Abstract
Adventitious rooting is a quantitative genetic trait regulated by both environmental and endogenous factors. To better understand the physiological and molecular basis of adventitious rooting, we took advantage of two classes of Arabidopsis thaliana mutants altered in adventitious root formation: the superroot mutants, which spontaneously make adventitious roots, and the argonaute1 (ago1) mutants, which unlike superroot are barely able to form adventitious roots. The defect in adventitious rooting observed in ago1 correlated with light hypersensitivity and the deregulation of auxin homeostasis specifically in the apical part of the seedlings. In particular, a clear reduction in endogenous levels of free indoleacetic acid (IAA) and IAA conjugates was shown. This was correlated with a downregulation of the expression of several auxin-inducible GH3 genes in the hypocotyl of the ago1-3 mutant. We also found that the Auxin Response Factor17 (ARF17) gene, a potential repressor of auxin-inducible genes, was overexpressed in ago1-3 hypocotyls. The characterization of an ARF17-overexpressing line showed that it produced fewer adventitious roots than the wild type and retained a lower expression of GH3 genes. Thus, we suggest that ARF17 negatively regulates adventitious root formation in ago1 mutants by repressing GH3 genes and therefore perturbing auxin homeostasis in a light-dependent manner. These results suggest that ARF17 could be a major regulator of adventitious rooting in Arabidopsis.
INTRODUCTION
Adventitious root formation is a complex process that is affected by multiple endogenous factors, including phytohormones, and environmental factors, such as wounding and light. The molecular mechanisms by which adventitious root formation is regulated are still poorly understood. Auxin plays a central role (Blakesley, 1994) and may interact with other endogenous factors or environmental stimuli, such as light. It was shown that auxin and light act antagonistically on the development of adventitious roots in Eucalyptus saligna and E. globulus (Fett-Neto et al., 2001). Recently, Niemi et al. (2005) showed that light sources with different spectra could affect adventitious root and mycorrhyza formation in Scots pine (Pinus sylvestris) in vitro. Arabidopsis thaliana serves as a valuable model system for dissecting the molecular mechanisms involved in the control of adventitious root initiation by diverse environmental signals. For example, King and Stimart (1998) have shown that several ecotypes of A. thaliana differ in their capacity to produce adventitious roots on the hypocotyl in response to auxin and that low and high rooting responses might be controlled by several genes acting independently in an additive-dominant manner. More recently, Konishi and Sugiyama (2003) identified temperature-sensitive mutants of Arabidopsis altered in adventitious rooting.
To gain further insight into the interaction between light and auxin in the regulation of adventitious rooting, we took advantage of two classes of mutants, superroot1 (sur1) and sur2 and argonaute1 (ago1), that we described previously (Boerjan et al., 1995; Delarue, 1996; Bohmert et al., 1998; Delarue et al., 1998; Camus, 1999). sur1 and sur2 are auxin overproducers that spontaneously develop adventitious roots on the hypocotyl as a consequence of increased endogenous auxin levels (Boerjan et al., 1995; Delarue et al., 1998). Although SUR2 is primarily involved in indole glucosinolate production, SUR1 is apparently required for the production of all glucosinolates in Arabidopsis (Barlier et al., 2000; Bak et al., 2001; Mikkelsen et al., 2004).
The ago1 mutant was first identified as a leaf developmental mutant (Bohmert et al., 1998). AGO1 is the founding member of a gene family that is conserved among eukaryotes (Bohmert et al., 1998), the members of which play a crucial role in the regulation of posttranscriptional gene silencing and related mechanisms (Fagard et al., 2000; Hammond et al., 2001; Carmell et al., 2002; Morel et al., 2002). AGO proteins are also called PPD proteins, because they all retain the conserved PAZ and PIWI domains (Cerutti et al., 2000). These proteins have been shown in both Drosophila melanogaster and human cells to be core components of the RNA-induced silencing complex, which targets mRNA for degradation using a microRNA (miRNA) as a guide (Hutvagner and Zamore, 2002; Ishizuka et al., 2002). Recently, it was shown that the role of the AGO1 gene in the miRNA pathway and its own regulation by this particular pathway are crucial for plant development (Vaucheret et al., 2004). Kidner and Martienssen (2004) reported that the leaf polarity defect observed in ago1 could be explained by an abnormal distribution of miRNAs targeting PHABULOSA and PHAVOLUTA transcription factors, which are known to control leaf polarity in plants. It was also shown that the steady state levels of several transcription factor targets of miRNAs were increased in rosette leaves of strong and weak alleles of ago1 (Vaucheret et al., 2004).
During our further characterization of the phenotype conferred by ago1, we discovered that, unlike the root, the apical part of ago1 mutants displayed resistance to auxin-mediated hypocotyl elongation and a defect in adventitious root formation in response to auxin. This prompted us to investigate a potential interaction between light and auxin in the regulation of adventitious rooting using an allelic series of ago alleles and ago sur double mutants. The data presented here demonstrate that the defect in adventitious root formation in ago1 mutants correlates with an alteration of auxin homeostasis and a hypersensitivity to light. We show that the mRNA of Auxin Response Factor17 (ARF17) accumulates in the hypocotyls of ago1 and demonstrate that deregulation of ARF17 expression and, as a consequence, GH3 gene expression at least in part explain the adventitious root phenotype of ago1 mutants. Thus, we conclude that AGO1, through its action on the regulation of ARF17 expression, regulates genes involved at the cross talk between auxin and light signaling during adventitious root development.
RESULTS
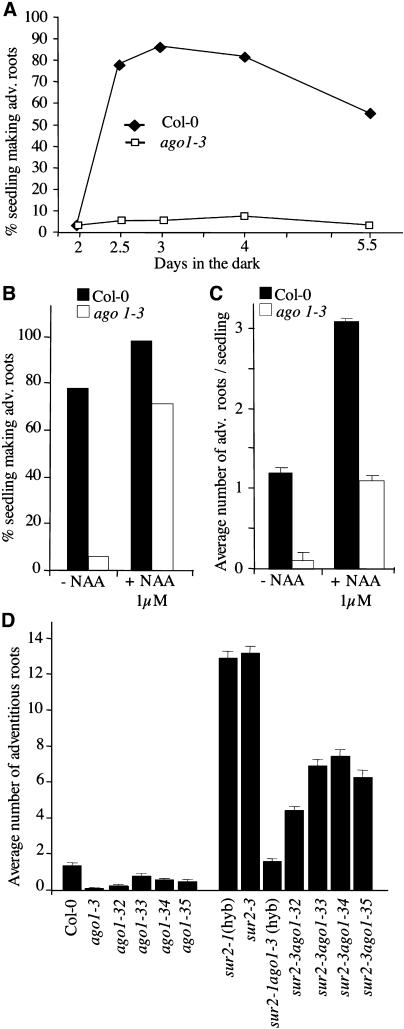
The experiments described in this article were done using one null and four hypomorphic ago1 mutants. The strong phenotype of ago1-3 has been described previously (Bohmert et al., 1998; Camus, 1999). In this study, we confirmed by protein gel blot analysis and sequencing that ago1-3 is a null allele (see Supplemental Table 1 and Supplemental Figure 1A online). The four new hypomorphic mutants (ago1-32 to ago1-35) were identified in a screen for mutants displaying a phenotype similar to that of other previously described hypomorphic allele mutants (Morel et al., 2002). ago1-32 shows a weak phenotype similar to that of ago1-26, whereas ago1-33, ago1-34, and ago1-35 are similar to ago1-27 (Morel et al., 2002). These mutants can grow in soil and, except for ago1-32, are fertile.
The Apical Part of ago1 Seedlings Is Specifically Impaired in Auxin Response
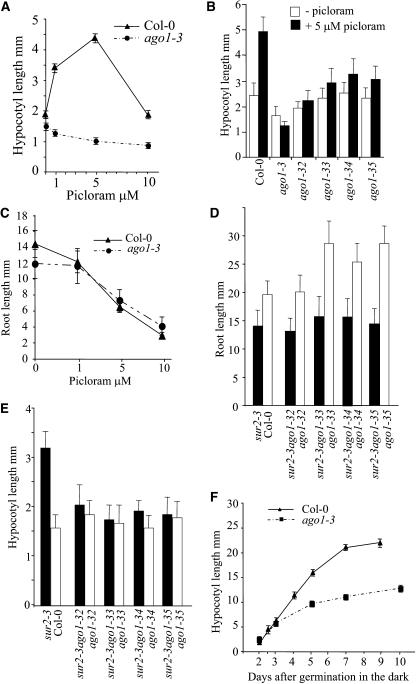
When wild-type seedlings were germinated on media containing increasing concentrations of picloram, an auxin-type herbicide (Hansen and Grossmann, 2000), their hypocotyls showed a maximum size at a concentration of 5 μM (Figure 1A). Unlike the wild type, ago1-3 plants showed no elongation of the hypocotyl when grown under the same conditions (Figure 1A). This defect of hypocotyl elongation in response to auxin was confirmed with the four weak allele mutants (Figures 1B and 2A). However, the root growth of the ago1-3 null mutant was inhibited normally on media containing different concentrations of either picloram (Figure 1C) or the auxins naphthylacetic acid (1-NAA) or indoleacetic acid (IAA) (data not shown). To investigate whether ago1 hypocotyl elongation was resistant to the increased endogenous content of auxin, double mutants between the different ago1 alleles and the sur2 auxin overproducer were produced. The sur2-1 ago1-3(hyb) double mutant is in a hybrid genetic background between Columbia (Col-0) and Wassilewskija (Ws) because it comes from a cross between a homozygote sur2-1 plant in the Ws ecotype and a heterozygote for the ago1-3 mutation in the Col-0 background. This hybrid genetic background will be referred to as (hyb) in the rest of the article. sur2 ago1 root had the same length as the sur2 root and was shorter than roots of the wild type or single ago1 mutants (Figure 1D) and therefore responded normally to increased endogenous levels of auxin. Conversely, the double mutant hypocotyl remained short (Figures 1E and 2B). The apical part of 8-d-old light-grown sur2-1 ago1-3(hyb) double mutants contained twice as much free IAA than did the wild type (C. Sorin, K. Ljung, J.D. Bussell, G. Sandberg, and C. Bellini, unpublished data), indicating that the ago1-3 mutation indeed induced resistance to increased levels of endogenous auxin that in the wild type were shown to induce elongation of the hypocotyl (Gray et al., 1998). On the contrary, when grown at high auxin concentrations, which inhibit wild-type hypocotyl elongation, the ago1-3 mutant was as sensitive as the wild type and showed reduction of hypocotyl elongation (Figures 2C and 2D). We also examined whether the ago1 hypocotyl could elongate under other growth conditions. Both the null and the weak allele mutants were grown on medium containing concentrations of gibberellic acid that promote hypocotyl elongation in the wild type. All ago1 alleles analyzed responded to gibberellic acid and elongated in the same proportion as the wild type (see Supplemental Figure 2 online). When grown in the dark, plants representing the weak alleles showed no mutant phenotype (see Figure 8). In the dark, the null allele ago1-3 was shorter than the wild type but was almost 10 times longer than in the light. The growth rate of ago1-3 followed that of the wild type for the first 3 d after germination, but then it decreased (Figure 1F).
Figure 1.
The Apical Part but Not the Root of ago1 Mutants Is Resistant to Auxin.
(A) Hypocotyl length of wild-type and ago1-3 siblings grown in vitro on increasing concentrations of picloram, 8 d after germination in the light.
(B) Hypocotyl length of wild-type and different ago1 mutants grown in vitro in the presence or absence of 5 μM picloram, 8 d after germination in the light.
(C) Root length of wild-type and ago1-3 siblings grown on increasing concentrations of picloram, 8 d after germination in the light.
(D) and (E) Root (D) and hypocotyl (E) length of Col-0 seedlings and siblings from a plant homozygous for the sur2-3 mutation and heterozygous for the different weak alleles of ago1, 8 d after germination in the light.
(F) Hypocotyl length of wild-type and ago1-3 siblings grown in vitro in the dark. The hypocotyl was measured at different time points.
Error bars indicate sd.
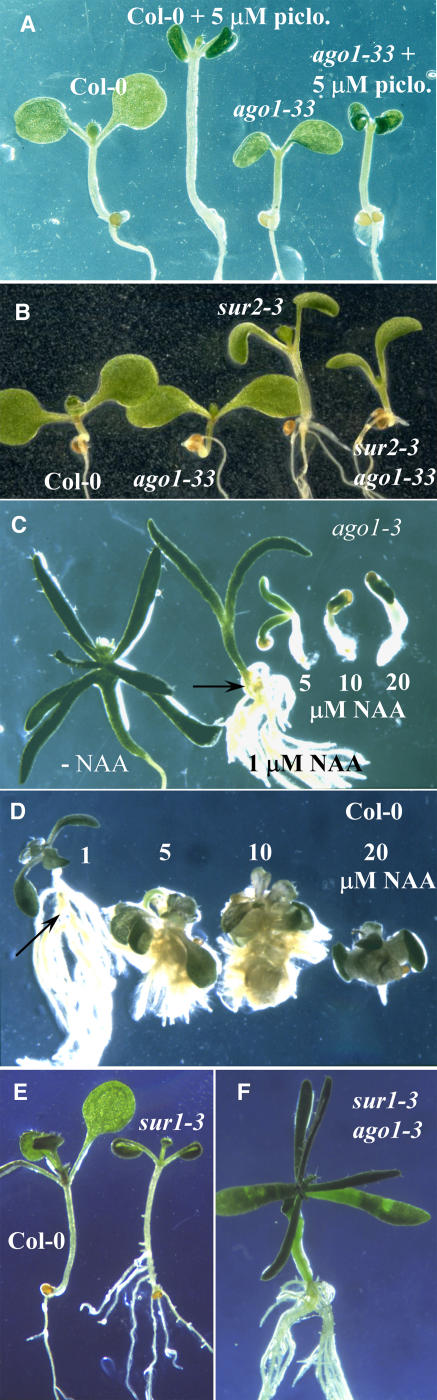
Figure 2.
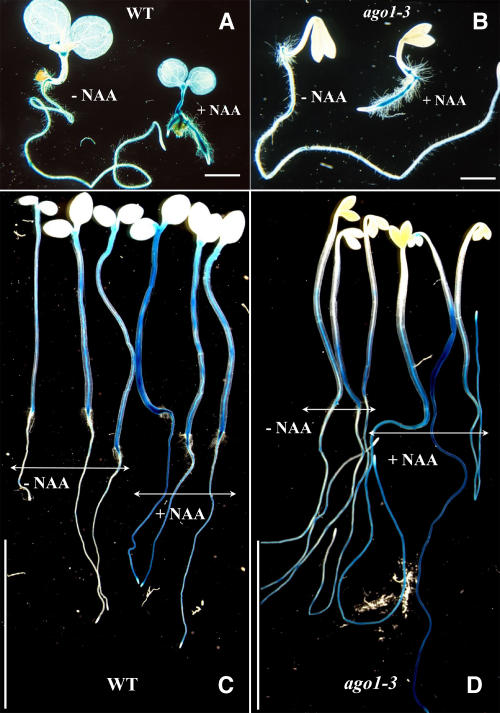
Auxin Resistance in the Apical Part Is Associated with a Defect in Adventitious Root Formation.
(A) Eight-day-old wild-type and ago1-33 siblings germinated and grown in the light in the presence or absence of 5 μM picloram.
(B) Wild-type and ago1-33 siblings (left) and sur2-3 and ago1-33 sur2-3 siblings (right). Seedlings were germinated and grown in the light for 8 d.
(C) and (D) Three-week-old ago1-3 (C) and wild-type (D) siblings grown in the light on media without or with increasing concentrations of 1-NAA. Arrows indicate the hypocotyl/root junction.
(E) Wild-type Col-0 (left) and sur1-3 (right) seedlings grown for 15 d in vitro.
(F) One-month-old sur1-3 ago1-3 double mutant.
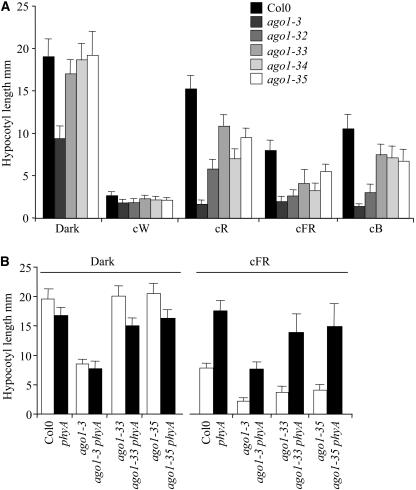
Figure 8.
ago1 Is Hyperresponsive to Light.
(A) Hypocotyl length of the different ago1 mutants grown in various light conditions. Hypocotyl length was measured on wild-type and mutant siblings from the different alleles grown in vitro under different light conditions for 8 d. cW, constant white light (150 μE·m−2·s−1); cR, constant red light (9 μE·m−2·s−1); cFR, constant far-red light (0.25 μE·m−2·s−1); cB, constant blue light (3.7 μE·m−2·s−1). Error bars indicate sd.
(B) phyA is epistatic to ago1 in far-red light. Hypocotyl length of Col-0, ago1-3, phyA, phyA ago1-3, phyA ago1-33, and phyA ago1-35, 8 d after germination either in the dark or in cFR. Error bars indicate sd.
To test whether, similar to the other auxin-resistant mutants (Pickett et al., 1990; Wilson et al., 1990; Timpte et al., 1995; Leyser et al., 1996), ago1-3 showed cross-resistance to cytokinin or ethylene, the mutant was grown on a medium containing either cytokinin or 1-aminocyclopropane-1-carboxylic acid, an ethylene precursor. Like the wild type, ago1-3 responded normally to both hormones (see Supplemental Figure 3 online). The double mutant between ago1-3 and the cytokinin overproducer amp1 (Chaudhury et al., 1993) showed a clear additive effect of both mutations (see Supplemental Figures 3B and 3C online).
Based on all of these results, we conclude that ago1 represents a new class of auxin-resistant mutants in which the hypocotyl is specifically resistant to auxin-mediated elongation.
ago1-3 Is Altered in Adventitious Root Formation but Not in Lateral Root Development
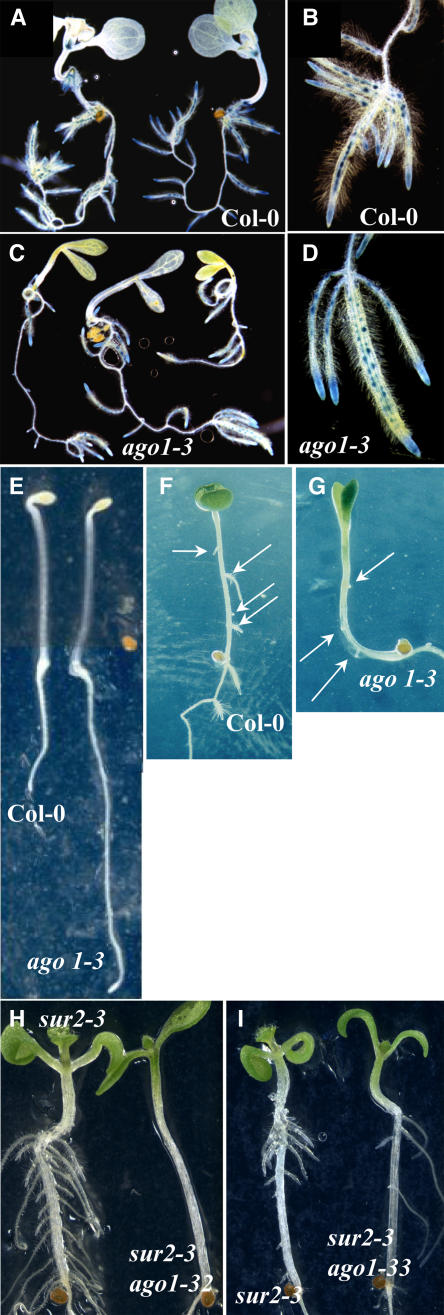
Because only the apical part of ago1 seedlings was resistant to auxin, we analyzed their capacity to produce adventitious roots either in response to exogenous auxin or in the auxin overproducer sur1 or sur2 background. When germinated and grown in the light in the presence of auxin, ago1-3 seedlings, unlike wild-type seedlings, were unable to develop adventitious roots on the hypocotyl (Figures 2C and 2D). Double mutants between ago1-3 and the auxin overproducer sur1-3 were also unable to produce adventitious roots from the hypocotyl (Figures 2E and 2F). We previously showed that adventitious roots in the hypocotyl initiate from the pericycle cells adjacent to the xylem poles, similar to lateral roots (Boerjan et al., 1995). Therefore, we checked whether ago1-3 mutant roots were able to initiate and develop lateral roots in response to exogenous auxin. We used the CYCB1:uidA promoter fusion as a reporter gene to monitor lateral root formation. Indeed, CYCB1 is one of the earliest genes expressed in the pericycle cells that will develop into a lateral root (Beeckman et al., 2001). As shown in Figures 3A to 3D, ago1-3 was able to initiate and develop lateral roots in response to 1 μM 1-NAA in a similar way to the wild type. These results indicate that, although adventitious and lateral roots develop from pericycle cells, ago1 is specifically impaired in adventitious root formation.
Figure 3.
Adventitious Root Initiation, but Not Lateral Root Initiation, Is Affected in ago1-3.
(A) to (D) Siblings from a heterozygous ago1-3 plant expressing the CycB1:uidA marker gene were germinated and grown in the light for 5 d on a medium without auxin and then transferred to a medium containing 1 μM NAA for 6 d.
(A) and (B) Wild type.
(C) and (D) ago1-3.
(E) Siblings from a heterozygous ago1-3 parent plant were germinated and grown in the dark for 2.5 d before transfer to the light.
(F) Col-0 could make up to four adventitious roots after etiolation and transfer to the light for 1 week on a medium containing 1 μM 1-NAA. Arrows indicate adventitious roots.
(G) ago1-3 siblings could very rarely develop up to three adventitious roots in the same conditions as in (F). Arrows indicate adventitious roots.
(H) and (I) sur2-3 and sur2-3 ago1 etiolated siblings after 1 week in the light. ago1-32 is the intermediate allele mutant (H), and ago1-33 is one of the weakest alleles (I).
When Primarily Etiolated, ago1-3 Seedlings Can Develop a Few Adventitious Roots in Response to Auxin
Because wild-type hypocotyl can spontaneously develop adventitious roots when it has been etiolated in the dark, and also because sur1 and sur2 have a longer hypocotyl than wild-type seedlings, we wondered whether the defect in adventitious rooting from ago1 hypocotyls was linked to its defect in elongation in the light in response to auxin. Mutant and wild-type seedlings were grown in the dark for different periods of time, and hypocotyl length was measured before they were transferred to the light for 1 week. After being etiolated for 2.5 d in the dark, both wild-type and mutant plants had an average hypocotyl length of 5 mm (Figure 1F), and most of wild-type seedlings developed at least one adventitious root when transferred to the light for 1 week (Figure 4A). The proportion of mutant seedlings able to develop at least one adventitious root remained extremely low irrespective of hypocotyl size (Figure 4A). This result suggested that a defect in hypocotyl elongation was not sufficient to explain the defect in adventitious rooting. Therefore, we tested the capacity of the ago1-3 mutant to develop adventitious roots in response to exogenous auxin after hypocotyl elongation in the dark. Mutant and wild-type siblings were etiolated for 2.5 d before transfer to the light (Figure 3E). They were then transferred onto a medium without auxin or containing 1 μM 1-NAA. Addition of auxin significantly increased the proportion of ago1-3 seedlings developing one or more adventitious roots (Figures 3F, 3G, 4B, and 4C). Figures 3F and 3G illustrate the fact that wild-type and ago1-3 seedlings could sometime make up to four and three adventitious roots, respectively, when transferred onto auxin-containing medium after etiolation. Nevertheless, this remained a rare event, as ago1-3 showed an average of one adventitious root and the wild type in the same conditions had three adventitious roots (Figure 4C). However, weak allele mutants etiolated for 2.5 d before transfer to the light on a medium containing 1 μM 1-NAA produced almost as many adventitious roots as the wild type (data not shown).
Figure 4.
Auxin Stimulates Adventitious Roots on Etiolated ago1 Seedlings but Never as in the Wild Type.
Emergent adventitious roots were scored 7 d after transfer to the light. At least 40 seedlings were used for each data point. This was repeated on three independent biological replicates.
(A) Siblings from a heterozygous ago1-3 mother plant were germinated and grown in the dark for different times, then transferred to the light. The proportion of seedlings forming one or more adventitious roots was determined after 1 week in the light.
(B) Proportion of wild-type and mutant seedlings forming one or more adventitious roots after transfer to the light for 1 week on a medium with or without 1-NAA.
(C) Average number of adventitious roots formed on wild-type or mutant seedlings in the absence or presence of 1-NAA. Error bars indicate se.
(D) Average number of adventitious roots formed on Col-0, sur2, the five ago1 mutants, and the five sur2 ago1 double mutants after etiolation and transfer to the light for 1 week. Error bars indicate se.
Adventitious roots were also scored in etiolated seedlings of the different sur2 ago1 double mutants at 7 d after transfer to the light. The sur2-1 ago1-3(hyb) mutant developed as many adventitious roots as the wild type in the absence of auxin but never more (Figure 4D). Double mutants with the weak alleles developed more adventitious roots but never as many as the single sur2 mutant in the same conditions (Figures 3H, 3I, and 4D).
The sur2-1 ago1-3(hyb) Double Mutant Can Initiate Adventitious Roots in the Dark
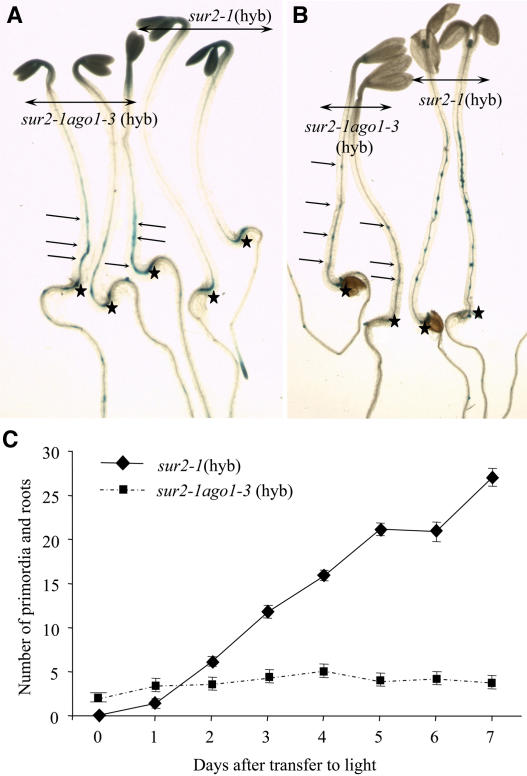
To check whether the results described above were linked to a defect in the initiation of adventitious roots or to a blockage of the subsequent development of initiated adventitious roots, we monitored the expression of the CycB1:uidA marker gene in the sur2-1 ago1-3(hyb) double mutant. Seedlings were etiolated in the dark to obtain 5-mm hypocotyls before transfer to the light for 1 to 7 d. Figures 5A and 5C show that sur2-1(hyb) did not express the reporter gene at 2.5 d after germination in the dark in the hypocotyl but clearly showed adventitious root initiation at 1 d after transfer to the light (Figure 5C), which regularly increased with time (Figures 5B and 5C). Unexpectedly, the sur2-1 ago1-3(hyb) double mutant already expressed the reporter gene in hypocotyl pericycle cells before being transferred to the light (Figures 5A and 5C). In contrast with sur2-1(hyb), though, no more primordia were initiated on the hypocotyl even several days after transfer to the light (Figures 5B and 5C). These results indicate that light is required for the induction of adventitious root initiation and development in sur2-1(hyb) mutants but has an inhibitory effect in ago1-3 sur2-1(hyb) double mutants. Because adventitious roots in ago1-3 sur2-1(hyb) seedlings were initiated in the dark and stopped after transfer to the light, the ago1-3 mutation may alter light regulatory pathways.
Figure 5.
sur2 ago1 Double Mutants Initiate Adventitious Roots in the Dark.
(A) and (B) CycB1:uidA expression in etiolated siblings of sur2-1 and sur2-1 ago1-3(hyb), 2.5 d after germination in the dark (A) and after transfer to the light for 3 d (B). Arrows indicate GUS staining in the sur2-1 ago1-3(hyb) hypocotyl. Stars indicate the junction between the hypocotyl and the root.
(C) CycB1:uidA expression in etiolated siblings of sur2-1 and sur2-1 ago1-3(hyb) was monitored in seedlings germinated and grown in the dark for 2.5 d and transferred to light for 1 to 7 d. This allowed identification and scoring of the initiation of very early primordia as well as older emerging roots. Error bars indicate se.
Auxin Homeostasis Is Altered in the Apical Part of ago1-3 Seedlings but Not in the Root
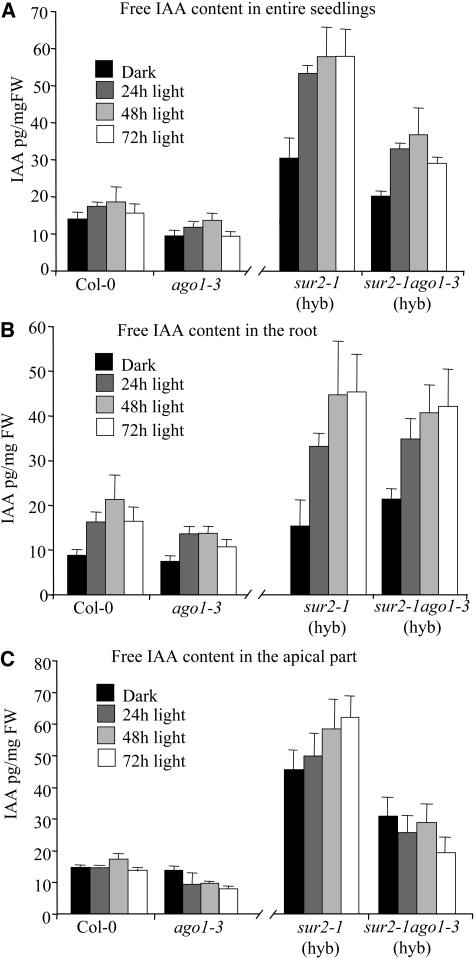
The fact that sur2 ago1 double mutants developed fewer adventitious roots than sur2 mutants under the same growth conditions suggested that the ago1 mutation could affect endogenous auxin levels. Therefore, we determined the endogenous content of free IAA in the different genotypes. Seedlings of the wild type, ago1-3, sur2-1(hyb), and sur2-1 ago1-3(hyb) were grown in the dark for 2.5 d and then transferred to the light for 24, 48, or 72 h. Figure 6A shows that ago1-3 entire seedlings had a slightly lower free IAA content than the wild type either in the dark or after transfer to the light (P < 0.05). A more pronounced effect of the ago1-3 mutation could be observed in the auxin-overproducing sur2 background. The double mutant sur2-1 ago1-3(hyb) had a reduction in free IAA compared with the single sur2-1(hyb) mutant (P < 0.01), although the level remained higher than in the wild type (P < 0.01). Interestingly, when IAA was quantified in the root only, no differences between sur2-1(hyb) and sur2-1 ago1-3(hyb) were detected (Figure 6B). In the dark, the ago1-3 root had the same auxin content as the wild-type root, and no clear differences were observed after transfer to the light (Figure 6B). On the contrary, when free IAA was quantified in the apical part of the seedlings (hypocotyl plus cotyledons), the endogenous content in the ago1-3 mutant was similar to the wild-type level in the dark, but it decreased after transfer to the light (P < 0.01). This effect was even more striking in the sur2-1(hyb) background. The free IAA content clearly increased in the apical part of sur2-1(hyb) after transfer to the light (P < 0.05) but decreased in that of the sur2-1 ago1-3(hyb) double mutant (P < 0.05) (Figure 6C).
Figure 6.
Free IAA Content in Col-0, ago1-3, sur2-1(hyb), and ago1-3 sur2-1(hyb) Seedlings.
The endogenous free IAA level was measured in seedlings germinated and grown for 2.5 d in the dark, then transferred to the light for 24, 48, or 72 h. FW, fresh weight.
(A) Entire seedlings. Free IAA content was lower in ago1-3 than in the wild type (P < 0.5). The sur2-1 ago1-3(hyb) double mutant contained significantly more auxin than the wild type (P < 0.01) but less than sur2-1(hyb) (P < 0.01).
(B) Root. The auxin content increased in all the genotypes after transfer to the light. No significant difference between ago1-3 and Col-0 or sur2-1(hyb) and sur2-1 ago1-3(hyb) could be detected.
(C) Apical part (cotyledons plus hypocotyl). The auxin content decreased significantly in ago1-3 seedlings after transfer to the light (P < 0.01) and was significantly lower than that in the wild type after 72 h in the light (P < 0.01). The auxin content increased significantly in sur2-1(hyb) after transfer to the light (P < 0.05) but decreased in sur2-1 ago1-3(hyb) (P < 0.05).
Three biological replicates were used for each data point. Error bars indicate sd. A t test was performed according to http://graphpad.com/quickcalcs/ttest1.cfm.
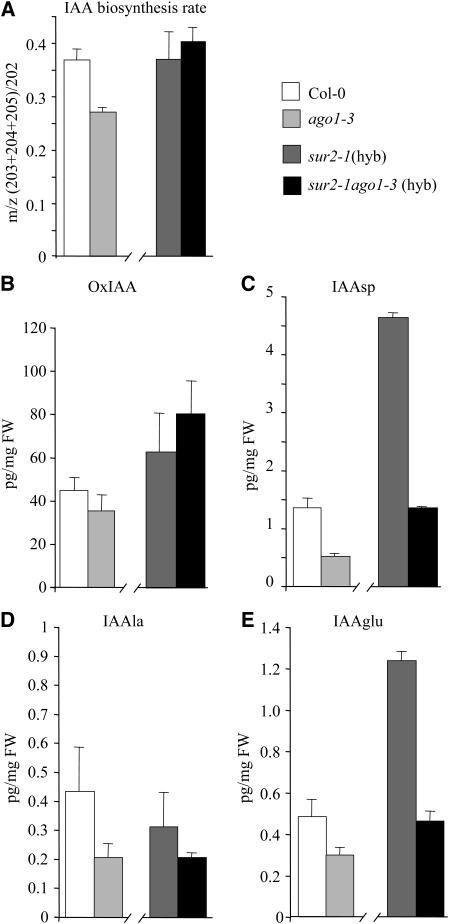
The IAA biosynthesis rate was measured in the apical part of the different genotypes at 72 h after transfer to the light by monitoring the incorporation of deuterium via de novo synthesis of IAA, as described previously (Ljung et al., 2001). Although the biosynthesis rate was low at that stage of development, incorporation of deuterium could be measured, indicating that some IAA was synthesized. The rate of synthesis was slightly lower but significantly different in the ago1-3 mutant compared with the wild type (P < 0.05); nevertheless, no difference was detected between the auxin overproducer sur2-1 mutant and the sur2-1 ago1-3(hyb) double mutant (Figure 7A). Similar results were obtained with entire seedlings (data not shown). By contrast, incorporation of deuterium was almost not detectable in the root of both genotypes (data not shown), indicating that, at that stage of development, the auxin was synthesized mainly in the apical part. These results indicate that the AGO1 gene might be required for the regulation of auxin biosynthesis. Nevertheless, the decrease of auxin content in sur2-1 ago1-3(hyb) cannot be explained by a lower biosynthesis rate compared with sur2-1, suggesting that ago1-3 may be altered in other pathways that regulate auxin homeostasis. Thus, we measured the endogenous contents of several IAA metabolites in the different genotypes and showed that the levels of the amide conjugates IAAsp (P < 0.01), IAAla (P < 0.05), and IAAglu (P < 0.05) were lower in ago1-3 and ago1-3 sur2-1(hyb) compared with Col-0 and sur2-1(hyb), respectively (Figures 7C to 7E), suggesting an inhibition of the IAA conjugation pathway in the ago1-3 background. The level of oxoindole-3-acetic acid was not statistically different at that stage of development between ago1-3 and Col-0 seedlings or in the ago1-3 sur2-1(hyb) double mutant compared with sur2-1(hyb) siblings (Figure 7B). Nevertheless, because the decrease of IAA content in sur2-1 ago1-3(hyb) compared with sur2-1(hyb) cannot be explained by a reduction of IAA biosynthesis or increased conjugation, further analyses are needed to check for a potential activation of the catabolic pathway in ago1-3. These results suggest that AGO1 influences the overall regulation of auxin homeostasis in the apical part of Arabidopsis seedlings.
Figure 7.
IAA Biosynthesis Rate and Level of IAA Metabolites in Col-0, ago1-3, sur2-1(hyb), and ago1-3 sur2-1(hyb) Seedlings.
(A) IAA biosynthesis rate in the apical part of seedlings (hypocotyl plus cotyledons), 72 h after transfer to the light. Four replicates were used for each data point. Error bars indicate sd.
(B) to (E) Quantification of IAA metabolites. Measurements were performed on entire seedlings grown for 2.5 d in the dark for oxoindole-3-acetic acid (B), N-(indole-3-acetyl)-Asp (C), N-(indole-3-acetyl)-Ala (D), and N-(indole-3-acetyl)-Glu (E). Four replicates were used for each data point. Error bars indicate sd. FW, fresh weight.
ago1 Is Hyperresponsive to Light
Although the ago1-3 mutant did not show a characteristic deetiolated phenotype (short hypocotyl, open and expanded cotyledons), some aspects of its phenotype in the dark, such as the shorter hypocotyl and longer primary root (Figure 3E), suggest a defect in light perception or in light regulatory pathways. Indeed, ago1-3 had a root that was almost three times longer than the wild-type root for the same hypocotyl size at 2.5 d after germination in the dark (Figure 3B). This root length was equivalent to that of ago1-3 or wild-type roots when the seedlings were germinated and grown in the light for 7 d (data not shown). The different ago1 alleles were grown under different monochromatic light conditions. They were grown under low-fluence constant red light (cR) (9 μE·m−2·s−1), low-fluence constant blue light (cB) (4 μE·m−2·s−1), or very low-fluence constant far-red light (cFR) (0.25 μE·m−2·s−1) for 7 d. In these conditions, the wild type was shorter than when it was grown in the dark, but its hypocotyl still elongated. The ago1-3 mutant did not elongate at all in any light conditions except in the dark (Figure 8A), suggesting a general upregulation of light regulatory pathways. The weak allele mutants were all shorter than the wild type in cR, cB, and cFR (Figure 8A). The weakest allele mutants displayed similar elongation of the hypocotyl in cB but showed more heterogeneity in cR and cFR. Three of the hypomorphic mutants, ago1-32, ago1-33, and ago1-34, were almost as short as the null mutant ago1-3 in very low-fluence cFR. These results suggested that ago1 mutants were hypersensitive to light. That four of them were considerably shorter in cFR suggested a deregulation in the phytochrome A (PHYA)–dependent pathway. To test this hypothesis, we generated double mutants with phyA. As shown in Figure 8B, we observed a clear epistasy of the phyA mutation on ago1-3, ago1-33, and ago1-35 in cFR. These results strongly suggest that at least the light regulatory pathways mediated by PHYA are upregulated in ago1.
Expression of GH3 Genes Is Downregulated in ago1-3
We have analyzed the expression of several auxin-inducible genes in the ago1-3 mutant. Using RNA gel blot experiments, we have checked the expression of six auxin-inducible Aux/IAA genes (IAA1, IAA4, IAA7, IAA14, IAA17, and IAA19) (Abel et al., 1995; Rouse et al., 1998; Nagpal et al., 2000; Fukaki et al., 2002; Tatematsu et al., 2004). None of the six genes was upregulated in ago1-3 compared with the wild type in the absence of auxin (data not shown). All of them were induced by auxin, although the induction in ago1-3 was weaker in the case of IAA7 and IAA14 (data not shown).
The expression patterns of four auxin-inducible reporter genes were also analyzed. IAA2:uidA-, DR5:uidA-, SAUR-AC:uidA-, and GH3:uidA-expressing lines were crossed with heterozygous ago1-3 plants. Wild-type and mutant plants were grown for 1 week on medium without auxin or containing 1 μM 1-NAA. Upon analysis of histochemical β-glucuronidase (GUS) staining, we observed the same expression pattern of DR5:uidA, IAA2:uidA, or SAUR-AC:uidA as well as the same response to auxin treatment between wild-type and ago1-3 plants. By contrast, GH3:uidA expression was induced in the root and in the hypocotyl of the wild type when grown in the presence of 1 μM 1-NAA but never in the hypocotyl of ago1-3 mutants, which, however, showed a normal induction in the root (Figures 9A and 9B). In the absence of auxin, light-grown wild-type and ago1-3 seedlings showed similar patterns of expression in the root at the points of lateral root initiation. Nevertheless, in contrast with the wild type, the apical meristem was rarely stained in 1-week-old, light-grown ago1-3 seedlings (Figures 9A and 9B). Because these differences between the wild type and ago1-3 were observed in light-grown seedlings, we checked the conditions used for adventitious root induction. Seedlings were first etiolated for 2.5 d and then transferred to long-day conditions for 2 d. After 44 h, they were transferred in liquid culture medium in the presence or absence of 10 μM 1-NAA and kept in the same growth conditions for 4 h, then stained overnight for GUS expression. In the absence of auxin, GH3:uidA was expressed in all hypocotyls of wild-type seedlings, whereas in ago1-3, expression was restricted to the bottom third of the hypocotyl. The roots of nontreated wild-type or ago1-3 seedlings mainly expressed GH3:uidA at the site of lateral root initiation. As described previously, the ago1-3 root is longer than the wild-type root and initiates several lateral roots in the dark, which could explain the greater GUS expression in roots of etiolated ago1-3 compared with the wild type. When seedlings where treated with 10 μM 1-NAA, GH3:uidA was induced all along the root of both wild-type and ago1-3 siblings (Figures 9C and 9D). GH3:uidA expression was also stronger in hypocotyls of both the wild type and ago1-3 after auxin treatment (Figures 9C and 9D). Nevertheless, expression of the reporter gene still was not induced in the upper part of ago1-3 hypocotyls (Figure 9D). These results suggest that the regulation of GH3-like gene expression could be altered in ago1-3.
Figure 9.
Expression of the GH3:uidA Reporter Gene in ago1-3 Seedlings.
(A) and (B) GH3:uidA expression in wild-type (A) and ago1-3 (B) seedlings geminated and grown in the light for 8 d in the absence or presence of 1 μM 1-NAA. Bars = 2 mm.
(C) and (D) GH3:uidA expression in wild-type (C) and ago1-3 (D) seedlings that were etiolated for 2.5 d in the dark before transfer to the light for 44 h. Seedlings were then transferred to liquid culture medium without or with 10 μM 1-NAA for 4 h in the same growth conditions. GUS staining was overnight. From left to right in each panel: three nontreated seedling and three seedlings treated with NAA. Bars = 6 mm.
The mRNA of the Auxin Response Factor ARF17 Accumulates in the Hypocotyl of ago1-3
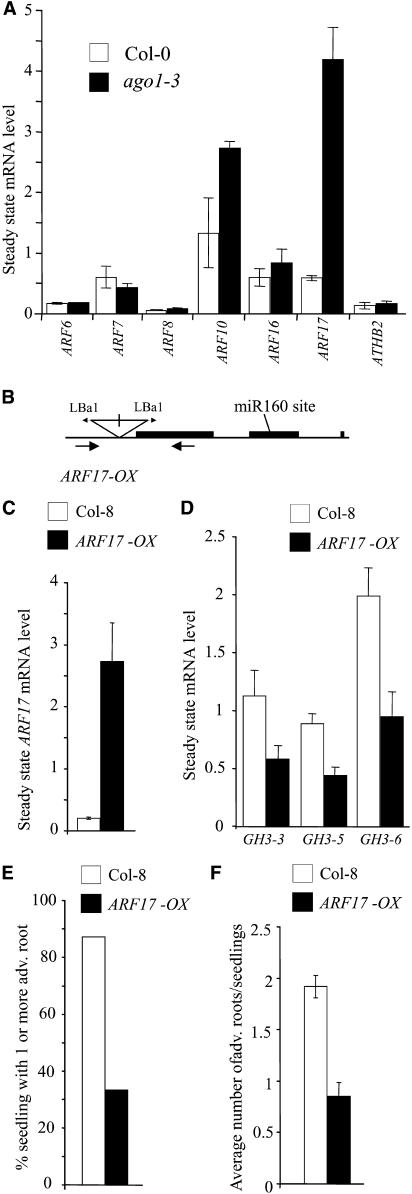
It was recently shown that ago1 mutants accumulate several miRNA targets, including two auxin response factors, ARF8 and ARF17, in rosette leaves (Vaucheret et al., 2004). Five ARF genes were identified as potential targets for miRNAs: ARF6, ARF8, ARF10, ARF16, and ARF17 (Rhoades et al., 2002). To address whether any of these genes could be involved in AGO-modified auxin responses during adventitious root formation, we analyzed their expression in wild-type and ago1-3 hypocotyls that had been etiolated for 2.5 d in the dark and transferred to the light for 48 h. A significant sevenfold increase in the steady state level of ARF17 mRNA was detected in the hypocotyl of the ago1-3 mutant (Figure 10A). Moreover, the ARF10 mRNA showed a twofold increase, and no significant difference was detected for ARF6, ARF8, or ARF16. Additionally, no difference in expression was detected for either ARF7/NPH4 or ATHB2, analyzed as transcription factors putatively not targeted by miRNAs and because they were shown previously to act in the cross talk between light and auxin signaling pathways (Stowe-Evans et al., 2001; Morelli and Ruberti, 2002).
Figure 10.
The Auxin Response Factor ARF17 Represses GH3 Genes and Adventitious Root Formation.
(A) Total RNAs were extracted from hypocotyls of Col-0 and ago1-3 siblings etiolated in the dark for 2.5 d and then transferred for 48 h into the light. The indicated mRNAs were quantified by real-time quantitative PCR using primers surrounding putative miRNA cleavage sites. ATHB2 and ARF7 were used as controls not targeted by miRNAs. Expression for each gene was normalized to that of ACTIN2. Error bars indicate se of two independent biological replicates.
(B) Scheme of the ARF17 OX T-DNA insertion line SALK 062511. Black boxes represent CDS, lines represent introns, untranslated regions, or promoters. The positions of the inverted repeat T-DNA insertion and left border primers (Lba1) are indicated. Approximate positions of the genomic primers used in genotyping are indicated with arrows below the transcript.
(C) Relative abundance of ARF17 transcript in ARF17 OX and Col-8 hypocotyls etiolated for 2.5 d and exposed to light for 2 d. Expression for the gene was normalized to that of ACTIN2. Quantification was made by real-time quantitative PCR using primers surrounding the putative miRNA cleavage site. Error bars indicate se of two independent biological replicates.
(D) Relative abundance of GH3-3, GH3-5, and GH3-6 transcripts in ARF17 OX and Col-8 hypocotyls etiolated for 2.5 d and exposed to light for 2 d. Quantification was performed using semiquantitative RT-PCR as described in Methods. Expression for the genes was normalized to that of 18S rRNA. Error bars indicate se of three independent RT-PCR replicates (P < 0.01). These experiments were repeated on two independent biological replicates.
(E) Proportion of wild-type and ARF17 OX siblings developing one or more adventitious roots after etiolation and transfer to the light for 1 week.
(F) Average number of adventitious roots formed on ARF17 OX and Col-8 after etiolation and transfer to the light for 1 week. Error bars indicate se (P < 0.01; n > 30). Observations were done on three independent biological replicates.
ARF17 Represses the Expression of GH3 Genes and Negatively Regulates Adventitious Root Formation
ARFs bind auxin response elements present in the promoter of auxin-inducible genes such as Aux/IAA, SAUR, and GH3 and either repress or activate their transcription (Tiwari et al., 2003). Although repression activity has not been demonstrated for ARF17, its sequence is more closely related to the repressor group of ARFs (Tiwari et al., 2003). Therefore, we suggest that the overexpression of ARF17 in ago1-3 could negatively regulate the expression of several GH3-related genes and in this way repress adventitious root formation. To test this hypothesis, we characterized a SALK line containing a T-DNA insertion in the promoter region of the ARF17 gene. Initial RT-PCR on the line SALK 062511 indicated the presence of an ARF17 transcript in T-DNA homozygotes. The molecular characterization of the insertion indicated the presence of two T-DNAs as inverted repeats ∼200 bp 5′ of the coding sequence (CDS) and ∼100 bp upstream of the start of transcription (Figure 10B). The pROK vector used in the generation of SALK lines (Alonso et al., 2003) carries a Cauliflower mosaic virus 35S promoter oriented toward the left border, which was likely to induce the expression of ARF17. Real-time PCR was performed as described above and indicated that the line was in fact an overexpresser. The abundance of ARF17 transcript was increased 7 times with respect to wild-type siblings in 12-d-old entire seedlings (data not shown) and ∼13 times in hypocotyls etiolated for 2.5 d followed by 2 d in the light (Figure 10C). Importantly, because the primers bound the miRNA target sequence in ARF17, the excess transcript is not immediately degraded through miRNA processes and is able to accumulate. As this line has retained kanamycin resistance (see http://signal.salk.edu/tdna_FAQs.html), a single T-DNA insertion locus was confirmed by screening the segregation of selfed progeny derived from heterozygous plants on kanamycin-supplemented medium (data not shown). Homozygote ARF17 OX plants did not show any obvious phenotypic difference compared with wild-type siblings: they grew normally in soil and were fully fertile (data not shown). To verify the hypothesis that ARF17 could repress GH3 expression, we analyzed the transcript abundance for GH3-3, GH3-5 (AtGH3a), and GH3-6 (DFL1) in hypocotyls etiolated for 2.5 d followed by 2 d in the light. All three genes were significantly repressed in ARF17 OX (Figure 10D). Because the expression of these three genes was correlated to the adventitious root number (C. Sorin, L. Negroni, T. Balliau, H. Corti, M.P. Jacquemot, M. Daventure, G. Sandberg, M. Zivy, and C. Bellini, unpublished data), we checked whether ARF17 OX was altered in the development of adventitious roots. Indeed, although the adult plant did not have an obvious phenotype, ARF17 OX produced fewer adventitious roots than did wild-type siblings after etiolation and transfer to the light for 1 week (Figures 10E and 10F). These results suggest that ARF17 could negatively regulate adventitious root formation by repressing GH3 gene expression.
DISCUSSION
ago1 Mutants Represent a New Class of Auxin-Resistant Mutants Altered in Their Capacity to Develop Adventitious Roots
We have demonstrated that the null and weak ago1 allele mutants analyzed in this study are specifically resistant to auxin in the apical part of the seedling. Indeed, they were not able to elongate in the presence of picloram, which is known to stimulate hypocotyl elongation in the wild type (Delarue et al., 1998). Nevertheless, ago1 was able to respond to auxin concentrations that inhibit hypocotyl elongation. The root responded normally to all of the auxins tested, suggesting that the auxin resistance was restricted to the apical part. Because the hypomorphic mutants display the same auxin resistance in the hypocotyl as the null mutant, this cannot be attributed to the pleiotropic developmental phenotype of the null mutant. Therefore, ago1 differs from other auxin-resistant mutants described to date, such as axr1, aux1, axr2, axr3, and axr4, which were selected for the resistance of the root to inhibitory concentrations of auxin (Estelle and Somerville, 1987; Pickett et al., 1990; Wilson et al., 1990; Hobbie and Estelle, 1995; Leyser et al., 1996). These auxin-resistant mutants, except axr4, also display cross-resistance to other hormones, such as cytokinin and ethylene. This was not the case for ago1 mutants, which responded normally to exogenous cytokinin, ethylene, and gibberellic acid. Although ago1-3 has an altered developmental phenotype, it is still able to behave like the wild type in various growth conditions. The defect in hypocotyl elongation in the presence of picloram is not attributable to a general problem of elongation, because the ago1 mutants were able to elongate in the light when grown on medium containing gibberellic acid or in the dark. Together, our results show that the apical part of ago1 is specifically resistant to auxin-mediated hypocotyl elongation.
This auxin resistance is also characterized by a reduced number of adventitious roots in the hypocotyl in response to auxin. Unlike the hypocotyl, the roots of the ago1-3 null mutant were still able to normally initiate lateral roots in response to exogenous auxin, as shown by the normal induction and expression of the CycB1:uidA reporter gene in the roots of mutant seedlings. These results strongly support the hypothesis that different regulatory pathways control lateral root and adventitious root initiation, although both root types initiate from pericycle cells. A mutation in AGO1 clearly uncouples these pathways.
We further demonstrated that the defect in adventitious rooting is not related to defective hypocotyl elongation, because ago1-3, unlike the wild type, was barely able to produce adventitious roots irrespective of the size of its hypocotyl. However, when seedlings were first etiolated and then transferred to the light on medium containing 1 μM 1-NAA, most of the ago1-3 seedlings could make at least one adventitious root, but the average number of adventitious roots always remained lower than in the wild type in the presence of auxin. This was also observed in the sur2 ago1 double mutant, indicating that ago1 plants were able to initiate adventitious roots under certain conditions. This means that the pericycle cells in the hypocotyl were still able to reenter the cell cycle in an ago1 background and that the inhibition occurred at a different level. Indeed, when the expression of the CycB1:uidA reporter gene was monitored in sur2-1(hyb) and sur2-1 ago1-3(hyb), we observed that, unlike sur2-1, the double mutant had already initiated adventitious roots at 2.5 d after germination in the dark. Nevertheless, the number of adventitious roots did not increase after transfer to the light, unlike in sur2-1 seedlings. These results could be explained by two hypotheses. (1) The ago1-3 mutation modifies the endogenous auxin content in the double mutant sur2-1 ago1-3(hyb), leading to a reduction or a blockage of adventitious rooting. This, however, would not explain the initiation of adventitious roots in the sur2-1 ago1-3(hyb) hypocotyl in the dark. (2) The ago1-3 mutation could modify light regulatory pathways in such a way that it affects auxin homeostasis and has an inhibitory effect on adventitious root initiation and development in ago1 or sur2 ago1. To further test the latter hypothesis, we analyzed the behavior of ago1 mutants in different light conditions and measured the endogenous content of IAA and IAA metabolites in the different genotypes.
Light Regulatory Pathways Are Upregulated in ago Mutants
Although the hypomorphic mutants did not show any significant phenotype in the dark, the null allele mutant ago1-3 had a shorter hypocotyl and a longer root than did the wild type. This observation supported the hypothesis of a potential deregulation of light perception and signaling, which was further tested by growing ago1 mutants under different light conditions (white, cR, cB, and cFR light). Under all conditions tested, the null allele mutant ago1-3 remained as short as in white light, and all of the weak alleles displayed a significant inhibition of hypocotyl growth compared with the wild type under low fluences of cR, cB, and cFR (P < 0.01). The different weak allele mutants did not show exactly the same behavior. Thus, it should be possible to classify the different ago1 hypomorphic alleles according to their behavior in different light conditions and possibly to identify alleles more specifically affecting one or the other pathway. If such alleles could be identified, they could be used in global approaches, such as transcriptome analysis, to identify candidate genes potentially involved in the different light regulatory pathways. Three of the hypomorphic mutants were almost as short as the null allele mutant ago1-3 in extremely low-fluence cFR, suggesting that a deregulation of the PHYA-dependent pathway might account for part of the phenotype of these ago1 mutants. Analysis of the double mutants with phyA showed a phyA epistasy on ago1, confirming that the PHYA-dependent signaling pathways were upregulated in ago1 mutants. Double mutants with other mutants affected in light perception or signaling are currently being analyzed to determine the relative contribution of the different pathways to the regulation of adventitious rooting.
ago1-3 Disrupts Auxin Homeostasis in the Apical Part of the Seedlings
Measurements of the endogenous level of free IAA showed that it was lower in ago1-3 and ago1-3 sur2-1(hyb) entire seedlings than in their respective controls Col-0 and sur2-1(hyb). Interestingly, free IAA content was shown to decrease in the apical part of ago1-3 and ago1-3 sur2-1(hyb) after transfer to the light, unlike in the root, where the auxin content was the same as in the respective controls Col-0 and sur2-1(hyb). In ago1-3, this could be explained by a lower auxin biosynthesis rate compared with that in the wild type. This result indicated that the AGO1 gene might regulate genes involved in the regulation of auxin biosynthesis. Nevertheless, the effect of ago1-3 on the auxin biosynthesis rate could not be detected in an auxin overproducer background, and no differences in auxin biosynthesis were detected in ago1-3 sur2-1(hyb) compared with sur2-1(hyb). Therefore, the lower level of free IAA in the apical part of the ago1-3 sur2-1(hyb) double mutant in the dark and its decrease after transfer of the seedlings to the light cannot be explained only by the defect in biosynthesis. This suggested that AGO1 has a more general role in the regulation of auxin homeostasis and might also be needed, directly or indirectly, for the regulation of auxin conjugation and/or catabolism. Indeed, the level of IAA conjugates was reduced in ago1-3 and ago1-3 sur2-1(hyb), indicating that IAA conjugation is downregulated in ago1-3. No significant increase in the level of oxoindole-3-acetic acid (one of the primary catabolites) could be detected in ago1-3 sur2-1(hyb) compared with sur2-1(hyb), suggesting that auxin catabolism might not be affected, although results observed with ago1-3 sur2-1(hyb) suggest that further analysis is required. Together, these results allow for the conclusion that AGO1 regulates, directly or indirectly, genes that control different aspects of auxin homeostasis in Arabidopsis.
ARF17 and GH3 Might Control Adventitious Rooting by Modulating IAA Homeostasis in a Light-Dependent Manner
Expression of auxin-inducible genes was analyzed in the ago1-3 mutant. No significant difference between the wild type and ago1-3 was detected, except for the GH3:uidA fusion. Unlike in the wild type, this reporter gene was not induced in the ago1-3 mutant hypocotyls when the seedlings were grown in the light in the presence of auxin. Nevertheless, when seedlings were first etiolated before transfer to the light, expression of GH3:uidA was detected in both wild-type and ago1-3 hypocotyls. However, in ago1-3, the expression pattern was different from that in the wild type and restricted to the bottom part of the hypocotyl. Auxin treatment increased the expression of GH3:uidA in both wild-type and ago1-3 siblings, but the expression pattern was not modified in the ago1-3 hypocotyl. This indicated that exogenous NAA could not induce GH3:uidA expression in the upper part of ago1-3 hypocotyl. These results suggested a potential deregulation of the expression of endogenous GH3 genes in ago1-3. This hypothesis was confirmed by results that we recently obtained by analyzing, through two-dimensional gel electrophoresis, the protein profile of the mutant hypocotyl (C. Sorin, L. Negroni, T. Balliau, H. Corti, M.P. Jacquemot, M. Daventure, G. Sandberg, M. Zivy, and C. Bellini, unpublished data). The expression of three GH3 proteins, GH3-3 (Staswick et al., 2002), GH3-5 (AtGH3a) (Tanaka et al., 2002), and GH3-6 (DFL1) (Nakazawa et al., 2001), was positively correlated with adventitious root formation, and they accumulated in the hypocotyls of sur2-1 but not ago1-3 sur2-1(hyb) (C. Sorin, L. Negroni, T. Balliau, H. Corti, M.P. Jacquemot, M. Daventure, G. Sandberg, M. Zivy, and C. Bellini, unpublished data). Because the endogenous auxin level in sur2-1 ago1-3(hyb) was at least twice the wild-type level, the downregulation of GH3 genes in an ago1 background is not strictly related to the auxin content. Interestingly, it has been shown that GH3-related proteins can adenylate, in vitro, several phytohormones, such as jasmonate, IAA, and salicylic acid (Staswick et al., 2002). GH3-3, GH3-5, and GH3-6 genes are induced by auxin (Hagen and Guilfoyle, 2002), and the encoded proteins belong to subgroup II of GH3 proteins that were reported to adenylate IAA in vitro (Staswick et al., 2002). Staswick et al. (2005) reported that six recombinant GH3 proteins, including the three listed above, could produce, in vitro, auxin conjugates with several amino acids and that a DFL1-overexpressing line contained an increased level of IAA-Asp. Based on these observations, we conclude that the reduction in auxin conjugates in an ago1-3 background is probably attributable to the reduced expression of GH3 genes. Some GH3 genes are regulated by both light and auxin (Hsieh et al., 2000; Nakazawa et al., 2001; Tanaka et al., 2002; Takase et al., 2003), suggesting that they could act at the cross talk of auxin and light signaling pathways. Based on our results and those described in the literature, we propose that an abnormal regulation of several GH3-related genes in ago1 mutants alters auxin homeostasis in a light-dependent manner. Here, we also show that among five ARFs that are targeted for degradation by a miRNA, only ARF17 mRNA accumulates to a high level in the hypocotyl of ago1-3. ARF17 belongs to the auxin response factor family that is represented by 23 members in Arabidopsis (Hagen and Guilfoyle, 2002). Because ARF17 lacks the characteristic protein–protein interaction domains present in the majority of ARFs and that are necessary for the interaction with other ARFs and Aux/IAA proteins (Kim et al., 1997; Ulmasov et al., 1999; Guilfoyle and Hagen, 2001), it is unlikely that ARF17 interacts either with other ARFs or with Aux/IAA proteins. ARFs bind auxin response elements present in the promoter of auxin-inducible genes such as Aux/IAA, SAUR, and GH3 and either repress or activate their transcription (Tiwari et al., 2003). Although repression activity has not been demonstrated for ARF17, its sequence is more closely related to the repressor group of ARFs (Tiwari et al., 2003). Therefore, the overexpression of ARF17 in the ago1-3 mutant could negatively regulate the expression of several GH3-related genes. This was confirmed by the characterization of an ARF17-overexpressing line, which showed a clear reduction in the expression of GH3-3, GH3-5 (AtGH3a), and GH3-6 (DFL1). ARF17 OX also produced significantly fewer adventitious roots than did the wild type, confirming the correlation between adventitious rooting and the expression level of GH3 genes observed in our proteomic experiments (C. Sorin, L. Negroni, T. Balliau, H. Corti, M.P. Jacquemot, M. Daventure, G. Sandberg, M. Zivy, and C. Bellini, unpublished data). Therefore, we conclude that the overexpression of ARF17 in ago1 is likely to be at least partially responsible for the defect in adventitious root formation.
Although at a lower level than ARF17, ARF10 also accumulates in ago1-3. ARF10 is also a potential repressor of auxin-inducible genes, and it is phylogenetically related to ARF17 and ARF16 (Okushima et al., 2005). Thus, it will be interesting to analyze whether its accumulation also contributes to the auxin-related phenotype conferred by ago1-3 and what could be the relative contribution of ARF10 and ARF17 in the control of adventitious root development.
Interestingly, it was shown recently that ARF8, which belongs to the activator group of ARFs, could positively regulate the expression of three GH3 genes, including GH3-5 and GH3-6, and in this way possibly modulate auxin homeostasis (Tian et al., 2004). Although ARF8 is targeted by a miRNA, its expression is not modified in the hypocotyl of ago1-3. We suggest that ARF17 is likely to compete with ARF8 in the regulation of GH3 gene expression.
We have demonstrated that the defect in adventitious rooting of ago1 mutants correlates with a perturbation of auxin homeostasis in the apical part of the seedling and the upregulation of light signaling pathways. Because ago1-3 is affected in biosynthesis and conjugation and potentially in degradation processes, we conclude that AGO1 is required for the proper regulation of auxin homeostasis in the apical part of the plant. We have also shown that expression of several GH3 genes is likely to be repressed by the auxin response factor ARF17 in the hypocotyl of the ago1-3, where the defect in auxin homeostasis was observed. Therefore, we suggest that ARF17 and GH3 could be involved in the control of adventitious root formation. Because several GH3 genes were shown to act in the cross talk between light and auxin signaling pathways, we suggest that ARF17 could be a major regulator of adventitious root formation by repressing GH3 genes and therefore modulating auxin homeostasis in a light-dependent manner.
METHODS
Plant Material and Growth Conditions
The origins of the different Arabidopsis thaliana ago1 mutants are described in the supplemental data online. sur1-3, sur2-1, and sur2-3 mutants have been described (Boerjan et al., 1995; Delarue et al., 1998; Barlier et al., 2000). A SALK insertion line (ecotype Col-8) with a T-DNA insertion in the promoter region of the ARF17 gene was obtained from the Nottingham Arabidopsis Stock Centre (reference number SALK 062511). Seeds from the phyA-211 mutant were obtained through the Nottingham Arabidopsis Stock Centre (reference number N6223). The GH3:uidA construct, which contains the promoter of the soybean GH3 gene (Hagen et al., 1991), was a gift from T. Guilfoyle (University of Missouri, Columbia, MO). The production of Arabidopsis transgenic lines was described by Delarue (1996). The SAUR-AC1:uidA- (Gil and Green, 1996), IAA2:uidA-, and DR5:uidA-expressing lines were provided by P. Gil (Michigan University, East Lansing, MI), A. Marchant (UPSC, Sweden), and B. Scheres (Utrecht University, The Netherlands), respectively. The CycB1:uidA-expressing line was provided by D. Inzé (VIB, Gent, Belgium). Heterozygous plants segregating the ago1-3 mutation were crossed with the different transgenic lines. Wild-type and ago1-3 seedlings expressing the marker genes were selected in F2 progeny.
Double mutants were obtained by crossing either heterozygous plants for ago1-3 or ago1-32 alleles or homozygous ago1-33, ago1-34, or ago1-35 plants with heterozygous sur1-3 (Col-0) or homozygous sur2-1 (Ws), sur2-3 (Col-0), or phyA-211 (Col-0). Double mutants were selected in the F3 generation. Because ago1-3 and sur2-1 are in two different ecotypes, the double mutant ago1-3 sur2-1, the background of which is a mix of Ws and Col-0, was always compared with its siblings, which we called sur2-1(hyb) to avoid confusion.
For seed production and crosses, the plants were grown in a greenhouse. Seeds were sown on soil, and seedlings were transferred into individual pots 10 d after germination. Plants were grown under 16 h of light with 10 to 15°C night temperature and 20 to 25°C day temperature.
For in vitro culture, seedlings were grown as described previously (Santoni et al., 1994). The conditions in the controlled-environment chambers were as follows: 150 μE·m−2·s−1 irradiance provided by BIOLUX fluorescent tubes, 16 h of light, 60% relative humidity, 15°C night temperature, and 20°C day temperature. 1-NAA was dissolved in ethanol. Picloram was dissolved in DMSO.
For dark growth conditions, Petri dishes were wrapped with four layers of aluminum foil and placed vertically.
For experiments in different light qualities, the plates were placed under continuous irradiance at a constant temperature of 20°C. Blue light (460 nm, 3.7 μE·m−2·s−1) was provided by TLD tubes 36W/18 (Philips, Montpellier, France) filtered by a Plexiglas layer (blauw number 627; Rohm and Haas, Philadelphia, PA). Red light (660 nm, 9 μE·m−2·s−1) was provided by light-emitting diodes (NLS01 number 9600; Nijssen, Utrecht, The Netherlands). Far-red light (700 to 760 nm, 0.25 μE·m−2·s−1) was provided by True-Lite fluorescent tubes (Bio-Elektrik, Leinburg, Germany) filtered by a layer of Plexiglas (Rohm and Haas).
Histochemical GUS Assays
Histochemical assays for GUS expression were performed as described previously (Mollier et al., 1995).
Hypocotyl and Root Measurements
Hypocotyl and root measurements were performed as described by Gendreau et al. (1997). At least 30 seedlings were used for each data point. All measurements were done on two independent biological replicates. Error bars in the figures indicate sd.
Scoring of Adventitious Roots
Seedlings were etiolated in the dark for 2.5 d and then transferred to the light. Emergent adventitious roots were scored at 7 d after transfer to the light. At least 40 seedlings were used for each data point. This was repeated on three independent biological replicates.
Adventitious root primordia and emerging adventitious roots were scored in sur2-1 ago1-3(hyb) and sur2-1 expressing the CycB1:uidA reporter gene. GUS staining was performed before counting blue dots (very early primordia) and emerging roots on the hypocotyls using a stereomicroscope. For each data point, 30 seedlings were analyzed. The experiment was repeated three times for time point 0 (i.e., seedlings grown in the dark only).
Molecular Characterization of the arf17 Insertion Line
The line SALK 062511 (Alonso et al., 2003) carries a T-DNA insertion ∼200 bp 5′ of the CDS and ∼100 bp upstream of the start of transcription. The flanking sequence tag for this line (http://signal.salk.edu/cgi-bin/tdnaexpress) runs upstream into the promoter. PCRs of genomic DNA performed using the SALK T-DNA primer Lba1 (http://signal.salk.edu/tdnaprimers.html) in combination with the ARF17 genomic primers ARF17-1S-LF (5′-CGAGGGATAAGCACAAAAACATGA-3′) and ARF17-1S-RR (5′-CCGTTGTTAGCAAGTGACGCC-3′) indicated the presence of two T-DNAs as inverted repeats. Sequencing across the left border toward ARF17-1S-RR confirmed the insertion point and the presence of the inverted repeat (Figure 10B). The pROK vector used to generate SALK lines (Alonso et al., 2003) carries the Cauliflower mosaic virus 35S promoter oriented toward the left border. The presence of the intact 35S promoter was confirmed by sequencing.
PCR Experiments
Hypocotyls were collected from mutant and wild-type siblings segregating from heterozygous parents that had been grown in the dark for 2.5 d, then in the light for 2 d. Two independent biological replicate experiments were performed. RNA was extracted from approximately 50 mg of hypocotyls using the RNAqueous kit (Bio-Rad, Hercules, CA). cDNAs were made using the iScript kit (Bio-Rad).
Real-time RT-PCR template quantification was performed using a Bio-Rad iCycler with iQ SYBR Green Supermix (Bio-Rad). This followed a two-step protocol: 95°C for 3 min, followed by 50 cycles of denaturation at 95°C for 30 s and annealing/extension at 60°C for 45 s. Melt curves were derived after each amplification by increasing the temperature in 0.5°C increments from 55 to 95°C. Reactions were performed in triplicate for each sample. Acceptable standard curves were those for which 1 ≥ E ≥ 0.85 and r2 ≥ 0.985, where E is the PCR efficiency and r2 is the correlation coefficient obtained with the standard curve.
Primers for At3g18780/ACTIN2, At5g37020/ARF8, and At1g77850/ARF17 are given by Vazquez et al. (2004). Other primers were as follows: At1g30330/ARF6 (5′-TGCGAAGCGAGCTTGCTC-3′ and 5′-GCTCACAAACTCCGGCCAAG-3′); At5g20730/ARF7 (5′-AGAACTCAATCTTTTGGTGTC-3′ and 5′-CGTTTTTGCACCTTTGTATAAG-3′); At2g28350/ARF10 (5′-ACAATGGCGGTGGCGAGTC-3′ and 5′-GATGGTGATCCGAAGAGTTGTTGAG-3′); At4g30080/ARF16 (5′-AAGCCCGTTAAGCTCTGTTC-3′ and 5′-GGAGGAGGAGGTGGTCTATTC-3′); and At4g16780/ATHB2 (5′-ACATGAGCCCACCCACTAC-3′ and 5′-GAAGAGCGTCAAAAGTCAAGC-3′). Quantification of each gene was normalized to ACTIN2.
Semiquantitative RT-PCR was performed using as internal standard the 18S rRNA primers/competimers (Ambion, Austin, TX). PCR amplification was performed using Taq DNA polymerase (New England Biolabs, Beverly, MA). For each gene-specific primer pair, the optimum annealing temperature and linear amplification range were predetermined using 0.012 μg of RNA per 10 μL of PCR assay. The sequences of the gene-specific primers used for RT-PCR were as follows: At2g23170/GH3-3 (5′-AAGTTTGTGCGGAGGAAGAA-3′ and 5′-AAAGCGGGCTGAAGTGTGT-3′); At4g27260/GH3-5 (5′-AATGCCAACAATCGAAGAGG-3′ and 5′-CTTGCACTCAAATTCCACGA-3′); and At5g54510/GH3-6 (5′-CCTATGCTGGGCTTTACAGG-3′ and 5′-ACCAGGGGACCATTTAGGAC-3′). The PCR products were resolved on a 1.5% agarose gel and stained with ethidium bromide. The PCR products were then quantified by scanning densitometry. A t test was performed according to http://graphpad.com/quickcalcs/ttest1.cfm.
Analysis of Endogenous IAA Content
Seedlings were grown in the dark for 2.5 d and then transferred to the light for 24, 48, or 72 h. Entire seedlings, the apical part (cotyledons plus hypocotyl), or roots from several seedlings were pooled to obtain an average of 15 mg of fresh weight.
Samples were extracted, purified, and analyzed by gas chromatography–selected reaction monitoring–mass spectrometry as described previously (Edlund et al., 1995). Calculation of the isotopic dilution factors was based on the addition of 50 pg [13C6]IAA/mg tissue. A t test was performed according to http://graphpad.com/quickcalcs/ttest1.cfm.
Quantitative Analysis of IAA Conjugates and Catabolites
Seedlings were grown in the dark for 2.5 d. Entire seedlings were pooled to obtain 30 to 70 mg of fresh weight. Samples were extracted, purified, and analyzed by liquid chromatography–multiple reaction monitoring–mass spectrometry as described previously (Kowalczyk and Sandberg, 2001). A t test was performed according to http://graphpad.com/quickcalcs/ttest1.cfm.
Deuterium-Feeding Experiments
Seedlings were grown in the dark for 2.5 d and then transferred to the light for 48 h. Then, entire seedlings, apical parts, or roots were transferred to liquid culture medium (Santoni et al., 1994) with or without 30% deuterated water for 20 h. The samples were then weighed and frozen in liquid nitrogen. The extraction and purification were performed as described previously (Ljung et al., 2001). Calculation of isotopic dilution was based on the addition of 50 pg [13C6]IAA/mg tissue. Analysis by gas chromatography–high-resolution mass spectrometry was done at a resolution of at least 10,000, and isotopomers of the base peaks of methylated and trimethylsilylated IAA were measured with mass-to-charge ratios of 202.105, 203.112, 204.118, 205.124, and 208.125. Corrections were incorporated for the contribution of natural isotopic abundances to mass-to-charge ratios of 203 to 205. The incorporation of deuterium into the IAA molecule was then calculated. Corrections for background were made by analyzing the samples from the control plants grown without deuterated water. A t test was performed according to http://graphpad.com/quickcalcs/ttest1.cfm.
Supplementary Material
Acknowledgments
The authors thank Roger Granbom for technical assistance and Rishikesh Bhalerao, Ove Nilsson, Alan Marchant, Arsenio Villarejo, and Markus Grebe for discussions and critical reading of the manuscript. This work was supported by the Commission of the European Communities (Contract ERBIO4CT960689), the Institut National de la Recherche Agronomique (C.B. and H.V.), the Swedish Natural Sciences Research Council (G.S.), and the Swedish Foundation for Strategic Research (C.B.). C.S. and I.C. were the recipients of PhD fellowships from the Ministère de la Recherche et de l'Enseignement Supérieur. G.G. is currently the recipient of a PhD fellowship from the Ministère de la Recherche et de l'Enseignement Supérieur.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Catherine Bellini (catherine.bellini@genfys.slu.se).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031625.
References
- Abel, S., Nguyen, D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Bak, S., Tax, F.E., Feldmann, K.A., Galbraith, D.W., and Feyereisen, R. (2001). CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeckman, T., Burssens, S., and Inze, D. (2001). The peri-cell-cycle in Arabidopsis. J. Exp. Bot. 52, 403–411. [DOI] [PubMed] [Google Scholar]
- Blakesley, D. (1994). Auxin metabolism and adventitious root initiation. In Biology of Adventitious Root Formation, T.D. Davis and B.E. Haissig, eds (New York: Plenum Press), pp. 143–154.
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Vanonckelen, H., Vanmontagu, M., and Inze, D. (1995). superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus, I. (1999). ARGONAUTE d'Arabidopsis thaliana Définit une Famille de Gènes Conservés chez les Eucaryotes: Impliqués dans le Développement. PhD dissertation (Paris: Université Pierre et Marie Curie).
- Carmell, M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. (2002). The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16, 2733–2742. [DOI] [PubMed] [Google Scholar]
- Cerutti, L., Mian, N., and Bateman, A. (2000). Domains in gene silencing and cell differentiation proteins: The novel PAZ domain and redefinition of the Piwi domain. Trends Biochem. Sci. 25, 481–482. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Letham, S., Craig, S., and Dennis, E.S. (1993). Amp1-A mutant with high cytokinin levels and altered embryonic pattern, faster vegetative growth, constitutive photomorphogenesis and precocious flowering. Plant J. 4, 907–916. [Google Scholar]
- Delarue, M. (1996). Approches Génétiques et Physiologiques du Développement d'Arabidopsis thaliana: Caratérisation des mutants cristal et superroot. PhD dissertation (Paris: Université Pierre et Marie Curie).
- Delarue, M., Prinsen, E., Vanonckelen, H., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostasis. Plant J. 14, 603–611. [DOI] [PubMed] [Google Scholar]
- Edlund, A., Eklof, S., Sundberg, B., Moritz, T., and Sandberg, G. (1995). A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 108, 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fett-Neto, A.G., Fett, J.P., Veira Goulart, L.W., Pasquali, G., Termignoni, R.R., and Ferreira, A.G. (2001). Distinct effects of auxin and light on adventitious root development in Eucalyptus saligna and Eucalyptus globulus. Tree Physiol. 21, 457–464. [DOI] [PubMed] [Google Scholar]
- Fukaki, H., Tameda, S., Masuda, H., and Tasaka, M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29, 153–168. [DOI] [PubMed] [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Hofte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil, P., and Green, P.J. (1996). Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: The 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 15, 1678–1686. [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 9, 7187–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle, T., and Hagen, G. (2001). Auxin response factors. J. Plant Growth Regul. 20, 281–291. [Google Scholar]
- Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. [PubMed] [Google Scholar]
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hansen, H., and Grossmann, K. (2000). Auxin-induced ethylene triggers abscisic acid biosynthesis and growth inhibition. Plant Physiol. 124, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7, 211–220. [DOI] [PubMed] [Google Scholar]
- Hsieh, H.L., Okamoto, H., Wang, M., Ang, L.H., Matsui, M., Goodman, H., and Deng, X.W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hutvagner, G., and Zamore, P.D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. [DOI] [PubMed] [Google Scholar]
- Ishizuka, A., Siomi, M.C., and Siomi, H. (2002). A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 16, 2497–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428, 81–84. [DOI] [PubMed] [Google Scholar]
- Kim, J., Harter, K., and Theologis, A. (1997). Protein-protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 94, 11786–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.J., and Stimart, D.P. (1998). Genetic analysis of variation for auxin-induced adventitious root formation among eighteen ecotypes of Arabidopsis thaliana L. Heynh. J. Hered. 89, 481–487. [DOI] [PubMed] [Google Scholar]
- Konishi, M., and Sugiyama, M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130, 5637–5647. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, M., and Sandberg, G. (2001). Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127, 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Leyser, H.M.O., Pickett, F.B., Dharmaseri, S., and Estelle, M. (1996). Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 10, 403–413. [DOI] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J. 28, 465–474. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Naur, P., and Halkier, B.A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. [DOI] [PubMed] [Google Scholar]
- Mollier, P., Montoro, P., Delarue, M., Bechtold, N., Bellini, C., and Pelletier, G. (1995). Promoterless gusA expression in a large number of Arabidopsis thaliana transformants obtained by the in planta infiltration method. C. R. Acad. Sci. Paris 318, 465–474. [Google Scholar]
- Morel, J.B., Godon, C., Mourrain, P., Beclin, C., Boutet, S., Feuerbach, F., Proux, F., and Vaucheret, H. (2002). Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli, G., and Ruberti, I. (2002). Light and shade in the photocontrol of Arabidopsis growth. Trends Plant Sci. 7, 399–404. [DOI] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa, M., Yabe, N., Ichikawa, T., Yamamoto, Y.Y., Yoshizumi, T., Hasunuma, K., and Matsui, M. (2001). DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J. 25, 213–221. [DOI] [PubMed] [Google Scholar]
- Niemi, K., Julkunen-Tiitto, R., Tegelberg, R., and Haggman, H. (2005). Light sources with different spectra affect root and mycorrhiza formation in Scots pine in vitro. Tree Physiol. 25, 123–128. [DOI] [PubMed] [Google Scholar]
- Okushima, Y., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, F.B., Wilson, A.K., and Estelle, M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94, 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Rouse, D., Mackay, P., Stirnberg, P., Estelle, M., and Leyser, O. (1998). Changes in auxin response from mutations in an AUX/IAA gene. Science 279, 1371–1373. [DOI] [PubMed] [Google Scholar]
- Santoni, V., Bellini, C., and Caboche, M. (1994). Use of two-dimensional protein-pattern analysis for the characterization of Arabidopsis thaliana mutants. Planta 192, 557–566. [Google Scholar]
- Staswick, P.E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M.T., Maldonado, M.C., and Suza, W. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17, 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe-Evans, E.L., Luesse, D.R., and Liscum, E. (2001). The enhancement of phototropin-induced phototropic curvature in Arabidopsis occurs via a photoreversible phytochrome A-dependent modulation of auxin responsiveness. Plant Physiol. 126, 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase, T., Nakazawa, M., Ishikawa, A., Manabe, K., and Matsui, M. (2003). DFL2, a new member of the Arabidopsis GH3 gene family, is involved in red light-specific hypocotyl elongation. Plant Cell Physiol. 44, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Tanaka, S., Mochizuki, N., and Nagatani, A. (2002). Expression of the AtGH3a gene, an Arabidopsis homologue of the soybean GH3 gene, is regulated by phytochrome B. Plant Cell Physiol. 43, 281–289. [DOI] [PubMed] [Google Scholar]
- Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M.K., Harper, R.M., Liscum, E., and Yamamoto, K.T. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, C., Muto, H., Higuchi, K., Matamura, T., Tatematsu, K., Koshiba, T., and Yamamoto, K.T. (2004). Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light conditions. Plant J. 40, 333–343. [DOI] [PubMed] [Google Scholar]
- Timpte, C., Lincoln, C., Pickett, F.B., Turner, J., and Estelle, M. (1995). The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 8, 561–569. [DOI] [PubMed] [Google Scholar]
- Tiwari, S.B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.K., Picket, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222, 377–383. [DOI] [PubMed] [Google Scholar]
NOTE ADDED IN PROOF
- The word hybrid is misused in this article. A hybrid is an F1 product. The double mutant sur2-1 ago1-3 is indeed in a recombinant genotype between Col-0 and Ws. Hybrid should be understood here as recombinant genotype.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.