Abstract
Regulation of the IκBα and IκBβ proteins is critical for modulating NF-κB-directed gene expression. Both IκBα and IκBβ are substrates for cellular kinases that phosphorylate the amino and carboxy termini of these proteins and regulate their function. In this study, we utilized a biochemical fractionation scheme to purify a kinase activity which phosphorylates residues in the amino and carboxy termini of both IκBα and IκBβ. Peptide microsequence analysis by capillary high-performance liquid chromatography ion trap mass spectroscopy revealed that this kinase was the DNA-dependent protein kinase catalytic subunit (DNA-PKcs). DNA-PK phosphorylates serine residue 36 but not serine residue 32 in the amino terminus of IκBα and also phosphorylates threonine residue 273 in the carboxy terminus of this protein. To determine the biological relevance of DNA-PK phosphorylation of IκBα, murine severe combined immunodeficiency (SCID) cell lines which lack the DNA-PKcs gene were analyzed. Gel retardation analysis using extract prepared from these cells demonstrated constitutive nuclear NF-κB DNA binding activity, which was not detected in extracts prepared from SCID cells complemented with the human DNA-PKcs gene. Furthermore, IκBα that was phosphorylated by DNA-PK was a more potent inhibitor of NF-κB binding than nonphosphorylated IκBα. These results suggest that DNA-PK phosphorylation of IκBα increases its interaction with NF-κB to reduce NF-κB DNA binding properties.
NF-κB comprises a family of proteins including p50, p52, p65 or RelA, p100, p105, and c-Rel which regulate the expression of a variety of cellular and viral genes (reviewed in references 7, 75, and 79). Each of these proteins contains a region known as the Rel homology domain which is critical for the DNA binding and dimerization properties of these proteins. One of the major regulatory mechanisms which control NF-κB activity is the unique cellular localization of different members of this family. In unstimulated cells, p65 or RelA is nearly exclusively localized in the cytoplasm (4–6, 13, 34), but it translocates to the nucleus upon treatment of the cells with a variety of inducers such as phorbol esters, interleukin 1, and tumor necrosis factor alpha (TNF-α) (43, 73). RelA dimerizes with other NF-κB family members (7, 75, 79) and activates gene expression via its potent transactivation domain (8, 67, 70). Thus, cellular proteins which regulate the nuclear translocation of NF-κB are critical for the control of NF-κB activation of viral and cellular genes.
The IκB proteins constitute a group of cytoplasmic proteins that bind to NF-κB and sequester these proteins in the cytoplasm by preventing their nuclear localization. A number of different IκB proteins have been identified including IκBα, IκBβ, IκBγ (reviewed in reference 79), and IκBɛ (80). IκBα (41) and IκBβ (76) are the best studied of these regulatory proteins. Treatment of cells with a variety of agents such as phorbol esters, TNF-α, and UV irradiation results in the degradation of IκBα and IκBβ and the nuclear translocation of NF-κB (12, 17, 43, 73). IκB present in the nucleus terminates the induction process in response to TNF-α and other activators (2, 3, 60).
IκBα and IκBβ have distinct functional domains. For example, the N terminus and the ankyrin repeats of IκBα are required for the cytoplasmic regulation of NF-κB while C-terminal sequences are required to regulate NF-κB function in the nucleus (60). The activity of IκB is regulated by its phosphorylation state. The C termini of the IκBα and IκBβ proteins contain PEST domains with serine and threonine residues that are phosphorylated by cellular kinases which regulate the intrinsic stability of these proteins (10, 11, 25, 57, 61, 66, 81). In addition, the amino termini of these proteins each contain two closely spaced serine residues that are also capable of being phosphorylated by cellular kinases (16, 17, 28, 32, 77). Serine residues at positions 32 and 36 of IκBα (16, 17, 28, 32, 77) and 19 and 23 of IκBβ (62) are phosphorylated when cells are treated with various agents such as TNF-α and phorbol esters. Phosphorylation of these residues leads to their ubiquitination and proteasome-mediated degradation (1, 23, 24, 28, 32, 58, 69, 77). Mutations of these amino-terminal serine residues in IκBα and IκBβ prevent the degradation of these proteins upon treatment of cells with TNF-α or phorbol esters and inhibit the nuclear translocation of NF-κB (16, 28, 62, 77).
Biochemical fractionation has been performed to identify cellular kinases that are capable of phosphorylating IκBα. A protein complex migrating at approximately 700 kDa is capable of phosphorylating IκBα on serine residues 32 and 36, resulting in IκBα degradation by the proteasome (24, 51). Two related kinases isolated from a similar-size complex, IKKα and IKKβ, phosphorylate serine residues 32 and 36 in IκBα (27, 63, 65, 83, 85). Another kinase, RSK1, also phosphorylates the amino terminus of IκBα (71). In contrast to IKKα and IKKβ, RSK1 phosphorylates IκBα exclusively on serine residue 32. Cellular kinases are also capable of phosphorylating the carboxy terminus of IκBα. For example, casein kinase II phosphorylates serine and threonine residues in the carboxy terminus of IκBα (10, 20, 58, 59, 82). Mutation of these carboxy-terminal serine and threonine residues increases the steady-state levels of IκBα (57, 58). These results suggest that multiple cellular kinases are capable of phosphorylating IκBα and regulating different aspects of its function.
To further characterize cellular kinases that phosphorylate IκBα, we utilized biochemical fractionation and peptide sequence analysis by ion trap mass spectrometry (MS). This analysis demonstrated that the catalytic subunit of DNA-dependent protein kinase, DNA-PKcs, phosphorylated specific residues in the amino and carboxy termini of IκBα and IκBβ. We also investigated the regulation of IκBα and NF-κB in equine and murine severe combined immunodeficiency (SCID) cell lines deficient in DNA-PKcs. SCID cells, which lack the DNA-PKcs gene, exhibited constitutive nuclear NF-κB DNA binding activity, in contrast to control cell lines. DNA-PK phosphorylation of IκBα increased its ability to inhibit NF-κB DNA binding properties. These studies suggest that IκB is a target for DNA-PK phosphorylation and that this phosphorylation regulates the DNA binding properties of NF-κB in the nucleus.
MATERIALS AND METHODS
Plasmid construction.
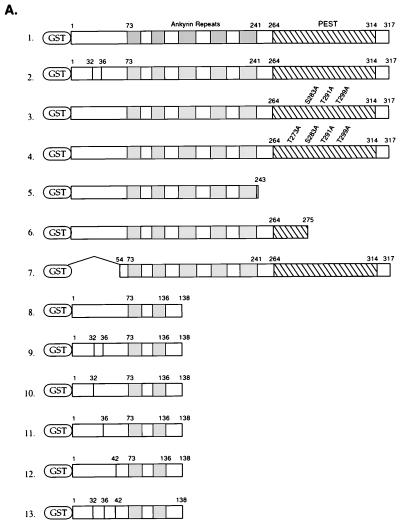
Full-length wild-type IκBα cDNA and IκBα cDNA with mutations at the codons for serine residues 32 and 36 in a pCMV4 vector (gift from Dean Ballard) (16) were digested with BamHI and HindIII and cloned into the pGEX-KG vector at BamHI-HindIII sites. A deletion of the carboxy-terminal 74 amino acids of IκBα, including the PEST domain, was accomplished by digesting the IκBα cDNA with BamHI and HincII and cloning the resulting fragment into pGEX-KG cut with BamHI and SmaI. Another deletion of the C terminus of wild-type IκBα was accomplished by digesting the IκBα cDNA with BamHI and SacI and cloning the fragment into pGEX-KG at the BamHI-SacI site to produce an IκBα cDNA encoding the amino-terminal 138 amino acids of IκBα. These truncated IκBα cDNAs were then utilized to make single-stranded M13 DNA templates for mutating either serine residue 36, serine residue 32, or both serine residues to alanine or tyrosine residue 42 to phenylalanine. The oligonucleotides used for the site-directed mutagenesis mutations were as follows: for changing serine residue 32 to alanine, 5′-GAC CGC CAC GAC GCC GGC GGC CTG GAC TCC-3′; for changing serine residue 36 to alanine, 5′GCG GCC TGG ACG CCA TGA AAGA-3′; and for changing tyrosine residue 42 to phenylalanine, 5′-CGA GGA GTT CGA GCA GAT GG-3′. Finally, fragments extending from amino acids 1 to 53 for both wild-type IκBα and IκBα with mutations at serine residues 32 and 36 were generated by BamHI and XhoI digestion of these cDNAs and cloning of the resulting cDNA fragments into pGEX-KG.
The IκBβ mutants were constructed by digesting the pCMV4-IκBβ cDNA (62) (gift from Dean Ballard) with NcoI and cloning this full-length IκBβ coding sequence into pGEX-KG. This glutathione S-transferase (GST)-IκBβ cDNA was used with a quick-change site-directed mutagenesis kit (Stratagene) to mutate serine residues 19 and 23 to alanine by using the oligonucleotide 5′-CGA TGA ATG GTG CGA CGG CGG CCT GGG CGCTC TAGGTCC CGA CG-3 and its complementary oligonucleotide. The oligonucleotide 5′-ACC TGA CGA CGA GGA CTA AAA GCT TAG CCC TTG CA-3′ and its complementary oligonucleotide were used to delete 54 amino acid residues at the C terminus of IκBβ.
IκBα site-directed mutants (with mutations S283A, T291A, and/or T299A) were prepared by using a quick-change site-directed mutagenesis kit (Stratagene) to mutate serine residue 283 and threonine residues 291 and 299 to alanine with oligonucleotides 5′-ATG CTG CCA GAG GCT GAG GAT GAG GAG-3′, 5′-GGA GAG CTA TGA CGC AGA GTC AGA GT-3′, and 5′-TTC ACG GAG TTC GCA GAG GAC GA-3′ and the complementary oligonucleotides, respectively. IκBα proteins with mutations T273A and T293A were constructed by using IκBα templates containing the triple mutant (mutations S283A, T291A, and T299A) and oligonucleotides 5′-GCA GCT GGG CCA GCT GGC ACT AGA AAA CCT-3′ and 5′-TAT GAC GCA GAG GCA GAG TTC ACG GAG-3′, respectively.
Adenovirus constructs containing wild-type and mutant IκBα were constructed by inserting the IκBα wild-type cDNA cut with ClaI and SmaI into pBlueScriptSK. An XbaI-KpnI IκBα cDNA fragment was then inserted into the pAC/CMVplpa plasmid to generate pAC/CMV-IκBα wt. The pCMV4-IκBα construct containing mutations at codons for serine residues 32 and 36 was digested with ClaI and SmaI and cloned into pAC/CMV plasmid.
Expression of bacterially produced GST-IκBα proteins.
The wild-type and mutant IκBα pGEX-KG constructs were transformed into Escherichia coli BL21 DE3. Cultures (400 ml) of E. coli were grown to an optical density at 600 nm of 0.6 to 0.8 and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h. Cells were pelleted, resuspended in buffer A (20 mM HEPES [pH 7.9], 400 mM NaCl, 5 mM dithiothreitol [DTT], 50 mM mannitol, 10 mM sodium ascorbate, 10% glycerol, 0.1 mM EDTA, 0.1% Nonidet P-40 [NP-40], 1 mM phenylmethylsufonyl fluoride [PMSF]), mildly sonicated, and centrifuged. The supernatant was incubated with 0.5 ml of glutathione agarose matrix (Sigma) for 1 h at 4°C. The matrix was then washed four times with buffer A and two times with buffer B (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40, 5 mM DTT, 1 mM PMSF). These GST fusion proteins were eluted off the matrix by 5 mM glutathione and then were fractionated through Q-Sepharose columns with a 0.1 to 0.5 M KCl gradient to remove contaminating DNA.
Cell culture and protein extraction.
Wild-type equine fibroblasts (CRL6288) and murine NIH 3T3 cells were purchased from the American Type Culture Collection ATCC. Equine SCID fibroblasts cell line 1863 (82) and murine SCID fibroblast cell line SF19 (39) were described previously. The parental (CB-17), SCID (SCID/St), and SCID-complemented (100E and 50D) cell lines have been previously described (49). HeLa spinner cells were maintained in supplemented Eagle minimal essential media with 5% newborn calf serum at a density of 4 × 105 cells/ml. Tetradecanoyl phorbol acetate (TPA) (Sigma; 50 ng/ml) and/or ionomycin (Calbiochem; 2 μM) was added to either HeLa, 3T3, or SF19 cells as outlined below.
To obtain cytoplasmic proteins, cells were washed with cold phosphate-buffered saline (PBS; pH 7.2), resuspended in buffer C (10 mM HEPES [pH 7.5], 0.1 mM EDTA, 10 mM KCl, 1 mM DTT, 50 mM NaF, 1 mM sodium orthovanadate, 5 μM okadaic acid, 0.5 mM PMSF, 5% glycerol, 10 μg of leupeptin and aprotinin per ml), and incubated on ice for 15 min. At the end of incubation, 1/10 volume of 10% NP-40 was added. Cells were vortexed for 30 s and then subjected to Eppendorf centrifugation for 30 s. Supernatants were collected as cytoplasmic proteins. The protein concentrations of the resultant supernatants containing cytoplasmic protein were determined by the Bradford assay using Bio-Rad reagent.
Preparation of recombinant adenoviruses.
The production of recombinant adenovirus utilized previously described methodology (21, 36). The pAC/CMVplpa plasmids encoding either wild-type IκBα or IκBα with mutations at serine residues 32 and/or 36 were cotransfected with the pJM17 adenovirus plasmid into 293 cells (adenovirus E1a-transformed human embryonic kidney cells). Viruses from 293 cell supernatants of cultures showing a complete cytopathic effect were purified by cesium chloride banding. Virus titers were determined by plaque assay using serial dilution (38). The pAC/CMVplpa, pJM17, and control recombinant replication-defective adenoviruses containing β-galactosidase were obtained from P. Nissen. Adenovirus infection of murine cell lines was performed by using a multiplicity of infection of 10 for 24 h.
Gel electrophoresis mobility shift assay.
Nuclei from NIH 3T3, SF19 (murine SCID fibroblasts), CRL6288 (normal equine fibroblast), and 1863 (equine SCID fibroblasts) cells and the murine cell lines CB-17, SCID/St, 100E, and 50D were resuspended in buffer containing 20 mM HEPES (pH 7.5), 50 mM KCl, 300 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1 mg of PMSF per ml, 10% glycerol, 10 μg of leupeptin, 10 μg of aprotinin per ml and extracted at 4°C on a rocker for 30 min, followed by Eppendorf centrifugation at 14,000 rpm for 5 min. The supernatants were collected, and 3 μg of each nuclear extract was incubated with 0.1 pmol of 32P-labeled double-stranded κB binding site oligonucleotide (5′-GCTGGGGACTTTC-3′) or SP1 binding site oligonucleotide (5′-ATTCGATCGGGGCGGGGCGAGC-3′) in buffer containing 1 μg of poly (dI-dC), 1 μg of bovine serum albumin, 10 mM HEPES (pH 7.9), 0.5 mM DTT, 0.1 mM EDTA, 60 mM KCl, 0.2 mM PMSF, 5 mM MgCl2, and 12% glycerol at room temperature for 15 min. Samples were analyzed by 5% native polyacrylamide gel electrophoresis (PAGE) followed by autoradiography.
For IκBα inhibition experiments, bacterially produced IκBα protein was cleaved from the GST moiety with thrombin, followed by the addition of PMSF. This IκBα protein was then phosphorylated by purified DNA-PK by using cold ATP. The nonphosphorylated and phosphorylated IκBα proteins were added to the gel retardation reaction mixture 30 min prior to the addition of the 32P-labeled probe. The recombinant p50 and p65 NF-κB proteins were produced in baculovirus expression vectors and purified by nickel agarose chromatography.
Immunocytochemistry and confocal microscopy.
Cells, including HeLa, 50D, and 100E cells, were cultured on coverslips, washed with PBS, and fixed with methanol at −20°C for 6 min. The cells were washed twice with PBS, blocked for 30 min in PBS containing 0.5% gelatin and 0.25% bovine serum albumin, and incubated for 1 h at room temperature with one of the following primary antibodies: a monoclonal antibody directed against DNA-PKcs, a rabbit polyclonal antibody directed against a carboxy-terminal peptide of IκBα (C-21; Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.), or a rabbit polyclonal antibody directed against a carboxy-terminal peptide of IκBβ p65 (C-20; Santa Cruz Biotechnology, Inc.). The cells were washed three times with 0.2% gelatin in PBS and incubated for 1 h with a goat anti-mouse fluorescein isothiocyanate (FITC) conjugate or a goat anti-rabbit FITC conjugate (Jackson ImmunoResearch, West Grove, Pa.). Samples were washed three times and then mounted in a solution of 1 mg of p-phenylenediamine per ml in 90% glycerol. The preparations were examined on a MRC1024 laser scanning confocal microscope (Bio-Rad, Microscience Division, Cambridge, Mass.) equipped with a 15-mW air-cooled krypton-argon laser (Ion Laser Technology, Salt Lake City, Utah) as a light source. The images were constructed from gray scale confocal fluorescence images with Adobe software.
Immunoprecipitation of DNA-PKcs.
Monoclonal antibodies 42-26, 25-4, and 18-2 directed against DNA-PKcs (19, 49) or flag monoclonal antibody (Kodak) was added to column fractions containing DNA-PKcs and incubated at 4°C for 1 h. Protein G-Sepharose was then added for 1 h, and the beads were washed three times with buffer D (20 mM HEPES [pH 7.9], 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 10% glycerol) and one time with buffer containing 12 mM HEPES (pH 7.9), 0.6 mM EDTA, 0.6 mM DTT, 6% glycerol, 60 mM KCl, and 7.5 mM MgCl2. Kinase reactions were then performed.
In vitro kinase assay.
Kinases which bound to GST-IκBα fusion proteins and which were able to phosphorylate IκBα were assayed by a modification of the previously described methods (44). Bacterially expressed GST-IκBα proteins were bound to glutathione agarose matrix (Sigma). HeLa S100 cells or column fractions were incubated with GST-IκBα-bound matrix in buffer containing 20 mM HEPES (pH 7.9), 100 mM KCl, 10% glycerol, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF at 4°C for 1 h. The matrix was washed twice with buffer containing 50 mM Tris (pH 8.0), 120 mM NaCl, 0.5% NP-40, 5 mM DTT, and 1 mM PMSF and once with the kinase reaction buffer (50 mM Tris [pH 7.4], 5 mM MnCl2, 5 mM DTT). The kinase assay was then performed in the kinase reaction buffer with the addition of 4 μM ATP, 10 μCi of [γ-32P]ATP, 5 mM NaF, 1 mM sodium orthovanadate, 40 μM MgCl2, a protease inhibitor cocktail (Boehringer Mannheim), and 1 mM PMSF at 30°C for 15 min. Similar results were seen when 2.5 μM okadaic acid was added to the kinase assay mixtures. The matrix was pelleted by centrifugation, 20 μl of 2× sodium dodecyl sulfate-PAGE (SDS-PAGE) sample buffer was added, and the samples were heated at 95°C for 5 min. The samples were resolved with SDS–12% polyacrylamide gels followed by autoradiography.
Western blot analysis.
Mono S fractions from the purification scheme for IκBα kinases were mixed with an equal volume of 2× SDS-PAGE sample buffer, boiled for 5 min, and resolved by SDS–6% PAGE. Following electrophoresis, the gel was transferred at 60 V to a Hybond-C nitrocellulose membrane (Amersham) for 8 h at 4°C. Affinity-purified goat antibody directed against DNA-PKcs (Santa Cruz Biotechnology, Inc.; SC-1551) at a 1:2,000 dilution was used as the primary antibody in the Western blot analysis. Donkey anti-goat immunoglobulin G horseradish peroxidase (Santa Cruz Biotechnology, Inc.; SC-2020) was used as the secondary antibody to detect the DNA-PKcs by enhanced chemiluminiscence reagents (ECL kit; Amersham).
Twenty to thirty micrograms of cytoplasmic proteins isolated from the NIH 3T3 and SF19 cells (39) was loaded on an SDS–10% PAGE gel and transferred to nitrocellulose membranes. The membranes were used in Western blot analysis with polyclonal rabbit antibody directed against IκBα (Santa Cruz Biotechnology, Inc.; SC-371) at a dilution of 1:3,000 or the p89 subunit of TFIIH (Santa Cruz Biotechnology, Inc.; SC-230) at a dilution of 1:5,000 before incubation with a second antibody and development with enhanced chemiluminescence.
Purification of IκBα kinases.
All the purification steps were conducted at 4°C. HeLa S100 extract (3 g) was prepared from 150 liters of cells (1011 cells) by the method of Dignam (29) and dialyzed against 0.1 M KCl buffer D (20 mM HEPES [pH 7.9], 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT). The extract was then fractionated on a phosphocellulose column which was washed with 150 ml of 0.1 M KCl buffer D, and the kinase activity was eluted with 100 ml of buffer D containing 0.3 M or 1.0 M KCl. The IκBα kinase activity was assayed with GST-IκBα fusion proteins, which extended from amino acid 1 to 138 and contained either wild-type sequences or sequences with mutations of serine residues 32 and 36, that were bound to glutathione agarose beads. The 0.3 M KCl fraction which contained the majority of the kinase activity was dialyzed against 0.05 M KCl buffer D and fractionated on a Q-Sepharose column (1 by 2 cm) which was eluted with 1.0 M KCl in buffer D. The active kinase fractions were pooled, loaded on a Superdex 200 column (26/60; Pharmacia), and eluted with 0.05 M KCl in buffer D. The active fractions were then pooled and fractionated with a heparin agarose column (1 by 3 cm). After the column was washed with 0.05 M KCl in buffer D, it was eluted with a 20-ml linear gradient of 0.05 to 0.8 M KCl in buffer D. The fractions containing the peak kinase activity were pooled and dialyzed against 0.05 M KCl in buffer D and were fractionated on a Mono Q fast protein liquid chromatography (FPLC) column (Pharmacia) with a linear gradient of 0.05 to 1.0 M KCl in buffer D. The fractions active for kinase activity were then pooled and dialyzed against 0.05 M KCl in buffer D and fractionated by using a Mono S FPLC column (Pharmacia) with a linear gradient of 0.05 to 0.6 M KCl. The fractions containing the peak of kinase activity were dialyzed against 0.1 M KCl in buffer D, aliquoted, frozen in liquid nitrogen, and stored at −80°C.
Peptide sequencing by ion trap MS of components of the IκBα kinase.
Purified fractions containing the IκBα kinase activity were pooled, loaded onto an SDS–6% polyacrylamide gel, and subjected to electrophoresis in Tris-glycine buffer. Coomassie brilliant blue-stained gel slices containing a 400-kDa protein species that correlated with the peak of kinase activity for IκBα were excised and subjected to in gel reduction, S-carboxyamidomethylation, and tryptic digestion (Promega). Sequence information on the digestion products was determined by capillary (Monitor C18 column [0.5 by 150 mm]; Michrom BioResources) reverse-phase chromatography, coupled to the electrospray ionization source of a quadruple ion trap mass spectrometer (Finnigan LCQ, San Jose, Calif.). The instrument was programmed to acquire successive sets of three-scan modes consisting of full-scan MS over the m/z range 395 to 1,118 amu, followed by two data-dependent scans on the most abundant ion in that full scan. These data-dependent scans allowed the automatic acquisition of high-resolution (zoom scan) spectra to determine charge state and exact mass and MS/MS spectra for peptide sequence information. Interpretation of the resulting MS/MS spectra was facilitated by searching the NCBI nr and dbest databases with the algorithm Sequest (31) and was then confirmed manually. The peptide sequences TVGALQVLGTEAQSSLK, LLLQGEADQSLLTFIDK, and SLGPPQGEEDSVPR had sequence identity with the DNA-dependent protein kinase catalytic subunit (40).
RESULTS
Identification of cellular kinases that phosphorylate IκBα.
To characterize cellular kinases that phosphorylate IκBα, we prepared S100 extract from both untreated and TPA-treated HeLa cells (29). The amino-terminal 138 amino acids of IκBα were fused to GST so that kinases that phosphorylated the amino terminus of IκBα could be detected. A GST-IκBα fusion protein containing mutations of serine residues 32 and 36 was also assayed in an attempt to differentiate kinases with specificity for these residues (16, 17, 28, 32, 77). Following binding of cellular proteins to GST-IκBα coupled to glutathione agarose beads, the beads were extensively washed and kinase reactions were performed, followed by SDS-PAGE and autoradiography.
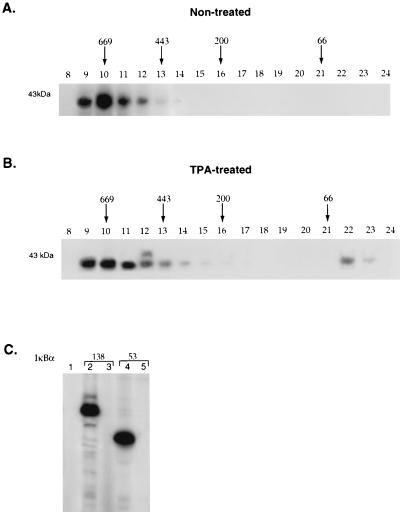
S100 extract from either untreated or TPA-treated HeLa cells was initially fractionated on a phosphocellulose column that was washed with buffer containing 0.1 M KCl and eluted with either 0.3 M KCl or 1.0 M KCl. The vast majority of the IκBα kinase activity which eluted in the 0.3 M KCl eluate was next fractionated on a Superdex 200 column. Superdex 200 fractionation of proteins isolated from untreated HeLa cells demonstrated one major peak of activity which migrated between 600 and 700 kDa (Fig. 1A). The phorbol ester-treated HeLa extract contained a similar amount of kinase activity in the 700-kDa fraction from the Superdex 200 column in addition to a second peak of IκBα kinase activity that migrated with markers of approximately 40 kDa (Fig. 1B). The specificity of kinases present in the 700-kDa fraction isolated from either untreated or TPA-treated HeLa cells was next determined. Kinases present in the 700-kDa fraction from both untreated (Fig. 1C) and TPA-treated (data not shown) extracts were able to phosphorylate a GST-IκBα fusion protein with serine residues at positions 32 and 36 (Fig. 1C, lanes 2 and 4) but not a GST-IκBα fusion protein with mutations of these serine residues (Fig. 1C, lanes 3 and 5).
FIG. 1.
Constitutive and inducible cellular kinases that phosphorylate the amino terminus of IκBα. S100 extract was prepared from HeLa cells that were either untreated (A) or treated with 50 ng of phorbol ester (TPA) per ml (B) for 1 h. The extract was fractionated on a phosphocellulose column, washed with 0.1 M KCl, and eluted with 0.3 M KCl. Each of these extracts was concentrated on a Q-Sepharose column and then fractionated on a Superdex 200 column. The column fractions from the Superdex 200 column were then assayed for their ability to bind and phosphorylate a GST-IκBα fusion protein truncated at amino acid 138. The positions of the molecular weight markers in the Superdex 200 column fractions are indicated. (C) Fraction 19 from untreated HeLa S100 extract that eluted from the mono Q column was assayed for its ability to phosphorylate 0.5 μg of either GST (lane 1), a GST-IκBα fusion protein truncated at amino acid 138 that contained wild-type or mutant sequences at serine residues 32 and 36 (lanes 2 and 3, respectively), and a GST-IκBα fusion protein truncated at amino acid 53 containing wild-type or mutant sequences at serine residues 32 and 36 (lanes 4 and 5, respectively). Similar results were obtained with TPA-treated extract.
Purification of a cellular kinase that phosphorylates the amino terminus of IκBα.
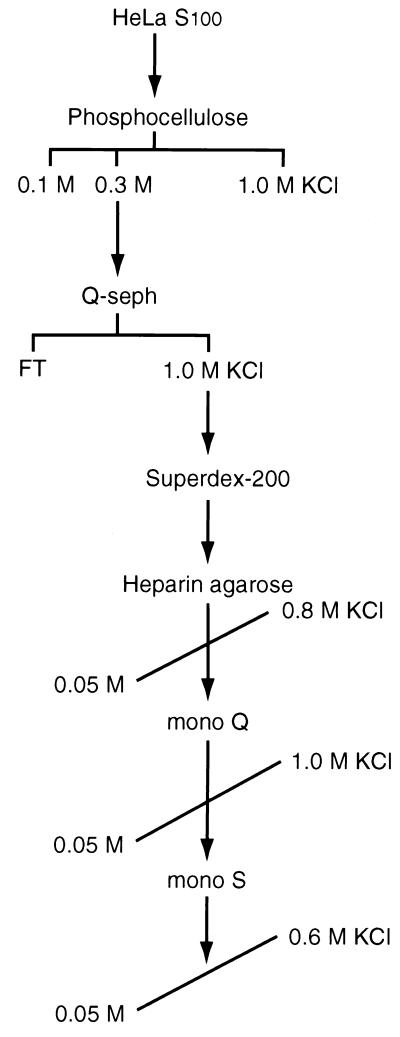
To purify kinases present in the 700-kDa fraction, S100 extract was prepared from 150 liters of untreated HeLa cells (Fig. 2). The purification scheme included elution of active kinase fractions from a phosphocellulose column with 0.3 M KCl, concentration of the sample on a Q-Sepharose column, and fractionation on a Superdex 200 column. The kinase activity that eluted in the 600- to 700-kDa Superdex 200 FPLC fraction was then further purified by heparin agarose, mono Q FPLC, and mono S FPLC chromatography. The fractions from the final mono S column were subjected to SDS-PAGE and Coomassie staining and were assayed for their ability to phosphorylate IκBα.
FIG. 2.
Fractionation of IκBα kinase activity. A schematic of the purification scheme used to purify cellular kinases that phosphorylate the amino terminus of IκBα is shown. S100 extract was obtained from untreated HeLa cells and fractionated on a phosphocellulose column washed with buffer containing 0.1 M KCl and eluted with 0.3 M KCl. This fraction was applied to a Q-Sepharose (Q-seph) column and eluted with 1.0 M KCl, followed by fractionation on a Superdex 200 column. Proteins with kinase activity for the 138 amino-terminal residues of IκBα were then fractionated on heparin agarose, mono Q FPLC, and mono S FPLC and eluted with KCl gradients as indicated. FT, column flowthrough.
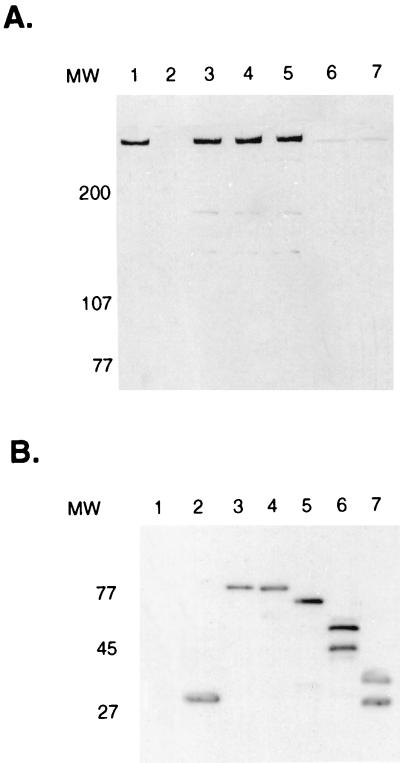
Coomassie staining of a polyacrylamide gel containing fractions from the mono S FPLC column revealed the presence of an approximately 400-kDa species which was most abundant in mono S fractions 20, 21, and 22 (Fig. 3A). The presence of this species correlated with the peak of IκBα kinase activity in these same column fractions (Fig. 3B). The presence of the 400-kDa species was associated with the peak of IκBα kinase activity in five different HeLa S100 preparations (data not shown). Following SDS-PAGE and Coomassie staining, the 400-kDa species was excised from the gel and subjected to gel tryptic digestion (42). The resultant mixture was analyzed by capillary high-performance liquid chromatography (HPLC) ion trap MS which allowed the acquisition of on-line MS/MS spectra for peptide sequence information. After manual and computer-assisted interpretation (31), peptide sequences that corresponded to the amino acid sequences TVGALQVLGTEAQSSLLK, LLLQGEADQSLLTFIDK, and SLGPPQGEEDSUPR in the DNA-dependent protein kinase catalytic subunit were obtained (40). The 90-kDa protein seen in the same column fractions as DNA-PK was also analyzed by peptide microsequence analysis and revealed a novel peptide sequence of unknown function (50).
FIG. 3.
Purified DNA-dependent protein kinase phosphorylates the amino terminus of IκBα. (A) Fractions (17 to 25) from the final mono S FPLC column were subjected to SDS-PAGE and Coomassie staining. (B) Kinase assays were also performed with these column fractions by using a GST-IκBα fusion protein extending from amino acids 1 to 138. (C) These same column fractions were also analyzed by Western blotting using polyclonal goat antibody directed against the DNA-dependent protein kinase catalytic subunit.
Western blot analysis with specific antibodies directed against DNA-PKcs demonstrated that the 400-kDa protein present in column fractions 20, 21, and 22 that correlated with the peak of IκBα kinase activity was in fact DNA-PKcs (Fig. 3C). Casein kinase II, which phosphorylates the C terminus of IκBα (10, 57, 61, 81), was not present in fractions containing DNA-PK, as determined by Western blot analysis with specific antibodies directed against this protein (data not shown). Western blot analysis was also used to analyze whether the Ku antigens (Ku86 and Ku70) copurified with DNA-PK in our IκBα kinase preparation (37, 74). This analysis indicated that low levels of the Ku antigens relative to that of DNA-PKcs were present and that there was increased amounts of Ku70 relative to Ku86 (data not shown). The significance of this finding remains to be determined.
DNA-PK phosphorylation of IκBα.
DNA-PK is a serine/threonine kinase which is a member of a family of protein kinases known as phosphatidylinositol 3 (PI-3) kinases that contain a conserved kinase domain and include ATM, FRAP, and FRP1 (26, 40, 68). DNA-PK can be detected in cellular extracts prepared from both the nucleus and the cytoplasm, although the majority of this activity is found in the nucleus (19, 54). This kinase comprises two subunits, which include a regulatory subunit that is the DNA end-binding heterodimer Ku and catalytic subunit DNA-PKcs. The kinase activity of DNA-PK is stimulated by the addition of double-stranded DNA and inhibited by the addition of wortmannin (40). DNA-PK phosphorylates a number of substrates including Jun (9), DNA replication factor A (18), p53 (33, 54, 56), the RNA polymerase II C-terminal domain (CTD) (30, 64), simian virus 40 T-antigen (22), hsp90 (19, 54), SP1 (46), Oct1 (35), and the serum response factor (59).
Several of these substrates including hsp90, p53, simian virus 40 T-antigen, Jun, and the serum response factor are phosphorylated by DNA-PK on serine and threonine residues which are followed by glutamine residues (reviewed in reference 53). However, the RNA polymerase II CTD, Oct-1, and SP1 are phosphorylated by DNA-PK but do not contain the above peptide sequence motif. DNA-PK phosphorylates both DNA-bound factors such as SP1, p53, T-antigen, and Jun and other substrates such as hsp90 and casein that are not bound to DNA. Finally, DNA-PK autophosphorylates DNA-PKcs, Ku70, and Ku86 (20, 54), and the autophosphorylation of DNA-PKcs inactivates its kinase activity (20).
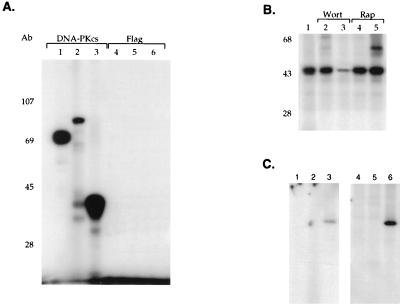
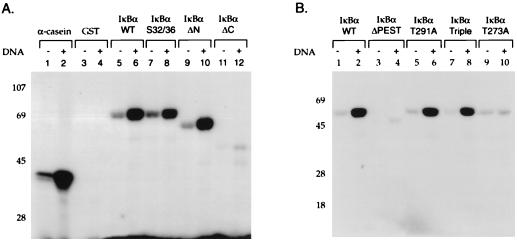
Although our results indicated that the presence of DNA-PKcs correlated with IκBα kinase activity, it was critical to demonstrate that DNA-PK itself was able to phosphorylate IκBα. A monoclonal antibody directed against DNA-PKcs was used to immunoprecipitate the kinase from the mono S column fractions and assay its ability to phosphorylate IκBα, IκBβ, and α-casein (which is a known substrate for DNA-PK [19, 54]). The unrelated flag monoclonal antibody was used as a control antibody in these immunoprecipitation studies. The fractions immunoprecipitated with antibody to DNA-PKcs were able to efficiently phosphorylate IκBα, IκBβ, and α-casein (Fig. 4A, lanes 1 to 3). In contrast, immunoprecipitation with the control antibody did not result in phosphorylation of these different substrates (Fig. 4A, lanes 4 to 6). These data support a role for DNA-PKcs in phosphorylating both IκBα and IκBβ.
FIG. 4.
Antibody directed against DNA-PK immunoprecipitates IκBα kinase activity. (A) Full-length GST-IκBα (lane 1), GST-IκBβ (lane 2), and α-casein (lane 3) were used in in vitro kinase assays with mono S column fractions immunoprecipitated with either the specific monoclonal antibody 42-26 directed against DNA-PKcs (lanes 1 to 3) or the unrelated flag monoclonal antibody (lanes 4 to 6), followed by SDS-PAGE and autoradiography. (B) In vitro kinase assays were performed with GST-IκBα truncated at amino acid 138 and the mono S column fractions containing DNA-PK (lane 1) in the presence of 50 or 250 nM wortmannin (Wort) (lanes 2 and 3, respectively) or 50 or 250 nM rapamycin (Rap) (lanes 4 and 5, respectively). (C) Immunoprecipitation of DNA-PK activity was performed with monoclonal antibodies (42-26 and 25-4) directed against DNA-PKcs. Cytoplasmic extracts prepared by either the Dignam method (lanes 1 to 3) or the rapid-lysis method (lanes 4 to 6) were immunoprecipitated with these antibodies and analyzed in in vitro kinase assays without substrate (lanes 1 and 4), with GST (lanes 2 and 5), or with GST-IκBα (lanes 3 and 6).
Wortmannin is an inhibitor of many members of the PI-3 kinase family including DNA-PK (40). Wortmannin will inhibit DNA-PK kinase activity at concentrations of approximately 250 nM, while it will inhibit other PI-3 kinases at much lower concentrations ∼5 nM (40). Next, we assayed whether the addition of wortmannin inhibited the IκBα kinase activity present in our mono S column fractions. The addition of wortmannin inhibited the ability of kinases present in the mono S column fraction to phosphorylate IκBα (Fig. 4B, lanes 2 and 3). In contrast, the addition of another kinase inhibitor, rapamycin, did not prevent the phosphorylation of IκBα (Fig. 4B, lanes 4 and 5). Finally, we demonstrated that the presence of DNA-PK in cytoplasmic extract was not dependent on the method of preparation. Immunoprecipitation with a monoclonal antibody directed against DNA-PKcs demonstrated that cytoplasmic extract prepared by both the standard S100 preparation procedure (29) and the rapid-lysis procedure (51) resulted in the presence of DNA-PK that was capable of phosphorylating IκBα (Fig. 4C, lanes 3 and 6). These results indicate that DNA-PK is able to specifically phosphorylate IκBα.
DNA-PK phosphorylates both the amino and carboxy termini of IκBα.
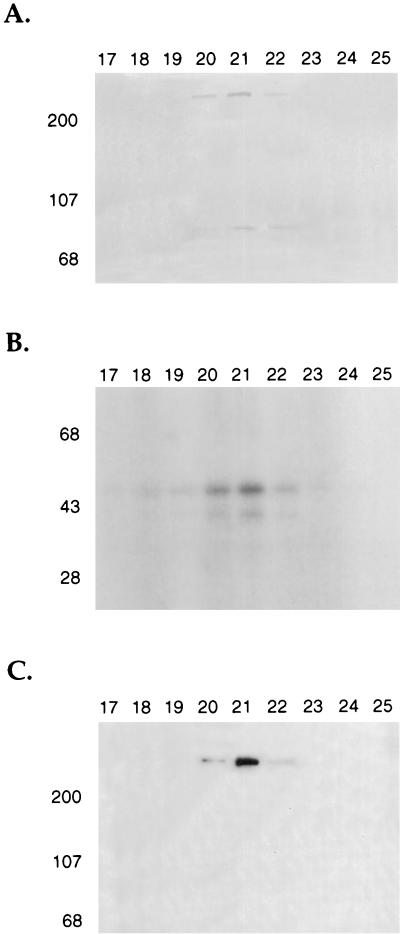
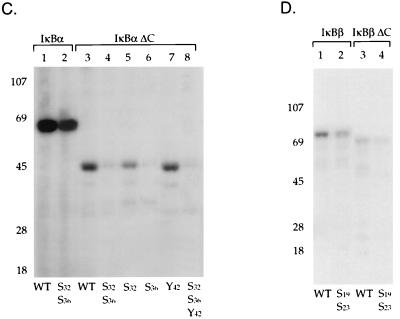
We next characterized the ability of a highly purified DNA-PK prepared by a previously described purification scheme (19) to phosphorylate GST fusion proteins containing wild-type and mutant IκBα (Fig. 5A) and IκBβ (Fig. 5B) proteins. Purified DNA-PK and our mono S column fractions containing DNA-PK gave equivalent specificities for the IκBα and IκBβ substrates (19). Numerous studies have demonstrated that serine and threonine phosphorylation by DNA-PK is highly dependent on the presence of free DNA ends (19, 54). Purified DNA-PK phosphorylated α-casein, and the degree of this phosphorylation was greatly enhanced by the addition of sonicated double-stranded salmon sperm DNA (Fig. 6A, lanes 1 and 2). There was no phosphorylation of GST by DNA-PK (Fig. 6A, lanes 3 and 4). The GST fusion protein containing the full-length IκBα protein was phosphorylated by DNA-PK, and this phosphorylation was stimulated by the addition of double-stranded DNA (Fig. 6A, lanes 5 and 6). Mutations of serine residues 32 and 36 in IκBα only slightly decreased IκBα phosphorylation (Fig. 6A, lanes 7 and 8). An IκBα protein with its 53 N-terminal amino acids deleted was strongly phosphorylated by DNA-PK (Fig. 6A, lanes 9 and 10), indicating significant C-terminal phosphorylation of IκBα by DNA-PK. A deletion of the carboxy-terminal 74 amino acids of IκBα markedly decreased its phosphorylation (Fig. 6A, lanes 11 and 12). These results were consistent with predominant DNA-PK phosphorylation of the carboxy terminus of IκBα.
FIG. 5.
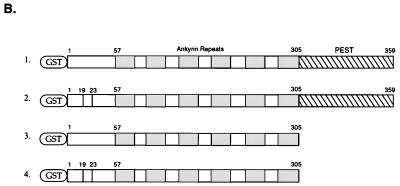
Schematic structure of IκBα and IκBβ fusion constructs. (A) GST fusion proteins were constructed with wild-type IκBα (lane 1), IκBα with mutations of serine residues 32 and 36 to alanine (lane 2), IκBα with mutations of serine residue 283 and threonine residues 291 and 299 to alanine (lane 3), IκBα with mutations of threonine residues 273, 291, and 299 and serine residue 283 to alanine (lane 4), IκBα with residues 244 to 317 deleted (lane 5), IκBα with residues 275 to 317 deleted (lane 6), IκBα with its amino-terminal 53 amino acids deleted (lane 7), IκBα truncated at amino acid 138 (lane 8), this construct with mutations of serine residues 32 and 36 to alanine (lane 9), serine residue 32 to alanine (lane 10), serine residue 36 to alanine (lane 11), tyrosine residue 42 to phenylalanine (lane 12), or serine residues 32 and 36 to alanine and tyrosine residue 42 to phenylalanine (lane 13). (B) GST fusion proteins were constructed with the full-length IκBβ (lane 1), this same construct having mutations of serine residues 19 and 23 to alanine (lane 2), IκBβ truncated at amino acid 305 (lane 3), and this protein with serine residues 19 and 23 changed to alanine (lane 4). The positions of the ankyrin repeats, the PEST domain, and serine and/or tyrosine residues that were mutated are shown.
FIG. 6.
Specificity of DNA-PK phosphorylation of IκBα and IκBβ. (A) Kinase assays were performed with purified DNA-PK and substrates including α-casein (lanes 1 and 2), GST (lanes 3 and 4), or GST fusion proteins containing wild-type IκBα (lanes 5 and 6), this construct with a mutation of serine residues 32 and 36 to alanine (lanes 7 and 8), IκBα with amino acids 1 to 53 deleted (lanes 9 and 10), or IκBα with amino acids 139 to 317 deleted (lanes 11 and 12). The kinase assays were performed in the absence (−) or presence (+) of sheared salmon sperm DNA (0.5 μg), as indicated. (B) GST fusion proteins containing either wild-type IκBα (lanes 1 and 2), IκBα (amino acids [aa] 1 to 275) with the majority of the PEST domain deleted (lanes 3 and 4), full-length IκBα with a mutation of threonine residue 291 to alanine (lanes 5 and 6), IκBα with mutations of serine residue 283 and threonine residues 291 and 299 to alanine (lanes 7 and 8), or IκBα with threonine residues 273, 291, and 299 and serine residue 283 changed to alanine (lanes 9 and 10) were analyzed in kinase assays in either the absence (−) or presence (+) of double-stranded DNA and purified DNA-PK. (C) GST fusion proteins containing either wild-type IκBα (lane 1), full-length IκBα with serine residues 32 and 36 changed to alanine (lane 2), IκBα (aa 1 to 138) with a deletion of aa 139 to 317 (lane 3), this same construct with serine residues 32 and 36 mutated to alanine (lane 4), IκBα (aa 1 to 138) with serine residue 32 changed to alanine (lane 5), IκBα (aa 1 to 138) with serine residue 36 changed to alanine (lane 6), IκBα (aa 1 to 138) with tyrosine residue 42 changed to phenylalanine (lane 7), and IκBα (aa 1 to 138) with mutations of serine residues 32 and 36 to alanine and tyrosine 42 to phenylalanine (lane 8) were analyzed in kinase assays in the presence of double-stranded DNA and purified DNA-PK. (D) GST fusion proteins containing wild-type IκBβ (lane 1), this same construct with serine residues 19 and 23 mutated to alanine (lane 2), a truncated IκBβ construct (aa 1 to 305) (lane 3), or a truncated IκBβ construct with serine residues 19 and 23 changed to alanine (lane 4) were also analyzed in kinase assays with double-stranded DNA and purified DNA-PK.
Next, we mapped sites in the carboxy terminus of IκBα that were potential sites for DNA-PK phosphorylation. The carboxy terminus of IκBα is critical for DNA-PK-dependent phosphorylation, as reflected in the major reduction of IκBα phosphorylation seen as a result of the deletion of its carboxy-terminal 42 amino acids (Fig. 6B, lanes 1 to 4). Casein kinase II has been demonstrated to phosphorylate the carboxy terminus of IκBα at serine residues 283 and 289 and threonine residues 291 and 299 (57, 61). Mutations of serine residue 283, threonine residue 291, and threonine residue 299 to alanine eliminated the casein kinase II phosphorylation of IκBα (57) (data not shown). However, DNA-PK was able to phosphorylate this IκBα mutant in addition to an IκBα mutant with a mutation at threonine 291, which has been reported to markedly decrease casein kinase II phosphorylation (57) (Fig. 6B, lanes 5 to 8). Mutation of additional serine and threonine residues in the carboxy terminus of IκBα extending between amino acids 251 and 317 revealed that only the mutation of threonine residue 273 markedly reduced DNA-PK phosphorylation of IκBα (Fig. 6B, lanes 9 and 10). Thus, different residues in the carboxy terminus of IκBα are critical for phosphorylation by DNA-PK and casein kinase II.
The ability of DNA-PK to phosphorylate residues in the amino terminus of IκBα was next analyzed. Mutation of serine residues 32 and 36 in wild-type IκBα resulted in a slight decrease in IκBα phosphorylation (Fig. 6D, lanes 1 and 2). Since DNA-PK also phosphorylates the carboxy terminus of IκBα, a truncated IκBα protein that contained only the amino-terminal 138 amino acids of this protein was next assayed. DNA-PK was able to phosphorylate this truncated GST-IκBα protein (Fig. 6C, lane 3), but phosphorylation of an IκBα protein containing mutations at both serine residues 32 and 36 was significantly reduced (Fig. 6C, lane 5). Next, we determined whether both of these amino-terminal serine residues were phosphorylated by DNA-PK. Mutation of serine residue 32 did not significantly decrease the ability of DNA-PK to phosphorylate IκBα (Fig. 6C, lane 6), while mutation of serine residue 36 markedly decreased DNA-PK phosphorylation of IκBα (Fig. 6C, lane 7). Mutation of tyrosine residue 42, which has also been demonstrated to be a site of IκBα phosphorylation by src-like kinases, did not alter DNA-PK phosphorylation (45). Finally, an IκBα protein containing mutations at serine residues 32 and 36 and tyrosine residue 42 was not phosphorylated by DNA-PK (Fig. 6C, lane 8). These results indicated that serine 36 was the predominant site in the amino terminus of IκBα that was phosphorylated by DNA-PK.
Next we characterized the ability of DNA-PK to phosphorylate IκBβ (62, 76). DNA-PK was able to phosphorylate a GST fusion protein containing full-length IκBβ (Fig. 6D, lane 1). An IκBβ protein with mutations of serine residues 19 and 23 (62) exhibited a slight decrease in phosphorylation by DNA-PK (Fig. 6D, lane 2). Next, we determined whether DNA-PK could phosphorylate GST fusion proteins containing the amino-terminal 305 amino acids of IκBβ, which lack the PEST domain. DNA-PK was able to phosphorylate this GST-IκBβ protein (Fig. 6D, lane 3), while an IκBβ protein containing mutations of serine residues 19 and 23 exhibited decreased phosphorylation by DNA-PK (Fig. 6D, lane 4). These results are consistent with the ability of DNA-PK to phosphorylate both the N and C termini of IκBα and IκBβ.
Interactions between DNA-PK and IκBα.
Since our purification scheme was based on the ability of kinases to bind to IκBα, we next characterized the domains of IκBα that were involved in interactions between these proteins. S100 extract was prepared from HeLa cells, and the presence of DNA-PK in this extract was determined by Western blot analysis (Fig. 7A, lane 1). Thus, we could determine whether DNA-PK present in the S100 extracts was able to directly interact with IκBα. The S100 extract was incubated with either GST alone (Fig. 7A, lane 2) or a variety of GST fusion proteins containing different portions of IκBα. These included GST fusion proteins with wild-type IκBα (Fig. 7A, lane 3), an IκBα protein having mutations of serine residues 32 and 36 (Fig. 7A, lane 4), an IκBα protein with its amino-terminal 53 amino acids deleted (Fig. 7A, lane 5), and a GST-IκBα protein containing either its amino-terminal 138 (Fig. 7A, lane 6) or 53 (Fig. 7A, lane 7) amino acids.
FIG. 7.
DNA-PK associates with IκBα. (A) HeLa S100 extract was incubated with a variety of GST-IκBα fusion proteins bound to glutathione agarose beads, followed by Western blot analysis with DNA-PKcs antibody. HeLa S100 extract with 50% of the input shown (lane 1) was incubated with either GST alone (lane 2), GST–wild-type IκBα (lane 3), GST-IκBα with mutations at serine residues 32 and 36 (lane 4), GST-IκBα with its amino terminal 53 amino acids deleted (lane 5), GST-IκBα containing the amino-terminal 138 amino acids of IκBα (lane 6), or GST-IκBα containing the amino terminal 53 amino acids of IκBα (lane 7). Western blot analysis was performed with a goat polyclonal antibody directed against DNA-PKcs. (B) The GST-IκBα fusion proteins used in panel A were subjected to SDS-PAGE and Western blot analysis with antibody directed against GST.
Following incubation of S100 with each of these GST fusion proteins, Western blot analysis was performed with specific antibodies directed against DNA-PKcs. This analysis indicated that DNA-PKcs was able to bind to IκBα proteins with intact ankyrin repeats and PEST domains (Fig. 7A, lanes 3 to 5). However, there was a marked decrease in the interaction of DNA-PKcs with IκBα proteins in which the majority of the ankyrin repeats were deleted (Fig. 7A, lanes 6 and 7). Western blot analysis revealed similar quantities of each of the GST-IκBα fusion proteins used in these assays (Fig. 7B). We noted that the degree of interaction between DNA-PKcs and IκBα was greatly diminished with more highly purified preparations of DNA-PK (data not shown). These results suggested that the interactions between IκBα and DNA-PKcs likely required the IκBα ankyrin repeats and could be mediated by additional cellular factors that are associated with DNA-PKcs.
SCID cells exhibit constitutive nuclear levels of NF-κB.
DNA-PK is critical in the process of DNA double-strand break repair. Cells deficient in DNA-PK are hypersensitive to ionizing radiation and to other agents which induce double-strand DNA breaks (14, 55). DNA-PK is also required in resolving double-strand breaks generated in lymphocyte precursors during V(D)J rearrangement (15, 49, 72). Germ line mutations in either chain of the Ku heterodimer or in DNA-PKcs result in the SCID phenotype (15, 49, 72). Recent data suggest that DNA-PK may play a role in traversing checkpoints after cell cycle arrest (52).
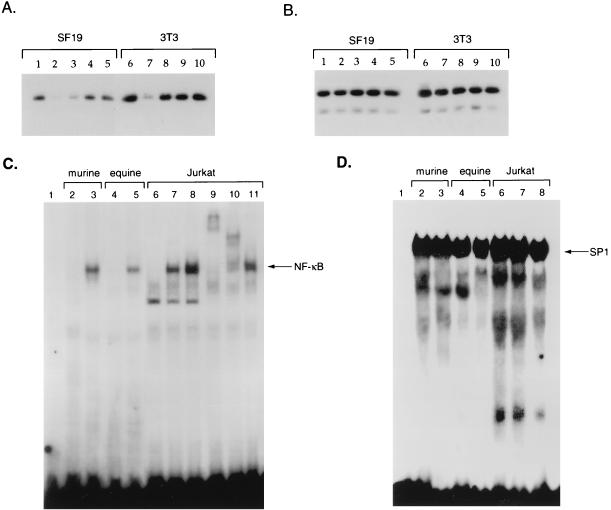
To address whether DNA-PK has a role in the regulation of IκBα protein levels and NF-κB nuclear translocation, we used the murine NIH 3T3 fibroblast cell line and the SCID mouse fibroblast cell line SF19, which is deficient in DNA-PK activity (39). Western blot analysis with DNA-PKcs antibody confirmed that 3T3 cells contained DNA-PKcs and that this protein was absent from SF19 cells (data not shown). Next we analyzed IκBα protein levels in cytoplasmic extracts prepared for these cells either prior to treatment with phorbol esters and ionomycin or following treatment with these inducers of IκBα degradation. Western blot analysis with rabbit polyclonal antibody indicated that the IκBα levels were approximately twofold lower in SF19 cells (Fig. 8A, lane 1) than in 3T3 cells (Fig. 8A, lane 6). Upon treatment of SF19 cells with TPA and ionomycin, there was a rapid decrease in IκBα protein levels at 15 min posttreatment followed by a restoration of IκBα protein levels, which increased to pretreatment levels between 45 and 60 min posttreatment (Fig. 8A, lanes 1 to 5). Upon treatment of 3T3 cells with TPA and ionomycin, a marked decrease in IκBα levels also occurred at 15 min posttreatment followed by a restoration of IκBα protein levels, which returned to pretreatment levels between 30 and 60 min posttreatment (Fig. 8A, lanes 6 to 10). As a control, Western blot analysis was performed with antibody directed against the p89 subunit of TFIIH (Fig. 8B). These results indicated that there were slight reductions in the levels of IκBα protein in cells lacking DNA-PKcs but that degradation of IκBα by agents that stimulate the nuclear translocation of NF-κB was not defective in SCID cells.
FIG. 8.
IκBα and NF-κB regulation in SCID cells. Cytoplasmic extracts were prepared from either the SCID cell line SF19 (lanes 1 to 5) or NIH 3T3 cells (lanes 6 to 10) that were untreated (lanes 1 and 6) or treated with TPA and ionomycin for 15 (lanes 2 and 7), 30 (lanes 3 and 8), 60 (lanes 4 and 9), or 90 (lanes 5 and 10) min. Western blot analysis was performed with 20 μg of each of these extracts by using rabbit polyclonal antibody directed against either IκBα (A) or the p89 subunit of TFIIH (B). Nuclear extracts were prepared from NIH 3T3 cells (lane 2), SF19 cells (lane 3), the wild-type equine cell line CRL6288 (lane 4), and the equine SCID cell line 1863 (lane 5) and subjected to gel retardation analysis with either an oligonucleotide containing an NF-κB (C) or an SP1 (D) binding site, followed by autoradiography. An analysis of probe alone is also shown (lane 1). For panel C, nuclear extract was prepared from Jurkat cells that were not treated (lane 6) or treated with TPA and ionomycin for 10 (lane 7) or 30 (lane 8) min. Antibody to the RelA (p65) (lane 10) or p50 (lane 10) subunits of NF-κB or CBP (lane 11) was added to the gel retardation assay mixtures for supershift analysis.
Next, we determined whether NF-κB DNA binding properties differed in nuclear extract prepared from SF19 and NIH 3T3 cells. Gel retardation analysis with a 32P-labeled NF-κB DNA probe was performed with nuclear extract prepared from untreated 3T3 and SF19 cells. There was no binding to the NF-κB probe in 3T3 nuclear extract (Fig. 8C, lane 2), while constitutive DNA binding of NF-κB was noted with SF19 nuclear extract (Fig. 8C, lane 3). To confirm that nuclear levels of NF-κB were indeed higher in cells lacking DNA-PK, we also prepared nuclear extract from control equine fibroblast cell line CRL6288 and equine SCID fibroblast cell line 1863 (82). Gel retardation analysis again confirmed constitutive NF-κB binding activity in nuclear extract prepared from the equine SCID cells, but not the control equine fibroblast cell line (Fig. 8C, lanes 4 and 5). Nuclear extracts prepared from untreated Jurkat cells (Fig. 8C, lane 6) and Jurkat cells treated with TPA and ionomycin (Fig. 8C, lanes 7 and 8) were also assayed in gel retardation assays with the NF-κB probe. This analysis demonstrated that the mobility of the NF-κB gel-retarded complex in Jurkat nuclear extract was the same as that seen with extracts prepared from the murine and equine SCID cell lines. Antibodies to either RelA (Fig. 8C, lane 9) or the p50 subunit of NF-κB (Fig. 8C, lane 10), when added to the TPA- and ionomycin-treated Jurkat nuclear extract supershifted the gel-retarded complex, indicating its specificity. Antibody to the coactivator protein CBP was added as a control to this gel retardation assay and did not alter the mobility of the NF-κB complex (Fig. 8C, lane 11). Finally, to standardize these nuclear extracts, we compared the amounts of SP1 protein prepared from these cell lines by using gel retardation analysis. These data indicated that the amounts of SP1 in the different cell lines were similar (Fig. 8D, lanes 1 to 8). These results indicate that SCID cell lines which lack DNA-PKcs have constitutive nuclear NF-κB DNA binding activity.
Increases in IκBα levels decrease NF-κB DNA binding properties in SCID cells.
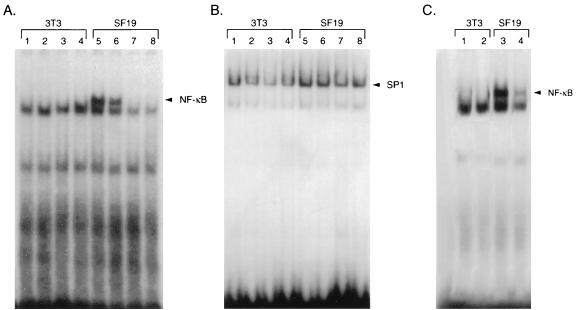
To determine whether constitutive NF-κB DNA binding in SCID cells could be modulated by overexpression of IκBα, we utilized recombinant adenoviruses containing either flag-tagged wild-type IκBα or a dominant-negative IκBα protein containing mutations of serine residues 32 and 36. A recombinant adenovirus expressing the β-galactosidase gene was used as a control. Western blot analysis with a flag monoclonal antibody confirmed the expression of the wild-type and the mutant IκB proteins upon infection of SF19 and 3T3 cells by the recombinant adenoviruses (data not shown). Each of these adenoviruses was then used to infect 3T3 and SF19 cells, and nuclear extract prepared 24 h later was analyzed by gel retardation analysis with an NF-κB oligonucleotide probe. Nuclear extract prepared from SF19 cells but not from 3T3 cells (Fig. 9A, lanes 1 and 5) revealed constitutive NF-κB DNA binding. A faster-mobility nonspecific gel-retarded species was present in these nuclear-extract preparations, and its mobility did not change with the addition of antibodies to p50 or p65, in contrast to the results seen with the slower-mobility species (data not shown). Adenoviruses expressing β-galactosidase did not alter the mobility of the NF-κB-specific gel-retarded species in SF19 extract (Fig. 9A, lanes 2 and 6). In contrast, infection with adenoviruses containing either wild-type IκBα (Fig. 9A, lanes 3 and 7) or a mutant IκBα with mutations at serine residues 32 and 36 (Fig. 9A, lanes 4 and 8) reduced NF-κB DNA binding in SF19 cells. There was no change in SP1 binding in the 3T3 and SF19 nuclear extracts infected with these different adenoviruses (Fig. 9B). The adenovirus construct containing wild-type IκBα was also assayed in SF19 and NIH 3T3 cells, and similar IκBα protein stability was found (data not shown).
FIG. 9.
Overexpression or decreased degradation of IκBα reduces NF-κB DNA-binding activity in SCID cells. A wild-type IκBα cDNA or an IκBα cDNA with mutations of serine residues 32 and 36 to alanine were cloned into adenovirus. These viruses and a control adenovirus containing the β-galactosidase gene were used to infect NIH 3T3 and SF19 cells at a multiplicity of infection of 10 and harvested 24 h later. Nuclear extracts were prepared from mock- and adenovirus-infected cells and subjected to gel retardation analysis with oligonucleotides containing NF-κB (A) or SP1 (B) binding sites. Gel retardation analysis was performed with nuclear extracts isolated from uninfected cells (lanes 1 and 5), cells infected with adenovirus containing the β-galactosidase gene (lanes 2 and 6), cells infected with adenovirus containing wild-type IκBα (lanes 3 and 7), and cells infected with adenovirus containing IκBα with serine residues 32 and 36 mutated to alanine (lanes 4 and 8). (C) NIH 3T3 (lanes 1 and 2) and SF19 (lanes 3 and 4) cells were treated with 50 μM TPCK (Sigma) for 45 min. Nuclear extracts were prepared from nontreated cells (lanes 1 and 3) and TPCK-treated cells (lanes 2 and 4) and subjected to gel retardation analysis with a double-stranded NF-κB binding oligonucleotide.
Finally, we determined whether the addition of the serine protease inhibitor TPCK (N-tosyl-l-phenylalanyl chloromethyl ketone), which prevents IκBα protein degradation (28, 32, 43), could alter the constitutive NF-κB DNA binding properties in SF19 nuclear extract. Following a 45-min treatment with 50 μM TPCK, nuclear extract was prepared from 3T3 and SF19 cells (Fig. 9C, lanes 1 to 4) and subjected to gel retardation analysis. TPCK was able to reduce NF-κB DNA binding in nuclear extract prepared from SF19 cells (Fig. 9C, lanes 3 and 4). The addition of TPCK did not alter SP1 binding in either 3T3 or SF19 cells (data not shown). Whether the ability of TPCK to prevent IκB degradation is related to its ability to inhibit protease and/or kinase activity is unclear (28, 32, 43). These results indicate that increases in IκBα protein reduce constitutive NF-κB DNA binding properties in SCID cells.
DNA-PK regulates NF-κB DNA binding properties.
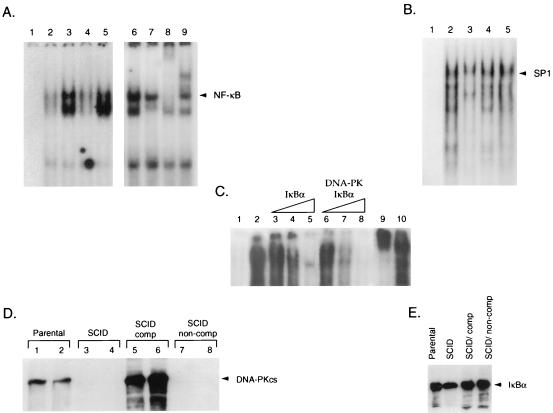
Although we demonstrated that SCID cells exhibited constitutive nuclear NF-κB DNA binding, it was important to determine whether the lack of DNA-PKcs was responsible for this phenotype. Nuclear extract was prepared from the parental murine cell line, CB-17, a SCID cell line (SCID-St), and SCID-St cells complemented with portions of human chromosome 8 either containing the DNA-PKcs gene (100E) or lacking this gene (50D). Gel retardation analysis indicated that NF-κB binding was markedly increased in extract prepared from SCID-St cells (Fig. 10A, lane 3) compared to that seen in extract from the CB-17 cells (Fig. 10A, lane 2). Nuclear extract prepared from the DNA-PKcs-complemented 100E cells had reduced NF-κB binding activity (Fig. 10A, lane 4) compared to nuclear extract prepared from 50D cells which lack DNA-PKcs (Fig. 10A, lane 5). SP1 binding levels were similar in nuclear extract prepared from each of these cells (Fig. 10B). This indicated that the lack of DNA-PKcs led to constitutive nuclear NF-κB DNA binding activity.
FIG. 10.
NF-κB DNA-binding activity in SCID and DNA-PKcs-complemented cells. (A) Nuclear extracts were prepared from CB-17 (parental), SCID-St (SCID), 100E (DNA-PKcs-complemented SCID cells), and 50D (non-DNA-PKcs-complemented SCID cells) cells and subjected to gel retardation analysis using NF-κB oligonucleotides. Results for probe alone (lane 1) and nuclear extract prepared from CB-17 (lane 2), SCID-St (lane 3), 100E (lane 4), and 50D (lane 5) cells are shown. Nuclear extract prepared from SCID-St cells (lane 6) was analyzed in these gel retardation assays in the presence of preimmune serum (lane 7), antibody directed against RelA (lane 8), or the NF-κB p50 subunit (lane 9). (B) Gel retardation analysis with an SP1 probe either alone (lane 1) or in the presence of nuclear extract prepared from CB-17 (lane 2), SCID-St (lane 3), 100E (lane 4), or 50D (lane 5) cells was performed. (C) The recombinant NF-κB subunits p50 and p65 (RelA) were purified following baculovirus expression and analyzed by gel retardation with NF-κB binding site oligonucleotides. Probe alone (lane 1) and the p50-p65 proteins without the addition of IκBα (lane 2) are shown, as are p50-p65 with 50, 100, or 200 ng of either purified IκBα (lanes 3 to 5, respectively) or DNA-PK-phosphorylated IκBα (lanes 6 to 8, respectively), which was added to the p50-p65 proteins approximately 30 min prior to the addition of the NF-κB probe. The results for p50-p65 proteins in the presence of antibody directed against p65 (RelA) (lane 9) or CBP (lane 10) are also indicated. (D) Western blot analysis was performed with cytoplasmic (odd-numbered lanes) or nuclear (even-numbered lanes) extracts prepared from CB-17 (lanes 1 and 2), SCID-St (lanes 3 and 4), 100E (lanes 5 and 6), and 50D (lanes 7 and 8) cells with a monoclonal antibody directed against DNA-PKcs (42-26). (E) Western blot analysis was performed on cytoplasmic extracts prepared from CB-17, SCID-St, 100E, and 50D cells with polyclonal antibody directed against IκBα.
To determine the potential mechanism by which DNA-PK was able to modulate NF-κB binding, we performed gel retardation analysis with recombinant p50 and p65 NF-κB proteins produced in baculovirus. Purified IκBα alone and IκBα phosphorylated by DNA-PK were incubated with the p50 and p65 proteins prior to the addition of a 32P-labeled NF-κB oligonucleotide probe. Kinase assays with [γ-32P]ATP demonstrated that the IκBα protein was phosphorylated by DNA-PK under these conditions (data not shown). Gel retardation analysis indicated that unphosphorylated IκBα (Fig. 10C, lanes 3 to 5) was a weaker competitor for p50-p65 binding to the NF-κB probe than the DNA-PK-phosphorylated IκBα (Fig. 10C, lanes 6 to 8). Antibody to p65 but not CBP supershifted the p50-p65 complex bound to the NF-κB oligonucleotide, indicating the specificity of this binding (Fig. 10C, lanes 9 and 10). These results suggested that DNA-PK phosphorylation of IκBα increases its interaction with NF-κB and thus reduces NF-κB DNA binding properties.
Western blot analysis demonstrated that DNA-PKcs was present in extract prepared from the parental and SCID cells complemented with a portion of human chromosome 8 containing the DNA-PKcs gene (Fig. 10D, lanes 1, 2, 5, and 6). However, DNA-PKcs was absent from SCID cells and SCID cells lacking the portion of human chromosome 8 that contained the DNA-PKcs gene (Fig. 10D, lanes 3, 4, 7, and 8). In addition, Western blot analysis confirmed that the levels of IκBα in these cell lines were similar although there was a slight but reproducible reduction of IκBα levels in the SCID cell line (Fig. 10E). These results indicate that DNA-PK phosphorylation of IκBα regulates NF-κB DNA binding properties rather than significantly altering IκBα levels.
DNA-PK and IκBα distribution in SCID and DNA-PKcs-complemented SCID cells.
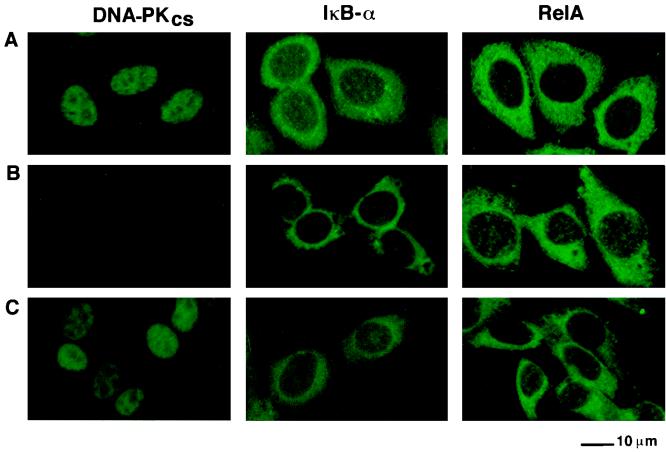
We next determined the intracellular localization of DNA-PK in HeLa cells, murine SCID cells lacking the DNA-PKcs gene (50D) and murine SCID cells complemented with a portion of human chromosome 8 which contains DNA-PKcs (100E). Immunofluorescence staining with monoclonal antibody 25-4 directed against human DNA-PKcs followed by analysis using confocal microscopy indicated that DNA-PKcs was present predominantly in the nuclei of HeLa (Fig. 11A) and 100E cells (Fig. 11C). As expected, no DNA-PKcs-specific nuclear staining was observed in the SCID mouse cell line (50D) which is deficient in both mouse and human DNA-PKcs (Fig. 11B).
FIG. 11.
Subcellular localization of DNA-PKcs, IκBα, and the RelA subunit of NF-κB. HeLa (A) 50D (B), and 100E (C) cells were stained by immunofluorescence with a monoclonal antibody directed against DNA-PKcs (25-4) (left panel), a polyclonal antibody directed against IκBα (C-21) (middle panel), or a polyclonal antibody directed against RelA (C-20) (right panel), followed by goat anti-mouse FITC or goat anti-rabbit FITC conjugate. Preparations were analyzed by confocal microscopy.
Next we determined whether the lack of DNA-PKcs in the SCID mouse 50D cells correlated with specific changes in the intracellular distribution of IκBα and the RelA or p65 subunit of NF-κB. IκBα was present in both the cytoplasm and the nuclei of HeLa and 100E cells (Fig. 11A and C). IκBα staining was slightly increased in the cytoplasm of 50D SCID cells, but there was reduced IκBα staining in the nuclei of these cells compared to that observed in the nuclei of 100E and HeLa cells. The RelA or p65 subunit of NF-κB was present predominantly in the cytoplasm of HeLa and 100E cells (Fig. 11A and C), whereas it was present in the cytoplasm and in increased amounts in the nuclei of 50D cells (Fig. 11B). Thus, the absence of DNA-PKcs in the nuclei of SCID mouse cells correlated with decreased nuclear staining of IκBα and with the presence of increased amounts of RelA in the nuclei of these cells.
DISCUSSION
In this study, we characterized cellular kinases that phosphorylate IκBα. Using a biochemical fractionation scheme, we isolated a cellular kinase, DNA-PK, that phosphorylated serine residue 36 and threonine residue 273 in IκBα. Threonine residue 273 in IκBα appeared to be a predominant site of phosphorylation by DNA-PK. However, the relative effects of phosphorylation of each of these residues on IκBα function were not addressed in this study. It is possible that phosphorylation of each of these residues may alter IκBα function by different mechanisms or that phosphorylation of both of these sites is required to alter NF-κB binding. Alternatively, only the predominant carboxy-terminal phosphorylation of IκB by DNA-PK may be critical for the regulation of NF-κB DNA binding properties.
The biological relevance of DNA-PK phosphorylation of IκB was demonstrated by an analysis of SCID cell lines in which DNA-PK activity is absent (15, 49, 72). SCID cell lines from different species including murine and equine species were found to have constitutive nuclear NF-κB DNA binding activity when analyzed in gel retardation assays. This was in contrast to SCID cell lines complemented with the DNA-PKcs gene (49) in which there was marked reduction in NF-κB DNA binding activity. These results and gel retardation assays with unphosphorylated and DNA-PK-phosphorylated IκBα suggest that DNA-PK phosphorylation of IκB enhances its ability to associate with NF-κB and inhibit NF-κB DNA binding properties.
Since DNA-PKcs is predominantly localized to the nucleus (19, 54), it likely phosphorylates IκB in the nucleus. However, we cannot rule out the possibility that small quantities of DNA-PK may be present in the cytoplasm and phosphorylate IκBα in the cytoplasm. The ability of double-stranded DNA to markedly stimulate DNA-PK phosphorylation of IκBα would also be consistent with a role for DNA-PK phosphorylation of IκB in the nucleus. Immunofluorescence studies demonstrate the presence of IκBα in both the cytoplasm and the nuclei of HeLa and SCID cells complemented with DNA-PKcs, with reduced levels of IκBα seen in the nuclei of SCID cells. Previous studies indicate that in some cell lines IκBα is present in both the nucleus and cytoplasm and that its role in the nucleus may be to inhibit NF-κB-mediated gene expression and terminate the response to inducers such as TNF-α (2, 3, 60). Increases in IκB phosphorylation by DNA-PK may inhibit NF-κB DNA binding activity in the nucleus by increasing associations between IκBα and NF-κB (78, 84).
Decreases in IκBα levels in the nuclei of SCID cells could be due to the fact that nonphosphorylated IκB is preferentially transported to the cytoplasm compared to phosphorylated IκBα. IκBα can shuttle between the nucleus and the cytoplasm to promote the efficient nuclear export of NF-κB complexes (3). This function is mediated by a nuclear export sequence in the C terminus of IκBα that is homologous to the previously described export signal found in the human immunodeficiency virus type 1 Rev protein. It is intriguing that this signal is located between amino acids 266 and 277 in IκBα, which includes the threonine residue (residue 273) that is phosphorylated by DNA-PK (3). Thus, it is possible that the phosphorylation of IκBα by DNA-PK leads to enhanced nuclear accumulation of IκBα and more potent inhibition of NF-κB DNA binding. DNA-PK phosphorylation of serine residue 36 may also lead to higher levels of IκBα in the nucleus by blocking the ability of inducible kinases such as IKKα and IKKβ to phosphorylate the amino terminus of IκB, which may lead to its degradation in the nucleus.
IκBα is subject to phosphorylation by a number of different kinases. These include kinases that are inducible by mitogenic stimuli, such as IKKα and IKKβ, that phosphorylate the amino terminus of IκBα and lead to its degradation by the proteasome (27, 63, 65, 83, 85). In addition, a phorbol ester-inducible kinase, RSK1, that specifically phosphorylates IκBα on serine residue 32 has been identified (71). Whether RSK1 in conjunction with DNA-PK can phosphorylate both serine residues 32 and 36, similar to the actions of IKKα and IKKβ, and thereby induce IκBα degradation remains to be determined. In addition to amino-terminal phosphorylation of IκBα, carboxy-terminal phosphorylation of IκBα by casein kinase II has been demonstrated (10, 11, 57, 61, 66). The four residues in the carboxy terminus of IκBα that are phosphorylated by casein kinase II are distinct from the predominant threonine residue that is phosphorylated by DNA-PK. The phosphorylation of IκBα by casein kinase II is likely involved in regulating its constitutive stability (11, 57, 61). Our results suggest that DNA-PK phosphorylation of IκBα is at least partially distinct from that of casein kinase II and that DNA-PK phosphorylation does not significantly alter IκBα protein stability. However, it is intriguing to note that casein kinase II phosphorylation of the C-terminal PEST domain of IκBβ but not of IκBα markedly increases the ability of IκBβ to inhibit NF-κB DNA binding properties (25, 78). Thus, DNA-PK and casein kinase II can phosphorylate both IκBα and IκBβ, but these kinases may have differential effects on IκBα and IκBβ interactions with NF-κB.
IκBα is a modular protein in which different domains are subject to regulation by different protein kinases. Inducible kinases, such as IKKα and IKKβ, which are activated by TNF-α and interleukin 1 lead to phosphorylation of the N terminus of IκBα and the degradation of IκBα (27, 63, 65, 83, 85). Constitutive phosphorylation of IκB by casein kinase II regulates basal IκBα protein stability (11, 57, 61). In this study, we demonstrate that DNA-PK phosphorylates both the amino and carboxy termini of IκBα and IκBβ. Further analysis will be required to determine the relevance of each of these phosphorylation sites. It is important to note that another member of the PI-3 kinase family, the product of the ataxia-telangiectasia gene (ATM) (68), has also been demonstrated to phosphorylate IκBα (47). Furthermore, an IκBα protein with its amino terminus deleted can prevent the marked radiosensitivity seen in ataxia-telangiectasia cells (48). Our results suggest that IκBα is regulated not only by kinases that lead to its degradation but by kinases that alter its interactions with NF-κB. This is likely critical in the control of the expression of genes involved in regulating cellular growth and proliferation.
ACKNOWLEDGMENTS
We thank Sharon Johnson for the preparation of the manuscript; Shane Fults for the preparation of the figures; and R. Robinson, and K. Pierce for expertise in the digestion and mass spectrometry. We thank Dean Ballard for providing the IκBα and IκBβ cDNAs and Cordula Kirchgessner for providing SCID and DNA-PKcs-complemented SCID cell lines.
This work was supported by grants from the NIH, the Veterans Administration, and the Robert Welch Foundation.
Li Liu and Youn-Tae Kwak contributed equally to this work.
REFERENCES
- 1.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1995;92:10599–10603. doi: 10.1073/pnas.92.23.10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle P A, Baltimore D. IκB: a specific inhibitor of the NF-κB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P A, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle P A, Baltimore D. A 65-kD subunit of active NF-kappaB is required for inhibition of NF-kappaB by I kappaB. Genes Dev. 1989;3:1689–1698. doi: 10.1101/gad.3.11.1689. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle P A, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 8.Ballard D W, Dixon E P, Peffer N J, Bogerd H, Doerre S, Stein B, Greene W C. The 65-kDa subunit of human NF-κB functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc Natl Acad Sci USA. 1992;89:1875–1879. doi: 10.1073/pnas.89.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bannister A J, Gottlieb T M, Kouzarides T, Jackson S P. c-Jun is phosphorylated by the DNA-dependent protein kinase in vitro; definition of the minimal kinase recognition motif. Nucleic Acids Res. 1993;21:1289–1295. doi: 10.1093/nar/21.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barroga C F, Stevenson J K, Schwarz E M, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II. Proc Natl Acad Sci USA. 1995;92:7637–7641. doi: 10.1073/pnas.92.17.7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beauparlant P, Lin R, Hiscott J. The role of the C-terminal domain of IκBα in protein degradation and stabilization. J Biol Chem. 1996;271:10690–10696. doi: 10.1074/jbc.271.18.10690. [DOI] [PubMed] [Google Scholar]
- 12.Beg A A, Finco T S, Nantermet P V, Baldwin A S J. Tumor necrosis factor and interleukin-1 lead to phosphorylation and loss of IκBα: a mechanism for NF-κB activation. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequences of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 14.Biederman K A, Sun J, Giacia A J, Tosto L M, Brown J M. scid mutation in mice confers hypersensitivity to ionizing radiation and a deficiency in DNA double-strand break repair. Proc Natl Acad Sci USA. 1991;88:1394–1397. doi: 10.1073/pnas.88.4.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 16.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown K, Gerstberger S, Carlson L, Fransozo G, Siebenlist U. Control of IκBα proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 18.Brush G S, Anderson C W, Kelly T J. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc Natl Acad Sci USA. 1994;91:12520–12524. doi: 10.1073/pnas.91.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter T, Vancurova I, Sun I, Lou W, DeLeon S. A DNA-activated protein kinase from HeLa cell nuclei. Mol Cell Biol. 1990;10:6460–6471. doi: 10.1128/mcb.10.12.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan D W, Lees-Miller S P. The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J Biol Chem. 1996;271:8936–8941. doi: 10.1074/jbc.271.15.8936. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Willingham T, Margraf L R, Schreiber-Agus N, DePhinho R A, Nisen P D. Effects of the MYC oncogene antagonist, MAD, on proliferation, cell cycling and the malignant phenotype of human brain tumour cells. Nat Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y-R, Lees-Miller S P, Tegtmeyer P, Anderson C W. The human DNA-activated protein kinase phosphorylates simian virus 40 T antigen at amino- and carboxy-terminal sites. J Virol. 1991;65:5131–5140. doi: 10.1128/jvi.65.10.5131-5140.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, Hagler J, Palombella V J, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 24.Chen Z J, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 25.Chu Z-L, McKinsey T A, Liu L, Qi X, Ballard D W. Basal phosphorylation of the PEST domain in IκBβ regulates its functional interaction with the c-rel proto-oncogene product. Mol Cell Biol. 1996;16:5974–5984. doi: 10.1128/mcb.16.11.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cimprich K A, Shin T B, Keith C T, Schreiber S L. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 28.DiDonato J A, Mercurio F, Karin M. Phosphorylation of IκBα precedes but is not sufficient for its dissociation from NF-κB. Mol Cell Biol. 1995;15:1302–1311. doi: 10.1128/mcb.15.3.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dvir A, Stein L Y, Calore B L, Dynan W S. Purification and characterization of a template-associated protein kinase that phosphorylates RNA polymerase II. J Biol Chem. 1993;268:10440–10447. [PubMed] [Google Scholar]
- 31.Eng J K, McCormick A L, Yates J R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 32.Finco T S, Beg A A, Baldwin A S J. Inducible phosphorylation of IκBα is not sufficient for its dissociation from NF-κB and is inhibited by protease inhibitors. Proc Natl Acad Sci USA. 1994;91:11884–11888. doi: 10.1073/pnas.91.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fiscella M, Ullrich S J, Zambrano N, Shields M T, Lin D, Lees-Miller S P, Anderson C W, Mercer W E, Appella E. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- 34.Ganchi P, Sun S-C, Greene W C, Ballard D W. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giffin W, Torrance H, Rodda D J, Prefontaine G G, Pope L, Hache R J G. Sequence-specific DNA binding by Ku autoantigen and its effects on transcription. Nature. 1996;380:265–268. doi: 10.1038/380265a0. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Foix A, Coats W, Baque S, Alam T, Gerard R, Newgard C. Adenovirus-mediated transfer of the muscle glycogen phosphorylase gene into hepatocytes confers altered regulation of glycogen metabolism. J Biol Chem. 1992;267:25129–25134. [PubMed] [Google Scholar]
- 37.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 38.Graham F, Preved L. Manipulation of adenovirus vectors. In: Murray E, editor. Methods in molecular biology. Jersey City, N.J: Humana Press, Inc.; 1991. pp. 109–128. [DOI] [PubMed] [Google Scholar]
- 39.Harrington J, Hsieh C-L, Gerton J, Bosma G, Lieber M R. Analysis of the defect in DNA end joining in the murine scid mutation. Mol Cell Biol. 1992;12:4758–4768. doi: 10.1128/mcb.12.10.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Lees-Miller S P, Anderson C W, Jackson S P. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 41.Haskill S, Beg A A, Tompkins S M, Morris J S, Yurochko A D, Sampson-Johannes A, Mondal K, Ralph P, Baldwin A S. Characterization of an immediate-early gene induced in adherent monocytes that encodes a kappa B-like activity. Cell. 1991;65:1281–1289. doi: 10.1016/0092-8674(91)90022-q. [DOI] [PubMed] [Google Scholar]
- 42.Hellman U, Wernstedt C, Gonez J, Heldin C H. Improvement of an in gel digestion procedure for the micro-preparation of internal protein fragments for amino acid sequencing. Anal Biochem. 1995;224:451–455. doi: 10.1006/abio.1995.1070. [DOI] [PubMed] [Google Scholar]
- 43.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben N Y, Baeuerle P A. Rapid proteolysis of I kappa B-alpha is necessary for activation of transcription factor NF-kappa B. Nature. 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 44.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein and UV-responsive protein kinase that binds and potentiates c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 45.Imbert V, Rupec R A, Livolsi A, Pahl H L, Traenckner E B-M, Mueller-Dieckmann C, Farahifar D, Rossi B, Auberger P, Baeuerle P A, Peyron J-F. Tyrosine phosphorylation of IκB-α activates NF-κB without proteolytic degradation of IκB-α. Cell. 1996;86:787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 46.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of SP1 by a DNA dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 47.Jung M, Kondratyev A, Lee S A, Dimtchev A, Dritschillo A. ATM gene product phosphorylates I kappa B-alpha. Cancer Res. 1997;57:24–27. [PubMed] [Google Scholar]
- 48.Jung M, Zhang Y, Lee S, Dritschilo A. Correction of radiation sensitivity in ataxia-telangiectasia cells by a truncated IκBα. Science. 1995;268:1619–1621. doi: 10.1126/science.7777860. [DOI] [PubMed] [Google Scholar]
- 49.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 50.Lane W S, Galat A, Harding M W, Schreiber S L. Complete amino acid sequence of the FK506 and rapamycin binding protein, FKBP, isolated from calf thymus. J Protein Chem. 1991;10:151–160. doi: 10.1007/BF01024778. [DOI] [PubMed] [Google Scholar]
- 51.Lee F S, Hagler J, Chen Z J, Maniatis T. Activation of the IκBα kinase complex by MEKK1, a kinase of the JNK pathway. Cell. 1997;88:213–222. doi: 10.1016/s0092-8674(00)81842-5. [DOI] [PubMed] [Google Scholar]
- 52.Lee S E, Mitchell R A, Cheng A, Hendrickson E A. Evidence for DNA-PK-dependent and -independent DNA double-strand break repair pathways in mammalian cells as a function of the cell cycle. Mol Cell Biol. 1997;17:1425–1433. doi: 10.1128/mcb.17.3.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lees-Miller S P. The DNA-dependent protein kinase, DNA-PK: 10 years and no ends in sight. Biochem Cell Biol. 1996;74:503–512. doi: 10.1139/o96-054. [DOI] [PubMed] [Google Scholar]
- 54.Lees-Miller S P, Chen Y-R, Anderson C W. Human cells contain a DNA-activated protein kinase that phosphorylates simian virus 40 T antigen, mouse p53, and the human Ku autoantigen. Mol Cell Biol. 1990;10:6472–6481. doi: 10.1128/mcb.10.12.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 56.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin R, Beauparlant P, Makris C, Meloche S, Hiscott J. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol Cell Biol. 1996;16:1401–1409. doi: 10.1128/mcb.16.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y-C, Brown K, Siebenlist U. Activation of NF-κB requires proteolysis of the inhibitor IκB-α: signal-induced phosphorylation of IκB-α alone does not release active NF-κB. Proc Natl Acad Sci USA. 1995;92:552–556. doi: 10.1073/pnas.92.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S-H, Ma J-T, Yueh A Y, Lees-Miller S P, Anderson C W, Ng S-Y. The carboxyl-terminal transactivation domain of human serum response factor contains DNA-activated protein kinase phosphorylation sites. J Biol Chem. 1993;268:21147–21154. [PubMed] [Google Scholar]
- 60.Luque I, Gelinas C. Distinct domains of IκBα regulate c-Rel in the cytoplasm and in the nucleus. Mol Cell Biol. 1998;18:1213–1224. doi: 10.1128/mcb.18.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKinsey T A, Brockman J A, Scherer D C, Al-Murrani S W, Green P L, Ballard D W. Inactivation of IκBβ by the Tax protein of human T-cell leukemia virus type 1: a potential mechanism for constitutive induction of NF-κB. Mol Cell Biol. 1996;16:2083–2090. doi: 10.1128/mcb.16.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 64.Peterson S R, Dvir A, Anderson C W, Dynan W S. DNA binding provides a signal for phosphorylation of the RNA polymerase II heptapetide repeats. Genes Dev. 1992;6:426–438. doi: 10.1101/gad.6.3.426. [DOI] [PubMed] [Google Scholar]
- 65.Regnier C H, Song H Y, Gao X, Goeddel D V, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez M S, Michalopoulos I, Arenzana-Seisdedos F, Hay R T. Inducible degradation of IκBα in vitro and in vivo requires the acidic C-terminal domain of the protein. Mol Cell Biol. 1995;15:2413–2419. doi: 10.1128/mcb.15.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruben S M, Narayanan R, Klement J F, Chen C-H, Rosen C A. Functional characterization of the NF-κB p65 transcriptional activator and an alternatively spliced derivative. Mol Cell Biol. 1992;12:444–454. doi: 10.1128/mcb.12.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle D A, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali S R, Simmons A, Clines G A, Sartiel A, Gatti R A, Chessa L, Sanal O, Lavin M F, Jaspers N G J, Taylor A M R, Arlett C F, Miki T, Wesissman S, Lovett M, Collins F S, Shiloh Y A. A single ataxia-telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 69.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmitz M L, Baeuerle P A. The p65 subunit is responsible for the strong transcription activating potential of NF-kappa B. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schouten G J, Vertegaal A C O, Whiteside S T, Israel A, Toebes M, Dorsman J C, van der Eb A, Zantema A. IκBα is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. EMBO J. 1997;16:3133–3144. doi: 10.1093/emboj/16.11.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P. Gene for the catalytic subunit of the human DNA-activated protein kinase maps to the site of the XRCC7 gene on chromosome 8. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun S-C, Ganchi P, Ballard D W, Greene W C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 74.Suwa A, Hirakata M, Takeda Y, Jesch S A, Mimori T, Hardin J A. DNA-dependent protein kinase (Ku protein-p350 complex) assembles on double-stranded DNA. Proc Natl Acad Sci USA. 1994;91:6904–6908. doi: 10.1073/pnas.91.15.6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 76.Thompson J E, Phillips R J, Erdjument-Bromage H, Tempst P, Ghosh S. IκBβ regulates the persistent response in a biphasic activation of NF-κB. Cell. 1995;80:573–582. doi: 10.1016/0092-8674(95)90511-1. [DOI] [PubMed] [Google Scholar]
- 77.Traenckner E B M, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκBα on serines 32 and 36 controls IκBα proteolysis and NF-κB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tran K, Merika M, Thanos D. Distinct functional properties of IκBα and IκBβ. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verma I M, Stevenson J K, Schwartz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 80.Whiteside S T, Epinat J, Rice N R, Israel A. I kappa B epsilon, a novel member of the IκB family, controls RelA and cRel NF-κB activity. EMBO J. 1997;16:1413–1426. doi: 10.1093/emboj/16.6.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Whiteside S T, Ernst M K, LeBail O, Laurent-Winter C, Rice N, Israel A. N- and C-terminal sequences control degradation of MAD3/IκBα in response to inducers of NF-κB activity. Mol Cell Biol. 1995;15:5339–5345. doi: 10.1128/mcb.15.10.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiler R, Leber R, Moore B B, VanDyke L F, Perryman L E, Meek K. Equine severe combined immunodeficiency: a defect in V(D)J recombination and DNA-dependent protein kinase activity. Proc Natl Acad Sci USA. 1995;92:11485–11489. doi: 10.1073/pnas.92.25.11485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 84.Zabel U, Baeuerle P A. Purified human IκB can rapidly dissociate the complex of the NF-kappa B transcription factor with its cognate DNA. Cell. 1990;61:255–265. doi: 10.1016/0092-8674(90)90806-p. [DOI] [PubMed] [Google Scholar]
- 85.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]