Abstract
Interferon establishes an antiviral state in numerous cell types through the induction of a set of immediate-early response genes. Activation of these genes is mediated by phosphorylation of latent transcription factors of the STAT family. We found that infection of primary foreskin fibroblasts with human cytomegalovirus (HCMV) causes selective transcriptional activation of the alpha/beta-interferon-responsive ISG54 gene. However, no activation or nuclear translocation of STAT proteins was detected. Activation of ISG54 occurs independent of protein synthesis but is prevented by protein tyrosine kinase inhibitors. Further analysis revealed that HCMV infection induced the DNA binding of a novel complex, tentatively called cytomegalovirus-induced interferon-stimulated response element binding factor (CIF). CIF is composed, at least in part, of the recently identified interferon regulatory factor 3 (IRF3), but it does not contain the STAT1 and STAT2 proteins that participate in the formation of interferon-stimulated gene factor 3. IRF3, which has previously been shown to possess no intrinsic transcriptional activation potential, interacts with the transcriptional coactivator CREB binding protein, but not with p300, to form CIF. Activating interferon-stimulated genes without the need for prior synthesis of interferons might provide the host cell with a potential shortcut in the activation of its antiviral defense.
Alpha interferon (IFN-α) and IFN-β are unique among the continuously growing superfamily of cytokines in their ability to confer resistance to viral infection (20, 36). The synthesis of IFN-α and IFN-β is induced at the transcriptional level after a cell encounters virus or double-stranded RNA (dsRNA) (16, 46). The subsequent secretion of the newly produced interferons and their binding to a common cell surface receptor results in the induction of a set of immediate-early response genes (12, 21, 24–26, 32, 44, 47). The activation of these interferon-stimulated genes (ISGs) represents the first step towards the development of an antiviral state. Control over ISGs is exerted by an IFN-α/β-activated transcription factor complex termed interferon-stimulated gene factor 3 (ISGF3), which binds to a common enhancer element referred to as the interferon-stimulated response element (ISRE) (10, 14, 23, 27, 43). ISGF3 is formed through the interaction of the DNA binding subunit ISGF3γ (p48) and the regulatory component ISGF3α (14, 28, 48), which itself is composed of two members of the STAT (signal transducers and activators of transcription) family of transcription factors, STAT1 and STAT2 (14, 15, 43). Both STAT proteins become tyrosine phosphorylated in response to IFN-α/β stimulation, which enables their nuclear translocation and DNA binding (8, 10, 13, 41). Transcriptionally active STAT1 has been shown to be a requirement for the antiviral and antiproliferative effects of IFN-α/β (5, 11, 31). The phosphorylation of STAT1 and STAT2 is mediated through the action of two related tyrosine kinases, Jak1 and Tyk2, which are enzymatically activated in response to IFN-α/β stimulation (35, 49).
Human cytomegalovirus (HCMV), a member of the betaherpesvirus family, is a prevalent pathogen which not only poses a major health threat to immunocompromised individuals but also accounts for the majority of virus-mediated birth defects (34). Previously, it was shown that several human viruses can inhibit the interferon-mediated activation of cellular genes that participate in the antiviral defense (1, 7, 17, 18, 42). During adenovirus infection, it appears that the protein encoded by the E1A gene interferes with the DNA-binding ability of ISGF3, resulting in the transcriptional suppression of the cellular ISGs (17, 18, 42). Since HCMV is able to complement the growth of an adenovirus E1A mutant (45), we were interested in determining whether HCMV infection could also alter the expression of the IFN-α/β-regulated immediate-early response genes. Surprisingly, we found that HCMV infection per se resulted in a robust transcriptional activation of the ISRE-controlled ISG54 gene and that this activation occurred in the absence of de novo protein synthesis. Furthermore, we identified a novel HCMV-induced putative transcription factor complex. Biochemical characterization suggests that it is composed of a recently described new member of the interferon regulatory factor (IRF) family and the transcriptional coactivator CREB binding protein (CBP).
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFFs) were maintained in Dulbecco modified Eagle medium (Irvine Scientific) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin (Irvine Scientific). HCMV Towne was obtained from the American Type Culture Collection, propagated as previously described, and stored at −80°C in Eagle minimal essential medium with 1% dimethyl sulfoxide and 10% FBS. HCMV infections were performed at a multiplicity of infection (MOI) of 5 PFU per cell. Mock infections were performed with media conditioned on actively growing cells for 2 days. Conditioned media were adjusted to 1% dimethyl sulfoxide and stored at −80°C until use.
Reagents.
Cycloheximide (CHX), genistein, and staurosporine were obtained from Sigma Chemical Co. and were used at 30 μg/ml, 100 μg/ml, and 50 ng/ml, respectively. The immunogen used to generate the anti-IRF3 antibody was human IRF3 amino acids 107 to 208 fused to glutathione S-transferase (pGEX2T; Pharmacia).
Whole-cell extracts.
HFFs were infected, mock infected, or stimulated as described above. At different times postinfection or poststimulation, the cells were scraped, pelleted, washed once with phosphate-buffered saline (PBS), and subsequently lysed on ice for 10 min in lysis buffer containing 20 mM HEPES (pH 7.4), 100 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 1 mM vanadate, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride. Lysates were vortexed and centrifuged at 15,000 × g for 10 min at 4°C. Protein concentration was measured by the Bio-Rad protein assay. Selected extracts were treated with 3 mM N-ethylmaleimide (NEM) for 20 min at room temperature and then quenched with 20 mM dithiothreitol on ice for 10 min.
Immunoprecipitation and immunoblotting.
Cellular extracts were subjected to immunoprecipitation with an antibody against the C terminus of Stat1 for 2 h prior to the addition of protein G-Sepharose beads (Pharmacia Biotech, Inc.) and incubation for an additional hour. The beads were pelleted at 15,000 × g for 2 min and washed three times with ice-cold lysis buffer (1 ml). Immunoprecipitates were boiled in sodium dodecyl sulfate sample buffer and resolved by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis. After transfer to Immobilon (Millipore), the blots were probed with a monoclonal antibody against Stat1 (Transduction Laboratories) and anti-phosphoStat1 antibodies (New England Biolabs). Reactive proteins were detected with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Amersham).
RNase protection assay analysis.
HFFs were cultured and infected as described above. Total RNA was isolated with TRIzol Reagent (Gibco BRL). 32P-labeled antisense riboprobes were generated by transcription of the linearized plasmid in vitro by using T7 or SP6 RNA polymerase (Promega). Labeled riboprobe and 10 μg of RNA were incubated in hybridization buffer {4:1 formamide and 5× stock; 5× stock was 200 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.4], 2 M NaCl, 5 mM EDTA} overnight at 56°C before digestion with T1 RNase (Gibco BRL) for 1 h at 37°C. After phenol extraction and ethanol precipitation, protected fragments were solubilized in RNA loading buffer (98% formamide, 10 mM EDTA [pH 8], bromophenol blue [1 mg/ml], xylene cyanol [1 mg/ml]), boiled for 2 min, and subjected to electrophoresis on a 4.5% polyacrylamide-urea gel.
In vitro transcription-translation reactions.
ISGF3γ, IRF1, and IRF2 were in vitro translated with rabbit reticulocyte lysate (Promega) according to the manufacturer’s instructions.
Immunofluorescence.
HFFs were seeded onto glass coverslips in 6-well plates and incubated overnight at 37°C under 5% CO2 in Dulbecco modified Eagle medium supplemented with 10% FBS. The cells were then treated with medium alone or 1,000 U of IFN-β per ml for 30 min or else infected with HCMV for 6 h. After treatment, cells were rinsed once with PBS and once with 1× PB (100 mM PIPES [pH 6.8], 2 mM MgCl2, 2 mM EGTA). Cells were fixed in methanol for 6 min at room temperature. Nuclei were permeabilized by incubating with 0.5% Nonidet P-40–PB for 10 min at room temperature. After one rinsing with PBS, the fixed cells were blocked in 10% goat serum in PBS for 35 min at room temperature. After blocking, the cells were incubated for 50 min at room temperature with an anti-Stat1 antibody (Transduction Laboratories). Cells were rinsed four times for 5 min in PBS, incubated with a goat anti-mouse Cy3-conjugated secondary antiserum for 40 min at room temperature, and mounted after rinsing on glass slides with 50% glycerol-PBS.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed with a 32P-end-labeled probe corresponding to the ISRE of the ISG15 promoter (5′ GATCCATGCCTCGGGAAAGGGAAACCGAAACTGAAGCC 3′). Equal amounts of protein were incubated with poly(dI-dC) and labeled oligonucleotides in ISRE binding buffer (40 mM KCl, 20 mM HEPES [pH 7.0], 1 mM MgCl2, 0.1 mM EGTA, 0.5 mM dithiothreitol, 4% Ficoll, 0.02% Nonidet P-40). Extracts incubated with the ISRE probe were in some instances supplemented with in vitro-translated ISGF3γ (Promega). Electrophoresis was performed by 6% nondenaturing TBE polyacrylamide gel electrophoresis, and the gels were dried and subjected to autoradiography. For competition experiments, unlabeled oligonucleotides were added to the reaction mixture prior to the addition of the labeled oligonucleotides. The oligonucleotide used as a competitor corresponded to either the ISG15-ISRE or the IFN-γ response region (GRR) sequence in the promoter of the high-affinity FcγRI receptor (5′ AATTAGCATGTTTCAAGGATTTGAGATGTATTTCCCAGAAAAG 3′). For supershift experiments, whole-cell extracts were incubated on ice with the specified antiserum for 1 h at 4°C prior to the addition of the labeled oligonucleotide.
RESULTS
HCMV infection activates transcription of the IFN-α/β-regulated ISG54 gene in the absence of protein synthesis.
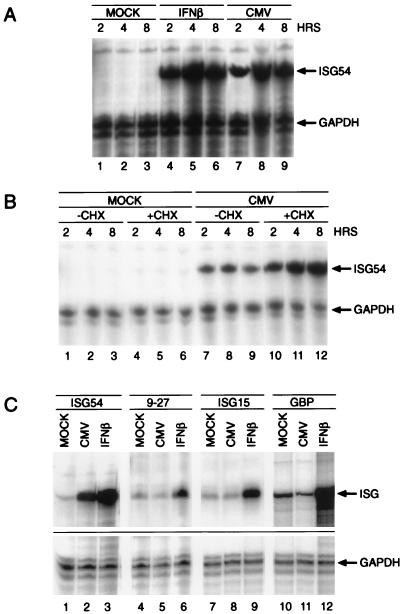
Activation of IFN-α/β-induced immediate-early response genes carrying the ISRE enhancer sequence requires the assembly of the ISGF3 transcription factor complex. Interference with the DNA binding capability of ISGF3 by the adenoviral E1A gene product prevents the transcriptional induction of cellular ISGs. Since HCMV is able to complement an adenovirus E1A mutant in this respect, we proceeded to determine whether infection with HCMV would result in a similar pattern of transcriptional suppression. HFFs were either treated with 1,000 U of IFN-β per ml or infected with HCMV Towne at an MOI of 5 PFU per cell, and total cellular RNA was then isolated at 2, 4, and 8 h after infection. We then performed RNase protection assays with a probe corresponding to the human ISG54 gene. Surprisingly, infection of the cells with HCMV was sufficient to induce a significant activation of the ISG54 gene (Fig. 1A, lanes 7 to 9). In order to investigate whether the induction of ISG54 by HCMV occurred in the absence of protein synthesis as suggested by the rapid kinetics of the transcriptional activation, we repeated the experiments in the presence of the translation inhibitor CHX.
FIG. 1.
Transcriptional activation of ISG54 by HCMV. (A) Primary human fibroblasts were incubated for the indicated times with either 1,000 U of IFN-β per ml (lanes 4 to 6) or 5 MOI of HCMV (lanes 7 to 9) or left untreated (lanes 1 to 3). Total cytoplasmic RNA was isolated, and the amounts of ISG54 and GAPDH mRNAs were determined by RNase protection assay with specific radiolabeled antisense RNA probes. (B) Experiments were performed as described for panel A, except that 50 μg of CHX per ml was present 30 min prior to and during the infection (compare lanes 7 to 9 with lanes 10 to 12). (C) Cells were infected for 6 h in the presence of 50 μg of CHX per ml, and total RNA was isolated as described above. RNase protection assays were performed with specific antisense probes corresponding to the ISG54, 9-27, ISG15, and GBP genes. GAPDH was used as an internal standard to ensure that equal amounts of RNA were loaded.
As shown in Fig. 1B (lanes 7 to 9 versus lanes 10 to 12), CHX pretreatment did not prevent HCMV-induced ISG54 transcription. In fact, an increased accumulation of message was observed, as was the case after stimulation with IFN-β in the presence of CHX (data not shown). To determine if the observed transcriptional activation after HCMV infection is restricted to the ISG54 gene or if other ISRE-containing genes are affected, we proceeded with the analysis of additional IFN-α/β-regulated immediate-early response genes such as the 9-27 (Fig. 1C, lanes 4 to 6), ISG15 (lanes 7 to 9), and GBP (lanes 10 to 12) genes. Unexpectedly, other than ISG54, HCMV infection failed to significantly activate the ISGs analyzed (Fig. 1C, lanes 5, 8, and 11).
Effects of protein kinase inhibitors on HCMV-mediated gene expression.
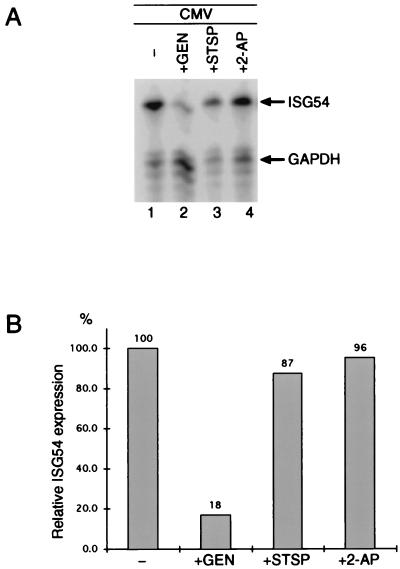
Since IFN-α/β-mediated gene expression requires the activity of tyrosine and serine-threonine kinases (13, 50), we wanted to determine whether the same was true for HCMV-induced activation of ISG54. Cells were preincubated with the kinase inhibitor staurosporine (Fig. 2A, lane 3) and the tyrosine-specific kinase inhibitor genistein (lane 2) for 30 min prior to infection with HCMV. Whereas genistein pretreatment resulted in an 82% inhibition of ISG54 expression, the effects of staurosporine were less significant (13% inhibition), indicating that protein tyrosine phosphorylation is an essential step in HCMV-induced gene transcription. In addition, we tested the PKR-specific inhibitor 2-aminopurine since this kinase is known to catalyze virus infection-dependent protein phosphorylation. Induction of ISG54 was unaffected by 2-aminopurine (Fig. 2A, lane 4).
FIG. 2.
Protein kinase activity is required for HCMV-mediated ISG54 expression. (A) Cells were infected for 6 h as described for Fig. 1 in the presence or absence of 100 μg of genistein per ml (lanes 2 and 1, respectively), 50 ng of staurosporine per ml (lane 3), or 50 μg of 2-aminopurine per ml (lane 4), and ISG54 and GAPDH mRNAs were analyzed by RNase protection assays as for Fig. 1 CHX (50 μg/ml) was present during the entire infection. (B) Radioactivity of the protected ISG54 and GAPDH mRNAs was quantitated with a Bio-Rad PhosphorImager. After normalization for GAPDH expression levels, the quantities of ISG54 were calculated as percentages of the amounts observed in the absence of the kinase inhibitors. GEN, genistein; STSP, staurosporine; 2-AP, 2-aminopurine.
HCMV infection initiates the assembly of a novel ISRE binding complex.
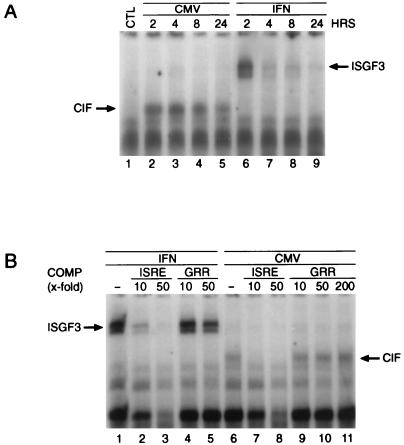
The ISRE enhancer element has been shown to be necessary and sufficient for the transcriptional activation of the ISGs (38, 39). To determine if HCMV causes the protein synthesis-independent formation of ISGF3 or another DNA binding complex, we performed EMSAs with the ISG15 and the ISG54 ISREs as probes. In contrast to IFN-β (Fig. 3A, lanes 6 to 9), HCMV infection did not initiate the assembly of STAT1, STAT2, and p48 into the ISGF3 complex. However, it did lead to the protein synthesis-independent appearance of an HCMV-inducible ISRE binding factor of distinct mobility, which we termed cytomegalovirus-induced ISRE-binding factor (CIF; Fig. 3A, lanes 2 to 5). These results suggest that CIF preexists in a latent, inactive form in the cell and is activated by a virus-induced, posttranslational modification. To verify that the ISRE binding of CIF occurred in a sequence-specific manner, we performed competition analysis with unlabeled oligonucleotides corresponding either to the ISRE or to the GRR, a distinct STAT1-binding sequence located in the promoter of the high-affinity FcγRI receptor. As shown in Fig. 3B, unlabeled ISRE at a 10-fold molar excess competed appropriately for CIF binding (lanes 7 and 8), whereas the addition of an up to 200-fold molar excess of GRR as a competitor DNA had no effect (lanes 9 to 11). Although ISG54 was induced to a much greater extent than the ISG15 gene, we were unable to detect by EMSA any significant differences in the binding characteristics of CIF to the two ISRE probes (data not shown). This suggests that additional factors or subtle differences in the specific ISRE sequence contribute to the differences in transcriptional induction.
FIG. 3.
HCMV infection induces a new ISRE binding complex. (A) Cells were infected at an MOI of 5 with HCMV (lanes 2 to 5) or were stimulated with 1,000 U of IFN-β per ml (lanes 6 to 9) for the indicated time intervals in the presence of 50 μg of CHX per ml. Whole-cell extracts were prepared, and EMSAs were performed with end-labeled ISRE as a probe. (B) To confirm binding specificity, EMSA was performed as described for panel A, except that a 10-fold (lanes 2, 4, 7, and 9), 50-fold (lanes 3, 5, 8, and 10), or 200-fold (lane 11) excess of unlabeled ISRE (lanes 2, 3, 7, and 8) or GRR (lanes 4, 5, 9, 10, and 11) was present during the binding reaction. COMP, competitor.
Role of STAT proteins in HCMV infection.
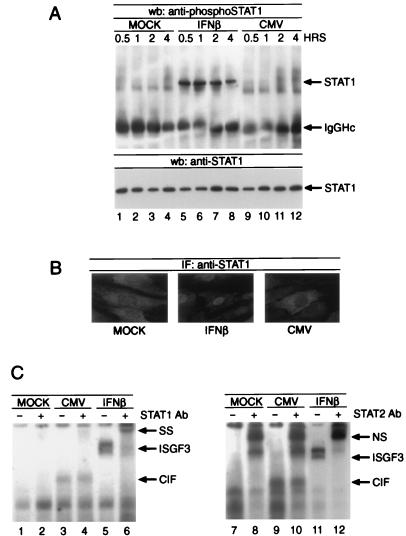
Activation of STAT proteins by tyrosine phosphorylation causes their nuclear translocation and facilitates DNA binding (23, 27). In order to investigate whether HCMV could cause tyrosine phosphorylation of STAT1, we performed immunoprecipitations followed by Western blot analysis with an anti-phosphoSTAT1 antibody that specifically recognizes STAT1 after its phosphorylation on Tyr701. STAT1 was immunoprecipitated from whole-cell lysates derived from cells that were either left untreated, stimulated with 1,000 U of IFN-β per ml, or infected with HCMV at an MOI of 5 PFU per cell for the indicated times. Although STAT1 was clearly tyrosine phosphorylated in response to IFN-β (Fig. 4A, lanes 5 to 8), no such modification was observed after viral infection (lanes 9 to 12). Loading of identical amounts of STAT1 protein was confirmed by Western blotting with a monoclonal anti-STAT1 antibody. To explore the possibility that HCMV infection might initiate STAT1 translocation in the absence of tyrosine phosphorylation, we subjected HFF cells to immunohistochemical staining utilizing a monoclonal antibody against STAT1. Whereas IFN-β treatment resulted in a robust nuclear presence of STAT1 (Fig. 4B, center panel), no translocation was observed during 6 h of HCMV infection (Fig. 4B, right panel). Since it appeared feasible that STAT1 or STAT2 might participate in the formation of CIF, even in an unphosphorylated form, we also executed supershift experiments on CIF- and ISGF3-containing extracts with anti-STAT1 (Fig. 4C, left panel) and anti-STAT2 (right panel) antibodies. As expected, both sera were able to supershift the ISGF3 complex (Fig. 4C, lanes 6 and 12); however, CIF migration was not affected (lanes 4 and 10). Taken together, these results clearly demonstrate that the STAT proteins known to bind the ISRE do not participate in the formation of CIF.
FIG. 4.
Role of STAT proteins in HCMV infection. (A) Cells were infected or stimulated as described for Fig. 3 for the times indicated, and whole-cell lysates were subject to immunoprecipitation by anti-STAT1 antibody. Western blotting was performed with a polyclonal phosphospecific STAT1 antibody (upper panel) or a monoclonal antibody against STAT1 to confirm equal protein loading (lower panel). (B) Cells were left untreated (left panel), stimulated with 1,000 U of IFN-β per ml (center panel), or infected with HCMV (right panel). Immunostaining was performed with a monoclonal antibody against STAT1 and a Cy3-conjugated secondary antiserum. (C) Extracts shown in Fig. 3 were used to perform EMSA supershift experiments with anti-STAT1 (left panel, lanes 2, 4, and 6)- or anti-STAT2 (right panel, lanes 8, 10, and 12)-specific antiserum. NS, nonspecific complex due to nonspecific interaction of the STAT2 antiserum with the probe; SS, supershifted complex; Ab, antibody; wb, Western blot; IF, immunofluorescence.
Comparison of CIF with the IRF family.
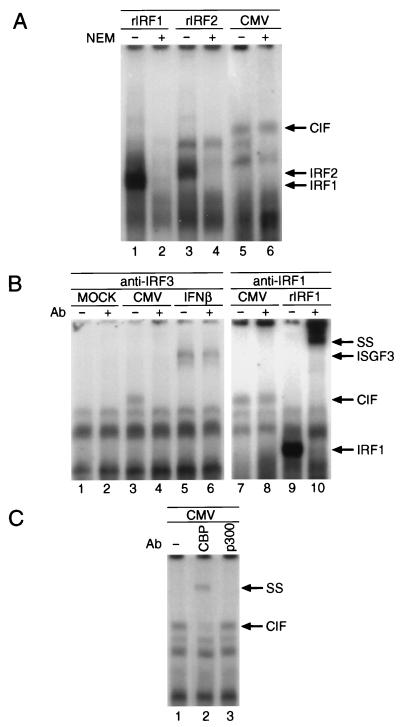
Several proteins in addition to STAT1, STAT2, and p48 have been shown to bind to the ISRE enhancer, where they function either as transcriptional activators or suppressors. Among them are several members of the IRF family (2, 6, 19, 30, 33, 51, 52) and the interferon consensus sequence binding protein ICSBP (22, 37). For some of the ISRE binding proteins it has been reported that their DNA binding activity is decreased by protein synthesis inhibition (e.g., VIBP) (4). In contrast, CIF activation was found to be enhanced in the presence of CHX (data not shown). This also suggests that IRF1 and IRF2 are not involved in the formation of CIF since their binding to the ISRE appears to be regulated through the abundance of those proteins rather than through a posttranslational modification. Another useful approach for distinguishing DNA binding proteins is their sensitivity towards alkylation by NEM, which in many cases eliminates protein-DNA interactions (9, 27). Therefore, we subjected a CIF-containing extract as well as in vitro-translated IRF1 or IRF2 to treatment with 3 mM NEM and analyzed for ISRE binding by EMSA. As shown in Fig. 5A, NEM exposure resulted in a complete loss of IRF1 and IRF2 binding to the ISRE (lanes 2 and 4, respectively), whereas CIF binding was not affected (lane 6). Since the target of NEM in the ISGF3 complex is the DNA binding component ISGF3γ (p48), the addition of exogenous, in vitro-translated ISGF3γ to the NEM-treated ISGF3-containing extract restored the binding of ISGF3 but did not alter the appearance of CIF (data not shown).
FIG. 5.
Characterization of CIF binding and composition. (A) In vitro-translated IRF1 (lanes 1 and 2), IRF2 (lanes 3 and 4), or a post-HCMV infection whole-cell lysate were subjected to alkylation with 3 mM NEM (lanes 2, 4, and 6) prior to analysis by EMSA. (B) Supershift analysis was performed as described for Fig. 4C, except that anti-IRF3 antibody was used in the experiments (lanes 2, 4, and 6). Antiserum against IRF1 (lanes 8 and 10) was used to demonstrate the specificity of the observed supershift of CIF by IRF3 antibody (lane 4). Supershifts with in vitro-translated IRF1 were performed to ensure the reactivity of the IRF1 antiserum (lane 9 versus lane 10). (C) CIF-containing extracts (lane 1) were incubated for 2 h with antiserum against either CBP (lane 2) or p300 (lane 3) prior to analysis by EMSA. rIRF1, in vitro-translated IRF1; rIRF2, in vitro-translated IRF2; SS, supershift; Ab, antibody.
Recently, a new member of the IRF family, IRF3, has been identified. Recombinant IRF3 is able to bind to the ISRE constitutively but has no intrinsic transactivation capabilities. Rather, it was suggested that it may require assembly with other coactivators to induce gene expression. In order to determine whether IRF3 participates in the formation of CIF, we performed supershift experiments with anti-IRF3 antisera. As shown in Fig. 5B, anti-IRF3 antibody caused the specific supershift of CIF (lane 4), whereas no effect was observed on the mobility of ISGF3 (lane 6) or in vitro-translated IRF1 or IRF2 (data not shown). To confirm the specificity of the immune reaction, anti-IRF1 antibody was used as a control serum. As expected, anti-IRF1 antisera did not influence the migration of CIF (lane 7 versus 8) but did supershift in vitro-translated IRF1 (lane 9 versus 10). Interestingly, the mobilities of CIF and recombinant IRF1 and IRF2 differ (compare Fig. 5A, lanes 1, 3, and 5) despite the similar sizes and structural features of the three IRF family members, suggesting that an additional protein(s) besides IRF3 might contribute to the assembly of CIF.
Since IRF3 does not possess intrinsic transactivation potential (2), we decided to determine whether the transcriptional coactivators CBP and p300 (3, 53) would participate in the formation of CIF to form a transcriptionally active complex. Experiments with supershifting antibodies specific for either CBP or p300 (40) revealed that CBP, but not p300, participates in the formation of CIF (Fig. 5C, lanes 2 versus 3).
DISCUSSION
IFN-α/β-mediated transcription of immediate-early response genes is an integral part of the cellular defense mechanism against viral infection. The concept of the interferon system is based on the transcriptional induction of the interferon genes upon viral infection. This results in the production and the release of newly synthesized IFN-α/β, which acts upon the neighboring cells or in an autocrine manner to induce the expression of ISGs. Activation of the Jak-STAT pathway is then required to establish an antiviral state in response to IFN-α/β. Unfortunately, this process causes a delayed response to the virus threat, since it requires the synthesis of IFN-α/β in order to produce cellular defensive actions. However, evidence exists for alternative pathways that regulate the transcription of ISGs under the control of the ISRE enhancer in a more timely fashion and independent of protein synthesis.
Previous reports have demonstrated that infection of cells with a mutant, E1A-deficient adenovirus can cause the activation of ISRE-containing genes. Wild-type adenovirus fails to induce ISG transcription because the E1A gene product interferes with the DNA binding of the ISGF3 transcription complex. Vesicular stomatitis virus infection of L929 cells also results in the formation of an ISRE binding complex referred to as VIBP. However, the formation of VIBP was prevented by inhibition of protein synthesis. Similar to virus infection, transfection of dsRNA is able to induce the binding of two distinct factors, DRAF1 and DRAF2, to the ISRE and to activate transcription of ISGs.
We describe the formation of a new ISRE binding factor, CIF, that occurs after infection of primary human fibroblasts with wild-type HCMV. CIF differs in a number of aspects from known ISRE binding proteins in that activation of CIF is paralleled by the transcriptional induction of ISG54, and both events are independent of protein synthesis. In contrast, VIBP assembly requires protein synthesis. The DRAF complexes display mobilities different from that of CIF in EMSAs. Furthermore, dsRNA transfection results in activation of ISG15 and ISG54 to comparable levels. On the other hand, CMV infection causes predominantly the expression of ISG54, without the upregulation of ISG15, 9-27, or GBP mRNA. Although we do not expect that ISG54 is the only HCMV-induced ISRE-containing gene, the observed specificity in gene expression is additional evidence that the activation of ISG54 by HCMV is not due to an autocrine stimulation by newly synthesized interferon, since IFN-α/β is able to activate all of the genes tested. Since we found that CIF bound the ISG15-ISRE to the same extent or better than the ISG54-ISRE, we believe that either additional proteins or subtle differences in the ISRE sequence account for the differences in transcriptional activation. It is notable in this context that the IP10 gene, whose promoter contains an ISRE that differs from the ISG54-ISRE in only two bases, is not significantly induced in response to IFN-α/β (29).
In our efforts to classify the proteins that compose CIF, we identified a recently described new member of the interferon regulatory factor family, IRF3, of unknown cellular function, as a component of CIF. Recombinant IRF1 and IRF2, which are of similar size and share a significant degree of homology with IRF3, display a higher mobility in EMSAs compared to CIF, suggesting that additional proteins contribute to the assembly of CIF. IRF3, which in contrast to IRF1 lacks transactivating potential, binds the ISRE sequence constitutively as a recombinant protein (2). IRF3 was found to support ISRE-dependent transcription in cotransfection and overexpression experiments, but it fails to induce transcription by itself (2). These facts suggested the possibility that IRF3 participates in the formation of a multicomponent transcription complex, perhaps functioning as a DNA binding component similar to the role of ISGF3γ in the assembly of the ISGF3 complex. Indeed, there are striking similarities between CIF and ISGF3 in that both complexes preexist in a latent form in the cell and are activated in the absence of protein synthesis, and their formation is sensitive to the presence of tyrosine kinase inhibitors but not of 2-aminopurine. Further analysis of the ISRE binding complex revealed that the transcriptional coactivator CBP participates in the formation of CIF, thereby contributing to the generation of a complex that is capable of initiating transcription.
In summary, our results show that HCMV infection results in the protein synthesis-independent activation of a novel cellular ISRE binding complex termed CIF. This putative transcription factor complex appears to selectively mediate the transcription of ISG54. Although we were able to establish IRF3 and CBP as components of CIF, the possibility that additional unidentified proteins might participate in CIF assembly remains. This hypothesis is supported by the fact that despite the abrogation of CIF formation by inhibition of tyrosine kinase activity, we were thus far unable to detect any tyrosine phosphorylation of either IRF3 or CBP. Since CIF is induced in the absence of protein synthesis, it is possible that the complex contains proteins which are part of the virion particle and are introduced into the cell upon viral entry. Alternatively, they may be cellular proteins that are activated upon viral contact or entry. Experiments are currently in progress to address this question.
Activating ISGs without the need for prior de novo synthesis of interferons might provide the host cell with a potential shortcut in the activation of its antiviral defense. Subsequent production and release of interferons by the host cell might be more beneficial to neighboring uninfected cells by protecting them from viral invasion. Alternatively, the restricted induction of ISG54 might be advantageous for the virus and its replication cycle. The ongoing further characterization of CIF will allow us to identify the underlying mechanism of CIF activation and its cellular function.
ACKNOWLEDGMENTS
We thank Andrew Larner and Keiko Ozata for their generous gifts of STAT1 and STAT2 and of the IRF1 and IRF2 antisera, respectively. We are also grateful to Robert Rickert for a critical reading of the manuscript. Antisera to CBP and p300 were generously provided by Pier Lorenzo Puri.
K.M. is a recipient of a fellowship from the Markey Foundation. This work was supported by NIH grants CA34729 (D.S.) and CA50773 (N.R.) and NIH training grant AI07384 (S.R.).
REFERENCES
- 1.Ackrill A M, Foster G R, Laxton C D, Flavell D M, Stark G R, Kerr I M. Inhibition of the cellular response to interferons by the products of the adenovirus type 5 E1A oncogene. Nucleic Acids Res. 1991;19:4387–4393. doi: 10.1093/nar/19.16.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-α. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 4.Bovelenta C, Lou J, Kanno Y, Park B-K, Thornton A M, Colligan J E, Schubert M, Ozato K. Vesicular stomatitis virus infection induces a nuclear DNA-binding factor specific for the interferon-stimulated response element. J Virol. 1995;69:4173–4181. doi: 10.1128/jvi.69.7.4173-4181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromberg J, Horvath C, Wen Z, Schreiber R, Darnell J. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon α and interferon γ. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cha Y, Deisseroth A B. Human interferon regulatory factor 2 gene. Intron-exon organization and functional analysis of 5′-flanking region. J Biol Chem. 1994;269:5279–5287. [PubMed] [Google Scholar]
- 7.Colamonici O R, Domanski P, Sweitzer S M, Larner A, Buller R M L. Vaccinia virus B18R gene encodes a type 1 interferon-binding protein that blocks interferon α transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 8.David M. Transcription factors in interferon signaling. Pharmacol Ther. 1995;65:149–161. doi: 10.1016/0163-7258(94)00050-d. [DOI] [PubMed] [Google Scholar]
- 9.David M, Larner A C. Activation of transcription factors by interferon alpha in a cell-free system. Science. 1992;257:813–815. doi: 10.1126/science.1496402. [DOI] [PubMed] [Google Scholar]
- 10.David M, Romero G, Zhang Z Y, Dixon J E, Larner A C. In vitro activation of the transcription factor ISGF3 by IFNα involves a membrane-associated tyrosine phosphatase and kinase. J Biol Chem. 1993;268:6593–6599. [PubMed] [Google Scholar]
- 11.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 12.Finbloom D S, Larner A C. Induction of early response genes by interferons, interleukins, and growth factors by the tyrosine phosphorylation of latent transcription factors: implications for chronic inflammatory diseases. Arthritis Rheum. 1995;38:877–889. doi: 10.1002/art.1780380702. [DOI] [PubMed] [Google Scholar]
- 13.Fu X-Y. A transcription factor with SH2 and SH3 domains is directly activated by an interferon-α induced cytoplasmic protein tyrosine kinase(s) Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 14.Fu X-Y, Kessler D S, Veals S A, Levy D E, Darnell J E., Jr ISGF-3, the transcriptional activator induced by IFN-α, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E J. The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of endogenous IFN-α and IFN-β genes by a regulatory transcription factor. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 17.Gutch M, Reich N. Repression of the interferon signal transduction pathway by the adenovirus E1A oncogene. Proc Natl Acad Sci USA. 1991;88:7913–7917. doi: 10.1073/pnas.88.18.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutch M, Reich N C. Response of the interferon signal transduction pathway by the adenovirus E1A oncogene. Proc Natl Acad Sci USA. 1991;88:7913–7917. doi: 10.1073/pnas.88.18.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 20.Isaacs A, Lindenmann J. Virus interference: the interferon. Proc R Soc Lond Ser B. 1957;147:258–267. [PubMed] [Google Scholar]
- 21.Kalvakolanu D V R, Sen G C. Differentiation-dependent activation of interferon-stimulated gene factors and transcription factor NF-κB in mouse embryonal carcinoma cells. Proc Natl Acad Sci USA. 1993;90:3167–3171. doi: 10.1073/pnas.90.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno Y, Kozak C A, Schindler C, Driggers P H, Ennist D L, Gleason S L, Darnell J E J, Ozato K. The genomic structure of the murine ICSBP gene reveals the presence of the gamma interferon-responsive element, to which an ISGF3α subunit (or similar) molecule binds. Mol Cell Biol. 1993;13:3951–3963. doi: 10.1128/mcb.13.7.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler D S, Veals S A, Fu X-Y, Levy D S. Interferon-α regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4:1753–1765. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- 24.Khan K D, Shuai K, Lindwall G, Maher S E, Darnell J E J, Bothwell A L M. Induction of the Ly-6A/E gene by interferon α/β and γ requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc Natl Acad Sci USA. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larner A C, Chaudhuri A, Darnell J E. Transcriptional induction by interferon: new protein(s) determine the extent and length of the induction. J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 26.Larner A C, Jonak G, Cheng Y-S E, Korant B, Knight E, Darnell J E. Transcriptional induction of two genes in human cells by β interferon. Proc Natl Acad Sci USA. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy D E, Kessler D S, Pine R, Darnell J E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 28.Levy D E, Lew D J, Kessler D S, Darnell J E., Jr Synergistic interaction between interferon-α and interferon-γ through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990;9:1105–1111. doi: 10.1002/j.1460-2075.1990.tb08216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luster A D, Unkeless J C, Ravetch J V. γ-Interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 30.Matsuyama T, Grossman A, Mittruecker H-W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 32.Mirkovitch J, Decker T, Darnell J E., Jr Interferon induction of gene transcription analyzed by in vivo footprinting. Mol Cell Biol. 1992;12:1–9. doi: 10.1128/mcb.12.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 34.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 35.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur A G, Barbieri G, Withuhn B A, Schindler C, Pellegrini S, Wilks A F, Ihle J N, Stark G R, Kerr I M. The protein tyrosine kinase JAK1 complements defects in the interferon-α/β and -γ signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 36.Muller U, Steinhoff U, Reis L F, Hemmi S, Pavlovic J, Zinkernagel R M, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 37.Nelson N, Marks M S, Driggers P H, Ozato K. Interferon consensus sequence-binding protein, a member of the interferon regulatory factor family, suppresses interferon-induced gene transcription. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pine R, Darnell J E., Jr In vivo evidence of interaction between interferon-stimulated gene factors and the interferon-stimulated response element. Mol Cell Biol. 1989;9:3533–3537. doi: 10.1128/mcb.9.8.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porter A C, Chernajovsky Y, Dale T C, Gilbert C S, Stark G R, Kerr I M. Interferon response element of the human 6-16 gene. EMBO J. 1988;7:85–92. doi: 10.1002/j.1460-2075.1988.tb02786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qureshi S A, Salditt-Georgieff M, Darnell J E., Jr Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated gene factor 3. Proc Natl Acad Sci USA. 1995;92:3829–3833. doi: 10.1073/pnas.92.9.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reich N, Pine R, Levy D, Darnell J E. Transcription of interferon-stimulated genes is induced by adenovirus particles but is suppressed by E1A gene products. J Virol. 1987;62:114–119. doi: 10.1128/jvi.62.1.114-119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staeheli P, Danielson P, Haller O, Sutcliffe J G. Transcriptional activation of the mouse Mx gene by type I interferon. Mol Cell Biol. 1986;6:4770–4774. doi: 10.1128/mcb.6.12.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tevethia M J, Spector D J. Complementation of an adenovirus 5 immediate early mutant by human cytomegalovirus. Virology. 1984;137:428–431. doi: 10.1016/0042-6822(84)90236-8. [DOI] [PubMed] [Google Scholar]
- 46.Thanos D, Maniatis T. The high mobility group protein HMG 1(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 47.Tomita Y, Cantell K, Kuwata T. Effects of human γ interferon on cell growth, replication of virus and induction of 2′-5′ oligoadenylate synthetase in three human lymphoblastoid cell lines and K562 cells. Int J Cancer. 1982;30:161–165. doi: 10.1002/ijc.2910300206. [DOI] [PubMed] [Google Scholar]
- 48.Veals S A, Santa Maria T, Levy D E. Two domains of ISGF3γ that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol Cell Biol. 1993;13:196–206. doi: 10.1128/mcb.13.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velazquez L, Fellous M, Stark G R, Pellegrini S. A protein tyrosine kinase in the interferon α/β signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 50.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 51.Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani Y, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. A novel interferon regulatory factor family transcription factor, ICSAT/Pip/LSIRF, that negatively regulates the activity of interferon-regulated genes. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu-Lee L, Hrachovy J A, Stevens A M, Schwarz L A. Interferon-regulatory factor 1 is an immediate-early gene under transcriptional regulation by prolactin in Nb2 T Cells. Mol Cell Biol. 1990;10:3087–3094. doi: 10.1128/mcb.10.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath K, Darnell J. Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]