Abstract
The DNA genome of a hepatitis B virus is generated by reverse transcription of the RNA pregenome. Replication initiation does not involve a nucleic acid primer; instead, the hepadnavirus P protein binds to the structured RNA encapsidation signal ɛ, from which it copies a short DNA primer that becomes covalently linked to the enzyme. Using in vitro-translated duck hepatitis B virus (DHBV) P protein, we probed the secondary structure of the protein-bound DHBV ɛ RNA (Dɛ) and observed a marked conformational change compared to free Dɛ RNA. Several initiation-competent mutant RNAs with a different free-state structure were similarly altered, whereas a binding-competent but initiation-deficient variant was not, indicating the importance of the rearrangement for replication initiation and suggesting a mechanistic coupling to encapsidation.
Hepatitis B viruses (hepadnaviruses) are small enveloped viruses that replicate by reverse transcription of an RNA intermediate. Despite this general similarity to other retroelements and retroviruses, many steps in their replication are unique (reviewed in references 28 and 30). For instance, extracellular virions contain DNA rather than RNA, and viral DNA does not usually integrate into the host genome. Major differences also exist in the mechanisms for encapsidation of genomic RNA and reverse transcriptase (RT). The RT (P protein), rather than the Gag-equivalent core protein, is responsible for specific RNA packaging (3); in contrast to retroviral RTs, P protein is synthesized as a separate entity (34), and so it lacks a Gag-like domain for mediating its coincorporation into nascent capsids. Furthermore, reverse transcription is initiated by protein priming (39) rather than by a nucleic acid primer, such as the host tRNAs used by retroviruses.
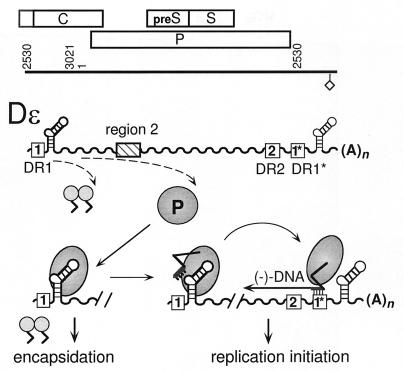
Key to these processes in hepatitis B viruses is the recognition and binding, by P protein, of a stem-loop structure close to the 5′ end of the pregenomic RNA; based on its initially recognized function as an encapsidation signal, it has been termed ɛ (Fig. 1) (16). Formation of this ribonucleoprotein (RNP) complex is required for the generation of replication-competent nucleocapsids (3) and apparently involves cellular chaperones, including Hsp90 (14) and p23 (15). Moreover, ɛ also acts as replication origin, as shown for duck hepatitis B virus (DHBV) (37, 38) and human hepatitis B virus (HBV) (29). The 5′-terminal nucleotide of minus-strand DNA becomes covalently linked to a Tyr residue in the unique terminal protein domain of P protein (see references 41 and 43 for DHBV and reference 21 for HBV) and, templated by an internal region of ɛ, is extended to a 3- or 4-nucleotide (nt) DNA primer (see Fig. 3A). After this priming reaction, the primer/P-protein complex translocates to another cis element, the direct repeat DR1 (Fig. 1). On the terminally redundant pregenomic RNA, ɛ and DR1 are present in two copies, but only 5′ ɛ and 3′ DR1 are active in minus-strand DNA synthesis (33). Hence, hepadnavirus first-strand DNA synthesis is discontinuous, as in retroviruses, but the initiation mechanism is quite different from that in retroviruses. The principal element defining the retroviral DNA initiation site is the 3′ end of a host tRNA held at a fixed position by base pairing to the complementary primer binding site on the genomic RNA (reviewed in references 24 and 25). While nucleic acid priming is used by almost all known retroelements (reviewed in reference 23), in hepatitis B viruses the information for exact start site selection must rely on the specific structure of the P-protein–ɛ complex.
FIG. 1.
Functional roles of the ɛ-P interaction. The straight line represents a linear version of the circular DHBV genome, the diamond represents the polyadenylation signal, and the open rectangles represent the three major open reading frames. Numbers are nucleotide positions. The wavy lines represent the terminally redundant RNA pregenome, which also serves as mRNA for the capsid (C) and the P protein (small and large circles). Dɛ is shown as a symbolic hairpin, and the direct-repeat elements DR1, DR2, and DR1* are shown as boxes. Region 2 is an as yet not well-defined second element required for DHBV pregenome encapsidation (7). Binding of P protein to 5′ Dɛ triggers the addition of dimeric capsid protein subunits and hence nucleocapsid assembly; in addition, replication is initiated by P copying 3 to 4 nt of the Dɛ bulge into a DNA primer that is covalently linked to the P protein. The temporal order, if any, of these events has not yet been established. The entire complex is then translocated to DR1*, and the primer is extended to form minus-strand DNA.
FIG. 3.
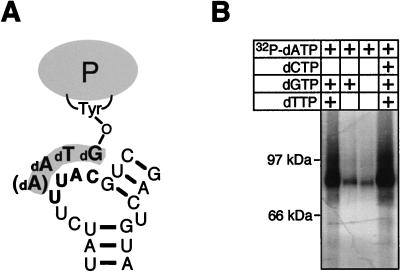
Authentic enzymatic activity of the immobilized and purified ɛ-P complex. (A) Schematic drawing of the DNA primer covalently linked to Tyr 96 of the DHBV P protein. Nucleotides in the bulge region of Dɛ serve as the template. (B) The immobilized and purified ɛ-P complex exhibits sequence-specific priming activity. Priming reactions were performed in the presence of [α-32P]dATP plus various combinations of unlabeled dNTPs as indicated in the figure, and samples were analyzed by sodium dodecyl sulfate-PAGE. Strong labeling of P protein was dependent on both dGTP and dTTP (left lane), in agreement with the synthesis of a primer with an authentic sequence.
For the P protein of DHBV, but not that of HBV, the priming reaction and limited primer elongation can be artificially reconstituted by in vitro translation of the protein in rabbit reticulocyte lysate (39), which provides the necessary chaperones. In contrast to the in vivo situation, Dɛ may be present at either end of the P mRNA used to program the lysate (cis priming) or may be added as a separate molecule (trans priming) (40). Despite this relaxed position specificity, all available evidence suggests that the in vitro reaction faithfully mimics the first step of hepadnavirus reverse transcription. By contrast, the DNA polymerase activity of HBV P protein expressed from recombinant baculoviruses (20) is also seen in the absence of ɛ (21).
Previous analyses of free in vitro-transcribed hepadnavirus ɛ RNAs (see references 19 and 31 for HBV and reference 4 for DHBV) had shown that their overall secondary structures are similar: they consist of a lower and an upper stem, separated by a bulged region of about 6 nt, and an apical loop of similar size (for Dɛ, see Fig. 4C). Mutational analyses with transfected cells (see references 19 and 31 for HBV and reference 32 for DHBV) demonstrated that these general structural features are important for the function of ɛ in encapsidation and replication, since gross alterations were not tolerated. Combining secondary-structure analyses and functional tests in the in vitro system of a panel of Dɛ variants, we have recently defined several additional determinants for a productive interaction between RNA and protein (6). While some of the data were compatible with a simple lock-and-key recognition mechanism, data for two classes of mutants suggested that, instead, a dynamic induced-fit mechanism operates during formation of an initiation-competent RNP complex: first, the ɛ signal of the related avian heron hepatitis B virus (HHBV), but not that of HBV (32), interacts productively with the DHBV P protein despite significant differences in primary sequence and apical secondary structure (4); similarly, we identified an initiation-competent Dɛ point mutant (L5) whose apical structure, in the free state, clearly differs from that of Dɛ (6). Hence, within certain limits, differently structured RNAs may adopt a new and probably similar structure once complexed with the protein. Second, we (6) and others (32) identified Dɛ mutants that physically bind to P protein in a competitive fashion but do not support the priming reaction. This phenotype might be explained if the corresponding variants, after the initial binding step, were unable to undergo a subsequent structural rearrangement required for the formation of a replication-competent RNP complex.
FIG. 4.
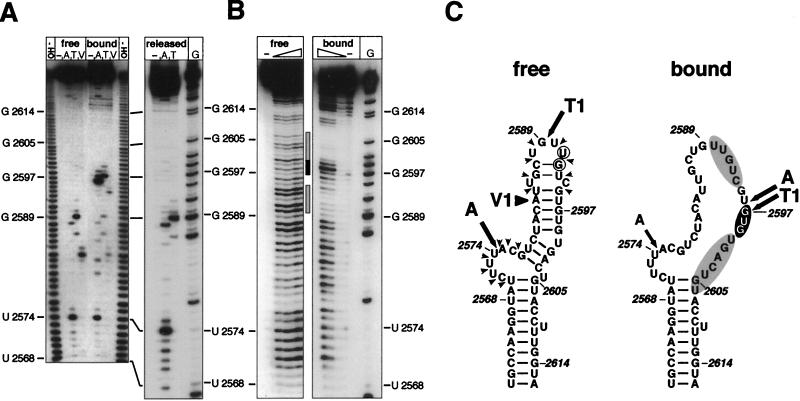
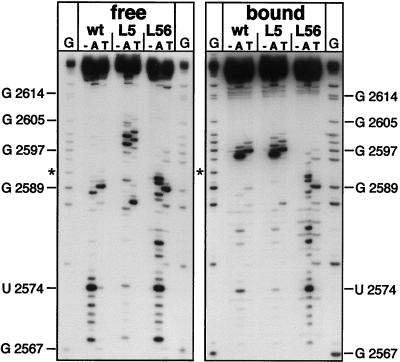
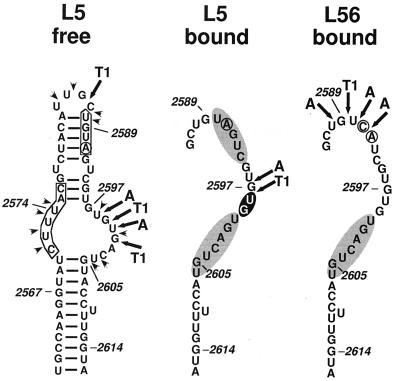
Secondary-structure analysis of free and P-protein-bound Dɛ RNA. (A) Nuclease probing. (Left) Free and P-protein-bound 5′-32P-labeled wt Dɛ RNA was subjected to limited digestion with RNase A (A), RNase T1 (T), and V1 nuclease (V), and the products were analyzed by denaturing PAGE. The minus signs represent an undigested control; a reference nucleotide ladder obtained by partial alkaline hydrolysis of 5′-32P-labeled wt Dɛ RNA is shown in lanes OH−. Right (“released”), the structural analysis of wt Dɛ RNA dissociated from the ɛ-P complex is shown. G represents a G-sequencing track obtained by digestion of wt Dɛ RNA with RNase T1 under denaturing conditions. (B) Lead probing. Free and P-protein-bound 5′-32P-labeled wt Dɛ RNA was treated with increasing amounts of lead acetate, and the products were analyzed by denaturing PAGE. The hypersensitive region of Dɛ in the ɛ-P complex is indicated by a solid bar, and regions showing reduced sensitivity are labeled with shaded bars. (C) Comparison of experimental nucleotide accessibilities in free and P-protein-bound Dɛ RNA with the secondary-structure model. Major sites accessible to single-strand-specific nucleases are indicated by arrows (A, RNase A; T1, RNase T1); The major V1 nuclease cleavage site is labeled by a large arrowhead (V1). Pb2+ cleavages in free RNA are denoted by small arrowheads. Lead cleavage sites enhanced in the ɛ-P complex have a black background, and regions protected from lead cleavage are shaded in gray. Numbers represent nucleotide positions. Nucleotides mutated in the Dɛ variants L5 (U2591A) and L56 (U2591C, G2592A) are encircled.
To obtain direct evidence for the proposed induced-fit mechanism, we used a recently established procedure to isolate the small amounts of the Dɛ-P protein complex from the reticulocyte lysate (5). Then we directly compared the secondary structures of free and P-protein-bound Dɛ RNAs. Our data demonstrate that the apical structure of Dɛ in the complex is drastically different from that in the free state. The priming-competent variant L5, although different from wild-type (wt) Dɛ in the free state, adopted essentially the same new structure as wt Dɛ in the complex. Very similar alterations were observed for three further priming-competent RNAs but not for a variant that is capable of physical but not productive binding to P protein. These data confirm the proposed induced-fit model and further support the notion that the interaction between ɛ and P protein is a highly dynamic process that eventually results in the proper positioning of the hepadnavirus RT on its template for correct initiation of DNA synthesis.
MATERIALS AND METHODS
Plasmid constructs.
Plasmid pDɛ1 (4) contains the ɛ sequence of DHBV16 between a HindIII site and a ClaI site downstream of a T7 promoter and served as template for in vitro transcription of wt Dɛ RNA. Plasmids encoding mutant Dɛ RNAs were obtained by replacing the HindIII-ClaI cassette of pDɛ1 with synthetic or PCR-generated DNA duplexes containing corresponding nucleotide exchanges. His6-tagged DHBV P protein was expressed from plasmid pT7AMVpol16H6 (5), which contains the complete P-protein open reading frame plus an insertion of six His residues between codons 2 and 3.
Dɛ RNA synthesis.
Mutant and wt Dɛ RNAs were obtained by in vitro transcription (T7 MEGAshortscript kit; Ambion, Austin, Tex.) of the corresponding plasmids after linearization with ClaI. The transcripts are nominally 76 nt long and contain the DHBV sequence from positions 2557 to 2624 plus short terminally vector-derived sequences as previously described (4). For 5′ 32P labeling, in vitro transcripts were dephosphorylated with calf intestinal alkaline phosphatase and rephosphorylated with T4 polynucleotide kinase in the presence of [γ-32P]ATP (5,000 Ci/mmol). Labeled transcripts (specific activity, 2 × 105 cpm/pmol) were gel purified, precipitated with isopropanol, and resuspended in TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 7.5]) to a final concentration of 5 μM.
In vitro generation and isolation of the RNP complex.
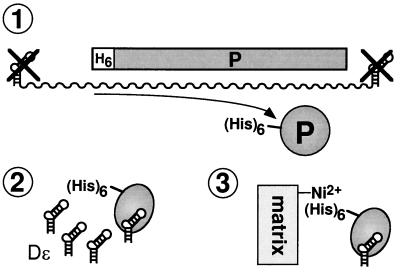
His6-tagged DHBV P protein was produced by in vitro translation in rabbit reticulocyte lysate. The RNA coding for P protein was obtained by in vitro transcription from plasmid pT7AMVpol16H6 linearized with AflII (DHBV position 2526). This P RNA ends exactly with the stop codon of the P ORF and does not contain any ɛ sequences. Usually, in vitro translation was performed in a total volume of 50 μl with 30 μl of rabbit reticulocyte lysate and 2.5 to 5 μg of P RNA. After a 30-min incubation at 30°C, 5′-end-labeled Dɛ RNA was added to a final concentration of 0.8 to 1 μM and the mixture underwent further incubation at 30°C for 60 to 90 min. The RNP complex was separated from free Dɛ RNA by immobilized-metal affinity chromatography (IMAC) essentially as previously described (5). In brief, the samples were mixed with 1 volume of Ni2+-nitrilotriacetic acid (NTA)-agarose (Qiagen, Hilden, Germany) in 400 μl of binding buffer (0.1 M sodium phosphate [pH 7.5], 150 mM NaCl, 0.1% Nonidet P-40, 20 mM imidazole, 100 μg of yeast tRNA per ml) and gently shaken for 90 min at 4°C. The beads were washed twice with 1 ml of binding buffer and at least twice with 1 ml of TMK buffer (50 mM Tris-Cl [pH 7.5], 40 mM KCl, 10 mM MgCl2, 100 μg of yeast tRNA per ml) until less than 10% of the remaining radioactivity was located in the supernatant as detected by Cerenkov scintillation counting. The total yield of P-protein-immobilized 32P-labeled Dɛ RNA was in the range of 105 cpm (500 fmol of RNA).
Probing of the RNP complex.
Aliquots of about 2 × 104 cpm of the Ni2+-NTA resin containing the isolated, immobilized RNP complex as described above were each mixed with 100 μl of TMK buffer and subjected to probing with nuclease or lead at 4°C. Nuclease probing was performed by treating the samples with RNase A (specific for unpaired pyrimidines; 1 ng per reaction), RNase T1 (specific for unpaired G residues; 2 U per reaction), or V1 nuclease (specific for double-stranded or “structured” regions; 14 mU per reaction) for 30 min with rocking. For Pb2+ probing, lead acetate (1 M) was added to final concentrations ranging from 10 to 100 mM. The samples were rocked for 90 min, and the reactions were stopped by adding EDTA to a final concentration of 20 mM to 110 mM. Both nuclease- and lead-treated samples were subsequently subjected to phenol extraction and ethanol precipitation of the RNA from the aqueous phase. Reaction products were analyzed by denaturing polyacrylamide gel electrophoresis (PAGE) on 8 to 10% polyacrylamide gels. G-specific RNA sequencing was performed by RNase T1 digestion under denaturating conditions (7 M urea, 20 mM sodium acetate, 1 mM EDTA [pH 5.0]) at 55°C.
To determine the structure of free Dɛ RNA under equivalent conditions, control reactions were carried out in parallel. For this purpose, in vitro translation reaction mixtures without template RNA were incubated with Ni2+-NTA resin and binding buffer. After the resin was washed several times, 5′-end-labeled Dɛ RNA was added and aliquots containing about 2 × 104 cpm were probed as described above.
Probing of Dɛ RNA dissociated from the RNP complex.
The RNP complex containing 5′-end-labeled wt Dɛ RNA and His6-tagged DHBV P protein was purified by IMAC as described above. After 1 volume of TMK buffer, unlabeled wt Dɛ RNA to a final concentration of 0.5 μM, and 1 U of RNasin per μl were added to the resin, the sample was incubated for 90 min at 30°C. The supernatant was removed, and the resin was washed once with 1.5 volumes of TMK. The supernatants were pooled. TMK was added to both the supernatant and the resin fractions to 300 μl, and aliquots of 100 μl containing free and complexed Dɛ RNA, respectively, were probed with RNase A and RNase T1 as described above.
In vitro priming activity of the RNP complex.
The RNP complex containing radiolabeled wt Dɛ RNA and His6-tagged DHBV P protein was immobilized on Ni2+-NTA resin as described above and separated from unbound wt Dɛ RNA by washing the beads twice with binding buffer and twice with 1× priming buffer (41) supplemented with yeast tRNA (100 μg/ml). Aliquots were taken and incubated with 10 μCi of [α-32P]dATP (3,000 Ci/mmol) and various combinations of unlabeled deoxynucleoside triphosphates (dNTPs; final concentration, 25 μM) in a total volume of 40 μl at 37°C for 1 h. After the resin was washed with 1 ml of priming buffer, P protein was eluted by adding imidazole to a final concentration of 200 mM and the extent of 32P labeling of P protein was analyzed by reducing sodium dodecyl sulfate-PAGE on 7.5% acrylamide gels.
RESULTS
Efficient separation of free and P-protein-bound Dɛ RNA.
The major drawback of the in vitro translation system is the very limited yield of P protein (in the range of 1 ng, or 10 fmol, per μl in the in vitro translation reaction). The protein binds Dɛ RNA with an apparent Kd in the range of 100 nM (4); the maximal priming signal, measured as the extent of [32P]dNMP labeling of the P protein, is obtained at about 1 μM (1 pmol per μl) Dɛ. For a structural analysis of the protein-bound RNA, it was therefore essential to isolate as much as possible of the complex and to completely remove free Dɛ RNA. We have recently shown that an N-terminally His6-tagged P protein is priming competent and can be specifically immobilized on Ni2+-NTA resin (5). This system (outlined in Fig. 2) efficiently separated binding-competent RNAs from an approximately 100-fold excess of input RNA molecules (5). To prove that the immobilized RNP complex is functionally authentic, we performed a priming reaction in the presence of [32P]dATP. If directed by the authentic template region in the Dɛ bulge, incorporation of dAMP into the primer [5′-GTA(A)-3′] (Fig. 3A) should depend on the presence of dGTP and dTTP. Indeed, labeling of the complex, from which most of the endogenous dNTPs in the lysate had been removed during the immobilization procedure, was strongly increased by dGTP plus dTTP but not by dGTP alone. No major further increase was observed when dCTP was also present (Fig. 3B). The amount of 32P-labeled P protein in the supernatant was below the detection limit, indicating that the P-protein molecules were enzymatically active while being bound to the Ni2+-containing resin. Hence, the immobilized His6-P protein–Dɛ complex has the same set of features previously described for the wt RNP in solution (39).
FIG. 2.
In vitro assembly and purification of the ɛ-P RNP complex. N-terminally His6-tagged DHBV P protein was expressed by in vitro translation in rabbit reticulocyte lysate from an RNA template lacking Dɛ sequences (step 1). Upon addition of in vitro-transcribed Dɛ RNA the ɛ-P complex was formed (step 2) and separated from free Dɛ RNA by IMAC (step 3).
A major structural rearrangement of Dɛ RNA upon binding to P protein.
Previous secondary-structure analyses of free Dɛ RNA (4) had shown that the most prominent signals obtained with the single-strand specific RNases A and T1 were U2574 and G2589, which are indicative of the bulge and the loop, respectively; the presence of a base-paired apical stem was demonstrated by cleavage in this region by the structure-specific V1 nuclease (Fig. 4C). Here we used the same enzymes to probe the structure of 5′-end-labeled wt Dɛ RNA bound to immobilized His6-P protein. To minimize potential effects of DNA primer synthesis, all the experiments were performed in the absence of exogenously added dNTPs. As a control for a possible influence of the Ni2+-NTA matrix and residual lysate components, we first reexamined the structure of free Dɛ RNA in the presence of Ni2+-NTA resin preincubated with reticulocyte lysate. No significant difference to the previously reported data was observed (Fig. 4A, lanes “free”). By contrast, the nuclease cleavage pattern of P-protein-bound RNA was dramatically different from that of free Dɛ RNA (lanes “bound”). Most striking was a series of new, neighboring product bands generated by RNases A and T1 in a region that is completely inaccessible to these nucleases in free RNA and that corresponds to right half of the upper stem (Fig. 4C). In contrast to these sites with enhanced reactivity, a strong reduction of the signal corresponding to G2589 in the loop and, less pronounced, at U2574 in the bulge was observed (see lanes wt in Fig. 5, where this reduction is more clearly seen). In addition, the V1 signal between the bulge and the loop in free RNA was substantially weakened.
FIG. 5.
Enzymatic secondary-structure analysis of Dɛ mutants L5 and L56 compared with wt Dɛ in their free state and bound to P protein. For further explanations and abbreviations, see the legend to Fig. 4. G-sequencing tracks on the left side of both panels (“free” and “bound”) were obtained from wt Dɛ RNA, and those on the right side were obtained from mutant L56 RNA. Note that a band representing G2592 (labeled with an asterisk) appears only with wt Dɛ, since L56 contains an A at that position.
A loss of signal intensity could be due either to shielding of the RNA by protein or to the intrinsic structure of the RNA; however, the appearance of new products in a previously inaccessible region demonstrates unequivocally that the structures of free and protein-bound RNA are different. In view of the secondary-structure model (Fig. 4C), this implies that several base pairs in the upper stem are opened. The notion that this structural difference is induced by RNA binding to the protein is supported by the following experiment. We incubated the immobilized RNP complex with an excess of unlabeled Dɛ RNA and examined the structures of the liberated molecules in parallel with those of molecules still bound to P protein. While the nuclease pattern of the bound fraction remained unchanged (results not shown), that of the RNA dissociated from the complex was virtually identical to that of free RNA (Fig. 4A, lanes “released”). These data indicate that the conformation of Dɛ RNA in the RNP complex is not stable in the absence of P protein and, upon dissociation, is rapidly reconverted into the structure adopted by the bulk of free Dɛ RNA molecules.
Pb2+ probing confirms the nuclease data and suggests specific contacts between protein and Dɛ RNA.
At high concentrations, Pb2+ cleaves RNA preferentially at unpaired residues and therefore, because of its smaller size, can be used to probe the RNA conformation with higher resolution than is possible with the bulky nucleases (10, 13). In addition, nucleotides in direct contact with protein should be protected from cleavage, resulting in footprint signals (9, 22). When free Dɛ RNA was incubated with increasing concentrations of Pb2+, cleavage occurred predominantly in the unpaired bulge and in the apical loop region, as expected (Fig. 4B, lanes “free”). A distinctly different pattern was obtained with Dɛ RNA in the RNP complex (lanes “bound”). A region in the upper stem around U2598 became highly sensitive, in accordance with the hypersensitive region defined by using the nucleases. A slightly enhanced reactivity was also detected around A2581, i.e., in the opposite strand of the upper stem. By contrast, the two regions immediately flanking the hypersensitive site (U2590 to C2594 and U2600 to U2606) were strongly protected from cleavage. In the structural model for free Dɛ RNA, they correspond to the 3′ half of the apical loop and the region opposite the bulge (Fig. 4C, “bound”). Taken together, these data are fully compatible with and extend the results of nuclease probing. They support the interpretation that the upper stem of Dɛ, upon binding to P protein, is at least partially opened and that this process is accompanied by specific protein-RNA contacts.
Priming-competent RNA variants with different free-state structures adopt similar new conformations to wt Dɛ in the complex.
To further substantiate the induced-fit mechanism suggested by the above data, we first took advantage of a previously identified point mutant of Dɛ, L5, that carries a single U-to-A exchange at nt 2591 in the apical loop (Fig. 4C) (6). In the free RNA, this nucleotide exchange results in a strikingly different nuclease pattern in the apical half of the structure (Fig. 5). For a compatible secondary-structure model, see Fig. 7. Nonetheless the variant binds to P protein and supports the priming reaction with at least half the efficiency of wt Dɛ (6). If the structure of Dɛ when bound by P protein is important for initiation of DNA synthesis, one would expect L5 RNA to adopt a very similar conformation. Reprobing free L5 RNA in the presence of Ni2+-containing resin preincubated with lysate did not reveal differences from the previously reported data. The bulge indicator nucleotide at U2574 and various signals at positions corresponding to the right half of the upper Dɛ stem (U2598 to G2601) that distinguish the variant from wt Dɛ were clearly visible (Fig. 5, lanes L5). In the complex, however, L5 RNA produced a new pattern that was nearly identical to that of protein-bound wt Dɛ. The same new hypersensitive sites appeared, and, moreover, the distinctive signals typical for free L5 RNA were strongly reduced. We conclude that in the protein-bound state, the structures of the two RNAs are very similar. That the bulk of molecules of the two RNA species have a different structure in the free state further supports the notion that the common new structure is induced by binding to the protein.
FIG. 7.
Comparison of experimental nucleotide accessibilities in free L5 RNA and in P-protein-bound L5 and L56 RNA with their secondary-structure models. (Left) Model of free L5 RNA. For comparison, nucleotides corresponding to the 6-nt bulge and the 4-nt apical loop of wt Dɛ are boxed. Nucleotides different from wt Dɛ are encircled. (Middle and right) Models of L5 and L56 RNA in the P-protein-bound state. Only the right half of each structure is shown. The lead-hypersensitive region of L5 has a black background. Regions significantly protected from lead cleavage in the ɛ-P complex have a gray background.
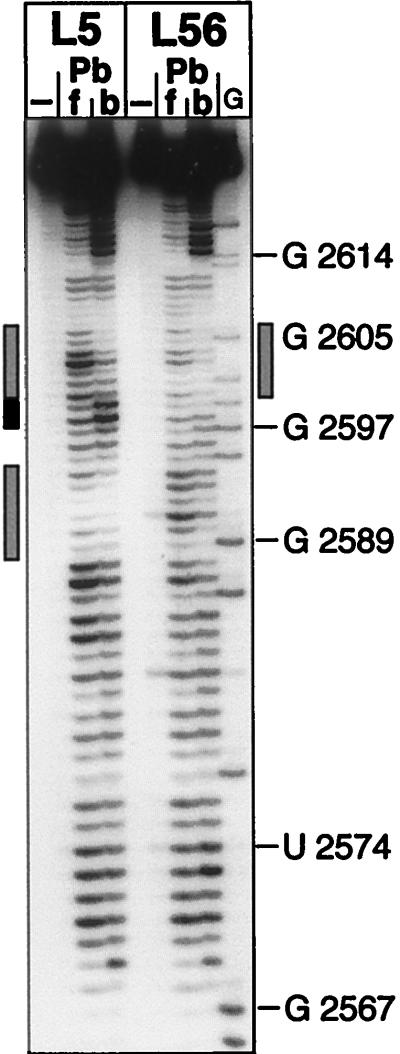
Further evidence for the similarity between the complex-specific structures of L5 and wt Dɛ was obtained by Pb2+ probing (Fig. 6). The most intense signals in free L5 RNA were at positions corresponding to single-stranded regions in the secondary-structure model shown in Fig. 7. In the complex, a prominent enhancement of the signals at positions U2598 and G2599 and strongly decreased signals in the adjacent regions were observed (schematically indicated in Fig. 7). These data are completely congruent with those obtained for wt Dɛ.
FIG. 6.
Lead probing of Dɛ mutants L5 and L56 in their free state (lanes f) and bound to P protein (lanes b). Free and P-protein-bound 5′-32P-labeled wt Dɛ RNA was treated with lead acetate (30 mM final concentration), and the products were analyzed by denaturing PAGE. The hypersensitive region of L5 is indicated by a solid bar, and regions showing reduced sensitivity are labeled with shaded bars. Numbers represent nucleotide positions.
To further corroborate the functional relevance of the structural alteration, we analyzed the P-protein-bound forms of several other RNAs (Fig. 8). A naturally occurring variant is the Hɛ signal of HHBV (35). It interacts productively with DHBV P protein despite seven nucleotide exchanges relative to Dɛ that do not allow for extensive base pairing in the upper stem (4) and result in a much more open apical structure (Fig. 8B). Consequently, many more positions are accessible to single-strand-specific nucleases in the free state (Fig. 8A, lanes Hɛ f). In the complex, however, the RNase A signals at C2596 and U2597, i.e., at the identical positions to those in Dɛ, were clearly enhanced (lanes Hɛ b). By contrast, the intensities at several other positions were decreased; this occurred most visibly between G2599 and U2606, i.e., overlapping with a region that in bound Dɛ is protected from Pb2+ attack. Next, we analyzed the artificial variant V3 (6) that bears six nucleotide exchanges relative to both Dɛ and Hɛ; its P-protein binding and priming phenotype is similar to that of Hɛ (6). The most prominent change between the free and bound form (lanes V3) was the increased RNase A signal intensity at C2595 and C2597, i.e., essentially the same region as in wt Dɛ (U2596, G2597). The minor 1-nt shift is most probably related to the specific sequence of the variant: unlike its counterpart in Dɛ, the A residue at position 2596 is not a substrate for RNase A or T1, while the intensity of C2595 is probably enhanced by the following A2596, as is commonly observed for the sequence pyrimidine-A (18). These data also indicate that the specific nature of the nucleotides in the hypersensitive region is not important.
FIG. 8.
Enzymatic secondary-structure analysis of Hɛ and Dɛ mutant V3 in their free state (lanes f) and in the ɛ-P complex (lanes b). (A) Nuclease probing. Hypersensitive sites are marked with arrowheads. For further explanations and abbreviations, see the legend to Fig. 4A. (B) Location of hypersensitive cleavage sites in the ɛ-P complex with respect to the secondary-structure model of Hɛ. Nucleotides different between Hɛ and wt Dɛ are encircled. The sequence of V3 differs at six positions from that of Hɛ and is denoted inside the large apical loop. Hypersensitive RNase A sites of P-protein-bound Hɛ and V3 RNA are indicated by arrows labeled Hɛ and V3.
Hence, three additional RNAs that interact productively with DHBV P protein all undergo a distinct conformational alteration in the ɛ-P complex similar to wt/Dɛ, despite their different primary sequences and free-state secondary structures. Essentially the same results were obtained with another Dɛ point mutant, G2586A. It carries a 1-nt exchange with respect to wt Dɛ just 5′ of the apical loop that results in an approximately fivefold-lower P-protein binding and priming efficiency (data not shown). These data further support the importance of a specific new RNA structure in a priming-competent hepadnavirus initiation complex.
A priming-incompetent Dɛ variant with a wt-like structure in the free state is not significantly altered when bound to P protein.
Binding of ɛ RNA to P protein is absolutely required for the template-directed priming reaction but not sufficient per se, as shown by the behavior of Dɛ variants that bind to P protein but do not support the priming reaction (6, 32); their competition with wt Dɛ suggests that they principally use the same binding site. An example is variant L56, which contains a 2-nt exchange in the loop (U2591C, G2592A; Fig. 4C). In our hands, L56 RNA bound to DHBV P protein with about one third the efficiency of wt Dɛ but elicited a priming signal of only about 3% of that of wt Dɛ (data not shown), in accord with previously reported data (32). In view of the above-described results, this priming defect might plausibly be related to an inability to undergo the conformational rearrangement observed for the priming-competent RNAs. Nuclease probing of free L56 RNA produced a pattern similar to that of wt Dɛ (Fig. 5, lanes L56). Importantly, however, this pattern changed only slightly upon binding to P protein. No hypersensitive sites were present in the region of the upper stem. Furthermore, no major signal reduction was seen in the loop region. The trivial explanation that the L56-P complex is unstable and what has been analyzed was free RNA can be ruled out, since no substantial dissociation of the L56-P complex during incubation in probing buffer was detectable (results not shown).
These results were corroborated by Pb2+ probing (Fig. 6, lanes L56). The free-state cleavage pattern, which was similar to that of free wt Dɛ RNA, changed only slightly in the complex. The most obvious alteration was a signal reduction between nt 2600 and 2606, i.e., corresponding to the second prominent footprint opposite the bulge region in Dɛ and L5 RNA (Fig. 7). However, no significant signal enhancement was seen in the region that is hypersensitive in complexed Dɛ and L5 RNA, and, at most, a weak protection occurred between the loop and the hypersensitive region. These data suggest that L56 RNA is trapped in a nonproductive complex with P protein.
DISCUSSION
Replication initiation in hepatitis B viruses employs a complex and highly unusual mechanism. In contrast to most DNA polymerases and reverse transcriptases, first-strand DNA synthesis does not rely on extending a given 3′ end of a nucleic acid primer whose position on the template strand is determined by Watson-Crick base pairing; rather, a specific tyrosine OH group of the P protein is used to anchor the first nucleotide of the minus DNA strand. Hence, the information for exactly positioning the hepadnavirus RT on the initiation site must rely on the interaction with its cognate RNA element ɛ. Its internal position on the template strand distinguishes it from other systems that do not require nucleic acid primers, like RNA replicases or the exceptional RT of the Mauriceville retroplasmid (8); it also differs from the protein-primed DNA replication mechanism used, e.g., by adenovirus (17) and phage phi 29 (26), where the initiation sites are usually confined to the 3′ ends of the template strands. This underscores the importance of structural information on the initiation complex for understanding the mechanism of hepatitis B virus replication.
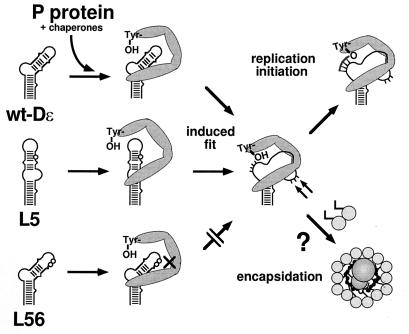
Previous analyses involving the in vitro translation system had shown a general correlation between preservation of the stem-loop structure and the priming ability of various Dɛ mutants (4, 6). However, several RNA variants did not seem to fit into this simple scheme. This led us to propose that formation of a functional initiation complex might require structural alterations in the RNA; these would be induced upon complex formation and would serve to properly juxtapose the template region to the active site and the Tyr-anchor residue of the enzyme (6). The data presented in this study fully support this model, which is summarized in Fig. 9.
FIG. 9.
Model for replication initiation complex formation in hepatitis B viruses. Free wt and mutant Dɛ RNAs are shown as schematic stem-loops representing their most likely secondary structures. Positions of nucleotides exchanged in the mutants L5 and L56 are symbolized by circles. P protein is represented by the U-shaped gray object. Binding of the protein to wt Dɛ and L5 RNA, probably via regions at the base of the bulge and in the vicinity of the apical loop, induces a distinct conformational change in the RNA and the reverse transcriptase. As a result, the primer template region, corresponding to the 3′-half of the bulge in free wt Dɛ RNA, becomes properly oriented with respect to the active site of the enzyme and the anchoring Tyr residue, and primer synthesis commences. L56 RNA binds to P protein but is unable to undergo this conformational shift, possibly because the loop mutations disturb the apical interaction with the protein. Formation of the initiation complex could provide a specific binding site for core protein dimers (connected circles) and hence could trigger encapsidation.
Both nuclease and Pb2+ probing revealed striking differences in the apical part of Dɛ upon complex formation. Most evident was the appearance of a set of new cleavage sites in a region that, in the free RNA, corresponds to the center of the right half of the upper stem (Fig. 4C). Three adjacent nucleotides at this new site were recognized by RNase A, T1 and Serratia marcescens nuclease (results not shown); a clear enhancement was also obvious with Pb2+. All the available data are in accord with the double-strandedness of this region in the free RNA: there are no signals with single-strand-specific nucleases, but the opposite strand is cleaved by V1 nuclease, which recognizes helical regions. Hence, these data demonstrate a remarkable structural difference between free and bound RNA.
In addition, several signals that are prominent in the free state were clearly reduced in the protein-bound RNA. Examples are G2589 in the loop and, most strikingly, the almost complete loss of the Pb2+ signals flanking the hypersensitive site. Such inaccessibility in the complex could be caused by intramolecular RNA structure or by shielding by the bound protein. We favor the second notion because protection in the loop (Fig. 4C) correlated with functional activity of the RNAs as template for P protein and mutations in this region can have a severe impact on P-protein binding and priming (6, 32), indicating that they are important for proper interaction. Furthermore, the immobilized complex probably contains additional proteins, including a dimer of Hsp90 (14) and p23 (15). Although the location of these RNP components with respect to P protein and Dɛ RNA remains to be elucidated, they may at least contribute to shielding the RNA target from the cleavage reagents. This view is further supported by the reduced nuclease signals in protein-bound Hɛ in the same region because there the excess pyrimidines are incompatible with formation of an extended new Watson-Crick paired structure.
The functional relevance of the structural alteration in protein-bound Dɛ RNA is strongly supported by the very similar changes observed for the four other priming-competent RNAs Hɛ, L5, V3, and G2586A. In the complex, all displayed essentially the same hypersensitive site as wt Dɛ, and reduced intensities were observed in the same regions. With the priming-deficient variant L56, by contrast, no hypersensitive site was generated in the complex and the only evident difference in the bound state was protection against Pb2+ cleavage in the region opposite the bulge. Notably, this corresponds to just one of the two regions that appear shielded in the priming-competent RNAs. This strict correlation between an altered conformation of the RNA in the protein-bound state and its template qualities provides strong evidence that functional RNA templates have a distinct structure in the complex that serves to properly position P protein over the initiation site.
Our results also favor an induced-fit mechanism over the recruitment of properly preformed conformers. First, in many probing experiments with free Dɛ RNA, we never observed significant signals in the complex-specific hypersensitive region, and native gel electrophoresis of free Dɛ and L5 gave no hints to the existence of such conformers at measurable concentrations (6). Second, L56 RNA has a free-state structure similar to that of Dɛ and apparently binds to P with that structure. Third, Dɛ released from the complex is indistinguishable from free Dɛ RNA, indicating that without the protein, the free-state structure is much more stable. Finally, recent data from RNA-protein complexes that are amenable to biophysical investigation suggest that such structural rearrangements are probably the rule rather than the exception (reviewed in reference 12).
For instance, interaction of human immunodeficiency virus type 1 Tat protein with TAR RNA induces a local conformational change in the bulge region of TAR that repositions the functional groups on the bases and the phosphate backbone that are critical for specific intermolecular recognition (1). This transition involves unpaired bulge residues which are stacked within the helix and become looped out upon complex formation. The structural changes induced by the binding of U1A protein to a stem-loop structure in the 3′-untranslated region of its pre-mRNA are mostly limited to the ordering of the flexible RNA single-stranded loop against the protein beta-sheet surface (2). In view of these data, the conformational alteration within Dɛ, involving the opening of several base pairs, is rather substantial.
Notably, the P protein itself also appears to be structurally altered upon RNA binding, as suggested by protease sensitivity assays (36). Specific proteolytic fragments correlated with binding to priming-competent Dɛ RNAs. Hence, it is likely not only that ɛ acts as a passive substrate that is molded to functional form by the enzyme but also that the RNA, in turn, is actively involved in altering the structure of the protein into an active, initiation-competent state. Clear-cut evidence for such a folding function of RNA has recently been obtained for the interactions of bacteriophage λ boxB RNA with N protein (27) and of Escherichia coli 4.5S RNA with Ffh protein (42). For hepadnaviruses, the strict interdependence of the principal viral replication components would rigidly limit reverse transcription to the correct template and hence protect the host from potentially deleterious unrestricted reverse transcription (11). Probably, further conformational changes occur during and after synthesis of the ɛ-templated primer in preparing P protein for the template switch to DR1*. In vivo, these dynamic alterations might be aided by the cellular chaperones that are associated with the Dɛ-P protein complex (15).
In summary, our data are fully compatible with a dynamic induced-fit model for the formation of a replication-competent hepatitis B virus initiation complex (Fig. 9). Key features are the requirement for a certain structure in the free state of the RNA that is recognized by P protein but, in addition, the ability of an RNA to undergo, together with the protein, the conformational rearrangements necessary for conversion into an active state. Protection of nucleotides opposite the bulge and previous mutational analyses (5, 40) suggest that the base of the bulge contains an important element for the initial recognition of ɛ by P protein. RNAs like wt Dɛ and L5 are then converted into a distinct priming-active conformation; this second step might involve bases in the loop region that are not protected in the inactive L56 RNA.
An intriguing speculation that can be inferred from these data is that formation of the complex-specific structure of Dɛ creates the actual packaging signal, perhaps involving the new nuclease-hypersensitive region (Fig. 9). Binding of P protein to ɛ is required, in an as yet poorly understood reaction, for recruitment of core protein to the complex and thus for capsid assembly (3, 28). It is not sufficient in itself, however, as shown by the packaging defect of variant L56 in the context of DHBV pgRNA (32). Preliminary data indicate that all three other binding-competent but priming-deficient RNAs are also severely encapsidation deficient. A structurally mediated coupling between replication and encapsidation might serve as a quality control to ensure the packaging of only the RNAs that are suited as templates for new DNA genomes.
ACKNOWLEDGMENTS
We thank Heinz Schaller and two unknown grant reviewers for stimulating discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB229) and, initially, grant Na154/3-1. We also acknowledge financial support by the Center for Clinical Research 1 (ZKF1) of the University Hospital Freiburg.
REFERENCES
- 1.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24:3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allain F H, Gubser C C, Howe P W, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interactions determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Schaller H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 1992;11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck J, Bartos H, Nassal M. Experimental confirmation of a hepatitis B virus (HBV) ɛ-like bulge-and-loop structure in avian HBV RNA encapsidation signals. Virology. 1997;227:500–504. doi: 10.1006/viro.1996.8329. [DOI] [PubMed] [Google Scholar]
- 5.Beck J, Nassal M. A sensitive procedure for mapping the boundaries of RNA elements binding in vitro translated proteins defines a minimal hepatitis B virus encapsidation signal. Nucleic Acids Res. 1996;24:4364–4366. doi: 10.1093/nar/24.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck J, Nassal M. Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J Virol. 1997;71:4971–4980. doi: 10.1128/jvi.71.7.4971-4980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvert J, Summers J. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J Virol. 1994;68:2084–2090. doi: 10.1128/jvi.68.4.2084-2090.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, Lambowitz A M. De novo and DNA primer-mediated initiation of cDNA synthesis by the mauriceville retroplasmid reverse transcriptase involve recognition of a 3′ CCA sequence. J Mol Biol. 1997;271:311–332. doi: 10.1006/jmbi.1997.1185. [DOI] [PubMed] [Google Scholar]
- 9.Ciesiolka J, Lorenz S, Erdmann V A. Structural analysis of three prokaryotic 5S rRNA species and selected 5S rRNA ribosomal protein complexes by means of Pb(II) induced hydrolysis. Eur J Biochem. 1992;204:575–581. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- 10.Ciesiolka J, Michalowski D, Wrzesinski J, Krajewski J, Krzyzosiak W J. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J Mol Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 11.Dhellin O, Maestre J, Heidmann T. Functional differences between the human LINE retrotransposon and retroviral reverse transcriptases for in vivo reverse transcription. EMBO J. 1997;16:6590–6602. doi: 10.1093/emboj/16.21.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frankel A D, Smith C A. Induced folding in RNA protein recognition: more than a simple molecular handshake. Cell. 1998;92:149–151. doi: 10.1016/s0092-8674(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 13.Gornicki P, Baudin F, Romby P, Wiewiorowski M, Kryzosiak W, Ebel J P, Ehresmann C, Ehresmann B. Use of lead(II) to probe the structure of large RNA’s. Conformation of the 3′ terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989;6:971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Seeger C. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc Natl Acad Sci USA. 1996;93:1060–1064. doi: 10.1073/pnas.93.3.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Toft D O, Seeger C. Hepadnavirus assembly and reverse transcription require a multicomponent chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Junker-Niepmann M, Bartenschlager R, Schaller H. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 1990;9:3389–3396. doi: 10.1002/j.1460-2075.1990.tb07540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King A J, van der Vliet P C. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 19.Knaus T, Nassal M. The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 1993;21:3967–3975. doi: 10.1093/nar/21.17.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanford R E, Notvall L, Beames B. Nucleotide priming and reverse transcriptase activity of hepatitis B virus polymerase expressed in insect cells. J Virol. 1995;69:4431–4439. doi: 10.1128/jvi.69.7.4431-4439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanford R E, Notvall L, Lee H, Beames B. Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J Virol. 1997;71:2996–3004. doi: 10.1128/jvi.71.4.2996-3004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lentzen G, Moine H, Ehresmann C, Ehresmann B, Wintermeyer W. Structure of 4.5S RNA in the signal recognition particle of Escherichia coli as studied by enzymatic and chemical probing. RNA. 1996;2:244–253. [PMC free article] [PubMed] [Google Scholar]
- 23.Levin H L. It’s prime time for reverse transcriptase. Cell. 1997;88:5–8. doi: 10.1016/s0092-8674(00)81851-6. [DOI] [PubMed] [Google Scholar]
- 24.Litvak S, Sarih-Cottin L, Fournier M, Andreola M, Tarrago-Litvak L. Priming of HIV replication by tRNA(Lys3): role of reverse transcriptase. Trends Biochem Sci. 1994;19:114–118. doi: 10.1016/0968-0004(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 25.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendez J, Blanco L, Salas M. Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16:2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mogridge J, Legault P, Li J, Van Oene M D, Kay L E, Greenblatt J. Independent ligand-induced folding of the RNA-binding domain and two functionally distinct antitermination regions in the phage lambda N protein. Mol Cell. 1998;1:265–275. doi: 10.1016/s1097-2765(00)80027-1. [DOI] [PubMed] [Google Scholar]
- 28.Nassal M. Hepatitis B virus morphogenesis. Curr Top Microbiol Immunol. 1996;214:297–337. doi: 10.1007/978-3-642-80145-7_10. [DOI] [PubMed] [Google Scholar]
- 29.Nassal M, Rieger A. A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J Virol. 1996;70:2764–2773. doi: 10.1128/jvi.70.5.2764-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nassal M, Schaller H. Hepatitis B virus replication—an update. J Viral Hepat. 1996;3:217–226. doi: 10.1111/j.1365-2893.1996.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 31.Pollack J R, Ganem D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. J Virol. 1993;67:3254–3263. doi: 10.1128/jvi.67.6.3254-3263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollack J R, Ganem D. Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J Virol. 1994;68:5579–5587. doi: 10.1128/jvi.68.9.5579-5587.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieger A, Nassal M. Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J Virol. 1996;70:585–589. doi: 10.1128/jvi.70.1.585-589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlicht H J, Radziwill G, Schaller H. Synthesis and encapsidation of duck hepatitis B virus reverse transcriptase do not require formation of core-polymerase fusion proteins. Cell. 1989;56:85–92. doi: 10.1016/0092-8674(89)90986-0. [DOI] [PubMed] [Google Scholar]
- 35.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavis J E, Ganem D. Evidence for activation of the hepatitis B virus polymerase by binding of its RNA template. J Virol. 1996;70:5741–5750. doi: 10.1128/jvi.70.9.5741-5750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavis J E, Perri S, Ganem D. Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J Virol. 1994;68:3536–3543. doi: 10.1128/jvi.68.6.3536-3543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang G H, Seeger C. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol. 1993;67:6507–6512. doi: 10.1128/jvi.67.11.6507-6512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang G H, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71:663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 40.Wang G H, Zoulim F, Leber E H, Kitson J, Seeger C. Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J Virol. 1994;68:8437–8442. doi: 10.1128/jvi.68.12.8437-8442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weber M, Bronsema V, Bartos H, Bosserhoff A, Bartenschlager R, Schaller H. Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J Virol. 1994;68:2994–2999. doi: 10.1128/jvi.68.5.2994-2999.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng N, Gierasch L M. Domain interactions in E. coli SRP: Stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]
- 43.Zoulim F, Seeger C. Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J Virol. 1994;68:6–13. doi: 10.1128/jvi.68.1.6-13.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]