Abstract
Human genome-wide association studies (GWAS) suggest a functional role for central glutamate receptor signaling and plasticity in body weight regulation. Here, we use UK Biobank GWAS summary statistics of body mass index (BMI) and body fat percentage (BF%) to identify genes encoding proteins known to interact with postsynaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-d-aspartate (NMDA) receptors. Loci in/near discs large homolog 4 (DLG4) and protein interacting with C kinase 1 (PICK1) reached genome-wide significance (P < 5 × 10−8) for BF% and/or BMI. To further evaluate the functional role of postsynaptic density protein-95 (PSD-95; gene name: DLG4) and PICK1 in energy homeostasis, we used dimeric PSD-95/disc large/ZO-1 (PDZ) domain–targeting peptides of PSD-95 and PICK1 to demonstrate that pharmacological inhibition of PSD-95 and PICK1 induces prolonged weight-lowering effects in obese mice. Collectively, these data demonstrate that the glutamate receptor scaffolding proteins, PICK1 and PSD-95, are genetically linked to obesity and that pharmacological targeting of their PDZ domains represents a promising therapeutic avenue for sustained weight loss.
Scaffolding proteins PICK1 and PSD-95 are genetically linked to obesity and therapeutic targets for sustained weight loss.
INTRODUCTION
The global prevalence of obesity has tripled since 1975, and it is estimated that >2 billion people will be living with overweight or obesity by 2030 (1, 2). Obesity is a risk factor for several severe noncommunicable diseases, including type 2 diabetes, cardiovascular disease, and cancer (3, 4). The development of long-acting glucagon-like peptide 1 receptor (GLP-1R) agonists and combined GLP-1R and glucose-dependent insulinotropic polypeptide (GIP) receptor co-agonists represents much-needed breakthroughs in obesity treatment (5, 6), yet these drugs are faced with a series of challenges. In addition to compliance issues, a large number of patients fail to reach a weight loss of >5% and substantial weight regain is seen for most patients after treatment cessation (7). Thus, it is clear that in parallel with the broad commercialization of incretin-based therapies, there is a growing need for medications with a different mode of action.
Over the past decade, genome-wide association studies (GWAS) have revealed numerous biological pathways that might contribute to obesity pathogenesis (8). Notably, many of the genes and pathways identified through genetic studies are enriched or exclusively expressed in the brain, emphasizing a key role of the central nervous system (CNS) in the pathobiological mechanisms underlying obesity (9). Genes relating to glutamatergic signaling and postsynaptic plasticity that are linked to α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-d-aspartate (NMDA) receptors are particularly enriched in genetic studies of obesity, pointing to an important role of the glutamatergic neurotransmitter system in the homeostatic mechanisms governing energy balance. Although incompletely mapped, a role for glutamate receptor–linked synaptic plasticity has also been identified in rodent studies related to energy metabolism (10–14). However, these pathways have yet to be carefully scrutinized for therapeutic utility in the context of obesity treatment.
Rodent studies confirm the potential weight-lowering benefits of disrupting NMDA receptor signaling (15–17). In addition, there is evidence supporting the idea that antagonizing AMPA receptors could mitigate food-motivated behavior (18, 19). However, AMPA and NMDA receptors are ubiquitously expressed throughout the CNS, and chemical modulators of these receptors are typically marred with off-target effects (20, 21). To circumvent these, scientists have begun exploring therapeutic targeting of intracellular receptor protein complexes to attain precise interference of postsynaptic receptor signaling (22). Both AMPA and NMDA receptors directly interact with intracellular scaffolding proteins, which participate in the anchoring and trafficking of glutamate receptors at the membrane (23). Many of these scaffolding proteins contain postsynaptic density protein-95 (PSD-95)/disc large/ZO-1 (PDZ) domains (23). PDZ domain–containing proteins support the formation and stability of postsynaptic glutamate receptor complexes and are emerging as promising targets for a variety of CNS disorders such as stroke and neuropathic pain (22, 24, 25). However, the involvement of this class of proteins in obesity pathogenesis and their therapeutic potential for weight loss have not been investigated.
RESULTS
Genetic screen of glutamate receptor–interacting PDZ domain–containing proteins
Genetic studies provide a unique opportunity to identify novel targets for obesity in free-living populations (26). Aiming to identify druggable targets for obesity treatment, we searched for proteins that are composed of at least one PDZ domain and that are known to engage in protein-protein interactions with glutamatergic AMPA or NMDA receptors. We identified six proteins that fulfill these criteria: glutamate receptor–interacting protein 1 (GRIP1), GRIP2, protein interacting with C kinase 1 (PICK1), PSD-95 [gene name discs large homolog 4 (DLG4)], synapse-associated protein 97 (SAP97; gene name DLG1), and SAP102 (gene name DLG3) (Fig. 1A). To investigate associations of genetic variants in or near the six genes with obesity phenotypes, we screened all autosomal variants within a ±500-kb region around the transcription start site for association with body mass index (BMI) and body fat percentage (BF%) in the UK Biobank. The DLG3 gene was excluded because of its location on the X chromosome, for which no relevant GWAS data on BMI or BF% are currently available. The DLG4 and PICK1 regions showed a genome-wide significant (P < 5 × 10−8) association with BF% and DLG4 also with BMI, whereas no genome-wide significant association was identified in the GRIP1, GRIP2, DLG1, or DLG2 gene regions (Fig. 1B and table S1). Accordingly, the loci around DLG4 and PICK1 were selected for phenome-wide association study (pheWAS) analysis.
Fig. 1. Overview of PDZ domain–containing proteins and the associations of genetic variants in the corresponding gene regions with BF% and BMI.
(A) PDZ domain–containing proteins that interact with glutamatergic AMPA or NMDA receptors: GRIP1, GRIP2, PICK1, PSD-95 (DLG4), SAP97 (DLG1), and SAP102 (DLG3). (B) Genetic variants in the PICK1 (rs17752670) and DLG4 (rs2242449) loci reached genome-wide significance [shown as −log10(P)] for association with BF% and BMI.
Phenome-wide associations of the DLG4 and PICK1 loci with obesity traits
We assessed the genetic link between the DLG4 and PICK1 loci and obesity by investigating phenome-wide associations with anthropometric traits in the UK Biobank (Figs. 2A and 3A). We identified three linkage disequilibrium (LD) blocks (r2 < 0.1) in the DLG4 locus, of which two (lead variants rs2242449 and rs144129583) contained a genome-wide significant association with BF% (Fig. 2, A and B). The rs2242449 variant was also genome-wide significantly associated with BMI (P < 4.1 × 10−9). Both rs2242449 and rs144129583 showed an association with body fat traits and not fat-free mass or height (Fig. 2C). The rs2242449 variant is an expression quantitative trait locus (eQTL) for DLG4 in the dorsolateral prefrontal cortex (DLPFC) (P = 1.4 × 10−7) (Fig. 2C), whereas no eQTL association was found for rs144129583.
Fig. 2. Human SNPs in the 1-Mb DLG4 region on chromosome 17 are associated with BF%.
(A) PheWAS for anthropometric traits in unrelated British individuals from the UK Biobank. The illustrated SNPs (x axis) are lead SNPs for the presented anthropometric traits (y axis). The highlighted SNPs rs2242449 and rs144129583 are lead SNPs for BF% in the locus. Icon colors are used to distinguish between the traits. (B) LD map of lead SNPs for anthropometric traits. Three LD blocks are identified in the locus chr17:6608115-7608115. LD blocks 1 and 3 show associations with BF% [rs2242449 (BF%): P = 7.22 × 10−9, β = 0.011, rs144129583 (BF%): P = 2.19 × 10−8, β = 0.022]. The strongest associations in LD blocks 2 and 3 are with height. (C) rs2242449 is associated with DLG4 transcript level in brain DLPFC tissue (P = 1.4 × 10−7) and located in an intron of DLG4. eQTL data were derived from Open Targets Genetics (https://genetics.opentargets.org/).
Fig. 3. Human SNPs in the 1-Mb PICK1 region on chromosome 22 are associated with BF%.
(A) PheWAS for anthropometric traits in unrelated British individuals from the UK Biobank. The illustrated SNPs (x axis) are lead SNPs for the presented anthropometric traits (y axis). The highlighted SNPs rs17752670 and rs4821764 are lead SNPs for BF%. Icon colors are used to distinguish between the traits. (B) LD map of lead SNPs for anthropometric traits. Two LD blocks are identified in the locus chr22:37962013-38962013 that are independently associated with BF% (rs17752670: P = 9.59 × 10−9, β = −0.013, rs4821764: P = 9.42 × 10−18, β = −0.016). (C) PICK1 is located in between the lead SNPs, and eQTL analysis from Open Target Genetics (https://genetics.opentargets.org/) reveals significant association of both lead SNPs with PICK1 transcript level in blood.
The PICK1 locus revealed two independent signals (r2 < 0.1) genome-wide significantly associated with BF% and fat mass traits, but not with BMI or fat-free mass (Fig. 3, A and B). The lead single-nucleotide polymorphisms (SNPs), rs17752670 and rs4821764, are eQTLs for PICK1 in the blood (rs17752670: P = 3.3 × 10−106 and rs4821764: P = 4.9 × 10−12) (Fig. 3C). Together, the genetic data suggest a connection between both DLG4 and PICK1 and obesity, possibly mediated via different pathways (central versus peripheral, respectively), as indicated by the eQTL associations of the lead SNPs.
Pharmacological inhibition of PSD-95 lowers body weight in obese mice
PSD-95, the protein product of the DLG4 gene, is a highly abundant component of the postsynaptic density (27). PSD-95 is involved in the regulation of NMDA and AMPA (via the protein stargazin) receptor trafficking and stabilization mediated via its PDZ domains (28, 29). The physiological and pharmacological role of PSD-95 in energy balance regulation remains elusive, and it has not yet been investigated as a drug target for weight loss. To probe the functional relevance of the link between DLG4 and obesity, we explored whether pharmacological targeting of PSD-95 lowers body weight in diet-induced obese (DIO) mice. We have previously developed a potent dimeric peptide–based inhibitor of PSD-95 (UCCB01-147; Fig. 4, A and B) for the treatment of ischemic stroke with the aim of uncoupling NMDA receptor activity from nitric oxide synthesis (30, 31). Here, we demonstrate that daily subcutaneous injections of UCCB01-147 dose-dependently lower body weight and food intake in DIO mice, and show that 25 mg kg−1 appears to be well tolerated for chronic dosing (fig. S1, A to G). Next, we assessed the effects of 14 days of repeated UCCB01-147 dosing on energy balance, glycemic control, and plasma lipid markers in DIO mice (Fig. 4C). We observed a 9% reduction in body weight, which was accompanied by a decrease in food intake (Fig. 4, D to G). The reduction in body weight was reflected in a selective loss of fat mass (Fig. 4H). Acute treatment with UCCB01-147 did not influence blood glucose levels assessed over a 24-hour period (Fig. 4, I and J). Glucose tolerance was unchanged after 7 days of treatment (fig. S1, H and I) but was improved following 14 days of treatment (Fig. 4, K and L). Plasma insulin and HOMA-IR score were not affected by UCCB01-147 treatment (Fig. 4M and fig. S1J), while plasma cholesterol and triglycerides were elevated following 14 days of treatment (Fig. 4, N and O). The changes in plasma lipids were not reflected in hepatic transcriptional changes in genes related to inflammation, glucose, or lipid metabolism (fig. S1K).
Fig. 4. Pharmacological inhibition of the PSD-95/nNOS/NMDA receptor complex reduces adiposity in DIO mice.
(A) Mechanism of action of PSD-95 inhibitor UCCB01-147 disrupting the PSD-95/nNOS/NMDA receptor complex. (B) Chemical structure of UCCB01-147. (C) Experimental design of the study in which DIO male C57BL/6J mice were treated with once-daily subcutaneous (s.c.) injections of UCCB01-147 (25 mg kg−1; n = 10 mice and n = 7 cages) or vehicle (isotonic saline, n = 9 mice and n = 6 cages) over 14 days. (D) Change in body weight. (E) Total change in body weight. (F) Cumulative food intake. (G) Total food intake. (H) Change in body composition. (I) Compound tolerance test on day 0. (J) Area under the curve of (I). (K) Intraperitoneal glucose tolerance test on day 14. (L) Area under the curve of (K). (M) Plasma insulin levels. (N) Plasma cholesterol levels. (O) Plasma triglyceride levels. (P) Chemical structure of UCCB01-144AA, a double alanine mutant with abolished potency at PSD-95. DIO male C57BL/6J mice were treated with once-daily subcutaneous injections of UCCB01-147 (25 mg kg−1; n = 6 mice and n = 6 cages), UCCB01-144AA (25 mg kg−1; n = 6 mice and n = 6 cages), or vehicle (isotonic saline, n = 6 mice and n = 6 cages) over 5 days. (Q) Change in body weight. (R) Total change in body weight. (S) Cumulative food intake. (T) Total food intake. Data were analyzed by two-tailed unpaired t test (E, G, H, J, and L to O), one-way analysis of variance (ANOVA), multiple comparison, Bonferroni post hoc test (R and T), and two-way ANOVA, multiple comparisons, Bonferroni post hoc test (D, F, I, K, Q, and S). Data represent means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Asterisks denote comparison between UCCB01-147 and UCCB01-144AA (Q and S).
To ensure that the UCCB01-147–induced weight loss is not driven by off-target effects, we engineered an inactive UCCB01-147 analog by introducing two alanine mutations to the pentapeptide, resulting in a peptide with comparable chemical structure but with essentially no binding to PSD-95 (UCCB01-144AA; Fig. 4P) (32). Next, we conducted a head-to-head comparison between equimolar dosing of UCCB01-147 and UCCB01-144AA to assess their weight-lowering efficacies in DIO mice. In contrast to UCCB01-147, treatment with UCCB01-144AA did not affect body weight or food intake (Fig. 4, Q to T), indicating that the weight-lowering effects of UCCB01-147 are mediated by PSD-95 inhibition. This notion was corroborated in a follow-up study in which we assessed the weight loss efficacy of a less potent monomeric peptide–based PSD-95 inhibitor, nerinetide (NA-1) (fig. S1, L and M) (25, 33). Equimolar dosing of NA-1 reduced body weight and food intake with less efficacy than UCCB01-147 in DIO mice (fig. S1, N to Q). Together, these results emphasize that pharmacological PSD-95 disruption is a promising strategy for obesity treatment.
Pharmacological inhibition of PICK1 lowers body weight in obese mice
The PDZ domain of PICK1 interacts with the C termini of the GluA2 and GluA3 subunits of AMPA receptors to regulate receptor trafficking (34). To study the functional role of PICK1 in energy homeostasis, we developed a lipidated variant of a selective dimeric PICK1 peptide inhibitor (mPD5) that interferes with PICK1-dependent phosphorylation of AMPA receptors (Fig. 5, A and B) (35). We demonstrate that mPD5 dose-dependently lowers body weight in obese mice and identify 56 mg kg−1 to be well tolerated for chronic studies (fig. S2, A to G). Subsequently, we conducted a 14-day study with once-daily subcutaneous dosing and demonstrated that mPD5 treatment induced a 9.8% vehicle-corrected weight loss (Fig. 5, C to E). The weight loss was reflected in a selective loss of fat mass and coincided with a reduction in food intake (Fig. 5, F to H). Global knockout of PICK1 impairs glucose tolerance (36), and overexpression of PICK1 in diabetic db/db mice improves glucose tolerance (37). In line with this, we observed that acute treatment with mPD5 transiently increased blood glucose levels but that chronic treatment improved glucose tolerance (Fig. 5, I to L, and fig. S2, H and I). The improvement in glucose tolerance was not reflected in changes in HOMA-IR score, plasma insulin or triglyceride levels but was accompanied by significantly lower levels of circulating cholesterol (Fig. 5, M to O, and fig. S2J). Except for a subtle increase in Il-1b mRNA levels, hepatic gene expression of targets relating to inflammation, glucose, and lipid metabolism was unaltered in response to chronic mPD5 treatment (fig. S2K).
Fig. 5. Pharmacological inhibition of PICK1 reduces adiposity in DOI mice.
(A) Mechanism of action of mPD5 inhibiting the PICK1/AMPA receptor complex. (B) Chemical structure of mPD5. (C) Study in which DIO male C57BL/6J mice were treated with once-daily subcutaneous injections of mPD5 (56 mg kg−1; n = 9 mice and n = 5 cages) or vehicle (isotonic saline, n = 9 mice and n = 5 cages) over 14 days. (D) Change in body weight. (E) Total change in body weight. (F) Cumulative food intake. (G) Total food intake. (H) Change in body composition. (I) Compound tolerance test on day 0. (J) Area under the curve of (I). (K) Intraperitoneal glucose tolerance test on day 14. (L) Area under the curve of (K). (M) Plasma insulin levels. (N) Plasma cholesterol levels. (O) Plasma triglyceride levels. (P) Study in which DIO male wild type or global PICK1 knockout C57BL/6J mice were treated with once-daily subcutaneous injections of mPD5 (56 mg kg−1; wild type: n = 4 mice and n = 4 cages; PICK1 knockout: n = 6 mice and n = 6 cages) or isotonic saline (wild type: n = 4 mice and n = 4 cages; PICK1 knockout: n = 6 mice and n = 6 cages) over 7 days. (Q) Change in body weight. (R) Total change in body weight. (S) Cumulative food intake. Data were analyzed by two-tailed unpaired t test (E, G, H, J, and L to O), one-way ANOVA, multiple comparisons, Bonferroni post hoc test (R), and two-way ANOVA, multiple comparisons, Bonferroni post hoc test (D, F, I, K, Q, and S). Data represent means ± SEM; *P < 0.05, **P < 0.01, and ****P < 0.0001. Asterisks denote the comparison between the mPD5-treated wild-type and PICK1 knockout groups (Q and S).
To confirm the target specificity of mPD5, we conducted a 7-day weight loss study in high-fat diet–fed global PICK1 knockout mice (Fig. 5P). We found that the body weight– and food intake–lowering effects of mPD5 observed in DIO wild type littermate mice were completely abolished in the PICK1 knockout mice, underscoring that the appetite-suppressing and weight-lowering effects of mPD5 are mediated via PICK1 (Fig. 5, Q to S).
Metabolic characterization of PSD-95 and PICK1 targeting inhibitors
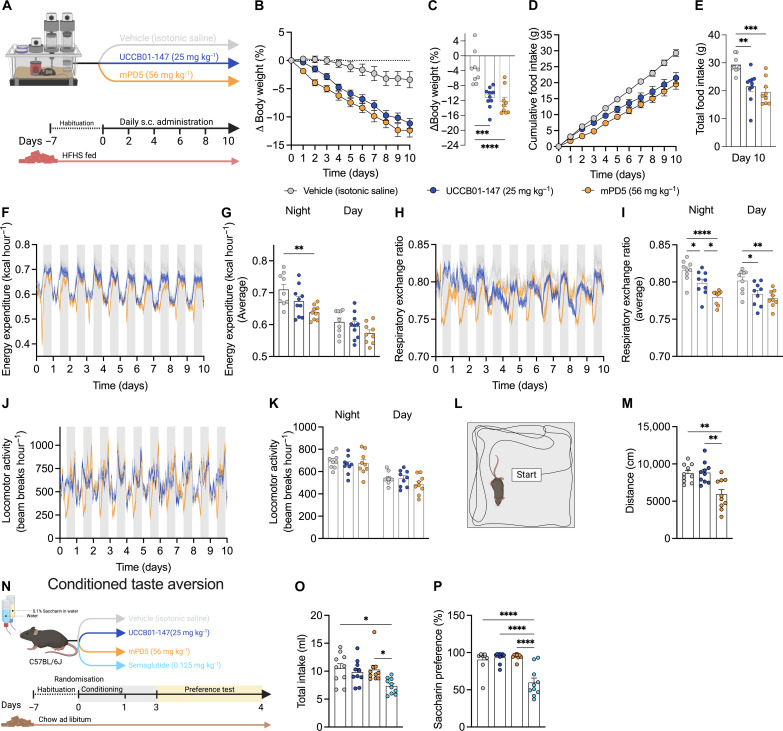
We report that the weight-lowering benefits of both UCCB01-147 and mPD5 are linked to reduced food intake. To understand whether changes in energy expenditure also contribute to the anti-obesity effect of these drug candidates, we used indirect calorimetry (Fig. 6A and fig. S3, A to G). In agreement with our previous results, we observed that both UCCB01-147 and mPD5 evoked a vehicle-corrected weight loss of ~10% over a 10-day treatment period reflected in a reduced food intake (Fig. 6, B to E). A numerical reduction in energy expenditure was observed for both compounds, and this reduction was statistically significant for mPD5-treated mice during the active phase relative to vehicle (Fig. 6, F and G). Unexpectedly, we found an increase in Dio2 mRNA in brown adipose tissue (BAT) following treatment with both UCCB01-147 and mPD5, and UCCB01-147 also increased Ppargc1a in BAT and Ucp1 in inguinal white adipose tissue (fig. S3, H to K). Both compounds reduce respiratory exchange ratio indicative of an increased utilization of fat as a metabolic substrate (Fig. 6, H and I). Total movement was not altered by UCCB01-147 or mPD5; however, a closer examination suggests a transient decrease in locomotion in the initial hours following mPD5 administration (Fig. 6, J and K). This was corroborated by a movement phenotype in the open-field test (Fig. 6, L and M). It is not uncommon for weight loss agents to elicit a hypolocomotive phenotype, occasionally associated with aversion. To explore that hypothesis, we conducted a conditioned taste aversion study using the two-bottle saccharin preference test paradigm (Fig. 6N and fig. S3, L to Q). In contrast to the positive control, semaglutide (30 nmol kg−1), a long-acting GLP-1 receptor agonist, mPD5, and UCCB01-147 did not reduce saccharin consumption or total liquid consumption compared to vehicle (Fig. 6, O and P). These data suggest that both UCCB01-147 and mPD5 lower body weight through a nonaversive reduction in food intake. Additional research is required to comprehend the transient impact on movement observed after mPD5 administration.
Fig. 6. Metabolic phenotyping of UCCB01-147 and mPD5.
(A) Indirect calorimetry study in which DIO male C57BL/6J mice were treated with once-daily subcutaneous injections of UCCB01-147 (25 mg kg−1; n = 10 mice and n = 10 cages), mPD5 (56 mg kg−1; n = 9 mice and n = 9 cages), or vehicle (isotonic saline, n = 9 mice and n = 9 cages) over 10 days in metabolic cages. (B) Change in body weight. (C) Total change in body weight. (D) Cumulative food intake. (E) Total food intake. (F) Longitudinal curves of energy expenditure. (G) Average energy expenditure during dark and light periods. (H) Longitudinal curves of respiratory exchange ratio. (I) Average respiratory exchange ratio during the dark and light periods. (J) Longitudinal curves of locomotor activity. (K) Average locomotor activity during the dark and light periods. (L) Open field test in which lean chow-fed C57BL/6J mice were treated with a single subcutaneous injection of UCCB01-147 (25 mg kg−1; n = 10 mice), mPD5 (56 mg kg−1; n = 10 mice), or vehicle (isotonic saline, n = 10 mice). (M) Distance traveled. (N) Conditioned taste aversion study in which lean chow-fed C57BL/6J mice were dosed with a single subcutaneous injection of UCCB01-147 (25 mg kg−1; n = 10 mice), mPD5 (56 mg kg−1; n = 10 mice), semaglutide (30 nmol kg−1; n = 10 mice), or vehicle (isotonic saline, n = 10 mice). (O) Total drinking volume. (P) Saccharin preference. Data were analyzed by one-way ANOVA, multiple comparisons, Bonferroni post hoc test (C, E, G, I, K, M, O, and P) and two-way ANOVA, multiple comparisons, Bonferroni post hoc test (B and D). Data represent means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Effects of pharmacological PSD-95 and PICK1 inhibition on brain activity and signaling
To map the global neuronal activity signatures following acute treatment with mPD5 and UCCB01-147, we used light sheet microscopy and c-Fos immunohistochemistry enabling quantitative whole-brain c-Fos expression profiles (Fig. 7A) (38, 39). mPD5 administration, but not UCCB01-147 administration, gave rise to a pronounced induction of whole-brain c-Fos expression compared to vehicle (Fig. 7, B and C). Volcano plot visualization revealed that mPD5 powerfully engages several brain regions involved in appetite regulation, i.e., subregions of the brainstem [area postrema (AP), dorsal motor nucleus of the vagus nerve (DMX), and nucleus of the solitary tract (NTS)] and the multiple hypothalamic subnuclei including the arcuate nucleus (ARC) and paraventricular hypothalamic nucleus (PVN) (Fig. 7, D to G). In contrast, UCCB01-147 gave rise to subtle changes in c-Fos expression compared to vehicle, and no significant increase in neuronal activity was observed in canonical appetite-regulating brain regions (Fig. 7, E, F, and H). It is crucial to emphasize that the absence of a distinct whole-brain c-Fos response following the administration of UCCB01-147 does not negate the possibility of a central mode of action. Instead, it suggests that interference with neuronal PSD-95 does not trigger a neuronal activity pattern conducive to c-Fos induction. While it cannot be ruled out that the compounds may exert some effects peripherally, the probability of major peripheral effects of UCCB01-147 appears low, considering the low expression of PSD-95 (Dlg4) in peripheral organs (fig. S4, A and B). Conversely, Pick1 mRNA can be detected in several peripheral tissues (fig. S4, A and B), yet the widespread neuronal c-Fos signal underscores that mPD5 engages multiple brain circuits.
Fig. 7. Whole brain quantification of c-Fos expression in response to UCCB01-147 or mPD5 treatment.
(A) Experimental design of study with whole-brain 3D mapping and quantification of c-Fos induction in response to acute treatment with mPD5 (56 mg kg−1; n = 8 mice), UCCB01-147 (25 mg kg−1; n = 8 mice), or vehicle (isotonic saline, n = 8 mice) in lean C57BL/6J mice. (B) Heatmaps of c-Fos density using spherical averaging (350 μm diameter). (C) Total c-Fos–positive cells in response to treatments. (D) Volcano plot of brain regions regulated for mPD5 relative to vehicle-treated mice. Light red indicates a log2 fold change >0 and Benjamini-Hochberg–adjusted P < 0.05. Light blue indicates a log2 fold change <0 and Benjamini-Hochberg–adjusted P < 0.05. (E) Volcano plot of brain regions regulated for UCCB01-147 relative to vehicle-treated mice. Light red indicates a log2 fold change >0 and Benjamini-Hochberg–adjusted P < 0.05. Light blue indicates a log2 fold change <0 and Benjamini-Hochberg–adjusted P < 0.05. (F) Heatmap of c-Fos activity in brain regions involved in body weight and appetite regulation. Data represent log2 fold change relative to the vehicle group average. (G) Top 20 regulated brain areas in response to acute mPD5 (56 mg kg−1) treatment represented as log2 fold change relative to the vehicle group average. (H) Top 20 regulated brain areas in response to acute UCCB01-147 (25 mg kg−1) treatment represented as log2 fold change relative to the vehicle group average. Data represent mean ± SEM; ****P < 0.0001. For brain region abbreviations, please refer to table S2.
Given the key role for the hypothalamus in central control of body weight and food intake, we performed RNA sequencing on hypothalamic samples obtained from DIO mice treated for 5 days with UCCB01-147, mPD5, or vehicle (fig. S5, A and B). Whereas treatment with mPD5 resulted in significant transcriptional regulation in the hypothalamus, UCCB01-147 did not alter gene expression beyond the adjusted threshold for statistical significance (fig. S5, C and D). Conversely, when assessing enrichment of functional terms, we noted that UCCB01-147 affected transcriptional signatures relating to extracellular matrix, angiogenesis, and synaptic function (fig. S5E). For mPD5, we observed enrichment of functional terms associated with cellular energy metabolism and mitochondrial bioenergetics (fig. S5F).
Region-specific effects on brain proteome following UCCB01-147 and mPD5 treatment
To further interrogate the neuronal underpinnings underlying the weight-lowering effects of mPD5 and UCCB01-147, we conducted mass spectrometry (MS)–based analyses of the proteome of six brain regions from DIO mice treated with vehicle, mPD5, or UCCB01-147 for 10 days (Fig. 8A and fig. S6, A to C). Principal components analysis (PCA) revealed a clear separation for brain regions as well as treatment groups (Fig. 8B and fig. S7, A to D). Using a threshold of false discovery rate–corrected P value < 0.05 and log2 fold change > 1.5, UCCB01-147 treatment resulted in differential expression of between 6 proteins in the least affected region (hypothalamus) and 260 proteins in the most affected region [nucleus accumbens (NAc)], when compared to vehicle (Fig. 8, C and F). In contrast, mPD5 treatment resulted in differential regulation of between 176 proteins in the least affected region (hypothalamus) and 364 proteins in the most affected region (brainstem), when compared to vehicle (Fig. 8, D and F). Although there was minimal overlap among brain regions in terms of proteins responding to UCCB01-147 treatment (Fig. 8C), there were 35 proteins that exhibited similar regulation across all brain regions when comparing mPD5 treatment to vehicle treatment (Fig. 8, D and E). Aligning with the three-dimensional (3D) c-Fos imaging and transcriptomics data, UCCB01-147 had subtle effects on the hypothalamic proteome. Instead, UCCB01-147 powerfully affected the proteome in the brainstem and in the NAc and, to a lesser degree, the cerebellum and the ventral midbrain. While the largest overlap of differentially expressed proteins (95 in total) between UCCB01-147 and mPD5 was detected in the NAc (Fig. 8F and fig. S8, A to J), the region with the most regulation between treatments was the brainstem (Fig. 8, G and H). When scrutinizing the most differentially regulated proteins (Fig. 8I), filtered for an absolute log2 fold change >1.5 and z-scored row-wise, distinct patterns of protein clusters are evident. It is important to highlight that a negative z score does not necessarily imply a reduction but can also represent that the fold change is the lowest among all the comparisons. Protein cluster analyses revealed increased protein expression in response to UCCB01-147 treatment (cluster 3), increased protein expression in the midbrain in response to both UCCB01-147 and mPD5 treatment (cluster 5), the highest fold change in the brainstem (cluster 6), the highest fold change in the midbrain with mPD5 and NAc with both UCCB01-147 and mPD5 treatments (cluster 7), and the highest fold change across regions with mPD5 treatment (cluster 10) (Fig. 8I). Overrepresentation analysis of clusters compared to the total identified proteome revealed Gene Ontology (GO) terms related to acetylcholine receptor binding (cluster 3), blood coagulation and inflammation (clusters 5 and 10), cellular response to retinoic acid and protein tyrosine kinase activity (cluster 6), and glutamatergic synapse and neuron spine (cluster 7) (Fig. 8J). Finally, upon assessing known PICK1 and PSD-95/Dlg4 interactors (identified with String PPIs), we see mostly a decrease in interacting proteins across regions and treatments, perhaps reflective of the prolonged treatment exposure (Fig. 8K). However, notable exceptions include a pervasive effect of UCCB01-147 in NAc to increase the expression of proteins that interact with PSD-95. A similar scenario is observed in the brainstem with the difference that UCCB01-147 in this region increases the expression of PICK1 interactors, including PICK1 protein in itself. This might reflect that PSD-95 can influence AMPA receptor trafficking through stargazin (40).
Fig. 8. Pharmacological PSD-95 and PICK1 inhibition differentially influence the brain proteome in a region-specific manner.
(A) Schematic anatomical location of dissected brain regions following 10 days of treatment. (B) PCA of all samples from different brain regions and treatment conditions. (C) UpSet plot for the number of unique and shared proteins that are differentially regulated with either UCCB01-147 (D) or mPD5 treatment compared to vehicle across brain regions. (E) Heatmap of log2 fold change (logFC) for the 35 proteins that respond similarly across brain regions with mPD5 treatment compared to vehicle. (F) Venn diagrams comparing similar or unique proteins that are differentially regulated with mPD5 or UCCB01-147 treatment. (G) Volcano plots for proteins logFC with UCCB01-147 (H) or mPD5 treatment compared to vehicle in the brainstem. (I) Heatmap of row-wise z-scored logFCs clustered by row for all proteins with an adjusted P < 0.05 and an absolute LogFC > 1.5. (J) Overrepresentation analysis for enriched gene ontologies (CC, BP, and MF) within each cluster with the total proteome set as background. (K) LogFC to either mPD5 or UCCB01-147 compared to vehicle in each brain region for significantly regulated proteins that are also interactors with PICK1 and PSD-95.
The widespread impact on the proteome beyond the hypothalamus and the brainstem led us to investigate the effects of UCCB01-147 and mPD5 on sucrose preference as an indicator of hedonic consumption. We noted a nonstatistically significant 18% reduction in sucrose preference following the administration of both compounds, with mPD5 exhibiting the most pronounced effect (P = 0.08) (fig. S9, A to D). Further investigation into the effects of targeting PSD-95 and PICK1 on the motivational aspects of feeding presents an interesting avenue for future research.
Pharmacological inhibition of PSD-95 and PICK1 yields sustained weight loss effects in obese mice
Pharmacological interference with glutamate receptors can induce lasting neuroplastic effects by diminishing excitatory neurotransmission, fostering changes in neural circuits, and possibly eliciting sustained disease-modifying benefits. To evaluate the long-term metabolic effects of mPD5 and UCCB01-147 treatment, we conducted a 13-day treatment study followed by a 29-day wash-out/weight recovery period. In this study, both compounds elicited significant weight-lowering benefits over the 13-day treatment period (fig. S10, A to C). Consistent with prior studies, weight loss for both compounds was solely reflected in a reduction of body fat mass (fig. S10D). Notably, although the half-life of both PDZ domain–interfering compounds is in the range of minutes to hours (35), the weight loss was sustained for ~10 days following treatment cessation. Four weeks after treatment cessation, mice treated with mPD5 had still not caught up with the body weight of the vehicle-treated group (fig. S10B), suggesting that PICK1 interference might have enduring effects on body weight. In a follow-up study, the sustained weight-lowering effects of mPD5 were assessed against the GLP-1 receptor agonist, liraglutide (fig. S10, E to G). After treatment cessation, mPD5-treated mice exhibited a continued weight loss for 3 days and sustained the lost weight for approximately 8 days. In contrast, liraglutide-treated mice promptly initiated weight regain upon treatment cessation, recovering at a faster rate than mPD5-treated mice. By day 35, liraglutide-treated mice equaled the weight of the vehicle-treated group, while mPD5-treated mice maintained a significantly lower weight than the vehicle-treated group (fig. S10H). To address potential confounding factors linked to differences in weight loss between liraglutide-treated mice and mPD5-treated mice, we conducted a study comparing mPD5-treated mice with a group subjected to caloric restriction to achieve equivalent weight loss (fig. S10, I and J). Upon cessation of calorie restriction and mPD5 treatment, the groups immediately diverged in weight regain. Consistent with previous observations, mPD5-treated mice exhibited a subdued weight regain after drug cessation, but 16 days into the recovery period, the groups had reconciled. The initial separation in weight regain was reflected in a robust counterregulatory hyperphagia in the calorie-restricted group (fig. S10K). In contrast, the mPD5 group returned to consuming the same amount of calories as the control mice, showing no compensatory behavior.
DISCUSSION
PICK1 and PSD-95 have previously been identified as opportunistic targets for the pharmacological treatment of neuropathic pain and stroke, respectively (33, 41). In the present study, we integrate human genomic data with in vivo pharmacology in rodents to identify and functionally validate the implication of PSD-95 and PICK1 in body weight regulation. We demonstrate that dimeric peptide inhibitors of PICK1 and PSD-95 drive a selective reduction in body weight and fat mass and that weight loss is sustained following treatment cessation. These findings encourage further preclinical assessment of targeting glutamate receptor scaffolding proteins for the treatment of obesity.
GWAS have linked AMPA and NMDA receptor biology to BMI (9, 42). By a targeted genetic association screen, we identified two independent loci near PICK1 and two loci near DLG4 for association with BF%. We replicate a previously identified locus and highlight an additional, independent BF% locus near PICK1 (43). Similarly, we replicate a previously reported BMI association for the DLG4 locus (44) and report a further independent signal associated with both BMI and BF% in the same gene region. The DLG4 region also contains a genome-wide significant locus for birth weight (44, 45). However, the birth weight lead SNP is only moderately correlated with the BF% lead SNP we identified (r2 ~ 0.4), suggesting that distinct gene regulatory mechanisms may be implicated in the regulation of prenatal growth and adult obesity.
The effectiveness of the PICK1 inhibitor mPD5 in lowering weight could be attributed to a decrease in food intake and was reflected in an exclusive reduction in body fat mass. The brain imaging data indicate that mPD5 interacts with canonical feeding regions in the hypothalamus, including ARC and PVN, as well as in the brainstem, such as DMX and NTS. Subsequent proteomics data further revealed that mPD5 appears to be signaling throughout the CNS and regulate proteins linked to synaptic function and immune responses. We observed no aversive response to mPD5, highlighting the disparity from incretin-based compounds. Even more intriguing, the sustained weight-lowering benefits observed after treatment cessation suggest that PICK1-related interference with AMPA receptor signaling offers neuromodulatory advantages for maintaining weight loss, extending beyond explanations provided by pharmacokinetic data. While the integrated c-Fos and proteomics data offer valuable mechanistic insights, more work is needed to pinpoint the causal site of action of mPD5 for weight loss. A deeper understanding of the mode of action of mPD5 could guide further medicinal chemistry refinement, with the goal of achieving more specific central actions. Such refinement may also alleviate the temporal sedative effects associated with mPD5 treatment.
While treatment with the PSD-95 inhibitor, UCCB01-147, echoed many of the key metabolic benefits of mPD5, such as anorectic effects and exclusive loss of fat mass, we did observe noticeable differences between the two molecules. Most apparent was the absence of widespread c-Fos induction in response to acute treatment in the brain. This could imply two nonexclusive possibilities: UCCB01-147 may exert its effects through distinct brain sites with subtle impacts, and/or interference with the NMDA receptor/PSD-95/neuronal nitric oxide synthase (nNOS) complex might not trigger neuronal c-Fos expression. Given the minimal peripheral expression of PSD-95, it is challenging to envision that the weight-lowering benefits are not centrally mediated. This notion also aligns with the pronounced changes in proteins relating to glutamatergic signaling in NAc and in the brainstem following UCCB01-147 treatment. Evaluation of the role of nNOS in the weight-lowering benefits of UCCB01-147 would be an intriguing avenue for future investigations. In context, determining which aspects of NMDA receptor signaling are retained in the context of PSD-95 inhibition is important and could perhaps aid in further refinement of targeting this pathway for weight loss.
The data presented here propose PSD-95 and PICK1 as therapeutic targets for weight loss. As we progress with the preclinical work focusing on the anti-obesity benefits, it is crucial that we in parallel interrogate the physiological implication of these scaffolding proteins in energy homeostasis and feeding behavior. Rodent studies have noted reduced PSD-95 protein expression in the hippocampus and prefrontal cortex following high-fat diet feeding (46–48). However, while a functional role for NMDA receptor signaling in hypothalamic hunger regulation is emerging (49, 50), the involvement of PSD-95 remains elusive. On the basis of studies in transgenic rodent models, PICK1 has been implicated in glycemic control and deficiency of PICK1 is associated with leanness (36, 51, 52). Of consideration, insights from incretin biology and pharmacology studies underscore that loss-of-function models do not always accurately predict physiological function and pharmacological potential (53). Another crucial consideration when advancing the scrutiny of the involvement of glutamate receptor scaffolding proteins in weight homeostasis and feeding behavior is to carefully distinguish and dissociate these aspects from those linked more broadly to emotional valence, where AMPA and NMDA receptors are known to be involved (54). In terms of directionality, disrupting NMDA receptor signaling holds clinical relevance for the treatment of Alzheimer’s disease and treatment-resistant depression (55, 56). In light of these considerations, a PSD-95 inhibitor has the potential to yield pleiotropic benefits, transcending its metabolic impact. Consequently, peptide inhibitors of PSD-95 are presently undergoing clinical evaluation for stroke treatment, demonstrating a favorable safety profile for human use (25, 31, 57, 58).
In conclusion, we reveal an association between PICK1 and DLG4 (PSD-95) loci and human anthropometric traits, highlighting the importance of further functional investigations into the roles of PSD-95 and PICK1 in body weight homeostasis and obesity pathogenesis. Complementing this, we demonstrate that pharmacological inhibition of PSD-95 or PICK1 effectively reduces body weight and fat mass in mouse models of obesity. Consequently, we propose that these PDZ domain–containing postsynaptic glutamate receptor scaffolding proteins represent a previously unknown drug target class for the treatment of obesity.
MATERIALS AND METHODS
PheWAS analysis
We used publicly available GWAS summary results from the UK Biobank (http://nealelab.is/uk-biobank/) to identify genetic variants associated with BF% and BMI with ±500 kb of the transcription start site of genes/proteins containing a PDZ domain and that interact with AMPA and NMDA receptors. The UK Biobank involves in-depth genetic and phenotypic characterization of over 500,000 individuals aged 40 to 69 years of age and living in the United Kingdom. We performed LD clumping to identify lead SNPs (MAF > 0.01) in the BF% and BMI-associated loci using the pruning algorithm implemented in PLINK v.1.9 (59) within a 1-Mb window and r2 < 0.1 in the 1000Genome Phase 1 reference panel. Next, we binned the SNPs based on the P value thresholds P < 5 × 10−8 (genome-wide significance), P < 1 × 10−6, and P < 1 × 10−5. We retrieved eQTL associations for the identified lead SNPs from the online database Open Target Genetics (https://genetics.opentargets.org/) (60–63). To implement a pheWAS for the DLG4 and PICK1 loci, we clustered the UK Biobank traits into categories that describe the nature of the trait (based on ICD-11 standards, if applicable). We only considered the category “anthropometric traits” in the present analyses. The sample size for the different anthropometric traits ranged between 106,251 and 336,935 individuals.
Animals
Eight-week-old C57BL/6J mice (Janvier, France) were maintained on a high-fat diet (D12331; Research Diets, New Brunswick, NJ, USA) for a minimum of 3 months to facilitate the progressive development of obesity. Mice were kept on a 12-hour light-dark cycle in a humidity-controlled (45 to 65%) and temperature-controlled environment (21° to 23°C) with ad libitum access to high-fat diet and water until initiation of experiments. All in vivo experiments were carried out in accordance with regulations regarding the care and use of experimental animals and under the approval of the Danish Animal Experimentation Inspectorate (2018-15-0201-01457, 2023-15-0201-01442, and 2023-15-0201-01530).
Drugs
Compounds mPD5, UCCB01-147, and UCCB01-144AA were commercially generated (WuXi AppTec, Shanghai, China). All drugs were diluted in isotonic saline and dosed subcutaneously. Solutions containing mPD5 or UCCB01-147 were pH-adjusted to pH 7.4. Liraglutide was purchased at a pharmacy (Victoza, Novo Nordisk, Bagsværd, Denmark), and semaglutide was obtained through the Novo Nordisk compound sharing service (Semaglutide, Novo Nordisk, Bagsværd, Denmark).
In vivo pharmacology studies
DIO C57BL/6J mice were single- or double-housed for all studies and had ad libitum access to high-fat diet. Mice were assigned to the test group or control group based on body weight, ensuring that the average body weight of each group was the same. Unless otherwise stated, mice were sham-injected with isotonic saline 3 days preceding the start of a study.
Study 1 (UCCB01-147 dose-response study)
DIO mice (average body weight 45 g, single-housed) were weighed 3 days before the first injection, and body composition was measured by magnetic resonance imaging (EchoMRI, Echo Medical System LLC, USA). DIO mice were subcutaneously injected with either UCCB01-147 (25 mg kg−1; n = 7 mice and n = 7 cages), UCCB01-147 (75 mg kg−1; n = 7 mice and n = 7 cages), or isotonic saline (n = 6 mice and n = 6 cages) daily for 9 days. Injections were performed 1 to 3 hours before the onset of the dark phase. On the seventh day, body composition was measured again in the morning and mice were subjected to a glucose tolerance test in the afternoon. Food intake and body weight were measured daily for the duration of the experiment. Two mice in the 75 mg kg−1 UCCB01-147 group were excluded from analyses because of adverse effects (sluggish movement and hypothermia). One mouse in the vehicle group was excluded because of excessive weight loss.
Study 2 (UCCB01-147 indirect calorimetry)
DIO mice (average body weight 42 g, single-housed) were acclimatized to indirect calorimetry cages for 1 week and sham-injected with isotonic saline 5 days before the start of the study. Mice were injected with UCCB01-147 (25 mg kg−1; n = 8 mice and n = 8 cages) or isotonic saline (n = 8 mice and n = 8 cages) daily for 7 days. Injections were performed 2 hours before the onset of the dark phase. Food intake and body weight were measured daily independently of the measurements taken by the automatic system (TSE System, Germany). Respiratory exchange ratio (volume of CO2 production:volume of O2 consumption), locomotor activity, food intake, and water intake were automatically measured every 20 min for the duration of the study. One mouse in the vehicle group was excluded because of poor health (visual inspection).
Study 3 (NA-1 weight loss)
DIO mice (average body weight 55 g, single-housed) were subcutaneously injected with UCCB01-147 (25 mg kg−1; n = 8 mice and n = 8 cages), a molar-matched dose of NA-1 (16.3 mg kg−1; n = 8 mice and n = 8 cages), or isotonic saline (n = 8 mice and n = 8 cages) 2 hours before the onset of the dark phase. Food intake and body weight were measured daily for the duration of the study. One vehicle mouse and one UCCB01-147–treated mouse were excluded during the study because of poor health (visual inspection).
Study 4 (comparative study of UCCB01-147 and UCCB01-144AA)
DIO mice (average body weight 49 g, single-housed) were injected once daily with UCCB01-147 (25 mg kg−1; n = 6 mice and n = 6 cages), UCCB01-144AA (25 mg kg−1; n = 6 mice and n = 6 cages, negative control), or isotonic saline (n = 6 mice and n = 6 cages) for 5 days. Injections were performed approximately 2 hours before the onset of the dark phase. Food intake and body weight were measured daily for the duration of the study.
Study 5 (mPD5 dose-response study, single-housed)
DIO mice (average body weight 51 g) were weighed 3 days before the first injection, and body composition was measured by magnetic resonance imaging (EchoMRI, Echo Medical System LLC, USA). Mice were subcutaneously injected once daily with mPD5 (19 mg kg−1; n = 6 mice and n = 6 cages), mPD5 (56 mg kg−1; n = 6 mice and n = 6 cages), or isotonic saline (n = 6 mice and n = 6 cages) for 7 days. Injections were performed 1 to 3 hours before the onset of the dark phase. On the seventh day, body composition was measured in the morning and mice underwent a glucose tolerance test in the afternoon. Food intake and body weight were measured daily for the duration of the study.
Study 6 (evaluation of weight loss recovery of UCCB01-147 and mPD5 relative to liraglutide)
DIO mice (average body weight 48 g; double-housed) were weighed and had body composition measured 2 days before the first injection (EchoMRI, Echo Medical System LLC, USA). Mice were subcutaneously injected with mPD5 (19 mg kg−1; n = 8 mice), mPD5 (56 mg kg−1; n = 8 mice and n = 4 cages), liraglutide (0.09 mg kg−1; n = 8 mice and n = 4 cages), or isotonic saline (n = 8 mice and n = 4 cages) daily for 9 days. Injections were performed 1 to 3 hours before the onset of the dark phase. Body weight was measured daily during pharmacological treatment and until day 21, whereafter body weight was monitored less frequently up until day 35.
Study 7 (UCCB01-147 versus mPD5 comparative study)
DIO mice (average body weight 53 g; double-housed) were weighed 3 days before the first injection, and body composition was measured (EchoMRI, Echo Medical System LLC, USA). Mice were treated with once-daily subcutaneous injections of mPD5 (56 mg kg−1; n = 8 mice and n = 4 cages), UCCB01-147 (25 mg kg−1; n = 8 mice and n = 4 cages), or isotonic saline (n = 8 mice and n = 4 cages) for 13 days. Injections were performed 1 to 3 hours before the onset of the dark phase. Body composition was measured again 13 days after the first injection. Body weight was measured daily for the duration of the pharmacological treatment period and at indicated time points thereafter to track recovery up until day 42.
Study 8 (UCCB01-147 weight loss study)
DIO mice (average body weight 60 g, single- and double-housed) were grouped 3 days before study start based on body weight such that each experimental group had approximately the same average body weight, and their body composition was determined by magnetic resonance imaging (EchoMRI, Echo Medical System LLC, USA). DIO mice were treated with once-daily subcutaneous injections of either UCCB01-147 (25 mg kg−1; n = 10 mice and n = 7 cages) or isotonic saline (n = 9 mice and n = 6 cages) for 14 days. Injections were performed 1 to 3 hours before the onset of the dark phase. On the first day of the study, a compound tolerance test was conducted, measuring blood glucose levels at time points 0, 0.5, 1, 2, 6, and 24 hours after administration of the first dose with a handheld glucometer (Contour XT, Bayer). On day 14, body composition was measured in the morning and mice were subjected to an intraperitoneal glucose tolerance test in the afternoon, measuring blood glucose levels at time points 0, 15, 30, 60, and 120 min after challenging the mice with an intraperitoneal glucose injection. Food intake and body weight were measured daily for the duration of the experiment.
Study 9 (mPD5 weight loss study)
DIO mice (with an average body weight of 60 g, double-housed) underwent weighing 3 days before the initial injection to make experimental groups based on body, and their body composition was assessed using magnetic resonance imaging (EchoMRI, Echo Medical System LLC, USA). Subsequently, DIO mice were treated with once-daily subcutaneous injections of either mPD5 (56 mg kg−1; n = 9 mice and n = 5 cages) or isotonic saline (n = 9 mice and n = 5 cages) for 14 days. Injections were administered 1 to 3 hours before the start of the dark phase. On the first day of the study, a compound tolerance test was conducted, measuring blood glucose levels at time points 0, 0.5, 1, 2, 6, and 24 hours after administration of the first dose using a handheld glucometer (Contour XT, Bayer). On day 14, body composition was measured in the morning, and in the afternoon, mice underwent an intraperitoneal glucose tolerance test, measuring blood glucose levels at time points 0, 15, 30, 60, and 120 min after the administration of an intraperitoneal glucose injection. Daily measurements of food intake and body weight were recorded throughout the experiment. One mouse and one cage were excluded from the mPD5 treatment group because of sickness. One mouse was excluded from the vehicle group because of constipation.
Study 10 (mPD5 and UCCB01-147 indirect calorimetry)
DIO mice (average body weight 60 g; single-housed) were acclimatized to metabolic cages (16-channel Promethion, Sable Systems International, NV, USA) for 7 days before initiation of the study. Oxygen consumption (VO2), carbon dioxide production (VCO2), respiratory exchange ratio, energy expenditure (in kilocalories per hour), and locomotor activity (in centimeters per second) were recorded every 15 min. Water was available ad libitum throughout the study period, and two pellets of high-fat diet were provided every day and the old ones were removed. On the day of initiation of the study, mice were divided into three experimental groups with similar average body weights and assigned to receive once-daily subcutaneous injections of mPD5 (56 mg kg−1; n = 10 mice and n = 10 cages), UCCB01-147 (25 mg kg−1; n = 10 mice and n = 10 cages), or vehicle (isotonic saline, n = 10 mice and n = 10 cages) for 10 days. Body weight and food intake were measured manually each day at the time of injection, and dosing was carried out at a volume of 5 μl g−1. Mice assigned to the different experimental groups were randomly distributed across two systems to correct for potential space-related bias. Raw data were analyzed using the online tool CalR (version 1.3, www.calrapp.org) and visualized using GraphPad Prism. One mouse in the vehicle group and one mouse in the mPD5 group were excluded because of constipation. One mouse in UCCB01-147 was excluded from analysis of locomotor activity and respiratory exchange ratio (outlier).
Study 11 (weight loss maintenance of mPD5 relative to mice receiving calorie restriction to match the weight loss trajectory)
DIO male C57BL/6J mice (average body weight 56 g; double-housed) were sham-injected 3 days before initiation of the study. Mice were grouped on the basis of body weight such that each experimental group had the same average body weight. The experimental groups were assigned to receive treatment with either mPD5 (56 mg kg−1; n = 8 mice and n = 4 cages), calorie restriction to match the weight loss of mPD5-treated mice (n = 8 mice and n = 4 cages), or vehicle (isotonic saline; n = 8 mice and n = 4 cages) for 11 days. Injections were performed approximately 2 hours before the onset of the dark phase. For the 11-day treatment period, food intake and body weight were measured daily at the time of injection, while food intake and body weight were measured once every 2 days during the recovery phase. One mouse was euthanized during the study in the 56 mg kg−1 mPD5 group due to sickness.
Study 12 (mPD5 PICK1 KO study)
DIO male wild-type and global PICK1 knockout mice (wild type, average body weight 36 g; global PICK knockout, average body weight 34 g; single-housed) were sham-injected 3 days before the study started. Mice were grouped on the basis of body weight such that each group had approximately the same average body weight and was assigned to receive once-daily subcutaneous injections of mPD5 (56 mg kg−1; wild type; n = 4 mice, n = 4 cages; global PICK1 knockout; n = 6 mice and n = 6 cages) or isotonic saline (wild type; n = 4 mice and n = 4 cages; global PICK1 knockout; n = 6 mice and n = 6 cages) for 7 days. Injections were performed approximately 2 hours before the onset of the dark phase. Food intake and body weight were measured daily at the time of injection.
Study 13 (mPD5 and UCCB01-147 open field test)
Locomotor activity in response to treatment with mPD5 or UCCB01-147 was evaluated using an open-field test. Lean male C57BL/6J mice were habituated in the procedure room 7 days before initiation of the experiment. The experiment was conducted by giving a single subcutaneous administration of mPD5 (56 mg kg−1 n = 10 mice), UCCB01-147 (25 mg kg-1; n = 10 mice), or vehicle (isotonic saline, n = 10 mice) 1 hour before recording their movement via ceiling-mounted Logitech C920 Pro cameras (1080 × 1080 pixels, 30 frames per second, Logitech software). Mice were divided into four groups based on body weight, and their movement was recorded for 30 min. Each run was conducted with one mouse from each treatment group and with run-to-run alternation so that treatments were equally distributed across all chambers. Movement traces and quantification of locomotor activity (velocity and distance traveled) were obtained using Noldus EthoVision XT software (Noldus, The Netherlands).
Study 14 (mPD5 and UCCB01-147 conditioned taste aversion relative to semaglutide)
A week before the study started (day –7), lean male C57BL/6J mice (average body weight 26 g, single-housed) were single-housed with ad libitum access to a chow diet, and one bottle of water (food and water were placed in the cage lid so that there was room for two water bottles) in a scantainer. The position of the water bottle was alternated daily between sides to minimize the development of any side preference. The animals were weighed and handled daily from day −3. On day −3, mice were exposed to a bottle containing 0.1% saccharin-flavored water with high palatability followed by subcutaneous administration of mPD5 (56 mg kg−1; n = 10 mice), UCCB01-147 (25 mg kg−1; n = 10 mice and n = 10 cages), semaglutide (30 nmol kg−1; n = 10 mice and n = 10 cages), or vehicle (isotonic saline, n = 10 mice and n = 10 cages). Three days after the first dosing, mice were exposed to a two-bottle taste preference test, i.e., a voluntary choice between tap water and the 0.1% saccharin solution. Saccharin and water intake was monitored for 24 hours followed by preference determination.
Study 15 (UCCB01-147 conditioned taste aversion)
An identical protocol to the one described for study 14 was applied, treating lean male C57BL/6J mice (average body weight 28 g) with subcutaneous injections of UCCB01-147 (25 mg kg-1; n = 10 mice and n = 10 cages) and vehicle (isotonic saline, n = 10 mice and n = 10 cages) as experimental groups.
Study 16 (mPD5 conditioned taste aversion)
An identical protocol to the one described for study 14 was applied, treating lean male C57BL/6J mice (average body weight 27 g) with subcutaneous injections of mPD5 (56 mg kg−1; n = 10 mice and n = 10 cages) and vehicle (isotonic saline, n = 10 mice and n = 10 cages) as experimental groups.
Study 17 (mPD5 and UCCB01-147 whole-brain c-Fos imaging)
DIO male C57BL6J mice (average body weight 45 g, single-housed) were injected with a single acute injection of mPD5 (56 mg kg−1; n = 8 mice), UCCB01-147 (25 mg kg-1; n = 8 mice), or vehicle (isotonic saline, n = 6 mice). Two hours after, mice were deeply anesthetized with pentobarbital and perfused intracardially with phosphate-buffered saline (PBS) and heparin (15,000 IU liter−1) for 2 min followed by neutral-buffered formalin (10%, Sigma-Aldrich, Denmark) for 5 min. Brains were harvested and postfixed overnight in neutral-buffered formalin at 4°C.
Study 18 (mPD5 and UCCB01-147 MS-based proteomics of hypothalamus, brainstem, nucleus accumbens, prefrontal cortex, cerebellum, and midbrain)
DIO mice (average body weight 53 g; double-housed) were sham-injected 3 days before initiation of the study. Mice were treated with once-daily subcutaneous injections of mPD5 (56 mg kg−1; n = 8 mice and n = 4 cages), UCCB01-147 (25 mg kg−1; n = 8 mice and n = 4 cages), or vehicle (isotonic saline, n = 8 mice and n = 4 cages) for 10 days. Injections were performed 2 hours before the onset of the dark phase. Food intake and body weight were measured daily at the time of injection for the duration of the experiment. On day 10, mice were euthanized by cervical dislocation and the brains were harvested and microdissected. Brain areas of interest were isolated using a coronal brain matrix. After isolation, tissue was snap-frozen and subsequently analyzed using liquid chromatography–MS/MS (LC-MS/MS) by the Proteomics Research Infrastructure at the University of Copenhagen.
Study 19 (bulk RNA sequencing study of hypothalami)
DIO mice (average body weight 53 g, double-housed) were injected with mPD5 (56 mg kg-1; n = 8 mice), UCCB01-147 (25 mg kg-1; n = 8 mice), or isotonic saline (n = 8 mice) daily for 5 days. On experimental days 1 to 4, injections were performed 2 hours before the onset of the dark phase. Body weight and food intake were measured daily. On the morning of the fifth experimental day, mice were injected and sacrificed 2 hours later. Whole hypothalami were frozen on dry ice in preparation for RNA isolation.
Study 20 (sucrose preference test)
DIO mice (average body weight 41 g, single-housed) were habituated in a scantainer for 7 days with ad libitum access to high-fat, high-sucrose diet, one bottle of tap water, and one bottle with a solution of 3% sucrose in tap water. Animal were treated with daily sham injections from day 0 to 2, and their food, water, and sucrose intake was measured at the time of injection. On day 2, mice were randomized on the basis of body weight, such that each group had the same average body weight, and assigned to receive once-daily subcutaneous treatment with mPD5 (56 mg kg−1; n = 9 mice and n = 9 cages), UCCB01-147 (25 mg kg−1; n = 9 mice and n = 9 cages), or isotonic saline (n = 9 mice and n = 9 cages) for 2 days. Sucrose and water intake was monitored daily followed by preference determination.
Study 21 (gene expression profiles of Pick1, Dlg4, AMPA, and NMDA receptor subunits)
Naïve lean mice (n = 6 mice) were euthanized by decapitation, and tissues were harvested and frozen immediately after dissection on dry ice (hypothalamus, white adipose tissue, BAT, heart, kidney, muscle, liver, and colon). The tissue was subjected to gene expression profiling, with the protocol outlined in the “Gene expression analysis (qPCR)” section.
Tissue clearing and whole-brain immunolabeling
Whole-brain clearing and staining was inspired by the iDISCO+ protocol (64). All the steps were performed at room temperature with gentle shaking unless otherwise specified. All the buffers were supplemented with 0.01% sodium azide (Sigma-Aldrich) to prevent bacterial and fungal growth. For delipidation and decolorization purposes, perfused brains were incubated with a 25% Quadrol [N,N,N′,N′-tetrakis(2-hydroxypropyl)ethylenediamine] solution in water at 37°C for 3 days. Then, the samples were transferred to a 5% ammonium solution in water (no wash in between) and incubated at 37°C for 1 day. Next, the samples were washed once in water for approximately 10 min before an increasing series of methanol (MeOH) (VWR) dilutions in water (washes of 1 hour in MeOH 20%, 40%, 60%, 80%, and twice 100%). Once dehydrated, samples were incubated overnight in a solution containing a 66% dichloromethane (DCM) (Sigma-Aldrich) in MeOH. Samples were then washed a few times in 100% MeOH throughout the day and bleached overnight at 4°C in a 5% hydrogen peroxide (Sigma-Aldrich) solution in MeOH (the samples were brought to 4°C before transferring them to the bleaching solution). Rehydration was done by incubating the samples in MeOH 60%, 40%, and 20% (1 hour each wash). After MeOH pretreatment, samples were washed in PBS twice for 15 min and 1 hour in PBS containing 0.2% Triton X-100 (Sigma-Aldrich) and further permeabilized by a 24-hour incubation at 37°C in permeabilization solution composed of 20% dimethyl sulfoxide (Sigma-Aldrich) and 2.3% glycine (Sigma-Aldrich) in PBS-T. To start the immunostaining, samples were first blocked with 0.2% gelatin (Sigma-Aldrich) in PBS-T for 24 hours at 37°C, and the same blocking buffer was used to prepare antibody solutions. The primary antibody against c-Fos (Cell Signaling Technology, no. 2250, 1:3000) was incubated for 11 days at 37°C with gentle shaking, then washed in PBS-T for 2 days (the wash solution was changed five times), and finally incubated for 8 days with secondary antibodies conjugated to an Alexa Fluor 647 fluorophore (Jackson ImmunoResearch, no. 711-605-152, 1:1000). After immunostaining, the samples were washed in PBS-T for 3 days (the wash solution was changed five times) and postfixed with 2% paraformaldehyde in PBS overnight at 4°C before the final dehydration. For the final dehydration, the samples were passed through a MeOH/water increasing concentration series: 20%, 40%, 60%, 80%, and 100% (1 hour each), followed by a wash in 66% DCM—33% MeOH overnight. MeOH was washed out with two final washes in DCM 100% (30 min each), and finally, the samples were transparized and stored in ethyl cinnamate (Sigma-Aldrich) for at least 3 days, at room temperature and in the dark, until imaging took place.
Light sheet microscopy
The images were acquired using a Zeiss LS7 Scanning Gaussian-beam Lightsheet Microscope with orthogonal light paths for illumination and detection. Two 5× illumination objectives [Carl Zeiss, numerical aperture (NA) 0.1] were used to generate dual-side illumination of the sample, together with an EC Plan-NEO 5× detection objective (Carl Zeiss, NA 0.16, 10.5-mm working distance). Images were acquired with two PCO Edge 4.2 M sCMOS cameras using Zeiss Zen Black software. The laser line used to capture the autofluorescence signal in the sample was 488 and 638 nm for the c-Fos–specific channel. To refine the emission signal, additional filters were used, i.e., BP (band-pass) 505–545 and LP (long-pass) 660 for the corresponding excitation.
Whole-brain c-Fos quantification
The tiled images were first stitched with a custom Python script using Terastitcher (65). The stitched images were then processed using the ClearMap 2 software (66). This enabled the generation of voxel maps depicting Fos cell densities and region-based cell count statistics. Briefly, the images underwent background removal processing. Local maxima were then detected to identify the initial cell locations. The locations served as seeds for a watershed algorithm, facilitating the estimation of cell volumes. Cells were filtered on the basis of their volume to eliminate smaller, artifactual maxima. Elastix (67) (available at https://elastix.lumc.nl) was used to align the autofluorescence images for each brain image to a reference atlas, and the coordinates of the filtered cells were transformed to a reference coordinate space (68). For the construction of voxel maps, each filtered cell was represented as a sphere of 350-μm diameter. Region- and voxel-wise statistics between the experimental groups were performed using the SciPy implementation of the Mann-Whitney U test. Last, we created volcano plots to represent the regional counts statistically.
Intraperitoneal glucose tolerance test
An intraperitoneal glucose tolerance test was performed on day 9 of studies 1 and 6. Mice were fasted for 5 hours before receiving a single intraperitoneal injection of glucose (2 g/kg) dissolved in isotonic saline. Blood glucose was measured at time points 0, 15, 30, 60, and 120 min after injection using a handheld glucometer (Contour XT, Bayer).
Gene expression analysis (qPCR)
BAT, inguinal white adipose tissue, and liver from mouse studies 8 and 9 were quickly dissected, frozen on dry ice, and stored at −80°C. Hypothalamus, inguinal white adipose tissue, BAT, heart, kidney, quadriceps, liver, and colon tissues were harvested from naïve mice. For RNA extraction, all tissues were homogenized in TRIzol reagent (QIAzol Lysis Reagent, Qiagen, 79306) with a stainless-steel bead (Qiagen, 69989) using TissueLyser II (Qiagen, 85300) set at 30 Hz for 3 min. Total RNA was isolated according to the manufacturer’s protocol using QIAwave RNA Mini Kit (Qiagen, 74534), RNA was quantified, and RNA quality was assessed using NanoDrop 2000 (Thermo Fisher Scientific, ND-2000). Total RNA was then converted to cDNA by mixing with FS buffer (Thermo Fisher Scientific, 18080-044), dithiothreitol (Thermo Fisher Scientific, 18080-044), and random primers (Sigma-Aldrich, 11034731001) followed by incubation at 70°C for 3 min in a thermal cycler (Eppendorf Mastercycler Pro) for first-strand synthesis. dNTP (Thermo Fisher Scientific, R0192), RNaseOUT (Thermo Fisher Scientific, 10777019), and SuperScript III (Thermo Fisher Scientific, 18080-044) were subsequently added for cDNA synthesis using a thermal cycler (Eppendorf Mastercycler Pro) with the following steps: 5 min at 25°C, 60 min at 50°C, and 15 min at 70°C. cDNA was diluted 1:20 for hypothalamus and 1:100 for all other tissues and stored at −20°C. Gene expression was analyzed by quantitative polymerase chain reaction (qPCR). cDNA, primers, and PrecisionPLUS qPCR Master Mix with SYBRgreen (Primer Design) were mixed in a 384-well plate and incubated in a LightCycler (Roche, LightCycler 480 II) using the following steps: 2 min at 95°C, 45 cycles of 60 s at 60°C, and, last, a melting curve analysis was performed. Primers are listed in table S3.
Transcriptomic analysis
Total RNA was isolated using RNeasy mini kit (Qiagen, no. 74106) according to the manufacturer’s protocol. Messenger RNA sequencing libraries were prepared using the Illumina TruSeq Stranded mRNA protocol (Illumina). Poly-A–containing mRNAs were purified by poly-T attached magnetic beads and fragmented, and cDNA was synthesized using SuperScript III reverse transcriptase (Thermo Fisher Scientific). cDNA was adenylated to prime for adapter ligation, and after a clean-up using AMPure beads (Beckman Coulter), DNA fragments were amplified using PCR followed by a final clean-up. Libraries were quality-controlled using a fragment analyzer instrument (Agilent Technologies) and subjected to 52–base pair (bp) paired-end sequencing on NovaSeq 6000 (Illumina). A total of 2.7 billion reads were generated.
Fastq files were first generated using bcl2fastq2 v. 2.20.0. The nf-core/rnaseq-pipeline v. 3.8 with the flags --with_UMI and --umitools_bc_pattern NNNNNNNN set was then used to align the reads to the GRCm38 primary assembly and produce corresponding gene counts (48). A multidimensional scaling plot was used to detect outlying samples. The gene expression of potential outliers was compared to the rest of the samples to look for evidence of inflammation or tissue contamination. Two samples were excluded because of likely contamination from surrounding tissue.
Differential expression testing
Genes with low expression were filtered on the basis of a threshold of 25 counts over at least five samples. The count matrix was analyzed using DESeq2 to perform count normalization and differential expression analyses between treatment groups. Benjamini-Hochberg correction was used. DESeq2 analysis was performed incorporating a design considering both the treatment and extraction pool. Pairwise differential expression results were generated for UCCB01-147 versus vehicle and mPD5 versus vehicle comparisons. The results were annotated with gene symbols using the org.Mm.eg.db database. Volcano plots were generated to visualize differentially expressed genes using the R package gglot2. Genes with absolute log2FoldChange < 2.5 were considered, and points were color-coded on the basis of adjusted P values (Padj): light orange for Padj < 0.05 and log2FoldChange < 0, dark orange for Padj < 0.05 and log2FoldChange < 0, and gray for Padj ≥ 0.05.
Functional enrichment analyses
Gene set enrichment analysis (GSEA) was performed to identify enriched pathways in each comparison. Gene lists were extracted from the DESeq2 results and ranked on the basis of log10(Padj)*log2FoldChange, and GSEA was conducted using the gseGO function from the clusterProfiler package (69). gseGO parameters were set to OrgDb = org.Mm.eg.db, ont = “ALL,” keyType = “ENSEMBL,” minGSSize = 10, maxGSSize = 300, and pvalueCutoff = 0.05. GSEA results were visualized using GraphPad Prism version 10.
MS-based proteomics
Sample preparation
Brain tissue samples were lysed with lysis buffer [1% (w/v) sodium deoxycholate, 100 mM tris-HCl (pH 8.5), and phosphatase inhibitor cocktail (Roche)] using the BeatBox tissue disruptor (Preomics) with 15 cycles of 30-s BeatBox high setting followed by 2-min incubation on ice. Samples were incubated for 10 min at 95°C followed by 10 min of BeatBox in medium setting. Samples were reduced with 5 mM (final concentration) TCEP for 15 min at 55°C, alkylated with 20 mM (final concentration) CAA for 30 min at room temperature, and digested adding trypsin/LysC at a 1:50 enzyme/protein ratio. Peptides were cleaned up using iST PSI desalting plates (Preomics).
LC-MS (DIA)
Peptides were separated on a 110-cm mPAC HPLC column (Thermo Fisher Scientific) with a Vanquish Neo HPLC system (Thermo Fisher Scientific) coupled through a nano-electrospray source to a Tribrid Ascend mass spectrometer (Thermo Fisher Scientific) with a nonlinear gradient of 1 to 50% buffer B (0.1% formic acid and 80% acetonitrile) at a flow rate of 300 nl/min over 120 min. Spray voltage was set to 2200 (V) on a 10-μm-inner-diameter fused silica emitter (Evosep). The column temperature was kept at 50°C. Samples were acquired using a DIA data acquisition method, where the Tribrid mass spectrometer was performing a full scan [120-K resolution, scan range 390 to 1010 mass/charge ratio (m/z), auto maximum injection time, AGC target 100%], followed by 75 DIA windows (8 m/z each) of MS/MS scans in the Orbitrap analyzer (scan range 400 to 1000 m/z, 15-K resolution, 27-ms maximum injection time, AGC target 100%). Normalized HCD collision energy (%) was set to 30, and 1 m/z DIA windows overlap was allowed.
Bioinformatics
The ProteinGroup file from Spectronaut was loaded into R Studio V.4.2.1. Raw intensities were log2-transformed and filtered to ensure at least 50% quantification in each condition (brain region/treatment). The data were then median-scaled, and differential abundance analysis was performed using LIMMA (70). Treatment comparisons (UCCB01-147 versus vehicle, mPD5 versus vehicle) within each brain region were extracted. The eBayes function was then used for the empirical smoothening of SEs and the moderation of t statistics. A Benjamini-Hochberg–corrected P value of <0.05 and a fold change of 1.5 were set as the significance cutoffs. PCA was conducted on the complete proteome observations using the prcomp function (stats package). Heatmaps were made in Pheatmap package (1.0.12), and the number of clusters was determined by measuring the within-cluster sum of squares, average silhouette width, and Dunn index. Within-cluster overrepresentation analysis was conducted with the clusterProfiler (71) package (4.6.2) using the enrichGO function with universe set to all proteins with gene names identified, all ontologies specified, pvalueCutoff < 0.05, and qvalueCutoff < 0.2.
Statistical analyses
Statistical analyses were performed in GraphPad Prism version 9; statistical tests used are described in respective figure legends. All data are presented as means ± SEM, and all P values ≤ 0.05 were considered statistically significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Acknowledgments
We thank L. Sachs for technical assistance. We acknowledge the Single-Cell Omics Platform and the Rodent Metabolic Phenotyping Platform at the Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR) for technical expertise and support. Mass spectrometry analyses were performed by the Proteomics Research Infrastructure (PRI) at the University of Copenhagen (UCPH), supported by the Novo Nordisk Foundation (NNF) (grant agreement number NNF19SA0059305).
Funding: This work was supported by a research grant from the Novo Nordisk Foundation (grant number: NNF18CC0033668 to N.F.), research grants from the Lundbeck Foundation (fellowship R238-2016-2859), and the Novo Nordisk Foundation support CC (grant numbers: NNF17OC0026114 and NNF22OC0073778). K.L.M. is supported by the Lundbeck Foundation (R344-2020-1063) and by the Novo Nordisk Foundation (NNF21OC0070287). The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center, based at the University of Copenhagen, Denmark, and partially funded by an unconditional donation from the Novo Nordisk Foundation (www.cbmr.ku.dk) (grant number NNF18CC0034900).
Author contributions: N.F., J.P., and C.C. conceptualized the research. S.M. performed the pheWAS analyses. K.S. and A.B. developed and synthesized UCCB01-147. K.L.M. developed and synthesized mPD5. J.P., N.F., A.S.B.-R., A.J., R.C.A., and C.V.M. conducted in vivo pharmacology studies. J.P., A.J., T.T., G.S., G.H., and S.E.J. conducted whole-brain c-Fos expression study. N.K.K., J.K.L., J.P., M.A., A.J., C.S., J.H.S., and A.S.D. conducted proteomic studies. N.F. and C.V.M. conducted RNA sequencing study. All authors provided input for in vivo studies. J.P., N.F., S.M., T.O.K., and C.C. wrote the manuscript. All authors contributed to the editing and proofreading of the final draft.
Competing interests: C.C. and J.P. are cofounders of Ousia Pharma ApS, a biotech company developing therapeutics for treatment of obesity. A.B. and K.S. are cofounders of Avilex Pharma, a biotech company developing neuroprotectants for the treatment of stroke. K.L.M. is a co-inventor of a filed patent application disclosing the lipidated dimeric peptides, their usage, and their extended utilization (WO2021/176094; filing date,3 June 2020). The patent application is filed by the University of Copenhagen and is licensed to Zyneyro ApS. K.L.M. is a cofounder of Zyneyro ApS, a biotech company developing drugs for the treatment of chronic pain. A.S.B.-R. is an employee of Novo Nordisk A/S. The other coauthors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data have been submitted to GEO and are publicly available as of the date of publication (accession number: GSE221549). The scripts used to analyze the RNA-seq data are available at Zenodo (10.5281/zenodo.10276328). The mass spectrometry–based proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier (PXD047515).
Supplementary Materials
This PDF file includes:
Tables S1 to S3
Figs. S1 to S10
REFERENCES AND NOTES
- 1.Kelly T., Yang W., Chen C. S., Reynolds K., He J., Global burden of obesity in 2005 and projections to 2030. Int. J. Obes. 32, 1431–1437 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Bauer U. E., Briss P. A., Goodman R. A., Bowman B. A., Prevention of chronic disease in the 21st century: Elimination of the leading preventable causes of premature death and disability in the USA. Lancet 384, 45–52 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Must A., Spadano J., Coakley E. H., Field A. E., Colditz G., Dietz W. H., The disease burden associated with overweight and obesity. JAMA 282, 1523–1529 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Lauby-Secretan B., Scoccianti C., Loomis D., Grosse Y., Bianchini F., Straif K.; International Agency for Research on Cancer Handbook Working Group , Body fatness and cancer—Viewpoint of the IARC Working Group. N. Engl. J. Med. 375, 794–798 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilding J. P. H., Batterham R. L., Calanna S., Davies M., Van Gaal L. F., Lingvay I., McGowan B. M., Rosenstock J., Tran M. T. D., Wadden T. A., Wharton S., Yokote K., Zeuthen N., Kushner R. F.; STEP 1 Study Group , Once-weekly semaglutide in adults with overweight or obesity. N. Engl. J. Med. 384, 989–1002 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Jastreboff A. M., Aronne L. J., Ahmad N. N., Wharton S., Connery L., Alves B., Kiyosue A., Zhang S., Liu B., Bunck M. C., Stefanski A.; SURMOUNT-1 Investigators , Tirzepatide once weekly for the treatment of obesity. N. Engl. J. Med. 387, 205–216 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Wilding J. P. H., Batterham R. L., Davies M., Van Gaal L. F., Kandler K., Konakli K., Lingvay I., McGowan B. M., Oral T. K., Rosenstock J., Wadden T. A., Wharton S., Yokote K., Kushner R. F.; STEP 1 Study Group , Weight regain and cardiometabolic effects after withdrawal of semaglutide: The STEP 1 trial extension. Diabetes Obes. Metab. 24, 1553–1564 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claussnitzer M., Susztak K., Gaining insight into metabolic diseases from human genetic discoveries. Trends Genet. 37, 1081–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locke A. E., Kahali B., Berndt S. I., Justice A. E., Pers T. H., Day F. R., Powell C., Vedantam S., Buchkovich M. L., Yang J., Croteau-Chonka D. C., Esko T., Fall T., Ferreira T., Gustafsson S., Kutalik Z., Luan J., Mägi R., Randall J. C., Winkler T. W., Wood A. R., Workalemahu T., Faul J. D., Smith J. A., Zhao J. H., Zhao W., Chen J., Fehrmann R., Hedman Å. K., Karjalainen J., Schmidt E. M., Absher D., Amin N., Anderson D., Beekman M., Bolton J. L., Bragg-Gresham J. L., Buyske S., Demirkan A., Deng G., Ehret G. B., Feenstra B., Feitosa M. F., Fischer K., Goel A., Gong J., Jackson A. U., Kanoni S., Kleber M. E., Kristiansson K., Lim U., Lotay V., Mangino M., Leach I. M., Medina-Gomez C., Medland S. E., Nalls M. A., Palmer C. D., Pasko D., Pechlivanis S., Peters M. J., Prokopenko I., Shungin D., Stančáková A., Strawbridge R. J., Sung Y. J., Tanaka T., Teumer A., Trompet S., van der Laan S. W., van Setten J., Van Vliet-Ostaptchouk J. V., Wang Z., Yengo L., Zhang W., Isaacs A., Albrecht E., Ärnlöv J., Arscott G. M., Attwood A. P., Bandinelli S., Barrett A., Bas I. N., Bellis C., Bennett A. J., Berne C., Blagieva R., Blüher M., Böhringer S., Bonnycastle L. L., Böttcher Y., Boyd H. A., Bruinenberg M., Caspersen I. H., Chen Y.-D. I., Clarke R., Warwick Daw E., de Craen A. J. M., Delgado G., Dimitriou M., Doney A. S. F., Eklund N., Estrada K., Eury E., Folkersen L., Fraser R. M., Garcia M. E., Geller F., Giedraitis V., Gigante B., Go A. S., Golay A., Goodall A. H., Gordon S. D., Gorski M., Grabe H.-J., Grallert H., Grammer T. B., Gräßler J., Grönberg H., Groves C. J., Gusto G., Haessler J., Hall P., Haller T., Hallmans G., Hartman C. A., Hassinen M., Hayward C., Heard-Costa N. L., Helmer Q., Hengstenberg C., Holmen O., Hottenga J.-J., James A. L., Jeff J. M., Johansson Å., Jolley J., Juliusdottir T., Kinnunen L., Koenig W., Koskenvuo M., Kratzer W., Laitinen J., Lamina C., Leander K., Lee N. R., Lichtner P., Lind L., Lindström J., Lo K. S., Lobbens S., Lorbeer R., Lu Y., Mach F., Magnusson P. K. E., Mahajan A., Mc Ardle W. L., Lachlan S. M., Menni C., Merger S., Mihailov E., Milani L., Moayyeri A., Monda K. L., Morken M. A., Mulas A., Müller G., Müller-Nurasyid M., Musk A. W., Nagaraja R., Nöthen M. M., Nolte I. M., Pilz S., Rayner N. W., Renstrom F., Rettig R., Ried J. S., Ripke S., Robertson N. R., Rose L. M., Sanna S., Scharnagl H., Scholtens S., Schumacher F. R., Scott W. R., Seufferlein T., Shi J., Smith A. V., Smolonska J., Stanton A. V., Steinthorsdottir V., Stirrups K., Stringham H. M., Sundström J., Swertz M. A., Swift A. J., Syvänen A.-C., Tan S.-T., Tayo B. O., Thorand B., Thorleifsson G., Tyrer J. P., Uh H.-W., Vandenput L., Verhulst F. C., Vermeulen S. H., Verweij N., Vonk J. M., Waite L. L., Warren H. R., Waterworth D., Weedon M. N., Wilkens L. R., Willenborg C., Wilsgaard T., Wojczynski M. K., Wong A., Wright A. F., Zhang Q.; The Life Lines Cohort Study, Brennan E. P., Choi M., Dastani Z., Drong A. W., Eriksson P., Franco-Cereceda A., Gådin J. R., Gharavi A. G., Goddard M. E., Handsaker R. E., Huang J., Karpe F., Kathiresan S., Keildson S., Kiryluk K., Kubo M., Lee J.-Y., Liang L., Lifton R. P., Ma B., Mc Carroll S. A., Mc Knight A. J., Min J. L., Moffatt M. F., Montgomery G. W., Murabito J. M., Nicholson G., Nyholt D. R., Okada Y., Perry J. R. B., Dorajoo R., Reinmaa E., Salem R. M., Sandholm N., Scott R. A., Stolk L., Takahashi A., Tanaka T., va’nt Hooft F. M., Vinkhuyzen A. A. E., Westra H.-J., Zheng W., Zondervan K. T.; The ADIPOGen Consortium; The AGEN-BMI Working Group; The CARDIOGRAMplus CD Consortium; The CKDGen Consortium; The GLGC, The ICBP; The MAGIC Investigators; The Mu THER Consortium; The MIGen Consortium; The PAGE Consortium; The Repro Gen Consortium; The GENIE Consortium; The International Endogene Consortium, Heath A. C., Arveiler D., Bakker S. J. L., Beilby J., Bergman R. N., Blangero J., Bovet P., Campbell H., Caulfield M. J., Cesana G., Chakravarti A., Chasman D. I., Chines P. S., Collins F. S., Crawford D. C., Cupples L. A., Cusi D., Danesh J., de Faire U., den Ruijter H. M., Dominiczak A. F., Erbel R., Erdmann J., Eriksson J. G., Farrall M., Felix S. B., Ferrannini E., Ferrières J., Ford I., Forouhi N. G., Forrester T., Franco O. H., Gansevoort R. T., Gejman P. V., Gieger C., Gottesman O., Gudnason V., Gyllensten U., Hall A. S., Harris T. B., Hattersley A. T., Hicks A. A., Hindorff L. A., Hingorani A. D., Hofman A., Homuth G., Hovingh G. K., Humphries S. E., Hunt S. C., Hyppönen E., Illig T., Jacobs K. B., Jarvelin M.-R., Jöckel K.-H., Johansen B., Jousilahti P., Wouter Jukema J., Jula A. M., Kaprio J., Kastelein J. J. P., Keinanen-Kiukaanniemi S. M., Kiemeney L. A., Knekt P., Kooner J. S., Kooperberg C., Kovacs P., Kraja A. T., Kumari M., Kuusisto J., Lakka T. A., Langenberg C., Marchand L. L., Lehtimäki T., Lyssenko V., Männistö S., Marette A., Matise T. C., Mc Kenzie C. A., Knight B. M., Moll F. L., Morris A. D., Morris A. P., Murray J. C., Nelis M., Ohlsson C., Oldehinkel A. J., Ong K. K., Madden P. A. F., Pasterkamp G., Peden J. F., Peters A., Postma D. S., Pramstaller P. P., Price J. F., Qi L., Raitakari O. T., Rankinen T., Rao D. C., Rice T. K., Ridker P. M., Rioux J. D., Ritchie M. D., Rudan I., Salomaa V., Samani N. J., Saramies J., Sarzynski M. A., Schunkert H., Schwarz P. E. H., Sever P., Shuldiner A. R., Sinisalo J., Stolk R. P., Strauch K., Tönjes A., Trégouët D.-A., Tremblay A., Tremoli E., Virtamo J., Vohl M.-C., Völker U., Waeber G., Willemsen G., Witteman J. C., Carola Zillikens M., Adair L. S., Amouyel P., Asselbergs F. W., Assimes T. L., Bochud M., Boehm B. O., Boerwinkle E., Bornstein S. R., Bottinger E. P., Bouchard C., Cauchi S., Chambers J. C., Chanock S. J., Cooper R. S., de Bakker P. I. W., Dedoussis G., Ferrucci L., Franks P. W., Froguel P., Groop L. C., Haiman C. A., Hamsten A., Hui J., Hunter D. J., Hveem K., Kaplan R. C., Kivimaki M., Kuh D., Laakso M., Liu Y., Martin N. G., März W., Melbye M., Metspalu A., Moebus S., Munroe P. B., Njølstad I., Oostra B. A., Palmer C. N. A., Pedersen N. L., Perola M., Pérusse L., Peters U., Power C., Quertermous T., Rauramaa R., Rivadeneira F., Saaristo T. E., Saleheen D., Sattar N., Schadt E. E., Schlessinger D., Slagboom P. E., Snieder H., Spector T. D., Thorsteinsdottir U., Stumvoll M., Tuomilehto J., Uitterlinden A. G., Uusitupa M., van der Harst P., Walker M., Wallaschofski H., Wareham N. J., Watkins H., Weir D. R., Wichmann H.-E., Wilson J. F., Zanen P., Borecki I. B., Deloukas P., Fox C. S., Heid I. M., O.’onnell J. R., Strachan D. P., Stefansson K., van Duijn C. M., Abecasis G. R., Franke L., Frayling T. M., Mc Carthy M. I., Visscher P. M., Scherag A., Willer C. J., Boehnke M., Mohlke K. L., Lindgren C. M., Beckmann J. S., Barroso I., North K. E., Ingelsson E., Hirschhorn J. N., Loos R. J. F., Speliotes E. K., Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan B. P., Roberts J. L., Fogarty K. V., Reynolds K. A., Jonas J. M., Hudson J. I., Memantine in the treatment of binge eating disorder: An open-label, prospective trial. Int. J. Eat. Disord. 41, 520–526 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Huang X. T., Yang J. X., Wang Z., Zhang C. Y., Luo Z. Q., Liu W., Tang S. Y., Activation of N-methyl-D-aspartate receptor regulates insulin sensitivity and lipid metabolism. Theranostics 11, 2247–2262 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suyama S., Ralevski A., Liu Z. W., Dietrich M. O., Yada T., Simonds S. E., Cowley M. A., Gao X. B., Diano S., Horvath T. L., Plasticity of calcium-permeable AMPA glutamate receptors in Pro-opiomelanocortin neurons. eLife 6, e25755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krashes M. J., Koda S., Ye C., Rogan S. C., Adams A. C., Cusher D. S., Maratos-Flier E., Roth B. L., Lowell B. B., Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J. Clin. Invest. 121, 1424–1428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oginsky M. F., Goforth P. B., Nobile C. W., Lopez-Santiago L. F., Ferrario C. R., Eating ‘Junk-Food’ produces rapid and long-lasting increases in NAc CP-AMPA receptors: Implications for enhanced cue-induced motivation and food addiction. Neuropsychopharmacology 41, 2977–2986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith K. L., Rao R. R., Velazquez-Sanchez C., Valenza M., Giuliano C., Everitt B. J., Sabino V., Cottone P., The uncompetitive N-methyl-D-aspartate antagonist memantine reduces binge-like eating, food-seeking behavior, and compulsive eating: Role of the nucleus accumbens shell. Neuropsychopharmacology 40, 1163–1171 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng S. N., Yan Y. H., Zhu T. L., Ma B. K., Fan H. R., Liu Y. M., Li W. G., Li F., Long-term NMDAR antagonism correlates weight loss with less eating. Front. Psych. 10, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Popik P., Kos T., Zhang Y., Bisaga A., Memantine reduces consumption of highly palatable food in a rat model of binge eating. Amino Acids 40, 477–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.T. L. Fetterly, A. M. Catalfio, C. R. Ferrario, Effects of junk-food on food-motivated behavior and NAc glutamate plasticity; insights into the mechanism of NAc calcium-permeable AMPA receptor recruitment. bioRxiv 540977 [Preprint] (2023); 10.1101/2023.05.16.540977. [DOI] [PMC free article] [PubMed]
- 19.Hettes S. R., Gonzaga W. J., Heyming T. W., Nguyen J. K., Perez S., Stanley B. G., Stimulation of lateral hypothalamic AMPA receptors may induce feeding in rats. Brain Res. 1346, 112–120 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Lipton S. A., Failures and successes of NMDA receptor antagonists: Molecular basis for the use of open-channel blockers like memantine in the treatment of acute and chronic neurologic insults. NeuroRx 1, 101–110 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogawski M. A., Revisiting AMPA receptors as an antiepileptic drug target. Epilepsy Curr. 11, 56–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen N. R., Calyseva J., Fernandes E. F. A., Luchow S., Clemmensen L. S., Haugaard-Kedstrom L. M., Stromgaard K., PDZ domains as drug targets. Adv. Ther. 2, 1800143 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim E., Sheng M., PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771–781 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Sorensen A. T., Rombach J., Gether U., Madsen K. L., The scaffold protein PICK1 as a target in chronic pain. Cell 11, 1255 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill M. D., Goyal M., Menon B. K., Nogueira R. G., McTaggart R. A., Demchuk A. M., Poppe A. Y., Buck B. H., Field T. S., Dowlatshahi D., van Adel B. A., Swartz R. H., Shah R. A., Sauvageau E., Zerna C., Ospel J. M., Joshi M., Almekhlafi M. A., Ryckborst K. J., Lowerison M. W., Heard K., Garman D., Haussen D., Cutting S. M., Coutts S. B., Roy D., Rempel J. L., Rohr A. C., Iancu D., Sahlas D. J., Yu A. Y. X., Devlin T. G., Hanel R. A., Puetz V., Silver F. L., Campbell B. C. V., Chapot R., Teitelbaum J., Mandzia J. L., Kleinig T. J., Turkel-Parrella D., Heck D., Kelly M. E., Bharatha A., Bang O. Y., Jadhav A., Gupta R., Frei D. F., Tarpley J. W., McDougall C. G., Holmin S., Rha J. H., Puri A. S., Camden M. C., Thomalla G., Choe H., Phillips S. J., Schindler J. L., Thornton J., Nagel S., Heo J. H., Sohn S. I., Psychogios M. N., Budzik R. F., Starkman S., Martin C. O., Burns P. A., Murphy S., Lopez G. A., English J., Tymianski M.; ESCAPE-NA1 Investigators , Efficacy and safety of nerinetide for the treatment of acute ischaemic stroke (ESCAPE-NA1): A multicentre, double-blind, randomised controlled trial. Lancet 395, 878–887 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Hinney A., Korner A., Fischer-Posovszky P., The promise of new anti-obesity therapies arising from knowledge of genetic obesity traits. Nat. Rev. Endocrinol. 18, 623–637 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J., Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics 5, 1158–1170 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kornau H. C., Schenker L. T., Kennedy M. B., Seeburg P. H., Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269, 1737–1740 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Chen L., Chetkovich D. M., Petralia R. S., Sweeney N. T., Kawasaki Y., Wenthold R. J., Bredt D. S., Nicoll R. A., Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature 408, 936–943 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Sommer J. B., Bach A., Mala H., Stromgaard K., Mogensen J., Pickering D. S., In vitro and in vivo effects of a novel dimeric inhibitor of PSD-95 on excitotoxicity and functional recovery after experimental traumatic brain injury. Eur. J. Neurosci. 45, 238–248 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Bach A., Clausen B. H., Moller M., Vestergaard B., Chi C. N., Round A., Sorensen P. L., Nissen K. B., Kastrup J. S., Gajhede M., Jemth P., Kristensen A. S., Lundstrom P., Lambertsen K. L., Stromgaard K., A high-affinity, dimeric inhibitor of PSD-95 bivalently interacts with PDZ1-2 and protects against ischemic brain damage. Proc. Natl. Acad. Sci. U.S.A. 109, 3317–3322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kucharz K., Sondergaard Rasmussen I., Bach A., Stromgaard K., Lauritzen M., PSD-95 uncoupling from NMDA receptors by Tat- N-dimer ameliorates neuronal depolarization in cortical spreading depression. J. Cereb. Blood Flow Metab. 37, 1820–1828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bach A., Clausen B. H., Kristensen L. K., Andersen M. G., Ellman D. G., Hansen P. B. L., Hasseldam H., Heitz M., Ozcelik D., Tuck E. J., Kopanitsa M. V., Grant S. G. N., Lykke-Hartmann K., Johansen F. F., Lambertsen K. L., Stromgaard K., Selectivity, efficacy and toxicity studies of UCCB01-144, a dimeric neuroprotective PSD-95 inhibitor. Neuropharmacology 150, 100–111 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Dev K. K., Nishimune A., Henley J. M., Nakanishi S., The protein kinase C alpha binding protein PICK1 interacts with short but not long form alternative splice variants of AMPA receptor subunits. Neuropharmacology 38, 635–644 (1999). [DOI] [PubMed] [Google Scholar]
- 35.L. Jensen, mPD5, an efficacious PICK1 inhibitor for treating chronic pain. bioRxiv 530471 [Preprint] (2023); 10.1101/2023.03.03.530471. [DOI]
- 36.Holst B., Madsen K. L., Jansen A. M., Jin C., Rickhag M., Lund V. K., Jensen M., Bhatia V., Sorensen G., Madsen A. N., Xue Z., Moller S. K., Woldbye D., Qvortrup K., Huganir R., Stamou D., Kjaerulff O., Gether U., PICK1 deficiency impairs secretory vesicle biogenesis and leads to growth retardation and decreased glucose tolerance. PLOS Biol. 11, e1001542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qiu H., Ma L., Feng F., PICK1 attenuates high glucose-induced pancreatic beta-cell death through the PI3K/Akt pathway and is negatively regulated by miR-139-5p. Biochem. Biophys. Res. Commun. 522, 14–20 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Renier N., Wu Z., Simon D. J., Yang J., Ariel P., Tessier-Lavigne M., iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Hansen H. H., Perens J., Roostalu U., Skytte J. L., Salinas C. G., Barkholt P., Thorbek D. D., Rigbolt K. T. G., Vrang N., Jelsing J., Hecksher-Sorensen J., Whole-brain activation signatures of weight-lowering drugs. Mol. Metab. 47, 101171 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bats C., Groc L., Choquet D., The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 53, 719–734 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Christensen N. R., De Luca M., Lever M. B., Richner M., Hansen A. B., Noes-Holt G., Jensen K. L., Rathje M., Jensen D. B., Erlendsson S., Bartling C. R., Ammendrup-Johnsen I., Pedersen S. E., Schonauer M., Nissen K. B., Midtgaard S. R., Teilum K., Arleth L., Sorensen A. T., Bach A., Stromgaard K., Meehan C. F., Vaegter C. B., Gether U., Madsen K. L., A high-affinity, bivalent PDZ domain inhibitor complexes PICK1 to alleviate neuropathic pain. EMBO Mol. Med. 12, e11248 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turcot V., Lu Y., Highland H. M., Schurmann C., Justice A. E., Fine R. S., Bradfield J. P., Esko T., Giri A., Graff M., Guo X., Hendricks A. E., Karaderi T., Lempradl A., Locke A. E., Mahajan A., Marouli E., Sivapalaratnam S., Young K. L., Alfred T., Feitosa M. F., Masca N. G. D., Manning A. K., Medina-Gomez C., Mudgal P., Ng M. C. Y., Reiner A. P., Vedantam S., Willems S. M., Winkler T. W., Abecasis G., Aben K. K., Alam D. S., Alharthi S. E., Allison M., Amouyel P., Asselbergs F. W., Auer P. L., Balkau B., Bang L. E., Barroso I., Bastarache L., Benn M., Bergmann S., Bielak L. F., Bluher M., Boehnke M., Boeing H., Boerwinkle E., Boger C. A., Bork-Jensen J., Bots M. L., Bottinger E. P., Bowden D. W., Brandslund I., Breen G., Brilliant M. H., Broer L., Brumat M., Burt A. A., Butterworth A. S., Campbell P. T., Cappellani S., Carey D. J., Catamo E., Caulfield M. J., Chambers J. C., Chasman D. I., Chen Y. I., Chowdhury R., Christensen C., Chu A. Y., Cocca M., Collins F. S., Cook J. P., Corley J., Corominas Galbany J., Cox A. J., Crosslin D. S., Cuellar-Partida G., D'Eustacchio A., Danesh J., Davies G., Bakker P. I. W., Groot M. C. H., Mutsert R., Deary I. J., Dedoussis G., Demerath E. W., Heijer M., Hollander A. I., Ruijter H. M., Dennis J. G., Denny J. C., Di Angelantonio E., Drenos F., Du M., Dube M. P., Dunning A. M., Easton D. F., Edwards T. L., Ellinghaus D., Ellinor P. T., Elliott P., Evangelou E., Farmaki A. E., Farooqi I. S., Faul J. D., Fauser S., Feng S., Ferrannini E., Ferrieres J., Florez J. C., Ford I., Fornage M., Franco O. H., Franke A., Franks P. W., Friedrich N., Frikke-Schmidt R., Galesloot T. E., Gan W., Gandin I., Gasparini P., Gibson J., Giedraitis V., Gjesing A. P., Gordon-Larsen P., Gorski M., Grabe H. J., Grant S. F. A., Grarup N., Griffiths H. L., Grove M. L., Gudnason V., Gustafsson S., Haessler J., Hakonarson H., Hammerschlag A. R., Hansen T., Harris K. M., Harris T. B., Hattersley A. T., Have C. T., Hayward C., He L., Heard-Costa N. L., Heath A. C., Heid I. M., Helgeland O., Hernesniemi J., Hewitt A. W., Holmen O. L., Hovingh G. K., Howson J. M. M., Hu Y., Huang P. L., Huffman J. E., Ikram M. A., Ingelsson E., Jackson A. U., Jansson J. H., Jarvik G. P., Jensen G. B., Jia Y., Johansson S., Jorgensen M. E., Jorgensen T., Jukema J. W., Kahali B., Kahn R. S., Kahonen M., Kamstrup P. R., Kanoni S., Kaprio J., Karaleftheri M., Kardia S. L. R., Karpe F., Kathiresan S., Kee F., Kiemeney L. A., Kim E., Kitajima H., Komulainen P., Kooner J. S., Kooperberg C., Korhonen T., Kovacs P., Kuivaniemi H., Kutalik Z., Kuulasmaa K., Kuusisto J., Laakso M., Lakka T. A., Lamparter D., Lange E. M., Lange L. A., Langenberg C., Larson E. B., Lee N. R., Lehtimaki T., Lewis C. E., Li H., Li J., Li-Gao R., Lin H., Lin K. H., Lin L. A., Lin X., Lind L., Lindstrom J., Linneberg A., Liu C. T., Liu D. J., Liu Y., Lo K. S., Lophatananon A., Lotery A. J., Loukola A., Luan J., Lubitz S. A., Lyytikainen L. P., Mannisto S., Marenne G., Mazul A. L., McCarthy M. I., McKean-Cowdin R., Medland S. E., Meidtner K., Milani L., Mistry V., Mitchell P., Mohlke K. L., Moilanen L., Moitry M., Montgomery G. W., Mook-Kanamori D. O., Moore C., Mori T. A., Morris A. D., Morris A. P., Muller-Nurasyid M., Munroe P. B., Nalls M. A., Narisu N., Nelson C. P., Neville M., Nielsen S. F., Nikus K., Njolstad P. R., Nordestgaard B. G., Nyholt D. R., O'Connel J. R., O'Donoghue M. L., Olde Loohuis L. M., Ophoff R. A., Owen K. R., Packard C. J., Padmanabhan S., Palmer C. N. A., Palmer N. D., Pasterkamp G., Patel A. P., Pattie A., Pedersen O., Peissig P. L., Peloso G. M., Pennell C. E., Perola M., Perry J. A., Perry J. R. B., Pers T. H., Person T. N., Peters A., Petersen E. R. B., Peyser P. A., Pirie A., Polasek O., Polderman T. J., Puolijoki H., Raitakari O. T., Rasheed A., Rauramaa R., Reilly D. F., Renstrom F., Rheinberger M., Ridker P. M., Rioux J. D., Rivas M. A., Roberts D. J., Robertson N. R., Robino A., Rolandsson O., Rudan I., Ruth K. S., Saleheen D., Salomaa V., Samani N. J., Sapkota Y., Sattar N., Schoen R. E., Schreiner P. J., Schulze M. B., Scott R. A., Segura-Lepe M. P., Shah S. H., Sheu W. H., Sim X., Slater A. J., Small K. S., Smith A. V., Southam L., Spector T. D., Speliotes E. K., Starr J. M., Stefansson K., Steinthorsdottir V., Stirrups K. E., Strauch K., Stringham H. M., Stumvoll M., Sun L., Surendran P., Swift A. J., Tada H., Tansey K. E., Tardif J. C., Taylor K. D., Teumer A., Thompson D. J., Thorleifsson G., Thorsteinsdottir U., Thuesen B. H., Tonjes A., Tromp G., Trompet S., Tsafantakis E., Tuomilehto J., Tybjaerg-Hansen A., Tyrer J. P., Uher R., Uitterlinden A. G., Uusitupa M., Laan S. W., Duijn C. M., Leeuwen N., van Setten J., Vanhala M., Varbo A., Varga T. V., Varma R., Velez Edwards D. R., Vermeulen S. H., Veronesi G., Vestergaard H., Vitart V., Vogt T. F., Volker U., Vuckovic D., Wagenknecht L. E., Walker M., Wallentin L., Wang F., Wang C. A., Wang S., Wang Y., Ware E. B., Wareham N. J., Warren H. R., Waterworth D. M., Wessel J., White H. D., Willer C. J., Wilson J. G., Witte D. R., Wood A. R., Wu Y., Yaghootkar H., Yao J., Yao P., Yerges-Armstrong L. M., Young R., Zeggini E., Zhan X., Zhang W., Zhao J. H., Zhao W., Zhao W., Zhou W., Zondervan K. T.; CHD Exome+ Consortium; EPIC-CVD Consortium; ExomeBP Consortium; Global Lipids Genetic Consortium, GoT2D Genes Consortium; EPIC InterAct Consortium; INTERVAL Study; ReproGen Consortium; T2D-Genes Consortium; MAGIC Investigators; Understanding Society Scientific Group, Rotter J. I., Pospisilik J. A., Rivadeneira F., Borecki I. B., Deloukas P., Frayling T. M., Lettre G., North K. E., Lindgren C. M., Hirschhorn J. N., Loos R. J. F., Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 50, 26–41 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y., Day F. R., Gustafsson S., Buchkovich M. L., Na J., Bataille V., Cousminer D. L., Dastani Z., Drong A. W., Esko T., Evans D. M., Falchi M., Feitosa M. F., Ferreira T., Hedman A. K., Haring R., Hysi P. G., Iles M. M., Justice A. E., Kanoni S., Lagou V., Li R., Li X., Locke A., Lu C., Magi R., Perry J. R., Pers T. H., Qi Q., Sanna M., Schmidt E. M., Scott W. R., Shungin D., Teumer A., Vinkhuyzen A. A., Walker R. W., Westra H. J., Zhang M., Zhang W., Zhao J. H., Zhu Z., Afzal U., Ahluwalia T. S., Bakker S. J., Bellis C., Bonnefond A., Borodulin K., Buchman A. S., Cederholm T., Choh A. C., Choi H. J., Curran J. E., de Groot L. C., De Jager P. L., Dhonukshe-Rutten R. A., Enneman A. W., Eury E., Evans D. S., Forsen T., Friedrich N., Fumeron F., Garcia M. E., Gartner S., Han B. G., Havulinna A. S., Hayward C., Hernandez D., Hillege H., Ittermann T., Kent J. W., Kolcic I., Laatikainen T., Lahti J., Leach I. M., Lee C. G., Lee J. Y., Liu T., Liu Y., Lobbens S., Loh M., Lyytikainen L. P., Medina-Gomez C., Michaelsson K., Nalls M. A., Nielson C. M., Oozageer L., Pascoe L., Paternoster L., Polasek O., Ripatti S., Sarzynski M. A., Shin C. S., Narancic N. S., Spira D., Srikanth P., Steinhagen-Thiessen E., Sung Y. J., Swart K. M., Taittonen L., Tanaka T., Tikkanen E., van der Velde N., van Schoor N. M., Verweij N., Wright A. F., Yu L., Zmuda J. M., Eklund N., Forrester T., Grarup N., Jackson A. U., Kristiansson K., Kuulasmaa T., Kuusisto J., Lichtner P., Luan J., Mahajan A., Mannisto S., Palmer C. D., Ried J. S., Scott R. A., Stancakova A., Wagner P. J., Demirkan A., Doring A., Gudnason V., Kiel D. P., Kuhnel B., Mangino M., McKnight B., Menni C., O’Connell J. R., Oostra B. A., Shuldiner A. R., Song K., Vandenput L., van Duijn C. M., Vollenweider P., White C. C., Boehnke M., Boettcher Y., Cooper R. S., Forouhi N. G., Gieger C., Grallert H., Hingorani A., Jorgensen T., Jousilahti P., Kivimaki M., Kumari M., Laakso M., Langenberg C., Linneberg A., Luke A., McKenzie C. A., Palotie A., Pedersen O., Peters A., Strauch K., Tayo B. O., Wareham N. J., Bennett D. A., Bertram L., Blangero J., Bluher M., Bouchard C., Campbell H., Cho N. H., Cummings S. R., Czerwinski S. A., Demuth I., Eckardt R., Eriksson J. G., Ferrucci L., Franco O. H., Froguel P., Gansevoort R. T., Hansen T., Harris T. B., Hastie N., Heliovaara M., Hofman A., Jordan J. M., Jula A., Kahonen M., Kajantie E., Knekt P. B., Koskinen S., Kovacs P., Lehtimaki T., Lind L., Liu Y., Orwoll E. S., Osmond C., Perola M., Perusse L., Raitakari O. T., Rankinen T., Rao D. C., Rice T. K., Rivadeneira F., Rudan I., Salomaa V., Sorensen T. I., Stumvoll M., Tonjes A., Towne B., Tranah G. J., Tremblay A., Uitterlinden A. G., van der Harst P., Vartiainen E., Viikari J. S., Vitart V., Vohl M. C., Volzke H., Walker M., Wallaschofski H., Wild S., Wilson J. F., Yengo L., Bishop D. T., Borecki I. B., Chambers J. C., Cupples L. A., Dehghan A., Deloukas P., Fatemifar G., Fox C., Furey T. S., Franke L., Han J., Hunter D. J., Karjalainen J., Karpe F., Kaplan R. C., Kooner J. S., McCarthy M. I., Murabito J. M., Morris A. P., Bishop J. A., North K. E., Ohlsson C., Ong K. K., Prokopenko I., Richards J. B., Schadt E. E., Spector T. D., Widen E., Willer C. J., Yang J., Ingelsson E., Mohlke K. L., Hirschhorn J. N., Pospisilik J. A., Zillikens M. C., Lindgren C., Kilpelainen T. O., Loos R. J., New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nat. Commun. 7, 10495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrington N. M., Beaumont R. N., Horikoshi M., Day F. R., Helgeland O., Laurin C., Bacelis J., Peng S., Hao K., Feenstra B., Wood A. R., Mahajan A., Tyrrell J., Robertson N. R., Rayner N. W., Qiao Z., Moen G. H., Vaudel M., Marsit C. J., Chen J., Nodzenski M., Schnurr T. M., Zafarmand M. H., Bradfield J. P., Grarup N., Kooijman M. N., Li-Gao R., Geller F., Ahluwalia T. S., Paternoster L., Rueedi R., Huikari V., Hottenga J. J., Lyytikainen L. P., Cavadino A., Metrustry S., Cousminer D. L., Wu Y., Thiering E., Wang C. A., Have C. T., Vilor-Tejedor N., Joshi P. K., Painter J. N., Ntalla I., Myhre R., Pitkanen N., van Leeuwen E. M., Joro R., Lagou V., Richmond R. C., Espinosa A., Barton S. J., Inskip H. M., Holloway J. W., Santa-Marina L., Estivill X., Ang W., Marsh J. A., Reichetzeder C., Marullo L., Hocher B., Lunetta K. L., Murabito J. M., Relton C. L., Kogevinas M., Chatzi L., Allard C., Bouchard L., Hivert M. F., Zhang G., Muglia L. J., Heikkinen J.; EGG Consortium, Morgen C. S., van Kampen A. H. C., van Schaik B. D. C., Mentch F. D., Langenberg C., Luan J., Scott R. A., Zhao J. H., Hemani G., Ring S. M., Bennett A. J., Gaulton K. J., Fernandez-Tajes J., van Zuydam N. R., Medina-Gomez C., de Haan H. G., Rosendaal F. R., Kutalik Z., Marques-Vidal P., Das S., Willemsen G., Mbarek H., Muller-Nurasyid M., Standl M., Appel E. V. R., Fonvig C. E., Trier C., van Beijsterveldt C. E. M., Murcia M., Bustamante M., Bonas-Guarch S., Hougaard D. M., Mercader J. M., Linneberg A., Schraut K. E., Lind P. A., Medland S. E., Shields B. M., Knight B. A., Chai J. F., Panoutsopoulou K., Bartels M., Sanchez F., Stokholm J., Torrents D., Vinding R. K., Willems S. M., Atalay M., Chawes B. L., Kovacs P., Prokopenko I., Tuke M. A., Yaghootkar H., Ruth K. S., Jones S. E., Loh P. R., Murray A., Weedon M. N., Tonjes A., Stumvoll M., Michaelsen K. F., Eloranta A. M., Lakka T. A., van Duijn C. M., Kiess W., Korner A., Niinikoski H., Pahkala K., Raitakari O. T., Jacobsson B., Zeggini E., Dedoussis G. V., Teo Y. Y., Saw S. M., Montgomery G. W., Campbell H., Wilson J. F., Vrijkotte T. G. M., Vrijheid M., de Geus E., Hayes M. G., Kadarmideen H. N., Holm J. C., Beilin L. J., Pennell C. E., Heinrich J., Adair L. S., Borja J. B., Mohlke K. L., Eriksson J. G., Widen E. E., Hattersley A. T., Spector T. D., Kahonen M., Viikari J. S., Lehtimaki T., Boomsma D. I., Sebert S., Vollenweider P., Sorensen T. I. A., Bisgaard H., Bonnelykke K., Murray J. C., Melbye M., Nohr E. A., Mook-Kanamori D. O., Rivadeneira F., Hofman A., Felix J. F., Jaddoe V. W. V., Hansen T., Pisinger C., Vaag A. A., Pedersen O., Uitterlinden A. G., Jarvelin M. R., Power C., Hypponen E., Scholtens D. M., Lowe W. L. Jr., Smith G. D., Timpson N. J., Morris A. P., Wareham N. J., Hakonarson H., Grant S. F. A., Frayling T. M., Lawlor D. A., Njolstad P. R., Johansson S., Ong K. K., McCarthy M. I., Perry J. R. B., Evans D. M., Freathy R. M., Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 51, 804–814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Helgeland O., Vaudel M., Sole-Navais P., Flatley C., Juodakis J., Bacelis J., Koloen I. L., Knudsen G. P., Johansson B. B., Magnus P., Kjennerud T. R., Juliusson P. B., Stoltenberg C., Holmen O. L., Andreassen O. A., Jacobsson B., Njolstad P. R., Johansson S., Characterization of the genetic architecture of infant and early childhood body mass index. Nat. Metab. 4, 344–358 (2022). [DOI] [PubMed] [Google Scholar]
- 46.Hao S., Dey A., Yu X., Stranahan A. M., Dietary obesity reversibly induces synaptic stripping by microglia and impairs hippocampal plasticity. Brain Behav. Immun. 51, 230–239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lizarbe B., Soares A. F., Larsson S., Duarte J. M. N., Neurochemical modifications in the hippocampus, cortex and hypothalamus of mice exposed to long-term high-fat diet. Front. Neurosci. 12, 985 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy P., Martinelli I., Moruzzi M., Maggi F., Amantini C., Micioni Di Bonaventura M. V., Cifani C., Amenta F., Tayebati S. K., Tomassoni D., Ion channels alterations in the forebrain of high-fat diet fed rats. Eur. J. Histochem. 65, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu T., Kong D., Shah B. P., Ye C., Koda S., Saunders A., Ding J. B., Yang Z., Sabatini B. L., Lowell B. B., Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron 73, 511–522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grzelka K., Wilhelms H., Dodt S., Dreisow M. L., Madara J. C., Walker S. J., Wu C., Wang D., Lowell B. B., Fenselau H., A synaptic amplifier of hunger for regaining body weight in the hypothalamus. Cell Metab. 35, 770–785 e775 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Mao Z., Huang J., Xia J., PICK1 is essential for insulin production and the maintenance of glucose homeostasis. Mol. Biol. Cell 29, 587–596 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Backe M. B., Andersen R. C., Jensen M., Jin C., Hundahl C., Dmytriyeva O., Treebak J. T., Hansen J. B., Gerhart-Hines Z., Madsen K. L., Holst B., PICK1-deficient mice maintain their glucose tolerance during diet-induced obesity. J. Endocr. Soc. 7, bvad057 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller T. D., Finan B., Bloom S. R., D'Alessio D., Drucker D. J., Flatt P. R., Fritsche A., Gribble F., Grill H. J., Habener J. F., Holst J. J., Langhans W., Meier J. J., Nauck M. A., Perez-Tilve D., Pocai A., Reimann F., Sandoval D. A., Schwartz T. W., Seeley R. J., Stemmer K., Tang-Christensen M., Woods S. C., DiMarchi R. D., Tschop M. H., Glucagon-like peptide 1 (GLP-1). Mol. Metab. 30, 72–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanacora G., Treccani G., Popoli M., Towards a glutamate hypothesis of depression: An emerging frontier of neuropsychopharmacology for mood disorders. Neuropharmacology 62, 63–77 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R., Reddy P. H., Role of glutamate and NMDA receptors in Alzheimer's disease. J. Alzheimers Dis. 57, 1041–1048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serafini G., Howland R. H., Rovedi F., Girardi P., Amore M., The role of ketamine in treatment-resistant depression: A systematic review. Curr. Neuropharmacol. 12, 444–461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aarts M., Liu Y., Liu L., Besshoh S., Arundine M., Gurd J. W., Wang Y. T., Salter M. W., Tymianski M., Treatment of ischemic brain damage by perturbing NMDA receptor- PSD-95 protein interactions. Science 298, 846–850 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Bach A., Chi C. N., Pang G. F., Olsen L., Kristensen A. S., Jemth P., Stromgaard K., Design and synthesis of highly potent and plasma-stable dimeric inhibitors of the PSD-95-NMDA receptor interaction. Angew. Chem. Int. Ed. Engl. 48, 9685–9689 (2009). [DOI] [PubMed] [Google Scholar]
- 59.Chang C. C., Chow C. C., Tellier L. C., Vattikuti S., Purcell S. M., Lee J. J., Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghoussaini M., Mountjoy E., Carmona M., Peat G., Schmidt E. M., Hercules A., Fumis L., Miranda A., Carvalho-Silva D., Buniello A., Burdett T., Hayhurst J., Baker J., Ferrer J., Gonzalez-Uriarte A., Jupp S., Karim M. A., Koscielny G., Machlitt-Northen S., Malangone C., Pendlington Z. M., Roncaglia P., Suveges D., Wright D., Vrousgou O., Papa E., Parkinson H., MacArthur J. A. L., Todd J. A., Barrett J. C., Schwartzentruber J., Hulcoop D. G., Ochoa D., McDonagh E. M., Dunham I., Open Targets Genetics: Systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fairfax B. P., Humburg P., Makino S., Naranbhai V., Wong D., Lau E., Jostins L., Plant K., Andrews R., McGee C., Knight J. C., Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 343, 1246949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Wijst M., de Vries D. H., Groot H. E., Trynka G., Hon C. C., Bonder M. J., Stegle O., Nawijn M. C., Idaghdour Y., van der Harst P., Ye C. J., Powell J., Theis F. J., Mahfouz A., Heinig M., Franke L., The single-cell eQTLGen consortium. Elife 9, e52155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.GTEx Consortium , Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Renier N., Adams E. L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A. E., Kadiri L., Umadevi Venkataraju K., Zhou Y., Wang V. X., Tang C. Y., Olsen O., Dulac C., Osten P., Tessier-Lavigne M., Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bria A., Iannello G., TeraStitcher—A tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13, 316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kirst C., Skriabine S., Vieites-Prado A., Topilko T., Bertin P., Gerschenfeld G., Verny F., Topilko P., Michalski N., Tessier-Lavigne M., Renier N., Mapping the fine-scale organization and plasticity of the brain vasculature. Cell 180, 780–795 e725 (2020). [DOI] [PubMed] [Google Scholar]
- 67.Shamonin D. P., Bron E. E., Lelieveldt B. P., Smits M., Klein S., Staring M.; Alzheimer's Disease Neuroimaging Initiative , Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer's disease. Front. Neuroinform. 7, 50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perens J., Salinas C. G., Skytte J. L., Roostalu U., Dahl A. B., Dyrby T. B., Wichern F., Barkholt P., Vrang N., Jelsing J., Hecksher-Sorensen J., An optimized mouse brain atlas for automated mapping and quantification of neuronal activity using iDISCO+ and light sheet fluorescence microscopy. Neuroinformatics 19, 433–446 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G., Wang L. G., Han Y., He Q. Y., clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., Smyth G. K., limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., G., Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3
Figs. S1 to S10