Summary
A comprehensive understanding of neuronal diversity and connectivity is essential for understanding the anatomical and cellular mechanisms that underlie functional contributions. With the advent of single-cell analysis, growing information regarding molecular profiles leads to the identification of more heterogeneous cell types. Therefore, the need for additional orthogonal recombinase systems is increasingly apparent, as heterogeneous tissues can be further partitioned into increasing numbers of specific cell types defined by multiple features. Critically, new recombinase systems should work together with pre-existing systems without cross-reactivity in vivo. Here, we introduce novel site-specific recombinase systems based on ΦC31 bacteriophage recombinase for labeling multiple cell types simultaneously and a novel viral strategy for versatile and robust intersectional expression of any transgene. Together, our system will help researchers specifically target different cell types with multiple features in the same animal.
eTOC Blurb
Jeong et al. created a new viral technique, ΦC31/pSIO vector, to target specific neuronal cell types without cross-reactivity with Cre− and Flp recombinases. These easy-to-use viral tools for robust and effective intersectional expression of numerous transgenes in neurons with multiple characteristics will help the functional understanding of diverse brain circuitry.
Introduction
Defining the function of specific cellular ensembles within multicellular biological systems has remained a major thrust of modern biological research 1–3. Especially, the complexity of the nervous system is not only due to a large number of neurons but also to the enormous number of synapses through which these neurons are connected. A more fascinating aspect of this complexity is specificity. Each neuron in the brain has a very specific pattern of projections and connections. Elucidating this specificity is the major goal of many neuroscientists and will provide important frameworks for understanding brain function.
During past decades, substantial progress has been made toward this goal through the development of molecular and viral tools for visualizing and manipulating specific cells 4–7. Various transgenic mouse lines expressing Cre or Flp recombinases in specific cell types have been developed 6,8–11 and several viral tools for targeting neurons have also been introduced 2,12,13. The Cre and Flp recombinase systems have been the most widely used approaches to synergistically target specific transgene expressions because of their incredibly low cross-reactivity in vivo 4–6. A typical application of these recombinases for viral-mediated gene delivery uses the DIO (Double-flanked Inverted Open reading frame) or FLEX (FLip EXcision) system to express a transgene in a Cre or Flp recombinase-dependent manner 4,7,14. However, with the advent of single-cell analysis methods, more information on the molecular profiling of the brain will lead to the identification of more heterogeneity 15,16. Even with the increased number of available transgenic mice expressing Cre, and Flp recombinases in specific cell types, it is obvious that new recombinases and their counterparts are needed for targeting additional cell types, especially with viral-mediated gene delivery. Unfortunately, because of cross-reactivity and low efficiency, several efforts to develop these new recombinase and reporter pairs have not been fully successful for viral-mediated gene delivery in vivo 4. Thus, here, we developed the novel recombinase and reporter pairs that can be used together with Cre and Flp in a minimal cross-reactivity for viral-mediated gene delivery.
Moreover, recently, sophisticated studies have revealed significant cellular heterogeneity, even in seemingly homogenous tissue. The advent of single-cell analysis methods has enabled in-depth molecular profiling of the tissue, leading to the identification of a substantial amount of heterogeneity 15,17–22. For instance, single-cell transcriptomics and electrophysiological characterization of cholecystokinin (CCK)-expressing neurons in the brain have identified at least five distinct subpopulations of these neurons 23. Another set of studies on dopaminergic neurons in the midbrain area suggest that it can be classified into subtypes ranging from 5 to 10 subtypes 16,24–27. Furthermore, genetically similar cell types can have anatomically distinct properties such as different axonal target areas in the brain 2,28–33. This suggests that neurons can be identified not only by multiple genetic markers but also by a number of other features, including anatomical properties. Thus, to effectively study the heterogeneity of cells, more tools capable of defining a growing number of individual cellular populations beyond Cre and Flp recombinase-mediated gene delivery are immensely needed. For example, viral or molecular tools to label neurons expressing both Cre− or Flp recombinase in the same animals were introduced. Several reporter mice line allow us to achieve the labeling of Cre− and Flp recombinase expressing neurons8, but the brain-area specific and developmental stage-specific labeling of both Cre− and Flp recombinase expressing neurons is not possible. In addition to that, labeling neurons expressing only Cre− not Flp or vice versa has not been reliable with transgenic reporter mice. Other efforts have been made for this purpose, such as INTRSECT (INTronic Recombinase Sites Enabling Combinatorial Targeting)4,34 as well as, tTARGIT (tTA-driven Recombinase-Guided Intersectional Targeting)35, but the efficiency of labeling and the expression level of some transgenes are not high enough for some applications. Especially, so far, only a few transgenes have been tested for reliable expression. Here, in addition to the novel recombinase system, we develop easily applicable viral tools for the robust and efficient intersectional expression of various transgenes, which will provide valuable tools for anatomical and functional understanding of neural circuitry.
Results
PhiC31- and Bxb1-dependent viral-mediated gene delivery
The bacteriophage ΦC31 and Bxb1 recombinases, also known as integrases, are distinct from Cre and Flp recombinases, and are in a different class of recombinases 36–39. Several studies have successfully utilized ΦC31 to deliver genes into the specific region of genome in C. elegans and in Drosophila40,41. While Cre or Flp recombinases recognize identical and palindromic target sites, the target sites for ΦC31 and Bxb1 recombinases, attB and attP sequences are different from each other (Fig. 1A). Thus, unlike the DIO or FLEX system, the recombination between attB and attP sites results in chimeric attL and attR sequences, which cannot be further recombined (Fig. 1B and C). Using this feature, we designed two vectors, pSIO (phiC31-mediated Single-flanked Inverted Open reading frame) and bSIO (bxb1-mediated Single-flanked Inverted Open reading frame) for ΦC31 and Bxb1 recombinases-dependent transgene expression, respectively (Fig. 1C). We then examined the efficiency and the cross-reactivity for the Cre, Flp, ΦC31, and Bxb1 recombinase systems. Our results indicate that these four recombinases show comparable efficiency with no detectable cross-reactivity in HEK293T cells (Fig. 1D, Supplementary Fig. 1A). The co-transfection of HEK293T cell with a plasmid expressing ΦC31 together with mCherry (ΦC31-IRES-mCherry) and a plasmid expressing EGFP in ΦC31-dependent manner (pSIO-EGFP) showed that EGFP expression was observed in the cell expressing mCherry, indicating the specific expression of EGFP in ΦC31-expressing cells (Supplementary Fig. 1B). Moreover, we can use separate expression vectors for each of the four recombinase systems – cDIO (cre-mediated DIO), fDIO (flp-mediated DIO), pSIO, and bSIO – to simultaneously express different transgenes. These vectors show high efficiency without any cross-reactivity, indicating that our systems can be used to express different transgenes in four differentially defined cell populations simultaneously (Fig. 1D). To further validate the efficacy of the ΦC31/pSIO or Bxb1/bSIO recombinase system in vivo, we generated an adeno-associated virus (AAV) expressing ΦC31 or Bxb1 (AAV-EF1α-ΦC31 or AAV-EF1α-Bxb1) and AAV expressing mScarlet-I in ΦC31- or Bxb1-dependent manner (AAV-EF1α-pSIO-mScarlet-I or AAV-EF1α-bSIO-mScarlet-I) under EF1α promoter. With the ΦC31/pSIO system, we were able to induce strong transgene expression (mScarlet-I) in the hippocampal dentate gyrus (DG; n = 3 animals) area without any cross-reactivity with other recombinase systems (n = 3 for Cre recombinase alone; n = 2 for Flp recombinase alone; Fig. 1E). However, with the Bxb1/bSIO system, we found that the bSIO-mScarlet-I vector itself showed a slight leaky expression in vivo (n = 3 animals) (Supplementary Fig. 2A). Thus, we decided to focus on the in vivo application of the ΦC31/pSIO system. To compare the efficacy of gene transport using the ΦC31/pSIO system to other recombinases, we co-injected AAVs expressing different fluorescent proteins with different recombinase pairings (Cre/cDIO, Flp/fDIO, C31/pSIO). Injections of a similar amount of viral particle for each recombinase pair resulted in the compatible level of different fluorescent protein production (Supplementary Fig. 2B).
Figure. 1: Simultaneous use of multiple orthogonal recombinases to deliver different transgenes in various cell types.
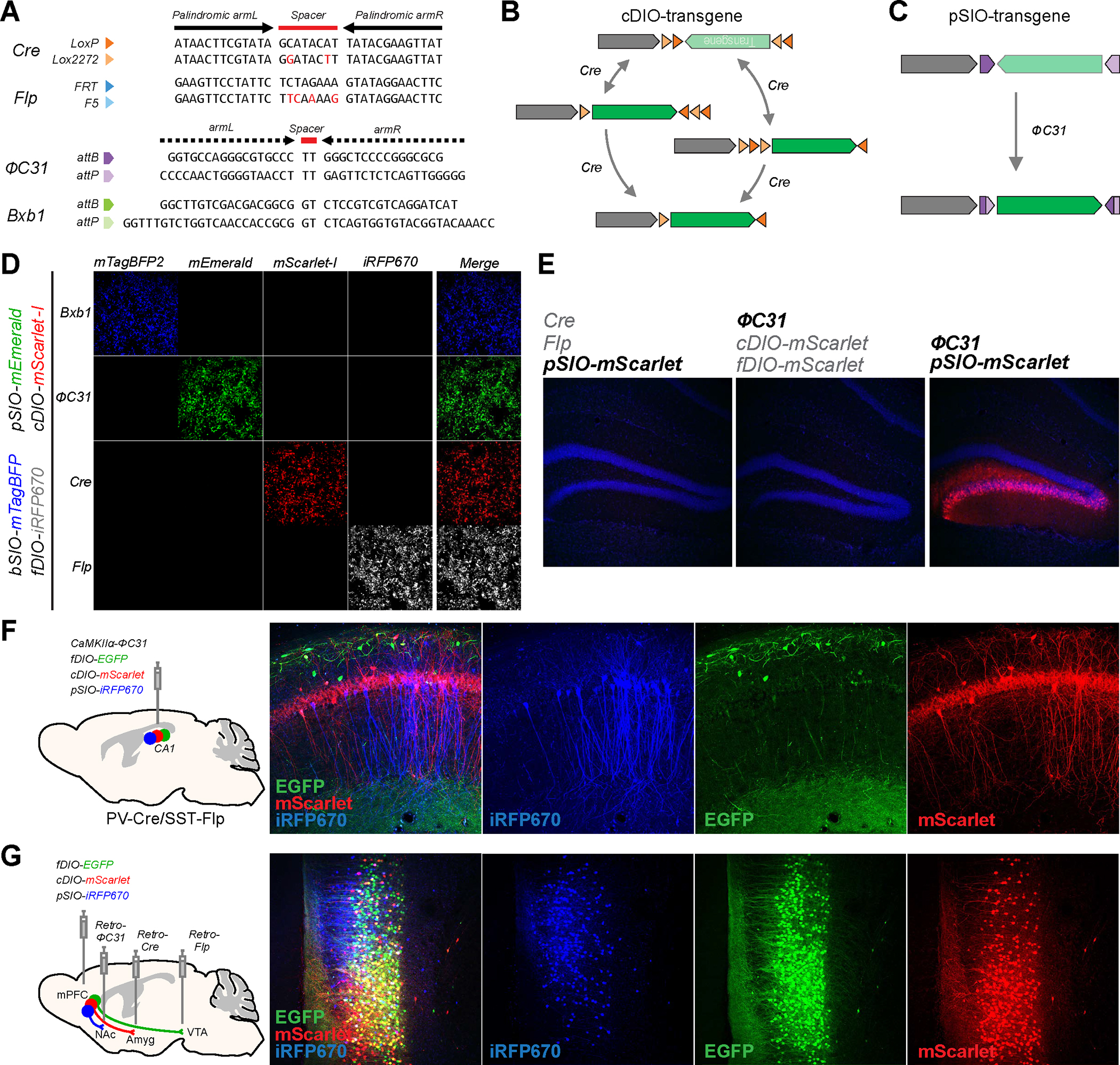
A, Sequence of the target sites for four recombinases used in cDIO, fDIO, bSIO, and pSIO vectors. The Cre and Flp recognition sites contain 13-bp palindromic sequences (indicated by arrows) flanking a 8-bp non-symmetric core sequence (indicated as spacer). The PhiC31 and Bxb1 recognition sites contain non-palindromic recognition sequences (indicated by dashed arrow) flanking 2-bp spacer. B and C, Design of cDIO, pSIO, and the possible flow of recombination to express the transgene. D, Specificity of the differential transgenes expression when bSIO, pSIO, cDIO, and fDIO vectors are delivered to the same cells. The plasmids with bSIO-mTagBFP2, pSIO-mEmerald, cDIO-mScarlet-I, fDIO-iRFP670, and a single recombinase expressing plasmid (total 5 plasmids) were co-transfected in HEK293T cells. Each row shows different fluorescence channels from the same cells imaged. E, Specificity of ΦC31/pSIO system in vivo. Combinations of AAVs as indicated were co-injected in the DG. F, Usage of cDIO, fDIO, and pSIO to express three different transgenes in three populations defined by promoter activities. AAV-CaMKIIα-ΦC31, AAV-EF1α-fDIO-EGFP, AAV-EF1α-cDIO-mScarlet-I, and AAV-EF1α-pSIO-iRFP670 were co-injected into CA1 of PV-Cre/SST-Flp transgenic mice. The lower titer of AAV-CaMKIIα-ΦC31 was used to restrict the iRFP670 expression in a sparse population for better visualization of the cell morphology. iRFP670 is pseudocolored in blue. G, Usage of cDIO, fDIO, and pSIO vectors to express three different transgenes in three populations defined by separate anatomical connectivity. AAV-EF1α-fDIO-EGFP, AAV-EF1α-cDIO-mScarlet-I, and AAV-EF1α-pSIO-iRFP670 were co-injected into mPFC, and retroAAV-Flp, retroAAV-Cre, and retroAAV-ΦC31 were injected into VTA, BLA, and NAc, respectively. d-g, Scale bar = 50 μm.
To demonstrate the application of having an additional recombinase, we used Cre, Flp, and ΦC31 recombinases to target three genetically different neuronal populations in the mouse brain. First, we applied the ΦC31 recombinases to the transgenic mice expressing Cre and Flp recombinases under the control of the parvalbumin (PV) and somatostatin (SST) promoters, respectively (PV-Cre/SST-Flp). We injected AAV expressing ΦC31 recombinase under CaMKIIα promoter (AAV-CaMKIIα-ΦC31), along with AAV-EF1α-fDIO-EGFP, AAV-EF1α-cDIO-mScarlet-I, and AAV-EF1α-pSIO-iRFP670 into hippocampal CA1 area of PV-Cre/SST-Flp mice (n =2 animals) (Fig. 1F). This approach enables us to express three different fluorescent proteins, EGFP, mScarlet-I, and iRFP670 in SST+, PV+, and CaMKIIα+ neurons, respectively, in the hippocampal CA1. In a separate set of mice, we also injected retrogradely-transducing AAV (retroAAV) expressing ΦC31 (retroAAV- EF1α-ΦC31) into the dorsolateral striatum (DLS) of PV-Cre/SST-Flp mice and injected AAV-EF1α-fDIO-EGFP, AAV-EF1α-cDIO-mScarlet-I, and AAV-EF1α-pSIO-iRFP670 into the primary motor cortex (M1) (Supplementary Fig. 2C). With this approach, we were able to express EGFP and mScarlet-I in SST+ and PV+ neurons, respectively, and iRFP670 in DLS-projecting motor cortical neurons to differentiate three distinct neuronal populations in M1, two defined by genetic markers and one defined by the anatomical property (Supplementary Fig. 2C).
Next, we validated this new recombinase combination system for separating neuronal populations with different neural pathways. Using engineered viral tracing strategies, recent developments in modern neuroscience have revealed that adjacent neurons in the same brain region can have distinct neuroanatomical connectivity profiles, convey different information streams, and contribute uniquely to brain function 28,29,33. Therefore, we tested whether our new recombinase ΦC31 can expand the viral tracing toolkit and provide a robust means to map out different neural pathways.
Since cortical pyramidal neurons are projecting to other subcortical brain regions distinct and collateral manners 42,43, we decided to label medial prefrontal cortical (mPFC) neurons projecting into three different brain areas 44–46. We injected AAV-EF1α-fDIO-EGFP, AAV-EF1α-cDIO-mScarlet-I, and AAV-EF1α-pSIO-iRFP670 into the mPFC, and injected retroAAV-Flp, retroAAV-Cre, and retroAAV-ΦC31 into major targets of the mPFC, ventral regimental area (VTA), basolateral amygdala (BLA), and nucleus accumbens (NAc), respectively (n = 3 animals) (Fig. 1G). In this way, distinct mPFC neurons were labeled with three different fluorescent proteins depending on the projection target areas. Notably, although we detected neurons that expressed one of the three fluorescent proteins, a significant fraction of labeled neurons expressed multiple fluorescent proteins (Fig 1G) – suggesting that these neurons form collateralizing axonal fibers to multiple target areas 43. Together, these results clearly demonstrate that the ΦC31/pSIO recombinase system can be used with other recombinases to specifically express transgenes in differentially defined populations in vivo.
Generating knock-in mice expressing PhiC31 in specific neuronal cell types
To expand the use of the ΦC31/pSIO system, we decided to generate knock-in mice expressing ΦC31 in specific neuronal populations. Using the CRISPR-directed gene targeting method (Easi-CRISPR; See Method) 47, we generated two knock-in mice, where recombinase expression is mediated by 2A sequence from a bicistronic transcript, one expressing ΦC31 under SERT (Serotonin Transporter)/Slc6a4 promoter to label serotonergic neurons (SERT-P2A-PhiC31), and one expressing ΦC31 under VGluT2 (Vesicular glutamate transporter 2)/Slc17a6 promoter to label subsets of glutamatergic neurons (VGluT2-P2A-PhiC31) (Fig. 2).
Figure. 2: Generation of transgenic animals expressing ΦC31 in specific cellular populations.
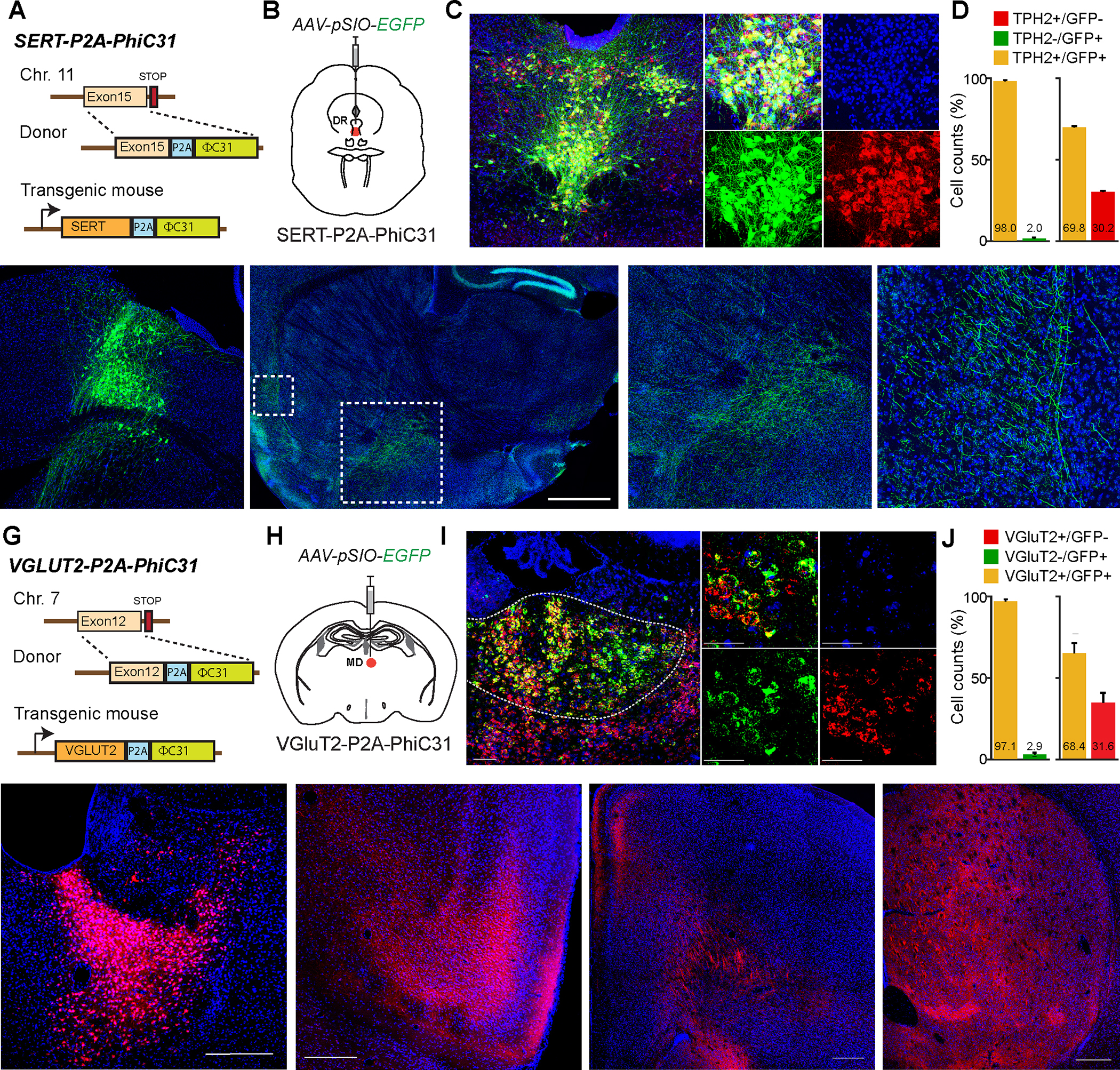
A, Schematic showing the insertion site of ΦC31 into SERT gene to target serotonergic neurons. B, Schematic showing the injection of AAV-EF1α-pSIO-EGFP into DR (dorsal raphe) of SERT-P2A-ΦC31 mice. C, Fluorescent image showing that EGFP expression in Tryptophan hydroxylase (TPH)-positive neurons. D, The quantification of all EGFP- and TPH-postive neurons (n = 593) in the injection areas show that more than 98% of EGFP-positive neurons are co-localized with TPH immunostaining. E, Fluorescent image showing the EGFP-expressing SERT-positive neurons in DR. F, Images showing the axonal projections of serotonergic neurons projecting to multiple brain areas including amygdala (Amy), ventral pallidum (VP), lateral orbitofrontal cortex (LO), and so on. G, Schematic showing the insertion site of ΦC31 into VGLUT2 gene to target subsets of glutamatergic neurons. H, Schematic showing the injection of AAV-EF1α-pSIO-EGFP into MD (mediodorsal thalamus) of VGluT2-P2A-ΦC31 mice. I, Fluorescent image showing that EGFP-expressing neurons in MD are expressing VGluT2 mRNA. J, The quantification of the fluorescent in situ hybridization (FISH) results showed that more than 97% of EGFP-positive neurons are co-localized with VGluT2 mRNA (n = 1038). K, The image showing the injection sites of AAV-EF1α-pSIO-mScarlet-I into MD of VGLUT2-P2A-ΦC31. L-N, Images showing the axonal projections of VGluT2-positive MD neurons labeled by mScarlet-I in anterior insular cortex (AIC; L), anterior cingulate cortex (ACC; M), striatum (STR; N), and so on. ACC; anterior cingulate cortex, AIC; anterior insular cortex, CL; central lateral thalamus, GP; globus pallidus, Hb; habenula, IL; infralimbic cortex, MPO; medial preoptic area, PrL; prelimbic cortex, STR; striatum, VP; ventral pallidum.
To validate the delivery of transgenes, EGFP, in serotonergic neurons, we injected AAV-hSyn-pSIO-EGFP into dorsal raphe (DR) of SERT-P2A-PhiC31 mice (Fig 2A, B). The EGFP expression was highly colocalized with serotonergic neuronal marker, Tryptophan hydroxylase (TPH) (n = 3 animals) (Fig 2C). We found that more than 98% of EGFP-positive neurons are TPH-positive, suggesting that the combination of SERT-P2A-PhiC31 knockin mouse and pSIO-mediated gene delivery can specifically target serotonergic neurons in DR (Fig. 2D). We also counted all EGFP-positive and TPH-positive neurons in the areas of injection and found that around ~70% TPH positive neurons were labelled with our viral injection (See Methods). We must note that the efficacy can vary based on the serotype, titers, and volume of the viruses. It is also possible that the level of SERT expression in certain TPH-positive neurons can be low as reported 48.
To examine the specificity and expression level of the transgenes further, we again injected AAV-hSyn-pSIO-EGFP into DR of SERT-P2A-PhiC31 mice (Fig. 2E). We found that the EGFP-positive axonal fibers of EGFP in regions such as amygdala, lateral orbital cortex, and so on (Fig. 2F), suggesting that the SERT-P2A-PhiC31 mice and pSIO vector-mediated gene expressions align well with the previously documented patterns of the serotonergic projections 49. This indicates that SERT-P2A-PhiC31 mice and pSIO-dependent gene delivery can be a valuable addition to investigate the roles of monoamine neurotransmitters.
In a similar manner, to achieve the EGFP expression in VGluT2 neurons, we injected AAV-hSyn-pSIO-EGFP into the mediodorsal thalamus (MD) of VgluT2-P2A-PhiC31 mice (Fig. 2G, H). Using fluorescent in situ hybridization against VgluT2 and EGFP, we validated the EGFP expression in VgluT2-positive neurons (n = 3 animals) (Fig. 2I, J). We found that even at the mRNA level of expression, more than 97% of EGFP-positive neurons express VGluT2 mRNA, indicating that VGluT2-P2A-PhiC31 can be used to efficiently label VGLUT2-positive neurons (Fig. 2J). We also counted all EGFP-positive and VgluT2 mRNA-positive neurons in the areas of injection and found that around ~70% VgluT2-positive neurons were labelled with our viral injection (See Methods). Again, we have to note that the efficacy can vary based on the serotype, titers, volume of the viruses, and so on.
When we injected AAV-hSyn-pSIO-mScarlet-I into the MD of VGluT2-P2A-PhiC31 mice (Fig. 2K), we could detect the strong fluorescent protein expression of axonal fibers of MD neurons in their target areas including anterior insular cortex, anterior cingulate cortex, and striatum (Fig. 2L to 2N), indicating that VgluT2-P2A-PhiC31 mice and pSIO-dependent gene system can deliver the efficient expression to detect the projection patterns of the VgluT2-postive neurons.
PhiC31-dependent expression of various functional transgenes in mouse brain
The introduction of novel recombinases that are orthogonal to Cre and Flp is essential for the manipulating and monitoring different cell types in the brain. To deliver various transgenes expressing molecular tools for manipulating and monitoring the activity of specific neurons, and tracing specific neuronal populations using the ΦC31/pSIO system, we designed several versions of pSIO vectors (Key Resource Table). First, we generated the several vectors expressing different optogenetic tools with the ΦC31/pSIO system. We simply replace mScarlet-I or EGFP in our pSIO vector with a number of optogenetics tools, including ChR2-EYFP, oChIEF-Citrine, ChrimsonR-tdTomato, and NpHR-EYFP 50–53. Then, we injected AAV-pSIO vectors expressing these tools into several different areas of the brains (Fig 3A to 3D). Two to three weeks later, we performed a whole-cell patch clamp recording of neurons expressing optogenetic tools that can be visualized by fluorescent proteins tagged with opsins (Fig. 3A to 3D). We found that the illumination of specific wavelength of light for each optogenetic tool can successfully manipulate the excitability of neurons expressing these tools, demonstrating the efficacy of optogenetic tool delivery with ΦC31/pSIO system (Fig. 3A to 3D).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP (Goat) | Abcam | AB5450; RRID:AB_304897 |
| Anti-GFP (Rabbit) | Millipore | AB3080; RRID:AB_91337 |
| Anti-TPH (Tryptophan hydroxylase) | Abcam | AB5295; RRID:AB_2207555 |
| Anti-Goat Alexa 488 | Life Technology | A11055; RRID:AB_2534102 |
| Anti-Goat Alexa 647 | Life Technology | A21447; RRID:AB_2535864 |
| Anti-Goat Rabbit 568 | Life Technology | A10042; RRID:AB_2534017 |
| Anti-Goat Rabiit 488 | Life Technology | A32790; RRID:AB_2762833 |
| RNAscope Probes | ||
| Probe for EGFP | Advanced Cell Diagnostics, Inc. | 31917 |
| Probe for Mm-Slc17a6 | Advanced Cell Diagnostics, Inc. | 400281 |
| Bacterial and Virus Strains | ||
| EnvA-pseudotyped, glycoprotein- deleted rabies virus; EnvA-RVΔG-eGFP | Lim Lab | N/A |
| Adeno-associated virus-DJ; various | Lim Lab | N/A |
| retroAAV | Lim Lab | N/A |
| Chemicals | ||
| Fluoromount-G | Thermo Fisher Scientific | 00–4959-52 |
| Experimental Models: Cell Lines | ||
| AAV-293 cells | Agilent | 240073 |
| HEK293T cells | ATCC | CRL-3216 |
| B7GG cells | Ed Callaway Lab | N/A |
| BHK-EnvA cells | Ed Callaway Lab | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: VGLUT2-Cre | The Jackson Laboratory | Stock#: 028863, RRID:IMSR_JAX:028863 |
| Mouse: PV-Cre | The Jackson Laboratory | Stock#: 017320, RRID:IMSR_JAX:017320 |
| Mouse: SST-Flp | The Jackson Laboratory | Stock#: 031629, RRID:IMSR_JAX:031629 |
| Mouse: PV-Cre/SST-Flp | Lim Lab | Newly generated for this study |
| Mouse: VGLUT2-P2A-PhiC31 | Lim Lab | Newly generated at Salk Transgenic Core for this study |
| Mouse: SERT-P2A-PhiC31 | Lim Lab | Newly generated at Salk Transgenic Core for this study |
| Mouse: C57BL/6J | The Jackson Laboratory | JAX: 000664; RRID: IMSR_JAX:000664 |
| Recombinant DNA for virus | ||
| pAAV-EF1α-Cre+ Flp+-PhiC31 | This paper | N/A |
| pAAV- EF1α-Cre+ Flp+-tTA2 | This paper | N/A |
| pAAV-CamKII-PhiC31 | This paper | N/A |
| pAAV-EF1α-fDIO-mScarlet-I | This paper | N/A |
| pAAV-hSyn-pSIO-mScarlet-I | This paper | N/A |
| pAAV-EF1α-pSIO-mEmerald | This paper | N/A |
| pAAV-EF1α-fDIO-iRFP670 | This paper | N/A |
| pAAV-EF1α-fDIO-EGFP | This paper | N/A |
| pAAV-EF1α-cDIO-mCherry | a gift from Bryan Roth | RRID:Addgene_500462 |
| pAAV-EF1α-cDIO-mScarlet-I | This paper | N/A |
| pAAV-EF1α-Bxb 1 | This paper | N/A |
| pAAV-EF1α-bSIO-mScarlet-I | This paper | N/A |
| pAAV-EFlα-bSIO-mTagBFP2 | This Paper | N/A |
| pAAV-EF1α-Cre+ Flp−-mTagBFP2 | This paper | N/A |
| pAAV-EF1α-Cre− Flp+-iRFP670 | This paper | N/A |
| pAAV-EFlα-Cre+ Flp−_v0.5 -mTagBFP2 | This paper | N/A |
| pAAV-EF1α-Cre− Flp+_v0.5-iRFP670 | This paper | N/A |
| pAAV-EF1α-Cre+ Flp+-Bxb1 (293) | This paper | N/A |
| pAAV-EF1α-Cre+ Flp+-Bxb1 (468) | This paper | N/A |
| pAAV-TRE3G-pSIO-mScarlet-I | This paper | N/A |
| pAAV-EF1α-Cre+ Flp−-iRFP670myc | This paper | N/A |
| pAAV-EFlα-Cre− Flp+-mEmerald | This paper | N/A |
| pAAV-EF1α-pSIO-jGCaMP8m | This paper | N/A |
| pAAV-EFlα-pSIO-oPBG | This paper | N/A |
| pAAV-hSyn-pSIO-mRuby-2A-TVA | This paper | N/A |
| pAAV-EFlα-pSIO-EGFP | This paper | N/A |
| pAAV-EF1α-pSIO-ChR2-EYFP | This paper | N/A |
| pAAV-EF1α-pSIO-ChrimsonR- tdTomato | This paper | N/A |
| pAAV-EF1α-pSIO-oChIEF-Citrine | This paper | N/A |
| pAAV-EF1α-pSIO-NpHR-EYFP | This paper | N/A |
| pAAV-EF1α-pSIO-hM3Dq-mCherry | This paper | N/A |
| pAAV-EF1α-pSIO-hM4Di-mCherry | This paper | N/A |
| pAAV-EF1α-mCherry-IRES-Cre | Fenno et al.4 | RRID:Addgene_55632 |
| pAAV-EF1α-mCherry-IRES-Flp | Fenno et al.4 | RRID:Addgene_55634 |
| pAAV-EF1α-mCherry-IRES-PhiC31 | This paper | N/A |
| Software and Algorithms | ||
| Photoshop 2022 | Adobe | N/A |
| Illustrator 2022 | Adobe | N/A |
| FIJI (ImageJ) | NIH | N/A |
| Clampfit 10.4 | Molecular Devices | N/A |
| pClamp 10.4 | Molecular Devices | N/A |
Figure. 3: Expression of various transgenes with the ΦC31/pSIO system.
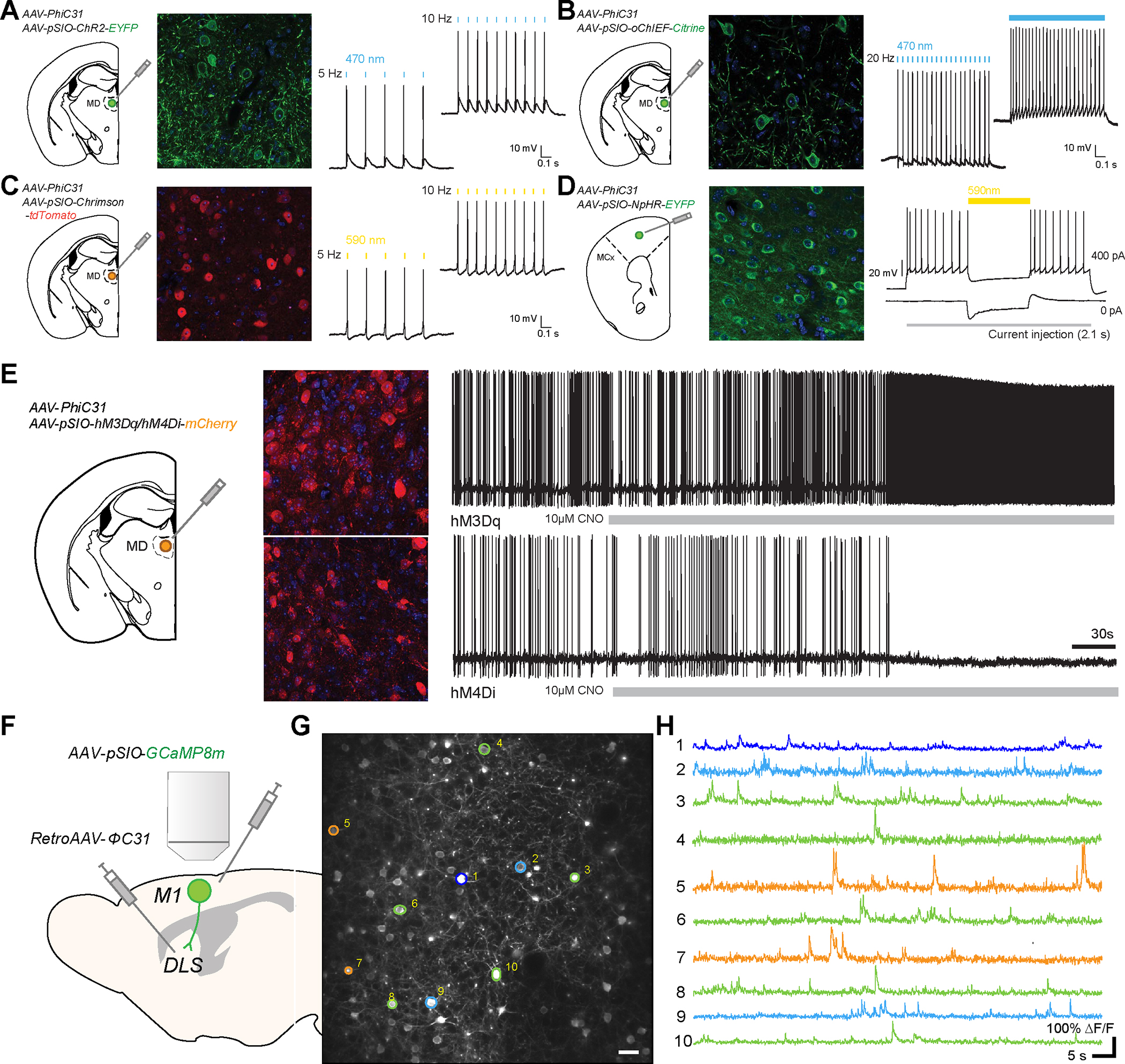
A, Schematic diagram showing the injection of AAV-EF1α-pSIO-ChR2-EYFP and AAV-EF1α-ΦC31 into mediodorsal thalamus (MD) (left). Confocal image showing the expression of ChR2-EYFP (middle). The validation of the functional ChR2 expressed with ΦC31/pSIO system, showing the action potential with light stimulation (470 nm) (right). B, Schematic diagram showing the injection of AAV-EF1α-pSIO-oChIEF-Citrine and AAV-EF1α-ΦC31 into mediodorsal thalamus (MD) (left). Confocal image showing the expression of oChIEF-Citrine (middle). The validation of the functional oChIEF expressed with ΦC31/pSIO system, showing the action potential with light stimulation (470 nm) (right). C, Schematic diagram showing the injection of AAV-EF1α-pSIO-ChrimsonR-tdTomato and AAV-EF1α-ΦC31 into mediodorsal thalamus (MD) (left). Confocal image showing the expression of ChrimsonR-tdTomato (middle). The validation of the functional ChrimsonR expressed with ΦC31/pSIO system, showing the action potential with light stimulation (590 nm) (right). D, Schematic showing the injection of AAV-EF1α-pSIO-NpHR3.0-EYFP and AAV-CamKII- ΦC31 into the prelimbic cortex (PrL) (left). Confocal image showing the expression of NpHR3.0-EYFP in PrL (middle). The validation of the NpHR3.0 expressed with ΦC31/pSIO system, showing the reduced the current injection (100 pA, 200 pA)-induced action potential with light stimulation (590 nm) (right). E, Schematic diagram showing the injection of AAV-EF1α-pSIO-hM3Dq-mCherry or AAV-EF1α-pSIO-hM4Di-mCherry together with AAV-EF1α-ΦC31 into mediodorsal thalamus (MD) (left). Confocal image showing the expression of hM3Dq-mCherry and hM4Di-mCherry (middle). The change of neuronal excitability of neurons expressing hM3Dq or hM4Di after the bath application of CNO (right). F, Diagram showing the two-photon imaging of primary motor cortical (M1) neurons projecting to dorsolateral striatum (DLS). To express GCaMP8m in this population, we injected AAV- AAV-EF1α-pSIO-GcaMP8m into M1 and retroAAV- ΦC31 into DLS. G, Two-photon samples images of M1 neurons expressing GcaMP8m. Scale bar = 25 μm. H, Samples calcium activity traces of M1 neurons in G.
Next, we designed vectors expressing chemogenetic tools (AAV-EF1α-pSIO-hM3Dq-mCherry and AAV-EF1α-pSIO-hM4Di-mCherry) with ΦC31/pSIO system. Two to three weeks after the injection of these viruses into the MD thalamic nuclei, we performed the whole cell patch clamp recording from mCherry-labelled neurons. We found that the bath application of clozapine-N-oxide (CNO) increased or decreased the excitability of neurons expressing hM3Dq or hM4Di, respectively (Fig. 3E)
To test the delivery of a genetically encoded calcium indicator, jGCaMP8m54, with the ΦC31/pSIO system, we generated an AAV vector expressing jGCaMP8m in a ΦC31-dependent manner (AAV-EF1α-pSIO-jGCaMP8m) (Fig. 3F). To examine the efficacy of this vector system, we injected retroAAV-ΦC31 into the dorsal lateral striatum (DLS) and injected AAV-EF1α-pSIO-jGCaMP8m into primary motor cortex (M1). A month after the injections of viruses, we performed two-photon imaging of GCaMP8m labeled neurons in M1 (Fig. 3F). As in Fig. 3G, we found that we were able to image the calcium activities of M1 neurons projecting to DLS even a month after the injection.
Evaluating the application of PhiC31/pSIO system for the in vivo application using the terminal optogenetic stimulation
To test the in vivo functionality of the transgene expression with ΦC31/pSIO system, we decided to examine the behavioral changes induced by optogenetic stimulation. It has been well-documented that the activation of VTA glutamatergic projections to LHb is involved in aversive behaviors 55,56. Thus, we tested whether a VGluT2-P2A-PhiC31 and pSIO-dependent gene expression system could reproduce the same behavioral effects. We injected AAV-EF1α-pSIO-ChR2-EYFP or AAV-EF1α-pSIO-EGFP into the ventral tegmental area (VTA) of VGluT2-P2A-PhiC31 mice, then implanted optic fibers into the lateral habenula (LHb) (Fig. 4A). Three weeks after the injections, we examined whether the optogenetic stimulation of axonal terminals of VTA glutamatergic neurons can induce conditioned place aversion (CPA) (Fig. 4B). Indeed, we observed that the stimulation of VTA glutamatergic axon fibers expressing ChR2 could elicit a strong aversion to the stimulated chamber (Fig. 4C to 4E). This result indicates that the ΦC31/pSIO system is capable of delivering a level of ChR2 expression sufficient for the optogenetic stimulation of axon terminals. The axon tracing results of MD neurons (Fig. 2G) and the behavioral tasks with the terminal stimulation (Fig 4) with ΦC31/pSIO system suggest that ΦC31/pSIO system is novel recombinase system that is suitable for orthogonal use in vivo alongside Cre/cDIO and Flp/fDIO systems.
Figure 4. Optogenetic stimulation of axonal fibers of VTA VGluT2 neurons in LHb induces conditioned place aversion.
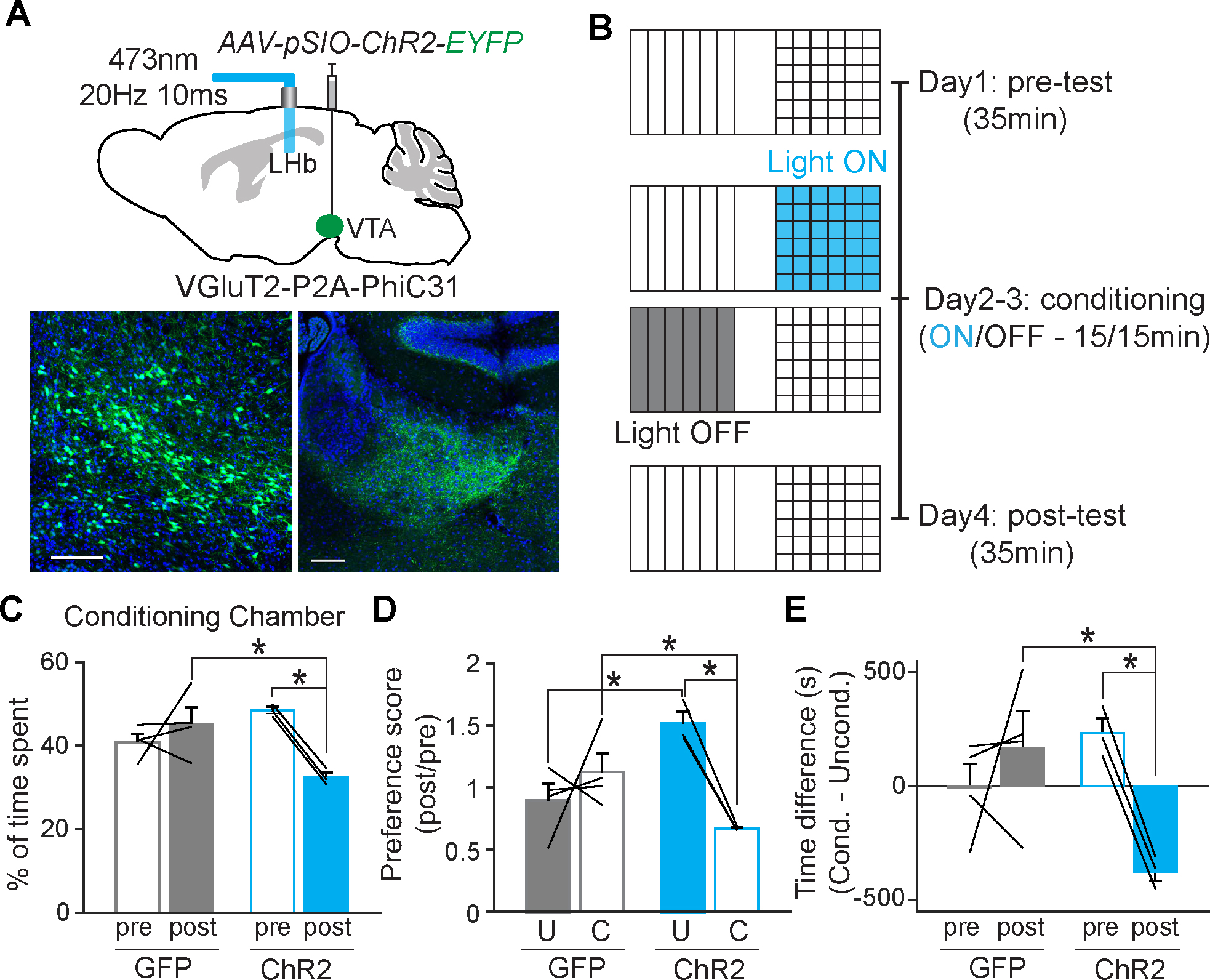
A, Schematic diagram of the injection of AAV-pSIO-ChR2-EYFP into the VTA of VGluT2-P2A-PhiC31, and the optic fiber implantation to stimulate the axonal fibers of VTA VgluT2 neurons in lateral habenula (LHb) (top). Confocal images of VTA GluT2 neurons expressing ChR2-EYFP and axonal fibers in LHb (bottom). B, Diagram showing the timeline of conditioned place preference/aversion (CPP/CPA) using optogenetic stimulation. C, The percentage of time spent in the conditioning chamber where animals received optogenetic stimulation. Two-way repeated measure ANOVA, Holm-sidak multiple comparison, GFP; n = 4, ChR2; n =3, For pre vs. post within ChR2 group; t = 3.309; P = 0.021, For GFP vs. ChR2 within post group; t = 3.384; P = 0.009. D, The preference scores for the unconditioned chamber and conditioned chamber after the optogenetic stimulation. Two-way repeated measure ANOVA, Holm-sidak multiple comparison, GFP; n = 4, ChR2; n =3, For unconditioned vs conditioned within ChR2 group; t = 3.230; P = 0.023, For GFP vs ChR2 within conditioned; t =2.547; P = 0.046, For GFP vs. ChR2 within conditioned; t = 3.453; P = 0.015. E, The difference in time spent in the conditioning chamber before and after the optogenetic stimulation. Two-way repeated measure ANOVA, Holm-sidak multiple comparison, GFP; n = 4, ChR2; n =3, For pre vs. post within ChR2 group; t = 3.041; P = 0.029, For GFP vs. ChR2 within post group; t = 3.217; P = 0.010.
PhiC31-mediated intersectional gene delivery into cells expressing both Cre and Flp recombinase.
Recent single-cell sequencing studies have increasingly demonstrated that it is necessary to define cell populations by multiple features 17,19,23,24,30. Although the list of transgenic and knock-in animals expressing Cre− or Flp recombinase for marking specific neuronal types is growing, it is possible that many neurons defined by one molecular marker can be further differentiated by another molecular marker or specific features. Thus, viral strategies to efficiently and reliably deliver various transgenes in either neurons expressing both Cre and Flp (Cre+/Flp+), neurons expressing Cre, but not Flp (Cre+/Flp−), and/or neurons expressing Flp, but not Cre (Cre−/Flp+) separately are needed. Previously, the INTRSECT method was developed to achieve this 4. However, the efficiency of labeling and the expression level of different transgenes with this method have not been fully validated, so only a few transgenes have been useful to express intersectionally with this approach so far 4,5. In particular, the INTRSECT approach necessitates locating the intron insertion site and screening for transgenic expression for every gene to be delivered in order to express a new transgene. The screening process often proves difficult. Moreover, unwanted intron sequences are present in the expressed transgenes when this strategy is used.
To meet this challenge, we have designed a two-vector system. In one vector, a partial fragment of the ΦC31 sequence is inverted using the fDIO configuration, and the whole sequence is inverted with the cDIO configuration (Fig. 5A). Thus, only when both the Cre and Flp recombinases are expressed, the two fragments of the ΦC31 recombinase are oriented correctly to permit the expression of functional ΦC31 with a short Flp target site (F3) inserted within the recombinase (Supplementary Fig. 3A and 3B). This vector, named Cre+Flp+-ΦC31 can, in turn, control the expression of a transgene in the pSIO vector. In combination with the vector expressing a transgene in a ΦC31-dependent manner (pSIO vector), intact transgenes can be expressed in Cre+/Flp+ cells. (Fig. 5A). As intended, we found that the combination of Cre+Flp+-ΦC31 and pSIO-mScarlet-I vectors showed specific expression of mSclaret-I only when both Cre and Flp were expressed together in HEK293T cells (Fig. 5B). Furthermore, the injection of AAV-EF1α-Cre+Flp+-ΦC31 and AAV-EF1α-pSIO-mScarlet-I into hippocampal DG produces a strong expression of mScarlet-I only when both Cre and Flp recombinases were expressed together, demonstrating the specificity and efficacy of this system in vivo (n = 3 animals per each condition) (Fig. 5C). We also applied the same strategy using Bxb1 instead of ΦC31 recombinase, to generate Cre+Flp+-Bxb1 and found that this vector system shows high specificity but has lower efficiency compared to Cre+Flp+-ΦC31 in HEK293T cells (Supplementary Fig. 3C). Since we also found the leaky expression from bSIO vectors in vivo (Supplementary Figure 2A), we decided to focus on the Cre+Flp+-ΦC31 system.
Figure. 5: Two vectors strategy for expressing a transgene in cells expressing both Cre and Flp recombinases.
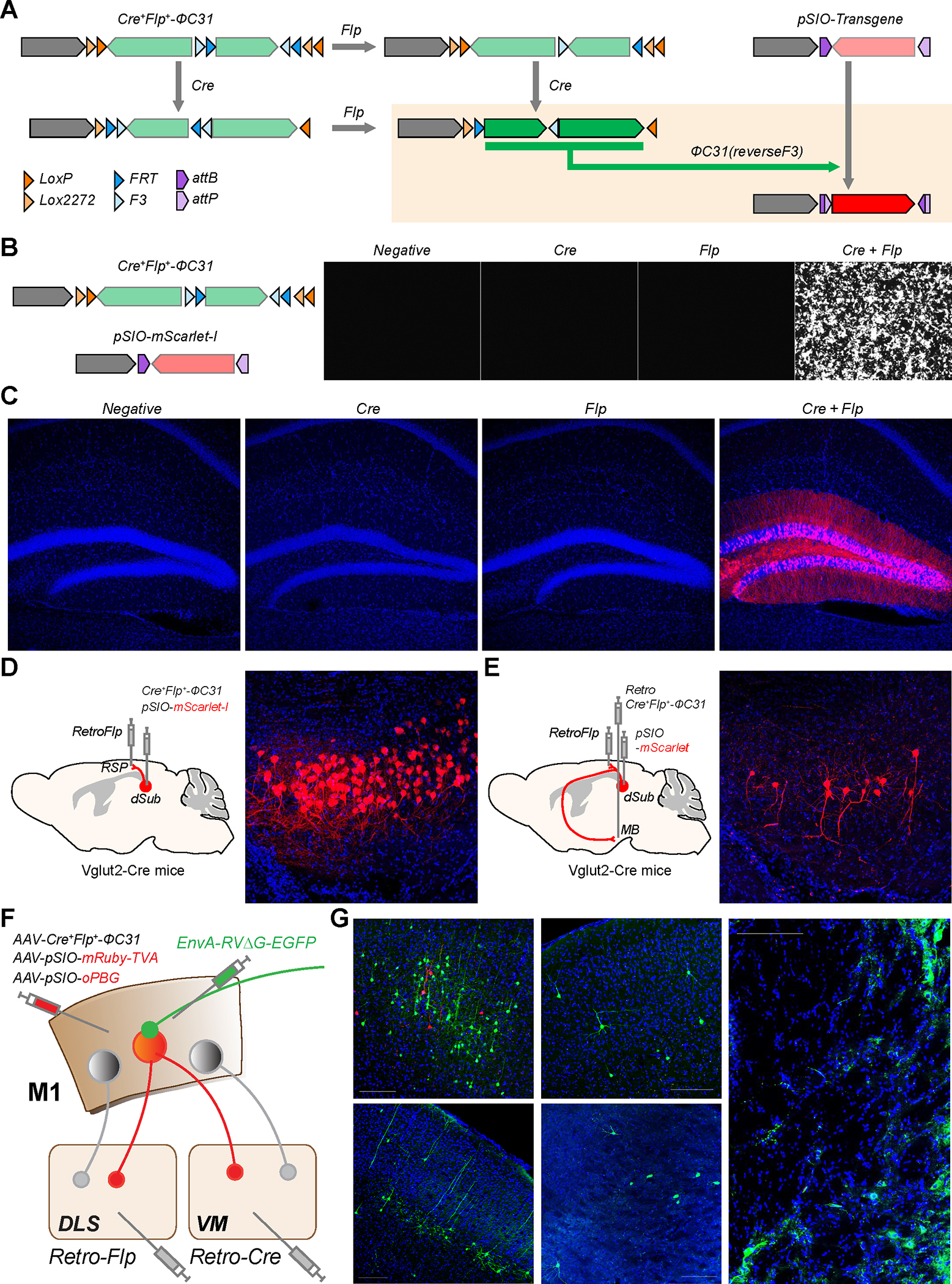
A, Design of Cre+Flp+-ΦC31 and the results of recombination on Cre+Flp+-ΦC31 and pSIO vector in the presence of Cre and/or Flp recombinases. B, Specificity of Cre+Flp+-ΦC31 and pSIO vector strategy in HEK293T cells. Cre+Flp+-ΦC31, pSIO-mScarlet-I, and Cre and/or Flp expressing plasmids were co-transfected. C, Specificity of Cre+Flp+-ΦC31 and pSIO vector strategy in vivo. AAV-EF1α-Cre+Flp+-ΦC31, AAV-EF1α-pSIO-mScarlet-I, and a combination of AAV-Cre and/or AAV-Flp were co-injected into DG. Scale bar = 50 μm D, Usage of Cre+Flp+-ΦC31 and pSIO vector strategy to express the transgene in cell population defined by two factors. AAV-EF1α-Cre+Flp+-ΦC31 and AAV-EF1α-pSIO-mScarlet-I were co-injected into dSub, and retroAAV-Flp was injected into RSP of VGLUT2-Cre transgenic mice. E, Usage of Cre+Flp+-ΦC31 and pSIO vector system to express the transgene in cell population defined by three factors. AAV-EF1α-pSIO-mScarlet-I, retroAAV-Flp, and retroAAV-EF1α-Cre+Flp+-ΦC31 were injected into dSub, RSP, and MB, respectively, of VGLUT2-Cre transgenic mice. d, e, Scale bar = 100 μm. F, Schematic showing a strategy for examining inputs to M1 neurons projecting to both DLS and VM. To express TVA and oPBG in M1 neurons projecting to both DLS and VM, we injected AAV-EF1α-Cre+Flp+-ΦC31, AAV-EF1α-pSIO-mRuby-TVA, and AAV-EF1α-pSIO-oPBG into M1, then injected retroAAV-Cre and retroAAV-Flp into VM and DLS, respectively. G, Image showing the injection sites showing starter cells (red) in M1 (top left), and inputs from other brain areas. cM1; contralateral side of M1, SS; somatosensory cortex, TH; motor thalamus, GPe; globus pallidus, external. Scale bar = 50 μm.
To further test the leakiness and specificity of Cre+Flp+-ΦC31/pSIO combination, we injected AAV-EF1α-Cre+Flp+-ΦC31and AAV-EF1α-pSIO-EGFP into the medial prefrontal cortex (mPFC) of VGluT1-Cre and VGAT-Flp mice, respectively. Three weeks after the injection, we found no leaky expression in the injected areas, suggesting that the Cre+Flp+-ΦC31/pSIO combination show very low level of leaky expression when only Cre or Flp recombinases are present (Supplementary Fig. 3D).
As noted earlier, neurons can also be defined using multiple features including both genetic markers and axonal target areas. For example, neurons in the dorsal subiculum (dSub) heterogeneously express vesicular glutamate transporters VGLUT1 or VGLUT2 and have distinct contributions to contextual memory formation and long-term memory storage 57,58. In addition to that, only a fraction of these neurons project to the retrosplenial cortex (RSP). Using our strategy, we successfully expressed a target transgene only in dSub VGLUT2+ neurons which project to the RSP by injecting AAV-EF1α-Cre+Flp+-ΦC31 and AAV-EF1α-pSIO-mScarlet-I in the dSub of VGLUT2-Cre transgenic mice and retroAAV-Flp into RSP (Fig. 5D). Moreover, we could apply an additional layer of specificity to target even more specific cell populations by delivering Cre+Flp+-ΦC31 in the retroAAV. We injected AAV-EF1α-pSIO-mScarlet-I in the dSub of VGLUT2-Cre transgenic mice and retroAAV-Flp and retroAAV-EF1α-Cre+Flp+-ΦC31 into RSP and mammillary body (MB), respectively 59. With this strategy, we found that mScarlet-I is strongly expressed in a subpopulation of VGLUT2+ dSub neurons that collateralize axonal projections to both RSP and MB (Fig. 5E). These results demonstrate that our Cre+Flp+ system can be used to express the transgene in cells defined by two or three features.
As well as features of output projection patterns, neurons may have distinct brain-wide inputs 28,29,33. Thus, tools to map inputs to specific cell types defined by multiple features are necessary. Delivering ΦC31 recombinase in neurons expressing both Cre and Flp recombinase can expand the tool to deliver any transgenes into Cre+/Flp+ neurons. Thus, to examine the brain-wide inputs to Cre+/Flp+ neurons which are defined by multiple features, we generated vectors expressing TVA receptors and rabies virus glycoprotein (RVG) in a ΦC31-dependent manner (AAV-hSyn-pSIO-mRuby-TVA and AAV-EF1α-pSIO-oPBG) 60,61. Using these vectors, we aimed to map inputs to M1 neurons projecting to both DLS and ventromedial thalamus (VM). We injected retroAAV-Cre and retroAAV-Flp into the VM and DLS, respectively, and injected AAV-EF1α-Cre+Flp+-ΦC31, AAV-hSyn-pSIO-mRuby-TVA, and AAV-EF1α-pSIO-oPBG into the M1. Two weeks later we injected a rabies virus, EnvA-RVΔG-EGFP, which targets TVA-expressing neurons, into M1 (Fig. 5F). With this approach, we found that we were able to label brain-wide inputs, including motor thalamus (TH), contralateral cortex, globus pallidus, and so on, to specific M1 neurons projecting to both the DLS and VM (Fig. 5G). Altogether, Cre+Flp+-ΦC31 system enables us to distinguish neural populations by their input/output connectivity features.
Overall, together with the pSIO vectors expressing various molecular tools (Fig. 3; Key Resource Table), we believe that we can provide novel approach to deliver intact genes into Cre+/Flp+ neurons easily and efficiently.
Delivery of different transgenes in cells expressing a different combination of Cre and Flp recombinases
In addition to the intersectional expression of transgenes in Cre+/Flp+ neurons, targeting cells that express Cre recombinase but not Flp recombinase or vice versa: Cre+/Flp− or Cre−/Flp+ cells, respectively, is also important to understand the cell type-specific roles of neurons. To achieve transgene expression in Cre+/Flp− cells, it is possible that Flp target sites can be placed upstream and downstream of the transgene in the cDIO configuration (Fig. 6A and Supplementary Fig. 4A). Cre recombination inverts the direction of the transgene, but the presence of Flp will excise the transgene regardless of Cre recombination – thus, preventing transgene expression. Although this approach is beginning to be used in the field 62, critical confounding issue, when simultaneously using Cre+/Flp+, Cre+/Flp−, and Cre−/Flp+ vectors, is that recombination can also occur between vectors with identical target sites, leading to significant false positives.
Figure. 6: Vector strategies for the delivery of different transgenes in cells expressing a different combination of Cre and Flp recombinases.
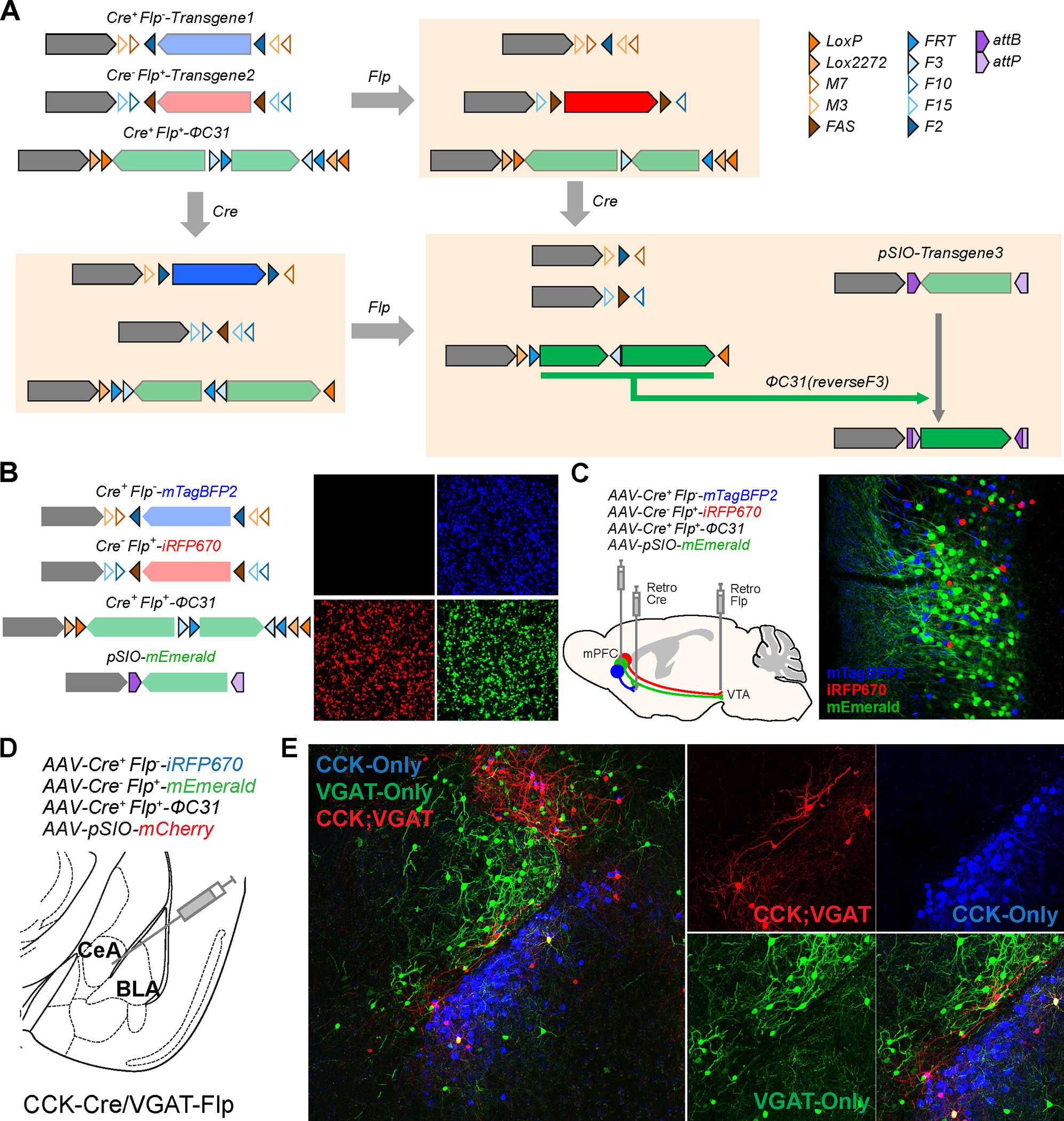
A, Design of Cre+Flp−, Cre−Flp+, and Cre+Flp+ vectors and the results of recombination. B, Exclusive expression of three transgenes in cells expressing a different combination of recombinases. Cre+Flp−-mTagBFP, Cre−Flp+-iRFP670, Cre+Flp+-ΦC31, pSIO-mEmerald, and Cre and/or Flp expressing plasmids were co-transfected in HEK293T cells. iRFP670 is pseudocolored in red. Scale bar = 50 μm. C, Simultaneous usage of Cre+Flp−, Cre−Flp+, and Cre+Flp+ vector strategies in vivo. AAV-EF1α-Cre+Flp−-mTagBFP2, AAV-EF1α-Cre−Flp+-iRFP670, AAV-EF1α-Cre+Flp+-ΦC31, and AAV-EF1α-pSIO-mEmerald were co-injected into mPFC, retroAAV-Cre and retroAAV-Flp were injected into NAc and VTA, respectively. Scale bar = 100 μm. D, Schematic of the injection of AAV-EF1α-Cre+Flp−-iRFP670, AAV-EF1α-Cre−Flp+-mEmerald, AAV-EF1α-Cre+Flp+-ΦC31, and AAV-EF1α-pSIO-mScarlet into the amygdala (at the border between the central amygdala (CeA) and basolateral amygdala (BLA)) of CCK-Cre/VGAT-Flp mice. We injected a mix of 150 nl of each virus (total of 600 nl). E, Confocal images showing overall amygdala areas injected (left) and images of CeA and BLA (right) with high magnification, showing that we could differential CCK-only, VGAT-only, and CCK- and VGAT expressing neurons with different fluorescent proteins.
For example, when we use several vectors containing overlapping and identical target sites for Cre or Flp recombinases (Cre+Flp−_v0.5-mTagBFP, Cre−Flp+_v0.5-iRFP670, Cre+Flp+-ΦC31; Supplementary Figure 4A), we found that the unwanted inter-vector recombination between identical Cre or Flp recognition sites happened, leading to the expression of all transgenes from all vectors transfected – indicating that these strategies cannot be reliably used together (Supplementary Figure 4B). Therefore, it is crucial to use different variants of target recombination site to avoid undesired inter-vector recombination that may lead to inappropriate transgene expression 62. To overcome this issue, we used the combination of different recognition sites for Cre and Flp recombinase. We have designed a new version of Cre+/Flp− and Cre−/Flp+ that contains completely exclusive sets of target sites (M7, M3, and FAS for Cre recombinase; F10, F15, F2 for Flp recombinase; Figure 6A and Supplementary Figure 4C) without any overlap among the vectors 63–67. With these new vector designs, we can exclusively and simultaneously express different transgenes in distinct cell populations containing different recombinase combinations in culture cells (Cre+/Flp−, Cre−/Flp+, and Cre+/Flp+; Fig. 6B).
Next, we tested whether the combination of Cre+/Flp+, Cre+/Flp−, and Cre−/Flp+ vectors can exclusively express different transgenes in vivo. To differentiate mPFC cells that project either to the NAc or VTA only, or to both target areas, we injected AAV-EF1α-Cre+Flp−-mTagBFP2, AAV-EF1α-Cre−Flp+-iRFP670, AAV-EF1α-Cre+Flp+-ΦC31, and AAV-EF1α-pSIO-mEmerald into the mPFC, and injected retroAAV-Cre and retroAAV-Flp into NAc and VTA, respectively (Fig. 6C). Three weeks after the injection, we found that the three different fluorescent proteins are exclusively expressed in three distinct populations: mPFC neurons projecting to both the NAc and VTA express mEmerald, those projecting only to the NAc express mTagBFP2, and those projecting only to the VTA express iRFP670 (Fig. 6C). We found no mEmerald expression in mTagBFP2 (0 cells in 246 mTagBFP2-expressing cells from 3 animals) or iRFP670 (0 cells in 187 iRFP670 expressing cells from 3 animals)-positive neurons. The TagBFP2-postive- and iRFP670-postive cells show no overwrapping expression at all. We should point out that this is the experiment showing a proof of principle. The results may differ depending on the virus titer and injection sites. At least in this experiment when we achieved a stable and robust expression of Cre and Flp recombinase using viral-mediated gene delivery, our vector designs can successfully separate three different neuronal populations by expressing different fluorescent proteins.
We further validated this strategy using transgenic mouse lines, in which brain area has molecular heterogeneity of well-known markers. It is reported that both basolateral amygdala (BLA) and central amygdala (CeA) have cholecystokinin (CCK)-positive neurons, and CCK neurons in BLA are glutamatergic and CCK neurons in CeA are GABAergic 68. Moreover, CeA area has non-CCK-positive GABAergic neurons. Thus, we tested whether our strategy can differentiate distinct CCK-positive GABAergic neurons, CCK-negative GABAergic neurons, and CCK-positive non-GABAergic neurons in the amygdala. We injected AAV-EF1α-Cre+Flp−-iRFP670, AAV-EF1α-Cre−Flp+-mEmerald, AAV-EF1α-Cre+Flp+-ΦC31, and AAV-EF1α-pSIO-mCherry into the border between the central amygdala (CeA) and basolateral amygdala (BLA) area of CCK-Cre::VGAT-Flp transgenic mice (Fig. 6D). Indeed, we found CCK-positive but non-GABAergic neurons (Cre+/Flp−) labeled with iRFP670 signals in the BLA, CCK-negative GABAergic neurons (Cre−/Flp+) with mEmerald in the CeA. We could also observe that CCK-positive GABAergic neurons (Cre+/Flp+) in both BLA and CeA were labeled with mCherry with a very small number of mCherry signals in iRFP670-positive neurons (8 cells in 276 iRFP670-positive neurons from 3 animals) and in mEmerald-positive neurons (11 cells in 312 mEmerald-positive neurons from 3 animals) (Fig. 6E). Although it is difficult to track the expression level and timing of recombinases in transgenic animals, we believe there is a possibility of minor leaky or non-specific expression of Cre and Flp recombinases in the mouse line we used because a small amount of short-term expression of recombinases is required for inducing recombination in viral vectors. These results suggest that our new multiplexed recombinases strategy can contribute to the understanding of heterogeneous neural circuit function in the same brain area in the same animal.
The intersectional gene delivery into cells expressing Cre, Flp, and ΦC31 recombinase
As the number of specific neuronal types is growing, the specific delivery of transgene into neurons possessing multiple features can be a future challenge. Thus, we extended our approach to develop a method to express transgenes in cells simultaneously expressing Cre, Flp, and ΦC31 recombinase; Cre+/Flp+/ΦC31+ neurons. The tetracycline-dependent transcription factor, tTA2s, from the Tet-Off system comprises two components, the DNA binding domain Tet repressor (TetR) domain and the transcription activation domain (AD) 69. Similar to the Cre+Flp+-ΦC31 vector design, the two components of the tTA2s are inverted separately and the peptide translated from the Flp target site was positioned between TetR and AD, creating Cre+Flp+-tTA2. The tTA2s with the peptide insertion are induced when both Cre and Flp are expressed. The ΦC31 is further required to invert the direction of the transgene in the pSIO vector driven by Tet Response Element promoter (pTRE), another component of the Tet-Off system which needs tTA2s for its expression. Therefore, the transgene expression requires the presence of Cre, Flp, and ΦC31 in the same cell (Fig. 7A). We confirmed that this strategy was successfully applied in HEK293T cells with high specificity (Fig. 7B). Next, to test this strategy in vivo, we injected AAV-EF1α-Cre+Flp+-tTA2s and AAV-pTRE-pSIO-mScarlet-I, as well as a combination of either AAV-Cre, AAV-Flp, and/or AAV-ΦC31 into the DG. We found that the transgene was strongly expressed when all three of Cre, Flp, and ΦC31 were expressed together (n = 3 animals per each group) (Fig. 7C).
Figure. 7: Two vectors strategy for expressing a transgene in cells with Cre, Flp, and ΦC31 expression.
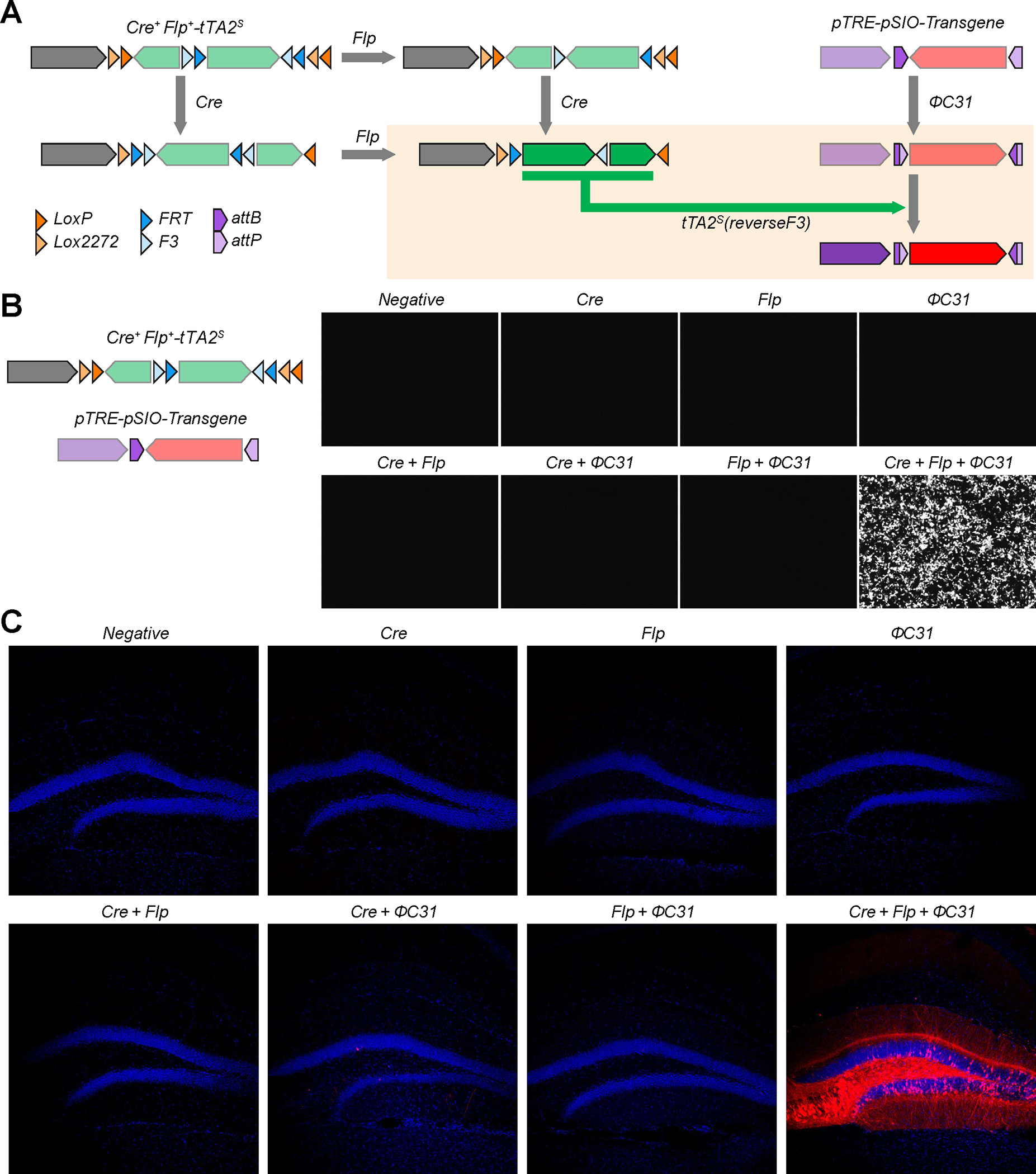
A, Design of vector strategy for Cre+Flp+ΦC31+ and the results of recombination. Cre and Flp recombinases induce the expression of tTA2s on the Cre+Flp+-tTA2s vector, while ΦC31 inverts the direction of the transgene in the pTRE-pSIO-transgene vector. tTA2s can drive transcription of the TRE promoter. B, Specificity of Cre+Flp+-tTA2s and pTRE-pSIO vector strategy in HEK293T cells. Cre+Flp+-tTA2s, pTRE-pSIO-mScarlet-I, and Cre, Flp, and/or ΦC31 expressing plasmids were co-transfected. C, Specificity of Cre+Flp+-tTA2s and pTRE-pSIO vector strategy in vivo. AAV-EF1α-Cre+Flp+-tTA2s, AAV-pTRE-pSIO-mScarlet-I, and combination of AAV-Cre, AAV-Flp, and/or AAV-ΦC31 were co-injected into DG. Scale bar = 50 μm
DISCUSSION
In the present study, we introduce novel recombinase system, ΦC31/pSIO system, that can be used together with Cre and Flp recombinases with high efficiency with minimal cross-reactivity for viral-mediated gene delivery as well as for knock-in mice to target specific cell types. In addition, we have designed new viral vector strategies that can express any intact transgenes in cells expressing any combination of Cre and Flp recombinases (e.g. Cre+/Flp+, Cre+/Flp−, and Cre−/Flp+). As these vectors are uniquely designed to contain no overlapping target sites among them, Cre+/Flp+, Cre+/Flp−, and Cre−/Flp+ vectors can be used together to express different transgenes without undesired crosstalk, providing an invaluable tool to study the interaction of specific cell types in highly heterogeneous tissue. Furthermore, we expanded our vector toolkit by designing a system to specifically mediate transgene expression in cells expressing all three recombinases: Cre, Flp, and ΦC31.
Although previous attempts to achieve the viral-mediated expression of specific transgenes in both Cre− and Flp−recombinase expressing neurons have been made 4–6, these strategies possess several drawbacks. Strategies using stop cassette flanked by recombinase target sites in addition to DIO configuration risks leaky transgene expression as the stop cassette strategy has been demonstrated to have a much higher background than the DIO strategy, especially, for viral-mediated gene delivery 6,7. Another strategy, termed INTRSECT, directly splits the transgene into multiple fragments and inverts the direction of these fragments 4,5,34. Although elegant, this strategy requires extensive designing and testing to confirm specificity, functionality, and efficiency for every new transgene, which significantly limits its versatility. Because of these limitations, only a few transgenes were validated for intersectional expression after the introduction of this strategy. Even in these validated transgenes, there are unwanted intron sequences within the open reading frame. We thought that this can be a major limitation because it will be very challenging for researchers to keep up with the use of novel optogenetic tools or activity indicators in the future. Moreover, this approach also precludes the expression of transgenes that may be hard to split into multiple fragments such as very small transgenes or non-coding RNAs, including microRNAs and microRNA-based shRNAs.
Our system is designed to express ΦC31 in neurons expressing both Cre− and FLP recombinases, specifically, and then to express the transgenes in a ΦC31-dependent manner. Because of the high efficiency of ΦC31 recombinase as Cre and Flp recombinases (Fig. 1, Supplementary Fig. 1), we achieved a robust and specific expression of many transgenes in Cre+ Flp+ neurons with our design. Therefore, our method will eliminate the difficult screening procedure required by the INTERSECT method to identify intron sites for the optimal expression of each transgene via Cre− and Flp−dependent recombinases. Since the original gene without intron sites will be expressed, the expression levels and functionality of transgenes will be uniform, eliminating the need for screening configurations for optimal expression, and providing researchers with a simple method to deliver different transgenes for various applications. Indeed, we showed that we can express various optogenetic and chemogenetic tools and GCaMP8m in a ΦC31-dependent manner, and express TVA and oPBG in Cre+/Flp+ neurons with this strategy, indicating that we can easily express various tools in Cre+/Flp+ neurons with our strategy in vivo. (Fig. 5). Our new suite of viral vector strategies described here is flexible, such that any intact transgene can be easily placed into the vector with a simple cloning process. This will allow the researcher to design the experiment to intersectionally express any transgenes without a challenging optimization process. Taken together, our suite of tools greatly improves and expands the recombinase toolkit by providing new orthogonal recombinases and modular viral vectors that can be used together with high efficiency and specificity without unwanted crosstalk between different recombinase systems.
We also designed vectors that can express different transgenes into distinct subpopulations of neurons simultaneously (e.g. Cre+ Flp+, Cre+ Flp−, Cre− Flp+). We showed that our unique design has very little cross-expression when we introduce the robust and stable expression of Cre and Flp recombinases in vivo and in cell culture (Fig. 6). However, we have to point out that there should be some variability based on the individual experimental application with different transgenic mouse line and viral tools. It has been reported anecdotally that the expression level of Cre and Flp recombinases in certain transgenic or knock-in mice can vary depending on brain regions and developmental stages, and it is difficult to measure and compare the expression level of recombinases directly in vivo. This variability in recombinase expression can produce non-specific expression with any viral-mediate gene delivery system. Our system should have similar issues. Thus, even though we have tested and validated the specificity and efficacy of our system in cell culture, viral-mediated delivery of recombinases in vivo, and in several transgenic mouse lines, further modification of the systems can be still necessary based on the experimental application.
In this study, we also generated a viral strategy to express tTA2 in both Cre− and Flp expressing neurons, for the reliable intersectional expression of transgenes. One advantage of using tTA is that we can control the timing of expression through the administration of doxycycline 70. The tTA expression, in the absence of doxycycline, will constitutively activate the expression of transgenes, but in the presence of doxycycline, the expression of transgenes will be interrupted. Thus, by controlling the timing and duration of doxycycline administration, we will be able to control the expression of transgenes in specific neural populations at specific times during experimental procedures. Using a tTA-dependent and ΦC31-dependent gene expression strategy, we also generated a reliable tool to express any transgenes in Cre−, Flp− and ΦC31-dependent manner. This approach will provide an additional instrument to control the expression of transgenes in specific neuronal populations and time.
With the advent of single-cell analysis methods, an increasing number of novel cell types are being reported recently. This identification of the cellular heterogeneity in the brain opens up a new direction in the field, understanding the roles of individual neurons as well as the meaning of heterogeneity in brain function 71–73. However, the molecular tools necessary to define the increasing numbers of novel cell types have not been fully available. Here, we developed and validated a viral strategy to target specific neuronal cell types using a novel recombinase combination, ΦC31/pSIO vector, without the cross-reactivity with Cre− and Flp recombinases. We develop easily applicable viral tools for the robust and efficient intersectional expression of various transgenes in neurons with 2 or 3 different features, which will provide valuable tools for the anatomical and functional understanding of heterogeneous neural circuitry.
STAR METHODS (Summary)
RESOURCES AVAILABILITY
Lead Contact
All reagents detailed in this manuscript are freely available for academic use. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Byung Kook Lim (bklim@ucsd.edu)
Materials availability
DNA constructs and viruses generated by the authors will be distributed to other research investigators upon request. All DNA constructs will be uploaded to ADDGENE (see Key Resources Table) upon the acceptance of the manuscript for publication. The mouse lines, SERT-P2A-PhiC31 and VGluT2-P2A-PhiC31, will be submitted for deposition at the Jackson Laboratory.
Data and code availability
The data and raw images that support the findings from this study are available from the Lead Contact upon request. No stand-alone code was generated for this project.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All procedures for the use of mice were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Diego. Mice were maintained on a 12/12 hours light/dark cycle with regular mouse chow and water available ad libitum. PV-Cre, SST-Flp, and VGLUT2-Cre homozygous transgenic mice were obtained from the Jackson Laboratory (008069, 028579, and 016963, respectively). PV-Cre/SST-Flp transgenic mice were produced by crossing PV-Cre homozygous mice with SST-Flp homozygous mice. Heterozygous VGluT2-Cre were produced by crossing VGLUT2-Cre mice with C57BL/6J. CCK-Cre/VGAT-Flp transgenic mice were produced by crossing CCK-Cre homozygous mice (Jackson Lab: 012706) with VGAT-Flp (Jackson lab: 029591) homozygous mice. VGluT2-P2A-PhiC31 and SERT-P2A-PhiC31 were newly generated in this study and back-crossed multiple times with C57BL6 before use.
METHOD DETAILS
Viral vectors
All plasmids were constructed using either restriction enzyme reaction/ligation or NEBuilder cloning kit. DNA for codon-optimized PhiC31 was a gift from Philippe Soriano (Addgene plasmid # 13795; http://n2t.net/addgene:13795; RRID:Addgene_13795). DNA for codon-optimized Bxb1 was a gift from Orion Weiner (Addgene plasmid # 119901; http://n2t.net/addgene:119901; RRID:Addgene_119901). All the vectors and sequence information will be deposited to addgene.
All AAV vectors used in this study were packaged as AAV serotype DJ and generated as previously described. In brief, AAV vectors were produced by calcium phosphate transfection of HEK293T cells with three plasmids: an AAV vector plasmid carrying target constructs, an AAV helper plasmid (pHelper, Agilent), and AAV rep-cap helper plasmid (pRC-DJ, a gift from M. Kay). 72 hours post-transfection, the cells were collected and lysed by three freeze-thaw cycles. Viral particles were purified by an iodixanol step-gradient ultracentrifugation followed by buffer exchange and concentration using a 100-kDa molecular cutoff ultrafiltration device (Millipore). The genomic titers of each AAV were determined by quantitative polymerase chain reaction. The AAV vectors were combined/diluted in PBS to working concentration.
Cell culture
HEK293T cells were cultured in DMEM supplemented with 10% FBS. Plasmids were transfected as a mixture by calcium phosphate transfection in 24-well plates and fluorescence images were taken by Olympus cellSens. For confocal imaging of Fig. 1c, 3b, and Supplementary Fig. 4, cells were cultured on Poly-D-Lysine treated round coverslips. After fixation with 4% paraformaldehyde (PFA), the coverslips were mounted on a slide glass using ProLong™ Glass Antifade Mountant (ThermoFisher Scientific). Images were acquired by Olympus FluoView FV1200 confocal microscope.
Generation of SERT-P2A-PhiC31 and VGluT2-P2A-PhiC31 knock-in mice
Sert-P2A-PhiC31o knock-in mice were created using Easi-CRISPR method47 at the Salk Transgenic Core Facility. To generate the Sert-P2A-PhiC31o knock-in mice, mouse zygotes were microinjected with “Cas9 protein: Sert-sgRNA1: Sert-sgRNA2: Sert-ssDNA-HDR template” ribonucleoprotein (RNP) mixtures at concentrations of 20ng/uL:10ng/uL:10ng/uL;10ng/ul. Injected zygotes were implanted into B6D2F1 pseudo-pregnant females. Successful knock-in pups were detected by PCR genotyping and confirmed by sequencing data. Cas9 protein was purchased from IDT (Alt-R® S.p. HiFi Cas9 Nuclease V3, 1081060). Sert-sgRNA1 (sequence: ACCAACGGAAAUUCCGUGUG) and Sert-sgRNA2 (sequence: AUCAGCUUGGCUUAGAGGGG) were purchased from Synthego. Sert-ssDNA-HDR template (2131 bp) was purchased from IDT (Megamer™ Single-Stranded DNA Fragments).
VGluT2-P2A-PhiC31o knock-in mice were created using Easi-CRISPR method47 at the Salk Transgenic Core Facility. To generate the VGluT2-P2A-PhiC31o knock-in mice, mouse zygotes were microinjected with “Cas9 protein: VGluT2-sgRNA1: VGluT2-sgRNA2: VGluT2-ssDNA-HDR template” ribonucleoprotein (RNP) mixtures at concentrations of 20ng/uL:10ng/uL:10ng/uL;10ng/ul. Injected zygotes were implanted into B6D2F1 pseudo-pregnant females. Successful knock-in pups were detected by PCR genotyping and confirmed by sequencing data. Cas9 protein was purchased from IDT (Alt-R® S.p. HiFi Cas9 Nuclease V3, 1081060). VGluT2-sgRNA1 (sequence: GCAAGACGCGUACACCUAUA) and VGluT2-sgRNA2 (sequence: AUCGUUAUGAAUAAUCAUCU) were purchased from Synthego. VGluT2-ssDNA-HDR template (2128 bp) was purchased from IDT (Megamer™ Single-Stranded DNA Fragments).
Stereotaxic injection
All AAV injections were conducted on adult mice at age of 2 months or more. Mice were anesthetized with ketamine (100 mg/kg) and dexmedetomidine (1 mg/kg) and placed in a stereotaxic instrument (David Kopf Instruments). Stereotaxic coordinates for each injection site were as followed; DG (AP−2.0, ML+1.5, DV−2.1), CA1 (AP−2.0, ML+1.5, DV−1.75), mPFC (AP+1.9, ML+0.3, DV−2.75), VTA (AP−3.0, ML+0.5, DV−4.4), BLA (AP−1.3, ML+2.9, DV−4.6), NAc (AP+1.3, ML+0.5, DV−4.5), dSub (AP−3.2, ML+1.8, DV−1.7), RSP (AP-2.5, ML+0.25, DV−0.95), MB (AP−2.8, ML+0.3, DV−5.25).
In most experiments, the working titers for retro AAVs were 1~21012 GC/mL and the injection volume was 200~400 nl. However, we do have some modifications in the specific sets of experiments. For Figure 1F, the titer for AAV-CaMKIIα-ΦC31 was 2×108 GC/mL to sparsely label pyramidal CA1 neurons, and all other titers were 1~2×1011 GC/mL. For Figure 5F–G, the titer for the rabies virus was 1.4×109 GC/mL. For the experiment in Figure 6D–E, we injected a mix of 150 nl of each virus (total of 600 nl). Thus, the particle amount for the individual virus injected is lower than in other experiments. For the experiment in Supplement Figure 2B, we injected a mix of 80 nl of each virus (total 480 nl).
Brain preparation and confocal imaging
3 weeks after AAV injection, the mice were perfused with Saline followed by 4% PFA. Perfused brains were fixed with 4% PFA overnight at 4°C and the PFA was replaced by PBS after the fixation. Brains were sliced into 50μm sections by Vibratome and mounted in ProLong™ Glass Antifade Mountant. Slices that have mTagBFP2 expression were immunostained with rabbit Anti-tRFP antibody (Evrogen) and Goat anti-Rabbit IgG Alexa Fluor 568 (ThermoFisher Scientific) before mounting. Images were acquired by Olympus FluoView FV1200 confocal microscope.
Optogenetic stimulation-induced conditioned place avoidance (CPA) test
To stimulate the axonal terminals of VTA glutamatergic neurons in LHb, we injected AAV-EF1α-pSIO-ChR2-EYFP into the VTA, then implanted optic fibers into LHb. Two weeks after the surgery, mice were subjected to the conditioned place avoidance test in the three-chamber apparatus (68.58 (w) × 22.86 (l) × 25.4 (h) cm). Two side chambers of identical dimensions (26.7 × 22.86 cm) were configured differently, one with black and white patterns on the walls and a rough floor material, and the other with black and white squares on the walls and a smooth floor material. On day one, mice were placed in the center chamber for five minutes before being allowed to freely investigate the entire apparatus for thirty minutes (pre-test) to establish a preference baseline. On days 2 and 3, mice were confined to one of the side chambers for 15 min under the optogenetic stimulation (473nm laser, 5mW, 20Hz, 10ms, Opto Engine LLC) and consecutively to the other side of the chamber without optical stimulation for another 15 min. Optic stimulation in left or right chambers and stimulation treatment order were counterbalanced across mice. Optic stimulation in the left or right chambers and the order of stimulation treatments were randomized among rodents. On day four, identical to day one, mice were placed in the center chamber for five minutes and allowed to explore the complete apparatus for thirty minutes (post-test). Using a camcorder, locomotor activities were captured and analyzed using the EthoVision XT 13 software (Noldus Information Technology).
Immunohistochemistry and cell counting
Mice were anesthetized with the 1–2% isoflurane and then perfused with phosphate buffered saline (PBS) followed by 4% Paraformaldehyde (PFA). The dissected brains were fixed with 4% PFA overnight at 4°C and then sliced into 40 μm coronal sections using a Vibratome (Leica VT1000S). For immunostaining, brain slices were incubated overnight at 4°C with primary antibodies in a PBS solution containing 0.3% Triton X-100, followed by 1 hour at room temperature with secondary antibodies in a PBS solution containing 0.3% Triton X-100. Images were acquired by a confocal microscope using an 20x objective (N.A. 0.75) (Leica Stellaris 5). Cell counting was performed using Fiji (ImageJ).
Multiplex fluorescent in situ hybridization
Mice (VGluT2-P2A-PhiC31) that underwent surgeries for targeting glutamatergic neurons in mediodorsal thalamus (MD) were deeply anesthetized with isoflurane and transcardially perfused with ice-cold PBS. The brains were extracted, submerged in embedding medium (Tissue-Tek O.C.T.; Sakura), and frozen with 2-methylbutane (Sigma) chilled with dry ice in 70% ethanol. The frozen brain blocks were stored at −20°C for at least one day, then sliced with a cryostat microtome (Thermo-Fisher) to obtain 20-μm coronal sections. For each brain, 8 sections regularly sampled across the entire dorsal striatum were mounted on SuperFrost Plus slides (Fisher Scientific) and processed exactly as described in the RNAscope assay online protocol (ACD; Advanced Cell Diagnostics). We used probe against EGFP mRNA to label the PhiC31 expressing MD neurons. Probes against VGluT2 mRNAs were used to determine whether the labeled MD neurons were VGluT2-expressing glutamatergic neurons. All probes were purchased from ACD. Because the labeling of eGFP mRNA was the strongest, we assigned the Atto-647 secondary probe to the EGFP probe to prevent the bleed-through of the fluorescent signal into neighboring channels during imaging. Images of the MD were acquired through a 20x objective (N.A. 0.75) on a confocal microscope (Leica Stellaris 5). All eGFP-labeled neurons were manually quantified.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics
Statistical analyses for Figure 4 were performed with GraphPad Prism 6 or MATLAB R2015b. Final samples sizes were estimated using power analysis for a subset of behavioral experiments. Comparisons between two groups with normally distributed data were made using the Two-way repeated measure ANOVA, Holm-sidak multiple comparison.
Supplementary Material
Highlight.
The novel recombinase system, ΦC31/pSIO system, for viral-mediated gene delivery
Creation of two transgenic mouse lines for cell-type specific expression of ΦC31
Two vectors strategy to express any transgenes interjectionally in Cre+/Flp+ neurons
Strategy for the temporal expression of transgenes in specific neuronal populations.
ACKNOWLEDGEMENTS
We thank Dr. Tom Hnasko as well as Lim lab members for the helpful discussion. We thank J. Lee, S. Mo and Y. Lee for testing our viruses in vivo. This work was supported by NIH grant R01NS121231, R01DA049787, U01MH114829, and NSF grant (IOS-2022241). M.J., Q.W., and J.-H. C. was supported by an NIH T32 training grant (5T32NS007220). D.H.S is supported by the National Research Foundation of Korea (NRF-2018R1A5A2023879).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information consists of Figures S1–S4.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Kuhlman SJ, and Huang ZJ (2008). High-resolution labeling and functional manipulation of specific neuron types in mouse brain by Cre-activated viral gene expression. PLoS One 3, e2005. 10.1371/journal.pone.0002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo L, Callaway EM, and Svoboda K (2008). Genetic dissection of neural circuits. Neuron 57, 634–660. 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian X, and Zhou B (2021). Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. J Biol Chem 296, 100509. 10.1016/j.jbc.2021.100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods 11, 763–772. 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenno LE, Ramakrishnan C, Kim YS, Evans KE, Lo M, Vesuna S, Inoue M, Cheung KYM, Yuen E, Pichamoorthy N, et al. (2020). Comprehensive Dual- and Triple-Feature Intersectional Single-Vector Delivery of Diverse Functional Payloads to Cells of Behaving Mammals. Neuron 107, 836–853 e811. 10.1016/j.neuron.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. (2015). Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958. 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sohal VS, Zhang F, Yizhar O, and Deisseroth K (2009). Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 459, 698–702. 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480 e422. 10.1016/j.cell.2018.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerfen CR, Paletzki R, and Heintz N (2013). GENSAT BAC cre-recombinase driver lines to study the functional organization of cerebral cortical and basal ganglia circuits. Neuron 80, 1368–1383. 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, and Gerfen CR (2007). Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci 27, 9817–9823. 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang ZJ, and Zeng H (2013). Genetic approaches to neural circuits in the mouse. Annu Rev Neurosci 36, 183–215. 10.1146/annurev-neuro-062012-170307. [DOI] [PubMed] [Google Scholar]
- 12.Callaway EM, and Luo L (2015). Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci 35, 8979–8985. 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimidschstein J, Chen Q, Tremblay R, Rogers SL, Saldi GA, Guo L, Xu Q, Liu R, Lu C, Chu J, et al. (2016). A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat Neurosci 19, 1743–1749. 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, and Ghyselinck NB (2003). A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21, 562–565. 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 15.Iourov IY, Vorsanova SG, and Yurov YB (2012). Single cell genomics of the brain: focus on neuronal diversity and neuropsychiatric diseases. Curr Genomics 13, 477–488. 10.2174/138920212802510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030 e1016. 10.1016/j.cell.2018.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cadwell CR, Palasantza A, Jiang X, Berens P, Deng Q, Yilmaz M, Reimer J, Shen S, Bethge M, Tolias KF, et al. (2016). Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat Biotechnol 34, 199–203. 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Teichmann SA, and Meyer KB (2018). From Tissues to Cell Types and Back: Single-Cell Gene Expression Analysis of Tissue Architecture. Annu Rev Biomed Da S 1, 29–51. 10.1146/annurev-biodatasci-080917-013452. [DOI] [Google Scholar]
- 19.Gupta RK, and Kuznicki J (2020). Biological and Medical Importance of Cellular Heterogeneity Deciphered by Single-Cell RNA Sequencing. Cells 9. 10.3390/cells9081751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz-Manchado AB, Gonzales CB, Zeisel A, Munguba H, Bekkouche B, Skene NG, Lonnerberg P, Ryge J, Harris KD, Linnarsson S, and Hjerling-Leffler J (2018). Diversity of Interneurons in the Dorsal Striatum Revealed by Single-Cell RNA Sequencing and PatchSeq. Cell Rep 24, 2179–+. 10.1016/j.celrep.2018.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Papalexi E, and Satija R (2018). Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18, 35–45. 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 22.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 23.Fuzik J, Zeisel A, Mate Z, Calvigioni D, Yanagawa Y, Szabo G, Linnarsson S, and Harkany T (2016). Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat Biotechnol 34, 175–183. 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hook PW, McClymont SA, Cannon GH, Law WD, Morton AJ, Goff LA, and McCallion AS (2018). Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am J Hum Genet 102, 427–446. 10.1016/j.ajhg.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e519. 10.1016/j.cell.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poulin JF, Gaertner Z, Moreno-Ramos OA, and Awatramani R (2020). Classification of Midbrain Dopamine Neurons Using Single-Cell Gene Expression Profiling Approaches. Trends Neurosci 43, 155–169. 10.1016/j.tins.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiklova K, Bjorklund AK, Lahti L, Fiorenzano A, Nolbrant S, Gillberg L, Volakakis N, Yokota C, Hilscher MM, Hauling T, et al. (2019). Single-cell RNA sequencing reveals midbrain dopamine neuron diversity emerging during mouse brain development. Nat Commun 10, 581. 10.1038/s41467-019-08453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knowland D, Lilascharoen V, Pacia CP, Shin S, Wang EH, and Lim BK (2017). Distinct Ventral Pallidal Neural Populations Mediate Separate Symptoms of Depression. Cell 170, 284–297 e218. 10.1016/j.cell.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilascharoen V, Wang EH, Do N, Pate SC, Tran AN, Yoon CD, Choi JH, Wang XY, Pribiag H, Park YG, et al. (2021). Divergent pallidal pathways underlying distinct Parkinsonian behavioral deficits. Nat Neurosci 24, 504–515. 10.1038/s41593-021-00810-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin R, Wang R, Yuan J, Feng Q, Zhou Y, Zeng S, Ren M, Jiang S, Ni H, Zhou C, et al. (2018). Cell-type-specific and projection-specific brain-wide reconstruction of single neurons. Nat Methods 15, 1033–1036. 10.1038/s41592-018-0184-y. [DOI] [PubMed] [Google Scholar]
- 31.Sorensen SA, Bernard A, Menon V, Royall JJ, Glattfelder KJ, Desta T, Hirokawa K, Mortrud M, Miller JA, Zeng H, et al. (2015). Correlated gene expression and target specificity demonstrate excitatory projection neuron diversity. Cereb Cortex 25, 433–449. 10.1093/cercor/bht243. [DOI] [PubMed] [Google Scholar]
- 32.Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, et al. (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78. 10.1038/s41586-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pribiag H, Shin S, Wang EH, Sun F, Datta P, Okamoto A, Guss H, Jain A, Wang XY, De Freitas B, et al. (2021). Ventral pallidum DRD3 potentiates a pallido-habenular circuit driving accumbal dopamine release and cocaine seeking. Neuron 109, 2165–2182 e2110. 10.1016/j.neuron.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenno LE, Mattis J, Ramakrishnan C, and Deisseroth K (2017). A Guide to Creating and Testing New INTRSECT Constructs. Curr Protoc Neurosci 80, 4 39 31–34 39 24. 10.1002/cpns.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sabatini PV, Wang J, Rupp AC, Affinati AH, Flak JN, Li C, Olson DP, and Myers MG (2021). tTARGIT AAVs mediate the sensitive and flexible manipulation of intersectional neuronal populations in mice. Elife 10. 10.7554/eLife.66835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond CS, and Soriano P (2007). High-Efficiency FLP and Phi C31 Site-Specific Recombination in Mammalian Cells. Plos One 2. ARTN e162 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, and Calos MP (2001). Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol 21, 3926–3934. 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turan S, and Bode J (2011). Site-specific recombinases: from tag-and-target- to tag-and-exchange-based genomic modifications. Faseb J 25, 4088–4107. 10.1096/fj.11-186940. [DOI] [PubMed] [Google Scholar]
- 39.Xu ZY, Thomas L, Davies B, Chalmers R, Smith M, and Brown W (2013). Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. Bmc Biotechnol 13. Artn 87 10.1186/1472-6750-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang FJ, Chen CN, Chang T, Cheng TW, Chang NC, Kao CY, Lee CC, Huang YC, Hsu JC, Li J, et al. (2022). phiC31 integrase for recombination-mediated single-copy insertion and genome manipulation in Caenorhabditis elegans. Genetics 220. 10.1093/genetics/iyab206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateman JR, Lee AM, and Wu CT (2006). Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173, 769–777. 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lui JH, Nguyen ND, Grutzner SM, Darmanis S, Peixoto D, Wagner MJ, Allen WE, Kebschull JM, Richman EB, Ren J, et al. (2021). Differential encoding in prefrontal cortex projection neuron classes across cognitive tasks. Cell 184, 489–506 e426. 10.1016/j.cell.2020.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Au-Young SM, Shen H, and Yang CR (1999). Medial prefrontal cortical output neurons to the ventral tegmental area (VTA) and their responses to burst-patterned stimulation of the VTA: neuroanatomical and in vivo electrophysiological analyses. Synapse 34, 245–255. . [DOI] [PubMed] [Google Scholar]
- 45.Sesack SR, Deutch AY, Roth RH, and Bunney BS (1989). Topographical organization of the efferent projections of the medial prefrontal cortex in the rat: an anterograde tract-tracing study with Phaseolus vulgaris leucoagglutinin. J Comp Neurol 290, 213–242. 10.1002/cne.902900205. [DOI] [PubMed] [Google Scholar]
- 46.Anastasiades PG, and Carter AG (2021). Circuit organization of the rodent medial prefrontal cortex. Trends Neurosci 44, 550–563. 10.1016/j.tins.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, et al. (2017). Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18, 92. 10.1186/s13059-017-1220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang KW, Ochandarena NE, Philson AC, Hyun M, Birnbaum JE, Cicconet M, and Sabatini BL (2019). Molecular and anatomical organization of the dorsal raphe nucleus. Elife 8. 10.7554/eLife.46464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Friedmann D, Xiong J, Liu CD, Ferguson BR, Weerakkody T, DeLoach KE, Ran C, Pun A, Sun Y, et al. (2018). Anatomically Defined and Functionally Distinct Dorsal Raphe Serotonin Sub-systems. Cell 175, 472–487 e420. 10.1016/j.cell.2018.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gradinaru V, Thompson KR, and Deisseroth K (2008). eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol 36, 129–139. 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8, 1263–1268. 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 52.Lin JY, Lin MZ, Steinbach P, and Tsien RY (2009). Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J 96, 1803–1814. 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nat Methods 11, 338–346. 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Rozsa M, Liang Y, Bushey D, Wei Z, Zheng J, Reep D, Broussard GJ, Tsang A, Tsegaye G, et al. (2023). Fast and sensitive GCaMP calcium indicators for imaging neural populations. Nature 615, 884–891. 10.1038/s41586-023-05828-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mondoloni S, Mameli M, and Congiu M (2022). Reward and aversion encoding in the lateral habenula for innate and learned behaviours. Transl Psychiatry 12, 3. 10.1038/s41398-021-01774-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Root DH, Mejias-Aponte CA, Qi J, and Morales M (2014). Role of glutamatergic projections from ventral tegmental area to lateral habenula in aversive conditioning. J Neurosci 34, 13906–13910. 10.1523/JNEUROSCI.2029-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wozny C, Beed P, Nitzan N, Possnecker Y, Rost BR, and Schmitz D (2018). VGLUT2 Functions as a Differential Marker for Hippocampal Output Neurons. Front Cell Neurosci 12, 337. 10.3389/fncel.2018.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamawaki N, Corcoran KA, Guedea AL, Shepherd GMG, and Radulovic J (2019). Differential Contributions of Glutamatergic Hippocampal-->Retrosplenial Cortical Projections to the Formation and Persistence of Context Memories. Cereb Cortex 29, 2728–2736. 10.1093/cercor/bhy142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kinnavane L, Vann SD, Nelson AJD, O’Mara SM, and Aggleton JP (2018). Collateral Projections Innervate the Mammillary Bodies and Retrosplenial Cortex: A New Category of Hippocampal Cells. eNeuro 5. 10.1523/ENEURO.0383-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, Conzelmann KK, Young JA, and Callaway EM (2007). Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647. 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim EJ, Jacobs MW, Ito-Cole T, and Callaway EM (2016). Improved Monosynaptic Neural Circuit Tracing Using Engineered Rabies Virus Glycoproteins. Cell Rep 15, 692–699. 10.1016/j.celrep.2016.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saunders A, Johnson CA, and Sabatini BL (2012). Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front Neural Circuits 6, 47. 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Langer SJ, Ghafoori AP, Byrd M, and Leinwand L (2002). A genetic screen identifies novel non-compatible loxP sites. Nucleic Acids Res 30, 3067–3077. 10.1093/nar/gkf421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee G, and Saito I (1998). Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216, 55–65. 10.1016/s0378-1119(98)00325-4. [DOI] [PubMed] [Google Scholar]
- 65.Missirlis PI, Smailus DE, and Holt RA (2006). A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination. BMC Genomics 7, 73. 10.1186/1471-2164-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlake T, and Bode J (1994). Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry 33, 12746–12751. 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 67.Turan S, Kuehle J, Schambach A, Baum C, and Bode J (2010). Multiplexing RMCE: versatile extensions of the Flp-recombinase-mediated cassette-exchange technology. J Mol Biol 402, 52–69. 10.1016/j.jmb.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 68.Shen CJ, Zheng D, Li KX, Yang JM, Pan HQ, Yu XD, Fu JY, Zhu Y, Sun QX, Tang MY, et al. (2019). Cannabinoid CB1 receptors in the amygdalar cholecystokinin glutamatergic afferents to nucleus accumbens modulate depressive-like behavior. Nat Med 25, 337–349. 10.1038/s41591-018-0299-9. [DOI] [PubMed] [Google Scholar]
- 69.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, and Hillen W (2000). Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci U S A 97, 7963–7968. 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gossen M, and Bujard H (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A 89, 5547–5551. 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guan J, Wang Y, Lin Y, Yin Q, Zhuang Y, and Ji G (2020). Cell Type-Specific Predictive Models Perform Prioritization of Genes and Gene Sets Associated With Autism. Front Genet 11, 628539. 10.3389/fgene.2020.628539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McKenzie AT, Wang MH, Hauberg ME, Fullard JF, Kozlenkov A, Keenan A, Hurd YL, Dracheva S, Casaccia P, Roussos P, and Zhang B (2018). Brain Cell Type Specific Gene Expression and Co-expression Network Architectures. Sci Rep-Uk 8. ARTN 8868 10.1038/s41598-018-27293-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deco G, Kringelbach ML, Arnatkeviciute A, Oldham S, Sabaroedin K, Rogasch NC, Aquino KM, and Fornito A (2021). Dynamical consequences of regional heterogeneity in the brain’s transcriptional landscape. Sci Adv 7. 10.1126/sciadv.abf4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and raw images that support the findings from this study are available from the Lead Contact upon request. No stand-alone code was generated for this project.


