Abstract
Rap1p localization factor 4 (RLF4) is a Saccharomyces cerevisiae gene that was identified in a screen for mutants that affect telomere function and alter the localization of the telomere binding protein Rap1p. In rlf4 mutants, telomeric silencing is reduced and telomere DNA tracts are shorter, indicating that RLF4 is required for both the establishment and/or maintenance of telomeric chromatin and for the control of telomere length. In this paper, we demonstrate that RLF4 is allelic to NMD2/UPF2, a gene required for the nonsense-mediated mRNA decay (NMD) pathway (Y. Cui, K. W. Hagan, S. Zhang, and S. W. Peltz, Mol. Cell. Biol. 9:423–436, 1995, and F. He and A. Jacobson, Genes Dev. 9:437–454, 1995). The NMD pathway, which requires Nmd2p/Rlf4p together with two other proteins, (Upf1p and Upf3p), targets nonsense messages for degradation in the cytoplasm by the exoribonuclease Xrn1p. Deletion of UPF1 and UPF3 caused telomere-associated defects like those caused by rlf4 mutations, implying that the NMD pathway, rather than an NMD-independent function of Nmd2p/Rlf4p, is required for telomere functions. In addition, telomere length regulation required Xrn1p but not Rat1p, a nuclear exoribonuclease with functional similarity to Xrn1p (A. W. Johnson, Mol. Cell. Biol. 17:6122–6130, 1997). In contrast, telomere-associated defects were not observed in pan2, pan3, or pan2 pan3 strains, which are defective in the intrinsic deadenylation-dependent decay of normal (as opposed to nonsense) mRNAs. Thus, loss of the NMD pathway specifically causes defects at telomeres, demonstrating a physiological requirement for the NMD pathway in normal cell functions. We propose a model in which the NMD pathway regulates the levels of specific mRNAs that are important for telomere functions.
Telomeres, the ends of linear chromosomes, have multiple functions within the cell. Telomeres stabilize chromosomes by preventing end-to-end fusions (62, 63, 78, 85). Telomerase, an enzyme necessary for the maintenance of telomeric DNA, is inactive in most somatic cells and is activated in many malignant cells (reviewed in references 3, 8, and 25). Furthermore, the lack of telomerase activity in somatic cells limits the life span of human fibroblasts (5). In addition, genes adjacent to telomeres are often packaged in an inaccessible heterochromatin-like structure (23) that results in epigenetic transcriptional repression termed telomeric silencing or telomere position effect. Telomeric DNA is composed of simple short repeats in most organisms, and telomere length is controlled by the actions of multiple gene products (reviewed in reference 58).
In the yeast Saccharomyces cerevisiae, the most abundant telomere-binding protein, repressor activator protein (Rap1p) (9, 53, 80), binds double-stranded telomere repeats and is required for both telomeric silencing (43, 83) and telomere length regulation (59, 61, 83). In wild-type cells, Rap1p localizes to a small number of foci located primarily near the nuclear periphery (21, 38), which colocalize with telomeric DNA (21). A number of mutations, including those in SIR genes, result in an altered localization of Rap1p (11) and likely reflect changes in the structure of telomeric chromatin (19, 21).
To identify additional proteins involved in telomere function, we performed a genetic screen using circular plasmids that included both telomeric and centromeric DNA (TEL+CEN plasmids). TEL+CEN plasmids are highly unstable due to antagonism between the telomere and centromere sequences (18). We selected for mutant strains in which this TEL+CEN antagonism was lost (i.e., TEL+CEN plasmids were stabilized). As a secondary screen, we analyzed the nuclear distribution of Rap1p in these mutant strains by indirect immunofluorescence microscopy. To date, we have identified six genes that, when mutated, perturb the localization of Rap1p. These genes are termed Rap1p localization factor (RLF) genes (17, 19, 40). The present paper describes the characterization of RLF4. In addition to the stabilization of TEL+CEN plasmids and the altered localization of Rap1p, rlf4 strains are defective in telomeric silencing and have shortened telomeres. Cloning of RLF4 identified rlf4-1 as a new allele of NMD2/UPF2, a gene encoding a component of the pathway required for decay of mRNAs that prematurely terminate translation (12, 28).
This unique nonsense-mediated decay (NMD) pathway targets nonsense messages for rapid degradation. Targets of NMD include mRNAs containing nonsense or frameshift mutations (24, 32, 48, 49, 54, 69), transcripts with short upstream open reading frames (ORFs) (12, 68, 70, 72), and inefficiently spliced, intron-containing RNAs that enter the cytoplasm (29, 32). Nonsense mRNAs are stabilized in cells containing suppressor tRNAs, indicating that NMD targeting is closely linked to premature translation termination (4, 24, 54, 70). The NMD pathway requires three elements: a translational arrest, cis-acting sequences within the 5′ proximal two-thirds to three-quarters of the message, and trans-acting factors such as the Upf/Nmd proteins. The Upf/Nmd proteins include Upf1p (13, 49, 71), Nmd2p/Upf2p (12, 28), and Upf3p (46, 47, 49). Because Upf1p and Nmd2p, as well as Nmd2p and Upf3p, physically interact (27) and because single and double mutations in any of the UPF genes inhibit mRNA decay to the same extent, the NMD proteins clearly operate together in the NMD pathway (2, 12).
In the present paper, we demonstrate that RLF4 is allelic to NMD2. Like nmd2/rlf4 strains, both upf1 and upf3 strains have telomeric defects. Furthermore, strains lacking Xrn1p, the cytoplasmic exoribonuclease that degrades mRNAs targeted by the NMD pathway, also have short telomeres, suggesting that NMD and UPF genes affect telomere function via Xrn1p-mediated mRNA decay. Our results indicate that wild-type telomeric chromatin function and telomere length control require the NMD pathway specifically, rather than mRNA decay in general.
MATERIALS AND METHODS
Yeast and Escherichia coli strains and plasmids.
The genotypes of the yeast strains used in this study are listed in Table 1. E. coli XL1-blue (Stratagene, Inc.) was used for all standard plasmid preparations. Yeast cultures were grown overnight at 30°C in either minimal (SDC) or rich (YPAD) medium with 2% glucose, unless otherwise stated.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| W303 Strains | ||
| YJB447 | MATα ade2-1 ura3-1 his3 leu2-3,112 trp1 can1-100 VII-L URA3-TEL | A. Lustig |
| YJB1274 | Matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 nmd2::HIS3 | 28 |
| YJB1324 | Mata ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 upf1::HIS3 | 27 |
| YJB1471 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 nmd2::HIS3 | This study |
| YJB2239 | Matα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 nmd2::HIS3/+ est1::URA3/+ VR-ADE2-TEL | This study |
| YJB2263 | Mata ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 VR-ADE2-TEL | This study |
| YJB2262 | YJB2263 nmd2::HIS3 | This study |
| YJB1680 | MATα ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 VII-L URA3-TEL | This study |
| YJB2297 | YJB1680 upf1::HIS3 | This study |
| YJB2299 | YJB1680 nmd2::HIS3 | This study |
| YJB2739 | MATa ade2-1 his3-11,15 leu2-3 trp1-1 ura3-1 can1-100 | 7 |
| YJB2735 | YJB2739 pan2::LEU2 | 7 |
| YJB2736 | YJB2739 pan3::HIS3 | 7 |
| YJB2740 | YJB2739 pan2::LEU2 pan3::HIS3 | 7 |
| YJB2827 | MATα ade2-1 his3-11,15 leu2-3 trp1-1 ura3-1 can1-100 VII-L URA3-TEL | This study |
| YJB2823 | YJB2827 pan2::LEU2 | This study |
| YJB2824 | YJB2827 pan3::HIS3 | This study |
| YJB2826 | YJB2827 pan2::LEU2 pan3::HIS3 | This study |
| S150-2B strains | ||
| YJB276 | MATa leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ [cir+] | S. Enomoto |
| YJB492 | MATα leu2-3,112 ura3-52 [trp1-289]b his3Δade2Δ VII-L URA3-TEL [cir0] | S. Enomoto |
| YJB539 | MATa leu2-3,112 ura3-52 [trp1-289] his3Δ ade2Δ rlf4-1 VII-L URA3-TEL [cir0] | This study |
| YJB1222 | YJB539 [p108] | This study |
| YJB1260 | YJB539 [pRACUT1] | This study |
| YJB1275 | YJB1260 [pRS315-NMD2] | This study |
| YJB2541 | YJB1260 [pRS315-NMD2] | This study |
| YJB1338 | MATα leu2-3,112 ura3-52 [trp1-289] his3Δ ade2Δ rlf4-1 VII-L URA3-TEL [cir0] | This study |
| YJB1593 | MATa leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ cyh2 [cir+] nmd2::HIS3 [pRACUT1] | This study |
| YJB1594 | MATα leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ cyh2 [cir+] nmd2::HIS3 [pRACUT1] | This study |
| YJB2763 | MATa leu2-3,112 ura3-52 trp1-289 his3Δ ade2Δ cyh2 [cir+] nmd2::HIS3 | This study |
| Other | ||
| YJB2793 (=CH1305) | MATa ade2 ade3 leu2 ura3 lys2 can1 Gal+ | 42 |
| YJB2794 (=RDKY1977) | MATa ade2 ade3 leu2 ura3 lys2 can1 Gal+xrn1Δ | 34 |
| YJB2796 (=FY23) | Mata ura3-52 leu2Δ1 trp1Δ63 | 1 |
| YJB2797 (=DAT1-17) | FY23 his3Δ200 rat1-1 | 1 |
| YJB2889 | Matα his4-38 SUF1-1 leu2-1,3 trp1-Δ1 lys1-1 ura3-52 | 46 |
| YJB2890 | YJB2889 upf3Δ1::TRP1 | 46 |
| YJB2891 | YJB2890 [pLS17 = pUPF3] | 46 |
| YJB2892 | YJB2890 [pRS316] | 81 |
Brackets around plasmids indicate that they are extrachromosomal. Boldface indicates relevant genotypes.
Brackets indicate that the mutation is suppressed.
The plasmid pRACUT1 contains an autonomously replicating sequence (ARS), the ADE2 gene, centromere sequence from CEN IV, URA3, and telomere repeat sequence (18). pRS315-NMD2(X-S) contains the full-length NMD2 gene on an XbaI-SalI fragment (28), cloned into the pRS315 vector backbone (81). NMD2 was disrupted in the S150-2B background by transforming YJB276 with SacI/SalI-digested pBs-nmd2::HIS3 (28). This disruption replaces 444 bp of the promoter and 1,286 bp of the coding sequence of NMD2 with an XbaI-ClaI fragment containing the HIS3 gene. Deletion of NMD2 in YJB276-nmd2::HIS3 was confirmed by Southern blot hybridization, and original disruptants were back-crossed to YJB1260. Strains YJB1593 and YJB1594 are His+ spores from the resulting diploid.
URA3 was integrated adjacent to the left telomere of chromosome VII at the ADH4 locus (VII-L URA3-TEL) (23) and was introduced into strains containing nmd2::HIS, upf1::HIS, pan2::LEU2, pan3::HIS, and pan2::LEU2 pan3::HIS3, all in the W303 background, by standard genetic crosses.
Cloning of RLF4.
YJB539 (rlf4-1) cannot grow on 5-fluoro-orotic acid (5-FOA) due to defective silencing of the integrated URA3 gene at the left end of chromosome VII (VII-L URA3-TEL) (23). YJB539 was transformed (20) with a YCP50-LEU2 library (41), plated onto SDC minus Leu to select for transformants, and then replica plated onto SDC plus 5-FOA to select for restoration of silencing at telomeres. 5-FOA-resistant colonies were then streaked onto SDC minus Leu, SDC minus uracil, and SDC plus 5-FOA plates to identify colonies of cells that contained a library plasmid, the VII-L URA3-TEL marker, and were consistently 5-FOA resistant. Of ∼125,000 colonies screened, 124 colonies met the criteria described above. Plasmids from these candidates were rescued into E. coli (XL1-blue) cells. Plasmid p108 contained an ∼11-kb insert and conferred 5-FOA resistance on YJB539.
Southern analysis.
Telomere repeat probes were prepared by end labeling oligonucleotide 238 (CACCACACCCACACACCACCACACCCACACACCACCACAC), which is identical to 2.5 repeats of the telomere template sequence in TLC1. DNA was denatured and transferred (60) to a nylon membrane (Micron Separations, Inc.). Total genomic DNA was electrophoresed overnight in 1% agarose with Tris-acetate buffer. Blots were hybridized at 65°C in QuikHyb (Stratagene, Inc.) according to the manufacturer’s instructions and then washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 65°C for 20 to 30 min and 0.5× SSC–0.1% SDS at 65°C for 10 min.
Northern analysis.
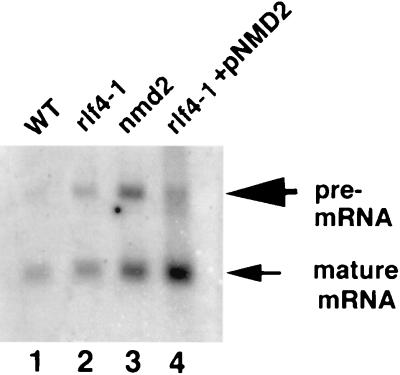
Total RNA was prepared from mid-log-phase cultures by a glass bead method (22). RNA was subjected to electrophoresis in 4% formaldehyde–1% agarose gels and transferred (60) to a nylon membrane (Micron Separations, Inc.). CYH2 probe was prepared by random priming (Oligolabelling kit; Pharmacia, Inc.) of an ∼0.5-kb EcoRI-HindIII fragment from pGEM-4Z-CYH2 (28). The blots were incubated with the CYH2 probe at 42°C in hybridization solution containing 50% formamide, 5× Denhardt’s solution, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.1-mg/ml single-stranded herring sperm DNA, 0.2% SDS, and 5.5% dextran sulfate. The blots were washed at 42°C in 2× SSC–0.1% SDS for 20 to 30 min and analyzed on a Molecular Dynamics Storm 840 PhosphorImager with Image Quant version 1.11 software. Ratios were determined by dividing the corrected volumes of pre-mRNA signal by the corrected volumes of mature mRNA signal for each individual strain.
Antisera and indirect immunofluorescence microscopy.
Indirect immunofluorescence was performed with anti-Rap1p antiserum exactly as described previously (19).
RESULTS
Telomeric chromatin is altered in rlf4-1 mutants.
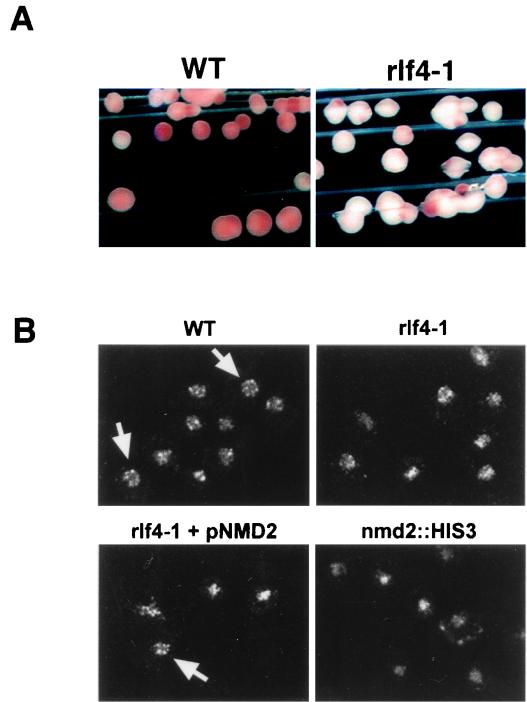
In S. cerevisiae, telomere repeats, like centromere sequences, stabilize circular plasmids (52). In contrast, circular plasmids containing both telomere and centromere sequences exhibit TEL+CEN antagonism, i.e., they are highly unstable (18). Genes that affect telomeric chromatin (e.g., RAP1, SIR2, SIR3, and SIR4) also affect the stability of TEL and TEL+CEN plasmids (18, 52), suggesting that telomeric DNA on plasmids has some of the properties of chromosomal telomeres. The rlf4-1 mutation was identified in a genetic screen (18, 19) for mutants in which TEL+CEN antagonism was lost, i.e., TEL+CEN plasmids were stabilized. When pRACUT1, a TEL+CEN plasmid carrying ADE2, was transformed into an ade2Δ strain, the instability of the TEL+CEN plasmid could be visualized by the predominance of red sectors in the colonies (Fig. 1A). In contrast, ade2Δ strains carrying the rlf4-1 allele gave rise to colonies with predominantly white sectors, indicating that the TEL+CEN plasmid was quite stable (i.e., antagonism was lost) (Fig. 1A).
FIG. 1.
rlf4-1 mutant strain exhibits reduced TEL+CEN antagonism and altered localization of Rap1p. (A) TEL+CEN antagonism assay. WT (YJB276) and rlf4-1 (YJB539) strains were transformed with pRACUT1. The left panel is a transformant of pRACUT1 into YJB276, and the right panel is a transformant of pRACUT1 into YJB539. Transformants were streaked on complete medium to assay loss of pRACUT1, which is indicated by the red. (B) Anti-Rap1p indirect immunofluorescence assay. Images are confocal laser micrographs of the equatorial Z section of each nucleus. WT (YJB492), rlf4-1 (YJB1260), nmd2::HIS3 (YJB1274), and rlf4-1 + pNMD2 (YJB1275) strains are shown. Arrows point to nuclei with characteristic WT Rap1p distribution.
The localization of Rap1p within yeast nuclei, which is thought to reflect the state of telomeric chromatin (19, 21), was altered in rlf4-1 strains. In wild-type (WT) strains, Rap1p stains as a small number of discrete foci that localize primarily near the nuclear periphery (19, 38) (Fig. 1B). In contrast, in most rlf4-1 cells, the Rap1p foci appeared less discrete, such that it was difficult to count the exact number of foci within a single nucleus. Diffuse Rap1p staining within the central portion of the nuclei was often evident in rlf4-1 cells (Fig. 1B). Alterations in Rap1p localization in other strains with defects in telomeric chromatin function have been observed (e.g., sir3, sir4, and rlf2 strains) (19, 21).
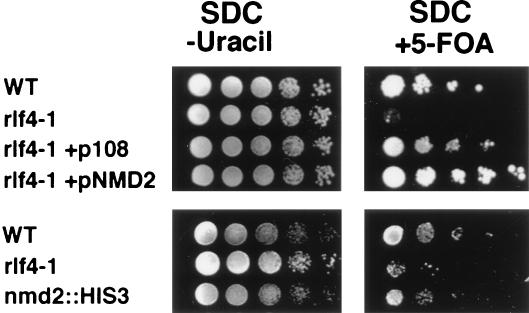
Genes inserted adjacent to telomeres are silenced in S. cerevisiae (23). Telomeric silencing was measured by determining the proportion of cells in the population that repressed a telomeric copy of URA3 by plating the cells on 5-FOA, which kills cells expressing URA3 (23). In WT cells, VII-L URA3-TEL is subject to epigenetic silencing; more than half of the cells in the population expressed URA3 and thus could grow on medium lacking uracil; yet a significant proportion (1 to 50%) of the cells also grew on SDC containing 5-FOA, indicating that the URA3 gene was silenced (Fig. 2). In the rlf4-1 strain, a very small proportion (10−4 to 10−5) of the cells silenced VII-L URA3-TEL (Fig. 2). Thus, almost all of the cells were unable to grow on 5-FOA, indicating that RLF4 is required for telomeric silencing.
FIG. 2.
Telomeric silencing assay. rlf4-1 mutant strain exhibits reduced telomeric silencing. (Top panels) WT (YJB492), rlf4-1 (YJB1338), rlf4-1 + p108 (YJB1222), and rlf4-1 + pNMD2 (YJB2541); (bottom panels) WT (YJB492), rlf4-1 (YJB1260), and nmd2::HIS3 (YJB2763). 1:10 serial dilutions of fresh overnight cultures were spotted onto the indicated media and grown for 48 h at 30°C. Reduced growth on SDC containing 5-FOA indicates reduced silencing of VII-L URA3-TEL. Growth on SDC plates (not shown) was similar to growth on SDC plates lacking uracil.
Telomere length control is altered in rlf4-1 mutants.
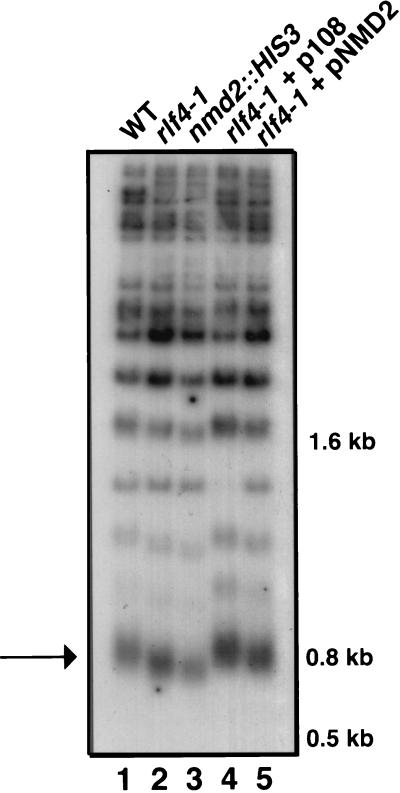
In wild-type S. cerevisiae cells, the length of telomere repeat DNA is maintained within a narrow size range (79). Although the average telomere lengths are slightly different in different strain backgrounds, in most strains telomere repeats (TG1–3/C1–3A) are ∼250 to 350 bp. At least 15 genes that affect average telomere length have been identified (reviewed in references 56, 58, and 86). We measured average telomere length by digesting chromosomal DNA with PstI, which digests ∼520 bp from the telomere proximal end of Y′ repeats, which are present at ∼50% of the chromosome ends (57). Digestion of the WT strain released an ∼800-bp terminal fragment (Fig. 3, arrow), indicating that telomere repeats are an average of ∼280 bp in the S150-2B strain background (Fig. 3, lane 1). In rlf4-1 strains, the terminal PstI fragment was ∼700 bp (Fig. 3, lane 2), indicating that the telomere repeat tract was ∼100 bp shorter than the telomere repeat tract in the parental strain. An ∼100-bp reduction in telomere length was consistently observed in all rlf4-1 progeny and always cosegregated with the TEL+CEN antagonism and telomeric silencing phenotypes of rlf4-1 strains. Taken together, the phenotypes of rlf4-1 strains indicate that both telomeric chromatin function and telomere length control are altered in rlf4-1 strains.
FIG. 3.
rlf4/nmd2 mutants exhibit shortened telomere tracts. Total genomic DNA was digested with PstI and separated in 1% agarose. The arrow points to the terminal PstI fragment of ∼800 bp, which includes ∼520 bp of the Y′ TAS element and ∼180 to 280 bp of telomere repeat sequence TG1–3/C1–3A. Other bands are terminal X element fragments and/or internal X and Y′ fragments. Lanes 1 to 5, WT (YJB492), rlf4-1 (YJB1338), nmd2::HIS3 (YJB2763), rlf4-1 + p108 (YJB1222), and rlf4-1 + pNMD2 (YJB2541), respectively.
Cloning of RLF4.
RLF4 was cloned by complementation of the telomeric silencing defect in rlf4-1 strains (see Materials and Methods). One genomic clone, p108, restored TEL+CEN antagonism (data not shown), telomeric silencing (Fig. 2), and WT telomere length (Fig. 3, lane 4) to the rlf4-1 strain YJB539.
Restriction and DNA sequence analysis of the insert in p108 indicated that the plasmid contained an ∼11-kb fragment of chromosome VIII including at least portions of four ORFs: ORF76W, NMD2, ORF78W, and IRE1. A plasmid copy of NMD2 [(pRS315-NMD2(X-S) (28)], which is hereafter called pNMD2, was sufficient to restore TEL+CEN antagonism, telomeric silencing, Rap1p localization, and telomere length control to YJB539 (Fig. 1, 2, and 3; data not shown). Furthermore, nmd2::HIS3 strains had defects in telomeric chromatin function and telomere length control like those observed in the rlf4-1 strain; TEL+CEN plasmids were stabilized (data not shown), Rap1p appeared more diffuse (Fig. 1B), telomeric silencing was reduced (Fig. 2), and telomeric DNA was shorter than that in the isogenic WT strain (Fig. 3, lane 3), suggesting that NMD2 was the gene on p108 responsible for the suppression of rlf4-1 phenotypes.
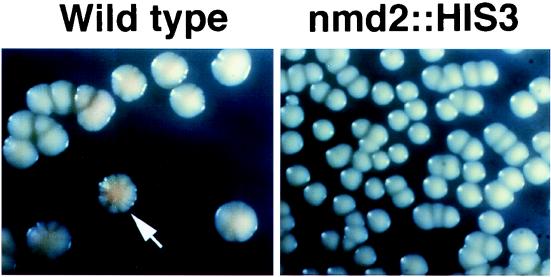
Mutations in genes encoding components of the NMD pathway cause steady-state accumulation of a number of RNAs that normally have very short half-lives (reviewed in reference 32). One of these messages is PPR1, a positive regulator of URA3 (55). Thus, it was possible that derepression of the VII-L URA3-TEL in YJB539 occurred due to upregulation by Ppr1p, rather than by reduced telomeric silencing. To distinguish between these possibilities, we monitored the expression of an ADE2 gene integrated adjacent to the right end of chromosome V (V-R ADE2-TEL) (23) in a strain lacking the normal chromosomal copy of ADE2. The ADE2 gene is not upregulated by Prp1p (82) or by mutation of NMD2 (49a). Red and white colony sectoring was used to monitor the status of the telomeric ADE2 gene. In these strains, expression of ADE2 leads to the cells being colored white and lack of ADE2 expression results in cells being colored red. WT and nmd2::HIS3 tetrad progenies that carried V-R-ADE2-TEL were isolated from the heterozygous diploid YJB2239. WT (NMD2) progeny gave rise to red-white sectored colonies (Fig. 4), which are characteristic of the normal epigenetic silencing of telomere-adjacent genes (23). In contrast, nmd2::HIS3 progeny gave rise to solid white colonies (Fig. 4), which are indicative of extensive derepression of the telomeric ADE2 gene. Thus, despite the possible upregulation of the URA3 gene in nmd2/rlf4 strains, telomeric silencing is perturbed in these strains due to alteration of the telomeric chromatin.
FIG. 4.
Silencing of a telomere-proximal V-R ADE2-TEL gene is lost in an nmd2::HIS3 mutant strain. Fresh overnight cultures of WT (YJB2263) and nmd2::HIS3 (YJB2262) strains were diluted 1:105 in sterile water and then plated on complete medium to assay expression of V-R ADE2-TEL. Red sectors indicate silencing of V-R ADE2-TEL, and white sectors indicate expression of V-R ADE2-TEL.
A rlf4-1 strain is defective in nonsense-mediated mRNA decay.
Because the nmd2::HIS3 strain had defects in telomeric functions similar to those of rlf4-1 strains, we asked whether an rlf4-1 strain had defects in nonsense-mediated mRNA decay similar to those of an nmd2::HIS3 strain. To ensure that genetic background did not contribute to any differences observed, we disrupted NMD2 in the S150-2B background (see Materials and Methods) and compared mRNA stabilities between isogenic WT, rlf4-1, and nmd2::HIS3 strains.
The CYH2 transcript is a poorly spliced RNA, and the CYH2 pre-mRNA accumulates in strains defective in nonsense-mediated mRNA decay (29). We prepared total RNA and asked whether CYH2 pre-mRNA accumulated in the rlf4-1 strain as it did in the nmd2::HIS3 strain by analyzing Northern blots with a CYH2 probe (Fig. 5). The ratio of CYH2 pre-mRNA to mature mRNA was approximately twofold higher in the rlf4-1 strain than the ratio of pre-mRNA to mature mRNA (0.31) in the WT strain (Fig. 5, compare lanes 1 and 2). Introduction of pNMD2 into the rlf4-1 strain (Fig. 5, lane 4) reduced the ratio of pre-mRNA to mRNA to 0.27, suggesting that the aberrant accumulation of pre-mRNA in the rlf4-1 strain was due to loss of Nmd2p function.
FIG. 5.
A rlf4-1 strain exhibits defective nonsense-mediated mRNA decay. Total RNA was isolated from mid-log-phase cells and separated in 1% agarose–4% formaldehyde. The Northern blot was probed with an ∼500 bp radiolabeled fragment of CHY2 from pGEM-4Z-CYH2.
rlf4-1 is an allele of NMD2.
Since the mutant phenotypes of rlf4-1 and nmd2::HIS3 were similar in all cases, and since pNMD2 complemented all phenotypes of rlf4-1 strains, we performed genetic linkage analysis to ask whether rlf4-1 and nmd2::HIS3 were allelic. Haploid rlf4-1 strain YJB1260 was crossed to haploid nmd2::HIS3 strain YJB1593, the resulting diploid was sporulated, and TEL+CEN antagonism was scored in the tetrad progeny with pRACUT1. In 10 complete tetrads and more than 60 other individual spores, no restoration of TEL+CEN antagonism was observed, which is consistent with the idea that rlf4-1 and nmd2::HIS3 are allelic.
We then determined the nature of the nmd2 allele from the rlf4-1 mutant strain by PCR amplification and subsequent DNA sequencing of overlapping segments covering the complete NMD2 coding sequence. Comparison of the rlf4-1 allele to the NMD2 sequence in the GenBank database revealed that the AAG lysine codon at amino acid 304 of NMD2 had been changed to a UAG stop codon in rlf4-1. Thus, the predicted product of rlf4-1 includes only the N-terminal one-third of the Nmd2p peptide sequence. These data, together with the phenotypic similarities of the rlf4-1 and nmd2::HIS3 alleles, indicate that RLF4 and NMD2 are allelic.
Telomere functions require the NMD pathway.
Nmd2p is a member of the NMD pathway that targets nonsense messages for exoribonucleolytic decay (27, 28). We asked whether the telomeric defects of rlf4 strains were caused by loss of NMD function or rather by loss of a putative NMD-independent function of Nmd2p. If the telomeric defects of nmd2 strains are due to inactivation of the NMD pathway, then we expected that mutation of genes encoding other members of this pathway, UPF1 and UPF3, would result in similar telomeric defects.
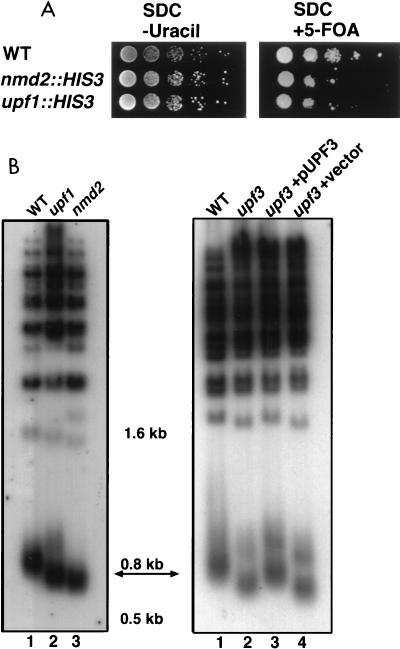
We generated an upf1 strain containing VII-L URA3-TEL in the W303 background and assayed telomeric silencing in this strain. The silencing defect caused by an nmd2::HIS3 mutation is less severe in this genetic background (∼10- to 100-fold reduction in 5-FOA resistance [Fig. 6A]) than in S150-2B (∼100- to 1,000-fold reduction in 5-FOA resistance [Fig. 2]). Similar to an nmd2::HIS3 strain, the upf1 VII-L URA3-TEL strain exhibited an ∼100-fold reduction in 5-FOA resistance relative to the WT strain, suggesting a modest silencing defect (Fig. 6A). Thus, telomeric chromatin is disrupted to the same degree in upf1 and nmd2 strains.
FIG. 6.
upf1 and upf3 strains exhibit telomeric defects. (A) upf1 telomeric silencing assay. Serial dilutions (1:10) of fresh overnight cultures were spotted onto the indicated media and grown for 48 h at 30°C. Reduced growth on SDC containing 5-FOA indicates reduced silencing of VII-L URA3-TEL. WT (YJB1680), nmd2::HIS3 (YJB2299), and upf1::HIS3 (YJB2297) are shown. Growth on SDC plates (not shown) was similar to growth on SDC plates lacking uracil. (B) upf1 and upf3 telomere length assay. Total genomic DNA was digested with PstI and separated in 1% agarose. The arrow points to the terminal PstI fragment of ∼800 bp, which includes ∼520 bp of the Y′ TAS element and ∼180 to 280 bp of telomere repeat sequence TG1–3/C1–3A. (Left panel) Lanes 1 to 3, WT (YJB447), upf1::HIS3 (YJB1324), and nmd2::HIS3 (YJB1274), respectively; (right panel) lanes 1 to 4 WT (YJB2889), upf3::TRP1 (YJB2890), upf3::TRP1 + pUPF3 (YJB2891), and upf3::TRP1 + vector (YJB2892), respectively.
We also assayed telomere length in both an upf1 strain and an upf3 strain. Like telomeres in nmd2 strains, the bulk of the telomeres in the upf1 and upf3 strains was ∼100 bp shorter than that in the isogenic WT strains (Fig. 6B). upf1 and upf3 strains had terminal PstI restriction fragments that were similar in size to those of nmd2 mutants and were significantly shorter than those of the isogenic WT strains. Addition of plasmid-born UPF3 to the upf3 strain restored normal telomere length regulation (Fig. 6B, right panel, lanes 2 and 3). Taken together, the silencing defect in the upf1 strain and the telomere length reduction in the upf1 and upf3 strains suggest that it is the NMD pathway, rather than Nmd2p alone, that contributes to telomere functions in WT cells.
A strain deficient in cytoplasmic exoribonucleolytic activity also has short telomeres.
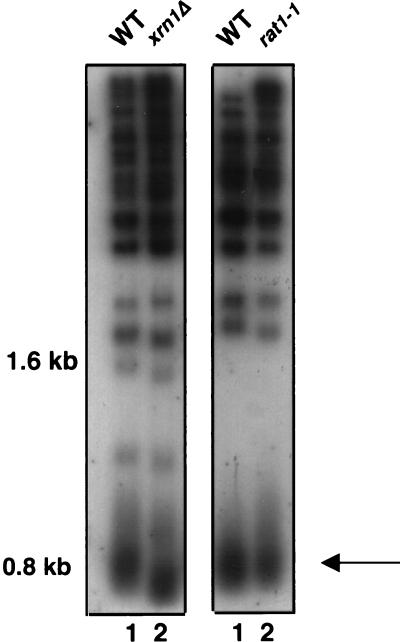
The NMD pathway could contribute to telomere functions by mediating the decay of specific mRNAs. Alternatively, this pathway could perform a novel function at telomeres that is unrelated to its role in mRNA decay. To distinguish between these possibilities, we analyzed average telomere lengths in strains lacking the major exoribonuclease (Xrn1p) required for cytoplasmic RNA decay (44, 65, 67). If the decay of mRNAs targeted by the NMD pathway is responsible for the telomere-associated phenotypes of upf1, nmd2, and upf3 mutants, then Xrn1p should also contribute to telomere functions. Consistent with the hypothesis that telomeric defects arise in an rlf4/nmd2 strain due to defective nonsense-mediated mRNA decay, an xrn1Δ strain (34) had telomeres ∼100 bp shorter than those in the isogenic WT strain (Fig. 7, left panel, lane 2).
FIG. 7.
An xrn1Δ strain, but not a rat1-1 strain, exhibits defective telomere length control. Total genomic DNA was digested with PstI and separated in 1% agarose. The arrow points to the terminal PstI fragment of ∼800 bp, which includes ∼520 bp of the Y′ TAS element and ∼180 to 280 bp of telomere repeat sequence TG1–3/C1–3A. (Left panel) Lanes 1 and 2, WT (YJB2793) and xrn1Δ (YJB2794), respectively; (right panel), lanes 1 and 2, WT (YJB2796) and rat1-1 (YJB2797), respectively. All strains were grown at 30°C.
Xrn1p, which is exclusively cytoplasmic (30), is functionally similar to Rat1p, a nuclear exoribonuclease that performs an essential function in S. cerevisiae. Localization of Rat1p to the cytoplasm allows it to complement an xrn1Δ mutation (33, 73). Telomeres are, by definition, nuclear in S. cerevisiae, because the nuclear membrane remains intact throughout the cell cycle. Because telomeres are nuclear, we asked whether Rat1p might play a role in telomere function by analyzing telomere length in a rat1-1 strain. In contrast to the xrn1Δ strain, the rat1-1 strain did not exhibit any significant alteration in the length of the terminal PstI fragment when cells were grown at permissive (23°C) and semipermissive (30 and 33°C) temperatures (Fig. 7 and data not shown). Since Xrn1p and Rat1p can be functionally redundant yet differ in cellular localization (33), the finding that the xrn1Δ strain had a defect in telomere length control and that the rat1-1 strain did not suggests that telomere defects arise due to perturbation of mRNA degradation by a cytoplasmic exoribonuclease rather than due to mRNA degradation by a nuclear exoribonuclease. These results also support the hypothesis that the telomere defects of an rlf4/nmd2 strain occur due to inactivation of the NMD pathway, which targets mRNAs for degradation by Xrn1p.
Telomere functions are not perturbed in strains deficient in the decay of normal messages.
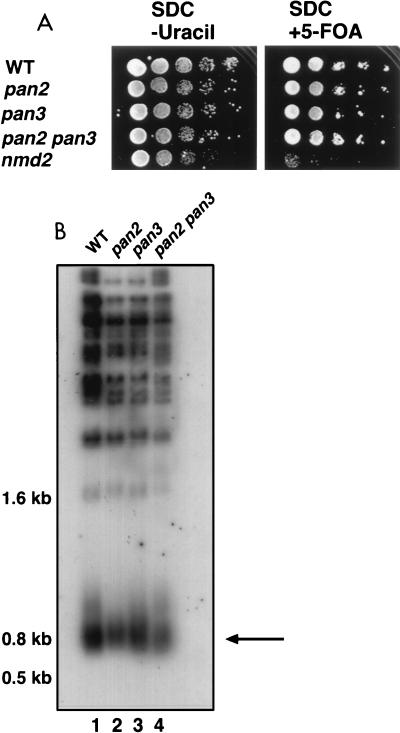
Like the NMD pathway, the intrinsic mRNA decay pathway relies on Xrn1p activity for final exoribonucleolytic activity (44, 65, 67). However, the intrinsic mRNA decay pathway requires progressive deadenylation followed by 5′ decapping, whereas the NMD pathway bypasses the deadenylation requirement for mRNA decay (14, 65, 66, 76). Poly(A) nuclease (PAN) deadenylates mRNA in a poly(A)-binding-protein-dependent manner (6, 77). To ask if it is intrinsic mRNA decay or NMD pathway-specific decay that is required for telomere functions, we analyzed pan2 and pan3 mutants. PAN2 and PAN3 encode two subunits of the PAN nuclease, and pan2 and pan3 mutant strains accumulate mRNAs with average poly(A) tails longer than those of WT strains (6, 7). We asked whether strains that might be hampered in the processes of normal mRNA turnover would exhibit defects in telomeric functions similar to those of strains deficient in nonsense-mediated mRNA decay. We analyzed telomeric silencing and telomere length control in strains carrying deletions of PAN2 and PAN3. Neither the pan2 strain, the pan3 strain, nor a double mutant pan2 pan3 strain exhibited a reduction in telomeric silencing relative to an isogenic WT strain (Fig. 8A). Similarly, telomere length control in a pan2 strain, a pan3 strain, and a pan2 pan3 double mutant strain was indistinguishable from that of the isogenic WT strain (i.e., terminal telomere tracts were not shortened [Fig. 8B]). Thus, mutants with defects in the deadenylation process required for efficient poly(A) shortening-dependent mRNA turnover do not exhibit altered telomeric chromatin or telomere length control. These results are consistent with the idea that the NMD pathway specifically, rather than the intrinsic pathway that targets mRNA for degradation, is required for normal telomere function.
FIG. 8.
pan mutants do not exhibit telomere defects. (A) Telomeric silencing assay. Serial dilutions (1:10) of fresh overnight cultures were spotted onto the indicated media and grown for 48 h at 30°C. WT (YJB2827), pan2::LEU2 (YJB2823), pan3::HIS3 (YJB2824), pan2::LEU2 pan3::HIS3 (YJB2826), and nmd2::HIS3 (YJB1593) are shown. Growth on SDC plates (not shown) was similar to growth on SDC plates lacking uracil. (B) Telomere length assay. Total genomic DNA was digested with PstI and separated in 1% agarose. The arrow points to the terminal PstI fragment of ∼800 bp, which includes ∼520 bp of the Y′ TAS element and ∼180 to 280 bp of telomere repeat sequence TG1–3/C1–3A. Lanes 1 to 4, WT (YJB2739), pan2::LEU2 (YJB2735), pan3::HIS3 (YJB2736), and pan2::LEU2 pan3::HIS3 (YJB2740), respectively.
DISCUSSION
rlf4-1 is an allele of the nonsense-mediated decay gene NMD2.
RLF4, which was isolated by virtue of its defects in telomere-associated phenotypes, is allelic with NMD2/UPF2 by several criteria. First, NMD2 complemented all of the mutant phenotypes of an rlf4-1 strain. Second, nmd2 and rlf4 strains exhibited similar telomere-associated phenotypes as well as a similar defect in nonsense-mediated decay. Third, genetic linkage analysis indicated that rlf4-1 and nmd2::HIS3 cosegregate. Finally, the DNA sequence of rlf4-1 indicated that it is a nonsense allele that would encode only the first one-third of Nmd2p. The phenotypic similarities of rlf4-1 and the nmd2::HIS3 deletion allele suggest that the rlf4-1 allele is effectively a null allele of NMD2.
How does NMD2/RLF4 affect telomere functions?
NMD2/RLF4 encodes a protein necessary for nonsense-mediated mRNA decay (12, 28). The NMD pathway targets mRNAs for rapid turnover when the translational machinery encounters a premature termination codon during translation (reviewed in references 32, 69, and 70). NMD2/RLF4 is one of three genes, the other two being UPF1 and UPF3, that are required for nonsense-mediated decay. Like nmd2/rlf4 strains, upf1 and upf3 strains had telomere-associated defects (Fig. 6), suggesting that the NMD pathway, rather than an NMD-independent function of Nmd2/Rlf4p, is required for normal telomere function.
We envision two general models to explain why the NMD pathway proteins might be required for telomere functions: a direct model and an indirect model. In the direct model, NMD proteins have a second function at telomeres in the nucleus. In the indirect model, the NMD pathway affects telomere function by altering the stability of one or more specific mRNAs that are important for telomere chromatin function and/or telomere length control. A number of genes, such as RAP1 and RIF1, affect both telomere length control and telomeric silencing (24a, 59). It is also possible that the NMD pathway affects telomere functions even more indirectly, by altering the level of an mRNA that encodes a protein that then, in turn, regulates telomere-related genes. Nmd2p has a putative bipartite nuclear localization sequence that is required for function (26, 28). Upf3p also has predicted nuclear localization sequences and nuclear export sequences (46, 79a). Thus, at least two of the NMD proteins may have an opportunity to interact directly with telomeres, at least transiently.
To begin to distinguish between the direct and indirect models of NMD protein involvement in telomere function, we asked whether Xrn1p is required for telomere function. XRN1 has been isolated from a wide range of biochemical and genetic screens and is also known as KEM1, SEP1, DST2, RAR5, and SKI1 (15, 36, 37, 39, 84). We found that, like upf1, nmd2, and upf3 mutants, the xrn1 mutant had short telomeres (Fig. 7, left panel). Our work confirms a previous report that telomeres are shorter in xrn1/kem1 strains (51). Unlike the previous report, we found that xrn1 mutants did not display a senescence phenotype (data not shown); rather, in keeping with the observations of others (16, 33, 36, 45), xrn1 cells exhibited a slow-growth phenotype. The fact that Xrn1p is exclusively cytoplasmic (30, 33) supports the indirect model that Xrn1p (and Upf1p, Nmd2p, and Upf3p) affects telomeres via message degradation.
A second exoribonuclease in S. cerevisiae is encoded by the essential gene RAT1 (1, 33, 35). Rat1p localizes predominantly to the nucleus; yet this nuclear localization appears saturable, such that multicopy expression of RAT1 results in both nuclear and cytoplasmic localization of Rat1p and partially restores the slow growth and RNA turnover phenotypes of an xrn1Δ strain (33, 73). In contrast to the xrn1 mutant, the rat1-1 mutant had normal telomere length control (Fig. 7), suggesting that, despite the similarities between Xrn1p and Rat1p, the NMD proteins do not affect telomere functions via nuclear Rat1p activity. This result is again consistent with the indirect model that NMD proteins are required for telomere function because they target specific mRNAs for decay by Xrn1p.
Telomere functions could be indirectly affected by all mRNA decay pathways or by the NMD pathway specifically. To distinguish between these possibilities, we analyzed the telomere-associated phenotypes of pan strains which are deficient in the poly(A)-binding-protein-dependent PAN. PAN is the only characterized deadenylase in yeast, and poly(A) tail deadenylation is a prerequisite for targeting of a message for the intrinsic pathway of mRNA turnover (6, 7, 14, 77). Pan2p and Pan3p interact physically, and pan2 and pan3 mutant strains have mRNA populations with average poly(A) tail lengths longer than those of WT strains (7). The fact that neither a pan2, a pan3, nor a pan2 pan3 strain exhibited reduced telomeric silencing or shortened telomere tracts (Fig. 8) is consistent with the hypothesis that it is the NMD pathway specifically, rather than intrinsic mRNA decay, that is required for telomere function. Because some (inefficient) deadenylation occurs in strains lacking Pab1p, it is possible that other Pab1p-independent PANs exist in yeast (7, 10, 76), and, thus, we cannot exclude the possibility that a Pab1p-independent PAN may contribute to telomere function. Despite this caveat, we propose the model that the levels of mRNAs important for telomere functions are inappropriately increased in upf and nmd mutants, leading to defects in telomeric silencing and length control. This model implies that in WT cells, the level of these mRNAs is regulated by the NMD pathway. While our data support this indirect model, we cannot rule out the possibility that the NMD proteins have a second, more direct, role in the nucleus that is independent of their cytoplasmic function and of the function of Rat1p and Xrn1p.
Is there a role for the nonsense-mediated decay pathway in regulating normal rather than nonsense mRNAs?
Sequence features that the NMD pathway recognizes include upstream ORFs (uORFs) and frameshifts. Interestingly, a number of messages encoding telomere-associated proteins include uORFs or frameshifts. For example, TEL2, EST1, and EST2 (genes required for telomerase activity and/or telomere length control) contain clusters of uORFs within 100 bp of the initiation codons (50, 74), and EST3 requires a frameshift for translation of a full-length functional protein (64). Neither uORFs nor frameshifts alone are sufficient to ensure that mRNAs containing them are targeted by the NMD pathway (71). For example, the GCN4 mRNA, which is regulated by a mechanism involving four short uORFs, is not targeted for decay by the NMD pathway (reviewed in reference 31). Analysis of specific mRNA levels and mRNA half-lives will be required to determine whether the NMD pathway specifically and directly regulates the expression of genes that encode proteins known to be required for telomeric silencing and/or telomere length control.
The NMD pathway has been presumed to target defective rather than normal mRNAs for degradation (70). The telomere-associated phenotypes of upf1, nmd2, upf3, and xrn1 mutants implicate the NMD pathway in the regulation of normal mRNAs involved in telomere function. The half-lives of mRNAs are thought to be governed by a number of competing stabilizing and destabilizing forces (reviewed in references 32, 70 and 75). Upf-mediated decay may be one of the destabilizing forces that contribute to the regulation of mRNA half-life for one or more normal mRNAs important for telomere functions.
ACKNOWLEDGMENTS
We thank Maryam Gerami-Nejad for technical assistance and David Gartner and Mark Sanders of the University of Minnesota Imaging Center for assistance with digital imaging. We thank Feng He, Allen Jacobson, Alan Sachs, and Michael Culbertson for providing strains and plasmids. We thank Steve Johnston, Cathy Asleson, Michael Lelivelt, and Jeff Dahlseid for critical reading of the manuscript and many helpful suggestions.
This work was supported by a grant from the National Institutes of Health (GM38636) to J.B.
ADDENDUM IN PROOF
Recently, CTF13, a gene important for centromere function, was found to be regulated, indirectly, by the NMD pathway (J. N. Dahlseid, J. Puziss, R. L. Shirley, A. L. Atkin, P. Hieter, and M. R. Culbertson, Genetics, in press).
REFERENCES
- 1.Amberg D C, Goldstein A L, Cole C N. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- 2.Atkin A L, Schenkman L R, Eastham M, Dahlseid J N, Lelivelt M J, Culbertson M R. Relationship between yeast polyribosomes and Upf proteins required for nonsense mRNA decay. J Biol Chem. 1997;272:22163–22172. doi: 10.1074/jbc.272.35.22163. [DOI] [PubMed] [Google Scholar]
- 3.Autexier C, Greider C W. Telomerase and cancer: revisiting the telomere hypothesis. Trends Biochem Sci. 1996;21:387–391. [PubMed] [Google Scholar]
- 4.Belgrader P, Maquat L E. Nonsense but not missense mutations can decrease the abundance of nuclear mRNA for the mouse major urinary protein, while both types of mutations can facilitate exon skipping. Mol Cell Biol. 1994;14:6326–6336. doi: 10.1128/mcb.14.9.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. . (See comments.) [DOI] [PubMed] [Google Scholar]
- 6.Boeck R, Tarun S J, Rieger M, Deardorff J A, Muller-Auer S, Sachs A B. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem. 1996;271:432–438. doi: 10.1074/jbc.271.1.432. [DOI] [PubMed] [Google Scholar]
- 7.Brown C E, Tarun S Z, Jr, Boeck R, Sachs A B. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryan T M, Reddel R R. Telomere dynamics and telomerase activity in in vitro immortalized human cells. Eur J Cancer. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 9.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caponigro G, Parker R. Multiple functions for the poly(A) binding protein in mRNA decapping and deadenylation in yeast. Genes Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- 11.Cockell M, Palladino F, Laroche T, Kyrion G, Liu C, Lustig A J, Gasser S M. The carboxy termini of Sir4 and Rap1 affect Sir3 localization: evidence for a multicomponent complex required for yeast telomeric silencing. J Cell Biol. 1995;129:909–924. doi: 10.1083/jcb.129.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui Y, Hagan K W, Zhang S, Peltz S W. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 13.Culbertson M R, Underbrink K M, Fink G R. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 15.Dykstra C C, Hamatake R K, Sugino A. DNA strand transfer protein β from yeast mitotic cells differs from strand transfer protein alpha from meiotic cells. J Biol Chem. 1990;265:10968–10973. [PubMed] [Google Scholar]
- 16.Dykstra C C, Kitada K, Clark A B, Hamatake R K, Sugino A. Cloning and characterization of DST2, the gene for DNA strand transfer protein beta from Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2583–2592. doi: 10.1128/mcb.11.5.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto, S., E. L. Chamberlain, M. Gerami-Nejad, and J. Berman. Unpublished observations.
- 18.Enomoto S, Longtine M S, Berman J. TEL+CEN antagonism on plasmids involves telomere repeat sequence tracts and gene products that interact with chromosomal telomeres. Chromosoma. 1994;103:237–250. doi: 10.1007/BF00352248. [DOI] [PubMed] [Google Scholar]
- 19.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 20.Gietz R D, Schiestl R H. Transforming yeast with DNA. Methods Mol Cell Biol. 1995;5:255–269. [Google Scholar]
- 21.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser S M. The clustering of telomeres and colocalization with Rap1, Sir3 and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschling, D. E. 1998. http://www.fhcrc.org/∼gottschling/rprep.html.
- 23.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 24.Gozalbo D, Hohmann S. Nonsense suppressors partially revert the decrease of the mRNA level of a nonsense mutant allele in yeast. Curr Genet. 1990;17:77–79. doi: 10.1007/BF00313252. [DOI] [PubMed] [Google Scholar]
- 24a.Hardy C F J, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 25.Harley C B, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr Opin Genes Dev. 1995;5:249. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 26.He F, Brown A H, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]
- 27.He F, Brown A H, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 29.He F, Peltz S W, Donahue J L, Rosbash M, Jacobson A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc Natl Acad Sci USA. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heyer W D, Johnson A W, Reinhart U, Kolodner R D. Regulation and intracellular localization of the Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol Cell Biol. 1995;15:2728–2736. doi: 10.1128/mcb.15.5.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinnebusch A G. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- 32.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 33.Johnson A W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol Cell Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson A W, Kolodner R D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol Cell Biol. 1995;15:2719–2727. doi: 10.1128/mcb.15.5.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kenna M, Stevens A, McCammon M, Douglas M G. An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol Cell Biol. 1993;13:341–350. doi: 10.1128/mcb.13.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J, Ljungdahl P O, Fink G R. kem mutations affect nuclear fusion in Saccharomyces cerevisiae. Genetics. 1990;126:799–812. doi: 10.1093/genetics/126.4.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kipling D, Tambini C, Kearsey S E. rar mutations which increase artificial chromosome stability in Saccharomyces cerevisiae identify transcription and recombination proteins. Nucleic Acids Res. 1991;19:1385–1391. doi: 10.1093/nar/19.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein F, Laroche T, Cardenas M E, Hofmann J F-X, Schweizer D, Gasser S M. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolodner R, Evans D H, Morrison P T. Purification and characterization of an activity from Saccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc Natl Acad Sci USA. 1987;84:5560–5564. doi: 10.1073/pnas.84.16.5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konkel L M, Enomoto S, Chamberlain E M, McCune-Zierath P, Iyadurai S J, Berman J. A class of single-stranded telomeric DNA-binding proteins required for Rap1p localization in yeast nuclei. Proc Natl Acad Sci USA. 1995;92:5558–5562. doi: 10.1073/pnas.92.12.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouprina N, Kroll E, Bannikov V, Bliskovsky V, Gizatullin R, Kirillov A, Shestopalov B, Zakharyev V, Hieter P, Spencer F, Larionov V. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kranz J E, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyrion G, Liu K, Liu C, Lustig A J. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7:1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 44.Larimer F W, Hsu C L, Maupin M D, Stevens A. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene. 1992;120:51–57. doi: 10.1016/0378-1119(92)90008-d. [DOI] [PubMed] [Google Scholar]
- 45.Larimer F W, Stevens A. Disruption of the gene XRN1, coding for a 5′→3′ exoribonuclease, restricts yeast cell growth. Gene. 1990;95:85–90. doi: 10.1016/0378-1119(90)90417-p. [DOI] [PubMed] [Google Scholar]
- 46.Lee B-S, Culbertson M R. Identification of an additional gene required for eukaryotic nonsense mRNA turnover. Proc Natl Acad Sci USA. 1995;92:10354–10358. doi: 10.1073/pnas.92.22.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S I, Umen J G, Varmus H E. A genetic screen identifies cellular factors involved in retroviral-1 frameshifting. Proc Natl Acad Sci USA. 1995;92:6587–6591. doi: 10.1073/pnas.92.14.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leeds P, Peltz S W, Jacobson A, Culbertson M R. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 49.Leeds P, Wood J M, Lee B S, Culbertson M R. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Lelivelt, M., and M. R. Culbertson. Personal communication.
- 50.Lendvay T S, Morris D K, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Z-P, Gilbert W. The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell. 1994;77:1083–1092. doi: 10.1016/0092-8674(94)90447-2. [DOI] [PubMed] [Google Scholar]
- 52.Longtine M S, Enomoto S, Finstad S, Berman J. Yeast telomere repeat sequence (TRS) improves circular plasmid segregation, and TRS plasmid segregation involves the RAP1 gene product. Mol Cell Biol. 1992;12:1997–2009. doi: 10.1128/mcb.12.5.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Longtine M S, Wilson N M, Petracek M E, Berman J. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr Genet. 1989;16:225–239. doi: 10.1007/BF00422108. [DOI] [PubMed] [Google Scholar]
- 54.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Losson R, Lacroute F. Cloning of a eukaryotic regulatory gene. Mol Gen Genet. 1981;184:394–399. doi: 10.1007/BF00352511. [DOI] [PubMed] [Google Scholar]
- 56.Louis E J. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- 57.Louis E J, Borts R H. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics. 1995;139:125–136. doi: 10.1093/genetics/139.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowell J E, Pillus L. Telomere tales: chromatin, telomerase, and telomere function in Saccharomyces cerevisiae. Cell Mol Life Sci. 1998;54:32–49. doi: 10.1007/s000180050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 60.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 61.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 62.McClintock B. Cytological observations of deficiencies involving known genes, translocations and an inversion in Zea mays. M Agric Exp Stn Res Bull. 1931;163:1–30. [Google Scholar]
- 63.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morris D K, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 65.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 66.Muhlrad D, Decker C J, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 68.Oliveira C C, McCarthy J E G. The relationship between eukaryotic translation and mRNA stability. A short upstream open reading frame strongly inhibits translational initiation and greatly accelerates mRNA degradation in the yeast Saccharomyces cerevisiae. J Biol Chem. 1995;270:8936–8943. doi: 10.1074/jbc.270.15.8936. [DOI] [PubMed] [Google Scholar]
- 69.Peltz S W, Brown A H, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 70.Peltz S W, He F, Welch E, Jacobson A. Nonsense-mediated mRNA decay in yeast. Prog Nucleic Acids Res Mol Biol. 1994;47:271–298. doi: 10.1016/s0079-6603(08)60254-8. [DOI] [PubMed] [Google Scholar]
- 71.Peltz S W, Trotta C, He G, Brown A, Donahue J, Welch E, Jacobson A. Identification of the cis-acting sequences and trans-acting factors involved in nonsense-mediated mRNA decay. In: Tuite M, McCarthy J, Brown A, Sherman F, editors. Protein synthesis and targeting in yeast. Berlin, Germany: Springer-Verlag; 1993. pp. 1–10. [Google Scholar]
- 72.Pinto I, Na J G, Sherman F, Hampsey M. cis- and trans-acting suppressors of a translation initiation defect at the cyc1 locus of Saccharomyces cerevisiae. Genetics. 1992;132:97–112. doi: 10.1093/genetics/132.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poole T L, Stevens A. Comparison of features of the RNase activity of 5′-exonuclease-1 and 5′-exonuclease-2 of Saccharomyces cerevisiae. Nucleic Acids Symp Ser. 1995;33:79–81. [PubMed] [Google Scholar]
- 74.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 76.Sachs A B, Davis R W. The poly(A) binding protein is required for poly(A) shortening and 60S ribosomal subunit-dependent translation initiation. Cell. 1989;58:857–867. doi: 10.1016/0092-8674(89)90938-0. [DOI] [PubMed] [Google Scholar]
- 77.Sachs A B, Deardoff J A. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 78.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 79.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79a.Shirley, R. L., M. J. Lelivelt, L. R. Schenkman, J. N. Dahlseid, and M. R. Culbertson. Personal communication.
- 80.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 81.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stotz A, Muller P P, Linder P. Regulation of the ADE2 gene from Saccharomyces cerevisiae. Curr Genet. 1993;24:472–480. doi: 10.1007/BF00351708. [DOI] [PubMed] [Google Scholar]
- 83.Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci USA. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tishkoff D X, Rockmill B, Roeder G S, Kolodner R D. The sep1 mutant of Saccharomyces cerevisiae arrests in pachytene and is deficient in meiotic recombination. Genetics. 1995;139:495–509. doi: 10.1093/genetics/139.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 86.Zakian V A. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu Rev Genet. 1996;30:141–172. doi: 10.1146/annurev.genet.30.1.141. [DOI] [PubMed] [Google Scholar]