Abstract
Human leukosialin (CD43) is expressed in a cell lineage-specific as well as a differentiation stage-specific fashion. The leukosialin promoter, made up of an Sp1 binding site and a sequence similar to that of an initiator, possesses high transcriptional potential. Previous data have demonstrated that the leukosialin gene is down-regulated in nonproducing cells by DNA methylation. In this paper the repressive mechanism of DNA methylation in expression systems is reported. In vitro DNA methylation with SssI (CpG) methylase of leukosialin-chloramphenicol acetyltransferase (CAT) constructs drastically reduced transcriptional activities in stable transfection systems with the human HeLa and Jurkat cell lines. On the other hand, the transcriptional repression by in vitro methylation was less pronounced in Drosophila melanogaster cells, which lack genomic methylation. In these cells, Sp1 could transactivate equally well both the unmethylated and methylated leukosialin promoter. In order to test whether one of the methyl-CpG-binding proteins, MeCP2, is responsible for transcriptional repression of the leukosialin gene, I isolated the human MeCP2 cDNA (encoding 486 amino acid residues) and expressed it in Drosophila cells. I found that MeCP2 substantially inhibited Sp1-activated transcription when the leukosialin promoter was methylated. The level of repression was directly proportional to the amount of MeCP2 expression vector transfected. Analysis of C-terminal deletion mutants of MeCP2 showed that repressive activity of Sp1 transactivation is localized to the N-terminal region consisting of amino acid residues 1 to 193, which encompass the methyl-binding domain. These results suggest that interference with Sp1 transactivation by MeCP2 is an important factor in the down-regulation of leukosialin gene expression by DNA methylation.
DNA methylation of the C-5 position of cytosine within CpG dinucleotides plays a fundamental role in regulating gene expression in vertebrates (2). It has been demonstrated that two types of mechanisms are involved in silencing genes by DNA methylation. CpG methylation itself down-regulates transcription by preventing the binding of transcription factors to their recognition sequences (3, 9, 38). As an indirect mechanism, DNA methylation suppresses transcription through repressor molecules which can bind to methylated CpG. To date, the methyl-binding proteins MeCP1 (6, 11, 32) and MeCP2 (29, 33, 36) and the histone H1 (21, 52) have been identified as such repressors. Also in the category of an indirect mechanism is gene inactivation by alteration in chromosome structure (22, 23). A relationship between methyl-CpG repression and nuclear architecture has also been suggested by the finding that a Mar-binding protein, which is implicated in loop domain formation of chromatin, is essentially identical to MeCP2 (53). Indirect mechanisms seem to be more general strategies to suppress gene activities. Recently, the significance of methyl-binding proteins in biological processes was demonstrated when a knockout of the MeCP2 gene resulted in a defect in mouse embryonic development (48). In this study, an indirect mechanism of transcriptional repression by DNA methylation was investigated with a tissue-specific gene.
Human leukosialin (CD43) is a major sialoglycoprotein on the surfaces of hematopoietic cells. It has been demonstrated that this molecule plays a role in signal transduction as well as cell adhesion (15). The expression of leukosialin is regulated in a cell lineage-specific as well as a differentiation stage-specific manner. Leukosialin is present in T lymphocytes, granulocytes, monocytes, platelets, and hematopoietic stem cells but is absent in erythrocytes (8, 13, 18, 51). In an erythroid cell lineage, its expression is observed only at an early stage of differentiation and then decreases during cell maturation (4). Transcription from the TATA-less promoter of the leukosialin gene (25) is mediated by the transcription factor Sp1, which binds to the GGGTGG motif located about 40 bp upstream from the transcription start site (26). The transcription initiation sequence, which fits into the consensus sequence of an initiator (45, 46), is likely responsible for the basal level of transcription (28). This regulatory region is ubiquitously functional in mammalian cells, providing transcriptional activity comparable to that of the cytomegalovirus enhancer and promoter (54), which is one of the strongest transcriptional regulatory elements in mammalian cells (5). A previous study demonstrated that DNA methylation plays a pivotal role in leukosialin gene expression (28). There is a high positive correlation between gene activity and the demethylation state of the 5′ region of the leukosialin gene in various human cell lines and tissues. The DNA methyltransferase inhibitor 5-azacytidine was able to induce expression of the endogenous leukosialin gene in nonexpressing cells. In addition, in vitro DNA methylation of the 5′ region drastically reduced transcriptional activity in a transient-expression system. Thus, transcriptional regulation of the leukosialin gene is constitutively achieved by alterations in DNA methylation, and leukosialin gene expression provides an excellent system to study how DNA methylation regulates tissue-specific expression of a gene.
Here I tested the effect of in vitro DNA methylation of leukosialin reporter constructs in expression systems. I analyzed the influence of DNA methylation in a stable-transfection system with human cells. Next, I used Drosophila melanogaster cells, which lack genome methylation (37, 50), as recipient cells. Although Drosophila cells possess a general transcriptional machinery with functional homologs in mammalian cells (1, 19, 20), the transcription factor Sp1 is absent in Drosophila cells (10). In addition, it was shown previously that the leukosialin promoter requires exogenous Sp1 for transcriptional activation in Drosophila cells (26). Taking advantage of these facts, I first investigated the influence of CpG methylation on Sp1 transactivation of the leukosialin promoter in transient-expression assays with these cells. I then examined the effect of methyl-binding proteins on transcription from the methylated promoter. For this purpose, I isolated the human MeCP2 cDNA and expressed it in Drosophila cells. Using a cotransfection system, I show that MeCP2 strongly represses Sp1 transactivation when the promoter is methylated.
MATERIALS AND METHODS
Cell culture.
Human epithelial HeLa cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, 500 U of penicillin per ml, and 100 μg of streptomycin per ml. Human leukemia Jurkat cells (T lymphocytic) were maintained in RPMI 1640 medium with 20% FCS and the same supplements in a humidified 5% CO2 atmosphere. Drosophila Schneider cell line 2 (SL2), derived from Drosophila embryos (43) (kindly provided by R. Evans), was cultured in Schneider’s Drosophila medium (GIBCO BRL) with 10% FCS and 2 mM glutamine.
In vitro DNA methylation.
Leukosialin-chloramphenicol acetyltransferase (CAT) plasmids were constructed as described previously (25). pCAT-Basic (Promega) is a promoterless CAT vector utilized for leukosialin CAT construction. pCAT-Control (Promega) is an expression vector in which the CAT gene is under the control of the simian virus 40 (SV40) enhancer and promoter. These CAT plasmid DNAs were treated with SssI (CpG) methylase or HpaII methylase (New England Biolabs) in the presence (methylated) or absence (unmethylated) of 5 mM S-adenosylmethionine. After phenol extraction and ethanol precipitation, the methylated DNAs were treated with HpaII restriction enzyme. By electrophoresis on agarose gels, equal amounts of closed circular DNAs (more than 80% of total DNA) were observed between methylated and unmethylated plasmids.
Isolation of human methyl-CpG-binding protein MeCP2 cDNA and construction of MeCP2 expression plasmids.
A partial rat MeCP2 DNA fragment was obtained by PCR (40) of rat brain cDNA (Clontech) with primers based on sequences reported for rat MeCP2 (29). With rat MeCP2 DNA as a probe, a human leukemia cell line HL60 cDNA plasmid library (Invitrogen) was screened under low-stringency conditions. Seven positive clones were isolated, and the largest was sequenced by the dideoxy chain termination method (42). Drosophila expression plasmids of intact and C-terminal deletion mutants of MeCP2 were constructed as follows. Various MeCP2 cDNA fragments were generated by PCR with the largest insert as the template. The 5′ primer 5′TTTGAATTCAGAATACACCTTGCTTCTGT3′ contains the sequence from −43 to −23 relative to the translation start site, attached to an EcoRI recognition sequence and three thymine residues. This primer was used for all constructs. The 3′ primers contain 24 nucleotides derived from various coding regions and the 3′ untranslated region of MeCP2, preceded by three thymine residues, a BamHI recognition sequence, and a stop codon. The PCR products were digested with EcoRI and BamHI restriction enzymes and cloned into the EcoRI and BamHI cloning sites of the actin 5C vector (A5C) (kindly provided by R. M. Evans), an expression vector that drives a high level of transcription in Drosophila cells (49). As a control plasmid, the coding region of human glycophorin, an erythroid membrane protein, was amplified by PCR from the glycophorin A cDNA (27) and cloned into A5C. The fidelity of each construct was confirmed by sequencing with an ABI PRISM 377 DNA sequencer (Perkin-Elmer).
DNA transfection and CAT assay.
For stable-expression assays with HeLa cells, equimolar amounts of CAT constructs, equivalent to 10 μg of pCAT-Basic, were cotransfected into HeLa cells with 2 μg of the neomycin selection marker pcDNAINeo (Invitrogen) by a lipofection method (14). The DNA complex with Lipofectin (GIBCO BRL) was incubated with 106 cells in Opti-MEM (GIBCO BRL) for 5 h, and the transfection was stopped by the addition of two times the volume of DMEM with 10% FCS. After 48 h, the medium was changed to DMEM containing 400 μg of G418 per ml and cultivation was continued for 3 weeks. Approximately 50 to 120 colonies visible on each plate were trypsinized and pooled for replating onto a 100-mm-diameter dish. After cultivation, 5 × 106 cells were subjected to the CAT assay described by Gorman et al. (16) with a slight modification. For the stable-expression assay with Jurkat cells, amounts of the leukosialin CAT constructs equivalent to those described above were cotransfected into 106 Jurkat cells with 2 μg of pcDNAINeo (Invitrogen) by the calcium phosphate method (17). After 5 h, the medium was changed to RPMI 1640 with 20% FCS, and after 48 h, the cells were cultivated in medium containing 1 mg of G418 per ml. Every 3 to 4 days, half of the medium was slowly aspirated by pipette to maintain the cells on the plate and fresh medium containing G418 was added. After 2 weeks, approximately 10 to 30 clumped cells were visible. Cultivation was continued until the total number of cells exceeded 2 × 107. G418-resistant Jurkat cells (5 × 106) were subjected to the CAT assay described above. For the transient-expression assay with Drosophila SL2 cells, 4 μg of leukosialin CAT constructs was cotransfected with either 0.5 μg of an Sp1 expression plasmid, pPacSp1 (10) (kindly provided by J. T. Kadonaga), or an insertless expression vector, A5C. The effects of MeCP2 and its C-terminal deletion mutants were tested by adding 0.5 μg of expression constructs to the above-described DNA mixture. Cells (106) were transfected by the calcium phosphate method (17), and after 48 h, CAT activities were determined.
Southern blot hybridization.
High-molecular-weight genomic DNAs were prepared from stably transfected cells by an NaDodSO4-proteinase K treatment, following phenol-chloroform extraction as described previously (24). Ten-microgram samples of genomic DNAs were digested with MspI or HpaII restriction enzymes, resolved on a 1.5% agarose gel, and transferred to a Nytran nylon filter (Schleicher & Schuell). For the probe, a 560-bp MspI fragment of the 5′ region was prepared from the leukosialin genomic clone LeuS-2 (25) and labeled with [α-32P]dCTP. The blot was prehybridized, hybridized, and washed as described previously (41).
Fluorescence microscopy.
A fusion protein containing the N-terminal 70 amino acid residues of MeCP2 was produced with the glutathione S-transferase gene system (47). A rabbit was immunized three times with 1 mg of fusion protein in Freund’s complete adjuvant, and polyclonal antibodies were purified on protein A-agarose (GIBCO BRL). Drosophila SL2 cells were transfected with expression constructs of MeCP2 and its C-terminal deletion mutants by the calcium phosphate method (17). Two days later, cells were fixed with 3.7% formaldehyde for 30 min and permeabilized with 0.5% Triton X-100 for 20 min. After being blocked in phosphate-buffered saline containing 4 mg of bovine serum albumin per ml, transfected cells were incubated with the above-described polyclonal anti-MeCP2 antibodies in a 1:100 dilution for 2 h at room temperature. After several washings with phosphate-buffered saline, the cells were incubated with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Sigma) for 1 h at room temperature. The cells were washed and observed under an Olympus Fluoview laser microscope. The nuclear regions of the cells were identified by staining the cells with the DNA-binding dye propidium iodide (Molecular Probes).
Nucleotide sequence accession number.
The sequence of the 1,669-bp MeCP2 cDNA insert was reported to GenBank with the accession no. L37298.
RESULTS
Constitutive repression of the promoter of leukosialin-CAT constructs in stably transfected human cell lines after in vitro methylation of CpG sites.
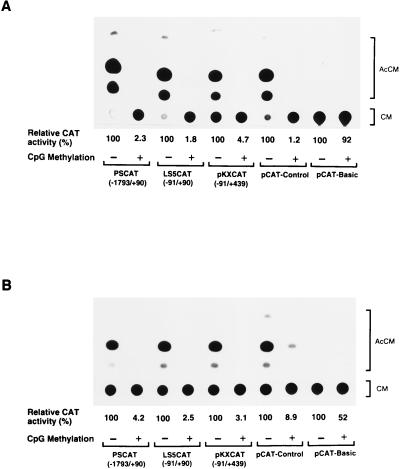
In a previous study, the effect of in vitro methylation at CpG sites of the 5′ region of the leukosialin gene was analyzed by CAT assay in a transient-transfection system (28). These results showed that the transcriptional activity of the 5′ region was substantially reduced by CpG methylation. Since transient-transfection assays require high copy numbers of nonintegrated transgenes, I employed a stable-expression system in order to more faithfully reproduce in vivo gene expression. In addition, it has been demonstrated that Sp1 binding sites play a key role in the maintenance of a methylation-free CpG island in a housekeeping gene, the adenine phosphoribosyltransferase (APRT) gene (7, 30). Since the leukosialin promoter contains an Sp1 binding site, I tested whether this site could demethylate the methylated-leukosialin CAT constructs and induce transcriptional activity in a stable-expression system. First, the effect of in vitro methylation on the transcriptional activity of leukosialin CAT constructs was analyzed in non-leukosialin-expressing HeLa cells. For this purpose, I used three leukosialin CAT constructs: PSCAT(−1793/+90), LS5CAT(−91/+90), and pKXCAT(−91/+439) (25). These constructs were methylated in vitro with SssI (CpG) methylase and cotransfected with pcDNAINeo into HeLa cells. G418-selected cells were subjected to the CAT assay as described in Materials and Methods. The results are presented in Fig. 1A. All three constructs showed a significant reduction in transcriptional activity resulting from CpG methylation in stably transfected HeLa cells. On the other hand, unmethylated constructs showed considerable CAT activities, and these results were consistent with those observed in transient-transfection assays (28). An approximately 20- to 50-fold reduction in activity by CpG methylation was observed with these constructs. Similar repression of transcriptional activity was observed in pCAT-Control, where the CAT gene is under the control of the SV40 enhancer and promoter (Fig. 1A).
FIG. 1.
Effect of in vitro DNA methylation on transcriptional activities of the 5′ regions of the leukosialin gene exogenously introduced and stably maintained in leukosialin-expressing and non-leukosialin-expressing cells. (A) CAT constructs were in vitro methylated with SssI (CpG) methylase. Methylated or unmethylated constructs were cotransfected with pcDNAINeo into non-leukosialin-expressing HeLa cells. After G418 selection, visible colonies were pooled and G418-resistant cells were subjected to the CAT assay described in Materials and Methods. pCAT-Basic is a promoterless vector used for the leukosialin CAT constructs. pCAT-Control possesses the SV40 enhancer and promoter. Relative CAT activities compared with that of each unmethylated construct are presented. The values are averages of results from three independent experiments. (B) The same experiment was conducted with leukosialin-expressing Jurkat cells. G418-resistant cells were obtained as described in Materials and Methods, and cellular extracts were subjected to the CAT assay. AcCM, acetylchloramphenicol; CM, chloramphenicol.
Next, I investigated the effect of methylation on transcriptional activity in a leukosialin-expressing hematopoietic cell line, Jurkat. The constructs described above were cotransfected with pcDNAINeo into Jurkat cells, and G418-resistant cells were subjected to the CAT assay. The results were similar to those seen with stably transfected HeLa cells, showing that methylation greatly reduced transcriptional activity in these leukosialin-expressing cells (Fig. 1B). In this case, the transcriptional activity of pCAT-Control was less affected by methylation than that of HeLa cells. Transcriptional activation was not observed even in the leukosialin-expressing cells despite the fact that the Sp1 element plays a dominant role in leukosialin gene expression.
Methylation states of the 5′ regulatory regions, methylated in vitro and stably transfected in HeLa cells.
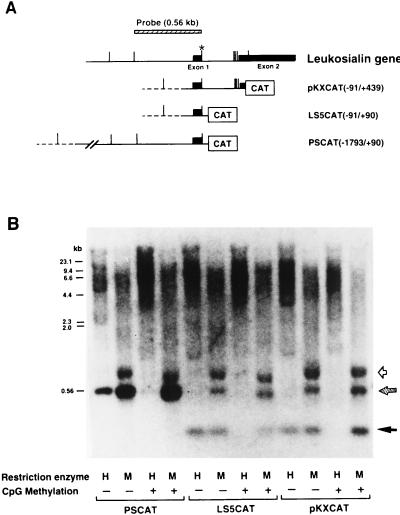
In order to determine whether repression of transcriptional activity of methylated-leukosialin constructs was due to a continuously methylated state of the regulatory region, I performed Southern blot hybridization analyses of genomic DNAs from stably transfected HeLa cells. Methylation-sensitive HpaII and insensitive MspI restriction enzymes were used for digestion. Maps of transfected constructs and the probe used for the analysis are shown in Fig. 2A. A previous study showed that MspI digestion produces two hybridizing bands in HeLa cells (28). One is a 0.56-kb DNA sequence between two CCGG sites at −493 and +68 (Fig. 2B). The other band is a 0.8-kb DNA, which appears to be generated by the polymorphic difference of the CCGG sequence at +68. These two bands were only faintly visible following HpaII digestion of genomic DNA from untransfected HeLa cells, which shifted bands to higher molecular weights. Genomic DNA derived from HeLa cells stably transfected with unmethylated PSCAT(−1793/+90) showed hypomethylation of the regulatory region of the transfected gene, which produced a 0.56-kb HpaII band (Fig. 2B). Stable transfection with methylated PSCAT did not produce the 0.56-kb band but produced higher-molecular-size smeared bands, indicating that the regulatory region of the transfected gene is still methylated. Similar results were obtained with LS5CAT(−91/+90) and pKXCAT(−91/+439). With these constructs, the CCGG site at +68 was present in the constructs but the upstream site at −493 was not. Therefore, MspI digestion showed a lower band, one generated by cleavage of the sequence in the CAT vector (pCAT-Basic) (Fig. 2B). Without methylation, this band was observed following HpaII digestion, but with CpG methylation of these constructs, the band shifted to higher molecular weights. Thus, the methylation states of the regulatory regions in the sequences of the transfected constructs were largely maintained in HeLa cells. Therefore, demethylation of the region surrounding the Sp1 element, which has been observed in the APRT gene in ES cells and in transgenic mice (7, 30), was not detected in this study.
FIG. 2.
Methylation states of the 5′ regions of the leukosialin gene exogenously introduced and stably maintained in non-leukosialin-expressing HeLa cells. (A) Schematic representations of the leukosialin gene and leukosialin CAT constructs. The exons are depicted by filled boxes, and introns and the 5′ flanking regions are depicted with horizontal lines. The vector sequences are depicted with dotted lines. MspI (CCGG) sites are shown with vertical bars. The asterisk indicates the polymorphic site, where an MspI site is lost in one allele of HeLa cells. The MspI DNA fragment (560 bp) used for a hybridization probe is presented at the top. (B) Genomic DNAs (10 μg) from HeLa cells stably transfected with CpG-methylated- or unmethylated-leukosialin CAT constructs were digested with HpaII (H) or MspI (M) and separated by 1.5% agarose gel electrophoresis. The blotted filter was hybridized with the 560-bp DNA fragment of the 5′ region of the leukosialin gene shown in panel A. The hatched arrow indicates the position of the fragments produced by the endogenous gene as well as the exogenously introduced gene. The filled arrow indicates the signal produced by the exogenously introduced gene. The open arrow indicates the position of a polymorphic fragment of the endogenous leukosialin gene of HeLa cells.
Sp1 can transactivate the leukosialin promoter in Drosophila cells even when the promoter is methylated.
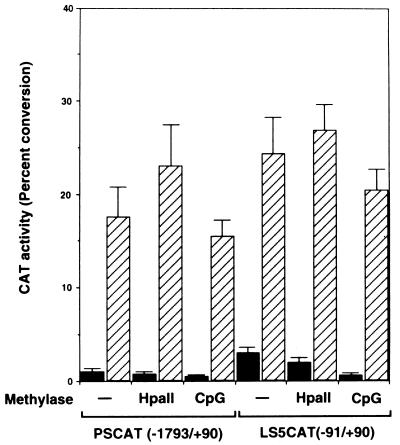
The above results also suggest that endogenous methyl-binding proteins might participate in constitutive repression of transcription from a methylated template. Thus, the level of repressive activity appears to be high in human cells. In order to determine what factors are involved in repression of transcriptional potential of the leukosialin gene by DNA methylation, it is crucial to employ a transfection system that uses host cells deficient in or with low numbers of endogenous methyl-CpG repressors. Since Drosophila cells lack genomic methylation (37, 50), it is likely that methyl-binding proteins are present at very low levels in these cells. Therefore, I chose Drosophila Schneider cell line 2 (SL2) cells as recipient cells to perform a transient-expression assay. The effect of in vitro methylation of the leukosialin CAT constructs PSCAT and LS5CAT was tested in Drosophila SL2 cells. The results are shown in Fig. 3. In agreement with a previous observation (26), the leukosialin regulatory region could confer a weak transcriptional activity without cotransfection of the Sp1 expression vector pPacSp1. This activity might represent the basal transcriptional level conferred by an initiator-like element (26). Cotransfection of pPacSp1 led to an approximately 10-fold increase in promoter activity from the unmethylated-leukosialin promoter. Basal transcriptional activities in the absence of Sp1 were decreased by DNA methylation with HpaII and SssI (CpG) methylase. Notably, a 10-fold reduction in transcriptional activity was observed in LS5CAT when the promoter was methylated with SssI (CpG) methylase. Therefore, the basal transcriptional activity was inhibited by DNA methylation in Drosophila SL2 cells. In contrast, cotransfection of pPacSp1 significantly increased transcriptional activity from the methylated promoter. In the presence of Sp1, the transcriptional activities of the CpG methylated promoter were enhanced to the 60 to 70% level of unmethylated promoter activities. It is noteworthy that the HpaII methylated promoter produced higher transcriptional activity than the unmethylated promoter in the presence of Sp1. These results indicate that Sp1 can transactivate the leukosialin promoter in Drosophila SL2 cells even when the promoter is methylated.
FIG. 3.
Transactivation of methylated-leukosialin promoter activity by Sp1 in Drosophila cells. The CAT constructs were methylated in vitro with HpaII or SssI (CpG) methylase. Methylated or unmethylated CAT constructs (4 μg) were transfected with 0.5 μg of A5C (filled bars) or with 0.5 μg of pPacSp1 (hatched bars) into Drosophila SL2 cells. After 48 h, the cellular extract was subjected to the CAT assay. CAT activity is presented as a percentage of the level of conversion of the acetylated form. Each value is the mean and standard deviation of results from three independent experiments.
Coexpression of human MeCP2 cDNA reduced transcriptional activity from the methylated promoter in Drosophila cells.
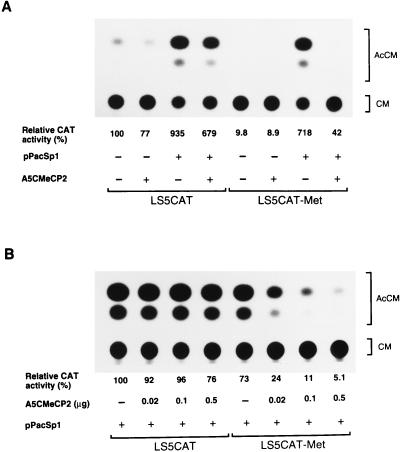
In order to determine whether down-regulation of the leukosialin gene by DNA methylation is mediated through methyl-CpG-binding proteins, the effect of human MeCP2 on transcriptional activity was tested with Drosophila SL2 cells. In this experiment, I isolated human MeCP2 cDNAs from a HL60 cDNA library by using a rat cDNA as a probe. The largest insert, 1,669 bp (GenBank accession no. L37298), has an open reading frame encoding a product of 486 amino acid residues, which is 94% identical to the rat MeCP2 (29). The MeCP2 cDNA was cloned into A5C, which provided high expression in Drosophila cells driven by the A5C promoter (49), and the expression vector was designated A5CMeCP2. The effect of MeCP2 on basal and Sp1-activated transcription was examined by cotransfection. The results of transient-transfection experiments are presented in Fig. 4A. Cotransfection of A5CMeCP2 resulted in only a slight decrease in the transcriptional activity of the unmethylated LS5CAT in the presence or absence of Sp1. Similarly, in the absence of Sp1, addition of A5CMeCP2 had a slight effect on the transcriptional activity of methylated LS5CAT. On the other hand, in the presence of Sp1, the transcriptional activity of methylated LS5CAT was sharply reduced by addition of A5CMeCP2. In this case, MeCP2 decreased Sp1-activated transcription of the leukosialin promoter by 12-fold. The level of repression is directly proportional to the amount of MeCP2 expression vector transfected (Fig. 4B). This repression by MeCP2 is specific for the methylated CpG promoter, since the addition of A5CMeCP2 had little influence on the transcriptional activity of the unmethylated promoter. These results indicate that MeCP2 represses Sp1 transactivation when the promoter is methylated.
FIG. 4.
Repressive effect of MeCP2 on Sp1-activated transcription of the methylated-leukosialin promoter in Drosophila cells. (A) The LS5CAT(−91/+90) construct was methylated in vitro with SssI (CpG) methylase. Methylated or unmethylated LS5CAT (4 μg) was transfected with or without the Sp1 expression plasmid pPacSp1 (0.5 μg) and with or without the MeCP2 expression plasmid A5CMeCP2 (0.5 μg) into Drosophila SL2 cells. The total amount of transfected DNAs was adjusted to 5 μg by adding A5C. Relative CAT activities compared with those of unmethylated-LS5CAT transfection are presented. Values are averages of results from three independent experiments. (B) Effect of A5CMeCP2 concentration on the transcriptional repression of LS5CAT(−91/+90) in Drosophila SL2 cells. Methylated or unmethylated LS5CAT construct (4 μg) was cotransfected with pPacSp1 (0.5 μg) and various amounts of A5CMeCP2 (0.02 to 0.5 μg). The total amount of transfected DNA was adjusted to 5 μg by adding A5C. Relative CAT activities are shown as described for panel A. AcCM, acetylchloramphenicol; CM, chloramphenicol; Met, methylated.
The MBD of MeCP2 is required for repression of Sp1-activated transcription.
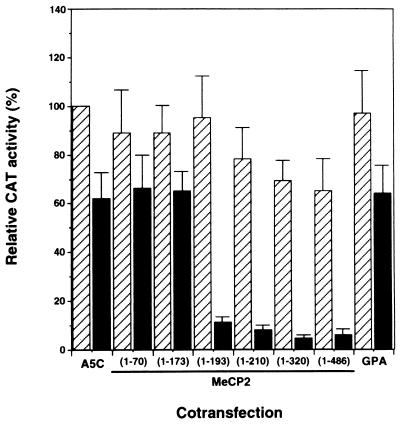
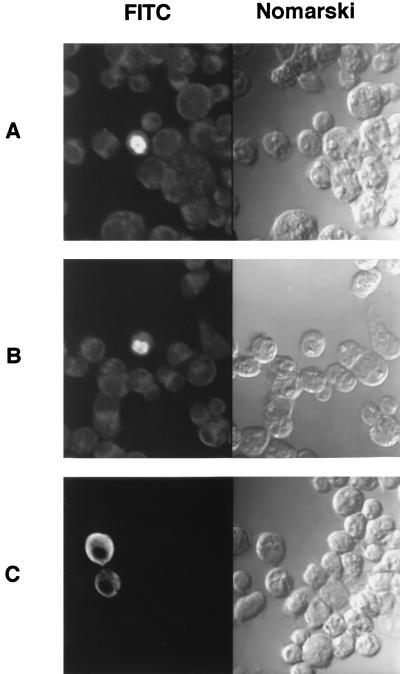
To localize the region of MeCP2 responsible for repression of Sp1 transactivation, C-terminal deletion mutants of MeCP2 cDNA were generated and cloned into A5C as described in Materials and Methods. These constructs were introduced into Drosophila SL2 cells with pPacSp1 and methylated or unmethylated LS5CAT. Figure 5 shows the results of cotransfection experiments with C-terminal deletion mutants. Significant reduction of transcriptional activity from the methylated promoter was observed in the constructs bearing DNA encoding at least the N-terminal 193 amino acid residues. The transcriptional activities of the methylated constructs were reduced more than sixfold compared with those of unmethylated constructs. In these constructs transcriptional repression was observed apparently when the CAT construct was methylated. When an expression vector bearing DNA encoding nonnuclear protein glycophorin A, which is a membrane protein found in human erythrocytes, was cotransfected, the transcriptional level was reduced to a much smaller extent by CpG methylation. The minimal region of repressive activity includes a methyl-binding domain (MBD) (amino acid residues 78 to 162) (34). Further deletion resulted in the loss of repressor activity. The mutant form of MeCP2 containing amino acid residues 1 to 173 and an MBD, however, did not show repressive activity. To discover the cellular localization of C-terminal deletion mutants, immunofluorescence analyses of transfected SL2 cells were conducted with rabbit anti-MeCP2 antibodies against the MeCP2 fusion protein representing amino acids 1 to 70 (Fig. 6). The mutant containing the N-terminal 193 amino acid residues exhibited exclusive nuclear distribution (Fig. 6B), which was verified by staining with propidium iodide (data not shown). The nuclear accumulation of this mutant is comparable to that of full-length MeCP2 (Fig. 6A). On the other hand, the mutant containing the N-terminal 173 amino acid residues predominantly showed cytoplasmic staining (Fig. 6C). Therefore, it is likely that this mutant lacks a nuclear localization signal (NLS), which may be located between amino acid residues 173 and 193. These results indicate that the MBD of MeCP2 is required for suppression of Sp1 transactivation and the resulting decrease in leukosialin gene activity by CpG methylation.
FIG. 5.
Effect of a carboxy-terminal deletion of MeCP2 on transcriptional repression of the methylated-leukosialin promoter. Methylated (filled bars) or unmethylated (hatched bars) LS5CAT(−91/+90) construct (4 μg) was cotransfected with pPacSp1 (0.5 μg) and an expression plasmid (0.5 μg) bearing genes encoding various C-terminal deletion mutations of MeCP2 into Drosophila SL2 cells. Amino acid residues of MeCP2 encoded in the mutants are shown in parentheses. A5C is the insertless expression vector. GPA is a control plasmid in which the cDNA of human glycophorin A, an erythroid membrane protein, is cloned into the A5C vector. Relative CAT activities compared with that of unmethylated LS5CAT cotransfected with pPacSp1 and A5C are presented. Values are the means and standard deviations of results from three independent experiments.
FIG. 6.
Localization of MeCP2 and its C-terminal deletion mutants expressed in Drosophila cells. SL2 cells were transiently transfected with the A5C expression plasmid bearing DNA encoding full-length MeCP2 (A), the N-terminal 193 amino acid residues of MeCP2 (B), or the N-terminal 173 amino acid residues of MeCP2 (C). The cells were stained with rabbit polyclonal antibodies against the MeCP2 fusion protein representing amino acids 1 to 70 and imaged by fluorescence microscopy and with Nomarski optics. FITC, fluorescein isothiocyanate.
DISCUSSION
The leukosialin gene is expressed in tissue-specific and differentiation stage-specific manners. Previous results indicated that the strong promoter composed of an Sp1 binding site and a sequence similar to that of an initiator was regulated by DNA methylation (28). Therefore, leukosialin gene regulation provides an excellent system to understand how DNA methylation is involved in tissue-specific gene expression. The effect of DNA methylation on the transcriptional activity of the leukosialin promoter was tested in a stable-transfection system with expressing Jurkat and nonexpressing HeLa cells. In both cell lines, transcriptional activities were drastically suppressed by CpG methylation of leukosialin CAT constructs. A similar reduction was also observed in transient-transfection systems with HeLa cells (28) and Jurkat cells (data not shown). Southern blot hybridization of genomic DNAs from stably transfected cells showed that the exogenously introduced leukosialin promoter maintained the same methylation state as that of transfected constructs. Recent studies have demonstrated that an Sp1 binding site is involved in the demethylation of the CpG island of the APRT gene (7, 30). However, demethylation of the methylated leukosialin promoter was not observed in the stable-expression system despite the presence of an Sp1 binding site. It is likely that demethylation requires dramatic phenotypic changes in cells leading to differentiation or that demethylation occurs in the cells having such potential. In fact, demethylation of the CpG island was successful in embryonic stem cells (7).
Human cells appear to have a strong repressive activity mediated by methyl-binding proteins. Therefore, in order to determine what kinds of methyl-CpG repressors are involved in silencing a methylated gene, host cells deficient in such repressors were required for transfection experiments. In this study Drosophila SL2 cells were adopted as recipient cells. It is known that the Drosophila genome does not exhibit detectable amounts of DNA methylation (37, 50). Therefore, it was assumed that Drosophila cells do not possess significant DNA methylation capacity. In addition, the general transcriptional machineries of Drosophila and mammals are thought to be highly conserved (1, 19, 20). Thus, it is presumed that Drosophila cells are deficient in methyl-binding proteins and provide a suitable system to investigate the effect of methylation on the transcription of a mammalian gene. Furthermore, it is known that Drosophila SL2 cells lack Sp1 activity (10). Taking advantage of these facts, I tested the effect of methylation on Sp1-activated transcription of the leukosialin gene. In this study, I observed a substantial decrease in transcriptional activity in the absence of Sp1 when the leukosialin CAT construct was methylated. This inhibition might be due to binding of ubiquitous nuclear factor(s) such as the histone H1. However, the binding affinity of the repressive factor appeared weak because Sp1 expression could transactivate equally well both the methylated promoter and the unmethylated promoter. A similar observation was also reported by Rhodes et al. (39), who showed that the basal transcriptional level of the methylated α1(I) collagen promoter was considerably repressed in SL2 cells and that Sp1 could transactivate the methylated promoter. Thus, Sp1 may exclude such repressors from the methylated promoter and enhance transcription. In this regard, it has been demonstrated that Sp1 can replace the histone H1 repressor that binds to a regulatory sequence and activate transcription (12). Since this evidence was obtained with the Drosophila system, it is likely that the increases in transcriptional activities observed following cotransfection with an Sp1 expression vector resulted from this mechanism. In Drosophila cells, therefore, endogenous suppressive proteins exist but do not function as strong repressors for CpG methylated genes.
In this study the effect of MeCP2 on transcription from a methylated promoter was tested. A human MeCP2 cDNA obtained from an HL60 cDNA library was 88% identical to the rat cDNA at the nucleotide level and 94% identical at the amino acid level (29). Expression of human MeCP2 in Drosophila cells significantly reduced Sp1-activated transcription from the methylated-leukosialin promoter. The basal transcriptional level and Sp1-activated transcription of the unmethylated promoter were minimally influenced by cotransfection of A5CMeCP2. In this cotransfection experiment it was difficult to determine whether the transcriptional activity of the methylated-leukosialin promoter in the absence of Sp1 was affected by MeCP2, because the transcriptional activity of the methylated-CAT construct without Sp1 is so low. Therefore, the question of whether the basal transcriptional level of the leukosialin gene is reduced by MeCP2 will require further analysis. The repressive effect of MeCP2 on Sp1 transactivation was positively correlated with the concentration of cotransfected A5CMeCP2 in the methylated DNA, but increasing the amount of A5CMeCP2 produced only minimal repression of transcriptional activity of the unmethylated promoter. Thus, these results demonstrate that the repressive activity of MeCP2 to a methylated template can be reconstituted in Drosophila cells.
C-terminal deletion analysis of MeCP2 revealed repressive activities in constructs bearing DNA encoding at least the N-terminal amino acid residues 1 to 193, which encompass the MBD. This observation was not consistent with the previously obtained result that repressive activity is localized to a region between amino acid residues 207 and 310 of mouse MeCP2 (36). This discrepancy may result from the different systems used for the analysis. However, using an in vitro transcription system, Nan et al. also found that activity of the methylated adenovirus late promoter was suppressed by mutants containing only the MBD (36). A previous study localized the NLS to amino acid residues 255 to 271 of mouse MeCP2 (35), but my immunofluorescence analyses indicated an additional NLS located between amino acid residues 173 and 193. Although this NLS was found not to be active in mammalian cells, it may be functional in Drosophila cells to facilitate transport into the nucleus. When these findings are considered together, it appears that the MBD of MeCP2 is sufficient for the repression of Sp1 transactivation. Although the present study employed in vitro methylation of the whole CAT constructs, the CAT gene and vector sequences seemed to be less effective targets for this repression. Recent study of the transient expression of ρ-globin CAT constructs, which had CpG methylation in the various components of the plasmid, showed that promoter methylation is a major mediator in the suppression of transcription (44). At present it is not clear whether MeCP2 binding to methylated CpG can interfere with Sp1 binding to the leukosialin promoter. The Sp1 binding site of the leukosialin promoter is a GGGTGG in core sequence and is not a CpG methylation site (25). Therefore, a methylated-CpG site adjacent to the Sp1 site available for MeCP2 binding is responsible for suppression of Sp1-activated transcription. Further studies are required to define whether binding of MeCP2 to methylated CpG blocks cis-regulatory sequences and prevents the access of Sp1 and/or the basal transcriptional machinery of RNA polymerase II. Structurally, the leukosialin promoter is not rich in CpG sites, and probably the number of those sites is not sufficient for interaction with a different methyl-CpG-binding protein, MeCP1, which requires at least 12 CpG sites for binding (32). Therefore, MeCP2 might be an important factor that strongly suppresses the leukosialin gene when it is methylated. Further studies are needed to reveal whether weak methyl-binding repressors such as histone H1 also contribute to down-regulation of the methylated-leukosialin gene in human cells. Recently, it was demonstrated that the Sp1 knockout mouse showed significantly reduced expression of MeCP2 (31). Therefore, it is possible that Sp1 transactivation and transcriptional repression by MeCP2 are coupled regulatory mechanisms in higher-order eukaryotes and that these two types of regulation also contribute to the epigenetic mechanism of DNA methylation required for proper gene expression.
ACKNOWLEDGMENTS
I thank M. Fukuda and H. M. Ranney for helpful discussions, J. T. Kadonaga for providing the pPacSp1 plasmid, R. M. Evans for providing the A5C vector, and S. Iki and S. Yoshizumi for technical assistance.
REFERENCES
- 1.Ahearn J M, Bartolomei M S, West M L, Cisek L J, Corden J L. Cloning and sequence analysis of the mouse genomic locus encoding the largest subunit of RNA polymerase II. J Biol Chem. 1987;262:10695–10705. [PubMed] [Google Scholar]
- 2.Antequera F, Bird A P. Molecular biology and biological significance. In: Jost J P, Saluz H P, editors. DNA methylation. Basel, Switzerland: Brickhauser Verlag; 1993. pp. 169–185. [Google Scholar]
- 3.Bednarik D P, Duckett C, Kim S U, Perez V L, Griffis K, Guenther P, Folks T M. DNA CpG methylation inhibits binding of NF-κB proteins to the HIV-1 long terminal repeat cognate DNA motifs. New Biol. 1991;3:969–976. [PubMed] [Google Scholar]
- 4.Bettaib A, Farance F, Mitjavila M T, Mishal Z, Dokhelar M C, Tursz T, Breton-Gorius J, Vainchenker W, Kieffer N. Use of a monoclonal antibody (GA3) to demonstrate lineage restricted O-glycosylation on leukosialin during terminal erythroid differentiation. Blood. 1988;71:1226–1233. [PubMed] [Google Scholar]
- 5.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckensyein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 6.Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell. 1991;64:1123–1134. doi: 10.1016/0092-8674(91)90267-3. [DOI] [PubMed] [Google Scholar]
- 7.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson S R, Fukuda M. Isolation and characterization of leukosialin, a major sialoglycoprotein on human leukocytes. J Biol Chem. 1986;261:12779–12786. [PubMed] [Google Scholar]
- 9.Comb M, Goodman H W. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 11.Cross S H, Meehan R R, Nan X, Bird A. A component of the transcriptional repressor MeCP1 shares a motif with DNA methyltransferase and HRX proteins. Nat Genet. 1997;16:256–259. doi: 10.1038/ng0797-256. [DOI] [PubMed] [Google Scholar]
- 12.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 13.Dyer M J S, Hunt S V. Committed T-lymphocyte stem cells of rats characterized by surface W3/13 antigen and radiosensitivity. J Exp Med. 1981;154:1164–1177. doi: 10.1084/jem.154.4.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felgner P L, Gadek T R, Holm M, Roman R, Chan H W, Wenz M, Northrop J P, Ringold G M, Danielsen M. Lipofectin: a high efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukuda M. Leukosialin, a major O-glycan-containing sialoglycoprotein defining leukocyte differentiation and malignancy. Glycobiology. 1991;1:347–356. doi: 10.1093/glycob/1.4.347. [DOI] [PubMed] [Google Scholar]
- 16.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Gulley M L, Ogata L C, Thorson J A, Dailey M O, Kemp J D. Identification of murine Pan-T cell antigen which is also expressed during the terminal phases of B cell differentiation. J Immunol. 1988;140:3751–3757. [PubMed] [Google Scholar]
- 19.Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- 20.Heiermann R, Pongs O. In vitro transcription with extracts of nuclei of Drosophila embryos. Nucleic Acids Res. 1985;13:2709–2730. doi: 10.1093/nar/13.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jost J-P, Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo to methylated nonspecific DNA sequences. Proc Natl Acad Sci USA. 1992;89:9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kass S U, Pruss D, Wolffe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 23.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 24.Kudo S, Fukuda M. Structural organization of glycophorin A and B genes: glycophorin B gene evolved by homologous recombination at Alu repeat sequences. Proc Natl Acad Sci USA. 1989;86:4619–4623. doi: 10.1073/pnas.86.12.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudo S, Fukuda M. A short, novel promoter sequence confers the expression of human leukosialin, a major sialoglycoprotein on leukocytes. J Biol Chem. 1991;266:8483–8489. [PubMed] [Google Scholar]
- 26.Kudo S, Fukuda M. Transcriptional activation of human leukosialin (CD43) gene by Sp1 through binding to a GGGTGG motif. Eur J Biochem. 1994;223:319–327. doi: 10.1111/j.1432-1033.1994.tb18997.x. [DOI] [PubMed] [Google Scholar]
- 27.Kudo S, Onda M, Fukuda M. Characterization of glycophorin A transcripts: control by the common erythroid-specific promoter and alternative usage of different polyadenylation signals. J Biochem. 1994;116:183–192. doi: 10.1093/oxfordjournals.jbchem.a124492. [DOI] [PubMed] [Google Scholar]
- 28.Kudo S, Fukuda M. Tissue-specific transcriptional regulation of human leukosialin (CD43) gene is achieved by DNA methylation. J Biol Chem. 1995;270:13298–13302. doi: 10.1074/jbc.270.22.13298. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 30.Macleod D, Charlton J, Mullis J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 31.Marin M, Karis A, Visser P, Grosveld F, Philipsen S. Transcription factor Sp1 is essential for early embryonic development but dispensable for cell growth and differentiation. Cell. 1997;89:619–628. doi: 10.1016/s0092-8674(00)80243-3. [DOI] [PubMed] [Google Scholar]
- 32.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpG. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 33.Meehan R R, Lewis J D, Bird A P. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nan X, Meehan R R, Bird A. Dissection of the methyl-CpG binding domain from the chromosome protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 37.Patel C V, Gopinathan K P. Determination of trace amount of 5-methylcytosine in DNA by reverse-phase high-performance liquid chromatography. Anal Biochem. 1987;164:164–169. doi: 10.1016/0003-2697(87)90381-2. [DOI] [PubMed] [Google Scholar]
- 38.Prendergast G C, Lawe D, Ziff E B. Association of Myn, the murine homolog of max, with c-myc stimulates methylation-sensitive DNA binding and ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes K, Rippe R A, Umezawa A, Nehls M, Brenner D A, Breindl M. DNA methylation represses the murine α1(I) collagen promoter by an indirect mechanism. Mol Cell Biol. 1994;14:5950–5960. doi: 10.1128/mcb.14.9.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 44.Singal R, Ferris R, Little J A, Wang S Z, Ginder G D. Methylation of the minimal promoter of an embronic globin gene silences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smale S T, Baltimore D. The “Initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 46.Smale S T, Schmit M C, Berk A J, Baltimore D. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci USA. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 48.Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 49.Thummel C S, Boulet A M, Lipshitz H D. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- 50.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 51.Vargas-Cortes M, Axelsson B, Larsson A, Berzins T, Perlman P. Enhancement of human spontaneous cell-mediated cytotoxicity by a monoclonal antibody against the large sialoglycoprotein (CD43) on peripheral blood lymphocytes. Scand J Immunol. 1988;27:661–671. doi: 10.1111/j.1365-3083.1988.tb02399.x. [DOI] [PubMed] [Google Scholar]
- 52.Weintraub H. Histone-H1-dependent chromatin super-structures and the suppression of gene activity. Cell. 1984;38:17–27. doi: 10.1016/0092-8674(84)90522-1. [DOI] [PubMed] [Google Scholar]
- 53.Weitzel J M, Buhrmester H, Strätling W H. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol Cell Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu A, Kudo S, Fukuda M. A novel expression vector composed of a regulatory element of the human leukosialin-encoding gene in different types of mammalian cells. Gene. 1995;160:283–286. doi: 10.1016/0378-1119(95)00206-l. [DOI] [PubMed] [Google Scholar]