Abstract
The hydrolytic activity of the ATP synthase in bovine mitochondria is inhibited by a protein called IF1, but bovine IF1 has no effect on the synthetic activity of the bovine enzyme in mitochondrial vesicles in the presence of a proton motive force. In contrast, it has been suggested based on indirect observations that human IFI inhibits both the hydrolytic and synthetic activities of the human ATP synthase and that the activity of human IF1 is regulated by the phosphorylation of Ser-14 of mature IF1. Here, we have made both human and bovine IF1 which are 81 and 84 amino acids long, respectively, and identical in 71.4% of their amino acids and have investigated their inhibitory effects on the hydrolytic and synthetic activities of ATP synthase in bovine submitochondrial particles. Over a wide range of conditions, including physiological conditions, both human and bovine IF1 are potent inhibitors of ATP hydrolysis, with no effect on ATP synthesis. Also, substitution of Ser-14 with phosphomimetic aspartic and glutamic acids had no effect on inhibitory properties, and Ser-14 is not conserved throughout mammals. Therefore, it is unlikely that the inhibitory activity of mammalian IF1 is regulated by phosphorylation of this residue.
Keywords: mitochondria, ATP synthase, regulation, inhibitor protein IF1, unidirectional inhibition
In 1963, Pullman and Monroy isolated from bovine mitochondria a low molecular weight protein, now known as IF1, or inhibitor of F1-ATPase, that, as its name implies, inhibits ATP hydrolysis by the F1-catalytic domain of the ATP synthase (1). They demonstrated that in the presence of either of the electron donors NADH or succinate, bovine submitochondrial particles (SMPs), which are everted vesicles of the inner mitochondrial membrane with the F1-catalytic domain of the membrane bound ATP synthase extending into the surrounding milieu, generate a proton motive force (pmf) across the vesicular membrane, and that these vesicles were able to make ATP from ADP and phosphate. However, the addition of bovine IF1 at a single concentration did not uncouple oxidative phosphorylation (1) and therefore it was concluded that bovine IF1 is a unidirectional inhibitor of ATP hydrolysis by the ATP synthase, without effect on the ability of the enzyme to carry out ATP synthesis. Direct single observations of this unidirectional inhibitory activity were made subsequently with purified bovine ATP synthase coreconstituted into phospholipid vesicles with bacteriorhodopsin (2). On provision of light, together with ADP, Mg2+, and phosphate, the bacteriorhodopsin generated a transmembrane pmf to drive the synthesis of ATP by the ATP synthase, but, addition of IF1 supplied at a single concentration, did not inhibit ATP synthesis. Also, when a pmf is generated in mitochondrial membrane vesicles, IF1 disengages from the ATP synthase complex (3), and disengagement from F1 occurs effectively only in the presence of ADP and phosphate with the enzyme’s rotor turning in the synthetic direction (4).
Bovine IF1 is a basic protein of 84 amino acids (5), and the active form is dimeric with the monomers associated via an antiparallel α-helical coiled-coil in their C-terminal regions, and the N-terminal inhibitory regions extending in opposite directions (Fig. 1A) (6, 7). In the free solution, these inhibitory regions are intrinsically disordered (8, 9), but in crystals of bovine IF1, they are folded into an α-helix (10). In the process of inhibiting the hydrolytic activity of the ATP synthase, each intrinsically disordered inhibitory region interacts initially with the most open of the three catalytic interfaces of the enzyme, the "empty" interface. Then, concomitant with the consecutive hydrolysis of two ATP molecules in each F1-domain, the catalytic interfaces close and the inhibitory regions become progressively structured and increasingly enveloped by the enzyme, leading to the final fully inhibited state where each of the inhibitory regions is bound in the "DP" (or diphosphate) interface (10). In this fully inhibited state, each of the inhibitory regions from residues 1 to 13 remains disordered and occupies the central cavity surrounding the α-helical coiled-coil region of the γ-subunit in the central stalk of the enzyme's rotor. Residues 14 to 18 form a short α-helix and together with an extended region from residues 19 to 20 they interact with the α-helical coiled-coil region of the γ-subunit. The extended region is followed by an α-helix from residues 21 to 81. This α-helix includes the bound inhibitory region from residues 21 to 46, which extends unbroken via residues 47 to 81 from the surface of the F1-catalytic domain. Residues 47 to 81 provide the C-terminal dimerization region (7).
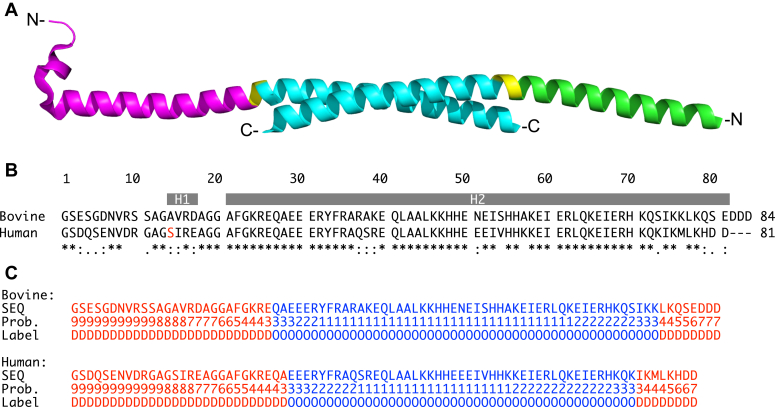
Figure 1.
Structure of bovine IF1and sequence conservation in the bovine and human proteins.A, on the left is shown, the structure of the monomer of bovine IF1 made of 84 amino acids derived from the inhibitory region (residues 11–46 magenta) in the crystal structure of monomeric bovine IF1 bound to bovine F1-ATPase (PDB 4TT3) and the dimerization region (blue) from the structure in crystals of isolated dimeric bovine IF1 (PDB 1GMJ). The dimerization regions of the two monomers are associated via an antiparallel α-helical coiled-coil (residues 49–81). The right-hand monomer is taken from the same crystal structure of the isolated dimer, with the inhibitory region colored green. The left-hand monomer was resolved from residues 11 to 82, and the right-hand monomer from residues 20 to 79. The yellow regions in each monomer are residues Lys-47 and His-48, found between the inhibitory region and the interacting domain of the coiled-coil. In free solution, the two inhibitory regions are intrinsically disordered (9), and become ordered, as shown in the left-hand monomer, on binding to the F1-catalytic domain of ATP synthase (10). B, comparison of the sequences of bovine and human IF1. Sequence conservation is shown above where asterisks, colons, and periods, respectively, indicate identical, strongly conserved, and weakly similar residues. Human residue Ser-14 (red) has been proposed to be reversibly phosphorylated (13). The gray boxes H1 (residues 14–17) and H2 (residues 21–81) indicate the positions of α-helical regions in the crystal structure of bovine IF1 (7). C, intrinsically disordered regions (D, red) of bovine and human IF1 predicted with SPOT-Disorder 2 (57) with the probability scores (Prob.). Regions predicted to be ordered (O) are blue.
Until now, the biochemical properties of the closely related human IF1 have not been studied in vitro, and the assumption that human IF1 has a unidirectional inhibitory effect on the human ATP synthase, inhibiting only ATP hydrolysis and not synthesis, has been challenged (11, 12). Thus, it has been asserted that in mitochondria within human cells, IF1 inhibits both ATP hydrolysis and synthesis and moreover that reversible phosphorylation of residue Ser-14 of mature IF1, referred to previously as residue Ser-39 of the mitochondrial precursor protein, regulates both hydrolytic and synthetic inhibitory activities by preventing IF1 from binding to the ATP synthase during either ATP hydrolysis or ATP synthesis (13). The evidence for the inhibition of mitochondrial ATP synthesis by IF1 has been questioned previously (14). Here, we have studied the effects of both bovine and human IF1 on ATP hydrolysis and ATP synthesis by the ATP synthase in bovine SMPs. For this purpose, we have adopted an established method for assaying ATP (15) and have used this technique to follow the generation of ATP and to explore the impact on this process of various inhibitors, including bovine and human IF1 and mutant forms added at a range of molar excesses. The mutant forms of human IF1 include those where Ser-14 has been substituted by the phosphomimetic residues, aspartic acid and glutamic acid.
Results
Comparison of features of human and bovine IF1
The sequences of bovine and human IF1 are identical in 71.4% of their amino acids and conservatively substituted in a further 13.1% (Fig. 1B). In the inhibitory region, 71.1% of amino acids are identical and a further 17.8% are highly conserved and in the coiled-coil region observed in the bovine dimeric protein (residues 44–84), 73.2% of amino acids are identical in the human ortholog and an additional 7.3% are substituted conservatively. Moreover, the residues of bovine IF1 that are involved in binding it to the bovine enzyme are identical, except for residues 11, 15, and 17 where bovine serine, valine, and aspartic acid residues, respectively, are conservatively substituted by human glycine, isoleucine, and glutamic acid. The amino acid residues that bind bovine IF1 to the F1-domain have been identified by systematic mutation and kinetic analysis (16). They interact predominantly with specific residues in the C-terminal domain of the β-subunit, with another residue in the C-terminal domain of the adjacent α-subunit and with three others in the α-helical coiled-coil region of the γ-subunit (see Table S1). These interacting residues are identical in the human subunits (Table S1), and the sequences of the same three α-, β-, and γ-subunits are identical in 98.4%, 98.6%, and 92.3% of their amino acid residues, respectively (Fig. S1). Another feature of the bovine inhibitor proteins is that its N-terminal region is intrinsically disordered (10), and the same region is predicted to be so in the human protein (Fig. 1C). Therefore, it is reasonable to conclude that bovine and human IF1 molecules have closely similar structures and properties and that they bind to their cognate F1-domains in an essentially identical fashion. Thus, the inhibitory properties of human IF1 studied, as they are here, with the more readily available and sufficiently abundant ATP synthase in bovine SMPs, illuminate the binding properties of human IF1 with the human ATP synthase.
ATP synthase content of bovine SMPs
In order to ensure that the investigations of the inhibitory properties included experimental conditions similar to physiological circumstances, the content of ATP synthase in bovine SMPs was determined by absolute quantitative mass spectrometry (MS) using stable isotopically labeled peptide standards. It was found to be 0.48 nmol/mg n-dodecyl-β-D-maltoside extract (Table S2), with corresponding molar ratios for the α-, e- and OSCP-subunits of 3.0:0.90:1.2, close to the values 3:1:1 expected from the structure of the enzyme complex (17). The range of molar excesses of IF1 employed in the experiments below includes the range of estimated physiological levels, but also extends beyond it (see Table S3).
Design of inhibitor proteins
All four versions of the bovine inhibitor proteins that were studied contained the mutation Y33W to aid spectroscopic quantitation of the protein. This conservative substitution has no deleterious effect on the inhibitory activity of IF1 (16). These four versions of bovine IF1 were as follows: the full-length dimeric protein BovIF1(1–84)-Y33W, where the ATP hydrolytic inhibitory activity of dimers is regulated by both pH and concentration of cations; BovIF1(1–84)-Y33W-H49K, where the mutation H49K makes the protein constitutively active in the inhibition of ATP hydrolysis from pH values below 8 (18); the monomeric inhibitors BovIF1(1–62)-Y33W and BovIF1(1–62)-Y33W-H49K, without and with the mutation H49K, respectively. The eight versions of the human inhibitor protein included two versions of the full-length dimeric WT protein, HumIF1(1–81)-WT and HumIF1(1–81)-Y33W, plus the constitutively active forms HumIF1(1–81)-H49K and HumIF1(1–81)-Y33W-H49K. In addition, four forms of human IF1 were prepared to investigate the possible role of phosphorylation of residue Ser-14 in the regulation of the hydrolytic and synthetic activities of the protein, all with the mutation Y33W. The four versions, where Ser-14 has been replaced by the phosphomimetic residues aspartic acid and glutamic acid, are HumIF1(1–81)-S14D-Y33W, HumIF1(1–81)-S14D-Y33W-H49K, HumIF1(1–81)-S14E-Y33W and HumIF1(1–81)-S14E-Y33W-H49K. All fourteen proteins were produced by bacterial overexpression, with full-length bovine proteins prepared by a combination of cation and anion exchange chromatography and the other twelve by affinity purification and proteolytic removal of affinity tags. Their purities were validated by SDS-PAGE and their sequences were checked by MS (Figs. S2 and S3, and Table S4).
Characterization of bovine SMPs
ATP synthesis by bovine SMPs was demonstrated by generating a pmf with either NADH or succinate, in the presence of ADP and phosphate. This synthetic activity was abolished by the separate addition of the uncoupling agent carbonyl cyanide 4-(trifluoromethoxy)-phenylhydrazone and by oligomycin, rotenone, and antimycin, specific inhibitors of ATP synthase, complex I and complex III, respectively (Fig. S4). As expected, the addition of the inhibitors BovIF1(1–84)-Y33W, BovIF1(1–84)-Y33W-H49K, BovIF1(1–62)-Y33W and BovIF1(1–62)-Y33W-H49K, over a range of concentrations, had similar effects in inhibiting ATP hydrolysis (Fig. 2). In contrast, addition of the same proteins had no significant effect on ATP synthesis, whereas in control samples lacking the inhibitor protein, ATP synthesis was abolished by the addition of oligomycin (Fig. 2).
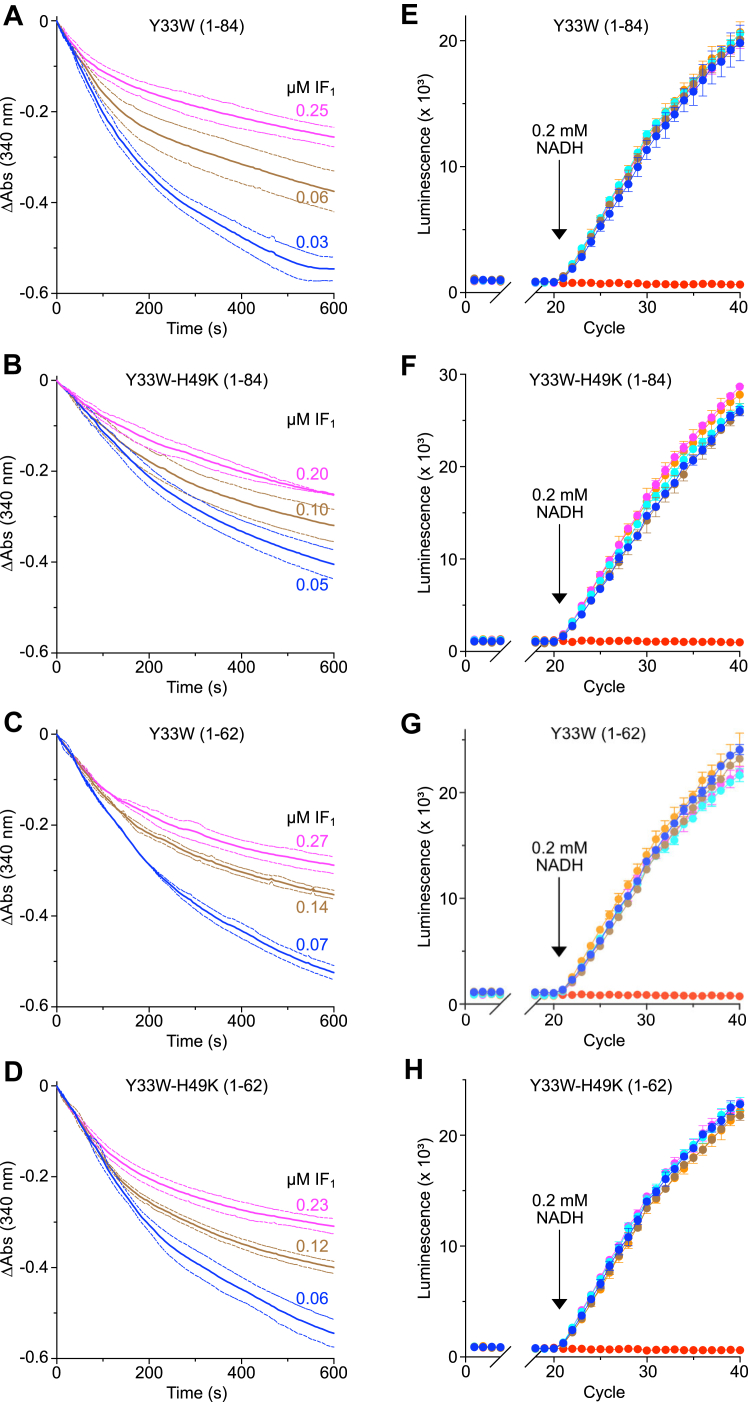
Figure 2.
Inhibition of ATP hydrolysis and the continued generation of ATP in the presence of mutated forms of bovine IF1.A–D, inhibition of ATP hydrolysis and (E–H), effects on ATP synthesis in bovine SMPs by increasing concentrations of mutant forms of bovine IF1. A and E, IF1(1–84)-Y33W, (B and F), IF1(1–84)-Y33W-H49K; (C and G), IF1(1–62)-Y33W; (D and H), IF1(1–62)-Y33W-H49K. A–D, measurements were made in triplicate, except for B 0.20 μM where duplicate values were used, average values were corrected to an initial value of zero, and ± SD shown with bounding dashed lines (Files S1–S4). In E–H, luminescence signals are given in relative light units; ( ), SMPs only; (
), SMPs only; ( ), (
), ( ), and (
), and ( ), SMPs in the presence of inhibitor proteins in molar ratios of 0.5, 2.5, 5, and 10, respectively, with respect to the quantity of ATP synthase; (
), SMPs in the presence of inhibitor proteins in molar ratios of 0.5, 2.5, 5, and 10, respectively, with respect to the quantity of ATP synthase; ( ), in the presence of oligomycin (10 μg/ml). Background luminescence levels were established for 20 measurement cycles, and ATP generation was initiated at cycle 20 by the addition of NADH (0.2 mM). Measurements were made in triplicate and average values ± SD are shown (Files S5-S8). SMP, submitochondrial particle.
), in the presence of oligomycin (10 μg/ml). Background luminescence levels were established for 20 measurement cycles, and ATP generation was initiated at cycle 20 by the addition of NADH (0.2 mM). Measurements were made in triplicate and average values ± SD are shown (Files S5-S8). SMP, submitochondrial particle.
Effects of human inhibitor proteins on the hydrolytic and synthetic activities of bovine ATP synthase
The four inhibitor proteins HumIF1(1–81), HumIF1(1–81)-Y33W, HumIF1(1–81)-H49K, and HumIF1(1–81)-Y33W-H49K all inhibited ATP hydrolysis by bovine SMPs effectively. Constitutively active forms with the mutation H49K were the most potent, and at a concentration of 0.8 μM they inhibited hydrolysis as effectively as 1 μM oligomycin (Figs. 3 and 4). However, all four of these human inhibitor proteins had no significant effect on ATP synthesis (Figs. 3 and 4). Indeed, at the higher concentrations employed, the human inhibitor proteins had a stimulatory effect on ATP synthesis, presumably arising from the inhibition of the low levels of ATP hydrolysis by ATP synthase in a small fraction of leaky SMPs that are unable to maintain a pmf or possibly from uncoupled partially formed ATP synthase complexes that arise as intermediates during the process of assembly of the enzyme (19, 20). Similar stimulatory effects on ATP synthesis were especially evident, for example, in the presence of large molar excesses of BovIF1(1–84)-Y33W (Fig. S5). In the presence of HumIF1(1–81)-H49K, the rate of ATP synthesis calculated from the initial slope of the continuous assay was 0.414 μmol min−1 mg−1 bovine SMPs and 0.434 μmol min−1 mg−1 SMPs from the quench assay (Fig. 4). As the ATP synthase content of SMPs is 0.48 nmol mg−1 (Table S2), the calculated rate of synthesis of ATP approaches 15 molecules s−1 enzyme−1, which corresponds to the enzyme's rotor turning at ca. five revolutions per second, assuming that all ATP synthase complexes are active.
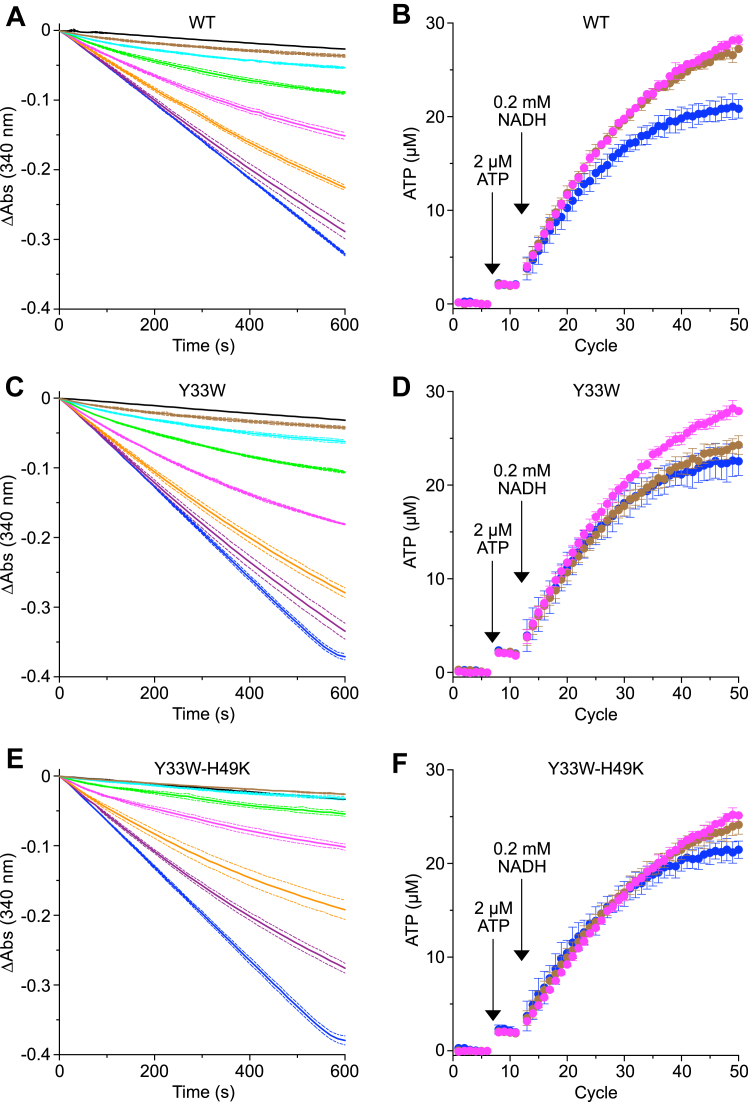
Figure 3.
Effect of human IF1and mutant forms on the activity of ATP synthase.Left hand panels (A), (C), and (E), ATP hydrolysis by bovine SMPs inhibited by increasing concentrations of (A), HumIF1(1–81); (C), HumIF1(1–81)-Y33W; (E), HumIF1(1–81)-Y33W-H49K. ( ), no inhibitor; (—), plus 1 μM oligomycin. IF1 concentrations: (
), no inhibitor; (—), plus 1 μM oligomycin. IF1 concentrations: ( ), 0.05 μM; (
), 0.05 μM; ( ), 0.1 μM; (
), 0.1 μM; ( ), 0.2 μM; (
), 0.2 μM; ( ), 0.4 μM; (
), 0.4 μM; ( ), 0.8 μM; (
), 0.8 μM; ( ), 1.6 μM. Measurements were made in triplicate, and the average values are shown, corrected to an initial value of zero, and ± SD shown as bounding dashed lines (Files S9–S11). Right hand panels (B), (D), and (F), ATP synthesis by bovine heart SMPs coupled to NADH oxidation, in the presence or absence of (B), HumIF1(1–81)-WT; (D), HumIF1(1–81)-Y33W; (F), HumIF1(1–81)-Y33W-H49K. ATP was measured by a luminescence continuous real-time assay with a luciferase-luciferin reagent. The luminescence signal was calibrated with 2 μM ATP, and the background signal before addition of ATP was subtracted. ATP synthesis was initiated by the addition of NADH (0.2 mM). (
), 1.6 μM. Measurements were made in triplicate, and the average values are shown, corrected to an initial value of zero, and ± SD shown as bounding dashed lines (Files S9–S11). Right hand panels (B), (D), and (F), ATP synthesis by bovine heart SMPs coupled to NADH oxidation, in the presence or absence of (B), HumIF1(1–81)-WT; (D), HumIF1(1–81)-Y33W; (F), HumIF1(1–81)-Y33W-H49K. ATP was measured by a luminescence continuous real-time assay with a luciferase-luciferin reagent. The luminescence signal was calibrated with 2 μM ATP, and the background signal before addition of ATP was subtracted. ATP synthesis was initiated by the addition of NADH (0.2 mM). ( ), no inhibitor; (
), no inhibitor; ( ), 0.5 μM IF1; (
), 0.5 μM IF1; ( ), 5 μM IF1. IF1 at 5 μM provides an IF1:ATP synthase molar ratio of 393:1. Data points are the average signal ± SD, n = 4 wells (Files S12–S14). SMP, submitochondrial particle.
), 5 μM IF1. IF1 at 5 μM provides an IF1:ATP synthase molar ratio of 393:1. Data points are the average signal ± SD, n = 4 wells (Files S12–S14). SMP, submitochondrial particle.
Figure 4.
Impact of constitutively active human IF1on ATP hydrolysis and ATP synthesis by bovine SMPs.A, ATP hydrolysis inhibited by increasing concentrations of constitutively active human IF1(1–81)-H49K. ( ), no inhibitor; (—), plus 1 μM oligomycin. IF1 at various concentrations; (
), no inhibitor; (—), plus 1 μM oligomycin. IF1 at various concentrations; ( ), 0.05 μM; (
), 0.05 μM; ( ), 0.1 μM; (
), 0.1 μM; ( ), 0.2 μM; (
), 0.2 μM; ( ), 0.4 μM; (
), 0.4 μM; ( ), 0.8 μM; (
), 0.8 μM; ( ), 1.6 μM. Measurements were made in triplicate, and the traces represent the average values corrected to an initial value of zero, and ± SD shown with bounding dashed lines (File S15). B–D, ATP synthesis by bovine SMPs coupled to NADH oxidation, in the presence or absence of human IF1(1–81)-H49K. ATP synthesis was initiated by the addition of NADH (0.2 mM). In (B) and (C), ATP synthesis was measured with a luminescence continuous real-time assay, and in (D), by a quench assay with luciferase-luciferin reagent. In (B), the concentrations of human IF1(1–81)-H49K were as follows: (
), 1.6 μM. Measurements were made in triplicate, and the traces represent the average values corrected to an initial value of zero, and ± SD shown with bounding dashed lines (File S15). B–D, ATP synthesis by bovine SMPs coupled to NADH oxidation, in the presence or absence of human IF1(1–81)-H49K. ATP synthesis was initiated by the addition of NADH (0.2 mM). In (B) and (C), ATP synthesis was measured with a luminescence continuous real-time assay, and in (D), by a quench assay with luciferase-luciferin reagent. In (B), the concentrations of human IF1(1–81)-H49K were as follows: ( ), no inhibitor; (
), no inhibitor; ( ), 0.2 μM; (
), 0.2 μM; ( ), 1 μM; (
), 1 μM; ( ), 5 μM; (
), 5 μM; ( ), 10 μM; (
), 10 μM; ( ), 20 μM. A concentration of 20 μM IF1 provides a molar excess of ca. 1600:1 with respect to the quantity of ATP synthase. Background luminescence levels were established for ten measurement cycles. N = 4 wells, and data points correspond to the average signal ± SD (File S16). In (C), the luminescence signal was calibrated with 2 μM ATP before the addition of NADH, and the background signal before addition of ATP was subtracted. (
), 20 μM. A concentration of 20 μM IF1 provides a molar excess of ca. 1600:1 with respect to the quantity of ATP synthase. Background luminescence levels were established for ten measurement cycles. N = 4 wells, and data points correspond to the average signal ± SD (File S16). In (C), the luminescence signal was calibrated with 2 μM ATP before the addition of NADH, and the background signal before addition of ATP was subtracted. ( ), no inhibitor; (
), no inhibitor; ( ), 5 μM IF1(1–81)-H49K. The data points are the average signal ± SD, n = 4 wells (File S17). D, ATP synthesis determined with the quench assay. (
), 5 μM IF1(1–81)-H49K. The data points are the average signal ± SD, n = 4 wells (File S17). D, ATP synthesis determined with the quench assay. ( ), no inhibitor; (
), no inhibitor; ( ), 5 μM IF1(1–81)-H49K, corresponding to an IF1:ATP synthase molar ratio of ca. 700:1. Data points are the average signal ± SD, n = 3 (File S18). In (C), a linear regression (
), 5 μM IF1(1–81)-H49K, corresponding to an IF1:ATP synthase molar ratio of ca. 700:1. Data points are the average signal ± SD, n = 3 (File S18). In (C), a linear regression ( ) was applied to the initial rate (25 s) of ATP synthesis in the presence of IF1 H49K, and in (D) to the rate over 240 s. The rates of ATP synthesis (magenta) in μmol min−1 mg−1 were calculated from the slopes. SMP, submitochondrial particle.
) was applied to the initial rate (25 s) of ATP synthesis in the presence of IF1 H49K, and in (D) to the rate over 240 s. The rates of ATP synthesis (magenta) in μmol min−1 mg−1 were calculated from the slopes. SMP, submitochondrial particle.
Are the inhibitory properties of IF1 regulated by phosphorylation?
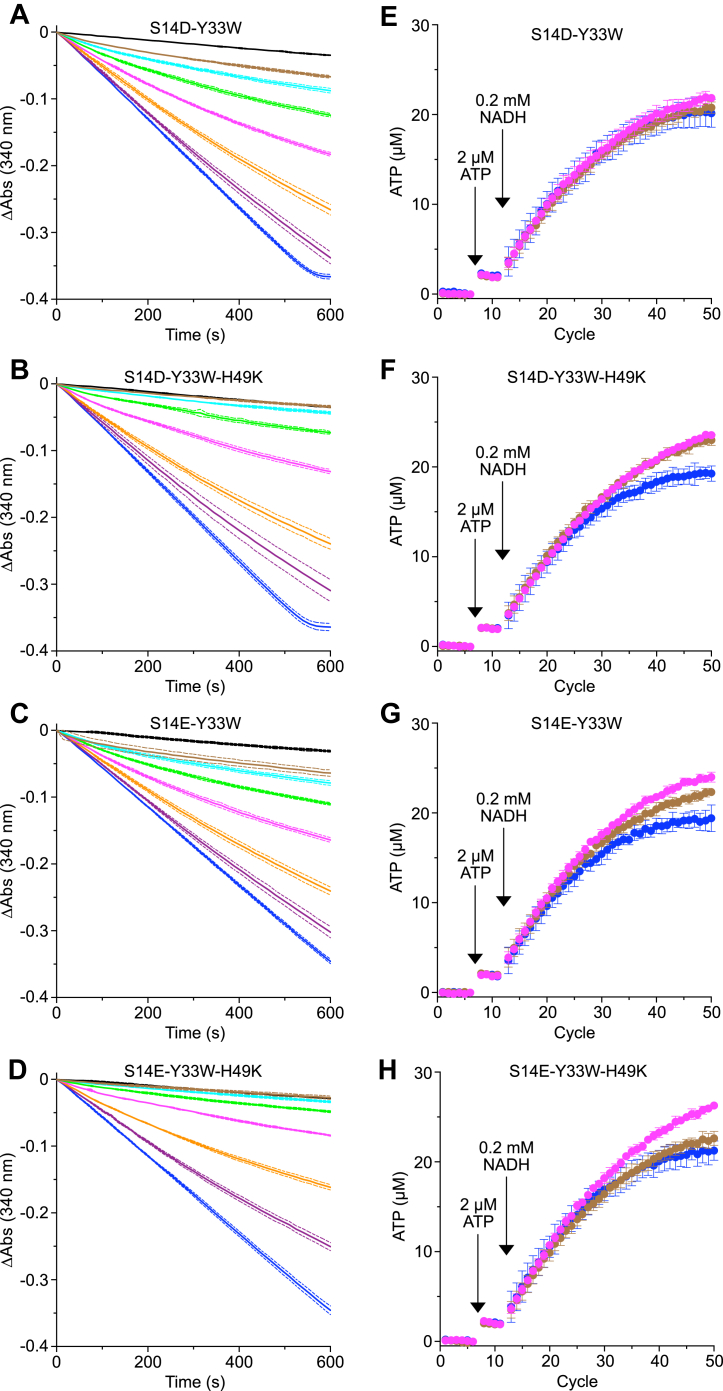
The proposal that the inhibitory activity of human IF1 is regulated by the phosphorylation of residue Ser-14 (13) was investigated with the four inhibitor proteins, HumIF1(1–81)-S14D-Y33W, HumIF1(1–81)-S14E-Y33W, and constitutively active forms HumIF1(1–81)-S14D-Y33W-H49K and HumIF1(1–81)-S14E-Y33W-H49K. All four were as effective inhibitors of ATP hydrolysis as the inhibitor protein containing Ser-14, and none of them inhibited ATP synthesis (Fig. 5). On this basis, it is concluded that the inhibitory activity of human IF1 on ATP hydrolysis is unlikely to be regulated by phosphorylation of Ser-14, and these experiments provide further evidence that human IF1 does not inhibit ATP synthesis.
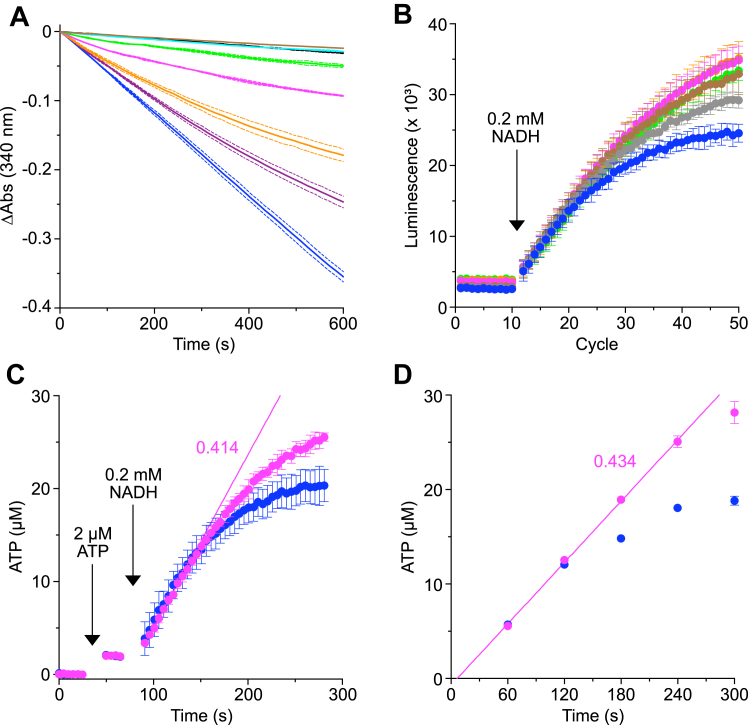
Figure 5.
Impact of phosphomimetic mutant forms of human IF1on the hydrolytic and synthetic activities of bovine ATP synthase.A–D, the impacts on ATP hydrolysis. E–H, the impacts on ATP synthesis. The following inhibitors were employed. In (A) and (E), IF1(1–81)-S14D-Y33W; in (B) and (F), IF1(1–81)-S14D-Y33W-H49K; in (C) and (G), IF1(1–81)-S14E-Y33W; in (D) and (H), IF1(1–81)-S14E-Y33W-H49K. In (A–D), inhibition of ATP hydrolysis by bovine SMPs by increasing concentrations of IF1. ( ), 0.05 μM; (
), 0.05 μM; ( ), 0.1 μM; (
), 0.1 μM; ( ), 0.2 μM; (
), 0.2 μM; ( ), 0.4 μM; (
), 0.4 μM; ( ), 0.8 μM; (
), 0.8 μM; ( ), 1.6 μM. (
), 1.6 μM. ( ) and (—), controls with no inhibitor and 1 μM oligomycin, respectively. Measurements were made in triplicate, and traces correspond to the average values corrected to an initial value of zero, with ± SD shown as bounding dashed lines (Files S19–S22). In (E–H), ATP synthesis by bovine heart SMPs coupled to NADH oxidation, measured by a luminescence continuous real-time assay and with a luciferase-luciferin reagent. The luminescence signal was calibrated with 2 μM ATP, and the background signal before addition of ATP was subtracted. ATP synthesis was initiated by the addition of NADH (0.2 mM). (
) and (—), controls with no inhibitor and 1 μM oligomycin, respectively. Measurements were made in triplicate, and traces correspond to the average values corrected to an initial value of zero, with ± SD shown as bounding dashed lines (Files S19–S22). In (E–H), ATP synthesis by bovine heart SMPs coupled to NADH oxidation, measured by a luminescence continuous real-time assay and with a luciferase-luciferin reagent. The luminescence signal was calibrated with 2 μM ATP, and the background signal before addition of ATP was subtracted. ATP synthesis was initiated by the addition of NADH (0.2 mM). ( ), no inhibitor; (
), no inhibitor; ( ), 0.5 μM IF1; (
), 0.5 μM IF1; ( ), 5 μM IF1. A concentration of 5 μM IF1 provides a molar ratio of ca. 400:1 with respect to the ATP synthase. Data points are the average signal ± SD, n = 4 wells (Files S23–S26). SMP, submitochondrial particle.
), 5 μM IF1. A concentration of 5 μM IF1 provides a molar ratio of ca. 400:1 with respect to the ATP synthase. Data points are the average signal ± SD, n = 4 wells (Files S23–S26). SMP, submitochondrial particle.
Discussion
Fundamental properties of IF1
Three fundamental properties of the free bovine dimeric inhibitor that are likely to underlie its biological role in regulating the ATP synthase in both bovine and human mitochondria have been established previously by in vitro experimentation. The first such property is that the N-terminal portion of the inhibitory region from residue 1 to about residue 45 of bovine IF1 is intrinsically disordered (8, 9, 10). The N-terminal region of the human protein is predicted to be disordered also up to around residue 28, as it is also in the bovine protein (Fig. 1C), and, given that the disordered region of bovine IF1 has been demonstrated experimentally to extend up to around residue 45, the sequence conservation implies that it is likely to do so also in the human protein. It is noteworthy that a property of many intrinsically disordered proteins is that they bind to more than one biological target (21, 22). This characteristic raises the possibility that in addition to binding to the ATP synthase, IF1 binds to other as yet uncharacterized sites. This feature may help to explain some of the many diverse roles that have been connected to IF1 (see below).
A second fundamental property of mammalian IF1 is that the active inhibitor is dimeric, (see Fig. 1A). Previously, it has been demonstrated that each of the inhibitory regions in dimeric bovine IF1 can bind simultaneously to an F1-catalytic domain (23). However, the recent structure of the intact dimeric ATPase shows that the two F1-heads are too far apart to permit the dimeric inhibitor to straddle between the two catalytic domains in the isolated dimer (17). In the inner membranes of mitochondria, the dimers self-associate in long rows on the tips of the cristae (24, 25), and IF1 dimers can span across the dimer–dimer interface between adjacent F1-domains and such complexes have been observed by cryo-EM (26, 27). Although the biological significance of these inhibited complexes is currently unclear, they may participate in the formation of the variety of structures that the cristae can assume. Cells both overexpressing or lacking IF1 are reported to have altered cristae (28, 29, 30, 31).
A third fundamental property is that the inhibitory activity of bovine IF1 is influenced by both the pH and the ionic strength of cations of the surrounding milieu (6, 9). Thus, at around pH 8 and above, and at low concentrations of cations, the dimeric IF1 is inactive because the dimers self-associate and form oligomers of dimers where the inhibitory regions are occluded. At pH values of around 7 and below, and/or at high concentrations of cations, IF1 consists of free and active dimers, where the N-terminal inhibitory regions can bind to ATP synthase complexes and prevent the hydrolysis of ATP. This in vitro inactivation-activation process provides the basis for a possible mechanism of the physiological regulation of the activity of IF1. Thus, for example, the inactive inhibitor could be activated from inactive oligomers by a decrease in the pH of the mitochondrial matrix, such as occurs during ischemia (32). It could also be activated by an increased concentration of Ca2+ ions in the mitochondrial matrix either accompanying cell signalling events or by the cytoplasmic overload of Ca2+ ions and their subsequent uptake into the mitochondrial matrix, the opening of the mitochondrial permeability transition pore, disruption mitochondrial membranes, and necrotic cell death (33).
The experiments described here have confirmed and further defined a fourth fundamental property of IF1, namely that it is a unidirectional inhibitor of ATP hydrolysis only, with no direct effect on the synthesis of ATP. Kobayashi et al. (4) have shown that the physical restoration of rotation in the F1 domain from an IF1 inhibited state depended upon both the direction of application of the force and the nature of the bound substrate. Thus, it required both the manipulation of the γ-subunit in the synthetic direction and the presence of bound ADP and Pi before significant levels of rotation could be observed. These experiments also suggested that the re-establishment of rotation was accompanied by the release of bound IF1. Moreover, as shown here, there was no support for the proposal that the inhibitory activity of IF1 is regulated by phosphorylation of Ser-14 (13), as the replacement of this residue by either of the phosphomimetic residues aspartate or glutamate had no effect on the inhibitory properties of IF1. To a first approximation, the carboxylate of an aspartate or glutamate residue provides a point negative charge to mimic the point negative charge of a phosphoserine, but unlike a dianionic phosphate monoester, it has one negative charge only. Also, the aspartate and glutamate residues are smaller and have a different geometry from phosphoserine. Hence, although the introduction of an aspartate or a glutamate often mimics the function of phosphoserine; for examples see (34, 35), it may not necessarily do so (36). However, crucially, it should also be noted that Ser-14 of IF1 is not strictly conserved in mammals and other vertebrates (Fig. S6), as might be expected if the residue were involved in a basic regulatory mechanism. For example, in cattle, pigs, horses, African bush elephants, and bottle-nose dolphins it is replaced by an alanine residue (Fig. S6). Recent work also refutes the proposed inhibition of ATP synthesis by IF1 in cell cultures, and the regulation of IF1 by PKA-dependent phosphorylation (37). Therefore, on the basis of the experimental evidence and evolutionary considerations, on balance it is unlikely that the activity of mammalian IF1 is regulated in vivo by the reversible phosphorylation of Ser-14.
The structural basis of the unidirectionality of inhibition is not understood currently, but it could stem from two sources: the catalytic F1-domain, the inhibitor protein IF1, or both. While the structure of the catalytic domain of the enzyme in hydrolytic mode is well known (38, 39), there is no detailed structural information about the enzyme in synthetic mode. In synthetic mode, the rotor turns in a counter-clockwise sense (as viewed perpendicular to the plane of the mitochondrial membrane with the mitochondrial matrix above), and the torque of the rotor is driven by the pmf, whereas in hydrolytic mode, the rotor turns in the opposite sense and rotation is driven by the binding and release of substrates and products from the catalytic domain (38, 39). Thus, it seems possible, even likely, that the detailed structure of the enzyme will differ in these two modes, with perhaps the most significant differences being found in the structure of membrane extrinsic part of the rotor (made of single copies of the γ-, δ-, and ε-subunits) and especially of the elongated α-helical structure of the γ-subunit with which the N-terminal region of IF1 interacts in hydrolytic inhibition. Thus, the reversal of the direction of rotation with accompanying changes in the catalytic interfaces themselves can be envisaged to release any inhibitor bound previously in hydrolytic mode and prevent the inhibitor from (re)binding during synthetic mode. Removal of the unstructured residues 1 to 12 of IF1 did not affect the inhibitory activity significantly, but further truncation of IF1 up to residue 22 resulting in removal of the short α-helix and the N-terminal region of the long α-helix (Fig. 1) resulted in weaker binding of the truncated versions of IF1 to F1-ATPase (16), accompanied by a significant loss of the rotational and directional-dependent activation of hydrolytic activity (4).
Roles of IF1
Many biological roles have been ascribed to IF1 (reviewed in ref (14)), and, for example, its roles as a factor in the process of assembly of ATP synthase clearly relate to its inhibition of partially formed intermediates that are capable of ATP hydrolysis but not synthesis (19, 20). Also, its association with dimeric ATP synthase complexes in influencing the formation of the mitochondrial cristae points to the local inhibition of ATP hydrolysis. In addition, it is accepted that following the dissipation of the mitochondrial pmf and lowered matrix pH associated with ischaemia that IF1 is a reversible noncompetitive inhibitor of ATP hydrolysis by mitochondrial ATP synthase. More recently it was shown that altering the levels of IF1 affects cellular and mitochondrial Ca2+ handling (40) and may be unrelated to the interaction of IF1 with ATP synthase (41). Perhaps the most widely discussed and least understood role is the association of IF1 with various forms of cancer (14, 42). It has been observed that IF1 is expressed at elevated levels in many forms of the disease, and it has been suggested to play a role in the switch of the cancer cells to a more aerobic glycolytic metabolism (43, 44, 45). However, the molecular mechanisms underlying this pro-oncogenic role remain obscure. These suggestions that IF1 inhibits ATP synthesis are based on indirect observations, and not, as here, by direct biochemical analysis. It remains possible that the intrinsically disordered region of IF1 leads it to bind not only to the mitochondrial ATPase but also to other, as yet unidentified, targets, and that this property may provide an explanation of the apparently contradictory suggestions that have been made about the properties of the protein.
Experimental procedures
Numbering of IF1 proteins
The 25 amino acid N-terminal mitochondrial import sequences of bovine and human IF1 are removed during import of the proteins into the organelle, and the mature proteins are numbered from 1 to 84 and 1 to 81, respectively. In mitochondria, both mature proteins have a ragged N-terminus with forms starting at residues −1, 1, 2, and 3 (46, 47, 48).
Protein estimation
The concentration of proteins in solution were determined with bicinchoninic acid (BCA, Thermo Fisher Scientific). In experiments with bovine IF1 containing the mutation Y33W, protein concentrations were estimated from the UV absorbance at 280 nm.
Bovine SMPs
Bovine mitochondria from approximately half a heart, prepared as described before (49), were resuspended in SMP buffer (10 mM 3-(N-Morpholino)propanesulphonic acid, pH 7.5, and 250 mM sucrose) and centrifuged (11,300g, 12 min, 4 °C). The pellets were resuspended in 30 ml of SMP buffer with a Dounce homogenizer. The pH of the suspension was adjusted to 9 with 2.5 M Tris base, stirred at 4 °C for 10 min, and then diluted with SMP buffer (30 ml), and centrifuged (38,000g, 14 min, 4 °C). The pellet was resuspended in SMP buffer (20 ml), and the washing procedure was repeated twice more. The suspension of washed mitochondria in 40 ml of SMP buffer, plus bovine cytochrome c (final concentration 100 μM) was kept at 4 °C for 1 h. Magnesium sulfate was added (final concentration 15 mM), and the suspension was sonicated ten times at 150 W for 15 s at 4 °C, with 1 min intervening pauses, with a Q700 sonicator (QSonica; probe diameter 0.5 inches). Then the suspension was centrifuged (24,700g, 20 min), and the supernatant was recentrifuged (74,700g, 30 min, 4 °C). The resultant pellet of SMPs was resuspended in SMP buffer, homogenized, and stored at −80 °C.
ATP synthase content of SMPs
Bovine SMPs (41 mg/ml) were diluted to 10 mg/ml and extracted for 15 min at 4 °C with buffer containing 0.1 M ammonium bicarbonate, pH 8, 0.2 mM neutral tris(2-carboxyethyl)-phosphine) (TCEP, Sigma), and 1% (w/v) n-dodecyl-β-D-maltoside (final concentration). The sample was centrifuged (14,000g, 15 min, 4 °C), and proteins in the supernatant at a concentration of 1 mg/ml, estimated with BCA, were digested for 2 to 2.5 h at 37 °C with trypsin (sequencing grade from Roche; trypsin:protein, 1:100, w/w) in the presence of 0.1 M ammonium bicarbonate, 0.2 mM TCEP, and 0.5% (w/v) sodium deoxycholate (50). Then digestion was continued for a further 18 h at 37 °C in the presence of a second portion of trypsin and finally in the presence of a third portion of trypsin for 1.5 to 2 h. The concentrations of synthetic peptides containing stable isotopes (Table S5; Cambridge Research Biochemicals) dissolved in 10% (v/v) aqueous acetonitrile were determined by amino acid analysis (Protein and nucleic acid facility, Department of Biochemistry, University of Cambridge). A known amount of this internal reference standard was added to the digest (51), plus an equal volume of ethyl acetate saturated with water and containing formic acid (ca. 4%, v/v) (50). The sample was vortexed, then centrifuged (14,000g, 2 min, 18 °C) to separate the phases. Portions of the lower aqueous phase containing the peptides was fractionated on a C18 reverse phase column in a Proxeon EASY-nLC1000, eluted with a gradient of acetonitrile containing 0.1% (v/v) formic acid, with the column effluent coupled directly to a Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific). Peptides were fragmented by collision-induced dissociation with nitrogen. Peak areas for the labeled and endogenous peptides (charge state 2+) were calculated from the extracted ion chromatograms of the parent ions with Xcalibur (Thermo Fisher Scientific) with an m/z tolerance of 5 ppm, and used to determine the subunit ratio and absolute amount of ATP synthase (Fig. S7, Table S6). Labeled peptides gave a linear signal over the analytical range of 31.3 to 2000 fmol (Fig. S8).
Overexpression of WT and mutant forms of bovine and human IF1
Bovine and human IF1 variants were constructed with the mutation Y33W to facilitate protein quantitation by absorbance at 280 nm and used as a calibration standard for protein estimations by the BCA method. The mutation H49K was introduced into the bovine protein by site directed mutagenesis with the forward primer agaaacacaaggaaaatgagatctctcatcatgcaaaggag and the reverse primer tcattttccttgtgtttcttcaaggcggccagctgttctttag, where the bold letters indicate the mutated codon. The primers were designed to anneal to the same sequence on the opposite strands of the plasmid. The entire plasmid was amplified by PCR, and template DNA was digested with the restriction enzyme Dpn I. The sequences were cloned into the expression vector pRun and were confirmed by DNA sequencing. The DNA sequence for human IF1 encoded the intact protein (residues 1–81) plus the N-terminal sequence MSHHHHHHSAENLYFQ. Specific mutations were introduced into the coding sequence of the human protein with the NEBaseChanger kit (New England Biolabs). Expression plasmids encoding WT and mutated forms of bovine and human IF1 were transformed into cells of Escherichia coli C41 (DE3). Cells were grown in 2xTY medium at 37 °C until the A600 reached 0.6 absorbance units. Then expression of recombinant proteins was induced with IPTG (final concentration 600 μM). Cells expressing bovine proteins were grown at 25 °C for 18 h, centrifuged (6500g, 10 min), resuspended in buffer containing 10 mM Tris–HCl, pH 8, 1 mM EDTA, and complete EDTA-free protease inhibitors (Sigma-Aldrich), and broken with a Constant Cell disruptor (Constant Cell). Subsequent steps were carried out at 4 °C. Broken cells were centrifuged (265,000g, 45 min), and the supernatant was dialyzed for 18 h against buffer A (10 mM Tris–HCl, pH 8, and 1 mM EDTA). The solution was applied to a HiTrap SP HP cation exchange column (5 ml; GE Healthcare). The inhibitor protein eluted from the column at a salt concentration of 350 mM on a linear gradient of NaCl from 0 to 1 M. Fractions containing the inhibitor were dialyzed against buffer A for 18 h and then applied to a HiTrap Q HP column (5 ml; GE Healthcare). On a linear gradient of NaCl from 0 to 1 M, the inhibitor protein eluted at 300 mM. Purified IF1 was exchanged into ultrapure water on a PD-10 column (GE Healthcare). Cells expressing human IF1 and variants were grown at 37 °C for ca. 4 h, centrifuged (2700g, 10 min, 4 °C), washed in cold buffer A consisting of 20 mM Tris–HCl, pH 7.4, 0.1 M NaCl, 10% (v/v) glycerol and containing cOmplete protease inhibitors (Sigma-Aldrich), and centrifuged (3900g, 10 min, 4 °C). Cells were resuspended in cold buffer A containing cOmplete EDTA-free protease inhibitors and broken, on ice, with a Q700 sonicator equipped with sonic probe 0.75 inches in diameter. Broken cells were centrifuged (10,845g, 15 min, 4 °C), and imidazole (5 M, pH 8) was added to the supernatant to give a final concentration of 20 mM. The sample was mixed by rotation at 4 °C for 1 h with nickel-nitrilotriacetic acid agarose resin (Qiagen). Then the resin with IF1 bound via the His-tag was collected by settling, or centrifugation (27g, 1 min, 4 °C), washed with buffer A plus 20 mM imidazole, and finally IF1 was eluted with buffer A containing 0.5 M imidazole. The eluted IF1 was dialyzed in a 3.5 kDa cut-off Slide-A-Lyzer cassette (Thermo Fisher Scientific) into buffer A at 4 °C. The His-tag was removed in the dialysis cassette by addition of tobacco etch virus (TEV) protease (ca. 0.1 mg/ml, final concentration) and neutralized 2 mM TCEP and incubation with stirring at 30 °C for 3 h in buffer A. The cleaved sample was then kept for 1 h at 4 °C in the presence of the nickel-nitrilotriacetic acid agarose and 10 mM imidazole. The unbound IF1 was dialyzed at 4 °C against Milli-Q water in a 3.5 kDa cut-off Slide-A-Lyzer cassette.
Overexpression and purification of C terminally truncated bovine IF1
DNA-encoding residues 1 to 62 of bovine IF1 with an N-terminal hexa-histidine tag, followed by a site for TEV protease, as described above for human IF1, was cloned into pRun. The point mutation H49K was introduced by site directed mutagenesis and the protein was expressed as described above for intact bovine IF1. The bacterial pellet was resuspended in His buffer A (20 mM Tris, pH 7.4, 0.1 M NaCl, and 10 mM imidazole) plus one cOmplete EDTA-free protease inhibitor tablet (Roche). Cells were broken with a constant cell disruptor and then centrifuged (234,000g, 45 min, 4 °C). The supernatant was applied to a HisTrap column (5 ml, GE Healthcare), which was eluted with a gradient of imidazole from 10 to 500 mM. The purified protein was dialyzed against His buffer A plus TCEP in the presence of TEV protease for 18 h at 18 °C to cleave the His-tag. The free His-tag was removed by affinity chromatography and the truncated forms were exchanged into ultrapure water.
Characterization of inhibitor proteins
The purity of inhibitor proteins was examined by SDS-PAGE and staining with Coomassie R250. Stained gels were imaged with an Epson Perfection V850 Pro flatbed scanner. Intact molecular masses were measured by electrospray MS in a Q-Trap 4000 (ABSciex) instrument, operated in MS mode, and calibrated with a mixture of myoglobin and trypsinogen. Reconstructed molecular masses were calculated with Peakview (Sciex).
Inhibition of ATP hydrolysis by SMPs
The ATP hydrolase activity of SMPs was measured in the presence of various inhibitor proteins with a coupled ATP-generating system (52). The inhibition of the ATP hydrolysis by endogenous ADP bound to SMPs was alleviated by incubation for 10 min at room temperature with an equal volume of stripping buffer (20 mM Tris–HCl, pH 7.4, 50% glycerol v/v, and 4 mM EDTA). The stripped SMPs were added to the assay mix consisting of 50 mM Tris–HCl, pH 7.4, 50 mM KCl, 2 mM MgCl2, 0.14 or 0.2 mM NADH, 1 μM rotenone or piericidin A, pyruvate kinase (20 μg/ml), and lactate dehydrogenase (10 μg/ml), to a final concentration of ca. 4 to 20 μg/ml, depending on the measured uninhibited activity. The assay mix (180 μl, 37 °C) was dispensed into a 96-well plate (Corning) warmed to 37 °C. The assay was initiated by the addition of 20 μl of a solution containing 10 mM phosphoenolpyruvate, 20 mM ATP, and the requisite inhibitor protein to give a final assay volume of 200 μl. The oxidation of NADH was monitored from the UV absorbance at 340 nm for 10 min at 37 °C with a SpectraMax M2 plate reader (Molecular Devices). Measurements were made in triplicate.
ATP synthesis by SMPs
In some experiments, the synthesis of ATP was followed by a continuous real-time assay (53) and in others by a quench assay. In both assays, bovine SMPs were added to the buffer consisting of 20 mM Tris-phosphate, pH 7.4, and 5 mM MgCl2, plus 20 or 100 μM of the adenylate kinase inhibitor, P1,P5-di(adenosine-5′)pentaphosphate, and 100 μM ADP. The reaction mixture was warmed to 37 °C. The final concentration of ATP synthase in the assay solution was 0.5 to 1.5 pmol per 100 μl.
The continuous real-time assay
The luciferase reagent from ATP bioluminescence kit CLS II (Roche) was prepared according to the manufacturer’s instructions, and 1 μl was added to each 100 μl of assay solution containing the bovine SMPs dispensed in wells in opaque white 96-well plates (Corning or PerkinElmer) at 37 °C. Luminescence was measured with a ClarioStar plate reader (BMG Labtech). Typically, a single row of 12 wells was read simultaneously, with three samples in quadruplicate, distributed in a repeating pattern from the outside to the centre to help offset the time difference across the row. Background luminescence values were established over 10 to 20 measurement cycles (5 or 7 s with a measurement interval time of 0.1 s). Then a pmf was generated by the addition of either 200 μM NADH or 4 mM succinate, and the synthesis of ATP was followed during a further 20 to 40 cycles. In assays conducted in the presence of bovine and human IF1 and mutants thereof, the inhibitor proteins were preincubated with SMPs in the assay mixture for 2 to 3 min before initiating ATP synthesis by the generation of a pmf. Control assays were performed in the presence of the respiratory inhibitors oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, rotenone, or antimycin A. Measurements were made in triplicate or quadruplicate.
The quench assay
The assay was performed in microcentrifuge tubes instead of in the wells of 96-well plates, in the absence of the luciferase reagent, but otherwise under the same conditions as described above. Samples (10 μl) were taken from the reaction mixture at 1 min intervals and added to 4% (w/v) trichloroacetic acid (40 μl), and mixed 20 s later with 1 M Tris–HCl, pH 8 (250 μl), at room temperature. The luciferase reagent (30 μl) was added to the mixture and three portions (100 μl) were transferred into wells in an opaque white 96-well plate. The luminescence signal was measured in a ClarioStar plate reader at room temperature. The instrument was calibrated with standard samples of ATP.
Data availability
All data are available within the paper and Supporting information, except for MS data deposited to the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD045079.
Supporting information
This article contains supporting information (10, 55, 56).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Mr P. Sharratt (Department of Biochemistry, University of Cambridge) for performing amino acid analysis, and Dr J. He (Medical Research Council Mitochondrial Biology Unit, Cambridge) for the advice in the design and sequencing of the human IF1 gene products.
Author contributions
J. E. W. conceptualization; J. E. W. supervision; J. E. W. funding acquisition; J. E. W. project administration; J. E. W. methodology; J. E. W. writing-original draft; J. C., I. N. W., C. J. W., S. D., I. M. F., and J. E. W. formal analysis; J. C., I. N. W., C. J. W., S. D., I. M. F., and investigation; J. C. and I. N. W. visualization.
Funding and additional information
The work was supported by the Medical Research Council, United Kingdom via Program Grant MR/M009858/1, Project Grant MR/V009672/1, and Grant MC_UU_00015/8 (to J. E. W.). C. J. W. received support from Peterhouse, University of Cambridge.
Reviewed by members of the JBC Editorial Board. Edited by Phillip A. Cole
Footnotes
Present address for Charlotte J. Wright: Tree of Life, Wellcome Sanger Institute, Cambridge CB10 1SD, United Kingdom.
Supporting information
References
- 1.Pullman M.E., Monroy G.C. A naturally occurring inhibitor of mitochondrial adenosine triphosphatase. J. Biol. Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 2.Runswick M.J., Bason J.V., Montgomery M.G., Robinson G.C., Fearnley I.M., Walker J.E. The affinity purification and characterization of ATP synthase complexes from mitochondria. Open Biol. 2013;3 doi: 10.1098/rsob.120160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippe G., Sorgato M.C., Harris D.A. The binding and release of the inhibitor protein are governed independently by ATP and membrane potential in ox-heart submitochondrial vesicles. Biochim. Biophys. Acta. 1988;933:12–21. doi: 10.1016/0005-2728(88)90051-5. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi R., Ueno H., Okazaki K.I., Noji H. Molecular mechanism on forcible ejection of ATPase inhibitory factor 1 from mitochondrial ATP synthase. Nat. Commun. 2023;14:1682. doi: 10.1038/s41467-023-37182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker J.E., Gay N.J., Powell S.J., Kostina M., Dyer M.R. ATP synthase from bovine mitochondria: sequences of imported precursors of oligomycin sensitivity conferral protein, factor 6, and adenosinetriphosphatase inhibitor protein. Biochemistry. 1987;26:8613–8619. doi: 10.1021/bi00400a018. [DOI] [PubMed] [Google Scholar]
- 6.Cabezon E., Butler P.J., Runswick M.J., Walker J.E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 7.Cabezón E., Runswick M.J., Leslie A.G., Walker J.E. The structure of bovine IF1, the regulatory subunit of mitochondrial F-ATPase. EMBO J. 2001;20:6990–6996. doi: 10.1093/emboj/20.24.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon-Smith D.J., Carbajo R.J., Yang J.C., Videler H., Runswick M.J., Walker J.E., Neuhaus D., et al. Solution structure of a C-terminal coiled-coil domain from bovine IF1: the inhibitor protein of F1 ATPase. J. Mol. Biol. 2001;308:325–339. doi: 10.1006/jmbi.2001.4570. [DOI] [PubMed] [Google Scholar]
- 9.Boreikaite V., Wicky B.I.M., Watt I.N., Clarke J., Walker J.E. Extrinsic conditions influence the self-association and structure of IF1, the regulatory protein of mitochondrial ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 2019;116:10354–10359. doi: 10.1073/pnas.1903535116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bason J.V., Montgomery M.G., Leslie A.G.W., Walker J.E. Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11305–11310. doi: 10.1073/pnas.1411560111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Bermúdez J., Cuezva J.M. The ATPase Inhibitory Factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochim. Biophys. Acta. 2016;1857:1167–1182. doi: 10.1016/j.bbabio.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Domínguez-Zorita S., Romero-Carramiñana I., Cuezva J.M., Esparza-Moltó P.B. The ATPase inhibitory factor 1 is a tissue-specific physiological regulator of the structure and function of mitochondrial ATP synthase: a closer look into neuronal function. Front. Physiol. 2022;13 doi: 10.3389/fphys.2022.868820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Bermúdez J., Sánchez-Aragó M., Soldevilla B., Del Arco A., Nuevo-Tapioles C., Cuezva J.M. PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H+-ATP synthase. Cell Rep. 2015;12:2143–2155. doi: 10.1016/j.celrep.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 14.Solaini G., Sgarbi G., Baracca A. The F1Fo-ATPase inhibitor, IF1, is a critical regulator of energy metabolism in cancer cells. Biochem. Soc. Trans. 2021;49:815–827. doi: 10.1042/BST20200742. [DOI] [PubMed] [Google Scholar]
- 15.Lemasters J.J., Hackenbrock C.E. Adenosine triphosphate: continuous measurement in mitochondrial suspension by firefly luciferase luminescence. Biochem. Biophys. Res. Commun. 1973;55:1262–1270. doi: 10.1016/s0006-291x(73)80030-0. [DOI] [PubMed] [Google Scholar]
- 16.Bason J.V., Runswick M.J., Fearnley I.M., Walker J.E. Binding of the inhibitor protein IF1 to bovine F1-ATPase. J. Mol. Biol. 2011;406:443–453. doi: 10.1016/j.jmb.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spikes T.E., Montgomery M.G., Walker J.E. Structure of the dimeric ATP synthase from bovine mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23519–23526. doi: 10.1073/pnas.2013998117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schnizer R., Van Heeke G., Amaturo D., Schuster S.M. Histidine-49 is necessary for the pH-dependent transition between active and inactive states of the bovine F1-ATPase inhibitor protein. Biochim. Biophys. Acta. 1996;1292:241–248. doi: 10.1016/0167-4838(95)00208-1. [DOI] [PubMed] [Google Scholar]
- 19.He J., Ford H.C., Carroll J., Douglas C., Gonzales E., Ding S., et al. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2018;115:2988–2993. doi: 10.1073/pnas.1722086115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J., Carroll J., Ding S., Fearnley I.M., Montgomery M.G., Walker J.E. Assembly of the peripheral stalk of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2020;117:29602–29608. doi: 10.1073/pnas.2017987117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babu M.M., Kriwacki R.W., Pappu R.V. Structural biology. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 22.Uversky V.N., Oldfield C.J., Dunker A.K. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J. Mol. Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 23.Cabezón E., Arechaga I., Jonathan P., Butler G., Walker J.E. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J. Biol. Chem. 2000;275:28353–28355. doi: 10.1074/jbc.C000427200. [DOI] [PubMed] [Google Scholar]
- 24.Dudkina N.V., Sunderhaus S., Braun H.P., Boekema E.J. Characterization of dimeric ATP synthase and cristae membrane ultrastructure from Saccharomyces and Polytomella mitochondria. FEBS Lett. 2006;580:3427–3432. doi: 10.1016/j.febslet.2006.04.097. [DOI] [PubMed] [Google Scholar]
- 25.Davies K.M., Strauss M., Daum B., Kief J.H., Osiewacz H.D., Rycovska A., et al. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu J., Zhang L., Zong S., Guo R., Liu T., Yi J., et al. Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1. Science. 2019;364:1068–1075. doi: 10.1126/science.aaw4852. [DOI] [PubMed] [Google Scholar]
- 27.Pinke G., Zhou L., Sazanov L.A. Cryo-EM structure of the entire mammalian F-type ATP synthase. Nat. Struct. Mol. Biol. 2020;27:1077–1085. doi: 10.1038/s41594-020-0503-8. [DOI] [PubMed] [Google Scholar]
- 28.Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., et al. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Faccenda D., Nakamura J., Gorini G., Dhoot G.K., Piacentini M., Yoshida M., et al. Control of mitochondrial remodeling by the ATPase inhibitory factor 1 unveils a pro-survival relay via OPA1. Cell Rep. 2017;18:1869–1883. doi: 10.1016/j.celrep.2017.01.070. [DOI] [PubMed] [Google Scholar]
- 30.Weissert V., Rieger B., Morris S., Arroum T., Psathaki O.E., Zobel T., et al. Inhibition of the mitochondrial ATPase function by IF1 changes the spatiotemporal organization of ATP synthase. Biochim. Biophys. Acta Bioenerg. 2021;1862 doi: 10.1016/j.bbabio.2020.148322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domínguez-Zorita S., Romero-Carramiñana I., Santacatterina F., Esparza-Moltó P.B., Simó C., Del-Arco A., et al. IF1 ablation prevents ATP synthase oligomerization, enhances mitochondrial ATP turnover and promotes an adenosine-mediated pro-inflammatory phenotype. Cell Death Dis. 2023;14:413. doi: 10.1038/s41419-023-05957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouslin W., Broge C.W. Regulation of mitochondrial matrix pH and adenosine 5’-triphosphatase activity during ischemia in slow heart-rate hearts. Role of Pi/H+ symport. J. Biol. Chem. 1989;264:15224–15229. [PubMed] [Google Scholar]
- 33.Duchen M.R. Mitochondria and Ca2+ in cell physiology and pathophysiology. Cell Calcium. 2000;28:339–348. doi: 10.1054/ceca.2000.0170. [DOI] [PubMed] [Google Scholar]
- 34.Thorsness P.E., Koshland D.E. Inactivation of isocitrate dehydrogenase by phosphorylation is mediated by the negative charge of the phosphate. J. Biol. Chem. 1987;262:10422–10425. [PubMed] [Google Scholar]
- 35.Martin I., Kim J.W., Lee B.D., Kang H.C., Xu J.C., Jia H., et al. Ribosomal protein s15 phosphorylation mediates LRRK2 neurodegeneration in Parkinson’s disease. Cell. 2014;157:472–485. doi: 10.1016/j.cell.2014.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paleologou K.E., Schmid A.W., Rospigliosi C.C., Kim H.Y., Lamberto G.R., Fredenburg R.A., et al. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J. Biol. Chem. 2008;283:16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sgarbi G., Righetti R., Del Dotto V., Grillini S., Giorgio V., Baracca A., et al. The pro-oncogenic protein IF1 does not contribute to the Warburg effect and is not regulated by PKA in cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 2023;1870 doi: 10.1016/j.bbadis.2023.166879. [DOI] [PubMed] [Google Scholar]
- 38.Abrahams J.P., Leslie A.G.W., Lutter R., Walker J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 39.Walker J.E. The ATP synthase: the understood, the uncertain and the unknown. Biochem. Soc. Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 40.Faccenda D., Gorini G., Jones A., Thornton C., Baracca A., Solaini G., et al. The ATPase Inhibitory Factor 1 (IF1) regulates the expression of the mitochondrial Ca2+ uniporter (MCU) via the AMPK/CREB pathway. Biochim. Biophys. Acta Mol. Cell Res. 2021;1868 doi: 10.1016/j.bbamcr.2020.118860. [DOI] [PubMed] [Google Scholar]
- 41.Pavez-Giani M.G., Sánchez-Aguilera P.I., Bomer N., Miyamoto S., Booij H.G., Giraldo P., et al. ATPase inhibitory factor-1 disrupts mitochondrial Ca2+ handling and promotes pathological cardiac hypertrophy through CaMKIIδ. Int. J. Mol. Sci. 2021;22:4427. doi: 10.3390/ijms22094427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esparza-Moltó P.B., Cuezva J.M. The role of mitochondrial H+-ATP synthase in cancer. Front. Oncol. 2018;8:53. doi: 10.3389/fonc.2018.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luciaková K., Kuzela S. Increased content of natural ATPase inhibitor in tumor mitochondria. FEBS Lett. 1984;177:85–88. doi: 10.1016/0014-5793(84)80986-2. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez-Cenizo L., Formentini L., Aldea M., Ortega A.D., García-Huerta P., Sánchez-Aragó M., et al. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J. Biol. Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez-Aragó M., Formentini L., Martínez-Reyes I., García-Bermudez J., Santacatterina F., Sánchez-Cenizo L., et al. Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis. 2013;2:e46. doi: 10.1038/oncsis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Runswick M.J., Walker J.E., Gibson B.W., Williams D.H. The frayed N-terminal of the inhibitor protein of bovine mitochondrial F1-ATPase. Biochem. J. 1986;235:515–519. doi: 10.1042/bj2350515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu G., Shin S.B., Jaffrey S.R. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19310–19315. doi: 10.1073/pnas.0908958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaca Jacome A.S., Rabilloud T., Schaeffer-Reiss C., Rompais M., Ayoub D., Lane L., et al. N-terminome analysis of the human mitochondrial proteome. Proteomics. 2015;15:2519–2524. doi: 10.1002/pmic.201400617. [DOI] [PubMed] [Google Scholar]
- 49.Smith A.L. Preparation, properties, and conditions for assay of mitochondria: Slaughterhouse material, small-scale. Methods Enzymol. 1967;10:81–86. [Google Scholar]
- 50.Masuda T., Tomita M., Ishihama Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008;7:731–740. doi: 10.1021/pr700658q. [DOI] [PubMed] [Google Scholar]
- 51.Gerber S.A., Rush J., Stemman O., Kirschner M.W., Gygi S.P. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pullman M.E., Penefsky H.S., Datta A., Racker E. Partial resolution of the enzymes catalyzing oxidative phosphorylation. I. Purification and properties of soluble dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 1960;235:3322–3329. [PubMed] [Google Scholar]
- 53.Lemasters J.J., Hackenbrock C.R. Continuous measurement and rapid kinetics of ATP synthesis in rat liver mitochondria, mitoplasts and inner membrane vesicles determined by firefly-luciferase luminescence. Eur. J. Biochem. 1976;67:1–10. doi: 10.1111/j.1432-1033.1976.tb10625.x. [DOI] [PubMed] [Google Scholar]
- 54.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Pancrazio F., Mavelli I., Isola M., Losano G., Pagliaro P., Harris D.A., et al. In vitro and in vivo studies of FoF1 ATP synthase regulation by inhibitor protein IF1 in goat heart. Biochim. Biophys. Acta. 2004;1659:52–62. doi: 10.1016/j.bbabio.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 56.Esparza-Moltó P.B., Nuevo-Tapioles C., Chamorro M., Nájera L., Torresano L., Santacatterina F., et al. Tissue-specific expression and post-transcriptional regulation of the ATPase inhibitory factor 1 (IF1) in human and mouse tissues. FASEB J. 2019;33:1836–1851. doi: 10.1096/fj.201800756R. [DOI] [PubMed] [Google Scholar]
- 57.Hanson J., Paliwal K.K., Litfin T., Zhou Y. SPOT-Disorder2: Improved protein intrinsic disorder prediction by ensembled deep learning. Genomics Proteomics Bioinformatics. 2019;17:645–656. doi: 10.1016/j.gpb.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available within the paper and Supporting information, except for MS data deposited to the ProteomeXchange Consortium via the PRIDE (54) partner repository with the dataset identifier PXD045079.