Abstract
A genetic screen to isolate gene products required for vacuolar morphogenesis in the yeast Saccharomyces cerevisiae identified VAM7, a gene which encodes a protein containing a predicted coiled-coil domain homologous to the coiled-coil domain of the neuronal t-SNARE, SNAP-25 (Y. Wada and Y. Anraku, J. Biol. Chem. 267:18671–18675, 1992; T. Weimbs, S. H. Low, S. J. Chapin, K. E. Mostov, P. Bucher, and K. Hofmann, Proc. Natl. Acad. Sci. USA 94:3046–3051, 1997). Analysis of a temperature-sensitive-for-function (tsf) allele of VAM7 (vam7tsf) demonstrated that the VAM7 gene product directly functions in vacuolar protein transport. vam7tsf mutant cells incubated at the nonpermissive temperature displayed rapid defects in the delivery of multiple proteins that traffic to the vacuole via distinct biosynthetic pathways. Examination of vam7tsf cells at the nonpermissive temperature by electron microscopy revealed the accumulation of aberrant membranous compartments that may represent unfused transport intermediates. A fraction of Vam7p was localized to vacuolar membranes. Furthermore, VAM7 displayed genetic interactions with the vacuolar syntaxin homolog, VAM3. Consistent with the genetic results, Vam7p physically associated in a complex containing Vam3p, and this interaction was enhanced by inactivation of the yeast NSF (N-ethyl maleimide-sensitive factor) homolog, Sec18p. In addition to the coiled-coil domain, Vam7p also contains a putative NADPH oxidase p40phox (PX) domain. Changes in two conserved amino acids within this domain resulted in synthetic phenotypes when combined with the vam3tsf mutation, suggesting that the PX domain is required for Vam7p function. This study provides evidence for the functional and physical interaction between Vam7p and Vam3p at the vacuolar membrane, where they function as part of a t-SNARE complex required for the docking and/or fusion of multiple transport intermediates destined for the vacuole.
The intracellular compartments of eukaryotic cells are largely defined by the composition of the resident proteins contained therein. Thus, efficient biosynthetic trafficking of these resident proteins must be maintained to ensure the integrity and functionality of individual organelles. The acidified vacuole of the budding yeast Saccharomyces cerevisiae is one such organelle. Analogous to the lysosome of mammalian cells, the vacuole functions in a variety of cellular processes, including macromolecular degradation, metabolite storage, and cytosolic ion and pH homeostasis (41, 45). Thus, vacuolar function requires an influx of resident proteins such as hydrolytic proteases, lipases, and transporters. These proteins traffic via vesicle-mediated transport reactions that require appropriate cargo selection and vesicle budding from donor membranes, followed by the docking and fusion of the transport intermediates with the correct target organelle. Several genetic screens have been designed to identify mutants defective in vacuolar protein sorting (vps) (4, 68), vacuolar protease activity (pep) (40), and vacuolar morphogenesis (vam) (84). Through these screens, over 40 complementation groups have been identified that display defects in the delivery of proteins from the trans-Golgi network to the vacuole. These complementation groups have been further classified (classes A through F) with respect to their specific morphological and biochemical phenotypes (5, 63).
Biochemical investigation of protein transport in numerous experimental systems has identified several proteins proposed to be involved in the docking and fusion of transport vesicles with their target membranes (30, 66, 67, 78). These proteins have been found to be highly homologous among the various systems, suggesting that the docking and fusion of transport vesicles utilizes a conserved mechanism (11, 23). Namely, members of the syntaxin, synaptobrevin-VAMP, Sec1p, and Rab families have been shown to function in vesicular transport at multiple stages of the secretory pathway. In neuronal cells, the transmembrane proteins synaptobrevin (v-SNARE) and syntaxin (t-SNARE) associate with donor and target membranes, respectively (8, 10, 79). According to the SNARE hypothesis, complementary pairing between synaptobrevin and syntaxin provides, in part, the specificity required for the targeting of cargo to the appropriate destination (76). Stable interaction between synaptobrevin and syntaxin requires an additional t-SNARE molecule, SNAP-25 (synapse-associated protein of 25 kDa), that forms a t-SNARE complex with syntaxin (31, 32, 58, 75). This ternary interaction ensures the docking stability and specificity necessary for subsequent fusion to occur. Sec1p and Rab family members are suggested to regulate the formation and/or activity of these SNARE interactions (56, 66, 67). After the coupling of v- and t-SNAREs, N-ethylmaleimide-sensitive factor (NSF) and soluble NSF attachment proteins (SNAPs) catalyze the disassembly of SNARE complexes, allowing fusion of the transport vesicle with the target membrane (16, 75, 76). Alternatively, it has also been proposed that the NSF-SNAP complex functions in priming SNAREs via ATP hydrolysis prior to docking (50, 51, 55, 80).
Several of the VPS gene products have been found to be members of the conserved syntaxin, Sec1p, and Rab families. The syntaxin homolog Pep12p (9), Sec1p family member Vps45p (17, 60), and Rab GTPase Vps21p (35) are believed to direct the transport of vacuolar proteins from the trans-Golgi to an endosomal compartment (9, 15). Genetic and biochemical studies have also identified a set of VPS gene products that mediate trafficking at a late stage of vacuolar protein transport. These gene products include Vam3p (20, 77, 83), Vps33p (6, 65), and Ypt7p (86), which share homology with syntaxin, Sec1p, and Rab family members, respectively. Additionally, the yeast NSF homolog Sec18p (21) has been shown to function both in Golgi-to-endosome transport with Pep12p (15) and in vacuole-to-vacuole fusion with Ypt7p and Vam3p (50, 55, 80). The prevalence of these SNARE complex members suggests that the mechanisms of docking and fusion are conserved in the vacuolar protein sorting pathway.
The VAM7 gene was originally identified in a screen for mutants defective in vacuolar assembly and morphogenesis (84). Cells with the VAM7 gene deleted lack normal vacuoles and, instead, accumulate numerous vesicular structures that contain vacuolar proteins (82). Furthermore, Vam7p has been determined to contain an amino-terminal NADPH oxidase p40phox (PX) domain (61) and a carboxy-terminal heptad repeat homologous to the coiled-coil domain of SNAP-25 family members (85). Here, we report the function of the VAM7 gene product in vacuolar protein sorting by characterizing a temperature-conditional allele of VAM7 (vam7tsf). Analysis of the vam7tsf mutant indicates that Vam7p functions at a late stage in vacuolar delivery of multiple proteins that transit via distinct biosynthetic pathways. Inactivation of the vam7tsf gene product results in the accumulation of numerous aberrant membrane compartments which likely correspond to unfused transport intermediates. Localization experiments suggest that a fraction of Vam7p associates with vacuolar membranes. Genetic and physical analysis determined that Vam7p functionally interacts with the vacuolar t-SNARE Vam3p. Our results suggest that Vam7p functions at the vacuole as part of a t-SNARE complex in a manner analogous to SNAP-25 in synaptic vesicle trafficking.
MATERIALS AND METHODS
Strains and media.
The S. cerevisiae strains used in these studies are listed in Table 1. Yeast strains were grown in standard yeast extract-peptone-dextrose (YPD), yeast extract-peptone-fructose (YPF), or synthetic dextrose (SD) media supplemented with appropriate amino acids (72). Yeast transformations were done by the lithium acetate method (39) with single-stranded carrier DNA. Standard bacterial medium (52) was supplemented with 100 μg of ampicillin per ml for plasmid retention. Escherichia coli transformations with the XL1Blue strain (14) were done as previously described (28).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 | 65a |
| BHY10 | SEY6210; leu2-3,112::pBHY11(CPY-Inv LEU2) | 35 |
| TKSY32 | BHY10; pep12-60 vam7Δ::HIS3 | This study |
| TKSY43 | SEY6210; vam7Δ::HIS3 | This study |
| TKSY44 | BHY10; vam7Δ::HIS3 | This study |
| TKSY48 | SEY6210; sec18-1 vam7Δ::HIS3 | This study |
| TKSY49 | SEY6210; vam7Δ::HIS3 vam3Δ::LEU2 + pVAM3.416 | This study |
| TKSY50 | SEY6210; vam7Δ::HIS3 vam3Δ::LEU2 + pVAM3-6.416 | This study |
| CBY9 | BHY10; pep12-60 | 15 |
| CBY42 | SEY6210; sec18-1 vam3Δ::HIS3 | C. Burd |
| EGY1181-10 | SEY6210; sec18-1 | E. Gaynor |
| TDY2 | SEY6210; vam3Δ::LEU2 | 20 |
DNA methods.
Recombinant DNA manipulations were performed by standard methods (49). Restriction and modification enzymes were purchased from Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and New England Biolabs (Beverly, Mass.). DNA sequencing was done by the UCSD CFAR Core facility with the ABI Prism BigDye and Amplitaq DNA polymerase FS. Gels were analyzed with a Perkin-Elmer–Applied Biosystems 373XL DNA sequencer. The plasmid pYVQ706 containing the VAM7 open reading frame was a generous gift from Yoh Wada (University of Tokyo, Tokyo, Japan). Its construction is described elsewhere (82). Plasmids pTKS30 and pTKS23 were generated by subcloning the 1.8-kb NsiI-HindIII fragment (containing the entire VAM7 ORF) of pYVQ706 into the PstI-HindIII polylinker sites of pRS414 and pRS424 (73), respectively. A PCR-based HIS3 disruption fragment was constructed with primers containing overhangs of the first and last 50 bases of the VAM7 coding sequence (7). The amplified DNA fragment was isolated and transformed into SEY6210 or BHY10. The resulting His+ colonies were isolated and confirmed for insertion of the HIS3 gene into the VAM7 locus by PCR of genomic DNA. Furthermore, this procedure was used to generate other vam7Δ strains described in Table 1. Plasmid pTKS43ts-167 containing a temperature-conditional allele of vam7 (vam7-167) was constructed by PCR-based mutagenesis and gapped-plasmid repair (53). Primers complementary to the 5′ or 3′ ends of the VAM7 coding sequence were used to mutagenize VAM7 under limiting dATP conditions. The resulting PCR product was purified and cotransformed with PstI-AatII gapped pTKS30. Candidates were isolated for temperature-conditional secretion of a CPY-invertase fusion protein (57). The GFP-Vam7p fusion protein was constructed by PCR with oligonucleotides introducing an in-frame BamHI site at the start ATG. The PCR fragment was cloned into the BglII-SalI sites of the pRS416 version of pGOGFP (18) to generate plasmid pTKS35. PX domain point mutants were made by using the gene SOEing method (38, 87) and gapped-plasmid repair. Point mutations were recovered from yeast cells and confirmed by sequencing for the desired mutations. Plasmids pVAM3.414, pVAM3.416, pVAM3.426, pVAM3-6.416, and pPEP12.426 were previously described (20).
Preparation of antiserum against Vam7p.
To construct the glutathione S-transferase (GST)-Vam7p fusion protein, the 943-base XbaI-HindIII fragment of VAM7 was inserted into PGEX-KG (Pharmacia LKB, Uppsala, Sweden). The resulting fusion protein containing the C-terminal 182 amino acids of Vam7p was expressed in E. coli XL1Blue and isolated by affinity binding to glutathione-coupled Sepharose. After being eluted from the Sepharose, the GST-Vam7p fusion protein was further purified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroelution. Purified protein was then used to immunize New Zealand White rabbits as previously described (36). Crude antiserum was affinity purified by binding to cyanogen bromide-coupled GST-Vam7p (29). Vam7p was detected in yeast cell lysates by immunoblotting and enhanced chemiluminescence (ECL) detection as previously described (2).
Metabolic labeling and immunoprecipitations.
To examine the biosynthetic trafficking of vacuolar proteins, yeast cells were radiolabeled as previously described (2, 35, 36). Immunoprecipitations of ALP, CPS, and CPY were done accordingly (19, 36). Aminopeptidase I (API) immunoprecipitations were performed as previously described (20). API antibody was a generous gift from Dan Klionsky (University of California, Davis, Calif.) (43). Antibodies to ALP, CPY, CPS, Pep12p, and Vam3p have been previously described (9, 19, 20, 42, 44).
Subcellular fractionation.
Subcellular fractionation experiments with Vam7p were done as previously described with modifications. Unlabeled spheroplasts made from wild-type cells (SEY6210) were lysed, and fractions were generated as described previously (24). Each fraction was detected by immunoblotting and ECL as described previously (2). For temperature-shift experiments, spheroplasts from EGY1181-10 were radiolabeled at 26°C with EXPRE35S35S label (NEN/DuPont) for 35 min and chased for 10 min with unlabeled methionine, cysteine, and yeast extract to final concentrations of 5 mM, 1 mM, and 0.2%, respectively. After 10 min, cells were either shifted to 38°C or remained at 26°C for an additional 30 min. After the temperature shift, cells were harvested, fractions were generated, and Vam7p was immunoprecipitated as described previously (24). Accudenz density gradients were prepared as described earlier (3). The 13,000 × g pellet fraction was generated from unlabeled spheroplasts as described above and resuspended in 1 ml of lysis buffer (0.2 M sorbitol; 50 mM potassium acetate; 20 mM HEPES, pH 6.8; 2 mM EDTA) containing 60% Accudenz A.G. (Accurate Chemical & Scientific Corporation, Westbury, N.Y.). A 2-ml portion of 55% Accudenz A.G. was then layered on top of the resuspended pellet, followed by an additional 2 ml of 35% Accudenz A.G. After a 14-h spin at 200,000 × g, the top 3 ml, the remaining 2 ml, and the sediment were individually harvested, precipitated with 10% trichloroacetic acid (TCA), washed with acetone, and detected by immunoblotting. The top 3 ml was designated the float fraction, and the remaining 2 ml was designated the nonfloat fraction.
Microscopy.
Nomarski optics analysis and fluorescence microscopy of the green fluorescent protein (GFP)-Vam7p fusion protein were done as described previously (18). For electron microscopy (EM) analysis, vam7 mutants were prepared as described earlier (20, 64).
Native immunoprecipitations.
Appropriate yeast strains were grown in YPD or SD supplemented with 2% Casamino Acids and appropriate amino acids to mid-log phase. Six units of optical density at 600 nm (OD600; 1 OD600 unit ≈ 108 cells) were harvested, spheroplasted, and allowed to recover for 10 min at 26°C. Accordingly, the cells were then shifted to 38°C or retained at 26°C for 1 h and then lysed in 0.5% Triton X-100, 0.2 M sorbitol, 50 mM potassium acetate, 20 mM HEPES (pH 6.8), and 2 mM EDTA and homogenized as described previously (24) at 5 OD600 units/ml. The homogenate was extracted on ice for 10 min and then centrifuged at 13,000 × g for 10 min. One milliliter of extract was transferred to a new Eppendorf tube and incubated with 5 μl of affinity-purified anti-Vam7p polyclonal antibody for 1 h at 4°C. Protein A-Sepharose beads (Pharmacia) were added, and the mixture was allowed to incubate for an additional 1 h. After incubation, bound protein was washed six times with 1 ml of lysis buffer containing 0.1% Triton X-100. Supernatant was completely removed from the beads with a Hamilton syringe and immunoprecipitated proteins were eluted with SDS-PAGE protein sample buffer. Two OD units were loaded, resolved by SDS-PAGE, and detected by immunoblotting and ECL.
RESULTS
A vam7tsf mutant displays defects in the maturation of multiple vacuolar proteins and accumulates aberrant membranous compartments.
Mutations in a specific subset of VPS and VAM genes cause an accumulation of vacuolar proteins in numerous small, prevacuolar compartments (5, 63, 84). Not surprisingly, many of the VAM genes overlap with the VPS genes, including VPS41/VAM2 and VAM6/VPS39 (54). A previous study determined that vam7Δ mutants also display a similar aberrant vacuolar morphology (82). In complementation studies, we found that the VAM7 gene complemented vps43 mutants, indicating that VAM7 and VPS43 correspond to the same locus. To address the primary role for VAM7, a temperature-conditional allele of VAM7 was generated by error-prone PCR mutagenesis as previously described (see Materials and Methods). One mutant, vam7-167, was characterized in detail. DNA sequencing analysis of the vam7-167 mutant identified two amino acid changes: leucine 134 to proline and leucine 287 to proline. Western blotting revealed that mutant Vam7p expression in the vam7-167 strain was found to be similar to that observed for wild-type Vam7p at both permissive and nonpermissive temperatures for 30 min (data not shown). Henceforth, this allele will be denoted as vam7tsf (temperature-sensitive for function).
The biosynthetic trafficking of vacuolar hydrolases such as carboxypeptidase Y (CPY) can be monitored by posttranslational modifications that correspond to transport through distinct compartments of the secretory and vacuolar protein sorting pathways (45). Precursor CPY (p1CPY) is first translocated into the endoplasmic reticulum (ER), where it receives core glycosyl modifications. Upon transport to the Golgi complex, the p1 precursor is further mannosylated to produce the p2 form (p2CPY). Like CPY, the type II vacuolar transmembrane hydrolases alkaline phosphatase (ALP) and carboxypeptidase S (CPS) are both transported via the secretory pathway to the vacuole as inactive precursors (pALP and pCPS, respectively). Both CPY and CPS are delivered from the Golgi to the vacuole via an endosomal compartment (15, 19), while ALP traffics through an alternative Golgi-to-vacuole pathway (18). Upon reaching the vacuole, the precursors are proteolytically cleaved to generate the active, mature forms (mCPY, mCPS, and mALP) of these enzymes.
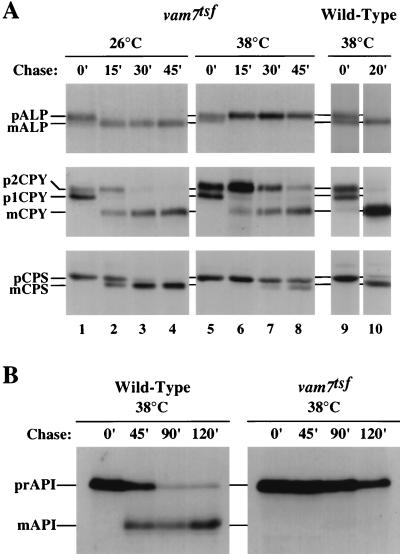
To determine the phenotypic consequences of Vam7p inactivation, the processing of CPY, ALP, and CPS was assayed in vam7tsf cells by pulse-chase analysis. The vam7tsf mutant cells were grown to logarithmic phase at 26°C and then incubated at either 26°C or 38°C for 10 min. Each culture was then pulse-labeled for 10 min with [35S]cysteine and [35S]methionine and chased with unlabeled cysteine and methionine for 0, 15, 30, or 45 min. Lysates were produced and immunoprecipitated with antibodies to ALP, CPS, and CPY, separated by SDS-PAGE, and analyzed by autoradiography. As shown in Fig. 1A, vacuolar hydrolases were rapidly matured in the vam7tsf mutant incubated at 26°C with kinetics nearly identical to those for wild-type cells. In contrast, the vam7tsf mutant incubated at 38°C displayed a complete block in the maturation of ALP and severe kinetic defects in the processing of CPY and CPS. The processing of CPY was significantly slower in vam7tsf cells (processing half-time, t1/2 ≈ 30 to 40 min) than in wild-type cells incubated at 38°C (t1/2 ≈ 5 to 10 min). Similarly, the processing of CPS in the vam7tsf mutant (t1/2 ≈ 50 to 60 min) was severely impaired compared to that in wild-type cells at 38°C (t1/2 ≈ 10 to 15 min). Immunoprecipitation of extracellular p2CPY revealed that only a small amount (5%) was secreted by vam7tsf cells at 38°C after 45 min of chase (Table 2), suggesting that the p2CPY accumulated in an intracellular compartment.
FIG. 1.
Vacuolar protein sorting in vam7tsf mutant cells. (A) TKSY43 (vam7Δ) cells transformed with either complementing plasmid (pTKS30) or plasmid containing a temperature-sensitive-for-function (tsf) allele of vam7 (pTKS43ts-167) were incubated at either the permissive (26°C) or the nonpermissive (38°C) temperature for 10 min and then labeled with [35S]cysteine and [35S]methionine for 10 min. Cells were subsequently chased with unlabeled cysteine and methionine and harvested at the time points indicated. ALP, CPY, and CPS were immunoprecipitated, resolved by SDS-PAGE, and visualized by autoradiography. CPS samples were treated with endoglycosidase H prior to electrophoresis. ER-modified and Golgi-modified CPYs are denoted by p1CPY and p2CPY, respectively. Other precursor (p) and mature (m) forms of vacuolar proteins are indicated. (B) API delivery in vam7tsf cells. TKSY43 (vam7Δ) cells transformed with either complementing plasmid (pTKS30) or plasmid containing the vam7tsf allele (pTKS43ts-167) were incubated at the nonpermissive temperature (38°C) for 10 min and then labeled with [35S]cysteine and [35S]methionine for 10 min. Cells were subsequently chased with unlabeled cysteine and methionine and harvested at the indicated time points. API was recovered from lysates by immunoprecipitation, resolved by SDS-PAGE, and visualized by autoradiography. Precursor API (prAPI) and mature API (mAPI) are indicated.
TABLE 2.
CPY maturation and mislocalization in vam7tsf mutanta
| Strain | Temp (°C) | Fraction
|
||
|---|---|---|---|---|
| Intracellular
|
Extracellular
|
|||
| % Mature | % Precursor | % Precursor | ||
| Wild-type | 38 | >95 | <2 | <2 |
| vam7tsf | 26 | >95 | <2 | <2 |
| vam7tsf | 38 | 70 | 25 | 5 |
Strains were incubated at the indicated temperature for 10 min, labeled with [35S]cysteine and [35S]methionine for 10 min, and chased with unlabeled cysteine and methionine for 45 min. Cells were then spheroplasted, and samples were centrifuged to generate intracellular and extracellular fractions. CPY was immunoprecipitated and quantified by autoradiography.
The vacuolar hydrolase API has been shown to traffic from the cytoplasm to the vacuole through macroautophagy (1, 43, 70, 71). In the vacuole, precursor API (prAPI) is converted to mature API (mAPI). Previously, we determined that Vam3p function is required for the delivery of API to the vacuole (20). To determine if Vam7p function is also required for the transport of API to the vacuole, pulse-chase analysis was carried out on wild-type and vam7tsf cells. At 26°C, in both wild-type and vam7tsf cells, API was matured to its 50-kDa vacuolar form with a processing half-time of ca. 2 h (data not shown). In wild-type cells at 38°C, the majority of API was matured by 120 min (Fig. 1B). In contrast, in vam7tsf cells at the nonpermissive temperature, API remained solely in its precursor 61-kDa form after 120 min of chase. Thus, Vam7p function is required for the transport and maturation of API.
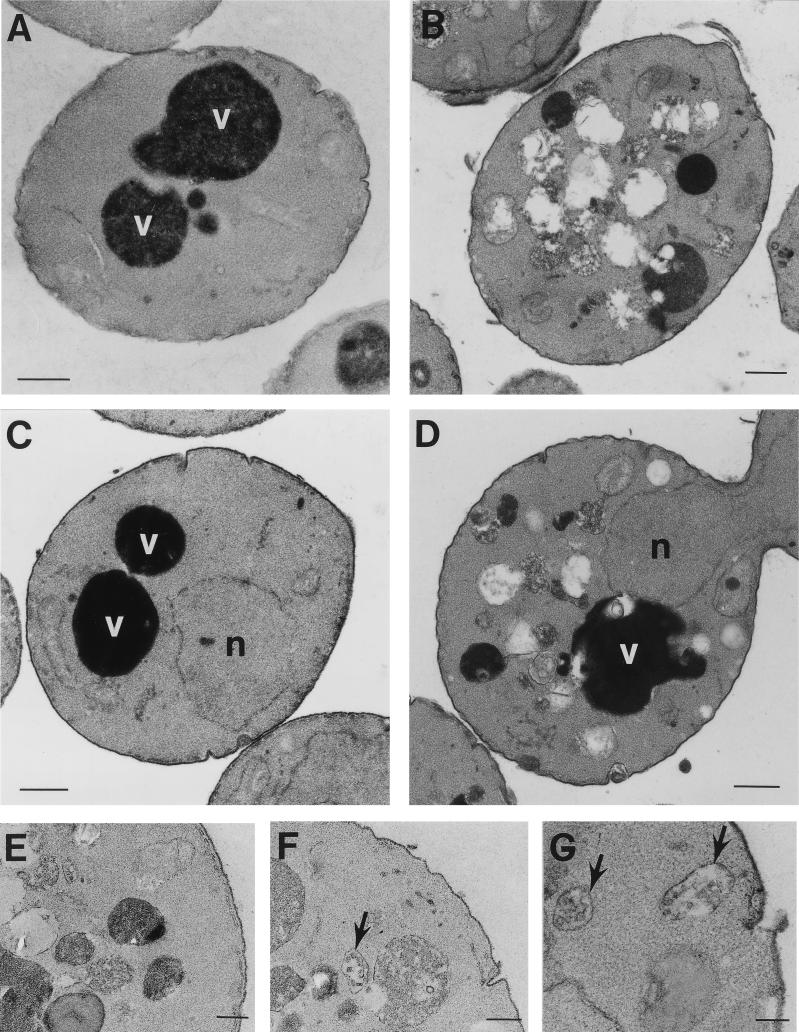
Whereas wild-type cells normally have one to three large, electron-dense vacuoles and a cytoplasm relatively free of membrane compartments (Fig. 2A), ultrastructural examination of vps41/vam2Δ (54, 62), ypt7/vam4Δ (86), vam6/vps39Δ (54), and vam3Δ (77, 83) mutants revealed an absence of normal vacuoles. By EM these deletion mutants were shown to contain numerous aberrant membrane compartments that were 200 to 600 nm in diameter. Examination of the vam7Δ mutant by EM revealed that it displays an aberrant morphology (Fig. 2B). Prominent vacuoles were not observed in vam7Δ cells. Instead, vam7Δ cells accumulated numerous smaller, membrane-enclosed compartments.
FIG. 2.
Ultrastructural analysis of vam7Δ and vam7tsf mutants. A cross-section of (A) wild-type (SEY6210) and (B) vam7Δ cells grown at 30°C viewed by EM. Mutant cells containing the vam7tsf allele (TKSY43 plus pTKS43ts-167) were incubated at the permissive temperature (26°C) or the nonpermissive temperature (38°C) for 3 h (C and D, respectively) prior to preparation for EM analysis. vam7tsf cells at the nonpermissive temperature accumulated novel compartments in addition to a prominent electron-dense vacuolar compartment. Enlargements of novel compartments (E) accumulated after 3 h at the nonpermissive temperature, including those that resemble multi-vesicular bodies (F and G, arrows) also seen in vam3tsf and vps41tsf mutants. n, nucleus; v, vacuole. Bars: A to D, 500 nm; E to G, 200 nm.
To assess the immediate consequences of inactivating Vam7p on vacuolar morphology, vam7tsf cells were examined by EM. Previous studies have determined that the vps41tsf (19) and vam3tsf mutants (20) maintained relatively intact vacuoles following 3 h at the nonpermissive temperature. Thus, it appeared that the structural integrity of the vacuole was retained, suggesting that the primary functions of these gene products are not in vacuolar maintenance. Additionally, these mutants accumulated numerous small, novel membrane compartments, which may represent unfused transport intermediates. Inactivation of the vam7tsf gene product also resulted in the accumulation of aberrant membranous compartments that were similar to compartments that accumulated in the vam7Δ mutant. After 3 h at the nonpermissive temperature, the vam7tsf mutant contained numerous clusters of novel membranous compartments (Fig. 2D and E). Interestingly, some of these compartments appeared to resemble multivesicular bodies (Fig. 2F and G) similar to those found in vam3tsf (20) and vps41tsf (19) mutants. Furthermore, after 3 h at the nonpermissive temperature, intact vacuoles still remained, indicating that vacuolar integrity was not compromised. Importantly, the morphology of the vam7tsf mutant incubated at the permissive temperature (Fig. 2C) resembled that of wild-type cells. The results of these phenotypic analyses are consistent with the VAM7 gene product functioning in vacuolar delivery of biosynthetic cargoes.
Vam7p associates with vacuolar membranes.
To identify the VAM7 gene product, a polyclonal antiserum was raised against a fusion protein containing GST fused to the carboxy-terminal 182 amino acids of Vam7p. The resulting antiserum was affinity purified and specifically recognized a protein of ca. 35-kDa in wild-type cells that was not observed with preimmune sera or in vam7Δ cells (data not shown). This closely corresponds to the predicted molecular mass of 38.6 kDa for the product of the VAM7 open reading frame.
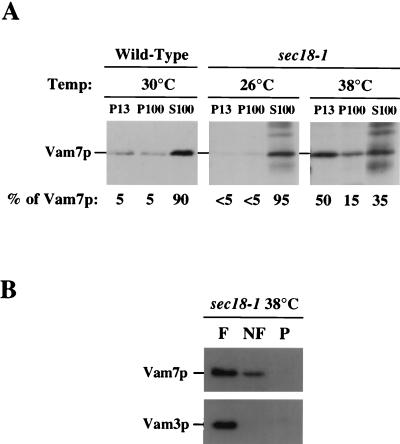
The localization of Vam7p was determined by using differential centrifugation with wild-type cells. Lysates from wild-type cells were initially fractionated by centrifugation at 13,000 × g to generate the pellet (P13) and supernatant (S13) fractions. The S13 fraction was then subjected to another centrifugation at 100,000 × g to produce additional pellet (P100) and supernatant (S100) fractions. Vam7p was predominantly found in the S100 fraction (ca. 90% of total Vam7p; Fig. 3A), suggesting that Vam7p is largely a soluble protein. Small amounts (ca. 5% each) of Vam7p were found in the P13 and P100 fractions, consistent with Vam7p associating with a membrane fraction or a large protein complex.
FIG. 3.
Subcellular localization of the VAM7 gene product. (A) SEY6210 (wild-type) spheroplasts (5 OD600 units) were lysed and fractionated by differential centrifugation (see Materials and Methods), generating P13, P100, and S100 fractions. Fractions were TCA-precipitated, and proteins from 2 OD600 units were resolved by SDS-PAGE. Resolved proteins were then transferred to nitrocellulose, immunoblotted with anti-Vam7p antibody, and visualized by ECL fluorography. EGY118-110 (sec18-1) spheroplasts were labeled with [35S]cysteine and [35S]methionine at the permissive temperature (26°C) for 30 min and chased for 15 min. Cells then remained at the permissive temperature or were shifted to the nonpermissive temperature (38°C) for an additional 30 min. After the temperature shift, cells were harvested and fractionated as described above. TCA-precipitated lysates were immunoprecipitated with anti-Vam7p antibody, revolved by SDS-PAGE, and visualized by autoradiography. (B) Accudenz gradient analysis of Vam7p from the P13 fraction of inactivated sec18-1 cells. The P13 fraction from EGY118-110 (sec18-1) cells incubated at the nonpermissive temperature (38°C) was resuspended in lysis buffer containing 60% Accudenz and loaded at the bottom of a 60%–55%–35% Accudenz step gradient. After centrifugation at 200,000 × g for 14 h, floating (F), nonfloating (NF), and pellet (P) fractions were harvested and analyzed by Western blotting.
A previous report identified a carboxy-terminal domain in Vam7p that is homologous to the coiled-coil domain of SNAP-25 (85). In neuronal cells, NSF catalyzes the dissociation of SNARE complexes which include SNAP-25 (74–76). In yeast cells, the NSF homolog Sec18p also has been shown to mediate the dissociation of SNARE complexes, as inactivation of the conditional sec18-1 allele results in the stabilization of SNARE complexes (15, 74, 80). To determine if Vam7p associates with membranes in a Sec18p-dependent manner, subcellular fractionation was performed on the sec18-1 strain under conditions where Sec18-1p was inactive (nonpermissive temperature and absence of Mg2+-ATP). The distribution of Vam7p in sec18-1 cells at the permissive temperature (26°C) was nearly identical to its distribution in wild-type cells (Fig. 3A). In striking contrast, when sec18-1 cells were incubated at the nonpermissive temperature (38°C) for 30 min, ca. 50% of Vam7p shifted from the S100 fraction to the P13 fraction.
It is possible that the sec18-1-induced shift of Vam7p to the P13 fraction may be due to protein aggregation and not membrane association. Thus, gradient analysis was performed to confirm the Vam7p membrane association (3). The P13 fraction from sec18-1 cells incubated at the nonpermissive temperature was resuspended with 60% Accudenz lysis buffer and loaded on the bottom of an Accudenz step gradient. After 14 h of centrifugation, samples were divided into three fractions. The top fraction (F) contained membrane-associated floating material, while the load fraction contained nonfloating (NF) material. The pellet (P) fraction corresponded to pelletable material not associated with membranes. Identification of Vam7p by Western blotting determined that the majority of Vam7p from the P13 fraction was at the top of the gradient (Fig. 3B). Additionally, the vacuolar transmembrane protein, Vam3p, was found to float to the top of the gradient. Thus, the inactivation of Sec18p enhances the association of Vam7p with a membrane compartment.
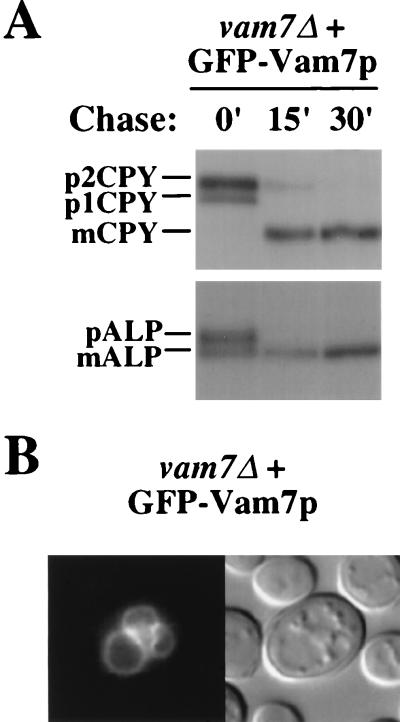
As an alternative approach to determine the subcellular localization of Vam7p, GFP was fused to the amino terminus of Vam7p. Pulse-chase analysis of vam7Δ with a single-copy (CEN) plasmid containing the coding sequence for the GFP-Vam7p fusion revealed that ALP and CPY matured with wild-type kinetics (Fig. 4A), indicating that the GFP-Vam7p fusion complements the vam7Δ phenotype. Additionally, GFP-Vam7p distributed identically to chromosomal Vam7p by subcellular fractionation with approximately twofold-higher expression compared to native Vam7p (data not shown). By fluorescence microscopy, the GFP-Vam7p fusion protein was detected both in the cytoplasm and on vacuolar membranes (Fig. 4B). This result suggests that Vam7p peripherally associates with vacuolar membranes possibly through transient interactions, which may be partially disrupted when cells are lysed during fractionation experiments.
FIG. 4.
Localization of the GFP-Vam7p fusion protein. (A) Complementation of the CPY and ALP sorting defects of vam7Δ mutants with the GFP-Vam7p fusion protein. TKSY43 (vam7Δ) cells harboring a single-copy (CEN) plasmid (pTKS35) encoding the GFP-Vam7p fusion protein were labeled with [35S]cysteine and [35S]methionine for 10 min at 30°C and chased with unlabeled cysteine and methionine for the indicated times. CPY and ALP were immunoprecipitated and then visualized by SDS-PAGE and autoradiography. (B) Nomarski optics image (left panel) and fluorescence localization (right panel) of GFP-Vam7p in TKSY43 (vam7Δ) cells containing the GFP-Vam7p fusion protein (pTKS35).
VAM7 interacts with the putative vacuolar t-SNARE VAM3.
The VAM3 gene product has been shown to be required at a late step in the transport of multiple proteins to the vacuole. Similar to vam7tsf mutants, a vam3tsf strain displays defects in the transport of ALP, CPS, CPY, and API to the vacuole (20). Vam3p localizes to vacuolar membranes, and its homology to syntaxin suggests that it functions as a t-SNARE in protein transport to the vacuole (20, 77, 83). Similarities in phenotypes between vam3 and vam7 mutants together with their homologies to syntaxin and SNAP-25, respectively, suggest that they may function together at a common step in the VPS pathway.
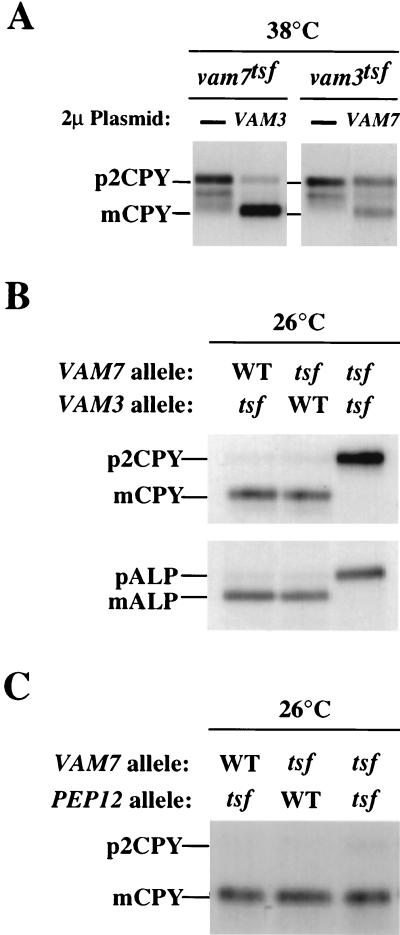
To determine whether VAM3 and VAM7 genetically interact, multicopy plasmids (2μm) overexpressing Vam3p or Vam7p were transformed into vam7tsf or vam3tsf cells, respectively. These strains were then analyzed by pulse-chase immunoprecipitation assays for suppression of protein transport defects at the nonpermissive temperature. vam3tsf or vam7tsf cells transformed with 2μm vector alone exhibited the same protein-sorting defects at the nonpermissive temperature as had the vam3tsf or vam7tsf cells (Fig. 5A). Overexpression of Vam7p partially suppressed the CPY missorting defects in vam3tsf cells, while overexpression of Vam3p significantly rescued the temperature-conditional CPY sorting defect of the vam7tsf mutant. Furthermore, a strain harboring both the vam7tsf and vam3tsf mutations was produced to test for synthetic vacuolar-protein-sorting defects at 26°C, the permissive temperature for the individual tsf mutants. Pulse-chase analysis of the double mutant revealed that while single vam3tsf or vam7tsf mutants displayed no defects in the maturation of both ALP and CPY, the double vam3tsf vam7tsf mutant displayed a strong defect in the processing of ALP and CPY (Fig. 5B). As a control experiment, we conducted genetic tests with VAM7 and PEP12. The PEP12 gene product functions as an endosomal t-SNARE required for the transport of CPY from the trans-Golgi to the endosome (9, 15, 19). A pep12tsf vam7tsf double mutant did not display a synthetic CPY sorting defect (Fig. 5C). Additionally, overexpression of Pep12p did not suppress the CPY transport defect of the vam7tsf mutant (data not shown). These results show that VAM3 and VAM7 specifically interact and function together at a late stage of the vacuolar-protein-sorting pathway. As the vam7tsf mutant contains a mutation in the heptad repeat homologous to SNAP-25 (leucine 287 to proline), it is possible that the phenotypes associated with the double vam7tsf vam3tsf mutant are due to an abrogation in the coiled-coil interaction between Vam7tsfp and Vam3tsfp.
FIG. 5.
Genetic interactions between VAM7 and VAM3. (A) Suppression of vam7tsf and vam3tsf mutant cells. TKSY43 (vam7Δ) or TDY2 (vam3Δ) containing tsf alleles of either vam7 (pTKS43ts-167) or vam3 (pVAM3.6-416), respectively, were transformed with a multicopy vector (2μm) containing no insert, VAM7 (pTKS23), or VAM3 (pVAM3.426) as indicated. Cells grown at 26°C were harvested, incubated at the permissive or nonpermissive temperature for 10 min, and labeled with [35S]cysteine and [35S]methionine for 10 min. After the labeling, cells were chased with unlabeled cysteine and methionine for 30 min and harvested by TCA precipitation. Lysates from these cells were immunoprecipitated with the indicated antibodies and analyzed as previously described. (B) Synthetic interactions between vam7tsf and vam3tsf. Double-mutant strains (vam3Δ vam7Δ) harboring the vam3tsf allele (pTKS30 and pVAM3.6-416), the vam7tsf allele (pTKS43ts-167 and pVAM3-416), or both tsf alleles (pTK43ts-167 and pVAM3.6-416) were incubated at 26°C for 10 min and labeled with [35S]cysteine and [35S]methionine for an additional 10 min. After the labeling, the cells were chased with unlabeled cysteine and methionine for 45 min, harvested, and assayed for vacuolar protein sorting as described above. (C) Double-mutant cells (pep12-60 vam7Δ) were transformed with CEN plasmids containing VAM7 (pTKS30) or the vam7tsf allele (pTKS43ts-167). Single- and double-mutant strains were assayed for CPY sorting as described above.
Vam3p coimmunoprecipitates with Vam7p.
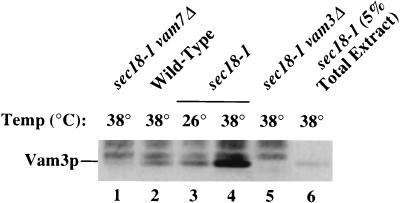
Our genetic findings suggest that Vam7p physically interacts with Vam3p in a complex on vacuolar membranes. In addition, the data suggest that this interaction may be enhanced by inactivation of the SEC18 gene product. In order to determine whether Vam7p may be in a complex that contains Vam3p, Vam7p was immunoprecipitated under native conditions with detergent extracts from wild-type and sec18-1 yeast cells. After immune complexes were pelleted with protein A-Sepharose beads, bound protein was washed with 0.1% Triton X-100 and then eluted from the beads with sample buffer. Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by immunoblotting with anti-Vam3p or anti-Vam7p antibodies (Fig. 6). When compared to a control fraction (lane 6), ca. 10 to 15% of total Vam3p coimmunoprecipitated with Vam7p in wild-type cells incubated at 38°C or sec18-1 cells incubated at 26°C (lanes 2 and 3). In striking contrast, in sec18-1 cells incubated at 38°C, the amount of Vam3p that coimmunoprecipitated with Vam7p increased ca. fivefold (lane 4). Importantly, Vam3p did not coimmunoprecipitate with Vam7p in extracts made from sec18-1 vam7Δ or sec18-1 vam3Δ strains incubated at 38°C (lanes 1 and 5). The interaction between Vam7p and Vam3p was also specific, as Pep12p did not coimmunoprecipitate with Vam7p in extracts made from sec18-1 cells incubated at 38°C (data not shown). Consistent with results from the genetic studies, these coimmunoprecipitation experiments demonstrate that Vam7p is in a complex that contains Vam3p.
FIG. 6.
Native immunoprecipitation of Vam3p with Vam7p. Spheroplasts (5 OD600 equivalents) from the following strains were generated and incubated at either 26 or 38°C as indicated for 1 h: TKSY48 (sec18-1 vam7Δ), SEY6210 (wild type), EGY118-110 (sec18-1), and CBY42 (sec18-1 vam3Δ). Spheroplasts were then lysed with 0.5% Triton X-100 and incubated with anti-Vam7p antibody for 1 h at 4°C, followed by the addition of protein A-Sepharose for an additional 1 h. Bound immunocomplexes were washed with 0.1% Triton X-100 and eluted from beads with SDS-PAGE sample buffer. Two-OD600-unit samples were loaded onto SDS-PAGE gels, resolved by electrophoresis, and transferred to nitrocellulose paper. Vam3p and Vam7p were immunoblotted with anti-Vam3p and -Vam7p antibody, respectively, and visualized by ECL fluorography. Of the total sec18-1 38°C extract, 5% was TCA precipitated and loaded as an indication of the amount of total Vam3p bound to Vam7p.
Mutations in the PX domain of Vam7p result in synthetic defects with the vam3tsf mutant.
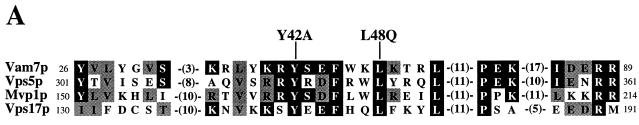
Recently, an extensive database search revealed that Vam7p contains a domain that is present in a number of proteins with diverse functions (61). This domain, termed the PX domain, characterizes a family of proteins that includes the p40phox and p47phox subunits of the NADPH oxidase complex, CPK-like phosphatidylinositol 3-kinases, and proteins involved in vesicular trafficking, including the SNX-1 family members (47) and the yeast proteins Mvp1p (22), Vps5p (37), and Vps17p (46). No function has been determined for this domain, nor has any requirement for the PX domain in a specific protein been identified. The region of the PX domain in Vam7p with highest homology to Vps5p, Mvp1p, and Vps17p spans amino acids 26 to 89 (Fig. 7A). To determine whether the PX domain is critical for Vam7p function, two point mutations in codons of conserved amino acids were made: tyrosine 42 to alanine (vam7Y42A) and leucine 48 to glutamine (vam7L48Q).
FIG. 7.
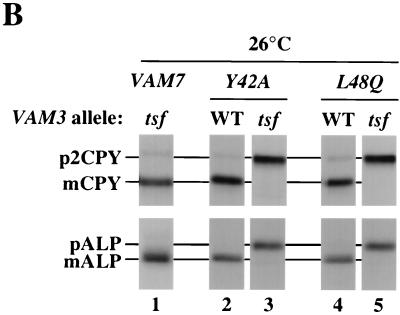
Vacuolar trafficking defects of VAM7 PX domain mutants with the vam3tsf mutation. (A) The PX domain of Vam7p aligned with the PX domains of three yeast proteins involved in vesicular trafficking are shown. The regions of highest homology between the four PX domains are indicated by the numbers on the right and left of the sequence. Gap lengths are indicated in parentheses. Amino acid residues of at least two other proteins that are conserved with the PX domain of Vam7p are highlighted in black. Amino acid residues that are similar to Vam7p PX domain residues are indicated in gray. The amino acids that were changed (Y42A and L48Q) are denoted in boldface above the Vam7p sequence. (B) Synthetic interactions between vam7 PX and vam3tsf mutations. Standard pulse-chase analysis was conducted on vam3Δ vam7Δ double mutants harboring CEN plasmids containing wild-type VAM7 (pTKS30), mutant VAM7 encoding tyrosine 42-to-alanine (Y42A) or leucine 48-to-glutamine (L48Q) mutations, and wild-type VAM3 (pVAM3.416) or vam3tsf (pVAM3-6.416) alleles. Double mutants were labeled for 10 min and chased for 30 min at 26°C.
To determine whether point mutations in the PX domain affect Vam7p function, the vam7Y42A and vam7L48Q mutants were assayed for the trafficking of CPY and ALP to the vacuole by pulse-chase immunoprecipitation experiments as described above (Fig. 7B). Both the vam7Y42A and vam7L48Q mutants did not display any significant defects in ALP or CPY trafficking after 30 min of chase at 26°C (lanes 2 and 4, respectively). However, when the PX mutations were combined with the vam3tsf mutation, a strong synthetic phenotype was observed. Both vam7Y42A vam3tsf and vam7L48Q vam3tsf double mutants displayed a complete block in ALP and CPY trafficking after 30 min of chase at 26°C (lanes 3 and 5, respectively). In contrast, the vam3tsf mutant at 26°C displayed normal, rapid processing of its vacuolar hydrolases (lane 1). As the mutations in the vam7tsf allele map outside of the PX domain, results from these experiments suggest that the mutations in the PX domain also cause defects in Vam7p function, possibly by altering its interaction with Vam3p or some other component of the SNARE complex.
DISCUSSION
The experiments described here investigate the role of Vam7p in vacuolar protein sorting. Previous reports suggested that Vam7p functions in vacuolar morphogenesis (82) and identified an amino-terminal PX domain of unknown function (61) and a carboxy-terminal heptad repeat with homology to the coiled-coil of SNAP-25 (85). Analysis of the vam7tsf mutant (double mutant; leucine 134 to proline and leucine 287 to proline) revealed that, like the vacuolar syntaxin homolog Vam3p (20), Vam7p functions in the delivery of multiple proteins that all converge at the vacuole via distinct biosynthetic pathways. Furthermore, examination of the vam7tsf mutant incubated at the nonpermissive temperature by EM revealed the accumulation of numerous small aberrant membranous compartments, suggesting that the docking and/or fusion of transport intermediates to the vacuole was impaired. Localization experiments determined that a portion of Vam7p associates with vacuolar membranes. The large cytoplasmic pool and the absence of any consensus palmitoylation motif or shift in the electrophoretic mobility of the membrane-associated fraction suggest that Vam7p is not modified with palmitate as is SNAP-25 (33). However, transient palmitoylation cannot be ruled out. Genetic studies demonstrated that VAM7 functionally interacts with VAM3. Consistent with these observations, Vam7p was found to physically associate in a complex that contains Vam3p and this interaction was enhanced by inactivation of Sec18p. This suggests that Vam7p may be a component of a t-SNARE complex analogous to syntaxin–SNAP-25 complexes at the plasma membrane of neuronal cells. Lastly, mutations in the PX domain of Vam7p resulted in synthetic protein sorting defects when expressed in the vam3tsf mutant. Together, the data presented here suggest that Vam7p functions in conjunction with Vam3p, possibly as part of a t-SNARE complex on vacuolar membranes to mediate the docking and fusion of multiple transport intermediates from distinct biosynthetic pathways.
Vam7p regulates a late step in vacuolar protein trafficking.
Initial analysis of the VAM7 gene suggested it functions in the maintenance of vacuolar morphology but not in vacuolar protein sorting (84). Examination of vam7Δ mutants revealed that these mutants lack normal vacuoles and instead accumulate numerous abnormal membrane compartments. Analysis of CPY by Western blotting in vam7Δ mutants found CPY predominantly in its mature form (82). However, steady-state analyses of this kind can be misleading, as slow processing of vacuolar precursors can occur in nonvacuolar compartments. For example, nonvacuolar processing has been observed in Class E vps mutants which develop a proteolytically competent prevacuolar compartment (2, 59). Pulse-chase and EM analysis of the vam7tsf mutant suggests that Vam7p functions primarily in protein trafficking. After temperature inactivation, while vacuolar integrity and inheritance remain normal, newly synthesized p2CPY immediately accumulates and is slowly processed to the mature form only after prolonged chase. Additionally, the bulk of p2CPY is not secreted, indicating that it is sorted away from the late secretory pathway at the trans-Golgi but may accumulate in transport intermediates, such as prevacuolar endosomes. This phenotype has also been observed in vps41tsf (19) and vam3tsf mutants (20). The common phenotypes associated with these vam and vps mutants suggest that they function together to regulate a late step in vacuolar transport.
Vam7p as a component of a vacuolar SNARE complex.
Sequence similarities between some of the VPS gene products and proteins implicated in the docking of transport vesicles to target membranes in mammalian systems suggest that they share conserved functions. The identification of syntaxin homologs, Sec1p family members, and Rab GTPases that are required in vacuolar protein sorting implies that transport within the VPS pathway is mediated by mechanisms common to other vesicle-mediated transport steps. In neuronal cells, docking is mediated by the pairing of synaptobrevin (v-SNARE) on the vesicle membrane with the t-SNAREs syntaxin and SNAP-25 on the target membrane to form the stable complex necessary for fusion events (31, 32, 58, 76). Analysis of the Vam7p sequence identified a region (amino acids 253 to 313) predicted to form coiled-coil interactions that shares homology to SNAP-25 (85), suggesting that it may function in a similar manner.
In addition to conserved coiled-coil domains, SNAP-25-like molecules are defined by a distinct set of characteristics. SNAP-25 family members, including the yeast plasma membrane t-SNARE Sec9p, are characterized by their role in docking and fusion, localization at target membranes, and interactions with SNARE molecules (12, 13, 31, 32, 69, 76). Additionally, physical interaction between SNAP-25 and syntaxin is enhanced in the presence of nonhydrolyzable ATP analogs or by inactivation of NSF or Sec18p (13, 32, 75, 76). As shown here, Vam7p fulfills these criteria. Phenotypic analysis of the vam7tsf mutant identified the intracellular accumulation of vacuolar precursors and aberrant membranous compartments, suggesting that the targeting and/or fusion of transport intermediates was disrupted. A fraction of Vam7p localizes on vacuolar membranes. Vam7p interacts with the vacuolar t-SNARE Vam3p, suggesting that together they may form part of a t-SNARE complex. Genetic and physical analyses further determined that inactivation of the SEC18 gene product resulted in enhanced association of Vam7p with a Vam3p-containing complex. Based on these observations, it can be proposed that the Vam7p functions like SNAP-25 in a vacuole-specific SNARE complex.
A previous study of Sec18p function in the VPS pathway provided evidence for a late stage in the delivery of CPY from the Golgi to the vacuole that does not require Sec18p activity (25). This study identified the processing of a small kinetic pool of p2CPY to mCPY that occurred even after temperature inactivation of the sec18-1 gene product. Homology between components of known SNARE complexes to VPS gene products and results presented here suggest that Sec18p may regulate a vacuolar SNARE complex that is required for biosynthetic transport. One possible explanation for this apparent discrepancy is suggested by results from in vitro studies of vacuole-to-vacuole fusion. The in vitro vacuole-to-vacuole homotypic fusion reaction exhibits requirements for the same transport factors as those that have been shown to function in biosynthetic trafficking to the vacuole, including Vam3p (55), Ypt7p (26, 50), and Sec18p (27, 51). Although vacuole-to-vacuole homotypic fusion and biosynthetic transport intermediate-to-vacuole heterotypic fusion are not necessarily considered to be equivalent reactions, the mechanisms utilized in docking and fusion may be similar. The in vitro studies have suggested that ATP hydrolysis by Sec18p functions in activating SNARE components at a step prior to docking (27, 51, 55). Thus, it is possible that the Sec18p-independent delivery of a small pool of p2CPY to the vacuole occurred via SNARE complexes that had been activated prior to shifting the sec18-1 mutant cells to the nonpermissive temperature.
To date, no v-SNARE that mediates vacuolar protein trafficking at a late step has been identified. In vitro reconstitution of vacuolar inheritance by homotypic vacuole-to-vacuole fusion assays has identified the v-SNARE Nyv1p gene product as functioning with Vam3p in vacuole-to-vacuole fusion (55, 80). The v-SNARE Vti1p has been suggested to function in the transport of CPY from the Golgi to the endosome (48, 81). Interestingly, Vti1p has also been recently reported to coimmunoprecipitate with Vam3p (34). Thus, Vti1p may be the v-SNARE that functions with Vam7p and Vam3p in targeting biosynthetic traffic to the vacuole. Further genetic and biochemical studies with Vti1p, Vam3p, and Vam7p will be necessary to demonstrate the coassembly of all three proteins in a stable SNARE complex.
The PX domain is required for Vam7p function.
The PX domain is common to a diverse set of proteins, including two subunits of the NADPH oxidase complex, p40phox and p47phox, the CPK family of phosphatidylinositol 3-kinases, and phospholipase D proteins (61). The PX domains of these family members range in size, with highest homology in a region spanning ca. 60 amino acids. PX domains commonly contain a short proline-rich region, although it is not found in Vam7p. Interestingly, PX domains are also prominent in proteins implicated in vesicular trafficking, including the mammalian protein SNX1, which has been shown to interact with the cytosolic tail of the EGF receptor (47), and the yeast proteins Vps5p, Vps17p, Mvp1p, and Vam7p. The role of the PX domain has not been defined. However, it has been suggested to function in protein-protein interactions. Consistent with this idea, Vps5p and Vps17p have been shown to physically interact (37), although it is not known if their PX domains mediate this interaction. Synthetic interactions with the vam3tsf mutation suggest that the PX domain of Vam7p may contribute to or regulate the interaction between these two proteins or it may mediate interactions with other components of the docking and fusion machinery. Further genetic and physical interaction studies will provide a greater understanding of PX domain function in Vam7p and possibly other PX domain-containing proteins.
Model for Vam7p as a SNAP-25-like molecule in the docking and fusion of transport intermediates to the vacuole.
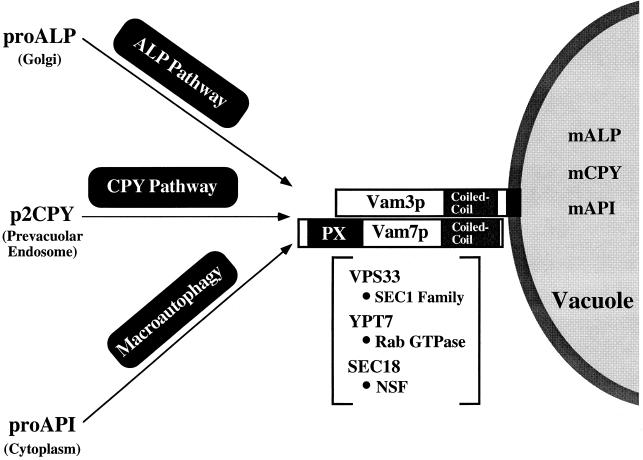
Based on the genetic and biochemical studies described here and an examination of protein transport in other systems, the specific roles for gene products involved in the late events of vacuolar protein sorting can be proposed (Fig. 8). Docking and/or fusion of prevacuolar transport intermediates from at least three distinct biosynthetic pathways to the vacuole requires the SNAP-25 family member Vam7p and the syntaxin homolog Vam3p. These interactions may be regulated by the Rab GTPase, Ypt7p (84, 86), and the VPS33 gene product, the SEC1 homolog shown to genetically interact with VAM3 (20). Lastly, transport to the vacuole may require Sec18p to activate and dissociate SNARE complexes, allowing for the docking and/or fusion of transport intermediates with the vacuole. Further investigation of components of the Vam3p SNARE complex will be required to understand the precise function of Vam7p. Screens to identify proteins that physically and genetically interact with the PX domain of Vam7p, such as the class C VPS genes that also function late in the pathway (65), will provide insight into the function of this domain and its involvement in docking and/or fusion mechanisms. Results from these examinations should help in understanding the molecular mechanisms that direct the specific docking and fusion of transport intermediates with the appropriate target membrane in both yeast and other eukaryotes.
FIG. 8.
Model depicting genes involved in docking and/or fusion at the vacuole. Three vacuolar proteins proceed to the vacuole via distinct biosynthetic pathways, indicated above the arrows. Vam7p and Vam3p function as a t-SNARE complex in the docking and/or fusion of transport intermediates to the vacuole. Other genes that are proposed to regulate the docking and fusion of transport intermediates to the vacuole are enclosed by brackets. The homologies between these VPS genes and proteins implicated in vesicular docking and fusion are indicated (•). The PX domain and coiled-coil domains of Vam7p and Vam3p are also schematically diagrammed.
ACKNOWLEDGMENTS
We thank Y. Wada for generously providing plasmids and strains and for sharing unpublished results; D. Klionsky for providing the API antiserum; G. Odorizzi and R. Aroian for assistance with the fluorescence microscopy and Delta Vision software; C. Hofeditz for EM analysis (Core B headed by M. Farquhar of Program Project Grant CA58689); E. Gaynor for providing strains; and members of the Emr lab, especially B. Wendland, C. Burd, and M. Babst, for reagents, helpful discussions and critical reading of the manuscript.
This work was supported by a grant from the NIH (CA58689 to S.D.E.). T.K.S. is a member of the Biomedical Sciences Graduate Program. S.D.E. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Baba M, Osumi M, Scott S V, Klionsky D J, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst M, Sato T K, Banta L M, Emr S D. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babst M, Wendland B, Estepa E J, Emr S D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosomal function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bankaitis V A, Johnson L M, Emr S D. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banta L M, Robinson J S, Klionsky D J, Emr S D. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banta L M, Vida T A, Herman P K, Emr S D. Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol Cell Biol. 1990;10:4638–4649. doi: 10.1128/mcb.10.9.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumert M, Maycox P R, Navone F, DeCamilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO J. 1989;8:379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becherer K A, Rieder S, Emr S D, Jones E W. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett M K, Garcia-Arraras J E, Elferink L A, Peterson K, Fleming A M, Hazuka C D, Scheller R H. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 11.Bennett M K, Scheller R H. The molecular machinery for secretion is conserved from yeast to neurons. Proc Natl Acad Sci USA. 1993;90:2559–2563. doi: 10.1073/pnas.90.7.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasi J, Chapman E R, Link E, Binz T, Yamasaki S, De Camilli P, Sudhof T C, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- 13.Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 14.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–379. [Google Scholar]
- 15.Burd C G, Peterson M, Cowles C R, Emr S D. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clary D O, Griff I C, Rothman J E. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- 17.Cowles C R, Emr S D, Horazdovsky B F. Mutations in the VPS45 gene, a SEC1 homologue, result in vacuolar protein sorting defects and accumulation of membrane vesicles. J Cell Sci. 1994;107:3449–3459. doi: 10.1242/jcs.107.12.3449. [DOI] [PubMed] [Google Scholar]
- 18.Cowles C R, Odorizzi G, Payne G S, Emr S D. The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell. 1997;91:109–118. doi: 10.1016/s0092-8674(01)80013-1. [DOI] [PubMed] [Google Scholar]
- 19.Cowles C R, Snyder W B, Burd C G, Emr S D. An alternative Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darsow T, Rieder S E, Emr S D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eakle K A, Bernstein M, Emr S D. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ekena K, Stevens T H. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol Cell Biol. 1995;15:1671–1678. doi: 10.1128/mcb.15.3.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- 24.Gaynor E C, te Heesen S, Graham T R, Aebi M, Emr S D. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham T R, Emr S D. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas A, Wickner W. Homotypic vacuole fusion requires Sec17p (yeast alpha-SNAP) and Sec18p (yeast NSF) EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D L. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1988. [Google Scholar]
- 30.Hay J C, Scheller R H. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Sudhof T C, Niemann H. Synaptic vesicle membrane fusion complex: action of clostridial neurotoxins on assembly. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J. 1994;14:2317–2325. doi: 10.1002/j.1460-2075.1995.tb07226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess D T, Slater T M, Wilson M C, Skene J H. The 25 kDa synaptosomal-associated protein SNAP-25 is the major methionine-rich polypeptide in rapid axonal transport and a major substrate for palmitoylation in adult CNS. J Neurosci. 1992;12:4634–4641. doi: 10.1523/JNEUROSCI.12-12-04634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holthuis J C M, Nichols B J, Dhruvakumar S, Pelham H R B. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horazdovsky B F, Busch G R, Emr S D. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297–1309. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horazdovsky B F, Emr S D. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem. 1993;268:4953–4962. [PubMed] [Google Scholar]
- 37.Horazdovsky B F, Seaman M N J, Mclaughlin S A, Yoon S-H, Emr S D. A sorting nexin-1 homologue, Vps5p, forms a complex with Vps17p and is required for recycling the vacuolar protein-sorting receptor. Mol Biol Cell. 1997;8:1529–1541. doi: 10.1091/mbc.8.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horton R M, Cai Z L, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 39.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones E W. Proteinase mutants of Saccharomyces cerevisiae. Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones E W, Webb G C, Hiller M A. Biogenesis and function of the yeast vacuole. In: Pringle J R, Broach J R, Jones E W, editors. Molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 363–470. [Google Scholar]
- 42.Klionsky D J, Banta L M, Emr S D. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klionsky D J, Cueva R, Yaver D S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klionsky D J, Emr S D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klionsky D J, Herman P K, Emr S D. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohrer K, Emr S D. The yeast VPS17 gene encodes a membrane associated protein required for the sorting of soluble vacuolar hydrolases. J Biol Chem. 1993;268:559–569. [PubMed] [Google Scholar]
- 47.Kurten R C, Cadena D L, Gill G N. Enhanced degradation of EGF receptors by a sorting nexin, SNX1. Science. 1996;272:1008–1010. doi: 10.1126/science.272.5264.1008. [DOI] [PubMed] [Google Scholar]
- 48.Lupashin V V, Waters M G. t-SNARE activation through transient interaction with a Rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- 49.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 50.Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136:307–317. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- 52.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 53.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 54.Nakamura N, Hirata A, Ohsumi Y, Wada Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- 55.Nichols B J, Ungermann C, Pelham H R B, Wickner W T, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- 56.Novick P, Brennwald P. Friends and family: the role of Rab GTPases in vesicular traffic. Cell. 1993;75:597–601. doi: 10.1016/0092-8674(93)90478-9. [DOI] [PubMed] [Google Scholar]
- 57.Paravicini G, Horazdovsky B F, Emr S D. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35 null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol Biol Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pevsner J, Hsu S C, Braun J E, Calakos N, Ting A E, Bennett M K, Scheller R H. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- 59.Piper R C, Cooper A A, Yang H, Stevens T H. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J Cell Biol. 1995;131:603–618. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piper R C, Whitters E A, Stevens T H. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Eur J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- 61.Ponting C P. Novel domains in NADPH oxidase subunits, sorting nexins, and PtdIns 3-kinases: binding partners of SH3 domains? Protein Sci. 1996;5:2353–2357. doi: 10.1002/pro.5560051122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radisky D C, Snyder W B, Emr S D, Kaplan J. Identification of VPS41, a gene required for vacuolar trafficking and the assembly of the yeast high affinity iron transport system. Proc Natl Acad Sci USA. 1997;94:5662–5666. doi: 10.1073/pnas.94.11.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rieder S E, Banta L M, Kohrer K, McCaffery J M, Emr S D. Multilamellar endosome like compartment accumulates in the yeast vps28. Mol Biol Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rieder S E, Emr S D. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a.Robinson J S, Klionsky D J, Banta L M, Emr S D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rothman J E. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 67.Rothman J E, Sollner T H. Throttles and dampers: controlling the engine of membrane fusion. Science. 1997;276:1212–1213. doi: 10.1126/science.276.5316.1212. [DOI] [PubMed] [Google Scholar]
- 68.Rothman J H, Stevens T H. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 69.Schiavo G, Rosetto O, Catsicas S, Polverino de Laureto P, Das Gupta B R, Benfenati F, Montecucco C. Identification of the nerve terminal targets of botulinum neurotoxin serotypes A, D, and E. J Biol Chem. 1993;268:23784–23787. [PubMed] [Google Scholar]
- 70.Scott S V, Baba M, Ohsumi Y, Klionsky D J. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scott S V, Hefner-Gravink A, Morano K A, Noda T, Ohsumi Y, Klionsky D J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sherman F, Fink G R, Lawrence L W. Methods in yeast genetics: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1979. [Google Scholar]
- 73.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sogaard M, Tani K, Ye R R, Geromanos S, Tempst P, Kirchhausen T, Rothman J E, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- 75.Sollner T, Bennett M K, Whiteheart S W, Scheller R H, Rothman J E. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 76.Sollner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 77.Srivastava A, Jones E W. Pth1/Vam3p is the syntaxin homolog at the vacuolar membrane of Saccharomyces cerevisiae required for the delivery of vacuolar hydrolases. Genetics. 1998;148:85–98. doi: 10.1093/genetics/148.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sudhof T C. The synaptic vesicle cycle: a cascade of protein protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 79.Trimble W S, Cowan D M, Scheller R H. VAMP-1: a synaptic vesicle-associated integral membrane protein. Proc Natl Acad Sci USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ungermann C, Nichols B J, Pelham H R B, Wickner W. A vacuolar v-t-SNARE complex, the predominant form in vivo and on isolated vacuoles, is disassembled and activated for docking and fusion. J Cell Biol. 1998;140:61–69. doi: 10.1083/jcb.140.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.von Mollard G F, Nothwehr S F, Stevens T H. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wada Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. II. VAM7, a gene for regulating morphogenic assembly of the vacuoles. J Biol Chem. 1992;267:18671–18675. [PubMed] [Google Scholar]
- 83.Wada Y, Nakamura N, Ohsumi Y, Hirata A. Vam3p, a new member of syntaxin related proteins, is required for vacuolar assembly in the yeast Saccharomyces cerevisiae. J Cell Sci. 1997;110:1299–1306. doi: 10.1242/jcs.110.11.1299. [DOI] [PubMed] [Google Scholar]
- 84.Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- 85.Weimbs T, Low S H, Chapin S J, Mostov K E, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wichmann H, Hengst L, Gallwitz D. Endocytosis in yeast: evidence for the involvement of a small GTP binding protein (Ypt7p) Cell. 1992;71:1131–1142. doi: 10.1016/s0092-8674(05)80062-5. [DOI] [PubMed] [Google Scholar]
- 87.Yon J, Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989;17:4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]