Abstract
Although a surprisingly large number of genes affect yeast telomeres, in most cases it is not known if their products act directly or indirectly. We describe a one-hybrid assay for telomere binding proteins and use it to establish that six proteins that affect telomere structure or function but which had not been shown previously to bind telomeres in vivo are indeed telomere binding proteins. A promoter-defective allele of HIS3 was placed adjacent to a chromosomal telomere. Candidate proteins fused to a transcriptional activation domain were tested for the ability to activate transcription of the telomere-linked HIS3 gene. Using this system, Rif1p, Rif2p, Sir2p, Sir3p, Sir4p, and Cdc13p were found to be in vivo telomere binding proteins. None of the proteins activated the same reporter gene when it was at an internal site on the chromosome. Moreover, Cdc13p did not activate the reporter gene when it was adjacent to an internal tract of telomeric sequence, indicating that Cdc13p binding was telomere limited in vivo. The amino-terminal 20% of Cdc13p was sufficient to target Cdc13p to a telomere, suggesting that its DNA binding domain was within this portion of the protein. Rap1p, Rif1p, Rif2p, Sir4p, and Cdc13p activated the telomeric reporter gene in a strain lacking Sir3p, which is essential for telomere position effect (TPE). Thus, the telomeric association of these proteins did not require any of the chromatin features necessary for TPE. The data support models in which the telomere acts as an initiation site for TPE by recruiting silencing proteins to the chromosome end.
Telomeres, the protein-DNA complexes at the ends of eukaryotic chromosomes, are essential for chromosome stability in yeast (69). Telomeres protect chromosomes from degradation and end-to-end fusions and allow their complete replication by providing a substrate for the enzyme telomerase. In addition, in some organisms, including Saccharomyces cerevisiae (23), telomeres are a specialized site for gene expression because transcription of genes near telomeres is reversibly repressed, a phenomenon called telomere position effect (TPE).
Telomeric DNA typically consists of a simple, repeated sequence with the strand running 5′ to 3′ toward the end of the molecule being rich in G residues. For example, telomeres in Saccharomyces consist of 300 ± 75 bp of C1-3A/TG1-3 DNA (71). In some organisms, the G-rich strand extends beyond the end of the molecule to form a single-stranded overhang or G tail. Because these extensions are found in organisms from yeast (83) to vertebrates (50, 52, 88), they are probably a general feature of eukaryotic chromosomes.
Telomeric DNA from yeast (86) to humans (77) is assembled into a nonnucleosomal, protein-DNA complex, the telosome. The duplex C1-3A/TG1-3 DNA binding protein Rap1p is the major protein in the yeast telosome. Anti-Rap1p antibodies specifically immunoprecipitate telomeric C1-3A/TG1-3 tracts from chromosomes and linear plasmids (14, 86). Rap1p is also associated with the telomeres of meiotic chromosomes (37). Mutations in RAP1 affect telomere length and TPE. However, Rap1p also binds to many nontelomeric sites, where it can act as either a transcriptional repressor or a transcriptional activator (reviewed in reference 89).
The proteins Rif1p and Rif2p were identified by their ability to interact with the carboxyl terminus of Rap1p in a two-hybrid system (28, 85). Loss of either protein causes telomere lengthening, a phenotype exacerbated by simultaneous loss of both proteins (85). Several models can explain these phenotypes: Rif1p and Rif2p could be telosomal proteins, nucleases, telomerase inhibitors, or transcriptional regulators of Rap1p-mediated transcription.
Proteins that bind to the single-stranded G tail on telomeres were first identified in the ciliated protozoan Oxytricha by virtue of their unusual property of remaining bound to DNA in 2 M salt (24). Because the Oxytricha proteins bind to the terminal G tail, unlike Rap1p, their binding is telomere limited. Although there is no evidence for salt-stable telomere binding proteins in Saccharomyces (87), genetic data suggest that Saccharomyces has a telomere-limited binding protein(s) that affects TPE and prevents end-to-end fusions (84). Gel shift analysis reveals that there are multiple yeast proteins that bind specifically to single-stranded TG1-3 DNA in vitro (39, 79). However, several of these single-stranded TG1-3 binding proteins also bind RNA (39, 79), and in several cases, their mutation or overexpression does not affect telomeres (39). Cdc13p also binds single-stranded TG1-3 DNA in vitro but not RNA or fully duplex telomeric DNA (40, 59). Moreover, Cdc13p affects both telomere length (25) and the susceptibility of telomeric DNA to the cell cycle-specific degradation (19) that occurs at the end of S phase (82). In addition, cells with the cdc13-2 allele have the same phenotype as cells lacking telomerase (59), yet extracts from cdc13-2 cells have normal telomerase activity in vitro (42). These data can be explained if Cdc13p binds telomeres in vivo and affects their accessibility to both nucleases and telomerase. However, immunoprecipitation experiments similar to those used to detect Rap1p at telomeres (14) do not reveal a physical association of Cdc13p with telomeric DNA (41a).
SIR2, SIR3, and SIR4 are essential for TPE (3) as well as for transcriptional silencing at internal loci (66, 73). Although all three proteins are chromatin associated in vivo (30, 75), none is thought to bind DNA. Rather, the Sir proteins bind chromatin by binding histones and/or by homotypic or heterotypic interactions with each other (29, 30, 53, 54, 56, 75). Sir3p also interacts directly with Rap1p (56). Chromatin cross-linking reveals that Sir2p, Sir3p, and Sir4p are associated with the transcriptionally silent chromatin near telomeres (30, 75). Cytological studies show that Rap1p and the three Sir proteins are concentrated in discrete foci near the nuclear periphery and that these foci colocalize with subtelomeric DNA (21, 22). The association of Sir proteins with subtelomeric chromatin and with Rap1p foci at the nuclear periphery is disrupted by mutations that eliminate TPE (12, 21, 22, 29, 30, 75). Thus, there is compelling evidence that the Sir proteins affect silencing by association with subtelomeric DNA, but there is no direct evidence that the Sir proteins are telosomal proteins.
A surprisingly large number of proteins, in addition to those listed above, affect yeast telomeres (see Discussion). We sought to develop a general method that could be used to determine whether these activities are a consequence of physical association with telomeres. As an alternative to cytological and biochemical approaches, we developed a one-hybrid system for use as a genetic assay for telomere binding proteins. In a one-hybrid system, the binding site of interest is placed immediately upstream of a reporter gene, a library that encodes protein fragments fused to a transcriptional activation domain is introduced, and fusion proteins that recognize the binding site are identified by the ability to activate the reporter gene. Any plasmid that activates transcription encodes a peptide that is potentially a sequence-specific DNA binding protein or is associated via protein-protein interactions with a sequence-specific DNA binding protein. In the one-hybrid system described in this paper, the cis-acting site is a fully functional chromosomal telomere, whereas in previous one-hybrid systems, the DNA binding sites were tested outside their normal chromosomal context (15, 16, 38, 81). Because the binding site is a chromosomal telomere, all proteins normally associated with telomeres should be present, and any interactions detected by the system are likely to be functionally relevant. Using this one-hybrid system, we show that Rap1, Sir2, Sir3, Sir4, Rif1, Rif2, and Cdc13 proteins are telomere binding proteins in vivo.
MATERIALS AND METHODS
All experiments were done in S. cerevisiae strains derived from YM701 (MATa ura3-52 his3-200 ade2-101 lys2-801 trp1-901 tyr1; from M. Johnston). The HIS-Tel strain was constructed by transforming YM701 with PvuII-digested pYAHISTEL, which inserts the promoter-defective allele of HIS3 and URA3 at ADH4 and deletes the ∼20 kb of DNA distal of ADH4. The HIS3-URA3 fragment in pYAHISTEL was obtained from p601 (2). pYAHISTEL also contained 71 bp of C1-3A/TG1-3 DNA immediately 5′ to HIS3, which served as a substrate for telomere formation in yeast. The C1-3A DNA was obtained as a 100-bp blunt-ended XhoI-HindIII fragment from pYTCA-1X (67). The 5.2-kb PvuII fragment also contained the last 594 bp of the ADH4 open reading frame (ORF) (starting at the XbaI site; isolated from pYA4-2 [61]) next to the URA3 gene that was used to target the fragment to chromosome VII-L. The HIS-Int strain was constructed by digesting plasmid pYAHU with PvuII and transforming YM701. pYAHU was constructed by inserting the ∼3.7-kb PvuII-SmaI fragment from p601 (2) into MscI-digested pYA4-2. pYACAHIS, used to make HIS-Int-CA, was constructed in a three-fragment ligation. The telomere tract for the ligation was obtained by cutting pCT300 (∼300-bp C1-3A EcoRI fragment of pYLPV in the BamHI/SalI site of the cloning vector pVZ1) with PvuII and NarI, followed by partial digestion with EcoRI to liberate a ∼550-bp fragment containing 276 bp of C1-3A/TG1-3 DNA as well as polylinker sequences and most of the lacI gene. A ligation was performed with the 550-bp fragment containing telomeric DNA, the 5.8-kb MscI-XbaI fragment from pYA4-2, and the 3.6-kb EcoRI-XbaI fragment from p601. pYACAHIS was digested with XmnI prior to transformation. In the final step in constructing HIS-Tel, HIS-Int, and HIS-Int-CA strains, yeast transformants were selected on media lacking uracil.
The sir3Δ strains were constructed by using pNO3, which eliminated almost the entire SIR3 ORF. To construct pNO3, plasmid pKL3 (a kind gift from R. Sternglanz), which contains the SIR3 ORF, ∼400 bp of DNA 5′ to the gene, and ∼1 kb 3′ of the gene, was used. Plasmid pKL3 was partially digested with EcoRI and HpaI in such a way as to delete ∼200 bp of DNA 5′ to the gene, as well as the entire ORF with the exception of the last 26 bp. The ∼4.2-kb vector was left with ∼200 bp of DNA from the 5′ end of SIR3 and ∼1 kb from the 3′ end. The 4.5-kb LYS2 gene was excised from pTD27 (a kind gift from T. Davis) with EcoRI and PvuII and ligated into the digested vector to make pNO3. Plasmid pNO3 was digested with PvuII prior to transformation. The sir3::LYS2 strains were constructed by transforming with EcoRI-digested pJR317 (35). In sir3::LYS2, LYS2 is inserted at the XhoI site in SIR3, after amino acid 961 of the 978-amino-acid Sir3p (35). Cells with sir3::LYS2 lack TPE (3) as well as HM silencing (35), phenotypes confirmed for the sir3::LYS2 allele in the YM701 background. Sir3p was not detectable by Western analysis of extracts from sir3::LYS2 cells with a Sir3p polyclonal antiserum (generously provided by L. Pillus) that readily detected Sir3p in a wild-type extract (data not shown).
To construct plasmids for expressing fusion proteins (Fig. 1B), fragments of genes were inserted in frame into the EcoRI cloning site in pJG4-5 (27) and pRF4-6NL (a kind gift from R. Finley). When pJG4-5 carries an insert, it expresses a polypeptide that is a fusion of the inserted gene with the B42 activation domain, the hemagglutinin (HA) tag, and a nuclear localization signal. B42 is an Escherichia coli sequence that activates transcription in yeast (49). pRF4-6NL is very similar to pJG4-5 but lacks the B42 activation domain. For each fusion protein, two independent clones from E. coli were assayed. RAP1 was tested as a 1.4-kb NruI-BglI fragment (from plasmid D123 [72]). The RIF1 constructs contained a 2.7-kb XmnI-HindIII fragment from pCH450 (28). The RIF2 constructs were made by PCR amplifying the entire ORF from pBS/RIF2 (85), using Taq polymerase (Boehringer Mannheim). EcoRI and XhoI sites engineered into the PCR product allowed insertion of RIF2 into EcoRI-XhoI-digested pJG4-5 or pRF4-6NL. SIR3 constructs contained a 1.6-kb EcoRI fragment consisting of nucleotides 1313 to 2911 of SIR3 from pKL3 (a pUC19-based plasmid from R. Sternglanz containing SIR3). The entire ORF of SIR4 was PCR amplified from genomic DNA by using Vent polymerase (New England Biolabs, Beverly, Mass.). EcoRI and XhoI sites engineered into the PCR product allowed insertion of SIR4 into EcoRI-XhoI-digested pJG4-5 or pRF4-6NL. The CDC13 gene product was assayed both as full-length protein and as three separate polypeptides. The plasmid for expressing full-length CDC13 was made by isolating the ∼3-kb NcoI-SalI fragment from pTHA-CDC13 (40) and inserting it into EcoRI-XhoI-digested pJG4-5. Portions of CDC13 were cloned as EcoRI fragments from nucleotides 1 to 754, 755 to 1525, and 1526 to 2941 from pVZ-CDC13 (contains a 3-kb SalI-BamHI fragment from pTHA-CDC13 in the BamHI-SalI site of pVZ1). Two different constructs were tested for EST1: the 2.1-kb HincII fragment which contains almost the entire ORF (constructed by B. Balakumaran) and the carboxyl-terminal EcoRI-HincII fragment, both of which were isolated from pVZ-EST1 (made by A. Wolf). The TEL2 insert, which expressed amino acids 27 to 688 of Tel2p, was cloned as an NruI-SalI restriction fragment obtained from pT2Na (68). NDJ1 constructs contained the entire 352-amino-acid ORF and were made by PCR amplification from genomic DNA, using Vent polymerase (New England Biolabs). XhoI sites engineered into the PCR product allowed insertion of NDJ1 into XhoI-digested pJG4-5 or pRF4-6NL. EST2 was cloned by S.-C. Teng, using PCR amplification of the entire 884-amino-acid ORF, from genomic DNA with High Fidelity polymerase (Boehringer Mannheim).
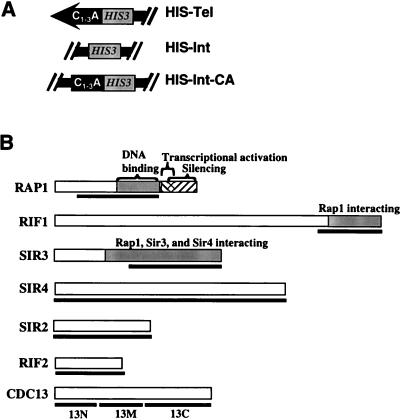
FIG. 1.
Schematic representation of strains and proteins. (A) Chromosomal context of reporter genes. Strains with HIS-Tel contained the promoter-defective allele of HIS3 integrated with its TATA element ∼50 bp from the start of the telomeric C1-3A/TG1-3 tract at the chromosome VII-L telomere. HIS3 was inserted within ADH4 in such a way that the ∼20 kb of DNA that is normally distal of ADH4 was deleted (23). HIS-Int contains the same HIS3 allele integrated at the same site within ADH4 but without deletion of distal DNA such that the reporter gene was ∼20 kb from the chromosome VII-L telomere. HIS-Int-CA was the same as HIS-Int except that it contained a ∼276-bp tract of C1-3A/TG1-3 DNA ∼50 bp distal to the HIS3 promoter. For all three strains, there is a copy of URA3 proximal to HIS3 that served as the selectable marker during transformation. (B) Structures of proteins that tested positive in the one-hybrid assay. Bars below the proteins indicate regions of the proteins tested in the studies described here. The 462 amino acids of the 827-amino-acid Rap1p that were tested contain the DNA binding domain but not the endogenous transcriptional activation domain. For Rif1p, 367 amino acids of the 1,915-amino-acid protein were expressed in the fusion protein, which included the region that interacts with Rap1p by two-hybrid analysis (28). For Sir3p, the carboxyl 532 amino acids of the 978-amino-acid protein were expressed. By two-hybrid analysis, amino acids 307 to 978 of Sir3p are sufficient for its interaction with Rap1p, Sir3p, and Sir4p (56). By in vitro analysis, the region of Sir3p sufficient for Sir4p interaction was delimited further (amino acids 622 to 978 [75]). Fusion proteins contained the entire ORF for Sir4p (1,358 amino acids), Sir2p (563 amino acids), and Rif2p (395 amino acids). The 924-amino-acid Cdc13p was tested both as a full-length fusion protein and as three fusion proteins containing 251 amino acids (Cdc13N-Actp), 257 amino acids (Cdc13M-Actp), or 416 amino acids (Cdc13C-Actp).
To select for expression of the HIS3 reporter gene, cells were grown to stationary phase at 30°C (∼2 days) in liquid medium lacking tryptophan and containing 3% raffinose to avoid glucose repression of fusion protein expression. Five microliters of cells was then spotted in 10-fold serial dilutions onto 3% Gal-His plates containing 3-amino-1,2,4-triazole (3-AT) (Sigma, St. Louis, Mo.) or 3% Gal-Trp control plates. The concentration of 3-AT was determined empirically for each experiment because different lots of 3-AT differed in strength. Moreover, the effective concentration of 3-AT in plates decreased over time. In many cases where the fusion protein being tested did not activate HIS-Tel, cells carrying only the vector and thus expressing a 104-amino-acid polypeptide grew better on test plates than cells expressing the test protein (e.g., proteins without activation domain in Fig. 1B [Rif1p and Rif2p] or Fig. 4A [Cdc13Np]).
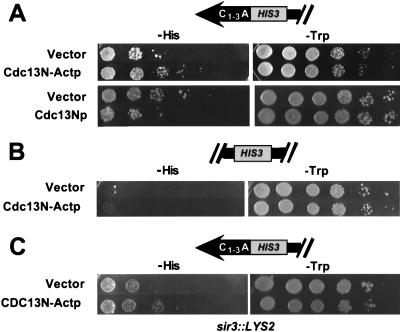
FIG. 4.
The amino-terminal portion of Cdc13p activated HIS-Tel. The amino-terminal 251 amino acids of Cdc13p (or vector alone) were expressed as a fusion protein in HIS-Tel with or without the transcriptional activation domain (A), in HIS-Int with the activation domain (B), or in sir3::LYS2 HIS-Tel with the activation domain (C). Cells were spotted in 10-fold serial dilutions on test plates that select for the telomere interaction (left) or control plates (right). Test plates in panels A and B had no 3-AT; test plates in panel C contained 50 mM 3-AT. Plates were incubated at 30°C for 5 (test plates, A and B), 3 (control plates), or 13 (test plates, C) days.
Expression of proteins was verified by Western analysis using a monoclonal mouse anti-HA antibody (Boehringer Mannheim). Samples were prepared essentially as described previously (39, 40). Proteins were electrophoretically separated on 7.8% acrylamide gels. Gels were blotted to nitrocellulose (39, 40). The primary antibody for detection was anti-HA antibody diluted 1:100 (Amersham Corp., Arlington Heights, Ill.). The secondary antibody was horseradish peroxidase-conjugated anti-mouse (Amersham Corp.).
RESULTS
The telomere one-hybrid system: general considerations.
We constructed strains in which a promoter-defective allele of HIS3 (2) was inserted immediately adjacent to the left telomere of chromosome VII (Fig. 1A, HIS-Tel). In this strain, transcription of the HIS3 reporter gene was necessary for growth on plates lacking histidine. Because the HIS3 allele supports a very low level of basal transcription, HIS-Tel cells grew poorly on plates lacking histidine (data not shown). HIS3 was chosen as a reporter gene because a competitive inhibitor of His3p, 3-AT, can be used to select for cells expressing different levels of His3p. The more 3-AT in the medium, the more His3p is required for growth (2).
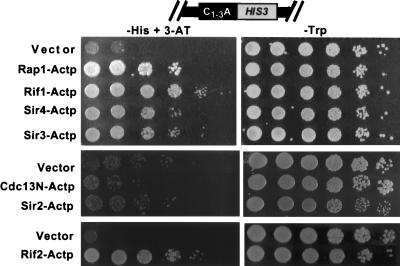
Hybrid proteins consisting of a candidate telomere binding protein fused to the B42 transcriptional activation domain (49) were expressed under the control of a galactose-inducible promoter in the HIS-Tel strain from extrachromosomal plasmids carrying the TRP1 gene. If the fusion protein binds telomeres in vivo, it might activate transcription of the telomeric HIS3 gene and allow improved growth on galactose plates lacking histidine (hereafter referred to as test plates). In some cases, the entire candidate protein was present in the fusion protein, whereas in others, portions of the candidate protein were expressed (Fig. 1B). Fusion proteins also contained an HA tag that was used in Western blot analysis to ensure that the fusion proteins were expressed. For each fusion protein, at least two independent yeast transformants were tested. Each fusion protein was tested a minimum of six times on at least 2 different days. Transcriptional activation was assessed by plating 10-fold serial dilutions of each strain on test plates (Fig. 2, left panels). To demonstrate that similar numbers of cells were plated for all strains, the same dilutions were also plated on galactose medium that contained histidine but lacked tryptophan (Fig. 2, right panels). The control plates selected for plasmid maintenance and induced expression of the fusion proteins but did not require expression of HIS-Tel.
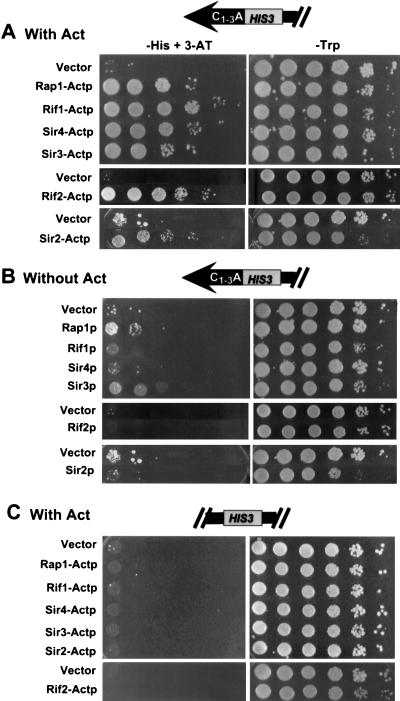
FIG. 2.
Rap1, Rif1, Sir4, Sir3, Rif2, and Sir2 fusion proteins activated HIS3 at a telomere (HIS-Tel) but not at an internal site on the chromosome (HIS-Int). HIS-Tel (A and B) or HIS-Int (C) cells expressing various proteins either fused to the activation domain (A and C) or without an activation domain (B) were spotted in 10-fold serial dilutions onto test plates that selected for the telomere interaction (left) or control plates (right). Here and in all other figures, test and control plates both contained galactose to induce expression of the fusion proteins. Here and in all subsequent figures, cells presented in the same photograph were assayed on the same plate (i.e., the vector control was always on the same plate as the protein being tested). Each fusion protein was tested in a range of 3-AT concentrations. In this and other figures, the 3-AT concentration showing the greatest growth difference between the control (vector alone) and the cells expressing the fusion protein is shown. In panels A and B, the concentration of 3-AT was 5 (Sir2), 20 (Rif2), or 10 (all other fusion proteins) mM. In panel C, plates had no 3-AT except for Rif2-Actp, which had 20 mM 3-AT. The HIS3 gene in HIS-Int was functional since cells containing vector alone often formed a few colonies at the highest dilution. Consistent with published reports (31), expression of some fusion proteins inhibited growth (see, especially, Sir2p with or without activation domain, control plates). Plates were incubated at 30°C for 4 (control plates), 8 (5 mM 3-AT), 7 (10 mM 3-AT), or 6 (20 mM 3-AT) days.
Several controls were used to establish that fusion proteins that tested positive in the telomere one-hybrid system did so by binding to the chromosome VII-L telomere. To establish that transcriptional activation depended on the fusion proteins, cells with the fusion proteins were also tested on glucose medium, where expression of the fusion proteins is low or absent. Activation by the fusion protein was always compared to activation by the vector alone, which produced a 104-amino-acid polypeptide consisting of the activation domain, a nuclear localization signal, and the HA tag. In addition, each candidate protein was also tested without an activation domain. Overexpression of Sir4p (12) or the carboxyl terminus of Rap1p (84) decreases TPE, presumably because the proteins titrate factors important for TPE away from telomeres. If a fusion protein activated the HIS3 reporter gene in trans by reducing TPE, it should also activate when expressed without its activation domain. Finally, specificity for the telomere was established by determining if fusion proteins activated the HIS3 reporter gene when it was inserted 20 kb from the left telomere of chromosome VII (Fig. 1A, HIS-Int).
To determine if a telomere one-hybrid system can detect telomere binding proteins, we first examined the behavior of the known telomere binding protein Rap1p. When HIS-Tel contained vector alone (Fig. 2A, left panel, Vector), a few colonies grew in the spot containing the most cells. However, expression of Rap1-Actp, which contained the DNA binding region of Rap1p (Fig. 1B) fused to the transcriptional activation domain, caused a ∼1,000-fold increase in plating efficiency on test plates compared to cells containing vector alone (Fig. 2A, left, Rap1-Actp). This growth was dependent on protein expression because activation was not seen on plates lacking galactose (data not shown). Rap1-Actp did not activate the same HIS3 allele when the gene was 20 kb from the telomere (HIS-Int [Fig. 2C, left, Rap1-Actp]). Without an activation domain, Rap1p supported a very modest increase in colony-forming ability compared to cells carrying vector alone, indicating that activation was not due to relief of TPE (Fig. 2B, left, Rap1p). Rap1p-Actp also activated HIS-Tel in a sir3 strain (Fig. 3, left, Rap1-Actp). This result provided definitive evidence that the activating effect of Rap1-Actp on HIS-Tel was not due to its decreasing TPE since TPE is eliminated in a sir3 strain (3). Taken together, these results indicate that a protein known to bind telomeres in vivo, Rap1p, behaves as predicted in this one-hybrid system, thus defining a new in vivo telomere binding assay for S. cerevisiae.
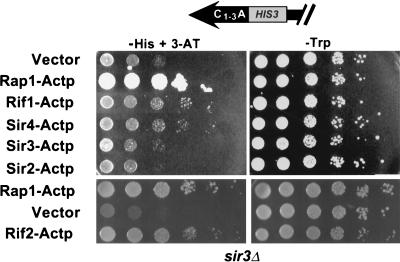
FIG. 3.
Rap1p, Rif1p, Sir4p, and Rif2p interactions with a telomere did not require Sir3p. HIS-Tel sir3Δ strains carrying vector alone (top rows) or expressing various fusion proteins were spotted in 10-fold serial dilutions onto test plates that select for the telomere interaction (left; 50 mM 3-AT) or control plates (right). Plates were incubated at 30°C for 2 (control plates, top), 4 (control plates, bottom), 6 (test plates, bottom), or 10 (test plates, top) days.
Sir3, Sir4, Sir2, Rif1, and Rif2 fusion proteins activate the telomeric reporter gene.
Next, other proteins that affect telomeres but which had not been shown to interact directly with telomeres were tested. Fusion proteins containing either 367 amino acids from the carboxyl terminus of Rif1p, 532 amino acids from the carboxyl terminus of Sir3p (Fig. 1B), or full-length Sir2p, Sir4p, or Rif2p were made and subjected to telomere one-hybrid analysis.
Each of the fusion proteins activated HIS-Tel, supporting a ∼200-fold (Sir3-Actp and Sir2-Actp), ∼1,000-fold (Sir4-Actp), or ∼10,000-fold (Rif1-Actp and Rif2-Actp) increase in plating efficiency on test plates compared to cells containing vector alone (Fig. 2A, left). None of these fusion proteins activated HIS-Int (Fig. 2C, left). Each of these fusion proteins showed no increase in colony-forming ability compared to vector alone when expressed without an activation domain (Fig. 2B, left). (Although the Sir3 peptide without the fused activation domain generated a haze of cells at several dilutions, these cells never generated colonies.) In addition, Rif1-Actp, Rif2-Actp, and Sir4-Actp activated HIS-Tel in a sir3 strain (Fig. 3; discussed in more detail below). We conclude that Sir2p, Sir3p, Sir4p, Rif1p, and Rif2p, like Rap1p, bind chromosomal telomeres in vivo.
The telomere one-hybrid assay is specific for telomere binding proteins.
To ensure that the activation of HIS3 was not a general property of fusion proteins, a random collection of 20 genomic fragments fused to the transcriptional activation sequence was tested in the HIS-Tel strain. Each of these fusion proteins was negative in the telomere one-hybrid assay, as was RNA polymerase II subunit B4 (33), which is not suspected of having any telomere function (data not shown).
Several other proteins that affect telomeres in vivo were also tested (see Materials and Methods for structures of these fusion proteins). Est1p binds single-stranded TG1-3 DNA in vitro (79). However, since Est1p has a higher affinity in vitro for RNA than for single-stranded TG1-3 DNA and binds telomerase RNA in vivo (41, 74), it may function by interacting with telomerase RNA. Est2p is the presumed catalytic subunit of yeast telomerase (43). Ndj1p/Tam1p localizes to the ends of meiotic chromosomes (11, 13). Tel2p affects both telomere length (48) and TPE (68) and binds C1-3A/TG1-3 DNA in vitro (37a). Although Western blot analysis showed that each of these fusion proteins was expressed, none activated HIS-Tel (data not shown).
Binding of Rap1p, Rif1p, Rif2p, and Sir4p but not Sir2p to the telomere does not require endogenous Sir3p.
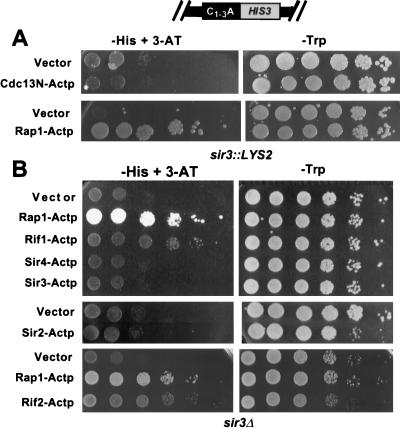
The telomere one-hybrid system can be used not only to determine if a given protein binds telomeres in vivo but also to assess requirements for binding. For example, since the carboxyl-terminal regions of Sir3p and Rif1p were positive in the one-hybrid system, these regions (Fig. 1B) must contain the necessary information for telomere localization. The one-hybrid system can also be used to determine if specific proteins influence the binding of other proteins to the telomere. As a first step in this type of analysis, we determined if each of the proteins giving positive results in the one-hybrid system in a wild-type strain could activate the reporter gene in a HIS-Tel sir3 strain.
Since Sir3p is required for TPE (3), constitutive transcription of HIS-Tel should be higher in sir3 strains. As anticipated, HIS-Tel sir3 cells carrying vector alone efficiently generated colonies on test plates containing <50 mM 3-AT. Thus, HIS-Tel sir3 cells containing candidate fusion proteins were tested on medium containing 50 mM 3-AT, the lowest concentration of 3-AT at which the HIS-Tel sir3 strain containing vector alone did not generate colonies efficiently (Fig. 3, left). On test plates containing 50 mM 3-AT, HIS-Tel sir3 cells expressing Rap1-Actp, Rif1-Actp, Sir4-Actp, and Rif2-Actp generated colonies more efficiently than cells carrying vector alone, whereas Sir3-Actp and Sir2-Actp did not (Fig. 3, left panels). However, cells expressing Rif1-Actp and Sir4-Actp grew slowly on test plates. This low growth rate was not due to an inhibitory effect of high 3-AT concentrations on cell growth since cells expressing Rap1-Actp or Rif2-Actp grew well on these plates (Fig. 3). These data show that binding of Rap1p, Rif1p, Rif2p, and Sir4p to the telomere does not require Sir3p but that the association of Rif1p and Sir4p with the telomere is altered in the absence of Sir3p. One possibility is that Sir3p stabilizes the association of these proteins with the telosome.
Since Sir3-Actp, a fusion protein that lacks the amino-terminal 45% of Sir3p, failed to activate in the sir3 strain, it probably binds telomeres by interacting with endogenous, full-length Sir3p. Indeed, the carboxyl terminus of Sir3p interacts by two-hybrid analysis with full-length Sir3p (56). Others have shown that elimination of or even simple fusions to the amino terminus of Sir3p eliminate Sir3p silencing function (10, 47, 51). We infer from the one-hybrid data that alteration of the amino terminus of Sir3p affects TPE by preventing Sir3p association with telomeres. Telomere association of Sir2-Actp, a fusion containing full-length Sir2p, also required Sir3p, a result consistent with the finding that E. coli-synthesized Sir2p and Sir3p interact in vitro (54).
The amino-terminal region of Cdc13p interacts with telomeres in vivo.
To determine if Cdc13p is an in vivo telomere binding protein, we first tested full-length Cdc13p. Full-length Cdc13p with or without an activation domain activated HIS-Tel, suggesting that overexpression of full-length Cdc13p affects TPE (data not shown). Next we expressed portions of Cdc13p as fusion proteins (Fig. 1B). Cdc13N-Actp contained the first 251 amino acids of the 924-amino-acid Cdc13p, Cdc13M-Actp contained the middle 257 amino acids, and Cdc13C-Actp contained the carboxyl-terminal 416 amino acids. Cdc13M-Actp did not activate HIS-Tel (data not shown). The C-terminal region showed weak activity, but removing the activation domain did not eliminate this activity, suggesting that the C-terminal region reduced TPE in trans (data not shown). However, HIS-Tel cells expressing Cdc13N-Actp generated colonies ∼50-fold better than vector alone (Fig. 4A). This activity was completely dependent on the activation domain (Fig. 4A) and did not occur when HIS3 was 20 kb from the telomere (HIS-Int [Fig. 4B]). Cdc13N-Actp also activated HIS-Tel ∼50-fold in a sir3 strain (Fig. 4C). Thus, the activation of HIS-Tel by Cdc13N-Actp was not due to an effect on TPE. We conclude that the N-terminal domain of Cdc13p associates with telomeres in vivo.
Although Rap1p, Sir2p, Sir3p, Sir4p, Rif1p, and Rif2p bind internal tracts of telomeric DNA, Cdc13p binding is telomere limited.
Since Cdc13p binds single-stranded TG1-3 DNA in vitro, it is possible that Cdc13N-Actp activated HIS-Tel by binding to the single-stranded TG1-3 tails thought to be present at the ends of all chromosomes. If this model is correct, Cdc13N-Actp should not bind internal tracts of C1-3A/TG1-3 DNA. To test this possibility, a strain called HIS-Int-CA was constructed. This strain was the same as HIS-Int except that it contained a 276-bp tract of C1-3A/TG1-3 DNA immediately distal to HIS3 (Fig. 1B). Since many organisms, including yeast (46, 80), have stretches of telomeric DNA at internal sites on the chromosome, binding to internal tracts of telomeric DNA is also likely to be biologically relevant.
Proteins like Rap1p that bind duplex C1-3A/TG1-3 DNA or proteins like Rif1p and Rif2p that bind to the telomere via protein-protein interactions are expected to activate HIS-Int-CA. Indeed, Rap1-Actp, Rif1-Actp, Rif2-Actp, Sir3-Actp, and Sir4-Actp all activated HIS-Int-CA efficiently, allowing colony growth at dilutions 100- to 10,000-fold higher than with vector alone (Fig. 5, left). However, neither Cdc13N-Actp nor Sir2-Actp activated HIS-Int-CA (Fig. 5). Since Cdc13N-Actp activated HIS-Tel in a sir3 strain (Fig. 4C) but Sir2-Actp did not (Fig. 3), binding of Cdc13N-Actp to a telomere required neither Sir2p nor Sir3p. The failure of Cdc13N-Actp to activate HIS-Int-CA cannot be attributed to the absence of any of the other known telomere binding proteins since Rap1-Actp, Rif1-Actp, Rif2-Actp, Sir3-Actp, and Sir4-Actp activated HIS-Int-CA (Fig. 5). These data suggest that Cdc13N-Actp failed to activate HIS-Int-CA because its in vivo substrate is a single-stranded TG1-3 tail that is not present at internal tracts of C1-3A/TG1-3 DNA.
FIG. 5.
Rap1p, Rif1p, Rif2p, Sir3p, and Sir4p, but not Cdc13p or Sir2p, bind an internal tract of telomeric DNA in a wild-type strain. The indicated fusion proteins were expressed in HIS-Int-CA cells. Cells were spotted in 10-fold serial dilutions on control plates (right) or test plates (left) containing 10 (top), 0 (middle), or 20 (bottom) mM 3-AT. Control plates were incubated for 4 days, and test plates were incubated for 6 (top and bottom) or 16 (middle) days.
The fusion proteins were also tested in a sir3 HIS-Int-CA strain (Fig. 6, left). Only Rap1-Actp, Rif1-Actp, and Rif2-Actp activated in this strain. Thus, although Sir4p can bind to the telomere in the absence of Sir3p (Fig. 3), Sir3p is required for Sir4p binding to internal tracts of C1-3A/TG1-3 DNA (Fig. 6B).
FIG. 6.
Sir4p requires Sir3p to bind an internal tract of C1-3A/TG1-3 DNA. The indicated fusion proteins were expressed in HIS-Int-CA sir3 strains. Cells were spotted in 10-fold serial dilutions onto test plates (left) or control plates (right). The 3-AT concentration was 35 (A) or 50 (B) mM. Control plates were incubated for 4 days, and test plates were incubated for 17 (A) or 13 (B) days.
DISCUSSION
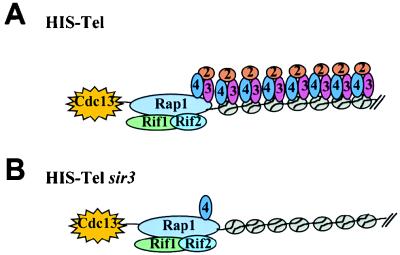
This paper describes a one-hybrid system that provides an in vivo assay for telomere binding proteins. In this scheme, a reporter gene with a minimal promoter was positioned immediately adjacent to a chromosomal telomere (HIS-Tel [Fig. 1A]). Candidate telomere binding proteins fused to a transcriptional activation domain were tested for the ability to activate the reporter gene. Because activation was assayed in living cells and because the reporter gene was next to a chromosomal telomere, proteins that were positive in the system are very likely to be biologically relevant. The versatility of the telomere one-hybrid system was demonstrated by the fact that proteins that localize to telomeres by direct binding to telomeric DNA (Rap1p and Cdc13p) or by protein-protein interactions (Rif1p, Rif2p, Sir2p, Sir3p, and Sir4p) gave positive signals. The one-hybrid data on telomere-associated proteins combined with earlier information on the distribution of proteins in subtelomeric chromatin (30, 75) in both wild-type and sir3 cells are summarized in Fig. 7.
FIG. 7.
Schematic representation of proteins associated with chromosomal telomeres and subtelomeric regions in vivo in a wild-type (A) or sir3 (B) strain. The physical presence of Rap1p at telomeres in vivo was first shown in reference 13 and confirmed herein. The presence of Sir2p, Sir3p, and Sir4p on subtelomeric nucleosomes is from references 30 and 75. The ovals labeled 2, 3, and 4 represent Sir2p, Sir3p, and Sir4p. Although one molecule of each telomere binding protein is indicated, nothing is known about the stoichiometry of telomere binding proteins except that there are 10 to 20 molecules of Rap1p per telomere (20, 87).
Since none of the fusion proteins activated the reporter gene at a nontelomeric site (HIS-Int [Fig. 2C and 4B), activation was due not to their recognizing HIS3 DNA or to their having a general affinity for nucleosomes. Although subtelomeric nucleosomes have several features such as hypoacetylated histones that are characteristic of silent chromatin, these modifications are lost in a sir3 strain (7, 55). Likewise, Sir2p, Sir3p, and Sir4p bind subtelomeric nucleosomes in wild-type cells, but the association of all three Sir proteins with subtelomeric chromatin is eliminated in a sir3 strain (30, 75). Rap1p, Sir4p, Rif1p, Rif2p, and Cdc13p activated HIS-Tel even in a sir3 strain (Fig. 3 and 4C). Given that TPE and the chromatin features associated with TPE are not present in sir3 cells, this activation cannot be attributed to these proteins binding subtelomeric nucleosomes by virtue of their having silencing specific modifications that would be absent at HIS-Int. Thus, these proteins must activate HIS-Tel by association with the telomere itself (Fig. 7).
Given that genes next to yeast telomeres are subject to TPE, it is perhaps surprising that transcription can be used to monitor telomere binding. Previous studies identified a ∼100-bp region between the telosome and subtelomeric nucleosomes that is highly accessible in vivo to both the dam methylase (86) and nucleases (69). To maximize transcription of the reporter gene, its TATA box was positioned within this nuclease-hypersensitive region. Since more 3-AT was needed to prevent growth of cells carrying vector alone in sir3 compared to wild-type cells (Fig. 3 and 4C), transcription of the reporter genes was reduced by TPE despite the favorable position of the TATA box. However, since TPE affects basal, not activated, transcription (23, 65), the activating nature of the fusion proteins was able to overcome the repressive effects of TPE.
The one-hybrid system was specific for telomere binding proteins in that most fusion proteins, including several that affect telomeres, were negative in this system. However, the fusion proteins that were positive in the system did not exhibit identical activation behaviors. Since 3-AT is a competitive inhibitor of His3p, the concentration of 3-AT at which a strain can grow is a rough indicator of the amount of His3p it produces. By this criterion, different fusion proteins generated different amounts of His3p. For example, wild-type Rap1-Actp-expressing HIS-Tel cells were able to grow at very high concentrations of 3-AT, whereas cells expressing Sir2-Actp were not (data not shown). In addition, the fraction of HIS-Tel cells able to grow on test plates varied depending on which fusion protein was expressed. For example, in the wild-type HIS-Tel strain, virtually all cells expressing Rif1-Actp or Rif2-Actp grew on test plates, whereas only ∼2% of the Rap1-Actp-expressing cells formed colonies (although the Rap1-Actp-expressing colonies grew faster than Rif1-Actp-expressing cells) (Fig. 2A). However, in the sir3 strain, ∼100% of the Rap1-Actp-expressing cells grew on test plates (Fig. 3), probably because Sir3p, a protein involved in transcriptional repression, in some way masks the Rap1p transcriptional activation domain. Several factors probably contribute to the number of positive cells as well as to their growth rate, including the ability of the fusion protein to compete with the endogenous protein for telomere binding, the number of copies of the protein at the telomere, the position of its acidic activation domain within the multiprotein-DNA complex, the stability of its association, and the fraction of the cell cycle during which it is telomere bound. Hence, a negative result in the one-hybrid assay does not rule out telomere association. For example, Est2p, the catalytic subunit of telomerase (42), surely interacts with telomeres in vivo yet was negative in the one-hybrid assay (data not shown). Probably, the Est2p interaction with the telomere is too transient or too infrequent (or both) to give a positive signal in the one-hybrid system.
Biochemical and cytological data demonstrate that Sir2, Sir3, and Sir4p are components of subtelomeric chromatin (12, 22, 29, 30, 60, 75). Although genetic evidence suggests that Sir3p is also a telosomal protein (44, 47), the one-hybrid data shown here provide the first direct evidence that the three Sir proteins are integral components of the telosome (summarized in Fig. 7). The association of the three Sir proteins with the telomere provides direct support for models in which C1-3A/TG1-3 DNA acts as an initiation site for silencing by recruiting silencing proteins to the telomere itself. Our results also demonstrate that requirements for Sir4p binding are different at the telomere (Fig. 3) than at both subtelomeric nucleosomes (75) and internal tracts of C1-3A/TG1-3 DNA (Fig. 6B) since Sir4p association was Sir3p independent only at the telomere. Thus, there must be something special about the telomere that facilitates Sir4p binding to C1-3A/TG1-3 DNA in the absence of Sir3p, such as the presence of Cdc13p or the higher concentration of Rap1p that results from telomere-telomere interactions (21). Either of these explanations might also explain why Sir2p was detectable at telomeric (Fig. 2) but not internal (Fig. 5) tracts of telomeric DNA.
This paper provides the first direct evidence that Rif1p and Rif2p are telosomal proteins (Fig. 2). Their presence at the telomere in combination with the telomere lengthening seen in their absence (28, 85) supports the idea that their binding limits the access of telomeric DNA to telomerase. That telosomal proteins might limit telomere replication was first proposed from the paradoxical result that adding extra telomeres or internal tracts of C1-3A/TG1-3 DNA results in telomere elongation (67). Telosomal proteins that limit telomere replication are probably widespread, as the human telomere binding protein TRF1 also inhibits telomere lengthening (78).
Our results also provide the first direct evidence that Cdc13p is a telosomal protein (summarized in Fig. 7). Cdc13N-Actp, which contained the amino-terminal ∼20% of Cdc13p, activated the reporter gene at a telomere (Fig. 4A) but not at an internal stretch of telomeric DNA (Fig. 5). Since other telomere binding proteins activated HIS-Int-CA (Fig. 5), the failure of Cdc13N-Actp to activate in this strain was not due to the absence of these proteins at the internal C1-3A/TG1-3 tract. These data can be explained if Cdc13N-Actp binds single-stranded TG1-3 DNA in vivo as it does in vitro (40, 59). In a gel shift assay, Cdc13p binds single-stranded TG1-3 DNA and C1-3A/TG1-3 with a 5- to 9-base single-stranded TG1-3 tail but does not bind duplex C1-3A/TG1-3 DNA (40). Since the amino-terminal 251 amino acids of Cdc13p were sufficient for telomere binding in vivo (Fig. 1B), this region of the protein is likely to contain the Cdc13p DNA binding domain. In earlier experiments, we were unable to detect association of HA-tagged Cdc13p with telomeres by an immunoprecipitation approach (41a), perhaps because its telomeric association was lost during extract preparation. This difference emphasizes the benefits of a sensitive in vivo assay for telomere binding.
In addition to the proteins tested here, there are many others that affect yeast telomeres in vivo or bind telomeric DNA in vitro, including the products of TEL1 (48), HDF1 (63), HDF2 (5, 26), STN1 (25), RAD50 (36), MRE11 (4), XRS2 (4), PIF1 (70), KEM1/SEP1 (45), CDC17/POL1 (9), RFC1/CDC44 (1), EST4 (57), GAL11 (76), WTM1, WTM2, and WTM3 (62), SAS2 (64), HST3 and HST4 (6), CAC1 (17, 32, 55), CAC2 and CAC3 (32), TOP3 (34), TBF1 (8), and SET1 (58). To understand their mechanism of action, it is important to determine if they are physically associated with telomeres and, if so, to establish requirements for telomere binding. The one-hybrid system described here is a relatively simple in vivo assay that can be applied to any candidate telomere binding protein or used as a screen for new telosomal proteins. In addition, these methods should be applicable to other organisms, including human cells in culture (18), where a reporter gene can be positioned next to a chromosomal telomere.
ACKNOWLEDGMENTS
We thank past and present members of the Zakian lab for recombinant DNA, B. Balakumaran and A. Taggart for helpful discussions, and especially J.-J. Lin for sharing information prior to publication. We thank C. Freudenreich, E. Monson, A. Taggart, and S.-C. Teng for their comments on the manuscript. We also thank R. Brent, R. Finley, and other members of the Brent lab for reagents and advice, L. Pillus for sharing information about Sir3p, and L. Breeden for hospitality during the course of some of this work.
This research was supported by NIH grant GM43255. M.K.A. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute. B.D.B. was supported in part by NIH training grant AG00057 administered by the Pathology Department at the University of Washington.
REFERENCES
- 1.Adams A K, Holm C. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4614–4620. doi: 10.1128/mcb.16.9.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexandre C, Grueneberg D A, Gilman M Z. Studying heterologous transcription factors in yeast. Methods Companion Methods Enzymol. 1993;5:147–155. [Google Scholar]
- 3.Aparicio O M, Billington B L, Gottschling D E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 4.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 8.Brigati C, Kurtz S, Balderes D, Vidali G, Shore D. An essential yeast gene encoding a TTAGGG repeat-binding protein. Mol Cell Biol. 1993;13:1306–1314. doi: 10.1128/mcb.13.2.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson M J, Hartwell L. CDC 17: an essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- 10.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 11.Chua P R, Roeder G S. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 12.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad M N, Dominguez A M, Dresser M E. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 14.Conrad M N, Wright J H, Wolf A J, Zakian V A. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell. 1990;63:739–750. doi: 10.1016/0092-8674(90)90140-a. [DOI] [PubMed] [Google Scholar]
- 15.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 16.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 17.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 18.Farr C, Fantes J, Goodfellow P, Cooke H. Functional reintroduction of human telomeres into mammalian cells. Proc Natl Acad Sci USA. 1991;88:7006–7010. doi: 10.1073/pnas.88.16.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilson E, Roberge M, Giraldo R, Rhodes D, Gasser S M. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J Mol Biol. 1993;231:293–310. doi: 10.1006/jmbi.1993.1283. [DOI] [PubMed] [Google Scholar]
- 21.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B K, Grunstein M, Gasser S M. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 24.Gottschling D E, Zakian V A. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 25.Grandin N, Reed S I, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 26.Gravel S, Larrivee M, Labrecque P, Wellinger R J. Yeast Ku as a regulator of chromosomal DNA end structure. Science. 1998;280:741–744. doi: 10.1126/science.280.5364.741. [DOI] [PubMed] [Google Scholar]
- 27.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 28.Hardy C F, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 29.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 30.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 31.Holmes S G, Rose A B, Steuerle K, Saez E, Sayegh S, Lee Y M, Broach J R. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 33.Khazak V, Sadhale P P, Woychik N A, Brent R, Golemis E A. Human RNA polymerase II subunit hsRPB7 functions in yeast and influences stress survival and cell morphology. Mol Biol Cell. 1995;6:759–775. doi: 10.1091/mbc.6.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim R A, Caron P R, Wang J C. Effects of yeast DNA topoisomerase III on telomere structure. Proc Natl Acad Sci USA. 1995;92:2667–2671. doi: 10.1073/pnas.92.7.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimmerly W J, Rine J. Replication and segregation of plasmids containing cis-acting regulatory sites of silent mating-type genes in Saccharomyces cerevisiae are controlled by the SIR genes. Mol Cell Biol. 1987;7:4225–4237. doi: 10.1128/mcb.7.12.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kironmai K M, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cell. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 37.Klein F, Laroche T, Cardenas M E, Hofmann J F, Schweizer D, Gasser S M. Localization of RAP1 and topoisomerase II in nucleic and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Kota R, Runge K W. The yeast telomere length regulator TEL2 encodes a protein that binds to telomeric DNA. Nucleic Acids Res. 1998;26:1528–1535. doi: 10.1093/nar/26.6.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J J, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- 39.Lin J J, Zakian V A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994;22:4906–4913. doi: 10.1093/nar/22.23.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J-J, Zakian V A. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 41a.Lin, J.-J., and V. A. Zakian. Unpublished results.
- 42.Lingner J, Cech T R, Hughes T R, Lundblad V. Three ever shorter telomerase (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingner J, Hughes T R, Shevchenko A, Mann M, Lundblad V, Cech T R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 44.Liu C, Lustig A J. Genetic analysis of Rap1p/Sir3p interactions in telomeric and HML silencing in Saccharomyces cerevisiae. Genetics. 1996;143:81–93. doi: 10.1093/genetics/143.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Lee A, Gilbert W. Gene disruption of a G4-DNA-dependent nuclease in yeast leads to cellular senescence and telomere shortening. Proc Natl Acad Sci USA. 1995;92:6002–6006. doi: 10.1073/pnas.92.13.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Louis E J, Naumova E S, Lee A, Naumov G, Haber J E. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lustig A J, Liu C, Zhang C, Hanish J P. Tethered Sir3p nucleates silencing at telomeres and internal loci in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2483–2495. doi: 10.1128/mcb.16.5.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lustig A J, Petes T D. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 50.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 51.Marcand S, Buck S W, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- 52.McElligott R, Wellinger R J. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 54.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monson E K, de Bruin D, Zakian V A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 57.Morris D K, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 58.Nislow C, Ray E, Pillus L. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol Biol Cell. 1997;8:2421–2436. doi: 10.1091/mbc.8.12.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 60.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S M. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 61.Paquin C E, Williamson V M. Ty insertions at two loci account for most of the spontaneous antimycin A resistance mutations during growth at 15°C of Saccharomyces cerevisiae strains lacking ADH1. Mol Cell Biol. 1986;6:70–79. doi: 10.1128/mcb.6.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pemberton L F, Blobel G. Characterization of the Wtm proteins, a novel family of Saccharomyces cerevisiae transcriptional modulators with roles in meiotic regulation and silencing. Mol Cell Biol. 1997;17:4830–4841. doi: 10.1128/mcb.17.8.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Porter S E, Greenwell P W, Ritchie K B, Petes T D. The DNA-binding protein Hdf1p (a putative Ku homologue) is required for maintaining normal telomere length in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:582–585. doi: 10.1093/nar/24.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reifsnyder C, Lowell J, Clarke A, Pillus L. Yeast SAS silencing genes and human genes associated with AML and HIV-1 Tat interactions are homologous with acetyltransferases. Nat Genet. 1996;14:42–49. doi: 10.1038/ng0996-42. [DOI] [PubMed] [Google Scholar]
- 65.Renauld H, Aparicio O M, Zierath P D, Billington B L, Chhablani S K, Gottschling D E. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- 66.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Runge K W, Zakian V A. Introduction of extra telomeric DNA sequences into Saccharomyces cerevisiae results in telomere elongation. Mol Cell Biol. 1989;9:1488–1497. doi: 10.1128/mcb.9.4.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 70.Schulz V P, Zakian V A. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 71.Shampay J, Szostak J W, Blackburn E H. DNA sequences of telomeres maintained in yeast. Nature. 1984;310:154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 72.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 73.Stavenhagen J B, Zakian V A. Internal tracts of telomeric DNA act as silencers in Saccharomyces cerevisiae. Genes Dev. 1994;8:1411–1422. doi: 10.1101/gad.8.12.1411. [DOI] [PubMed] [Google Scholar]
- 74.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki Y, Nishizaw M. The yeast GAL11 protein is involved in regulation of the structure and the position effect of telomeres. Mol Cell Biol. 1994;14:3791–3799. doi: 10.1128/mcb.14.6.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tommerup H, Dousmanis A, de Lange T. Unusual chromatin in human telomeres. Mol Cell Biol. 1994;14:5777–5785. doi: 10.1128/mcb.14.9.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 79.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 80.Walmsley R M, Chan C S M, Tye B-K, Petes T D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 81.Wang M M, Reed R R. Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature. 1993;364:121–126. doi: 10.1038/364121a0. [DOI] [PubMed] [Google Scholar]
- 82.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 83.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 84.Wiley E, Zakian V A. Extra telomeres, but not internal tracts of telomeric DNA, reduce transcriptional repression at Saccharomyces telomeres. Genetics. 1995;139:67–79. doi: 10.1093/genetics/139.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 86.Wright J H, Gottschling D E, Zakian V A. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 87.Wright J H, Zakian V A. Protein-DNA interactions in soluble telosomes from Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:1454–1460. doi: 10.1093/nar/23.9.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright W E, Tesmer V M, Huffman K E, Levene S D, Shay J W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zakian V A. Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]