Abstract
Background
Lung cancer metastasis to the brain presents significant clinical challenges. Therefore, elucidating its underlying mechanisms and characterizing its transcriptomic landscape is essential for developing therapeutic interventions.
Methods
We analyzed two distinct single-cell RNA sequencing datasets of lung cancer metastasis to analyze the evolutionary trajectory of brain metastatic tumors. In addition, a systematic comparison of cell-cell interaction between tumor cells and lymphocytes was conducted within primary and brain metastatic tumors.
Results
The brain metastatic tumors showed greater transcriptomic changes (reflected by a higher pseudotime) than tumors in the lymph nodes and primary tumors. Furthermore, our investigation has not only revealed specific shared ligand-receptor pairs in both mLN and mBrain, exemplified by the interaction between SPP1 and CD99 in T cells, but has also unveiled a diverse array of ligand-receptor pairs exclusive to the mBrain. Notably, this includes distinctive pairs such as APP and IL1 observed specifically in myeloid cells.
Conclusion
The distinct microenvironment in the brain may influence the observed transcriptomic changes in tumors, emphasizing the significance of the specific environment in determining tumor behavior and therapeutic response.
Keywords: Lung cancer brain metastasis, Single-cell RNA sequencing, Tumor microenvironment, Therapeutic response
1. Introduction
Lung cancer remains the foremost cause of cancer-related mortality globally. Its rapid metastasis, even in early stages, to various organs poses a significant challenge to targeted therapies, thereby diminishing the efficacy of systemic treatments [1]. Over the past decade, advances in understanding lung cancer's molecular biology have led to the development of novel and effective therapies, significantly enhancing patient survival rates [2]. Recently, immunotherapies have emerged as a focal point in non-small cell lung cancer (NSCLC) treatment, with ongoing research into multiple potential influencing factors [[3], [4], [5], [6]]. This progress is predominantly attributed to breakthroughs in NSCLC treatment.
Several researchers are investigating the mechanisms underlying the aggressive nature of lung cancer [7,8]. Current research focuses on understanding how lung cancer cells resist or even thrive under hypoxia [9], how immune cells can be manipulated by tumor cells, and the diverse pathways involved in the migration of tumor cells. The findings suggest that lung cancer metastasis is a complex phenomenon and multiple pathways are implicated in the process. Here, we focused on NSCLC from its primary site to the most frequent sites of metastasis: the brain, bone, and liver. We investigated the pathology, molecular pathways, and genetic characteristics of lung cancer metastasis to these organs. Understanding these complex mechanisms is crucial to develop effective therapeutic interventions for this fatal disease.
Single-cell RNA sequencing (scRNA-seq) can be used to detect tumor heterogeneity and examine gene expression patterns in tissues. This technology has enabled researchers to identify novel biomarkers and eludicate the complex heterogeneity of NSCLC tissues [[10], [11], [12]]. Nevertheless, the mechanisms of lung cancer metastasis and heterogeneity in metastatic tissues at the single-cell level have not been completely elucidated [13]. Moreover, cellular heterogeneity and shifts in the microenvironment between primary lung and metastatic tumors need to be further explored. In this study, we aimed to elucidate the differences and similarities between primary and metastatic tumors of NSCLC at the single-cell level.
2. Materials and methods
Single-cell RNA sequencing data collection and processing We obtained two distinct scRNA-seq datasets on lung cancer metastasis from the GSE131907 [14] and GSE123904 [15] databases. We assessed the quality of raw data and filter out poor-quality cells to ensure high-quality cells for downstream analysis. We restricted our analysis to include only cells derived from normal lung tissue (nLung), tumor lung tissue (tLung), metastatic lymph nodes (mLN), and metastatic brain (mBrain) tissue. Next, all the data were integrated using the “NormalizeData,” “FindVariableFeatures,” and “ScaleData” functions of Seurat v4. Principal component analysis (PCA) was then performed to reduce the dimensions of the integrated data, and the harmony approach was used to identify the anchor genes shared across the different batches. Uniform manifold approximation and projection (UMAP) dimensional reduction analysis [16] was performed on the scaled matrix (with only the most variable genes) using the first 30 PCA components to obtain a two-dimensional representation of the cell states. Cell clustering was then performed using the ‘FindClusters’ function, which implements the shared nearest neighbor modularity optimization-based clustering algorithm with a resolution of 0.6. The cell types were annotated manually based on the original annotation after detecting the top hits for each cluster.
2.1. Trajectory analysis
The differentiation of the pseudotime of a lineage between sample groups can provide valuable information on disease progression, such as the transition from a healthy to a diseased state or from the primary tumor to metastasis. We used Monocle 3 to infer cellular trajectories and determine the pseudotime of cells in different groups [17]. Seurat was used to create a dimensional-reduced cell matrix (UMAP) and group labels for each cell type to generate the specific input format for Monocle 3. We set the normal lung as the beginning point of the trajectory to infer the expected differentiation trajectory for each cell type. This approach enabled us to compare the pseudotime of cells between different sample groups and identify potential differences in disease progression and metastatic potential.
3. Detection of cluster-specific genes
The differentially expressed gene (DEG) analysis between different cell clusters was conducted using the Seurat “FindAllMarkers” function based on the Wilcoxon rank-sum test. The genes expressed in more than 25% of the cells with log2 fold change >0.5 compared with the background and a false discovery rate <0.001 were defined as DEGs for each cluster. In addition, DEGs among normal tissues, primary tumors, and metastasis tumors for each cell type were analyzed using the “FindMarkers” function. All the parameters were set as default.
3.1. Functional enrichment analysis
Functional enrichment analyses of DEGs were conducted by hypergeometric test using the WEB-based GEne SeT AnaLysis Toolkit (WebGestalt R v0.4.4) [18]. All human protein-coding genes were used as the background gene sets. The Benjamini–Hochberg procedure was used for multiple test corrections [19]. The pathways with an adjusted p-value (FDR) of <0.05 were considered significant.
3.1.1. Cell–cell communication analysis
We employed an R package called CellChat [20] to identify and visualize differential cell-to-cell interactions among nLung, tLung, mLN, and mBrain. We selectively used 2021 pre-validated ligand–receptor (L–R) interactions as a priori network information. For each L-R pair, we then calculated their information flow strength and communication probability between different cell groups by using the functions computeCommunProb, computeCommunProbPathway, and aggregateNet with standard parameters. We used a one-sided permutation test (n = 100) for each L–R pair to predict differential intercellular communication pathways among nLung, tLung, mLN, and mBrain groups. This test randomly permutes the group labels of cells and then recalculates the communication probability between the two cell groups [20]. The interactions with a p-value <0.05 were considered statistically significant.
3.2. Survival analysis
We conducted survival analysis using the bladder cancer cohort from The Cancer Genome Atlas. Additionally, we used several online resources for pan-cancer and expression analyses of targets of differentially expressed/edited microRNAs. These resources were UALCAN, CbioPortal, and the Gene Expression Profiling Interactive Analysis (GEPIA) tool (v1.0 and v2.0) [21,22]. Survival and co-expression analyses were performed using the GEPIA web interface (GEPIA (Gene Expression Profiling Interactive Analysis) (cancer-pku.cn)).
3.3. Statistical analysis
All the results are shown as the mean ± standard deviation. For parametric data, the Student's t-test was utilized to assess the statistical differences between two groups. For nonparametric data analysis, the Wilcoxon test was used. All statistical analyses were conducted using GraphPad Prism 8.0.2 and SPSS 22.0. Statistical significance was established at p < 0.05.
4. Results
4.1. Metastatic tumor shows a higher pseudotime than primary tissue
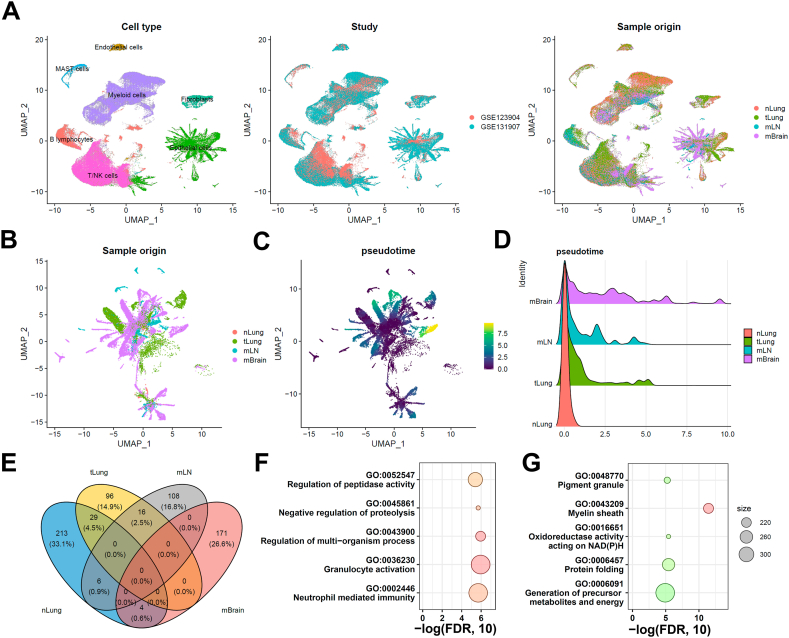
We investigated the diversity of cells and cellular changes in the samples to compare the cellular heterogeneity and microenvironment changes in primary lung tumors and metastatic tumors. First, we reprocessed the scaled gene-barcode matrices, removed low-quality cells, and conducted standard clustering analysis using Seurat. Then, we identified 160,904 high-quality single cells grouped into seven major cell clusters, namely T/NK cells, B lymphocytes, myeloid cells, mast cells, fibroblasts, endothelial cells, and epithelial cells (Fig. 1A). The UMAP analysis confirmed that batch effects were minimized across different studies. Next, we assessed the proportions of major cell types across different sample sites and found few fibroblast and endothelial cells in the mLN group (Fig. S1). Moreover, we observed an increased proportion of T/NK cells in the primary lung tumor, whereas a decreased proportion of T/NK cells was observed in the mLN and mBrain groups. We also observed an increased proportion of B-lymphocytes in the lung tumor, specifically in the mLN group. In contrast, the proportion of myeloid cells decreased in both lung primary and metastatic tumors compared with normal lung tissue. Finally, we also observed an increase in the proportion of epithelial cells in the tumor lung tissue (both primary tumor and metastasis group), suggesting the crucial role of epithelial cells in the development and progression of lung cancer.
Fig. 1.
Single-cell RNA-seq integrative analysis in lung cancer metastasis. (A) UMAP projection of all the cells grouped by cell clusters (n = 7), studies, or sample origin sites. (B) UMAP projection of tumor cells grouped by different origin sites. (C) Pseudotime analysis of tumor cells. (D) Pseudotime distribution among the normal lung tissue (nLung), tumor lung tissue (tLung), metastatic lymph nodes (mLN), and metastatic brain (mBrain) groups. (E) Comparative analysis of differentially expressed genes in tumor cells. Functional pathway enrichment analysis of (F) mLN-specific differentially expressed genes and (G) mBrain-specific differentially expressed genes.
Therefore, we selected all the epithelial cells for further analysis to understand the cellular changes during lung cancer metastasis (Fig. 1B). The pseudotime analysis was performed using the Monocle 3 software to investigate the developmental trajectory of the epithelial cells [17]. All the normal epithelial cells were selected as the root. Our results indicated that the metastatic tumor in the mBrain group (Fig. 1C and D) had a longer pseudotime compared with the metastatic tumor in the mLN group and the primary tumor in the tLung group. This finding suggests a potential transcriptomic heterogeneity within these tumors in different tissues. Further investigations into the underlying mechanisms of this heterogeneity could provide novel insights into the metastatic process.
We next compared DEGs among nLung, tLung, mLN, and mBrain groups. Collectively, 96, 108, and 171 genes were differentially expressed (log2 fold change >0.5 and adjusted p-value <0.001) in tLung, mLN, and mBrain groups, respectively (Fig. 1E). The pathway enrichment analysis was also performed on the epithelial cells from the mLN and mBrain groups. Epithelial cells in the mLN group were enriched in pathways related to granulocyte activation, regulation of multi-organism, and neutrophil-mediated immunity. In contrast, the epithelial cells in the mBrain group were enriched in pathways related to the myelin sheath, oxidoreductase activity acting on NAD(P)H, and protein folding, indicating different cellular states and functions in different metastatic sites (Fig. 1F-G). Recently, a pan-cancer analysis showed that the cancer cell states correlate with their interaction with the tumor microenvironment (TME) [23]. Therefore, we compared the differential gene expression patterns in the TMEs of the primary tumors and the respective TMEs at different sites of metastasis.
4.1.1. Interactions between T cell and tumor cell in metastatic vs. non-metastatic cancer cells
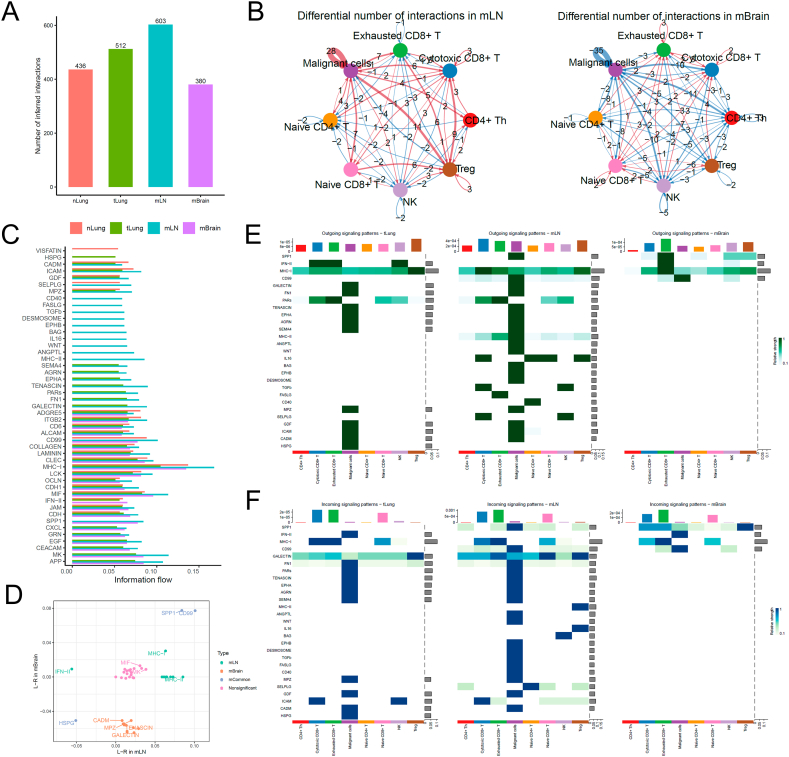
We first re-clustered and fine-annotated the T-cell subtypes and identified their seven subtypes, namely CD4+ Th, cytotoxic CD8+ T, exhausted CD8+ T, naive CD4+ T, naive CD8+ T, NK cell, and Treg. Then, we used an R package called CellChat [20] to infer the differential communication patterns (one-sided permutation test, see Methods) between T cells and tumor malignant cells at nLung, tLung, mLN, and mBrain sites. The overall cellular interactions between T cells and malignant cells increased in the tLung and mLN groups, whereas these interactions decreased in the metastatic brain (mBrain) group (Fig. 2A). This may happen because the blood–brain barrier restricts the presence of T cells in the brain tissue, or microglial cells limit the infiltration of T cells in the brain (Fig. 2B). Notably, differential communication patterns between mLN and tLung and mBrain and tLung groups revealed a consistent pattern (Fig. 2C).
Fig. 2.
Cell–cell communication analysis between T cells and malignant cells in tumor. (A) Bar plot shows the total number of the inferred cell–cell communication networks among nLung, tLung, mLN, and mBrain groups. (B) Circle plots summarize the number of differentially expressed interactions between mLN and tLung or mBrain and tLung groups. (C) Top representative differentially expressed ligand–receptor signaling pathways among the four groups. (D) Scatterplot shows common or specific signaling pathways between T cells and malignant cells in the mLN (x-axis) and mBrain (y-axis) groups. (E, F) Visualization of cell–cell communication between different cell groups for incoming and outgoing signals.
We further examined the information flow for each pair of ligand–receptor signaling pathways and observed differences in the contributions of immune and inflammatory pathways among different sites (Fig. 2C). We created a diagonal plot to illustrate the most significant differential ligand–receptor (L–R) interactions in mLN and mBrain metastatic versus primary sites (Fig. 2D). The strength of cellular interaction between SPP1 and CD99 increased in both mLN and mBrain groups, whereas the ligand-receptor interactions involving HSPG decreased in both groups. Additionally, we observed several site-specific communication patterns. For instance, the expression of MHC-I/II signaling pathways increased, whereas that of the IFN-II signaling pathway decreased in the mLN group. Furthermore, CADM, MPZ, TENASCIN, and GALECTIN signaling pathways were decreased in the mBrain group. Further, we created two heatmaps to compare the differential number of interactions or interaction strengths (Fig. 2E and F).
4.1.2. B cell to tumor cell interactions in metastatic vs. non-metastatic cancer cells
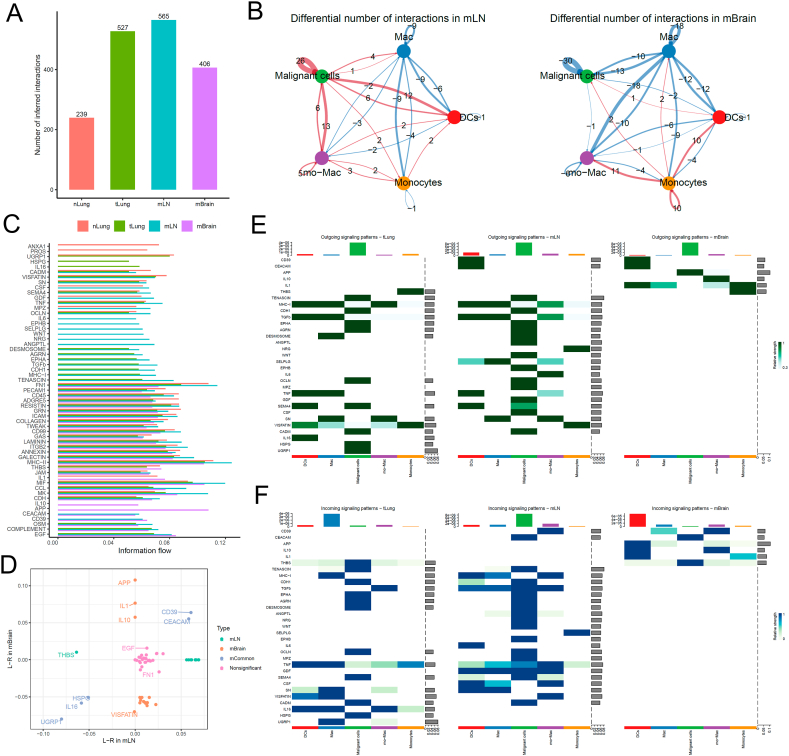
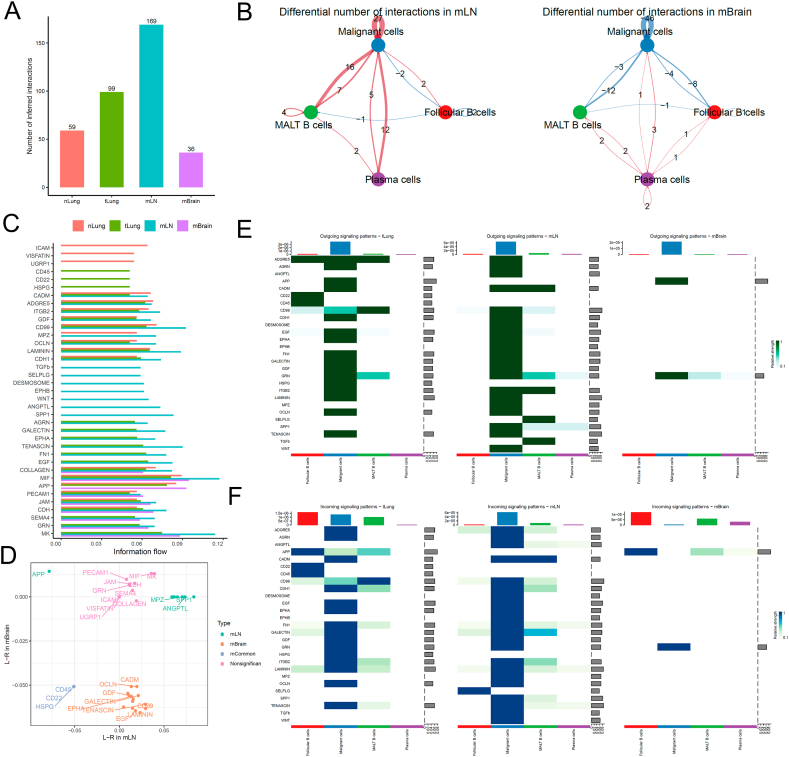
We re-clustered and fine-annotated the B-cell subtypes and identified their three subtypes, namely follicular B, MALT B, and plasma cells. The overall cellular interactions between B cells and malignant cells were similar to those between T cells and malignant cells (Fig. 3A). The interactions between B cells and malignant cells, specifically those between MALT B and malignant cells, increased in the tLung and mLN groups but decreased in the mBrain group (Fig. 3B). Then, we examined the information flow for each pair of ligand–receptor signaling pathways among different sites (Fig. 3C and D). We observed an increased strength of B-cell interactions between MPZ, SPP1, and ANGPTL in only mLN group, and the ligand-receptor HSPG, CD22, and CD45 decreased in both mLN and mBrain groups. Notably, any mBrain-specific signaling pathway was not observed with B cells. However, multiple cell surface, cell adhesion, and migration-related signaling pathways, including EGF, LAMININ, CD99, EPHA, GALECTIN, GDF, OCLN, and CADM, were significantly decreased in the mBrain group. Fig. 3E and F presents a detailed comparison of the differential number of interactions or interaction strengths for B cells and malignant cells.
Fig. 3.
Cell–cell communication analysis between B cells and malignant cells in tumor. (A) Bar plot showing the total number of the inferred cell–cell communication networks among nLung, tLung, mLN, and mBrain groups. (B) Circle plots summarize the number of differential expressed interactions between mLN and tLung or mBrain and tLung groups. (C) Top representative differentially expressed L–R signaling pathways among the four groups. (D) Scatterplot shows common or specific signal pathways between B cells and malignant cells in the mLN (x-axis) and mBrain (y-axis) groups. (E, F) Detailed visualization of cell–cell communication between different cell groups for incoming and outgoing signals.
4.1.3. Myeloid to tumor cell interactions in metastatic vs. non-metastatic cancer cells
We re-clustered and fine-annotated myeloid cells and identified their four subtypes, namely dendritic cells (DCs), macrophage, monocyte-macrophage, and monocytes. The overall cellular interactions between myeloid cells and malignant cells in the tumor tissues were higher than in the normal tissues (Fig. 4A). However, the interactions in the tLung and mLN groups were stronger than in the mBrain group (Fig. 4B). The information flow for each pair of ligand–receptor signaling pathways showed an increased strength of cellular interaction between CD39 and CEACAM in both mLN and mBrain groups, and the ligand-receptors HSPG, IL16, and UGRP1 were decreased in both groups (Fig. 4C). In addition, three mBrain-specific signaling pathways, including IL1, IL10, and APP were found (Fig. 4D), which may play a role in promoting the growth and survival of cancer cells in the brain. Overall, our analysis revealed a landscape of cell–cell interactions across various metastatic tumor sites in lung cancer.
Fig. 4.
Cell–cell communication analysis between myeloid and malignant cells in tumor. (A) Bar plot showing the total number of the inferred cell–cell communication networks among nLung, tLung, mLN, and mBrain groups. (B) Circle plots summarize the number of differential expressed interactions between mLN and tLung or between mBrain and tLung groups. (C) Top representative differentially expressed L-R signaling pathways among the four groups. (D) Scatterplot shows common or specific signal pathways between myeloid and malignant cells in mLN (x-axis) and mBrain (y-axis) groups. (E, F) Detailed visualization of cell–cell communication between different cell groups for incoming and outgoing signals.
5. Discussion
We conducted a comprehensive analysis of metastatic lung cancer at the single-cell level using public databases. We used pseudotime and cell communication analyses and identified for the first time that tumors from different metastatic regions have distinct evolutionary patterns. These distinct evolutionary patterns can be attributed to differences in the TMEs, especially the proportion of immune cells. Our findings suggest that the metastatic tumor in the mBrain group has undergone more transcriptomic alterations (a higher pseudotime) than the metastatic tumor in the mLN and the primary tumor in the tLung group. Another study further supported the idea that metastases have unique genomic and epigenetic characteristics by providing an integrated genomic and DNA methylation analysis of advanced NSCLC with brain metastases. The study identified differences in somatic mutations, copy number variants, and DNA methylation profiles between primary lung tumors and brain metastatic tissue [24]. These diversity across tumors from various tissues validates the concept that the TME is pivotal in modulating the genetic and epigenetic profile of the tumor [25]. Each tissue provides a unique microenvironment, potentially impacting tumor cell activities such as growth, invasion, and therapeutic resistance [26]. Moreover, the distance the tumor travels may also influence its evolutionary status.
Further, we compared primary, mLN, and mBrain tumors and observed significant changes in tumor-related immune cell populations. The overall number of T, B, and myeloid cells in the brain was less than that in the lymph node and primary tumors due to the blood–brain barrier. However, we found that these immune cells interact with brain cancer cells, and significant differences were observed among the four groups. The interactions involving B cells were the least, whereas the number of interactions involving myeloid and T cells was similar. Zhang et al. [27] also suggested that brain metastasis environment is significantly immunosuppressed compared to the primary lung tumor due to reduced T-cell and B-cell abundance/activity and increased neutrophil infiltration. This suggests a distinct immune landscape in brain metastases.
Exhausted T cells show a significant increase in the signaling pathways involving MHC-I, SPP1, and IFN-II. SPP1 is closely related to PD-L1; its increase can mediate macrophage polarization and facilitate immune escape in lung adenocarcinoma [28]. IFN-II may be the prime activator of CD8+ T-cell exhaustion to restrict antitumor immunity [29]. Notably, both the activated signals implied the immune escape mechanism of brain metastatic tumors, which was align with other researches in this field [27,30].
DCs showed the most robust interactions with other cell types and modulated immune-related signaling pathways, such as the IL-10 and APP pathways, through both incoming and outgoing signals. IL10 is an anti-inflammatory cytokine that can suppress the immune response, and it can be beneficial in limiting inflammation and tissue damage in certain contexts. However, IL-10 can promote tumor growth and metastasis by suppressing the ability of the immune system to detect and eliminate cancer cells. It has indicated that targeting tumors with IL-10 can inhibit DC-induced apoptosis of CD8+ T cells [31]. APP is a transmembrane protein involved in cell adhesion, signaling, and the regulation of neuronal survival. A class of enzymes called secretases cleavesAPP to produce various fragments, including beta-amyloid, which is the main component of the plaques found in the brains of patients with Alzheimer's disease. In lung cancer brain metastasis, APP may be involved in promoting cancer cell survival and proliferation [32]. Lung cancer cells that metastasize to the brain express high levels of APP, and blocking the APP pathway can inhibit the growth and survival of these cells. One proposed mechanism for the role of APP in lung cancer brain metastasis speculates its interaction with a protein called APLP2, which is also involved in cell adhesion and survival [33,34]. APLP2 is upregulated in lung cancer brain metastasis, and its interaction with APP may promote cancer cell survival and migration to the brain [35].
While utilizing the same datasets (GSE131907 and GSE123904), our study employed a unique analytical approach, leading to findings that diverge in specific L-R interactions and pathways when compared to the original reports. Our approach revealed distinct patterns of cellular communication and pathway activation that were not as emphasized in the original studies. By doing so, our research corroborates and expands upon the findings of the referenced articles, providing a more comprehensive and nuanced understanding of the cellular and molecular dynamics in metastatic lung adenocarcinoma.
In summary, this study demonstrates that metastatic tumors in the mBrain group exhibited more extensive transcriptomic alterations than those in the mLN group and primary tLung tumors, as evidenced by increased pseudotime in scRNA-seq analysis. This variation in evolutionary status likely results from the unique microenvironments of different tissue sites. A critical advancement of this research is the elucidation of lung cancer cell adaptation and evolution during disease progression and in response to therapy. Additionally, the study highlights distinct gene expression profiles at various stages of tumor progression and metastasis, contributing to a refined understanding of lung cancer. These findings emphasize the importance of considering the specific TME and suggest a future focus on translating molecular insights into clinical applications. This may include the development of targeted therapies and personalized medicine strategies and the identification of therapeutic targets to counteract resistance. Furthermore, integrating scRNA-seq with other omics data, such as genomic and proteomic data, promises a more comprehensive understanding of lung cancer biology.
However, the study has limitations. The datasets (GSE131907 and GSE123904) may have constraints regarding sample size, diversity, and representativeness. The methodology's exclusion of paired sample analysis from the same patients limits insights into individual cancer progression. Additionally, the lack of experimental validation of computational findings presents a significant limitation; these theoretical results require further empirical verification through in vitro or in vivo methods to fully understand the dynamic nature of cancer progression.
Overall, this research is crucial for understanding tumor behavior and response to therapy, guiding future research towards developing novel therapies for brain metastatic tumors, particularly by targeting unique immune evasion signals characteristic of brain metastasis.
6. Ethical approval and consent to participate
Not applicable.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA (http://portal.gdc.cancer.gov/) and GEO (www.ncbi.nlm.nih.gov/) under the accession number GSE131907 and GSE123904.
CRediT authorship contribution statement
Xiaoyuan Wang: Writing – original draft, Formal analysis, Conceptualization. Hao Liang: Writing – review & editing, Formal analysis, Data curation. Xiaoli Tang: Writing – review & editing, Data curation. Xiaodong Ling: Supervision. Yingnan Yang: Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to the members of Yang laboratory for their valuable comments. We also thank the support of Harbin Medical University Cancer Hospital.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e27071.
Contributor Information
Xiaodong Ling, Email: xiaodongling@hrbmu.edu.cn.
Yingnan Yang, Email: yangyingnanltx@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Hirsch F.R., et al. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 2.Zappa C., Mousa S.A. Non-small cell lung cancer: current treatment and future advances. Transl. Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizzo A. Identifying optimal first-line treatment for advanced non-small cell lung carcinoma with high PD-L1 expression: a matter of debate. Br. J. Cancer. 2022;127(8):1381–1382. doi: 10.1038/s41416-022-01929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mollica V., et al. The impact of ECOG performance status on efficacy of immunotherapy and immune-based combinations in cancer patients: the MOUSEION-06 study. Clin. Exp. Med. 2023;23(8):5039–5049. doi: 10.1007/s10238-023-01159-1. [DOI] [PubMed] [Google Scholar]
- 5.Santoni M., et al. The impact of gender on the efficacy of immune checkpoint inhibitors in cancer patients: the MOUSEION-01 study. Crit. Rev. Oncol. Hematol. 2022;170 doi: 10.1016/j.critrevonc.2022.103596. [DOI] [PubMed] [Google Scholar]
- 6.Rizzo A., et al. Impact of Proton pump inhibitors and histamine-2-receptor antagonists on non-small cell lung cancer immunotherapy: a systematic review and meta-analysis. Cancers. 2022;14(6) doi: 10.3390/cancers14061404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang B., et al. Analysis of risk factors and gene mutation characteristics of different metastatic sites of lung cancer. Cancer Med. 2022;11(1):268–280. doi: 10.1002/cam4.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomuleasa C., et al. [The fundamental mechanisms of metastatic spread and chemotherapy resistance in lung cancer] Pneumologia. 2011;60(2):99–103. [PubMed] [Google Scholar]
- 9.Salem A., et al. Targeting hypoxia to improve non–small cell lung cancer outcome. JNCI: J. Natl. Cancer Inst. 2018;110(1):14–30. doi: 10.1093/jnci/djx160. [DOI] [PubMed] [Google Scholar]
- 10.Hao D., et al. The single-cell immunogenomic landscape of B and plasma cells in early-stage lung adenocarcinoma. Cancer Discov. 2022;12(11):2626–2645. doi: 10.1158/2159-8290.CD-21-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q., et al. Molecular profiling of human non-small cell lung cancer by single-cell RNA-seq. Genome Med. 2022;14(1):1–18. doi: 10.1186/s13073-022-01089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sinjab A., et al. Resolving the spatial and cellular architecture of lung adenocarcinoma by multiregion single-cell sequencing. Cancer Discov. 2021;11(10):2506–2523. doi: 10.1158/2159-8290.CD-20-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sultana A., et al. Single-cell RNA-seq analysis to identify potential biomarkers for diagnosis, and prognosis of non-small cell lung cancer by using comprehensive bioinformatics approaches. Translational Oncology. 2023;27 doi: 10.1016/j.tranon.2022.101571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim N., et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat. Commun. 2020;11(1):2285. doi: 10.1038/s41467-020-16164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laughney A.M., et al. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat. Med. 2020;26(2):259–269. doi: 10.1038/s41591-019-0750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McInnes L., Healy J., Saul N., Grossberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3(1):861. [Google Scholar]
- 17.Trapnell C., et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y., et al. WebGestalt Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 20.Jin S., et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandrashekar D.S., et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z., et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkley D., et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 2022;54(8):1192–1201. doi: 10.1038/s41588-022-01141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y., et al. Integrated genomic and DNA methylation analysis of patients with advanced non-small cell lung cancer with brain metastases. Mol. Brain. 2021;14(1):176. doi: 10.1186/s13041-021-00886-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H., Han G., Chen J. Heterogeneity of tumor immune microenvironment in malignant and metastatic change in LUAD is revealed by single-cell RNA sequencing. Aging (Albany NY) 2023;15(12):5339. doi: 10.18632/aging.204752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu X., et al. Single-cell omics traces the heterogeneity of prostate cancer cells and the tumor microenvironment. Cell. Mol. Biol. Lett. 2023;28(1):1–16. doi: 10.1186/s11658-023-00450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q., et al. The spatial transcriptomic landscape of non-small cell lung cancer brain metastasis. Nat. Commun. 2022;13(1):5983. doi: 10.1038/s41467-022-33365-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Du W., Chen Z., Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp. Cell Res. 2017;359(2):449–457. doi: 10.1016/j.yexcr.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 29.Lukhele S., et al. The transcription factor IRF2 drives interferon-mediated CD8+ T cell exhaustion to restrict anti-tumor immunity. Immunity. 2022;55(12):2369–2385. e10. doi: 10.1016/j.immuni.2022.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wischnewski V., et al. Phenotypic diversity of T cells in human primary and metastatic brain tumors revealed by multiomic interrogation. Nat. Can. (Ott.) 2023;4(6):908–924. doi: 10.1038/s43018-023-00566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao J., et al. Targeting tumors with IL-10 prevents dendritic cell-mediated CD8+ T cell apoptosis. Cancer Cell. 2019;35(6):901–915. e4. doi: 10.1016/j.ccell.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Mikaeili Namini A., et al. An in silico comparative transcriptome analysis identifying hub lncRNAs and mRNAs in brain metastatic small cell lung cancer (SCLC) Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-22252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., et al. Expression of amyloid precursor-like protein 2 (APLP2) in glioblastoma is associated with patient prognosis. Folia Neuropathol. 2018;56(1):30–38. doi: 10.5114/fn.2018.74657. [DOI] [PubMed] [Google Scholar]
- 34.Sliker B.H., et al. Beta 2-microglobulin regulates amyloid precursor-like protein 2 expression and the migration of pancreatic cancer cells. Cancer Biol. Ther. 2019;20(6):931–940. doi: 10.1080/15384047.2019.1580414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McHenry P.R., Prosperi J.R. Proteins found in the triple-negative breast cancer secretome and their therapeutic potential. Int. J. Mol. Sci. 2023;24(3):2100. doi: 10.3390/ijms24032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA (http://portal.gdc.cancer.gov/) and GEO (www.ncbi.nlm.nih.gov/) under the accession number GSE131907 and GSE123904.