Abstract
Although the splicing of transcripts from most eukaryotic genes occurs in a constitutive fashion, some genes can undergo a process of alternative splicing. This is a genetically economical process which allows a single gene to give rise to several protein isoforms by the inclusion or exclusion of sequences into or from the mature mRNA. CD44 provides a unique example; more than 1,000 possible isoforms can be produced by the inclusion or exclusion of a central tandem array of 10 alternatively spliced exons. Certain alternatively spliced exons have been ascribed specific functions; however, independent regulation of the inclusion or skipping of each of these exons would clearly demand an extremely complex regulatory network. Such a network would involve the interaction of many exon-specific trans-acting factors with the pre-mRNA. Therefore, to assess whether the exons are indeed independently regulated, we have examined the alternative exon content of a large number of individual CD44 cDNA isoforms. This analysis shows that the downstream alternatively spliced exons are favored over those lying upstream and that alternative exons are often included in blocks rather than singly. Using a novel in vivo alternative splicing assay, we show that intron length has a major influence upon the alternative splicing of CD44. We propose a kinetic model in which short introns may overcome the poor recognition of alternatively spliced exons. These observations suggest that for CD44, intron length has been exploited in the evolution of the genomic structure to enable tissue-specific patterns of splicing to be maintained.
Alternative pre-mRNA splicing allows a single gene to direct the synthesis of several structurally and functionally distinct protein isoforms. A number of patterns of alternative splicing have been described and include the use of alternative 5′ or 3′ splice sites, mutually exclusive use of exons, failure to remove introns, and the inclusion or skipping of whole exons (16, 25, 33). In some cases, alternative splicing results in dramatic changes in the polypeptide chain either by altering the transcriptional frame or by leading to premature termination. In other cases, the changes are more subtle, leading to the addition or deletion of one or more protein domains by exon skipping.
A particularly noteworthy example is the case of the lymphocyte homing receptor CD44, which is expressed on lymphocytes and at a variety of nonlymphoid sites and is believed to act primarily as an adhesion molecule with an affinity for extracellular matrix components, including hyaluronic acid, collagen, and fibronectin (22, 35, 43). It is perhaps one of the most impressive examples of alternative splicing yet described because the central 10 of a total of 20 exons can be included or skipped to produce over 1,000 potential isoforms (Fig. 1) (31, 32, 41). The most frequently found isoform is the so-called hematopoietic variant (CD44H), in which all of the alternative exons are omitted. Isoforms containing alternative exons have been estimated to amount to 1 to 10% of all CD44 isoforms. Distinct functions for some of these isoforms have been described. Isoforms containing exon V6 promote tumor metastasis (13) and are upregulated upon lymphocyte activation so that they can augment immune responses (1). Alternative splicing may also regulate the affinity of CD44 for hyaluronic acid, its major ligand (2, 4). CD44 isoforms containing exon V3 also promote tumor metastasis (4) and carry modified glycosaminoglycans, which can bind growth factors (5). The CD44 gene is large, spanning more than 50 kb of genomic DNA. This size has made the study of CD44 splicing difficult, although cell fusion experiments have indicated the existence of dominant trans-acting factors promoting exon inclusion (23).
FIG. 1.
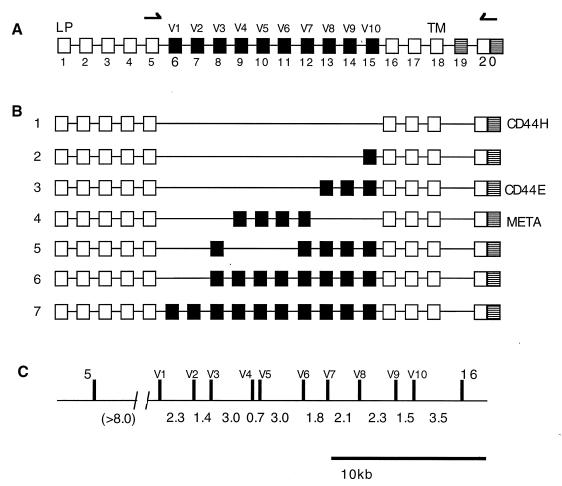
Alternative splicing of CD44. (A) Map of the CD44 gene. Exon 1 encodes the leader peptide (LP), and exon 18 encodes the transmembrane domain (TM). Exons 6 to 15 correspond to the alternatively spliced exons (V1 to V10). Exon 19 encodes a short cytoplasmic domain (32). (B) Examples of some previously observed isoforms of CD44: 1, basic, or hematopoietic, variant CD44H; 2, V10 only; 3, epithelial variant (35); 4, META-1 variant (13). (C) Scale map of the genomic locus encompassing the alternatively spliced exons. Intron sizes are given in kilobases. The exon 5-V1 intron size has not been directly measured but is at least 8 kb.
The development of in vitro and in vivo splicing systems has allowed many of the principal mechanisms governing constitutive and alternative splicing to be elucidated. These mechanisms can be divided into cis-acting sequence elements within the pre-mRNA and trans-acting protein factors. Alternatively spliced exons often have suboptimal splice sites, and mutation of these toward consensus at the 5′ and 3′ sites, polypyrimidine tracts, and branch points increases the utilization of alternatively spliced exons (8, 9, 11). A further sequence element, the exon recognition element (ERE) (24, 34, 37, 40), was recently defined. EREs are frequently purine rich and are thought to interact with the serine-arginine (SR) family of pre-mRNA splicing factors in a sequence-specific fashion. Mutation of a 5′ splice site not only abolishes splicing of the downstream intron but also inhibits removal of the upstream intron, leading to skipping of an internal exon (38, 42). These observations have led to the exon definition model of splicing, in which it is proposed that splicing factors binding to the 5′ splice site recruit other factors to define the exon as a unit for splicing (30).
Many of these analyses have been performed with in vitro splicing reactions and have allowed the minimal intron and exon sizes compatible with splicing to be delineated (7, 44). However, because these systems do not allow the examination of RNAs of more than some hundreds of nucleotides in size, they shed little light onto the functional significance of the very large introns, often kilobases in length, found in many vertebrate genes. In this report, we first analyze splicing patterns of individual CD44 transcripts in normal mouse tissues to determine some basic rules governing the repertoire of CD44 isoforms. Using a novel in vivo alternative splicing assay, we demonstrate a dramatic effect of intron length upon CD44 alternative splicing. As introns are shortened, exon inclusion increases, suggesting a kinetic proximity model for the splicing of alternative exons. Alternatively spliced exons are frequently included in contiguous blocks, a fact which may suggest that all of these exons may not each have a separate function. Instead, they may act as stuffers, extending the N-terminal hyaluronic acid binding domain from the cell surface.
MATERIALS AND METHODS
Plasmid construction.
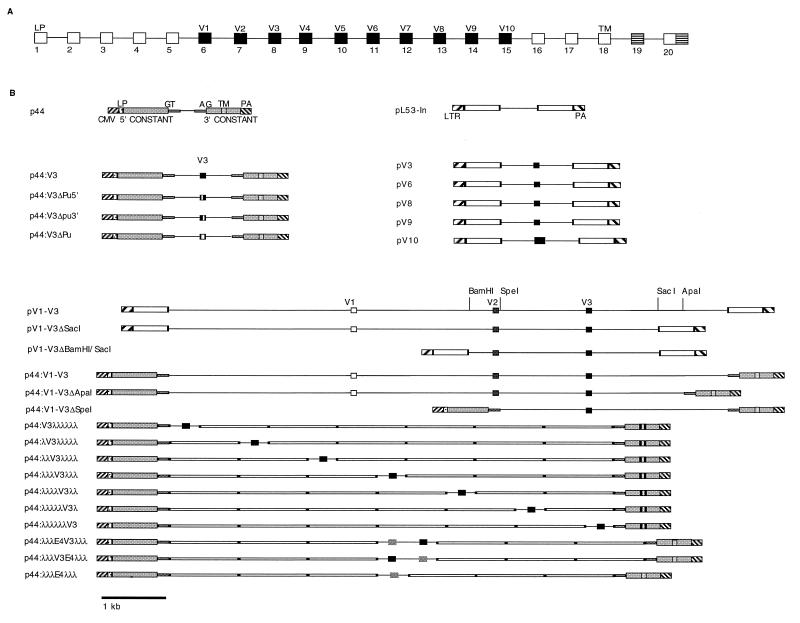
All plasmids are shown in Fig. 2. p3-10cDNA, a CD44 cDNA which contains all the alternatively spliced exons from V3 and V10, was a gift from David Jackson (18). pL53-In was a gift from Michael Reth (3). A 9-kb genomic fragment encompassing CD44 exons V1 to V3 was cloned into pL53-In from a yeast artificial chromosome (YAC) clone encompassing the CD44 locus to form pV1-V3 (32). Sequencing of the introns surrounding the alternatively spliced exons allowed the design of oligonucleotides to amplify cassettes containing single CD44 alternative exons flanked by 200 bp of intron. These products were cloned into pL53-In to give pV3, pV6, pV8, pV9, and pV10 and resequenced to ensure the absence of mutations introduced by amplification.
FIG. 2.
Plasmid constructs. (A) Exon map of the CD44 gene. (B) Diagrammatic representation of the constructs used in this work. pV3, pV6, pV8, pV9, and pV10 were constructed in pL53-In. For p44 and its derivatives, shaded areas represent the 5′ and 3′ constant regions of human CD44, consisting of exons 1 to 5 and exons 16 to 18 or 20, respectively. CMV, cytomegalovirus promoter; LTR, Rous sarcoma virus long terminal repeat promoter; LP, leader peptide; GT, start of intron following exon 5; AG, end of intron prior to exon 15; TM, transmembrane domain; PA, polyadenylation signal.
The CD44 splicing vector p44 was constructed in two halves. A PCR product encompassing exons 1 to 5 derived from cDNA was fused to a genomic PCR product encompassing exon 5 followed by 200 bp of intron. This product was cloned into pCDNA3 (Invitrogen). A similar fusion of genomic exon 16 and its flanking 3′ intron was made with cDNA for exons 16 to 20. This product was cloned into the 3′ end of the polylinker of pCDNA3 to create the full-length CD44 splicing vector. CD44 expression from this construct was verified by immunofluorescence of transiently transfected COS-7 cells with an anti-CD44 monoclonal antibody and by reverse transcription (RT)-PCR. Derivatives of this basic construct were made by insertions of genomic and bacteriophage lambda stuffer fragments into the multiple cloning site. The stuffer fragments were amplified from phage lambda (nucleotides 23448 to 24415) and cloned directionally on either side of V3 to form the series p44:V3λλλλλλ to p44:λλλλλλV3. Constitutive exon 4 with 200 bp of flanking intron was amplified from genomic DNA and introduced upstream or downstream of V3 in the construct p44:λλλV3λλλ to form p44:λλλE4V3λλλ and p44:λλλV3E4λλλ. Modifications of the purine-rich element within V3 were introduced by PCR with modified oligonucleotides. In all of these constructs, the donor and acceptor exons were flanked by 200 bp of their natural introns and the alternatively spliced exon cassettes were likewise flanked by at least 200 bp of their natural introns.
Amplification of murine CD44.
Normal mouse tissue RNAs (kidney, liver, stomach, spleen, small and large intestines, and skeletal muscle) were transcribed separately with avian myeloblastosis virus reverse transcriptase. The cDNAs were amplified separately in a 35-cycle PCR with a pair of primers flanking the alternatively spliced region (FCD44: ACC CCA GAA GGC TAC ATT TTG CAC; RCD44: TTC CGG GTC TCG TCA GCT GTC ATA). Products from this amplification were TA cloned into pBluescript KS−. Replica filters from the transformation plate were hybridized to two probes: first, an oligonucleotide which hybridizes to all CD44 isoforms, and second, a cDNA probe containing all the alternatively spliced exons (V1 to V10). Colonies which contained alternatively spliced exons were picked and placed in grids in 96-well plates. Replicas of these were transferred to nylon membranes and probed with oligonucleotides specific for each alternative exon (31) in order to establish their precise exon composition.
Transfections.
COS-7 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum in 10-cm tissue culture plates. At 50% confluence, the cells were transiently transfected with 10 μg of purified plasmid DNA by use of either Lipofectin (Gibco-BRL) or the DEAE-dextran method. Cells were harvested 48 h after transfection and stained for fluorescence-activated cell sorter (FACS) analyses. RNA from the remaining cells was extracted with RNAzol B (Cinna Biotecx Laboratories).
Antibodies and FACS analysis.
Anti-human CD44–phycoerythrin conjugate, which detects all isoforms of CD44, was purchased from Sigma. Anti-CD44V3 and anti-CD44V6 were gifts from David Jackson. Fluorescein isothiocyanate-conjugated Fc-specific goat anti-mouse immunoglobulin G was purchased from DAKO and preabsorbed with mouse serum before use in double staining. FACS analysis was carried out with a FACSCalibur apparatus driven from the CellQuest software package (Becton-Dickinson).
cDNA synthesis and RT-PCR.
RNA (10 μg) was used in cDNA synthesis with avian myeloblastosis virus reverse transcriptase. Synthesis was primed with oligo(dT) for amplification of preproinsulin or by use of a primer annealing to 3′-untranslated sequences in pCDNA3 (CAG TCG AGG CTG ATC AGC GAG CT) to prevent the priming of endogenous CD44 transcripts. Five percent of the cDNA was used in a 30-cycle PCR amplification with Taq polymerase. Oligonucleotide primer pairs which anneal to the donor and acceptor exons of rat preproinsulin (CCC GGA ATT CTG GAA CCG CGA G and AGC AGA TCT TGG TGC AGC ACT GAT CCA) or CD44 exons 5 and 16, flanking the alternatively spliced region (AGT GAA AGG AGC AGC ACT TCA GG and TCA GAT CCA TGA GTG GTA TGG GAC), were chosen. Reaction products were resolved by electrophoresis on 1% agarose or 6% polyacrylamide gels. DNA was transferred to Hybond N+ (Amersham) and probed with γ-32P-end-labelled oligonucleotides specific for the donor rat preproinsulin exon or CD44 exon 5. Autoradiograms were examined by laser densitometry for quantitation.
RESULTS
CD44 exons are not chosen at random.
CD44 contains 10 alternatively spliced exons arranged in a central tandem array which, by free inclusion or exclusion, could produce 1,024 isoforms. However, sequencing of a number of alternatively spliced isoforms in our laboratory suggested that some isoforms are represented more commonly than others. To confirm these initial observations, we performed a systematic survey of CD44 isoforms expressed in seven normal mouse tissues (kidney, liver, stomach, spleen, small and large intestines, and skeletal muscle). CD44 was amplified from mouse cDNA with primers directed to exons 5 and 20 (Fig. 1), which lie on opposite sides of the alternatively spliced region (Fig. 3A, panel 1). These products were cloned, and the exon composition of about 1 to 2,000 clones from each tissue was determined by oligonucleotide probing with primers for each of the 10 alternatively spliced exons.
FIG. 3.
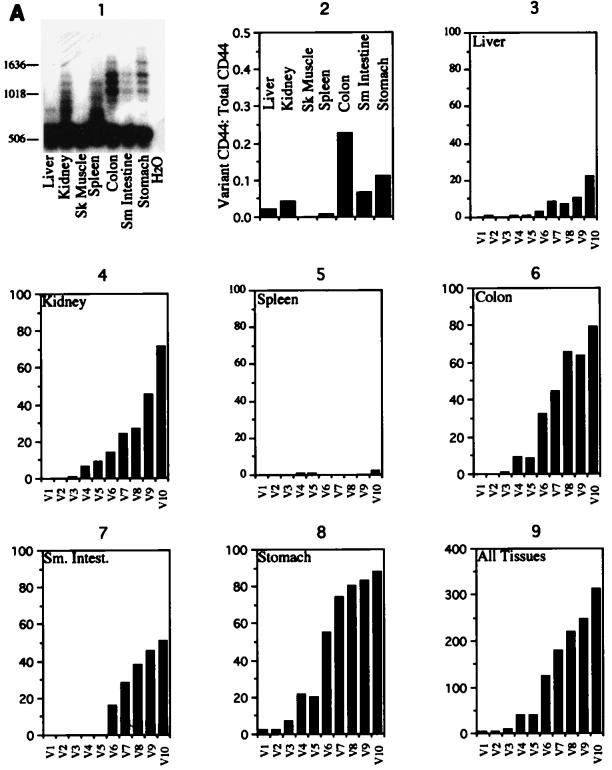
Murine CD44 splicing patterns. (A) CD44 was amplified from normal mouse tissue cDNAs and, following Southern transfer, probed with an oligonucleotide which detects all CD44 isoforms (panel 1). A total of 1,000 to 2,000 CD44-containing clones from each tissue were analyzed. The majority of these clones were CD44H, containing no alternative exons (panel 2). The exon compositions of 328 alternative exon-containing variants were determined (panels 3 to 8) (liver [n = 28], kidney [n = 77], spleen [n = 4], colon [n = 80], small (Sm) intestine (Intest) [n = 51], and stomach [n = 88]). No alternatively spliced isoforms were observed in skeletal (Sk) muscle. (B) Schematic of the alternative exon compositions of alternatively spliced cDNA clones included in this study. The numbers at the left represent alternative exons V1 to V10. Exons scored as present are indicated by black dots.
The cloned isoforms were examined in two ways. First, a general survey of alternative exon inclusion frequencies revealed that CD44H is the predominant variant (Fig. 3A, panel 2). The percentage of variants which contained the alternatively spliced exons varied from 24% in the colon to 0% in skeletal muscle. In addition, in isoforms which contained alternative exons, there was a clear hierarchy of expression, with the most 3′ alternative exons being included more frequently than the 5′ exons (Fig. 3A, panels 3 to 8). Thus, for a combination of all the tissues examined, V10 was observed approximately 100 times more frequently than V1 (Fig. 3A, panel 9). Second, we examined the alternative exon content of each alternatively spliced isoform. These data indicated that alternative exons were not chosen independently of each other. If an alternative exon was selected, it was most likely to be included together with all the alternative exons found downstream. Thus, isoforms such as variant 5 (Fig. 1B), which included an alternatively spliced exon, skipped the next, and then included others downstream, were comparatively rare, accounting for 16, 12, 14, and 18% of the alternatively spliced isoforms from the colon, stomach, kidney, and liver, respectively. Only one isoform (containing V5 and V7) in the kidney included, skipped, included, and then skipped exons again. The exon compositions of the alternatively spliced CD44 isoforms are shown in Fig. 3B.
Although it was possible that this strategy favored the amplification of shorter isoforms, previous analyses comparing PCR and immunoprecipitation showed the PCR to be representative (20, 21). Furthermore, the patterns of splicing observed, i.e., a bias toward the use of the most 3′ alternative exons together with a propensity to include exons in contiguous blocks, would not result from a bias toward preferential amplification of shorter cDNAs.
Minigenes containing large genomic fragments of CD44 are spliced faithfully in vivo.
We chose to study the control of splicing of exon V3, which has been shown to bind growth factors and promote tumor metastasis. V3 is used infrequently in normal tissues but is expressed by many tumor cell lines (15, 19). Genomic fragments of human CD44 were cloned into the exon trap vector pL53-In (3) between donor and acceptor exons from the rat preproinsulin gene (Fig. 2). Splicing was assayed by RT-PCR following transient transfection into COS-7 cells, which show almost exclusively CD44H (no alternatively spliced exons) and no expression of exon V3 in RT-PCR analysis (data not shown).
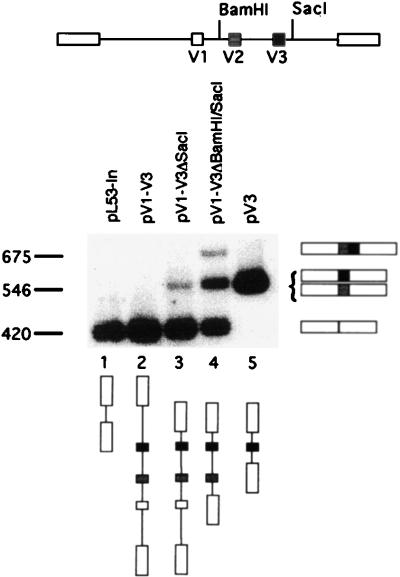
A 9-kb fragment of the human CD44 gene containing exons V1 to V3 was cloned into pL53-In to generate pV1-V3 (this and all other plasmid constructs are drawn to scale in Fig. 2). When transfected into COS-7 cells, this construct mimicked the COS-7 endogenous pattern of CD44 splicing, in which none of the alternatively spliced exons were used (Fig. 4, lane 2). The 5′ and 3′ introns flanking V3 were 1.5 and 4 kb, respectively. This construct was large, so to allow study of the cis-acting sequence elements controlling the splicing of V3, we constructed a minigene, pV3. This construct has a 500-bp fragment containing V3 flanked on each side by 300 bp of intron (100 bp from the preproinsulin vector and 200 bp from CD44 itself). To our surprise, when the introns were reduced to 300 bp, V3 became included constitutively (Fig. 4, lane 5). Smaller deletions made in the 9-kb clone pV1-V3 also led to an increase in V3 inclusion, with some inclusion of V2 (Fig. 4, lanes 3 and 4).
FIG. 4.
Shortening of introns activates the splicing of V3. RT-PCR analysis of splicing from the heterologous reporter pL53-In is shown. Lane 1, no insert; lane 2, 9-kb genomic fragment of CD44 with exons V1 to V3; lanes 3 to 5, shortening of the introns surrounding V3. Fragment sizes are indicated in base pairs. The fragment migrating at 550 bp in lane 3 was shown by Southern analysis to contain either exon V3 or exon V2.
Construction of a CD44 reporter minigene.
The upregulation of V3 inclusion observed when the surrounding introns were shortened could be explained in a variety of ways. First, splicing could be a function of intron length. Second, negative regulatory sequences within the introns could have been deleted. Finally, the result might have been an artifact generated by the chimeric construct in which human CD44 exons were flanked by rat preproinsulin exons.
To test these possibilities, we designed a CD44 reporter in which exons 1 to 5 and 16 to 20 are fused as cDNA (designated p44; Fig. 2). Exons 5 and 16 flank the alternatively spliced region of the CD44 gene (Fig. 1), so these are included with 200 bp of their flanking intron sequences to form the donor and acceptor sites, respectively, in this construct. Genomic fragments from CD44 can be cloned between these exons. This construct produces a pre-mRNA which, when spliced, encodes a full-length CD44 open reading frame, allowing the protein to reach the cell surface. This fact allows splicing to be assayed in vivo by FACS analysis as well as by RT-PCR. An antibody to an epitope encoded by alternatively spliced exon V3 is used in combination with an antibody which detects all isoforms of CD44. Therefore, in a double-staining procedure, the ratio of V3 staining to total CD44 staining reflects the degree of inclusion of V3 in alternatively spliced transcripts.
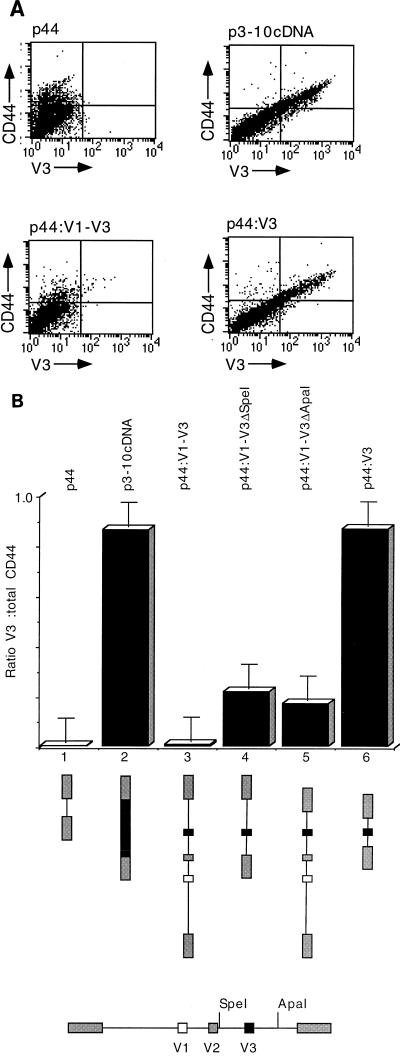
Figure 5A shows total CD44 staining plotted on the y axis and V3 staining plotted on the x axis. Cells positive for both appear in the upper right quadrant, and cells in which CD44 skips V3 appear in the upper left quadrant. Cells which appear in the lower left quadrant are untransfected cells. Construct p44 is spliced to form CD44H and shows only total CD44 staining (upper left quadrant). Conversely, p3-10cDNA, which is a cloned cDNA containing all exons from V3 to V10, shows approximately 100% double staining, as expected (upper right quadrant).
FIG. 5.
In vivo splicing with a CD44 reporter. (A) Scatter plots from FACS analyses of V3 splicing from p44, which has no insert; p3-10cDNA, a CD44 cDNA control containing all exons from V3 to V10; p44:V3, V3 surrounded by short, 400-bp introns; and p44:V1-V3, containing a 9-kbp genome DNA fragment encompassing exons V1 to V3. Total CD44 is shown on the y axis; V3 is shown on the x axis. Cells positive for both are in the upper right quadrant; V3-negative, CD44-positive cells (V3 skipping) are in the upper left quadrant; and cells negative for both (untransfected) are in the lower left quadrant. (B) Histogram illustrating the data from panel A and the results obtained with the 9-kb p44V1-V3 genomic insert and two restriction fragments shortening the introns surrounding V3. Bars represent V3-positive cells expressed as a ratio to total CD44-positive cells.
When the genomic fragment containing exons V1 to V3 was introduced into p44, the results were similar to those obtained with the preproinsulin reporter. p44:V1-V3 is spliced in the same manner as endogenous COS-7 CD44, leading to skipping of V3. Shortening the natural introns surrounding V3 by use of convenient restriction sites in the upstream intron (p44:V1-3ΔSpeI) or downstream intron (p44:V1-3ΔApaI) led to an increase in the inclusion of V3. V3 was included constitutively when the flanking introns were shortened to 400 bp. These data are given in scatter plots in Fig. 5A and are represented graphically in Fig. 5B.
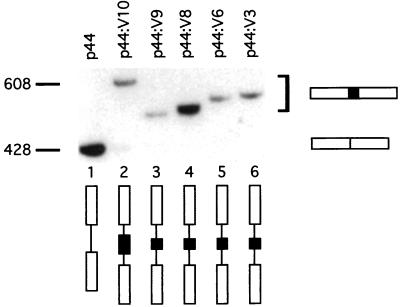
To test whether this result was unique to exon V3 or was applicable to other CD44 alternatively spliced exons, we made constructs with pL53-In and p44 in which the introns flanking exons V6, V8, V9, and V10 were shortened to approximately 400 bp (pV6, pV8, pV9, and pV10, respectively; Fig. 2). The splicing of these constructs was analyzed by RT-PCR (Fig. 6) and by FACS analysis for p44:V6 and p44:V3 by use of antibodies directed to epitopes encoded by these exons (data not shown). In each case, shortening of the introns had the same effect upon inclusion of these exons, which are normally skipped in COS-7 CD44. Splicing of these exons became constitutive, except for pV10, in which a minor fraction of transcripts skipped the exon (Fig. 6, lane 2).
FIG. 6.
Alternative exons flanked by short introns are spliced constitutively. RT-PCR analysis of splicing from several single-exon reporter constructs surrounded by short, 400-bp introns in pL53-In is shown. Fragment sizes are indicated in base pairs.
Splicing of exon V3 is strictly controlled by intron length.
Since shortening of the introns surrounding a range of CD44 alternative exons promoted exon inclusion, we chose to examine whether splicing efficiency was a function of intron length. To this end, we created artificial introns by using a prokaryotic stuffer sequence. A series of constructs were created by cloning multiple copies of a 1-kb section of phage lambda DNA directionally into either 5′ or 3′ introns of construct p44:V3 to make seven constructs, p44:V3λλλλλλ to p44:λλλλλλV3. Each construct contained six repeats of this stuffer fragment surrounding exon V3 and flanked by 200 bp of the natural CD44 intron sequence on each side (Fig. 2).
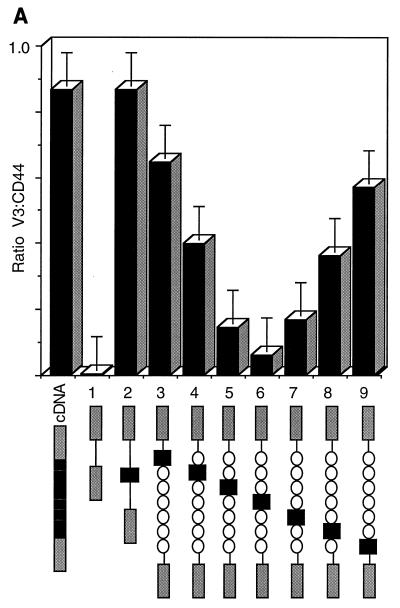
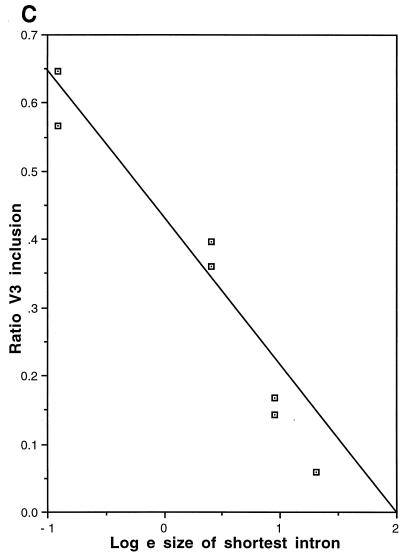
These constructs were transfected into COS-7 cells, and the results were analyzed by FACS analysis and RT-PCR (Fig. 7A and B). When V3 lay at either end of the stuffer DNA (Fig. 7A and B, p44:V3λλλλλλ [lane 4] or p44:λλλλλλV3 [lane 10]), the shortest intron was 400 bp and the longest was 6.4 kbp. In this position, such as in lane 2, where both introns were short (p44:V3), V3 was included almost constitutively. There appeared to be no polarity to the effect, as it was seen equally in constructs with either a short 5′ intron or a short 3′ intron. As V3 was moved toward the center of the stuffer fragment, its inclusion was reduced in a titratable fashion. When V3 was surrounded by 3.5 kbp of introns (p44:λλλV3λλλ; lane 7), it became almost completely lost, mimicking the splicing of the endogenous COS-7 CD44 gene. The reduction in CD44 splicing appeared to be exponential, and a plot of percentage of V3 inclusion versus the natural logarithm of the length of the shortest intron demonstrated this relationship (Fig. 7C). The results obtained from the FACS analysis mirrored those obtained by RT-PCR; laser densitometry provided confirmation that RT-PCR was quantitative in its representation of CD44 splicing.
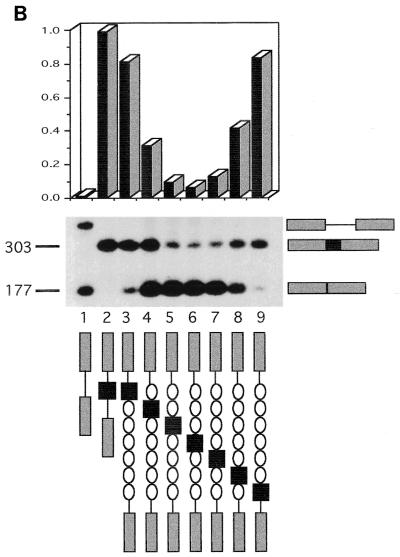
FIG. 7.
Effect of intron size on alternative splicing of V3. (A) FACS analysis of splicing is shown. Black box, V3; gray boxes, donor and acceptor sequences; open circles, 1-kb lambda DNA repeats. Column designated cDNA, control cDNA (V3 to V10); column 1, empty vector; column 2, p44:V3 (V3 with short, 400-bp introns); columns 3 to 9, V3 moved sequentially across a 6-kbp lambda stuffer segment. (B) RT-PCR analysis of the same transfectants is shown. The ratio of V3 to total CD44, as measured by laser densitometry, is illustrated graphically. Fragment sizes are indicated in base pairs. (C) V3 inclusion is exponentially related to intron length.
Constitutive exons can overcome the restraint of long introns.
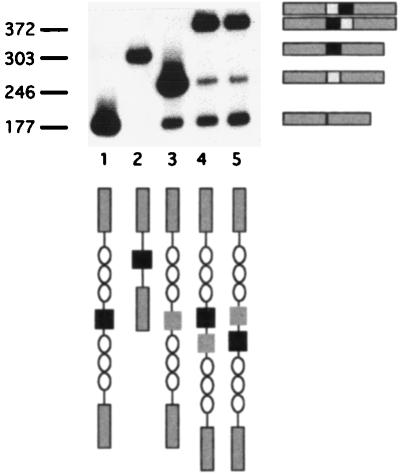
Many vertebrate genes which have introns many tens of kilobases in length have been described, implying that the splicing of constitutive exons in these genes cannot be limited by flanking intron length. We cloned exon 4, a constitutive exon from CD44, into the center of the lambda stuffer fragment to form p44:λλλE4λλλ. This construct contains exon 4 surrounded by 200 bp of its own introns and 3 kbp of lambda stuffer DNA on each side. Exon 4 was chosen because it does not interrupt the translational frame of CD44, so we could avoid potential interference from nonsense codon-mediated decay (26) and exon skipping mediated by nonsense codon-containing exons (17).
Because no antibody which is specific for epitopes encoded by exon 4 exists, the results were examined only by RT-PCR. As expected, constitutive exon 4 was spliced faithfully in this construct and was included in 95% of transcripts, while in construct p44:λλλV3λλλ, exon V3 was skipped (Fig. 8, lanes 3 and 1, respectively). To confirm the requirement of proximity to a constitutive exon, we introduced V3 either 400 bp upstream (p44:λλλE4V3λλλ) or 400 bp downstream (p44:λλλV3E4λλλ) of constitutive exon 4. In both of these constructs, V3 and exon 4 lay in the middle of the 6-kb lambda stuffer fragment, yet V3 splicing was efficiently rescued by its proximity to the constitutive exon.
FIG. 8.
V3 splicing is rescued by proximity to a “strong” exon. Exon 4 of CD44 (light gray boxes) was cloned on either side of V3 (dark gray boxes) in the p44:λλλV3λλλ construct, and data were obtained by RT-PCR. Fragment sizes are indicated in base pairs.
V3 contains a purine-rich element which is essential for splicing.
Examination of the sequences of many of the alternatively spliced CD44 exons reveals purine-rich stretches consistent with exon recognition elements. V3 contains the sequence TGAAGAAAATGAAGATGAAAGAGACAGA, encoding a stretch of acidic amino acids. This sequence was mutated to a pyrimidine-rich stretch by replacement with serine codons TCT and TCC. We made three constructs in which the 5′ half (p44:V3ΔPu5′; TCTTCCTCTTCCTCTGATGAAAGAGACAGA), the 3′ half (p44:V3ΔPu3′; AATGAAGAAAATGAATCTTCCTCTTCCTCT), or both halves (p44:V3ΔPu; TCTTCCTCTTCCTCTTCTTCCTCTTCCTCT) of the purine-rich stretch were mutated; boldfacing indicates the mutated sequence. These mutations were made in p44:V3, which has two short introns flanking V3 and constitutively includes V3 (see above).
Both the 5′ and the 3′ mutations reduced exon inclusion from approximately 100 to 70%, and the double mutation completely abolished V3 inclusion (Fig. 9). SR proteins are known to bind purine-rich EREs and have been shown to enhance the splicing of some alternatively spliced exons both in vitro and by cotransfection in vivo (6, 27). Despite the clear dependence of V3 splicing on the integrity of a purine-rich splicing enhancer, we were unable to enhance V3 inclusion in the construct p44:λλλV3λλλ by cotransfection with known SR proteins SRp20, SF2, SC35, SRp30c, SRp40, and SRp55 (data not shown).
FIG. 9.
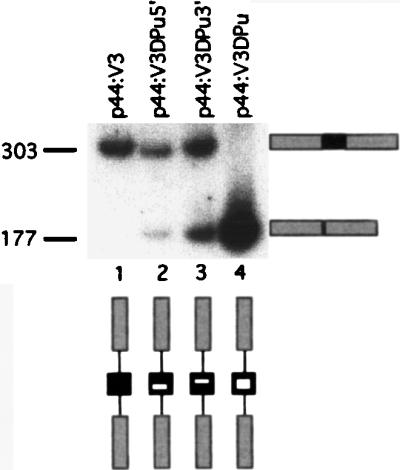
Mutation of a putative ERE affects V3 splicing. RT-PCR analysis of splicing from constructs in which the purine-rich stretch of V3 was replaced by pyrimidines is shown. Lane 1, unmutated pV3; lanes 2 and 3, mutation of 5′ and 3′ halves, respectively; lane 4, double mutant. Fragment sizes are indicated in base pairs.
DISCUSSION
Influence of intron length upon alternative splicing.
A number of fundamental rules governing splicing have been elucidated by use of in vitro and in vivo splicing systems. Because of difficulties in cloning, mutating, and transcribing large tracts of DNA, most of these analyses have concentrated upon the definition of minimal intron and exon sizes and the effects of mutations in and around exons (7, 44). There has been no systematic analysis of the function of the large introns, often many kilobases, found in many vertebrate genes. Some hints of an effect of intron length on alternative splicing have come from shortening of the intron preceding the transmembrane domain in the immunoglobulin genes. Although this analysis was complicated by competition between the processes of polyadenylation and splicing, it does seem that shorter introns promote splicing to generate isoforms including a transmembrane domain (29).
Mutational inactivation of the 5′ splice site represses splicing not only to the mutated site but also of the upstream intron. Thus, the splicing machinery often fails to remove an apparently normal intron upstream of the mutated 5′ splice site (38, 42). This observation led to the exon definition model of splicing, which proposes that splicing factors assembled on both 5′ and 3′ splice sites communicate with each other to define an exon for splicing (30). This model holds true for most vertebrate genes, in which, as a general rule, exons are relatively small (<300 bp) and introns are large (14). However, in invertebrates, such as Drosophila melanogaster, the situation is reversed, so that in many cases, large exons are flanked by small introns (14, 28). In this situation, a rule of intron definition seems to hold, in which the unit of recognition for splicing is the intron (39). Thus, the splicing machinery appears to define units for splicing based upon the shortest distance between two splice sites. There does appear to be a reciprocal relationship between intron and exon sizes, such that shortened introns can compensate for a reduction in splicing efficiency caused by larger exons (36).
Our results demonstrate a relationship between intron length and splicing of a variety of CD44 alternatively spliced exons. This effect was examined closely for exon V3, which is skipped in isoforms present in most normal tissues but is included in a number of tumors. We showed that the splicing of V3 is reduced as intron length is increased. One interpretation of these results is that the effects can be explained by intron, rather than exon, definition, similar to that observed in invertebrates. However, this is probably not the case, since V3 is not a long exon (126 bp) and the shortest intron used in these experiments was 500 bp, too large for intron definition (39) and within the range of average sizes for mammalian genes in which exon definition is believed to operate.
Based upon our observations, we propose a kinetic probability model for splicing in CD44, which may have applicability to other vertebrate genes. Thus, constitutive exons which have strong splice sites are well defined. Contact between two such exons results in a high probability of productive spliceosome formation. Conversely, alternatively spliced exons are normally poorly defined and, upon contact with well-defined constitutive exons, have a low probability of productive spliceosome formation. The definition of such exons may be promoted by the presence of factors, such as SR proteins either singly or in combination, which may bind in a sequence-specific manner to EREs (24, 34, 37, 40) or other, as-yet-undefined enhancer elements. It has been shown that the binding of such factors improves exon recognition, due in part to the enhancement of the binding of U1-snRNP to the alternative 5′ splice site (10). We propose that intron length determines the frequency of contact between two exons such that long introns result in infrequent contact and short introns result in a proportionately higher frequency of contact. In this way, a short intron may overcome the disadvantage of a poorly defined alternative exon by giving rise to an increase in the likelihood of contact between it and its neighbor, increasing the probability of spliceosome formation. This model predicts that the probability of contact and hence splicing will decrease exponentially with intron length. This prediction is supported by our observation on the reduction in V3 splicing as introns are artificially enlarged. Furthermore, since mRNA secondary structures have been shown to affect the choice of alternative 5′ splice sites (12), another effect of intron enlargement is the additional potential for secondary and tertiary structures in the pre-mRNA which could obscure the already poorly defined exon.
CD44 isoforms are not generated at random, and this property may be influenced by intron length.
Our analysis of CD44 alternative splicing in the normal mouse led to three observations. First, and as had been previously thought, the most frequently found isoform (90 to 95%) is CD44H, which skips all the alternatively spliced exons. Second, in other isoforms there is a clear hierarchy of expression of the alternatively spliced exons, so that V10 is found most frequently and V1 is found least frequently. Third, detailed examination of these isoforms shows that once a 5′ alternatively spliced exon is chosen, it frequently is followed by all the alternatively spliced exons lying downstream. Eighty percent of the isoforms containing alternatively spliced exons are of this type, and isoforms containing a single exon are rare. These rules are not hard and fast, and several isoforms which contain single, centrally placed exons, such as V6 in isolation, have been reported, particularly from lymphoid tissues (1). However, overall the isoforms reported by others are in keeping with our findings.
The genomic structure of CD44 is highly conserved between humans and mice. In particular, the lengths of the introns in the alternatively spliced region are almost identical (32, 41). All the introns in this region are 1.5 to 3.5 kb, with two exceptions, the intron between V4 and V5, which is approximately 0.5 kb, and the intron preceding the first alternatively spliced exon, V1. This intron is at least 8 kb in length (Fig. 1A), and because we have so far been unable to span this intron with cloned genomic DNA, this figure may represent a conservative estimate.
The intron length model predicts that the splicing of alternative exons is enhanced by proximity to a constitutive exon. Exon V10 is the alternatively spliced exon located most proximal to a constitutive exon (3.5 kb) and would therefore be predicted to be found most frequently. As mentioned above, the most 5′ of the alternatively spliced exons, V1, is preceded by a very long intron, a fact which would be predicted to reduce its splicing efficiency. Apart from the intron preceding V1, all the other introns are in the range of 0.5 to 3.5 kb. Thus, once V10 is selected, its neighbor, V9, becomes the most proximal exon and is favored above the others. Similarly, V8 is enhanced by the selection of V9, and so on. At any point, however, the splicing can jump to the next constitutive exon, exon 5, terminating alternative exon inclusion. This model explains the finding that the frequency of exon expression falls in a hierarchy from V10 to V1. It also explains why 80% of isoforms contain a contiguous string of exons downstream of the first selected exon. If a contiguous string of exons is included, then the effective intron sizes are kept to a minimum, whereas if one or more exons in the string are skipped, then a much larger intron, formed by the sum of intron and exon lengths to be excluded, has to be overcome.
Further evidence of the strength of this model comes from an examination of isoforms containing exons V4 and V5. These two exons are separated by a short, 0.5-kb intron. Because of their close proximity, the intron length model would predict that if V5 is used, then V4 would be likely to follow. Indeed, V4 is included in 86% of transcripts containing V5. V3 is separated from V4 by a 3-kb intron, and only 14% of isoforms containing V4 also contain V3, again consistent with the intron length model.
In this paper, we have examined the splicing patterns of the CD44 gene and have demonstrated a clear hierarchy and connectivity in the patterns of alternative splicing. We have also demonstrated the influence of intron length upon alternative splicing, with short introns favoring exon inclusion. From these observations, we propose a kinetic model for the alternative splicing of CD44 in which short introns promote more frequent collisions between a poorly defined alternatively spliced exon and a neighboring constitutive exon, thus enhancing splicing efficiency.
Alternative splicing has been demonstrated in many genes, and in many instances, such as CD44, has been shown to have functional sequelae for the individual cell and organism. Phylogenetic comparisons show many of the spliceosomal components to be highly conserved among species as diverse as humans, mice, yeast, Drosophila, and Caenorhabditis elegans. In contrast, many alternatively spliced mammalian genes, such as the cell surface receptors CD44 and CD45, are found only in higher vertebrates and not in invertebrates. This fact implies that the alternative splicing of these genes has evolved within a predetermined background of splicing factors. In other words, genes may well have evolved to suit the available splicing factors rather than that the splicing factors have evolved to meet the splicing needs of the genes. To develop alternative splicing in a gene, evolution may mutate sequences in and around the exons to either enhance or reduce exon definition and hence splicing efficiency. This process is necessarily constrained by the requirement to conserve the primary coding potential. A complementary and perhaps more powerful mechanism for this objective is the modulation of intron length. Subtle changes in intron length can result from unequal meiotic crossover, and such events do not disrupt the primary sequence of the gene. A number of alternatively spliced genes, such as CD44 and CD45, have alternative exons surrounded by introns of 1 to 4 kb, which may be the optimum distance over which this effect operates. If these observations hold true for other large, alternatively spliced genes, then intron length could allow rapid fine-tuning of alternative splicing to comply with phylogenetically ancient patterns of splicing factor expression. Put another way, we propose that intron length is modulated so as to place an alternative exon within a window so that its expression may be modulated by tissue-specific patterns of trans-acting protein factors.
ACKNOWLEDGMENTS
We thank David Jackson for the gift of monoclonal antibodies and the CD44 p3-10cDNA construct, Magda Peblanski for extensive assistance with FACS analyses, and Jon Frampton for critical reading of the manuscript.
This work was supported by the Wellcome Trust, the Centre Nationale de la Recherche Scientifique (CNRS), and the Arthritis and Rheumatism Council. M.V.B. was the recipient of a travelling fellowship under the E. U. Human Capital and Mobility Program.
Martyn V. Bell and Alison E. Cowper contributed equally to this work.
REFERENCES
- 1.Arch R, Wirth K, Hofmann M, Ponta H, Matzku S, Herrlich P, Zoller M. Participation in normal immune responses of a metastasis-inducing splice variant of CD44. Science. 1992;257:682–685. doi: 10.1126/science.1496383. [DOI] [PubMed] [Google Scholar]
- 2.Aruffo A, Stamenkovic I, Melnick M, Underhill C B, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990;61:1303–1313. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 3.Auch D, Reth M. Exon trap cloning: using PCR to rapidly detect and clone exons from genomic DNA fragments. Nucleic Acids Res. 1990;18:6743–6744. doi: 10.1093/nar/18.22.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolazzi A, Jackson D, Bennett K, Aruffo A, Dickinson R, Shields J, Whittle N, Stamenkovic I. Regulation of growth and dissemination of a human lymphoma by CD44 splice variants. J Cell Sci. 1995;108:1723–1733. doi: 10.1242/jcs.108.4.1723. [DOI] [PubMed] [Google Scholar]
- 5.Bennett K L, Jackson D G, Simon J C, Tanczos E, Peach R, Modrell B, Stamenkovic I, Plowman G, Aruffo A. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J Cell Biol. 1995;128:687–698. doi: 10.1083/jcb.128.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caceres J F, Stamm S, Helfman D M, Krainer A R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 7.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominski Z, Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992;12:2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski Z, Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992;12:2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASf. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eperon L P, Estibeiro J P, Eperon I C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986;324:280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- 12.Eperon L P, Graham I R, Griffiths A D, Eperon I C. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 13.Gunthert U, Hofmann M, Rudy W, Reber S, Zoller M, Haussmann I, Matzku S, Wenzel A, Ponta H, Herrlich P. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991;65:13–24. doi: 10.1016/0092-8674(91)90403-l. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins J D. A survey on intron and exon lengths. Nucleic Acids Res. 1988;16:9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann M, Rudy W, Zoller M, Tolg C, Ponta H, Herrlich P, Gunthert U. CD44 splice variants confer metastatic behavior in rats: homologous sequences are expressed in human tumor cell lines. Cancer Res. 1991;51:5292–5297. [PubMed] [Google Scholar]
- 16.Horowitz D S, Krainer A R. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 17.Hull J, Shackleton S, Harris A. The stop mutation R553X in the CFTR gene results in exon skipping. Genomics. 1994;19:362–364. doi: 10.1006/geno.1994.1070. [DOI] [PubMed] [Google Scholar]
- 18.Jackson D G, Bell J I, Dickinson R, Timans J, Shields J, Whittle N. Proteoglycan forms of the lymphocyte homing receptor CD44 are alternatively spliced variants containing the v3 exon. J Cell Biol. 1995;128:673–685. doi: 10.1083/jcb.128.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson D G, Buckley J, Bell J I. Multiple variants of the human lymphocyte homing receptor CD44 generated by insertions at a single site in the extracellular domain. J Biol Chem. 1992;267:4732–4739. [PubMed] [Google Scholar]
- 20.Jackson D G, Schenker T, Waibel R, Bell J I, Stahel R A. Expression of alternatively spliced forms of the CD44 extracellular-matrix receptor on human lung carcinomas. Int J Cancer Suppl. 1994;8:110–115. doi: 10.1002/ijc.2910570724. [DOI] [PubMed] [Google Scholar]
- 21.Jackson D G, Screaton G R, Bell M V, Dickinson R, Timans J, Shields J, Whittle N, Collins I, Fawcett J, Simmons D, Bell J I. 5th International Human Leucocyte Differentiation Antigen Workshop Handbook. Oxford, United Kingdom: Oxford University Press; 1994. Structure of the CD44 gene and its expression during lymphocyte activation; pp. 1727–1729. [Google Scholar]
- 22.Jalkanen S, Jalkanen M. Lymphocyte CD44 binds the COOH-terminal heparin-binding domain of fibronectin. J Cell Biol. 1992;116:817–825. doi: 10.1083/jcb.116.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konig H, Moll J, Ponta H, Herrlich P. Trans-acting factors regulate the expression of CD44 splice variants. EMBO J. 1996;15:4030–4039. [PMC free article] [PubMed] [Google Scholar]
- 24.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T. Mechanisms of alternative splicing. Science. 1991;251:33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- 26.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 27.Mayeda A, Helfman D M, Krainer A R. Modulation of exon skipping and inclusion by heterogeneous nuclear ribonucleoprotein A1 and pre-mRNA splicing factor SF2/ASF. Mol Cell Biol. 1993;13:2993–3001. doi: 10.1128/mcb.13.5.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mount S M, Burks C, Hertz G, Stormo G D, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson M L, Perry R P. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci USA. 1986;83:8883–8887. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Screaton G R, Bell M V, Bell J I, Jackson D G. The identification of a new alternative exon with highly restricted tissue expression in transcripts encoding the mouse Pgp-1 (CD44) homing receptor. Comparison of all 10 variable exons between mouse, human, and rat. J Biol Chem. 1993;268:12235–12238. [PubMed] [Google Scholar]
- 32.Screaton G R, Bell M V, Jackson D G, Cornelis F B, Gerth U, Bell J I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci USA. 1992;89:12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith C W, Patton J G, Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- 34.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stamenkovic I, Aruffo A, Amiot M, Seed B. The hematopoietic and epithelial forms of CD44 are distinct polypeptides with different adhesion potentials for hyaluronate-bearing cells. EMBO J. 1991;10:343–348. doi: 10.1002/j.1460-2075.1991.tb07955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterner D A, Carlo T, Berget S M. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 38.Talerico M, Berget S M. Effect of 5′ splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990;10:6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talerico M, Berget S M. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian M, Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 41.Tolg C, Hofmann M, Herrlich P, Ponta H. Splicing choice from ten variant exons establishes CD44 variability. Nucleic Acids Res. 1993;21:1225–1229. doi: 10.1093/nar/21.5.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treisman R, Proudfoot N J, Shander M, Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982;29:903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- 43.Wayner E A, Carter W G, Piotrowicz R S, Kunicki T J. The function of multiple extracellular matrix receptors in mediating cell adhesion to extracellular matrix: preparation of monoclonal antibodies to the fibronectin receptor that specifically inhibit cell adhesion to fibronectin and react with platelet glycoproteins Ic-IIa. J Cell Biol. 1988;107:1881–1891. doi: 10.1083/jcb.107.5.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wieringa B, Hofer E, Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984;37:915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]