Abstract
Objective:
To identify, in myometrial stem/progenitor cells, the presumptive cell of origin for uterine fibroids, substrates of Mediator-associated CDK8/19 kinase, which is known to be disrupted by uterine fibroid driver mutations in Mediator subunit MED12.
Design:
Experimental study.
Setting:
Academic research laboratory.
Patient(s):
Women undergoing hysterectomy for uterine fibroids.
Intervention(s):
Stable isotopic labeling of amino acids in myometrial stem/progenitor cell culture (SILAC) coupled with chemical inhibition of CDK8/19 and downstream quantitative phosphoproteomics and transcriptomic analyses.
Main Outcome Measure(s):
High-confidence Mediator kinase substrates identified by SILAC-based quantitative phosphoproteomics were determined using an empirical Bayes analysis and validated orthogonally by in vitro kinase assay featuring reconstituted Mediator kinase modules comprising wild-type or G44D mutant MED12 corresponding to the most frequent uterine fibroid driver mutation in MED12. Mediator kinase-regulated transcripts identified by RNA sequencing were linked to Mediator kinase substrates by computational analyses.
Result(s):
A total of 296 unique phosphosites in 166 proteins were significantly decreased (≥ twofold) upon CDK8/19 inhibition, including 118 phosphosites in 71 nuclear proteins representing high-confidence Mediator kinase substrates linked to RNA polymerase II transcription, RNA processing and transport, chromatin modification, cytoskeletal architecture, and DNA replication and repair. Orthogonal validation confirmed a subset of these proteins, including CUX1 and FOXK1, to be direct targets of MED12-dependent CDK8 phosphorylation in a manner abrogated by the most common uterine fibroid driver mutation (G44D) in MED12, implicating these substrates in disease pathogenesis. Transcriptome-wide profiling of Mediator kinase-inhibited myometrial stem/progenitor cells revealed alterations in cell cycle and myogenic gene expression programs to which Mediator kinase substrates could be linked directly. Among these, CUX1 is an established transcriptional regulator of the cell cycle whose corresponding gene on chromosome 7q is the locus for a recurrent breakpoint in uterine fibroids, linking MED12 and Mediator kinase with CUX1 for the first time in uterine fibroid pathogenesis. FOXK1, a transcriptional regulator of myogenic stem cell fate, was found to be coordinately enriched along with kinase, but not core, Mediator subunits in myometrial stem/progenitor cells compared with differentiated uterine smooth muscle cells.
Conclusion(s):
These studies identify a new catalog of pathologically and biologically relevant Mediator kinase substrates implicated in the pathogenesis of MED12 mutation-positive uterine fibroids, and further uncover a biochemical basis to link Mediator kinase activity with CUX1 and FOXK1 in the regulation of myometrial stem cell fate.
Keywords: MED12 mutations, mediator kinase, myometrial stem/progenitor cells, phosphoproteomics, uterine fibroids
Uterine fibroids (UFs; leiomyomas) are benign monoclonal neoplasms of the myometrium (MM) and are the most frequent reproductive tumors in women worldwide (1). Although benign, these tumors are nonetheless associated with significant morbidity; they are the primary indicator for hysterectomy and a major cause of gynecologic and reproductive dysfunction, ranging from menorrhagia and pelvic pain to infertility, recurrent miscarriage, and preterm labor (2, 3). The annual US health care costs associated with UFs have been estimated at $5.9 to $34.9 billion (4), rendering UFs a significant public health and financial burden.
Current treatment options for UFs rely on medical or surgical intervention strategies designed, respectively, to manage clinical symptoms in the short-term or provide long-term remedial benefit (5, 6). Recent efforts to improve the clinical utility of medical therapies, including strategies to enhance efficacy and improve tolerability, such as the use of gonadotropin-releasing hormone receptor antagonists to reduce heavy menstrual bleeding and pain associated with UFs, have led to several promising results (7, 8). However, the use of these and other agents that target the hypothalamic-pituitary-gonadal axis is constrained by the negative collateral impact of induced hypoestrogenicity. Accordingly, no long-term, noninvasive treatment option currently exists for UFs, and greater insight regarding tumor etiology will be key to the development of more effective medical therapies.
The prevailing model of UF pathogenesis involves the genetic transformation of a single MM stem cell (SC) into a tumor-initiating stem cell (UF SC) that seeds and sustains monoclonal tumor growth characterized by an increase in cell size and number, as well as abundant extracellular matrix production, under the influence of endocrine, autocrine, and paracrine growth factor and hormone receptor signaling (2, 9, 10). Most of the genetic drivers thought to be dominantly responsible for cell transformation have been identified. Among these, mutations in genes encoding the RNA polymerase II (Pol II) transcriptional Mediator subunit MED12 are by far the most prevalent, accounting for approximately 70% of UFs (11). A proportionally smaller fraction of tumors are thought to arise from genetic alterations leading to overexpression of HMGA2 (approximately 20%), disruption of the COL4A5-COL4A6 locus (approximately 3%), or biallelic loss of fumarate hydratase (approximately 2%) (12–14).
All UF-linked mutations in MED12 reside exclusively in exons 1 or 2 and feature predominantly missense alterations and less frequently small in-frame deletions or insertions. Among the former, the overwhelming majority are missense mutations that lead to substitutions at three highly conserved MED12 amino acids: Leu-36, Gln-43, and Gly-44 (11, 15). Importantly, molecular genetic analyses in human tumors and gene-targeting studies in mice strongly support the notion that UF-linked MED12 mutations are drivers of tumorigenesis (14, 16, 17).
Mediator is a multiprotein signal processor through which regulatory information conveyed by gene-specific transcription factors is transduced to RNA Pol II (16). MED12, along with MED13, CCNC, and CDK8 (or its close paralog CDK19), comprise a four-subunit ‘‘kinase’’ module that variably associates with a 26-subunit Mediator core (16, 18). Notably, the kinase module is a major ingress of signal transduction through Mediator, and CDK8/19 kinase activity has been shown to be required for nuclear transduction of signals instigated by multiple oncogenic pathways with which MED12 is biochemically and genetically linked, suggesting a close functional relationship between MED12 and CCNC-CDK8 (16, 19). Confirming this prediction, we and others have previously reported that MED12 activates CCNC-dependent CDK8/19 in Mediator (20–22). Mechanistically, we showed that this occurs via direct stabilization of the CDK8 activation (T)-loop by MED12 residues recurrently mutated in UFs, suggesting that the UF driver mutations alter T-loop conformation and disrupt CCNC-CDK8 kinase activity (18). We confirmed this and showed that pathogenic mutations in MED12 disrupt CDK8/19 kinase activity both in vitro and, more importantly, in clinically relevant patient fibroids (18, 21–23). Collectively, these findings identify a common molecular defect associated with UF-linked mutations in MED12 and implicate aberrant Mediator-associated CDK8/19 kinase activity in the pathogenesis of UFs. Nonetheless, the identity of biologically relevant Mediator kinase substrates and their pathogenic role in UF development have not been established.
In this study, we used quantitative phosphoproteomics coupled with a highly specific chemical inhibitor of CDK8/19 to identify Mediator kinase substrates in MM stem/progenitor cells, the presumptive cells of origin for UFs. We identified a catalog of high-confidence Mediator kinase targets linked to RNA metabolism, transcription, chromatin modification, and DNA replication and repair, including proteins previously implicated in UFs as well as those with no previously established disease associations. Orthogonal validation revealed a subset of these high-confidence targets to be direct substrates of Mediator kinase in vitro in a manner abrogated by the pre-dominant UF driver mutation in MED12. Transcriptome-wide expression profiling revealed Mediator kinase-dependent regulation of cell cycle and myogenic gene expression programs to which validated Mediator kinase substrates could be linked directly. Among these, CUX1 is an established transcriptional regulator of the cell cycle whose corresponding gene on chromosome 7q is the locus for a recurrent breakpoint in UFs (24–28). FOXK1, with established links to myogenic stem cell fate (29–32), is shown here to be coordinately enriched along with kinase, but not core, Mediator subunits in MM stem/progenitor cells compared with differentiated uterine smooth muscle cells. Together, these studies forge new links between Mediator kinase, which is known to be disrupted by UF driver mutations in MED12, and downstream targets in MM stem/progenitor cells and further impute a biochemical basis to link Mediator kinase activity with CUX1 and FOXK1 in the regulation of myometrial stem cell fate.
MATERIALS AND METHODS
Myometrial Side Population Isolation
Myometrial samples were obtained from two women undergoing surgery for symptomatic UFs at the University of Texas Health Science Center San Antonio after providing informed consent. Patient 1 was a 43-year-old Hispanic woman who received norethindrone before surgery. Patient 2 was a 45-year-old Hispanic woman who received medroxyprogester-one acetate (Depo-Provera) 2 months before surgery. The University of Texas Health Science Center San Antonio Institutional Review Board approved the protocol for recovery of surgical specimens. Protocols for tissue dissociation and isolation of the MM side population (SP) cell fraction have been described (33) and are detailed in the supplemental materials and methods, available online.
Cell Culture
Myometrial SP cells and myometrial non-SP cells were cultured in Mesenchymal Basal Stem Cell Medium (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS; Gibco) or DMEM:F12 medium (Thermo Fisher Scientific) with 10% FBS (Gibco), respectively, under hypoxic (2% O2, 5% CO2) or normoxic (20% O2, 5% CO2) conditions as specified. All cell cultures were supplemented with 0.1% antibiotic/antiMYCotic (Thermo Fisher Scientific).
Stable Isotope Labeling and Inhibitor Treatment
Myometrial SP cells were cultured in SILAC medium (Thermo Fisher Scientific) supplemented with 10% dialyzed FBS (Cambridge Isotopes), 0.1% antibiotic/antiMYCotic (Thermo Fisher Scientific), and either Arg8/Lys10 (heavy) or Arg0/Lys0 (light) isotopically labeled amino acids. The cells were metabolically labeled for five passages in either heavy or light medium before 45 minutes of treatment with 100 nM CCT251545 or dimethyl sulfoxide (DMSO) and subsequent cell extraction and sample preparation for phosphoproteomics analysis. Three replicate samples were prepared; samples 1 and 2 had light cells treated with vehicle and heavy cells treated with inhibitor, and sample 3 had heavy cells treated with vehicle and light cells treated with inhibitor.
Quantitative Phosphoproteomics and Data Analysis
Protocols were performed as described in detail (Supplemental materials, available online) using a Nanoflow UPLC:Ultimate 3000 nano UHPLC system (Thermo Fisher Scientific) and a Q Exactive HF Orbitrap LC-MS/MS system (Thermo Fisher Scientific). Raw MS files were analyzed and searched against a human protein database using Maxquant (1.6.2.6).
Antibodies, Plasmids, and Protein Expression and Purification
Detailed descriptions of antibodies as well as plasmids and procedures used to generate and purify recombinant Mediator kinase subunits and substrates are provided in the supplemental materials. Human Flag-CDK8 wild-type (WT) and kinase-dead (D173A), HIS-CYCLIN C, CBP-MED13, HA-MED12 WT, and G44D proteins were expressed in High Five insect cells using the baculovirus expression system, as previously described (22, 34).
In Vitro Kinase Assay
Assays were performed as described previously (21, 22), with additional experimental details outlined in the supplemental materials.
RNA Sequencing, Validation, and Computational Analyses
RNA from MM SP cells treated without (DMSO) or with 100 nM CCT251545 for 3 hours (three independent replicate experiments) was processed for library construction and sequencing, as well as validation by reverse transcriptase quantitative polymerase chain reaction (RT-qPCR), as described in detail in the supplemental materials. Data normalization and differential expression were performed using R package DESeq2, version 1.32. Gene set enrichment, gene ontology, and transcription factor-binding site analyses were performed as described in the supplemental materials.
RESULTS
Identification of High-Confidence Mediator Kinase Substrates in MM Stem/Progenitor Cells Using SILAC-Based Quantitative Phosphoproteomics
To identify Mediator kinase substrates in MM stem/progenitor cells, dissociated single-cell suspensions from MM of women undergoing hysterectomy for symptomatic UFs were processed for stem/progenitor cell isolation using the ‘‘side population (SP)’’ method (35). This flow cytometric-based approach has been used successfully to identify and sort somatic stem cells from multiple tissue types, including MM, based on their enhanced ability to efflux intracellular Hoechst dye via the ATP-binding cassette (ABC) family of transporter proteins (36). Previously, SP cells isolated from human MM using this approach were shown to be undifferentiated, multipotent, and mesenchymal-derived, as expected for smooth muscle stem cells/progenitors (33, 36, 37). Indeed, we confirmed that the MM SP cells isolated in this study were mesenchymal-derived (CD90+) and undifferentiated (expressing low levels of nuclear hormone receptors and smooth muscle markers) (Supplemental Fig. 1, available online). Hereafter, we refer to these cells as MM SP (myometrium side population or stem/progenitor-enriched) cells.
We used SILAC (Stable Isotope Labeling of Amino Acids in Cell culture) coupled with downstream phosphoproteomic analysis to identify Mediator kinase substrates in MM SPs (Fig. S2A). To this end, patient-derived MM SPs were cultured in the presence of heavy (Lys8/Arg10) or light (Lys0/Arg0) isotopically labeled amino acids before treatment without (DMSO) or with the highly specific and professionally credentialed CDK8/19 chemical inhibitor CCT251545 (38, 39). Mechanistically, this type I binding inhibitor was shown to occupy the CDK8 ATP-binding site while concomitantly promoting reinsertion of the CDK8 C-terminus into the ligand-binding site; this unique binding mode is presumed to underlie the high selectivity of CCT251545 for CDK8/19 (>100-fold selectivity over 291 other profiled kinases) (39, 40). The labeling experiments were performed in triplicate and included a label swap in which the inhibitor treatment protocol in replicates 1 and 2 (light and heavy isotopically labeled cells treated with vehicle and inhibitor, respectively) was reversed in replicate 3. To increase the probability of identifying direct Mediator kinase targets, we used short-term (45-minute) treatment with CDK8/19 inhibitor, which we validated as sufficient to confer ≥90% suppression of Mediator kinase activity, as measured by the level of γ-interferon-stimulated STAT1 S727 phosphorylation (Supplemental Fig. 2B), a validated biomarker of cellular Mediator kinase function (39, 41). Cell lysates from control (DMSO) and inhibitor-treated cells were mixed 1:1 (based on total protein concentration) before phosphopeptide enrichment using iron immobilized metal affinity chromatography and subsequent analysis by LC-MS/MS. Notably, a portion of the unenriched protein lysate was also processed by LC-MS/MS to permit normalization of samples for total protein levels, thus ensuring that inhibitor-dependent changes in measured phosphorylation levels were not attributable to alterations in protein abundance.
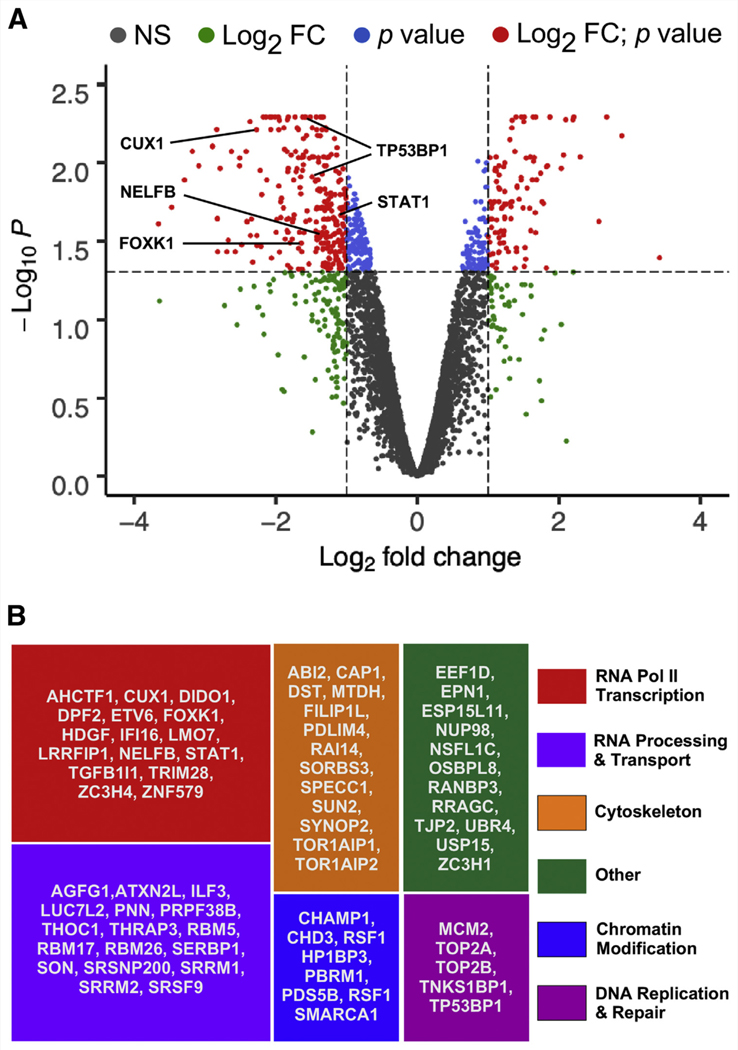
In total, we identified 6,679, 7,273, and 7,939 phosphosites corresponding to 2,570, 2,707, and 2,832 different proteins, respectively, that were measurably altered in samples 1, 2, and 3 upon CDK8/19 inhibition (Supplemental Table 1 and Supplemental Fig. 3). To identify ‘‘high-confidence’’ Mediator kinase-dependent phosphosites, we filtered these data using the following criteria for quantified inhibitor-dependent changes in the normalized phosphorylation levels of each unique phosphosite: a quantified change must occur in at least two of three replicate samples; a quantified change must exceed a minimal threshold of ≥ twofold; and a quantified change must meet a minimal significance threshold of P<.05 as determined by an empirical Bayes statistical approach (41, 42). Application of these criteria produced a circumscribed list of 420 Mediator kinase-dependent phosphosites corresponding to 237 different proteins. Among these, the vast majority (approximately 70%, or 296 phosphosites in 166 proteins) were decreased, as expected from short-term inhibitor treatment designed to identify direct Mediator kinase targets (Fig. 1A and Supplemental Table 2). Nonetheless, 124 phosphosites in 71 proteins were increased upon CDK8/19 inhibition (Fig. 1A and Supplemental Table 2), revealing indirect, albeit Mediator kinase-dependent, targets indicative of rapid changes in phosphorylation-dependent signaling following Mediator kinase inhibition. Of the 166 proteins showing decreased phosphorylation levels upon Mediator kinase inhibition, 71 are reported to be nuclear in their cellular localization. We consider these 71 proteins, encompassing 118 phosphosites (Supplemental Table 3), to be ‘‘high-confidence’’ targets of direct phosphorylation by Mediator kinase that is itself restricted to the interphase nucleus. The remaining 95 cytosolic proteins likely are indirect targets of Mediator kinase and/or direct targets capable of transient localization to the interphase nucleus or targets during mitosis, in which nuclear membrane dissolution could remove a barrier to their otherwise restricted sequestration from Mediator. Notably, the cytosolic phosphoproteins identified as Mediator kinase-dependent in this study showed significant enrichment for microtubule-binding, actin cytoskeletal organization, and cadherin-mediated cell-cell adhesion (Supplemental Fig. 4A), suggesting a possible and heretofore unappreciated role for Mediator kinase in the regulation of contractile force production. Regarding Mediator kinase-dependent nuclear phosphoproteins deemed high-confidence substrates, functional categorization and gene ontology analysis revealed these 71 gene products to be significantly enriched for biologic functions related to RNA polymerase II transcription, RNA processing and transport, cytoskeletal architecture, chromatin regulation and modification, and DNA replication and repair (Figs. 1B and Supplemental Fig. 4B), suggesting roles for Mediator kinase beyond its well-established function in transcription.
FIGURE 1.
Identification of high-confidence Mediator kinase substrates in myometrial (MM) side population (SP) cells. (A) Volcano plot of statistically significant phosphosite changes (empirical Bayes analysis) in MM SP cells after 45 minutes of treatment with CCT251545. Mediator kinase-dependent phosphosites considered ‘‘high-confidence’’ are those undergoing an inhibitor-dependent change in abundance of ≥ twofold with P<.05 (red dots). Putative direct Mediator kinase phosphosites (upper left quadrant) are those reduced in abundance upon Mediator kinase inhibition. Phosphopeptides in validated Mediator kinase substrates are indicated. NS = not significant. (B) Functional categorization of high-confidence Mediator kinase substrates.
Among the 71 high-confidence Mediator kinase substrates, significantly enriched phosphosite motifs were identified using iceLOGO, revealing a consensus Ser/Pro motif and a preference for Pro and Asp residues at the –2 and –1 sites relative to the prime Ser phosphorylation site (Supplemental Fig. 4C). This analysis confirms an apparent CDK8/19 site-specific preference for PX(S/T) motifs within substrates identified in previous studies (41, 43, 44). Furthermore, as reported previously for CDK8 (41), we observed no overrepresentation of basic residues positioned C-terminal to the phosphosite, distinguishing the CDK8/19 consensus motif from that of other cyclin-dependent kinases (45).
MED12-Dependent CDK8/19 Substrate Phosphorylation Is Compromised by UF Driver Mutation G44D
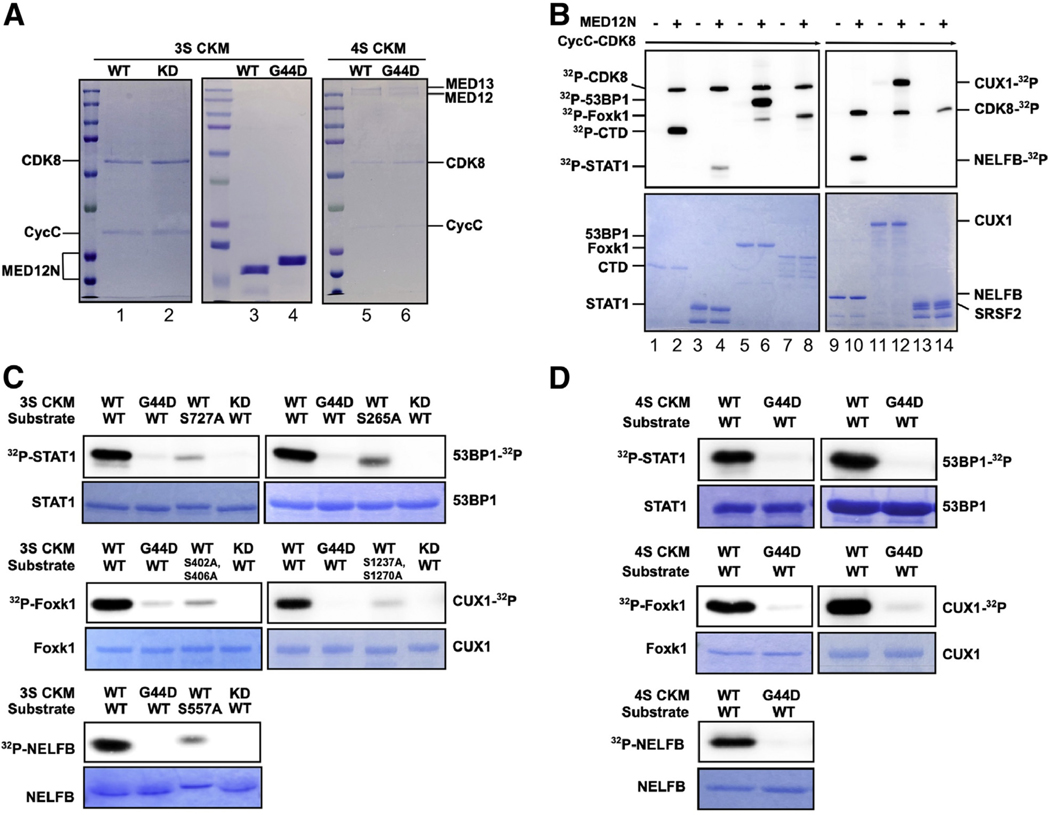
To validate CDK8/19-dependent phosphoproteins as direct substrates of Mediator kinase, we selected a subset of six mapped phosphosites in five different proteins for further analysis in vitro. Among these, two proteins and their corresponding phosphosites, STAT1 S727 and TP53BP1 S265, were previously identified as Mediator kinase targets in other cellular contexts (39, 41), substantiating our phosphoproteomics approach in MM SP cells and also providing positive controls for site-specific Mediator kinase activity in our validation assays. An additional three proteins carrying four phosphosites (CUX1 S1237, FOXK1 S416 and S420 [mouse equivalents S402 and S406], and NELFB S557), none of which have previously been linked with Mediator kinase, were chosen for further analysis based on their established associations with UFs (CUX1 [25, 46]), muscle stem cell biology (FOXK1 [29, 32]), or RNA Pol II transcriptional elongation (NELFB [47, 48]) with which Mediator kinase itself has been implicated (16, 19). For each of the five candidate substrates tested, purified recombinant WT or phosphomutant (Ser to Ala) derivatives were assessed for their abilities to serve as direct Mediator kinase substrates in vitro. We also tested a recombinant protein fragment corresponding to three tandem copies of the heptapeptide repeat sequence present in the RNA Pol II C-terminal domain (CTD), a reported Mediator kinase substrate, providing an additional positive control for Mediator kinase activity in our system (21–23). Finally, we included a negative control protein, SRSF2, which did not meet the criteria for consideration as a statistically significant high-confidence Mediator kinase substrate by SILAC analysis. We used two different sources of Mediator kinase for substrate phosphorylation analyses. First, a three-subunit partial Mediator kinase module was reconstituted from baculovirus-expressed and purified human CCNC-CDK8 and an Escherichia coli-expressed recombinant MED12 protein fragment spanning amino acids 1 to 100 (Fig. 2A). Previously, we showed that MED12 (1–100) is sufficient to bind and activate CCNC-CDK8 through CDK T-loop stabilization (18, 22). Second, we used the intact four-subunit kinase module reconstituted from purified baculovirus coexpressed human MED13, MED12, CCNC, and CDK8 subunits (Fig. 2A).
FIGURE 2.
Validation of select Mediator kinase substrates. (A) Purified three-subunit and four-subunit CDK8 kinase modules (3S CKM and 4S CKM, respectively) analyzed by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. For 3S CKM, CDK8 wild-type (WT) and kinase-dead (KD) (lanes 1 and 2), as well as MED12 (1–100) WT and G44D mutant derivatives (lanes 3 and 4), are indicated. For 4S CKM, MED12 WT and G44D mutant derivatives (lanes 5 and 6) are indicated. (B) In vitro kinase assays were performed using CCNC-CDK8 WT without (–) or with (+) MED12 (1–100) WT along with each of the indicated substrates (RNA Pol II [polymerase II] C-terminal domain [CTD] [lanes 1 and 2], STAT1 [lanes 3 and 4], TP53BP1 [lanes 5 and 6], Foxk1 [lanes 7 and 8], NELFB [lanes 9 and 10], CUX1 [lanes 11 and 12], or SRSF2 [lanes 13 and 14]). Kinase reactions were resolved by SDS-PAGE and subjected to Phosphoimager analysis to detect phosphorylated (32P) substrates (top panels) or Coomassie blue staining to visualize input substrates (bottom panels). 32P-CDK8 indicates autophosphorylation of CDK8, which is stimulated by MED12 (1–100). RNA Pol II CTD and SRSF2 are positive and negative controls for Mediator kinase activity, respectively. (C and D) In vitro kinase assays were performed using 3S CKM (C) or 4S CKM (D) and either WT or phosphomutant substrates bearing Ser to Ala substitution mutations at Mediator kinase-dependent phosphosites mapped by SILAC analysis. Where indicated, kinase reactions were performed using 3S CKM or 4S CKM reconstituted with CDK8 WT or CDK8 KD as well as MED12 WT or G44D mutant derivatives. Kinase reactions were resolved by SDS-PAGE and analyzed as described in (B).
Consistent with our previous observations (18, 22), CCNC-CDK8 alone exerted minimal kinase activity toward the RNA Pol II CTD, which was otherwise greatly stimulated by the addition of MED12 (1–100), confirming MED12 as a potent activator of CCNC-CDK8 (Fig. 2B, lanes 1 and 2). Notably, MED12 (1–100) also greatly stimulated CCNC-CDK8 kinase activity toward all five test substrates, confirming each to be a direct Mediator kinase substrate in vitro (Fig. 2B, lanes 3–12). As expected on the basis of the results of our phosphoproteomics profiling, SRSF2 was not detectably phosphorylated by CCNC-CDK8, either alone or in complex with MED12 (1–100) (Fig. 2B, lanes 13 and 14), thus revealing substrate selectivity to be an important feature recapitulated in these in vitro Mediator kinase assays. Notably, the ability of MED12 to stimulate CCNC-CDK8-dependent phosphorylation of all five test substrates in these assays was effectively abrogated by a debilitating substitution mutation (D173A) in the CDK8 active site (22), confirming that the stimulatory impact of MED12 in this setting is mediated exclusively through CDK8, as opposed to an unrelated kinase possibly present at undetectable levels in our purified reconstituted system (Fig. 2C). Importantly, phosphorylation of all five test substrates by Mediator kinase in vitro was significantly diminished by Ala substitution mutations at each of their corresponding Mediator kinase-dependent phosphosite Ser residues mapped in MM SP cells (Fig. 2C). Together, these analyses confirm direct and site-specific substrate phosphorylation by Mediator kinase in vitro and thus validate our phosphoproteomics analysis in MM SP cells.
Previously, we showed that UF driver mutations in MED12 disrupt Mediator kinase activity directed toward the RNA Pol II CTD (18, 21–23). To determine if this pathogenic impairment extends to biologically relevant substrates identified in this study, we compared WT MED12 (1–100) and its G44D mutant derivative, corresponding to the most prevalent MED12 UF driver mutation (11, 16), for their abilities to stimulate CCNC-CDK8-dependent phosphorylation of the five substrates validated above. Compared with WT MED12, mutant G44D was significantly impaired in its ability to stimulate CCNC-CDK8-dependent phosphorylation of all five substrates (Fig. 2C), confirming the deleterious impact of this dominant UF driver mutation on Mediator kinase function across a range of its natural substrates.
To confirm these observations in a more biologically relevant context, we purified intact Mediator kinase modules from insect cells coexpressing all four kinase module subunits (MED13, CCNC, CDK8, and either WT or G44D mutant MED12) and then compared WT and mutant MED12-containing kinase modules for their abilities to directly phosphorylate all five test substrates in vitro. Comparative analyses of WT and G44D mutant MED12-containing Mediator kinase modules revealed the latter to be significantly impaired in its ability to phosphorylate all five substrates tested (Fig. 2D). Together, these results newly identify STAT1, TP53BP1, CUX1, FOXK1, and NELFB to be direct substrates of Mediator kinase in a manner largely abrogated by pathogenic MED12 mutant G44D, thus revealing Mediator kinase disruption to be a general defining feature of MED12 UF driver mutations, with a potential broad impact on the Mediator kinase-dependent phosphoproteome.
Mediator Kinase Substrates Are Functionally Linked with Mediator Kinase-Regulated Genes in MM SP Cells
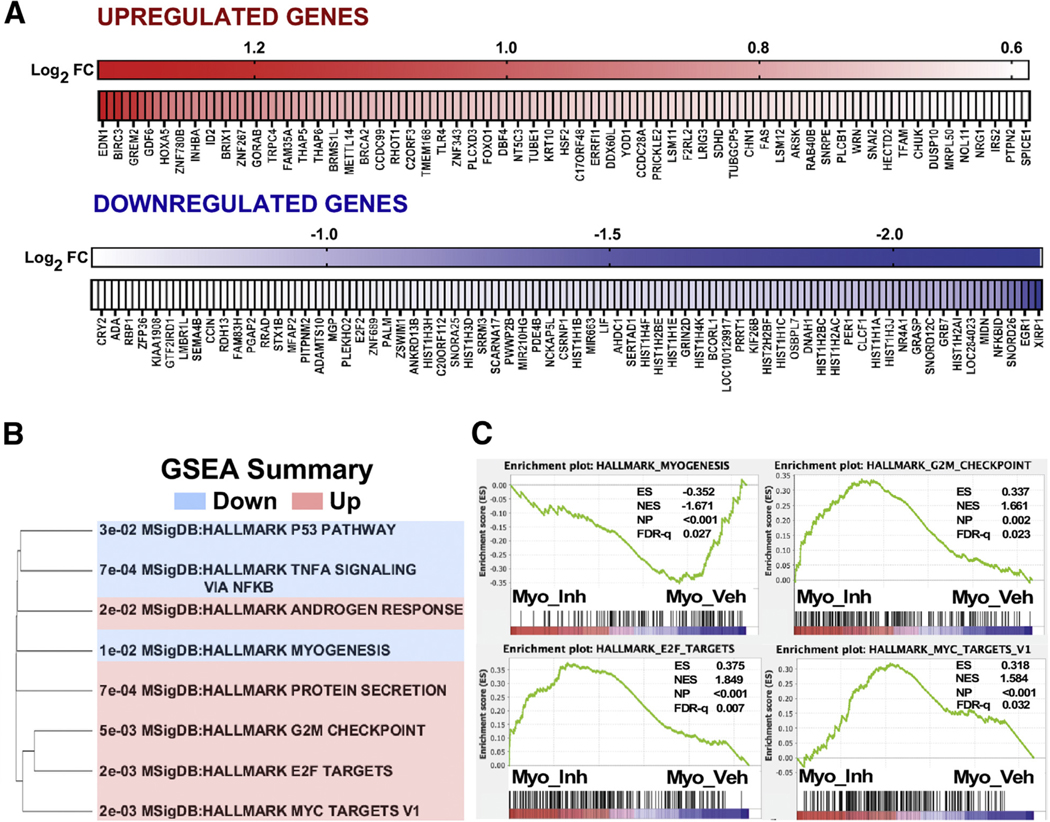
To examine the impact of Mediator kinase inhibition on global gene expression, we performed RNA sequencing in MM SP cells 3 hours after treatment with DMSO or CCT251545, which permitted us to capture primarily direct, as opposed to indirect, gene expression changes arising from short-term Mediator kinase inhibition. Using relatively standard criteria (minimum 1.5-fold change; P<.05), we identified 401 genes (123 up and 278 down) that were differentially expressed as a function of CCT254515 treatment (Fig. 3A, Supplemental Table 4, and Supplemental Fig. 5). Gene set enrichment analysis linked Mediator kinase activity with Hallmark myogenesis, E2F, MYC, and cell cycle checkpoint pathways, among others, suggesting a possible role for Mediator kinase in the regulation of MM SP cell growth and differentiation (Fig. 3B and C).
FIGURE 3.
Mediator kinase regulates myogenic and cell growth-related gene expression programs in myometrial side population (MM SP) cells. (A) Heat maps of 401 genes differentially expressed (FC≥1.5-fold; P<.05) in MM SP cells upon 3 hours of CCT251545 treatment, including both up-regulated (n = 123) and down-regulated (n=278) genes. Only a subset of genes are labeled. (B and C) Hallmark gene sets enriched in Mediator kinase-inhibited MM SP cells. GSEA =XX.
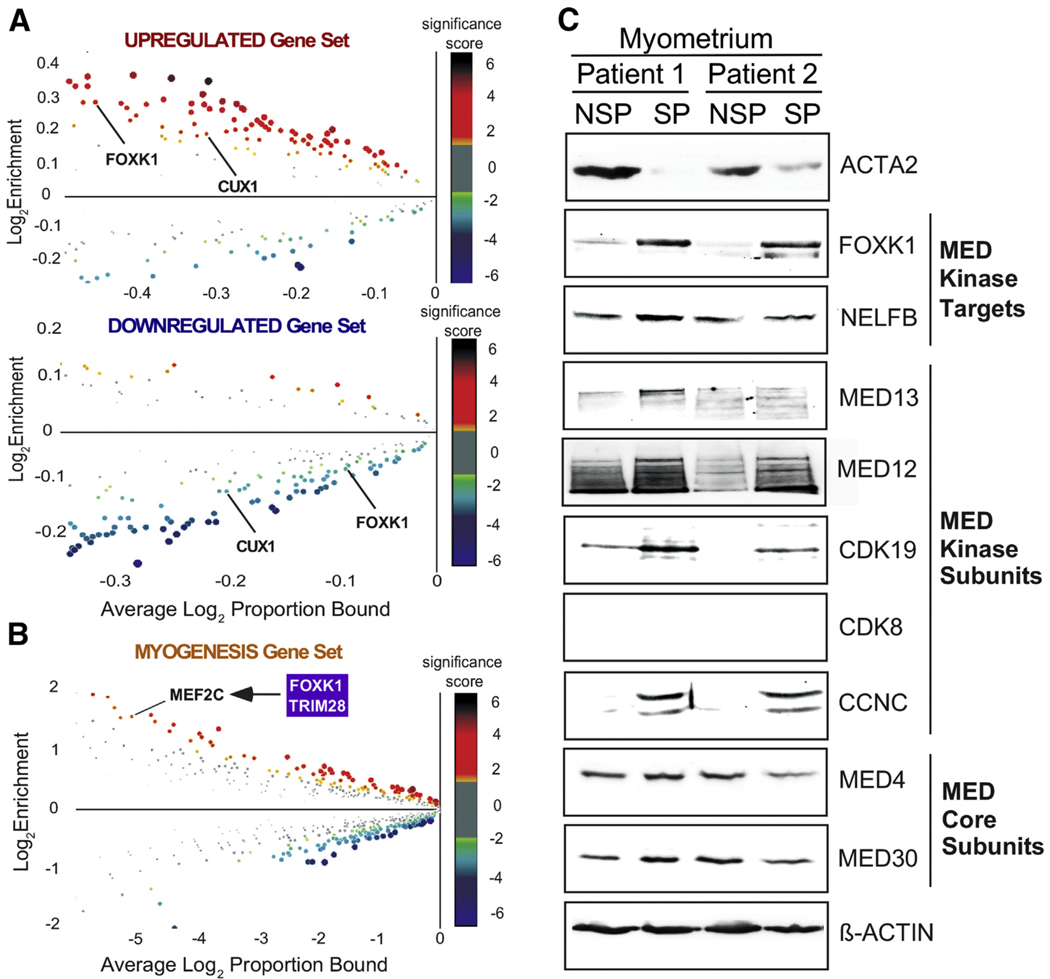
Because a number of Mediator kinase substrates identified in this study are transcription factors (Fig. 1B), we explored possible functional connections between CDK8/19 inhibitor-dependent alterations in gene expression and transcription factor activity. To this end, we searched promoter sequences (±2 Kb from the transcription start site) of the 401 genes differentially expressed as a function of CCT251545 treatment for enriched (or depleted) transcription factor-binding sites using CiiiDER (49). This analysis yielded several intriguing links with Mediator kinase substrates. For example, the binding sites for Mediator kinase substrates FOXK1 and CUX1 were found to be enriched or depleted within the promoters of genes that were up- or down-regulated, respectively, by Mediator kinase inhibition (Fig. 4A), implicating FOXK1 and CUX1 as downstream transcriptional effectors of Mediator kinase in MM SP cells. Furthermore, FOXK1, through a mechanism independent of its own DNA-binding activity, was previously shown to interact with and regulate the function of the myogenic transcription factor MEF2C (32), whose cognate binding site was enriched in Mediator kinase-regulated Hallmark myogenesis genes (Fig. 4B). In addition, TRIM28/KAP1, identified in this study as a Mediator kinase substrate, is an established transcriptional cofactor for MEF2, and, notably, TRIM28 S473, identified in this study as a Mediator kinase-dependent phosphosite (Supplemental Table 3), was previously shown to function as a phosphorylation switch for muscle stem cell differentiation (50). We also Observed enrichment of transcription factor-binding sites for genes with altered expression in CCT251545-treated MM SCs, including EGR1, FOXO1, and E2F2 (Supplemental Fig. 6). Notably, Mediator kinase substrates STAT1, YBX1, Q27 and ILF3 are established co/regulators of EGR1 (51–53), whereas Mediator kinase substrates STAT1, ETV6, and TRIM28 are known to regulate FOXO1 (54–56). Finally, Mediator kinase substrate CUX1, implicated in UF pathogenesis, is an established regulator of E2F2 in regulation of cell cycle gene transcription (27). Together, these findings reveal multiple functional connections between Mediator kinase substrates and Mediator kinase-dependent gene expression programs in MM SP cells, indicative of transcriptional regulation through Mediator kinase activity.
FIGURE 4.
Mediator kinase-regulated genes are linked functionally with Mediator kinase substrates in myometrial side population (MM SP) cells. (A) Ciiider enrichment plots for 401 Mediator kinase-regulated genes, including up-regulated and down-regulated gene sets. Plots show the enrichment (proportional ratio of promoter regions bound by transcription factors [indicated by dots]) and average log proportion bound. Dot size and color reflect the log2 P value (significance score). Significance scores greater than or less than zero indicate overrepresentation or underrepresentation of transcription factor-binding sites, respectively, in promoter regions of queried genes. Note that binding sites for Mediator kinase substrates FOXK1 and CUX1 (indicated) are overrepresented and underrepresented in up-regulated and down-regulated gene sets, respectively. (B) Ciiider enrichment plot for Hallmark myogenesis gene set. Note that binding sites for MEF2, a Mediator kinase-regulated gene that physically and functionally interacts with Mediator kinase substrates FOXK1 and TRIM28, are enriched in the myogenic gene set. (C) Whole cell lysates of paired SP and non-SP fractions from two different patient-derived myometria were processed by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblot analysis using antibodies specific for Mediator kinase substrates (FOX1, NELFB), Mediator kinase module subunits (MED13, MED12, CDK8/CDK19, CCNC), Mediator core subunits (MED4, MED30), α-smooth muscle actin (ACTA2), and β-ACTIN (loading control). Paired SP and non-SP fractions were isolated from uteri bearing MED12 mutation-negative (patient 1) or MED12 mutation-positive (patient 2) uterine fibroids. Note that FOXK1, along with kinase, but not core, Mediator subunits is enriched in undifferentiated MM SP cells compared with differentiated non-SP (NSP) (ACTA2-expressing) cells. MED = Mediator.
FOXK1 Is Enriched Along with Mediator Kinase Components in MM SCs
Among the Mediator kinase substrates validated in this study, FOXK1 is of particular interest, given its established roles in myogenic stem cell fate and biologic processes known to be regulated by Mediator kinase. Murine Foxki 1 has been shown to be required for activation and proliferation of myogenic progenitors while repressing myogenic differentiation (29, 30, 32). Furthermore, FOXK1 is known to regulate Wnt/β-catenin signaling and aerobic glycolysis, two cellular programs also controlled by Mediator kinase (57–62). These observations, coupled with our identification of FOXK1 as a direct target of Mediator kinase, suggest a possible basis to link Mediator kinase activity directly with FOXK1 in MM stem/progenitor cell biology. To begin to investigate this possibility, we assessed the expression profile of FOXK1 and individual Mediator kinase subunits in SP (stem/progenitor-enriched) and non-SP (differentiated) cells derived from patient MM. This analysis revealed coordinate enrichment of FOXK1 and Mediator kinase module, but not core Mediator, subunits in patient-derived MM SP cells compared with differentiated non-SP cells (Fig. 4C). Because SP and non-SP cells are typically cultured under hypoxic and normoxic conditions, respectively, we repeated these analyses using cells cultured under identical conditions of hypoxia. We observed similar results (Supplemental Fig. 7), indicating that enriched expression of FOX1 and Mediator kinase in SP vs. non-SP cells likely reflects the distinct differentiation status of the cells as opposed to a differential hypoxic response. This suggests an important function for Mediator kinase and FOXK1 in MM stem/progenitor cell biology.
DISCUSSION
Our previous discovery that UF driver mutations in MED12 alter CDK8 T-loop conformation and compromise CCNC-CDK8 kinase activity suggests that disruption of Mediator kinase function is a contributing factor in UF pathogenesis. In this study, we sought to identify biologically relevant Mediator kinase substrates in MM stem/progenitor cells to further explore this hypothesis. Using SILAC in conjunction with CDK8/19 chemical inhibition and quantitative phosphoproteomics, we identified the minimal Mediator kinase-dependent phosphoproteome in MM stem/progenitor cells, including both direct and indirect downstream targets. Application of stringent criteria regarding the magnitude and significance of inhibitor-dependent changes in protein phosphorylation levels across multiple experiments revealed 118 phosphosites in 71 nuclear proteins that we consider high-confidence Mediator kinase substrates. Among these, we validated five as direct targets of Mediator kinase in vitro in a manner abrogated by the most common UF driver mutation in MED12, implicating these substrates in disease pathogenesis. Further, transcriptome-wide expression profiling revealed Mediator kinase activity to be prominently linked with cell cycle and myogenic gene expression programs to which Mediator kinase targets, most notably CUX1 and FOXK1, could be linked directly.
CUX1 is a homeodomain transcription factor whose corresponding gene is commonly deleted in a UF subset characterized by recurrent structural aberrations affecting chromosome 7q22 (25, 46, 63). Notably, loss of heterozygosity at 7q22 has been observed in UFs as well as breast and ovarian cancers (46, 64, 65). Furthermore, CUX1 has been identified as a haploinsufficient tumor suppressor in acute myeloid leukemia, and inactivating CUX1 mutations are found at low frequencies in multiple tumor types (66, 67). Together, these observations suggest that CUX1 may function as a tumor suppressor. Biologically, CUX1 is implicated in a broad range of physiological processes, including regulation of cell proliferation and death, cell differentiation, and tissue development (68). Notably, previous studies have revealed that CUX1 is highly expressed in undifferentiated cells and appears to play an important role in the regulation of stem cell fate. For example, CUX1 is highly expressed in long-term hematopoietic stem cells compared with short-term hematopoietic stem cells and myeloid progenitors, and CUX1 depletion in vivo triggered expansion of myeloid and short-term hematopoietic stem cells and myeloid progenitors (69). Thus, CUX1 appears to control hematopoietic stem cell line-age specification and differentiation by maintaining quiescence and suppressing proliferation and self-renewal of hematopoietic stem cells. At the molecular level, CUX1 is known to regulate cell cycle gene transcription through both E2F-dependent and E2F-independent mechanisms, thereby ensuring proper temporal controls on key cell cycle phase transitions. For example, as the DNA-binding component of the multiprotein HINF-D transcription complex, CUX1, through an E2F-independent mechanism, represses histone gene transcription in midlate S phase, thus restricting the timing of histone gene expression to early S phase, and also activates mitotic regulators in late G1/S phase before their requirement for cytokinesis in M phase (26, 28). On the other hand, CUX1 has also been shown to physically interact and functionally cooperate with E2F in transcriptional activation of non-histone cell-cycle-regulated genes (27). It is therefore notable that among Mediator kinase-regulated genes identified in this study, replication-dependent histone genes and E2F-dependent genes were prominently enriched, and CUX1-binding sites were found to be significantly overrepresented in their respective promoter regions. Thus, Mediator kinase could represent an important regulator of CUX1-dependent cell-cycle gene expression in MM SP cells.
FOXK1, a member of the Forkhead Box K transcription factor family, is implicated in a variety of biologic processes relevant to UF pathogenesis. For example, FOXK1 promotes muscle progenitor cell proliferation and represses myogenic differentiation (31, 32), activates the Wnt/β-catenin pathway to promote cell growth (62), regulates metabolism to favor aerobic glycolysis (61), and interacts with TP53BP1 (identified in this study as a Mediator kinase substrate) to dictate the choice of DNA repair pathway following DNA double-strand breaks (70). The role of FOXK1 in the regulation of both Wnt/β-catenin signaling and myogenic cell fate is particularly notable. For example, the Wnt/β-catenin pathway has been shown to be crucial for the renewal of myometrial stem cells and their differentiation into smooth muscle, and further, targeted overexpression of constitutively active β-catenin in the uterine mesenchyme of mice promotes uterine fibroid tumors (2, 9, 71). Moreover, Wnt/β-catenin-stimulated TGFβ3 induces cell proliferation and extracellular matrix deposition, hallmark features of UFs (9, 71). It is therefore notable that we and others have previously implicated Mediator kinase in the regulation of canonical Wnt/β-catenin signaling (16, 57, 60). Regarding FOXK1 and myogenesis, genetic studies revealed an important role for FOXK1 in the regulation of myogenic progenitor cell fate. Thus, Foxk1-deficient mice were shown to exhibit severely impaired skeletal muscle regeneration accompanied by reduced muscle stem cell numbers, impaired stem cell activation, and perturbed stem cell cycle kinetics (29). Cell biologic and biochemical analyses further revealed that Foxk1 promotes muscle stem cell proliferation and inhibits myogenic differentiation by repressing the transcriptional activities of Foxo4 and Mef2, respectively (32). Together, these studies unequivocally implicate FOXK1 in the regulation of muscle stem cell fate and skeletal muscle regeneration. Although the role of FOXK1 in the control of myometrial stem cell fate is unknown, we nonetheless found FOXK1 to be coordinately enriched along with kinase, but not core, Mediator subunits in myometrial stem/progenitor cells relative to differentiated smooth muscle cells, suggesting a comparatively important functional role for FOXK1 in the stem/progenitor cell pool. Furthermore, in MM SP cells, the presence of FOXK1-binding sites in Mediator kinase-regulated genes was significantly correlated with their higher expression, while binding sites for transcription factor MEF2, with which FOXK1 functionally interacts, were enriched in the promoters of Mediator kinase-dependent Hallmark myogenic genes. Accordingly, FOXK1 could function as a key downstream effector of Mediator kinase in the regulation of myometrial stem cell fate, a possibility consistent with an emerging role for Mediator kinase in stem cell biology.
In this regard, recent work has implicated Mediator kinase activity directly in the regulation of stem cell plasticity and fate determination. For example, a developmentally programmed reduction in CDK8 expression is known to be associated with naive pluripotency during animal development in vivo, and chemical inhibition of CDK8/19 kinase activity has been shown sufficient to revert primed pluripotent stem cells to a naive pluripotent state in vitro (72, 73). Furthermore, CDK8, through a mechanism dependent upon c-MYC, has been shown to promote cancer stem cell self-renewal and tumorigenicity in colon and brain cancers (74, 75). These findings are germane to the pathogenesis of UFs, since the vast majority of these tumors are presumed to arise from genetic transformation of myometrial stem/progenitor cells through somatic mutations in MED12 that disrupt CDK8/19 kinase activity. We hypothesize that Mediator kinase disruption as a consequence of pathogenic MED12 mutations alters myometrial stem cell fate through deregulation of key substrates, including CUX1 and FOXK1, that control critical cell growth and myogenic gene expression programs. Further studies will be required to confirm this hypothesis and clarify the role of CUX1, FOXK1, and additional Mediator kinase substrates in the regulation of myometrial stem cell fate and tumor formation.
Finally, we note that many Mediator kinase substrates identified in this study participate in biologic processes other than transcription, including DNA replication and repair. This finding indicates a broader function for Mediator beyond its well-established role in transcription, and further identifies additional Mediator kinase-dependent pathways susceptible to dysregulation as a consequence of UF driver mutations in MED12. Notably, recent work has shown that MED12 mutation-positive UF stem cells accumulate high levels of unrepaired DNA double-strand breaks accompanied by reduced expression of DNA damage response and repair genes (76). Conceivably, altered phosphorylation dynamics of Mediator kinase substrates, including direct DNA repair effectors or transcriptional regulators of DNA damage response genes, could contribute to the enhanced DNA damage load observed in MED12 mutation-positive UF stem cells. Notably, we observed that Mediator kinase-inhibited MM SPs showed significant enrichment of Hallmark ‘‘G2/M checkpoint’’ and ‘‘p53 pathway’’ gene sets, consistent with elevated levels of DNA damage in these cells. Future studies will seek to clarify whether and how Mediator kinase activity contributes to the DNA damage response in myometrial stem cells and whether its disruption by MED12 mutations contributes to genomic instability and UF formation.
CONCLUSION
In summary, we report a new catalog of Mediator kinase substrates in myometrial stem/progenitor cells, including transcription factors CUX1 and FOXK1, that we identify as candidate downstream effectors of Mediator kinase in the control of cell growth and myogenic gene expression programs. Aberrant substrate phosphorylation and altered downstream signaling as a consequence of MED12 mutations that impair CDK8/19 kinase activity could perturb myometrial stem cell fate, contributing to fibrotic transformation and UF formation.
Supplementary Material
Acknowledgments:
We thank Drs. Rong Li (George Washington University), Alain Nepvue (McGill University), Kuniyoshi Iwabuchi (Kanazawa University), and Beatrice Eymin (Institute for Advanced Biosciences) for kind gifts of the plasmids for NELFB, CUX1, TP53BP1, and SRSF2, respectively.
L.B. has nothing to disclose. S.K. has nothing to disclose. R.S. has nothing to disclose. L.H. has nothing to disclose. J.B. is a former employee of The Institute of Cancer Research, which operates a Rewards to Inventors scheme and has a commercial interest in the development of inhibitors of the Wnt pathway, CDK8/19, and other cyclin-dependent kinases, with intellectual property licensed to Merck KGaA and Cyclacel Pharmaceuticals. J.B. holds equity in NeoPhore Ltd. S.F.C. has nothing to disclose. K.L.T. has nothing to disclose. T.G.B. reports receiving grants from the National Institutes of Health during the conduct of the study.
Supported by US Department of Health and Human Services, National Institutes of Health grants 1R01HD087417 and 1R01HD094378 (to T.G.B.) and Cancer Research UK grants C309/A11566, C368/A6743, and A368/A7990 (to J.B.). L.B. was supported by a Cancer Prevention Research Institute of Texas Research Training Award (RP170345) and an Institutional Research and Academic Career Development Award (IRACDA) K12GM111726.
REFERENCES
- 1.Doherty L, Mutlu L, Sinclair D, Taylor H. Uterine fibroids: clinical manifestations and contemporary management. Reprod Sci 2014;21:1067–92. [DOI] [PubMed] [Google Scholar]
- 2.Bulun SE. Uterine fibroids. N Engl J Med 2013;369:1344–55. [DOI] [PubMed] [Google Scholar]
- 3.Stewart EA, Laughlin–Tommaso SK, Catherino WH, Lalitkumar S, Gupta D, Vollenhoven B. Uterine fibroids. Nat Rev Dis Primers 2016;2:16043. [DOI] [PubMed] [Google Scholar]
- 4.Cardozo ER, Clark AD, Banks NK, Henne MB, Stegmann BJ, Segars JH. The estimated annual cost of uterine leiomyomata in the United States. Am J Obstet Gynecol 2012;206:211.e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartels CB, Cayton KC, Chuong FS, Holthouser K, Arian SE, Abraham T, et al. An evidence-based approach to the medical management of fibroids: a systematic review. Clin Obstet Gynecol 2016;59:30–52. [DOI] [PubMed] [Google Scholar]
- 6.Giuliani E, As-Sanie S, Marsh EE. Epidemiology and management of uterine fibroids. Int J Gynaecol Obstet 2020;149:3–9. [DOI] [PubMed] [Google Scholar]
- 7.Al-Hendy A, Lukes AS, Poindexter AN 3rd, Venturella R, Villarroel C, Critchley HOD, et al. Treatment of uterine fibroid symptoms with relugolix combination therapy. N Engl J Med 2021;384:630–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch SE, Mayer DC. Oriahnn: new drug approved for treating heavy menstrual bleeding in women with uterine fibroids. Ann Pharmacother 2021:1–9. [DOI] [PubMed]
- 9.Commandeur AE, Styer AK, Teixeira JM. Epidemiological and genetic clues for molecular mechanisms involved in uterine leiomyoma development and growth. Hum Reprod Update 2015;21:593–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkafas H, Qiwei Y, Al-Hendy A. Origin of uterine fibroids: conversion of myometrial stem cells to tumor-initiating cells. Semin Reprod Med 2017;35:481–6. [DOI] [PubMed] [Google Scholar]
- 11.Makinen N, Mehine M, Tolvanen J, Kaasinen E, Li Y, Lehtonen HJ, et al. MED12, the mediator complex subunit 12 gene, is mutated at high frequency in uterine leiomyomas. Science 2011;334:252–5. [DOI] [PubMed] [Google Scholar]
- 12.Markowski DN, Holzmann C, Bullerdiek J. Genetic alterations in uterine fibroids—a new direction for pharmacological intervention? Expert Opin Ther Targets 2015;19:1485–94. [DOI] [PubMed] [Google Scholar]
- 13.Mehine M, Kaasinen E, Heinonen HR, Makinen N, Kampjarvi K, Sarvilinna N, et al. Integrated data analysis reveals uterine leiomyoma subtypes with distinct driver pathways and biomarkers. Proc Natl Acad Sci U S A 2016;113:1315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehine M, Makinen N, Heinonen HR, Aaltonen LA, Vahteristo P. Genomics of uterine leiomyomas: insights from high-throughput sequencing. Fertil Steril 2014;102:621–9. [DOI] [PubMed] [Google Scholar]
- 15.Croce S, Chibon F. MED12 and uterine smooth muscle oncogenesis: state of the art and perspectives. Eur J Cancer 2015;51:1603–10. [DOI] [PubMed] [Google Scholar]
- 16.Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol 2015;50:393–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mittal P, Shin YH, Yatsenko SA, Castro CA, Surti U, Rajkovic A. Med12 gain-of-function mutation causes leiomyomas and genomic instability. J Clin Invest 2015;125:3280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li YC, Chao TC, Kim HJ, Cholko T, Chen SF, Li G, et al. Structure and non-canonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module. Sci Adv 2021;7:eabd4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fant CB, Taatjes DJ. Regulatory functions of the Mediator kinases CDK8 and CDK19. Transcription 2019;10:76–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol 2009;29:650–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park MJ, Shen H, Spaeth JM, Tolvanen JH, Failor C, Knudtson JF, et al. Oncogenic exon 2 mutations in Mediator subunit MED12 disrupt allosteric activation of cyclin C-CDK8/19. J Biol Chem 2018;293:4870–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turunen M, Spaeth JM, Keskitalo S, Park MJ, Kivioja T, Clark AD, et al. Uterine leiomyoma-linked MED12 mutations disrupt mediator-associated CDK activity. Cell Rep 2014;7:654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park MJ, Shen H, Kim NH, Gao F, Failor C, Knudtson JF, et al. Mediator kinase disruption in MED12-mutant uterine fibroids from Hispanic women of South Texas. J Clin Endocrinol Metab 2018;103:4283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramdzan ZM, Nepveu A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer 2014;14:673–82. [DOI] [PubMed] [Google Scholar]
- 25.Schoenmakers EF, Bunt J, Hermers L, Schepens M, Merkx G, Janssen B, et al. Identification of CUX1 as the recurrent chromosomal band 7q22 target gene in human uterine leiomyoma. Genes Chromosomes Cancer 2013; 52:11–23. [DOI] [PubMed] [Google Scholar]
- 26.Seguin L, Liot C, Mzali R, Harada R, Siret A, Nepveu A, et al. CUX1 and E2F1 regulate coordinated expression of the mitotic complex genes Ect2, MgcRacGAP, and MKLP1 in S phase. Mol Cell Biol 2009;29:570–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truscott M, Harada R, Vadnais C, Robert F, Nepveu A. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol Cell Biol 2008;28:3127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, et al. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: a mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proc Natl Acad Sci U S A 1996;93:11516–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawke TJ, Jiang N, Garry DJ. Absence of p21CIP rescues myogenic progenitor cell proliferative and regenerative capacity in Foxk1 null mice. J Biol Chem 2003;278:4015–20. [DOI] [PubMed] [Google Scholar]
- 30.Meeson AP, Hawke TJ, Graham S, Jiang N, Elterman J, Hutcheson K, et al. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells 2004;22:1305–20. [DOI] [PubMed] [Google Scholar]
- 31.Shi X, Garry DJ. Sin3 interacts with Foxk1 and regulates myogenic progenitors. Mol Cell Biochem 2012;366:251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Wallis AM, Gerard RD, Voelker KA, Grange RW, DePinho RA, et al. Foxk1 promotes cell proliferation and represses myogenic differentiation by regulating Foxo4 and Mef2. J Cell Sci 2012;125:5329–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ono M, Qiang W, Serna VA, Yin P, Coon JSt, Navarro A, et al. Role of stem cells in human uterine leiomyoma growth. PLoS One 2012;7:e36935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H, Spaeth JM, Kim NH, Xu X, Friez MJ, Schwartz CE, et al. MED12 mutations link intellectual disability syndromes with dysregulated GLI3-dependent Sonic Hedgehog signaling. Proc Natl Acad Sci U S A 2012;109:19763–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golebiewska A, Brons NH, Bjerkvig R, Niclou SP. Critical appraisal of the side population assay in stem cell and cancer stem cell research. Cell Stem Cell 2011;8:136–47. [DOI] [PubMed] [Google Scholar]
- 36.Ono M, Maruyama T, Masuda H, Kajitani T, Nagashima T, Arase T, et al. Side population in human uterine myometrium displays phenotypic and functional characteristics of myometrial stem cells. Proc Natl Acad Sci U S A 2007;104:18700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono M, Kajitani T, Uchida H, Arase T, Oda H, Nishikawa-Uchida S, et al. OCT4 expression in human uterine myometrial stem/progenitor cells. Hum Reprod 2010;25:2059–67. [DOI] [PubMed] [Google Scholar]
- 38.Chemical Probes Portal. Available at: https://www.chemicalprobes.org/cct251545. Accessed.
- 39.Dale T, Clarke PA, Esdar C, Waalboer D, Adeniji-Popoola O, Ortiz-Ruiz MJ, et al. A selective chemical probe for exploring the role of CDK8 and CDK19 in human disease. Nat Chem Biol 2015;11:973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mallinger A, Crumpler S, Pichowicz M, Waalboer D, Stubbs M, Adeniji-Popoola O, et al. Discovery of potent, orally bioavailable, small-molecule inhibitors of WNT signaling from a cell-based pathway screen. J Med Chem 2015;58:1717–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poss ZC, Ebmeier CC, Odell AT, Tangpeerachaikul A, Lee T, Pelish HE, et al. Identification of Mediator kinase substrates in human cells using cortistatin A and quantitative phosphoproteomics. Cell Rep 2016;15:436–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Margolin AA, Ong SE, Schenone M, Gould R, Schreiber SL, Carr SA, et al. Empirical Bayes analysis of quantitative proteomics experiments. PLoS One 2009;4:e7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 2009;139:757–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, et al. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 2013;38:250–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malumbres M Cyclin-dependent kinases. Genome Biol 2014;15:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng WR, Scherer SW, Koutsilieris M, Huizenga JJ, Filteau F, Tsui LC, et al. Loss of heterozygosity and reduced expression of the CUTL1 gene in uterine leiomyomas. Oncogene 1997;14:2355–65. [DOI] [PubMed] [Google Scholar]
- 47.Hewitt SC, Li R, Adams N, Winuthayanon W, Hamilton KJ, Donoghue LJ, et al. Negative elongation factor is essential for endometrial function. FASEB J 2019;33:3010–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet 2013; 47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gearing LJ, Cumming HE, Chapman R, Finkel AM, Woodhouse IB, Luu K, et al. CiiiDER: a tool for predicting and analysing transcription factor binding sites. PLoS One 2019;14:e0215495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev 2015;29:513–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lasham A, Mehta SY, Fitzgerald SJ, Woolley AG, Hearn JI, Hurley DG, et al. A novelEGR-1dependentmechanismforYB-1modulationofpaclitaxelresponse in a triple negative breast cancer cell line. Int J Cancer 2016;139:1157–70. [DOI] [PubMed] [Google Scholar]
- 52.Schiavone D, Avalle L, Dewilde S, Poli V. The immediate early genes Fos and Egr1 become STAT1 transcriptional targets in the absence of STAT3. FEBS Lett 2011;585:2455–60. [DOI] [PubMed] [Google Scholar]
- 53.Wu TH, Shi L, Lowe AW, Nicolls MR, Kao PN. Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3. PLoS One 2019;14:e0216042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fiorentino L, Cavalera M, Menini S, Marchetti V, Mavilio M, Fabrizi M, et al. Loss of TIMP3 underlies diabetic nephropathy via FoxO1/STAT1 interplay. EMBO Mol Med 2013;5:441–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ibarra IL, Hollmann NM, Klaus B, Augsten S, Velten B, Hennig J, et al. Mechanistic insights into transcription factor cooperativity and its impact on protein-phenotype interactions. Nat Commun 2020;11:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santoni de Sio FR, Barde I, Offner S, Kapopoulou A, Corsinotti A, Bojkowska K, et al. KAP1 regulates gene networks controlling T-cell development and responsiveness. FASEB J 2012;26:4561–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature 2008; 455:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galbraith MD, Andrysik Z, Pandey A, Hoh M, Bonner EA, Hill AA, et al. CDK8 kinase activity promotes glycolysis. Cell Rep 2017;21:1495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He L, Gomes AP, Wang X, Yoon SO, Lee G, Nagiec MJ, et al. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol Cell 2018;70:949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim S, Xu X, Hecht A, Boyer TG. Mediator is a transducer of Wnt/betacatenin signaling. J Biol Chem 2006;281:14066–75. [DOI] [PubMed] [Google Scholar]
- 61.Sukonina V, Ma H, Zhang W, Bartesaghi S, Subhash S, Heglind M, et al. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature 2019;566:279–83. [DOI] [PubMed] [Google Scholar]
- 62.Wang W, Li X, Lee M, Jun S, Aziz KE, Feng L, et al. FOXKs promote Wnt/betacatenin signaling by translocating DVL into the nucleus. Dev Cell 2015;32: 707–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon NS, Rong Zeng W, Premdas P, Santaguida M, Berube G, Nepveu A. Expression of N-terminally truncated isoforms of CDP/CUX is increased in human uterine leiomyomas. Int J Cancer 2002;100:429–32. [DOI] [PubMed] [Google Scholar]
- 64.Neville PJ, Thomas N, Campbell IG. Loss of heterozygosity at 7q22 and mutation analysis of the CDP gene in human epithelial ovarian tumors. Int J Cancer 2001;91:345–9. [DOI] [PubMed] [Google Scholar]
- 65.Zeng WR, Watson P, Lin J, Jothy S, Lidereau R, Park M, et al. Refined mapping of the region of loss of heterozygosity on the long arm of chromosome 7 in human breast cancer defines the location of a second tumor suppressor gene at 7q22 in the region of the CUTL1 gene. Oncogene 1999;18:2015–21. [DOI] [PubMed] [Google Scholar]
- 66.McNerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood 2013;121:975–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wong CC, Martincorena I, Rust AG, Rashid M, Alifrangis C, Alexandrov LB, et al. Inactivating CUX1 mutations promote tumorigenesis. Nat Genet 2014; 46:33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hulea L, Nepveu A. CUX1 transcription factors: from biochemical activities and cell-based assays to mouse models and human diseases. Gene 2012; 497:18–26. [DOI] [PubMed] [Google Scholar]
- 69.An N, Khan S, Imgruet MK, Gurbuxani SK, Konecki SN, Burgess MR, et al. Gene dosage effect of CUX1 in a murine model disrupts HSC homeostasis and controls the severity and mortality of MDS. Blood 2018; 131:2682–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang M, Feng X, Pei G, Srivastava M, Wang C, Chen Z, et al. FOXK1 participates in DNA damage response by controlling 53BP1 function. Cell Rep 2020;32:108018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanwar PS, Lee HJ, Zhang L, Zukerberg LR, Taketo MM, Rueda BR, et al. Constitutive activation of beta-catenin in uterine stroma and smooth muscle leads to the development of mesenchymal tumors in mice. Biol Reprod 2009;81:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lynch CJ, Bernad R, Martinez-Val A, Shahbazi MN, Nobrega-Pereira S, Calvo I, et al. Global hyperactivation of enhancers stabilizes human and mouse naive pluripotency through inhibition of CDK8/19 Mediator kinases. Nat Cell Biol 2020;22:1223–38. [DOI] [PubMed] [Google Scholar]
- 73.Martinez-Val A, Lynch CJ, Calvo I, Ximenez-Embun P, Garcia F, Zarzuela E, et al. Dissection of two routes to naive pluripotency using different kinase inhibitors. Nat Commun 2021;12:1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, et al. CDK8 maintains tumor dedifferentiation and embryonic stem cell pluripotency. Cancer Res 2012;72:2129–39. [DOI] [PubMed] [Google Scholar]
- 75.Fukasawa K, Kadota T, Horie T, Tokumura K, Terada R, Kitaguchi Y, et al. CDK8 maintains stemness and tumorigenicity of glioma stem cells by regulating the c–MYC pathway. Oncogene 2021;40:2803–15. [DOI] [PubMed] [Google Scholar]
- 76.Prusinski Fernung LE, Al-Hendy A, Yang Q. A preliminary study: human fibroid Stro-1+/CD44+ stem cells isolated from uterine fibroids demonstrate decreased DNA repair and genomic integrity compared to adjacent myometrial Stro-1+/CD44+ cells. Reprod Sci 2019;26:619–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.