Summary
Primary open-angle glaucoma (POAG), a leading cause of irreversible blindness globally, shows disparity in prevalence and manifestations across ancestries. We perform meta-analysis across 15 biobanks (of the Global Biobank Meta-analysis Initiative) (n = 1,487,441: cases = 26,848) and merge with previous multi-ancestry studies, with the combined dataset representing the largest and most diverse POAG study to date (n = 1,478,037: cases = 46,325) and identify 17 novel significant loci, 5 of which were ancestry specific. Gene-enrichment and transcriptome-wide association analyses implicate vascular and cancer genes, a fifth of which are primary ciliary related. We perform an extensive statistical analysis of SIX6 and CDKN2B-AS1 loci in human GTEx data and across large electronic health records showing interaction between SIX6 gene and causal variants in the chr9p21.3 locus, with expression effect on CDKN2A/B. Our results suggest that some POAG risk variants may be ancestry specific, sex specific, or both, and support the contribution of genes involved in programmed cell death in POAG pathogenesis.
Keywords: glaucoma, genetics, GWAS, disease disparity, transcriptomic, genetic interactions, cross-ancestry, disease prediction, PRS, pleiotropy
Graphical abstract

Highlights
-
•
GWAS of diverse POAG data identify novel loci including sex/ancestry-specific signals
-
•
PRS for POAG have differential ancestry-specific pattern of associations in EHR
-
•
Unique inter-loci interactions contribute to disease etiology
-
•
Results point to contribution of programmed cell death genes in pathogenesis of POAG
Lo Faro et al. perform the largest and most diverse POAG GWAS to date and identify novel sex- and ancestry-specific associations. They further perform extensive statistical analysis implicating vascular and cancer-related genes in disease etiology.
Introduction
Glaucoma is a complex eye disease characterized by a progressive loss of optic nerve (ON) fibers, which manifests initially as visual field loss, and if untreated ultimately leads to irreversible blindness.1 Primary open-angle glaucoma (POAG) represents the most prevalent type of glaucoma. POAG affects the trabecular meshwork (TM), the inner retina, where retinal ganglion cell axons form the ON, and, presumably due to transsynaptic degeneration, the visual pathways, including the visual cortex.2,3 High intraocular pressure (IOP) is a major risk factor identified in POAG patients.4 Other risk factors are advanced age and positive family history, and difference in prevalence has been shown for POAG across ethnicities and sex.5,6,7 Moreover, there is disparity in POAG clinical presentations and outcome across ancestries.8,9
Through genome-wide association studies (GWASs), significant progress has been made in understanding the genetic pathophysiology of glaucoma in humans. In addition, animal models have also provided a valuable resource for understanding the biological mechanisms.10 However, there is still a lack of understanding of the underlying pathologic mechanisms in POAG, limiting the development of specific interventions in patients.
Several studies have found that POAG is associated with a variety of cardiovascular diseases and vascular risk factors.11,12,13,14,15,16,17,18,19,20,21 In a previous large multi-ancestry GWAS,22 results from gene-enrichment analysis have implicated perturbation of molecular mechanisms in the vascular system that contribute to blood vessel morphogenesis, vasculature development, and regulation of endothelial cell proliferation. Analysis in large electronic health records (EHRs), coupled with a zebrafish model system, showed association of reduced genetically predicted expression of a gene that encodes for glutamate receptor GRIK5, which potentially determines blood vessel numbers, integrity in the eye, and increased vascular permeability, with comorbid vascular and eye diseases.23 However, no study has previously performed detailed exploration of the genetics that underlies the potential vascular connection with POAG across ancestries. Large population-based and clinical-based biobanks offer an opportunity to do large-scale in silico investigations to elucidate common molecular systemic pathways between POAG and vascular systems. Furthermore, knowledge gained from large biobanks and combined with eQTL human data from the GTEx project (https://gtexportal.org/) allow a deeper exploration on how the interaction occurs in humans between the genes identified in GWAS, such as SIX6 and CDKN2A/B.24,25
The Global Biobank Meta-analysis Initiative (GBMI) is a collaborative group that currently involves 19 biobanks from 12 countries spanning 4 different continents: North America (Canada, USA), East Asia (China, Japan), Europe (Iceland, Estonia, Finland, the Netherlands, Norway, Scotland, UK), and Australia. Each GBMI-affiliated biobank has paired genetic and phenotypic data collated through different types of electronic health data, such as self-report data from questionnaires, billing codes, doctors’ narrative notes, and death registry for >2.1 million individuals representing diverse ancestries: African ancestry individuals, Admixed Americans, Central or South Asians, East Asians, Europeans, and Middle Eastern. A detailed description of each biobank is found in Zhou et al.26
In this study, we conducted a large-scale meta-analysis of POAG GWAS in 15 GBMI biobanks from 6 ancestries (n = 1,487,441). Then, we merged our data with two previously published GWASs, and the combined dataset represents the largest and most diverse POAG study to date. We then leveraged sophisticated statistical methods to identify unique molecular actors across ancestries.
Results
Discovery of novel ancestry-specific POAG loci
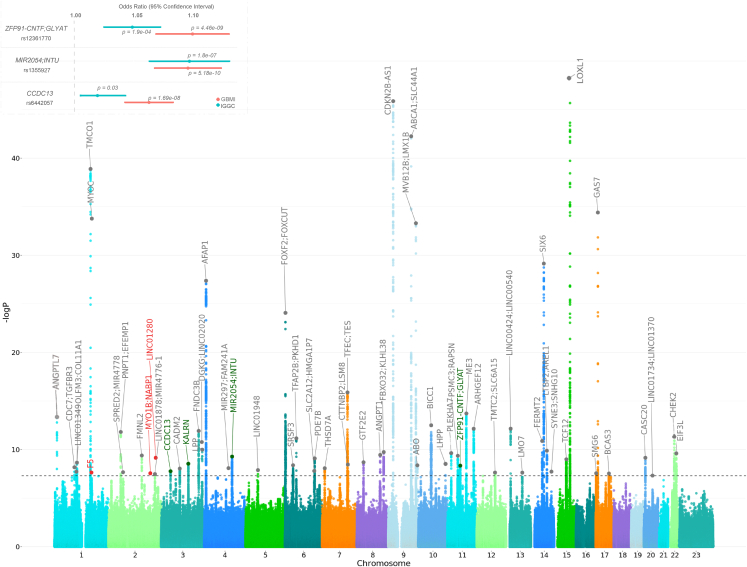
We report here a multi-ancestry genome-wide association meta-analysis study of 26,848 POAG cases and 1,460,593 controls from 15 GBMI biobanks. A total of 62 loci that reach the genome-wide significant threshold were identified (Figures 2, S6, S7, and S8; Table S3). Five of these loci were novel, and encompass the genes F5, RPL37A-LINC01280, ZFP91-CNTF-GLYAT, CCDC13, and MIR2054-INTU (Table 1). Of these five loci, the latter three and only ZFP91-CNTF-GLYAT were observed at subgenome-wide significance levels in IGGC (International Glaucoma Genetics Consortium) multi-ethnic and GGLAD (Genetics of Glaucoma in People of African Descent) summary data, respectively (Figure 2, correlation between the effect sizes was high for IGGC r = 0.71, and low for GGLAD r = 0.12).22,27 ZFP91-CNTF-GLYAT variant is within the same locus of rs574982531_OR10W1-OR5B17, identified in females in Gharakhrani et al., in sex-stratified analysis, whose sentinel SNP is ∼133 kb upstream.22 A locus defined here as ANGPTL7-MTOR encompasses previously reported rare variants and corresponds to rs143038218_UBIAD1 locus.28,29 In addition, a Bayesian cross-ancestry meta-analysis using Meta-Regression of Multi-AncEstry Genetic Association of GBMI data, identified two additional loci, one of which is a novel locus specific to African ancestry (rs77136907_MYO1B;NABP1, p = 2.74e−08; inverse-variance meta-analysis p = 2.99e−07) (Table S7).26,30 Three previously identified loci, rs11024102_PLEKHA7, rs58812088_ FNDC3B, and rs3825942_LOXL1, might be due to other glaucoma subtypes (Table S7). Therefore, we cannot rule out potential effects of non-POAG glaucoma for these signals i.e., especially due to primary angle closure glaucoma (PACG) in East Asian populations, where this glaucoma subtype is more common,6,31,32,33 or from biobanks where phenotyping was based on self-reporting (Table S1). However, some of the loci are potentially pleiotropic for POAG and PACG/exfoliation syndrome (XFS). For example, rs993471_COL11A1, previously reported in Choquet and co-workers, attained genome-wide statistical significance in combined meta-analysis and is associated with PACG.34,35 While variants in LTBP2 are reported in the IGGC-POAG GWAS22 and independently in patients with POAG and in patients with XFS using candidate gene sequencing analysis.36
Figure 2.
Genome-wide association study of POAG: Manhattan plot and effect size comparison of novel variants in GBMI and IGGC
The x axis is the position on each chromosome and the y axis is the –log10 p value from the GWAS for each SNP. The black line demarcates the threshold for genome-wide significance (p = 5.0e−8). The four replicated novel regions and the nearest genes that reach the threshold for significance are indicated in green, the potentially novel regions and the nearest genes are indicated in red. Previously reported genes are in gray. Details of all genome-wide significant signals are in Table S3. Inset: the effect size comparison of significant novel single nucleotide polymorphism in GBMI and. p indicates the p value of each variant.
Table 1.
Novel loci significantly associated with POAG (p < 5.0e−8) in the GBMI
| Chr | Bp | Effect allele | Non-effect allele | SNP | Gene | Function | No. of cases | No. of controls | MAF | Beta | SE | p | Hp |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 191,664,446 | C | G | rs77136907 | MYO1B; NABP1 | intergenic | 3,134 | 641,377 | 0.001 | 0.58 | 0.13 | 2.74e−08 | 0.00004 |
| 2 | 216,590,253 | T | C | rs12476634 | LINC01280; RPL37A | ncRNA, exonic | 2,538 | 484,482 | 0.001 | 1.04 | 0.16 | 6.91e−10 | 0.03 |
| 3 | 42,753,558 | A | G | rs6442057 | CCDC13 | intronic | 26,848 | 1,460,599 | 0.26 | 0.06 | 0.01 | 1.69e−08 | 0.7 |
| 4 | 127,549,411 | T | C | rs1355927 | MIR2054; INTU | intergenic | 26,848 | 1,460,599 | 0.07 | 0.09 | 0.01 | 5.18e−10 | 0.1 |
| 11 | 58,666,759 | C | A | rs12361770 | ZFP91-CNTF; GLYAT | intergenic | 17,789 | 1,213,831 | 0.15 | 0.09 | 0.01 | 4.46e−09 | 0.4 |
Chr, chromosome; Bp, base pair position (hg38); SNP, single-nucleotide polymorphism; MAF, minor allele frequency; SE, standard error; p, p value; Hp, heterogeneity p value.
We further performed meta-analysis of GBMI and public datasets from IGGC and GGLAD data, generating the largest and most diverse GWAS to date. We combined the European-specific IGGC meta-analysis data (GCST90011767), the GLADD meta-analysis (GCST009245), and the GBMI data in which we excluded FinnGen (a subset present in IGGC of European ancestry), BioME from African ancestry (a subset present in GGLAD), and UKBB-Africa (a subset present in GGLAD) cohorts. We refer to GBMI-IGGC-GGLAD for this meta-analysis (Table S1). Here, additional novel loci that encompass the following prioritized genes LOC654841, KBTBD8, ADGRL3, DDIT4L, INTU, HMGXB3, KCNK5, MAD1L1, APPL2-KCCAT198, CATSPERB, OR5B12-OR5B21, and FENDRR attained genome-wide significance (Table S7; Figures S11 and S12). Five of the genes in these novel loci are involved in cardiovascular conditions and six in cancer processes.37,38,39,40,41,42,43,44,45,46,47,48,49 In GBMI-IGGC-GGLAD meta-analysis, two GBMI novel loci, RPL37A-LINC01280 and MIR2054;INTU, maintained the genome-wide significance level (RPL37A-LINC01280 p = 4.16e−9; MIR2054;INTU p = 1.54e−9). While the other three GBMI novel loci, CCDC13, MYO1B;NABP1, and ZFP91-CNTF;GLYAT, were attenuated to subgenome-wide significance level (CCDC13 p = 6.20e−8; MYO1B;NABP1 p = 2.77e−6; ZFP91-CNTF;GLYAT p = 1.69e−6) (Tables S3 and S7). In GBMI-IGGC-GGLAD, where the GBMI_FinnGen cohort was excluded to avoid overlap, the lead SNP rs1469837390 in the F5 locus dropped out.
Identification of novel sex-specific loci
In the GBMI dataset, we performed sex-stratified analyses to identify any variants that demonstrated sex-specific association or effect size heterogeneity. We identified one low-frequency African-specific novel association in females rs116625313_PRKG2;RASGEF1B (females p = 2.85e−8, beta = 1.52 vs. males p = 0.35, beta = −0.59), (Table S4). In addition, four novel loci with POAG association specific to males were identified (Table S5) with low-frequency lead variants, three of which are African specific in the GBMI African ancestry cohorts (2%–6% in population frequencies): rs111739240_TMEM167B, rs17057880_MIR3142HG-ATP10B, and rs114598725_ARMC4, and one is male non-Finnish European specific, rs150385013_LINC02024-LOC105374060 (>0.1% population frequencies, Table S5).
We next searched for coding variants at associated loci that explain the association signal and therefore implicate a likely functional gene. The novel rs111739240_TMEM167B variant is in linkage disequilibrium (LD) with a proxy variant, rs17641032 (r2 = 0.7 in 1000 Genomes Project Africans) at 7%, 5%, and 3% frequencies in European, African, and East-Asian populations, respectively. However, the proxy rs17641032 variant is associated with POAG in African ancestry BioVU male subjects (African ancestry individual cases n = 69, p = 5.05e−4) but not in European ancestry subjects (European cases, n = 213, p = 0.892), confirming African-specific association of this locus. The proxy rs17641032 variant is a GTEx eQTL for TMEM167B and AMIGO1.25 In GTEx tissue with the largest sample size (n = 706), skeletal muscle, the proxy variant rs17641032 is associated with expression changes for three genes in both sexes (TMEM167B p = 0.00027, CELSR2 p = 0.0086, and AMPD2 p = 0.041; Table S8). However, only CELSR2 has male-specific expression changes at this variant (males p = 0.02, females p = 0.21) in GTEx skeletal muscle tissue (Figure S11). In addition, transcriptome-wide association study (TWAS)-PheWAS analysis revealed associations of the CELSR2 with traits/phenotypes within the endocrine/metabolic (lipid traits) and circulatory groups with hyperlipidemia and angina pectoris, respectively, as the top phenotype associations (Table S24).
In addition, 14 of the loci that have association signals in combined sex meta-analysis have significant albeit attenuated signals, only in either male (3 loci) or females (11 loci) in sex-stratified analysis (Table S4 and S5). Four of these loci (near CADM2, DGKG, KALRN, and ARHGEF12) show significant effect size differences between males and females based on the Cochran heterogeneity test (Tables S4 and S5; Figure S10). All four genes prioritized using TWAS that are near lead variants for the four loci that show significant heterogeneity between males and females (CADM2, DGKG, KALRN, and ARGHEF2) have also been shown to have sex-biased expression patterns.50,51
We examined differences in POAG risk between sex in BioVU participants who are ≥40 years old and self-identify as European Americans (EA) or African Americans (AA). In 12,755 POAG cases, in an overall total of 1,372,397 BioVU participants, there was higher odds of POAG in AA males relative to females (odds ratio [OR] = 1.15; 95% CI: 1.06–1.24; p = 3.85e−4) while the risk was marginally lower in males than females in EA (OR = 0.96; 95% CI: 0.92–1; p = 0.048).
Ancestry-specific loci and heterogeneity in cross-ancestry effect sizes
To evaluate consistency in the cross-ancestry effect sizes, we estimated the correlation between beta values of independent genome-wide significant SNPs between European, African, and Asian populations in the GBMI dataset. As expected, the highest correlation was between the European and Asian ancestry (Pearson correlation coefficient [r] = 0.87), while correlation between African with other continental populations was much lower, probably due to the small African samples in GBMI study: European and African ancestry (r = 0.34), and African and Asian ancestry (r = 0.25) (Figures S6, S7, and S8). However, three novel loci and two previously identified loci showed heterogeneity in effect sizes between ancestries and biobanks. We infer these variants to be ancestry specific. We define ancestry-specific variants using the following metrics: (1) the lead GWAS variant in the statistically significant locus and any other variants in LD is only observed in association with the trait in a particular ancestry, (2) where the LD pattern is complex, the LD variant is only observed in the ascertained ancestry as gleaned from gnomAD and 1000 genomes databases, (3) further, for variants that are found in very low frequencies in admixed populations, besides the original ascertained source population, the variant should be absent from presumed main ancestral continental populations for the admixed populations in 1000 genomes data.52 We therefore classified the following variants as ancestry specific: the novel rs12476634_LINC01280, rs77136907_MYO1B;NABP1 loci (African and Hispanic Americans), the novel rs1469837390_F5 (Europeans), the well-known rs74315329_MYOC (Europeans), and rs147660927_ANGPTL7-MTOR28 (Northern Europeans) (Tables S3 and S7).26 These loci that we classify as either northern European or African specific have similar patterns that are highly specific to the two ancestries described here.53,54 In fact, the lead variants in northern-European-specific loci (rs1469837390_F5 and rs147660927_ANGPTL7-MTOR) are only reported in Finnish and non-Finnish European individuals in gnomAD, while the lead variants in African-specific loci (rs12476634_LINC01280 and rs77136907_MYO1B;NABP1) are observed in this study and in the gnomAD/1000 genome databases in individuals of African and Hispanic Americans, who are generally African admixed.55,56,57 In addition, the five sex-stratified association signals reported were ancestry specific: ATPB10, TMEM167B and ARMC4, PRKG2;RASGEF1B (African specific), and LINC02024;LOC105374060 (European specific) (Tables S4 and S5). The five sex- and ancestry-specific loci reported show substantial difference in minor allele frequencies of the lead variants in biobanks tested (Tables S6). Post-hoc power analysis indicates that our study is powered to detect these sex-specific signals (supplemental information). Association with POAG for three of the five sex- and ancestry-specific loci identified in GBMI were further replicated in additional biobanks: ATPB10 (African specific, p = 0.028) in PMBB, PRKG2;RASGEF1B (African specific, p = 0.0336), and LINC02024;LOC105374060 (European specific, p = 0.00536) in All of US dataset (Tables S6). However, considering the small sample sizes in this study, further studies are necessary to confirm the role and effect of ancestry- and sex-specific variants identified here.
The well-known MYOC p.Gln368Ter variant, which hitherto had not been defined as ancestry specific, shows association in GBMI European ancestry biobanks only (the variant was not imputed in African and Asian ancestry biobanks).58 The variant is the only missense mutation among >100 coding variants in the MYOC gene so far identified as glaucoma causing that has been implicated in late-onset POAG,59,60,61,62,63,64,65,66 and shows evidence of founder effect in European ancestry individuals.59,67,68,69 In addition, >92% (289/314) carriers of this allele in the latest version of the gnomAD database are of European ancestry, and non-European populations that the allele has been reported in have a history of admixture with European populations. Furthermore, in 1000 genomes, this variant is present in Europeans and an individual of South Asian ancestry sampled in the UK.53,54 The variant has not been reported in continental African populations and those observed in AA individuals are probably due to admixture.53,54,70
Overall, the ancestry-specific loci and those that show heterogeneity between the ancestry might be driving most of the overall heterogeneity observed between the biobanks (Figure S1).26 Phenotypic heterogeneity might also be a contributing factor with all self-reporting biobanks clustering together (Figure S1) relative to those that used ICD codes to identify cases.
Enrichment analyses prioritize vascular and cell proliferation mechanisms
To identify the functional roles of POAG-associated variants and which tissues are mediating the genetic effects, we performed enrichment analyses with DEPICT using GBMI-IGGC-GGLAD summary statistics (see methods). DEPICT prioritized 101 co-regulated genes, of which 43 (43%) have been previously reported for POAG and 58 (57%) that were novel (Table S9). Nearly 50% (49 genes) of the genes identified here were reported to be associated with vascular traits including cardiac diseases and arterial stiffness measurement, while nearly a third (34 genes) were associated with cancer, including cutaneous melanoma, keratinocyte carcinoma, breast carcinoma, prostate carcinoma, and lung carcinoma (Table S9). The gene-enrichment analysis was performed using the statistically significant 110 biological pathways identified (p < 3.45e−6) (Table S10). Among the top and common enrichment signals were biological pathways involved in vascular-related blood vessel development/morphogenesis, angiogenesis, and epithelial cell proliferation (Table S10). In addition, several biological pathways crucial in cell development and proliferation, such as the IGF1 subnetwork, insulin-like growth factor binding, and Wnt signaling were significantly enriched (Table S10). Tissue/cell-type enrichment analysis prioritized 10 significant tissues and cell types, with musculoskeletal and cardiovascular systems being the most represented (Table S11).
TWAS analyses identify novel associations
TWAS conducted in the GBMI-IGGC-GGLAD meta-analysis summary statistics identified 18 gene-trait associations (p < 2.5e−6, Bonferroni correction for mean 20,000 genes tested) in the GTEx tissues chosen in this study (methods; Table S12). Most of these genes (16/18) showed associations in at least one other tissue among the additional 22 other GTEx tissues potentially relevant to ocular diseases (Tables S12–S15). The JTI model generated most genes and gene-trait associations among the three cis prediction models (Tables S14 and S15; Figure S13). However, most of the gene-trait associations in JTI overlap with what was obtained using PrediXcan and UTMOST models (Figure S13). Using the three cis models made it possible to have a comprehensive TWAS prioritization of gene-trait associations. For example, one additional gene-trait association (rs1704654_CCDC88C;CATSPERB) that transects novel loci identified in GBMI-IGGC-GGLAD was prioritized using PrediXcan and UTMOST models but not the most robust JTI (Table S15). TWAS results of EyeGEx retinal data using a modified version PrediXcan models’ pipeline used for GTEx data (STAR Methods)71 were largely consistent with those in GTEx tissues with all the five genes (GAS7, ACP2, CLIC5-ENPP5, and LAMTOR3) prioritized in retinal tissues as statistically significant signals (Table S13).
Overall, the 253 gene-trait associations across the EyeGEx retinal tissue and 23 GTEx tissues were estimated to define the 90% credible set at the locus via probabilistic fine-mapping. One hundred and sixteen of the gene-trait associations in the 90% credible set intersect with 57 out of the 109 GWAS loci identified. A total of 15 of these gene-trait associations were novel, 9 of which were unique to GBMI (Tables S14 and S15). One hundred and fifty-six gene-trait associations intersect 64 previously known loci, while 86 did not intersect with any of the genome-wide significant risk loci and correspond to 68 loci with subgenome-wide GWAS signals (TWAS “loci”) that will potentially attain significance in a more powered GWAS (Figure S13; Tables S14 and S15). For example, LAMTOR3, implicated in cellular proliferation during tissue homeostasis, had previously been reported using TWAS (p = 1.5e−7) but an associated GWAS signal in Gharahkhani and co-workers was at subgenome-wide significance level22,72; while the GWAS signal in our more powered larger meta-analysis attained genome-wide significance (p = 8.6e−48). Another example that exemplifies this is SEC31B (SEC31 homolog B, COPII coat complex component), a gene on chromosome 10, a novel significant TWAS association signal in three GTEx brain tissues using the JTI model (Table S14). The SNP located near this gene, rs11190559, was the strongest signal (p = 2.4e−5) with 47 other variants in the locus with association signal with p < 1e−4 (Tables S14 and S15). The SEC31B gene has been reported in the context of 2 anomalous pigmentary syndromes with ocular manifestations; retinitis pigmentosa 37 and Hermansky-Pudlak syndrome 1.73 A GWAS variant in the vicinity of the gene is associated with heart rate.74 Moreover, loss-of-function mutations in SEC31B promote colorectal cancer metastasis and a rare form of anemia.75,76
Seven of the novel TWAS loci were unique to PrediXcan and/or UTMOST models (Figure S13; Table S15). Overall, nearly two-thirds of the genes cumulatively prioritized using the three cis TWAS models (181/271) have vascular-related and/or cell senescence/proliferation functional roles or have been implicated in vascular or neoplastic diseases. A fifth of these vascular and cancer-related genes are cilia-related genes (Table S16).
We confirmed that all but four TWAS-prioritized genes (14/18; Table S14) that transect novel loci associated with POAG are robustly expressed in all eye tissues based on the publicly available Ocular Tissue Database (OTD).77,78 However, the three genes missing in the OTD, ABHD18, ACKR2, and LAMTOR3, have been shown to be expressed in mouse and pig retina.79,80
PRS prediction performance and effect of POAG liability across EHR
Prediction performance of the leave-biobank-out GBMI-IGGC-GGLAD POAG meta-analysis as discovery GWAS in five biobanks with African, Asian, and European ancestry subjects, as estimated by R2 on the liability scale, ranged from 0.0166 (95% CI: 0.01, 0.025) for Lifelines to 0.0484 (95% CI: 0.042, 0.056) for UKBB (Figure 3A). However, consistent with previous findings, performance was lower for the two non-European ancestry populations (Figure 3A).81 Results for the more balanced case-control ratio of ∼1:4 in BioVU and Lifelines showed an improvement in the R2 (Figure S14).82,83,84,85 Similarly, European ancestry subjects with polygenic risk scores (PRSs) in the highest risk decile of the PRS distribution had 2.1- to 4-fold higher odds of POAG compared with those in the mid decile (Lifelines: 95% CI: 1.8, 2.24; UKBB: 95% CI: 3.34, 4.79). In African ancestry individuals, the OR between the highest and mid decile polygenic risk was slightly lower but significant only in the UKBB (2.3-fold [95% CI: 1.21, 4.97]) while in BBJ East-Asian samples it is 1.72-fold (95% CI: 1.59, 1.85) (Figure 3B). Furthermore, the PRSs were robustly associated with POAG phecode across five biobanks (Tables S17–S23).
Figure 3.
Assessment of prediction performance and odds ratio in polygenic risk scores for POAG across multiple biobanks
(A) Prediction performance of GBMI-IGGC-GGLAD and IGGC POAG meta-analysis across five biobanks. The proportion of variance in POAG explained by PRS (Nagelkerke’s R2 in liability scale) is reported on the y axis. The source dataset used to estimate the marginal effect size of SNPs are listed in parentheses for each biobank. The PRSs of POAG were generated using PRScs auto. Performance of UKBB and BBJ using IGGC was not checked because data from the two biobanks are part of IGGC meta-analysis. AFR, African ancestry; EAS, East Asian ancestry; EUR, European ancestry.
(B) The odds ratio (OR) between top decile and mid-decile PRS POAG across five biobanks. The dashed line indicates OR = 1. The averaged OR was calculated using the inverse-variance weighted method. PRS was stratified into deciles with the mid-decile (40%–60%) used as the reference group. The discovery dataset used to estimate the marginal effect size of SNPs are listed in parentheses for each biobank. AFR, African ancestry; EAS, East Asian ancestry; EUR, European ancestry.
(C) Proportion of phecode groups associated with polygenic risk generated from leave-biobank-out POAG GBMI-IGGC-GGLAD meta-analysis across three biobanks. Asterisks indicate groups that are significantly enriched.
In general, summary statistics from IGGC yielded better predictions (R2) and ORs across the five biobanks and the GLGS cohort relative to GBMI data (Figures 3A, 3B, and S14).81 Moreover, there was no significant improvement in predictions when using the larger GBMI-IGGC-GGLAD meta-analysis summary data as a source relative to IGGC (Figure 3A). This could be due to phenotype heterogeneity and potentially unbalanced case-control ratio in GBMI compared with IGGC. These were exemplified by elevated prediction observed for the more accurately phenotyped GLGS cohort with balanced case-control ratio compared with equally balanced target data in BioVU and Lifelines (Figure S14).
As expected, the genetic risk for POAG was associated with ocular traits, but also with other phenotypes, such as circulatory, neoplasm, and musculoskeletal traits, across the five biobanks (Figures 3C; Tables S17–S23). Additional association signals with circulatory traits and neoplasms were observed in African (e.g., UKBB_AA_circulatory, Fisher’s test p = 0.0376) and European ancestry (e.g., BioVU_neoplasm p = 0.0002) cohorts (Figure 3C). We then explored POAG comorbidity pattern across circulatory codes (n = 171: ≥100 cases) in a total of 1,968,903 BioVU subjects of African (n = 273,379) and European ancestry (n = 1,695,524), after excluding all individuals with other eye codes (phecode 360–379) and correcting for age and sex. African ancestry individuals who have POAG have significantly higher odds of comorbidity with circulatory codes relative to European ancestry individuals (OR = 2.63; 95% CI: 1.66–4.16; p = 3.4e−5).
Interaction between the SIX6 and CDKN2B-AS1 loci
We confirmed that the haplotype containing the rs33912345_SIX6 risk allele (C allele) has reduced SALRNA1 and SIX6 expression relative to the haplotype containing the reference allele (Figures 4A and 4B), potentially indicating that the missense rs33912345_SIX6 variant contributes to etiology of POAG by downregulating the expression of the genes in the SIX6 locus. To further study if there is an association signal in the SIX6 locus independent of the rs33912345_SIX6 missense variant, we rebuilt the prediction model in skeletal muscle tissue by excluding all variants in LD with the missense variant (retaining those with r2 < 0.1). We found weak or no association signals with either SIX6 variants (p = 0.041 vs. p = 5.93e−15 for original model) or SALRNA1 (p = 0.78 vs. p = 1.96e−32) with LD-constrained rebuilt PrediXcan and UTMOST models, respectively, indicating that all the association signals detected in the locus in the original gene models are attributable to the rs33912345_SIX6 missense variant. Similarly, all the chr9p21.3 association signals observed with POAG are mainly attributable to the exonic rs1008878_CDKN2B-AS1 variant (Figure S15).
Figure 4.
Genetic correlations and expression effects in GTEx skeletal muscle tissue: insights into SIX6 and CDKN2B-AS1 loci interactions and pheWAS
(A) Correlations in measured gene expressions in GTEx skeletal muscle tissue between genes in the SIX6 and CDKN2B-AS1 loci.
(B) Effect of rs33912345 variants in GTEx-measured gene expressions in skeletal muscle tissue (n = 716). Difference in GTEx-measured gene expressions of SIX6 locus genes, SIX6 and SALRNA1, between (i) individuals who carry the rs33912345C causal allele (represented here by proxy variant rs7493429, r2 = 0.74) versus those with the wild-type allele and (ii) different genotype causal/wild-type combination. p values (p) are based on t test (i) and ANOVA tests (ii).
(C) Effects of genetic interaction between rs33912345_SIX6 proxy variant and rs10811650_CDKN2B-AS1 on CDKN2B-AS1 locus gene expressions, (i) Expression pattern in individuals who carry wild-type rs33912345_SIX6 proxy variant in combination with rs10811650_CDKN2B-AS1 genotypes in brain cortex, (ii) expression pattern in individuals who carry rs33912345_SIX6 proxy variant causal allele in combination with rs10811650_CDKN2B-AS1 genotypes in brain cortex, and (iii) expression pattern in individuals who carry homozygous rs33912345_SIX6 proxy variant in combination with rs10811650_CDKN2B-AS1 genotypes in skeletal muscle. p values (p) are based on ANOVA tests.
(D) GReX-PheWAS for categorizing phenome-wide associations for CDKN2A and CDKN2B genetically regulated expressions in BioVU and UKBB. (i) Miami plot of TWAS Z scores (y axis) across phenotypes, colored by phecode group. The dotted gray line shows the significance threshold for Benjamini-Hochberg FDR correction and phenotypes are labeled if the association passes Bonferroni correction. (ii) Boxplots of –log10 Benjamini-Hochberg FDR-adjusted p values of genetical GTAs across nine phenotype groups. The dotted gray line shows FDR-adjusted p value of 0.05.
We next explored the interaction between rs33912345_SIX6 proxy variant and CDKN2 genes and their potential consequences on the expression pattern on chr9p21.3 genes. We first confirmed that there was correlation between expression levels of SIX6 and chr9p21.3 loci TWAS prioritized genes in GTEx data (Figures 4A, S16, S17, and S18). We also observed interactions between the two loci in association with POAG and vascular-related traits (Phecodes 411, 411.1–411.9) in BioVU (Table S25). We further found that the proxy variant has significant effect on expression of CDKN2A and CDKN2B via interaction with chr9p21.3 top GWAS variants that are associated with vascular and neoplastic traits in the GWAS catalog (rs10811650 and rs2891168). The eQTL effect of the chr9p21.3 variants disappears in the presence of the rs33912345 proxy causal allele (Figures 4Ci and 4Cii). However, in the presence of the homozygous rs33912345_SIX6 proxy, the eQTL effect is observed for those with homozygous causal alleles for the local variants with dramatic reduction in expression for CDKN2B, and increase in expression for CDKN2A (Figures 4Ciii, S16, S17, and S18).
We explored potential biological consequences of the interaction between the two loci by performing TWAS-PheWAS for the two CDKN2B-AS1 loci genes in the UKBB (n = 396,618) and BioVU (n = 59,805) cohorts. We applied the best-performing model with the highest prediction ability estimated from the cross-validation for each gene among PrediXcan, UTMOST, and JTI.86,87,88 For the two chr9p21.3 genes, the best-performing models in brain cortex tissue were JTI (CDKN2A, r2 = 0.0257) and UTMOST (CDKN2B, r2 = 0.0413). We performed GReX-PheWAS for the 2 chr9p21.3 genes in the same 2 cohorts, followed by meta-analysis of the 2 PheWAS (n = 456,423) across 731 traits and diseases, grouped into 17 categories. The GReX-PheWAS analysis revealed associations for CDKN2A/B in the CDKN2B-AS1 locus with glaucoma and enrichment for phenotypes of the circulatory and neoplasm groups, with coronary atherosclerosis and skin cancers as the top phenotype associations (Figure 4D). These phenotypes include traits that are a result of senescence (upregulation) and cell proliferations (downregulation). We also detected associations with lipid disorders, which have been implicated in both these two groups, consistent with the defined functions of the two genes, suggesting that an etiology that connects these traits is a balance between cell proliferation and senescence.89,90
Discussion
In this study, we aimed to determine the genetic mechanisms underlying POAG. We leveraged a suite of statistical tools to analyze a combination of massive GBMI genome-wide discovery resources, previously published GWASs, and publicly available GTEx data. In the GBMI GWAS meta-analysis for POAG across 15 biobanks (n = 1,487,441) we identified a total of 62 risk loci, of which six were novel (F5, MYO1B;NABP1, RPL37A-LINC01280, CCDC13, MIR2054-INTU, ZFP91-CNTF-GLYAT) and three of these were reported in IGGC at subgenome-wide significance levels.22 The novel associations encompass loci near genes INTU-MIR2054, ZFP91-GLYAT-CNTF, and CCDC13, with only the latter not prioritized by TWAS.91,92,93,94 The larger GBMI-IGGC-GGLAD meta-analysis revealed additional novel associations in the loci that encompass LOC654841, KBTBD8, ADGRL3, DDIT4L, HMGXB3, KCNK5, MAD1L1, APPL2-KCCAT198, CATSPERB, OR5B12-OR5B21, and FENDRR genes. In the GBMI-IGGC-GGLAD, TWAS prioritized 6 novel POAG susceptibility genes that correspond to 5 of the 12 additional novel loci and all have functional roles or have been previously implicated in vascular and/or neoplastic-related pathologies: COL4A495, MFF96 (corresponding to the LOC654841 locus), LAMTOR397,98 (corresponding to the DDIT4L locus), HMGXB3,99,100 ALDH1L2101 (corresponding to the APPL2-KCCAT198 locus), and CCDC88C102,103,104 (corresponding to the CATSPERB locus). By virtue of being gene based and a more well-powered analysis than GWAS, TWAS identified potentially 68 additional novel loci that are at subgenome-wide GWAS association.75,76 In fact, 11 of these loci are confirmed by an independent multi-trait analysis of larger data,105 which is consistent with our assertion that, as more POAG cases are collected over time, these signals will be detected at genome-wide significant level.
Review of literature on functional roles of the genes that fall within the loci in this study, coupled with enrichment, genetic correlation, and PheWAS analyses, all point POAG risk to potential link to vascular and proliferation mechanisms. Two-thirds of all the TWAS-prioritized POAG-associated genes identified in this study have been previously associated with vascular-related traits and/or implicated in carcinoma and act as tumor suppressors. In addition, all the genes that are near or regulated by the sex-associated novel loci are also implicated with risk of vascular-related diseases and neoplasms.51,106,107,108,109,110,111,112 We performed enrichment analyses in which cell proliferation and angiogenesis pathways were significantly represented.
We further explored pleiotropy of genetic risk for POAG across the phenome in five biobanks, and confirmed association of POAG genetic risk burden with vascular and neoplastic-related traits. Interestingly, consistent with our findings, analysis of genetic drug discovery/repurposing for the GBMI, POAG GWAS identified four molecules approved for treatment of vascular-related conditions and two additional molecules approved for neoplasm-related conditions.113 In addition, an anti-hyperlipidemic drug initially developed for the treatment of coronary artery disease, probucol, was prioritized.113 In summary, our results reinforce the role of vascular processes in glaucoma. The results also highlight pathways related to cell proliferation mechanisms that have not been reported in previous GWASs. Therefore, these results add more insights into understanding glaucoma pathogenesis.
Previous studies showed disparity in POAG prevalence across ancestries and sexes. While non-genetic factors are likely to drive many of the observed health disparities characterized to date, differences in some underlying genetics (including differences in frequency among populations, unique unobserved rare variants, and/or difference in effect sizes at common SNPs) play a significant role in differential disease etiology.114 In the GBMI analysis, on both sexes we observed two novel loci (rs1469837390_F5 in FinnGen; rs12476634_LINC01280 in African and Hispanic Americans) and two known POAG loci (rs74315329_MYOC in Europeans; rs147660927_ANGPTL7-MTOR in HUNT, FinnGen, EstBB, Lifelines, QSKIN) that showed ancestry-specific effects. Sex-stratified association analyses in GBMI identified an additional three African-specific novel loci that were associated with POAG in males only: ATPB10, TMEM167B, and ARMC4, and one African-specific novel locus PRKG2;RASGEF1B that is associated with POAG in females only. PRKG2, the closest gene to the lead female-associated variant, is induced by estrogen and progesterone.115 Interestingly, estrogen has been reported as having protective effects in glaucoma.116 Fourteen additional loci show significant effect size differences between ancestries, three of which also have significant difference in effect between males and females. Genes prioritized using TWAS that are near the lead variants for the loci that show effect difference between the sexes have sex-biased gene expression patterns.50,51 We further performed in silico validation of the novel male-associated African-specific TMEM167B locus and showed that the variant is a male-specific regulator of CELSR2. CELSR2 has been previously reported to be differentially expressed between males and females and associated with cardiovascular traits.51 ARMC4 has also been shown to be differentially expressed between males and females.51,107,108
Results from PRS PheWAS across the BioVU, UKBB, and EstBB also suggest that genetic risk burdens of POAG are associated with vascular-related traits, especially in GBMI African ancestry, and neoplasms in GBMI European ancestry populations. Moreover, in BioVU we observed higher risk of POAG comorbidity with vascular-related traits in African ancestry relative to individuals of European ancestry. These potentially indicate unique shared biology between POAG and circulatory traits in African ancestry populations.117,118,119 Consistent with previous findings,120 our study also shows sex-related disparity in POAG in African ancestry. Larger epidemiological and genetic studies in individuals of different ancestries will clarify the role of GWAS signals identified here or other additional signals in differential risk burden between ancestries and sexes.121
We performed extensive in silico validation of the SIX6 and CDKN2B-AS1 loci, using GTEx and EHR data across BioVU and UKBB. In our study we confirmed evidence of significant interaction between rs33912345_SIX6 and causal variants in the chr9p21.3 region, with concomitant effect on expression of CDKN2A and CDKN2B genes in the CDKN2B-AS1 locus. These effects are particularly enhanced between homozygous rs33912345_SIX6 and homozygous causal variants in chr9p21.3. However, in contrast to previous studies in mouse models,122,123,124,125 our analysis in GTEx muscle tissue indicates that CDKN2B might be the causal gene in humans because it undergoes expected expression reduction, while expression of CDKN2A increases. In a mouse model with elevated IOP, the SIX6 missense variant was shown to increase the risk of glaucoma-associated vision loss by disrupting the development of the neural retina by upregulating the CDKN2A expression, leading to a reduced number of retinal ganglion cells.122,123,124,125 Our results highlight a regulatory mechanism for rs33912345_SIX6, which aligns to previously reported regulatory roles for this variant. Carnes and co-authors reported that patients who have the rs33912345_SIX6 CC genotype (homozygous for the risk allele) have a significantly thinner retinal nerve fiber layer (a glaucoma endophenotype) than patients homozygous with non-risk allele.122 Furthermore, a Brazilian population study reported an additive association effect of an independent chromosome 16 homozygous rs1362756 in the SALL1 gene in the presence of rs33912345_SIX6 CC genotype, but not with heterozygous or wild-type genotypes.126 This suggests that the trans-effect of the rs33912345_SIX6 variant in POAG risk is complex and potentially involves multiple loci and genes. In addition, TWAS-PheWAS analysis for the two chr9p21.3 locus genes revealed associations with circulatory and neoplasm traits. Thus, the SIX6/CDKN2A/B association pattern observed here potentially reflects the balance between tumor suppression, senescence (apoptosis, autophagic), and cell proliferation or tumorigenesis. Collectively, our results implicate programmed cell death as the underlying common mechanism behind the shared genes. These findings suggest that apoptosis triggered by the ON degeneration in POAG disease may play a protective role in various neoplasms by promoting programmed death of cancer cells and vice versa. Future analyses will clarify the trans-effect of the rs33912345_SIX6 with other POAG loci in the genome in the context of glaucoma etiology.
By integrating evidence from vascular and cell proliferation mechanisms that emerged from our results, we suggest that primary cilia may potentially be a common link between vascular diseases and glaucoma pathophysiology. The novel associated genes identified in the GBMI GWAS, CCDC13, CNTF, and INTU, the gene CELSR2 with male-specific association in Africans, and more than 40 other genes prioritized using TWAS, have been found to be linked to primary cilia, a microtubule-based cellular structure located on the surfaces of vertebrate cells.127,128,129 In addition, the other male-specific associations, ATP10B and ARMC4 genes, are cilia-related genes implicated in ocular, vascular, and neoplastic traits.107,130,131,132,133,134,135 Primary cilia are crucial in normal function of the smooth muscles and play important roles in cell proliferation mechanisms.136 In fact, primary cilia, through their dysfunction, contribute to cancer via interference in signaling pathways, such as the Wnt signaling.136,137 Some investigations have suggested a direct link between Wnt signaling and glaucoma, since Wnt signaling is involved in the regeneration of the ON after injury and also in IOP regulation.102,138,139 Primary cilia on vascular endothelium have been proposed to play a critical role in the regulation of vascular barrier.136 The vascular barrier controls the exchange of molecules and ions between blood and tissues and prevents dangerous substances from entering the tissues and causing damage.137,140 Primary cilia are present in the eye, i.e., retinal pigment epithelium, and they carry out sensory function through various signaling pathways.140,141 Loss of primary cilia is associated with several pathologies that have anomalies in these mechanisms, such as retinopathy and Leber congenital amaurosis.142,143 In the eye, the TM also contains primary cilia and these structures change with IOP. When the IOP is elevated, primary cilia shorten, promoting the expression of tumor necrosis factor α and transforming growth factor β (TGF-β), perhaps initiating mechanisms leading to glaucoma.144 Recent studies have shown that one of the first genes linked to glaucoma, MYOC-producing myocilin, is present in the base of primary cilia and its expression affects pathways related to ciliary signaling, such as TGF-β.145,146 This suggests that ciliary pathways are involved in the secretion of myocilin, of which accumulation in the TM of mutant MYOC cause open-angle glaucoma.147 Investigating the mechanisms that influence specific primary cilia functionality will contribute to understanding the role of this structure in the pathogenesis of glaucoma, and eventually to develop novel drugs and therapies.
Strengths and limitations of the study
In this study, there are several methodological strengths as well as limitations. The combined dataset represents the largest multi-ancestry genetic study conducted in POAG and thus powered to detect novel associations. Furthermore, the meta-analysis with two other available datasets and the vast resources from different biobanks afforded an opportunity to explore genetic interactions between likely causal variants of glaucoma, and to conduct in silico investigation of the potential shared biology between POAG, cancer, and vascular traits. On the other hand, using the two larger published GWASs on POAG, it did not allow us to perform replications on the identified novel loci.
Several issues that are inherent in a biobank collaborative context, such as GBMI, might lead to a conservative signal detection. POAG phenotyping methods in GBMI were different across the biobanks (i.e., phecode mapping, or a combination of physicians’ diagnosis, glaucoma medications, or self-reporting). In addition, some of the phenotyping was for glaucoma and not specifically POAG, potentially introducing biological heterogeneity. Even though this is not a major issue in biobanks with a majority of European and African ancestry subjects where POAG is the predominant glaucoma subtype, the signals detected from Asians will also probably be from PACG, where this subtype predominates.6,31,32,33 In addition, signals from other minor glaucoma subtypes (such as XFS) might also introduce some noise. In this study, we showed that only three previously identified loci were potentially signals from other subtypes—and all other loci, including novel ones, were detected in other published data. Moreover, despite using powerful approaches to mitigate the effect of unbalanced case-control sampling inherent in a biobank context, the attenuating effect of inappropriate controls will persist, making our signal detection conservative.26,148
Furthermore, even though this analysis used multi-ancestry meta-analysis, the subjects are still predominantly of European ancestry. We believe that recruitment of cohorts that include ethnic minorities will improve knowledge transferability and health equity. Finally, even though we did extensive in silico validation using available data and gleaned broad biological pathways that might be implicated in POAG, we did not perform functional validation of the novel genes identified in this study. Therefore, other in vitro studies using relevant human tissues and in vivo using model organisms are still necessary to define the biological role of these genes in POAG and potential link to vascular and neoplastic mechanisms.
Conclusion and future directions
Our study identified large effect ancestry and sex-specific loci associated with POAG. A larger genetic study in individuals of more diverse ancestries will clarify the role of GWAS signals identified here. Moreover, there is a need for more studies to ascertain the magnitude of inter-loci interactions and the roles of the SIX6 missense variant in the overall interaction landscape. Furthermore, the role of these interactions and differences in disease burden for POAG and vascular/cancer-related traits among ancestries need to be elucidated. In summary, by integrating evidence from vascular and cell proliferation mechanisms that emerged from our results, we suggest that primary cilia may potentially be involved in glaucoma pathophysiology. Further investigations are needed to elucidate their role and mechanisms in glaucoma.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Association results | This paper | https://www.globalbiobankmeta.org/resources |
| LDlinkR | 192 | https://github.com/CBIIT/LDlinkR |
| gnomAD | 53 | https://gnomad.broadinstitute.org/ |
| 1000 Genomes | 52 | https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/ |
| GTEx version 8 | 25 | https://gtexportal.org/home |
| Software and algorithms | ||
| Joint-tissue imputation | 88 | https://github.com/gamazonlab/MR-JTI |
| PrediXcan | 86 | https://github.com/hakyimlab/PrediXcan |
| UTMOST | 87 | https://github.com/Joker-Jerome/UTMOST |
| DEPICT | 164 | https://www.nature.com/articles/ncomms6890 |
| PRS-CS | 173 | https://www.nature.com/articles/s41467-019-09718-5 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Jibril Hirbo (jibril.hirbo@vumc.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The all-biobank meta-analysis results and plots (including ancestry/sex-stratified and cross-ancestry meta-analyses) are available for downloading at https://www.globalbiobankmeta.org/resources and for browsing at the browser http://results.globalbiobankmeta.org.
-
•
Scripts used for quality control, meta-analysis, and summary of results are available at https://github.com/globalbiobankmeta and deposited at https://zenodo.org/badge/latestdoi/295461030.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Method details
Association testing
The overall design of this study is reported in Figure 1. As part of the GBMI, a large-scale multi-ancestry meta-analysis of genome-wide association studies was conducted including a total of 26,848 cases and 1,460,593 controls from 15 global biobanks across six ancestries (Table S1, Figures S1, and S2). Phenotype definition, biobank specific quality control and standardized GWAS was performed by each contributing biobank. POAG phenotyping was done using any of the three approaches: phecode mapping (BioVU, UKBB, HUNT, MGI and CCPM), ICD9/ICD10 codes (BioMe, FinnGen, EstBB, MGB, deCODE) and either one or a combination of physicians’ diagnosis, glaucoma medications or self-reporting (BBJ, TWB, Lifelines, GS and QSkin) (Table S1).149 Potential effect of heterogeneity in phenotyping was checked by comparing a subset of manually reviewed and phecode defined patients in BioVU with an independent traditionally phenotyped cohort (Supplementary information, Table S2).
Figure 1.
Workflow of this study
A total of 15 biobanks joined the GBMI POAG meta-analysis (n = 1,487,441: 26,848 cases, 1,460,593 controls), where phenotyping was harmonized across biobanks, biobank-specific quality control was performed and standardized genome-wide association study (GWAS) was conducted. This was followed by a meta-analysis of GBMI, IGGC of European ancestry, and GGLAD (n = 1,478,037: 46,325 cases, 1,431,712 controls). On the Meta-GBMI multi-ethnic biobanks summary data, we performed functional impact, enrichment analysis, transcriptome-wide association study (TWAS), and fine-mapping. Polygenic risk scores (PRSs) for POAG were constructed from the leave-biobank-out GBMI-IGGC-GGLAD meta-analysis summary statistics with PRS-CS and validated in six biobanks (BBJ, BioVU, Estonian, GLGS, Lifelines, and UKBB). PRSs were then tested for association using phenome-wide association studies (PheWAS) across four biobanks. Then, to confirm and interpret our results, we examined the expression effects of missense variants in SIX6-CDKN2B-AS1 and TMEM167B loci in GTEx data. MAC, minor allele count.
Meta-analysis was performed followed by the variant-level quality control for each biobank by flagging markers with different allele frequencies compared to gnomAD and excluding markers with imputation quality score <0.3.26,54 Sex-specific association analysis was also conducted across 9 biobanks in 7,916 cases and 269,105 controls (males) and across 10 biobanks in 9,538 cases and 342,870 controls (females) (Tables S4—S5). The number of independent loci and number of top hits for each biobank contributing to GBMI are reported in Figure S5.
To increase the power to discover additional variants associated with POAG, a meta-analysis was performed combining GBMI (n = 1,259,040), International Glaucoma Genetics Consortium (IGGC) of European ancestry (n = 192,702) and Genetics of Glaucoma in People of African Descent (GGLAD) (n = 26,295) summary statistics, excluding any biobank cohorts from GBMI that overlap with the two other datasets in our analysis (FinGenn, BioME and UKBB Africans). Fixed-effect meta-analyses based on inverse-variance weighting were performed. We obtained a total number of cases of 46,325 and number of controls of 1,431,712 (Table S7).22,27 We further performed ancestry specific meta-analysis of the data. We defined genome-wide significant loci by iteratively spanning the ±500 kb region around the most significant variant and merging overlapping regions until no genome-wide significant variants were detected within ±500 kb. The most significant variant in each locus was selected as the index variant. To identify independent variants, clumping was performed in PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) using r2 < 0.05 as linkage disequilibrium (LD) metric threshold, physical distance of 500 kb, 1000 Genomes Project Europeans as LD reference panel and the significance threshold for index SNPs set at P < 1x10−5.150 Polymorphisms were considered significantly associated with POAG if genome-wide significant level (p value<5.0e−8) was attained. Quantile-quantile (Q-Q) and Manhattan plots were generated to visualize the results.
Single variant replication analyses were performed in three biobanks that had at least 50 POAG cases: Penn Medicine Biobank (PMBB), Colorado Center for Personalized Medicine (CCPM) and All of US (Table S6).151,152,153 PMBB and CCPM are part of the GBMI that joined the consortium after initial analysis of the data and have been previously described and use same phenotyping and GWAS protocols as other GBMI biobanks.154 All of Us Research Program is an NIH initiative that plans to enroll an observational cohort of diverse group of at least 1 million persons in the United States.154 We used April 2023 v7 Curated Data Repository (CDR – vers. 2022Q4R9) with 7/1/2022 cutoff date for participant EHR data (n = >413k, >245k of which have short read WGS data). In All of US, we defined cases as those with ICD9-365.11,365.12, ICD10-H40.11, H40.12, and SNOMED CT 77075001, 50485007 codes, while controls are >18 years old with no eye related ICD9/10 and SNOMED CT codes. Individuals were stratified based on All of US predicted ancestry, and precomputed PCs 1–10 (provided by All-of-US) included as part of the covariates. Sex specific association analysis was performed using REGENIE in PMBB and CCPM, while Wald logistic regression tests using HAIL in All of US with POAG as the outcome and variants as the predictor performed, adjusting for age and the first ten PCs as covariates. For comparison purposes, regression analysis on all data in each biobank was performed adding sex to the covariates for combined sex analysis.
Checking for potential for phenotype heterogeneity in BioVU samples
POAG was ascertained in the BioVU dataset using phecodes in VUMC electronic health records. All subjects (n = 144,017) with an ophthalmology examination code (CPT = 92002, 92004, 92012, or 92014) were selected using at least two instances of POAG phecode that covers ICD-9 and/or ICD-10 codes 365.11, H40.11XX, 365.12, H40.12XX (n = 17,824) while controls exclude those with codes for eye diseases (360–379, H00-H59, E08.3, E09.3, E10.3, E11.3, E13.3, Q10.XX-Q15.XX, n = 53,919). The groups were then filtered to include only subjects with genotyping data from Illumina MEGA-Array.
The de-identified electronic health records of the glaucoma subjects were then reviewed to confirm the diagnoses. The patients’ names, addresses, other forms of identification, and the treating physicians’ names were removed. Records which were available included the problem list stating POAG or NTG, medication lists, clinical notes including detailed ophthalmic examinations, ophthalmic operative notes, correspondence letters and discharge summaries. Additional high-level confirmation included an anterior segment examination without secondary-glaucoma evidence, and open angles by gonioscopy.
Only those subjects (n = 1040) who had POAG reported in any of the above categories by the treating physicians were included as cases in the manual review. Any subject, who was coded for POAG without supporting confirmation or who had contradictory information in the medical record, was excluded to minimize inclusion errors. Supporting information including: reported peripheral vision loss, intraocular pressure, glaucomatous optic nerve appearance, glaucoma surgeries and use of glaucoma medications was reviewed and noted. A total of 716 individuals who had MEGA array genotyped have their EHR manually curated. A total of 220 out of the 716 individuals conflicted with manually curated records of which 172 are those with only a single ICD9+ICD10 mention of POAG. A total of 48 phecode assignations conflicted with manually curated records: 5 cases were missed by phecode phenotyping, 4 were pigmentary glaucoma, 7 POAG suspect, 9 pseudoexfoliation syndromes (XFS) and 23 primary angle closure glaucoma (PACG) (Table S2). Using any mention of ICD9 or ICD10 POAG had a far much higher number of conflicts (114) with manually curated information. Thus conservatively, we estimate that ∼1.2% (∼320) and ∼3.2% (∼560) might potentially be XFS and PACG cases, respectively, among the total GBMI 26,848 POAG cases.
Therefore, as further quality check step, we compared our GBMI-IGGC-GGLAD meta-analysis signals with well powered GWASs for XFS and PACG, and determined that three of the loci might be attributed to these two glaucoma subtypes: 1) The rs3825942_LOXL1 loci that is the main XFS signal and previously reported in GWAS of POAG in a recent study that include Biobank level data155,156 2) The rs11024102_PLEKHA7 has been associated only with PACG and glaucoma in East Asian ancestry individuals, while, 3) the lead variant in rs58812088_ FNDC3B in our study is just ∼12kb away from and in high LD (r2 = 0.7) with PACG lead variant in rs16856870_FNDC3B locus.35,157 However, three loci: rs993471_COL11A1, rs2276035_ARHGEF12, rs12150284_GAS7 have previously been shown to be POAG-PACG shared loci.34,35
Additional cohort included in risk prediction was from Groningen Longitudinal Glaucoma Study (GLGS). The GLGS consisted of Dutch individuals with a diagnosis of POAG. The original GLGS cohort has been described in detail by Heeg et al. (2005).158 After the inclusion of the initial cohort in 2000–2001, the GLGS continued recruiting new participants during follow-up. We included glaucoma patients who visited the outpatient department of the University Medical Center Groningen in 2015 and who gave written informed consent for a blood sample for genetic analyses. In the GLGS, glaucoma patients had to show glaucomatous visual field loss in at least one eye. For glaucomatous visual field loss, two consecutive tests had to be abnormal in at least one eye, after an initial test that was discarded to reduce the influence of learning. Defects had to be compatible with glaucoma and without any other explanation. Those with pseudoexfoliative or pigment dispersion glaucoma or a history of angle-closure or secondary glaucoma were excluded.8 In GLGS, genomic DNA was extracted from the peripheral blood, using Gentra Systems Purogene chemistry. The genotyping was done using the Illumina Infinium Global Screening Array (GSA) MultiEthnic Disease beadchip version, which contains 692,367 markers.139,159
To check if difference in ancestry and sex-specific associations in our study was potentially correlated with difference in POAG prevalence, cases in European and African American was estimated in 3,414,079 BioVU patients who are ≥40 years older and self-identify as either European Americans (EA, n = 1,218,124; 555,132 males and 662,825 females) or African Americans (AA, n = 154,273; 66,691 males and 87,541 females).
Comorbidity with circulatory related codes (phecodes = 394–459) in a total of 1,968,903 individuals with ≥2 instances of mention of the relevant code excluding individuals with any mention of eye codes (phecodes = 360–379) (AA, n = 273,379; 1,549 cases and 271,830 controls; and EA, n = 1695,524; 5,446 cases and 1,690,078 controls) was inferred by performing logistic regression analysis conditioned on sex and age as covariates. Odd ratio comparison was done using epitools in R.
Post-hoc power analysis for ancestry and sex specific novel loci
For the SNPs identified in the GBMI analyses carried out in males of the two African cohorts, a post hoc power analysis, based on a likelihood ratio test framework, was performed in the combined dataset where the variants were observed (BioVU and BioMe cohorts), using the “genpwr” R package. For the SNP rs111739240_TMEM167B, as input parameters, we set a MAF of 0.025, for an odds ratio of 6, a sample size of N = 7,860, and number of cases = 155, a case rate of 0.019, and a standard GWAS significance of p value of 5E−8 (Table S5). For the SNP rs78909751_ARMC4, as input parameters, we set a MAF of 0.032, for an odds ratio of 5.41 (N = 7,860, cases = 155, case rate of 0.019, p value of 5E−8 (Table S5). Based on this power analysis and under an additive model, for both variants, we had 80% of the power of detection with a sample size of more than 4,617 individuals.
SNPs and gene annotations
Significant polymorphisms were annotated with the gene inside whose transcript-coding region they are located, or alternatively, the nearest gene. In addition, the polymorphic sites were functionally annotated using ANNOVAR.160 Exonic SNPs were investigated further using SNPnexus to uncover non-synonymous variants.161 Damaging effect of non-synonymous SNP on protein structure and function was investigated using the Sorting Intolerant From Tolerant and Polymorphism Phenotyping scoring tools.162,163
Enrichment analysis
To identify the functional roles and tissue specificity of the associated variants, we performed gene prioritization and tissue- and gene-set enrichment analyses using DEPICT (Tables S9—S11) in which we prioritized POAG-associated genes using a co-regulation-based method across multiple different tissues.164 The tool assesses the potential role of genes independent of the presence of an eQTL, making it possible to test more genes.
For the gene set enrichment analysis, gene expression data from 77,840 samples was used to predict gene function for all genes in the genome based on similarities in gene expression. In DEPICT, the probability of a gene being a member of a gene set was estimated based on their co-functionality to prioritize the most likely causal genes. A total of 14,461 reconstituted gene sets was generated which represent a set of biological annotations: Gene Ontology (GO) gene sets, REACTOME gene sets, Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets, InWeb protein-protein interactions, and Mouse Genetics Initiative gene sets (MP). Bonferroni correction was applied for multiple comparisons of 14,461 independent tests (p < 0.05/14,461).
For tissue enrichment, microarray data from 37,427 human tissues of 209 Medical Subject Heading (MeSH) annotations from Affymetrix HGU133a2.0 platform microarrays was used to identify genes with high expression in different cells and tissues.
TWAS and fine-mapping analyses
We used three gene-based methods, PrediXcan, joint tissue imputation (JTI) and unified test for molecular signatures (UTMOST) for correlating the genetic component of gene expression with phenotype.86,87,88 PrediXcan estimates gene expression weights by training a linear prediction model in a reference sample with both gene expression and SNP genotype data.165 UTMOST and JTI methods borrow information across different tissues transcriptomes, leveraging shared genetic regulation, to improve prediction performance in a tissue-dependent manner.88
There is only one ocular tissue in GTEx data with available corresponding genotype data needed to build TWAS models, so we used RNA-seq and genotype data from peripheral retina and proxy tissues that have biological functions that we inferred to be crucial in visual perception. These proxy tissues include: vascular tissues (artery and heart tissues) that are crucial in production and drainage of aqueous humor (defects in this system can cause glaucoma), tissues of the nervous systems, which the eye is considered its extension of, and the liver tissue because it is a pivotal metabolic center.166,167,168 We consider the brain cortex as the most relevant GTEx tissue since the visual cortex is located there.
For the three models, gene expression prediction models were trained for 23 different human nervous, vascular and liver tissues (Tables S11—S12) using GTEx v8 data.169 The corresponding genotype data were imputed using the University of Michigan Imputation Server, with 1000 Genomes Project (phase 3 ver. 5) as a reference panel.170,171 Models with non-zero weights that met set significance criteria (FDR< 0.05 from the cross-validation in each tissue) were retained in the database. For each model, there was also a corresponding file with covariance data for the SNPs in each model. The three models were applied to the GBMI-IGGC-GGLAD GWAS meta-analysis summary results (n = 1,478,037: 46,325 cases and 1,431,712 controls).
Publicly available RNAseq data for eye tissues by Ratnapriya et al., (EyeGEx) have no associated data from other tissues and thus we only generated Predixcan models. To analyze RNA-seq data from peripheral retina tissues, gene expression values for all samples from retina tissue were normalized: 1) genes with expression below ≤6 reads (unnormalized) in ≤20% of samples read counts were excluded, 2) were normalized between samples using TMM, and 3) expression values for each gene inverse normal transformed across samples. We used PLINK to compute genotype principal components (PCs) based on 5,473,093 SNPs after pruning variants in pairwise LD.150 After regressing out all the covariates associated with the expression data (age, sex,os_od, mgs_level, rin, postmortem_interval_hrs), and ten first PCs, residuals were used to train PrediXcan model. Expression prediction models were trained using an elastic net model, excluding SNPs with >10% missingness, MAF <0.01 and deviation from Hardy-Weinberg across all 405 samples. These filtering steps led to a total of 7,732,396 SNPs. The prediction performance was evaluated by the correlation between the predicted and observed expression levels in 5-fold cross-validation. A total of 2807 genes with Pearson correlation r > 0.1 and p < 0.05 were considered as "predictable genes" and used in subsequent analysis. We presume that the mechanisms that underlie complex ocular conditions like POAG (like other complex diseases) can span tissue types across the human body, and thus we believe that focusing on just ocular tissues can lead to misleading conclusions.
Fine-mapping of TWAS association signals was done using FOCUS, a probabilistic gene-level fine-mapping method, to define credible sets of genes that explain the expression-trait signal at any given locus.172 The default non-informative priors implemented in FOCUS were used to estimate the posterior inclusion probability and a 90% credible set of genes at a given locus.
Polygenic risk scores (PRS) and pleiotropy
PRS for POAG were constructed from the leave-biobank-out GBMI-IGGC-GGLAD meta-analysis summary statistics in six different biobanks, BioVU (n = 85,615), UK Biobank (n = 370,088), Biobank Japan or BBJ (n = 178,726), Lifelines (n = 14,930), Estonian Biobank (n = 53,821), and glaucoma cohort GLGS159 (n = 3,739), with PRS-CS-auto option, and the best performing European reference panel (based on R2) of either 1KG Phase 3 or UK Biobank was used to estimate LD.53,173 In the target samples, genotypes were filtered for SNPs using the following criteria: minor allele frequency< 0.01, missing genotype rate< 0.05, filters out variants which have Hardy-Weinberg equilibrium exact test p value < 1e−6, and exclude individuals with missing data <0.1. Only unrelated individuals with genetic relatedness less than 0.05 were retained. We used EHR/or self-reported health conditions (depending on the data availability in each biobank) in a total of 617,565 individuals across five biobanks (excluding GLGS that was used to check the effect in a more balanced case-control cohort) to explore pleiotropy of genetic risk for POAG. We then evaluated the predictive performance of the PRSs generated using leave-biobank-out meta-analysis results in the six biobanks. PRSs were then tested for association with 17 disease categories using a phenome-wide association study (PheWAS).
Expression effects of missense variants (male-specific rs74113753 & trans-ancestry rs33912345)
Variants in SIX6 and CDKN2B-AS1 loci (chr9p21.3) have been associated with POAG,174,175,176,177,178,179,180,181 related risk factors and endophenotypes, such as peripapillary retinal nerve fiber layer,182 optic nerve degeneration,178,183 and vertical cup-disc ratio (VCDR)184,185,186,187,188,189,190 across all ancestries. The rs33912345_SIX6 missense variant (p.His141Asn) is the lead variant in the SIX6 locus, outside the DNA binding site and it is speculated to affect the ability of the SIX6 gene to interact with other transcription factors and cofactors.125 The variant rs33912345 is also Expression quantitative trait locus (eQTL) in GTEx for several genes within the SIX6 locus.169 The SIX6 and the CDKN2B-AS1 loci are known to be associated with POAG174,175,176,177,178,179,180,181 and cardiovascular disease.191 Studies in both glaucoma models and cell lines indicated that SIX6 missense variants interact with genes in the chr9p21.3 locus.124,125 However, no comprehensive analysis of this interaction has been done in human genetic and transcriptomic data.
In the SIX6 locus, the effect of the missense rs33912345_SIX6 variant on expression patterns of genes SIX6 and the nearby long non-coding RNA, SALRNA1, on genes CDKN2A and CDKN2B (CDKN2B-AS1 locus) were determined by performing regression analysis of residuals of the GTEx normalized gene expression levels. For each tissue, the possible confounders (sex, platform, first five principal components, and Probabilistic Estimation of Expression Residuals [PEER] factors)88 have been accounted for in the analysis. Since the missense variant (rs33912345_SIX6) did not pass the quality control filters, we used a proxy SNP, rs7493429_SIX6, which was in high LD with the missense variant (r2 = 0.724, D’ = 0.99), determined using R package LDlinkR (https://github.com/CBIIT/LDlinkR).192
We evaluated the effect of the variant rs33912345_SIX6 in the SIX6 locus on trait association to variants in CDKN2B-AS1 locus in four steps. We first checked the differences in the gene expressions of SIX6 and SALRNA1 measured in skeletal muscle tissues (the only tissues with GTEx expression data remaining after correcting for confounders) between the haplotype background that has the reference and the alternate allele. Secondly, we rebuilt the muscle skeletal genes expression model while excluding any variants that were in LD with the rs33912345_SIX6 missense variant (r2 > 0.1). The PrediXcan and UTMOST muscle tissue models were the best performing gene models for SIX6 (PrediXcan r2 = 0.131, compared to JTI r2 = 0.120) and SALRNA1 (UTMOST r2 = 0.024 compared to PrediXcan r2 = 0.016), respectively. Thirdly, we evaluated the Pearson correlations in gene expressions between genes in the SIX6 and CDKN2B-AS1 loci. We then checked for statistical interaction between the SIX6 sentinel SNP and two genome-wide significant variants from the CDKN2B-AS1 locus, with traits that represent groups in LD with glaucoma association in the loci. The chr9p21.3 POAG associated GWAS variants are in LD in European ancestry individuals (r2 > 0.1) with more than 140 GWAS catalog variants in the ∼260 kb cluster associated with vascular,191 carcinoma, and neurological traits.193 We used two variants in the locus that are associated with a whole range of the cardiovascular and cancer related traits; rs2891168_CDKN2B-AS1 (coronary artery disease, myocardial infarction, and beta blocking agent use measurement),44,194,195,196,197,198 and rs10811650_CDKN2B-AS1 (breast cancer, melanoma, and hair color).135,199,200,201 We finally determined if these interactions had effects on expression patterns of CDKN2A & CDKN2B in GTEx data brain cortex tissue. Mean expression difference between allele combinations was assessed using Student’s t test and ANOVA (Table S24).
To determine phenotypic consequences of the SIX6-CDKN2B-AS1 loci interactions, and African male-specific missense variant, we further performed TWAS-PheWAS for the two chr9p21.3 genes and CELSR2 gene (that have male-specific expression association with the missense variant) in summary statistics from the UKBB (n = 396,618) and BioVU (n = 59,805), followed by meta-analysis of the two PheWAS (n = 456,423) across 731 traits and diseases grouped into 17 categories (Table S23).148,202
We also evaluated the effect of African-specific missense variant rs74113753_TMEM167B using proxy rs17641032_TMEM167B variant (−862bp, r2 = 0.699, D’ = 1, 1000 genomes)192 on measured gene expression residuals for a total of 24 genes in a 500 kb window either side of the variant that passed quality filters in GTEx muscle skeletal tissues (n = 706) in all the samples and sex-stratified set (males = 469, females = 237).
Acknowledgments
We would like to acknowledge Dr. Anand Swaroop (NEI) for sharing genotyped data (for EyeGEx RNA Seq data at GTEx) to build PrediXcan eye tissue model, Melinda Aldrich for critical review on an earlier version of the manuscript. We also would like to acknowledge the organizing committee of the International Common Disease Alliance for intellectual contributions on the setup of the GBMI as a nascent activity to the larger effort. We thank Daniel King from the Hail team and Sam Bryant from the Stanley Center Data Management team for helping with the Google bucket setup and data sharing and Sinead Chapman for helping in paper submission.
Jibril Hirbo is supported by P50MD017347, and through grant 5U01HG011720-03 (to N.J.C.) and R35HG010718 (to E.R.G.), E.R.G. is supported by the National Institutes of Health (NIH) Awards R35HG010718, R01HG011138, and R01GM140287, and NIH/NIA AG068026. V.L.F. was supported by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 675033 (EGRET plus). Additional funding was provided by the Rotterdamse Stichting Blindenbelangen (grant no. B20150036). K.J. was supported by the Joseph Ellis Family and William Black Research Funds, and an Unrestricted Departmental Grant to the Vanderbilt Eye Institute from Research to Prevent Blindness, Inc., NY.
We acknowledge all the participants in the study and biobanks that were involved in generating POAG summary data: BioBank Japan (Yukinori Okada, Koichi Matsua, and Masahiro Kanai), BioMe (Ruth Loos, Judy Cho, Eimear Kenny, Michael Preuss, and Simon Lee), BioVU (Nancy Cox and Jibril Hirbo), Colorado Center for Personalized Medicine (Kathleen Barnes, Michelle Daya, and Chris Gignoux), deCODE Genetics (Kári Stefánsson and Unnur þorsteinsdóttir), Estonian Biobank (Andres Metspalu, Reedik Mägi, Tõnu Esko, and Priit Palta), FinnGen (Aarno Palotie, Mark Daly, Samuli Ripatti, Mitja Kurki, and Juha Karjalainen), Generation Scotland (Caroline Hayward and Riccardo Marioni), HUNT (Kristian Hveem, Cristen Willer, Sarah Graham, Ben Brumpton, and Brooke Wolford), Lifelines (Esteban Lopera, Serena Sanna, and Harold Snieder), Michigan Genomics Initiative (Sebastian Zoellner, Michael Boehnke, Lars Fritsche, and Anita Pandit), Taiwan Biobank (Yen-Feng Lin, Yen-Chen Feng, and Hailiang Huang), and UK Biobank (Konrad Karczewski and Alicia Martin).
Author contributions
Study design, V.L.F., W.Z., N.J.C., and J.H.; data collection/contribution, V.L.F., S.S., N.J.C., J.H., Y.W., E.L.-M., K.L., M.K., Y.O., P.S., P.P., A.R.M., N.M.J., L.G., S.S.V., A.V., D.J.R., J.S., K.C., C.G., and GBMI; data analysis, V.L.F., A.B., W.Z., D.Z., P.S., and J.H.; writing, V.L.F. and J.H.; revision, V.L.F., A.B., W.Z., D.Z., Y.W., K.L., M.K., E.L.-M., P.S., P.P., R.T., X.Z., S.N., S.S., I.M.N., H.S., I.S., A.R.M., M.A.B., C.W., S.M., N.I., E.R.G., N.M.J., K.J., N.J.C., and J.H. All authors provided critical inputs in interpretation of the data and approved the final version of the manuscript.
Declaration of interests
E.R.G. received an honorarium from the journal Circulation Research of the American Heart Association as a member of the Editorial Board. S.M. is a co-founder and holds stock in Seonix Pty Ltd.
Published: February 20, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2024.101430.
Supplemental information
References
- 1.Quigley H.A., Broman A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta N., Ly T., Zhang Q., Kaufman P.L., Weinreb R.N., Yücel Y.H. Chronic ocular hypertension induces dendrite pathology in the lateral geniculate nucleus of the brain. Exp. Eye Res. 2007;84:176–184. doi: 10.1016/j.exer.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Saccà S.C., Pascotto A., Camicione P., Capris P., Izzotti A. Oxidative DNA Damage in the Human Trabecular Meshwork. Arch. Ophthalmol. 2005;123:458–463. doi: 10.1001/archopht.123.4.458. [DOI] [PubMed] [Google Scholar]
- 4.Nemesure B., Honkanen R., Hennis A., Wu S.Y., Leske M.C., Barbados Eye Studies Group Incident Open-angle Glaucoma and Intraocular Pressure. Ophthalmology. 2007;114:1810–1815. doi: 10.1016/j.ophtha.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Gramer E., Grehn F. Springer Science & Business Media; 2012. Pathogenesis and Risk Factors of Glaucoma. [Google Scholar]
- 6.Tham Y.-C., Li X., Wong T.Y., Quigley H.A., Aung T., Cheng C.-Y. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Zetterberg M. Age-related eye disease and gender. Maturitas. 2016;83:19–26. doi: 10.1016/j.maturitas.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Bonnemaijer P.W.M., Lo Faro V., Sanyiwa A.J., Hassan H.G., Cook C., GIGA study group. Van de Laar S., Lemij H.G., Klaver C.C.W., Jansonius N.M., Thiadens A.A.H.J. Differences in clinical presentation of primary open-angle glaucoma between African and European populations. Acta Ophthalmol. 2021;99:e1118–e1126. doi: 10.1111/aos.14772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyari F., Abdull M.M., Bastawrous A., Gilbert C.E., Faal H. Epidemiology of glaucoma in Sub-Saharan Africa: Prevalence, incidence and risk factors. Middle East Afr. J. Ophthalmol. 2013;20:111–125. doi: 10.4103/0974-9233.110605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evangelho K., Mastronardi C.A., de-la-Torre A. Experimental Models of Glaucoma: A Powerful Translational Tool for the Future Development of New Therapies for Glaucoma in Humans—A Review of the Literature. Medicina. 2019;55:280. doi: 10.3390/medicina55060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emre M., Orgül S., Gugleta K., Flammer J. Ocular blood flow alteration in glaucoma is related to systemic vascular dysregulation. Br. J. Ophthalmol. 2004;88:662–666. doi: 10.1136/bjo.2003.032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlson M.E., de Moraes C.G., Link A., Wells M.T., Harmon G., Peterson J.C., Ritch R., Liebmann J.M. Nocturnal Systemic Hypotension Increases the Risk of Glaucoma Progression. Ophthalmology. 2014;121:2004–2012. doi: 10.1016/j.ophtha.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeganathan V.S.E., Wong T.Y., Foster P.J., Crowston J.G., Tay W.T., Lim S.C., Saw S.-M., Tai E.S., Aung T. Peripheral artery disease and glaucoma: the singapore malay eye study. Arch. Ophthalmol. 2009;127:888–893. doi: 10.1001/archophthalmol.2009.136. [DOI] [PubMed] [Google Scholar]
- 14.Broadway D.C., Drance S.M. Glaucoma and vasospasm. Br. J. Ophthalmol. 1998;82:862–870. doi: 10.1136/bjo.82.8.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flammer J., Orgül S., Costa V.P., Orzalesi N., Krieglstein G.K., Serra L.M., Renard J.-P., Stefánsson E. The impact of ocular blood flow in glaucoma. Prog. Retin. Eye Res. 2002;21:359–393. doi: 10.1016/s1350-9462(02)00008-3. [DOI] [PubMed] [Google Scholar]
- 16.Grunwald J.E., Piltz J., Hariprasad S.M., DuPont J. Optic nerve and choroidal circulation in glaucoma. Invest. Ophthalmol. Vis. Sci. 1998;39:2329–2336. [PubMed] [Google Scholar]
- 17.Flammer J. The vascular concept of glaucoma. Surv. Ophthalmol. 1994;38(Suppl):S3–S6. doi: 10.1016/0039-6257(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 18.Drance S., Anderson D.R., Schulzer M., Collaborative Normal-Tension Glaucoma Study Group Risk factors for progression of visual field abnormalities in normal-tension glaucoma. Am. J. Ophthalmol. 2001;131:699–708. doi: 10.1016/s0002-9394(01)00964-3. [DOI] [PubMed] [Google Scholar]
- 19.Cursiefen C., Wisse M., Cursiefen S., Jünemann A., Martus P., Korth M. Migraine and tension headache in high-pressure and normal-pressure glaucoma. Am. J. Ophthalmol. 2000;129:102–104. doi: 10.1016/s0002-9394(99)00289-5. [DOI] [PubMed] [Google Scholar]
- 20.Ko F., Boland M.V., Gupta P., Gadkaree S.K., Vitale S., Guallar E., Zhao D., Friedman D.S. Diabetes, Triglyceride Levels, and Other Risk Factors for Glaucoma in the National Health and Nutrition Examination Survey 2005-2008. Invest. Ophthalmol. Vis. Sci. 2016;57:2152–2157. doi: 10.1167/iovs.15-18373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wise L.A., Rosenberg L., Radin R.G., Mattox C., Yang E.B., Palmer J.R., Seddon J.M. A prospective study of diabetes, lifestyle factors, and glaucoma among African-American women. Ann. Epidemiol. 2011;21:430–439. doi: 10.1016/j.annepidem.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gharahkhani P., Jorgenson E., Hysi P., Khawaja A.P., Pendergrass S., Han X., Ong J.S., Hewitt A.W., Segrè A.V., Rouhana J.M., et al. Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries. Nat. Commun. 2021;12:1258. doi: 10.1038/s41467-020-20851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unlu G., Gamazon E.R., Qi X., Levic D.S., Bastarache L., Denny J.C., Roden D.M., Mayzus I., Breyer M., Zhong X., et al. GRIK5 Genetically Regulated Expression Associated with Eye and Vascular Phenomes: Discovery through Iteration among Biobanks, Electronic Health Records, and Zebrafish. Am. J. Hum. Genet. 2019;104:503–519. doi: 10.1016/j.ajhg.2019.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramdas W.D., van Koolwijk L.M.E., Lemij H.G., Pasutto F., Cree A.J., Thorleifsson G., Janssen S.F., Jacoline T.B., Amin N., Rivadeneira F., et al. Common genetic variants associated with open-angle glaucoma. Hum. Mol. Genet. 2011;20:2464–2471. doi: 10.1093/hmg/ddr120. [DOI] [PubMed] [Google Scholar]
- 25.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W., Kanai M., Wu K.-H.H., Humaira R., Tsuo K., Hirbo J.B., Wang Y., Bhattacharya A., Zhao H., Namba S., et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human diseases. medRxiv. 2021 doi: 10.1016/j.xgen.2022.100192. Preprint at. 10.11.19.21266436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genetics of Glaucoma in People of Genetics of Glaucoma in People of African Descent GGLAD Consortium. Hauser M.A., Allingham R.R., Aung T., Van Der Heide C.J., Taylor K.D., Rotter J.I., Wang S.-H.J., Bonnemaijer P.W.M., Williams S.E., et al. Association of Genetic Variants With Primary Open-Angle Glaucoma Among Individuals With African Ancestry. JAMA. 2019;322:1682–1691. doi: 10.1001/jama.2019.16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanigawa Y., Wainberg M., Karjalainen J., Kiiskinen T., Venkataraman G., Lemmelä S., Turunen J.A., Graham R.R., Havulinna A.S., Perola M., et al. Rare protein-altering variants in ANGPTL7 lower intraocular pressure and protect against glaucoma. PLoS Genet. 2020;16:e1008682. doi: 10.1371/journal.pgen.1008682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khawaja A.P., Cooke Bailey J.N., Wareham N.J., Scott R.A., Simcoe M., Igo R.P., Jr., Song Y.E., Wojciechowski R., Cheng C.-Y., Khaw P.T., et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 2018;50:778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mägi R., Horikoshi M., Sofer T., Mahajan A., Kitajima H., Franceschini N., McCarthy M.I., COGENT-Kidney Consortium T2D-GENES Consortium. Morris A.P., Morris A.P. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allison K., Patel D., Alabi O. Epidemiology of Glaucoma: The Past, Present, and Predictions for the Future. Cureus. 2020;12:e11686. doi: 10.7759/cureus.11686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapetanakis V.V., Chan M.P.Y., Foster P.J., Cook D.G., Owen C.G., Rudnicka A.R. Global variations and time trends in the prevalence of primary open angle glaucoma (POAG): a systematic review and meta-analysis. Br. J. Ophthalmol. 2016;100:86–93. doi: 10.1136/bjophthalmol-2015-307223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMonnies C.W. Glaucoma history and risk factors. J. Opt. 2017;10:71–78. doi: 10.1016/j.optom.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choquet H., Paylakhi S., Kneeland S.C., Thai K.K., Hoffmann T.J., Yin J., Kvale M.N., Banda Y., Tolman N.G., Williams P.A., et al. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat. Commun. 2018;9:2278. doi: 10.1038/s41467-018-04555-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khor C.C., Do T., Jia H., Nakano M., George R., Abu-Amero K., Duvesh R., Chen L.J., Li Z., Nongpiur M.E., et al. Genome-wide association study identifies five new susceptibility loci for primary angle closure glaucoma. Nat. Genet. 2016;48:556–562. doi: 10.1038/ng.3540. [DOI] [PubMed] [Google Scholar]
- 36.Jelodari-Mamaghani S., Haji-Seyed-Javadi R., Suri F., Nilforushan N., Yazdani S., Kamyab K., Elahi E. Contribution of the latent transforming growth factor-β binding protein 2 gene to etiology of primary open angle glaucoma and pseudoexfoliation syndrome. Mol. Vis. 2013;19:333–347. [PMC free article] [PubMed] [Google Scholar]
- 37.Du L., Li C.-R., He Q.-F., Li X.-H., Yang L.-F., Zou Y., Yang Z.-X., Zhang D., Xing X.-W. Downregulation of the ubiquitin ligase KBTBD8 prevented epithelial ovarian cancer progression. Mol. Med. 2020;26:96. doi: 10.1186/s10020-020-00226-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamada Y., Yasukochi Y., Kato K., Oguri M., Horibe H., Fujimaki T., Takeuchi I., Sakuma J. Identification of 26 novel loci that confer susceptibility to early-onset coronary artery disease in a Japanese population. Biomed. Rep. 2018;9:383–404. doi: 10.3892/br.2018.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kan Z., Jaiswal B.S., Stinson J., Janakiraman V., Bhatt D., Stern H.M., Yue P., Haverty P.M., Bourgon R., Zheng J., et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 40.Simonson B., Subramanya V., Chan M.C., Zhang A., Franchino H., Ottaviano F., Mishra M.K., Knight A.C., Hunt D., Ghiran I., et al. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci. Signal. 2017;10:eaaf5967. doi: 10.1126/scisignal.aaf5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams S., Bateman A., O’Kelly I. Altered expression of two-pore domain potassium (K2P) channels in cancer. PLoS One. 2013;8:e74589. doi: 10.1371/journal.pone.0074589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pardo L.A., Stühmer W. The roles of K channels in cancer. Nat. Rev. Cancer. 2014;14:39–48. doi: 10.1038/nrc3635. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Baron C.P., Jonsson P., Thomas C., Dryer S.E., Williams C. The Two-Pore Domain Potassium Channel KCNK5: Induction by Estrogen Receptor α and Role in Proliferation of Breast Cancer Cells. Mol. Endocrinol. 2011;25:1326–1336. doi: 10.1210/me.2011-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Harst P., Verweij N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukasaki K., Miller C.W., Greenspun E., Eshaghian S., Kawabata H., Fujimoto T., Tomonaga M., Sawyers C., Said J.W., Koeffler H.P. Mutations in the mitotic check point gene, MAD1L1, in human cancers. Oncogene. 2001;20:3301–3305. doi: 10.1038/sj.onc.1204421. [DOI] [PubMed] [Google Scholar]
- 46.Barbieri M., Esposito A., Angellotti E., Rizzo M.R., Marfella R., Paolisso G. Association of genetic variation in adaptor protein APPL1/APPL2 loci with non-alcoholic fatty liver disease. PLoS One. 2013;8:e71391. doi: 10.1371/journal.pone.0071391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma X.-W., Ding S., Ma X.-D., Gu N., Guo X.-H. Genetic variability in adapter proteins with APPL1/2 is associated with the risk of coronary artery disease in type 2 diabetes mellitus in Chinese Han population. Chin. Med. J. 2011;124:3618–3621. [PubMed] [Google Scholar]
- 48.Li Y., Zhang W., Liu P., Xu Y., Tang L., Chen W., Guan X. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. OncoTargets Ther. 2018;11:1403–1412. doi: 10.2147/OTT.S149511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J., Du W. LncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein. OncoTargets Ther. 2019;12:4287–4295. doi: 10.2147/OTT.S195853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bianconi E., Casadei R., Frabetti F., Ventura C., Facchin F., Canaider S. Sex-Specific Transcriptome Differences in Human Adipose Mesenchymal Stem Cells. Genes. 2020;11:909. doi: 10.3390/genes11080909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oliva M., Muñoz-Aguirre M., Kim-Hellmuth S., Wucher V., Gewirtz A.D.H., Cotter D.J., Parsana P., Kasela S., Balliu B., Viñuela A., et al. The impact of sex on gene expression across human tissues. Science. 2020;369:eaba3066. doi: 10.1126/science.aba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.1000 Genomes Project Consortium. Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. G.P., and The 1000 Genomes Project Consortium (2010). A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., et al. A global reference for human genetic variation (2015) Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daya M., Rafaels N., Brunetti T.M., Chavan S., Levin A.M., Shetty A., Gignoux C.R., Boorgula M.P., Wojcik G., Campbell M., et al. Association study in African-admixed populations across the Americas recapitulates asthma risk loci in non-African populations. Nat. Commun. 2019;10:880. doi: 10.1038/s41467-019-08469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belbin G.M., Cullina S., Wenric S., Soper E.R., Glicksberg B.S., Torre D., Moscati A., Wojcik G.L., Shemirani R., Beckmann N.D., et al. Toward a fine-scale population health monitoring system. Cell. 2021;184:2068–2083.e11. doi: 10.1016/j.cell.2021.03.034. [DOI] [PubMed] [Google Scholar]
- 57.Vishnu A., Belbin G.M., Wojcik G.L., Bottinger E.P., Gignoux C.R., Kenny E.E., Loos R.J.F. The role of country of birth, and genetic and self-identified ancestry, in obesity susceptibility among African and Hispanic Americans. Am. J. Clin. Nutr. 2019;110:16–23. doi: 10.1093/ajcn/nqz098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Craig J.E., Han X., Qassim A., Hassall M., Cooke Bailey J.N., Kinzy T.G., Khawaja A.P., An J., Marshall H., Gharahkhani P., et al. Multitrait analysis of glaucoma identifies new risk loci and enables polygenic prediction of disease susceptibility and progression. Nat. Genet. 2020;52:160–166. doi: 10.1038/s41588-019-0556-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fingert J.H., Héon E., Liebmann J.M., Yamamoto T., Craig J.E., Rait J., Kawase K., Hoh S.T., Buys Y.M., Dickinson J., et al. Analysis of myocilin mutations in 1703 glaucoma patients from five different populations. Hum. Mol. Genet. 1999;8:899–905. doi: 10.1093/hmg/8.5.899. [DOI] [PubMed] [Google Scholar]
- 60.Allingham R.R., Wiggs J.L., De La Paz M.A., Vollrath D., Tallett D.A., Broomer B., Jones K.H., Del Bono E.A., Kern J., Patterson K., et al. Gln368STOP myocilin mutation in families with late-onset primary open-angle glaucoma. Invest. Ophthalmol. Vis. Sci. 1998;39:2288–2295. [PubMed] [Google Scholar]
- 61.Wiggs J.L., Allingham R.R., Vollrath D., Jones K.H., De La Paz M., Kern J., Patterson K., Babb V.L., Del Bono E.A., Broomer B.W., et al. Prevalence of mutations in TIGR/Myocilin in patients with adult and juvenile primary open-angle glaucoma. Am. J. Hum. Genet. 1998;63:1549–1552. doi: 10.1086/302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Angius A., Spinelli P., Ghilotti G., Casu G., Sole G., Loi A., Totaro A., Zelante L., Gasparini P., Orzalesi N., et al. Myocilin Gln368stop mutation and advanced age as risk factors for late-onset primary open-angle glaucoma. Arch. Ophthalmol. 2000;118:674–679. doi: 10.1001/archopht.118.5.674. [DOI] [PubMed] [Google Scholar]
- 63.Hewitt A.W., Mackey D.A., Craig J.E. Myocilin allele-specific glaucoma phenotype database. Hum. Mutat. 2008;29:207–211. doi: 10.1002/humu.20634. [DOI] [PubMed] [Google Scholar]
- 64.Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scelsi H.F., Barlow B.M., Saccuzzo E.G., Lieberman R.L. Common and rare myocilin variants: Predicting glaucoma pathogenicity based on genetics, clinical, and laboratory misfolding data. Hum. Mutat. 2021;42:903–946. doi: 10.1002/humu.24238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alward W.L.M., van der Heide C., Khanna C.L., Roos B.R., Sivaprasad S., Kam J., Ritch R., Lotery A., Igo R.P., Jr., Cooke Bailey J.N., et al. Myocilin Mutations in Patients With Normal-Tension Glaucoma. JAMA Ophthalmol. 2019;137:559–563. doi: 10.1001/jamaophthalmol.2019.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baird P.N., Richardson A.J., Mackey D.A., Craig J.E., Faucher M., Raymond V. A common disease haplotype for the Q368STOP mutation of the myocilin gene in Australian and Canadian glaucoma families. Am. J. Ophthalmol. 2005;140:760–762. doi: 10.1016/j.ajo.2005.04.043. [DOI] [PubMed] [Google Scholar]
- 68.Baird P.N., Craig J.E., Richardson A.J., Ring M.A., Sim P., Stanwix S., Foote S.J., Mackey D.A. Analysis of 15 primary open-angle glaucoma families from Australia identifies a founder effect for the Q368STOP mutation of myocilin. Hum. Genet. 2003;112:110–116. doi: 10.1007/s00439-002-0865-5. [DOI] [PubMed] [Google Scholar]
- 69.Faucher M., Anctil J.-L., Rodrigue M.-A., Duchesne A., Bergeron D., Blondeau P., Côté G., Dubois S., Bergeron J., Arseneault R., et al. Founder TIGR/myocilin mutations for glaucoma in the Québec population. Hum. Mol. Genet. 2002;11:2077–2090. doi: 10.1093/hmg/11.18.2077. [DOI] [PubMed] [Google Scholar]
- 70.Sale M.M., FitzGerald L.M., Kagame K., Erdmann I., Craig J.E., Dickinson J.L., Cooper R.L. Investigation of the prevalence of the myocilin Q368STOP mutation in Ugandan glaucoma patients. Ophthalmic Genet. 2002;23:67–69. doi: 10.1076/opge.23.1.67.2204. [DOI] [PubMed] [Google Scholar]
- 71.Ratnapriya R., Sosina O.A., Starostik M.R., Kwicklis M., Kapphahn R.J., Fritsche L.G., Walton A., Arvanitis M., Gieser L., Pietraszkiewicz A., et al. Retinal transcriptome and eQTL analyses identify genes associated with age-related macular degeneration. Nat. Genet. 2019;51:606–610. doi: 10.1038/s41588-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Teis D., Taub N., Kurzbauer R., Hilber D., de Araujo M.E., Erlacher M., Offterdinger M., Villunger A., Geley S., Bohn G., et al. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 2006;175:861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N., Ding Y.U., Yu T., Li J., Shen Y., Wang X., Fu Q., Shen Y., Huang X., Wang J. Causal variants screened by whole exome sequencing in a patient with maternal uniparental isodisomy of chromosome 10 and a complicated phenotype. Exp. Ther. Med. 2016;11:2247–2253. doi: 10.3892/etm.2016.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van den Berg M.E., Warren H.R., Cabrera C.P., Verweij N., Mifsud B., Haessler J., Bihlmeyer N.A., Fu Y.-P., Weiss S., Lin H.J., et al. Discovery of novel heart rate-associated loci using the Exome Chip. Hum. Mol. Genet. 2017;26:2346–2363. doi: 10.1093/hmg/ddx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yehia L., Niazi F., Ni Y., Ngeow J., Sankunny M., Liu Z., Wei W., Mester J.L., Keri R.A., Zhang B., Eng C. Germline Heterozygous Variants in SEC23B Are Associated with Cowden Syndrome and Enriched in Apparently Sporadic Thyroid Cancer. Am. J. Hum. Genet. 2015;97:661–676. doi: 10.1016/j.ajhg.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang C., Chen N., Li X., Lu D., Hou Z., Li Y., Jin Y., Gu J., Yin Y. Mutations in the coat complex II component SEC23B promote colorectal cancer metastasis. Cell Death Dis. 2020;11:157. doi: 10.1038/s41419-020-2358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wagner A.H., Anand V.N., Wang W.-H., Chatterton J.E., Sun D., Shepard A.R., Jacobson N., Pang I.-H., Deluca A.P., Casavant T.L., et al. Exon-level expression profiling of ocular tissues. Exp. Eye Res. 2013;111:105–111. doi: 10.1016/j.exer.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bryan J.M., Fufa T.D., Bharti K., Brooks B.P., Hufnagel R.B., McGaughey D.M. Identifying core biological processes distinguishing human eye tissues with precise systems-level gene expression analyses and weighted correlation networks. Hum. Mol. Genet. 2018;27:3325–3339. doi: 10.1093/hmg/ddy239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M., et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 80.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 81.Wang Y., Namba S., Lopera E., Kerminen S., Tsuo K., Läll K., Kanai M., Zhou W., Wu K.-H., Favé M.-J., et al. Global biobank analyses provide lessons for computing polygenic risk scores across diverse cohorts. medRxiv. 2021 doi: 10.1016/j.xgen.2022.100241. Preprint at. 11.18.21266545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328–3329. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y., Guo J., Ni G., Yang J., Visscher P.M., Yengo L. Theoretical and empirical quantification of the accuracy of polygenic scores in ancestry divergent populations. Nat. Commun. 2020;11:3865. doi: 10.1038/s41467-020-17719-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., et al. GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hu Y., Li M., Lu Q., Weng H., Wang J., Zekavat S.M., Yu Z., Li B., Gu J., Muchnik S., et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat. Genet. 2019;51:568–576. doi: 10.1038/s41588-019-0345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou D., Jiang Y., Zhong X., Cox N.J., Liu C., Gamazon E.R. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat. Genet. 2020;52:1239–1246. doi: 10.1038/s41588-020-0706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang H., Wang Y., Guan L., Chen Y., Chen P., Sun J., Gonzalez F.J., Huang M., Bi H. Lipidomics reveals carnitine palmitoyltransferase 1C protects cancer cells from lipotoxicity and senescence. J. Pharm. Anal. 2021;11:340–350. doi: 10.1016/j.jpha.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Millner A., Atilla-Gokcumen G.E. Lipid Players of Cellular Senescence. Metabolites. 2020;10:339. doi: 10.3390/metabo10090339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang L., Hauser E.R., Shah S.H., Pericak-Vance M.A., Haynes C., Crosslin D., Harris M., Nelson S., Hale A.B., Granger C.B., et al. Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am. J. Hum. Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ji J.-Z., Elyaman W., Yip H.K., Lee V.W.H., Yick L.-W., Hugon J., So K.-F. CNTF promotes survival of retinal ganglion cells after induction of ocular hypertension in rats: the possible involvement of STAT3 pathway. Eur. J. Neurosci. 2004;19:265–272. doi: 10.1111/j.0953-816x.2003.03107.x. [DOI] [PubMed] [Google Scholar]
- 93.Pease M.E., Zack D.J., Berlinicke C., Bloom K., Cone F., Wang Y., Klein R.L., Hauswirth W.W., Quigley H.A. Effect of CNTF on retinal ganglion cell survival in experimental glaucoma. Invest. Ophthalmol. Vis. Sci. 2009;50:2194–2200. doi: 10.1167/iovs.08-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shpak A.A., Guekht A.B., Druzhkova T.A., Kozlova K.I., Gulyaeva N.V. Ciliary neurotrophic factor in patients with primary open-angle glaucoma and age-related cataract. Mol. Vis. 2017;23:799–809. [PMC free article] [PubMed] [Google Scholar]
- 95.Xu S., Xu H., Wang W., Li S., Li H., Li T., Zhang W., Yu X., Liu L. The role of collagen in cancer: from bench to bedside. J. Transl. Med. 2019;17:309. doi: 10.1186/s12967-019-2058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Youle R.J., van der Bliek A.M. Mitochondrial Fission, Fusion, and Stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finak G., Bertos N., Pepin F., Sadekova S., Souleimanova M., Zhao H., Chen H., Omeroglu G., Meterissian S., Omeroglu A., et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat. Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 98.Karnoub A.E., Dash A.B., Vo A.P., Sullivan A., Brooks M.W., Bell G.W., Richardson A.L., Polyak K., Tubo R., Weinberg R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 99.Chiappetta G., Manfioletti G., Pentimalli F., Abe N., Di Bonito M., Vento M.T., Giuliano A., Fedele M., Viglietto G., Santoro M., et al. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int. J. Cancer. 2001;91:147–151. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1033>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 100.Zhou X., Chada K. HMGI family proteins: architectural transcription factors in mammalian development and cancer. Keio J. Med. 1998;47:73–77. doi: 10.2302/kjm.47.73. [DOI] [PubMed] [Google Scholar]
- 101.Krupenko S.A., Krupenko N.I. ALDH1L1 and ALDH1L2 Folate Regulatory Enzymes in Cancer. Adv. Exp. Med. Biol. 2018;1032:127–143. doi: 10.1007/978-3-319-98788-0_10. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez J.M., Jr. Existence of the canonical Wnt signaling pathway in the human trabecular meshwork. Invest. Ophthalmol. Vis. Sci. 2012;53:6972. doi: 10.1167/iovs.12-10985. [DOI] [PubMed] [Google Scholar]
- 103.Wang W.-H., McNatt L.G., Pang I.-H., Millar J.C., Hellberg P.E., Hellberg M.H., Steely H.T., Rubin J.S., Fingert J.H., Sheffield V.C., et al. Increased expression of the WNT antagonist sFRP-1 in glaucoma elevates intraocular pressure. J. Clin. Invest. 2008;118:1056–1064. doi: 10.1172/JCI33871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang X., Huai G., Wang H., Liu Y., Qi P., Shi W., Peng J., Yang H., Deng S., Wang Y. Mutual regulation of the Hippo/Wnt/LPA/TGF-β signaling pathways and their roles in glaucoma (Review) Int. J. Mol. Med. 2018;41:1201–1212. doi: 10.3892/ijmm.2017.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han X., Gharahkhani P., Hamel A.R., Ong J.S., Rentería M.E., Mehta P., Dong X., Pasutto F., Hammond C., Young T.L., et al. Large-scale multitrait genome-wide association analyses identify hundreds of glaucoma risk loci. Nat. Genet. 2023;55:1116–1125. doi: 10.1038/s41588-023-01428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tan J., Che Y., Liu Y., Hu J., Wang W., Hu L., Zhou Q., Wang H., Li J. CELSR2 deficiency suppresses lipid accumulation in hepatocyte by impairing the UPR and elevating ROS level. Faseb. J. 2021;35:e21908. doi: 10.1096/fj.202100786RR. [DOI] [PubMed] [Google Scholar]
- 107.Hjeij R., Lindstrand A., Francis R., Zariwala M.A., Liu X., Li Y., Damerla R., Dougherty G.W., Abouhamed M., Olbrich H., et al. ARMC4 mutations cause primary ciliary dyskinesia with randomization of left/right body asymmetry. Am. J. Hum. Genet. 2013;93:357–367. doi: 10.1016/j.ajhg.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Onoufriadis A., Shoemark A., Munye M.M., James C.T., Schmidts M., Patel M., Rosser E.M., Bacchelli C., Beales P.L., Scambler P.J., et al. Combined exome and whole-genome sequencing identifies mutations in ARMC4 as a cause of primary ciliary dyskinesia with defects in the outer dynein arm. J. Med. Genet. 2014;51:61–67. doi: 10.1136/jmedgenet-2013-101938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Samani N.J., Braund P.S., Erdmann J., Götz A., Tomaszewski M., Linsel-Nitschke P., Hajat C., Mangino M., Hengstenberg C., Stark K., et al. The novel genetic variant predisposing to coronary artery disease in the region of the PSRC1 and CELSR2 genes on chromosome 1 associates with serum cholesterol. J. Mol. Med. 2008;86:1233–1241. doi: 10.1007/s00109-008-0387-2. [DOI] [PubMed] [Google Scholar]
- 110.Jiang L., Zhang X., Xiang C., Geradts J., Wei Q., Liang Y., Huang H., Xu J.-F. Differential cellular localization of CELSR2 and ING4 and correlations with hormone receptor status in breast cancer. Histol. Histopathol. 2018;33:835–842. doi: 10.14670/HH-11-979. [DOI] [PubMed] [Google Scholar]
- 111.Yamada Y., Toyota M., Hirokawa Y., Suzuki H., Takagi A., Matsuzaki T., Sugimura Y., Yatani R., Shiraishi T., Watanabe M. Identification of differentially methylated CpG islands in prostate cancer. Int. J. Cancer. 2004;112:840–845. doi: 10.1002/ijc.20335. [DOI] [PubMed] [Google Scholar]
- 112.Xu M., Zhu S., Xu R., Lin N. Identification of CELSR2 as a novel prognostic biomarker for hepatocellular carcinoma. BMC Cancer. 2020;20:313. doi: 10.1186/s12885-020-06813-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Namba S., Konuma T., Wu K.-H., Zhou W., Okada Y., Global Biobank Meta-analysis Initiative A practical guideline of genomics-driven drug discovery in the era of global biobank meta-analysis. medRxiv. 2021 doi: 10.1016/j.xgen.2022.100190. Preprint at. 12.03.21267280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cole B.S., Gudiseva H.V., Pistilli M., Salowe R., McHugh C.P., Zody M.C., Chavali V.R.M., Ying G.S., Moore J.H., O’Brien J.M. The Role of Genetic Ancestry as a Risk Factor for Primary Open-angle Glaucoma in African Americans. Invest. Ophthalmol. Vis. Sci. 2021;62:28. doi: 10.1167/iovs.62.2.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y., Yan L., Liu J., Cui S., Qiu J. cGMP-dependent protein kinase II determines β-catenin accumulation that is essential for uterine decidualization in mice. Am. J. Physiol. Cell Physiol. 2019;317:C1115–C1127. doi: 10.1152/ajpcell.00208.2019. [DOI] [PubMed] [Google Scholar]
- 116.Fotesko K., Thomsen B.S.V., Kolko M., Vohra R. Girl Power in Glaucoma: The Role of Estrogen in Primary Open Angle Glaucoma. Cell. Mol. Neurobiol. 2022;42:41–57. doi: 10.1007/s10571-020-00965-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Salowe R., Salinas J., Farbman N.H., Mohammed A., Warren J.Z., Rhodes A., Brucker A., Regina M., Miller-Ellis E., Sankar P.S., et al. Primary Open-Angle Glaucoma in Individuals of African Descent: A Review of Risk Factors. J. Clin. Exp. Ophthalmol. 2015;6:450. doi: 10.4172/2155-9570.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Siesky B., Harris A., Racette L., Abassi R., Chandrasekhar K., Tobe L.A., Behzadi J., Eckert G., Amireskandari A., Muchnik M. Differences in ocular blood flow in glaucoma between patients of African and European descent. J. Glaucoma. 2015;24:117–121. doi: 10.1097/IJG.0b013e31829d9bb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.The Advanced Glaucoma Intervention Study (AGIS): 3. Baseline characteristics of black and white patients (1998). Ophthalmology 105, 1137–1145. [DOI] [PubMed]
- 120.Khachatryan N., Pistilli M., Maguire M.G., Salowe R.J., Fertig R.M., Moore T., Gudiseva H.V., Chavali V.R.M., Collins D.W., Daniel E., et al. Primary Open-Angle African American Glaucoma Genetics (POAAGG) Study: gender and risk of POAG in African Americans. PLoS One. 2019;14:e0218804. doi: 10.1371/journal.pone.0218804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ye X., She X., Shen L. Association of sex with the global burden of glaucoma: an analysis from the global burden of disease study 2017. Acta Ophthalmol. 2020;98:e593–e598. doi: 10.1111/aos.14330. [DOI] [PubMed] [Google Scholar]
- 122.Carnes M.U., Liu Y.P., Allingham R.R., Whigham B.T., Havens S., Garrett M.E., Qiao C., Katsanis N., Wiggs J.L., et al. NEIGHBORHOOD Consortium Investigators Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS Genet. 2014;10:e1004372. doi: 10.1371/journal.pgen.1004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Teotia P., Van Hook M.J., Wichman C.S., Allingham R.R., Hauser M.A., Ahmad I. Modeling Glaucoma: Retinal Ganglion Cells Generated from Induced Pluripotent Stem Cells of Patients with SIX6 Risk Allele Show Developmental Abnormalities. Stem Cell. 2017;35:2239–2252. doi: 10.1002/stem.2675. [DOI] [PubMed] [Google Scholar]
- 124.Iglesias A.I., Springelkamp H., van der Linde H., Severijnen L.-A., Amin N., Oostra B., Kockx C.E.M., van den Hout M.C.G.N., van IJcken W.F.J., Hofman A., et al. Exome sequencing and functional analyses suggest that SIX6 is a gene involved in an altered proliferation–differentiation balance early in life and optic nerve degeneration at old age. Hum. Mol. Genet. 2014;23:1320–1332. doi: 10.1093/hmg/ddt522. [DOI] [PubMed] [Google Scholar]
- 125.Skowronska-Krawczyk D., Zhao L., Zhu J., Weinreb R.N., Cao G., Luo J., Flagg K., Patel S., Wen C., Krupa M., et al. P16INK4a Upregulation Mediated by SIX6 Defines Retinal Ganglion Cell Pathogenesis in Glaucoma. Mol. Cell. 2015;59:931–940. doi: 10.1016/j.molcel.2015.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vasconcellos J.P.C., Silva Y.C.O., Rodrigues T.A.R., da Silva F.C., Alves M., Araki M.V.R., Schimiti R.B., Costa V.P., Melo M.B. Association of SALL1 rs1362756 and SIX1/SIX6 rs33912345 variants with POAG in a Brazilian population. Invest. Ophthalmol. Vis. Sci. 2020;61:1250. [Google Scholar]
- 127.Staples C.J., Myers K.N., Beveridge R.D.D., Patil A.A., Howard A.E., Barone G., Lee A.J.X., Swanton C., Howell M., Maslen S., et al. Ccdc13 is a novel human centriolar satellite protein required for ciliogenesis and genome stability. J. Cell Sci. 2014;127:2910–2919. doi: 10.1242/jcs.147785. [DOI] [PubMed] [Google Scholar]
- 128.Yang N., Leung E.L.-H., Liu C., Li L., Eguether T., Jun Yao X.-J., Jones E.C., Norris D.A., Liu A., Clark R.A., et al. INTU is essential for oncogenic Hh signaling through regulating primary cilia formation in basal cell carcinoma. Oncogene. 2017;36:4997–5005. doi: 10.1038/onc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nobbio L., Fiorese F., Vigo T., Cilli M., Gherardi G., Grandis M., Melcangi R.C., Mancardi G., Abbruzzese M., Schenone A. Impaired expression of ciliary neurotrophic factor in Charcot-Marie-Tooth type 1A neuropathy. J. Neuropathol. Exp. Neurol. 2009;68:441–455. doi: 10.1097/NEN.0b013e31819fa6ba. [DOI] [PubMed] [Google Scholar]
- 130.Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Steggink L.C., Boer H., Meijer C., Lefrandt J.D., Terstappen L.W.M.M., Fehrmann R.S.N., Gietema J.A. Genome-wide association study of cardiovascular disease in testicular cancer patients treated with platinum-based chemotherapy. Pharmacogenomics J. 2021;21:152–164. doi: 10.1038/s41397-020-00191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tan P. Intrinsic Subtypes of Gastric Cancer, Based on Gene Expression Pattern, Predict Survival and Respond Differently to Chemotherapy. Sci. Tech. Rep. 2011 doi: 10.4016/32808.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liang Y., Jiang L., Zhong X., Hochwald S.N., Wang Y., Huang L., Nie Q., Huang H., Xu J.-F. Discovery of Aberrant Alteration of Genome in Colorectal Cancer by Exome Sequencing. Am. J. Med. Sci. 2019;358:340–349. doi: 10.1016/j.amjms.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 134.Boutin T.S., Charteris D.G., Chandra A., Campbell S., Hayward C., Campbell A., Hinds D., Hinds D., et al. 23andMe Research Team, , . UK Biobank Eye & Vision Consortium Insights into the genetic basis of retinal detachment. Hum. Mol. Genet. 2020;29:689–702. doi: 10.1093/hmg/ddz294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kichaev G., Bhatia G., Loh P.-R., Gazal S., Burch K., Freund M.K., Schoech A., Pasaniuc B., Price A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019;104:65–75. doi: 10.1016/j.ajhg.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ma N., Zhou J. Functions of Endothelial Cilia in the Regulation of Vascular Barriers. Front. Cell Dev. Biol. 2020;8:626. doi: 10.3389/fcell.2020.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pala R., Jamal M., Alshammari Q., Nauli S.M. The Roles of Primary Cilia in Cardiovascular Diseases. Cells. 2018;7:233. doi: 10.3390/cells7120233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hackam A.S. The Wnt signaling pathway in retinal degenerations. IUBMB Life. 2005;57:381–388. doi: 10.1080/15216540500137586. [DOI] [PubMed] [Google Scholar]
- 139.Lo Faro V., Ten Brink J.B., Snieder H., Jansonius N.M., Bergen A.A. Genome-wide CNV investigation suggests a role for cadherin, Wnt, and p53 pathways in primary open-angle glaucoma. BMC Genom. 2021;22:590. doi: 10.1186/s12864-021-07846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun C., Zhou J., Meng X. Primary cilia in retinal pigment epithelium development and diseases. J. Cell Mol. Med. 2021;25:9084–9088. doi: 10.1111/jcmm.16882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.May-Simera H.L., Wan Q., Jha B.S., Hartford J., Khristov V., Dejene R., Chang J., Patnaik S., Lu Q., Banerjee P., et al. Primary Cilium-Mediated Retinal Pigment Epithelium Maturation Is Disrupted in Ciliopathy Patient Cells. Cell Rep. 2018;22:189–205. doi: 10.1016/j.celrep.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dilan T.L., Singh R.K., Saravanan T., Moye A., Goldberg A.F.X., Stoilov P., Ramamurthy V. Bardet-Biedl syndrome-8 (BBS8) protein is crucial for the development of outer segments in photoreceptor neurons. Hum. Mol. Genet. 2018;27:283–294. doi: 10.1093/hmg/ddx399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang W., Li L., Su Q., Gao G., Khanna H. Gene Therapy Using a miniCEP290 Fragment Delays Photoreceptor Degeneration in a Mouse Model of Leber Congenital Amaurosis. Hum. Gene Ther. 2018;29:42–50. doi: 10.1089/hum.2017.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou P., Zhou J. The Primary Cilium as a Therapeutic Target in Ocular Diseases. Front. Pharmacol. 2020;11:977. doi: 10.3389/fphar.2020.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Takahashi H., Noda S., Mashima Y., Kubota R., Ohtake Y., Tanino T., Kudoh J., Minoshima S., Oguchi Y., Shimizu N. The myocilin (MYOC) gene expression in the human trabecular meshwork. Curr. Eye Res. 2000;20:81–84. [PubMed] [Google Scholar]
- 146.Itakura T., Peters D.M., Fini M.E. Glaucomatous MYOC mutations activate the IL-1/NF-κB inflammatory stress response and the glaucoma marker SELE in trabecular meshwork cells. Mol. Vis. 2015;21:1071–1084. [PMC free article] [PubMed] [Google Scholar]
- 147.O’Brien E.T., Ren X., Wang Y. Localization of myocilin to the golgi apparatus in Schlemm’s canal cells. Invest. Ophthalmol. Vis. Sci. 2000;41:3842–3849. [PubMed] [Google Scholar]
- 148.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Scholtens S., Smidt N., Swertz M.A., Bakker S.J.L., Dotinga A., Vonk J.M., van Dijk F., van Zon S.K.R., Wijmenga C., Wolffenbuttel B.H.R., Stolk R.P. Cohort Profile: LifeLines, a three-generation cohort study and biobank. Int. J. Epidemiol. 2015;44:1172–1180. doi: 10.1093/ije/dyu229. [DOI] [PubMed] [Google Scholar]
- 150.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Verma A., Damrauer S.M., Naseer N., Weaver J., Kripke C.M., Guare L., Sirugo G., Kember R.L., Drivas T.G., Dudek S.M., et al. The Penn Medicine BioBank: Towards a Genomics-Enabled Learning Healthcare System to Accelerate Precision Medicine in a Diverse Population. J. Personalized Med. 2022;12:1974. doi: 10.3390/jpm12121974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wiley L.K., Shortt J.A., Roberts E.R., Lowery J., Kudron E., Lin M., Mayer D.A., Wilson M.P., Brunetti T.M., Chavan S., et al. Building a Vertically-Integrated Genomic Learning Health System: The Colorado Center for Personalized Medicine Biobank. medRxiv. 2022 doi: 10.1101/2022.06.09.22276222. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.All of Us Research Program Investigators. Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E. The “All of Us” Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhou W., Kanai M., Wu K.-H.H., Rasheed H., Tsuo K., Hirbo J.B., Wang Y., Bhattacharya A., Zhao H., Namba S., et al. Global Biobank Meta-analysis Initiative: Powering genetic discovery across human disease. Cell Genom. 2022;2:100192. doi: 10.1016/j.xgen.2022.100192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Aung T., Ozaki M., Lee M.C., Schlötzer-Schrehardt U., Thorleifsson G., Mizoguchi T., Igo R.P., Jr., Haripriya A., Williams S.E., Astakhov Y.S., et al. Genetic association study of exfoliation syndrome identifies a protective rare variant at LOXL1 and five new susceptibility loci. Nat. Genet. 2017;49:993–1004. doi: 10.1038/ng.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vithana E.N., Khor C.-C., Qiao C., Nongpiur M.E., George R., Chen L.-J., Do T., Abu-Amero K., Huang C.K., Low S., et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat. Genet. 2012;44:1142–1146. doi: 10.1038/ng.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Heeg G.P., Blanksma L.J., Hardus P.L.L.J., Jansonius N.M. The Groningen Longitudinal Glaucoma Study. I. Baseline sensitivity and specificity of the frequency doubling perimeter and the GDx nerve fibre analyser. Acta Ophthalmol. Scand. 2005;83:46–52. doi: 10.1111/j.1600-0420.2005.00423.x. [DOI] [PubMed] [Google Scholar]
- 159.Lo Faro V., Nolte I.M., ten Brink J.B., Snieder H., Jansonius N.M., Bergen A.A., Lifelines Cohort Study, Lifelines Cohort Study (1AD). Mitochondrial genome study identifies association between primary open-angle glaucoma and variants in MT-CYB, MT-ND4 genes and haplogroups. Front. Genet. 0. 2021;12:781189. doi: 10.3389/fgene.2021.781189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Oscanoa J., Sivapalan L., Gadaleta E., Dayem Ullah A.Z., Lemoine N.R., Chelala C. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update) Nucleic Acids Res. 2020;48:W185–W192. doi: 10.1093/nar/gkaa420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pers T.H., Karjalainen J.M., Chan Y., Westra H.-J., Wood A.R., Yang J., Lui J.C., Vedantam S., Gustafsson S., Esko T., et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Friedland A.B. Relationship between arterial pulsations and intraocular pressure. Exp. Eye Res. 1983;37:421–428. doi: 10.1016/0014-4835(83)90017-9. [DOI] [PubMed] [Google Scholar]
- 167.Guidoboni G., Sacco R., Szopos M., Sala L., Verticchio Vercellin A.C., Siesky B., Harris A. Neurodegenerative Disorders of the Eye and of the Brain: A Perspective on Their Fluid-Dynamical Connections and the Potential of Mechanism-Driven Modeling. Front. Neurosci. 2020;14:566428. doi: 10.3389/fnins.2020.566428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chan J.W., Chan N.C.Y., Sadun A.A. Glaucoma as Neurodegeneration in the Brain. Eye Brain. 2021;13:21–28. doi: 10.2147/eb.s293765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Website Michigan Imputation Server. https://imputationserver.sph.umich.edu/.
- 171.Delaneau O., Marchini J., 1000 Genomes Project Consortium. 1000 Genomes Project Consortium Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 2014;5:3934. doi: 10.1038/ncomms4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ge T., Chen C.-Y., Ni Y., Feng Y.-C.A., Smoller J.W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Bailey J.N.C., Loomis S.J., Kang J.H., Allingham R.R., Gharahkhani P., Khor C.C., Burdon K.P., Aschard H., Chasman D.I., Igo R.P., Jr., et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat. Genet. 2016;48:189–194. doi: 10.1038/ng.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Osman W., Low S.-K., Takahashi A., Kubo M., Nakamura Y. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum. Mol. Genet. 2012;21:2836–2842. doi: 10.1093/hmg/dds103. [DOI] [PubMed] [Google Scholar]
- 176.Springelkamp H., Iglesias A.I., Mishra A., Höhn R., Wojciechowski R., Khawaja A.P., Nag A., Wang Y.X., Wang J.J., Cuellar-Partida G., et al. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum. Mol. Genet. 2017;26:438–453. doi: 10.1093/hmg/ddw399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gharahkhani P., Burdon K.P., Fogarty R., Sharma S., Hewitt A.W., Martin S., Law M.H., Cremin K., Bailey J.N.C., Loomis S.J., et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat. Genet. 2014;46:1120–1125. doi: 10.1038/ng.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Nakano M., Ikeda Y., Tokuda Y., Fuwa M., Omi N., Ueno M., Imai K., Adachi H., Kageyama M., Mori K., et al. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PLoS One. 2012;7:e33389. doi: 10.1371/journal.pone.0033389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Li Z., Allingham R.R., Nakano M., Jia L., Chen Y., Ikeda Y., Mani B., Chen L.-J., Kee C., Garway-Heath D.F., et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum. Mol. Genet. 2015;24:3880–3892. doi: 10.1093/hmg/ddv128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fan B.J., Wang D.Y., Pasquale L.R., Haines J.L., Wiggs J.L. Genetic Variants Associated with Optic Nerve Vertical Cup-to-Disc Ratio Are Risk Factors for Primary Open Angle Glaucoma in a US Caucasian Population. Invest. Ophthalmol. Vis. Sci. 2011;52:1788–1792. doi: 10.1167/iovs.10-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ramdas W.D., van Koolwijk L.M.E., Ikram M.K., Jansonius N.M., de Jong P.T.V.M., Bergen A.A.B., Isaacs A., Amin N., Aulchenko Y.S., Wolfs R.C.W., et al. A Genome-Wide Association Study of Optic Disc Parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cheng C.-Y., Allingham R.R., Aung T., Tham Y.-C., Hauser M.A., Vithana E.N., Khor C.C., Wong T.Y. Association of common SIX6 polymorphisms with peripapillary retinal nerve fiber layer thickness: the Singapore Chinese Eye Study. Invest. Ophthalmol. Vis. Sci. 2014;56:478–483. doi: 10.1167/iovs.14-15863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W., et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cao D., Jiao X., Liu X., Hennis A., Leske M.C., Nemesure B., Hejtmancik J.F. CDKN2B polymorphism is associated with primary open-angle glaucoma (POAG) in the Afro-Caribbean population of Barbados, West Indies. PLoS One. 2012;7:e39278. doi: 10.1371/journal.pone.0039278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Shiga Y., Akiyama M., Nishiguchi K.M., Sato K., Shimozawa N., Takahashi A., Momozawa Y., Hirata M., Matsuda K., Yamaji T., et al. Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum. Mol. Genet. 2018;27:1486–1496. doi: 10.1093/hmg/ddy053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Mori K., Nakano M., Tokuda Y., Ikeda Y., Ueno M., Sotozono C., Kinoshita S., Tashiro K. Stronger Association of CDKN2B-AS1 Variants in Female Normal-Tension Glaucoma Patients in a Japanese Population. Invest. Ophthalmol. Vis. Sci. 2016;57:6416–6417. doi: 10.1167/iovs.16-20417. [DOI] [PubMed] [Google Scholar]
- 187.Ng S.K., Burdon K.P., Fitzgerald J.T., Zhou T., Fogarty R., Souzeau E., Landers J., Mills R.A., Casson R.J., Ridge B., et al. Genetic Association at the 9p21 Glaucoma Locus Contributes to Sex Bias in Normal-Tension Glaucoma. Invest. Ophthalmol. Vis. Sci. 2016;57:3416–3421. doi: 10.1167/iovs.16-19401. [DOI] [PubMed] [Google Scholar]
- 188.Chen Y., Hughes G., Chen X., Qian S., Cao W., Wang L., Wang M., Sun X. Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population. Invest. Ophthalmol. Vis. Sci. 2015;56:2595–2600. doi: 10.1167/iovs.14-16269. [DOI] [PubMed] [Google Scholar]
- 189.Taylor K.D., Guo X., Zangwill L.M., Liebmann J.M., Girkin C.A., Feldman R.M., Dubiner H., Hai Y., Samuels B.C., Panarelli J.F., et al. Genetic Architecture of Primary Open-Angle Glaucoma in Individuals of African Descent: The African Descent and Glaucoma Evaluation Study III. Ophthalmology. 2019;126:38–48. doi: 10.1016/j.ophtha.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rong S.S., Lu S.Y., Matsushita K., Huang C., Leung C.K.S., Kawashima R., Usui S., Tam P.O.S., Young A.L., Tsujikawa M., et al. Association of the SIX6 locus with primary open angle glaucoma in southern Chinese and Japanese. Exp. Eye Res. 2019;180:129–136. doi: 10.1016/j.exer.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 191.Zhao W., Smith J.A., Mao G., Fornage M., Peyser P.A., Sun Y.V., Turner S.T., Kardia S.L.R. The cis and trans effects of the risk variants of coronary artery disease in the Chr9p21 region. BMC Med. Genom. 2015;8:21. doi: 10.1186/s12920-015-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Myers T.A., Chanock S.J., Machiela M.J. LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front. Genet. 2020;11:157. doi: 10.3389/fgene.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Koyama S., Ito K., Terao C., Akiyama M., Horikoshi M., Momozawa Y., Matsunaga H., Ieki H., Ozaki K., Onouchi Y., et al. Population-specific and trans-ancestry genome-wide analyses identify distinct and shared genetic risk loci for coronary artery disease. Nat. Genet. 2020;52:1169–1177. doi: 10.1038/s41588-020-0705-3. [DOI] [PubMed] [Google Scholar]
- 195.Nikpay M., Goel A., Won H.-H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Hartiala J.A., Han Y., Jia Q., Hilser J.R., Huang P., Gukasyan J., Schwartzman W.S., Cai Z., Biswas S., Trégouët D.A., et al. Genome-wide analysis identifies novel susceptibility loci for myocardial infarction. Eur. Heart J. 2021;42:919–933. doi: 10.1093/eurheartj/ehaa1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wu Y., Byrne E.M., Zheng Z., Kemper K.E., Yengo L., Mallett A.J., Yang J., Visscher P.M., Wray N.R. Genome-wide association study of medication-use and associated disease in the UK Biobank. Nat. Commun. 2019;10:1891. doi: 10.1038/s41467-019-09572-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 199.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Matsunaga H., Ito K., Akiyama M., Takahashi A., Koyama S., Nomura S., Ieki H., Ozaki K., Onouchi Y., Sakaue S., et al. Transethnic Meta-Analysis of Genome-Wide Association Studies Identifies Three New Loci and Characterizes Population-Specific Differences for Coronary Artery Disease. Circ. Genom. Precis. Med. 2020;13:e002670. doi: 10.1161/CIRCGEN.119.002670. [DOI] [PubMed] [Google Scholar]
- 201.Landi M.T., Bishop D.T., MacGregor S., Machiela M.J., Stratigos A.J., Ghiorzo P., Brossard M., Calista D., Choi J., Fargnoli M.C., et al. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat. Genet. 2020;52:494–504. doi: 10.1038/s41588-020-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The all-biobank meta-analysis results and plots (including ancestry/sex-stratified and cross-ancestry meta-analyses) are available for downloading at https://www.globalbiobankmeta.org/resources and for browsing at the browser http://results.globalbiobankmeta.org.
-
•
Scripts used for quality control, meta-analysis, and summary of results are available at https://github.com/globalbiobankmeta and deposited at https://zenodo.org/badge/latestdoi/295461030.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.