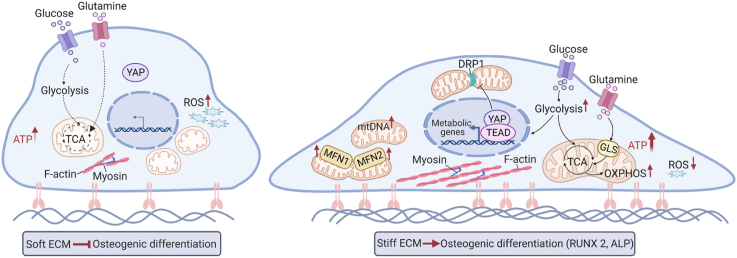
Abstract
The biophysical factors of biomaterials such as their stiffness regulate stem cell differentiation. Energy metabolism has been revealed an essential role in stem cell lineage commitment. However, whether and how extracellular matrix (ECM) stiffness regulates energy metabolism to determine stem cell differentiation is less known. Here, the study reveals that stiff ECM promotes glycolysis, oxidative phosphorylation, and enhances antioxidant defense system during osteogenic differentiation in MSCs. Stiff ECM increases mitochondrial fusion by enhancing mitofusin 1 and 2 expression and inhibiting the dynamin-related protein 1 activity, which contributes to osteogenesis. Yes-associated protein (YAP) impacts glycolysis, glutamine metabolism, mitochondrial dynamics, and mitochondrial biosynthesis to regulate stiffness-mediated osteogenic differentiation. Furthermore, glycolysis in turn regulates YAP activity through the cytoskeletal tension-mediated deformation of nuclei. Overall, our findings suggest that YAP is an important mechanotransducer to integrate ECM mechanical cues and energy metabolic signaling to affect the fate of MSCs. This offers valuable guidance to improve the scaffold design for bone tissue engineering constructs.
Keywords: Matrix stiffness, Metabolic regulation, YAP, Stem cell fate, Cellular mechanics
Graphical abstract
Highlights
-
•
Glycolysis is the main source of ATP for ECM stiffness-induced osteogenic differentiation in MSCs.
-
•
Stiff ECM promotes mitochondrial fusion and biosynthesis.
-
•
YAP acts as a mechan-matabolic sensor to regulate osteogenic capacity of MSCs.
-
•
ECM stiffness-induced glycolysis regulates YAP via cellular mechanics.
1. Introduction
The properties of the physiological environment can be simulated by biomaterials to modulate cell behaviors. Microenvironments trigger biochemical signaling through mechanotransduction, which allows cells to sense, respond, and adapt to their physical surroundings. Mesenchymal stem cells (MSCs) have been widely used in studies of tissue regeneration and stem cell therapy [1]. Their homeostatic maintenance, self-renewal, and cell fate decisions have been known to be regulated by the stem cell microenvironment, namely the stem cell niche. Extracellular matrix (ECM) stiffness is one of the critical factors regulating the fate of stem cells [2]. Cells can detect and react to ECM stiffness by adhesive ligands [3]. It has been reported that stiff ECM promotes osteogenesis, while soft ECM induces adipogenesis of MSCs [4]. Furthermore, ECM stiffening increases cell spreading and actomyosin contractility and enhances the expression of RUNX2, Smad, β-catenin, Yes-associated protein (YAP) [5], and TRPV4 [6] to promote osteogenic differentiation [7]. New insights into the mechanism of substrate stiffness-mediated differentiation can contribute to guiding MSC fate in therapeutically desirable directions.
The ability of the metabolism to guide the fate of stem cells has received significant attention in the last decade [8]. It has demonstrated that cell metabolism not only contributes to energy production but also plays critical roles in stem cell proliferation and lineage commitment [9,10]. It has been shown that embryonic stem cells transfer anaerobic glycolysis into the mitochondrial oxidative metabolism to meet the energy requirement of cardiac differentiation [11]. During the differentiation of pluripotent stem cells (PSCs), mitochondrial fusion proteins, MFN1 and 2, and optic atrophy 1 are increased to activate oxidative metabolism [12]. It has been reported that biomaterials can be designed to regulate cellular metabolism to promote tissue regeneration, which is through the delivery of metabolic factors such as citrate and glutamine [13,14]. Whereas how biomaterial properties affect energy metabolism to regulate cell fate remains elusive.
Notably, recent studies have indicated the linkage of metabolic responses to ECM stiffness. Soft ECM increases DRP1- and MIEF1/2-dependent mitochondrial fission to produce more reactive oxygen species (ROS) in breast cancer cells [15]. ECM stiffness-induced disassembly of stress fiber triggers high glycolytic rates, which enable cells to adapt to the mechanical properties of the tissue microenvironment [16]. These studies suggest that biomaterial stiffness-mediated energy metabolism is essential to regulate cell behavior and fate. YAP is a well-known mechanotransducer that is activated by mechanical cues and ECM stiffness [17]. YAP regulates a variety of biological behaviors, including stem cell growth and differentiation [18]. ECM stiffening can promote the proliferation of endothelial cells through YAP/TAZ-dependent glutaminolysis and anaplerosis in pulmonary hypertension [19]. Moreover, cell migration of hepatocellular carcinoma cells can be accelerated by a stiffer ECM, which involves YAP-mediated aerobic glycolysis [20]. Nevertheless, little is known about the interplay of YAP and metabolism in ECM stiffness-induced osteogenesis.
Here, our results show that stiff ECM could promote the osteogenic differentiation of MSCs by regulating metabolic processes such as glycolysis, OXPHOS, mitochondrial dynamics, and glutaminolysis. Moreover, metabolism processes can also regulate cellular mechanics. Glycolysis is involved in F-actin polymerization, myosin contractility, and paxillin formation. YAP provides a mechanistic link between mechanotransduction, metabolism, and osteogenic differentiation. Taken together, our work highlights that the engineered ECM properties can regulate YAP-mediated energy metabolism to enhance the osteogenesis of MSCs, which inspires innovations in biomaterials for improved bone regeneration.
2. Results
2.1. ECM stiffness regulates osteogenic differentiation and ATP production in MSCs
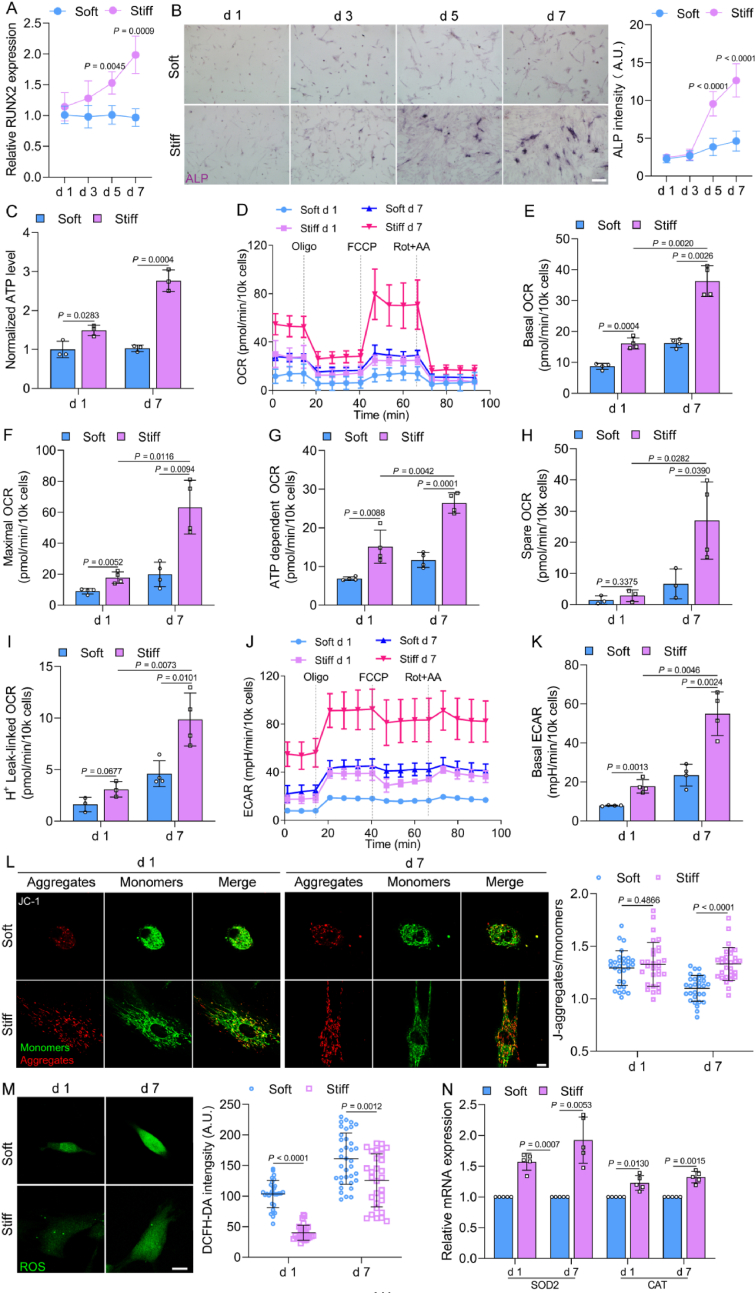
MSCs were seeded on soft (4.47 kPa) and stiff (40 kPa) ECM with normal growth medium for 1–7 days. The osteogenic differentiation of MSCs was promoted on a stiff ECM, with an increase of osteogenic marker RUNX2 expression and alkaline phosphatase (ALP) after 5 and 7 days (Fig. 1A and B), consistent with previous studies [5,21]. To examine the changes in bioenergetics, we measured ATP production on days 1 and 7. More ATP (two-fold higher) was generated in cells cultured on a stiff ECM compared with on a soft ECM (Fig. 1C). Bioenergetic profiling was performed by Seahorse analyses. The basal oxygen consumption rate (OCR), maximal OCR, and ATP-dependent OCR of cells on a stiff ECM were higher than those cultured on a soft ECM on days 1 and 7 (Fig. 1 D–G). Moreover, spare capacity OCR and proton leak OCR were both increased on day 7 on the stiff ECM (Fig. 1H and I). Likewise, the basal extracellular acidification rate (ECAR) was increased on the stiff ECM (Fig. 1J and K), indicating that stiff ECM promote the OXPHOS and glycolysis of MSCs. The mitochondrial membrane potential (ΔΨm) on the stiff ECM increased after 7 days (Fig. 1L). In addition, the ROS level on the stiff ECM was lower than that on the soft ECM (Fig. 1M), which was consistent with the increase in the gene expression of superoxide dismutase 2 (SOD2) and catalase (CAT) on the stiff ECM (Fig. 1N). These results indicate that ECM stiffness, in the absence of a mineralization-inducing medium, regulates osteogenic differentiation, which was accompanied by increased energy production and oxidative stress resistance in MSCs.
Fig. 1.
ECM stiffness promotes osteogenic differentiation of MSCs with increased ATP production. (A) RUNX2 gene expression of MSCs on soft and stiff ECM for 1–7 days (n = 4 for each group). (B) Left, representative images of ALP staining in MSCs for 1–7 days, scale bars, 200 μm. Right, quantification of ALP intensity (left to right, n = 12, 12, 10, 14, 19, 15, 14, 13). (C) Normalized ATP levels of MSCs on days 1 and 7 (n = 3 for each group). (D) Oxygen consumption rate (OCR) was determined by Seahorse in MSCs on days 1 and 7 (n = 4 for each group). (E–I) Quantification of individual parameters for basal OCR (E), maximal OCR (F), ATP dependent respiration (G), spare OCR (H), and H+ leak-linked OCR (I). (J and K) Extracellular acidification rate (ECAR) of MSCs (J) and quantification of basal ECAR (K, n = 4 for each group). (L) Mitochondrial membrane potential (ΔΨm) of MSCs on soft and stiff ECM on days 1 and 7. Left, representative images of JC-1 staining in MSCs, scale bars, 10 μm. Right, the ratio of J-aggregates to monomers (n = 30 for each group). (M) Levels of ROS in MSCs. Left, representative images, scale bars, 25 μm. Right, quantification of intensity (left to right, n = 30, 30, 36, 30). (N) The gene expression levels of SOD2 (n = 5 for each group) and CAT (n = 5 for each group) in MSCs. Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from unpaired two-tailed Student's t-test, except for ROS intensity d 1 in M (Mann-Whitney test).
2.2. ECM stiffness regulates metabolic modulation in MSCs
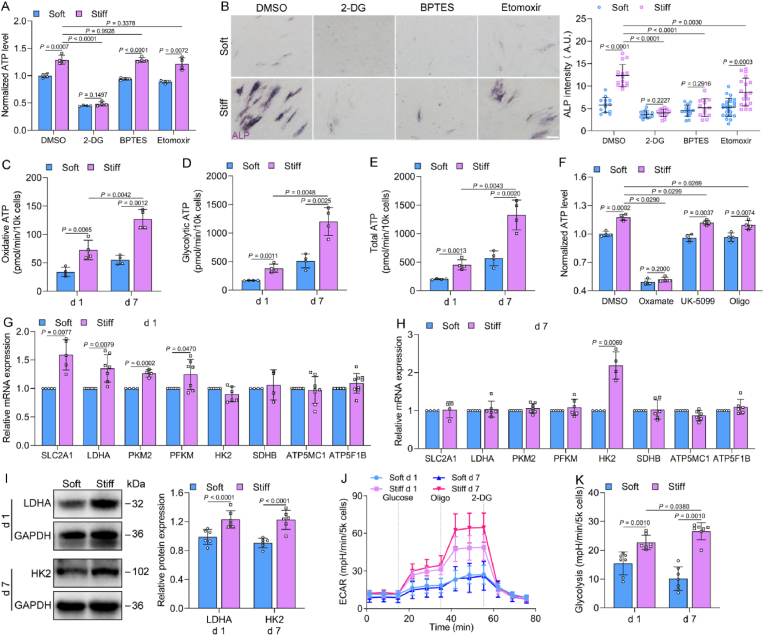
To determine the contribution of glucose, glutamine, and fatty acids to ATP production, their utilization was individually inhibited by 2-DG, BPTES, and etomoxir, respectively, and then ATP production was measured after 1 day. The inhibitor 2-DG was decreased, whereas BPTES and etomoxir barely affected ATP production (Fig. 2A), indicating that the primary energy source is glucose for MSCs cultured on a stiff ECM. These three inhibitors all suppressed ALP expression on the stiff ECM on day 7 (Fig. 2B), suggesting that glucose, glutamine, and fatty acids are vital for the stiffness-induced osteogenic differentiation of MSCs.
Fig. 2.
ECM stiffness enhanced metabolic activities in MSCs. (A) Intracellular ATP levels of MSCs on soft and stiff ECM on day 1 (n = 4 for each group). Cells were treated with inhibitors of glucose (100 mM 2-DG), glutamine (20 μM BPTES), and fatty acids (200 μM etomoxir). (B) Left, representative images of ALP staining in MSCs treated with the inhibitors after 7 days, scale bars, 200 μm. Right, quantification of ALP intensity (left to right, n = 13, 15, 14, 15, 15, 13, 24, 19). (C–E) Theoretical ATP production from OXPHOS, glycolysis, and total ATP by Seahorse analysis in MSCs on soft and stiff ECM on days 1 and 7 (n = 4 for each group). (F) Intracellular ATP levels of MSCs on day 1 (n = 4 for each group). Cells were treated with the inhibitors of lactate dehydrogenase (100 mM Oxamate), mitochondrial pyruvate carrier (100 mM UK-5099), and ATP synthase (2 mM Oligomycin, Oligo). (G and H) Metabolic gene expression analysis of MSCs on days 1 (G, n = 5 for SLC2A1, 7 for LDHA, 4 for PKM2, 7 for HK2, 7 for PFKM, 4 for SDHB, 8 for ATP5MC1, 9 for ATP5F1B) and 7 (H, n = 4 for SLC2A1, 6 for LDHA, 7 for PKM2, 6 for HK2, 4 for PFKM, 6 for SDHB, 6 for ATP5MC1, 6 for ATP5F1B). (I) Left, immunoblots for the protein expression of glycolytic enzymes in MSCs on days 1 and 7. Right, quantification of LDHA and HK2 protein expression (n = 6 for each group). (J and K) Extracellular acidification rate (ECAR) was determined by Seahorse in MSCs on days 1 and 7 (J) and quantification of glycolytic rates (K, n = 7 for each group). Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from unpaired two-tailed Student's t-test (A–I) or Mann-Whitney test (K).
We calculated theoretical ATP production from OXPHOS or glycolysis was increased on the stiff ECM [22]. The ATP produced from OXPHOS or glycolysis was increased with ∼32% and ∼57% increases on the stiff ECM on day 7 compared to day 1 (Fig. 2C and E). Moreover, the ATP produced from glycolysis was greater than 80% of the total amount produced (Fig. 2D and E). To further verify the specific energy-production pathways of ATP production, lactate dehydrogenase, mitochondrial pyruvate carrier, and mitochondrial ATP synthase were inhibited by oxamate, UK-5099, and oligomycin, respectively. Oxamate reduced the intracellular ATP levels by approximately 50%, whereas ATP levels were slightly decreased by UK-5099 and oligomycin (Fig. 2F). These data demonstrate that glycolysis is the main pathway for energy generation in MSCs cultured on a stiff ECM.
Furthermore, the genes related to glycolysis, such as glucose transporter 1 (SLC2A1), lactate dehydrogenase A (LDHA), pyruvate kinase M2 (PKM2), and phosphofructokinase (PFKM) were upregulated on the stiff ECM on day 1 (Fig. 2G). Interestingly, only hexokinase 2 (HK2) was upregulated in genes associated with glycolysis on a stiff ECM on day 7 (Fig. 2H), which was consistent with the increase of HK2 from RNA sequencing (RNA-seq) analysis (Fig. S3 A). RNA-seq analysis showed that there were no significant changes in genes of the TCA cycle and the mitochondrial electron transport chain (ETC) complexes on the stiff ECM compared to the soft ECM on day 7 (Fig. S3 B–G). Additionally, the protein expression of glycolytic enzymes LDHA and HK2 were also upregulated on the stiff ECM on day 1 and day 7, respectively (Fig. 2I). The measurement of ECAR indicated that the glycolytic rates were dramatically increased on the stiff ECM compared to the soft ECM on days 1 and 7 (Fig. 2J and K). These findings support the increased glycolysis associated with the osteogenic differentiation of MSCs on a stiff ECM.
2.3. ECM stiffness regulates mitochondrial dynamics and biogenesis in MSCs
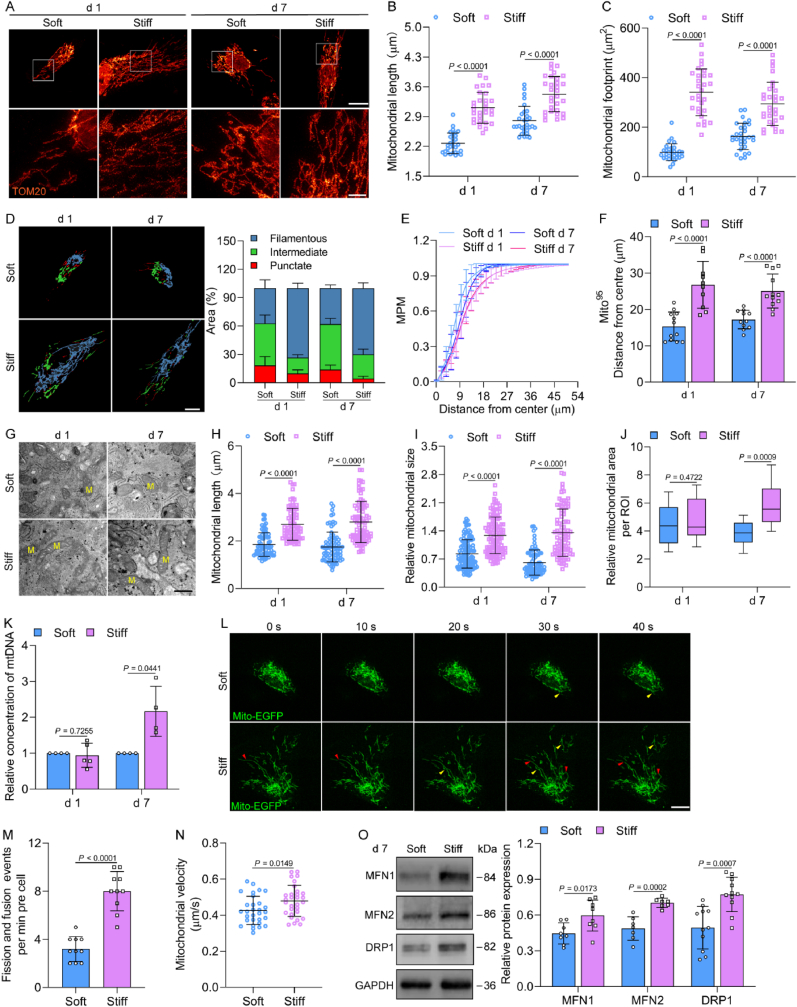
The differentiation of stem cells is an energy-demanding process. Mitochondria are the main energy powerhouse in cells. The metabolites of glycolysis enter mitochondria to fuel the TCA cycle and OXPHOS [23]. The super-resolution imaging and confocal results of mitochondria indicated that the mitochondrial length and footprint increased in a stiffness-dependent manner from 1 to 40 kPa (Fig. 3 A–C, Fig. S4A). This observation was also confirmed through unbiased machine learning algorithms in which cells displayed filamentous mitochondria on a stiff ECM (Fig. 3D). However, ECM stiffness greater than 80 kPa could not regulate mitochondrial length and footprint (Fig. S4A). Furthermore, the mitochondrial probability map (MPM) shifted to the left and the mito95 value decreased on the soft ECM (Fig. 3E and F), which suggested there were fewer perinuclear mitochondria on the stiff than on the soft ECM. The transmission electron microscopy results showed elongated mitochondria with well-developed cristae, and the mitochondrial size was increased on the stiff ECM, as confirmed on days 1 and 7 (Fig. 3 G–I). The mitochondrial area per region-of-interest (ROI) was increased on the stiff ECM (Fig. 3J). The RNA-seq results indicated that a mitochondrial biosynthesis-related gene, mitochondrial transcription factor A (TFAM), was upregulated on the stiff ECM (Fig. S4B). The mitochondrial DNA (mtDNA) copy number on the stiff ECM was also elevated (Fig. 3K). These data revealed that mitochondria appeared more elongated and fused on the stiff ECM in MSCs, which was accompanied by increased mitochondrial biogenesis.
Fig. 3.
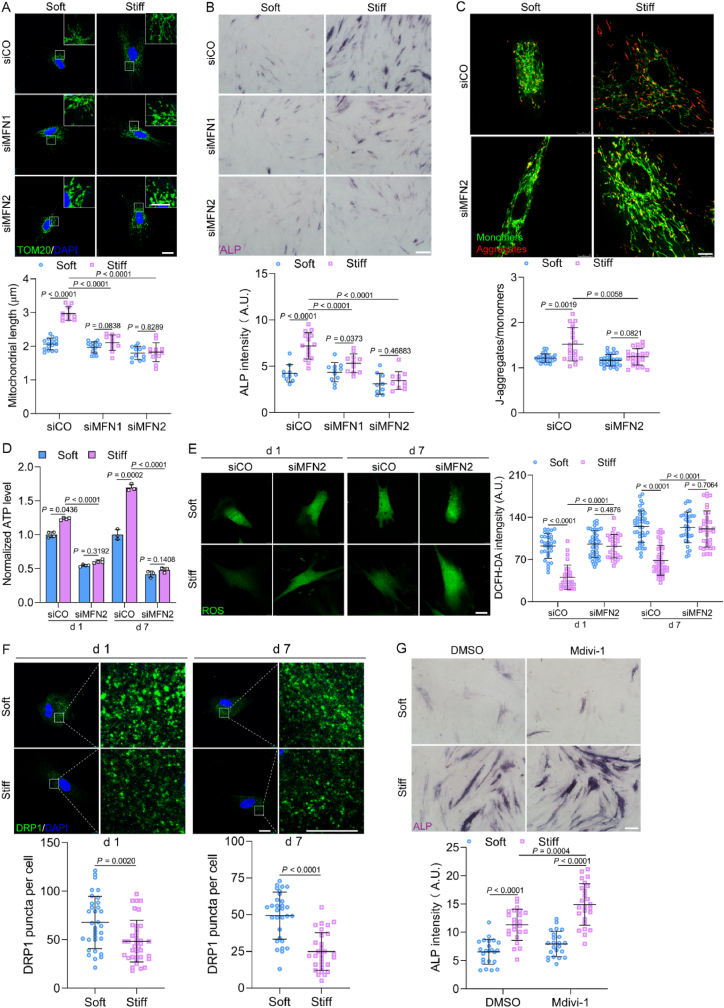
ECM stiffness regulates mitochondrial dynamics and biogenesis. (A–C) Mitochondrial morphology in MSCs on soft and stiff ECM on days 1 and 7. Representative STED images (top) and high magnification of white boxed regions (bottom) were shown (A), scale bars, 20 μm and 4 μm. Mitochondrial length (B, n = 30 for each group) and mitochondrial footprint (C, n = 30 for each group) were analyzed. (D) Left, representative images of classification of mitochondrial morphology, scale bars, 20 μm. Right, quantification of punctate, intermediate, and filamentous mitochondria (n = 10 for each group). (E) Mitochondrial probability map (MPM) was plotted for the accumulation of mitochondria in MSCs (n = 10 for each group). (F) Mito95 values (95th percentile) were calculated (left to right, n = 12, 10, 11, 12). (G–J) Transmission electron micrographs of mitochondria. Representative images of mitochondria (G), scale bars, 500 nm. The average mitochondrial length (H, left to right, n = 74, 74, 79, 83), relative mitochondrial size (I, left to right, n = 120, 116, 79, 88), and relative mitochondrial area per ROI (J, left to right, n = 11, 14, 10, 13) were quantified. (K) The mtDNA copy number was detected by qPCR (n = 4 for each group). (L) Time-lapse frames of mitochondrial fission and fusion. Red arrow, fission event; yellow arrow, fusion event, scale bars, 15 μm. (M) Quantification of fission and fusion events per minute per cell and (N) mitochondrial velocity (n = 30 for each group). (O) Left, immunoblots for mitochondrial fission and fusion protein expression in MSCs on day 7. Right, quantification of MFN1, MFN2 and DRP1 protein expression (n = 8 for MFN1, 7 for MFN2, 11 for DRP1). Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from Mann-Whitney test (B,C,F,H,I) or unpaired two-tailed Student's t-test (J,K,M−O).
To visualize mitochondrial dynamics, MSCs were transfected with mito-EGFP. The results of time-lapse imaging demonstrated that fission/fusion and motility were promoted by the stiff ECM in MSCs (Fig. 3 L–N, Movie S1). MFN1, MFN2, and DRP1 are key regulators of mitochondrial dynamics and their expression was barely affected by stiffness on day 1 (Fig. S5A), but increased on the stiff ECM on day 7 (Fig. 3O). To investigate their roles in stiffness-mediated osteogenic differentiation, MFN1 or 2 was successfully inhibited to reduce the mitochondrial fusion in MSCs (Fig. 4 A, Figs. S5B and C). The ALP expression was decreased by siMFN1 or 2, especially siMFN2 at day 7 (Fig. 4B). The knockdown of MFN2 also eliminated the variations that the ECM stiffness caused in terms of ΔΨm, ATP, and ROS levels (Fig. 4C–E), indicating that MFN2 is vital to energy synthesis and anti-oxidative capability, which promotes the osteogenic differentiation of MSCs. Next, we detected the fission factor DRP1 puncta, the active form of DRP1 [24]. Notably, the number of DRP1 puncta was decreased on the stiff ECM (Fig. 4F), which was consistent with the results on the elongated and fused mitochondria. The inhibition of DRP1 by Mdivi-1 enhanced ALP expression of MSCs cultured on the stiff ECM (Fig. 4G). Collectively, our data indicate that stiff ECM promoted mitochondrial fusion, which was mediated by increasing MFN1 and MFN2 expression and inhibiting DRP1 activity and both contributed to the osteogenic differentiation of MSCs.
Fig. 4.
ECM stiffness regulates osteogenic differentiation of MSCs through mitochondrial dynamics. (A) Top, representative images of mitochondrial morphology in MSCs with knockdown of MFN1 (siMFN1) and MFN2 (siMFN2), scale bar, 20 μm; inset scale bar, 10 μm. Bottom, quantification of mitochondrial length (left to right, n = 16, 10, 15, 11, 12, 12). (B) Top, Representative images of ALP staining in MSCs with knockdown of MFN1 (siMFN1) and MFN2 (siMFN2) for 7 days, scale bars, 200 μm. Bottom, quantification of ALP intensity (left to right, n = 10, 15, 11, 11, 9, 11). (C) ΔΨm in MSCs with knockdown of MFN2 (siMFN2) on stiff and soft ECM on day 7. Top, representative images of JC-1 staining, scale bars, 10 μm. Bottom, the ratio of J-aggregate to monomers (n = 26, 19, 30, 23). (D) Levels of intracellular ATP in MSCs with knockdown of MFN2 (siMFN2) on days 1 and 7 (n = 4 for 1 d, 3 for 7 d). (E) Levels of ROS of MSCs with knockdown of MFN2 on days 1 and 7. Left, representative images, scale bars, 25 μm. Right, quantification of intensity (n = 31 for siCO 1 d, 41 for siMFN2 1 d, 30 for siCO 7 d, and siMFN2 7 d). (F) DRP1 puncta were detected in MSCs on days 1 and 7. Top, Representative images of DRP1 puncta. Scale bar, 20 μm; enlarged inset scale bar, 10 μm. Bottom, the number of total DRP1 puncta (left to right, n = 33, 38, 30, 30). (G) Top, representative images of ALP staining in MSCs with DRP1 inhibitor Mdivi-1 (25 μM), scale bars, 200 μm. Bottom, quantification of ALP intensity (left to right, n = 23, 24, 23, 25). Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from Mann-Whitney test (F) or unpaired two-tailed Student's t-test (A–E,G).
2.4. YAP involves in stiffness regulates metabolism in MSCs
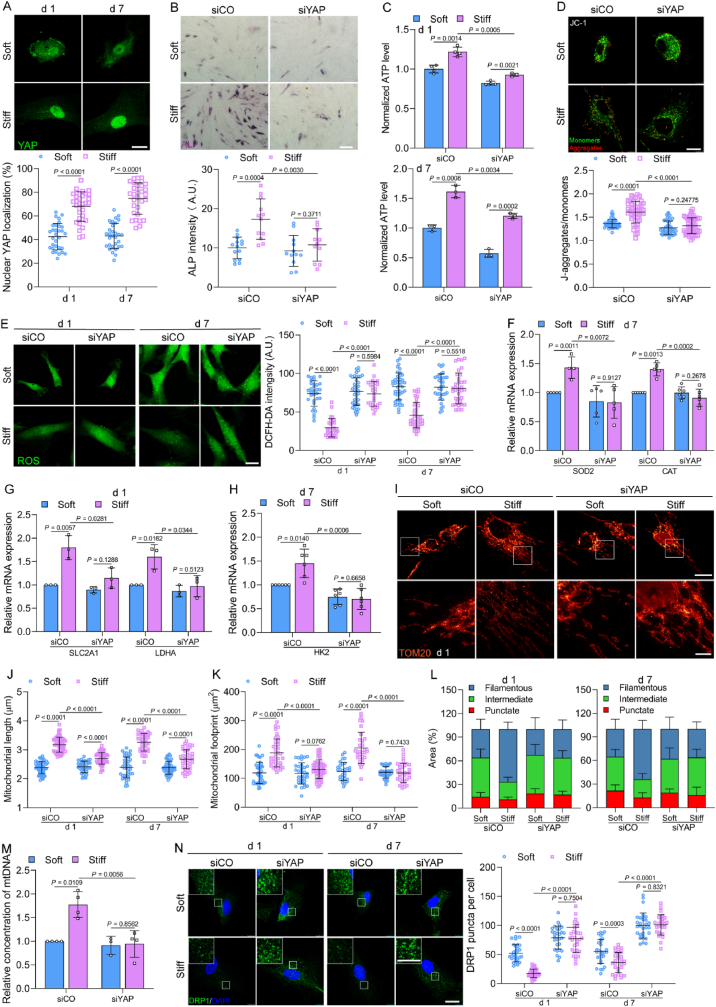
Our study and previous studies have indicated that stiff ECM can enhance cytoskeletal tension and focal adhesion (FA) formation, as well as upregulate YAP (Fig. 5A, Fig. S6 A–L) [21,25,26]. In addition, stiff ECM increased YAP activation by inducing actin polymerization and actomyosin contractility (Figs. S6L and M). We first validated successful YAP inhibition using immunofluorescence (Fig. S7A). The knockdown of YAP suppressed stiff-induced osteogenic differentiation (Fig. 5B), consistently with previous studies [5,27]. We further investigated their role in the stiffness-mediated regulation of MSCs metabolism. The knockdown of YAP inhibited ATP production, decreased ΔΨm, elevated ROS production, and suppressed SOD2 and CAT gene expression in MSCs cultures on a stiff ECM (Fig. 5 C–F). Furthermore, the inhibition of YAP reduced SLC2A1, LDHA, and HK2 gene expression on the stiff ECM (Fig. 5G and H). Our data indicate that stiff ECM enhance energetics through YAP and show that YAP is instrumental to the regulation of ATP production, glycolysis, and the antioxidant capacity of MSCs cultured on a stiff ECM.
Fig. 5.
ECM stiffness promotes metabolism by YAP in MSCs. (A) Top, representative immunofluorescence of YAP in MSCs on soft and stiff ECM on days 1 and 7, scale bar, 20 μm. Bottom, quantification of nuclear YAP localization (n = 31 for 1 d, 30 for 7 d). (B) Top, representative images of ALP staining in MSCs with knockdown of YAP (siYAP) for 7 days, scale bars, 200 μm. Bottom, quantification of ALP intensity (left to right, n = 14, 12, 12, 11). (C) Intracellular ATP levels of MSCs transfected with siCO or siYAP on days 1 (top) and 7 (bottom) (n = 4 for each group 1 d, 3 for each group 7 d). (D) ΔΨm in MSCs with knockdown of YAP (siYAP) on day 7. Top, representative images of JC-1 staining, scale bars, 20 μm. Bottom, the ratio of J-aggregates to monomers (n = 44, 46, 43, 50). (E) Levels of ROS in MSCs with depleted YAP (siYAP). Left, representative images, scale bars, 20 μm. Right, quantification of intensity (left to right, n = 31, 28, 41, 28, 42, 40, 33, 35). (F) The gene expression levels of SOD2 and CAT of MSCs transfected with siCO or siYAP on soft and stiff ECM on day 7 (left to right, n = 5, 4, 5, 5, 6, 5, 6, 6). (G and H) Metabolic gene expression analysis of MSCs with knockdown of YAP (siYAP) on days 1 (G, n = 3 for each group) and 7 (H, n = 6 for each group). (I–K) Mitochondrial morphology of MSCs with knockdown of YAP (siYAP) on soft and stiff ECM. Representative STED images (top) and high magnification of white boxed regions (bottom) were shown on day 1 (I), scale bars, 20 μm and 4 μm. Mitochondrial length (J, left to right, n = 47, 50, 36, 56, 41, 39, 33, 47) and mitochondrial footprint (K, left to right, n = 40, 37, 33, 47, 30, 30, 32, 34) was calculated on days 1 and 7. (L) Classification of mitochondrial morphology of MSCs transfected with siCO or siYAP on days 1 and 7 (n = 7 for each group). (M) Quantification of mtDNA copy number in MSCs with depleted YAP (siYAP) on day 7 (n = 4 for each group). (N) DRP1 puncta were detected in MSCs transfected with siCO or siYAP on days 1 and 7. Left, Representative images of DRP1 puncta, scale bar, 20 μm; inset scale bar, 10 μm. Right, the number of total DRP1 puncta (n = 35 for each group). Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from Mann-Whitney test (A,D and E) or unpaired two-tailed Student's t-test (B,C,F–H,J,K,M,N).
To investigate the role of YAP in the regulation of the mitochondrial network by ECM stiffness, we knocked down YAP to examine mitochondrial morphology and dynamics. Mitochondrial fusion was inhibited with decreased length and footprint after silencing YAP in MSCs cultured on a stiff ECM (Fig. 5 I–K, Fig. S7B). Thus, there was a decrease in filamentous mitochondria and an increase in intermediate mitochondria (Fig. 5L, Fig. S7C). Moreover, the mtDNA content was suppressed by the knockdown of YAP in MSCs cultured on a stiff ECM (Fig. 5M), indicating that YAP was involved in stiffness-induced mitochondrial dynamics and biogenesis. However, siYAP did not change the expression levels of MFN1, MFN2, and DRP1 (Fig. S7D); instead, it increased the number of DRP1 puncta (Fig. 5N), demonstrating that YAP enhances mitochondrial fusion via the inhibition of DRP1 activity on a stiff ECM.
2.5. YAP-mediated ECM stiffness affects glutamine metabolism of osteogenesis
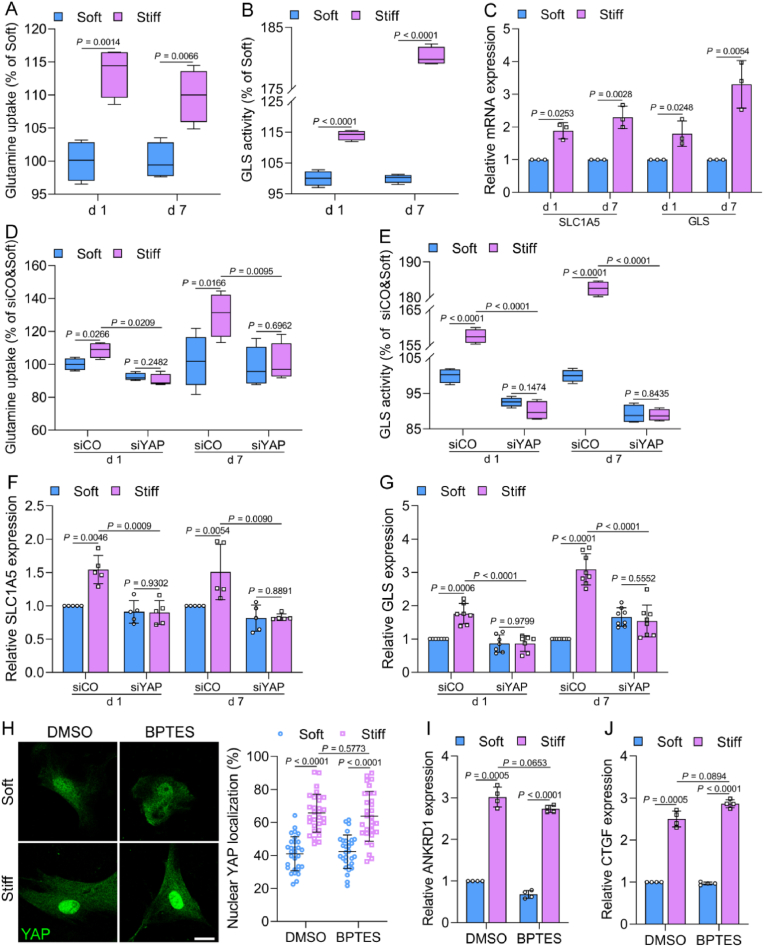
Emerging evidences suggest that although glutamine did not provide as much energy as glucose, it is vital to bone formation and homeostasis [28]. Our data showed that ECM stiffness-induced osteogenic differentiation was reduced by the inhibition of glutamine metabolism (Fig. 2B). Glutamine consumption and glutaminase (GLS) activity were increased in MSCs cultured on a stiff ECM on days 1 and 7 (Fig. 6A and B). Furthermore, the gene expression of SLC1A5 (a glutamine transporter) and GLS was upregulated by the stiff ECM (Fig. 6C). Moreover, we evaluated whether ECM stiffness-induced glutamine metabolism was also regulated by YAP. The knockdown of YAP inhibited glutamine uptake and GLS activity (Fig. 6D and E), and the gene expression levels of SLC1A5 and GLS were decreased in MSCs cultured on the stiff ECM (Fig. 6F and G). However, the YAP nuclear localization and the expression of YAP targets were not affected by the inhibition of glutamine consumption on the stiff ECM (Fig. 6 H–J). These data indicate that stiff ECM could regulate glutamine metabolism to contribute to osteogenesis, and YAP is indispensable for this process.
Fig. 6.
ECM stiffness regulates glutamine metabolism of osteogenesis by YAP. (A) Glutamine uptake and (B) GLS activity in MSCs on soft and stiff ECM on days 1 and 7 (n = 4 for each group). (C) The gene expression of SLC1A5 and GLS in MSCs on days 1 and 7 (n = 3 for each group). (D and E) Glutamine uptake (D, n = 4 for 1 d, 6 for 7 d) and GLS activity (E, n = 4 for each group) in MSCs with knockdown of YAP (siYAP) on days 1 and 7. (F and G) The gene expression levels of SLC1A5 (F, n = 5 for each group) and GLS (G, n = 7 for 1 d, 8 for 7 d) in MSCs transfected with siCO or siYAP on days 1 and 7. (H) Left, representative immunofluorescence of YAP in MSCs treated with 20 μM BPTES for 6 h, scale bar, 25 μm. Right, quantification of nuclear YAP localization (n = 30 for each group). (I and J) The gene expression levels of ANKRD1 (I) and CTGF (J) in MSCs cultured with 20 μM BPTES for 6 h (n = 4 for each group). Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from unpaired two-tailed Student's t-test.
2.6. Glycolysis regulates cell mechanics to affect YAP activity
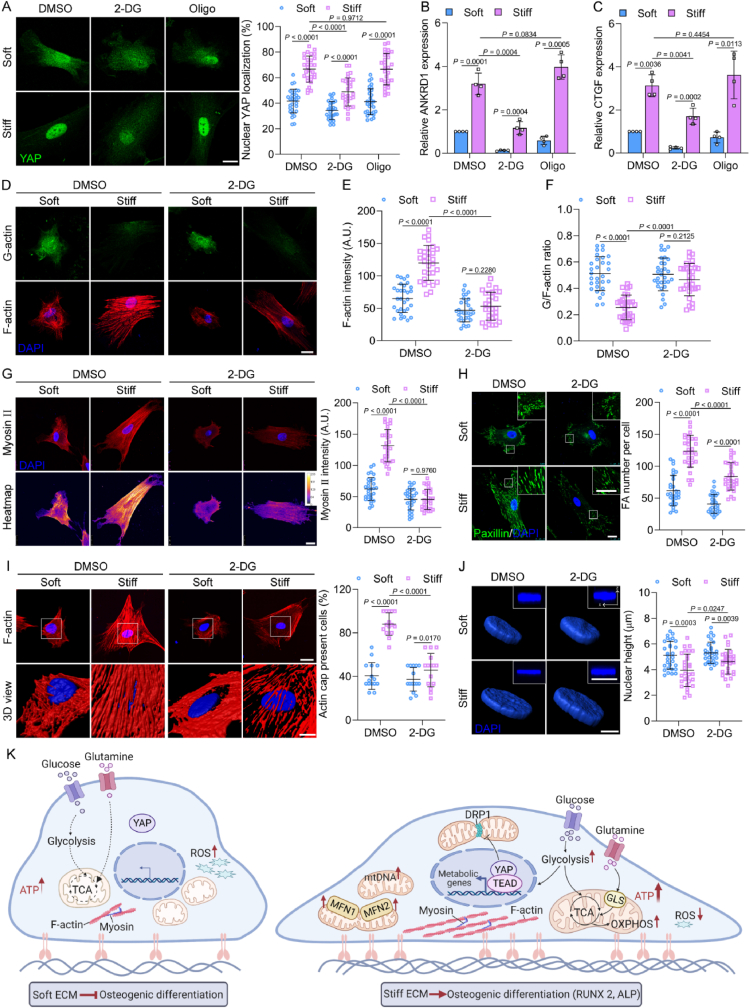
As our data showed that YAP regulates ECM stiffness-induced metabolism, especially glycolysis, we wondered whether ECM stiffness-induced metabolism could regulate YAP. The YAP nuclear localization, expression, and target genes were substantially decreased by the inhibition of glycolysis by 2-DG (Fig. 7 A–C, Figs. S8A and B). However, YAP activity was not affected by the inhibition of OXPHOS with oligomycin (Fig. 7 A–C). Moreover, disruption of MFN1 or MFN2 did not regulate the nuclear localization of YAP, whereas reduced the gene expression of YAP targets (Fig. S9 A–C).
Fig. 7.
Glycolysis regulates YAP activity via cell mechanoresponsive. (A) Left, representative immunofluorescence of YAP in MSCs treated by 100 mM 2-DG or 2 μM oligomycin (Oligo) for 6 h, scale bar, 25 μm. Right, quantification of nuclear YAP localization (n = 30 for each group). The gene expression levels of ANKRD1 (B) and CTGF (C) in MSCs with 100 mM 2-DG or 2 μM oligomycin (Oligo) for 6 h (n = 4 for each group). (D–F) G-actin and F-actin in MSCs treated with 100 mM 2-DG for 6 h, Representative images (D), scale bar, 25 μm. Quantification of F-actin intensity (E, n = 30 for each group) and G/F actin ratio (F, n = 30 for each group). (G) Left, representative images and heatmap of myosin II in MSCs treated with 100 mM 2-DG for 6 h, scale bar, 25 μm. Right, quantification of myosin II intensity (n = 30 for each group). (H) Left, representative images of paxillin in MSCs treated with 100 mM 2-DG for 6 h, scale bar, 25 μm; inset scale bar, 20 μm. Right, quantification of FA number per cell (n = 30 for each group). (I) Left, representative images of apical actin stress fibers (top) and high magnification and three-dimensional (3D) reconstruction of white boxed regions (bottom), scale bars, 30 μm and 10 μm. Right, quantification of the actin cap formation (n = 15 for each group). (J) Left, reconstructed DAPI images in MSCs treated with 100 mM 2-DG for 6 h, scale bar, 10 μm; inset scale bar, 20 μm. Right, quantification of nuclear height (n = 30 for each group). (K) Schematic illustration represents ECM stiffness-driven metabolism in regulating osteogenic differentiation of MSCs through YAP. Data from at least three independent experiments. All graphs showed mean ± s.d. Statistical significance was derived from unpaired two-tailed Student's t-test.
To further investigate the underlying mechanism, we examined the effect of glycolysis on the Hippo pathway, as well as F-actin and myosin dynamics. Treatment with 2-DG depressed YAP expression but did not affect the phosphorylation of YAP or LATS (Fig. S8 A–D). Furthermore, 2-DG decreased F-actin intensity and increased G/F-actin levels in MSCs cultured on the stiff ECM (Fig. 7 D–F). Similarly, myosin II and the number and length of FA were suppressed by 2-DG (Fig. 7G and H, Fig. S10). As YAP nuclear translocation was related to nuclear flattening [29], the results indicated that 2-DG inhibited actin cap numbers (Fig. 7I), which increased the nuclear height of MSCs cultured on the stiff ECM (Fig. 7J), explaining why YAP was transported into nuclei. Overall, glycolysis is coupled to cell mechanics, which in turn regulates YAP localization and activity.
3. Discussion
Mechanical cues generated by the ECM can regulate stem cell fate and behavior. Emerging evidence has indicated that metabolism is vital to stem cell fate decision [10,30,31]. However, it is unclear how mechanical cues produced by the ECM are integrated with metabolic responses in stem cells. Our findings demonstrate that ATP production was increased, and the function of the antioxidant defense system was enhanced to decrease the effects of ROS accumulation on stiff ECM. It has been reported that undifferentiated PSCs rely more on glycolysis than OXPHOS for energy production, and switch to oxidative metabolism during differentiation [12,32]. However, our results show that both glycolysis and OXPHOS were enhanced during the osteogenic differentiation of MSCs on a stiff ECM. The contribution of glycolysis to ATP production was more than 80% which is consistent with a previous study [33]. Nevertheless, there is no agreement among researchers on the metabolic switch during osteogenesis. During the osteogenic differentiation of MSCs in a mineralization-inducing medium, OXPHOS was activated, while glycolysis remained unchanged [34] or decreased [35]. A recent study showed that glycolysis continuously increases during the osteogenesis of osteoblast, while OXPHOS increases first and then decreases [36]. Our study reveals a mode of ECM stiffness-induced metabolism switch during osteogenic differentiation in MSCs. Additionally, rate-limiting enzymes are involved in stiffness-induced glycolysis in a duration-dependent manner. Glycolytic enzymes, such as PFKM and PKM2 were upregulated rapidly on the first day of ECM stiffness-induced osteogenic differentiation, while the expression level of HK2 increased after 7 days of culture, thereby contributing to glycolysis.
Mitochondria are not only cellular energy hubs, they are also central regulators of stem cell fate decisions [37]. However, it needs to be further elucidated on how mitochondrial dynamics regulate stem cell differentiation, not to mention their role in ECM stiffness-induced differentiation. Our results indicate that stiff ECM promotes mitochondrial fusion by increasing MFN1 and MFN2 expression and inhibiting DRP1 activity. MFN1, especially MFN2 and DRP1 are involved in stiffness-regulated the osteogenic differentiation of MSCs. Mitochondrial fusion provides sufficient energy, maintains membrane potential, and mitigates oxidative damage [38], which are critical for the osteogenic differentiation of MSCs [39]. Mineralization-inducing medium increased mitochondrial fusion during MSC osteogenesis, which was correlated with increased MFN2 expression, except that the expression levels of MFN1 and DRP1 did not change [40]. Therefore, our results represent that in the absence of a mineralization-inducing medium, MSCs could respond to changes in ECM stiffness to regulate metabolic processes and mitochondrial dynamics to promote osteogenic differentiation.
We explored the underlying mechanism of ECM stiffness in the regulation of metabolism in the osteogenic differentiation of MSCs. YAP is a mechanotransducer, and regulator of cellular energy status [41]. Previous studies have demonstrated that the loss of YAP impairs glycolysis in renal cell carcinoma cells [42] or breast cancer cells [43]. Nevertheless, the roles of YAP in the regulation of mechanical cues and metabolism remain largely unexplored. Our findings show that YAP could regulate the expression levels of SLC2A1, LDHA, and HK2 in cells cultured on a stiff ECM to mediate glycolysis. YAP also played a role to regulate DRP1 activity and mitochondrial biosynthesis on a stiff ECM. A recent study has reported that YAP enhances phosphocreatine production by increasing the expression of cytoplasmic creatine kinase in pancreatic cancer cells cultured on a stiff ECM [44]. Thus, our results shed light on the role of YAP in ECM stiffness-induced metabolic remodeling.
Glutamine metabolism has been reported to be critical for stem cell fate specification, even though its contribution to the energy supply is less significant [28,45]. Recent work has reported that ECM stiffness can stabilize microtubules by regulating glutamine metabolism, which allows breast cancer cells to adapt to mechanical cues [46]. Our results indicate that stiff ECM increase glutamine uptake and GLS activity in MSCs. Thus, ECM stiffness controls the replenishment of glutamine. Mechanistically, YAP is involved in the metabolism of glutamine on a stiff ECM, which promotes the osteogenic differentiation of MSCs. Furthermore, a previous study has demonstrated that ECM stiffness promotes glutamate synthesis by upregulating GLS through the YAP/TAZ signaling pathway in squamous cell carcinoma [47]. Likewise, YAP/TAZ-dependent glutaminolysis can affect endothelial cell growth and migration in a model of pulmonary hypertension-induced ECM stiffening [19].
Notably, the metabolism can in turn regulate YAP [41,48]. The inhibition of glycolysis decreased YAP/TAZ transcriptional activity in breast cancer cells [49]. However, it is unknown whether ECM stiffness-induced metabolic responses can regulate YAP activity in MSCs. Our results indicate that the inhibition of glycolysis, not the inhibition of OXPHOS, impaired YAP transcriptional activity. We also concluded that YAP was regulated by glycolysis which was independent of the Hippo pathway, but dependent on cell mechanics. ECM stiffness has been reported to increase the nuclear localization of YAP through the regulation of nuclear pores [29]. Our findings reveal that the inhibition of glycolysis impaired cytoskeletal tension, which in turn reduced nuclear flattening on a stiff ECM, which might be a reason for the reduction in YAP nuclear import.
4. Conclusion
In summary, our findings reveal that ECM stiffness can regulate metabolism to contribute to the osteogenic differentiation of MSCs. YAP is a component of a feedback mechanism that maintains favorable metabolic substrates and mechanics, which determine the fate of MSCs (Fig. 7K). These findings provide an effective metabolic target for cell therapies based on stem cells and also provide new insights for advanced biomaterial design in the field of regenerative medicine.
5. Experimental section
5.1. Cell culture
Human MSCs were purchased from Nuwacell (RC02003, Nuwacell, Hefei, China). This study conformed to the standards in the Declaration of Helsinki. All procedures were approved by the Biological and Medical Ethics Committee of Beihang University (IRB No. BM20210097). The cells were cultured in α-MEM (12000022, Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (Amresco, Solon, OH, USA) at 37 °C and 5% CO2. The medium was changed every three days.
5.2. ECM preparation
Polyacrylamide (PA) gels were prepared according to a previous study [50]. PA gels with different Young's moduli were obtained by adjusting the ratio of acrylamide and diacrylamide in aqueous solution and induced to polymerize on amino-silanated coverslips by adding N,N,N′,N′-tetramethylethylenediamine (TEMED) and ammonium persulphate. Next, 0.2 mg ml−1 sulfosuccinimidyl-6-(4′-azido-2′-nitrophenylamino) hexanoate (sulfo-SANPAH, ProteoChem, Hurricane, UT, USA) was used to covalently bind proteins to the polyacrylamide ECM, which was activated by ultraviolet light at 365 nm for 20 min. PA gels were washed with 50 mM HEPES (pH 8.5), and coated with 50 μg mL−1 fibronectin (356008, Corning Life Sciences, NY, USA) overnight at 4 °C. PA gels were washed two times with phosphate-buffered saline (PBS) before seeding the cells. The elastic modulus of PA gels was confirmed by a universal testing machine (EZ-LX, Shimadzu, Kyoto, Japan) at strain rate of 5 mm min−1, and the resluts were showed in Fig. S1. The CCK8 assays showed that PA gels were not cytotoxic to MSCs (Fig. S2).
5.3. Quantitative real-time polymerase chain reaction
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and cDNA was synthesized using a reverse transcription kit (Takara, Otsu, Japan) according to the manufacturer's instructions. The primers for quantitative real-time polymerase chain reaction (qPCR) were synthesized by Sangon Biotech (Shanghai, China) and listed in Table S1. qPCR was carried out with a two-step SYBR Green qPCR kit (Takara, Otsu, Japan). The thermocycler (QuantStudio 1, Thermo Fisher Scientific, Waltham, MA, USA) program was as follows: 95 °C for 30 s, followed by 40 cycles of 95 °C for 10 s and 56 °C for 34 s, followed by 55 °C–95 °C for the generation of a melting curve. The quantification of relative gene expression was performed by the 2−ΔΔCT method, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a reference gene for data normalization. The mtDNA copy number was quantified as a ratio of COX1 mtDNA/GAPDH DNA.
5.4. Alkaline phosphatase staining
For alkaline phosphatase (ALP) staining, the cells were seeded on gels housed in plates and cultured in growth medium for 1, 3, 5, and 7 days. Cells were fixed with 4% paraformaldehyde for 30 min, and washed three times with PBS. BCIP/NBT solution (Beyotime, Shanghai, China) was added to the cells, and the plates were incubated in the dark. Each sample was imaged by a microscope (Olympus, Tokyo, Japan), and the intensity of ALP staining was analyzed with ImageJ software (http://imagej.net/ImageJ) to quantify the amount of ALP expression. In certain experiments, 500 μM 2-DG (D0051, Tokyo Chemical Industry, Tokyo, Japan), 10 μM BPTES (M5015, Abmole, Houston, TX, USA), or 200 μM etomoxir (S8244, Selleck, Houston, TX, USA), or 25 μM Mdivi-1 (HY-15886, MedChemExpress, Monmouth Junction, NJ, USA) were incubated with the cells as described.
5.5. ATP measurement
The intracellular ATP level was measured using an enhanced ATP assay kit (Beyotime, Shanghai, China). Cells were lysed with lysis buffer on ice, and then centrifuged at 12000 g for 5 min at 4 °C to collect the supernatant. Aliquots of 40 μl of the supernatant were mixed rapidly with 100 μl of ATP detection reagent in a white-walled 96-well plate, and the relative light unit (RLU) values were measured with a luminometer (Thermo Fisher Scientific). The ATP concentration of each sample was calculated using the standard curve. The ATP measurements were normalized to the protein concentrations. In certain experiments, 100 mM 2-DG, 20 μM BPTES, 200 μM etomoxir, 2 μM Oligomycin (A606700-0005, BBI Life Sciences, Shanghai, China), 100 mM oxamate (M9783, Abmole), 100 μM UK-5099 (M7455, Abmole), 1 μM cytochalasin D (MB0784, Meilunbio, Dalian, China), and 50 μM blebbistatin (MB5060, Meilunbio, Dalian, China) were incubated with cells for 1 h, and then the intracellular ATP levels were measured.
5.6. Measurement of the mitochondrial membrane potential
The mitochondrial membrane potential (ΔΨm) was measured by JC-1 staining (T3168, Thermo Fisher Scientific). The J-aggregates fluoresce red and indicate high ΔΨm, while the monomers fluoresce green and indicate low ΔΨm. Cells were incubated with 1 μM JC-1 for 20 min at 37 °C and washed with pre-warmed PBS. Images were rapidly captured with a confocal microscope (Leica SP8, Wetzlar, Germany), and the density of the J-aggregates (red, Ex = 535 nm) and the monomers (green, Ex = 485 nm) were quantified by ImageJ software.
5.7. ROS measurement
Intracellular ROS levels were detected with the cell-permeable probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, HY-D0940, MedChemExpress). Cells were incubated with 5 μM DCFH-DA in the dark at 37 °C for 30 min and wash two times with pre-warmed PBS. Images were rapidly acquired with a confocal microscope, and the fluorescence intensity was quantified by ImageJ software.
5.8. Seahorse assay
Cells were digested from the PA gels with trypsin and seeded on an XF96 plate (Agilent, Santa Clara, CA, USA) at a density of 10,000 cells per well. The complete Seahorse medium consisted of 1 g l−1 glucose (G7528, Sigma-Aldrich, St. Louis, MO, USA), 2 mM glutamine (G8540, Sigma-Aldrich) and 1 mM sodium pyruvate (S8636, Sigma-Aldrich) in Seahorse XF base medium (102353-100, Agilent, Santa Clara, CA, USA) with pH 7.4. Cells were incubated at 37 °C for 1 h after changing the medium to 175 μl of complete Seahorse medium in a CO2-free incubator. The inhibitors 2 μM oligomycin (ab141829, Abcam, Cambridge, UK), 2 μM carbonyl cyanide-4 phenylhydrazone (FCCP, C2920, Sigma-Aldrich), and 1 μM rotenone (R8875, Sigma-Aldrich)/antimycin (ab141904, Abcam) were injected into the port of the sensor cartridge for the detection of oxygen consumption rate (OCR). The 10 mM glucose, 2 μM oligomycin, and 2 μM 2-DG (D8375, Sigma-Aldrich), were injected into the port of the sensor cartridge for the detection of extracellular acidification rate (ECAR). The cell culture plate and sensor cartridge were run on the Seahorse XF96 bioanalyzer (Agilent) to detect OCR and ECAR.
5.9. RNA-sequencing
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Library construction and transcriptome sequencing were performed by Novogene (Beijing, China). The sequencing library was generated using the NEBNext Ultra™ RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA), and sequenced on the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Clean reads were mapped to genes in GRCh38 genome by HISAT2 2.0.5 software [51]. Differential expression analysis was performed with DESeq2 1.16.1 package [52]. Enrichment analysis Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway were performed using the clusterProfiler 3.4.4 tool [53]. The gene heatmaps and enrichment dot bubble were draw using http://www.bioinformatics.com.cn/.
5.10. Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 20 min at room temperature, and washed three times with PBS. Cells were permeabilized with 0.2% Triton X-100 for 7 min, washed three times and then blocked with 5% BSA for 30 min. Cells were rinsed three times with PBS, immediately followed by incubation with the primary antibodies (a list of specific antibodies is provided in Table S2) overnight at 4 °C. The antibodies were removed, and the cells were washed with PBS three times. Subsequently, the cells were incubated with secondary antibodies or phalloidin (1:1000, A34055, Thermo Fisher Scientific) for 1 h at room temperature and protected from light. F- and G-actin were visualized with phalloidin and DNase I 488 (1:500, D12371, Thermo Fisher Scientific), which were incubated with cells for 1 h. Finally, the cells were washed three times and stained with DAPI (1:2000, D9542, Sigma-Aldrich) for 5 min. Cell images were captured by a confocal microscope, and analyzed using Fiji software (http://fiji.sc/Fiji). Image z-stacks of nuclei were collected with a step size of 0.3 μm and reconstructed using Imaris 9.0.1 software.
5.11. Super-resolution imaging
Cells were stained as described as above, and the primary antibody for mitochondrial immunostaining was TOM20 (1:200, sc-17764, Santa Cruz Biotechnology). The secondary antibody was Abberior STAR RED (640 nm, 1:150, Abberior, NanoTag). Super resolution imaging was performed on the Abberior STED system (Abberior Instruments, Göttingen, Germany) equipped with an Olympus 100 × UPLXAPO 100XO 1.45 NA oil immersion objective.
5.12. Mitochondrial morphology analysis
Mitochondrial length, footprint and distribution were detected using Fiji software (MiNA and Fiji plugin, http://fiji.sc/Fiji). Mitochondrial morphology was further classified as punctate, intermediate, and filamentous by machine-learning [54]. The percentage area of each category in each image was calculated and then compared among the groups. The distribution of mitochondria in the perinuclear region was detected from the cell center or MPM [55]. The shift of the curve to the left indicated that the mitochondria accumulated towards the center of the cell. The mito95 value was defined the distance from the center of the cell to the location containing 95% of the mitochondria.
5.13. Transmission electron microscopy
Cells were fixed with 2% paraformaldehyde and 2.5% glutaraldehyde in PBS at 4 °C for 24 h, and then washed with PBS three times at room temperature for 15 min each time. After washing, the samples were post-fixed with 1% osmium acid and 1.5% potassium ferricyanide for 1 h and rinsed with ultrapure water three times for 15 min each time. Samples were stained with 1% uranyl acetate overnight, followed by washing with ultra-pure water three times for 15 min each time. Stained cells were dehydrated in an ice box through a series of ethanol and propylene oxide baths, followed by infiltration with EMbed-812 resin and embedding at 60 °C. Samples were imaged with a transmission electron microscope (H-7650, Hitachi, Tokyo, Japan), and data analysis was performed using ImageJ software.
5.14. Visualization of mitochondria in living cells
The mEmerald-TOMM20-N-10 plasmid was a gift from Michael Davidson (Addgene plasmid 54282, Watertown, MA, USA). The plasmid was transfected into cells using Lipofectamine 3000 reagent (L3000075, Thermo Fisher Scientific). The transfection reagent was removed and replaced with normal complete medium after 6 h transfection. Two days after transfection, the imaging of living cells was performed by a spinning disk confocal microscope (Dragonfly, Andor, Belfast, UK) at 37 °C with 5% CO2. Fluorescence images were collected with a 40 × 1.30 NA oil immersion objective at 10 s intervals for 3–5 min. The quantitation of mitochondrial movement was acquired by ImageJ as previously described [56].
5.15. Western blotting
Cells were lysed with RIPA buffer (Beyotime) containing protease inhibitors (04693132001, complete EDTA-free, Roche, Basel, Switzerland), and the supernatant was collected by centrifugation at 4 °C. The concentrations of proteins were quantified using the BCA assay kit (P0010, Beyotime, Shanghai, China), and 30 μg of protein from each sample was separated on a 12% SDS-PAGE gel. The proteins were transferred to a PVDF membrane, which was blocked in 5% bovine serum albumin (BSA) for 2 h at room temperature. The PVDF membrane was incubated with the primary antibodies (a list of specific antibodies is provided in Table S2) overnight at 4 °C. Secondary antibody conjugated to horseradish peroxidase was incubated with the PVDF membrane for 1 h at room temperature, and target proteins were detected by enhanced chemiluminescence (ECL) reagent (Applygen, Beijing, China). The bands were imaged by an imaging system (Sagecreation, Beijing, China), and quantified with Quantity One software (Bio-Rad, Hercules, CA, USA).
5.16. Transfections
The small interfering RNAs (siRNAs) for YAP, MFN1 and MFN2 were synthesized by GenePharma (GenePharma, Shanghai, China), and the sequences of the siRNAs are listed in Table S3. The day before transfection, the medium was changed to complete medium without penicillin and streptomycin. When cells reached 80% confluence, 10 nM siRNA was diluted in Opti-MEM (31985062, Thermo Fisher Scientific), and transfected into cells with RNAiMAX (13778150, Thermo Fisher Scientific). Cells were used for subsequent experiments after 48 h of transfection, and the transfection efficiency was assessed by western blotting.
5.17. Glutamine assay
The glutamine level was measured from the medium using a glutamine enzyme immunoassay (EIA) kit (TW210, TW-REAGENG, Shanghai, China). Cells were cultured in the same volume of normal medium containing 2 mM glutamine. The uptake of glutamine by cells was inversely proportional to the concentration of glutamine in the medium. The supernatant was collected by centrifugation at 1000 g for 5 min, and the enzyme-linked immunosorbent assay was performed according to the manufacturer's instructions. The optical density (OD) was measured with a plate reader (Varioskan Flash Multimode Reader, Thermo Fisher Scientific), and the glutamine level was normalized to cell number.
5.18. Glutaminase activity assay
The glutaminase activity of cells was measured using the glutaminase (GLS) activity assay kit (BC1445, Solarbio) according the manufacturer's instructions. Cells cultured on gels were digested and sonicated at 32% power for 3 min (3 s per time, 7 s per interval) in an ice bath. Extracts were centrifuged at 12000 g for 15 min at 4 °C, and the supernatants were combined to the reaction solution. The absorbance at 630 nm was recorded for each sample against a water blank, and the enzyme activity was calculated. Glutaminase activity was normalized to cell number.
5.19. Statistical analysis
Statistical analysis was performed using SPSS 25.0 software, and the graphs were generated by GraphPad Prism 9.0.0 software. All data represented mean ± standard deviation (SD). The normal distribution of all data was assessed using the Shapiro-Wilk test (significance value of 0.05). If the data passed the normality test, the significance of means between two comparable groups was determined using the unpaired two‐tailed Student's t-test; if not, statistical significance of two comparable groups was determined using the Mann-Whitney test. Multiple comparisons of normally distributed datasets were performed using One-way ANOVA with Dunnet's test. The P values were given in the Fig. legends, and P < 0.05 was considered to be statistically significant. Fig. 7 was created with BioRender.com.
Data availability statement
Data supporting the findings of this study are available in the article or from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the Biological and Medical Ethics Committee of Beihang University.
CRediT authorship contribution statement
Jing Na: Writing – original draft, Visualization, Validation, Methodology, Investigation, Data curation, Conceptualization. Zhijie Yang: Validation, Methodology, Data curation. Qiusheng Shi: Validation, Methodology. Chiyu Li: Software, Methodology. Yu Liu: Validation, Methodology. Yaxin Song: Visualization, Software. Xinyang Li: Visualization, Software. Lisha Zheng: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization. Yubo Fan: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China [grant numbers 32171310, 11972067, U20A20390, 11827803, 12332019].
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.02.003.
Contributor Information
Lisha Zheng, Email: lishazheng@buaa.edu.cn.
Yubo Fan, Email: yubofan@buaa.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Yin J.Q., Zhu J., Ankrum J.A. Manufacturing of primed mesenchymal stromal cells for therapy. Nat. Biomed. Eng. 2019;3:90–104. doi: 10.1038/s41551-018-0325-8. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher D.A., Mullins R.D. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Z., Guo S.S., Fassler R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang C., Tibbitt M.W., Basta L., Anseth K.S. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 6.Lee H.P., Stowers R., Chaudhuri O. Volume expansion and TRPV4 activation regulate stem cell fate in three-dimensional microenvironments. Nat. Commun. 2019;10:529. doi: 10.1038/s41467-019-08465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S.B., Kim J.K., Lee G., Kim D.H. Mechanical properties of materials for stem cell differentiation. Adv Biosyst. 2020;4 doi: 10.1002/adbi.202000247. [DOI] [PubMed] [Google Scholar]
- 8.Ly C.H., Lynch G.S., Ryall J.G. A metabolic roadmap for somatic stem cell fate. Cell Metabol. 2020;31:1052–1067. doi: 10.1016/j.cmet.2020.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Schell J.C., Wisidagama D.R., Bensard C., Zhao H., Wei P., Tanner J., Flores A., Mohlman J., Sorensen L.K., Earl C.S., Olson K.A., Miao R., Waller T.C., Delker D., Kanth P., Jiang L., DeBerardinis R.J., Bronner M.P., Li D.Y., Cox J.E., Christofk H.R., Lowry W.E., Thummel C.S., Rutter J. Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat. Cell Biol. 2017;19:1027–1036. doi: 10.1038/ncb3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yucel N., Wang Y.X., Mai T., Porpiglia E., Lund P.J., Markov G., Garcia B.A., Bendall S.C., Angelo M., Blau H.M. Glucose metabolism drives histone acetylation landscape transitions that dictate muscle stem cell function. Cell Rep. 2019;27:3939–3955. doi: 10.1016/j.celrep.2019.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung S., Dzeja P.P., Faustino R.S., Perez-Terzic C., Behfar A., Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat. Clin. Pract. Cardiovasc. Med. 2007;4(Suppl 1):S60–S67. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisowski P., Kannan P., Mlody B., Prigione A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018;19 doi: 10.15252/embr.201745432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C., Tian X., Kim J.P., Xie D., Ao X., Shan D., Lin Q., Hudock M.R., Bai X., Yang J. Citrate-based materials fuel human stem cells by metabonegenic regulation. Proc. Natl. Acad. Sci. U S A. 2018;115:E11741–E11750. doi: 10.1073/pnas.1813000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang R., Zhang X., Liu J., Li X., Zhou D., Luan S. Functional gelatin hydrogel scaffold with degraded-release of glutamine to enhance cellular energy metabolism for cartilage repair. Int. J. Biol. Macromol. 2022;221:923–933. doi: 10.1016/j.ijbiomac.2022.09.039. [DOI] [PubMed] [Google Scholar]
- 15.Romani P., Nirchio N., Arboit M., Barbieri V., Tosi A., Michielin F., Shibuya S., Benoist T., Wu D., Hindmarch C.C.T., Giomo M., Urciuolo A., Giamogante F., Roveri A., Chakravarty P., Montagner M., Cali T., Elvassore N., Archer S.L., De Coppi P., Rosato A., Martello G., Dupont S. Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance. Nat. Cell Biol. 2022;24:168–180. doi: 10.1038/s41556-022-00843-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J.S., Burckhardt C.J., Lazcano R., Solis L.M., Isogai T., Li L., Chen C.S., Gao B., Minna J.D., Bachoo R., DeBerardinis R.J., Danuser G. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature. 2020;578:621–626. doi: 10.1038/s41586-020-1998-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Totaro A., Panciera T., Piccolo S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 2018;20:888–899. doi: 10.1038/s41556-018-0142-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moya I.M., Halder G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019;20:211–226. doi: 10.1038/s41580-018-0086-y. [DOI] [PubMed] [Google Scholar]
- 19.Bertero T., Oldham W.M., Cottrill K.A., Pisano S., Vanderpool R.R., Yu Q., Zhao J., Tai Y., Tang Y., Zhang Y.Y., Rehman S., Sugahara M., Qi Z., Gorcsan J., 3rd, Vargas S.O., Saggar R., Saggar R., Wallace W.D., Ross D.J., Haley K.J., Waxman A.B., Parikh V.N., De Marco T., Hsue P.Y., Morris A., Simon M.A., Norris K.A., Gaggioli C., Loscalzo J., Fessel J., Chan S.Y. Vascular stiffness mechanoactivates YAP/TAZ-dependent glutaminolysis to drive pulmonary hypertension. J. Clin. Invest. 2016;126:3313. doi: 10.1172/JCI86387. 3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Q.P., Luo Q., Deng B., Ju Y., Song G.B. Stiffer matrix accelerates migration of hepatocellular carcinoma cells through enhanced aerobic glycolysis via the MAPK-YAP signaling. Cancers. 2020;12:490. doi: 10.3390/cancers12020490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 22.Mookerjee S.A., Gerencser A.A., Nicholls D.G., Brand M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 2017;292:7189–7207. doi: 10.1074/jbc.M116.774471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakrabarty R.P., Chandel N.S. Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell. 2021;28:394–408. doi: 10.1016/j.stem.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kraus F., Roy K., Pucadyil T.J., Ryan M.T. Function and regulation of the divisome for mitochondrial fission. Nature. 2021;590:57–66. doi: 10.1038/s41586-021-03214-x. [DOI] [PubMed] [Google Scholar]
- 25.Xie J., Bao M., Hu X., Koopman W.J.H., Huck W.T.S. Energy expenditure during cell spreading influences the cellular response to matrix stiffness. Biomaterials. 2021;267 doi: 10.1016/j.biomaterials.2020.120494. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M., Sun Q., Liu Y., Chu Z., Yu L., Hou Y., Kang H., Wei Q., Zhao W., Spatz J.P., Zhao C., Cavalcanti-Adam E.A. Controllable ligand spacing stimulates cellular mechanotransduction and promotes stem cell osteogenic differentiation on soft hydrogels. Biomaterials. 2021;268 doi: 10.1016/j.biomaterials.2020.120543. [DOI] [PubMed] [Google Scholar]
- 27.Brusatin G., Panciera T., Gandin A., Citron A., Piccolo S. Biomaterials and engineered microenvironments to control YAP/TAZ-dependent cell behaviour. Nat. Mater. 2018;17:1063–1075. doi: 10.1038/s41563-018-0180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu Y., Newman H., Shen L., Sharma D., Hu G., Mirando A.J., Zhang H., Knudsen E., Zhang G.F., Hilton M.J., Karner C.M. Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metabol. 2019;29:966–978. doi: 10.1016/j.cmet.2019.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elosegui-Artola A., Andreu I., Beedle A.E.M., Lezamiz A., Uroz M., Kosmalska A.J., Oria R., Kechagia J.Z., Rico-Lastres P., Le Roux A.L., Shanahan C.M., Trepat X., Navajas D., Garcia-Manyes S., Roca-Cusachs P. Force triggers YAP nuclear entry by regulating transport across nuclear pores. Cell. 2017;171:1397–1410. doi: 10.1016/j.cell.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 30.Shapira S.N., Christofk H.R. Metabolic regulation of tissue stem cells. Trends Cell Biol. 2020;30:566–576. doi: 10.1016/j.tcb.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodríguez-Colman M.J., Schewe M., Meerlo M., Stigter E., Gerrits J., Pras-Raves M., Sacchetti A., Hornsveld M., Oost K.C., Snippert H.J., Verhoeven-Duif N., Fodde R., Burgering B.M. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature. 2017;543:424–427. doi: 10.1038/nature21673. [DOI] [PubMed] [Google Scholar]
- 32.van Gastel N., Carmeliet G. Metabolic regulation of skeletal cell fate and function in physiology and disease. Nat. Metab. 2021;3:11–20. doi: 10.1038/s42255-020-00321-3. [DOI] [PubMed] [Google Scholar]
- 33.Lee W.C., Ji X., Nissim I., Long F. Malic enzyme couples mitochondria with aerobic glycolysis in osteoblasts. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shum L.C., White N.S., Mills B.N., Bentley K.L., Eliseev R.A. Energy metabolism in mesenchymal stem cells during osteogenic differentiation. Stem Cells Dev. 2016;25:114–122. doi: 10.1089/scd.2015.0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C.T., Shih Y.R., Kuo T.K., Lee O.K., Wei Y.H. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cell. 2008;26:960–968. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- 36.Suh J., Kim N.K., Shim W., Lee S.H., Kim H.J., Moon E., Sesaki H., Jang J.H., Kim J.E., Lee Y.S. Mitochondrial fragmentation and donut formation enhance mitochondrial secretion to promote osteogenesis. Cell Metabol. 2023;35:345–360. doi: 10.1016/j.cmet.2023.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Barzegari A., Aaboulhassanzadeh S., Landon R., Gueguen V., Meddahi-Pelle A., Parvizpour S., Anagnostou F., Pavon-Djavid G. Mitohormesis and mitochondrial dynamics in the regulation of stem cell fate. J. Cell. Physiol. 2022;237:3435–3448. doi: 10.1002/jcp.30820. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda Y., Shirakabe A., Brady C., Zablocki D., Ohishi M., Sadoshima J. Molecular mechanisms mediating mitochondrial dynamics and mitophagy and their functional roles in the cardiovascular system. J. Mol. Cell. Cardiol. 2015;78:116–122. doi: 10.1016/j.yjmcc.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren L., Chen X., Chen X., Li J., Cheng B., Xia J. Mitochondrial dynamics: fission and fusion in fate determination of mesenchymal stem cells. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.580070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forni M.F., Peloggia J., Trudeau K., Shirihai O., Kowaltowski A.J. Murine mesenchymal stem cell commitment to differentiation is regulated by mitochondrial dynamics. Stem Cell. 2016;34:743–755. doi: 10.1002/stem.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibar C., Irvine K.D. Integration of hippo-YAP signaling with metabolism. Dev. Cell. 2020;54:256–267. doi: 10.1016/j.devcel.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White S.M., Avantaggiati M.L., Nemazanyy I., Di Poto C., Yang Y., Pende M., Gibney G.T., Ressom H.W., Field J., Atkins M.B., Yi C. YAP/TAZ inhibition induces metabolic and signaling rewiring resulting in targetable vulnerabilities in NF2-deficient tumor cells. Dev. Cell. 2019;49:425–443. doi: 10.1016/j.devcel.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng X., Han H., Liu G.P., Ma Y.X., Pan R.L., Sang L.J., Li R.H., Yang L.J., Marks J.R., Wang W., Lin A. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017;36:3325–3335. doi: 10.15252/embj.201797609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papalazarou V., Zhang T., Paul N.R., Juin A., Cantini M., Maddocks O.D.K., Salmeron-Sanchez M., Machesky L.M. The creatine-phosphagen system is mechanoresponsive in pancreatic adenocarcinoma and fuels invasion and metastasis. Nat. Metab. 2020;2:62–80. doi: 10.1038/s42255-019-0159-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu V., Roy I.J., Torres A., Jr., Joly J.H., Ahsan F.M., Graham N.A., Teitell M.A. Glutamine-dependent signaling controls pluripotent stem cell fate. Dev. Cell. 2022;57:610–623. doi: 10.1016/j.devcel.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torrino S., Grasset E.M., Audebert S., Belhadj I., Lacoux C., Haynes M., Pisano S., Abelanet S., Brau F., Chan S.Y., Mari B., Oldham W.M., Ewald A.J., Bertero T. Mechano-induced cell metabolism promotes microtubule glutamylation to force metastasis. Cell Metabol. 2021;33:1342–1357. doi: 10.1016/j.cmet.2021.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Bertero T., Oldham W.M., Grasset E.M., Bourget I., Boulter E., Pisano S., Hofman P., Bellvert F., Meneguzzi G., Bulavin D.V., Estrach S., Feral C.C., Chan S.Y., Bozec A., Gaggioli C. Tumor-stroma mechanics coordinate amino acid availability to sustain tumor growth and malignancy. Cell Metabol. 2019;29:124–140. doi: 10.1016/j.cmet.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santinon G., Pocaterra A., Dupont S. Control of YAP/TAZ activity by metabolic and nutrient-sensing pathways. Trends Cell Biol. 2016;26:289–299. doi: 10.1016/j.tcb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Enzo E., Santinon G., Pocaterra A., Aragona M., Bresolin S., Forcato M., Grifoni D., Pession A., Zanconato F., Guzzo G., Bicciato S., Dupont S. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tse J.R., Engler A.J. Preparation of hydrogel substrates with tunable mechanical properties. Curr. Protoc. Cell Biol. 2010 doi: 10.1002/0471143030.cb1016s47. (Chapter 10) Unit 10.16. [DOI] [PubMed] [Google Scholar]
- 51.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 52.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen K.H., Dasgupta A., Lin J., Potus F., Bonnet S., Iremonger J., Fu J., Mewburn J., Wu D., Dunham-Snary K., Theilmann A.L., Jing Z.C., Hindmarch C., Ormiston M.L., Lawrie A., Archer S.L. Epigenetic dysregulation of the dynamin-related protein 1 binding partners MiD49 and MiD51 increases mitotic mitochondrial fission and promotes pulmonary arterial hypertension: mechanistic and therapeutic implications. Circulation. 2018;138:287–304. doi: 10.1161/CIRCULATIONAHA.117.031258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopez-Domenech G., Covill-Cooke C., Ivankovic D., Halff E.F., Sheehan D.F., Norkett R., Birsa N., Kittler J.T. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. EMBO J. 2018;37:321–336. doi: 10.15252/embj.201696380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fehrenbacher K.L., Yang H.C., Gay A.C., Huckaba T.M., Pon L.A. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr. Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available in the article or from the corresponding author on reasonable request.