Abstract
In this study, we examined the effect of acetylation of the NH2 tails of core histones on their binding to nucleosomal DNA in the absence or presence of bound transcription factors. To do this, we used a novel UV laser-induced protein-DNA cross-linking technique, combined with immunochemical and molecular biology approaches. Nucleosomes containing one or five GAL4 binding sites were reconstituted with hypoacetylated or hyperacetylated core histones. Within these reconstituted particles, UV laser-induced histone-DNA cross-linking was found to occur only via the nonstructured histone tails and thus presented a unique tool for studying histone tail interactions with nucleosomal DNA. Importantly, these studies demonstrated that the NH2 tails were not released from nucleosomal DNA upon histone acetylation, although some weakening of their interactions was observed at elevated ionic strengths. Moreover, the binding of up to five GAL4-AH dimers to nucleosomes occupying the central 90 bp occurred without displacement of the histone NH2 tails from DNA. GAL4-AH binding perturbed the interaction of each histone tail with nucleosomal DNA to different degrees. However, in all cases, greater than 50% of the interactions between the histone tails and DNA was retained upon GAL4-AH binding, even if the tails were highly acetylated. These data illustrate an interaction of acetylated or nonacetylated histone tails with DNA that persists in the presence of simultaneously bound transcription factors.
DNA in the cell nucleus exists in the form of chromatin. Chromatin structure is quite complex, and several different levels of chromatin packaging have to be perturbed in order for transcription factors to gain access to their binding sites within regulatory DNA sequences (12). In this study, we examined the binding of transcription factors to the first level of chromatin organization, the nucleosome. The nucleosome is the basic chromatin subunit; it consists of a DNA fragment about 180 to 200 bp long wrapped around an octamer of core histones, two each of H2B, H2A, H3, and H4. As demonstrated earlier, the histone octamer represents a tripartite assembly with an overall shape of a cylindrical wedge and a centrally located tetramer, (H3-H4)2, flanked by two H2A-H2B dimers (6). The surface of the histone octamer has 12 periodically located, binary structural motifs that permit the docking of DNA on the octamer; discovery of this structure yielded a model for the nucleosome in which the NH2 termini emerge at alternating sides of the DNA (7). This model is in excellent agreement with the recently resolved crystal structure of the nucleosome core particle at 2.8 Å (45). The histone tails are located external to the core particle and are subject to acetylation, a posttranslational modification which is believed to be involved in transcriptional regulation (29, 30, 43). Until the late 1980s, only indirect circumstantial evidence existed to support the originally proposed hypothesis of Allfrey et al. (3) that acetylation remodels chromatin structure and facilitates transcription (2, 35). The first direct link between histone acetylation and transcriptionally active chromatin became apparent with the development of an immunochemical procedure for the fractionation of chromatin by use of an antibody which specifically recognizes hyperacetylated histones (29, 30). The use of the same fractionation scheme with an antibody specific for H4 showed that both transcriptional silencing of the yeast mating type cassette and telomere silencing are accompanied by a strong decrease in H4 acetylation levels (13). Thus, all of the above studies strongly suggest a close relationship between histone acetylation and transcription.
How can histone acetylation, a posttranscriptional modification of the most abundant proteins within the cell nucleus, be important in transcriptional regulation? At least two different scenarios can be envisaged. In the first model, the reduction of the lysine-positive charges within the histone NH2 tails could perturb or even abolish their interaction with DNA, hence loosening the nucleosome and higher-order chromatin structure (26, 43). This scenario would allow easier transcription factor binding and thus facilitate transcription (43, 75). This hypothesis has become very popular, with recent discoveries suggesting “targeted histone acetylation”: it was found that components of the basal transcriptional machinery, transcription coactivators (11, 14, 15, 47, 51, 77) and transcription corepressors (1, 32, 39, 40), possess intrinsic histone acetyltransferase or histone deacetylase activities. Thus, recruitment of such coactivators or corepressors by transcription factors to specific DNA sequences may determine the acetylation status of core histones and consequently “open” (upon histone acetylation and subsequent removal of histone tails from their interaction with DNA) or return (upon histone deacetylation) the nucleosomes to their repressive state (72). In this way, transcription factor binding and transcription itself will, respectively, be promoted or inhibited by targeted histone acetylation.
In the alternative model, the acetylation of histones was viewed as a signal for the binding (elimination) of other factors (65). This model is essentially based on the use of antibodies which recognize specific acetylated lysine residues of histone H4 (37). For example, antibodies against specific H4 lysines gave a characteristic distribution pattern in polytene chromosomes from larval salivary glands of some chironomid insects (63, 64). Interestingly, the female inactive X chromosome was not immunolabeled with the different antibodies used (64). Immunolabeling of human metaphase chromosomes with antibodies against the most highly acetylated forms of H4 also showed a specific labeling pattern (38). In summary, the immunofluorescence studies carried out with these antibodies demonstrated that constitutive, centric heterochromatin and facultative heterochromatin in mammalian cells contained underacetylated forms of H4, while acetylated H4 was preferentially located in regions enriched in coding DNA (38, 65). This specificity of localization of differentially acetylated H4 forms was suggested to act as a signal for other factors (65).
In this study, we focused on the fate of the histone NH2 tails in nucleosome particles reconstituted with hyperacetylated histones and on the binding of the chimeric GAL4-AH transcription factor. To this end, we used a unique combination of UV-induced laser cross-linking together with immunochemical and molecular biology techniques. We found that the association of the histone tails with nucleosomal DNA is both dynamic and persistent, surviving both histone acetylation and GAL4-AH binding.
MATERIALS AND METHODS
Preparation of DNA probes.
The 180- and 150-bp DNA fragments containing the five centered GAL4 binding sites were generated by PCR amplification from plasmid pG5H as previously described (76). The 154-bp probe with a single GAL4 binding site at 32 bp from the ends was prepared from plasmid pBEND401G1 by digestion with SalI and MluI (17).
32P labeling of the 154-bp probe was carried out with T4 polynucleotide kinase. The 180- and 150-bp probes were labeled either by T4 polynucleotide kinase treatment or by PCR, when a higher specific activity was needed. The PCR mixture contained dATP, dGTP, and dTTP each at a concentration of 200 μM, dCTP at 100 μM, and 5 μl of [α-32P]dCTP (3,000 Ci/mmol; ICN). The fragments were amplified by numerous cycles and, after separation on a native 8% polyacrylamide (acrylamide/bisacrylamide, 29:1)–1× Tris-borate-EDTA (TBE) gel, were excised from the gel and electroeluted (71). The quantity of the probe was determined either fluorimetrically or by comparison with a DNA mass ladder (Gibco-BRL) on ethidium bromide-stained agarose gels.
Oligonucleosome and histone isolation.
Linker histone-depleted oligonucleosomes were prepared from chicken erythrocyte nuclei. Chicken erythrocyte nuclei isolated as described previously (46) were digested with micrococcal nuclease (5 U per 50 μg of DNA) in 10 mM Tris-HCl [pH 7.8]–50 mM NaCl–1 mM CaCl2–0.5 mM phenylmethylsulfonyl fluoride for 45 min at 37°C. The digestion was stopped by the addition of EDTA to a final concentration of 5 mM, the digested material was dialyzed for 6 to 8 h against 0.25 mM EDTA, and oligonucleosomes were recovered in the supernatant after centrifugation of the lysed nuclei on a bench-top centrifuge for 10 min. Oligonucleosomes were depleted from H1 and H5 linker histones and from nonhistone proteins by fractionation in a 10 to 30% sucrose gradient containing 0.65 M NaCl (19). The peak fraction, containing essentially mononucleosomes and small amounts of dinucleosomes, was dialyzed against 100 mM NaCl, divided into aliquots, and frozen at −80°C.
Chicken core histones were isolated from the oligonucleosomes by overnight extraction with HCl (21). Highly hyperacetylated histone octamers were isolated from HeLa cells grown in butyrate as described by Ausio and van Holde (8). Briefly, butyrate-grown HeLa cell chromatin was fractionated in the presence of divalent cations to obtain fractions enriched in hyperacetylated histones. Six milligrams of the hyperacetylated chromatin fraction (fraction a) was dialyzed against 0.633 M NaCl–0.1 M potassium phosphate–1 mM dithiothreitol–5 mM sodium butyrate (pH 6.7). The dialyzed chromatin was loaded onto a hydroxylapatite column (1.5 by 15 cm), and linker histones were eluted with 100 to 120 ml of the above-described buffer. Elution of hyperacetylated core histones was performed with the same buffer but containing 1 M NaCl. The eluted histones were concentrated and kept frozen at −80°C until use.
The extent of histone acetylation was assessed on acid-urea-Triton gels (18). The acetylated histones obtained in this way contained an average of 17 acetyl groups per histone octamer.
Digestion with trypsin.
Tailless nucleosomes were obtained by trypsin digestion. Briefly, 150 μl of nucleosomes (150 μg/ml) in 50 mM Tris-HCl (pH 7.5)–100 mM NaCl was incubated at 37°C with trypsin (Sigma) at a ratio of 1 μg of trypsin/25 μg nucleosomes. At various times after the beginning of the digestion, aliquots were removed and transferred to separate tubes, and the digestion was stopped by the addition of diisopropylfluorophosphate (Sigma) at a final concentration of 0.01%. The extent of trypsin digestion was checked by sodium dodecyl sulfate (SDS)–18% polyacrylamide gel electrophoresis (42).
Transcription factor GAL4-AH purification and nucleosome reconstitution.
The chimeric transcription factor GAL4-AH, containing the DNA binding and dimerization domains of GAL4 linked to an artificial 15-amino-acid putative amphipathic helix, was purified as described previously (44).
Reconstitution of nucleosomes containing the 32P-labeled fragment with one or five GAL4 binding sites was carried out by either the histone octamer transfer method (74) or salt dialysis as described by Vettese-Dadey et al. (70). For octamer transfer, 3 μg of donor nucleosomes was mixed with 30 ng of 32P-labeled probe in 1 M NaCl–10 mM Tris-HCl (pH 8.0)–1 mM EDTA in a final volume of 50 μl and incubated for 20 min at 37°C. The reaction mixtures were serially diluted to 0.9, 0.7, 0.5, and 0.3 M NaCl with dilution buffer (50 mM HEPES [pH 7.5], 1 mM EDTA [pH 8.0]) and incubated at each dilution step for 20 min at 30°C. Finally, the reaction mixtures were brought to 0.1 M NaCl with 10 mM Tris-HCl (pH 7.5)–1 mM EDTA (pH 8.0)–20% glycerol and incubated for 30 min at 30°C.
For salt dialysis nucleosome reconstitution, 2 to 3 μg of core histones was mixed with 2.1 μg of carrier thymus DNA and 50 to 100 ng of 32P-labeled DNA probe in 2 M NaCl–10 mM Tris-HCl (pH 8.0)–1 mM EDTA (pH 8.0)–10 mM β-mercaptoethanol–1 mg of bovine serum albumin (BSA) per ml in a total volume of 100 μl. The reaction mixtures were incubated for 15 to 30 min at room temperature, transferred to dialysis tubing, and dialyzed at 4°C against 10 mM Tris-HCl (pH 8.0)–1 mM EDTA (pH 8.0)–10 mM β-mercaptoethanol containing 1.2, 1.0, 0.8, and 0.6 M NaCl. Each dialysis step was carried out for 2 h. Finally, the reconstituted material was dialyzed overnight against 10 mM Tris-HCl (pH 8.0)–1 mM EDTA (TE). The reconstituted nucleosomes were analyzed on a 4% native polyacrylamide (acrylamide/bisacrylamide, 19:1)–0.5× TBE gel. Under optimal conditions, more than 85–90% of the 32P-labeled fragment was usually nucleosome reconstituted.
Binding reactions.
Nucleosomes reconstituted by octamer transfer or by salt dialysis were incubated with increasing concentrations of diluted GAL4-AH (stock solution, 2 mg/ml, diluted in 10 mM HEPES [pH 7.5]–100 mM KCl–10 mM ZnCl2–5 mM dithiothreitol–1 mg of BSA per ml). Final reaction mixtures were brought to 20 μl with binding buffer (20 mM HEPES [pH 7.5], 50 mM KCl, 5% glycerol, 2 mM dithiothreitol, 1 mM ZnCl2, 1 mg of BSA per ml) and incubated for 30 min at 30°C. The binding of GAL4-AH was analyzed on a 4% polyacrylamide–0.5× TBE gel at 4°C and a constant amperage of 8 mA. The binding reactions were quantified by use of a PhosphorImager and Image Quant Software (Molecular Dynamics). UV laser irradiation of the GAL4-AH-bound nucleosomes was performed immediately after completion of the binding reactions.
Preparation of antibodies.
Antibodies against core histones H2A, H2B, and H4 were prepared by injecting rabbits with histone-RNA complexes essentially as described previously (4). All antibodies were immunospecifically purified from sera by use of respective antigens conjugated to CNBr-Sepharose 4B (Pharmacia Biotech, Inc.).
Immunoblotting.
Histones were separated by electrophoresis in SDS–18% polyacrylamide gels (42). The proteins were transferred to nitrocellulose filters (Amersham) by electroblotting in 12.5 mM Tris-HCl (pH 8.3)–125 mM glycine–0.05% SDS–20% methanol for 1 h at a constant amperage of 200 mA. The electroblotted proteins were stained with 0.2% India ink in phosphate-buffered saline (PBS) supplemented with 0.2% Tween 20. After protein visualization, the filters were rinsed with PBS and blocked for 1 h in 10% nonfat dry milk–0.3% Tween 20–PBS. The filters were rinsed with PBS and overlaid with affinity-purified antibodies in PBS–10% fetal calf serum–0.2% Tween 20. After incubation for 1 h with gentle shaking at room temperature, the filters were washed three times with PBS–0.5 M NaCl–0.5% Triton X-100 and twice with PBS–0.5 M NaCl, each washing step lasting 10 min. The filters were incubated for 1 h with peroxidase-conjugated secondary antibody and, after extensive washing as described above, developed by use of an ECL kit (Amersham).
UV laser irradiation.
UV laser irradiation was carried out with a single 5-ns pulse from the fourth harmonics (266 nm) of a Surelite II (Continuum) Nd:YAG laser. The pulse energy was measured with a calibrated pyroelectrical detector (Ophir Optronics Ltd.) by use of an 8% deviation beam splitter. The electrical signal from the detector was transmitted to a computer for further processing. The sample (usually 20 μl) was irradiated in a 0.65-ml siliconized Eppendorf tube. The size of the laser beam was adjusted by means of a set of circular diaphragms to perfectly fit the surface area of the sample. Special care was taken to avoid air bubbles in the sample solution.
Quantitative estimation of protein covalently linked to DNA.
The total amount of protein cross-linked to DNA was determined by repeated phenol extractions. After irradiation, 20 μl of sample was mixed with 130 μl of TE and 100 μl of TE-saturated phenol. The solution was then vortexed and centrifuged for 3 min in a bench-top centrifuge, the aqueous phase was carefully recovered, and the phenol phase was extracted three more times with 200 μl of TE. The aqueous phases were pooled, and 100 μl of phenol was added. After the addition of 730 μl of TE to the phenol-phase fraction, the quantities of labeled DNA in both the phenol and the aqueous phases were measured by Cerenkov counting. The cross-linking yield was calculated as the ratio of phenol counts to phenol plus aqueous counts after subtraction of the background counts (irradiated DNA in the absence of protein). The quantum efficiency was calculated by dividing the cross-linking yield by the number of photons absorbed by a nucleotide base.
Immunoslot assay.
The cross-linking of individual histones was estimated by a slot immunoassay (48). The covalent histone-DNA complexes in the reconstituted nucleosomes were separated from the non-cross-linked proteins through preformed CsCl gradients. The gradients were fractionated, and the fractions containing the peak of DNA and covalent histone-DNA complexes were pooled. Five micrograms of the cross-linked material (measured as the amount of DNA) was dotted onto nitrocellulose filters, and the presence of individual histones was detected by the protocol described in the Immunoblotting section (see above).
Immunoprecipitation.
The immunoprecipitation of individual covalent histone-DNA complexes was performed essentially as described by Moss et al. (48). Fifty microliters of IgGsorb (The Enzyme Center, Malden, Mass.) was resuspended in 0.5 ml of 1% BSA–0.25 mg of laser-irradiated Escherichia coli DNA per ml in PBS and shaken for 1 h at room temperature to block sites of nonspecific absorption. The pellet obtained after centrifugation for 30 s in a bench-top centrifuge was resuspended in 0.5 ml of a mixture consisting of the specific antibody, the irradiated 32P-labeled reconstituted particles (corresponding to about 3 μg of DNA), and 200 μg of carrier-irradiated nucleosomes. The ratio of antibody to 32P-labeled core particles plus carrier-irradiated nucleosomes was 1:2.5 in antibody buffer (50 mM HEPES [pH 7.5], 2 M NaCl, 0.1% SDS, 1% Triton X-100, 1% deoxycholate, 5 mM EDTA, 0.1% BSA). The suspension was shaken for 2 to 3 h at room temperature and washed five times with antibody buffer and three times with rinse buffer (50 mM HEPES [pH 7.5], 0.15 M NaCl, 5 mM EDTA). The suspension was centrifuged for 30 s between each wash. The amounts of immunoprecipitated individual histone-DNA complexes were measured by Cerenkov counting.
RESULTS
Histones within reconstituted nucleosomes are efficiently cross-linked to DNA upon UV laser irradiation.
UV irradiation with conventional sources does not induce detectable amounts of core histone-DNA cross-linking (53). However, high-intensity UV laser irradiation of nuclei and chromatin leads to efficient histone-DNA cross-linking, and individual histones within the covalent protein-DNA complexes can be visualized with the help of specific antibodies (4, 20, 49). Histone-DNA cross-linking is essentially determined by the biphotonic mechanism of protein-DNA cross-linking operating in the presence of high-intensity laser irradiation (4, 33, 53). We sought to determine the efficiency of histone-DNA cross-linking after nucleosome reconstitution procedures.
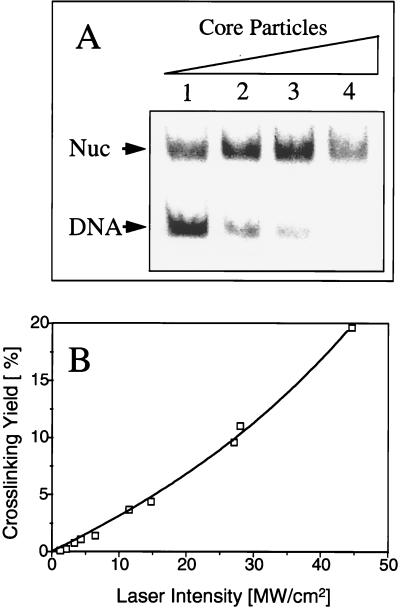
Nucleosomes were efficiently reconstituted on a 32P-labeled DNA probe by the octamer transfer method, as shown in Fig. 1A. As shown in Fig. 1B, under these conditions, irradiation of the reconstituted nucleosomes with a single 5-ns laser pulse led to significant cross-linking of the histones with the labeled DNA. Moreover, the dependence of the cross-linking yield on the laser intensity (the dose-response curve) fit perfectly with a theoretical curve for a biphotonic reaction (see also Fig. 4A and B). These results confirmed our previous data on the biphotonic mechanism of protein-DNA cross-linking induced by high-intensity laser irradiation (4, 5).
FIG. 1.
Dose-response curve for reconstituted nucleosomes. (A) Nucleosome particles are efficiently reconstituted by octamer transfer. Thirty nanograms of the 32P-labeled 180-bp probe DNA containing five centered GAL4 binding sites was reconstituted into nucleosome cores by the octamer transfer method. The mobilities of the nucleosome core (Nuc) and the naked DNA are indicated. The concentrations of donor nucleosomes were 0.1 μg (lane 1), 0.5 μg (lane 2), 1 μg (lane 3), and 3 μg (lane 4). (B) UV laser-induced histone-DNA cross-linking proceeds via a biphotonic mechanism. Reconstituted nucleosomes were irradiated with a single 266-nm laser pulse at different intensities. The amount of cross-linked 32P-labeled DNA was measured by the phenol extraction procedure and plotted against the laser intensity. The experimental points were computer fitted to reflect two-quantum processes (4, 5).
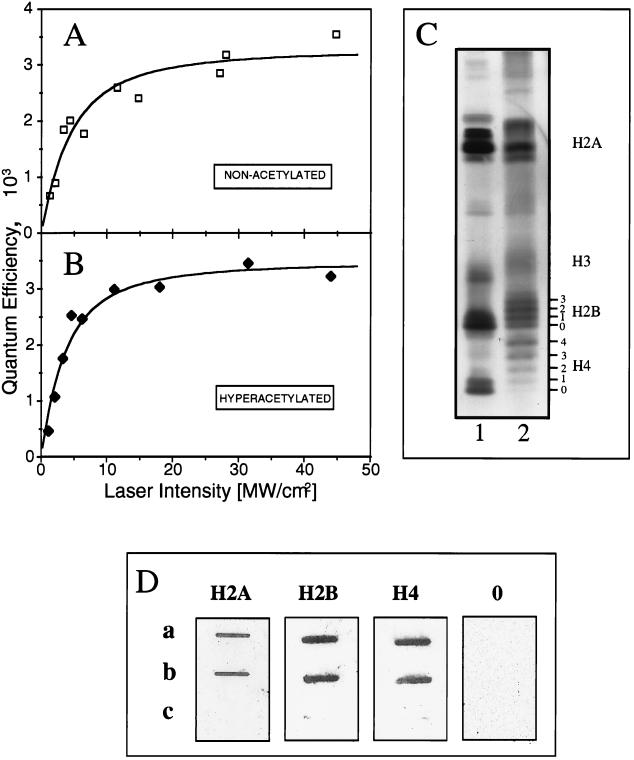
FIG. 4.
Hypoacetylated and hyperacetylated nucleosomes are cross-linked with the same efficiency in 100 mM NaCl. 32P-labeled 180-bp probe DNA was reconstituted in nucleosomes with control, hypoacetylated, or hyperacetylated histones (17 acetyl groups per histone octamer). Samples, in a solution of 100 mM NaCl, were irradiated with single 266-nm laser pulses at different intensities, and the yield of cross-linked DNA was measured by the phenol extraction procedure. (A and B) Dose-response curves for hypoacetylated (A) and hyperacetylated (B) nucleosomes. (C) Acid-urea-Triton gel electrophoresis of hypoacetylated (lane 1) and hyperacetylated (lane 2) histones used for reconstitution. The number of acetylated groups is indicated by numbers 0 to 4. (D) Immunoslot assay of the reaction of antibodies to H2A, H2B, and H4 and preimmune IgG (0) with covalent histone-DNA complexes isolated after centrifugation in CsCl gradients of irradiated nucleosomes containing hypoacetylated (a) and hyperacetylated (b) histones; c, immunoslot assay of the material from control (hypoacetylated), nonirradiated particles after centrifugation in CsCl. The experiment was carried out as described in the legend to Fig. 3B.
UV laser-induced cross-linking of histones to DNA within reconstituted nucleosomes occurs via their NH2 tails only.
The structure of the histone core octamer has been determined by X-ray crystallography to a resolution of 3.1 Å (6, 7). All histone chains contain a folded motif, the histone fold, and an externally located NH2-terminal tail region which possesses a large number of positively charged residues. The histone folds contain many sites of interactions with nucleosomal DNA (45). In addition, the positively charged histone tails also interact with DNA; however, these domains seem to be devoid of any regular structure when not bound to nucleosomal DNA (10). Our earlier immunochemical data indicated that in native chromatin, laser-induced histone-DNA cross-linking was achieved essentially via the NH2 tails (59). In order to determine whether this is also the case for reconstituted nucleosomes, we carried out two types of experiments.
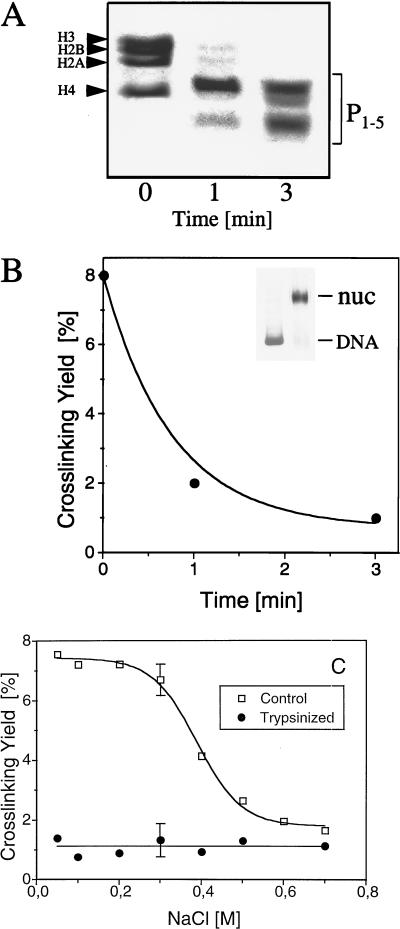
In the first set of experiments, we removed the NH2 tails of donor nucleosomes by trypsin digestion and used the truncated nucleosomes for octamer transfer reconstitution. The reconstituted particles, containing trypsin-truncated histones, were irradiated, and the total amount of cross-linked histones was compared to that of nucleosomes with native histones. The kinetics of trypsin digestion of donor nucleosomes are shown in Fig. 2A. Under these conditions, 1 min of digestion partially removed the NH2 tails, while 3 min was sufficient for their complete elimination. At the same time, the histone fold domains (peptides P1–5 in Fig. 2A; see also reference 66) remained intact. The reconstitution of particles with truncated nucleosomes was as efficient as that of non-trypsin-digested native nucleosomes (compare Fig. 1A and the inset of Fig. 2B; see also references 9 and 69). However, the efficiency of histone cross-linking within reconstituted nucleosomes containing trypsin-truncated (i.e., without histone NH2 tails) core histones was decreased to insignificant levels (Fig. 2B). These data strongly suggest that the cross-linking of histones to DNA in reconstituted particles occurred via the histone NH2 tails.
FIG. 2.
Core histones are cross-linked to DNA via their NH2 tails. (A) Electrophoresis (18% polyacrylamide–SDS) of histones isolated from donor nucleosomes digested with trypsin for the indicated times. P1–5, trypsin-resistant peptides of the core histones, designated as described by van Holde (67). (B) Dependence of the yield of cross-linked DNA on reconstituted nucleosomes containing trypsinized histones. Nucleosomes were reconstituted under optimal conditions (3 μg of donor nucleosomes for 30 ng of 32P-labeled 180-bp probe DNA) by using as donors either native nucleosomes or nucleosomes digested with trypsin for 1 or 3 min. Each sample was irradiated with a single 266-nm laser pulse at a laser intensity of 25 MW/cm2. The amount of cross-linked DNA was measured by the phenol extraction method, and the yield of cross-linked DNA was plotted against the time of trypsin digestion of donor nucleosomes. The inset represents the mobilities of the naked 180-bp DNA fragment and of reconsti- tuted particles (nuc) determined by using as donors nucleosomes digested with trypsin for 3 min. (C) NaCl concentration dependence of the yield of cross-linked DNA on nucleosome particles reconstituted with native histones or with histones from donor nucleosomes that had been digested with trypsin for 3 min. Both samples at different NaCl concentrations were irradiated with a single laser pulse (25 MW/cm2), and the dependence of the yield of cross-linked DNA (measured by phenol extraction) on NaCl concentration was determined. Each experimental point represents the average of four independent experiments. Because the errors of measurements at different ionic strengths were found to be essentially the same, for simplicity error bars are shown for only two experimental points.
This conclusion was further supported by the dependence of histone cross-linking on the concentration of NaCl (Fig. 2C). In the cross-linking reactions, increasing NaCl concentrations above 0.3 M led to a pronounced decrease in the efficiency of cross-linking, which became insignificant at 0.5 to 0.6 M NaCl. Increasing NaCl concentrations above 0.3 M NaCl released the NH2 tails from their interactions with DNA (16, 73). Moreover, direct contact between proteins and DNA is necessary in order for UV laser-induced cross-linking to occur (UV light is a “zero-length” cross-linking agent). Thus, the lack of cross-linking above 0.5 to 0.6 M NaCl was most likely due to the release of the NH2 tails from DNA at these salt concentrations. These data are in agreement with our earlier conclusion that cross-linking is achieved via histone NH2 tails. This conclusion is also supported by the lack of cross-linking for tailless nucleosomes within the range of 0.1 to 0.7 M NaCl (Fig. 2C).
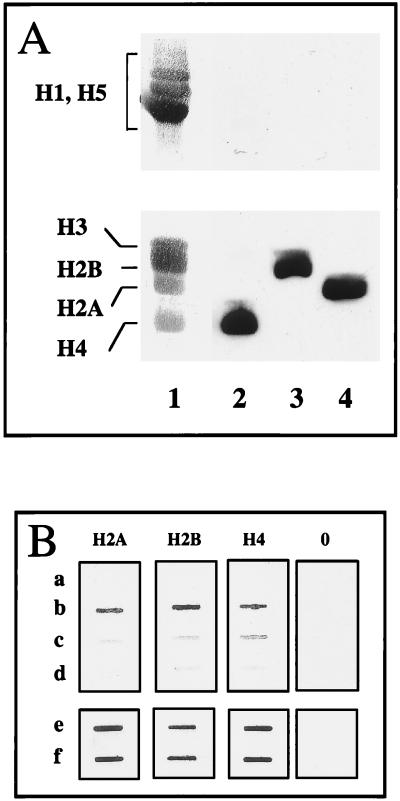
In the second experimental approach, nucleosomes were reconstituted from purified native histones. These reconstituted particles were digested with trypsin and irradiated with the laser, and the covalent histone-DNA complexes were purified from the non-cross-linked proteins on CsCl gradients. The amount of individual histones cross-linked to DNA within the purified complexes was estimated by an immunoslot assay with highly specific antibodies. The specificity of the antibodies is illustrated in Fig. 3A. If the NH2 tails were responsible for histone-DNA cross-linking, we expected to observe a disappearance of the cross-linked histones within the CsCl-purified protein-DNA complexes isolated from irradiated and trypsin-digested (tailless) nucleosomes. This is because of the fact that the non-cross-linked histone fold domains are dissociated from DNA during centrifugation in CsCl gradients. Figure 3B illustrates that antibody detection of histones H2A, H2B, and H4 in the CsCl fractions indeed was dependent on the presence of the NH2 tails of each histone (i.e., lost by trypsin digestion). Since each antibody reacted with the histone fold domains in the absence of the histone tails and the presence of histones in the CsCl fractions was dependent on UV laser-induced cross-linking (Fig. 3B), these results clearly demonstrate that the histone-DNA cross-linking was achieved via the nonstructured tails.
FIG. 3.
Immunochemical evidence for selective histone NH2 tail-DNA cross-linking induced by UV laser irradiation. (A) Specificity of the histone antibodies used. Hen erythrocyte histones were separated by 18% polyacrylamide–SDS gel electrophoresis, electroblotted, and stained with India ink (lane 1) or reacted with immunopurified antibodies against H4 (lane 2), H2B (lane 3), and H2A (lane 4). (B) Immunoslot assay for the presence of core histones in cross-linked protein-DNA complexes obtained upon irradiation of nucleosomes. Nucleosomes containing 180 bp of DNA were reconstituted by histone octamer transfer by using as donors either native nucleosomes or nucleosomes digested with trypsin for 1 or 3 min. The samples were irradiated with identical doses, and the cross-linked histone-DNA complexes were separated from the free histones on CsCl gradients. The CsCl gradients were fractionated, and the fractions containing the DNA peak were pooled. Five micrograms (measured as DNA) from each pooled sample was dotted on a nitrocellulose filter and reacted with antibodies against H2A, H2B, and H4 and preimmune IgG (0). a, Nonirradiated particles; b, irradiated particles; c and d, irradiated particles containing 1- and 3-min trypsin-digested histones, respectively; e and f, control slots showing the reaction of the antibodies with nucleosomes containing native or 3-min trypsin-digested histones.
Hyperacetylated NH2 tails of core histones interact at an efficiency similar to that of hypoacetylated tails with nucleosomal DNA at nearly physiological ionic strengths.
Histone acetylation is a posttranslational modification that correlates strongly with the transcriptional regulation of numerous genes (for recent reviews, see references 27, 54, 57, and 72). However, the precise role of this modification remains unclear (28, 65, 72). Acetylation occurs on specific lysine residues within the core histone NH2 tails (41, 67). A widely accepted hypothesis is that histone acetylation, by reducing the positive charge of histone tails, releases them from their interaction with DNA. This is thought to result in the observed enhanced transcription factor binding to nucleosomes containing acetylated histones (43, 70, 72). Since we have shown that UV laser irradiation induces histone-DNA cross-linking via the core histone NH2 tails only, this method appears to be an ideal tool for directly studying the interaction of hyperacetylated histone tails with DNA. To this end, we reconstituted nucleosomes by salt dialysis using highly hyperacetylated core histones (17 acetyl groups per histone octamer; Fig. 4C) isolated by a special fractionation procedure as described previously (8, 26). Nucleosome reconstitution with the hyperacetylated core histones was as efficient as reconstitution with the hypoacetylated core histones (data not shown).
Hyperacetylated and control (hypoacetylated) reconstituted nucleosomes were UV laser irradiated at different intensities, and the yield of cross-linking was calculated. The dose-response curves for both preparations at 0.1 M NaCl are shown in Fig. 4A and B. Both dependencies were essentially identical. Thus, at 0.1 M NaCl, hyperacetylated NH2 tails interacted as closely as hypoacetylated tails with nucleosomal DNA. This conclusion was further confirmed by the immunochemical data presented in Fig. 4D. The reactions of specific antibodies against individual core histones with covalent histone-DNA complexes isolated from irradiated nucleosomes containing hypoacetylated and hyperacetylated histones showed the same intensities.
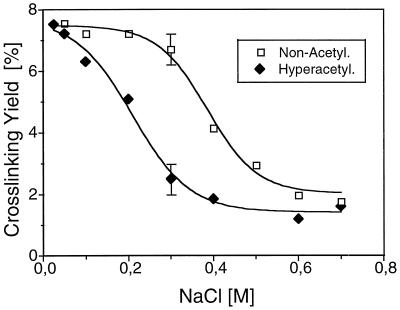
The efficiencies of histone cross-linking of nucleosomes containing hyperacetylated versus hypoacetylated histones differed at a higher ionic strength (Fig. 5). Increasing the NaCl concentration resulted in decreased cross-linking efficiency, reaching a plateau of insignificant cross-linking for both preparations. However, for hyperacetylated particles, this effect was observed at lower NaCl concentrations. The binding of histone NH2 tails to DNA is essentially electrostatic in nature (16, 67), and increasing NaCl concentrations affect electrostatic interactions. Thus, these data indicate a weaker interaction of hyperacetylated histone NH2 tails with nucleosomal DNA at a higher ionic strength.
FIG. 5.
UV laser-induced cross-linking detects some perturbations in the histone NH2 tail-DNA interactions in nucleosomes containing hyperacetylated histones. Particles containing 180 bp of DNA and reconstituted with hypoacetylated (□) or hyperacetylated (⧫) histones were irradiated with a single laser pulse (25 MW/cm2) at different ionic strengths (50 to 700 mM NaCl), and the yield of cross-linked DNA was measured. Data derived from three independent experiments are presented as a graph of the percentage of cross-linking DNA yield versus NaCl concentration. For simplicity, the error bars at one NaCl concentration only are shown.
The NH2 tails of core histones and GAL4-AH transcription factors coexist on the same nucleosomal DNA.
We demonstrated that at nearly physiological ionic strengths, the hyperacetylation of histone NH2 tails only slightly perturbs their interaction with DNA. Next we asked whether transcription factor binding affects the interaction of the histone NH2 tails with DNA. To answer this question, we carried out immunoprecipitation experiments (Fig. 6). As a model system, we used nucleosomal templates containing 180 and 150 bp of DNA with five centered GAL4 binding sites, because a perturbation, if induced, should be more apparent in a particle containing multiple bound transcription factors. The 180-bp particle contains almost 40 bp of linker DNA and, since the sites of DNA interaction with the histone NH2 tails are not well known (7, 45), it is possible that some of the histone tails might interact with the linker DNA. If this is the case, the binding of GAL4-AH should not affect the histone tail-DNA interactions, since the GAL4 binding sites are located within the nucleosomal DNA and not the linker DNA. The use of the linkerless 150-bp particle overcomes this problem, since it allows for studying the effect of an interaction of the histone NH2 tails with nucleosomal DNA only.
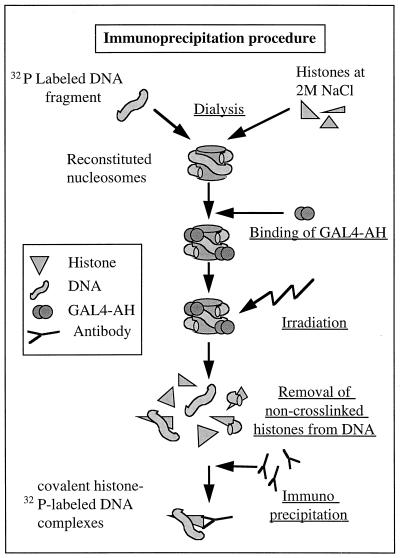
FIG. 6.
Schematic presentation of the experimental strategy for studying the effect of five nucleosome-bound GAL4-AH molecules on histone NH2 tail-DNA cross-linking efficiency. For experimental details, see Materials and Methods.
Briefly, 32P-labeled particles, with or without five bound GAL4-AH molecules, were UV laser irradiated with identical doses, and the covalent histone-DNA complexes were immunoprecipitated with highly specific antibodies against individual histones (Fig. 3A). The precipitation was carried out under conditions in which the non-cross-linked histones were completely removed from the DNA (for details, see Materials and Methods). A comparison of the amounts (measured as 32P counts) of precipitated individual histone-DNA complexes for the different samples allowed us to judge the efficiency of histone NH2 tail-DNA cross-linking in the presence or absence of bound GAL4-AH dimers. This comparison provided a measure of the effect of GAL4-AH binding on histone tail interactions with DNA.
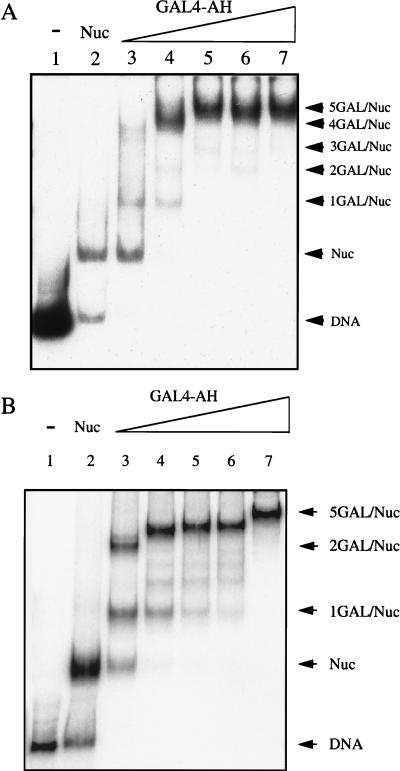
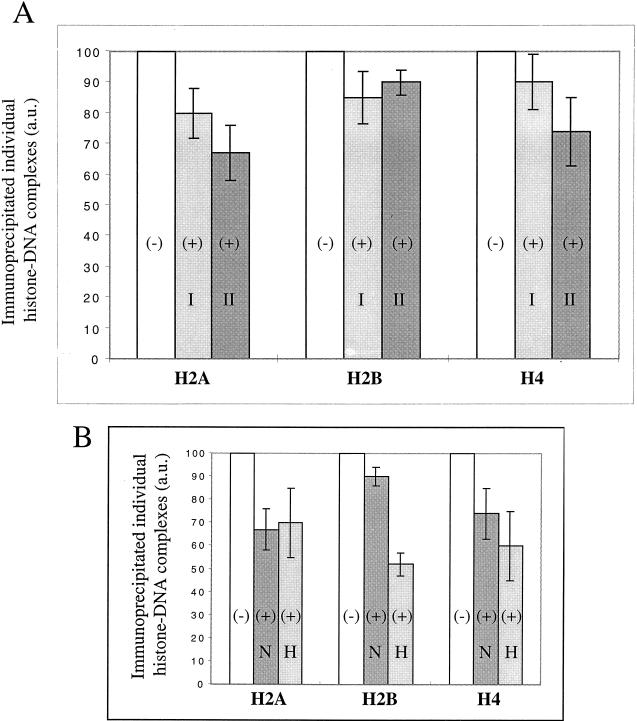
Initially, we performed experiments to determine the optimal conditions for the saturation of GAL4-AH on all five sites on the reconstituted particles (Fig. 7). Once these conditions were determined, we irradiated the samples and carried out the immunoprecipitation experiments. The immunoprecipitation data are presented in Fig. 8. As shown in Fig. 8A, the cross-linking yield for individual histones in hypoacetylated 180- and 150-bp particles containing five GAL4-AH dimers was decreased compared to that in samples lacking bound GAL4-AH, suggesting that some perturbation in histone tail-DNA interactions occurred. However, this perturbation was found to be relatively small, the highest level being observed for the 180-bp particle H2A (33% cross-linking yield decrease).
FIG. 7.
Titration of reconstituted nucleosomes with GAL4-AH. (A) Reconstituted particles containing the 180-bp DNA fragment and bearing five central GAL4-AH binding sites were incubated without (lane 2) or with increasing amounts of GAL4-AH (lanes 3 to 7). The GAL4-AH-bound nucleosomes were separated from the unbound nucleosomes on a 4% polyacrylamide gel. An autoradiogram of the gel is shown. The concentrations of GAL4-AH used were as follows: 0 (lane 2), 25 nM (lane 3), 51 nM (lane 4), 154 nM (lane 5), 309 nM (lane 6), and 515 nM (lane 7). The arrows Lane 1 consists of the loaded DNA fragment used in the reconstitution. The arrows indicate the positions of the different nucleosome complexes. (B) Same as panel A but for particles containing the 150-bp DNA fragment.
FIG. 8.
Binding of five GAL4-AH molecules does not substantially affect the histone NH2 tail-DNA interactions in reconstituted particle preparations containing either hypoacetylated or hyperacetylated histones. (A) 32P-labeled hypoacetylated 150-bp (I) and 180-bp (II) DNA particles containing or not containing five GAL4-AH molecules were laser irradiated at identical doses, and the covalent complexes containing individual cross-linked histones were immunoprecipitated with specific antihistone antibodies (see Materials and Methods for details). The amount of immunoprecipitated DNA was measured by Cerenkov counting. A histogram showing the percentage of immunoprecipitated individual covalent histone-DNA complexes in the presence of five nucleosome-bound GAL4-AH molecules relative to that in the absence of bound GAL-AH is shown. +, presence of five GAL4-AH factors; −, absence of these factors. The results are averaged over three independent experiments with each of the antibodies used. a.u., arbitrary units. (B) Same as panel A but with 180-bp DNA particles reconstituted with either hypoacetylated or hyperacetylated histones. N, particles containing hypoacetylated histones; H, particles containing hyperacetylated histones. The data represent average values from several experiments. For hypoacetylated nucleosomes, six, four, and five independent immunoprecipitations were carried out with antibodies against H2A, H2B, and H4, respectively. The results for hyperacetylated nucleosomes are averaged over three independent experiments with each of the antibodies used.
Histones H2A and H4 of the hyperacetylated 180-bp particle containing 10 bound GAL4-AH molecules were cross-linked with essentially the same efficiency as those of the GAL-AH-bound hypoacetylated particle, while H2B cross-linking was affected (about a 50% decrease in the cross-linking yield, compared to 10% in the hypoacetylated particle; Fig. 8B). Thus, histone hyperacetylation may be responsible for the selective perturbation of H2B NH2 tail-DNA interactions within hyperacetylated nucleosome particles bound to GAL4-AH.
The binding of five GAL4-AH dimers to both hypo- and hyperacetylated nucleosomes did not completely perturb the interaction of histone NH2 tails with nucleosomal DNA. In all cases, the reduction in cross-linking efficiency was twofold or less with GAL4-AH binding. This finding raises the possibility that the hyperacetylation of histones may have a similar magnitude of effect on GAL4-AH binding to nucleosomal DNA. To test this idea, we reconstituted nucleosome particles with hypoacetylated and hyperacetylated histones containing one or five GAL4 binding sites and studied quantitatively the binding of GAL4-AH to both types of particles. The results of these studies are presented in Fig. 9. The binding of GAL4-AH was enhanced 2 to 2.5 times in hyperacetylated nucleosomes. This effect was found to be more apparent for the template with one GAL4 binding site. Thus, the hyperacetylation of histones had only a modest effect on GAL4-AH binding to nucleosomal DNA, as expected from the cross-linking data given above. These results are in excellent agreement with the previously published data of Vettese-Dadey et al. (70), who reported that only the most highly acetylated histone, H4, substantially affected binding, which was more apparent for the basic helix-loop-helix (bHLH) protein USF than for GAL4-AH.
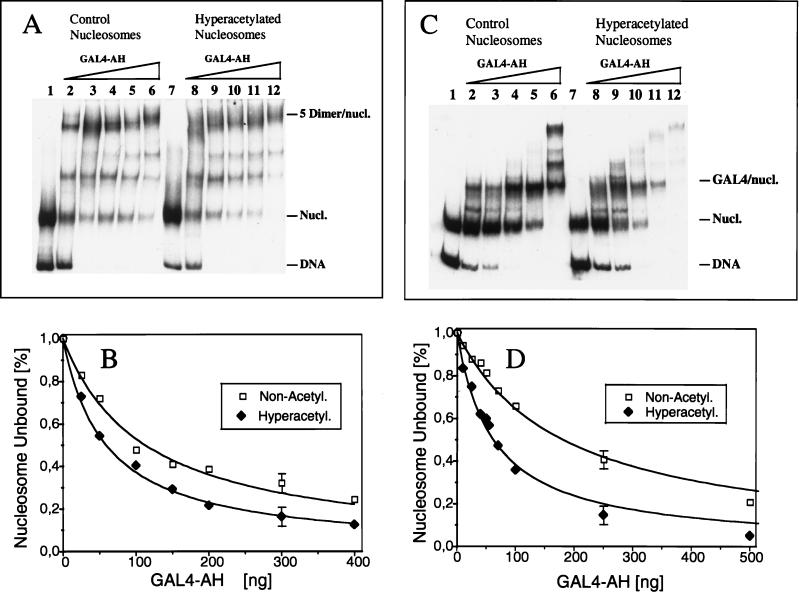
FIG. 9.
Binding of GAL4-AH to reconstituted nucleosomes containing hypoacetylated or hyperacetylated core histones. (A) 32P-labeled 180-bp probe DNA containing five GAL4 binding sites was reconstituted in nucleosome cores with either hypoacetylated (lanes 1 to 6) or hyperacetylated (lanes 7 to 12) core histones and allowed to react with increasing amounts of GAL4-AH. The mobilities of the naked DNA and the nucleosome complexes (Nucl.) are indicated. The concentrations of GAL4-AH used in these experiments were 0 (lanes 1 and 7), 22 nM (lanes 2 and 8), 68 nM (lanes 3 and 9), 90 nM (lanes 4 and 10), 135 nM (lanes 5 and 11), and 180 nM (lanes 6 and 12). (B) Graphic representation of data from representative experiments shown in panel A. The decrease in unbound nucleosome bands versus GAL4-AH concentrations is shown. Quantification was performed with the help of a PhosphorImager. The percentage of unbound nucleosomes was calculated as the ratio of the nucleosome band signal to the radioactivity signal of the whole lane □, hypoacetylated histones; ⧫, hyperacetylated histones. (C) An experiment similar to that in panel A was carried out but with a 154-bp probe containing a single GAL4 binding site. The concentrations of GAL4-AH used were 0 (lanes 1 and 7), 22 nM (lanes 2 and 8), 45 nM (lanes 3 and 9), 110 nM (lanes 4 and 10), 220 nM (lanes 5 and 11), and 440 nM (lanes 6 and 12). (D) Dependence of the percentage of unbound nucleosomes on GAL4-AH concentrations for the data presented in panel C. The measurements were performed as described for panel B.
DISCUSSION
Transcription regulation in eukaryotes requires the coordinated binding of numerous basal and specific transcription factors (60). This binding, however, is impeded by nucleosomes in the presence of transcription factor cognate DNA sequences (22, 52). The histone core octamer binds to nucleosomal DNA with a high affinity; thus, the resulting nucleosomal complex is very stable (6, 7). Thus, in many instances transcription factors have to overcome the nucleosome barrier in order to gain access to their DNA binding sequences. It should be noted that different transcription factors bind to nucleosomal DNA with different levels of affinity relative to naked DNA (12, 52). It has been proposed that core histone NH2 tails play an important role in hindering the interaction of transcription factors with nucleosomal DNA (43, 72). Accordingly, the hyperacetylation of lysine residues within core histone tails may substantially weaken histone NH2 tail-DNA interactions by displacing the tails away from the DNA. This situation in turn may facilitate the binding of transcription factors to DNA (43, 70). To test this model, in this work we examined the effect of both histone hyperacetylation and transcription factor binding on the interaction of histone NH2 tails with nucleosomal DNA by a combination of UV laser-induced cross-linking and molecular biology techniques.
UV laser-induced histone-DNA cross-linking within reconstituted nucleosomes.
We showed that histone-DNA cross-linking within reconstituted nucleosomes is achieved via core histone NH2 tails in two different sets of experiments. In the first set, we demonstrated that no histone-DNA cross-links were induced upon laser irradiation of nucleosomes reconstituted with trypsin-truncated (tailless) histones (Fig. 2B). In agreement with this interpretation, the reduction of histone tail-DNA interactions by rising ionic strength (16) led to a manifold decrease in the efficiency of cross-linking (Fig. 2C). In the alternative set of experiments, we first cross-linked the histones in reconstituted particles and then digested the histone NH2 tails with trypsin. The digestion of the tails released the histones from the DNA upon centrifugation in CsCl gradients (Fig. 3B), demonstrating again that the UV laser-induced cross-linking was accomplished solely via the N-terminal region of the histone molecule. This conclusion is in agreement with our previous data on laser-induced histone-DNA cross-linking in native chromatin (59). It should be noted that the efficiencies of histone NH2 tail cross-linking and the respective interactions of the histone NH2 tails with the DNA, unlike those for the C-terminal domain of H2A (66), were essentially the same for mononucleosomes and for high-molecular-weight chromatin (3a, 59). Thus, our experimental model, a nucleosome containing 180 bp of DNA, is a representative one for studying histone NH2 tail interactions with DNA.
Why is histone cross-linking achieved via the NH2 tails only? One possible explanation is the following. In order for the photochemical reaction resulting in cross-linking to occur, two events have to take place at the same time: (i) very close protein-DNA contact and (ii) a favorable orientation of both chromophores, the DNA base and the amino acid residue. Some of the amino acid residues from the histone fold-domain are in close contact with DNA (45). However, the histone folds are organized in a relatively rigid structure in the histone octamer, in contrast to their NH2 tails, which seem to be flexible (16). Thus, flexibility of the histone tails may present favorably oriented amino acids for efficient protein-DNA cross-linking.
Interaction of histone NH2 tails with DNA within reconstituted nucleosomes containing hyperacetylated histones.
The fact that UV histone-DNA cross-linking occurs exclusively via the NH2 tails provides an approach to examine the effect of histone acetylation on the interaction of NH2 tails with DNA. Since histone acetylation is restricted to the NH2 tails and since direct contact between the tails and nucleosomal DNA is needed for cross-linking to occur (53), the cross-linking yield is a direct measure of the extent of the histone tail-DNA interactions. To this end, we reconstituted nucleosomes by the salt dialysis method using highly hyperacetylated histone octamers (17 acetyl groups per histone octamer) (Fig. 4C) and compared their efficiency for cross-linking with that of particles containing hypoacetylated histones (Fig. 4 A and B and Fig. 5). The cross-linking yields for both samples at 100 to 150 mM NaCl were very similar, demonstrating that at these nearly physiological ionic strengths, the NH2 tails were not released from nucleosomal DNA upon histone acetylation. However, raising the ionic strength led to an earlier decrease in the cross-linking yield for hyperacetylated nucleosomes (Fig. 5). Thus, bearing in mind that the histone NH2 tail interaction is essentially electrostatic (16), these data reflect weakening in the interaction between the hyperacetylated histone tails and DNA without their release from the DNA at physiological ionic strengths, in contrast to previous models (for a review, see reference 72). This conclusion is further enhanced by recent in vivo data showing that the hyperacetylated histones of actively transcribed ribosomal genes can be cross-linked to DNA by use of UV laser irradiation (49). The above finding is also supported by data on chemically induced histone-DNA cross-linking (performed partially via the NH2 tails) which indicated changes in hyperacetylated histone NH2 tail-DNA interactions but not their total displacement from the DNA (23).
Reconstituted oligonucleosome complexes with the same highly hyperacetylated histones as those used in the present work were studied in the past with the help of sedimentation and electron microscopy techniques (26). Both types of analysis showed that at nearly physiological ionic strengths (100 to 150 mM NaCl), the hyperacetylated oligonucleosomes remained in an extended conformation, in contrast to their hypoacetylated counterparts. Thus, the weakening of hyperacetylated histone NH2 tail-DNA interactions detected in this study obviously affects the compaction of the nucleosomal filament.
Binding of GAL4-AH to reconstituted nucleosomes containing hypoacetylated and hyperacetylated histones.
We also addressed the question of whether the weakened hyperacetylated histone-DNA interactions affected transcription factor binding and if these histone-DNA interactions were in turn affected by transcription factor binding. To this end, we studied the interactions of the chimeric transcription factor GAL4-AH with hypoacetylated and hyperacetylated nucleosomes, since GAL4-AH can invade nucleosomes with relatively small changes in affinity relative to naked DNA (12, 52). For both samples, saturation of nucleosomes with five GAL4-AH dimers led to a decrease in core histone cross-linking efficiency, as judged by immunoprecipitation with specific antibodies against individual core histones (Fig. 8). However, interactions of the NH2 tails with DNA were still detected after GAL4-AH binding (greater than 50% for hyperacetylated H2B and H4 and 70% for H2A). Since the five GAL4 sites covered 90 bp of the core particle DNA, this result clearly demonstrates that in both hypoacetylated and hyperacetylated nucleosomes, the histone NH2 tails remained associated with DNA, which was simultaneously bound by GAL4-AH. Interestingly, the effect of GAL4-AH binding on the interactions of the H2B tail with DNA was largely acetylation dependent (a fivefold greater reduction in cross-linking). However, this result was somehow not surprising, considering the position occupied by the histone H2B tails in the core particle (7, 45).
It should be noted that we have no data on the fate of the histone H3 tails within reconstituted nucleosomes containing bound GAL4-AH transcription factors. Although we raised antibodies against histone H3, these antibodies were found not to function at the high ionic strengths (see Materials and Methods) necessary for specific immunoprecipitation of individual covalent histone-DNA complexes. However, our antibodies functioned in immunoblotting, and by using them we found the same efficiencies of cross-linking of hyperacetylated and hypoacetylated H3 tails to nucleosomal DNA in an immunoslot assay (data not shown).
Based on the above data, hyperacetylation of histones should not be expected to play a dramatic role in the enhancement of GAL4-AH binding. In fact, this was found to be the case: the binding of GAL4-AH was enhanced 2 to 2.5 times for hyperacetylated nucleosomes (Fig. 9). The increased affinity of GAL4-AH for its binding sites on hyperacetylated nucleosomes was more apparent for one GAL4 binding site template. Recently, Vettese-Dadey et al. (70) demonstrated that GAL4-AH binding to nucleosomes containing acetylated histones was modestly stimulated. By using a gel retardation assay coupled to immunochemical techniques, these authors were able to show that only core particles containing the most highly acetylated forms of histones had the highest affinity for GAL4-AH. This affinity was found to be two to three times higher than that for hypoacetylated particles. These results fully agree with the data presented here for nucleosomes containing highly hyperacetylated histones. Interestingly, Vettese-Dadey et al. (70) observed stronger effects of H4 acetylation on the binding of the bHLH protein USF (70). It will be interesting to determine in future studies if USF has a stronger effect on the DNA binding of histone tails. It has also been recently shown that for transcription factors which involve a large DNA binding domain, such as TFIIIA, acetylation does not have any effect on the efficiency of their binding to nucleosomal DNA (34).
Histone hyperacetylation and transcription.
Previous studies on the effect of histone acetylation on transcription factor binding to nucleosomes have proposed that acetylation may release histone tails from DNA, thereby stimulating transcription factor access (reviewed in reference 72). This model was based in part on the observation that removal of the histone tails with trypsin (43, 69) similarly stimulated transcription factor access to nucleosomal DNA. However, the present study provides a different view of the dynamic interactions of transcription factors and histone tails with nucleosomal DNA, based upon two crucial observations. First, our data clearly demonstrate that both hypoacetylated and hyperacetylated histone NH2 tails bound to nucleosomal DNA. Hyperacetylation of histone tails weakened histone tail-DNA binding but did not abolish the interaction. Second, greater than 50% of the NH2 tail-DNA interactions persisted during the occupancy of 90 bp of nucleosomal DNA by GAL4-AH dimers (saturation of the five GAL4 binding sites) within either hypoacetylated or hyperacetylated particles. Thus, our data indicate that the histone tails remained associated with nucleosomal DNA when acetylated and also when the nucleosomal DNA was cooccupied by DNA binding transcription factors. These data argue against a simple model in which histone tails are mere inhibitors of transcription factor access through mutually exclusive binding. Instead, these data support a more dynamic role of histone tails and their acetylation in enhancing factor access and in transcription regulation.
It is becoming increasingly clear that transcription may be regulated by histone NH2 tails indirectly, since they can be the target for repressor proteins (31, 56). For example, Edmonson et al. (24) showed that the in vitro binding of the NH2 tails of histones H4 and H3 to the transcription repressor Ssn6-Tup1 complex is negatively regulated by histone acetylation. Furthermore, mutations within the NH2 tails of histones H3 and H4 which abolish Tup1-histone binding led to in vivo enhancement of transcription by more than one order of magnitude. However, even if enhanced transcription factor binding and displacement of repressor proteins operate synergistically via histone acetylation, this activity can account for only a portion of the several hundredfold enhancement of transcription observed in vivo; consequently, other activities, such as those of chromatin remodeling factors (36, 55, 61, 62, 68), activator proteins (60), and so forth, must participate in the activation process. Recently, an example demonstrating the complexity of the activation of transcription was the finding that tumor suppressor p53 can be acetylated by its coactivator, p300 (which until recently was thought to have a specific histone acetyltransferase activity only), resulting in a remarkable enhancement of its binding to DNA and activation of its biochemical function (28).
The omnipresent histones.
In this study, we demonstrated that the binding of five GAL4-AH dimers to DNA (90 bp of DNA occupancy) in both hypoacetylated and hyperacetylated nucleosomes results in a weak alteration of the histone NH2 tail-DNA interaction. At the same time, the saturation of the five GAL4 binding sites induces a massive disruption of folded histone domain-DNA interactions (52, 74). These findings suggest that in vivo, during the process of transcription, histones might not leave DNA but instead might remain anchored to it (25, 49, 50, 58) through their NH2 tails (49, 50). Indeed, Nacheva et al. (50), by using chemical cross-linking with histone NH2 tails, showed that the actively transcribed hsp70 gene (this gene does not have nucleosome organization when actively transcribed) contained histones in amounts comparable to those of the nonactive gene. However, when a procedure for cross-linking via the histone folded domains was used, no histones were found on the hsp70 gene, indicating that histone folded domain-DNA interactions were disrupted. The second line of evidence came from the study of Mutskov et al. (49). These authors, by using histone NH2 tail UV laser-induced cross-linking, demonstrated the presence of hyperacetylated histones on the coding sequences of actively transcribed, nonnucleosomally organized rat ribosomal genes. All of these data clearly show that actively transcribed genes are covered with histones and that the histones remain attached to the DNA via their NH2 tails.
ACKNOWLEDGMENTS
We thank E. Moudrianakis and S. Khochbin for helpful and stimulating discussions as well as for careful reading of the manuscript.
This work was supported by grants from CNRS, INSERM (contract 4E006B), and Region Rhône-Alpes (project Emergence).
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin-3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Allegra P, Sterner R, Clayton D F, Allfrey V G. Affinity chromatographic purification of nucleosomes containing transcriptionally active DNA sequences. J Mol Biol. 1987;196:379–388. doi: 10.1016/0022-2836(87)90698-x. [DOI] [PubMed] [Google Scholar]
- 3.Allfrey V G, Faulkner R, Mirsky A E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Angelov, D., V. Mutskov, and S. Dimitrov. Unpublished data.
- 4.Angelov D, Stefanovsky V, Dimitrov S I, Russanova V, Pashev I. A picosecond UV laser induced protein-DNA crosslinking in reconstituted nucleohistones, nuclei and whole cells. Nucleic Acids Res. 1988;16:4525–4538. doi: 10.1093/nar/16.10.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelov D, Berger M, Cadet J, Marion C, Spassky A. High-intensity ultraviolet laser probing of nucleic acids. Trends Photochem Photobiol. 1994;3:643–663. [Google Scholar]
- 6.Arents G, Burlingame R W, Wang B-C, Love W E, Moudrianakis E N. The nucleosomal core histone octamer at 3.1A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arents G, Moudrianakis E N. DNA protein interactions in chromatin and the structure of the histone octamer. In: Sarma H, Sarma M, editors. Structural biology: the state of the art. Proceedings of the Eighth Conversation. Albany: State University of New York; 1994. pp. 93–108. [Google Scholar]
- 8.Ausio J, van Holde K. Histone hyperacetylation: its effect on nucleosome conformation and stability. Biochemistry. 1986;22:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 9.Ausio J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone tails in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 10.Banéres J-L, Martin A, Parello J. The N tails of histones H3 and H4 adopt a highly structured conformation in the nucleosome. J Mol Biol. 1997;273:503–508. doi: 10.1006/jmbi.1997.1297. [DOI] [PubMed] [Google Scholar]
- 11.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 12.Beato M, Eisfeld K. Transcription factor access to chromatin. Nucleic Acids Res. 1997;25:3559–3563. doi: 10.1093/nar/25.18.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Transcriptional silencing in yeasts is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 14.Brownell J, Allis C D. An activity gel assay detects a single catalytically active histone acetyltransferase subunit in Tetrahymena macronuclei. Proc Natl Acad Sci USA. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brownell J, Zhou J, Ranally T, Kobayashi R, Edmonson D, Roth S, Allis C D. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 16.Cary P D, Moss T, Bradbury E M. High resolution proton-magnetic-resonance studies of chromatin core particles. Eur J Biochem. 1978;89:475–482. doi: 10.1111/j.1432-1033.1978.tb12551.x. [DOI] [PubMed] [Google Scholar]
- 17.Chen H, Li B, Workman J L. A histone DNA-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor-induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dimitrov S, Wolffe A P. Fine resolution of histones by two-dimensional polyacrylamide gel electrophoresis: developmental implications. Methods Companion Methods Enzymol. 1997;12:57–61. doi: 10.1006/meth.1997.0447. [DOI] [PubMed] [Google Scholar]
- 19.Dimitrov S I, Russanova V, Pashev I. The globular domain of histone H5 is internally located in the 30 nm chromatin fiber: an immunochemical study. EMBO J. 1987;6:2387–2392. doi: 10.1002/j.1460-2075.1987.tb02516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dimitrov S I, Stefanovsky V, Karagyozov L, Angelov D A, Pashev I G. The enhancers and promoters of the Xenopus laevis ribosomal spacer are associated with histones upon active transcription of the ribosomal genes. Nucleic Acids Res. 1990;18:6393–6397. doi: 10.1093/nar/18.21.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dimitrov S I, Almouzni G, Dasso M, Wolffe A. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 1993;160:214–227. doi: 10.1006/dbio.1993.1299. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov S I, Wolffe A P. Chromatin and nuclear assembly: experimental approaches toward the reconstitution of transcriptionally active and silent states. Biochim Biophys Acta. 1995;1260:1–13. doi: 10.1016/0167-4781(94)00182-3. [DOI] [PubMed] [Google Scholar]
- 23.Ebralidze K K, Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Nucleosomal structure at hyperacetylated loci probed in nuclei by DNA-histone crosslinking. Nucleic Acids Res. 1993;21:4734–4738. doi: 10.1093/nar/21.20.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edmonson D G, Smith M M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 25.Ericsson C, Grossbach U, Bjorkroth B, Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990;60:73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Ramirez M, Rocchini C, Ausio J. Modulation of chromatin folding by histone acetylation. J Biol Chem. 1995;270:17923–17928. doi: 10.1074/jbc.270.30.17923. [DOI] [PubMed] [Google Scholar]
- 27.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 29.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1989;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone acetylation co-maps with generalized DNase I sensitivity in the chicken β-blobin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 32.Heinzel T, Lavinsky R M, Mullen T-M, Söderström M, Laherty C D, Torchia J, Yang W-M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 33.Hockensmith J W, Kubasek W L, Worachek W R, Evertsz E M, von Hippel P H. DNA-protein interactions. Methods Enzymol. 1991;208:211–236. doi: 10.1016/0076-6879(91)08015-a. [DOI] [PubMed] [Google Scholar]
- 34.Howe L, Ausio J. Nucleosome translational position and not histone acetylation determines TFIIA binding to nucleosomal Xenopus laevis 5S rRNA genes. Mol Cell Biol. 1998;18:1156–1162. doi: 10.1128/mcb.18.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ip Y T, Jackson V, Meyer J, Chalkley R. The separation of transcriptionally engaged genes. J Biol Chem. 1988;263:14044–14052. [PubMed] [Google Scholar]
- 36.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 37.Jeppesen P, Mitchell A, Turner B M, Perry P. Antibodies to defined histone epitopes reveal variations in chromatin conformation and underacetylation of centric heterochromatin in human metaphase chromosomes. Chromosoma. 1992;101:322–332. doi: 10.1007/BF00346011. [DOI] [PubMed] [Google Scholar]
- 38.Jeppesen P, Turner B M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 39.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 40.Khochbin S, Wolffe A P. The origin and utility of histone deacetylases. FEBS Lett. 1997;419:157–160. doi: 10.1016/s0014-5793(97)01423-3. [DOI] [PubMed] [Google Scholar]
- 41.Kuo M-H, Brownell J A, Sobel R E, Ranalli T, Cook R G, Edmonson D G, Roth S Y, Allis C D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- 42.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 43.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 44.Lin Y S, Carey M, Ptachne M, Green M R. How different transcription activators can cooperate. Cell. 1988;54:659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- 45.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 46.Mirzabekov A D, Pruss D V, Ebralidze K K. Chromatin superstructure-dependent crosslinking with DNA of the histone H5 residues Thr1, His25 and His62. J Mol Biol. 1989;211:479–491. doi: 10.1016/0022-2836(90)90366-T. [DOI] [PubMed] [Google Scholar]
- 47.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani I, Allis C D. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;86:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 48.Moss T, Dimitrov S I, Houde D. UV-laser crosslinking of proteins to DNA. Methods Companion Methods Enzymol. 1997;11:225–234. doi: 10.1006/meth.1996.0409. [DOI] [PubMed] [Google Scholar]
- 49.Mutskov V J, Russanova V, Dimitrov S I, Pashev I. Histones associated with non-nucleosomal rat-ribosomal genes are acetylated, while those bound to nucleosome organized gene copies are not. J Biol Chem. 1996;271:11852–11857. doi: 10.1074/jbc.271.20.11852. [DOI] [PubMed] [Google Scholar]
- 50.Nacheva G A, Guschin D Y, Preobrazhenskaya O V, Karpov V L, Ebralidze K K, Mirzabekov A D. Changes in the pattern of histone binding to DNA upon transcription activation. Cell. 1989;58:27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetytransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 52.Owen-Hughes T, Workman J L. Experimental analysis of chromatin function in transcriptional control. Crit Rev Euk Gene Expression. 1994;4:403–441. [PubMed] [Google Scholar]
- 53.Pashev I G, Dimitrov S I, Angelov D. Laser-induced protein-DNA crosslinking. Trends Biochem Sci. 1991;16:323–326. doi: 10.1016/0968-0004(91)90133-g. [DOI] [PubMed] [Google Scholar]
- 54.Pazin M J, Kadonaga J T. What’s up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 55.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 56.Roth S Y, Shimuzu M, Johnson L, Grunstein M, Simpson T. Stable nucleosome positioning and complete repression by the yeast a2 repressor are disrupted by amino-terminal mutations in histone H4. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 57.Roth S Y, Allis C D. Histone acetylation and chromatin assembly: a single escort, multiple dances? Cell. 1996;87:5–8. doi: 10.1016/s0092-8674(00)81316-1. [DOI] [PubMed] [Google Scholar]
- 58.Solomon M J, Larsen P L, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 59.Stefanovsky V, Dimitrov S I, Russanova V R, Angelov D, Pashev I G. Laser-induced crosslinking of histones to DNA in chromatin and core particles: implications in studying histone-DNA interactions. Nucleic Acids Res. 1989;23:10069–10081. doi: 10.1093/nar/17.23.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 61.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by the binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 62.Tsukiyama T, Daniel C, Tamkun G, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 63.Turner B M, Franchi L, Wallace H. Islands of acetylated H4 in polytene chromosomes and their relationship to chromatin packaging and transcriptional activity. J Cell Sci. 1990;96:335–346. doi: 10.1242/jcs.96.2.335. [DOI] [PubMed] [Google Scholar]
- 64.Turner B M, Birley A J, Lavender J. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 65.Turner B M, O’Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 66.Usachenko S I, Bavykin S G, Gavin I M, Bradbury E M. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc Natl Acad Sci USA. 1994;91:6845–6849. doi: 10.1073/pnas.91.15.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Holde K. Chromatin. Berlin, Germany: Springer-Verlag KG; 1988. [Google Scholar]
- 68.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 69.Vettese-Dadey M, Walter P, Chen H, Juan L J, Workman J L. Role of the histone amino termini in the facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 71.Volgelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 73.Walker I O. Differential dissociation of histone tails from core chromatin. Biochemistry. 1984;23:5622–5628. doi: 10.1021/bi00318a037. [DOI] [PubMed] [Google Scholar]
- 74.Walter P P, Owen-Hughes T A, Côté J, Workman J L. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol Cell Biol. 1995;15:6178–6187. doi: 10.1128/mcb.15.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolffe A P, Pruss D. Targeted chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 76.Workman J L, Kingston R E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992;258:1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- 77.Yang H J, Ogryzko V, Nishikawa J, Howard B, Nakatani Y. Ap300/CBP-associated factor that competes with the adenoviral oncoprotein E1a. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]