Abstract
Objective
This study sought to explore the clinical value of matrix metalloproteinases 12 (MMP12) in multiple cancers, including lung adenocarcinoma (LUAD).
Methods
Using >10,000 samples, this retrospective study demonstrated the first pan-cancer analysis of MMP12. The expression of MMP12 between cancer groups and their control groups was analyzed using Wilcoxon rank-sum tests. The clinical significance of MMP12 expression in multiple cancers was assessed using receiver operating characteristic curves, Kaplan–Meier curves, and univariate Cox analysis. A further LUAD-related analysis based on 4565 multi-center and in-house samples was performed to verify the findings regarding MMP12 in pan-cancer analysis partly.
Results
MMP12 mRNA is highly expressed in 13 cancers compared to their controls, and the MMP12 protein level is elevated in some of these cancers (e.g., colon adenocarcinoma) (P < .05). MMP12 expression makes it feasible to distinguish 21 cancer tissues from normal tissues (AUC = 0.86). A high MMP12 expression is a prognosis risk factor in eight cancers, such as adrenocortical carcinoma (hazard ratio >1, P < .05). The elevated MMP12 expression is also a prognosis protective factor in breast-invasive carcinoma and colon adenocarcinoma (hazard ratio <1, P < .05). Some pan-cancer findings regarding MMP12 are verified in LUAD—MMP12 expression is upregulated in LUAD at both the mRNA and protein levels (P < .05), has the potential to distinguish LUAD with considerable accuracy (AUC = .91), and plays a risk prognosis factor for patients with the disease (P < .05).
Conclusions
MMP12 is highly expressed in most cancers and may serve as a novel biomarker for the prediction and prognosis of numerous cancers.
Keywords: expression, prognosis, immune microenvironment, standardized mean difference, area under the curve
Introduction
Cancer has gradually become one of the leading causes of human illness and mortality. International cancer statistics indicate that there were approximately 19 million new cancer cases worldwide and more than 10 million cancer deaths in 2020. 1 Targeted therapy offers greater promise than traditional approaches (e.g., surgery) in treating several cancers. 2 Imatinib provides a noteworthy example of its effectiveness in targeting the BCR-ABL fusion gene, which has led to a significant improvement in the 10-year survival rate for patients with chronic myeloid leukemia (from less than 50% to approximately 80%). 3 Cancer cells expressing PD-L1 can circumvent immune response inhibition by activating the interaction between PD-1/PD-L1, which promotes immune evasion; PD-1 inhibitors, such as nivolumab, can effectively block this interaction and suppress immune evasion and provide benefits to patients with non-small-cell lung cancer. 4 These examples highlight the immense potential value of targeted therapy for treating cancers. However, the presence of drug resistance and the occurrence of side effects associated with current medications underscore the need for researchers to prioritize the exploration of novel cancer markers.5,6 Furthermore, a lack of valuable markers for many cancers also limits the development of targeted therapy. Therefore, it is crucial to explore markers that potentially play vital roles in multiple cancers.
Among numerous potential tumor markers, matrix metalloproteinase 12 (MMP12) has garnered increasing attention from researchers. MMP12, a metalloproteinase secreted by macrophages, has been found to be aberrantly expressed in various tumors, affecting tumor progression and prognosis through multiple mechanisms.7-9 In liver hepatocellular carcinoma (LIHC), upregulation of MMP12 has been associated with tumor growth and progression by promoting angiogenesis, ultimately resulting in a poorer prognosis for patients. 10 In lung adenocarcinoma (LUAD), MMP12 protein expression levels are significantly higher in tumor tissues than control lung tissues. Knocking down MMP12 or inhibiting its expression can suppress the proliferation and invasion of LUAD cells, possibly by affecting the expression of vascular endothelial growth factor and the epithelial-mesenchymal transition.11,12 In renal cell carcinoma and esophageal squamous cell carcinoma, patients with increased MMP12 expression have a significantly worse prognosis compared to those with low MMP12 expression.13,14 Furthermore, MMP12 has been proposed as a target for tumor immunosuppression and immune checkpoints. 15 These studies shed light on the pro-tumorigenic role of MMP12 and its potential therapeutic value. However, some studies have also suggested anti-tumor effects of MMP12 in colorectal cancer: knocking out MMP12 leads to the accumulation of M2 macrophages (which predominantly exhibit pro-cancer effects) in the tumor microenvironment, thereby promoting the growth of colorectal tumors. 16 In ovarian cancer, high levels of MMP12 mRNA have been associated with better overall survival (OS). 17 Therefore, previous research indicates that MMP12 plays an important role in various tumors, but its clinical significance may not be consistent across different types of cancer. Closing this research gap is needed to identify the pan-cancer clinical significance of MMP12.
We analyzed MMP12’s cancer cell effects, mRNA and protein expression, immune effects, clinical significance, and potential mechanisms of MMP12 in 33 cancer tissues and 21 normal tissues by collecting data from the DepMap Portal, Xena database, and Clinical Proteomic Tumor Analysis Consortium database. Additionally, we used data from The Cancer Genome Atlas (TCGA), Genotype-Tissue Expression, ArrayExpress, and Gene Expression Omnibus to analyze MMP12’s expression in LUAD and combined those data with our in-house data to explore the role of MMP12 in LUAD as well as its important pan-cancer role, thereby providing a potential target of cancer immunotherapy.
Materials and Methods
This retrospective study was approved by the medical ethics review committee of the First Affiliated Hospital of Guangxi Medical University (No. 6 Shuangyong Road, Nanning, China) on October 26, 2021, with the approval number 2021(KY-E-246). Informed consent was signed by all patients involved in the in-house data. The personal identification information of the patients included in this study has been removed, and it is not possible to ascertain the identity of the patients through the information provided in this article. The reporting of the study adheres to REMARK guidelines. 18
Collecting MMP12-related Expression and Prognosis Data Across Cancers
The DepMap Portal includes data on numerous cell types. We collected data on RNA interference (n of samples = 494) to analyze the essential roles of MMP12 in multiple cancer cells. An RNA interference score of less than 0 indicates that MMP12 is essential for a specific cancer cell.
The Xena database collects data on tumors and their normal samples from various datasets such as TCGA. TCGA data were extracted from the Xena database to analyze MMP12’s mRNA expression among 33 tumor tissue types (n of samples = 8305) and 21 normal tissue types (n of samples = 671). The Clinical Proteomic Tumor Analysis Consortium database contains MMP12 protein level data in various cancers from the Proteomic Data Commons. The protein level data from this database were collected to detect the difference in MMP12 protein levels between breast-invasive carcinoma (BRCA), colon adenocarcinoma (COAD), head and neck squamous cell carcinoma (HNSCC), and uterine corpus endometrial carcinoma (UCEC) and their control samples.19-21
Clinical parameters were collected from the Xena database, including patients’ ages, genders, and cancer stages as defined by the American Joint Committee on Cancer. The cancer patients’ prognosis data were also acquired from the Xena database, including OS, disease-specific survival, progression-free interval, and disease-free interval. The define details of clinical endpoints can be seen in previous research. 22
The Prediction Effect and Prognosis Value of MMP12 for Cancers
The area under the curve (AUC) size of receiver operating characteristic (ROC) curves was applied to determine the ability of MMP12 expression to differentiate tumor tissues and normal tissues. We plotted the summary ROC (sROC) to evaluate the overall ability to discriminate between tumor tissues and normal tissues. Univariate Cox regression and Kaplan–Meier curves revealed differences in prognosis of patients with various expression levels of MMP12. The high-MMP12 and low-MMP12 groups were identified using a cutoff value determined by the “survminer” software package.
We also examined the role of MMP12 expression in the immune environment and signaling pathways. Three types of data, including neoantigen count, 23 tumor mutational burden (TMB), and microsatellite instability (MSI), were acquired from Sanger Box (v3.0). 24 We also revealed the regulation of MMP12 expression on six types of patient’s immune cells, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells; the infiltration level data of these cells were obtained from TIMER. 25 Immune environment data based on the ESTIMATE algorithm 26 were acquired from Sanger Box (v3.0); they contained three types of scores, including immune, stromal, and ESTIMATE scores. Using the clusterProfiler package, 27 Kyoto Encyclopedia of Genes and Genomes 28 signaling pathways that MMP12 may participate in multiple cancers were determined with gene set enrichment analysis; those signaling pathways with the P-value <.05 were included.
Validation of MMP12 mRNA Expression in LUAD
To validate the MMP12 expression in LUAD at mRNA levels, LUAD-related public datasets were collected from several public databases: TCGA, Genotype-Tissue Expression, ArrayExpress, and Gene Expression Omnibus. The data retrieval strategy for datasets was “(mRNA or gene) AND lung AND (adenocarcinoma OR [non-small cell]).” The inclusion criteria for datasets and their samples were as follows: (1) the samples were sourced from humans; (2) samples from the LUAD group were obtained from pathologically diagnosed LUAD tissues or cells; (3) samples from the control group were obtained from pathologically diagnosed normal lung tissues or cells; (4) the dataset had complete mRNA expression data. The exclusion criteria were as follows: (1) incomplete mRNA expression data; (2) duplicate samples in various datasets. Ultimately, 59 datasets were included in this study and merged to 29 datasets based on the same platform (e.g., GPL10558). Details of the 59 datasets are listed in Supplementary Material 1.
Validation of MMP12 Protein Expression in LUAD
To verify MMP12 expression in LUAD at the level of proteins, in-house tissue microarrays (LUC1021, LUC1502, and LUC481) were purchased from Fanpu Biotech (Guilin, China), including 64 LUAD samples and 24 non-LUAD control samples. The inclusion of samples was as follows: (1) samples were sourced from human LUAD tissues and non-LUAD control lung tissues; (2) samples were pathologically identified; (3) samples were collected from the patients who signed the informed consent. These samples were used in an immunohistochemistry experiment.
We conducted the immunohistochemistry experiment following the instructions of the reagent manufacturer. We used a .01 M citrate buffer solution (pH = 6.0) to wash the dewax and repaired tissue slides to extract the antigen, and we used 3% H2O2 to deactivate the endogenous peroxidase. We used the rabbit anti-human MMP12 monoclonal antibody (ab137444, Abcam, UK) in a 1:100 dilution to incubate the prepared tissue slides at 37°C for 30 min. In contrast, we incubated the negative control slides in phosphate buffer overnight. We added a secondary antibody, horseradish peroxidase (D-3004-15, Changdao Biotechnology Co. Ltd., Shanghai, China), to the tissue slides, which were then kept at room temperature (approximately 25°C) for 25 minutes and finally stained with 3,3’-diaminobenzidine for 10 minutes.
The dehydrated and sealed slides were used to assess the degree of MMP12 protein expression under microscopy. Positive and negative anti-MMP12 antibody staining showed diverse colors (brown granules in the nucleus and/or cytoplasm for the positive staining and blue particles for the negative). All specimens of the tissue microarrays were evaluated for positive cells in five randomly selected regions. In the visual field, the anti-MMP12 body staining intensity score was indicated by integers from 0 through 3, representing no staining, light staining, moderate staining, and strong staining. For positive cells, integers from 0 through 4 represented <5%, 5%–25%, 26%–50%, 51%–75%, and >75%, respectively, in the visual field. The final score (i.e., the product of the intensity score and the positive cells score) represents the MMP12 protein level in the LUAD and control tissues.
Validation of the Potential Clinical Value of MMP12 in LUAD
The AUC values of the ROC curves and an sROC curve were applied to determine the ability of MMP12 expression to differentiate LUAD samples and control specimens. We used the Kaplan–Meier curve to evaluate the prognosis differences between LUAD individuals with a high MMP12 and those with a low MMP12 expression. The “survminer” package was employed to determine the high-MMP12 and low-MMP12 groups.
Statistical Analysis
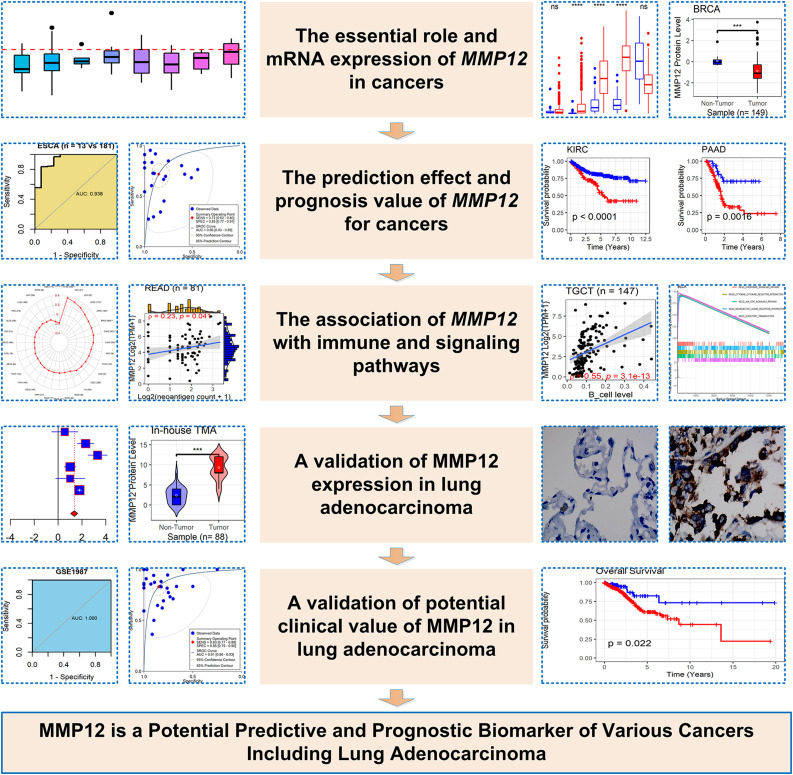
We conducted Wilcoxon rank-sum tests to explore the differences in MMP12 expression between cancer groups and their control groups (e.g., LUAD vs control). The method was also used to detect MMP12 expression differences in patients with various clinical parameters. Using the “meta” package, differences in MMP12 expression level between the LUAD group and the non-LUAD group were evaluated with a standardized mean difference (SMD). We used Begg’s test to evaluate publication bias, and the P-value of less than .1 indicated significant publication bias. 29 All correlation analyses in this study were done with Spearman’s rank correlation coefficient. The sROC curve was produced using Stata (v15.0), and the remaining calculations were conducted in R (v4.1.0). Figure 1 shows the overall framework design of this research.
Figure 1.
Research overflow of this study.
Results
Pan-Cancer MMP12 Expression Level
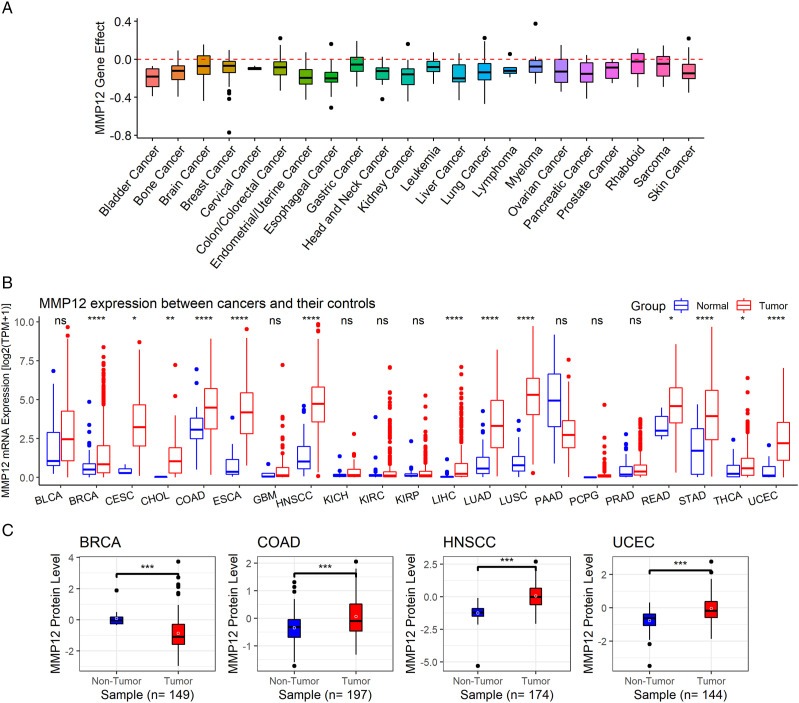
MMP12 is essential for various cancers, particularly esophageal cancer (ESCA) and liver cancer (Figure 2A). MMP12 mRNA overexpression compared to normal tissues was observed in the cancer tissues of 13 cancer types (P < .05), including BRCA, COAD, ESCA, HNSCC, LIHC, LUAD, UCEC, cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), and thyroid carcinoma (THCA) (Figure 2B). Furthermore, at the protein level, MMP12 expression was higher in COAD, HNSCC, and UCEC than in normal tissues, while the opposite result was found in BRCA (P < .05) (Figure 2C).
Figure 2.
Essential role of MMP12, MMP12 mRNA expression, and MMP12 protein levels in cancers. Panel A: Identification of essential roles of MMP12 for multiple cancers. Panel B: The differential expression of MMP12 mRNA between cancers and controls; P-value was based on the Wilcoxon rank-sum test with a false discovery rate. Panel C: The differential levels of MMP12 protein between cancers and controls. nsP ≥ .05; *P < .05; **P < .01; ***P < .001; ****P < .0001.
Clinical Significance of Pan-Cancer MMP12 Expression
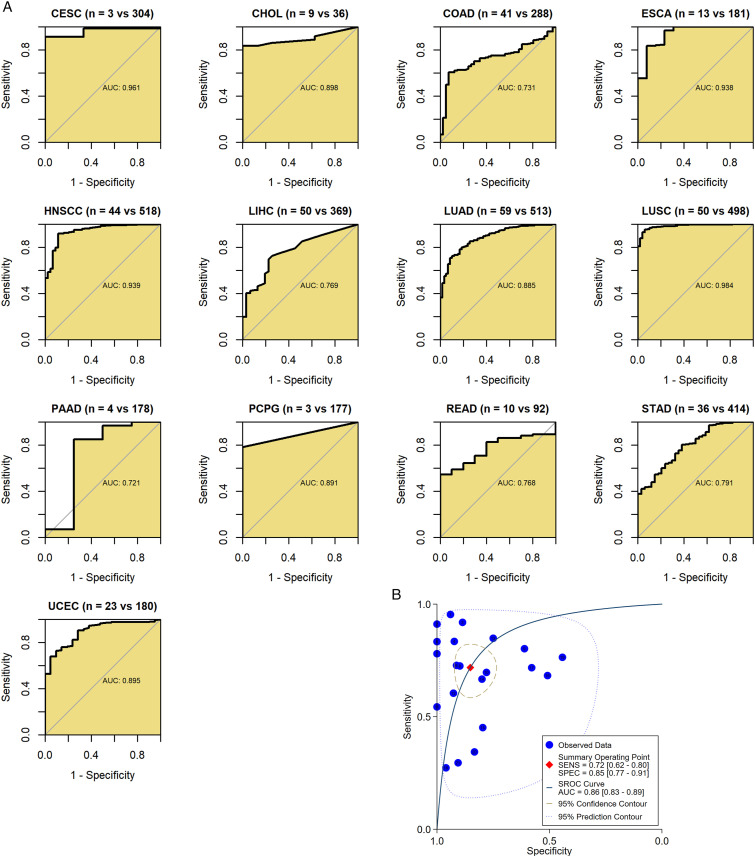
We drew ROC curves to identify the pan-cancer clinical significance of MMP12. The results show that 13 of 21 cancer types have an AUC value above .7 (Figure 3A), indicating that MMP12 has a conspicuous ability to distinguish these cancer tissues from normal tissues. In addition, the AUC in sROC was .86, indicating that MMP12 has a good ability to distinguish 21 cancer tissues from normal tissues (Figure 3B).
Figure 3.
Ability of MMP12 to differentiate the tumor tissue from the control tissue. Panel A: MMP12 can accurately distinguish cancer tissues from control tissues in some cancers. Panel B: MMP12 can well distinguish cancers from controls in 21 cancer types.
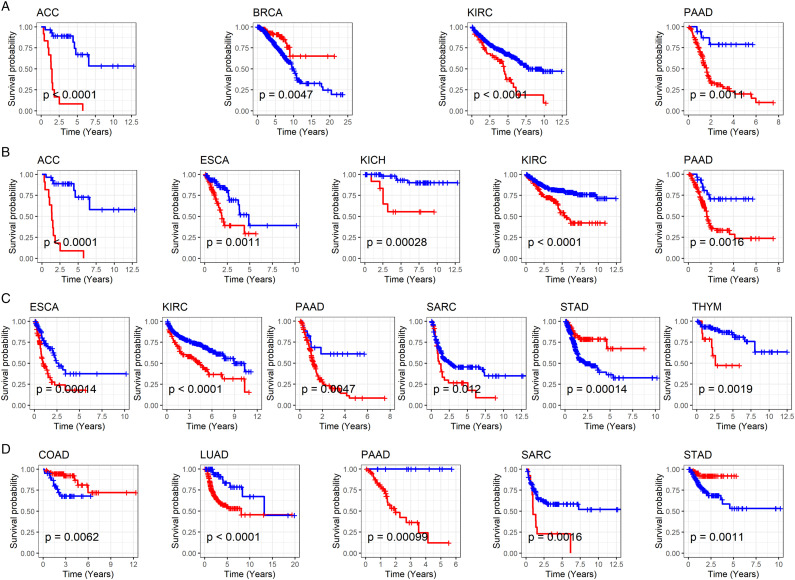
Univariate Cox analysis showed that high MMP12 expression predicts a poor OS (hazard ratio [HR] > 1, P < .05) in adrenocortical carcinoma (ACC), kidney renal clear cell carcinoma (KIRC), and pancreatic adenocarcinoma (PAAD) and a favorable OS in BRCA (HR < 1, P < .05) (Table 1). For disease-specific survival, high MMP12 expression was associated with poor clinical outcomes in ACC, ESCA, kidney chromophobe (KICH), KIRC, and PAAD (HR > 1, P < .05) (Table 1). In addition, high expression of MMP12 was associated with a poor progression-free interval in ESCA, KIRC, PAAD, sarcoma (SARC), and thymoma (THYM) (HR > 1, P < .05) and favorable progression-free interval in STAD (HR < 1, P < .05) (Table 2). Upregulation of MMP12 in LUAD, PAAD, and SARC was associated with a decreased disease-free interval (HR > 1, P < .05), while elevated expression of MMP12 was relevant to an increased disease-free interval in COAD and STAD (HR < 1, P < .05) (Table 2). Finally, we used Kaplan–Meier curves to test MMP12 expression and prognosis in cancer patients, which validated the above results (P < .05) (Figure 4).
Table 1.
Relation of MMP12 Expression With Overall Survival (OS) and Disease-specific Survival (DSS) of Cancer Patients.
| Cancer (sample) | OS HR a | P-value | Cancer (sample) | DSS HR | P-value |
| ACC (41) | 1.555 | .002 | ACC (39) | 6.396 | <.001 |
| BLCA (416) | .975 | .455 | BLCA (401) | .972 | .496 |
| BRCA (1143) | .879 | .031 | BRCA (1115) | .926 | .324 |
| CESC (273) | 1.072 | .253 | CESC (272) | 1.062 | .380 |
| CHOL (40) | .988 | .949 | CHOL (38) | 1.045 | .809 |
| COAD (317) | .920 | .184 | COAD (302) | .920 | .339 |
| DLBC (44) | 1.035 | .865 | DLBC (44) | 1.210 | .488 |
| ESCA (188) | 1.099 | .119 | ESCA (185) | 1.217 | .007 |
| GBM (114) | .888 | .373 | GBM (106) | .895 | .429 |
| HNSCC (554) | .974 | .437 | HNSCC (526) | .959 | .347 |
| KICH (77) | 2.440 | .055 | KICH (77) | 3.341 | .017 |
| KIRC (500) | 1.282 | <.001 | KIRC (486) | 1.303 | <.001 |
| KIRP (240) | 1.007 | .979 | KIRP (238) | 1.147 | .576 |
| LGG (205) | 1.118 | .721 | LGG (201) | 1.175 | .606 |
| LIHC (326) | 1.121 | .086 | LIHC (317) | 1.078 | .390 |
| LUAD (549) | 1.042 | .230 | LUAD (515) | 1.005 | .913 |
| LUSC (515) | 1.000 | .989 | LUSC (457) | .957 | .396 |
| MESO (67) | 1.001 | .993 | MESO (49) | .913 | .528 |
| OV (382) | .947 | .337 | OV (354) | .946 | .369 |
| PAAD (176) | 1.146 | .034 | PAAD (170) | 1.159 | .037 |
| PCPG (93) | 1.408 | .425 | PCPG (93) | 1.240 | .718 |
| PRAD (518) | .615 | .410 | PRAD (516) | .961 | .951 |
| READ (101) | .939 | .664 | READ (95) | .885 | .596 |
| SARC (219) | 1.063 | .336 | SARC (214) | 1.082 | .240 |
| SKCM (94) | 1.118 | .521 | SKCM (94) | 1.212 | .317 |
| STAD (406) | .944 | .111 | STAD (384) | .915 | .065 |
| TGCT (127) | 1.647 | .123 | TGCT (127) | 1.670 | .129 |
| THCA (503) | 1.273 | .226 | THCA (497) | .255 | .127 |
| THYM (106) | 1.180 | .842 | THYM (106) | 2.488 | .301 |
| UCEC (178) | .915 | .472 | UCEC (176) | 1.066 | .657 |
| UCS (54) | 1.225 | .106 | UCS (52) | 1.203 | .164 |
| UVM (33) | 1.417 | .082 | UVM (33) | 1.480 | .054 |
aNotes: hazard ratio.
Table 2.
Relation of MMP12 Expression With Progression-free Interval (PFI) and Disease-free Interval (DFI) of Cancer Patients.
| Cancer (sample) | PFI HR a | P-value | Cancer (sample) | DFI HR | P-value |
|---|---|---|---|---|---|
| ACC (40) | 1.008 | .965 | ACC (20) | 165.846 | .223 |
| BLCA (415) | 1.003 | .928 | BLCA (189) | .999 | .995 |
| BRCA (1142) | .973 | .632 | BRCA (975) | .988 | .873 |
| CESC (276) | .960 | .507 | CESC (173) | .858 | .147 |
| CHOL (40) | .884 | .450 | CHOL (29) | 1.019 | .910 |
| COAD (314) | .934 | .247 | COAD (121) | .684 | .020 |
| DLBC (43) | .930 | .697 | DLBC (26) | .366 | .119 |
| ESCA (185) | 1.189 | .002 | ESCA (91) | 1.219 | .055 |
| GBM (113) | .991 | .947 | HNSCC (134) | 1.202 | .072 |
| HNSCC (553) | .993 | .839 | KICH (36) | .027 | .538 |
| KICH (77) | 2.011 | .089 | KIRC (112) | .764 | .582 |
| KIRC (489) | 1.202 | .009 | KIRP (133) | 1.279 | .463 |
| KIRP (237) | 1.048 | .810 | LGG (52) | 1.083 | .894 |
| LGG (204) | .660 | .207 | LIHC (274) | 1.056 | .532 |
| LIHC (326) | 1.061 | .328 | LUAD (331) | 1.160 | .003 |
| LUAD (545) | 1.037 | .259 | LUSC (316) | .950 | .423 |
| LUSC (515) | .981 | .638 | MESO (11) | 1.987 | .085 |
| MESO (65) | .966 | .783 | OV (191) | .967 | .632 |
| OV (382) | .916 | .087 | PAAD (71) | 1.382 | .001 |
| PAAD (175) | 1.127 | .047 | PCPG (77) | 2.092 | .469 |
| PCPG (92) | 1.444 | .084 | PRAD (367) | .534 | .125 |
| PRAD (518) | 1.155 | .330 | READ (32) | 1.185 | .620 |
| READ (100) | .962 | .760 | SARC (129) | 1.223 | .002 |
| SARC (216) | 1.160 | .003 | STAD (252) | .864 | .033 |
| SKCM (93) | 1.125 | .455 | TGCT (101) | 1.004 | .967 |
| STAD (409) | .903 | .009 | THCA (357) | 1.409 | .110 |
| TGCT (125) | .992 | .923 | UCEC (126) | .729 | .077 |
| THCA (502) | 1.086 | .575 | UCS (26) | .894 | .766 |
| THYM (106) | 2.175 | .029 | |||
| UCEC (178) | .899 | .313 | |||
| UCS (54) | 1.211 | .103 | |||
| UVM (32) | 1.186 | .389 |
aNotes: hazard ratio.
Figure 4.
Relation of MMP12 expression with overall survival (A), disease-specific survival (B), progression-free interval (C), and disease-free interval (D) of cancer patients. The red and blue curves represent the high-MMP12 expression group and low-MMP12 expression group, respectively.
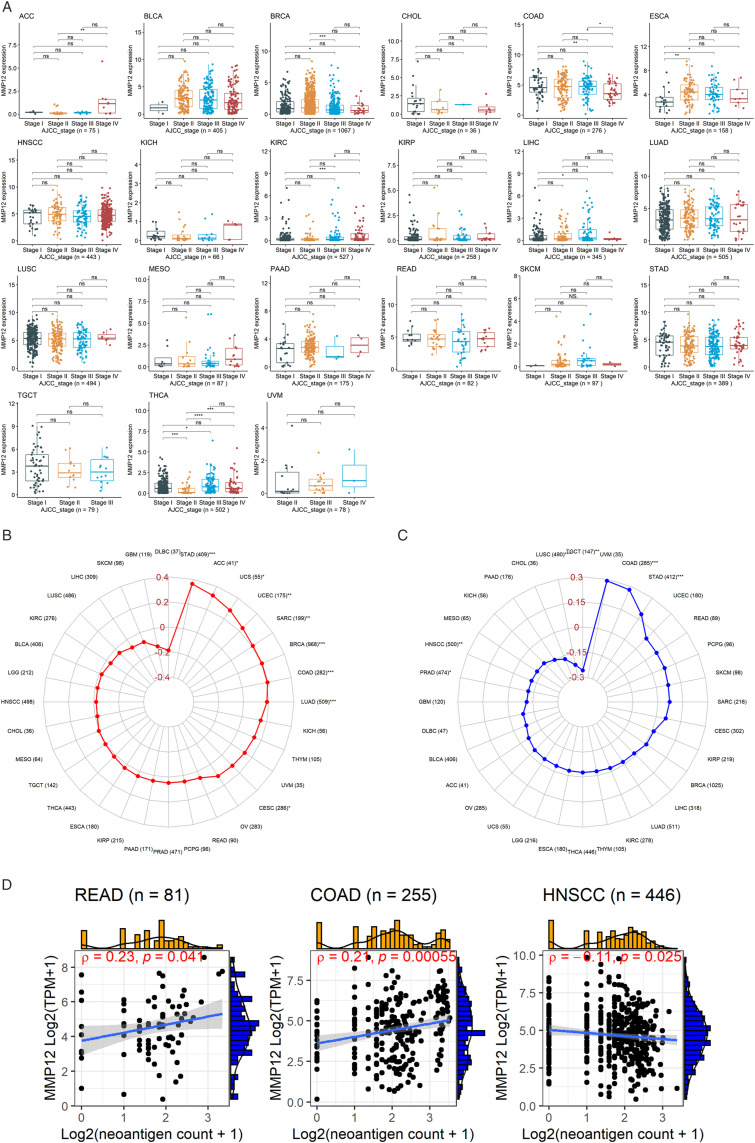
MMP12 expressed variously among the cancer staging levels of 21 cancer types. MMP12 expression was at high levels in the advanced stages of several cancers, including ACC, ESCA, KIRC, LIHC, and THCA (P < .05) (Figure 5A). In contrast, it expressed at low levels in terminal cancers, including BRCA and COAD (P < .05) (Figure 5A). Other cancers, including bladder urothelial carcinoma (BLCA), CHOL, HNSCC, KICH, kidney renal papillary cell carcinoma (KIRP), LUAD, LUSC, mesothelioma (MESO), PAAD, READ, skin cutaneous melanoma (SKCM), STAD, testicular germ cell tumor (TGCT), and uveal melanoma (UVM), showed little difference in MMP12 expression between various stages (Figure 5A). Furthermore, we observed that MMP12 expression did not differ significantly by age and gender in most cancers (Supplementary Materials 2 and 3).
Figure 5.
Relation of MMP12 expression with tumor stages (panel A), tumor mutational burden (panel B), microsatellite instability (panel C), and neoantigen (panel D) of cancer patients. *P < .05; **P< .01; ***P < .001.
Relationship Between the Expression of Immune Gene MMP12 and Genomic Heterogeneity
TMB is related to the number of tumor cell mutations; a high level of TMB can induce the body to produce neoantigens and cause more immune cells to play a role in immune recognition. 30 MMP12 expression was positively correlated with TMB (P < .05) in STAD, ACC, uterine carcinosarcoma (UCS), UCEC, SARC, BRCA, COAD, LUAD, and CESC (Figure 5B).
Microsatellites, because they are short-chain repetitive DNA sequences, are prone to MSI when they are affected by mismatch repair. 31 The expression of MMP12 was positively correlated with COAD and STAD but negatively correlated with prostate adenocarcinoma, HNSCC, LUSC, and TGCT (P < .05) (Figure 5C).
DNA damage increases neoantigens on the surface of tumor cells, which benefits immune cells in recognizing and killing tumor cells. 32 Our research found a weak correlation between MMP12 expression and cancer neoantigens in READ and COAD (P < .05) (Figure 5D).
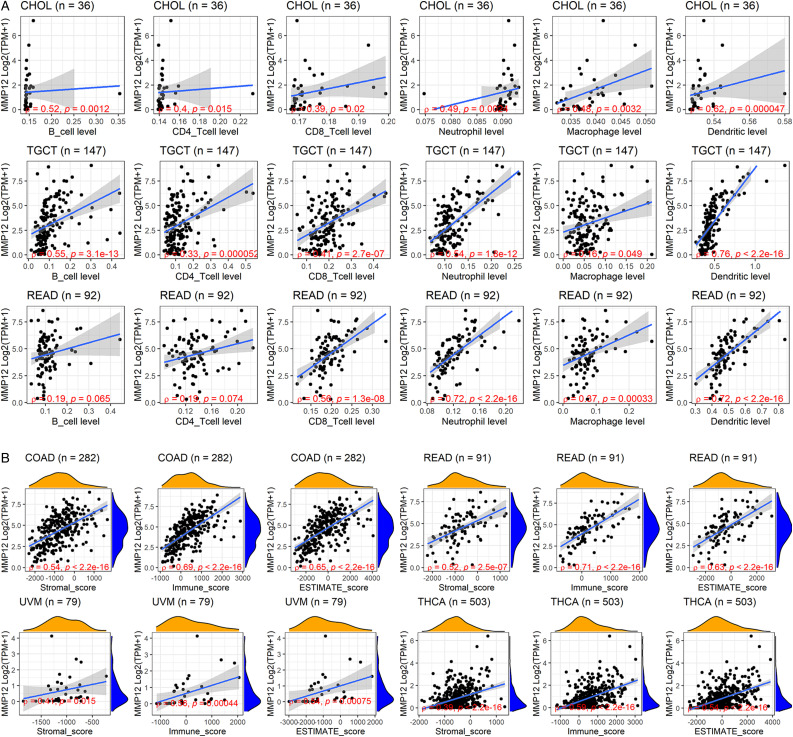
Correlation Assessment of MMP12 Expression and the Immune Microenvironment
MMP12 expression was correlated with the degree of six types of immune cell infiltration in various cancers (Figure 6A and Supplementary Material 4). Notably, MMP12 expression was significantly associated with the infiltration levels of B cells and dendritic cells in CHOL, TGCT, and READ (P < .05) (Figure 6A). Moreover, the expression levels of MMP12 were relevant to the immune microenvironment (Figure 6B and Supplementary Material 5). Among THCA, COAD, READ, and UVM, MMP12 expression had the strongest relationship with the stromal score in COAD, and the relationship of MMP12 expression with immune and ESTIMATE scores was conspicuous in READ and COAD (Figure 6B).
Figure 6.
Relation of MMP12 expression and infiltration levels of immune cells. Panel A: TIMER algorithm; B: ESTIMATE algorithm.
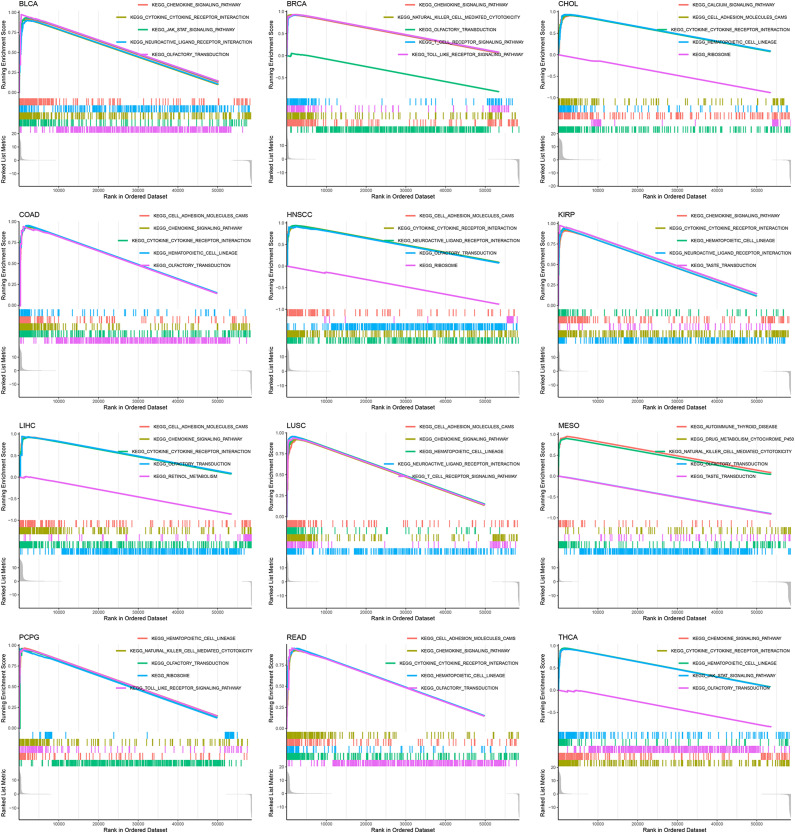
MMP12 Expression and Potential Signaling Pathways
The results of gene enrichment analysis show that MMP12 may participate in 31 potential molecular mechanisms in 33 cancers. The analysis results of 12 cancers (BLCA, BRCA, CHOL, COAD, HNSCC, KIRP, LIHC, LUSC, MESO, pheochromocytoma and paraganglioma, READ, and THCA) suggest that MMP12 is associated with at least five signaling pathways, indicating that MMP12 is likely to regulate the occurrence and development of cancer through these pathways, such as the cytokine–cytokine receptor interaction and the chemokine signaling pathway (Figure 7 and Supplementary Material 6).
Figure 7.
Potential signaling pathways of MMP12 may affect multiple cancers.
Overall Expression Level of MMP12 in LUAD
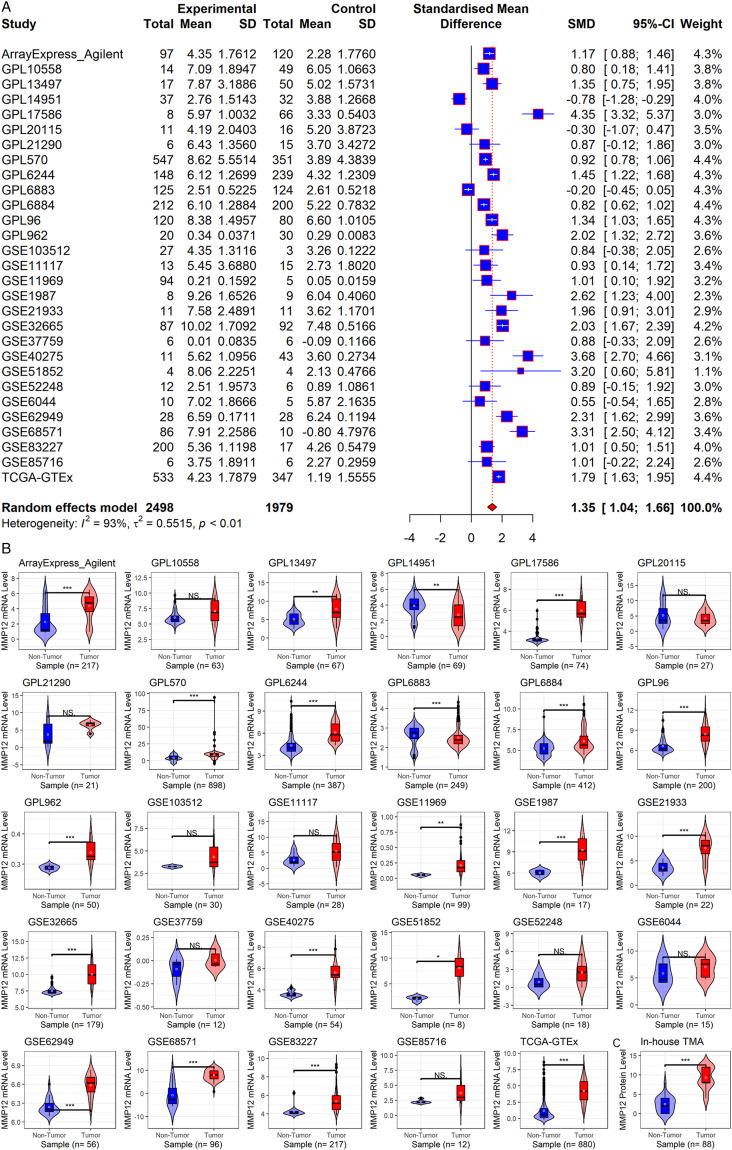
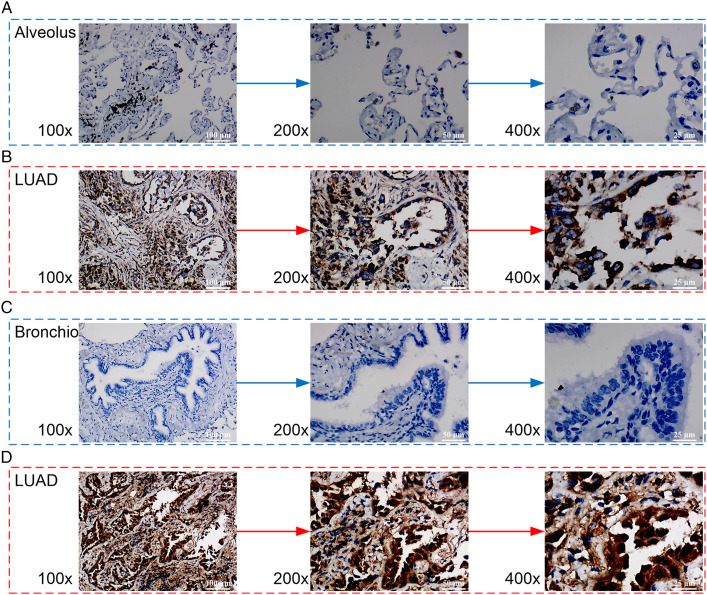
To further explore the findings regarding MMP12 in pan-cancer analysis, we examined the comprehensive expression level of MMP12 in LUAD. Among the 29 collected datasets, MMP12 mRNA was upregulated in LUAD (SMD = 1.35; 95% CI [1.04–1.66]) (Figure 8A), and no significant publication bias was found using Begg’s test (P > .1, Supplementary Material 7). A Wilcoxon rank-sum test also revealed overexpression of MMP12 mRNA in LUAD (P < .05) (Figure 8B). In addition, using in-house tissue microarrays, no positive MMP12 protein staging was found in the alveoli and bronchi of the control tissue (Figures 9A and C). However, a high level of MMP12 protein was observed in the LUAD tissue (Figures 9B and D), which was confirmed by the further Wilcoxon rank-sum test (P < .05) (Figure 8C).
Figure 8.
MMP12 mRNA and protein levels in LUAD. Panel A: MMP12 mRNA expression forest plot in LUAD and control tissues. Panel B: Violin plots of MMP12 mRNA expression in each dataset. Panel C: The violin plot of MMP12 protein levels. The P-value is calculated based on the Wilcoxon rank-sum test. NSP ≥ .05; *P < .05; **P< .01; ***P < .001.
Figure 9.
Microscopic images of MMP12 protein levels in control tissues (panels A and C) and LUAD tissues (panels B and D). The numerical value in the bottom left corner of each image represents the magnification of the microscope. The white numerical value in the lower right corner of each image represents the scale.
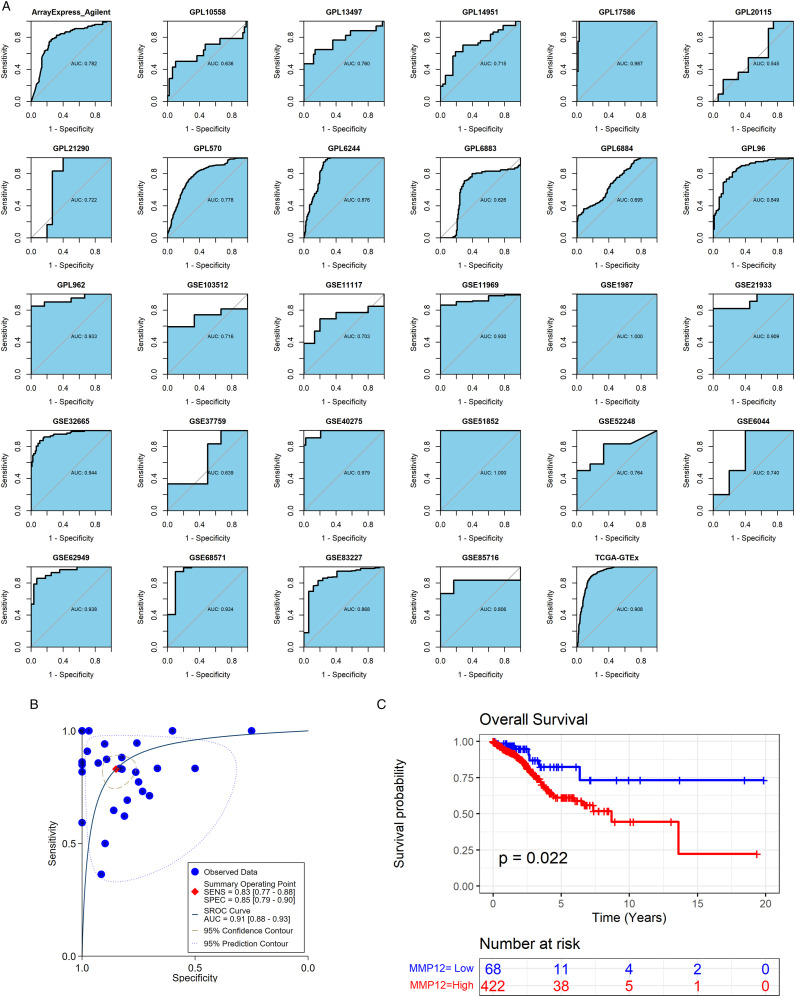
Clinical Value of MMP12 in LUAD
Among the 29 included datasets, ROC curves showed that MMP12 mRNA expression in LUAD exceeded moderate accuracy in 19 datasets (AUC >.75) (Figure 10A). The sROC analysis revealed that MMP12 mRNA expression accurately distinguished LUAD from non-LUAD (sensitivity = .83, specificity = .85; AUC = .91) (Figure 10B). Furthermore, using OS curves, a lower MMP12 expression in LUAD patients tended to predict a good prognosis (P = .022) (Figure 10C).
Figure 10.
Clinical value of MMP12 in LUAD. Panels A–B: MMP12 can well distinguish LUAD from controls. Panel C: Kaplan–Meier curves of the relation of MMP12 expression with overall survival of LUAD patients.
Discussion
Cancer seriously threatens human health as one of the leading causes of death, so it is meaningful to explore novel markers for identifying the disease status and prognosis of cancer patients. 33 As an immune gene, MMP12 can regulate inflammatory responses and play an essential role in specific cancers. 34 However, there was a dearth of a comprehensive pan-cancer analysis of MMP12.
This study used numerous multi-center samples and multiple approaches to explore the pan-cancer expression, clinicopathology, potential mechanisms, and clinical significance of MMP12. MMP12 was differently expressed in various cancers, and its expression level was related to TMB, MSI, neoantigen counts, and immune microenvironments in some cancers. The relationship between MMP12 expression and the potential molecular mechanism, prognosis, and clinical significance was also investigated in various cancers. Furthermore, we analyzed the expression of MMP12 in LUAD to support pan-cancer research by using public database datasets and in-house tissue microarrays to comprehensively determine that MMP12 mRNA and protein expression are upregulated in LUAD.
MMP12 is highly expressed in various cancers and plays a critical role in these diseases. Studies report that elevated MMP12 expression represents a poor prognosis for BRCA and that the gene was positively correlated with neutrophils and dendritic cells. 35 In terms of COAD, overexpressed MMP12 promotes tumors and is associated with a poor OS of patients. 36 In HNSCC, increasing MMP12 expression promotes cancer cell proliferation, migration, invasion, and the metastasis of cancer cells. 37 Overexpressed MMP12 mRNA may promote the progression and metastasis of ESCA, LIHC, and LUSC and is associated with a poor prognosis in cancer patients.25,38,39 Additionally, a high expression of MMP12 mRNA in LUAD promotes lymph node metastasis. 12 Similarly, our study detected that MMP12 is differently expressed in various cancers and that MMP12 is associated with patients’ cancer status and prognosis in several cancers. Briefly, in regard to expression between cancer and normal tissues, MMP12 mRNA is overexpressed in 13 cancers (CESC, etc.), and higher MMP12 protein levels are observed in COAD, HNSCC, and UCEC. Some of these findings have not previously been reported. For instance, our study for the first time describes an elevated MMP12 expression in UCEC, including mRNA and protein levels, indicating the novelty of our findings. Notably, our study found that, in contrast to MMP12 mRNA expression, MMP12 protein levels are lower in BRCA than in control tissues; this may be related to various factors, such as translation-level regulation and post-translational modification regulation,40-42 which requires further experimental verification. Such a phenomenon also indicates that investigations of gene expression should at least focus on both mRNA and protein levels rather than on only one of them. Regarding cancer status prediction, our study indicates that MMP12 makes it feasible to distinguish 21 cancer tissues from normal tissues. For prognosis, MMP12 expression is a prognosis risk factor in multiple cancers, including ACC, ESCA, KICH, KIRC, LUAD, PAAD, SARC, and THYM. Notably, MMP12 expression may be a prognosis protective factor for patients with BRCA and STAD; this may be attributed to the beneficial effects of MMP12 on macrophage development and suppression of angiogenesis in these two types of cancer, leading to its anti-tumor function.16,43 This indicates that the clinical significance may not be consistent across different types of cancer. Altogether, elevated MMP12 expression may serve as a potential predictive and prognostic biomarker for various cancers.
MMP12 may act as a potential target gene for immunotherapy in cancers. Previously, highly expressed MMP12 has been identified as an immune gene promoting the proliferation of immune cells (e.g., B cells and dendritic cells), and the gene stimulates the host immune system to cause immune responsiveness.44,45 TMB and MSI are considered predictive biomarkers for immunotherapy, as they may contribute to the generation of neoantigens and thus stimulate the host immune system to recognize and clear neoantigens in the immune microenvironment.32,46,47 In our study, MMP12 expression is related to TMB, MSI, and neoantigen numbers in various cancers (e.g., STAD and COAD), implying its participation in the immune microenvironment. Such a conclusion is also supported by the correlation (mainly positive) of MMP12 expression with several immune cells (e.g., B cells and dendritic cells) and immune scores (e.g., ESTIMATE algorithm scores). Moreover, MMP12 may participate in the occurrence and development of cancers through molecular signaling pathways, such as the cytokine–cytokine receptor interaction and the chemokine signaling pathway as has been verified in colorectal cancer and breast cancer.48,49 The abovementioned findings suggest that MMP12 may have the potential to act as a tumor marker in tumor immunotherapy.
Some pan-cancer findings regarding MMP12 are verified in LUAD. With relatively small samples (n = 52), Lv et al. determined that MMP12 protein levels are higher in LUAD tissues than in normal tissues, and they report that MMP12 can promote the proliferation and growth of cancer cells and increase their invasiveness. 12 Employing a large sample (n = 4565) from multiple centers and in house, we identified the upregulation of MMP12 expression in LUAD at both the mRNA and protein levels. We also show for the first time that MMP12 mRNA expression has the potential to distinguish LUAD with considerable accuracy. Furthermore, MMP12 expression serves as a risk prognosis factor for patients with LUAD. Thus, MMP12 may play an important role in LUAD as a predictive and prognostic biomarker. Based on this, a promising application of MMP12 in LUAD involves detecting MMP12 mRNA expression levels during pathological diagnosis of potential patients with LUAD; this approach may contribute to the evaluation of patient prognosis.
This study has a few limitations. For example, for various cancers, we need to collect more samples to determine MMP12 expression at the protein level. We also need to include more clinicopathological parameters to explore whether other clinicopathological variables may impact our results. More in vivo and in vitro experiments are needed to investigate the pan‐cancer mechanisms of MMP12. In addition, the pan-cancer examination of the relationship between prognosis and MMP12 expression was based on retrospective data; thus, prospective studies are needed for further validation.
Conclusion
This study comprehensively explores MMP12 in multiple cancers. MMP12 is highly expressed in most cancers. The gene may serve as a novel biomarker for the prediction and prognosis of numerous cancers.
Supplemental Material
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Acknowledgments
The authors thank Guangxi Key Laboratory of Medical Pathology for its technical support in computational and clinical pathology. The results shown in the study are in part based upon data generated by the DepMap Portal (https://depmap.org/portal/), Xena database (https://xena.ucsc.edu/), Proteomic Data Commons (https://pdc.cancer.gov), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/), and TCGA Research Network (www.cancer.gov/tcga).
Appendix.
Abbreviations
- ACC
adrenocortical carcinoma
- AUC
area under the curve
- BLCA
bladder urothelial carcinoma
- BRCA
breast-invasive carcinoma
- CESC
cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
cholangiocarcinoma
- COAD
colon adenocarcinoma
- DLBC
lymphoid neoplasm diffuse large b-cell lymphoma
- ESCA
esophageal carcinoma
- GBM
glioblastoma multiforme
- HNSCC
head and neck squamous cell carcinoma
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KICH
kidney chromophobe
- KIRC
kidney renal clear cell carcinoma
- KIRP
kidney renal papillary cell carcinoma
- LGG
brain lower grade glioma
- LIHC
liver hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- LUSC
lung squamous cell carcinoma
- MESO
mesothelioma
- MMP12
matrix metalloproteinase 12
- OS
overall survival
- OV
ovarian serous cystadenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PCPG
pheochromocytoma and paraganglioma
- PRAD
prostate adenocarcinoma
- READ
rectum adenocarcinoma
- ROC
receiver operating characteristic
- SARC
sarcoma
- SKCM
skin cutaneous melanoma
- sROC
summary receiver operating characteristic
- SMD
standardized mean difference
- STAD
stomach adenocarcinoma
- OS
overall survival
- TCGA
The Cancer Genome Atlas
- TGCT
testicular germ cell tumors
- THCA
thyroid carcinoma
- THYM
thymoma
- TMB
tumor mutation burden
- MSI
microsatellite instability
- UCEC
uterine corpus endometrial carcinoma
- UCS
uterine carcinosarcoma
- UVM
uveal melanoma.
Authors’ Contributions: Study design and draft writing: Guo-Sheng Li, Yu-Xing Tang, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen. Data acquisition: Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, and He-Qing Huang. Data analysis and interpretation: Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, and Rong-Quan He. All authors were involved in reading and revising the draft and approved the final version for publication.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was funded by Guangxi Medical High-level Key Talents Training “139” Program (2020), Guangxi Zhuang Autonomous Region Medical Health Appropriate Technology Development and Application Promotion Project (S2020031), Guangxi Higher Education Undergraduate Teaching Reform Project (2022JGA146 and 2021JGA142), Guangxi Educational Science Planning Key Project (2021B167), and Guangxi Medical University Key Textbook Construction Project (Gxmuzdjc2223).
Supplemental Material: Supplemental material for this article is available online.
Ethical Statement
Ethical Approval
This study was approved by the medical ethics review committee of the First Affiliated Hospital of Guangxi Medical University (No. 6 Shuangyong Road, Nanning, China) on October 26, 2021, with the approval number 2021(KY-E-246). Informed consent was signed by all patients involved in the in-house data.
ORCID iD
Gang Chen https://orcid.org/0000-0003-2402-2987
Data Availability Statement
The data that support the findings of pan-cancer analyses are available in public databases, including DepMap Portal (https://depmap.org/portal/), Xena database (https://xenabrowser.net/datapages), Proteomic Data Commons (https://pdc.cancer.gov), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/), and TCGA Research Network (www.cancer.gov/tcga). Data on in-house tissue samples used during the current study are available from the corresponding author upon reasonable request.
References
- 1.Xia C, Dong X, Li H, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl) 2022;135(5):584-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, Pirollo KF, Chang EH. Tumor-targeted p53-gene therapy enhances the efficacy of conventional chemo/radiotherapy. J Control Release. 2001;74(1-3):115-128. [DOI] [PubMed] [Google Scholar]
- 3.Waarts MR, Stonestrom AJ, Park YC, Levine RL. Targeting mutations in cancer. J Clin Invest 2022(8):132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lahiri A, Maji A, Potdar PD, et al. Lung cancer immunotherapy: progress, pitfalls, and promises. Mol Cancer 2023;22(1):40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H, Wang L, Bernards R. Rational combinations of targeted cancer therapies: background, advances and challenges. Nat Rev Drug Discov. 2023;22(3):213-234. [DOI] [PubMed] [Google Scholar]
- 6.Zhong L, Li Y, Xiong L, et al. Small molecules in targeted cancer therapy: advances, challenges, and future perspectives. Signal Transduct Target Ther. 2021;6(1):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banerjee S, Lo WC, Majumder P, et al. Multiple roles for basement membrane proteins in cancer progression and EMT. Eur J Cell Biol. 2022;101(2):151220. [DOI] [PubMed] [Google Scholar]
- 8.Marcos-Jubilar M, Orbe J, Roncal C, et al. Association of SDF1 and MMP12 with atherosclerosis and inflammation: clinical and experimental study. Life 2021;11(5):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan PP, Ding WY, Wang P. The roles of prostaglandin F2 in regulating the expression of matrix metalloproteinase-12 via an insulin growth factor-2-dependent mechanism in sheared chondrocytes. Signal Transduct Target Ther. 2018;3:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo ZY, Jiang LP. Matrix metalloproteinase 12 (MMP12) as an adverse prognostic biomarker of vascular invasion in hepatic cell carcinoma. Eur Rev Med Pharmacol Sci. 2022;26(7):2238-2249. [DOI] [PubMed] [Google Scholar]
- 11.Hung WY, Lee WJ, Cheng GZ, et al. Blocking MMP-12-modulated epithelial-mesenchymal transition by repurposing penfluridol restrains lung adenocarcinoma metastasis via uPA/uPAR/TGF-beta/Akt pathway. Cell Oncol. 2021;44(5):1087-1103. [DOI] [PubMed] [Google Scholar]
- 12.Lv FZ, Wang JL, Wu Y, Chen HF, Shen XY. Knockdown of MMP12 inhibits the growth and invasion of lung adenocarcinoma cells. Int J Immunopathol Pharmacol. 2015;28(1):77-84. [DOI] [PubMed] [Google Scholar]
- 13.Beres B, Yusenko M, Peterfi L, Kovacs G, Banyai D. Matrix metalloproteinase 12 is an independent prognostic factor predicting postoperative relapse of conventional renal cell carcinoma - a short report. Cell Oncol (Dordr). 2022;45(1):193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han F, Zhang S, Zhang L, Hao Q. The overexpression and predictive significance of MMP-12 in esophageal squamous cell carcinoma. Pathol Res Pract. 2017;213(12):1519-1522. [DOI] [PubMed] [Google Scholar]
- 15.Huang H, Hu Y, Guo L, Wen Z. Integrated bioinformatics analyses of key genes involved in hepatocellular carcinoma immunosuppression. Oncol Lett. 2021;22(6):830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Zhang X, Liu Q, et al. Knocking out matrix metalloproteinase 12 causes the accumulation of M2 macrophages in intestinal tumor microenvironment of mice. Cancer Immunol Immunother. 2020;69(8):1409-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng L, Qian J, Zhu F, Wu F, Zhao H, Zhu H. The prognostic values of matrix metalloproteinases in ovarian cancer. J Int Med Res. 2020;48(1):300060519825983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krug K, Jaehnig EJ, Satpathy S, et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell 2020;183(5):1436-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, Chen L, Savage SR, et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell. 2021;39(3):361-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dou Y, Kawaler EA, Cui Zhou D, et al. Proteogenomic characterization of endometrial carcinoma. Cell 2020;180(4):729-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li GS, Huang HQ, Liang Y, et al. BCAT1: A risk factor in multiple cancers based on a pan-cancer analysis. Cancer Med. 2022;11(5):1396-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen W, Song Z, Zhong X, et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Commentary iMeta 2022;1(3):e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Ren H, Peng R, Li Y, Ming L. Identification of key genes associated with progression and prognosis for lung squamous cell carcinoma. PeerJ. 2020;8:e9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353-D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088-1101. [PubMed] [Google Scholar]
- 30.Jardim DL, Goodman A, de Melo Gagliato D, Kurzrock R. The challenges of tumor mutational burden as an immunotherapy biomarker. Cancer Cell. 2021;39(2):154-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De' Angelis GL, Bottarelli L, Azzoni C, et al. Microsatellite instability in colorectal cancer. Acta Biomed. 2018;89(9-S):97-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schumacher TN, Scheper W, Kvistborg P. Cancer neoantigens. Annu Rev Immunol 2019;37:173-200. [DOI] [PubMed] [Google Scholar]
- 33.Alsharif F. Discovering the use of complementary and alternative medicine in oncology patients: A systematic literature review. Evid Based Complement Alternat Med. 2021;2021:6619243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia N, Xia L, Zhang WF, Zhou FX. [Immune-related genes and their determined immune cell microenvironment to predict the prognosis of gastric adenocarcinoma]. Zhonghua Yi Xue Za Zhi. 2022;102(12):840-846. [DOI] [PubMed] [Google Scholar]
- 35.Cheng T, Chen P, Chen J, Deng Y, Huang C. Landscape Analysis of matrix metalloproteinases unveils key prognostic markers for patients with breast cancer. Front Genet. 2021;12:809600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klupp F, Neumann L, Kahlert C, et al. Serum MMP7, MMP10 and MMP12 level as negative prognostic markers in colon cancer patients. BMC Cancer 2016;16:494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saleem Z, Shaikh AH, Zaman U, et al. Estimation of salivary matrix metalloproteinases- 12 (MMP- 12) levels among patients presenting with oral submucous fibrosis and oral squamous cell carcinoma. BMC Oral Health. 2021;21(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, Wang JM, Luo JH, et al. Airway epithelial integrin beta4-deficiency exacerbates lipopolysaccharide-induced acute lung injury. J Cell Physiol. 2021;236(11):7711-7724. [DOI] [PubMed] [Google Scholar]
- 39.He MK, Le Y, Zhang YF, et al. Matrix metalloproteinase 12 expression is associated with tumor FOXP3(+) regulatory T cell infiltration and poor prognosis in hepatocellular carcinoma. Oncol Lett 2018;16(1):475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell 2016;165(3):535-550. [DOI] [PubMed] [Google Scholar]
- 41.Kokkinidis M, Glykos NM, Fadouloglou VE. Catalytic activity regulation through post-translational modification: the expanding universe of protein diversity. Adv Protein Chem Struct Biol. 2020;122:97-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das T, Shin SC, Song EJ, Kim EE. Regulation of deubiquitinating enzymes by post-translational modifications. Int J Mol Sci. 2020;21(11):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margheri F, Serrati S, Lapucci A, et al. Systemic sclerosis-endothelial cell antiangiogenic pentraxin 3 and matrix metalloprotease 12 control human breast cancer tumor vascularization and development in mice. Neoplasia 2009;11(10):1106-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates AM, Fischer CL, Abhyankar VP, et al. Matrix metalloproteinase response of dendritic cell, gingival epithelial keratinocyte, and T-cell transwell Co-cultures treated with Porphyromonas gingivalis Hemagglutinin-B. Int J Mol Sci. 2018;19(12):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JT, Pamir N, Liu NC, et al. Macrophage metalloelastase (MMP12) regulates adipose tissue expansion, insulin sensitivity, and expression of inducible nitric oxide synthase. Endocrinology. 2014;155(9):3409-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li R, Han D, Shi J, et al. Choosing tumor mutational burden wisely for immunotherapy: A hard road to explore. Biochim Biophys Acta Rev Cancer. 2020;1874(2):188420. [DOI] [PubMed] [Google Scholar]
- 47.Chan TA, Yarchoan M, Jaffee E, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 2019;30(1):44-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Cheng L, Li C, Zhang C, Wang L, Zhang J. Identification of tumor microenvironment-related prognostic genes in colorectal cancer based on bioinformatic methods. Sci Rep. 2021;11(1):15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hernandez L, Magalhaes MA, Coniglio SJ, Condeelis JS, Segall JE. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res. 2011;13(6):R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Supplemental Material for MMP12 is a Potential Predictive and Prognostic Biomarker of Various Cancers Including Lung Adenocarcinoma by Guo-Sheng Li, Yu-Xing Tang, Wei Zhang, Jian-Di Li, He-Qing Huang, Jun Liu, Zong-Wang Fu, Rong-Quan He, Jin-Liang Kong, Hua-Fu Zhou, and Gang Chen in Cancer Control.
Data Availability Statement
The data that support the findings of pan-cancer analyses are available in public databases, including DepMap Portal (https://depmap.org/portal/), Xena database (https://xenabrowser.net/datapages), Proteomic Data Commons (https://pdc.cancer.gov), Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/), and TCGA Research Network (www.cancer.gov/tcga). Data on in-house tissue samples used during the current study are available from the corresponding author upon reasonable request.