Summary
Molecular mechanisms underlying quantitative variations of pathogenicity remain elusive. Here, we identified the Xanthomonas campestris XopJ6 effector that triggers disease resistance in cauliflower and Arabidopsis thaliana. XopJ6 is a close homolog of the Ralstoniapseudosolanacearum PopP2 YopJ family acetyltransferase. XopJ6 is recognized by the RRS1-R/RPS4 NLR pair that integrates a WRKY decoy domain mimicking effector targets. We identified a XopJ6 natural variant carrying a single residue substitution in XopJ6 WRKY-binding site that disrupts interaction with WRKY proteins. This mutation allows XopJ6 to evade immune perception while retaining some XopJ6 virulence functions. Interestingly, xopJ6 resides in a Tn3-family transposon likely contributing to xopJ6 copy number variation (CNV). Using synthetic biology, we demonstrate that xopJ6 CNV tunes pathogen virulence on Arabidopsis through gene dosage-mediated modulation of xopJ6 expression. Together, our findings highlight how sequence and structural genetic variations restricted at a particular effector gene contribute to bacterial host adaptation.
Subject areas: Classification Description, Genetics, Bacteriology, Molecular plant pathology
Graphical abstract

Highlights
-
•
Xanthomonas campestris XopJ6 effector protein is a YopJ family acetyltransferase
-
•
XopJ6 avirulence activity triggers immunity in Arabidopsis and cauliflower accessions
-
•
A XopJ6 natural variant evades recognition by the RRS1-R/RPS4 NLR pair
-
•
xopJ6 copy number variation modulates pathogen virulence on Arabidopsis
Classification Description: Genetics; Bacteriology; Molecular plant pathology
Introduction
During the co-evolution of plants and pathogens, plants have developed a complex two-tiered immune system that allows the recognition of both conserved pathogen-associated molecular patterns (PAMPs) and pathogen-specific secreted molecules (effectors).1,2 PAMP recognition by cell surface receptors triggers the activation of defense responses known as PAMP-triggered immunity (PTI).3 These responses include rapid bursts of reactive oxygen species (ROS) and Ca2+, activation of mitogen-activated and Ca2+-dependent protein kinase signaling cascades, production of different phytohormones, and an extensive transcriptional reprogramming.4,5 Adapted pathogens can overcome PTI by secreting effectors able to subvert the plant immunity and physiology to the pathogen’s benefit.6 In turn, plants have evolved intracellular immune receptors able to recognize some of these effectors to potentiate PTI.7,8,9 This later response known as effector-triggered immunity (ETI) is often associated with the hypersensitive response (HR), a form of host-induced programmed cell death to prevent the pathogen proliferation and spread.10 Therefore, effectors constitute double-edged swords that confer either strong advantages or disadvantages for bacterial multiplication, depending on the host plant genotype.
Many gram-negative bacterial pathogens are able to translocate their type III effector (T3E) proteins directly into the host cells using specialized type III secretion systems (T3SS).11 The YopJ family of T3E proteins, named after the Yersinia spp. outer protein J, is one of the most widely distributed T3E in both animal- and plant-associated bacteria.12 This T3E family is characterized by the presence of a conserved protease-like catalytic triad,13 required for acetyltransferase activity of several YopJ members from both animal14,15,16 and plant pathogens.17,18,19,20,21 These include the Ralstonia pseudosolanacearum PopP2 effector, which exerts its virulence functions by acetylating defensive WRKY transcription factors (TFs) to prevent WRKY binding to DNA and the subsequent activation of defense-related genes.22,23 In resistant Arabidopsis accessions, PopP2 is recognized by a pair of NOD-like immune receptors (NLRs) RRS1-R (RESISTANCE TO RALSTONIA SOLANACEARUM1) and RPS4 (RESISTANCE TO PSEUDOMONAS SYRINGAE4) with an N-terminal TIR (Toll-Interleukin-1 Receptor) signaling domain.24 RRS1-R and RPS4 cooperate genetically and molecularly in resistance to different pathogens.25,26,27,28,29,30,31 RRS1-R effectively intercepts PopP2-mediated disabling of defensive WRKY TFs thanks to a C-terminal WRKY decoy domain. Acetylation of RRS1-R by PopP2 activates the receptor complex and ETI.22,23
Xanthomonas campestris pv. campestris (Xcc) is the causal agent of black rot on wild and cultivated Brassicaceae, including the Arabidopsis thaliana model species.32 Xcc possesses a T3SS essential for pathogenicity to deliver a repertoire of 18–28 T3E proteins.33 The contribution of few Xcc T3Es to virulence has been reported although the underlying molecular mechanisms remain largely unknown.34,35,36,37,38,39,40 To date, the only Xcc T3E with known avirulence activity is AvrAC/Xanthomonas outer protein AC (XopAC) on Arabidopsis41 and Matthiola incana.42 In Arabidopsis, XopAC is recognized by the intracellular immune receptor ZAR1 (HOPZ-ACTIVATED RESISTANCE 1)/RKS1 (RESISTANCE-RELATED KINASE 1) complex that perceives uridylylation of PBS1-LIKE PROTEIN 2 (PBL2) triggered by XopAC.43,44 Upon activation, the ZAR1/RKS1/PBL2 complex assembles into a pentameric resistosome which functions as a calcium channel in the plasma membrane mediating immune signaling and cell death.44,45
In this study, we identified the Xcc XopJ6 effector as a close homolog of PopP2 from the YopJ family of acetyltransferases. We found that xopJ6 confers avirulence to Xcc in both cauliflower and Arabidopsis. In addition, we showed that XopJ6 targets WRKY TFs by acetylation, similar to PopP2, highlighting a conserved virulence strategy for disease establishment across bacterial phytopathogens. Consistent with PopP2 recognition mechanism, XopJ6 is recognized by the RRS1-R/RPS4 immune receptor complex with RRS1-R functioning as a sensor of XopJ6 acetyltransferase activity. By exploring the natural diversity of Xcc genomes, we found a single-amino acid variant of XopJ6 which can evade RRS1-R/RPS4-mediated recognition while retaining some virulence activity. In addition, we observed a variation in copy number of xopJ6 in Xcc strains, which is positively correlated with both XopJ6 expression in vitro and Xcc virulence in Arabidopsis. These results highlight how subtle variations in sequence or expression of a single T3E gene can quantitatively tune bacterial pathogenicity and determine the outcome of the interaction.
Results
Copy number variation (CNV) and extreme sequence conservation at the xopJ6 loci
To investigate the virulence of Xcc on cauliflower (Brassica oleracea var. botrytis cv. Clovis), 37 different Xcc strains were inoculated in leaf mesophyll by infiltration. Of all the strains tested, seven phylogenetically related strains (CN02, CN03, CN06, CN08, CN12, CN17, and CN18), as well as the CN07 strain, caused an HR-like collapse of the tissues 24 h post-inoculation (Figures 1A–1C). To identify the Xcc effector gene triggering this cell death response, we screened for a positive correlation between the ability/inability to cause HR (strains CN02, CN03, CN06, CN08, CN12, CN17, and CN18 versus strains 8004, ATCC33913, CN14, CN15, and CN16) and the presence/absence of genes as inferred from their genomic sequences (Table S1). This approach yielded 42 candidate genes including xopJ6, a T3E gene that encodes a putative YopJ family acetyltransferase (IPR005083, Table S2). In silico analyses of Xcc genomic sequences evidenced one (CN03, CN06, CFBP4954, CN13, and CFBP6865), two (CN02, CN07, CN12, and CN17), three (CN08), or four (CN18) xopJ6 copies per genome (Figure 1A). The exact copy number of xopJ6 was experimentally confirmed by Southern blot for all strains except for CN18 (Figure S1A): indeed, four xopJ6 copies could be identified in CN18 genome sequence versus at least two by Southern analysis. All xopJ6 sequences were identical between and within strains except for strains CN13, CFBP4954, and CFBP6865. xopJ6CN13 and xopJ6CFBP4954 differed from the consensus sequence by only two SNPs (T214G and C1146A) leading to Cys72Gly and Asn382Lys substitutions in the XopJ6 protein (Figure S2). xopJ6CFBP6865 sequence harbored both the T214G SNP and an insertion by a 1.2 kb-long IS3-family insertion sequence (IS) at position 232–233 (Figure S2A). The most remarkable features of xopJ6 loci are both extreme sequence conservation and CNV. For the sake of simplicity, we used xopJ6CN06 as xopJ6 reference sequence unless otherwise specified.
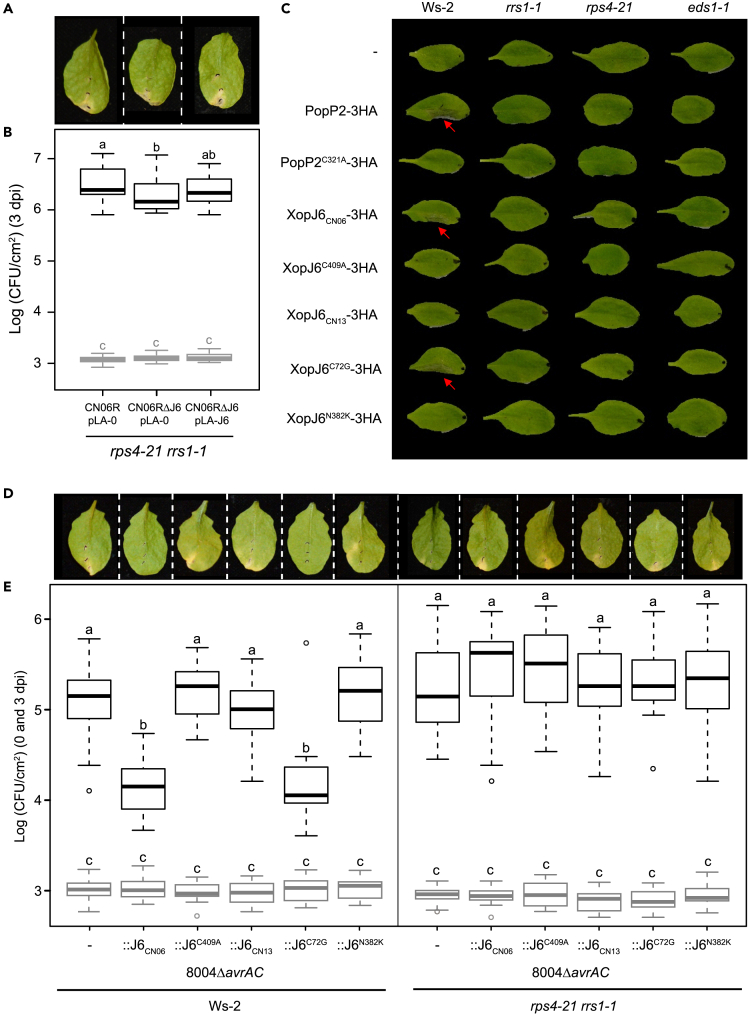
Figure 1.
xopJ6 gene confers avirulence in Arabidopsis and cauliflower
(A) Phylogeny of X. campestris pv. campestris strains (Xcc) as described.39 Red and blue dots indicate the presence of xopAC and xopJ6, respectively. Alleles not able to confer avirulence are indicated by empty dots. The number of blue dots indicates the number of xopJ6 copies. The question mark indicates that the number of xopJ6 copies is uncertain.
(B) Tissue collapse (TC) observed on cauliflower 24h after infiltration with Xcc strains at 108 CFU/mL presented in panel A (-: indicates no collapse; the experiment was repeated 2 times with similar results) and median disease index (DI) on Arabidopsis thaliana (Ath) Col-0, Ws-2 and Sf-2 accessions 7 days after wound inoculation with bacterial suspensions at 108 CFU/mL (from Figure S1B; at least three independent experiments, 4 plants per replicate, 4 leaves per plant; color code: white, no symptom; yellow, faint chlorosis; orange, extended chlorosis; red, necrosis).
(C) Collapse observed on cauliflower leaves 24h after infiltration with Xcc CN06R rifampicin-resistant strain, xopJ6 deletion mutant (CN06RΔJ6) and the complemented strain (CN06RΔJ6:1301-J6) at 108 CFU/mL.
(D) Bacterial population in individual cauliflower hydathodes six days after dip inoculation of various Xcc rifampicin-resistant strains, corresponding xopJ6-deleted strains (ΔJ6 for strains with one copy and ΔΔJ6 for strain with two copies) and the CN06 complemented strain (CN06RΔJ6:1301-J6) with bacterial suspensions at 108 CFU/mL.
(E) Disease symptoms observed 10 days after wound inoculation of Arabidopsis Ws-2 leaves with Xcc CN06R rifampicin-resistant strain and xopJ6 deletion mutant carrying an empty pLAFR6 plasmid (−) or containing the xopJ6 gene (+J6) with bacterial suspensions at 108 CFU/mL. Pictures are representative of the median disease index.
(F) Bacterial populations at 0 dpi (in gray, at least 20 values per strain) and 3 dpi (in black, 27 values per strain) after infiltration of Arabidopsis Ws-2 leaves with same strains as in panel E with bacterial suspensions at 105 CFU/mL. For panels D, E and F, at least three independent experiments were performed and combined and different letters indicate statistical groups determined using nonparametric Kruskal-Wallis or HSD tests (p value < 0.01).
xopJ6 is a T3E gene located within a Tn3-family transposon
xopJ6 contains a cis-regulatory element (PIP-box, TTCG-N16-TTCG, −147 to −123 bp from start codon) bound by the master hrp regulatory protein HrpX.46 Using an epitope-tagged XopJ6-c-Myc placed under the control of its own promoter (Table S1), we could confirm that accumulation of XopJ6 is dependent on both HrpG and HrpX regulators (Figure S3A) in both rich and hrp-inducing media.47
xopJ6 in Xcc is located in a highly conserved 9-kb region flanked by 37-bp inverted repeats (IRs) which also contains genes coding for a Tn3-family transposase (IPR002513), a tyrosine recombinase (IPR020876), and a hypothetical protein (Figure S3B). Such genetic element, typical of Tn3-family transposons,48 was registered as Tn6714 in The Transposon Registry.49 The chromosomal copies of Tn6714 were found as simple or composite transposons in three insertion sites located within a 80-kb chromosomal region of Xcc (Figure S3C). A dynamic and parsimonious scenario implying insertions, duplications, and losses is proposed to explain the variation in copy number of xopJ6 in Xcc (Figure S3D).
Single Tn6714 copies were found in CN13 and CFBP6865 on 186-kb and 62-kb contigs, respectively, corresponding to the observed plasmid sizes in those two strains (195 kb in CN1333 and 63 kb in CFBP686550). The exact location of Tn6714CFBP4954 could not be determined. xopJ6 is thus expressed from either chromosomal or plasmid-borne copies of Tn6714.
xopJ6 is an avirulence gene on cauliflower and Arabidopsis thaliana
The presence of xopJ6 was correlated with the ability to trigger HR on cauliflower cv. Clovis for the 37 Xcc strains tested except for strains CN13, CFBP4954, and CFBP6865 which failed to induce any collapse (Figures 1A and 1B). To test for possible xopJ6-mediated HR, a xopJ6 deletion mutant was constructed in CN06R, a rifampicin-resistant derivative of CN06 strain that expresses a consensus xopJ6 sequence. This CN06RΔJ6 deletion mutant failed to cause HR and could be complemented with xopJ6CN06 sequence under the control of a strong constitutive tac promoter (CN06RΔJ6:1301-J6, Figure 1C). Upon dip inoculation, multiplication of CN06RΔJ6 in hydathodes was increased by 10-fold as compared to wild-type and CN06RΔJ6:1301-J6 strains (Figure 1D). Comparable xopJ6-mediated avirulence activity was observed in strains CN03R and CN12R, but not strain CN13R which expresses a xopJ6 allele with both T214G and C1146A SNPs (Figure 1D). These results demonstrate that xopJ6 encodes for an avirulence protein recognized in cauliflower cv. Clovis.
To study possible xopJ6 avirulence functions in Arabidopsis, pathogenicity assays were performed on accessions Col-0, Ws-2, and Sf-2 with all the wild-type Xcc strains by wound inoculation (Figures 1B and S1B). Interestingly, strains causing xopJ6-mediated HR on cauliflower do not cause disease in Ws-2 and Sf-2, indicating that xopJ6 is also recognized in Arabidopsis. This was further confirmed in Ws-2 by wound inoculation with strains CN06R, CN06RΔJ6, and the complemented strain. CN06RΔJ6 was the only strain able to cause disease (Figure 1E) with up to 10-fold higher multiplication in the mesophyll as compared to wild-type and complemented strains (Figure 1F). Altogether, these results demonstrate that xopJ6 behaves as an avirulence gene on both cauliflower and Arabidopsis.
XopJ6 belongs to the YopJ family of acetyltransferases and is very closely related to Ralstonia pseudosolanacearum PopP2
XopJ6 is a protein of 565 residues that belongs to the YopJ family of bacterial acetyltransferases, composed of evolutionary-conserved T3E proteins from a wide variety of animal and plant bacterial pathogens as well as nitrogen-fixing rhizobia.12 A phylogenetic analysis revealed that XopJ6 does not fall with the Xanthomonas group II homologs but into the R. pseudosolanacearum group V with PopP2 being its closest homolog (Figure 2A). Group V is closest to YopJ homologs from animal pathogens (group I).
Figure 2.
XopJ6 from Xanthomonas campestris pv. campestris is a close homolog to PopP2 from Ralstonia pseudosolanacearum
(A) Phylogenetic tree of 25 members of bacterial effectors of the YopJ family. The position of XopJ6CN06 and its close ortholog PopP2GMI1000 is highlighted in red. Neighbor-joining tree with bootstrap support values indicated above each node. The scale bar indicates the evolutionary distance in number of amino acid substitutions per site computed using the Poisson correction method. Division in groups is based on a previous report.12
(B) Pairwise sequence alignment of PopP2GMI1000 and XopJ6CN06. ClustalW2 alignment using the BLOSUM cost matrix with open and extension gap penalties of 10 and 0.1, respectively. The background color represents identical (black) or similar (gray) residues. The amino acids forming the conserved catalytic triad13 are framed in orange and the residues involved in binding to acetyl-CoA, InsP6 co-factor, and RRS1-R in yellow, green, and blue, respectively.20,51 The two amino acid substitutions found in natural variants of XopJ6 (C72G and N382K) are indicated by a red dot (see Figure S2B).
XopJ6 and PopP2 share 35.4% overall identity with a good conservation of the acetyltransferase domain52 (residues 232–565 and 151–488, respectively [Figure 2B]). Known residues driving PopP2 biological activities are conserved in XopJ6CN0620,51: catalytic triad (PopP2H260/D279/C321, H346/D365/C409 in XopJ6), residues important for interaction with WRKY TFs (PopP2E284/D292/N296, E370/D378/N382 in XopJ6), a lysine residue targeted by PopP2 autoacetyltransferase activity (PopP2K383, K471 in XopJ6), and residues involved in binding with host co-factor Inositol-6-Phosphate (InsP6) (PopP2R416, R505 in XopJ6) and with acetyl-coenzyme A (PopP2K316/S389/F446, K404/S477/F530 in XopJ6) (Figure 2B). Such conservation suggests that XopJ6CN06 is an acetyltransferase that could modify WRKY TFs by acetylation, as previously shown for PopP2.22,23
The Arabidopsis RRS1-R/RPS4 NLR pair mediates XopJ6 recognition
In Arabidopsis, PopP2 is recognized by the RRS1-R/RPS4 NLR pair. We tested whether this NLR pair was also involved in XopJ6 recognition by inoculating the rps4-21 rrs1-1 double mutant with CN06R strain. In contrast to Ws-2 plants (Figures 1E and 1F), CN06R and CN06RΔJ6 strains caused similar disease index (Figures 3A and S4A) and multiply to similar levels in the rps4-21 rrs1-1 mutant (Figure 3B). This suggests that RRS1-R and/or RPS4 are required for recognition of XopJ6 and resistance to strain CN06R. These results are consistent with RRS1-R/RPS4 conferring resistance to Xcc CFBP6943,53 a strain which contains at least one copy of xopJ6 (Figure S1A).
Figure 3.
XopJ6N382K escapes recognition by RRS1-R and RPS4 in Ws-2
(A) Pictures representative of the median disease index observed at 10 days after wound inoculation of Arabidopsis Ws-2 rps4-21 rrs1-1 mutant with bacterial suspensions at 108 CFU/mL. Strains are described in panel B.
(B) Bacterial populations at 0 dpi (in gray, at least 20 values per strain) and 3 dpi (in black, 27 values per strain) after infiltration of Arabidopsis Ws-2 rps4-21 rrs1-1 mutant with Xcc CN06R rifampicin-resistant strain or xopJ6-deleted (ΔJ6) strain carrying an empty plasmid (pLA-0) or containing xopJ6 (pLA-J6) with bacterial suspensions at 105 CFU/mL. Three independent experiments were performed and combined.
(C) Composite image of hypersensitive response (HR) observed 48 h post-infiltration of Arabidopsis wild-type Ws-2 accession or rrs1-1, rps4-21, and eds1-1 mutants with Pseudomonas fluorescens Pf0-1 (OD600 = 0.2) delivering the indicated effector protein. Red arrows indicate leaves showing HR. This experiment was performed between two to three times with similar results.
(D) Pictures representative of the median disease index observed 10 days after wound inoculation of Arabidopsis Ws-2 wild-type accession or rps4-21 rrs1-1 mutant with bacterial suspensions at 108 CFU/mL. Strains are described in panel E.
(E) Bacterial populations at 0 dpi (in gray, 17 values per strain) and 3 dpi (in black, 27 values per strain) after infiltration of Arabidopsis Ws-2 wild-type accession or rps4-21 rrs1-1 mutant with Xcc 8004ΔavrAC strains carrying a genomic copy of xopJ6 (:J6) with various mutations (C409A, C72G and/or N382K) with bacterial suspensions at 108 CFU/mL. Three independent experiments were performed and combined. Panels B and E: statistical groups were determined using nonparametric Kruskal-Wallis test (p value < 0.01) and are indicated by different letters. See also Figure S4.
To further investigate the genetic requirement of RRS1-R and RPS4 in XopJ6 recognition, we used the Pseudomonas fluorescens (Pf0-1)-mediated effector delivery assay in wild-type Ws-2 and the mutant lines rrs1-1 and rps4-2154 (Figure 3C). The eds1-1 mutant was also inoculated since EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1) is necessary for RRS1-R/RPS4 function.55,56,57 The amount of triple hemagglutinin (HA)-tagged effector proteins produced by Pf0-1 was verified by immunoblot (Figure S4B). After 24 h, XopJ6 triggered a cell death response in Ws-2 similar to PopP2, whereas no HR was observed on rrs1-1, rps4-21, and eds1-1 mutant lines. Moreover, as observed with the PopP2C321A catalytic mutant, a XopJ6C409A variant mutated in its putative main catalytic Cys409 residue was unable to trigger HR on Ws-2, suggesting that XopJ6 is an acetyltransferase whose activity is responsible for activation of RRS1-R/RPS4-dependent immunity. Unlike AvrRps4 recognition that involves a second R gene pair, RRS1B/RPS4B,58 full inhibition of XopJ6-triggered HR in single rrs1-1 and rps4-21 mutants, indicates that, as previously observed with PopP2, XopJ6 recognition in Ws-2 does not seem to require the RRS1B/RPS4B gene pair.
These results were further confirmed by gain-of-function experiments in Xcc strain 8004 which does not naturally carry xopJ6. xopJ6 was expressed in the 8004ΔavrAC mutant strain to avoid avrAC-mediated ETI in Ws-2.43,44 Strain 8004ΔavrAC::J6CN06 was avirulent on Ws-2 (Figures 3D and S4C), and its multiplication was reduced by 10-fold compared to strain 8004ΔavrAC (Figure 3E), confirming that xopJ6 is responsible for the avirulence observed on this accession. This plant response depends on XopJ6 acetyltransferase activity since strain 8004ΔavrAC::J6C409A expressing XopJ6-C409A catalytic mutant causes disease and multiplies on Ws-2 leaves similarly to strain 8004ΔavrAC (Figures 3D, 3E, and S4C). When delivered by the Xcc CN06RΔJ6 strain, PopP2 also triggered a reduction in bacterial multiplication in Arabidopsis Ws-2 accession (Figure S4D). However, contrary to XopJ6CN06, no cell death was observed on the cauliflower cv. Clovis in response to PopP2 (Figure S4E), suggesting a very specific mechanism for XopJ6 recognition in cauliflower.
XopJ6 physically interacts with RRS1-R and AtWRKY22 in planta
Based on the strong similarities between PopP2 and XopJ6 acetyltransferase domains, we hypothesized that XopJ6 also triggers activation of RRS1-R/RPS4 through manipulation of RRS1-R WRKY domain. XopJ6 physical interaction with RRS1-R WRKY domain was first demonstrated using a yeast two-hybrid assay. While full-length BD-XopJ6 fusion protein was auto-active in yeast, XopJ6 catalytic unit (residues 224 to 565) used as bait (BD-XopJ6224-565) specifically interacted with AD-RRS1-RCterm, like PopP2 (Figures 4A and S5A). In order to probe for XopJ6/RRS1-R interaction in vivo, FRET-FLIM (Förster resonance energy transfer by fluorescence lifetime imaging microscopy) experiments were conducted. XopJ6CN06 fused to the cyan fluorescent protein (XopJ6CN06-CFP) and RRS1-R fused to the yellow fluorescent protein (RRS1-R-YFP) were transiently co-expressed in N. benthamiana (Figure S5B) and served as FRET donor and acceptor, respectively. XopJ6CN06-CFP showed a nucleocytoplasmic localization whereas RRS1-R-YFP was restricted to the nucleus (Figure 4B). The average CFP lifetime in nuclei expressing XopJ6CN06-CFP alone was higher than in nuclei where both XopJ6CN06-CFP and RRS1-R-YFP could be detected indicative of a physical interaction (Table 1; Figure 4C). Similar results were obtained with AtWRKY22 (Table 1; Figures 4C and 4D) indicating that XopJ6 can interact with WRKY TFs, like PopP2. Finally, like PopP2,20 Xop6C409A catalytic mutant also interacted with RRS1-R and AtWRKY22 (Table 1; Figures 4C and 4D). No FRET could be detected between XopJ6-CFP and YFP alone (Table 1; Figure S5C) further confirming the specificity of the observed XopJ6-WRKY domain interactions.
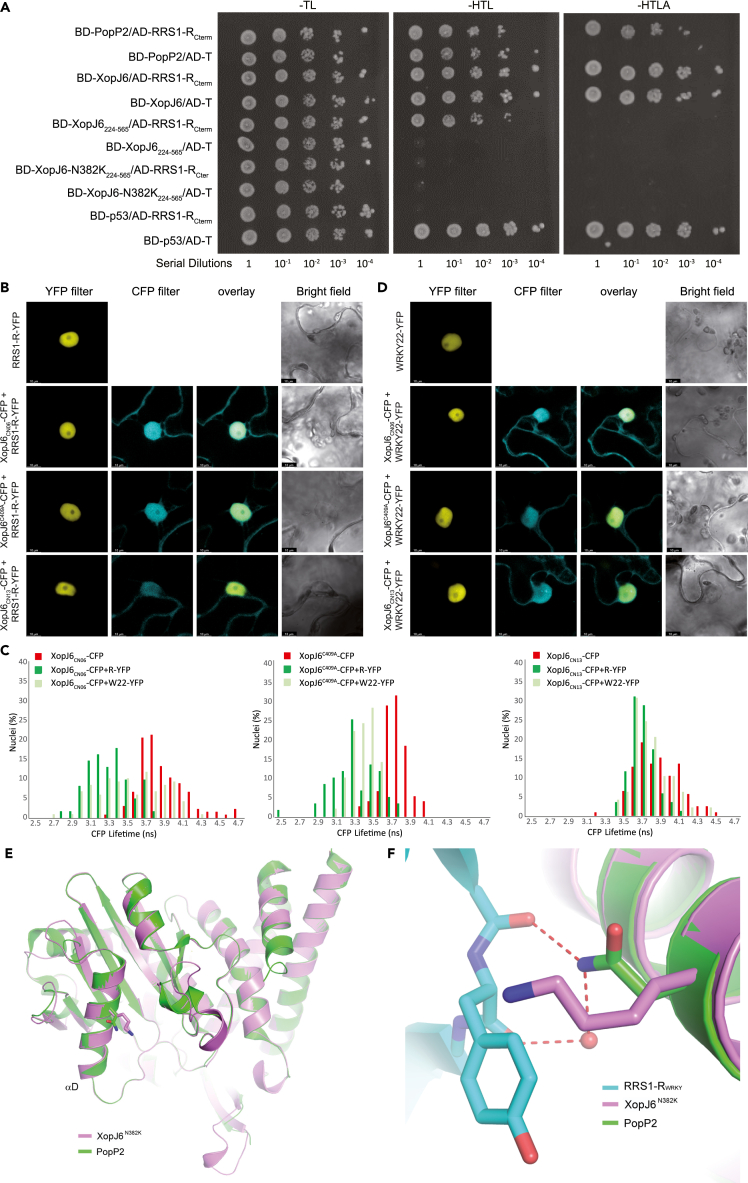
Figure 4.
Natural substitution of the Asn-382 residue with a Lysine (N382K) in XopJ6CN13 prevents physical interaction of XopJ6 with RRS1-R and AtWRKY22 in vivo
(A) The XopJ6 full-length protein or its catalytic unit (residues 224 to 565, designated by XopJ6224-565) containing or not the N382K substitution were used as baits (BD-XopJ6224-565 and BD-XopJ6-N382K224-565, respectively). RRS1-RCterm fused to the Gal4 activating domain (AD) was used as prey (AD-RRS1-RCterm). Full-length PopP2 was used as a positive control bait protein (BD-PopP2) for testing interaction with AD-RRS1-RCterm. The murine p53 protein was used as negative control (BD-p53) for testing interactions with AD-RRS1-RCterm and as a positive control for interaction with the SV40 large T-antigen (AD-T). Full-length XopJ6 causes autoactivation of yeast reporter genes. Yeast transformants were grown on non-selective SD/-Trp/-Leu (SD-TL) medium or selective medium SD/-His/-Trp/-Leu/(SD-HTL) or SD/-His/-Trp/-Leu/-Ade (SD-TLHA). Plates were photographed three days after plating. This experiment was repeated three times with similar results. See Figure S5A.
(B and D) Confocal images of N. benthamiana epidermal cells showing nucleocytoplasmic localization of XopJ6 and its variants co-expressed either with RRS1-R-YFP (B) or AtWRKY22-YFP (D). This experiment was performed three times with similar results.
(C) CFP lifetime distribution of XopJ6CN06, XopJ6C409A and XopJ6CN13 C-terminally fused with CFP in plant nuclei and expressing or not either RRS1-R-YFP or AtWRKY22-YFP. See Figures S5B and S5C.
(E) Superposition of the structures of PopP2 (PDB code 5W3X) and XopJ6 generated by homology modeling. The substrate recognition helix (αD in PopP2) is labeled. The side chains of PopP2-N296 and XopJ6-N382K are shown in stick presentation.
(F) XopJ6-N382K mutation on the interface between PopP2 and RRS1-RWRKY. The hydrogen bonds are depicted as dashed lines, and the water molecule is shown as red sphere.
Table 1.
XopJ6CN06 and XopJ6C409A, but not XopJ6CN13, physically interact in planta with RRS1-R and AtWRKY22 as measured by FRET-FLIM
| Donor | Acceptor | T(ns)a | Δt (ps)b | SEc | semd | Ne | E(%)f | p valueg |
|---|---|---|---|---|---|---|---|---|
| XopJ6CN06-CFP | – | 3.8475 | – | 0.2541 | 0.0218 | 135 | ||
| XopJ6CN06-CFP | RRS1-R-YFP | 3.2694 | 578 | 0.2288 | 0.0293 | 61 | 15.02 | 7.17x10−35 |
| XopJ6CN06-CFP | AtWKRY22-YFP | 3.4824 | 365 | 0.3413 | 0.0314 | 118 | 9.49 | 3.59x10−19 |
| XopJ6CN06-CFP | YFP | 4.0041 | −156 | 0.2572 | 0.0319 | 65 | −4.07 | 6.81 x10−5 |
| XopJ6C409A-CFP | – | 3.7244 | – | 0.1384 | 0.0158 | 77 | ||
| XopJ6C409A-CFP | RRS1-R-YFP | 3.2726 | 452 | 0.2542 | 0.0328 | 60 | 12.13 | 2.89x10−26 |
| XopJ6C409A-CFP | AtWKRY22-YFP | 3.3641 | 360 | 0.1275 | 0.0180 | 50 | 9,67 | 3.34x10−29 |
| XopJ6CN13-CFP | – | 3,8062 | – | 0.2329 | 0.0205 | 129 | ||
| XopJ6CN13-CFP | RRS1-R-YFP | 3.6427 | 163 | 0.1748 | 0.0184 | 90 | 4.29 | 5.27x10−8 |
| XopJ6CN13-CFP | AtWKRY22-YFP | 3.8303 | −24 | 0.2816 | 0.0276 | 50 | −0.63 | 0.47495 |
| XopJ6CN13-CFP | YFP | 3.9187 | −112 | 0.2516 | 0.0356 | 50 | −2.96 | 0.00509 |
The statistical test used was two sided. The lifetime measurements were carried out from two to three independent expression assays performed in N. benthamiana (leaf samples were taken between 36 and 48 hours after infiltration with A. tumefaciens).
Mean lifetime, T, in nanoseconds (ns). For each nucleus, average fluorescence decay profiles were plotted and fitted with exponential function using a non-linear square estimation procedure and the mean lifetime was calculated according to τ = Σ αiti² / Σ αiti with I(t) = Σ αi e-t/ti.
Δt=τD – τDA (in ps).
Standard error.
Standard error of the mean.
Total number of measured nuclei.
% FRET efficiency: E=1 – (τDA/τD).
p value of the difference between the donor lifetimes in the absence and in the presence of acceptor (Student’s t test).
The WRKY DNA-binding domain is a substrate of XopJ6 acetyltransferase activity
To determine whether RRS1-R WRKY domain (RRS1-RCterm) and AtWRKY22 behave as XopJ6 substrates, both proteins were C-terminally fused to the enhanced GFP (RRS1-RCterm-eGFP and AtWRKY22-eGFP) and transiently co-expressed in Nicotiana benthamiana with WT PopP2 or XopJ6CN06, or their corresponding catalytic mutants. Consistent with previous reports,20 immunoblotting with anti-GFP and anti-Ac-K antibodies showed Lys acetylation of immuno-purified RRS1-RCterm-GFP and AtWRKY22-GFP with PopP2, but no acetylation was observed with XopJ6 (Figure S5D).
Acetylation was further examined in E. coli by coexpressing RRS1-RCterm or AtWRKY22 N-terminally and C-terminally fused with glutathione S-transferase (GST) and hexahistidine (His6), respectively, with His6-PopP2 or His6-XopJ6. A truncated form of XopJ6 lacking its first 223 residues (His6-XopJ6224-565) comparable to PopP2 acetyltransferase domain (PopP2149-488)20 was used to improve His6-XopJ6 solubility. Immunoblotting with anti-His6 and anti-Ac-K antibodies of the GST-purified proteins revealed the presence of acetylated RRS1-RCterm and AtWRKY22 only in presence of PopP2 but not with XopJ6224-565 (Figure S5E).
The acetylation status of purified GST-tagged proteins was determined by mass spectrometry-based analysis. Since GST-RRS1-RC-term-His6 co-expressed with XopJ6 variants could not be purified as efficiently as GST-AtWRKY22-His6, this analysis was restricted to the latter. With a coverage of almost 50% of AtWRKY22, acetylation of Lys-139 could be detected upon expression with both PopP2 and XopJ6, but not with XopJ6C409A catalytic mutant (Table S3; Table S4). This Lys-139 resides in the WRKY domain heptad (WRKYGQK139) of AtWRKY22, a canonical DNA-binding domain found in all WRKY TFs.59 PopP2-dependent acetylation of this critical lysine residue was previously reported to inhibit binding of WRKY TF to DNA.22 Together, these data indicate that XopJ6 targets defensive WRKY TFs by acetylation, but probably less efficiently than PopP2, as evidenced by the lack of detection of acetylated forms of RRS1-RCterm and AtWRKY22 co-expressed with active XopJ6 (Figures S5D and S5E).
The N382K substitution in XopJ6CN13 natural variant disrupts physical interaction with WRKY TFs and evades recognition by RRS1-R/RPS4
Strains CN13, CFBP4954, and CFBP6865 are unable to induce collapse on cauliflower cv. Clovis (Figures 1A and 1B), suggesting that their XopJ6 variants have impaired avirulence activity. The virulent Xcc strain 8004 expressing xopJ6CN13 also failed to trigger HR on cauliflower cv. Clovis (Figure S4F). In Arabidopsis Ws-2 plants, XopJ6CN13 delivered by Pf0-1 did not trigger a cell death response (Figure 3C) and Xcc 8004ΔavrAC::J6CN13 strain reached bacterial populations comparable to those of the untransformed virulent 8004ΔavrAC strain (Figures 3D and 3E). Thus, the XopJ6CN13 natural variant lacks avirulence activity in two different host plants likely through evasion of RRS1-R/RPS4-mediated recognition in Arabidopsis.
FRET-FLIM measurements in N. benthamiana using XopJ6CN13-CFP fusion as a donor demonstrate that XopJ6CN13, while showing a nucleocytoplasmic localization similar to that of wild-type XopJ6 and XopJ6-C409A, is unable to physically interact with RRS1-R or AtWRKY22 (Figures 4B–4D; Table 1). This finding is consistent with the inability of XopJ6CN13 to trigger activation of RRS1-R/RPS4-mediated disease resistance in Ws-2 (Figures 3C–3E). XopJ6CN13 variant differs from XopJ6CN06 by two residue substitutions, C72G and N382K (Figure 2B). To determine whether either of these two substitutions allows XopJ6CN13 to evade immune recognition, the avirulence activity of XopJ6C72G and XopJ6N382K was examined in Ws-2 upon delivery by Pf0-1 (Figure 3C) or Xcc (Figures 3D, 3E, and S4C). Pf0-1-mediated delivery of XopJ6CN13 and XopJ6N382K did not trigger any cell death response in Ws-2, in contrast to XopJ6CN06 and XopJ6C72G variants. Similarly, Xcc strains 8004ΔavrAC::J6CN13 and 8004ΔavrAC::J6N382K were virulent on Ws-2 in contrast to 8004ΔavrAC::J6CN06 and 8004ΔavrAC::J6C72G. Finally, strains 8004:J6CN13 and 8004:J6N382K failed to cause HR on cauliflower cv. Clovis in contrast to strains 8004:J6CN06 and 8004:J6C72G (Figure S4F). Together, these results demonstrate the critical role of the N382K substitution for XopJ6CN13 immune evasion of Xcc in both cauliflower cv. Clovis and Arabidopsis Ws-2.
The tri-dimensional structure of PopP2 alone and in complex with RRS1-R C-terminal WRKY domain51 was used to model XopJ6 yielding a structure very similar to PopP2 (Figure 4E). XopJ6CN06 N382 residue corresponds to PopP2-N296, a residue located in a loop mediating physical interaction with RRS1-R WRKY domain.51 By overlapping the XopJ6 and PopP2-RRS1-RC-term structures, the N382K substitution in XopJ6CN13 might not only lead to the loss of two hydrogen bonds that help to recognize the main chain of RRS1-R WRKY domain K1217 and Y1218 (within the conserved WRK1217Y1218GQK heptad) but also introduce a steric collapse with the side chain of Y1218 (Figures 4E and 4F). This modeling could explain why the N382K substitution in XopJ6 catalytic unit abolished the interaction with RRS1-RCterm in yeast two-hybrid assay (Figure 4A) and why the XopJ6CN13 variant cannot physically interact with either AtWRKY22 or RRS1-R in planta (Table 1; Figure 4C), allowing Xcc strains encoding XopJ6CN13 variant (strains CN13 and CFBP4954) to evade recognition (Figures 1B, 3C–3E, S4C, and S4F).
XopJ6 plays a role in Xcc virulence and contributes to PTI suppression
We tested if XopJ6CN06 could contribute to bacterial virulence on Arabidopsis lacking RRS1-R/RPS4 immune receptors. Indeed, bacterial multiplication of the xopJ6 deletion mutant in CN06R strain was significantly reduced on rps4-21 rrs1-1 compared to wild-type strain (Figure 3B). In the susceptible Col-0 accession that is unable to recognize PopP2,27,28 pathogenicity and bacterial growth of strains CN06RΔJ6 and CN03RΔJ6 were significantly reduced compared to corresponding wild-type strains (Figures 5A, 5B, and S6A). Pathogenicity could only be partially complemented for CN06RΔJ6 strain. In strain CN13R, XopJ6CN13 also contributes to pathogenicity but not to bacterial multiplication in planta (Figures 5A, 5B, and S6A).
Figure 5.
XopJ6 is a virulence determinant for Xcc and contributes to PTI attenuation
(A) Pictures representative of the median disease index observed 10 days after wound inoculation of Arabidopsis Col-0 accession with bacterial suspensions at 108 CFU/mL. Strains are described in panel B.
(B) Bacterial populations at 0 dpi (in gray, at least 22 values per strain) and 3 dpi (in black, 48 values per strain) after infiltration of Arabidopsis Col-0 accession with Xcc CN06R, CN03R and CN13R rifampicin-resistant strains, corresponding xopJ6 deletion mutants (ΔJ6) and for CN06, the complemented strain (CN06RΔJ6:1301-J6) with bacterial suspensions at 105 CFU/mL. At least three independent experiments were performed and combined. Statistical groups were determined using the nonparametric Kruskal-Wallis test (p value < 0.01) and are indicated by different letters.
(C) Composite image showing that both XopJ6CN06 and XopJ6CN13 delivered by Pf0-1 allow for the subsequent HopQ1-mediated cell death. in N. benthamiana. Suspensions of Pf0-1 wild-type strain or Pf0-1-expressing XopJ6CN06-3HA, XopJ6C409A-3HA or XopJ6CN13-3HA were infiltrated into N. benthamiana leaves (white dotted line). After 7 h, a suspension of P. syringae DC3000 was infiltrated in an overlapping area (red dotted line). Values in brackets indicate the frequency of tissue collapse at 48 h post-inoculation in overlapping areas of leaves from three independent experiments (8 leaves per replicate).
(D) ROS-burst assays in response to flg22 treatment in N. benthamiana leaves transiently expressing XopJ6 variants (XopJ6CN06-3HA, XopJ6C409A-3HA or XopJ6CN13-3HA). For each condition, ROS production was normalized to the total luminescence produced in the second half of the same leaf expressing the GFP-3HA protein and treated with 100 nM flg22. Three independent experiments were performed and combined (mean ± SD; n = 16 leaf discs per condition in a given experiment). Letters a and b denote statistical significance (p value <0.05, one-way ANOVA with Bonferroni post-test). See also Figures S6B and S6C.
We then tested whether XopJ6CN06 and XopJ6CN13 were both able to suppress basal immune response. In a PTI suppression assay in N. benthamiana, we tested if a non-pathogenic bacterium delivering a putative PTI-suppressing effector can interfere with effector-triggered cell death caused by an avirulent bacterial pathogen such as Pseudomonas syringae pv. tomato DC3000.60 If the non-pathogenic bacterium does not express a PTI-suppressing effector, Pst-mediated cell death is compromised by the strong PTI response. In the case of PTI suppression triggered by a type III effector protein, Pst-mediated cell death should be observed. XopJ6CN06, catalytically inactive XopJ6C409A, and XopJ6CN13 were delivered as 3HA-tagged proteins in plant cells by the non-pathogenic Pf0-1 strain. Infiltration with DC3000 triggered a HopQ1-mediated cell death response only with prior delivery of either XopJ6CN06 or XopJ6CN13, but not with XopJ6C409A, suggesting that XopJ6 acetyltransferase activity contributes to PTI suppression (Figures 5C and S6B). This result was further confirmed with the ability of active XopJ6CN06 and XopJ6CN13, but not XopJ6C409A, to alter flg22-induced ROS production in N. benthamiana (Figures 5D and S6C). Together, these observations indicate that, like XopJ6CN06, XopJ6CN13, although unable to interact with WRKY TFs, contributes to suppression of ROS production, suggesting that this natural XopJ6 variant would exert its virulence functions without interfering with the defensive WRKY TFs.
CNV of xopJ6 modulates Xcc pathogenicity in Arabidopsis
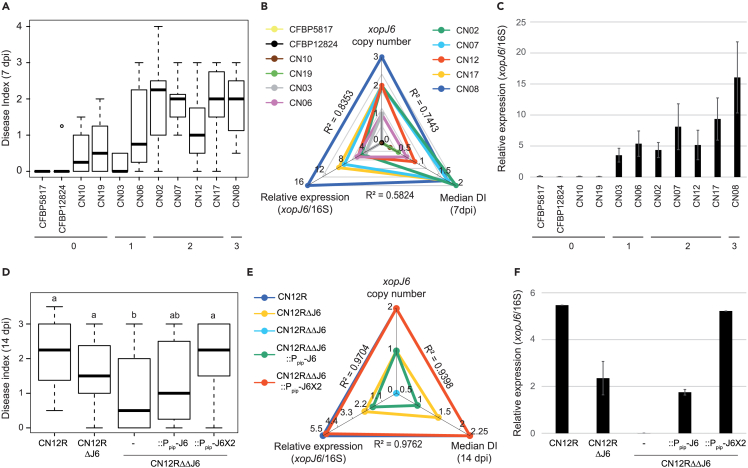
The presence of multiple identical copies of xopJ6 in wild Xcc strains was intriguing (Figures 1A and S1A) and prompted us to check whether CNV of xopJ6 could have a quantitative contribution to bacterial pathogenicity. We tested whether the number of xopJ6 copies correlates with the pathogenicity of natural Xcc strains virulent on Col-0, i.e., not expressing xopAC. Strain CN18 was excluded from this analysis due to ambiguous xopJ6 copy numbers inferred from genomic sequence data and Southern blot analyses (Figure S1A). At seven days post-inoculation, Xcc aggressiveness was positively correlated with xopJ6 copy number (R2 = 0.74, Figures 6A and 6B). xopJ6 expression level determined in vitro in hrp-inducing medium was also positively correlated with xopJ6 copy number (R2 = 0.83, Figures 6B, 6C, and S7). Together, these data suggest that CNV in xopJ6 could modulate Xcc pathogenicity on Arabidopsis by tuning xopJ6 expression levels.
Figure 6.
Copy number variation of xopJ6 modulates xopJ6 expression level and pathogen virulence on Col-0
(A and D) Disease index observed after wound inoculation of Col-0 leaves with Xcc strains at 108 CFU/mL (A) DI at 7 dpi for different wild-type Xcc strains that do not contain a xopAC allele able to confer avirulence on Col-0 and excluding CN18 for which the number of xopJ6 copies is uncertain (from Figure S1A). (D) DI at 14 dpi for Xcc CN12R rifampicin-resistant wild-type strain, deleted from one or two xopJ6 copies and complemented with one or two xopJ6 copies under its native promoter (Ppip). At least 3 independent experiments were performed (4 plants, 4 leaves per replicate) and combined using the median DI for each plant. Statistical groups were determined using nonparametric Kruskal-Wallis test (p value < 0.01) and are indicated by different letters.
(B and E) Spider charts showing the correlation between the number of xopJ6 copies, the relative expression of xopJ6 and the median disease index in various Xcc strains (B) and in Xcc CN12R strains described in panel D (E). The correlation coefficients between each character are indicated.
(C and F) Relative expression of xopJ6 in various Xcc strains (C) and in Xcc CN12R strains described in panel D (F). RT-qPCR were performed on RNA samples extracted after growth in MME minimal medium. Relative expression was calculated as the ΔCp between xopJ6 and the reference gene 16S rRNA. Three independent experiments were performed and combined. Numbers indicated below the histograms A and C correspond to the number of xopJ6 copies. See also Figure S6D.
To investigate the contribution of xopJ6 CNV on Xcc pathogenicity, we engineered serial deletions of the two xopJ6 copies in strain CN12R (CN12RΔJ6 and CN12RΔΔJ6). The double xopJ6 deletion mutant CN12RΔΔJ6 was then used for genomic complementation with one or two copies of xopJ6 under the control of its native promoter (CN12RΔΔJ6:552-J6 and CN12RΔΔJ6:552-J6x2 strains). xopJ6 copy number, xopJ6 expression in hrp-inducing medium, and pathogenicity were again highly correlated with each other (0.94 < R2 values < 0.97, Figures 6D–6F). Bacterial multiplication of CN12RΔΔJ6 was restored by complementation with either one or two copies of xopJ6 while xopJ6 overexpression controlled by a tac promoter (CN12RΔΔJ6:1301-J6 strain) resulted in 3- to 4-fold overexpression of xopJ6 relative to wild-type CN12R strain and conferred significantly higher bacterial growth, demonstrating that increase in xopJ6 expression level promotes bacterial virulence (Figure S6D; Table S5). By contrast, overexpression of xopJ6CN13 in CN12RΔΔJ6 (CN12RΔΔJ6:1301-J6CN13 strain) only partially restored pathogenicity to wild-type levels (Figure S6D; Table S5), suggesting that XopJ6CN13 exhibits a lower virulence activity than XopJ6. Collectively, these results demonstrate that variations of xopJ6 copy number modulate the aggressiveness and multiplication of a Xcc strain and indicate that changes in copy number of effector genes are exploited by natural Xcc strains to fine-tune their virulence on host plants.
Discussion
This work reveals how distinct genetic variations within the xopJ6 effector gene of Xcc enable this bacterial pathogen to evade immune recognition and modulate its virulence on its hosts. XopJ6 represents an additional member of the YopJ family of acetyltransferases that are widely distributed in both animal and plant bacterial pathogens. xopJ6 confers avirulence in both Arabidopsis and cauliflower, and variation in xopJ6 copy number potentiates Xcc virulence in a gene dosage-dependent manner, demonstrating how CNV can contribute to bacterial pathogenicity.
xopJ6 is one of the few known avirulence genes of Xcc in Arabidopsis, along with xopAC and xopAM.33,41 Surprisingly, we could not identify strains harboring both ETI-inducing xopAC and xopJ6 genes (Figure 1A). This intriguing result might reflect the distribution pattern of the matching resistance genes in the host population for these Xcc strains, which were originally isolated from infected Brassica crop plants or seeds.33,61
XopJ6 recognition is mediated by the well-characterized RRS1-R/RPS4 NLR pair, consistent with XopJ6 being a close homolog of PopP2. This recognition likely depends on the acetylation of RRS1-R WRKY decoy domain, as reported for PopP2 recognition (Table S3).22 Because Xcc is a natural pathogen of Arabidopsis,62 this study possibly identifies a native RRS1-R/RPS4 immune function compared to the detection of R. pseudosolanacearum PopP2 and Pseudomonas syringae pv. pisi AvrRps420,25,27,63 which are likely not naturally occurring Arabidopsis pathogens. In addition, no avrRps4 homolog has been identified in Xcc or the Xanthomonas genus. However, the Xanthomonads-restricted T3E XopS from X. campestris pv. euvesicatoria was recently shown to interact with and stabilize the WRKY40 TF in pepper.64
XopJ6 recognition in cauliflower confers immunity against Xcc inside hydathodes and therefore represents the first Xcc avirulence determinant reported in a Brassica crop. Interestingly, XopJ6 recognition mechanisms likely differ between Arabidopsis and cauliflower: there are no Brassica oleracea RRS1-R/RPS4 orthologs coding for NLR receptors with an integrated WRKY domain in the available B. oleracea genome sequences. Such genes were only described in the Arabidopsis thaliana close relatives A. lyrata and Capsella rubella.58 Consistent with PopP2 not being recognized in the cauliflower commercial hybrid cv. Clovis (Figure S4E), the genetic characterization of xopJ6-mediated resistance in this cultivar should reveal a yet-unknown recognition mechanism.
Natural variation in xopJ6 reflects Xanthomonas adaptation to evade host recognition
Compelling evidences demonstrate the importance of natural variations (SNPs, insertions or deletions [indels], recombinations) in avirulence genes as a strategy to evade NLR-mediated recognition by the host immunity and gain host adaptation.33,65,66,67 Here, we identified a natural variant of XopJ6 that evades recognition in cauliflower and Arabidopsis thanks to a single N382K substitution (Figures 3D, 3E, S4C, and S4F). In Arabidopsis, this substitution abolishes RRS1-R/RPS4-mediated immunity by disrupting the interaction between XopJ6 and RRS1-R WRKY domain (Figure 4). In PopP2, the corresponding residue N296 is located in the substrate recognition α-helix (αD) making direct contact with RRS1-R WRKY domain.51 A triple PopP2E284A/D292A/N296A mutant containing substitutions in critical residues of this αD helix is unable to acetylate RRS1-R WRKY domain and consequently fails to elicit ETI in Ws-2 plants.51 Homology modeling of XopJ6N382K acetyltransferase domain based on the crystal structure of PopP2 predicts substantial steric crashes induced by the N382K substitution, preventing interaction with RRS1-R WRKY domain (Figures 4D and 4E). The natural selection of a XopJ6 variant capable of evading immune surveillance by RRS1-R/RPS4 illustrates the ongoing arms race between Xcc and its host plants. It would thus be interesting to mine the natural genetic diversity of Arabidopsis and cauliflower NLRs for extended recognition capabilities of the XopJ6CN13 variant. This would pave the way for engineering NLR variants with optimized integrated domains allowing a more efficient and broader sensing of pathogen effectors.
XopJ6 virulence functions go beyond the manipulation of WKRY TFs
The XopJ6CN13 variant, though unable to interact with WRKY TFs, retains some of its immune-suppressing function (Figure 5) while evading recognition by RRS1-R/RPS4 (Figures 3 and 4). This indicates that XopJ6 virulence functions are likely not restricted to the targeting of defensive WRKY TFs. We could not identify any natural PopP2 variants within the αD′ substrate recognition helix among the different publicly available R. pseudosolanacearum genomes, suggesting that targeting of defensive WRKY TFs by PopP2 represents a major virulence strategy which cannot be easily dispensed with by R. pseudosolanacearum. Besides WRKY TFs, PopP2 is known to interact with several other host targets including the cysteine protease RD1968 and Phytoalexin Deficient 4 (PAD4), an important immune regulator in basal and NLR-triggered immunity and whose acetylation by PopP2 affects plant immunity.69 Investigation of XopJ6CN06 and XopJ6CN13 proximal proteomes in Arabidopsis and in cauliflower could thus uncover key virulence targets relevant for Xcc pathogenicity and plant immunity.
Are xopJ6 and popP2 the result of convergent evolution?
Both PopP2 and XopJ6 proteins share sequence similarities with YopJ homologs from animal pathogens (Figure 2A). The evolutionary history of these two genes is unclear. xopJ6 and popP2 GC contents (53% and 60%, respectively) differ from the rest of Xcc and R. pseudosolanacearum genomes (65% and 67%, respectively) suggesting recent acquisitions of these genes by horizontal gene transfer (HGT).70 The presence of xopJ6 in a transposon could have facilitated such HGT. However, xopJ6 sequence homology to popP2 is mostly restricted to the acetyltransferase domain, indicating that both genes diverged significantly. Both genes may also have originated from two distinct yopJ homologs of bacterial pathogens and evolved acetylation of plant-specific WRKY TFs independently in Xanthomonas and Ralstonia.
Is CNV a common strategy to tune pathogenicity?
xopJ6 expression is tightly controlled by the master regulators HrpG and HrpX as expected for T3E genes (Figure S3A).71,72,73 The timely and coordinated expression of the T3 secretion machinery and its T3E genes is known to be critical for the outcome of the interaction with the hosts. Yet, little is known about the natural diversity of T3E gene expression levels and their biological importance in bacterial pathogens colonizing animal and plant hosts. Such diversity could result from promoter polymorphisms or CNV of strictly- or near-identical T3E genes as observed for xopJ6 in Xcc. Such CNV is very likely driven by the multiple linked insertions of a Tn3-like transposon carrying xopJ6 in Xcc chromosome and possibly by insertion in native plasmids whose copy numbers relative to the chromosome are unknown (Figures S3C and S3D).
Importantly, the number of xopJ6 copies does not only correlate with its expression levels but also determines Xcc pathogenicity on susceptible Arabidopsis accession (Figures 6 and S6D; Table S5). This remarkable correlation even holds on the collection of natural Xcc strains tested. Rare examples of duplicated T3E genes have been reported in pathogenic bacteria such as two identical copies of xopZ in X. oryzae pv. oryzae (Xoo) strain PXO99A as part of a large 212-kb genomic duplication.74 Yet, single xopZ mutants are not significantly impaired in pathogenicity on rice in contrast to a double mutant indicating that a single copy of xopZ would be sufficient to promote pathogenicity in the conditions tested. Modulation of pathogen virulence through CNV of effector genes has been reported in oomycetes and nematodes, although in both cases CNV does not refer to identical gene copies.75,76 Such scenarios rather refer to duplication events leading to later functional diversification so that each gene copy might serve slightly distinct functions. At this stage, it is difficult to determine whether modulation of T3E gene expression level reflects an evolutionary strategy commonly used by pathogenic bacteria to fine-tune their pathogenicity. Yet, CNV of T3E genes should now be easier to identify based on genomic sequences assembled from long-read technologies. In parallel, determining how CNVs of T3E genes are distributed among natural bacterial strains that differ in origin, host range, or aggressiveness should provide insights into how this source of genetic variation enables a pathogen to better adapt to its hosts.
Limitations of the study
Several questions remain unanswered in this study: How is the effector protein XopJ6 recognized in Brassica crops where the RRS1-R/RPS4 NLR pair is absent? What are the non-WRKY targets of XopJ6? Are CNV or gene dosage common strategies used by parasitic microbes of plants and animals to tune aggressiveness on their hosts? These questions will require further investigations.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GFP from mouse IgG1k (clones 7.1 and 13.1) | Roche | Cat#11814460001; RRID:AB_390913 |
| anti-HA-HRP, High affinity | Roche | Cat#12013819001; RRID:AB_390917 |
| acetylated-Lysine Mouse mAb (Ac-K-103) | Cell Signaling | Cat#9681; RRID:AB_331799 |
| anti-His6-HRP | Roche | Cat#11965085001; RRID:AB_514487 |
| goat anti-mouse IgG(H + L)-HRP | Bio-Rad | Cat#170-6516; RRID:AB_11125547 |
| goat anti mouse IgG2a-HRP | Bio-Rad | Cat#STAR133P; RRID:AB_1102655 |
| c-Myc Monoclonal Antibody (9E10) | Thermo Fisher Scientific | Cat#MA1-980; RRID:AB_558470 |
| Chemicals, peptides | ||
| Glutathione Sepharose 4G | Sigma-Aldrich | Cat#GE17-0756-01 |
| GFP-Trap® Agarose | Chromotek | Cat#gta-20 |
| Protease inhibitor cocktail | Sigma-Aldrich | Cat#P9599 |
| Clarity™ Western ECL substrate | Bio-Rad | Cat#1705060 |
| PageRuler™ Prestained Protein Ladder, 10 to 180 kDa | Thermo Fisher Scientific | Cat#26617 |
| 7.5% Mini-PROTEAN® TGX™ Gels | Bio-Rad | Cat#4561026 |
| Gentamicin sulfate | Duchefa Biochemie | Cat#1405-41-0 |
| Carbenicillin disodium | Duchefa Biochemie | Cat#4800-94-6 |
| Spectinomycin pentahydrate | Duchefa Biochemie | Cat#22189-32-8 |
| Chloramphenicol | Duchefa Biochemie | Cat#56-75-7 |
| Kanamycine sulfate monohydrate | Duchefa Biochemie | Cat#25389-94-0 |
| Tetracyclin hydrochloride | Sigma-Aldrich | Cat#T7660 |
| Rifampicin | Sigma-Aldrich | Cat#R3501 |
| Pimaricin (DELVOCID® Instant) | DSM | Cat#00226 |
| 3′,5′-Dimethoxy-4′-hydroxyacetophenone (acetosyringone) | Sigma-Aldrich | Cat#D134406 |
| Tween® 20 | Sigma-Aldrich | Cat#P9416 |
| Tween® 80 | Sigma-Aldrich | Cat#P1754 |
| Sodium butyrate | Sigma-Aldrich | Cat#303410 |
| alpha32P-dCTP | Hartmann Analytic | Cat#FP-105 |
| Random Primer 6 | BioLabs | Cat#S1230S |
| Transcriptor Reverse Transcriptase | Roche | Cat#3531287001 |
| p-Nitrophenyl-β-D-glucuronide, PNPG | Sigma-Aldrich | Cat#N1627 |
| Lysozyme | Merck | Cat#1.05281 |
| Triton™ X-100 | Sigma-Aldrich | Cat#X100 |
| N-Lauroylsarcosine sodium salt | Sigma-Aldrich | Cat#L5125 |
| Wizard® SV Gel and PCR Clean-Up System | Promega | Cat#A9281 |
| Wizard® Plus SV Minipreps DNA Purification System | Promega | Cat#A1340 |
| Wizard® Genomic DNA Purification Kit | Promega | Cat#A1120 |
| RNeasy Mini Kit | QIAGEN | Cat#74106 |
| Turbo DNase | Invitrogen | Cat#AM1907 |
| Amersham Rediprime II Random DNA Labeling System | Cytiva | Cat#RPN1633 |
| IllustraTM ProbeQuantTM G-50 Micro Columns | Cytiva | Cat#28-9034-08 |
| Light Cycler 480 SYBR Green I Master | Roche Diagnostics | Cat#04887352001 |
| Critical commercial assays/components | ||
| Gateway™ BP Clonase™ II Enzyme mix | Thermo Fisher Scientific | Cat#11789020 |
| Gateway™ LR Clonase™ II Enzyme mix | Thermo Fisher Scientific | Cat#11791100 |
| Gateway™ pDONR™207 Vector | Invitrogen | N/A |
| Gateway™ pDONR™221 Vector | Invitrogen | N/A |
| PrimeStar® Max DNA polymerase | Takarabio | Cat#R045A |
| Phusion™ High-Fidelity DNA Polymerase | Thermo Fisher Scientific | Cat#F630 |
| GoTaq® DNA polymerase | Promega | Cat#M3001 |
| T4 DNA Ligase | Promega | Cat#M1801 |
| Taq DNA ligase | BioLabs | Cat#M0208 |
| T5 exonuclease | BioLabs | Cat#M0363 |
| Deposited data | ||
| Raw and deposited data (Mendeley) | This paper | https://doi.org/10.17632/k3gzy92c77.1 |
| MS-based proteomic data | This paper | PXD038104 (Pride) |
| Essential equipments | ||
| FLUOStar® Omega | BMG Labtech | N/A |
| Tissue Lyser MM 400 grinder | Retsch | Cat#20.745.0001 |
| ChemiDoc™ imaging system | Bio-Rad | N/A |
| Stratalinker® UV crosslinker | Stratagene | Cat#400075 |
| Molecular Dynamics Storm® 840 system | Molecular Dynamics | N/A |
| Storage Phosphor Screen | Molecular Dynamics | N/A |
| LightCycler® 480 II | Roche Diagnostics | N/A |
| NanoDrop™ Lite | Thermo Fischer Scientific | Cat#ND-LITE |
| Gene Pulser Xcell Electroporation Systems | Bio-Rad | Cat#1652660 |
| Experimental models: Organisms/strains | ||
| Bacterial and yeast strains used in this study, see Table S1. | ||
| Nicotiana benthamiana | N/A | |
| Arabidopsis thaliana Col-0 | N/A | |
| Arabidopsis thaliana Sf-2 | N/A | |
| Arabidopsis thaliana Ws-2 | N/A | |
| Arabidopsis thaliana rps4-21 (Ws-2) | N/A | |
| Arabidopsis thaliana rrs1-1 (Ws-2) | N/A | |
| Arabidopsis thaliana eds1-1 (Ws-2) | N/A | |
| Arabidopsis thaliana rps4-21 rrs1-1 (Ws-2) | N/A | |
| Capsicum annuum cv Early Calwonder (ECW) | N/A | |
| Capsicum annuum cv ECW-10R | N/A | |
| Brassica oleracea var Botrytis cv Clovis F1 | Vilmorin | Cat#351496 |
| Recombinant DNA | ||
| Plasmids used in this study are listed in Table S1 | ||
| Oligonucleotides | ||
| Primers used in this study are listed in Table S6 | ||
| Softwares | ||
| R | On-line | https://www.r-project.org/ |
| TAU_POGRAPHY-FLIM software v. 3.1 | On-line | https://trigenotoul.com/ |
| Mascot (version 2.7.0.1, Matrix Science) | Matrix Science Inc. | http://www.matrixscience.com/ |
| Proline | On-line | http://proline.profiproteomics.fr/#downloads. |
| Geneious 11.1.4 | Dotmatics | https://www.geneious.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents used in this study should be directed to and will be fulfilled by the lead contact, Laurent Deslandes (laurent.deslandes@inrae.fr).
Material availability
This study did not generate any unique reagent.
Data and code availability
-
•
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
-
•
Unprocessed photos of immunoblots, southern blots and HR presented in this study have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
MS-based proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository77 with the dataset identifier PXD038104.
Experimental models and study participant details
Bacterial strains and growth conditions
The Xanthomonas campestris pv. campestris (Xcc), Escherichia coli, Pseudomonas fluorescens (Pf0-1) and Agrobacterium tumefaciens strains and plasmids used in this study are listed in Table S1.
Xcc cells were grown on MOKA rich medium (Yeast Extract 4 g/L, Casamino acids 8 g/L, K2HPO4 2 g/L, MgSO4.7H2O 0.3 g/L)78 or MME minimal medium (K2HPO4 10.5 g/L, 4.5 g/L, (NH4)2SO4 1 g/L, Casamino acids 0.15%, MgSO4 1mM)79 at 28°C. Antibiotics were used at the concentration of 50 μg/mL for rifampicin and kanamycin, and 5 μg/mL for gentamicin and tetracycline. Pimaricin (30 μg/mL) was added in plates used for internal growth curves. Spontaneous rifampicin-resistant Xcc strains were selected by plating on MOKA rifampicin plates 10 mL of an overnight culture in MOKA. After three days at 28°C, two individual clones were isolated.
Pf0-1 strains were grown at 28°C on King’s B medium (Peptone 20 g/L, Glycerol 10 g/L, K2HPO4 1.5 g/L, MgSO4.7H2O 1.5 g/L, pH7.2). Antibiotics were used at the concentration of 30 μg/mL for chloramphenicol, 5 μg/mL for tetracycline and 15 μg/mL for gentamicin.
A. tumefaciens strains were grown in liquid YEB medium (yeast extract 10 g/L, peptone 10 g/L, NaCl 5 g/L, pH7.0) supplemented with adequate antibiotics for 16 h at 28°C under shaking.
E. coli strains were grown on Luria-Bertani rich medium Peptone 10 g/L, Yeast Extract 5 g/L, NaCl 10 g/L),80 at 37°C. Antibiotics were used at the concentrations of 50 μg/mL for ampicillin and kanamycin, 5 μg/mL for gentamicin and tetracycline and 40 μg/mL for spectinomycin.
Plant material and growth conditions
Arabidopsis thaliana Sf-2, Col-0 and Ws-2 plants, and Ws-2 null mutants (rps4-21, rrs1-1, rps4-21 rrs1-1 and eds1-1) were grown in short days (8-h light) at 22°C (60% relative humidity, 125 μE m−2.s−1 fluorescent illumination) for four weeks.
Nicotiana benthamiana plants were sown on soil and grown at 24°C under long day photoperiod (16-h light) with 60% relative humidity. Leaves of 4–5 weeks-old plants were used for Agrobacterium tumefaciens-mediated transient expression for immunoblotting, co-localization studies or FRET-FLIM assays.
Cauliflower (Brassica oleracea var. botrytis cv. Clovis, Vilmorin) plants were grown for four weeks in a greenhouse (14-h light, 24°C–28°C during the day, 22°C–26 °C at night).
Methods details
Construction of Xanthomonas campestris pv. campestris deletion mutants
xopJ6 deletion mutants were constructed using the sacB system.81 Amplicons of ∼500 bp located upstream and downstream of the region to delete were obtained by PCR using specific oligonucleotides (Table S6, LN809-812) and cloned into the MCS of pΔ13, a goldengate compatible derivative of pK18mobsacB plasmid.33 The sequence of all cloned amplicons was verified by Sanger sequencing.
Plasmids from E. coli donor strains were introduced in Xcc recipient starins by triparental mating as described82 using pRK2073 as a helper plasmid. Km-resistant, Rif-resistant and sucrose-susceptible transconjugants were cultured overnight in liquid MOKA-Rif and plated on MOKA-Rif plates containing 10% sucrose. Deletion events were screened in sucrose-resistant, Rif-resistant and Km-susceptible clones by colony-PCR using LN850 and LN813 oligonucleotides (Table S6). For each deletion, two individual positive clones were selected and isolated.
Complementation plasmids
The gene corresponding to xopJ6 with 5′ and 3′ UTR (330 bp before the start codon and 109 bp after the stop codon) was amplified by PCR using the oligonucleotides xopJ6a.m.-Eco and xopJ6AV-Hind (Table S6) and Xcc CN06 genomic DNA as template. PCR amplicon was cloned into the multiple cloning site (MCS) of the pLAFR6 plasmid linearized with EcoRI and HindIII restriction enzymes.83
The 3myc-tagged version of xopJ6 with its 5′ UTR was amplified using the oligonucleotides LN850 and LN851 (Table S6) and cloned into the goldengate compatible pBRM-P plasmid84 (Table S1).
Plasmidic DNA was prepared using Wizard Plus SV Minipreps DNA Purification System (Promega) and sequenced using Eurofins Genomics facilities.
Plasmids were introduced in Xcc by triparental mating as described.82 Clones were tested by PCR for the presence of the plasmid using J6-seq2 and J6-seq3rev oligonucleotides (Table S6). For each complementation, two positive clones were selected and isolated.
Chromosomal integration of xopJ6 in Xcc 8004
Gene corresponding to xopJ6 with 5′ and 3′ UTR (330 bp before the start codon and 109 bp after stop codon) was amplified by PCR using xopJ6a.m.-Eco and xopJ6AV-Hind oligonucleotides (Table S6). WT xopJ6 and xopJ6CN13 genes were amplified from Xcc CN06 and Xcc CN13 genomic DNA, respectively. PCR amplicons were cloned into the MCS of the pBluescript KS (Stratagene) giving rise to pBS-xopJ6 and pBS-xopJ6CN13 plasmids respectively. The pBS-xopJ6C72G plasmid (transition T214G) was constructed by sub-cloning the EcoRI-NcoI fragment of the pBS-xopJ6CN13 plasmid into the pBS-xopJ6 plasmid. Similarly, the pBS-xopJ6N382K plasmid (transition C1146A) was constructed by sub-cloning the NcoI-HindIII fragment of pBS-xopJ6CN13 plasmid into the pBS-xopJ6 plasmid. The pBS-xopJ6C409A plasmid (transitions T1225G and G1226C) was constructed using DpnI-based mutagenesis as described85 with XopJ6-DpnI-fwd and XopJ6-DpnI-rev oligonucleotides (Table S6). After PCR amplification with Phusion High-Fidelity DNA Polymerase (Thermofisher) and pBS-xopJ6 plasmid as matrix (18 cycles 30 s 98°C, 30 s 60°C, 5 min 72°C), amplicon was digested with DpnI (Promega) for 1 h at 37°C and transformed by electroporation in E. coli TG1 competent cells.
Clones obtained were sequenced and EcoRI-HindIII fragments of pBS-xopJ6, pBS-xopJ6CN13, pBS-xopJ6C72G, pBS-xopJ6N382K, and pBS-xopJ6C409A plasmids were sub-cloned into the pK18mobsacB derivative pCZ1013 plasmid (Table S1) giving rise to pCZ1013-xopJ6, pCZ1013-xopJ6CN13, pCZ1013-xopJ6C72G, pCZ1013-xopJ6N382K, and pCZ1013-xopJ6C409A, respectively (Table S1).
Plasmids obtained were introduced in Xcc 8004 and 8004 ΔxopAC strains by triparental mating and genomic integration of xopJ6 was verified by PCR using J6-seq2 and J6-seq3-rev oligonucleotides (Table S6).
Chromosomal integration of xopJ6 in Xcc CN strains
Integration regions cloned into the pCZ1013 plasmid (XC_0133 to XC_0135) are not conserved in CN strains. We therefore used two other pK18mobsacB derivatives, pCZ1301 Corresponds to pK18-GI,86 and pEL552, both with integrative regions located between XC_3385 and XC_3386. To construct pEL552, oligonucleotides 1301_modif_NotI and 1301_modif_BamHI were hybridised to form an adapter that was ligated into pCZ1301 digested with BamHI and NotI, leading to the deletion of a 265 bp fragment that contain the tac promoter. xopJ6 and xopJ6CN13 were subcloned into pCZ1301 and pEL552 from pBS-xopJ6 and pBS-xopJ6CN13, respectively, using EcoRI and HindIII.
Two copies of xopJ6 were cloned by isothermal assembly87 in pEL552 digested by HindIII. The two xopJ6 copies were amplified with Phusion High-Fidelity DNA Polymerase (ThermoFisher) using J6_left_copy_AM_fw_552 and J6_left_copy_AV_rev oligonucleotides (from 330 bp before the start codon to 32 bp after the stop codon, Table S6) for the first copy of xopJ6 and J6_right_copy_AM_fw and J6_right_copy_AV_rev_552 oligonucleotides (from 75 bp before the start codon (contains the RBS site) to 32 bp after stop codon, Table S6) for the second copy of xopJ6 and with pBS-xopJ6 as template. PCR products were purified using Wizard SV Gel and PCR Clean-Up System (Promega) and cloned into pEL552 linearized by HindIII during 1 h at 50°C in presence of Phusion High-Fidelity DNA Polymerase (ThermoFisher), T5 exonuclease (BioLabs) and Taq DNA Ligase (BioLabs). Ligation products were transformed in E. coli TG1 and clones obtained were tested by colony PCR using J6-seq2 and J6-seq3-rev oligonucleotides to detect the presence of xopJ6 and J6-seq2 and J6-seq0-rev oligonucleotides (Table S6) to confirm the presence of 2 copies of xopJ6.
Plasmids were sequenced and then introduced in Xcc CN06R, and/or CN12R strains by triparental mating (same protocol as for deletion). Integration of xopJ6 was verified by PCR using J6-seq2 and J6-seq3-rev oligonucleotides to detect the presence of xopJ6 and J6-seq2 and J6-seq0-rev oligonucleotides (Table S6) to confirm the presence of two copies of xopJ6.
Plasmid constructions for Pf0-1 delivery
The different XopJ6 variants (full length and truncated clones) were amplified by PCR from Xcc CN06 strain genomic DNA using PrimeSTAR Max DNA polymerase (Takara Bio). The sequences corresponding to XopJ6-C72G and XopJ6-N382K were generated by two-step PCR. All PCR products flanked with attB1 and attB2 recombination sites were recombined in pENTR plasmid to generate the relevant pENTR clones. All DNA constructs were verified by Sanger sequencing. The inserts cloned in pENTR vectors were then recombined in pDEST vectors via LR reaction (Invitrogen). For in planta expression of epitope tagged proteins, the desired pENTRY plasmids were subjected to an LR reaction with (i) pAM-PAT-35S-GWY-3HA, or -CFP or (ii) pBIN-35S-GWY-3HA, -YFP or -CFP. Oligonucleotides used are listed in Table S6.
In vitro growth curves of Xcc
In vitro bacterial growth was analyzed for each Xcc strain derivative and compared to the corresponding wild-type (Figure S7). To this end, growth curves were generated using the FLUOStar Omega apparatus (BMG Labtech, Offenburg, Germany) with four independent replicates as described.88 For each strain, two independent overnight pre-cultures in MOKA rich medium were washed in MME minimal medium. Growth rates were measured using 96-well flat-bottom microtiter plates containing 200 μL of MOKA or MME inoculated at an optical density OD600nm of 0.15 with two technical replicates for each preculture. The microplates were shaken continuously at 700 rpm using the linear shaking mode and OD600nm was measured every 5 min for 24 h.
Pathogenicity assays on arabidopsis
Pathogenicity assays were conducted on four week old plants grown on Jiffy pots in a growth chamber at 22°C, with a 9-h light period and a light intensity of 192 μmol/m2/s.
Wound inoculations were performed on four fully extended leaves per plant, four plants per strain per replicate, by piercing the central vein three times (from the middle to the tip of the leaf) using a needle (1.2 × 38mm) dipped in a bacterial inoculum at 108 CFU/mL in 1 mM MgCl2.89 After inoculation, trays were covered with a lacerated transparent Saran film and moved in a growth chamber with 70% humidity, 22°C, 8-h light period. Disease development on leaves was scored at different days after inoculation using the following disease index score: 0: no symptom; 1: chlorosis at the inoculation point; 2: extended chlorosis; 3: necrosis; 4: leaf death.
Mesophyll infiltrations were performed on four fully extended leaves per plant, four plants per strain per replicate using 1 mL needleless syringes as described90 with a bacterial inoculum at 105 CFU/mL in 1 mM MgCl2. To determine in planta bacterial population densities during infection, we performed Internal Growth Curves (IGC).78 After inoculation, trays were covered with a lacerated transparent food conservation film and moved in a growth chamber with 70% humidity, 22°C, 8 h light period. Leaf discs were sampled at zero and three days after inoculation using a Biopsy punch (diameter 0.65 cm, surface area 0.33 cm2) and ground individually using a Tissue Lyser MM 400 grinder (Retsch), twice 30 s at a frequency of 30 Hz with two glass beads (diameter 4 mm) in 200 μL of sterile water. The homogenates were serially diluted in sterile water and 5 μL drops were spotted three times for dilutions from non-diluted to 10−1 for T0 and from 10−1 to 10−4 at 3dpi on MOKA plates supplemented with rifampicin and pimaricin. Plates were incubated at 28°C for 48h and colonies were enumerated in spots containing 1 to 30 colonies. Bacterial densities in leaves were calculated as log CFU/cm2.
Cauliflower infection assays
Pathogenicity assays were conducted on the second true leaf of four week old Brassica oleracea cv Botrytis var Clovis-F1 plants (Vilmorin) grown in a greenhouse.
Hypersensitive response assays were performed by mesophyll infiltration using 1 mL needleless syringe and a bacterial inoculum at 108 CFU/mL of 1 mM MgCl2. After inoculation, trays were moved in a growth chamber with 70% humidity, 22°C, 8-h light period. Collapse was observed 24 h later.
Bacterial populations in hydathodes were determined after dip-inoculation. For that purpose, the second leaf was dipped for 20 s in a bacterial solution at 108 CFU/mL containing 0.5% Tween 80 in 1 mM MgCl2 as described.86,91 After six days, hydathodes were sampled using a Biopsy punch (diameter, 1.5 mm; surface area, 1.77 mm2), individually ground in 1.2 mL deepwell plates containing 5–6 glass beads (2 mm diameter) and 200 μL of sterile water. Bacterial solutions were plated as described above except that dilutions from non-diluted to 10−5 were spotted on plates.
Pseudomonas fluorescens-mediated delivery of effector proteins in Arabidopsis
For Pf0-1-mediated delivery, PopP2 and XopJ6 variant sequences were introduced by LR recombination in pBBR1-AvrRps4prom-GWY-3HA destination vector that allows the expression, under the control of AvrRps4 promoter sequence (504 bp of promoter sequence PCR-amplified from avrRps4Pip151 genomic clone), of proteins tagged in the C-terminus end with 3-HA epitope tag.22 For hypersensitive response assay, freshly grown Pf0-1 strains on King’s B agar plates containing appropriate antibiotics (tetracycline 5 μg/mL, chloramphenicol 30 μg/mL, and gentamicin 15 μg/mL) were harvested in 10 mM MgCl2 and washed two times in the same buffer. The final bacterial density was adjusted to OD600nm = 0.2. Four week old plants were placed in a humid environment 14 h before infiltration to facilitate inoculation. Leaves were hand-infiltrated with 1 mL needleless syringes and kept in a growth chamber with 70% humidity, 22°C, 8-h light period for 24–48 h. Arabidopsis protein samples were prepared from Ws-2 leaves, 7 h after infiltration with Pf0-1. For each sample, eight leaf discs (7 mm2) were ground in liquid nitrogen and total proteins were extracted using the procedure described for proteins transiently expressed in N. benthamiana and analyzed by western-blot.
PTI suppression assays
For suppression of PTI elicited by Pf0-1 in N. benthamiana, bacterial suspensions of Pf0-1 transformed with the different pBBR-derived constructs at OD600nm = 0.2 in 10 mM MgCl2 were infiltrated into fully expanded leaves of N. benthamiana. PTI was allowed to develop during 7 h, after which a P. syringae pv. tomato DC3000 suspension at OD600nm = 0.02 was infiltrated into an overlapping area. Tissue collapse in the overlapping area was evaluated two days after infiltration. To confirm the expression of 3HA-tagged effector proteins delivered by Pf0-1, four leaf discs of N. benthamiana were harvested 7 h after infiltration of a bacterial suspensions (OD600nm = 0.2 in 10 mM MgCl2) and proteins were extracted and analyzed by western-blot.
ROS burst assays
ROS production was quantified as described.92 Four weeks-old N. benthamiana leaves were infiltrated with A. tumefaciens carrying pBIN plasmids with corresponding genes (Table S1). XopJ6 variants (XopJ6CN06-3HA, XopJ6C409A-3HA or XopJ6CN13-3HA) were transiently expressed in one-half of an N. benthamiana leaf while the second half was expressing GFP-3HA as an internal control. 24 h post-infiltration, 16 leaf discs of 4 mm2 were harvested for each condition and individually incubated overnight in a 96-well plate (OpiPlate-96, PerkinElmer) containing 100 μL of millipure water. For ROS production measurements, water was replaced by an elicitor mix (50 nM flg22, 100 μM Luminol, 20 μg/mL HorseRadish Peroxidase) and relative luminescence was quantified in each well every 60 s during 1 h, using a Pekin Elmer luminometer. For each condition, ROS production was normalized to the total luminescence produced by the GFP-3HA control expressed in the same leaf (flg22-treated).
Southern blot analysis
Genomic DNA was extracted using the Wizard Genomic DNA Purification Kit (Promega) following the supplier’s instructions for Gram-negative bacteria from the pellet of 2 mL of overnight grown bacteria in MOKA rich medium previously washed in water. Six μg of genomic DNA digested overnight at 37°C with BamHI and HindIII were fractionated on a 1% agarose gel in TAE 1X (40 mM Tris-acetate, 1 mM EDTA, pH 8.3) during 15 h at 20 V. After denaturation treatment in the gel (10 min incubation in 0.25 N HCl, 30 min in 0.5 M NaOH −1.5 M NaCl and 30 min in 3 M NaAc pH 5.5), DNA was transferred to nylon membrane (Hybond N+) in SSC 20X buffer (0.3 M sodium citrate in 3M NaCl). After UV crosslink of DNA to the membrane (Stratalinker UV crosslinker, automatic settings), the membrane was pre-hybridized in Church buffer (1% BSA, 1 mM EDTA, 7% SDS, 0.25 M NaPi pH 7.2) 3h at 65°C. In parallel, an internal xopJ6 amplicon obtained by PCR using J6-seq2 and J6-seq3rev oligonucleotides (Table S6) and pBS-J6 as matrix (Table S1) was purified with the Wizard SV10 mM Gel and PCR Clean-Up System (Promega). 30 ng of the purified PCR product in 50 μL of 10 mM Tris pH8, 1 mM EDTA was labeled with alpha32 P-dCTP (Hartmann Analytic) using the Amersham Rediprime II Random DNA Labeling System (Cytiva) following supplier’s instructions. The probe was purified using Illustra ProbeQuant G-50 Micro Columns (Cytiva) and denatured by addition of 1 volume of 0.2 M NaOH. Then, 200 μL of the probe (specific to nucleotides 973 to 1537 of xopJ6) was added to the pre-hybridized membrane and incubated overnight at 65°C. The membrane was then washed three times in 0.1X SSC 0.1% SDS for 20 min at room temperature, dried, used to expose a Storage Phosphor Screen and imaged using a Molecular Dynamics Storm 840 system.
RNA extraction and real-time quantitative PCR
After an overnight preculture in MOKA rich medium, bacteria were centrifuged and washed in MME minimal medium. Three mL of MME medium were inoculated at 2.5 108 CFU/mL and after 6 h of growth at 28°C, 2 mL were centrifuged and the pellets were frozen and stored at −20°C. Total RNA were extracted using the RNeasy Mini Kit (Qiagen) with modification of the first step: frozen pellets were resuspended in 100 μL of 10 mM Tris pH8 - 1 mM EDTA containing lysozyme (few milligrams resuspended in 3 mL), mixed and incubated 5 min at room temperature. After addition of 350 μL of RLT buffer containing 1% β-mercaptoethanol, samples were passed at least 10 times through a 1 mL syringe with a needle (1.2 × 38mm) and centrifuged for 3 min at maximum speed. Next steps were performed following supplier’s instructions. Total RNA was quantified using a NanoDrop Lite (Thermo Fischer Scientific) and four μg were treated with Turbo DNase (Invitrogen) following supplier’s instructions. Absence of genomic DNA was verified by PCR using xopJ6 specific oligonucleotides (J6seq2 and J6seq3rev, Table S6). Total RNA was quantified using a NanoDrop Lite (Thermo Fischer Scientific) and 1 μg was reverse-transcribed using Transcriptor Reverse Transcriptase (Roche) and Random Primer 6 (BioLabs) for 30 min at 55°C.
Real-time quantitative PCR (RT-qPCR) was performed on a LightCycler 480 II machine (Roche Diagnostics), using Light Cycler 480 SYBR Green I Master (Roche Diagnostics) and 10-fold diluted cDNA. Oligonucleotides used for RT-qPCR are listed in Table S6. The crossing point (Cp) for each sample was calculated by the second derivative maximum using the LightCycler 480 software version SW 1.5.1.62 (Roche Diagnostics). Relative expression was calculated as the ΔCp between xopJ6 (Xcc_CN06-G2_01.2135 in CN06 Xcc strain) and the reference gene 16S rRNA (XC_4393).
Agrobacterium-mediated transient expression assays in N. benthamiana
For transient expression in N. benthamiana leaf epidermal cells, A. tumefaciens strains were grown in liquid YEB medium containing appropriate antibiotics for 16 h at 28°C under shaking. Cells were harvested by centrifugation and resuspended in infiltration medium (10 mM MES pH5.6, 10 mM MgCl2, 150 μM acetosyringone) at OD600nm = 0.25. For co-expression, bacterial suspension carrying individual constructs were mixed in a 1:1 ratio (Total OD600nm = 0.5). After incubation at room temperature for 1 h, bacteria were infiltrated into the leaves of four week old N. benthamiana plants using a needleless syringe. Plants were incubated for 36–48 h in growth chambers under controlled conditions before harvesting leaf disc samples.
Protein extraction, immunoprecipitation and immunoblotting
Xcc protein samples were prepared from bacterial cells grown for 6 h in MOKA or MME media inoculated at 1.25 108 CFU/mL or 2.5 108 CFU/mL, respectively. Pelleted bacteria from 1.4 mL were resuspended in 60 μL of 2x Laemmli buffer 0.125M Tris pH 6.8, 4% SDS, 20% glycerol, 0.2 M DTT,93 and after 10 min denaturation at 95°C, 15 μL of total bacterial cell extracts were loaded on 7.5% Mini-PROTEAN TGX Gels (Bio-Rad). Quality of the protein electrophoresis was visualised using the stain-free option of the ChemiDoc imaging system (Bio-Rad).
N. benthamiana protein samples were prepared from leaf discs harvested 48 h after Agrobacterium-mediated transformation and homogenised in 1 mL of ice-cold IP Buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 10 mM EDTA, 2 mM DTT, 10 mM sodium butyrate, 1x Plant protease cocktail inhibitor (SIGMA), 0.1% Triton). The extract was centrifuged at 13000 g for 2 min at 4°C and 50 μL of the supernatant (crude extract) was denatured for 3 min at 95°C in 50 μL of Laemmli buffer (2X). For immunoprecipitation of GFP-tagged proteins, the remaining supernatant was transferred in a new tube and incubated with 8 μL of GFP-trap agarose beads (Chromotek) for 1 h at 4°C under stirring. Beads were washed twice with 800 μL of IP2 buffer and subsequently denatured for 3 min at 95°C in 50 μL of Laemmli buffer (2X) before analysing the IPs by immunoblotting (SDS-PAGE). Transferred proteins were visualised by Ponceau S red staining. Membranes were blocked in a 2% milk TBST (Tris Buffer Saline-Tween-20; 50 mM Tris-HCl pH7.5, 150 mM NaCl, and 0.2% Tween 20) solution before immunodetection.
The following primary antibodies were used in this study: monoclonal c-myc antibody (9E10, Thermofisher Scientific; dilution 1:10000), anti-Acetylated Lysine (Ac-K-103, Cell Signaling Technology; dilution 1:2000), anti-HA-HRP (3F10; Roche; dilution 1:5000), anti-GFP (mouse monoclonal; Roche; dilution 1:3000), anti-His6-HRP (mouse monoclonal; Roche; dilution 1:25000). The appropriate horseradish peroxidase-conjugated secondary antibody was applied to the membranes: goat anti-mouse IgG-HRP (Bio-Rad; dilution 1:10000) for detection of anti-GFP; goat anti-mouse IgG2a-HRP (Bio-Rad; dilution 1:5000 for detection of anti-Ac-K antibody). Immunodetections were performed using Clarity Western ECL substrate reagent (Bio-Rad).
Bacterial acetylation assay
Single colonies of Rosetta (DE3) E. coli cells (Novagen) co-transformed with derivatives of the pCDF-GST-gene-His6 and pETDuet-1-gene destination vectors were grown during overnight and then diluted (1/20) in 50 mL of LB medium supplemented with carbenicillin (50 μg/mL), spectinomycin (50 μg/mL) and chloramphenicol (30 μg/mL) at 37°C. At an OD600nm of 0.6, protein expression was induced with 250 μM IPTG (isopropyl-β-thiogalactopyranoside) during 3 h at 28°C. Pelleted cells were concentrated 10 times in PBS (phosphate-buffered saline, pH 8.0) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma Aldrich), 10 mM Sodium butyrate, 0.1% Triton, 1 mM EDTA, and 2.5 mM DTT) and lysed using a French press. After centrifugation, supernatants were incubated with 30 μL of pre-equilibrated Glutathione Sepharose 4B (GE Healthcare) for 2 h at 4°C with rotation. Sepharose beads were recovered by centrifugation and then washed three times with buffer for 10 min at 4°C with rotation. Purified GST-tagged proteins were denatured in 100 μL of Laemmli buffer (2X) for 3 min at 95°C. Proteins samples were separated by SDS-PAGE (Bio-Rad) and analyzed by immunoblot.
Fluorescence microscopy
Colocalisation assays were performed as described.20 Briefly, A. tumefaciens strains carrying CFP- and YFP-tagged constructs were co-infiltrated in the leaves of four weeks-old N. benthamiana plants. Leaves were sampled for imaging between 36 and 48 h post-infiltration. CFP and YFP fluorescence was observed by laser confocal microscopy (TCS SP2-AOBS and TCS SP8-AOBS, Leica, Germany) using a water-immersed objective (HC PL APO 63x/1.20 W or HC FLUOTAR 25x/0.95 W VISIR). A laser excited CFP fluorescence at λ = 458 nm and detected in the 470–510 nm range. YFP was excited at λ = 514 nm and detected in 525–585 nm range. Images were acquired sequentially with the software Leica LCS version 2.61 and LAS X version 3.57.
FRET-FLIM data acquisition
Fluorescence lifetime measurements were performed in time domain using a streak camera. The light source is a 439 nm pulsed laser diode (PLP-10, Hamamatsu, Japan) delivering ultrafast picosecond pulses of light. All images were acquired with a 60x oil immersion lens (plan APO 1.4 N.A., IR) mounted on an inverted microscope (Eclipse TE2000E, Nikon, Japan). The fluorescence emission is directed back into the detection unit through a short pass filter and a band-pass filter (483/32 nm). The detector is a streak camera (Streakscope C10627, Hamamatsu Photonics, Japan) coupled to a fast and high-sensitivity CCD camera (model C8800-53C, Hamamatsu). For each acquisition, average fluorescence decay profiles were plotted and lifetimes were estimated by fitting data with exponential function using a non-linear least-squares estimation procedure.94 Fluorescence lifetime of the donor (CFP) was experimentally measured in the presence and absence of the acceptor (YFP). FRET efficiency (E) was calculated by comparing the lifetime of the donor in the presence (τDA) or absence (τD) of the acceptor: E = 1-(τDA)/(τD). Statistical comparisons between control (donor) and assay (donor + acceptor) lifetime values were performed by Student’s t test.
Yeast two-hybrid
Constructs for yeast two-hybrid analysis (PopP2, XopJ6, XopJ6224-565, XopJ6-N382K224-565 and RRS1-RCterm) were prepared in the MatchMaker GAL4 two-hybrid system (Clontech) using the pGBG-GWY (BD fusion) and pGAD-GWY (AD fusion) vectors to express bait and prey proteins, respectively. Co-expression of BD-p53 and AD-T (Clontech) was used as positive control on selective medium. Bait and prey proteins were tagged with a c-Myc and an HA epitope tag, respectively. Bait and prey constructs were introduced in AH109 Saccharomyces cerevisae strain (Clontech) according to the manufacturer’s instructions. Transformed cells were plated on SD-TL medium and incubated 3 days at 30°C. Colonies were then dropped by dilution gradient on different media: non-selective (-TL) and selective (-TLH/-TLHA) and incubated 3 days at 30°C. Protein expression in yeast was evaluated following incubation of cultures under shaking at 30°C overnight in SD-TL media. Cultures were processed for protein extraction according to a lithium acetate (Liac)/NaOH pre-treatment protocol as previously described (Zhang et al., 2011). Samples were denatured 3 min at 95°C in Laemmli buffer (2X) before separation by SDS-PAGE and immunoblotting with anti-c-Myc-HRP and anti-HA-HRP conjugated antibodies.
Sample preparation for proteomic analysis
Recombinant GST-tagged proteins were expressed in Rosetta (DE3) E. coli cells (Novagen) co-transformed with pDUET- and pCDF-GST-based plasmids. Briefly, for each combination, bacteria were precultured in 10 mL liquid LB medium with appropriate antibiotics (carbenicillin (50 μg/mL) and spectinomycin (50 μg/mL)) at 37°C for 12 h under constant shaking (180 rpm). The precultures were diluted in 50 mL liquid LB medium (ration 1:25, with antibiotics) at 37°C under shaking (180 rpm). At OD600nm = 0.6, protein expression was induced with 0.2 mM IPTG for 4 h at 28°C under shaking. Cultures were centrifuged (6500 rpm, 20 min, 4°C) and pelleted cells were concentrated 10 times in PBS (phosphate-buffered saline, pH 8.0) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma Aldrich), 10 mM Sodium butyrate, 0.1% Triton, 1 mM EDTA, and 2.5 mM DTT) and lysed using a French press. After centrifugation, supernatants were incubated with 30 μL of pre-equilibrated Glutathione Sepharose 4B (GE Healthcare) for 2 h at 4°C with rotation. Sepharose beads were recovered by centrifugation and then washed three times with buffer for 10 min at 4°C with rotation. Pulled-down GST-tagged proteins were denaturated in 50 μL of of Laemmli buffer (2X) containing 25 mM dithiothreitol (DTT) at 95°C for 5 min, and cysteines were alkylated by the addition of 90 mM iodoacetamide for 30 min at room temperature. Protein samples were loaded onto a 10% SDS-polyacrylamide gel and subjected to short electrophoresis. After Quick Coomassie Blue (Generon) staining of the gel, gel slices were excised, washed twice with 50 mM ammonium bicarbonate-acetonitrile (1:1, v/v) and then washed once with acetonitrile. Proteins were in-gel digested by the addition of 60 μL of a solution of modified sequencing grade trypsin in 25 mM ammonium bicarbonate (20 ng/μL, sequence grade, Promega, Charbonnières, France). The mixture was incubated at 37°C overnight. The resulting peptides were extracted from the gel by one round of incubation (15 min, 37°C) in 1% formic acid–acetonitrile (40%) and two rounds of incubation (15 min each, 37°C) in 1% formic acid–acetonitrile (1:1). The extracted fractions were air-dried. Tryptic peptides were resuspended in 14 μL of 2% acetonitrile and 0.05% trifluoroacetic acid for further MS analysis.
Nano LC-MS/MS analysis
Peptides mixtures were analyzed by nano-LC-MS/MS using nanoRS UHPLC system (Dionex, Amsterdam, The Netherlands) coupled to a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Five microliters of each sample were loaded on a C18 pre-column (5 mm × 300 μm; Thermo Fisher) at 20 μL/min in 5% acetonitrile, 0.05% trifluoroacetic acid. After 5 min of desalting, the pre-column was switched on line with the analytical C18 column (15 cm × 75 μm; Reprosil C18 in-house packed) equilibrated in 95% of solvent A (5% acetonitrile +0.2% formic acid in water) and 5% of solvent B (80% acetonitrile +0.2% formic acid in water). Peptides were eluted using a 5–50% gradient of B during 105 min at a 300 nL/min flow rate. The Q-Exactive Plus was operated in data-dependent acquisition mode with the Xcalibur software. Survey scan MS spectra were acquired in the Orbitrap on the 350–1500 m/z range with the resolution set to a value of 70 000. The ten most intense ions per survey scan were selected for HCD fragmentation, and the resulting fragments were analyzed in the Orbitrap with the resolution set to a value of 17 500. Dynamic exclusion was used within 30s to prevent repetitive selection of the same peptide.
Bioinformatics analysis of nanoLC-MS/MS-based proteomic data
Acquired MS and MS/MS data were searched with Mascot (version 2.7.0, http://matrixscience.com) against a custom-made database containing all interest protein sequences and contaminants protein sequences.
The search included methionine oxydation, protein N-terminal threonine (T), serine (S) and lysine (K) acetylation, as variable modifications, and carbamidomethylation of cysteine as fixed modifications. Trypsin P was chosen as the enzyme and 3 missed cleavages were allowed. The mass tolerance was set to 10 ppm for the precursor ion and to 20 mmu for fragment ions. Raw MS signal extraction of identified peptides was performed across different samples. Validation of identifications was performed through a false-discovery rate set to 1% at protein and peptide-sequence match level, determined by target-decoy search using the in-house-developed software Proline software version 2.1 (http://proline.profiproteomics.fr/). Then, peptide identifications were grouped into proteins. Acetylated and no acetylated ratio of peptides containing the K319 residue of AtWRKY22 lying in the conserved WRKY heptad were calculated. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD038104.
Protein modeling
Homology models of XopJ6 were obtained on SWISS-MODEL, an automated web-based homology modeling server (http://swissmodel.expasy.org/workspace/). The original and mutated amino acid sequences of XopJ6 were uploaded to the server. The crystal structure of PopP2-IP6 (PDB code 5W3T) was selected as template.
Multiple sequence alignment and phylogeny reconstruction
The amino acid sequences of several members of the YopJ/HopZ/AvrRxv family were retrieved from the NCBI database. The GenBank accession numbers of the full-length proteins are as follows: AAG39033 for AvrXv4 (Xanthomonas euvesicatoria), CAF32331 for PopP1 (Ralstonia pseudosolanacearum), AAA27595 for AvrRxv (X. euvesicatoria), CAC16700 for HopZ2 (Pseudomonas syringae), AAD39255 for AvrBsT (X. euvesicatoria), ABM32744 for Aave2166 (Acidovorax citrulli), NP_443964 for NopJ (Sinorhizobium fredii), EKG29639 for HopZ4 (P. syringae), WP_011347382 for XopJ5 (X. campestris), ABM33278 for Aave2708 (A. citrulli), AAF63400 for ORFB (Erwinia amylovora), AAF71492 for HopZ3 (P. syringae), AAR02168 for HopZ1a (P. syringae), WP_004661226 for HopZ1b (P. syringae), AAL84243 for HopZ1c (P. syringae), CAF32358 for PopP3 (R. pseudosolanacearum), CAD13849 for RipAE (R. pseudosolanacearum), CAD15839 for RipJ (R. pseudosolanacearum), CAD14570 for PopP2 (R. pseudosolanacearum), XopJ6CN06 (X. campestris), AAT08443 for VopA (Vibrio parahaemolyticus), NP_710166 for AopP (Aeromonas salmonicida), AAL21745 for AvrA (Salmonella enterica), AKN09807 for YopJ (Yersinia pestis) and AAN37537 for YopP (Y. enterocolitica). The sequences were aligned using MAFFT version 7.017 implemented in Geneious version 7.1.9 (Biomatter Ltd.). The FFT-NS-ix1000 algorithm was used with the BLOSUM62 scoring matrix, a gap open penalty of 1.53 and an offset value of 0.123. This multiple sequence alignment was used to reconstruct the phylogenetic tree using MEGA X95 with the Neighbor-joining method calculating the genetic distances with the Jukes-Cantor substitution model and running a 1,000 replicates-bootstrap resampling test.
In silico characterization of transposon
The identification of transposon-like flanking inverted repeats was performed using xopJ6-containing genomic regions from available Xcc genomic sequences as query with the “Inverted Repeats Finder” algorithm version 2.2 and parameters by default https://tandem.bu.edu/cgi-bin/irdb/irdb.exe,.96
Quantification and statistical analysis
For FRET-FLIM analyses, significance of the difference between the donor lifetimes in the absence and in the presence of acceptor was assessed by a Student’s t test. Mass spectrometry based quantitative proteomics to assess acetylated residues on peptides parameters regarding precursor and fragment mass error tolerances were set respectively at 10 ppm and 0.6 Da for LTQ-Orbitrap Velos Pro data, and 10 and 20 ppm for Q-Exactive Plus data. The Proline software was used for the compilation, grouping, and filtering of the results (conservation of rank 1 peptides, peptide length ≥6 amino acids, peptide score ≥25, and minimum of one specific peptide per identified protein group) as described.97 Proline was then used for compilation, grouping and spectral counting-based comparison of the protein groups identified in the different samples. The acetylation sites were considered accurate when their localisation probability was above 75%.
For disease symptoms scoring and bacterial growth in planta, results were analyzed and plotted with R software. Boxplot representations are as follows: middle bar: median; box limit: upper and lower quartiles; extremes: minimum and maximum values. Dots indicate outliers. Statistical groups were determined using nonparametric Kruskal-Wallis or HSD tests and are indicated by different letters.
For RT-qPCR data, mean ΔCt were calculated from three technical replicates for two to three biological replicates (independently grown sets of bacteria). The mean relative expression from the biological replicates +/− SEM was represented in barplots. ΔCt values were used to assess the significance of the difference in ΔCt between the different genetic lines. A one-way ANOVA was applied when the assumptions about the underlying distribution of the data were fulfilled, or a kruskal-test when they were not.
Acknowledgments
J.S.L. and M.E. were funded by a PhD grant from the French Ministry of National Education and Research. M.G.-F.’s PhD grant was obtained from the French Laboratory of Excellence project (TULIP ANR-10-LABX-41; ANR-11-IDEX-0002-02) and INRAE. The authors benefited from the COST action FA1208 SUSTAIN and CA16107 EuroXanth. L.D. was supported by two research grants from the Agence Nationale de la Recherche (ANR15-CE20-0016-01and ANR-18-CE20-0015). L.D.N. was supported by a research grant from the Agence Nationale de la Recherche (ANR-18-CE20-0020-01). This work was supported by the French Laboratory of Excellence project ‘‘TULIP’’ (ANR-10-LABX-41; ANR-11-IDEX-0002-02), the "École Universitaire de Recherche (EUR)" TULIP-GS (ANR-18-EURE-0019), the Région Occitanie, European funds (Fonds Européens de DEveloppement Régional, FEDER), Toulouse Métropole, and the French Ministry of Research with the Investissement d'Avenir Infrastructures Nationales en Biologie et Santé program (ProFI, Proteomics French Infrastructure project, ANR-10-INBS-0008, and FBI, France BioImaging infrastructure project, ANR-10-INBS-0004). Z.-M.Z. was supported by the National Natural Science Foundation of China (32171184). We thank Baptiste Mayjonade for his help in genomic DNA preparation and Fabrice Roux for helpful discussions at the beginning of the project.
Author contributions
E.L., M.G.-F., M.E., C.V., J.S.L., C.Pouzet, A.J., C.G., C.Pichereaux, Z.-M.Z., S.C., and L.D. performed experiments. E.L., M.G.-F., L.D., and L.D.N. designed the experiments and wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published: February 15, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109224.
Contributor Information
Laurent Deslandes, Email: laurent.deslandes@inrae.fr.
Laurent D. Noël, Email: laurent.noel@inrae.fr.
Supplemental information
References
- 1.Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 2.Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Macho A.P., Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol. Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 4.Boller T., Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 5.Li B., Meng X., Shan L., He P. Transcriptional Regulation of Pattern-Triggered Immunity in Plants. Cell Host Microbe. 2016;19:641–650. doi: 10.1016/j.chom.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toruño T.Y., Stergiopoulos I., Coaker G. Plant-Pathogen Effectors: Cellular Probes Interfering with Plant Defenses in Spatial and Temporal Manners. Annu. Rev. Phytopathol. 2016;54:419–441. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 8.Yuan M., Ngou B.P.M., Ding P., Xin X.F. PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 2021;62:102030. doi: 10.1016/j.pbi.2021.102030. [DOI] [PubMed] [Google Scholar]
- 9.Ngou B.P.M., Ahn H.K., Ding P., Jones J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021;592:110–115. doi: 10.1038/s41586-021-03315-7. [DOI] [PubMed] [Google Scholar]
- 10.Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol. Plant Pathol. 2019;20:1163–1178. doi: 10.1111/mpp.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büttner D., Bonas U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 2010;34:107–133. doi: 10.1111/j.1574-6976.2009.00192.x. [DOI] [PubMed] [Google Scholar]
- 12.Ma K.W., Ma W. YopJ Family Effectors Promote Bacterial Infection through a Unique Acetyltransferase Activity. Microbiol. Mol. Biol. Rev. 2016;80:1011–1027. doi: 10.1128/MMBR.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orth K., Xu Z., Mudgett M.B., Bao Z.Q., Palmer L.E., Bliska J.B., Mangel W.F., Staskawicz B., Dixon J.E. Disruption of Signaling by Yersinia Effector YopJ, a Ubiquitin-Like Protein Protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 14.Jones R.M., Wu H., Wentworth C., Luo L., Collier-Hyams L., Neish A.S. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 15.Mukherjee S., Keitany G., Li Y., Wang Y., Ball H.L., Goldsmith E.J., Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 16.Trosky J.E., Liverman A.D.B., Orth K. Yersinia outer proteins: Yops. Cell Microbiol. 2008;10:557–565. doi: 10.1111/j.1462-5822.2007.01109.x. [DOI] [PubMed] [Google Scholar]
- 17.Cheong M.S., Kirik A., Kim J.G., Frame K., Kirik V., Mudgett M.B. AvrBsT acetylates Arabidopsis ACIP1, a protein that associates with microtubules and is required for immunity. PLoS Pathog. 2014;10:e1003952. doi: 10.1371/journal.ppat.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J., Manning A.J., Wolfgeher D., Jelenska J., Cavanaugh K.A., Xu H., Fernandez S.M., Michelmore R.W., Kron S.J., Greenberg J.T. Acetylation of an NB-LRR Plant Immune-Effector Complex Suppresses Immunity. Cell Rep. 2015;13:1670–1682. doi: 10.1016/j.celrep.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma K.W., Jiang S., Hawara E., Lee D., Pan S., Coaker G., Song J., Ma W. Two serine residues in Pseudomonas syringae effector HopZ1a Are required for acetyltransferase activity and association with the host Co-factor. New Phytol. 2015;208:1157–1168. doi: 10.1111/nph.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tasset C., Bernoux M., Jauneau A., Pouzet C., Brière C., Kieffer-Jacquinod S., Rivas S., Marco Y., Deslandes L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi S., Prokchorchik M., Lee H., Gupta R., Lee Y., Chung E.H., Cho B., Kim M.S., Kim S.T., Sohn K.H. Direct acetylation of a conserved threonine of RIN4 by the bacterial effector HopZ5 or AvrBsT activates RPM1-dependent immunity in Arabidopsis. Mol. Plant. 2021;14:1951–1960. doi: 10.1016/j.molp.2021.07.017. [DOI] [PubMed] [Google Scholar]
- 22.Le Roux C., Huet G., Jauneau A., Camborde L., Trémousaygue D., Kraut A., Zhou B., Levaillant M., Adachi H., Yoshioka H., et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 23.Sarris P.F., Duxbury Z., Huh S.U., Ma Y., Segonzac C., Sklenar J., Derbyshire P., Cevik V., Rallapalli G., Saucet S.B., et al. A Plant Immune Receptor Detects Pathogen Effectors that Target WRKY Transcription Factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 24.Griebel T., Maekawa T., Parker J.E. NOD-like receptor cooperativity in effector-triggered immunity. Trends Immunol. 2014;35:562–570. doi: 10.1016/j.it.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Hinsch M., Staskawicz B. Identification of a new Arabidopsis disease resistance locus, RPS4, and cloning of the corresponding avirulence gene, avrRps4, from Pseudomonas syringae pv. pisi. Mol. Plant Microbe Interact. 1996;9:55–61. doi: 10.1094/mpmi-9-0055. [DOI] [PubMed] [Google Scholar]
- 26.Gassmann W., Hinsch M.E., Staskawicz B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 27.Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deslandes L., Olivier J., Theulieres F., Hirsch J., Feng D.X., Bittner-Eddy P., Beynon J., Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc. Natl. Acad. Sci. USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- 30.Williams S.J., Sohn K.H., Wan L., Bernoux M., Sarris P.F., Segonzac C., Ve T., Ma Y., Saucet S.B., Ericsson D.J., et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- 31.Birker D., Heidrich K., Takahara H., Narusaka M., Deslandes L., Narusaka Y., Reymond M., Parker J.E., O'Connell R. A locus conferring resistance to Colletotrichum higginsianum is shared by four geographically distinct Arabidopsis accessions. Plant J. 2009;60:602–613. doi: 10.1111/j.1365-313X.2009.03984.x. [DOI] [PubMed] [Google Scholar]
- 32.Vicente J.G., Holub E.B. Xanthomonas campestris pv. campestris (cause of black rot of crucifers) in the genomic era is still a worldwide threat to brassica crops. Mol. Plant Pathol. 2013;14:2–18. doi: 10.1111/j.1364-3703.2012.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guy E., Genissel A., Hajri A., Chabannes M., David P., Carrère S., Lautier M., Roux B., Boureau T., Arlat M., et al. Natural Genetic Variation of Xanthomonas campestris pv. campestris Pathogenicity on Arabidopsis Revealed by Association and Reverse Genetics. mBio. 2013;4:e00538-12. doi: 10.1128/mBio.00538-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J.M. A Xanthomonas uridine 5'-monophosphate transferase inhibits plant immune kinases. Nature. 2012;485:114–118. doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 35.Kay S., Boch J., Bonas U. Characterization of AvrBs3-like effectors from a Brassicaceae pathogen reveals virulence and avirulence activities and a protein with a novel repeat architecture. Mol. Plant Microbe Interact. 2005;18:838–848. doi: 10.1094/MPMI-18-0838. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y., Li T., Xu T., Tang Z., Guo J., Cai Y. Multiple Xanthomonas campestris pv. campestris 8004 type III effectors inhibit immunity induced by flg22. Planta. 2020;252:88. doi: 10.1007/s00425-020-03484-1. [DOI] [PubMed] [Google Scholar]
- 37.Liu L., Wang Y., Cui F., Fang A., Wang S., Wang J., Wei C., Li S., Sun W. The type III effector AvrXccB in Xanthomonas campestris pv. campestris targets putative methyltransferases and suppresses innate immunity in Arabidopsis. Mol. Plant Pathol. 2017;18:768–782. doi: 10.1111/mpp.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan L., Rong W., Luo H., Chen Y., He C. The Xanthomonas campestris effector protein XopDXcc8004 triggers plant disease tolerance by targeting DELLA proteins. New Phytol. 2014;204:595–608. doi: 10.1111/nph.12918. [DOI] [PubMed] [Google Scholar]
- 39.Denancé N., Szurek B., Doyle E.L., Lauber E., Fontaine-Bodin L., Carrère S., Guy E., Hajri A., Cerutti A., Boureau T., et al. Two ancestral genes shaped the Xanthomonas campestris TAL effector gene repertoire. New Phytol. 2018;219:391–407. doi: 10.1111/nph.15148. [DOI] [PubMed] [Google Scholar]
- 40.Yan X., Tao J., Luo H.L., Tan L.T., Rong W., Li H.P., He C.Z. A type III effector XopLXcc8004 is vital for Xanthomonas campestris pathovar campestris to regulate plant immunity. Res. Microbiol. 2019;170:138–146. doi: 10.1016/j.resmic.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Xu R.Q., Blanvillain S., Feng J.X., Jiang B.L., Li X.Z., Wei H.Y., Kroj T., Lauber E., Roby D., Chen B., et al. AvrACXcc8004, a type III effector with a leucine-rich repeat domain from Xanthomonas campestris pathovar campestris confers avirulence in vascular tissues of Arabidopsis thaliana ecotype Col-0. J. Bacteriol. 2008;190:343–355. doi: 10.1128/JB.00978-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubrow Z.E., Carpenter S.C.D., Carter M.E., Grinage A., Gris C., Lauber E., Butchachas J., Jacobs J.M., Smart C.D., Tancos M.A., et al. Cruciferous weed isolates of Xanthomonas campestris yield insight into pathovar genomic relationships and genetic determinants of host- and tissue-specificity. Mol. Plant Microbe Interact. 2022;35:791–802. doi: 10.1094/MPMI-01-22-0024-R. [DOI] [PubMed] [Google Scholar]
- 43.Guy E., Lautier M., Chabannes M., Roux B., Lauber E., Arlat M., Noël L.D. xopAC-triggered Immunity against Xanthomonas Depends on Arabidopsis Receptor-Like Cytoplasmic Kinase Genes PBL2 and RIPK. PLoS One. 2013;8:e73469. doi: 10.1371/journal.pone.0073469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G., Roux B., Feng F., Guy E., Li L., Li N., Zhang X., Lautier M., Jardinaud M.F., Chabannes M., et al. The Decoy Substrate of a Pathogen Effector and a Pseudokinase Specify Pathogen-Induced Modified-Self Recognition and Immunity in Plants. Cell Host Microbe. 2015;18:285–295. doi: 10.1016/j.chom.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Bi G., Su M., Li N., Liang Y., Dang S., Xu J., Hu M., Wang J., Zou M., Deng Y., et al. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell. 2021;184:3528–3541.e12. doi: 10.1016/j.cell.2021.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Koebnik R., Krüger A., Thieme F., Urban A., Bonas U. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J. Bacteriol. 2006;188:7652–7660. doi: 10.1128/JB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wengelnik K., Van den Ackerveken G., Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol. Plant Microbe Interact. 1996;9:704–712. doi: 10.1094/mpmi-9-0704. [DOI] [PubMed] [Google Scholar]
- 48.Nicolas E., Lambin M., Dandoy D., Galloy C., Nguyen N., Oger C.A., Hallet B. The Tn3-family of Replicative Transposons. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0060-2014. [DOI] [PubMed] [Google Scholar]
- 49.Tansirichaiya S., Rahman M.A., Roberts A.P. The Transposon Registry. Mob. DNA. 2019;10:40. doi: 10.1186/s13100-019-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bellenot C., Carrère S., Gris C., Noël L.D., Arlat M. Genome Sequences of 17 Strains from Eight Races of Xanthomonas campestris pv. campestris. Microbiol. Resour. Announc. 2022;11:e0027922. doi: 10.1128/mra.00279-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z.M., Ma K.W., Gao L., Hu Z., Schwizer S., Ma W., Song J. Mechanism of host substrate acetylation by a YopJ family effector. Nat. Plants. 2017;3:17115. doi: 10.1038/nplants.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Z.M., Ma K.W., Yuan S., Luo Y., Jiang S., Hawara E., Pan S., Ma W., Song J. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat. Struct. Mol. Biol. 2016;23:847–852. doi: 10.1038/nsmb.3279. [DOI] [PubMed] [Google Scholar]
- 53.Debieu M., Huard-Chauveau C., Genissel A., Roux F., Roby D. Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol. Plant Pathol. 2016;17:510–520. doi: 10.1111/mpp.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sohn K.H., Segonzac C., Rallapalli G., Sarris P.F., Woo J.Y., Williams S.J., Newman T.E., Paek K.H., Kobe B., Jones J.D.G. The nuclear immune receptor RPS4 is required for RRS1SLH1-dependent constitutive defense activation in Arabidopsis thaliana. PLoS Genet. 2014;10:e1004655. doi: 10.1371/journal.pgen.1004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhattacharjee S., Halane M.K., Kim S.H., Gassmann W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science. 2011;334:1405–1408. doi: 10.1126/science.1211592. [DOI] [PubMed] [Google Scholar]
- 56.Huh S.U., Cevik V., Ding P., Duxbury Z., Ma Y., Tomlinson L., Sarris P.F., Jones J.D.G. Protein-protein interactions in the RPS4/RRS1 immune receptor complex. PLoS Pathog. 2017;13:e1006376. doi: 10.1371/journal.ppat.1006376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 58.Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015;6:6338. doi: 10.1038/ncomms7338. [DOI] [PubMed] [Google Scholar]
- 59.Rushton P.J., Somssich I.E., Ringler P., Shen Q.J. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 60.Badel J.L., Piquerez S.J.M., Greenshields D., Rallapalli G., Fabro G., Ishaque N., Jones J.D.G. In planta effector competition assays detect Hyaloperonospora arabidopsidis effectors that contribute to virulence and localize to different plant subcellular compartments. Mol. Plant Microbe Interact. 2013;26:745–757. doi: 10.1094/MPMI-06-12-0154-R. [DOI] [PubMed] [Google Scholar]
- 61.He Y.Q., Zhang L., Jiang B.L., Zhang Z.C., Xu R.Q., Tang D.J., Qin J., Jiang W., Zhang X., Liao J., et al. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biol. 2007;8:R218. doi: 10.1186/gb-2007-8-10-r218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kniskern J.M., Traw M.B., Bergelson J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol. Plant Microbe Interact. 2007;20:1512–1522. doi: 10.1094/MPMI-20-12-1512. [DOI] [PubMed] [Google Scholar]
- 63.Sohn K.H., Hughes R.K., Piquerez S.J., Jones J.D.G., Banfield M.J. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proc. Natl. Acad. Sci. USA. 2012;109:16371–16376. doi: 10.1073/pnas.1212332109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raffeiner M., Üstün S., Guerra T., Spinti D., Fitzner M., Sonnewald S., Baldermann S., Börnke F. The Xanthomonas type-III effector XopS stabilizes CaWRKY40a to regulate defense responses and stomatal immunity in pepper (Capsicum annuum) Plant Cell. 2022;34:1684–1708. doi: 10.1093/plcell/koac032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J., Chen L., Lu X., Peng Q., Zhang Y., Yang J., Zhang B.Y., Yang B., Waletich J.R., Yin W., et al. Natural allelic variations provide insights into host adaptation of Phytophthora avirulence effector PsAvr3c. New Phytol. 2019;221:1010–1022. doi: 10.1111/nph.15414. [DOI] [PubMed] [Google Scholar]
- 66.Herbers K., Conrads-Strauch J., Bonas U. Race-specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature. 1992;356:172–174. [Google Scholar]
- 67.Bentham A.R., Petit-Houdenot Y., Win J., Chuma I., Terauchi R., Banfield M.J., Kamoun S., Langner T. A single amino acid polymorphism in a conserved effector of the multihost blast fungus pathogen expands host-target binding spectrum. PLoS Pathog. 2021;17:e1009957. doi: 10.1371/journal.ppat.1009957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bernoux M., Timmers T., Jauneau A., Brière C., de Wit P.J.G.M., Marco Y., Deslandes L. RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell. 2008;20:2252–2264. doi: 10.1105/tpc.108.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huh S.U. PopP2 interacts with PAD4 in an acetyltransferase activity-dependent manner and affects plant immunity. Plant Signal. Behav. 2021;16:2017631. doi: 10.1080/15592324.2021.2017631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salanoubat M., Genin S., Artiguenave F., Gouzy J., Mangenot S., Arlat M., Billault A., Brottier P., Camus J.C., Cattolico L., et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 71.Luneau J.S., Cerutti A., Roux B., Carrère S., Jardinaud M.F., Gaillac A., Gris C., Lauber E., Berthomé R., Arlat M., et al. Xanthomonas transcriptome inside cauliflower hydathodes reveals bacterial virulence strategies and physiological adaptations at early infection stages. Mol. Plant Pathol. 2022;23:159–174. doi: 10.1111/mpp.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Noël L., Thieme F., Nennstiel D., Bonas U. cDNA-AFLP analysis unravels a genome-wide hrpG-regulon in the plant pathogen Xanthomonas campestris pv. vesicatoria. Mol. Microbiol. 2001;41:1271–1281. doi: 10.1046/j.1365-2958.2001.02567.x. [DOI] [PubMed] [Google Scholar]
- 73.Teper D., Pandey S.S., Wang N. The HrpG/HrpX Regulon of Xanthomonads-An Insight to the Complexity of Regulation of Virulence Traits in Phytopathogenic Bacteria. Microorganisms. 2021;9:187. doi: 10.3390/microorganisms9010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Song C., Yang B. Mutagenesis of 18 type III effectors reveals virulence function of XopZ(PXO99) in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 2010;23:893–902. doi: 10.1094/MPMI-23-7-0893. [DOI] [PubMed] [Google Scholar]
- 75.Castagnone-Sereno P., Mulet K., Danchin E.G.J., Koutsovoulos G.D., Karaulic M., Da Rocha M., Bailly-Bechet M., Pratx L., Perfus-Barbeoch L., Abad P. Gene copy number variations as signatures of adaptive evolution in the parthenogenetic, plant-parasitic nematode Meloidogyne incognita. Mol. Ecol. 2019;28:2559–2572. doi: 10.1111/mec.15095. [DOI] [PubMed] [Google Scholar]
- 76.Qutob D., Tedman-Jones J., Dong S., Kuflu K., Pham H., Wang Y., Dou D., Kale S.D., Arredondo F.D., Tyler B.M., Gijzen M. Copy number variation and transcriptional polymorphisms of Phytophthora sojae RXLR effector genes Avr1a and Avr3a. PLoS One. 2009;4:e5066. doi: 10.1371/journal.pone.0005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Blanvillain S., Meyer D., Boulanger A., Lautier M., Guynet C., Denancé N., Vasse J., Lauber E., Arlat M. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One. 2007;2:e224. doi: 10.1371/journal.pone.0000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arlat M., Gough C.L., Barber C.E., Boucher C., Daniels M.J. Xanthomonas campestris contains a cluster of hrp genes related to the larger hrp cluster of Pseudomonas solanacearum. Mol. Plant Microbe Interact. 1991;4:593–601. doi: 10.1094/mpmi-4-593. [DOI] [PubMed] [Google Scholar]
- 80.Miller J. Cold Spring Harbor Laboratory Press; 1992. A Short Course in Bacterial Genetics. [Google Scholar]
- 81.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 82.Turner P., Barber C., Daniels M. Behaviour of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 1984;195:101–107. doi: 10.1007/BF00332731. [DOI] [Google Scholar]
- 83.Bonas U., Stall R.E., Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. Mol. Gen. Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 84.Szczesny R., Jordan M., Schramm C., Schulz S., Cogez V., Bonas U., Büttner D. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol. 2010;187:983–1002. doi: 10.1111/j.1469-8137.2010.03312.x. [DOI] [PubMed] [Google Scholar]
- 85.Zheng L., Baumann U., Reymond J.L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cerutti A., Jauneau A., Auriac M.-C., Lauber E., Martinez Y., Chiarenza S., Leonhardt N., Berthomé R., Noël L.D. Immunity at Cauliflower Hydathodes Controls Systemic Infection by Xanthomonas campestris pv campestris. Plant Physiol. 2017;174:700–716. doi: 10.1104/pp.16.01852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 88.Boulanger A., Déjean G., Lautier M., Glories M., Zischek C., Arlat M., Lauber E. Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 2010;192:1487–1497. doi: 10.1128/JB.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Meyer D., Lauber E., Roby D., Arlat M., Kroj T. Optimization of pathogenicity assays to study the Arabidopsis thaliana-Xanthomonas campestris pv. campestris pathosystem. Mol. Plant Pathol. 2005;6:327–333. doi: 10.1111/j.1364-3703.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 90.Lummerzheim M., de Oliveira D., Castresana C., Miguens F.C., Louzada E., Roby D., Van Montagu M., Timmerman B. Identification of Compatible and Incompatible Interactions Between Arabidopsis thaliana and Xanthomonas campestris pv. campestris and Characterization of the Hypersensitive Response. Mol. Plant Microbe Interact. 1993;6:532–544. doi: 10.1094/MPMI-6-532. [DOI] [Google Scholar]
- 91.Luneau J.S., Baudin M., Quiroz Monnens T., Carrère S., Bouchez O., Jardinaud M.F., Gris C., François J., Ray J., Torralba B., et al. Genome-wide identification of fitness determinants in the Xanthomonas campestris bacterial pathogen during early stages of plant infection. New Phytol. 2022;236:235–248. doi: 10.1111/nph.18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sang Y., Macho A.P. Analysis of PAMP-Triggered ROS Burst in Plant Immunity. Methods Mol. Biol. 2017;1578:143–153. doi: 10.1007/978-1-4939-6859-6_11. [DOI] [PubMed] [Google Scholar]
- 93.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 94.Camborde L., Jauneau A., Brière C., Deslandes L., Dumas B., Gaulin E. Detection of nucleic acid-protein interactions in plant leaves using fluorescence lifetime imaging microscopy. Nat. Protoc. 2017;12:1933–1950. doi: 10.1038/nprot.2017.076. [DOI] [PubMed] [Google Scholar]
- 95.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gelfand Y., Rodriguez A., Benson G. TRDB--the Tandem Repeats Database. Nucleic Acids Res. 2007;35:D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bouyssié D., Hesse A.M., Mouton-Barbosa E., Rompais M., Macron C., Carapito C., Gonzalez de Peredo A., Couté Y., Dupierris V., Burel A., et al. Proline: an efficient and user-friendly software suite for large-scale proteomics. Bioinformatics. 2020;36:3148–3155. doi: 10.1093/bioinformatics/btaa118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
-
•
Unprocessed photos of immunoblots, southern blots and HR presented in this study have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
MS-based proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository77 with the dataset identifier PXD038104.