Abstract
Glucocorticoid receptor (GR) and octamer transcription factors 1 and 2 (Oct-1/2) interact synergistically to activate the transcription of mouse mammary tumor virus and many cellular genes. Synergism correlates with cooperative DNA binding of the two factors in vitro. To examine the molecular basis for these cooperative interactions, we have studied the consequences of protein-protein binding between GR and Oct-1/2. We have determined that GR binds in solution to the octamer factor POU domain. Binding is mediated through an interface in the GR DNA binding domain that includes amino acids C500 and L501. In transfected mammalian cells, a transcriptionally inert wild-type but not an L501P GR peptide potentiated transcriptional activation by Oct-2 100-fold above the level that could be attained in the cell by expressing Oct-2 alone. Transcriptional activation correlated closely with a striking increase in the occupancy of octamer motifs adjacent to glucocorticoid response elements (GREs) on transiently transfected DNAs. Intriguingly, GR–Oct-1/2 binding was interrupted by the binding of GR to a GRE. We propose a model for transcriptional cooperativity in which GR–Oct-1/2 binding promotes an increase in the local concentration of octamer factors over glucocorticoid-responsive regulatory regions. These results reveal transcriptional cooperativity through a direct protein interaction between two sequence-specific transcription factors that is mediated in a way that is expected to restrict transcriptional effects to regulatory regions with DNA binding sites for both factors.
The initiation of transcription of genes by RNA polymerase II is usually controlled through complex transcriptional regulatory regions containing multiple binding sites for sequence-specific upstream transcription factors (74, 88). Overall transcriptional responsiveness is determined by cooperative and competitive interactions between the DNA-bound factors that are often dependent upon the precise juxtaposition of DNA binding sites (44). The regulatory potential of individual transcription factors can be influenced indirectly through the manipulation of DNA and/or chromatin structure (6, 8, 55, 88, 91). The precise arrangement of the transcription factor binding sites is also important for the cooperative interactions of sequence-specific transcription factors with transcriptional coactivators and the basal transcription machinery (43, 53, 62, 80, 94).
In many instances, transcriptional cooperativity also correlates with direct protein-protein interactions between individual sequence-specific transcription factors. Often, protein-protein contacts between heterologous factors stabilize DNA binding or alter the sequence specificity of binding. Thus, DNA-bound serum response factor is required for the formation of a ternary complex which includes serum response factor accessory protein 1 (SAP-1) (90); the sequence-specific DNA binding of DNA-dependent protein kinase is mediated through its association with DNA-bound Ku autoantigen (30), and the interaction of herpes simplex virus protein 16 (VP16) with host cell factor and the POU homeodomain protein octamer transcription factor 1 (Oct-1) alters the DNA sequence specificity of Oct-1 DNA binding (42, 98). Indeed, the sequence-specific DNA binding of several homeodomain proteins has been demonstrated to depend on stabilizing interactions with other sequence-specific transcription factors, including Exd, Ubx, Pbx, Hox, Ftz-F1, Ear3, and Arp1 (16, 34, 36, 71, 95, 96, 105).
There have also been a number of recent reports of sequence-specific transcription factors effectively functioning as transcriptional coactivators to potentiate activation of transcription by binding to other DNA-bound factors without directly contacting DNA themselves. For example, the glucocorticoid receptor (GR) has been demonstrated to act as a coactivator of Stat5 in prolactin-responsive cells (82). Similarly, GR and NF-IL6 appear to interact in a manner that allows each factor to potentiate transcriptional activation by the other from either a glucocorticoid response element (GRE) or an NF-IL6 DNA recognition sequence (63). The atypical homeodomain protein prospero enhances the DNA binding of the homeodomain proteins Dfd and HoxA5 in the absence of prospero DNA binding sites (35). In contrast, protein-protein binding can also restrict transcriptional activation potential. Thus, the Stat5-GR interaction appears to repress the activation of transcription through a GRE and prospero inhibits DNA binding by Eve (35, 82).
GR is a ligand-activated nuclear hormone receptor (7) that exists in the cytoplasms of unstimulated cells in an inactive complex with heat shock proteins and immunophilins (72). Hormone binding induces dissociation of the receptor-heat shock protein complex and rapid transfer of the free, transcriptionally active receptor to the nucleus, where it binds with high affinity to GREs (5, 83) as a homodimer to regulate transcription through interactions with transcriptional coactivators such as steroid receptor coactivator and CREB binding protein (14, 82). DNA binding of each GR monomer is mediated through two Cys4 zinc fingers that are highly conserved within the nuclear receptor superfamily (48).
Studies of the mouse mammary tumor virus (MMTV) promoter-proximal regulatory region have revealed two ways in which GR can influence the ability of other sequence-specific transcription factors to regulate transcription (7). This region of the MMTV long terminal repeat (LTR) encodes binding sites for nuclear factor 1 (NF1) (9, 11, 59) and octamer transcription factors (8, 10, 89) in addition to a complex GRE (15, 78). However, transcription from the MMTV promoter-proximal regulatory region is tightly controlled by hormone, with NF1 and Oct-1 having little effect on transcription in the absence of steroid (12, 65). Prior to stimulation, the MMTV LTR is packaged into a series of precisely positioned or phased nucleosomes that appear to specifically exclude NF1 from the MMTV promoter (2, 3, 64, 67, 70, 102). Upon hormone treatment, the binding of liganded GRs to the GRE is proposed to initiate a remodelling of the chromatin that makes the promoter accessible to NF1 and leads to the occupancy of the MMTV NF1 binding site in vivo (3, 8, 21, 70). Support for this hypothesis comes from a series of in vitro experiments which have demonstrated that GR and NF1 bind to the MMTV LTR and activate transcription independently on chromatin-free templates (8, 40). Further, on transiently transfected plasmid DNAs, which are not precisely packaged into chromatin, NF1 binding to the MMTV LTR occurs constitutively and is not affected by GR (3, 50).
In contrast, DNA binding and the activation of MMTV transcription by GR and Oct-1 have been shown to be highly cooperative both in vivo and in vitro (8, 91). Although the MMTV octamer motifs occur in vivo in linker DNA between two nucleosomes (91), and thus are expected to be more accessible than the NF1 binding sites, binding to the octamer motifs in vivo was also observed to be strictly hormone dependent (91). The binding of Oct-1 to the MMTV LTR in vitro was also dependent on GR (8). Specifically, addition of GR decreased the concentration of octamer factor required to saturate the octamer motifs in in vitro footprinting experiments (8). Further, transcriptional regulation of chromatin-free DNA templates by Oct-1 and GR was highly cooperative. Preliminary experiments have indicated that GR and Oct-1 may bind directly in solution through an interaction requiring the Oct-1 homeodomain (45). However, this interaction does not appear sufficient for either GR or Oct-1 to act as a transcriptional coactivator to potentiate transcription in the absence of octamer motifs or GREs. Rather, there is some evidence to suggest that GR may antagonize the activation of transcription by Oct-1 from regulatory regions that lack GR binding sites (45, 101).
Octamer transcription factors bind through their POU domains (100) to an 8-bp DNA recognition sequence known as an octamer motif (27, 97). The POU domain is a bipartite DNA binding domain (DBD) comprised of a homeodomain and a POU-specific helix-turn-helix motif (41). Binding of the two domains to DNA is highly cooperative. Oct-2 is highly similar in its POU domain to Oct-1 but has a distinct transcriptional regulatory potential (86) and does not interact with VP16 (46). Oct-1 and Oct-2 are constitutively active in cells in which they are expressed. Oct-1 is a ubiquitous protein, while Oct-2 is expressed mainly in lymphoid cells (20, 85). Like Oct-1, Oct-2 appears to activate transcription synergistically with GR (101). Furthermore, this transcriptional synergism also seems to result, at least in part, from the cooperative binding of GR and Oct-2 to DNA (101).
In this study, we have investigated the mechanism of transcriptional synergism between GR and Oct-1 or Oct-2 (Oct-1/2) by examining the molecular basis for their direct binding. Our results indicate that the POU domains of Oct-1 and Oct-2 bind to the DBD of GR in solution. We identified two point mutations in the DNA-contact α-helix in the C-terminal zinc finger of GR that individually prevented Oct-1/2 binding to GR in vitro and in vivo. The result of GR–Oct-1/2 binding in transiently transfected cells was a striking recruitment of the octamer factor to binding sites adjacent to GR binding sites, and this recruitment correlated directly with a dramatic increase in reporter gene transcription. Indeed, recruitment of Oct-2 to a promoter by a GR peptide unable to significantly activate transcription on its own resulted in transcriptional activation approximately 100-fold higher than could otherwise be achieved by ectopic expression of Oct-2 alone. Intriguingly, addition of a GRE to protein incubations was sufficient to disrupt preformed GR-Oct complexes. Our data support a mechanism for transcriptional cooperativity in which GR directly recruits Oct-1/2 to glucocorticoid-responsive promoters. However, the nature of the GR–Oct-1/2 interaction seems to ensure that Oct-1 and Oct-2 do not act as transcriptional coactivators of DNA-bound GR on promoters lacking octamer motifs.
MATERIALS AND METHODS
Plasmids.
The plasmids pGALO (77), pNLVP16 (23), and pG5E1BCAT (54) have been previously described, as have the GR and Oct plasmids GRWT (pRDN93) (58), X795, X616, X568 (76), and pCGNOCT-2 (86). Most of the plasmids containing point mutations in the GR DBD have also been described previously (79) as has the MMTV CAT reporter plasmid pHCWT (13). Plasmids used for in vitro translation include pSP6Luc (Promega) and pET11dCREB (24). The plasmid encoding the G504R point mutation (pT7G504R) was constructed by substituting the XhoI/PstI (blunted) GR fragment from pVAG504R (69) into pT7X556 (29) (XhoI/SacI [blunted]), to produce a plasmid that expresses an RNA encoding amino acids (aa) 407 to 523 of rat GR under the control of the T7 promoter. The plasmid with an L501P mutation in the wild-type (WT) GR full-length sequence (GRWT) was constructed by a site-directed mutagenesis strategy and then subcloned into p6RGR (66). Vectors expressing GRWT and GR with the L501P mutation (GRL501P) with N-terminal c-myc tags (pMTG-GR and pMTG-GRL501P) were created by cloning the BglII/SmaI 350-bp fragment from pCRIIMTG (a gift from J. Bell, University of Ottawa) and the MscI/BamHI GR fragments from p6RGR into pTL2 (provided by M. Petkovich, Queen’s University) digested with BglII and BamHI. GRWT, GRL501P, and GRC500Y DBDs fused to the Gal4 DBD were constructed by inserting NdeI/SacI fragments from the appropriate pT7-X556 vector (79) in frame into pGALO 3′ to the Gal4 DBD. The plasmid expressing the Oct-1 POU domain fused to the VP16 transcriptional activation domain was prepared by cloning a PCR-amplified Oct-1 fragment encoding aa 265 to 444 into pNLVP16. pGSTOct-2POU was cloned by inserting a PCR-amplified DNA fragment encoding aa 186 to 367 of Oct-2 (20) into pGEX 2T (Pharmacia).
The plasmids used in in vivo footprinting experiments were prepared by subcloning oligonucleotides encoding a consensus GRE (HindIII/BamHI DNA fragment from pTKCAT-GRE [93]) adjacent to the MMTV distal octamer motif (pBSGREOCTd) (−61 to −51; 5′-GATCC ACCTT ATTTA CATAA GCA-3′; BamHI/HindIII), a Gal4 DNA binding site adjacent to the histone H2B octamer motif (pBSGALOCT) (5′-CGGAG TACTG TCCTC CGGTA CCTGT ATGCA AAT-3′) and a consensus Gal4 DNA binding site adjacent to an intracisternal A particle (IAP) enhancer core binding site (pBSGALIAP) (5′-CGGAG TACTG TCCTC CGGTA CCCTG CGCAT GTG-3′) into the HindIII site (GREOCTd) or the SmaI site (GALOCTd and GALIAP) of pBluescript (Stratagene).
The reporter plasmids for transient transfection were constructed by multimerizing oligonucleotides prior to insertion into pG5E1BCAT. An oligonucleotide containing four copies of the distal octamer motif (described above) was blunted and inserted into the XbaI (blunted) site of pG5E1BCAT. pG54XOctmt was produced by inserting two copies of a synthetic oligonucleotide containing two mutant octamer motifs (5′-CTAGA GCTTC GGCAA ATAAG GTGGC TTCGG CAAAT AAGGT GT-3′) into the XbaI site.
Cell culture, transfections, and chloramphenicol acetyltransferase (CAT) assays.
CHOK1 cells (American Type Culture Collection, Manassas, Va.) were maintained in α-minimal essential medium supplemented with 10% fetal bovine serum (Life Technologies, Burlington, Vt.). HeLa and Sf7 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum.
Transfections for the selection of stable cell lines were performed with Lipofectamine (Life Technologies) (10 μl per 15-cm-diameter dish). Cells were incubated with Lipofectamine-DNA for 6 h, and transfection was stopped by the addition of fetal bovine serum to 10%. Selection with 400 μg of G418 per ml was initiated 24 h after transfection.
For CAT analysis, transfections (30) were performed with DEAE dextran as previously described (12) (2 μg of CAT reporter, 3 μg Rous sarcoma virus β-galactosidase (β-Gal), and 10 μg of total DNA) or with Lipofectamine (10 μl per 60-mm-diameter dish). Highly sheared salmon sperm DNA was used to equalize the DNA loadings between experiments where necessary. Equivalent results were obtained by both techniques. Following transfection, the cells were incubated overnight. The medium was changed the next morning, and cells were harvested 24 to 48 h later. CAT assays were performed as previously described, and parallel β-Gal assays were used to normalize results for variations in transfection efficiencies. Each experiment was repeated in duplicate a minimum of three separate times. The error bars in the figures reflect the standard errors of the means of all repetitions. The levels of expression of the various Gal and Gal-GR constructs employed in these assays were quantified by Western blot analysis with a Gal4 antibody (Santa Cruz Biotechnologies, Santa Cruz, Calif.) and a Bio-Rad model GS-525 phosphorimager. In all instances, transfections were performed with amounts of Gal4 DBD (GalO)-, Gal-GRWT-, and Gal-GRL501P-expressing plasmids that expressed equal levels of the proteins.
Transfection of cells for in vivo footprinting assays was performed with CaPO4 exactly as previously described (3), with 1.5 μg of footprinting template, and with 1.5 μg of GR, Gal-GR, and Oct-2 expression plasmids, as indicated in the figures, in DNA mixtures supplemented to 5 μg with highly sheared salmon sperm DNA. Cells were harvested for footprinting 24 h after transfection. In experiments with full-length GRWT cells were treated with 10−6 M dexamethasone (Dex) or vehicle for 15 min prior to harvesting.
Finally, transfections to prepare nuclear extracts for electrophoretic mobility shift assays (EMSA) were performed with Lipofectamine (15 μl per 60-mm-diameter dish). Cells were harvested 48 h after transfection.
In vitro binding to GST fusion proteins.
Glutathione S-transferase (GST) fusion proteins were expressed and purified on glutathione-Sepharose essentially as described previously (46). GR peptides were in vitro translated in the presence of [35S]methionine with the coupled transcription-translation TNT reticulocyte lysate system (Promega). Steroid binding was accomplished by addition of 10−6 M Dex to the in vitro-translated GR for at least 2 h at 4°C followed by incubation at 25°C for 25 min as previously described (25). The association of GR with heat shock proteins was stabilized by the addition of 20 mM MoNa2O4 (49). The levels of binding of the in vitro-translated GRs to the GST fusion proteins were determined essentially as described previously (46). Briefly, equal amounts of in vitro-translated proteins were incubated with 0.5 μg of immobilized GST fusion protein in 200 μl of binding buffer (20 mM HEPES [pH 7.9], 60 mM KCl, 12% glycerol, 1.5 mM EDTA, 1 mM dithiothreitol [DTT], 0.15 mM phenylmethylsulfonyl fluoride, 0.1% Nonidet P-40) for 90 min at 4°C. Following four washes with binding buffer, the proteins retained on the affinity matrix were eluted in sodium dodecyl sulfate (SDS) sample buffer and resolved by SDS-polyacrylamide gel electrophoresis (PAGE). Levels of binding were compared to those of 10% of the in vitro-translated proteins added to the incubations.
In oligonucleotide competition experiments, following the 90-min incubation at 4°C, 100 ng of oligonucleotides encoding a GRE (5′-ACAGT TCGAC ATAGA ACAAA CTGTT CTTAA AAGGT ACCCA-3′), an IAP enhancer core (28) (5′-CTGCG CATGT GCCAA GGGTA TCTTA TGACT-3′), a Ku antigen DNA binding site from the C3H strain of MMTV (51) (5′-AGCTT GTCTC AAGAA GAAAA AGACG AGAG-3′), or highly sheared calf thymus (CT) DNA was added and the incubations were continued for a further 20 min at room temperature. Subsequently, samples were washed and processed as in the standard assay.
Peroxide treatment of the GR.
The GR DBD peptide X556 was expressed in BL21 (DE3) and partially purified as previously described (29). X556 was approximately 10% pure, as estimated by Coomassie blue staining of SDS-polyacrylamide gels.
In vitro-translated X616 and bacterially expressed X556 were treated with 20 mM H2O2 in the presence of 50 mM EDTA for 1 h at 4°C followed by neutralization for 15 min with catalase as previously described (39, 87). Restoration of zinc was attempted by treatment with 0.1 M DTT in the presence of 250 μM ZnCl2 (29).
Immunoprecipitations and Western blot analysis.
Western blotting was performed with Immobilon-P membranes (Millipore), and proteins were visualized by enhanced chemiluminescence (Amersham).
Cell lines stably expressing WT and L501P GRs with N-terminal c-myc tags were prepared by cotransfecting pMTG-GRWT or pMTG-GRL501P and pSV2NEO into murine Sf7 fibroblasts and selecting stable colonies by treatment with G418. Western blot analysis of whole-cell extracts with myc-tag antibody 9E10 identified two cell lines, Sf7 myc-GR and Sf7 myc-GRL501P, expressing nearly equal levels of GRWT and GRL501P as determined by phosphorimager analysis. Binding assays were performed as previously described with 9E10 and whole-cell extracts prepared 1 h following treatment of the cells with 10−6 M Dex and full-length 35S-labelled in vitro-translated Oct-1/2. Following vigorous washing, phosphorimager quantification of SDS-polyacrylamide gels was used to compare specific binding by myc-GR-containing cell extracts to that of an extract prepared from parental Sf7 cells that do not express myc-tagged GR. myc-GR loading in each experiment was confirmed by Western blot analysis.
In vivo footprinting of transiently transfected plasmids.
Nucleus preparation and λ exonuclease analysis were performed essentially as described previously (61). A total of 1.5 × 105 nuclei were used for each λ exonuclease analysis. Restriction enzyme digestion of the nuclei was used to create specific entry points for exonuclease digestion. Nuclei from cells transfected with pHCWT were digested with BamHI, pBSGREOCTd was digested with SmaI, and pBSGALOCT and pBSGALIAP were digested with XhoI. Each sample was simultaneously digested with 100 U of restriction enzyme and 15 U of λ exonuclease (Life Technologies) for 15 min at 30°C. Reactions were stopped with SDS-EDTA, and the DNAs were purified by proteinase K digestion and phenol-chloroform extraction.
Linear PCR extension (25 cycles) to reveal exonuclease pausing was performed with primers extending from +74 to +52 of the MMTV LTR for plasmid pHCWT, a T3 polymerase promoter primer for pBSGALOCT, and a T7 polymerase promoter primer for pBSGALIAP. Reactions were carried out as described previously (61) with Taq DNA polymerase (Perkin-Elmer) in the buffer supplied by the manufacturer. Products were resolved on 6% polyacrylamide–7 M urea gels.
Nuclear extracts and EMSA.
Nuclear extracts from transiently transfected cells were prepared by a micropreparation procedure (1) from confluent 60-mm-diameter plates. In all extracts used for EMSA, levels of transiently expressed proteins were first verified by Western blotting. 32P-labelled oligonucleotides for GALOCTd and GALIAP have been described above. The GALmtOCT oligonucleotide had a 2-bp mutation (underlined) that is said to eliminate Oct-1/2 binding (19) (5′-CGGAG TACTG TCCTC CGGTA CCTGT CGGCA AATAA GGT-3′). Nuclear extracts (1 μg) were preincubated alone or in combination for 4 h at 4°C. This incubation was followed by addition of 2 ng of 32P-labelled oligonucleotides, 1 μg of poly(dI-dC), 1 μg of bovine serum albumin, and 100 ng of competitor oligonucleotides, as indicated in the figures, for 20 min at room temperature in 12 mM HEPES (pH 7.9)–12% glycerol–60 mM NaCl–0.12 mM EDTA. Protein-DNA complexes were resolved on 4% polyacrylamide (acrylamide-bisacrylamide [39.5:1]) gels in 0.5× Tris-borate-EDTA.
An EMSA with bacterially expressed GRX556 treated as described above was performed with the 32P-labelled 42-bp GRE oligonucleotide from pTKCAT GRE (93) in the same buffer for 20 min at room temperature followed by PAGE as described above.
RESULTS
The GR DBD directs binding of GR to the Oct-1 POU domain in vitro.
To begin a detailed examination of the potential for the GR to bind directly to Oct-1/2, we expressed the POU domain of Oct-1 as a GST fusion protein (46). A previous report implicating the POU homeodomain of Oct-1 in GR binding and the lack of similarity between Oct-1 and Oct-2 outside of the POU domain suggested this region as the likely target for GR binding (45, 86). In a first series of experiments, we tested the ability of in vitro-translated rat GR fragments to bind to the GST–Oct-1 POU domain fusion protein (Fig. 1). In vitro-translated GR is incorporated into a heat shock protein-containing multiprotein complex that closely mimics the complexed state of unliganded GR in the cytoplasm (22). In addition to stabilizing the ligand binding domain in a conformation able to accept ligand, association of GR with these accessory proteins masks DNA binding, nuclear localization, and other functions of GR (72). Addition of the synthetic steroid Dex to in vitro-translated receptor results in the release of the heat shock proteins from the liganded receptor (22).
FIG. 1.
Specific binding of the GR to the POU domain of Oct-1 in vitro is abrogated by C-terminal truncation into the DBD. (A) Schematic presentation of in vitro-translated rat GR fragments used in panels B and C. The positions of the DBD, ligand binding domain, and heat shock protein binding domains of GR are diagrammed. (B) In vitro-translated Dex-treated GR, CREB, and firefly luciferase (Luc.) were tested for binding to GST–Oct-1 POU by a pulldown assay. Levels of binding (lanes 1 to 3) were compared to those of 10% of the receptors added to the incubations (lanes 4 to 6). The migrations of molecular mass markers are indicated to the left of the autoradiographs. (C) In vitro-translated GR fragments were tested for binding to GST–Oct-1 POU. GRWT and X795, which contain intact ligand binding domains and which were complexed with heat shock proteins in the absence of ligand, were tested for Oct-1 POU binding in both the presence (+) and absence (−) of bound steroid (Dex). SDS-polyacrylamide gels were exposed equally to compare specific levels of binding (lanes 1 to 6) to those of 10% of the in vitro-translated proteins added to the incubation (lanes 7 to 12). (D) In vitro-translated full-length and C-terminally deleted GRs produced by transcription of a restricted DNA template were tested for binding to immobilized recombinant GST–Oct-1 POU. Bound untreated and Dex-treated full-length receptors are shown in lanes 1 and 2, respectively, while the C-terminal truncations are shown in lanes 3 to 5. Levels of binding of proteins are compared to levels of binding of 10% of the input proteins shown in lanes 6 to 10.
As expected, in vitro-translated, liganded rat GR was observed to bind directly to GST–Oct-1 POU (Fig. 1B, lane 1). Binding appeared to be specific, as neither in vitro-translated CREB nor luciferase bound to GST–Oct-1 POU (Fig. 1B, lanes 2 and 3). Further, GR was not retained by GST alone and binding was completely resistant to the addition of ethidium bromide (47, 73). Finally, binding of GR to GST–Oct-1 POU appeared to require the dissociation of heat shock proteins induced by ligand, as binding was undetectable in the absence of added steroid (Fig. 1C, lanes 1 and 2).
Preliminary mapping of the GST–Oct-1 POU binding domain on GR indicated that deletion of the N- or C-terminal domain of the receptor had no significant effect on POU domain binding but that omission of the heat shock protein binding domain in the C terminus of GR rendered binding constitutive (Fig. 1C, lanes 3 to 6). Exactly the same results were obtained for the binding of GRWT and GR fragments to a GST–Oct-2 POU domain fusion protein (73). Binding assays with additional GR constructs mapped the C-terminal boundary for GST–Oct-1 POU binding between aa 494 and 523 of the GR DBD (Fig. 1D).
To analyze the interaction of the GR DBD to the POU domain of Oct-1 in more detail, the effect of specific GR DBD point mutations on the binding of in vitro-translated GR peptides to GST–Oct-1 POU was assessed (Fig. 2). Two of the peptides tested, containing C500Y and L501P mutations that map within the DNA-contact α-helix of the second zinc finger of the GR DBD, failed to bind GST–Oct-1 POU (Fig. 2B, lanes 9 and 10). By contrast, several other mutations, including cysteine mutations that disrupt zinc coordination (C460Y, C492R, and C495Y), had no effect on GST–Oct-1 POU binding. Repetition of the binding assay with WT and C500Y GR fragments purified from bacteria verified that binding was direct and did not require additional factors present in the reticulocyte lysate (73).
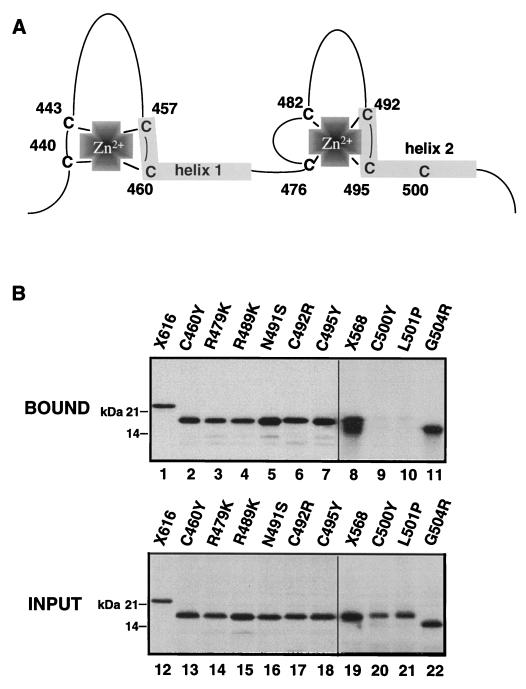
FIG. 2.
The mutations C500Y and L501P in the GR DBD eliminate binding to GST–Oct-1 POU in vitro. (A) Schematic summary of the DNA binding domain of the rat GR DBD showing the position of the zinc-coordinating cysteines and the two α-helices that contact DNA. (B) The in vitro-translated GRWT DBD (aa 407 to 568) (lanes 8 and 19) and specific point mutation-containing peptides (all of which were in the aa 407 to 556 GR backbone except G504R, which was in the aa 407 to 523 backbone) (lanes 2 to 11) were tested for binding to GST–Oct-1 POU (lanes 2 to 11) in comparison to that of X616 (lanes 1 and 12), as described for Fig. 1.
The GR DBD zinc requirements for Oct-1 binding were examined further by removing zinc from the GR DBD by treatment with hydrogen peroxide in the presence of EDTA (39, 87) (Fig. 3). This treatment completely eliminated sequence-specific DNA binding of a bacterially expressed GR DBD fragment (Fig. 3A, lanes 3 and 4). Approximately 10% of the DNA binding activity of GR could subsequently be restored by DTT treatment in the presence of 250 μM zinc chloride (lanes 5 and 6). The same treatments had no discernible effect on the binding of the in vitro-translated GR DBD to GST–Oct-1 POU (Fig. 3B, lanes 1 to 3). Taken together, these results indicate that while coordination of zinc, and thus the zinc-stabilized tertiary structure of the GR DBD, did not appear to be required for binding to GST–Oct-1 POU, the DNA-contact α-helix within the second GR zinc finger contained at least two determinants, Cys500 and Leu501, required for Oct-1 POU domain binding.
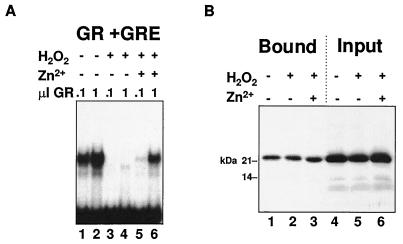
FIG. 3.
Removal of zinc from the GR DBD has no effect on binding to GST–Oct-1 POU. (A) The effects of peroxide-EDTA treatment, which strips the coordinated zinc from the GR DBD, and the subsequent partial reconstitution of zinc coordination on the binding of bacterially expressed X556 to a GRE was evaluated by EMSA. (B) Effects of the same treatment described for panel A on the binding of in vitro-translated X616 to GST–Oct-1 POU.
Point mutations in the GR DBD prevent the binding of GR to Oct-1/2 in vivo.
To determine the involvement of residues in the α-helix extending from the second finger of the GR DBD in the binding of full-length GR to WT Oct-1/2 in solution, cellular extracts were prepared from murine Sf7 fibroblasts stably transfected to express similar levels of full-length WT and L501P GRs with N-terminal c-myc tags. The binding of full-length, in vitro-translated, Oct-1 and Oct-2 to the GRs in extracts prepared 1 h after glucocorticoid hormone treatment was determined by examining the amounts of Oct-1 and Oct-2 immunoprecipitated with an antibody to the c-myc tag (Fig. 4). Both Oct-1 and Oct-2 were efficiently immunoprecipitated with GRWT, but neither protein was detected above background in GRL501P immunoprecipitates. Thus, both Oct-1 and Oct-2 were observed to bind full-length, liganded GR and this binding was completely abrogated by the L501P point mutation in the GR DBD.
FIG. 4.
An L501P substitution in full-length GRWT eliminates binding to full-length WT Oct-1 and Oct-2. GRWT (lanes 2 and 6) and GRL501P (lanes 3 and 7) with N-terminal c-myc tags in nuclear extracts of stably transfected CHOK1 fibroblasts were tested for binding to in vitro-translated full-length Oct-2 (A) and Oct-1 (B) by immunoprecipitation with anti-myc antibody 9E10. Binding to immunoprecipitated control extracts lacking myc-tagged GR is shown in lanes 4 and 8. Levels of binding were compared to those of 10% of input Oct-2 and Oct-1 (lanes 1 and 5, respectively) and were quantified by exposure of the SDS-polyacrylamide gels to a storage phosphor screen. The relative counts are displayed beneath the lanes. (C) A Western blot of the immunoprecipitated GRWT, GRL501P, and control nuclear extracts with BUGR2, a GR-specific antibody, demonstrates that equal amounts of the two receptor forms were immunoprecipitated from the extracts.
In a third test of GR-Oct binding, we examined the ability of the Oct-1 POU domain to bind to the GR DBD in two hybrid experiments in mammalian cells (Fig. 5). Because L501P and C500Y mutations in the GR DBD interrupt DNA binding by GR as well as binding to Oct-1/2, a Gal4 DBD was added to GR DBD constructs (aa 407 to 556) to distinguish transcriptional effects resulting from the loss of GR-octamer protein binding from effects due to the abrogation of the binding of GR to DNA. Transcription was monitored from a reporter gene with Gal4 DNA binding sites immediately adjacent to a minimal adenovirus E1B promoter.
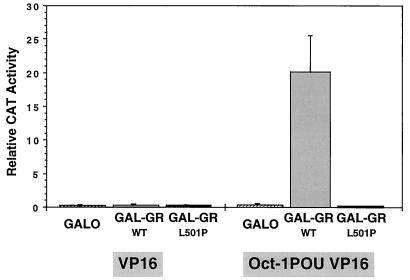
FIG. 5.
Two-hybrid analysis of the binding of the GR DBD to the Oct-1 POU domain in CHO cells as reflected by the activation of pG5E1bCAT transcription. Gal-GRWT, Gal-GRL501P, or the Gal4 DBD (GalO) were coexpressed with the VP16 activation domain alone (left) or tethered to the POU domain of Oct-1 (right). The means and standard errors of the means of results of five independent transfections are shown. Western blot analysis indicated that GalO, Gal-GRWT, and Gal-GRL501P were expressed at the same level (73). Similar results were also obtained with HeLa cells (73).
The Gal4 DBD (GalO) and Gal4 DBD-GR DBD fusion proteins (Gal-GRWT and Gal-GRL501P) coexpressed with the acidic activation domain of VP16 in CHO cells had no effect on E1B expression. Similarly, expression of a construct with the Oct-1 POU domain fused to the activation domain of VP16 failed to induce transcription when the construct was coexpressed with GalO. E1B expression was activated over 80-fold, however, when Gal-GRWT was coexpressed with Oct-1–VP16. By contrast, Gal-GRL501P expressed at similar levels was completely unable to induce E1B transcription. The same result was obtained with Gal-GRC500Y (52). Essentially the same results also were obtained with full-length Oct-2 (see below). These results confirmed that the GR DBD interacted with the POU domains of Oct-1 and -2 in the nucleus in a way that was completely sensitive to C500Y and L501P mutations in the GR DBD.
GR recruits octamer factors to adjacent octamer motifs in vivo.
DNA binding experiments have shown that, in vitro, GR decreases the concentration of octamer factor required to saturate binding sites adjacent to GREs (8). Further, we have demonstrated that the levels of occupancy of the MMTV promoter octamer motifs by Oct-1 in vivo on chromatin closely paralleled the levels of binding of GR to the MMTV GREs (91). However, in this instance, GR binding to the MMTV promoter induced a rearrangement of chromatin structure that may have promoted Oct-1 binding by increasing the accessibility of the octamer motifs. Thus, it was not possible in these experiments to distinguish the extent to which occupancy of the octamer motifs resulted from the cooperativity in DNA binding observed in vitro between Oct-1 and GR.
One way in which to distinguish direct effects on DNA binding from indirect effects resulting from the remodelling of the chromatin structure is to examine the binding of transcription factors to transiently transfected DNAs that fail to adopt a regular chromatin structure in the absence of DNA replication (75). This approach has previously been used to support the hypothesis that the binding of NF1 to the MMTV promoter is dependent solely on chromatin remodelling (3). Therefore, in order to dissociate rearrangements in chromatin structure from cooperativity in DNA binding, we examined the effect of GR on the binding of Oct-2 with a series of transiently transfected DNA templates (Fig. 6).
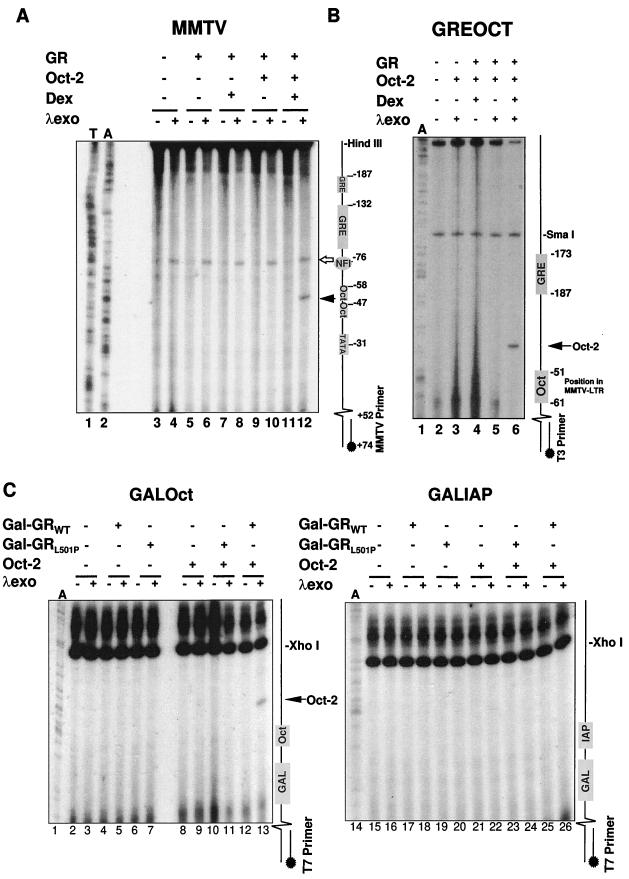
FIG. 6.
GR binding recruits Oct-2 to octamer motifs adjacent to GR binding sites in the nucleus. (A) Nuclei prepared from CHO cells transfected with the MMTV promoter construct pHCWT, GR, and/or Oct-2 expression plasmids and treated with 10−6 M Dex or vehicle for 15 min were restricted with HindIII and digested with λ exonuclease (λexo) as indicated. Digestion was revealed by linear PCR extension of a primer extending from +74 to +52 of the MMTV LTR, and pause sites were positioned relative to A and T sequencing tracks amplified with the same primer. The Dex-, GR-, and Oct-2-specific λ pause site is indicated by the solid arrow, while the constitutive pause site generated by NF1 is indicated by the open arrow. The positions of transcription factor binding sites in the MMTV LTR are indicated schematically to the right of the autoradiogram. (B) Nuclei prepared from CHO cells transfected with pBluescript containing the strong GRE and octamer motif sequences from the MMTV LTR separated by 29 nucleotides (8) and GR and/or Oct-2 expression plasmids and treated with 10−6 M Dex or vehicle for 15 min were restricted with SmaI and digested with λ exonuclease as indicated. Digestion was revealed by linear PCR extension of a T3 polymerase primer, and pause sites were positioned relative to an A sequencing track amplified with the same primer. The positions of the octamer motif sequence and GRE sequences in the MMTV LTR are summarized schematically. The Dex-, GR-, and Oct-2-specific λ pause site is indicated by the arrow. (C) Nuclei prepared from CHO cells transfected with pBluescript containing either a Gal4 binding site separated by 8 nucleotides from the octamer motif sequence from the MMTV LTR (left) or a nonspecific oligonucleotide encoding an IAP enhancer core (right) along with Gal-GRWT, Gal-GRL501P, and/or Oct-2 expression plasmids were restricted with XhoI and digested with λ exonuclease as indicated. Digestion was revealed by linear PCR extension of a T7 polymerase primer, and pause sites were positioned relative to an A sequencing track amplified with the same primer. The positions of the octamer motif-IAP sequence and of the Gal4 sequence are summarized schematically. The Gal-GRWT-, Oct-2-, and octamer motif-dependent specific λ pause site is indicated by the arrow. Western blots of cellular extracts verified that Gal-GRWT and Gal-GRL501P were expressed to the same levels (73).
Sequence-specific binding of transcription factors to DNA in the cell can be detected directly in nuclei by exonuclease digestion of restricted DNA (60, 61, 103). Many DNA-bound transcription factors, including NF1, provide a barrier to the exonuclease, forcing a pause in digestion that can be subsequently viewed by primer extension or PCR. Because binding is revealed as a positive signal, this method is particularly useful for footprinting sequence-specific binding to transiently transfected plasmid DNAs which rarely become saturated with factors due to what are often very high copy numbers of plasmids in individual cells.
The MMTV promoter contains a complex GRE, an NF1 binding site and two adjacent octamer motifs (17, 92). Binding to the NF1 site can be detected by λ exonuclease digestion of transiently transfected templates, but GR binding does not impede λ exonuclease sufficiently to be revealed as a pause site (61). Further, NF1 binding to the transiently transfected MMTV DNA is known to be constitutive and unaffected by steroid (3) and can thus be used as an internal control for the binding of other factors like Oct-1/2 to the promoter.
In the first instance, we examined transcription factor binding to the MMTV LTR in plasmid pHCWT (Fig. 6A). In the absence of steroid, a λ exonuclease pause site immediately upstream of the NF1 binding site on the MMTV promoter was readily detected by linear PCR, both in the presence and in the absence of full-length GRWT (lanes 4 and 6). As expected, the intensity of this pause site was unaffected by Dex treatment in the presence of full-length GRWT or coexpressed Oct-2 (lanes 8, 10, and 12). By contrast, no pausing adjacent to the octamer motifs in the MMTV LTR was detectable at this level of amplification in the absence of steroid hormone, even upon ectopic expression of Oct-2 (lanes 4, 6, 8, and 10). Upon hormone treatment, however, a strong Oct-2-dependent pause site was detected (lane 12). Somewhat unexpectedly, this pause site mapped exactly between the two MMTV octamer motifs at −49 (lane 12) rather than near −60, adjacent to the upstream octamer motif.
To confirm that this pause site did indeed reflect the binding of Oct-2 to the MMTV LTR, and to show that Oct-2 binding was not dependent on any other factor that might bind to the MMTV promoter but GR, we repeated the footprinting with a promoterless pBluescript construct into which the distal MMTV octamer motif (−61 to −51) was cloned adjacent to the strong single GRE from the MMTV LTR (−187 to −172) (Fig. 6B). On this plasmid, a single steroid- and Oct-2-dependent pause site was detected immediately adjacent to the octamer motif (lane 6). We therefore conclude that steroid-activated GR can promote the binding of Oct-2 to octamer motifs adjacent to GREs on plasmid DNA in the cell nucleus. This experiment also indicated that the promotion of Oct-2 DNA binding was not sensitive to the orientations and relative positioning of the GREs and octamer motifs, as both the spacing between and the orientations of the MMTV GRE and octamer motif were altered in the pBluescript plasmid in comparison to those of the natural promoter.
To evaluate the effect of the L501P mutation, which disrupts GR–Oct-1/2 protein-protein binding, on the binding of Oct-2 to octamer motifs in the nucleus, we examined the ability of Gal4 DBD-GR DBD fusion proteins to promote the binding of Oct-2 to a histone H2B octamer motif cloned into pBluescript adjacent to a single Gal4 DNA binding site (Fig. 6C). Interestingly, expression of the Gal-GR proteins alone did not result in a exonuclease pause site adjacent to the Gal4 DNA binding site (lanes 5 and 7), suggesting that binding of the Gal4 DBD to DNA, like binding of GR to DNA, does not impede progress of the λ exonuclease. However, coexpression of Gal-GRWT and Oct-2 again led to the appearance of an exonuclease pause site immediately adjacent to the octamer motif (lane 13). Oct-2 DNA binding was completely sensitive to the L501P mutation in GR, as coexpression of Gal-GRL501P to the same levels as Gal-GRWT (73) was completely unable to promote Oct-2-dependent exonuclease pausing (lane 11). Further, no pause was detected on templates in which a nonspecific oligonucleotide had been substituted for the octamer motif (lanes 14 to 26). Together, these results suggest that the L501P-sensitive binding of GR to Oct-2 in the cell strongly promotes the binding of Oct-2 to octamer motifs adjacent to binding sites for GR. Further, the DNA binding of Oct-2 was not dependent on the sequence-specific binding of GR to DNA, as Gal-GRWT appeared to be as effective in recruiting Oct-2 from a Gal4 DNA binding site as full-length GRWT was from a GRE.
Recruitment potentiates the activation of transcription by the octamer factors to levels that cannot otherwise be attained in the cell.
The ability of many transcription factors, including Oct-1/2, to activate transcription is usually directly proportional to their affinities for individual response elements and to their concentrations in the nucleus. Thus, when joined to minimal promoters, weak octamer factor binding sites direct the activation of transcription by Oct-1/2 more poorly than high-affinity binding sites (19). When considered in this context, the dramatic L501P-sensitive increase of Oct-2 DNA binding we observed with GR might be expected to dramatically potentiate the activation of transcription by Oct-2 from promoters with octamer motifs and GR binding sites linked in cis. Further, this effect might be expected to account for a significant proportion of the transcriptional cooperativity observed between GR and Oct-1/2 on such promoters (8, 101).
We examined the potential of GR–Oct-1/2 binding to promote transcriptional activation from consensus, high-affinity octamer motifs (19) linked in cis to GR binding sites in transient transfection assays (Fig. 7). In these experiments, the use of transcriptionally inert Gal DBD-GR DBD fusion proteins allowed us to focus specifically on transcription induced by Oct-2 and by an Oct-1–VP16 fusion protein, in the absence of the N- and C-terminal GR transcriptional activation functions. Further, linking the GR DBD to a Gal4 DBD enabled us to evaluate the consequences of the L501P GR mutation, which abrogated both GR–Oct-1/2 binding and the promotion of Oct-2 binding to transiently transfected plasmids.
FIG. 7.
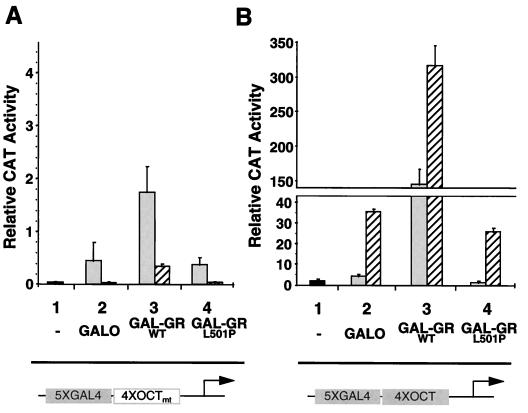
Gal-GRWT dramatically potentiates octamer motif-dependent activation of transcription by Oct-2 and Oct-1–VP16. The interaction of full-length Oct-2 (shaded bars) and Oct-1–VP16 (hatched bars) with Gal-GR fusion proteins was examined with reporter plasmids containing five Gal4 binding sites 5′ to four mutated (A) or WT (B) octamer motifs upstream of a minimal E1B promoter and CAT gene. The Gal4 DBD alone (GalO) or Gal-GR fusion proteins were coexpressed with Oct-2 (shaded bars) or Oct-1–VP16 (hatched bars). The black bars show activity in the absence of ectopically expressed proteins (lanes 1). Expression of GalO alone had no significant effect on CAT activity generated from the octamer mutant reporter construct (52).
Transcription was recorded from two reporter constructs derived from the original G5E1BCAT plasmid used in the two-hybrid experiments described above. The first construct contained four copies of a consensus octamer motif oligonucleotide inserted between the five Gal4 motifs and the E1B promoter (G54xOctE1B). The second construct contained four copies of a related oligonucleotide with two substitutions in the octamer motif that have been previously described as eliminating Oct-1/2 DNA binding and transcriptional induction (G54xOctmtE1B) (19). Under our conditions in CHO cells, this previously reported mutation eliminated transcriptional activation by Oct-1–VP16, and was 90 to 95% effective in abrogating the induction of transcription by Oct-2 (Fig. 7).
Coexpression of Oct-2 or Oct-1–VP16 with the Gal4 DBD (GalO) or Gal-GRL501P had no additional effect on the activity derived from the reporter gene with the mutant octamer motifs (Fig. 7A). However, when Gal-GRWT was coexpressed with Oct-1–VP16 or Oct-2, reporter activity was induced 5-fold (Oct-2) to 10-fold (Oct-1–VP16). Interestingly, while a strong activation of reporter gene expression still existed, the 95-bp increase in separation between the Gal4 DNA binding sites and the E1B minimal promoter reduced the activation of transcription by Oct-1–VP16 by approximately eightfold (compare Fig. 7A with Fig. 5). This reduction suggested that proximity to the E1B promoter was important for translating two-hybrid GR–Oct-1–VP16 binding into activation of transcription.
For the E1B construct containing WT octamer motifs (Fig. 7B), expression of Oct-1–VP16 or Oct-2 alone or together with GalO or Gal-GRL501P resulted in a 10- to 25-fold induction of transcription over the level obtained from the reporter with the mutant octamer motifs (lanes 2 and 4; note the difference in scale from that of Fig. 7A). Coexpression with Gal-GRWT further potentiated the induction of CAT activity 10-fold (Oct-1–VP16) to 100-fold (Oct-2). Thus, the striking recruitment of Oct-2 to octamer motifs by Gal-GRWT was directly reflected by a very strong increase in octamer factor-dependent transcription. By contrast, coexpression of GalO or the Gal-GRs had no significant effect on the activation of E1B transcription by Oct-2 from an E1B CAT reporter gene containing the octamer motifs but from which the Gal4 binding sites had been removed (52).
The experiments described above were performed at levels of Oct-2 and Oct-1–VP16 expression that maximally activated transcription of the G54xOctE1B reporter gene in the absence of coexpressed GR. Therefore, our results indicated that GR-mediated recruitment of the octamer factors not only increased the efficiency of the use of limiting quantities of octamer factor in the cell but actually increased octamer motif-dependent transcription beyond the level that could otherwise be attained in the cell, regardless of the amount of octamer factor expressed.
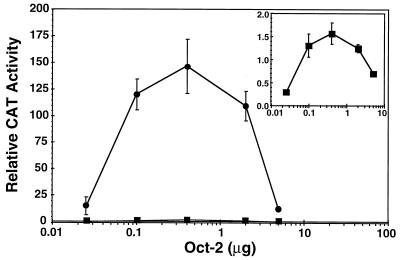
This finding is illustrated further by the results shown in Fig. 8. Transfection of increasing amounts of the Oct-2 expression vector in the presence of Gal-GRL501P yielded a curve for transcriptional induction consistent with rising concentrations of Oct-2 at first increasing transcription but eventually saturating the system to effect transcriptional squelching (31) at high concentrations (Fig. 8, inset). The shape of the curve obtained for the dose-dependent induction of the G54xOctE1B promoter when increasing amounts of Oct-2 were coexpressed with Gal-GRWT was strikingly similar. Notably, squelching was observed at the same Oct-2 concentration in both instances. However, across the entire range of Oct-2 concentrations, coexpression of Gal-GRWT resulted in a consistent 100-fold increase in Oct-2-dependent E1B transcription.
FIG. 8.
GR binding potentiates the activation of transcription by Oct-2 far above the level that can be obtained by ectopic expression of Oct-2 alone. CAT activity from the E1B reporter with five Gal4 binding sites and four octamer motifs was determined upon cotransfection of increasing amounts of Oct-2 expression plasmid in the presence of a fixed amount of a Gal-GRWT (circles) or Gal-GRL501P (squares) expression vector. The results with Gal-GRL501P are shown on an enlarged scale in the inset. CAT activity from experiments with increasing amounts of Oct-2 transfected alone or in the presence of GalO reached a plateau of activation (∼5 relative CAT units) by 400 ng of Oct-2, with a slow decline thereafter to background levels (∼0.3 relative CAT units) (data not shown). The data represent the means and standard errors of the means of results from three to five independent experiments.
Binding of GR to a GRE destabilizes association with Oct-1/2.
Despite the strength of the two-hybrid effect we observed with the GR DBD tethered to DNA through the Gal4 DBD, previous authors have demonstrated that Oct-1 and Oct-2 do not appear to enhance the activation of transcription through a GRE in the absence of octamer motifs (101). Similarly, we observed that expression of Oct-2 had no effect on the ability of the Gal-GR fusion protein to activate transcription from a GRE (52). These results suggested the intriguing possibility that the binding of GR to DNA might be exclusive of the continued interaction with Oct-1/2 that we have observed in solution.
To examine how GR binding to DNA might affect GR–Oct-1/2 binding, we first examined the effect of various DNAs on GR–Oct-1/2 binding in a GST pulldown assay (Fig. 9A). In vitro-translated Dex-liganded GR was preincubated with GST–Oct-1 POU beads under standard binding conditions to allow complex formation to reach equilibrium. Subsequently, the GR–Oct-1 complex was challenged with various DNAs. The preformed GR–Oct-1 complexes were unaffected by the addition of highly sheared CT DNA or two different nonspecific oligonucleotides to the binding assay (lanes 3 to 5; compare with Fig. 1). However, the addition of an oligonucleotide encoding a GRE following the equilibration of GR-Oct binding completely disrupted the preformed GR–Oct-1 POU complexes. Thus, GR–Oct-1 binding appeared to be highly sensitive to the exposure of the GR to a GRE.
FIG. 9.
The binding of GR to Oct-1 POU in solution is sensitive to the presence of a GRE. (A) Following the incubation of GR with GST–Oct-1 POU, binding was challenged with a consensus GRE (lane 2), highly sheared CT DNA (lane 3), or nonspecific oligonucleotides encoding an IAP enhancer core (lane 4) or a binding site for the Ku autoantigen on the C3H MMTV LTR (lane 5). GST–Oct-1 POU binding was determined by exposure of an SDS-polyacrylamide gel and compared to that of 10% of the in vitro-translated GR added to the incubation. (B) EMSA with nuclear extracts prepared from untransfected CHO cells and cells transfected with expression plasmids for Gal-GRWT, Gal-GRL501P, or Oct-2. The radiolabelled oligonucleotide employed contained a single Gal4 binding site and an octamer motif. Nuclear extracts were preincubated prior to the addition of the labelled probe and competitor DNAs. Incubation was continued for a further 20 min prior to PAGE. The individual combinations of nuclear extracts and unlabelled competitor oligonucleotides (added at a 100-fold molar excess) are summarized at the top of the panel.
To examine the relationship between GR-Oct protein-protein and protein-DNA binding in more detail, mobility shift assays were performed with nuclear extracts prepared from cells transfected to overexpress high levels of either Gal-GRWT, Gal-GRL501P, or Oct-2 (Fig. 9B). The Gal-GR fusion proteins and Oct-2 formed specific complexes with an oligonucleotide containing both an octamer motif and a Gal4 binding site (lanes 2 to 4). By chance, the Oct-2 and the Gal-GR peptides formed complexes on the Gal-octamer oligonucleotide that migrated similarly. Further, the abundance of the Gal-GR peptides and Oct-2 in these extracts was such that the binding of endogenous Oct-1 to the oligonucleotide could not be detected even with five times the maximum amount of nuclear extract used in each incubation (lane 1). WT and L501P Gal-GR binding was competed by excess Gal4 oligonucleotide but not by a consensus GRE or octamer motif (lanes 5 to 10). Similarly, Oct-2 binding was not affected by a GRE or Gal4 binding site but was competed by an unlabelled octamer motif oligonucleotide (lanes 11 to 13). When Gal-GRWT was preincubated together with Oct-2 prior to the addition of the labelled oligonucleotide, only a single, lower-mobility Gal–GRWT–Oct-2 DNA ternary complex was detected, even though the bound complex contained less than 5% of the total labelled oligonucleotide added to the incubation (lane 14). The lack of higher-mobility complexes representing the binding of Gal-GRWT or Oct-2 alone to DNA suggests that, under these binding conditions, nearly all of the GR and Oct-2 was associated with heterocomplexes in solution and that the GR–Oct-2 complexes bound coordinately to DNA. However, when Gal-GRL501P was preincubated with Oct-2, only the higher-mobility complexes containing individually bound Gal-GRL501P and Oct-2 were detected (lane 17).
Formation of the Gal–GRWT–Oct-2 low-mobility complex was specifically sensitive to the addition of unlabelled GRE-containing oligonucleotide. Addition of the GRE together with the radiolabelled Gal4-octamer motif oligonucleotide following preincubation of Oct-2–Gal–GRWT completely prevented formation of the lower-mobility ternary complex (lane 15). By contrast, the nonspecific IAP oligonucleotide had no detectable effect on the formation of the low-mobility complex (lane 16). Thus, in two separate assays, GR–Oct-1/2 binding appeared to be not just inhibited but actually disrupted by addition of a GRE to incubations containing preformed GR–Oct-1/2 complexes.
DISCUSSION
We have examined the ability of GR to bind directly to Oct-1/2 and have investigated the consequences of this protein-protein interaction for DNA binding and transcriptional activation by Oct-1/2. Our results indicate that the binding of the GR DBD to the POU domains of Oct-1 and Oct-2 dramatically potentiates the ability of the octamer factors to activate transcription in a promoter-specific manner by encouraging the occupancy of octamer motifs adjacent to GREs. Interestingly, GR–Oct-1/2 binding appeared to be incompatible with the binding of GR to a GRE. These data suggest a mechanism for transcriptional cooperativity between GR and Oct-1/2 in which protein-protein interactions between GR and the octamer factors occur in a way designed to restrict transcriptional cooperativity to promoters containing DNA binding sites for both factors.
The formation of a complex between the DNA binding domain of GR and the POU domain of Oct-1 was evidenced in several different assays. GST pulldown experiments demonstrated that full-length GR could bind specifically to the POU domains of Oct-1 and Oct-2. Binding was localized to the GR DBD and occurred in a manner that was independent of zinc coordination, as neither point mutation of zinc-coordinating cysteines or treatment of the zinc fingers with H2O2 influenced POU domain binding in vitro. However, individual mutation of two amino acids in the DNA-contact helix of finger 2 of the GR DBD (56), C500Y and L501P, did block GR–Oct-1/2 binding in vitro and in two-hybrid assays in mammalian cells. Further, the L501P mutation was sufficient to prevent the binding of full-length GR to full-length Oct-1/2 in immunoprecipitation experiments and abrogated the coordinate binding of GR and Oct-2 to DNA in EMSA.
These results suggest that a surface in the GR DBD that is distinct from, but that overlaps, the DNA binding interface of the DBD, mediates specific binding to Oct-1/2 in the cell. Overlap between the two interfaces appears to be limited, as several point mutations that interfered with GR DNA binding and/or DNA-dependent dimerization (R479K, R489K, and N491S), had no effect on POU domain binding. Further, although the mutations C500Y and L501P interfere with POU domain binding, crystallographic (56) and nuclear magnetic resonance (4) analysis of the GR DBD places these amino acids on the surface of the second DNA-contact α-helix of the GR DBD that is oriented inwards, towards the core of the DBD. It therefore seems unlikely that either amino acid directly contacts the POU domain. We suggest that it is more probable that the C500Y and L501P mutations force a change in the tertiary structure of the GR DBD that alters the configuration of the POU binding surface in a way that results in a loss of binding.
Many protein-protein interactions have also been mapped to the POU domains of Oct-1 and -2 and to the homeodomain of other homeobox proteins that affect either DNA binding site selection or the transcriptional response of these factors (18, 32, 98). The two best-characterized interactions with octamer factors are those with VP16 (42, 46) and the coactivator Bob1 (also known as Oca-B and OBF-1) (33, 57, 84), with binding to VP16 being specific to Oct-1. In additional binding studies, we have verified that GR can bind specifically to the homeodomains of Oct-1 and Oct-2 (99a). Further, in our experiments, a point mutation in the homeodomain that has previously been reported to distinguish VP16 binding (46) has no effect on GR binding (data not shown). Therefore, it seems that while the presence of a POU-specific domain increases the specificity and affinity of DNA sequence recognition by POU homeodomain proteins, additional interactions, including that observed here with GR, may still be required to direct and increase the binding of these factors to octamer motifs in the cell.
Truncation of GR from the C terminus indicated that the potential to bind Oct-1/2 was lost upon deletion into the DBD. Perhaps surprisingly, preliminary binding and two-hybrid studies attempting to delimit the N-terminal requirements for the binding of GR to Oct-1/2 suggest that the entire GR DBD may be required (99). Thus, although the zinc-coordinated tertiary structure of the GR zinc fingers was not required for octamer factor binding, other determinants within this region, or the overall folding of the DBD, are likely to prove necessary for Oct-1/2 binding.
It is interesting that the GR DBD can engage in protein-protein interactions with a number of other sequence-specific transcription factors, including AP-1, NF-κB, NF-IL6, Stat5, and Nur77 (37, 63, 68, 81, 82, 104). Moreover, these interactions are usually mediated by the DBD of the other factor as well. By contrast, interactions between GR and other factors that do not bind DNA, transcriptional coactivators or heat shock protein 90, for example (14, 38), occur outside the DBD. In vitro DNA binding experiments suggest that DBD-DBD interactions can modify the DNA binding potential of GR and/or its partner. Thus, GR binding to AP-1 modifies the DNA binding potentials of both factors, repressing binding to simple motifs in favor of composite elements (26). Similarly, GR binding to NF-κB and Nur77 represses the levels of DNA binding of these factors (37, 68). By contrast, binding to Stat5 represses GR DNA binding (82).
The consequences of the binding of GR to Oct-1/2 appear to be distinct from those of other protein-protein interactions with the GR DBD described to date. Unlike Stat5, AP-1, NF-κB, and Nur77 binding, association with Oct-1/2 is not expected to impair or modify GR DNA binding. Furthermore, the C500Y and L501P mutations that we have identified have not been reported to disrupt the binding of GR to the other factors. However, as no direct comparison has been made, a more detailed analysis of GR–Oct-1/2 binding requirements will be needed to clarify the distinction between this interaction and other protein binding events mediated by the GR DBD.
In vitro footprinting of the DNA binding of GR and Oct-1/2 indicated that GR binding to the MMTV LTR decreased the concentration of Oct-1 required to saturate the MMTV octamer motifs (8). Our results indicate that GR has a similar ability to promote the binding of Oct-1/2 to the MMTV octamer motifs on plasmid DNA in the cell. They also indicate that this recruitment of Oct-1/2 to the MMTV LTR is dependent upon L501P-sensitive GR–Oct-1/2 binding. Exonuclease footprinting of specific nuclear factor binding to a DNA template containing a single GRE and an MMTV octamer motif showed that Oct-2-dependent occupancy of the octamer motifs was strongly induced by liganded GR and did not involve any additional sequence-specific or general transcription factors. The same potentiation of binding was observed when the GR DBD was tethered to DNA by a Gal4 DNA binding site adjacent to the octamer motif. Further, in this instance, binding to the octamer motif was completely sensitive to the L501P mutation in the GR DBD, which abrogated GR–Oct-1/2 binding in other assays. The implication of these results is that the steroid-dependent occupancy of octamer motifs in the MMTV LTR observed in vivo on chromatin is likely to depend to a large extent on the association of GR and Oct-1/2. This possibility is in direct contrast to NF1 binding to the MMTV LTR, which appears to be entirely dependent on the remodelling of the chromatin structure (91).
Interestingly, the Oct-2- and GR-dependent pause site that we observed on the MMTV LTR mapped at −49, exactly between the two octamer motifs. An explanation for the location of this pause is not obvious as GR–Oct-2-dependent binding to a subcloned distal octamer motif (−61 to −51) was able to impede digestion by the exonuclease. However, this result is exactly consistent with exonuclease pausing previously reported in in vivo footprinting of MMTV LTR chromatin that also placed a steroid-dependent pause site at −49 (21). At the time, this pause was ascribed to basal transcription factor binding to the MMTV TATA box. Our results now suggest that it is more likely that this pause reflects the binding of octamer factors to the MMTV LTR.
While our results agree with almost all of the in vitro and in vivo GR–Oct-1/2 DNA binding data obtained to date, they at first glance appear to conflict with the results of a previous report in which Oct-1 binding to the transiently transfected MMTV LTR was not detected to increase in response to hormone treatment (50). However, in these experiments, exonuclease III was the processive enzyme employed in the footprinting. A subsequent report demonstrated that the −49 pause site, and thus Oct-1/2 binding, can be detected only with λ exonuclease (61).
The extent to which the promotion of octamer factor DNA binding by GR affected transcriptional activation by Oct-1/2 was examined in transient transfection assays. The use of truncated GR-Gal4 DBD fusion proteins unable to activate reporter gene expression when they were expressed alone allowed us to directly correlate octamer factor DNA binding with transcriptional activation. With the simple reporter system employed, the L501P-sensitive promotion of octamer factor DNA binding directly correlated with an increase in the transcriptional activation potential of Oct-1/2 of close to 2 orders of magnitude. This transcriptional effect was entirely dependent upon the presence of Gal4 DNA binding sites in the reporter gene promoter. Therefore, we propose that binding to GR provides a strong selective advantage for the activation of transcription by octamer factors from glucocorticoid-responsive promoters. Furthermore, these results suggest one way in which transcriptional activation by a constitutively active nuclear factor can be made dependent on inducible signals.
An unusual feature of the potentiation of Oct-1/2 DNA binding by GR was that the GR–Oct-1/2 association in solution was at least significantly weakened by the binding of GR to a GRE. In both GST pulldown experiments and EMSA, the addition of a GRE but not nonspecific DNAs resulted in the disruption of GR–Oct-1/2 binding. This result has not previously been reported for other instances of cooperative DNA binding of transcription factors. One obvious potential advantage to weakening the link between GR and Oct-1/2 following the binding of GR to DNA would be a tighter control of gene expression by reducing the potential for Oct-1/2 to influence the transcription of GR-responsive genes lacking octamer motifs. Thus, Oct-1 and Oct-2 are not expected to act as transcriptional coactivators of GR, despite their ability to bind in solution. A potential disadvantage of this effect is predicted to be the inability of GR to recruit Oct-1/2 to octamer motifs following the DNA binding of GR. However, as demonstrated in our experiments and by in vivo footprinting experiments performed with cells expressing normal levels of GR and Oct-1 (91), this limitation does not appear to be a major detriment to recruiting Oct-1/2 to the MMTV promoter.
Two potential ways in which GR–Oct-1/2 binding in solution may lead to the simultaneous binding of GR and Oct-1/2 to promoters are summarized schematically in Fig. 10. In the first instance (Fig. 10A), association of GR and Oct-1/2 DBDs in solution occurs in a manner which leaves the GR DBD free to recognize and initiate binding to GREs (step i). However, binding of GR to the GRE is incompatible with continued association with Oct-1/2 and leads to the release of the octamer factors from GR (step ii). When release occurs within transcriptional regulatory regions containing octamer motifs, Oct-1 and -2 binding is encouraged through an increase in their local concentrations (step iii). In the absence of an octamer motif, however, the octamer factor is not retained. One additional advantage of this mechanism is that it minimizes the requirement for precise juxtaposition of the GREs and octamer motifs. While we have not performed a rigorous evaluation of this possibility, the promotion of Oct-2 binding to transiently transfected plasmid DNAs by GR was not observed to be sensitive to the spacing between, or the orientations of, the GREs and octamer motifs used in this study.
FIG. 10.
Two possible mechanisms for the recruitment of Oct-1/2 by GR to octamer motifs adjacent to GREs. (A) Upon treatment with steroid, liganded GR is transported into the nucleus, where it can form a complex with Oct-1/2 through an interface that involves the zinc fingers of GR and the POU domain of Oct-1/2 (step i). Mutations L501P and C500Y in the GR DBD specifically abrogate octamer factor binding. Although the binding to Oct-1/2 requires determinants in the GR DBD, GR complexed with Oct-1/2 retains the ability to recognize and bind to a GRE (step ii). Binding of GR to a GRE is preferred to, and destabilizes, binding to Oct-1/2. During the course of GR DNA binding, the interaction between GR and Oct-1/2 is weakened in a manner that leads to the release of the octamer factor from GR in vitro. In the absence of nearby octamer motifs, the octamer factor is not retained. However, in the presence of an adjacent octamer motif, the release of Oct-1/2 directly promotes octamer motif binding (step iii). (B) The second mechanism is similar to that shown in panel A, except that the associated GR and octamer factors bind coordinately to transcriptional regulatory regions containing GREs and octamer motifs linked in cis. Coordinate binding may potentially occur in preference to binding by the individual factors to regulatory regions with binding sites for only GR or Oct-1/2.
A second possibility is that on transcriptional regulatory regions containing both octamer motifs and GREs, associated GR and Oct-1/2 initiate DNA binding at the same time (Fig. 10B). This joint binding would be a more direct way to facilitate octamer factor binding and is supported by our results demonstrating that Gal-GR fusion proteins also promoted the binding of Oct-2 adjacent to Gal4 binding sites. However, another report that full-length GR represses the ability of Oct-1 to bind to octamer motifs in the absence of GREs (45) suggests that GR may still need to release the octamer factor for DNA binding to occur. Interestingly, in both the previous study and in preliminary experiments that we have performed (73), the GR DBD alone is unable to repress octamer factor DNA binding in vitro or to repress octamer factor-dependent transcription in transfection experiments. Thus, it seems that the repression of octamer factor DNA binding by full-length GR depends on determinants outside the minimal region of GR required for octamer factor binding in solution. Finally, while not indicated in Fig. 10, neither model for recruitment of Oct-1/2 to DNA by GR specifically excludes the possibility of continued, but fundamentally altered, interaction between GR and Oct-1 or Oct-2 when both factors are bound to their respective response elements.
One approach that is expected to provide additional insight into the mechanism through which GR–Oct-1/2 association promotes the binding of Oct-1/2 to DNA would be to examine how complex formation affects the kinetics of Oct-1/2 DNA binding. For example, continued association of GR and Oct-1/2 following DNA binding might be expected to decrease the rate of dissociation of Oct-1/2 from octamer motifs. By contrast, the release of Oct-1/2 into solution adjacent to octamer motifs is expected to increase the on rate for Oct-1/2 DNA binding.
Finally, our results suggest that the recruitment of Oct-1/2 to DNA by GR plays a major role in promoting the occupancy of octamer motifs in the MMTV and other glucocorticoid-responsive promoters in vivo on chromatin templates. However, in order to directly evaluate this prediction, it will be necessary to identify mutations in the GR DBD that block Oct-1/2 binding without affecting the binding of GR to DNA or the ability of GR to promote the rearrangement of chromatin.
ACKNOWLEDGMENTS
We thank K. Yamamoto and W. Herr for providing us with many of the GR and Oct-1/2 plasmids required to complete this work. We also thank P. Sassone-Corsi for the gift of the CREB plasmid, E. Fearon for the mammalian two-hybrid plasmids, J. Bell for pCRIIMTG, and M. Petkovitch for pTL2. The critical comments of Y. Lefebvre and M. Ekker on the manuscript are particularly appreciated.
This work was funded by a grant from the Medical Research Council of Canada (MRC) to R.J.G.H. R.J.G.H. is a Scholar of the MRC and the Cancer Research Society Inc. M.E.L. is a Junior Fellow of the National Cancer Institute of Canada. C.S.-P. holds an MRC-Arthritis Society postdoctoral fellowship, and J.M.W. was given an L. Siminovitch postdoctoral training award. G.G.P. was funded through an MRC graduate studentship.
REFERENCES
- 1.Andrews N C, Faller D V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archer T K, Cordingley M G, Wolford R G, Hager G L. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–698. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer T K, Lefebvre P, Wolford R G, Hager G L. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–1576. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 4.Baumann H, Paulsen K, Kovacs H, Berglund H, Wright A P H, Gustafsson J-Å, Härd T. Refined solution structure of the glucocorticoid receptor DNA-binding domain. Biochemistry. 1993;32:13463–13471. doi: 10.1021/bi00212a011. [DOI] [PubMed] [Google Scholar]
- 5.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 6.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beato M, Herrlich P, Schütz G. Steroid receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 8.Brüggemeier U, Kalff M, Franke S, Scheidereit C, Beato M. Ubiquitous transcription factor OTF-1 mediates induction of the MMTV promoter through synergistic interaction with hormone receptors. Cell. 1991;64:565–572. doi: 10.1016/0092-8674(91)90240-y. [DOI] [PubMed] [Google Scholar]
- 9.Brüggemeier U, Rogge L, Winnacker E L, Beato M. Nuclear factor I acts as a transcription factor on the MMTV promoter but competes with steroid hormone receptors for DNA binding. EMBO J. 1990;9:2233–2239. doi: 10.1002/j.1460-2075.1990.tb07393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buetti E. Stably integrated mouse mammary tumor virus long terminal repeat DNA requires the octamer motifs for basal promoter activity. Mol Cell Biol. 1994;14:1191–1203. doi: 10.1128/mcb.14.2.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buetti E, Kühnel B, Diggelmann H. Dual function of a nuclear factor I binding site in MMTV transcription regulation. Nucleic Acids Res. 1989;17:3065–3078. doi: 10.1093/nar/17.8.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cato A C B, Miksicek R, Schütz G, Arnemann J, Beato M. The hormone regulatory element of mouse mammary tumor virus mediates progesterone induction. EMBO J. 1986;5:2237–2240. doi: 10.1002/j.1460-2075.1986.tb04490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cato A C B, Skroch P, Weinmann J, Butkeraitis P, Ponta H. DNA sequences outside the receptor-binding sites differently modulate the responsiveness of the mouse mammary tumour virus promoter to various steroid hormones. EMBO J. 1988;7:1403–1410. doi: 10.1002/j.1460-2075.1988.tb02957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 15.Chalepakis G, Arnemann J, Slater E, Bruller H J, Gross B, Beato M. Differential gene activation by glucocorticoids and progestins through the hormone regulatory element of mouse mammary tumor virus. Cell. 1988;53:371–382. doi: 10.1016/0092-8674(88)90157-2. [DOI] [PubMed] [Google Scholar]
- 16.Chan S K, Jaffe L, Capovilla M, Botas J, Mann R S. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 17.Chavez S, Beato M. Nucleosome-mediated synergism between transcription factors on the mouse mammary tumor virus promoter. Proc Natl Acad Sci USA. 1997;94:2885–2890. doi: 10.1073/pnas.94.7.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleary M A, Herr W. Mechanisms for flexibility in DNA sequence recognition and VP16-induced complex formation by the Oct-1 POU domain. Mol Cell Biol. 1995;15:2090–2100. doi: 10.1128/mcb.15.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-1: activation of a transcriptionally silent motif through Oct-1 and VP-16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 20.Clerc R, Corcoran L, LeBowitz H, Baltimore D, Sharp P. The B-cell specific Oct-2 protein contains a POU box and homeo-box-type domains. Genes Dev. 1988;2:1570–1581. doi: 10.1101/gad.2.12a.1570. [DOI] [PubMed] [Google Scholar]
- 21.Cordingley M G, Riegel A T, Hager G L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell. 1987;48:261–270. doi: 10.1016/0092-8674(87)90429-6. [DOI] [PubMed] [Google Scholar]
- 22.Dalman F C, Bresnick E H, Patel P D, Perdew G H, Watson S J, Jr, Pratt W B. Direct evidence that the glucocorticoid receptor binds to hsp90 at or near the termination of receptor translation in vitro. J Biol Chem. 1989;264:19815–19821. [PubMed] [Google Scholar]
- 23.Dang C V, Barrett J, Villa-Garcia M, Resar L M, Kato G J, Fearon E R. Intracellular leucine zipper interactions suggest c-Myc hetero-oligomerization. Mol Cell Biol. 1991;11:954–962. doi: 10.1128/mcb.11.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Groot R P, Delmas V, Sassone-Corsi P. DNA bending by transcription factors CREM and CREB. Oncogene. 1994;9:463–468. [PubMed] [Google Scholar]
- 25.Denis M, Poellinger L, Wilstom A-C, Gustafsson J-Å. Requirement of hormone for thermal conversion of the glucocorticoid receptor to the DNA-binding state. Nature. 1988;333:686–688. doi: 10.1038/333686a0. [DOI] [PubMed] [Google Scholar]
- 26.Diamond M I, Miner J N, Yoshinaga S K, Yamamoto K R. c-Jun and c-Fos levels specify positive or negative glucocorticoid regulation from a composite GRE. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 27.Falkner F G, Zachau H G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984;310:71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- 28.Falzon M, Kuff E L. A variant binding sequence for transcription factor EBP-80 confers increased promoter activity on a retroviral long terminal repeat. J Biol Chem. 1990;265:13084–13090. [PubMed] [Google Scholar]
- 29.Freedman L P, Luisi B F, Korszun Z R, Basavappa R, Sigler P B, Yamamoto K R. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- 30.Giffin W, Torrance H, Rodda D J, Préfontaine G G, Pope L, Haché R J G. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 31.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 32.Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- 33.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 34.Guichet A, Copeland J W, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause H M, Ephrussi A. The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 35.Hassan B, Li L, Bremer K A, Chang W, Pinsonneault J, Vaessin H. Prospero is a panneural transcription factor that modulates homeodomain protein activity. Proc Natl Acad Sci USA. 1997;94:10991–10996. doi: 10.1073/pnas.94.20.10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayashi S, Scott M P. What determines the specificity of action of Drosophila homeo domain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- 37.Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf H J, Herrlich P, Cato A C. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howard K J, Holley S J, Yamamoto K R, Distelhorst C W. Mapping the HSP90 binding region of the glucocorticoid receptor. J Biol Chem. 1990;265:11928–11935. [PubMed] [Google Scholar]
- 39.Hutchison K A, Matic G, Czar M J, Pratt W B. DNA-binding and non-DNA-binding forms of the transformed glucocorticoid receptor. J Steroid Biochem Mol Biol. 1992;41:715–718. doi: 10.1016/0960-0760(92)90410-k. [DOI] [PubMed] [Google Scholar]
- 40.Kalff M, Gross B, Beato M. Progesterone receptor stimulates transcription of mouse mammary tumour virus in a cell-free system. Nature. 1990;344:360–362. doi: 10.1038/344360a0. [DOI] [PubMed] [Google Scholar]
- 41.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 42.Kristie T M, Sharp P A. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 43.Kurokawa R, Soderström M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 44.Kurokawa R, Yu V C, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld M G, Glass C K. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 45.Kutoh E, Strömstedt P-E, Poellinger L. Functional interference between the ubiquitous and constitutive octamer transcription factor 1 (OTF-1) and the glucocorticoid receptor by direct protein-protein interaction involving the homeo subdomain of OTF-1. Mol Cell Biol. 1992;12:4960–4969. doi: 10.1128/mcb.12.11.4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai J-S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeo domain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 47.Lai J-S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. Evolution of the nuclear receptor gene superfamily. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leach K L, Dahmer M K, Hammond N D, Sando J J, Pratt W B. Molybdate inhibition of glucocorticoid receptor inactivation and transformation. J Biol Chem. 1979;254:11884–11890. [PubMed] [Google Scholar]
- 50.Lee H L, Archer T K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994;14:32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J W, Moffitt P G, Morley K L, Peterson D O. Multipartite structure of a negative regulatory element associated with a steroid hormone-inducible promoter. J Biol Chem. 1991;266:24101–24108. [PubMed] [Google Scholar]
- 52.Lemieux, M. E., G. G. Préfontaine, L. Pope, and R. J. G. Haché. Unpublished data.
- 53.Li P, He X, Gerrero M R, Mok M, Aggarwal A, Rosenfeld M G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993;7:2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 54.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 55.Lu Q, Wallrath L L, Elgin S C. The role of a positioned nucleosome at the Drosophila melanogaster hsp26 promoter. EMBO J. 1995;14:4738–4746. doi: 10.1002/j.1460-2075.1995.tb00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 57.Luo X, Ikeda Y, Parker K L. A cell-specific nuclear receptor is essential for adrenal and gonadal development and sexual differentiation. Cell. 1994;77:481–490. doi: 10.1016/0092-8674(94)90211-9. [DOI] [PubMed] [Google Scholar]
- 58.Miesfeld R, Rusconi S, Godowski P J, Maler B A, Okret S, Wilkström A-C, Gustafsson J-Å, Yamamoto K R. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell. 1986;46:389–399. doi: 10.1016/0092-8674(86)90659-8. [DOI] [PubMed] [Google Scholar]
- 59.Miksicek R, Borgmeyer U, Nowock J. Interaction of the TGGCA-binding protein with upstream sequences is required for efficient transcription of mouse mammary tumor virus. EMBO J. 1987;6:1355–1360. doi: 10.1002/j.1460-2075.1987.tb02375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 61.Mymryk J, Archer T. Detection of transcription factor binding in vivo using lambda exonuclease. Nucleic Acids Res. 1994;22:4344–4345. doi: 10.1093/nar/22.20.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Näär A M, Boutin J M, Lipkin S M, Yu V C, Holloway J M, Glass C K, Rosenfeld M G. The orientation and spacing of core DNA-binding motifs dictate selective transcriptional responses to three nuclear receptors. Cell. 1991;65:1267–1279. doi: 10.1016/0092-8674(91)90021-p. [DOI] [PubMed] [Google Scholar]
- 63.Nishio Y, Isshiki H, Kishimoto T, Akira S. A nuclear factor for interleukin-6 expression (NF-IL6) and the glucocorticoid receptor synergistically activate transcription of the rat alpha 1-acid glycoprotein gene via direct protein-protein interaction. Mol Cell Biol. 1993;13:1854–1862. doi: 10.1128/mcb.13.3.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ostlund Farrants A-K, Blomquist P, Kwon H, Wrange Ö. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parks W P, Hubbell E S, Goldberg R J, O’Neill F J, Scolnick E M. High frequency variation in mammary tumor virus expression in cell culture. Cell. 1976;8:87–93. doi: 10.1016/0092-8674(76)90189-6. [DOI] [PubMed] [Google Scholar]
- 66.Pearce D, Yamamoto K R. Mineralocorticoid and glucocorticoid receptor activities distinguished by nonreceptor factors at a composite response element. Science. 1993;259:1161–1165. doi: 10.1126/science.8382376. [DOI] [PubMed] [Google Scholar]
- 67.Perlmann T, Wrange Ö. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988;7:3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Philips A, Maira M, Mullick A, Chamberland M, Lesage S, Hugo P, Drouin J. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol Cell Biol. 1997;17:5952–5959. doi: 10.1128/mcb.17.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Picard D, Yamamoto K R. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Piña B, Brüggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 71.Power S C, Cereghini S. Positive regulation of the vHNF1 promoter by the orphan receptors COUP-TFI/Ear3 and COUP-TFII/Arp1. Mol Cell Biol. 1996;16:778–791. doi: 10.1128/mcb.16.3.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pratt W B, Toft D O. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev. 1997;18:306–360. doi: 10.1210/edrv.18.3.0303. [DOI] [PubMed] [Google Scholar]
- 73.Préfontaine, G. G., and R. J. G. Haché. Unpublished data.
- 74.Pugh B. Mechanisms of transcription complex assembly. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 75.Reeves R, Gorman C M, Howard B. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 1985;13:3599–3615. doi: 10.1093/nar/13.10.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rusconi S, Yamamoto K R. Functional dissection of the hormone and DNA-binding activities of the glucocorticoid receptor. EMBO J. 1987;6:1309–1315. doi: 10.1002/j.1460-2075.1987.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sadowski I, Ma J, Trienzenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 78.Scheidereit C, Geisse S, Westphal H M, Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983;304:749–752. doi: 10.1038/304749a0. [DOI] [PubMed] [Google Scholar]
- 79.Schena M, Freedman L P, Yamamoto K R. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes Dev. 1989;3:1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- 80.Schüle R, Müller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 81.Schüle R, Rangarajan P, Kliewer S, Ransone L J, Bolado J, Yang N, Verma I M, Evans R M. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 82.Stöcklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]
- 83.Strahle U, Klock G, Schütz G. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci USA. 1987;84:7871–7875. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 85.Sturm R A, Das G, Herr W. The ubiquitous octamer-binding protein Oct-1 contains a POU domain with a homeo box subdomain. Genes Dev. 1988;2:1582–1599. doi: 10.1101/gad.2.12a.1582. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 87.Tienrungroj W, Meshinchi S, Sanchez E R, Pratt S E, Grippo J F, Holmgren A, Pratt W B. The role of sulfhydryl groups in permitting transformation and DNA binding of the glucorticoid recepetor. J Biol Chem. 1987;262:6992–7000. [PubMed] [Google Scholar]
- 88.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 89.Toohey M G, Lee J W, Huang M, Peterson D O. Functional elements of the steroid hormone-responsive promoter of mouse mammary tumor virus. J Virol. 1990;64:4477–4488. doi: 10.1128/jvi.64.9.4477-4488.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Treisman R, Marais R, Wynne J. Spatial flexibility in ternary complexes between SRF and its accessory proteins. EMBO J. 1992;11:4631–4640. doi: 10.1002/j.1460-2075.1992.tb05565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Truss M, Bartsch J, Schelbert A, Haché R J G, Beato M. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 1995;14:1737–1751. doi: 10.1002/j.1460-2075.1995.tb07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Truss M, Chalepakis G, Beato M. Interplay of steroid hormone receptors and transcription factors on the mouse mammary tumor virus promoter. J Steroid Biochem Mol Biol. 1992;43:365–378. doi: 10.1016/0960-0760(92)90071-p. [DOI] [PubMed] [Google Scholar]
- 93.Truss M, Chalepakis G, Slater E P, Mader S, Beato M. Functional interaction of hybrid response elements with wild-type and mutant steroid hormone receptors. Mol Cell Biol. 1991;11:3247–3258. doi: 10.1128/mcb.11.6.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Dijk M A, Murre C. Extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 96.van Dijk M A, Peltenburg L T, Murre C. Hox gene products modulate the DNA binding activity of Pbx1 and Pbx2. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]
- 97.Verrijzer C P, Alkema M J, van Weperen W W, Van Leeuwen H C, Strating M J, van der Vliet P C. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 1992;11:4993–5003. doi: 10.1002/j.1460-2075.1992.tb05606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walker S, Hayes S, O’Hare P. Site-specific conformational alteration of the oct-1 POU domain-DNA complex as the basis for differential recognition by vmw65 (VP16) Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 99.Walther, R., and R. J. G. Haché. Unpublished data.
- 99a.Wang, J. M., et al. Unpublished data.
- 100.Wegner M, Drolet D W, Rosenfeld M G. POU-domain proteins: structure and function of developmental regulators. Curr Opin Cell Biol. 1993;5:488–498. doi: 10.1016/0955-0674(93)90015-i. [DOI] [PubMed] [Google Scholar]
- 101.Wieland S, Döbbeling U, Rusconi S. Interference and synergism of glucocorticoid receptor and octamer factors. EMBO J. 1991;10:2513–2521. doi: 10.1002/j.1460-2075.1991.tb07791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wrange Ö, Carlstedt-Duke J, Gustafsson J Å. Stoichiometric analysis of the specific interaction of the glucocorticoid receptor with DNA. J Biol Chem. 1986;261:11770–11778. [PubMed] [Google Scholar]
- 103.Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984;309:229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- 104.Yang-Yen H F, Chambard J C, Sun Y L, Smeal T, Schmidt T J, Drouin J, Karin M. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 105.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon N, Pick L. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]