Abstract
Background
CD molecules plays a vital role in gastric cancer (GC). We used bioinformatics analysis methods to develop prognosis related CD molecules risk signature; On the other hand, we used the experiments to further explore the function and mechanism of differentially expressed prognostic CD molecules (TREM1) in GC.
Methods
Kaplan-Meier survival and univariate Cox regression analysis were used to evaluate the overall survival of CD molecule genes in gastric cancer. ROC curve and Kaplan-Meier curves were used to analyze the predictive value of CD molecule related genes risk signature by “survival and timeROC” R packages. GSEA, and Cibersortx software were used to analyze the functional enrichment. Finally, we verified the function and mechanism of TREM1 in GC by gene silencing and MAPK inhibitor (SB203580) in vitro and vivo.
Results
A total of 41 prognosis related risk factors in gastric cancer were identified based on CD molecules, including TREM1 and ect. The high-risk patients had higher risk score and shorter survival time. ROC curves revealed that this risk signature accurately predicted survival times of gastric cancer patients at the 1-, 2-, 3-, 4- and 5-year. The frequency of T cells follicular helper and NK cells activated were added in low-risk group. Next, differentially expressed prognostic CD molecules analysis revealed that TREM1 was identified as key genes in GC progression based on TCGA and GES158662 and GSE15459 datasets of GC. In vitro experiments, TREM1 silencing significantly inhibited GC cell proliferation and migration, induced cell apoptosis. GSEA revealed that TREM1 activated cancer related signaling pathway, including MAPK signaling pathway and ect. High expression of TREM1 was related Macrophages M2 and Mast cells resting in GC tissues. Moreover, knockdown of TREM1 inhibited tumor growth through downregulated MAPK signaling pathway in vivo.
Conclusion
These results identified that CD molecule related genes as a novel prognostic and diagnostic biomarker in gastric cancer. TREM1 acts as an oncogene role in GC by activated MAPK signaling pathway.
Keywords: CD molecules, Gastric cancer, TREM1, Prognosis, MAPK signaling pathway
1. Introduction
Gastric cancer is one of gastrointestinal malignant tumor with high morbidity and mortality [1]. Due to bad living habits and Helicobacter pylori infection, there were one million new cases and seven hundred thousand deaths worldwide in 2020 [2]. Because there are no obvious symptoms in the early stage of gastric cancer, many patients are diagnosed with advanced gastric cancer. Despite continuous progress in molecular diagnostic techniques, surgical and adjuvant therapy techniques, the overall survival rate of gastric cancer patients is still poor [3]. The 5-year survival rate is less than 20% in patients with advanced gastric cancer [4]. Therefore, it is particularly important to find the key genes and therapeutic targets that affect the prognosis of gastric cancer.
CD molecules are glycoproteins and glycolipids of the cell membrane surface, which mediate its interaction with antigen, with other components of the immune system [5]. CD molecules were used as a surface marker for cell identification and isolation, which was widely involved in the cell growth, differentiation, migration and activation [6]. CD molecule not only participate in recognizing antigens, capturing antigens, and promoting the interaction between immune cells and antigens or immune molecules, but also mediate the adhesion between immune cells and immune cells, and between immune cells and the matrix, which plays an key role in immune response, activation and effector stages [7]. More and more studies have confirmed that CD molecules are closely related with the progression of cancers, including gastric cancer, and expected to be a potential biomarker for the diagnosis and prognosis of gastric cancer [8,9]. We used the data of patients with gastric cancer from the public databases the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) to explore the clinical significance and function of prognosis related CD molecules and differentially expressed CD molecules in between gastric cancer tissues with normal tissues. We found key genes in the progression of Gastric cancer by differentially expressed CD molecules analysis based on TCGA and GEO dataset and further explored and validated the gene that may be therapeutic targets.
Bioinformatics analysis has revealed that protein-coding gene triggering receptors expressed by myeloid cells-1 (TREM1) was a key gene in gastric cancer progression. Previous study has reported that TREM1 was upregulated in most cancers and positively associated with poor prognosis, immune response, pro-tumor pathways [10]. TREM1 silencing significantly inhibited invasion and migration of liver cancer cells by mediating macrophage polarization [11]. TREM1 expression was associated with persistent DNA damage, and inflammation [12]. However, the understanding of oncogenic roles of TREM1 in gastric cancer remains incomplete.
In this study, Kaplan-Meier survival and univariate Cox regression analysis was used to evaluate the overall survival of CD molecules in gastric cancer. We then further explored the role of immune cell infiltration in gastric cancer. We identified the survival-related CD molecules in the context of GC and develop a prognostic signature for gastric cancer patients. Moreover, we identified that TREM1 was key gene in GC progression by differentially expressed CD molecules analysis based on TCGA and GES158662 and GSE15459 datasets of GC. Finally, we verified the function and mechanism of TREM1 in gastric cancer by gene silencing and MAPK inhibitor (SB203580) in vitro and vivo.
2. Materials and methods
2.1. Collection of gastric cancer datasets
Clinical data, count and Fragments Per Kilobase of transcript per Million (FPKM) data of 375 gastric cancer patients were acquired from UCSC Xena. A total of 422 CD molecules related genes were retrieved from the KEGG pathways as targeted genes set. GSE158662 dataset was downloaded from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158662), including 3 normal tissues sample and 3 gastric cancer sample. Next, GSE103236 dataset was downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE103236), including 9 normal tissues sample and 10 gastric cancer; GSE15459 dataset was downloaded to verify the KM survival curve of CD36 and TREM1 high/low expression in gastric cancer patients, including 200 primary gastric tumors.
2.2. Prognosis related CD molecules genes
Kaplan-Meier survival and univariate Cox regression analysis was used to evaluate the overall survival of CD molecule in gastric cancer. The “glmnet” R package (Version 4.1.1) was used to LASSO regression to prevent model overfitting. LASSO regression analysis was used to obtain the CD molecules related prognostic signature and the corresponding risk coefficients based on multivariate Cox regression analysis. The risk scores of gastric cancer patients were calculated by the regression coefficients of genes and their corresponding mRNA expressions. The patients with GC were divided into high-risk group and low-risk group based on the cutoff value of the median value of risk scores.
Kaplan-Meier curves was analyzed by using the “survival” R packages (Version 3.2.10). We used the “timeROC” R packages (1.18.0) to create a time dependent ROC curve to assess the risk score's efficacy in predicting the 1, 2, 3, 4 and 5-year survival of gastric cancer patients.
2.3. Cibersort analysis
The CIBERSORT [13] was used to analyze the abundance of 22 tumor immune infiltrating cell types in the tumor immune microenvironment (TME) of gastric cancer patient samples. The heatmap was used to show the differential abundance of 22 immune infiltrating cell. Wilcoxon test was used to analyze the statistical differences of high and low risk group.
2.4. Immune checkpoint molecules expression
The potential immunotherapeutic markers including 18 ICB-related genes were explored in high- and low risk groups by Wilcoxon test. The heatmap of ICB-related genes expression was drawn by R package of “pheatmap” (version 4.0.4).
2.5. The expression analysis of OS related CD molecule genes
The differential expression of overall survival (OS) related CD molecule genes in gastric cancer tissues and normal tissues were analyzed based log 2 |fold change| ≥1, P < 0.05.
2.6. Copy number alteration (CNA)
The copy number alteration frequency and type of OS related CD molecule genes were analyzed by cBioPortal online web (http://www.cbioportal.org/).
2.7. Gene set enrichment analysis (GSEA)
The genomic data of gastric patients in the TCGA database were divided into high expression of TREM1 group and low expression of TREM1 group. The hallmark and KEGG gene set was used for the enrichment analysis in GSEA v 4.1.0 software.
2.8. Cell culture
The gastric mucosal epithelial cell line (GES-1) and GC cell lines (AGC and HGC27) were obtained from the Global Bioresource Center (ATCC, USA). The cells were cultured in RPMI-1640 medium (Gibco, USA) added with 10% fetal bovine serum (FBS), 100 μg/mL streptomycin and 100 U/mL penicillin (Gibco, USA) and maintained in a humidified atmosphere of 5% CO2 at 37 °C.
2.9. Cell transfection
The shRNA sequences of TREM1 were purchased from Hangzhou guannan Biotechnology Co., LTD. For cell transfection, AGC and HGC27 were transfected with TREM1 shRNA using Lipofectamine™ 3000 (Invitrogen, USA) according to the manufacturer's instructions to establish stable cell lines. Transfection efficiency was detected by RT-qPCR assay. We used shRNA sequence targeting TREM1 (shRNA-1: 5′-CACCGGGTTCCGGTGTTCAACATTGCGAACAATGTTGAACACCGGAACCC-3′; shRNA-2: 5′-CACCGGTGTTCAACATTGTCATTCTCGAAAGAATGACAATGTTGAACACC-3′; and shRNA-3: 5′-CACCGGTCTTCTCTGTCCTGTTTGCCGAAGCAAACAGGACAGAGAAGACC-3′).
2.10. RT-qPCR assay
Total RNA was isolated by using TRIzol reagent (Thermo Fisher, Cat. #15596026) from GES-1, AGC and HGC27 cell lines. Total RNA was synthesized to cDNA by reverse transcription kit (Vazyme, Cat. #R211-01). The cDNA was amplified by RT‒qPCR using AceQ Universal SYBR qPCR Master Mix (Cat. No. A25742; Thermo Fisher Co., Ltd.) on an ABI 7500 PCR system (Thermo Fisher Scientific, Inc.). The cycling parameters used were 95 °C for 15s, 55–60 °C for 15s, and 72 °C for 15s for 45 cycles. Ct values were determined during the exponential amplification phase of real time PCR. The 2−△△Ct method was need to calculate relative expression levels in GC cell lines. Each experiment was performed in triplicate. The primer sequences were as follows:
TREM1, forward: 5′-GAACTCCGAGCTGCAACTAAA-3′;
TREM1, reverse:5′-TCTAGCGTGTAGTCACATTTCAC-3′
GAPDH, forward: 5′-CACAAGCAGAGTGCTGAAGGTG-3′;
GAPDH, forward: 5′-ACCACCCTGTTGCTGTAGCCAA-3′.
2.11. Cell proliferation assay
Cell counting kit-8(Cat. #GK10001-1) was used to detect the cell proliferation after cell transfection. In brief, logarithmic growth cells (1 × 105) were seeded into 96-well cell culture plates. 10 μL of CCK-8 solution was added at 0, 24, 48 and 72 h. The absorbance of each well was determined by enzyme label detection at 450 nm (Molecular Devices, USA). All assays were repeated at least three times.
2.12. Flow cytometric assay
Cell apoptosis was detected by using Annexin V- FITC/PI apoptosis detection kit. In brief, 500 μLbinding buffer was added the cells, and 5 μL Annexin V-FITC apoptosis detection kit (cat.no.556547; BD Biosciences) and 5 μL PI staining solution (cat. No. 556547; BD Biosciences) were added and stained the cells in the dark for 15 min. Next, the cell apoptotic rate was analyzed by flow cytometer (Becton Dickinson, USA). All assays were repeated at least three times. The cell apoptosis was analyzed by using FloJo software (BD Biosciences, USA).
2.13. Cell migration assay
Cell migration was analyzed by using 24-well transwell chambers (Cat. #3433). For migration assays, 1 × 105 cells were plated in the top chamber. After 24 h of culture, the chamber was token out and fixed with 10% neutral formalin. Wiping off the cells that do not pass through the chamber membrane with a cotton swab. The invasion cells were stained with crystal violet for 20 min. The migration cells were photographed under the microscope from randomly select four visual fields.
2.14. Tumor xenograft model
A total of 15 male athymic mice (BALB/c-nu/nu; 6 weeks old; 20–25 g) were purchased from shanghai Southern Model Biotechnology Co., LTD (2022-00). The Animal experiment protocol listed below has been reviewed and approved by Laboratory animal management ethics committee of ZPPH (Approval No. 20230514170023672211). Mice were divided into three group, including TREM1 shRNA group, control group, and TREM1 shRNA + MAPK inhibitor (SB203580) group. Each mouse was subcutaneously inoculated with AGS cell from differential treat. The tumor volume was sequentially recorded 0 days, 3 days, 7 days, 10 days, 13 days. 15 days, 18 days, and 21 days post administration. All experimental animals were killed by inhaling carbon monoxide gas. The tumor tissues were collected and tumor weight was measured.
2.15. Statistical analysis
SPSS 19.0 (IBM Corporation, Armonk, NY, USA) was used for statical analysis. The dates are expressed as means ± standard deviation. One way ANOVA test was used to compare the mean between multiple samples, The Student's t-test was used to compare the mean between two samples. P < 0.05 was considered statical significance.
3. Results
3.1. Identification of prognosis related risk factors in gastric cancer based on CD molecules
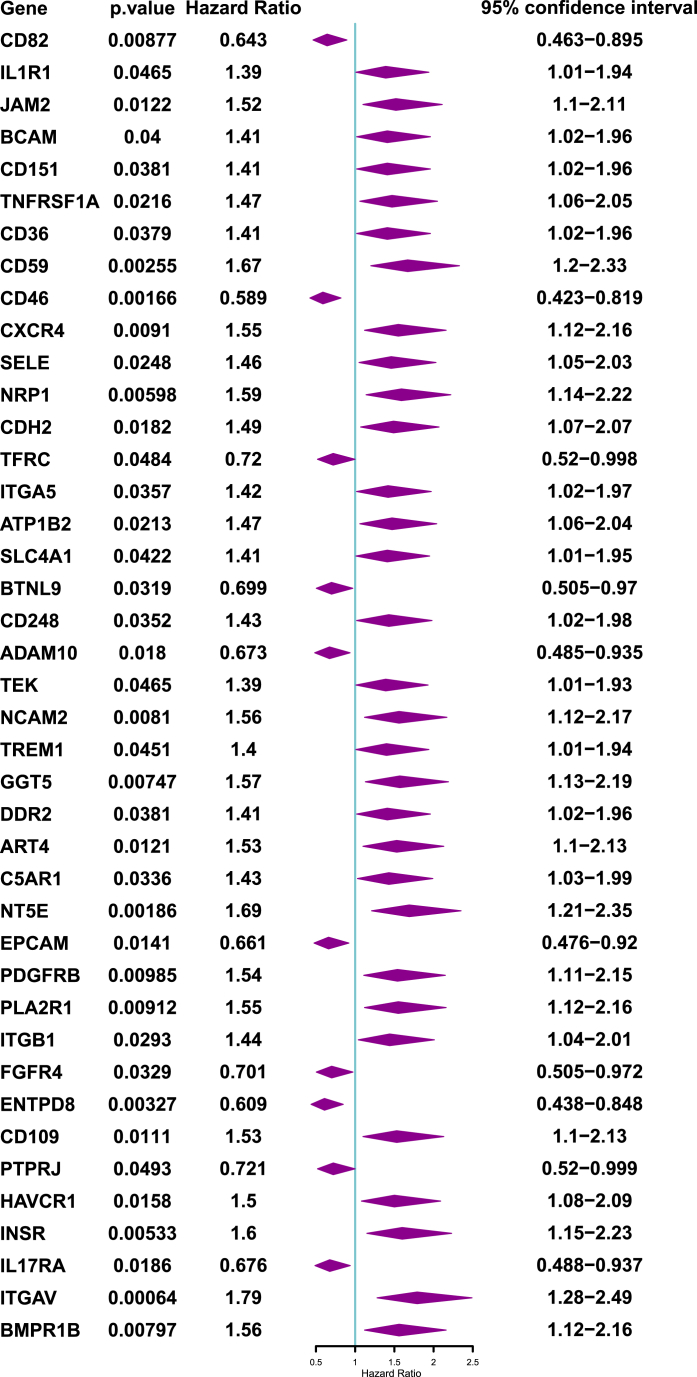
The RNA-seq data of the 422 CD molecules related genes was downloaded to explore the overall survival associated CD molecules by Kaplan-Meier curves analysis and univariate Cox regression analysis. A total of 41 prognosis related CD molecules were acquired from 422 CD molecules in gastric cancer (Supplemental Fig. 1). Among, the high expression of TREM1, TNFRSF1A, SLC4A1, TEK, BCAM, PLA2R1, PDGFRB, ATP1B2, ART4, HAVCR1, SELE, CDH2, IL1R1, CD248, INSR, NT5E, CXCR4, NCAM2, NRP1, JAM2, CD59, ITGB1, CD109, BMPR1B, GGT5, ITGAV, CD36, ITGA5, C5AR1, CD151, DDR2, and FGFR4 have shorter survival time than low expression group in gastric cancer (Supplemental Fig. 1A), the low expression of PTPRJ, BTNL9, CD46, ADAM10, ENTPD8, TFRC, IL17RA, EPCAM, CD82 group with gastric cancer patients have shorter survival time than high expression group (Supplemental Fig. 1B). Next, poor prognosis related risk factors were evaluated in gastric cancer by univariate Cox regression analysis from prognosis related CD molecules. As shown in Fig. 1, the overall survival univariate forest plot shown that CD82, CD46, TFRC, BTNL9, ADAM10, EPCAM, FGFR4, ENTPD8, PTPRJ, and IL17RA were protective factors for the poor prognosis of gastric cancer patients, IL1R1, JAM2, BCAM, CD151, TNFRSF1A, CD36, CD59, CXCR4, SELE, NRP1, CDH2, ITGA5, ATP1B2, SLC4A1, CD248, TEK, NCAM2, TREM1, GGT5, DDR2, ART4, C5AR1, NT5E, PLA2R1, ITGB1, CD109, HAVCR1, INSR, ITGAW and BMPR1B were harmful factors for the poor prognosis of gastric cancer patients. These CD molecules genes were risk factors affecting the prognosis of gastric cancer.
Fig. 1.
Univariate Cox regression analyses of CD molecules influencing overall survival in gastric cancer.
3.2. Construction and function analysis of CD molecules genes related prognostic model in gastric cancer
A total of 41 CD prognosis related genes were used for LASSO Cox regression. We used LASSO regression analysis to build a CD molecule related prognostic model (Supplemental Fig. 2A). By using multivariate Cox proportional HR analysis, ten CD molecules were found and used to create prognostic signature for patients OS (Supplemental Fig. 2B), including four low risk genes (IL17RA, PTPRJ, CD82 and ITGA5) and six high risk genes (CD151, ART4, CD59, PLA2R1 and NRP1) (Supplemental Fig. 2C). Each patient's risk value was computed and ranked in increasing order. The patients of the low-risk group was longer survival time. Next, the heat map showed that the expression of ten CD molecules, IL17RA, PTPRJ, CD82 and ITGA5 were high-expressed in the low-risk group, and CD161, ART4, CD59, PLA2R1 and NRP1 were high-expressed in the high-risk group than low-risk group (Supplemental Fig. 2D). To explore the predictive value of the signature, the Kaplan-Meier survival curves were analyzed. GC patients of high -risk group has shorter OS (Supplemental Fig. 2E). ROC analysis confirmed that the area under curve (AUC) was 0.649 at 1 year; 0.641 at 2 years, 0.717 at 3 years, 0.71 at 4 years, 0.734 at 5 years, respectively (Supplemental Fig. 2F), AUC values were more than 0.5 regardless of the predicted survival time at 1, 2, 3, 4, 5-year survival in the training set.
3.3. Comparison of the immune microenvironment and the immune checkpoint molecules expression of prognosis related CD molecular genes
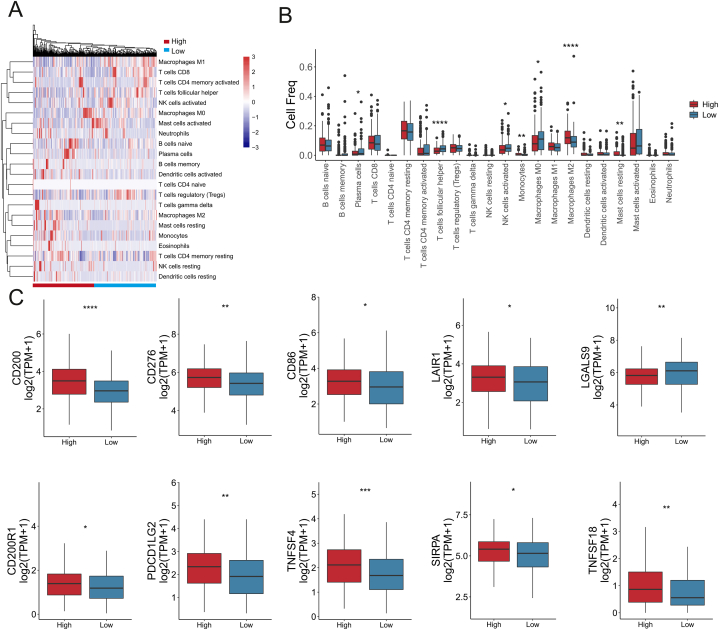
We employed CIBERSORT to analyze the difference of 22 immune cells in between high risk and low risk groups for each gastric cancer tissues. The heatmap of the tumor infiltrating immune cells expression was showed in high/low risk groups, red indicated high risk group and blue indicated low risk group (Fig. 2A). The composition of TIICs remained basically the same, mainly composed of T cells CD4 memory, Dendritic cells (DC), NK cells, Macrophages, and Mast cells. Wilcoxon test analysis showed Plasma cells, T cells follicular helper, NK cells activated, Macrophages M0, was higher infiltration, and Monocytes, Macrophages M2, and Mast cells resting were lower infiltration in the low-risk group than high-risk group (Fig. 2B).
Fig. 2.
The landscape of TME. (A) The heatmap of tumor infiltrating immune cells expression. Red indicates high risk group; Blue indicates low risk group. (B) Differentially analysis of 22 tumor infiltrating immune cells by Wilcoxon test analysis. (C) The expression of immune checkpoint molecules in low- and high -risk groups of gastric cancer patients. (*P < 0.05; **P < 0.01; ***P < 0.001).
Immune blocking checkpoint (ICB) related gene expression levels were correlated with therapeutic response of immune checkpoint inhibitors and targeted ICB checkpoints has emerged as promising strategy in cancers treatment [14,15]. To better explore the potential of CD molecules for predicting the response of gastric cancer patients to immunotherapy, we analyzed the expression of immune checkpoint molecules in low-and high-groups of gastric cancer. As shown in Fig. 2C, the expression of immunomodulators (CD200, CD276, CD86, LAIR1, CD200R1, PDCD1LG2, TNFSF4, SIRPA, and TNFSF18) were significantly reduced in low-risk group than high risk group of gastric cancer, the expression of LGALS9 was significantly increased in low-risk group. These results suggested that GC patients with high risk may benefit less from available immune blockade therapy.
3.4. The differential expression and copy number alteration of prognosis related CD molecules in gastric cancer
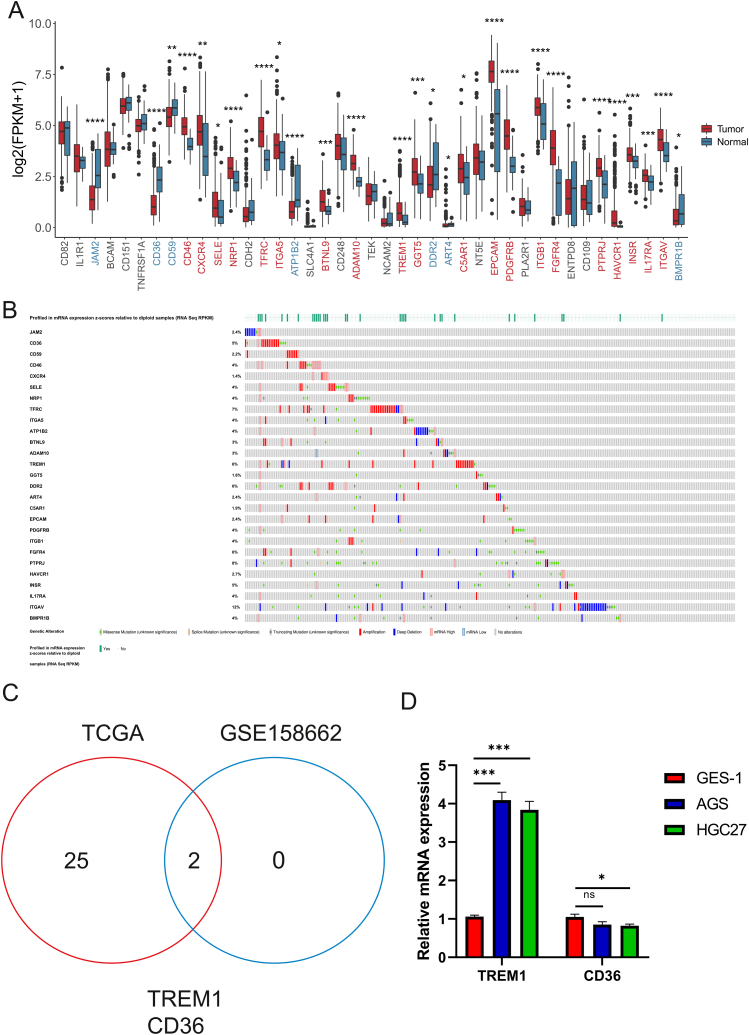
Next, we analyzed that the expression of 41 CD molecules genes in gastric cancer tissues and normal tissues. The expression of JAM2, CD36, CD59, ATP1B2, and ART4 were significantly downregulated in gastric cancer tissues compared with normal group, the expression level of CD46, CXCR4, SELE, NRP1, TFRC, ITGA5, BTNL6, ADAM10, TREM1, GGT5, C5AR1, EPCAM, PDGFRB, ITGB1, FGFR4, PTPRJ, HAVCR1, INSR, IL17RA, and ITGAV were significantly upregulated in gastric cancer tissues compared with normal group (Fig. 3A). cBioPortal shown that the CNA of CD molecules genes in gastric cancer (Fig. 3B). The amplification and CNA frequency of TREM1 (6%), TFRC (7%), FGFR4 (6%), PTPRJ (8%), ITGAV (12%) are significantly upregulated in tumor samples. To identify key candidate CD molecules involved in GC progression, we analyzed the GEO GSE158662 (Supplementary Fig. 3A) and GSE103236 (Supplementary Fig. 3B) gastric cancer RNA-seq datasets and screened 2 CD molecules that were significantly differentially expressed in GC compared with normal cancer. The expression of CD36 was significantly decreased in gastric cancer, and the expression of TREM1 was significantly increased in gastric cancer (Supplementary Figs. 3A and 3B). Venn diagram showed that a total of 2 CD molecules were differentially expressed in between gastric cancer tissues and normal tissues based on TCGA dataset and GEO of gastric cancer (Fig. 3C). Moreover, the survival time of CD36 high/low expression no significant difference in gastric cancer patients (Supplementary Fig. 3C). KM survival analysis of TREM1 high/low expression confirmed that the survival time of TREM1 high expression significantly shorter than TREM1 low expression in gastric patients (Supplementary Fig. 3D). Further, we detected the expression of TREM1 and CD36 in gastric mucosal epithelial cells (GES-1), and gastric cancer cells (AGS, HGC27). As shown in Fig. 3D, TREM1 expression was significantly upregulated in AGS and HGC27 cells lines compared with GES-1 cell, CD36 expression was significantly downregulated in HGC27 and no significantly downregulated in AGS compared with GES-1 cell. Therefore, we further explored the role of TREM1 by using the functional experiments of vivo and vitro based on CD molecules expression and prognosis analysis.
Fig. 3.
The expression and CNA of CD molecules genes in GC. (A) The differentially expressed CD molecule genes in tumor compared with normal group based on the GC-TCGA dataset, Red font indicates upregulated genes, Blue font indicates downregulated genes (*P < 0.05; **P < 0.01; ***P < 0.001). (B) copy number alteration frequency of CD molecule genes in gastric cancer. (C) Veen diagram shows total of 2 differentially expressed CD molecule genes in between GC tissue with normal tissues base on TCGA and GSE158662 datasets. (D) The mRNA expression of TREM1 and CD36 was detected in GES-1, AGS, HGC27 cells lines (*P < 0.05; ***P < 0.001).
3.5. Knockdown of TREM1 inhibited cell proliferation, migration and induced cell apoptosis in GC cells
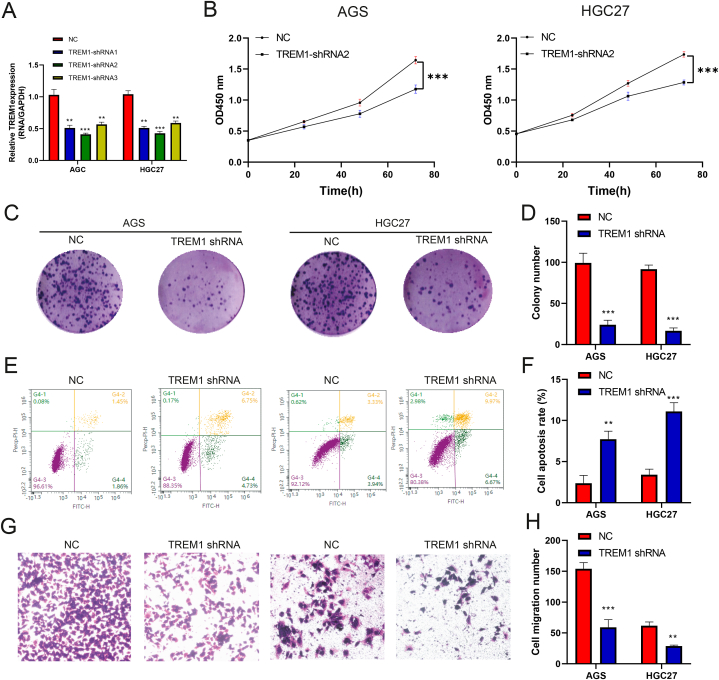
The role of TREM1 in GC cells is still unclear. We investigated the role of TREM1 in GC cells. First, RT-qPCR assays showed that TREM1 expression was significantly reduced in TREM1 shRNA group (Fig. 4A). Knockdown of TREM1 inhibited the proliferation of AGS and HGC27 cell lines, as shown by the CCK-8 assay (Fig. 4B). Colon formation assay results shown that knockdown of TREM1 decreased the cell growth number of AGS and HGC27 cell lines (Fig. 4C and D). Next, we analyzed the effect of TREM1 on the apoptosis of AGS and HGC27 cell lines using flow cytometry. We found that knockdown of TREM1 increased the cell apoptotic rates (Fig. 4E and F). In addition, transwell assay revealed that cell migration was reduced in the TREM1 shRNA group compared with the control group (Fig. 4G and H). Collectively, these results suggested that knockdown of TREM1 inhibited cell proliferation, migration and promoted cell apoptosis in GC cells.
Fig. 4.
Effects of TREM1 on the proliferation, migration and apoptosis of AGS and HGC27 cells. (A) TREM1 expression was analyzed by RT-qPCR assay. (B) CCK-8 assay detected the AGS and HGC27 cells proliferation. (C) Colon formation assay was used to detected cell growth number of AGS and HGC27 cells. (D) colony number was calculated. (E) Flow cytometry assay was used to detect the apoptosis of AGS and HGC27 cell lines. (F) Cell apoptosis rate was calculated. (G) Transwell assay was performed to explore the migration of AGS and HGC27 cell lines. (H) Cell migration number were calculated. Data are represented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
3.6. MAPK pathway was activated by TREM1 in gastric cancer
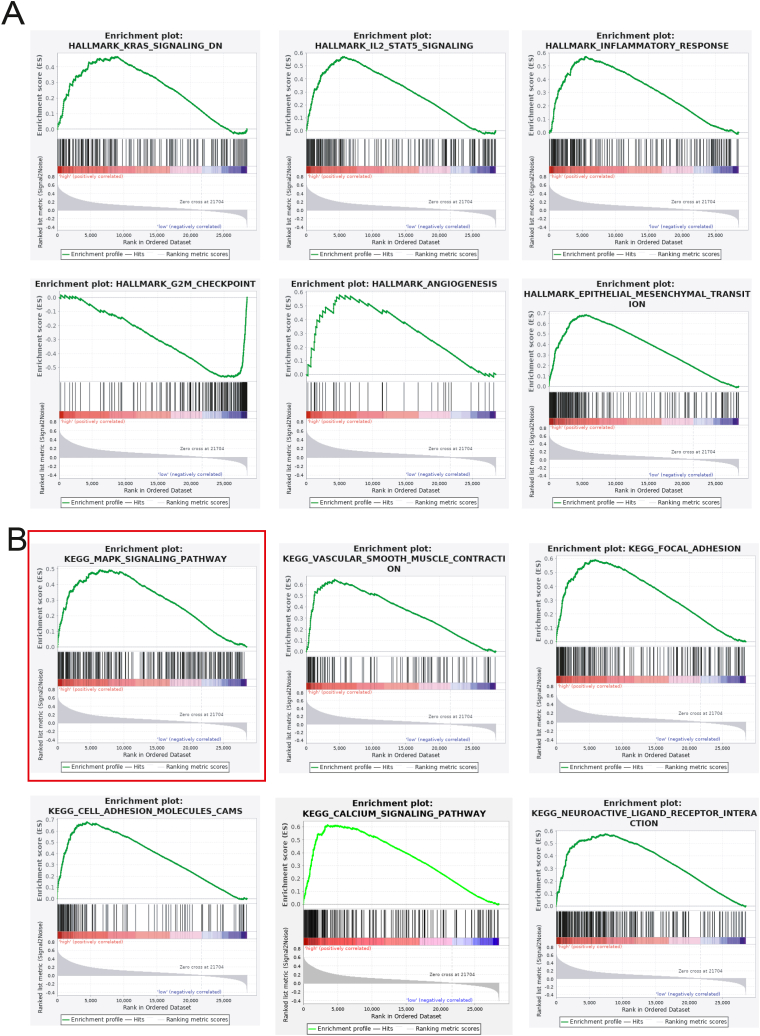
To further investigate the potential mechanism of TREM1 in gastric cancer, Gene set enrichment analysis was performed to explore the involved Hallmark and KEGG functional pathways of high/low expression of TREM1. The hallmark of the TREM1 high expression group is shown in Fig. 5A, including HALLMARK KRAS signaling, IL2-STAT5 signaling, Inflammatory response, G2M checkpoint, Angiogenesis, Epithelial mesenchymal transition. The KEGG pathways of the high expression of TREM1 are shown in Fig. 5B, including MARK signaling pathway, Vascular smooth muscle contraction, Focal adhesion, cell adhesion molecules, Calcium signaling pathway, Neuroactive ligand receptor interaction. High expression of TREM1 significantly activated MARK signaling pathway in gastric cancer by GSEA analysis. Mitogen-activated protein kinase (MAPK) signaling pathway is the basic pathway in mammalian cells, which is closely related to cell proliferation, differentiation, apoptosis, angiogenesis and other physiological activities. The abnormal activation of some proteins in the MAPK pathway is an important cause of various cancers including gastric cancer. Therefore, intervention of this pathway can be used as one of the strategies for tumor treatment.
Fig. 5.
Gene set enrichment analysis. (A) The HALLMARK pathway enrichment of high/low expression of TREM1 in gastric cancer. (B) The KEGG pathway enrichment of high/low expression of TREM1 in gastric cancer.
Immune cells in the tumor microenvironment (TME) play an essential role in tumor progression. We used Cibersort to analyze the relationship between TREM1 expression and immune cell infiltration. We found that low expression of TREM1 was related with plasma cells, T cells follicular helper, NK cells activated, Macrophages M0 immune cell infiltration; high expression of TREM1 was related with Monocytes, Macrophages M2, Mast cells resting immune cell infiltration (Supplementary Fig. 4).
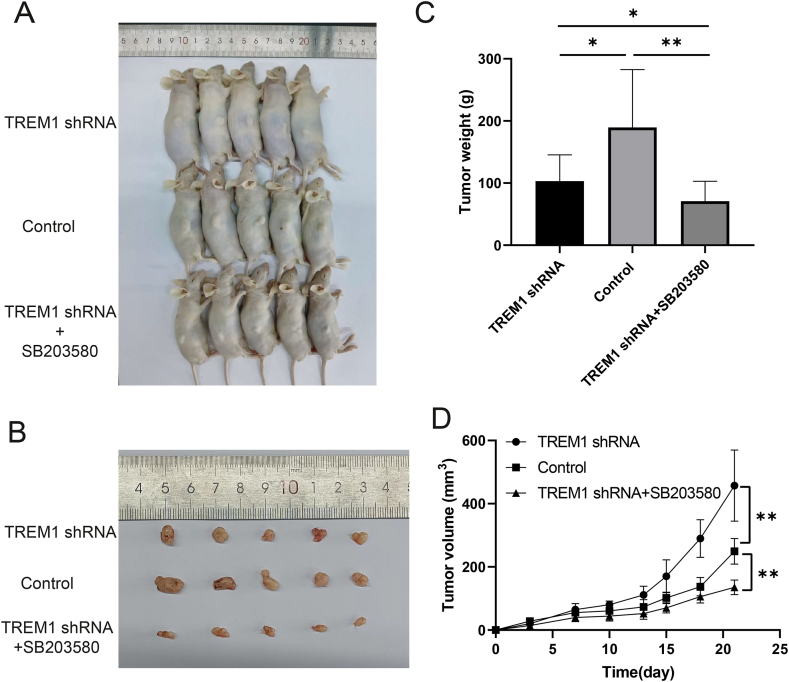
3.7. TREM1 inhibited tumor growth through downregulated MAPK signaling pathway in vivo
To understand the importance of the MAPK pathway in TREM1 medicated gastric cancer, we performed joint experiments using the MAPK inhibitor (SB2023580) + TREM1 shRNA in xenograft mouse models. As shown in Fig. 6A and B, the TREM1 shRNA group significantly suppressed tumor growth compared with control group, TREM1 shRNA + SB2023580 significantly inhibited tumor growth compared with TREM1 shRNA group and Control group, including tumor weight (Fig. 6C) and tumor volume (Fig. 6D). We further confirmed that MAPK signaling is responsible for the carcinogenic function of TREM1.
Fig. 6.
TREM1 silencing inhibited tumor growth and tumor volume by downregulated MAPK signaling pathway. (A and B) Tumor growth of TREM1 shRNA, Control, TREM1 shRNA + SB203580. (C) Tumor weight was measured. (D) Tumor volume was measured at 0, 3, 5, 7, 10, 13, 15, 18, 21 days. Data are presented as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
4. Discussion
CD molecules play an crucial role in the mechanism of tumor immune response, which involved in the occurrence of inflammation and the development and metastasis of tumor cells [16]. However, the value of CD molecules in evaluating the clinical prognosis of gastric cancer patients has not been reported. Kaplan-Meier curves analysis and univariate Cox regression analysis revealed that a total of 38 overall survival associated CD molecule genes in gastric cancer. Among, CD82, CD46, TFRC, BTNL9, ADAM10, EPCAM, FGFR4, ENTPD8, PTPRJ, and IL17RA were a protective factor for the poor prognosis of gastric cancer patients, IL1R1, JAM2, BCAM, CD151, TNFRSF1A, CD36, CD59, CXCR4, SELE, NRP1, CDH2, ITGA5, ATP1B2, SLC4A1, CD248, TEK, NCAM2, TREM1, GGT5, DDR2, ART4, C5AR1, NT5E, PLA2R1, ITGB1, CD109, HAVCR1, INSR, ITGAW and BMPR1B were harmful factors for the poor prognosis of gastric cancer patients.
NRP1 expression was upregulated in gastric cancer tissues and associated with the poor overall survival [17]. Low expression of PLA2R1 was associated with poor disease-free survival, upregulation of PLA2R1 inhibited thyroid cancer cell proliferation, invasion and migration by inhibiting the activation of the Wnt/β-catenin pathway [18]. EGFR/Wnt/β-catenin increased the CD55/CD59 expression play a key role in suppressing complement and CD8+T cell activation to achieve tumor immune escape and immune checkpoint blockade resistance [19]. CD151 was high expressed and associated with poor prognosis, high TNM stage, depth of invasion in GC patients [20]. High expression of CD73 (Ecto-5′-nucleotidase (NT5E)) was an independent predictor for poor prognostic factor for gastric cancer patients [21]. Integrin subunit alpha 5 (ITGA5) high expression was associated with poor prognosis and immune infiltration of gastrointestinal tumors patients [22]. High expression of IL-17RA was associated with GC patient's overall survival [23]. These CD molecules genes were independent risk factors for poor OS in gastric cancer.
Tumor microenvironment (TME) play a critical effect on tumors incidence and development [24]. CD molecules risk score is significantly correlated with the immune microenvironment of gastric cancer [25]. The expressed glycoproteins in immune infiltrates surface can be used as biomarkers for immune cells classification. CD molecules also affect the pro-tumor or anti-tumor activity of immune cells [26]. T cells follicular helper, NK cells activated, Macrophages M0, was higher infiltration, and Monocytes, Macrophages M2, and Mast cells resting in the low group were lower infiltration than high-risk group. More importantly, some scholars have proposed that targeted treatment of CD molecules can effectively improve the microenvironment of immunosuppression, which may provide a new window for tumor immunotherapy [27].
Next, TREM1 was identified differentially expressed Key CD molecules in between GC tissues with normal tissues. TREM1 is a member of the lg-like immunoregulatory receptor family and a major amplifier of innate immune responses [28]. TREM1 is clinically valuable diagnostic and prognostic biomarker in cancer [29]. Previous study has reported that downregulation of TREM1 suppressed invasion and migration of liver cancer cells by mediating macrophage polarization [11]. In this study, we found that TREM1 was upregulated in gastric cancer tissues compared with normal tissues, and high expression of TREM1 was associated with poor prognosis of gastric cancer patients. The amplification and CAN frequency of TREM1 was 6% in gastric cancer patients. In vitro, TREM1 silencing inhibited cell proliferation, migration and induced cell apoptosis in gastric cancer cells.
We used GSEA analysis to investigate the possible molecular pathways of TREM1 to better explore the underlying biological process. HALLMARK pathways revealed that TREM1 involved in cancer related pathway, including KRAS signaling [30], IL2-STAT5 signaling [31], Inflammatory response [32], G2M checkpoint [33], Angiogenesis and epithelial mesenchymal transition [34]. KEGG enrichment results revealed that TREM1 activated MAPK signaling pathway [35], Vascular smooth muscle contraction [36], focal adhesion [37], adhesion molecules, Calcium signaling pathway [38], and neuroactive ligand receptor interaction. Tumor associated cellular malignant phenotypes and pathways were significant enriched in high expression of TREM1 of gastric cancer. MAPK pathways plays an important role in the regulation of tumor cell proliferation and differentiation, and was closely related to cell apoptosis [39,40]. MAPK was highly expressed in gastric cancer, and could play a role in the occurrence and metastasis of gastric cancer [35]. It has the function of oncogene in gastric cancer [41]. Inhibition of MAPK pathway reduced the proliferation and metastasis of gastric cancer cells [42]. In addition, clinical anti-gastric cancer drugs such as cisplatin and gemcitabine induced apoptosis of cancer cells by MAPK dependent manner [43]. We found that TREM1 significantly activated MAPK signaling pathway in gastric cancer. We further explored that TREM1 silencing + SB203580 significantly inhibited tumor growth and tumor volume in vivo mice model. Therefore, TREM1 silencing combined with MAPK inhibitor (SB203580) provides a theoretical basis for development of new anti-tumor methods.
Our findings establish a novel prognosis signature based on survival related CD molecules in GC. These results identified that CD molecule related genes as a novel prognostic and diagnostic biomarker in gastric cancer. Further, we identified that TREM1 was key genes in GC progression. TREM1 silencing significantly inhibited cell proliferation, migration and induced cell apoptosis in GC cells. TREM1 acts as an oncogene role in GC by activated MAPK signaling pathway.
Our study had some limitations, we did not elucidate the molecular mechanism of TREM1 activates MAPK signaling pathway. We have not studied how the abnormal expression of TREM1 in GC. It is worthwhile to further investigate TREM1 upstream regulate factor in GC. We next explored that the molecular mechanism of TREM1 activated MAPK signaling pathway. The role of TREM1 in GC still needs to be further explored.
5. Conclusion
Prognosis related CD molecules genes were identified and associated with immune microenvironment and immune checkpoint molecules expression of gastric cancer, which may help to reveal the pathogenesis of gastric cancer and provide new ideas for its diagnosis and treatment. Importantly, TREM1 upregulation was associated with poor prognosis of gastric cancer patients. TREM1 promoted cell proliferation, migration and tumor growth in gastric cancer cells through activated MAPK signaling pathway.
Ethics declarations statements
The Animal experiment protocol listed below has been reviewed and approved by Laboratory animal management ethics committee of ZPPH (Approval No. 20230514170023672211).
Funding
This research was supported by The Health Bureau of Zhejiang province (No. 2022KY501 and No. 2024KY026).
Data availability statement
GSE158662, GSE103236, GSE15459 dataset was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/). Data will be made available on request.
CRediT authorship contribution statement
Long Chen: Writing – original draft, Conceptualization. Fen Huang: Validation, Formal analysis, Data curation. Xiaopan Luo: Visualization, Methodology, Investigation. Zan Chen: Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not application.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e26852.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Machlowska J., et al. Gastric cancer: epidemiology, risk factors, classification, genomic characteristics and treatment strategies. Int. J. Mol. Sci. 2020;21(11) doi: 10.3390/ijms21114012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smyth E.C., et al. Gastric cancer. Lancet. 2020;396(10251):635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 3.Xu Y., et al. The application of CA72-4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim. Biophys. Acta Rev. Canc. 2021;1876(2) doi: 10.1016/j.bbcan.2021.188634. [DOI] [PubMed] [Google Scholar]
- 4.Sawaki K., Kanda M., Kodera Y. Review of recent efforts to discover biomarkers for early detection, monitoring, prognosis, and prediction of treatment responses of patients with gastric cancer. Expet Rev. Gastroenterol. Hepatol. 2018;12(7):657–670. doi: 10.1080/17474124.2018.1489233. [DOI] [PubMed] [Google Scholar]
- 5.Zola H. Medical applications of leukocyte surface molecules--the CD molecules. Mol. Med. 2006;12(11–12):312–316. doi: 10.2119/2006-00081.Zola. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimura K., et al. Combined inhibition of PD-1/PD-L1, Lag-3, and Tim-3 axes augments antitumor immunity in gastric cancer-T cell coculture models. Gastric Cancer. 2021;24(3):611–623. doi: 10.1007/s10120-020-01151-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wattanawongdon W., Bathpho T.S., Tongtawee T. Co-expression of LGR5 and CD133 cancer stem cell predicts a poor prognosis in patients with gastric cancer. Turk. J. Gastroenterol. 2021;32(3):261–268. doi: 10.5152/tjg.2021.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv K., et al. Lymphocyte-activation gene 3 expression associates with poor prognosis and immunoevasive contexture in Epstein-Barr virus-positive and MLH1-defective gastric cancer patients. Int. J. Cancer. 2021;148(3):759–768. doi: 10.1002/ijc.33358. [DOI] [PubMed] [Google Scholar]
- 9.Yiming L., et al. CD133 overexpression correlates with clinicopathological features of gastric cancer patients and its impact on survival: a systematic review and meta-analysis. Oncotarget. 2015;6(39):42019–42027. doi: 10.18632/oncotarget.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X., et al. Overexpression of TREM1 is associated with the immune-suppressive microenvironment and unfavorable prognosis in pan-cancer. J. Inflamm. Res. 2023;16:1375–1391. doi: 10.2147/JIR.S398284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen M., et al. Downregulation of triggering receptor expressed on myeloid cells 1 inhibits invasion and migration of liver cancer cells by mediating macrophage polarization. Oncol. Rep. 2021;45(4) doi: 10.3892/or.2021.7988. [DOI] [PubMed] [Google Scholar]
- 12.Li X., et al. Persistent DNA damage and oncogenic stress-induced Trem1 promotes leukemia in mice. Haematologica. 2022;107(11):2576–2588. doi: 10.3324/haematol.2021.280404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman A.M., et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman A., Patel S.P., Kurzrock R. PD-1-PD-L1 immune-checkpoint blockade in B-cell lymphomas. Nat. Rev. Clin. Oncol. 2017;14(4):203–220. doi: 10.1038/nrclinonc.2016.168. [DOI] [PubMed] [Google Scholar]
- 15.Postow M.A., Callahan M.K., Wolchok J.D. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 2015;33(17):1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin H.S., Park Y. Hitting the complexity of the TIGIT-CD96-CD112R-CD226 axis for next-generation cancer immunotherapy. BMB Rep. 2021;54(1):2–11. doi: 10.5483/BMBRep.2021.54.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G., et al. Hypomethylated gene NRP1 is co-expressed with PDGFRB and associated with poor overall survival in gastric cancer patients. Biomed. Pharmacother. 2019;111:1334–1341. doi: 10.1016/j.biopha.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Xu Q., et al. Clinical importance of PLA2R1 and RASSF9 in thyroid cancer and their inhibitory roles on the Wnt/beta-catenin pathway and thyroid cancer cell malignant behaviors. Pathol. Res. Pract. 2022;238 doi: 10.1016/j.prp.2022.154092. [DOI] [PubMed] [Google Scholar]
- 19.Shao F., et al. Silencing EGFR-upregulated expression of CD55 and CD59 activates the complement system and sensitizes lung cancer to checkpoint blockade. Nat. Cancer. 2022;3(10):1192–1210. doi: 10.1038/s43018-022-00444-4. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y.M., et al. Overexpression of CD151 predicts prognosis in patients with resected gastric cancer. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0058990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu S., et al. NT5E is associated with unfavorable prognosis and regulates cell proliferation and motility in gastric cancer. Biosci. Rep. 2019;39(5) doi: 10.1042/BSR20190101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H., et al. ITGA5 is a prognostic biomarker and correlated with immune infiltration in gastrointestinal tumors. BMC Cancer. 2021;21(1):269. doi: 10.1186/s12885-021-07996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y.X., et al. Increased chemokine receptor IL-17RA expression is associated with poor survival in gastric cancer patients. Int. J. Clin. Exp. Pathol. 2015;8(6):7002–7008. [PMC free article] [PubMed] [Google Scholar]
- 24.Hinshaw D.C., Shevde L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wo Y.J., et al. The roles of CD38 and CD157 in the solid tumor microenvironment and cancer immunotherapy. Cells. 2019;9(1) doi: 10.3390/cells9010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roma-Rodrigues C., et al. Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 2019;20(4) doi: 10.3390/ijms20040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu L., et al. Assessment of the expression of the immune checkpoint molecules PD-1, CTLA4, TIM-3 and LAG-3 across different cancers in relation to treatment response, tumor-infiltrating immune cells and survival. Int. J. Cancer. 2020;147(2):423–439. doi: 10.1002/ijc.32785. [DOI] [PubMed] [Google Scholar]
- 28.Bosco M.C., Raggi F., Varesio L. Therapeutic potential of targeting TREM-1 in inflammatory diseases and cancer. Curr. Pharmaceut. Des. 2016;22(41):6209–6233. doi: 10.2174/1381612822666160826110539. [DOI] [PubMed] [Google Scholar]
- 29.Cioni B., et al. Androgen receptor signalling in macrophages promotes TREM-1-mediated prostate cancer cell line migration and invasion. Nat. Commun. 2020;11(1):4498. doi: 10.1038/s41467-020-18313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buscail L., Bournet B., Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17(3):153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 31.Lutz V., et al. IL18 receptor signaling regulates tumor-reactive CD8+ T-cell exhaustion via activation of the IL2/STAT5/mTOR pathway in a pancreatic cancer model. Cancer Immunol. Res. 2023;11(4):421–434. doi: 10.1158/2326-6066.CIR-22-0398. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y., et al. Cancer and ER stress: mutual crosstalk between autophagy, oxidative stress and inflammatory response. Biomed. Pharmacother. 2019;118 doi: 10.1016/j.biopha.2019.109249. [DOI] [PubMed] [Google Scholar]
- 33.Matheson C.J., Backos D.S., Reigan P. Targeting WEE1 kinase in cancer. Trends Pharmacol. Sci. 2016;37(10):872–881. doi: 10.1016/j.tips.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J.X., et al. LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene. 2018;37(20):2660–2675. doi: 10.1038/s41388-018-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Wu S., et al. ORAI2 promotes gastric cancer tumorigenicity and metastasis through PI3K/Akt signaling and MAPK-dependent focal adhesion disassembly. Cancer Res. 2021;81(4):986–1000. doi: 10.1158/0008-5472.CAN-20-0049. [DOI] [PubMed] [Google Scholar]
- 36.Riascos-Bernal D.F., et al. The FAT1 cadherin drives vascular smooth muscle cell migration. Cells. 2023;12(12) doi: 10.3390/cells12121621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paluch E.K., Aspalter I.M., Sixt M. Focal adhesion-independent cell migration. Annu. Rev. Cell Dev. Biol. 2016;32:469–490. doi: 10.1146/annurev-cellbio-111315-125341. [DOI] [PubMed] [Google Scholar]
- 38.Puri B.K. Calcium signaling and gene expression. Adv. Exp. Med. Biol. 2020;1131:537–545. doi: 10.1007/978-3-030-12457-1_22. [DOI] [PubMed] [Google Scholar]
- 39.Lee S., Rauch J., Kolch W. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int. J. Mol. Sci. 2020;21(3) doi: 10.3390/ijms21031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullah R., et al. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin. Cancer Biol. 2022;85:123–154. doi: 10.1016/j.semcancer.2021.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Jiang T., et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer. 2021;20(1):66. doi: 10.1186/s12943-021-01358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y., Fan D. Ginsenoside Rg5 induces G2/M phase arrest, apoptosis and autophagy via regulating ROS-mediated MAPK pathways against human gastric cancer. Biochem. Pharmacol. 2019;168:285–304. doi: 10.1016/j.bcp.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Zhong C., et al. LINC00665: an emerging biomarker for cancer diagnostics and therapeutics. Cells. 2022;11(9) doi: 10.3390/cells11091540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GSE158662, GSE103236, GSE15459 dataset was downloaded from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/). Data will be made available on request.