Abstract
Despite the improvements in clinical outcomes for DLBCL, a significant proportion of patients still face challenges with refractory/relapsed (R/R) disease after receiving first-line R-CHOP treatment. To further elucidate the underlying mechanism of R/R disease and to develop methods for identifying patients at risk of early disease progression, we integrated clinical, genetic and transcriptomic data derived from 2805 R-CHOP-treated patients from seven independent cohorts. Among these, 887 patients exhibited R/R disease within two years (poor outcome), and 1918 patients remained in remission at two years (good outcome). Our analysis identified four preferentially mutated genes (TP53, MYD88, SPEN, MYC) in the untreated (diagnostic) tumor samples from patients with poor outcomes. Furthermore, transcriptomic analysis revealed a distinct gene expression pattern linked to poor outcomes, affecting pathways involved in cell adhesion/migration, T-cell activation/regulation, PI3K, and NF-κB signaling. Moreover, we developed and validated a 24-gene expression score as an independent prognostic predictor for treatment outcomes. This score also demonstrated efficacy in further stratifying high-risk patients when integrated with existing genetic or cell-of-origin subtypes, including the unclassified cases in these models. Finally, based on these findings, we developed an online analysis tool (https://lymphprog.serve.scilifelab.se/app/lymphprog) that can be used for prognostic prediction for DLBCL patients.
Subject terms: Cancer genetics, Cancer genomics, Cancer genomics, Clinical genetics
Introduction
DLBCL is one of the most common and aggressive types of lymphoid malignancies, accounting for approximately 30% of all non-Hodgkin lymphomas. Despite its aggressiveness, the disease can be cured, with most patients achieving long-term remission following standard R-CHOP or similar regimens [1, 2]. Nevertheless, approximately one-third of patients experience refractory or relapsed (R/R) disease, and most relapses occur within the first few years [2, 3]. Salvage treatment followed by high-dose chemotherapy and autologous stem cell transplantation has been established as second-line treatment for younger patients with R/R disease, but only a minority of these patients achieve long-term remission [3–6]. Recently, CD19 CAR-T-cell therapy was approved in the US as a second-line treatment in DLBCL patients with R/R disease and as a third-line treatment in several other countries [7, 8]. Other agents also showed promising results, including bispecific antibodies, anti-PD1 antibodies, anti-CD19 antibodies and antibody–drug conjugates [9–11]. Methods for identifying patients who are likely to develop R/R disease within the first few years after diagnosis might benefit from these new therapies and thus are urgently needed to further improve the overall outcomes of DLBCL patients.
Clinically, the International Prognostic Index (IPI) is commonly used to predict disease risk [12, 13]. However, there has been a growing interest in developing more advanced prognostic tools that account for a range of clinical parameters, biomarkers, and gene expression profiling, incorporating immunohistochemical or molecular techniques [14–17]. Twenty years ago, two major subtypes of DLBCL based on cell-of-origin (COO), germinal center B-cell-like (GCB) DLBCL and activated B-cell-like (ABC) DLBCL, were identified by gene expression profiling, with the association of poor outcomes in ABC-DLBCL [18]. In addition, DLBCL patients with a dual rearrangement of MYC and BCL2, “double/triple-hit” lymphoma, were recognized as a high-grade category with a poor prognosis in the latest WHO and ICC classification [19, 20]. Recently, several studies have identified molecular subtypes associated with different patient outcomes based on genetic alterations (mutations, copy number variation (CNV) and selected translocations) [21–24]. Additionally, direct sequencing of R/R tumors has shown that mutations in several genes (TP53, KMT2D, CREBBP, NFKBIE, FOXO1, MS4A1) might be associated with therapeutic resistance [25]. However, the prognostic value of these genetic changes remains to be proven. Finally, gene expression signatures [26–29] or tumor cell states and ecosystems [30] based on transcriptomic analysis have been developed and used to predict overall survival (OS) in DLBCL patients. However, to date, there is still a lack of studies that compare or integrate genetic and transcriptomic features when evaluating patient outcomes.
To further understand the mechanism underlying drug resistance and disease progression, and to develop a molecular tool for identifying patients at risk for early R/R disease, here, we performed comprehensive genomic and transcriptomic analyses on diagnostic (untreated) tumors from 2805 R-CHOP-treated DLBCL patients. This included our own study cohort and other six published cohorts with clinical, DNA mutation, and/or gene expression data available [21–24, 31–33]. We characterized the clinical characteristics, mutation profile and gene expression pattern in 887 patients who developed R/R disease within two years and compared these data with those from 1918 patients who remained in remission at two years. Furthermore, we sought to establish a risk classifier capable of effectively predicting the treatment outcomes of DLBCL patients, especially those with early R/R disease.
Methods
Clinical and genetic data
This study included analyses of clinical and genetic data from seven DLBCL cohorts. The first cohort (our cohort) included 161 R-CHOP-treated DLBCL patients diagnosed in Sweden or China during 2001–2015 (Table S1A). The Swedish patients (n = 73) were diagnosed at Karolinska/Stockholm Country and Uppsala University Hospitals, and their samples were newly sequenced in this study. The Chinese patients (n = 88) have been described previously [34–37] and the data were reanalyzed here. DLBCL samples from patients with chronic hepatitis B virus infection were excluded from the current study due to the potential genetic differences observed in these tumors [35]. The performance of various sequencing platforms is summarized in Table S1B, and details of the data analysis, especially on somatic mutation calling and gene expression analysis are provided in the Supplemental Methods. The study was approved by the Institutional Review Boards of Tianjin Medical University Cancer Institute, Uppsala University and Karolinska Institutet.
Six published DLBCL cohorts with available clinical/DNA mutation/gene expression data were also included in the analysis (Table S2A, B) [21–24, 31–33]. Clinical data were collected from the original deposits, and information on LymphGen subtypes was available in five cohorts [21–24, 32, 33]. For the two remaining cohorts (our cohort/GSE117556), LymphGen subtypes were predicted using the LymphGen tool (https://llmpp.nih.gov/lymphgen/index.php).
Meta-analysis of DLBCL patients with poor and good outcomes treated with R-CHOP
Several criteria were used to enroll samples for meta-analysis: (1) DLBCL patients were treated with R-CHOP and key clinical data were available; (2) Sufficient PFS data were available to define two-year outcomes; (3) Either DNA mutation or gene expression data were available. Subsequently, individuals who experienced R/R disease within two years were characterized as having poor outcomes, whereas those who remained in remission at two years were considered to have good outcomes.
Differentially expressed genes (DEGs) and gene set enrichment analysis (GSEA)
The RNAseq datasets (our cohort, n = 108; Schmitz et al. [23], n = 219) and microarray datasets (GSE117556, n = 723; GSE181063, n = 326) [21, 31] were separately combined using the R package Limma to remove batch effects between datasets [38], generating an RNAseq-based dataset and a microarray-based dataset. DEGs between DLBCLs with poor and good outcomes were identified based on criteria of FDR q < 0.1 and fold change >1.2. Further details on data analysis and GSEA are provided in the Supplemental Methods.
Establishment and validation of a risk signature to predict R/R disease within two years
The above RNAseq and microarray datasets were further merged into a larger cohort (n = 1376), using the quantile normalization approach described previously to normalize cross-platform datasets [39]. Univariate Cox regression was performed to assess the association of PFS and gene expression levels in the entire cohort, identifying prognostic genes (p < 0.01). Subsequently, we randomly assigned samples to a discovery cohort (70%, n = 964) and a validation cohort (30%, n = 412). The LASSO algorithm was used to extract gene-expression risk signatures, using the gene expression levels of overlapping genes between the prognostic genes and the DEGs between the poor and good outcome groups as the input variable and the two-year outcome of each patient as the outcome variable (Fig. S1). In this process, the discovery cohort was further divided into a training cohort (80%, n = 772) and a test cohort (20%, n = 192). One thousand risk models were constructed by employing various random seeds in the discovery cohort. Subsequently, their performance was evaluated in the test cohort, selecting the model with the highest area under the curve (AUC) value and accuracy for predicting two-year outcomes as the optimal risk model. The validation cohort was then utilized to demonstrate the performance of the risk model. Furthermore, another two independent datasets were used to test the established risk model: validation cohort-2 (RNAseq, n = 49) [40], and validation cohort-3 (microarray, remaining sample of GSE181063, n = 484). Further details on data computing, risk score calculation, optimal threshold, and risk classification are provided in the Supplemental Methods.
Results
Clinical characteristics of different DLBCL cohorts
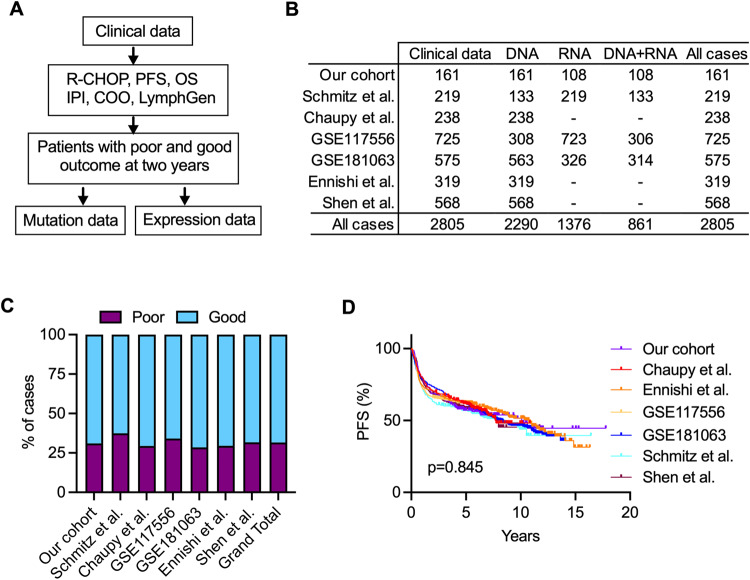
To investigate the characteristics of DLBCL patients with early R/R disease following R-CHOP treatment, we initially enrolled 161 DLBCL patients (our cohort; Table S1A). Of these, 50 (31%) experienced R/R disease within two years and were classified as having poor outcomes, whereas the remaining patients who were still in remission at two years, were categorized as having good outcomes. As expected, those with poor outcomes tended to be older, were more likely to have an ABC subtype, and had presented with more advanced disease at diagnosis and higher IPI scores than those with good outcomes (Table S3). To validate these findings, data from six published cohorts were compiled using the same enrollment criteria, comprising 2644 patients (Fig. 1A, B; Table S2A, B). Approximately 32% (range: 29%∼37%) of them experienced R/R disease within two years (Fig. 1C), and these patients displayed similar high-risk characteristics, despite variations in individual parameters among individual cohorts (Table S4). Moreover, the analysis demonstrated no statistically significant difference in PFS among all cohorts, as evidenced by the overlapping survival curves (Fig. 1D). Together, these results suggest that DLBCL patients with poor outcomes share common high-risk characteristics and exhibit similar clinical outcomes across different cohorts. Furthermore, this effort resulted in a cohort of 2805 uniformly R-CHOP-treated DLBCL patients, with 887 of them identified as having poor outcomes within two years. Additionally, we had access to genetic data for 2290 samples and transcriptomic data for 1376 samples (Fig. 1B; Table S2A), with 861 samples having both data types, enabling comprehensive integrated genomic and transcriptomic analyses.
Fig. 1. DLBCL cohorts used to identify patients with poor outcomes within two years following R-CHOP treatment.
A The workflow for identifying patients with poor and good outcomes in different cohorts. B An overview of clinical, DNA mutation, and gene expression data for patient cohorts included in this study. C The percentage of patients with poor outcomes within each cohort. D Kaplan–Meier survival analysis illustrating PFS in different cohorts. The p value was calculated by the log-rank test.
Mutation spectrum in DLBCL patients with poor outcomes
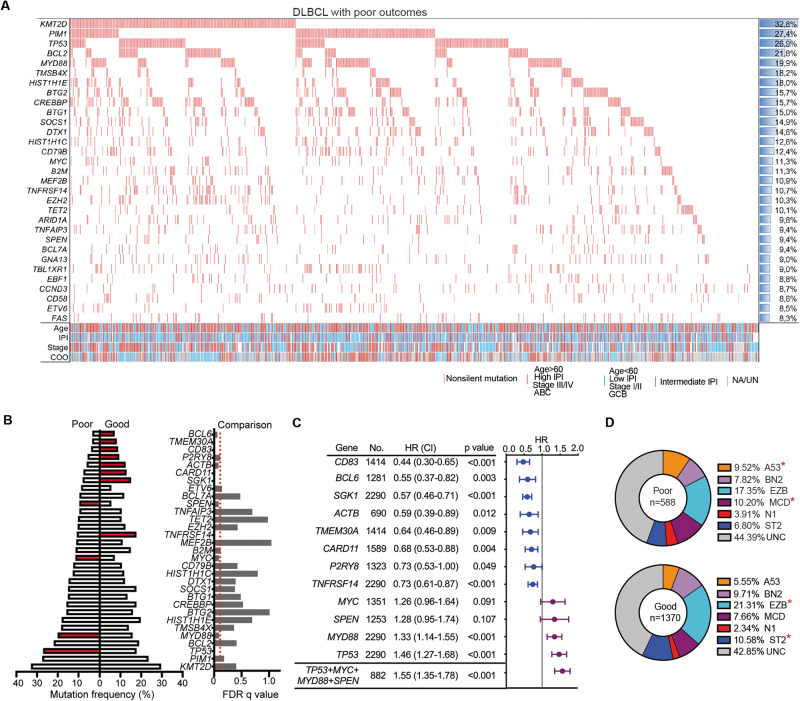
To investigate the mutation profiles of the 2290 DLBCL tumors, we focused on genes that were sequenced in all cohorts and specifically examined genes that were reported in at least three cohorts. This approach led to the identification of 99 genes for further analysis (Table S5A). Among these, 24 were found to be mutated in at least 10% of DLBCLs, including KMT2D, PIM1, TP53, BCL2, HIST1H1E, MYD88, CREBBP, and others (Table S5B). Many of these genes have been previously characterized as significant mutation targets in DLBCL [21–24, 35]. Moreover, the mutation frequencies of these 99 genes in the combined cohort exhibited a strong correlation (Pearson’s correlation coefficients; r value: 0.863–0.963) with those observed in the individual cohorts (Fig. S2A), indicating the consistency of mutation patterns across different cohorts. Furthermore, when examining the mutation frequencies of genes overlapped between the seven cohorts, we also observed a similar consistency (Fig. S2B; Table S5B).
Subsequently, a focused analysis was performed on the mutational profiles of DLBCL patients with poor outcomes in the combined cohort, consisting of 702 patients. We identified 57 genes that were affected by nonsilent mutations in at least 5% of the samples, with 20 of these being mutated in at least 10% of cases (Fig. 2A; Table S6). The most frequently mutated genes in DLBCL with poor outcomes were KMT2D (32.8%), PIM1 (27.4%), TP53 (26.9%), BCL2 (21.8%), MYD88 (19.9%), TMSB4X (18.2%), HIST1H1E (18.0%), BTG2 (15.7%), and others. We then compared the mutational profiles of tumors among patients with poor and good outcomes, revealing significant enrichment (q < 0.1) of mutations in four genes (TP53, MYD88, SPEN, MYC) and significant depletion of mutations in eight genes (CD83, BCL6, SGK1, ACTB, TMEM30A, CARD11, P2RY8, TNFRSF14) in DLBCLs with poor outcomes (Fig. 2B). The examination of these genes within the individual cohorts revealed a largely similar trend in mutation frequency within the poor outcome group across various cohorts (Fig. S3). Moreover, Cox regression analysis demonstrated that mutations in the four enriched genes were associated with worse PFS, whereas mutations in the eight depleted genes were associated with better PFS (Fig. 2C). Combined analysis of mutations in any of the four enriched genes (TP53 + MYD88 + SPEN + MYC) showed a slightly higher hazard ratio than that of individual genes (1.55 vs. 1.26–1.46; Fig. 2C). Finally, approximately 53% of the samples were classified into known LymphGen subtypes, and the A53 and MCD subtypes were significantly overrepresented in patients with poor outcomes, while the EZB and ST2 subtypes were enriched in patients with good outcomes (Fig. 2D). These findings indicate that a distinct mutation spectrum was associated with early R/R disease in DLBCL patients.
Fig. 2. Mutation pattern in tumors derived from DLBCL patients with poor outcomes following R-CHOP treatment.
A Catalog of the most frequently mutated genes in DLBCL tumors among patients with poor outcomes. Genes that affected by nonsilent mutations across more than three cohorts and observed in more than 8% of all patients (n = 702) were included. B Comparison of mutation frequencies between DLBCLs with poor and good outcomes using Fisher’s exact test, adjusted by the false discovery rate (FDR) q value (q < 0.1 was considered significant). The presented genes included those most significantly mutated between the two outcome groups and those with mutation frequencies greater than 10%. Genes with FDR q < 0.1 were indicated as red color. C Forest plots display the association between the mutation of individual/combined genes and PFS in the combined cohorts. D Donut chart illustrating the distribution of LymphGen DNA subtypes in patients with poor or good outcomes in the combined cohorts. Fisher’s exact test was used to compute p values. *p < 0.05; NA not available, UN unknown, HR hazard ratio, CI confidence interval, UNC unclassified.
Gene expression profiles in DLBCLs among patients with poor outcomes
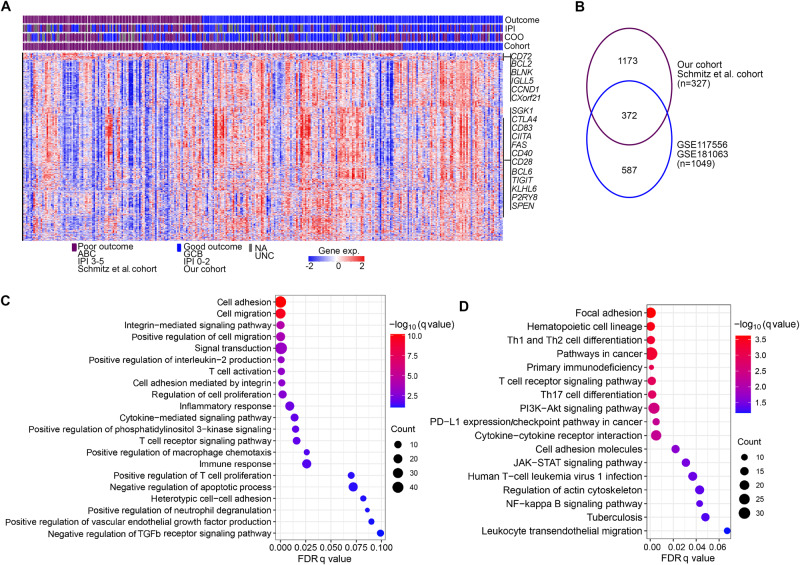
To compare the gene expression profiles between good and poor outcome groups, we first conducted analyses of DEGs in the RNAseq dataset (n = 327), encompassing 122 cases with poor outcomes (Fig. S4A). In this dataset, DEGs associated with poor outcomes were characterized by a small group of upregulated genes (n = 73) that included BCL2, BLNK, CXorf21, CD72, IGLL5, CCND2, TNFRSF8, NME1, BTLA and CD52, and a large group of downregulated genes (n = 1472) that included 49 genes that have been described to be frequently mutated in lymphoma [35] and immune checkpoint genes (CTLA4, TIGIT, CD80) (Fig. 3A; Table S7). Among the DEGs, four genes (SGK1, CD83, BCL6, P2RY8) were found to be less frequently mutated in DLBCLs with poor outcomes, whereas one gene (SPEN) was preferentially mutated in this group. Subsequently, we analyzed DEGs in the microarray-based dataset (n = 1049, Fig. S4B), including 336 cases with poor outcomes, and further compared them with those from the RNAseq-based dataset. This analysis revealed 372 overlapping genes (Fig. 3B), constituting 24% and 39% of the total DEGs in the respective cohorts. Notably, more than 98% (367) of these overlapping DEGs showed consistent upregulation or downregulation in both cohorts (Table S7), suggesting an overall consistency across different datasets and patient groups. We then performed GSEA of the overlapping DEGs and identified several significantly enriched pathways, including cell adhesion/migration/proliferation, PI3K, T-cell activation, T-cell receptor, NF-κB signaling, and PD-1 expression/checkpoint pathways (Fig. 3C, D).
Fig. 3. Gene expression pattern in tumors derived from DLBCL patients with poor and good outcomes at two years following R-CHOP treatment.
The analysis of differentially expressed genes (DEGs) was conducted separately in the RNAseq dataset (our cohort/Schmitz et al. cohort, n = 327) and the microarray dataset (GSE117556/GSE181063; n = 1049). A threshold of FDR q < 0.1 and fold change >1.2 was used to define DEGs from each dataset. A The heatmap illustrating DEGs between the two outcome groups in the RNAseq dataset. Each column represents a sample, ordered by outcome groups. Within each group, samples were ordered following the input of the data accordingly. Each row represents a gene. Selected genes that are frequently mutated in B-cell lymphomas or important for immune responses are highlighted in the figure. B The numbers of overlapping DEGs between the RNAseq dataset and the microarray dataset. GSEA analysis of the overlapping DEGs from (B) (C: Gene Ontology biological pathway, (D): Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway).
Establishment and validation of a risk signature to predict early R/R disease
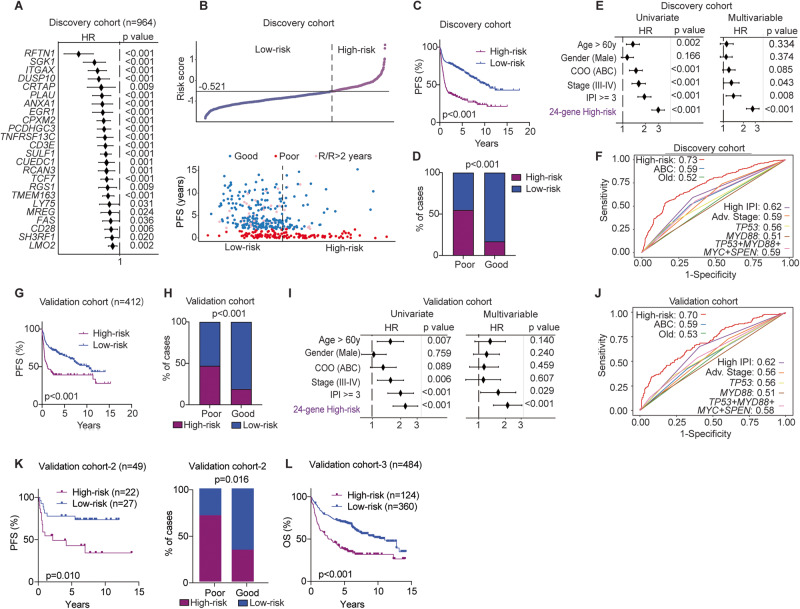
To establish a robust risk signature representative of early R/R DLBCL, we next employed a quantile normalization approach to integrate RNAseq-based and microarray-based datasets, creating a large, normalized dataset (Fig. S5; n = 1376). These samples were then randomly assigned as discovery and validation cohorts (70% and 30% respectively, Fig. S1). Univariate Cox regression was first performed to assess PFS and gene expression levels, identifying 656 prognostic genes from the entire cohort, with 242 of them overlapping with the 372 DEGs (Fig. 3B; Table S8). Using the expression levels of these 242 genes, and the two-year outcomes of each patient as inputs, the LASSO algorithm was subsequently employed to extract risk signatures in the discovery cohort (n = 964). Among 1000 established combinations, a 24-gene panel exhibiting the highest AUC value and accuracy in the test cohort in predicting two-year outcomes was selected as the optimal risk model/signature (Fig. 4A; Table S9). Notably, several of these genes play roles in relevant cellular processes/pathways, including BCR signaling (RFTN1), protein kinases (SGK1), apoptosis (FAS), cell differentiation markers (CD28, CD3E, ITGAX, LY75, TNFRSF13C/BAFFR), cytokines and growth factors (PLAU), and transcription factors (LMO2, EGR1, TCF7). Subsequently, using the 24-gene expression score calculated in each tumor, we categorized patients into high- and low-risk groups based on a threshold that provided an optimal trade-off between sensitivity and specificity in predicting two-year outcomes (Fig. 4B). High-risk DLBCL patients assigned by the 24-gene expression score exhibited significantly worse PFS, and more high-risk patients experienced poor outcomes within two years than those assigned to the low-risk group (Fig. 4C, D). Additionally, the 24-gene risk score displayed a positive predictive value (PPV) of 0.61 and a negative predictive value (NPV) of 0.79 in predicting two-year outcomes, with an overall accuracy of 74%. Multivariable analyses revealed that this 24-gene risk score served as an independent predictor (p < 0.001) of two-year prognosis, even after adjusting for key clinical risk factors (Fig. 4E). Moreover, ROC curve analysis demonstrated that the 24-gene scoring stratification outperformed the COO subtype, key clinical parameters, and various mutational statuses (TP53, MYD88, etc.), in predicting two-year outcomes (Fig. 4F). Furthermore, the inclusion of mutational status of the four preferentially mutated genes (individually or in combination) identified in the poor outcome patients did not enhance its predictive efficiency of early R/R DLBCL (Fig. S6A).
Fig. 4. Development and validation of an independent gene-expression signature to predict treatment outcomes in DLBCL patients.
The RNAseq and microarray datasets were merged into a larger cohort (n = 1376) using a quantile normalization approach. The samples were subsequently randomly divided into a discovery cohort (70%, n = 964; A–F) and a validation cohort (30%, n = 412; G–J). A Forest plots showing the association between the expression levels of the 24 genes and PFS within the discovery cohort. B The distribution of 24-gene expression scores in each DLBCL patient, and the correlation between PFS and risk groups in the discovery cohort. Patients were assigned to high- and low-risk groups based on the optimal threshold for the ROC curve, set at −0.521. Each dot represents one patient. C Kaplan–Meier survival analysis illustrating PFS between high- and low-risk groups in the discovery cohort. The p value was calculated by the log-rank test. D Bar plots showing the distribution of high- and low-risk patients within poor and good outcome groups in the discovery cohort. Fisher’s exact test was utilized to determine the p value. E Univariate and multivariable Cox regression analyses demonstrating the prognostic independence of the 24-gene-expression scores in the discovery cohort. Key clinical parameters such as age, subtype, stage, and IPI factors are included in the analysis. F ROC curves demonstrating the performance of different parameters in identifying DLBCL patients with two-year poor outcomes in the discovery cohort. AUC values are indicated. G Kaplan–Meier survival analysis illustrating PFS of the high- and low-risk groups in the validation cohort (n = 412). Patients were classified into high- and low-risk groups using the same threshold established in the discovery cohort (−0.521). The p value was calculated by the log-rank test. H Bar plots showing the distribution of high- and low-risk patients within poor and good outcome groups in the validation cohort. Fisher’s exact test was used to determine the p value. I Univariate and multivariable Cox regression analyses demonstrating the independent prognostic role of the 24-gene expression score in the validation cohort. J ROC curves demonstrating the performance of different parameters in identifying DLBCL patients with two-year poor outcomes in the validation cohort. K, L Two additional independent cohorts (RNAseq=49, CNP0001327; microarray=484, remaining samples of the GSE181063 cohort were only available for OS data) were used to evaluate the algorithm of the 24-gene risk score. HR hazard ratio, CI confidence interval, ROC receiver operating characteristic, AUC area under the curve, OS overall survival.
Consistent with the findings in the discovery cohort, we observed worse PFS and more patients experiencing poor outcomes among high-risk patients in the validation cohort (n = 412; Fig. 4G, H). The PPV, NPV and overall accuracy in predicting two-year outcomes in the validation cohort were 0.55, 0.75 and 70%, respectively, which were slightly lower than those observed in the discovery cohort. Multivariable analyses and ROC curves confirmed the independent predictive capability of the 24-gene risk score, with an AUC value of 0.7 for predicting two-year outcomes in the validation cohort (Fig. 4I, J). Moreover, combining mutation status (TP53, MYD88, SPEN, MYC) with the 24-gene risk score also failed to improve the prediction performance (Fig. S6B).
Subsequent evaluations, especially among samples with available double-hit and double-expressor status, further confirm its independent prognostic value (Fig. S7). Furthermore, the comparison of ROC curves with existing gene expression-based classifiers [27, 28, 41] demonstrated that our 24-gene risk score had the best performance in predicting two-year outcomes (Fig. S8). In addition, the results across individual cohorts revealed similar patterns in 24-gene expression, differences in PFS, and the distribution of patients in high- and low-risk groups (Fig. S9–10). Finally, the 24-gene score algorithm was successfully validated on two additional cohorts derived from different platforms (RNAseq, n = 49; microarray, n = 484) (Fig. 4K–L). These findings underscore the robustness and generalizability of the 24-gene expression score as an independent prognostic tool for predicting early disease progression in DLBCL.
The association of the 24-gene expression score and COO subtypes of DLBCL
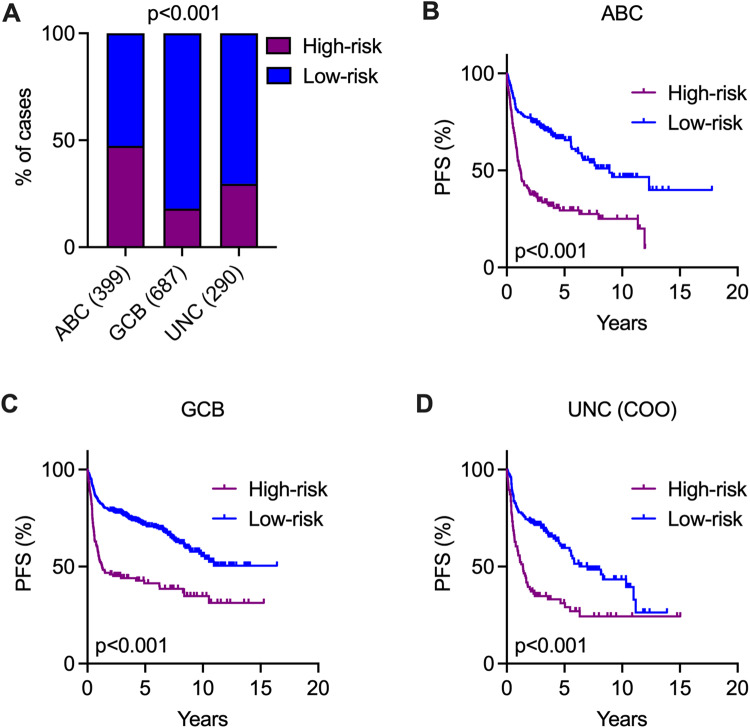
We next assessed the correlation between the 24-gene expression score and the COO subtypes, which are known predictors of DLBCL patient outcomes. Within the entire cohort (n = 1376), a notably higher proportion of ABC-DLBCL cases were categorized as high-risk, while a lower percentage of GCB-DLBCL and unclassified cases fell into the high-risk group (Fig. 5A). Moreover, patients classified as high-risk exhibited significantly poorer PFS in both ABC and GCB subtypes when compared to those in the low-risk group (Fig. 5B, C). Notably, the 24-gene risk score was also effective in identifying high-risk patients among the unclassified COO cases (Fig. 5D). Furthermore, these results were consistent across the discovery, validation, and individual cohorts, indicating no significant cohort-related bias in the analysis of combined data (Fig. S11–12). Together, our findings suggest that the 24-gene expression score can further stratify high-risk patients among COO subtypes, including those unclassified cases.
Fig. 5. Independent risk stratification by 24-gene expression scores in various COO subtypes of DLBCLs.
The analysis was performed on all samples with available gene expression data (n = 1376). A Bar plots showing the distribution of high- and low-risk patients in various COO subtypes. Fisher’s exact test was used to determine the p value. Kaplan–Meier survival analysis illustrating PFS between high- and low-risk groups in ABC-DLBCL (B), GCB-DLBCL (C) and UNC-DLBCL (D). COO cell-of-origin, UNC unclassified. The p value was calculated by the log-rank test. Independent analyses in the individual cohorts, discovery cohort and validation cohort are presented in Fig. S11–S12.
Use the 24-gene expression score to stratify high-risk patients within LymphGen subtypes
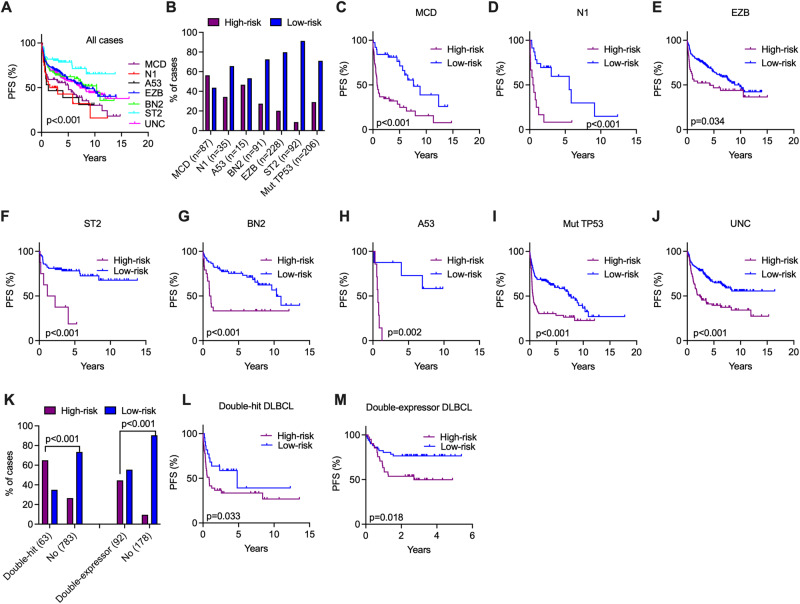
To investigate the potential relationship between the 24-gene expression score and DNA molecular subtypes in DLBCLs, we examined the performance of the risk scores across LymphGen subtypes. Among the 1376 samples with transcriptomic data, LymphGen subtype information was available for 956 samples. Consistent with previous studies [23, 24], the MCD, N1, and A53 subtypes in the entire cohort were associated with inferior survival, whereas the EZB, BN2, and ST2 subtypes had favorable outcomes (Fig. 6A). The distribution of high- and low-risk patients in these DNA subtypes is summarized in Fig. 6B and the 24-gene risk score was significantly effective in further stratifying high-risk patients across all individual DNA subtypes (Fig. 6C–H). Since the A53 subtype of LymphGen is mainly defined by TP53 mutations and CNVs and since CNV data were not available in three cohorts (our cohort/GSE117556/GSE181063), we conducted an independent analysis on DLBCLs with TP53 mutations (n = 206). Notably, patients carrying TP53 mutations who were assigned to the high-risk group had significantly worse PFS than those assigned to the low-risk group (Fig. 6I). Moreover, in those 284 unclassified cases, where LymphGen subtypes were not assigned and TP53 were not mutated (Table S2A), the 24-gene risk score was still able to effectively stratify high-risk patients (Fig. 6J). Furthermore, the 24-gene risk score also demonstrated its significant capability in assigning double-hits and double-expressors into the high-risk group (Fig. 6K), and further identifying high-risk patients within each subtype (Fig. 6L–M). Additionally, these analyses were conducted separately in the discovery and validation cohorts, and the results revealed relatively consistent results observed in the combined cohort (Fig. S13). Thus, the utility of the 24-gene expression score can be extended to risk stratification among the genetic subtypes.
Fig. 6. Individualized risk stratification by 24-gene expression score in LymphGen DNA subtypes.
All samples with available gene expression data and LymphGen DNA subtypes were combined for the analysis (n = 956). A Kaplan–Meier survival analysis showing the PFS in the individual LymphGen DNA subtypes. B Bar plots showing the distribution of high- and low-risk patients in the different DNA subtypes. C–J Kaplan–Meier survival analysis showing the PFS of high- and low-risk patients in the indicated DNA subtypes and the unclassified subtype. K Bar plots showing the distribution of high- and low-risk patients in the double-hits (n = 63) and double-expressors (n = 92) of MYC and BCL2. Among 1376 samples with 24-gene risk scores, 846 cases were evaluated for double-hit status, with 63 identified as double-hit. A total of 270 samples were assessed for double-expressor status, with 92 identified as double-expressors. Fisher’s exact test was used to compute p values. L, M Kaplan–Meier survival analysis showing the PFS of high- and low-risk patients in the indicated groups. UNC unclassified. For all Kaplan–Meier survival analyses, the p value was calculated by the log-rank test. Independent analyses in the discovery and validation cohorts are presented in Fig. S13.
Discussion
Despite recent advances in the characterization of the genome and transcriptome of DLBCL [21–24, 32–36, 42–46], including analysis at the single-cell level [44], our knowledge of patients with early R/R disease is still limited. Our study integrated data from seven cohorts comprising 2805 R-CHOP-treated patients, including 887 cases with poor treatment outcomes within two years. Our analyses revealed a distinct mutation pattern and gene expression profile in these patients with poor outcomes. Additionally, we established a 24-gene expression score as an independent prognostic predictor for early R/R disease in DLBCL patients. This risk score also demonstrated its effectiveness in further stratifying COO and genetic subtypes into high- and low-risk groups. Overall, our study provides valuable insights into understanding the molecular mechanisms underlying early R/R DLBCL and highlights the importance of incorporating gene expression profiling into clinical decision-making for DLBCL patients.
Various studies have previously utilized NGS technologies to describe genetic subtypes and prognostic models in DLBCL, including DNA mutation-based subtypes [21–24] and gene expression-based risk signatures [26–29] or prognostic tumor microenvironments [30]. However, the focus has been largely on OS, rather than specifically addressing features associated with early disease progression. Some studies have developed risk models by incorporating OS data from patients treated with CHOP and R-CHOP [27, 28], whereas some risk signatures were developed without validation cohorts or based on limited sample sizes [26, 29]. Moreover, none of these risk models/signatures attempted to integrate both genetic and transcriptomic data in their evaluation. Our study benefits from many patients with both genomic and transcriptomic data and the use of multiple independent cohorts to validate findings. Moreover, we defined early R/R disease based on a two-year cutoff, which is associated with the poorest outcomes [6, 47]. Notably, previous studies indicated that patients who survived and remained event-free for 24 months had a similar consecutive OS as the general population matched for sex and age [6, 48]. Our newly developed 24-gene classifier is primarily designed to optimize its effectiveness in predicting two-year outcomes, surpassing the performance of existing classifiers in this aspect. Moreover, the integration of the cross-platform normalization approach facilitates the merging of data from diverse platforms, generating a robust dataset for establishing a more reliable risk classifier. Finally, in light of our findings, we created an online analysis tool (accessible at https://lymphprog.serve.scilifelab.se/app/lymphprog) designed for user-friendly and efficient risk assessment, which lays the foundation for developing a practical algorithm with potential applications in clinical settings.
Four genes were preferentially mutated in DLBCL with poor outcomes. Of these, TP53, MYC, and MYD88 mutations have been associated with early R/R disease in DLBCLs in previous studies [25, 49–52], whereas SPEN is a novel candidate, and plays a role in negatively regulating NOTCH signaling. Studies have indicated that aberrant NOTCH signaling can contribute to drug resistance in various cancers [53], including DLBCL [54], suggesting that targeting the NOTCH pathway may hold promise as a therapeutic strategy for patients with early R/R disease. Conversely, the eight genes preferentially mutated in DLBCL with good outcomes align with their roles as markers for genetic subtypes associated with favorable prognosis, including BN2, ST2, and EZB [23, 24]. Loss-of-function TMEM30A mutations have been associated with increased B-cell signaling and enhanced drug uptake in DLBCLs, potentially contributing to a better treatment response [55]. Notably, the study also identified genes, such as SGK1 and CD83, where both higher mutation frequency and higher mRNA expression were linked to better treatment outcomes in DLBCL patients. This suggests that tumor cells with such features may exhibit increased sensitivity to R-CHOP treatment. However, although the identification of these differentially mutated genes may help to understand the mechanism underlying early R/R disease, the mutation status of individual genes appears to provide limited predictive value when compared to gene expression-based risk signatures.
We further demonstrated that our risk score algorithm shows superior performance in further stratifying high-risk patients from COO and DNA molecular subtypes. Importantly, it showed the capacity to further identify high-risk patients among the unclassified cases in these subtyping tools. While this 24-gene risk score may not identify all high-risk patients, possibly due to the considerable intratumor and intertumor heterogeneity of the disease, it shows promise in effectively predicting two-year outcomes in more than 70% of DLBCL patients following R-CHOP treatment. For high-risk patients identified by this classifier, intensified immunochemotherapy protocols, such as incorporating high-dose methotrexate [56], or R-double-CHOP [57], may be considered while awaiting more definitive evidence on new immunotherapies or combined therapies [58]. Moreover, close monitoring of high-risk patients during or after R-CHOP treatment is necessary. Tools such as monitoring measurable residual disease through cell-free tumor DNA analysis may help to track treatment response and disease status in these high-risk patients [59], facilitating timely interventions if needed.
In summary, leveraging data from multiple cohorts, our study identified mutated genes, altered pathways, and gene expression-based risk signatures associated with poor outcomes in DLBCL patients. However, despite combining several cohorts to achieve a relatively homogeneous dataset, considerable heterogeneities (sample resources, sequencing platforms/methods, variant callers, thresholds in identifying prognostic genes and DEGs) between acquired data types from individual cohorts may still introduce variability. Therefore, these findings may require further validation in additional cohorts. To further improve the predictive value of the 24-gene expression score, incorporating additional genetic/epigenetic features, such as genome-wide mutational signatures [37], CNVs, noncoding drivers [60], RNA editing [46], and tumor microenvironment, as well as the application of supplementary tools, such as proteomic, metabolic, and single-cell studies, may be considered.
Supplementary information
Acknowledgements
This work was supported by the Swedish Cancer Society, the Swedish Research Council, Radiumhemmets, the Center for Innovative Medicine, the Mayo Clinic and KI collaborative grant, the Knut and Alice Wallenberg Foundation, O. E. and Edla Johansson Foundation and the Guangdong Provincial Key Laboratory of Human Disease Genomics (2020B1212070028).
Author contributions
WR analyzed and interpreted the data and wrote the manuscript; HW, XL, MY, XY, and DL performed the bioinformatics analysis; SAO, MB, XW, KS, GE, RMA, BS, BEW, and HZ collected samples and clinical information; WR, and SAO interpreted the clinical information; KW supervised the bioinformatics analysis; KES, BEW and HZ were involved in study supervision; and QPH designed and supervised the study and wrote the manuscript.
Funding
Open access funding provided by Karolinska Institute.
Data availability
The datasets analyzed during the current study are available in the China National GeneBank Sequence Archive of China National GeneBank Database with accession Nos. CNP0001228 and CNP0001220, and in the National Center for Biotechnology Information (NCBI) database with accession No. PRJNA952204.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Weicheng Ren, Hui Wan, Sulaf Abd Own.
These authors jointly supervised this work: Kui Wu, Huilai Zhang, Björn Engelbrekt Wahlin, Karin E. Smedby, Qiang Pan-Hammarström.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-023-02120-7.
References
- 1.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:235–42. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 2.Harrysson S, Eloranta S, Ekberg S, Enblad G, Jerkeman M, Wahlin BE, et al. Incidence of relapsed/refractory diffuse large B-cell lymphoma (DLBCL) including CNS relapse in a population-based cohort of 4243 patients in Sweden. Blood Cancer J. 2021;11:9. doi: 10.1038/s41408-020-00403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarkozy C, Coiffier B. Primary refractory diffuse large B cell lymphoma in the rituximab era. Curr Opin Oncol. 2015;27:377–83. doi: 10.1097/CCO.0000000000000209. [DOI] [PubMed] [Google Scholar]
- 4.Coiffier B, Sarkozy C. Diffuse large B-cell lymphoma: R-CHOP failure-what to do? Hematol Am Soc Hematol Educ Program. 2016;2016:366–78. doi: 10.1182/asheducation-2016.1.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viardot A, Goebeler ME, Hess G, Neumann S, Pfreundschuh M, Adrian N, et al. Phase 2 study of the bispecific T-cell engager (BiTE) antibody blinatumomab in relapsed/refractory diffuse large B-cell lymphoma. Blood. 2016;127:1410–6. doi: 10.1182/blood-2015-06-651380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maurer MJ, Ghesquieres H, Jais JP, Witzig TE, Haioun C, Thompson CA, et al. Event-free survival at 24 months is a robust end point for disease-related outcome in diffuse large B-cell lymphoma treated with immunochemotherapy. J Clin Oncol. 2014;32:1066–73. doi: 10.1200/JCO.2013.51.5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locke FL, Miklos DB, Jacobson CA, Perales MA, Kersten MJ, Oluwole OO, et al. Axicabtagene ciloleucel as second-line therapy for large B-cell lymphoma. N. Engl J Med. 2022;386:640–54. doi: 10.1056/NEJMoa2116133. [DOI] [PubMed] [Google Scholar]
- 8.Sauter CS, Senechal B, Riviere I, Ni A, Bernal Y, Wang X, et al. CD19 CAR T cells following autologous transplantation in poor-risk relapsed and refractory B-cell non-Hodgkin lymphoma. Blood. 2019;134:626–35. doi: 10.1182/blood.2018883421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansell SM, Minnema MC, Johnson P, Timmerman JM, Armand P, Shipp MA, et al. Nivolumab for relapsed/refractory diffuse large B-cell lymphoma in patients ineligible for or having failed autologous transplantation: a single-arm, phase II study. J Clin Oncol. 2019;37:481–9. doi: 10.1200/JCO.18.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamadani M, Gopal AK, Pasquini M, Kim S, Qiu X, Ahmed S, et al. Allogeneic transplant and CAR-T therapy after autologous transplant failure in DLBCL: a noncomparative cohort analysis. Blood Adv. 2022;6:486–94. doi: 10.1182/bloodadvances.2021005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutchings M, Mous R, Clausen MR, Johnson P, Linton KM, Chamuleau MED, et al. Dose escalation of subcutaneous epcoritamab in patients with relapsed or refractory B-cell non-Hodgkin lymphoma: an open-label, phase 1/2 study. Lancet. 2021;398:1157–69. doi: 10.1016/S0140-6736(21)00889-8. [DOI] [PubMed] [Google Scholar]
- 12.International Non-Hodgkin’s Lymphoma Prognostic Factors, Project. A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 13.Biccler JL, Eloranta S, de Nully Brown P, Frederiksen H, Jerkeman M, Jorgensen J, et al. Optimizing outcome prediction in diffuse large B-cell lymphoma by use of machine learning and nationwide lymphoma registries: a nordic lymphoma group study. JCO Clin Cancer Inf. 2018;2:1–13. doi: 10.1200/CCI.18.00025. [DOI] [PubMed] [Google Scholar]
- 14.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359:2313–23. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alizadeh AA, Gentles AJ, Alencar AJ, Liu CL, Kohrt HE, Houot R, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118:1350–8. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–7. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Zhang E, Ma Z, Wu W, Kosinska A, Zhang X, et al. Enhancing virus-specific immunity in vivo by combining therapeutic vaccination and PD-L1 blockade in chronic hepadnaviral infection. PLoS Pathog. 2014;10:e1003856. doi: 10.1371/journal.ppat.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 19.Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, et al. The 5th edition of the world health organization classification of hematolymphoid tumours: lymphoid neoplasms. Leukemia. 2022;36:1720–48. doi: 10.1038/s41375-022-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campo E, Jaffe ES, Cook JR, Quintanilla-Martinez L, Swerdlow SH, Anderson KC, et al. The international classification of mature lymphoid neoplasms: a report from the clinical advisory committee. Blood. 2022;140:1229–53. doi: 10.1182/blood.2022015851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a haematological malignancy research network report. Blood. 2020;135:1759–71. doi: 10.1182/blood.2019003535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–90. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl J Med. 2018;378:1396–407. doi: 10.1056/NEJMoa1801445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic Implications. Cancer Cell. 2020;37:551–568 e514. doi: 10.1016/j.ccell.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rushton CK, Arthur SE, Alcaide M, Cheung M, Jiang A, Coyle KM, et al. Genetic and evolutionary patterns of treatment resistance in relapsed B-cell lymphoma. Blood Adv. 2020;4:2886–98. doi: 10.1182/bloodadvances.2020001696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merdan S, Subramanian K, Ayer T, Van Weyenbergh J, Chang A, Koff JL, et al. Gene expression profiling-based risk prediction and profiles of immune infiltration in diffuse large B-cell lymphoma. Blood Cancer J. 2021;11:2. doi: 10.1038/s41408-020-00404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan M, Yang P, Wang F, Luo X, Li B, Ding Y, et al. Whole transcriptome data analysis reveals prognostic signature genes for overall survival prediction in diffuse large B cell lymphoma. Front Genet. 2021;12:648800. doi: 10.3389/fgene.2021.648800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Yuan W, Zhang J, Li L, Zhang L, Zhang X, et al. Identification and validation of a prognostic prediction model in diffuse large B-cell lymphoma. Front Endocrinol (Lausanne) 2022;13:846357. doi: 10.3389/fendo.2022.846357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosquera Orgueira A, Diaz Arias JA, Cid Lopez M, Peleteiro Raindo A, Antelo Rodriguez B, Aliste Santos C, et al. Improved personalized survival prediction of patients with diffuse large B-cell Lymphoma using gene expression profiling. BMC Cancer. 2020;20:1017. doi: 10.1186/s12885-020-07492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steen CB, Luca BA, Esfahani MS, Azizi A, Sworder BJ, Nabet BY, et al. The landscape of tumor cell states and ecosystems in diffuse large B cell lymphoma. Cancer Cell. 2021;39:1422–37. doi: 10.1016/j.ccell.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sha C, Barrans S, Cucco F, Bentley MA, Care MA, Cummin T, et al. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J Clin Oncol. 2019;37:202–12. doi: 10.1200/JCO.18.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ennishi D, Jiang A, Boyle M, Collinge B, Grande BM, Ben-Neriah S, et al. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J Clin Oncol. 2019;37:190–201. doi: 10.1200/JCO.18.01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen R, Fu D, Dong L, Zhang MC, Shi Q, Shi ZY, et al. Simplified algorithm for genetic subtyping in diffuse large B-cell lymphoma. Signal Transduct Target Ther. 2023;8:145. doi: 10.1038/s41392-023-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Miranda NF, Georgiou K, Chen L, Wu C, Gao Z, Zaravinos A, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124:2544–53. doi: 10.1182/blood-2013-12-546309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren W, Ye X, Su H, Li W, Liu D, Pirmoradian M, et al. Genetic landscape of hepatitis B virus-associated diffuse large B-cell lymphoma. Blood. 2018;131:2670–81. doi: 10.1182/blood-2017-11-817601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georgiou K, Chen L, Berglund M, Ren W, de Miranda NF, Lisboa S, et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood. 2016;127:3026–34. doi: 10.1182/blood-2015-12-686550. [DOI] [PubMed] [Google Scholar]
- 37.Ye X, Ren W, Liu D, Li X, Li W, Wang X, et al. Genome-wide mutational signatures revealed distinct developmental paths for human B cell lymphomas. J Exp Med. 2021;218:e20200573. doi: 10.1084/jem.20200573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foltz SM, Greene CS, Taroni JN. Cross-platform normalization enables machine learning model training on microarray and RNA-seq data simultaneously. Commun Biol. 2023;6:222. doi: 10.1038/s42003-023-04588-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T, Liu H, Jiao L, Zhang Z, He J, Li L, et al. Genetic characteristics involving the PD-1/PD-L1/L2 and CD73/A2aR axes and the immunosuppressive microenvironment in DLBCL. J Immunother Cancer. 2022;10:e004114. [DOI] [PMC free article] [PubMed]
- 41.Zamani-Ahmadmahmudi M, Nassiri SM. Development of a reproducible prognostic gene signature to predict the clinical outcome in patients with diffuse large B-cell lymphoma. Sci Rep. 2019;9:12198. doi: 10.1038/s41598-019-48721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nie M, Du L, Ren W, Joung J, Ye X, Shi X, et al. Genome-wide CRISPR screens reveal synthetic lethal interaction between CREBBP and EP300 in diffuse large B-cell lymphoma. Cell Death Dis. 2021;12:419. doi: 10.1038/s41419-021-03695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nie M, Ren W, Ye X, Berglund M, Wang X, Fjorden K, et al. The dual role of CD70 in B-cell lymphomagenesis. Clin Transl Med. 2022;12:e1118. doi: 10.1002/ctm2.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye X, Wang L, Nie M, Wang Y, Dong S, Ren W, et al. A single-cell atlas of diffuse large B cell lymphoma. Cell Rep. 2022;39:110713. doi: 10.1016/j.celrep.2022.110713. [DOI] [PubMed] [Google Scholar]
- 45.Buhler MM, Martin-Subero JI, Pan-Hammarstrom Q, Campo E, Rosenquist R. Towards precision medicine in lymphoid malignancies. J Intern Med. 2022;292:221–42. doi: 10.1111/joim.13423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pecori R, Ren W, Pirmoradian M, Wang X, Liu D, Berglund M, et al. ADAR1-mediated RNA editing promotes B cell lymphomagenesis. iScience. 2023;26:106864. doi: 10.1016/j.isci.2023.106864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jakobsen LH, Bogsted M, Brown PN, Arboe B, Jorgensen J, Larsen TS, et al. Minimal loss of lifetime for patients with diffuse large B-cell lymphoma in remission and event free 24 months after treatment: a danish population-based study. J Clin Oncol. 2017;35:778–84. doi: 10.1200/JCO.2016.70.0765. [DOI] [PubMed] [Google Scholar]
- 48.Ekberg S, Jerkeman M, Andersson PO, Enblad G, Wahlin BE, Hasselblom S, et al. Long-term survival and loss in expectancy of life in a population-based cohort of 7114 patients with diffuse large B-cell lymphoma. Am J Hematol. 2018;93:1020–8. doi: 10.1002/ajh.25147. [DOI] [PubMed] [Google Scholar]
- 49.Morin RD, Assouline S, Alcaide M, Mohajeri A, Johnston RL, Chong L, et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin Cancer Res. 2016;22:2290–2300. doi: 10.1158/1078-0432.CCR-15-2123. [DOI] [PubMed] [Google Scholar]
- 50.Park HY, Lee SB, Yoo HY, Kim SJ, Kim WS, Kim JI, et al. Whole-exome and transcriptome sequencing of refractory diffuse large B-cell lymphoma. Oncotarget. 2016;7:86433–45. doi: 10.18632/oncotarget.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mareschal S, Dubois S, Viailly PJ, Bertrand P, Bohers E, Maingonnat C, et al. Whole exome sequencing of relapsed/refractory patients expands the repertoire of somatic mutations in diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2016;55:251–67. doi: 10.1002/gcc.22328. [DOI] [PubMed] [Google Scholar]
- 52.Greenawalt DM, Liang WS, Saif S, Johnson J, Todorov P, Dulak A, et al. Comparative analysis of primary versus relapse/refractory DLBCL identifies shifts in mutation spectrum. Oncotarget. 2017;8:99237–44. doi: 10.18632/oncotarget.18502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Li Y, Ahmad A, Azmi AS, Banerjee S, Kong D, et al. Targeting notch signaling pathway to overcome drug resistance for cancer therapy. Biochim Biophys Acta. 2010;1806:258–67. doi: 10.1016/j.bbcan.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hartert KT, Wenzl K, Krull JE, Manske M, Sarangi V, Asmann Y, et al. Targeting of inflammatory pathways with R2CHOP in high-risk DLBCL. Leukemia. 2021;35:522–33. doi: 10.1038/s41375-020-0766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ennishi D, Healy S, Bashashati A, Saberi S, Hother C, Mottok A, et al. TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma. Nat Med. 2020;26:577–88. doi: 10.1038/s41591-020-0757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leppa S, Jorgensen J, Tierens A, Meriranta L, Ostlie I, de Nully Brown P, et al. Patients with high-risk DLBCL benefit from dose-dense immunochemotherapy combined with early systemic CNS prophylaxis. Blood Adv. 2020;4:1906–15. doi: 10.1182/bloodadvances.2020001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurita D, Miura K, Nakagawa M, Ohtake S, Sakagami M, Uchino Y, et al. Dose-intensified CHOP with rituximab (R-Double-CHOP) followed by consolidation high-dose chemotherapies for patients with advanced diffuse large B-cell lymphoma. Int J Hematol. 2015;101:585–93. doi: 10.1007/s12185-015-1780-6. [DOI] [PubMed] [Google Scholar]
- 58.Morschhauser F, Feugier P, Flinn IW, Gasiorowski R, Greil R, Illes A, et al. A phase 2 study of venetoclax plus R-CHOP as first-line treatment for patients with diffuse large B-cell lymphoma. Blood. 2021;137:600–9. doi: 10.1182/blood.2020006578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurtz DM, Soo J, Co Ting Keh L, Alig S, Chabon JJ, Sworder BJ, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol. 2021;39:1537–47. doi: 10.1038/s41587-021-00981-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu H, Uuskula-Reimand L, Isaev K, Wadi L, Alizada A, Shuai S, et al. Candidate Cancer Driver Mutations in Distal Regulatory Elements and Long-Range Chromatin Interaction Networks. Mol Cell. 2020;77:1307–21. doi: 10.1016/j.molcel.2019.12.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed during the current study are available in the China National GeneBank Sequence Archive of China National GeneBank Database with accession Nos. CNP0001228 and CNP0001220, and in the National Center for Biotechnology Information (NCBI) database with accession No. PRJNA952204.