Abstract
In humans, there are two isoforms each of clathrin heavy chain (CHC17 and CHC22) and light chain (LCa and LCb) subunits, all encoded by separate genes. CHC17 forms the ubiquitous clathrin-coated vesicles that mediate membrane traffic. CHC22 is implicated in specialized membrane organization in skeletal muscle. CHC17 is bound and regulated by LCa and LCb, whereas CHC22 does not functionally interact with either light chain. The imbalanced interactions between clathrin subunit isoforms suggest a distinct evolutionary history for each isoform pair. Phylogenetic and sequence analysis placed both heavy and light chain gene duplications during chordate evolution, 510-600 million years ago. Genes encoding CHC22 orthologues were found in several vertebrate species, with only a pseudogene present in mice. Multiple paralogons surrounding the CHC genes (CLTC and CLTD) were identified, evidence that genomic or large-scale gene duplication produced the two CHC isoforms. In contrast, clathrin light chain genes (CLTA and CLTB) apparently arose by localized duplication, within 1-11 million years of CHC gene duplication. Analysis of sequence divergence patterns suggested that structural features of the CHCs were maintained after gene duplication, but new interactions with regulatory proteins evolved for the CHC22 isoform. Thus, independent mechanisms of gene duplication expanded clathrin functions, concomitant with development of neuromuscular sophistication in chordates.
Keywords: phylogenetic, membrane traffic, coated vesicle, CHC17, CHC22
Analysis of genetic evolution is informative for characterizing protein isoform diversification. Genomic analyses of the Hox gene clusters (1) and genes of the MHC (2) have revealed coevolution of proteins that function together, as well as mechanisms of gene duplication that facilitated sophistication of the vertebrate nervous and immune systems. Protein evolution in membrane traffic pathways, which also diversified for neuromuscular and immunological sophistication, has not been extensively analyzed. A single previous phylogenetic study analyzed the adaptor and COP proteins, both of which mediate interaction between receptors and membrane vesicle coats (3). These two related protein families apparently diverged early in eukaryotic evolution, as yeast and humans have identifiable orthologues of both gene families. Here we extend evolutionary analysis to the vesicle coat protein clathrin and find that clathrin gene duplication occurred later, in conjunction with chordate evolution.
In humans, there are two isoforms of clathrin heavy chain (CHC), encoded by genes CLTC and CLTD at genomic loci 17q23.2 for CHC17 (4) and 22q11.21 for CHC22 (5-8). The CHC17 isoform trimerizes to form a triskelion (three-legged molecule) with each leg bound by a regulatory light chain (LC) subunit (9). Clathrin triskelia self-assemble into a polyhedral protein coat attached to membrane vesicles by adaptor molecules that sequester receptors into the vesicle. The resulting coated vesicles sort proteins during endocytosis and organelle biogenesis. CHC17 is expressed ubiquitously in vertebrate tissues and a functional orthologue is present in all eukaryotic organisms analyzed. The CHC22 isoform is highly expressed in human skeletal muscle, with a low level detected in other tissues. CHC22 is concentrated at neuromuscular and myotendinous junctions, and its expression increases during myogenesis and muscle regeneration (10, 11). Thus, CHC22 appears to have a role distinct from CHC17 in specialized muscle membrane organization. CHC17 and CHC22 have a remarkably high protein sequence identity (85%), despite their evident differences in function. A further conundrum in CHC diversification was posed when the gene encoding CHC22 was not found (12) in the paralogous region on mouse chromosome 16 that corresponds to its location in humans. This finding suggested that CHC22 either arose from a human-specific gene duplication or was lost in mice.
In contrast to the CHC isoforms, the two vertebrate clathrin LCs, LCa and LCb, have more divergent sequences (60% identity), but their functions are more similar. LCa and LCb both bind and regulate CHC17, but do not functionally interact with CHC22 (10, 11). LCa and LCb are encoded on separate human chromosomes by genes CLTA and CLTB, at 9p13.3 and 5q35.2, respectively. Both have additional isoforms arising from neuron-specific splicing differences. LCs are implicated in regulation of clathrin assembly and function (9, 13) and the two LCs are expressed at characteristically different levels in different vertebrate tissues. This finding suggests that LCs control distinct tissue-specific functions of CHC17. Invertebrates and yeasts have a single LC gene (9), which apparently partners with the CHC17 functional orthologue in these species.
Evolutionary studies of duplicated genes indicate that large scale genomic duplications, perhaps as extensive as one or two rounds of whole genome duplication, occurred during chordate evolution (14). Local gene duplication has also occurred at a constant rate throughout evolutionary time and together with large scale duplication has contributed to the increased biological complexity leading to vertebrate evolution (15). The present study applies rigorous genome analysis to define the duplication mechanisms and the divergence rates by which the CHC and LC gene families evolved, gaining insight into the functional role of the resulting isoform diversity.
Materials and Methods
See Supporting Text, Tables 1-4, and Figs. 6-11, which are published as supporting information on the PNAS web site, for more detail.
Data Sets. CHC and LC amino acid data sets and paralogon data sets were assembled following literature and database searches. Amino acid sequences were used for all but one paralogon data set (YPEL1/YPEL2). Alignments were made by using Genetics Computer Group (16) pileup, vector nti suite 7.0 (InforMax), alignx or mafft (17). DNA and/or protein accession codes are noted in Tables 1-3.
Phylogenetic Analyses. Bayesian phylogenetic analyses used mrbayes3b4 (18). Sampling was with one cold chain and three heated chains (temperature of 0.2), which were run for 300,000 generations (500,000 for paralogons). Trees were sampled every 100th generation, and the first 350 trees (1,000 for paralogons) were discarded before a consensus tree was generated. Convergence occurred well before reaching these limits. Amino acid analyses used a poisson model (blossum for paralogons) and gamma distances (four categories). The nucleotide analysis (YPEL1/YPEL2 data set) used a codon-based GTR+G+I model of substitution. Trees were rooted at the midpoint. Parsimony analyses were with paup* version 4.0b10 (19), using the tree bisection-reconnection branch-swapping algorithm, heuristic searches and 1,000 replicates. Tree topologies obtained with the two methods were statistically compared by using the Templeton test with a parsimony model, as implemented in paup* version 4.0b10. In all analyses, the test failed to reject one of the two topologies (α = 0.05).
Divergence Time Estimations. Divergence times were estimated by using the Bayesian relaxed molecular clock approach with the multidistribute program package (20). Vertebrate and urochordate sequences that were complete or almost complete were analyzed. To root the tree, the LC data set also included Aplysia californica, and the CHC data set included three protostome sequences (Drosophila melanogaster, Anopheles gambiae, and Aedes aegyptii). The tree topologies for the divergence time analyses were generated by mrbayes3b4, with the same parameters as for the paralogon analysis. In both data sets, the root of the ingroup tree was set at 595 ± 32.5 million years ago (MYA), corresponding to the urochordate-vertebrate separation. An alternative older separation was explored by using 715 ± 92.5 MYA. Several internal nodes were constraints: the synapsid-diapsid split (306-332 MYA), the lissamphibian-amniote separation (338-385 MYA), and the Actinopterygian-Sarcopterygian split (>411 MYA). We also investigated the age difference between the CHC and LC duplications where the urochordate-vertebrate divergence time was constrained at the same value for both data sets. In this analysis the divergence time was set at the average of the values obtained with the CHC and LC data sets, with a small standard deviation (±1 million years).
Creation of Paralogue Maps. Paralogous regions identified by an automated analysis (ref. 21 and http://wolfe.gen.tcd.ie/dup) were used as a starting point. The genes surrounding CLTC or CLTA were searched by blast for the genomic location of the most closely related family members. For candidate paralogons, data sets were constituted and investigated by phylogenetic analysis as described above. For the CHC paralogon data set, the probability of six or seven paralogons being located within these sequences by chance was determined by using the calculation p = L2/N(K-1), using n = 26,588 as the estimated number of proteins in the human genome (22), where L = ∑ for all X from X = K - 2 to X = J - 2, and K = 6 or 7 paralogons within a continuous stretch of J genes.
Site-Specific Evolutionary Rate Analysis. Sequence fragments were removed before analysis with the program diverge (23). Because of gaps in the full-length CHC sequences and length differences between CHC22 and CHC17, 43 residues were excluded from the CHC analysis. For similar reasons, 77 residues were removed from the LC analysis, including 30 residues of the neuronal insert region. A de novo neighbor-joining tree was built within diverge, clades (LCa and LCb or CHC17 and CHC22) were selected, and posterior probabilities for functional divergence were estimated for each position (Table 4).
Results
Three approaches were used to investigate the phylogeny and evolution of clathrin genes. First, extensive phylogenetic trees were constructed and gene divergence times were calculated. Genes flanking the clathrin genes were then analyzed to identify the likely mechanism by which each gene family duplicated. Finally, calculations were made to assess the likelihood of individual amino acids contributing to the development of different functions within the CHC and LC gene families.
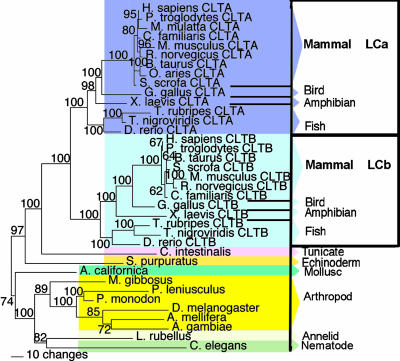
Clathrin Gene Families Duplicated During Chordate Evolution. Phylogenetic trees constructed for the proteins encoded by the CHC gene family, using Bayesian (Figs. 1, 6, and 7) or parsimony (data not shown) analysis, supported a common gene ancestor for CHC22 and CHC17. Both trees revealed that CLTD and CLTC genes are present in bony fishes (actinopterygians Takifugu rubripes and Tetraodon nigroviridis), but only a single CHC-encoding gene, orthologous to both CLTC and CLTD, was found in the genome of the urochordate tunicate sea squirt (Ciona intestinalis). Thus, the timing of CHC gene duplication coincides with the time frame in which the theorized en bloc genome duplications occurred during chordate evolution (24). Phylogenetic trees of the LC gene family constructed by either Bayesian (Figs. 2 and 8) or parsimony (data not shown) analysis showed a similar time frame for LC gene duplication. Using the Bayesian relaxed molecular clock approach (20), the CHC and LC gene duplications were calculated to occur between 510 and 600 MYA. The interval between the two duplication events did not exceed 11 million years (Fig. 9).
Fig. 1.
Bayesian phylogenetic tree of CHCs. Animal kingdom phylogenetic tree of CHC proteins, displaying clade probabilities (as percentages of 2,650 replications), with selected outlying species shown. The scale bar shows the number of sequence changes per branch length. D. rerio A, C, D, and E represent sequences found on chromosomes 10, 23, 9, and 11, and sequence B is unassigned in the D. rerio genome.
Fig. 2.
Bayesian phylogenetic tree of clathrin LCs. Animal kingdom phylogenetic tree for LC proteins, displaying clade probabilities (as percentages of 2,650 replications), with selected outlying species shown. The scale bar shows the number of sequence changes per branch length.
Full-length sequences and sequence fragments for CLTD gene homologues, predicted to encode CHC22, were found in several vertebrate genomes (Pan troglodytes, Canis familiaris, Gallus gallus, Xenopus laevis, Takifugu rubripes, Danio rerio, and Tetraodon nigroviridis). However, in the entire Mus musculus genome, only a partial CLTD gene (chromosome region 16A1 from 17541k to 17654k), surrounded by the same genes that flank human CLTD (Fig. 3), was found. The partial mouse gene was missing the 5′ end and most of the central exons. PCR-based sequencing of one exon near the 3′ end revealed that, for 16 mouse strains (representing three Mus species and three subspecies of Mus musculus), the exon contained stop codons in all three reading frames (Fig. 10). Thus, mouse CLTD-like genomic DNA would encode a nonfunctional pseudogene, confirming the absence of a functional murine CLTD gene (12).
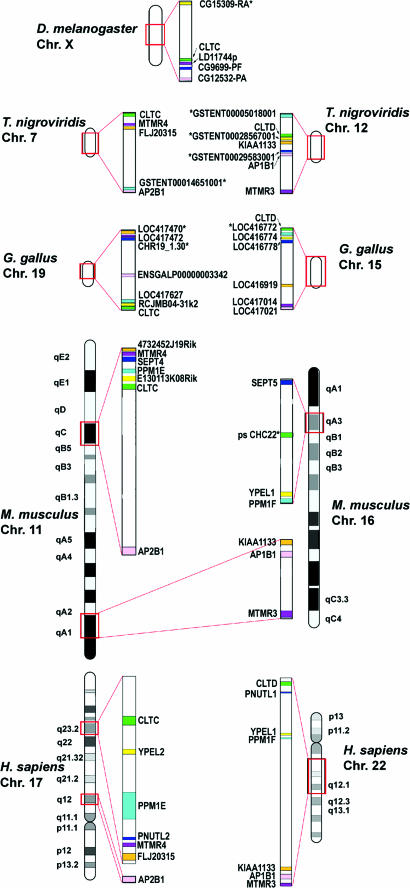
Fig. 3.
Paralogous chromosomal regions surrounding CHC genes. Human chromosomal regions 17q12-17q23.2 (Lower Left) and 22q11.21-22q12.1 (Lower Right) contain seven pairs of paralogons for genes encoding ring finger proteins FLJ20315/KIAA1133 (orange), dual-specificity phosphatases MTMR4/MTMR3 (purple), septins PNUTL2/PNUTL1 (blue), partner of pix proteins PPM1E/PPM1F (aqua), yippee-like proteins YPEL2/YPEL1 (yellow), β adaptins AP2B1 and AP1B1 (pink), and CHCs CLTC/CLTD (green). Orthologues of these paralogons (designated by the same colors as the human genes), mapped to one syntenic region in the D. melanogaster (Top), and two syntenic regions in the vertebrate species, are shown. Chromosomes from all species (Upper Left and Right) are on the same scale, with paralogous regions (boxed in red) expanded (Center). M. musculus 11qC is similar to human chromosome 17, whereas human chromosome 22 has orthologues divided between M. musculus 11qA1 and 16qA3. The M. musculus pseudogene similar to human CLTD (ps CHC22) is found on 16qA3, and no orthologue to CLTD is found elsewhere. Asterisks denote genes located by different methodology (see Supporting Text).
In the zebrafish (Danio rerio) genome, one full-length CHC-encoding gene (chromosome 11) and four fragments were found. If all of the fragments represent full-length genes, then zebrafish could have at least two CLTC orthologues (chromosome 10 and unmapped) and three CLTD orthologues (chromosomes 9, 11, and 23). This finding is consistent with previous observations that two zebrafish genes are frequently found per human gene (25), but will not be confirmed until the genome analysis is completed. Only two LC-encoding genes were found in zebrafish on chromosomes 1 and 14, sorting into the LCa and LCb clades, respectively.
In addition to CLTA and CLTB, two LC pseudogenes lacking introns, at 8p22 and 12p13.31, were found only in the human genome. Both LC pseudogenes LCps8 and LCps12 sorted within the LCa clade of the tree (data not shown). Thus, these pseudogenes were probably processed mRNAs reincorporated into the vertebrate genome some time after rodents diverged from the lineage giving rise to humans (≈90 MYA).
Clathrin Gene Families Duplicated by Distinct Mechanisms. To investigate the duplication mechanism for the clathrin genes, we searched for neighboring paralogous genes in the human genome. Genes were considered paralogons if they were homologues arising from duplications occurring in approximately the same time frame as the clathrin gene duplications (Fig. 11). The chromosomal regions surrounding CLTA and CLTB contained only one identifiable pair of paralogons, RING finger protein genes KIAA1100 and RNF38, each immediately adjacent to an LC gene. Lack of other local paralogons suggests the LC gene duplication was a localized event. By contrast, in the vicinity of the CHC genes there were five additional paralogons contained within 1 MB on chromosome 17 (comprising 17 genes) and within 11.2 MB on chromosome 22 (165 genes) (Fig. 3). It is unlikely (P < 0.001) that so many paralogons would group together by chance on either chromosome, indicating that they originated from a large-scale duplication. The five paralogous pairs near CLTC and CLTD, respectively, comprised genes encoding septins (PNUTL2 and PNUTL1, previously noted as paralogons; ref. 21), myotubularin-related FYVE-dual-specificity phosphatases [MTMR4 and MTMR3 (KIAA0371)], hypothetical RING finger proteins (FLJ20315 and KIAA1133), partner of PIX proteins (PPM1E and PPM1F), and yippee-like proteins (YPEL2 and YPEL1). Extension of the analyzed region on chromosome 17 to 23 MB (344 genes) added a sixth paralogon pair in the vicinity of CLTC and CLTD (P < 0.05 for a chance grouping). These paralogons (AP2B1 and AP1B1) encode subunits of clathrin-associated adaptors AP2 and AP1.
Many of the same paralogons that flank CLTC and CLTD in the human genome are syntenic in the equivalent regions of four additional genomes, mouse (M. musculus), chicken (G. gallus), green spotted pufferfish (T. nigrovividis), and fruit fly (D. melanogaster) (Fig. 3). The vertebrate genomes each have two syntenic groups of these paralogons (except in mouse, where one group is split into two distinct regions), and the fruit fly has a single syntenic group, strengthening the argument that the CLTC and CLTD gene duplication occurred during a large-scale, possibly genomic duplication event. Thus, although CHC and LC gene duplications occurred in the same time frame, they seem to have occurred by different mechanisms.
Residues Contributing to Functional Divergence of Clathrin Isoforms. Comparing two protein isoforms for the rate of amino acid evolution at each sequence position can be used to identify residues with potential to confer differential functions (26). Using diverge software (23), the CHC and LC gene families were analyzed to find amino acid residues that evolved at different rates in each pair of paralogues. The program generates a posterior probability that each residue in an aligned clade has functional divergence compared to its counterpart in the other clade.
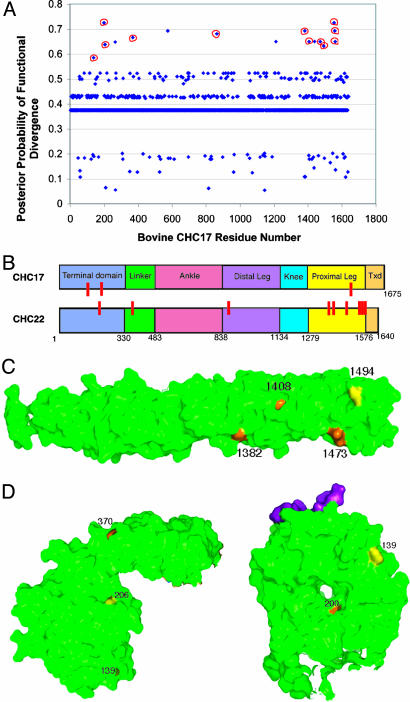
diverge analysis of full-length CHC sequences revealed 16 residues with a significant posterior probability of functional divergence (Fig. 4A and Table 4). Four of these residues appeared less conserved in one clade when sequence fragments from additional species were inspected and were therefore eliminated from further consideration. Three of the remaining 12 divergent residues (139, 200, and 206) localize to the N-terminal domain (TD), and one (370) is in the linker region that connects the globular TD to the extended linear domains of CHC repeat motifs, which make up the triskelion legs (27, 28) (Fig. 4B). Residue 864 in the distal leg segment was uniquely divergent in the entire central portion of the legs. The seven additional divergent residues localized to the CHC region involved in LC binding for CHC17 (comprising the C-terminal half of the proximal leg segment and the region involved in trimerization) (13). Of these seven residues, six were conserved in CHC22 but variable in CHC17, whereas S1494 was the opposite (Table 4). From the x-ray structures of CHC17 fragments (Figs. 4 C and D and 5C), it is seen that divergent residues 1382 and 1473 localize to the CHC17-LC interface, whereas 1408 and 1494 are on a perpendicular leg face.
Fig. 4.
Site-specific profile for CHC evolutionary rate changes. The posterior probability of functional divergence [P(S1|X)] was quantitated by using diverge at each amino acid site in an alignment of CHC protein sequences. (A) Posterior probabilities of functional divergence at each sequence position of CHC17 and CHC22. Sixteen residues had posterior probabilities >0.58, which was calculated to indicate significant divergence. The 12 residues circled in red (139, 200, 206, 370, 864, 1382, 1408, 1473, 1494, 1555, 1559, and 1561) are depicted in B, C, and D below. diverge parameters for CHC22/CHC17 are ThetaML = 0.384, AlphaML = 0.283, SE Theta = 0.092, LRT Theta = 17.41. (B) CHC domains are represented on this bar diagram, with the location of residues with significant posterior probabilities of divergence noted in red on the isoform where it is more conserved. The approximate boundaries of each domain are numbered according to residue position in CHC17. Txd, trimerization domain. (C) Residues with predicted functional divergence between CHC17 and CHC22 are mapped onto the crystallographic structure (green) of the CHC proximal leg (27). Residues conserved only in CHC17 are noted in yellow, and those conserved only in CHC22 are noted in orange. (D) Residues with predicted functional divergence between CHC17 and CHC22 are mapped onto the crystallographic structure (green) of the terminal domain with the “clathrin box” peptide bound in its groove noted in purple (31). Residues conserved only in CHC17 are noted in yellow, and those conserved only in CHC22 are noted in orange.
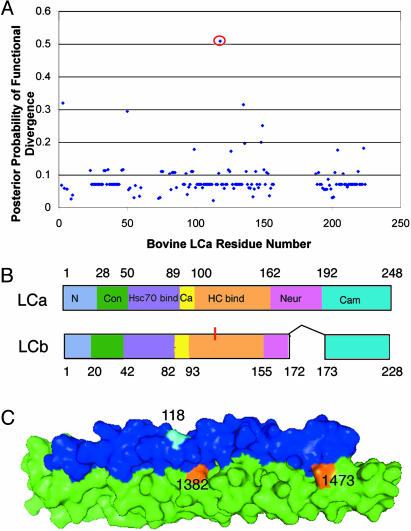
Fig. 5.
Site-specific profile for clathrin LC evolutionary rate changes. The posterior probability of functional divergence [P(S1|X)] was quantitated by using diverge at each amino acid site in an alignment of LC protein sequences. (A) Posterior probabilities calculated for each LC residue reveal only residue 118 as implicated in functional divergence between LCa and LCb by a calculated posterior probability >0.5. diverge parameters for LCa/LCb are ThetaML = 0.160, AlphaML = 0.471, SE Theta = 0.113, LRT Theta = 1.98. (B) Clathrin LC domains are represented on this bar diagram, with the position of 118 indicated in red on LCb, where it is conserved. The domains are as follows: N, N terminus of LCa; Con, sequence of 100% identity between all mammalian LCa and LCb sequences, which regulates clathrin assembly in vitro; Hsc70bind, binding site for Hsc70 on LCa; Ca, calcium-binding site shared by both LCs; HC bind, CHC-binding region shared by both LCs; Neur, region of neuronal inserts for both LCs; Cam, calmodulin-binding domain shared by both LCs (9). (C) LC-HC interface along the proximal leg (13) with only the interacting portions of each subunit shown. Heavy chain is in green and light chain is in blue, with residue 118 in cyan. Residues 1382 and 1473 are conserved in CHC22 and variable in CHC17 (see Fig. 4).
The diverge analysis of LCa and LCb sequences revealed only one residue with significant posterior probability of functional divergence (Fig. 5 A and B). The identified residue 118 is a conserved glutamate (E) in all LCb sequences and is A, Q, M, V, or E in LCa. This position is in the CHC17-binding region, but is opposite to the CHC17-binding interface (Fig. 5C) (13). Overall, diverge analyses of both the CHCs and LCs identified evolutionary differences that could create isoform-specific binding sites for accessory proteins.
Discussion
Analysis of clathrin gene families suggests that the two mammalian CHC isoforms evolved by large-scale (possibly genomic) duplication, whereas the two LC isoforms arose by local gene duplication. That different gene duplication mechanisms generated the CHC and LC isoforms correlates with the differential functions of the clathrin subunits. The LCs diversify tissue-specific regulation of CHC17, whereas duplication of the CHCs diversified their capacity for novel structural roles in different tissues. Interestingly, the time of the two gene duplications was close, with <11 million years separating the two events. Thus, both clathrin gene duplications contributed to the development of increasing complexity during chordate evolution 510-600 MYA through different mechanisms of gene diversification.
The evolutionary changes in CHC22 after CHC gene duplication indicate limited structural diversification of the two isoforms and selection for novel accessory protein-binding sites. Key features of the CHC17 structure were conserved in CHC22, consistent with CHC22 having a functional TD, linker, distal and proximal leg segments, and trimerization domain. Biochemical analysis confirms that CHC22 trimerizes (10). New binding sites for CHC22-specific accessory proteins at both termini were suggested by modeling the CHC22 sequence into the CHC17 structure. Seven divergent residues (six conserved in CHC22) localized to the C-terminal regions that would mediate LC binding in CHC17, suggesting an acquired function for this domain in CHC22. Divergent residues 1382 and 1473 are at a position that would disrupt potential LC binding and divergent residues 1408 and 1494, at the perpendicular face, could contribute to novel protein-binding sites, because these are predicted to be excluded from lattice contacts (28, 29). Consistent with this model, CHC22 does not bind LCs and binds sorting nexin 5 in this region (11). LCs acquire an α-helical structure only when bound to CHC17 (13). This structural plasticity can apparently support more sequence variation than observed for CHCs, allowing LCs to diversify for regulation of CHC17. The one LC residue position that displayed significantly divergent evolutionary rates between the two isoforms localized to a surface of LCb opposite to the CHC17 binding face. Conservation of this position in LCb suggests a previously unrecognized protein-binding site in this region that is already known to influence clathrin assembly (9).
In CHC22, divergent residue 139 localized to a groove between blades of the predicted TD β-propeller structure (30), adjacent to the groove that, in CHC17, binds a clathrin-box motif shared by CHC17-binding proteins (31). Combined with divergent TD residues 200 and 206 and linker residue 370, residue 139 could form a previously undefined CHC22 TD-binding site. Of interest is that human CHC17 polymorphisms also arise in the TD (residues 228 and 371), suggesting population diversity in CHC17 regulation. The only centrally located divergent residue of CHC22 (position 864, conserved in CHC22 and variable in CHC17) could influence interactions between the distal and proximal leg segments, according to its position in the recently modeled clathrin lattice (28, 29). The evolved acquisition of protein-binding sites for CHC22 likely specialized its role at neuromuscular and myotendinous junctions. Because of its structural conservation, we propose that CHC22 could contribute to organization of membrane proteins at these sites in a fashion analogous to CHC17's organization of receptors into transport vesicles.
The loss of a full-length CLTD orthologue in mice, but its continued presence in both birds and carnivores, suggests that the mouse mutation occurred in the rodent lineage <90 MYA. Initial analysis of the rat genome suggested a similar arrangement to the mouse genome and lack of a CLTD orthologue where expected. However, functional studies (11) indicate the presence of CHC22 protein in rat (but not mouse), so the rat CLTD orthologue may still appear upon refined analysis of the rat genome. Compared to other species, the mouse genome has rearranged extensively where the CLTD pseudogene fragment is located (32), possibly explaining its degeneration. Although mice are frequently used as models for human disease conditions, there are many murine features that reflect evolutionary divergence from humans (33-35). Thus, the loss of CHC22 in the mouse lineage should not be considered an indication of its general nonfunctionality. In fact, a comparison of mouse and human skeletal muscle, focusing on regions where CHC22 is concentrated, should reveal how mice have compensated for the loss of CHC22 and provide further insight into CHC22's tissue-specific function in human muscle.
It is notable that the six paralogons adjacent to CLTC and CLTD encode proteins with functions that might interact or influence those of CHC17 and CHC22. We speculate that it may not be coincidental that this group of genes remained near the clathrin genes, perhaps constituting a “membrane traffic” gene cluster. Such functional groupings have been found for other gene families, e.g., the Hox gene clusters (1) and the MHC (36, 37). The most obvious clathrin-related paralogons are the genes encoding the β subunits of the AP1 and AP2 adaptors. These subunits stimulate clathrin assembly and are part of the adaptors that link the clathrin coat to vesicle membranes and cargo (30). Many endocytic proteins, including the AP2 adaptor, bind to both inositol polyphosphates and CHC17, thereby controlling membrane vesicle budding (30). This activity could be influenced by adjacent myotubularins, one of which (encoded by MTMR3) is a known inositol lipid 3-phosphatase (38). RING finger proteins are E3 ubiquitin ligases, potentially creating cargo for association with CHC17 or CHC22. Septins participate in vesicle targeting and fusion and in cytokinesis, perhaps interacting with CHC17 at the mitotic spindle (39), where its function is unknown (40). PPM1F encodes a calmodulin kinase phosphatase (41) that could affect the interaction between calmodulin and neuronal clathrin-coated vesicles (9). Yippee-like protein (YPEL1-encoded) is associated with morphogenesis during cell development (42), a possible connection with CHC22 function in myogenesis. Finally, genes encoding epsin proteins, which are associated with clathrin-coated vesicles, are present close to the paralogous regions described here. Although these epsins are not strictly paralogons by the divergence criteria applied to the others, their presence may be significant with regard to interactions with locally encoded gene products. Further studies of the relationship between clathrins and the gene products of adjacent paralogons could well reveal some interesting interactions involved in membrane traffic.
In conclusion, patterns of divergence and identification of linked genes have suggested previously undescribed aspects of clathrin function that can be studied further. Thus, a look into the past provides a focus for the future, even for these relatively conserved clathrin gene families, indicating the potential of such analysis for other gene families arising from duplication of genes encoding “housekeeping” proteins.
Supplementary Material
Acknowledgments
We thank S. Newmyer for identifying LC sequences from some species. This work was supported by National Institutes of Health Grants GM-38093 (to F.M.B.), CA-09043 (to D.E.W.), and AI-31168 (to P.P.) and by fellowships from the Wellcome Trust and Muscular Dystrophy Association (to M.C.T.).
Author contributions: F.M.B. and P.P. designed research; D.E.W., L.A.-R., and M.C.T. performed research; D.E.W., L.A.-R., and J.D.W. analyzed data; and D.E.W., L.A.-R., P.P., and F.M.B. wrote the paper.
Abbreviations: CHC, clathrin heavy chain; LC, light chain; MYA, million years ago; TD, terminal domain.
References
- 1.Ferrier, D. E. & Minguillon, C. (2003) Int. J. Dev. Biol. 47, 605-611. [PubMed] [Google Scholar]
- 2.Yu, C. Y. (1998) Exp. Clin. Immunogenet. 15, 213-230. [DOI] [PubMed] [Google Scholar]
- 3.Schledzewski, K., Brinkmann, H. & Mendel, R. R. (1999) J. Mol. Evol. 48, 770-778. [DOI] [PubMed] [Google Scholar]
- 4.Dodge, G. R., Kovalszky, I., McBride, O. W., Yi, H. J., Chu, M., Saitta, B., Stokes, D. G. & Ioozo, R. V. (1991) Genomics 11, 174-178. [DOI] [PubMed] [Google Scholar]
- 5.Sirotkin, H., Morrow, B., DasGupta, R., Goldberg, R., Patanjali, S. R., Shi, G., Cannizzaro, L., Shprintzen, R., Weissman, S. M. & Kucherlapati, R. (1996) Hum. Mol. Genet. 5, 617-624. [DOI] [PubMed] [Google Scholar]
- 6.Kedra, D., Peyrard, M., Fransson, I., Collins, J. E., Dunham, I., Roe, B. A. & Dumanski, J. P. (1996) Hum. Mol. Genet. 5, 625-631. [DOI] [PubMed] [Google Scholar]
- 7.Gong, W., Emanuel, B. S., Collins, J., Kim, D. H., Wang, Z., Chen, F., Zhang, G., Roe, B. & Budarf, M. L. (1996) Hum. Mol. Genet. 5, 789-800. [DOI] [PubMed] [Google Scholar]
- 8.Long, K. R., Trofatter, J. A., Ramesh, V., McCormick, M. K. & Buckler, A. J. (1996) Genomics 35, 466-472. [DOI] [PubMed] [Google Scholar]
- 9.Brodsky, F. M., Chen, C. Y., Knuehl, C., Towler, M. C. & Wakeham, D. E. (2001) Annu. Rev. Cell Dev. Biol. 17, 517-568. [DOI] [PubMed] [Google Scholar]
- 10.Liu, S.-H., Towler, M. C., Chen, E., Chen, C.-Y., Song, W., Apodaca, G. & Brodsky, F. M. (2001) EMBO J. 20, 272-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Towler, M. C., Gleeson, P. A., Hoshino, S., Rahkila, P., Manalo, V., Ohkoshi, N., Ordahl, C., Parton, R. G. & Brodsky, F. M. (2004) Mol. Biol. Cell 15, 3181-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puech, A., Saint-Jore, B., Funke, B., Gilbert, D. J., Sirotkin, H., Copeland, N. G., Jenkins, N. A., Kucherlapati, R., Morrow, B. & Skoultchi, A. I. (1997) Proc. Natl. Acad. Sci. USA 94, 14608-14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, C. Y., Reese, M. L., Hwang, P. K., Ota, N., Agard, D. & Brodsky, F. M. (2002) EMBO J. 21, 6072-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van de Peer, Y. (2004) Nat. Rev. Genet. 5, 752-763. [DOI] [PubMed] [Google Scholar]
- 15.Gu, X., Wang, Y. & Gu, J. (2002) Nat. Genet. 31, 205-209. [DOI] [PubMed] [Google Scholar]
- 16.Genetics Computer Group (1994) Program Manual for the Wisconsin Package, Version 8 (Genetics Computer Group, Madison, WI).
- 17.Katoh, K., Misawa, K., Kuma, K. & Miyata, T. (2002) Nucleic Acids Res. 30, 3059-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 19.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, MA).
- 20.Thorne, J. L. & Kishino, H. (2002) Syst. Biol. 51, 689-702. [DOI] [PubMed] [Google Scholar]
- 21.McLysaght, A., Hokamp, K. & Wolfe, K. H. (2002) Nat. Genet. 31, 200-204. [DOI] [PubMed] [Google Scholar]
- 22.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 23.Gu, X. & Vander Velden, K. (2002) Bioinformatics 18, 500-501. [DOI] [PubMed] [Google Scholar]
- 24.Yaspo, M. L. (2001) Trends Mol. Med. 7, 494-501. [DOI] [PubMed] [Google Scholar]
- 25.Taylor, J. S., Braasch, I., Frickey, T., Meyer, A. & Van de Peer, Y. (2003) Genome Res. 13, 382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu, X. (2003) Genetica 118, 133-141. [PubMed] [Google Scholar]
- 27.Ybe, J. A., Brodsky, F. M., Hofmann, K., Lin, K., Liu, S. H., Chen, L., Earnest, T. N., Fletterick, R. J. & Hwang, P. K. (1999) Nature 399, 371-375. [DOI] [PubMed] [Google Scholar]
- 28.Fotin, A., Cheng, Y., Sliz, P., Grigorieff, N., Harrison, S. C., Kirchhausen, T. & Walz, T. (2004) Nature 432, 573-579. [DOI] [PubMed] [Google Scholar]
- 29.Wilbur, J. H., P. K. & Brodsky, F. M. (2005) Traffic 6, 346-350. [DOI] [PubMed] [Google Scholar]
- 30.Owen, D. J., Collins, B. M. & Evans, P. R. (2004) Annu. Rev. Cell Dev. Biol. 20, 153-191. [DOI] [PubMed] [Google Scholar]
- 31.ter Haar, E., Harrison, S. C. & Kirchhausen, T. (2000) Proc. Natl. Acad. Sci. USA 97, 1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botta, A., Lindsay, E. A., Jurecic, V. & Baldini, A. (1997) Mamm. Genome 8, 890-895. [DOI] [PubMed] [Google Scholar]
- 33.Maass, A., Konhilas, J. P., Stauffer, B. L. & Leinwand, L. A. (2002) Cold Spring Harbor Symp. Quant. Biol. 67, 409-415. [DOI] [PubMed] [Google Scholar]
- 34.Coughlin, S. R. (2000) Nature 407, 258-264. [DOI] [PubMed] [Google Scholar]
- 35.Biassoni, R., Cantoni, C., Pende, D., Sivori, S., Parolini, S., Vitale, M., Bottino, C. & Moretta, A. (2001) Immunol. Rev. 181, 203-214. [DOI] [PubMed] [Google Scholar]
- 36.Abi-Rached, L., Gilles, A., Shiina, T., Pontarotti, P. & Inoko, H. (2002) Nat. Genet. 31, 100-105. [DOI] [PubMed] [Google Scholar]
- 37.Kasahara, M., Yawata, M. & Suzuki, T. (1999) in Major Histocompatibility Complex: Evolution, Structure, and Function, ed. Kasahara, M. (Springer, Tokyo), pp. 27-44.
- 38.Walker, D. M., Urbe, S., Dove, S. K., Tenza, D., Raposo, G. & Clague, M. J. (2001) Curr. Biol. 11, 1600-1605. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto, C. T., McKinney, J. & Jeng, Y. Y. (2000) Am. J. Physiol. 279, C369-C374. [DOI] [PubMed] [Google Scholar]
- 40.Kartmann, B. & Roth, D. (2000) J. Cell Sci. 114, 839-844. [DOI] [PubMed] [Google Scholar]
- 41.Harvey, B. P., Banga, S. S. & Ozer, H. L. (2004) J. Biol. Chem. 279, 24889-24898. [DOI] [PubMed] [Google Scholar]
- 42.Farlie, P., Reid, C., Wilcox, S., Peeters, J., Reed, G. & Newgreen, D. (2001) Genes Cells 6, 619-629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.