Targeting the DNA/RNA helicase DHX9 induces the accumulation of double-stranded RNA and R-loops in small cell lung cancer cells, enhancing immunotherapy response and opening new therapeutic opportunities in immunologically ‘cold’ tumors.
Abstract
Activating innate immunity in cancer cells through cytoplasmic nucleic acid sensing pathways, a phenomenon known as “viral mimicry,” has emerged as an effective strategy to convert immunologically “cold” tumors into “hot.” Through a curated CRISPR-based screen of RNA helicases, we identified DExD/H-box helicase 9 (DHX9) as a potent repressor of double-stranded RNA (dsRNA) in small cell lung cancers (SCLC). Depletion of DHX9 induced accumulation of cytoplasmic dsRNA and triggered tumor-intrinsic innate immunity. Intriguingly, ablating DHX9 also induced aberrant accumulation of R-loops, which resulted in an increase of DNA damage–derived cytoplasmic DNA and replication stress in SCLCs. In vivo, DHX9 deletion promoted a decrease in tumor growth while inducing a more immunogenic tumor microenvironment, invigorating responsiveness to immune-checkpoint blockade. These findings suggest that DHX9 is a crucial repressor of tumor-intrinsic innate immunity and replication stress, representing a promising target for SCLC and other “cold” tumors in which genomic instability contributes to pathology.
Significance:
One promising strategy to trigger an immune response within tumors and enhance immunotherapy efficacy is by inducing endogenous “virus-mimetic” nucleic acid accumulation. Here, we identify DHX9 as a viral-mimicry-inducing factor involved in the suppression of double-stranded RNAs and R-loops and propose DHX9 as a novel target to enhance antitumor immunity.
See related commentary by Chiappinelli, p. 389.
This article is featured in Selected Articles from This Issue, p. 384
INTRODUCTION
Activating innate immunity in cancer cells is a potent means by which cold (“immune-desert” or “immune-excluded”) tumors can be turned hot (“immune-inflamed,” in which T cells and other immune cells are positioned near the tumor cells), enhancing responsiveness to immune-checkpoint blockade (ICB) therapies (1–5). One promising strategy to trigger an innate immune response within tumors and boost cancer immunotherapy is by inducing the intracellular accumulation of endogenous “virus-mimetic” nucleic acids, including double-stranded RNAs (dsRNA) and double-stranded DNAs (dsDNA; refs. 6–9). These nucleic acids are sensed by the host innate immune system as evidence of viral replication and provoke an immediate and robust IFN-driven antiviral response (10, 11). Such an antiviral response is potently immunogenic, and viral-mimicry-inducing therapies have shown very promising results in preclinical models (6, 9–13) and in clinical trials (14, 15).
Small cell lung cancer (SCLC), the most lethal type of lung cancer, is a classic example of a cold tumor. Despite having one of the highest mutational burdens because of its strong association with tobacco smoking, SCLC is characterized by a reduced antigen presentation and an immunologically desert tumor microenvironment (TME; refs. 16–18). Despite the addition of ICB therapy to standard platinum-based chemotherapy in first-line treatment, SCLC remains a devastating disease and only a minority of SCLC patients derive lasting benefit from these treatments (19–22). Recent studies analyzing murine and human SCLC tumors suggest the existence of an “inflamed” SCLC subtype characterized by high expression of immune-related genes and human leukocyte antigens (HLA) and exhibiting the greatest benefit from ICB therapy among all the SCLC subtypes (9, 23, 24). This emphasizes the urgent need to identify novel therapeutic targets able to enhance antitumor immunity and ultimately sensitize these cold tumors to immunotherapies.
Although SCLCs have been uniformly treated with DNA-damaging platinum-based chemotherapy, recent studies have demonstrated the potential of replication stress inducers as a therapeutic strategy for SCLC tumors (12, 13, 25), including inhibitors of poly (ADP-ribose) polymerase (PARP) and checkpoint kinase 1 (CHK1). These inhibitors block the DNA damage response (DDR) pathway to induce replication stress and cell death in cancer cells whose genome is unstable because of mutations in DDR pathway genes and/or dysregulated cell-cycle progression. Recently, PARP, CHK1, and WEE1 inhibitors have been reported to work in synergy with PD-1/PD-L1 blockade in SCLC (26, 27) as well as triple-negative breast cancer (28) and ovarian cancer (29). PARP or CHK1 inhibition induces cyclic GMP-AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway activation, which senses cytoplasmic DNA generated by DNA damage, activating tumor-intrinsic innate immunity (26).
In the current study, we undertook a discovery effort to identify critical regulators of dsRNA in SCLC cells through a curated CRISPR-based screen of RNA helicases and identified the DExD/H-box helicase 9 (DHX9) as a promising strategy by which viral mimicry can be triggered in immunologically cold tumors. DHX9 is an abundant RNA/DNA helicase capable of unwinding both RNA and DNA duplexes, as well as more complex nucleic acid structures (30, 31). Its functions include regulation of transcription, RNA processing and transport, and maintenance of genomic stability (32). We demonstrate that DHX9 suppresses the accumulation of dsRNAs as well as R-loops (DNA/RNA hybrids), and its depletion leads not only to an antiviral immune response but also to DNA replication stress and DNA damage in SCLC cells. DHX9 deletion causes a dramatic decrease in cancer cell viability in vitro and promotes increased immunogenicity in mouse models of SCLC, significantly enhancing ICB responsiveness. These findings provide the first description of the role of DHX9 on tumor immunity and genomic instability, and the identification of a novel viral mimicry-inducing strategy to enhance antitumor immunity and boost cancer immunotherapy in immunologically cold tumors, such as SCLC.
RESULTS
DHX9 Suppresses dsRNA Accumulation in SCLCs
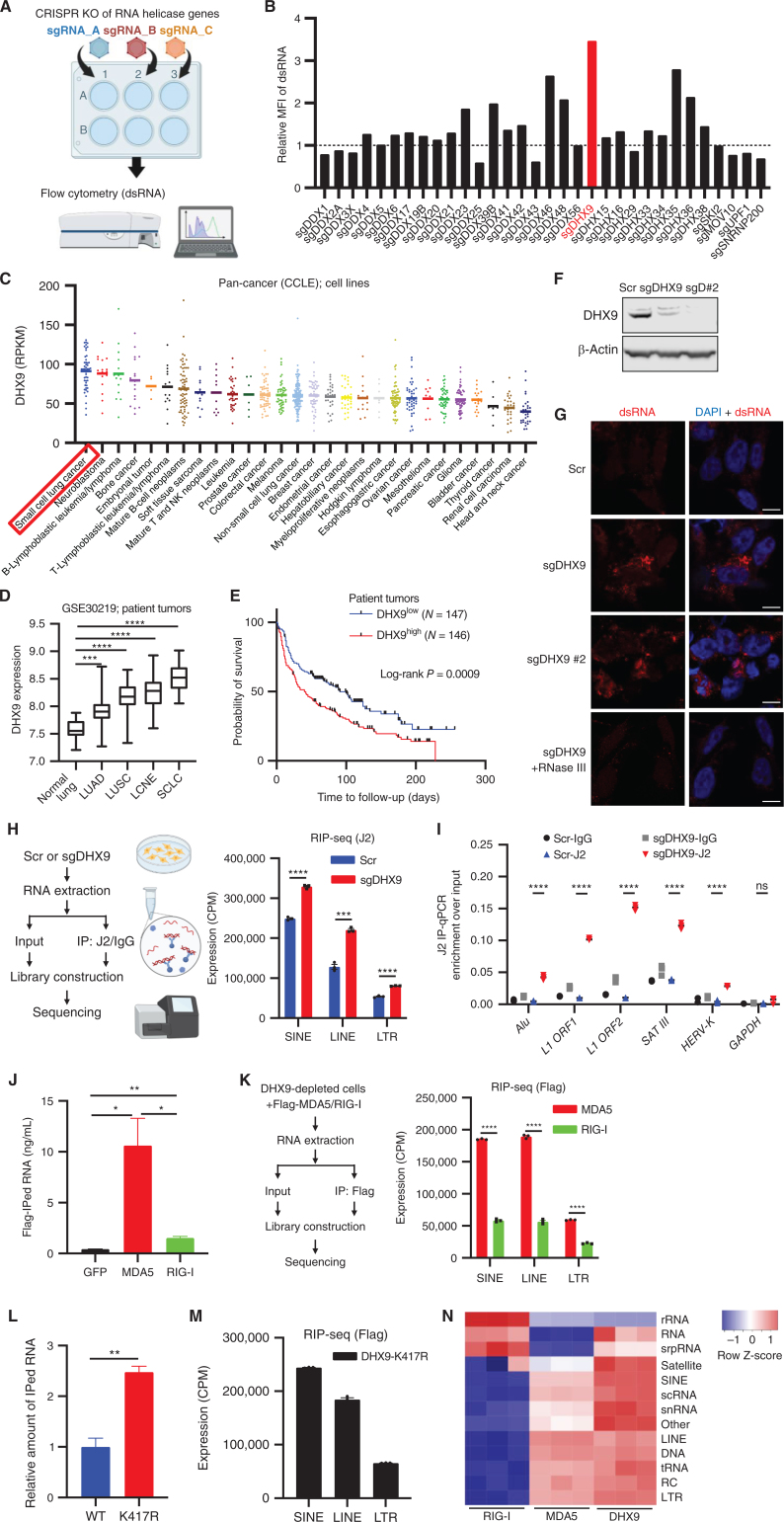
Previous studies have revealed that some RNA helicases have the ability to unwind dsRNA structures (31, 33, 34). Thus, we hypothesized that targeting RNA helicases would be a novel and efficient strategy to induce innate immune response through dsRNA accumulation in cold tumors. To identify RNA helicases whose functions are critical for dsRNA unwinding in SCLC cells, we performed a curated flow cytometry-based CRISPR screen that detected levels of intracellular dsRNA in SCLC cells (Fig. 1A and B). We included sgRNAs targeting 32 different genes (Supplementary Table S1) that have been reported to work as RNA helicases and to contribute to RNA-related functions including splicing, nuclear export, cytoplasmic transport, translation regulation, and mRNA degradation (35). We used the H446 SCLC cell line because it showed the potential to strongly increase endogenous dsRNA after treatment with decitabine, a DNA-methyltransferase inhibitor reported to induce dsRNA accumulation in cancer cells (ref. 10; Supplementary Fig. S1A). Among the top hit candidates of the screen (Fig. 1B), sgRNA-mediated depletion of the DExD/H-box helicase 9 (DHX9) significantly increased dsRNA. Intriguingly, the tumor type with the highest expression of DHX9 levels among all the tumor types registered in the Cancer Cell Line Encyclopedia (CCLE) database was SCLC (Fig. 1C). Indeed, most SCLC cell lines exhibited high expression of DHX9 at the protein level (Supplementary Fig. S1B). To investigate the relevance of DHX9 in human SCLC and other tumor types, we interrogated transcriptomic data from public cancer data sets, including The Cancer Genome Atlas (TCGA). Importantly, DHX9 overexpression was observed in many human lung cancer types compared with normal lung, and SCLC showed the strongest DHX9 expression among all the lung cancer subtypes (Fig. 1D), consistent with the CCLE analysis (Fig. 1C). Together, these results suggest that DHX9 would be a promising target in SCLC. Next, we sought to investigate the relevance of DHX9 expression on patient survival. Kaplan–Meier analysis of lung tumor patients, as well as other tumor types including ovarian and breast cancers, revealed that high DHX9 expression was associated with poor patient prognosis (Fig. 1E; Supplementary Fig. S1C), which is consistent with previous reports on prostate cancer and hepatocellular carcinoma patients (36, 37).
Figure 1.
DHX9 suppresses double-stranded RNA (dsRNA) accumulation in SCLCs. A, Schematic of the screen to identify critical regulators of dsRNA. Created with BioRender.com. B, Result of the dsRNA regulator screen. Relative mean fluorescence intensity (MFI) of dsRNA level in H446 cells depleted of RNA helicase genes was compared. C, DHX9 mRNA expression was profiled in 28 cancer types. The expression data of cancer cell lines (CCLE) were downloaded from cBioPortal, and the cell lines were subgrouped based on the information from the Depmap database (sample_info.csv, “Subtype”). In case “Subtype” information is not available, “primary_disease” was used for subgrouping. D, Analysis of DHX9 expression in indicated lung cancer (patient tumor) subtypes and normal lung. Data were downloaded from the GEO database (GSE30219). Normal lung tissue (N = 14), LUAD (N = 85), LUSC (N = 61), LCNE (N = 56), SCLC (N = 20). Bars indicate the min and max values. E, Survival curve analysis of lung tumor patients. Data were downloaded from the GEO database (GSE30219). F, Immunoblot (IB) of DHX9 protein in Scramble, sgDHX9, and sgDHX9 #2 H446 cells. G, Immunofluorescence images of dsRNA (red) staining of Scramble or sgDHX9 cells (treated w/wo RNase III). Nuclei were counterstained with DAPI. Scale bar = 10 μm. H, Schematic (left) and result (right) of J2-RIP-seq analysis. Expression levels of specific retrotransposon classes (SINE, LINE, LTR) in Scramble or sgDHX9 cells are summarized (n = 3). CPM, counts per million. I, Result of RIP-qRT-PCR analysis of the indicated retrotransposon elements (n = 3). 36B4 was used as a reference. J, RNA amounts that were pulled down with Flag antibody were compared among Flag-GFP-, Flag-MDA5-, and Flag-RIG-I-expressing cells. K, Schematic (left) and result (right) of sequencing analysis of RNA pulled down with Flag antibody. Expression levels of specific retrotransposon classes (SINE, LINE, LTR) are summarized (n = 3). CPM: counts per million. L, Relative RNA amounts that were pulled down with Flag antibody were compared between 3xFlag-DHX9-WT- and 3xFlag-DHX9-K417R-expressing cells. M, Sequencing analysis of RNA pulled down with Flag antibody. Expression levels of specific retrotransposon classes (SINE, LINE, LTR) are summarized (n = 3). CPM: counts per million. N, Heat map of Flag-RIP-seq results comparing DHX9, MDA5, and RIG-I bound RNA species (n = 3). rRNA: ribosomal RNA, srpRNA: signal recognition particle RNA, scRNA: small conditional RNA, snRNA: small nuclear RNA, tRNA: transfer RNA, RC: rolling circle, RNA: other RNA repeats, DNA: DNA repeat elements. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student t test (D, H, K, and L), log-rank test (E), one-way ANOVA (J), two-way ANOVA followed by the Tukey multiple comparisons test (I).
Recently, DHX9 has been demonstrated to bind directly to short interspersed nuclear element (SINE) Alu elements, which could be a source of dsRNA and circular RNA (circRNA; ref. 38). This finding may implicate DHX9 as an important mediator of cellular dsRNA. To validate the finding of the screen, we depleted DHX9 by two different sgRNA sequences (sgDHX9 and sgDHX9 #2) in H446 cells (Fig. 1F) and tested dsRNA accumulation by immunofluorescence (IF) staining using a dsRNA-specific antibody (J2 antibody). Cells lacking DHX9 showed an accumulation of cytoplasmic dsRNA when stained with J2 antibody. Furthermore, the J2 signal was efficiently diminished by dsRNA-specific RNase III treatment, suggesting that the J2 antibody is specifically recognizing dsRNAs (Fig. 1G). We depleted DHX9 in additional SCLC cell lines (H196, H82, and DMS-114) and confirmed the accumulation of dsRNA by intracellular flow cytometry in all the SCLC cell lines tested (Supplementary Fig. S1D and S1E).
Previous studies reported that the sources of immunogenic dsRNAs generated in cancer cells treated with certain epigenetic inhibitors were various endogenous retroviral element (ERE) subtypes, including SINEs, long interspersed nuclear elements (LINE), and long terminal repeats (LTR; refs. 10, 39, 40). To identify the genomic sources of dsRNA accumulated in DHX9-depleted cells, we used the J2 antibody to immunoprecipitate dsRNAs in DHX9-depleted SCLC cells and performed RNA Immunoprecipitation Sequencing (RIP-seq). Sequencing J2-enriched RNAs showed that the dsRNAs derived from LINE and SINE families were strongly increased in sgDHX9 cells (Fig. 1H), indicating that DHX9 plays an important role in unwinding LINE- and SINE-derived dsRNA structures in SCLC cells. Also, dsRNAs from LTR families were significantly increased in sgDHX9 cells (Fig. 1H; Supplementary Fig. S1F). In addition, J2-RIP experiments followed by quantitative RT-PCR (qRT-PCR) analysis confirmed that DHX9 depletion increased the dsRNAs derived from various ERE subfamilies, including SINEs (Alu), LTR-ERVs (HERV-K), and satellite repeats (SAT III), as well as LINEs (L1 ORF; Fig. 1I).
We next wanted to understand which nucleic acid sensors were involved in the recognition of the dsRNAs accumulated in DHX9-depleted cells, as well as the specific genomic sources of dsRNAs recognized by these sensors. Because both MDA5 and RIG-I are key cytosolic pattern recognition receptors that recognize dsRNA (40), we performed RIP-seq analysis in DHX9-depleted H446 cells that stably expressed either Flag-tagged MDA5 or RIG-I. Interestingly, we found that RNA levels in MDA5 pulldowns were significantly higher than RIG-I pulldowns (Fig. 1J). More notably, sequencing results showed that dsRNA derived from LINEs and SINEs, which were also J2-enriched RNAs from sgDHX9 cells (Fig. 1H), were mainly bound to MDA5. This suggests that MDA5 might be a major contributor of the dsRNA-sensing pathway in DHX9-depleted cells (Fig. 1K).
To assess whether the helicase activity of DHX9 is required for suppressing dsRNAs in this model, we stably transduced H446 cells with Flag-tagged DHX9-WT or DHX9-K417R helicase dead mutant (Supplementary Fig. S1G) and performed RIP. Of note, we detected a significant increase of RNAs pulled down by DHX9-K417R helicase dead mutant, suggesting an impaired dsRNA unwinding capacity when compared with DHX9-WT (Fig. 1L) and supporting the idea that DHX9 helicase activity contributes to the suppression of dsRNA accumulation. Furthermore, sequencing RNAs from DHX9-K417R pulldowns demonstrated that SINEs and LINEs, which were enriched in sgDHX9 cells (Fig. 1H), can directly bind to DHX9 (Fig. 1M), further indicating these are targets of DHX9's helicase activity. Finally, heat map analysis of RIP-seq results showed that the enrichment pattern of RNA species bound to DHX9-K417R was similar to MDA5 but not to RIG-I, further supporting the idea that dsRNAs that are sensed by MDA5 (>100 bp) are the main targets of DHX9 (Fig. 1N). Taken together, these results demonstrate that DHX9 suppresses ERE-derived dsRNA accumulation by unwinding dsRNA in SCLC cells.
DHX9 Depletion Induces IFN Response in SCLCs
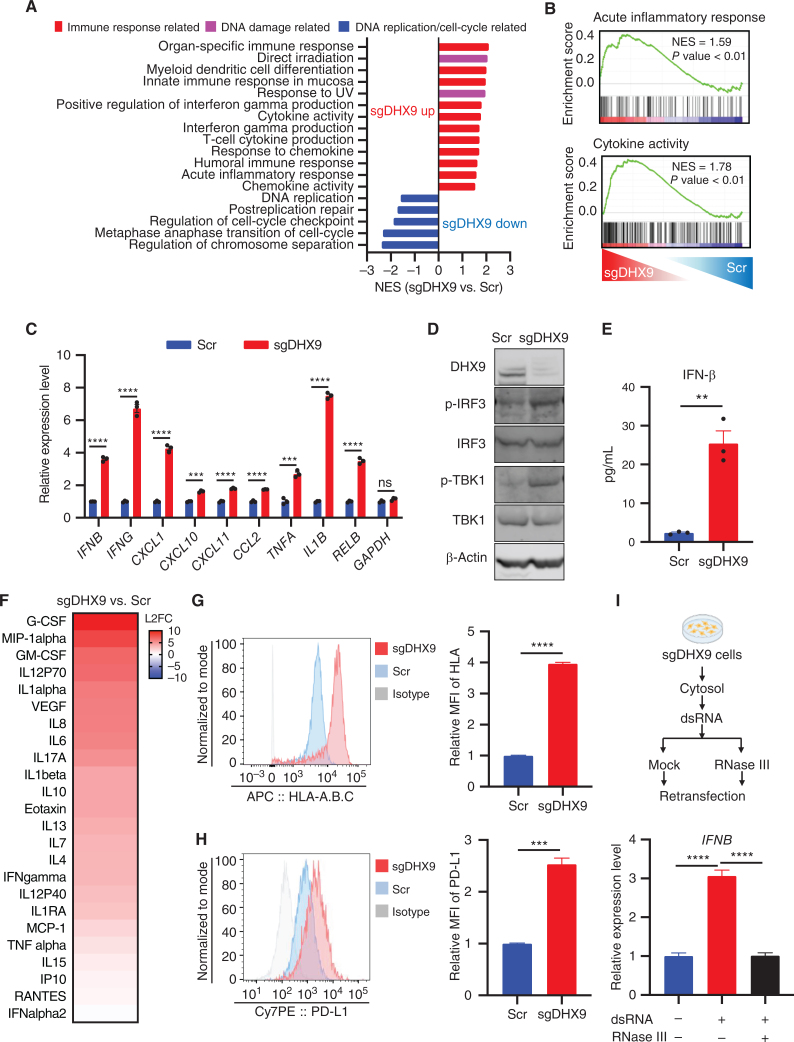
To investigate the biological impact of dsRNA accumulation in SCLC cells, we performed RNA sequencing (RNA-seq) on Scramble versus sgDHX9 SCLC cells. Consistently, gene set enrichment analysis (GSEA) revealed that DHX9 depletion induced the upregulation of gene-expression pathways associated with immune and inflammatory responses and cytokine activity (Fig. 2A and B), and sgDHX9 cells showed increased expression of many interferon stimulated genes (ISG) by RNA-seq (Supplementary Fig. S2A). We validated by qRT-PCR that multiple ISGs (IFNB, CXCL10, CXCL11, and CCL2), as well as nuclear factor kappa B (NF-kB)-responsive genes (TNFA, IL1B, and RELB) were strongly induced by DHX9 loss, indicating the activation of an antiviral transcription program in response to DHX9 depletion (Fig. 2C; Supplementary Fig. S2B; Supplementary Table S2). sgDHX9 cells also showed increased levels of phospho (p-)IRF3 and p-TBK1 (Fig. 2D), markers of innate immunity able to activate a type I IFN response. Indeed, we were able to readily detect a significant increase in the secretion of IFNβ into the culture medium after DHX9 depletion (Fig. 2E), as well as the secretion of multiple other cytokines and chemokines detected by Luminex multiplex assay (Fig. 2F; Supplementary Fig. S2C).
Figure 2.
DHX9 depletion induces IFN response. A, Gene sets significantly upregulated and downregulated are shown, based on GSEA result. Immune response–related gene sets are in red, DNA damage–related gene sets are in pink, and DNA replication/cell-cycle–related gene sets are in blue. B, GSEA with C5 (ontology) gene sets based on RNA-seq results of sgDHX9 vs. Scramble cells. C, qRT-PCR analysis of the immune-related genes comparing Scramble and sgDHX9 H196 cells (n = 3). 36B4 was used as a reference. D, Immunoblot (IB) of the indicated proteins in Scramble and sgDHX9 H196 cells. E, ELISA of human IFNβ protein in conditioned medium from Scramble and sgDHX9 H196 cells. F, Log2 fold change (FC) of cytokine/chemokine differences of sgDHX9 H196 compared with Scramble. The cytokine/chemokine levels were quantified with Proteome Profiler Human Cytokine Array Kit. G and H, Flow cytometry analysis of HLA-A.B.C (G) or PD-L1 (H) expression on the cell surface of Scramble and sgDHX9 H196 cells. Data are representative of three independent experiments (left). Mean fluorescence intensity (MFI) was quantified by FlowJo (right; n = 3). I, Schematic (top) and result (bottom) of qRT-PCR analysis of IFNB gene in H196 cells treated with cytoplasmic dsRNA (n = 3). 36B4 was used as a reference. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student t test (C, E, G, and H), one-way ANOVA (I).
To further explore the potential impact of DHX9 loss on antitumor immunity, we analyzed cell-surface expression of HLA-A, -B, and -C on SCLC cells by flow cytometry, revealing a strong induction of these MHC class I molecules after DHX9 depletion (Fig. 2G; Supplementary Fig. S2D). Furthermore, DHX9 loss also induced cell-surface PD-L1 expression (Fig. 2H; Supplementary Fig. S2E) which has been reported to be minimally expressed in more than 80% of SCLC tumors (41). Next, to evaluate the immunogenic capacity of dsRNAs produced after DHX9 loss, we pulled down dsRNAs from DHX9-depleted cells using the J2 antibody and transfected these dsRNAs into recipient cells. Importantly, transfection of dsRNA isolated from DHX9 depleted cells significantly activated IFNB expression in recipient cells, whereas digestion with RNase III (which cleaves dsRNA), completely canceled this effect (Fig. 2I), further suggesting that dsRNAs are responsible for the IFN response induced by DHX9 depletion. Together, these results indicate that DHX9 depletion activates dsRNA-sensing pathways and provokes the hypothesis that targeting DHX9 may provide unexplored strategies to activate antitumor immune pathways in SCLC.
DHX9 Depletion Causes R-loop Accumulation, DNA Damage, and CGAS–STING Pathway Activation
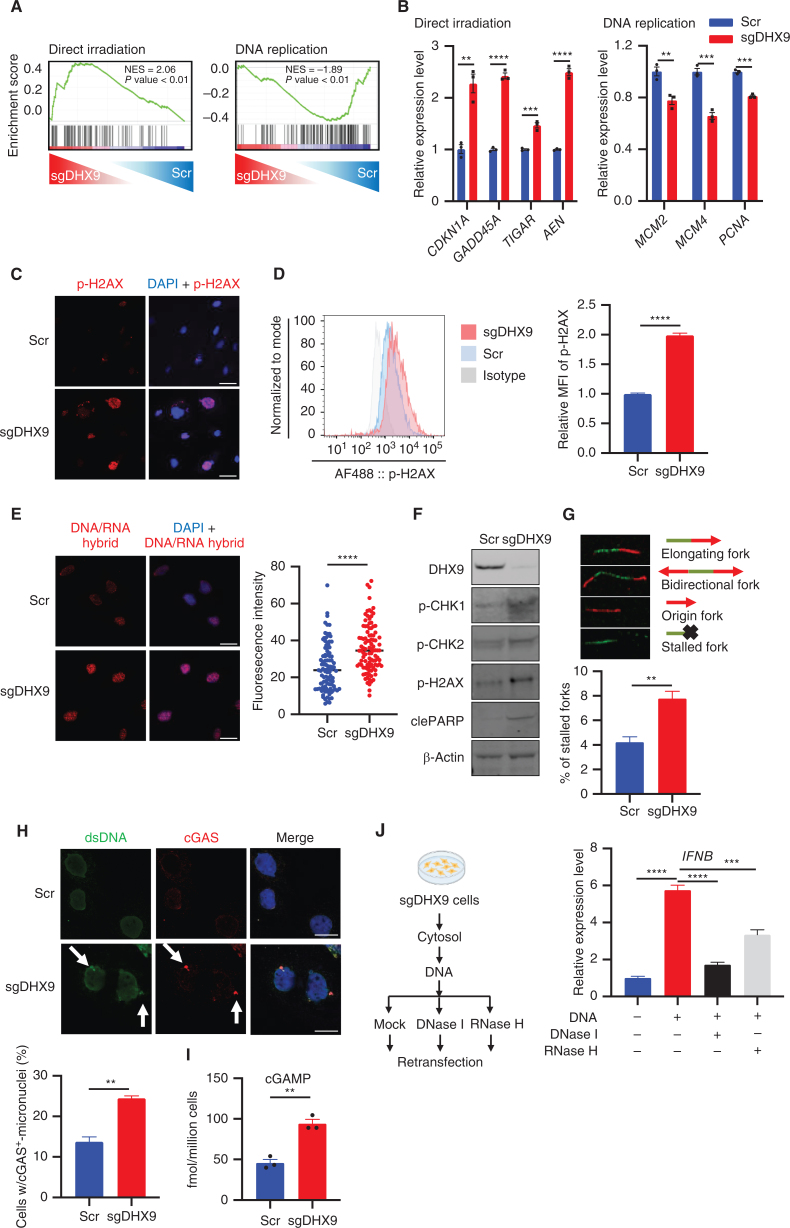
Intriguingly, we found that DHX9 depletion led not only to an increase in dsRNAs and an IFN response but also to an upregulation of DNA damage–associated gene signatures and downregulation of cancer-associated pathways when examined by analysis of RNA-seq data from DHX9-depleted SCLC cells (Figs. 2A and 3A). We validated by qRT-PCR in multiple DHX9-depleted cells that genes involved in DNA damage were upregulated, whereas cancer-associated genes, such as genes related to DNA replication, were downregulated (Fig. 3B; Supplementary Fig. S3A). This finding suggests that conflicts in completing DNA replication in sgDHX9 cells might result in DNA damage. Indeed, IF and flow cytometry analysis showed that the DNA double-stranded breaks (DSB) marker p-H2AX was strongly induced in multiple DHX9-depleted cells (Fig. 3C and D; Supplementary Fig. S3B).
Figure 3.
DHX9 depletion causes R-loop accumulation, DNA damage, and cGAS–STING pathway activation. A, GSEA with C2 (curated) gene sets, based on RNA-seq results of sgDHX9 vs. Scramble cells. B, qRT-PCR analysis of the direct irradiation response and replication-related genes comparing Scramble and sgDHX9 H196 cells (n = 3). 36B4 was used as a reference. C, Immunofluorescence images of p-H2AX (red) staining of Scramble and sgDHX9 H196 cells. Scale bar = 50 μm. D, Flow cytometry analysis of intracellular p-H2AX levels in Scramble and sgDHX9 H196 cells. Data are representative of three independent experiments (left). Mean fluorescence intensity (MFI) was quantified by FlowJo (right; n = 3). E, Immunofluorescence images of DNA/RNA hybrid (red) staining of Scramble and sgDHX9 H196 cells (left) and quantification of fluorescence intensity (right; 150 cells were counted per group, n = 3). Scale bar = 50 μm. F, Immunoblot (IB) of the indicated proteins in Scramble and sgDHX9 H196 cells. G, DNA fiber assay of Scramble and sgDHX9 H196 cells. The percentage of stalled forks over the total number of different replication structures was measured (>150 labeled forks were counted per group, n = 3). H, Immunofluorescence images of dsDNA (green) and cGAS (red) staining of Scramble and sgDHX9 H196 cells (left) and quantification of cells with cGAS+-micronuclei (150 cells were counted per group, n = 3). Scale bar = 25 μm. I, ELISA of human cGAMP protein in Scramble and sgDHX9 H196 cells. J, Schematic (left) and result (right) of qRT-PCR analysis of IFNB gene in H196 cells treated with cytoplasmic DNA (n = 3). 36B4 was used as a reference. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student t test (B, D, E, G, H, and I), one-way ANOVA (J).
Previous studies reported that DHX9 is involved in unwinding R-loops or DNA/RNA hybrid structures (42, 43), and excessive R-loop formation can trigger genomic instability and replication stress by impairing replication fork progression and inducing DSBs (44, 45), suggesting that aberrant R-loop accumulation might be a source of DNA damage in DHX9-depleted cells. To test this hypothesis, we first compared the distribution and strength of R-loops (stained with S9.6 DNA/RNA hybrid antibody) in Scramble versus sgDHX9 cells. Consistent with previous findings, we observed an increased accumulation of R-loop structures in sgDHX9 cells, which were efficiently degraded by RNase H treatment (Fig. 3E; Supplementary Fig. S3C). As we expected, loss of DHX9 also activated the DNA damage checkpoint pathway in H196, H82, and H1048 cells, as indicated by increased p-CHK1 and p-CHK2, and apoptosis, as indicated by cleaved PARP as well as p-H2AX (Fig. 3F; Supplementary Fig. S3D). To test whether the accumulation of R-loops is directly responsible for genomic instability and DNA damage, SCLC cells were transfected with a vector expressing RNase H1, which preferentially degrades the RNA portion of the R-loops. Exogenous RNase H1 expression suppressed the increase of p-CHK2, p-H2AX, and cleaved PARP in sgDHX9 cells, supporting the idea that genomic instability and DNA damage in cells deficient for DHX9 were caused by accumulation of R-loops (Supplementary Fig. S3E). Next, we sought to determine the impact of DHX9 depletion on DNA replication stress and we performed the DNA fiber assay, which enables us to analyze replication fork progression and processivity on single DNA molecules visualized by immunofluorescence (42, 46). We found that the proportion of stalled forks significantly increased in DHX9-depleted cells (Fig. 3G), indicating that R-loop accumulation in cells deficient for DHX9 triggers replication stress, which likely contributes to genomic instability.
Next, we asked whether the helicase activity of DHX9 was required for the unwinding of DNA/RNA hybrids in our model. Consistent with previous results, the transfection of siDHX9-3′UTR decreased the cell growth of H446 cells, whereas the expression of DHX9-WT but not K417R helicase dead mutant rescued the effects of DHX9 KD on cell viability (Supplementary Fig. S3F and S3G). Of note, we also found that DHX9-WT but not K417R mutant rescued the increase of DNA/RNA hybrids induced after DHX9 loss, suggesting that helicase activity of DHX9 is required to unwind DNA/RNA hybrid structures (Supplementary Fig. S3H).
The reduced expression of replication factors in sgDHX9 cells (Fig. 3B) raised the question of whether stalled replication forks might result from insufficient replication proteins. However, we found that exogenous expression of RNase H1, which resolves R-loops, decreased the number of stalled forks in sgDHX9 cells (Supplementary Fig. S3I). This further supports the idea that the accumulation of R-loops contributes to fork stalling. In addition, because key DNA repair genes were not upregulated in sgDHX9 cells and to gain more insight into this mechanism, we reviewed RNA-seq data comparing sgDHX9 and Scr cells and found that senescence-related genes were upregulated in DHX9-depleted cells (Supplementary Fig. S3J and S3K). Consistent with this, secretion of cytokines related to aging/senescence (IL1β, IL6, IL10, TNFα, and MCP-1) was strongly increased in sgDHX9 cells (Fig. 2F). As reported in previous studies (47, 48), DNA damage might result in cellular senescence, and consequently DNA replication/repair genes might be downregulated. Based on these reports, DHX9-depleted cells might experience a senescence-like phenotype because of DNA damage, which can potentially contribute to the suppression of some key DNA repair genes.
Replication stress inducers including PARP and CHK1 inhibitors have been reported to activate innate immune responses through the cGAS–STING pathway in SCLC cells (26). Thus, we next interrogated whether DHX9 loss-induced replication stress can contribute to the activation of the cGAS–STING pathway and found that DHX9 depletion increased the number of cells with micronuclei that stained positive for cGAS and dsDNA compared with Scramble cells (Fig. 3H). To measure cGAS activation, we then quantified by ELISA the levels of the second messenger, 2′3′-cyclic GMP-AMP (cGAMP). This assay revealed that DHX9 depletion also elicited a substantial increase in cGAMP production (Fig. 3I), suggesting an activation of the cGAS–STING pathway. Next, to examine the immunogenic capacity of cytosolic DNAs produced after DHX9 loss, we transfected cytoplasmic DNAs extracted from DHX9-depleted cells into recipient cells. Notably, transfection of cytoplasmic DNA isolated from DHX9-depleted cells significantly activated IFNB expression in recipient cells (Fig. 3J). Intriguingly, RNase H, which selectively degrades RNA strands in DNA/RNA hybrids, partially rescued IFNB induction caused by DNA transfection, whereas DNase I, which degrades DNA in the forms of dsDNA, ssDNA, and DNA/RNA hybrids, almost completely canceled this effect. This suggests that cytoplasmic DNA/RNA hybrids as well as dsDNAs accumulated after DHX9 depletion contribute to the induction of an IFN response in SCLC cells (Fig. 3J). Altogether, these data indicate that the R-loops generated after DHX9 loss in SCLC cells promote DNA replication stress and DSBs, which likely contribute to the activation of an innate immune response.
DHX9 Loss Triggers IFN Signaling through dsRNA and dsDNA Antiviral Sensing Pathways in SCLC Cells
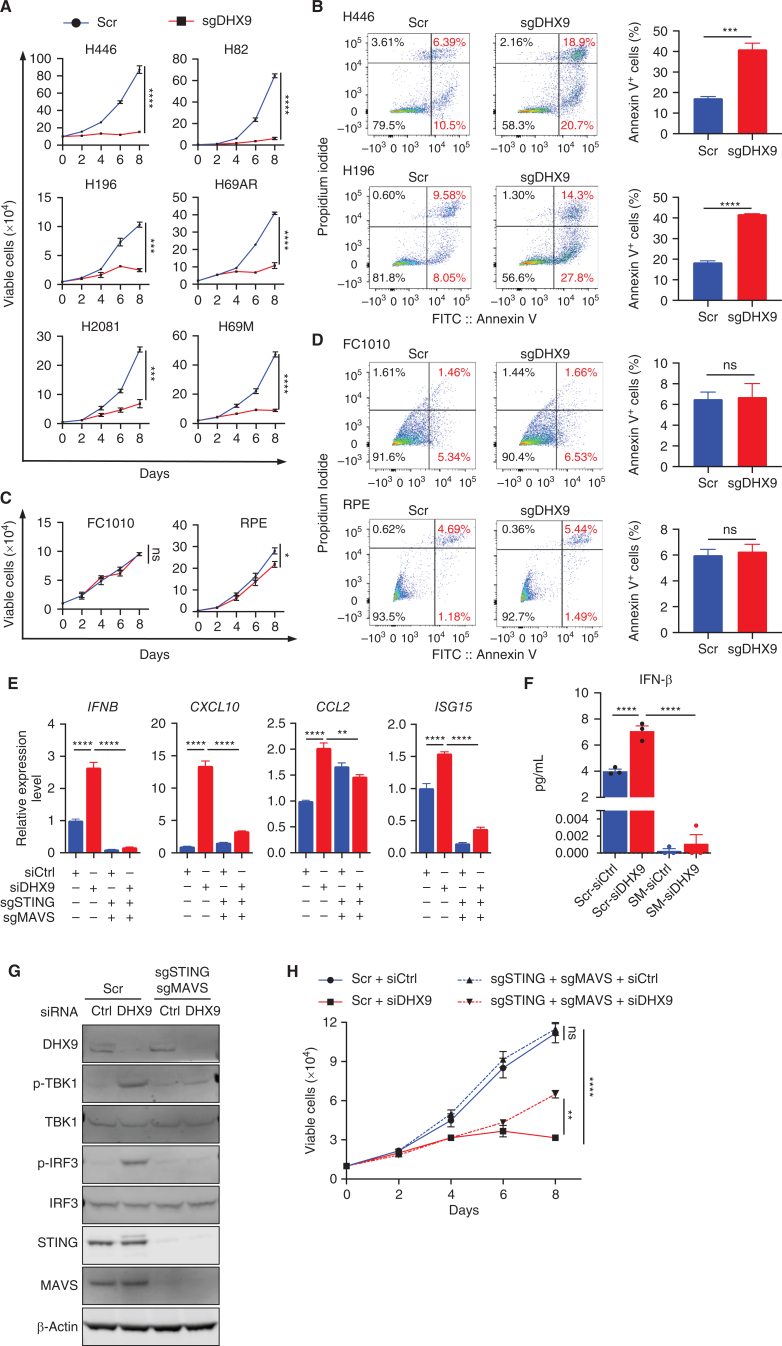
Based on the data above and that both innate immune responses and genomic instability can promote growth arrest and cell death, we next evaluated the effects of DHX9 depletion on SCLC cell proliferation and viability. We found a dramatic decrease in cell proliferation in all SCLC cells tested, including chemoresistant SCLC cells as well as increased apoptosis (Fig. 4A and B; Supplementary Fig. S4A and S4B). Notably, DHX9 depletion had minimal effects on the proliferation and apoptosis of normal cells (Fig. 4C and D) and did not induce R-loop accumulation nor increase the expression of p-CHK2, p-H2AX, or cleaved PARP, supporting cancer cell–specific effects (Supplementary Fig. S4C and S4D). Most SCLCs lack functional TP53 and RB1, which are known to be drivers of genomic instability and replication stress (49–51), positioning DHX9 as a promising target and vulnerability to selectively accelerate replication stress-induced DNA damage in those tumors.
Figure 4.
DHX9 loss triggers dsRNA and dsDNA antiviral sensing pathways and IFN signaling in SCLC cells. A, Growth curves of the indicated SCLC cell lines (n = 3). B, Flow cytometry analysis of apoptotic (annexin V+) cells in Scramble and sgDHX9 H196 and H446 cells. Data are representative of three independent experiments (left). Quantification of apoptotic cells is shown (right; n = 3). C, Growth curves of the indicated normal cell lines (n = 3). D, Flow cytometry analysis of apoptotic (annexin V+) cells in Scramble and sgDHX9 FC1010 and RPE cells. Data are representative of three independent experiments (left). Quantification of apoptotic cells is shown (right; n = 3). E, qRT-PCR analysis of the immune-related genes comparing Scramble and sgSTING + sgMAVS H196 cells transfected with siCtrl or siDHX9 (n = 3). 36B4 was used as a reference. F, ELISA of human IFNβ protein in conditioned medium from Scramble and sgSTING + sgMAVS H196 cells, transfected with siCtrl or siDHX9 (n = 3). SM: sgSTING + sgMAVS. G, Immunoblot (IB) of the indicated proteins in Scramble and sgSTING + sgMAVS H196 cells transfected with siCtrl or siDHX9. H, Growth curves of Scramble and sgSTING + sgMAVS H196 cells, transfected with siCtrl or siDHX9 (n = 3). Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student t test (A, B, C, and D), two-way ANOVA followed by Tukey multiple comparisons tests (E, F, and H).
DHX9 depletion caused dsRNA and dsDNA accumulation in SCLC cells (Fig. 1G; Supplementary Fig. S1E; Fig. 3H), which could induce activation of dsRNA- or dsDNA-sensing pathways. Thus, to determine whether dsRNA- or dsDNA-sensing pathways are responsible for inducing antiviral innate immunity after DHX9 depletion, we performed CRISPR-mediated depletion of MAVS and STING combined with transfection of siCtrl or siDHX9. Notably, double-KO (dKO) of MAVS and STING was able to abrogate the induction of multiple ISGs (Fig. 4E) and IFNβ protein secretion (Fig. 4F) observed after DHX9 depletion as well as completely abrogate the phosphorylation of TBK1 and IRF3 induced by DHX9 loss (Fig. 4G). To systematically investigate which specific cytosolic nucleic acid sensing pathways are activated by DHX9 loss, we depleted multiple nucleic acid sensors/mediators (cGAS, STING, MDA5, RIG-I, MAVS, PKR, TLR3, and TLR9) and tested IFNB and CXCL10 expression after DHX9 loss (Supplementary Fig. S4E). Depletion of cGAS or STING strongly suppressed IFNB and CXCL10 induction upon DHX9 loss, and depletion of MDA5 or MAVS also partially rescued this phenotype, whereas depletion of other sensors had no significant effects. These results indicate that although the cGAS–STING pathway might have a major contribution to the IFN response, the dsRNA-sensing pathway (MDA5-MAVS) also plays a significant role. This further suggests that both cytoplasmic dsRNA and dsDNA generated upon DHX9 depletion contribute to the downstream induction of antiviral and interferon pathways in SCLC cells.
To evaluate the contribution of cytoplasmic dsRNA-/dsDNA-sensing pathways to cell proliferation upon DHX9 loss, we depleted MAVS and STING individually and as dKO and tested the effects of DHX9 loss on cell proliferation (Fig. 4H; Supplementary Fig. S4F). Codepletion of MAVS and STING partially rescued cell growth defect as well as depletion of each gene individually. Consistent with this, the depletion of type I interferon signaling through IFNAR1 also partially rescued the growth defect caused by DHX9 loss (Supplementary Fig. S4G and S4H). Together, these data suggest a role of dsRNA- and dsDNA-sensing pathways on the antiviral immune response and cell death induced by DHX9 depletion. However, the observed partial rescue of cell proliferation also suggests that there may be redundancy in dsRNA/dsDNA-recognition pathways or that other pathways might contribute to DHX9 depletion-induced cell death.
CRISPR Screen Identifies Modulators of Sensitivity and Resistance to DHX9 Loss
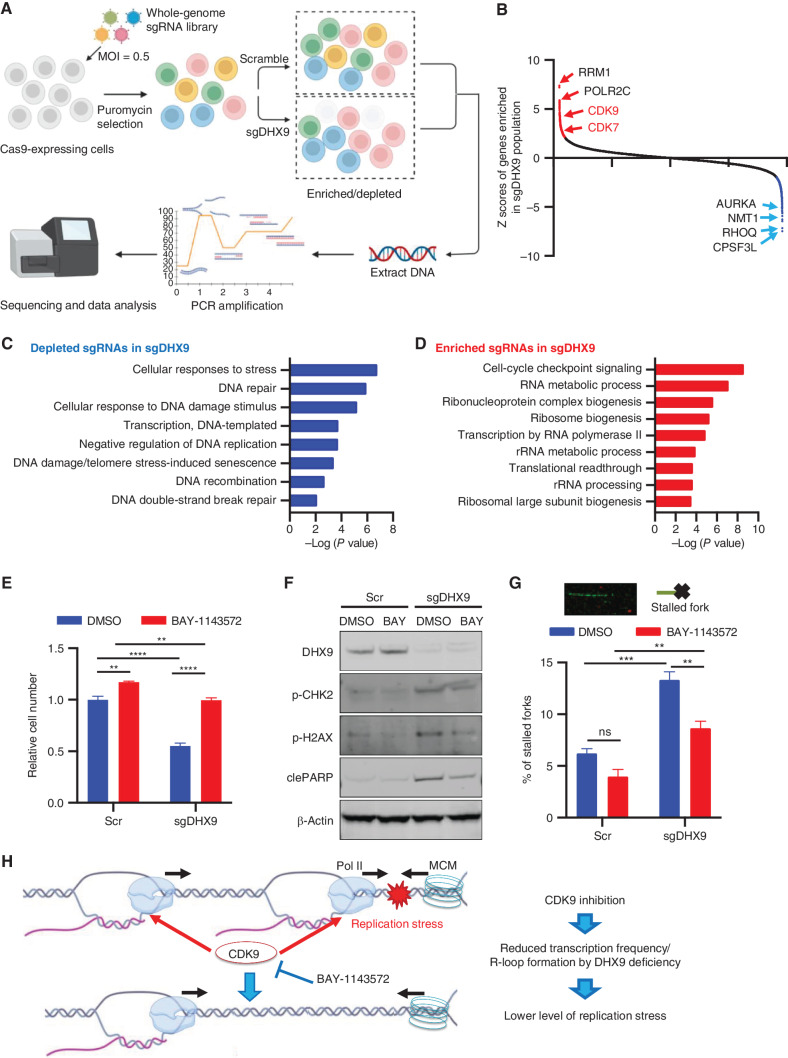
To interrogate the mechanisms by which DHX9 loss induces cell death in SCLC, we next performed a genome-wide, in vitro pooled CRISPR screen in SCLC cells to identify the genes whose deletion would rescue or increase the lethality observed following DHX9 depletion. Briefly, cells were transduced with the Brunello Human CRISPR Knockout Pooled Library (52) and selected in the presence of puromycin for efficient lentivirus transduction. Then, cells were transduced with either Scramble control or sgDHX9 vectors. Cells were further propagated, and genomic DNA was extracted to identify sgRNAs enriched and depleted in DHX9-depleted cells compared with control (Fig. 5A). This CRISPR screen identified multiple genes whose ablation resulted in resistance or sensitivity to DHX9 loss-induced lethality, and consequent enriched or depleted sgRNA representation in sgDHX9 cells (Fig. 5B).
Figure 5.
CRISPR screen identifies modulators of sensitivity and resistance to DHX9 loss. A, Schematic of the genome-wide CRISPR screening method to reveal regulators of DHX9 loss-related cell death. Created with BioRender.com. B, Top-rated enriched and depleted sgRNAs from the genome-wide CRISPR screening are summarized. C and D, Gene ontology analysis of sgRNA targeted depleted (C) and enriched (D) genes in sgDHX9 population. E, Relative cell number of Scramble and sgDHX9 H82 cells treated with DMSO or 0.5 μmol/L BAY-1143572. Luminescence of CellTiter-Glo was detected on day 5 after seeding (n = 3). F, Immunoblot (IB) of the indicated proteins in Scramble and sgDHX9 H82 cells treated with DMSO or 0.5 μmol/L BAY-1143572. G, DNA fiber assay of Scramble and sgDHX9 H82 cells treated with DMSO or 0.5 μmol/L BAY-1143572. The percentage of stalled forks over the total number of different replication structures was measured (>150 labeled forks were counted per group, n = 3). H, Schematic model of growth rescue effect by CDK9 inhibition in DHX9-depleted cells. Created with BioRender.com. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA followed by Tukey multiple comparisons test (E and G).
As expected, gene ontology (GO) analysis of the list of genes targeted by sgRNAs depleted in sgDHX9 cells revealed gene signatures related to cellular stress, DNA damage, and DNA repair, indicating that loss of these genes increased the sensitivity of SCLC cells to DHX9 loss-dependent cell death (Fig. 5C). Indeed, siRNA knockdown (KD) of AURKA, NMT1, and RHOQ genes included in the top hits accelerated cell death caused by siDHX9 (Supplementary Fig. S5A). Conversely, the screen also identified genes whose ablation resulted in resistance to DHX9 loss-induced lethality (sgRNAs enriched in DHX9-depleted population). Functional annotation of these genes revealed a striking enrichment for factors that are involved in the RNA metabolic process, transcription, ribosome biogenesis, and cell-cycle checkpoint signaling (Fig. 5D). One of the top sgRNAs enriched in DHX9-depleted cells were sgRNAs targeting CDK9 (Fig. 5B), a kinase essential for transcriptional elongation (53). sgRNAs targeting CDK7 also involved in transcription elongation by phosphorylating CDK9 were also enriched in the DHX9-depleted population (Fig. 5B). Importantly, conflicts between transcription and DNA replication represent a significant cause of replication stress (45). These conflicts may result from the cotranscriptional occurrence of R-loops, which could impede the progression of DNA replication forks (42, 54). Interestingly, it has been previously reported that inhibition of CDK9 decreased replication stress and DNA damage caused by R-loops, enabling DNA replication in cells treated with inhibitors or siRNAs against certain chromatin modifiers (55). Similarly, another study showed that RNA transcription is altered in the absence of p53, inducing increased replication stress and genomic instability that can be rescued by pharmacologic inhibition of transcription through CDK inhibitors (56). This raises the possibility that conflicts with transcription might cause replication stress when DHX9 is depleted. This may also explain why sgRNAs targeting CDK9 were able to rescue DHX9 depletion-induced lethality in our screen. Thus, ongoing transcription and perhaps the resulting R-loop formation might represent a prerequisite for impaired DNA replication upon DHX9 depletion leading to replication stress, DNA damage, and ultimately cell death. Importantly, this might also explain why SCLC cells are particularly vulnerable to DHX9 deletion, as almost all SCLC tumors have loss-of-function mutations in TP53 and RB1 genes, which are linked with replication stress and genomic instability (49–51). We propose that a major source of replication stress in SCLC cells might arise from conflicts in transcription, as has been shown to occur in p53-deficient cells (56). Thus, therapeutic strategies that exacerbate this stress could selectively kill SCLC cells by replicative damage.
To validate the CRISPR screen results, we initially examined the protein levels of CDK9 and CDK7 in H82 cells using the same sgRNAs used in the screen. We observed a reduction in both proteins, although these sgRNAs did not achieve complete elimination of CDK9 and CDK7 protein levels (Supplementary Fig. S5B). Given the essential nature of CDK9 and CDK7, this partial reduction in their protein levels provides an explanation for the enrichment of sgRNAs targeting these essential genes in our screen. We also evaluated the impact of DHX9 depletion on cell viability in sgCDK9 and sgCDK7 cells and confirmed that the depletion of CDK9 and CDK7 partially rescued the effects of DHX9 loss (Supplementary Fig. S5C). These findings align with the results of the CRISPR screen, in which we observed an enrichment of sgRNAs targeting CDK9 and CDK7. Consistent with this, siRNAs targeting CDK9 as well as CDK7 also recovered cell growth of DHX9 KD H82 cells (Supplementary Fig. S5D). Importantly, we found that CDK9 inhibition with a potent and highly selective CDK9 inhibitor (BAY-1143572) rescued the decrease in cell proliferation and diminished the induction of p-CHK2, p-H2AX, and cleaved PARP observed after DHX9 depletion (Fig. 5E and F), although CDK9 inhibition decreased cell growth of FC1010 normal fibroblast cells (Supplementary Fig. S5E). A higher concentration of BAY-1143572 inhibited the SCLC cell growth as well (Supplementary Fig. S5F), consistent with previous reports (57). By performing the DNA fiber assay, we found that the proportion of stalled forks significantly increased in DHX9-depleted cells and that CDK9 inhibition greatly reduced the proportion of stalled forks both in sgDHX9 and control cells (Fig. 5G).
Given that CDK9 inhibition could affect transcription globally, which may indirectly contribute to the rescuing effects, we performed dot-blot experiments using the S9.6 antibody to directly evaluate R-loop accumulation in DHX9-depleted cells after CDK9 inhibition. Consistent with previous results, we observed a significant increase of R-loops in sgDHX9 cells, which was partially rescued by CDK9 inhibition (Supplementary Fig. S5G). Furthermore, we directly measured the gene-specific R-loop level using qDRIP (quantitative differential DNA/RNA immunoprecipitation; Supplementary Figs. S5H–S5P; ref. 58). Of note, CDK9 inhibitor treatment alleviated R-loop build-up at multiple promoter regions, including Gadd45b, RhoB (direct DHX9-targeted genes; ref. 59), and ThumpD2 (Supplementary Figs. S5N–S5P). Additionally, we confirmed that CDK9 inhibition had no significant effects on the expression levels of key nucleic acid sensors or mediators, excluding any potential impact of CDK9 inhibition in pathways involved in dsRNA/dsDNA sensing (Supplementary Fig. S5Q). These findings indicate that a major source of replication stress in SCLC cells arises from transcription-associated damage, which is further induced by DHX9 depletion (Fig. 5H).
Together, these results illuminate a novel mechanism of DHX9 loss-induced cell death in cancer cells and position DHX9 as a promising therapeutic target for SCLC and other tumor types, whereas genomic instability and replication stress contribute to pathology.
DHX9 Depletion Decreases Tumor Growth, Induces Immune Cell Infiltration, and Enhances Response to ICB Therapy
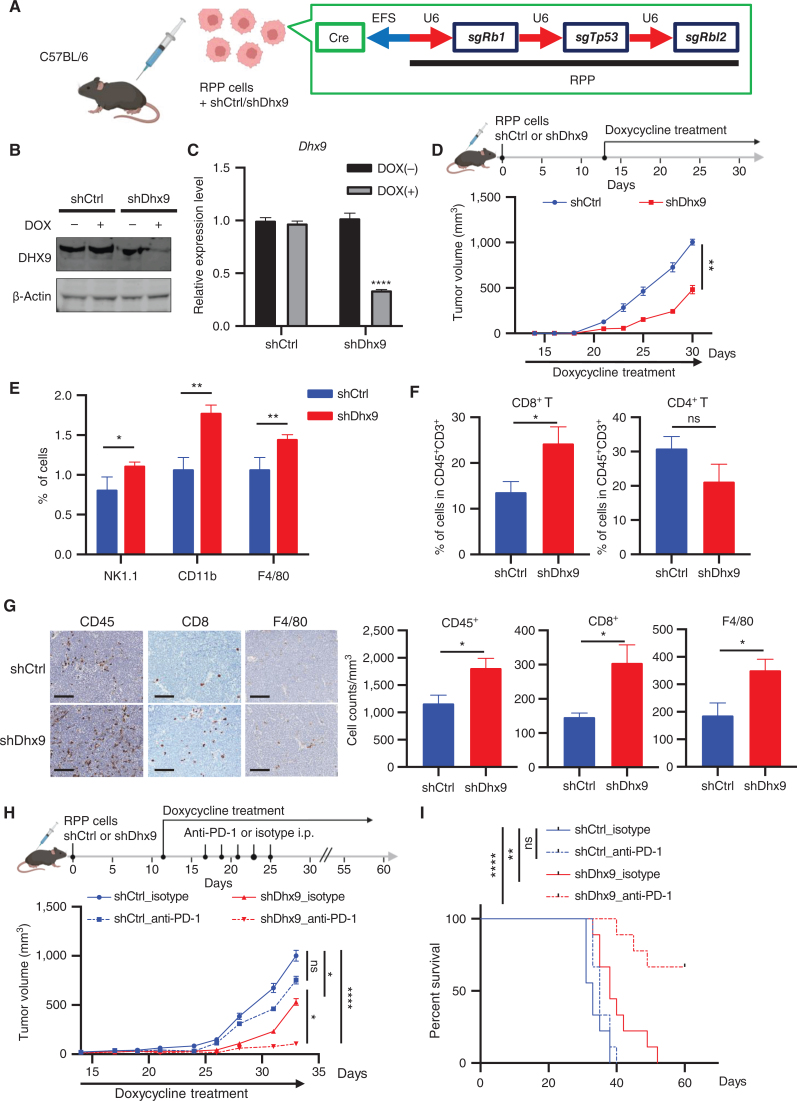
The induction of immune transcriptional programs by DHX9 loss (Fig. 2A–C) suggests the hypothesis that such immune signaling may also impact the tumor microenvironment and provoke antitumor immune responses. To test this hypothesis, we generated RPP cells (a murine SCLC cell line generated by triple knockout of Tp53, Rb1, and Rbl2 genes; refs. 24, 60) stably expressing shRNA targeting either control (shCtrl) or DHX9 (shDhx9) under control of a Tet-inducible promoter, into the flanks of C57BL/6 mice (Fig. 6A). When tumors became palpable, the mice were fed with doxycycline water to induce DHX9 KD (Fig. 6B and C). We found that DHX9 KD significantly decreased tumor volume and weight (Fig. 6D; Supplementary Fig. S6A), whereas the body weight of mice was not affected by the treatment (Supplementary Fig. S6B). To better understand the in vivo immune consequences of DHX9 depletion in SCLC, we evaluated tumor infiltration of immune cells by flow cytometry analysis and observed a striking increase of immune cells expressing NK-1.1, CD11b, and F4/80 (Fig. 6E). In addition, intratumoral cytotoxic CD8+ T cells were significantly increased in shDhx9 tumors, indicating a functional immune response, whereas CD4+ T cells did not change significantly (Fig. 6F; Supplementary Fig. S6C; Supplementary Table S6). IHC analysis also confirmed enhanced infiltration of CD8+ T cells as well as F4/80+ macrophages in DHX9-depleted tumors (Fig. 6G). Thus, these data demonstrate that DHX9 KD enhances immunogenicity in vivo potentially bypassing a major pathway of cancer cell immune evasion. To further assess whether immune cells are responsible for the reduced growth of DHX9-depleted tumors, we first evaluated the effects of DHX9 depletion in immunodeficient (NSG) mice using the same murine SCLC cell line (RPP cells) to replicate the same experimental conditions. We found that whereas DHX9 depletion decreased tumor volume in NSG mice (Supplementary Fig. S6D), this effect on tumor growth was smaller when compared with the effects of DHX9 depletion observed in immunocompetent mice (Fig. 6D). These results further suggest that both DNA damage and immune infiltration induced by DHX9 depletion contribute to the reduced tumor growth. Furthermore, given that CD8+ T cells and NK cells were significantly increased in DHX9-depleted tumors (Fig. 6E and F), we then performed the same in vivo experiment as in Fig. 6D and tested the effects of CD8+ T-cell depletion and NK-cell depletion using CD8a antibody and NK1.1 antibody, respectively. Although we found that CD8+ T-cell depletion slightly rescued the decrease in tumor growth observed after DHX9 loss, the observed partial rescue also suggests that other immune cell types might contribute to the reduced tumor growth observed in DHX9-depleted tumors (Supplementary Fig. S6E). Together, these results suggest that immune cells as well as intracellular replication stress contribute to the reduced growth of DHX9 KD tumors.
Figure 6.
DHX9 depletion decreases tumor growth, induces immune cell infiltration, and enhances response to ICB therapy. A, Schematic of in vivo tumor growth assay. RPP cells, which were transduced with DOX-inducible shCtrl or shDhx9 vector, were transplanted into C57BL/6 mice. Created with BioRender.com. B and C, Immunoblot (IB; B) and qRT-PCR analysis (C) of DHX9 expression in shCtrl and shDhx9 RPP cells treated w/wo DOX. D, Tumor growth curves of shCtrl and shDhx9 RPP tumors (n = 6). E, Flow cytometry quantification of the indicated infiltrating immune cells in shCtrl and shDhx9 RPP tumors. Each population was analyzed by FlowJo (n = 4). F, Flow cytometry quantification of infiltrating CD8+ T cells and CD4+ T cells of CD45+CD3+ cells in shCtrl and shDhx9 RPP tumors (n = 6). G, Representative IHC images of indicated infiltrating immune cells in shCtrl and shDhx9 RPP tumors (left) and quantification (n = 6; right). Scale bar = 100 μm. H, Tumor growth curves of shCtrl and shDhx9 RPP tumors treated with isotype control or anti–PD-1 antibody (n = 9). I, Survival curves for mice in H. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired Student t test (C, D, E, F, and G), two-way ANOVA followed by Tukey multiple comparisons test (H) and log-rank test (I).
As DHX9 loss induced IFN response and increased intratumoral infiltration of cytotoxic CD8+ T cells in SCLC tumors, we next sought to determine whether DHX9 depletion potentiates ICB-based immunotherapy in the SCLC syngeneic model. To accomplish this, we treated immunocompetent C57BL/6-bearing RPP cells expressing DOX-inducible shCtrl or shDhx9 with either anti–PD-1 or isotype IgG control antibodies. We excluded anti–CTLA-4 antibodies because based on multiple clinical trials there is currently no role for anti–CTLA-4 antibodies in the treatment of SCLC (61–63). Notably, although control RPP tumors were not very sensitive to PD-1/PD-L1 blockade, consistent with a previous report (26), DHX9 depletion dramatically enhanced the sensitivity of RPP tumors to anti–PD-1 therapy, resulting in increased mouse survival (Fig. 6H and I). To validate our findings in a second in vivo model, we generated KP3 cells (mouse SCLC cell line derived from the RP GEMM model; refs. 64, 65) stably expressing DOX-inducible shCtrl or shDhx9 and engrafted them into the flanks of immunocompetent recipient mice (Supplementary Fig. S6F). Similarly, DHX9 loss also enhanced the sensitivity of RP tumors to anti–PD-1 therapy, resulting in increased mouse survival (Supplementary Fig. S6G and S6H). Again, we observed a striking increase of CD8+ T cells in shDhx9 tumors (Supplementary Fig. S6I and S6J). Taken together, these data suggest that DHX9 loss in SCLC tumors induces a robust antitumor immunity, which may ultimately lead to improved ICB treatment outcomes in immunologically cold tumors.
DHX9 Is Negatively Correlated with Immune Signatures and Associated with Poor Clinical Outcomes in Cancer Patient Data Sets
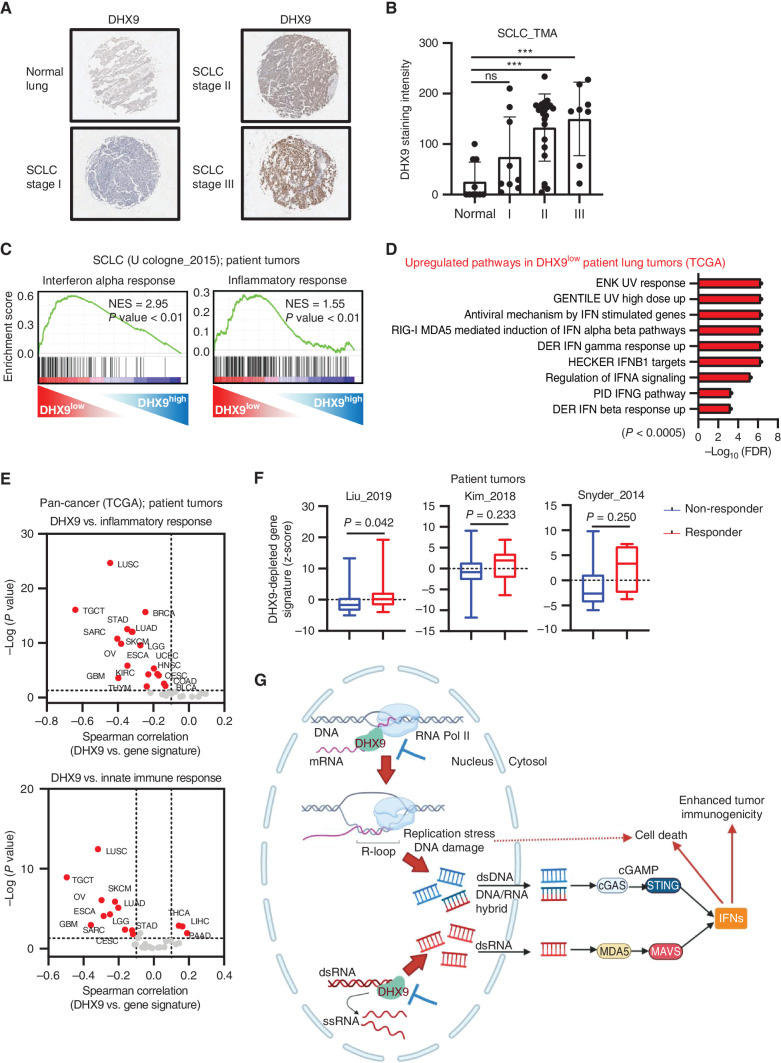
To investigate whether DHX9 is associated with human tumorigenesis, we tested DHX9 protein expression by IHC in a human SCLC tissue microarray (TMA), including 40 SCLC tumor samples and normal lung controls (Fig. 7A). Consistent with the public transcriptomic data (Fig. 1D), SCLC tumors exhibited higher staining intensity of DHX9 than normal counterparts (Fig. 7B). Notably, DHX9 expression increased in higher tumor stages, supporting the association of high DHX9 expression and poor patient prognosis in SCLC patients.
Figure 7.
DHX9 is associated with poor clinical outcomes in cancer patient data sets. A, Representative IHC images of DHX9 expression in SCLC tumors and normal lung tissue. B, Quantification of IHC images of DHX9 in A. Normal lung tissue (N = 10), stage I (N = 9), stage II (N = 23), stage III (N = 8). C, GSEA with H (hallmark) gene sets, based on RNA-seq results of 81 SCLC patient tumors (N = 40 DHX9low vs. N = 41 DHX9high). Data were downloaded from cBioPortal (U Cologne, Nature 2015; ref. 51). D, Gene ontology analysis of genes upregulated in DHX9low lung patient tumors. Data were downloaded from TCGA. E, Correlation analysis between DHX9 expression level and z-scores of the indicated gene sets in different tumor types of TCGA (patient tumors). F, Boxplots of DHX9-depleted signature z-scores in nonresponder and responder of patients treated with ICB therapy to anticipate the potential clinical relevance of targeting DHX9 in immunotherapy settings. The DHX9-depleted gene signature was prepared based on genes upregulated in sgDHX9 SCLC cells when compared with Scramble and includes the two most prominent features of DHX9-depleted cells: DDR and immune response. G, Schematic model of antitumor effects caused by DHX9 inhibition. Created with BioRender.com. Data represent mean ± SEM. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA (B) and unpaired Student t test (F).
We next interrogated transcriptomic data from a human SCLC data set (51) to determine whether DHX9 expression is associated with the tumor immune microenvironment. GSEA revealed that DHX9 expression inversely correlated with gene signatures involved in IFN-alpha response as well as inflammatory response (Fig. 7C), confirming our results from in vitro and in vivo models. Indeed, many of the IFN-stimulated genes and NF-kB–responsive genes were tended to be highly expressed in the DHX9low group (Supplementary Fig. S7A). We also interrogated TCGA to explore the broader relevance of DHX9 across human lung cancers and GO analysis revealed that gene signatures involved in IFN-related pathways were strongly upregulated in DHX9 low-expressing lung cancer tumors when compared with DHX9 high-expressing tumors (Fig. 7D), suggesting that DHX9 is a crucial repressor of tumor-intrinsic innate immunity in human lung tumors. DHX9 expression was also inversely correlated with DDR, consistent with activation of apoptosis and DNA damage checkpoint pathway in sgDHX9 cells in vitro (Fig. 3F). In addition, pan-cancer analysis exhibited a strong negative correlation between DHX9 expression and gene signatures of inflammatory response and innate immune response in a variety of tumor types (Fig. 7E). Consistent with this, GSEA of pan-cancer RNA-seq data also showed an upregulation of interferon-alpha response and inflammatory response in DHX9low tumors (Supplementary Fig. S7B).
To anticipate the clinical relevance of targeting DHX9 for immunotherapy, we created a DHX9-depleted gene signature (Supplementary Table S3), including genes that were upregulated in sgDHX9 cells based on RNA-seq analysis and related to the two most prominent features of DHX9-depleted cells: DDR and immune response. Given that this signature is expected to be upregulated when DHX9 is inhibited in cancer cells, we wanted to evaluate its clinical relevance in immunotherapy settings. Thus, we assessed the potential predictive value of DHX9 inhibition across public data sets of patients treated with anti–PD-1-based or anti–CTLA-4-based ICB therapy (66–68). Notably, we found that high levels of the DHX9-depleted gene signature were positively associated with ICB response across most of the data sets, suggesting that DHX9 inhibition may sensitize cancers to immunotherapy (Fig. 7F). In addition, the estimated immune-score of the tumor microenvironment in DHX9low tumors tended to be higher than DHX9high tumors (Supplementary Fig. S7C). Thus, these data provide evidence that DHX9 inhibition could sensitize tumors to ICB therapies, including immunologically cold tumors such as SCLC (Fig. 7G).
DISCUSSION
Induction of innate immune responses in tumor cells has been gathering attention as a promising strategy to enhance ICB therapy response (10, 11, 26), especially in immunologically cold tumors. Here, we demonstrate that the RNA helicase DHX9 functions as a repressor of innate immune signaling and replication stress in cancer cells through the unwinding of dsRNA and R-loop structures.
Our study revealed that depletion of DHX9 induces immunogenic dsRNAs, causing tumor cell–intrinsic antiviral signaling, and alters transcription and DNA replication in cancer cells to induce the formation of R-loops, compromising genome stability and cell viability. Notably, triggering these tumor-intrinsic events greatly improves ICB treatment outcomes in immunologically cold tumors, such as SCLC. Although prior studies have shown the potential of DHX9 to unwind dsRNA (38, 69), DNA/RNA hybrids (31, 70), and other more complex nucleic acid structures (32), our findings strongly suggest that dsRNAs and R-loops derived from DHX9 depletion directly contribute to induction of innate immune responses and replication stress in cancer cells, highlighting its potential as novel target to enhance antitumor immunity and boost cancer immunotherapy. Recently, a study reported by Crossley and colleagues demonstrated that DNA/RNA hybrids derived from R-loops are sensed by cGAS and TLR3 and activate an innate immune response and apoptosis (71), which is further supported by the data presented here, suggesting that R-loop–inducing therapies might be a therapeutic strategy to effectively enhance antitumor immunity while concurrently inducing DNA damage and compromising cell viability in cancer cells.
Almost all SCLCs exhibit inactivation of TP53 and RB1, both of which are key regulators of DNA damage and cell-cycle checkpoint (49–51). Intriguingly, although genomic instability in SCLC is among the highest in all the cancer types (72), the tumor microenvironment tends to be immune desert (16), which might explain the reason why SCLCs are relatively unresponsive to ICB therapies and remain as a recalcitrant disease (20). Thus, an urgent need exists to identify novel targets to improve outcomes of SCLC patients, who have what is considered the most lethal type of lung cancer. Increasing evidence has now emerged indicating that targeting components of the DDR pathway, including PARP, CHK1, or WEE1, might be an effective strategy with significant antitumor effects in SCLC preclinical models (12, 13, 25). Importantly, recent studies showed that DDR inhibitors potentiate ICB therapies in SCLC preclinical models by evoking beneficial IFN responses in the tumor microenvironment through accumulating cytosolic DNA and activating cGAS–STING and STAT1 pathways (26, 27). Intriguingly, we found that DHX9 depletion in SCLC cells triggers innate immune signaling and DNA damage through dsRNA and R-loop production, at least partially generated from various EREs and genomic instability, representing a more robust viral mimicry-inducing factor and a vulnerability preferentially in cancer cells under replication stress.
A major reason explaining the poor response of cold tumors to immunotherapy is the lack of an immunogenic tumor microenvironment, often linked to decreased CD8+ T-cell abundance and defective IFN signaling (73). Our data in immunocompetent SCLC mouse models indicate that the tumor-intrinsic effects of DHX9 depletion not only induced a significant decrease in tumor growth but also led to the recruitment of multiple immune cells into the TMA, including CD8+ T cells, dramatically sensitizing SCLC tumors to anti–PD-1 immunotherapy. These findings strongly suggest that targeting DHX9 could potentially convert immunologically cold tumors to hot while concurrently promoting cancer cell death.
Results from this study support efforts aimed at designing DHX9 inhibitors to exploit its therapeutic potential as anticancer therapy and to improve responsiveness to immunotherapy in immunologically cold tumors. Although DHX9 is reported to be essential during embryonic development, additional studies using conditional knockdown systems showed that reduced levels of DHX9 in adult mice did not cause any deleterious effects at the organismal level, although DHX9 loss was lethal to tumor cells (74). Consistent with these data, our findings indicate that DHX9 depletion has minimal or no effects on the proliferation of normal cells, which also emphasizes the contribution of intrinsic replication stress to DHX9 loss-related cell death in SCLC cells and suggests that targeting DHX9 might be an effective and realistic therapeutic strategy to treat cancers.
Given that DHX9 is broadly expressed and plays a central role in diverse cell homeostasis processes, potential consequences of direct DHX9 inhibition need to be carefully evaluated in future studies. A recent study reported that DHX9 is required for a productive CD8+ T-cell response against viral infection, protecting T cells from apoptosis and regulating genes that are necessary for effector T-cell differentiation independent of its nuclear sensor role (75). Based on these observations, it might be argued that systemic treatment with DHX9 inhibitors could restrict the antitumor activity of certain immune cells. However, we and others have shown that DHX9 is overexpressed in certain cancers, including SCLC (Fig. 1D; refs. 76–78), and that rapidly dividing tumor cells that are under replication stress are heavily vulnerable to DHX9 depletion compared with normal cells, highlighting the potential for a therapeutic window in which tumor growth control could be achieved with limited toxicity. Importantly, the fact that DHX9 depletion not only impacts cancer cell survival but also potentiates antitumor immune responses to ICB therapy in SCLC models might open a new range of therapeutic possibilities that will expand the breadth and depth of response to cancer immunotherapies. Nevertheless, further research should be conducted to fully evaluate the systemic consequences of DHX9 inhibition.
In summary, our study identified DHX9 as a novel vulnerability in immunologically cold tumors, especially in those whose genome is unstable, such as SCLC. We show that DHX9 is a crucial suppressor of dsRNA and R-loop accumulation, and the genetic perturbation of DHX9 leads to innate immune response activation as well as DNA replication stress and DNA damage. Moreover, we found that these tumor-intrinsic effects can turn cold tumors into hot, dramatically enhancing ICB responsiveness and highlighting DHX9 as a potential novel target to boost cancer immunotherapy in immunologically cold tumors.
METHODS
Cell Lines and Cell Culture
The human SCLC cell lines NCI-H69, NCI-H69AR, NCI-H841, SHP-77, NCI-H187, NCI-H345, NCI-H524, NCI-H1048, and NCI-H82 were obtained from the laboratory of Dr. Joan Albanell and were authenticated following short tandem repeat (STR) genotyping. The RPP SCLC mouse cell line was obtained from the laboratory of Dr. Matthew Oser. The KP3 (RP) SCLC mouse cell line was obtained from the laboratory of Dr. Julien Sage. RPE cells were obtained from the laboratory of Dr. Johnathan R. Whetstine, and FC1010 primary fibroblast cells were isolated in the FCCC tissue culture facility (from Dr. Hossein Borghaei) and were authenticated following STR genotyping. NCI-H196, NCI-H446, NCI-H1436, NCI-H2081, Colo668, NCI-H1694, NCI-H841, DMS-114, and HEK293T cells were obtained from the American Type Culture Collection and used for all experiments before reaching 10 passages. NCI-H69M was established from NCI-H69, as previously reported (79). All cells were routinely tested for Mycoplasma and found to be free of contamination.
H69, H69M, H69AR, H82, H446, H196, SHP-77, H187, H345, H524, H1436, H2081, Colo668, H1694, H841, H1048, and DMS-114 were cultured in RPMI-1640 containing 10% fetal bovine serum (FBS, HyClone), 2.5 g/L glucose, and 1× penicillin/streptomycin (10,000 U/mL; Thermo Fisher Scientific). HEK293T was maintained in Dulbecco's Modified Eagles Medium (DMEM) containing 10% FBS and 1× penicillin/streptomycin. RPP and KP3 mouse cell lines were cultured in RPMI-1640 containing 10% FBS, 1× penicillin/streptomycin, and HITES (1× insulin–transferrin–selenium, 10 nmol/L β-estradiol, and 10 nmol/L hydrocortisone). RPE was cultured in DMEM/F-12, 10% FBS, 2.5 mmol/L L-glutamine, 15 mmol/L HEPES, 0.5 mmol/L sodium pyruvate, and 0.01 mg/mL hygromycin. FC1010 was cultured in RPMI-1640 with 15% FBS, 2 mmol/L L-glutamine, 1 mmol/L sodium pyruvate, and 1× penicillin/streptomycin. All cells were maintained in a humidified incubator with 5% CO2 at 37°C. Subtypes of SCLC cell lines are summarized in Supplementary Table S4.
CRISPR-Cas9 Gene-Editing Vectors
Target sequences of DHX9, MAVS, STING, IFNAR1, and DNA/RNA sensors and RNA helicases (for screen) for CRISPR interference were designed using the single-guide RNA (sgRNA) designer (http://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). Sequences of sgRNA for human DHX9 were 5′-GGGGTAGAATCTGATACCGA-3′ (sgDHX9) and 5′-CAAAACATTATACTGGCATG-3′ (sgDHX9 #2). sgRNA sequences of human MAVS and STING were 5′-ACTGGAGCAGATGATAGGCT-3′ and 5′-GGTACCGGGGCAGCTACTGG-3′, respectively. sgRNA sequence of human IFNAR1 was 5′-GTACATTGTATAAAGACCAC-3′. sgRNA sequences of RNA helicase genes and DNA/RNA sensor genes are listed in Supplementary Table S1. sgRNA from the Gecko library v2 was used as a dummy sgRNA (5′-ATCGTTTCCGCTTAACGGCG-3′) for Scramble control (Scr). LentiCRISPR v2 vectors were cloned as previously described (80, 81). Lentiviral plasmids were transduced into HEK293T cells along with pMD2.G and psPAX2 using X-treme Gene 9 DNA Transfection Reagent (Roche; no. XTG9-RO) according to the manufacturer's instructions. The 48-hour supernatant of transduced HEK293T cells was collected and filtered with a 0.45 μm filter.
Transduction of SCLC Cells with Lentiviral Vectors
Culture supernatant from HEK293T cells containing virus particles was applied to SCLC cell lines, with 8 μg/mL polybrene (Santa Cruz Biotechnology; no. sc-134220). Cells were centrifuged with virus at 2,000 rpm at 37°C for 2 hours. The cells were incubated at 37°C in 5% CO2 for 24 hours, and then the media were replaced with complete RPMI-1640. Virus-infected cells were selected for 48 hours using 1.0 μg/mL puromycin (Gibco) or 5.0 μg/mL blasticidin (Gibco), from 48 hours after infection.
siRNA Transfection
DHX9 siRNAs (no. s4020, s4021) were purchased from Thermo Fisher Scientific, with s4021 used in most experiments. The siRNA targeting the 3′UTR of DHX9 was purchased from Horizon (no. A-009950-16-0005). A nonspecific control siRNA duplex (siCtrl) was purchased from Thermo Fisher Scientific (Silencer Select Negative Control No. 1 siRNA, no. 4390844). Other siRNAs used in this study included AURKA (no. s197), NMT1 (no. s9602), RHOQ (no. s23825), CDK9 (no. s2835), and CDK7 (no. s2829; Thermo Fisher Scientific). siRNAs were transfected using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific; no. 13778500) according to the manufacturer's instructions.
IHC Staining
Mouse tumor tissues were collected and fixed in 10% neutral buffered formalin for 24 hours, dehydrated, and embedded in paraffin. Hematoxylin and eosin (H&E)-stained sections were used for morphologic evaluation purposes and 5-μm unstained sections for IHC studies.
IHC staining was carried out on a VENTANA Discovery XT automated staining instrument (Ventana Medical Systems) using VENTANA reagents according to the manufacturer's instructions. Briefly, slides were deparaffinized using EZ Prep solution (no. 950-102) for 16 minutes at 72°C. Epitope retrieval was accomplished with CC1 solution (EDTA, pH 9.0.; no. 950-224) at high temperature (e.g., 95–100°C) for 32 minutes. Rabbit primary antibodies: anti-mouse CD8 (1:50, Cell Signaling Technology; no. 98941), anti-mouse CD45 (1:50, Cell Signaling Technology; no. 70257), and F4/80 (1:800, Cell Signaling Technology; no. 70076) tittered with a TBS antibody diluent into user fillable dispensers for use on the automated stainer. The immune complex was detected using the Ventana OmniMap anti-Rabbit detection kit (no. 760-4311) and developed using the VENTANA ChromMap DAB detection kit (no. 760-159) according to the manufacturer's instructions. Slides were then counterstained with hematoxylin II (no. 790-2208) for 8 minutes, followed by Bluing reagent (no. 760-2037) for 4 minutes.
The slides were then dehydrated with ethanol series, cleared in xylene, and mounted. As a negative control, the primary antibody was replaced with normal rabbit IgG to confirm the absence of specific staining.
Quantitative Image Analysis
Immunostained slides were scanned using an Aperio ScanScope CS 5 slide scanner (Aperio). Scanned images were then viewed with Aperio's image viewer software (ImageScope, version 11.1.2.760, Aperio). Selected regions of interest were outlined manually by a pathologist. The positive percentage score for CD8, CD45, and F4/80 was quantified using the Aperio V9 algorithm.
Immunoblotting
Protein was extracted from cell lines with Pierce RIPA Buffer (Thermo Fisher Scientific; no. 89900) and quantified by Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; no. 23225). Protein extracts were subjected to polyacrylamide gel electrophoresis using the 4%–12% NuPAGE gel system (Invitrogen) and transferred to PVDF membranes (Millipore). Transferred protein was immunoblotted using antibodies against TBK1 (no. 3013), S172 pTBK1 (no. 5483), IRF3 (no. 4302), S396 pIRF3 (no. 4947), S345 pCHK1 (no. 2348), T68 pCHK2 (no. 2197), S139 pH2AX (no. 9718), cleaved PARP (no. 5625), STING (no. 13647), MAVS (no. 3993), CDK9 (no. 2316), CDK7 (no. 2090), β-actin (no. 3700; Cell Signaling Technology), mouse DHX9 (no. ab26271), IFNAR1 (no. ab124764), V5 tag (no. ab27671; Abcam), and human DHX9 (no. sc-137232; Santa Cruz Biotechnology) after blocking with LICOR Blocking Buffer (LICOR; no. 927-60001).
Secondary antibodies were purchased from LICOR Biosciences: IRDye 800CW Goat anti-Mouse IgG (H+L) (no. 926-32210), IRDye 680RD anti-Rabbit IgG (H+L) (no. 926-68071), and IRDye 800CW Goat anti-Rabbit IgG (H+L) (no. 926-32211). LICOR Antibody Diluent (LICOR; no. 927-65001) was used to dilute primary and secondary antibodies. Phospho-specific antibodies were diluted in CanGet Signal Immunoreaction Enhancer Solutions (TOYOBO; no. NKB-101) 1 (for primary) and 2 (for secondary). Imaging of blots was performed using the LICOR Odyssey system.
Immunocytochemistry
Cells were plated on BioCoat Culture Slide (Corning; no. 354630) after trypsinization and incubated overnight. To detect expression of proteins in nuclei and micronuclei, cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes and permeabilized with 0.5% Triton X-100 for 15 minutes. After blocking with MAXblock Blocking Medium (Active Motif; no. 15252) for 1 hour at 37°C, cells were stained overnight at 4°C with primary antibodies and for 1 hour at room temperature with secondary antibodies. Coverslips were mounted with ProLong Gold Antifade reagent with DAPI (Invitrogen; no. P36935), and immunofluorescent visualization of nuclei was counterstained with DAPI in the reagent. Immunofluorescence was detected using a Leica SP8 confocal microscope and analyzed with ImageJ software. Antibody against dsRNA (J2; 1:200, no. 10010200) was purchased from SCICONS, and S139 pH2AX (1:1,000, no. 9718) and cGAS (1:100, no. 79978) were purchased from Cell Signaling Technology, and dsDNA antibody (1:100, no. ab27156) was from Abcam. For RNase III treatment, cells were treated with 20 U/mL of RNase III for 30 minutes at 37°C before fixation.
To detect DNA/RNA hybrid, cells were fixed with ice-cold, 100% methanol for 20 minutes at −20°C and permeabilized with 0.5% Triton X-100 for 15 minutes. Cells were incubated with antibody against DNA/RNA hybrid (Sigma-Aldrich; no. MABE1095, clone S9.6) at 1:50 dilution overnight at 4°C, followed by secondary donkey anti-mouse IgG (H+L) conjugated with Alexa Fluor 594 at 1:500 dilution for 1 hour at room temperature. For RNase H treatment, cells were incubated with 120 U RNase H (Takara Bio; no. 2150A) for 4 hours in RNase H buffer (40 mmol/L Tris-HCl pH8.0, 4 mmol/L MgCl2, 1 mmol/L dithiothreitol, 4% glycerol, and 0.003% BSA) before immunocytochemistry assay.
ELISA
IFNβ ELISA (R&D Systems; no. DIFNB0) and 2′,3′-Cyclic GAMP (cGAMP) ELISA (Arbor Assays; no. K067-H1) kits were used according to the manufacturer's instructions. Conditioned media from cells cultured for 72 hours after seeding (for IFNβ) and cell lysates (for cGAMP) were collected and analyzed. For cells treated with siRNAs, 72-hour-culture conditioned media from the cells were collected at day 6 after transfection.
For cytokine array assay, the Proteome Profiler Human Cytokine Array Kit (R&D Systems; no. ARY005B) was used according to the manufacturer's instructions. Conditioned media were collected 72 hours after seeding the cells.
Flow Cytometry Analysis
For detecting proteins on the cell membrane, collected cells were washed with PBS and stained with anti–PD-L1 (BioLegend; no. 329718, isotype control: no. 400232) and anti–HLA-A, B, C (BioLegend; no. 311410, isotype control: no. 400220) antibodies diluted with PBS containing 2% FBS at 2 μg/mL. Stained cells were analyzed on a BD LSR II Flow Cytometer, and fluorescence levels were compared with isotype control antibodies. Data were analyzed using the FlowJo software (TreeStar). Dead cells were excluded by staining with PI or Zombie NIR Fixable Viability Kit (BioLegend; no. 423106).
For intracellular flow cytometry, collected cells were fixed with 4% formaldehyde in PBS and permeabilized with 0.1% Triton X-100 in PBS. The cells were then incubated with primary antibodies, followed by secondary antibodies. The primary antibody against dsRNA (J2; 1:200, no. 10010200) was purchased from SCICONS, and S139 pH2AX (1:1,000, no. 9718) was from Cell Signaling Technology. Isotype controls, normal mouse IgG2a (no. ab18413), and rabbit IgG (no. ab172730) were purchased from Abcam. Secondary donkey anti-mouse IgG (H+L) conjugated with Alexa Fluor 488 (no. A32766) and donkey anti-rabbit IgG (H+L) conjugated with Alexa Fluor 488 (no. A32790) were purchased from Thermo Fisher Scientific. Dead cells were excluded by prefixation staining with Zombie NIR Fixable Viability Kit.
Murine SCLC GEMM, Cell Line Derivation, and Tumor Implantation Studies
The RPP-631 (RPP) SCLC mouse cell lines were established in the laboratory of Dr. Matthew G. Oser (Dana-Farber Cancer Institute, Boston, MA, USA), which were originally derived from SCLC tumors that were generated in LSL-Cas9 C57BL/6 mice that were intratracheally injected with AAV that encode Cre-recombinase and sgRNAs targeting Trp53, Rb1, and Rbl2 (RPP) genes (60). Histopathology of the tumor from which the cell lines were derived showed small cell lung cancer.
To generate the syngeneic mouse tumor model, 8.0 × 106 RPP cells, which stably express SMARTvector Tet-inducible Dhx9 shRNA (horizon; no. V3SM11253) or control shRNA (horizon; no. VSC11652), were subcutaneously implanted into the flank of C57BL/6 mice or NSG mice after mixing with Matrigel (Corning; no. 354234) at 1:1 ratio. To generate RP tumors, 1.0 × 106 KP3 cells (a kind gift from Dr. Julien Sage) were subcutaneously implanted into the flank of B6129SF1 mice after mixing with Matrigel at a 1:1 ratio. Doxycycline water (5% sucrose with 2 mg/mL of doxycycline; Sigma) was provided to all the groups to induce knockdown of the Dhx9 gene once palpable tumor formation was confirmed. The doxycycline water was changed every other day. Tumor size was measured every 2–3 days by digital caliper. Tumor volumes were calculated using the formula: volume = (length × width2)/2. Both female and male C57BL/6 mice (Jackson Laboratories) of 6–8 weeks old were used for the transplantation studies.
Antibody Treatment, Tumor Collection, and Survival Analysis
Mice were euthanized with CO2 and their tumors were quickly extracted, washed in PBS, and minced using a sterilized razor blade. For tumor IHC staining of CD8, CD45, and F4/80 and flow cytometry analysis of infiltrated immune cells, all mice were euthanized on day 32 after inoculation of RPP cells. Collected tumor tissues were fixed in 10% phosphate-buffered formaldehyde for IHC and were processed for flow cytometry analysis using the Tumor Dissociation Kit, mouse (Miltenyi; no. 130-096-730) and gentleMACS Dissociator (Miltenyi; no. 130-093-235) according to the manufacturer's instructions, followed by washing and filtering with a 70-μm cell strainer.
For combination treatments, mice were administered 200 μg rat IgG2a isotype control (Bio X Cell; no. BP0089) or anti–PD-1 antibody (Bio X Cell; no. BP0146) via i.p. injection on days 17, 19, 21, 23, and 25 after inoculation of RPP cells. Doxycycline water was provided once palpable tumor formation was confirmed to induce knockdown of the Dhx9 gene. Tumor size was measured every 2–3 days by digital caliper, and mouse survival was monitored with tumor volume exceeding 1,000 mm3, weight loss >15%, and decreasing behavioral conditions considered as endpoints.
For depletion of CD8 T cells or NK cells, C57BL/6 mice were treated with 10 mg/kg anti-CD8a antibody (Bio X Cell; no. BP0061) or anti-NK1.1 antibody (Bio X Cell; no. BP0036) twice per week.
Quantitative PCR with Reverse Transcription
Total RNAs were extracted using the RNeasy Mini Kit (Qiagen; no. 74106) according to the manufacturer's instructions. A total 1 μg of extracted RNA was used to generate cDNA with the SuperScript III First-Strand Synthesis SuperMix for RT-qPCR kit (Thermo Fisher Scientific; no. 18080-044). qRT-PCR of the indicated genes (Supplementary Table S2) was performed using Power SYBR Green PCR Master Mix (Applied Biosystems; no. 4367659) and the Applied Biosystems QuantStudio 6 Pro Real-Time PCR System and software. The relative expression was normalized with the expression of the housekeeping genes 36B4 (for human cells) or Actb (for mouse cells) and analyzed with the –ΔΔCt relative quantification method.
RNA-seq
Total RNAs were extracted and purified using the RNeasy Mini Kit (Qiagen; no. 74106) from NCI-H196, NCI-H446, and NCI-H82 cells, which were infected with lentivirus containing Scramble or sgDHX9 vector at day 7 after selection. Using a 2100 Bioanalyzer RNA 6000 Nano assay (Agilent), the quality of RNA was assessed. RNA concentration was measured using a Qubit 2.0 Fluorometer (Life Technologies). Illumina sequencing libraries were constructed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) and sequenced on Illumina NovaSeq 6000 by pair-end sequencing with a read length of 2 × 150 bp by Novogene.
Expression levels for each gene were quantified from the sequencing data using Kallisto (82). The data were then summarized using the tximport package (ver. 1.18.0) of R software (ver. 4.0.3) and RStudio (RStudio), and scaledTPM counts were used for further analysis as expression values. GSEA was performed to identify gene signatures that are upregulated and downregulated in sgDHX9 cells compared with Scramble or DHX9low tumors compared with DHX9high tumors (for data from database).
RIP-seq
Scramble or sgDHX9 H446 cells (2.0 × 107) were harvested, and RNA immunoprecipitation (RIP) was conducted using a Magna RIP RNA-Binding Protein Immunoprecipitation kit (Sigma-Aldrich; no. 17-700) according to the manufacturer's instructions. Briefly, cell pellets were lysed in RIP lysis buffer, followed by incubation with RIP buffer containing magnetic beads conjugated with J2 (SCICONS; no. 10010200) or isotype control (Abcam; no. ab18413) antibody at 4°C overnight. Samples were then incubated with proteinase K, and immunoprecipitated RNAs were recovered by phenol:chloroform:isoamyl alcohol purification. RNA was quantified using a QuantiFluor RNA System (Promega; no. E3310) and assessed for quality with the 2100 Bioanalyzer RNA 6000 Nano assay (Agilent) before library generation. Ribosomal RNA (rRNA) was removed using NEBNext rRNA Depletion Kit v2 (NEB; no. E7400L) according to the manufacturer's instructions. For pulling down of MDA5 and RIG-I bound RNA, we transfected pcDNA3-Flag-MDA5 and pcDNA3-Flag-RIG-I expression vector, respectively, to DHX9-depleted cells 2 days before collecting RNA. Anti-Flag (DYKDDDDK)-tag antibody (Invitrogen; no. MA1-91878) was used for IP. For pulling down of DHX9-bound RNA, we prepared pCDH-3xFlag-DHX9-K417R–expressing H446 cells and performed IP using anti-Flag (DYKDDDDK)-tag antibody.
Illumina sequencing libraries were prepared by Novogene, with the NEBNext Ultra II RNA Library Prep Kit for Illumina (NEB) according to the manufacturer's instructions. Libraries were analyzed for insert size distribution using the 2100 Bioanalyzer RNA 6000 Nano assay (Agilent). Libraries were quantified using the Qubit 2.0 Fluorometer (Life Technologies) and sequenced on Illumina NovaSeq 6000 by pair-end sequencing with a read length of 2 × 150 bp, by Novogene.
For analysis of RE expression from RIP-seq data, the trimmed FASTQ reads by TrimGalore (version 0.6.4) were mapped against H. sapiens UCSC hg19 using the Bowtie 2 (version 2.3.5) alignment software (83). The counts of RE were calculated by RepEnrich2 (84) using default settings and were normalized by one million reads as CPM (counts per million). In this software, reads mapping uniquely to the genome are assigned to subfamilies of repetitive elements based on their degree of overlap to “RepeatMasker” annotated genomic instances of each repetitive element subfamily. On the other hand, reads mapping to multiple locations are separately mapped to repetitive element assemblies—referred to as repetitive element psuedogenomes. By combining the counts from uniquely mapping reads and multimapping reads, “RepEnrich2” keeps track of all repetitive elements that every read aligns to and systematically estimates enrichment from all mapping reads. The heat map was visualized using the “ggplot2” package of R software (version 4.0.5).
RIP-qRT-PCR
Scramble or sgDHX9 H446 cells (5.0 × 106) were harvested, and cytoplasmic fractions were extracted using the Nuclear Extract Kit (Active Motif; no. 40010) according to the manufacturer's instructions. To isolate RNA, an equal volume of 70% ethanol was added to the cytoplasmic fractions, and then, according to the manufacturer's instructions, purification was performed using the RNeasy Plus Mini Kit (Qiagen; no. 74106). The total RNA was dissolved with 38 μL RNase-free H2O. Then 2 μL total RNA was used as input and the remainder was divided into 2 tubes. In all, 2 μg of J2 antibody (SCICONS; no. 10010200) and mouse control IgG2a (Abcam; no. ab18413) were conjugated to 20 μL protein G agarose (Millipore; no. 16-266) per pulldown by rotation overnight at 4°C. To digest single-stranded RNA, 1 μL of RNase A (Sigma-Aldrich; no. R6513) was added to each tube and then mixed with 1 mL IP buffer (50 mmol/L Tris-HCl [pH 7.4], 125 mmol/L NaCl, 1 mmol/L EDTA, 0.1% Triton X-100). The RNA samples were incubated with antibody-conjugated protein G agarose beads overnight at 4°C. Beads were washed with IP buffer three times and then incubated in 50 μL proteinase K digestion solution [1× TE, 100 mmol/L NaCl, 1% SDS, and 1 μL of 20 mg/mL Proteinase K solution (Thermo Fisher Scientific; no. AM2546)] for 20 minutes at 45°C to isolate RNA. After centrifugation, 50 μL of the supernatant was added to 300 μL Buffer RLT Plus from the RNeasy Plus Mini Kit (QIAGEN; no. 74106) to purify RNA. The final product containing dsRNA was denatured for 5 minutes at 95°C, followed by reverse transcription using qScript cDNA SuperMix (Quantabio; no. 95048), which contains both random primers and oligo(dT) primers. qRT-PCR was performed using the primers listed in Supplementary Table S2 by Applied Biosystems QuantStudio 6 Pro Real-Time PCR System and software.
DNA Fiber Assay
Cells were pulse-labeled with 25 μmol/L ldU (Sigma-Aldrich; no. I7125) for the first 30 minutes, followed by 250 μmol/L CIdU (Sigma-Aldrich; no. C6891) for 30 minutes. The cells were trypsinized and resuspended in PBS then diluted to the concentration of 1.0 × 105 to 1.0 × 106 cells/mL. At the end of an APS-coated glass slide (Matsunami; no. SUAPS1190), 2 μL of cell suspension was placed. After air-drying for 8 minutes, 7 μL of fiber lysis solution (200 mmol/L Tris-HCl [pH 7.5], 50 mmol/L EDTA, 0.5% SDS) was pipetted on top of the cell suspension and mixed gently. Cell lysis proceeded for 5 minutes. The slides were tilted at 15° to allow the DNA to spread down to the bottom of the slide. Slides were air-dried for 15 minutes and fixed in methanol/acetic acid (3:1). After washing with distilled water, DNA was denatured in 2.5 M HCl for 80 minutes. The slides were washed with PBS three times and blocked in 5% BSA in PBS for 1 hour. After blocking, the slides were incubated with primary antibodies, anti-IdU (BD; no. 347580), and anti-CldU (Abcam; no. ab6326) and followed by secondary antibodies, donkey anti-mouse IgG (H+L) conjugated with Alexa Fluor 488 (Thermo Fisher Scientific; no. A32766), and donkey anti-rat IgG (H+L) conjugated with Alexa Fluor 594 (Thermo Fisher Scientific; no. A21209).
Proliferation Assay
Cells were seeded in 12-well plates at low density (2,500–10,000 cells/well) and cultured in complete RPMI-1640 medium in a humidified incubator with 5% CO2 at 37°C. After 2, 4, 6, and 8 days, cells were harvested and counted.
Transfection of Cytoplasmic dsRNA/DNA
Cytoplasmic fraction was extracted from DHX9-depleted cells (4.0 × 106) using the Nuclear Extract Kit (Active Motif; no. 40010) according to the manufacturer's instructions. Cytoplasmic DNA was extracted using the Gel Extraction Kit (QIAGEN; no. 28704) after treatment of RNase A/T1 Mix (Thermo Fisher Scientific; no. EN0551). To isolate RNA, an equal volume of 70% ethanol was added to the cytoplasmic fractions, and purification was performed using the RNeasy Plus Mini Kit (QIAGEN; no. 74106). Pulldown of dsRNA was performed as detailed in the “RIP-qRT-PCR” section.
For the cytoplasmic DNA retransfection assay, extracted cytoplasmic DNA was digested with 1 μL DNase I (Thermo Fisher Scientific; no. EN0521), 1 μL RNase H (NEB; no. M0523S), or 1 μL H2O (mock) in 10 μL reaction mixture for 60 minutes at 37°C prior to retransfection into H196 cells with Lipofectamine 3000 (Thermo Fisher Scientific; no. L3000001). For cytoplasmic dsRNA retransfection, purified dsRNA was digested with 1 μL RNase III (Thermo Fisher Scientific; no. AM2290) or 1 μL H2O (mock) in 5 μL reaction mixture for 60 minutes at 37°C prior to retransfection. After 24 hours, RNA was isolated using the RNeasy Plus Mini Kit.
DHX9 Expression Vectors
The wild-type and helicase dead mutant (K417R) human DHX9 cDNAs were cloned into pCDH-3xFLAG-GFP-puroR vector (a kind gift from Dr. Lu Chen) with the AscI/PacI sites, generating pCDH-3xFLAG-DHX9 (WT/K417R)-puroR. For rescue experiments, siRNA targeting DHX9-3′UTR was transfected to knockdown-only endogenous DHX9.
R-loop Detection by Dot Blot
For R-loop (DNA/RNA hybrid) detection by dot blot, DNA samples were prepared using the DRIP protocol, as previously reported (85). Briefly, cells were lysed in 1.6 mL TE buffer containing 50 μL of 20% SDS and 5 μL of 20 mg/mL proteinase K at 37°C overnight. Extracted DNA was phase separated using phenol/chloroform/isoamyl alcohol (25:24:1), precipitated with ethanol, and resuspended in TE buffer. Then, purified DNA was digested using a cocktail of restriction enzymes (HindIII, SspI, EcoRI, BsrGI, and XbaI; 30 U each), treated with RNase A (10 μg/mL) and RNase III (2U), and again purified by phenol/chloroform/isoamyl alcohol (25:24:1). DNA was spotted on a nitrocellulose membrane, blocked with LICOR Blocking Buffer, and incubated with antibody against DNA/RNA hybrid (Sigma-Aldrich; no. MABE1095, clone S9.6) dsDNA (Abcam; no. ab27156). Secondary antibody IRDye 800CW Goat anti-Mouse IgG (H+L) (LICOR; no. 926-32210) was used to detect the signal. Imaging of blots was performed using the LICOR Odyssey system.
Generation and Characterization of Synthetic Spike-in R-loop
Synthetic R-loop spike-in was generated according to (86). Briefly, E. coli genomic DNA was isolated by phenol-chloroform extraction. A 320 bp of DNA was PCR amplified with L286F (T7-promoter bearing) and L286R (Supplementary Table S2) and purified using DNA Clean and Concentrator-5 (Zymo Research; no. D4014). In vitro transcription reactions were carried out at 37°C for 1 hour with the HiScribe T7 High Yield RNA Synthesis Kit (NEB; no. E2040S) with 200 ng of template DNA. RNA was purified using the Direct-zol RNA Miniprep kit (Zymo Research; no. R2052). RNA was checked by running a 7% UREA-PAGE gel. Complementary L286 ssDNA was synthesized by IDT (Supplementary Table S2), and the DNA/RNA hybrid was reconstituted with a molar ratio of 1:5 in the NEB Buffer 2.1. The reconstituted hybrid is fractionated in 1.8% agarose gel, from which the RNase H sensitive species was excised and eluted following a gel-crushing method (87). The purified R-loop was quantitated by agarose gel electrophoresis followed by SYBR green II staining. R-loop is further characterized by an R-loop EMSA assay containing 0.55 nmol/L of R-loop and S9.6 antibody (0.11 and 0.55 nmol/L). The reaction was incubated at 37°C for 30 minutes and fractioned in a 5% native PAGE gel visualized by SYBR green II staining.
Quantitative DNA-RNA Immunoprecipitation-qPCR with Spike-Ins
qDRIP was performed as per the method detailed by (86). Briefly, H82 cells were pelleted at 500 × g for 5 minutes, followed by resuspension in 4 mL of DPBS and a gently layered 3 mL of Ficoll Paque Plus (Cytiva; no. 17144003). This was centrifuged at 400 × g for 40 minutes at room temperature to separate live cells. In total, 750,000 trypan-blue-negative cells per experimental condition were resuspended in 1.6 mL of TE buffer, to which 5 μL of a 16.7 fM of synthetic E. coli DNA/RNA hybrid L286 was added as spike-in control (see below). Cells were incubated in 50 μL of 20% SDS and 5 μL of Proteinase K (20 mg/mL, Thermo Fisher Scientific; no. EO0491) at 37°C for 3 hours, followed by phenol-chloroform extraction and ethanol precipitation. The DNA was then sonicated to an average fragment length of 300 bp using the QSonica 800R3 (no. Q800R3-110) at 40% Amplitude, 15 seconds ON or OFF alternating for a total of 7 minutes ON time. The resulting fragment distribution was analyzed using High Sensitivity Tapestation D1000 screentape (Agilent; no. 5067-5584). Immunoprecipitation was performed with the S9.6 antibody (Kerafast; no. Kf-Ab01137-23.0) and DynaGreen Protein A/G Magnetic Beads (Thermo Fisher Scientific; no. 80106G). RNase H treatment (NEB; no. M0297S) was carried out at 37°C overnight. The qPCR reactions were then conducted using 2× qRT-PCR Brilliant III SYBR Master Mix (Agilent; no. 600886) on the QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific; 4485691). The results of these reactions were analyzed using R version 4.3.1.
Viability Assay
Cells were plated in 96-well plates at low density (2,000–8,000 cells/well) and cultured in complete RPMI-1640 medium in a humidified incubator with 5% CO2 at 37°C for 96 hours. Luminescent values of CellTiter-Glo Cell Viability assay (Promega; no. G7571) were obtained by the CLARIOstar Plus Microplate Reader and software (BMG Labtech).
CRISPR Screen and Analysis
On day 0 (day of infection), ∼7.0 × 107 (∼1,000 cells/sgRNA) H82 cells, which stably express Cas9, were resuspended in complete media with 10% FBS, 8 μg/mL polybrene at a concentration of 1.0 × 106 cells/mL in 50 mL conical tubes, and the Human Brunello CRISPR knockout pooled library was added at an MOI of 0.3. The cells were then distributed onto low-adherence 6-well plates at a density of 2.0 × 106 cells per well, and the plates were centrifuged at 2,000 rpm for 2 hours. The following day (day 1), the virus was removed by changing media, and the cells were transferred to low-adherence 10-cm plates at a concentration of 0.4 × 106 cells/mL. On day 4, the cells were plated in fresh media in the presence of 1 μg/mL puromycin and were selected for 72 hours.
Following completion of puromycin selection (day 7), resistant cells were cultured with fresh media and grown in complete media until day 13. On day 13, 2.5 × 107 cells were mixed with lentivirus containing Scramble or sgDHX9, with 8 μg/mL polybrene. The cells were then distributed onto low-adherence 6-well plates at a density of 2.0 × 106 cells per well, and the plates were centrifuged at 2,000 rpm for 2 hours. The following day (day 14), the virus was washed away, and the cells were transferred to low-adherence 10-cm plates. Until day 23, the cells were grown in complete media, keeping a minimum of 2.5 × 107 cells per group. The screen was performed in 3 biological replicates.
After completion of the screen, genomic DNA (gDNA) was isolated using Blood and Cell Culture DNA Midi Kit (Qiagen; no. 13343) according to the manufacturer's protocol. To attach sequencing adaptors and barcode samples, PCR of gDNA was performed using Ex Taq DNA Polymerase (TaKara; no. RR001) as previously described (52). Samples were purified with Agencourt AMPure XP SPRI beads according to the manufacturer's instructions (Beckman Coulter; no. A63880). Purified DNA samples were sequenced on a Nextseq2000 (Illumina). The sgRNA read count and hits calling were analyzed by MAGeCK v0.5.7 algorithm (88). Each gene symbol of enriched or depleted sgRNAs in DHX9-loss population was mapped to GO resources (http://www.geneontology.org/) to rank GO terms.
Human Brunello CRISPR knockout pooled library (52) was a gift from Drs. David Root and John Doench (Addgene; no. 73178).
Pan-cancer Analysis of TCGA Data Set
TCGA pan-cancer gene expression with patient annotation data sets were retrieved from the Genomic Data Commons of the National Cancer Institute (https://gdc.cancer.gov/about-data/publications/pancanatlas). Signature scores of different tumor types were calculated in GSVA using the “z-score” method (89). Spearman correlation and multiple testing corrections were done in R software (ver. 4.0.3).
Data Availability
The RNA-seq data are deposited in the GEO repository (Accession Number: GSE244103). We also registered raw data of RIP-seq experiments in the GEO repository [Accession Number: GSE244104 (J2), GSE247217 (Flag)]. Data generated from the CRISPR screen are provided in Supplementary Table S5. All other data and materials can be requested from the corresponding author.
Statistical Analyses
All graphs depict mean ± SEM unless otherwise indicated. Tests for differences between two groups were performed using two-tailed unpaired Student t test or Mann–Whitney test. Two-way analysis of variance (ANOVA) was performed where applicable using the Tukey multiple comparison test. Values of P < 0.01–0.05 (*), P < 0.001–0.01 (**), P < 0.001–0.0001 (***), or P < 0.0001 (****) were considered significant. GraphPad Prism7 was used for statistical analysis of experiments, data processing, and presentation.
Study Approval
All animal experiments were performed under protocols approved by the Institutional Animal Care and Use Committee at FCCC.
Supplementary Material
sgRNA sequences
PCR primer sequences
DHX9 depleted signature
SCLC cell line information
CRISPR screen results
Flow cytometry settings
DHX9 suppresses ERE-derived dsRNA accumulation in SCLCs
Induction of IFN response after DHX9 loss in SCLC cell lines
DNA damage and R-loop accumulation in DHX9 depleted SCLC cells
DHX9 depletion decreases cell viability and induces dsRNA and dsDNA antiviral sensing pathways in SCLC cells
CDK9 inhibition rescues the DHX9 depletion effect on cell survival by reducing replication stress
DHX9 loss decreases tumor growth, induces immune cell infiltration and enhances response to immunotherapy
DHX9-low tumors are associated with interferon and inflammatory signatures
Acknowledgments
The authors thank Dr. David A. Barbie (Dana-Farber Cancer Institute, Boston, MA, USA) for critically reading the manuscript. The GEMM-derived SCLC cell line RPP was kindly provided by Dr. Matthew G. Oser (Dana-Farber Cancer Institute, Boston, MA, USA). The GEMM-derived SCLC cell line KP3 (RP) was kindly provided by Dr. Julien Sage (Stanford University, Stanford, CA, USA). The authors thank Mengmeng Li (Fox Chase Cancer Center, Philadelphia, PA, USA) for statistical consulting. This work was supported by the Lung Cancer Research Foundation (I. Cañadas), the G. Harold and Leila Y. Mathers Foundation (I. Cañadas), the W.W. Smith Charitable Trust (I. Cañadas) and the Bucks County Chapter of the Board of Associates of Fox Chase Cancer Center (I. Cañadas). Additional funding was provided by a Grant-in-Aid for a Japan Society for Promotion of Science (JSPS) fellowship to T. Murayama (202160224). This work was also supported by NIH grants R35GM150538 (to L. Chen) and T32GM142606 training fellowship (to X. Jiang).
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
T. Murayama reports funding was provided for work outside the submitted work by a Grant-in-Aid for a Japan Society for Promotion of Science (JSPS) fellowship (202160224; http://www.jsps.go.jp/english/e-pd/index.html). R.M. Prasad reports a patent for 18/043,548 pending. E.R. Gerstein reports grants from NIH/NIGMS during the conduct of the study. L. Chen reports grants from NIH/NIGMS during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
T. Murayama: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J. Nakayama: Data curation, software, formal analysis, validation, investigation, visualization, methodology. X. Jiang: Methodology. K. Miyata: Data curation, software, formal analysis, validation, visualization, methodology. A.D. Morris: Investigation, visualization, methodology. K.Q. Cai: Resources, formal analysis, investigation, visualization, methodology. R.M. Prasad: Validation, methodology. X. Ma: Validation, methodology. A. Efimov: Resources, methodology. N. Belani: Methodology, writing–review and editing. E.R. Gerstein: Investigation, methodology. Y. Tan: Resources, investigation, methodology. Y. Zhou: Data curation, software, formal analysis, visualization, methodology. W. Kim: Data curation, software, methodology. R. Maruyama: Data curation, methodology. K.S. Campbell: Resources, methodology. L. Chen: Funding acquisition, investigation, visualization, methodology. Y. Yang: Data curation, formal analysis, validation, visualization, methodology. S. Balachandran: Conceptualization, formal analysis, writing–review and editing. I. Cañadas: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Demaria O, Cornen S, Daeron M, Morel Y, Medzhitov R, Vivier E. Harnessing innate immunity in cancer therapy. Nature 2019;574:45–56. [DOI] [PubMed] [Google Scholar]
- 2. Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti–PD-1 immunotherapy. Cell 2017;170:1109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ishizuka JJ, Manguso RT, Cheruiyot CK, Bi K, Panda A, Iracheta-Vellve A, et al. Loss of ADAR1 in tumours overcomes resistance to immune checkpoint blockade. Nature 2019;565:43–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ribas A, Medina T, Kummar S, Amin A, Kalbasi A, Drabick JJ, et al. SD-101 in combination with pembrolizumab in advanced melanoma: results of a phase Ib, multicenter study. Cancer Discov 2018;8:1250–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J, Huang D, Saw PE, Song E. Turning cold tumors hot: from molecular mechanisms to clinical applications. Trends Immunol 2022;43:523–45. [DOI] [PubMed] [Google Scholar]
- 6. Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature 2020;588:169–73. [DOI] [PubMed] [Google Scholar]
- 7. Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 2018;174:549–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen JZ, Qiu Z, Wu Q, Finlay D, Garcia G, Sun D, et al. FBXO44 promotes DNA replication-coupled repetitive element silencing in cancer cells. Cell 2021;184:352–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 2018;24:1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 2015;162:974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 2015;162:961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res 2017;77:3870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Von Hoff DD, Rasco DW, Heath EI, Munster PN, Schellens JHM, Isambert N, et al. Phase I study of CC-486 alone and in combination with carboplatin or nab-paclitaxel in patients with relapsed or refractory solid tumors. Clin Cancer Res 2018;24:4072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DiNardo CD, Jonas BA, Pullarkat V, Thirman MJ, Garcia JS, Wei AH, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med 2020;383:617–29. [DOI] [PubMed] [Google Scholar]
- 16. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity 2020;52:17–35. [DOI] [PubMed] [Google Scholar]
- 17. Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Antonia SJ, López-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883–95. [DOI] [PubMed] [Google Scholar]
- 20. Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 2018;379:2220–9. [DOI] [PubMed] [Google Scholar]
- 21. Liu SV, Reck M, Mansfield AS, Mok T, Scherpereel A, Reinmuth N, et al. Updated overall survival and PD-L1 subgroup analysis of patients with extensive-stage small-cell lung cancer treated with atezolizumab, carboplatin, and etoposide (IMpower133). J Clin Oncol 2021;39:619–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gadgeel SM, Pennell NA, Fidler MJ, Halmos B, Bonomi P, Stevenson J, et al. Phase II study of maintenance pembrolizumab in patients with extensive-stage small cell lung cancer (SCLC). J Thorac Oncol 2018;13:1393–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gay CM, Stewart CA, Park EM, Diao L, Groves SM, Heeke S, et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021;39:346–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mahadevan NR, Knelson EH, Wolff JO, Vajdi A, Saigi M, Campisi M, et al. Intrinsic immunogenicity of small cell lung carcinoma revealed by its cellular plasticity. Cancer Discov 2021;11:1952–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sen T, Tong P, Diao L, Li L, Fan Y, Hoff J, et al. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res 2017;23:6239–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov 2019;9:646–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Taniguchi H, Caeser R, Chavan SS, Zhan YA, Chow A, Manoj P, et al. WEE1 inhibition enhances the antitumor immune response to PD-L1 blockade by the concomitant activation of STING and STAT1 pathways in SCLC. Cell Rep 2022;39:110814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017;23:3711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 2019;79:311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–65. [DOI] [PubMed] [Google Scholar]
- 31. Lee CG, Chang KA, Kuroda MI, Hurwitz J. The NTPase/helicase activities of Drosophila maleless, an essential factor in dosage compensation. EMBO J 1997;16:2671–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget 2016;7:42716–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choi H, Kwon J, Cho MS, Sun Y, Zheng X, Wang J, et al. Targeting DDX3X triggers antitumor immunity via a dsRNA-mediated tumor-intrinsic type I interferon response. Cancer Res 2021;81:3607–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem 2002;277:12810–5. [DOI] [PubMed] [Google Scholar]
- 35. Bourgeois CF, Mortreux F, Auboeuf D. The multiple functions of RNA helicases as drivers and regulators of gene expression. Nat Rev Mol Cell Biol 2016;17:426–38. [DOI] [PubMed] [Google Scholar]
- 36. Chellini L, Pieraccioli M, Sette C, Paronetto MP. The DNA/RNA helicase DHX9 contributes to the transcriptional program of the androgen receptor in prostate cancer. J Exp Clin Cancer Res 2022;41:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi F, Cao S, Zhu Y, Yu Q, Guo W, Zhang S. High expression of DHX9 promotes the growth and metastasis of hepatocellular carcinoma. J Clin Lab Anal 2021;35:e24052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aktas T, Avsar Ilik I, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 2017;544:115–9. [DOI] [PubMed] [Google Scholar]
- 39. Chen R, Ishak CA, De Carvalho DD. Endogenous retroelements and the viral mimicry response in cancer therapy and cellular homeostasis. Cancer Discov 2021;11:2707–25. [DOI] [PubMed] [Google Scholar]
- 40. Sadeq S, Al-Hashimi S, Cusack CM, Werner A. Endogenous double-stranded RNA. Noncoding RNA 2021;7:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu S, Jia M, Li Y, Sun PL, Gao H. Differential expression of PD-L1 in central and peripheral and TTF1-positive and -negative small-cell lung cancer. Front Med 2020;7:621838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Patel PS, Abraham KJ, Guturi KKN, Halaby MJ, Khan Z, Palomero L, et al. RNF168 regulates R-loop resolution and genomic stability in BRCA1/2-deficient tumors. J Clin Invest 2021;131:e140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DHX9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep 2018;23:1891–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Crossley MP, Bocek M, Cimprich KA. R-loops as cellular regulators and genomic threats. Mol Cell 2019;73:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Petermann E, Lan L, Zou L. Sources, resolution and physiological relevance of R-loops and RNA-DNA hybrids. Nat Rev Mol Cell Biol 2022;23:521–40. [DOI] [PubMed] [Google Scholar]
- 46. Murayama T, Takeuchi Y, Yamawaki K, Natsume T, Li M, Marcela RN, et al. MCM10 compensates for Myc-induced DNA replication stress in breast cancer stem-like cells. Cancer Sci 2021;112:1209–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Collin G, Huna A, Warnier M, Flaman JM, Bernard D. Transcriptional repression of DNA repair genes is a hallmark and a cause of cellular senescence. Cell Death Dis 2018;9:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res 2007;35:7417–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Egloff S. CDK9 keeps RNA polymerase II on track. Cell Mol Life Sci 2021;78:5543–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gorthi A, Romero JC, Loranc E, Cao L, Lawrence LA, Goodale E, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature 2018;555:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Klusmann I, Wohlberedt K, Magerhans A, Teloni F, Korbel JO, Altmeyer M, et al. Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc Natl Acad Sci U S A 2018;115:E11311–E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yeo CQX, Alexander I, Lin Z, Lim S, Aning OA, Kumar R, et al. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep 2016;15:132–46. [DOI] [PubMed] [Google Scholar]
- 57. Lucking U, Scholz A, Lienau P, Siemeister G, Kosemund D, Bohlmann R, et al. Identification of Atuveciclib (BAY 1143572), the first highly selective, clinical PTEFb/CDK9 inhibitor for the treatment of cancer. ChemMedChem 2017;12:1776–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Crossley MP, Bocek MJ, Hamperl S, Swigut T, Cimprich KA. qDRIP: a method to quantitatively assess RNA-DNA hybrid formation genome-wide. Nucleic Acids Res 2020;48:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yuan W, Al-Hadid Q, Wang Z, Shen L, Cho H, Wu X, et al. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res 2021;49:8573–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oser MG, Sabet AH, Gao W, Chakraborty AA, Schinzel AC, Jennings RB, et al. The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev 2019;33:1718–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Paz-Ares L, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022;7:100408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Owonikoko TK, Park K, Govindan R, Ready N, Reck M, Peters S, et al. Nivolumab and Ipilimumab as maintenance therapy in extensive-disease small-cell lung cancer: CheckMate 451. J Clin Oncol 2021;39:1349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ready NE, Ott PA, Hellmann MD, Zugazagoitia J, Hann CL, de Braud F, et al. Nivolumab monotherapy and nivolumab plus ipilimumab in recurrent small cell lung cancer: results from the CheckMate 032 randomized cohort. J Thorac Oncol 2020;15:426–35. [DOI] [PubMed] [Google Scholar]
- 64. Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nat Med 2011;17:1504–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nishiga Y, Drainas AP, Baron M, Bhattacharya D, Barkal AA, Ahrari Y, et al. Radiotherapy in combination with CD47 blockade elicits a macrophage-mediated abscopal effect. Nat Cancer 2022;3:1351–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu D, Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Arnon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med 2019;25:1916–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. [DOI] [PubMed] [Google Scholar]
- 68. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lin YC, Yu YS, Lin HH, Hsiao KY. Oxaliplatin-induced DHX9 phosphorylation promotes oncogenic circular RNA CCDC66 expression and development of chemoresistance. Cancers 2020;12:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chakraborty P, Huang JTJ, Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat Commun 2018;9:4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Crossley MP, Song C, Bocek MJ, Choi JH, Kousorous J, Sathirachinda A, et al. R-loop-derived cytoplasmic RNA-DNA hybrids activate an immune response. Nature 2023;613:187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iams WT, Porter J, Horn L. Immunotherapeutic approaches for small-cell lung cancer. Nat Rev Clin Oncol 2020;17:300–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene 2016;35:5093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jiao A, Sun C, Wang X, Lei L, Liu H, Li W, et al. DExD/H-box helicase 9 intrinsically controls CD8(+) T cell-mediated antiviral response through noncanonical mechanisms. Sci Adv 2022;8:eabk2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wei X, Pacyna-Gengelbach M, Schluns K, An Q, Gao Y, Cheng S, et al. Analysis of the RNA helicase A gene in human lung cancer. Oncol Rep 2004;11:253–8. [PubMed] [Google Scholar]
- 77. Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, et al. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics 2014;7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liu S, He L, Wu J, Wu X, Xie L, Dai W, et al. DHX9 contributes to the malignant phenotypes of colorectal cancer via activating NF-kappaB signaling pathway. Cell Mol Life Sci 2021;78:8261–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Canadas I, Rojo F, Taus A, Arpi O, Arumi-Uria M, Pijuan L, et al. Targeting epithelial-to-mesenchymal transition with Met inhibitors reverts chemoresistance in small cell lung cancer. Clin Cancer Res 2014;20:938–50. [DOI] [PubMed] [Google Scholar]
- 80. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014;343:84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 83. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Criscione SW, Zhang Y, Thompson W, Sedivy JM, Neretti N. Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 2014;15:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sanz LA, Chedin F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA-RNA immunoprecipitation and high-throughput sequencing. Nat Protoc 2019;14:1734–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Crossley MP, Cimprich KA. Quantitative DNA-RNA immunoprecipitation sequencing with spike-ins. Methods Mol Biol 2022;2528:381–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sun Y, Sriramajayam K, Luo D, Liao DJ. A quick, cost-free method of purification of DNA fragments from agarose gel. J Cancer 2012;3:93–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sgRNA sequences
PCR primer sequences
DHX9 depleted signature
SCLC cell line information
CRISPR screen results
Flow cytometry settings
DHX9 suppresses ERE-derived dsRNA accumulation in SCLCs
Induction of IFN response after DHX9 loss in SCLC cell lines
DNA damage and R-loop accumulation in DHX9 depleted SCLC cells
DHX9 depletion decreases cell viability and induces dsRNA and dsDNA antiviral sensing pathways in SCLC cells
CDK9 inhibition rescues the DHX9 depletion effect on cell survival by reducing replication stress
DHX9 loss decreases tumor growth, induces immune cell infiltration and enhances response to immunotherapy
DHX9-low tumors are associated with interferon and inflammatory signatures
Data Availability Statement
The RNA-seq data are deposited in the GEO repository (Accession Number: GSE244103). We also registered raw data of RIP-seq experiments in the GEO repository [Accession Number: GSE244104 (J2), GSE247217 (Flag)]. Data generated from the CRISPR screen are provided in Supplementary Table S5. All other data and materials can be requested from the corresponding author.