Abstract
Molecular beacons (MBs) are stem–loop DNA probes used for identifying and reporting the presence and localization of nucleic acid targets in vitro and in vivo via target-dependent dequenching of fluorescence. A drawback of conventional MB design is present in the stem sequence that is necessary to keep the MBs in a closed conformation in the absence of a target, but that can participate in target binding in the open (target-on) conformation, giving rise to the possibility of false-positive results. In order to circumvent these problems, we designed MBs in which the stem was replaced by an orthogonal DNA analog that does not cross-pair with natural nucleic acids. Homo-DNA seemed to be specially suited, as it forms stable adenine-adenine base pairs of the reversed Hoogsteen type, potentially reducing the number of necessary building blocks for stem design to one. We found that MBs in which the stem part was replaced by homo-adenylate residues can easily be synthesized using conventional automated DNA synthesis. As conventional MBs, such hybrid MBs show cooperative hairpin to coil transitions in the absence of a DNA target, indicating stable homo-DNA base pair formation in the closed conformation. Furthermore, our results show that the homo-adenylate stem is excluded from DNA target binding, which leads to a significant increase in target binding selectivity.
INTRODUCTION
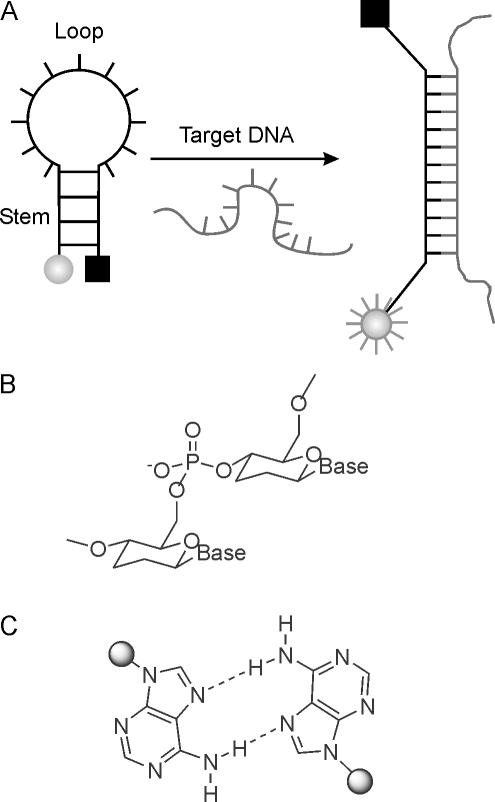
The post-genomic era continuously demands for novel and improved DNA recognition probes with high sensitivity and selectivity for use in fundamental biomedical studies, disease diagnosis, functional genomics and drug discovery. A particularly useful concept for diagnosing single stranded DNA or RNA with high mismatch discrimination is that of the molecular beacons (MBs), first described by Tyagi and Kramer (1). MBs are single-stranded oligonucleotide hybridization probes that form a stem–loop structure. The loop contains a probe sequence that is designed to be complementary to a DNA-target, and the stem is formed by complementary arm sequences that are located on either side of the probe. A fluorophore is covalently linked to the end of one arm and a quencher to the end of the other arm. In the absence of a target, the MB is closed and fluorescence is quenched. However, after hybridization to a nucleic acid target sequence the stem of the MB melts and fluorescence is emitted owing to increased distance between fluorophore and quencher (Figure 1A).
Figure 1.
(A) Working principle of an MB. In the absence of a DNA target, the MB is in its closed hairpin state, and fluorescence is quenched due to close proximity of fluorophore and quencher (left). Upon target binding to the loop region, the hairpin stem melts and fluorescence is emitted as a consequence of the increased distance of fluorophore and quencher; (B) chemical structure of homo-DNA; (C) structure of the reversed Hoogsteen A–A base pair in homo-DNA duplexes.
MB probes display higher target selectivity compared with linear probes, owing to their structurally well-defined hairpin loop state in the absence of a target (2). The MB concept has been developed and exploited extensively in the past for the detection of single nucleotide polymorphisms (3), for visualizing the distribution and transport of mRNAs in living cells (4), as probes for reporting enzymatic activity (5), as well as for a variety of other applications (6). An updated overview on literature on MBs can be found in the Internet (http://www.molecular-beacons.org).
Although MBs are very sensitive to mismatches in the probe-target region, their flanking arm sequences, containing 4–5 nt each, are prone to fortuitously participate in DNA-target binding, thus constituting a potential source of false-positive results. A solution to this problem exists in theory by replacing the stem part of an MB by an oligonucleotidic pairing system that is orthogonal and consequently does not cross-pair with DNA or RNA. Known pairing systems with such properties are, for example, enantio-DNA (L-DNA) (7–11), hexopyranosyl RNA (pRNA) (12–15) or homo-DNA (16–19).
In the approach described here, we focused on homo-DNA (Figure 1B) for the stem part of MBs. Homo-DNA, a homolog of natural DNA, was shown before by Eschenmoser and co-workers (18) to form stable, antiparallel duplexes with itself, without cross-pairing to natural DNA. Moreover, homo-DNA can easily be prepared by standard automated phosphoramidite chemistry and is thus fully compatible with DNA synthesis. Particularly intriguing is the property of homo-DNA oligoadenylates to form A–A self-pairs of the reversed Hoogsteen type (Figure 1C) with thermal stabilities that roughly match that of natural Watson–Crick base pairs. Exploiting this property reduces the number of necessary building blocks for the arms of an MB to only one. A further advantage is the expected higher enzymatic stability owing to the modifications of either end of an MB with unnatural homo-DNA building blocks.
On the basis of UV- and fluorescence-melting curves, we show here that the stems in homo-DNA MBs do not interact with flanking target DNA sequences while those of conventional MBs do and thus give rise to false-positive results. In addition, we confirmed this significantly increased binding selectivity by a DNA microarray experiment as a mimic of a more complex cDNA environment.
MATERIALS AND METHODS
Synthesis of oligonucleotides
Oligonucleotides were prepared on the 0.2 or 1.3 μmol scale on a commercial DNA-synthesizer using standard phosphoramidite chemistry on DABSYL- or DABCYL-CPG (Glen Research) solid support. Standard DNA phosphoramidite building blocks were from Glen Research. Homo-DNA A-phosphoramidite was prepared as described previously (19). The MBs FMB1–5, containing fluorescein at the 5′ end, were prepared by reaction with 6-FAM amidite, (Glen Research) in the last synthesis cycle. Cy5 containing MBs (Cy5MB1–7) were prepared by using 5′-amino-modifier C3-TFA phosphoramidite (Glen Research) as the last synthetic step, followed by post-synthetic labeling (vide infra). The coupling time was extended to 6 min for non-standard phosphoramidites. 5-(ethylthio)-1H-tetrazole (0.25 M in CH3CN) was used as the activator. The cleavage of oligonucleotides from solid supports and the cleavage of all protecting groups were carried out under standard conditions (concentrated NH3 at 55°C for 17 h).
Post-synthetic labeling of 5′-amino-modified oligonucleotides with Cy5 mono NHS-ester (Amersham Biosciences) was carried out at room temperature for 1 h in 0.1 M carbonate buffer (pH 8.5) according to the manufacturer's protocol. MBs were purified by reverse high-performance liquid chromatography on a NUCLEOSIL 300–305 C18 column (Macherey-Nagel). The MBs were desalted over SepPak (Waters) or evaporated (4×) from 500 μl of H2O and characterized by ESI−-MS (Table 2).
Table 2.
MBs and reference sequences prepared and investigated as well as their ESI− mass-spectrometric characterization
| Molecular beacons | (M − H)− calculated | (M − H)− found | |
|---|---|---|---|
| FMB1 | FAM-GCGCGAAGTTAAGACCTATGCGCGC-Dabsyl | 8726.9 | 8727.2 |
| FMB2 | FAM-CGCGAAGTTAAGACCTATGCGCG-Dabsyl | 8107.6 | 8108.5 |
| FMB3 | FAM-TCGCGAAGTTAAGACCTATGCGCGA-Dabsyl | 8725.0 | 8725.4 |
| FMB4 | FAM-aaaaAAGTTAAGACCTATGaaaa-Dabsyl | 8252.6 | 8254.2 |
| FMB5 | FAM-aaaaaAAGTTAAGACCTATGaaaaa-Dabsyl | 8906.4 | 8906.8 |
| Cy5MB1 | Cy5-NH2C3-CCGGTGGGTTGCTTTGAAGCCGG-Dabcyl | 8364.0 | 8362.7 |
| Cy5MB2 | Cy5-NH2C3-CCGGTGGGTTGCATTGAAGCCGG-Dabcyl | 8373.0 | 8373.3 |
| Cy5MB3 | Cy5-NH2C3-CCGCTGGGTTGCATTGAAGGCGG-Dabsyl | 8409.0 | 8409.7 |
| Cy5MB4 | Cy5-NH2C3-GCGGTGGGTTGCATTGAAGCCGC-Dabcyl | 8373.0 | 8373.8 |
| Cy5MB5 | Cy5-NH2C3-GCGCTGGGTTGCATTGAAGGCGC-Dabcyl | 8373.0 | 8373.5 |
| Cy5MB6 | Cy5-NH2C3-aaaaTGGGTTGCTTTGAAGaaaa-Dabsyl | 8544.4 | 8544.9 |
| Cy5MB7 | Cy5-NH2C3-aaaaTGGGTTGCATTGAAGaaaa-Dabsyl | 8553.4 | 8554.0 |
| Cy5Ref | Cy5-NH2C3-CTTTTGAGCGTGAGC | 5375.0 | 5373.9 |
Upper case letters, 2′-deoxynucleotides; lower case letters, homo-DNA nucleotides; bold, probe region; italics, stem region; bold underline, mismatched base.
UV-melting curves
Oligonucleotides were mixed in a 1:1 stoichiometry using the UV-extinction coefficients of natural oligodeoxynucleotides. UV-melting curves were recorded on a Cary 3E UV/vis spectrophotometer (Varian) at 260 and 280 nm in 50 mM KCl, 10 mM Tris and 3.5 mM MgCl2, pH 8.0 at a duplex concentration of 2 μM. Consecutive heating–cooling–heating cycles in the temperature interval of 15–85°C with a linear gradient of 0.5°C/min were applied. Heating and cooling ramps were superimposable in all cases, indicating equilibrium conditions.
Fluorescence spectra
Steady-state fluorescence emission spectra of the fluorescein-labeled MBs FMB1–5 (Table 2) were recorded at 517 nm upon excitation at 490 nm with a bandwidth of 5 nm for the excitation and emission by using a Cary Eclipse Fluorescence spectrophotometer (Varian). The concentration of MBs and targets was 200 nM in 50 mM KCl, 10 mM Tris and 3.5 mM MgCl2, pH 8.0. Concentrations of MBs were calculated by using the approximation that 1 OD260/ml = 33 μg oligonucleotide. A quartz cuvette with a volume capacity of 3000 μl was used for the measurements. Every sample was denatured and annealed at room temperature overnight before measurement. Fluorescence intensity of MB/target duplexes was measured between 15 and 85°C by heating at a rate of ∼0.5°C/min. The maximum intensity (at 517 nm) was taken for the fluorescence-temperature profiles.
Incubation of microarrays
Hybridization to the Escherichia coli K12 starter V2 array (MWG-Biotech, Germany) was carried out with 100 nM (120 μl) of Cy5-labeled MBs Cy5MB1–7, together with an equal amount of reference oligonucleotide Cy5Ref (Table 2) in hybridization buffer (MWG-Biotech) at 15°C for 2 h. Subsequently, the slides were washed successively with 30 mM sodium citrate, 300 mM NaCl, 0.1% SDS (2×), 15 mM sodium citrate, 150 mM NaCl (1×) and 7.5 mM sodium citrate, 75 mM NaCl (1×), 110 ml each, at 4°C for 5 min. The slide was dried by centrifugation (Heraeus-Christ, Labofuge III) at room temperature for 5 min at 1200 r.p.m.
Imaging and quantification
Microarrays were scanned at 10 μm resolution with a ScanArray 4000 (Packard Biochip Technologies) scanning laser confocal fluorescence microscope. The emitted fluorescence signal was detected by a photomultiplier tube (PMT) at 670 nm. For all microarray experiments, the laser power and PMT gain were calibrated to the fluorescence intensity of the reference oligonucleotide Cy5Ref, set to 100%. The fluorescent signals were analyzed by quantifying the pixel density (intensity) of each spot using QuantArray (Version 3, Packard Bioscience). The local background signal was automatically subtracted from the hybridization signal of each separate spot, and then the mean signal intensity of each spot was used for data analysis. The relative fluorescence Frel was calculated by Frel = FSpot/FRef × 100, where FSpot is the absolute fluorescence intensity of each spot and FRef the absolute intensity of Cy5Ref.
RESULTS
To determine the effect of one or two additional base pairs flanking a potential probe sequence on thermal stability, we first measured Tms on a linear probe sequence, simulating a 15mer target region and one additional stem base of a MB on either side (Table 1, X,Y). These probes were hybridized to potential targets T7–11 with a T-overhang and either a full, A–T match or an A–A mismatch in the center of the probe. The bases opposite X and Y on the probes were varied. In the case of the probe with X,Y = G,C it appeared that the mismatch in the center of the probe reduced the Tm by ∼9°C in the absence of any attractive interaction between X,Y and the targets. However, one or two additional base pairs flanking the probe region led to a recovery of the Tm to values that almost reached that of the fully matched duplex, despite the presence of a central A–A mismatch. This clearly shows that 1–2 additional base pairs to the flanking tract of a probe sequence can thermally compensate for a mismatch in the probe region. The same experiments with a hybrid probe containing homo-A nucleotides in positions X and Y showed no increase in Tm for all combinations of natural bases in the target, indicating the absence of attractive interactions between homo-A and opposing natural nucleotides at the junction site and thus supporting the concept.
Table 1.
Tm data (260 nm) of a linear probe with homo-DNA or DNA cap-nucleotides and the corresponding matched or mismatched targets T7–11, showing in some cases additional interactions with the cap-nucleotides of the probe
| 5′-XXAAGTTAAGACCTATGY-3′ | Tm (°C)a | ||
|---|---|---|---|
| X,Y = a,a | X,Y = G,C | ||
| T7 | 3′-TTTTTTTCAATTCTGGATACTTTTT-5′ | 51.6 | 50.1 |
| T8 | 3′-TTTTTTTCAATTCAGGATACTTTTT-5′ | 42.3 | 41.2 |
| T9 | 3′-TTTTTTTCAATTCAGGATACGTTTT-5′ | 42.5 | 48.0 |
| T10 | 3′-TTTTCTTCAATTCAGGATACGTTTT-5′ | 43.4 | 50.7 |
| T11 | 3′-TTTTATTCAATTCAGGATACATTTT-5′ | 42.2 | 41.7 |
Upper case letters, 2′-deoxynucleotides; lower case letters, homo-DNA nucleotides; bold, target sequence; bold italics, junction site; bold underline, mismatch.
aDuplex concentration: 2 μM in 50 mM KCl, 10 mM Tris and 3.5 mM MgCl2, pH 8.0.
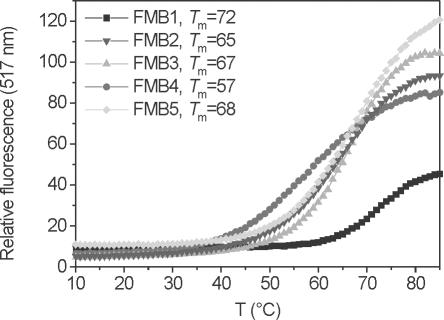
The next step was to determine whether MBs with a homo-DNA stem are able to form hairpin structures. For this purpose, we prepared the MBs FMB1–5 that were fluorescein labelled at their 5′ end and that contained a Dabsyl quencher at their 3′ end (Table 2). FMB1 and FMB3 were equipped with a natural 5 bp DNA stem with a G–C or an A–T base pair terminating the stem, while MB FMB4,5 contained a 4 or 5 bp homoadenine DNA stem. Thermal denaturation profiles (Figure 2) showed, for all cases, sigmoidal melting with Tms between 57 and 72°C. Comparison of the Tms of FMB3 and FMB4 clearly reveals similar thermal stability of the two hairpin structures, indicating similar thermal stability of a pentahomoadenosyl-DNA stem to an unmodified DNA stem with four G–C and one A–T pair at the end. Residual fluorescence in the closed form was similar for all MBs, indicating similar quenching efficiency. In the melted form, the relative fluorescence signal was again of similar magnitude except for the case of FMB1, in which the ∼2-fold reduced signal intensity is most likely due to quenching by the dG-nucleotide to which the fluorophore was attached. Thus, homo-DNA and conventional MBs seem to have a similar dynamic signal to background ratios.
Figure 2.
Fluorescence-melting curves and corresponding Tms of the MBs FMB1–5 in the absence of a DNA target. The concentrations of MBs were 200 nM in 50 mM KCl, 10 mM Tris and 3.5 mM MgCl2, pH 8.0. The monophasic transitions reflect hairpin to random coil transitions.
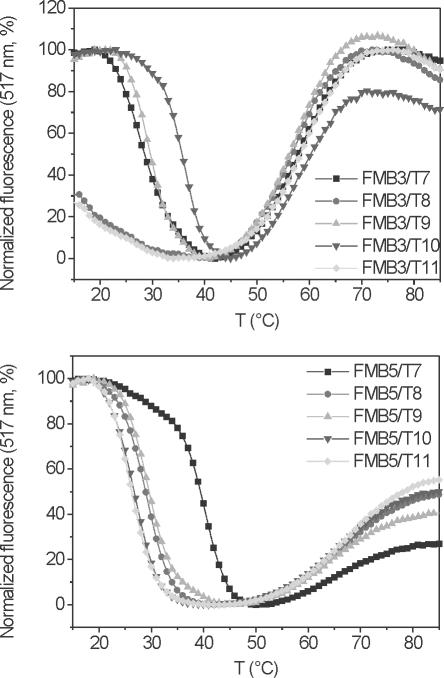
A comparison of target binding selectivity of homo-DNA and conventional MBs was subsequently performed with FMB3 and FMB5, due to the similar thermal stabilities of their stems. Fluorescence-melting profiles were recorded after incubation with targets T7–T11 (Figure 3). The melting profiles typically showed high relative fluorescence at low temperatures, where the targets are bound to the loop and the hairpin is molten. Upon increasing the temperature, fluorescence is quenched owing to loss of the target and concomitant hairpin formation of the MB. The second transition occurring at higher temperature is associated with an increase in fluorescence and corresponds to the melting of the hairpin to a random coil MB structure. Thus, the transition at lower temperature is indicative for target binding. It becomes clear that the conventional MB FMB3 binds best and strongest to the target T10, containing a mismatch in the center and two matching base pairs to the stem region (Figure 3, top and Table 3). The target T9, containing a central mismatch and only one additional G–C base pair to the stem still shows equal affinity to the MB as its real target T7, containing no mismatch and no flanking assistance from pairing of the target to the stem. These results clearly show that stem participation in natural DNA MBs occurs and can severely obstruct sequence analysis.
Figure 3.
Fluorescence-melting curves of MBs FMB3 (top) and FMB5 (bottom) in the presence of the DNA targets T7–11. The concentration of MBs and targets were 200 nM each in 50 mM KCl, 10 mM Tris and 3.5 mM MgCl2, pH 8.0. The transition at lower temperature corresponds to DNA-target melting from the MBs and concomitant hairpin formation. The transition at higher temperature reflects the hairpin to random coil transition of the MBs.
Table 3.
Tm data [fluorescence-melting curves (°C)] for dissociation of targets T7–11 from the MBs FMB3 and FMB5
| Target | FMB3 | FMB5 |
|---|---|---|
| T7 | 29 | 40 |
| T8 | <15 | 29 |
| T9 | 30 | 29 |
| T10 | 35 | 27 |
| T11 | <15 | 27 |
Experimental conditions as indicated in Figure 3.
The situation is different with the homo-DNA MB FMB5 (Figure 3, bottom), in which only the fully matched target T7 binds strongly to the MB while all other targets show lower Tms, irrespective of the nature of the flanking sequence. The relative discrimination of binding to target T7 and T8 (match versus mismatch in the probe region) of the conventional MB FMB3 seems to be higher than that of the homo-DNA MB FMB5 (Table 3). Also, we note that the Tms for target binding are generally higher in the case of FMB5 relative to FMB3, despite equal stability of the loop–stem structure of the two MBs.
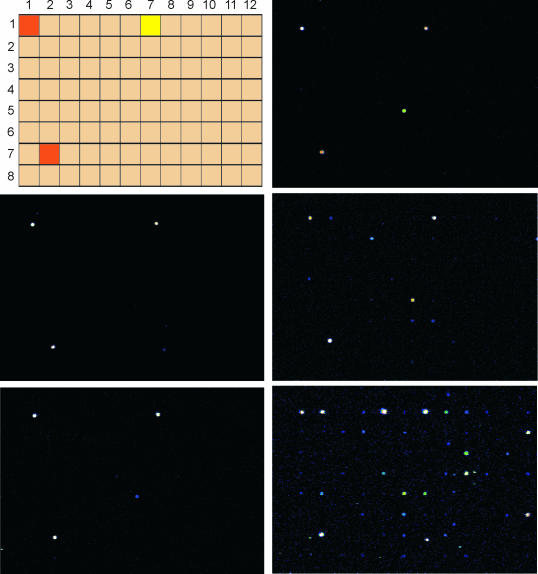
In order to prove the increased sequence selectivity of homo-DNA MBs over conventional MBs, we decided to probe target selectivity on a DNA array. The commercial E.coli K12 starter V2 array from MWG-Biotech was chosen for this purpose. It contains 94 different 50mer oligodeoxynucleotides (Figure 4, top left), of which B3734 was chosen as the target, as it is represented twice on the slide. Oligonucleotide Ara5 was targeted by the Cy5-labeled, linear reference oligonucleotide Cy5Ref that was used as an internal standard for loading control.
Figure 4.
Top left: relative arrangement of the 96 oligonucleotides on the E.coli K12 starter V2 array from MWG-Biotech. The location of the target oligodeoxynucleotide B3734 that appears in duplicate on the slide is depicted in orange, the reference oligonucleotide Ara5 that is complementary to Cy5Ref is in yellow. Fluorescence microscope pictures after hybridization and washing under non-stringent (15°C) conditions: center left, Cy5MB6/Cy5Ref; bottom left, Cy5MB7/Cy5Ref; top right, Cy5MB1/Cy5Ref; center right, Cy5MB2/Cy5Ref, bottom right, Cy5MB4/Cy5Ref.
MBs Cy5MB1–7, containing a 15mer loop sequence that is complementary to the central part of B3734, were equipped with either a 4 bp homo-DNA stem (CyMB6 and 7) or a 4 bp natural CG-stem with different sequence motifs (CyMB1–5). Again, Cy5MBs contained a fluorescence quencher at the 3′ end and a Cy5 dye at the 5′ end, linked via a C3-linker element. Cy5MB1 and Cy5MB6 were complete matches to the target while Cy5MB2–5 and Cy5MB7 contained a mismatch at position 9 of the probe sequence. The arrays were incubated with reference oligonucleotide Cy5Ref and each of the Cy5MB1–7 separately, using hybridization conditions of low stringency (15°C). This was explicitly chosen to work out the differences of the intrinsic binding properties of the two MB-systems. Figure 4 highlights fluorescence microscope pictures of the arrays for selected cases, and Figure 5 shows quantitative binding data to the most prominent binding sites.
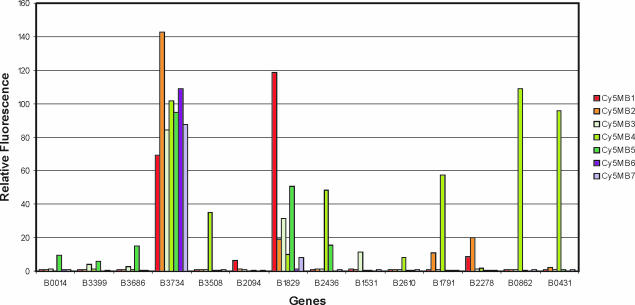
Figure 5.
Quantitative data of binding of the Cy5MB1–7 to the target B3734 and the 13 most important off-target sites in the array experiment. The data are referenced to the linear oligonucleotide Cy5Ref, the binding of which to target Ara5 was set to 100%.
It appears that the fully matched homo-DNA MB Cy5MB6 only binds to its two target sites while that containing one mismatch (Cy5MB7) shows one additional off-target binding site (Figure 5, target B1829) to which it binds with low affinity (∼10%). Thus, under the given non-stringent conditions, where no single mismatch discrimination can be expected, a very well behaved binding pattern occurs with off-target binding being essentially absent.
The situation becomes different with MBs having a natural CCGG stem with or without a mismatch in the probe sequence. Already, Cy5MB1 with no mismatch bound even stronger (∼120%) to B1829 compared with its real target B3734 (∼70%). The situation becomes even worse with Cy5MB2 having the central mismatch. This probe bound appreciably to at least 10 off-target sequences (Figure 4) and, for an unknown reason, even stronger (∼140%) to its target B3734 than Cy5MB1, having no mismatch (Figure 5). This stays in contrast to the MB with the homo-DNA stem, where only one very weak (<10%) off-target binding site was found. Permutation of the stem nucleotides in the conventional MBs while leaving the mismatch in the center of the probe (Cy5MB3–5) made participation of stem binding even clearer. Interestingly, sequence alignment of CyMB2–5 to the off-targets found in the array, based on Watson–Crick binding (see Supplementary Material), led to no rational explanation of the observed affinities.
DISCUSSION
MBs are extremely useful hybridization probes that can report the presence of a DNA or RNA analyte in vitro and its localization in vivo displaying a high signal to background ratio and enhanced specificity compared with linear oligonucleotide probes. In addition, MBs can be color coded by using appropriate fluorophores as reporter units, which offers an additional degree of freedom in analysis. However, when working with cDNA libraries as targets or in vivo, considerable care has to be taken in the design of the hairpin arms. In typical MBs, these arms consist of up to two-thirds of the number of nucleotides involved in target recognition and are thus a potential source of false-positive results owing to participation in binding to the targets. Optimization of MB technology in the past resulted in the use of 2′-O-Me-RNA MBs in order to increase biostability and affinity of the MBs to RNA and to abolish RNase H degradation of the targets at the same time (20). To better control the role of the stems in target binding, shared-stem MBs were designed in which at least one arm of the stem is part of the recognition sequence (21). But in these cases, the increased selectivity of conventional MBs to their targets, relative to linear oligonucleotides, is lost. This increased selectivity is due to the reference state of the unbound MB being a hairpin and not a random coil structure. Thus, novel design strategies for improved selectivity better maintain this structural feature. A very recent approach in circumventing random binding of the stem part after target recognition involved the use of stems that are attached in an inverted fashion to the probe region, via 3′–3′ and 5′–5′ junctions (22). Although this measure increases selectivity, it is certainly not optimal, as stem participation is not principally excluded in this case also.
The use of an oligonucleotidic pairing system that is orthogonal to DNA and RNA for stem design, as described here, is conceptually simple and straightforward. It confers higher biostability (especially exonuclease stability) to MBs and can, as reported here, reduce the number of necessary building blocks for stem components to one. This renders MB synthesis easy and economic. The obtained results with the homo-DNA MBs are of interest in several aspects. Compared with conventional MBs, those with a homo-DNA stem show significantly enhanced target selectivity by abolishing secondary attractive interactions between stem and target. Neither with defined linear hybrid oligonucleotides, nor with homo-DNA MBs was any contribution of homo-DNA to target binding observed. The signal-to-background ratios of the homo-DNA MBs FMB4,5 are similar to that of the conventional FMB2,3. This can be deduced from the fluorescence-melting profiles without target, as well as from the melting experiments with the different targets T7–11 (Figures 2 and 3). The intrinsic selectivity of loop-target binding (match versus mismatch in the loop region) in absence of possible secondary interactions via the arm sequences (targets T7/T8) shows a slight advantage for the conventional MB FMB3 relative to the homo-DNA MB FMB5. Whether the higher target discrimination of FMB3 is merely owing to the generally lower Tms for target binding (Table 3) or is intrinsic to the DNA stem structure remains unclear at this point and calls for a thermodynamic analysis of the sequential transitions depicted in Figure 3. Finally, the fact that no other nucleoside than homoadenosine is required for homo-DNA stem design obliterates the problem of fluorescence quenching by a neighboring guanine base that has to be taken care of in conventional MB design (e.g. FMB1).
The use of non-natural oligonucleotidic pairing systems in biotechnology is appealing and only little explored. Among the few examples are L-DNA that has been used in the past as aptamers (spiegelmers) for selective binding of biopolymers (23), pyranosyl RNA as a nanosystem (24) and the 3′-deoxy variant of pRNA (pDNA) as a scaffold for tuning aptamer and ribozyme activity (25). The work described here contains the first application of homo-DNA in nucleic acid biotechnology.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr Eric Grüter and Prof. R. Häner for helpful advice with the DNA array experiments. Financial support from the Swiss National Science Foundation (grant no: 200020-107692) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the University of Bern.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet G., Tyagi S., Libchaber A., Kramer F.R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl Acad. Sci. USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyagi S., Bratu D.P., Kramer F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 4.Bratu D.P., Cha B.J., Mhlanga M.M., Kramer F.R., Tyagi S. Visualizing the distribution and transport of mRNAs in living cells. Proc. Natl Acad. Sci. USA. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundu L.M., Burgdorf L.T., Kleiner O., Batschauer A., Carell T. Cleavable substrate containing molecular beacons for the quantification of DNA-photolyase activity. Chembiochem. 2002;3:1053–1060. doi: 10.1002/1439-7633(20021104)3:11<1053::AID-CBIC1053>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 6.Tan W., Wang K., Drake T.J. Molecular beacons. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 7.Garbesi A., Capobianco M.L., Colonna F.P., Tondelli L., Arcamone F., Manzini G., Hilbers C.W., Aelen J.M.E., Blommers M.J.J. L-DNAs as potential antimessenger oligonucleotides: a reassessment. Nucleic Acids Res. 1993;21:4159–4165. doi: 10.1093/nar/21.18.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damha M.J., Giannaris P.A., Marfey P., Reid L.S. Oligonucleotides containing unnatural L-2′-deoxyribose. Tetrahedron Lett. 1991;32:2573–2576. [Google Scholar]
- 9.Damha M.J., Giannaris P.A., Marfey P. Antisense L/D-oligonucleotide chimeras: nuclease stability, base-pairing properties, and activity at directing Ribonuclease H. Biochemistry. 1994;33:7877–7885. doi: 10.1021/bi00191a015. [DOI] [PubMed] [Google Scholar]
- 10.Urata H., Ogura E., Shinohara K., Ueda Y., Akagi M. Synthesis and properties of mirror-image DNA. Nucleic Acids Res. 1992;20:3325–3332. doi: 10.1093/nar/20.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashley G.W. Modeling, synthesis and hybridization properties of (L)-ribonucleic acid. J. Am. Chem. Soc. 1992;114:9731–9736. [Google Scholar]
- 12.Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 13.Beier M., Reck F., Wagner T., Krishnamurthy R., Eschenmoser A. Chemical etiology of nucleic acid structure: comparing pentopyranosyl-(2′–4′) oligonucleotides with RNA. Science. 1999;283:699–703. doi: 10.1126/science.283.5402.699. [DOI] [PubMed] [Google Scholar]
- 14.Pitsch S., Krishnamurthy R., Bolli M., Wendeborn S., Holzner A., Minton M., Lesueur C., Schönvogt I., Jaun B., Eschenmoser A. Pyranosyl-RNA (‘p-RNA’): base-pairing selectivity and potential to replicate. Helv. Chim. Acta. 1995;78:1621–1635. [Google Scholar]
- 15.Pitsch S., Wendeborn S., Jaun B., Eschenmoser A. Why pentose- and not hexose-nucleic acids? Part VII. Pyranosyl-RNA (‘p-RNA’) Helv. Chim. Acta. 1993;76:2161–2183. [Google Scholar]
- 16.Eschenmoser A., Dobler M. Warum Pentose und nicht Hexose-Nucleinsäuren? Teil I. Helv. Chim. Acta. 1992;75:218–259. [Google Scholar]
- 17.Otting G., Billeter M., Wüthrich K., Roth H.-J., Leumann C., Eschenmoser A. Warum Pentose und nicht Hexose-Nucleinsäuren? Teil IV, Homo-DNS: 1H-,13C-,31P- und 15N NMR spektroskopische Untersuchungen von ddGlc (A-A-A-A-A-T-T-T-T-T) in wässriger Lösung. Helv. Chim. Acta. 1993;76:2701–2756. [Google Scholar]
- 18.Hunziker J., Roth H.-J., Böhringer M., Giger A., Diederichsen U., Göbel M., Krishnan R., Jaun B., Leumann C., Eschenmoser A. Warum Pentose und nicht Hexose-Nucleinsäuren?, Teil III, Oligo(2′,3′-dideoxy-β-d-glucopyranosyl)-nucleotide (Homo-DNS): Paarungseigenschaften. Helv. Chim. Acta. 1993;76:259–352. [Google Scholar]
- 19.Böhringer M., Roth H.-J., Hunziker J., Göbel M., Krishnan R., Giger A., Schweizer B., Schreiber J., Leumann C., Eschenmoser A. Warum Pentose und nicht Hexose-Nucleinsäuren? Teil II, Oligonucleotide aus 2′,3′-dideoxy-β-d-glucopyranosylbausteinen (‘Homo-DNS’): Herstellung. Helv. Chim. Acta. 1992;75:1416–1477. [Google Scholar]
- 20.Tsourkas A., Behlke M.A., Bao G. Hybridization of 2′-O-methyl and 2′-deoxy molecular beacons to RNA and DNA targets. Nucleic Acids Res. 2002;30:5168–5174. [PMC free article] [PubMed] [Google Scholar]
- 21.Tsourkas A., Behlke M.A., Bao G. Structure–function relationships of shared-stem and conventional molecular beacons. Nucleic Acids Res. 2002;30:4208–4215. doi: 10.1093/nar/gkf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne K.A. Sequence-specific, self-reporting hairpin inversion probes. J. Am. Chem. Soc. 2005;127:1989–1994. doi: 10.1021/ja046369w. [DOI] [PubMed] [Google Scholar]
- 23.Eulberg D., Buchner K., Maasch C., Klussmann S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: identification of a biostable substance P antagonist. Nucleic Acids Res. 2005;33:e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eschenmoser A., Miculka C., Windhab N. Preparation and uses of non-helical supramolecular nanosystems based on pyranosyl-nucleotides. Patent WO1997EP06907 19971210(WO9825953) 1998
- 25.Ackermann D., Wu X., Pitsch S. 3-Deoxyribopyranose (4′→2′)-oligonucleotide duplexes (p-DNA duplexes) as substitutes of RNA hairpin structures. Helv. Chim. Acta. 2002;85:1463–1478. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.