Summary
Dysregulation of normal transcription factor activity is a common driver of disease. Therefore, the detection of aberrant transcription factor activity is important to understand disease pathogenesis. We have developed Priori, a method to predict transcription factor activity from RNA sequencing data. Priori has two key advantages over existing methods. First, Priori utilizes literature-supported regulatory information to identify transcription factor-target gene relationships. It then applies linear models to determine the impact of transcription factor regulation on the expression of its target genes. Second, results from a third-party benchmarking pipeline reveals that Priori detects aberrant activity from 124 single-gene perturbation experiments with higher sensitivity and specificity than 11 other methods. We applied Priori and other top-performing methods to predict transcription factor activity from two large primary patient datasets. Our work demonstrates that Priori uniquely discovered significant determinants of survival in breast cancer and identified mediators of drug response in leukemia.
Subject areas: Gene network, Molecular mechanism of gene regulation, Biocomputational method, Biological constraints
Graphical abstract

Highlights
-
•
Priori predicts transcription factor activity using prior regulatory information
-
•
Priori detects perturbed transcription factors better than 11 other methods
-
•
Priori uniquely identified FOXA1 activity as a determinant of survival in BIDC
-
•
Priori nominated FOXO1 activity as a mediator of venetoclax sensitivity in AML
Gene network; Molecular mechanism of gene regulation; Biocomputational method; Biological constraints
Introduction
The coordinated expression and activity of transcription factors are fundamental mechanisms in establishing and maintaining cell identity and function. Transcription factors bind to cis-regulatory DNA sequences, including promoters and enhancers, and modulate gene transcription.1,2 Dysregulation of these normal transcription factor functions frequently contributes to the development of a pathogenic cell phenotype.3,4 Abnormal transcription factor activity can result from mutations in the putative cis-regulatory DNA binding sequences or in the transcription factors themselves. Recent studies have highlighted the importance of aberrant expression of pathogenetic transcription factors as drivers of disease.4 For example, MYC, which is important for cellular growth and proliferation, is the most frequently amplified oncogene. Elevated levels of MYC has been shown to promote tumorigenesis in a variety of tissue types.4 In tumor cells expressing high levels of MYC, the transcription factor accumulates in cis-regulatory regions of genes associated with cellular proliferation and growth, resulting in transcriptional amplification of MYC’s gene regulatory network and, subsequently, abnormal cellular proliferation.5,6 Therefore, detection of abnormal transcription factor activity is valuable for better understanding the mechanisms that underly disease pathogenesis.
Gene expression profiling, including RNA sequencing (RNA-seq), is commonly used to monitor dynamic changes in transcription factors and their gene regulatory networks. Initial studies to infer transcription factor activity only used transcription factor gene expression as a proxy for activity.7,8,9 However, this approach has several shortcomings. Gene expression is only an indirect measurement of protein activity due to the complex mechanisms controlling protein synthesis and degradation.10,11,12 Additionally, feedback loops may alter the expression of transcription factors in response to their regulatory activity.4,13,14,15 Reliable predictions of transcription factor activity, therefore, cannot be limited to evaluating transcription factor expression alone.
An alternative approach to inferring transcription factor activity is to assess the expression of downstream target genes.7,8,9 This approach has two major benefits. First, evaluation of hundreds or thousands of downstream targets instead of a single transcription factor likely improves the prediction robustness. While some of these targets may be context-specific, analyzing them in aggregate likely improves the prediction generalizability across many contexts. Second, as target gene expression is downstream of transcription factor control, these signatures are expected to reflect the actual transcriptional impact more accurately. Therefore, accounting for the downstream impact of transcription factors on its gene regulatory networks is likely important for activity inference.
Multiple methods have been developed to quantify transcription factor activity from gene expression data. These approaches can be grouped based on how they select gene expression features. Methods like Univariate Linear Model (ULM) and Multivariate Linear Model (MLM) use every gene in a dataset, nominating transcription factors as a covariate that best estimates the expression of all other genes.16 However, these methods develop activity signatures using genes that may not have a true biological relationship to the transcription factor of interest. Gene set approaches like Over-Representation Analysis (ORA), Fast Gene Set Enrichment (FGSEA), Gene Set Variation Analysis (GVSA), and AUCell infer activity using sets of published transcription factor target genes or target genes curated by experts.17,18,19,20 While gene set methodologies are simple and popular, they are susceptible to the quality and comparability of gene set signatures.21 Network inference approaches, including Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe), infer gene regulatory networks based on the covariance of transcription factors and its putative targets.22,23 This process is not completely unsupervised, however, as ARACNe requires a user-defined list of transcription factors in order to infer gene regulatory networks.22,23 The same group that developed ARACNe also created Virtual Inference of Protein-activity by Enriched Regulon analysis (VIPER) to infer transcription factor activity from ARACNe gene expression signatures.24 The challenge with these approaches is deconvoluting combinatorial regulation, where the expression of a target gene is controlled by multiple transcription factors. While some of these methods, including VIPER, have an option to correct for this, it remains difficult to infer transcriptional networks as there are many possible solutions that can explain the underlying data.7,25 While these methods deploy various techniques to generate activity scores using the expression of downstream target genes, most do not select their target gene features from literature-supported transcriptional relationships.
Recent studies have highlighted that grounding predictions using transcription factor activity methods remains challenging.25,26,27,28,29 A rigorous evaluation of widely used transcription factor algorithms demonstrated that most methods do not robustly detect perturbed transcription factors.20 Despite this, there are precision medicine clinical studies that use inferred transcription activity from bulk RNA-seq as a marker to guide clinical decisions. While there is an increasing number of single cell and spatial -omic modalities available to clinical researchers, these studies as well as many larger cohorts and clinical trials most commonly use markers identified from bulk sequencing of RNA or DNA. Therefore, it is critical to develop methods that can robustly detect aberrant transcription factor activity from primary patient bulk RNA-seq data.
Here, we propose an approach that uses prior, peer-reviewed biological information to infer transcription factor activity called Priori. Our method has two major advantages over the existing methods. First, Priori identifies transcription factor target genes using carefully extracted transcriptional regulatory networks from Pathway Commons.30,31 This resource continually collects information on biological pathways including molecular interactions, signaling pathways, regulatory networks, and DNA binding. Pathway Commons currently contains data from 22 high-quality databases with over 5,700 detailed pathways and 2.4 million interactions. Using the transcriptional relationships from Pathway Commons, Priori fits linear models to the expression of transcription factors and their target genes. These models allow Priori to understand the impact and direction of transcription factor regulation on its known target genes. Second, comparison with a third-party benchmarking workflow reveals that Priori detects aberrant transcription factor activity from 124 gene perturbation experiments with higher sensitivity and specificity than 11 other methods. We applied Priori and three other methods nominated from the benchmarking workflow to generate activity scores from two large primary patient datasets, TCGA-BRCA and Beat AML. We demonstrate that Priori can be deployed to discover significant predictors of survival in breast cancer as well as identify mediators of drug response in leukemia from primary patient samples that were not robustly detected using the other methods.
Results
Priori uses prior biological information to infer transcription factor activity
For each transcription factor in an RNA-seq dataset, Priori generates an activity score (Figure 1A). The activity score is a weighted, aggregate statistic that reflects the impact and direction of transcription factor regulation on its target genes. Priori first identifies the known target genes for each transcription factor in an RNA-seq dataset from Pathway Commons (or another network provided by the user).30,31 Priori then assigns weights to each target gene by correlating the target gene expression to its transcription factor (Equation 5). To identify the targets that are most impacted by transcription factor regulation, Priori separates the up- and down-regulated genes by their transcription factor-target gene weights and ranks their expression (Equation 6). These ranks are subsequently scaled by multiplying it with the transcription factor-target gene weights. The activity score for each transcription factor is then calculated by summing the weighted ranks (Equation 7). With this single-component model, researchers can use Priori to predict transcription factor activity from RNA-seq data.
Figure 1.
Overview of the Priori methodology and benchmarking workflow
(A) Priori generates an activity score for each transcription factor in an RNA-seq dataset. Priori first extracts the downstream target genes for each transcription factor from Pathway Commons. Priori then calculates weights for each target gene by correlating the expression of each transcription factor to its target genes. Priori then ranks the absolute expression of all genes in the dataset and scales these ranks by the transcription factor-target gene weights. The summation of the weighted target gene ranks is the transcription factor activity score.
(B) Schematic overview of the benchmarking workflow. We generated transcription factor activity scores for each method using normalized RNA-seq counts following single-gene knockdown or over-expression. Priori, along with other methods that use prior information, generated activity scores with transcriptional relationships from Pathway Commons. AUROC and AUPRC values were calculated for each down-sampling permutation. 100 down-sampling permutations were performed to compare an equal number of perturbed and unperturbed genes.
In order to compare how well Priori detects aberrant transcription factor activity to other methods, we used a third-party benchmarking workflow called decoupleR (Figure 1B).20 The decoupleR workflow facilitated an unbiased, common evaluation scheme to determine how often each method correctly identified transcription factors that have been knocked-down or over-expressed from RNA-seq data, resembling the pathologic disruption of normal transcription factor regulation.32 Using this workflow, we generated transcription factor activity scores for 16 methods, including Priori, using the default parameters (Table 1). Several of these methods were developed by the authors of the decoupleR workflow, including the standard, normalized (Norm), and corrected (Corr) versions of Weighted Mean (WMEAN), Weighted Sum (WSUM), Univariate Decision Tree (UDT), and Multivariate Decision Trees (MDT).20 These authors also developed a normalized version of FGSEA. For methods that use prior information, we generated activity scores using the Pathway Commons transcriptional relationships. We ranked the transcription factor activity scores for each experiment and compared the ranks of perturbed and unperturbed transcription factors. We evaluated how often the perturbed transcription factor activity score was among the top activity scores in each experiment. The authors of the decoupleR pipeline defined this threshold by the number of perturbed transcription factors in the dataset. Since the number of unperturbed transcription factors vastly outnumbered the perturbed transcription factors, we implemented a down-sampling strategy. For each down-sampling permutation, we calculated area under the precision recall curve (AUPRC) and receiver operating characteristic (AUROC) metrics. The results of this third-party benchmarking workflow allowed us to objectively compare the sensitivity and specificity of Priori to detect perturbed transcription factor activity to 11 other methods.
Table 1.
Transcription factor activity methods evaluated using the decoupleR benchmarking workflow
| Feature Selection | Method | Acronym | Normalized Method | Corrected Method | Reference |
|---|---|---|---|---|---|
| All features | Univariate Linear Model | ULM | – | – | Teschendorff and Wang npj genomic medicine (2020)16 |
| All features | Multivariate Linear Model | MLM | – | – | Teschendorff and Wang npj genomic medicine (2020)16 |
| Gene set | Over Representation Analysis | ORA | – | – | Badia-i-Mompel et al. Bioinformatic Advances (2022)20 |
| Gene set | Fast Gene Set Enrichment | FGSEA | Norm FGSEA | – | Korotkevich et al. bioRxiv (2021)17 |
| Gene set | Gene Set Variation Analysis | GVSA | – | – | Hänzelmann et al. BMC Bioinformatics (2013)18 |
| Gene set | AUCell | – | – | – | Aibar et al. Nature Methods (2017)19 |
| Inferred networks | Virtual Inference of Protein-activity by Enriched Regulon analysis | VIPER | – | – | Alvarez et al. Nature Genetics (2016)24 |
| Curated gene networks | Weighted Mean | WMEAN | Norm WMEAN | Corr WMEAN | Badia-i-Mompel et al. Bioinformatic Advances (2022)20 |
| Curated gene networks | Weighted Sum | WSUM | Norm WSUM | Corr WSUM | Badia-i-Mompel et al. Bioinformatic Advances (2022)20 |
| Curated gene networks | Univariate Decision Tree | UDT | – | – | Badia-i-Mompel et al. Bioinformatic Advances (2022)20 |
| Curated gene networks | Multivariate Decision Trees | MDT | – | – | Badia-i-Mompel et al. Bioinformatic Advances (2022)20 |
ID: Norm = normalized; Corr = corrected.
Priori identifies perturbed transcription factors with greater sensitivity and specificity than other methods
The decoupleR workflow evaluates transcription factor activity methods using a curated dataset from Holland et al.32 This dataset includes RNA-seq data from 94 knockdown and 30 over-expression experiments of 62 different transcription factors (Table S1). Before using this dataset to compare transcription factor activity methods, we wanted to evaluate whether transcription factor knockdown resulted in decreased normalized expression and over-expression resulted in increased normalized expression. Moreover, since Priori and other methods use the transcription factor expression to generate activity scores, we wanted to understand the extent to which transcription factor expression alone could predict gene perturbation status. To assess this, we compared the normalized expression of perturbed transcription factors to unperturbed genes. We found that the normalized expression of knocked-down transcription factors was significantly less than unperturbed genes (Figure S1A). While over-expressed transcription factors were associated with a higher normalized expression, it was not significantly different than unperturbed genes (Figure S1B). This data indicates that while transcription factor expression is a reasonable indicator of gene perturbation, other features are needed to confidently predict aberrant activity.
Using the decoupleR workflow, we generated activity scores for 16 transcription factor activity methods using RNA-seq data from the 124 gene perturbation experiments. To understand the patterns in predicted activity across the methods, we correlated the activity scores (Figure 2A). The Priori activity scores were dissimilar from the other methods, indicating that Priori identified a unique pattern of transcription factor activity. To assess how well the methods detected perturbed transcription factors, we calculated AUPRC and AUROC metrics for each down-sampling permutation (Figures 2B–2D; Table S2). Priori had a greater AUPRC and AUROC values than the other methods across all experiments. ULM, Norm WMEAN, Norm WSUM, and VIPER were the next closest methods by AUPRC and AUROC. Collectively, these experiments show that Priori detects perturbed transcription factors with greater sensitivity and specificity than other methods.
Figure 2.
Priori detects aberrant transcription factor activity with improved sensitivity and specificity
(A) Using the decoupleR workflow, transcription factor activity scores were generated using the perturbation dataset. Spearman correlation of the activity scores for each method.
(B) Activity scores were generated using Pathway Commons transcriptional relationships. Mean AUPRC and AUROC values across the 100 down-sampling permutations for each method.
(C and D) The distribution of (C) AUPRC and (D) AUROC values across the 100 down-sampling permutations from (B). Error bars represent the SEM.
(E and F) Activity scores were generated using (E) DoRothEA or (F) OmniPath transcriptional relationships. Mean AUPRC and AUROC values across the 100 down-sampling permutations for each method.
While our analyses showed that Priori can accurately identify perturbed transcription factors, we wanted to understand the extent to which prior information from other datasets impacted its performance. Pathway Commons is one of several large databases that curates transcriptional relationships. DoRothEA is a comprehensive resource, which assembles transcription factor-target gene relationships from ChIP-seq peaks, inferred regulatory networks (including ARACNe), transcription factor binding motifs, and literature-curated resources.33 OmniPath, on the other hand, integrates intra- and inter-cellular signaling networks in addition to transcriptional relationships from 100 different resources, including DoRothEA and Pathway Commons.34 To evaluate the impact of prior information on each method, we used the decoupleR workflow to generate activity scores using transcription factor-target gene interactions from DoRothEA or OmniPath instead of Pathway Commons. We used the default parameters for each method, including a lower limit of 15 downstream targets for the Priori analyses. We found that in both instances, Priori had higher AUPRC and AUROC values than the other methods (Figures 2E, 2F, and S1C‒S1F; Tables S3 and S4). Priori exhibited similar AUPRC and AUROC values when using transcriptional relationships from DoRothEA, OmniPath, or Pathway Commons. However, Priori had the highest mean AUPRC and AUROC values using the Pathway Commons transcriptional relationships. Our analyses demonstrated that regardless of the prior transcriptional relationship information, Priori detected perturbed transcription factor activity with improved sensitivity and specificity.
We evaluated how transcriptional relationships from Pathway Commons, DoRothEA, and.
OmniPath affected the performance of each method. However, these databases do not provide the appropriate information for methods that were designed to generate activity scores from networks with signed edges weighted by likelihood, like VIPER. While decoupleR does not allow for networks with signed edges, we wanted to understand how VIPER’s performance changed with ARACNe-generated networks with likelihood-weighted edges.20 The authors of ARACNe have published cancer-type-specific networks trained on TCGA RNA-seq datasets.23 We identified the cancer types and organ sites associated with each cell line tested in the perturbation dataset (Figures S2A‒S2C; Table S5). With decoupleR, we generated VIPER activity scores using any TCGA ARACNe network that was trained on a cancer type that was also evaluated in the perturbation dataset. We observed that VIPER had greater AUPRC and AUROC values when using the transcriptional relationships from Pathway Commons than those from the TCGA ARACNe networks (Figure S2D). ARACNe can also be used to reverse engineer gene regulatory networks from an RNA-seq dataset.23 Using the new implementation of ARACNe, ARACNe-AP, we inferred transcriptional relationships using the perturbation RNA-seq data. We observed improved AUPRC and AUROC values when VIPER used ARACNe-AP transcriptional relationships as prior information (Figure S2E). However, these AUPRC and AUROC values were still not greater than Priori using transcriptional relationships from Pathway Commons.
Priori’s predictions are robust to noise
We demonstrated that Priori detects perturbed transcription factors with improved sensitivity and specificity, particularly when it used Pathway Commons transcriptional relationships as prior information. However, we wanted to understand how robust these predictions are to noise. For methods that use prior information, the major sources of noise are introduced in the RNA-seq data and the prior transcriptional relationships. First, to evaluate the effect of noisy gene expression data, we introduced increasing amounts of zero-centered, Gaussian-distributed noise to the Holland et al. perturbation dataset. We evaluated the accuracy of perturbed transcription factor predictions using the decoupleR pipeline as described above. We observed a drop in AUPRC and AUROC in most methods when more than one standard deviation of gaussian-centered noise was introduced (Figures S3A and S3B; Table S6). Notably, Priori could still accurately identify perturbed transcription factors even when five standard deviations of noise was added. Second, to understand the impact of noisy prior information, we gradually removed the number of transcription factor-target gene relationships from the prior Pathway Commons network. We observed that Priori and other methods that use prior information had similar AUPRC and AUROC values as long as 20% of the network was retained (Figures S3C and S3D; Table S6). It was unclear, however, whether this pattern was due to the diminished number of transcriptional relationships or to the masking of true relationships. To test this, we randomized the target genes in the Pathway Commons by sampling with replacement any feature in the gene expression dataset. We observed a drop in prediction accuracy when more than 60% of the target genes were randomized (Figures S3E and S3F; Table S6). These analyses demonstrate that Priori is robust to artificial noise introduced to the gene expression data and can accurately identify perturbed transcription factors as long as 60% of the Pathway Commons network is retained.
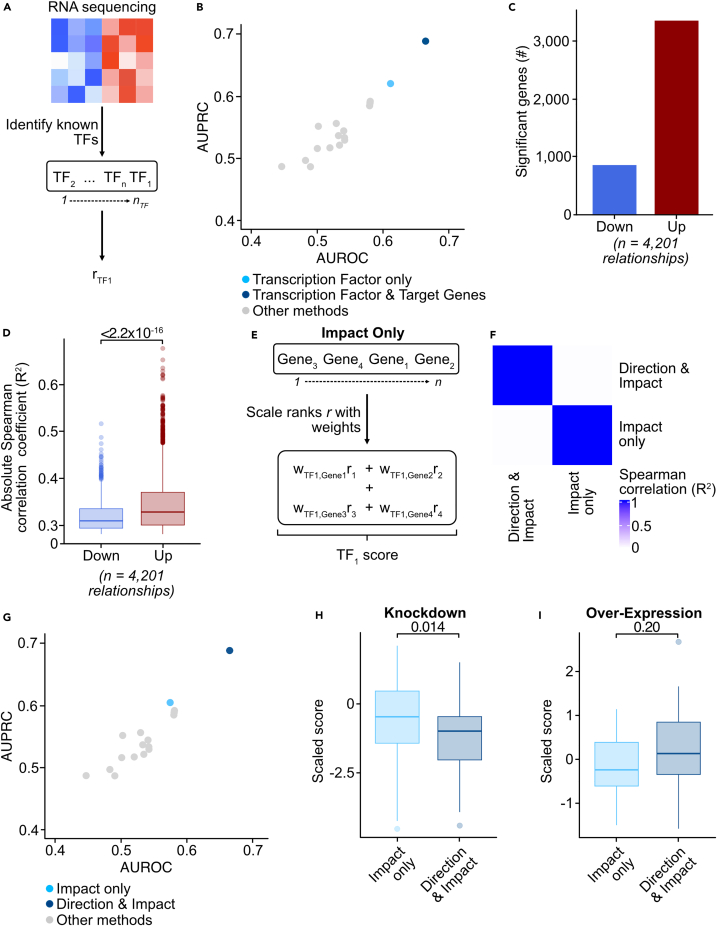
Priori’s improved performance is due to evaluating the direction of transcriptional regulation
We evaluated how Priori’s prediction accuracy was affected by artificial noise. However, it was unclear what enabled Priori to detect aberrant activity better than other methods. We previously demonstrated that transcription factor expression is a reasonable indicator of gene perturbation. We wanted to understand the extent to which assessment of transcription factor expression enabled Priori to detect perturbed transcription factors. We designed a variant of Priori that only uses transcription factor expression to infer transcription factor activity (Figure 3A). Like Priori, this method first identifies known transcription factors in an RNA-seq dataset from Pathway Commons. While this alternative method also deploys rank-based analyses, the activity score is the normalized rank of the expression of a given transcription factor relative to others in a dataset (Equation 8). We used the decoupleR benchmarking pipeline to evaluate how well this alternative method detects aberrant transcription factor activity using known transcription factors from Pathway Commons. While this alternative method had a higher AUROC and AUPRC than the other methods, Priori still detected perturbed transcription factors with higher sensitivity and specificity (Figure 3B). These data imply that while assessment of transcription factor expression is important for detection of aberrant activity, performance is improved when target gene expression is evaluated as well.
Figure 3.
Evaluation of the direction and impact of transcriptional regulation is critical for Priori to detect aberrant transcription factor activity
(A) Schematic showing how transcription factor activity scores were generated using transcription factor expression only (in contrast to both transcription factor and target gene expression as shown in Figure 1A). The alternative method first identified transcription factors in the perturbation dataset from Pathway Commons. The method then ranks the transcription factors by expression and reports the normalized rank as the activity score.
(B) Activity scores were generated using the method outlined in (A). Transcriptional relationships from Pathway Commons were used as prior information. Mean AUPRC and AUROC values across the 100 down-sampling permutations. Mean AUPRC and AUROC values from the methods in Figure 2B are also shown.
(C) Priori identified transcription factor target genes in the perturbation dataset using Pathway Commons transcriptional relationships. The expression of transcription factors and their target genes were evaluated using Spearman correlation. Statistical significance was determined using the Spearman correlation p value with an FDR post-test correction. The Spearman correlation coefficient was used to determine down-regulated (R2 < 0) and up-regulated target genes (R2 > 0).
(D) Absolute Spearman correlation coefficient of the expression of transcription factors and their down-regulated or up-regulated target genes. Statistical significance was determined by a two-sided Student’s t test. Error bars represent the SEM.
(E) Schematic showing how Priori was altered to assess only the impact of transcriptional regulation (in contrast to both direction and impact of regulation as shown in Figure 1A).
(F) Using the decoupleR workflow, transcription factor activity scores were generated using the perturbation dataset. Spearman correlation of the activity scores for each method.
(G) Activity scores were generated using the method outlined in (E). Transcriptional relationships from Pathway Commons were used as prior information. Mean AUPRC and AUROC values across the 100 down-sampling permutations for Priori using only the impact of transcriptional relationships. Mean AUPRC and AUROC values from the methods in Figure 2B are also shown.
(H and I) Priori activity scores using absolute and relative transcriptional relationships were z-transformed. Distribution of scaled scores for perturbed transcription factors across all (H) knockdown and (I) over-expression experiments. Statistical significance was determined by a two-sided Student’s t test. Error bars represent the SEM.
While Pathway Commons does not provide signed relationships, we designed Priori to infer the impact and direction of transcription factor regulation on its target genes. Priori correlates the expression of transcription factors and their known target genes. Priori subsequently assigns the impact and direction of transcriptional regulation as the sign and coefficient of the Spearman correlation, respectively. Using the sign of the Spearman correlation, we observed that Priori inferred 2,507 more significant up-regulated targets than down-regulated target genes in the perturbation dataset (Figure 3C). Using the coefficient of the Spearman correlation, we observed that Priori predicted that transcription factors had a significantly greater impact on its up-regulated targets than its down-regulated target genes (Figure 3D). These analyses suggest the relevance of evaluating the regulatory direction of transcription factors on their target genes.
To understand how assessment of the direction of transcriptional regulation is important for detecting aberrant transcription factor activity, we created another variant of Priori that only evaluates the absolute impact of transcriptional regulation (Figure 3E). Like Priori, this method deploys a rank-based analysis to evaluate the expression of transcription factors and their target genes (Equation 9). However, this alternative method does not delineate target genes by their direction of transcriptional regulation, which is indicated by the sign of the Spearman correlation. We used the decoupleR pipeline to generate activity scores using the Pathway Commons transcriptional relationships as prior information. These activity scores were highly dissimilar to the scores generated by Priori that evaluated both the direction and impact of transcriptional regulation (R2 = 0.013; Figure 3F). Moreover, when Priori only uses the impact of transcriptional regulation to detect perturbed transcription factors, its AUPRC and AUROC values are less than when it uses both the direction and impact (Figure 3G). Accounting for direction of transcriptional regulation likely allows Priori to detect knocked-down and over-expressed transcription factors. We observed an expected decrease in scaled scores across all knockdown experiments when Priori evaluates both the direction and impact of transcriptional regulation (Figure 3H). Scaled scores less than zero indicate that the activity of a transcription factor is down-regulated compared to all other perturbed and unperturbed transcription factors in the dataset (and vice versa). These scaled scores are significantly less than the impact-only method. Consistently, Priori predicted an expected increase in activity to over-expressed transcription factors (Figure 3I). While these scores are not statistically different from the scores from the impact-only method, the mean predicted activity from the impact-only scores was less than zero. Overall, these analyses demonstrate that assessment of the direction and impact of transcriptional regulation allows Priori to detect aberrant transcription factor activity with improved sensitivity and specificity.
FOXA1 transcription factor activity is a significant determinant of survival among patients with invasive breast ductal carcinoma
Since we demonstrated that Priori identifies perturbed regulators with greater sensitivity and specificity than other methods, we sought to determine whether Priori could be used to understand transcription factor drivers of disease. We used Priori to generate transcription factor activity scores for 637 patients with invasive breast ductal carcinoma (BIDC) from the TCGA-BRCA cohort.35 To understand the impact of prior information on these scores, we generated scores using transcriptional relationships not only from Pathway Commons, but from DoRothEA and Omnipath as well. In addition, we compared these predicted scores to activity scores from the three top methods identified in the decoupleR benchmark analysis: VIPER, ORA, and Norm WMEAN. Regardless of the prior information, the Priori scores from these patients clustered by breast cancer subtypes (Figures 4A and S4A‒S4C). Like in the Holland et al. perturbation dataset, we observed more up-regulated than down-regulated target genes (Figure S4D). However, we did not observe a clear separation of breast cancer subtypes when using activity scores from the other methods (Figures S4E‒S4G). BIDC is classified into three molecular subtypes: luminal, HER2, and basal cancers.36,37,38,39,40 The most common types, luminal and basal breast cancers, are distinguished by hormone receptor expression. Luminal breast cancers express estrogen and progesterone receptors, whereas basal breast cancers do not. Unsupervised clustering of the Priori Pathway Commons scores revealed that the predicted activity of transcription factors that regulate the expression of estrogen receptors (ESR1) and progesterone receptors (NR3C1) were decreased in the basal breast cancer cluster (Cluster 2; Figure 4B). Moreover, Cluster 2 had decreased predicted GATA3 activity, which is critical to luminal cell specification, as well as increased predicted MYC activity, which is consistent with previous studies.36,41,42,43 These analyses show that Priori transcription factor activity scores delineated primary BIDC patients by their molecular subtype.
Figure 4.
FOXA1 transcription factor activity is a significant determinant of survival for patients with BIDC
(A) Priori scores were generated from RNA-seq of 637 patients with BIDC. UMAP dimensional reduction and projection of Priori scores. Dots are colored by the breast cancer molecular subtype.
(B) Unsupervised hierarchical clustering of Priori scores generated in (A).
(C) Mean absolute difference of Priori scores from patients in the clusters 1 and 2 defined in (B).
(D) Distribution of FOXA1 Priori scores among patients in clusters 1 and 2 defined in (B). Statistical significance was determined by a two-sided Student’s t test. Error bars represent the SEM.
(E‒G) Kaplan-Meier survival analysis of patients grouped by (E) molecular subtype, (F) FOXA1 Priori scores, or (G) FOXA1 normalized gene expression counts. Patients among the top 90% of Priori scores or counts were grouped into “High” and those in the bottom 10% were grouped into “Low”. Statistical significance was determined by a log rank Mantel-Cox test.
(H) Differential gene expression network enrichment between clusters defined in (B). Select significantly enriched nodes are shown.
While the transcription factors associated with luminal and basal breast cancer are likely important in distinguishing BIDC patient samples by their molecular subtype, the greatest difference in predicted Priori activity between the two clusters was FOXA1 (Figures 4C and 4D; Table 2; Table S7). FOXA1 is a forkhead protein associated with mammary gland development. While the difference in FOXA1 activity was significantly different between the patient clusters defined by the DoRothEA and OmniPath Priori scores, its activity was much less pronounced between the clusters defined by the other methods (Figures S5A‒S5E; Table 2; Table S7). Evaluation of the greatest difference in predicted activity between the other networks and methods nominated transcription factors other than FOXA1. While FOXA1 was the second greatest difference in predicted activity between the Priori DoRothEA and OmniPath scores, these methods instead nominated ESR1 (Figures S5F and S5G). Notably, FOXA1 is a known regulator of ESR1.44 The other methods nominated different transcription factors, including OR10H2 by VIPER, ACTL6A by ORA, and TGFβ2 by Norm WMEAN (Figures S5H‒S5J; Table 2; Table S7). Together, these analyses show that we were able nominate drivers of basal and luminal breast cancer with Priori, identifying that luminal cancer samples are associated with high predicted FOXA1 activity.
Table 2.
The top 5 differences in predicted transcription factor activity between BIDC primary patient sample clusters
| Method | Network | Transcription factor | Absolute difference in activity (Cluster 1 vs. 2) |
|---|---|---|---|
| Priori | Pathway Commons | FOXA1 | 2.382 |
| ESR1 | 2.324 | ||
| PAX2 | 2.235 | ||
| GATA3 | 2.226 | ||
| XBP1 | 2.173 | ||
| Priori | DoRothEA | ESR1 | 2.209 |
| FOXA1 | 2.099 | ||
| GATA3 | 2.008 | ||
| HOXB13 | 2.007 | ||
| MYOD1 | 2.002 | ||
| Priori | OmniPath | ESR1 | 2.279 |
| FOXA1 | 2.271 | ||
| GATA2 | 2.236 | ||
| GATA3 | 2.199 | ||
| TFAP2C | 2.027 | ||
| VIPER | ARACNe BRCA | OR10H2 | 3.2 |
| PBRM1 | 3.136 | ||
| SAP130 | 2.979 | ||
| ELOB | 2.79 | ||
| ZIC2 | 2.783 | ||
| ORA | Pathway Commons | ACTL6A | 1.394 |
| RUVBL1 | 1.394 | ||
| RUVBL2 | 1.394 | ||
| KAT5 | 1.154 | ||
| TRRAP | 1.048 | ||
| Norm WMEAN | Pathway Commons | TGFB2 | 62.63 |
| RHOA | 48.07 | ||
| EDNRA | 46.81 | ||
| PTHLH | 32.89 | ||
| LEP | 22.77 |
To understand the clinical impact of transcription factor activity in breast cancer, we evaluated survival differences among the patients in the BIDC cohort. Patients grouped by their molecular subtypes demonstrated no significant difference in survival (Figure 4E). When we grouped patients by predicted FOXA1 Priori activity scores, patients with low FOXA1 activity had a significantly decreased chance of survival, which is consistent with previous reports (Figure 4F).45 We did not observe a survival difference when patients were instead grouped by FOXA1 expression or by the FOXA1 activity scores generated by Priori using DoRothEA or OmniPath prior networks or the other methods (Figures 4G and S6). Additionally, we did not see a survival difference in the transcription factor drivers nominated by Priori using DoRothEA or OmniPath prior networks or the other methods (Figures S7A‒S7E). ORA predicted that ACTL6A activity was either 1.91 or 3.30 in the vast majority of samples (52.7% and 42.7%, respectively; Figure S7F). As a result, normal quantiles could not be calculated, so all samples were included in the survival analysis. These data suggest that high FOXA1 transcription factor activity is protective of survival in BIDC.
In order to understand how FOXA1 activity may mediate survival in BIDC, we generated a differential gene regulatory network between the two patient clusters identified by Priori using Pathway Commons transcriptional relationships (Figure 4H).30,31,46 This analysis suggests two molecular mechanisms that distinguish luminal samples in Cluster 1 and basal samples in Cluster 2. First, we observed down-regulation of a positive feedback loop of ESR1, FOXA1, XBP1, NF1, and FOS in the basal Cluster 2 samples (Table S8).47 This is consistent with basal breast cancer, which is characterized by repression of estrogen receptor encoded by ESR1. Additionally, this analysis also shows that MYC downregulates FOXA1 cell cycle target genes, CCND1 and CDKN1B, in basal breast cancer.48 Downregulation of cell proliferation is a known mechanism of chemotherapy resistance in basal breast cancer.49 These analyses nominate putative targets in the FOXA1 network that may regulate survival in BIDC.
FOXO1 transcription factor activity mediates venetoclax resistance in acute myeloid leukemia
Aberrant transcription factor activity is also an important regulator of drug resistance in multiple tumor types.3,50,51,52,53 As we have shown that Priori nominates transcription factor regulation associated with breast cancer survival, we wanted to understand whether Priori could also be used to identify mediators of drug sensitivity. Since ex vivo drug screening data are not available in the TCGA datasets, we calculated Priori scores for 859 patients with acute myeloid leukemia (AML) from the Beat AML cohort.54,55 This dataset provides paired baseline RNA-seq and ex vivo drug sensitivity data. Once again, we generated activity scores with Priori using transcriptional relationships from Pathway Commons, DoRothEA, and Omnipath as well three of the top methods from the decoupleR benchmark analysis (VIPER, ORA, and Norm WMEAN). Consistent with the Holland et al. perturbation and TCGA-BRCA datasets, we observed more up-regulated than down-regulated target genes (Figure S8A). To nominate transcription factors mediators of drug sensitivity, we correlated predicted transcription factor activity scores from each method to drug response. We found 11,075 significant inhibitor-transcription factor activity relationships using Priori, 2,934 of which were also identified when using Priori scores that were generated from DoRothEA or OmniPath prior networks (Figures 5A and S8B; Table S9). In contrast, only 192 of Priori Pathway Commons relationships were identified by VIPER, ORA, and Norm WMEAN (Figure S8C; Table S9). VIPER likely identified the most significant inhibitor-transcription factor activity relationships (29,682) because it generated scores for more transcription factors than the other methods (1,363; Figure S8D). Among the strongest correlations from Priori was predicted FOXO1 transcription factor activity with venetoclax resistance (R2 = −0.5895; Figure 5B). This relationship was the 33rd, 16th, and 10th highest correlations among Priori scores that used Pathway Commons, DoRothEA, or OmniPath as prior networks, respectively (Figures S8E and S8F). Venetoclax resistance is more highly correlated with predicted FOXO1 activity than FOXO1 expression alone (R2 = −0.499; Figure 5C). Notably, while FOXO1 Norm WMEAN activity scores were directly proportional to venetoclax resistance, the VIPER and ORA activity scores were anti-correlated to venetoclax resistance (Figures S8G‒S8I). These findings nominate FOXO1 activity as a mediator of venetoclax activity in AML.
Figure 5.
FOXO1 is a critical mediator of response to venetoclax in AML
(A) Priori scores generated from RNA-seq of 859 patients with AML. Spearman correlation of Priori scores and ex vivo drug response AUC data.
(B and C) Spearman correlation of ranked venetoclax AUC and ranked (B) FOXO1 Priori scores or (C) FOXO1 normalized counts. Statistical significance was determined using the Spearman correlation p value with an FDR post-test correction.
(D and E) THP-1 cells were transduced with lentiviral particles harboring expression cassettes for hSpCas9 and a non-targeting or FOXO1 guide RNA. Cells were cultured for 3 days along a 7-point curve with venetoclax. Cell viability was assessed by CellTiter Aqueous colorimetric assay. ns = not significant; ∗ = p < 0.05, ∗∗ = p < 0.01, ∗∗∗ = p < 0.001, ∗∗∗∗ = p < 0.0001.
Venetoclax induces cancer cell death by restoration of intrinsic mitochondrial apoptosis. Venetoclax blocks BCL2 from sequestering factors that activate pro-apoptotic BCL2 family proteins, such as BAX.56 In mantle cell lymphoma (MCL), it has been reported that genomic regions of BAX and multiple other pro-apoptotic BCL2 family proteins are bound by FOXO1.57 The authors further demonstrated that disruption of FOXO1 activity sensitized MCL cell lines to venetoclax. However, the relationship between FOXO1 activity and venetoclax resistance has not been investigated in AML. Prior work has shown that monocytic AML is intrinsically resistant to venetoclax-based therapy.58 Given the results from our analysis of transcription factor activity in AML patients, we wanted to understand the extent to which FOXO1 knockdown could sensitize monocytic AML to venetoclax treatment. We used CRISPR-Cas9 to knock-out FOXO1 in THP-1 cells, a cell line model of monocytic leukemia (Figure S9). Both FOXO1 CRISPR guides significantly increased venetoclax sensitivity in this cell line model of monocytic AML, suggesting FOXO1 is an important mediator of venetoclax sensitivity (Figures 5D and 5E). These findings were consistent with the predictions from Priori (regardless of prior network) and Norm WMEAN, demonstrating how Priori can be used to detect transcription factor mediators of drug resistance.
Discussion
We have developed Priori, a computational algorithm that infers transcription factor activity using prior biological knowledge. The results from a third-party, unbiased benchmarking workflow demonstrate that Priori detects perturbed transcription factors with higher sensitivity and specificity than 11 other methods. Our analyses show that while accounting for transcription factor expression aids Priori in detecting perturbed transcription factors, Priori’s improved performance over other methods is likely due to assessment of the direction and impact of transcription factor regulation on their target genes. Using Priori, we identified FOXA1 activity as a regulator of survival in BIDC and nominated important downstream targets that may contribute to this survival difference.45 Importantly, there was no significant survival difference among patients that were stratified using FOXA1 scores generated by VIPER, ORA, Norm WMEAN, or even Priori that used DoRothEA or OmniPath as prior information. Moreover, we used Priori to nominate transcription factor regulators of drug sensitivity in AML. We found that predicted FOXO1 activity by Priori is highly associated with venetoclax resistance. We validated these findings in a cell line model of monocytic AML, which is resistant to venetoclax.58 While Priori (regardless of prior information) and Norm WMEAN predicted that FOXO1 activity was associated with venetoclax resistance, VIPER and ORA FOXO1 activity scores were significantly associated with venetoclax sensitivity.
Priori leverages the Pathway Commons resource to identify known gene regulatory networks in gene expression data. While using these relationships enables Priori to ground its findings in peer-reviewed literature, this may limit the discovery of novel transcriptional relationships. Analyses from other groups suggest that prior-based method tend to replicate prior information.59 Indeed, the analysis of BIDC patient samples revealed that Priori was able to identify known transcription factor drivers of BIDC from several biological datasets, including Pathway Commons, DoRothEA, and OmniPath. Our findings also showed that Priori detected a relationship between FOXA1 activity and BIDC survival. While it has been shown that high FOXA1 expression is associated with improved outcomes in patients with estrogen receptor-positive disease, these findings reveal a novel relationship between FOXA1 activity in estrogen receptor-positive and receptor-negative disease.45,60,61,62 Our analyses provide a novel mechanistic hypothesis that is ready for experimental investigation.
Pathway Commons integrates publicly available RNA, DNA, and protein data sourced from a variety of tissue types. However, Pathway Commons is not designed to curate tissue-specific transcription factor gene regulatory networks. Cell context influences regulatory interactions between transcription factors and their downstream target genes.4,63 The single-gene perturbation experiments, whose gene expression data we used to evaluate the transcription factor activity methods, were performed in numerous cell types. The small size of this dataset precluded a definitive evaluation of tissue-context as a determinant of method performance. However, we designed Priori to allow researchers to include their own regulatory networks for context-specific evaluation of their experiments. Overall, Priori should generate robust predictions that are generalizable across many cellular contexts.
In our study, we used RNA-seq data from large clinical cohorts to evaluate Priori. While we demonstrated that Priori can be used to identify determinants of survival and mediators of drug response in this context, more investigation is needed to understand Priori’s ability to predict transcription factor activity from smaller scale experiments. Notably, in a different study, we have applied a preliminary version of Priori to investigate the mechanism of combined FLT3 and LSD1 inhibition in FLT3-ITD AML.64 We used Priori to identify important determinants of the drug combination response and showed that predicted activity of a putative drug combination target, MYC, decreased in six patient samples. Since Priori scores are normalized across all samples analyzed within the same run, we expect that Priori has more power to identify differences in transcription factor activity among larger patient cohorts. Researchers can contextualize Priori scores from smaller scale experiments by generating scores from the large and small cohort RNA-seq data in the same run. Of course, this is only possible if the RNA-seq data has been consistently normalized and batch effects have been mitigated. We encourage researchers to only compare Priori scores that have been generated in the same run.
In conclusion, results from this study showed that our transcription factor activity method, Priori, detects perturbed transcription factors with improved sensitivity and specificity over other commonly used methods. Using Priori, we found that predicted FOXA1 activity is a significant determinant of survival in BIDC. We nominated putative FOXA1 targets that may be important for this survival difference. Lastly, we found that predicted FOXO1 activity is highly correlated with venetoclax resistance. We validated these findings in vitro using cell line model of AML that is intrinsically resistant to venetoclax.
Limitations of the study
The decoupleR benchmarking workflow facilitated a robust and unbiased comparison of numerous transcription factor activity methods.20 Employing the dataset curated by Holland et al., the workflow reported average AUROC and AUPRC values across all down-sampling runs as well as the transcription factor activity scores generated for each run.32 However, the workflow did not report the positively identified perturbed transcription factors for each run. Consequently, our ability to investigate the distinctive characteristics of experiments (e.g., cell type, functional genetic technique, perturbed transcription factor) that might have influenced each method’s predictions was constrained. Further efforts are warranted to include these reporting metrics, enabling a more comprehensive understanding of the factors underlying Priori’s proficiency in identifying perturbed transcription factor activity.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Holland et al. perturbation dataset (RNA-seq) | Holland et al.32 | Used in this study: https://zenodo.org/records/8368697/files/holland_rna_expr.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://zenodo.org/record/5645208/files/rna_expr.rds?download=1 |
| Holland et al. perturbation dataset (metadata) | Holland et al.32 | Used in this study: https://zenodo.org/records/8368697/files/holland_rna_meta.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://zenodo.org/record/5645208/files/rna_expr.rds?download=1 |
| TCGA PanCancer Atlas Breast Invasive Carcinoma (RNA-seq) | TCGA35 | Used in this study: https://zenodo.org/records/8368697/files/tcga_brca_normalized_counts.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://www.cbioportal.org/study/summary?id=brca_tcga_pan_can_atlas_2018 |
| TCGA PanCancer Atlas Breast Invasive Carcinoma (metadata) | TCGA35 | Used in this study: https://zenodo.org/records/8368697/files/tcga_brca_metadata.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://www.cbioportal.org/study/summary?id=brca_tcga_pan_can_atlas_2018 |
| Beat AML (RNA-seq) | Bottomly et al.55 | Used in this study: https://zenodo.org/records/8368697/files/beataml_rna_expr.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://github.com/biodev/beataml2.0_data/raw/main/beataml_waves1to4_norm_exp_dbgap.txt |
| Beat AML (Inhibitor AUC values) | Bottomly et al.55 | Used in this study: https://zenodo.org/records/8368697/files/beataml_inhibitor_auc.tsv Database: https://doi.org/10.1101/2022.12.16.520295 Original source: https://github.com/biodev/beataml2.0_data/raw/main/beataml_probit_curve_fits_v4_dbgap.txt |
| Software and algorithms | ||
| Priori | This study | Github: https://github.com/ohsu-comp-bio/regulon-enrichment https://doi.org/10.5281/zenodo.10553601 |
| decoupleR version 2.0.0 (fork for this study) | Badia-i-Mompel et al.20 | Github: https://github.com/ohsu-comp-bio/decoupleR https://doi.org/10.5281/zenodo.10553605 |
| decoupleRBench version 0.1.0 (fork for this study) | Badia-i-Mompel et al.20 | Github: https://github.com/ohsu-comp-bio/decoupleRBench https://doi.org/10.5281/zenodo.10553607 |
| decoupleR workflow | This study | Github: https://github.com/ohsu-comp-bio/decoupler_workflow https://doi.org/10.5281/zenodo.10553605 |
| Pathway Commons | Rodchenkov et al.31 | https://www.pathwaycommons.org/ |
| Patterns | Babur et al.65 | https://code.google.com/archive/p/biopax-pattern/ |
| Univariate Linear Model (ULM) version 1.0.0 | Teschendorff and Wang16 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-ulm.R |
| Multivariate Linear Model (MLM) version 1.0.0 | Teschendorff and Wang16 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-mlm.R |
| Over Representation Analysis (ORA) version 1.0.0 | Badia-i-Mompel et al.20 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-ora.R |
| Fast Gene Set Enrichment (FGSEA) version 1.20.0 | Korotkevich et al.17 | https://github.com/ctlab/fgsea/ |
| Gene Set Variation Analysis (GVSA) version 1.42.0 | Hänzelmann et al.18 | https://bioconductor.org/packages/3.17/bioc/html/GSVA.html |
| AUCell version 1.16.0 | Aibar et al.19 | https://github.com/aertslab/AUCell |
| Virtual Inference of Protein-activity by Enriched Regulon analysis (VIPER) version 1.28.0 | Alvarez et al.24 | https://www.bioconductor.org/packages/release/bioc/html/viper.html |
| Weighted Mean (WMEAN) version 1.0.0 | Badia-i-Mompel et al.20 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-wmean.R |
| Weighted Sum (WSUM) version 1.0.0 | Badia-i-Mompel et al.20 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-wsum.R |
| Univariate Decision Tree (UDT) version 1.0.0 | Badia-i-Mompel et al.20 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-udt.R |
| Multivariate Decision Trees (MDT) version 1.0.0 | Badia-i-Mompel et al.20 | https://github.com/ohsu-comp-bio/decoupleR/blob/master/R/statistic-mdt.R |
| OmnipathR version 3.6.0 | Turei et al.34 | https://www.bioconductor.org/packages/release/bioc/html/OmnipathR.html |
| dorothea version 1.10.0 | Garcia-Alonso et al.33 | https://bioconductor.org/packages/release/data/experiment/html/dorothea.html |
| ARACNE-AP version 1.0 | Lachmann et al.23 | https://github.com/califano-lab/ARACNe-AP |
| arcane.networks version 1.18.0 | Lachmann et al.23 | https://bioconductor.org/packages/release/data/experiment/html/aracne.networks.html |
| Seurat version 4.4.0 | Hafemeister, Satija et al.66 | https://github.com/satijalab/seurat |
| CausalPath version 1.8.0 | Babur et al.46 | https://github.com/PathwayAndDataAnalysis/causalpath |
| Geneious Prime | Dotmatics | https://www.geneious.com/ |
| ICE CRISPR Analysis Tool | Synthego | https://ice.synthego.com/#/ |
| Experimental models: Cell lines | ||
| THP-1 | ATCC | Cat# TIB-202 |
| Lenti-X 293T cells | Clontech | Cat# 632180 |
| Chemicals, peptides, and recombinant proteins | ||
| RPMI 1640 Medium | Gibco | Cat# 11875093 |
| HyClone Characterized FBS | Cytiva Life Sciences | Cat# SH30071.04 |
| GlutaMAX | Gibco | Cat# 35050079 |
| 2-Mercaptoethanol | Sigma Aldrich | Cat# M6250 |
| HighPrep PCR Clean-up System | MagBio Genomics | Cat# AC-60001 |
| Recombinant DNA | ||
| FOXO1 gRNA in pLentiCRISPR v2 backbone (#1) | Genscript | Sequence: GCTCGTCCCGCCGCAACGCG |
| FOXO1 gRNA in pLentiCRISPR v2 backbone (#2) | Genscript | Sequence: ACAGGTTGCCCCACGCGTTG |
| Non-targeting pLentiCRISPR v2 | Addgene | Cat# 169795 |
| psPAX2 | Addgene | Cat# 12260 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Emek Demir (demire@ohsu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The sources of the datasets supporting the current study are presented in the key resources table and the method details section.
-
•
All original and forked code have been deposited at GitHub and are publicly available as of the date of publication.
-
•
Any additional information required to re-analyze the data reported in this paper or reproduce the results is available from the lead contact upon request.
Experimental model and study participant details
Cell lines
THP-1 cells (DSMZ) were cultured in RPMI (Gibco) supplemented with 10% fetal bovine serum (FBS, HyClone), 2 mM GlutaMAX (Gibco), 100 units/mL Penicillin, 100 μg/mL Streptomycin (Gibco), and 0.05 mM 2-Mercaphightoethanol (Sigma Aldrich). Sex of THP-1 cells is male. All cells were cultured at 5% CO2 and 37°C. Cell lines were tested monthly for mycoplasma contamination.
Method details
Priori algorithm
Pre-processing
Priori normalizes and scales the input RNA-seq data prior to downstream analysis. Priori first filters out counts with a standard deviation less than 0.1 (controlled by the thresh_filter parameter). Priori linearly shifts the remaining counts by the minimum value and then log2 normalizes them (xgene). Priori then scales the normalized counts (zgene) using one of four methods: “standard”
| (Equation 1) |
(where the mean and standard deviation of all normalized counts are indicated by and , respectively), “robust”
| (Equation 2) |
(where the median normalized counts is indicated by and the interquartile range is indicated by ), “minmax”
| (Equation 3) |
(where the minimum and maximum normalized counts are indicated by and , respectively), or “quant” where the normalized counts are scaled using the inverse of the cumulative distribution function, F(x).67
| (Equation 4) |
Priori defaults to the “standard” scaling function.
Network
Priori uses known gene regulatory networks to predict transcription factor activity from RNA-seq data. By default, Priori extracts transcriptional relationships from the Pathway Commons database to generate activity scores.30,31 Users can also generate Priori scores using other gene regulatory networks with the regulon parameter. The user-defined network must specify the transcription factor (Regulator) and their downstream target genes (Target). Pathway relationships in Pathway Commons are represented with the BioPAX language.30,68 BioPAX abstracts major pathway relationships, including gene regulatory networks, into a standardized format. However, BioPAX representations cannot be interpreted directly. In order to identify the gene regulatory networks encoded in Pathway Commons, we extracted transcription factors and their primary targets using Patterns.65 Using the extracted network, Priori removes transcription factors with less than 15 downstream targets. Users can control the number of targets with the regulon_size parameter.
Activity scores
Once the network is prepared, Priori generates an activity score for each transcription factor in an RNA-seq dataset. To calculate the activity score for a transcription factor (), Priori first calculates weights for its target genes (). The target gene weight is the product of the F-statistic () and the Spearman correlation coefficient () of the transcription factor and its target gene expression:
| (Equation 5) |
where i represents the range of transcription factors in a dataset, j represents the range of target genes for a given transcription factor , and r represents the rank of the scaled counts relative to all features in the dataset. The non-negative, log2-normalized counts are used to calculate the F-statistic.
Priori first uses the transcription factor-target gene weights to determine the direction of regulation. Priori defines k down-regulated targets among all j target genes as those with . Priori also identifies l up-regulated targets among all j target genes as those with . Priori then ranks the scaled target counts for each transcription factor grouped by their direction of regulation ( or ). Priori weighs the ranks of the scaled counts using the target gene weight :
| (Equation 6) |
The resulting activity score for a given transcription factor () is the summation of the weighted ranks:
| (Equation 7) |
where downregulated genes are scaled by −1 prior to summation. Finally, the activity scores for each transcription factor are z-transformed relative to all other transcription factors and then again to all other samples in the dataset.
Alternative method: Evaluation of transcription factor expression only
We wanted to understand the extent to which assessment of transcription factor expression enabled Priori to detect perturbed transcription factors. We designed an alternative method that only uses the transcription factor expression to infer transcription factor activity. To infer the activity for a transcription factor , this alternative method first ranks the transcription factor scaled counts relative to other transcription factors in the dataset, . The metho then scales these ranks by the total number of transcription factors, , and normalizes them using a normal distribution:
| (Equation 8) |
Alternative method: Impact of transcriptional regulation only
We wanted to understand the extent to which assessment of transcriptional regulation on target genes enabled Priori to detect perturbed transcription factors. We designed an alternative method that only uses the impact of transcriptional regulation to infer transcription factor activity. The alternative method uses the same values of the weighted ranks ( or ) as calculated above. However, the resulting activity score for a given transcription factor () is the summation of the weighted ranks, but the down-regulated genes are not scaled by −1 prior to summation:
| (Equation 9) |
The activity scores for each transcription factor are z-transformed relative to all other transcription factors and then again to all other samples in the dataset.
Benchmarking workflow
Pre-processing
The decoupleR benchmarking workflow has been previously described.20 The Holland et al. normalized gene expression counts and metadata for were downloaded Zenodo (https://zenodo.org/record/5645208).20 The normalized RNA-seq data was linearly shifted by the minimum value so all values were non-negative.32
Transcription factor activity scores and p Values
With the decoupleR workflow (version 2.0.0), we generated transcription factor activity scores for 11 methods including Priori: AUCell (version 1.16.0), FGSEA (version 1.20.0), GSVA (version 1.42.0), MDT (version 1.0.0), ORA (version 1.0.0), UDT (version 1.0.0), ULM (version 1.0.0), VIPER (version 1.28.0), WMEAN (version 1.0.0), and WSUM (version 1.0.0). decoupleR also generated normalized transcription factor activity scores for FGSEA, WMEAN, and WSUM as well as corrected scores for WMEAN and WSUM.20 We generated transcription factor activity scores for each method using the default parameters. OmniPath (downloaded using OmipathR package version 3.6.0) and DoRothEA (downloaded using Dorothea package version 1.10.0), which assign confidence scores to their transcriptional relationships, were filtered for high confidence relationships (A, B, or C). We also calculated p values for the Priori activity scores using a Student’s two-sided t-test with an FDR post-test correction.
Area under the receiver operating characteristic and precision recall curves
Using the decoupleRBench package (version 0.1.0), we generated AUROC and AUPRC values. Briefly, we ranked the absolute value of the activity scores from the decoupleR workflow for each experiment.20 The activity scores were ranked separately for each method. We determined whether the perturbed transcription factors were among the top “n” scores. “n” was defined as the number of unique perturbed transcription factors in the dataset. There are 62 unique transcription factors in the Holland et al. perturbation dataset, which is the dataset that we used to compare the prediction accuracy of Priori to other methods.32 Therefore, a true positive is assigned to a method whose perturbed transcription factor ranks among the top 62 activity scores. For methods that use the Pathway Commons transcriptional relationships (a total of 610 transcription factors), this is the top 10.2% of features. As the number of unperturbed transcription factors in the dataset substantially outnumbered the perturbed factors, we deployed a down-sampling strategy to compare an equal number. We calculated AUROC and AUPRC values for 100 down-sampling permutations.
ARACNe network
In order to better understand whether Priori’s performance advantages depend on the design of its input transcription factor network, we generated VIPER activity scores using transcriptional relationships from TCGA ARACNe networks and an alternative network using ARACNe-AP.23 We downloaded the TCGA ARACNe networks using the arcane.networks package (version 1.18.0). For the ARACNe-AP network, we computed it from the Holland et al. perturbation dataset.32 ARACNe-AP requires a list of transcription factors in order to generate a gene regulatory network. We used a list of transcription factors from the Alvarez et al. 2016 publication that was provided by Dr. Mariano Alvarez on September 25, 2019.24 We excluded transcription factors that were not present in the Holland et al. dataset, resulting in an input list of 1,726 transcription factors. We ran ARACNe-AP (version 1.0, created with java 1.8.0_171-b11) with 100 bootstraps, --p-value = 1E-8, and –random seeds = TRUE.23 The consolidated interactome included 1,726 transcription factors and 302,444 interactions.
Noise
In order to investigate the robustness transcription factor activity scores to noise, we artificially altered the input data, including the gene expression data from the Holland et al. perturbation dataset and the transcriptional relationships from Pathway Commons, to each method. First, we added zero-centered, Gaussian-distributed noise to the gene expression data using the stats package (R base version 4.1.3). We increased the amount of noise by altering the standard deviation of the Gaussian distribution. Next, we evaluated activity scores by artificially altering the Pathway Commons transcriptional relationships. We tested this in two ways. First, we randomly removed transcription factor-target gene pairs in the Pathway Commons prior network using the stats package (R base version 4.1.3). Second, we randomized the transcription factor-target gene pairs by sampling genes in the Holland et al. dataset with replacement. We replaced these selected genes as the target genes of randomly selected transcription factors using the stats package (R base version 4.1.3).
Data analysis
Benchmarking workflow
Custom scripts were used to evaluate the results of the decoupleR benchmarking workflow. In order to compare the scores across the different methods, we z-transformed the transcription factor activity scores. We calculated the Spearman correlation between the activity scores and p values of each method.
TCGA BRCA
The normalized gene expression counts and metadata from the Breast Invasive Carcinoma (TCGA, PanCancer Atlas) study were downloaded from cBioPortal (https://www.cbioportal.org).35,69 Priori scores were generated from the normalized gene expression counts linearly shifted by the minimum value using the same Pathway Commons, DoRothEA, or OmniPath relationships that were evaluated in the decoupleR benchmarking workflow. VIPER scores were generated using the ARACNe BRCA network, --pleiotropy = TRUE, and --eset.filter = FALSE.23,24 Norm WMEAN scores were generated using Pathway Commons transcriptional relationships, --times = 100, --sparse = TRUE, and --randomize_type = rows.20 ORA scores were generated using Pathway Commons transcriptional relationships, --n_up = 300, --n_down = 300, --n_background = 20000, and --with_ties = TRUE. Custom scripts were used to exclude patients without basal or luminal BIDC. The Seurat package (version 4.4.0) was used to perform dimensional reduction on the Priori scores by PCA and UMAP.66 Survival data was analyzed for significance using a log rank Mantel-Cox test. We assigned patients to “High” or “Low” groups depending on whether they were among the top 90% or bottom 10% of the value of interest. Differential gene expression networks were generated from the normalized gene expression data using CausalPath (version 1.18.0).46 An FDR threshold of 0.001 was used to evaluate significant relationships following 100 permutations.
Beat AML
The normalized gene expression counts and inhibitor AUC values from the Beat AML study were downloaded from GitHub (https://github.com/biodev/beataml2.0_data).54,55 Priori scores were generated from the normalized gene expression counts of baseline AML patient samples from the Beat AML cohort using the same Pathway Commons, DoRothEA, or OmniPath relationships that were evaluated in the decoupleR benchmarking workflow. These counts were also linearly shifted by the minimum value. VIPER scores were generated using a trained ARACNe-AP network, --pleiotropy = TRUE, and --eset.filter = FALSE.23,24 For the ARACNe-AP network, we computed it from the normalized RNA-seq data from Beat AML.54,55 ARACNe-AP requires a list of transcription factors in order to generate a gene regulatory network. We used a list of transcription factors from the Alvarez et al. 2016 publication that was provided by Dr. Mariano Alvarez on September 25, 2019.24 We excluded transcription factors that were not present in the Holland et al. dataset, resulting in an input list of 1,408 transcription factors. We ran ARACNe-AP (version 1.0, created with java 1.8.0_171-b11) with 100 bootstraps, --p-value = 1E-8, and --random seeds = TRUE.23 The consolidated interactome included 1,402 transcription factors and 320,632 interactions. Norm WMEAN scores were generated using Pathway Commons transcriptional relationships, --times = 100, --sparse = TRUE, and --randomize_type = rows.20 ORA scores were generated using Pathway Commons transcriptional relationships, --n_up = 300, --n_down = 300, --n_background = 20000, and --with_ties = TRUE. Custom scripts were used to exclude patients without a diagnosis of AML or those with a prior myeloproliferative neoplasm. Priori scores and single-inhibitor drug AUC values on the same patient sample were evaluated using a Spearman correlation. Significant correlations were those with an FDR <0.05.
Cell culture
CRISPR
Two FOXO1 CRISPR guide RNAs in a pLentiCRISPR v2 backbone were obtained from GenScript.70 The target sequence for guide RNA (gRNA) 1 is GCTCGTCCCGCCGCAACGCG and the sequence for gRNA 2 is ACAGGTTGCCCCACGCGTTG. Additionally, a non-targeting CRISPR guide RNA (target sequence CCTGGGTTAGAGCTACCGCA) generated by scrambling the target sequence of LentiCRISPRv2-ACTB-C1 in a pLentiCRISPR v2 backbone was obtained from Addgene (Cat# 169795). Lentivirus was produced by transfecting Lenti-X 293T cells (Clontech) with the SMARTvector transfer plasmid and packaging/pseudotyping plasmids. psPAX2 was a gift from Didier Trono (Addgene plasmid #12260; http://n2t.net/addgene:12260; RRID:Addgene_12260). The supernatant containing lentivirus was collected after 48 h of culture and filtered with a 0.45 μm filter. THP-1 cells were transduced with virus via spinnoculation in the presence of polybrene. Transduced cells were selected with 1 μg/mL puromycin to produce a stable cell line.
CRISPR validation
FOXO1 knockdown was validated using Tracking of Indels by Decomposition (TIDE).71 Briefly, cellular DNA was PCR-amplified using primers upstream (sequence AAGTAGGGCACGCTCTTGAC) and downstream (sequence CGTTCCCCCAAATCTCGGAC) of the FOXO1 gRNA target sequences. The primers were designed in Geneious Prime and synthesized by Integrated DNA Technologies. Paramagnetic beads were used to purify the PCR DNA fragments (MagBio Ref #AC-60001) and subsequently sequenced by EuroFins. Inference of CRISPR Edits (ICE) was performed using the Synthego web tool (https://ice.synthego.com/#/).
Drug sensitivity assay
Cells were cultured for 72 h along a 7-point dose curve with venetoclax. Cell viability was assessed by CellTiter Aqueous colorimetric assay.
Quantification and statistical analysis
Values are represented as the mean and error bars are the SEM unless otherwise stated. Python and R were used to perform statistical analyses. Significance was tested using two-sided Student’s t test. Correlations were performed using Spearman’s rank method. Statistical significance in the survival analyses was determined by a log rank Mantel-Cox test. Where appropriate, p values were adjusted for repeated testing using the Benjamini–Hochberg method.
Acknowledgments
W.M.Y. received funding from the NIH National Cancer Institute (1 F30CA278500-01A1), E.D. received funding from the National Institutes of Health (5U2CCA233280-03, 5U01CA224012–02, U24CA264007) and O.B. received funding from the NIH National Heart, Lung, and Blood Institute (R01HL146549) for this work. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We would like to thank all of the patients for their precious time and donation of samples supporting this research. We appreciate the OHSU core facilities ExaCloud Cluster Computational Resource and the Advanced Computing Center for their assistance. We would also like to thank Michal R. Grzadkowski for his advice in the early stages of Priori’s development.
Author contributions
W.M.Y.: conceptualization, software, formal analysis, validation, investigation, visualization, methodology, writing-original draft, writing-review and editing. J.E.: conceptualization, software, formal analysis, validation, investigation, visualization, methodology, writing-review and editing. H.H.: conceptualization, formal analysis, investigation, writing-review and editing. J.S.: formal analysis, investigation, writing-review and editing. O.N.: conceptualization, investigation, writing-review and editing. Ö.B.: conceptualization, investigation, writing-review and editing. T.P.B.: conceptualization, resources, supervision, funding acquisition, validation, investigation, writing-review and editing. E.D.: conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-review and editing. The co-first authors may identify themselves as lead authors in their respective CVs.
Declaration of interests
W.M.Y. is a former employee of Abreos Biosciences, Inc. and was compensated in part with common stock options. Pursuant to the merger and reorganization agreement between Abreos Biosciences, Inc. and Fimafeng, Inc., W.M.Y. surrendered all of his common stock options in 03/2021. T.P.B. has received research support from AstraZeneca, Blueprint Medicines as well as Gilead Sciences and is the institutional PI on the FRIDA trial sponsored by Oryzon Genomics. The authors certify that all compounds tested in this study were chosen without input from any of our industry partners. The other authors do not have competing interests, financial or otherwise.
Published: February 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109124.
Supplemental information
References
- 1.Fuda N.J., Ardehali M.B., Lis J.T. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spitz F., Furlong E.E.M. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 3.Bushweller J.H. Targeting transcription factors in cancer — from undruggable to reality. Nat. Rev. Cancer. 2019;19:611–624. doi: 10.1038/s41568-019-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee T.I., Young R.A. Transcriptional Regulation and Its Misregulation in Disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional Amplification in Tumor Cells with Elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R., et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khatri P., Sirota M., Butte A.J. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. PLoS Comput. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen T.-M., Shafi A., Nguyen T., Draghici S. Identifying significantly impacted pathways: a comprehensive review and assessment. Genome Biol. 2019;20:203. doi: 10.1186/s13059-019-1790-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Essaghir A., Toffalini F., Knoops L., Kallin A., van Helden J., Demoulin J.-B. Transcription factor regulation can be accurately predicted from the presence of target gene signatures in microarray gene expression data. Nucleic Acids Res. 2010;38:e120. doi: 10.1093/nar/gkq149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Sousa Abreu R., Penalva L.O., Marcotte E.M., Vogel C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel C., Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koussounadis A., Langdon S.P., Um I.H., Harrison D.J., Smith V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015;5 doi: 10.1038/srep10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiełbasa S.M., Vingron M. Transcriptional Autoregulatory Loops Are Highly Conserved in Vertebrate Evolution. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benito J., Zheng H., Ng F.S., Hardin P.E. Transcriptional feedback loop regulation, function and ontogeny in Drosophila. Cold Spring Harb. Symp. Quant. Biol. 2007;72:437–444. doi: 10.1101/sqb.2007.72.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornstein C., Winter D., Barnett-Itzhaki Z., David E., Kadri S., Garber M., Amit I. A negative feedback loop of transcription factors specifies alternative dendritic cell chromatin states. Mol. Cell. 2014;56:749–762. doi: 10.1016/j.molcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teschendorff A.E., Wang N. Improved detection of tumor suppressor events in single-cell RNA-Seq data. npj Genom. Med. 2020;5:43. doi: 10.1038/s41525-020-00151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korotkevich G., Sukhov V., Budin N., Shpak B., Artyomov M.N., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021;2 doi: 10.1101/060012. Preprint at. [DOI] [Google Scholar]
- 18.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14:7–15. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.-C., Geurts P., Aerts J., et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badia-i-Mompel P., Vélez Santiago J., Braunger J., Geiss C., Dimitrov D., Müller-Dott S., Taus P., Dugourd A., Holland C.H., Ramirez Flores R.O., Saez-Rodriguez J. decoupleR: ensemble of computational methods to infer biological activities from omics data. Bioinform. Adv. 2022;2:vbac016. doi: 10.1093/bioadv/vbac016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung J.-H., Yang T.-H., Hu Z., Weng Z., DeLisi C. Gene set enrichment analysis: performance evaluation and usage guidelines. Brief. Bioinform. 2012;13:281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Margolin A.A., Nemenman I., Basso K., Wiggins C., Stolovitzky G., Dalla Favera R., Califano A. ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context. BMC Bioinf. 2006;7:S7–S15. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachmann A., Giorgi F.M., Lopez G., Califano A. ARACNe-AP: gene network reverse engineering through adaptive partitioning inference of mutual information. Bioinformatics. 2016;32:2233–2235. doi: 10.1093/bioinformatics/btw216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvarez M.J., Shen Y., Giorgi F.M., Lachmann A., Ding B.B., Ye B.H., Califano A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen C., Fleming K., Prendergast N., Rubio R., Emmert-Streib F., Bontempi G., Haibe-Kains B., Quackenbush J. Inference and validation of predictive gene networks from biomedical literature and gene expression data. Genomics. 2014;103:329–336. doi: 10.1016/j.ygeno.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walhout A.J.M. What does biologically meaningful mean? A perspective on gene regulatory network validation. Genome Biol. 2011;12:109. doi: 10.1186/gb-2011-12-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barbuti R., Gori R., Milazzo P., Nasti L. A survey of gene regulatory networks modelling methods: from differential equations, to Boolean and qualitative bioinspired models. J. Membr. Comput. 2020;2:207–226. doi: 10.1007/s41965-020-00046-y. [DOI] [Google Scholar]
- 28.Fernald G.H., Capriotti E., Daneshjou R., Karczewski K.J., Altman R.B. Bioinformatics challenges for personalized medicine. Bioinformatics. 2011;27:1741–1748. doi: 10.1093/bioinformatics/btr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yngvadottir B., Macarthur D.G., Jin H., Tyler-Smith C. The promise and reality of personal genomics. Genome Biol. 2009;10:237. doi: 10.1186/gb-2009-10-9-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami E.G., Gross B.E., Demir E., Rodchenkov I., Babur Ö., Anwar N., Schultz N., Bader G.D., Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodchenkov I., Babur O., Luna A., Aksoy B.A., Wong J.V., Fong D., Franz M., Siper M.C., Cheung M., Wrana M., et al. Pathway Commons 2019 Update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 2020;48:D489–D497. doi: 10.1093/nar/gkz946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holland C.H., Tanevski J., Perales-Patón J., Gleixner J., Kumar M.P., Mereu E., Joughin B.A., Stegle O., Lauffenburger D.A., Heyn H., et al. Robustness and applicability of transcription factor and pathway analysis tools on single-cell RNA-seq data. Genome Biol. 2020;21:36. doi: 10.1186/s13059-020-1949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Alonso L., Holland C.H., Ibrahim M.M., Turei D., Saez-Rodriguez J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019;29:1363–1375. doi: 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Türei D., Valdeolivas A., Gul L., Palacio-Escat N., Klein M., Ivanova O., Ölbei M., Gábor A., Theis F., Módos D., et al. Integrated intra- and intercellular signaling knowledge for multicellular omics analysis. Mol. Syst. Biol. 2021;17 doi: 10.15252/msb.20209923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger A.C., Korkut A., Kanchi R.S., Hegde A.M., Lenoir W., Liu W., Liu Y., Fan H., Shen H., Ravikumar V., et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690–705.e9. doi: 10.1016/j.ccell.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sørlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perou C.M., Jeffrey S.S., van de Rijn M., Rees C.A., Eisen M.B., Ross D.T., Pergamenschikov A., Williams C.F., Zhu S.X., Lee J.C., et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc. Natl. Acad. Sci. USA. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenton J.D., Carey L.A., Ahmed A.A., Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J. Clin. Oncol. 2005;23:7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 39.Tamimi R.M., Baer H.J., Marotti J., Galan M., Galaburda L., Fu Y., Deitz A.C., Connolly J.L., Schnitt S.J., Colditz G.A., Collins L.C. Comparison of molecular phenotypes of ductal carcinoma in situand invasive breast cancer. Breast Cancer Res. 2008;10:R67–R69. doi: 10.1186/bcr2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorlie T., Tibshirani R., Parker J., Hastie T., Marron J.S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouros-Mehr H., Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev. Dyn. 2006;235:3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouros-Mehr H., Slorach E.M., Sternlicht M.D., Werb Z. GATA-3 Maintains the Differentiation of the Luminal Cell Fate in the Mammary Gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sotiriou C., Neo S.-Y., McShane L.M., Korn E.L., Long P.M., Jazaeri A., Martiat P., Fox S.B., Harris A.L., Liu E.T. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc. Natl. Acad. Sci. USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seachrist D.D., Anstine L.J., Keri R.A. FOXA1: A Pioneer of Nuclear Receptor Action in Breast Cancer. Cancers. 2021;13:5205. doi: 10.3390/cancers13205205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Babur Ö., Luna A., Korkut A., Durupinar F., Siper M.C., Dogrusoz U., Vaca Jacome A.S., Peckner R., Christianson K.E., Jaffe J.D., et al. Causal interactions from proteomic profiles: Molecular data meet pathway knowledge. PATTER. 2021;2 doi: 10.1016/j.patter.2021.100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., Iliopoulos D., Zhang Q., Tang Q., Greenblatt M.B., Hatziapostolou M., Lim E., Tam W.L., Ni M., Chen Y., et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508:103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Green A.R., Aleskandarany M.A., Agarwal D., Elsheikh S., Nolan C.C., Diez-Rodriguez M., Macmillan R.D., Ball G.R., Caldas C., Madhusudan S., et al. MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br. J. Cancer. 2016;114:917–928. doi: 10.1038/bjc.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nedeljković M., Damjanović A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer—How We Can Rise to the Challenge. Cells. 2019;8:957. doi: 10.3390/cells8090957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia-Alonso L., Iorio F., Matchan A., Fonseca N., Jaaks P., Peat G., Pignatelli M., Falcone F., Benes C.H., Dunham I., et al. Transcription Factor Activities Enhance Markers of Drug Sensitivity in Cancer. Cancer Res. 2018;78:769–780. doi: 10.1158/0008-5472.CAN-17-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alessandrini F., Pezzè L., Menendez D., Resnick M.A., Ciribilli Y. ETV7-Mediated DNAJC15 Repression Leads to Doxorubicin Resistance in Breast Cancer Cells. Neoplasia. 2018;20:857–870. doi: 10.1016/j.neo.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neel D.S., Bivona T.G. Resistance is futile: overcoming resistance to targeted therapies in lung adenocarcinoma. npj Precis. Oncol. 2017;1:3. doi: 10.1038/s41698-017-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matkar S., Sharma P., Gao S., Gurung B., Katona B.W., Liao J., Muhammad A.B., Kong X.-C., Wang L., Jin G., et al. An Epigenetic Pathway Regulates Sensitivity of Breast Cancer Cells to HER2 Inhibition via FOXO/c-Myc Axis. Cancer Cell. 2015;28:472–485. doi: 10.1016/j.ccell.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyner J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. Functional genomic landscape of acute myeloid leukaemia. Nature. 2018;562:526–531. doi: 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bottomly D., Long N., Schultz A.R., Kurtz S.E., Tognon C.E., Johnson K., Abel M., Agarwal A., Avaylon S., Benton E., et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell. 2022;40:850–864.e9. doi: 10.1016/j.ccell.2022.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mihalyova J., Jelinek T., Growkova K., Hrdinka M., Simicek M., Hajek R. Venetoclax: A new wave in hematooncology. Exp. Hematol. 2018;61:10–25. doi: 10.1016/j.exphem.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 57.Brown F., Hwang I., Sloan S., Hinterschied C., Helmig-Mason J., Long M., Youssef Y., Chan W.K., Prouty A., Chung J.H., et al. PRMT5 Inhibition Promotes FOXO1 Tumor Suppressor Activity to Drive a Pro-Apoptotic Program That Creates Vulnerability to Combination Treatment with Venetoclax in Mantle Cell Lymphoma. Blood. 2021;138:681. doi: 10.1182/blood-2021-153733. [DOI] [Google Scholar]
- 58.Pei S., Pollyea D.A., Gustafson A., Stevens B.M., Minhajuddin M., Fu R., Riemondy K.A., Gillen A.E., Sheridan R.M., Kim J., et al. Monocytic Subclones Confer Resistance to Venetoclax-Based Therapy in Patients with Acute Myeloid Leukemia. Cancer Discov. 2020;10:536–551. doi: 10.1158/2159-8290.CD-19-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hawe J.S., Saha A., Waldenberger M., Kunze S., Wahl S., Müller-Nurasyid M., Prokisch H., Grallert H., Herder C., Peters A., et al. Network reconstruction for trans acting genetic loci using multi-omics data and prior information. Genome Med. 2022;14:125. doi: 10.1186/s13073-022-01124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perou C.M., Sørlie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 61.Lupien M., Eeckhoute J., Meyer C.A., Wang Q., Zhang Y., Li W., Carroll J.S., Liu X.S., Brown M. FoxA1 Translates Epigenetic Signatures into Enhancer-Driven Lineage-Specific Transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hurtado A., Holmes K.A., Ross-Innes C.S., Schmidt D., Carroll J.S. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacNeil L.T., Pons C., Arda H.E., Giese G.E., Myers C.L., Walhout A.J.M. Transcription Factor Activity Mapping of a Tissue-Specific In Vivo Gene Regulatory Network. Cels. 2015;1:152–162. doi: 10.1016/j.cels.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yashar W.M., Curtiss B.M., Coleman D.J., VanCampen J., Kong G., Macaraeg J., Estabrook J., Demir E., Long N., Bottomly D., et al. Disruption of the MYC Super-Enhancer Complex by Dual Targeting of FLT3 and LSD1 in Acute Myeloid Leukemia. Mol. Cancer Res. 2023;21:631–647. doi: 10.1158/1541-7786.MCR-22-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Babur Ö., Aksoy B.A., Rodchenkov I., Sümer S.O., Sander C., Demir E. Pattern search in BioPAX models. Bioinformatics. 2014;30:139–140. doi: 10.1093/bioinformatics/btt539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 68.Demir E., Cary M.P., Paley S., Fukuda K., Lemer C., Vastrik I., Wu G., D’Eustachio P., Schaefer C., Luciano J., et al. BioPAX – A community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The sources of the datasets supporting the current study are presented in the key resources table and the method details section.
-
•
All original and forked code have been deposited at GitHub and are publicly available as of the date of publication.
-
•
Any additional information required to re-analyze the data reported in this paper or reproduce the results is available from the lead contact upon request.