Abstract
Plants acclimate to variations in light intensity by changing the antenna size of photosystems. This acclimation allows them to undergo efficient photosynthesis and creates a protective strategy to minimize photodamage. Chlorophyll b synthesis by chlorophyllide a oxygenase (CAO) is a key regulatory step in the control of antenna size. Recently, we found that higher plant CAOs consist of three domains (A, B, and C domains) and confirmed that the C domain possesses catalytic function. To investigate the function of the A domain, we fused various combinations of these three domains with green fluorescent protein (GFP) and introduced them into Arabidopsis thaliana. When a full-length CAO-GFP fusion protein was introduced into a chlorophyll b–less chlorina1-1 mutant, chlorophyll b accumulated to almost the same levels as in the chlorophyll b–containing Columbia wild type, but the CAO-GFP could not be detected by immunoblotting. By contrast, when a GFP-C domain fusion was introduced into chlorina1-1 or Columbia wild type, a large amount of GFP-C domain protein accumulated and the chlorophyll a/b ratio decreased drastically from 3.6 to 2.2 in Columbia wild type. When an A domain-GFP was introduced into Columbia wild type, A domain-GFP levels were very low. Conversely, a large amount of the protein accumulated when it was introduced into the chlorina1-1 mutant. These results indicate that the A domain may sense the presence of chlorophyll b and regulate the accumulation of CAO protein in the chloroplasts.
INTRODUCTION
The first step in photosynthesis is the capture of light energy by the photosynthetic pigments in light-harvesting systems, which are composed of core and peripheral antenna complexes (Green and Durnford, 1996). The core complexes of photosystems I and II contain P700-chlorophyll a-protein complex and CP43/CP47 as internal light-harvesting antenna, respectively, and these complexes contain chlorophyll a and β-carotene as the photosynthetic pigments (Alfonso et al., 1994; Ben-Shem et al., 2003). On the other hand, depending on the photosynthetic organism, peripheral antenna complexes contain a variety of pigments, such as phycobilins, chlorophyll b, and fucoxanthin (Green and Durnford, 1996; Ting et al., 2002).
In chlorophytes and prochlorophytes, chlorophyll b is a pigment of the peripheral antenna complexes (Green and Durnford, 1996; La Roche et al., 1996), which harvest light energy at ∼470 and 650 nm. Light energy at these specific wavelengths within the light spectrum is not efficiently absorbed by chlorophyll a. Thus, organisms that use chlorophyll b in their antenna systems are capable of harvesting a wider range of light energy. In addition to this light-harvesting function, chlorophyll b also plays an important role in the regulation of photosynthetic antenna size. The antenna size of photosystem II in green plants is determined by the amount of the light-harvesting complex associated with core complexes of photosystem II (LHCII) (Jansson, 1994). LHCII levels are well correlated with chlorophyll b synthesis (Bailey et al., 2001). In a previous study, LHCII levels increased when chlorophyll b synthesis was activated by feeding 5-aminolevulinic acid, a precursor of chlorophylls (Tanaka et al., 1994). LHCII levels were low in chlorophyll b–less mutants (Peter and Thornber, 1991; Härtel et al., 1996; Espineda et al., 1999), and the ratio of chlorophyll to reaction center in photosystem II was ∼100 in chlorophyll b–less mutants compared with 300 in wild-type plants (Polle et al., 2000). When plants were grown under low light intensities, chlorophyll b synthesis was enhanced and antenna size increased (Bailey et al., 2001).
As a result of the observation that chlorophyll b is distributed exclusively in LHCs, it is known that chlorophyll b synthesis is critical for LHC formation. However, if chlorophyll b is overproduced, excess LHC or free chlorophyll b would accumulate and induce photodamage. Chlorophyll b levels are determined by two reactions: the synthesis of chlorophyll b by chlorophyllide a oxygenase (CAO) (Tanaka et al., 1998) and the reconversion of chlorophyll b to chlorophyll a by the chlorophyll cycle (Ohtsuka et al., 1997). Because a defect in CAO results in chlorophyll b–less or –deficient phenotypes, there are no other pathways capable of bypassing the CAO reaction (Tanaka et al., 1998; Espineda et al., 1999). Furthermore, CAO is considered the only enzyme responsible for chlorophyll b synthesis. This conclusion was validated with biochemical studies using recombinant proteins, which revealed that CAO carries out two-step oxygenation reactions and converts chlorophyllide a to chlorophyllide b without any other proteins except reduced ferredoxin (Oster et al., 2000). These results suggest that CAO enzyme activity is important for chlorophyll b synthesis. Chlorophyll a/b ratios are reported to correlate with CAO mRNA levels in Arabidopsis thaliana (Harper et al., 2004) and Dunaliella salina (Masuda et al., 2002, 2003) during acclimation to different light intensities, indicating that chlorophyll b synthesis is at least partly regulated by the level of CAO mRNA. However, the chlorophyll a/b ratio was only slightly decreased (from 2.85 to 2.65) in CAO-overexpressing plants (Tanaka et al., 2001), which suggests that CAO activity is regulated at the post-transcriptional level.
Chlorophyll b exists in prochlorophytes and chlorophytes and is synthesized by CAO (Tomitani et al., 1999), although the gene(s) for chlorophyll b synthesis has not been identified in Prochlorococcus (Hess et al., 2001). Recently, we isolated a full-length sequence that encodes a CAO gene from Prochlorothrix hollandica (PhCAO) (Nagata et al., 2004). Comparisons of the PhCAO gene sequence with those of higher plants revealed that the mature sequences of Arabidopsis CAO (AtCAO) and Oryza sativa CAO (OsCAO) consist of three domains (Nagata et al., 2004). The N-terminal conserved sequence (A domain; 134 amino acids) is found in AtCAO and OsCAO but not in PhCAO. On the contrary, the conserved C-terminal sequence is common to all CAO sequences (C domain; 336 or 343 amino acids). The A domain and the C domain are linked by a less conserved sequence, the B domain, which consists of 30 amino acids. The C domain has a Rieske center and nonheme iron binding motifs (Tomitani et al., 1999) and catalyzes the conversion of chlorophyll a to chlorophyll b without the help of the A domain (Nagata et al., 2004). These characteristics indicate that the A domain plays a role in the regulation of CAO activity in higher plants.
To isolate and reveal the functions of the individual domains, we introduced genes for the various CAO domains into Arabidopsis. When the complete CAO gene was overexpressed, CAO protein levels were below a detectable level and the chlorophyll a/b ratio was decreased only slightly. However, when the gene corresponding to the C domain was introduced into Arabidopsis, a large amount of C domain accumulated and the chlorophyll a/b ratio was reduced drastically. In addition, we also determined that the A domain reduced CAO protein levels only when chlorophyll b accumulated. Based on these results, we propose that the A domain is involved in sensing the presence of chlorophyll b and functions to destabilize CAO proteins.
RESULTS
The N-Terminal 56 Residues Contain a Transit Peptide Sequence
Using cyanobacteria as a model system, Nagata et al. (2004) reported that AtCAO consists of three domains (Figure 1A) and demonstrated that the C domain alone can catalyze the conversion of chlorophyll a to chlorophyll b. To assess the functions of the A domains in higher plants, we constructed transgenic Arabidopsis plants that constitutively overexpress full-length or truncated CAO proteins lacking their A or C domain (Figure 1B). These constructs were ectopically overexpressed under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Green fluorescent protein (GFP) was fused to CAO domains so that the amount and localization of the transgene products could be readily monitored by microscopic analyses (Niwa et al., 1999).
Figure 1.
Domain Structures of the AtCAO and GFP Proteins and the CAO-GFP Fusion Proteins.
Nagata et al. (2004) reported the domain structure of AtCAO, assuming that the N-terminal 36 residues exist as a transit peptide. We used the N-terminal 56 residues to guide the fusion proteins into chloroplasts (see Results). A, A domain; B, B domain; C, C domain; GFP, sGFP (S65T); tp, N-terminal 56 residues of AtCAO.
(A) Domain structures of the AtCAO and GFP proteins.
(B) Structures of the CAO-GFP fusion proteins.
Because CAO is a chloroplast protein, it was important that the transgene constructs be designed to encode a transit peptide that would facilitate import specific for chloroplasts. To date, the exact transit peptide sequence for CAO has not been determined experimentally; thus, we predicted the transit peptide region in the CAO sequence according to the methods of Konishi et al. (1993) and Emanuelsson et al. (2000). Konishi et al. (1993) proposed that the transit peptides of higher plants have a common secondary β-turn/β-sheet/α-helix structure. In the CAO sequence, computer-assisted analysis predicted that the N-terminal 50 residues fold into this same structure. A neural network–based method for cleavage site predictions developed by Emanuelsson et al. (2000) predicted that the transit sequence is cleaved after the first 36 residues. Furthermore, the N-terminal 56 residues were not highly conserved between AtCAO and OsCAO. Thus, we hypothesized that the N-terminal 56 residues contain a transit peptide that would facilitate import into chloroplasts.
As a method to confirm that the N-terminal 56 residues of CAO sufficiently guided the transgene products to the chloroplasts, we analyzed transgenic plants that overexpressed the N-terminal 56 residues (tp) fused to GFP (tp-GFP; Figure 1B). GFP signals in the transgenic tp-GFP plants were observed exclusively in the chloroplasts (Figure 2C). By contrast, GFP constructs that lacked the N-terminal 56 residues (GFP) did not target to chloroplasts (data not shown). Based on these experiments, the N-terminal 56 residues were used to guide transgene products to chloroplasts in the experiments that followed (Figure 1B).
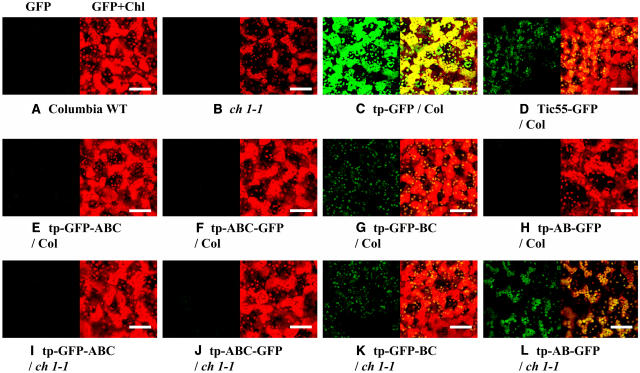
Figure 2.
Chloroplast Targeting of the CAO-GFP Fusion Proteins.
Fluorescence at 680 nm (chlorophylls) and 522 nm (GFP) of rosette leaves were monitored separately by confocal microscopy with a ×40 objective lens. GFP (left in each panel) and merged fluorescent (GFP+Chl; right in each panel) images are indicated. Chlorophyll and GFP fluorescence are indicated by red and green coloring, respectively. Note that the overlaps of green and red are indicated by yellow in the merged images. All fluorescence was recorded with an equal laser intensity. Chlorophyll b–less phenotypes ([B] and [L]) have less chlorophyll fluorescence compared with that in chlorophyll b–containing phenotypes. Bars = 50 μm.
(A) Columbia wild type (WT), a nontransformant of the Columbia wild type.
(B) ch1-1, a nontransformant of ch1-1.
(C) tp-GFP/Col, a transgenic Columbia wild type overexpressing the N-terminal 56 residues of CAO fused to GFP.
(D) Tic55-GFP/Col, a transgenic Columbia wild type overexpressing full-length Tic55 fused to GFP at its C terminus.
(E) tp-GFP-ABC/Col, a transgenic Columbia wild type overexpressing full-length CAO fused to GFP at its N terminus.
(F) tp-ABC-GFP/Col, a transgenic Columbia wild type overexpressing full-length CAO fused to GFP at its C terminus.
(G) tp-GFP-BC/Col, a transgenic Columbia wild type overexpressing the C domain of CAO fused to GFP.
(H) tp-AB-GFP/Col, a transgenic Columbia wild type overexpressing the A domain of CAO fused to GFP.
(I) tp-GFP-ABC/ch1-1, a transgenic ch1-1 mutant overexpressing full-length CAO fused to GFP at its N terminus.
(J) tp-ABC-GFP/ch1-1, a transgenic ch1-1 mutant overexpressing full-length CAO fused to GFP at its C terminus.
(K) tp-GFP-BC/ch1-1, a transgenic ch1-1 mutant overexpressing the C domain of CAO fused to GFP.
(L) tp-AB-GFP/ch1-1, a transgenic ch1-1 mutant overexpressing the A domain of CAO fused to GFP.
Full-Length CAO-GFP Constructs Complemented a Chlorophyll b–Less Mutant Phenotype of chlorina1-1
Before constructing transgenic plants that overexpressed CAO proteins lacking A or C domains, we produced transgenic plants that overexpress the entire CAO sequence fused to GFP. This experiment was designed to determine whether an intact native CAO protein is required for chlorophyll b biosynthesis. It has been reported that either C- or N-terminal GFP fusion influences protein functions in different manners (Kosemund et al., 2000). Thus, we tested the effect of GFP fusion to either the C or the N terminus (tp-ABC-GFP or tp-GFP-ABC; Figure 1B) on chlorophyll b biosynthesis and intracellular CAO localization. The constructs (tp-ABC-GFP or tp-GFP-ABC) were introduced into a Columbia background chlorophyll b–less mutant, chlorina1-1 (ch1-1). This mutant contains a 31-bp deletion in the intrinsic CAO gene and is rendered unable to synthesize chlorophyll b (Oster et al., 2000). Because ch1-1 lacks the ability to synthesize chlorophyll b, any chlorophyll b accumulation resulting from transformation with our various gene constructs would allow us to individually assess the contribution of transgene products to chlorophyll b synthesis.
Overexpression of the CAO-GFP fusion proteins complemented the chlorophyll b–less phenotype of ch1-1. Specifically, transgenic ch1-1 plants overexpressing the full-length CAO fused to GFP at the C terminus (tp-ABC-GFP) or the N terminus (tp-GFP-ABC) showed similar chlorophyll a/b ratios of 3.39 or 3.57, respectively (Table 1). These values were similar to that of the Columbia wild type (3.68). Total chlorophyll levels per fresh weight of rosette leaves of ch1-1 mutant were ∼25% of Columbia wild-type levels; however, chlorophyll levels were rescued by overexpression of either tp-ABC-GFP or tp-GFP-ABC (Table 1). Therefore, we conclude that GFP fusion to either the C or the N terminus of CAO does not interfere with chlorophyll b biosynthesis. Overexpression of the full-length CAO fused to GFP proteins (tp-ABC-GFP or tp-GFP-ABC) slightly decreased the chlorophyll a/b ratio in the Columbia wild-type background (Table 1). These results indicate that overexpression of the CAO gene by the CaMV 35S promoter only slightly increased the rate of chlorophyll b biosynthesis.
Table 1.
Chlorophyll Accumulation in the Transformants
| Chlorophyll a/b Ratio
|
Total Chlorophyll (nmol/mg Fresh Weight)
|
|||||
|---|---|---|---|---|---|---|
| Arabidopsis Transformants | Average | sd | Samples | Average | sd | Samples |
| Columbia wild type (nontransformant) | 3.68 | 0.13 | 5 | 1.97 | 0.29 | 6 |
| ch1-1 (nontransformant) | na | na | 3 | 0.47 | 0.07 | 6 |
| Full length | ||||||
| tp-ABC-GFP/Col | 2.95 | 0.23 | 7 | 2.00 | 0.22 | 8 |
| tp-GFP-ABC/Col | 3.13 | 0.43 | 9 | 2.10 | 0.07 | 4 |
| tp-ABC-GFP/ch1-1 | 3.39 | 0.17 | 3 | 1.89 | 0.27 | 4 |
| tp-GFP-ABC/ch1-1 | 3.57 | 0.28 | 9 | 1.89 | 0.23 | 4 |
| Domains A+B | ||||||
| tp-AB-GFP/Col | 3.64 | 0.13 | 13 | 2.18 | 0.20 | 8 |
| tp-AB-GFP/ch1-1 | na | na | 4 | 0.88 | 0.01 | 4 |
| Domains B+C | ||||||
| tp-GFP-BC/Col | 2.24 | 0.10 | 3 | 2.06 | 0.16 | 5 |
| tp-GFP-BC/ch1-1 | 2.29 | 0.30 | 5 | 1.96 | 0.09 | 4 |
| Additional controls | ||||||
| tp-GFP/Col | 3.72 | 0.08 | 5 | 2.24 | 0.19 | 4 |
The names of the transgenic plants are described in the legend to Figure 2. na, not analyzed.
As a method to determine intracellular and intraorganelle localization, transgenic plants overexpressing the full-length CAO-GFP fusions (tp-ABC-GFP or tp-GFP-ABC) were examined by confocal microscopy. However, GFP signals were not observed in any of the full-length CAO-GFP plants that regained chlorophyll b–synthesizing capacity (Figures 2E, 2F, 2I, and 2J). The ABC-GFP and GFP-ABC proteins were not detected by immunoblotting with either anti-GFP or anti-CAO antibodies in transgenic plants overexpressing mRNA for the full-length CAO-GFP fusion (Figures 3A, 3B, and 4). The results of confocal microscopy and immunoblot analysis suggest that a low level accumulation of CAO protein was sufficient for normal chlorophyll b biosynthesis.
Figure 3.
Immunoblot Analysis of the CAO-GFP Fusion and Truncated CAO Proteins in Transgenic Plants.
Equal fresh weights (2.5 mg) of rosette leaves were subjected to SDS-PAGE, and the CAO-GFP fusion or CAO proteins were detected using anti-GFP (A) or anti-CAO ([B] and [C]) antibodies. Gray arrowheads at left indicate the estimated size of CAO (∼56 kD), and black arrowheads indicate specific CAO signals detected by anti-CAO antibodies.
(A) and (B) The names of the transgenic plants are described in the legend to Figure 2.
(C) tp-ABC/ch1-1, a transgenic ch1-1 mutant overexpressing full-length CAO; tp-BC/ch1-1, a transgenic ch1-1 mutant overexpressing CAO without the A domain; tp-C/ch1-1, a transgenic ch1-1 mutant overexpressing CAO without the A and B domains.
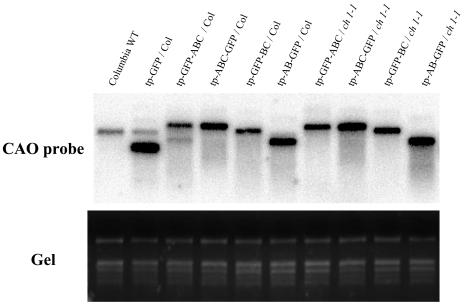
Figure 4.
RNA Gel Blot Analysis of the Transgene mRNA.
Five micrograms of total RNA extracted from rosette leaves of the transgenic plants was separated on a formaldehyde-agarose gel. RNA was transferred onto a nylon membrane and hybridized with a 32P-labeled full-length CAO gene–specific probe. rRNA bands separated on a gel were shown by ethidium bromide staining to demonstrate equal loading of samples (Gel). The names of the transgenic plants are described in the legend to Figure 2.
Effects of A Domain Removal from CAO on Chlorophyll b and Protein Levels
We constructed transgenic plants that overexpressed the C domain of CAO fused to GFP (tp-GFP-BC; Figure 1B) in the Columbia wild type and ch1-1 mutants. The chlorophyll a/b ratios of these transgenic Columbia wild-type and ch1-1 plants were 2.24 and 2.29, respectively. These values were significantly lower than that of the full-length CAO-GFP–overexpressing plants (Table 1). These results indicate that the C domain alone is sufficient for chlorophyll b biosynthesis and that the presence of the A domain in the CAO protein has a contrasting function, inhibiting chlorophyll b biosynthesis. Transgenic tp-GFP-BC mRNA levels were similar to those of tp-ABC-GFP or tp-GFP-ABC transgenic plants (Figure 4). The significantly decreased chlorophyll a/b ratio observed in the transgenic tp-GFP-BC plants was not attributed to enhanced CAO mRNA accumulation. Total chlorophyll levels per fresh weight of rosette leaves in transgenic plants (tp-GFP-BC) were almost the same as in the Columbia wild type (Table 1). These results suggest that the overexpression of CAO mRNAs changed chlorophyll a/b ratios but did not alter total chlorophyll levels.
In contrast with mRNA levels, a significant difference in CAO-GFP fusion protein levels was observed in the tp-GFP-BC and the full-length CAO-GFP (tp-ABC-GFP and tp-GFP-ABC) transgenic plants. In tp-GFP-BC transgenic plants, confocal microscopy detected GFP fluorescence in chloroplasts (Figures 2G and 2K) and anti-GFP and anti-CAO antibodies detected significant protein accumulation at the predicted molecular size (∼70 kD; Figures 3A and 3B). However, in tp-ABC-GFP or tp-GFP-ABC transgenic plants, transgenic protein accumulation was below a detectable level by confocal microscopy (Figures 2E, 2F, 2I, and 2J) and immunoblot analysis (Figures 3A and 3B). These results indicate that increased chlorophyll b accumulation in the tp-GFP-BC plants was a result of increased CAO protein accumulation and suggest that the A domain may suppress CAO protein accumulation.
These results were confirmed in the transgenic plants that lacked GFP (Figure 3C). When the complete CAO (tp-ABC), CAO without the A domain (tp-BC), or CAO without the A and B domains (tp-C) genes were introduced into ch1-1 mutants, the chlorophyll b–less mutant phenotype was rescued. Immunoblot analysis could not detect CAO proteins with the Columbia wild type or even in the tp-ABC–overexpressing lines. Weak bands of ∼56 kD were observed, and it was concluded that these bands were not CAO. This conclusion was reached because identical bands were also observed in the ch1-1 mutant and in ch1-1 background transgenic plants. The ch1-1 mutant has a defective CAO gene and therefore cannot produce appropriately sized CAO protein. On the other hand, a large amount of CAO proteins accumulated when the A domain was removed (tp-BC and tp-C). These data suggest that the accumulation of CAO proteins was not affected by GFP but was regulated by the A domain. We also observed a slight increase in CAO accumulation when the B domain was removed (Figure 3C). Although these results are reproducible, further experimentation is warranted and necessary to clarify the physiological function of the B domain.
Localization of the GFP-BC Protein
The GFP signals in tp-GFP-BC plants were observed by confocal microscopy at peripheral and internal regions of chloroplasts (Figure 5D). We constructed transgenic plants that overexpress the envelope-specific protein, Tic55 (Caliebe et al., 1997), with a C-terminal fusion to GFP (Tic55-GFP). The GFP signals of the Tic55-GFP proteins were targeted to chloroplasts (Figure 2D) and showed a ring-like distribution pattern around the chloroplasts (Figure 5B). The GFP signals of GFP-BC in chloroplasts were similar to those of Tic55-GFP (Figure 5D). Additionally, the GFP signals of GFP-BC proteins inside the chloroplasts almost overlapped with chlorophyll fluorescence. The signals inside the chloroplasts formed a gradation pattern, and stronger and weaker patches of fluorescence were evident (Figure 5D).
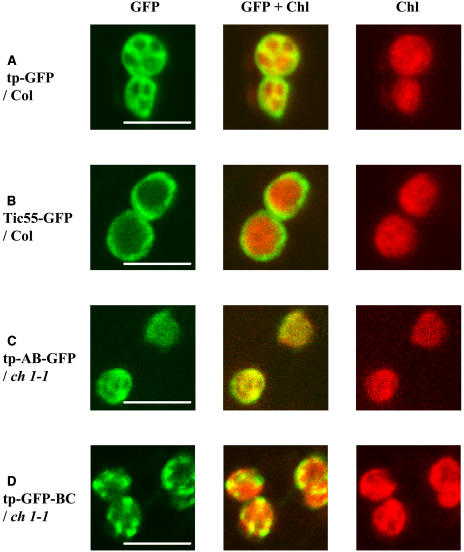
Figure 5.
GFP Fluorescence Pattern of the CAO-GFP Fusion Proteins in Chloroplasts.
Fluorescence at 680 nm (chlorophylls) and 522 nm (GFP) of chloroplasts were monitored separately by confocal microscopy with a ×100 oil-immersion objective lens. Chlorophyll (Chl) and GFP fluorescence are indicated by red and green coloring, respectively. Note that the overlaps of green and red are indicated by yellow in the merged images. To optimize the images, the intensities of actinic laser and gain levels of fluorescence were changed in each sample. Bars = 5 μm. The names of the transgenic plants are described in the legend to Figure 2.
Chloroplasts were isolated from tp-GFP-BC and Tic55-GFP transgenic plants, and envelope and thylakoid membranes were fractionated by sucrose density gradient ultracentrifugation. Tic55-GFP was detected in envelope fractions and Lhcb proteins were found exclusively in fractions associated with thylakoid membranes (Figures 6A and 6B). These results indicate that chloroplast envelope and thylakoid membranes were effectively separated by sucrose gradient separation. By contrast, GFP-BC was found in both envelope and thylakoid fractions (Figure 6A). These results were consistent with those obtained with confocal microscopy and strongly suggest that the GFP-BC proteins are localized on envelope and thylakoid membranes.
Figure 6.
Localization of GFP-BC Proteins in Chloroplast Membranes.
Stroma, envelope, and thylakoid fractions were isolated from chloroplasts harvested from transgenic plants. An equal amount of protein from each fraction was separated by SDS-PAGE, and the proteins were transferred onto a polyvinylidene difluoride membrane. S, stroma; E, envelope; T, thylakoids. The names of the transgenic plants are described in the legend to Figure 2.
(A) Proteins (0.5 μg) were subjected to immunoblotting using anti-GFP antibodies, and localization of GFP fusion proteins was analyzed.
(B) Proteins (0.17 μg) were subjected to immunoblotting using anti-Lhcb antibodies.
Chlorophyll b Feedback Regulates CAO Protein Accumulation
Considering the inhibitory function of the A domain on CAO accumulation, it is possible to speculate that when chlorophyll b synthesis is in excess, the A domain decreases CAO protein levels. However, if the transgene constructs contain a catalytic C domain, chlorophyll b is inevitably synthesized in both Columbia wild-type and ch1-1 mutant backgrounds and the effects of chlorophyll b on protein accumulation cannot be examined. To overcome this problem, we compared the accumulation of the fusion protein of the A domain and GFP (tp-AB-GFP; Figure 1B) in the presence (Columbia wild-type background) or absence (ch1-1 background) of chlorophyll b.
Chlorophyll b did not accumulate in the transgenic ch1-1 plants, and overexpression of the tp-AB-GFP gene did not change the chlorophyll a/b ratio in transgenic Columbia wild-type plants (Table 1). GFP signals of tp-AB-GFP were found in chloroplasts in the transgenic ch1-1 background (Figure 2L). In contrast with the ch1-1 background, no GFP signal was observed in the Columbia wild-type background (Figure 2H). These results were also supported by immunoblotting with anti-GFP antibodies. Significant amounts of the transgene products were observed in the ch1-1 background, whereas little transgene product was observed in the Columbia wild-type background (∼45 kD; Figure 3A). With the exception of the weak signals of AB-GFP, similar results were obtained with anti-CAO antibodies (Figure 3B). It is likely that this was attributable to the weak reactivity of the anti-CAO antibodies against the A domain (see Methods). These results indicate that the presence of the A domain prevents protein accumulation only when chlorophyll b is accumulated.
Immunoblot analysis with tp-AB-GFP plants showed that the A domain downregulated the accumulation of AB-GFP in the presence of chlorophyll b. However, tp-AB-GFP mRNA levels were almost the same in both Columbia wild-type and ch1-1 backgrounds (Figure 4). To clarify whether chlorophyll b regulates the accumulation of AB-GFP at the translational level, we investigated the distribution of tp-AB-GFP mRNA in polysome gradients (Figure 7A). RNA gel blot analysis of tp-AB-GFP mRNA showed that almost all of the mRNA associated with ribosomes in both Columbia wild-type and ch1-1 backgrounds. There was no significant difference in the distribution profiles between the two plants, which indicates that chlorophyll b does not influence the translation activity of tp-AB-GFP mRNA (Figures 7A and 7B). Lhcb1 mRNA distributed to polysome and nonpolysome fractions (fraction 2 in Figures 7A and 7B), indicating that a part of Lhcb1 mRNA was not translated. However, tp-AB-GFP mRNA distributed only to polysomal fractions in both the Columbia wild type and ch1-1 (Figure 7B). This specific distribution may be caused by the Tobacco mosaic virus Ω sequence in tp-AB-GFP mRNA, which increases translational efficiency.
Figure 7.
Association of tp-AB-GFP mRNA with Polysomes in the Transgenic Plants.
(A) Total extracts of the rosette leaves of tp-AB-GFP/Col and tp-AB-GFP/ch1-1 were fractionated on sucrose gradients. The gradients were fractionated into 10 fractions consisting of equal volumes. An equal proportion of the RNA purified from each fraction was separated on a formaldehyde-agarose gel, and rRNA bands separated on a gel were shown by ethidium bromide staining to confirm equal loading of samples (Gel). RNA was transferred onto a nylon membrane and hybridized with a 32P-labeled full-length CAO or Lhcb1 probe. Fractions are numbered from top to bottom of the gradient.
(B) Distribution profiles of tp-AB-GFP and Lhcb1 mRNA that associated to polysomes. The relative quantification data of signal intensities were measured using NIH Image software (http://rsb.info.nih.gov/nih-image/).
The names of the transgenic plants are described in the legend to Figure 2.
DISCUSSION
The A Domain Regulates CAO Accumulation
Although we were unable to detect CAO proteins with our anti-CAO antibodies in either Columbia wild-type or full-length CAO-GFP–overexpressing (tp-ABC-GFP or tp-GFP-ABC) plants, chlorophyll b was actively synthesized, indicating that low levels of CAO are sufficient for chlorophyll b accumulation at normal levels. By contrast, when the CAO sequence lacking the A domain–encoding region (tp-GFP-BC) was introduced into Arabidopsis, a large amount of GFP-BC accumulated. With the exceptions of the low light–adapted Prochlorococcus (Urbach et al., 1998) and CAO-overexpressing cyanobacteria (Xu et al., 2001), whose chlorophyll a/b ratios were less than 1.0, the chlorophyll a/b ratio decreased drastically in this study (Espineda et al., 1999; Bailey et al., 2001; Tanaka et al., 2001). These results indicate that the A domain reduces CAO protein accumulation and therefore controls the production of chlorophyll b. It is possible that this might be the reason why CAO overexpression resulted in a decreased chlorophyll a/b ratio of only 0.2 (Tanaka et al., 2001).
The A Domain Senses the Presence of Chlorophyll b and Regulates CAO Protein Levels
When the tp-AB-GFP gene was introduced into a chlorophyll b–less ch1-1 mutant, a large amount of AB-GFP accumulated despite the presence of the A domain, whereas the accumulation of AB-GFP was very low when the same construct was introduced into the Columbia wild type. These data suggest that the A domain senses the presence of chlorophyll b and regulates CAO accumulation. It is possible that chlorophyll b and the A domain regulate the accumulation of CAO on multiple levels. Specifically, the translation, the import into the chloroplasts, and the stabilization of CAO proteins may be affected by chlorophyll b and the A domain. Translational regulation by chlorophyll b is not likely, because chlorophyll b is located in the chloroplasts, whereas CAO proteins are synthesized by cytoplasmic ribosomes. Furthermore, polysome analysis showed that the translation was not affected by chlorophyll b. The second possibility is that the import of CAO into chloroplasts is inhibited by chlorophyll b. It has been reported that protochlorophyllide a reductase accumulates on the chloroplast envelope when import into the chloroplasts is inhibited (Reinbothe et al., 1996; Kim and Apel, 2004). We also observed the accumulation of GFP in the cytoplasmic spaces when GFP was not transported into the chloroplasts. These results indicate that if the import of CAO is inhibited, the proteins accumulate in either cytoplasmic spaces or on the chloroplast envelopes. However, we did not observe the accumulation of tp-AB-GFP or full-length CAO-GFP in either the cytoplasmic spaces or on the chloroplast envelopes in chlorophyll b–containing plants. Based on these results, we propose that the proteins with the A domain (full-length CAO, GFP-ABC, ABC-GFP, and AB-GFP) are degraded in the chloroplasts after or during the transport into chloroplasts (Figure 8). According to this hypothesis, it is possible that the A domain has two functions: to sense the presence of chlorophyll b and to destabilize CAO proteins.
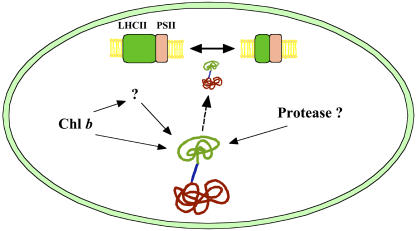
Figure 8.
Regulation of CAO Protein Accumulation in the Chloroplasts.
CAO consists of three domains, A, B, and C. The A and B domains are not involved in the catalytic function, and only the C domain is sufficient for the conversion of chlorophyll a to chlorophyll b. The A domain regulates the accumulation of CAO by sensing the presence of chlorophyll b. When the chlorophyll b level is low or absent, CAO is stable in the chloroplasts and chlorophyll b synthesis is accelerated. However, when a large amount of chlorophyll b accumulates, the A domain senses the presence of chlorophyll b and destabilizes CAO, resulting in the inhibition of chlorophyll b synthesis. Chloroplast protease(s) might be involved in the degradation of CAO proteins. By this mechanism, CAO levels are maintained at the required levels and the activity of chlorophyll b synthesis is regulated. Finally, synthesized chlorophyll stabilizes LHCII and regulates photosynthetic antenna size. The A, B, and C domains of CAO are illustrated in green, blue, and red, respectively. PSII, photosystem II.
There are two possible mechanisms for the A domain to sense the presence of chlorophyll b. The first would involve a direct interaction of chlorophyll b with the A domain. The A domain does not have a catalytic function but it does contain a putative chlorophyll binding motif (ExxNxR; Eggink and Hoober, 2000). According to this direct interaction mechanism, chlorophyll b binds to the A domain and destabilizes the CAO proteins. The second possibility is that the A domain senses the presence of chlorophyll b indirectly. Chlorophyll b induces the formation of LHC and enlarges the antenna size of the photosystems (Tanaka et al., 1994; Bailey et al., 2001). It is possible that these effects might induce changes in the photosynthetic activity and redox state of the chloroplast. The A domain senses these changes and regulates the accumulation of the CAO proteins. Further experimentation is required to clarify the interaction between chlorophyll b and the A domain.
The involvement of a domain regulating protein stability was reported previously with auxin/indoleacetic acid (Aux/IAA) proteins, which are involved in the auxin signaling pathway. Domain II of the Aux/IAA proteins (13 amino acids) is essential for the rapid degradation of Aux/IAA proteins, and the deletion of this domain resulted in Aux/IAA protein accumulation (Ramos et al., 2001). This rapid degradation is important for the transient increase in protein levels. The degradation processes in Aux/IAA proteins require a ubiquitin system (Gray et al., 2001). However, ubiquitin has not been reported previously in chloroplasts (Beers et al., 1992). Therefore, it is possible that CAO might use a different system for its degradation. The importance of the N-terminal sequence for protein degradation in chloroplasts was also reported with LHCII (Yang et al., 2000). LHCII became stable toward protease when the N-terminal sequence was deleted. Although in vitro experiments showed that the N-terminal sequence destabilizes LHCII, LHCII is the most abundant protein in the thylakoid membranes, whereas CAO protein is present at extremely low levels. Thus, we speculate that the LHC degradation system differs from that of CAO and that some novel mechanisms function for the control of CAO accumulation. The A domain destabilizes not only the C domain of CAO but also GFP if these proteins are fused to the A domain. These data allow us to conclude that if a protein acquires an A domain–like sequence, the protein can be regulated at the post-translational level. At present, we have found no proteins that exhibit sequence similarity with the A domain. However, we speculate that some sequences are involved in the regulation of protein accumulation in chloroplasts in the same manner as the A domain, because post-translational regulation is known to be especially important in chloroplasts.
Physiological Functions of the A Domain
One of the physiological functions of the A domain is to maintain CAO at a low level. This is important because low CAO levels are sufficient for the chlorophyll b synthesis that is required for LHC formation. If the chlorophyll a/b ratio decreases below 2 as a result of CAO protein accumulation, either an excessive amount of LHC will accumulate or chlorophyll b will not properly localize in the photosystems. This phenomenon would occur because LHC is the only chlorophyll b binding complex. Damage or impairment of the photosystems might be induced in CAO-overproducing plants. Therefore, it is possible that the A domain might function to keep CAO at an adequate level (Figure 8).
We reported that PhCAO does not contain sequences corresponding to the A and B domains of AtCAO but only contains sequences similar to the C domain with a short C-terminal extension (Nagata et al., 2004). Considering that the CAOs of Chlamydomonas and Dunaliella have long N-terminal extensions, the A domain must be acquired in the eukaryotic stage. It is interesting that the LHC superfamily is also found only in eukaryotic photosynthetic organisms (Green and Durnford, 1996). One possible function of the A domain of eukaryotic CAOs is to interact with the LHC during the assembly of chlorophyll and LHCII apoproteins (Hoober and Eggink, 2001). This also indicates that the A domain has other functions besides the regulation of protein accumulation. Further studies are required to clarify the function of the A domain.
Localization of the CAO Proteins
Using GFP fluorescence as a marker, we could not identify CAO localization in the chloroplasts. This was likely attributable to the fact that transgenic protein levels were too low and GFP fluorescence was not observed when the full-length CAO was used. However, we found that the GFP signal of GFP-BC localized on the edge and inner spaces of the chloroplasts. In addition, fractionation experiments with chloroplasts indicated that GFP-BC localized on thylakoid and envelope membranes. These results are consistent with the previous report that CAO exists on both chloroplast envelopes and thylakoid membranes (Eggink et al., 2004). The GFP fluorescent pattern of AB-GFP was similar to that of GFP in chloroplasts (Figures 5A and 5C). Therefore, in contrast with GFP-BC, it is possible that the AB domain has no role in dictating protein localization and that the C domain determines the localization of CAO proteins. This CAO localization is consistent with previous reports that chlorophyllide a, a substrate of CAO, is synthesized in the chloroplast envelope (Joyard et al., 1990) and that chlorophyll a bound to the chlorophyll–protein complexes in the thylakoid membranes is converted to chlorophyll b during reorganization of the photosystems (Tanaka and Tsuji, 1982; Ohtsuka et al., 1997). Determination of the exact localization of CAO by immunoelectron microscopy and identification of the proteins that interact with CAO are necessary to understand the regulatory mechanisms of protein accumulation in the chloroplasts.
METHODS
Plant Materials
Arabidopsis thaliana plants were grown at 23°C under continuous light conditions (white fluorescent light at 60 to 90 μmol·m−2·s−1). Seeds of the Columbia wild-type strain and the ch1-1 mutant were germinated in soil and grown for 4 to 6 weeks in pots. Seeds of the transgenic Arabidopsis were germinated on plates containing half-strength MS medium (2.3 g/L; Wako Pure Chemical Industries, Osaka, Japan), 0.7% (w/v) agar, and 50 μg/mL kanamycin. Transgenic plants successfully grown for 1 week were transferred into soil and continuously grown for 4 to 6 weeks. After 4 to 6 weeks, rosette leaves of the plants were harvested and used for the experiments.
Plasmid Construction
A sGFP (S65T) overexpression system containing the CaMV 35S promoter, the Tobacco mosaic virus Ω sequence, and the nopaline synthase terminator (from Y. Niwa, University of Shizuoka, Shizuoka, Japan) (Niwa et al., 1999) was incorporated into the multicloning site of a pGreenII-0029 plasmid at EcoRI and HindIII (from R.P. Hellens and P. Mullineaux, John Innes Centre, Colney, UK) (Hellens et al., 2000). This plasmid was used to construct the transformation plasmid vectors containing the transgenes. The sGFP (S65T) gene in this plasmid was substituted by transgenes (full-length or truncated AtCAO cDNA sequences fused to sGFP [S65T] gene; Figures 1A and 1B) at the SalI and NotI sites. The DNA sequence integrity of the entire coding region was verified by sequencing.
Transformation Techniques
A series of transformation plasmids were introduced into Agrobacterium tumefaciens strain C58 (GV2260) by electroporation. Arabidopsis plants (Columbia wild type and ch1-1 mutants) were transformed by infiltration with Agrobacterium cells containing the series of transformation plasmids. Primary Arabidopsis transformants were selected on plates containing half-strength MS medium (2.3 g/L), 0.7% (w/v) agar, and 50 μg/mL kanamycin.
RNA Gel Blot Analysis
Total RNA was extracted from Arabidopsis rosette leaves using an RNeasy RNA extraction kit (Qiagen, Hilden, Germany). Total RNA (5 μg) was denatured with formamide/formaldehyde and fractionated on a 1.0% (w/v) agarose–18% (v/v) formaldehyde gel. The gel was subsequently blotted onto Hybond-XL membranes (Amersham Biosciences, Piscataway, NJ), and RNA was cross-linked to these membranes by UV radiation. The membranes were hybridized with a specific full-length radiolabeled CAO probe and washed for 20 min at 60°C with 2× SSC buffer (1× SSC buffer is 16.65 mM NaCl and 16.65 mM sodium citrate) containing 0.1% (w/v) SDS and then for 20 min at 60°C with 1× SSC buffer containing 0.1% (w/v) SDS. Signals were developed using a FUJI BAS-1500 system (Fuji Film, Tokyo, Japan).
Polysome Analysis
Polysomes were isolated as described previously by Barkan (1998). Frozen rosette leaves (0.3 g) were ground in liquid nitrogen, and 1 mL of polysome extraction buffer (0.2 M Tris-HCl, pH 9.0, 0.2 M KCl, 35 mM MgCl2, 12.5 mM EGTA, 0.2 M sucrose, 1% [w/v] Triton X-100, 2% [w/v] polyoxyethylene-20-cetyl ether, 100 mM β-mercaptoethanol, 0.5 mg/mL heparin, 100 μg/mL chloramphenicol, and 25 μg/mL cycloheximide) was added. Leaf material was ground further until thawed and placed on ice for 10 min. Nuclei and insoluble materials were pelleted by centrifugation for 5 min at 15,000 rpm. Sodium deoxycholate was added to the supernatant to a final concentration of 0.5% (w/v), and the sample was placed on ice for 5 min. Remaining insoluble material was subsequently pelleted by centrifugation at 15,000 rpm for 15 min. The supernatant (0.4 mL) was layered onto 3.5-mL sucrose gradients that were prepared as described by Barkan (1998) and centrifuged with a RPS-56T swing rotor (Hitachi Koki, Tokyo, Japan) in a Himac CP70G ultracentrifuge at 45,000 rpm for 65 min at 4°C. Four hundred microliters of each fraction was collected by gentle pipetting from the top of the gradient and added to 50 μL of a solution consisting of 5% SDS and 0.2 M EDTA, pH 8.0. RNA was purified from each fraction by phenol:chloroform:isoamyl alcohol (25:24:1) extraction and subsequent ethanol precipitation. Finally, RNA samples were resuspended in 20 μL of TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA), and 5 μL of each fraction was examined by RNA gel blot analysis.
Immunoblot Analysis
Ten milligrams of rosette leaves was homogenized with 100 μL of extraction buffer (50 mM Tris, pH 6.8, 2 mM EDTA, 10% [w/v] glycerol, 2% [w/v] SDS, and 6% [v/v] 2-mercaptoethanol), and the homogenate samples were centrifuged at 10,000g for 5 min. The supernatants (25 μL) was subjected to 10% (w/v) acrylamide SDS-PAGE separation, and the resolved proteins were blotted onto a Hybond-P membrane (Amersham Biosciences). Anti-GFP rabbit antibodies (diluted 1:5000; Invitrogen, Carlsbad, CA) or anti-CAO rabbit antibodies (diluted 1:1000) were used to detect CAO or GFP transferred proteins. Anti-rabbit IgG linked to horseradish peroxidase (Amersham Biosciences) was used as a secondary antibody specific for both primary antibodies. Cross-reactive protein bands were developed using an ECL plus protein gel blot analysis kit (Amersham Biosciences). The anti-CAO antibodies were raised against a polypeptide corresponding to the Tyr-120 to Val-516 sequence of CAO protein in our laboratory.
Purification of Envelope and Thylakoid Membranes from Chloroplasts
Envelope and thylakoid membranes were isolated from chloroplasts of Arabidopsis plants as described by Block et al. (2002). The leaves were homogenized in a blender in 250 mL of ice-cold buffer containing 0.45 M sorbitol, 20 mM Tricine/NaOH, pH 8.4, 10 mM EDTA, 10 mM NaHCO3, and 0.1% BSA. A chloroplast pellet was obtained by centrifugation at 1500g for 3 min. The chloroplasts were disrupted in 10 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.6, 4 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride. Chloroplast subfractions were separated on a step gradient of 2.0, 0.93, and 0.6 M sucrose in buffer R [10 mM 3-(N-morpholino)-propanesulfonic acid, pH 7.6, and 4 mM MgCl2] by ultracentrifugation at 70,000g for 1 h. The chloroplast envelope fraction was collected at the interface between the 0.6 and 0.93 M sucrose layers, and thylakoids were collected at the interface between the 0.93 and 2.0 M sucrose layers.
Confocal Microscopy
Fluorescence images were recorded on a MRC 1024 confocal system (Bio-Rad Laboratories, Hercules, CA) and an Axioplan fluorescence microscope equipped with an Achroplan ×40 objective lens and an α Plan-Fluar ×100 oil-immersion objective lens (Carl Zeiss, Jena, Germany). An argon laser (25 mW) was used to generate an excitation source at 488 nm, and GFP and chlorophyll fluorescence were recorded at 522 and 680 nm, respectively. The images were processed with Adobe Photoshop 4.0 software (Adobe, San Jose, CA).
HPLC Analysis
Rosette leaves were homogenized in liquid nitrogen, and pigments were extracted with 100% acetone. The extracts were then centrifuged at 15,000g for 5 min, and the supernatant was subjected to HPLC. Pigments were separated on an octadecyl column (Shim-pack CLC-ODS column, 6.0 × 150 mm; Shimadzu, Kyoto, Japan) using methanol or methanol:acetone (70:30) as the elution agent at a flow rate of 1.7 mL/min. Elution profiles were monitored by measuring at 650 nm. Chlorophyll content was quantified from the chromatographic peak areas after calibration of the chromatographic response with known quantities of the relevant pigments (Juntec, Atsugi, Japan).
Acknowledgments
This work was supported in part by a Grant-in-Aid (15370015) to A.T. from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Ayumi Tanaka (ayumi@pop.lowtem.hokudai.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.031518.
References
- Alfonso, M., Montoya, G., Cases, R., Rodriguez, R., and Picorel, R. (1994). Core antenna complexes, CP43 and CP47, of higher plant photosystem II: Spectral properties, pigment stoichiometry, and amino acid composition. Biochemistry 33, 10494–10500. [DOI] [PubMed] [Google Scholar]
- Bailey, S., Walters, R.G., Jansson, S., and Horton, P. (2001). Acclimation of Arabidopsis thaliana to the light environment: The existence of separate low light and high light responses. Planta 213, 794–801. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297, 38–57. [Google Scholar]
- Beers, E.P., Moreno, T.N., and Callis, J. (1992). Subcellular localization of ubiquitin and ubiquitinated proteins in Arabidopsis thaliana. J. Biol. Chem. 267, 15432–15439. [PubMed] [Google Scholar]
- Ben-Shem, A., Frolow, F., and Nelson, N. (2003). Crystal structure of plant photosystem I. Nature 426, 630–635. [DOI] [PubMed] [Google Scholar]
- Block, M.A., Tewari, A.K., Albrieux, C., Maréchal, E., and Joyard, J. (2002). The plant S-adenosyl-l-methionine:Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur. J. Biochem. 269, 240–248. [DOI] [PubMed] [Google Scholar]
- Caliebe, A., Grimm, R., Kaiser, G., Lübeck, J., Soll, J., and Heins, L. (1997). The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 16, 7342–7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggink, L.L., and Hoober, J.K. (2000). Chlorophyll binding to peptide maquettes containing a retention motif. J. Biol. Chem. 275, 9087–9090. [DOI] [PubMed] [Google Scholar]
- Eggink, L.L., LoBrutto, R., Brune, D.C., Brusslan, J., Yamasato, A., Tanaka, A., and Hoober, J.K. (2004). Synthesis of chlorophyll b: Localization of chlorophyllide a oxygenase and discovery of a stable radical in the catalytic subunit. BMC Plant Biol. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Espineda, C.E., Linford, A.S., Devine, D., and Brusslan, J.A. (1999). The AtCAO gene, encoding chlorophyll a oxygenase, is required for chlorophyll b synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 10507–10511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Green, B.R., and Durnford, D.G. (1996). The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714. [DOI] [PubMed] [Google Scholar]
- Harper, A.L., von Gesjen, S.E., Linford, A.S., Peterson, M.P., Faircloth, R.S., Thissen, M.M., and Brusslan, J.A. (2004). Chlorophyllide a oxygenase mRNA and protein levels correlate with the chlorophyll a/b ratio in Arabidopsis thaliana. Photosynth. Res. 79, 149–159. [DOI] [PubMed] [Google Scholar]
- Härtel, H., Lokstein, H., Grimm, B., and Rank, B. (1996). Kinetic studies on the xanthophyll cycle in barley leaves (influence of antenna size and relations to nonphotochemical chlorophyll fluorescence quenching). Plant Physiol. 110, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens, R.P., Edwards, E.A., Leyland, N.R., Bean, S., and Mullineaux, P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42, 819–832. [DOI] [PubMed] [Google Scholar]
- Hess, W.R., Rocap, G., Ting, C.S., Larimer, F., Stilwagen, S., Lamerdin, J., and Chisholm, S.W. (2001). The photosynthetic apparatus of Prochlorococcus: Insights through comparative genomics. Photosynth. Res. 70, 53–71. [DOI] [PubMed] [Google Scholar]
- Hoober, J.K., and Eggink, L.L. (2001). A potential role of chlorophylls b and c in assembly of light-harvesting complexes. FEBS Lett. 489, 1–3. [DOI] [PubMed] [Google Scholar]
- Jansson, S. (1994). The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta 1184, 1–19. [DOI] [PubMed] [Google Scholar]
- Joyard, J., Block, M., Pineau, B., Albrieux, C., and Douce, R. (1990). Envelope membranes from mature spinach chloroplasts contain a NADPH:protochlorophyllide reductase on the cytosolic side of the outer membrane. J. Biol. Chem. 265, 21820–21827. [PubMed] [Google Scholar]
- Kim, C., and Apel, K. (2004). Substrate-dependent and organ-specific chloroplast protein import in planta. Plant Cell 16, 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi, T., Maruta, Y., Shinohara, K., and Watanabe, A. (1993). Transit peptides of thylakoid luminal proteins: The sites of stromal processing are conserved among higher plants. Plant Cell Physiol. 34, 1081–1087. [Google Scholar]
- Kosemund, K., Geiger, I., and Paulsen, H. (2000). Insertion of light-harvesting chlorophyll a/b protein into the thylakoid: Topographical studies. Eur. J. Biochem. 267, 1138–1145. [DOI] [PubMed] [Google Scholar]
- La Roche, J., van der Staay, G.W.M., Partensky, F., Ducret, A., Aebersold, R., Li, R., Golden, S.S., Hiller, R.G., Wrench, P.M., Larkum, A.W.D., and Green, B.R. (1996). Independent evolution of the prochlorophytes and green plant chlorophyll a/b light-harvesting proteins. Proc. Natl. Acad. Sci. USA 93, 15244–15248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T., Polle, J.E.W., and Melis, A. (2002). Biosynthesis and distribution of chlorophyll among the photosystems during recovery of the green alga Dunaliella salina from irradiance stress. Plant Physiol. 128, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T., Tanaka, A., and Melis, A. (2003). Chlorophyll antenna size adjustments by irradiance in Dunaliella salina involve coordinate regulation of chlorophyll a oxygenase (CAO) and Lhcb gene expression. Plant Mol. Biol. 51, 757–771. [DOI] [PubMed] [Google Scholar]
- Nagata, N., Satoh, S., Tanaka, R., and Tanaka, A. (2004). Domain structures of chlorophyllide a oxygenase of green plants and Prochlorothrix hollandica in relation to catalytic functions. Planta 218, 1019–1025. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18, 455–463. [DOI] [PubMed] [Google Scholar]
- Ohtsuka, T., Ito, H., and Tanaka, A. (1997). Conversion of chlorophyll b to chlorophyll a and the assembly of chlorophyll with apoproteins by isolated chloroplasts. Plant Physiol. 113, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster, U., Tanaka, R., Tanaka, A., and Rüdiger, W. (2000). Cloning and functional expression of the gene encoding the key enzyme for chlorophyll b biosynthesis (CAO) from Arabidopsis thaliana. Plant J. 21, 305–310. [DOI] [PubMed] [Google Scholar]
- Peter, G.F., and Thornber, J.P. (1991). Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-proteins. J. Biol. Chem. 266, 16745–16754. [PubMed] [Google Scholar]
- Polle, J.E.W., Benemann, J.R., Tanaka, A., and Melis, A. (2000). Photosynthetic apparatus organization and function in the wild type and a chlorophyll b-less mutant of Chlamydomonas reinhardtii: Dependence on carbon source. Planta 211, 335–344. [DOI] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, O., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe, S., Reinbothe, C., Neumann, D., and Apel, K. (1996). A plastid enzyme arrested in the step of precursor translocation in vivo. Proc. Natl. Acad. Sci. USA 93, 12026–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., and Tsuji, H. (1982). Calcium-induced formation of chlorophyll b and light-harvesting chlorophyll a/b-protein complex in cucumber cotyledons in the dark. Biochim. Biophys. Acta 680, 265–270. [Google Scholar]
- Tanaka, A., Ito, H., Tanaka, R., Tanaka, N.K., Yoshida, K., and Okada, K. (1998). Chlorophyll a oxygenase (CAO) is involved in chlorophyll b formation from chlorophyll a. Proc. Natl. Acad. Sci. USA 95, 12719–12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, A., Tanaka, Y., and Tsuji, H. (1994). Preferential accumulation of apoproteins of the light-harvesting chlorophyll a/b protein complex in greening barley leaves treated with 5-aminolevulinic acid. Planta 192, 92–97. [Google Scholar]
- Tanaka, R., Koshino, Y., Sawa, S., Ishiguro, S., Okada, K., and Tanaka, A. (2001). Overexpression of chlorophyllide a oxygenase (CAO) enlarges the antenna size of photosystem II in Arabidopsis thaliana. Plant J. 26, 365–373. [DOI] [PubMed] [Google Scholar]
- Ting, C.S., Rocap, G., King, J., and Chisholm, S.W. (2002). Cyanobacterial photosynthesis in the oceans: The origins and significance of divergent light-harvesting strategies. Trends Microbiol. 10, 134–142. [DOI] [PubMed] [Google Scholar]
- Tomitani, A., Okada, K., Miyashita, H., Matthijs, H.C.P., Ohno, T., and Tanaka, A. (1999). Chlorophyll b and phycobilins in the common ancestor of cyanobacteria and chloroplasts. Nature 400, 159–162. [DOI] [PubMed] [Google Scholar]
- Urbach, E., Scanlan, D.J., Distel, D.L., Waterbury, J.B., and Chisholm, S.W. (1998). Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46, 188–201. [DOI] [PubMed] [Google Scholar]
- Xu, H., Vavilin, D., and Vermaas, W. (2001). Chlorophyll b can serve as the major pigment in functional photosystem II complexes of cyanobacteria. Proc. Natl. Acad. Sci. USA 98, 14168–14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D.-H., Paulsen, H., and Andersson, B. (2000). The N-terminal domain of the light-harvesting chlorophyll a/b-binding protein complex (LHCII) is essential for its acclimative proteolysis. FEBS Lett. 466, 385–388. [DOI] [PubMed] [Google Scholar]