Abstract
The DPB11 gene, which genetically interacts with DNA polymerase II (ɛ), one of three replicative DNA polymerases, is required for DNA replication and the S phase checkpoint in Saccharomyces cerevisiae. To identify factors interacting with Dbp11, we have isolated sld (synthetically lethal with dpb11-1) mutations which fall into six complementation groups (sld1 to -6). In this study, we characterized SLD2, encoding an essential 52-kDa protein. High-copy SLD2 suppressed the thermosensitive growth defect caused by dpb11-1. Conversely, high-copy DPB11 suppressed the temperature-sensitive growth defect caused by sld2-6. The interaction between Sld2 and Dpb11 was detected in a two-hybrid assay. This interaction was evident at 25°C but not at 34°C when Sld2-6 or Dpb11-1 replaced its wild-type protein. No interaction between Sld2-6 and Dpb11-1 could be detected even at 25°C. Immunoprecipitation experiments confirmed that Dpb11 physically interacts with Sld2. sld2-6 cells were defective in DNA replication at the restrictive temperature, as were dpb11-1 cells. Further, in dpb11-1 and sld2-6 cells, the bubble-shaped replication intermediates formed in the region of the autonomously replicating sequence reduced quickly after a temperature shift. These results strongly suggest the involvement of the Dpb11-Sld2 complex in a step close to the initiation of DNA replication.
Eukaryotic chromosomal DNA is replicated exactly once in the S phase of the cell cycle. This is regulated mainly in the initiation step of DNA replication. In Saccharomyces cerevisiae, chromosomal DNA replication is initiated at a restricted region, the autonomously replicating sequence (ARS) (reviewed in references 14 and 49). The six-subunit origin recognition complex binds the ARS throughout the cell cycle (9, 16), and the prereplicative complexes (pre-RC) assemble on the ARS at the end of mitosis (16). Six Mcm family proteins, Mcm2 to -7, which have a conserved amino acid sequence and interact with one another to form large complexes (32, 53), are the components of the pre-RC. Their loading onto the ARS depends on the origin recognition complex and Cdc6 protein (2, 18, 34, 52). Cdc45, which interacts with the Mcm proteins, also functions in the initiation of chromosomal DNA replication (2, 15, 24, 26, 28, 36, 40, 41, 56, 57). For the initiation of DNA replication, Cdc28/Clb5 or Cdc28/Clb6 and Cdc7/Dbf4 protein kinases are required and phosphorylate the pre-RC components (17, 33, 42).
It has been suggested that at the G1/S phase boundary, DNA replication enzymes, including DNA polymerases, are recruited to the pre-RC to initiate both leading- and lagging-strand synthesis (2). In S. cerevisiae, three DNA polymerases, I (α), II (ɛ), and III (δ), are known to be essential for chromosomal DNA replication (reviewed in reference 51). DNA polymerase I (Pol I) is the best candidate for initiation of synthesis on both strands because it associates with the DNA primase complex. That is, DNA primase synthesizes RNA, and Pol I synthesizes a short DNA strand by using the RNA primer. Subsequently, Pol II and Pol III can elongate the DNA strand by using the short DNA fragment as a primer.
Pol II is purified as a complex of 256-, 80-, 34-, 30-, and 29-kDa polypeptides (22); the 256-kDa subunit is the catalytic subunit, but the functions of the other subunits are not yet known. The 256- and 80-kDa subunits are encoded by POL2 and DPB2, respectively, and are essential for chromosomal DNA replication (3, 5, 38). Because loss of the 34- and 30-kDa subunits, both of which are encoded by DPB3, increased the spontaneous mutation frequency, it has been suggested that Dpb3 is required for fidelity of DNA synthesis (4).
To identify factors interacting with Pol II, we isolated a multicopy suppressor, DPB11, of dpb2-1 (6). High-copy DPB11 also suppressed the growth defect caused by pol2-11 and pol2-12 mutations, which lie in the region corresponding to the C-terminal domain of Pol2 (6). The C-terminal domain of Pol2 is important for holding other subunits (38), and it has been suggested that this domain plays a role in the function of the S phase checkpoint (39). The amino acid sequence of Dpb11 is similar to that of Cut5/Rad4 of Schizosaccharomyces pombe, which is required for onset of S phase and the cell cycle checkpoint (44–46). Both the Dpb11 and Cut5/Rad4 proteins have two pairs of BRCT repeats, which have been suggested to be a domain for interaction between proteins (11, 13). The phenotype of the thermosensitive dpb11-1 mutant and genetic evidence suggest that Dpb11 interacts with the Pol II complex and is required for DNA replication and the S phase checkpoint (6).
To further understand the Dpb11 function, we have tried to identify factors interacting with Dpb11 by isolation of sld (synthetically lethal with dpb11-1) mutants. So far, we have been able to isolate six complementation groups of sld mutants. In this paper, we describe a new gene, SLD2, that is essential for cell growth. Genetic and biochemical analyses of the SLD2 gene suggested that complex formation between Dpb11 and Sld2 is essential for chromosomal DNA replication.
MATERIALS AND METHODS
Microorganisms.
Yeast strains are listed in Table 1. Escherichia coli DH5α (47) was used for plasmid propagation.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| W303-1A/B | MATa/α diploid, cross of W303-1A and W303-1B | R. Rothstein |
| HTY2-1 | MATα ade2 ade3 his3 leu2 trp1 ura3 | K. Tanaka |
| YHA410 | MATa ade1 bar1Δ his2 leu2 trp1 ura3 | |
| YHA411 | YHA410 dpb11-1 | |
| YYK1 | MATα dpb11-1 ade2 ade3 leu2 trp1 ura3 | Segregant from HTY2-1 × YHA411 |
| YYK2 | YYK1[pYK1] | |
| YHA400 | MATa ade5-1 his7 leu2-3,112 trp1-289 ura3-52 dpb11Δ::LEU2[YEp195DPB11] | |
| YYK3 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sld2Δ::LEU2[YEp195SLD2] | |
| YYK4 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sld2Δ::LEU2[YEp195SLD2] | |
| YYK5 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sld2Δ::LEU2[YCp22sld2-6] | |
| YYK6 | MATa ade2-1 bar1Δ::URA3 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 sld2Δ::LEU2[YCp22SLD2] | |
| YYK7 | YYK5 bar1Δ::URA3 | |
| SS111-2-11 | MATa pol2-11 ade2-101 can1 his3 gal2 trp1-289 tyr1 ura3-52 | J. Campbell |
| YHA9 | MATa dpb2-1 ade5-1 his3-Δ200 trp1-289 ura3-52 can1 | |
| L40 | MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ | |
| CB001 | MATa leu2 trp1 ura3 prb pep4::URA3 |
Plasmids.
pYK1 and YEp181-DPB11 were constructed by subcloning the 3.0-kb SalI DPB11 DNA fragment (6) into the SalI site of YEp352ADE3 (obtained from K. Tanaka, Osaka University) or YEplac181 (20), respectively. YCp111DPB3 was constructed by subcloning the 2.3-kb HaeII DPB3 fragment (4), after filling in with T4 DNA polymerase, into the SmaI site of YCplac111 (20). pBS(SK+)-SLD2, YCp111SLD2, YCp22SLD2, YCp33SLD2, and YEp195SLD2 were constructed by subcloning the 3.9-kb HindIII SLD2 DNA fragment (Fig. 1A) into the HindIII site of pBS(SK+), YCplac111, YCplac22, YCplac33, or YEplac195 (20), respectively. The YEp213-based chromosomal DNA library (25) was used for cloning the SLD genes.
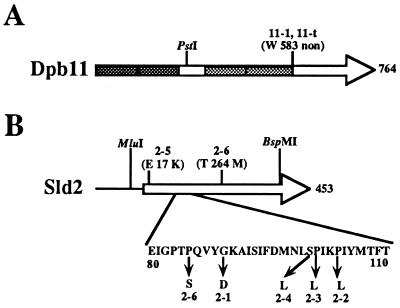
FIG. 1.
Locations of mutation sites in the DPB11 and SLD2 genes. The amino acid substitution is shown for each mutant allele. (A) A mutation site in the DPB11 gene. In the dpb11-1 allele, the G at nucleotide 1748 (nucleotide 1 is A in the first ATG of the ORF) was replaced by A. 11-t indicates dpb11-t, which was constructed by deleting the region corresponding to C-terminal portion after the stop codon at nucleotide 1784. Each pair of shaded boxes indicates the repeated BRCT (13) sequences. (B) Mutation sites in the SLD2 gene. Nucleotide substitutions occurring in each mutant allele are as follows: A for G at nucleotide 266 (nucleotide 1 is A in the first ATG of the ORF) in sld2-1, T for C at nucleotide 311 in sld2-2, T for C at nucleotide 302 in sld2-3, T for C at nucleotide 299 in sld2-4, A for G at nucleotide 49 in sld2-5, and T for C at nucleotides 253 and 791 in sld2-6.
Genetic methods and manipulation of DNA.
General genetic methods were as described previously (48). YYK2 was treated with 1% ethyl methanesulfate for 60 min in 0.2 M phosphate buffer (pH 8.0) containing 2% glucose before being plated on YPD plates. Manipulation of DNA was as described by Sambrook et al. (47).
Disruption of the SLD2 gene.
The genomic sequence between the MluI and BspMI sites was removed from the pBS(SK+)SLD2 plasmid and replaced by the 1.6-kb LEU2 fragment isolated from YDp-L (10) (Fig. 1B). The LEU2-disrupted genomic fragment was subsequently removed from the plasmid and introduced into the W303 diploid. Southern blot analysis was performed on the Leu+ transformants to confirm that one copy of the endogenous SLD2 gene was successfully disrupted.
Isolation of the sld2-6 allele.
The diploid strain containing the disrupted SLD2 gene was transformed with YEp195SLD2, and the resultant Ura+ transformants were sporulated and dissected. One Ura+ Leu+ segregant, YYK3, was used for further study. YCp22SLD2 was treated with hydroxylamine as described previously (3) and used for transformation of YYK3. About 6,000 transformants grown at 25°C on Ura− Trp− plates were replica plated to one set of 5-fluoro-orotic acid (5-FOA) plates and incubated at 25 and 37°C. One clone showed temperature-sensitive growth. The plasmid (YCpsld2-6) was recovered and retransformed into YYK3 to confirm the temperature-sensitive phenotype. The resultant strain, YYK5, showed thermosensitivity and was used for further analysis.
Determination of mutation sites.
The dpb11-1 mutation site was determined by sequencing YCp22dpb11-1, which had been isolated as a thermosensitive allele by plasmid shuffling (6). The sld2-6 mutation site was determined by sequencing YCp22sld2-6, which had been isolated as a thermosensitive allele by plasmid shuffling. To identify the sld2-1, -2, -3, -4, and -5 mutation sites, each mutation allele was amplified by PCR with genomic DNA isolated from the respective mutant as the template and then sequenced. All sequencing was performed with customized primers by using the PRISM dye terminator cycle sequencing ready reaction kit (ABI) according to the manufacturer’s instructions.
Synchronization of yeast cells.
In order to facilitate the synchronization of cells, bar1 derivatives were constructed by replacing the endogenous gene with a URA3 insertion mutant allele by the one-step gene replacement method (43). Cultures of yeast cells were grown to log phase (2 × 106 to 3 × 106 cells/ml) and then arrested with 30 ng of α-factor (Peptide Institute Inc., Osaka, Japan) per ml at 25°C for 4 h. Thereafter, α-factor was removed by centrifuging the cells at low speed. The cells were then resuspended in fresh YPD medium containing 100 μg of pronase per ml at various temperatures.
Measurement of DNA content.
The DNA concentration was measured as described previously (50). Cells were arrested with α-factor and released at 37°C. At each time point, 108 cells were collected, washed with 1 ml of water, and resuspended in 1 ml of ice-cold 5% trichloroacetic acid. Cells were incubated on ice for 15 min and washed with 1 ml of ice-cold water three times. Thereafter, cells were resuspended in 100 μl of water and mixed with 200 μl of diphenylamine reagent (1.5% [wt/vol] diphenylamine and 2.75% [vol/vol] sulfuric acid in glacial acetic acid) and incubated at 100°C for 15 min. The absorbance at 595 nm was measured, and the amount of DNA was calculated with reference to the absorbance of standard DNA.
Two-hybrid analysis.
PCR-amplified DNA from the SLD2 or sld2-6 gene was cloned into pBTM116 (8) to allow production of LexA-Sld2 or LexA–Sld2-6 fusion protein. PCR-amplified DNA from the DPB11 or dpb11-t gene was cloned into pACT2 (7) to allow production of Gal4-Dpb11 or Gal4–Dpb11-t fusion protein. The dpb11-t gene was amplified by PCR, with one of the oligonucleotides containing a stop codon corresponding to Trp583. Plasmids were introduced into yeast strain L40, and the transformants were selected on medium lacking tryptophan, uracil, and leucine. Colonies thus isolated were patched onto medium lacking the same amino acids. When grown, the yeast cells were replicated to filter paper (Whatman no. 50). The filter paper was frozen in liquid nitrogen, and the color was developed with Z buffer (10 mM KCl, 1 mM MgSO4, Na-PO4, pH 7.0) containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside).
Immunoprecipitation.
Immunoprecipitation was carried out as described previously (28). Cells were harvested by centrifugation, washed once in ice-cold water, and finally resuspended in 0.5 ml of lysis buffer (10% glycerol, 50 mM Tris [pH 7.5], 1 mM EDTA, 0.15 M NaCl, 0.2% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, complete protein inhibitor [Boehringer Mannheim]). Cells were broken by vortexing with glass beads. Lysates were clarified by centrifugation in a microcentrifuge for 10 min at 4°C. Protein extracts (0.5 ml; diluted to 2 mg/ml in cold lysis buffer) were adsorbed against 50 μl (50% [vol/vol] slurry) of protein A-agarose beads (Sigma) by mixing on a rotating wheel for 30 min at 4°C. The beads were pelleted, and the supernatant was recovered and mixed for 2 h with monoclonal antibody 12CA5 followed by a further 2-h incubation with 50 μl of fresh protein A-agarose beads. The immune complex was recovered after the beads were washed five times with 1 ml of cold lysis buffer. The precipitated samples were boiled for 5 min in the presence of sodium dodecyl sulfate (SDS) and 2-mercaptoethanol and subjected to SDS-polyacrylamide gel electrophoresis. The separated proteins were transferred to Immobilon P filters (Millipore) by electroblotting. The filters were blocked in 5% skim milk powder in TTBS (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 0.05% Tween 20) for 1 h, probed with primary antibodies for 2 h, and then incubated with alkaline phosphatase-conjugated secondary antibodies for 1 h. Filters were washed once for 15 min and three times for 5 min each in TTBS after both the primary and secondary antibody incubations. Detection was with an Alkaline Phosphatase Conjugate Substrate Kit (Bio-Rad).
2D gel analysis.
Yeast cell samples were mixed with an equal volume of toluene stop solution (95% ethanol, 3% toluene, 20 mM Tris-HCl, pH 7.5), followed immediately by the addition of 0.5 M EDTA to a final concentration of 10 mM (31). Total DNA was prepared by CsCl gradient centrifugation and digested with PstI. The replication intermediates were enriched by benzoylated naphthoylated DEAE (BND)-cellulose chromatography (29). Neutral-neutral two-dimensional (2D) gels were run, blotted, and probed (12).
RESULTS
Isolation of synthetically lethal mutations with dpb11-1.
The combination of mutations in genes coding for interacting proteins often results in lethality at the permissive condition for one of the single mutations (21). Thus, we screened for mutations which are lethal in combination with the dpb11-1 mutation to identify factors interacting with Dpb11. The YYK2 (dpb11-1 ade2 ade3 ura3) strain harboring pYK1 (DPB11 ADE3 URA3) was mutagenized and grown on YPD plates at 25°C. If YYK2 loses pYK1 during growth, the colonies show a white or pinkish color. However, cells having an additional sld (synthetically lethal with dpb11-1) mutation will form red colonies, because cells cannot grow without pYK1.
Among 50,000 colonies, 14 mutant clones which exhibited red colonies were obtained. These mutant strains were crossed with YHA400 {Δdpb11 ura3[YEp195 DPB11 (URA3)]}, and the resultant diploids were streaked onto 5-FOA plates and incubated at 25°C. Three diploid clones could not grow on 5-FOA plates, suggesting that a mutation lethal at the permissive temperature occurred in the DPB11 gene. One diploid clone grew very slowly on the 5-FOA plate, although the mutation was recessive. This clone has not been analyzed further. The remaining 10 clones were crossed with a wild-type strain and sporulated, and the resultant asci were dissected. The segregation patterns of the spore clones showed that each mutant strain has a single sld mutation. These sld mutations did not confer any detectable growth defect at 16, 25, 30, or 37°C. This result suggested that the synthetic lethality caused by the sld and dpb11-1 mutations is not from the simple addition of two defective mutations and is caused by a defect in a specific interaction.
To clone the SLD genes, we introduced a chromosomal DNA library into each mutant clone and replica plated the transformants onto 5-FOA plates to select colonies which could lose the DPB11 plasmid and retain the SLD gene. We isolated five different DNA clones from the mutant strains. The restriction maps of these five DNA clones differed from that of DPB11. The region of each DNA clone essential for complementation was delimited and sequenced from both ends. The S. cerevisiae genomic database revealed one open reading frame (ORF) for each DNA clone. We named these ORFs SLD1, -2, -3, -4, and -5. To confirm that each mutation occurred in the corresponding SLD gene, we mapped the mutation site of the SLD gene on the chromosome. One mutant strain could not be complemented by those SLD genes, and we named the sld mutation in this strain sld6-1. Thus, there were six complementation groups (Table 2).
TABLE 2.
sld complementation groups
| Mutation | Allelic mutation | Mutation allele(s) | Essentiala |
|---|---|---|---|
| sld1 | dpb3 | -1 | − |
| sld2 | -1, -2, -3, -4, -5 | + | |
| sld3 | -1 | + | |
| sld4 | cdc45 | -1 | + |
| sld5 | -1 | + | |
| sld6 | -1 | NDb |
+ and −, essential or nonessential for the cell growth, respectively.
ND, not determined.
SLD1 was identical to DPB3, which encodes the third-largest subunit of Pol II (4), and SLD4 was identical to CDC45, which is required for the initiation of chromosomal DNA replication (24, 26, 28, 41, 56, 57). SLD2, SLD3, and SLD5 were new genes. In this study, we further analyzed the SLD2 gene. The analysis of the SLD3 and SLD5 genes will be described elsewhere.
Sld2 is essential for cell growth.
The SLD2 gene corresponds to the YKL108w ORF (19), which encodes a 52-kDa protein. The deduced amino acid sequence has no significant similarities with the known proteins in GenBank.
One of the copies of the SLD2 gene was disrupted in a diploid strain by replacement with the LEU2 gene, and the resultant strain was sporulated and dissected. All of 20 tetrads examined showed two viable and two lethal spores. All viable spores clones were Leu−, indicating that the SLD2 gene is essential for cell growth. Furthermore, microscopic observation revealed that all of the inviable spores exhibited a dumbbell shape.
To understand the function of SLD2, we isolated a thermosensitive sld2-6 mutation by the plasmid-shuffling method (see Materials and Methods). The sld2-6 cells could grow at 25°C but not at 34°C. At the restrictive temperature, 80% of the sld2-6 cells arrested with a dumbbell shape with the nucleus adjacent to the isthmus between mother and daughter cells (data not shown), which is the typical terminal morphology for mutants defective in DNA replication.
Mutation sites of dpb11 and sld2.
To localize the region important for the interaction between Dpb11 and Sld2, the mutation sites of the dpb11-1 and sld2-1, -2, -3, -4, -5, and -6 alleles were determined as described in Materials and Methods. The dpb11-1 mutation was found at nucleotide 1748 (nucleotide 1 is the A in the first ATG of the ORF) and was a G-to-A alteration which changed a tryptophan codon to a nonsense codon (Fig. 1A). Thus, dpb11-1 encodes a truncated 66-kDa protein. To confirm that the truncation of Dpb11 caused the thermosensitive growth phenotype, the region corresponding to the C-terminal portion of the protein was deleted from the gene beyond the stop codon at nucleotide 1784 (dpb11-t) and transferred to the chromosome. dpb11-t cells showed a thermosensitive growth phenotype, as did dpb11-1 cells.
The sld2-1, -2, -3, -4, and -6 mutations clustered in a region corresponding to the N-terminal region of the protein (amino acids [aa] 85 to 104) (Fig. 1B). Thus, this region might be important for the interaction between Dpb11 and Sld2. However, the amino acid sequence in this region does not have any characteristic features. The sld2-6 allele has two C-to-T alterations occurring at nucleotide 253 (corresponding to a change from P to S at aa 85) and at nucleotide 791 (corresponding to a change from T to M at aa 264). We do not know which mutation in the sld2-6 mutant is responsible for the thermosensitivity.
Genetic interactions between Sld2 and Dpb11.
The sld2-1, -2, -3, -4, and -5 mutations were isolated as those which cannot be combined with the dpb11-1 mutation at 25°C, suggesting that Dpb11 interacts with Sld2. A mutation occurring in one subunit of a complex is often suppressed by increased dosage of another subunit. The SLD2 gene on a high-copy-number plasmid was introduced into a dpb11-1 mutant, and high-copy DPB11 was introduced into the newly isolated sld2-6 mutant cells. Both strains harboring YEpDPB11 or YEpSLD2 could grow at 35.5°C, while they cannot grow without those plasmids (Fig. 2A and B). The suppression was dependent on the copy number of these genes, since YCpDPB11 and YCpSLD2 suppressed the growth defect more weakly than YEpDPB11 and YEpSLD2 (Fig. 2A and B). Thus, Dpb11 interacts with Sld2 genetically.
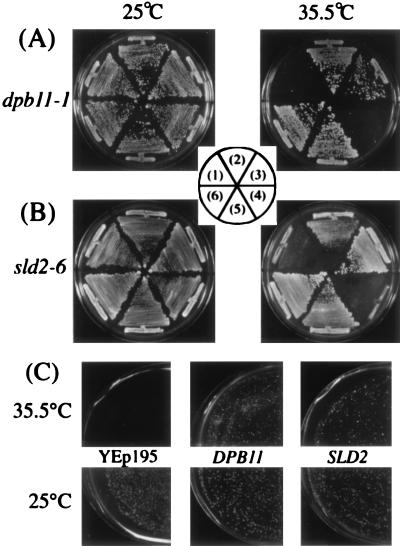
FIG. 2.
Suppression of temperature-sensitive growth of dpb11-1, sld2-6, and pol2-11 mutants. (A) YHA411 (dpb11-1) cells harboring YCplac22 (1), YCp22DPB11 (2), YCp22SLD2 (3), YEplac195 (4), YEp195DPB11 (5), and YEp195SLD2 (6) plasmids were streaked onto YPD plates and incubated at the indicated temperature for 3 days. (B) YYK5 (sld2-6) cells harboring YCplac33 (1), YCp33DPB11 (2), YCp33SLD2 (3), YEplac195 (4), YEp195DPB11 (5), and YEp195SLD2 (6) plasmids were streaked onto YPD plates and incubated at the indicated temperature for 3 days. (C) SS111-2-1 (pol2-11) cells harboring YEp195SLD2, YEp195DPB11, or YEplac195 were spread on the plates after appropriate dilution and incubated at the indicated temperature for 3 days.
We previously isolated DPB11 as a multicopy suppressor of the thermosensitive growth phenotype of dpb2-1 cells, whose mutation is in the second largest subunit of Pol II. This gene also suppresses the phenotype of pol2-11 cells, whose mutation is in the largest subunit of Pol II. Furthermore, we showed that dpb11-1 is synthetically lethal with pol2-11 (6). If Sld2 forms a complex with Dpb11, high-copy SLD2 might also suppress the growth defect of dpb2-1 or pol2-11. As shown in Fig. 2C, high-copy SLD2 suppressed the growth defect of pol2-11 weakly but not that of dpb2-1 (data not shown). To investigate the synthetic lethality between sld2-6 and a mutation in POL2, YYK4 was crossed with a pol2-11 strain and sporulated, and the asci were dissected. The resultant strain, bearing pol2-11, sld2Δ::LEU2, and YEplac195SLD2, was transformed with YCpsld2-6, and the transformants were streaked onto 5-FOA plates. No transformant could grow on 5-FOA plates, indicating that an sld2-6 mutation cannot be combined with a pol2-11 mutation. Taken together, these results suggest that a complex of Dpb11 and Sld2 interacts with Pol II.
Physical interaction between Sld2 and Dpb11.
To determine whether Dpb11 physically interacts with Sld2, two-hybrid analysis was employed. The ORF of SLD2 was fused to the LexA binding domain of pBTM116 (8), and the ORF of DPB11 was fused to the Gal4 activation domain of pACT2 (7). The resultant two plasmids could complement the thermosensitive growth of dpb11-1 or Δsld2 cells, respectively, indicating that the fused gene in each case is functional. They were introduced into L40 cells, and expression of the lacZ gene reporter was examined. As shown in Fig. 3, the lacZ gene was expressed, suggesting that Sld2 interacts with Dpb11.
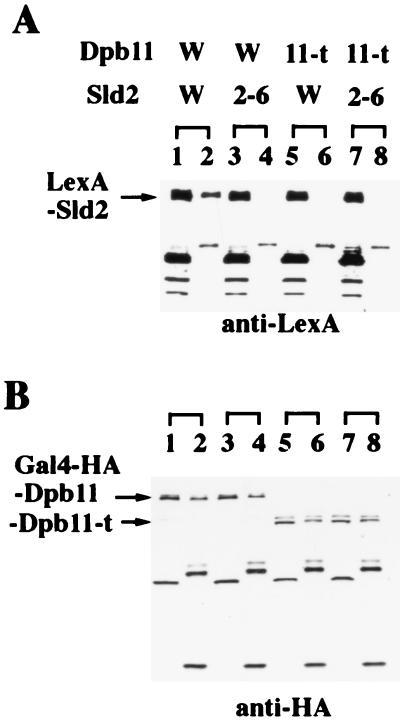
FIG. 3.
Physical interaction between Dpb11 and Sld2. V, Sld2, and Sld2-6 in the column headed LexABD denote plasmids that express LexA, LexA-Sld2, and LexA-Sld2-6 proteins, respectively. V, Dpb11, and Dpb11-t in the column headed Gal4AD denote plasmids that express Gal4, Gal4-Dpb11, and Gal4-Dpb11-t proteins, respectively. Transformants of L40 carrying each pair of the plasmids were assayed for β-galactosidase activity by colony color with X-Gal.
dpb11-t and sld2-6 were cloned on pACT2 and pBTM116, respectively. L40 cells harboring various combinations of the pACT2 and pBTM116 derivatives were constructed and incubated at 25 and 34°C. As shown in Fig. 3, the expression level of the lacZ gene in cells harboring the wild-type DPB11 and sld2-6 or harboring dpb11-t and SLD2 was almost the same as that in cells harboring the wild-type pair at 25°C but was reduced at 34°C. Moreover, cells harboring dpb11-t and sld2-6 did not express the lacZ gene even at 25°C. These results suggest that Sld2 and Dpb11 interact physically and that the mutations in SLD2 and DPB11 reduce the interaction.
To examine the interaction between Sld2 and Dpb11 more directly, immunoprecipitation was employed. Cells harboring the LexA-Sld2 and Gal4-HA-Dpb11 plasmids were lysed, and the proteins were precipitated with a monoclonal antibody against hemagglutinin (HA). The precipitates were analyzed by SDS-polyacrylamide gel electrophoresis followed by Western blotting with antibodies against HA or LexA. As shown in Fig. 4, the HA antibody precipitated Sld2 as well as Dpb11. Cells harboring Gal4–HA–Dpb11-t and LexA-Sld2 plasmids were lysed and mixed with HA antibody. The HA antibody precipitated Dbp11-t but not Sld2. Similarly, Sld2-6 in cells harboring Gal4-HA-Dpb11 and LexA–Sld2-6 was not coprecipitated with Dpb11 by HA antibody.
FIG. 4.
Coimmunoprecipitation of Dpb11 and Sld2. Dpb11 W and Dpb11 11-t, plasmids that express Gal4-HA-Dpb11 and Gal4–HA–Dpb11-t truncated proteins, respectively. Sld2 W and Sld2 2-6, plasmids that express LexA-Sld2 and LexA–Sld2-6 mutant proteins, respectively. Proteins extracted from yeast strain CB001 expressing each pair of the plasmids were precipitated with an anti-HA monoclonal antibody (12CA5). The protein extracts and the precipitates were analyzed for the presence of LexA-Sld2 or LexA–Sld2-6 probed with anti-LexA serum (A) and for the presence of Gal4-HA-Dpb11 or Gal4–HA–Dpb11-t probed with anti-HA monoclonal antibody (12CA5) (B). In lanes 1, 3, 5, and 7, the soluble protein extracts before immunoprecipitation were applied. In lanes 2, 4, 6, and 8, the immunoprecipitates were applied.
These results suggest that temperature-sensitive mutations are defective in the interaction between Dpb11 and Sld2 and further suggest that the synthetic lethality of dpb11-1 with sld2-6 is caused by the lack of interaction between Dpb11 and Sld2.
sld2-6 cells are defective in DNA replication, as are dpb11-1 cells.
Previously, we showed by flow cytometric analysis that dpb11-1 cells are defective in the progression of S phase at the restrictive temperature. We further analyzed DNA replication in dpb11-1 and sld2-6 cells. dpb11-1 and sld2-6 cells were arrested with α-factor and released at 37°C. Total DNA was measured by the diphenylamine method (50) after the temperature shift-up. dpb11-1 and sld2-6 cells did not increase their DNA contents at 37°C, unlike wild-type cells, although mutant cells started budding at 37°C in a manner similar to that for the wild type (Fig. 5), suggesting that Dpb11 and Sld2 are required for DNA replication.
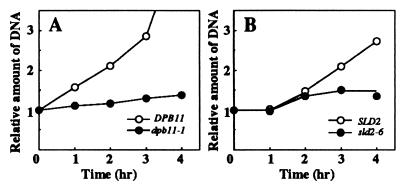
FIG. 5.
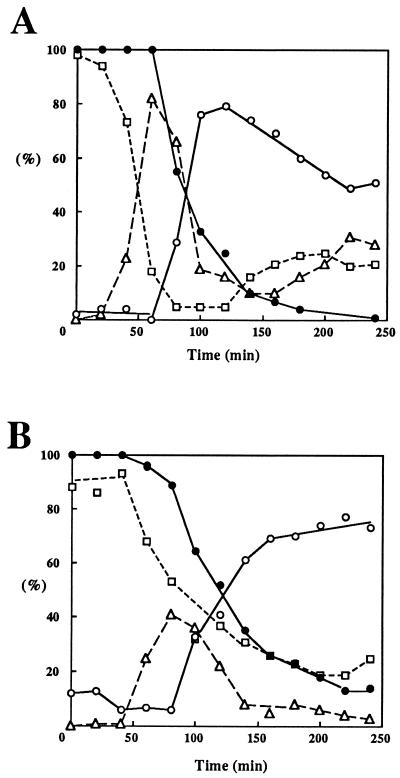
DNA replication in dpb11-1 and sld2-6 cells. (A) YHA410 (DPB11) and YHA411 (dpb11-1) cells were arrested with α-factor at 25°C and then released at 37°C as described in Materials and Methods. Aliquots were withdrawn at the indicated times to measure total DNA by the diphenylamine method (50). (B) YYK6 (SLD2) and YYK7 (sld2-6) cells were synchronized, and the total DNA at 37°C was measured as for panel A. The delay of DNA synthesis in YYK6 and YYK7 is probably caused by delayed release from α-factor (see Fig. 6).
dpb11-1 cells lose viability quickly after a temperature shift-up (6). To determine the point at which the cells start losing viability, the viability of synchronized cells was examined. As shown in Fig. 6, both dpb11-1 and sld2-6 cells started losing viability when cells having a large bud appeared. These results suggest that both mutant cells lose viability during S phase.
FIG. 6.
Viability of dpb11-1 and sld2-6 cells after release from α-factor arrest at the restrictive temperature. YHA411 (dpb11-1) (A) and YKK4 (sld2-6) (B) cells were arrested by α-factor treatment at 25°C and then released at 37°C. Aliquots were withdrawn at the indicated times to determine cell morphology and cell number. Cells were spread on YPD plates and incubated at 25°C for 3 days. Colonies grown on YPD plates were scored. Symbols: □, cells with no bud; ▵, cells with a small bud; ○, cells with a large bud; •, viability of cells.
The fact that dpb11-1 and sld2-6 mutant cells showed similar defects in DNA replication and loss of viability at the same point during the cell cycle is consistent with the conclusion that Dpb11 and Sld2 form a complex and participate in the same process.
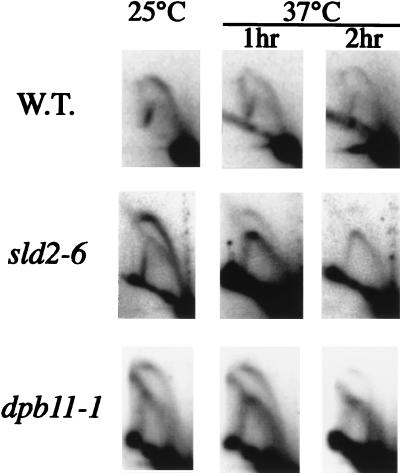
To examine the DNA replication defect in dpb11-1 and sld2-6 cells in detail, formation of replication intermediates in an active replicator, ARS306 (56), was analyzed by the 2D gel method (12). dpb11-1 and sld2-6 cells, as well as wild-type cells, were grown to log phase at the permissive temperature and were shifted to the restrictive temperature for 1 or 2 h before harvest. At both 25 and 37°C, the wild-type strain gave a clear bubble arc signal, indicating that replication initiates in the ARS306 region and that forks pass through the PstI site used for digestion of chromosomal DNA (Fig. 7). However, a complete fork arc in addition to a bubble arc was observed in both dpb11-1 and sld2-6 cells even at 25°C. The bubble arc signals further decreased after the shift to 37°C. This can be explained by the reduced amount of DNA synthesis initiated from ARS306 and the replication forks moved in from other replication origins. Similar results were also obtained for ARS501 and ARS1 (data not shown). Furthermore, no replication intermediate was observed when dpb11-1 and sld2-6 cells were arrested with α-factor and released at the restrictive temperature (data not shown). These results showed that Dpb11 and Sld2 work together in a step close to the initiation of DNA replication.
FIG. 7.
Neutral-neutral 2D gel analysis of the chromosomal ARS306 locus in wild-type (W.T.) (YHA410), dpb11-1 (YHA411), and sld2-6 (YYK5) strains. Cells were grown and harvested at 25°C or shifted to 37°C for 1 or 2 h prior to harvest. DNA was digested with PstI and probed with the 0.5-kb genomic fragment containing ARS306 (55).
To further examine the function of Dpb11 and Sld2 in the progression of S phase after DNA replication initiates, we tested whether Dpb11 and Sld2 execute any function after a hydroxyurea (HU) block, because HU is an inhibitor of ribonucleotide reductase that causes replication forks to stall. Wild-type or mutant cells were arrested with α-factor and released to YPD medium containing 0.2 M HU at 25°C. Cells were then released from the HU block at 37°C, and nuclear division was examined under an epifluorescence microscope after nuclei were stained with 4′,6-diamino-2-phenylindole. As shown in Table 3, most of the sld2-6 and dpb11-1 cells showed one nucleus at 80 min after release from HU, while more than 50% of wild-type cells had two nuclei. Further incubation of sld2-6 cells did not increase the population of cells having two nuclei. In the case of dpb11-1 cells, the nucleus divided abnormally because of the loss of the checkpoint, as described previously (6). Since dpb11-1 and sld2-6 cells arrested in M phase by nocodazole and released at the restrictive temperature could divide, this result suggests that Dpb11 and Sld2 function in the S phase after the HU block.
TABLE 3.
Nuclear division after release from an HU block
| Strain | % of cells having two nucleia at:
|
||
|---|---|---|---|
| 0 min | 80 min | 240 min | |
| YYK6 | 2 | 52 | NDb |
| YYK7 (sld2-6) | 8 | 6 | 5 |
| YHA410 | 0 | 58 | ND |
| YHA411 (dpb11-1) | 5 | 8 | 52c |
Cells were synchronized with α-factor, released to YPD medium containing 0.2 M HU at 25°C for 90 min, and then transferred to 37°C. After 30 min of incubation, cells were washed and released from the HU block at 37°C. At the indicated time after release from HU, nuclei were observed.
ND, not determined.
Most of these cells have two nuclei with the abnormal morphology described previously (6).
The transcript level of SLD2 fluctuates and peaks at the G1/S boundary.
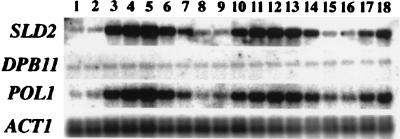
The upstream DNA sequence of the SLD2 gene has one MluI cell cycle box, which functions in cell cycle-dependent transcription (1, 30). Therefore, the transcript level of SLD2 during the cell cycle was examined. The cells were synchronized by elutriation (54), and RNA was extracted at various times during the cell cycle.
As shown in Fig. 8, the transcript level of SLD2 fluctuated with the cell cycle and peaked at the G1/S phase boundary, similarly to the transcript level of POL1, while the DPB11 transcript level was constant. The increase of the transcript level of SLD2 at the G1/S boundary is consistent with an important function at this point in the cell cycle.
FIG. 8.
The transcript level of the SLD2 gene is regulated during the cell cycle and peaks at the G1/S boundary. G1 cells obtained by centrifugal elutriation were released into fresh medium. Samples were withdrawn every 20 min, and total RNA was extracted for Northern blot analysis. The blot was probed with 32P-labelled fragments of the SLD2, DPB11, POL1, and ACT1 genes.
DISCUSSION
Several lines of evidence suggest that Dpb11 and Sld2 form a complex and are required for chromosomal DNA replication in S. cerevisiae. First, sld2 mutations were synthetically lethal with dpb11-1. Second, the growth defect of sld2-6 cells is suppressed by high-copy DPB11, and the growth defect of dpb11-1 cells is suppressed by high-copy SLD2. Third, two-hybrid analysis showed an interaction between Dpb11 and Sld2. Thermosensitive Dpb11-1 and Sld2-6 are defective in the interaction, and this interaction was dependent on the temperature. Fourth, LexA-Sld2 was coprecipitated with HA-Dpb11 by antibody against HA. Fifth, the phenotypes of dpb11 and sld2 are similar.
The mutations that occurred in SLD2 were localized to a region corresponding to a 20-aa stretch, a region that must be important for the interaction of Sld2 with Dpb11. Although we could not find any significant homology in Sld2 with known protein sequences, there is a target site, SPIK, for phosphorylation by Cdc28 kinase (37). In Dbp11, deletion of C-terminal amino acids abolishes the interaction with Sld2 at the restrictive temperature. Since the truncated Dpb11-1 still has two pairs of BRCT repeats, the removal of the C terminus may change the conformation of Dpb11 at high temperature. Although we have not determined the region in Dpb11 that is important for the interaction with Sld2, BRCT repeats are candidates for the interaction domain.
The Dbp11-Sld2 complex is required for DNA synthesis, because dpb11-1 and sld2-6 mutants are defective in DNA replication at the restrictive temperature. dpb11-1 and sld2-6 cells showed reduced signal intensity of a bubble arc in 2D gel electrophoresis, suggesting that Dpb11-Sld2 functions in an early step of DNA replication. Although the 2D gel analysis cannot discriminate between initiation and an early step of elongation, Dpb11 seems to play a role in an early step of elongation by the following reasons. First, dpb11-1 cells showed a stability of ARS plasmids similar to that of wild-type cells (our unpublished observation), whereas ARS plasmids are unstable in cdc6, mcm, orc and cdc45 mutants, which are defective in initiation of DNA replication (23, 24, 27, 28, 35, 53, 56). Second, Dpb11 and Sld2 are required for the progression of S phase after release from an HU block (Table 3). Therefore, Dpb11-Sld2 may function in the first step of elongation by interacting with initiation proteins.
As the Dpb11-Sld2 complex also interacts with Pol II which is recruited to the pre-RC on the ARS at initiation of DNA replication (2), it is conceivable that Dpb11-Sld2 brings Pol II onto the pre-RC and thus that Dpb11-Sld2 is essential for the transition from the initiation to the elongation step in DNA replication. So far, we have not succeeded in detecting actual complexes including Dpb11 and Sld2 in cell extracts without overexpression of Dbp11 and Sld2. This may be due to low abundance and unstable properties of Dpb11 and Sld2. The actual complex may consist of Dpb11, Sld2, and other Sld proteins and may connect Pol II to Mcm proteins. Thus, it seems likely that the dpb11-1 and sld2-6 mutations disrupt this large complex and thus confer a defect in DNA replication. However, we cannot rule out other possibilities, such as that the Dpb11-Sld2 complex is directly involved in the initiation step or that they are accessary factors of Pol II.
Dpb11-Sld2 may function in the transition from the initiation to the elongation step of DNA replication and may move with Pol II in the replication fork. Thus the Dpb11-Sld2 complex may monitor any defect in DNA replication that might occur during the initiation and subsequent elongation steps. This could be important for a checkpoint function. While both dpb11-1 and sld2-6 cells lose viability quickly after a temperature shift, sld2-6 cells did not show any significant sensitivity to UV, methyl methanesulfonate, or HU, unlike other checkpoint mutants, including dpb11-1 cells. Moreover, the terminal phenotype of sld2-6 cells after 4 h at the restrictive temperature was different from that of dpb11-1 cells. After the temperature shift, the dpb11-1 cells first showed a dumbbell shape with one nucleus near the isthmus; the nucleus was divided aberrantly, and then some of cells divided (6). The sld2-6 cells arrested with a dumbbell shape and one nucleus near the isthmus, but the nucleus did not divide. This is not caused by residual activity of Sld2-6, because the SLD2 disruptant cells also showed a dumbbell shape, whereas about 30% of the DPB11 disruptant cells divided aberrantly like the dpb11-1 cells at the restrictive temperature (our unpublished observation). Because Dpb11 is still intact in sld2-6 mutant cells, Dpb11 may interact with other proteins in a checkpoint control. Dpb11-1 may also lose the interaction with other checkpoint proteins in addition to Sld2.
The transcript level of SLD2 fluctuates in the cell cycle and peaks at the G1/S boundary, whereas the transcript level of DPB11 is constant during the cell cycle (Fig. 8). Although we have not examined the amounts of Dpb11 and Sld2 proteins during the cell cycle, the amount of the Dpb11-Sld2 complex may peak at the G1/S boundary. This is consistent with a function for Dpb11-Sld2 in an early step of DNA replication.
Our previous study showed that dpb11-1 is synthetically lethal with either dpb2-1 or pol2 (6). In this study, we screened sld mutations and classified them into six complementation groups. However, we obtained no sld mutations in DPB2 or POL2. Mutations in DPB2 and POL2 were isolated by in vitro mutagenesis and the plasmid-shuffling method (3, 5). Therefore, a mutation in these genes may be difficult to isolate, or it may simply be that the sld screening has not yet been saturated.
ACKNOWLEDGMENTS
We thank S. J. Elledge, K. Tanaka, N. Ogawa, and K. Shirahige for yeast strains and plasmids, H. Iwasaki and H. Shinagawa for LexA antibodies, and L. H. Johnston for critical reading of the manuscript.
This study was supported by grants from Ministry of Education, Science, Culture and Sports, Japan.
REFERENCES
- 1.Andrews B J, Mason S W. Gene expression and the cell cycle: a family affair. Science. 1993;261:1543–1544. doi: 10.1126/science.8372349. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 3.Araki H, Hamatake R K, Johnston L H, Sugino A. DPB2, the gene encoding DNA polymerase II subunit B, is required for chromosome replication in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:4601–4605. doi: 10.1073/pnas.88.11.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araki H, Hamatake R K, Morrison A, Johnson A L, Johnston L H, Sugino A. Cloning DPB3, the gene encoding the third subunit of DNA polymerase II of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:4867–4872. doi: 10.1093/nar/19.18.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki H, Ropp P A, Johnson A L, Johnston L H, Morrison A, Sugino A. DNA polymerase II, the probable homolog of mammalian DNA polymerase ɛ, replicates chromosomal DNA in the yeast Saccharomyces cerevisiae. EMBO J. 1992;11:733–740. doi: 10.1002/j.1460-2075.1992.tb05106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araki H, Leem S-H, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(ɛ) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1997;283:141–156. doi: 10.1016/s0076-6879(97)83013-3. [DOI] [PubMed] [Google Scholar]
- 8.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 9.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origin of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 10.Berben G, Dumont J, Gilliquet V, Bolle P-A, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 11.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 12.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 13.Callebaut I, Mornon J-P. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 14.Campbell J L, Newlon C S. Chromosomal DNA replication. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces cerevisiae: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 41–146. [Google Scholar]
- 15.Dalton S, Hopwood B. Characterization of Cdc47p-minichromosome maintenance complex in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley J F X, Cocker J H, Dowell S J, Harwood J, Rowley A. Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle. J Cell Sci Suppl. 1995;19:67–72. doi: 10.1242/jcs.1995.supplement_19.9. [DOI] [PubMed] [Google Scholar]
- 17.Diffley J F X. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 18.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dujon B, Alexandraki D, Andre B, Ansorge W, Baladron V, Ballesta J P G, et al. Complete DNA sequence of yeast chromosome XI. Nature. 1994;369:371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- 20.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 21.Guarente L. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 1993;9:362–366. doi: 10.1016/0168-9525(93)90042-g. [DOI] [PubMed] [Google Scholar]
- 22.Hamatake K R, Hasegawa H, Clark A B, Bebenek K, Kunkel T A, Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- 23.Hardy C F J. Characterization of an essential Orc2p-associated factor that plays a role in DNA replication. Mol Cell Biol. 1996;16:1832–1841. doi: 10.1128/mcb.16.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardy C F J. Identification of Cdc45p, an essential factor required for DNA replication. Gene. 1997;187:239–246. doi: 10.1016/s0378-1119(96)00761-5. [DOI] [PubMed] [Google Scholar]
- 25.Hasegawa H, Sakai A, Sugino A. Isolation, DNA sequence and regulation of a new cell division cycle gene from the yeast Saccharomyces cerevisiae. Yeast. 1989;5:509–524. doi: 10.1002/yea.320050610. [DOI] [PubMed] [Google Scholar]
- 26.Hennesy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 27.Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huberman J A, Spotila L D, Nawotka K A, El-Assouli S M. The in vivo replication origin of the yeast 2μm plasmid. Cell. 1987;51:473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 30.Johnston L H, Lowndes N F. Cell cycle control of DNA synthesis in budding yeast. Nucleic Acids Res. 1992;20:2403–2410. doi: 10.1093/nar/20.10.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston L H, Williamson D H. An alkaline sucrose gradient analysis of the mechanism of nuclear DNA synthesis in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1978;164:217–225. doi: 10.1007/BF00267387. [DOI] [PubMed] [Google Scholar]
- 32.Lei M, Kawasaki Y, Tye B K. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loo S, Fox C A, Rine J, Kobayashi R, Stillman B, Bell S. The origin recognition complex in silencing, cell cycle progression, and DNA replication. Mol Biol Cell. 1995;6:741–756. doi: 10.1091/mbc.6.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moir D, Stewart S E, Osmond B C, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: properties and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno S, Nurse P. Substrates for p34cdc2: in vivo veritas? Cell. 1990;61:549–551. doi: 10.1016/0092-8674(90)90463-o. [DOI] [PubMed] [Google Scholar]
- 38.Morrison A, Araki H, Clark A B, Hamatake R K, Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1991;62:1143–151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 39.Navas T A, Zhou Z, Elledge S J. DNA polymerase ɛ links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 40.Newlon C S. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- 41.Owens J C, Detweiler C S, Li J L. CDC45 is required for conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci USA. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piatti S, Böhm T, Cocker J H, Diffley J F X, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 43.Rothstein R J. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 44.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saka Y, Fantes P, Sutani T, McInerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 49.Stillman B. Initiation of chromosomal DNA replication in eukaryotes: lessons from lambda. J Biol Chem. 1994;269:7047–7050. [PubMed] [Google Scholar]
- 50.Sudden R E, Krizus A. Correction factors for the diphenylamine test for deoxyribonucleic acid in yeast. Microbios. 1985;43:233–243. [PubMed] [Google Scholar]
- 51.Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 53.Tye B-K. The MCM2-3-5 proteins: are they replication licensing factor? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 54.White J H M, Barker D G, Nurse P, Johnston L H. Periodic transcription as a means of regulating gene expression during the cell cycle: contrasting modes of expression of DNA ligase genes in budding and fission yeast. EMBO J. 1986;5:1705–1709. doi: 10.1002/j.1460-2075.1986.tb04414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu J, Newlon C S, Huberman J A. Localization of a DNA replication origin and termination zone on chromosome III of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4733–4741. doi: 10.1128/mcb.12.10.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]