Abstract
The eukaryotic single-stranded DNA binding protein replication protein A (RPA) participates in major DNA transactions. RPA also interacts through its middle subunit (Rpa2) with regulators of the cell division cycle and of the response to DNA damage. A specific contact between Rpa2 and nascent simian virus 40 DNA was revealed by in situ UV cross-linking. The dynamic attributes of the cross-linked DNA, namely, its size distribution, RNA primer content, and replication fork polarity, were determined. These data suggest that Rpa2 contacts the early DNA chain intermediates synthesized by DNA polymerase α-primase (RNA-DNA primers) but not more advanced products. Possible signaling functions of Rpa2 are discussed, and current models of eukaryotic lagging-strand DNA synthesis are evaluated in view of our results.
The single-stranded DNA (ssDNA) binding protein (SSB) replication protein A (RPA) participates in eukaryotic DNA replication (16, 58–60), DNA excision repair (12), and homologous recombination (22, 32). In addition, RPA communicates with regulators of the cell division cycle and of the response to DNA damage (9, 30, 41, 42). RPA is a heterotrimer, containing subunits of 70, 29 to 34, and 11 to 14 kDa (Rpa1, -2, and -3, respectively), all of which are conserved among eukaryotes and essential for viability in Saccharomyces cerevisiae (6, 7, 16, 34, 58, 59). Although functionally analogous to prokaryotic SSBs (31), RPA differs from them in its subunit complexity, in being modified by cellular regulators, and in its mode of template binding (59).
RPA’s roles in DNA replication are inferred primarily from studying simian virus 40 (SV40) DNA replication in systems reconstituted in vitro and its binding interactions with defined ssDNA templates and other replication proteins (13). Such data portray RPA as a component of a ternary primosome complex whose other members are a replication initiator-helicase (the viral large T antigen) and DNA polymerase (Pol) α-primase. The primosome assembles at the SV40 replication origin (ori), melts ori duplex DNA, and primes the leading DNA strands at this site. However, it also continues to unwind parental DNA and to prime lagging-strand intermediates throughout the replication cycle (11, 14, 33, 35, 36, 44, 47, 52, 56). In these processes, RPA facilitates parental DNA unwinding, stabilizes the exposed template, and attracts other replication proteins.
The relevant functions of individual RPA subunits are understood in part. SSB activity has been localized to a central, ∼300-amino-acid region of Rpa1 fit to anchor RPA to the template strand (3, 19, 21, 43, 48). The ssDNA portion in direct contact with Rpa1 may be as small as 8 nucleotides (nt), although the stable, occluded binding site for RPA has been estimated to be closer to 30 nt (1–3, 26). Rpa2 and -3 form a stable complex that binds Rpa1, probably via Rpa3 (21, 25, 48). Each of the two smaller subunits features a single SSB motif found twice in Rpa1. Hence, the subunits may also bind ssDNA (43). In fact, within RPA, Rpa2 interacts with the primer end of a synthetic primed template (28). Conceivably, the weak DNA binding potential of the smaller subunits is bolstered at the replication fork by additional protein-DNA as well as protein-protein interactions of the primosome.
DNA damage-responsive and cell cycle-regulated protein kinases phosphorylate the N-terminal domain of Rpa2 (4, 8, 15, 20, 40, 61). DNA damage-induced phosphorylation occurs neither in human ataxia telangiectasia cells lacking ATM (30) nor in S. cerevisiae with a mutation in the ATM homologue MEC1 (9). Since MEC1 halts S-phase progression in response to replicative lesions (41, 42), the attendant phosphorylation of Rpa2 likely mediates the requisite switch between replicative and repair modes of DNA synthesis. Likewise, phosphorylation of Rpa2 by the G1/S checkpoint cyclin-dependent kinase 2 may be related to entry into S phase (46). Investigating the function of Rpa2 at the replication fork may provide clues to the relation between the phosphorylation of this subunit and the modulation of DNA synthesis.
To address this issue, Rpa2’s interaction with nascent DNA was monitored within replicating SV40 chromosomes by protein-DNA UV cross-linking. The data indicate that Rpa2 contacts an early intermediate produced by Pol α-primase (RNA-DNA primer [10, 37, 39, 53], henceforth RDP) but does not contact more-advanced products. This observation suggests a link between the replicative and signaling functions of RPA and provides a new vantage point from which to evaluate current models of eukaryotic lagging-DNA-strand synthesis.
MATERIALS AND METHODS
Materials.
Monoclonal antibodies (MAbs) specific to Rpa1 and Rpa2, namely, anti-SSB70A, anti-SSB70B, anti-SSB70C, and anti-SSB34A (25), as well as rabbit polyclonal antibodies raised against Rpa3, were received from Jerard Hurwitz, Memorial Sloan-Kettering Cancer Center, New York, N.Y. Sara Lavi, Tel Aviv University, also provided the anti-Rpa1 and anti-Rpa2 MAbs. Protease inhibitor cocktail tablets (Complete Mini) were purchased from Boehringer Mannheim. Other materials were as described previously (62).
DNA-protein UV cross-linking in replicating SV40 chromosomes.
Nuclear monolayers from SV40-infected CV-1 cells were prepared essentially as described previously (62). However, Complete Mini tablets, used according to the manufacturer’s instructions, replaced the protease inhibitor cocktail. The nuclear monolayers were gently agitated at 30°C for the times indicated in the figures with, per 100-mm-diameter plate, 600 to 1,200 μl of the replication mixture (50 mM K-acetate, 5 mM MgCl2, 2 mM dithiothreitol [DTT], 100 μg of leupeptin per ml, 0.05% Nonidet P-40, 2 μM [each] dGTP and dCTP, 20 μM bromodeoxyuridine triphosphate [BrdUTP], 0.5 μM [α-32P]dATP [500 Ci/mmol], 2 mM ATP, and 20 μM concentrations of other radioactive nucleoside triphosphates [rNTPs]). In the chase mixtures, 100 μM dATP and 1 mM dTTP replaced the radioactive and photoreactive deoxyNTPs (dNTPs) and the nuclear monolayers were further incubated as indicated in the figures. When nascent DNA was labeled in the RNA primer moiety, the corresponding replication mixture contained 0.5 μM [α-32P]UTP (3,000 Ci/mmol), 2 mM ATP, 20 μM concentrations of the three other rNTPs, 20 μM BrdUTP, 2 μM concentrations of the other dNTPs, and 200 μg of α-amanitin per ml. To end the reaction, the overlaying mixture was removed and the plates were chilled on ice. The plates were then placed on a model TFX-20M transilluminator (Vilber-Lourmat), providing a radiation spectrum with a peak at 312 nm. After irradiation for 6 to 10 min at 4°C, the nuclear monolayers were washed with 1 ml of 5 mM K-acetate–0.5 mM MgCl2–2 mM DTT–30 mM K-HEPES buffer (pH 7.4). Viral DNA and cross-linked proteins were extracted from the nuclei with sodium dodecyl sulfate (SDS)-NaCl buffer (23). Subsequently, the photolabeled proteins were extracted with phenol and precipitated from the phenol phase and interphase with acetone (62). Where indicated below, the acetone precipitate was resuspended in 250 μl of DNase buffer (40 mM Tris-HCl [pH 8.0], 6 mM MgCl2, 1.5 mM CaCl2) containing 0.4 mg of bovine serum albumin (BSA) per ml, the protease inhibitors, and 10 U of DNase I. Following incubation for 30 min at 37°C, an equal volume of 2× denaturation buffer (100 mM Tris-HCl [pH 7.5], 1% SDS, 140 mM β-mercaptoethanol) was added and the mixture was boiled for 10 min. Proteins and covalently linked nascent DNA were separated from free DNA by a second extraction with aqueous phenol and precipitated as described above. When characterization of the cross-linked DNA was intended, the first acetone precipitate was dissolved in 500 μl of 1× denaturation buffer without prior DNase I digestion. The photolabeled proteins were dissociated from the replicating viral DNA by boiling for 10 min, phenol extracted, and precipitated with acetone as described above. Reference protein markers for Western blot analysis were provided by DNase I-digested nonradioactive nuclear extracts prepared essentially as described above but without cross-linking.
To isolate photolabeled RPA heterotrimer, pulse-labeling of nascent DNA and UV cross-linking were performed with isolated replicating SV40 chromatin, essentially as described previously (62) but with some modifications. Complete Mini tablets replaced protease inhibitors in all solutions used for tagging and processing the cross-linked chromatin. The concentration and specific activity of [α-32P]dATP were as described above. Following pulse-labeling, the mixtures were transferred into a polystyrene culture plate and irradiated as described above. The plate was then washed twice with 300 μl of ice-cold low-salt buffer (20 mM HEPES-Na [pH 7.8], 5 mM K-acetate, 0.5 mM MgCl2, 0.5 mM DTT) followed by centrifugation at 220,000 × g for 90 min through a 200-μl 10% glycerol cushion in low-salt buffer. The chromatin pellet was suspended in 250 μl of DNase buffer, digested as described above with DNase I, and prepared for native immunoprecipitation by dilution with 5 volumes of 50 mM Tris-HCl (pH 7.5)–150 mM NaCl–1% Nonidet P-40. The resultant suspension was clarified by centrifugation at 4°C and 10,000 × g for 30 min.
Immunochemical detection of photolabeled RPA subunits.
The photolabeled proteins of the acetone precipitate were dissolved in 100 μl of 1× denaturation buffer and denatured by boiling for 10 min. They were then partially renatured by diluting the samples with 9 volumes of ice-cold washing buffer (50 mM Tris-HCl, [pH 7.5], 150 mM NaCl) containing 0.5% Nonidet P-40 and the protease inhibitors. Myeloma immunoglobulin G (IgG; 10 μg) used for preclearing was adsorbed to 4 μg of protein A-Sepharose via 20 μg of rabbit anti-mouse IgG in 300 μl of PBS (10 mM potassium phosphate buffer, [pH 7.4], 0.15 M NaCl) containing 1% BSA. The washed beads were incubated with the photolabeled protein mixtures for 3 h at 4°C on a rotator. Rabbit anti-mouse IgG was also used to enhance binding to protein A-Sepharose of the anti-RPA MAbs except anti-SSB70B and the polyclonal anti-Rpa3 antibodies that were adsorbed as such. Anti-SSB34A, anti-SSB70A, and anti-SSB70B hybridoma supernatants were used in amounts of 1, 2, and 2 ml, respectively. Purified anti SSB70C was used at 12 μg, the anti-Rpa3 antiserum was used at 4 to 10 μl, and both were adsorbed to protein A-Sepharose in 300 μl of PBS-BSA as described above. The precleared preparation was supplemented with, as the carrier, 1 mM dATP or UTP and 100 μg of denatured salmon sperm DNA per ml. This mixture was added to the protein A-antibody beads, and the suspension was rotated for 6 to 16 h at 4°C. The beads were rinsed four times with washing buffer containing 0.2% Nonidet P-40. Antibody-antigen conjugates were released by heating the suspension at 95°C for 10 min in 20 μl of 2× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and separated on SDS–10% (if not otherwise indicated) polyacrylamide gels. The gels were blotted on a nitrocellulose membrane or dried. Radioactivity was monitored and quantified by phosphorimaging (Fujix Bas 1000; Fuji) or autoradiography and densitometry (ScanJet 3p; Hewlett-Packard). TINA software (Raytest Isotopenmessgeräte GmbH), compatible with the TINA-PCBAS and TIFF files of the phosphorimager and scanner, respectively, was applied in both cases. These tools were also used for the densitometry of the enhanced-chemiluminescence (ECL) autoradiograms. The cross-linking intensity (CI) is the densitometric signal of the photolabeled protein normalized to the ECL signal of the immunoprecipitated counterpart. The relative labeling of RDP is the radioactivity in the 10- to 35-nt region divided by that in the 10- to 250-nt region representing the entire spectrum of Okazaki fragments and their precursors.
Photolabeled RPA heterotrimer was immunopurified from the cross-linked and DNase I-digested replicating SV40 chromatin. Following preclearing with a nonspecific antibody, as described above, native immunoprecipitation was performed with each of the three anti-Rpa1 MAbs (anti-SSB70A, anti-SSB70B, and anti-SSB70C) as well as with anti-SSB34A. The immunoprecipitates were released from protein A beads by heating the beads in 2× sample buffer as described above followed by washing in 200 μl of denaturation buffer. The supernatants were combined and extracted with phenol, and the proteins were precipitated from the phenol phase and interphase with acetone, as described above. The precipitates were denatured at 100°C for 10 min in 20 μl of SDS-PAGE sample buffer and separated on a gradient gel of SDS–8 to 15% polyacrylamide. Positions of RPA subunits were determined by sequential probing of an immunoblot with antibodies directed against each subunit. Photolabeled derivatives were detected by autoradiography.
Sizing cross-linked DNA chains.
Gel portions containing photolabeled proteins of interest were excised, swollen in 10 mM Tris-HCl (pH 7.5)–0.1 mM EDTA, and ground. The slurry was suspended in 300 μl of a similar buffer containing 1% SDS and 100 μg of proteinase K per ml and incubated for 2 h at 37°C. A comparable gel portion was extracted without protease to detect any free DNA migrating with the photolabeled conjugate on the SDS-polyacrylamide gel. The released DNA was mixed with 5 μg of carrier plasmid DNA or yeast tRNA and extracted with phenol, precipitated with ethanol, dissolved in formamide, and separated on a denaturing 12% polyacrylamide–7 M urea gel in 25 mM Tris-borate buffer, pH 8.3. After autoradiography and densitometric scanning, the profile of the unproteolyzed control was subtracted from that of the proteolyzed sample to yield the net profile of DNA released from the photolabeled protein.
Determination of replication fork polarity.
Replication fork polarity was determined by gel mobility retardation of the Rpa2 conjugate and by dot blot hybridization of the DNA released from it. In the first analysis, immunoprecipitated, photolabeled Rpa2, taken from half of a 100-mm-diameter plate, was dissolved in 25 μl of SDS-PAGE sample buffer containing 3 μg of either ssDNAs of pairs of M13mp10 clones encoding the lagging or leading SV40 template strands (38) or vector DNA. Following heating to 100°C for 1 min and slow cooling to 30°C, the mixtures were separated by SDS-PAGE. The radioactive species were detected by autoradiography. In the second analysis, DNA was released from the Rpa2 conjugate and purified as described above. It, along with an isolated RDP control, was then hybridized to dot blots, each containing 1 μg of ssDNA of one of the four template clones described above or of the vector DNA.
RESULTS
Interaction of Rpa2 with nascent SV40 DNA.
Mammalian replication proteins can be detected in situ by combining DNA-protein UV cross-linking with immunopurification (62). This protocol was adapted to monitor possible interactions between RPA and nascent SV40 DNA and eventually characterize the cross-linked DNA. Briefly, viral chromosomes replicating within nuclear monolayers or in a soluble fraction were pulse-labeled from photoreactive DNA precursor (5-BrdUTP) and radioactive dNTP or rNTP precursors. Following UV irradiation of the replication mixture, SV40 DNA and any cross-linked proteins were extracted. The photolabeled proteins were separated from the bulk nascent DNA, before or after DNase I digestion, depending on whether only their detection was intended (Fig. 1 to 3) or their detection as well as a characterization of the cross-linked DNA was required (Fig. 4 to 6).
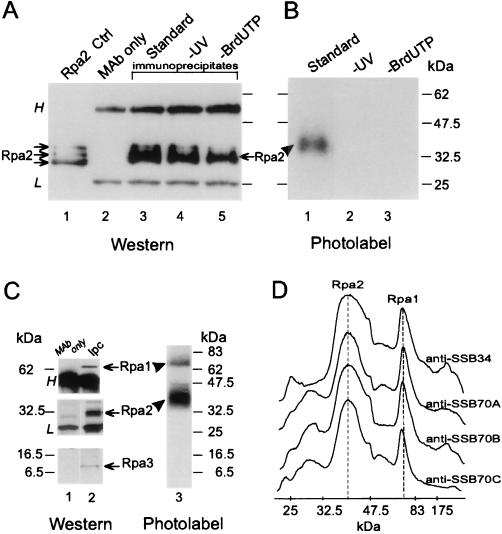
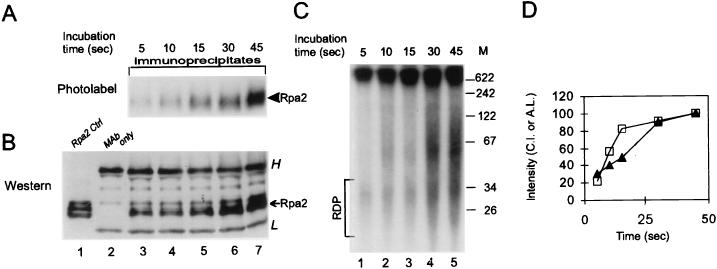
FIG. 1.
UV cross-linking of Rpa2 with nascent SV40 DNA. Proteins were photolabeled with nascent DNA within replicating SV40 chromosomes pulse-labeled by BrdUTP and [α-32P]dATP for 90 s. Following DNase I digestion, Rpa2 was immunopurified and detected by immunoblotting and autoradiography, as described in Materials and Methods. (A) Immunoblot probed with anti-SSB34A. Proteins extracted from a nonlabeled control nuclear monolayer (lane 1), antibody alone (lane 2), the immunoprecipitates of cross-linked proteins derived from the standard reaction mixture (lane 3), a nonirradiated control (lane 4), or a control with dTTP instead of BrdUTP (lane 5) are shown. (B) Autoradiogram of proteins immunoprecipitated from the standard mixture (lane 1), a nonirradiated control (lane 2), or the control with dTTP instead of BrdUTP (lane 3). (C) Immunoprecipitation of the photolabeled RPA heterotrimer. RPA was immunoprecipitated under native conditions from the photolabeled protein mixture derived from replicating SV40 chromatin with anti-SSB34. After separation by SDS-PAGE, individual RPA subunits were detected by immunoblotting and photolabeled derivatives were detected by autoradiography. (D) Densitometric tracings of photolabeled RPA precipitated under native conditions by the indicated MAbs and resolved by SDS-PAGE as described for panel C. H and L indicate heavy and light Ig chains, respectively. The arrows and arrowheads point to the ECL signal of Rpa2 and the photolabeled derivative, respectively. Ctrl, control; Ipc, immunoprecipitated cleared Rpa.
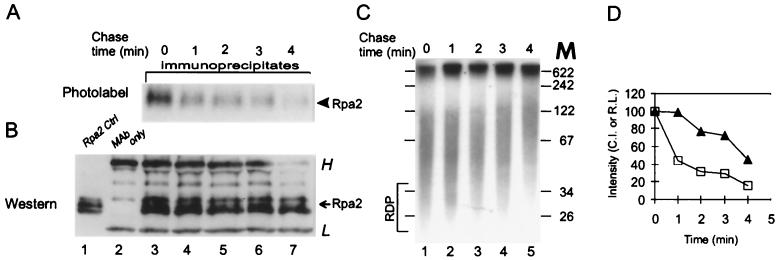
FIG. 3.
Pulse-chase kinetics of Rpa2’s photolabel. SV40 DNA pulse-labeled for 90 s with BrdUTP and [α-32P]dATP was chased with dTTP and nonlabeled dATP as indicated. After UV cross-linking, Rpa2 was immunopurified and detected as described in Materials and Methods and in the legend of Fig. 1. (A) Autoradiogram; (B) immunoblot; (C) patterns of nascent SV40 DNA during chase; (D) Rpa2 CI (open squares) and relative labeling (R.L.) of the RDP fraction (filled triangles) versus chase time. The photolabeled protein samples shown in panel A were isolated from the bulk (90%) of the reaction mixture, and the blot showing them was exposed for a week. The total nascent DNA shown in panel C represents 10% of the reaction mixture, and the gel containing it was exposed for 16 h. H and L indicate heavy and light Ig chains, respectively. The arrow points to the ECL signal of Rpa2; the arrowhead points to the photolabeled derivative. M, DNA size markers (in nucleotides).
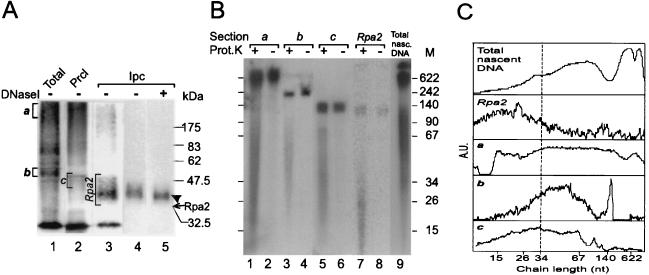
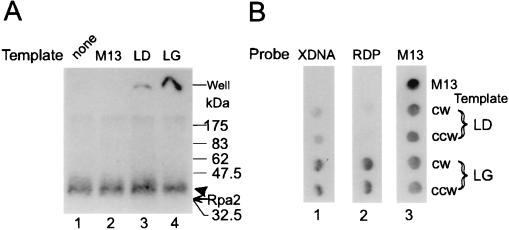
FIG. 4.
Sizing of nascent-DNA chains cross-linked to Rpa2. (A) Photolabeled Rpa2 not treated with DNase I was isolated from replication mixtures that had been pulse-labeled for 90 s as detailed in Materials and Methods. Crude photolabeled protein mixture (Total; lane 1); photolabeled proteins of the preclearing precipitate (Prcl; lane 2); immunoprecipitated cleared photolabeled Rpa2 (Ipc; lane 3); and nitrocellulose blots of the Rpa2 conjugate separated by SDS-PAGE, before or after DNase I digestion (lanes 4 and 5, respectively), are shown. (B) Portions of the bracketed gel sections a to c and Rpa2 (from panel A, lanes 1 to 3) were incubated with either proteinase K (Prot.K) (lanes 1, 3, 5, and 7) or buffer only (lanes 2, 4, 6, and 8). DNA was phenol extracted from the gel sections and separated by denaturing polyacrylamide-urea gel electrophoresis. Lane 9 contains an aliquot of nascent SV40 DNA (Total nasc. DNA) of the same replication mixture. The arrow marks the position of the ECL signal of Rpa2, and the arrowhead marks the position of the photolabeled derivative M, DNA size markers (in nucleotides). (C) Densitometric tracings of the total and released DNA preparations of panel B. The profile of the total DNA is shown. Net profiles of the DNA populations released from the indicated sections of panel A were obtained by subtracting the profile of an unproteolyzed sample (even-numbered lanes in panel B) from the original profile of the matched proteolyzed sample (odd-numbered lanes). However, residual spikes of large chains remained in the net profiles in sections b and c. These spikes coincide with the free-DNA contaminants and probably result from an excess of proteolyzed over unproteolyzed sample in the paired samples. The size distributions of chains in profiles a to c were estimated by setting their half-maximal heights as lower and upper borders. A.U., arbitrary absorption units.
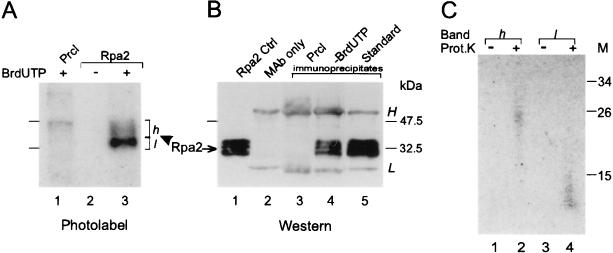
FIG. 6.
Cross-linking of Rpa2 to nascent DNA labeled in the RNA primer moiety. Protein-DNA conjugates radiolabeled from [α-32P]UTP were prepared and detected by autoradiography and Western blotting, and the cross-linked DNAs were sized, as detailed in Materials and Methods. (A) Autoradiogram of a preclearance (Prcl) immunoprecipitate (lane 1), an Rpa2 immunoprecipitate of a control replication mixture containing dTTP instead of BrdUTP (lane 2), or the standard mixture (lane 3). (B) Immunoblot probed with the anti-Rpa2 MAb. Shown are proteins extracted from a nonlabeled control (Cntrl) nuclear monolayer (lane 1), antibody alone (lane 2), a preclearance immunoprecipitate (lane 3), and Rpa2 immunoprecipitates from reaction mixtures without (lane 4) or with (lane 5) BrdUTP. (C) Nascent RNA-DNA released by proteolysis from photolabeled Rpa2 species of bands h and l (from panel A, lane 3) was resolved by gel electrophoresis (lanes 2 [band h] and 4 [band l]) along with respective unproteolyzed controls (lanes 1 and 3). h and l designate slow- and fast-migrating photolabeled Rpa2 species, respectively. H and L indicate heavy and light Ig chains, respectively. Prot.K proteinase K; M, protein size markers (in thousands). The arrow indicates the ECL signal of Rpa2, and the arrowhead indicates at the photolabeled derivative.
Rpa2 was immunopurified from the denatured photolabeled protein mixture with the cognate MAb anti-SSB34A and then visualized by Western blotting with the same antibody (Fig. 1A). Photolabeled derivatives were detected by autoradiography (Fig. 1B). Autoradiography revealed a radioactive product migrating between the 32.5- and 47-kDa size markers (lane 1), above the positions of the Rpa2 species detected by immunoblotting in the original protein mixture (Fig. 1A, lane 1) and the immunoprecipitates (lanes 3 to 5). Similar retardation, by the equivalent of 6 to 8 kDa, has been observed with a cross-linked 17-nt adduct (28). The radioactive Rpa2 derivative was seen neither in the nonirradiated control (Fig. 1B, lane 2) nor in that containing dTTP instead of BrdUTP (lane 3), indicating covalent linkage of DNA and protein. To compare the photolabeling potentials of the three RPA subunits, the intact RPA heterotrimer was precipitated under native conditions from the photolabeled protein mixture derived from replicating SV40 chromatin. In this case, the three anti-Rpa1 MAbs (anti-SSB70A, anti-SSB70B, and anti-SSB70C) and anti-SSB34A were used in separate immunoprecipitation attempts. After resolution of the individual subunits by denaturing gel electrophoresis, they were detected by immunoblotting and autoradiography. This process indicated that while all three subunits were precipitated, Rpa2 received the bulk of the photolabel; about 20% appeared in an Rpa1 derivative and none appeared in Rpa3 (Fig. 1C). The bias in favor of Rpa2 was observed regardless of the subunit specificity of the immunoprecipitating antibody (Fig. 1D).
Dynamics of the interaction between Rpa2 and nascent DNA.
The maturation kinetics of the nascent-DNA chains and the dynamics of the interaction between Rpa2 and nascent DNA were compared in continuous-labeling and pulse-chase experiments. Rpa2’s CI leveled off within ∼30 s (Fig. 2A, B, and D), similar to the rate at which the absolute labeling of the RDP fraction reached a steady-state value (Fig. 2C and D) (38). Moreover, Rpa2’s CI decayed with a half-life of ∼1 min (Fig. 3A, B, and D), faster than the conversion of RDPs into larger products (Fig. 3C and D). These data suggest that Rpa2 contacts RDPs and dissociates from them before further processing.
FIG. 2.
Behavior of Rpa2’s photolabel in continuous labeling. Protein-DNA conjugates were prepared after the indicated pulse-labeling times. Rpa2 was then immunopurified and detected by autoradiography and Western blot analysis, essentially as described in Materials and Methods and in the legend of Fig. 1. (A) Autoradiogram of photolabeled Rpa2; (B) immunoblot; (C) pattern of nascent-SV40-DNA chains from aliquots of the same replication mixtures; (D) Rpa2 CI (open squares) and absolute labeling (A.L.) of the RDP fraction (filled triangles) versus labeling time. H and L indicate heavy and light Ig chains, respectively. The arrow points to the ECL signal of Rpa2; the arrowhead points to the photolabeled derivative. Ctrl, control. M, DNA size markers (in nucleotides).
Size distribution of nascent DNA cross-linked to Rpa2.
To size the nascent-DNA chains cross-linked to Rpa2, a gel portion containing the original conjugate not treated with DNase I (Fig. 4A, lane 3) was digested with proteinase K. The released DNA was extracted with phenol and separated by denaturing gel electrophoresis, which revealed a major group of products ranging between ∼10 and 35 nt and another group ranging between ∼120 and 140 nt (Fig. 4B, lane 7). The latter, also extracted without proteolysis (Fig. 4B, lane 8), represented free DNA that was not covalently linked to Rpa2 but that migrated with the photolabeled protein in an SDS-polyacrylamide gel. The residual radioactivity seen in Fig. 4A, lane 3, between the well and the defined photolabeled Rpa2 band did not represent Rpa2 cross-linked to longer DNA chains. Blotting onto nitrocellulose removed this residue (lane 4), showing it to be free DNA not covalently bound to protein. Moreover, identical baseline intensities of the clarified region were obtained whether or not the Rpa2 conjugates had been subjected to digestion with DNase I (compare lanes 4 and 5). Moreover, the DNase I treatment only slightly changed the width, intensity, and position of the photolabeled Rpa2 band, consistent with the shortness of the original cross-linked chains (Fig. 4B, lane 7). Figure 4C compares the net scanned profile of the DNA released from Rpa2 (the difference between the densitometric tracings of lanes 7 and 8) to the profile of the original nascent DNA (derived from Fig. 4B, lane 9). While RDP-sized chains (area bordered by the 34-nt marker) constituted the bulk of DNA linked to Rpa2, they exhibited about 20% of the radioactivity of the “Okazaki zone” of total nascent DNA. We also determined the sizes of DNAs released from anonymous proteins found in the original mixture of photolabeled proteins (Fig. 4A, lane 1) or the preclearing precipitate (Fig. 4A, lane 2). Inspection of the respective autoradiograms (Fig. 4B, lanes 1 to 6) and derived net profiles yielded distributions of approximately 15 to 200, 30 to 80, and 15 to 70 nt (Fig. 4C). Considered together, these data indicated that selective binding of RDP-like chains is a distinctive property of Rpa2.
Lagging-strand polarity of DNA chains cross-linked to Rpa2.
The fork polarity of the DNA cross-linked to Rpa2 was determined by hybridization to SV40 ssDNA probes, representing the lagging or leading template strands, cloned in the M13 phage vector (38). Photolabeled Rpa2 not treated with DNase I was incubated under hybridization conditions with either lagging- or leading-strand probes or with the vector DNA, and the mixtures were resolved by SDS-PAGE. The nonhybridized conjugate and the hybridized form, expected at the well, were detected by autoradiography. As shown, the lagging-strand probes retained a considerable fraction of the radioactivity at the well, with the result that the intensity of the upper portion of photolabeled Rpa2 band was reduced (Fig. 5A, lane 4). The reason for the preferential retention seems to be the presence of the longer and better-hybridizing RDPs in this portion. In contrast, the leading-strand template retained only a residual fraction of the radioactivity at the well, about one-fifth of that seen with the lagging-strand template, and hardly reduced the intensity of the Rpa2 band (lane 3). In a complementary experiment, DNA released from photolabeled Rpa2 was hybridized to the individual probes on dot blots. In this case, a three- to fourfold bias in favor of the lagging-strand templates was found (Fig. 5B, lane 1). However, an RDP fraction purified directly from the replication mixture (Fig. 2C) hybridized to the same dots with a bias greater than 10-fold in favor of the lagging-strand templates (Fig. 5B, lane 2). The smaller bias seen with the Rpa2 conjugate and with the DNA released from it is attributed to increased contamination by fragments of long nascent chains due to the requisite higher specific radioactivity employed (500 versus 100 Ci/mmol) and longer isolation procedure. An alternative possibility, that Rpa2 also contacted leading chains, seems less likely since conjugates that were cross-linked to long nascent-DNA chains, expected at the top of the original SDS-polyacrylamide gel, were not evident.
FIG. 5.
Determination of fork polarity of DNA cross-linked to Rpa2. (A) Rpa2 cross-linked to intact nascent-DNA chains was immunopurified and hybridized to pairs of M13-SV40 ssDNA clones of leading (LD)- or lagging (LG)-strand fork polarity. The mixtures were resolved by SDS-PAGE and autoradiographed as described in Materials and Methods and the legend of Fig. 4. Lanes: 1, not hybridized; 2, vector; 3, mSVBH10 and mSVTB10 (clockwise replication fork [CW] and counterclockwise replication fork [CCW] leading-strand templates, respectively); 4, mSVBH11 and mSVTB11 (CW and CCW lagging-strand templates, respectively). (B) Nascent DNA was released by proteolysis of photolabeled Rpa2 (from Fig. 4B, lane 7). Following extraction from the gel, it was hybridized to dot blots containing the indicated leading- and lagging-strand probes (blot 1), along with a purified RDP fraction from Fig. 2C (blot 2) or vector DNA labeled by random priming (blot 3). XDNA, DNA released from the cross-linked Rpa2-DNA conjugate; RDP, RDP fraction; M13, vector DNA. The arrow indicates the position of the ECL signal of Rpa2, and the arrowhead indicates the photolabeled derivative.
Nascent DNA cross-linked to Rpa2 is covalently linked to RNA.
The presence of RNA primers in the nascent DNA cross-linked to Rpa2 was established by pulse-labeling the replicating DNA from [α-32P]UTP. Subsequent UV cross-linking and immunopurification revealed photolabeling of Rpa2 in the standard mixture but not in a control containing dTTP instead of BrdUTP (Fig. 6A, compare lanes 2 and 3). Hence, the radioactively labeled RNA was linked to Rpa2 through the cross-linked DNA moiety. Photolabeled in this manner, Rpa2 was resolved into two bands. The more intense, designated l, traveled near the major ECL signal of free Rpa2. The slower and fainter band (h) was retarded by the equivalent of ∼8 kDa and resembled in this regard Rpa2 photolabeled with a radioactive DNA precursor (Fig. 4A). Exhaustive proteolysis released from bands h and l chains of 20 to 30 and 10 to 15 nt, respectively (Fig. 6C, lanes 2 and 4). This difference may account, at least in part, for the difference in the electrophoretic mobilities of the parental photolabeled protein species h and l. Although the cross-linked nascent-DNA chains radiolabeled in the RNA moiety were also within the RDP size range, they appeared shorter, on average, than those radiolabeled in the DNA moiety (Fig. 4B, lane 7). This discrepancy is attributed to accentuation of the shorter chains in the number distribution due to radiolabeling of the fixed-size RNA moiety as opposed to enhancement of the longer chains in the weight distribution due to radiolabeling of the variable DNA moiety. Moreover, in the RNA radiolabeling mode, Rpa2 may be photolabeled as soon as the first photoreactive deoxynucleoside monophosphate (dNMP) residue is incorporated into the growing RDP; in the DNA radiolabeling mode, both the photoreactive and radioactive dNMPs are required.
DISCUSSION
Within the replicating SV40 chromosome, Rpa2 contacts RDPs, the products of Pol α-primase but not more advanced intermediates. This conclusion is based on the preferred cross-linking of Rpa2 (over Rpa1) to nascent SV40 DNA, kinetic attributes of the nascent-DNA chains cross-linked in situ to Rpa2, the size distribution of these cross-linked nascent-DNA chains, RNA primer content, and lagging-strand polarity (Fig. 1 to 6). These data also reinforce previous conclusions about the potential of Rpa2 to interact with DNA (28, 43), suggest a link between replicative and signaling functions of RPA, and provide a new vantage point from which to examine lagging-DNA-strand synthesis (45). Below, we elaborate on these issues.
Rpa2 may contact the growing ends of RDPs.
SSB activity was originally localized to Rpa1 (59), but recent evidence indicates that the smaller RPA subunits may also be endowed with this property. Philipova et al. have detected in the essential domains of Rpa2 and -3 a single copy of an SSB motif present twice in Rpa1 (43). They have also noted the weak SSB activity of Rpa2, as have others with the Rpa2-Rpa3 complex (46a). What could this SSB activity signify? Results of X-ray diffraction of a cocrystal containing the DNA binding domain of Rpa1 and an octanucleotide ligand indicate that the C-proximal domain of Rpa1 points toward the growing end of the primer strand (3). Since the C domain binds Rpa2 and -3 (18), Rpa2 may itself be near the primer end. In fact, Rpa2 has been cross-linked to the 3′ end of the primer strand of an RPA-primed template complex, and this reaction depended on the ability of Rpa1 to bind the template (28).
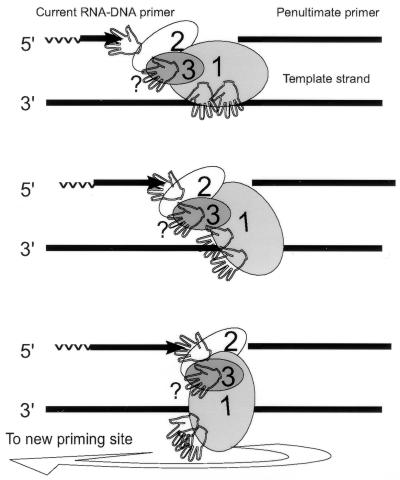
In what follows, we try to integrate the above facts and various attributes of the DNA cross-linked to Rpa2 (Fig. 2 to 6) into a preliminary viewpoint about the position and function of this subunit at the replication fork (Fig. 7). Accordingly, Rpa2 is situated to bind with its SSB motif a few dNMP residues at the 3′ end of the growing RDP. To maintain this position during RDP elongation, Rpa2 translocates with the growing end. Upon RDP completion, the contact is disrupted and RPA recycles to a new priming site. Our observations that support this scheme are, first, that the on and off rates of the contact between Rpa2 and nascent DNA matched the turnover kinetics of RDP (Fig. 2 and 3) and, second, that Rpa2 cross-linked with the entire spectrum of RDPs but not more advanced intermediates (Fig. 4 and 6).
FIG. 7.
Proposed contacts of RPA with DNA during the RDP cycle.
The ability of Rpa2 to interact with the 3′ end of the primer strand justifies reevaluation of RPA’s site size estimates (1, 2, 26, 27). Namely, different parts of model oligonucleotides used to determine this parameter can mimic different parts of a primed template, especially when RPA is limiting and the ligand is large enough to accommodate more than one subunit. Related to this may be the enhanced hyperphosphorylation of Rpa2 by the DNA-dependent protein kinase when RPA binds its ligand in the large-site mode (1). Presumably, contact of Rpa2 with a portion of the ligand under these conditions renders this subunit a better substrate for the kinase.
How can the SSB motif of Rpa2 recognize the primer end if the latter exists in duplex configuration? One possibility is that such recognition is enabled by spontaneous or RPA-assisted fraying of the duplex (17, 49). Nucleotide misincorporation and stalling of the fork also induce unwinding of the primer end, shifting it from the Pol to the proofreading site in certain Pols (54). However, because Pol α lacks a proofreader (55), it is tempting to speculate that associated Rpa2 compensates for this deficiency, acting as a potential sensor of the disabled primer end.
Rpa1 also cross-linked the primer strand, albeit more weakly than Rpa2. This interaction was observed both in SV40 chromosomes replicating in solution (Fig. 1C and D) or in nuclear monolayers (data not shown) and within a complex of purified RPA and a synthetic primed template (28). In contrast, when purified RPA interacts with single-stranded oligonucleotides, the affinity of Rpa1 to the ligand is orders of magnitude greater than that of Rpa2 (59). Therefore, the weaker binding of Rpa1 to the primer strand may reflect inadvertent interaction of RPA with traces of nascent DNA dissociated from the replicating chromosomes. Alternatively, Rpa1 does interact with the primer strand, either upstream or downstream of the gap RPA occupies, but this interaction is much weaker than that with the template strand. The apparent failure of Rpa3 to cross-link nascent DNA (Fig. 1C) may indicate that its SSB motif is idle, at least in ongoing replication. Alternatively, Rpa3 interacts with the primer strand under different circumstances or interacts with the template strand. To address these problems, it will be necessary to characterize all binding interactions of the three subunits with model primed templates and more complex systems that support the DNA transactions in which RPA partakes.
Possible signaling functions of Rpa2 within and beyond the replication fork.
Interaction with the growing primer end may qualify Rpa2 as a signal transducer within the replication apparatus, adapting the RPA heterotrimer itself or associated replication proteins to changes induced by DNA chain growth. RPA may be adjusted in this manner to the diminishing ssDNA template, being switched, perhaps, between its different binding modes (2) and/or induced to translocate to the next priming site. Rpa2 may also relay the growth signal to the associated Pol α-primase (5), permitting its progress along the template (51, 57) and/or cycling to a new priming site. Discontinuous translocation of the transcribing Escherichia coli RNA Pol has been proposed (inchworm model [51]), and an analogous mechanism may also benefit DNA Pol (57), allowing them to advance along rather than around their template. The strong pause site in RDP synthesis, accentuated by ATP depletion (39), hints that Pol α-primase translocates in such a manner.
Interaction of Rpa2 with the growing end of the primer and the reported interactions of Rpa2 with regulatory protein kinases (40, 59) implicate this subunit also in transduction of signals beyond the replication apparatus. For example, Rpa2 may report the condition of a disabled primer end to DNA damage-responsive checkpoints. Alternatively, signals received from checkpoints may be transmitted by Rpa2 to an associated replicative or repair protein and, in turn, shift the synthetic apparatus between the replicative and repair modes (29a). Although there is no evidence that Rpa2 phosphorylation inhibits ongoing SV40 DNA replication in experimental systems reconstituted in vitro (20, 29), this possibility remains to be explored under conditions closer to the physiological. The use of cellular replication systems, which are more responsive to checkpoints, may also be revealing. Such studies should also address the possible connection between the status of the primer end and Rpa2 phosphorylation.
Evaluation of models for lagging-DNA-strand synthesis.
Models of eukaryotic lagging-DNA-strand synthesis termed “initiation zone” and “nested discontinuity” have been described previously (38, 39, 45). Their current versions share the notion that RDPs produced by Pol α-primase are further processed by PCNA-dependent Pol δ. At ori, RDPs are extended into leading DNA strands. Those synthesized later are incorporated into Okazaki fragments of up to ∼200 nt (10, 37–39, 44, 45, 50, 53). It is with regard to the last step that the two models digress. In the first model, an Okazaki fragment arises by continuous elongation of a single RDP; in the second model, a contiguous array of RDPs matures into an Okazaki fragment in a repair-like process (38, 39). Hence, each model predicts a different population of nascent-DNA chains facing Rpa2: the entire spectrum of Okazaki fragment intermediates in the first model and only RDPs in the second. Selective cross-linking of RDPs to Rpa2 (Fig. 2 to 4 and 6) is consistent with the second model. Further support is lent to that model by the high intrinsic affinity and low cooperativity of RPA binding to the ssDNA target (27). These binding properties, which distinguish RPA from prokaryotic counterparts, hint that, in vivo, the eukaryotic protein encounters templates comprising a single or only a few binding sites. In fact, in nucleotide excision DNA repair, RPA interacts with a template of up to ∼30 nt (24). A similar portion may be available at the onset of the RDP cycle, according to the nested discontinuity model.
ACKNOWLEDGMENTS
We thank Jerard Hurwitz and Sara Lavi for providing anti-RPA antibodies, Marc Wold and Olga Lavrik for making results available prior to publication, and Mark Wold and Rolf Knippers for critical readings of the manuscript.
This work was supported by grants from the U.S.-Israel Binational Science Foundation, the Israel Cancer Society, and the German-Israeli Foundation for Research and Development to G.K.
REFERENCES
- 1.Blackwell L J, Borowiec J A, Mastrangelo I A. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol Cell Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell L J, Borowiec J A. Human replication protein A binds single-stranded DNA in two distinct complexes. Mol Cell Biol. 1994;14:3993–4001. doi: 10.1128/mcb.14.6.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochkarev A, Pfuetzner R A, Edwards A M, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- 4.Boubnov N V, Weaver D T. scid cells are deficient in Ku and replication protein A phosphorylation by the DNA-dependent protein kinase. Mol Cell Biol. 1995;15:5700–5706. doi: 10.1128/mcb.15.10.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun K A, Lao Y, He Z, Ingles C J, Wold M S. Role of protein-protein interactions in the function of replication protein A (RPA): RPA modulates the activity of DNA polymerase alpha by multiple mechanisms. Biochemistry. 1997;36:8443–8454. doi: 10.1021/bi970473r. [DOI] [PubMed] [Google Scholar]
- 6.Brill S J, Stillman B. Yeast replication factor-A functions in the unwinding of the SV40 origin of DNA replication. Nature. 1989;342:92–95. doi: 10.1038/342092a0. [DOI] [PubMed] [Google Scholar]
- 7.Brill S J, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- 8.Brush G S, Anderson C W, Kelly T J. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc Natl Acad Sci USA. 1994;91:12520–12524. doi: 10.1073/pnas.91.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brush G S, Morrow D M, Hieter P, Kelly T J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock P A, Seo Y S, Hurwitz J. Initiation of simian virus 40 DNA synthesis in vitro. Mol Cell Biol. 1991;11:2350–2361. doi: 10.1128/mcb.11.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins K L, Kelly T J. The effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coverley D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Requirement for replication protein SSB in human excision repair. Nature. 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 13.DePamphilis M L. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 14.Dornreiter I, Erdile L F, Gilbert I U, Von Winkler D, Kelly T J, Fanning E. Interaction of DNA polymerase α-primase with cellular replication protein A and SV40 T antigen. EMBO J. 1992;11:769–776. doi: 10.1002/j.1460-2075.1992.tb05110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dutta A, Din S, Brill S J, Stillman B. Phosphorylation of replication protein A: a role for cdc2 kinase in G1/S regulation. Cold Spring Harbor Symp Quant Biol. 1991;56:315–324. doi: 10.1101/sqb.1991.056.01.038. [DOI] [PubMed] [Google Scholar]
- 16.Fairman M P, Stillman B. Cellular factors required for multiple stages of SV40 replication in vitro. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgaki A, Hübscher U. DNA unwinding by replication protein A is a property of the 70 kDa subunit and is facilitated by phosphorylation of the 32 kDa subunit. Nucleic Acids Res. 1993;21:3659–3665. doi: 10.1093/nar/21.16.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomes X V, Wold M S. Structural analysis of human replication protein A. Mapping functional domains of the 70-kDa subunit. J Biol Chem. 1995;270:4534–4543. doi: 10.1074/jbc.270.9.4534. [DOI] [PubMed] [Google Scholar]
- 19.Gomes X V, Wold M S. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry. 1996;35:10558–10568. doi: 10.1021/bi9607517. [DOI] [PubMed] [Google Scholar]
- 20.Henricksen L A, Carter T, Dutta A, Wold M S. Phosphorylation of human replication protein A by the DNA-dependent protein kinase is involved in the modulation of DNA replication. Nucleic Acids Res. 1996;24:3107–3112. doi: 10.1093/nar/24.15.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henricksen L A, Umbricht C, Wold M S. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 22.Heyer W D, Rao M R, Erdile L F, Kelly T J, Kolodner R D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990;9:2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 24.Huang J C, Svoboda D L, Reardon J T, Sancar A. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci USA. 1992;89:3664–3668. doi: 10.1073/pnas.89.8.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenny M K, Schlegel U, Furneaux H, Hurwitz J. The role of human single stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J Biol Chem. 1990;13:7693–7700. [PubMed] [Google Scholar]
- 26.Kim C, Paulus B F, Wold M S. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 27.Kim C, Wold M S. Recombinant human replication protein A binds to polynucleotides with low cooperativity. Biochemistry. 1995;34:2058–2064. doi: 10.1021/bi00006a028. [DOI] [PubMed] [Google Scholar]
- 28.Lavrik O, Nasheuer H-P, Weisshart K, Wold M S, Prasad R, Beard W A, Wilson S H, Favre A. Subunits of human replication protein A are crosslinked by photoreactive primers synthesized by DNA polymerases. Nucleic Acids Res. 1998;26:602–607. doi: 10.1093/nar/26.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S H, Kim D K. The role of the 34-kDa subunit of human replication protein A in simian virus 40 DNA replication in vitro. J Biol Chem. 1995;270:12801–12807. doi: 10.1074/jbc.270.21.12801. [DOI] [PubMed] [Google Scholar]
- 29a.Lee S H, Kim D, Drissi R. Human xeroderma pigmentosum group A protein interacts with human replication protein A and inhibits DNA replication. J Biol Chem. 1995;270:21800–21805. doi: 10.1074/jbc.270.37.21800. [DOI] [PubMed] [Google Scholar]
- 30.Liu V F, Weaver D T. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohman T M, Ferrari M E. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 32.Longhese M P, Plevani P, Lucchini G. Replication factor A is required in vivo for DNA replication, repair, and recombination. Mol Cell Biol. 1994;14:7884–7890. doi: 10.1128/mcb.14.12.7884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melendy T, Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 replication. J Biol Chem. 1993;268:3389–3395. [PubMed] [Google Scholar]
- 34.Mitsis P G, Kowalczykowski S C, Lehman I R. A single-stranded DNA binding protein from Drosophila melanogaster: characterization of the heterotrimeric protein and its interaction with single-stranded DNA. Biochemistry. 1993;32:5257–5266. doi: 10.1021/bi00070a038. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y, Hurwitz J. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J Biol Chem. 1993;268:11008–11017. [PubMed] [Google Scholar]
- 36.Murakami Y, Wobbe C R, Weissbach L, Dean F B, Hurwitz J. Role of DNA polymerase α and DNA primase in simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1986;83:2869–2873. doi: 10.1073/pnas.83.9.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nethanel T, Kaufmann G. Two DNA polymerases may be required for synthesis of the lagging DNA strand of simian virus 40. J Virol. 1990;64:5912–5918. doi: 10.1128/jvi.64.12.5912-5918.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nethanel T, Reisfeld G, Dinter-Gottlieb G, Kaufmann G. An Okazaki piece of simian virus 40 may be synthesized by ligation of shorter precursor chains. J Virol. 1988;62:2867–2873. doi: 10.1128/jvi.62.8.2867-2873.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nethanel T, Zlotkin T, Kaufmann G. Assembly of simian virus 40 Okazaki pieces from DNA primers is reversibly arrested by ATP depletion. J Virol. 1992;66:6634–6640. doi: 10.1128/jvi.66.11.6634-6640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niu H, Erdjument-Bromage H, Pan Z Q, Lee S H, Tempst P, Hurwitz J. Mapping of amino acid residues in the p34 subunit of human single-stranded DNA-binding protein phosphorylated by DNA-dependent protein kinase and Cdc2 kinase in vitro. J Biol Chem. 1997;272:12634–12641. doi: 10.1074/jbc.272.19.12634. [DOI] [PubMed] [Google Scholar]
- 41.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 42.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 43.Philipova D, Mullen J R, Maniar H S, Lu J, Gu C, Brill S J. A hierarchy of SSB protomers in replication protein A. Genes Dev. 1996;10:2222–2233. doi: 10.1101/gad.10.17.2222. [DOI] [PubMed] [Google Scholar]
- 44.Prelich G, Stillman B. Coordinated leading and lagging DNA strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 45.Salas M, Miller T J, Leis J, DePamphilis M L. Mechanisms for priming DNA synthesis. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 131–176. [Google Scholar]
- 46.Santocanale C, Neecke H, Longhese M P, Lucchini G, Plevani P. Mutations in the gene encoding the 34 kDa subunit of yeast replication protein A cause defective S phase progression. J Mol Biol. 1995;254:595–607. doi: 10.1006/jmbi.1995.0641. [DOI] [PubMed] [Google Scholar]
- 46a.Sibenaller Z A, Sorensen B, Wold M S. The 32- and 14-kDa subunits of replication protein A are responsible for species-specific interactions with ssDNA. Biochemistry. 1998;37:12496–12506. doi: 10.1021/bi981110+. [DOI] [PubMed] [Google Scholar]
- 47.Stahl H, Dröge P, Knippers R. DNA helicase activity of SV40 large tumor antigen. EMBO J. 1986;5:1939–1944. doi: 10.1002/j.1460-2075.1986.tb04447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stigger E, Dean F, Hurwitz J, Lee S H. Reconstitution of functional human single-stranded DNA-binding protein from individual subunits expressed by recombinant baculoviruses. Proc Natl Acad Sci USA. 1994;91:579–583. doi: 10.1073/pnas.91.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treuner K, Ramsperger U, Knippers R. Replication protein A induces the unwinding of long double-stranded DNA regions. J Mol Biol. 1996;259:104–112. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 50.Tsurimoto T, Melendy T, Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990;346:534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- 51.Uptain S M, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 52.Waga S, Bauer G, Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994;269:10923–10934. [PubMed] [Google Scholar]
- 53.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 54.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 55.Wang T S-F. Cellular DNA polymerases. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 461–493. [Google Scholar]
- 56.Wiekowski M, Dröge P, Stahl H. Monoclonal antibodies as probes for a function of large T antigen during the elongation process of simian virus 40 DNA replication. J Virol. 1987;61:411–418. doi: 10.1128/jvi.61.2.411-418.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wlassoff W A, Dymshits G M, Lavrik O I. A model for DNA polymerase translocation: worm-like movement of DNA within the binding cleft. FEBS Lett. 1996;390:6–9. doi: 10.1016/0014-5793(96)00479-6. [DOI] [PubMed] [Google Scholar]
- 58.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Replication of simian virus 40 origin-containing DNA with purified components. Proc Natl Acad Sci USA. 1986;85:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wold M S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 60.Wold M S, Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zernik-Kobak M, Vasunia K, Connelly M, Anderson C W, Dixon K. Sites of UV-induced phosphorylation of the p34 subunit of replication protein A from HeLa cells. J Biol Chem. 1997;272:23896–23904. doi: 10.1074/jbc.272.38.23896. [DOI] [PubMed] [Google Scholar]
- 62.Zlotkin T, Kaufmann G, Jiang Y, Lee M Y W T, Uitto L, Syväoja J, Dornreiter I, Fanning E, Nethanel T. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–2305. [PMC free article] [PubMed] [Google Scholar]