Abstract
The protein tyrosine phosphatase SHP-1 is a critical regulator of macrophage biology, but its detailed mechanism of action remains largely undefined. SHP-1 associates with a 130-kDa tyrosyl-phosphorylated species (P130) in macrophages, suggesting that P130 might be an SHP-1 regulator and/or substrate. Here we show that P130 consists of two transmembrane glycoproteins, which we identify as PIR-B/p91A and the signal-regulatory protein (SIRP) family member BIT. These proteins also form separate complexes with SHP-2. BIT, but not PIR-B, is in a complex with the colony-stimulating factor 1 receptor (CSF-1R), suggesting that BIT may direct SHP-1 to the CSF-1R. BIT and PIR-B bind preferentially to substrate-trapping mutants of SHP-1 and are hyperphosphorylated in macrophages from motheaten viable mice, which express catalytically impaired forms of SHP-1, indicating that these proteins are SHP-1 substrates. However, BIT and PIR-B are hypophosphorylated in motheaten macrophages, which completely lack SHP-1 expression. These data suggest a model in which SHP-1 dephosphorylates specific sites on BIT and PIR-B while protecting other sites from dephosphorylation via its SH2 domains. Finally, BIT and PIR-B associate with two tyrosyl phosphoproteins and a tyrosine kinase activity. Tyrosyl phosphorylation of these proteins and the level of the associated kinase activity are increased in the absence of SHP-1. Our data suggest that BIT and PIR-B recruit multiple signaling molecules to receptor complexes, where they are regulated by SHP-1 and/or SHP-2.
Protein tyrosine phosphatases (PTPs) and protein tyrosine kinases (PTKs) control the level of tyrosyl phosphorylation on cellular proteins (reviewed in references 38, 52, and 56). Although many substrates for PTKs have been identified, the specific targets of individual PTP family members, along with the consequences of protein dephosphorylation for cellular physiology, remain largely unknown. The finding that some PTPs possess SH2 domains suggested that these molecules might be recruited to specific phosphotyrosyl (pY) sites (via their SH2 domains), where their PTP domains could catalytically amplify or terminate phosphotyrosine-mediated signals (14, 37). Two vertebrate SH2-containing PTPs (SHPs), SHP-1 and SHP-2 (1), have been cloned and characterized (reviewed in reference 37). Despite their structural similarity, SHP-1 and SHP-2 appear to possess distinct biological functions (reviewed in references 3, 36 to 38, 52, and 56). SHP-2 is expressed ubiquitously and typically transmits positive (i.e., signal-enhancing) signals downstream of receptor tyrosine kinases, cytokine receptors, and multichain immune recognition receptors, although recent work suggests that it may have negative (i.e., signal-attenuating) effects in some pathways (33, 54). In contrast, SHP-1 is expressed predominantly in hematopoietic cells, where it negatively regulates multiple growth factor and cytokine signaling pathways (reviewed in references 36, 38 and 56).
Our current understanding of the role played by SHP-1 in hematopoietic cell signaling has been enhanced greatly by the study of mice harboring two different mutations in the SHP-1 gene, the so-called motheaten (me/me) or motheaten viable (mev/mev) mutations (50, 58). The me allele has a single base pair deletion within SHP-1, which leads to abnormal splicing and the generation of a premature stop codon, such that me/me mice are effectively null for SHP-1 protein. The mev mutation also leads to aberrant splicing with the generation of two aberrant forms of SHP-1 with disrupted catalytic domains: one with a 5-amino-acid deletion and the other with a 23-amino-acid insertion. Based on the crystal structure of PTP-1B (2), both mutant proteins produced by the mev mutation would be expected to show markedly compromised substrate binding and/or catalytic ability. Accordingly, mev/mev mice have been shown to exhibit substantially decreased SHP-1 catalytic activity (27, 58). Both me/me and mev/mev mice exhibit multiple abnormalities in hematopoietic cell development and function. The most notable of these abnormalities, and the cause of early death in these animals, is the expansion and inappropriate activation of macrophages and granulocytes (reviewed in references 3 and 37).
Previous work from several laboratories has implicated SHP-1 in the regulation of the colony-stimulating factor 1 receptor (CSF-1R) signal transduction pathway. CSF-1, also known as macrophage colony-stimulating factor, is the major growth factor directing the proliferation, differentiation, and survival of cells of the mononuclear phagocyte lineage ex vivo and in vivo (reviewed in reference 51); hence, abnormal regulation of this pathway could contribute to the increased number of macrophages observed in me/me and mev/mev mice. Stimulation of the BAC1.2F5 macrophage cell line with CSF-1 results in tyrosine phosphorylation of SHP-1 (10, 59, 61). Although the CSF-1R itself probably phosphorylates SHP-1, association between SHP-1 and the CSF-1R has not been detected (10, 59, 61). More importantly, primary bone marrow-derived macrophages (BMM) from me/me mice are hypersensitive to the mitogenic effects of CSF-1, suggesting that SHP-1 negatively regulates CSF-1R signaling (10). The molecular details by which this regulation is effected and, in particular, the direct regulators and targets of SHP-1 remain largely unknown. A potentially attractive target is the CSF-1R itself, since it is hyperphosphorylated on tyrosine in me/me BMM (10). However, in the absence of a direct SHP-1–CSF-1R association, it is unclear how SHP-1 gains access to the CSF-1R. Other possible targets in this pathway include a group of Grb-2 SH3 domain-binding proteins that are hyperphosphorylated on tyrosine in me/me BMM (10). These proteins are likely to be recruited to tyrosyl-phosphorylated SHP-1 by virtue of its ability to bind the SH2 domains of Grb-2 (10).
Previously, we reported that SHP-1 constitutively coimmunoprecipitates with a 130-kDa tyrosine-phosphorylated species in murine BMM and some macrophage cell lines (10). This interaction, mediated via the SH2 domains of SHP-1, suggested that the 130-kDa species might be a regulator and/or substrate of SHP-1; thus, its identification and characterization are crucial to further understanding the mechanism of signal regulation by SHP-1 in macrophages. Previous attempts to identity this protein by immunoblotting eliminated c-Cbl, p130Cas, Jak2, Fak, Fps/Fes, GTPase-activating protein, Pyk2, SH2-containing inositol phosphatase (SHIP), vinculin, and the interleukin-3 receptor beta chain (10, 55a).
Here we show that the 130-kDa band actually consists of two transmembrane glycoproteins of different core sizes. We identify these proteins as a SIRP (signal-regulatory protein) family member known as BIT (brain immunoglobulin [Ig] like molecule with tyrosine-based activation motifs) (48) and PIR-B/p91A (22, 28), a protein that shares sequence similarity with human killer cell inhibitory receptors (KIRs), the Fcα receptor, and murine gp49B1. BIT, but not PIR-B, coimmunoprecipitates with the CSF-1R, suggesting that one of its functions may be to direct SHP-1 to the CSF-1R complex. However, other roles also seem likely. Our results indicate that BIT and PIR-B are direct substrates of SHP-1 in vivo. In addition, BIT and PIR-B coimmunoprecipitate with the same two, as yet unidentified, tyrosyl phosphoproteins, as well as an associated PTK activity. Tyrosyl phosphorylation of these proteins and the level of the associated PTK activity are enhanced in the absence of functional SHP-1. These data suggest that BIT and PIR-B recruit SHP-1 and other signaling molecules to the plasma membrane for the regulation of growth factor receptors and perhaps other macrophage signaling complexes.
MATERIALS AND METHODS
Cell culture.
BAC1.2F5 (35), a simian virus 40 large-T-antigen-immortalized cell line that retains CSF-1 dependence and many other properties of primary murine BMM, was maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and 20% conditioned media from L cells (LCM) as a source of CSF-1. Primary BMM from 10- to 20-day-old C3HeB/FeJLe-a/a-Hcphme me/me, C57BL/6J-Hcphme-v mev/mev (Jackson Laboratory, Bar Harbor, Maine), or normal littermate mice were prepared and cultured as described previously (10). We also generated new, early-passage cell lines from mev/mev and normal mice (N and meV) by immortalizing primary BMM by using the retrovirus pZip-Tex (4), which directs expression of simian virus 40 large T antigen. For infections, a 1:1 mixture of medium and supernatant from confluent Psi 2 cells expressing Zip-Tex virus was added to a 10-cm-diameter dish of subconfluent primary BMM for 4 h at 37°C. Fresh medium was added, and the cells were passaged for several days. Clones were picked following dilution, and their growth was monitored in the absence and presence of LCM. Clones that retained dependence on LCM were expanded and used for this study.
SHP-1 bacterial expression constructs.
Constructs directing the expression of glutathione S-transferase (GST) fusion proteins containing wild-type SHP-1 and a Cys453→Ser (C453S) mutation have been described elsewhere (42). The SHP-1 Asp419→Ala mutation (D419A) was generated by PCR-mediated mutagenesis. Briefly, the vector pGEX-2T-SHP-1, containing wild-type SHP-1 (44), was used as a template with primer pairs 5′-CACCCGAGAGGTGGAGAAAGGC-3′ plus 5′-GACCCCATGGGCGGGCCAG-3′ (codon 419 underlined) and 5′-GGGGCACTCCTAGGTTCAGGTTG-3′ plus 5′CTGGCCCGCCCATGGGGTC-3′ to generate 216- and 633-bp products, respectively. These products were used as templates in a third reaction with the 5′/3′ primer set (5′-CACCCGAGAGGTGGAGAAAGGC-3′ and 5′-GGGGCACTCCTAGGTTCAGGTTG-3′), and the product was digested with BglII and AvrII and subcloned into BglII/AvrII-cut pGEX-2T-SHP-1. The Arg459→Met (R459M) mutant was generated by using a similar strategy with the 3′/5′ primer set described above and the overlapping mutagenic oligonucleotides 5′-GGTGCCTGTCATGCCGATGC-3′ (codon 459 underlined) and 5′-GCATCGGCATGACAGGCACC-3′. The final PCR product was AvrII/BglII digested and ligated into AvrII/BglII-digested pGEX-2T-SHP-1. An SHP-1 mutant in which residues 451 to 475 are deleted was generated; this mutant (ΔP) lacks the PTP catalytic domain signature motif (V451HCSAG) and thus should be catalytically inactive and unable to bind substrates. Primer pairs for each reaction were 5′-CACCCGAGAGGTGGAGAAAGGC-3′ plus 5′-GTCACAGTCCAGGCCCTTGATGATGGGCCCTGCGTGA-3′ (underlined sequence denotes overlap) and 5′-AAGGGCCTGGACTGTGACATTG-3′ plus 5′-GGGGCACTCCTAGGTTCAGGTTG-3′. The resulting overlapping fragments were used as template with the 5′/3′ primer set (see above), and the product was digested and subcloned into AvrII/BglII-digested pGEX-2T-SHP-1. Transformants (in Escherichia coli DH5α) were screened for the ability to express the appropriately sized fusion protein following isopropylthio-β-d-galactoside (IPTG) induction, and the SHP-1 sequence in plasmids isolated from positive clones was confirmed by automated dideoxynucleotide sequencing.
Antibodies.
Affinity-purified antibodies directed against full-length SHP-1 (FL antibodies) or its C terminus (CTM antibodies) have been described previously (10, 31). Unless otherwise stated, FL antibodies, which predominantly recognize N-terminal determinants, were used for immunoprecipitations. Polyclonal antibodies against C-terminal peptides of SHP-1 and SHP-2, respectively, were purchased from Santa Cruz Biotechnology, Inc. Anti-SHP-1 monoclonal antibodies (MAbs), used for immunoblotting, were purchased from Transduction Laboratories, and antiphosphotyrosine (anti-pTyr) MAb 4G10 was from Upstate Biotechnology, Inc. Rabbit polyclonal anti-CSF-1R antibodies were the generous gift of Martine Roussel (St. Jude’s Cancer Research Center, Memphis, Tenn.) or were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti-mitogen-activated protein kinase (MAPK) antibodies were a gift from John Blenis (Harvard Medical School, Boston, Mass.). Polyclonal antisera against rat SHPS-1 and murine PIR-B/p91A were generated as described below.
Cloning of rat SHPS-1 and murine PIR-B.
Full-length SHPS-1 (for SH2 phosphatase substrate 1)/SIRPα1 was cloned by reverse transcription (RT)-PCR from rat brain total RNA by oligo(dT) priming, using primers 5′-CGGAATTCGCCATGGAGCCCGCCGGC-3′ (translation start site underlined) and 5′-CGGAATTCCCATTCACTTCCTCTGGAC-3′ (stop codon underlined). The generated fragment was cut with EcoRI and cloned into the vector pBS (Stratagene).
Murine PIR-B was cloned by its ability to interact with SHP-2 in a modified yeast two-hybrid system (29). The N- and C-terminal SH2 domains of SHP-2 were PCR amplified by using primers 5′-GAGGAGGTCGACCATCGCGGAGATGGTTTCACCCA-3′ and 5′-GAGAGAGTCGACTCAAGCATTTATACGAGTCGTGTT-3′. The PCR product was gel purified, cut with SalI, and cloned into the unique SalI site of pBTM/PDGFR. This modified bait plasmid includes the tyrosine kinase domain of the platelet-derived growth factor receptor driven from the ADH1 promoter. Expression of the platelet-derived growth factor receptor kinase results in tyrosyl phosphorylation of the target fusion proteins in yeast. Screening of the yeast two-hybrid system was carried out as described previously (29), using a VP16 cDNA library derived from EML cells (57). The PIR-B insert in VP16 was excised with BamHI (5′) and EcoRI (3′), gel purified, and used to probe an EML cell full-length cDNA library (29). Two full-length PIR-B clones were sequenced in both directions. The two clones, 4.1 and 20.1, were identical to the published sequences for p91A (22) and PIR-B1 (28).
Purification of GST fusion proteins.
GST fusion proteins were expressed and affinity purified by using glutathione-agarose by following standard techniques, with some minor modifications. Typically, 300-ml cultures of particular pGEX clones in DH5α were induced at an optical density at 600 nm of 0.7 to 1.0 with 0.1 mM IPTG. Cells were grown at 37°C for 3 h, pelleted, and lysed in a mixture containing 1% Triton X-100, 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, phenylmethylsulfonyl fluoride (20 μg/ml), and lysozyme (0.5 mg/ml). The lysate was sonicated, and a soluble fraction was prepared by centrifugation at 100,000 × g for 30 min at 4°C. Soluble material was incubated with glutathione-agarose beads (Sigma) for 2 h at 4°C, and the beads were washed extensively with lysis buffer followed by 50 mM Tris-HCl (pH 7.4)–150 mM NaCl.
Polyclonal antibody generation.
To generate polyclonal antisera against rat SHPS-1, the SHPS-1 cytoplasmic domain (amino acids 403 to 511) was subcloned by PCR from a pBS vector containing the full-length cDNA (see above), using primers 5′-ACGGAATTCAGGCCAAGGGCTCAACTTCTTCC-3′ and 5′-CGGAATTCCCATTCACTTCCTCTGGAC-3′. The resulting fragment was cut with EcoRI and ligated into EcoRI-digested pGEX-2T (Pharmacia). The GST fusion protein was expressed and purified as described above and then eluted from glutathione-agarose beads with 20 mM reduced glutathione (pH 7.4), dialyzed extensively, and concentrated in a Centricon-30 concentrator (Amicon). Rabbits were immunized with 400 μg of purified protein in complete Freund’s adjuvant and boosted every 4 weeks with 400 μg of protein in incomplete Freund’s adjuvant. First bleeds were made after the second boost, and titers were determined by immunoblotting of macrophage whole-cell lysates.
To prepare antiserum against PIR-B, a VP16 plasmid containing the cytoplasmic domain of PIR-B (amino acids 675 to 840) was cut with BamHI (5′) and EcoRI (3′), gel purified, and cloned into the BamHI and EcoRI sites of pGEX-3X (Pharmacia). GST–PIR-B fusion protein was affinity purified from E. coli BL21 lysates and eluted from glutathione-agarose beads as described above. Rabbits were immunized and boosted with 500 μg of purified GST–PIR-B fusion protein as described above. Test bleeds were collected after the second through fourth boosts, and titers were determined by immunoblotting of FDC-P1 cell lysates.
Immunoprecipitations and immunoblotting.
To prepare whole-cell extracts, cells were washed in phosphate-buffered saline containing 2 mM sodium orthovanadate and lysed on plates in Nonidet P-40 (NP-40) lysis buffer (1% NP-40, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM sodium orthovanadate, protease inhibitor cocktail [final concentrations, 10 μg of leupeptin, 1 μg of aprotinin, 1 μg of pepstatin, 1 μg of antipain, and 20 μg of phenylmethylsulfonyl fluoride per ml]). Lysates were vortexed and incubated on ice for 15 min prior to immunoprecipitations. For cell fractionation, cells were homogenized in hypotonic lysis buffer (20 mM HEPES [pH 7.4], 5 mM KCl, 1 mM MgCl2, 1 mM EDTA, 2 mM sodium orthovanadate, protease inhibitor cocktail). The crude nuclear pellet (P1) was obtained following centrifugation at 600 × g for 5 min. The resultant supernatant was subjected to centrifugation at 100,000 × g for 30 min, yielding crude microsomes (P100) and an S100 fraction. The P1 and P100 fractions were washed in hypotonic lysis buffer, resuspended in NP-40 lysis buffer, and precleared. Samples of each of these precleared fractions, representing the same number of cell equivalents, were subjected to immunoprecipitations with the indicated antibodies. For all other immunoprecipitations, precleared whole cell lysates were normalized for protein by using a bicinchoninic acid protein assay reagent kit (Pierce).
Immunoprecipitations were carried out by adding the appropriate antibodies plus protein A-Sepharose beads and incubating at 4°C for 2 to 3 h. Immune complexes were washed with NP-40 lysis buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto Immobilon-P membranes (Millipore). Immunoblots were blocked with 5% bovine serum albumin in TBST (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.05% Tween 20) for 1 h, incubated for 1 h with primary antibodies in TBST, washed three times for 10 min each in TBST, and then incubated for 1 h with horseradish peroxidase-conjugated secondary antibodies (Amersham). Following further washes, immunoblots were visualized by using enhanced chemiluminescence reagents (Amersham).
Lectin and GST fusion protein binding assays.
For lectin binding assays, anti-SHP-1 immune complexes from 2 × 107 BAC1.2F5 cells were boiled for 2 min in 200 μl of 0.5% SDS–1% β-mercaptoethanol. A 40-μl aliquot of the supernatant was removed as a control, while the remainder was diluted 10-fold in lectin binding buffer (1% NP-40, 50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.2 mM sodium orthovanadate, 2 mM MnCl2, 1 mM CaCl2). The sample was split into four and incubated for 1 h at 4°C with 0.25 mg of either Arachis hypogaea (peanut) lectin, Lens culinaris (lentil) lectin, concanavalin A, or wheat germ agglutinin. All lectins were cross-linked to agarose beads and were purchased from Sigma. The beads were washed in binding buffer, and bound protein was analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting.
For GST fusion protein binding assays, BAC1.2F5 cells were treated for 15 min with 100 μM pervanadate (100 μM sodium orthovanadate plus 100 μM hydrogen peroxide in medium) and then lysed in NP-40 lysis buffer (pH 7.4) containing 5 mM iodoacetic acid (to alkylate and inactivate endogenous PTPs) and 1 mM sodium orthovanadate. Following a 20-min incubation at 4°C, dithiothreitol was added to 15 mM to inactivate any unreacted iodoacetic acid. Lysates were precleared and then incubated for 2 h with 10 μg of either GST alone, GST–SHP-1(WT [wild type]), GST–SHP-1(C453S) or GST–SHP-1(D419A) bound to glutathione-agarose beads. Beads were washed in NP-40 lysis buffer and then incubated for 30 min at room temperature in 100 mM HEPES (pH 8.0) containing 1 mM, 20 mM, or no sodium orthovanadate. Eluates were removed, the beads were washed once in 100 mM HEPES, and bound proteins were analyzed by SDS-PAGE and anti-pTyr immunoblotting.
Deglycosylation of proteins in SHP-1 immunoprecipitates.
Anti-SHP-1 immunoprecipitates were denatured by boiling in 0.4% SDS–1% β-mercaptoethanol for 2 min. Samples were diluted fourfold with endoglycosidase buffer (50 mM sodium phosphate [pH 7.2], 0.66% NP-40, 2 mM sodium orthovanadate, plus protease inhibitor cocktail) and treated with 5 μl (1 U) of protein N-endoglycosidase F (endo F) (PNGase F; Boehringer Mannheim) for 1 h at 37°C or left untreated. Samples were then boiled in sample buffer and analyzed by SDS-PAGE and anti-pTyr immunoblotting.
RT-PCR of SIRP family proteins.
Total RNA was isolated by the guanidium isothiocyanate-CsCl centrifugation method (32). First-strand cDNA was synthesized from 5 μg of total RNA with the Superscript preamplification system (Gibco BRL), using oligo(dT) primers. Fifteen percent of this cDNA was used as template in PCRs with the following primer pairs: for full-length SIRP family clones, 5′-TACAGTGAATTCTCAGCCGCGGCCCATGGAGC-3′ and 5′-AGCATGAGAATTCCATTCACTTCCTCTGGAC-3; for cloning of the 270-bp fragment corresponding to the least conserved region of the SIRP family proteins (residues 37 to 122 of murine SHPS-1), 5′-CAGAAGTGAAGGTGACTCAG-3′ and 5′-TTCACACAGTAGTAGGTACC-3. PCR fragments were subcloned by blunt-end ligation into pUC19, and 10 random clones of each PCR product were subjected to automated sequencing.
Vaccinia virus expression system.
Recombinant vaccinia viruses encoding wild-type SHP-1 and an epitope-tagged version of the SHP-1 C453S mutant and the pSC-65 vaccinia virus vector used to generate recombinant viruses have been described previously (6). To subclone the D419A, R459M, and ΔP mutants of SHP-1 into a vaccinia virus vector (pSC-66), the mutants were first excised from the appropriate pGEX-2T vector with EcoRI and ligated into the vector pBS-KS+ (Stratagene). Mutant DNAs were then excised with SalI and NotI and subcloned into pSC-66 that had been digested with SalI and NotI. For infections, 2 × 107 BAC1.2F5 cells were deprived of CSF-1 for 12 h and then were left uninfected or were infected with each virus (5 PFU/cell) for 6 h at 37°C. Control and infected cells were stimulated with CSF-1 for 1 min or left unstimulated, washed extensively with RPMI, lysed, and subjected to immunoprecipitation with anti-SHP-1 CTM or anti-SHPS-1 antibodies. Immunoprecipitates were analyzed by SDS-PAGE and anti-pTyr immunoblotting. SHP-1 and BIT protein levels in whole-cell lysates and immunoprecipitates were analyzed by immunoblotting with the appropriate antibodies.
Immune complex kinase assays and V8 endoprotease partial peptide mapping.
BIT or PIR-B immunoprecipitates were washed once with low-salt buffer (30 mM HEPES [pH 7.4], 10 mM MnCl2, 0.2 mM sodium orthovanadate) and then incubated with 20 μl of autokinase buffer (30 mM HEPES [pH 7.4], 10 mM MnCl2, 0.2 mM sodium orthovanadate, 10 μM [γ-32P]ATP [final specific activity, 5 μCi/mmol]) for various times, as indicated. Reactions were terminated with sample buffer and analyzed by SDS-PAGE, anti-pTyr immunoblotting, and autoradiography. For V8 mapping, in vitro-labeled 55- and 120-kDa bands from anti-SHPS-1 and anti-PIR-B immunoprecipitates were excised from SDS-polyacrylamide gels and rehydrated in a 1:4 dilution of stacking buffer containing 1 mM EDTA. Gel slices were inserted into the wells of a 15% polyacrylamide gel (containing 1 mM EDTA) and overlaid with 0, 10, 50, or 200 ng of endoprotease Glu-C (V8 protease; Boehringer Mannheim) in 1:4 stacking buffer. Cleavage products were resolved and then analyzed by autoradiography.
RESULTS
Characterization of SHP-1-associated P130.
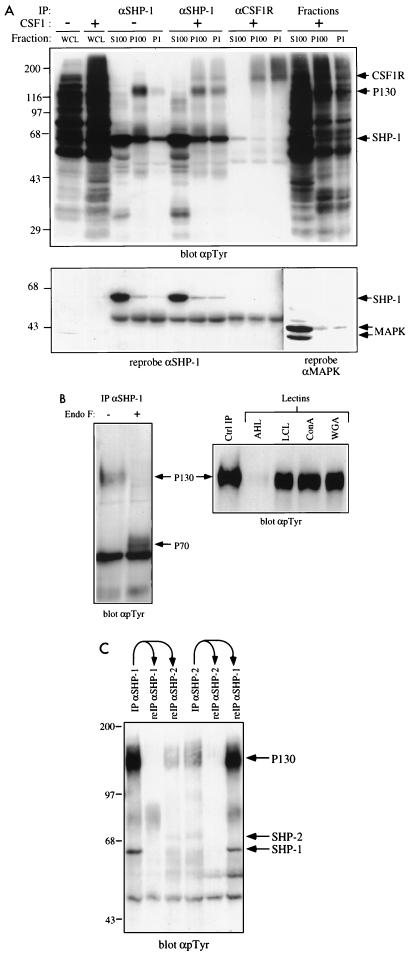
Previously, we observed a broad, 130-kDa phosphotyrosyl band (P130) in SHP-1 immunoprecipitates from BMM and BAC1.2F5 cell lysates. To further characterize P130, we first determined its intracellular location. SHP-1 immunoprecipitates were prepared from P1 (crude nuclear/cytoskeletal), P100 (microsomal), and S100 (cytoplasmic) fractions, generated from starved or CSF-1-stimulated (1 min) cells (Fig. 1A). In starved cells, the SHP-1–P130 complex localized mostly to the P100 fraction, although a small fraction (<5%) was found in the P1 fraction. In contrast, SHP-1 immunoblotting revealed that greater than 90% of the SHP-1 is cytosolic. Thus, P130 is associated with a small pool of membrane-associated SHP-1. This membrane-associated pool of SHP-1 also is more highly tyrosyl phosphorylated than the SHP-1 residing in the cytoplasmic compartment (S100). Upon CSF-1 stimulation, the SHP-1–P130 complex was found in both the P1 and P100 fractions. Some CSF-1R molecules also partitioned into the P1 fraction upon CSF-1 stimulation (Fig. 1A). The increase in SHP-1–P130 complex and CSF-IR in the P1 fraction most likely represents changes in cytoskeletal organization that occur upon CSF-1 stimulation, with consequent alteration of the cell fractionation properties of macrophages, rather than translocation into the nucleus. Interestingly, upon CSF-1 stimulation, a small amount of a tyrosyl-phosphorylated band with a molecular size of ∼160 kDa also could be detected in SHP-1 immunoprecipitates. This band comigrated with the CSF-1R (Fig. 1A), suggesting that SHP-1–P130 may associate transiently with the CSF-1R (see below).
FIG. 1.
P130 is a membrane-localized glycoprotein and associates with both SHP-1 and SHP-2. (A) SHP-1–P130 complex localizes to the membrane compartment. BAC1.2F5 cells were starved of CSF-1 for 20 h and then stimulated with 2,000 U of CSF-1 per ml for 1 min (+) or left unstimulated (−). Cells were fractionated as described in Materials and Methods, and aliquots of the P1, P100, and S100 fractions were immunoprecipitated (IP) with anti-SHP-1 (αSHP-1) or anti-CSF-1R antibodies. Immune complexes were analyzed by SDS-PAGE (10% gel) and anti-pTyr immunoblotting. The anti-SHP-1 antibodies used for this panel are weakly cross-reactive with SHP-2, accounting for the 70-kDa phosphotyrosyl species; SHP-1-specific reagents are used in all subsequent experiments. Whole-cell lysates (WCL) and each fraction also were analyzed. Blots were reprobed with an anti-SHP-1 MAb to test for protein levels, and blots of each fraction were reprobed with anti-MAPK to monitor contamination of the P1 and P100 fractions with soluble cytoplasmic proteins. The positions of migration of Gibco BRL protein molecular size standards are shown in kilodaltons at the left. (B) P130 is a glycoprotein. Anti-SHP-1 immunoprecipitates from randomly growing BAC1.2F5 cells were treated with endo F as described in Materials and Methods. Deglycosylated (+) and untreated (−) samples were analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting (left panel). Lectin binding assays (right panel) were carried out as described in Materials and Methods, using A. hypogaea lectin (AHL), L. culinaris lectin (LCL), concanavalin A (ConA), or wheat germ agglutinin (WGA). Bound proteins were analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting. An anti-SHP-1 immunoprecipitate was loaded as a control (Ctrl IP). (C) P130 can coimmunoprecipitate with SHP-1 and SHP-2. Specific antibodies to SHP-1 and SHP-2 were used in consecutive immunoprecipitations from the lysates of a randomly growing normal macrophage cell line (N). Bound proteins were analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting. The migration of molecular size standards is indicated in kilodaltons at the left.
The location of P130 within membrane fractions, together with its broad appearance on SDS-PAGE, suggested that it might be a glycoprotein. To test this possibility, we first treated SHP-1 immunoprecipitates from BAC1.2F5 cells with endo F. Endo F treatment caused a shift in the apparent molecular weight of the 130-kDa species, indicating that P130 is a glycoprotein with a core polypeptide size of around 70 kDa (Fig. 1B, left panel). The resulting 70-kDa core protein migrated as a doublet, perhaps reflecting differentially phosphorylated and/or O-glycosylated forms of the protein. Further evidence that P130 is a glycoprotein was provided by lectin binding experiments. SHP-1 immunoprecipitates were eluted at low pH, and the neutralized eluate was incubated with four different agarose-linked lectins. L. culinaris lectin, concanavalin A, and wheat germ agglutinin were found to bind P130, whereas P130 did not bind to peanut lectin (Fig. 1B, right panel). This analysis suggests that P130 from BAC1.2F5 cells does not contain terminal galactose-β(1-3)N-acetylgalactosamine, the disaccharide residue preferentially recognized by peanut lectin, whereas it probably contains α-mannose and β-N-acetyl-d-glucosamine.
We also examined whether P130 could bind to SHP-2. Using SHP-2 C-terminal peptide-specific antibodies, which do not cross-react with SHP-1, we observed tyrosyl-phosphorylated P130 in immunoprecipitates from lysates of asynchronous BAC1.2F5 cells (Fig. 1C, lane 4). The SHP-2-associated P130 species had biochemical (e.g., same core molecular size after endo F treatment) and immunological properties similar to those of the SHP-1-associated P130 (data not shown), suggesting that both SHPs associate with the same tyrosyl-phosphorylated glycoprotein. However, the SHP-1–P130 and SHP-2–P130 complexes appear to be distinct, as revealed by sequential immunodepletion experiments (Fig. 1C). Under conditions where SHP-1 (and hence the SHP-1-associated P130) was immunodepleted from lysates by using SHP-1-specific antibodies (lanes 1 and 2), SHP-2-associated P130 was still immunoprecipitated from the cleared lysate (lane 3). Conversely, P130 coimmunoprecipitated with SHP-1 from lysates cleared of SHP-2-associated P130 (lane 4 to 6). In addition to demonstrating the existence of separate SHP-P130 complexes, these data suggest that more P130 associates with SHP-1 than with SHP-2 in randomly growing macrophages.
Characterization and identification of P130 in primary BMM.
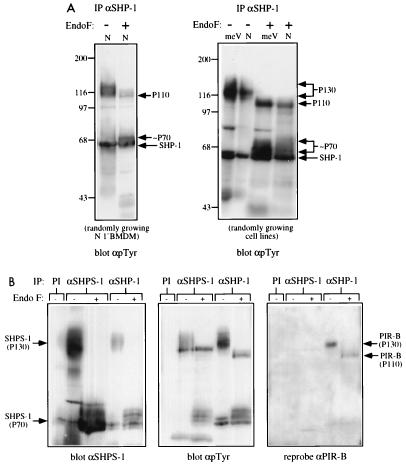
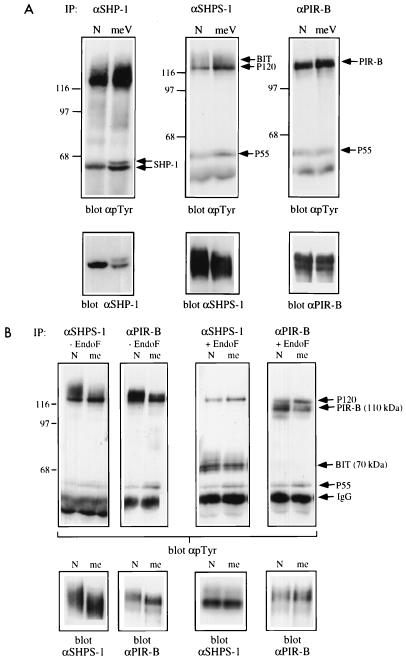
Although BAC1.2F5 cell P130 appeared to consist of a single tyrosyl-phosphorylated glycoprotein, different results were obtained upon analysis of normal primary BMM or two newly derived macrophage cell lines. These N and meV lines were generated by infecting BMM from normal and mev/mev mice, respectively, with ZipTex retrovirus and screening for cell clones that retained CSF-1 dependence (see Materials and Methods). Upon endo F treatment of SHP-1 immunoprecipitates from primary BMM or from either of these cell lines, a tyrosyl-phosphorylated core protein of 110 kDa was apparent in addition to the 70-kDa doublet observed in BAC1.2F5 cells (Fig. 2A). Interestingly, both the 70- and 110-kDa core proteins appeared hyperphosphorylated in the meV line compared with the N line (see below).
FIG. 2.
P130 consists of two glycoproteins, one a SIRP family member and the other PIR-B/p91A. (A) P130 consists of two distinct glycoproteins in primary BMM and cell lines derived from them. SHP-1 immunoprecipitates (IP) from randomly growing normal primary BMM (left panel) or N and meV cell lines (right panel) were treated with endo F (+) or left untreated (−). Deglycosylated proteins were analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting. The migration of molecular size standards is indicated in kilodaltons at the left. (B) Identification of the two glycoproteins as an SHPS-1-related protein and PIR-B/p91A. Preimmune (PI), anti-SHP-1, and anti-SHPS-1 immunoprecipitates from the meV cell line were treated with endo F (+) or left untreated (−) and then immunoblotted with anti-SHPS-1 (left panel) and anti-pTyr (middle panel) antisera. The anti-pTyr blot was stripped and reprobed with anti-PIR-B antisera (right panel).
The core molecular weights of these two SHP-1-associating proteins were similar to those of two recently cloned, novel membrane glycoproteins. The 70-kDa core protein was reminiscent of SHPS-1 (18, 39), a transmembrane glycoprotein that contains three extracellular Ig domains and a cytoplasmic domain with four potential ITIMs (immune receptor tyrosine-based inhibitory motifs) capable of binding SHPs or SHIP (reviewed in references 5 and 8). SHPS-1 becomes tyrosyl phosphorylated and associates with SHP-2 in CHO cells and fibroblast cell lines in response to various mitogens and upon cell adhesion (18). It also associates with SHP-1 in v-src-transformed rat fibroblasts overexpressing SHP-1. SHPS-1 (also called SIRPα1) is a member of the SIRP family (26); a protein cloned independently and termed BIT (48) is another member of this family (see below). The 110-kDa core protein had properties reminiscent of the myeloid cell- and B-cell-specific glycoprotein PIR-B (for paired Ig-like receptor B) (28), also known as p91A (22). PIR-B also is an ITIM-containing Ig superfamily member and is distantly related to human KIRs, which have been previously shown to associate with SHP-1 (6, 17, 40).
To test whether the two associating proteins that comprise P130 were SHPS-1 and PIR-B, we generated rabbit polyclonal antibodies directed against GST–SHPS-1 and GST–PIR-B fusion proteins. Given the strong sequence similarity between SIRPs that possess cytoplasmic domains (26), anti-SHPS-1 antibodies should cross-react with most, if not all, family members. The anti-SHPS-1 antisera reacted with P130 and its 70-kDa deglycosylated form in anti-SHP-1 immunoprecipitates prepared from early-passage immortalized macrophage cell lines, BAC1.2F5 cells, and primary BMM (Fig. 2B and data not shown). Thus, at least a portion of P130 consists of a SIRP-related protein. The amount of anti-SHPS-1-reactive 130-kDa protein in anti-SHP-1 immunoprecipitates was a small fraction of that present in anti-SHPS-1 immunoprecipitates, indicating that only a relatively minor percentage of total SHPS-1 was in a complex with SHP-1 under steady-state growth conditions (Fig. 2B, left panel). Conversely, the amount of tyrosyl-phosphorylated P130 was greater in SHP-1 immunoprecipitates than in SHPS-1 immunoprecipitates (Fig. 2B, middle panel). Therefore, SHP-1 basally associates with a small but highly phosphorylated pool of the total SHPS-1-related protein.
Reprobing the same blot with anti-PIR-B antisera revealed an immunoreactive band in anti-SHP-1 immunoprecipitates at 130 kDa in the absence of endo F and at 110 kDa following deglycosylation (Fig. 2B, right panel). Thus, SHPS-1 and PIR-B both associate basally with SHP-1 in primary macrophages and in newly generated macrophage cell lines. We failed to detect PIR-B in SHP-1 immunoprecipitates from BAC1.2F5 cells (data not shown), consistent with our earlier observation that only a single deglycosylated core protein could be detected in SHP-1 immunoprecipitates from this cell line (Fig. 1B). The absence of SHP-1-associated PIR-B in BAC1.2F5 cells is likely to reflect the very low levels of PIR-B protein expressed in these cells (data not shown). Our failure to detect PIR-B in anti-SHPS-1 immunoprecipitates or SHPS-1 in anti-PIR-B immunoprecipitates (Fig. 2B) argues that SHP-1 forms distinct complexes with each of these binding proteins.
Since SHPS-1 is one of a family of molecules (SIRPs) (26), we asked whether the component of P130 that was immunoreactive with anti-SHPS-1 antibodies represented a single protein or several SIRP family members. Although the known SIRPs show a high degree of overall sequence similarity, a region within the first Ig domain of these proteins is the most divergent. To determine which SIRP family member(s) are expressed in murine macrophages and thus were candidates for the SHPS-1-related protein that coimmunoprecipitates with SHP-1, we used primers flanking this divergent region to prime RT-PCRs for SIRP-related cDNA fragments. A single PCR fragment was generated (data not shown). Sequence analysis of multiple independent clones of this fragment revealed a single sequence (data not shown), which was identical to murine BIT (PID accession no. D85785) and distinct from murine SHPS-1 (PID accession no. D87967). Using primers flanking the BIT translation start and stop codons in RT-PCRs, we also obtained multiple independent full-length BIT cDNAs from macrophage mRNA. The same primer sets, when used in RT-PCRs with NIH 3T3 cell RNA, generated clones that were distinct from murine BIT but were identical to murine SHPS-1 (data not shown). Taken together, our data suggest that macrophages express mainly (if not entirely) a single SIRP (murine BIT). By inference, we conclude that the anti-SHPS-1-reactive band that associates with SHP-1 (and SHP-2) is largely, if not entirely, comprised of BIT. Although our anti-SHPS-1 and anti-PIR-B antibodies are not capable of immunodepleting BMM lysates of their respective antigens, based on the fraction of each of the two proteins immunodepleted by each antibody, and the absence of phosphotyrosyl bands other than the 70- and 110 kDa species following endo F treatment, we conclude that BIT and PIR-B are the only two major proteins contributing to P130.
BIT is in a complex with the CSF-1R in murine macrophages.
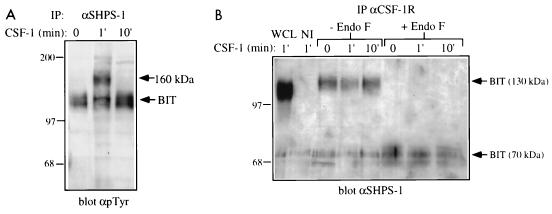
Our previous studies suggested that the CSF-1R may be a substrate for SHP-1 (10). However, the CSF1-R does not associate directly with the SH2 domains of SHP-1 (10, 59, 61) (see the introduction), and so it remained unclear how SHP-1 might access the CSF-1R. In our initial studies characterizing the SHP-1-associated P130 species, we noted the appearance of a weak tyrosyl-phosphorylated band with a molecular size of ∼160 kDa in SHP-1 immunoprecipitates from CSF-1-stimulated cells. This 160-kDa phosphotyrosyl protein comigrated with the tyrosyl-phosphorylated CSF-1R (Fig. 1A), raising the possibility that P130 associates with the CSF-1R, thereby delivering SHP-1 to the receptor.
To test this possibility, we first examined anti-SHPS-1 immunoprecipitates from starved and CSF-1-stimulated primary BMM. Upon 1 min of CSF-1 stimulation, a broad 160-kDa tyrosyl-phosphorylated protein appeared in anti-SHPS-1 immunoprecipitates (Fig. 3A); this protein coimmunoprecipitated with the tyrosyl-phosphorylated CSF-1R (data not shown). This association was transient, with no coimmunoprecipitating band detectable 10 min after CSF-1 stimulation (Fig. 3A). When anti-CSF-1R immunoprecipitates were immunoblotted for BIT (using anti-SHPS-1 antibodies), broad bands were detected at 130 and ∼70 kDa after endo F treatment (Fig. 3B). These data suggest that BIT and CSF-1R are associated in macrophages, since the bands were not detectable in nonimmune antiserum immunoprecipitates (Fig. 3B). We were unable to detect the CSF-1R by immunoblotting of anti-SHPS-1 immunoprecipitates, most likely because of the poor sensitivity of our anti-CSF-1R antibodies in immunoblot assays. Interestingly, BIT and the CSF-1R were present in a complex independent of stimulation, although (as expected) tyrosyl-phosphorylated CSF-1R was detectable only in anti-SHPS-1 immunoprecipitates following CSF-1 stimulation (compare Fig. 3A and B). Much more tyrosyl-phosphorylated CSF-1R was detected in anti-SHPS-1 immunoprecipitates than in anti-SHP-1 immunoprecipitates (data not shown). Since our anti-SHP-1 antibodies immunodeplete SHP-1 from macrophage lysates, whereas our anti-SHPS-1 antibodies remove only ∼60% of BIT (data not shown), these data suggest that the CSF-1R may form a direct complex with BIT.
FIG. 3.
Association of BIT with the CSF-1R. (A) Transient association and phosphorylation of a ∼160-kDa protein in BIT immunoprecipitates (IP). BAC1.2F5 cells were starved of CSF-1 for 22 h and then stimulated with 2,000 U of CSF-1 per ml for the indicated times or left unstimulated (0). BIT was immunoprecipitated from these lysates by using an anti-SHPS-1 antiserum, and bound proteins were analyzed by SDS-PAGE (8% gel) and anti-pTyr immunoblotting. The migration of molecular size standards is indicated in kilodaltons at the left. (B) BIT is associated constitutively with the CSF-1R. The CSF-1R was immunoprecipitated from the same set of lysates and treated with endo F or left untreated. The 1-min lysate was also immunoprecipitated with a nonimmune serum (NI). The presence of BIT in these immunoprecipitates was assessed by immunoblotting with the anti-SHPS-1 antiserum, which is able to recognize murine BIT. Whole-cell lysate (WCL) also was tested as a positive control. The migration of molecular size standards is indicated in kilodaltons at the left.
BIT and PIR-B are substrates of SHP-1.
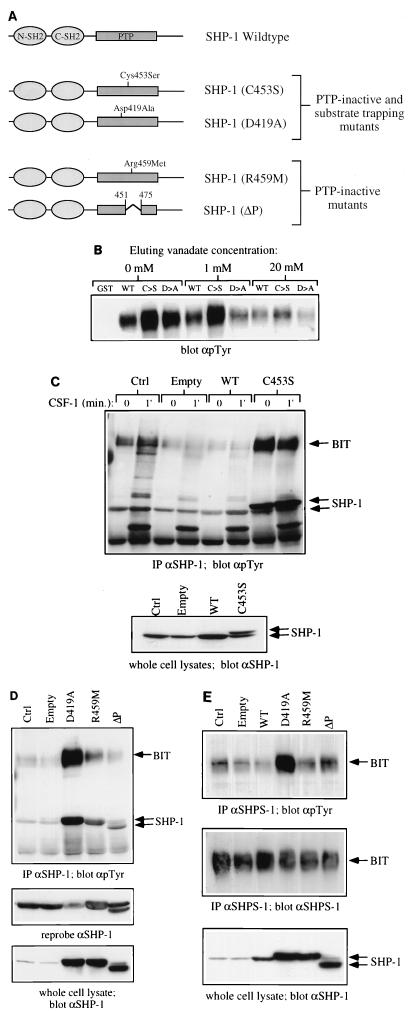
It has been proposed that SHPS-1 and possibly other SIRP family members are SHP-2 substrates as well as binding proteins (18, 26). In addition, Dos (the daughter of sevenless gene product), which binds to the Drosophila homolog of SHP-2, Csw (the corkscrew gene product) (16, 43), has been reported to be a Csw substrate (23, 46). These findings suggest that SHP-binding proteins may also be SHP substrates. To determine whether BIT and/or PIR-B might be substrates for SHP-1, we first treated SHP-1 immunoprecipitates containing BIT and PIR-B with recombinant GST–SHP-1. Tyrosyl phosphorylation of both proteins was reduced upon SHP-1 treatment, indicating that SHP-1 can dephosphorylate these proteins in vitro (data not shown). BIT and PIR-B bind to SHP-1 via the latter’s SH2 domains. However, if either of the 130-kDa proteins is an SHP-1 substrate, it should also have affinity for the SHP-1 PTP domain. To distinguish between SH2 domain and PTP domain binding, we compared the abilities of phosphotyrosyl proteins to bind to GST fusion proteins of wild-type SHP-1 with two catalytically inactive mutants (D419A and C453S) (Fig. 4A). By analogy with enzymological and structural studies of other PTP family members (reviewed in reference 13), such as PTP-1B (2, 15, 25) and PTP-PEST (19), these SHP-1 mutants would be expected to lose all (C453S) or nearly all (D419A) catalytic activity while retaining the ability to bind substrates. Such so-called substrate-trapping mutants have been used to identify in vivo substrates for several PTPs (15, 19; reviewed in references 13, 38, and 56). The tyrosyl-phosphorylated 130-kDa phosphotyrosyl species bound better to the C453S and D419A mutants than to wild-type SHP-1 (Fig. 4B, lanes 1 to 4). This suggests an interaction with both the catalytic cleft and the SH2 domains of SHP-1. Incubation with the active site competitor vanadate resulted in release of bound protein, confirming that binding can occur at the active site as well as the SH2 domains. Moreover, vanadate elution of P130 from the D419A mutant was more efficient than from the C453S (Fig. 4B, lanes 5 to 10). Since vanadate forms a covalent bond with the catalytic cysteinyl residue of PTPs (24, 25), it would be expected to have higher affinity for the D419A mutant (which retains this cysteinyl residue) than for the C453S mutant. Vanadate also was more effective at eluting P130 from the D419A mutant compared to wild-type SHP-1. Most likely, this observation can be explained by known features of the PTP catalytic mechanism: upon substrate binding, the so-called WPD loop (which, in SHP-1, contains D419) moves by nearly 10 Å toward the active-site cysteinyl residue, thus allowing D419 to serve as the general acid in catalysis. In the wild-type enzyme, this would lead to charge repulsion between the incoming general acid aspartyl residue and the negatively charged vanadate. This repulsive interaction is missing in the D419A mutant, making it likely that it would have a higher affinity for vanadate. Notably, a similar explanation has been proposed to explain the greater ability of D→A mutants to serve as substrate traps (reviewed in references 13, 38, and 56). No detectable amounts of P130 bound to either wild-type or mutant versions of the isolated PTP domain of SHP-1 (data not shown), suggesting that the SH2 domain(s) are necessary for P130 recruitment.
FIG. 4.
BIT and PIR-B are substrates of SHP-1 in macrophages. (A) Schematic representation of wild-type and mutant forms of SHP-1 used in this study. The properties of the PTP-inactive substrate-trapping and PTP-inactive nontrapping mutants are described in detail in the text. (B) P130 complex binds to substrate trapping mutants of SHP-1. GST fusion protein binding assays from BAC1.2F5 cell lysates were carried out as described in Materials and Methods, using 10 μg of either GST alone, GST–SHP-1(WT), GST–SHP-1(C453S) (C→S) or GST-SHP-1(D419A) (D→A). Bound proteins were eluted with 1 and 20 mM sodium orthovanadate or left untreated (0 mM). The amount of P130 remaining in the complex was analyzed by SDS-PAGE and anti-pTyr immunoblotting. (C) BIT binds preferentially to SHP-1(C453S) in vaccinia virus-infected BAC1.2F5 cells. Starved cells were either left uninfected (Ctrl) or infected with empty vaccinia virus (Empty), recombinant virus encoding wild-type SHP-1 (WT), or epitope-tagged SHP-1(C453S) (C→S) as described in Materials and Methods. Control and infected cells were stimulated with CSF-1 for 1 min or left unstimulated (0). Lysates were prepared and immunoprecipitated with anti-SHP-1 CTM antibodies, and immunoprecipitates were analyzed by SDS-PAGE and anti-pTyr immunoblotting (top panel). SHP-1 protein levels were analyzed in whole-cell lysates by immunoblotting with anti-SHP-1 MAb (lower panel). (D) Comparison of SHP-1-associated BIT phosphorylation in cells infected with phosphatase dead mutants of SHP-1. The experiment in Figure 4D was repeated with recombinant viruses for the expression of SHP-1 mutants D419A, R459M, and ΔP. SHP-1 immunoprecipitates from uninfected and virus infected BAC1.2F5 cells were analyzed by anti-pTyr immunoblotting. Levels of SHP-1 in immune complexes (middle panel) and whole-cell lysates (lower panel) were checked by reprobing and blotting with anti-SHP-1 MAb. (E) BIT phosphorylation in recombinant vaccinia virus-infected cells. BIT was immunoprecipitated from starved, uninfected and virus-infected BAC1.2F5 cells by using the anti-SHPS-1 antiserum, which is able to recognize murine BIT. Relative BIT tyrosine phosphorylation was analyzed by anti-pTyr blotting (top panel) and anti-SHPS-1 blotting (middle panel). The level of expression of each recombinant protein was checked by anti-SHP-1 blotting of whole-cell lysates (lower panel).
More compelling evidence that BIT is an SHP-1 substrate was provided by examining the effects of expressing these mutants in BAC1.2F5 cells (which do not express significant levels of PIR-B), using a vaccinia virus expression system. If BIT is an SHP-1 substrate, trapping mutants should bind more tyrosyl-phosphorylated BIT than either wild-type SHP-1 or catalytically impaired mutants that lose substrate binding capacity (nontrapping mutants) (Fig. 4A). By binding to tyrosyl-phosphorylated substrates, trapping mutants also might be expected to protect the tyrosyl-phosphorylated target from dephosphorylation by other cellular enzymes with tyrosine phosphatase activity (in vivo PTP protection assay). In the case of cells infected with vaccinia virus, these other enzymes include VH-1, a highly active dual-specificity phosphatase encoded in the vaccinia virus genome (21, 30). Infection with the parental (empty) vaccinia virus resulted in a dramatic decrease (up to 80 to 90%) in total cellular tyrosyl phosphorylation (data not shown), along with an equally dramatic decrease in SHP-1-associated tyrosyl-phosphorylated BIT (Fig. 4C), presumably reflecting the promiscuous action of VH-1. Cells infected with a recombinant vaccinia virus directing the expression of wild-type SHP-1 showed even lower levels of tyrosyl-phosphorylated BIT associated with SHP-1 (Fig. 4C, WT). However, cells expressing the C453S trapping mutant showed enhanced BIT tyrosyl phosphorylation (Fig. 4C, C453S). Notably, the level of tyrosyl-phosphorylated BIT associated with SHP-1 in C453S-infected cells was substantially higher than that found in control, uninfected cells, even though the level of expression of the mutant protein was essentially equal to that of the endogenous protein (Fig. 4C, bottom panel). Thus, our data suggest that tyrosyl-phosphorylated BIT is protected from dephosphorylation in C453S-expressing cells.
The ability of the C453S mutant to protect BIT from dephosphorylation by endogenous PTPs (including VH-1) could be due either to increased levels of its SH2 domains (given the ability of SH2 domains to protect their targets from phosphatase action [47]) and/or to direct protection afforded by the catalytically inactive but substrate-binding PTP domain. Cells overexpressing wild-type SHP-1 have increased amounts of its SH2 domains and increased total PTP activity; thus, the failure of overexpressed wild-type SHP-1 to cause increased BIT tyrosyl phosphorylation does not rule out SH2 domain protection. To distinguish between SH2 domain- and inactive PTP domain-mediated protection, we used two other catalytically inactive SHP-1 mutants, which would be expected to lose substrate binding ability (Fig. 4A). ΔP is an internal deletion in the PTP domain spanning the active-site cysteine. R459M should disrupt an interaction known to be critical for binding phosphate (2, 53); the analogous mutation in PTP-1B leads to a marked increase in Km, consistent with a significant decrease in affinity for binding substrates, along with a dramatic decrease in catalytic activity (15). Expression of nontrapping mutants should permit SH2 but not PTP domain-mediated protection.
Indeed, cells expressing the D419A trapping mutant exhibited a far greater increase in tyrosyl-phosphorylated BIT than those expressing either the R459M or ΔP mutant (Fig. 4D, top panel). These differences were not due to the levels of immunoprecipitated SHP-1 (middle panel) or the levels of expression of the various SHP-1 mutants (lower panel). Notably, cells expressing either R459M or ΔP displayed significant protection of tyrosyl-phosphorylated BIT compared to empty virus-infected cells. This level of protection presumably was afforded by the increased levels of the SHP-1 SH2 domains in the mutant-expressing cells and emphasizes the importance of distinguishing between SH2 and PTP domain protection when evaluating the effects of SHP trapping mutants (see Discussion). The effects of the various SHP-1 mutants on BIT tyrosyl phosphorylation were not restricted to SHP-1-associated BIT but rather affected the whole cellular pool of tyrosyl-phosphorylated BIT. This was revealed by comparing BIT tyrosyl phosphorylation in anti-SHPS-1 immunoprecipitates (Fig. 4E). In agreement with the previous experiments, BIT tyrosyl phosphorylation was markedly higher in cells expressing the D419A trapping mutant compared to wild-type, R459M, or ΔP forms of SHP-1. These in vivo protection assay data, together with the in vitro binding/vanadate elution experiments, strongly suggest binding of BIT to the PTP active site and, by inference, indicate that BIT is a bona fide substrate of SHP-1 in macrophages in vivo. Unfortunately, the same analysis could not be easily applied to PIR-B, since we fail to detect significant levels of this protein in BAC1.2F5 cells. Nevertheless, other data (see below) suggest that PIR-B also is an SHP-1 substrate.
Tyrosyl phosphorylation of BIT and PIR-B in mev/mevand me/meBMM.
Further evidence that BIT and PIR-B are in vivo substrates of SHP-1 was provided by studies of BMM from meν/meν mice. The forms of SHP-1 expressed in meν/meν mice have greatly reduced catalytic activity due to mutations within the PTP domain (see the introduction). Targets of SHP-1 in vivo would be expected to be hyper-phosphorylated in meν/meν BMM. Notably, however, the meν/meν forms of SHP-1 retain the potential to bind tyrosyl phosphoproteins via their SH2 domains; i.e., they can provide SH2 domain-mediated protection against dephosphorylation by other cellular PTPs. Thus, the meν/meν forms of SHP-1 should behave like the R459M and ΔP nontrapping mutants of SHP-1. To test these predictions, we examined the tyrosyl phosphorylation status of BIT and PIR-B in BMMs from meν/meν mice and their normal littermates. Consistent with the notion that these proteins are in vivo substrates of SHP-1, both were found to be hyperphosphorylated in meν/meν BMM (Fig. 5A).
FIG. 5.
BIT and PIR-B tyrosine phosphorylation in N, meV, and me/me primary BMM. (A) BIT and PIR-B are hyperphosphorylated in primary BMM from meν/meν mice. The relative levels of tyrosine phosphorylation of BIT and PIR-B in anti-SHP-1, anti-SHPS-1 (which recognizes murine BIT), and anti-PIR-B immunoprecipitates (IP) from N and meV primary BMM were compared by anti-pTyr immunoblotting (upper panels) and by reprobing and immunoblotting for protein levels (lower panels). The migration of molecular size standards is indicated in kilodaltons at the left. (B) BIT and PIR-B are hypophosphorylated in primary BMM from meν/meν mice. As in panel A, relative tyrosine phosphorylation levels in anti-SHPS-1 and anti-PIR-B immunoprecipitates from normal and meν/meν primary BMM were compared by anti-pTyr immunoblotting (upper panels) and immunoblotting for protein levels (lower panels). In addition, immunoprecipitates were endo F-treated to facilitate the visualization of BIT and PIR-B (right-hand panels). The migration of molecular size standards is indicated in kilodaltons at the left.
We reported previously that the 130-kDa phosphotyrosyl proteins associated with SHP-1 were not SHP-1 substrates. This conclusion was based on the finding that the tyrosyl phosphorylation levels of P130 recovered by binding to a GST-SH2 domain fusion protein of SHP-1 were comparable in normal and me/me BMM (10); notably, meν/meν lysates were not analyzed in our earlier studies. To resolve this apparent contradiction, we directly assessed BIT and PIR-B tyrosyl phosphorylation in me/me BMM. Consistent with our previous work, both proteins consistently showed a lower level of tyrosyl phosphorylation in me/me BMM (Fig. 5B). This decrease in BIT and PIR-B tyrosyl phosphorylation was most apparent when immunoprecipitates of each of these proteins were treated with endo F to separate the 130-kDa proteins from an associated tyrosyl-phosphorylated 120-kDa protein (see below). Although at first glance these findings appear to conflict with the idea that BIT and PIR-B are SHP-1 substrates, it is likely that the loss of SH2 domain protection in me/me cells allows dephosphorylation of BIT and PIR-B by other cellular PTPs, resulting in the observed hypophosphorylation. Such a model would predict that different tyrosyl phosphorylation sites on BIT and PIR-B may be preferred substrates for different PTPs (see Discussion).
BIT and PIR-B associate with additional phosphotyrosyl proteins in macrophages.
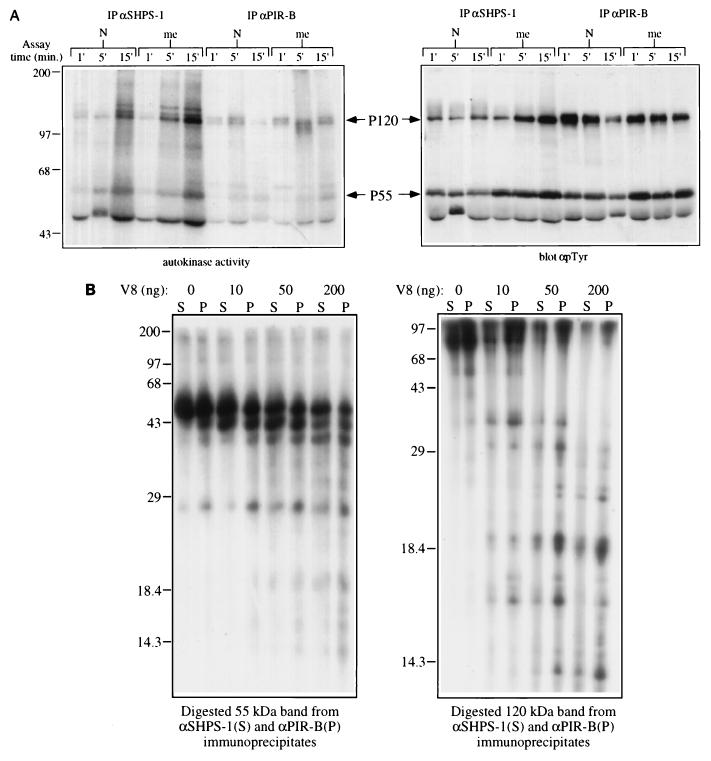
Two non-endo F-sensitive phosphotyrosyl proteins, with apparent molecular sizes of 120 kDa (P120) and 55 kDa (P55), consistently coimmunoprecipitated with BIT and PIR-B (Fig. 5). The P120 protein in particular was much more easily visualized following endo F treatment of BIT or PIR-B immunoprecipitates (Fig. 5B). Interestingly, in contrast to BIT and PIR-B, these proteins were hyperphosphorylated in both me/me and meν/meν primary BMM; hence, they are possible substrates of SHP-1. Alternatively, association of these proteins with BIT and PIR-B may be regulated by BIT and PIR-B tyrosyl phosphorylation. Attempts to identify P55 and P120 by using a battery of antibodies directed against similarly sized proteins have been unsuccessful. Immunoblotting experiments indicate that P55 is unlikely to be Hck, Fgr, Lyn, or p55Shc, whereas P120 does not appear to be Fak, Pyk2, c-Abl, c-Cbl, or p130Cas. BIT and PIR-B immunoprecipitates also contained an associated kinase activity (Fig. 6A, left panel). Interestingly, the major bands labeled in the in vitro kinase assay comigrated with P55 and P120 (Fig. 6A, right panel). Phosphoamino acid analysis revealed that the associated kinase activity(ies) was predominantly a PTK(s) (data not shown). Thus, P55, P120, or both may possess PTK activity. Interestingly, me/me BMM exhibit enhanced BIT- and PIR-B-associated PTK activity, consistent with the notion that SHP-1 regulates the activity of the BIT and PIR-B-associated PTK. To determine whether the same two proteins coimmunoprecipitated with BIT and PIR-B, we subjected the 32P-labeled 120- and 55-kDa bands generated by in vitro kinase assay to partial V8 endoprotease digestion and mapping (Cleveland analysis). Separation of the proteolytic products, followed by autoradiography, confirmed that the BIT and PIR-B coimmunoprecipitating P55 and P120 proteins were identical (Fig. 6B). We conclude that at least two additional phosphotyrosyl proteins, one or both of which may be a PTK, are associated with both BIT and PIR-B and that these proteins represent additional potential targets of SHP-1.
FIG. 6.
Characterization of BIT- and PIR-B-associated proteins. (A) BIT and PIR-B associate with a PTK activity in primary BMM. BIT and PIR-B from normal and me/me primary BMM were assayed for autokinase activity for the times indicated and analyzed by autoradiography (left panel) and anti-pTyr immunoblotting (right panel). Assay conditions are described in Materials and Methods. The migration of molecular size standards is indicated in kilodaltons at the left. IP, immunoprecipitation. (B) V8 endoprotease mapping of BIT- and PIR-B-associated P55 and P120 proteins. The autophosphorylated 55- and 120-kDa proteins from radiolabeled anti-SHPS-1 and anti-PIR-B immune complexes were excised from gels and subjected to in gel treatment with the indicated amounts of V8 protease as described in Materials and Methods. Labeled digests were analyzed by SDS-PAGE (15% gel) and autoradiography. The migration of molecular size standards is indicated in kilodaltons at the left of each panel.
DISCUSSION
Perhaps the most important physiological functions of SHP-1 are to regulate macrophage proliferation and activation. Mice with absent (me/me) or defective (mev/mev) SHP-1 have increased numbers of abnormally activated macrophages, which accumulate in multiple tissues, most notably the lungs. There, together with activated neutrophils, they cause a fatal interstitial pneumonia (reviewed in references 3 and 37). Although earlier work demonstrated intrinsic (i.e., cell-autonomous) signaling defects in me/me BMM (10, 12, 50), including dysregulation of the alpha interferon (12) and CSF-1R (10) signaling pathways, the molecular details by which regulation is normally effected remain largely unknown. Defining major SHP-1 binding proteins and/or substrates in macrophages should provide key insights into SHP-1 regulation and action.
In this report, we have shown that P130, the major phosphotyrosyl protein associated with SHP-1 in BMM, is comprised of two transmembrane glycoproteins, PIR-B and BIT. Some macrophage cell lines (e.g., BAC1.2F5) express low levels of PIR-B and consequently have only SHP-1–BIT complexes. BIT and PIR-B also associate with SHP-2, as well as two other, as yet unidentified phosphotyrosyl proteins (P120 and P55) and a PTK activity, which targets P120 and P55 in vitro and may be intrinsic to one or both of them. P120 and P55 may be recruited to BIT and PIR-B so that they can be regulated by SHP-1. Contrary to our previous prediction (10), experiments with substrate-trapping and nontrapping mutants of SHP-1 and analyses of meν/meν mice clearly indicate that BIT and PIR-B are SHP-1 substrates as well as SHP-1-binding proteins. Despite these shared properties, however, BIT and PIR-B probably have distinct functions in macrophages. BIT, for example, is present in a complex with the CSF-1R and may target SHP-1 to activated receptor complexes; as yet we have no evidence that PIR-B can play a similar role.
The SIRP family, of which SHPS-1 and BIT are members, is a group of highly similar glycoproteins containing Ig domains, some of which also have intracellular segments with multiple tyrosyl residues (26). SIRPs were initially defined as SHP-2-binding proteins. When SHP-1 was overexpressed ectopically in a rat fibroblastic cell line (18), it could form a complex with SHPS-1; however, the physiological significance of this interaction remained unclear. Our results indicate that SIRP interactions with SHP-1 do, in fact, occur in primary cells under physiological levels of expression of both proteins. Interestingly, one region within SIRP ectodomains is the most variable. Such a hypervariable region might serve as a ligand-binding domain and/or a domain that functions as the ligand for another cell surface molecule. Intriguingly, a recent study showed that purified BIT could promote cerebral cortical neurite extension (48), suggesting that BIT may be a ligand for an undefined neuronal receptor. It will be important to search for potential BIT ligands on cells that interact with macrophages in vivo. Alternatively, BIT may bind an extracellular matrix component. The constitutive tyrosyl phosphorylation of BIT may be thus misleading; it could simply reflect the fact that our experiments were performed with adherent macrophages.
PIR-B (also called p91A) is similar in topography to SIRPs, although it contains three more Ig domains and its sequence is only distantly related. Unlike SIRPs, which are widely expressed (18, 26), PIR-B expression is restricted to myeloid and B cells (22). PIR-B had been predicted to bind either or both of the SHPs (22, 28). We now have shown that such interactions do exist under physiologically relevant conditions in macrophages; similar interactions also take place in B cells (35a). The structure of PIR-B predicts it also may serve as a ligand and/or a receptor. Although distantly related to FcαR and KIRs, PIR-B does not bind to immune complexes, the ligands for Fc receptors (22), nor has it been shown to bind to major histocompatibility complex antigens, the ligands for KIRs (5). Given their divergence in sequence and structure, the BIT (and SIRPs in general) and PIR-B ectodomains probably interact with distinctly different classes of proteins.
In contrast, the cytoplasmic domains of BIT and PIR-B may deliver similar types of downstream signals. Both proteins bind SHP-1 and SHP-2 in BMM, but immunodepletion experiments clearly establish that the SHP-2–BIT and SHP-1–BIT complexes are distinct (Fig. 1C). Consistent with these findings, tyrosyl residues within the BIT (Y436 and Y477) and PIR-B (Y794) cytoplasmic domains conform to the ITIM consensus (6, 7). ITIMs, which consist of I/V-X-pY-X-X-L/V/I, are binding sites for SHP-1, SHP-2, and the inositol monophosphatase SHIP (for reviews, see references 5, 8, 49, and 55). Preliminary studies indicate that PIR-B also binds to SHIP (9a); similar studies are under way for BIT. The reason why the SHP-1–BIT (SHP-1–PIR-B) and SHP-2–BIT (SHP-2–PIR-B) complexes are distinct could be that the two SHPs compete for the same binding site(s). Alternatively, the two SHPs may bind to distinct sites on BIT and/or PIR-B. In that case, each SHP could, in principle, dephosphorylate the other’s binding site(s), resulting in mutually exclusive complexes. This model would be consistent with previous reports indicating that SHPS-1/SIRPα1 is an SHP-2 substrate in nonhematopoietic cells, together with our demonstration that BIT is an SHP-1 substrate in macrophages (Fig. 4 and 5). Mapping of the binding sites for the SHPs on BIT and PIR-B and tryptic phosphopeptide analysis of BIT and PIR-B in normal and me/me BMM should resolve these issues.
The physiological functions of BIT and PIR-B complexes remain to be determined. Other ITIM-containing proteins (e.g., CD22 and KIR) deliver inhibitory signals to multichain immune recognition receptors (for reviews, see references 5, 11, 36, 49, and 56). Studies with SHP-1-deficient DT40 B cells strongly suggest that KIR-mediated inhibition requires SHP-1 (41). By analogy, SIRPs also may play an inhibitory function. Overexpression of SHPS-1/SIRPα1 was reported to inhibit growth factor-induced mitogenic signaling in epithelial and fibroblastic cell lines (26). Since fibroblasts lack SHP-1 (34, 44, 60), SIRP-mediated inhibition would have to be mediated by SHP-2 or another SIRP-binding protein. However, it remains unclear whether these overexpression studies reflect a bona fide physiological function for this SIRP (or SIRPs in general) or a gratuitous effect of the overexpressed protein. Indeed, our data suggest that SIRPs may compete with other SHP-binding proteins (e.g., P97/P100 [9, 20, 55b]).
Nevertheless, several lines of evidence suggest that the BIT–SHP-1 complex regulates CSF-1R signaling, possibly by targeting SHP-1 to the CSF-1R. BMM lacking SHP-1 are hypersensitive to CSF-1 and display aberrant CSF-1R tyrosyl phosphorylation (10). The CSF-1R associates constitutively with BIT (Fig. 3B). However, it is difficult to detect tyrosyl-phosphorylated BIT in CSF-1R immunoprecipitates. This could reflect greater sensitivity of anti-SHPS-1 antibodies compared with anti-pTyr antibodies, but it is more likely that most of the BIT associated with the CSF-1R (especially prior to stimulation) is not tyrosyl phosphorylated. This is consistent with the small percentage of BIT that is tyrosyl phosphorylated (Fig. 2B), whereas the BIT signal in anti-CSF-1R immunoprecipitates (which do not immunodeplete CSF-1R under our conditions) is quite strong (Fig. 3B). Unphosphorylated, CSF-1R-associated BIT would not be able to bind SHP-1; thus, SHP-1 would not be expected to regulate basal (i.e., unstimulated) CSF-1R tyrosyl phosphorylation, in agreement with our previous studies (10). Upon CSF-1 stimulation, we expect that CSF-1R-associated BIT becomes tyrosyl phosphorylated, either directly by the CSF-1R or by a CSF-1R-activated PTK. This leads to recruitment of SHP-1 to the complex and CSF1-R dephosphorylation. Consistent with this model, a band with a size identical to the CSF-1R is detected in both BIT (Fig. 3A) and SHP-1 (Fig. 1) immunoprecipitates only upon CSF-1 stimulation. The kinetics of this transient association are consistent with the rapid dephosphorylation of the CSF-1R normally observed upon CSF-1 stimulation (Fig. 1A and 3A, reference 10, and data not shown). Consistent with the predictions of this model, CSF1-stimulated CSF-1R tyrosyl phosphorylation is increased and sustained in me/me BMM (10). Even in the complete absence of SHP-1, the CSF-1R is tyrosyl dephosphorylated eventually. This could be due to the action of BIT-bound SHP-2 (Fig. 1C), since overexpression of SHPS-1/SIRPα1 in fibroblastic cells (which lack SHP-1) antagonizes transformation by v-fms, the viral oncogene derived from the CSF-1R (26). However, if SHP-2 subserves this function, the distinct hematopoietic phenotypes of mice lacking SHP-1 (me/me mice) (reviewed in references 3 and 37) and embryonal stem cells expressing defective SHP-2 (markedly diminished hematopoiesis) (45) argue that SHP-1–BIT and SHP-2–BIT complexes are biologically nonequivalent. Our failure to detect CSF-1R–PIR-B complexes argues for distinct functions for PIR-B–SHP-1 and BIT–SHP-1 complexes. Given the similarity of PIR-B to KIR, gp49B1, and FcαR, an intriguing possibility is that BIT regulates receptor tyrosine kinase signaling, whereas PIR-B may negatively regulate other receptor signaling systems (e.g., macrophage multichain immune recognition receptors such as Fc receptors).
Several lines of evidence clearly establish BIT as an SHP-1 substrate. SHP-1 dephosphorylates BIT in vitro. BIT binds to substrate-trapping mutants of SHP-1 in vitro and is preferentially eluted by the active site competitor vanadate (Fig. 4B). Compelling evidence that BIT is an in vivo target of SHP-1 is provided by our finding that trapping mutants of SHP-1 bind substantially more tyrosyl-phosphorylated BIT than nontrapping mutants (Fig. 4C to E). Notably, BIT is more tyrosyl phosphorylated in cells expressing nontrapping mutants of SHP-1 than in empty virus-infected cells (Fig. 4D and E). Presumably, the increased concentration of the SHP-1 SH2 domains, which bind to tyrosyl-phosphorylated BIT (10), increases BIT phosphorylation by protecting these sites from other PTPs, including the vaccinia virus protein VH-1. This SH2 protection is distinct from active site PTP domain protection and presents a special problem for examining potential targets of SHPs. Our results show that only when trapping and nontrapping mutants are compared can increased tyrosyl phosphorylation of a potential SHP-1 target be interpreted confidently. Consistent with the effects of nontrapping mutants of SHP-1 in BAC1.2F5 cells, BIT is hyperphosphorylated in meν/meν mice (Fig. 5A). The effect of the SHP-1 mutants on PIR-B tyrosyl phosphorylation could not be evaluated in BAC1.2F5 cells, which lack significant PIR-B expression, and we have found it difficult to infect primary BMM with vaccinia viruses. Nevertheless, the hyperphosphorylation of PIR-B in meν/meν BMM (Fig. 5A) suggests that PIR-B is likely to be an SHP-1 substrate.
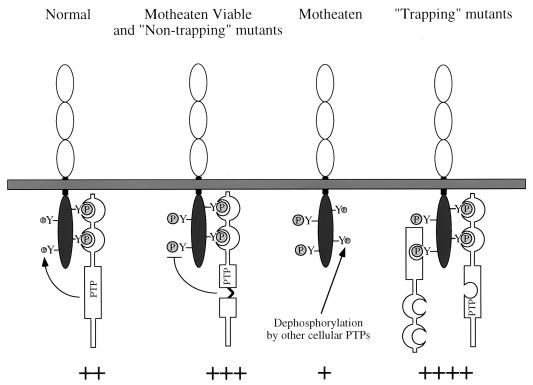
An apparent paradox was posed by the finding that levels of BIT and PIR-B tyrosyl phosphorylation are decreased in me/me BMM (Fig. 5B). We suspect that the explanation for this paradox is that the sites on BIT and PIR-B bound by SHP-1 are distinct from those that it dephosphorylates (Fig. 7). In normal BMM, SHP-1 is bound to these proteins via its SH2 domains, while its PTP domain dephosphorylates the other sites. Tyrosyl-phosphorylated BIT (and PIR-B) in such cells represents BIT (PIR-B) phosphorylated mainly on its SHP-1 binding sites. In meν/meν BMM, or in the presence of nontrapping SHP-1 mutants, the sites to which the SH2 domains of SHP-1 bind are still occupied and thus protected from dephosphorylation (SH2 protection), while the other tyrosines accumulate phosphate because they cannot be dephosphorylated by defective SHP-1. In me/me mice, however, sites normally occupied by the SHP-1 SH2 domains are now accessible to dephosphorylation by other PTPs. Additional phosphate may accumulate on the site(s) normally dephosphorylated by SHP-1, but there still could be a net decrease in overall BIT/PIR-B phosphorylation if, under normal conditions, the stoichiometry of phosphorylation of the sites bound by the SH2 domains is higher than those normally targeted by the catalytic domain (Fig. 7). Notably, BIT (PIR-B) tyrosyl phosphorylation in cells expressing trapping mutants of SHP-1 might be expected to be even higher than in meν/meν BMM, due to an additional PTP domain protection of sites normally targeted by SHP-1 (Fig. 7). This model argues that SHP-1 has preference not only for specific phosphotyrosyl proteins but for specific phosphorylation sites on these proteins. Moreover, it implies that multiple PTPs target BIT and PIR-B. Proof of this hypothesis will require phosphotryptic mapping studies and identification of other PTPs that target BIT and PIR-B. Notably, however, we have shown that SHP-2 associates with both of these proteins. In addition, recent experiments indicate that BIT and, to a lesser extent, PIR-B are hyperphosphorylated in CD45-deficient BMM (55b), raising the possibility that they are substrates for this receptor PTP as well.
FIG. 7.
Model of BIT and PIR-B phosphorylation in normal, meν/mev, and me/me BMM. In normal cells, SHP-1 can dephosphorylate certain sites while affording other sites protection through its SH2 domains. In meν/meν cells, SHP-1 still is capable of protecting sites via its SH2 domains but is unable to dephosphorylate substrate sites. Thus, BIT and PIR-B will be hyperphosphorylated in motheaten viable (+++) compared to normal (++) cells. In this way, the PTP-inactive nontrapping mutants of SHP-1 would resemble the meν/meν forms of SHP-1. In me/me cells, there is no SH2 domain protection, and these sites are dephosphorylated by other cellular PTPs. Although substrate sites would be hyperphosphorylated as in meν/meν cells, the SH2 domain-binding sites would be less phosphorylated than normal, resulting in an overall decrease in BIT and/or PIR-B tyrosyl phosphorylation (+). BIT and PIR-B tyrosyl phosphorylation in cells expressing trapping mutants of SHP-1 might be expected to be even higher than in meν/meν BMM (++++), due to an additional PTP domain protection of sites normally targeted by SHP-1.
Together, our results suggest that BIT and PIR-B function, at least in part, to target SHP-1 (and potentially SHP-2 and SHIP) to various membrane receptor complexes, but they probably have a more complicated role in macrophage signal transduction. Likewise, although the ability of SHP-1 to dephosphorylate BIT and PIR-B might be part of an autoregulatory and/or homeostatic mechanism to control the level of SHP-1 associated with these proteins, it seems more likely that SHP-1-mediated dephosphorylation has the more subtle and sophisticated function of controlling signaling complexes assembled on BIT and PIR-B. Such signaling complexes may include P55 and P120 and/or the associated PTK activity. Since tyrosyl phosphorylation of these proteins also is enhanced (and the associated PTK activity is increased) in me/me BMM, they may be substrates of SHP-1 and/or be phosphorylated by PTKs that are SHP-1 substrates. Further work will be required to identify these proteins, their mechanism of association with BIT and PIR-B, their biological function, and their importance in regulating the normal physiological functions that are disrupted in me/me mice.
ACKNOWLEDGMENTS
We thank Alana O’Reilly and Sue Miller for helpful comments on the manuscript, and we thank Susan Cohen for secretarial assistance.
This work was supported by National Institutes of Health grants PO1-DK50654 and R01 CA49152 (to B.G.N.) and R01-CA40987 (to L.R.R.). H.G. was supported by NRSA grant CA72144 and M.J.S.N. was supported by NRSA grant CA65048 from the National Institutes of Health. J.F.T. is a Fellow of the Leukemia Society of America.
REFERENCES
- 1.Adachi M, Fisher E H, Ihle J, Imai K, Jirik F, Neel B, Pawson T, Shen S-H, Thomas M, Ullrich A, Zhao Z. Mammalian SH2-containing protein tyrosine phosphatases. Cell. 1996;85:15. doi: 10.1016/s0092-8674(00)81077-6. [DOI] [PubMed] [Google Scholar]
- 2.Barford D, Flint A J, Tonks N K. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 3.Bignon J S, Siminovitch K A. Identification of PTP1C mutation as a genetic defect in motheaten and viable motheaten mice: a step toward defining the roles of protein tyrosine phosphatases in the regulation of hemopoietic cell differentiation and function. Clin Immunol Immunopathol. 1994;73:168–179. doi: 10.1006/clin.1994.1185. [DOI] [PubMed] [Google Scholar]
- 4.Brown M, McCormack M, Zinn K G, Farrell M P, Bikel I, Livingston D M. A recombinant murine retrovirus for simian virus 40 large T cDNA transforms mouse fibroblasts to anchorage-independent growth. J Virol. 1986;60:290–293. doi: 10.1128/jvi.60.1.290-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burshtyn, D. N., and E. O. Long. Regulation through inhibitory receptors: lessons from natural killer cells. Trends Cell Biol., in press. [DOI] [PubMed]
- 6.Burshtyn D N, Scharenberg A M, Wagtmann N, Rajagopalan S, Berrada K, Yi T, Kinet J-P, Long E O. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitory receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burshtyn D N, Yang W, Yi T, Long E O. A novel phosphotyrosine motif with a critical amino acid at position -2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 8.Cambier J C. Inhibitory receptors abound? Proc Natl Acad Sci USA. 1997;94:5993–5995. doi: 10.1073/pnas.94.12.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlberg K, Rohrschneider L R. Characterization of a novel tyrosine phosphorylated 100 kD protein that binds to tyrosyl phosphorylated SHP-2 and the 85 kDa subunit of PI3′-kinase in response to M-CSF and other cytokines. J Biol Chem. 1997;272:15943–15940. doi: 10.1074/jbc.272.25.15943. [DOI] [PubMed] [Google Scholar]
- 9a.Carlberg, K., and L. Rohrschneider. Unpublished data.
- 10.Chen H E, Chang S, Trub T, Neel B G. Regulation of colony-stimulating factor 1 receptor signaling in murine macrophages by the SH2-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;16:3685–3697. doi: 10.1128/mcb.16.7.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cyster J G, Goodnow C C. Tuning antigen receptor signaling by CD22: integrating cues from antigens and the microenvironment. Immunity. 1997;6:509–517. doi: 10.1016/s1074-7613(00)80339-8. [DOI] [PubMed] [Google Scholar]
- 12.David M, Chen H E, Goelz S, Larner A C, Neel B G. Differential regulation of the alpha/beta interferon-stimulated Jak/Stat pathway by the SH2 domain-containing tyrosine phosphatase SHPTP1. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denu J E, Stuckey J A, Saper M A, Dixon J E. Form and function in protein dephosphorylation. Cell. 1996;87:361–364. doi: 10.1016/s0092-8674(00)81356-2. [DOI] [PubMed] [Google Scholar]
- 14.Feng G S, Pawson T. Phosphotyrosine phosphatases with SH2 domains: regulators of signal transduction. Trends Genet. 1994;10:54–58. doi: 10.1016/0168-9525(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 15.Flint A J, Tiganis T, Barford D, Tonks N K. Development of “substrate trapping” mutants to identify physiological substrates of protein-tyrosine phosphatases. Proc Natl Acad Sci USA. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman R M, Jr, Plutzky J, Neel B G. Identification of a human src-homology 2 (SH2) containing tyrosine phosphatase: a putative homolog of Drosophila corkscrew. Proc Natl Acad Sci USA. 1992;89:11239–11243. doi: 10.1073/pnas.89.23.11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry A M, Lanier L L, Weiss A. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med. 1996;184:295–300. doi: 10.1084/jem.184.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, Kasuga M. A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garton A J, Flint A J, Tonks N K. Identification of p130casas a substrate for the cytosolic protein tyrosine phosphatase PTP-PEST. Mol Cell Biol. 1996;16:6408–6418. doi: 10.1128/mcb.16.11.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu H, Griffin J D, Neel B G. Characterization of two SHP-2-associated binding proteins and potential substrates in hematopoietic cells. J Biol Chem. 1997;272:16421–16430. doi: 10.1074/jbc.272.26.16421. [DOI] [PubMed] [Google Scholar]
- 21.Guan K, Broyles S S, Dixon J E. A tyr/ser protein phosphatase encoded by vaccinia virus. Nature. 1991;350:359–362. doi: 10.1038/350359a0. [DOI] [PubMed] [Google Scholar]
- 22.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohomori H, Takai T. Molecular cloning of a novel murine cell-surface glycoprotein homologous to killer cell inhibitory receptors. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 23.Herbst R, Carroll P M, Allard J D, Schilling J, Raabe T, Simon M A. Daughter of Sevenless is a substrate of the phosphotyrosine phosphatase corkscrew and functions during Sevenless signaling. Cell. 1996;85:899–909. doi: 10.1016/s0092-8674(00)81273-8. [DOI] [PubMed] [Google Scholar]
- 24.Huyer G, Liu S, Kelly J, Moffat J, Payette P, Kennedy B, Tsaprailis G, Gresser M J, Ramachandran C. Mechanism of inhibition of protein-tyrosine phosphatases by vanadate and pervanadate. J Biol Chem. 1997;272:843–851. doi: 10.1074/jbc.272.2.843. [DOI] [PubMed] [Google Scholar]
- 25.Jia Z, Barford D, Flint A J, Tonks N K. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 26.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. A family of proteins that inhibit signaling though tyrosine kinase receptors. Nature. 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 27.Kozlowski M, Mlinaric-Rascan I, Feng G S, Shen R, Pawson T, Siminovitch K A. Expression and catalytic activity of the tyrosine phosphatase PTP1C is severely impaired in motheaten and viable motheaten mice. J Exp Med. 1993;178:2157–2163. doi: 10.1084/jem.178.6.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubagawa H, Burrows P, Cooper M D. A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lioubin M N, Algate P A, Tsai S, Carlberg K, Aebersold R, Rohrschneider L R. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 30.Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenz U, Ravichandran K S, Pei D, Walsh C T, Burakoff S J, Neel B G. Lck-dependent tyrosyl phosphorylation of the phosphotyosine phosphatase SH-PTP1 in murine T cells. Mol Cell Biol. 1994;14:1824–1834. doi: 10.1128/mcb.14.3.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Marengere L E M, Waterhouse P, Duncan G S, Mittrucker H-W, Feng G-S, Mak T W. Regulation of T cell receptor signaling by tyrosine phosphatase SYP association with CTLA-4. Science. 1996;272:1170–1173. doi: 10.1126/science.272.5265.1170. [DOI] [PubMed] [Google Scholar]
- 34.Matthews R J, Bowne D B, Flores E, Thomas M L. Characterization of hematopoietic intracellular protein tyrosine phosphatases: description of a phosphatase containing an SH2 domain and another enriched in proline-, glutamic acid-, serine-, and threonine-rich sequences. Mol Cell Biol. 1992;12:2396–2405. doi: 10.1128/mcb.12.5.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan C, Pollard J W, Stanley E R. Isolation and characterization of a cloned growth factor-dependent macrophage cell line, BAC1.2F5. J Cell Physiol. 1987;130:420–427. doi: 10.1002/jcp.1041300316. [DOI] [PubMed] [Google Scholar]
- 35a.Nadler, M. J. S., J. F. Timms, K. Carlberg, L. Rohrschneider, and B. G. Neel. Unpublished data.
- 36.Neel B G. Role of phosphatases in lymphocyte activation. Curr Opin Immunol. 1997;9:405–420. doi: 10.1016/s0952-7915(97)80088-x. [DOI] [PubMed] [Google Scholar]
- 37.Neel B G. Structure and function of SH2-domain containing tyrosine phosphatases. Semin Cell Biol. 1993;4:419–432. doi: 10.1006/scel.1993.1050. [DOI] [PubMed] [Google Scholar]
- 38.Neel B G, Tonks N K. Protein tyrosine phosphatases in signal transduction. Curr Opin Cell Biol. 1997;9:193–204. doi: 10.1016/s0955-0674(97)80063-4. [DOI] [PubMed] [Google Scholar]
- 39.Noguchi T, Matozaki T, Fujioka Y, Yamao T, Tsuda M, Takada T, Kasuga M. Characterization of a 115-kDa protein that binds to SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in Chinese hamster ovary cells. J Biol Chem. 1996;271:27652–27658. doi: 10.1074/jbc.271.44.27652. [DOI] [PubMed] [Google Scholar]
- 40.Olcese L, Lang P, Vely F, Cambiaggi A, Marguet D, Blery M, Hippen K L, Biassoni R, Moretta A, Moretta L, Cambier J, Vivier E. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTP1D protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 41.Ono M, Okada H, Bolland S, Yanagi S, Kurosaki T, Ravetch J V. Deletion of SHIP or SHP-1 reveals two distinct pathways for inhibitory signaling. Cell. 1997;90:293–301. doi: 10.1016/s0092-8674(00)80337-2. [DOI] [PubMed] [Google Scholar]
- 42.Pei D, Lorenz U, Klingmuller U, Neel B G, Walsh C T. Intramolecular regulation of protein tyrosine phosphatase SH-PTP1: a new function for Src homology 2 domains. Biochemistry. 1994;33:15483–15493. doi: 10.1021/bi00255a030. [DOI] [PubMed] [Google Scholar]
- 43.Perkins L A, Johnson M R, Melnick M B, Perrimon N. The non-receptor protein tyrosine phosphatase Corkscrew functions in multiple receptor tyrosine kinase pathways in Drosophila. Dev Biol. 1996;180:63–81. doi: 10.1006/dbio.1996.0285. [DOI] [PubMed] [Google Scholar]
- 44.Plutzky J, Neel B G, Rosenberg R D. Isolation of a novel SRC homology 2 (SH2) containing tyrosine phosphatase. Proc Natl Acad Sci USA. 1992;89:1123–1127. doi: 10.1073/pnas.89.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qu C K, Shi Z Q, Shen R, Tsai F Y, Orkin S H, Feng G S. A deletion mutation in the SH2-N domain of SHP-2 severely suppresses hematopoietic cell development. Mol Cell Biol. 1997;17:5499–5507. doi: 10.1128/mcb.17.9.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raabe T, Riesgo-Escovar J, Liu X, Bausenwein B S, Deak P, Maroy P, Hafen E. DOS, a novel pleckstrin homology domain-containing protein required for signal transduction between sevenless and Ras1 in Drosophila. Cell. 1996;85:911–920. doi: 10.1016/s0092-8674(00)81274-x. [DOI] [PubMed] [Google Scholar]
- 47.Rotin D, Margolis B, Mohammadi M, Daly R, Daum G, Li N, Fischer E, Burgess W, Ullrich A, Schlessinger J. SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C-g. EMBO J. 1992;11:559–567. doi: 10.1002/j.1460-2075.1992.tb05087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sano S, Ohnishi H, Omori A, Hasegawa J, Kubota M. BIT, an immune antigen receptor-like molecule in the brain. FEBS Lett. 1997;411:327–334. doi: 10.1016/s0014-5793(97)00724-2. [DOI] [PubMed] [Google Scholar]
- 49.Scharenberg A, Kinet J. The emerging field of receptor-mediated inhibitory signaling: SHP or SHIP? Cell. 1996;87:961–964. doi: 10.1016/s0092-8674(00)81790-0. [DOI] [PubMed] [Google Scholar]
- 50.Shultz L D, Schweitzer P A, Rajan T V, Yi T, Ihle J N, Matthews R J, Thomas M L, Beier D R. Mutations at the murine motheatenlocus are within the hematopoietic cell protein phosphatase (HCPH) gene. Cell. 1993;73:1445–1454. doi: 10.1016/0092-8674(93)90369-2. [DOI] [PubMed] [Google Scholar]
- 51.Stanley E R. Action of the colony stimulating factor, CSF-1. Ciba Found Symp. 1986;118:29–41. doi: 10.1002/9780470720998.ch3. [DOI] [PubMed] [Google Scholar]
- 52.Streuli M. Protein tyrosine phosphatases in signaling. Curr Opin Cell Biol. 1996;8:182–188. doi: 10.1016/s0955-0674(96)80064-0. [DOI] [PubMed] [Google Scholar]
- 53.Stuckey J A, Schubert H L, Fauman E B, Zhang Z Y, Dixon J E, Saper M A. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature. 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- 54.Symes A, Stahl N, Reeves S A, Farruggella T, Servidel T, Gearan T, Yancopoulos G, Fink J S. The protein tyrosine phosphatase SHP-2 negatively regulates ciliary neurotrophic factor induction of gene expression. Curr Biol. 1997;7:687–700. doi: 10.1016/s0960-9822(06)00298-3. [DOI] [PubMed] [Google Scholar]
- 55.Thomas M L. Of ITAMs and ITIMs: turning on and off the B cell antigen receptor. J Exp Med. 1995;181:1953–1956. doi: 10.1084/jem.181.6.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Timms, J. F., H. Chen, and B. G. Neel. Unpublished data.
- 55b.Timms, J. F., and B. G. Neel. Unpublished data.
- 56.Tonks N K, Neel B G. From form to function: signaling by protein tyrosine phosphatases. Cell. 1996;87:365–368. doi: 10.1016/s0092-8674(00)81357-4. [DOI] [PubMed] [Google Scholar]
- 57.Tsai S, Bartelmez S, Sitnicka E, Collins S. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 1994;8:2831–2841. doi: 10.1101/gad.8.23.2831. [DOI] [PubMed] [Google Scholar]
- 58.Tsui H W, Siminovitch K A, deSouza L, Tsui F W L. Motheaten and viable motheatenmice have mutations in the haematopoietic cell phosphatase gene. Nat Genet. 1993;4:124–129. doi: 10.1038/ng0693-124. [DOI] [PubMed] [Google Scholar]
- 59.Yeung Y, Berg K L, Pixley F J, Angeletti R H, Stanley E R. Protein tyrosine phosphatase-1C is rapidly phosphorylated in tyrosine in macrophages in response to colony stimulating factor-1. J Biol Chem. 1992;267:23447–23450. [PubMed] [Google Scholar]
- 60.Yi T, Cleveland J L, Ihle J N. Protein tyrosine phosphatase containing SH2 domains: characterization, preferential expression in hematopoietic cells, and localization to human chromosome 12p12-13. Mol Cell Biol. 1992;12:836–846. doi: 10.1128/mcb.12.2.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yi T, Ihle J N. Association of hematopoietic cell phosphatase with c-Kit after stimulation with c-Kit ligand. Mol Cell Biol. 1993;13:3350–3358. doi: 10.1128/mcb.13.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]