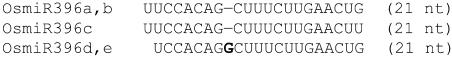Abstract
MicroRNAs (miRNAs) are a growing family of small noncoding RNAs that downregulate gene expression in a sequence-specific manner. The identification of the entire set of miRNAs from a model organism is a critical step toward understanding miRNA-guided gene regulation. Rice (Oryza sativa) and Arabidopsis thaliana, two plant model species with fully sequenced genomes, are representatives of monocotyledonous and dicotyledonous flowering plants, respectively. Thus far, experimental identification of miRNAs in plants has been confined to Arabidopsis. Computational analysis based on conservation with known miRNAs from Arabidopsis has predicted 20 families of miRNAs in rice. To identify miRNAs that are difficult to predict in silico or not conserved in Arabidopsis, we generated three cDNA libraries of small RNAs from rice shoot, root, and inflorescence tissues. We identified 35 miRNAs, of which 14 are new, and these define 13 new families. Thirteen of the new miRNAs are not conserved in Arabidopsis. Four of the new miRNAs are conserved in related monocot species but not in Arabidopsis, which suggests that these may have evolved after the divergence of monocots and dicots. The remaining nine new miRNAs appear to be absent in the known sequences of other plant species. Most of the rice miRNAs are expressed ubiquitously in all tissues examined, whereas a few display tissue-specific expression. We predicted 46 genes as targets of the new rice miRNAs: 16 of these predicted targets encode transcription factors, and other target genes appear to play roles in diverse physiological processes. Four target genes have been experimentally verified by detection of miRNA-mediated mRNA cleavage. Our identification of new miRNAs in rice suggests that these miRNAs may have evolved independently in rice or been lost in other species.
INTRODUCTION
MicroRNAs (miRNAs) are short, endogenous noncoding RNAs found in animals (Lee et al., 1993; Wightman et al., 1993; Lagos-Quintana et al., 2001, 2002, 2003; Lau et al., 2001; Lee and Ambros, 2001; Mourelatos et al., 2002; Ambros et al., 2003a; Aravin et al., 2003; Dostie et al., 2003; Grad et al., 2003; Lim et al., 2003a, 2003b), plants (Llave et al., 2002a; Mette et al., 2002; Park et al., 2002; Reinhart et al., 2002; Palatnik et al., 2003; Floyd and Bowman, 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004a, 2004b; Adai et al., 2005), and the Epstein-Barr virus (Pfeffer et al., 2004). In both animals and plants, the majority of the miRNA genes exist as independent transcriptional units, and they are transcribed by RNA polymerase II into long primary transcripts (termed pri-miRNAs) (Bartel, 2004; Kurihara and Watanabe, 2004; Parizotto et al., 2004). In animals, pri-miRNAs are trimmed in the nucleus to generate ∼70-nucleotide miRNA precursors (pre-miRNAs) with fold-back structures by a multiprotein complex called microprocessor, in which Drosha (an RNase III–like enzyme) and Pasha (a double-stranded RNA binding protein) are critical components (Lee et al., 2003; Denli et al., 2004). The pre-miRNAs are exported to the cytoplasm and subsequently cleaved by another RNase III–like enzyme called Dicer to generate mature miRNAs (Bernstein et al., 2001). However, the Arabidopsis thaliana genome does not appear to encode a Drosha ortholog, and it seems that the plant nuclear-localized Dicer homolog is likely to have Drosha function (Kurihara and Watanabe, 2004). Many miRNAs are conserved between species—often over wide evolutionary distances. For example, AthmiR166 is conserved in all lineages of land plants, including bryophytes, lycopods, ferns, and seed plants (Floyd and Bowman, 2004), and the Caenorhabditis elegans miRNA, let-7, is conserved in human, Drosophila melanogaster, and 11 other bilateral animals (Pasquinelli et al., 2000); but others are only conserved between more closely related species such as C. elegans and C. briggsae (Ambros et al., 2003a; Bartel, 2004). miRNAs downregulate the expression of specific mRNA targets, either by directing the cleavage of mRNAs or interfering with translation (Carrington and Ambros, 2003; Ambros, 2004; Bartel, 2004).
miRNAs have been identified by cloning and by computational approaches tailored to the key features of lin-4 and let-7, the two founding members of miRNAs from C. elegans, which include a fold-back hairpin RNA precursor coupled with evolutionary conservation (Ambros et al., 2003b). It was estimated that miRNA genes represent 1% of the expressed genome in complex organisms such as worms, flies, and humans (Lai, 2003; Lim et al., 2003b; Bartel, 2004). However, recent computational predictions have raised the number of miRNAs significantly in primates by comparative analysis of the human, mouse, and rat genomes (Berezikov et al., 2005). The identification of the entire set of miRNAs and their target genes from model organisms is of fundamental importance to understand regulatory networks and gene silencing mechanisms.
Rice (Oryza sativa) is the world's most important crop, as measured by the portion of calories it provides to the human diet. It is an established model system for monocots that includes all cereals. Rice is the only monocot species with a fully sequenced genome. The availability of the complete genome sequence of rice allowed the in silico identification of 20 families of rice miRNAs based on conservation of sequences with Arabidopsis miRNAs (Park et al., 2002; Reinhart et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004a; Adai et al., 2005). In addition to finding conserved miRNAs, cloning approaches revealed Arabidopsis miRNAs that are not conserved in rice. At least four well-characterized Arabidopsis miRNAs, miR158, miR161, miR163, and miR173, do not have homologs in rice (Jones-Rhoades and Bartel, 2004). Another miRNA (miR403) has been found to be conserved between Arabidopsis and Populus, whereas its counterpart could not be identified in rice (Sunkar and Zhu, 2004). Recently, evidence was shown that the nonconserved miR161 and miR163 from Arabidopsis may have evolved by inverted duplication of their target genes (Allen et al., 2004). Additionally, Berezikov et al. (2005) have predicted lineage-specific miRNAs in mammalian and nonmammalian animal species. Taken together, these observations support the notion that rice may express monocot- and/or rice-specific miRNAs.
This study was undertaken to identify new miRNAs that are difficult to predict in silico and verify previously predicted miRNAs from rice. Sequencing of small RNA libraries and subsequent analysis led to the identification of 14 new miRNAs. These new miRNAs from rice form 14 families, 13 of which are new and not present in Arabidopsis. Furthermore, we confirmed the existence of 15 of the 20 conserved families of miRNAs that were predicted previously. Based on sequence complementarity to miRNAs, we were able to predict 46 rice genes as putative targets of the new miRNAs. These predicted targets include not only transcription factors but also other genes involved in diverse physiological processes.
RESULTS
Identification of 13 New Families of Rice miRNAs
Because the available computational approaches can only identify miRNAs that are conserved between Arabidopsis and rice, a cloning approach was employed to identify rice miRNAs that may not be conserved or may have atypical features (Dugas and Bartel, 2004). To this end, we generated three independent small RNA libraries from rice in the size range of 18 to 26 nucleotides, from the shoots and roots of seedlings and inflorescence tissues of adult plants (O. sativa spp japonica cv Nipponbare). Small RNAs were isolated by size fractionation, ligated to 5′ and 3′ adapters, cloned, and sequenced. A total of ∼10,000 clones were sequenced (approximately one-third from each library), of which ∼5000 small cDNA sequences were between 18 and 26 nucleotides in length. The remaining sequences had either shorter fragments or self-ligated adapters. BLASTN searches revealed that 97% of these sequences have at least one match in the rice nuclear genome sequence version 3 annotated by The Institute for Genomic Research (TIGR) (www.tigr.org). The remaining 3% did not have a match and were not analyzed further. The lack of a match of these sequences may be due to unfinished regions in the rice genome sequence, sequencing errors, or other possibilities. Several clones were mapped to chloroplast or mitochondrial genomes and may represent either degradation or possibly regulatory products of organellar RNAs. The largest class of cloned RNAs represents fragments of abundant noncoding RNAs (rRNA, tRNA, small nuclear RNA, and small nucleolar RNA) as determined by BLASTN searches against the Rfam database. A small fraction represents mRNA breakdown products from rice. The remaining sequences constitute miRNAs (Tables 1 and 2) and endogenous small interfering RNAs (siRNAs) (data not shown). For 95% of the endogenous siRNAs, we could not detect their expression on small RNA gel blots.
Table 1.
New Rice miRNAs Identified by Cloning
| miRNA | Sequence (5′→3′) | RNA Blot | Size (nt) | No. of Loci | Chr. | Fold-Back (5′/3′) | Intergenic/Intron/Exon | Osi | Zm | At | Pt |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR390 | AAGCUCAGGAGGGAUAGCGCC (2) | + | 21 | 1 | 3 | 5′ | Intergenic | + | + | + | + |
| miR396d | UCCACAGGCUUUCUUGAACUG (10) | + | 21 | 2 | 4 | 5′ | Intergenic | + | + | − | − |
| miR396e | 2 | 5′ | Intergenic | ||||||||
| miR435 | UUAUCCGGUAUUGGAGUUGA (2) | + | 20 | 1 | 3 | 3′ | 11669.m03227_Intron_7(s) | + | − | − | − |
| miR436 | UGAGAGAGUGCACUUUCUCCC (2) | + | 21 | 1 | 2 | 5′ | J023035E19 (s) | + | − | − | − |
| miR437 | AAAGUUAGAGAAGUUUGACUU (2) | + | 21 | 1 | 2 | 3′ | 11668.m02729_Intron_15(s) | + | + | − | − |
| miR438 | UUCCCACGCGUUAUAGUGAAA (1) | + | 21 | 1 | 6 | 5′ | Intergenic | + | − | − | − |
| miR439a | UGUCGAACCGCG GUUGUUCGA (1) | + | 21 | 10 | 1 | 3′ | Intergenic | + | − | − | − |
| miR439b | 10 | 3′ | Intergenic | ||||||||
| miR439c | 1 | 3′ | Intergenic | ||||||||
| miR439d | 3 | 3′ | Intergenic | ||||||||
| miR439e | 7 | 3′ | Intergenic | ||||||||
| miR439f | 8 | 3′ | Intergenic | ||||||||
| miR439g | 8 | 3′ | Intergenic | ||||||||
| miR439h | 6 | 3′ | Intergenic | ||||||||
| miR439i | 9 | 3′ | Intergenic | ||||||||
| miR439j | 10 | 3′ | 11676.m02626_Intron_1(s) | ||||||||
| miR440 | AGUGUCUCCUGAUGAUCGGGACAA (2) | + | 24 | 1 | 11 | 3′ | 11687.m01522_Intron_9(s) | + | − | − | − |
| miR441a | UACCAUCAAUAUAAAUGUGGGAAA (3) | + | 24 | 3 | 1 | 3′ | Intergenic | + | − | − | − |
| miR441b | 3 | 3′ | Intergenic | ||||||||
| miR441c | 7 | 3′ | Intergenic | ||||||||
| miR442 | UGACGUGUAAAUUGCGAGACGAAU (2) | + | 24 | 1 | 4 | 5′ | 11670.m05347_Intron_1(s) | + | − | − | − |
| miR443 | AUCACAAUACAAUAAAUCUGGA (2) | + | 22 | 1 | 3 | 5′ | intergenic | + | − | − | − |
| miR444 | UUGCUGCCUCAAGCUUGCUGC (3) | + | 21 | 1 | 8 | 3′ | J033125N22 (s) | + | + | − | − |
| miR445a | UAAAUUAGUGUAUAAACAUCCGAU (2) | + | 24 | 9 | 5 | 3′ | Intergenic | + | + | − | − |
| miR445b | 6 | 3′ | Intergenic | ||||||||
| miR445c | 12 | 3′ | Intergenic | ||||||||
| miR445d | 3 | 3′ | Intergenic | ||||||||
| miR445e | 7 | 3′ | Intergenic | ||||||||
| miR445f | 3 | 3′ | Intergenic | ||||||||
| miR445g | 6 | 3′ | Intergenic | ||||||||
| miR445h | 3 | 3′ | 11669.m00786_Intron_2(as) | ||||||||
| miR445i | 6 | 3′ | 11680.m00766_Intron_1(s) | ||||||||
| miR446 | CAUCAAUAUGAAUAUGGGAAAUGG (2) | + | 24 | 1 | 6 | 3′ | Intergenic | + | − | − | − |
Frequency of cloning is indicated in parentheses after the miRNA sequences. Chromosomal (Chr.) position is indicated. miRNA location in the predicted fold-back structure is specified (5′ or 3′ arm). miRNAs that are conserved in other plants are indicated. Osi, O. sativa subspecies indica; Zm, Z. mays; At, A. thaliana; Pt, P. trichocarpa. nt, nucleotides.
Table 2.
Experimental Verification of Previously Predicted miRNAs in Rice
| Gene | miRNA Sequence (5′→3′) | Cloning Frequencies in Libraries |
|---|---|---|
| OsmiR156a-j | UGACAGAAGAGAGUGAGCAC | Shoot (12), root (5) |
| OsmiR156k,l | UGACAGAAGAGAGUGAGCACA | Shoot (3), root (1), inflorescence (1) |
| OsmiR 159a,b | UUUGGAUUGAAGGGAGCUCU | Shoot (3), inflorescence (2) |
| OsmiR159c | UUGGAUUGAAGGGAGCUCUGC | Shoot (2) |
| OsmiR160 | UGCCUGGCUCCCUGUAUGCCA | Shoot (1) |
| OsmiR164 | AUGGAGAAGCAGGGCACGUGCA | Shoot (3), inflorescence (3) |
| OsmiR166a-f | UCGGACCAGGCUUCAUUCCCC | Shoot (2), inflorescence (1) |
| OsmiR167a,b,c | UGAAGCUGCCAGCAUGAUCUA | Shoot (2) |
| OsmiR167d-I | UGAAGCUGCCAGCAUGAUCUG | Root (1) |
| OsmiR168a | UCGCUUGGUGCAGAUCGGGAC | Shoot (14), root (3), inflorescence (5) |
| OsmiR169b,c | CAGCCAAGGAUGACUUGCCGG | Shoot (2) |
| OsmiR169f,g | UAGCCAAGGAUGACUUGCCUA | Shoot (2), root (1) |
| OsmiR169 h-m | UAGCCAAGGAUGACUUGCCUG | Shoot (1) |
| OsmiR171a-f | UGAUUGAGCCGUGCCAAUAUC | Shoot (1), root (1), inflorescence (2) |
| OsmiR171g | GAGGUGAGCCGUGCCAAUAUC | Root (1) |
| OsmiR172a | AGAAUCUUGAUGAUGCUGCAU | Shoot (2), inflorescence (2) |
| OsmiR393 | UCCAAAGGGAUCGCAUUGAUC | Inflorescence (1) |
| OsmiR397b | UUAUUGAGUGCAGCGUUGAUG | Shoot (1), root (1) |
| OsmiR398 | UGUGUUCUCAGGUCGCCCCUG | Shoot (1) |
| OsmiR399a | UGCCAAAGGAGAAUUGCCCUG | Shoot (1) |
| OsmiR408 | CUGCACUGCCUCUUCCCUGGC | Root (1) |
miRNAs were distinguished from endogenous siRNAs on the basis of the ability of the miRNA surrounding sequences to adopt a hairpin structure (see Supplemental Figure 1 online). This analysis revealed that we had cloned 35 rice miRNAs. We also found one small RNA sequence that corresponds to OsmiR399g*. Sequence similarity searches against the central miRNA registry (http://www.sanger.ac.uk/Software/Rfam/mirna/search.shtml) showed that 14 of the miRNAs are new (Table 1). The remaining 21 (belonging to 15 families) were identical with previously predicted miRNAs in rice (Table 2). The newly identified 14 miRNAs correspond to 34 loci. These new miRNAs belong to two predominant size classes: 21 and 24 nucleotides in length (Figure 1). Nine of the 14 newly identified miRNAs begin with a 5′ uridine, which is a characteristic feature of miRNAs (Table 1). All 14 new miRNAs are perfectly conserved in Indica rice (O. sativa spp indica).
Figure 1.
Size Distribution of New miRNAs Cloned from Rice.
One of the newly identified miRNAs is represented by two genomic loci, OsmiR396d and OsmiR396e, and is a member of previously predicted OsmiR396 family in rice (Table 1, Figure 2). OsmiR396d was represented by 10 clones in our libraries and differed slightly in sequence from that of predicted OsmiR396 (Figure 2). The predicted OsmiR396 has three genomic loci (OsmiR396a, b, and c) and is represented by two members (Jones-Rhoades and Bartel, 2004). OsmiR396d differs from these two members by the presence of an additional nucleotide G between positions 8 and 9. Using a specific probe, we detected the expression of miR396d in rice and maize (Zea mays) but not in Arabidopsis (Figure 7A). Although cross-hybridization often occurs between members in the same miRNA family, this is prevented by the presence of an additional nucleotide in the middle of miR396d. Consistent with the absence of a signal in the Arabidopsis RNA gel blots, the miR396d sequence is not present in the Arabidopsis genome. OsmiR396d sequence and the secondary structure of its precursor sequences are conserved in barley (Hordeum vulgare), another monocotolydonous plant (data not shown).
Figure 2.
OsmiR396d Is a Unique Member of the Previously Predicted OsmiR396 Family in Rice.
Sequence alignment of miR396d and predicted members of OsmiR396 family. Bold letter represents the additional nucleotide (nt) in the new miRNA.
Figure 7.
Expression Patterns of New Rice miRNAs That Are Conserved in Another Monocot (Maize) or in Dicot (Arabidopsis) and Monocot (Maize).
The tRNA and 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls. Labeled RNA oligonucleotide was used as a size marker, and the position was indicated. nt, nucleotides.
Our sequence analysis indicated that we also identified a new miRNA that is conserved between monocots and dicots. We identified miR390 in rice through cloning, whereas the Arabidopsis counterpart was predicted through recent computational approaches (Bonnet et al., 2004; Wang et al., 2004b). miR390 is represented by one member with one locus in rice (Table 1), whereas in Arabidopsis and Populus, it is represented by two members with three and six loci, respectively (see Supplemental Figure 2 online).
Genomic Organization of the New Rice miRNAs
Genomic locations of the new miRNA genes in rice are shown in Table 1. The 14 newly identified miRNAs correspond to 34 loci. Hairpin structures can be predicted for all these 34 loci using miRNA surrounding sequences (see Supplemental Figure 1 online). Ten of these are encoded by single copy miRNA genes, whereas the other four (miR439, miR396d, miR441, and miR445) have multiple loci in the genome (Table 1). The exact origins of miRNAs corresponding to multiple genomic loci cannot be assigned unambiguously, and some of the loci could be pseudogenes. Our analysis of the genomic positions of the new miRNA genes shows that the majority localizes to intergenic regions (25 out of 34 loci). However, seven correspond to introns of protein-coding genes in either the sense (6) or antisense (1) orientation (Table 1). Our characterization of intronic origins of miRNAs was based on the latest annotation of the O. sativa spp japonica genome (version 3.0). Two miRNAs (miR439 and miR445) map to both intergenic and intronic locations. Two (miR435 and miR440) are derived from introns only. Another two miRNAs (miR436 and miR444) originate from the exons of protein-coding genes in the sense polarity (Table 1).
Biogenesis of OsmiR436 and OsmiR444
In general, most of the 20- to 25-nucleotide mature miRNAs are processed from a 70- to 300-nucleotide precursor, forming a hairpin structure that contains mature miRNA in either of its arms. Two miRNAs, miR436 and miR444, were mapped to the exons of the protein-coding genes J023035E19 (AK120922) and J033125N22 (AK103332), respectively, in the sense polarity (Table 1). The existence of these processed transcripts is supported by expression data (Kikuchi et al., 2003). Both of the precursor transcripts can form hairpin structures, and the miRNAs were detected on small RNA gel blots as discrete bands, suggesting that these are not nonspecific degradation products.
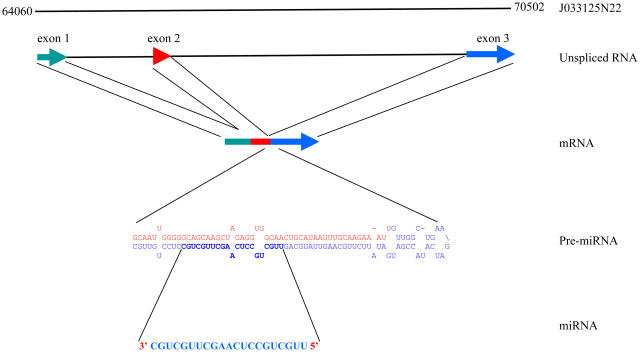
The biogenesis of OsmiR444 and OsmiR436 is unusual because the fold-back structure could not be predicted directly from the genomic sequence surrounding the miRNA. A hairpin structure can be predicted for a processed transcript (part of exons 2 and 3 sequences) but not with the genomic sequence, which suggests that the presence of an intron prevented the identification of a fold-back structure in the genomic locus of miR444 (Figure 3). The mature miR444 resides in the 3rd exon of the gene J033125N22. The open reading frame (ORF) of this gene is predicted to code for an unknown protein of 50 amino acids. It is possible though that the processed mRNA is just a pri-miRNA444 transcript and does not code for a protein.
Figure 3.
Schematic Representation of the Biogenesis of OsmiR444.
The hairpin structure requires parts of exon 2 and exon 3 of the host transcript J033125N22.
miR444 is conserved in monocots such as wheat (Triticum aestivum), barley, maize, sorghum (Sorghum bicolor), and sugarcane (Saccharum officinarum) but not in Arabidopsis (Figure 4A). The precursor sequences from all these plants can form a hairpin structure (Figure 4B). Unlike the situation in rice, the corresponding miRNA precursor fold-back structures can be predicted from the unspliced genomic sequences from the available sequences of other monocots.
Figure 4.
The miR444 Family Is Conserved in Monocots.
(A) Alignment of miR444 sequence from rice with the predicted homologs in wheat, barley, maize, sorghum, and sugarcane.
(B) Predicted fold-back structures of miR444 precursors from rice, wheat, barley, maize, sorghum, and sugarcane.
Similarly, OsmiR436 also resides in the same polarity of a processed transcript (J023035E19), and only the processed transcript can form a hairpin structure. The mature miR436 resides in the 3rd exon of the gene J023035E19. The predicted fold-back structure requires a very long part (720 nucleotides; exons 3 to 9) of the processed transcript (Figure 5) because of the presence of stem-loop structures protruding from the 3′ arm of the hairpin structure.
Figure 5.
Predicted Fold-Back Structure of OsmiR436 Precursor.
The fold-back structure was predicted with use of a 720-bp processed transcript. Protruding stem-loops in the 3′ arm of the hairpin are indicated by the slashes (\).
Expression Patterns of Rice miRNAs
The tissue- and development-specific expression of miRNAs might provide clues about their physiological function. In a wide range of organisms, many miRNAs have been found to be differentially expressed at different developmental stages, cell types, and tissues (Lee and Ambros, 2001; Lagos-Quintana et al., 2002; Aravin et al., 2003; Houbaviy et al., 2003). Several Arabidopsis miRNAs are expressed ubiquitously, whereas the expression of many others is regulated by development and shows preferential accumulation in certain tissues (Llave et al., 2002a; Park et al., 2002; Reinhart et al., 2002; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004). To assist with the determination of the function of the new rice miRNAs, we examined their expression in different organs and developmental stages (Figures 6 and 7).
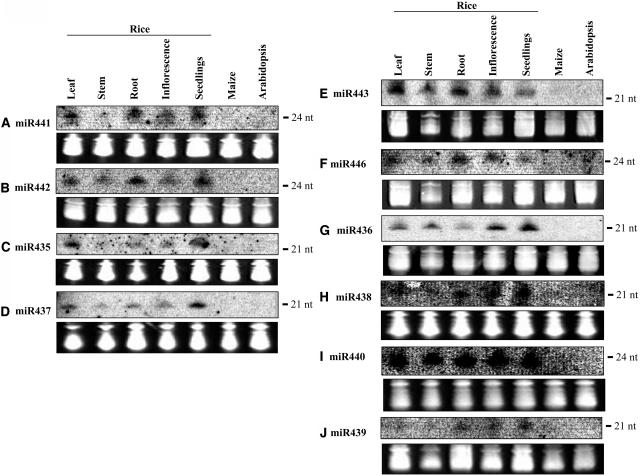
Figure 6.
Expression Patterns of miRNAs Cloned from Rice.
RNA gel blots of total RNA isolated from different tissues were probed with labeled oligonucleotides. The blots also included RNA from maize and Arabidopsis. The tRNA and 5S rRNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls. Labeled RNA oligonucleotide was used as a size marker, and the position was indicated. nt, nucleotides.
The expression patterns of miR441 and miR442 are similar: moderate expression in leaves, roots, and young seedlings and weaker expression in stems and inflorescences (Figures 6A and 6B). miR435 and miR437 also displayed similar expression patterns: moderate expression in leaves and young seedlings and weaker expression in other tissues tested (Figures 6C and 6D). miR443 and miR446 seem to be strongly expressed in leaves and roots and moderately in stem and inflorescence tissues (Figures 6E and 6F). miR436 is expressed in all tissues tested but the levels are higher in inflorescence and young seedlings (Figure 6G). miR438 expression was moderate in leaves, root, inflorescence, and young seedlings (Figure 6H). miR440 is uniformly and abundantly expressed in all rice tissues (Figure 6I), whereas miR439 shows very weak expression in seedlings and is barely detectable in other tissues (Figure 6J).
OsmiR396d showed strong and ubiquitous expression in all tissues, although the expression in roots was relatively lower (Figure 7A). miR445 was strongly expressed in mature stems but barely detected in leaf and inflorescence tissues (Figure 7B). OsmiR444 showed uniform expression in all rice tissues examined (Figure 7C). All three miRNAs (miR396d, miR444, and miR445) were found to be expressed in maize, although the signal was weaker, possibly because of pooled RNA from different tissues of maize (Figures 7A to 7C).
Some miRNAs displayed tissue-specific or developmental stage–specific expression patterns. Particularly interesting expression was observed for miR445 and miR390, which showed preferential expression in stems and roots, respectively (Figures 7B and 7D).
To help determine whether a miRNA is conserved within monocots, its expression was analyzed in another monocot, maize. The analysis showed that three miRNAs (miR396d, miR444, and miR445) are conserved and expressed in maize (Figures 7A to 7C). The presence of miR396d and miR445 miRNAs in other monocots is supported by their expression in maize, and miR444 by sequence and expression. In addition, miR437 sequence and conserved precursor fold-back structures are present in maize, sugarcane, and sorghum (see Supplemental Figure 3 online). The absence of sequence and expression of these four miRNAs in Arabidopsis suggests that they may be specific to monocots. As an example of conserved miRNAs between dicots and monocots, we tested the expression of a newly identified and conserved miRNA, miR390, in addition to the previously reported and conserved OsmiR156 and OsmiR171 miRNAs in rice, maize, and Arabidopsis (Figures 7D to 7F). In addition to the expected size, the OsmiR171 probe also hybridized to a slightly larger (∼23 bp) small RNA species in maize but not in rice or Arabidopsis, indicating that this larger species is specific to maize (Figure 7E). The remaining nine new rice miRNAs appear not to be conserved between plant species because they have no counterparts in other known plant sequences and their expression could not be detected in maize or Arabidopsis.
In summary, the RNA gel blot analysis confirmed the expression and sizes of 14 newly identified miRNAs in rice (Figures 6 and 7). The majority is expressed ubiquitously in all tissues.
Predicted Targets
Prediction of plant miRNA targets has been facilitated by their extensive sequence complementarity (Rhoades et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004b; Adai et al., 2005). Regulatory targets can be more confidently predicted for conserved miRNAs because complementary sites often are conserved across species boundaries. To identify the potential targets of our newly identified miRNAs, we used the miRNA sequences to search the rice mRNA sequences for antisense hits with the PATSCAN program (Dsouza et al., 1997). Based on transcriptome analysis in Arabidopsis transgenic plants overexpressing miRNAs, Weigel and colleagues (Schwab et al., 2005) devised a set of rules for predicting miRNA targets. These criteria include allowing one mismatch in the region complementary to nucleotides 2 to 12 of the miRNA but not at the cleaving site (10 and 11 nucleotides), and three additional mismatches were permitted between 12- and 21-nucleotide positions, but no more than two continuous mismatches within this region. Adopting these rules in predicting newly identified miRNA targets in rice, we allowed one mismatch between the positions 1 to 9 nucleotides from the 5′ end of miRNA and no mismatches between positions 10 and 11, and another two mismatches were allowed between positions 12 and 21/24. Gaps and mismatches are commonly seen in known animal and plant miRNA-mRNA basepairing interactions that are known to lead to cleavage or attenuation of translation (Aukerman and Sakai, 2003; Palatnik et al., 2003; Chen, 2004; Jones-Rhoades and Bartel, 2004). By applying the above rules, our analysis led to the prediction of 46 genes as putative targets for 11 new miRNAs in rice (Table 3). Predicted targets and their complementarity with the new miRNAs are provided in Supplemental Table 1 online. The number of predicted targets per miRNA varied greatly, from 1 to 15. Four of the miRNAs (miR435, miR443, miR444, and miR445) each has only one predicted target. We were unable to predict targets for the remaining three miRNAs (miR438, miR440, and miR442) by applying these criteria. To evaluate the false positive rates of our target predictions, we performed the same searches with 100 randomized sequences for each miRNA. The length and composition of the miRNAs were maintained in the randomized sets, and the searches were performed with the same mismatch settings (Rhoades et al., 2002; Jones-Rhoades and Bartel, 2004). According to these results (Table 3), the hit frequency with the authentic miRNAs is in most cases more than five times higher and relative.
Table 3.
Predicted Targets of Newly Identified Rice miRNAs
| miRNA | Target Gene | Target Site | Target Protein | Count | Mean | sd |
|---|---|---|---|---|---|---|
| miR390 | 11668.m00935(1); 11674.m03433(3); 11687.m03320(3) | ORF | Leu-rich repeat proteins | 3 | 0.24 | 0.7 |
| miR396d | 11668.m04403(1); 11668.m04580(1); 11669.m04718(1); 11670.m04997(1); 11670.m04998(1); 11680.m00165(1); 11680.m00999(1); 11669.m05248(1); 11670.m02258(1); 11687.m03206(1); 11686.m02909(1); 11687.m03206(2); 11670.m04996(1); 11680.m00166(1); 11670.m04758(1) | ORF | GRL transcription factors | 15 | 0.19 | 0.6 |
| miR435 | 11687.m03448(3) | ORF | F-box protein | 1 | 0.75 | 1.3 |
| miR436 | 11686.m04227(0); 11686.m04226(1) | ORF | Unknown proteins | 2 | 0.07 | 0.3 |
| miR437 | 11680.m04605(1); 11680.m04606(1) | ORF | Glu receptor proteins | 2 | 0.22 | 0.6 |
| miR439 | 11667.m02576 (3 target sites with 0, 2, and 4 mismatches) | ORF | Dirigent-like protein | 4 | 0.15 | 0.5 |
| 11680.m01869(1); 11668.m02918(3) | ORF | Unknown proteins | ||||
| 11673.m02474(3) | ORF | Retrotransposon | ||||
| miR441 | 11669.m05042(3) | ORF | Unknown protein | 10 | 0.00 | 0.0 |
| 11667.m06876(2); 11674.m00346(2); 11674.m00582(2); 11680.m01945(2); | 3′UTR | Unknown proteins | ||||
| 3′UTR | Unknown proteins | |||||
| 11682.m04930(3); 11687.m00089(3) 11682.m03939(3); | 3′UTR | Unknown proteins | ||||
| 11668.m04341(2) | 3′UTR | F-box protein | ||||
| 11686.m03687(3) | 3′UTR | UDP-glucoronosyl and UDP-glucosyltransferase | ||||
| miR443 | 11687.m04039(3) | ORF | Unknown protein | 1 | 0.13 | 0.4 |
| miR444 | 11668.m04852(0) | ORF | MADS box transcription factor | 1 | 0.19 | 0.6 |
| miR445 | 11680.m05074(3) | 5′UTR | RNA binding protein | 1 | 0.00 | 0.0 |
| miR446 | 11668.m04341(2) | 3′UTR | F-box protein | 6 | 0.01 | 0.1 |
| 11674.m00582(2); 11680.m01945(2); 11682.m04930(3); 11682.m03939(3) | 3′UTR | Unknown proteins | ||||
| 3′UTR | Unknown protein | |||||
| 11680.m01730(2) | 3′UTR | Unknown plant | ||||
| Specific protein |
For each potential target gene, the number of mismatches between the miRNA and mRNA is indicated in parentheses. Annotated rice mRNAs were also searched for sites complementary to randomized miRNAs (100 repeats), applying the same criteria that were used for predicting targets. The mean of the hit frequency of the randomized data and their standard deviations are recorded.
In animals, all known miRNA target sites were found in 3′ untranslated regions (UTRs) of protein coding genes, whereas in plants they are only occasionally in the 3′UTRs but are predominantly in the coding regions (Rhoades et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004b; Adai et al., 2005). In plants, they also have been predicted to reside in 5′UTRs (Sunkar and Zhu, 2004). Recently, miRNAs have also been predicted to target ORFs in humans (Lewis et al., 2005). Consistent with the earlier findings in Arabidopsis, 30 of our predicted target genes in rice have target sites in their ORFs. Fifteen genes have their predicted target sites in 3′UTR and only one in the 5′UTR.
Both miR444 and its predicted target, a MADS box transcription factor gene, are conserved in other monocots, such as wheat, maize, barley, and sugarcane, but not in Arabidopsis (data not shown). OsmiR396d, a new member of the miR396 family, is expected to target the growth-regulating factors (GRFs) (Jones-Rhoades and Bartel, 2004). miR396d displays near perfect complementarity with 15 of these GRF genes (Table 3; see Supplemental Table 1 online). The complementary sites of miR396d are highly conserved in GRF genes of sorghum, maize, barley, and sugarcane.
The predicted target of miR439, 11667.m02576, a dirigent-like protein gene, was found to have three complementary sites within its ORF (Figure 8). These three sites are very closely spaced and separated by gaps of 11 and 9 nucleotides. One target site corresponding to the positions 567 to 588 is perfectly complementary to the miRNA. The other two target sites correspond to the positions 599 to 619 and 628 to 648, with two and four mismatches, respectively (Figure 8). The two target sites corresponding to positions 567 to 588 and 628 to 648 are in frame, and there is partial amino acid sequence conservation between these two target sites (Figure 8). The presence of three target sites within one ORF has not been seen before and thus is unique among miRNA targets predicted thus far.
Figure 8.
OsmiR439 Is Predicted to Target Three Sites within the ORF of Its Target Gene, 11667.m02576.
Numbers represent the position of target sites in the ORF. Amino acid sequence corresponding to the target nucleotide sequence is shown.
It appears that our predicted targets play roles not only in development but also in diverse physiological processes. Sixteen of the predicted targets of two miRNAs (miR396d and miR444) are transcription factors (GRFs and MADS box transcription factors), whereas the remaining 30 predicted targets of nine miRNAs appear to play roles in a broad range of physiological processes and include protein kinases, F-box proteins, dirigent-like protein, Glu receptor-like proteins, RNA binding protein, retrotransposons, and 17 other proteins with unknown function (Table 3).
Identification of miRNA-Guided Cleavage of Target mRNAs in Rice
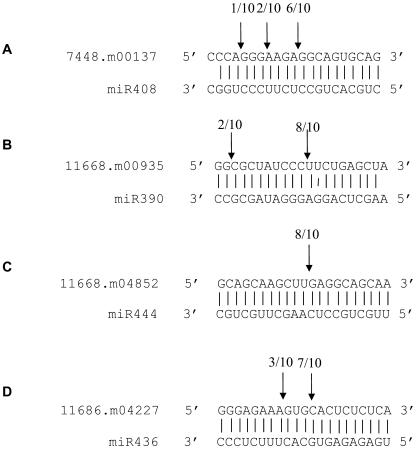
miRNAs negatively regulate target genes through miRNA-directed cleavage within the region of complementarity or interfering with translation (Carrington and Ambros, 2003; Ambros, 2004; Bartel, 2004; Dugas and Bartel, 2004; Mallory and Vaucheret, 2004). Most of the Arabidopsis miRNAs have been shown to guide the cleavage of target mRNAs (Llave et al., 2002b; Kasschau et al., 2003; Palatnik et al., 2003; Tang et al., 2003; Xie et al., 2003; Floyd and Bowman, 2004; Jones-Rhoades and Bartel, 2004; Mallory et al., 2004a, 2004b; Wang et al., 2004b). To test whether the predicted miRNA targets in rice can also be cleaved, we used an RNA ligase-mediated 5′ rapid amplification of cDNA ends (5′ RACE) procedure (Llave et al., 2002b) to map the cleavage sites in the predicted target genes from rice. We performed the 5′ RACE assays on four predicted target genes: two were representatives of targets of conserved miRNAs (11668.m00935 targeted by OsmiR390 and 7448.m00137 targeted by OsmiR408), one is a MADS box factor (11668.m04852) that is a predicted target of OsmiR444 as a representative of a monocot-specific miRNA target, and the other target served as a representative of rice-specific miRNA target (11686.m04227 targeted by OsmiR436). All four predicted targets were found to have specific cleavage sites corresponding to the miRNA complementary sequences (Figures 9A to 9D). In all these cases, the most common 5′ end of the mRNA fragments mapped to the nucleotides that pair with the 10th miRNA nucleotides from their 5′ ends.
Figure 9.
Identification of miRNA-Guided Cleavage Products of Target Genes in Rice.
(A) mRNA 7448.m00137.
(B) mRNA 11668.m00935.
(C) mRNA 11668.m04852.
(D) mRNA 11686.m04227.
Mapping of cleavage sites was done by RNA ligase-mediated 5′ RACE. Partial mRNA sequences from target genes were aligned with miRNAs. Numbers indicate the fraction of cloned PCR products terminating at different positions.
Experimental Verification of Previously Predicted miRNAs in Rice
With the exception of a member of the miR171 family, none of the predicted miRNAs in rice have been verified experimentally. The miR171b homolog in rice has been cloned in a recent study (Wang et al., 2004a). Sequence similarity searches show that 21 of the cloned miRNAs (in 15 families) in this study had been predicted in rice (Table 2). These include OsmiR156a, OsmiR156k, OsmiR159a/b, OsmiR159c, OsmiR160, OsmiR164, OsmiR166a-f, OsmiR167a-c, OsmiR167d-I, OsmiR168a, OsmiR169b/c, OsmiR169f/g, OsmiR169 h-m, OsmiR171a-f, OsmiR171g, OsmiR172a, OsmiR393, OsmiR397, OsmiR398, OsmiR399a, and OsmiR408 (Table 2). We noticed that all predicted members of four miRNA families—OsmiR169 (three members), OsmiR156 (two members), OsmiR159 (two members), and OsmiR171 (two members)—have appeared in our libraries (Table 2). Thus, our cloning supports the expression of all members of these four miRNA families in rice.
The frequency of cloning varies highly among miRNAs. Our analysis indicates that OsmiR168a is the most abundant miRNA in rice, which was represented by 22 clones in the library. Of the 22 clones, 14, 5, and 3 came from shoot, root, and inflorescence libraries, respectively. The OsmiR168 family was represented by two members (168a and 168b) that differ slightly in nucleotide sequence. All 22 clones belong to OsmiR168a and none to miR168b, which suggests that only miR168a is abundantly expressed, and miR168b is much lower in abundance or its expression may be limited to specific cells or tissues. An equally abundant miRNA family is the OsmiR156 family, with multiple (12) loci in the rice genome, which appeared 22 times in our sequencing. Seventeen clones corresponded to the OsmiR156a-j loci, whereas the other five clones belong to another homolog, miR156k/l. Slightly less abundant miRNAs are members of the OsmiR396 and OsmiR169 families, each represented by 10 and 9 clones, respectively. Interestingly, none of the 10 clones represented the predicted member of the OsmiR396 family; rather, all these 10 clones corresponded to the new member, OsmiR396d, identified in this study. Other miRNAs, OsmiR159, OsmiR164, OsmiR166, OsmiR167, OsmiR169, OsmiR171, OsmiR172, and OsmiR397, were cloned only a few times (two to eight), whereas the rest (OsmiR160, OsmiR393, OsmiR398, OsmiR399, and OsmiR408) appeared only once.
In addition to the 14 new miRNAs presented in Table 1, we also identified five other small RNAs (see Supplemental Table 2 online) encoded by the rice genome and have some features of miRNAs (derived from hairpin RNA precursors) (see Supplemental Figure 4 online). However, we are unable to designate them as miRNAs at this time because they could not be detected in the RNA gel blot analysis. Because of the lack of genetic tools in rice for validating miRNA biogenesis, such as mutants defective in Dicer-like genes, future research is required to confidently designate these small RNAs as either miRNAs or siRNAs.
DISCUSSION
Monocot-Specific and Rice-Specific miRNAs
In animals, most miRNAs are conserved across species boundaries (Ambros, 2004; Bartel, 2004). The strict conservation of miRNAs observed might suggest that the interactions between these miRNAs and their targets constitute essential processes and could play evolutionarily conserved roles (Pasquinelli et al., 2000). However, a few miRNAs appear to be species specific (Ambros et al., 2003a). miRNAs that are restricted to one or a few species may be implicated in species- or clade-specific functions. The identification of several miRNAs in Arabidopsis that are not conserved in rice suggests that these may play specific roles in Arabidopsis. These observations strongly supported the hypothesis that rice may also have a unique set of miRNAs. Thus far, 20 families of miRNAs discovered in Arabidopsis are predicted to be conserved in rice (Park et al., 2002; Reinhart et al., 2002; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004b; Adai et al., 2005). Of these, we have experimentally verified the expression of 15 families of miRNAs in rice. To a certain extent, the 75% coverage of predicted miRNAs in our libraries may reflect the extent of saturation of our miRNA cloning. Of the 14 new rice miRNAs we identified, only one (miR390) is conserved in both Arabidopsis and maize, whereas four others (miR396d, miR437, miR444, and miR445) are conserved in other monocots, such as maize, barley, sugarcane, and sorghum but not in Arabidopsis, which suggests that these latter ones are specific to monocots, and the remaining nine miRNAs are likely specific to rice. All of the new miRNAs are conserved in another rice subspecies, Indica (Table 1). This is expected given the extensive sequence colinearity between these two rice subspecies (Feng et al., 2002).
Compared with computational approaches, direct cloning has the advantage of identifying not only nonconserved miRNAs, but also atypical miRNAs. Cloning of miRNAs in rice resulted in the identification of an additional member (from the OsmiR396d and OsmiR396e loci) of the OsmiR396 family. The presence of an extra nucleotide in the middle of a miRNA from the same family was previously unknown. The identification of a new member of the OsmiR396 family with the added nucleotide in the miRNA sequences suggests that future computational approaches should incorporate this structural feature in their prediction strategies. It is interesting that this new member is missing from the Arabidopsis miR396 family.
Unusual Processing and Ubiquitous Expression Patterns of Rice miRNAs
Although the majority of the new miRNAs are mapped to intergenic regions, a considerable fraction maps to the introns, whereas two miRNAs (OsmiR436 and OsmiR444) map to exons of protein-coding genes. The expression of exon- and intron-derived miRNAs in the sense polarity is most likely driven by the promoters of the surrounding genes, whereas those in the antisense polarity within the introns are likely to be expressed as independent genes.
The structural analysis of full-length transcripts of pre-miRNA transcripts of miR172 revealed the presence of introns, and the processed transcripts are 5′ capped and polyadenylated (Aukerman and Sakai, 2003). A fold-back structure can be readily predicted for the AthmiR172 precursor from genomic sequences because the hairpin precursor resides in one exon. Nevertheless, this observation suggests that certain miRNA precursor sequences can enter the spliceosomal pathway. Consistent with this, we found that one of the newly identified miRNA precursors (OsmiR444) requires part of exons 2 and 3 of a protein-coding gene for hairpin formation. This shows that some miRNA precursors undergo processing and that processed transcripts are necessary for the prediction of the hairpin structure. Similarly, a hairpin structure for miR436 could only be predicted from a processed transcript, which suggests the presence of introns within the hairpin sequence. These findings raise the possibility that some other miRNA transcripts might also have introns within the hairpin precursor sequences, and it would be impossible to predict these miRNAs if the sequences of processed transcripts are not available.
Although the expression of several miRNAs from Arabidopsis are shown to be constitutive and ubiquitous, many miRNAs exhibit preferential expression in a temporal or tissue-specific manner (Llave et al., 2002a; Park et al., 2002; Reinhart et al., 2002; Aukerman and Sakai, 2003; Chen, 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004), whereas the expression of others is modulated by abiotic stress (Sunkar and Zhu, 2004) or nutritional status, such as sulfur starvation (Jones-Rhoades and Bartel, 2004). Most of the previously reported miRNAs were shown to be abundantly expressed in inflorescence tissues in Arabidopsis (Llave et al., 2002a; Park et al., 2002; Reinhart et al., 2002; Sunkar and Zhu, 2004). However, the expression of most of the newly identified rice miRNAs does not show higher expression in inflorescence tissues. In fact, with the exception of OsmiR436, all others seem to have lower levels of expression in the inflorescence compared with other tissues. Additionally, most of the newly identified rice miRNAs are ubiquitously expressed and few showed tissue-specific expression.
Predicted Targets Include Transcription Factors and Others with a Broad Range of Biological Functions
More than one-third of the genes in the human genome were predicted as miRNA targets, and these targets appear to be involved in a wide range of biological functions (Lewis et al., 2005). Previously reported miRNAs in Arabidopsis have targets that are predominantly transcription factors or parts of the RNA interference machinery, such as Dicer-like-1 and Argonaute1 (Xie et al., 2003; Vaucheret et al., 2004). More recently identified miRNAs in Arabidopsis may target transcripts that encode proteins involved in diverse physiological processes (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Wang et al., 2004b; Adai et al., 2005).
We were able to predict 46 genes as potential targets of 11 newly identified miRNAs in rice. Transcription factors (GRL and MADS box gene) represented one-third of the predicted targets of miRNAs in this study. The remaining 30 predicted targets appear to play roles in diverse physiological processes. For instance, F-box proteins that regulate diverse cellular processes, including cell cycle regulation, circadian rhythm, flower development, and hormonal signal transduction (Kuroda et al., 2002), are predicted targets of three miRNAs (miR435, miR441, and miR446). OsmiR439 is predicted to target a dirigent-like protein (Table 3). Thus far, dirigent proteins and their homologs have only been reported in vascular plants (Davin and Lewis, 2000). Although the precise role of these proteins is not known, they seem to function in the lignification in vascular plants (Burlat et al., 2001). Glu receptor proteins are probable targets of OsmiR437 (Table 3). Ionotropic Glu receptors function in animals as Glu-gated nonselective cation channels. Recent evidence suggests that plant Glu-receptor-like genes do encode nonselective cation channels, but these channels are not gated by amino acids; therefore, their roles in plants remain a mystery (Davenport, 2002). OsmiR390 is predicted to target three Leu-rich repeat protein kinases (Table 3). Fifteen unknown proteins are also predicted targets of newly identified miRNAs (Table 3). The identification of miRNAs that target genes with unknown functions provides a unique tool to probe the functions of these unknown genes.
The MADS box protein family is represented by >70 and >100 genes in rice and Arabidopsis, respectively. No miRNA that targets the MADS box transcription factors has been identified previously, either in Arabidopsis or rice. In this study, we found a miRNA (miR444) that targets a MADS box transcription factor gene in rice, and both the miRNA and the target sites in the MADS box genes are conserved in monocots. MADS domain proteins in plants were first identified as regulators of floral organ identity and have since been found to control additional developmental processes, such as meristem identity, root development, fruit dehiscence, and flowering time (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Riechmann and Meyerowitz, 1997; Theissen et al., 2000). miR444 precursors can be found in other monocot species, such as maize, wheat, barley, and sugarcane, from expression databases. The lack of a miR444 homolog and of the conserved target site in a MADS box gene(s) from Arabidopsis strongly suggests that this miRNA-mediated regulation of MADS box genes is conserved only in monocots. Future manipulation of OsmiR444 level and of the target MADS box genes should help unravel the importance of these interactions in rice and in monocots in general.
To our knowledge, no plant or animal miRNA has been previously predicted to have multiple target sites in one ORF. In the case of OsmiR439, one target site is perfectly complementary and the other two target sites have two and four mismatches, including in positions 9 and 11 from the 5′ end of the miRNA. The mismatch at position 11 may suggest that the interaction does not lead to cleavage because perfect matches at positions 10 and 11 are important for cleavage of the target (Jones-Rhoades and Bartel, 2004; Schwab et al., 2005). However, more recently it is shown that a mismatch at position 11 reduces the cleavage rate but still can be tolerated in vivo (Mallory et al., 2004b). The identification of miR439, with three complementary target sites within the ORF of a target gene (Figure 8), suggests that the cooperative regulation of one gene at three different sites by one miRNA may be important to downregulate the mRNA to the required level. Alternatively, despite its perfect complementarity with the target gene at one of the target sites, this miRNA might be involved in translational repression for which more than one target site may be necessary. Or one target site may be involved in cleavage and the others may be important for translational repression. Future functional studies with introduced silent mutations in the target sites may reveal the importance of three target sites in one ORF and may shed light on potentially unique regulatory mechanisms.
miRNA-Guided Cleavage of Target mRNAs in Rice
The cleavage of target mRNAs appears to be the predominant mode of gene regulation by plant miRNAs (Llave et al., 2002b; Kasschau et al., 2003; Palatnik et al., 2003; Tang et al., 2003; Xie et al., 2003; Jones-Rhoades and Bartel, 2004; Mallory et al., 2004a, 2004b; Vaucheret et al., 2004). Target mRNA fragments resulting from miRNA-guided cleavage are characterized by a 5′ phosphate group, and cleavage occurs near the middle of the basepairing interaction region with the miRNA (Llave et al., 2002b). We were able to map the cleavage sites for four of the predicted targets, and the results support that these are genuine miRNA targets in rice.
OsmiR408 was cloned in this study (Table 2). Previously, we reported the identification of miR408 from Arabidopsis (Sunkar and Zhu, 2004). miR408 was predicted to target an mRNA encoding a plantacyanin (7448.m00137). Its target site is conserved in several plant species (Sunkar and Zhu, 2004). In this study, we mapped the miR408 cleavage site in the rice plantacyanin transcript using a modified 5′ RACE procedure. The result validated plantacyanin as a genuine target of OsmiR408. The precise function of plantacyanins in plants is unknown; however, they have been proposed to function in cell-to-cell signaling, lignin formation, and stress responses (Romo et al., 2001; Kim et al., 2003). Our ability to map the cleavage products that correspond to the OsmiR390 complementary site in a Ser/Thr/Tyr protein kinase (11668.m00935) confirms that this kinase is indeed a miRNA target. This is a demonstration that protein kinases can be targeted by miRNAs. We also provide evidence that a rice MADS box transcription factor (11668.m04852) is a genuine target of OsmiR444. One of the predicted targets (11686.m04227, an unknown protein) of miR436 is also confirmed by the cleavage analysis.
In summary, our cloning study led to the discovery of 14 new rice miRNAs, of which only one (miR390) is conserved in Arabidopsis. At least four of the miRNAs (miR396d, miR437, miR444, and miR445) appear to be limited to monocots, suggesting monocot-specific miRNA-dependent regulatory processes. It is possible that more of the new miRNAs are present in other monocots whose sequence information is not available. It is also possible that more of these miRNAs may be conserved in maize but cannot be detected with the rice-specific probes because of possible subtle sequence differences. The putative monocot-specific miRNAs may have evolved after the divergence of monocots and dicots 200 million years ago, or they may have been lost in dicots.
METHODS
Cloning of miRNAs from Rice
Total RNA was isolated separately from shoots and roots of 4-week-old young seedlings and inflorescence of adult rice (Oryza sativa spp japonica cv Nipponbare) plants using TRIzol (Bethesda Research Laboratories, Life Technologies, Gaithersburg, MD) according to the manufacturer's instructions. Cloning of miRNAs was performed as described (Sunkar and Zhu, 2004). In brief, small RNAs from 18 to 26 nucleotides were size fractionated, purified, and ligated sequentially to 5′ and 3′ RNA/DNA chimeric oligonucleotide adapters. Reverse transcription was performed after ligation with the adapters, followed by PCR amplification. The resulting PCR products were cloned and transformed into competent cells. Plasmids were isolated from individual colonies and sequenced. The sequences were subsequently processed and used for BLAST analysis against the O. sativa spp japonica sequences in the TIGR database and other plant sequences in the National Center for Biotechnology Information (NCBI) database.
Sequence Analysis and Prediction of Fold-Back Structures
Automated base calling of raw sequence traces and vector removal was performed with the PHRED and CROSS MATCH programs from Ewing and Green (1998). All trimmed sequences longer than 16 nucleotides were used to search the Rfam (www.sanger.ac.uk/Software/Rfam) database with BLASTN (Altschul et al., 1997). This step allowed the removal of most non-siRNA and non-miRNA species in the data set. Putative origins for the remaining sequences were identified by BLASTN searches against the intergenic and intron sequences using the latest annotation 3.0 of O. sativa spp japonica from TIGR (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/). Candidates with perfect matches against these genomic data sets were used for fold-back secondary structure prediction with the mfold program (Zuker, 2003). Three different combinations of upstream and downstream sequences spanning the putative miRNA origins were used for the predictions: (1) 400 bp upstream and 20 to 30 bp downstream, (2) the same length combination but in reverse order, and (3) 200 bp upstream and 200 bp downstream. In the case of OsmiR444 and OsmiR436, only processed transcript sequences were used to predict the hairpin structure. OsmiR436 hairpin prediction required 700 nucleotides.
To identify putative target sequences, all predicted and cloned coding sequences and UTR sequences from O. sativa (TIGR all.cdna set) were searched with PatScan (Dsouza et al., 1997). The following parameters were used for these pattern searches, all referring to the 5′ end of the miRNAs in antisense orientation: not more than one mismatch between positions 1 and 9, no mismatch between positions 10 and 11, and not more than two mismatches for the rest of the sequence. False positive rates of predicted targets were estimated with the same search strategy using 100 randomized samples for each miRNA. The retrieved miRNA/target site pairs were ranked and scored by aligning them with the Needleman-Wunsch global alignment program from EMBOSS (Rice et al., 2000). Global cross-species conservations of miRNAs were determined by BLASTN searches against the nucleotide and EST databases from NCBI. The results were parsed with a Perl script that scored only those pairwise BLAST alignments (high similarity pair) as hits when they were not more than two positions shorter than the query sequence and 95% identical. More detailed conservations of miRNAs and target sequences within the genome sequences of Populus trichocarpa, O. sativa spp indica, and Arabidopsis thaliana were determined with the same BLASTN and PatScan strategy used for the japonica host. Weaker conserved origins in the genomic sequences of Arabidopsis and P. trichocarpa were identified with PatScan (maximal two mismatches) and subsequent fold-back structure predictions.
RNA Gel Blot Analysis
Total RNA was isolated from leaf, stem, root, and inflorescence tissues of adult plants as well as from 4-week-old young rice seedlings. Total RNA was also isolated from pooled maize (Zea mays) tissues (kernels, pericarp, endosperm, scutellum, coleoptile, embryo, silk, and young seedlings) and 3-week-old Arabidopsis seedlings using TRIzol. One hundred micrograms of total RNA was loaded per lane, resolved on a denaturing 15% polyacrylamide gel, and transferred electrophoretically to Hybond-N+ membranes (Amersham Biosciences, Buckinghamshire, UK). Hybridization and washings were performed as described (Sunkar and Zhu, 2004). The membranes were briefly air dried and then exposed to a phosphor imager.
5′ RACE
Total RNA from 4-week-old rice seedlings was extracted with Trizol reagent. Poly(A)+ mRNA was purified from total RNA with use of the Poly A kit (Promega, Madison, WI). RNA ligase-mediated 5′ RACE was performed using the GeneRacer kit (Invitrogen Life Technologies, Carlsbad, CA). The GeneRacer RNA Oligo adapter was directly ligated to mRNA (100 ng) without calf intestinal phosphatase and tobacco acid pyrophosphatase treatment. Gene-specific primers were designed and used for cDNA synthesis. Initial PCR was performed with the GeneRacer 5′ primer and gene-specific primers (7448.m00137, 11668.m00935, 11668.m04852, and 11686.m04227). Nested PCR was performed with 1 μL of the initial PCR reaction, GeneRace 5′ nested primer, and gene-specific internal primers. After amplification, 5′ RACE products were gel purified and cloned, and at least 10 independent clones were sequenced.
Supplementary Material
Acknowledgments
We thank Rebecca Stevenson for assistance in growing the rice plants and Shou-Wei Ding for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01GM0707501 and National Science Foundation Grant IBN-0212346 to J.-K.Z. P.J. was supported in part by a BOYSCAST fellowship from DST, the Government of India, and ICAR (New Delhi, India).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031682.
References
- Adai, A., Johnson, C., Mlotshwa, S., Archer-Evans, S., Manocha, V., Vance, V., and Sundaresan, V. (2005). Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res. 15, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, E., Xie, Z., Gustafson, A.M., Sung, G.-H., Spatafora, J.W., and Carrington, J.C. (2004). Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 36, 1282–1290. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Thomas, L.M., Alejandro, A.S., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V. (2004). The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Ambros, V., et al. (2003. b). A uniform system for microRNA annotation. RNA 9, 277–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros, V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. (2003. a). MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13, 807–818. [DOI] [PubMed] [Google Scholar]
- Aravin, A., Lagos-Quintana, M., Yalcin, A., Zavalon, M., Marks, D., Snyder, B., Gaasterland, T., Meyer, J., and Tuschl, T. (2003). The small RNA profile during Drosophila melanogaster development. Dev. Cell 5, 337–350. [DOI] [PubMed] [Google Scholar]
- Aukerman, M.J., and Sakai, H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-Like target genes. Plant Cell 15, 2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Berezikov, E., Guryev, V., van de Belt, J., Wienholds, E., Plasterk, R.H.A., and Cuppen, E. (2005). Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120, 21–24. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and de Peer, Y.V. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101, 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlat, V., Kwon, M., Davin, L.B., and Lewis, N.G. (2001). Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. [DOI] [PubMed] [Google Scholar]
- Chen, X. (2004). A microRNA as translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen, E.S., and Meyerowitz, E.M. (1991). The war of the whorls: Genetic interactions controlling flower development. Nature 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Davenport, R. (2002). Glutamate receptors in plants. Ann. Bot. (Lond) 90, 549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davin, L.B., and Lewis, N.G. (2000). Dirigent proteins and dirigent sites explain the mystery of specificity of radical precursor coupling in lignan and lignin biosynthesis. Plant Physiol. 123, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli, A.H., Tops, B.B.J., Plasterk, R.H.A., Ketting, R.F., and Hannon, G.J. (2004). Processing of primary microRNAs by the microprocessor complex. Nature 432, 231–234. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Mourelatos, Z., Yang, M., Sharma, A., and Dreyfuss, G. (2003). Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza, M., Larsen, N., and Overbeek, R. (1997). Searching for patterns in genomic data. Trends Genet. 13, 497–498. [DOI] [PubMed] [Google Scholar]
- Dugas, D.V., and Bartel, B. (2004). MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7, 512–520. [DOI] [PubMed] [Google Scholar]
- Ewing, P., and Green, B. (1998). Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- Feng, Q., et al. (2002). Sequence and analysis of rice chromosome 4. Nature 420, 316–320. [DOI] [PubMed] [Google Scholar]
- Floyd, S.K., and Bowman, J.L. (2004). Ancient microRNA target sequences in plants. Nature 428, 485–486. [DOI] [PubMed] [Google Scholar]
- Grad, Y., Aach, J., Hayes, G.D., Reinhart, B.J., Church, G.M., Ruvkun, G., and Kim, J. (2003). Computational and experimental identification of C. elegans microRNAs. Mol. Cell 11, 1253–1263. [DOI] [PubMed] [Google Scholar]
- Houbaviy, H.B., Murray, M.F., and Sharp, P.A. (2003). Embryonic stem cell-specific microRNAs. Dev. Cell 5, 351–358. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.J., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799. [DOI] [PubMed] [Google Scholar]
- Kasschau, K., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. (2003). P1/HC-Pro, a viral suppressor of RNA silencing interferes with Arabidopsis development and miRNA function. Dev. Cell 4, 205–217. [DOI] [PubMed] [Google Scholar]
- Kikuchi, S., et al. (2003). Collection, mapping and annotation of over 28,000 cDNA clones from japonica rice. Science 301, 376–379. [DOI] [PubMed] [Google Scholar]
- Kim, S., Mollet, J.C., Dong, J., Zhang, K., Park, S.Y., and Lord, E.M. (2003). Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 100, 16125–16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 101, 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K., and Matsui, M. (2002). Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol. 43, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Meyer, J., Borkhardt, A., and Tuschl, T. (2003). New microRNAs from mouse and human. RNA 9, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. (2002). Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. (2003). MicroRNAs: Runts of the genome assert themselves. Curr. Biol. 13, R925–R936. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., and Ambros, V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., and Kim, V.N. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Burge, C.B., and Bartel, D.P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Glasner, M.E., Yekta, S., Burge, C.B., and Bartel, D.P. (2003. a). Vertebrate microRNA genes. Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Weinstein, E.G., Abdelhakim, A., Yekta, S., Jones-Rhoades, M.W., Burge, C.B., and Bartel, D.P. (2003. b). The microRNAs of Caenorhabditis elegans. Genes Dev. 17, 991–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M., and Carrington, J.C. (2002. a). Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002. b). Cleavage of Scarecrew-like mRNA targets directed by a class of Arabidopsis microRNA. Science 297, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Dugas, D.V., Bartel, D.P., and Bartel, B. (2004. a). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Reinhart, B.J., Jones-Rhoades, M.W., Tang, G., Zamore, P.D., Barton, M.K., and Bartel, D.P. (2004. b). MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 23, 3356–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, A.C., and Vaucheret, H. (2004). MicroRNAs: Something important between the genes. Curr. Opin. Plant Biol. 7, 120–125. [DOI] [PubMed] [Google Scholar]
- Mette, M.F., van der Winden, J., Matzke, M., and Matzke, A.J. (2002). Short RNAs can identify new candidate transposable element families in Arabidopsis. Plant Physiol. 130, 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. (2002). miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik, J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Parizotto, E.A., Dunoyer, P., Rahm, N., Himber, C., and Vionnet, O. (2004). In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A.E., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408, 86–89. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S., Zavolan, M., Grasser, F.A., Chien, M., Russo, J.J., Ju, J., John, B., Enright, A.J., Marks, D., Sander, C., and Tuschl, T. (2004). Identification of virus-encoded microRNAs. Science 304, 734–736. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Jones-Rhoades, M.W., Bartel, B., and Bartel, D.P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M., Reinhart, B., Lim, L., Burge, B., Bartel, B., and Bartel, D. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., and Bleasby, A. (2000). EMBOSS: The European molecular biology open software suite. Trends Genet. 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1997). MADS domain proteins in plant development. Biol. Chem. 378, 1079–1101. [PubMed] [Google Scholar]
- Romo, S., Labrador, E., and Dopico, B. (2001). Water stress-regulated gene expression in Cicer arietinum seedlings and plants. Plant Physiol. Biochem. 39, 1017–1026. [Google Scholar]
- Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., and Weigel, D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell, in press. [DOI] [PubMed]
- Sunkar, R., and Zhu, J.K. (2004). Novel and stress regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. (2003). A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen, G., Becker, A., Di Rosa, A., Kanno, J., Kim, J.T., Munster, T., Winter, K.U., and Saedler, H. (2000). A short history of MADS box genes in plants. Plant Mol. Biol. 42, 115–149. [PubMed] [Google Scholar]
- Vaucheret, H., Vazquez, F., Crete, P., and Bartel, D.P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.-F., Zhou, H., Chen, Y.-Q., Luo, Q.-J., and Qu, L.-H. (2004. a). Identification of 20 new miRNAs from Oryza sativa. Nucleic Acids Res. 32, 1688–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X.J., Reyes, J.L., Chua, N.H., and Gaasterland, T. (2004. b). Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol. 5, R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., and Meyerowitz, E.M. (1994). The ABCs of floral homeotic genes. Cell 78, 203–209. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- Xie, Z., Kasschau, K.D., and Carrington, J.C. (2003). Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.