Abstract
The Drosophila homeodomain protein Even-skipped (Eve) is a transcriptional repressor, and previous studies have suggested that it functions by interfering with the basal transcription machinery. Here we describe experiments indicating that the mechanism of Eve repression involves a direct interaction with the TATA binding protein (TBP) that blocks binding of TBP-TFIID to the promoter. We first compared Eve activities in in vitro transcription systems reconstituted with either all the general transcription factors or only TBP, TFIIB, TFIIF30, and RNA polymerase II. In each case, equivalent and very efficient levels of repression were observed, indicating that no factors other than those in the minimal system are required for repression. We then show that Eve can function efficiently when its recognition sites are far from the promoter and that the same regions of Eve required for repression in vivo are necessary and sufficient for in vitro repression. This includes, in addition to an Ala-Pro-rich region, residues within the homeodomain. Using GAL4-Eve fusion proteins, we demonstrate that the homeodomain plays a role in repression in addition to DNA binding, which is to facilitate interaction with TBP. Single-round transcription experiments indicate that Eve must function prior to TBP binding to the promoter, suggesting a mechanism whereby Eve represses by competing with the TATA box for TBP binding. Consistent with this, excess TATA box-containing oligonucleotide is shown to specifically and efficiently disrupt the TBP-Eve interaction. Importantly, we show that Eve binds directly to TFIID and that this interaction can also be disrupted by the TATA oligonucleotide. We conclude that Eve represses transcription via a direct interaction with TBP that blocks TFIID binding to the promoter.
Regulation of transcription occurs by multiple distinct mechanisms, which can involve repressive as well as activating interactions between regulatory proteins and a variety of targets. Recent studies have identified a large number of proteins capable of repressing transcription, and evidence supporting a number of mechanisms has been presented (for reviews, see references 13 and 22). One way to distinguish different types of repressors is to consider two classes: those that function by influencing chromatin structure and those that interact with components of the transcriptional machinery. But even within these divisions there appear to be multiple different modes of repression, and a current challenge is to understand the underlying mechanisms.
A number of repressor proteins are now known or suspected to function by altering chromatin (reviewed in reference 26). One class consists of a number of sequence-specific DNA binding proteins that recruit to the template, via interacting corepressors, a histone deacetylase (reviewed in reference 36). This pathway, conserved from yeast cells to humans, suggests a satisfying though unproven mechanism: deacetylation of histones could allow tighter histone-DNA interactions, blocking the access of transcription factors to the promoters. Other repressors, including the SIR proteins in yeast cells (e.g., see reference 15), the Polycomb-group proteins in Drosophila and other metazoans (reviewed in references 35 and 38), and the TUP1-SSN6 corepressor complex in yeast cells (reviewed in reference 42), also appear to function by stabilizing chromatin structure, likely by interactions with histones. TUP1-SSN6, which is recruited to a number of different promoters by various transcription factors, is notable because it appears able to function both by influencing nucleosomal structure (see, for example, reference 5) and by establishing a repressive interaction with a component(s) of the basal transcription machinery (reference 40 and references therein). It is possible and perhaps likely that many repressors will utilize multiple mechanisms to ensure the silencing of target genes.
Many repressors function by interacting directly with other transcription factors. Certain of these employ a quenching mechanism by which the DNA-bound repressor directly interferes with the activity of an activator bound nearby (see references 13, 22, and 28 for further discussion). This mechanism may be particularly important for genes with complex promoter regions, allowing independent regulation of individual enhancer elements (reviewed in reference 9). Another class of repressors, called direct repressors, are thought to function by contacting components of the basal transcription machinery. One well-studied example is Dr1-DRAP1, a heterodimer conserved from yeast cells to humans (25). This protein appears to be a global repressor of transcription, as it targets promoters not by DNA binding but instead by an interaction with the general transcription factor, the TATA binding protein (TBP). In vitro experiments indicate that Dr1-DRAP1 does not interfere with TBP-DNA interaction but instead prevents the association of other general transcription factors, i.e., TFIIA and/or TFIIB (e.g., see references 32 and 51). The adenovirus E1A protein can also repress transcription from several promoters, likely through a direct interaction with TBP (45). Mot1 is an ATP-dependent global repressor in yeast cells that is also thought to function through TBP, in this case by dissociating it from DNA, although whether Mot1 directly contacts TBP is not known (1).
Several sequence-specific DNA binding proteins have also been suggested to function through interactions with general transcription factors. For example, the Drosophila Krüppel protein can interact in vitro with the small subunit of TFIIE (43) and several proteins can bind TBP. These include the unliganded thyroid hormone receptor (6) and two homeodomain proteins, the Drosophila Even-skipped protein (Eve) (50) and the mouse Msx1 protein (53). Human MDM2, which is recruited to promoters by the p53 protein, can repress basal transcription in vitro and has been shown to interact with both TBP and TFIIE (48). Although in many of these cases there is evidence that the protein-protein interactions detected are relevant to repression, the mechanisms are unclear and for several, alternative modes of repression have been proposed. For example, unliganded thyroid hormone receptor (and other nuclear receptors) have been shown to function, at least in part, by the histone deacetylase recruitment mechanism described above (16, 33). Krüppel may also function by quenching (29, 54), and Eve has been suggested to interfere indirectly with TBP (TFIID) binding to the promoter (2).
Eve is a primary pair-rule gene that plays a critical role in Drosophila embryogenesis, and genetic experiments have been consistent with Eve acting negatively in most instances (31). Eve was first suggested to function as a transcriptional repressor as determined by transient-transfection assays (10, 20). In vitro transcription experiments supported this idea (3) and suggested that Eve functions to interfere with an early step in assembly of the preinitiation complex (23). Subsequent transfection experiments also suggested that Eve functions as a direct repressor of basal transcription (11). This study also defined an Ala-Pro-rich repression domain, which is characteristic of repression regions found in several other repressors (13). How Eve actually functions is not known, although as mentioned above two models have been proposed. In one (2, 47), a form of cooperative DNA binding, probably mediated by residues encompassing the Ala-Pro-rich repression domain, is proposed to allow recognition of nonspecific DNA sites surrounding the promoter, thereby interfering with TFIID binding. In another (30, 50), it is suggested that the interaction between Eve and TBP, which requires the Eve repression domain, interferes in some way with TFIID function. Although both models focus on a step involving TFIID, the mechanisms proposed are very different.
Here we present experiments that provide new insights into how Eve represses transcription. The data strongly support the notion that the Eve-TBP interaction is responsible for repression and provide evidence for a novel mechanism by which this interaction directly interferes with TATA box binding by TBP-TFIID.
MATERIALS AND METHODS
Recombinant plasmids.
A Drosophila alcohol dehydrogenase (Adh) proximal promoter fragment (−40 to 10, relative to the RNA start site) was inserted into the SacI site of pC2AT (44) to construct the G-less cassette vector, Adh40/10-G380. Different in vitro transcription templates were made by inserting Eve binding site(s) at appropriate positions in Adh40/10-G380. Full-length cDNAs of Drosophila TBP, TFIIB, and TFIIF30 were cloned into the bacterial expression vector PET-his3a. PET-hisEve (wild type), PET-hisEveABCD, PET-hisEveABF, PET-hisEveABEF, and PET-hisEveBC2D2 were generated by subcloning appropriate DNA fragments from the PET-3a vector (50) into PET-his3a. PET-GAL4-EveCDEF-H6 and PET-GAL4-EveBCDEF-H6 were constructed from corresponding in vivo expression vectors (provided by M. Um) by subcloning into PET-23d (Novagen). Two Eve homeodomain mutant expression plasmids: PET-GAL4-EveΔN-BCDEF-H6 and PET-GAL4-EveΔC-BCDEF-H6 were made by PCR amplification of appropriate DNA fragments to create in-frame deletions, and PET-hisEve-BΔN was made by a similar strategy. Vectors encoding Eve proteins for in vitro transcription-translation were made by subcloning appropriate DNA fragments into the PET-3a vector. All plasmids were sequenced to confirm their identities. Detailed information concerning plasmid constructs is available upon request.
Protein purification.
RNA polymerase II (Pol II) was purified from HeLa nuclear extract pellets by the method of Reinberg and Roeder (41), which involved DE-52, Sephadex A-25 and phosphocellulose chromatography. General transcriptional factors TFIIA and TFIIE-TFIIF-TFIIH were partially purified from HeLa nuclear extract as described by Chiang et al. (4). Flag epitope-tagged TFIID was kindly provided by Hui Ge (National Institutes of Health). Bacterial strain BL21 was transformed with different PET expression plasmids and induced with isopropyl-β-d-thiogalactopyranoside as described by Um et al. (50). For his-dTFIIB and his-dTFIIF30, cell lysates were loaded onto a Ni2+ agarose column, and proteins were eluted with 200 mM imidazole (Qiagen). Purification of his-dTBP and GAL4-Eve-H6 fusion proteins was similar except that cell lysates were first passed through DEAE-CL6B, and flowthroughs were loaded on a Ni2+ column. His-Eve (wild type) and other Eve derivatives were purified under denaturing conditions according to standard protocol (Qiagen) and renatured by step dialysis. All recombinant proteins were dialyzed against buffer BC-100 (20 mM Tris [pH 7.9], 100 mM KCl, 20% glycerol, 0.05% Nonidet P-40 [NP-40], 0.1 mM EDTA, 5 mM dithiothreitol [DTT] and 0.5 mM phenylmethylsulfonyl fluoride).
In vitro transcription.
Each transcription reaction mixture (25 μl) contained 60 to 80 mM KCl, 12% glycerol (vol/vol), 25 mM HEPES-KOH (pH 8.2), 3 mM MgCl2, 5 mM DTT, bovine serum albumin (0.5 mg/ml), 4 U of RNasin (Promega), 0.1 mM 3′-o-methyl-GTP, 0.25 mM ATP and UTP, 25 μM CTP, 5 μCi of [α-32P]CTP, and 100 ng of the indicated supercoiled plasmid templates. In the complete system, each reaction mixture contained 100 ng of HeLa Pol II, 300 ng of TFIIA fraction, 10 ng of recombinant his-dTFIIB, 50 ng of Flag epitope-tagged holo-TFIID, 500 ng of TFIIE-TFIIF-TFIIH fraction, and the indicated amounts of purified Eve proteins. In the minimal system, each reaction mixture contained 100 ng of HeLa RNA Pol II, 50 ng of His-dTBP, 40 ng of His-dTFIIB, 48 ng of His-dTFIIF30, and the indicated amounts of Eve proteins. In both assays, the amount of each factor used was optimized such that they were all saturating relative to Pol II. After incubation at 30°C for 1 h, reactions were stopped by adding 100 μl of stop solution (20 mM EDTA, 1% sodium dodecyl sulfate [SDS], and 200 mM NaCl) and 125 μl of 2 M ammonium acetate. RNA was extracted with phenol-chloroform, precipitated with ethanol, analyzed by 6% polyacrylamide–urea gel electrophoresis, and visualized by autoradiography. Quantitation of labeled RNA was performed with a PhosphorImager (Molecular Dynamics).
For single-round transcription experiments, templates were first incubated with the indicated proteins (TBP or Eve) at 21°C for 20 min; other proteins were then added to reaction mixtures for another 20 min of incubation to allow preinitiation complexes to assemble. Sarkosyl (final concentration 0.018% [wt/vol]) and ribonucleotide 5′ triphosphates were added to reaction mixtures to start the transcription. After 30 min, reactions were stopped and analyzed as described above.
Gel mobility shift assays.
The DNA fragments indicated in the text were end labeled with Klenow polymerase in the presence of [α-P32]dATP. Binding reactions were carried out in a volume of 20 μl in 10 mM Tris (pH 7.9), 10% (vol/vol) glycerol, 100 mM KCl, 1 mM DTT, 0.05% NP-40, bovine serum albumin (0.3 mg/ml), poly(dC-dI) (5 μg/ml) and contained 0.25 ng of labeled DNA and the indicated amounts of Eve protein. After incubation at 30°C for 30 min, reaction mixtures were loaded onto a 10% polyacrylamide gel (40:1, acrylamide to bisacrylamide) containing 0.25 × TBE buffer (11). Electrophoresis was carried out at 200 V for 2 h.
Protein-protein interaction assays.
Glutathione S-transferase (GST) fusion protein binding assays were performed as described by Um et al. (50) with minor modification. DNA fragments of 16 bp were made by annealing two synthesized complementary DNA oligonucleotides. 35S-methionine-labeled Eve proteins were produced by in vitro transcription, which was followed by translation in reticulocyte lysate (Promega). In vitro-translated Eve proteins (1 μl) or purified recombinant His-tagged Eve proteins (200 ng) were incubated in 40-μl reaction mixtures with agarose-GST-TBP (2 μg) complexes in the absence or presence of the indicated amount of DNA fragment. After 2 h of incubation at room temperature, beads were washed and bound proteins were eluted with 20 mM reduced glutathione and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). In vitro-labeled Eve proteins were visualized by fluorography, and His-Eve proteins were detected by Western blotting carried out as described by Kohtz et al. (27), except that anti-Eve antibodies were used (7).
For the experiments examining the interaction between TFIID and Eve (see Fig. 8C), 6 μg of anti-Flag M2 monoclonal antibody (IBI/Kodak) was incubated with 5 μl of protein A Sepharose beads in 100 μl of IP buffer (20 mM Tris [pH 7.9], 500 mM NaCl, 0.1% NP-40) for 1 h at room temperature. After the beads were washed, they were incubated with 1 μg of Flag epitope-tagged TFIID in 100 μl of IPB buffer (13 mM HEPES [pH 7.9], 28 mM [NH4]2SO4, 100 mM KCl, 0.5 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride, 0.1% Tween 20) at room temperature for 3 h. Then 25 ng of purified His-tagged Eve was added to the tubes containing either bead-antibody-TFIID complexes or controls lacking TFIID, with or without the adenovirus major late (ML) TATA DNA fragment. After 3 h of incubation at room temperature, beads were washed and proteins were eluted three times with 100 μl of 0.2 M glycine (pH 2.5). Eluted proteins were precipitated with trichloroacetic acid, dissolved in sample buffer, and loaded onto a polyacrylamide-SDS gel. Immunoblotting was performed as described above.
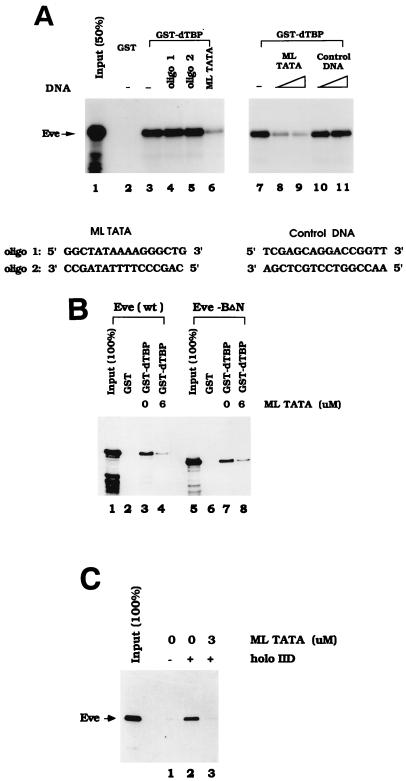
FIG. 8.
TATA sequence specifically inhibits the interaction between TBP and Eve. (A) 35S-labeled Eve was produced by in vitro transcription and translation and then incubated with either GST (lane 2) or GST-dTBP (lanes 3 to 11) in the absence (lanes 2, 3, and 7) or presence of various DNA oligonucleotides at concentrations of 10 μM (lanes 6, 8, and 10), 15 μM (lanes 9 and 11) or 20 μM (lanes 4 and 5). The sequences of ML TATA and the control DNA are shown below the panel. (B) His-tagged Eve (wt) or Eve-BΔN (200 ng) was incubated with either 2 μg of GST or 2 μg of GST-dTBP in the absence (lanes 2, 3, 6, and 7) or presence (lanes 4 and 8) of ML TATA at a concentration of 6 μM. The eluted Eve proteins were detected by Western blotting with anti-Eve antibodies. (C) Recombinant Eve-wt (25 ng) was incubated with either protein A Sepharose beads plus anti-Flag antibodies (lane 1) or 1 μg of Flag epitope-tagged TFIID bound to protein A Sepharose beads via anti-Flag antibodies (lanes 2 and 3). Then 3 μM ML TATA was added to one of these reaction mixtures (lane 3). Bound proteins were eluted and detected by Western blotting with anti-Eve antibodies.
RESULTS
Eve represses transcription in both fully and minimally reconstituted in vitro systems.
It has been shown previously that recombinant Eve protein purified from Escherichia coli can repress transcription in vitro in a binding site-dependent manner in Drosophila embryo nuclear extracts (3, 23) and in a reconstituted system (2). We extended this approach to examine the mechanism of Eve repression in more detail. In all experiments, Eve was tagged with six His residues at its N terminus, purified from E. coli by Ni2+ agarose beads under denaturing conditions, and then renatured (e.g., see Fig. 4A). In an initial experiment, in vitro transcription was reconstituted with TFIIA, IIE, IIF, and IIH partially purified from HeLa nuclear extract (4), recombinant Drosophila TFIIB, purified HeLa TFIID (4, 8), and RNA Pol II purified from HeLa nuclear extract pellets (41). For templates, the core sequence from the Drosophila Adh proximal promoter (−40 to 10, relative to the RNA start site [49]) was cloned into G-less cassette templates (44). Two templates were used in most experiments, one with three Eve binding sites (NP3) 49 bp upstream of the Adh core promoter and a 380-bp G-less sequence (NP3-Adh40/10-G380) and the other with the same Adh TATA sequence but without Eve binding sites and containing a 280-bp G-less cassette (Adh40/10-G280), which served as a control template (see Fig. 1A).
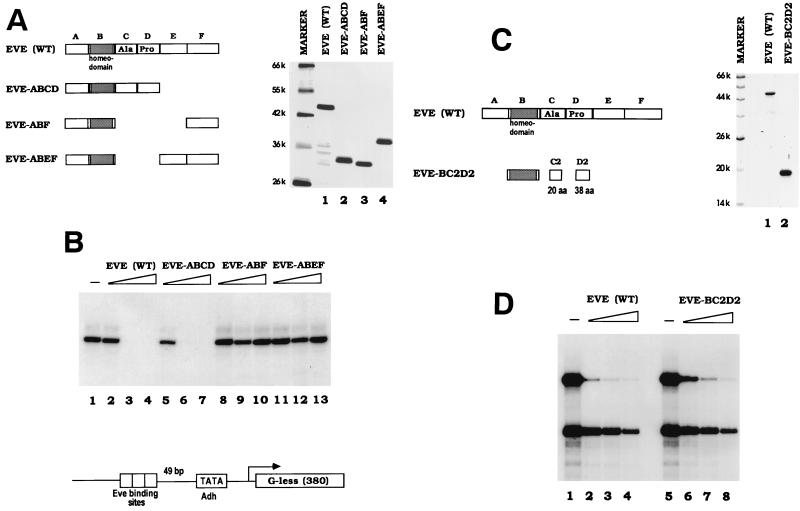
FIG. 4.
The Eve repression domain is required for repression in vitro. (A) Silver staining of a protein gel containing 500 ng of the Eve derivatives used in the transcription assays shown in panel B. The numbers at the left indicate the molecular weights of protein markers (k, thousand). A schematic diagram of the Eve proteins is shown on the left. Full-length Eve is divided into six regions according to the study by Han and Manley (11). (B) In vitro transcription assays in the minimal system. Reaction mixtures contained 0, 50, 125, or 250 nM concentrations of the indicated Eve derivative. The template used in the experiments, NP3-Adh40/10-G380, is diagrammed at the bottom of the panel. (C) Coomassie blue staining of a protein gel containing wild-type (WT) Eve and Eve-BC2D2 (2 μg). The diagram on the left depicts the subdivision of the Eve repression regions, as defined by Han and Manley (11). The sizes of molecular weight markers are indicated (k, thousand). (D) Eve (WT) and Eve-BC2D2 were analyzed for repression activity in the minimal transcriptional system. Reaction mixtures contained 0, 60, 120, or 240 nM concentrations of the indicated protein. The two templates used here are the same as in Fig. 1.
FIG. 1.
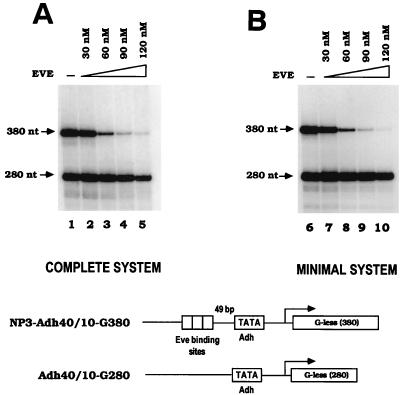
Eve efficiently represses in vitro transcription in reconstituted systems. (A) In vitro transcription assays were performed to examine Eve activity in a complete system. Each transcription reaction mixture contained purified HeLa RNA Pol II and Flag-tagged TFIID; partially purified HeLa TFIIA, TFIIE, TFIIF, and TFIIH; recombinant Drosophila TFIIB; and the indicated concentrations of purified recombinant Eve. Two G-less cassette templates were used in the reactions. NP3-Adh40/10-G380, with three Eve binding sites (NP3) 49 bp upstream of Drosophila Adh proximal promoter, was the test template, and Adh40/10-G280 was the control template. 32P-labeled RNA products were analyzed by electrophoresis and autoradiography. (B) Eve represses in vitro transcription reconstituted by a minimal set of factors. Transcription from Drosophila Adh promoter was reconstituted with HeLa Pol II and purified His-tagged Drosophila recombinant general transcriptional factors (dTFIIB, dTBP, and dTFIIF30). NP3-Adh40/10-G380 and Adh40/10-G280 served as templates, and the indicated concentrations of purified Eve were also included in reaction mixtures. A schematic diagram of two templates used in the transcription reactions is shown at the bottom of the figure.
In vitro transcription with the above components was carried out as described in Materials and Methods. When purified recombinant Eve was added to reaction mixtures, transcription from the template with Eve binding sites (NP3-Adh40/10-G380) was strongly and preferentially repressed in a concentration-dependent manner (Fig. 1A). The maximum binding site-dependent repression, defined as the fold repression obtained with NP3-Adh40/10-G380 divided by the fold repression obtained with Adh40/10-G280, was 20- to 25-fold. These findings are largely consistent with previous in vitro studies by Austin and Biggin (2), and the high efficiency of repression we detected is comparable to that observed previously in transient-transfection assays (11).
To extend these results, we wished to determine whether efficient repression could be detected in a simpler reconstituted transcription system. It is well known, for example, that activated transcription is not detected when TBP is substituted for TFIID (e.g., see reference 39), and it has been reported that Eve repression is at least fivefold less efficient when TBP replaces TFIID (2). We took advantage of studies of Tyree et al. (49), which showed that transcription from the Adh promoter fragment employed above could be obtained with only four factors: TBP, TFIIB, TFIIF30 (RAP30), and Pol II. The three Drosophila general factors were all His-tagged and purified from E. coli, and Pol II was again purified from HeLa nuclear extract pellets. Consistent with the results of Tyree et al. (49), efficient transcription was detected with this collection of factors and the Adh promoter–G-less cassette plasmids described above (Fig. 1B). We then tested whether Eve could repress transcription reconstituted by these four proteins in a binding site-dependent manner. As shown in Fig. 1A, increasing concentrations of Eve were added to reaction mixtures, and the relative amounts of transcription from the two templates were determined. The data (Fig. 1B) shows that Eve in fact repressed transcription reconstituted by these factors in a strong, binding site-dependent manner. At an Eve concentration of 120 nM, 36-fold repression was achieved from the template with Eve binding sites, whereas transcription from the control template was only slightly repressed (1.4-fold), resulting in 25-fold binding site-dependent repression. At higher Eve concentrations, moderate repression of the control template, also observed in the fully reconstituted system and previously by others (2, 23), was detected (results not shown). Most importantly, however, for each Eve concentration tested, binding site-dependent repression levels were equivalent in the complete and minimal systems. This finding indicates that components of the transcription machinery other than TBP, TFIIB, TFIIF30, and Pol II are unnecessary for full Eve repression in vitro. (Essentially identical levels of binding site-dependent repression were observed with preparations of Pol II purified to homogeneity, thus ruling out the presence of a corepressor in the Pol II preparation [results not shown].) Transcription mediated by this minimal set of general factors thus provides a relatively simple system to dissect the Eve repression mechanism.
Requirements for binding site-dependent repression by Eve.
In previous studies of Eve repression, reporter templates have always contained multiple Eve binding sites. To test whether the copy number of Eve binding sites affects Eve activity, we compared the repressive activity of Eve on the transcription of templates with different numbers of Eve binding sites upstream of the promoter. In vitro transcription assays were performed with minimal factors as described above. Besides NP3-Adh40/10-G380, two other templates, each with a single copy of an Eve binding site (NP; TCAATTAAATGA) 32 bp upstream of the Adh minimal promoter in either orientation, were also used. The data (Fig. 2) show that transcription from both templates with the single Eve binding site was not repressed at all, while transcription from NP3-Adh40/10-G380 was again efficiently inhibited. Increasing the number of Eve binding sites (to six) or inverting the three sites did not affect repression (data not shown). Given that each NP element binds two molecules of Eve (18), six Eve molecules should be bound to the NP3-containing templates. The binding site requirements for Eve repression are very similar to the requirements for activation by many transcriptional activators. In the case of Eve, this reflects in part cooperative DNA binding, as Eve binds significantly more tightly to a DNA fragment containing three binding sites than to a fragment with only a single site (results not shown).
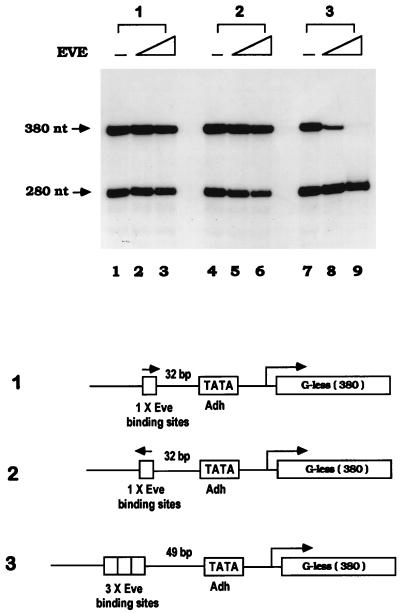
FIG. 2.
Repression by Eve requires multiple binding sites. In vitro transcription assays were carried out with the minimal set of general transcriptional factors (dTBP, dTFIIB, and dTFIIF30) and RNA Pol II as described in Fig. 1B. Three templates with different numbers of Eve binding sites were used in the reactions, along with the control template Adh40/10-G280. Then 0, 60, or 120 nM purified recombinant Eve was added to each reaction mixture. Templates are diagrammed at the bottom of the figure.
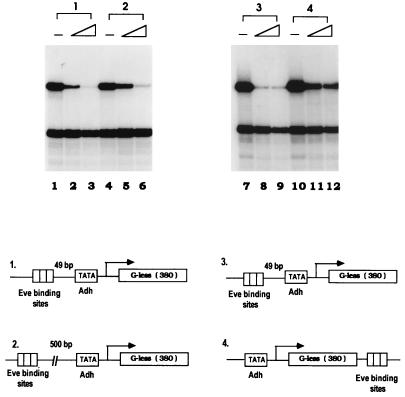
We next wished to determine whether Eve could function when its binding sites were moved to sites distant from the promoter. Although it is well established that some repressors can function at a considerable distance (see reference 9 for a review), Eve was essentially inactive in our previous transfection experiments when its binding sites were situated 500 bp upstream of the promoter (11), and previous in vitro experiments with Eve have all used templates with binding sites near the promoter (2, 3, 23). Indeed, to our knowledge it has not been established that a direct repressor can function when its sites are distant from the promoter. To address this, two transcription templates were constructed, one with three Eve binding sites 500 bp upstream of the Adh TATA sequence and another with three sites 400 bp downstream of the promoter (as indicated in Fig. 3). In vitro transcription assays were carried out with the minimal set of factors as described above. Strikingly, when Eve sites were situated 500 bp upstream, efficient repression nearly indistinguishable from that of the control was detected (Fig. 3, panels 1 and 2). For the template with Eve binding sites 400 bp downstream of the TATA sequence, transcription was also significantly repressed, although less efficiently than was observed with the control (Fig. 3, panels 3 and 4). These results establish that Eve can indeed efficiently repress transcription when its binding sites are located a considerable distance from the promoter. Why this was not observed in our previous transfection experiments is not clear, but it may reflect differences in the basal promoters employed.
FIG. 3.
Eve can repress in vitro transcription when its binding sites are distant from the promoter. In vitro transcription from Drosophila Adh proximal promoter was reconstituted with recombinant dTBP, dTFIIB, dTFIIF30, and Pol II. Three Eve binding sites (NP3) were inserted at the indicated positions in the template, Adh40/10-G380, which are shown by the diagram at the bottom of the figure. Transcription assays were performed in the absence of Eve (lanes 1, 4, 7, and 10) or in the presence of Eve at concentrations of 60 nM (lanes 2, 5, 8, and 11) or 120 nM (lanes 3, 6, 9, and 12).
The Eve repression domain is required for repression in vitro.
Our previous work defined an Ala-Pro-rich region in Eve, located just C terminal to the homeodomain, that is essential for repression in transfection assays (11) and for TBP binding (50) but not for DNA binding (11, 30). We next wished to determine whether this region is required for repression in vitro. To this end, three additional Eve derivatives were expressed in E. coli and purified as described above (see Fig. 4A), and their repression activities were determined. As shown in Fig. 4B, Eve-ABCD, in which the C-terminal 130 residues of Eve (EF) were deleted, retained full activity. This derivative was previously found to function like full-length Eve in transfection and TBP binding assays (11, 50). In sharp contrast, two other Eve derivatives, Eve-ABF and Eve-ABEF, which lack the defined repression domain, were completely inactive, indicating that the Ala-Pro-rich region is essential for transcriptional repression in vitro. In previous studies, the Eve repression domain CD was further subdivided to produce fragments that have various degrees of repressive activity in transfection assays (11). C2D2 (Fig. 4C), called the minimal repression domain, contains the majority of Ala-Pro residues and imparts to the homeodomain nearly full repression and TBP binding activities (11, 50). We prepared another His-tagged Eve derivative, Eve-BC2D2, which contains only the homeodomain plus the minimal repression domain, and tested its repressive activity. As shown in Fig. 4D, the 135-residue Eve-BC2D2 protein functioned as a strong binding site-dependent repressor, with activity comparable to that of full-length Eve. As expected, gel shift assays indicated that BC2D2 bound DNA with an affinity similar to that of wild-type Eve (data not shown). Taken together, our data show that Eve’s ability to repress in vitro transcription correlates completely both with repression activity in vivo and with TBP binding.
Sequences within the Eve homeodomain are required for repression in vitro.
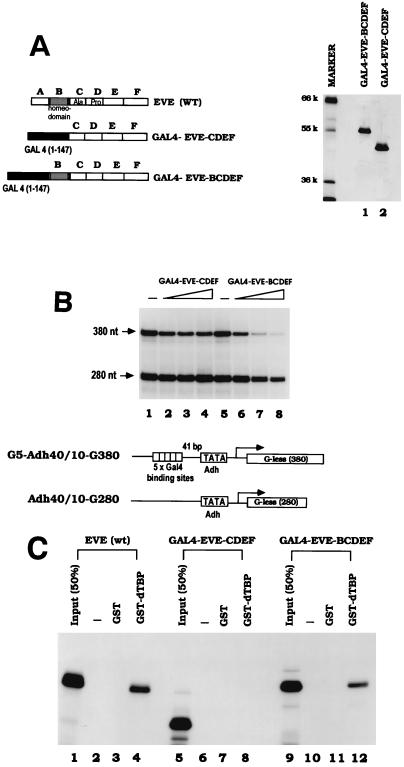
The above results, together with our previous studies, confirm the important role of the Ala-Pro-rich region in Eve-mediated repression and are entirely consistent with its functioning by interacting with TBP. Our previous results also showed that the homeodomain is required for interaction with TBP and can be required for optimal repression in transfection assays even in the context of a heterologous DNA binding domain (30, 50). However, in this case it was hard to exclude the possibility that the homeodomain’s role in repression reflects DNA and not TBP binding. This possibility may be strengthened by the fact that the homeodomain consensus binding site is very A-T rich, and evidence that the homeodomain protein Engrailed can repress transcription in vitro by binding to the TATA box and competing with TFIID has been presented (34). We therefore set out to examine more carefully the role of the Eve homeodomain in repression by utilizing GAL4-Eve fusion proteins in minimal in vitro transcription reactions. We first constructed and purified two such fusion proteins: GAL4-EveCDEF and GAL4-EveBCDEF (Fig. 5A; domain B corresponds to the homeodomain). Five GAL4 binding sites were placed 41 bp upstream of the Adh minimal promoter, and in vitro transcription was performed as described above with templates lacking or containing these sites. As shown in Fig. 5B, GAL4-EveCDEF had almost no repression activity, whereas GAL4-EveBCDEF significantly repressed transcription in a GAL4 binding site-dependent manner. Repression efficiency was about 50% the level observed with Eve itself. Gel shift assays indicated that both GAL4 fusion proteins bound GAL4 binding sites with the same affinity (data not shown). These results indicate that the homeodomain plays a role in repression in addition to DNA binding site recognition, although they do not by themselves indicate what this role actually is.
FIG. 5.
The Eve homeodomain contains residues required for repressing transcription and binding TBP. (A) Analysis of purified GAL4-Eve fusion proteins by SDS-PAGE. Proteins (500 ng) were visualized by silver staining. The molecular weights of protein markers are indicated on the left (k, thousand). A schematic diagram of the proteins is also shown. (B) In vitro transcription assays were performed with the minimal system and purified GAL4-Eve fusion proteins (as shown in Fig. 5A). Reaction mixtures contained 0, 70, 105, or 140 nM concentrations of the indicated GAL4-Eve protein. The template, G5-Adh40/10-G380, contained five GAL4 binding sites upstream of Adh promoter. A schematic diagram of the two templates used in the transcription reactions is shown. (C) [35S]methionine-labeled Eve (wt) and the two GAL4-Eve fusion proteins were produced by in vitro transcription and translation, and equal amounts (Input) of Eve proteins were incubated with either 2 μg of GST or 2 μg of GST-dTBP linked to glutathione-agarose beads. After an extensive washing, bound proteins were eluted with glutathione, resolved by electrophoresis, and visualized by autoradiography.
So far, we have observed a perfect correlation among Eve derivatives with respect to repression activity and TBP binding. To test the TBP binding ability of the GAL4-Eve fusion proteins, GST binding experiments were carried out. GST and GST-dTBP were purified from E. coli and immobilized on glutathione agarose beads. [35S]methionine-labeled GAL4- EveCDEF, GAL4-EveBCDEF, and wild-type Eve were produced by in vitro transcription and translation, and aliquots were mixed with the appropriate beads. Proteins, after an extensive washing, were eluted with buffer containing 20 mM glutathione, resolved by SDS-PAGE, and subjected to fluorography. The data obtained (Fig. 5C) show that GAL4-EveBCDEF specifically bound to GST-dTBP with almost the same efficiency as full-length Eve, while GAL4-EveCDEF did not interact detectably. These data indicate that the Eve homeodomain is required for the Eve-TBP interaction within the context of GAL4 fusion proteins. Once again, repression activity correlates with TBP binding.
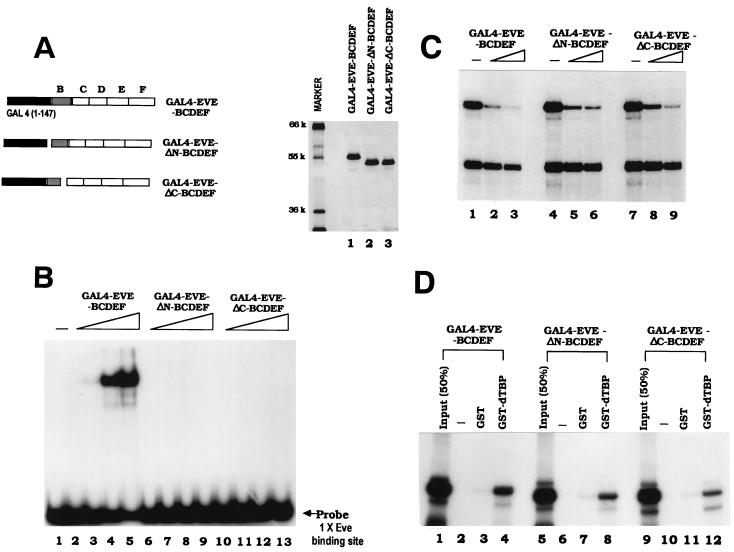
The activity of the GAL4-Eve BCDEF protein provided an opportunity to determine whether the role of the homeodomain in repression involves a function in addition to DNA binding, i.e., TBP binding. Based on the crystal structure of an Eve homeodomain-DNA complex, both the N-terminal arm and the C-terminal helix contribute to DNA recognition (18). We made two Eve homeodomain mutants based on this structure. In one (GAL4-EveΔN-BCDEF) the 8-amino-acid N-terminal arm was deleted, and in the other (GAL4-EveΔC- BCDEF) the 20-amino-acid C-terminal helix was deleted. These two GAL4 fusion proteins were purified from E. coli, as shown in Fig. 6A. The DNA binding activities of GAL4-Eve-BCDEF, GAL4-EveΔN-BCDEF, and GAL4-EveΔC-BCDEF were tested in gel shift assays with a 20-bp DNA fragment containing a single Eve binding site. The results (Fig. 6B) indicate that the DNA binding activity of both GAL4-Eve homeodomain mutants was completely abolished, establishing that as expected both the N-terminal arm and the C-terminal helix of the Eve homeodomain are required for DNA binding. Identical results were observed with a DNA fragment containing three Eve binding sites (NP3). When GAL4 binding sites were used as probes, both homeodomain mutants bound DNA with the same affinity as had GAL4-EveBCDEF (data not shown).
FIG. 6.
GAL4-Eve mutant proteins repress transcription and bind TBP. (A) Silver staining of a protein gel containing purified GAL4 Eve proteins (500 ng), which are also diagrammed on the left. The molecular weights of protein markers are indicated to the left of the gel (k, thousand). (B) DNA binding activity of the GAL4-Eve proteins in Fig. 6A was examined by gel shift assays. The DNA probe used in the experiment was an end-labeled 20-bp DNA fragment containing a single Eve binding site. Reaction mixtures contained 0, 4, 16, 64, or 256 nM concentrations of the indicated GAL4-Eve protein. Conditions for the gel shift assays are described in Materials and Methods. (C) The GAL4-Eve fusion proteins were tested for transcriptional repressive activity at concentrations of 100 nM (lanes 2, 5, and 8) or 200 nM (lanes 3, 6, and 9). G5-Adh40/10-G380 and Adh40/10-G280 were the templates used in the assays. (D) Binding of in vitro translated 35S-labeled GAL4-Eve fusion proteins to GST-dTBP. Equal amounts of the indicated GAL4-Eve proteins were incubated with either 2 μg of GST or 2 μg of GST-dTBP bound to glutathione-agarose beads. Protein complexes were eluted and analyzed as for Fig. 5.
We next compared the repression activities of the GAL4 fusion proteins in in vitro transcription assays by using the minimal system and DNA templates described above. Both GAL4-EveΔN-BCDEF and GAL4-EveΔC-BCDEF retained GAL4 binding site-dependent repressive activity, a result comparable to that observed with GAL4-EveBCDEF (Fig. 6C). These data establish that residues within the Eve homeodomain contribute to repression by a mechanism distinct from DNA binding. To determine whether the two homeodomain mutants retained the ability to interact with TBP, these two proteins, together with GAL4-Eve-BCDEF, were produced by in vitro transcription and translation, and binding experiments with purified GST-TBP were performed as described above. The results (Fig. 6D) indicate that both proteins retained almost full TBP binding activity, correlating with their activity in transcriptional repression. Together, these results establish that residues within the Eve homeodomain play a role in repression in addition to DNA binding and further support the notion that this is to facilitate binding to TBP.
Preincubation of TBP and template can prevent Eve repression.
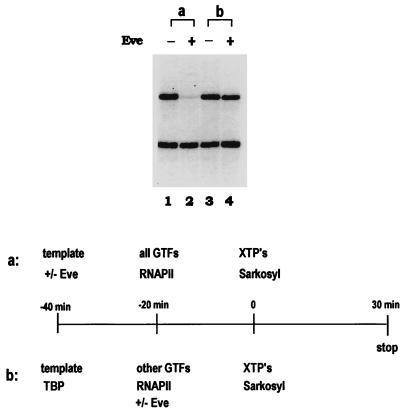
The data described above and previously (30, 50) argue strongly that an interaction with TBP is essential for Eve-mediated repression. But how this interaction actually represses transcription has not been addressed. Two general models can be suggested. In one, the Eve-TBP interaction interferes with TBP-TFIID binding to the TATA box, while in the other it blocks a subsequent step in preinitiation complex assembly. To distinguish between these, we carried out order-of-addition and preincubation experiments under conditions that allowed only a single round of transcription by using the anionic detergent Sarkosyl (14, 24). In the presence of Sarkosyl, transcription from already-assembled preinitiation complexes can occur but no new preinitiation complexes can form, limiting transcription to a single round. In the minimal transcription system used here, 0.018% (wt/vol) Sarkosyl was found to block preinitiation complex assembly but to allow transcription from preassembled factors (data not shown). Transcription reactions were performed with the Eve binding site-containing and the control templates described above. When the templates were preincubated first in the presence or absence of Eve and then with the transcription factors, followed by addition of XTPs and Sarkosyl, Eve strongly and specifically repressed transcription from the template containing Eve binding sites (Fig. 7, panel a). In contrast, if the templates were first preincubated with TBP, followed by addition of the other factors plus or minus Eve, no repression was observed (Fig. 7, panel b). These results suggest that Eve does not function by blocking a step subsequent to TBP binding and instead provide strong support for the idea that Eve functions by interfering with TBP (or TFIID; see below) binding to the promoter. We note that a similar conclusion was reached by Austin and Biggin (2), who used a related approach.
FIG. 7.
Preincubation of TBP and template prevents Eve repression. Single-round transcription assays were performed in the presence of 0.018% (wt/vol) Sarkosyl with the minimal set of transcription factors. The transcription templates were NP3-Adh40/10-G380 and Adh40/10-G280. Reaction mixtures contained 0 or 120 nM Eve. The detailed experimental procedure is depicted at the bottom of the figure.
TATA sequence specifically inhibits the interaction between TBP and Eve.
The data presented above provides evidence that the interaction between Eve and TBP results in repression by interfering with TBP binding to the promoter. This could be envisioned as establishing a competition between Eve and the TATA box for binding TBP. A prediction of such a model is that an excess of TATA-containing DNA should be able to compete the TBP-Eve interaction we described here and previously. Indeed, our observation that the addition of ethidium bromide to binding reaction mixtures actually enhanced this interaction (50) is consistent with this model. To probe the possible existence of such a competition directly, oligonucleotides containing the adenovirus ML TATA sequence (shown in Fig. 8A, bottom) were synthesized, purified, and annealed. Eve was produced by in vitro transcription and translation, GST and GST-dTBP were immobilized on beads, and binding experiments were performed as described above. Figure 8A shows that the 16-bp ML TATA DNA (at a concentration of 10 μM) significantly inhibited Eve binding to GST-dTBP. However, binding was not affected when the same concentration of each individual oligonucleotide (i.e., 20 μM) was added to reaction mixtures, indicating that only double-stranded ML DNA can inhibit the interaction between Eve and TBP. To exclude the possibility that any DNA fragment can block the Eve-TBP interaction, we tested a control 16-bp oligonucleotide, one previously shown not to interact with TBP (37), in binding assays. The data presented in Fig. 8A indicate that this oligonucleotide had no effect on the Eve-TBP interaction, whereas two concentrations of the ML TATA DNA (10 and 15 μM) resulted in increasing inhibition of the interaction between the two proteins.
To extend these results, we wished to confirm that the disruption of Eve-TBP binding by the TATA oligonucleotide was not an indirect effect that was reflecting, for example, an interaction with some component in the reticulocyte lysate, and also to provide evidence that the inhibition results from an interaction of the DNA with TBP and not with the Eve homeodomain. To this end, two recombinant Eve derivatives purified from E. coli, wild-type Eve and a mutant containing the ΔN homeodomain mutation, described above in the context of the GAL4-Eve fusion proteins, were used in binding reactions with GST-TBP in the presence or absence of the TATA oligonucleotide. Bound proteins were eluted with glutathione and detected by Western blotting with anti-Eve antibodies. The results (Fig. 8B) indicate that both Eve derivatives interacted strongly and specifically with GST-TBP and that in each case binding was inhibited by the TATA oligonucleotide. Together, these results indicate the existence of a competition between Eve and TATA-containing DNA for binding to TBP, which supports the hypothesis that Eve-mediated repression involves the inhibition of DNA binding by TBP.
We presented data here that Eve represses transcription equivalently in a fully reconstituted system (containing TFIID) and in the minimal system (containing TBP) used in the majority of our experiments. Because of this, we believe it is reasonable to assume that the mechanism we have elucidated with TBP also applies to TFIID-mediated transcription. But to provide direct support for this, we performed binding experiments similar to those just described, except we used TFIID in place of TBP. To this end, Flag epitope-tagged TFIID was purified from HeLa cells (4) and immobilized with anti-Flag antibodies bound to protein A Sepharose beads. These beads and control beads lacking TFIID were incubated with purified Eve in the presence or absence of 3 μM TATA oligonucleotide. After being washed, bound proteins were eluted and again detected by Western blotting with anti-Eve antibodies. The results (Fig. 8C) indicate that Eve bound very efficiently to TFIID and that, as with TBP, binding was disrupted by the TATA-containing oligonucleotide. These findings confirm that the Eve repression mechanism suggested by our data also applies to natural, TFIID-mediated transcription.
DISCUSSION
We have presented evidence indicating that the Drosophila Eve protein represses transcription by directly binding TBP in such a way that it interferes with or blocks the interaction between TBP-TFIID and DNA. Given that transcription was strongly and equally repressed by Eve in transcription reactions reconstituted with either all of the general transcription factors or only TBP, TFIIB, RAP30, and Pol II and that transcription in the minimal system was resistant to repression following preincubation of TBP and DNA, it is highly unlikely that any of the general transcription factors other than TBP are significant targets of Eve repression. It is conceivable that Eve, like several other characterized repressors, may function in vivo by more than one mechanism. However, the efficiency of the TBP-mediated repression described here suggests that this is an important aspect of Eve activity. It is also notable that the fully reconstituted system contained primarily human factors, while the minimal system was reconstituted largely with Drosophila proteins. This indicates that this mechanism of repression is likely evolutionarily conserved, at least from flies to humans, which is consistent with the fact that Eve binds equivalently to human and Drosophila TBP (50).
Our data indicate that the Eve homeodomain plays an important role in repression in addition to DNA binding, which is to facilitate interaction with TBP. It is now well established that homeodomains can participate in specific protein-protein interactions. Perhaps the best understood is the association of the viral activator VP16 with the POU domain protein Oct-1 (reviewed in reference 17). Elegant mutational studies revealed that exposed surface residues in the central region of the Oct-1 homeodomain are essential for interaction. This corresponds to the region of the Eve homeodomain that our deletion analysis suggested is necessary for both TBP binding and repression, although there is currently no evidence that the details of the two interactions are related. Perhaps more relevant to Eve repression is the murine Msx1 protein, in which specific residues in the N-terminal arm of the homeodomain were shown to be essential for TBP binding and repression but not DNA binding (53). This is analogous to the situation with Eve, except that the N-terminal arm is dispensable for the Eve-TBP interaction, so the exact role of the homeodomain in the two cases must be distinct. Indeed, several previous studies have shown that fusion proteins containing sequences encompassing the Ala-Pro-rich repression region (but not the homeodomain) fused to heterologous DNA binding domains are capable of repression in several different assays (2, 11, 21, 47). In our previous experiments, which involved transient transfections with plasmids encoding GAL4 fusion proteins similar to those analyzed here (11), we subsequently showed that the requirement of the homeodomain was promoter specific: repression of a promoter containing a TATA box required the homeodomain, while an initiator-containing promoter did not (30). Taken together, these findings suggest that the homeodomain does not always play an essential role in Eve-mediated repression. However, the studies described here indicate that it can be required in a defined in vitro transcription system and that its function extends beyond DNA binding. Whether residues within the homeodomain directly contact TBP or play an indirect role not involving specific contacts will require additional studies.
We initially characterized two repressors in transient-transfection assays, Eve and Engrailed (En) (11, 12). Although both contain Ala-rich motifs and displayed some similarities in activity in these assays, it is now clear that their mechanisms are distinct. This was suggested initially by one difference in their behavior in transfection assays, which was that Eve could repress basal (i.e., non-activated) expression whereas En could not. Using in vitro binding assays, we were also unable to detect an interaction between En and TBP (unpublished data). More recently, genetic and biochemical experiments have shown that an interaction between the En repression domain and a previously described corepressor, Groucho, can be required for En-mediated repression in vivo. In contrast, the Eve repression domain does not interact with Groucho and Eve-mediated repression is Groucho independent (21). The global repressor Dr1 also contains an Ala-rich domain that is essential for repression, and recent studies have shown that this region also interacts with TBP (52). However, since Dr1 blocks a step subsequent to DNA binding, the Eve-TBP and Dr1-TBP interactions are functionally distinct. The Drosophila Krüppel protein likewise contains an Ala-rich repression domain, which is located at its N terminus. Although its precise mode of action is not known, it is likely distinct from those just described and may involve a quenching mechanism (29). (Note that Krüppel also contains a C-terminal repression domain, one not Ala-rich, that appears to function by interacting with the small subunit of TFIIE [43].) Thus, while the presence of an Ala-rich region in a transcription factor may be diagnostic of a repression domain, it does not suggest a clear mechanism, since four Ala-rich repression domains appear to employ four distinct mechanisms. Many other repressors, of course, contain repression domains that are not Ala-rich (see reference 13 for a review), and it appears that repression motifs are as diverse in composition and function as are activation domains.
An important conclusion of our studies is that Eve represses transcription by preventing a stable TBP-TFIID and DNA interaction. Consistent with this, Austin and Biggin showed that template-bound Eve could block binding of TFIID to a TATA box on the same DNA (2). Based on this and other data, these authors also concluded that Eve functions by preventing TFIID from binding the promoter but proposed a mechanism different than that indicated by our data. Specifically, they provided evidence that sequences C terminal to the Eve homeodomain (likely corresponding to the Ala-Pro-rich repression domain) could impart to a heterologous DNA binding domain the ability to recognize low-affinity, nonspecific sites in the promoter region as a result of cooperative DNA binding nucleated by high-affinity sites located upstream of the TATA box. It was proposed that this nucleoprotein structure occluded the TATA box, preventing recognition by TBP-TFIID. Although it is attractive to consider that this “cooperative blocking” mechanism might function in conjunction with the repressive Eve-TBP interaction we have described to ensure strong repression, there are reasons to question the general significance of cooperative blocking. DNase footprinting studies by Hoey et al. (19), who used several DNA probes, did not detect any differences in the binding properties of Eve derivatives containing or lacking sequences C terminal to the homeodomain, and we have obtained essentially identical results with gel mobility shift assays (30; unpublished data). Johnson and Krasnow (23) measured template occupancy by Eve in nuclear extracts under conditions resulting in transcriptional repression. Using DNase footprinting assays, they found no indication of an Eve-DNA interaction outside of the Eve binding sites and over a distance of at least 100 bp downstream of the TATA box. Together, these results indicate that the Eve repression domain does not influence DNA binding by the homeodomain and that Eve can repress transcription without making DNA contacts outside of its binding sites. Thus, it seems that additional experiments are necessary to evaluate the generality of the cooperative blocking mechanism.
How does the Eve-TBP interaction prevent stable association of TBP-TFIID with the promoter? One possibility is based on observations suggesting that free TBP-TFIID exists as dimers, which must dissociate to bind DNA (e.g., see reference 46). Perhaps Eve binding to TBP prevents this required dissociation. However, by this view it is difficult to explain the observed DNA binding site dependence of Eve repression and also the ability of the TATA oligonucleotide to compete with the Eve and TBP-TFIID interaction. We favor instead a mechanism we refer to as the goalkeeper model. By this model, Eve (more precisely, multiple molecules of Eve) needs to be bound to the DNA in the vicinity (i.e., within several 100 bp) of the promoter (the “goal”) in order to fend off “shots” (i.e., TFIID). The multiple Eve molecules required would create a “hydraheaded goalkeeper,” whereby multiple repression domains could significantly increase Eve’s effectiveness. For example, they could not only allow a lower-affinity Eve-TBP interaction to compete successfully with TFIID-TATA binding but also simultaneously block TATA binding by more than one TFIID molecule. What prevents Eve from interacting with TFIID in solution and thereby nonspecifically inhibiting transcription? We suggest that the requirement for DNA binding by Eve reflects the relatively low affinity of the Eve-TBP interaction such that in solution the interaction, which would be with only a single Eve molecule, is sufficiently weak so as not to affect promoter binding by TFIID. In keeping with this, preincubation of Eve and TBP does not interfere with subsequent transcription from a template lacking Eve binding sites (unpublished data). In contrast, if Eve molecules are bound to DNA near the promoter, then repeated transient interactions could effectively block TFIID binding, leading to transcriptional repression. The most straightforward way Eve might accomplish this is via direct contact between the Eve repression domain and the concave DNA binding surface of TBP, but additional studies are required to address more precisely the exact nature of the repressive interaction.
ACKNOWLEDGMENTS
We are grateful to M. Um, K. Han, J. T. Kadonaga, and H. Ge for providing plasmids, to A. S. Manoukian for providing anti-Eve antibodies, and to H. Ge for providing Flag epitope-tagged HeLa TFIID and for helpful discussions. We thank K. Murthy for valuable advice on protein purification. L. Ge is thanked for excellent technical assistance.
This work is supported by National Institutes of Health grant GM 37971.
REFERENCES
- 1.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 2.Austin R J, Biggin M D. A domain of the even-skipped protein represses transcription by preventing TFIID binding to a promoter: repression by cooperative blocking. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggin M D, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- 4.Chiang C M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein-containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 6.Fondell J D, Brunel F, Hisatake K, Roeder R G. Unliganded thyroid hormone receptor alpha can target TATA-binding protein for transcriptional repression. Mol Cell Biol. 1996;16:281–287. doi: 10.1128/mcb.16.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the Even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 9.Gray S, Levine M. Transcriptional repression in development. Curr Opin Cell Biol. 1996;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- 10.Han K, Levine M S, Manley J L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989;56:573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- 11.Han K, Manley J L. Transcriptional repression by the Drosophila Even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- 12.Han K, Manley J L. Functional domains of the Drosophila Engrailed protein. EMBO J. 1993;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 14.Hawley D K, Roeder R G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 15.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davis J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 17.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch J A, Aggarwal A K. Structure of the Even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity. EMBO J. 1995;14:6280–6291. doi: 10.1002/j.1460-2075.1995.tb00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoey T, Warrior R, Manak J, Levine M. DNA-binding activities of the Drosophila melanogaster even-skipped protein are mediated by its homeo domain and influenced by protein context. Mol Cell Biol. 1988;8:4598–4607. doi: 10.1128/mcb.8.11.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaynes J B, O’Farrell P H. Activation and repression of transcription by homeodomain-containing proteins that bind a common site. Nature. 1988;336:744–749. doi: 10.1038/336744a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson A D. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 23.Johnson F B, Krasnow M A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeodomain proteins. Genes Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- 24.Kadonaga J T. Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J Biol Chem. 1990;265:2624–2631. [PubMed] [Google Scholar]
- 25.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingston R E, Bunker C A, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 27.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 28.Levine M, Manley J L. Transcriptional repression of eukaryotic promoters. Cell. 1989;59:405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- 29.Licht J D, Ro M, English M A, Grossel M, Hansen U. Selective repression of transcriptional activators at a distance by the Drosophila Krüppel protein. Proc Natl Acad Sci USA. 1993;90:11361–11365. doi: 10.1073/pnas.90.23.11361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manley J L, Um M, Li C, Ashali H. Mechanisms of transcriptional activation and repression can both involve TFIID. Philos Trans R Soc Lond B Biol Sci. 1996;351:517–526. doi: 10.1098/rstb.1996.0050. [DOI] [PubMed] [Google Scholar]
- 31.Manoukian A S, Krause H M. Concentration-dependent activities of the Even-skipped protein in Drosophila embryos. Genes Dev. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- 32.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 33.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 34.Ohkuma Y, Horikoshi M, Roeder R G, Desplan C. Engrailed, a homeodomain protein, can repress in vitro transcription by competition with the TATA box-binding protein transcription factor IID. Proc Natl Acad Sci USA. 1990;87:2289–2293. doi: 10.1073/pnas.87.6.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlando V, Paro R. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr Opin Genet Dev. 1995;5:174–179. doi: 10.1016/0959-437x(95)80005-0. [DOI] [PubMed] [Google Scholar]
- 36.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 37.Peterson M G, Tanese N, Pugh B F, Tjian R. Functional domains and upstream activation properties of cloned human TATA binding protein. Science. 1990;248:1625–1630. doi: 10.1126/science.2363050. [DOI] [PubMed] [Google Scholar]
- 38.Pirrotta V. Chromatin complexes regulating gene expression in Drosophila. Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- 39.Pugh B F, Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 40.Redd M J, Arnaud M B, Johnson A D. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 41.Reinberg D, Roeder R G. Factors involved in specific transcription by mammalian RNA polymerase II. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 42.Roth S Y. Chromatin-mediated transcriptional repression in yeast. Curr Opin Genet Dev. 1995;5:168–173. doi: 10.1016/0959-437x(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 43.Sauer F, Fondell J D, Ohkuma Y, Roeder R G, Jäckle H. Control of transcription by Krüppel through interaction with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- 44.Sawadogo M, Roeder R G. Factors involved in specific transcription by human RNA polymerase II: analysis of a rapid and quantitative in vitro assay. Proc Natl Acad Sci USA. 1985;82:4394–4398. doi: 10.1073/pnas.82.13.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song C Z, Loewenstein P M, Toth K, Tang Q, Nishikawa A, Green M. The adenovirus E1A repression domain disrupts the interaction between the TATA binding protein and the TATA box in a manner reversible by FTIIB. Mol Cell Biol. 1997;17:2186–2193. doi: 10.1128/mcb.17.4.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taggart A K, Pugh B F. Dimerization of TFIID when not bound to DNA. Science. 1996;272:1331–1333. doi: 10.1126/science.272.5266.1331. [DOI] [PubMed] [Google Scholar]
- 47.TenHarmsel A, Austin R J, Savenelli N, Biggin M D. Cooperative binding at a distance by even-skipped protein correlates with repression and suggests a mechanism of silencing. Mol Cell Biol. 1993;13:2742–2752. doi: 10.1128/mcb.13.5.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thut C J, Goodrich J A, Tjian R. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 1997;11:1974–1986. doi: 10.1101/gad.11.15.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyree C M, George C P, Lira-DeVito L M, Wampler S L, Dahmus M E, Zawel L, Kadonaga J T. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- 50.Um M, Li C, Manley J L. The transcriptional repressor Even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]
- 52.Yeung K C, Kim S, Reinberg D. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol Cell Biol. 1997;17:36–45. doi: 10.1128/mcb.17.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Catron K M, Abate-Shen C. A novel role for the Msx-1 homeodomain in transcriptional regulation: residues in the N-terminal arm mediate TATA binding protein interaction and transcriptional repression. Proc Natl Acad Sci USA. 1996;93:1764–1769. doi: 10.1073/pnas.93.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley J L. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991;5:254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]