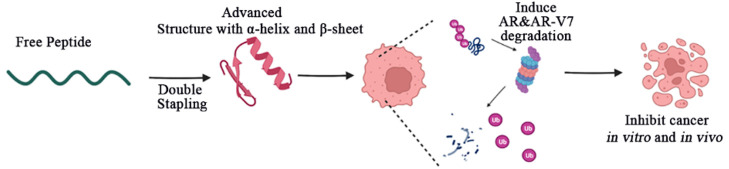
Abstract
Peptide drugs offer distinct advantages in therapeutics; however, their limited stability and membrane penetration abilities hinder their widespread application. One strategy to overcome these challenges is the hydrocarbon peptide stapling technique, which addresses issues such as poor conformational stability, weak proteolytic resistance, and limited membrane permeability. Nonetheless, while peptide stapling has successfully stabilized α-helical peptides, it has shown limited applicability for most β-sheet peptide motifs. In this study, we present the design of a novel double-stapled peptide capable of simultaneously stabilizing both α-helix and β-sheet structures. Our designed double-stapled peptide, named DSARTC, specifically targets the androgen receptor (AR) DNA binding domain and MDM2 as E3 ligase. Serving as a peptide-based PROTAC (proteolysis-targeting chimera), DSARTC exhibits the ability to degrade both the full-length AR and AR-V7. Molecular dynamics simulations and circular dichroism analysis validate the successful constraint of both secondary structures, demonstrating that DSARTC is a “first-in-class” heterogeneous-conformational double-stapled peptide drug candidate. Compared to its linear counterpart, DSARTC displays enhanced stability and an improved cell penetration ability. In an enzalutamide-resistant prostate cancer animal model, DSARTC effectively inhibits tumor growth and reduces the levels of both AR and AR-V7 proteins. These results highlight the potential of DSARTC as a more potent and specific peptide PROTAC for AR-V7. Furthermore, our findings provide a promising strategy for expanding the design of staple peptide-based PROTAC drugs, targeting a wide range of “undruggable” transcription factors.
Keywords: double-stapled peptide, α-helix and β-sheet structures, PROTAC, AR-V7, prostate cancer
Introduction
Peptide drugs have historically faced challenges, including issues such as poor blood stability and limited cell membrane penetration.1,2 However, recent advancements in peptide drug delivery systems and peptide modification chemistry have revitalized interest in the clinical application of peptide drugs.3−6 Notably, progress in peptide modification chemistry has opened up new avenues, with peptide stapling emerging as a highly promising strategy to enhance peptide stability and improve their ability to penetrate cell membranes.7−11 Among various peptide stapling chemistries, the all-hydrocarbon peptide stapling triggered by solid-phase ring-closing metathesis (RCM) reaction has garnered significant attention.12,13 Hydrocarbon-stapled peptides hold great promise as a therapeutic modality for a range of diseases, including cancer, viral infections, and autoimmune disorders.14−17 They offer several advantages over traditional peptide molecules and large protein molecules, including enhanced target specificity, improved cell permeability, and increased oral bioavailability.18−20 Remarkably, ALRN-6924, a representative stapled peptide drug, has progressed into clinical trials for the treatment of advanced solid tumors and lymphomas.21−23
Notably, hydrocarbon-stapled peptides have primarily been utilized to stabilize α-helical secondary conformations of linear peptides.6,24−27 Such peptide motifs commonly occur at the interacting interfaces of protein–protein interactions (PPIs).26,28,29 However, when it comes to other “undruggable” transcription factors, peptides tend to adopt alternative secondary structures, such as β-sheet conformations.30−32 Unfortunately, hydrocarbon peptide stapling has shown limited applicability for stabilizing β-sheet structures. Although there have been reports of stapled peptides based on nonhelical or β-hairpin motifs,33,34 the successful use of hydrocarbon peptide stapling to stabilize β-sheet structures remains rare. Therefore, there is an urgent need to develop β-sheet-stabilized stapled peptides in order to expand the scope of the peptide stapling strategy and target a broader range of “undruggable” molecules.
AR-V7 is a truncated form of the androgen receptor (AR) protein lacking the ligand-binding domain (LBD), which is responsible for binding to androgens like testosterone and dihydrotestosterone.35,36 Its role in the development and progression of castration-resistant prostate cancer (CRPC), a form of prostate cancer resistant to androgen deprivation therapy (ADT), has been extensively studied.37,38 Despite significant advancements in the use of small molecules targeting AR-V7 in recent years, no drugs specifically targeting AR-V7 have been approved for clinical use thus far. This unmet clinical need underscores the necessity for alternative approaches to effectively target AR-V7. In our previous proof-of-concept study, we developed a peptide PROTAC (proteolysis-targeting chimera) drug which could target AR-V7,39 which includes both α-helical and β-sheet peptide motifs in sequence. In this context, we renamed the AR DBD targeting peptide PROTAC to ARTC. The α-helical motif targeted the E3 ligase MDM2, while the β-sheet motif targeted the AR binding domain. Employing a nanoparticle-based delivery technique, ARTC demonstrated favorable characteristics for binding to and degrading AR-V7.39 These findings position ARTC as a peptide-based PROTAC drug candidate for AR and AR-V7.
Peptide-based PROTAC drugs have emerged as a promising alternative solution for various targets.40−44 In order to enhance the clinical potential of ARTC, we propose utilizing a peptide double-stapling strategy to stabilize both the α-helical and β-sheet peptide motifs. Previous studies have demonstrated that double-stapled peptides can exhibit significantly improved proteolytic stability and enhanced biological activities compared to their linear counterparts.15 However, the application of hydrocarbon stapling to simultaneously constrain both helix and sheet structures within a single peptide has not been explored.
In our study, we designed a double-stapled peptide named DSARTC (double-stapled AR targeting peptide PROTAC), which specifically targets the androgen receptor (AR) DNA binding domain (DBD) and MDM2 E3 ligase. This research represents the first instance of hydrocarbon stapling being utilized to stabilize the β-sheet secondary conformation of a peptide, resulting in a double-stapled peptide that incorporates both helix and sheet structures. DSARTC, as the “first-in-class” heterogeneous-conformational double-stapled peptide, has shown remarkable improvements in secondary structures, stability, and membrane permeability compared to the linear ARTC peptide. Additionally, as a stapled-peptide-based PROTAC, DSARTC demonstrates potent degradation capabilities for both AR and AR-V7. Moreover, the peptide has exhibited significant inhibitory effects on prostate cancer cell proliferation both in vitro and in vivo.
Our study not only contributes to the field of peptide-based PROTACs targeting AR-V7 but also presents a novel design concept for long-sequence peptide drugs. Our approach demonstrates that peptide stapling techniques should not be confined solely to stabilizing α-helical structures. Instead, they can be extended to encompass other secondary structures, thereby significantly broadening the druggability and application potential of peptide drugs. By exploring the stabilization of α-helix and β-sheet structures through hydrocarbon stapling, our research highlights the versatility of peptide stapling strategies. This finding opens up new possibilities for the design and development of long-sequence peptide drugs that target a wide range of disease-associated proteins and “undruggable” targets. The ability to stabilize diverse secondary structures with stapling techniques expands the toolkit available for peptide drug design, ultimately increasing the chances of success in developing therapeutically valuable peptide-based interventions.
Results and Discussion
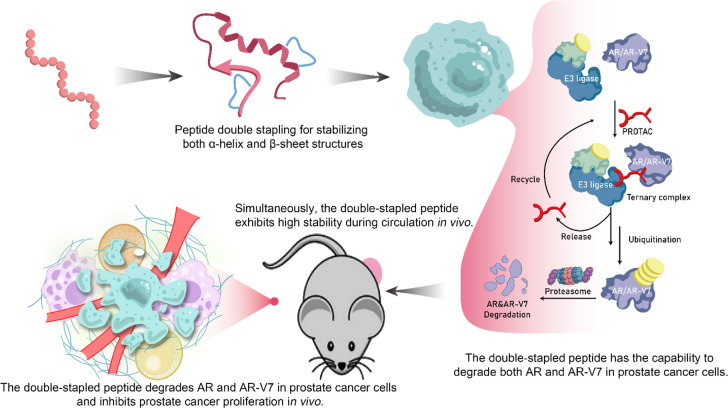
The development of peptide-based drugs is often hindered by inherent limitations such as limited stability and poor membrane penetration, which can greatly impact their therapeutic efficacy. In order to address these challenges, we implemented stapled peptide modification on our previously designed peptide PROTAC sequence that targets AR-V7. This modification approach enhances the stability of the peptide and improves its membrane penetration ability, ultimately augmenting its therapeutic potential. The resulting AR-targeting peptide degradation drug, DSARTC, incorporating staple peptide modifications, demonstrates the ability to effectively degrade intracellular AR and AR-V7, consequently leading to the inhibition of prostate cancer cell proliferation. By overcoming the limitations of stability and membrane penetration, our modified peptide PROTAC drug exhibits promising prospects for therapeutic intervention (Scheme 1).
Scheme 1. Utilization of Double-Stapled Peptide Chemistry in the Development of the Peptide PROTAC Drug Has Resulted in the Acquisition of Sophisticated Structural Features, Including the Simultaneous Incorporation of α-Helix and β-Sheet Motifs.
These structural modifications contribute to the notable enhancement of stability and improved membrane penetration capabilities of the stapled peptide PROTAC drug. Significantly, the stapled peptide PROTAC drugs have demonstrated the capacity to induce the degradation of both AR and AR-V7 in prostate cancer cells. Also, the stapled peptide PROTAC drugs effectively impede the progression of prostate cancer in vitro and in vivo. These findings highlight the potential of stapled peptide PROTAC drugs as a promising therapeutic strategy for the treatment of prostate cancer, offering a new avenue for combating this disease.
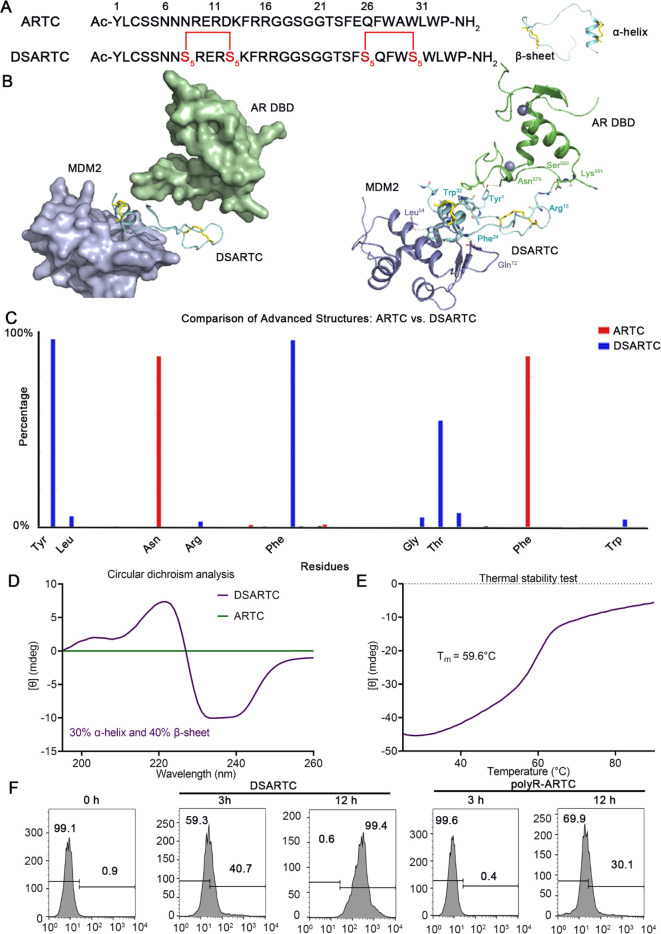
To accurately design the staple sites, we initiated simulations of the free linear peptide, termed ARTC, and its trimer structure with the AR DBD and MDM2. As shown in Figure S1, ARTC exhibited binding to the MDM2 region in an α-helical structure, while maintaining a β-sheet structure with AR DBD. Consequently, the staple modification of this peptide PROTAC presented a challenge, as it required stabilizing both the α-helix and β-sheet structures. To optimize the double-stapling sites in the peptide structure, we first calculated the binding of free peptide ARTC with the AR DBD and MDM2 at various residue locations. As shown in Figure S2, Arg9, Arg11, and Arg15 played a critical role in its binding with AR DBD, whereas Glu10 and Asp12 increased the binding energy. Thus, we choose Asn8–Asp12 as the first stapling sites for sheet structural stabilization. In another motif that is bound with MDM2, we also selected the negative residue pair Glu25–Ala29 for helical enhancement. Taken together, we designed DSARTC by replacing Asn8–Asp12 at the N-terminus and Glu25–Ala29 at the C-terminus with S5–S5 non-natural paired residues using double-peptide all-hydrocarbon stapling strategy (Figure 1A). Next, we successfully synthesized this double-stapled peptide, as presented in Figure S3. With rink amide resin as the support, the first half of the linear peptide until Lys13 was assembled using solid-phase peptide synthesis (SPPS). The first staple ring was formed through the first RCM reaction in the presence of Grubbs’ first-generation catalyst. Then, the remaining DSARTC was assembled, and the second staple was introduced using a similar RCM procedure. The crude double-stapled peptide was cleaved from the resin and purified by reversed-phase high-performance liquid chromatography (RP-HPLC) and verified by HPLC (>95%) and high-resolution mass spectrometry (HR-MS) (Figure S4).
Figure 1.
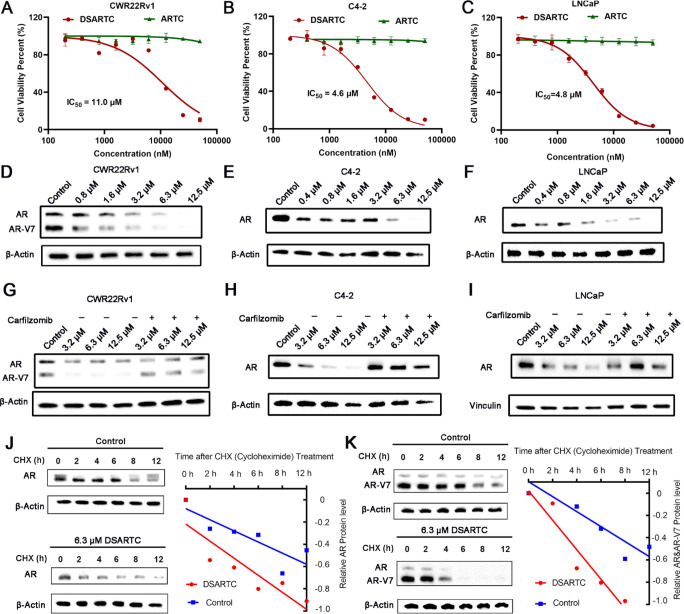
In silico design and structural characterization of DSARTC. (A) amino acid sequences of ARTC and DSARTC and the cartoon structure shown of DSARTC. Double-stapled peptide DSARTC incorporated both α-helix and β-sheet motifs within one peptide sequence. (B) Simulated complex structure of androgen receptor DNA binding domain (AR DBD) and MDM2 induced by the PROTAC peptide. The yellow sticks represent the staple structure, and the green dashes represent the hydron bonds. (C) Secondary structure analysis comparing ARTC and DSARTC by MD simulations. (D) Secondary structure analysis of DSARTC and free peptide using circular dichroism (CD). (E) Thermal stability analysis of DSARTC using CD. (F) Cellular uptake analysis comparing ARTC and polyR-DSARTC by flow cytometry at a used concentration of 5 μM in C4-2 cell lines.
We performed structural modeling analysis of the trimer structure of DSARTC complexed with AR DBD and MDM2. As shown in Figure 1B, DSARTC bound with the target proteins similar to the linear counterpart ARTC, consisting of the helical and sheet motifs. Tyr1 and Arg15, Phe24, and Trp28 can form hydrogen bonds with the AR DBD and MDM2, respectively. We next conducted a comprehensive analysis of the kinetic trajectory for free peptide ARTC and DSARTC, using molecular dynamic simulations. Specifically, we focused on the kinetic trajectory ranging from 500 to 1000 ns, evaluating the probability of advanced folded structure formation for each peptide type. This analysis allowed us to determine the most effective stapled peptide modification strategy that promoted advanced folding of the peptide structure, thus enhancing its stability and druggability. As illustrated in Figure 1C, we observed a higher frequency of advanced folding for peptides with staple modifications at Tyr1, Phe14, Thr22, and Trp28 sites. Simultaneously, simulation results have demonstrated that the selected binding site of the peptide does not affect the interaction between the peptide drug and the target protein. This finding ensures the high affinity between DSARTC and the AR DBD. To validate the predicted structure, we performed circular dichroism (CD) measurements. The CD results confirmed that DSARTC exhibited a significantly higher α-helix content and β-sheet structure content compared to the free peptide (Figure 1D). Additionally, thermal stability experiments supported the robust stability of DSARTC (Figure 1E). Furthermore, tests assessing serum stability and enzyme degradation stability demonstrated that the dual-stapled product, DSARTC, exhibited enhanced resistance to serum and enzyme degradation compared to the free peptide (Figures S5A and S5B). These findings highlight the improved in vivo circulation ability of DSARTC. To verify the superior cell membrane penetration ability of DSARTC compared to ARTC, we employed confocal microscopy to examine the drug’s membrane penetration. Polyarginine was introduced into the linear peptide ARTC, resulting in polyR-ARTC as the control. As illustrated in Figures 1F and S6, DSARTC exhibited a gradual entry into the cancer cells over time, whereas polyR-ARTC failed to penetrate the cell membrane and exert its function.
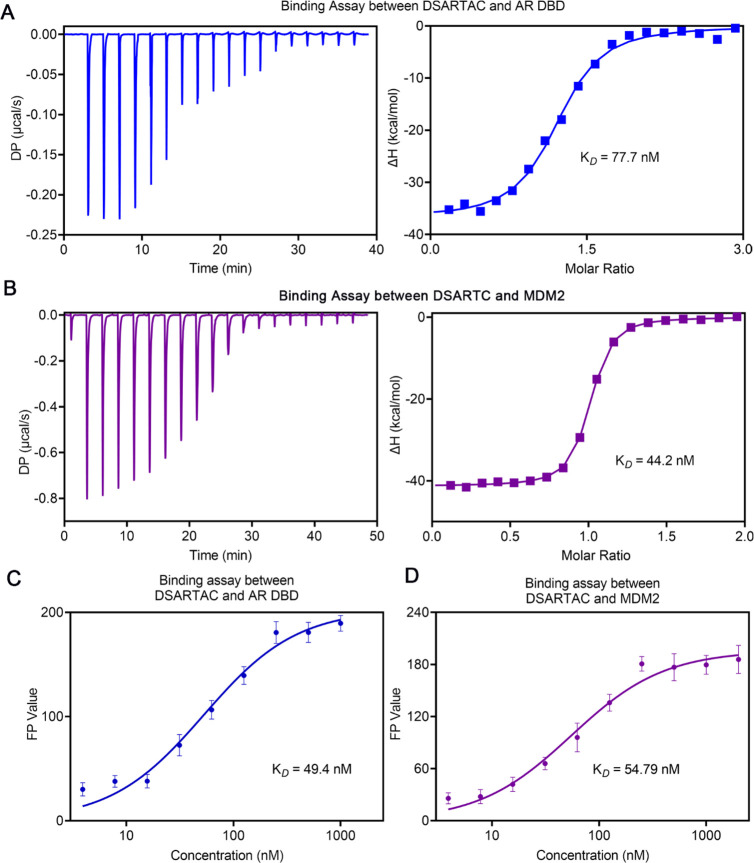
To ensure that the double-stapled peptide modification did not affect the binding of the peptide to the AR DBD and MDM2, we performed binding assays. The goal was to assess whether the stapling modifications altered the binding affinities of the peptide compared to the nonstapled peptide. Affinity constants of DSARTC with AR DBD and MDM2 were determined using techniques such as isothermal titration calorimetry (ITC) and fluorescence polarization (FP). The results, as depicted in Figures 2A to 2D, demonstrated that DSARTC maintained a high binding affinity to both AR DBD and MDM2, similar to the nonstapled peptide. These findings indicate that double-stapled peptide modification did not significantly affect the binding interactions between DSARTC and its target proteins. Thus, the stapling modifications did not compromise the peptide’s ability to bind to AR DBD and MDM2, ensuring its continued efficacy in targeting these proteins. To assess the potential of DSARTC in enhancing the combination of AR and MDM2 within cells, we conducted co-immunoprecipitation (Co-IP) experiments. These experiments aimed to investigate whether DSARTC could facilitate the interaction between endogenous AR and MDM2. The results obtained from the Co-IP experiments demonstrated that DSARTC effectively increased the interaction between endogenous AR and MDM2 (Figure S7). This outcome suggests that DSARTC has the capability to recruit MDM2 to the AR, which may lead to increased degradation of both AR and AR-V7. The enhanced interaction between AR and MDM2 further supports the potential therapeutic utility of DSARTC in promoting the degradation of AR and its variants.
Figure 2.
Affinity detection for DSARTC to MDM2 and AR DBD: (A) binding assays between DSARTC and AR DBD by ITC; (B) binding assays between DSARTC and MDM2 by ITC; (C) binding assay between DSARTC and AR DBD by fluorescence polarization; (D) binding assay between DSARTC and MDM2 by fluorescence polarization.
In order to evaluate the antiproliferative activity of DSARTC, we treated prostate cancer cell lines including C4-2, LNCaP, and CWR22Rv1 with various concentrations of DSARTC. As depicted in Figures 3A–C and S8, we observed a dose-dependent inhibitory effect on cell growth upon treatment with DSARTC, while the nonstapled free peptide ARTC did not exhibit any significant efficacy. The half-maximal degradation concentration (DC50) of DSARTC on AR and AR-V7 in CWR22Rv1 cells is 2.45 and 1.23 μM, respectively. The DC50 of DSARTC on AR in C4-2 and LNCaP cells is 2.67 and 1.87 μM, respectively. This indicates that the double-stapled peptide modification enhances the potency of the peptide PROTAC drug, leading to improved inhibition of prostate cancer cell proliferation. To further investigate the capability of DSARTC to induce AR and AR-V7 degradation, we conducted immunoblot analyses of AR and AR-V7. The results demonstrated that DSARTC effectively induced the degradation of both full-length AR and AR-V7 in a dose-dependent manner (Figures 3D–F). To confirm that DSARTC operates through the proteasome pathway, we performed experiments utilizing proteasome inhibitors. As illustrated in Figures 3G–I, the use of proteasome inhibitors, such as carfilzomib, significantly reversed the degradation of both full-length AR and AR-V7 induced by DSARTC. This observation suggests that the DSARTC-induced degradation of AR and AR-V7 is dependent on the proteasome pathway. Protein half-life experiments have additionally demonstrated that DSARTC exerts its effects by accelerating protein degradation while mitigating the influence of protein synthesis (Figures 3J,K). These findings further support the notion that DSARTC functions as a typical PROTAC drug, triggering targeted protein degradation through the proteasome pathway. As we mentioned below, one of the major advantages of peptide PROTAC is the bigger interface with target protein so peptide PROTAC drug could target proteins that are difficult to target by small molecules. As shown in Figure S9, ARV-110 is the most advanced clinical AR targeting PROTAC drug only for AR length but could not degrade AR-V7. To further investigate the impact of DSARTC on signaling pathways in prostate cancer cells, we conducted a reverse phase protein array analysis following DSARTC treatment in CWR22Rv1 cells. As depicted in Figure S10, numerous proteins were affected, including a significant decrease in the levels of eIF4G and AMPK. These proteins are known to be involved in the downstream signaling pathway of AR, confirming the specificity of DSARTC for its target. Furthermore, through GO and KEGG enrichment analysis (Figures S11–S13), we observed that DSARTC exerts effects on multiple cancer signaling pathways and influences immune-related pathways, such as inflammatory mediation. These findings highlight the therapeutic potential of DSARTC in prostate cancer cells, further supporting its promising role in prostate cancer treatment.
Figure 3.
DSARTC has demonstrated promising potential in inducing AR and AR-V7 degradation as well as inhibiting prostate cancer cells. Detection of cell viability in CWR22Rv1 (A), C4-2 (B), and LNCaP (C) cell lines following treatment with varying concentrations of DSARTAC drugs. Immunoblotting analysis of AR and AR-V7 protein levels in CWR22Rv1 (D), C4-2 (E), and LNCaP (F) cell lines following treatment with varying concentrations of DSARTAC drugs. Immunoblotting analysis of AR and AR-V7 protein levels in the presence of carfilzomib, in CWR22Rv1 (G), C4-2 (H), and LNCaP (I) cell lines following treatment with varying concentrations of DSARTAC drugs. IB analysis of WCLs from C4-2 cells (J) and CWR22Rv1 cells (K) treated with CHX for the indicated times, with or without 6.3 μM DSARTC drug treatment.
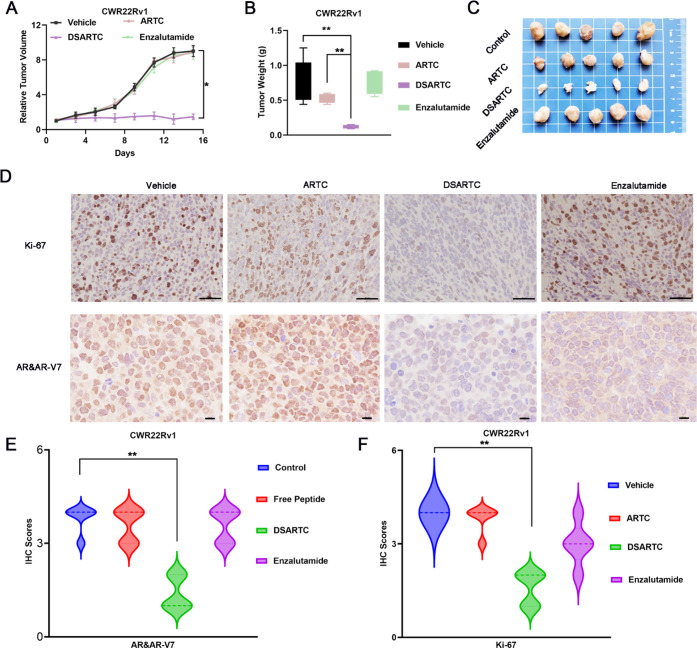
To evaluate the therapeutic potential of DSARTC in vivo, a subcutaneous xenograft mouse model of prostate cancer was established using enzalutamide-resistant CWR22Rv1 cells expressing both AR and AR-V7. The mice were divided into four treatment groups: PBS, free peptide, DSARTC, and enzalutamide. The drugs were administered via intraperitoneal injection every 2 days for a period of 2 weeks, and ARTC, DSARTC, and enzalutamide were administered at a dose of 10 mg/kg. The results of the in vivo efficacy study demonstrated that the free peptide had no significant effect on tumor growth. However, treatment with DSARTC significantly inhibited tumor growth in the CWR22Rv1 xenograft models, as observed in Figures 4A–C. On the other hand, enzalutamide did not show any effect on tumor growth in this model. Histopathological analysis using hematoxylin and eosin (H&E) staining and Ki-67 staining revealed a significant presence of apoptotic tumor cells in the DSARTC-treated tissues (Figure 4D). Additionally, immunohistochemical (IHC) analysis of Ki-67 as well as AR and AR-V7 in each treatment group further confirmed that DSARTC effectively induced the degradation of AR and AR-V7 in vivo (Figures 4E,F). Safety tests, including assessments of body weight and biochemical tests evaluating kidney and liver functions, have provided evidence that DSARTC exhibits no significant toxicity to the liver and kidneys at the administered dosages (Figure S14). Overall, the animal experiments demonstrated the ability of DSARTC to inhibit the proliferation of prostate cancer cells in vivo. These findings highlight the therapeutic potential of the double-stapled peptide and its application prospects in the treatment of prostate cancer.
Figure 4.
In vivo potency evaluation of DSARTC. (A) Tumor growth curves of CWR22Rv1 xenografts in nude mice (n = 5 per group) are presented as the mean ± SD values. The drugs were administered via intraperitoneal injection every 2 days for a period of 2 weeks; ARTC, DSARTC, and enzalutamide were administered at a dose of 10 mg/kg. Statistical analysis was performed using the t test, with ∗ indicating statistical significance at p < 0.05. (B) The average weight of tumors excised from each group of mice at the end of the drug treatment (n = 5) was analyzed using the t test, with ∗∗ indicating statistical significance at p < 0.01. (C) Photos of CWR22Rv1 tumors excised at the end of the experiment after different drug treatments. (D) The excised tumors from the control, free peptide, DSARTC, and enzalutamide groups were subjected to the IHC assay for Ki-67 staining, a marker of tumor cell growth, as well as for AR and AR-V7 staining, measuring the levels of AR and AR-V7 protein in the tumor cells. The scale bar was set to 73 μm. Statistical analysis of the IHC scores for AR and AR-V7 (E) and Ki-67 (F) on CWR22Rv1 tumors was conducted after treatment with different drugs. IHC intensity was scored as follows: 4 for highly positive, 3 for positive, 2 for minimally positive, and 1 for negative.
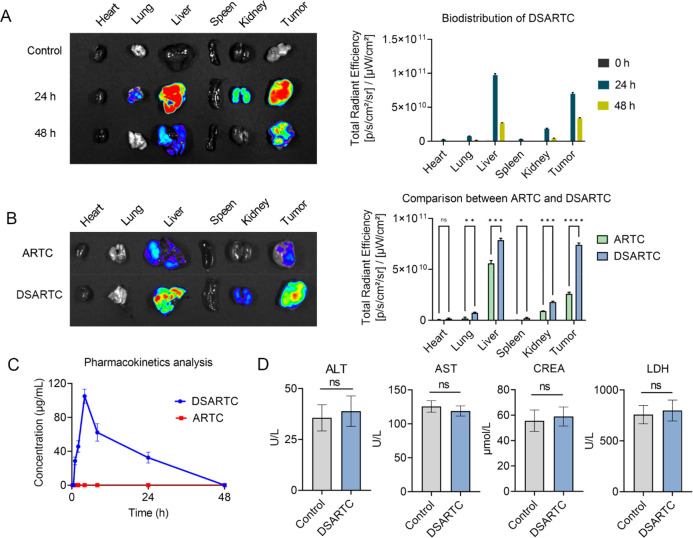
To assess the in vivo circulation stability of DSARTC, we conducted in vivo distribution and metabolism experiments. The results demonstrated that DSARTC exhibits prolonged retention at the tumor site, which is crucial for its long-lasting therapeutic effect. In Figure 5A, the data indicate that DSARTC remains present at the tumor site for more than 48 h, indicating its ability to persist in the target area for an extended period of time. This prolonged tumor retention provides a solid foundation for the drug to exert its therapeutic effects over a longer duration. Furthermore, compared with the free peptide, DSARTC exhibited significantly longer circulation time and enhanced tumor retention (Figure 5B). This indicates that the stapled peptide modification improves the stability and circulation characteristics of DSARTC in vivo, which is advantageous for its therapeutic efficacy. To further evaluate the pharmacokinetics character of DSARTC, LC-MS was used to evaluate the blood drug concentration of DSARTC in mice at different time points after intraperitoneal injection. As shown in Figure 5C, the Cmax of DSARTC is 104 μg/mL, the AUC of DSARTC is 1275.7 μg·h/mL, and the t1/2 of DSARTC is 13.9 h, whereas ARTC is rapidly degraded and undetectable in the blood. These findings highlight the favorable in vivo circulation stability of DSARTC, further supporting its potential as a promising therapeutic agent for prostate cancer treatment. To further assess the potential application of DSARTC, an acute toxicity analysis was conducted. A dosage of 30 mg/kg of DSARTC, three times the standard concentration, was administered for this analysis. Serum samples were collected to detect levels of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatine kinase (CREA), and lactate dehydrogenase (LDH), commonly utilized indicators of kidney and liver toxicity. As depicted in Figure 5D, the results indicate that DSARTC exhibited no significant signs of kidney or liver toxicity at this triple dose. This additional analysis further reinforces the favorable biosafety profile of DSARTC.
Figure 5.
Pharmacokinetics and safety analysis of DSARTC. (A) Biodistribution of DSARTC at different time points in CWR22RV1 xenograft models. (B) Comparison of biodistribution between free peptide and DSARTC in CWR22RV1 xenograft models. (C) Plasma concentration–time curve analysis of DSARTC and ARTC. (D) Acute toxicity analysis of DSARTC.
Conclusion
Through the incorporation of a stabilized β-sheet using hydrocarbon peptide stapling, we have successfully developed the “first-in-class” heterogeneous-conformational double-stapled peptide, DSARTC. This innovative approach represents the first report on the utilization of β-sheet modification within a peptide, showcasing the potential of hydrocarbon peptide stapling for alternative structural stabilization. By incorporating a stabilized β-sheet through hydrocarbon peptide stapling instead of the conventional α-helix, we have introduced novel possibilities in the realm of peptide stapling-assisted conformational constraining. This study marks the first report on the utilization of β-structure-modified peptide PROTAC drugs, showcasing their potential in the field. Furthermore, DSARTC stands out as the inaugural double-stapled peptide capable of simultaneously stabilizing both α-helix and beta-sheet structures.
These findings demonstrate that the incorporation of a β-structure modification not only enhances the binding affinity of the peptide PROTAC drug to the target protein but also improves the proteasomal degradation of the target protein. This bears particular significance for challenging targets such as AR-V7, which plays a critical role in the resistance observed in current prostate cancer therapies. The presented staple peptide PROTAC strategy not only addresses castration-resistant prostate cancer (CRPC) but also holds promise for targeting other traditionally deemed “undruggable” targets. The in vitro and in vivo experiments conducted in this study provide robust evidence supporting the efficacy of DSARTC as a potential therapeutic option for CRPC patients with enzalutamide resistance. Overall, this research significantly contributes to the field of PROTAC drug development, offering valuable insights and potential solutions for future investigations of cancer therapy.
Methods
Materials
Rink amide MBHA resin was obtained from Nankai Hecheng Science & Technology Co. Ltd. All amino acids, reagents, and solvents were sourced from Qir biochem or Titan Scientific Co. Ltd. The following antibodies were used: anti-AR (sc7305) and anti-GAPDH (sc-47724) from Santa Cruz; antivinculin antibody (V-4505), anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), secondary anti-mouse antibody, and secondary anti-rabbit antibody from Sigma-Aldrich. CCK8 reagents were purchased from Dojindo Laboratories.
Stapled Peptide Structure Simulation and Dynamic Analysis
The structures of MDM2 (PDB: 6Y4Q) and AR DBD (PDB: 1r4i) were picked as the initial models. The topology and coordinate files were constructed for the protein–peptide system using Amber’s leap. The peptide was first built using the ESMFold based on the natural peptide sequence. The stapled portion was then modified and bonded correctly. Force field files were generated using Ambertools. The ff14SB force field was used for the protein, the Generalized Amber Force Field 2 (gaff2) for the nonstandard amino acid, and the TIP3P model for water. The energy of the system was minimized using OpenMM to remove any clashes. The particle mesh Ewald (PME) method was used for long-range electrostatics with a 10 Å cutoff. The L-BFGS minimizer was used for 2000 steps. The system was heated and equilibrated with the NVT ensemble. The Langevin integrator was used with a 2 fs time step, The temperature was raised from 0 to 310 K over 200 ps. The system was equilibrated in the NPT ensemble. The Monte Carlo barostat was used to maintain 1 bar pressure with an anisotropic box scale. The Langevin integrator was used for temperature control as before. The system was equilibrated for 2 ns. A production MD simulation was run in the NPT ensemble. The simulation was run for 500 ns, saving frames every 10 ps. The trajectory was analyzed. The MMPBSA method was used to calculate the binding free energies of important amino acids. The DSSP program was used to calculate the β-sheet content of the stapled peptide. A short molecular docking simulation was performed to minimize this complex by 500 ps at constant pressure and temperature with a time step of 1 fs. Nonstandard amino acid parameters were determined using tleap, which is a program in AmberTools22. A cutoff of 10 Å was used for nonbonded interactions, and long-range electrostatic interactions were treated with the particle mesh Ewald method. All figures were drawn by using PyMOL.
Synthesis of Double-Stapled Peptides
Conventional solid-phase peptide synthesis (SPPS) was employed for the assembly of linear peptides. Rink amide MBHA resin (333 mg) was swollen in dichloromethane for 20 min at room temperature. The Fmoc group of the amino acid was deprotected using 20% piperidine in DMF (5 min × 2). Fmoc-AA-OH (0.5 mmol, 5 equiv), DIC (1 mmol, 10 equiv), and oxyme (0.5 mmol, 5 equiv) were dissolved in NMP (6 mL) and added to the resin, followed by shaking at 60 °C for 20 min to couple the first amino acid. The resin was then washed with DMF and DCM. Deprotection, washing, and coupling steps were repeated until the coupling of the third S5 amino acid to the resin. Notably, Fmoc-S5-OH and subsequent amino acids required 2 h for coupling. The resin was subjected to RCM reaction at room temperature using Grubbs’ first-generation catalyst (10 mM) in 1,2-dichloroethane (6 mL) for 2 h (2 cycles). Deprotection, washing, and coupling steps were repeated, until the last amino acid was attached to the resin. After deprotection, a solution of acetic anhydride and pyridine (1:1, 6 mL) was added to the resin. The second round of RCM reaction was performed using Grubbs’ first-generation catalyst (10 mM) in 1,2-dichloroethane (6 mL). Finally, the crude peptides were cleaved from the resin using cocktail B reagent (88% TFA, 2% TIPS, 5% water, and 5% phenols) for 4 h. The crude peptides were precipitated by adding cold diethyl ether to the solution followed by centrifugation.
HPLC and MS
The purified peptides were obtained using a preparative HPLC system equipped with a YMC-Pack ODS-AQ column (250 × 20 mm2, S-5 μm, 12 nm) with a flow rate of 20 mL/min. For analysis, an analytic HPLC system with a Waters “XBridge C18” column (4.6 × 150 mm2, 5 μm particle size) was used, operating at a flow rate of 1.0 mL/min. The gradient elution method involved a linear increase from 5% to 65% of solution B over a period of 5–30 min. The column temperature was maintained at a maximum of 30 °C, and the absorption was monitored at 214 nm. The mobile phase consisted of solution A (0.1% TFA in water) and solution B (0.1% TFA in MeCN). Finally, each peptide was identified using HR-Q-TOF-MS analysis conducted on an Agilent 6538 UHD Accurate Mass Q-TOF mass spectrometer.
ITC (Isothermal Titration Calorimetry)
ITC is a well-established technique utilized for determining the binding affinity between proteins and ligands. The signal measured during this process corresponds to the heat released upon interaction of the two reactants. MDM2 and AR-V7 proteins were obtained via E. coli expression and subsequently purified by using affinity chromatography. Prior to commencing ITC experiments, both the sample cells and injection syringe were meticulously cleaned following the manufacturer’s protocol. The sample cell was rinsed several times with 2 mL of buffer solution. Subsequently, the sample cell was loaded with 0.2 mL of protein solution (all proteins maintained a concentration of approximately 20 μM). Care was taken to prevent bubble formation during this loading process. Simultaneously, the syringe was filled with 0.04 mL of peptide solution (all peptides maintained concentrations around 200 μM). The ITC experiments were conducted at a temperature of 25 °C. An initial injection volume of 0.4 μL was administered, followed by subsequent injections maintained at a desired volume of 2 μL for 19 injections. Upon completion of the experiment, the acquired ITC data underwent analysis by using dedicated software provided by the instrument.
Cell Culture
LNCap, C4-2, and CWR22Rv1 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS).
Cell Viability Assay
Cells were seeded at a density of 4 × 103 cells per well in 96-well plates with three independent samples plated in duplicate for each concentration. After 24 h of treatment with free peptide or DSARTC, cell viability assays were conducted. Following a 2 h incubation with CCK-8 reagent, the absorbance values at 450 nm were measured using a microplate reader.
Immunoblotting
For immunoblotting analysis, cells were lysed in EBC buffer (50 mM Tris (pH, 7.5), 120 mM NaCl, and 0.5% NP-40) supplemented with protease inhibitors. The protein concentration in cell lysates was determined using a Pierce bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher). Equal amounts of the total protein were loaded onto SDS-PAGE gels for separation. The separated proteins were then transferred to poly(vinylidene fluoride) (PVDF) membranes and incubated with specific primary antibodies overnight at 4 °C. After incubation with an HRP-conjugated secondary antibody for 1 h at room temperature, the protein immunoreactive signals were detected.
Drug Uptake Analysis by Confocal and Flow Cytometer
Before DSARTC uptake was analyzed using confocal microscopy and a flow cytometer, DSARTC underwent initial labeling with fluorescein-Cy3. This labeling process involved the reaction of Cy3 esterified by NHS with the peptide drug’s amino group. For the confocal assay, C4-2 cells were seeded onto confocal plates. Following a 24 h incubation period, the cells were exposed to 5 μM of the drug at various time points, and subsequent confocal observations were conducted. DAPI was utilized for nuclear staining. In the flow assay, C4-2 cells were seeded in 6-well plates and allowed to attach for 24 h. Following this, the cells were exposed to 5 μM of the drug at different time intervals. Subsequently, the cells were digested and fixed to facilitate the flow analysis.
Immunoprecipitation Assays
For immunoprecipitation (IP) assays, a total of 3 mg of cell lysate was incubated with anti-Protein A/G-anti-AR beads for 4 h at 4 °C in the absence or presence of DSARTC. After incubation, the cell lysates were washed with EBC buffer to remove nonspecific binding. The proteins bound to the beads were then extracted by boiling at 95 °C for 5 min. The extracted proteins were separated by SDS-PAGE gels, transferred to PVDF membranes, and incubated with specific primary antibodies overnight at 4 °C. Following this, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature, and the protein immunoreactive signals were detected.
Evaluation of Antitumor Activity in CWR22Rv1 Xenograft Model
The treatment of four groups, each comprising 5 mice, commenced on day 1 once the tumors had reached a palpable size of approximately 30 mm3. In each medication treatment group, mice received injections every other day, spanning a total treatment period of 3 weeks. The mice were distributed across four treatment groups: PBS, free peptide, DSARTC, and enzalutamide. The drugs were administered via intraperitoneal injection every 2 days for a duration of 2 weeks, with ARTC, DSARTC, and enzalutamide administered at a dose of 10 mg/kg. Tumor dimensions were measured using calipers to determine length and width, and tumor volume was calculated using the formula tumor volume (V) = length × width2/2. Immunohistochemistry (IHC) staining images underwent analysis and scoring using the ImageJ profiler.
Pharmacokinetics Analysis
The objective of this study is to ascertain the pharmacokinetic (PK) profiles of DSARTC subsequent to intraperitoneal administration in male C57BL/6J mice. Animal feeding control measures will be implemented, wherein mice intended for subcutaneous (SC) administration will undergo an overnight fasting period, followed by feeding after a 4 h collection period. Upon drug injection, approximately 0.03 mL of blood will be collected at each specified time point and centrifuged at 4000g for 5 min in a 4 °C centrifuge. The resultant plasma samples were stored in a freezer at −75 ± 15 °C until analysis. The desired serial concentrations of working solutions were achieved by diluting stock solution of analyte with 0.1% formic acid and 50% methyl alcohol in DMSO solution. 20 μL of working solutions (5, 10, 20, 50, 100, 200, 500, 1000, 5000, and 10000 ng/mL) were added to 20 μL of the blank C57BL/6J mouse plasma to achieve calibration standards of 5–10000 ng/mL (5, 10, 20, 50, 100, 200, 500, 1000, 5000, and 10000 ng/mL) in a total volume of 40 μL. Four quality control samples at 10, 20, 50, 500, and 8000 ng/mL for plasma were prepared independently of those used for the calibration curves. These QC samples were prepared on the day of analysis in the same way as calibration standards. 40 μL of standards, 40 μL of QC samples, and 40 μL of unknown samples (20 μL of SD rat plasma with 20 μL of blank solution) were added to 150 μL of methyl alcohol containing an IS mixture for precipitating protein. Then the samples were vortexed for 30 s. After centrifugation at 4 °C and 4000 rpm for 15 min, the supernatant was diluted 2 times with 0.1% formic acid and 5% acetonitrile in water solution. 15 μL of diluted supernatant was injected into the LC/MS/MS system for quantitative analysis. Pharmacokinetic calculations were performed using WinNonlin software. Notably, the pharmacokinetic analysis was conducted by PHARMARON Inc.
Acute Toxicity Analysis of DSARTC
Acute toxicity analysis of DSARTC was conducted using male C57BL/6J mice. For this analysis, a dosage of 30 mg/kg of DSARTC, three times the standard concentration, was administered every other day. Blood samples were obtained from the mice, and subsequent serum samples were isolated to assess levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatine kinase (CREA), and lactate dehydrogenase (LDH).
Ethical Approval
All experimental procedures involving animals were conducted in accordance with institutional guidelines and approved by the Laboratory Animal Center and Biomedical Ethics Committee of Xi’an Jiaotong University. The study was assigned approval number 2020(G-135).
Statistics
The data are presented as the mean ± SD and represent triplicate samples. Statistical analyses were performed by using GraphPad Prism software. Differences among multiple groups and the effects of treatment in both in vivo and in vitro studies were assessed by using the nonparametric Kruskal–Wallis test for multiple comparisons and the t test for two-group comparisons. A p value of 0.05 or less was considered statistically significant for single comparisons.
Acknowledgments
The reverse phase protein array analysis was supported by FynnBio@.
Glossary
Abbreviations
- AR
androgen receptor
- PROTAC
proteolysis-targeting chimera.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacsau.3c00795.
Simulated structure, synthesis schematic, and synthesis results of DSARTC, along with the stability assessment and cellular uptake confocal analysis of DSARTC; experiments conducted to investigate the augmentation of AR binding to MDM2 facilitated by DSARTC within cells; analysis of proteomic changes of CWR22rv1 cells after DSARTC treatment (PDF)
Author Contributions
B.M. and D.L. contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was partially supported by the National Key R&D Program of China (2023YFC3404100), the National Natural Science Foundation of China (No. 81925028 and 82002717), the Shanghai Rising-Star Program and the Natural Science Foundation of Shaanxi Province of China (2021ZDLSF02-15), and the Institutional Foundation of the First Affiliated Hospital of Xi’an Jiaotong University (2021QN-14).
The authors declare no competing financial interest.
Supplementary Material
References
- Muttenthaler M.; King G. F.; Adams D. J.; Alewood P. F. Trends in peptide drug discovery. Nat. Rev. Drug Discovery 2021, 20 (4), 309–325. 10.1038/s41573-020-00135-8. [DOI] [PubMed] [Google Scholar]
- Henninot A.; Collins J. C.; Nuss J. M. The current state of peptide drug discovery: back to the future?. Journal of medicinal chemistry 2018, 61 (4), 1382–1414. 10.1021/acs.jmedchem.7b00318. [DOI] [PubMed] [Google Scholar]
- Yadav A. R.; Mohite S. K. Recent advances in protein and peptide drug delivery. Research journal of pharmaceutical dosage forms and technology 2020, 12 (3), 205–212. 10.5958/0975-4377.2020.00035.X. [DOI] [Google Scholar]
- Patil S.; Vhora I.; Amrutiya J.; Lalani R.; Misra A. Role of nanotechnology in delivery of protein and peptide drugs. Current pharmaceutical design 2015, 21 (29), 4155–4173. 10.2174/1381612821666150901095722. [DOI] [PubMed] [Google Scholar]
- Zorzi A.; Deyle K.; Heinis C. Cyclic peptide therapeutics: past, present and future. Curr. Opin. Chem. Biol. 2017, 38, 24–29. 10.1016/j.cbpa.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Li X.; Chen S.; Zhang W.-D.; Hu H.-G. Stapled helical peptides bearing different anchoring residues. Chem. Rev. 2020, 120 (18), 10079–10144. 10.1021/acs.chemrev.0c00532. [DOI] [PubMed] [Google Scholar]
- Chu Q.; Moellering R. E.; Hilinski G. J.; Kim Y.-W.; Grossmann T. N.; Yeh J. T.-H.; Verdine G. L. Towards understanding cell penetration by stapled peptides. MedChemComm 2015, 6 (1), 111–119. 10.1039/C4MD00131A. [DOI] [Google Scholar]
- Tan Y. S.; Lane D. P.; Verma C. S. Stapled peptide design: principles and roles of computation. Drug Discovery Today 2016, 21 (10), 1642–1653. 10.1016/j.drudis.2016.06.012. [DOI] [PubMed] [Google Scholar]
- Bird G. H.; Mazzola E.; Opoku-Nsiah K.; Lammert M. A.; Godes M.; Neuberg D. S.; Walensky L. D. Biophysical determinants for cellular uptake of hydrocarbon-stapled peptide helices. Nat. Chem. Biol. 2016, 12 (10), 845–852. 10.1038/nchembio.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y.; Fois G.; Flores J. R.; Tuvim M. J.; Zhou Q.; Yang K.; Leitz J.; Peters J.; Zhang Y.; Pfuetzner R. A.; Esquivies L.; Jones P.; Frick M.; Dickey B. F.; Brunger A. T. Inhibition of calcium-triggered secretion by hydrocarbon-stapled peptides. Nature 2022, 603 (7903), 949–956. 10.1038/s41586-022-04543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Geng H.; Zhang Q.; Liu Q.; Xie M.; Sun C.; Li W.; Lin H.; Jiang F.; Wang T.; Wu Y.-D.; Li Z. An in-tether chiral center modulates the helicity, cell permeability, and target binding affinity of a peptide. Angew. Chem. 2016, 128 (28), 8145–8149. 10.1002/ange.201602806. [DOI] [PubMed] [Google Scholar]
- Ueda A.; Makura Y.; Kakazu S.; Kato T.; Umeno T.; Hirayama K.; Doi M.; Oba M.; Tanaka M. E-selective ring-closing metathesis in α-helical stapled peptides using carbocyclic α, α-disubstituted α-amino acids. Org. Lett. 2022, 24 (4), 1049–1054. 10.1021/acs.orglett.1c04256. [DOI] [PubMed] [Google Scholar]
- Ali A. M.; Atmaj J.; Van Oosterwijk N.; Groves M. R.; Dömling A. Stapled peptides inhibitors: a new window for target drug discovery. Computational and structural biotechnology journal 2019, 17, 263–281. 10.1016/j.csbj.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A. R.; Maity A.; Chakraborty S.; Chakrabarti R. Computational design of stapled peptide inhibitor against SARS-CoV-2 receptor binding domain. Peptide Science 2022, 114 (5), e24267 10.1002/pep2.24267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.; Li X.; Li Y.; Yuan X.; Geng C.; Gao S.; Li J.; Ma B.; Wang Z.; Lu W.; Hu H.-G. Design of stapled peptide-based PROTACs for MDM2/MDMX atypical degradation and tumor suppression. Theranostics 2022, 12 (15), 6665. 10.7150/thno.75444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M.; Cong W.; Peng H.; Qing J.; Shen H.; Tang Y.; Geng C.; Chen S.; Zou Y.; Zhang W.-D.; Hu H.-G.; Li X. Stapled peptides targeting SARS-CoV-2 spike protein HR1 inhibit the fusion of virus to its cell receptor. J. Med. Chem. 2021, 64 (23), 17486–17495. 10.1021/acs.jmedchem.1c01681. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Deng Q.; Zhao H.; Xie M.; Chen L.; Yin F.; Qin X.; Zheng W.; Zhao Y.; Li Z. Development of stabilized peptide-based PROTACs against estrogen receptor α. ACS Chem. Biol. 2018, 13 (3), 628–635. 10.1021/acschembio.7b00985. [DOI] [PubMed] [Google Scholar]
- Verdine G. L.; Hilinski G. J. All-hydrocarbon stapled peptides as synthetic cell-accessible mini-proteins. Drug Discovery Today: Technologies 2012, 9 (1), e41–e47. 10.1016/j.ddtec.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Xie X.; Gao L.; Shull A. Y.; Teng Y. Stapled peptides: providing the best of both worlds in drug development. Future Medicinal Chemistry 2016, 8 (16), 1969–1980. 10.4155/fmc-2016-0102. [DOI] [PubMed] [Google Scholar]
- Cong W.; Shen H.; Liao X.; Zheng M.; Kong X.; Wang Z.; Chen S.; Li Y.; Hu H.; Li X. Discovery of an orally effective double-stapled peptide for reducing ovariectomy-induced bone loss in mice. Acta Pharmaceutica Sinica B 2023, 13, 3770. 10.1016/j.apsb.2023.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F.; Saleh M. N.; Infante J. R.; Goel S.; Falchook G. S.; Shapiro G.; Chung K. Y; Conry R. M.; Hong D. S.; Wang J. S.-Z.; Steidl U.; Walensky L. D.; Guerlavais V.; Payton M.; Annis D. A.; Aivado M.; Patel M. R. Phase I trial of a novel stapled peptide ALRN-6924 disrupting MDMX-and MDM2-mediated inhibition of WT p53 in patients with solid tumors and lymphomas. J. Clin Oncol 2017, 35, 2505. 10.1200/JCO.2017.35.15_suppl.2505. [DOI] [Google Scholar]
- Saleh M. N.; Patel M. R.; Bauer T. M.; Goel S.; Falchook G. S.; Shapiro G. I.; Chung K. Y.; Infante J. R.; Conry R. M.; Rabinowits G.; Hong D. S.; Wang J. S.; Steidl U.; Walensky L. D.; Naik G.; Guerlavais V.; Vukovic V.; Annis D. A.; Aivado M.; Meric-Bernstam F. Phase 1 trial of ALRN-6924, a dual inhibitor of MDMX and MDM2, in patients with solid tumors and lymphomas bearing wild-type TP53. Clin. Cancer Res. 2021, 27 (19), 5236–47. 10.1158/1078-0432.CCR-21-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallman D. A.; Borate U.; Cull E. H.; Donnellan W. B.; Komrokji R. S.; Steidl U. G.; Corvez M. M.; Payton M.; Annis D. A.; Pinchasik D.; Aivado M.; Verma A. Phase 1/1b study of the stapled peptide ALRN-6924, a dual inhibitor of MDMX and MDM2, as monotherapy or in combination with cytarabine for the treatment of relapsed/refractory AML and advanced MDS with TP53 wild-type. Blood 2018, 132, 4066. 10.1182/blood-2018-99-118780. [DOI] [Google Scholar]
- Morgan D. C.; McDougall L.; Knuhtsen A. G.; Jamieson A. Development of Bifunctional, Raman Active Diyne-Girder Stapled α-Helical Peptides. Chem.—Eur. J. 2023, 29 (41), e202300855 10.1002/chem.202300855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K.; Yin F.; Zhou Z.; Lian C.; Liu Y.; Sun C.; Li W.; Zhang J.; Li Z. Directional assembly of a stapled α-helical peptide. Chem. Commun. 2019, 55 (70), 10484–10487. 10.1039/C9CC04591K. [DOI] [PubMed] [Google Scholar]
- Chen H.; Zhan M.; Liu J.; Liu Z.; Shen M.; Yang F.; Kang Y.; Yin F.; Li Z. Structure-Based Design, Optimization, and Evaluation of Potent Stabilized Peptide Inhibitors Disrupting MTDH and SND1 Interaction. J. Med. Chem. 2022, 65 (18), 12188–99. 10.1021/acs.jmedchem.2c00862. [DOI] [PubMed] [Google Scholar]
- Jiang Y.; Zhang W.; Yang F.; Wan C.; Cai X.; Liu J.; Zhang Q.; Li Z.; Han W. Molecular design of stapled pentapeptides as building blocks of self-assembled coiled coil-like fibers. Science Advances 2021, 7 (4), eabd0492 10.1126/sciadv.abd0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarsson Y.; Jemth P. Affinity and specificity of motif-based protein-protein interactions. Curr. Opin. Struct. Biol. 2019, 54, 26–33. 10.1016/j.sbi.2018.09.009. [DOI] [PubMed] [Google Scholar]
- Russell R. B.; Alber F.; Aloy P.; Davis F. P.; Korkin D.; Pichaud M.; Topf M.; Sali A. A structural perspective on protein-protein interactions. Curr. Opin. Struct. Biol. 2004, 14 (3), 313–24. 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Ferrie J. J.; Karr J. P.; Tjian R.; Darzacq X. Structure”-function relationships in eukaryotic transcription factors: The role of intrinsically disordered regions in gene regulation. Mol. Cell 2022, 82 (21), 3970–84. 10.1016/j.molcel.2022.09.021. [DOI] [PubMed] [Google Scholar]
- Bokhovchuk F.; Mesrouze Y.; Meyerhofer M.; Fontana P.; Zimmermann C.; Villard F.; Erdmann D.; Kallen J.; Scheufler C.; Velez-Vega C.; Chène P. N-terminal β-strand in YAP is critical for stronger binding to scalloped relative to TEAD transcription factor. Protein Sci. 2023, 32 (1), e4545 10.1002/pro.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M. F.; Moore C. D.; Schulz K. N.; Alberto O.; Donague G.; Harrison M. M.; Zhu H.; Zaret K. S. Structural features of transcription factors associating with nucleosome binding. Molecular cell 2019, 75 (5), 921–32. 10.1016/j.molcel.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvarajan V.; Tram N. D.; Xu J.; Ngen S. T.; Koh J.-J.; Teo J. W.; Yuen T.-Y.; Ee P. L. R. Stapled β-Hairpin Antimicrobial Peptides with Improved Stability and Activity against Drug-Resistant Gram-Negative Bacteria. J. Med. Chem. 2023, 66, 8498. 10.1021/acs.jmedchem.3c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J. R.; Lampkin B. J.; Abakah C.; Moyer A.; Miao J.; Deprey K.; Cerulli R. A.; Lin Y.-S.; Baleja J. D.; Baker D.; Kritzer J. A. Stapled β-Hairpins Featuring 4-Mercaptoproline. J. Am. Chem. Soc. 2021, 143 (37), 15039–15044. 10.1021/jacs.1c04378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uo T.; Plymate S. R.; Sprenger C. C. The potential of AR-V7 as a therapeutic target. Expert Opinion on Therapeutic Targets 2018, 22 (3), 201–216. 10.1080/14728222.2018.1439016. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Wu D.; Thomas-Ahner J. M.; Lu C.; Zhao P.; Zhang Q.; Geraghty C.; Yan P. S.; Hankey W.; Sunkel B.; Cheng X.; Antonarakis E. S.; Wang Q.-E.; Liu Z.; Huang T. H.-M.; Jin V. X.; Clinton S. K.; Luo J.; Huang J.; Wang Q. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (26), 6810–6815. 10.1073/pnas.1718811115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z.; Li J.; Liu Y.; Shi Z.; Xuan Z.; Yang K.; Xu C.; Bai Y.; Fu M.; Xiao Q.; Sun H.; Shao C. The crucial role of AR-V7 in enzalutamide-resistance of castration-resistant prostate cancer. Cancers 2022, 14 (19), 4877. 10.3390/cancers14194877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yu C.; Shao Z.; Xia X.; Hu T.; Kong W.; He X.; Sun W.; Deng Y.; Liao Y.; Huang H. Selective degradation of AR-V7 to overcome castration resistance of prostate cancer. Cell death & disease 2021, 12 (10), 857. 10.1038/s41419-021-04162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B.; Fan Y.; Zhang D.; Wei Y.; Jian Y.; Liu D.; Wang Z.; Gao Y.; Ma J.; Chen Y.; Xu S.; Li L. De Novo Design of an Androgen Receptor DNA Binding Domain-Targeted peptide PROTAC for Prostate Cancer Therapy. Advanced Science 2022, 9 (28), 2201859. 10.1002/advs.202201859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Luo Q.; Wang Y.; Liang H.; Wang Y.; Hou Z.; Wan C.; Wang Y.; Liu Z.; Ye Y.; Zhu Li.; Wu J.; Yin F.; Li Z. Targeted Biomolecule Regulation Platform: A Split-and-Mix PROTAC Approach. J. Am. Chem. Soc. 2023, 145 (14), 7879–7887. 10.1021/jacs.2c12824. [DOI] [PubMed] [Google Scholar]
- Jin J.; Wu Y.; Chen J.; Shen Y.; Zhang L.; Zhang H.; Chen L.; Yuan H.; Chen H.; Zhang W.; Luan X. The peptide PROTAC modality: a novel strategy for targeted protein ubiquitination. Theranostics 2020, 10 (22), 10141. 10.7150/thno.46985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.; Jiao Z.; Hou Z.; Wang R.; Lian C.; Xing Y.; Luo Q.; An Y.; Yang F.; Wang Y.; Sha X.; Ruan Z.; Ye Y.; Liu Z.; Li Z.; Yin F. Selective Protein of Interest Degradation through the Split-and-Mix Liposome Proteolysis Targeting Chimera Approach. J. Am. Chem. Soc. 2023, 145 (40), 21860–21870. 10.1021/jacs.3c05948. [DOI] [PubMed] [Google Scholar]
- Liao H.; Li X.; Zhao L.; Wang Y.; Wang X.; Wu Y.; Zhou X.; Fu W.; Liu L.; Hu H.-G.; Chen Y.-G. A PROTAC peptide induces durable β-catenin degradation and suppresses Wnt-dependent intestinal cancer. Cell discovery 2020, 6 (1), 35. 10.1038/s41421-020-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Zhang M.; Liu S.; He Z.; Wang R.; Liang M.; An Y.; Jiang C.; Song C.; Ning Z.; Yin F.; Huang H.; Li Z.; Ye Y. Targeting UBE2C for degradation by bioPROTACs based on bacterial E3 ligase. Chin. Chem. Lett. 2023, 34 (4), 107732. 10.1016/j.cclet.2022.08.012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.