Abstract
Voltage-gated ion channels, whose first identified function was to generate action potentials, are divided into subfamilies with numerous members. The family of voltage-gated proton channels (HV) is tiny. To date, all species found to express HV have exclusively one gene that codes for this unique ion channel. Here we report the discovery and characterization of three proton channel genes in the classical model system of neural plasticity, Aplysia californica. The three channels (AcHV1, AcHV2, and AcHV3) are distributed throughout the whole animal. Patch-clamp analysis confirmed proton selectivity of these channels but they all differed markedly in gating. AcHV1 gating resembled HV in mammalian cells where it is responsible for proton extrusion and charge compensation. AcHV2 activates more negatively and conducts extensive inward proton current, properties likely to acidify the cytosol. AcHV3, which differs from AcHV1 and AcHV2 in lacking the first arginine in the S4 helix, exhibits proton selective leak currents and weak voltage dependence. We report the expansion of the proton channel family, demonstrating for the first time the expression of three functionally distinct proton channels in a single species.
Keywords: Aplysia californica, mollusc, patch-clamp, proton, voltage-gating
Introduction
The discovery of the voltage-gated proton channel dates back to 1982 [1]. The gene HVCN1 (Hydrogen voltage-gated channel 1) was identified 24 years later in 2006 by two groups simultaneously [2,3]. Surprisingly, each of over two dozen species evaluated to date has exclusively one gene. HV channels have diverse functions such as proton extrusion from the cytosol [4], charge compensation during the respiratory burst [5,6], B-cell receptor signalling [7], sperm maturation [8], and pH regulation [9–11]. Two variants of human HV1 are expressed. An isoform lacking the first 20 amino acids is initiated by an alternative start codon [7]. This short isoform exhibits twice slower activation, an enhanced phosphorylation response, and is enriched in malignant B cells [12]. Post-translational cleavage produces yet another isoform [13]. These two isoforms are derived from one single HVCN1 gene. Homology studies of the human genome reveal a second proton channel candidate, c15orf27 (TMEM266). This protein is expressed in human brain but does not conduct protons [14–16]. A quite different proton selective channel, OTOP or otopetrin, which we detected in the Aplysia genome, lacks voltage gating [17]. All species tested so far, including mammals, insects [18], fish [19], algae [20], fungi [21], sea urchin [22], and protists [23,24] possess exactly one or no HVCN1 gene. Here, we report the first electrophysiological description of three voltage-gated proton channels expressed from distinct individual genes, AcHV1, AcHV3 and AcHV3, in a single species, the classical model organism of neuronal plasticity, Aplysia californica.
Results
Aplysia HV channel paralogs
Increasing the radius of our genomic search from mammals to more distantly related species, we searched for high-probability proton channel candidates in several molluscan genomes. The recently annotated genome of Aplysia californica surprisingly yielded two sequences that met the criteria for HV channels: four transmembrane regions, a selectivity filter in S1 and a voltage-sensor with a highly conserved tryptophan residue in the S4 signature sequence, RxWRxxR [24]. Intriguingly, the sequences of both Aplysia channels differ significantly from each other. Although the short AcHV1 gene has only five exons, AcHV2 is much larger and possesses 10 exons (Fig. S1). Interestingly, we observed another channel candidate gene AcHV3, with 10 exons (Fig. S1), in which the first arginine above the tryptophan in the S4 signature sequence is missing. Moreover, a proline is localized directly N-terminal to the tryptophan. No HV channel with these two sequence anomalies has been described or reported to function. A preliminary analysis of the transcriptome and genome of octopus/cephalopods, oyster/bivalvia, and other gastropods also showed the presence of genes similar to the three AcHV (Fig. S2) suggesting that these types of HV channel populate different channel families in the mollusc phylum. Figure 1 presents an overview of the preliminary protein structure of the AcHV1, AcHV2 and AcHV3 transmembrane domains. Figure S3 shows an alignment of the transmembrane domains.
Fig. 1.

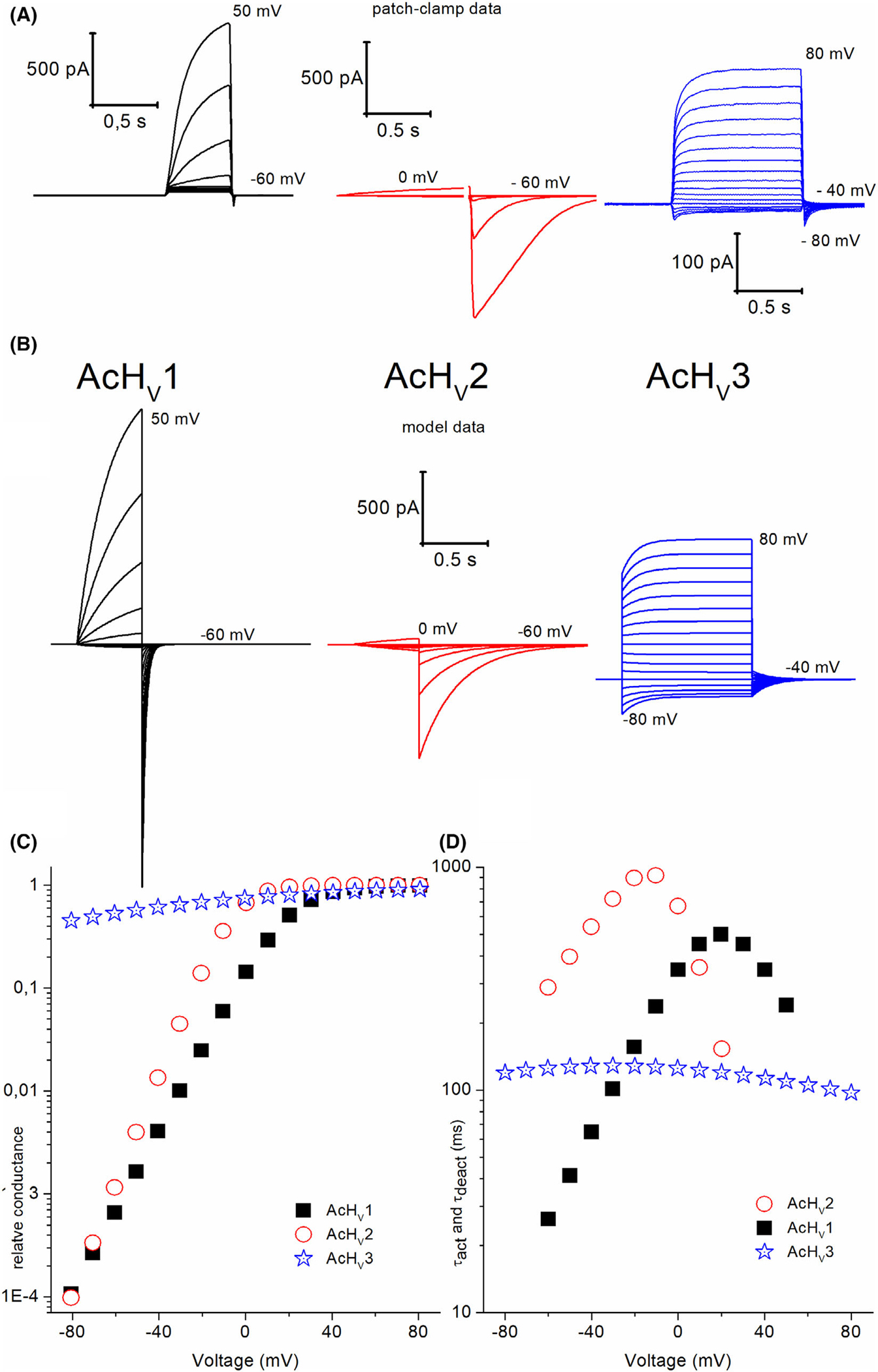
Structural models, alignment, and patch-clamp data of the three Aplysia californica HVs. (A) (i): Whole-cell patch-clamp measurement of AcHV1 showing outward proton currents. Pulses were applied from −60 to 40 mV in 10 mV steps. Measurement was taken at pHo = pHi = 6.5. The measured Vrev was ~ +3 mV. (ii) Zoom-in of dashed area from left showing an activation threshold potential (Vthres) of +10 mV. (iii) Family of pulses in a whole-cell measurement of AcHV2. Currents activate at potentials negative to Vrev allowing inward proton fluxes. Pulses were applied from −40 to 0 mV in 10 mV steps. The measurement was taken at pHo = pHi = 6.5, and Vrev was ~ −5 mV. (iv) Zoom-in of dashed area from left showing activation of proton currents at −30 mV. (v) Whole-cell current family of AcHV3 showing pulses from −90 to +100 mV in 10 mV increments. (vi) Magnification of the pulses around the holding potential of −40 mV, Vrev = 0 mV (dashed area left). The recording conditions are pHo = pHi = 7.0. (vii) Current–voltage plot from the three cells shown in (A). Maximal currents were obtained by a single exponential fitting of the activation kinetics and at different length pulses to avoid proton depletion. For AcHV3 peak, inward current during the pulse was defined as maximal inward current. (Viii) Comparison of activation of AcHV1 and AcHV2 represented in a threshold potential-reversal potential plot. Solid lines exhibit linear fittings of data for each channel in a pH range from 4.5 to 8.0. The total of cells analysed in each gene product was 19 and n = 60. Dashed line represents equality between Vrev and Vthres. Slopes for each channel appear almost identical which translates to the same voltage dependence of gating. However, threshold of activation is markedly more negative in AcHV2. AcHV1 presents an activation of Vthres = 0.776Vrev + 4.5 mV, meanwhile AcHV2 activation is described as Vthres = 0.767Vrev − 19.8 mV. We were unable to identify Vthres of AcHV3. (ix) GFP transfected tsA201 cell measured in pHo = 7, pHi = 5.5. Pulses from −90 to +70 mV showed no identifiable voltage or time-dependent conduction. (B) Structural models of the transmembrane domains of the AcHV1, AcHV2 and AcHV3 channel proteins in the closed state generated by homology modelling. The transmembrane helices are depicted as ribbons (from the N- to the C-terminus): S1 in red, S2 in grey, S3 in yellow, and S4 in blue. The three arginine residues in S4 thought to move outward upon membrane depolarization, the tryptophan residue in S4, and the aspartate residue in S1 required for proton selectivity are shown as sticks. Extracellular membrane loops are shown. Constructed by MODELLER9 [70]. (C) Sketch of the AcHV channels, in all channels S1 contains an aspartate. The proton channel signature sequence RxWRxxR is only partially conserved in AcHV3. (D) Alignment of the S4 helix in Aplysia HV [or AcHV] channels. Accession numbers XM_005100609, XM_005093050, XM_005094218, respectively, CLUSTALW used [25].
The three Aplysia californica channels have distinct biophysical properties
AcHV1
The first identified Aplysia HV channel, AcHV1, may be classified as a typical HV channel (Fig. 1). With 281 amino acids, it is comparable in size to the human HV1 channels (273 amino acids). N- and C-terminal domains are 50 and 100 amino acids in length, respectively, and loops between the transmembrane regions are small (~ 8–17 amino acids; Fig. S4). The S1 transmembrane helix possesses a typical aspartate selectivity filter [14], and the voltage sensor in S4 includes the canonical positively charged RxWRxxR motif [24]. In addition, several conserved negatively charged residues within S1, S2 and S3 are present, as well as a histidine residue at the beginning of S2 forming a putative Zn2+-binding site [2,26–28]. We cloned AcHV1 into pQBI-fC3 plasmid, which was also used for expression of human and insect HV1 channels [2,18,29]. In the construct, green fluorescent protein (GFP) is fused to the N-terminus of the channel. Distinct fluorescence of the membrane was observed in tsA201 cells (Fig. 2). Recorded under tight-seal, whole-cell conditions, AcHV1 exhibited reliable stable membrane currents (Fig. 3A) with an average maximal conductance of 1.5 ± 0.9 nSpF−1 (mean ± SD, n = 19), and capacity 11.4 ± 4.4 pF. AcHV1 has exquisite proton selectivity. The reversal potential of AcHV1 from tail currents [30] followed the Nernst potential for H+ almost perfectly (Fig. 3D). AcHV1 conductance was calculated from extrapolation of single exponential fits to the activating currents, producing gH-V plots (Fig. 3B). Pulse length was varied from 0.25 to 8 s to determine the time constant (τact) accurately, while minimizing H+ depletion. The gH-V plots of AcHV1 shifts by 43–45 mV per unit pHo are determined by two methods: plotting the voltage of first visible current (Vthres) against the reversal potential (Vrev; Fig. 3E) and by analysing the shift of the voltage at which gH was 10% maximal per change in ΔpH (Fig. 3F). As illustrated in Fig. 3C, τact in AcHV1 shifted with pHo, but also slowed at low pHo (τact increases with decreased pHo). Interestingly, the activation kinetics of AcHV1 is much faster than human, insect, or other mammalian HV1, but comparable to that of snails Helisoma trivolvis HtHV1 [31] and Lymnaea stagnalis LsHV1 [32]. In inside-out patch measurements, AcHV1 appears to respond to changes in pHi with a weaker voltage shift (Fig. 4B), as also observed in HtHV1. The more archetypical AcHV1 is expressed in 10 of the 12 investigated tissues. It is missing in the gut and the radular muscle. Interestingly, in human tissue hHV1 is expressed in the lung at levels comparable to all other tissues [2]; the expression of AcHV1 in the gills of Aplysia californica is comparatively low (Fig. S5). The voltage dependence of gating of τact in the recording shown was around 22 mV per e-fold change. The voltage dependence of gating of τdeact ranged also at 22 mV per e-fold change. Zinc inhibited the current (Fig. 5), and comparable to the proton channel of Nicoletia phytophila [29], Zn2+ slowed the activation time constant. We used the limiting slope technique to determine the gating charges of the recording shown in Fig. 3. In symmetrical pH conditions, we obtained 5.7 elementary charges.
Fig. 2.
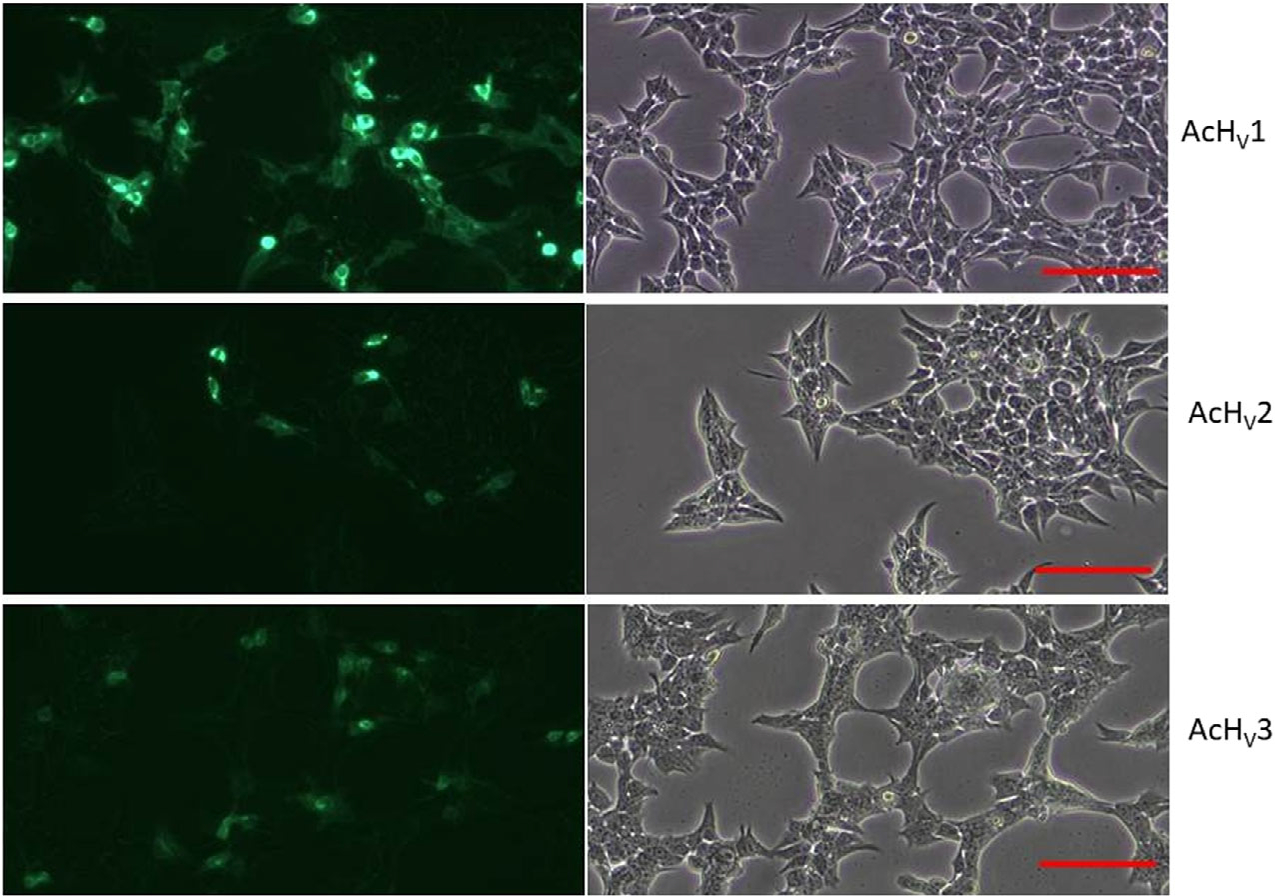
GFP tagged AcHV1–3 transfected tsA201 in fluorescence and transmission light. Direct comparison of the transfection of tsA201 cells with either AcHV1,2,3. One microlitre plasmid DNA of equal concentration was used. Calibration bar ≈ 100 μm. n = 3.
Fig. 3.
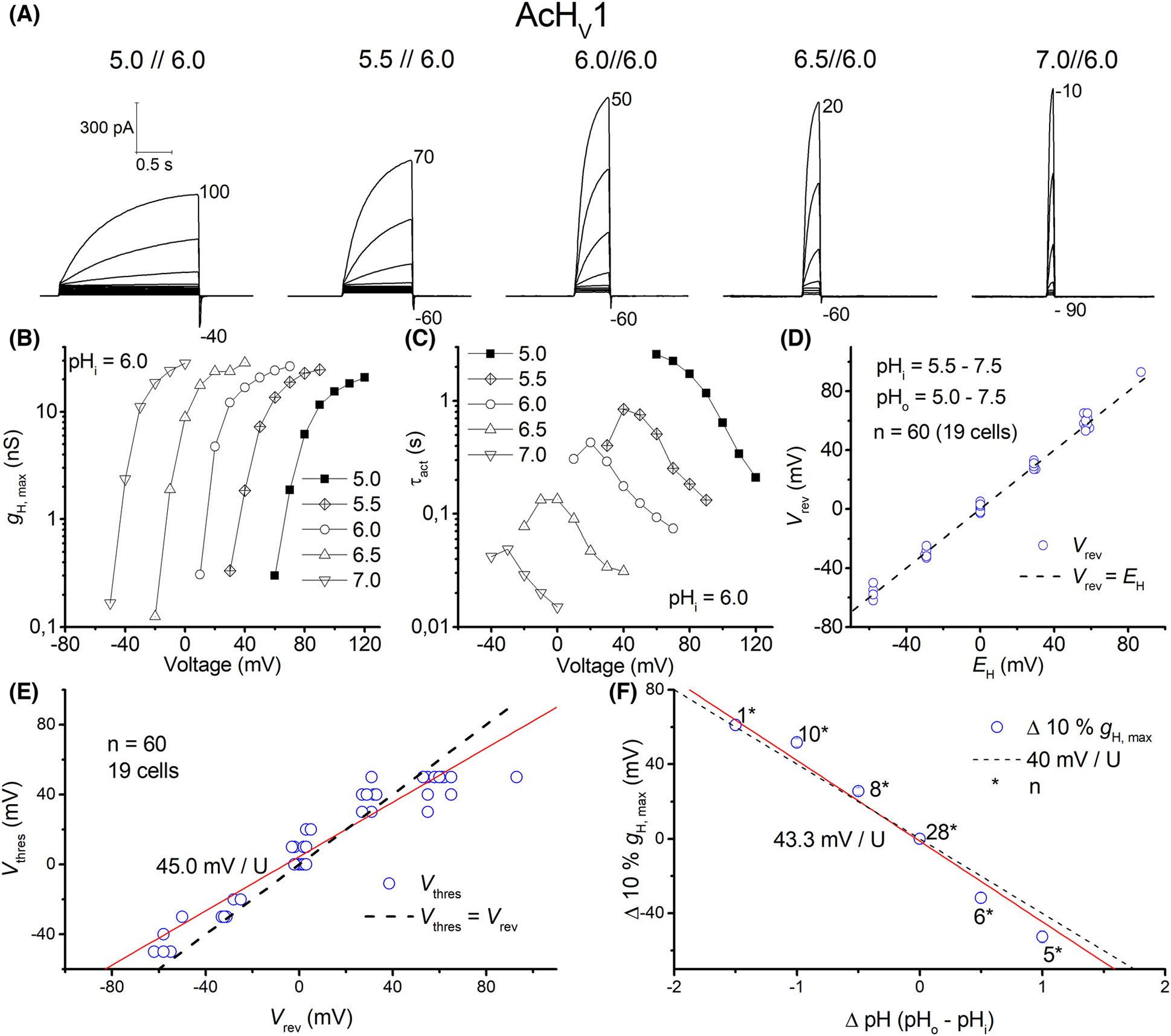
AcHV1 is a typical voltage-gated proton channel. (A) Whole-cell currents in the same cell with pHo ranging from 5–7 at pHi 6.0, in 10 mV increments, pulse length varied with pHo. Holding potential as depicted below the family. (B) Conductance-voltage plot of the current families in (A). (C) Typical speeding of activation kinetics with higher pHo. (D) Reversal potential (Vrev) measurements in 19 cells plotted against EH, showing proton selectivity of AcHV1. (E) Threshold-voltage plot; red line shows changes in Vthres according to Vrev, with a slope 45.0 mV per unit pH. Dashed line shows Vthres equal to Vrev. (F) 10% of maximal conductance vs. voltage shows 43.3 mV shift per unit pH. Dashed line shows the slope of 40 mV per U pH. Numbers with asterisks show number of recordings.
Fig. 4.
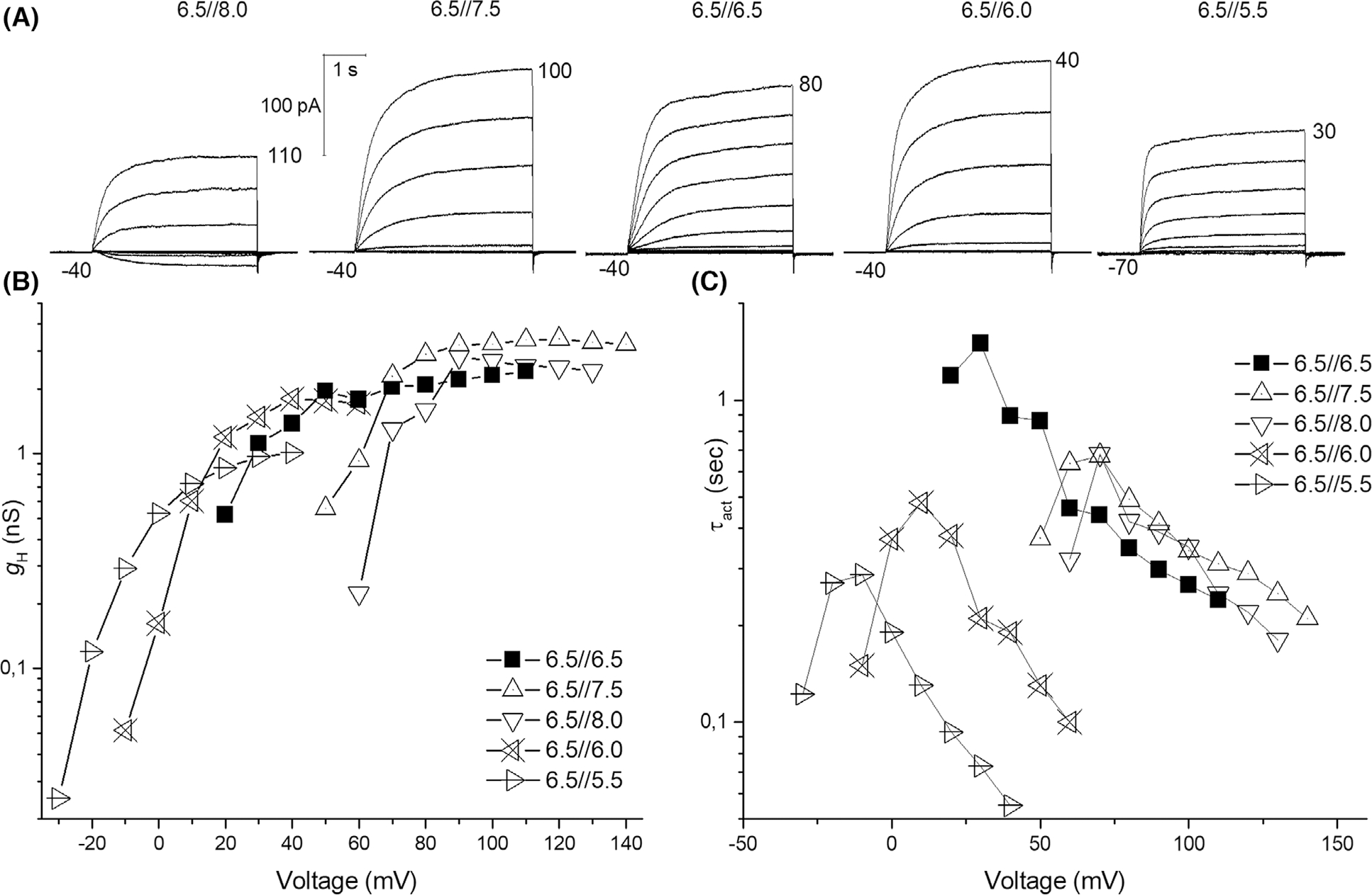
Inside-out patch of AcHV1 shows pHi dependence. (A) Current families in the same inside-out patch at five different pHi. Increments were 10 mV; holding potentials were −40 mV except in the last family where it was −70 mV. Inward currents were seen exclusively at pH 8.0//6.5. (B) Conductance-voltage plot of the current families in (A). (C) Activation kinetics show bell-shaped dependence on voltage.
Fig. 5.
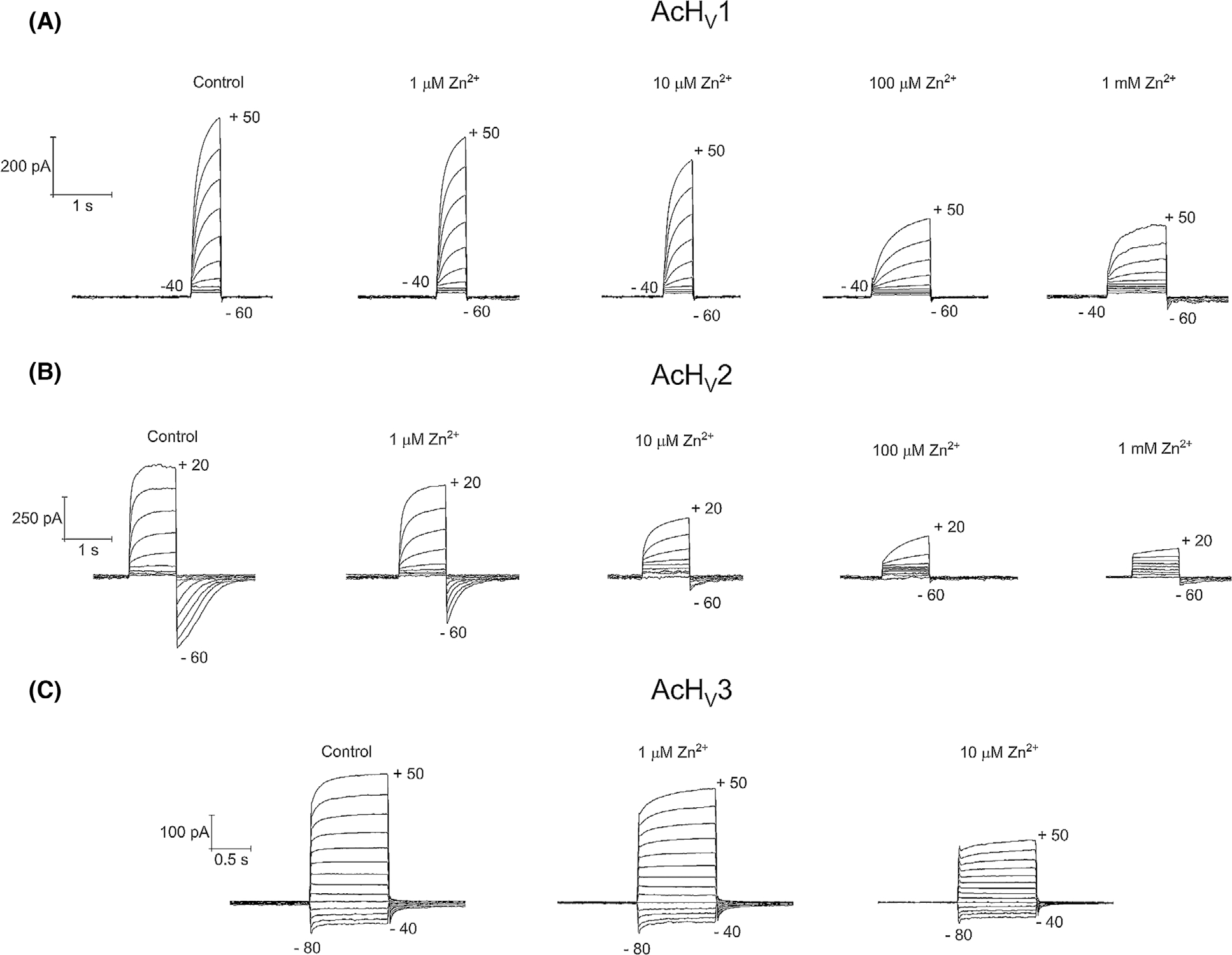
Zinc inhibition of the three proton channel paralogs. (A) AcHV1 current families at the shown zinc concentrations with an increment of 10 mV from −40 to 50 mV and a holding potential of −60 mV. (B) AcHV2 families at the shown zinc concentrations with an increment of 10 mV from −60 to 50 mV and a holding potential of −60 mV, pH 6.5//6.5. (C) AcHV3 current families at the shown zinc concentrations with an increment of 10 mV from −80 to 50 mV and a holding potential of −40 mV, pH 7.0//7.0.
AcHV2
With 339 amino acids, AcHV2 is significantly longer than AcHV1 (Fig. 1), mainly due to a larger extracellular loop between S1 and S2. The 79 residues in the S1–S2 loop possess 14 histidine residues (organized in pairs throughout the loop). Preliminary molecular dynamics (MD) simulations up to several hundreds of nanoseconds show no tendency towards a defined tertiary structure of the loop. The N- and C-terminal domains (70–90 amino acids) are within the normal range for proton channels (Fig. S4). Like AcHV1, AcHV2 has a typical residue for proton selectivity (aspartate in S1), the voltage-sensing RxWRxxR motif, and a histidine at the external part of S2 for Zn2+ binding. The expression of AcHV2 GFP fusion protein showed distinct plasma membrane fluorescence staining (Fig. 2). The average maximal conductance of tsA201 cells transfected with AcHV2 was 2.1 ± 0.8 nSpF−1, and the capacity was 9.0 ± 5.2 pF (n = 19). In contrast to AcHV1 and nearly all other HV, AcHV2 exhibits inward currents at symmetrical pH (Figs 6A and 1A). AcHV1 exhibited inward currents only with a large outward pH gradient, at pH 8.0//6.5 in inside-out patches (Fig. 4). AcHV2 was proton selective, agreeing nearly perfectly with the Nernst potential (Fig. 6D). Single exponential fits of current determined τact and were extrapolated to obtain IH and to calculate gH (Fig. 6B). Pulse lengths were varied from 0.25 to 30 s. To our surprise, pHo dependence of gating of AcHV2 was identical to AcHV1, 44 mV per unit pH (Fig. 6E,F). Activation kinetics (τact) exhibited clear pHo dependence and became faster with increasing pHo (Fig. 6C). The very negative threshold of activation distinguishes AcHV1 and AcHV2. While the primary task of mammalian proton channels is proton extrusion, AcHV2 conducts inward currents that would acidify the cell, but perhaps more importantly, that would depolarize the membrane. Thus, AcHV2 might adopt tasks typically ascribed to voltage-gated sodium or calcium channels by conducting action potentials. In dinoflagellates, HV1 conducts action potentials that trigger the bioluminescent flash [23,24]. By sensing pHo, AcHV2 could be part of the well-known respiratory pumping reflex of Aplysia triggered by low pHo [33,34] or the excitation of Aplysia neurons by lower pH [35]. The pHi dependence of AcHV2 was studied in inside-out patches (Fig. 7). The AcHV2 gH shifts around 40 mV per unit pHi with a suggested saturation at high pHi (Fig. 7), which resembles another snail Helix pomatia [36] and also occurs in hHV1 [37], but is in contrast to the compromised pHi dependence of AcHV1 and the snail channels HtHV1 [31,38] and LsHV1 [32]. The activation time constant of gating was around 15 mV per e-fold change under symmetrical pH conditions. The deactivation time constant appeared much slower, and from the specific recording shown in Fig. 3, we estimated around 31 mV per e-fold change. Gating charge determination via limiting slope method in symmetrical pH conditions revealed 5.3 elementary charges. Zinc inhibition measurements confirmed Zn2+ sensitivity of all three-channel paralogs (Fig. 5). AcHV2 was tested via mRNA analysis and RT-PCR quantification, showing very strong expression in Aplysia californica gut and gills. Expression was weak or absent in hepatopancreas, parapodia and radular muscle (Figs S5 and S6).
Fig. 6.
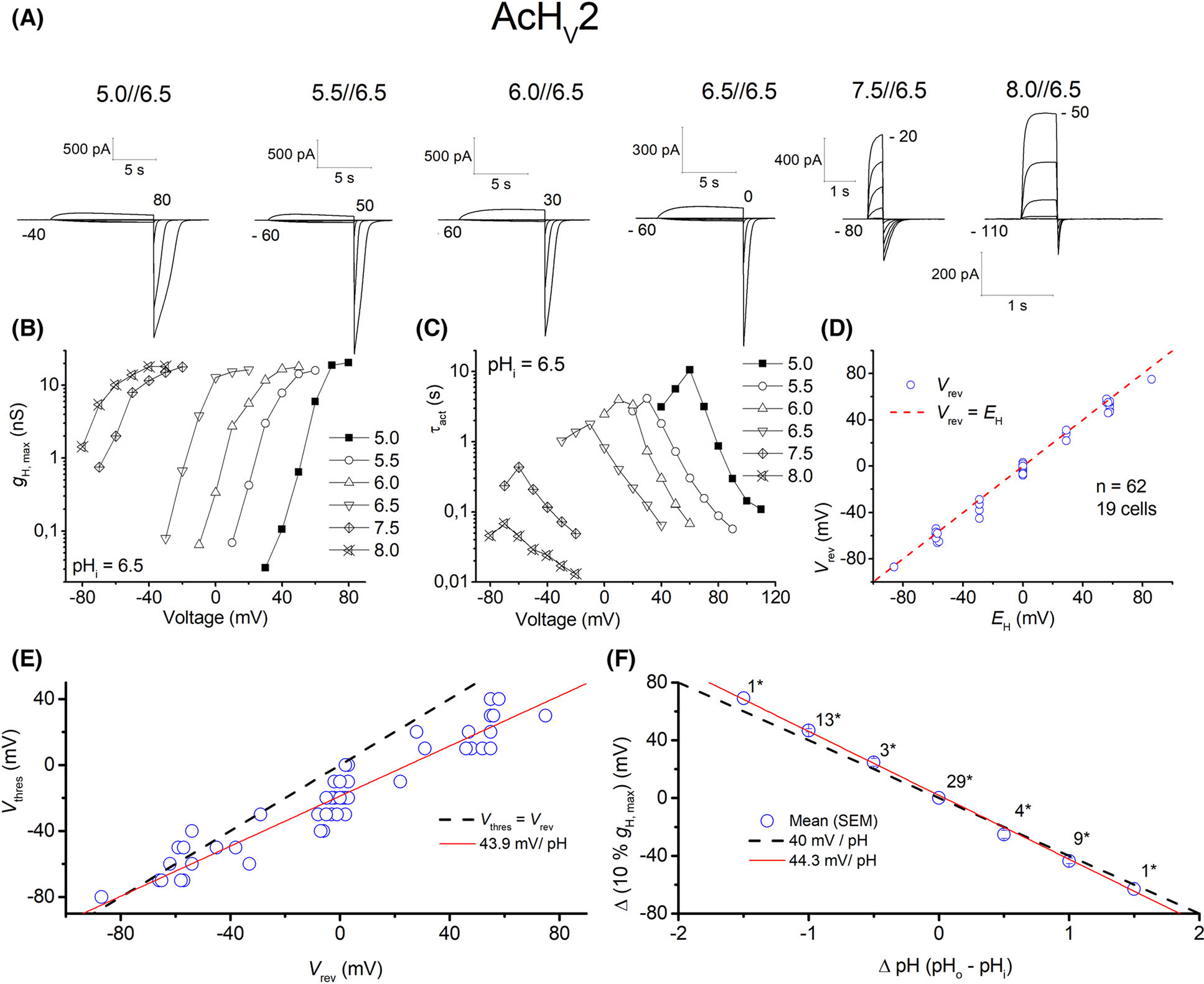
AcHV2 conducts inward proton currents. (A) Whole-cell currents in the same cell ranging from pHo 5–8 at pHi 6.5, in 10 mV increments, pulse length varied with pHo. Holding potential given below each family. (B) Conductance-voltage plot of the current families in (A). (C) Faster activation kinetics with higher pHo. (D) Reversal potential measurements of 19 cells plotted against EH, showing perfect proton selectivity of AcHV2. (E) Threshold-voltage plot; red line shows changes in Vthres according to reversal potential, with a slope 43.9 mV per unit pH. Dashed line shows Vthres equal to Vrev. (F) 10% of maximal conductance vs. voltage shows 44.3 mV shift per unit pH. Dashed line shows a slope of 40 mV/pH.
Fig. 7.
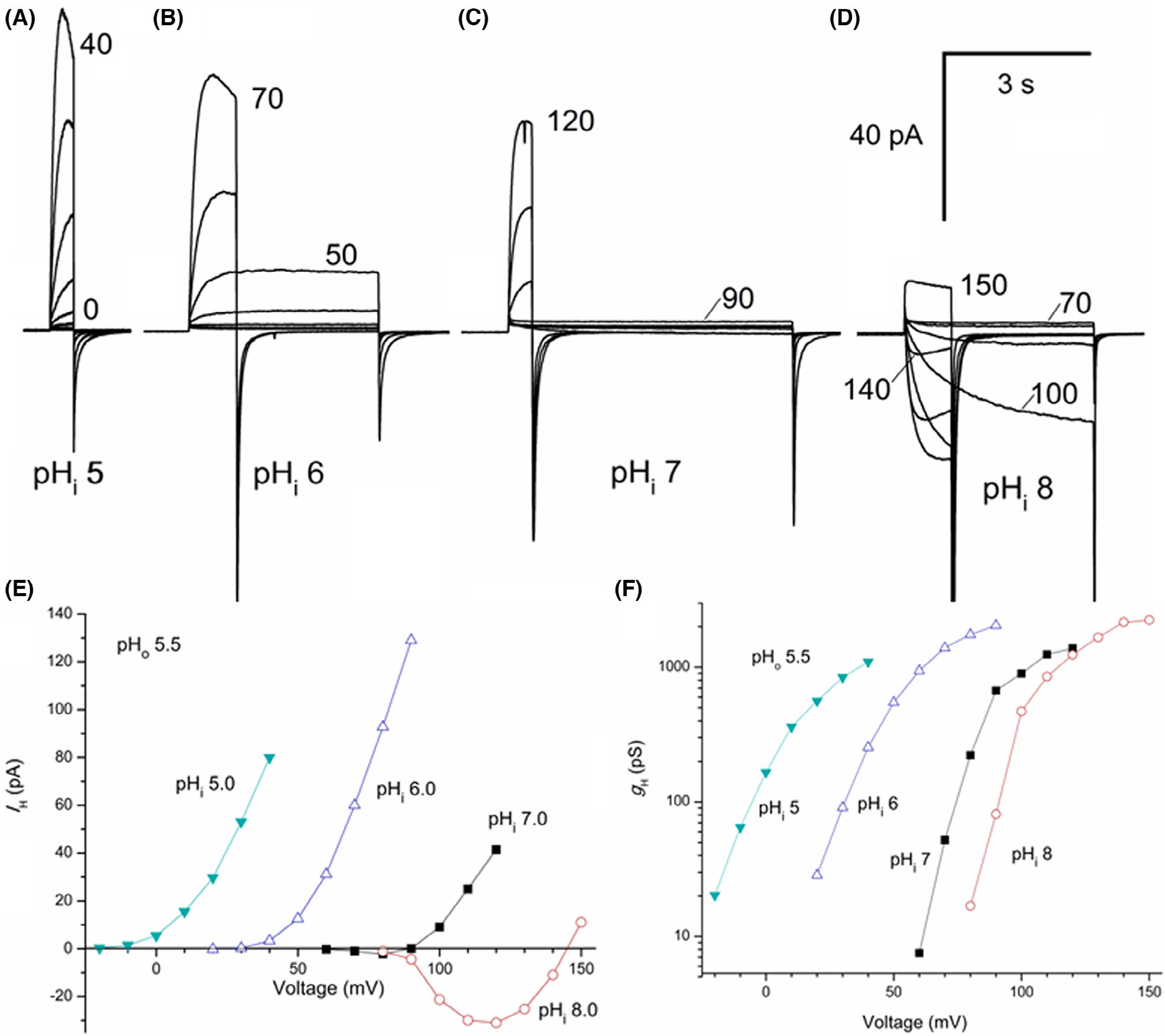
The sensitivity of AcHV2 channels to pHi determined in inside-out patches. (A–D) Families of currents all in the same inside-out patch with pHo 5.5 in the pipette solution were recorded in 10 mV increments up to the voltage indicated. In all cases, Vhold was −40 mV. Pulse duration was shortened for larger depolarizations to minimize proton depletion/accumulation. (E) Current–voltage curves for the families shown in (A–D). Currents were fitted with a single exponential and the extrapolated value or the peak current was plotted. (F) Conductance-voltage relationships for the same families.
AcHV3
The longest of the three channels is AcHV3 which contains 980 amino acids. The S1–S2 loop is 299 amino acids long (30% of the whole protein), lacks obvious organization, and includes 13 histidines. The N-terminus is short and contains 35 amino acids, while the C-terminus with 535 amino acids (54% of the whole protein) is the dominant portion of the channel. The C-terminus of all three paralogs had predicted coiled-coil motifs. The coiled-coil appeared shortly after the fourth transmembrane domain (prediction by https://waggawagga.motorprotein.de) [39]. A coiled-coil region in the C terminus of HV1 in other species is thought to stabilize dimerization [40–42]. Although the C-terminus contains several intracellular retention motifs, we were able to measure channel activity at the plasma membrane, potentially due to massive overexpression in tsA201, HEK-293, and COS-7 cells. Comparison of the localization of the three channels expressed in tsA201 cells is depicted in Fig. 2. The average maximal conductance of AcHV3 was hard to determine because of the overwhelming proton selective leak conductance [43] that we observed while measuring this channel. The leak conductance also introduced a novel phenomenon. Controlling the intracellular pH by the buffered pipette solution became challenging. AcHV3 (Fig. 8A) exhibited time-dependent currents with no detectable voltage dependence of the activation kinetics (n = 20; Fig. 8C). The voltage dependence of the gH was extraordinarily weak, in contrast with that of AcHV1 and AcHV2 (Figs 3B and 6B, respectively). The slope of the gH-V relationship was ~ 60 mV per e-fold change in AcHV3 over a wide voltage range, compared with ~ 4–6 mV per e-fold change in gH for AcHV1 and AcHV2. Therefore, in most recordings we did not reach a voltage at which the conductance was unmistakably maximal (Fig. 8B). Furthermore, many channels evidently remained open even at large negative holding potentials, due to the weak voltage dependence. However, the time-dependent currents of AcHV3 are smaller than the very robust time-dependent currents of AcHV1 and AcHV2. During solution changes of the external media, we observed a baseline current shift at the holding potential (Fig. 8D). AcHV1 and AcHV2 did not show such shifts during external pH changes. Solution exchange was a straightforward way to visualize the proton leak conductance. Addition of monovalent ions (Na+, K+, Cl−) to the bath solution had no effect on either leak or voltage dependent currents in AcHV3. AcHV3 showed in our mRNA analysis expression in every tested tissue. Furthermore, it was strongly expressed. However, our results regarding the amount of genomic contamination had an average of 20.5%. Therefore, we report the expression cautiously. Nevertheless, our findings were in good agreement with EST data, where clones encoding AcHV2 and AcHV3 were found in Aplysia juvenile CNS, but exclusively AcHV2 in pedal-pleural ganglia libraries (Fig. S2). Interestingly, orthologs to AcHV1 and AcHV2 were identified in an EST library of haemocyte cDNA in another mollusc, the oyster Crassostrea gigas, which may suggest connection to the mollusc immune system.
Fig. 8.
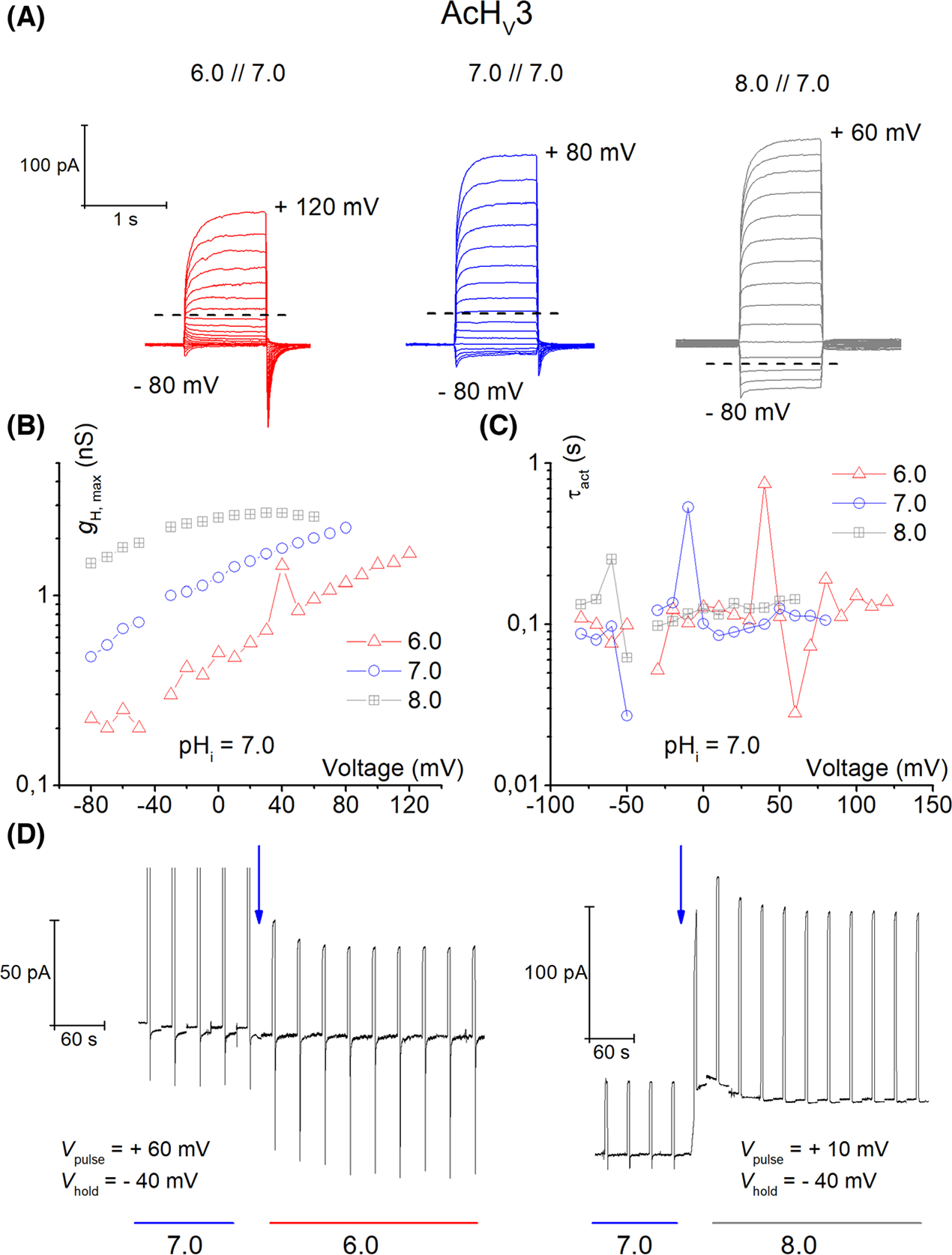
AcHV3 is proton selective and shows leak conductance of protons. (A) Typical whole-cell currents in the same cell ranging from pHo 6.0–8.0 at pHi 7.0, in 10 mV increments. Holding potential was −40 mV in each family. The dashed line shows the reversal potential. Whole-cell measurement from AcHV3 (n = 21). (B) Conductance-voltage plot of the current families in (A) (total conductance). (C) Activation kinetics from current families appears independent of pHo. (D) Proton leak currents are readily detectable by the shift in the baseline current at holding potential, during the solution exchange (blue arrow) to pHo 6 or pHo 8. Vhold in both exchanges is the same, but the test pulse voltage is different (as indicated).
AcHV1, AcHV2, and AcHV3 expressions depend minimally on animal age
Expression of AcHV mRNA is minimally dependent on age, as depicted in Fig. S7. While AcHV1 and AcHV3 express solely one mRNA isoform, we detected four different mRNA variants of AcHV2 (all encoding the same protein). Interestingly, exclusively AcHV2 expression increased recognizably but not significantly in older animals.
Generation of two-state kinetic models of AcHV1, AcHV2, and AcHV3
In this study, we describe three voltage-gated proton channels in one single species. The first two channels have biophysical properties comparable with those of voltage-gated proton channels in other species, but the third channel exhibits a considerable proton leak current. Aligning the sequences of the transmembrane domains of the three channels reveals that all of them possess an aspartate in the middle of S1 known as the selectivity filter (Fig. S3). However, the S4 segment reveals striking differences in the consensus sequence motif RxWRxxR (Fig. 1). The first R is missing in AcHV3, and the next position usually occupied by a hydrophobic leucine is occupied by a more hydrophilic proline [44]. To evaluate the functional consequences of the aberrant signature sequence, we generated a HH model of each channel.
The voltage-gated proton channel has at least one open and two closed states. These conclusions are based on the tail current kinetics and the obvious appearance of two distinct transitions between the states near to the threshold potential of activation (Fig. S8) [45]. Interestingly, the two components emerge also in trimers and tetramers [46]. However, neglecting the tail currents, it is possible to characterize most of the channel’s behaviour in a simple two-state model, which has one open and closed state. In this case, only one transition is needed; described by an activation rate constant equation and a deactivation rate constant equation. Here we used our experimental data to generate a HH model [47] for each AcHV1, AcHV2, and AcHV3. The aim is to compare the current of these channels at the same ΔpH. Figure 5 shows the three AcHV channel types compared to each other at symmetrical pH conditions. As mentioned above the currents appear almost identical to the measured proton currents (Fig. 9). However, tail currents kinetics of all modelled channels are appearing slower. This may be explained by the missing second closed state, which, if implemented, would drastically speed up the tail currents. However, AcHV3 is open at all displayed voltages and its activation and deactivation kinetics are weakly voltage dependent. In this situation, additional channels open during a step increase in voltage; therefore, we may be missing the full closed state to open state transition. Nevertheless, the simple two-state model reproduces the main features of the three channels.
Fig. 9.
AcHV1, AcHV2 and AcHV3 simple two-state model of each channel type. (A) Patch-clamp data displayed at symmetrical pH conditions taken from Figs 3, 6 and 8. (B) Currents of each AcHV type derived from the rate constant equations based on the measurements Figs 3, 6 and 8. Currents displayed in voltage increments of Δ10 mV. Two states are considered in the model. Therefore, the tail current kinetic is not identical to the patch-clamp recordings. AcHV3 model scaled has four times increased number of channels for better comparability. (C) Conductance voltage plot shows left shift of AcHV2. (D) Tail and activation time constants plotted in comparison to voltage.
Discussion
This is the first time that three different functional proton channel genes have been identified in a single species (Aplysia californica). Like all other HV1 channels, AcHV1, AcHV2, and AcHV3 are proton selective. AcHV1 and AcHV2 have evident pH-dependent voltage-gating. The most striking difference is that AcHV2 activates well negative to EH (Fig. S5), and thus, AcHV2 activity would depolarize, whereas AcHV1 activity would hyperpolarize the membrane at symmetrical pH. AcHV1 and AcHV2 are expressed in various tissues, in ganglia and parapodia they express together. Given its negative threshold, AcHV2 could generate a proton-mediated action potential that might contribute to the respiratory pumping reflex of Aplysia californica [33,34]. AcHV3 mRNA is present at substantial levels in all tested tissues. It thus appears to be more widespread throughout the body than the other AcHVs. However, our quantitative RNA analysis was potentially corrupted by genomic (DNA) contamination. Therefore, the apparent expression of AcHV3 in all tissues is to be carefully evaluated. It is most likely present in intracellular membranes in vivo; its C terminus has several cytoplasmic retention motifs, and proton leaks in plasma membranes of cells are typically deleterious. AcHV3 shows shallow voltage-dependent gating, which produces time-dependent outward currents that exhibit pH dependence. The voltage dependence is much weaker than that for AcHV1 and AcHV2. The limiting slope conductance of AcHV1 and AcHV2 is ~ 4–6 mV per e-fold change in voltage, whereas gH in AcHV3 changes e-fold in ~ 60 mV. The steepness of voltage-dependent gating reflects the amount of gating charge that moves across the membrane electrical field during channel opening [48,49]. For most voltage-gated ion channels the bulk of the gating charge moved is thought to reflect outward movement of cationic residues in S4, mostly Arg or Lys. Within this conceptual framework, the extremely weak voltage dependence of AcHV3 indicates that the aberrant signature sequence (LPWRxxR instead of RxWRxxR) most likely attenuates the voltage-dependent conformational change of S4.
Zinc is the most investigated voltage-gated proton channel inhibitor [27,50,51]. Zinc binds to the external part of the proton channel protein and prevents the channel from gating into an open configuration [28,52–55]. Furthermore, it has been shown that the ROS produced during the respiratory burst of phagocytes is massively reduced by zinc inhibition [6,56–58]. An initial investigation revealed that all three AcHVs are inhibited by zinc (Fig. 5). We recorded slowing of the time-dependent outward current and a decrease in tail currents. Both effects of zinc have been reported for other voltage-gated proton channels. Zinc inhibition was weaker for all three AcHVs than for mammalian channels.
AcHV2 acquires its distinctive inward currents by a simple reduction of voltage dependence of the rate constant equation for deactivation (Fig. 6). Additionally, its voltage dependence of activation is steeper than in AcHV1. Our model reproduced the currents reasonably. During the patch-clamp recordings of AcHV2, the main complications derived from the pronounced inward currents and the resulting tail currents. Acidification of the cytosol is the direct cause of this biophysical characteristic. Acidification of the cytosol exceeding the pH-buffering during the whole-cell measurements shifts the reversal potential towards a more negative value this changes the tail current shape visibly, best seen at the more acidic pHo (Fig. 6 A). The slower tail currents in the model compared to the patch-clamp currents are explained by the simple two-state model applied. The model of AcHV1 fitted reasonable the actual patch-clamp recordings but as for the AcHV2 model, the tail currents were slower than in the whole-cell recordings. Again, the simplistic model (C ⇋ O) cannot depict the two components of tail currents, it produces just a single component.
AcHV3 lacks R1 (position L429), the outermost arginine in S4, and has a proline (P430) before the equivalent of R2 in classical HVs (Fig. 1). Distinguishing it from all other known and electrophysiologically analysed native HV, AcHV3 exhibits proton-selective leak currents. The anomalous proton leak in AcHV3 could result from two main causes which may not be entirely separable: closed-channel proton leak or weak voltage dependence of gating. The lack of one of the three Arg in S4 might alter gating so that the channel does not close properly, such that the closed state leaks protons. However, no leak currents were reported in studies in which individual Arg in S4 of human or mouse HV1 were mutated to Ala [2,3]. Intriguingly, when outermost arginine (R1) was mutated to histidine, proton leak currents (through closed channels) were reported by Randolph et al. [59]. Here the principal hypothesis was that the histidine is able to shuttle protons across the membrane in the closed configuration comparable to results found in Shaker channels [60–62]. In a truncation study of the S4 segment of mouse HV1 (VSOP) [63], proton current was not observed if R1, R2 and R3 were missing, most likely making the channel non-functional by losing voltage dependence. If R3 was missing while R1 and R2 are still in place, the channel showed still voltage gating. R1 was not accessible by PEGylation assay, in the closed state of the proton channel, which suggests R1 has limited accessibility to the outer solution. Thus, a simple hydrophobic substitution (R to L) as it is the case in AcHV3 might not be the origin of proton leak current. However, substitution of R1 to a histidine in Kulleperuma et al. [64] was accessible by zinc from the external media. We recently evaluated the role of the hydrophobic gasket (HG) in human HV1 [43]. The HG consists of a ring of 3–4 hydrophobic amino acids near the middle of the transmembrane region of the voltage-sensing domains (S1–S4) of voltage-gating ion channels, including HV1 [43,65,66]. A variety of mutations in the HG designed to decrease its hydrophobicity, resulted in proton selective leak in apparently closed channels [43]. Similar manifestations of proton leak current were evident in AcHV3. This would indicate that a breach in the hydrophobic gasket potentially is the cause of the proton leak current in AcHV3. Or more simply one closed state does not close completely, while there is at least another close state most likely less voltage dependent.
Proline is known to be an alpha helix breaker [67] which might change the environment for S4 in AcHV3. Evidence consistent with the idea that replacing one S4 Arg impairs gating movement has been reported in CiHV1 [68]. The weak voltage dependence of gating of AcHV3, as seen in Fig. 8, supports the idea that the channel may not close completely in the range of voltages applied by us. This suggests that at the voltage applied not all channels in the measurement reach the closed state. S4 in AcHV3 has only two of the three cationic Arg, combined with its extremely weak voltage dependence, strongly suggesting that very little gating charge is moved. Clearly, gating in AcHV3 is grossly different from that of all other known HV channels.
Extreme negative voltages during voltage clamp measurements tend to disrupt the cell membrane. Holding the channel at a potential negative to EH results in sustained inward proton current that acidifies the cell, which is reflected by hyperpolarization of the reversal potential. During depolarizing voltage pulses, protons leave the cell and the reversal shifts positively due to depletion. Despite the high concentration of pH buffer in the pipette solution, the internal pH is poorly controlled, in the face of continuous proton leak. Therefore, we cannot determine the pH-dependent gating characteristics of AcHV3 or the absolute voltage shift of its conductance by the pH. However, as shown in Fig. 8, the channel shows extremely weak voltage dependence over a range of 200 mV both for the gH-V (Fig. 8B) and for τact-V (Fig. 8C). At each voltage applied, there was substantial proton selective conductance (Fig. 8B). However, our model of AcHV3 (Fig. 9) reproduced reasonably the measured patch-clamp current when we assumed the voltage dependence of AcHV3 was much less steep than that of the other two channels. The simple open and closed state model (C ⇋ O) did not simulate the tail currents perfectly in comparison to the actual patch-clamp measurements. If we introduce a second close state (C1 ⇋ C2 ⇋ O) into the model, the resulting currents fit the measurement better. We did not pursue the more complex model because the simple model suffices to show that the main difference between AcHV3 and the other two channels is likely the loss of voltage dependence of gating. Applying pH changes to our model resulted in currents comparable to those recorded from AcHV3 (Fig. S9). Although we cannot rule out a proton selective leak through closed AcHV3 channels, the shallow voltage dependence of AcHV3 may be according to “Occam’s razor” the simplest explanation for the unusual currents seen in AcHV3.
Despite the substantial differences in amino acid sequence among AcHV1, AcHV2, and AcHV3, (only 43% identity 1 vs. 2, 37% identity 1 vs. 3, 29% identity 2 vs. 3), we wondered if the difference in the S4 signature sequence might be responsible for the leak current in AcHV3. Molecular dynamics simulations suggest structural changes that affect the whole channel protein by comparing structurally AcHV1, AcHV2 and AcHV3 (Fig. 1). N-terminally from the Proline in AcHV3, the alpha helix unwinds this part widens in comparison to AcHV1 and AcHV2. The structural changes and the missing arginine are likely to impact on the voltage dependence of gating.
Summarizing our results, we found three genes coding for distinct voltage-gated proton channels in one species. All channels could be expressed at the plasma membrane and are proton selective. AcHV1 is a classically functioning voltage-gated proton channel with biophysical characteristics similar to the human voltage-gated proton channel. AcHV2 activates in a more negative voltage range, resulting in pronounced inward currents. In this respect, it resembles the voltage-gated proton channel from Karlodinium veneficum which is thought to mediate the action potential that trigger the flash in bioluminescent dinoflagellates [19]. AcHV3 is unique. Although it generates time-dependent, proton selective outward current, it exhibits pronounced proton selective currents over a wide voltage range. The expression of the three voltage-gated proton channels is extensive. In view of deleterious effects of proton leaks in plasma membranes, AcHV3 might be expressed mainly or exclusively in intracellular membrane in vivo. Aplysia californica is a classical model organism for neuroscience due to its huge size and the defined number of its neurons [69]. The diameter of the neurons (up to 1.1 mm) facilitates distinct assignment of specific neurons to specific tasks. The additional proton channels in Aplysia californica presumably exist to perform distinct as-yet-undetermined functions in addition to regulating the intracellular pH of these gigantic cells.
Materials and methods
Homology models
The homology models of the transmembrane domains (TMDs) of the AcHV1, AcHV2, and AcHV3 proteins were constructed by homology modelling to the TMD of the Ciona VSP (Ci-VSP) protein in the closed (down) state (PDB: 4G80) using modeller (release 9.18; [70]). First, the amino acid sequences of AcHV1, AcHV2, and AcHV3 were aligned to the amino acid sequence of Ci-VSP using the PSI/TM-Coffee web service of T-Coffee for transmembrane proteins [71]. The TMDs were found to have 54% (/29%) and 55% (/30%) sequence similarity (/identity) in the TM domains between AcHV1 and Ci-VSP, and between AcHV2 and Ci-VSP, respectively (S1–S2 loop not considered for percentage counting in the case of AcHV2; results not shown). The sequences are similar enough to construct accurate models of the AcHV1, AcHV2, and AcHV3 proteins based on the crystal structure of Ci-VSP as template. Then, the TMDs of the AcHV1, AcHV2, and AcHV3 proteins were modelled from the PSI/TM-Coffee alignments. For each of the proteins, 100 models were generated with modeller. For each protein, the five best models were selected based on the modeller objective function (molpdf scoring function) and refined using the web server for protein structure refinement 3drefine [72]. For each protein, the refined model with the best 3Drefine score was selected. The quality of the models was finally assessed using procheck [73].
Heterologous expression
For expression, the AcHV1, AcHV2, and AcHV3 genes were synthesized commercially (Eurofins/Genomics, Ebersberg, Germany); then, the synthesized DNA including a 5’ BamHI and 3’ EcoRI restriction site was cloned into a pEX-A2 plasmid. We subcloned the genes by restriction sites into a pQBI25-fC3 using 5’ BamHI and 3’ EcoRI restriction sites. Site-directed mutagenesis was performed using an overlapping PCR procedure. Clones were sequenced commercially to confirm mutations. GFP is fused physically to N-terminal side of each of the three channel proteins.
tsA201 (human kidney cell line) cells were grown to 85% confluency in 35 mm culture dishes. Cells were transfected with 1.3 μg plasmid DNA using polyethylenimine (Sigma, St. Louis, MO, USA). After 12 h at 37 °C in 5% CO2, cells were trypsinized and replated onto glass coverslips at low density for patch-clamp recording the same day and the next day. We selected green cells under fluorescence for recording. Whole-cell patch-clamp showed no other voltage- or time-dependent conductance under our recording conditions. tsA201 cells showed no native proton conductance without transfection induced expression of a protein by these cells. The level of expression of all mutants studied here was sufficiently high that potential contamination by native HV1 currents was negligible.
Aplysia rearing and sampling methods
Aplysia californica from the University of Miami National Resource for Aplysia were reared in fresh single-pass seawater at 13–15 °C from egg masses of wild-caught animals. Animals were fed an ad lib diet of Agardhiella subulata as previously described [74]. The animals used had mean live weight ± SD of 148 ± 46 g and were sexually mature.
Approximate 0.5 mL of haemolymph was drawn from the posterior sinus and preserved as whole blood in 0.5 mL of RNAlater (Sigma). Five millilitre of haemolymph was drawn from an additional three animals, and after centrifugation, the pellets were combined and preserved in 0.5 mL of RNAlater. Each animal, from which tissues were harvested, was anaesthetized by injection of 1/6th body weight (by volume) of isotonic MgCl2 and, after 5 min, euthanized by severing the connectives and removal of all nervous system ganglia. Ganglia and other tissues were immediately rinsed in artificial seawater (ASW: 417 mM NaCl, 10 mM KCl, 10 mM CaCl2, 55 mM MgCl2, and 15 mM HEPES-NaOH, pH 7.6), then minced, and each was placed in 0.5 mL RNAlater. All preserved haemolymph and tissue samples were refrigerated at 9.5 °C overnight and then moved to a −20 °C freezer until shipment to Germany on wet ice.
The tissues harvested were the paired cerebral, buccal, pleural, abdominal and pedal ganglia, rhinophore, and 20–50 mg portions of the I4 radular muscle, gill, parapodia, hepatopancreas, and gut directly adjacent to the buccal mass.
Electrophysiology
Whole-cell patch-clamp or excised patch recordings were done as described [75]: In brief, patch-clamp amplifiers EPC 10 (HEKA, Lambrecht, Germany) and Axopatch 200b (Molecular Devices, Sunnyvale, CA, USA) were used. Recordings were stored on hard discs and analysed with Origin (Origin 2017, Northampton, MA, USA). Patch pipettes were made from borosilicate capillaries GC 150TF-10 (Harvard Apparatus, Holliston, MA, USA) and pulled using a Flaming Brown automatic pipette puller P-1000 (Sutter Instruments, Novato, CA, USA). Pipettes were heat polished to a tip resistance ranging typically from 5–9 MΩ with pipette solutions used. Electrical contact with the pipette solution was achieved by a chloride silver wire and connected to the bath with an agar bridge made with Ringer’s solution. Seals were formed with Ringer’s solution (in mM 160 NaCl, 4.5 KCl, 2 CaCl2, 1 MgCl2, 5 HEPES, pH 7.4) in the bath, and the potential zeroed after the pipette was placed above the cell. Whole-cell solutions (pipette and bath) included 100 mM buffer close to its pKa with tetramethylammonium (TMA+) and methanesulfonate (CH3SO3−) as the main ions, 1 mm EGTA, and 1–2 mm Mg2+ with an osmolarity of 300 mOsmL−1. Buffers were HOMOPIPES at pH 4.5 and pH 5.0, MES at pH 5.5 and pH 6.0, Bis-Tris at pH 6.5, PIPES at pH 7.0, HEPES at pH 7.5, Tricine at pH 8.0, and CHES at pH 9.0. Resistance of the seals was usually > 3 GΩ. Currents are shown without correction for leak or liquid junction potentials. Data were collected between 11–23 °C. Currents were fitted to a rising exponential to obtain the activation time constant (τact). The proton conductance (gH) was calculated from the steady-state current (the fitted current extrapolated to infinite time) using reversal potentials (Vrev) measured in each solution in each cell. In these fits, the initial delay was ignored; the remaining current usually fitted a single exponential well. The reversal potential was measured by two methods. If the threshold was negative to the Vrev, it could be readily determined by the zero current. If Vrev was negative to Vthres, Vrev was determined with the tail current method. The magnitude of gH-V shift was determined by comparing the values of 10% of the maximal proton conductance, V(gH,max/10) or by the shift in Vthres. Overexpression of the channels in small cells resulted in large proton currents which removed enough protons from the cell to change pHi greatly. Proton channel gating kinetics depends strongly on pH; therefore, proton depletion is a significant source of error. To minimize this problem, families with different pulse lengths were applied. Longer pulses were used to determine pulses close to Vthres where τact is slow, while shorter pulses were used at more positive voltages. Zinc measurements were conducted as described [28].
Databank research
Database analysis
GenBank (nr), transcriptional survey sequences (TSA), and expressed sequence tags (EST) databases were screened with the TBLASTN algorithm for Aplysia sequences with significant homology to known HV1 sequences. The following criteria were used to identify putative Aplysia HV channels: (a) they possess four transmembrane regions (S1–S4), (b) they harbour a typical negatively charged (D/E) residue as selectivity filter in S1, (c) they possess a totally conserved tryptophan (W) residue in S4, (d) there are at least two positively charges residues (usually R) as putative voltage-sensor surrounding the W in S4. TSA coverage is shown in Fig. S10. Figure S11 shows amino acid sequences and TSA accession number. Preliminary cladogram is shown in Fig. S12.
Tissue distribution
Four animals were manually dissected, and tissue was stored in RNAlater (Qiagen, Hilden, Germany). Total RNA was prepared using RNeasy MiniKit (Qiagen) and reverse transcribed with Sensiscript reverse transcriptase (Qiagen). PCR analysis for the expression of AcHV1, AcHV2, and AcHV3 was conducted using AdvantageTaq polymerase mixture (TaKaRa, Saint-Germain-en-Laye, France) and a standard PCR protocol (56 °C annealing temperature). The following primers were used for amplification: AcHV1 forward 5’-AAAACCCTGCGAGAAACCTT-3′, AcHV1 reverse 5’-ATGATGAGAAGCCCAACACC-3′, AcHV2 forward 5’-GACTTTGACCTTTGCCCTAA-3′, AcHV2 reverse 5’-TATTCATGCCCAATTTGTTG-3′, AcHV3 forward 5’-CTTCCAATAGCAGGTTTCGT-3’, AcHV3 reverse 5’-CACAGTCAAAGGTGCTTCTG-3′. According to the identified database entries bands with a calculated 356 bp (AcHV1), 361 bp (AcHV2), 401 bp (AcHV3) were expected.
For a quantitative analysis of AcHV1–3 expression, the same primers and thermal protocols (see above) were used on a CFX Connect Real-Time System (Bio-Rad, Feld-kirchen, Germany) as a standard SybrGreen assay. For relative expression of the AcHV channels, the respective ΔCt value in direct comparison with the GAPDH expression in the same tissue was calculated. AcHV1 primers were intron spanning while AcHV2 + 3 were not. Genomic contamination of RNA samples cannot be excluded. As a control, qPCR calculations of tissue RNA samples with Reverse Transcriptase omitted in RT reaction were performed with the only exon-spanning primers for AcHV2 and AcHV3. The calculated relative amount of genomic DNA contamination on the total AcHV2/3 amount was between 32% and 4% (n = 6).
Modelling AcHV1 + 2 + 3
To model currents, presented rate constant equations were used for each channel (C ⇋ O).
| AcHV1: | α = 0.0083*e((100 + V)/22) | β = 4000*e(−(120 + V)/22) |
| AcHV2: | α = 1*e(V)/11 | β = 0.5*e(−V)/31 |
| AcHV3: | α = 6*e(V)/177 | β = 2*e(−V)/97 |
V, Voltage in mV.
Supplementary Material
Acknowledgements
We thank P. Drummer for excellent technical assistance. Supported by W. Lutz Stiftung to GC, Paulmanns Wunschkinder to CJ, NIH grant P40-OD010952 to LF, Kerscher’sche Stiftung to LN, NIH grant R35-GM126902 to TED.
Abbreviations
- AcHV1
Aplysia californica voltage-gated proton channel one
- GFP
green fluorescent protein
- gH-V
conductance voltage dependence
- HVCN1
hydrogen voltage-gated channel 1
- MD
molecular dynamics
- pHi
inside pH
- pHo
outside pH
- V rev
reversal potential
- ΔpH
difference between outside and inside pH
- τ act
time constant of activation
- τ deact
time constant of deactivation
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
Data availability statement
The data that supports the findings of this study are available in the figures of this manuscript and the supplementary material of this article.
References
- 1.Thomas RC, Meech RW. Hydrogen ion currents and intracellular pH in depolarized voltage-clamped snail neurones. Nature. 1982;299:826–8. 10.1038/299826a0 [DOI] [PubMed] [Google Scholar]
- 2.Ramsey IS, Moran MM, Chong JA, Clapham DE. A voltage-gated proton-selective channel lacking the pore domain. Nature. 2006;440:1213–6. 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006;312:589–92. 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- 4.Barish ME, Baud C. A voltage-gated hydrogen ion current in the oocyte membrane of the axolotl, Ambystoma. J Physiol. 1984;352:243–63. 10.1113/jphysiol.1984.sp015289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henderson LM, Chappell JB, Jones OT. Internal pH changes associated with the activity of NADPH oxidase of human neutrophils. Further evidence for the presence of an H+ conducting channel. Biochem J. 1988;251:563–7. 10.1042/bj2510563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeCoursey TE, Morgan D, Cherny VV. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 2003;422:531–4. 10.1038/nature01523 [DOI] [PubMed] [Google Scholar]
- 7.Capasso M, Bhamrah MK, Henley T, Boyd RS, Langlais C, Cain K, et al. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat Immunol. 2010;11:265–72. 10.1038/ni.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lishko PV, Botchkina IL, Fedorenko A, Kirichok Y. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 2010;140:327–37. 10.1016/j.cell.2009.12.053 [DOI] [PubMed] [Google Scholar]
- 9.Iovannisci D, Illek B, Fischer H. Function of the HVCN1 proton channel in airway epithelia and a naturally occurring mutation, M91T. J Gen Physiol. 2010;136:35–46. 10.1085/jgp.200910379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan D, Capasso M, Musset B, Cherny VV, Rios E, Dyer MJ, et al. Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc Natl Acad Sci USA. 2009;106:18022–7. 10.1073/pnas.0905565106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor AR, Chrachri A, Wheeler G, Goddard H, Brownlee C. A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol. 2011;9:e1001085. 10.1371/journal.pbio.1001085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hondares E, Brown MA, Musset B, Morgan D, Cherny VV, Taubert C, et al. Enhanced activation of an amino-terminally truncated isoform of the voltage-gated proton channel HVCN1 enriched in malignant B cells. Proc Natl Acad Sci USA. 2014;111:18078–83. 10.1073/pnas.1411390111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger TK, Fussholler DM, Goodwin N, Bonigk W, Muller A, Dokani Khesroshahi N, et al. Post-translational cleavage of Hv1 in human sperm tunes pH- and voltage-dependent gating. J Physiol. 2017;595:1533–46. 10.1113/JP273189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Musset B, Smith SM, Rajan S, Morgan D, Cherny VV, DeCoursey TE. Aspartate112 is the selectivity filter of the human voltage-gated proton channel. Nature. 2011;480:273–7. 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim IH, Hevezi P, Varga C, Pathak MM, Hong L, Ta D, et al. Evidence for functional diversity between the voltage-gated proton channel Hv1 and its closest related protein HVRP1. PLoS ONE. 2014;9:e105926. 10.1371/journal.pone.0105926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papp F, Lomash S, Szilagyi O, Babikow E, Smith J, Chang TH, et al. TMEM266 is a functional voltage sensor regulated by extracellular Zn2+. Elife. 2019;8:e42372. 10.7554/eLife.42372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu YH, Cooper AJ, Teng B, Chang RB, Artiga DJ, Turner HN, et al. An evolutionarily conserved gene family encodes proton-selective ion channels. Science. 2018;359:1047–50. 10.1126/science.aao3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaves G, Derst C, Jardin C, Franzen A, Musset B. Voltage-gated proton channels in polyneopteran insects. FEBS Open Bio. 2022;12:523–37. 10.1002/2211-5463.13361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratanayotha A, Kawai T, Higashijima SI, Okamura Y. Molecular and functional characterization of the voltage-gated proton channel in zebrafish neutrophils. Physiol Rep. 2017;5:e13345. 10.14814/phy2.13345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rangel-Yescas G, Cervantes C, Cervantes-Rocha MA, Suárez-Delgado E, Banaszak AT, Maldonado E, et al. Discovery and characterization of Hv1-type proton channels in reef-building corals. eLife. 2021;10:e69248. 10.7554/eLife.69248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C, Tombola F. Voltage-gated proton channels from fungi highlight role of peripheral regions in channel activation. Commun Biol. 2021;4:261. 10.1038/s42003-021-01792-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakata S, Miyawaki N, McCormack TJ, Arima H, Kawanabe A, Ozkucur N, et al. Comparison between mouse and sea urchin orthologs of voltage-gated proton channel suggests role of S3 segment in activation gating. Biochim Biophys Acta. 2016;1858:2972–83. 10.1016/j.bbamem.2016.09.008 [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez JD, Haq S, Bachvaroff T, Nowak KF, Nowak SJ, Morgan D, et al. Identification of a vacuolar proton channel that triggers the bioluminescent flash in dinoflagellates. PLoS ONE. 2017;12:e0171594. 10.1371/journal.pone.0171594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith SM, Morgan D, Musset B, Cherny VV, Place AR, Hastings JW, et al. Voltage-gated proton channel in a dinoflagellate. Proc Natl Acad Sci USA. 2011;108:18162–7. 10.1073/pnas.1115405108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson JD, Higgins DG, Gibson TJ. clustalw: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita K, Sakata S, Yamashita E, Fujiwara Y, Kawanabe A, Kurokawa T, et al. X-ray crystal structure of voltage-gated proton channel. Nat Struct Mol Biol. 2014;21:352–7. 10.1038/nsmb.2783 [DOI] [PubMed] [Google Scholar]
- 27.Musset B, Smith SM, Rajan S, Cherny VV, Sujai S, Morgan D, et al. Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J Physiol. 2010;588:1435–49. 10.1113/jphysiol.2010.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaves G, Bungert-Plumke S, Franzen A, Mahorivska I, Musset B. Zinc modulation of proton currents in a new voltage-gated proton channel suggests a mechanism of inhibition. FEBS J. 2020;287:4996–5018. 10.1111/febs.15291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaves G, Derst C, Franzen A, Mashimo Y, Machida R, Musset B. Identification of an HV 1 voltage-gated proton channel in insects. FEBS J. 2016;283:1453–64. 10.1111/febs.13680 [DOI] [PubMed] [Google Scholar]
- 30.Hodgkin AL, Huxley AF. The components of membrane conductance in the giant axon of Loligo. J Physiol. 1952;116:473–96. 10.1113/jphysiol.1952.sp004718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas S, Cherny VV, Morgan D, Artinian LR, Rehder V, Smith SME, et al. Exotic properties of a voltage-gated proton channel from the snail Helisoma trivolvis. J Gen Physiol. 2018;150:835–50. 10.1085/jgp.201711967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byerly L, Meech R, Moody W Jr. Rapidly activating hydrogen ion currents in perfused neurones of the snail, Lymnaea stagnalis. J Physiol. 1984;351:199–216. 10.1113/jphysiol.1984.sp015241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Croll RP. Sensory control of respiratory pumping in Aplysia californica. J Exp Biol. 1985;117:15–27. [Google Scholar]
- 34.Levy M, Achituv Y, Susswein AJ. Relationship between respiratory pumping and oxygen consumption in Aplysia depilans and Aplysia fasciata. J Exp Biol. 1989;141:389–405. [Google Scholar]
- 35.Brown AM, Berman PR. Mechanism of excitation of Aplysia neurons by carbon dioxide. J Gen Physiol. 1970;56:543–58. 10.1085/jgp.56.5.543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doroshenko PA, Kostyuk PG, Martynyuk AE. Transmembrane outward hydrogen current in intracellularly perfused neurones of the snail Helix pomatia. Gen Physiol Biophys. 1986;5:337–50. [PubMed] [Google Scholar]
- 37.Cherny VV, Morgan D, Musset B, Chaves G, Smith SM, DeCoursey TE. Tryptophan 207 is crucial to the unique properties of the human voltage-gated proton channel, hHV1. J Gen Physiol. 2015;146:343–56. 10.1085/jgp.201511456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherny VV, Morgan D, Thomas S, Smith SME, DeCoursey TE. Histidine(168) is crucial for DeltapH-dependent gating of the human voltage-gated proton channel, hHV1. J Gen Physiol. 2018;150:851–62. 10.1085/jgp.201711968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simm D, Hatje K, Kollmar M. Waggawagga: comparative visualization of coiled-coil predictions and detection of stable single alpha-helices (SAH domains). Bioinformatics. 2015;31:767–9. 10.1093/bioinformatics/btu700 [DOI] [PubMed] [Google Scholar]
- 40.Koch HP, Kurokawa T, Okochi Y, Sasaki M, Okamura Y, Larsson HP. Multimeric nature of voltage-gated proton channels. Proc Natl Acad Sci USA. 2008;105:9111–6. 10.1073/pnas.0801553105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SY, Letts JA, Mackinnon R. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc Natl Acad Sci USA. 2008;105:7692–5. 10.1073/pnas.0803277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tombola F, Ulbrich MH, Isacoff EY. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 2008;58:546–56. 10.1016/j.neuron.2008.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banh R, Cherny VV, Morgan D, Musset B, Thomas S, Kulleperuma K, et al. Hydrophobic gasket mutation produces gating pore currents in closed human voltage-gated proton channels. Proc Natl Acad Sci USA. 2019;116:18951–61. 10.1073/pnas.1905462116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–32. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 45.Cherny VV, Markin VS, DeCoursey TE. The voltage-activated hydrogen ion conductance in rat alveolar epithelial cells is determined by the pH gradient. J Gen Physiol. 1995;105:861–96. 10.1085/jgp.105.6.861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujiwara Y, Kurokawa T, Takeshita K, Nakagawa A, Larsson HP, Okamura Y. Gating of the designed trimeric/tetrameric voltage-gated H+ channel. J Physiol. 2013;591:627–40. 10.1113/jphysiol.2012.243006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Almers W Gating currents and charge movements in excitable membranes. Rev Physiol Biochem Pharmacol. 1978;82:96–190. 10.1007/BFb0030498 [DOI] [PubMed] [Google Scholar]
- 49.Sigg D, Bezanilla F. Total charge movement per channel. The relation between gating charge displacement and the voltage sensitivity of activation. J Gen Physiol. 1997;109:27–39. 10.1085/jgp.109.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahaut-Smith MP. The effect of zinc on calcium and hydrogen ion currents in intact snail neurons. J Exp Biol. 1989;145:455–64. [DOI] [PubMed] [Google Scholar]
- 51.Cherny VV, DeCoursey TE. pH-dependent inhibition of voltage-gated H+ currents in rat alveolar epithelial cells by Zn2+ and other divalent cations. J Gen Physiol. 1999;114:819–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jardin C, Chaves G, Musset B. Assessing structural determinants of Zn2+ binding to human HV1 via multiple MD simulations. Biophys J. 2020;118:1221–33. 10.1016/j.bpj.2019.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De La Rosa V, Bennett AL, Ramsey IS. Coupling between an electrostatic network and the Zn(2+) binding site modulates Hv1 activation. J Gen Physiol. 2018;150:863–81. 10.1085/jgp.201711822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiu F, Chamberlin A, Watkins BM, Ionescu A, Perez ME, Barro-Soria R, et al. Molecular mechanism of Zn2+ inhibition of a voltage-gated proton channel. Proc Natl Acad Sci USA. 2016;113:E5962–71. 10.1073/pnas.1604082113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jardin C, Ohlwein N, Franzen A, Chaves G, Musset B. The pH-dependent gating of the human voltage-gated proton channel from computational simulations. Phys Chem Chem Phys. 2022;24:9964–77. 10.1039/d1cp05609c [DOI] [PubMed] [Google Scholar]
- 56.Femling JK, Cherny VV, Morgan D, Rada B, Davis AP, Czirjak G, et al. The antibacterial activity of human neutrophils and eosinophils requires proton channels but not BK channels. J Gen Physiol. 2006;127:659–72. 10.1085/jgp.200609504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musset B, Cherny VV, DeCoursey TE. Strong glucose dependence of electron current in human monocytes. Am J Physiol Cell Physiol. 2012;302:C286–95. 10.1152/ajpcell.00335.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Droste A, Chaves G, Stein S, Trzmiel A, Schweizer M, Karl H, et al. Zinc accelerates respiratory burst termination in human PMN. Redox Biol. 2021;47:102133. 10.1016/j.redox.2021.102133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Randolph AL, Mokrab Y, Bennett AL, Sansom MS, Ramsey IS. Proton currents constrain structural models of voltage sensor activation. Elife. 2016;5:e18017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Starace DM, Bezanilla F. Histidine scanning mutagenesis of basic residues of the S4 segment of the shaker k+ channel. J Gen Physiol. 2001;117:469–90. 10.1085/jgp.117.5.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Starace DM, Bezanilla F. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 2004;427:548–53. 10.1038/nature02270 [DOI] [PubMed] [Google Scholar]
- 62.Starace DM, Stefani E, Bezanilla F. Voltage-dependent proton transport by the voltage sensor of the shaker K+ channel. Neuron. 1997;19:1319–27. 10.1016/s0896-6273(00)80422-5 [DOI] [PubMed] [Google Scholar]
- 63.Sakata S, Kurokawa T, Norholm MH, Takagi M, Okochi Y, von Heijne G, et al. Functionality of the voltage-gated proton channel truncated in S4. Proc Natl Acad Sci USA. 2010;107:2313–8. 10.1073/pnas.0911868107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulleperuma K, Smith SM, Morgan D, Musset B, Holyoake J, Chakrabarti N, et al. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J Gen Physiol. 2013;141:445–65. 10.1085/jgp.201210856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campos FV, Chanda B, Roux B, Bezanilla F. Two atomic constraints unambiguously position the S4 segment relative to S1 and S2 segments in the closed state of shaker K channel. Proc Natl Acad Sci USA. 2007;104:7904–9. 10.1073/pnas.0702638104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lacroix JJ, Hyde HC, Campos FV, Bezanilla F. Moving gating charges through the gating pore in a Kv channel voltage sensor. Proc Natl Acad Sci USA. 2014;111:E1950–9. 10.1073/pnas.1406161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Heijne G Proline kinks in transmembrane alpha-helices. J Mol Biol. 1991;218:499–503. 10.1016/0022-2836(91)90695-3 [DOI] [PubMed] [Google Scholar]
- 68.Gonzalez C, Rebolledo S, Perez ME, Larsson HP. Molecular mechanism of voltage sensing in voltage-gated proton channels. J Gen Physiol. 2013;141:275–85. 10.1085/jgp.201210857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moroz LL. Aplysia. Curr Biol. 2011;21:R60–1. 10.1016/j.cub.2010.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2016;54:5 6 1–5 6 37. 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Floden EW, Tommaso PD, Chatzou M, Magis C, Notredame C, Chang JM. PSI/TM-coffee: a web server for fast and accurate multiple sequence alignments of regular and transmembrane proteins using homology extension on reduced databases. Nucleic Acids Res. 2016;44:W339–43. 10.1093/nar/gkw300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharya D, Nowotny J, Cao R, Cheng J. 3Drefine: an interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016;44:W406–9. 10.1093/nar/gkw336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Laskowski RA, MacArthur MW, Thornton JM. PROCHECK: validation of protein-structure coordinates. In: CP Brock, T Hahn, H Wondratschek, U Müller, U Shmueli, E Prince, et al. , editors. International tables for crystallography. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2012. p. 684–7. [Google Scholar]
- 74.Gerdes R, Fieber LA. Life history and aging of captive-reared California Sea hares (Aplysia californica). J Am Assoc Lab Anim Sci. 2006;45:40–7. [PubMed] [Google Scholar]
- 75.Musset B, Cherny VV, Morgan D, Okamura Y, Ramsey IS, Clapham DE, et al. Detailed comparison of expressed and native voltage-gated proton channel currents. J Physiol. 2008;586:2477–86. 10.1113/jphysiol.2007.149427 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings of this study are available in the figures of this manuscript and the supplementary material of this article.


