Abstract
Activation of protein-encoding genes involves recruitment of an RNA polymerase II holoenzyme to promoters. Since the Srb4 subunit of the holoenzyme is essential for expression of most class II genes and is a target of at least one transcriptional activator, we reasoned that suppressors of a temperature-sensitive mutation in Srb4 would identify other factors generally involved in regulation of gene expression. We report here that MED6 and SRB6, both of which encode essential components of the holoenzyme, are among the dominant suppressors and that the products of these genes interact physically with Srb4. The recessive suppressors include NCB1 (BUR6), NCB2, NOT1, NOT3, NOT5, and CAF1, which encode subunits of NC2 and the Not complex. NC2 and Not proteins are general negative regulators which interact with TATA box binding protein (TBP). Taken together, these results suggest that transcription initiation involves a dynamic balance between activation mediated by specific components of the holoenzyme and repression by multiple TBP-associated regulators.
Expression of mRNA genes in eukaryotes involves the recruitment of RNA polymerase II and other general transcription factors to promoters (41, 51). Evidence that RNA polymerase II can be found associated with most of the general transcription factors and additional factors essential for initiation in vivo suggests that much of the transcription initiation apparatus can be recruited to promoters in a preassembled RNA polymerase II holoenzyme (6, 24, 28, 36, 43, 44, 55).
RNA polymerase II holoenzymes consist of RNA polymerase II and a subset of general transcription factors, together with Srb-Mediator proteins. Several lines of evidence indicate that the Srb-Mediator proteins are involved in the response to gene-specific activators. Truncation mutations of the C-terminal domain of the largest subunit of RNA polymerase II result in defects in activation (1, 14, 33, 53), and the Srb proteins were originally identified through genetic interactions with one such truncation mutant (19, 34, 57; reviewed in reference 29). The RNA polymerase II holoenzyme responds to the addition of transcriptional activators in vitro while purified polymerase and general factors alone do not (24, 28). The Srb-Mediator complex binds to the C-terminal domain and can be purified as a separate complex from holoenzyme. This purified Srb-Mediator complex is necessary to reconstitute the ability of a defined transcription system to respond to activators in vitro (19, 24). Activators have been shown to bind directly to the Srb-Mediator complex (19), and genetic and biochemical studies have identified the Srb4 subunit as a target of the well-studied acidic activator Gal4 (25).
Temperature-sensitive mutations in the essential Srb4 holoenzyme subunit can produce a rapid, general shutdown of mRNA synthesis, demonstrating that Srb4 is required for expression of most protein-encoding genes (58). Because essentially all of the Srb proteins are tightly associated with the holoenzyme in Saccharomyces cerevisiae cells, the Srb-containing holoenzyme likely functions in transcription initiation at most class II promoters in vivo.
To further investigate the role of Srb4 and the holoenzyme in transcriptional activation, we have isolated and characterized extragenic suppressors of the temperature-sensitive phenotype of a srb4-138 mutant. Srb4 normally has a positive role in transcription initiation, and the Srb4-138 mutation affects the function of the protein at the nonpermissive temperature (58). Suppressors of Srb4-138 must compensate for the reduced function of the mutant subunit and might therefore include mutations in other positive factors which increase their activity. The suppressors might also include mutations in negative factors which reduce their activity. Indeed, we have identified dominant and recessive suppressors of the temperature-sensitive phenotype of srb4-138 which occur in positive and negative regulators, respectively. The results described here support a model in which activation mediated by holoenzyme is repressed by general negative regulators associated with TATA box binding protein (TBP).
MATERIALS AND METHODS
Yeast manipulations.
Yeast strains and plasmids are listed in Table 1. Details of strain and plasmid constructions are available upon request. Yeast medium was prepared as described previously (57). Yeast transformations were done by a lithium acetate procedure (54). Plasmid shuffle techniques were performed as described previously (5) with 5-fluoro-orotic acid (5-FOA) as a selective agent against URA3 plasmids. Plasmids were recovered from yeast as described previously (20).
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| Z22 | MAT-a ura3-52 his3Δ200 leu2-3,112 |
| Z579 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [pCT127 (SRB4 LEU2 CEN)] |
| Z628 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [RY2882 (srb4-138 LEU2 CEN)] |
| Z804 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 ncb1-1 [RY2882 (srb4-138 LEU2 CEN)] |
| Z811 | MAT-α ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [RY7215 (srb4-138 URA3 CEN)] |
| Z828 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 ncb2-1 [RY2882 (srb4-138 LEU2 CEN)] |
| Z829 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 not1-10 [RY2882 (srb4-138 LEU2 CEN)] |
| Z830 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 not3-10 [RY2882 (srb4-138 LEU2 CEN)] |
| Z836 | MAT-α ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 [RY2882 (srb4-138 LEU2 CEN)] |
| Z837 | MAT-α ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 not1/URA3 [RY2882 (srb4-138 LEU2 CEN)] |
| Z838 | MAT-α ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 not3/URA3 [RY2882 (srb4-138 LEU2 CEN)] |
| Z847 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 SRB6-201 [RY2882 (srb4-138 LEU2 CEN)] |
| Z848 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 MED6-101 [RY2882 (srb4-138 LEU2 CEN)] |
| Z849 | MAT-a ura3-52 leu2-PET56 spt15Δ2 [YCp86 (SPT15 URA3 CEN)] |
| Z850 | MAT-a ura3-52 leu2-PET56 spt15Δ2 [RY7269 (SPT15 5′ FLAG tag LEU2 CEN)] |
| Z862 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 caf1-10 [RY2882 (srb4-138 LEU2 CEN)] |
| Z864 | MAT-a ura3-52 his3Δ200 leu2-3,112 srb4Δ2::HIS3 not5-10 [RY2882 (srb4-138 LEU2 CEN)] |
DNA methods.
DNA manipulations were performed as described previously (52). PCR amplifications were performed with Vent DNA polymerase (New England Biolabs) or Taq DNA polymerase (Perkin-Elmer) as described by the manufacturer.
Selection and analysis of srb4-138 suppressors.
Two-milliliter yeast extract-peptone-dextrose (YPD) cultures of the yeast strain Z628 were grown overnight at 30°C, plated at a density of 3 × 106 cells/plate, and placed at 36°C. Suppressors arose at a frequency of approximately one in 2 × 106 cells. One colony was picked from each plate, further colony purified, and subsequently retested for the ability to grow at 36°C.
To exclude intragenic revertants, the srb4-138 LEU2 plasmids were recovered from strains harboring suppressor mutations and transformed into Z811. Cells were streaked on 5-FOA to select against the URA3 version of srb4-138 and assayed for growth at 36°C on YPD, and those which grew were considered to have a suppressor mutation linked to the original plasmid-borne copy of srb4-138.
Dominant and recessive growth phenotypes were determined by mating the suppressors in the Z628 background to Z811 and assaying growth at 36°C on YPD. Diploids able to grow at 36°C contained a dominant suppressor. Diploids unable to grow at 36°C contained a recessive suppressor. To facilitate linkage analysis, the mating type of approximately half of the dominant suppressors and half of the recessive suppressors was switched by inducing expression of a plasmid-borne HO gene under the control of a galactose-inducible promoter.
Random spore analysis of the dominantly suppressive mutations was used to determine if two independent isolates were likely to contain mutations in the same gene. Haploids, each containing the srb4-138 mutation and an independently isolated suppressor mutation, were mated to each other to form diploids. These diploids were sporulated on plates, and a small quantity of spores was scraped off and shaken overnight at 30°C in 0.5 ml of 30 mM β-mercaptoethanol–100 ng of Zymolase 100 T (ICN) per ml. A total of 0.5 ml of 1.5% Nonidet P-40 and 0.4 g of glass beads were added, and the mixture was incubated on ice for 15 min. The suspension was then vortexed for 3 min, incubated on ice for 5 min, and vortexed for 2 min, and the glass beads were allowed to settle for 10 min at room temperature. The supernatant was removed and spun for 2 min, the pellet was washed once in water and then resuspended in water, and a portion was plated onto YPD. Approximately 50 of the haploid offspring were assayed for their ability to grow at 36°C. If all haploids were able to grow at 36°C, then the two suppressor isolates were assumed to contain mutations in the same gene.
Dominantly suppressive mutations were assayed for the ability to bypass the requirement for Srb4. Strains harboring dominant suppressors and carrying a LEU2 plasmid with srb4-138 were transformed with a URA3 version of srb4-138. Transformants were grown in synthetic complete Ura− Leu+ medium to permit loss of the LEU2 plasmid. The resultant strains were streaked on 5-FOA to select against the URA3-containing plasmid. Cells harboring dominant mutations could not survive on 5-FOA, indicating that there was still a requirement for srb4-138 even in the context of MED6-101 or SRB6-201.
Genetic complementation of the recessive alleles involved mating haploids, each containing the srb4-138 mutation and an independently isolated suppressor mutation, to form diploids and assessing the ability of these diploids to grow at 36°C. Diploids able to grow at 36°C were assumed to contain suppressor mutations in the same gene. Genomic clones of each complementation group were used to confirm the identity of each member of the complementation group and to identify additional members.
Cloning of dominant suppressors of srb4-138.
Genomic DNA clones containing MED6-101 and SRB6-201 were isolated by taking advantage of their ability to dominantly suppress the srb4-138 temperature-sensitive phenotype. Genomic DNA was isolated from strains containing the dominant suppressor alleles of MED6 and SRB6 (Z848 and Z847, respectively). Libraries were constructed in a yeast centromeric plasmid containing the URA3 gene as a selectable marker (57). These libraries were transformed into yeast cells containing srb4-138, and genomic clones were isolated from Ura+ transformants able to grow at 36°C. When necessary, the mutant genes were further subcloned.
Complementation analysis.
Complementation groups containing mutant alleles of NCB2, NOT1, NOT3, NOT5, and CAF1 were identified by transforming Z828 with a pCT3 plasmid containing wild-type NCB2 (pRY7212), Z829 with a YCP50 plasmid containing wild-type NOT1 (gift of M. Collart), Z830 with a pRS316 plasmid containing wild-type NOT3 (gift of M. Collart), Z864 with a pRS316 plasmid containing wild-type NOT5, and Z862 with a pRS316 plasmid containing wild-type CAF1 (RY7288). The resulting strains no longer grew at the nonpermissive temperatures, indicating that the suppression phenotype was reversed by the wild-type NCB2, NOT, and CAF1 genes. Confirmation that these represented the suppressor-containing genes was obtained through linkage analysis (NOT1 and NOT3) and gap repair (NCB2, NOT5, and CAF1).
Genetic linkage analysis.
The identities of not1 and not3 alleles as suppressors of srb4-138 were confirmed by genetic linkage analysis. The URA3 gene was integrated next to the NOT1 gene in Z836 with SacI-digested pES183 (gift of E. Shuster). The resulting strain, Z837, was mated with Z829. The resulting diploid strain was sporulated, and 20 tetrads were dissected. Analysis of the resulting spores showed that the temperature-sensitive phenotype always cosegregated with the Ura+ phenotype, indicating that the suppressor allele was tightly linked to the NOT1 gene. For NOT3, the URA3 gene was integrated next to the NOT3 gene in Z836 with EagI-digested pRS306 with NOT3 (gift of M. Collart). The resulting strain, Z838, was mated with Z830. The resulting diploid strain was sporulated, and 20 tetrads were dissected. Analysis of the resulting spores showed that the temperature-sensitive phenotype always cosegregated with the Ura+ phenotype, indicating that the suppressor allele was tightly linked to the NOT3 gene.
Sequence analysis.
Suppressors of the temperature-sensitive phenotype of srb4-138 were recovered by a plasmid gap repair technique (42). Gap-repaired plasmids carrying suppressor alleles of MED6, SRB6, NCB2, NOT5, and CAF1 were sequenced (Research Genetics). Suppressor alleles of NOT1 and NOT3 were obtained by PCR of genomic DNA from strains Z829 and Z830, respectively. PCR products were directly sequenced by Research Genetics.
Expression of recombinant Med6.
The MED6 open reading frame was cloned into baculoviral transfer vectors by PCR amplification of the gene with the plasmid pET-MED6 (31) and oligonucleotides 5′-GGAAGATCTATGAACGTGACACCGTTGGAT-3′ and 5′-TGCTCTAGATCATATGTAGTTTGGGGTGGA-3′. Recombinant baculoviruses were generated and used to infect Sf21 insect cells. Insect cell extracts were prepared as described previously (26).
Immunoprecipitation of Srb4, Med6, and Srb6.
Coimmunoprecipitation experiments were performed to test interactions of Med6 with various Srb proteins. An insect cell extract containing FLAG epitope-tagged Med6 or Srb4 was incubated with an extract containing an equimolar amount of untagged, recombinant Srb4, Srb6, or Med6 for 3 h on ice. Controls included the use of ovalbumin and the use of Med6 or Srb4 lacking FLAG epitope in the respective reactions. The anti-FLAG M2 antibody-coupled agarose beads (Eastman Kodak), equilibrated in the buffer MTB (19), were added to the reaction mixtures and incubated for 3 h at 4°C with constant agitation. Beads were precipitated and washed extensively with MTB. Proteins in the pellet were eluted by being boiled in sample buffer and analyzed by Western blotting. For the experiment shown in Fig. 4D, insect cell extracts containing those five recombinant proteins were prepared by coinfecting the cells with the recombinant baculoviruses at a multiplicity of infection of 5 to 10. Coimmunoprecipitations were performed as described above with anti-FLAG M2 antibody.
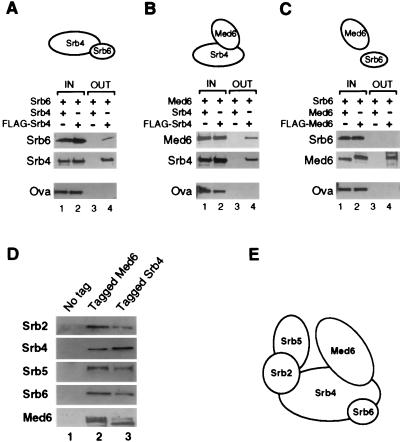
FIG. 4.
Med6 and Srb6 associate with Srb4. (A to C) Pairwise interactions of Med6 with Srb proteins. An insect cell extract containing one recombinant protein was incubated with an extract containing equimolar amounts of another recombinant protein which lacked (lanes 1 and 3) or contained (lanes 2 and 4) the FLAG epitope tag. Ovalbumin (Ova) was added to each reaction mixture to serve as a control for specific immunoprecipitation. The epitope-tagged Med6 or Srb4 and bound proteins were immunoprecipitated with anti-FLAG antibody. Fractions (1/10) of the load (IN) and all of the pellets (OUT) were analyzed by Western blotting with specific antibodies. A schematic interpretation of the binary interactions is presented at the top of each panel. (D) Insect cell extracts containing Med6 and Srb proteins were subjected to coimmunoprecipitation with anti-FLAG antibody. In the control reaction (lane 1, no tag), no tagged recombinant was included. In the other reactions, either Med6 (lane 2, tagged Med6) or Srb4 (lane 3, tagged Srb4) contained FLAG epitope tag. (E) A model depicting interactions between Med6 and dominant Srb proteins. This model assumes the stoichiometric association of the five proteins.
Antibody reagents.
A portion of Not1 (amino acids 1266 to 1442) was purified as a fusion to glutathione S-transferase (GST) from Escherichia coli DH5α according to previously published methods (56). The purified fusion protein was injected into rabbits to raise polyclonal antisera. Anti-polymerase II Western blotting analyses were performed with the mouse monoclonal antibody 8WG16. All other Western blot analyses were performed with rabbit polyclonal antisera. Anti-Spt3 antibody was the kind gift of J. Madison and F. Winston. Anti-TAFII90 antibody was the kind gift of J. Reese and M. Green.
GST-TBP affinity chromatography.
TBP affinity chromatography was performed as described previously (50) with the following modifications. To make whole-cell extract, yeast strain BJ926 was grown to an optical density of 3 in YPD at 30°C, harvested after being washed in 150 mM Tris acetate (pH 7.9)–50 mM potassium acetate, and stored at −80°C. Thawed cell pellet (130 g) was resuspended in 68 ml of 3× lysis buffer (450 mM Tris acetate [pH 7.9], 30% glycerol, 15 mM EDTA, 15 mM EGTA, 30 mM sodium fluoride, 1.8 mM sodium vanadate, 30 μM antipain-HCl, 15 mM benzamidine, 3 μg of aprotinin per ml, 3 μg of leupeptin per ml, 3 μg of pepstatin per ml, 0.25 mM phenylmethylsulfonyl fluoride [PMSF], 15 μM chymostatin). Cells were disrupted by bead beating for 20 cycles of 30 s of beating followed by 30 s of cooling in a stainless steel bead beater filled with 200 ml of 0.4- to 0.6-μm glass beads washed in 1× lysis buffer. After beating, dithiothreitol (DTT) and Na2S2O5 were added to 0.5 and 0.1 mM, respectively. The crude extract was centrifuged for 20 min at 10,000 rpm in a Sorvall GSA rotor. A one-ninth volume of 3 M (NH4)2SO4 (pH 7.9) was added slowly, and the mixture was stirred gently for 20 min and degassed. Ten percent polymin-P (1/100 volume) was added dropwise, and the extract was stirred gently for 20 min and degassed. The extract was centrifuged for 90 min at 42,000 rpm in a Ti 45 rotor (Beckman), and the supernatant (180 ml; 36 mg/ml) was frozen and stored at −80°C. Prior to use, the extract was thawed and dialyzed against buffer T(100) until the conductivity was equivalent to that of buffer T(150). Buffer T consists of 20 mM HEPES-KOH (pH 7.6); 10 mM magnesium acetate; 5 mM EGTA; 5 mM DTT; 20% glycerol; 0.5 μg each of leupeptin, pepstatin A, aprotinin, antipain-HCl, chymostatin, and bestatin per ml; 2 mM benzamidine-HCl; 0.5 mM PMSF; and potassium acetate added to the millimolar concentrations indicated in parentheses.
GST-yeast TBP and GST columns were prepared as described previously (50). Yeast whole-cell extract (∼100 mg) was diluted in buffer T(150) to a total volume of 30 ml. Fifteen milliliters of dilute extract was incubated with 1.0 ml of GST-yeast TBP or GST agarose at 4°C with rotation. Resin was collected by gentle centrifugation (1,000 × g, 1 min) and washed with 10 column volumes of buffer T(150). Bound proteins were eluted with 2 M potassium chloride. Peak fractions were pooled and dialyzed into buffer T(100).
Construction of FLAG-tagged TBP-containing yeast strain.
Plasmid RY7269 was constructed by PCR amplification with two sets of primers. The first set of primers generated a 1-kb fragment that incorporated the FLAG epitope behind the initial ATG of the open reading frame. This fragment was digested at the 5′ end with XhoI and at the 3′ end with Psp1406I (an endogenous site at nucleotide 14 of the TBP open reading frame). The second set of primers generated a 2-kb fragment including the TBP open reading frame and approximately 1 kb of 3′ downstream sequence. This fragment was digested at the 5′ end with Psp1406I and at the 3′ end with XmaI. The two digested fragments were then ligated into the LEU2 vector pRS315 digested with XhoI and XmaI. The resulting construct was transformed into yeast strain BYΔ2 (10), which has a genomic deletion of TBP covered by a wild-type copy of TBP on a URA3 plasmid. Selection against the URA3 plasmid with 5-FOA generated strain Z850 and confirmed that the tagged version of TBP was fully functional and able to complement the TBP deletion.
Immunoprecipitation of FLAG-TBP.
A crude fraction of yeast extract was prepared from yeast strains Z849 and Z850. Briefly, whole-cell extract was prepared as described previously (27). Whole-cell extract (50 mg) was diluted in buffer A(150) and passed over a 2-ml Bio-Rex 70 column equilibrated in buffer A(150). Buffer A consists of 20 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF, 1 mM benzamidine, and protease inhibitors as described above. The number in parentheses indicates the millimolar concentration of potassium acetate. The columns were washed with 20 column volumes of buffer BH(150) and eluted with 10 column volumes of buffer BH(300) and buffer BH(600). Buffer BH consists of 20 mM HEPES-KOH (pH 7.6), 1 mM EDTA, 10% glycerol, 0.5 mM PMSF, 1 mM benzamidine, and protease inhibitors as described above. The number in parentheses indicates the millimolar concentration of potassium acetate. Peak fractions of the BH(600) eluate were pooled and used for immunoprecipitations.
Approximately 100 μg of the BH(600) fraction was diluted with 4 volumes of buffer BH(0). Samples (approximately 1.5 ml each) were first cleared by incubation with 20 μl of anti-FLAG M1 affinity gel and then immunoprecipitated with 20 μl of anti-FLAG M2 affinity gel for 2 h at 4°C with rotation. Beads were collected by centrifugation at 8,000 × g and washed five times with BH buffer supplemented with various concentrations of potassium acetate. Bound proteins were eluted by being boiled for 1 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer without DTT. After centrifugation, additional sample buffer with DTT was added to the supernatant. Typically, 1/100 of the load and flowthrough and 1/5 of the eluate were loaded for Western blot analysis.
RESULTS
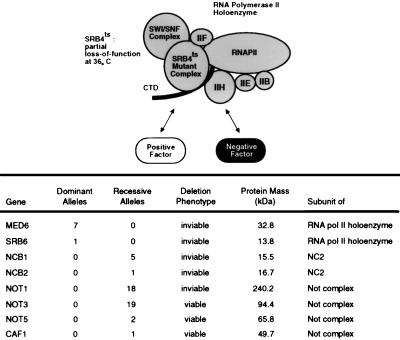
Fifty-four isolates were obtained in a genetic selection for suppressors of the temperature-sensitive phenotype of cells harboring srb4-138. Genetic analysis revealed that all 54 extragenic suppressor mutations occurred in eight genes (Fig. 1). Eight of the isolates were dominant suppressor mutations which occurred in two genes that encode components of the Srb-Mediator complex. The remaining 46 isolates were recessive suppressor mutations which occurred in six genes whose products are subunits of negative regulators.
FIG. 1.
Genetic suppressors of the temperature-sensitive Srb4-138 mutant RNA polymerase II holoenzyme. Conditional defects in the essential Srb4 subunit of the holoenzyme might be overcome by compensatory gain-of-function mutations in a general positive regulator or loss-of-function mutations in a global negative regulator. RNAPII and RNA polII, RNA polymerase II; CTD, C-terminal domain; ts, temperature sensitive.
MED6 and SRB6 alleles are dominant suppressors of srb4-138.
Genetic analysis of the 54 genetic suppressors indicated that eight had a dominant suppressor phenotype (Fig. 1). Linkage analysis of the eight dominant suppressors revealed that they fall into two groups. Group A consists of seven isolates, and group B consists of one isolate. Figure 2 shows the suppressor phenotype of one of the isolates from each group. To identify the gene represented in group A, a genomic DNA library was constructed from one of the dominant suppressor isolates, cells containing the srb4-138 temperature-sensitive mutation were transformed with the library, and recombinant DNA clones that suppressed the temperature-sensitive phenotype were isolated. The minimal fragment of genomic DNA with the suppressor phenotype was identified and sequenced and was found to encode a mutant form of MED6 (MED6-101). The MED6 dominant mutation is a G-to-T substitution at nucleotide 454, converting amino acid 152 from aspartic acid to tyrosine. To confirm that the suppressor mutation occurs in the MED6 gene, a gap repair method was used with plasmids lacking the MED6 open reading frame but retaining flanking DNA. Plasmids gap repaired from the suppressor strain conferred suppression while plasmids repaired from wild-type strains did not, confirming that the suppressor mutation occurs in MED6.
FIG. 2.
Dominant suppressors of srb4 temperature-sensitive mutant. Dominant mutations in MED6 or SRB6 suppress srb4-138. Shown are growth phenotypes of SRB4 cells and cells containing the srb4-138 mutation, either alone or with the MED6-101 or SRB6-201 mutation. Cells were spotted on YPD medium and incubated at 30 and 36°C for 2 days.
It is possible that the dominant suppressor mutations in MED6 eliminated a requirement for Srb4 function. To investigate this possibility, the MED6-101 allele was introduced into a strain with a genomic deletion of SRB4 covered by a CEN plasmid containing SRB4 and URA3, and the cells were plated on 5-FOA medium to select for cells that lost the plasmid (Fig. 3). No cells could be recovered on 5-FOA medium, indicating that some Srb4 function is essential for cell survival, even in the presence of MED6-101. These data suggest that Srb4-138 protein retains some function, even at temperatures that do not permit cell growth.
FIG. 3.
Dominant mutations in MED6 or SRB6 do not bypass a requirement for srb4-138. Shown are growth phenotypes of cells containing plasmid-borne copies of SRB4 or srb4-138, either alone or with the MED6-101 or SRB6-201 mutation. Cells were streaked on 5-FOA to select against URA3 versions of SRB4 or srb4-138 and incubated at 30°C for 2 days. Relevant genotypes are described as follows: 1, srb4Δ2::HIS3 [pCT127 (SRB4 LEU2 CEN)]; 2, srb4Δ2::HIS3 [pCT15 (SRB4 URA3 CEN)]; 3, srb4Δ2::HIS3 [pCT181 (srb4-138 LEU2 CEN)]; 4, srb4Δ2::HIS3 [pEG39 (srb4-138 URA3 CEN)]; 5, srb4Δ2::HIS3 MED6-101 [pEG39 (srb4-138 URA3 CEN)]; 6, srb4Δ2::HIS3 SRB6-201 [pEG39 (srb4-138 URA3 CEN)].
To identify the suppressor in group B, a genomic DNA library was prepared from cells containing this dominant mutation and transformed into cells containing the srb4-138 temperature-sensitive mutation. DNA clones that suppressed the temperature-sensitive phenotype were recovered and sequenced. The minimal fragment sufficient for the suppressor phenotype was found to contain a mutant allele of SRB6. The SRB6-201 dominant mutation is an A-to-C transversion at nucleotide 175, converting amino acid 59 from asparagine to histidine. Gap repair analysis confirmed that the suppressor mutation occurs in the SRB6 gene. As with the MED6 suppressor alleles, the dominant mutation in SRB6 was unable to bypass the requirement for some level of Srb4 function, as cells harboring SRB6-201 did not restore viability to cells with an SRB4 deletion (Fig. 3).
Med6 and Srb6 associate with Srb4.
Srb4, Med6, and Srb6 are components of the Srb-Mediator complex (19, 24, 31, 39). Srb4 and Srb6 are involved in similar functions in vivo (57, 58) and can form a complex in vitro (25). The observation that dominant mutations in MED6 can compensate for a partial loss of Srb4 function might reflect a physical interaction between Srb4 and Med6. We examined pairwise interactions between recombinant Srb4, Med6, and Srb6 proteins expressed in a baculovirus system (Fig. 4). Extracts containing FLAG epitope-tagged Srb4 or Med6 were incubated with extracts containing an equimolar amount of untagged Med6 or Srb6 protein. The epitope-tagged subunit was immunoprecipitated, and the pellet was analyzed by Western blotting for the untagged protein. Untagged protein was used in parallel reactions to control for specific immunopurification, and ovalbumin was added to each reaction mixture to control for nonspecific aggregation. The results confirmed previous evidence that Srb4 and Srb6 can form a complex (Fig. 4A) (25) and revealed that Srb4 and Med6 bind to one another in vitro (Fig. 4B). There were no detectable interactions between Med6 and Srb6 (Fig. 4C).
Srb2, Srb4, Srb5, and Srb6 form a complex in vitro (25). Figure 4D shows that Med6 binds to this Srb subcomplex. Extracts containing Srb2, Srb4, Srb5, Srb6, and Med6 proteins were incubated and immunoprecipitated with antibodies against epitope-tagged Srb4 or Med6. In both cases, all five proteins were coimmunoprecipitated (Fig. 4D, lanes 2 and 3). The genetic and biochemical data are consistent with the model for an Srb-Mediator subcomplex shown in Fig. 4E.
NCB1 and NCB2 loss-of-function mutations compensate for Srb4 defect.
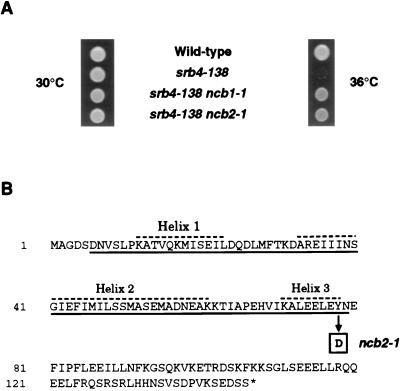
In addition to the eight dominant suppressors identified as alleles of MED6 and SRB6, 46 suppressors were characterized as recessive suppressors of srb4-138. We recently reported the identification of one of the recessive suppressors as NCB1 (BUR6), which encodes the large subunit of NC2 (13). Since NC2 is composed of two subunits, we tested whether any of the other complementation groups involved the NCB2 gene, which encodes the other subunit of this general negative regulatory factor. An isolate from each complementation group was transformed with a plasmid carrying a wild-type NCB2 gene, and the transformants were screened for the loss of the recessive suppressor phenotype. One group, consisting of a single isolate, showed this loss of suppression following transformation with the wild-type NCB2 gene. Subsequent gap repair analysis confirmed that a mutation in NCB2 was responsible for suppression. Thus, mutations in either subunit of yeast NC2 can cause suppression of srb4-138 (Fig. 5A). The suppressor allele (ncb2-1) was sequenced and found to affect the histone fold motif, which is important for the stable interaction of the two NC2 subunits (2, 4, 15, 38). The suppressor mutation is a T-to-G substitution at nucleotide 232, converting a highly conserved tyrosine to aspartic acid within the histone fold motif (Fig. 5B). This defect is similar to that identified for the suppressor mutation in the other subunit of NC2; the ncb1-1 mutation truncates the histone fold motif of this protein (13). Like NCB1, the NCB2 gene is essential (13, 23), and so the missense mutation must cause a partial functional defect in the small NC2 subunit.
FIG. 5.
Recessive mutations in either subunit of NC2 suppress srb4-138. (A) NC2 suppressors of the srb4 temperature-sensitive mutant. Shown are growth phenotypes of SRB4 cells and cells containing the srb4-138 mutation, either alone or in conjunction with the ncb1-1 or ncb2-1 mutation. Cells were spotted on YPD medium and incubated at 30 and 36°C for 2 days. (B) Sequence of NC2β. The suppressing mutation is indicated in boldface. The ncb2-1 recessive mutation is a T-to-G substitution at nucleotide 232, converting amino acid 78 from tyrosine to aspartic acid. The histone fold motif is underlined, and the helices of the motif are indicated by dashed lines.
NOT1, NOT3, NOT5, and CAF1 loss-of-function mutations compensate for Srb4 defect.
Since two of the recessive complementation groups define genes encoding a known negative regulator of transcription, we expected that additional negative regulators might be represented among the other complementation groups. Previous genetic and biochemical studies indicated that MOT1 (3, 11, 59), the NOT genes (8, 9, 40), and histones (18, 32, 49, 60) (reviewed in references 17, 62, and 63) all negatively regulate transcription. Consequently, representative isolates of the unidentified complementation groups were transformed with wild-type MOT1, NOT1, NOT2, NOT3, NOT4, NOT5, HTA1-HTB1, or HHT1-HHF1 and tested for viability at the restrictive temperature. Based on the loss of suppression of the srb4-138 phenotype when transformed with a copy of the wild-type gene, three complementation groups were found to represent recessive suppressor alleles of NOT1, NOT3, and NOT5 (Fig. 6). The identities of these suppressors were confirmed by linkage analysis or gap repair analysis. In addition, a disruption of the NOT3 gene also suppressed srb4-138, indicating that a loss-of-function mutation in NOT3 could alleviate the holoenzyme defect (data not shown). The suppressor alleles of these genes were sequenced, and the recessive mutations were identified. The not1-10 suppressor allele is a G-to-A substitution at nucleotide 5828 converting amino acid 1943 from glycine to aspartic acid. The not3-10 suppressor allele is a 19-bp duplication of nucleotides 1620 to 1638 that results in a frameshift and truncation of the protein. The not5-10 suppressor allele is a C-to-G mutation at nucleotide 1443 converting amino acid 481 from phenylalanine to leucine. As described for the previously identified ncb1-1 suppressor (13), suppressor alleles of NOT genes are able to rescue global transcriptional defects in poly(A) mRNA expression caused by srb4-138 at the restrictive temperature (data not shown).
FIG. 6.
Not complex suppressors of the srb4 temperature-sensitive mutant. Recessive mutations in NOT1, NOT3, NOT5, or CAF1 suppress srb4-138. Shown are growth phenotypes of SRB4 cells and cells containing the srb4-138 mutation, either alone or in conjunction with the not1-10, not3-10, not5-10, or caf1-10 mutation. Cells were spotted on YPD medium and incubated at 30 and 36°C for 2 days.
The Not proteins have recently been shown to associate with the Ccr4-Caf1 regulatory complex (35). Since three of the recessive complementation groups define NOT genes, we examined whether CCR4 or CAF1 was also represented among the recessive suppressors of srb4-138. The sole isolate of the last unidentified complementation group was transformed with wild-type CCR4 or CAF1 and tested for viability at the restrictive temperature. Based on the loss of suppression of the srb4-138 phenotype when transformed with a copy of the wild-type gene, this complementation group represented a recessive suppressor allele of CAF1 (Fig. 6). The identity of this suppressor was confirmed by gap repair analysis. The suppressor allele of this gene, caf1-10, was sequenced, and the recessive mutation was identified as an A-to-G substitution at nucleotide 739 that converts amino acid 247 from asparagine to aspartic acid. A disruption of CAF1 does not suppress the temperature-sensitive phenotype of srb4-138 (data not shown).
If the Not proteins are general negative regulators, then loss-of-function mutations in Not proteins might be expected to suppress defects due to at least some temperature-sensitive RNA polymerase II mutants. Indeed, in an independent selection for suppressors of temperature-sensitive mutations in the second largest subunit of RNA polymerase II, we have identified recessive suppressor mutations in NOT1 and NOT2 (29a).
NC2 and Not proteins associate with TBP.
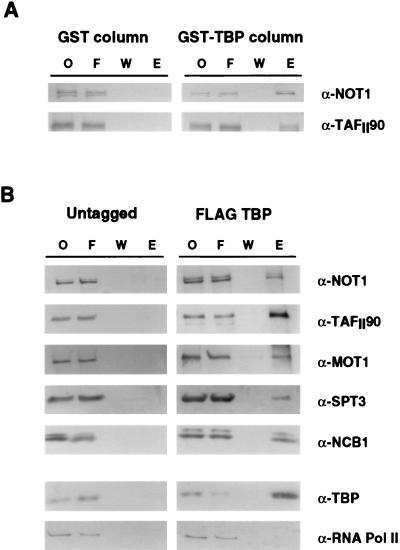
Med6 and Srb6 have previously been identified as components of the mediator subcomplex of RNA polymerase II holoenzyme (28, 31). Quantitative Western blot analysis revealed that NC2α, NC2β, Not1, and Not3 are not components of the holoenzyme (data not shown). Evidence that yeast NC2 binds TBP and represses transcription (13, 15, 16, 23) led us to investigate whether Not proteins also bind to TBP. Whole-cell extract was prepared and passed over a GST-TBP column, and various fractions were analyzed by Western blot analysis with anti-Not1 antibody. Under conditions in which TAFIIs are also retained, Not1 bound to the GST-TBP column (Fig. 7A).
FIG. 7.
TBP-associated proteins. (A) Western blot analysis of eluate from TBP affinity column eluates. Crude cell extract was passed over GST or GST-TBP columns. Bound proteins were eluted with 2 M KCl and probed with antibodies against Not1 and TafII90. Lanes indicated are onput (O), flowthrough (F), wash (W), and eluate (E). (B) Western blotting of immunoprecipitates from crude fractions of whole-cell extract derived from cells with (FLAG-TBP) or without (untagged) FLAG epitope-tagged TBP reveals that Not proteins interact, directly or indirectly, with TBP. Under these conditions, TBP also interacts with other proteins previously described as TBP-interacting factors, including TafII90, NC2, Spt3, and Mot1, but not RNA polymerase II. Lanes indicated are onput (O), flowthrough (F), wash (W), and eluate (E).
To confirm that Not’s interact with TBP, yeast TBP was epitope tagged at its N terminus with the FLAG epitope and immunoprecipitated from yeast cell extracts, and associated proteins were identified by Western blot analysis (Fig. 7B). TAFIIs, NC2, Mot1, and Spt3, each of which has previously been shown to bind TBP (3, 12, 13, 16, 45, 46, 50), were used as positive controls. Not1, TAFIIs, NC2, Mot1, and Spt3 were all found to coimmunopurify with TBP. In contrast, RNA polymerase II was not associated with the immunopurified TBP preparation.
DISCUSSION
Genetic selections can provide substantial new insights into the function of complex biological systems. Genetic and biochemical characterization of suppressors of RNA polymerase II mutations previously led us and others to the holoenzyme model. The isolation and characterization of eight genes found in a selection for suppressors of the srb4-138 allele provide additional insights into the holoenzyme components which are involved in transcription activation and the set of TBP regulators which appear to be general negative regulators of class II genes.
Functional interactions among holoenzyme subunits implicated in activation.
In principle, dominant suppressors of an srb4 temperature-sensitive mutant could reveal compensatory mutations in positive factors which are involved in class II gene expression. In fact, dominant mutations compensating for the srb4 mutation occurred in two genes whose products are also Srb-Mediator subunits, MED6 and SRB6. These two proteins are essential components of the holoenzyme and contribute to the response to activators in vivo and in vitro (19, 31, 57).
The functional interactions suggested by the genetic analysis are supported by physical interactions seen with recombinant Srb4, Srb6, and Med6. Srb4 interacts with both Med6 and Srb6 in vitro. Taken together, the genetic and biochemical results further refine our model for Srb subunit interactions within the holoenzyme (25) and extend it to incorporate Med6 (Fig. 4E).
General negative transcription factors associated with TBP.
Recessive mutations that suppress the srb4-138 defect occurred in the genes encoding both subunits of the negative regulator NC2. Genetic and biochemical evidence indicates that NC2 is a general negative regulator of transcription which is essential for yeast cell viability (13, 15, 16, 21–23, 37, 38, 47, 61, 64, 65). The protein represses transcription by binding to promoter-bound TBP and preventing the association of TFIIA and TFIIB during formation of the preinitiation apparatus (16, 22, 38).
Recessive suppressor mutations also occurred in genes encoding subunits of the Not complex. Previous experiments indicated that the Not protein complex can act as a negative regulator at several genes (8, 9). A recent report suggests that some components of this complex may have a positive role at certain genes, but it is not yet clear whether this role is direct (35). The evidence presented here indicates that Not proteins, like NC2, have a general negative regulatory function. Loss-of-function mutations in NOT1 can suppress defects in both SRB4 and RNA polymerase II subunit (RPB2) mutations. Similarly, loss-of-function mutations in NC2 suppress defects of both SRB4 and SRB6 mutations (13). The observation that mutations in NC2 or Not proteins can suppress defects in Srb and Rpb subunits of the RNA polymerase II holoenzyme indicates that NC2 and Not proteins contribute to a general level of transcriptional repression that must be overcome during transcription initiation by the holoenzyme.
While the mechanism of repression by NC2 involves TBP binding, the mechanism of repression by Not proteins has not been clear. We have found that Not1 associates with TBP, consistent with genetic evidence that Not mutations can relieve defects due to specific TBP mutations (7). Thus, Not proteins may be one of several factors, including NC2, that contribute to gene regulation by regulating TBP activity (30).
The balance between activation and repression.
Genetic analysis of suppressors of the srb4-138 mutation has revealed functional links between holoenzyme subunits involved in activation and two general negative regulators that associate with TBP. It is notable that recessive mutations in both NCB and NOT genes have previously been observed to compensate for defects in activation. The NOT genes were identified in a screen for suppressors of a defect in the GCN4 transcriptional activator (8, 9). A mutation in NCB1 (BUR6) can compensate for the loss of the upstream activating sequence in the SUC2 gene (47, 48). These results are consistent with the model that activators generally recruit holoenzymes in a manner that is dependent on Srb4, Srb6, and Med6 function and that NC2 and Not complexes generally inhibit transcription activation by this pathway.
ACKNOWLEDGMENTS
We thank P. Sharp, M. Green, V. Myer, and H. Madhani for advice and discussions. We thank F. Holstege, M. Collart, M. Green, Y.-J. Kim, J. Madison, J. Reese, K. Struhl, C. Wilson, and F. Winston for kind gifts of extracts, strains, plasmids, and antibodies. We thank A. S. Lee for technical assistance.
J.J.W. is a predoctoral fellow of the National Science Foundation. E.G.J. is a predoctoral fellow of the Howard Hughes Medical Institute. This work was supported by National Institutes of Health grants to R.A.Y.
REFERENCES
- 1.Allison L A, Ingles C J. Mutations in RNA polymerase II enhance or suppress mutations in GAL4. Proc Natl Acad Sci USA. 1989;86:2794–2798. doi: 10.1073/pnas.86.8.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arents G, Moudrianakis E N. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auble D T, Hansen K E, Mueller C G, Lane W S, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 4.Baxevanis A D, Landsman D. Histone and histone fold sequences and structures: a database. Nucleic Acids Res. 1997;25:272–273. doi: 10.1093/nar/25.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 6.Chao D M, Gadbois E L, Murray P J, Anderson S F, Sonu M S, Parvin J D, Young R A. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature. 1996;380:82–85. doi: 10.1038/380082a0. [DOI] [PubMed] [Google Scholar]
- 7.Collart M A. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol Cell Biol. 1996;16:6668–6676. doi: 10.1128/mcb.16.12.6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart M A, Struhl K. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 1993;12:177–186. doi: 10.1002/j.1460-2075.1993.tb05643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart M A, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. doi: 10.1101/gad.8.5.525. [DOI] [PubMed] [Google Scholar]
- 10.Cormack B P, Strubin M, Ponticelli A S, Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991;65:341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- 11.Davis J L, Kunisawa R, Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenmann D M, Arndt K M, Ricupero S L, Rooney J W, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- 13.Gadbois E L, Chao D M, Reese J C, Green M R, Young R A. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature. 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 15.Goppelt A, Meisterernst M. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 17.Grunstein M. Histone function in transcription. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 18.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 19.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Association of an activator with an RNA polymerase II holoenzyme. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman C S, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 21.Inostroza J A, Mermelstein F H, Ha I, Lane W S, Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992;70:477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Parvin J D, Shykind B M, Sharp P A. A negative cofactor containing Dr1/p19 modulates transcription with TFIIA in a promoter-specific fashion. J Biol Chem. 1996;271:18405–18412. doi: 10.1074/jbc.271.31.18405. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Na J G, Hampsey M, Reinberg D. The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA. 1997;94:820–825. doi: 10.1073/pnas.94.3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y J, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 25.Koh S S, Ansari A Z, Ptashne M, Young R A. An activator target in the RNA polymerase II holoenzyme. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 26.Koh S S, Hengartner C J, Young R A. Baculoviral transfer vectors for expression of flag(R) fusion proteins in insect cells. BioTechniques. 1997;23:622–627. doi: 10.2144/97234bm14. [DOI] [PubMed] [Google Scholar]
- 27.Koleske A J, Chao D M, Young R A. Purification of yeast RNA polymerase II holoenzymes. Methods Enzymol. 1996;273:176–184. doi: 10.1016/s0076-6879(96)73018-5. [DOI] [PubMed] [Google Scholar]
- 28.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 29.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 29a.Lee, T. Unpublished data.
- 30.Lee T, Young R A. Regulation of gene expression by TBP-associated proteins. Genes Dev. 1998;12:1398–1408. doi: 10.1101/gad.12.10.1398. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y C, Min S, Gim B S, Kim Y J. A transcriptional mediator protein that is required for activation of many RNA polymerase II promoters and is conserved from yeast to humans. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenfant F, Mann R K, Thomsen B, Ling X, Grunstein M. All four core histone N-termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 33.Liao S M, Taylor I C, Kingston R E, Young R A. RNA polymerase II carboxy-terminal domain contributes to the response to multiple acidic activators in vitro. Genes Dev. 1991;5:2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- 34.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 35.Liu H Y, Badarinarayana V, Audino D C, Rappsilber J, Mann M, Denis C L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998;17:1096–1106. doi: 10.1093/emboj/17.4.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 37.Meisterernst M, Roeder R G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991;67:557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- 38.Mermelstein F, Yeung K, Cao J, Inostroza J A, Erdjument-Bromage H, Eagelson K, Landsman D, Levitt P, Tempst P, Reinberg D. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 1996;10:1033–1048. doi: 10.1101/gad.10.8.1033. [DOI] [PubMed] [Google Scholar]
- 39.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberholzer U, Collart M A. Characterization of NOT5 that encodes a new component of the Not protein complex. Gene. 1998;207:61–69. doi: 10.1016/s0378-1119(97)00605-7. [DOI] [PubMed] [Google Scholar]
- 41.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 42.Orr-Weaver T L, Szostak J W, Rothstein R J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- 43.Ossipow V, Tassan J P, Nigg E A, Schibler U. A mammalian RNA polymerase II holoenzyme containing all components required for promoter-specific transcription. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 44.Pan G, Aso T, Greenblatt J. Interaction of elongation factors TFIIS and elongin A with a human RNA polymerase II holoenzyme capable of promoter-specific initiation and responsive to transcriptional activators. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 45.Poon D, Campbell A M, Bai Y, Weil P A. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 46.Poon D, Weil P A. Immunopurification of yeast TATA-binding protein and associated factors. J Biol Chem. 1993;268:15325–15328. [PubMed] [Google Scholar]
- 47.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recht J, Dunn B, Raff A, Osley M A. Functional analysis of histones H2A and H2B in transcriptional repression in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2545–2553. doi: 10.1128/mcb.16.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reese J C, Apone L, Walker S S, Griffin L A, Green M R. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994;371:523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- 51.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 54.Schiestl R H, Gietz R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 55.Shi X, Chang M, Wolf A J, Chang C H, Frazer-Abel A A, Wade P A, Burton Z F, Jaehning J A. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 57.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 58.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wade P A, Jaehning J A. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol Cell Biol. 1996;16:1641–1648. doi: 10.1128/mcb.16.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wan J S, Mann R K, Grunstein M. Yeast histone H3 and H4 N termini function through different GAL1 regulatory elements to repress and activate transcription. Proc Natl Acad Sci USA. 1995;92:5664–5668. doi: 10.1073/pnas.92.12.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White R J, Khoo B C, Inostroza J A, Reinberg D, Jackson S P. Differential regulation of RNA polymerases I, II, and III by the TBP-binding repressor Dr1. Science. 1994;266:448–450. doi: 10.1126/science.7939686. [DOI] [PubMed] [Google Scholar]
- 62.Wolffe A P. Transcription: in tune with the histones. Cell. 1994;77:13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 63.Wolffe A P. Histones, nucleosomes and the roles of chromatin structure in transcriptional control. Biochem Soc Trans. 1997;25:354–358. doi: 10.1042/bst0250354. [DOI] [PubMed] [Google Scholar]
- 64.Yeung K, Kim S, Reinberg D. Functional dissection of a human Dr1-DRAP1 repressor complex. Mol Cell Biol. 1997;17:36–45. doi: 10.1128/mcb.17.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeung K C, Inostroza J A, Mermelstein F H, Kannabiran C, Reinberg D. Structure-function analysis of the TBP-binding protein Dr1 reveals a mechanism for repression of class II gene transcription. Genes Dev. 1994;8:2097–2109. doi: 10.1101/gad.8.17.2097. [DOI] [PubMed] [Google Scholar]