ABSTRACT
Methylation modifications play pertinent roles in regulating gene expression and various biological processes. The silencing of the demethylase enzyme TET1 can affect the expressions of key oncogenes or tumour suppressor genes, thus contributing to tumour formation. Nonetheless, how TET1 affects the progression of cervical cancer is yet to be elucidated. In this study, we found that the expression of TET1 was significantly downregulated in cervical cancer tissues. Functionally, TET1 knockdown in cervical cancer cells can promote cell proliferation, migration, invasion, cervical xenograft tumour formation and EMT. On the contrary, its overexpression can reverse the aforementioned processes. Moreover, the autophagy level of cervical cancer cells can be enhanced after TET1 knockdown. Mechanistically, methylated DNA immunoprecipitation (MeDIP)-sequencing and MeDIP quantitative real-time PCR revealed that TET1 mediates the methylation of autophagy promoter regions. These findings suggest that TET1 affects the autophagy of cervical cancer cells by altering the methylation levels of NKRF or HIST1H2AK, but the specific mechanism needs to be investigated further.
KEYWORDS: Cervical cancer, TET1, autophagy, EMT
Introduction
Cervical cancer (CC) is one of the most prevalent gynaecological tumours and the fourth leading cause of female cancer-related mortality, with ~ 604,127 new cases of CC and 341,831 CC-related deaths reported worldwide [1]. Although the vaccine against cervical cancer has been available for more than 10 years, most women are still unvaccinated globally [2]. Therefore, clarifying the molecular mechanism of cervical cancer invasion and metastasis is immensely significant for improving patient prognosis and developing effective targeted drugs. Cervical cancer is mainly caused by human papillomavirus (HPV), but most HPV infections are transient and only some cells continue to remain infected [3]. This transient infection is insufficient to cause cervical epithelial transformation, and it is necessary to accumulate epigenetic aberrations later [4]. Furthermore, DNA methylation and histone abnormalities are necessary for the progression of cervical cancer [5]. Epigenetic changes have been observed to be related to the occurrence of cervical cancer. Related studies have shown that DNA methylase can regulate the invasion and metastasis of cervical cancer via different signal pathways [6,7]. However, few reports exist on the role of TET1 demethylation modification in the development of cervical cancer.
TET1, one of the main members of the TET family located at 10q21.3, was first identified in a patient with acute myeloid leukaemia who had chromosomal translocation [8]. TET1 contains three nuclear localization signals that indicate potential nuclear activity [9], which are predominantly expressed in foetal tissues, stem cells, and solid tumours [10]. TET1 is a highly active DNA cytosine oxygenase, which can be converted from 5-methylcytosine (5mc) to 5-hydroxymethylcytosine (5hmC) or compete with DNA methyltransferase, thereby causing passive demethylation and maintaining the unmethylated state of tumour suppressor genes [11,12]. TET1 abnormality is often closely linked to the occurrence and development of tumours, such as liver cancer, colorectal cancer, glioma, and acute leukaemia [13–16]. Many studies have shown that TET1 has varied functions in different malignant tumours. However, its role in cervical cancer remains unclear.
Autophagy is an evolutionarily conserved intracellular catabolic degradation process in which cytoplasmic macromolecules, aggregated proteins, damaged organelles, and pathogens are delivered to lysosomes and digested by hydrolases to release nucleotides, amino acids, fatty acids, sugars, and ATP, which are eventually cycled into the cytoplasmic matrix [17–22]. As type II programmed cell death, autophagy plays a crucial role in cancer, yet its dual role in cancer progression and inhibition remains controversial [23]. In most solid tumours, the dysregulation of TET1 often leads to tumorigenesis and causes alterations in a series of pathways, but how TET1 autophagy affects the occurrence and progression of cervical cancer is yet to be examined.
Therefore, this study aimed to investigate the changes in TET1 in cervical cancer and whether TET1 can affect cancer progression via autophagy.
Materials and methods
Clinical samples
We collected tissue samples from 15 cases of highly differentiated and poorly differentiated cervical carcinoma, as well as the corresponding paracancerous normal tissues, for the study of TET1, 5mc, and 5hmc expression. The average age of patients with normal cervical tissues was 50.70 ± 6.76 years, while the average age of patients with cervical cancer was 49.18 ± 9.63 years. There was no statistically significant difference in age between the two groups (p = 0.525), indicating comparability. All samples were collected from the Department of Pathology, Affiliated Hospital of Guizhou Medical University. All patients did not receive preoperative chemoradiotherapy. All samples were obtained with the informed consent of patients and approved for use by the Ethics Review Committee of the Guizhou Medical University. In addition, 4 pairs of cervical cancer tissues and adjacent normal cervical tissues were used to detect the TET1 expression by Western blotting.
IHC staining
The tissue slides were routinely treated by dewaxing and hydrating, followed by antigenic repair and treatment with 3% H2O2 to remove endogenous peroxidase and blocking with sheep serum. The corresponding antibodies TET1 (GeneTex 124,207, 1:500), 5mc (GeneTex 128,455, 1:200), 5hmc (GeneTex 629,765, 1:200), LC3 (Beyotime, AF5225, 1:200), and P62 (Beyotime, AF5312, 1:200) were added to the tissues and incubated overnight at 4°C. After washing thrice with PBS, the slides were stained first with the immunohistochemical kit (ZSGB-BIO, PV-9000) and then with haematoxylin, followed by differentiation with hydrochloric acid ethanol, washing with warm water, and sealing with neutral glue. Each sample was scored according to the proportion of positive-stained cells (proportional score, 0–4) and staining intensity (intensity score, 0–3). The staining index was calculated by multiplying the ratio score and intensity score.
Cell culture and plasmid construction
SiHa and HeLa cervical cancer cells were purchased from Nanjing Kepai Biological Technology Co., Ltd. (CBP60552 and CBP60232, respectively). The cells were cultured in DMEM medium supplemented with 10% foetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin and incubated in a constant temperature incubator containing 5% CO2 at 37°C.
The TET1-targeting plasmid was constructed by the LentiCRISPRv2 system, and the sgRNA of the targeted gene was designed using the CRISPR design tool (http://tools.genome Engineering.org). The following two pairs of sequences with higher scores were selected: Oligo-TET1-F1, caccGTCCAACAACAGCACCATGC, Oligo-TET1-R1, aaacCCCGTGCCAGTCAGAAGCCTC; Oligo-TET1-F2, caccGATCTGGAAGGCCTTGCATGT, Oligo-TET1-R2, aaacCCATGCAAGGCCTTCCAGATC. According to the sequencing results, the second sgRNA sequence was selected. The lentiviral packaging plasmids pSPAX2, pMD2G, and TET1 LentiCRISPRv2 expression plasmids were transfected into 293T cells with Lipofectamine3000. After culturing for 24–48 h, the virus solution was collected. In order to obtain the TET1-knockout HeLa and SiHa cervical cancer cell lines, 1 mL of the virus solution and 10 ng polybrene were added, while 1 μg/mL puromycin was added after 48 h, followed by the screening of the TET1-targeted cells.
In order to obtain TET1-overexpressed HeLa and SiHa cervical cancer, the TET1-overexpressed plasmid (Addgene 49,792) was transfected into the cervical cancer cells. After 48 h, 5 mg/mL G418 was added to the culture medium for 15 days to obtain stably overexpressed TET1 cells.
Cell viability assay
To evaluate the effect of 3 MA on SiHa cell viability, cells (5 × 103per well) were plated in 96-well plates and cultured in 100 μl of complete growth medium, and then treated with the indicated concentration of 3 MA(10 mM) for 24 h, 48 h and 72 h. Cell viability was determined using a Cell Counting Kit-8 (Beyotime,C0037). Briefly, 10 μl CCK-8 solution was added to each well, the plate was incubated for 2 h at 37°C, and the optical density was measured at 450 nm using a micro-plate reader (Bio-rad, USA).
Western blotting
Total cellular protein was extracted by cells lysis buffer RIPA, which was added to a protease inhibitor mixture. Next, 30 ug of protein was separated by SDS-PAGE, and the protein was transferred onto a polyvinylidene fluoride (PVDF) membrane by the wet transfer method, and the protein – antibody complex was detected by immunoassay(19). Primary antibodies used were as follows: TET1 (GeneTex, 1:1000), E-cadherin (GeneTex, 1:1000), Vimentin (GeneTex, 1:1000), N-cadherin (GeneTex, 1:1000), Beclin1 (GeneTex, 1:1000), LC3 (Beyotime, AF5225, 1:500), P62 (Beyotime, AF5312,1:500), and Atg5 (Beyotime, 1:500); GAPDH (Beyotime, AF0006, 1:1000). The second antibodies included HRP-goat anti-rabbit (Beyotime, AF0208, 1:1000) or HRP-goat anti-mouse (Beyotime, AF0216, 1:1000). Finally, the protein – antibody complexes were detected with an enhanced chemiluminescence (ECL) reagent.
RT-PCR
Total RNA was extracted with Trizol (Invitrogen, USA) reagent according to the manufacturer’s instructions. cDNA was synthesized by using the Beyort™ III cDNA first-strand synthesis premix. Then, cDNA amplification was performed by RT-PCR in Bio-Rad thermal cycling PCR instrument (Bio-Rad, USA). GAPDH was used as an internal reference. Table 1 lists the primers used in this study [24].
Table 1.
The primer sequence used in this study.
| Gene name | Primer type | Primer Sequence |
|---|---|---|
| GAPDH | Forward | GGAGCGAGATCCCTCCAAAAT |
| Reverse | GGCTGTTGTCATACTTCTCATGG | |
| E-cadherin | Forward | TACACTGCCCAGGAGCCAGA |
| Reverse | TGGCACCAGTGTCCGGATTA | |
| Vimentin | Forward | GACGCCATCAACACCGAGTT |
| Reverse | CTTTGTCGTTGGTTAGCTGGT | |
| Snail | Forward | GAGGCGGTGGCAGACTAG |
| Reverse | GACACATCGGTCAGACCAG | |
| NKRF | Forward | AGCCAGCTCAAATTGTAGTCT |
| Reverse | ACCTTATCTTGTTTGTCGCCA | |
| HIST1H2AK | Forward | TTTTGAGACAAGGCCTGGC |
| Reverse | CCTGGTAGGCGGAGTTTACA | |
| PGRMC2 | Forward | GCAGGATTAGCTGCTTCTTTTA |
| Reverse | TCAGGGAGATATGATTAGAGATAGAAT | |
| AHCYL2 | Forward | CCTGACCTTGTGATCCACCT |
| Reverse | TCACACCTGTAATCCCAGCA | |
| IFT27 | Forward | GAGCACTGATCACCATCGAA |
| Reverse | GCAAGGCACACATCACCTAA | |
| PARD3B | Forward | ATGGGTTGCACTAGCAGAGG |
| Reverse | CCCAGCTCTTCTTGCTCTGA | |
| CHSY1 | Forward | CCTCGTTACTTCTGGGTTGC |
| Reverse | CCATGCTTTCTGTGTGCAGT | |
| GRAMD3 | Forward | GCACTGCATTTGTTTGTTTTCA |
| Reverse | TGGATTTACTGCAGTGCCTTT | |
| INSIG2 | Forward | GGAGCTGAAATTGTGGCAATA |
| Reverse | GAATTCGGCTGTGAATCCAT | |
| PALB2 | Forward | TTTCCCATCCATGACTCAGA |
| Reverse | TCATGACCGTTGTTTGAACC | |
| ZNF558 | Forward | GGCAGAGCTGGTCTCTTCAG |
| Reverse | ACTGACTCCTGGGTTGATGG | |
| AHCYL2 | Forward | TACCGATACACAGCCATCCA |
| Reverse | AGAGTGGGAAAGGCAAAGTG | |
| ATXN10 | Forward | CTTACAATGGCCCTGCATTT |
| Reverse | CATCTGCATTTTCAGCAGGA | |
| PRKAG2 | Forward | CTTGGAGATGCAGGAGAAGG |
| Reverse | AGGTCTGGAGAAGCGGAGTT | |
| si-NKRF1 | sense | GGCUUGGUCUGGAUGUAGA |
| antisense | UCUACAUCCAGACCAAGCC |
Detection of the cell migration and invasion ability
The experimental cells were cultured in the logarithmic phase and inoculated into a 6-well plate at the density of 1 × 106 cells/well. The next day, cross lines were drawn in the middle of the plates using a 10 μL spear. Then, DMEM medium without FBS was added, and the cells were further cultured in an incubator at 37°C under a 5% CO2 atmosphere for 24 h. The cells were observed and photographed under an inverted microscope every day.
In the cell invasion test, 1 mg/L matrigel was added to the Transwell chamber and solidified overnight in an incubator at 37°C under a 5% CO2 atmosphere. The next day, the cells in the logarithmic phase were resuspended in a serum-free DMEM at the density of 5 × 105 cells/mL. Then, 200 μL of the cell suspension was added to each well in the upper chamber of Transwell, and 600 μL DMEM medium containing 10% FBS was added to the lower chamber (24-well plate). The cells were then cultured in an incubator at 37°C under a 5% CO2 atmosphere for 48 h. Then, the unmotivated cells and matrigel at the bottom of the upper chamber were gently wiped off with a wet cotton swab. After which, the Transwell chamber was fixed with 600 μL of methanol at room temperature for 20 min and dyed with 0.1% crystal violet at room temperature for 30 min. The migrated cells were observed and photographed under an inverted microscope; 5 fields of vision were randomly photographed for each membrane, and the number of cells in each field of vision was calculated [19,24].
Determination of cell proliferation and colony-formation ability
The cell proliferation was detected by RTCA assay (ACEA Biosciences Inc., USA). Briefly, the cells cultured to the logarithmic growth phase were digested, counted, and their concentration was adjusted to 1.25 × 104 cells/mL. Then, 2500 cells/well were added to the wells of the E-plate cell plate, and 5 duplicate wells were set for each group of cells. The E-plate cell plate with cells was placed into the RTCA instrument for monitoring and recording in accordance with the parameters set in the instrument instructions for the total monitoring time of 7 days.
In the colony formation experiment, the cells were inoculated into 6-well plates at the density of 200 cells/well. Each set of experiments was repeated thrice and cultured for 14 days. Then, the cell clones were fixed with methanol for 10 min and stained with 0.1% crystal violet. After 30 min, the colony formation was recorded and imaged with the Image Pro Plus, while the number of cell clones in each well was calculated.
Cell cycle detection
The cells in each group of the logarithmic phase were fixed overnight with 70% ethanol precooled on a 4°C ice bath and then stained with propidium iodide at 37°C for 30 min. The red fluorescence with a wavelength of 488 nm was detected by flow cytometry. Modfit.LT 4.1 software was used to analyse the DNA content in different cycles.
Tumor formation in nude mice
Female BALB/c nude mice (4 weeks old) were purchased from the Animal Research Center of Guizhou Medical University. SiHa cells were stably infected with lentivirus containing TET1-targeting sgRNA and TET1 cDNA to study the effect of TET1 on tumorigenesis in SiHa cells. To establish a subcutaneous tumour model, 1 × 107 cells/mouse were injected into the right side of 6-week-old female BALB/c nude mice. Tumour size was observed every 3 days starting from 1 week after inoculation. Mice were euthanized before the tumour volume reached 1,000 mm3, and the tumours were weighed. All animals were purchased through the Guizhou Medical University and approved by the Animal Ethics Committee of the Guizhou Medical University [24].
Transcriptome sequencing and database analysis
TET1cas9 and WT SiHa cells in 3 different passages were collected. The total RNA was extracted and sequenced by Shanghai Cloud-seq Biotech. Cuffdiff software was used to calculate and detect differentially expressed genes (mRNA) between two or two groups of samples. Fold change) ≥2.0, that is, log (FC) ≥1.0, p ≤ 0.05, and FPKM value in at least one (or a group of) samples ≥ 0.1 was set as the threshold of difference screening. Then, the differentially expressed genes were enriched by GO and KEGG [25] with the GGPLOT2 R packet. When using KEGG imagery please ensure that you cite this source in the appropriate Figure legend, as per the citation guidelines: www.kegg.jp/kegg/kegg1.html. For previous uses, the Kanehisa laboratory have happily provided permission.
The difference in the gene expression between cervical cancer and the normal control group was analysed by using the limma package of R software. The correlational analysis between TET1 and the autophagy gene was performed by using the KEGG (https://cistrome.shinyapps.io/timer/).(19)
MeDIP-seq and MeDIP-quantitative real-time PCR
MeDIP-Seq [19] was performed and analysed through Shanghai Cloud-seq Biotech, with 3 replicates in each group. Differential-methylated regions (DMRs) were identified by diffReps software. The default p < 0.0001 and fold change ≥ 2 were set as the threshold of differential methylation region. MeDIP-qPCR is a perfect combination of real-time quantitative PCR and MeDIP, which first interrupts the genome ultrasound into fragments of 200–1000 bp size [19]. The single-stranded DNA samples after heating denaturation were divided into two parts, one of which was added to an anti-5mc antibody (GeneTex 128,455) and the other was used as the total input DNA sample. The antibody complex of methylated DNA fragments in the sample was separated by affinity chromatography, while the remaining non-methylated DNA fragments in the sample were eluted. The purified methylated DNA fragments were then used as templates for real-time PCR. The primers used in the study are listed in Table 1.
Statistical analysis
The data are presented as the mean ±standard deviation (SD). Statistical analyses were conducted with the SPSS 19.0 software. Unpaired two-tailed Student’s t-tests were used to compare data between two groups. The Chi-square test was used to evaluate the statistical significance of differences in the IHC scores between cervical cancer and adjacent non-cancerous samples. p ≤ 0.05 were considered to indicate statistical significance.
Results
Downregulation of the protein level of TET1 in cervical cancer tissues
The expression of TET1 in normal cervical tissues and highly and poorly differentiated cervical cancer tissues was detected with immunohistochemistry and Western blot (Figure 1(a,b)). The results showed that the expression in cervical cancer tissues was significantly lower than that in adjacent normal tissues. TET1 was mainly expressed in the nucleus as brown – yellow particles. In cervical cancer tissues, TET1-positive cells were lightly stained and fewer in number, and TET1 expression in the poorly differentiated group was significantly lower than that in the highly differentiated group. In addition, we also analysed the expression of TET1 in cervical cancer through the TCGA database, and the results were consistent with the protein results(Figure 1(c)).
Figure 1.
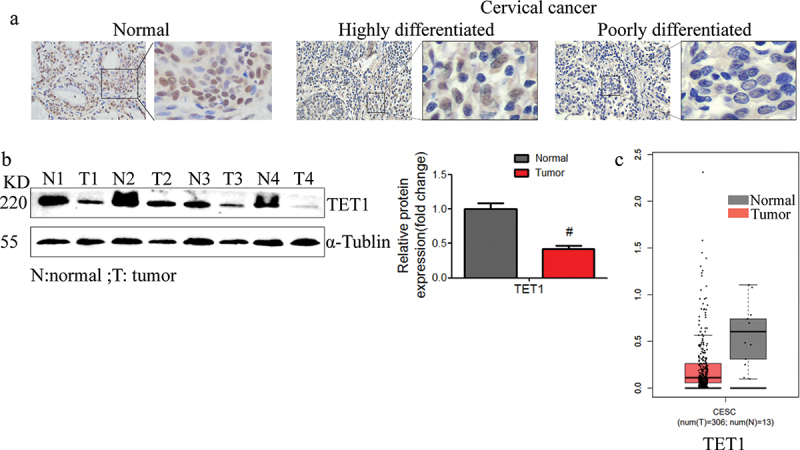
TET1 expression was downregulated in the cervical cancer tissues. (a) The expression of TET1 in the cervical cancer tissues with different degrees of differentiation; (b) The TET1 expression in cervical cancer and normal cervical tissues; (c) TCGA data indicating that TET1 was downregulated in cervical cancer.
Effect of TET1 knockdown and overexpression on the migration and invasion of cervical cancer cells
To investigate the role of TET1 in cervical cancer cells, the TET1 gene was targeted using CRISPR-Cas9 in SiHa and HeLa cells and TET1 was overexpressed in these cells (Figure 2(a,b)) The migration and invasion abilities of cervical cancer cells were significantly enhanced after TET1 knockdown and overexpression (Figure 2(c–g)).
Figure 2.
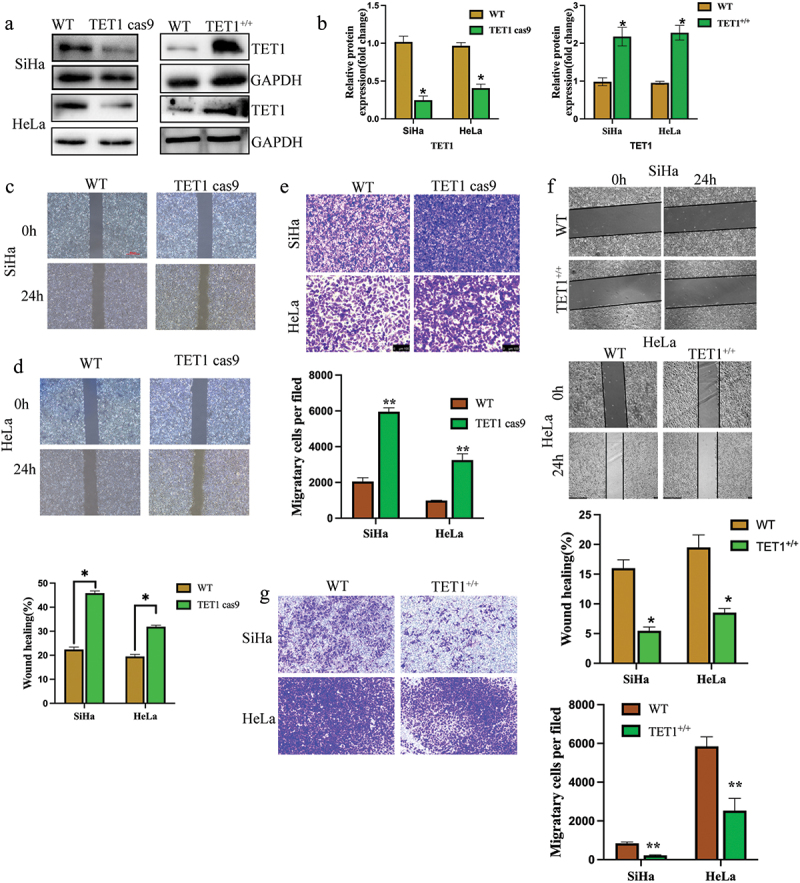
TET1 knockdown promoted the migration and invasion of cervical cancer cells.
(a,b) Verification of TET1 knockdown and overexpression in cervical cancer SiHa and HeLa cells; (c,d) The effect of TET1 knockdown on the migration of cervical cancer SiHa and HeLa cells was detected by scratch assay; (e) Transwell assay was performed to detect the effect of TET1 knockdown on the invasion ability of cervical cancer SiHa and HeLa cells; (f) The effect of TET1 overexpression on the migration of cervical cancer SiHa and HeLa cells was detected by scratch assay; (g) Transwell assay was performed to detect the effect of TET1 overexpression on the invasion ability of cervical cancer SiHa and HeLa cells.
Effect of TET1 on proliferation, clonal formation, and tumorigenesis of cervical cancer cells
The effect of TET1 on the clonal formation and proliferation of cervical cancer cells was investigated. The results showed that the number of colonies in the TET1cas9 SiHa and HeLa cell groups was 158 ± 17 and 143 ± 15, respectively, and those in the WT group were 61 ± 7 and 47 ± 5, respectively. Thus, the latter was significantly lower than the former (Figure 3(a)). However, the number of cell clones in the TET1 overexpression group was significantly lower than that in the WT group (Figure 3(b)). The cell cycle of cervical cancer SiHa cells was examined using flow cytometry. The findings revealed that after the deletion of the TET1 gene, the G0/G1 phase of SiHa cells decreased remarkably, but the S phase and G2/M phase increased significantly. These observations indicate that the DNA synthesis phase increased significantly, which accelerated cell growth (Figure 3(c)). The proliferation ability of cervical cancer cells was detected using RTCA, and no significant differences were perceived in the proliferation ability between the groups within 48 h of cell culture; however, the proliferation ability of the TET1cas9 group increased with the further increase in time (Figure 3(d)). In vivo experiments signified that compared with the HeLa cells in the WT group, the tumour volume of the TET1cas9 group was significantly increased, whereas that of the TET1 overexpression group was significantly decreased, which alludes that TET1 can inhibit tumorigenesis of cervical cancer cells in vivo (Figure 3(e)).
Figure 3.
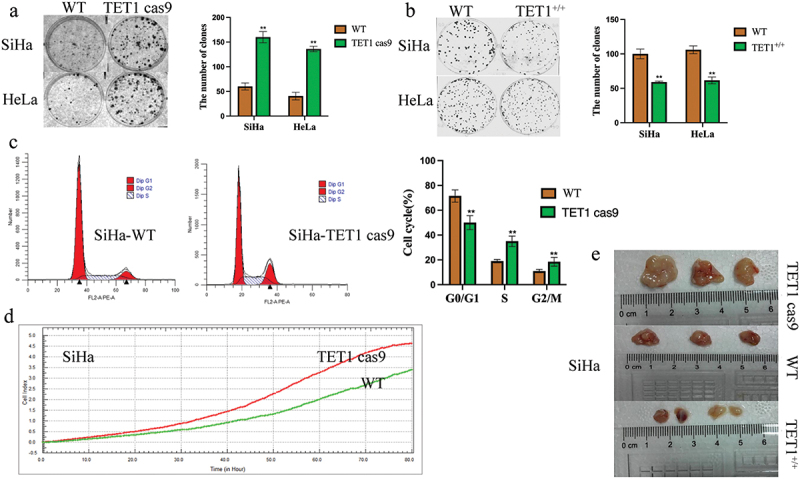
TET1 reduces the proliferation, clonal formation, and tumorigenesis of cervical cancer cells.
(a,b) The knockdown of TET1 promoted the colony formation of SiHa and HeLa cells; The overexpression of TET1 impaired the colony formation of SiHa and HeLa cells; (c,e) The effects of TET1 knockdown or overexpression on the proliferation of cervical cancer cells were detected by cell cycle(C), RTCA (d)and subcutaneous tumorigenesis(E)
Effect of TET1 on epithelial-mesenchymal transformation (EMT) of cervical cancer cells
The previous experimental results showed that TET1 knockdown in cervical cancer cells promoted the invasion and migration abilities of cervical cancer cells. Therefore, further studies were conducted, which revealed that after TET1 knockdown in SiHa cells, the epithelial cell indicator E-cadherin was significantly reduced, whereas N-cadherin, which are related to stromal cells, were significantly increased. The overexpression of TET1 had the opposite effects (Figure 4(a–d)). These findings suggest that TET1 could inhibit epithelial-mesenchymal transformation in cervical cancer cells.
Figure 4.
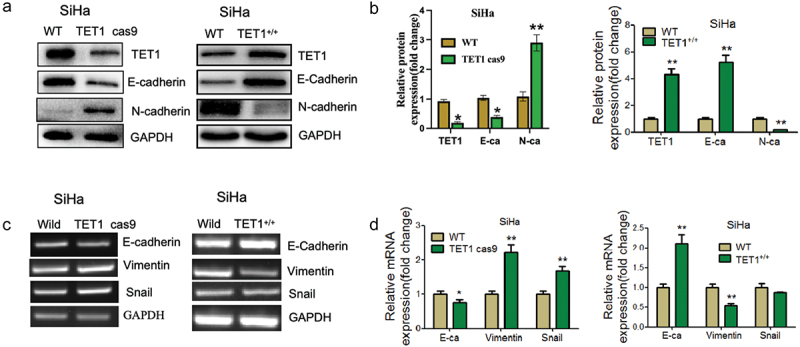
Effect of TET1 on epithelial-mesenchymal transformation (EMT) of cervical cancer SiHa cells.
(a,b) Western blotting was performed to detect the expression changes of EMT-related indicators after the knockdown or overexpression of TET1 in SiHa cells; (c,d) RT-PCR was performed to detect the expression changes of EMT-related indicators after the knockdown or overexpression of TET1 in SiHa cells.
Effect of TET1 on the biological functions of cervical cancer cells
TET1cas9 and WT SiHa cells were sequenced to analyse the differentially expressed genes and cellular functions related to TET1. KEGG analysis showed that the main signalling pathways that were upregulated in the TET1cas9 group were autophagy, mTOR, and RAP1, whereas the downregulated pathway mainly involved apoptosis (Figure 5(a,b)).
Figure 5.
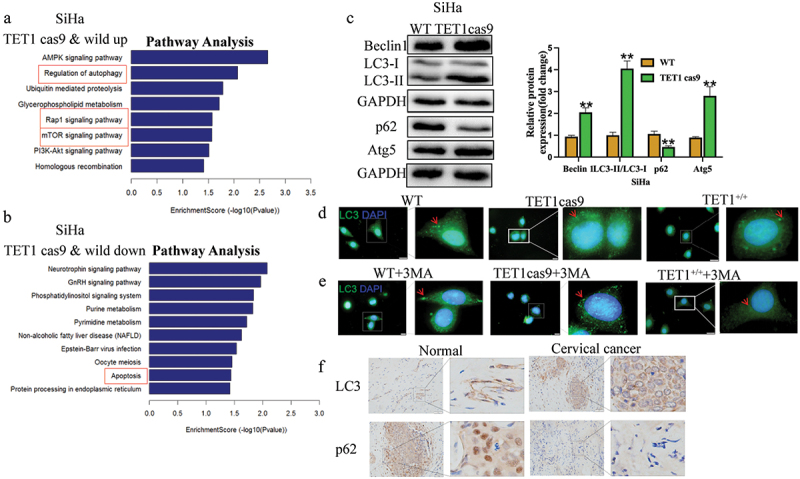
Effect of TET1 on the biological functions of cervical cancer cells.
(a) KEGG analysis revealed the signalling pathways associated with the upregulated activity of TET1 knockdown in SiHa cells when compared with that in wild-type cells; (b) KEGG analysis was performed to identify the signalling pathways associated with the downregulated activity of TET1 in SiHa cells when compared with that in wild-type cells. (c) Western blotting was performed to detect the autophagy indexes of SiHa cells after TET1 knockdown; (d,e) Immunofluorescence also showed that there were more autophagosomes in the TET1 cas9 group than in the WT group, while the TET1+/+ group had the least number of autophagosomes. Furthermore, immunofluorescence results show that knockdown of TET1 can attenuate the inhibitory effect of 3 MA on autophagy, while overexpression of TET1 can continue to inhibit autophagy in the presence of 3 MA; (f) Immunohistochemical detection of LC3 and P62 expression in cervical cancer and adjacent tissues.
Effect of TET1 on autophagy in cervical cancer cells
The effect of TET1 knockdown on the autophagy-related genes of SiHa cells was assessed using Western blotting. The levels of LC3II, Beclin1, and Atg5 in the TET1cas9 group were found to be significantly higher than those in the WT group, but the level of p62 was lower (Figure 5(c)). Immunofluorescence also showed that there were more autophagosomes in the TET1 cas9 group than in the WT group, while the TET1+/+ group had the least number of autophagosomes. Furthermore, immunofluorescence results show that knockdown of TET1 can attenuate the inhibitory effect of 3 MA on autophagy, while overexpression of TET1 can continue to inhibit autophagy in the presence of 3 MA(Figure 5(d,e)). Immunohistochemical detection of LC3 and P62 expressions in cervical cancer and adjacent tissues showed that the LC3 level was significantly higher in cervical cancer tissues than that in adjacent tissues, whereas the P62 level was significantly lower than that in the control group (Figure 5(f)). These results imply that TET1 May regulate the autophagy level in cervical cancer cells.
Effects of autophagy on TET1-mediated biological changes in cervical cancer cells
To further determine whether the effect of TET1 on the proliferation and migration of cervical cancer cells is regulated by autophagy, 3 MA (10 mM) was used to treat SiHa cells. The results indicated that the proliferation, invasion, and migration abilities of SiHa cells in the 3 MA group were significantly lower than those in the control group (Figure 6(a–c)). However, TET1 knockdown inhibited the effect of a 3 MA-induced decrease in invasion and migration abilities (Figure 6(d,e)).
Figure 6.
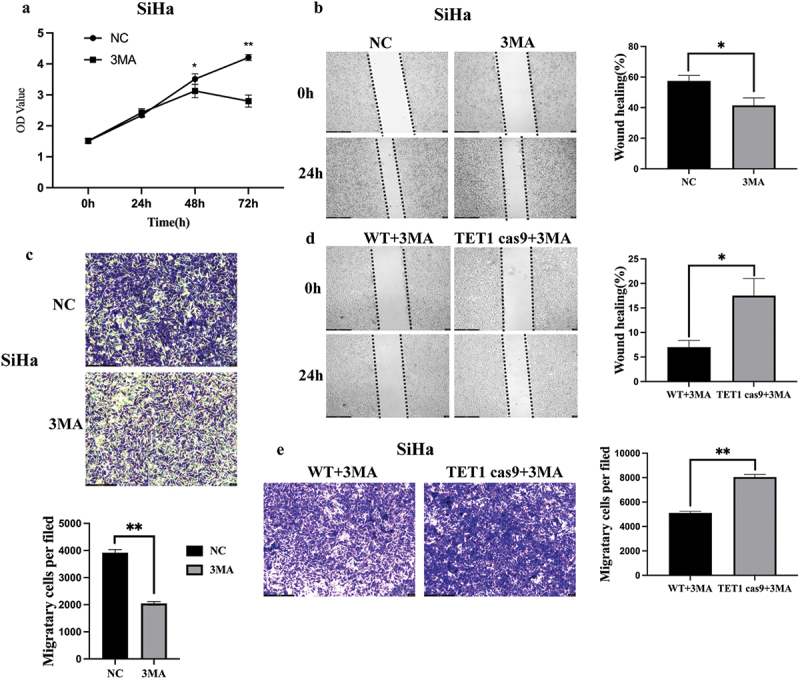
Effects of autophagy on TET1-mediated biological changes in cervical cancer cells.
(a – c) 3 MA to treat SiHa cells, and the results suggested that the proliferation, invasion, and migration of SiHa cells in the 3 MA group were significantly lower than those in the control group; (d,e) TET1 knockdown could inhibit this effect of a 3 MA-induced decrease in the invasion and migration abilities.
Effect of TET1 on gene methylation level and mRNA expression
As the main function of TET1 is demethylation, the expressions of 5mc and 5hmc were detected in highly and poorly differentiated tissue samples of cervical cancer using immunohistochemistry. The results showed that the expression of 5hmC was significantly lower than that of 5mc in poorly differentiated cervical cancer tissues (Figure 7(a,b)). Therefore, our data suggest that TET1 May affect cervical cancer progression by regulating the methylation levels. Furthermore, the MeDIP-Seq analysis of TET1cas9 and WT SiHa cells was performed, and the results were cross-checked with the transcriptome sequencing results. Compared with WT, 2009 methylation differentially enriched genes and 463 differentially expressed genes were detected in the TET1cas9 group. Examining the intersection of these differential genes revealed that compared with the WT group, there were 18 genes whose methylation was upregulated and expression was downregulated in the TET1cas9 group. Fourteen of these genes were selected for MeDIP-PCR verification, which indicated that compared with the WT group, the methylation levels of genes in the TET1cas9 group were significantly increased, except for PGRMC2, which implies that the sequencing results are reliable. Analysis of methylation enrichment regions of these genes showed that only the methylation differential enrichment regions of NKRF and HIST1H2AK were located in the promoter region. Further detection of these two genes with RT-PCR showed that compared with the WT group, the expression levels of these two genes were significantly decreased in the TET1cas9 group. Additionally, the correlation among TET1, NKRF, and HIST1H2AK was analysed using the TCGA database. The results signified that TET1 was positively correlated with the expressions of these two genes (Figure 7(c–g)) In order to observe the effect of NKRF on autophagy, we detected the expression changes of LC3B protein after knocking down NKRF. We observed increased expression of autophagy related protein LC3B after knocking down NKRF in SiHa(Figure 7(h,i)), indicating that NKRF may affect autophagy, and further indicating that TET1 May regulate autophagy through NKRF.
Figure 7.
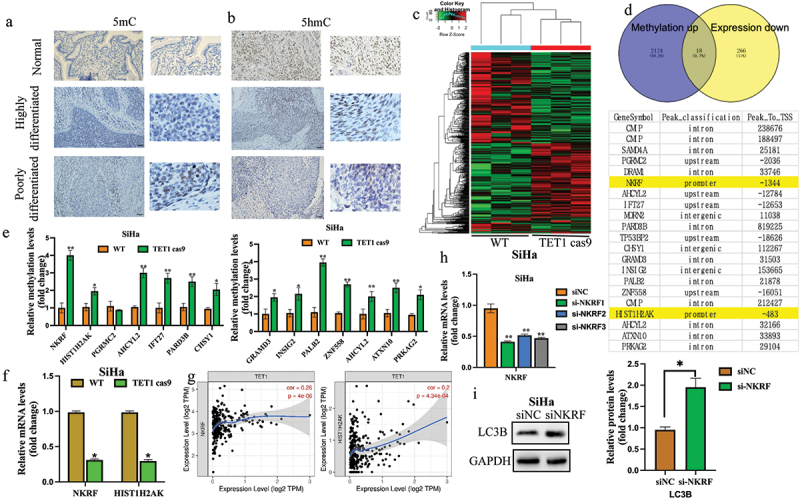
Effect of TET1 on the gene methylation level and mRNA expression.
(a,b) Immunohistochemistry was performed to detect the expression of 5 mC and 5 hmC in highly and poorly differentiated Normal cervical tissue cervical cancer tissues; (c,d) Genes with a high methylation level, but low mRNA expression and genes with a low methylation level but high mRNA expression were observed through methylation sequencing. Among them, only NKRF and HIST1H2AK were involved in the change of the promoter methylation level; (e,f) The specific methylation levels of the selected genes were detected by MEDIP-qPCR, and the NKRF and HIST1H2AK with upregulated promoter methylation and downregulated mRNA verified by q-PCR; (g) The correlation among TET1, NKRF, and HIST1H2AK was analysed with reference to the TCGA database. (h) The knockdown efficiency of NKRF was verified by RT-qPCR; (i) The expression of autophagy related protein LC3B was detected after NKRF knockdown.
Discussion
TET1 is a newly discovered demethylase that converts 5mc to 5hmC with the help of thymine DNA glycosylase and base excision repair, thereby initiating DNA demethylation and eventually producing 5-C [26]. Recent studies have shown that TET1-mediated DNA demethylation is widely involved in cell differentiation and tumour progression [27–29]. In addition, evidence that demethylation modification of TET1 is linked to the occurrence, proliferation, invasion, and metastasis of tumours is mounting [30]. This study confirmed the low expression of TET1 and the increased expression of autophagy-related proteins in cervical cancer tissues. TET1 can inhibit the proliferation, migration, and invasion of cervical cancer cells, which may be accomplished via regulation of the autophagy signalling pathway.
As a highly active DNA cytosine oxygenase, TET1 is believed to play a paramount role in maintaining the fidelity of DNA methylation patterns [14–16] and in sustaining the tumour suppressor genes in a nonmethylated state [17,18]. Low TET1 expression has been reported in various solid tumours, such as colorectal cancer, glioma, pancreatic cancer, and prostate cancer, and its decline is closely related to the increase in tumour malignancy [20–23]. In this study, TET1 was found to be lowly expressed in cervical cancer tissues. The proliferation, migration, and invasion abilities of cervical cancer cells were enhanced when the expression of TET1 was low, whereas this phenomenon was reversed when TET1 was overexpressed. All these findings indicate that TET1 also plays a role in the suppression of cervical cancer.
EMT is a process by which epithelial cells acquire mesenchymal characteristics [31]. In cancer, EMT is associated with tumour genesis, invasion, metastasis, and treatment resistance [32]. Therefore, in this study, we observed the effect of TET1 on EMT-related proteins to indicate the anti-tumour effect of TET1.
Autophagy plays vital role through autophagy-related (ATG) proteins in cancer [33]. The dual role of autophagy in cancer progression and inhibition has remained controversial so far. The numerous ATG proteins and their core complexes, including ULK1/2 kinase core complex, autophagy-specific class III PI3K complex, ATG9A trafficking system, ATG12, and LC3 ubiquitin-like conjugation systems, are responsible for multiple activities of the autophagy pathway and are involved in autophagy initiation, nucleation, elongation, maturation, fusion, and degradation [34–36].
Recent studies have revealed that TET1 May regulate various cellular functions and signalling pathways via demethylation and autophagy and participate in the occurrence and development of tumours. For example, Atg5 downregulates its expression via hypermethylation of its promoter, which is associated with the proliferation and carcinogenesis of melanoma cells [37]. Beclin-1 also plays an important role in autophagy and is considerably downregulated in breast cancer owing to DNA methylation [38]. Furthermore, Guo et al. found that TET1 plays an anticancer role by regulating autophagy in gliomas [39]. However, in cervical cancer, how TET1 ultimately participates in tumour genesis and development by influencing autophagy is yet to be established. In this study, the enrichment levels of 5mc and 5hmC were detected in clinical specimens, which showed that the level of 5hmC was significantly reduced in lowly differentiated cervical cancer tissues, whereas the level of 5mc was increased. TET1 May be involved in the progression of cervical cancer via demethylation. Transcriptome sequencing was further employed to screen the genes regulated by TET1 in cervical cancer cells. KEGG analysis revealed that the main signalling pathways affected by TET1 in SiHa cells were autophagy, MTOR, RAP1, PI3K/AKT, and apoptosis.
In addition, the TET1cas9 group of SiHa cells showed a higher level of autophagy compared to the WT group, and the autophagy level decreased after TET1 overexpression. Furthermore, SiHa WT, TET1cas9, and TET1+/+ cells were treated with the autophagy inhibitor 3 MA. Compared to the WT group, a decrease in autophagy levels was observed in SiHa cells after treatment with 3 MA, and knocking down or overexpressing TET1 could weaken or enhance the inhibitory effect of 3 MA on autophagy, as well as weaken the inhibitory effect on SiHa cell migration and invasion. These results suggest that TET1 May affect the migration and invasion of cervical cancer cells by regulating autophagy.
TET1cas9 and WT SiHa cells were analysed by MeDIP-Seq and cross-checked with transcriptome sequencing results. The results showed that 18 genes in TET1cas9 group had up-regulated methylation and down-regulated mRNA expression. Fourteen of these genes were selected for MeDIP-PCR verification. The results showed that compared with WT group, methylation levels of TET1cas9 genes were all increased except PGRMC2. However, only the methylation rich regions of NKRF and HIST1H2AK were located in the promoter region. Further RT-PCR was used to detect the expression of these two genes. Compared with WT group, the expression levels of these two genes in TET1 cas9 group were significantly reduced.These findings suggest that TET1 May influence the biological function of cervical cancer cells by regulating the methylation levels of the autophagy genes NKRF and HIST1H2AK, In addition, we observed increased expression of autophagy related protein LC3B after knockdown of NKRF, which further supported our view.
In summary, this study observed that TET1 can influence the level of autophagy, and even in the presence of autophagy inhibitors, it can still affect the migration and invasion of cervical cancer cells. Therefore, we speculate that TET1 May mediate the migration and invasion of cervical cancer cells by modulating autophagy. The findings have immense application value for the diagnosis, prognosis, and molecular targeted therapy of cervical cancer.
Acknowledgments
We are grateful to each of the authors who contributed to this study and to the National Natural Science Foundation of China (NSFC, grant number, 81960473), Guangxi Zhuang Autonomous Natural Science Foundation (grant number, 2018JJB140322), the project of science and technology innovation for Postgraduates of Hubei Medical College (grant number, YC2019013, YC2019018), the Supplemental Fund Supporting the NSFC Project from the Guiyang Science and Technology Bureau (grant number Zhuke (2017) 30–36), and the Guizhou Provincial Education Research Project (grant number KY (2021)) for supporting this study.
Funding Statement
The present study was supported by the National Natural Science Foundation of China (NSFC, grant number, 81960473), Guangxi Zhuang Autonomous Natural Science Foundation (grant number, 2018JJB140322), the project of science and technology innovation for Postgraduates of Hubei Medical College (grant number, YC2019013, YC2019018), the Supplemental Fund Supporting the NSFC Project from the Guiyang Science and Technology Bureau (grant number Zhuke (2017) 30–36), and the Guizhou Provincial Education Research Project (grant number KY (2021)).
Disclosure statement
The authors declare that they do not have financial or non-financial competing interests.
Author contributions
Concept presentation and experimental design: Yan Ding. Conduct of the experiment: Xiuying Chen, Jing Li, Ji Ren, Shan Wang, Yuxin Zhan. Data acquisition and analysis: Xiuying Chen, Yan Ding. Writing, reviewing and revising manuscripts: Ji Ren, Yan Ding. Study supervision: Yan Ding, Yujie Tan.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All the sequencing data generated by this study have been deposited in NCBI’s Gene Expression Omnibus(GEO) under accession number GSE236396 and GSE236315. Data used to determine the effect of TET1 on autophagy signalling pathways in SiHa cells were derived from KEGG(https://www.genome.jp/kegg/).
Ethics approval and informed consent
This study was approved by the Ethics Committee of the Guizhou Medical University. Clinical tissue samples are handled in strict accordance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent. All procedures involving animal use were approved by the Animal Ethics Committee of the Guizhou Medical University and were in accordance with the National Policy on Use of Laboratory Animals. And all methods were performed in accordance with the relevant guidelines and regulations of the Basel Declaration. And the study is reported in accordance with ARRIVE guidelines.
References
- [1].Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–15. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- [2].Maver PJ, Poljak M.. Progress in prophylactic human papillomavirus (HPV) vaccination in 2016: a literature review. Vaccine. 2018;36(36):5416–23. doi: 10.1016/j.vaccine.2017.07.113 [DOI] [PubMed] [Google Scholar]
- [3].Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17. doi: 10.1128/CMR.16.1.1-17.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7(1):11–22. doi: 10.1038/nrc2050 [DOI] [PubMed] [Google Scholar]
- [5].Su PH, Lai HC, Huang RL, et al. Paired box-1 (PAX1) activates multiple phosphatases and inhibits kinase cascades in cervical cancer. Sci Rep. 2019;9(1):9195. doi: 10.1038/s41598-019-45477-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gong Y, Wan JH, Zou W, et al. MiR-29a inhibits invasion and metastasis of cervical cancer via modulating methylation of tumor suppressor SOCS1. Future Oncol. 2019;15(15):1729–44. doi: 10.2217/fon-2018-0497 [DOI] [PubMed] [Google Scholar]
- [7].Hu Y, Wu F, Liu Y, et al. DNMT1 recruited by EZH2-mediated silencing of miR-484 contributes to the malignancy of cervical cancer cells through MMP14 and HNF1A. Clin Epigenetics. 2019;11(1):186. doi: 10.1186/s13148-019-0786-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu W, Wu G, Xiong F. et al. Advances in the DNA methylation hydroxylase TET1. Biomark Res. 2021. [cited 2021 Oct 16];9(1):76. doi: 10.1186/s40364-021-00331-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Greenberg MVC, Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- [10].Joshi K, Liu S, Breslin SJP, et al. Mechanisms that regulate the activities of TET proteins. Cell Mol Life Sci. 2022;79(7):363. doi: 10.1007/s00018-022-04396-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Charlton J, Jung EJ, Mattei AL, et al. Tets compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat Genet. 2020;52(8):819–27. doi: 10.1038/s41588-020-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu Y, Zhang R, Zhang X, et al. Curcumin may reverse 5-fluorouracil resistance on colonic cancer cells by regulating TET1-NKD-Wnt signal pathway to inhibit the EMT progress. Biomed Pharmacother. 2020;129:110381. doi: 10.1016/j.biopha.2020.110381 [DOI] [PubMed] [Google Scholar]
- [13].Thomson JP, Ottaviano R, Unterberger EB, et al. Loss of Tet1-associated 5-hydroxymethylcytosine is concomitant with aberrant promoter hypermethylation in liver cancer. Cancer Res. 2016;76(10):3097–108. doi: 10.1158/0008-5472.CAN-15-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Guo H, Zhu H, Zhang J, et al. TET1 suppresses colon cancer proliferation by impairing beta-catenin signal pathway. J Cell Biochem. 2019;120(8):12559–12565. doi: 10.1002/jcb.28522 [DOI] [PubMed] [Google Scholar]
- [15].Fu R, Ding Y, Luo J, et al. Ten-eleven translocation 1 regulates methylation of autophagy-related genes in human glioma. Neuroreport. 2018;29(9):731–8. doi: 10.1097/WNR.0000000000001024 [DOI] [PubMed] [Google Scholar]
- [16].Huang H, Jiang X, Li Z, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110(29):11994–9. doi: 10.1073/pnas.1310656110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6(4):463–477. doi: 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- [18].Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14(2):70–77. doi: 10.1016/j.tcb.2003.12.002 [DOI] [PubMed] [Google Scholar]
- [19].Fan J, Zhang Y, Mu J, et al. TET1 exerts its anti-tumor functions via demethylating DACT2 and SFRP2 to antagonize Wnt/β-catenin signaling pathway in nasopharyngeal carcinoma cells. Clin Epigenetics. 2018;10(1):103. doi: 10.1186/s13148-018-0535-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kang KA, Piao MJ, Hyun YJ, et al. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp Mol Med. 2019;51(4):1–14. doi: 10.1038/s12276-019-0238-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Neri F, Dettori D, Incarnato D, et al. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene. 2015;34(32):4168–76. doi: 10.1038/onc.2014.356 [DOI] [PubMed] [Google Scholar]
- [22].Debnath J, Gammoh N, Ryan KM. Autophagy and autophagy-related pathways in cancer. Nat Rev Mol Cell Biol. 2023;24(8):560–575. doi: 10.1038/s41580-023-00585-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu L, Wu Y, Li Q, et al. METTL3 promotes Tumorigenesis and metastasis through BMI1 m6A methylation in oral squamous cell carcinoma. Mol Ther. 2020;28(10):2177–2190. doi: 10.1016/j.ymthe.2020.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592. doi: 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wu J, Li H, Shi M, et al. TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/β-catenin signaling pathway to suppress EMT in pancreatic tumor cells. J Exp Clin Cancer Res. 2019;38(1):348. doi: 10.1186/s13046-019-1334-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pei YF, Tao R, Li JF, et al. Liu BY.TET1 inhibits gastric cancer growth and metastasis by PTEN demethylation and re-expression. Oncotarget. 2016;7(21):31322–31335. doi: 10.18632/oncotarget.8900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tsai KW, Li GC, Chen CH, et al. Reduction of global 5-hydroxymethylcytosine is a poor prognostic factor in breast cancer patients, especially for an ER/PR-negative subtype. Breast Cancer Res Treat. 2015;153(1):219–234. doi: 10.1007/s10549-015-3525-x [DOI] [PubMed] [Google Scholar]
- [29].Sjöström M, Zhao SG, Levy S, et al. The 5-hydroxymethylcytosine landscape of prostate cancer. Cancer Res. 2022;82(21):3888–3902. doi: 10.1158/0008-5472.CAN-22-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bao B, Teslow EA, Mitrea C, et al. Role of TET1 and 5hmC in an obesity-linked pathway driving cancer stem cells in triple-negative breast cancer. Mol Cancer Res. 2020;18(12):1803–1814. doi: 10.1158/1541-7786.MCR-20-0359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- [32].Goossens S, Vandamme N, Van Vlierberghe P, et al. EMT transcription factors in cancer development re-evaluated: beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868(2):584–591. doi: 10.1016/j.bbcan.2017.06.006 [DOI] [PubMed] [Google Scholar]
- [33].Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer. 2020. Jan 22;19(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Marsh T, Debnath J. Autophagy suppresses breast cancer metastasis by degrading NBR1. Autophagy. 2020;16(6):1164–1165. doi: 10.1080/15548627.2020.1753001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Klionsky DJ, Petroni G, Amaravadi RK, et al. Autophagy in major human diseases. EMBO J. 2021;40(19):e108863. doi: 10.15252/embj.2021108863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zeng J, Jing Y, Shi R, et al. Autophagy regulates biliary differentiation of hepatic progenitor cells through Notch1 signaling pathway. Cell Cycle. 2016;15(12):1602–10. doi: 10.1080/15384101.2016.1181234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2020;16(7):1296–1313. doi: 10.1080/15548627.2019.1671643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hill SM, Wrobel L, Ashkenazi A, et al. VCP/p97 regulates beclin-1-dependent autophagy initiation. Nat Chem Biol. 2021;17(4):448–455. doi: 10.1038/s41589-020-00726-x [DOI] [PubMed] [Google Scholar]
- [39].Fu R, Ding Y, Luo J, et al. TET1 exerts its tumour suppressor function by regulating autophagy in glioma cells. Biosci Rep. 2017;37(6). doi: 10.1042/BSR20160523 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request. All the sequencing data generated by this study have been deposited in NCBI’s Gene Expression Omnibus(GEO) under accession number GSE236396 and GSE236315. Data used to determine the effect of TET1 on autophagy signalling pathways in SiHa cells were derived from KEGG(https://www.genome.jp/kegg/).


