Abstract
Spatial transcriptomics (ST) technologies are rapidly becoming the extension of single-cell RNA sequencing (scRNAseq), holding the potential of profiling gene expression at a single-cell resolution while maintaining cellular compositions within a tissue. Having both expression profiles and tissue organization enables researchers to better understand cellular interactions and heterogeneity, providing insight into complex biological processes that would not be possible with traditional sequencing technologies. Data generated by ST technologies are inherently noisy, high-dimensional, sparse, and multi-modal (including histological images, count matrices, etc.), thus requiring specialized computational tools for accurate and robust analysis. However, many ST studies currently utilize traditional scRNAseq tools, which are inadequate for analyzing complex ST datasets. On the other hand, many of the existing ST-specific methods are built upon traditional statistical or machine learning frameworks, which have shown to be sub-optimal in many applications due to the scale, multi-modality, and limitations of spatially resolved data (such as spatial resolution, sensitivity, and gene coverage). Given these intricacies, researchers have developed deep learning (DL)-based models to alleviate ST-specific challenges. These methods include new state-of-the-art models in alignment, spatial reconstruction, and spatial clustering, among others. However, DL models for ST analysis are nascent and remain largely underexplored. In this review, we provide an overview of existing state-of-the-art tools for analyzing spatially resolved transcriptomics while delving deeper into the DL-based approaches. We discuss the new frontiers and the open questions in this field and highlight domains in which we anticipate transformational DL applications.
I. INTRODUCTION
Although multicellular organisms contain a common genome within their cells, the morphology and gene expression patterns of cells are largely distinct and dynamic. These differences arise from internal gene regulatory systems and external environmental signals. Cells proliferate, differentiate, and function in tissues while sending and receiving signals from their surroundings. These environmental factors cause cell fate to be highly dependent on the environment in which it exists. Therefore, monitoring a cell's behavior in the residing tissue is crucial to understanding cell function, as well as its past and future fate.1
Advancements in single-cell sequencing have transformed the genomics and bioinformatics fields. The advent of single-cell RNA sequencing (scRNAseq) has enabled researchers to profile gene expression levels of various tissues and organs, allowing them to create comprehensive atlases in different species.2–6 Moreover, scRNAseq enables the detection of distinct subpopulations present within a tissue, which has been paramount in discovering new biological processes, the inner workings of diseases, and effectiveness of treatments.7–14 However, high-throughput sequencing of solid tissues requires tissue dissociation, resulting in the loss of spatial information.15,16 To fully understand cellular interactions, data on tissue morphology and spatial information are needed, which scRNAseq alone cannot provide. The placement of cells within a tissue is crucial from the developmental stages (e.g., asymmetric cell fate of mother and daughter cells17) and beyond cell differentiation (such as cellular functions, response to stimuli, and tissue homeostasis18). These limitations would be alleviated by technologies that could preserve spatial information while measuring gene expression at the single-cell level.
Spatial transcriptomics (ST) provide an unbiased view of tissue organization crucial in understanding cell fate, delineating heterogeneity, and other applications.19 However, many current ST technologies suffer from lower sensitivities as compared to scRNAseq while lacking the single-cell resolution that scRNAseq provides.20 Targeted in situ technologies have tried to solve the issue of resolution and sensitivity but are limited in gene throughput and often require a priori knowledge of target genes.20 More specifically, in situ technologies [such as in situ sequencing (ISS),21 single-molecule fluorescence in situ hybridization (smFISH),22–24 targeted expansion sequencing,25 cyclic-ouroboros smFISH (osmFISH),26 multiplexed error-robust fluorescence in situ hybridization (MERFISH),27 sequential FISH (seqFISH+),28 and spatially resolved transcript amplicon readout mapping (STARmap)29] are typically limited to pre-selected genes that are on the order of hundreds, with the accuracy potentially dropping as more probes are added.29 We will refer to these methods as image-based techniques.
On the other hand, next generation sequencing (NGS)-based technologies [such as 10× Genomics' Visium and its predecessor,30,31 Slide-Seq,32 and high-definition spatial transcriptomics (HDST33)] barcode entire transcriptomes but have limited capture rates, and resolutions that are larger than a single cell34 (50–100 μm for Visium and 10 μm for Slide-Seq). Moreover, unlike image-based technologies, NGS-based methods allow for unbiased profiling of large tissue sections without necessitating a set of target genes.35,36 However, NGS-based technologies do not have a single-cell resolution, requiring cellular features to be inferred or related to the histological scale using computational approaches. Many current algorithms use traditional statistical or medical image processing frameworks that require human supervision,34,37,38 which is not ideal for large-scale analyses. Additionally, many algorithms are not generalizable across different sequencing platforms, which limits their utility and restricts multi-omics integration efforts.
Deep learning (DL) methods can use raw data to extract useful representations (or information) needed for performing a task, such as classification or detection.39 This quality makes this class of Machine Learning (ML) algorithms ideal for applications where the available data are large, higher-dimensional, and noisy, such as single-cell omics. DL models have been extensively used in scRNAseq studies (e.g., preprocessing,40,41 clustering,42,43 cell-type identification,44,45 and data augmentation46,47) and have shown to significantly improve upon traditional methods,10 suggesting the potential of such methods in ST analysis. Moreover, DL models can leverage multiple data sources, such as images and text data, to learn a set of tasks.48 Given that spatially resolved transcriptomics are inherently multi-modal (i.e., they consist of images and gene expression count data) and that downstream analysis consists of multiple tasks (e.g., clustering and cell-type detection), researchers have sought to develop ST-specific DL algorithms.
Spatially resolved transcriptomics have been utilized to unravel complex biological processes in many diseases (e.g., COVID-19,49,50 arthritis,51,52 cancer,31,33,53–55 Alzheimer's,56 diabetes,57,58 etc.). Continuous improvements and commercialization of ST technologies (such as 10×'s Visium) are resulting in wider use across individual labs. Therefore, scalable and platform-agnostic computational approaches are needed for the accurate and robust analysis of ST data. So far, DL methods have shown promising results in handling the scale and multi-modality of spatially resolved transcriptomics; however, DL-based models in this space remain nascent. Similar to scRNAseq analysis, we anticipate a suite of DL models to be developed in the near future to address many of the pressing challenges in the spatial omics field. This review aims to provide an overview of the current state-of-the-art (SOTA) DL models developed for ST analysis. Due to the potential and accessibility of NGS-based ST technologies, we primarily focus on methods and techniques developed for these technologies.
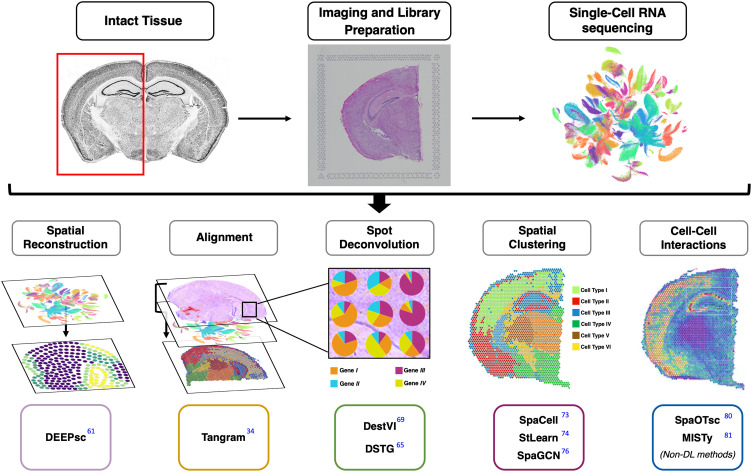
The remainder of this manuscript is organized as follows: We provide an overview of common scRNAseq and ST technologies in Sec. II, followed by a general description of common DL architectures used for ST analysis in Sec. III. Section IV is dedicated to the current DL methods developed for analyzing spatially resolved transcriptomics. In Sec. V, we conclude by discussing our outlook on the current challenges and future research directions in the ST domain. We provide an illustration of the main methods reviewed in this paper in Fig. 1 and provide the reader with a list of current SOTA methods for ST analysis in Table I. Given the pace of advancements in this field, the authors have compiled an online list of current DL methods for ST analysis on a dedicated repository (https://github.com/SindiLab/Deep-Learning-in-Spatial-Transcriptomics-Analysis), which will be maintained and continuously updated.
FIG. 1.
An overview of deep learning (and machine learning) methods for spatial transcriptomics presented in this review. In this work, we provide a brief background on related biological concepts, such as single-cell RNA sequencing (scRNAseq) and spatial transcriptomic (ST) technologies (Sec. II), followed by an overview of common deep learning architectures in Sec. III. We then dive deeper into specific machine learning techniques for spatial reconstruction (Sec. IV A), scRNAseq and ST alignment (Sec. IV B), ST spot deconvolution (Sec. IV C), spatial clustering (Sec. IV D), and cell–cell interaction (Sec. IV E). A more comprehensive list of the state-of-the-art methods for spatial transcriptomics is provided in Table I.
TABLE I.
A list of relevant methods for the analysis of spatial transcriptomics data. The italicized boldfaced methods are the ones that utilize deep learning (or elements closely aligned). We review these methods in depth in this paper.
| Category | Method | Year | Framework | Language | Software availability |
|---|---|---|---|---|---|
| Spatial reconstruction | Seurat59 | 2015 | Statistical | R | https://github.com/satijalab/seurat |
| novoSpaRc60 | 2019 | Optimization/statistical | Python | https://github.com/rajewsky-lab/novosparc | |
| DEEPsc 61 | 2021 | Machine learning | MATLAB | https://github.com/fmaseda/DEEPsc | |
| Alignment and integration | Spatial backmapping62 | 2015 | Scoring scheme | R | https://github.com/jbogp/nbt_spatial_backmapping |
| Tangram 34 | 2021 | Machine learning | Python | https://github.com/broadinstitute/Tangram | |
| GLUER 63 | 2021 | Machine learning | Python | https://github.com/software-github/GLUER | |
| Spot deconvolution | Stereoscope64 | 2020 | Statistical | Python | https://github.com/almaan/stereoscope |
| DSTG 65 | 2021 | Machine learning | Python, R | https://github.com/Su-informatics-lab/DSTG | |
| SPOTlight66 | 2021 | Machine learning | R | https://github.com/MarcElosua/SPOTlight | |
| RTCD67 | 2021 | Statistical | R | https://github.com/dmcable/RCTD | |
| SpatialDWLS68 | 2021 | Optimization/statistical | R | https://github.com/RubD/Giotto | |
| DestVI 69 | 2021 | Machine learning | Python | https://github.com/YosefLab/scvi-tools | |
| Cell2location70 | 2022 | Statistical | Python | https://github.com/BayraktarLab/cell2location | |
| CARD71 | 2022 | Machine learning | R, C++ | https://github.com/YingMa0107/CARD | |
| Spatial clustering | HMRF72 | 2018 | Statistical | R, Python, C | https://bitbucket.org/qzhudfci/smfishhmrf-py/ |
| SpaCell 73 | 2019 | Machine learning | Python | https://github.com/BiomedicalMachineLearning/SpaCell | |
| StLearn 74 | 2020 | Machine learning | Python | https://github.com/BiomedicalMachineLearning/stLearn | |
| BayesSpace75 | 2021 | Statistical | R, C++ | https://github.com/edward130603/BayesSpace | |
| SpaGCN 76 | 2021 | Machine learning | Python | https://github.com/jianhuupenn/SpaGCN | |
| Spatially variable genes identification | Trendsceek77 | 2018 | Statistial | R | https://github.com/edsgard/trendsceek |
| SpatialDE78 | 2018 | Statistical | Python | https://github.com/Teichlab/SpatialDE | |
| Spark79 | 2020 | Statistical | R, C++ | https://github.com/xzhoulab/SPARK | |
| Cell–cell communication | SpaOTsc80 | 2020 | Machine learning | Python | https://github.com/zcang/SpaOTsc |
| MISTy81 | 2020 | Machine learning | R | https://github.com/saezlab/mistyR | |
| Giotto82 | 2021 | Statistical | R | https://github.com/RubD/Giotto |
II. BIOLOGICAL BACKGROUND
A. Single-cell RNA sequencing (scRNAseq)
RNA sequencing (RNA-seq) provides comprehensive insight on cellular processes (such as identifying genes that are upregulated or downregulated, etc.). However, traditional bulk RNA-seq is limited to revealing the average expression from a collection of cells, and not disambiguation single-cell behavior. Thus, it is difficult to delineate cellular heterogeneity with traditional RNA-seq, which is a disadvantage since cellular heterogeneity has been shown to play a crucial role in understanding many diseases.83 Therefore, researchers have turned to single-cell RNA-seq (scRNAseq) in order to identify cellular heterogeneity within tissues. ScRNAseq technologies have been instrumental in the study of key biological processes in many diseases, such as cancer,84 Alzheimer's,85 cardiovascular diseases,86 etc. (see Ref. 83 for more details). RNA sequencing of cells at a single-cell resolution, scRNAseq, generally consists of four stages:
-
(i)
Isolation of single-cells and lysing: Cells are selected through laser microdissection, fluorescence-activated cell sorting (FACS), microfluidic/microplate technology (MT), or a combination of these methods,87 with MT being highly complementary to NGS-based technologies.88 MT encapsulates each single-cell into an independent microdroplet containing unique molecular identifiers (UMI), lysis buffer for cell lysis (to increase the capturing of as many RNA molecules as possible), oligonucleotide primers, and deoxynucleotide triphosphates (dNTPs) in addition to the cells themselves. Due to MT's higher isolation capacity, thousands of cells can be simultaneously tagged and analyzed, which is beneficial for large-scale scRNAseq studies.
-
(ii)
Reverse transcription: One challenge in RNA sequencing is that RNA cannot be directly sequenced from cells, and thus, RNA must first be converted to complementary DNA (cDNA).89 Although various technologies employ different techniques, the reverse transcription phase generally involves capturing mRNA using poly[T] sequence primers that bind to mRNA ploy[A] tail prior to cDNA conversion. Based on the sequencing platform, other nucleotide sequences are added to the reverse transcription; for example, in NGS protocols, UMIs are added to tag unique mRNA molecules so that it could be traced back to their originating cells, enabling the combination of different cells for sequencing.
-
(iii)
cDNA amplification: Given that RNA cannot be directly sequenced from cells, single-stranded RNAs must first be reverse transcribed to cDNA. However, due to the small amount of mRNA in cells, limited cDNA is produced, which is not optimal for sequencing. Therefore, the limited quantity of cDNA must be amplified prior to library preparation and sequencing.90 The amplification is often done by either PCR (an exponential amplification process with its efficiency being sequence dependent) or IVT (a linear amplification method that requires an additional round of reverse transcription of the amplified RNA) before sequencing.89,91 The final cDNA library consists of an adaptor-ligated sequencing library attached to each end.
-
(iv)
Sequencing library construction: Finally, every cell's tagged and amplified cDNA is combined for library preparation and sequencing similar to bulk RNA sequencing methods, followed by computational pipelines for processing and analysis.92
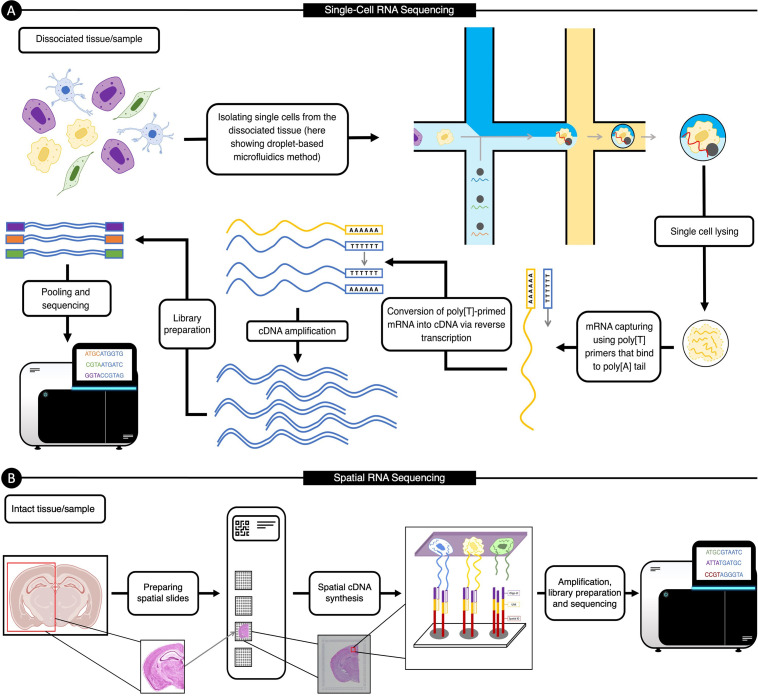
Figure 2(A) illustrates an example of the workflow for scRNAseq. For more details of each stage and various scRNAseq workflows, we refer the reader to Refs. 91 and 93–95.
FIG. 2.
Example of single-cell RNA sequencing and spatial transcriptomics workflows. (A) An illustration of droplet-based microfluidics single-cell RNA sequencing that consists of (1) dissociating a tissue or biological sample, (2) isolating single cells, unique molecular identifiers, and lysis buffer, (3) cell lysis, (4) mRNA capture and reverse transcription, (5) cDNA amplification, and (6) library construction. We describe this process in more detail in Sec. II A). (B) A visualization of the steps for next-generation sequencing-based spatial transcriptomic (as described in Sec. II B), which include (1) tissue preparation, staining, and imaging, (2) permeabilizing the tissue, (3) cDNA synthesis, amplification, and library construction, followed by (5) sequencing.
B. Spatial transcriptomics technologies
More recently, technologies that profile gene expression while retaining spatial information have emerged. These technologies are collectively known as spatial transcriptomics (ST). The various ST technologies provide different advantages and are chosen based on experimental factors, such as the size of tissue to be assayed, the number of genes to be probed, a priori knowledge of target genes, cost, etc. In general, ST technologies can be divided into two broad categories: imaging-based and next-generation sequencing (NSG)-based technologies. In this section, we provide an overview of popular techniques, with more emphasis on NGS-based approaches. For a more comprehensive and technical review of ST technologies, we refer the reader to Asp et al.,96 Rao et al.,20 and Moses and Pachter.97
1. Imaging-based technologies
Imaging-based technologies are broadly subdivided into in situ hybridization (ISH)-based methods, in situ sequencing (ISS)-based methods, or methods that borrow elements from both approaches. Unlike RNAseq methods described above, ISH and most ISS-based techniques require labeled probes. This means that the target genes must be known in advance, and, moreover, the number of genes that can be measured is limited.20
a. In situ hybridization (ISH)-based approaches
ISH-based methods aim to detect the absence or presence of target RNA (or DNA) sequences while localizing the information of the desired sequences to specific cells or chromosomal sites.98,99 ISH-based techniques use the labeled probes (usually made with DNA or RNA) that bind to desired sequences in fixed cells or tissue, therefore detecting the desired sequence through the hybridization of a complementary probe. The hybridized probes are then visualized through isotopic and nonisotopic (fluorescent and nonfluorescent) approaches.99 ISH-based techniques have been limited by the number of distinguishable transcripts; however, recent innovations have resulted in ample multiplexing capabilities.20
b. In situ sequencing (ISS)-based approaches
ISS-based approaches aim to sequence the RNA content of a cell in situ using DNA balls that amplify the RNA signals: RNA is first reverse transcribed to cDNA, followed by circular amplification (to generate a long single-stranded DNA that contains repeats of the target sequence) and sequencing.100 Although the transcript can be localized at subcellular resolution, micrometer- or nanometer-sized DNA balls are often used to amplify the signals to reach sufficient signal for imaging.96 Initially, the first ISS-based method101 used targeted padlock probes (a single-stranded DNA molecule containing regions complementary to the target cDNA) followed by sequence-by-ligation21 to detect desired genes. This method provided a subcellular resolution and an ability to detect single-nucleotide variants (SNVs). This ISS protocol is targeted and yields a detection efficiency of approximately 30%.102 Several recent ISS techniques build upon this approach and improve traditional ISS by introducing novel modifications to the experimental aspect of the protocol, as well as ways to mitigate the number of cells that can be discriminated simultaneously.103–105 For example, barcode in situ targeted sequencing (BaristaSeq)104 uses sequencing-by-synthesis and has led to increased read lengths and enabled higher throughput and cellular barcoding with improved detection efficiency compared to the initial ISS approach.104 Another ISS-based technique is spatially resolved transcript amplicon readout mapping (STARmap)29 that reduces noise and avoids the cDNA conversion complications by utilizing the improved padlock-probe and primer design; STARmap adds a second primer to target the site next to the padlock probe in order to circumvent the reverse transcription step. STARmap also uses advanced hydrogel chemistry and takes advantage of an error-robust sequencing-by-ligation method, resulting in detection efficiency that is comparable to scRNAseq methods (around 40%).29,96 Although most ISS approaches (including the ones mentioned here) are targeted, ISS-based methods could also be untargeted,25,106 but this typically leads to much lower sensitivity (around 0.005%) and molecular crowding, affecting the rolling-circle amplification bias.106,107
In imaging-based approaches, the generated image is segmented and processed to produce a cell-level gene-expression matrix. The gene-expression matrix is generated through processing the generated image(s), which can be done manually or automatically. However, given the biased and laborious nature of manual segmentation, there has been a shift toward designing general and automated techniques.108,109 The accurate and general automation of this process still remains a challenge, therefore motivating the application of recent machine learning and computer vision approaches to this field,108–111 which have shown improvements compared to the traditional methods.112 Although this manuscript focuses on methods for NGS-based technologies, many of those techniques (including the ones in Secs. IV C and IV D) can be extended to image-based technologies as well.
C. Next generation sequencing (NGS)-based technologies
Due to the unbiased capture of mRNA, NGS-based technologies can shed light on the known and unknown morphological features using only the molecular characterization of tissues.1 This unbiased and untargeted nature of NGS technologies makes them ideal for studying and exploring new systems,20 a major advantage compared to most image-based technologies that require target genes a priori. While NGS-based approaches differ in the specifics of the protocols, they all build on the idea of adding spatial barcodes before library preparation, which are then used to map transcripts back to the appropriate positions (known as spots or voxels). An example workflow of NGS-based spatial sequencing is depicted in Fig. 2(B). In this section, we provide a general overview of the four most common spatial transcriptomics technologies. For a more complete review of these technologies, we refer the readers to Refs. 20 and 96.
Ståhl et al.31 were the first to successfully demonstrate the feasibility of using NGS for spatial transcriptomics (this initial approach is often referred to as spatial transcriptomics). Their innovation was to add spatial barcodes prior to library preparation, enabling the mapping of expressions to appropriate spatial spots. More specifically, Ståhl et al. positioned oligo(dT) probes and unique spatial barcodes as microarrays of spots on the surface of slides. Next, fresh frozen tissue slices were placed on the microarray and processed to release mRNA (using enzymatic permeabilization), which was then hybridized with the probes on the surface of the slides. This approach consists of (i) collecting histological imaging [using standard fixation and staining techniques, including hematoxylin and eosin (H and E) staining] for investigating the morphological characteristics and (ii) sequencing the spatially barcoded cDNA to profile gene expressions. In the initial experiments, each slide consisted of approximately 1000 spots, each having a diameter of 100 μm with 200 μm center-to-center distance.1 This approach provides researchers with an unbiased technique for analyzing large tissue areas without the need for selecting target genes in advance.20,35,112
After the initial success of spatial transcriptomics, 10× Genomics subsequently improved the resolution (shrinking the spot diameters to 55 μm with 100 μm center-to-center distance) and sensitivity (capturing more than 104 transcripts per spot) of the approach and eventually commercializing it as Visium30,113) The development and commercialization of the spatial transcriptomics resulted in relatively rapid adoption across fields, such as cancer biology,55,114 developmental biology,115,116 and neuroscience.117,118 The histological imaging and gene expression profiling of Visium are similar to the initial approach: the staining and imaging of the tissues are through traditional staining techniques, including H and E staining for visualizing the tissue sections using a brightfield microscope and immunofluorescence staining to visualize protein detection in the tissue sections through a fluorescent microscope. Visium protocol allows for both fresh frozen (FF) or formalin-fixed paraffin-embedded (FFPE) tissues. For FF tissues, similar to Ståhl et al.,31 the tissue is permeabilized, allowing the release of mRNA, which hybridizes to the spatially barcoded oligonucleotides present on the spots. The captured mRNA then goes through a reverse transcription process that results in cDNA, which is then barcoded and pooled for generating a library.119 For FFPE tissues, tissue is permeabilized to release ligated probe pairs from the cells that bind to the spatial barcodes on the slide, and the barcoded molecules are pooled for downstream processing to library generation.119
Building on the spatial transcriptomics, Vickovic et al.33 proposed high-definition spatial transcriptomics (HDST) that improved the resolution to about 2 μm. Similar to the other approaches, HDST also employs specific barcodes ligated to beads that are coupled to a spot (prior to lysis), so that expressions are mapped to the tissue image. However, the innovation of HDST includes the use of 2 μm beads placed in hexagonal wells, enabling accurate compartmentalization and grouping of the biological materials in the experiment.33 Simultaneously, Rodriques et al.32 introduced SlideSeq that utilizes slides with randomly barcoded beads to capture mRNA, also increasing the resolution (to 10 μm) and sensitivity (500 transcripts per 54 bead) spatial-resolved sequencing compared to Ståhl et al.31 However, SlideSeq placed the barcoded beads in rubber and onto glass slides, as opposed to HDST's hexagonal beads, and determines the position of each random barcode by in situ-indexing.20,32
Despite the differences, all NGS-based technologies use spatial barcodes to tag released RNAs, which then go through conventional processes for sequencing similar to scRNAseq. After sequencing, the data are processed to construct the spatial location of each read (using the spatial barcode) and to construct a gene-expression matrix (mapping the reads to the genome to identify the transcript of origin). Given that most technologies have resolutions larger than a single cell (commonly having expression for 3–30 cells in each spot), the data processing and analysis procedures are relatively similar.
III. MACHINE LEARNING AND DEEP LEARNING BACKGROUND
With the technologies now defined, we next describe common Machine Learning (ML) methods used to analyze the ST data. In this section, we first provide a discussion of the algorithmic development of ML and deep learning (DL) models and then discuss common architectures used for spatially resolved transcriptomics (and scRNAseq data).
ML refers to a computer algorithm's ability to acquire knowledge by extracting patterns and features from the raw data.120 All ML algorithms depend on the data, which must be available before the methods can be used, and a defined mathematical objective. ML models' lifecycle consists of two phases, namely, training and evaluation. During training, ML algorithms analyze the data to extract patterns and adjust their internal parameters based on optimizing their objectives (known as loss function). In the evaluation (or inference) stage, the trained model makes predictions (or performs the task it was trained to do) on unseen data.
There are two main types of ML algorithms: supervised and unsupervised. An ML algorithm is considered to be unsupervised if it utilizes raw inputs without any labels to optimize its objective function (an example would be the K-Means clustering algorithm121). Conversely, if an algorithm uses both raw data and the associated labels (or targets) in training, then it is a supervised learning algorithm. Supervised learning is the most common form of ML.39 An example of supervised learning in scRNAseq analysis would be classifying cell subpopulations using prior annotations: this requires a labeled set of cell types for training (the available annotations), an objective function for calculating learning statistics (“teaching” the model), and testing data for measuring how well the model can predict the cell type (label) on data it has not seen before (i.e., generalizability of the model). Another common example of supervised learning is regression, where a model predicts continuous values as opposed to outputting labels or categorical values in classification. For supervised tasks, a model is trained on the majority of the data (known as training set) and then evaluated on held-out data (test set). Depending on the size of our dataset, there can also be a third data split known as a validation set, which is used to measure the performance of the model throughout training to determine early stopping:122 Early stopping is when we decide to stop the training of a model due to overfitting (or over-optimization) on the training set. Overfitting training data worsens the generalizability of the model on unseen data, which early stopping aims to avoid.122 In addition to supervised and unsupervised algorithms, there are also semi-supervised learning, where a model uses a mix of both supervised and unsupervised tasks, and self-supervised, where the computer algorithm generates new or additional labels to improve its training or to learn a new task.
Raw experimental data typically contains noise or other unwanted features, which present many challenges for ML algorithms. Therefore, it is often necessary to carefully preprocess data or to rely on domain-specific expertise in order to transform the raw data into some internal representation from which ML models can learn.39 Deep learning (DL) algorithms, however, aim to use only raw data to automatically extract and construct useful representations required for learning the tasks at hand. In a broad sense, DL models are able to learn from observations through constructing a hierarchy of concepts, where each concept is defined by its relation to simpler concepts. A graphical representation of the hierarchy of concepts (and learning) will consist of many layers, with many nodes and edges connecting the vertices, somewhat resembling humans' neural network. This graph is referred to as an artificial neural network (ANN). ANNs are composed of interconnected nodes (“artificial neurons”) that resemble and mimic our brains' neuronal functions. An ANN is considered to be a DL model if it consists of many layers—often more than three, hence being called deep.
Many tasks that humans perform can be viewed as mappings between sets of inputs and outputs. For example, humans can take a snapshot image of their surroundings (input) and detect the relevant objects (the outputs). DL, and more generally, Artificial Intelligence, aims to learn such mappings in order to model human-level intelligence. Mathematically, ANNs are universal function approximators, meaning that, theoretically, they can approximate any (continuous) function.123–125 Cybenko123 proved this result for a one-layer neural network with an arbitrary number of neurons (nodes) and a sigmoid activation function by showing that such architecture is dense within the space of continuous functions (this result has now been extended to ANNs with multiple layers124). While constructing arbitrarily-long single-layer ANNs is not possible, it has been shown that ANNs with many layers (deeper) generally learn faster and more reliably than ANNs with few wide (many neurons) layers.126 This has allowed researchers to employ deep networks for learning very complex functions through constructing simple non-linear layers that can transform the representation of each module (starting with the raw input) into a representation at a higher, slightly more abstract level.39
DL models' ability to approximate highly non-linear functions has revolutionized many domains of science, including computer vision,127–129 natural language processing,130–132 and bioinformatics.133–135 DL is becoming increasingly incorporated in many computational pipelines and studies, especially in genomics and bioinformatics, including scRNAseq and spatial transcriptomics analysis. In Secs. III A–III F, we provide a brief overview of essential deep learning architectures that have been used in spatial transcriptomics and scRNAseq analysis. In Fig. 3, we present illustrations of the architectures discussed in Secs. III A–III F. Note that for simplicity, we have categorized all graph convolution networks (GCNs)136 as DL models; this is because (i) GCNs can easily be extended to include more layers (deeper networks) and (ii) lack of other existing methods that incorporate some elements of DL. A more comprehensive description of each architecture can be found in the seminal textbook by Goodfellow et al.120
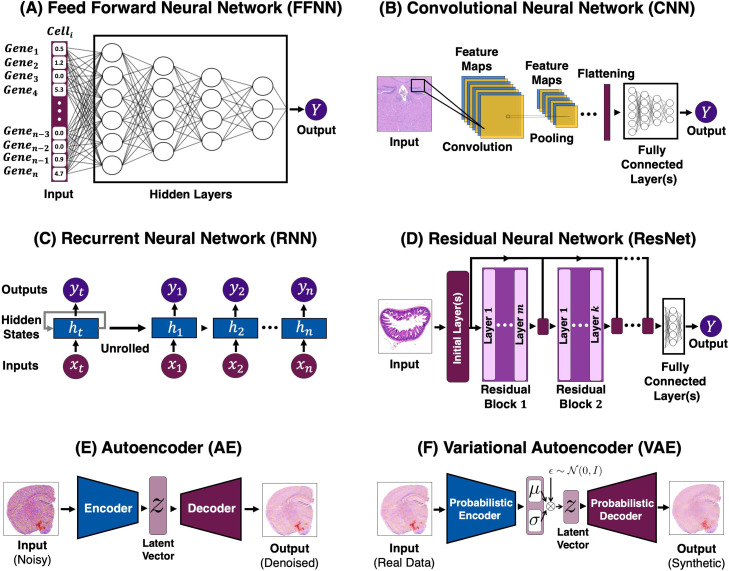
FIG. 3.
Examples of deep learning architectures. Models depicted in (a), (b), (c), and (d) are examples of supervised learning, and networks shown in (e) and (f) are unsupervised. (a) An example of an FFNN architecture with gene expression count as its input. (b) An example of CNN architecture, where the model passes the inputs through the three stages of a CNN (with non-linear activation not depicted) to extract features. Then, outputs are flattened and fed into a fully connected layer (or layers). (c) The general training flow of an RNN, with the unrolled version showing the time step-dependent inputs, hidden state, and outputs. The inputs to RNNs need to have a sequential structure (e.g., time-series data). (d) An illustration of an ResNet. In traditional ResNets, there are identity mappings (or skip connections) that pass the input of a residual block to its output (often through addition). (e) Here, we show the general architecture of a trained denoising AE in the inference stage, with a noisy histology slide as its input, yielding a denoised version of the input image. (6) A depiction of a traditional VAE in the inference stage. VAE's aim is to generate synthetic data that closely resembles the original input. This is done through regularizing the latent space of an AE with the use of a probabilistic encoder and decoder.
A. Feed forward neural network (FFNN)
FFNNs, the quintessential example of Artificial Neural Networks (ANNs), aim to approximate a function mapping a set of inputs to their corresponding targets [see Fig. 3(a)]. More specifically, given an input and a target , where , FFNNs aim to learn the optimal parameters such that . FFNNs are the building blocks of many more advanced architectures (e.g., convolutional neural networks) and, therefore, are of paramount importance in the field of ML.120 As mentioned previously, ANNs are universal function approximators, and they represent a directed acyclic graph of function compositions hierarchy within the network. Each layer of an FFNN, ( being the ith layer), is often a simple linear function: For example, we can have a linear function for outputting of the form Eq. (1), with weight parameters and a bias
| (1) |
However, a model composed of only linear functions can only approximate linear mappings. As such, we must consider non-linear activation functions to increase model capacity, enabling the approximation of complex non-linear functions. In the simplest case, neural networks (NNs) use an affine transform (controlled by learned parameters) followed by a non-linear activation function, which, theoretically, enables them to approximate any non-linear function.137 Moreover, we could compose many of such non-linear transformations to avoid infinitely wide-neural networks when approximating complex functions. However, in this context, finding a set of optimal functions ( ) is a practically impossible task. As such, we restrict the class of function that we use for to the following form in Eq. (2):
| (2) |
where superscript i enumerates the layers, is a non-linear activation function (usually a Rectified Linear Unit138), denotes the output of the layer (with indicating the input data), weights , and biases . Note that because of the dimensionality of the mapping, , and we must have a vector of biases . FFNNs are composed of such functions in chains; to illustrate, consider a three-layer neural network
| (3a) |
| (3b) |
| (3c) |
with representing the hidden states or hidden layers.
FFNNs find the optimal contribution of each parameter (i.e., weights and biases) by minimizing the desired objective. The goal is to generalize the task to data the model has never seen before (testing data). Although the non-linearity increases the capacity of FFNNs, it causes most objective functions to become non-convex. In contrast to convex optimization, non-convex loss functions do not have global convergence guarantees and are sensitive to the initial starting point (parameters of the network).139 Therefore, such optimization is often done through stochastic gradient descent (SGD) (or some variant of it). Moreover, given the sensitivity to initial values, weights are typically chosen to be small random values, with biases initialized to zero or small positive values.120,140,141
Almost all neural networks use iterative gradient descent (GD)-based optimizers to train. GD has three main variants, which differ in the amount of data utilized to calculate the gradients for updating the parameters. The classic GD variant, referred to as batch GD, uses all data points to make the updates to the parameters in one iteration. However, this approach is generally not feasible, since the amount of data required for training DL models almost never fit in memory.142 The second variant of GD is stochastic gradient descent (SGD) where the parameters are updated for every training datum. Computationally, it has been shown that the noise in SGD accelerates its convergence compared to batch GD, but SGD also has the possibility of overshooting, especially for highly non-convex optimization functions.142,143 The third variant, and the most frequently used one for deep learning, is mini-batch GD that updates the parameters for every batch of training data—if the batch size is one, then this variant is just SGD, and if batch size is the entire dataset, then it is equivalent to batch GD. Conventionally, optimization of NNs is done through gradient descent performed backward in the network, which itself consists of two components: a numerical algorithm for efficient computation of the chain rule for derivatives (backpropagation144) and a GD-based optimizer (e.g., Adam145 or AdaGrad146). The optimizer is an algorithm that performs gradient descent, while backpropagation is an algorithm for computing the expression for gradients during the backward pass of the model.
B. Convolutional neural network (CNN)
Learning from images, such as detecting edges and identifying objects, has been of interest for some time in computer science.147 Images contain a lot of information; however, only a small amount of that information is often relevant to the task at hand. For example, an image of a stained tissue contains both important information, namely, the tissue itself, and irrelevant pixels, such as the background. Prior to DL, researchers would hand-design a feature extractor to learn relevant information from the input. Much of the work had focused on the appropriate feature extractors for desired tasks (e.g., see the seminal work by Marr and Hildreth148). However, one of the main goals in ML is to extract features from raw inputs without hand-tuned kernels for feature extraction. CNNs149,150 are a specialized subset of ANNs that use the convolution operation (in at least one of their layers) to learn appropriate kernels for extracting important features beneficial to the task at hand. Mathematically, convolution between two functions f and w is defined as a commutative operation shown in the following equation:
| (4) |
Using our notation, we intuitively view convolution as the area under f(s) weighted by and shifted by x. In most applications, discrete functions are used. As an example, assume we have a 2D kernel K that can detect edges in a 2D image I with dimension m × n. Since I is discrete, we can use the discrete form of Eq. (4) for the convolution of I and K over all pixels
| (5) |
However, since there is less variation in the valid range of m and n (the dimensions of the image) and the operation is commutative, most algorithms implement Eq. (5) equivalently
| (6) |
Typical CNNs consist of a sequence of layers (usually three) which include a layer performing convolution, hence called a convolutional layer (affine transform), a detector stage (non-linear transformation), and a pooling layer. The learning unit of a convolutional layer is called a filter or kernel. Each convolutional filter is a matrix, typically of small dimensions (e.g., 3 × 3 pixels), composed of a set of weights that acts as an object detector, with the weights being continuously calibrated during the learning process. CNNs' objective is to learn an optimal set of filters (weights) that can detect the needed features for specific tasks (e.g., image classification). The result of convolution between the input data and the filter's weights is often referred to as a feature map [as shown in Fig. 3(b)]. Once a feature map is available, each value of this map is passed through a non-linearity (e.g., ReLU). The output of a convolutional layer consists of as many stacked feature maps as the number of filters present within the layer.
There are two key ideas behind the design of CNNs: First, in data with grid-like topology, local neighbors have highly correlated information. Second, equivariance to translation can be obtained if units at different locations share weights. In other words, sharing parameters in CNNs enabled the detection of features regardless of the locations where they appear in. An example of this would be detecting a car. In a dataset, a car could appear at any position in a 2D image, but the network should be able to detect it regardless of the specific coordinates.147 These design choices provide CNNs with three main benefits compared to other ANNs: (i) sparse interactions, (ii) shared weights, and (iii) equivariant representations.149
Another way of achieving equivariance to translation is to utilize pooling layers. Pooling decreases the dimension of learned representations and makes the model insensitive to small shifts and distortions.39 In the pooling layers, we use the outputs of the detector stage (at certain locations) to calculate a summary statistic for a rectangular window of values (e.g., calculating the mean of a 3 × 3 patch). There are many pooling operations, with common choices being max-pooling (taking the maximum value of a rectangular neighborhood), mean-pooling (taking the average), and L2 norm (taking the norm). In all cases, rectangular patches from one or several feature maps are inputted to the pooling layer, where semantically similar features are merged into one. CNNs typically have an ensemble of stacked convolution layers, non-linearity, and pooling layers, followed by fully connected layers that produce the final output of the network. The backpropagation of gradients through CNNs is analogous to FFNNs, enabling the model to learn an optimal set of filters for the task(s) at hand. CNNs have been effectively used in many applications in computer vision and time-series analysis and are being increasingly utilized for analysis of the ST data, since spatial-omics are multi-modal, with one of the modalities being images (as we discuss in Sec. IV).
C. Recurrent neural network (RNN)
Just as CNNs are specialized to process data with a grid-like topology, RNNs'144 special characteristics make them ideal for processing sequential data }, where denotes the ith element in the ordered sequence X. Examples of such sequence-like structures include time series and natural language. RNNs process sequential inputs one at a time and implicitly maintain a history of previous elements of the input sequence. We present an illustration of the conventional RNN architecture in Fig. 3(c). Similar to FFNNs or CNNs, RNNs can be composed of many layers, with each layer depending on the previous hidden state, , and a shared set of parameters, . A deep RNN with n hidden states can be expressed as follows:
| (7a) |
| (7b) |
| (7c) |
The idea behind sharing in RNN states is similar to CNNs: parameter sharing across different time points allows RNNs to generalize the model to sequences of variable lengths and share statistical strengths at different positions in time.120,151 Similar to FFNNs, RNNs learn by propagating the gradients of each hidden state's inputs at discrete times. This process becomes more intuitive if we consider the outputs of hidden units at various time iterations as if they were the outputs of different neurons in a deep multi-layer network. However, due to the sequential nature of RNNs, the backpropagation of gradients shrinks or grows at each time step, causing the gradients to potentially vanish or blow up. This fact and the inability to parallelize training at different hidden states (due to the sequential nature of RNNs) makes RNNs notoriously hard to train, especially for longer sequences.130,152 However, when these issues are averted (via gradient clipping or other techniques), RNNs are powerful models and gain state-of-the-art capabilities in many domains, such as natural language processing. The training challenges combined with the nature of scRNAseq data have resulted in fewer developments of RNNs for single-cell analysis. However, recently, some studies have used RNNs and long short-term memory153 (a variant of RNNs) for predicting cell types and cell motility (e.g., see Kimmel et al.154).
D. Residual neural network (RestNet)
As mentioned above, deep RNNs may suffer from vanishing or exploding gradients. Such issues can also arise in other deep neural networks as well, where gradient information could diminish as the depth increases (through approaches, such as batch normalization,155 aim to help with gradient issues). One way to alleviate vanishing gradients in very deep networks is to allow gradient information from successive layers to pass through, helping with maintaining information propagation even as networks become deeper. ResNets156 achieve this by skip (or residual) connections that add the input to a block (a collection of sequential layers) to its output. For an FFNN, consider function f in Eq. (2). Using the same notation as in Eq. (2), ResNet's inner layers take the form shown in the following equation:
| (8) |
The addition of , the input of the current layer [or the output of the th layer], to the current ith layer output is the skip or residual connection helps flow the information from the input deeper in the network, thus stabilizing training and avoiding vanishing gradient in many cases.156,157 Indeed, this approach can be contextualized within the traditional time integration framework for dynamical systems. For example, consider the following equation:
| (9) |
In the simplest case, this system can be discretized and advanced using and some scaled value of , or a combination of scaled values of . Forward Euler, perhaps the simplest time integrator, advances the solution as shown through the scheme in the following equation:
| (10) |
where h is a sufficiently small real positive value. ResNets use this idea to propose a different way of calculating the transformations in each layer, as shown in Eq. (8).
ResNets consist of residual blocks (also called modules), each of which contains a series of layers. For visual tasks, these blocks often consist of convolutional layers, followed by activation functions, with the skip connection adding the input information to the output of the residual blocks (as opposed to the individual layers inside). ResNets have different depths and architectures, with a number usually describing the depth of the model [e.g., ResNet50 means there are 50 layers (there are 48 convolution layers, one MaxPool, and one AveragePool layer)].
ResNets have transformed DL by enabling the training of very deep neural networks, setting the state-of-the-art performance in many areas, particularly in computer vision.156 The pre-trained ResNets on ImageNet dataset158 are widely used for transfer learning, where the network is either used as is or further fine-tuned on the specific dataset. Pre-trained ResNet models have also been used in spatial transcriptomics analysis, as we discuss later in this manuscript. (ImageNet is the standard dataset for benchmarking performance of machine learning algorithms in classification and object recognition. ImageNet contains more than 14 million hand-annotated images).
E. Autoencoder (AE)
AEs159,160 are neural networks that aim to reconstruct (or copy) the original input via a non-trivial mapping. Conventional AEs have an “hour-glass” architecture [see Fig. 3(e)] consisting of two networks: (i) an encoder network, , which maps an input to a latent vector where, ideally, z contains the most important information from x in a reduced space (i.e., ), (ii) the decoder network, , which takes z as the input and maps it back to , ideally, reconstructing x exactly; i.e., . AEs were traditionally used for dimensionality reduction and denoising, trained by minimizing a mean squared error (MSE) objective between the input data and the reconstructed samples (outputs of the decoder).
Over time, the AE framework has been generalized to stochastic mappings, i.e., probabilistic encoder–decoder mappings, and . A well-known example of such generalization is variational autoencoders (VAEs),161 where by using the same hour-glass architecture, one can use probabilistic encoders and decoders to generate new samples drawn from an approximated posterior. Both traditional AEs and VAEs have practical applications in many biological fields, have been used extensively in scRNAseq (see Ref. 10 for an overview of these models), and are becoming more frequently employed in spatial transcriptomics analysis, which we overview later in this work.
F. Variational autoencoder (VAE)
One can describe VAEs161 as AEs that regularize the encoding distribution, enabling the model to generate new synthetic data. The general idea behind VAEs is to encode the inputs as a distribution over the latent space, as opposed to a single point (which is done by AEs). More specifically, VAEs draw samples z from an encoding distribution, , and subsequently feed the sample through a differentiable generator network, obtaining . Then, x is sampled from a distribution . Moreover, VAEs utilize an approximate inference network (i.e., the encoder) to obtain z. With this approach, now is considered a decoder network, decoding z that comes from . VAEs can take advantage of gradient-based optimization for training through maximizing the variational lower bound, , associated with x. Figure 3(f) depicts the architecture of traditional VAEs.
Mathematically, we can express the objective function as in Eq. (11)
where denotes the entropy, and is the Kullback–Leibler divergence. The first term in Eq. (11a) is the joint log-likelihood of the hidden and visible variables under the approximate posteriors over the latent variables. The second term of Eq. (11a) is the entropy of the approximate posterior. This entropy term encourages the variational posterior to increase the probability mass on a range of z which could have produced x, as opposed to mapping to a one-point estimate of the most likely value.120
Compared to other generative models [e.g., generative adversarial networks (GANs)162], VAEs have desirable mathematical properties and training stability.120 However, they suffer from two major weaknesses: (i) classic VAEs create “blurry” samples (those that adhere to an average of the data points), rather than the sharp samples that GANs generate due to GANs' adversarial training. (ii) The other major issue with VAEs is posterior collapse: when the variational posterior and actual posterior are nearly identical to the prior (or collapse to the prior), which results in poor data generation quality.163 To alleviate these issues, different algorithms have been developed, which have been shown to significantly improve the quality of data generation.163–169 VAEs are used extensively for the analysis of single-cell RNA sequencing (see Erfanian et al.10), and we anticipate them to be applied to a wide range of spatial transcriptomics analyses as well.
IV. DEEP LEARNING MODELS FOR SPATIALLY RESOLVED TRANSCRIPTOMICS ANALYSIS
In Secs. IV A–IV E, we describe the use of ML and DL to problems emerging from spatial transcriptomics.
A. Spatial reconstruction
Prior to the advancement of spatial transcriptomics, several studies aimed to reconstruct spatial information using gene expression data, with most of these works using a statistical framework. As perhaps one of the most influential models in this space, Satija et al.59 introduced Seurat: a tool that utilized spatial reference maps constructed from a few landmark in situ patterns to infer the spatial location of cells from corresponding gene expression profiles (i.e., scRNAseq data). This approach showed promising results: Satija et al. tested Seurat's capabilities and performance on developing zebrafish embryo dataset (containing 851 cells) and a reference atlas constructed from colorigenic in situ data for 47 genes,59 confirming Seurat's accuracy with several experimental assays. Additionally, they showed that Seurat can accurately identify and localize rare cell populations. Satija et al. also demonstrated that Seurat was a feasible computational solution for handling stochastic noise in omics data and finding a correspondence between ST and scRNAseq data.
Although Seurat proved to be successful in some applications, it had the limitation of requiring spatial patterns of marker genes expression.60 To alleviate Seurat's limitations, newer methods that did not require spatial reference atlases were developed. A more recent and an influential model in this space is novoSpaRc,60 with the ability to infer spatial mappings of single cells de novo. For novoSpaRc, Nitzan et al.60 assume that cells that are closer to one another physically have similar gene expressions as well, therefore searching for spatial arrangement possibilities that place cells with similar expressions closer in space. Nitzan et al. formulate this search through a generalized optimal-transport problem for probabilistic embedding.
NovoSpaRc shows very promising results when it is applied to spatially reconstruct mammalian liver and intestinal epithelium, and embryos from fly and zebrafish from gene expression data.60 However, novoSpaRc (and similar models) use a generic framework and cannot be easily adapted to specific biological systems, which may be required given the vast diversity of biological processes and organisms. For this reason, many have utilized ML algorithms to specifically adapt to the biological system by learning from the data, as opposed to using pre-defined algorithms that remain unchanged. Indeed, we anticipate that DL models will soon play a salient role in spatial reconstruction of scRNAseq, given their ability to extract features from the raw data while remaining flexible across different applications. In this section, we review DEEPsc,61 a system-adaptive ML model that aims to impute spatial information onto non-spatial scRNAseq data.
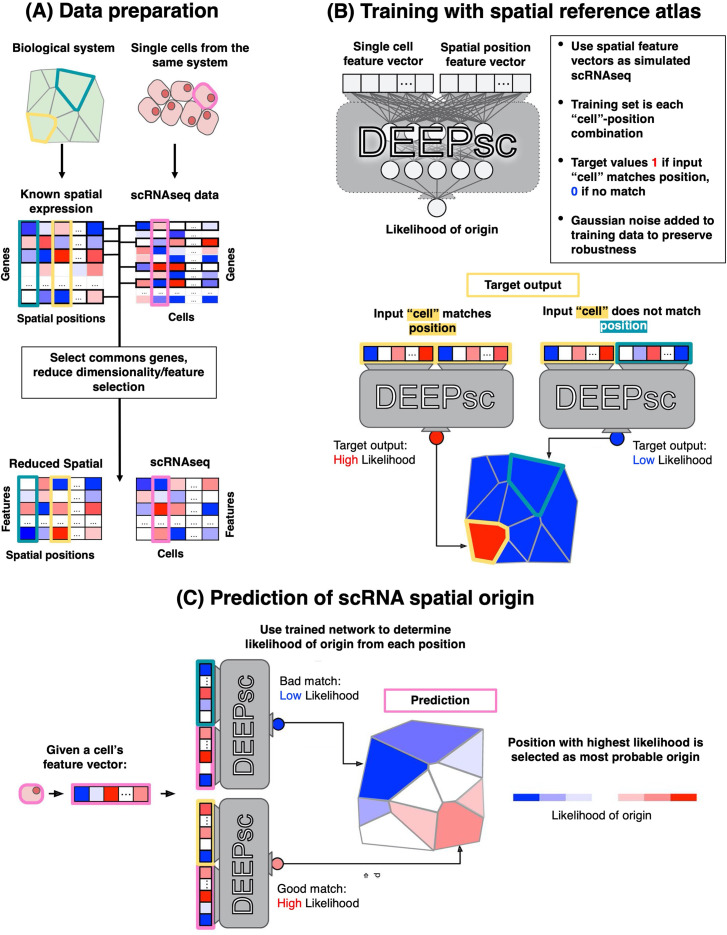
DEEPsc is a spatial reconstruction method that requires a reference atlas (see Fig. 4). This reference map can be expressed as a matrix , where is the number of spatial locations, and is the number of genes. Maseda et al. start by selecting common genes between Mspatial and the gene expression matrix, (where is the number of cells, and is the number of genes), resulting in a spatial matrix and an expression matrix , where is the common genes between the two matrices. Next, S is projected into a lower dimension using principal component analysis (PCA), and the same PCA coefficients are used to project E into these principal components. In the last step of processing, both matrices are normalized by their largest elements, resulting all elements of the matrices E and S to be in [0, 1]. Let us denote the normalized and PCA-reduced spatial and gene expression matrices as and , respectively.
FIG. 4.
An overview of DEEPsc training and inference. (a) Maseda et al. find the common genes in both spatial and scRNAseq data and perform dimensionality reduction on each data modality (with the final matrices having the same number of features). (b) During the training, DEEPsc uses spatial expression to “simulate” single-cell gene expression vectors. More specifically, every feature vector from the spatial expression is concatenated with all other vectors (labeled as “non-match”) and also itself (labeled as “match”) to form the input data to the neural network. (c) During inference, the scRNAseq feature vectors are concatenated with all spatial feature vectors, where the model should place a high probability for locations where the gene expression could have originated from. This figure was obtained from Maseda et al.61 [Front. Genet. 12, 348 (2021). Copyright 2021 Authors; licensed under a Creative Commons Attribution [CC-BY] license).
DEEPsc requires known spatial expression to learn the correct spatial positions, given the gene expression. More specifically, Maseda et al. construct training vectors of size (with N being the number of features preserved in the reduced matrix ). The first N elements of Inpij correspond to the spatial expression at the ith position, and the last N elements correspond to some position j in the reference atlas, including the position j = i. During training, DEEPsc's goal is produce the highest likelihood when (meaning that ) and assign low likelihood when . DEEPsc also adds Gaussian noise to posi (the first N elements of Inpij), which aims to preserve robustness and avoid overfitting. The addition of noise can lead to DEEPsc learning a complex non-linear mapping between the spatial positions in the reference atlas rather than a simple step-like function that activates when an exact match is inputted. During inference stage (i.e., after DEEPsc is trained), posi is replaced with the gene expression feature vector, which are the elements of , and the goal is to predict the likelihood of the expression vector being originated from all possible positions j.
DEEPsc's network is an FFNN with two hidden layers, with each , mapping to an , where y can be viewed as a likelihood that the input cell originated from the input spatial position.170 Given that for each training data Inpij there will be non-matches (labels of zero) and only one match for when j = i, the training labels will have many more zeros than ones. Therefore, Maseda et al. propose a nontraditional objective function that accounts for the imbalance between zeros and ones in the training data. This objective function is shown in the following equation:
| (12) |
where is the networks predicted outputs, and is the true target ( if it exactly matches, and otherwise). This allows DEEPsc to avoid producing trivially zero outputs, which is important given the sparsity of the data. Maseda et al. also employ strategies in data splitting which helps to account for the inherent sparsity in the targets.61 It is important to note that Maseda et al. also formulate a novel system-adaptive scoring scheme to evaluate the performance of DEEPsc using the spatial reference. However, the scoring scheme does not fall within the scope of this manuscript.
Maseda et al. apply DEEPsc for spatial imputation of four different biological systems (Zebrafish,59 Drosophila,171 Cortex,172 and Follicle173) achieving accuracy comparable to several existing models while having higher precision and robustness.61 DEEPsc also shows better consistency across the different biological systems tested, which can be attributed to its system-adaptive design. In addition, the authors attribute the performance and generalizability of DEEPsc to the use of FFNN (which have been noted before in other biological applications as well45,47) and the various strategies for robustness used during the training of DEEPsc. On the other hand, a weakness of DEEPsc is its training time, which depends non-linearly on the number of locations available. However, this issue can be potentially mitigated by considering a small subset of possible locations, or a more optimized design when training the model.
B. Alignment
Alignment in ST analysis refers to the process of mapping scRNAseq data to a physical domain while aiming to match the geometry with the available spatial data. As previously stated, NGS-based technologies suffer from limited capture rates and significant dropout174 (especially at higher resolutions). Before the use of DL in ST analysis, many computational approaches aimed to spatially reconstruct key marker genes scRNAseq data by assuming continuity in the gene space,60 or by leveraging local alignment information.59 Moreover, most techniques for alignment or deconvolution of spatial data either learned a program dictionary32 or estimated a probabilistic distribution of the data64 for the cell types at each spot. However, such approaches are not generalizable to all experimental settings, since finding the mapping of sparse or sporadically distributed genes to the spots is difficult and is error-prone due to dropouts.34
DL frameworks have the potential of providing robust models that can adapt to specific species or technologies while being generalizable to other datasets and platforms. The potential application of DL in the alignment of spatially resolved transcriptomics came to fruition recently through the work of Biancalani et al., called Tangram.34 Tangram is a framework that, among many of its capabilities, can align scRNAseq or single-nucleus(sn) RNAseq profiles to spatial data; for the sake of simplicity, we refer to both data types as scRNAseq, although there are differences between the two methods (see Ref. 175 for a systematic comparison of scRNAseq and snRNAseq approaches). Tangram aims to: (i) learn the transcriptome-wide spatial gene expression map at a single-cell resolution and (ii) relate the spatial information back to histological and anatomical data obtained from the same samples. Tangram's general workflow is to learn a mapping between the data modalities and then construct specific models for the downstream tasks (such as deconvolution, correcting low-quality data, etc.). We first summarize Tangram's alignment algorithm and then provide the applications in which DL models are utilized.
Tangram's general objective is to learn a spatial matrix describing the spatial alignments for the cells, with ncells and ngenes denoting the number of single-cells and the number of genes, respectively. Let the expression of gene k in cell i be denoted by , a non-negative value. Next, Tangram partitions (“voxelizes”) the spatial volume at the finest possible resolution (depending on the spatial technology) as a one-dimensional array. This allows Tangram to construct (1) a matrix , where Gjk is a non-negative value denoting the expression of gene k in voxel j, and (2) a cell-density vector , where is the cell density in voxel j (with the total density for each voxel summing to 1).
The learning of transcriptome-wide spatial gene expression map at a single-resolution happens through learning a mapping operator , where Mij denotes the probability of cell i being in voxel j. Moreover, given any matrix , each element of the operator M is assigned according the following equation:
| (13) |
ensuring that , i.e., assigning a probability distribution along the voxels using the well-known function. Biancalani et al. define an additional quantity, , which denotes the spatial gene expression as predicted by the operator M, and a vector where is the predicted cell density for each voxel j.
Given the preliminary quantities, we can now write Tangram's generic objective function as shown in the following equation:
| (14) |
where “ ” denotes the matrix slicing, and is the cosine similarity, defined as follows:
| (15) |
The objective function, Eq. (14), learns a proportional mapping of the genes to the voxels. Additionally, this loss function can be further modified to incorporate prior knowledge. Indeed, Biancalani et al. modify this to regularize for the learned density distributions and the cells contained within each voxel, as shown in the following equation:
| (16) |
where minimizing the divergence (first term) enforces that the learned density distribution and the expected distribution are similar, and the additional loss over the voxels (third term) penalizes the model if the predicted gene expression is not proportional to the expected gene expression. Biancalani et al. minimize the objective shown in Eq. (16) through gradient-based optimizers implemented in PyTorch.176,177 After optimizing Eq. (16), Tangram is able to map all scRNAseq profiles onto the physical space, thus performing the alignment. It is important to note that although Tangram learns a linear operator M, this mapping could be replaced with a deep neural network as well.
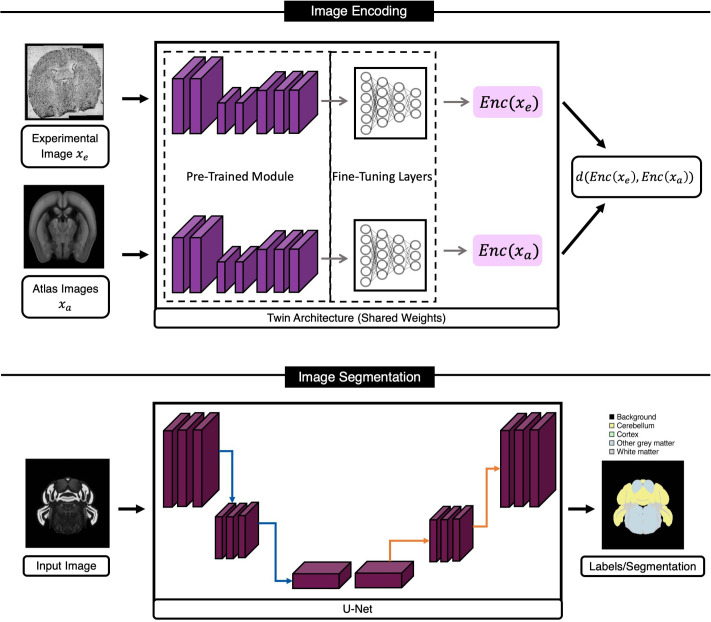
Tangram utilizes DL to integrate anatomical and molecular features, specifically for mouse brain images. To do so, the authors use an image segmentation network (U-Net178) in combination with a “Twin” network179 to produce segmentation masks of anatomical images, with both networks being CNNs. We present a general overview of these architectures in Fig. 5. The twin network uses DenseNet:180 a deep NNs that concatenates the outputs at each layer to propagate salient information to deeper layers in the network (refer to Sec. III D for the motivation behind such approaches). More specifically, Tangram uses a pre-trained DenseNet encoder (trained on ImageNet) to encode images and remove technical noise and artifacts. Biancalani et al. also add two additional layers to the pre-trained encoder, which map the outputs to a smaller latent space. The encoder of the twin network is fine-tuned on learning the prediction of spatial depth difference between two images: Two random images are inputted to the twin network with their spatial difference depth being the desired target output, dtrue. The network then tries to predict the depth, dpred, for all N inputs, ultimately comparing them against the corresponding true depth differences, dtrue (as shown in the following equation):
| (17) |
FIG. 5.
Tangram's DL framework for aligning/integrating histology and anatomical data with molecular data. Tangram's model for this task is a combination of encoding (using a twin network) and segmentation modules. The twin network learns a similarity metric for brain sections based on anatomical features in images, while the U-Net model is trained to segment five different classes on mouse brain images. This figure was recreated for this manuscript based on illustrations from Biancalani et al.34
The segmentation model of Tangram generates five custom segmentation masks (background, cortex, cerebellum, white matter, and other gray matter) that are compatible with the existing Allen ontology atlas. The segmentation model is a U-Net, which uses a pre-trained ResNet50 (Ref. 156) as its core. Finally, for each pixel in input images, the model's last layer (a softmax function) assigns a probability of belonging to one of the five segmentation classes. Tangram's segmentation model aims to optimize the superposition of the cross entropy and Jaccard index, as presented in the following equation:
| (18) |
with p denoting the model prediction and g referring to the ground truth image.
Biancalani et al. demonstrate that Tangram learns an accurate mapping between the spatial data and scRNAseq gene expression when applied to fine- or coarse-grained spatial atlases. The authors show that their approach works well across different technologies (namely, ISH, smFISH, Visium, STARmap, and MERFISH) at different resolutions and gene coverage and is able to learn a robust and accurate alignment mapping for the isocortex of the adult healthy mouse brain.34 While Tangram can offer different advantages based on spatial technology, it can produce consistent spatial mappings and overcome limitations in resolution or throughput, which is beneficial in many ST experiments and studies.
C. Spot deconvolution
One downside of using NGS-based technologies remains to be their resolution: Despite the recent technological advancements, most ST platforms (e.g., spatial transcriptomics, Visium, DBiT-seq,181 Nanostring GeoMx,182 and SlideSeq) do not have a single-cell resolution. The number of cells captured in each spot still varies based on the tissues [about 1–10 (Ref. 183)] and the technology used. On the other hand, we cannot assume that all cells within a spot are the same, due to the heterogeneity of the cells. Therefore, it is necessary to use computational approaches for inferring the cell types in each spot or voxel. Such estimations would be possible if there were a complementary scRNAseq dataset. The process of inferring the cellular composition of each spot is known as cell-type deconvolution. Deconvolution has been at the forefront of computational efforts, and it is important in building organ atlases.20,118,184 In fact, cell-type deconvolution is an existing procedure for inferring cell-type composition in RNAseq data using scRNAseq. However, methods developed for bulk RNAseq do not account for the spatial components of ST datasets and are therefore generally inadequate. Given that deconvolution is an existing practice in RNAseq studies, we will refer to the spatial deconvolution problem as spot deconvolution to distinguish between the traditional methods and the ones developed for ST analysis.
We divide spot deconvolution methods into three categories: (i) statistical methods, (ii) machine learning, and (iii) deep learning, with many of the current models falling into the first two categories. We now dive deeper into the two existing models that use DL for performing spot deconvolution.
1. DestVI
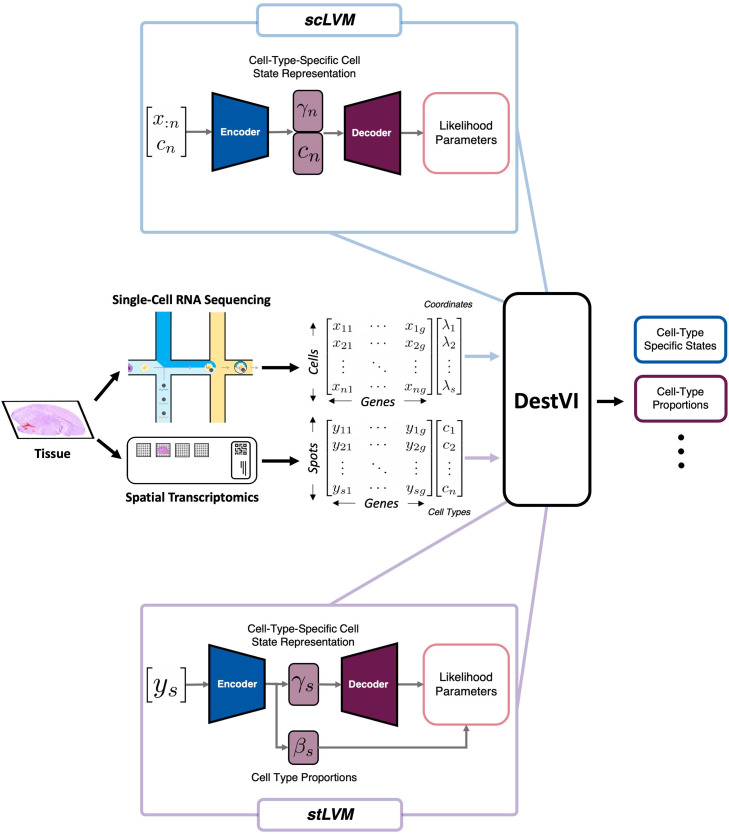
DestVI (deconvolution of spatial transcriptomics profiles using variation inference) is a Bayesian model for spot deconvolution. DestVI employs a conditional deep generative model (similar to scVI,185 a popular model for scRNAseq analysis) to learn cell-type profiles and continuous sub-cell-type variations, aiming to recover the cell-type frequency and the average transcriptions state of cell types at each spot. To do so, DestVI takes a pair of transcriptomics datasets as inputs: (i) a reference scRNA-seq data and (ii) a query spatial transcriptomics data (from the same samples). DestVI then outputs the expected proportion of cell types for every spot and continuous estimation of cell state for the cell types present in each spot, which can be viewed as the average state of the cell types in each spot, which Lopez et al. suggest as useful for downstream analysis and formulation of biological hypotheses.69
DestVI uses two different latent variable models (LVMs) for distinguishing cell-type proportions and delineating cell-type-specific sub-states (shown in Fig. 6). The first LVM is for single-cell data (therefore named scLVM) which assumes the counts follow a negative binomial (NB) distribution, which has shown to model RNAseq count data well.185–187 Specifically, Lopez et al. assume that for each gene g and cell n, the count of observed transcripts, xng, follows an NB distribution parameterized with : is a parameter that depends on the type assigned to the cell cn, the total number of detected molecules ln, and a low-dimensional latent vector γn (which Lopez et al. set ) that describes the variability of cell-type assignment to cell cn, and a neural network f parameters (in this case, a two-layer NN). The second parameter of the NB, pg, is optimized using variational Bayesian inference. We can summarize the assumptions for scLVM as shown in the following equation:
| (19) |
with the latent variable . Each cn (the annotations) are represented by a one-hot encoded vector, which is concatenated with γ to serve as the input of the NN f. Lopez et al. use a VAE to optimize for the marginal conditional likelihood .
FIG. 6.
Visualization of DestVI's computation workflow for spot deconvolution. DestVI uses information from both data modalities of the ST data (coordinates and scRNAseq). DestVI defines two latent variable models (LVMs) for each data modality: an LVM for modeling scRNAseq data (scLVM, shown at the top) and one that aims to model the ST data (stLVM, shown at the bottom). We describe each one in Sec. IV C. This figure was recreated for this manuscript based on illustrations from Lopez et al.69
Finally, for scLVM, a mean-field Gaussian distribution , parameterized by another two-layer NN g, is inferred for each cell which quantifies the cell state and the associated uncertainty. The NN g takes a concatenation of (i) the gene expression vector xn and (ii) the one-hot encoded cell annotations as its inputs. The network g outputs the mean and variance of the variational distribution for γn, obtained through optimizing Eq. (20)
| (20) |
where is the prior likelihood for γn. Similar to training any other VAE, the observations are split into mini-batches, and sampling from the variational distribution is done using the reparameterization trick described in Kingma and Welling.161 The computational workflow for scLVM is visualized in Fig. 6(b).
The second LVM aims to model the spatial transcriptomics data (hence called stLVM) with the assumption that the number of observed transcripts xsg at each spot s for each gene g follows an NB distribution. Additionally, Lopez et al. also assume that each spot has C(s) cells, with each cell n in a spot s being generated from the latent variables . For stLVM's NB distribution, the rate parameter , where αg is a correction factor for the gene-specific bias between spatial and scRNAseq data, lg is the overall number of molecules observed in each spot, and fg is an NN network with parameters . These assumptions and quantities allow Lopez et al. to formulate the total gene expression xsg as shown in the following equation:
| (21) |
Moreover, using a parameter to designate the abundance of every cell type in every spot, βsc, and NB's rate-shape parameterization property (see Aragón et al.188), Eq. (21) can be rewritten as follows:
| (22) |
supposing that cells from a given cell type c in a spot s must come from the same covariate .
The covariate in DestVI allows for the model to account for ST technology discrepancies by assuming various empirical priors (refer to Fig. 6). Lopez et al. simplify the problem of identifying every cell type in each spot to determine the density cell types, i.e., assuming that there cannot be significantly different cell states of the same cell types within a spot. Lopez et al. use a penalized likelihood method to infer point estimates for γc, α, and β. With the additional two strategies to stabilize the training of DestVI, the final objective function for stLVM consists of (i) the negative binomial likelihood, (ii) the likelihood of the empirical prior, and (iii) the variance penalization for α.
Lopez et al. use simulations to present DestVI's ability to provide higher resolution compared to the existing methods and estimate gene expression by every cell type in all spots. Furthermore, they show that DestVI is able to accurately deconvolute spatial organization when applied mouse tumor model. In the cases tested, Lopez et al. demonstrate that DestVI is capable of identifying important cell-type-specific changes in gene expression between different tissue regions or between conditions and that it can provide a high resolution and accurate spatial characterization of the cellular organization of tissues.
2. DSTG
Deconvoluting spatial transcriptomics data through the graph-based convolutional network (DSTG)65 is a recent semi-supervised model that employs graph convolutional networks (GCN)136 for spot deconvolution. DSTG utilizes scRNAseq to construct pseudo-ST data and then build a link graph that represents the similarity between all spots in both real and pseudo-ST data. The pseudo-ST is generated by combining scRNAseq transcriptomics of multiple cells to mimic the expression profiles at each spot; while the real ST data are unlabeled, the pseudo-ST has labels. To construct the link graph, DSTG first reduces the dimensionality of both real and pseudo data using canonical correlation analysis189 and then identifies mutual nearest neighbors.190 Next, a GCN is used on the link graph to propagate the real and pseudo-ST data into a latent space that is turned into a probability distribution of the cell compositions for each spot.
Song et al. form the link graph by taking the number of spots as the number of vertices, resulting in a graph , where denotes the number of spots, and E represents the edges between them. DSTG takes two inputs: (i) the adjacency matrix of graph G, represented by A, and (ii) a combination of both real and pseudo datasets , where m is the number of variable genes, and (the total number of spots in both datasets) with Spseudo and Sreal indicating the number of spots in the pseudo and real ST datasets, respectively. Next, Song et al. normalize the adjacency matrix (for efficient training of DSTG) using the diagonal degree of A, denoted by D, as shown in the following equation:
| (23) |
where (with I denoting the identity matrix). Given the two inputs, DSTG's graph convolution layers take the following form:
| (24) |
with denoting the ith hidden layer of DSTG, and being a non-linear activation function [in this case, ]. The output of DSTG is denoted by , the proportion of cell type at each spot . Song et al. design DSTG's architecture as shown in the following equation:
| (25) |
where k is the last layer, and is the predicted proportions at each spot in the pseudo and real data, denoted by Ypseudo and Yreal, respectively. It is important to note that Song et al. chose a GCN with three layers after performing an ablation study on the number of layers. Finally, DSTG is trained by optimizing the cross entropy loss
| (26) |
with denoting the label for true/predicted cell type t at spot s. Note that this constitutes a semi-supervised training for DSTG since only labels for the pseudo-ST are used in training, but the model will also learn to predict labels for the real dataset as well [refer to Eq. (25)].
Song et al. note that, compared to traditional approaches, DSTG provides three key advantages: (i) given that DSTG uses variable genes and a non-linear GCN, it allows for learning complex deconvolution mappings from the ST data. (ii) The weights assigned to the different cell types in the pseudo-ST and the semi-supervised scheme allow DSTG to identify key features that allow the model to learn the cellular composition in real data. (iii) DSTG's scalability and adaptability will be beneficial in ST analysis, given that the sequencing depth of the ST data is expected to increase. Song et al. show that DSTG consistently outperforms the benchmarked state-of-the-art model (SPOTlight) on both synthetic data and real data. More specifically, DSTG is evaluated on simulated data generated from PBMC where it shows high accuracy between the predicted cell compositions and the true proportions. Song et al. also find that DSTG's deconvolution of the ST data from complex tissues, including mouse cortex, hippocampus, and human pancreatic tumor slices, is consistent with the underlying cellular mixtures.65
D. Spatial clustering
Clustering allows the aggregation of data into subpopulations based on some shared metric of distance or “closeness.” In RNA sequencing studies, clustering is the first step of identifying cell clusters, often followed by laborious manual annotation (e.g., through identifying differentially expressed genes) or some automated workflows.191 Clustering has been a crucial step in many scRNAseq studies, often performed using graph-based community detection algorithms (such as Louvain192 or Leiden193) or more traditional methods (such as K-Means121). Although the scRNAseq techniques can be used in some ST studies (e.g., for multiplexed FISH data where a single-cell resolution is available), the result may be discontinuous or erroneous since the spatial coordinates have not been taken into account.76 Therefore, there is a need for ST-specific methods that can utilize both gene expression and histology data to produce clusters that are coherent, both in gene expression and physical space.
Recently, new frameworks for spatial clustering of the ST data have emerged which utilize both spatial and expression information available. Zhu et al.72 introduced a hidden-Markov random field (HMRF)-based method to model the spatial dependency of gene expression using both the sequencing and imaging-based single-cell transcriptomic profiling technologies. HMRF is a graph-based model used to model the spatial distribution of signals. Using the ST data, Zhu et al. create a grid where neighboring nodes are connected to each other. However, the spatial pattern cannot be observed directly (since it is “hidden”), and it must be inferred through observations that depend on the hidden states probabilistically. Similar to Zhu et al., BayesSpace75 employs a Bayesian formulation of HMRF and uses the Markov chain Monte Carlo (MCMC) algorithm to estimate the model parameters. Despite the ability of these methods to cluster voxels (or cells) into distinct subpopulations, these approaches suffer from the lack of versatility required to handle different modalities present in the ST data.76
With the emergence of newer technologies, the scale and variability within datasets are increasing, requiring more general and flexible models for accurate and robust analysis of these studies. A few ML-based approaches have been proposed to combat some of these mentioned challenges. Below, we review the ML-based methods that offer scalability and are generally more applicable to various experimental settings.
1. SpaCell
SpaCell73 is a double-stream DL framework that utilizes both histology images and the associated spot gene counts. For the histology data, Tan et al. first preprocess the images (removing low-quality images, stain normalization, normalizing the pixels using a z-transform, and removing background noise). Next, they split each histology image into tiles that contain one spot each (sub-images of 299 × 299 pixels). For the preprocessing of the count matrix (which contains the reads at each spot), Tan et al. follow traditional scRNAseq preprocessing workflows, including count normalization, removal of outlier genes, and cells with too few genes. After the preprocessing stage, each tile (containing the image of a spot) corresponds to a column in the count matrix (reads from the same spot). At this point, each image , and the count matrix M will be in a space. However, Tan et al. reduce the count matrix to only contain 2048 most variable genes at each spot, therefore resulting in a new count matrix . Let us denote the spot of as , which has a corresponding image xi.
In order to spatially cluster cells of the same type, both image and count data must be used. The first step in SpaCell is to pass on the spot images, xi, to a pre-trained ResNet50 (trained on ImageNet data) in order to output feature vectors, (each having the same dimensionality as columns of ). Next, to extract features from both modalities, SpaCell uses two separate AEs for the image feature vectors, , and the most-variable-genes counts, , with both AEs having the same latent dimension (we discuss the reason behind this later). Let us denote the AE for images as , and the gene counts AE as , with and indicating the encoder and decoders, respectively.
Given N spots, each AE in SpaCell aims to minimize three objective functions for their respective inputs [i.e., for and for ]: (i) the mean squared error (MSE) between the input and the output [shown in Eq. (27)], (ii) the KL divergence between the probability distributions for the input and constructed output of all N spots [denoted by p and q in Eq. (28), respectively], and (iii) binary cross entropy (BCE), shown in Eq. (29)
| (27) |
| (28) |
| (29) |
Once training has concluded, SpaCell encodes both images and gene counts, i.e., and , to be used for clustering. More specifically, clustering is performed on a matrix that is the concatenation of the latent vectors produced by each AE, . This is why the latent spaces of have the same dimension. After obtaining the concatenated matrix, the downstream clustering is performed using K-Means (which can be substituted for other algorithms as well). Through this procedure, SpaCell uses both data modalities and can produce clusters that are highly accurate when compared to the true clusters (annotated by pathologists).
2. StLearn
StLearn74 is a DL model that, among many of its capabilities, can perform segment (cluster) spatial regions based on their function and expression. StLearn's DL component lies within its spatial morphological gene expression (SME) normalization. The SME normalization aims to combine critical information from Hematoxylin and Eosin stained (H and E) tissue images and transcriptome-wide gene expression profiles to then take advantage of in downstream analysis, such as clustering, spatial trajectory inference, and cell–cell interaction.
The SME normalization procedure includes (i) spatial location: In order to use the spatial positions for selecting neighboring spot pairs, Pham et al. consider two spots si and sj as neighbors if the center-to-center Euclidean distance between the two spots, , is less than a specified distance r, i.e., . Pham et al. include all paired spots si and sj as the input to adjust for the gene expression of the center spot si. The next step in SME normalization is (ii) Morphological similarity: stLearn calculates the morphological similarity between spots using feature vectors produced by an ImageNet-pre-trained ResNet50. More specifically, all H&E images corresponding to each spot si are inputted to the ResNet50 model, producing a feature vector . Subsequently, stLearn performs PCA on each feature vector xi, resulting in reduced-dimension feature vectors . To calculate the morphological distance (MD) between two neighboring spots si and sj [according to the criterion defined in (i)], Pham et al. measure the cosine similarity between two reduced feature vectors [refer to Eq. (15) for the definition of cosine similarity]; this MD is shown in the following equation:
| (30) |
As the last step in SME normalization, the gene expression at each spot si is normalized using the MD distance, as shown in the following equation:
| (31) |
where GEi denotes raw gene expression counts at spot si, and n is the total number of neighbors identified for spot si. StLearn's SME normalization enables a smoothing of the expression data based on morphological similarity with the goal of improving clustering through grouping morphologically similar sections of the tissue together.
After the SME normalization step, stLearn's clustering algorithm consists of two steps: First, finding general global clusters (utilizing traditional clustering techniques) using SME-normalized data as its input, followed by local clustering that aims to identify spatially separated sub-clusters and merge spots that are collocated, resulting in spatially contiguous clusters. Pham et al. note that stLearn constitutes the first-of-its-kind method to use tissue morphology in normalizing gene expression, outperforming two competing approaches on two tested data (Seurat on Allen Mouse Brain Atlas194 and SpatialLIBD195 on human brain tissue sections196).74
3. SpaGCN
SpaGCN76 is a graph convolution network (GCN) that integrates both spatial information and histology images to perform spatial clustering. Using each spot as vertices, Hu et al. create a weighted undirected graph, , where is the total number of spots, and E is the set of edges with prescribed weights representing the similarity between the nodes. The weight of each of these edges is determined by (i) the distance between the two spots (nodes) that the edge connects and (ii) the associated histology information (in this case, pixel intensity). This means that two spots are deemed similar if they are physically close to one another, and they seem similar in the histology image.
In order to attribute the pixel information to each spot, Hu et al. use the mean RGB pixel intensity of each spot within a window of size 50 × 50 pixels. That is, given a spot s with physical coordinates (xs, ys) and pixel coordinates , SpaGCN calculates the mean and variance of all the pixels present within a 50 × 50 pixels centered at . Let psr, psg and psb denote the means, and refer to the variance for the red, green, and blue channels, respectively. SpaGAN then summarizes the pixel mean and variance information as a unified value, as shown in the following equation:
| (32) |
Furthermore, zs is rescaled using the mean and standard deviation of each coordinate (including the newly created z axis), with an additional scaling factor that can put more emphasis on histology data when needed. Let μz denote the mean of zs, and let be the standard deviation of with ; then, we can formulate the rescaling as the following:
| (33) |
where α denotes the scaling factor described previously (α = 1 by default).
Using the rescaled value in Eq. (33), the weight of each edge between two vertices s and k is calculated as shown in the following equation:
| (34) |
where l denotes the characteristic length scale, and d(s, k) is the traditional Euclidean distance, as shown in the following equation:
| (35) |
SpaGCN's network construction (and backpropagation) is similar to other GCNs, inspired by Kipf and Welling136 (for an overview of GCNs, refer to Sec. IV C 2). The network intakes the adjacency matrix A to represent the graph G, and a reduced-dimension representation of the gene expression matrix, which Hu et al. achieve using PCA with 50 principal components. The output of the GCN network is a matrix that includes combined information on histology, gene expression, and spatial position. SpaGCN then uses the output of the GCN to perform unsupervised clustering of the spatial data.
SpaGCN uses the Louvain algorithm (an iterative unsupervised clustering algorithm) on the output of GCN to initialize cluster centroids, with the number of clusters (controlled by Louvain's resolution parameter) being optimized on maximizing the Silhouette score197). The iterative updates are based on optimizing a metric that defines the distance between each spot and all cluster centroids using the t-distribution as a kernel. For a centroid cj, a total of N clusters, and the embedded point hi for spot i, this metric can be defined as the probability of assigning cell i to cluster j, as shown in the following equation:
| (36) |
Hu et al. further refine the clusters using an auxiliary target distribution [shown in Eq. (37)] that prefers spots assignments with the highest confidence and normalizes the centroid contribution to the overall loss function as the following:
| (37) |
Finally, spaGCN is trained by optimizing the divergence between the p and q distributions, as shown in the following equation:
| (38) |
Hu et al. demonstrate that SpaGCN can accurately identify spatial clusters that are consistent with manual annotations since SpaGCN utilizes information from both gene expression and histology. The authors perform spatial clustering with SpaGCN on the human dorsolateral prefrontal cortex, human primary pancreatic cancer, and multiple mouse tissue data, showing that SpaGCN performs consistently well, outperforming other state-of-the-art models (stLearn, BayesSpace, and Louvain). These results show the feasibility and potential of SpaGCN for clustering spatial-resolved transcriptomics.
E. Cell–cell interactions
Multicellular organisms depend on intricate cell–cell interactions (CCIs) which dictate cellular development, homeostasis, and single-cell functions.198 Unraveling such interaction within tissues can present unique insight on complex biological processes and disease pathogenesis.198–201 CCI has been investigated using both scRNAseq and RNAseq, wherein most approaches test for enrichment in ligand–receptor profiles in the expression data.202–204 However, the ST data can offer a more comprehensive view of CCI, since the distance traveled by ligand signal is crucial in determining the type of cell–cell signaling.183 Given the importance of CCI and the advantages that the ST data provide, several computational approaches for inferring cellular interactions using the ST data have been developed, such as SpaOTsc,80 Giotto,82 MISTy.81
SpaOTsc is a model that can be used in integrating scRNAseq data with spatial measurements, and in inferring cellular interactions in spatial-resolved transcriptomics data. SpaOTsc aims to estimate cellular interactions by analyzing the relationships between ligand–receptor pairs and their downstream genes. SpaOTsc formulates a spatial metric using the optimal transport algorithm, returning a mapping that contains the probability distribution of each scRNA-seq cell over a spatial region. SpaOTsc also utilizes a random forest in order to infer the spatial range of ligand–receptor signaling and subsequently remove the long-distance connections. Another approach is Giotto:82 Giotto is an extended and comprehensive toolbox designed for ST analysis and visualization, which includes a CCI model that calculates an enrichment score (the weighted mean expression of a ligand and the corresponding receptor in the two neighboring cells). Giotto then constructs an empirical null distribution by moving the locations for each cell type, subsequently calculating the corresponding statistical significance (P-value), and ordering the ligand–receptors pair-wise for all neighboring cells. Although the mentioned models have shown to discover simple cellular interactions, such approaches often fail to detect complex gene–gene interactions, which is essential in understanding many diseases. Though DL models for CCI are nascent, we anticipate such models to better learn complex interactions from the raw data.
V. CONCLUSIONS AND OUTLOOK
The ST field is rapidly growing, with new datasets and analysis pipelines released weekly. The innovations in biological methods will continue to spur creativity in algorithm development, with an emphasis on ML-based frameworks. Although the space of DL models for ST analysis is currently small, we anticipate the field to experience a paradigm shift toward deep-learned models. In this review, our goal was to provide readers with the necessary biological, mathematical, and computational background for understanding the existing approaches and expanding upon the current models to address the challenges posed by the ST domain.
In this manuscript, we provided an overview of current DL-based techniques for alignment and integration of the ST data, spatial clustering, spot deconvolution, inferring cell–cell communication, and approaches for reconstructing spatial coordinates using scRNAseq data (with limited or no spatial reference atlas). The DL methods we presented, in comparison to their conventional counterparts, offer accuracy and scalability advantages. However, DL methods are not always the preferred choice as they are computationally expensive and may lack biological interpretability. As more methods for ST analysis are developed, we believe that standard datasets for benchmarking new models as well as comprehensive accuracy and efficiency analysis of existing techniques will be of significant value to the field. Though the existing methods set the new state-of-the-art in their respective categories, room for improvements remains large. Among the ST downstream analyses, applications of DL algorithms for studying cell–cell communication and identification of spatially variable genes remain mostly underexplored. Given DL models' ability to extract sophisticated patterns from the raw data, we anticipate that DL approaches will prove useful in unraveling complex biological processes, aiding the efforts in identifying cellular interactions and highly variable genes in a spatial context.
Recent technological advancements have enabled researchers to utilize various single-cell omics sources to construct multi-omics datasets, providing comprehensive view of many diseases [e.g., COVID19 (Refs. 205 and 206) and cancer207] and developmental processes.208,209 As single-cell analysis enters the multi-omics age, the need for integrating the ST data with other single-cell sources will increase. Therefore, we expect an increase in ML-based frameworks for data integration and alignment, spearheaded by DL-based approaches. Additionally, due to the noise and multi-modality of the ST data, there exists an unmet need for methods that account for batch effects in spatial and gene expression data. Given the success of DL techniques for batch effect removal in scRNAseq, we foresee DL models being widely used for batch effect correction of spatially resolved transcriptomics data.
Despite the recentness of ST technologies, researchers have successfully used these technologies to generate spatially resolved cell atlases, providing new insight on a wide range of biological processes and organs.118,210–213 Such studies not only show the tremendous potential that ST technologies hold but also highlight the need for scalable and efficient analysis tools. The application of DL to ST analysis remains a rapidly evolving nascent domain, demonstrating promising great prospects in advancing the field of ST, and the integration of ST datasets with other omics data.
ACKNOWLEDGMENTS
We wish to acknowledge O. Davalos for offering fruitful discussions and providing useful insight on earlier versions of this work. We also would like to thank M. Hilscher and T. Buvoli for providing valuable feedback on earlier drafts of this manuscript. A.A.H. is supported by the National Human Genome Research Institute of the National Institutes of Health under Award No. F31HG012718 (National Research Service Award). Additionally, the authors received support from the National Institutes of Health (Nos. R15-HL146779 and R01-GM126548) and the National Science Foundation (No. DMS-1840265).
The Visium slide and its visualizations in Figs. 1 and 3 were accessed from 10× Genomics.214 The mouse brain histology image in Fig. 1 was taken from Ref. 215. The illustration of mouse brain in Fig. 2 was obtained from BioRender.216
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Author Contributions
A.A.H. and S.S.S. wrote the manuscript, edited the drafts, and conceptualized the figures, which A.A.H then created.
A. Ali Heydari: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Validation (equal); Visualization (equal); Writing – original draft (equal); Writing – review & editing (equal). Suzanne Sindi: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Funding acquisition (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Supervision (equal); Validation (equal); Visualization (equal); Writing – original draft (equal); Writing – review & editing (equal).
DATA AVAILABILITY
The data that support the findings of this study are available within the article.
References
- 1. Larsson L., Frisén J., and Lundeberg J., “ Spatially resolved transcriptomics adds a new dimension to genomics,” Nat. Methods 18, 15–18 (2021). 10.1038/s41592-020-01038-7 [DOI] [PubMed] [Google Scholar]
- 2. Packer J. S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J., Waterston R. H., and Murray J. I., “ A lineage-resolved molecular atlas of c. elegans embryogenesis at single-cell resolution,” Science 365, eaax1971 (2019). 10.1126/science.aax1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F., Huang D., Xu Y., Huang W., Jiang M., Jiang X., Mao J., Chen Y., Lu C., Xie J., Fang Q., Wang Y., Yue R., Li T., Huang H., Orkin S. H., Yuan G.-C., Chen M., and Guo G., “ Mapping the mouse cell atlas by microwell-seq,” Cell 172, 1091–1107 (2018). 10.1016/j.cell.2018.02.001 [DOI] [PubMed] [Google Scholar]
- 4. Schaum N., Karkanias J., Neff N. F., May A. P., Quake S. R., Wyss-Coray T., Darmanis S., Batson J., Botvinnik O., Chen M. B., Chen S., Green F., Jones R. C., Maynard A., Penland L., Pisco A. O., Sit R. V., Stanley G. M., Webber J. T., Zanini F., Baghel A. S., Bakerman I., Bansal I., Berdnik D., Bilen B., Brownfield D., Cain C., Chen M. B., Cho M., Cirolia G., Conley S. D., Demers A., Demir K., de Morree A., Divita T., du Bois H., Dulgeroff L. B. T., Ebadi H., Espinoza F. H., Fish M., Gan Q., George B. M., Gillich A., Genetiano G., Gu X., Gulati G. S., Hang Y., Hosseinzadeh S., Huang A., Iram T., Isobe T., Ives F., Jones R. C., Kao K. S., Karnam G., Kershner A. M., Kiss B. M., Kong W., Kumar M. E., Lam J. Y., Lee D. P., Lee S. E., Li G., Li Q., Liu L., Lo A., Lu W.-J., Manjunath A., May A. P., May K. L., May O. L., McKay M., Metzger R. J., Mignardi M., Min D., Nabhan A. N., Neff N. F., Ng K. M., Noh J., Patkar R., Peng W. C., Puccinelli R., Rulifson E. J., Sikandar S. S., Sinha R., Sit R. V., Szade K., Tan W., Tato C., Tellez K., Travaglini K. J., Tropini C., Waldburger L., van Weele L. J., Wosczyna M. N., Xiang J., Xue S., Youngyunpipatkul J., Zardeneta M. E., Zhang F., Zhou L., May A. P., Neff N. F., Sit R. V., Castro P., Croote D., DeRisi J. L., Stanley G. M., Webber J. T., Baghel A. S., Chen M. B., Espinoza F. H., George B. M., Gulati G. S., Kershner A. M., Kiss B. M., Kuo C. S., Lam J. Y., Lehallier B., Nabhan A. N., Ng K. M., Nguyen P. K., Rulifson E. J., Sikandar S. S., Tan S. Y., Travaglini K. J., van Weele L. J., Wang B. M., Wosczyna M. N., Yousef H., May A. P., Quake S. R., Stanley G. M., Webber J. T., Beachy P. A., Chan C. K. F., George B. M., Gulati G. S., Huang K. C., Kershner A. M., Kiss B. M., Nabhan A. N., Ng K. M., Nguyen P. K., Rulifson E. J., Sikandar S. S., Travaglini K. J., Wang B. M., Weinberg K., Wosczyna M. N., Wu S. M., Barres B. A., Beachy P. A., Chan C. K. F., Clarke M. F., Kim S. K., Krasnow M. A., Kumar M. E., Kuo C. S., May A. P., Metzger R. J., Neff N. F., Nusse R., Nguyen P. K., Rando T. A., Sonnenburg J., Wang B. M., Weissman I. L., Wu S. M., Quake S. R., T. T. M. Consortium, O. coordination, L. coordination, O. collection, processing, L. preparation, sequencing, C. data analysis, C. type annotation, W. Group, S. text writing Group, and P. investigators, “ Single-cell transcriptomics of 20 mouse organs creates a tabula muris,” Nature 562, 367–372 (2018). 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Regev A., Teichmann S. A., Lander E. S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., Clevers H., Deplancke B., Dunham I., Eberwine J., Eils R., Enard W., Farmer A., Fugger L., Gottgens B., Hacohen N., Haniffa M., Hemberg M., Kim S., Klenerman P., Kriegstein A., Lein E., Linnarsson S., Lundberg E., Lundeberg J., Majumder P., Marioni J. C., Merad M., Mhlanga M., Nawijn M., Netea M., Nolan G., Pe'er D., Phillipakis A., Ponting C. P., Quake S., Reik W., Rozenblatt-Rosen O., Sanes J., Satija R., Schumacher T. N., Shalek A., Shapiro E., Sharma P., Shin J. W., Stegle O., Stratton M., Stubbington M. J. T., Theis F. J., Uhlen M., van Oudenaarden A., Wagner A., Watt F., Weissman J., Wold B., Xavier R., and Yosef N., “ The human cell atlas,” Elife 6, e27041 (2017). 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davie K., Janssens J., Koldere D., De Waegeneer M., Pech U., Kreft Ł., Aibar S., Makhzami S., Christiaens V., Bravo González-Blas C., Poovathingal S., Hulselmans G., Spanier K. I., Moerman T., Vanspauwen B., Geurs S., Voet T., Lammertyn J., Thienpont B., Liu S., Konstantinides N., Fiers M., Verstreken P., and Aerts S., “ A single-cell transcriptome atlas of the aging drosophila brain,” Cell 174, 982–998 (2018). 10.1016/j.cell.2018.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Y., Wang D., Peng M., Tang L., Ouyang J., Xiong F., Guo C., Tang Y., Zhou Y., Liao Q., Wu X., Wang H., Yu J., Li Y., Li X., Li G., Zeng Z., Tan Y., and Xiong W., “ Single-cell RNA sequencing in cancer research,” J. Exp. Clin. Cancer Res. 40, 81 (2021). 10.1186/s13046-021-01874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Derakhshani A., Asadzadeh Z., Safarpour H., Leone P., Shadbad M. A., Heydari A., Baradaran B., and Racanelli V., “ Regulation of CTLA-4 and PD-l1 expression in relapsing-remitting multiple sclerosis patients after treatment with fingolimod, IFNβ-1α, glatiramer acetate, and dimethyl fumarate drugs,” J. Pers. Med. 11, 721 (2021). 10.3390/jpm11080721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kusnadi A., Ramírez-Suástegui C., Fajardo V., Chee S. J., Meckiff B. J., Simon H., Pelosi E., Seumois G., Ay F., Vijayanand P., and Ottensmeier C. H., “ Severely ill patients with COVID-19 display impaired exhaustion features in SARS-CoV-2–reactive cd8+ T cells,” Sci. Immunol. 6, eabe4782 (2021). 10.1126/sciimmunol.abe4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Erfanian N., Heydari A. A., Iañez P., Derakhshani A., Ghasemigol M., Farahpour M., Nasseri S., Safarpour H., and Sahebkar A., “ Deep learning applications in single-cell omics data analysis,” bioRxiv (2021). 10.1101/2022.02.28.482392 [DOI] [PubMed] [Google Scholar]
- 11. Park S. R., Namkoong S., Friesen L., Cho C.-S., Zhang Z. Z., Chen Y.-C., Yoon E., Kim C. H., Kwak H., Kang H. M., and Lee J. H., “ Single-cell transcriptome analysis of colon cancer cell response to 5-fluorouracil-induced DNA damage,” Cell Rep. 32, 108077 (2020). 10.1016/j.celrep.2020.108077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Su Y., Chen D., Yuan D., Lausted C., Choi J., Dai C. L., Voillet V., Duvvuri V. R., Scherler K., Troisch P., Baloni P., Qin G., Smith B., Kornilov S. A., Rostomily C., Xu A., Li J., Dong S., Rothchild A., Zhou J., Murray K., Edmark R., Hong S., Heath J. E., Earls J., Zhang R., Xie J., Li S., Roper R., Jones L., Zhou Y., Rowen L., Liu R., Mackay S., O'Mahony D. S., Dale C. R., Wallick J. A., Algren H. A., Zager M. A., Wei W., Price N. D., Huang S., Subramanian N., Wang K., Magis A. T., Hadlock J. J., Hood L., Aderem A., Bluestone J. A., Lanier L. L., Greenberg P. D., Gottardo R., Davis M. M., Goldman J. D., and Heath J. R., “ Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19,” Cell 183, 1479–1495 (2020). 10.1016/j.cell.2020.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iqbal F., Lupieri A., Aikawa M., and Aikawa E., “ Harnessing single-cell RNA sequencing to better understand how diseased cells behave the way they do in cardiovascular disease,” Arteriosclerosis Thrombosis Vasc. Biol. 41, 585–600 (2021). 10.1161/ATVBAHA.120.314776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heming M., Li X., Rauber S., Mausberg A. K., Borsch A.-L., Hartlehnert M., Singhal A., Lu I.-N., Fleischer M., Szepanowski F., Witzke O., Brenner T., Dittmer U., Yosef N., Kleinschnitz C., Wiendl H., Stettner M., and Horste G. M. Z., “ Neurological manifestations of COVID-19 feature t cell exhaustion and dedifferentiated monocytes in cerebrospinal fluid,” Immunity 54, 164–175 (2021). 10.1016/j.immuni.2020.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., Li N., Szpankowski L., Fowler B., Chen P., Ramalingam N., Sun G., Thu M., Norris M., Lebofsky R., Toppani D., Kemp D. W., Wong M., Clerkson B., Jones B. N., Wu S., Knutsson L., Alvarado B., Wang J., Weaver L. S., May A. P., Jones R. C., Unger M. A., Kriegstein A. R., and West J. A. A., “ Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex,” Nat. Biotechnol. 32, 1053–1058 (2014). 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Treutlein B., Brownfield D. G., Wu A. R., Neff N. F., Mantalas G. L., Espinoza F. H., Desai T. J., Krasnow M. A., and Quake S. R., “ Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq,” Nature 509, 371–375 (2014). 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barresi M. J. and Gilbert S. F., Developmental Biology ( Oxford University Press, 2019). [Google Scholar]
- 18. Dries R., Chen J., Del Rossi N., Khan M. M., Sistig A., and Yuan G.-C., “ Advances in spatial transcriptomic data analysis,” Genome Res. 31, 1706–1718 (2021). 10.1101/gr.275224.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noel T., Wang Q. S., Greka A., and Marshall J. L., “ Principles of spatial transcriptomics analysis: A practical walk-through in kidney tissue,” Front. Physiol. 12, 809346 (2022). 10.3389/fphys.2021.809346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao A., Barkley D., França G. S., and Yanai I., “ Exploring tissue architecture using spatial transcriptomics,” Nature 596, 211–220 (2021). 10.1038/s41586-021-03634-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ke R., Mignardi M., Pacureanu A., Svedlund J., Botling J., Wahlby C., and Nilsson M., “ In situ sequencing for RNA analysis in preserved tissue and cells,” Nat. Methods 10, 857–860 (2013). 10.1038/nmeth.2563 [DOI] [PubMed] [Google Scholar]
- 22. Codeluppi S., Borm L. E., Zeisel A., Manno G. L., van Lunteren J. A., Svensson C. I., and Linnarsson S., “ Spatial organization of the somatosensory cortex revealed by cyclic smFISH,” bioRxiv (2018). 10.1101/276097 [DOI] [PubMed] [Google Scholar]
- 23. Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., and Tyagi S., “ Imaging individual mRNA molecules using multiple singly labeled probes,” Nat. Methods 5, 877–879 (2008). 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Femino A. M., Fay F. S., Fogarty K., and Singer R. H., “ Visualization of single RNA transcripts in situ,” Science 280, 585–590 (1998). 10.1126/science.280.5363.585 [DOI] [PubMed] [Google Scholar]
- 25. Alon S., Goodwin D. R., Sinha A., Wassie A. T., Chen F., Daugharthy E. R., Bando Y., Kajita A., Xue A. G., Marrett K., Prior R., Cui Y., Payne A. C., Yao C.-C., Suk H.-J., Wang R., Yu C.-C. J., Tillberg P., Reginato P., Pak N., Liu S., Punthambaker S., Iyer E. P. R., Kohman R. E., Miller J. A., Lein E. S., Lako A., Cullen N., Rodig S., Helvie K., Abravanel D. L., Wagle N., Johnson B. E., Klughammer J., Slyper M., Waldman J., Jané-Valbuena J., Rozenblatt-Rosen O., Regev A., Consortium I., Church G. M., Marblestone A. H., Boyden E. S., Ali H. R., Sa'd M. A., Alon S., Aparicio S., Battistoni G., Balasubramanian S., Becker R., Bodenmiller B., Boyden E. S., Bressan D., Bruna A., Burger M., Caldas C., Callari M., Cannell I. G., Casbolt H., Chornay N., Cui Y., Dariush A., Dinh K., Emenari A., Eyal-Lubling Y., Fan J., Fatemi A., Fisher E., González-Solares E. A., González-Fernández C., Goodwin D., Greenwood W., Grimaldi F., Hannon G. J., Harris O., Harris S., Jauset C., Joyce J. A., Karagiannis E. D., Kovačević T., Kuett L., Kunes R., Yoldaş A. K., Lai D., Laks E., Lee H., Lee M., Lerda G., Li Y., McPherson A., Millar N., Mulvey C. M., Nugent F., O'Flanagan C. H., Paez-Ribes M., Pearsall I., Qosaj F., Roth A. J., Rueda O. M., Ruiz T., Sawicka K., Sepúlveda L. A., Shah S. P., Shea A., Sinha A., Smith A., Tavaré S., Tietscher S., Vázquez-García I., Vogl S. L., Walton N. A., Wassie A. T., Watson S. S., Weselak J., Wild S. A., Williams E., Windhager J., Whitmarsh T., Xia C., Zheng P., and Zhuang X., “ Expansion sequencing: Spatially precise in situ transcriptomics in intact biological systems,” Science 371, eaax2656 (2021). 10.1126/science.aax2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Codeluppi S., Borm L. E., Zeisel A., La Manno G., van Lunteren J. A., Svensson C. I., and Linnarsson S., “ Spatial organization of the somatosensory cortex revealed by osmFISH,” Nat. Methods 15, 932–935 (2018). 10.1038/s41592-018-0175-z [DOI] [PubMed] [Google Scholar]
- 27. Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., and Zhuang X., “ Spatially resolved, highly multiplexed RNA profiling in single cells,” Science 348, aaa6090 (2015). 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eng C.-H. L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G.-C., and Cai L., “ Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH+,” Nature 568, 235–239 (2019). 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang X., Allen W. E., Wright M. A., Sylwestrak E. L., Samusik N., Vesuna S., Evans K., Liu C., Ramakrishnan C., Liu J., Nolan G. P., Bava F.-A., and Deisseroth K., “ Three-dimensional intact-tissue sequencing of single-cell transcriptional states,” Science 361, eaat5691 (2018). 10.1126/science.aat5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.10x Genomics, see https://www.10xgenomics.com/spatial-transcriptomics for “ Spatial transcriptomics,” 2021.
- 31. Ståhl P. L., Salmén F., Vickovic S., Lundmark A., Navarro J. F., Magnusson J., Giacomello S., Asp M., Westholm J. O., Huss M., Mollbrink A., Linnarsson S., Codeluppi S., Borg Å., Pontén F., Costea P. I., Sahlén P., Mulder J., Bergmann O., Lundeberg J., and Frisén J., “ Visualization and analysis of gene expression in tissue sections by spatial transcriptomics,” Science 353, 78–82 (2016). 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 32. Rodriques S. G., Stickels R. R., Goeva A., Martin C. A., Murray E., Vanderburg C. R., Welch J., Chen L. M., Chen F., and Macosko E. Z., “ Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution,” Science 363, 1463–1467 (2019). 10.1126/science.aaw1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vickovic S., Eraslan G., Salmén F., Klughammer J., Stenbeck L., Schapiro D., Aijo T., Bonneau R., Bergenstråhle L., Navarro J., Gould J., Griffin G. K., Borg Å., Ronaghi M., Frisén J., Lundeberg J., Regev A., and Ståhl P. L., “ High-definition spatial transcriptomics for in situ tissue profiling,” Nat. Methods 16, 987–990 (2019). 10.1038/s41592-019-0548-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Biancalani T., Scalia G., Buffoni L., Avasthi R., Lu Z., Sanger A., Tokcan N., Vanderburg C. R., Segerstolpe Å., Zhang M., Avraham-Davidi I., Vickovic S., Nitzan M., Ma S., Subramanian A., Lipinski M., Buenrostro J., Brown N. B., Fanelli D., Zhuang X., Macosko E. Z., and Regev A., “ Deep learning and alignment of spatially resolved single-cell transcriptomes with tangram,” Nat. Methods 18, 1352–1362 (2021). 10.1038/s41592-021-01264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salmén F., Ståhl P. L., Mollbrink A., Navarro J., Vickovic S., Frisén J., and Lundeberg J., “ Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections,” Nat. Protoc. 13, 2501–2534 (2018). 10.1038/s41596-018-0045-2 [DOI] [PubMed] [Google Scholar]
- 36. Jemt A., Salmén F., Lundmark A., Mollbrink A., Fernández Navarro J., Ståhl P. L., Yucel-Lindberg T., and Lundeberg J., “ An automated approach to prepare tissue-derived spatially barcoded RNA-sequencing libraries,” Sci. Rep. 6, 37137 (2016). 10.1038/srep37137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tustison N. J., Cook P. A., Klein A., Song G., Das S. R., Duda J. T., Kandel B. M., van Strien N., Stone J. R., Gee J. C., and Avants B. B., “ Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements,” NeuroImage 99, 166–179 (2014). 10.1016/j.neuroimage.2014.05.044 [DOI] [PubMed] [Google Scholar]
- 38. Balakrishnan G., Zhao A., Sabuncu M. R., Guttag J., and Dalca A. V., “ VoxelMorph: A learning framework for deformable medical image registration,” IEEE Trans. Med. Imaging 38, 1788–1800 (2019). 10.1109/TMI.2019.2897538 [DOI] [PubMed] [Google Scholar]
- 39. LeCun Y., Bengio Y., and Hinton G., “ Deep learning,” Nature 521, 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 40. Eraslan G., Simon L. M., Mircea M., Mueller N. S., and Theis F. J., “ Single-cell RNA-seq denoising using a deep count autoencoder,” Nat. Commun. 10, 390 (2019). 10.1038/s41467-018-07931-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Badsha M. B., Li R., Liu B., Li Y. I., Xian M., Banovich N. E., and Fu A. Q., “ Imputation of single-cell gene expression with an autoencoder neural network,” Quant. Biol. 8, 78–94 (2020). 10.1007/s40484-019-0192-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li X., Wang K., Lyu Y., Pan H., Zhang J., Stambolian D., Susztak K., Reilly M. P., Hu G., and Li M., “ Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis,” Nat. Commun. 11, 2338 (2020). 10.1038/s41467-020-15851-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu J., Li X., Hu G., Lyu Y., Susztak K., and Li M., “ Iterative transfer learning with neural network for clustering and cell type classification in single-cell RNA-seq analysis,” Nat. Mach. Intell. 2, 607–618 (2020). 10.1038/s42256-020-00233-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shao X., Yang H., Zhuang X., Liao J., Yang P., Cheng J., Lu X., Chen H., and Fan X., “ scDEEPSort: A pre-trained cell-type annotation method for single-cell transcriptomics using deep learning with a weighted graph neural network,” Nucl. Acids Res. 49, e122 (2021). 10.1093/nar/gkab775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma F. and Pellegrini M., “ ACTINN: Automated identification of cell types in single cell RNA sequencing,” Bioinformatics 36, 533–538 (2020). 10.1093/bioinformatics/btz592 [DOI] [PubMed] [Google Scholar]
- 46. Heydari A. A., Davalos O. A., Zhao L., Hoyer K. K., and Sindi S. S., “ ACTIVA: Realistic single-cell RNA-seq generation with automatic cell-type identification using introspective variational autoencoders,” Bioinformatics 38, 2194–2201 (2022). 10.1093/bioinformatics/btac095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marouf M., Machart P., Bansal V., Kilian C., Magruder D. S., Krebs C. F., and Bonn S., “ Realistic in silico generation and augmentation of single-cell RNA-seq data using generative adversarial networks,” Nat. Commun. 11, 166 (2020). 10.1038/s41467-019-14018-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bayoudh K., Knani R., Hamdaoui F., and Mtibaa A., “ A survey on deep multimodal learning for computer vision: Advances, trends, applications, and datasets,” Visual Comput. 38, 2939–2970 (2021). 10.1007/s00371-021-02166-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Butler D., Mozsary C., Meydan C., Foox J., Rosiene J., Shaiber A., Danko D., Afshinnekoo E., MacKay M., Sedlazeck F. J., Ivanov N. A., Sierra M., Pohle D., Zietz M., Gisladottir U., Ramlall V., Sholle E. T., Schenck E. J., Westover C. D., Hassan C., Ryon K., Young B., Bhattacharya C., Ng D. L., Granados A. C., Santos Y. A., Servellita V., Federman S., Ruggiero P., Fungtammasan A., Chin C.-S., Pearson N. M., Langhorst B. W., Tanner N. A., Kim Y., Reeves J. W., Hether T. D., Warren S. E., Bailey M., Gawrys J., Meleshko D., Xu D., Couto-Rodriguez M., Nagy-Szakal D., Barrows J., Wells H., O'Hara N. B., Rosenfeld J. A., Chen Y., Steel P. A. D., Shemesh A. J., Xiang J., Thierry-Mieg J., Thierry-Mieg D., Iftner A., Bezdan D., Sanchez E., Campion T. R., Sipley J., Cong L., Craney A., Velu P., Melnick A. M., Shapira S., Hajirasouliha I., Borczuk A., Iftner T., Salvatore M., Loda M., Westblade L. F., Cushing M., Wu S., Levy S., Chiu C., Schwartz R. E., Tatonetti N., Rennert H., Imielinski M., and Mason C. E., “ Shotgun transcriptome, spatial omics, and isothermal profiling of SARS-CoV-2 infection reveals unique host responses, viral diversification, and drug interactions,” Nat. Commun. 12, 1660 (2021). 10.1038/s41467-021-21361-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Delorey T. M., Ziegler C. G. K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., Abbondanza D., Fleming S. J., Subramanian A., Montoro D. T., Jagadeesh K. A., Dey K. K., Sen P., Slyper M., Pita-Juárez Y. H., Phillips D., Biermann J., Bloom-Ackermann Z., Barkas N., Ganna A., Gomez J., Melms J. C., Katsyv I., Normandin E., Naderi P., Popov Y. V., Raju S. S., Niezen S., Tsai L. T. Y., Siddle K. J., Sud M., Tran V. M., Vellarikkal S. K., Wang Y., Amir-Zilberstein L., Atri D. S., Beechem J., Brook O. R., Chen J., Divakar P., Dorceus P., Engreitz J. M., Essene A., Fitzgerald D. M., Fropf R., Gazal S., Gould J., Grzyb J., Harvey T., Hecht J., Hether T., Jané-Valbuena J., Leney-Greene M., Ma H., McCabe C., McLoughlin D. E., Miller E. M., Muus C., Niemi M., Padera R., Pan L., Pant D., Pe'er C., Pfiffner-Borges J., Pinto C. J., Plaisted J., Reeves J., Ross M., Rudy M., Rueckert E. H., Siciliano M., Sturm A., Todres E., Waghray A., Warren S., Zhang S., Zollinger D. R., Cosimi L., Gupta R. M., Hacohen N., Hibshoosh H., Hide W., Price A. L., Rajagopal J., Tata P. R., Riedel S., Szabo G., Tickle T. L., Ellinor P. T., Hung D., Sabeti P. C., Novak R., Rogers R., Ingber D. E., Jiang Z. G., Juric D., Babadi M., Farhi S. L., Izar B., Stone J. R., Vlachos I. S., Solomon I. H., Ashenberg O., Porter C. B. M., Li B., Shalek A. K., Villani A.-C., Rozenblatt-Rosen O., and Regev A., “ COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets,” Nature 595, 107–113 (2021). 10.1038/s41586-021-03570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vickovic S., Schapiro D., Carlberg K., Lotstedt B., Larsson L., “ Korotkova M., Hensvold A. H., Catrina A. I., Sorger P. K., Malmstrom V., Regev A., and Ståhl P. L., Three-dimensional spatial transcriptomics uncovers cell type dynamics in the rheumatoid arthritis synovium,” bioRxiv (2020). 10.1101/2020.12.10.420463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carlberg K., Korotkova M., Larsson L., Catrina A. I., Ståhl P. L., and Malmstrom V., “ Exploring inflammatory signatures in arthritic joint biopsies with spatial transcriptomics,” Sci. Rep. 9, 18975 (2019). 10.1038/s41598-019-55441-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Berglund E., Maaskola J., Schultz N., Friedrich S., Marklund M., Bergenstråhle J., Tarish F., Tanoglidi A., Vickovic S., Larsson L., Salmén F., Ogris C., Wallenborg K., Lagergren J., Ståhl P., Sonnhammer E., Helleday T., and Lundeberg J., “ Spatial maps of prostate cancer transcriptomes reveal an unexplored landscape of heterogeneity,” Nat. Commun. 9, 2419 (2018). 10.1038/s41467-018-04724-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moncada R., Barkley D., Wagner F., Chiodin M., Devlin J. C., Baron M., Hajdu C. H., Simeone D. M., and Yanai I., “ Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas,” Nat. Biotechnol. 38, 333–342 (2020). 10.1038/s41587-019-0392-8 [DOI] [PubMed] [Google Scholar]
- 55. Ji A. L., Rubin A. J., Thrane K., Jiang S., Reynolds D. L., Meyers R. M., Guo M. G., George B. M., Mollbrink A., Bergenstråhle J., Larsson L., Bai Y., Zhu B., Bhaduri A., Meyers J. M., Rovira-Clavé X., Hollmig S. T., Aasi S. Z., Nolan G. P., Lundeberg J., and Khavari P. A., “ Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma,” Cell 182, 497–514 (2020). 10.1016/j.cell.2020.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen W.-T., Lu A., Craessaerts K., Pavie B., Sala Frigerio C., Corthout N., Qian X., Laláková J., Kuhnemund M., Voytyuk I., Wolfs L., Mancuso R., Salta E., Balusu S., Snellinx A., Munck S., Jurek A., Fernandez Navarro J., Saido T. C., Huitinga I., Lundeberg J., Fiers M., and De Strooper B., “ Spatial transcriptomics and in situ sequencing to study alzheimer's disease,” Cell 182, 976–991 (2020). 10.1016/j.cell.2020.06.038 [DOI] [PubMed] [Google Scholar]
- 57. Backdahl J., Franzén L., Massier L., Li Q., Jalkanen J., Gao H., Andersson A., Bhalla N., Thorell A., Rydén M., Ståhl P. L., and Mejhert N., “ Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin,” Cell Metab. 33, 1869–1882 (2021). 10.1016/j.cmet.2021.07.018 [DOI] [PubMed] [Google Scholar]
- 58. Theocharidis G., Thomas B. E., Sarkar D., Mumme H. L., Pilcher W. J. R., Dwivedi B., Sandoval-Schaefer T., Sîrbulescu R. F., Kafanas A., Mezghani I., Wang P., Lobao A., Vlachos I. S., Dash B., Hsia H. C., Horsley V., Bhasin S. S., Veves A., and Bhasin M., “ Single cell transcriptomic landscape of diabetic foot ulcers,” Nat. Commun. 13, 181 (2022). 10.1038/s41467-021-27801-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Satija R., Farrell J. A., Gennert D., Schier A. F., and Regev A., “ Spatial reconstruction of single-cell gene expression data,” Nat. Biotechnol. 33, 495–502 (2015). 10.1038/nbt.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nitzan M., Karaiskos N., Friedman N., and Rajewsky N., “ Gene expression cartography,” Nature 576, 132–137 (2019). 10.1038/s41586-019-1773-3 [DOI] [PubMed] [Google Scholar]
- 61. Maseda F., Cang Z., and Nie Q., “ DEEPsc: A deep learning-based map connecting single-cell transcriptomics and spatial imaging data,” Front. Genet. 12, 348 (2021). 10.3389/fgene.2021.636743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Achim K., Pettit J.-B., Saraiva L. R., Gavriouchkina D., Larsson T., Arendt D., and Marioni J. C., “ High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin,” Nat. Biotechnol. 33, 503–509 (2015). 10.1038/nbt.3209 [DOI] [PubMed] [Google Scholar]
- 63. Peng T., Chen G. M., and Tan K., “ Gluer: Integrative analysis of single-cell omics and imaging data by deep neural network,” bioRxiv (2021). 10.1101/2021.01.25.427845 [DOI] [Google Scholar]
- 64. Andersson A., Bergenstråhle J., Asp M., Bergenstråhle L., Jurek A., Navarro J. F., and Lundeberg J., “ Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography,” Commun. Biol. 3, 565 (2020). 10.1038/s42003-020-01247-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Song Q. and Su J., “ DSTG: Deconvoluting spatial transcriptomics data through graph-based artificial intelligence,” Briefings Bioinf. 22, bbaa414 (2021). 10.1093/bib/bbaa414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Elosua-Bayes M., Nieto P., Mereu E., Gut I., and Heyn H., “ SPOTlight: Seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes,” Nucl. Acids Res. 49, e50 (2021). 10.1093/nar/gkab043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cable D. M., Murray E., Zou L. S., Goeva A., Macosko E. Z., Chen F., and Irizarry R. A., “ Robust decomposition of cell type mixtures in spatial transcriptomics,” Nat. Biotechnol. 40, 517–526 (2021). 10.1038/s41587-021-00830-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dong R. and Yuan G.-C., “ SpatialDWLS: Accurate deconvolution of spatial transcriptomic data,” Genome Biol. 22, 145 (2021). 10.1186/s13059-021-02362-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lopez R., Li B., Keren-Shaul H., Boyeau P., Kedmi M., Pilzer D., Jelinski A., Yofe I., David E., Wagner A., Ergen C., Addadi Y., Golani O., Ronchese F., Jordan M. I., Amit I., and Yosef N., “ Destvi identifies continuums of cell types in spatial transcriptomics data,” Nat. Biotechnol. 40, 1360–1369 (2022). 10.1038/s41587-022-01272-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kleshchevnikov V., Shmatko A., Dann E., Aivazidis A., King H. W., Li T., Elmentaite R., Lomakin A., Kedlian V., Gayoso A., Jain M. S., Park J. S., Ramona L., Tuck E., Arutyunyan A., Vento-Tormo R., Gerstung M., James L., Stegle O., and Bayraktar O. A., “ Cell2location maps fine-grained cell types in spatial transcriptomics,” Nat. Biotechnol. 40, 661–671 (2022). 10.1038/s41587-021-01139-4 [DOI] [PubMed] [Google Scholar]
- 71. Ma Y. and Zhou X., “ Spatially informed cell-type deconvolution for spatial transcriptomics,” Nat. Biotechnol. 40, 1349–1359 (2022). 10.1038/s41587-022-01273-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu Q., Shah S., Dries R., Cai L., and Yuan G.-C., “ Identification of spatially associated subpopulations by combining scRNAseq and sequential fluorescence in situ hybridization data,” Nat. Biotechnol. 36, 1183–1190 (2018). 10.1038/nbt.4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tan X., Su A., Tran M., and Nguyen Q., “ SpaCell: Integrating tissue morphology and spatial gene expression to predict disease cells,” Bioinformaics 36, 2293–2294 (2020). 10.1093/bioinformatics/btz914 [DOI] [PubMed] [Google Scholar]
- 74. Pham D., Tan X., Xu J., Grice L. F., Lam P. Y., “ Raghubar A., Vukovic J., Ruitenberg M. J., and Nguyen Q., stLearn: Integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues,” bioRxiv (2020). 10.1101/2020.05.31.125658 [DOI] [Google Scholar]
- 75. Zhao E., Stone M. R., Ren X., Guenthoer J., Smythe K. S., Pulliam T., Williams S. R., Uytingco C. R., Taylor S. E. B., Nghiem P., Bielas J. H., and Gottardo R., “ Spatial transcriptomics at subspot resolution with BayesSpace,” Nat. Biotechnol. 39, 1375–1384 (2021). 10.1038/s41587-021-00935-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hu J., Li X., Coleman K., Schroeder A., Ma N., Irwin D. J., Lee E. B., Shinohara R. T., and Li M., “ SpaGCN: Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network,” Nat. Methods 18, 1342–1351 (2021). 10.1038/s41592-021-01255-8 [DOI] [PubMed] [Google Scholar]
- 77. Edsgard D., Johnsson P., and Sandberg R., “ Identification of spatial expression trends in single-cell gene expression data,” Nat. Methods 15, 339–342 (2018). 10.1038/nmeth.4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Svensson V., Teichmann S. A., and Stegle O., “ Spatialde: Identification of spatially variable genes,” Nat. Methods 15, 343–346 (2018). 10.1038/nmeth.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun S., Zhu J., and Zhou X., “ Statistical analysis of spatial expression patterns for spatially resolved transcriptomic studies,” Nat. Methods 17, 193–200 (2020). 10.1038/s41592-019-0701-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cang Z. and Nie Q., “ Inferring spatial and signaling relationships between cells from single cell transcriptomic data,” Nat. Commun. 11, 2084 (2020). 10.1038/s41467-020-15968-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tanevski J., Flores R. O. R., Gabor A., Schapiro D., and Saez-Rodriguez J., “ Explainable multi-view framework for dissecting intercellular signaling from highly multiplexed spatial data,” bioRxiv (2021). 10.1101/2020.05.08.084145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dries R., Zhu Q., Dong R., Eng C.-H. L., Li H., Liu K., Fu Y., Zhao T., Sarkar A., Bao F., George R. E., Pierson N., Cai L., and Yuan G.-C., “ Giotto: A toolbox for integrative analysis and visualization of spatial expression data,” Genome Biol. 22, 78 (2021). 10.1186/s13059-021-02286-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gupta R. K. and Kuznicki J., “ Biological and medical importance of cellular heterogeneity deciphered by single-cell RNA sequencing,” Cells 9, 1751 (2020). 10.3390/cells9081751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. van Galen P., Hovestadt V., Wadsworth M. H. II, Hughes T. K., Griffin G. K., Battaglia S., Verga J. A., Stephansky J., Pastika T. J., Lombardi Story J., Pinkus G. S., Pozdnyakova O., Galinsky I., Stone R. M., Graubert T. A., Shalek A. K., Aster J. C., Lane A. A., and Bernstein B. E., “ Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity,” Cell 176, 1265–1281 (2019). 10.1016/j.cell.2019.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Masuda T., Sankowski R., Staszewski O., Bottcher C., Amann L., Sagar, Scheiwe C., Nessler S., Kunz P., van Loo G., Coenen V. A., Reinacher P. C., Michel A., Sure U., Gold R., Grun D., Priller J., Stadelmann C., and Prinz M., “ Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution,” Nature 566, 388–392 (2019). 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- 86. Churko J. M., Garg P., Treutlein B., Venkatasubramanian M., Wu H., Lee J., Wessells Q. N., Chen S.-Y., Chen W.-Y., Chetal K., Mantalas G., Neff N., Jabart E., Sharma A., Nolan G. P., Salomonis N., and Wu J. C., “ Defining human cardiac transcription factor hierarchies using integrated single-cell heterogeneity analysis,” Nat. Commun. 9, 4906 (2018). 10.1038/s41467-018-07333-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gross A., Schoendube J., Zimmermann S., Steeb M., Zengerle R., and Koltay P., “ Technologies for single-cell isolation,” Int. J. Mol. Sci. 16, 16897–16919 (2015). 10.3390/ijms160816897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ma S., Murphy T. W., and Lu C., “ Microfluidics for genome-wide studies involving next generation sequencing,” Biomicrofluidics 11, 021501 (2017). 10.1063/1.4978426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hwang B., Lee J. H., and Bang D., “ Single-cell RNA sequencing technologies and bioinformatics pipelines,” Exp. Mol. Med. 50, 1–14 (2018). 10.1038/s12276-018-0071-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Haque A., Engel J., Teichmann S. A., and Lonnberg T., “ A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications,” Genome Med. 9, 75 (2017). 10.1186/s13073-017-0467-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Stark R., Grzelak M., and Hadfield J., “ RNA sequencing: The teenage years,” Nat. Rev. Genet. 20, 631–656 (2019). 10.1038/s41576-019-0150-2 [DOI] [PubMed] [Google Scholar]
- 92. van Dijk E. L., Auger H., Jaszczyszyn Y., and Thermes C., “ Ten years of next-generation sequencing technology,” Trends Genet. 30, 418–426 (2014). 10.1016/j.tig.2014.07.001 [DOI] [PubMed] [Google Scholar]
- 93. Zhang X., Li T., Liu F., Chen Y., Yao J., Li Z., Huang Y., and Wang J., “ Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems,” Mol. Cell 73, 130–142 (2019). 10.1016/j.molcel.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 94. Kolodziejczyk A. A., Kim J. K., Svensson V., Marioni J. C., and Teichmann S. A., “ The technology and biology of single-cell RNA sequencing,” Mol. Cell 58, 610–620 (2015). 10.1016/j.molcel.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 95. Huang X.-T., Li X., Qin P.-Z., Zhu Y., Xu S.-N., and Chen J.-P., “ Technical advances in single-cell RNA sequencing and applications in normal and malignant hematopoiesis,” Front. Oncol. 8, 582 (2018). 10.3389/fonc.2018.00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Asp M., Bergenstråhle J., and Lundeberg J., “ Spatially resolved transcriptomes—Next generation tools for tissue exploration,” BioEssays 42, 1900221 (2020). 10.1002/bies.201900221 [DOI] [PubMed] [Google Scholar]
- 97. Moses L. and Pachter L., “ Museum of spatial transcriptomics,” Nat. Methods 19, 534–546 (2022). 10.1038/s41592-022-01409-2 [DOI] [PubMed] [Google Scholar]
- 98. Rautenstrauss B. W. and Liehr T., FISH Technology ( Springer; Verlag, Berlin, 2002). [Google Scholar]
- 99. Jensen E., “ Technical review: In situ hybridization,” Anat. Rec. 297, 1349–1353 (2014). 10.1002/ar.22944 [DOI] [PubMed] [Google Scholar]
- 100. Hilscher M. M., Gyllborg D., Yokota C., and Nilsson M., “ In situ sequencing: A high-throughput, multi-targeted gene expression profiling technique for cell typing in tissue sections,” in In Situ Hybridization Protocols, edited by Nielsen B. S. and Jones J. ( Springer, New York, NY, 2020), pp. 313–329. [DOI] [PubMed] [Google Scholar]
- 101. Weinstein J. A., Regev A., and Zhang F., “ Dna microscopy: Optics-free spatio-genetic imaging by a stand-alone chemical reaction,” Cell 178, 229–241 (2019). 10.1016/j.cell.2019.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Larsson C., Grundberg I., Soderberg O., and Nilsson M., “ In situ detection and genotyping of individual mRNA molecules,” Nat. Methods 7, 395–397 (2010). 10.1038/nmeth.1448 [DOI] [PubMed] [Google Scholar]
- 103. Gyllborg D., Langseth C. M., Qian X., Choi E., Salas S. M., Hilscher M. M., Lein E. S., and Nilsson M., “ Hybridization-based in situ sequencing (HYBISS) for spatially resolved transcriptomics in human and mouse brain tissue,” Nucl. Acids Res. 48, e112 (2020). 10.1093/nar/gkaa792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen X., Sun Y.-C., Church G. M., Lee J. H., and Zador A. M., “ Efficient in situ barcode sequencing using padlock probe-based BaristaSeq,” Nucl. Acids Res. 46, e22 (2017). 10.1093/nar/gkx1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lee H., Salas S. M., Gyllborg D., and Nilsson M., “ Direct RNA targeted transcriptomic profiling in tissue using hybridization-based RNA in situ sequencing (HybRISS),” bioRxiv (2020). 10.1101/2020.12.02.408781 [DOI] [Google Scholar]
- 106. Lee J. H., Daugharthy E. R., Scheiman J., Kalhor R., Ferrante T. C., Terry R., Turczyk B. M., Yang J. L., Lee H. S., Aach J., Zhang K., and Church G. M., “ Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues,” Nat. Protoc. 10, 442–458 (2015). 10.1038/nprot.2014.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Lee J. H., “ Quantitative approaches for investigating the spatial context of gene expression,” WIREs Syst. Biol. Med. 9, e1369 (2017). 10.1002/wsbm.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Gudla P. R., Nakayama K., Pegoraro G., and Misteli T., “ Spotlearn: Convolutional neural network for detection of fluorescence in situ hybridization (fish) signals in high-throughput imaging approaches,” Cold Spring Harbor Symp. Quant. Biol. 82, 57–70 (2017). 10.1101/sqb.2017.82.033761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Partel G., Hilscher M. M., Milli G., Solorzano L., Klemm A. H., Nilsson M., and Wahlby C., “ Automated identification of the mouse brain's spatial compartments from in situ sequencing data,” BMC Biol. 18, 144 (2020). 10.1186/s12915-020-00874-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Pardo E., Morgado J. M. T., and Malpica N., “ Semantic segmentation of mFISH images using convolutional networks,” Cytometry, Part A 93, 620–627 (2018). 10.1002/cyto.a.23375 [DOI] [PubMed] [Google Scholar]
- 111. Frankenstein Z., Uraoka N., Aypar U., Aryeequaye R., Rao M., Hameed M., Zhang Y., and Yagi Y., “ Automated 3d scoring of fluorescence in situ hybridization (FISH) using a confocal whole slide imaging scanner,” Appl. Microsc. 51, 4 (2021). 10.1186/s42649-021-00053-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Littman R., Hemminger Z., Foreman R., Arneson D., Zhang G., Gómez-Pinilla F., Yang X., and Wollman R., “ Joint cell segmentation and cell type annotation for spatial transcriptomics,” Mol. Syst. Biol. 17, e10108 (2021). 10.15252/msb.202010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.x.Genomics, Visium Spatial Gene Expression Reagent Kits—User Guide, 2021.
- 114. Hunter M. V., Moncada R., Weiss J. M., Yanai I., and White R. M., “ Spatially resolved transcriptomics reveals the architecture of the tumor/microenvironment interface,” bioRxiv (2021). 10.1101/2020.11.05.368753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fawkner-Corbett D., Antanaviciute A., Parikh K., Jagielowicz M., Gerós A. S., Gupta T., Ashley N., Khamis D., Fowler D., Morrissey E., Cunningham C., Johnson P. R., Koohy H., and Simmons A., “ Spatiotemporal analysis of human intestinal development at single-cell resolution,” Cell 184, 810–826 (2021). 10.1016/j.cell.2020.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Asp M., Giacomello S., Larsson L., Wu C., Furth D., Qian X., Wardell E., Custodio J., Reimegård J., Salmén F., Osterholm C., Ståhl P. L., Sundstrom E., Åkesson E., Bergmann O., Bienko M., Månsson-Broberg A., Nilsson M., Sylvén C., and Lundeberg J., “ A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart,” Cell 179, 1647–1660 (2019). 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- 117. Batiuk M. Y., Tyler T., Mei S., Rydbirk R., Petukhov V., Sedmak D., Frank E., Feher V., Habek N., Hu Q., Igolkina A., Roszik L., Pfisterer U., Petanjek Z., Adorjan I., Kharchenko P. V., and Khodosevich K., “ Selective vulnerability of supragranular layer neurons in schizophrenia,” bioRxiv (2021). 10.1101/2020.11.17.386458 [DOI] [Google Scholar]
- 118. Ortiz C., Navarro J. F., Jurek A., Martin A., Lundeberg J., and Meletis K., “ Molecular atlas of the adult mouse brain,” Sci. Adv. 6, eabb3446 (2020). 10.1126/sciadv.abb3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.10x Genomics, see https://www.10xgenomics.com/products/spatial-gene-expression for “ Spatial gene expression,” 2022.
- 120. Goodfellow I., Bengio Y., and Courville A., Deep Learning ( The MIT Press, 2016). [Google Scholar]
- 121. Lloyd S., “ Least squares quantization in pcm,” IEEE Trans. Inf. Theory 28, 129–137 (1982). 10.1109/TIT.1982.1056489 [DOI] [Google Scholar]
- 122. Prechelt L., “ Early stopping-but when?,” in Proceedings of the Neural Networks: Tricks of the Trade, This Book is an Outgrowth of a 1996 NIPS Workshop ( Springer-Verlag, Berlin, Heidelberg, 1998), pp. 55–69. [Google Scholar]
- 123. Cybenko G., “ Approximation by superpositions of a sigmoidal function,” Math. Control Signals Syst. 2, 303–314 (1989). 10.1007/BF02551274 [DOI] [Google Scholar]
- 124. Scarselli F. and Chung Tsoi A., “ Universal approximation using feedforward neural networks: A survey of some existing methods, and some new results,” Neural Networks 11, 15–37 (1998). 10.1016/S0893-6080(97)00097-X [DOI] [PubMed] [Google Scholar]
- 125. Chen T. and Chen H., “ Universal approximation to nonlinear operators by neural networks with arbitrary activation functions and its application to dynamical systems,” IEEE Trans. Neural Networks 6, 911–917 (1995). 10.1109/72.392253 [DOI] [PubMed] [Google Scholar]
- 126. Uzair M. and Jamil N., “ Effects of hidden layers on the efficiency of neural networks,” in Proceedings of the IEEE 23rd International Multitopic Conference (INMIC) ( IEEE, 2020), pp. 1–6. [Google Scholar]
- 127. Krizhevsky A., Sutskever I., and Hinton G. E., “ ImageNet classification with deep convolutional neural networks,” in Advances in Neural Information Processing Systems, edited by Pereira F., Burges C. J. C., Bottou L., and Weinberger K. Q. ( Curran Associates, Inc., 2012), Vol. 25. [Google Scholar]
- 128. Zhu X., Lyu S., Wang X., and Zhao Q., “ Tph-yolov5: Improved yolov5 based on transformer prediction head for object detection on drone-captured scenarios,” arXiv:2108.11539 [cs.CV] (2021).
- 129. Tan M. and Le Q. V., “ Efficientnetv2: Smaller models and faster training,” arXiv:2104.00298 [cs.CV] (2021).
- 130. Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A. N., Kaiser L. u, and Polosukhin I., “ Attention is all you need,” in Advances in Neural Information Processing Systems, edited by Guyon I., Luxburg U. V., Bengio S., Wallach H., Fergus R., Vishwanathan S., and Garnett R. ( Curran Associates, Inc., 2017), Vol. 30. [Google Scholar]
- 131. Brown T. B., Mann B., Ryder N., Subbiah M., Kaplan J., Dhariwal P., Neelakantan A., Shyam P., Sastry G., Askell A., Agarwal S., Herbert-Voss A., Krueger G., Henighan T., Child R., Ramesh A., Ziegler D. M., Wu J., Winter C., Hesse C., Chen M., Sigler E., Litwin M., Gray S., Chess B., Clark J., Berner C., McCandlish S., Radford A., Sutskever I., and Amodei D., “ Language models are few-shot learners,” arXiv:2005.14165 [cs.CL] (2020).
- 132. Devlin J., Chang M., Lee K., and Toutanova K., “ BERT: Pre-training of deep bidirectional transformers for language understanding,” in Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies (NAACL-HLT), Minneapolis, MN, 2–7 June 2019, edited by Burstein J., Doran C., and Solorio T. ( Association for Computational Linguistics, 2019), Vol. 1 (Long and Short Papers), pp. 4171–4186. [Google Scholar]
- 133. Poplin R., Chang P.-C., Alexander D., Schwartz S., Colthurst T., Ku A., Newburger D., Dijamco J., Nguyen N., Afshar P. T., Gross S. S., Dorfman L., McLean C. Y., and DePristo M. A., “ A universal SNP and small-indel variant caller using deep neural networks,” Nat. Biotechnol. 36, 983–987 (2018). 10.1038/nbt.4235 [DOI] [PubMed] [Google Scholar]
- 134. Li H., Shaham U., Stanton K. P., Yao Y., Montgomery R. R., and Kluger Y., “ Gating mass cytometry data by deep learning,” Bioinformatics 33, 3423–3430 (2017). 10.1093/bioinformatics/btx448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., and Hassabis D., “ Highly accurate protein structure prediction with alphafold,” Nature 596, 583–589 (2021). 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Kipf T. N. and Welling M., “ Semi-supervised classification with graph convolutional networks,” in Proceedings of the 5th International Conference on Learning Representations (ICLR), Toulon, France, 24–26 April 2017 ( OpenReview.net, 2017). [Google Scholar]
- 137. Hornik K., “ Approximation capabilities of multilayer feedforward networks,” Neural Networks 4, 251–257 (1991). 10.1016/0893-6080(91)90009-T [DOI] [Google Scholar]
- 138. Nair V. and Hinton G. E., “ Rectified linear units improve restricted Boltzmann machines,” in Proceedings of the 27th International Conference on International Conference on Machine Learning (ICML) ( Omnipress, Madison, WI, 2010), pp. 807–814. [Google Scholar]
- 139. Nocedal J. and Wright S., Numerical Optimization ( Springer Science and Business Media, 2006). [Google Scholar]
- 140. Glorot X. and Bengio Y., “ Understanding the difficulty of training deep feedforward neural networks,” in Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, edited by Teh Y. W. and Titterington M. ( PMLR, Chia Laguna Resort, Sardinia, Italy, 2010), Vol. 9, pp. 249–256. [Google Scholar]
- 141. He K., Zhang X., Ren S., and Sun J., “ Delving deep into rectifiers: Surpassing human-level performance on imageNet classification,” in Proceedings of the IEEE International Conference on Computer Vision (ICCV) (2015). [Google Scholar]
- 142. Ruder S., “ An overview of gradient descent optimization algorithms,” arXiv:1609.04747 (2016).
- 143. Smith S., Elsen E., and De S., “ On the generalization benefit of noise in stochastic gradient descent,” in Proceedings of the 37th International Conference on Machine Learning, edited by De H. III and Singh A. ( PMLR, 2020), Vol. 119, pp. 9058–9067. [Google Scholar]
- 144. Rumelhart D. E., Hinton G. E., and Williams R. J., “ Learning representations by back-propagating errors,” Nature 323, 533–536 (1986). 10.1038/323533a0 [DOI] [Google Scholar]
- 145. Kingma D. P. and Ba J., “ Adam: A method for stochastic optimization,” in Proceedings of the 3rd International Conference on Learning Representations (ICLR), San Diego, CA, 7–9 May 2015, edited by Bengio Y. and LeCun Y. ( Conference Track Proceedings, 2015). [Google Scholar]
- 146. Duchi J., Hazan E., and Singer Y., “ Adaptive subgradient methods for online learning and stochastic optimization,” J. Mach. Learn. Res. 12, 2121–2159 (2011), see https://jmlr.org/papers/v12/duchi11a.html [Google Scholar]
- 147. Zhao Z.-Q., Zheng P., Xu S.-T., and Wu X., “ Object detection with deep learning: A review,” IEEE Trans. Neural Networks Learn. Syst. 30, 3212–3232 (2019). 10.1109/TNNLS.2018.2876865 [DOI] [PubMed] [Google Scholar]
- 148. Marr D. and Hildreth E., “ Theory of edge detection,” Proc. R. Soc. London, Ser. B 207, 187–217 (1980). 10.1098/rspb.1980.0020 [DOI] [PubMed] [Google Scholar]
- 149. LeCun Y., Boser B., Denker J. S., Henderson D., Howard R. E., Hubbard W., and Jackel L. D., “ Backpropagation applied to handwritten zip code recognition,” Neural Comput. 1, 541–551 (1989). 10.1162/neco.1989.1.4.541 [DOI] [Google Scholar]
- 150. LeCun Y. and Bengio Y., “ Convolutional networks for images, speech, and time series,” in The Handbook of Brain Theory and Neural Networks ( MIT Press, Cambridge, MA, 1998), pp. 255–258. [Google Scholar]
- 151.Note that if the model chose a separate parameter for each , for , then the model could not generalize to any inputs where (size of X is greater than n).
- 152. Kolen J. F. and Kremer S. C., “ Gradient flow in recurrent nets: The difficulty of learning longterm dependencies,” in A Field Guide to Dynamical Recurrent Networks ( IEEE, 2001), pp. 237–243. [Google Scholar]
- 153. Hochreiter S. and Schmidhuber J., “ Long short-term memory,” Neural Comput. 9, 1735–1780 (1997). 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 154. Kimmel J. C., Brack A. S., and Marshall W. F., “ Deep convolutional and recurrent neural networks for cell motility discrimination and prediction,” IEEE/ACM Trans. Comput. Biol. Bioinf. 18, 562–574 (2021). 10.1109/TCBB.2019.2919307 [DOI] [PubMed] [Google Scholar]
- 155. Ioffe S. and Szegedy C., “ Batch normalization: Accelerating deep network training by reducing internal covariate shift,” in Proceedings of the 32nd International Conference on Machine Learning, edited by Bach F. and Blei D. ( PMLR, Lille, France, 2015), Vol. 37, pp. 448–456. [Google Scholar]
- 156. He K., Zhang X., Ren S., and Sun J., “ Deep residual learning for image recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) ( IEEE, 2016), pp. 770–778. [Google Scholar]
- 157. Shah A., Kadam E., Shah H., Shinde S., and Shingade S., “ Deep residual networks with exponential linear unit,” in Proceedings of the Third International Symposium on Computer Vision and the Internet ( Association for Computing Machinery, New York, NY, 2016), pp. 59–65. [Google Scholar]
- 158. Deng J., Dong W., Socher R., Li L.-J., Li K., and Fei-Fei L., “ ImageNet: A large-scale hierarchical image database,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition (2009) pp. 248–255.
- 159. Ballard D. H., “ Modular learning in neural networks,” in Proceedings of the AAAI (AAAI, 1987), Vol. 1, pp. 279–284. [Google Scholar]
- 160. Hinton G. E., “ Connectionist learning procedures11 this chapter appeared in volume 40 of artificial intelligence in 1989, reprinted with permission of North-holland publishing. It is a revised version of technical report cmu-cs-87-115, which has the same title and was prepared in june 1987 while the author was at Carnegie Mellon University. the research was supported by contract n00014-86-k-00167 from the office of naval research and by grant ist-8520359 from the national science foundation,” in Machine Learning, edited by Kodratoff Y. and Michalski R. S. ( Morgan Kaufmann, San Francisco, CA, 1990), Chap. 20, pp. 555–610. [Google Scholar]
- 161. Kingma D. P. and Welling M., “ Auto-encoding variational bayes,” in Proceedings of the International Conference on Learning Representations (2014). [Google Scholar]
- 162. Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., Courville A., and Bengio Y., “ Generative adversarial nets,” in Advances in Neural Information Processing Systems, edited by Ghahramani Z., Welling M., Cortes C., Lawrence N., and Weinberger K. Q. ( Curran Associates, Inc., 2014), Vol. 27. [Google Scholar]
- 163. He J., Spokoyny D., Neubig G., and Berg-Kirkpatrick T., “ Lagging inference networks and posterior collapse in variational autoencoders,” in Proceedings of the International Conference on Learning Representations (2019). [Google Scholar]
- 164. Yang Z., Hu Z., Salakhutdinov R., and Berg-Kirkpatrick T., “ Improved variational autoencoders for text modeling using dilated convolutions,” in Proceedings of the International Conference on Machine Learning ( PMLR, 2017), pp. 3881–3890. [Google Scholar]
- 165. Heydari A. A. and Mehmood A., “ SRVAE: Super resolution using variational autoencoders,” Proc. SPIE 11400, 114000U (2020). 10.1117/12.2559808 [DOI] [Google Scholar]
- 166. Semeniuta S., Severyn A., and Barth E., “ A hybrid convolutional variational autoencoder for text generation,” in Proceedings of the Conference on Empirical Methods in Natural Language Processing ( Association for Computational Linguistics, Copenhagen, Denmark, 2017), pp. 627–637. [Google Scholar]
- 167. Heydari A. A., Thompson C. A., and Mehmood A., “ Softadapt: Techniques for adaptive loss weighting of neural networks with multi-part loss functions,” arXiv:1912.12355 (2019).
- 168. Tolstikhin I., Bousquet O., Gelly S., and Schoelkopf B., “ Wasserstein auto-encoders,” in Proceedings of the International Conference on Learning Representations (2018). [Google Scholar]
- 169. Daniel T. and Tamar A., “ SoftIntroVAE: Analyzing and improving the introspective variational autoencoder,” arXiv:2012.13253 [cs.LG] (2021).
- 170.The astute reader will note that although Maseda et al. refer to DEEPsc as DL model, the methods's two-layer FFNN is not considered deep model in most definitions.
- 171. Nikos K., Philipp W., Jonathan A., Anastasiya B., Salah A., Claudia K., Christine K., Nikolaus R., and Zinzen R. P., “ The drosophila embryo at single-cell transcriptome resolution,” Science 358, 194–199 (2017). 10.1126/science.aan3235 [DOI] [PubMed] [Google Scholar]
- 172. Tasic B., Menon V., Nguyen T. N., Kim T. K., Jarsky T., Yao Z., Levi B., Gray L. T., Sorensen S. A., Dolbeare T., Bertagnolli D., Goldy J., Shapovalova N., Parry S., Lee C., Smith K., Bernard A., Madisen L., Sunkin S. M., Hawrylycz M., Koch C., and Zeng H., “ Adult mouse cortical cell taxonomy revealed by single cell transcriptomics,” Nat. Neurosci. 19, 335–346 (2016). 10.1038/nn.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Joost S., Zeisel A., Jacob T., Sun X., Manno G. L., Lonnerberg P., Linnarsson S., and Kasper M., “ Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity,” Cell Syst. 3, 221–237 (2016). 10.1016/j.cels.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dropout refers to the scenario when a gene is observed at a moderate or high expression level in a subset of cells, but not detected in other cells.
- 175. Ding J., Adiconis X., Simmons S. K., Kowalczyk M. S., Hession C. C., Marjanovic N. D., Hughes T. K., Wadsworth M. H., Burks T., Nguyen L. T., Kwon J. Y. H., Barak B., Ge W., Kedaigle A. J., Carroll S., Li S., Hacohen N., Rozenblatt-Rosen O., Shalek A. K., Villani A.-C., Regev A., and Levin J. Z., “ Systematic comparison of single-cell and single-nucleus RNA-sequencing methods,” Nat. Biotechnol. 38, 737–746 (2020). 10.1038/s41587-020-0465-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.See https://pytorch.org/ for PyTorch is one of the most popular DL library in Python.
- 177. Paszke A., Gross S., Massa F., Lerer A., Bradbury J., “ Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L., Desmaison A., Kopf A., Yang E. Z., DeVito Z., Raison M., Tejani A., Chilamkurthy S., Steiner B., Fang L., Bai J., and Chintala S., Pytorch: An imperative style, high-performance deep learning library,” arXiv:1912.01703 (2019).
- 178. Ronneberger O., Fischer P., and Brox T., “ U-net: Convolutional networks for biomedical image segmentation,” in Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), edited by Navab N., Hornegger J., Wells W. M., and Frangi A. F. ( Springer International Publishing, Cham, 2015), pp. 234–241. [Google Scholar]
- 179. Koch G. R., “ Siamese neural networks for one-shot image recognition,” in Proceedings of the 32nd International Conference on Machine Learning (2015). [Google Scholar]
- 180. Huang G., Liu Z., Van Der Maaten L., and Weinberger K. Q., “ Densely connected convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) ( IEEE, 2017), pp. 2261–2269. [Google Scholar]
- 181. Liu Y., Yang M., Deng Y., Su G., Enninful A., Guo C. C., Tebaldi T., Zhang D., Kim D., Bai Z., Norris E., Pan A., Li J., Xiao Y., Halene S., and Fan R., “ High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue,” Cell 183, 1665–1681 (2020). 10.1016/j.cell.2020.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.See https://www.nanostring.com/ for information on Nanostring’s GeoMx technology.
- 183. Hu J., Schroeder A., Coleman K., Chen C., Auerbach B. J., and Li M., “ Statistical and machine learning methods for spatially resolved transcriptomics with histology,” Comput. Struct. Biotechnol. J. 19, 3829–3841 (2021). 10.1016/j.csbj.2021.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Garcia-Alonso L., Handfield L.-F., Roberts K., Nikolakopoulou K., Fernando R. C., “ Gardner L., Woodhams B., Arutyunyan A., Polanski K., Hoo R., Sancho-Serra C., Li T., Kwakwa K., Tuck E., Kleshchevnikov V., Tarkowska A., Porter T., Mazzeo C. I., van Dongen S., Dabrowska M., Vaskivskyi V., Mahbubani K. T., Park J.-E., Jimenez-Linan M., Campos L., Kiselev V., Lindskog C., Ayuk P., Prigmore E., Stratton M. R., Saeb-Parsy K., Moffett A., Moore L., Bayraktar O. A., Teichmann S. A., Turco M. Y., and Vento-Tormo R., “ Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro,” bioRxiv (2021). 10.1101/2021.01.02.425073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Lopez R., Regier J., Cole M. B., Jordan M. I., and Yosef N., “ Deep generative modeling for single-cell transcriptomics,” Nat. Methods 15, 1053–1058 (2018). 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Grun D., Kester L., and van Oudenaarden A., “ Validation of noise models for single-cell transcriptomics,” Nat. Methods 11, 637–640 (2014). 10.1038/nmeth.2930 [DOI] [PubMed] [Google Scholar]
- 187. Grønbech C. H., Vording M. F., Timshel P. N., Sønderby C. K., Pers T. H., and Winther O., “ scVAE: Variational auto-encoders for single-cell gene expression data,” Bioinformatics 36, 4415–4422 (2020). 10.1093/bioinformatics/btaa293 [DOI] [PubMed] [Google Scholar]
- 188. Aragón J., Eberly D., and Eberly S., “ Existence and uniqueness of the maximum likelihood estimator for the two-parameter negative binomial distribution,” Stat. Probab. Lett. 15, 375–379 (1992). 10.1016/0167-7152(92)90157-Z [DOI] [Google Scholar]
- 189. Kettenring J. R., “ Canonical analysis of several sets of variables,” Biometrika 58, 433–451 (1971). 10.1093/biomet/58.3.433 [DOI] [Google Scholar]
- 190. Haghverdi L., Lun A. T. L., Morgan M. D., and Marioni J. C., “ Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors,” Nat. Biotechnol. 36, 421–427 (2018). 10.1038/nbt.4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Pasquini G., Rojo Arias J. E., Schafer P., and Busskamp V., “ Automated methods for cell type annotation on SCRNA-seq data,” Comput. Struct. Biotechnol. J. 19, 961–969 (2021). 10.1016/j.csbj.2021.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Blondel V. D., Guillaume J.-L., Lambiotte R., and Lefebvre E., “ Fast unfolding of communities in large networks,” J. Stat. Mech. Theory Exp. 2008, P10008. 10.1088/1742-5468/2008/10/p10008 [DOI] [Google Scholar]
- 193. Traag V. A., Waltman L., and van Eck N. J., “ From Louvain to Leiden: Guaranteeing well-connected communities,” Sci. Rep. 9, 5233 (2019). 10.1038/s41598-019-41695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T.-M., Chi Chin M., Chong J., Crook B. E., Czaplinska A., Dang C. N., Datta S., Dee N. R., Desaki A. L., Desta T., Diep E., Dolbeare T. A., Donelan M. J., Dong H.-W., Dougherty J. G., Duncan B. J., Ebbert A. J., Eichele G., Estin L. K., Faber C., Facer B. A., Fields R., Fischer S. R., Fliss T. P., Frensley C., Gates S. N., Glattfelder K. J., Halverson K. R., Hart M. R., Hohmann J. G., Howell M. P., Jeung D. P., Johnson R. A., Karr P. T., Kawal R., Kidney J. M., Knapik R. H., Kuan C. L., Lake J. H., Laramee A. R., Larsen K. D., Lau C., Lemon T. A., Liang A. J., Liu Y., Luong L. T., Michaels J., Morgan J. J., Morgan R. J., Mortrud M. T., Mosqueda N. F., Ng L. L., Ng R., Orta G. J., Overly C. C., Pak T. H., Parry S. E., Pathak S. D., Pearson O. C., Puchalski R. B., Riley Z. L., Rockett H. R., Rowland S. A., Royall J. J., Ruiz M. J., Sarno N. R., Schaffnit K., Shapovalova N. V., Sivisay T., Slaughterbeck C. R., Smith S. C., Smith K. A., Smith B. I., Sodt A. J., Stewart N. N., Stumpf K.-R., Sunkin S. M., Sutram M., Tam A., Teemer C. D., Thaller C., Thompson C. L., Varnam L. R., Visel A., Whitlock R. M., Wohnoutka P. E., Wolkey C. K., Wong V. Y., Wood M., Yaylaoglu M. B., Young R. C., Youngstrom B. L., Yuan X. F., Zhang B., Zwingman T. A., and Jones A. R., “ Genome-wide atlas of gene expression in the adult mouse brain,” Nature 445, 168–176 (2007). 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- 195. Pardo B., Spangler A., Weber L. M., Page S. C., Hicks S. C., Jaffe A. E., Martinowich K., Maynard K. R., and Collado-Torres L., “ spatialLIBD: An R/bioconductor package to visualize spatially-resolved transcriptomics data,” BMC Genomics 23, 434 (2022). 10.1186/s12864-022-08601-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Maynard K. R., Collado-Torres L., Weber L. M., Uytingco C., Barry B. K., Williams S. R., Catallini J. L., Tran M. N., Besich Z., Tippani M., Chew J., Yin Y., Kleinman J. E., Hyde T. M., Rao N., Hicks S. C., Martinowich K., and Jaffe A. E., “ Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex,” Nat. Neurosci. 24, 425–436 (2021). 10.1038/s41593-020-00787-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Rousseeuw P. J., “ Silhouettes: A graphical aid to the interpretation and validation of cluster analysis,” J. Comput. Appl. Math. 20, 53–65 (1987). 10.1016/0377-0427(87)90125-7 [DOI] [Google Scholar]
- 198. Armingol E., Officer A., Harismendy O., and Lewis N. E., “ Deciphering cell–cell interactions and communication from gene expression,” Nat. Rev. Genet. 22, 71–88 (2021). 10.1038/s41576-020-00292-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Arneson D., Zhang G., Ying Z., Zhuang Y., Byun H. R., Ahn I. S., Gomez-Pinilla F., and Yang X., “ Single cell molecular alterations reveal target cells and pathways of concussive brain injury,” Nat. Commun. 9, 3894 (2018). 10.1038/s41467-018-06222-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Skelly D. A., Squiers G. T., McLellan M. A., Bolisetty M. T., Robson P., Rosenthal N. A., and Pinto A. R., “ Single-cell transcriptional profiling reveals cellular diversity and intercommunication in the mouse heart,” Cell Rep. 22, 600–610 (2018). 10.1016/j.celrep.2017.12.072 [DOI] [PubMed] [Google Scholar]
- 201. Cohen M., Giladi A., Gorki A.-D., Solodkin D. G., Zada M., Hladik A., Miklosi A., Salame T.-M., Halpern K. B., David E., Itzkovitz S., Harkany T., Knapp S., and Amit I., “ Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting,” Cell 175, 1031–1044 (2018). 10.1016/j.cell.2018.09.009 [DOI] [PubMed] [Google Scholar]
- 202. He B., Bergenstråhle L., Stenbeck L., Abid A., Andersson A., Borg Å., Maaskola J., Lundeberg J., and Zou J., “ Integrating spatial gene expression and breast tumour morphology via deep learning,” Nat. Biomed. Eng. 4, 827–834 (2020). 10.1038/s41551-020-0578-x [DOI] [PubMed] [Google Scholar]
- 203. Efremova M., Vento-Tormo M., Teichmann S. A., and Vento-Tormo R., “ CellPhoneDB: Inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes,” Nat. Protoc. 15, 1484–1506 (2020). 10.1038/s41596-020-0292-x [DOI] [PubMed] [Google Scholar]
- 204. Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., Fau C., Lacroix M., and Colinge J., “ SingleCellSignalR: Inference of intercellular networks from single-cell transcriptomics,” Nucl. Acids Res. 48, e55 (2020). 10.1093/nar/gkaa183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Stephenson E., Reynolds G., Botting R. A., Calero-Nieto F. J., Morgan M. D., Tuong Z. K., Bach K., Sungnak W., Worlock K. B., Yoshida M., Kumasaka N., Kania K., Engelbert J., Olabi B., Spegarova J. S., Wilson N. K., Mende N., Jardine L., Gardner L. C. S., Goh I., Horsfall D., McGrath J., Webb S., Mather M. W., Lindeboom R. G. H., Dann E., Huang N., Polanski K., Prigmore E., Gothe F., Scott J., Payne R. P., Baker K. F., Hanrath A. T., Schim van der Loeff I. C. D., Barr A. S., Sanchez-Gonzalez A., Bergamaschi L., Mescia F., Barnes J. L., Kilich E., de Wilton A., Saigal A., Saleh A., Janes S. M., Smith C. M., Gopee N., Wilson C., Coupland P., Coxhead J. M., Kiselev V. Y., van Dongen S., Bacardit J., King H. W., Baker S., Bradley J. R., Dougan G., Goodfellow I. G., Gupta R. K., Hess C., Kingston N., Lehner P. J., Matheson N. J., Owehand W. H., Saunders C., Smith K. G. C., Summers C., Thaventhiran J. E. D., Toshner M., Weekes M. P., Bucke A., Calder J., Canna L., Domingo J., Elmer A., Fuller S., Harris J., Hewitt S., Kennet J., Jose S., Kourampa J., Meadows A., O'Brien C., Price J., Publico C., Rastall R., Ribeiro C., Rowlands J., Ruffolo V., Tordesillas H., Bullman B., Dunmore B. J., Fawke S., Graf S., Hodgson J., Huang C., Hunter K., Jones E., Legchenko E., Matara C., Martin J., O'Donnell C., Pointon L., Pond N., Shih J., Sutcliffe R., Tilly T., Treacy C., Tong Z., Wood J., Wylot M., Betancourt A., Bower G., De Sa A., Epping M., Huhn O., Jackson S., Jarvis I., Marsden J., Nice F., Okecha G., Omarjee O., Perera M., Richoz N., Sharma R., Turner L., De Bie E. M. D. D., Bunclark K., Josipovic M., Mackay M., Michael A., Rossi S., Selvan M., Spencer S., Yong C., Ansaripour A., Mwaura L., Patterson C., Polwarth G., Polgarova P., Stefano G. d, Allison J., Butcher H., Caputo D., Clapham-Riley D., Dewhurst E., Furlong A., Graves B., Gray J., Ivers T., Kasanicki M., Gresley E. L., Linger R., Meloy S., Muldoon F., Ovington N., Papadia S., Phelan I., Stark H., Stirrups K. E., Townsend P., Walker N., Webster J., Rostron A. J., Simpson A. J., Hambleton S., Laurenti E., Lyons P. A., Meyer K. B., Nikolić M. Z., Duncan C. J. A., Smith K. G. C., Teichmann S. A., Clatworthy M. R., Marioni J. C., Gottgens B., Haniffa M., C. I. of Therapeutic Immunology, and I. D.-N. I. of Health Research (CITIID-NIHR) COVID-19 BioResource Collaboration, “ Single-cell multi-omics analysis of the immune response in COVID-19,” Nat. Med. 27, 904–916 (2021). 10.1038/s41591-021-01329-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Bernardes J. P., Mishra N., Tran F., Bahmer T., Best L., Blase J. I., Bordoni D., Franzenburg J., Geisen U., Josephs-Spaulding J., Kohler P., Kunstner A., Rosati E., Aschenbrenner A. C., Bacher P., Baran N., Boysen T., Brandt B., Bruse N., Dorr J., Drager A., Elke G., Ellinghaus D., Fischer J., Forster M., Franke A., Franzenburg S., Frey N., Friedrichs A., Fuß J., Gluck A., Hamm J., Hinrichsen F., Hoeppner M. P., Imm S., Junker R., Kaiser S., Kan Y. H., Knoll R., Lange C., Laue G., Lier C., Lindner M., Marinos G., Markewitz R., Nattermann J., Noth R., Pickkers P., Rabe K. F., Renz A., Rocken C., Rupp J., Schaffarzyk A., Scheffold A., Schulte-Schrepping J., Schunk D., Skowasch D., Ulas T., Wandinger K.-P., Wittig M., Zimmermann J., Busch H., Hoyer B. F., Kaleta C., Heyckendorf J., Kox M., Rybniker J., Schreiber S., Schultze J. L., and Rosenstiel P., “ Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19,” Immunity 53, 1296–1314 (2020). 10.1016/j.immuni.2020.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Zhang Z., Huang K., Gu C., Zhao L., Wang N., Wang X., Zhao D., Zhang C., Lu Y., and Meng Y., “ Molecular subtyping of serous ovarian cancer based on multi-omics data,” Sci. Rep. 6, 26001 (2016). 10.1038/srep26001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Lee J., Hyeon D. Y., and Hwang D., “ Single-cell multiomics: Technologies and data analysis methods,” Exp. Mol. Med. 52, 1428–1442 (2020). 10.1038/s12276-020-0420-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Welch J. D., Kozareva V., Ferreira A., Vanderburg C., Martin C., and Macosko E. Z., “ Single-cell multi-omic integration compares and contrasts features of brain cell identity,” Cell 177, 1873–1887 (2019). 10.1016/j.cell.2019.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Maniatis S., Aijo T., Vickovic S., Braine C., Kang K., Mollbrink A., Fagegaltier D., Andrusivová Ž., Saarenpaa S., Saiz-Castro G., Cuevas M., Watters A., Lundeberg J., Bonneau R., and Phatnani H., “ Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis,” Science 364, 89–93 (2019). 10.1126/science.aav9776 [DOI] [PubMed] [Google Scholar]
- 211. Wu S. Z., Al-Eryani G., Roden D. L., Junankar S., Harvey K., Andersson A., Thennavan A., Wang C., Torpy J. R., Bartonicek N., Wang T., Larsson L., Kaczorowski D., Weisenfeld N. I., Uytingco C. R., Chew J. G., Bent Z. W., Chan C.-L., Gnanasambandapillai V., Dutertre C.-A., Gluch L., Hui M. N., Beith J., Parker A., Robbins E., Segara D., Cooper C., Mak C., Chan B., Warrier S., Ginhoux F., Millar E., Powell J. E., Williams S. R., Liu X. S., O'Toole S., Lim E., Lundeberg J., Perou C. M., and Swarbrick A., “ A single-cell and spatially resolved atlas of human breast cancers,” Nat. Genet. 53, 1334–1347 (2021). 10.1038/s41588-021-00911-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Tyser R. C. V., Mahammadov E., Nakanoh S., Vallier L., Scialdone A., and Srinivas S., “ Single-cell transcriptomic characterization of a gastrulating human embryo,” Nature 600, 285–289 (2021). 10.1038/s41586-021-04158-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Zhang M., Eichhorn S. W., Zingg B., Yao Z., Cotter K., Zeng H., Dong H., and Zhuang X., “ Spatially resolved cell atlas of the mouse primary motor cortex by merfish,” Nature 598, 137–143 (2021). 10.1038/s41586-021-03705-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.See https://www.10xgenomics.com/products/spatial-gene-expression for the visuals on spatial transcriptomics.
- 215. Mousavi Z., Kourosh-Arami M., Mohsenzadegan M., and Komaki A., “ An immunohistochemical study of the effects of orexin receptor blockade on phospholipase c-b3 level in rat hippocampal dentate gyrus neurons,” Biotech. Histochem. 96, 191–196 (2021). 10.1080/10520295.2020.1778088 [DOI] [PubMed] [Google Scholar]
- 216.See https://biorender.com/ for accessing the mouse brain visual used in this manuscript.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available within the article.