Abstract
BACKGROUND:
SGLT2 (sodium-glucose cotransporter 2) inhibitors (SGLT2i) can protect the kidneys and heart, but the underlying mechanism remains poorly understood.
METHODS:
To gain insights on primary effects of SGLT2i that are not confounded by pathophysiologic processes or are secondary to improvement by SGLT2i, we performed an in-depth proteomics, phosphoproteomics, and metabolomics analysis by integrating signatures from multiple metabolic organs and body fluids after 1 week of SGLT2i treatment of nondiabetic as well as diabetic mice with early and uncomplicated hyperglycemia.
RESULTS:
Kidneys of nondiabetic mice reacted most strongly to SGLT2i in terms of proteomic reconfiguration, including evidence for less early proximal tubule glucotoxicity and a broad downregulation of the apical uptake transport machinery (including sodium, glucose, urate, purine bases, and amino acids), supported by mouse and human SGLT2 interactome studies. SGLT2i affected heart and liver signaling, but more reactive organs included the white adipose tissue, showing more lipolysis, and, particularly, the gut microbiome, with a lower relative abundance of bacteria taxa capable of fermenting phenylalanine and tryptophan to cardiovascular uremic toxins, resulting in lower plasma levels of these compounds (including p-cresol sulfate). SGLT2i was detectable in murine stool samples and its addition to human stool microbiota fermentation recapitulated some murine microbiome findings, suggesting direct inhibition of fermentation of aromatic amino acids and tryptophan. In mice lacking SGLT2 and in patients with decompensated heart failure or diabetes, the SGLT2i likewise reduced circulating p-cresol sulfate, and p-cresol impaired contractility and rhythm in human induced pluripotent stem cell–derived engineered heart tissue.
CONCLUSIONS:
SGLT2i reduced microbiome formation of uremic toxins such as p-cresol sulfate and thereby their body exposure and need for renal detoxification, which, combined with direct kidney effects of SGLT2i, including less proximal tubule glucotoxicity and a broad downregulation of apical transporters (including sodium, amino acid, and urate uptake), provides a metabolic foundation for kidney and cardiovascular protection.
Keywords: diabetes mellitus, gastrointestinal microbiome, heart, kidney, metabolome, plasma, proteome, sodium-glucose transporter 2 inhibitors, uremic toxins, urine
Clinical Perspective.
What Is New?
This is the first comprehensive omics analysis in response to an SGLT2 (sodium-glucose cotransporter 2) inhibitor (SGLT2i) with the goal to define early physiologic responses that establish a kidney and cardioprotective environment with primary studies being performed in a murine model and key findings confirmed in patients.
The SGLT2i dapagliflozin inhibits gut microbiome formation of uremic toxins, at least in part through an off-target effect.
Direct effects of the SGLT2i on the early proximal tubule are facilitated by a large SGLT2 interactome, which causes downregulation or inhibition of multiple apical transporters.
What Are the Clinical Implications?
SGLT2i can reduce the deleterious effects of uremic toxins on organs, such as the heart; a proposed off-target effect suggests that this occurs independent of kidney function.
The direct effect of SGLT2i on the early proximal tubule lowers glucotoxicity; effects through interacting transporters cause a broad functional remodeling of that nephron segment.
SGLT2i reduce the early proximal tubule uptake not only of sodium and sugar but of many other metabolites, which decreases cardiovascular risk factors such as hypertension, volume retention, and hyperuricemia.
SGLT2 (sodium-glucose cotransporter 2) is primarily expressed in the brush border of the early proximal tubule of the kidney, where the transporter is responsible for quantitative reabsorption of filtered glucose.1 Inhibitors of SGLT2 (SGLT2i) are an effective therapy to treat hyperglycemia in patients with type 2 diabetes, and these compounds also protect the heart and kidneys from failing. This includes patients with chronic or acute heart failure (HF), chronic or acute impairment in kidney function, and reduced or preserved cardiac ejection fraction, and is independent of hyperglycemia (ie, type 2 diabetes) and sex.2–6 As a consequence, SGLT2i constitute a pillar of therapy for HF with reduced or preserved ejection fraction7,8 and are guideline-directed in patients with chronic kidney disease irrespective of the presence of diabetes.9 Whereas an increased risk for ketoacidosis is a safety concern for the use of SGLT2i in patients with type 1 diabetes, and therefore, approval in this patient group is limited to a few countries, similar cardiovascular benefits and mechanisms are expected.10–13
Many effects and pleiotropic mechanisms have been purported for SGLT2i on the basis of large-scale and reductionist approaches, covering enhanced ketogenesis and fasting-like metabolic responses, renal and systemic hemodynamic effects, on- and off-target effects, direct effects on the heart, vasculature, and tubular cells, and many more.5,14–16 None of the studies, however, was successful in crystalizing a key mechanism of SGLT2i that is critical for cardiovascular and renal protection. This may be explained by 3 interrelated reasons. First, glucose reabsorption is a central function of the kidney. Given the central role of glucose metabolism across all tissues, however, understanding the functional integration of manipulating kidney glucose handling will likely require considering signals of metabolic crosstalk. Second, analyses—both omics studies and reductionist approaches—have mainly been performed at later disease stages, possibly reflecting improved tissue structure and integrity rather than primary physiologic and pathophysiologic mechanisms of SGLT2i. Third, many SGLT2i studies analyzed transcripts, but not proteins and their respective metabolites.
Therefore, this study aimed to shed light on physiologic mechanisms of SGLT2i. We hypothesized that substantial metabolic organ communication occurs in response to SGLT2i, and thus elected an integrated omics approach. To this end, we performed integrated metabolomics and proteomics analyses in all major metabolic organs, biofluids, and intestinal microbiota in nondiabetic mice and in the early phase of uncomplicated hyperglycemia in Akita mice, a genetic murine model of type 1 diabetes that shows protective effects in response to chronic SGLT2i17 (Figure 1A and 1B). Short-term exposure to SGLT2i for 1 week was studied with the goal to identify primary metabolic effects and communications that may set up a protective environment rather than studying consequences of improved organ function in response to long-term treatment. A high-fat Western diet (WD) was chosen to mimic the metabolic environment of a Western lifestyle. Dapagliflozin was chosen because of its documented cardioprotective and renoprotective properties in patients with and without diabetes.4 We identified several unanticipated metabolic signals (Figure 1C). In follow-up studies, we exposed human microbial culture to SGLT2i, used SGLT2 knockout (KO) mice to probe for off-target effects of SGLT2i, and performed translational validations in human kidney, patient plasma, and human induced pluripotent stem cell (hiPSC)–derived human engineered heart tissues (EHTs; Figure 1D). The generated data provide new insights on physical SGLT2 interactors and the SGLT2i-dependent proteome, metabolome, phosphoproteome, and metaproteome, and thereby a potential roadmap to novel and early mechanistic targets of SGLT2i, which may also facilitate metabolic drug development.
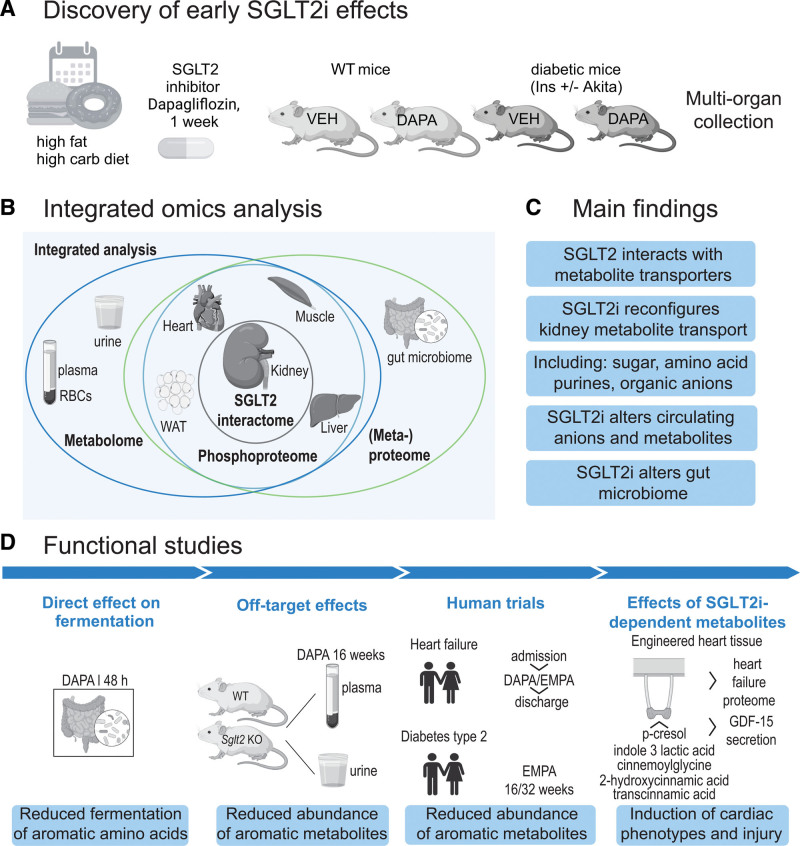
Figure 1.
Overview of study design and phenotypes. A, Overall discovery strategy. Wild-type (WT) and hyperglycemic type 1 diabetic Akita mice were put on a high-fat, high-carbohydrate Western diet for 4 weeks, followed by SGLT2i (sodium-glucose cotransporter 2 inhibitor), dapagliflozin (10 mg/kg of Western diet), or vehicle (repelleted Western diet) for 1 week. B, Organs and body fluids were snap-frozen and subjected to multi-omics analysis using mass spectrometry. SGLT2 interactome was determined from kidney. C, Main findings. D, Overview of functional follow-up studies and key findings. GDF15 indicates growth/differentiation factor 15; RBC, red blood cell; and WAT, white adipose tissue.
METHODS
Mouse SGLT2i Treatment Studies
All animal experiments were conducted in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and approved by the VA San Diego Healthcare System Institutional Animal Care and Use Committee. Nineteen 8-week-old C57BL/6J male mice (wild type [WT]; strain 000664; The Jackson Laboratory) and 16 diabetic C56BL/6-Ins2Akita/J male mice (Akita; strain 003548; The Jackson Laboratory) were randomly allocated for housing (4 or 5 mice per cage) and acclimatized for 2 weeks in the VA San Diego Healthcare System vivarium. The animals were then fed WD (42.7% kcal carbohydrate and 42% kcal fat; TD.88137; Envigo) for 4 weeks followed by 1 week of WD with added dapagliflozin (10 mg/kg diet; WT, n=10; Akita, n=8) or a repelleted WD diet (vehicle; WT, n=9; Akita, n=8). All animals were housed in the same room with a 12:12-hour light–dark cycle and free access to water. Female mice were not enrolled because female Akita mice do not develop robust hyperglycemia.18 At the time of sample collection, identities of collected samples were concealed by a numeric code.
Urine and Blood Collection and Organ Harvest
Blood glucose and body weight were measured 1 day before WD feeding, 1 day before starting the drug treatment, and at the end of the study. Spontaneous urine was collected by gently grabbing the mouse at the same time points as blood glucose and body weight measurement. At the end of study, mice were anesthetized by injection of a ketamine (Henry Schein Animal Health) and xylazine (Bimeda-MTC Animal Health) cocktail (100 mg/kg and 10 mg/kg, respectively; 10 μL/g IP) followed by blood collection by retro-orbital plexus puncture. The remaining blood was flushed out by cardiac perfusion of cold saline, and then organs (kidney, liver, heart, skeletal muscle, and white adipose tissue) were immediately harvested and frozen in liquid nitrogen and stored at −80°C for further analyses. In addition, feces from the cecum was collected for metaproteome analysis. Blood glucose level was determined with a glucometer (Contour; Bayer). Glucose concentration in urine was determined using Infinity Glucose Hexokinase Liquid Stable Reagent (TR15421; Thermo Fisher Scientific) and normalized to urine creatinine measured by the kinetics of the alkaline picrate Jaffe reaction (Infinity; Thermo Scientific). The organs from the same animals were used for proteomics, phosphoproteomics, and metabolomics, as well as metagenomics and metaproteomics, as outlined in the following.
Organ Proteomics and Phosphoproteomics
Protein Extraction
Tissue pieces (20–40 mg) were homogenized in 300 µL extraction buffer at 4°C by bead-beating in a bullet blender (Storm) for 3 minutes with amplitude 8 (2 steel beads per sample). Proteins were extracted in a buffer containing 6 M guanidinium hydrochloride, 10 mM HEPES (pH 8.5), 5 mM TCEP, and 10 mM chloroacetamide, supplemented with protease inhibitors (Complete; Roche), phosphatase inhibitors (PhosSTOP; Roche), and benzonase (200 U/mL). After boiling (10 minutes; 95°C), protein concentrations were determined by bicinchoninic acid assay. Samples were diluted to 1 M guanidinium hydrochloride and proteins were digested with trypsin at 37°C overnight at an enzyme:protein ratio of 1:50. Digestion was stopped by adding trifluoroacetic acid (TFA) to a final concentration of 1%. Peptides were desalted by reverse phase 5×C18 disks (Empore; Dr Maisch) StageTip solid phase extraction. Peptides were dried down and reconstituted in 100 mM tetraethylammonium bromide (pH 8.5). Peptide concentrations were determined by NanoDrop.
Tandem Mass Tag Labeling
Peptides were labeled with TMTpro 16-plex (Thermo Fisher Scientific) according to the manufacturer’s instructions, allowing direct comparisons between dapagliflozin (n=8) and vehicle (n=8) per organ (kidney, liver, heart, muscle, white adipose tissue, and gut microbiome) separately for each mouse strain (WT and Akita). In total, 12 TMTpro 16-plex labeling reactions, 1 batch per organ and genotype, were performed. Vehicle and dapagliflozin were labeled with tandem mass tag (TMT) label reagents (vehicle: 126, 127N, 127C, 128N, 130C, 131N, 131C, and 132N; dapagliflozin: 128C, 129N, 129C, 130N, 132C, 133N, 133C, and 134N). TMT labels were reconstituted in anhydrous acetonitrile for 5 minutes with occasional vortexing and added to the peptides. Depending on the organ, 10 to 20 µg peptides were labeled per sample. After 1 hour, labeling reactions were quenched with 5 µL 5% hydroxylamine for 15 minutes. From the combined peptides, 40 µg were set aside for proteome analysis; the remainder (120 to 200 µg) was used for phosphopeptide enrichment.
Phosphopeptide Enrichment
TMT-labeled peptides (120–200 µg) were desalted on C18 Sep-Pak columns. Dried-down peptides were reconstituted in loading buffer (80% acetonitrile, 1 M glycolic acid, and 5% TFA). To each sample, 500 µg equilibrated MagReSyn Ti-IMAC magnetic beads (ReSyn Biosciences) were added and incubated while shaking for 20 minutes. Beads were separated on a magnetic stand for 30 seconds. Phosphopeptides were washed for 2 minutes in loading buffer, followed by 2 washes in 80% acetonitrile, 1% TFA, and a final wash with 10% acetonitrile, 0.2% TFA. Enriched phosphopeptides were eluted in 3 rounds with 80 µL 1% ammonia for 20 minutes and were acidified with TFA. After drying down, peptides were loaded and desalted on C18 Stage Tips.
Reverse-Phase High-pH Peptide Fractionation
TMT-labeled peptides for proteome and phosphoproteome analysis were dried down, desalted on C18 Stage Tips, and stepwise eluted in 25 mM ammonium formate (pH 10) with increasing concentrations of acetonitrile (10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, 50%), resulting in 8 fractions per sample. Eluted peptides were dried down and reconstituted in 0.1% formic acid for mass spectrometry (MS) analysis.
MS Measurements
Tryptic peptides in 0.1% formic acid were analyzed on an UltiMate3000 rapid separation liquid chromatography (LC) system (Thermo Fisher Scientific) coupled to an Exploris 480 mass spectrometer including high-field asymmetric waveform ion mobility spectrometry pro interface (Thermo Fisher Scientific). Peptides were separated on a 2-column setup with a trap column (Acclaim PepMap 100 C18, 3 µm particle size, 2 cm×75 µm; Thermo Fisher Scientific) at a flow rate of 5 µL/min and an analytical column (Aurora 25 cm×75 µm; IonOpticks) at a flow rate of 400 nL/min using a 150-minute gradient with mobile phase A as 0.1% formic acid in LC-MS–grade water and mobile phase B as 0.1% formic acid in LC-MS–grade acetonitrile. The peptide gradient was run for 5 minutes at 2% B, followed by a linear gradient to 30% B over 120 minutes, at 126 minutes switched to 90% B, and finished with a column equilibration step from 136 to 150 minutes at 2% B. All MS measurements were recorded in data-dependent mode in positive polarity with 3 high-field asymmetric waveform ion mobility spectrometry control voltages: for proteome, −40, −55, −70; and for phosphoproteome, −40, −60, −80. Cycle times were 1 second for each control voltage. MS1 spectra were recorded in profile mode at 120 000 resolution within a scan range of 350 to 1350 m/z, with normalized automatic gain control target of 300%, RF lens of 40%, included charge states of 2 to 6, minimal intensity of 5×104, and auto maximum injection time. MS2 spectra were recorded in centroid mode at 45 000 resolution with a normalized higher-energy collisional dissociation collision energy of 32% (35% for phosphoproteome), an isolation window of 0.7 m/z, a normalized automatic gain control target of 75%, a fixed first mass at 110 m/z, and auto maximum injection time.
Data Analysis
Raw files were analyzed with Proteome Discoverer v2.4 (Thermo Fisher Scientific). Proteome and phosphoproteome were analyzed separately, with 1 analysis per organ combining WT and Akita. In total, 12 Proteome Discoverer searches were performed: 5 for proteome (kidney, liver, heart, muscle, fat), 5 for phosphoproteome (kidney, liver, heart, muscle, fat), and 2 for metaproteome (gut microbiome WT, gut microbiome Akita). Searches were performed with Sequest HT as search engine against a canonical reference murine database downloaded from UniProt on September 10, 2021 (55 336 sequences). Metaproteome analyses were performed with the same settings as proteome analyses except for an additional study-specific metagenomics-derived database, which was generated as described in the following. Methionine oxidation (+15.995 Da), protein N-terminus acetylation (+42.011 Da), protein methionine (+79.996 Da) loss (−131.04 Da), protein methionine loss with acetylation (−89.08 Da), and STY phosphorylation (+) for phosphoproteome searches only were set as dynamic modifications. TMT pro-labeled lysine (+304.207 Da), TMT pro-labeled peptide N-terminus (+304.207 Da), and cysteine carbamylation (+57.021 Da) were set as static modifications. Searches were performed with a precursor mass tolerance of 10 ppm and fragment tolerance of 0.2 Da. Two or 3 missed cleavages were allowed for proteome or phosphoproteome searches, respectively. Peptide and protein identifications were performed with a false discovery rate cutoff of 0.01 with a minimal peptide length of 6 amino acids. Reporter ion quantification included an integration tolerance of 20 ppm using the most confident centroid as integration method. Downstream analyses were performed in R. Statistical analysis was performed with the limma R package19 using Bayes moderated t test and including multiple testing correction by Benjamini-Hochberg, kinase substrate enrichment analysis with the KSEAapp R package,20 gene ontology enrichment analysis with STRINGdb and ClusterProfiler,21 and phospho motif visualization with ggseqlogo22; upset plots were generated with UpsetR23 and ggplot224 for general visualization.
Mouse and Human SGLT2 Interactome Analysis
Snap-frozen kidney cortices of 12-week-old mice (n=4, female, biologic replicates, 8 separate coimmunoprecipitations [4 with anti-SGLT2 and 4 with isotype control]) were homogenized in a tissue grinder using modified RIPA buffer containing 1% NP40, 150 mm NaCl, 0.25% sodium deoxycholate, 50 mm Tris (pH 7.5) supplemented with protease inhibitors (Complete, Roche), phosphatase inhibitors (PhosStop; Roche), and DNaseI (200 U/mL). For the human interactome analysis, the study was conducted in accordance with the ethical principles stated by the Declaration of Helsinki, and the study was approved by the local ethics committee of the chamber of physicians in Hamburg. Three cortex pieces from 1 human donor with nephrectomy were subject to lysis, as mentioned previously. Lysates were cleared by centrifugation for 15 minutes at 4°C at 18 000×g, and then 2 µg primary antibody was added. After 30 minutes rotation at room temperature, 100 µL protein A/protein G magnetic beads (Miltenyi) were added, and lysate was kept on rotation at 4°C for 1 hour. Prechilled µ columns (Miltenyi) in prechilled magnet (Miltenyi) were equilibrated with modified RIPA before adding the lysate. After 3 washes with wash buffer 1, containing 50 mM Tris (pH 7.5), 150 mM NaCl, 5% glycerol, and 0.05% NP40, and 5 washes with wash buffer 2, containing 50 mM Tris (pH 7.5) and 150 mM NaCl, column-bound proteins were partially digested for 30 minutes at room temperature with 25 µL digestion buffer containing 2 M urea, 12.5 mM Tris, 7.5 mM ammonium bicarbonate, 1 mM dithiothreitol, and 5 ng/µL trypsin. Peptides were eluted twice with 50 µL 2 M urea, 25 mM Tris, 7.5 mM ammonium bicarbonate, and 5 mM chloroacetamide, and fully digested overnight at room temperature in the dark. After acidification with TFA, peptides were desalted by C18 StageTip solid phase extraction,25 dried down in a vacuum concentrator (Labconco Centrivac), and reconstituted in 0.1% formic acid before LC-MS measurement. The antibodies used were rabbit anti-murine SGLT2 (20802; BiCell), specificity for SGLT2 previously confirmed using Sglt2 KO mice,26 rabbit anti-human SGLT2 (142195; Cell Signaling; amino terminus targeted), and, for isotype control, rabbit immunoglobulin G (Sigma). Bioinformatics of proteins was performed as for regular proteomics data. Human protein atlas27 version 23 was used and image and gene data were available from proteinatlas.org.
Metagenomics and Metaproteomics
DNA Extraction
To generate a project-specific reference database for metaproteomics, whole genome sequencing of the gut microbiome was performed. DNA was extracted from 4 cecal feces samples of the 4 experimental groups (WT vehicle and dapagliflozin, diabetic vehicle and dapagliflozin). DNA extraction of samples was done using the FastDNA Spin kit for Soil (MP Biomedicals). Biomass was homogenized in 1000 μL sodium phosphate buffer. Then, 500 μL sample, 480 μL sodium phosphate buffer, and 120 μL MT buffer were added to a Lysing Matrix E tube. Bead beating was performed at 6 m/s for 4 cycles of 40 seconds each. Gel electrophoresis using Tapestation 2200 and Genomic DNA Screentapes (Agilent) was used to validate product size and purity of a subset of DNA extracts. DNA concentration was measured using the Qubit dsDNA HS/BR Assay kit (Thermo Fisher Scientific).
Shotgun DNA Sequencing and Bioinformatics
Sequencing libraries were prepared using the NEBNext Ultra II DNA library prep kit for Illumina (New England Biolabs) following the manufacturer’s protocol. Library concentrations were measured in triplicate using the Qubit dsDNA HS kit and library size estimated using TapeStation with D1000 HS ScreenTape. The sequencing libraries were pooled in equimolar concentrations and diluted to 4 nM. The samples were paired-end sequenced (2×150 bp) on a NovaSeq 6000 platform (Illumina). Fastp software was used to filter Raw Tags to obtain high-quality sequencing data (Clean Tags). Host sequences were removed by alignment with host genome sequences using Bowtie2. Metagenomics-derived proteome database generation is described in the following. The proteomics data were searched against this generated database as mentioned in Data Analysis. Phylogenetic analysis was performed as previously described.28 Annotation of tryptophan and phenylalanine fermenting bacteria was annotated on species level–based previous work.29,30 Log2 fold change data were then plotted in a cumulative histogram.
Metagenomics-Derived Proteome Database Generation
Metaproteomics database assembly was performed by BMKGENE (Biomarker Technologies). Raw reads were filtered using Trimmomatic with a quality cutoff of 20 and reads shorter than 30 bp were discarded. The reads considered from the host were removed using DeconSeq version 0.4.331 and bowtie2 with the National Center for Biotechnology Information. The remaining high-quality reads of all samples were assembled by multiple megahits using Megahit V1.1.2,32 which makes use of succinct de Bruijn graphs. Contigs ≥300 bp were selected as the final assembly result. Results were evaluated by Quast33 and then contigs were used for further gene prediction and annotation.
Nonredundant Gene Set Construction
On the basis of the assembly results, MetaGeneMark version 3.26 (http://exon.gatech.edu/meta_gmhmmp.cgi) using default measures was used to predict the presence of open reading frames. Predicted genes from all samples were gathered together to form a large gene set. MMseqs2 version 11-e1a1c (https://github.com/soedinglab/mmseqs2) was used to construct the nonredundant gene set by setting 95% identity and 90% coverage of the gene with the longer sequences in the clustering.
Gene Abundance Calculation
Bowtie2 version 2.3.4 was used to map clean reads of each sample to the nonredundant gene set.34 MarkDuplicates in the Picard toolkits version 2.0.11 was used to remove the polymerase chain reaction duplicates in the reads, and HTSeq v0.9.1 was then used to calculate gene counts.35 Transcripts per kilobase of exon model per million mapped reads of the gene, calculated as [(gene count/gene length)×106/sum (gene count/gene length)], were used to normalize gene abundance. The abundance of genes was compared using the R program DESeq2 package.36 The differences were considered significant when the false discovery rate was <0.05.
Metaproteomics Sample Preparation
Samples for metaproteomics were obtained from the microbial suspension in PBS. Isolated bacteria were obtained by sequential centrifugation at 4°C. As a first step, the suspension was centrifuged at 800×g for 15 minutes to remove cells and particulate matter and then at 10 000×g for 15 minutes to pellet bacteria. Samples were prepared as described previously for tissue samples for proteomics.
Untargeted Metabolomics
Metabolite Extraction
A total of 10 mg of perfused kidney tissue was homogenized in a glass vial as previously described67 in 800 µL ice-cold methanol/acetonitrile/water (2:2:1; v/v). The homogenized sample was then sonicated at 4°C at maximum strength for 10 minutes and then kept at −20ºC for 2 hours to allow for complete protein precipitation. After that, 750 µL of the homogenized solution was transferred to a 1.5 mL Eppendorf vial, and the samples were centrifuged at 4ºC for 20 minutes. Supernatants were dried down in a speedvac (Labconco Centrivap) at 8°C. Then, samples were resuspended in 200 µL of H2O/ACN (1:1; v/v) at a volume of 20 µL/mg tissue. The reconstituted solution was sonicated in the ice bath for 10 minutes, vortexed, and spun down at 13 000 rpm for 15 minutes to remove any debris. Supernatant was subjected to LC-MS analysis. For quality-control purposes, a pooled sample of all experimental samples was prepared by using 5 µL metabolite solution from each sample. This quality control sample was used to monitor instrument and LC performance throughout the runs.
For plasma samples, 20 µL of plasma were extracted with 4× volume of ice-cold acetonitrile-methanol 1:1, followed by vortexing, incubation at −20ºC in the freezer, and precipitation of proteins by centrifugation at 16 000×g for 20 minutes. For urine analysis, urinary creatinine was measured by a commercially available assay (500701; Cayman). Equal amounts of urine (diluted according to urinary creatinine) were extracted with 4× volume of ice-cold acetonitrile-methanol 1:1, followed by vortexing and incubation at −20ºC in the freezer. Proteins were precipitated by centrifugation at 16 000×g for 20 minutes. The supernatant was dried down, and the urine was resuspended in 200 µL of acetonitrile:water 1:1.
Untargeted Metabolomics Analysis and MS
Untargeted LC-MS analysis for metabolomics was performed on a quadrupole time-of-flight instrument (Impact II; Bruker) as previously described.37,38 This was coupled with an ultrahigh-performance LC instrument (Bruker Elute; Bruker) or an Agilent Infinity 1290 ultrahigh-performance LC device (Agilent). To increase metabolome coverage and minimize ion suppression, a dual fractionation strategy was performed by using both reverse phase and hydrophilic liquid interaction chromatography (HILIC) on the same sample. For reverse phase, we used an Acquity BEH C18 column (1.0×100 mm, 1.7 μm particle size; Waters). For HILIC fractionation, we used an Acquity BEH amide (1.0×100 mm, 1.7 μm particle size; Waters). Both chromatographies were run at 150 μL/min. A binary buffer system of buffer A (0.1% formic acid) and buffer B (0.1% formic acid in acetonitrile) was used. The following gradient was used for reverse phase: 99% A for 1 minute, 1% A over 9 minutes. The following gradient was used for HILIC: 1% A for 1 minute, 35% A over 13 minutes, 60% A over 3 minutes, and held at 60% A for an additional 1 minute.
Calibration was performed by using sodium formate (post-run mass calibration). Data were acquired with quadrupole time-of-flight over an m/z range of 50 to 1000 Da in positive ion mode and negative ion mode (HILIC only). Electrospray source conditions were set as follows: end plate offset, 500 V; dry gas temperature, 200°C; drying gas, 6 L/min; nebulizer, 1.6 bar; and capillary voltage, 3500 V. The injection volume was 2 μL. For molecule identification, putative molecules of interest were fragmented using different collision energies (10, 20 eV) or ramp collision energies (20–50 eV) in data-dependent mode.
Untargeted Metabolomics Analysis: Data Processing, Metabolite Identification
Data were processed using XCMS39 as previously described.38 Metabolites with a q (false discovery rate) value <0.05 were considered. Post hoc corrected t tests were performed. Features with >1.5-fold change between the SGLT2i group and control were considered significantly changed and subjected to metabolite identification. Metabolite entities were confirmed by exact mass (±10 ppm), MS2 spectra match using METLIN product ions40 and in-source fragments,41 and retention time of authentic standards. Data were analyzed and plotted as described previously, using limma R package19 and ggplot2 for data visualization.
Cytoscape
Metabolite–proteome interactions were derived from STITCH using physical interactions only42 and further visualized in Cytoscape.42 Data were imported from metabolite and protein differential expression lists and connected by STITCH. The String package (installed version 1.6.0 in Cytoscape version 3.9.2) and stringify command were used to interconnect nodes between organs. Data were visualized through annotated doughnut-shaped terms. The edges represent protein interactions by stitch algorithm (experiments, databases, medium confidence). Proteins and metabolites without connections were removed to make the figure more legible.
Targeted Metabolomics
A total of 15 µL of serum samples or creatinine-normalized urine samples were extracted with 4× volume of ice-cold acetonitrile:methanol 1:1 v/v. Samples were vortexed and incubated at −20°C for 2 hours. Then, samples were spun down using a centrifuge at 16 000×g at 4° for 20 minutes. The supernatant extracts were transferred in 96-well plates and subjected to targeted metabolomics containing a list of uremic toxins as defined by Tanaka and colleagues.43
We used a triple-quadrupole (QQQ; Agilent Triple Quadrupole 6495C) coupled to an ultrahigh-pressure LC system (1290 Infinity; Agilent Technologies). Data acquisition was done with Agilent MassHunter Workstation Data Acquisition (version 10.1). For metabolite separation, a dual strategy using both polar (BEH amide) and aromatic (Phenylhexyl) stationary phases was used. For HILIC, a BEH amide column (2.1 µm; 1.7×100 mm; Waters) was used for separation. A total of 2 μL was injected. The binary buffer system consisting of buffer A and buffer B was used. The flow rate was 0.4 mL/min. Buffer A was 99.9% H2O and 0.1% formic acid with 20 mM NH4HCO2. Buffer B was 99.9% acetonitrile and 0.1% formic acid. The gradient with A:B ratios was as follows: 0 min, 5:95; 1.5 min, 5:95; 14 min, 35:65; 17 min, 55:45; 19 min, 55:45; 19.1 min, 5:95 off. Dynamic multireaction monitoring was used. The collision energies and product ions (MS2 or quantifier and qualifier ion transitions) were optimized. Electrospray ionization source conditions were set as follows: gas temperature, 120°C; gas flow, 13 L/min; nebulizer, 23 psi; sheath gas temperature, 400°C; sheath gas flow, 12 L/min; cap voltage, 1500 V (positive mode); and nozzle voltage, 500 V (positive mode). For the phenylhexyl separation using reverse phase, we used the following binary buffer system: buffer A was 0.1% formic acid in water, and buffer B was 0.1% formic acid in acetonitrile. We used a phenylhexyl CSH column (1.7 µm, 1.0×100 mm; Waters) for separation at a flow rate of 0.15 mL/min. The gradient was as follows: 0 minutes 99% A, 2.5 minutes 99% A, 8.0 minutes 80% A, 10.5 minutes 1% A, 12.5 minutes 1% A, 14 minutes 99% A, 15 minutes 99% A. The QQQ settings were as follows: gas temperature, 200°C; gas flow, 15 L/min; nebulizer, 25 psi; sheath gas temperature, 325°C; sheath gas flow, 9 L/min; cap voltage, 3000 V (positive mode); and nozzle voltage, 500 V (positive mode). The data were recorded in dynamic multiple reaction monitoring mode with 500 ms cycling time. Transition lists and retention times of the metabolites can be found in Excel File S10.
Targeted Metabolomics of the Dahl Salt-Sensitive Hypertensive Rat Model
Urine and serum were obtained from dapagliflozin- and vehicle-treated hypertensive animals as described in a previous study by Kravtsova et al44 and analyzed as described previously.
Microbiota Fermentation
Donor Recruitment and Fecal Sample Processing
Fresh fecal samples were collected from 3 healthy donors in July 2022. Anonymous sample collection and further processing are exempt from ethics approval by the National Scientific Committee (National Videnskabsetisks Komite). The donors were between 25 and 45 years of age, had regular eating patterns and bowel movements, and did not take any food supplements containing prebiotics or probiotics or any medication affecting gut transit or digestion during the 3 months preceding the sample donation. All donors provided written consent. Each fecal sample was transferred to a sealed bag containing a CO2 gas pack (BD GasPak EZ pouch system) immediately, as described,45 and was processed within 4 hours after defecation. By using fresh samples, a freeze/thaw bias was excluded. A fecal slurry (10% m/v) was prepared using anaerobic peptone water. All work was conducted in a Baker Ruskinn anaerobic bench to keep a strict anaerobic environment.
In vitro Batch Fermentations
To investigate the effect of dapagliflozin addition on microbiota composition and metabolic activity, in vitro batch fermentations were conducted using modified and anaerobically prepared MacFarlane as base medium.46 The medium contained (in g/L) 1.0 cellobiose, 1.0 xylan, 1.0 arabinogalactan, 0.5 inulin, 1.0 soluble starch, 3.0 amicase, 5.0 Bacto Tryptone, 1.5 meat extract, 4.5 yeast extract, 4.0 mucin, 0.005 hemin, 0.4 bile salt, 3.0 KH2PO4, 9.0 NaHCO3, 3.05 MgSO4, 0.5 CaCl2·2H2O, 1.0 MnCl2·4H2O, 0.025 FeSO4·7H2O, 0.5 ZnSO2·7H2O, 4.5 NaCl, and 4.5 KCl resuspended in MilliQ water. Tween 80 (1 mL) was added together with short-chain fatty acids for a final concentration of 33 mM acetate, 1 mM isobutyrate, 1 mM isovalerate, 1 mM valerate, and 9 mM propionate. After autoclavation, dapagliflozin dissolved in ethanol was added at 100 nM final concentration. Media were inoculated in triplicate with 0.1 mL of fecal slurry, creating 1% concentration. Samples were placed in a shaking incubator at 140 rpm at 37°C. Samples were collected after 0, 24, and 48 hours. Effects were compared with baseline for both dapagliflozin and control (vehicle) samples.
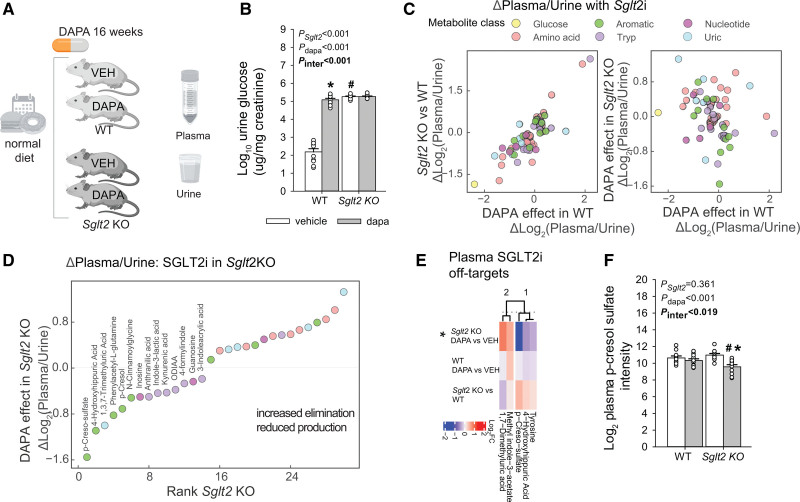
Sglt2 KO Mouse Study
The generation of Sglt2 KO mice on a C57BL6 background has been published.47 At 5 weeks of age, male Sglt2 WT and littermate KO mice, maintained on standard rodent diet (Envigo 7001), were switched to the same diet containing dapagliflozin (10 mg/kg diet) or repelleted diet (4 groups of mice). Blood glucose was measured in awake mice by tail snip at 9, 13, 17, and 21 weeks of age and mean values calculated per mouse. At 21 weeks of age, a spontaneous urine sample was collected from awake mice to determine urine glucose:creatinine ratios and for the performance of urine metabolomics. Blood was subsequently collected by retroorbital plexus puncture under isoflurane anesthesia for the performance of plasma metabolomics. Over the 48 hours before this final sample collection, food intake was determined with single housing of the mice in regular cages. All samples were collected in nonfasted mice between 9:00 and 11:00 am.
Human Serum Samples
CYCLE Cohort
For this study, data and biosamples of patients enrolled in the CYCLE study (Characterization of Phenotypes in Acute Heart Failure Patients) were used. All participants provided written informed consent and the study was performed in accordance with the Declaration of Helsinki and was approved by the local ethics committee (PV5983). CYCLE is an ongoing, prospective, observational study enrolling participants ≥18 years of age with acute decompensated HF, defined as at least 3 out of 6 typical signs and symptoms of HF (ie, orthopnea, paroxysmal nocturnal dyspnea, fatigue, pulmonary rales, peripheral edema, gut congestion); onset or worsening of these symptoms within the past 7 days; and NT-proBNP (N-terminal pro-B-type natriuretic peptide) ≥300 pg/mL. Patients with other medical conditions that could result in the previously described signs and symptoms as well as patients with malignancies or a life expectancy <12 months were excluded. An extensive clinical assessment and blood samples were obtained at baseline, after 3 and 5 days, and at discharge. The blood samples were collected using a standardized procedure and were stored at −80°C until measurement. Standardized echocardiography was performed at baseline and at discharge, and all participants were followed up by the local registry office to obtain mortality data.
GDF15 Measurement
GDF15 (growth/differentiation factor 15) was measured in patient serum in the CYCLE study using a commercially available ELISA following the manufacturer’s instructions (BMS2258; Invitrogen).
Randomized Controlled Trial in Diabetic Patients
The trial was approved by The Central Denmark Region Committees on Health Research Ethics (VEK approval 1-10-72-52-19) and The Danish Medicine Agency (LMS approval 2019032214).48 The trial was monitored by the local Good Clinical Practice Unit at Aarhus and Aalborg University Hospital and complied with the Declaration of Helsinki. All participants gave written informed consent. Biosamples from 60 patients receiving empagliflozin or matching placebo who were enrolled in SEMPA (Effect of Empagliflozin and Semaglutide on Cardio-Renal Target Organ Damage in Patients With Type 2 Diabetes: A Randomized Trial) were used (URL: https://www.clinicaltrialsregister.eu; Unique identifier: 2019-000781-38). The trial was a 32-week, partly placebo-controlled, partly open-label prospective study, originally designed to assess the effects of semaglutide, empagliflozin, or their combination on kidney oxygenation and arterial stiffness.48 The trial included 120 participants with type 2 diabetes (HbA1c >48 mmol/mol), age ≥50 years, with a high risk of cardiovascular disease (eg, a history of myocardial infarction or stroke or estimated glomerular filtration rate <60 mL·min·1.73 m2). Key exclusion criteria included estimated glomerular filtration rate <45 mL min 1.73 m2 and liver disease or a cardiovascular or cerebrovascular event within 90 days before randomization. Participants were randomly assigned to 1.0 mg subcutaneous semaglutide once weekly (Ozempic; Novo Nordisk A/S; 30 participants), 10 mg empagliflozin daily (Jardiance; Boehringer Ingelheim International; 30 participants), combination treatment (30 participants), or tablet placebo (30 participants) for 32 weeks. Outcomes were assessed at baseline, week 16, and week 32. Blood samples were collected using a standardized procedure and stored at −80°C until measurement.
hiPSC-Derived Engineered Heart Tissues
Human Induced Pluripotent Stem Cells
The in-house control hiPSC line (UKEi001-A) was registered at the European Human Stem Cell Registry (hPSCreg) and had previously been produced from dermal fibroblasts from human punch skin biopsies as described in previously published protocols.49 The use of this cell line was approved by the Ethical Committee of the University Medical Center Hamburg-Eppendorf (approval PV4798, 28.10.2014) with written informed consent.
Thawing, Passaging, and Culture of hiPSC
Cryopreserved hiPSC were thawed at 37°C for 2 to 3 minutes and then drawn up into a pipette containing 5 mL FTDA (DMEM, 2 mM L-glutamine, 5 mg/L transferrin/selenium, 0.1% human serum albumin, 1× lipid mix, 5 mg/L insulin, dorsomorphin, 30 ng/mL fibroblast growth factor [FGF; 100-18B; Peprotech], 2.5 ng/mL activin A, 0.5 ng/mL transforming growth factor–β), supplied by the ROCK-Inhibitor Y-27632 (1:1000; orb154626; Biorbyt). After centrifugation for 3 minutes at 800 rpm, the pellet was resuspended in FTDA containing 0.3 µL/mL FGF and 10 µM Y-27632. The cells were then cultured on Geltrex-coated (1:100; 14133-02; Gibco A) 6-well plates and incubated under hypoxic conditions at 37°C, 5% CO2, and 5% O2. The cells were fed daily with prewarmed FTDA, supplemented with 0.3 µL/mL FGF, and split twice a week when the cell layer was >90% confluent. For that purpose, before detachment with Accutase solution (supplemented with 10 µM Y; 07922; StemCell Technologies) for 5 to 10 minutes at 37°C, the wells were washed once with PBS. The cells were detached from the wells, transferred to falcons using a 1000-µL pipette, and washed twice with prewarmed FTDA supplemented with Y and FGF, followed by centrifugation at 800 rpm for 3 minutes. After removal of the supernatant, cells were resuspended in the same medium, counted, and densities of 50 000 to 70 000 cells/cm² (ie, 500 000 to 700 000 cells per well or 4.5 to 6.5×106 cells per T75 flask) were plated. Before enrolling cells into experiments, hiPSC were passaged 3 times.
Differentiation of hiPSC to hiPSC-Induced Cardiac Myocytes
The differentiation of hiPSC to cardiac myocytes was performed as previously described.50 In brief, confluent hiPSC, expanded in T75 flasks, were preincubated in FTDA supplemented with 10 µM Y-27632 for ≥1 hour at 37°C. The flasks were washed once with PBS and then dissociated with 0.5 mM EDTA in PBS into small cell clusters. The flasks were washed with Roswell Park Memorial Institute (RPMI; 21875-034; Gibco) and added to the cell suspension. After centrifugation at 800 rpm for 10 minutes, the pellet was resuspended in stage 0 medium (DMEM/F12, 2 mM L-glutamine, 5 mg/L transferrin, 5 µg/L selenium, 0.1% human serum albumin, 1× lipid mix, 5 mg/L insulin, 50 nM dorsomorphin, 2.5 ng/mL activin A, 0.5 ng/mL transforming growth factor–β1, 30 ng/mL FGF, 4 mg/mL polyvinyl alcohol, 10 µM Y-27632) and triturated 3 to 5 times with a serologic 10-mL pipette. Cells were counted in a Neubauer counting chamber and 30×106 cells per 100 mL stage 0 medium were transferred to prewarmed spinner flasks. The spinner flasks were then placed on a magnetic stirrer in the hypoxia incubator (37°C, 5% O2, 5% CO2, 95% relative humidity) under constant stirring at 40 rpm to induce formation of embryonic bodies (EBs).
After EB generation overnight in spinner flasks, mesoderm was induced in T175 flasks (83.3912.502; Sarstedt) for 3 days. For that purpose, Pluronic-coated (1% Pluronic in PBS [P2443; Sigma]) T175 flasks were washed twice with PBS and preincubated with Stage 1 medium (RPMI 1640, 4 mg/mL polyvinyl alcohol [P8136; Sigma], 5 mg/L transferrin, 5 µg/L selenium, 10 µM Y-27632, 1× lipid mix [L5146; Sigma], 250 µM polyacrylic acid, 2.5 ng/mL activin A [338-AC; R&D Systems], 4 mM BMP4 in 0.1% BSA [314-BP; R&D Systems], FGF 100 µg/mL in PBS/0.1% human serum albumin [05-720-1B; Biological Industries]). EB suspension from spinner flask was then transferred to T175 flask. After sedimentation of EBs, the supernatant was removed and all EBs were pooled in 1 flask; all other flasks were washed with RPMI 1640 (supplemented with 10 mM HEPES [91.05.4; Roth] and 4 mg/mL polyvinyl alcohol), followed by an additional washing step. Afterward, EB volume was calculated and 160 to 250 µL EB volume was distributed in 46 mL stage 1 medium. EBs were incubated for 3 days in the hypoxia incubator (37°C, 5% O2, 5% CO2, 95% relative humidity) with daily half-medium change with stage 1 medium.
After mesoderm induction in stage 1, differentiation of mesoderm progenitors to cardiac myocytes was performed during stage 2. Medium from T175 flasks was removed and flasks were washed twice with washing medium. After estimation of EB volume, 180 to 250 µL EB volume per flask was incubated in 43 mL FDM (RPMI 1640, 0.5% [v/v] Pen/Strep, 5 mg/L transferrin, 5 µg/L selenium, 1× lipid mix, 0.05% human serum albumin, 250 nM polyacrylic acid, 1 µM XAV, 10 µM Y-27632) with daily half-medium changes. After 3 days’ culture, medium was replaced by stage 2 medium supplemented with insulin (I9278 [Sigma]; RPMI 1640, 0.5 % [v/v] Pen/Strep, 1 µM XAV, 10 µM Y-27632, 10 mM HEPES, 500 µM 1-thioglycerol [MTG; M6146; Sigma], 2% B27 stock [0.156 mg insulin in 1 mL B27; I9278-5 mL; Sigma-Aldrich, Merck]) and incubated for another 4 days in same conditions and daily half-medium exchange. Next, medium was replaced by RDM (RPMI 1640, 0.5% [v/v] Pen/Strep, 10 µM Y-27632, 10 mM HEPES, 500 µM 1-thioglycerol [MTG], 2% B27 stock) and EBs were cultured for another 12 days as previously mentioned.
Dissociation of hiPSC-Derived Cardiac Myocytes
To receive single cardiac myocytes, 3-dimensional EBs were dissociated. Medium was removed from T175 flasks, followed by 2 intermittent washing steps for 5 minutes with HBSS without Ca2+ (14175-053; Gibco) and incubation with 12 mL collagenase solution/200 µL EB volume (HBSS without Ca2+, 1 mg/mL collagenase II [200 units/mg LS004176; Worthington], 10 mM HEPES, 10 µM Y-27632, 30 µM BTS [N-benzyl-p-toluene sulfonamide; B3082-25G; TCI]) for up to 3.5 hours at 37°C, 90% relative humidity, 5% CO2, 20% O2 until EBs started to disaggregate. The cells were then triturated by pipetting and incubated by the same amount of blocking buffer (RPMI 1640, 1% Pen/Strep, 6 µL DNase/mL) before centrifugation for 10 minutes at 100×g. The pellet was resuspended and gently triturated in prewarmed basic medium (RPMI 1640, 1% Pen/Strep). The dissociated hiPSC-derived cardiac myocytes were either directly used to generate EHT or frozen in 10% DMSO in fetal calf serum.
Generation of EHTs
Fibrin-based human EHTs were generated in agarose casting molds with transparent silicone posts as previously described.50,51 In brief, casting molds were generated in a 24-well plate using 2% agarose in PBS and spacers. Transparent silicone posts were placed into the casting molds and the master mix (1 million hiPSC cardiomyocytes, 5.6 µL 2× DMEM, 0.1% 10 mM Y-27632); before use, 2.5 µL/EHT fibrinogen (200 mg/mL in NaCl 0.9%; F4753 [Sigma]) and NKM added up to 97 µL/EHT (DMEM [F0415; Biochrom], 1% Pen/Strep, 10% horse serum [26050; Gibco], 1% of 200 mM glutamine [Gibco]) was prepared. For each EHT, 97 µL master mix was first pipetted into 3 µL of 3 U/mL thrombin (BP11101104; Biopur) and then into the agarose molds around the posts. The plate was then incubated at 37°C, 7% CO2, 40% O2, and 98% relative humidity for 1.5 to 2 hours until the fibrin was polymerized and EHTs were transferred into a new 24-well plate containing prewarmed culture medium (DMEM [Sigma], 1% Pen/Strep, 10% horse serum, 10 μg/mL insulin, 33 μg/mL aprotinin [A1153; Sigma]). The culture medium was changed 3 times each week and supplemented with 200 µM tranexamic acid (857653-50G; Sigma) for matrix stabilization. After 7 to 10 days, a beating pattern could be observed macroscopically, allowing video-optical contraction analysis.
Incubation of EHTs With Metabolites
On day 13 after the generation of the EHTs, the culture medium was deprived to 4% horse serum, followed by further deprivation of serum to serum-free medium (DMEM, 1% Pen/Strep, 10 µg/mL insulin, 33 µg/mL aprotinin, 2 mM L-glutamine, 5 ng/mL triiodothyronine T3 [IRMM469; Sigma], 50 ng/mL hydrocortisone [H0888; Sigma]) on day 14 of EHT culture. On day 15, 5 to 6 EHTs per group of hiPSC-derived cardiac myocytes were treated daily with 300 µM or 3 mM of either p-cresol (PCL) or indole-3-lactic acid (ILA) in DMSO, or DMSO as appropriate control for 5 days. As force in the 3-mM PCL group declined within hours of first treatment, after 72 hours, PCL was replaced with DMSO to investigate a potential functional rescue.
For proteomics analyses, 4 to 6 EHTs per group were treated continuously for 5 days starting from day 15 with 300 µM of PCL, ILA, cinnemoylglycine, 2-hydroxycinnamic acid, transcinnamic acid, or DMSO as vehicle control.
Video-optical analysis of EHTs for different parameters such as contractile force, frequency, and relaxation time to 80% were investigated as previously described.51 For functional analyses of the treated EHTs, short-term electrical pacing was applied as previously described.52 Biphasic pulses (4 ms per direction) at 2 V (electrical field strength 2 V/cm) and a frequency of 0.5 to 4 Hz with increasing increments of 0.5 Hz were applied. For analysis and representation of the data, Excel and Graph Pad Prism 6.0 were used.
Proteomic Analysis of EHT Tissue
Samples were prepared as described previously in the Proteomics section. C18 stage tip desalted peptides (unlabeled, unfractionated) dissolved in 0.1% formic acid were measured by LC-MS as described previously with some modifications. The peptide gradient used was 5 minutes at 2% B, reaching 8% B at 10 minutes, followed by a linear gradient to 25% B over 80 minutes, followed by a linear gradient to 35% B over 10 minutes, at 101 minutes switched to 90% B, and finished with a column equilibration step from 110 to 120 minutes at 2% B. All MS measurements for EHT samples were recorded in data-independent acquisition mode in positive polarity with high-field asymmetric waveform ion mobility spectrometry control voltages −45 and −65. MS1 spectra were recorded in profile mode at 120 000 resolution within a scan range of 380 to 1500 m/z, with normalized automatic gain control target of 300%, RF lens of 40%, and auto maximum injection time. MS2 spectra were recorded at 30 000 for 400 to 1000 Da with nonoverlapping isolation windows of 15 Da and normalized HCD collision energy of 28%. MS RAW files were analyzed by Spectronaut version 17 (Biognosys) using directDIA+ with the predefined factory settings. Trypsin/P was selected as digestion enzyme allowing for 2 missed cleavages, for peptides in the range of 7 to 57 amino acids. Cysteine carbamylation was set as fixed modification, methionine oxidation and protein N-terminal acetylation as variable modifications. For Pulsar searches, identification for PSM, peptide, and protein group were set to false discovery rate 0.01. Data were searched against a UniProt Homo sapiens reviewed database (downloaded March 4, 2023; 20 422 entries).
Western Blotting
Membrane fraction of whole kidney was prepared as previously described47 and protein concentration was determined using a Protein Assay Kit (Bio-Rad Laboratories). A total of 25 µg of protein per each lane was electrophoresed using a 7.5% Criterion TGX precast gel system (Bio-Rad Laboratories) and transferred to polyvinylidene fluoride membrane. After blocking with 5% nonfat dried milk in TBS-T buffer, the membrane was incubated with primary antibodies for overnight at 4oC and then with horseradish peroxidase–conjugated secondary antibodies for 1 hour at room temperature. The membrane incubated with enhanced chemiluminescence reagent was imaged on a Chemidoc Imaging System (Bio-Rad Laboratories), and images were analyzed with Image Lab Software, version 6.0.1 (Bio-Rad Laboratories). Primary antibodies used in the study were anti-OAT1 (organic anion transporter 1) antibody (rabbit polyclonal, 1:1000 [26574-1-AP; Proteintech]) and anti–β-actin antibody (mouse monoclonal, 1:1000 [3700; Cell Signaling Technology]). Secondary antibodies were anti-mouse immunoglobulin G horseradish peroxidase–linked antibody (1:3000 [7076; Cell Signaling Technology]) and anti-rabbit immunoglobulin G horseradish peroxidase–linked antibody (1:3000 [7074; Cell Signaling Technology]). Statistical analysis was performed by using 2-sided t test.
Data Integration
Interactive Data Exploration
The presented data were made explorable as an interactive web shiny application. To this end, result tables were read into R to create a summarized experiment by the R package SummarizedExperiment (version 1.26.1). The R packages iSEE53 (version 2.8.0) and iSEEu54 (version 1.8.0) were used to publish shiny applications for the phosphoproteome and proteome, respectively. Gene Ontology term enrichment results were registered as feature sets that map to included proteins. In the phosphoproteome application, proteins were registered as feature sets that combine all phosphosites of the protein.
Data Sharing
All proteomics data are deposited in the PRIDE ProteomeXchange data repository,55,56 accessible through http://www.ebi.ac.uk/pride using the following credentials: project name, Metabolic reconfiguration through SGLT2 inhibition improves metabolic waste excretion; project accession, PXD036817; project DOI, 10.6019/PXD036817; username, reviewer_pxd036817@ebi.ac.uk; and password, Wal8896C. The metaproteomics database is deposited under https://figshare.com/s/288394c518595ca0d1b2, reserved under doi 10.6084/m9.figshare.24160935.
RESULTS
Overview on the Strategy of Matched Integrative Interorgan-omics Discovery and Translation
One week after initiation of dapagliflozin or vehicle, we performed deep multi-omics profiling combining proteomics, phosphoproteomics, metabolomics, and metaproteomics approaches from the same animals across major metabolic organs (Figure 1A and 1B). Plasma glucose levels were decreased by dapagliflozin in Akita mice, but not in nondiabetic WT mice. Glucosuria was induced by dapagliflozin in WT mice, but no obvious further increase was observed for Akita mice (Figure S1A). Less glucose filtration in response to SGLT2i prevents net increase in glucosuria in Akita.17 Body weight did not change in any group by the short treatment (Figure S1A). Proteomics analysis quantified a total of 9501 proteins (8421 genes) and 10 744 phosphorylation sites in the kidney, liver, heart, muscle, and white adipose tissue (Excel Files S1 and S5). Of these, most of the significant SGLT2i-induced changes were observed in the kidney (Figure S1B). All the proteome data from all tissues in all organs are also explorable in a shiny server–based application at https://kidneyapp.shinyapps.io/SGLT2i_proteome.
SGLT2 Inhibition Reconfigures the Kidney Proteome
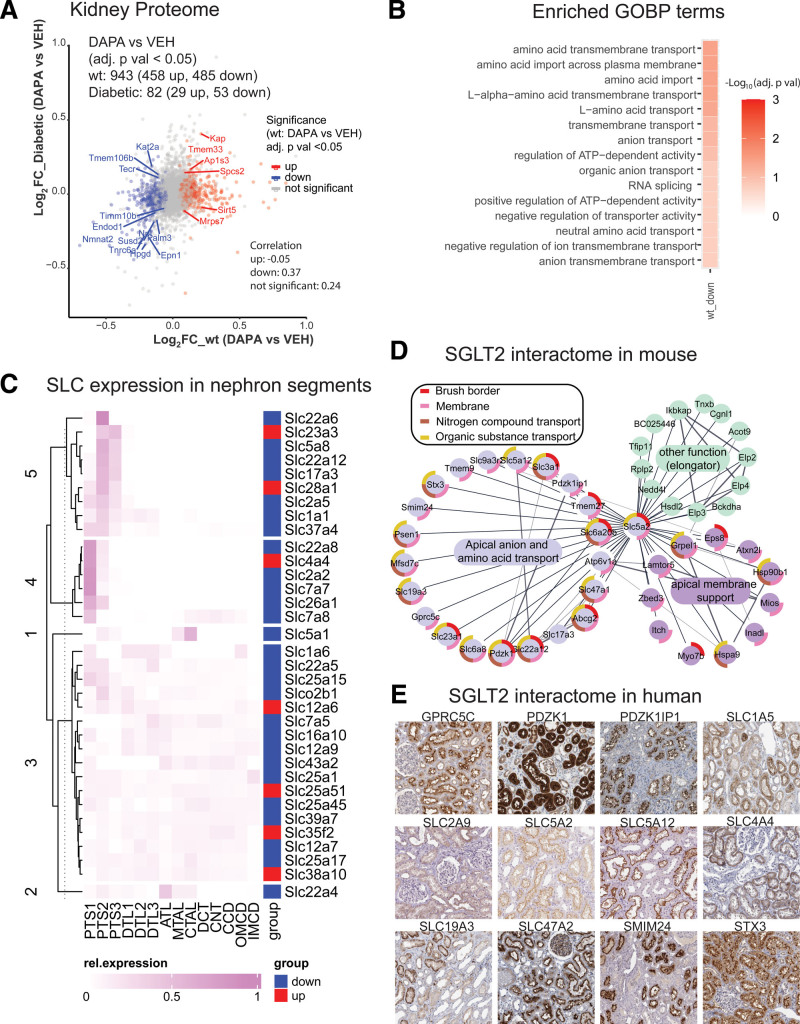
We quantified 6676 proteins in kidney cortex, with 6107 proteins for WT and 6207 proteins for Akita mice (Excel File S1). The analysis showed that the effect of SGLT2i on the kidney proteome was stronger in WT than in diabetic mice: In WT mice, 455 proteins were increased and 485 proteins were decreased by SGLT2i (Figure 2A). SGLT2i treatment changed fewer proteins in diabetic mice (29 increased and 53 decreased). Both groups showed significant alterations in kidney mitochondrial protein abundance, but only WT mice showed a robust upregulation in inner mitochondrial membrane proteins (Figure S2A). Moreover, only WT mice reacted to SGLT2i with downregulation of kidney proteins localized to the apical or basolateral membrane (Figure 2B) or proteins involved in mRNA splicing. Following up on the observed stronger changes in WT mice, Gene Ontology term enrichment (Excel File S2 and Figure S2A and S2B) indicated consistent alterations in transmembrane transport across a variety of functionally interconnected proteins (Figure S2B), with most transporters being downregulated by SGLT2i in WT mice. Clustering of differentially expressed proteins in WT mice on the basis of nephron segment–resolved RNA-sequencing data57 revealed that a large fraction of segment-specific protein changes by SGLT2i were localized to proximal tubule segments S1, S2, and S3 (clusters 1, 7, and 9; Excel File S3), consistent with a primary site of action in S1/2 proximal tubule.14 Among the 34 changed membrane transporters of the solute carrier group (SLC) in WT kidney, 27 were downregulated. Half of the SLCs displayed segment-specific expression for proximal tubule S1 and S2/3 segments (Figure 2C). The associated functions of the downregulated SLCs as well as their segment-specific expression according to the clustering in a protein metabolite interaction network are shown in Figure S2C.
Figure 2.
Integrated proteome/interactome analysis indicates broad remodeling of kidney metabolite transport by SGLT2i. A, Volcano plot quantification of proteomics analysis of kidney cortex. Overlapping significant proteins (false discovery rate <0.05) between wild-type (WT) and Akita mice upon SGLT2i (sodium-glucose cotransporter 2 inhibitor) treatment are labeled with their gene name. B, Enrichment of Gene Ontology Biological Process (GOBP) terms in WT kidney cortex (kidney cortex proteome was used as background). C, Nephron localization of altered transmembrane transporters of the solute carrier group (SLC). D, Mass spectrometry–based interactome analysis of mouse SGLT2i. SGLT2 interacts with membrane transporters that mediate the reabsorption or secretion of sodium, amino acids, urate, protons, anions, and multiple other compounds. E, Human tissue atlas of selected putative human SGLT2 interactors (source: The Human Proteome Atlas; https://www.proteinatlas.org).
SGLT2 Interacts With Multiple Apical Proximal Tubule Metabolite Transporters
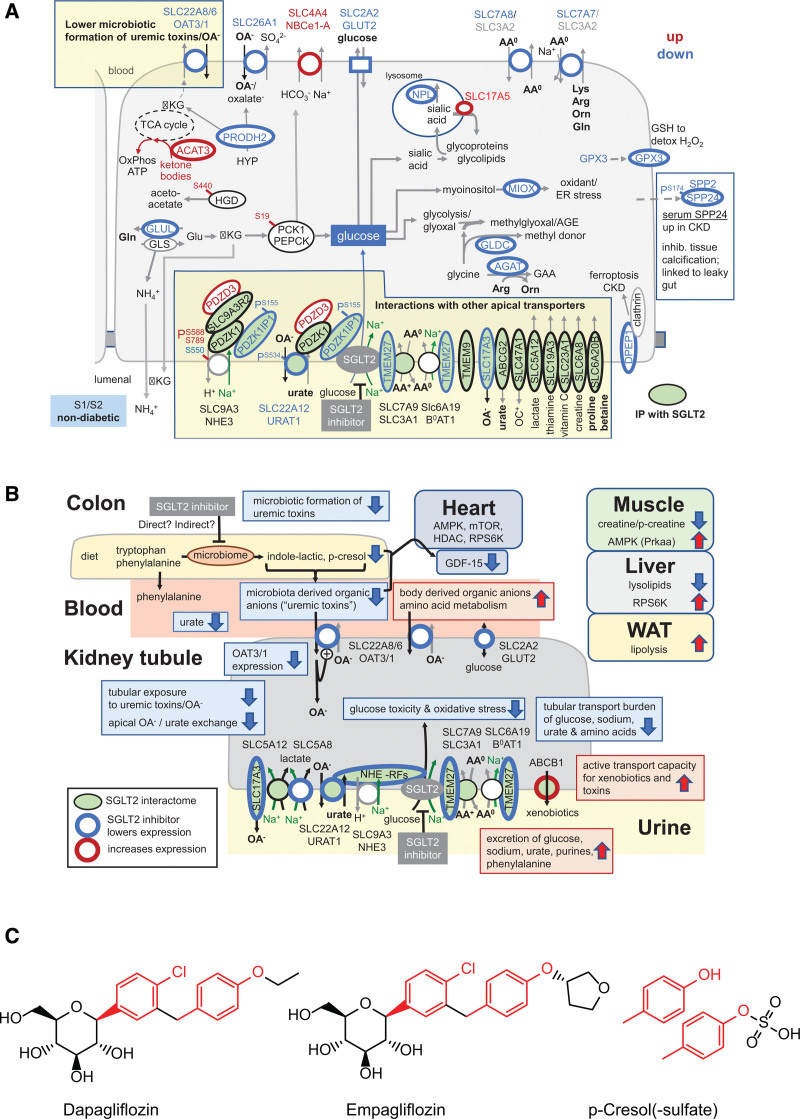
To determine whether the changed proteins interact directly with SGLT2, we performed interactome analysis of the SGLT2 transporter from native mouse kidneys. Using affinity purification with MS from native mouse kidney (Excel File S4), we found that SGLT2 interacted with sodium- and hydrogen transporter–regulating proteins (including Pdzk1ip1 [MAP17], Pdzk1 [NHERF3], and Slc9a3r2 [NHERF2]), as previously predicted by bioinformatics,58 but also with several metabolite transporters. Transporter substrates included creatine (Slc6a8), vitamin C (Slc23a1), urate (Slc22a12, Abcg2, Slc17a3), lactate and ketone bodies (Slc5a12), thiamine (Slc19a3), organic anion and drug detoxification (Slc17a3, Slc47a1 [MATE1]), and amino acids (Slc3a1, Slc6a20b, Tmem27; Figure 2D), many of which are linked to sodium cotransport. Four interacting proteins (urate transporter Slc22a12, amino acid transport modulator TMEM27, NHE3-interacting MAP17, and organic anion secretory transporter Slc17a3) were also significantly decreased by SGLT2i in WT mice in the proteome analysis. Other SGLT2-interacting proteins included TMEM9 (which facilitates assembly of lysosomal proton-transporting V-type ATPase and enhances lysosomal acidification and trafficking59), Atp6v1a (which has a role in acidification of eukaryotic intracellular organelles), Smim24, Stx3 (which has a role in cell polarity), Psen1, Mfsd7c (which switches mitochondrial ATP synthesis to thermogenesis in response to heme60), Lamtor5 (which has a role in mTOR signaling), and Gprc5c (which has a role in cellular effects of retinoic acid). To further confirm the relevance in human, we immunopurified SGLT2 from human kidney using a different antibody than in mice, followed by interactome analysis (Excel File S4). Consistent with the mouse results, the human SGLT2 interactome was associated with organic substance transport (24 proteins) and revealed copurification of SGLT2 with proteins related to NHE-RF, as well as urate and amino acid transporters. Whereas several proteins were identical in human and mouse SGLT2 interactome (Excel File S4), some were functionally similar (Slc47a1 in mouse and SLC47A2 in human are both drug excretion transporters; Slc22a12 in mouse and SLC2A9 in human are both urate transporters). Most of these protein transporters are immunolocalized to proximal tubule cells in the human kidney in the Human Protein Atlas (Figure 2E).
Thus, SGLT2i-dependent proteome and SGLT2 kidney interactome suggest that SGLT2i induces reconfiguration of early proximal tubule transport, consistent with the idea that SGLT2i interferes with the early proximal tubule transport of many substrates beyond glucose.1
SGLT2 Inhibition Changes the Global Phosphoproteome Landscape Including Metabolically Regulated Proteins
Phosphoproteomic investigation quantified a total of 10 744 phosphorylation sites across organs (Excel File S5). The entire phosphoproteome dataset is explorable (https://kidneyapp.shinyapps.io/SGLT2i_phosphoproteome). Altered protein phosphorylation by SGLT2i was primarily found in the WT kidney, similar to the findings of the proteome analysis. This included 343 phosphorylation sites, 165 with increased phosphorylation, and 178 with decreased phosphorylation (Figure S3A and Excel File S5). Functional enrichment analysis showed effects on GTPases, cytoskeleton, and RNA splicing in the kidney (Figure S3B and S3C and Excel File S2). Closer analysis of amino acid sequences surrounding phosphorylation sites revealed distinct alterations in WT mice (Figure S3D) but also in diabetic mice.
Comparing all analyzed organs, SGLT2i increased basophilic phosphorylation sites in the liver and white adipose tissue, whereas the heart showed a unique pattern of a decreased basophilic phosphorylation pattern (Figure S3D). In the nondiabetic and diabetic kidney, kinase-substrate enrichment analysis indicated that SGLT2i downregulated PRKAA1 (protein kinase AMP-activated catalytic subunit alpha 1; Figure S3E), a nutrient-controlled kinase primarily expressed in the distal nephron.57 Moreover, SGLT2i reduced kidney substrates of mTOR in nondiabetic mice and SGK1 in diabetic mice.61 Downregulation of mTORC1 mRNA has been found in response to SGLT2i along the tubular system in youth-onset type 2 diabetes.62 mTOR downregulation may reflect reduced glucotoxicity in S1/2 segments but energy-consuming upregulation of compensatory transport in downstream tubule segments. In contrast to the kidney, kinase substrate enrichment analysis suggested an increased RPS6 kinase activity by SGLT2i in liver (and to a lesser extent in heart), and a decreased activation of cGMP-dependent protein kinase (PRKG1) in the heart (Figure S3E). The most substantial proteome and phosphoproteome changes in liver and white adipose tissue are shown in Figure S4A and S4B. Closer examination of the SGLT2i-targeted phosphorylation sets in the heart revealed that a large fraction of altered regulatory phosphorylation sites were metabolically regulated sites and proteins, such as AMPK (5′-adenosine monophosphate–activated protein kinase), HDAC (histone deacetylase), Raptor, and other metabolic sensors in heart (Figure S3F). Many of the changes in the heart occurred in diabetic but also in nondiabetic mice, indicating effects in part independent of blood glucose levels. Overall, metabolic signaling dominated the phosphorylation landscape in the kidney and other organs, without singling out a single kinase.
SGLT2i-Dependent Metabolome Unravels Metabolite Signals Primarily in Plasma and Urine But Also in Other Organs
To determine whether the altered signaling profiles could, in part, be explained by metabolic signaling other than glucose, we performed untargeted metabolomics analysis in selected organs and biofluids. In total, 186 unique metabolites changed (322 summed for all organs), with 136 for WT mice and 96 for diabetic mice (corresponding to 201 for WT and 144 for Akita mice when summed for all organs). We found that SGLT2i chiefly acted on the abundance of metabolites in urine and plasma (Figure 3A), and the diversity of molecules was more extensive in nondiabetic versus diabetic mice. Consistent with initial phenotyping of the mice (Figure S1A), SGLT2i enhanced sugars (glucose) in the urine in WT mice without changing plasma levels, whereas SGLT2i in hyperglycemic Akita mice lowered plasma sugars (glucose) without a robust increase in glucosuria. SGLT2 administration broadly perturbed amino acid metabolism. SGLT2i decreased lipids in plasma in WT mice, but in liver, heart, and red blood cells in Akita mice. Reduced triacylglyceride and phosphoglyceride species were found in white adipose tissue in WT, and a decrease in creatine:creatine-phosphate ratios was observed in skeletal muscles particularly of diabetic animals (Figure S4C and S4D). An extensive catalogue of metabolic changes can be found in Excel File S6 and Figure S5.
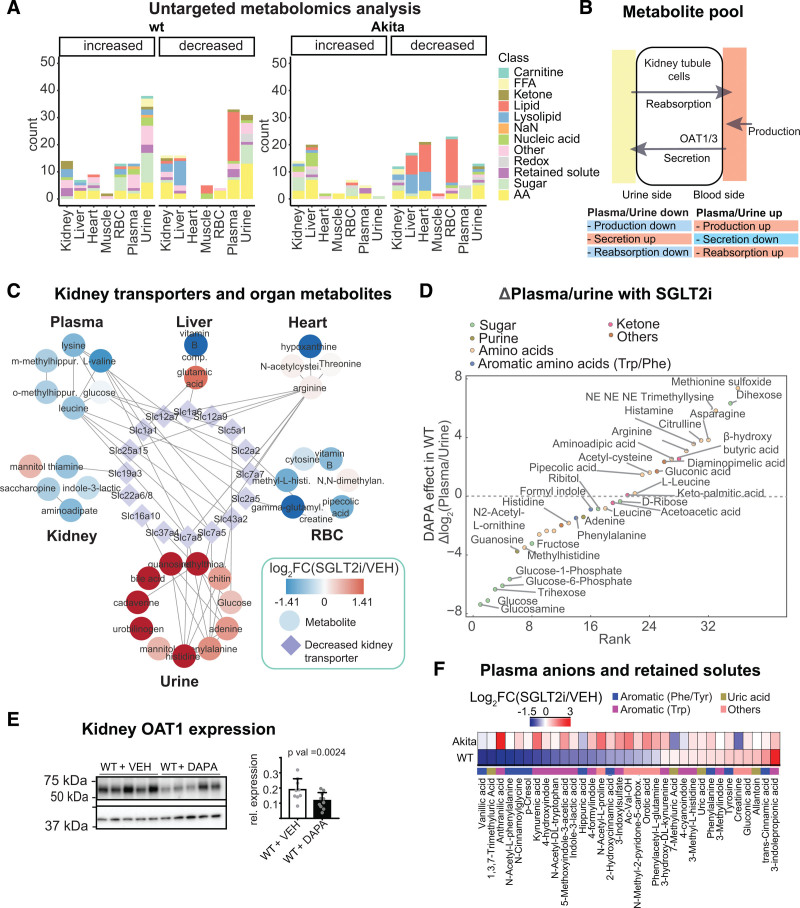
Figure 3.
Untargeted metabolome analysis reveals SGLT2i-induced alterations in amino acid and organic anion metabolism. A, Overview of untargeted metabolomics results in wild-type (WT) and diabetic Akita mice. Significant metabolites from >30 000 quantified features with breakdown of altered metabolites by organ and genotype. B, The metabolite pool is sensitive to changes in production as well as kidney secretion and reabsorption. C, Illustration of interorgan communication by linking solute carrier group (SLCs) downregulated in the kidney to enhanced urinary metabolites and depleted metabolites in other organs. D, SGLT2 (sodium-glucose cotransporter 2)–dependent changes in plasma:urine ratios indicate increased urinary excretion of glucose and glucose metabolites, and less secretion of substrates of the kidney organic anion transporters (see text for details). Urine metabolites were normalized to creatinine. E, Immunoblot analysis of OAT1 (organic anion transporter 1) in SGLT2i-treated WT mice shows reduction of OAT1 (Slc22a6; normalized to b-actin; 2-sided t test), consistent with proteomics results. F, Targeted analysis of microbiota-derived organic anions (uremic toxins) in the plasma of WT and diabetic Akita mice. RBC indicates red blood cell.
Downregulation in Kidney Transporters Can Contribute to SGLT2i Effects on Metabolites in Plasma, Urine, and Other Organs
SGLT2i changed the kidney proteome and phosphoproteome (Figure 2A through 2C and Figure S3A through S3C), and SGLT2 orchestrates other metabolite transporters (Figure 2D and 2E). Its inhibition induced prominent alterations in plasma and urine metabolites (Figure 3A). Thus, we hypothesized that renal proteomic reconfiguration could be partly responsible for the observed metabolite alterations. Several processes can alter the circulating metabolite composition of body fluids (Figure 3B). Changes in urinary excretion could occur if the body production or tubular transport is altered. In WT mice, 27 SLCs were decreased by SGLT2i in the kidney. Combining these with urinary metabolite profiles (each normalized to urinary creatinine) yielded a small number of potential links (Figure 3C). With a focus on metabolites increased by SGLT2i in urine and including the metabolite profiles of other organs in the analysis, SGLT2i effects could contribute to metabolite alterations in other organs (Figure 3C).
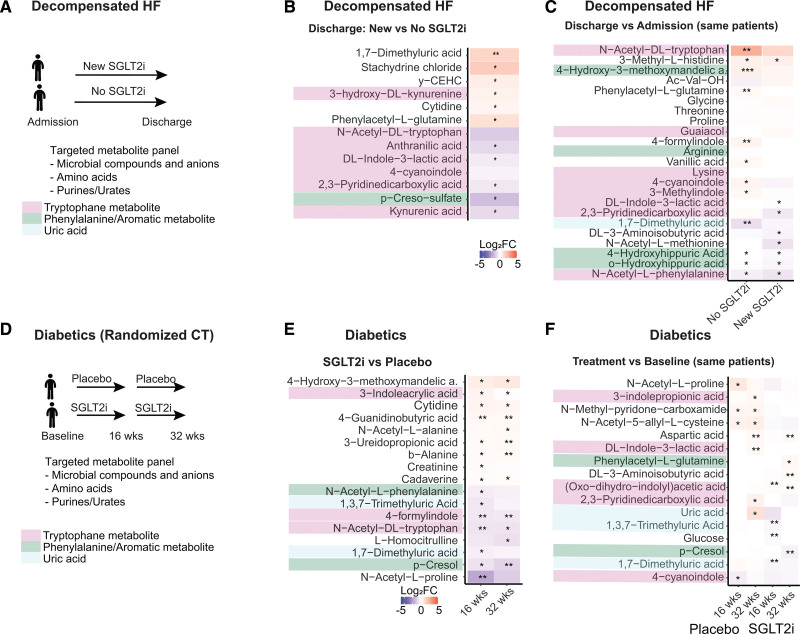
SGLT2i Reduced Plasma Levels of Gut-Derived Organic Anions (Uremic Toxins) Despite Lower Renal Secretory Transporter Expression
Next, we analyzed plasma over urine concentrations of metabolites that can reflect primary effects on kidney transport. As expected, the plasma:urine ratio for glucose and multiple other sugar metabolites decreased (Figure 3D), likely because of reduced tubular reabsorption. Metabolites with increased plasma:urine ratios were lysine metabolites, including diaminopimelic acid (a unique component of the cell wall of Gram-negative bacteria and a precursor of lysine), trimethyl lysine, pipecolic acid, and amino adipic acid (last oxidation product of lysine), a protective metabolite against obesity and diabetes in murine models.63 Many of the compounds with increased plasma:urine ratios in response to SGLT2i are substrates of the proximal tubule organic anion transporter OAT1 (Figure 3D): this included acetyl cysteine,64 citrulline,65 gluconate,66 and methionine,67 indicating that kidney secretion of these compounds may be reduced. This was corroborated by the finding of the proteome analysis that SGLT2i reduced OAT1 and OAT3 expression in WT, confirmed by immunoblot analysis of kidney membrane fractions (Figure 3E). At the same time, we observed that SGLT2i reduced plasma levels of so-called retained or uremic toxins (Figure 3A). To substantiate this finding, we performed a targeted analysis of serum metabolites that are known to be retained in kidney disease and are chiefly produced by the gut microbiome.43 SGLT2i decreased many of these uremic toxins in plasma of WT mice, including PCL, ILA, anthranilic acid, vanillic acid, and 3-indoxylsulfate (Figure 3F). In an independent nondiabetic animal model (hypertensive Dahl SS rats), the plasma:urine ratio of PCL sulfate and ILA was likewise markedly decreased by SGLT2i (Figure S4E). As noted previously, this cannot be explained by a downregulation of the kidney secretory capacity for uremic toxins (OAT1/OAT3) and suggested alteration of the formation of these compounds by SGLT2i.
SGLT2i Reshapes Intestinal Microbiota Fermentation of Amino Acids to Uremic Toxins
Many uremic toxins are produced by microbiota, and SGLT2i altered metabolites from uremic toxins, predominantly from phenylalanine and tryptophan. Thus, we hypothesized that SGLT2i affected the microbiota to alter production of these metabolites. To test this hypothesis, we analyzed matched cecal microbiomes using metagenomics-assisted metaproteomics (Figure S6A). In total, 6899 species were found by metagenomics. Translating this to the proteome, we quantified 14 621 proteins from these species (8249 for WT, 10 847 for Akita mice) with more robust effects of SGLT2i in WT versus Akita mice (Excel File S7 and Figure S1B). The phylogenetic overview summarizes the reconfiguration of the microbiota by SGLT2i in WT mice from phylum to family level (Figure 4A). On the genus level, SGLT2i prompted an increase in Akkermansia and Lachnoclostridium and a decrease in Acetatifactor and Bilophila (Figure S6B). Diversity analysis of the metaproteome revealed higher alpha diversity for diabetic mice after treatment, and beta diversity was significantly different for control and treated animals for both WT and diabetic mice (Figure S6C and S6D). Metaproteome analysis also covered proteins typical for the digestive tract, with most of them being downregulated by SGLT2i (34 down, 3 up; Excel File S7). As in the kidney, several of the decreased proteins were involved in nutrient transport, including Slc15a1 for oligopeptides, urate/xenobiotic secretory transporter Abcg2, and Slc27a4 for fatty acid import.
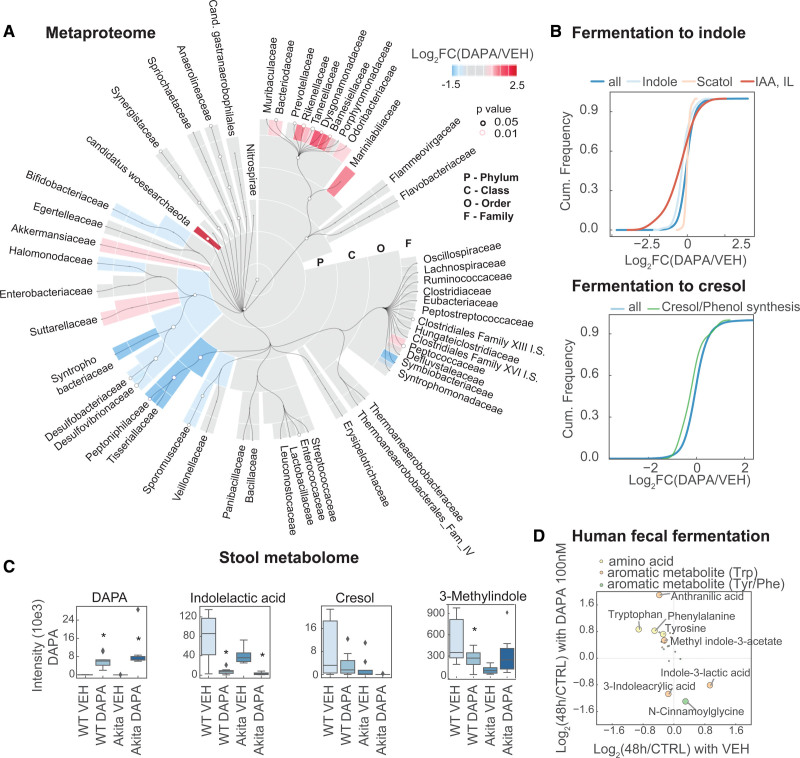
Figure 4.
Metaproteomics analysis demonstrates reshaping and attenuation of the microbiome fermentation of amino acids to uremic toxins by SGLT2i. A, Taxonomic analysis of metaproteome of gut microbiota with SGLT2i (sodium-glucose cotransporter 2 inhibitor) on family level. B, Cumulative histogram analysis of cresol- and phenol-producing bacteria species on metaproteome level, as well as tryptophan-metabolizing bacteria. C, Murine fecal metabolomics results of key metabolites. D, Targeted metabolomics of human fecal microbiota fermentation (48 hours) in the presence or absence of SGLT2i dapagliflozin (100 nM). Metabolites with >0.5-fold change (48 hours fermentation versus control) and P<0.05 between both conditions are labeled with an open circle (3 donors, with 3 fermentations each). IAA indicates indoleacrylic acid; and IL, indolelactate.
Because many of the plasma solutes altered by SGLT2i in WT mice were metabolites of aromatic amino acids, we analyzed the abundance of the bacteria that can metabolize these amino acids on the proteomics level. For indole-generating bacteria, we found a reduction of species that express tryptophanases to generate indole from tryptophan (indoleacrylic acid; Figure 4B). We also found a reduction of cresol and phenol builders, using aromatic amino acids such as phenylalanine as a substrate (Figure 4B). Examination of the stool metabolome revealed that dapagliflozin was present in the stool (Figure 4C). Metabolites from tryptophan and phenylalanine fermentation were decreased in these stool samples from dapagliflozin-treated mice (Figure 4C). To probe for a direct effect of dapagliflozin in the stool, we added the SGLT2i to human stool. After anaerobic fermentation, we found reduced indolelactic acid, a tryptophan metabolite, and cinnamoylglycine (Figure 4D); that is, compounds related to those reduced in plasma by SGLT2i in WT mice (Figure 3F). Addition of SGLT2i to human stool fermentation also reversed the disappearance of the parent molecule of these toxins, particularly the aromatic amino acids tryptophan, tyrosine, and phenylalanine (Figure 4D). This suggests that the physiologic effects of SGLT2i on the microbiota to generate fewer uremic toxins may also involve direct effects of SGLT2i on the microbiota.
Net Reduction of Uremic Toxin Metabolites by SGLT2i Is Independent of SGLT2
We then examined whether the reduced metabolites are independent of the presence of the SGLT2 protein, and whether these effects remain over a longer period. To distinguish the pure SGLT2 effect from an off-target effect, and in the absence of hyperglycemic confounders, we used the SGLT2 KO mouse model (Sglt2 KO).47 This mouse has a deletion of the Slc5a2 gene in exons 1 through 5, rendering the SGLT2 protein nonfunctional. We treated both Sglt2 KO and WT mice with SGLT2i (dapagliflozin) for 16 weeks on a normal diet, analyzed plasma and urine, and performed plasma targeted analysis using the panel of retained metabolites (Figure 5A). Sglt2 KO mice had lower blood glucose, higher glucose excretion, and higher food intake (Figure 5B and Figure S6E). In the KO mice, SGLT2i did not affect any of these measures. We calculated plasma:urine ratios (normalized to creatinine) to understand overall body handling in response to SGLT2i. The ratios for many metabolites were similarly changed in Sglt2 KO versus WT mice as they were by dapagliflozin versus vehicle treatment of WT mice (Figure 5C left panel), suggesting consequences of SGLT2 inhibition. Among others, this included a decline in the ratio for glucose, reflecting an increase in urine glucose elimination. As expected, absence of SGLT2 blocked the effect of dapagliflozin on glucose excretion (Figure 5C right panel). On the other hand, dapagliflozin significantly changed the ratio for a number of metabolites in Sglt2 KO mice, indicating an off-target effect (Figure 5D and Excel File S8). Ranking changes in the plasma:urine ratios revealed that dapagliflozin reduced the ratios in Sglt2 KO mice for multiple aromatic acid metabolites, including cresols and hippurates, followed by tryptophan metabolites. The plasma-level analysis identified 5 metabolites significantly changed by SGLT2i in the Sglt2 KO environment, including PCL sulfate and a modified hippuric acid (Figure 5E and 5F). This experiment suggests that SGLT2i treatment has off-target effects, including on cresols, hippurates, and tryptophan metabolites.
Figure 5.
Using Sglt2 KO mice to probe for metabolic off-target effects of SGLT2i. A, Study design. B, Quantification of glucosuria. C, Quantification of plasma:urine ratios in dapagliflozin-treated mice and Sglt2 knockout (KO) mice as compared with untreated wild-type (WT) mice. D, Rank of changes in plasma:urine ratios with SGLT2i (sodium-glucose cotransporter 2 inhibitor; all P<0.05). E, Analysis of off-target effects on mouse plasma, defined as significant changes in the presence of SGLT2i in Sglt2 KO mice. F, Quantification of p-cresol sulfate abundance in response to SGLT2i in both WT and Sglt2 KO mice. B and F, 2-way ANOVA to probe for a significant effect of Sglt2 genotype (Sglt2), dapagliflozin (dapa), or the interaction between the 2 factors (Pinteraction). If the interaction was statistically significant, then a pairwise multiple comparison procedure (Holm-Sidak method) identified the significant effects. *P<0.05 versus vehicle; #P<0.05 versus WT.
Metabolic Organ Communication Effects of SGLT2i Are Relevant in Humans
Given that the kidney SGLT2 interactome as well as the microbial response to SGLT2i were similar in mice and humans, we wondered whether SGLT2i would affect circulating solutes with cardiovascular relevance similarly in longitudinal data. Targeted metabolome analysis was performed on plasma samples derived from patients from 2 independent studies, targeting 80 organic anions and uremic toxins. In the first cohort of >40 patients, we analyzed the effect in a real-world setting of decompensated HF (Figure 6A). We found that SGLT2i significantly reduced or blunted the increase in plasma of several organic anions in patients with HF, but only when SGLT2i was added during the hospitalization (Figure 6B and 6C). This included several tryptophan metabolites, such as ILA, kynurenic acid, and acetyl-tryptophan, as well as the phenylalanine metabolites phenylacetyl glutamine and PCL sulfate. We also analyzed longitudinal data from a randomized controlled trial with SGLT2i (empagliflozin) in diabetic patients (Figure 6D). We found a reduction in plasma of dimethyluric acid and trimethyluric acid, a reduction of PCL, and blunted increase of indole metabolites, compared with baseline and compared with placebo (Figure 6E and 6F). The majority of the decreased solutes in both clinical studies was derived from purine metabolism and gut aromatic amino acid metabolism, consistent with the metabolic organ communication observed in mice.
Figure 6.
SGLT2i-induced reduction in uremic toxins translates to humans. A and D, Design of decompensated heart failure (HF) cohort and randomized controlled trial in patients with diabetes. B and C, Decompensated HF: heatmaps of SGLT2i (sodium-glucose cotransporter 2 inhibitor)–induced log2 fold changes for comparisons at discharge (B) and between admission and discharge (C). D, Randomized control trial of patients with diabetes: heatmaps of SGLT2i-induced log2 fold changes for SGLT2i versus placebo (E) and treatment versus baseline (F). SGLT2i treatment reduces uremic toxins originating from aromatic amino acid fermentation.
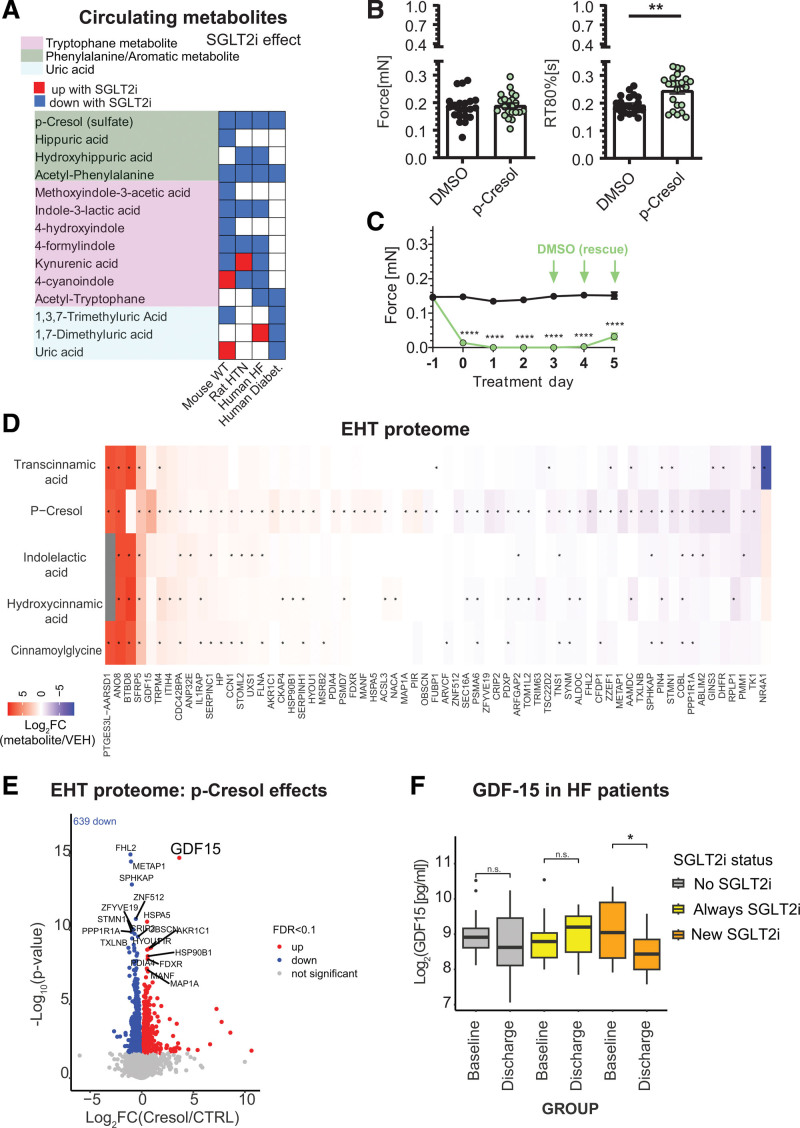
SGLT2i-Dependent Metabolite p-Cresol Induces Stress Signals in Human EHT
A consistent global trend was observed between different circulating metabolites in human, mouse (including Sglt2 KO mice), and rat studies (Figure 7A), with PCL sulfate or PCL consistently decreased by SGLT2i in all systems observed. To analyze this compound’s effect on human tissue, we exposed hiPSC EHTs to PCL. EHTs are 3-dimensional cultures of induced pluripotent stem cell–derived cardiac myocytes that can be phenotyped in terms of force, frequency, and rhythmic measures. We found that PCL altered relaxation time of cardiac myocytes at previously reported concentrations in patients (300 μM)68,69 (Figure 7B). Tenfold higher concentrations (low mM range) within hours significantly decreased the force and frequency (Figure 7C and Figure S7A), also when EHTs were frequency-controlled (Figure S7B). The effect of PCL on EHTs was partially reversible and rescued by washing out the compound (Figure 7C and Video S1). Indolelactic acid had no measurable effect on contractile measures in EHTs derived from 4 or 5 differentiation rounds (Figure S7C).
Figure 7.
Effects of SGLT2i-modulated uremic toxin, p-cresol, on human engineered heart tissue. A, Summary of metabolite changes in response to SGLT2i (sodium-glucose cotransporter 2 inhibitor) in mouse, rat, and human. B, Human engineered heart tissue (EHT) was exposed to either DMSO (black) or 300 µM p-cresol (PCL; green) for 5 days. Bar charts show force (mN), and relaxation time from peak to 80% relaxation (RT 80%; seconds) in spontaneously contracting EHTs. C, Time course of spontaneous force 1 day before exposure to 3 mM PCL (−1; green) or DMSO (black) until 5 days of daily treatment. After 3 days of treatment, PCL was replaced by DMSO (green arrows) for another 48 hours’ incubation (n=16–22 per group, derived from 4 [300 µM] and 5 [3 mM] independent rounds of cardiac differentiation unpaired, parametric T test [** = P<0.01 and **** = P<0.0001]). D and E, Proteomic effects of incubation of EHT with p-cresol (300 µM) or other SGLT2i-modulated metabolites for 5 days. F, GDF15 (growth/differentiation factor 15) in patients with heart failure (HF) with new SGLT2i treatment (patients from Figure 6A). FDR indicates false discovery rate; HTN, hypertension; and WT, wild type.
To further elucidate the mechanism, we performed proteomics analysis of EHT tissues treated with 300 μM of PCL, indole lactic acid, and 3 other aromatic metabolites (Figure 7D and Excel File S9). The most robust changes were observed in response to PCL (Figure 7D and Excel File S9). Proteins increasing across several metabolites included the heart disease–associated channel TRPM4 (transient receptor potential cation channel subfamily M member 4),70,71 the mechanosensor FLNA (filamin A), and the growth factor CCN1 (CYR61 [cysteine-rich 61]); reduced proteins included PPP1R1A (protein phosphatase 1 regulatory inhibitor subunit 1), the downregulation of which has been reported as hallmark in human HF.72 A unique feature of PCL-altered proteome was a reduction of cardiac sarcomere-associated proteins (Figure S7D), as well as a strong induction of the stress signal GDF15 (Figure 7E). Considering modulation of this pathway in patients with HF, we examined GDF15 levels. We found that circulating GDF15 was reduced in patients receiving SGLT2i for HF therapy (Figure 7F).
DISCUSSION
By performing a systematic analysis of SGLT2i treatment in vivo, we report a physiologic framework for cardiorenal benefits, involving reconfiguration by SGLT2i of the kidney (Figure 8A and Figure S8) and systemic metabolic communication (Figure 8B). Several SGLT2-dependent and off-target effects of SGLT2i on the kidney, heart, vessels, and other organs have been identified.14,73,74 Our study illuminates a central role of the microbiome and metabolic and early proximal tubule reconfiguration. Whereas several studies have explored the long-term effects on other tissues,75–77 we focused on relatively healthy mice at early time points to avoid dominating secondary omics signals from fibrosis, serum responses, and tissue decay that hamper mechanistic interpretation of omics data.78 SGLT2i had larger effects on metabolism and kidney reconfiguration in nondiabetic mice than in hyperglycemic Akita mice. This may in part reflect the greater effect of SGLT2i on glucosuria, calorie loss, and intracellular glucose of proximal tubules in nondiabetic animals.17
Figure 8.
Integrated views on the reconfiguration by SGLT2i of the early proximal tubule and systemic metabolic communication. A, Summary of findings and metabolic communication of SGLT2i (sodium-glucose cotransporter 2 inhibitor) in an early proximal tubule centric view in wild-type (WT) mice. Proteins with red lining were upregulated and proteins with blue lining were downregulated by SGLT2i. Proteins showing green filling have been shown to interact with SGLT2 in affinity purification analysis (see also Figure 2D). For further discussion, see text and Figure S8. B, Summary of findings and metabolic communication of SGLT2i on a systemic level in WT mice. C, Chemical structures of SGLT2 inhibitors dapagliflozin and empagliflozin and p-cresol. ACAT3 indicates acetyl-coenzyme A acetyltransferase 3; AGE, advanced glycation end products; AMPK, 5′-adenosine monophosphate–activated protein kinase; CKD, chronic kidney disease; ER, endoplasmic reticulum; GLUT2, glucose transporter 2; HGD, homogentisate 1,2-dioxygenase; NHE3, Na+/H+ exchanger 3; OAT1, organic anion transporter 1; PDZK1IP1, PDZK1 interacting protein 1; SLC, solute carrier; TMEM27, transmembrane protein 27; and WAT, white adipose tissue.
Two overarching mechanisms for renal protection emerge from our data. First, enzymes involved in the handling of glucose, oxidative stress, and cellular detoxification were decreased by SGLT2i, including MIOX (myo-inositol oxygenase) and GPX3 (glutathione peroxidase 3), both of which are primarily expressed in the early proximal tubule.57 Increase in MIOX has been implicated in oxidative stress and mitochondrial injury in the diabetic kidney,79 and its deficiency shields kidneys from tubulointerstitial injury by reducing glucose-induced oxidant and endoplasmic reticulum stress.80 Dapagliflozin also decreased GPX3, a secreted protein that binds specifically to basement membranes of mouse renal cortex tubule cells and detoxifies H2O2.81 Other early proximal tubule-specific protein changes are consistent with kidney protection, among these ACAT3 (acetyl-coenzyme A acetyltransferase 3), HGD (homogentisate 1,2-dioxygenase), SPP2 (secreted phosphoprotein 2), DPEP1 (dipeptidase 1), SLC4A4 (solute carrier family 4 member 4), and GLUT2 (glucose transporter 2)82 (Figure 8A; for more detail, see Excel File S1). Thus, evidence is presented on the protein level that SGLT2i reduces glucotoxicity and oxidative stress in the early proximal tubule.
SGLT2i affected more transporters and their substrates than anticipated. It is known that SGLT2i inhibits early proximal reabsorption not only of glucose but also in part of NaCl, bicarbonate, and fluid.83 As a consequence, SGLT2i can improve kidney cortex oxygenation84 and kidney outcome.85 The acute natriuretic and chronic blood pressure–lowering effects of SGLT2i have been linked to partial inhibition of the coexpressed Na+/H+ exchanger NHE3 (Na+/H+ exchanger 3).86,87 In interactome studies, we found that PDZK1IP1 (PDZK1 interacting protein 1; MAP17) interacts with SGLT2i, consistent with its activating role,88 alongside NHE regulating factors NHE-RF3 (Pdzk1) and NHE-RF2 (Slc9a3r2).89 Moreover, SGLT2i altered phosphorylation of these proteins (reduced Pdzk1ip1 S152 and S155 and NHE3 [Slc9a3] S550; increased NHE3 [Slc9a3] S588 and S789). In addition, we found that SGLT2 interacts with apical metabolite transporters in the early proximal tubule, including organic anions and cations, urate, creatine, lactate, thiamine, vitamin C, and neutral amino acids (Figures 2D and 2E and 8A). SGLT2i reduces several of these transporters. SGLT2 also interacts with and dapagliflozin reduced the expression of the ancillary protein TMEM27 (transmembrane protein 27; collectrin), a trafficking regulator for anionic (Slc1a1), neutral (Slc6a19), and cationic (Slc7a9) amino acid transporters. Some of these amino acids, such as phenylalanine, were consistently lost in the urine by SGLT2i. The apical transporters are coordinated to maintain tubular reabsorption in response to the daily changes in glomerular filtration rate and, thus, the tubular load of many substrates.1 Whereas this coupling makes physiologic sense, we show here that this makes a broad range of apical transporters sensitive to SGLT2i, so that SGLT2i not only lowers glucose uptake but more broadly reduces the oxygen-consuming transport burden of the early proximal tubule. The early proximal blockages would also protect the kidney by causing a more equal transport distribution along the nephron, potentially leading to a more favorable stress tolerance82 (Figure S8).
Two novel overarching mechanisms for cardiovascular protection by SGLT2i emerge from the study, both related to metabolic communication with the kidney. First, SGLT2i altered the excretion of many metabolites with cardiovascular relevance, such as purine metabolites. For instance, uric acid is a cardiovascular risk factor.90 Uricosuric effects of SGLT2i have been described in humans and mice with a potential role of the urate transporter URAT1 (urate transporter 1).91,92 This clinically relevant finding is mechanistically underpinned here by showing for the first time evidence for direct interaction of SGLT2i with URAT1 (Slc22a12), as well as reduced protein expression of URAT1 and reduced phosphorylation at S534 by SGLT2i. Urate, similar to many other organic anions, is transported through a machinery of more promiscuous transporters that perform tubular secretion. Results from 2 rodent models revealed that some organic anions accumulated in the blood in response to SGLT2i, indicating reduced tubular secretion (Figure 3 and Figure S4E). The organic anion transporters OAT1 and OAT3 (Slc22a8 and Slc22a6) mediate the basolateral uptake step in the proximal tubule for the overall secretion of organic anions, largely in exchange for α-ketoglutarate. Proteome quantification revealed reduced OAT1/3 protein expression by dapagliflozin in WT mice, as confirmed by Western blotting for OAT1 (Figure 3E). Moreover, OAT1/3 downregulation has potential for cardiovascular benefits. Accumulated OAT1/3 substrates included various anionic lysine metabolites with a potential link to cardiovascular disease,38 as well as acetyl-cysteine, which has antioxidant and anti-inflammatory properties in chronic kidney disease,93 and citrulline, the supplementation of which can lower blood pressure.94 OAT3 KO in mice is associated with lower blood pressure.95 SGLT2i decreased levels of circulating anions labeled as uremic toxins. Downregulation of OAT1/3—the major kidney excretion pathway for toxins and anions—would be expected to increase circulating levels of microbiota-derived uremic toxins, as observed for other anions, but the opposite effect was observed. The most likely explanation lies in the proposed gut–kidney axis as part of the remote sensing theory.96 Proximal tubule cells sense elevated levels of endogenous, gut microbiome–derived, metabolites, such as indoxyl sulfate (by means of the aryl-hydrocarbon receptor and epidermal growth factor receptor axis), which then induces their own renal secretion by upregulating OAT1.97 Reduced microbiome formation of these metabolites in response to SGLT2i would decrease OAT1/OAT3. This decrease, in turn, is expected to reduce the basolateral uptake and exposure of organic anions including nephro-toxic substances.26
Second, SGLT2i altered the microbiome in WT mice on the proteome level, mostly in a beneficial direction (Figure 4). The observed increases in Akkermansia, for instance, have previously been associated with responses to ketone bodies and amelioration of cardiovascular health problems such as hypertension.98 Increases in Desulfovibrio and reductions in Clostridiae are antagonized by the SGLT2 inhibitor on the proteome level and have been suggested to be causative for obesity in mice and humans.99 Moreover, SGLT2i reduced the microbiotic formation or circulating levels of multiple organic anions that are uremic toxins, among these PCL/PCL sulfate and ILA, including in patients (Figure 6). This class of compounds conveys renal and cardiovascular risk. Thus, evidence is presented that SGLT2i decreases microbiotic formation of uremic toxins; this potentially includes direct inhibitory effects on the microbiotic fermentation of amino acids (phenylalanine, tryptophan) as observed in human stool (Figure 4D). Animal studies confirm that this effect is at least in part independent of the presence of SGLT2, indicating an off-target effect (Figure 5): plasma levels of PCL sulfate are strongly reduced by SGLT2i in mice lacking SGLT2 (Figure 5C), and several other gut-derived tryptophan metabolites show a negative change in plasma:urine ratios (Figure 5D and 5E). Supporting the notion that SGLT2i also lower relevant cardiovascular toxic metabolites in humans, we found that circulating PCL100 and ILA101 levels were decreased in multiple patient populations newly treated with SGLT2i (Figure 6). Moreover, PCL directly and reversibly affected human EHT function and increased the stress-signal GDF15 in this preparation, whereas SGLT2i lowered circulating GDF15 levels in patients with HF (Figure 7). GDF15 is a heart-derived biomolecule,102 a cardiovascular biomarker candidate,103 and is sufficient to cause cardiac atrophy.104 Although our study was not designed to assess its biomarker potential, its potential instruction through circulating retained metabolites emphasizes its central role in body homeostasis and organ communication.102 In essence, SGLT2i could affect the microbiome formation of uremic toxins as an unanticipated off-target mechanism with protective potential on the heart and kidney. It is tempting to speculate that the aromatic part of the SGLT2i molecules examined here—which exhibits certain structural similarities to cresol (Figure 8C)—is responsible for this off-target effect.
SGLT2 inhibitors establish a favorable metabolic environment and functional state in the early proximal tubule, offering potential kidney and heart protection. Mechanisms are in part off-target and include the inhibition of gut microbiome–mediated uremic toxin formation. This is complemented by direct effects of SGLT2i on the early proximal tubule, which reduces glucotoxicity and—facilitated by a functionally coupled SGLT2 interactome—causes broad downregulation or inhibition of multiple apical transporters. These actions contribute to reduced kidney transport burden and oxygen consumption, and support natriuresis and urate excretion. Thus, SGLT2 inhibitors have the potential to positively influence blood pressure, volume retention, and hyperuricemia, ultimately benefiting cardiovascular health. The presented resource opens opportunities for a deeper understanding of SGLT2 inhibitor effects on metabolic, kidney, and heart function, and thus cardiovascular health.
ARTICLE INFORMATION
Acknowledgments
The authors thank Dr Trine Salomon, Stefan Gatzemeier, Mette Løbner, and Dr Angeliki Marietou for technical support and Drs Yi Yang and Liang Qiao (Department of Chemistry, Fudan University) for sharing code for metaproteome analysis.
Sources of Funding
Dr Rinschen was supported by the DFG (grants RI 2811/1-1 and RI 2811/2-1 and SFB1192-project B10), the Young Investigator Award from the Novo Nordisk Foundation (grant NNF19OC0056043), the Carlsberg Young Investigator fellowship, and the Aarhus University Research Foundation. This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 754513 and The Aarhus University Research Foundation (to Dr Rinschen). Dr Vallon was supported by the National Institutes of Health (grants R01DK112042, RF1AG061296, R01DK132690, and P30DK079337) and the Department of Veterans Affairs. Dr Zeller is supported by the DZHK (grant FKZ 81Z0710102, 2018–2025). Dr Fenton and Dr Rinschen were supported by the Novo Nordisk Foundation (grant NNF20SA0061466). Dr Staruschenko was supported by the National Institutes of Health (grant R35 HL135749). Dr Raabe was supported by the Studienstiftung des Deutschen Volkes. Dr Cuello was supported by the DZHK (grant FKZ 81Z0710115) and the DFG (grants CU53/5-1 and CU53/10-1). Dr Hoxha was supported by the Deutsche Forschungsgesellschaft (Heisenberg Programme). Dr Schwab received funding from the Aarhus University Research Foundation (grant AU FF-F-2020-7-2).
Disclosures
Dr Rinschen declares pending research funding from Novo Nordisk unrelated to this work. Over the past 12 months, Dr Vallon has served as a consultant for Lexicon and received speaker honoraria from AstraZeneca and grant support for investigator-initiated research from AstraZeneca, Boehringer Ingelheim, Gilead, Lexicon, Maze, Merck, and Novo-Nordisk. Dr Magnussen receives study-specific funding from the German Center for Cardiovascular Research (DZHK; Promotion of Women Scientists Programme; FKZ 81X3710112), the Deutsche Stiftung für Herzforschung, the Dr Rolf M. Schwiete Stiftung, NDD, and Loewenstein Medical unrelated to the current work. Dr Magnussen has received speaker fees from AstraZeneca, Novartis, Boehringer Ingelheim/Lilly, Bayer, Pfizer, Sanofi, Aventis, Apontis, and Abbott outside this work. Dr Dugourd and R. Fallegger report funding from Pfizer. Dr Saez-Rodriguez reports funding from GSK, Pfizer, and Sanofi and fees from Travere Therapeutics, Stadapharm, and Astex. Dr Hoxha served on advisory boards for Novartis, Morphosys AG, Sotio, and Argenx. The other authors declare no conflict of interest.
Supplemental Material
Figures S1–S8
Video S1
Excel Files S1–S10
Supplementary Material
Nonstandard Abbreviations and Acronyms
- ACAT3
- acetyl-coenzyme A acetyltransferase 3
- AMPK
- 5′-adenosine monophosphate–activated protein kinase
- CYCLE
- Characterization of Phenotypes in Acute Heart Failure Patients
- CYR61
- cysteine-rich 61
- DPEP1
- dipeptidase 1
- EB
- embryonic bodies
- EHT
- engineered heart tissue
- FGF
- fibroblast growth factor
- FLNA
- filamin A
- GDF15
- growth/differentiation factor 15
- GLUT2
- glucose transporter 2
- GPX3
- glutathione peroxidase 3
- HDAC
- histone deacetylase
- HF
- heart failure
- HGD
- homogentisate 1,2-dioxygenase
- HILIC
- hydrophilic liquid interaction chromatography
- hiPSC
- human induced pluripotent stem cell
- HSA
- human serum albumin
- ILA
- indole-3-lactic acid
- LC
- liquid chromatography
- LC-MS
- liquid chromatography mass spectrometry
- MIOX
- myo-inositol oxygenase
- MS
- mass spectrometry
- NHE3
- Na+/H+ exchanger 3
- NT-proBNP
- N-terminal pro-B-type natriuretic peptide
- OAT1
- organic anion transporter 1
- PCL
- p-cresol
- PDZK1IP1
- PDZK1 interacting protein 1
- PPP1R1A
- protein phosphatase 1 regulatory inhibitor subunit 1
- PRKAA1
- protein kinase AMP-activated catalytic subunit alpha 1
- RPMI
- Roswell Park Memorial Institute
- SGLT2
- sodium-glucose cotransporter 2
- SGLT2i
- sodium-glucose cotransporter 2 inhibitors
- SLC
- solute carrier
- SLC4A4
- solute carrier family 4 member 4
- SPP2
- secreted phosphoprotein 2
- TFA
- trifluoroacetic acid
- TMEM27
- transmembrane protein 27
- TMT
- tandem mass tag
- TRPM4
- transient receptor potential cation channel subfamily M member 4
- URAT1
- urate transporter 1
- WD
- Western diet
- WT
- wild type
V. Vallon and M.M. Rinschen contributed equally.
This manuscript was sent to Manuel Mayr, Guest Editor, for review by expert referees, editorial decision, and final disposition.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.123.065517.
For Sources of Funding and Disclosures, see page 881.
Circulation is available at www.ahajournals.org/journal/circ
Contributor Information
Young Chul Kim, Email: y8kim@vapop.ucsd.edu.
Søren Gullaksen, Email: soegul@clin.au.dk.
Benedikt Schrage, Email: b.schrage@uke.de.
Janice Raabe, Email: janice.raabe@hotmail.de.
Arvid Hutzfeldt, Email: a.hutzfeldt@uke.de.
Fatih Demir, Email: fatih.demir@biomed.au.dk.
Elina Kovalenko, Email: elinak@biomed.au.dk.
Aurelien Dugourd, Email: bf242@ad.uni-heidelberg.de.
Robin Fallegger, Email: robin.fallegger@uni-heidelberg.de.
Johannes Jaegers, Email: johannes.jaegers@directbox.com.
Qing Li, Email: qingli@bce.au.dk.
Olha Kravtsova, Email: okravtsova@usf.edu.
Maria Crespo-Masip, Email: chrysopoulou@biomed.au.dk.
Amelia Palermo, Email: apalermo@mednet.ucla.edu.
Robert A. Fenton, Email: robert.a.fenton@biomed.au.dk.
Elion Hoxha, Email: e.hoxha@uke.de.
Stefan Blankenberg, Email: s.blankenberg@uke.de.
Paulus Kirchhof, Email: p.kirchhof@uke.de.
Tobias B. Huber, Email: t.huber@uke.de.
Esben Laugesen, Email: esben.laugesen@clin.au.dk.
Tanja Zeller, Email: t.zeller@uke.de.
Maria Chrysopoulou, Email: chrysopoulou@biomed.au.dk.
Julio Saez-Rodriguez, Email: julio.saez@uni-heidelberg.de.
Thomas Eschenhagen, Email: t.eschenhagen@uke.uni-hamburg.de.
Alexander Staruschenko, Email: staruschenko@usf.edu.
Gary Siuzdak, Email: siuzdak@scripps.edu.
Clarissa Schwab, Email: schwab@bce.au.dk.
Friederike Cuello, Email: f.cuello@uke.de.
REFERENCES
- 1.Vallon V. Glucose transporters in the kidney in health and disease. Pflugers Arch. 2020;472:1345–1370. doi: 10.1007/s00424-020-02361-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herrington WG, Staplin N, Wanner C, Green JB, Hauske SJ, Emberson JR, Preiss D, Judge P, Mayne KJ, Ng SYA, et al. ; The EMPA-KIDNEY Collaborative Group. Empagliflozin in patients with chronic kidney disease. N Engl J Med. 2023;388:117–127. doi: 10.1056/NEJMoa2204233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anker SD, Butler J, Filippatos G, Ferreira JP, Bocchi E, Böhm M, Brunner-La Rocca H-P, Choi D-J, Chopra V, Chuquiure-Valenzuela E, et al. ; EMPEROR-Preserved Trial Investigators. Empagliflozin in heart failure with a preserved ejection fraction. N Engl J Med. 2021;385:1451–1461. doi: 10.1056/NEJMoa2107038 [DOI] [PubMed] [Google Scholar]
- 4.Heerspink HJL, Stefánsson BV, Correa-Rotter R, Chertow GM, Greene T, Hou F-F, Mann JFE, McMurray JJV, Lindberg M, Rossing P, et al. ; DAPA-CKD Trial Committees and Investigators. Dapagliflozin in patients with chronic kidney disease. N Engl J Med. 2020;383:1436–1446. doi: 10.1056/NEJMoa2024816 [DOI] [PubMed] [Google Scholar]
- 5.van der Aart–van der Beek AB, de Boer RA, Heerspink HJL. Kidney and heart failure outcomes associated with SGLT2 inhibitor use. Nat Rev Nephrol. 2022;18:294–306. doi: 10.1038/s41581-022-00535-6 [DOI] [PubMed] [Google Scholar]
- 6.Voors AA, Angermann CE, Teerlink JR, Collins SP, Kosiborod M, Biegus J, Ferreira JP, Nassif ME, Psotka MA, Tromp J, et al. The SGLT2 inhibitor empagliflozin in patients hospitalized for acute heart failure: a multinational randomized trial. Nat Med. 2022;28:568–574. doi: 10.1038/s41591-021-01659-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, Deswal A, Drazner MH, Dunlay SM, Evers LR, et al. 2022 AHA/ACC/HFSA guideline for the management of heart failure: executive summary: a report of the American College of Cardiology/American Heart Association joint committee on clinical practice guidelines. Circulation. 2022;145:e876–e894. doi: 10.1161/CIR.0000000000001062 [DOI] [PubMed] [Google Scholar]
- 8.Visseren FLJ, Mach F, Smulders YM, Carballo D, Koskinas KC, Bäck M, Benetos A, Biffi A, Boavida J-M, Capodanno D, et al. ; ESC National Cardiac Societies. 2021 ESC guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J. 2021;42:3227–3337. doi: 10.1093/eurheartj/ehab484 [DOI] [PubMed] [Google Scholar]
- 9.Buse JB, Wexler DJ, Tsapas A, Rossing P, Mingrone G, Mathieu C, D'Alessio DA, Davies MJ. 2019 update to: Management of hyperglycaemia in type 2 diabetes, 2018: a consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia. 2020;63:221–228. doi: 10.1007/s00125-019-05039-w [DOI] [PubMed] [Google Scholar]
- 10.Fattah H, Vallon V. The potential role of SGLT2 inhibitors in the treatment of type 1 diabetes mellitus. Drugs. 2018;78:717–726. doi: 10.1007/s40265-018-0901-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu H, Sridhar VS, Perkins BA, Rosenstock J, Cherney DZI. SGLT2 inhibition in type 1 diabetes with diabetic kidney disease: potential cardiorenal benefits can outweigh preventable risk of diabetic ketoacidosis. Curr Diab Rep. 2022;22:317–332. doi: 10.1007/s11892-022-01471-2 [DOI] [PubMed] [Google Scholar]
- 12.Hropot T, Battelino T, Dovc K. Sodium-glucose co-transporter-2 inhibitors in type 1 diabetes: a scoping review. Horm Res Paediatr. 2022;96:620–630. doi: 10.1159/000527653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupsa BC, Kibbey RG, Inzucchi SE. Ketones: the double-edged sword of SGLT2 inhibitors? Diabetologia. 2023;66:23–32. doi: 10.1007/s00125-022-05815-1 [DOI] [PubMed] [Google Scholar]
- 14.Vallon V, Verma S. Effects of SGLT2 inhibitors on kidney and cardiovascular function. Annu Rev Physiol. 2021;83:503–528. doi: 10.1146/annurev-physiol-031620-095920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Packer M. Critical reanalysis of the mechanisms underlying the cardiorenal benefits of SGLT2 inhibitors and reaffirmation of the nutrient deprivation signaling/autophagy hypothesis. Circulation. 2022;146:1383–1405. doi: 10.1161/CIRCULATIONAHA.122.061732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S, Wang Q, Christodoulou A, Mylonas N, Bakker D, Nederlof R, Hollmann MW, Weber NC, Coronel R, Wakker V, et al. Sodium glucose cotransporter-2 inhibitor empagliflozin reduces infarct size independently of sodium glucose cotransporter-2. Circulation. 2023;147:276–279. doi: 10.1161/CIRCULATIONAHA.122.061688 [DOI] [PubMed] [Google Scholar]
- 17.Vallon V, Gerasimova M, Rose MA, Masuda T, Satriano J, Mayoux E, Koepsell H, Thomson SC, Rieg T. SGLT2 inhibitor empagliflozin reduces renal growth and albuminuria in proportion to hyperglycemia and prevents glomerular hyperfiltration in diabetic Akita mice. Am J Physiol Renal Physiol. 2014;306:F194–F204. doi: 10.1152/ajprenal.00520.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gurley SB, Mach CL, Stegbauer J, Yang J, Snow KP, Hu A, Meyer TW, Coffman TM. Influence of genetic background on albuminuria and kidney injury in Ins2(+/C96Y) (Akita) mice. Am J Physiol Renal Physiol. 2010;298:F788–F795. doi: 10.1152/ajprenal.90515.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiredja DD, Koyutürk M, Chance MR. The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics. 2017;33:3489–3491. doi: 10.1093/bioinformatics/btx415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan Li, et al. clusterProfiler 40: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2:100141. doi: 10.1016/j.xinn.2021.100141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagih O. A versatile R package for drawing sequence logos. Bioinformatics. 2017;33:3645–3647. doi: 10.1093/bioinformatics/btx469 [DOI] [PubMed] [Google Scholar]
- 23.Conway JR, Lex A, Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickham H. ggplot2. Springer; 2009. doi: 10.1007/978-0-387-98141-3 [Google Scholar]
- 25.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- 26.Navarro Garrido A, Kim YC, Oe Y, Zhang H, Crespo-Masip M, Goodluck HA, Kanoo S, Sanders PW, Bröer S, Vallon V. Aristolochic acid-induced nephropathy is attenuated in mice lacking the neutral amino acid transporter B0AT1 (Slc6a19). Am J Physiol Renal Physiol. 2022;323:F455–F467. doi: 10.1152/ajprenal.00181.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 28.Long S, Yang Y, Shen C, Wang Y, Deng A, Qin Q, Qiao L. Metaproteomics characterizes human gut microbiome function in colorectal cancer. npj Biofilms Microbiomes. 2020;6:14. doi: 10.1038/s41522-020-0123-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saito Y, Sato T, Nomoto K, Tsuji H. Identification of phenol- and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol Ecol. 2018;94:fiy125. doi: 10.1093/femsec/fiy125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roager HM, Licht TR. Microbial tryptophan catabolites in health and disease. Nat Commun. 2018;9:3294. doi: 10.1038/s41467-018-05470-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmieder R, Edwards R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS One. 2011;6:e17288. doi: 10.1371/journal.pone.0017288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033 [DOI] [PubMed] [Google Scholar]
- 33.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Pyl PT, Huber W. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinschen MM, Palygin O, Guijas C, Palermo A, Palacio-Escat N, Domingo-Almenara X, Montenegro-Burke R, Saez-Rodriguez J, Staruschenko A, Siuzdak G. Metabolic rewiring of the hypertensive kidney. Sci Signal. 2019;12:eaax9760. doi: 10.1126/scisignal.aax9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinschen MM, Palygin O, El-Meanawy A, Domingo-Almenara X, Palermo A, Dissanayake LV, Golosova D, Schafroth MA, Guijas C, Demir F, et al. Accelerated lysine metabolism conveys kidney protection in salt-sensitive hypertension. Nat Commun. 2022;13:4099. doi: 10.1038/s41467-022-31670-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: a web-based platform to process untargeted metabolomic data. Anal Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guijas C, Montenegro-Burke JR, Domingo-Almenara X, Palermo A, Warth B, Hermann G, Koellensperger G, Huan T, Uritboonthai W, Aisporna AE, et al. METLIN: a technology platform for identifying knowns and unknowns. Anal Chem. 2018;90:3156–3164. doi: 10.1021/acs.analchem.7b04424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue J, Domingo-Almenara X, Guijas C, Palermo A, Rinschen MM, Isbell J, Benton HP, Siuzdak G. Enhanced in-source fragmentation annotation enables novel data independent acquisition and autonomous METLIN molecular identification. Anal Chem. 2020;92:6051–6059. doi: 10.1021/acs.analchem.0c00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhn M, von Mering C, Campillos M, Jensen LJ, Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/nar/gkm795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka H, Sirich TL, Plummer NS, Weaver DS, Meyer TW. An enlarged profile of uremic solutes. PLoS One. 2015;10:e0135657. doi: 10.1371/journal.pone.0135657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kravtsova O, Bohovyk R, Levchenko V, Palygin O, Klemens CA, Rieg T, Staruschenko A. SGLT2 inhibition effect on salt-induced hypertension, RAAS, and Na+ transport in Dahl SS rats. Am J Physiol Renal Physiol. 2022;322:F692–F707. doi: 10.1152/ajprenal.00053.2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bircher L, Schwab C, Geirnaert A, Greppi A, Lacroix C. Planktonic and sessile artificial colonic microbiota harbor distinct composition and reestablish differently upon frozen and freeze-dried long-term storage. mSystems. 2020;5:e00521–e00519. doi: 10.1128/mSystems.00521-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bircher L, Geirnaert A, Hammes F, Lacroix C, Schwab C. Effect of cryopreservation and lyophilization on viability and growth of strict anaerobic human gut microbes. Microb Biotechnol. 2018;11:721–733. doi: 10.1111/1751-7915.13265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallon V, Platt KA, Cunard R, Schroth J, Whaley J, Thomson SC, Koepsell H, Rieg T. SGLT2 mediates glucose reabsorption in the early proximal tubule. J Am Soc Nephrol. 2011;22:104–112. doi: 10.1681/ASN.2010030246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gullaksen S, Vernstrøm L, Sørensen SS, Ringgaard S, Laustsen C, Funck KL, Poulsen PL, Laugesen E. Separate and combined effects of semaglutide and empagliflozin on kidney oxygenation and perfusion in people with type 2 diabetes: a randomised trial. Diabetologia. 2023;66:813–825. doi: 10.1007/s00125-023-05876-w [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- 50.Breckwoldt K, Letuffe-Brenière D, Mannhardt I, Schulze T, Ulmer B, Werner T, Benzin A, Klampe B, Reinsch MC, Laufer S, et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat Protoc. 2017;12:1177–1197. doi: 10.1038/nprot.2017.033 [DOI] [PubMed] [Google Scholar]
- 51.Hansen A, Eder A, Bönstrup M, Flato M, Mewe M, Schaaf S, Aksehirlioglu B, Schwoerer AP, Uebeler J, Eschenhagen T. Development of a drug screening platform based on engineered heart tissue. Circ Res. 2010;107:35–44. doi: 10.1161/CIRCRESAHA.109.211458 [DOI] [PubMed] [Google Scholar]
- 52.Hirt MN, Boeddinghaus J, Mitchell A, Schaaf S, Börnchen C, Müller C, Schulz H, Hubner N, Stenzig J, Stoehr A, et al. Functional improvement and maturation of rat and human engineered heart tissue by chronic electrical stimulation. J Mol Cell Cardiol. 2014;74:151–161. doi: 10.1016/j.yjmcc.2014.05.009 [DOI] [PubMed] [Google Scholar]
- 53.Rue-Albrecht K, Marini F, Soneson C, Lun ATL. iSEE: interactive summarized experiment explorer. F1000Res. 2018;7:741. doi: 10.12688/f1000research.14966.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rue-Albrecht K, Soneson C, Marini F, Lun A, Stadler M. iSEEu: iSEE Universe; 2022. doi: 10.18129/B9.bioc.iSEEu [Google Scholar]
- 55.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, et al. The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L, Chou C-L, Knepper MA. A comprehensive map of mRNAs and their isoforms across all 14 renal tubule segments of mouse. J Am Soc Nephrol. 2021;32:897–912. doi: 10.1681/ASN.2020101406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wicik Z, Nowak A, Jarosz-Popek J, Wolska M, Eyileten C, Siller-Matula JM, von Lewinski D, Sourij H, Filipiak KJ, Postuła M. Characterization of the SGLT2 interaction network and its regulation by SGLT2 inhibitors: a bioinformatic analysis. Front Pharmacol. 2022;13:901340. doi: 10.3389/fphar.2022.901340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung Y-S, Jun S, Kim MJ, Lee SH, Suh HN, Lien EM, Jung H-Y, Lee S, Zhang J, Yang J-I, et al. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/β-catenin signalling. Nat Cell Biol. 2018;20:1421–1433. doi: 10.1038/s41556-018-0219-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Ivica NA, Dong T, Papageorgiou DP, He Y, Brown DR, Kleyman M, Hu G, Chen WW, Sullivan LB, et al. MFSD7C switches mitochondrial ATP synthesis to thermogenesis in response to heme. Nat Commun. 2020;11:4837. doi: 10.1038/s41467-020-18607-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lang F, Görlach A, Vallon V. Targeting SGK1 in diabetes. Expert Opin Ther Targets. 2009;13:1303–1311. doi: 10.1517/14728220903260807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaub JA, AlAkwaa FM, McCown PJ, Naik AS, Nair V, Eddy S, Menon R, Otto EA, Demeke D, Hartman J, et al. SGLT2 inhibitors mitigate kidney tubular metabolic and mTORC1 perturbations in youth-onset type 2 diabetes. J Clin Invest. 2023;133:e164486. doi: 10.1172/JCI164486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu W-Y, Shen Y, Zhu H, Gao J, Zhang C, Tang L, Lu S-Y, Shen C-L, Zhang H-X, Li Z, et al. 2-Aminoadipic acid protects against obesity and diabetes. J Endocrinol. 2019;243:111–123. doi: 10.1530/JOE-19-0157 [DOI] [PubMed] [Google Scholar]
- 64.Aslamkhan AG, Han Y-H, Yang X-P, Zalups RK, Pritchard JB. Human renal organic anion transporter 1-dependent uptake and toxicity of mercuric-thiol conjugates in Madin-Darby canine kidney cells. Mol Pharmacol. 2003;63:590–596. doi: 10.1124/mol.63.3.590 [DOI] [PubMed] [Google Scholar]
- 65.Nakakariya M, Shima Y, Shirasaka Y, Mitsuoka K, Nakanishi T, Tamai I. Organic anion transporter OAT1 is involved in renal handling of citrulline. Am J Physiol Renal Physiol. 2009;297:F71–F79. doi: 10.1152/ajprenal.90662.2008 [DOI] [PubMed] [Google Scholar]
- 66.Kaler G, Truong DM, Khandelwal A, Nagle M, Eraly SA, Swaan PW, Nigam SK. Structural variation governs substrate specificity for organic anion transporter (OAT) homologs: potential remote sensing by OAT family members. J Biol Chem. 2007;282:23841–23853. doi: 10.1074/jbc.M703467200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J Proteome Res. 2011;10:2842–2851. doi: 10.1021/pr200093w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Assis RP, Castro JFA, Gutierres VO, Arcaro CA, Brotto RS, Oliveira OMMF, Baviera AM, Brunetti IL. Effects of uremic solutes on reactive oxygen species in vitro model systems as a possibility of support the renal function management. BMC Nephrol. 2015;16:50. doi: 10.1186/s12882-015-0029-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu H-J, Yen C-H, Wu I-W, Hsu K-H, Chen C-K, Sun C-Y, Chou C-C, Chen C-Y, Tsai C-J, Wu M-S, et al. The association of uremic toxins and inflammation in hemodialysis patients. PLoS One. 2014;9:e102691. doi: 10.1371/journal.pone.0102691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng J, Zong P, Yan J, Yue Z, Li X, Smith C, Ai X, Yue L. Upregulation of transient receptor potential melastatin 4 (TRPM4) in ventricular fibroblasts from heart failure patients. Pflugers Arch. 2021;473:521–531. doi: 10.1007/s00424-021-02525-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guinamard R, Bouvagnet P, Hof T, Liu H, Simard C, Sallé L. TRPM4 in cardiac electrical activity. Cardiovasc Res. 2015;108:21–30. doi: 10.1093/cvr/cvv213 [DOI] [PubMed] [Google Scholar]
- 72.Wittköpper K, Fabritz L, Neef S, Ort KR, Grefe C, Unsöld B, Kirchhof P, Maier LS, Hasenfuss G, Dobrev D, et al. Constitutively active phosphatase inhibitor-1 improves cardiac contractility in young mice but is deleterious after catecholaminergic stress and with aging. J Clin Invest. 2010;120:617–626. doi: 10.1172/JCI40545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dyck JRB, Sossalla S, Hamdani N, Coronel R, Weber NC, Light PE, Zuurbier CJ. Cardiac mechanisms of the beneficial effects of SGLT2 inhibitors in heart failure: evidence for potential off-target effects. J Mol Cell Cardiol. 2022;167:17–31. doi: 10.1016/j.yjmcc.2022.03.005 [DOI] [PubMed] [Google Scholar]
- 74.De Pascalis A, Cianciolo G, Capelli I, Brunori G, La Manna G. SGLT2 inhibitors, sodium and off-target effects: an overview. J Nephrol. 2021;34:673–680. doi: 10.1007/s40620-020-00845-7 [DOI] [PubMed] [Google Scholar]
- 75.Shi J, Qiu H, Xu Q, Ma Y, Ye T, Kuang Z, Qu N, Kan C, Hou N, Han F, et al. Integrated multi-omics analyses reveal effects of empagliflozin on intestinal homeostasis in high-fat-diet mice. iScience. 2023;26:105816. doi: 10.1016/j.isci.2022.105816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Osataphan S, Macchi C, Singhal G, Chimene-Weiss J, Sales V, Kozuka C, Dreyfuss JM, Pan H, Tangcharoenpaisan Y, Morningstar J, et al. SGLT2 inhibition reprograms systemic metabolism via FGF21-dependent and -independent mechanisms. JCI Insight. 2019;4:e123130. doi: 10.1172/jci.insight.123130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Zhang Z, Li C, Tang D, Dai Y. Metabolomics study reveals the alteration of fatty acid oxidation in the hearts of diabetic mice by empagliflozin. Mol Omics. 2022;18:643–651. doi: 10.1039/d2mo00036a [DOI] [PubMed] [Google Scholar]
- 78.Rinschen MM, Saez-Rodriguez J. The tissue proteome in the multi-omic landscape of kidney disease. Nat Rev Nephrol. 2020;17:205–219. doi: 10.1038/s41581-020-00348-5 [DOI] [PubMed] [Google Scholar]
- 79.Zhan M, Usman IM, Sun L, Kanwar YS. Disruption of renal tubular mitochondrial quality control by myo-inositol oxygenase in diabetic kidney disease. J Am Soc Nephrol. 2015;26:1304–1321. doi: 10.1681/ASN.2014050457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma I, Deng F, Liao Y, Kanwar YS. Myo-inositol oxygenase (MIOX) overexpression drives the progression of renal tubulointerstitial injury in diabetes. Diabetes. 2020;69:1248–1263. doi: 10.2337/db19-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olson GE, Whitin JC, Hill KE, Winfrey VP, Motley AK, Austin LM, Deal J, Cohen HJ, Burk RF. Extracellular glutathione peroxidase (Gpx3) binds specifically to basement membranes of mouse renal cortex tubule cells. Am J Physiol Renal Physiol. 2010;298:F1244–F1253. doi: 10.1152/ajprenal.00662.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vallon V, Thomson SC. The tubular hypothesis of nephron filtration and diabetic kidney disease. Nat Rev Nephrol. 2020;16:317–336. doi: 10.1038/s41581-020-0256-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Layton AT, Vallon V. SGLT2 inhibition in a kidney with reduced nephron number: modeling and analysis of solute transport and metabolism. Am J Physiol Renal Physiol. 2018;314:F969–F984. doi: 10.1152/ajprenal.00551.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laursen JC, Søndergaard-Heinrich N, de Melo JML, Haddock B, Rasmussen IKB, Safavimanesh F, Hansen CS, Størling J, Larsson HBW, Groop P-H, et al. Acute effects of dapagliflozin on renal oxygenation and perfusion in type 1 diabetes with albuminuria: a randomised, double-blind, placebo-controlled crossover trial. EClinicalMedicine. 2021;37:100895. doi: 10.1016/j.eclinm.2021.100895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pruijm M, Milani B, Pivin E, Podhajska A, Vogt B, Stuber M, Burnier M. Reduced cortical oxygenation predicts a progressive decline of renal function in patients with chronic kidney disease. Kidney Int. 2018;93:932–940. doi: 10.1016/j.kint.2017.10.020 [DOI] [PubMed] [Google Scholar]
- 86.Onishi A, Fu Y, Patel R, Darshi M, Crespo-Masip M, Huang W, Song P, Freeman B, Kim YC, Soleimani M, et al. A role for tubular Na+/H+ exchanger NHE3 in the natriuretic effect of the SGLT2 inhibitor empagliflozin. Am J Physiol Renal Physiol. 2020;319:F712–F728. doi: 10.1152/ajprenal.00264.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Borges-Júnior FA, dos Santos DS, Benetti A, Polidoro JZ, Wisnivesky ACT, Crajoinas RO, Antônio EL, Jensen L, Caramelli B, Malnic G, et al. Empagliflozin inhibits proximal tubule NHE3 activity, preserves GFR, and restores euvolemia in nondiabetic rats with induced heart failure. J Am Soc Nephrol. 2021;32:1616–1629. doi: 10.1681/ASN.2020071029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coady MJ, Tarazi AE, Santer R, Bissonnette P, Sasseville LJ, Calado J, Lussier Y, Dumayne C, Bichet DG, Lapointe JY. MAP17 is a necessary activator of renal Na+/glucose cotransporter SGLT2. J Am Soc Nephrol. 2017;28:85–93. doi: 10.1681/ASN.2015111282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lanaspa MA, Giral H, Breusegem SY, Halaihel N, Baile G, Catalán J, Carrodeguas JA, Barry NP, Levi M, Sorribas V. Interaction of MAP17 with NHERF3/4 induces translocation of the renal Na/Pi IIa transporter to the trans-Golgi. Am J Physiol Renal Physiol. 2007;292:F230–F242. doi: 10.1152/ajprenal.00075.2006 [DOI] [PubMed] [Google Scholar]
- 90.Saito Y, Tanaka A, Node K, Kobayashi Y. Uric acid and cardiovascular disease: a clinical review. J Cardiol. 2021;78:51–57. doi: 10.1016/j.jjcc.2020.12.013 [DOI] [PubMed] [Google Scholar]
- 91.Novikov A, Fu Y, Huang W, Freeman B, Patel R, van Ginkel C, Koepsell H, Busslinger M, Onishi A, Nespoux J, et al. SGLT2 inhibition and renal urate excretion: role of luminal glucose, GLUT9, and URAT1. Am J Physiol Renal Physiol. 2019;316:F173–F185. doi: 10.1152/ajprenal.00462.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Suijk DLS, van Baar MJB, van Bommel EJM, Iqbal Z, Krebber MM, Vallon V, Touw D, Hoorn EJ, Nieuwdorp M, Kramer MMH, et al. SGLT2 inhibition and uric acid excretion in patients with type 2 diabetes and normal kidney function. Clin J Am Soc Nephrol. 2022;17:663–671. doi: 10.2215/CJN.11480821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cepaityte D, Leivaditis K, Varouktsi G, Roumeliotis A, Roumeliotis S, Liakopoulos V. N-acetylcysteine: more than preventing contrast-induced nephropathy in uremic patients: focus on the antioxidant and anti-inflammatory properties. Int Urol Nephrol. 2023; doi: 10.1007/s11255-022-03455-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.An P, Wan S, Luo Y, Luo J, Zhang X, Zhou S, Xu T, He J, Mechanick JI, Wu W-C, et al. Micronutrient supplementation to reduce cardiovascular risk. J Am Coll Cardiol. 2022;80:2269–2285. doi: 10.1016/j.jacc.2022.09.048 [DOI] [PubMed] [Google Scholar]
- 95.Vallon V, Eraly SA, Wikoff WR, Rieg T, Kaler G, Truong DM, Ahn S-Y, Mahapatra NR, Mahata SK, Gangoiti JA, et al. Organic anion transporter 3 contributes to the regulation of blood pressure. J Am Soc Nephrol. 2008;19:1732–1740. doi: 10.1681/ASN.2008020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nigam SK, Granados JC. A Biological basis for pharmacokinetics: the remote sensing and signaling theory. Clin Pharmacol Ther. 2022;112:456–460. doi: 10.1002/cpt.2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jansen J, Jansen K, Neven E, Poesen R, Othman A, van Mil A, Sluijter J, Sastre Torano J, Zaal EA, Berkers CR, et al. Remote sensing and signaling in kidney proximal tubules stimulates gut microbiome-derived organic anion secretion. Proc Natl Acad Sci USA. 2019;116:16105–16110. doi: 10.1073/pnas.1821809116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chakraborty S, Galla S, Cheng X, Yeo J-Y, Mell B, Singh V, Yeoh BS, Saha P, Mathew AV, Vijay-Kumar M, et al. Salt-responsive metabolite, β-hydroxybutyrate, attenuates hypertension. Cell Rep. 2018;25:677–689.e4. doi: 10.1016/j.celrep.2018.09.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petersen C, Bell R, Klag KA, Lee S-H, Soto R, Ghazaryan A, Buhrke K, Ekiz HA, Ost KS, Boudina S, et al. T cell-mediated regulation of the microbiota protects against obesity. Science. 2019;365:eaat9351. doi: 10.1126/science.aat9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harlacher E, Wollenhaupt J, Baaten CCFMJ, Noels H. Impact of uremic toxins on endothelial dysfunction in chronic kidney disease: a systematic review. Int J Mol Sci. 2022;23:531. doi: 10.3390/ijms23010531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Karu N, McKercher C, Nichols DS, Davies N, Shellie RA, Hilder EF, Jose MD. Tryptophan metabolism, its relation to inflammation and stress markers and association with psychological and cognitive functioning: Tasmanian Chronic Kidney Disease pilot study. BMC Nephrol. 2016;17:171. doi: 10.1186/s12882-016-0387-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang T, Liu J, McDonald C, Lupino K, Zhai X, Wilkins BJ, Hakonarson H, Pei L. GDF15 is a heart-derived hormone that regulates body growth. EMBO Mol Med. 2017;9:1150–1164. doi: 10.15252/emmm.201707604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wollert KC, Kempf T, Wallentin L. Growth differentiation factor 15 as a biomarker in cardiovascular disease. Clin Chem. 2017;63:140–151. doi: 10.1373/clinchem.2016.255174 [DOI] [PubMed] [Google Scholar]
- 104.Ozcan M, Guo Z, Valenzuela Ripoll C, Diab A, Picataggi A, Rawnsley D, Lotfinaghsh A, Bergom C, Szymanski J, Hwang D, et al. Sustained alternate-day fasting potentiates doxorubicin cardiotoxicity. Cell Metab. 2023;35:928–942.e4. doi: 10.1016/j.cmet.2023.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]