Abstract
Attention network theory proposes three distinct types of attention—alerting, orienting, and control—that are supported by separate brain networks and modulated by different neurotransmitters, that is, norepinephrine, acetylcholine, and dopamine. Here, we explore the extent of cortical, genetic, and molecular dissociation of these three attention systems using multimodal neuroimaging. We evaluated the spatial overlap between fMRI activation maps from the attention network test (ANT) and cortex‐wide gene expression data from the Allen Human Brain Atlas. The goal was to identify genes associated with each of the attention networks in order to determine whether specific groups of genes were co‐expressed with the corresponding attention networks. Furthermore, we analyzed publicly available PET‐maps of neurotransmitter receptors and transporters to investigate their spatial overlap with the attention networks. Our analyses revealed a substantial number of genes (3871 for alerting, 6905 for orienting, 2556 for control) whose cortex‐wide expression co‐varied with the activation maps, prioritizing several molecular functions such as the regulation of protein biosynthesis, phosphorylation, and receptor binding. Contrary to the hypothesized associations, the ANT activation maps neither aligned with the distribution of norepinephrine, acetylcholine, and dopamine receptor and transporter molecules, nor with transcriptomic profiles that would suggest clearly separable networks. Independence of the attention networks appeared additionally constrained by a high level of spatial dependency between the network maps. Future work may need to reconceptualize the attention networks in terms of their segregation and reevaluate the presumed independence at the neural and neurochemical level.
Keywords: Aceylcholine, Alerting, Attention, Attention Network Test, Control, Cortical Gene Expression, Dopamine, Multimodal Imaging, Network Neuroscience, Neuromodulation, Norepinephrine, Orienting
We examined the hypotheses derived from attention network theory, which suggest distinct neuromodulation of the alerting, orienting, and control attention networks. Through a comparison of attention network maps with relevant gene expression maps and receptor/transporter maps obtained from PET imaging, we found that these relationships were not supported.
1. INTRODUCTION
Attention prioritizes the processing of goal‐relevant over less relevant information which ensures adaptive behavior in information‐rich environments (Cowan, 1999; Posner & Fan, 2008). Attention network theory (ANT) distinguishes between three different types of attention: (i) alerting attention, which is the initiation of a state of heightened alertness in anticipation of upcoming stimuli; (ii) orienting attention, which is the shift of the attentional focus to prioritize information processing at a particular spatial location, and (iii) control attention, which is the selective amplification of relevant aspects of a stimulus when irrelevant information is present (Posner & Dehaene, 1994; Posner & Petersen, 1990). At the brain level, each type of attention is supported by distinct and distributed set of cortical and subcortical regions: the three attention networks alerting, orienting, and control (Petersen & Posner, 2012).
A core tenet of ANT is the presumed independence of the three attention networks. This independence is thought to arise from dissociating effects of large neuromodulatory systems: alerting is modulated by norepinephrine, orienting by acetylcholine, and control by dopamine (Posner & Rothbart, 2007). While the relevance of all three neuromodulators in attention is well established (Noudoost & Moore, 2011; Robbins, 1997), the evidence for the assumed dissociation and specificity in regard to the three attention networks is less conclusive. Experimental work with the ANT (Fan et al., 2002), a behavioral protocol to assess the efficiency of all three attention networks simultaneously, was unable to detect influences of the potent acetylcholine agonist nicotine on any of the three attention networks (see McCormick, 2022, for review). Furthermore, direct manipulations of the noradrenergic system by blocking the norepinephrine reuptake impact orienting attention but not alerting and control (Reynaud et al., 2019), while indirect measure of locus coeruleus activity suggest an involvement of norepinephrine in all three attention networks (Gabay et al., 2011; Geva et al., 2013). Only for dopamine, the evidence is more favorable, with clear demonstrations of increased dopamine release during task performance (Badgaiyan & Wack, 2011) and dissociating effects in patients with Parkinson's disease whose cardinal symptom is dopaminergic impairment (Yang et al., 2022). It needs to be noted, however, that many studies have used rather indirect approaches to the neuromodulators and that neuroimaging work has been able to detect neuromodulation at the brain level in the absence of behavioral effects (Ikeda et al., 2017; Thienel et al., 2009).
The distal causation of these neuromodulatory effects is likely of genetic origin (Green et al., 2008). In line with the behavioral genetics literature (Polderman et al., 2015), it has been shown that the behavioral efficiency of the attention networks is heritable (Fan et al., 2001). Several studies have attempted to implicate candidate polymorphisms on genes with direct relevance for the neuromodulatory systems in behavioral and neural markers of attention and have found some evidence for the hypothesized dissociation (Fan et al., 2003; Fossella et al., 2002; Rueda et al., 2005). The available evidence, however, is not conclusive (Green et al., 2008; Posner et al., 2014) and genetic association studies on candidate genes have been disputed more recently, despite their apparent face value (Border et al., 2019; Montag et al., 2020; S. R. Moore, 2017).
Given the paucity of supporting evidence for the molecular and genetic dissociation of the three attention systems in humans, we decided to revisit the hypothesis by utilizing two novel and complementary approaches: a comprehensive examination of brain‐wide gene transcriptomic activity and the expression of receptor and transporter molecules as discerned through molecular neuroimaging.
ANT theory's underpinnings predominantly rely on systemic pharmacological manipulations and exhibit some ambiguity in elucidating the precise mechanisms through which neuromodulatory signals affect the attention networks. All three proposed neuromodulatory systems share common characteristics, including the localized origin of synthesizing neurons in the brainstem and midbrain, and widespread target sites throughout the cortex and subcortical areas such as the basal ganglia and the thalamus, where these neurotransmitters exert their neuromodulatory influences at noradrenergic, cholinergic, and dopaminergic synapses. Consequently, the chemical neuromodulation of attention networks could occur through direct mechanisms, involving the modification or regulation of synaptic connections within the attention networks themselves, or through indirect pathways, influencing neural circuitry in regions such as the striatum and thalamus, which subsequently impact the attention networks via other neurotransmitters like glutamate or GABA (Ross & Van Bockstaele, 2021; Speranza et al., 2021; Tang et al., 2012).
Since ANT theory designates the cortical projection sites of ascending dopaminergic, cholinergic, and noradrenergic fibers as central hubs in the attention networks (Posner & Fan, 2008), it seems reasonable to begin by exploring the possibility of direct neuromodulation. Under this premise, we posit that the cortex‐wide activation profile of each attention network, as activated by the ANT (Fan et al., 2002; Fan et al., 2005), corresponds to the relative availability of pertinent receptor and transporter molecules (approach 1) and the relative expression of relevant genes in brain tissue (approach 2). Assuming that the molecular neuromodulation of attention networks occurs at the synaptic levels, we hypothesize that (1) brain regions that activate during alerting show higher availability of the norepinephrine transporter (NET) and higher expression of genes involved in norepinephrine binding, (2) that brain regions that activate during orienting show higher availability of nicotinergic acetylcholine receptors and vesilcular acetylcholine transporters, and a higher expression of genes involved in acetylcholine‐gated cation‐selective channel activity, and (3) that brain regions that activate during attention control show higher availability of the dopamine transporter (DAT) and dopamine D1 and D2 receptors, and higher expression of genes involved in dopamine binding. We expect stronger covariation between task‐evoked activity within each attentional domain and the mentioned gene sets as compared to randomly defined gene sets of equal size. We also expect stronger covariation between task‐evoked activity within each attentional domain and receptor/transporter availability as compared to random null models of the attention networks. Moreover, we expect specificity of the associations in a way that gene sets and receptor/transporter availability with presumed relevance for one attentional domain is more strongly related to this domain than to the other two domains. Finally, given the assumed independence of the three attention networks, we expect (4) that the genome‐wide transcriptomic signatures of the attention networks are uncorrelated.
To this end, we utilize a sample of healthy volunteers who completed the ANT during functional magnetic resonance imaging (Markett et al., 2022), publicly available maps of group‐level receptor and transporter distribution maps from positron emission tomography (Hansen, Shafiei, et al., 2022), and microarray measures for over 20,000 genes measured at 3702 locations in the brains of six donors as provided by the Allen Human Brain Atlas (AHBA) and curated by the abagen toolbox (Hawrylycz et al., 2012; Markello et al., 2021).
2. METHODS
2.1. Functional imaging data set
We used the publicly available (https://osf.io/st9ae/) group‐level attention network maps from our previous study (Markett et al., 2022). The maps were obtained from N = 78 healthy young volunteers (n = 35 female, n = 43 male, mean age M = 26.18, SD = 5.34) who completed the revised ANT (see next paragraph for details) during fMRI. All participants provided informed written consent and received remuneration. The study protocol adhered to the Declaration of Helsinki and was approved by a local ethics committee.
2.2. Attentional network test
The maps were derived by using the ANT (Xuan et al., 2016). The task followed a 4 × 2 design with the factors cueing condition (no cue, double cue, valid spatial cue, invalid spatial cue) and target (congruent flanker, incongruent flanker). Participants responded to a total of 288 trials, split into four sessions of 78 trials each. A typical trial sequence is shown in Figure 1. Throughout each trial, participants were instructed to maintain fixation on a central fixation cross. Target stimuli appeared in one out of two boxes presented laterally to the fixation cross and included five arrows pointing either to the left or to the right. Participants were instructed to indicate as fast and as accurate as possible via button press whether the most central of the five arrows pointed left or right while ignoring the four flanking arrows that could either point into the same (congruent) or opposite direction (incongruent). Shortly before target onset, one out of four different cues was presented that carried information that a target was about to appear (temporal double cue) or where the target was about to appear (spatial cue). Spatial cues were either valid (i.e., giving correct information on the target location) or invalid (i.e., pointing at a location where the target did not appear). Some targets were preceded by no cue as a baseline condition. Cues were presented for 100 ms, the onset asynchrony between cues and targets was 0, 400, or 800 ms, targets were presented for 500 ms with an additional response window of 1200 ms, and trials were spaced with a jittered interval of 4000 ms on average, systematically sampled from a distribution ranging from 2000 to 12,000 ms. There were 48 trials with double cue, 48 trials with invalid cue, 48 trials with no cue, and 144 trials with valid cues. We realized an equal proportion of congruent and incongruent targets (144 each). The combinations of cue‐target asynchronies, target location, and flanker types were counterbalanced for each cue condition. For more details on stimulus dimension and timing, we refer to our previous work (Markett et al., 2022).
FIGURE 1.
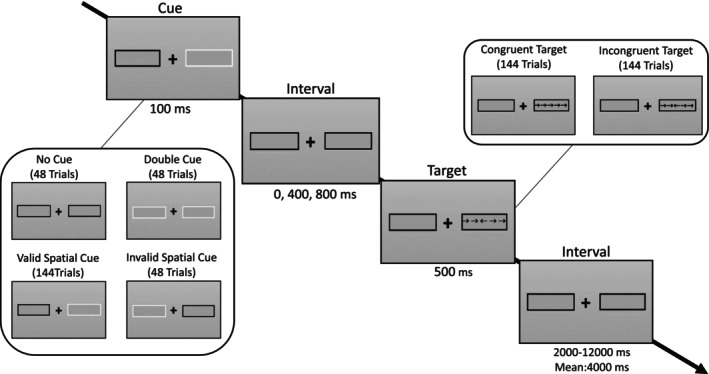
Overview of the stimuli and their timing within a standard trial sequence. Each trial commenced with a 100 ms display of one of three cues: no cue, a double cue, or a spatial cue. Following this, there was a cue‐target interval of 0, 400, or 800 ms, after which five arrows were presented for 500 ms as the target stimulus. Participants were required to indicate, through button presses, whether the central arrow pointed to the left or right. In half of the trials, the flanking arrows were congruent, while in the other half, they were incongruent. The time elapsed between the offset of the target and the onset of the subsequent cue was a jittered interval, with a mean duration of 4000 ms across trials and a range of 2000–12,000 ms. The targets appeared at either the cued position (in cases of valid spatial cues) or at an uncued position (in cases of invalid spatial cues). The experiment consisted of a total of 288 trials, organized across four separate runs.
2.3. Functional MRI acquisition and preprocessing
Brain images were collected on a 3T Siemens Prisma scanner equipped with a 32 channel had coil. We used the following pulse sequences from the HCP‐Lifespan project (Glasser et al., 2013; Harms et al., 2018): a T1‐weighted structural image (Multiecho MPRAGE, voxel size 0.8 mm, isotropic, time to repeat TR = 2.4 s, time to echo TE = 22 ms, flip angle 8°), a T2‐weighted structural image (SPACE, voxel size 0.8 mm isotropic, TR = 3.2 s, TE = 563 ms, flip angle 120°), and four runs of task fMRI, including two spin echo fieldmaps (A‐P and P‐A encoding) followed by BOLD fMRI (multiband echoplanar, 72 slices, TR = 800 ms, voxel size 2 mm isotropic, TE = 37 ms, flip angle 52°, A‐P encoding direction).
Images were preprocessed with the HCP minimal preprocessing pipelines (v4.1) using Freesurfer (v6.0) and FSL (6.0.1) under Linux Debian 10. Preprocessing scripts are described in Glasser et al. (2013) and can be obtained from Github (github.com/Washington-University/HCPpipelines).
T1‐ and T2‐weighted images were corrected for gradient distortions, aligned, brain extracted, bias field corrected, and registered to MNI space using nonlinear transformation. We then ran the images through HCP's Freesurfer pipeline with improved brain extraction, alignment, and adjustment of the white matter surface. Cortical surfaces were subsequently registered to template space based on cortical folding (MSMsulc, Robinson et al., 2018) and downsampled to the 32k_LR surface space. Functional images and the corresponding field maps were processed with the fMRIVolume pipeline which included correction for gradient distortions, motion, and EPI image distortions, followed by co‐registration with the T1 structural image and normalization to MNI volumetric space using MSMsulc. All transformations were applied in a single step. Following intensity normalization to their global 4D mean and masking, volume time series underwent additional processing using the fMRISurface pipeline. This processing involved creating individual CIFTI dense time series grayordinate files. During this step, we applied light volume and surface‐based smoothing, employing a Gaussian filter with a full width at half maximum of 4 mm.
2.4. Attention network maps
We conducted first‐level analyses in SPM12 (www.fil.ion.ucl.ac.uk/) using a general linear model. To facilitate the analysis, we converted surface images into “fakevolumetric” nifti‐images. Condition‐specific regressors were generated by convolving delta functions with SPM's canonical hemodynamic response function. Adhering to the ANT's design featuring four types of cues and two types of targets, we established separate regressors to represent the onsets of the following events: Congruent targets following double cues, congruent targets following valid cues, congruent targets following invalid cues, congruent targets following no cues, incongruent targets following double cues, incongruent targets following valid cues, incongruent targets following invalid cues, and incongruent targets following no cues. In addition to these eight regressors, we introduced an extra regressor for error trials, 12 regressors to account for the six head motion parameters and their temporal derivatives, and one constant per run. The three attention networks were operationalized through linearly weighted contrasts on estimated beta images from first‐level analyses in SPM12: the alerting network was obtained by contrasting the double cue minus the no cue condition across target conditions. The orienting network was defined as the validity effect by contrasting the invalid cue minus valid cue condition across targets. The control network was obtained by contrast incongruent minus congruent targets across cue conditions. Group‐level maps were obtained by submitting the individual contrast images, using the Sandwich Estimator (SwE) Toolbox for SPM12 (Guillaume et al., 2014). Specifically, we implemented a modified SwE procedure that included a type c small sample size correction and a wild bootstrapping procedure consisting of 999 bootstraps.
2.5. Cortical parcellation
Linking imaging data from different modalities (functional activation maps, PET maps, gene‐expression data) requires a brain parcellation as a common reference scheme. We utilized the Lausanne‐219 parcellation which is a high resolution derivative of Freesurfer's surface‐based Desikan‐Killiany‐Atlas (Cammoun et al., 2012; Desikan et al., 2006) with 219 cortical brain regions. PET‐maps were already available in this format. For the functional and gene‐expression analyses, we created our own group version of the Lausanne‐219 parcellation based on the T1‐weighted structural images acquired alongside the functional imaging data. Structural images were run through HCP's freesurfer pipeline (Glasser et al., 2013) and subsequently parcellated according to the Lausanne‐219 atlas with scripts distributed with the CATO toolbox (de Lange & van den Heuvel, 2021). All individual parcellations were subsequently aligned with fsaverage_32LR space and a group atlas was created by assigning each vertex to the most frequent Lausanne‐label across participants. We used this group atlas to extract region‐wise mean activation differences from the unthresholded second‐level contrast maps for each ANT contrast. For the gene‐expression analysis, we resampled the group parcellation to fsaverage5 space with 10k resolution for each hemisphere separately, as required by abagen. We complemented the cortical parcellation with the standard Freesurfer subcortical parcellation which includes seven bilateral regions.
2.6. Null model
Testing the spatial correspondence of different brain maps requires a null model that accounts for spatial non‐independence (Alexander‐Bloch et al., 2018). We evaluated statistical significance of the correlations between functional task activation and gene‐expression profiles and between task activations and receptor/transporter maps through a spatial permutation approach (the “spin test”) where we swapped the parcellation labels randomly while accounting for the intrinsic geometry of the cortex (Váša et al., 2018). We created the null model based on centroid coordinates for each cortical region after projecting the group‐level parcellation onto a sphere. For each permutation, coordinates were rotated around three axes with randomly generated angles. The same angles (with opposing signs) were used for both hemispheres to preserve hemispheric symmetry. To keep the alignment between rotated and unrotated parcellations intact, we matched each rotated to the closest unrotated region (minimum Euclidian distance), starting with the region that was most distant to all other regions on average and then progressing though all other regions in descending order. Code snippets for null model creations were taken from Github (github.com/frantisekvasa/rotate_parcellation).
2.7. Overlap of activation maps
We quantified the degree of functional overlap between the phenotypic activation maps (brain images) by calculating product–moment correlations between respective activation vectors. Statistical significance was determined via permutation testing based on the null model. We randomly rotated each activation map one million times to obtain permutated activation maps that account for spatial autocorrelations in the brain maps (see Section 2.5). For each pairwise comparison (alerting vs. control, alerting vs. orienting, and control vs. orienting), rotation was performed twice, that is, one activation vector (e.g., alerting) was rotated first and correlated with the observed activation vector (e.g., control) and vice versa. Thus, each observed correlation (e.g., alerting vs. control) was compared to two million expected correlations (alerting observed vs. control rotated, and alerting rotated vs. control observed).
2.8. Gene expression analyses
For gene expression analyses, we used abagen (Markello et al., 2021), an open‐source Python interface that enables integration of the AHBA (Hawrylycz et al., 2012) with neuroimaging data. The AHBA is an open‐access database containing gene expression and other microarray expression data collected from six human postmortem brains. abagen intends to improve workflows related to the AHBA in terms of transparency, reproducibility, and standardization. The tool enhances transcriptional data analyses as these are often accompanied by a wide range of processing options making results from different studies less comparable.
We utilized abagens workflow for correlated gene expression analyses, employing its default parameters. The detailed processing steps are documented in the abagen methods report (Appendix B).
This procedure involved uploading a surface brain atlas to generate a genes‐by‐regions expression matrix with a total of 15,632 incorporated genes.
2.9. Gene expression and attention networks
We correlated each of the three regions‐by‐activation vectors from the ANT conditions with the regions‐by‐genes expression matrix obtained from abagen, resulting in one product–moment correlation coefficient for each ANT condition by gene combination. We obtained corresponding p‐values through permutation tests with the rotated spatial null model (see Section 2.5). We performed one million rotations to determine significance with adequate accuracy even after stringent Bonferroni‐correction for multiple testing (0.05/15632 = 0.0000032). For each gene, we determined the relative frequency by which the absolute correlation coefficient after random permutation was equal to or larger than the absolute empirical (observed) correlation coefficients. To distinguish positive from negative correlations, the p‐values were transformed into z‐values under consideration of the empirical correlations' signs. In a last step, all genes were rank‐ordered based on these z‐values. Rank ties were resolved by inspecting the empirical correlation coefficient, allowing duplicate z‐values to be unambiguously assigned to a rank. These rank indices, each obtained by considering both the p‐value and empirical correlation coefficient with its sign, serve as input for the subsequent gene set enrichment analysis (GESA). In this analysis step, no thresholds were applied, and all genes, along with their respective rank indices, were preserved without exclusions.
2.10. Gene‐expression‐similarities of attention networks
We calculated pair‐wise correlations between the three vectors containing the association between each gene by ANT condition combination (see Section 2.8) to quantify gene‐expression‐similarities between the three attention networks. Corresponding p‐values were computed by randomly rotating the ANT activation maps relative to the gene expression maps (see Section 2.6) and calculating the proportion of pair‐wise correlation coefficients of genetic associations results that were at least as extreme as the original pair‐wise correlation coefficients. ANT activation maps were rotated in equal directions to preserve their phenotypic correlations. Under the assumption of no association between ANT activation maps and gene expression maps, the pair‐wise correlations of association results were expected to reflect, on average, the phenotypic correlations between ANT activation maps. In case of true associations between ANT activation maps and gene expression maps, genetic similarities (i.e., pair‐wise correlations of genetic association results) were expected to exceed the phenotypic correlation.
2.11. Gene set definition
We grouped the gene expression decoding results for each ANT conditions by their underlying molecular functions (MFs) to detect similarities between single genes on a higher level. A common tool for classifying such gene functions is the PANTHER Classification System (Mi et al., 2021). PANTHER allows functional sorting of proteins, based on various criteria such as signaling and metabolic pathways or external aspects of the Gene Ontology database. The Gene Ontology (Ashburner et al., 2000; Gene Ontology Consortium, 2021) provides biological information to PANTHER that can be divided into three domains: MFs, biological processes, and cellular components (Mi & Thomas, 2009). Given our hypotheses on local activity of dopamine, norepinephrine, and acetylcholine, we focused our analyses only MFs, that is, on all molecular activities that arise almost directly from one or more gene products, such as binding or transport activity. In contrast, biological processes represent activities superordinate to MFs, thus not describing actions on a local, individual level but rather general metabolic or physiological processes. Cellular components, on the other hand, describe cell structures (e.g., mitochondria, cytosol) instead of cell activity and therefore do not provide relevant information about the neurotransmitter system.
Further, the selection of MFs we hypothesize being involved in alerting, orienting, and control focused primarily on transmitter interaction rather than synthesis since regions of transmitter synthesis are relatively confined. We rather expect the experimental manipulation leading to increased postsynaptic activity in the target regions of the three transmitter systems.
Before performing statistical analyses, we preselected those three MFs (out of around 5000 available MF in the database) that best reflect neurotransmitter binding activity of dopamine, norepinephrine, and acetylcholine:
-
Dopamine binding (GO:0035240, 7 genes)
Binding to dopamine, a catecholamine neurotransmitter formed by aromatic‐l‐amino‐acid decarboxylase from 3,4‐dihydroxy‐l‐phenylalanine.
-
Norepinephrine binding (GO:0051380, 4 genes)
Binding to norepinephrine, (3,4‐dihydroxyphenyl‐2‐aminoethanol), a hormone secreted by the adrenal medulla and a neurotransmitter in the sympathetic peripheral nervous system and in some tracts of the CNS. It is also the biosynthetic precursor of epinephrine.
-
Acetylcholine‐gated cation‐selective channel activity (GO:0022848, 18 genes)
Selectively enables the transmembrane transfer of a cation by a channel that opens upon binding acetylcholine.
2.12. Gene set enrichment analysis
We used GSEA to assess whether genes belonging to the specified gene sets. Dopamine binding, norepinephrine binding, and acetylcholine‐gated cation‐selective channel activity showed stronger co‐expression with the attention network maps relative to all other expressed genes. Association results derived from correlating gene expression maps and ANT activation maps (see Section 2.8) were uploaded to PANTHER v17, including unique Gene IDs and their corresponding rank index. This index indicates the extent to which gene expression and fMRI activation spatially overlap in comparison to other genes. Identical tool settings and processing steps were applied to all gene lists (Appendix B). The results of these enrichment tests are lists of significantly over‐ or under‐represented MFs for each ANT contrast as well as information about all mapped genes for each MF. GSEA was primarily applied to evaluate whether the three hypothesized categories are statistically overrepresented. In addition, we explored the potential over‐ or under‐representation of other categories (Appendix D). However, it is crucial to interpret these exploratory findings with caution, considering the autocorrelations of gene expressions.
2.13. Analysis of subcortical areas
To gain a more comprehensive understanding of the overlap between brain images and gene expression patterns in subcortical regions, we extended our analytical approach, previously applied to the cortex (as described in Sections 2.6, 2.11), to subcortical areas, including the thalamus, caudate, putamen, pallidum, hippocampus, amygdala, and the accumbens area (Desikan‐Killiany brain parcellation).
For these subcortical regions, we performed an adapted spin test by generating permutations of all 5040 possible orders while considering the hemisphere as a fixed factor. This allowed us to determine pairwise significance of overlap in ANT activation. We correlated the observed activation vector of one ANT condition (e.g., alerting) with 5039 shuffled activation vectors of the other ANT condition (e.g., control), and vice versa. Similarly, we computed gene expression similarities (as described in Section 2.9) and conducted GSEA (as detailed in Section 2.11) for the subcortical regions.
We also utilized the Desikan‐Killiany brain parcellation to retrieve gene expression data for 15,632 genes from the AHBA within these seven subcortical brain regions.
2.14. PET data set
We obtained 19 publicly available group‐level maps of receptor and transporter availability in nine neurotransmitter systems (Hansen, Shafiei, et al., 2022), derived from PET imaging in 27 different samples with N = 1239 healthy participants in total and individual sample sizes ranging from n = 3 to n = 174. From the 19 maps, we selected six maps with direct relevance for the three hypothesized neurotransmitter systems: Norepinephrine (NET), acetylcholine (nicotinergic receptor a4b2 and vesicular acetylcholine transporter [vACht]), and dopamine (d1‐receptor, d2‐receptor, and DAT). All data were already provided in the Lausanne‐219 parcellation and can be accessed via github.com/netneurolab/hansen_receptors.
2.15. Molecular similarities of attention networks
Relationships between attention networks and PET maps were assessed through Pearson correlations. Corresponding p‐values were obtained through permutation testing with the spin test (5000 permutations each). We report all correlations with false‐discovery‐rate‐adjusted p‐values (q = .05).
2.16. Open science statement
Code for structural and functional preprocessing can be obtained from https://github.com/Washington-University/HCPpipelines. Group‐level data, decoding results, and analyses code can be obtained from github.com/schinhan/ant_genes. We have published other work on the same data set (Markett et al., 2022).
3. RESULTS
3.1. Overlap of activation maps
In a first step, we parcellated the three attention network maps (Figure 2a) into 219 ROIs (Figure 2b) to evaluate their functional overlap. These activation vectors of attention networks (Figure 2c) were positively intercorrelated: r = .546 (p = 3.5e‐06) for alerting versus control, r = .824 (p = 2e‐06) for alerting versus orienting, and r = .636 (p = 1e‐06) for control versus orienting. The p‐values indicate that, for each pairwise comparison, almost none of the randomly rotated brain maps displayed stronger correlations with the observed brain maps than the observed brain maps themselves (Figure 2d).
FIGURE 2.
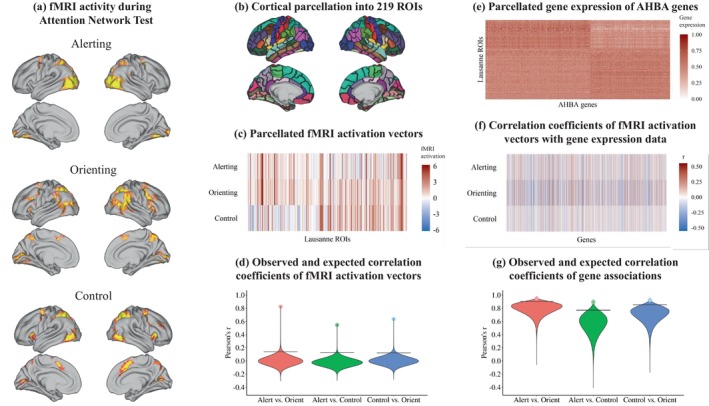
Outline of the step‐by‐step workflow for our analysis: Attention network maps (a) were parcellated into 219 regions using the Lausanne parcellation (b), generating one activation vector of length 219 for each attention network, displayed in panel (c). Functional overlap between attention network test (ANT) networks was evaluated by correlating the ANT brain activation vectors, displayed as dots in panel (d). Statistical significance of the correlations was evaluated by a spatial permutation test. The violins in panel (d) show the distributions of 2 million correlations obtained from spatially rotating the ANT activation vectors according to the spin‐test null model. Horizontal lines reflect the 95th quantiles of correlations expected under the null hypothesis. We then obtained regional gene expression data from the AHBA using the abagen pipeline. The matrix in panel (e) visualizes gene expression for a total of 15,632 genes, with values ranging from 0 to 1 across all 219 regions of the Lausanne parcellation. For visualization, the order of genes and ROIs was rearranged into two clusters. Associations between gene expression and ANT networks were evaluated by correlating the activation vectors from panel (c) with each of the 15,632 gene expression vectors in panel (e), yielding a correlation coefficient for each gene with the alerting, orienting, and control networks, displayed in panel (f). In a last step, we assessed the similarity between the gene expression associations of each attention network displayed in panel (f) by computing their pairwise correlation, shown as dots in panel (g). The violins show the distribution of two million correlations obtained from spatially rotating the ANT activation vectors (panel e) according to the spin‐test null model. Again, horizontal lines reflect the 95th quantiles of correlations expected under the null hypothesis.
Additionally, we computed Dice similarity coefficients (DSC) to offer a more comprehensive evaluation of spatial overlap. The DSC quantifies set overlap based on binary vectors. To determine whether a given brain region belonged to a particular attention network, we initially extracted mean activation for each task condition (e.g., incongruent flankers) in all 219 brain regions at the participant level. We then conducted t‐tests to identify regions significantly activated by the experimental manipulation (p < .05, FDR‐corrected). The resulting binary vectors were used to calculate DSC for all ANT comparisons. We obtained a DSC = .444 (p = .003) for alerting versus control, DSC = .583 (p = 8.15e‐05) for alerting versus orienting, and DSC = .592 (p = 1.10e‐04) for control versus orienting. In practical terms, this means that the pairwise similarity between ANT maps ranges from 44.4 to 59.2%. P‐values were obtained through the same spin test approach as used for the correlational overlap of ANT maps, involving two million permutations for each comparison.
In an exploratory endeavor, we aimed to identify which regions were primarily responsible for this high overlap between attention networks and where regional differences emerged. In Appendix C, we provide visualizations of regions displaying the greatest absolute differences in brain activation across all three comparisons within the ANT. This includes highlighting elements consistently activated across all three attention networks and those showing activation in just one or two of them.
3.2. Gene expression analyses
Second, we examined the spatial correspondence between gene expression and spatial activation of attention networks for each gene in comparison to all other genes. The gene expression matrix extracted from abagen is shown in Figure 2e. Correlation of the region‐by‐gene expression matrix with the spatial activation vectors of the ANT conditions yielded an ANT condition‐by‐gene association matrix (Figure 2f). Significance testing with one million permutations of the spatial null model revealed 3871 genes whose cortex‐wide expression patterns covaried with the activation maps (p < .05, FDR corrected) in alerting, 6905 genes in orienting, and 2556 genes in control.
Overall, the associations between the gene expression maps from abagen and the ANT maps exceed the expected associations under the null hypothesis since the observed permutation‐based p‐values strongly deviate from the expected p‐values (Figure 3a). In other words, the activation maps and gene expression map exhibit overall stronger overlaps than those resulting from random rotation of the activation maps.
FIGURE 3.
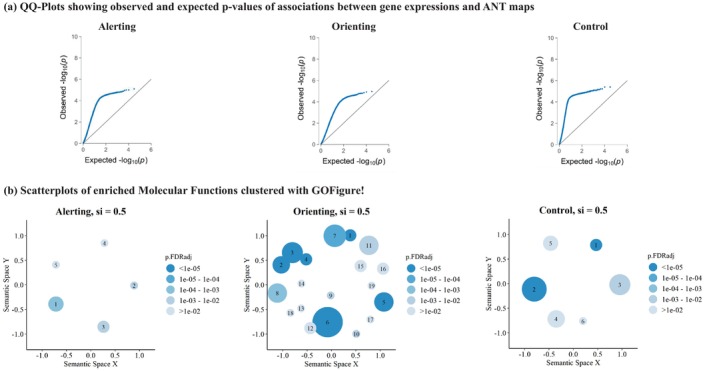
QQ‐plots in panel (a) display permutation‐based p‐values for all 15,632 genes. These p‐values were derived by associating the gene expression map from abagen with the ANT activation maps and comparing them to gene associations of 1 million rotated ANT activation maps. On the y‐axes, we present observed permutation‐based p‐values, plotted against the expected p‐value distribution under the null hypothesis on the x‐axes. Notably, leftward deflections (indicated by blue dots) from the projected null (diagonal line) signify an enrichment of low p‐values. GO‐Figure! plots in panel (b) summarize enriched MFs from GSEA. We have included all significant MFs (p < .05, FDR corrected) in the creation of these summary visualizations with GO‐Figure. Each cluster's color represents the p‐value of the selected representative, while the size of the circles indicates the number of MF assigned to the cluster. Spatial distances between clusters reflect their semantic similarity. For this analysis, we specified a maximum of 20 clusters, a default similarity cut‐off of si = .50, and restricted the analysis to molecular functions. The similarity cut‐off is a parameter between 0 and 1, determining the degree of semantic similarity necessary for two GO‐terms within the Gene Ontology to be assigned to the same cluster. Labels of the cluster's representatives are given in Table 2.
Association results of all expressed genes and the different ANT conditions are displayed in respective Manhattan plots (Appendix C, Figure C.1), with further details provided in Appendix S1 (Table B1). We note that Manhattan plots do not highlight specific genomic regions that contain accumulations of strong association p‐values as typically shown in genome‐wide association studies. In this expression‐based analysis, clusters of genes may span the whole genome and still overlap in their cortex‐wide expression patterns, producing “horizontal band of associations” with similar test‐statistics in Manhattan plots (e.g., see band of associations in ranging from −log10(p) = 4 to −log10(p) = 5).
To identify groups of genes with a relative enrichment of signals, we carried out GSEA, which implies that all genes (both significant and nonsignificant genes) were included along with their corresponding z‐values.
3.3. Gene‐expression‐similarities of attention networks
Next, we aimed to test whether the general gene expression patterns showed spatial overlap between the alerting, orienting, and control network. The empirical correlation between vectors containing the genetic associations of ANT maps, which, again, represent the correlation coefficients between activation maps and gene expression maps, aggregated to r = .893 (p = .0002) for alerting versus control, r = .948 (p = .0015) for alerting versus orienting, and r = .922 (p = .0008) for control versus orienting. By comparison, under the null model of no association between gene expression and ANT activation maps (operationalized through one million random rotations of the ANT activation maps), correlations of the gene association results derived for the three ANT conditions were lower and approximated the direct correlations of ANT maps (mean r = .554 for alerting vs. control, mean r = .785 for alerting vs. orienting, and mean r = .695 for control vs. orienting). The enhanced similarity in association results as indicated by higher‐than‐expected correlation coefficients suggests a shared systematic covariation between gene expression and ANT maps (Figure 2g).
3.4. Gene set enrichment analysis
GSEA were applied for all three ANT conditions to quantify the probability to which the genes associated with their hypothesized MFs are coincidentally or systematically ranked higher within the whole gene list. To do so, we uploaded association results of all 15,632 genes to PANTHER, of which 14,466 genes could be successfully annotated within the database. Consequently, we derived GSEA results from these 14,466 genes and excluded the unmapped gene IDs from subsequent analysis.
Table 1 shows enrichment analysis results (Mann–Whitney U test statistic) for all three MF defined a priori. Test statistics inform about how many genes were mapped to this MF and whether the MF is over‐ or underrepresented. No ANT contrast was significantly positively related to its matching MF: There was no evidence for positive enrichment of genes related to norepinephrine binding in alerting, nor for dopamine binding in control, nor for acetylcholine‐gated cation‐selective channel activity in orienting. Furthermore, there was also no positive enrichment for any pre‐defined MF and the other two ANT conditions. In contrast, we did find an enrichment toward the bottom extreme for norepinephrine binding and the control network, meaning that the distribution of values of this gene set (z‐scores derived from the permutation‐based p‐value and the sign of the observed correlation coefficient) was shifted toward smaller values relative to the overall list of genes, suggesting lower expression at activated regions.
TABLE 1.
GSEA results of MF defined a priori.
| ANT condition | Alerting | Control | Orienting | ||||
|---|---|---|---|---|---|---|---|
| Number | ± | p | ± | p | ± | p | |
| Dopamine binding | 6 | ‐ | .384 | ‐ | .739 | ‐ | .544 |
| Norepinephrine binding | 3 | ‐ | .057 | ‐ | .018 | ‐ | .054 |
| Acetylcholine‐gated cation‐selective channel activity | 10 | ‐ | .643 | ‐ | .252 | ‐ | .277 |
Note: Bold value indicates statistical significance.
TABLE 2.
GO‐terms functioning as representatives of clusters generated with GO‐figure (si = 0.50).
| ANT condition | GO‐terms with FDR‐adjusted p‐value |
|---|---|
| Alerting |
1. Signaling receptor binding (p adj = 2.62E‐04) 2. Structural constituent of ribosome (p adj = 1.27E‐03) 3. Active transmembrane transporter activity (p adj = 8.29E‐03) 4. Protein serine kinase activity (p adj = 2.52E‐02) 5. ATP binding (p adj = 3.56E‐02) |
| Orienting |
1. Structural constituent of ribosome (p adj = 1.58E‐09) 2. DNA binding (p adj = 9.24E‐07) 3. Double‐stranded DNA binding (p adj = 9.24E‐07) 4. Regulatory region nucleic acid binding (p adj = 1.99E‐06) 5. Transcription regulator activity (p adj = 2.16E‐06) 6. Signaling receptor binding (p adj = 2.77E‐06) 7. Protein serine kinase activity (p adj = 3.37E‐05) 8. ATP binding (p adj = 1.21E‐04) 9. Chromatin binding (p adj = 1.46E‐03) 10. Antioxidant activity (p adj = 5.76E‐03) 11. Transmembrane signaling receptor activity (p adj = 6.59E‐03) 12. Fatty acid binding (p adj = 1.05E‐02) 13. Copper ion binding (p adj = 1.68E‐02) 14. Glycosaminoglycan binding (p adj = 2.74E‐02) 15. Oxidoreductase activity, acting on the CH‐NH group of donors (p adj = 3.09E‐02) 16. Helicase activity (p adj = 3.67E‐02) 17. Metal ion transmembrane transporter activity (p adj = 3.67E‐02) 18. Calcium ion binding (p adj = 3.91E‐02) 19. Catalytic activity, acting on RNA (p adj = 4.70E‐02) |
| Control |
1. Structural constituent of ribosome (p adj = 2.72E‐08) 2. Signaling receptor binding (p adj = 6.89E‐06) 3. Transmembrane signaling receptor activity (p adj = 1.93E‐03) 4. Protein serine kinase activity (p adj = 1.07E‐02) 5. ATP binding (p adj = 1.03E‐02) 6. Copper ion binding (p adj = 3.96E‐02) |
In summary, the preselected MFs yielded no significant hits; nonetheless, we detected positive enrichment (p < .05, FDR‐corrected) for 4 other MFs in alerting (Table D.1, Appendix D), 41 MFs in orienting (Table D.2, Appendix D), and 8 MFs in control (Table D.3, Appendix D). Negative associations appeared for 4 MFs in alerting, 25 MFs in orienting, and 17 MFs in control. Among these hits, no MF was directly related to dopamine, acetylcholine, or norepinephrine (see Appendix D).
3.5. Summarizing GESA results
Following GSEA with Panther, we focused on MFs that were significantly associated with attention networks, filtered with a significance threshold of p < .05, FDR corrected. Given the potentially extensive and complex nature of these lists, we sought to enhance the interpretability of our enrichment results by employing GO‐Figure! (Reijnders & Waterhouse, 2021), an open‐source Python software designed to cluster lists of GO‐terms and reduce redundancies (Figure 3b).
Our process involved using the significant GO‐terms, along with their corresponding p‐values, as input for analysis. Pairwise similarities were computed among all inserted terms, following the common formula established by Lin (1998). MFs that exceeded a similarity score of si = .50 were grouped into clusters, as they were assumed to be highly functionally similar. For each cluster, we selected a representative MF based on several criteria, including the p‐value, the number of annotated proteins, and their relationships within the Gene Ontology database. In this selection process, parent terms were prioritized over child terms.
The representatives were then plotted in a two‐dimensional space based on the pairwise semantic similarity of the representatives. This numerical value reflects the extent of functional and hierarchical similarity within the Gene Ontology.
It is important to note that the enriched categories presented in GO‐Figure! may be subject to potential influences from autocorrelations of gene expressions, as discussed in Fulcher et al. (2021). This consideration raises the possibility of false‐positive results, even when accounting for spatial autocorrelation of brain activation with the spin test.
3.6. Analyses of subcortical areas
We extended our analyses of cortical ANT maps to subcortical maps. We applied the same analysis pipeline as described in Sections 3.1, 3.4, encompassing the calculation of overlap between ANT maps, assessment of gene expression similarities across different ANT conditions, and GSEA.
To determine significance, we employed permutation tests. In these tests, we computed correlations between randomly shuffled ANT maps and observed ANT maps or gene expression patterns of randomly shuffled maps. The resulting p‐values were derived by assessing how many of these randomly generated correlations exceeded the observed correlation coefficient, relative to the total number of permutations conducted. An illustrative depiction of this workflow, akin to Figure 2, is available in Appendix C.
In the subcortical regions, we found no statistically significant overlaps among the three attention network maps. The correlation coefficients were as follows: r = .556 (p = .506) for alerting versus control, r = .146 (p = .743) for alerting versus orienting, and r = .377 (p = .368) for control versus orienting comparisons. Similarly, no significant overlap was observed in gene expression patterns: r = .778 (p = .128) for alerting versus control, r = .426 (p = .241) for alerting versus orienting, and r = .238 (p = .522) for control versus orienting.
3.7. Attention networks and neurotransmitter systems measured by PET
Our complementary methodological approach involved the analysis of PET‐maps representing neurotransmitter receptors and transporters to evaluate their spatial overlap with the attention networks. Based on attention network theory, we expected brain regions with stronger activation during attention to correlate with the availability of receptor and transporter molecules within three neurotransmitter systems. As shown in Figure 4, however, empirical results were not in line with our hypotheses: The availability of the noradrenalin transporter did not match the alerting network, and the availability of dopamine D1 and D2 receptors and the DAT did not correspond with the control network. Also, the availability of the most abundant nicotinergic acetylcholine receptor did not correlate with the activation of the orienting network. The exploratory analyses on other neurotransmitter systems did not reveal significant relationships (see Figure 4).
FIGURE 4.
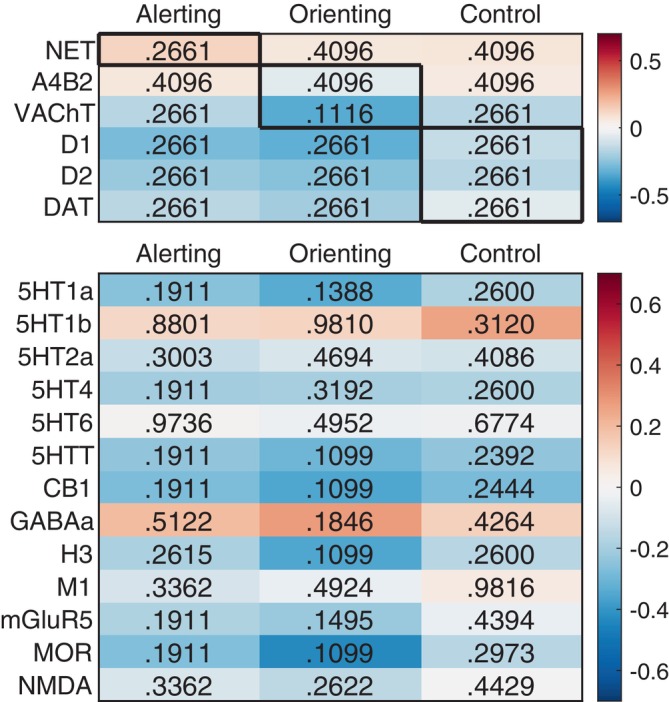
Correlations between neurotransmitter maps as revealed by PET‐imaging (rows) and attention networks (columns) based on the Lausanne‐219 parcellation. P‐values were obtained through permutation testing under consideration of the autocorrelations across brain regions. The upper part of the table shows the hypothesized relationships (bold‐framed cells) and the lower part of the table gives the results from the exploratory analysis. None of the correlations were significant at p < .05, FDR‐corrected.
4. DISCUSSION
Attention network theory states that three distinct attention networks are each modulated by a specific neurotransmitter: the alerting network by norepinephrine, the orienting network by acetylcholine, and the attention control network by dopamine. We sought to ascertain these hypotheses by assessing the molecular signatures of attention networks through a multimodal neuroimaging design: We compared fMRI activation maps from the ANT with cortex‐wide gene expression patterns and with the spatial distribution of neurotransmitter receptors and transporters as revealed by PET imaging.
If the three attention networks were modulated by distinct neurotransmitters as proposed by attention network theory, we would expect spatial correspondence between the networks' activation patterns on the one hand and the expression‐levels of genes related to transmitter binding as well as the availability of receptor and transporter molecules of the proposed neurotransmitter on the other hand.
4.1. No evidence for the hypothesized neuromodulators
We did not find any evidence for the suggested neuromodulatory separation for any of the three attention networks. Even though a substantial number of genes co‐expressed significantly with the attention networks, there was no enrichment of gene sets linked to the MFs of dopamine binding, norepinephrine binding, and acetylcholine‐gated channel activity. Regarding the spatial distribution of receptor and transporter molecules in the PET images, there was also no spatial correspondence with attention network in the hypothesized direction: the distribution of NET did not correlate with the activation of the alerting network, the distribution of a4b2 receptors did not correlate with activation of the orienting network, and neither DRD1 nor DRD2 availability correlated with the activation of the attention control network. Only the distribution of the vACht correlated with the activation of the orienting network, however, in the opposite direction, suggesting higher transporter availability outside the orienting network.
From this, we conclude that the specific hypotheses on distinct neuromodulation of the three attention networks do not hold, at least not at the level of transcriptomic activity and the availability of receptor or transporter molecules, and at least not in the general and straightforward manner as suggested by attention network theory. We will discuss implications against methodological questions further below.
4.2. No evidence for a transcriptomic separation of attention networks
Despite the unfavorable evidence for the a priori hypotheses, our exploratory analysis revealed that our design was at least in principle capable of detecting MFs in relationship with the attention networks. We found a substantial number of genes (3871 for alerting, 6905 for orienting, 2556 for control) whose cortex‐wide transcription co‐varied with the activation maps. Among a ranked list of all available genes, GESA further prioritized several MFs for all three attention networks. These included genes involved in the regulation of protein biosynthesis, phosphorylation, enzymatic activity, and receptor binding. The implication of such broad terms in all three networks is not surprising, given the high similarity of the co‐expression patterns across networks. Such broad terms, however, could also result from spatial autocorrelations of the gene expression maps (Fulcher et al., 2021), and should therefore be interpreted with caution.
In absolute numbers, we implicated most genes in the orienting network. The additional MFs prioritized here included genes involved in transcriptomic activity and regulation. Among all identified gene sets, the only gene set with direct relevance for a concrete neurotransmitter was glutamate receptor activity (GO:0008066). This gene set, however, was underrepresented among the associations for the control network. In sum, the exploratory analysis of the attention networks' transcriptomic signatures revealed a relatively broad set of associated MFs. Contrary to the claims of attention network theory, there was no clear distinction in the networks' transcriptomic profiles and no evidence for any given neurotransmitter system whose neuromodulatory activity might support the hypothesized separation of the networks at the molecular level.
4.3. Methodological considerations
The current approach is based on various assumptions of attention network theory. In the following, we will discuss the current finding against these assumptions and highlight some challenges within attention network theory that may prompt a need for reevaluation of its central principles.
One of the fundamental assumptions of attention network theory is the independence of the three attention networks. Independence refers to the idea that the three networks exhibit uncorrelated behavioral responses when manipulated, distinct activation patterns, and independent neuromodulatory influences (Petersen & Posner, 2012; Posner & Fan, 2008; Posner & Rothbart, 2007). While the behavioral independence of the three networks has been well‐documented (Callejas et al., 2004; Fan et al., 2002, 2009; Ishigami & Klein, 2010; Markett et al., 2022), our findings suggest that there is significant overlap between the cortical activation patterns of the three networks. This overlap limits the scope of the theory but does not necessarily rule out the possibility of separate neuromodulation which may still occur despite overlapping activation patterns. This, however, was not the case. The lack of independence in functional activation, transcriptomic activity, and receptor expression, impose significant constraints on the predictions of attention network theory.
Our approach also rests on the assumption that the three attention networks can be operationalized using activation maps from the ANT. This assumption is based on attention network theory, which views the ANT as the standard protocol for separating the networks and considers it capable of fully activating them (Fan et al., 2005; Fan & Posner, 2004). By keeping visual stimuli constant and by counterbalancing motor responses, the ANT is believed to reveal the extra neural effort required for different attention systems. But while the ANT's validity can be reasonably assumed, it is unclear whether the presumed transmitter systems modulate a given network as a whole (and not only selected regions within the network) and if activation across the network is proportional to the hypothesized neuromodulation. Such proportional relationship would imply uniform neuromodulation of the several processes captured by the ANT activation maps such as increased activation due to the prioritization of information processing in sensory cortices (Brefczynski & DeYoe, 1999; Kastner et al., 1999; Müller et al., 2003), increased activation due to the maintenance and allocation of the attentional focus in premotor cortex and the frontal eye fields (T. Moore et al., 2003; Thompson, 2005), and increased activity linked to the detection of increased demand for attentional resources in the anterior cingulate (Botvinick et al., 2004). Since attention network theory does not specify the mechanisms of neuromodulation, the current results do not necessarily refute the specific hypotheses. However, they raise doubts that the attention networks as currently operationalized are modulated as a whole, and indicate the need to amend the theory. This revision should include a specification of the relationship between attention networks and activation patterns, the functional neuroanatomy of each network, and the potential target regions of neuromodulatory influences.
Another fundamental assumption underlying our approach pertains to the translation of the proposed neuromodulation into three gene sets derived from the gene ontology, specifically encompassing MFs associated with transmitter binding. This assumption warrants careful examination from two distinct angles. First, can we reasonably infer that the selected transcriptomic markers adequately represent the cortical projection sites, and are the chosen MFs the most appropriate operationalization? Second, can we anticipate that neural activity patterns across the brain will indeed correlate with the relative expression of genes following neuromodulation?
Regarding the first question: The MFs selected are linked to transmitter binding, a process that characterizes neurotransmitter interaction with synaptic molecules. The gene products examined include key components such as the DAT, dopamine receptors, associated intracellular signaling molecules, various adrenergic receptors, and proteins forming subunits for the pentameric nicotinergic acetylcholine receptor. Our premise is that if the experimental manipulations within the ANT lead to the hypothesized increase in activity of dopaminergic neurons in the midbrain, noradrenergic neurons in the locus coeruleus, and cholinergic neurons in the basal forebrain, we should observe a neuromodulation of ongoing neural activity within their respective projection areas. Given that these projection areas are known to possess an abundance of the mentioned receptor and transporter molecules, this should also be reflected in their transcriptomic activity. If the hypothesized neuromodulation indeed acts directly on the attention networks, we would expect local parcel‐wise associations between transcriptomic activity and attention network activation.
This leads to the second question. The hypothesized direct neuromodulatory effects on the networks as a whole are not definitively established. Attention network theory does not explicitly outline the mechanisms of the presumed neuromodulation. While descriptions of the attention networks in the literature include the cortical target sites of these neurotransmitters (Posner & Fan, 2008), suggesting a direct effect, it is equally plausible that neuromodulation operates indirectly. For example, locus coeruleus activity and norepinephrine release influence attention by initiating broader adjustments in brain states, including alterations in cortical, subcortical, and autonomic activity (Ross & Van Bockstaele, 2021). Norepinephrine's influence extends to the thalamus, which subsequently modulates cortical activity during sensory processing (O'Donnell et al., 2012). Dopamine, as a neuromodulator, acts on striato‐thalamic‐cortical loops (Parent & Hazrati, 1995). Dopamine's modulation of medium spiny neurons in the striatum results in the disinhibition of thalamic glutamatergic neurons projecting to the cortex (Speranza et al., 2021). These striato‐thalamic‐cortical loops are further modulated by acetylcholine, particularly through the nicotinic receptor (Exley & Cragg, 2009). If neuromodulatory effects on the attention networks prove to be indirect, perhaps involving the basal ganglia or the thalamus, or being more locally confined to individual cortical targets, we may find little basis for assuming correlations between transcriptomic and neural activity across the entire brain.
Our present findings suggest that there is no detectable relationship between neural activation patterns elicited by the experimental manipulation of attention and receptor or transporter availability, or related transcriptomic activity. Given that the relationship between receptor/transporter availability and gene expression is not straightforward (Hansen et al., 2022), the absence of a relationship with either level of observation can be considered complementary evidence. Our exploratory analysis also did not reveal any other MF whose transcriptomic activity would suggest a comprehensive relationship between the activation maps and dopamine, norepinephrine, or acetylcholine. This may constrain the strong claim of network‐wide neuromodulation, but does not preclude neuromodulation through other mechanisms, such as more nuanced or dynamic neuromodulatory activity in brain stem or midbrain nuclei. In light of the mixed evidence from pharmacological studies with the ANT (Badgaiyan & Wack, 2011; McCormick, 2022; Reynaud et al., 2019; Thienel et al., 2009); however, it seems reasonable to revise attention network theory regarding the presumed neuromodulation of attention networks. The analysis of subcortical areas similarly failed to establish a clear relationship between ANT activations and molecular measures. Nevertheless, it is essential to recognize that subcortical nuclei are often relatively small and may not be adequately distinguished at the limited spatial resolution inherent to transcriptomic, molecular, and functional neuroimaging techniques (for an in‐depth discussion, please refer to Boeken et al., 2022). Future investigations may consider optimizing processing pipelines for subcortical areas or employing spatially constrained hypotheses to uncover more regionally specific effects.
Finally, our approach is based on the idea that group‐level data from three different sources can be combined in a correlational design on the grounds of a cortical parcellation. This methodological approach is widely applied when combing transcriptomic and neuroimaging data (Fornito et al., 2019; Hansen, Markello, et al., 2022; Seidlitz et al., 2020), probing mechanistic hypotheses in clinical neuroscience (Buckner et al., 2008; de Lange et al., 2019; Fornito & Bullmore, 2014; Zhou et al., 2012), and multimodal neuroimaging where the invasive nature of one of assessment methods precludes direct comparisons in the same participants (Scholtens et al., 2014; van den Heuvel et al., 2015). Despite the limitations of small sample sizes of postmortem brains, constraints in spatial resolution with PET imaging, and unaccounted individual variability, it is important to recognize that the AHBA microarray gene expression maps and group‐level receptor/transporter maps still represent state‐of‐the‐art techniques. But even though we used an established null model to reduce spurious influences on our test statistics (Alexander‐Bloch et al., 2018; Váša et al., 2018), it needs to be noted that the here presented relationships are correlational. The absence of direct psychopharmacological manipulation or experimental effects on neuromodulators limits conclusions on causality. Furthermore, it is essential to emphasize that our study's statistical approach, while yielding a nonsignificant correlation between the molecular markers and attention network maps, does not definitively conclude the absence of any spatial association. Instead, it suggests that the observed relationship is highly unlikely, given the general baseline similarity of the brain maps we analyzed. Our findings should be considered within the context of the data's nature and the inability to conduct equivalence testing. Further research utilizing alternative statistical methods may provide additional insights into the practical significance of the observed associations.
4.4. Implications for attention network theory
Since its first conception more than 30 years ago (Posner & Petersen, 1990), attention network theory has established itself as an influential account of how higher cognitive functions such as attention emerges from a network of distributed brain areas (Posner & Dehaene, 1994). By incorporating neuropsychological evidence (Fernandez‐Duque & Posner, 2001), modern neuroimaging (Fan et al., 2005; Markett et al., 2014; Petersen & Posner, 2012; Xuan et al., 2016), as well as developmental (Posner et al., 2014), genetic (Fan et al., 2001; Green et al., 2008), and pharmacological data (Marrocco & Davidson, 1998), attention network theory has not only stipulated hundreds of empirical investigations (Arora et al., 2020) but also achieved a level of sophistication that allows for specific hypotheses on the molecular signatures of attention networks. The present findings, however, indicate that some of these predictions do not hold in the proposed way. The ANT activation maps do neither align with the hypothesized distribution of receptor and transporter molecules nor with transcriptomic profiles that would suggest clearly separable networks along molecular lines. Separability and presumed independence of the attention networks is additionally constrained by a high level of spatial dependency between the network maps. Since attention network theory acknowledges interactions between the attention networks (Callejas et al., 2004; Fan et al., 2009; Xuan et al., 2016), it may be reasonable to reconceptualize the attention networks in terms of their segregation and integration. Future work will also need to readdress the different observational layers and specify how the functional activation maps relate to the underlying brain network (Betzel et al., 2016; Cole et al., 2016; Liu et al., 2022; Markett et al., 2022; Murphy et al., 2020), in order to reevaluate the presumed independence of attention networks at the neural and neurochemical level, and to specify the presumed neuromodulatory influences on alerting, orienting, and attentional control.
Supporting information
DATA S1: Supporting information.
DATA S2: Supporting information.
ACKNOWLEDGEMENTS
This work has been supported by a grant from the German Research Association, Grant/ Award Number: MA‐6792/3‐1 awarded to Sebastian Markett. Open Access funding enabled and organized by Projekt DEAL.
Schindler, H. , Jawinski, P. , Arnatkevičiūtė, A. , & Markett, S. (2024). Molecular signatures of attention networks. Human Brain Mapping, 45(3), e26588. 10.1002/hbm.26588
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available in ant_genes at https://github.com/schinhan/ant_genes. These data were derived from the following resources available in the public domain: Github; https://github.com/justinehansen/hansen_receptors-1; ABAGEN, https://github.com/rmarkello/abagen; and OSF, https://osf.io/st9ae/.
REFERENCES
- Alexander‐Bloch, A. F. , Shou, H. , Liu, S. , Satterthwaite, T. D. , Glahn, D. C. , Shinohara, R. T. , Vandekar, S. N. , & Raznahan, A. (2018). On testing for spatial correspondence between maps of human brain structure and function. NeuroImage, 178, 540–551. 10.1016/j.neuroimage.2018.05.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, S. , Lawrence, M. A. , & Klein, R. M. (2020). The attention network test database: ADHD and cross‐cultural applications. Frontiers in Psychology, 11, 388. 10.3389/fpsyg.2020.00388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C. A. , Blake, J. A. , Botstein, D. , Butler, H. , Cherry, J. M. , Davis, A. P. , Dolinski, K. , Dwight, S. S. , Eppig, J. T. , Harris, M. A. , Hill, D. P. , Issel‐Tarver, L. , Kasarskis, A. , Lewis, S. , Matese, J. C. , Richardson, J. E. , Ringwald, M. , Rubin, G. M. , & Sherlock, G. (2000). Gene ontology: Tool for the unification of biology. Nature Genetics, 25(1), 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badgaiyan, R. D. , & Wack, D. (2011). Evidence of dopaminergic processing of executive inhibition. PLoS One, 6(12), e28075. 10.1371/journal.pone.0028075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzel, R. F. , Gu, S. , Medaglia, J. D. , Pasqualetti, F. , & Bassett, D. S. (2016). Optimally controlling the human connectome: The role of network topology. Scientific Reports, 6(1), 30770. 10.1038/srep30770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeken, O. J. , Cieslik, E. C. , Langner, R. , & Markett, S. (2022). Characterizing functional modules in the human thalamus: Coactivation‐based parcellation and systems‐level functional decoding. Brain Structure and Function, 228, 1811–1834. 10.1007/s00429-022-02603-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border, R. , Johnson, E. C. , Evans, L. M. , Smolen, A. , Berley, N. , Sullivan, P. F. , & Keller, M. C. (2019). No support for historical candidate gene or candidate gene‐by‐interaction hypotheses for major depression across multiple large samples. American Journal of Psychiatry, 176(5), 376–387. 10.1176/appi.ajp.2018.18070881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinick, M. M. , Cohen, J. D. , & Carter, C. S. (2004). Conflict monitoring and anterior cingulate cortex: An update. Trends in Cognitive Sciences, 8(12), 539–546. 10.1016/j.tics.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Brefczynski, J. A. , & DeYoe, E. A. (1999). A physiological correlate of the “spotlight” of visual attention. Nature Neuroscience, 2(4), 370–374. 10.1038/7280 [DOI] [PubMed] [Google Scholar]
- Buckner, R. L. , Andrews‐Hanna, J. R. , & Schacter, D. L. (2008). The brain's default network: Anatomy, function, and relevance to disease. Annals of the New York Academy of Sciences, 1124, 1–38. 10.1196/annals.1440.011 [DOI] [PubMed] [Google Scholar]
- Callejas, A. , Lupiáñez, J. , & Tudela, P. (2004). The three attentional networks: On their independence and interactions. Brain and Cognition, 54(3), 225–227. 10.1016/j.bandc.2004.02.012 [DOI] [PubMed] [Google Scholar]
- Cammoun, L. , Gigandet, X. , Meskaldji, D. , Thiran, J. P. , Sporns, O. , Do, K. Q. , Maeder, P. , Meuli, R. , & Hagmann, P. (2012). Mapping the human connectome at multiple scales with diffusion spectrum MRI. Journal of Neuroscience Methods, 203(2), 386–397. 10.1016/j.jneumeth.2011.09.031 [DOI] [PubMed] [Google Scholar]
- Cole, M. W. , Ito, T. , Bassett, D. S. , & Schultz, D. H. (2016). Activity flow over resting‐state networks shapes cognitive task activations. Nature Neuroscience, 19(12), 1718–1726. 10.1038/nn.4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, N. (1999). An embedded‐processes model of working memory. In Miyake A. & Priti S. (Eds.), Models of working memory: Mechanisms of active maintenance and executive control (pp. 62–101). Cambridge University Press. [Google Scholar]
- de Lange, S. C. , Scholtens, L. H. , van den Berg, L. H. , Boks, M. P. , Bozzali, M. , Cahn, W. , Dannlowski, U. , Durston, S. , Geuze, E. , van Haren, N. E. M. , Hillegers, M. H. J. , Koch, K. , Jurado, M. Á. , Mancini, M. , Marqués‐Iturria, I. , Meinert, S. , Ophoff, R. A. , Reess, T. J. , Repple, J. , … van den Heuvel, M. P. (2019). Shared vulnerability for connectome alterations across psychiatric and neurological brain disorders. Nature Human Behaviour, 3(9), 988–998. 10.1038/s41562-019-0659-6 [DOI] [PubMed] [Google Scholar]
- de Lange, S. C. , & van den Heuvel, M. P. (2021). Structural and functional connectivity reconstruction with CATO—A connectivity analysis TOolbox [Preprint]. Neuroscience. 10.1101/2021.05.31.446012 [DOI] [PubMed] [Google Scholar]
- Desikan, R. S. , Ségonne, F. , Fischl, B. , Quinn, B. T. , Dickerson, B. C. , Blacker, D. , Buckner, R. L. , Dale, A. M. , Maguire, R. P. , Hyman, B. T. , Albert, M. S. , & Killiany, R. J. (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage, 31(3), 968–980. 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Exley, R. , & Cragg, S. J. (2009). Presynaptic nicotinic receptors: A dynamic and diverse cholinergic filter of striatal dopamine neurotransmission. British Journal of Pharmacology, 153(S1), S283–S297. 10.1038/sj.bjp.0707510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Fossella, J. , Sommer, T. , Wu, Y. , & Posner, M. I. (2003). Mapping the genetic variation of executive attention onto brain activity. Proceedings of the National Academy of Sciences of the United States of America, 100(12), 7406–7411. 10.1073/pnas.0732088100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , Gu, X. , Guise, K. G. , Liu, X. , Fossella, J. , Wang, H. , & Posner, M. I. (2009). Testing the behavioral interaction and integration of attentional networks. Brain and Cognition, 70(2), 209–220. 10.1016/j.bandc.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J. , McCandliss, B. , Fossella, J. , Flombaum, J. , & Posner, M. (2005). The activation of attentional networks. NeuroImage, 26(2), 471–479. 10.1016/j.neuroimage.2005.02.004 [DOI] [PubMed] [Google Scholar]
- Fan, J. , McCandliss, B. D. , Sommer, T. , Raz, A. , & Posner, M. I. (2002). Testing the efficiency and independence of attentional networks. Journal of Cognitive Neuroscience, 14(3), 340–347. 10.1162/089892902317361886 [DOI] [PubMed] [Google Scholar]
- Fan, J. , & Posner, M. (2004). Human attentional networks. Psychiatrische Praxis, 31, 210–214. 10.1055/s-2004-828484 [DOI] [PubMed] [Google Scholar]
- Fan, J. , Wu, Y. , Fossella, J. A. , & Posner, M. I. (2001). Assessing the heritability of attentional networks. BMC Neuroscience, 7, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Duque, D. , & Posner, M. I. (2001). Brain imaging of attentional networks in normal and pathological states. Journal of Clinical and Experimental Neuropsychology, 23(1), 74–93. 10.1076/jcen.23.1.74.1217 [DOI] [PubMed] [Google Scholar]
- Fornito, A. , Arnatkevičiūtė, A. , & Fulcher, B. D. (2019). Bridging the gap between connectome and transcriptome. Trends in Cognitive Sciences, 23(1), 34–50. 10.1016/j.tics.2018.10.005 [DOI] [PubMed] [Google Scholar]
- Fornito, A. , & Bullmore, E. T. (2014). Connectomics: A new paradigm for understanding brain disease. European Neuropsychopharmacology, 25, 733–748. 10.1016/j.euroneuro.2014.02.011 [DOI] [PubMed] [Google Scholar]
- Fossella, J. , Sommer, T. , Fan, J. , Wu, Y. , Swanson, J. M. , Pfaff, D. W. , & Posner, M. I. (2002). Assessing the molecular genetics of attention networks. BMC Neuroscience, 3(1), 14. 10.1186/1471-2202-3-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulcher, B. D. , Arnatkeviciute, A. , & Fornito, A. (2021). Overcoming false‐positive gene‐category enrichment in the analysis of spatially resolved transcriptomic brain atlas data. Nature Communications, 12(1), 2669. 10.1038/s41467-021-22862-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay, S. , Pertzov, Y. , & Henik, A. (2011). Orienting of attention, pupil size, and the norepinephrine system. Attention, Perception, & Psychophysics, 73(1), 123–129. 10.3758/s13414-010-0015-4 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium . (2021). The gene ontology resource: Enriching a GOld mine. Nucleic Acids Research, 49, D325–D334. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva, R. , Zivan, M. , Warsha, A. , & Olchik, D. (2013). Alerting, orienting or executive attention networks: Differential patters of pupil dilations. Frontiers in Behavioral Neuroscience, 7(45), 1–11. 10.3389/fnbeh.2013.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, M. F. , Sotiropoulos, S. N. , Wilson, J. A. , Coalson, T. S. , Fischl, B. , Andersson, J. L. , Xu, J. , Jbabdi, S. , Webster, M. , Polimeni, J. R. , Van Essen, D. C. , Jenkinson, M. , & WU‐Minn HCP Consortium . (2013). The minimal preprocessing pipelines for the human connectome project. NeuroImage, 80(C), 105–124. 10.1016/j.neuroimage.2013.04.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, A. E. , Munafò, M. R. , DeYoung, C. G. , Fossella, J. A. , Fan, J. , & Gray, J. R. (2008). Using genetic data in cognitive neuroscience: From growing pains to genuine insights. Nature Reviews Neuroscience, 9(9), 710–720. 10.1038/nrn2461 [DOI] [PubMed] [Google Scholar]
- Hansen, J. Y. , Markello, R. D. , Tuominen, L. , Nørgaard, M. , Kuzmin, E. , Palomero‐Gallagher, N. , Dagher, A. , & Misic, B. (2022). Correspondence between gene expression and neurotransmitter receptor and transporter density in the human brain. NeuroImage, 264, 119671. 10.1016/j.neuroimage.2022.119671 [DOI] [PubMed] [Google Scholar]
- Hansen, J. Y. , Shafiei, G. , Markello, R. D. , Smart, K. , Cox, S. M. L. , Nørgaard, M. , Beliveau, V. , Wu, Y. , Gallezot, J.‐D. , Aumont, É. , Servaes, S. , Scala, S. G. , DuBois, J. M. , Wainstein, G. , Bezgin, G. , Funck, T. , Schmitz, T. W. , Spreng, R. N. , Galovic, M. , … Misic, B. (2022). Mapping neurotransmitter systems to the structural and functional organization of the human neocortex. Nature Neuroscience, 25(11), 1569–1581. 10.1038/s41593-022-01186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, M. P. , Somerville, L. H. , Ances, B. M. , Andersson, J. , Barch, D. M. , Bastiani, M. , Bookheimer, S. Y. , Brown, T. B. , Buckner, R. L. , Burgess, G. C. , Coalson, T. S. , Chappell, M. A. , Dapretto, M. , Douaud, G. , Fischl, B. , Glasser, M. F. , Greve, D. N. , Hodge, C. , Jamison, K. W. , … Yacoub, E. (2018). Extending the human connectome project across ages: Imaging protocols for the lifespan development and aging projects. NeuroImage, 183, 972–984. 10.1016/j.neuroimage.2018.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz, M. J. , Lein, E. S. , Guillozet‐Bongaarts, A. L. , Shen, E. H. , Ng, L. , Miller, J. A. , van de Lagemaat, L. N. , Smith, K. A. , Ebbert, A. , Riley, Z. L. , Abajian, C. , Beckmann, C. F. , Bernard, A. , Bertagnolli, D. , Boe, A. F. , Cartagena, P. M. , Chakravarty, M. M. , Chapin, M. , Chong, J. , … Jones, A. R. (2012). An anatomically comprehensive atlas of the adult human brain transcriptome. Nature, 489(7416), 391–399. 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda, Y. , Funayama, T. , Tateno, A. , Fukayama, H. , Okubo, Y. , & Suzuki, H. (2017). Modafinil enhances alerting‐related brain activity in attention networks. Psychopharmacology, 234(14), 2077–2089. 10.1007/s00213-017-4614-9 [DOI] [PubMed] [Google Scholar]
- Ishigami, Y. , & Klein, R. M. (2010). Repeated measurement of the components of attention using two versions of the attention network test (ANT): Stability, isolability, robustness, and reliability. Journal of Neuroscience Methods, 190(1), 117–128. 10.1016/j.jneumeth.2010.04.019 [DOI] [PubMed] [Google Scholar]
- Kastner, S. , Pinsk, M. A. , De Weerd, P. , Desimone, R. , & Ungerleider, L. G. (1999). Increased activity in human visual cortex during directed attention in the absence of visual stimulation. Neuron, 22(4), 751–761. 10.1016/S0896-6273(00)80734-5 [DOI] [PubMed] [Google Scholar]
- Lin, D. (1998). Automatic retrieval and clustering of similar words. In 36th Annual Meeting of the Association for Computational Linguistics and 17th International Conference on Computational Linguistics (Vol. 2, pp. 768–774). [Google Scholar]
- Liu, Z.‐Q. , Vázquez‐Rodríguez, B. , Spreng, R. N. , Bernhardt, B. C. , Betzel, R. F. , & Misic, B. (2022). Time‐resolved structure‐function coupling in brain networks. Communications Biology, 5(1), 532. 10.1038/s42003-022-03466-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markello, R. D. , Arnatkeviciute, A. , Poline, J.‐B. , Fulcher, B. D. , Fornito, A. , & Misic, B. (2021). Standardizing workflows in imaging transcriptomics with the abagen toolbox. eLife, 10, e72129. 10.7554/eLife.72129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markett, S. , Nothdurfter, D. , Focsa, A. , Reuter, M. , & Jawinski, P. (2022). Attention networks and the intrinsic network structure of the human brain. Human Brain Mapping, 43(4), 1431–1448. 10.1002/hbm.25734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markett, S. , Reuter, M. , Montag, C. , Voigt, G. , Lachmann, B. , Rudorf, S. , Elger, C. E. , & Weber, B. (2014). Assessing the function of the fronto‐parietal attention network: Insights from resting‐state fMRI and the attentional network test: Assessing the function of the fronto‐parietal attention network. Human Brain Mapping, 35(4), 1700–1709. 10.1002/hbm.22285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrocco, R. T. , & Davidson, M. (1998). Neurochemistry of attention. In R. Parasuraman (Ed.), The attentive brain (pp. 35–50). The MIT Press. [Google Scholar]
- McCormick, C. R. (2022). Lifestyle factors and their impact on the networks of attention. Applied Cognitive Psychology, 36(1), 135–153. 10.1002/acp.3904 [DOI] [Google Scholar]
- Mi, H. , & Thomas, P. (2009). PANTHER pathway: An ontology‐based pathway database coupled with data analysis tools. Protein Networks and Pathway Analysis, 123–140. 10.1007/978-1-60761-175-2_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H. , Ebert, D. , Muruganujan, A. , Mills, C. , Albou, L.‐P. , Mushayamaha, T. , & Thomas, P. D. (2021). PANTHER version 16: A revised family classification, tree‐based classification tool, enhancer regions and extensive API. Nucleic Acids Research, 49(D1), D394–D403. 10.1093/nar/gkaa1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag, C. , Ebstein, R. P. , Jawinski, P. , & Markett, S. (2020). Molecular genetics in psychology and personality neuroscience: On candidate genes, genome wide scans, and new research strategies. Neuroscience & Biobehavioral Reviews, 118, 163–174. 10.1016/j.neubiorev.2020.06.020 [DOI] [PubMed] [Google Scholar]
- Moore, S. R. (2017). Commentary: What is the case for candidate gene approaches in the era of high‐throughput genomics? A response to Border and Keller (2017). Journal of Child Psychology and Psychiatry, 58(3), 331–334. 10.1111/jcpp.12697 [DOI] [PubMed] [Google Scholar]
- Moore, T. , Armstrong, K. M. , & Fallah, M. (2003). Visuomotor origins of covert spatial attention. Neuron, 40(4), 671–683. 10.1016/S0896-6273(03)00716-5 [DOI] [PubMed] [Google Scholar]
- Müller, N. G. , Bartelt, O. A. , Donner, T. H. , Villringer, A. , & Brandt, S. A. (2003). A physiological correlate of the “zoom lens” of visual attention. The Journal of Neuroscience, 23(9), 3561–3565. 10.1523/JNEUROSCI.23-09-03561.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, A. C. , Bertolero, M. A. , Papadopoulos, L. , Lydon‐Staley, D. M. , & Bassett, D. S. (2020). Multimodal network dynamics underpinning working memory. Nature Communications, 11(1), 3035. 10.1038/s41467-020-15541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noudoost, B. , & Moore, T. (2011). The role of neuromodulators in selective attention. Trends in Cognitive Sciences, 15(12), 585–591. 10.1016/j.tics.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, J. , Zeppenfeld, D. , McConnell, E. , Pena, S. , & Nedergaard, M. (2012). Norepinephrine: A neuromodulator that boosts the function of multiple cell types to optimize CNS performance. Neurochemical Research, 37(11), 2496–2512. 10.1007/s11064-012-0818-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent, A. , & Hazrati, L.‐N. (1995). Functional anatomy of the basal ganglia. I. The cortico‐basal ganglia‐thalamo‐cortical loop. Brain Research Reviews, 20(1), 91–127. 10.1016/0165-0173(94)00007-C [DOI] [PubMed] [Google Scholar]
- Petersen, S. E. , & Posner, M. I. (2012). The attention system of the human brain: 20 years after. Annual Review of Neuroscience, 35, 73–89. 10.1146/annurev-neuro-062111-150525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman, T. J. C. , Benyamin, B. , de Leeuw, C. A. , Sullivan, P. F. , van Bochoven, A. , Visscher, P. M. , & Posthuma, D. (2015). Meta‐analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics, 47(7), 702–709. 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- Posner, M. I. , & Dehaene, S. (1994). Attentional networks. Trends in Neurosciences, 17(2), 75–79. [DOI] [PubMed] [Google Scholar]
- Posner, M. I. , & Fan, J. (2008). Attention as an organ system. In Topics in integrative neuroscience: From Cells to Cognition (pp. 31–61). Cambridge: Cambridge University Press. 10.1017/CBO9780511541681.005 [DOI] [Google Scholar]
- Posner, M. I. , & Petersen, S. E. (1990). The attention system of the human brain. Annual Review of Neuroscience, 13(1), 25–42. 10.1146/annurev.ne.13.030190.000325 [DOI] [PubMed] [Google Scholar]
- Posner, M. I. , & Rothbart, M. K. (2007). Research on attention networks as a model for the integration of psychological science. Annual Review of Psychology, 58(1), 1–23. 10.1146/annurev.psych.58.110405.085516 [DOI] [PubMed] [Google Scholar]
- Posner, M. I. , Rothbart, M. K. , Sheese, B. E. , & Voelker, P. (2014). Developing attention: Behavioral and brain mechanisms. Advances in Neuroscience, 2014, 1–9. 10.1155/2014/405094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnders, M. J. M. F. , & Waterhouse, R. M. (2021). Summary visualizations of gene ontology terms with GO‐figure! Frontiers in Bioinformatics, 1, 6. 10.3389/fbinf.2021.638255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud, A. J. , Froesel, M. , Guedj, C. , Hassen, S, B. H. , Cléry, J. , Meunier, M. , Ben Hamed, S. , & Hadj‐Bouziane, F. (2019). Atomoxetine improves attentional orienting in a predictive context. Neuropharmacology, 150, 59–69. 10.1016/j.neuropharm.2019.03.012 [DOI] [PubMed] [Google Scholar]
- Robbins, T. W. (1997). Arousal systems and attentional processes. Biological Psychology, 45(1–3), 57–71. 10.1016/S0301-0511(96)05222-2 [DOI] [PubMed] [Google Scholar]
- Robinson, E. C. , Garcia, K. , Glasser, M. F. , Chen, Z. , Coalson, T. S. , Makropoulos, A. , Bozek, J. , Wright, R. , Schuh, A. , Webster, M. , Hutter, J. , Price, A. , Cordero Grande, L. , Hughes, E. , Tusor, N. , Bayly, P. V. , Van Essen, D. C. , Smith, S. M. , Edwards, A. D. , … Rueckert, D. (2018). Multimodal surface matching with higher‐order smoothness constraints. NeuroImage, 167, 453–465. 10.1016/j.neuroimage.2017.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J. A. , & Van Bockstaele, E. J. (2021). The locus coeruleus‐ norepinephrine system in stress and arousal: Unraveling historical, current, and future perspectives. Frontiers in Psychiatry, 11, 601519. 10.3389/fpsyt.2020.601519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda, M. R. , Rothbart, M. K. , McCandliss, B. D. , Saccomanno, L. , & Posner, M. I. (2005). From the cover: Training, maturation, and genetic influences on the development of executive attention. Proceedings of the National Academy of Sciences of the United States of America, 102(41), 14931–14936. 10.1073/pnas.0506897102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtens, L. H. , Schmidt, R. , de Reus, M. A. , & van den Heuvel, M. P. (2014). Linking macroscale graph analytical organization to microscale Neuroarchitectonics in the macaque connectome. Journal of Neuroscience, 34(36), 12192–12205. 10.1523/JNEUROSCI.0752-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidlitz, J. , Nadig, A. , Liu, S. , Bethlehem, R. A. I. , Vértes, P. E. , Morgan, S. E. , Váša, F. , Romero‐Garcia, R. , Lalonde, F. M. , Clasen, L. S. , Blumenthal, J. D. , Paquola, C. , Bernhardt, B. , Wagstyl, K. , Polioudakis, D. , de la Torre‐Ubieta, L. , Geschwind, D. H. , Han, J. C. , Lee, N. R. , … Raznahan, A. (2020). Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nature Communications, 11(1), 3358. 10.1038/s41467-020-17051-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speranza, L. , Di Porzio, U. , Viggiano, D. , De Donato, A. , & Volpicelli, F. (2021). Dopamine: The neuromodulator of long‐term synaptic plasticity, reward and movement control. Cells, 10(4), 735. 10.3390/cells10040735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y.‐Y. , Rothbart, M. K. , & Posner, M. I. (2012). Neural correlates of establishing, maintaining, and switching brain states. Trends in Cognitive Sciences, 16(6), 330–337. 10.1016/j.tics.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thienel, R. , Voss, B. , Kellermann, T. , Reske, M. , Halfter, S. , Sheldrick, A. J. , Radenbach, K. , Habel, U. , Jon Shah, N. , Schall, U. , & Kircher, T. (2009). Nicotinic antagonist effects on functional attention networks. The International Journal of Neuropsychopharmacology, 12(10), 1295. 10.1017/S1461145709990551 [DOI] [PubMed] [Google Scholar]
- Thompson, K. G. (2005). Neuronal basis of covert spatial attention in the frontal eye field. Journal of Neuroscience, 25(41), 9479–9487. 10.1523/JNEUROSCI.0741-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel, M. P. , Scholtens, L. H. , Barrett, L. F. , Hilgetag, C. C. , & de Reus, M. A. (2015). Bridging cytoarchitectonics and connectomics in human cerebral cortex. The Journal of Neuroscience, 35(41), 13943–13948. 10.1523/JNEUROSCI.2630-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váša, F. , Seidlitz, J. , Romero‐Garcia, R. , Whitaker, K. J. , Rosenthal, G. , Vértes, P. E. , Shinn, M. , Alexander‐Bloch, A. , Fonagy, P. , Dolan, R. J. , Jones, P. B. , Goodyer, I. M. , the NSPN consortium , Sporns, O. , & Bullmore, E. T. (2018). Adolescent tuning of association cortex in human structural brain networks. Cerebral Cortex, 28(1), 281–294. 10.1093/cercor/bhx249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan, B. , Mackie, M.‐A. , Spagna, A. , Wu, T. , Tian, Y. , Hof, P. R. , & Fan, J. (2016). The activation of interactive attentional networks. NeuroImage, 129, 308–319. 10.1016/j.neuroimage.2016.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J. , Pourzinal, D. , Rheinberger, T. , Copland, D. A. , McMahon, K. L. , Byrne, G. J. , & Dissanayaka, N. N. (2022). The attention network test in Parkinson and Lewy body disease: A systematic review. Cognitive and Behavioral Neurology, 35(1), 1–13. 10.1097/WNN.0000000000000292 [DOI] [PubMed] [Google Scholar]
- Zhou, J. , Gennatas, E. D. , Kramer, J. H. , Miller, B. L. , & Seeley, W. W. (2012). Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron, 73(6), 1216–1227. 10.1016/j.neuron.2012.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DATA S1: Supporting information.
DATA S2: Supporting information.
Data Availability Statement
The data that support the findings of this study are available in ant_genes at https://github.com/schinhan/ant_genes. These data were derived from the following resources available in the public domain: Github; https://github.com/justinehansen/hansen_receptors-1; ABAGEN, https://github.com/rmarkello/abagen; and OSF, https://osf.io/st9ae/.


