Abstract
Intra-articular adeno-associated virus (AAV) gene therapy has been explored as a potential strategy for joint diseases. However, concerns of low transduction efficacy, off-target expression, and neutralizing antibodies (Nabs) still need to be addressed. In this study, we demonstrated that AAV6 was the best serotype to transduce joints after screening serotypes 1 to 9. To develop a more effective AAV vector, a set of novel AAV capsids were rationally engineered. The mutant AAV62 created by swapping variable region I (VRI) of AAV2 into AAV6 induced a higher transduction efficiency per AAV genome copy number. To further investigate the roles of specific amino acids in the transduction of AAV62 and AAV6, we found out that AAV6D with the deletion of threonine at residue 265 induced a 2-fold higher transduction than AAV6, while the transduction efficiency from AAV6M with the mutation of alanine to glutamine at residue 263 was 10-fold lower. AAV6D efficiently transduced both synoviocytes and chondrocytes with low AAV genome copy numbers in other tissues and less Nab formation. This study demonstrates that novel AAV mutants with rational engineering may enhance joint transduction after intra-articular administration in mice, with the potential to evade AAV Nabs and minimize off-target effects in the liver.
Key words: AAV, engineering, joint, Nab, transduction, rational design
Graphical abstract

Li and colleagues’ study investigated the role of VRI of various serotypes in joint gene delivery through rational design, and developed a novel variant that achieves high local transduction and low neutralizing antibody (Nab) formation, with limited off-target effects. These findings offer potential clinical applications for optimized joint gene therapy.
Introduction
Joint diseases are one of the most common disorders, mostly resulting from inflammation or degeneration in the musculoskeletal system involving different joints of the body and heavily impact patients’ life quality and well-being.1 Those diseases present significant challenges in terms of treatments. In recent years, biologics have emerged as a promising alternative of disease-modifying antirheumatic drugs for treatment of joint diseases. These biologics target specific molecules to alleviate the underlying mechanisms of joint diseases. For instance, the application of proinflammatory cytokine antagonists, including interleukin inhibitors and tumor necrosis factor-α (TNF) inhibitors,2 has demonstrated successful reduction of inflammation in clinical trials. Furthermore, targeting B or T cells has been employed in clinics, such as rituximab3 and abatacept.4 Despite these advancements, achieving satisfactory outcomes still remains a challenge. The efficacy of these treatments often takes a few weeks to manifest, and multiple doses are typically required due to the short half-life of proteins.5 Additionally, the most common drug delivery method involves systemic treatment through intravenous infusion. While this approach can benefit multiple joints and other affected areas, it presents outstanding concerns such as off-target effects and difficulties in achieving the ideal concentration within the joint.5,6 Intra-articular injection is able to solve these outstanding concerns raised from systemic treatment. However, effective therapeutic outcome requires multiple intra-articular injections due to rapid clearance of drugs in joints and chronic condition in many joint diseases.
Intra-articular gene therapy, particularly with adeno-associated virus (AAV) vectors, has shown great potential for treatment of joint diseases.7 Compared with protein therapy with systemic treatment, intra-articular AAV gene therapy offers several distinct advantages, including long-term persistence8 and minimal off-target and low toxicity,7 and has been explored as a possible method to address the most severe problems in local joints such as arthritis.9
Recently, 13 AAV serotypes and more than 100 variants have been isolated in human and nonhuman primate tissue samples10 and developed as gene therapy vectors due to their broad tissue tropisms.11 Various wild-type serotypes8,12,13,14,15 have been explored for joint delivery, but there have been no thorough study and standard criteria on which serotype is the best option in vivo. Additionally, clinical trials revealed many limitations and unwanted effects from those serotypes. For intra-articular AAV gene delivery, three main obstacles still remain: transduction efficiency in the joint, potential risks of AAV vectors entering the blood and inducing off-target transduction into other tissues,16 and Nabs against AAV.17
To address these issues, AAV capsid engineering using direct revolution or rational design has become a popular tool to develop more potential AAV vectors.18,19 The AAV virion is composed of 60 subunits of capsid proteins with nine variable regions (VRs) defined between the conserved regions of serotypes. For rational design of novel AAV capsids, domain swapping, in which VRs from different serotypes are switched onto each other, provided insights into the roles of the VRs from different variants, and identified the domains that are critical for cell uptake and transduction.20 VRI has been reported to play an important role in AAV transduction, biodistribution, and Nab recognition in different tissues.21,22,23 Albright et al. reported that modification of VRI from AAVrh.10 into AAV1 decreased the vascular and hepatic transduction and enhanced CNS transduction through altered SIA footprints.23 Pei et al. isolated a mutant AAV LP2-10 with the substitution of residues 261–272 in VRI from AAV6 into AAV8, which exhibited a slightly lower liver transduction than AAV8.22 Moreover, Bowles et al. developed chimeric vector AAV2.5 with VRI modification, which showed a lower Nab level compared with the wild-type AAV2.24 However, there is limited information regarding the specific role of VRI in joint gene delivery.
In this study, we systematically evaluated the transduction efficiency of AAV serotypes 1–9 in the mouse knee joint, then we performed rational design to develop novel AAV capsids by VRI swapping among AAV2, 5, 6, and 8, and modification of a single amino acid in VRI of AAV6 to gain insight into the relationship between structure and AAV gene delivery efficacy. Our findings indicated that AAV62 exhibited better transduction efficiency out of all these mutants with VRI swapping after intra-articular injection. AAV62 induced a higher transduction efficiency per AAV genome copy number than AAV6, the best serotype to transduce the joints, with an altered Nab profile from AAV6. Additionally, we have identified a promising AAV6 mutant with deletion of one amino acid at residue 265 that demonstrated a higher transduction compared with AAV6 and AAV62 after intra-articular injection while generating fewer Nabs.
Results
AAV6 induced the highest transduction among AAV serotypes in joints
To verify the transduction efficiency of AAV serotypes in mouse joints, 5 × 109 vg of AAV encoding the luciferase gene (AAV/luc) were injected in both mouse knee joints at a volume of 5 μL in each joint. In vivo imaging was performed weekly from weeks 1 through 6 and at 5 months after AAV injection (Figure 1A). For all AAV vectors, luciferase expression generally increased in the knee joint during the first 2 weeks, and then remained stable after 3 weeks. Among all the serotypes, AAV6 induced the highest transduction followed by AAV1, AAV2, AAV5, AAV7, AAV8, and AAV9, while AAV3 and 4 showed the lowest transduction (Figure 1B, p < 0.05). Obvious luciferase expression was also detected in the livers of mice treated with intra-articular injection of AAV7, AAV8, and AAV9, but not in mice treated with the other serotypes (Figure 1A).
Figure 1.
Transduction efficiency with different AAV serotypes in the joints of mice
An amount of 5 × 109 vg of AAV/luc vectors from different serotypes were administered into the knee joints of C57BL/6 mice. Imaging was taken at the indicated time points. (A) The representative imaging at week 6 post AAV injection. (B) The transgene expression in joints with AAV serotypes at different time points. Data are shown as means ± SEM (n = 5). (C) In vitro analysis of luciferase activity in the cell lysate of AAV transduced joints. Mice were euthanized at week 6 after intra-articular injection and the joints were harvested for luciferase activity analyses. Data are shown as means ± SEM (n = 5). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05.
At week 6 post AAV intra-articular injection, the mice were killed, and their knee joints were collected and homogenized with 5x protein lysis buffer overnight at 4°C. In vitro luciferase activity was measured among AAV1-9/luc. Consistent with in vivo luciferase results, AAV6 showed the highest transduction (p < 0.05), followed by AAV5 and AAV2, with no significant difference observed between them (p > 0.05, Figure 1C).
AAV genome copy number in knee joints and livers from different serotypes
To investigate the efficiency of transduction into the joints from different AAV serotypes after intra-articular injection, we measured the viral genome copy number in AAV-treated knee joints. The results showed that the highest genome copy numbers were detected in AAV6-treated joints (1.73 ± 0.55), followed by AAV8, AAV7, and AAV1 (p < 0.05, Figure 2A). After normalizing luciferase activity per AAV genome by calculating the ratio of in vitro luciferase activity per gram of joint tissue to the vector copy number per diploid DNA genome, AAV6 still had the highest transgene expression per copy number, followed by AAV2, AAV5, and AAV1 (p < 0.05, Figure 2B). These findings indicate that AAV6 is the most efficient serotype to transduce the joint tissues.
Figure 2.
AAV copy number in mice knee and liver after injection of AAV1-9 at week 6
(A) AAV copy number in the knees. Six weeks after injection of 5 × 109vg AAV1-9/luc, AAV copy number was measured by qPCR using the primer of luciferase. Data are shown as means ± SEM (n = 4). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05. (B) The normalized transduction per copy number. In vitro luciferase intensity in each serotype was divided by copy number, respectively. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05. (C) AAV copy number in the liver. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05.
We also examined AAV copy numbers in the liver because the AAV genome in the liver serves as a parameter for studying the ability of AAV vectors to cross the local barrier and enter the bloodstream after intra-articular injection. The results showed that AAV2, AAV7, AAV8, and AAV9 exhibited higher viral copy numbers (0.023 ± 0.004, 0.029 ± 0.013, 0.1 ± 0.021, 0.11 ± 0.031, respectively) in the liver relative to the serotypes AAV1, AAV3, AAV4, AAV5, and AAV6, which were less than 0.02 per cell (Figure 2C, p < 0.05).
AAV is able to transduce both synoviocytes and chondrocytes in the joints
To determine which cell types in the joint were transduced by AAV/luc vectors from the serotypes of interest, specifically 2, 5, 6, and 8, the knee joints of mice were fixed and stained by rabbit anti-luciferase primary antibody at week 6 after intra-articular injection (Figure S1). All vectors were able to transduce 20%–30% of chondrocytes with no significant difference among serotypes 2, 5, 6, and 8 (22.3% ± 2.5%, 20% ± 4.7%, 25.7% ± 2.1%, and 30.3% ± 4.5%, respectively, p > 0.05). There was a similar transduction efficiency in synoviocytes with serotypes AAV2, 5, and 6, with transduction rates of 28.3% ± 4.5%, 25.0% ± 5.6% and 23.7% ± 3.8%, respectively. However, AAV8 induced much lower transduction in synoviocytes than the other serotypes with a transduction rate of 13.3% ± 2.8% (p < 0.05, Figure S1).
VRI swapping changed the AAV transduction profile in joints
AAV variable region I (VRI) has been shown to play a role in transduction efficiency and Nab profiles in previous studies.22 Our study showed that AAV6 is the best serotype for transducing the joint, as described above. In this next study, we explored whether the swapping of the VRI from other serotypes into the AAV6 capsid, or the VRI from AAV6 into other serotypes, impacted the transduction efficiency and immune profile. We selected a representative group for our investigation—AAV2, AAV5, AAV6, and AAV8, which are also among the most extensively investigated in other studies. First, we examined VP1, VP2, and VP3 expression in the AAV variants’ capsids. After the transfection of mutants’ plasmids into 293 cells, a western blot was performed on the cell lysates. As shown in Figure 3A, all three VPs were generated. Next, we made AAV/luc vectors from these plasmids and quantified the virus titer in both the supernatant and cell lysates using primers of the ITRs. The yield of AAV56, in which VRI of AAV5 was replaced from that of AAV6, was the lowest, with a titer of less than 1 × 107 vg/μL, and the highest yield of AAV vectors was AAV62. The other mutants produced virus vector yields similar to AAV6 (Figure 3B). Further, we administered AAV/luc vectors at a dose of 5 × 109 vg from these mutants and AAV6 into the knee joints of C57BL/6 mice via intra-articular injection. AAV56 and AAV65 were excluded from the subsequent experiments due to the low virus production in AAV56 and they are a paired group. At weeks 1 and 6 post AAV administration, imaging was carried out. At week 1, the highest transduction was observed with AAV6. At week 6, we found that AAV62 and AAV6 induced similar transduction efficiencies, which were higher than the other mutants. AAV68, which had VRI swapping of AAV8 into AAV6, experienced 9-fold lower transduction efficiency compared with AAV6 (p < 0.05, Figures 3C and 3D). However, AAV26 and AAV86, with the VRI swapping of AAV6 into AAV2 and AAV8, respectively, experienced enhanced joint transduction compared with AAV2 or AAV8. We collected the knee joints at week 6 post AAV injection from mice treated with AAV6 and the mutants AAV62, AAV26, AAV68, and AAV86 for luciferase analysis in vitro. Consistent with the results of in vivo imaging in mice, AAV62 and AAV6 demonstrated over 3-fold higher luciferase activity than AAV86, 68, and 26 in vitro (Figure 3E).
Figure 3.
Package and transduction of the novel AAV vectors with VRI swapping
(A) AAV capsid protein (VP1, VP2, VP3) in each novel virus. (B) Packaging ability in each novel virus after VRI swapping. Both supernatant and cell lysates were collected and AAV titers in supernatant and cell lysates were quantified respectively based on qPCR. Data are shown as means ± SEM (n ≥ 4). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05. (C) In vivo imaging of AAV6, 62, 65, 26, 68, and 86 at week 6. (D) In vivo luciferase intensity of AAV6, 62, 65, 26, 68, and 86 at weeks 1 and 6. Data are shown as means ± SEM (n = 4). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05. (E) In vitro luciferase assay at week 6. Data are shown as means ± SEM (n = 4). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05.
VRI swapping changed the AAV vector biodistribution after intra-articular injection
We analyzed the genome copy number in the joints of mice treated with AAV vectors by qPCR using primers specific for luciferase. Mice receiving AAV6 exhibited an over 5-fold higher copy number in the knee joints than those receiving AAV62 or AAV68, and a 2-fold higher copy number in the knee joints than those receiving AAV26 and AAV86 (Figure 4A). However, after normalization to AAV genome copy number in the joints, the highest transgene expression per copy number was found to be in mice injected with AAV62, followed by AAV68, AAV6, and AAV26, and the lowest being AAV86. AAV62 showed 8-fold and 15-fold higher transduction efficiency per copy number than AAV68 and AAV6, respectively, and over 50-fold higher transduction efficiency per copy number than AAV26 and AAV86 (Figure 4B). Next, we examined the AAV genome copy number in the liver, and the highest genome copy number was found in mice treated with AAV86 with ∼0.1 AAV genome per cell, followed by AAV68 and 26, with AAV62 and AAV6 having the lowest genome copy number in the liver, though the mean value of AAV62 is slightly higher than AAV6, there was no significant difference (p < 0.05, Figure 4C).
Figure 4.
AAV copy number in mice knee and liver after injection of AAV6, 62, 68, 26, and 86 at week 6
(A) AAV copy number of AAV6, 62, 68, 26, and 86 in the knee. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005. (B) The normalized transduction per copy number. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗∗∗∗p < 0.001. (C) AAV copy number in the liver. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
Cell tropisms of AAV mutants with VRI swapping in joints
After collecting the joints from mice treated with AAV6, AAV62, and AAV26 at week 6 post AAV injection for analysis of specific cell tropism, we found that the transduction rate in synoviocytes was similar for all three AAV mutants. However, a 1.5-fold lower transduction rate was observed in chondrocytes of mice treated with AAV26 compared with AAV 6 or 62. Specifically, the transduction percentages for chondrocytes were 24.3% ± 4.2%, 25.7% ± 2.1%, 14.7% ± 2.5%, in AAV62, AAV6, and AAV26, respectively (p < 0.05) (Figure S2).
AAV mutants with VRI swapping had different Nab patterns
It has been demonstrated that joint fluid also contains Nabs, which are able to block effective AAV transduction after intra-articular administration.25 We studied the Nab profile in blood after AAV intra-articular injection. Serum from AAV6-treated mice had a Nab titer of 1:300 to AAV6, and 1:100 of Nabs to AAV62 (Figure 5A). Similarly, serum from mice treated with AAV62 had a Nab titer of approximately 1:300 to AAV62, but only a Nab titer of approximately 1:100 to AAV6 (Figure 5B). Neither AAV62 nor AAV6 generated detectable Nabs against AAV26 and AAV2 (Figures 5A and 5B). On the other hand, AAV26 and AAV2 still induced moderate Nabs against AAV6 and AAV62, with Nab titers ranging from 1:10–100 (Figures 5C and 5D).
Figure 5.
Neutralizing antibodies comparison among AAV6, 62, 26, and 2/luc
(A–D) The AAV/luc vectors (including AAV6, AAV62, AAV26, and AAV2) were incubated with serial dilution of serum from mice injected with 5e9 vg of AAV6 (A), AAV62 (B), AAV26 (C), and AAV2 (D) for 1 h. The mixture of diluted serum and AAV was then added into Huh-7 cells at a multiplicity of infection of 2e3. After 48 h, the luciferase intensity was measured and AAV neutralization titer for each sample was determined by the serum dilution that reached 50% transduction compared with the result from the mixture of AAV vector with PBS. Data are presented as mean ± SD (n = 3).
Single amino acid change in AAV6 capsid VRI enhanced AAV transduction
There is only a two-amino acid difference between the VRIs of AAV2 and AAV6. Compared with AAV2, the VRI region of AAV6 differs by the insertion of one amino acid (threonine) at residue 265 of VP1 and the substitution of glutamine for alanine at residue 263. To study the role of two individual amino acids in AAV6 joint transduction, we designed two novel AAV capsids: the mutant AAV6D with a deletion of threonine at residue 265 and the mutant AAV6M with a mutation of glutamine to alanine at residue 263.
In order to avoid potential saturation from a high signal of luciferase expression, a dose of 1 × 109 vg of AAV/luc vectors was injected into the knee joints of mice, and imaging was performed at weeks 1 and 6 (Figure 6A). At week 1, the signal for all groups was relatively low, and there was no significant difference between different groups. At week 6, AAV6D induced 2-fold higher transgene expression than AAV6 and AAV62, while AAV6M induced around 10-fold lower transgene expression than AAV6 (Figure 6B).
Figure 6.
Transduction profile of AAV6D and AAV6M in vivo
(A) In vivo imaging of AAV6D, 6M, 6, and 62 at week 6. (B) In vivo luciferase intensity of AAV6D, 6M, 6, and 62 at week 1 and week 6. Data are shown as means ± SEM (n ≥ 4). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗∗∗∗p < 0.001. (C) In vitro luciferase assay at week 6. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗∗∗p < 0.005, ∗∗∗∗p < 0.001. (D) AAV genome copy number in the knee joint. Data are shown as means ± SEM (n ≥ 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. (E) The normalized luciferase expression per AAV genome copy number in the joints. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05. (F) AAV genome copy number in the liver. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons.
In agreement with the data in mice imaging, we detected 3.5-fold more luciferase activity in the cell lysates of knee joints treated with AAV6D compared with AAV6 (p < 0.05, Figure 6C). However, no significant difference of AAV genome copy numbers was found in the joints, even though the copy number between AAV6D and AAV6 was slightly higher with the value of 0.39 ± 0.18 and 0.38 ± 0.18 per cell, respectively. AAV6M and AAV62 showed relatively lower copy numbers, with values of 0.26 ± 0.15 and 0.21 ± 0.12 per cell (Figure 6D). After normalization with AAV gene copy number, we found that AAV6D and AAV62 induced 2-fold and 4-fold higher transgene expression per copy number than AAV6 in the joints, respectively (Figure 6E). The copy numbers in the liver were also more than 100 times lower than those of the knee joints, with no significant difference between AAV6 and the AAV mutants (AAV6D and AAV6M), indicating a limited capacity for crossing the local barrier and entering the bloodstream (Figure 6F).
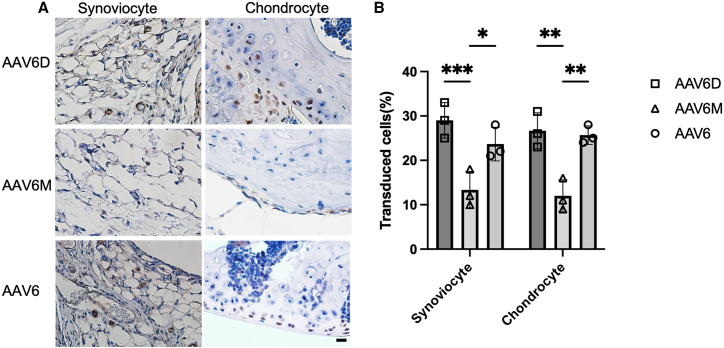
In addition, AAV6D demonstrated a comparable cell tropism to AAV6 (p > 0.05, Figures 7A and 7B), as it transduced 29.3% ± 4.5% of synoviocytes and 26.8% ± 4.1% of chondrocytes in the joints.
Figure 7.
Cell tropism in the mice knee joint after intra-articular injection of AAV6, AAV6D, or AAV6M
(A) Representative IHC staining of mice knee joints of AAV6, 6D, and 6M. Mice knee joints were fixed in 4% paraformaldehyde and were stained with rabbit anti-luciferase primary antibody. Positive cells were stained brown in color. (B) The percentages of positively stained cell in both synoviocytes and chondrocytes. The positively stained synoviocytes and chondrocytes were counted in 10 fields of view and averaged in each joint. Data are shown as means ± SEM (n = 3). Data were analyzed using one-way ANOVA followed by Bonferroni multiple comparison test for group comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.005.
Furthermore, we analyzed the Nab profiles in the blood of mice treated with intra-articular injection of AAV6 mutants. The administration of AAV6D generated more than 3-fold lower Nab titer in the injected mice compared with AAV6 (Figure S3). Additionally, we found that AAV6 Nabs were more than 3-fold less efficient in blocking AAV6D transduction than AAV6 (Figure S3).
Discussion
Local joint gene delivery is a promising strategy for achieving concentrated gene expression and fewer off-target side effects in other organs.26,27 Multiple serotypes of AAV have been explored intra-articularly in different species, such as mice, dogs, and horses. So far, several transgenes have been investigated for local gene therapy in arthritis by targeting the underlying molecular and cellular mechanisms that contribute to joint inflammation and damage. For example, clinical trials have shown that intra-articular injection of AAV expressing interleukin-1 receptor antagonist protein (IL-1Ra) or transforming growth factor β 1 (TGF-β1) can reduce pain and improve joint function in patients with osteoarthritis.28 In animal models of rheumatoid arthritis, intra-articular injection of AAV vectors encoding CTLA-47 or PD-L129 have been shown to reduce joint inflammation and improve joint function.
For intra-articular application of AAV vector-mediated gene delivery, there are still several challenges that need to be addressed. In particular, achieving high transduction efficiency, minimizing off-target effects, and avoiding Nabs in joint fluid are critical for successful AAV gene therapy. In order to optimize the capsid for the local joint gene delivery, this study compared the results of transduction in the mouse knee joints from AAV serotypes 1 to 9 and explored novel capsids by swapping VRI among AAV2, 5, 6, and 8, or modifying a single amino acid with either deletion at residue 265 (AAV6D) or mutation at residue 263 (AAV6M) on AAV6 capsid. Our study showed that AAV6D exhibited the strongest transduction efficiency with decreased Nab formation and limited off-target effects in the liver.
Our study indicates AAV6 demonstrated the overall best performance for joint gene therapy among AAV serotypes 1 to 9. Although AAV transduction in the joints has been studied, the information regarding the most suitable AAV serotype for joint gene therapy is still limited. Some of the previous studies only focused on a limited selection of serotypes, or in different experimental settings, ranging from in vitro cell lines, ex vivo human or equine joint samples, to in vivo mouse models. In the mouse studies, Kyostio-Moore et al. used an osteoarthritis mouse model and reported that the AAV1 capsid carrying the LacZ gene resulted in the most robust transduction of the joints and was the only serotype to transduce multiple cell types including chondrocytes, synovium, adipocytes, joint capsule, and skeletal muscle when compared with AAV2, 5, and 8.12 However, those studies did not apply AAV6. Yoon et al. compared AAV1–9 and AAV6.2, rh8, 10, 39, and 43 by injecting AAV/GFP in mice knee; they reported that AAV2 had the most positive transduced cells in both mouse knee cartilage and synovium, followed by AAV6.2 and AAV6.30 Nevertheless, they evaluated the transduction through counting the positive GFP cells per field, a method that relies more on qualitative analysis. In contrast, our study quantifies the total transgene expression and signal intensity utilizing luciferase reporter gene, which provides a more quantitative approach.
To verify the role of VRI in joint transduction, we have made several AAV mutants by swapping of VRI between AAV2, 5, 8 and AAV6. Substitution of VRI into AAV2 and AAV8 virions from AAV6 increased transduction. Swapping of AAV6 VRI from AAV8 decreased transduction in the joints. It was worth noting that AAV62 with substitution of VRI from AAV2 into AAV6 showed similar transduction to AAV6 at peak time points, but a lower transduction compared with AAV6 in the first week. This may contribute to a late virion cell and nuclear entry or capsid uncoating process. Similar to this finding, Clare et al. showed that AAV2 capsid exhibited a lag phase of intra-cellular trafficking before uncoating when compared with AAV6 and AAV8, resulting in a slower transduction.31 Interestingly, AAV62 showed a higher transduction per AAV genome copy number than AAV6. However, we compared the AAV binding efficiency using human synoviocytes and chondrocytes in vitro and found that there was no significant difference between AAV6 and AAV62 (data not shown). It is possible that the difference in transduction efficiency per AAV genome copy number between AAV6 and AAV62 may be attributed to the role of VRI in AAV trafficking, second-strand synthesis, and transcription of the input AAV genome.32 AAV62 may exhibit a slower transportation into the nucleus but a higher transcription than AAV6, which could potentially explain the variations in the presence of AAV genomes within cells but comparable transgene expression levels and transduced cells in our study.
It has been reported that certain amino acids in the AAV capsid VRI play important roles in transduction.33,34 Cabanes-Creus et al. has shown that insertion of an amino acid after residue 264 significantly increased AAV2 transduction efficiency in the muscle.34 Our previous work demonstrated that most AAV2 mutants with substitution of the amino acid at residue 265 from other 19 amino acids increased muscle transduction.35 In this study, AAV6D with just one amino acid deletion of threonine at 265 residue showed 2-fold higher transduction efficiency than AAV6. One potential mechanism may be due to the change of the stability of the intra-loop hydrogen bond network within VRI. This network is known to influence how the capsid interacts and binds with particles during various reactions. Specifically, during the capsid glycosylation, several motifs of the AAV capsid protein bind to glycans through hydrogen bond,36 which is essential for maintaining the proper conformation and function of the AAV. Hydrogen bond network has been found to be involved in capsid assembly, stability, cell entry, uncoating during cell entry, and endocytosis.37 Different serotypes have different numbers of hydrogen bonds in the VRI region. For example, AAV1 has four hydrogen bonds in the VRI region but AAV2 has two hydrogen bonds.38 The stability of the VRI loop can be reduced by deleting an amino acid residue involved in hydrogen bonding such as tyrosine or threonine,39 or substituting the amino acid residue with a residue that does not form a hydrogen bond. For example, AAV8 VRI only has one hydrogen bond, and it was reported that the deletion of T265 in AAV8, which is involved in the hydrogen bond, resulted in significantly increased transduction in the muscle and decreased transduction in the liver, while the deletion of the neighboring amino acid S266 did not impact transduction efficiency.38 Those findings indicate that the specific amino acid in VRI domain of AAV capsid plays an important role in the AAV life cycle and transduction profile.
The prevalence of pre-existing Nabs against AAV is high in humans, as Nabs against human-derived capsids have been detected in 40%–80% of human populations,17 especially for some common serotypes, such as AAV2 and AAV8, with cross-activity between serotypes.40 It has been demonstrated that Nabs can be detected in joint fluids in patients with joint disorders with a similar titer to that in circulation.41 AAV Nabs have been reported to potentially influence the efficacy of the gene therapy. Currently, several methods have been investigated to decrease Nab activity, including immunosuppression before AAV application,42 depletion of circulating IgG by IdeS or IdeZ43 or plasma apheresis, temporal blockage of all antibody isotypes with protein M,44 utilization of decoys that mimic the antigen and thus block the antibody binding,45 or capsid engineering.46 It has been demonstrated that VRI is a critical domain for Nab recognition.21,22,24
In this study, we only focused on Nab profiles in a mouse model by analyzing the Nab generated after immunization of the mice with AAV vectors, as it was not feasible to obtain human serum samples from patients treated with parental AAV serotypes. The newly formed Nab profile in a mouse model is a good indicator to study the level of immunogenicity induced by these variants, and their ability to evade the cross-reactivity of Nabs generated from other AAV variants. Both novel capsids AAV62 and AAV6D induce much lower Nab titers than AAV2 and AAV6 after immunizing the mice with those AAV vectors. Most importantly, these novel mutants have a higher capacity to evade Nabs generated from AAV2 or AAV6 immunization. This finding is consistent with our previous work, a single insertion of threonine at residue 265 on AAV2 decreased Nab titer and increased the ability to evade AAV2 Nabs.35 However, it is important to study the ability of AAV variants to evade pre-existing Nabs in comparison with parental AAV serotypes using human sera. In the future, we will collect serum samples from individuals and study the Nab profile for these AAV variants.
The most current gene therapy for joint diseases involves the direct delivery of immunomodulators into the joints.8,47 The expression of these immunomodulators in off-targeted tissues16,48 may induce unwanted side effects by interfering with systemic immune response. Additionally, the liver is the most common tissue transduced by AAV vectors after systemic administration.49 Recently, clinical trials have documented liver toxicity in patients with systemic AAV gene delivery.48,50 Therefore, it is imperative to apply for AAV vectors that have no or weak capacity to cross the joint barrier into the circulation to decrease the potential risk of liver toxicity and immune dysregulation. In this study, we did not detect any transgene expression in the liver of mice treated with intra-articular injection of AAV6 or its mutants AAV62 and AAV6D in spite of the presence of very low viral genome copy number. In contrast, AAV7, AAV8, and AAV9 induced a strong liver transduction after intra-articular injection. By swapping the VRI of AAV6 into AAV8, the mutant AAV86 showed significantly reduced liver transduction in mice compared with AAV8. This result is consistent with our previous finding that the mutant AAV LP2-10 with substitution of VRI from AAV6 into AAV8 had a decreased human hepatocyte transduction when compared with AAV8 in a chimeric mouse model with re-population of human hepatocytes.22 In this mouse model, AAV6 induced a lower human hepatocyte transduction than AAV8. These studies implicate that VRI plays a role in liver transduction and modification of VRI could potentially help mitigate off-target effects.
Our study primarily focuses on the role of VRI in joint transduction. However, VRI may not be the only VR that impacts joint transduction. There are nine VRs in the VP3 region of AAV that are distinct between serotypes; various VRs from different serotypes have been reported to play important roles in AAV functions. For example, Tenney et al. reported that swapping VR VII and IX from AAV8 into AAV2 can nearly match AAV8’s efficiency in transducing mouse liver.51 Gurda et al. reported that VRIV in AAV9 is involved in Nab binding, receptor binding, and transduction efficiency;52 they also identified that VRVIII plays a role in Nab escape and transduction in AAV2 and AAV8.53 Moreover, Ito et al. reported IV, VIII, and IX in AAV3 are important for reducing the Nab response.54 Yan et al. reported VRVI in AAV6 is critical for human airway epithelia transduction.55 Given the limited information regarding the specific role of various VRs in joint gene delivery have been investigated, in future, more could be explored to investigate the role of other VRs in joint transduction.
In conclusion, the AAV6 mutants described in this study expand the AAV vector pool available for joint gene delivery. The results from this study provide compelling evidence that the VRI from AAV6 plays a critical role in determining joint transduction efficiency and the activity of neutralizing antibodies. This study laid a foundation for potential clinical applications of the novel AAV capsids for joint gene delivery.
Materials and methods
Plasmid mutagenesis
To generate the mutated AAV variants, we synthesized the corresponding mutation sequences and cloned them into plasmids using two restriction enzymes (MfeI and Bsu36I for AAV6 cleavage, SwaI and EcoNI for AAV5 cleavage, EcoNI and BsiWI for AAV2 cleavage, and BsiWI and ApaI for AAV8 cleavage). Specifically, we created AAV62, 65, and 68 by swapping the VRI region of AAV2, AAV5, and AAV8 into AAV6, respectively. We also generated AAV26, 56, and 86 by swapping the VRI of AAV6 into AAV2, AAV5, and AAV8. To generate AAV6D, we deleted one amino acid, threonine, at position 265, while for AAV6M, we substituted one amino acid at position 263 from alanine to glutamine (Figure S4).
Recombinant AAV virus production
HEK-293 cells were cultured in Dulbecco’s Modified Eagle’s Medium with 10% fetal calf serum, 100 U mL−1 penicillin G and 100 μg mL−1 streptomycin at 37°C. Cells were routinely split 1:5 three times a week when they reached approximately 90% confluency.
To generate recombinant AAV viruses, the standard triple transfection method was used with the XX6-80 adenoviral helper plasmid, a packaging plasmid, and an ITR plasmid containing the luciferase gene with a strong, long-term, and ubiquitous CBA promoter. Both supernatant and cell lysates were subjected to a cesium chloride (CsCl) gradient ultracentrifugation to purify the AAV vectors. To quantify AAV titers, a quantitative real-time polymerase chain reaction (qPCR) was carried out using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) to detect the AAV sequence ITR, and primers for the ITR (forward: 5′- AAC ATG CTA CGC AGA GAG GGA GTG G -3′, reverse: 5′-CAT GAG ACA AGG AAC CCC TAG TGA TGG AG-3′) were designed and synthesized (Gene Script, NJ, USA). All RT-qPCRs included 40 cycles and a melt-curve. Serial dilutions of AAV with known titers were used as standards. Alkaline gel electrophoresis was also performed to verify AAV vector genome integrity; SYPRO Ruby protein gel stain (Thermo Fisher, Waltham, MA, USA) was used to confirm the presence of all the three AAV capsid viral proteins (VP1, VP2, and VP3). After AAV titer quantification, the viruses were diluted to the same concentration.
Animals
C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME, USA). All care and procedures were in accordance with the Guide for the Care and Use of Laboratory Animals and all procedures received prior approval by the University of North Carolina Institutional Animal Care and Usage Committee (UNC IACUC ID: 21–233.0). For the procedure of AAV injection, 7- to 8-week-old mice were anesthetized with isoflurane and 5 μL PBS containing 5 × 109 particles of AAV were injected into each knee joint. In each group, both of each mouse’s knee joints were injected with the same virus.
In vivo bioluminescence imaging
In vivo bioluminescence imaging was performed to assess the transduction efficiency of different AAV vectors in the knee joints of male C57BL/6 mice. The mice were anesthetized with 2.5% isoflurane and intraperitoneally injected with 100 μL luciferase substrate luciferin (150 mg/kg; Invitrogen, CA, USA). Bioluminescence imaging was performed 10 min after injection using a charge-coupled device camera (Berthold Technologies, Bad Wildbad, Germany) for 5 min. The photon signal was color-coded, with red presenting the highest intensity and blue the lowest. The signal intensities from the regions of interest were measured in total photon flux (photons/s per cm2).
In vitro luciferase assay
Six weeks after injection with AAV, the mice were euthanized and their joint tissues were harvested. Approximately 10 mg of each tissue was minced on ice and homogenized in 500 μL of 5X Passive Lysis Buffer (Promega, Madison, WI, USA) overnight at 4°C. For luminometric analysis, 25 μL of lysates and 100 μL of D-luciferin substrate (Promega, Madison, WI, USA) were mixed and transferred to 96-well plates. Total protein concentration in tissue lysates was determined using the BCA assay (Bio-Rad, Hercules, CA, USA).
AAV copy number
The biodistribution of AAV was determined by quantifying the AAV copy number in various organs. Knee and liver tissues were collected at 6 weeks post-injection. Approximately 25 mg of tissue was extracted from each organ and treated with proteinase K using a DNeasy kit (Qiagen, Hilden, Germany) to extract host and vector genomic DNA. To determine the AAV copy number per cell by qPCR assay, primers for the luciferase gene were designed and synthesized (forward: 5′- TGAGTACTTCGAAATGTCCGTTC-3′, reverse: 5′-GTATTCAGCCCATATCGTTTCAT-3′) (Gene Script, NJ, USA). The diploid genomes were calculated based on nanodrop. The normalized transduction per vector genome was determined by initially calculating the ratio of in vitro luciferase activity per gram of joint tissue to the vector copy number per diploid DNA genome for each sample. Then, using AAV6 as a baseline (set to 1), the expression levels of the other groups were normalized against AAV6 to obtain their relative ratios.
Detection of Nabs
For comparison of humoral immune response among AAV capsids, three serum samples from each group were collected from the mice 6 weeks after AAV injection. Huh-7 cells were seeded in a 48-well plate at a density of 105 cells/well in 200 μL of ex vivo medium (Lonza, Basel, Switzerland). The cells were cultured for 3–4 h at 37°C and allowed to adhere to the well. The sera were diluted as 1:3, 1:10, 1:33, 1:100, 1:333, 1:1000, and 1:3333 with PBS. Each diluted serum was then incubated with 5 μL of different AAV vectors (4 × 107 vg/μL) for 1 h at room temperature. The mixture of serum and AAV was then added into Huh-7 cells. After 48 h, cells were lysed by 5x protein lysis buffer (Promega, Madison, WI, USA) and mixed with 100 μL luciferin (Promega, Madison, WI, USA), and luciferase luminescence was measured at a wavelength of 562 nm. The Nab titer was determined based on 50% inhibition of signal intensity.
Tissue histopathology
At week 6, the mice were euthanized and the knee joints were collected by dissecting the femur and tibia 5 mm away from the knee joint. The harvested knees were then fixed in 4% paraformaldehyde and decalcified in 5% trichloroacetic acid for 7 days. They were then dehydrated, embedded in paraffin, and sectioned at 5 μm thickness. The sections were stained with hematoxylin and eosin staining and immunohistochemistry staining (IHC). To perform IHC, the sections were de-paraffinized, rehydrated, subjected to heat-induced antigen retrieval at 95°C for 8 min in 0.01 M sodium citrate, and cooled at room temperature for 25 min. Subsequently, the samples were incubated in 3% H2O2 in methanol for 20 min, blocked with 10% normal goat serum for 1 h, and incubated overnight at 4°C with rabbit anti-luciferase primary antibody (Sigma-Aldrich, St. Louis, MO, USA, 1/1,500 dilution). Negative controls were treated with 10% normal goat serum without the primary antibodies. Anti-rabbit antibody (abcam, Cambridge, UK, 1/500 dilution) was used as a secondary antibody. The color was developed using a Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA, USA) and DAB Substrate Kit (Vector Laboratories, Burlingame, CA, USA). Three knee joints were used in each group, and 10 fields of view per joint were evaluated. Positively stained cells in each field of view were counted by two blinded observers and averaged.
Statistical analysis
GraphPad Prism 9 software was used for statistical analysis. Data were shown as mean ± SD, and the box and whisker plots and line chart were used for descriptive statistics. Differences among each group were determined by one-way ANOVA or Student’s t test. Bonferroni and Sidak tests were used for multiple comparisons between groups. The significance level was set at 0.05. Based on the power analysis of our preliminary data using nQuery software, the power of mouse sample size was over 80% at a significance level of 0.05.
Data and code availability
The authors confirm that the data generated during this study are included in this article and its supplemental information. Raw data that support the findings of this study are available upon request from the corresponding author.
Acknowledgments
We would like to thank Drs. Matthew Hirsch, Richard Loeser, Roland Arnold, Shannon Wallet, Junjiang Sun, and Charles Askew who provided us with valuable suggestions, and Dr. Chuanan Zhang from UNC vector core for technical support of AAV virus production. We also acknowledge the UNC Biomedical Research Imaging Center (BRIC) Small Animal Imaging (SAI) facility for assistance with mouse imaging and UNC Animal Histopathology Core for histological services. This work was partly supported by National Institutes of Health grants R01HL144661, R01AI168242, R01HL151348. The graphical abstract was created with BioRender.com.
Author contributions
W.L. conducted the experiments and wrote the manuscript; S.F. and L.F. assisted with the experiments and data analysis; and C.L. designed the experiments and revised the paper. All authors reviewed, edited, and approved the final manuscript.
Declaration of interests
W.L., R.J.S., and C.L. are inventors on the patent application related to this work. C.L. is a cofounder of Bedrock Therapeutics, NabGen, GeneVentiv, and Astro, Inc. He has licensed patents by UNC and has received royalties from these startups and Asklepios Biopharmaceutical. R.J.S. is the founder and a shareholder at Asklepios Biopharmaceutical. He holds patents that have been licensed by the UNC to Asklepios Biopharmaceutical, for which he receives royalties. He has consulted for Baxter Health Care and has received payment for speaking.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2024.101211.
Supplemental information
References
- 1.Barbour K.E., Helmick C.G., Boring M., Brady T.J. Vital Signs: Prevalence of Doctor-Diagnosed Arthritis and Arthritis-Attributable Activity Limitation — United States, 2013–2015. MMWR Morb. Mortal. Wkly. Rep. 2017;66:246–253. doi: 10.15585/mmwr.mm6609e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arend W.P., Dayer J.-M. Cytokines and cytokine inhibitors or antagonists in rheumatoid arthritis. Arthritis Rheum. 1990;33:305–315. doi: 10.1002/art.1780330302. [DOI] [PubMed] [Google Scholar]
- 3.Bryant A., Moore J. Rituximab and its potential for the treatment of rheumatoid arthritis. Therapeut. Clin. Risk Manag. 2006;2:207–212. doi: 10.2147/tcrm.2006.2.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansen D.T.S.L., el Bannoudi H., Arens R., Habets K.L.L., Hameetman M., Huizinga T.W.J., Stoop J.N., Toes R.E.M. Abatacept decreases disease activity in a absence of CD4(+) T cells in a collagen-induced arthritis model. Arthritis Res. Ther. 2015;17:220. doi: 10.1186/s13075-015-0731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curtis J.R., Singh J.A. Use of biologics in rheumatoid arthritis: current and emerging paradigms of care. Clin. Therapeut. 2011;33:679–707. doi: 10.1016/j.clinthera.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphreys J., Hyrich K., Symmons D. What is the impact of biologic therapies on common co-morbidities in patients with rheumatoid arthritis? Arthritis Res. Ther. 2016;18:282. doi: 10.1186/s13075-016-1176-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Wang F., Wang B., Zhang J., Yu J.-Y. Intra-articular gene delivery of CTLA4-FasL suppresses experimental arthritis. Int. Immunol. 2012;24:379–388. doi: 10.1093/intimm/dxs041. [DOI] [PubMed] [Google Scholar]
- 8.Evans C.H., Ghivizzani S.C., Robbins P.D. Gene Delivery to Joints by Intra-Articular Injection. Hum. Gene Ther. 2018;29:2–14. doi: 10.1089/hum.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Theis K.A., Murphy L.B., Guglielmo D., Boring M.A., Okoro C.A., Duca L.M., Helmick C.G. Prevalence of Arthritis and Arthritis-Attributable Activity Limitation - United States, 2016-2018. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1401–1407. doi: 10.15585/mmwr.mm7040a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J., Hakobyan N., Valentino L.A., Feldman B.L., Samulski R.J., Monahan P.E. Intra-articular factor IX protein or gene replacement protects against development of hemophilic synovitis in the absence of circulating factor IX. Blood. 2008;112:4532–4541. doi: 10.1182/blood-2008-01-131417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salganik M., Hirsch M.L., Samulski R.J. 2017. Adeno-associated Virus as a Mammalian DNA Vector; p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyostio-Moore S., Bangari D.S., Ewing P., Nambiar B., Berthelette P., Sookdeo C., Hutto E., Moran N., Sullivan J., Matthews G.L., et al. Local gene delivery of heme oxygenase-1 by adeno-associated virus into osteoarthritic mouse joints exhibiting synovial oxidative stress. Osteoarthritis Cartilage. 2013;21:358–367. doi: 10.1016/j.joca.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Payne K.A., Lee H.H., Haleem A.M., Martins C., Yuan Z., Qiao C., Xiao X., Chu C.R. Single intra-articular injection of adeno-associated virus results in stable and controllable in vivo transgene expression in normal rat knees. Osteoarthritis Cartilage. 2011;19:1058–1065. doi: 10.1016/j.joca.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Q., Luo H., Zhou C., Yu H., Yao S., Fu F., Seeley R., Ji X., Yang Y., Chen P., et al. Comparative intra-articular gene transfer of seven adeno-associated virus serotypes reveals that AAV2 mediates the most efficient transduction to mouse arthritic chondrocytes. PLoS One. 2020;15 doi: 10.1371/journal.pone.0243359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boissier M.-C., Lemeiter D., Clavel C., Valvason C., Laroche L., Begue T., Bessis N. Synoviocyte infection with adeno-associated virus (AAV) is neutralized by human synovial fluid from arthritis patients and depends on AAV serotype. Hum. Gene Ther. 2007;18:525–535. doi: 10.1089/hum.2006.174. [DOI] [PubMed] [Google Scholar]
- 16.Fischer K.B., Collins H.K., Callaway E.M. Sources of off-target expression from recombinase-dependent AAV vectors and mitigation with cross-over insensitive ATG-out vectors. Proc. Natl. Acad. Sci. USA. 2019;116:27001–27010. doi: 10.1073/pnas.1915974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 19.Bartel M., Schaffer D., Büning H. Enhancing the clinical potential of AAV vectors by capsid engineering to evade pre-existing immunity. Front. Microbiol. 2011;2 doi: 10.3389/fmicb.2011.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker J., Fakhiri J., Grimm D. Fantastic AAV Gene Therapy Vectors and How to Find Them-Random Diversification, Rational Design and Machine Learning. Pathogens. 2022;11:756. doi: 10.3390/pathogens11070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govindasamy L., Padron E., McKenna R., Muzyczka N., Kaludov N., Chiorini J.A., Agbandje-McKenna M. Structurally Mapping the Diverse Phenotype of Adeno-Associated Virus Serotype 4. J. Virol. 2006;80:11556–11570. doi: 10.1128/JVI.01536-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei X., Shao W., Xing A., Askew C., Chen X., Cui C., Abajas Y.L., Gerber D.A., Merricks E.P., Nichols T.C., et al. Development of AAV Variants with Human Hepatocyte Tropism and Neutralizing Antibody Escape Capacity. Mol. Ther. Methods Clin. Dev. 2020;18:259–268. doi: 10.1016/j.omtm.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albright B.H., Simon K.E., Pillai M., Devlin G.W., Asokan A. Modulation of Sialic Acid Dependence Influences the Central Nervous System Transduction Profile of Adeno-associated Viruses. J. Virol. 2019;93:e00332-19. doi: 10.1128/JVI.00332-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowles D.E., McPhee S.W.J., Li C., Gray S.J., Samulski J.J., Camp A.S., Li J., Wang B., Monahan P.E., Rabinowitz J.E., et al. Phase 1 Gene Therapy for Duchenne Muscular Dystrophy Using a Translational Optimized AAV Vector. Mol. Ther. 2012;20:443–455. doi: 10.1038/mt.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kyostio-Moore S., Berthelette P., Cornell C.S., Nambiar B., Figueiredo M.D. Hyaluronic acid synthase-2 gene transfer into the joints of Beagles by use of recombinant adeno-associated viral vectors. AJVR (Am. J. Vet. Res.) 2018;79:505–517. doi: 10.2460/ajvr.79.5.505. [DOI] [PubMed] [Google Scholar]
- 26.Paulk N. Gene Therapy: It Is Time to Talk about High-Dose AAV: The deaths of two children with X-linked myotubular myopathy in the ASPIRO trial prompts a reexamination of vector safety. Genet. Eng. Biotechnol. News. 2020;40:14–16. doi: 10.1089/gen.40.09.04. [DOI] [Google Scholar]
- 27.Zincarelli C., Soltys S., Rengo G., Rabinowitz J.E. Analysis of AAV Serotypes 1–9 Mediated Gene Expression and Tropism in Mice After Systemic Injection. Mol. Ther. 2008;16:1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 28.Grässel S., Muschter D. Recent advances in the treatment of osteoarthritis. F1000Res. 2020;9 doi: 10.12688/f1000research.22115.1. F1000 Faculty Rev-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W., Sun J., Feng S.L., Wang F., Miao M.Z., Wu E.Y., Wallet S., Loeser R., Li C. Intra-articular delivery of AAV vectors encoding PD-L1 attenuates joint inflammation and tissue damage in a mouse model of rheumatoid arthritis. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1116084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon D.S., Lee K.-M., Cho S., Ko E.A., Kim J., Jung S., Shim J.-H., Gao G., Park K.H., Lee J.W. Cellular and Tissue Selectivity of AAV Serotypes for Gene Delivery to Chondrocytes and Cartilage. Int. J. Med. Sci. 2021;18:3353–3360. doi: 10.7150/ijms.56760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas C.E., Storm T.A., Huang Z., Kay M.A. Rapid Uncoating of Vector Genomes Is the Key toEfficient Liver Transduction with Pseudotyped Adeno-Associated VirusVectors. J. Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salganik M., Aydemir F., Nam H.-J., McKenna R., Agbandje-McKenna M., Muzyczka N. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J. Virol. 2014;88:1071–1079. doi: 10.1128/JVI.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Weigel-Van Aken K.A., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cabanes-Creus M., Navarro R.G., Liao S.H.Y., Baltazar G., Drouyer M., Zhu E., Scott S., Luong C., Wilson L.O.W., Alexander I.E., Lisowski L. Single amino acid insertion allows functional transduction of murine hepatocytes with human liver tropic AAV capsids. Mol. Ther. Methods Clin. Dev. 2021;21:607–620. doi: 10.1016/j.omtm.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li C., Diprimio N., Bowles D.E., Hirsch M.L., Monahan P.E., Asokan A., Rabinowitz J., Agbandje-McKenna M., Samulski R.J. Single Amino Acid Modification of Adeno-Associated Virus Capsid Changes Transduction and Humoral Immune Profiles. J. Virol. 2012;86:7752–7759. doi: 10.1128/JVI.00675-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer N.L., Chapman M.S. Adeno-associated virus (AAV) cell entry: structural insights. Trends Microbiol. 2022;30:432–451. doi: 10.1016/j.tim.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poudel L., Twarock R., Steinmetz N.F., Podgornik R., Ching W.-Y. Impact of Hydrogen Bonding in the Binding Site between Capsid Protein and MS2 Bacteriophage ssRNA. J. Phys. Chem. B. 2017;121:6321–6330. doi: 10.1021/acs.jpcb.7b02569. [DOI] [PubMed] [Google Scholar]
- 38.Warischalk, J.K. Uncovering structural components of the adeno-associated viral capsid that can be modified to improve clinical gene therapy outcomes. 142.
- 39.Castle M.J., Turunen H.T., Vandenberghe L.H., Wolfe J.H. Controlling AAV Tropism in the Nervous System with Natural and Engineered Capsids. Methods Mol. Biol. 2016;1382:133–149. doi: 10.1007/978-1-4939-3271-9_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kruzik A., Fetahagic D., Hartlieb B., Dorn S., Koppensteiner H., Horling F.M., Scheiflinger F., Reipert B.M., de la Rosa M. Prevalence of Anti-Adeno-Associated Virus Immune Responses in International Cohorts of Healthy Donors. Mol. Ther. Methods Clin. Dev. 2019;14:126–133. doi: 10.1016/j.omtm.2019.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cottard V., Valvason C., Falgarone G., Lutomski D., Boissier M.-C., Bessis N. Immune Response Against Gene Therapy Vectors: Influence of Synovial Fluid on Adeno-Associated Virus Mediated Gene Transfer to Chondrocytes. J. Clin. Immunol. 2004;24:162–169. doi: 10.1023/B:JOCI.0000019781.64421.5c. [DOI] [PubMed] [Google Scholar]
- 42.Prasad S., Dimmock D.P., Greenberg B., Walia J.S., Sadhu C., Tavakkoli F., Lipshutz G.S. Immune Responses and Immunosuppressive Strategies for Adeno-Associated Virus-Based Gene Therapy for Treatment of Central Nervous System Disorders: Current Knowledge and Approaches. Hum. Gene Ther. 2022;33:1228–1245. doi: 10.1089/hum.2022.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elmore Z.C., Oh D.K., Simon K.E., Fanous M.M., Asokan A. Rescuing AAV gene transfer from neutralizing antibodies with an IgG-degrading enzyme. JCI Insight. 2020;5 doi: 10.1172/jci.insight.139881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bryant D.H., Bashir A., Sinai S., Jain N.K., Ogden P.J., Riley P.F., Church G.M., Colwell L.J., Kelsic E.D. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 2021;39:691–696. doi: 10.1038/s41587-020-00793-4. [DOI] [PubMed] [Google Scholar]
- 45.Mingozzi F., Anguela X.M., Pavani G., Chen Y., Davidson R.J., Hui D.J., Yazicioglu M., Elkouby L., Hinderer C.J., Faella A., et al. Overcoming preexisting humoral immunity to AAV using capsid decoys. Sci. Transl. Med. 2013;5:194ra92. doi: 10.1126/scitranslmed.3005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gross D.-A., Tedesco N., Leborgne C., Ronzitti G. Overcoming the Challenges Imposed by Humoral Immunity to AAV Vectors to Achieve Safe and Efficient Gene Transfer in Seropositive Patients. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.857276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodrich L.R., Grieger J.C., Phillips J.N., Khan N., Gray S.J., McIlwraith C.W., Samulski R.J. scAAVIL-1ra dosing trial in a large animal model and validation of long-term expression with repeat administration for osteoarthritis therapy. Gene Ther. 2015;22:536–545. doi: 10.1038/gt.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matagne V., Borloz E., Ehinger Y., Saidi L., Villard L., Roux J.-C. Severe offtarget effects following intravenous delivery of AAV9-MECP2 in a female mouse model of Rett syndrome. Neurobiol. Dis. 2021;149 doi: 10.1016/j.nbd.2020.105235. [DOI] [PubMed] [Google Scholar]
- 49.Yang L., Jiang J., Drouin L.M., Agbandje-Mckenna M., Chen C., Qiao C., Pu D., Hu X., Wang D.-Z., Li J., Xiao X. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc. Natl. Acad. Sci. USA. 2009;106:3946–3951. doi: 10.1073/pnas.0813207106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colella P., Ronzitti G., Mingozzi F. Emerging Issues in AAV-Mediated In Vivo Gene Therapy. Mol. Ther. Methods Clin. Dev. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenney R.M., Bell C.L., Wilson J.M. AAV8 capsid variable regions at the two-fold symmetry axis contribute to high liver transduction by mediating nuclear entry and capsid uncoating. Virology. 2014;454–455:227–236. doi: 10.1016/j.virol.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 52.Gurda B.L., DiMattia M.A., Miller E.B., Bennett A., McKenna R., Weichert W.S., Nelson C.D., Chen W.J., Muzyczka N., Olson N.H., et al. Capsid Antibodies to Different Adeno-Associated Virus Serotypes Bind Common Regions. J. Virol. 2013;87:9111–9124. doi: 10.1128/JVI.00622-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurda B.L., Raupp C., Popa-Wagner R., Naumer M., Olson N.H., Ng R., McKenna R., Baker T.S., Kleinschmidt J.A., Agbandje-McKenna M. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J. Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ito M., Takino N., Nomura T., Kan A., Muramatsu S.I. Engineered adeno-associated virus 3 vector with reduced reactivity to serum antibodies. Sci. Rep. 2021;11:9322. doi: 10.1038/s41598-021-88614-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan Z., Lei-Butters D.C.M., Keiser N.W., Engelhardt J.F. Distinct transduction difference between adeno-associated virus type 1 and type 6 vectors in human polarized airway epithelia. Gene Ther. 2013;20:328–337. doi: 10.1038/gt.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data generated during this study are included in this article and its supplemental information. Raw data that support the findings of this study are available upon request from the corresponding author.