Abstract
The transcription factor Pax6 is essential for the development of the eyes and the central nervous system of vertebrates and invertebrates. Pax6 contains two DNA-binding domains; an N-terminal paired domain and a centrally located homeodomain. We have previously shown that the vertebrate paired-less isoform of Pax6 (Pax6ΔPD), and several other homeodomain proteins, interact with the full-length isoform of Pax6 enhancing Pax6-mediated transactivation from paired domain-DNA binding sites. By mutation analyses and molecular modeling we now demonstrate that, surprisingly, the recognition helix for specific DNA binding of the homeodomains of Pax6 and Chx10 interacts with the C-terminal RED subdomain of the paired domain of Pax6. Basic residues in the recognition helix and the N-terminal arm of the homeodomain form an interaction surface that binds to an acidic patch involving residues in helices 1 and 2 of the RED subdomain. We used fluorescence resonance energy transfer assays to demonstrate such interactions between Pax6 molecules in the nuclei of living cells. Interestingly, two mutations in the homeodomain recognition helix, R57A and R58A, reduced protein–protein interactions, but not DNA binding of Pax6ΔPD. These findings suggest a critical role for the recognition helix and N-terminal arm of the paired class homeodomain in protein–protein interactions.
INTRODUCTION
Pax6 is a highly conserved member of the Pax family of transcription factors that plays pivotal roles during embryogenesis in both vertebrates and invertebrates (1,2). Pax6 contains two DNA-binding domains. A flexible linker region separates the N-terminally located paired domain (PD) from the paired type homeodomain (HD). The region C-terminal to the HD is rich in proline, serine and threonine, and functions as a transcriptional activation domain (3–7). The zebrafish and human Pax6 proteins have an overall amino acid sequence identity of 97%, while invertebrate and mouse Pax6 proteins show more than 90% sequence identity in the PD (8). In vertebrates, Pax6 is expressed in the developing neuroretina and lens of the eye, in the nasal placode, in the pancreas, in the pituitary gland [reviewed in (8)] and in the pineal gland (9,10). Pax6 is essential for normal development of several organs, including the brain, the pancreas and the eye. The importance of Pax6 in development of the eye is illustrated by the fact that Pax6 induces ectopic eyes in flies and frogs upon misexpression (11–13). While the homozygous Pax6 mutation is lethal to mouse embryos, the heterozygous mutant gives the Small eye phenotype (14). In humans, heterozygous mutations cause aniridia and are also associated with Peters anomaly and other congenital eye disorders (15).
The paired class HDs are able to bind cooperatively to palindromic DNA sequences of the type TAAT(N)2–3ATTA, named P2 or P3 after the number of base pairs separating the two TAAT/ATTA palindromic core sequences (16). The HD of Pax6 binds preferentially to P3 sites (5). In contrast to the cooperative DNA binding/dimerization of other HDs (e.g. Hox), which relies on sequences extrinsic to the HD, the paired class HDs can rely entirely on the 60-amino acid-long HD to achieve cooperativity upon DNA binding (16).
Several isoforms of Pax6 have been reported (17–19). One of these is the 33/32 kDa Pax6ΔPD isoform that lacks the PD owing to the use of an internal start codon for translation between the PD and the HD (18). An alternatively spliced PD-less isoform of Pax6 has also been isolated from mouse brain cDNA (20). The Pax6ΔPD isoform is able to interact with full-length Pax6 thereby enhancing Pax6 transactivation from PD-binding sites in reporter gene assays (21). The HD of Pax6ΔPD can interact with both the HD and the PD of Pax6. We found that the HDs of a number of other homeodomain proteins are also able to interact with Pax6 (21). Several other proteins have been shown to bind to Pax proteins via the PD and/or the HD leading to either repression or activation in reporter gene assays (22–32).
Here, we have investigated the DNA-independent interactions between the PD and the HD of Pax6 and between the PD of Pax6 and the HD of the paired class homeodomain protein Chx10. Surprisingly, helix 3 of the HD, which is the recognition helix for specific DNA binding, also mediates the protein–protein interactions with the Pax6 PD. Basic residues on one side of helix 3 interact with acidic residues in the RED subdomain of the PD. R3 and R5 in the N-terminal arm of the HD also contribute to the HD–PD interaction. R57 and R58 in the Pax6 HD are important for the protein–protein interaction with the PD, but they are not important for DNA binding. Our results suggest that protein–protein interactions involving the DNA-binding domains of Pax6 and paired-type homeodomain proteins may be instrumental in regulating the activity of these transcription factors.
MATERIALS AND METHODS
Plasmid constructions
All Pax6 constructs used in this work are derived from the zebrafish Pax6.1 cDNA (33). The GST Pax6 HD, GST Pax6 RED, GST Chx10 HD, HA-Pax6, HA-Pax6ΔHD, HA-Pax6ΔPD HA-Pax6ΔPDΔHD, HA-Chx10, pCI-Pax6 and P6CON-LUC constructs have been described previously (12,21). To construct pGST-18L-HD, a 296 bp EcoRI–XhoI PCR fragment encoding 18 amino acids of the linker region and the homeodomain of zebrafish Pax6 (amino acids 211–309) was amplified from pCI-Pax6 using the primers zf.pax6-HD5.5′ and zf.pax6-HD5.3′ and inserted into the EcoRI–XhoI sites of pGEX-4T3. HA-Pax6ΔHDh2-3 was constructed by ligation of the PCR product obtained from HA-Pax6 using primers delB and delD. HA-Pax6ΔHDh2-3 was then used as template with 6ΔPD.HA.5 and 6.HA.3 as primers to construct HA-Pax6ΔPDΔh2-3 following the same procedure as described previously for HA-Pax6ΔPDΔHD (21). HA-Pax6ΔPDΔh3 was constructed similarly using delC and delD primers with HA-Pax6Δh2-3 as template. The delD, 6ΔPD.HA.5 and 6.HA.3 primers have been described previously (21). The 5xHDp3-LUC reporter plasmid was constructed by replacing the GAL4-binding sites in pG5E1b-LUC (34) with five copies of a homeodomain p3 site. Oligonucleotides PaxHD-P3.1 (5′-GATCCTCTAGATAATGCGATTAGCGTAG-3′) and PaxHD-P3.2 (5′-GATCCTACGCTAATCGCATTATCTAGAG-3′) were annealed and five copies were cloned into the BamHI site of pUC19 making pUC19-5xHDp3. The HDp3 sites were then cloned into the pG5E1b-LUC vector by ligation of a 150 bp SmaI–XbaI fragment from pUC19-5xHDp3 to HindIII(end-filled)–XbaI cut pG5E1b-LUC.
The yeast expression vector pSOS (Stratagene) contains human SOS (residues 1–1066), the constitutive ADH1-promoter and the LEU2 gene. To generate the bait plasmid pSOS-zfPax6-HD, a 222 bp BamHI–SacI PCR fragment encoding the homeodomain of zebrafish Pax6 (amino acids 222–296) was amplified from pCI-Pax6 using the primers zf-pax6-HD1-5′ and zf-pax6-HD1-3′ and inserted into the BamHI–SacI sites of pSOS. Expression of the SOS–zfPax6-HD fusion protein in transformed cdc25-2 yeast cells was confirmed by immunoblotting with an anti-SOS1 specific antibody (Transduction Laboratories). The pMYR expression vector (Stratagene) contains the v-Src myristoylation sequence, the inducible GAL1-promoter and the URA3 gene. The pMYR-zfPax6-HD target vector was constructed by inserting a 222 bp EcoRI–SalI fragment, encoding the zfPax6 homeodomain (amino acids 222–296), amplified from pCI-Pax6 by PCR using the primer pairs zf.pax6-HD2-5′ and zf-pax6-HD2-3′ and inserted into the EcoRI–SalI sites in pMYR. The pMYR–zfPax6-PD fusion construct was made by PCR amplification of the zebrafish Pax6.1 paired domain (amino acids 21–154) from pCI-Pax6 using the primer pairs zf-pax6-PD2-5′ and zf-pax6-PD2-3′. The resulting 402 bp SmaI–SalI fragment was ligated into the SmaI–SalI sites of pMYR. The pSOS and pMYR constructs were verified by sequencing using the primers 5′-SOS and 5′-MYR, respectively.
The Gateway cloning system (Invitrogen) was used to make expression constructs for FRET experiments. Pax6 cDNAs were initially subcloned into entry vectors, and subsequently transferred into the destination vectors pDEST-EYFP-N1 and pDEST-ECFP-C1 (35). pDEST-EYFP-N1 and pDEST-ECFP-C1 contain a C-terminal yellow fluorescent protein (YFP) and an N-terminal cyan fluorescent protein (CFP), respectively. To generate pENTR1A-Pax6ΔHD, a 1.0 kb XhoI–NotI PCR fragment encoding the zebrafish Pax6.1 cDNA sequence lacking the homeodomain was amplified from HA.Pax6ΔHD using the primers Flag P6.5′ and PAX6.3′NOTI, and inserted into the XhoI–NotI sites of pENTR1A. pENTR1A-Pax6 was constructed by inserting a 1.3 kb XhoI–NotI fragment, encoding the full-length Pax6.1 cDNA amplified from pCI-Pax6 by PCR using the primer pairs Flag P6.5′ and PAX6.3′NOTI, into the XhoI–NotI sites of pENTR1A. The pENTR2B-Pax6ΔPD construct was made by inserting a 900 bp EcoRI–NotI cut PCR fragment from HA.Pax6ΔPD, made with primers FlagDPD.5′ and PAX6.3′ NOTI, into the EcoRI–NotI sites of pENTR2B. To make pENTR11-Pax6, the full-length Pax6.1 coding sequence was amplified from pCI-Pax6 using Pfu polymerase and the primers Flag P6.5′ and P63′Ustopp, and the resulting 1.3 kb PCR product was inserted into the XhoI–NotI sites of pENTR11. pENTR3C-zfPax6HD was constructed by inserting a 200 bp fragment encoding the homeodomain from Pax6.1 cDNA into the EcoRI–XhoI sites of pENTR3C. The entry vector pENTR1A-Pax6ΔPD (R53A.R57A) was made by PCR amplification of a 900 bp fragment from HA.Pax6ΔPD (R53A.R57A) using the primer pairs zf.pax6.HD7-5′ and zf.pax6.TAD2-3′. The 900 bp Pax6.1 cDNA sequence lacking the paired domain was ligated into the BamHI–XhoI sites of pENTR1A. To make pENTR11-Pax6ΔPD, a 900 bp SalI–NotI fragment, encoding Pax6.1 without the paired domain, was amplified by PCR from HA.Pax6ΔPD using the primers zf.pax6.HD6-5′ and zf.pax6.TAD1-3′, and inserted into the SalI–NotI sites of pENTR11. The pENTR11-Pax6ΔHD (E101A.E112A.E120A.E128A) vector was constructed by PCR using the Pfu polymerase. A 1.0 kb Pax6.1 sequence lacking the homeodomain was amplified from HA.Pax6ΔHD (E101A.E112A.E120A.E128A) using the primers zf.pax6.PD7-5′ and zf.pax6.TAD1-3′, and ligated into the SalI–NotI sites of pENTR11. All constructs were verified by nucleotide sequencing with the GFP-C1 and GFP-N1 primers (Invitrogen) using the BigDye v3.1 sequencing kit (Applied Biosystems).
In vitro mutagenesis
PCR-mediated in vitro mutagenesis was performed according to the manufacturer's instructions for the Quick-Change Site-Directed Mutagenesis kit (Stratagene). Mutagenized constructs were verified by sequencing. The specific primers used for mutagenesis are shown in Table 1.
Table 1.
Sequences of primers used in this study
| Name | Sequence |
|---|---|
| Pax6 R53A.3′ | 5′-CCTCCATTTCGCTGCTCTGTTTGAGAACC-3′ |
| Pax6 R53A.5′ | 5′-GGTTCTCAAACAGAGCAGCGAAATGGAGG-3′ |
| Pax6 S50A.3′ | 5′-CGCTCTTCTGTTTGCGAACCAGACCTG-3′ |
| Pax6 S50A.5′ | 5′-GACGTCTGGTTCGCAAACAGAAGAGCG-3′ |
| Pax6 R58A.3′ | 5′-CTTTTCCTCCGCCCTCCATTTCGCTC-3′ |
| Pax6 R58A.5′ | 5′-GAGCGAAATGGAGGGCGGAGGAAAAG-3′ |
| Pax6 N51Q.3′ | 5′-CCATTTCGCTCTTCTCTGTGAGAACCAGACC-3′ |
| Pax6 N51Q.5′ | 5′-GGTCTGGTTCTCACAGAGAAGAGCGAAATGG-3′ |
| Pax6 R44A.3′ | 5′-GAACCAGACCTGTATTGCTGCTTCTGGTAAATC-3′ |
| Pax6 R44A.5′ | 5′-GATTTACCAGAAGCAGCAATACAGGTCTGGTTC-3′ |
| Pax6 Q46A.3′ | 5′-GTTTGAGAACCAGACCGCTATTCTTGCTTCTGG-3′ |
| Pax6 Q46A.5′ | 5′-CCAGAAGCAAGAATAGCGGTCTGGTTCTCAAAC-3′ |
| Pax6 N51A.3′ | 5′-CATTTCGCTCTTCTGGCTGAGAACCAGACC-3′ |
| Pax6 N51A.5′ | 5′-GGTCTGGTTCTCAGCCAGAAGAGCGAAATG-3′ |
| Pax6 Q46A.3′ | 5′-GTTTGAGAACCAGACCGCTATTCTTGCTTCTGG-3′ |
| Pax6 Q46A.5′ | 5′-CCAGAAGCAAGAATAGCGGTCTGGTTCTCAAAC-3′ |
| Pax6 E120A.3′ | 5′-CTGTCTCGGATTGCCCACGCGAAGATTGAC-3′ |
| Pax6 E120A.5′ | 5′-GTCAATCTTCGCGTGGGCAATCCGAGACAG-3′ |
| Pax6 E112A.3′ | 5′-GAAGATTGACGGACACGCCCTCTTGTACTG-3′ |
| Pax6 E112A.5′ | 5′-CAGTACAAGAGGGCGTGTCCGTCAATCTTC-3′ |
| Pax6 R57A.3′ | 5′-CTTTTCCTCCCTCGCCCATTTCGCTCTTC-3′ |
| Pax6 R57A.5′ | 5′-GAAGAGCGAAATGGGCGAGGGAGGAAAAG-3′ |
| Pax6 E128A.3′ | 5′-GTGCAGACCCCCGCTGATAGCAGC-3′ |
| Pax6 E128A.5′ | 5′-GCTGCTATCAGCGGGGGTCTGCAC-3′ |
| Chx10 R57A.3′ | 5′-CTTCTCCCTCTTCGCCCACTTGGCTCTG-3′ |
| Chx10 R57A.5′ | 5′-CAGAGCCAAGTGGGCGAAGAGGGAGAAG-3′ |
| Chx10 Q46A.3′ | 5′-CTGGAACCACACGCGTATCCTGTCTTCTG-3′ |
| Chx10 Q46A.5′ | 5′-CAGAAGACAGGATACGCGTGTGGTTCCAG-3′ |
| Chx10 R44A.3′ | 5′-CCACACCTGTATCGCGTCTTCTGGGAGC-3′ |
| Chx10 R44A.5′ | 5′-GCTCCCAGAAGACGCGATACAGGTGTGG-3′ |
| Chx10 Q50A.3′ | 5′-GCTCTGCGGTTCGCGAACCACACCTGTATCC-3′ |
| Chx10 Q50A.3′ | 5′-GGATACAGGTGTGGTTCGCGAACCGCAGAGC-3′ |
| Chx10 N51Q.3′ | 5′-CTTGGCTCTGCGTTGCTGGAACCACACCTG-3′ |
| Chx10 N51Q.5′ | 5′-CAGGTGTGGTTCCAGCAACGCAGAGCCAAG-3′ |
| Chx10 R53A.3′ | 5′-CCTCCACTTGGCTGCGCGGTTCTGGAACC-3′ |
| Chx10 R53A.5′ | 5′-GGTTCCAGAACCGCGCAGCCAAGTGGAGG-3′ |
| Chx10 R58A.5′ | 3′-CAGCACTTCTCCCTCGCCCTCCACTTGGC-3′ |
| Chx10 R58A.3′ | 3′-GCCAAGTGGAGGGCGAGGGAGAAGTGCTG-3′ |
| Pax6 R57Aon A53.3′ | 5′-CTTTTCCTCCCTCGCCCATTTCGCTGCTC-3′ |
| Pax6 R57Aon A53.5′ | 5′-GAGCAGCGAAATGGGCGAGGGAGGAAAAG-3′ |
| DelB | 5′-GTAGCTCGAGCTTTCAAACTCTTTTTCA-3′ |
| DelC | 5′-TGGTCTCGAGATTTTTGCAGCAAGTCTTTCTCG-3′ |
| zf.pax6-HD1.5′ | 5′-CTAGGATCCTGAGGCTTCAGCTTAAA-CGAAAAC-3′ |
| zf.pax6-HD1.3′ | 5′-TACGAGCTCGGC-TTGTCTTCTTTGATTTCTTAAC-3′ |
| zf.pax6-HD2.5′ | 5′-GCAGAATTCAGGCTTCAGCTTAAACGAAAACTG-3′ |
| zf.pax6-HD2.3′ | 5′-AGCGTCGACGGCTTGTCTTCTTTGA-TTTCTTAAC-3′ |
| zf.pax6-PD2.5′ | 5′-CGACCCGGGTGCAAAACAGTCACAGTGGAGTG-3′ |
| zf.pax6-PD2.3 | 5′-TGAGTCGACCTGTTGCTTTTCGCTAG-CCAGG-3′ |
| 5′-SOS | 5′-CCAAGACCAGGTACCATG-3′ |
| 5′-MYR | 5′-ACTACTAGCAGCTGTAATAC-3′ |
| zf.pax6-HD5.5′ | 5′-ACAGAATTCCTCCAATGGCGAGGACTCAGATG-3′ |
| zf.pax6-HD5.3′ | 5′-CGACTCGAGCTGCTGCTGATGGGTATGTGACT-3′ |
| zfHD R(-)7A.5′ | 5′-GATGAGACCCAAATGGCGCTTCAGCTTAAACG-3′ |
| zfHD R(-)7A.3′ | 5′-CGTTTAAGCTGAAGCGCCATTTGGGTCTCATC-3′ |
| zfHD K(-)3A.5′ | 5′-CAAATGAGGCTTCAGCTTGCACGAAAACTGCAAAGGAATC-3′ |
| zfHD K(-)3A.3′ | 5′-GATTCCTTTGCAGTTTTCGTGCAAGCTGAAGCCTCATTTG-3′ |
| zfHD R(-)2A.5′ | 5′-GAGGCTTCAGCTTAAAGCAAAACTGCAAAGGAATCG-3′ |
| zfHD R(-)2A.3′ | 5′-CGATTCCTTTGCAGTTTTGCTTTAAGCTGAAGCCTC-3′ |
| zfHD K(-)1A.5′ | 5′-CTTCAGCTTAAACGAGCACTGCAAAGGAATCGC-3′ |
| zfHD K(-)1A.3′ | 5′-GCGATTCCTTTGCAGTGCTCGTTTAAGCTGAAG-3′ |
| zfHD R(+)3A.5′ | 5′-CTTAAACGAAAACTGCAAGCGAATCGCACTTCTTTC-3′ |
| zfHD R(+)3A.3′ | 5′-GAAAGAAGTGCGATTCGCTTGCAGTTTTCGTTTAAG-3′ |
| zfHD R(+)5A.5′ | 5′-GAAAACTGCAAAGGAATGCCACTTCTTTCACACAAG-3′ |
| zfHD R(+)5A.3′ | 5′-CTTGTGTGAAAGAAGTGGCATTCCTTTGCAGTTTTC-3′ |
| zfHD K61A.5′ | 5′-GGAGGAGGGAGGAAGCGTTAAGAAATCAAAGAAG-3′ |
| zfHD K61A.3′ | 5′-CTTCTTTGATTTCTTAACGCTTCCTCCCTCCTCC-3′ |
| zfPD.E101A-5′ | 5′-GACTCCCGCGGTGGTCGGCAAAATTG-3′ |
| zfPD.E101A-3′ | 5′-CAATTTTGCCGACCACCGCGGGAGTC-3′ |
| zfPD.D123A-5′ | 5′-GTGGGCAATCCGAGCCAGGCTGCTAT-3′ |
| zfPD.D123A-3′ | 5′-ATAGCAGCCTGGCTCGGATTGCCCAC-3′ |
| P63′Ustopp | 5′-CTAGCGGCCGCTGTAGTCTGGGCCAGTA-3′ |
| FlagDPD.5′ | 5′-GCGGAATTCGGGCGCAGATGGCATGTAT-3′ |
| Pax6.3′NotI | 5′-ATAGCGGCCGCTCACTGTAGTCTGGGCCA-3′ |
| Flag P6.5′ | 5′-GACCTCGAGCAAAACAGTCACAGTGGAGTG-3′ |
| zf.pax6.HD7-5′ | 5′-AGCGGATCCGCATGTATGAAAAGCTGAGGATGCT-3′ |
| zf.pax6.TAD2-3′ | 5′-GACCTCGAGTCGCGTTCTGCCTGTAGTCTGGG-3′ |
| zf.pax6.HD6-5′ | 5′-CGAGTCGACTATGAAAAGCTGAGGATGCTGAACG-3′ |
| zf.pax6.TAD1-3′ | 5′-GACGCGGCCGCTCGCGTTCTGCCTGTAGTCTGGG-3′ |
| zf.pax6.PD7-5′ | 5′-CGAGTCGACAACAGTCACAGTGGAGTGAACCAG-3′ |
| GFP-N1-3′ | 5′-CCGTTTACGTCGCCGTCCAG-3′ |
| GFP-C1-5′ | 5′-GATCACATGGTCCTGCTGGA-3′ |
Glutathione S-transferase (GST) pull-down assays
GST fusion proteins purified from Escherichia coli LE392 or E.coli BL21-Star(DE3)pLysS (Invitrogen) extracts using glutathione–sepharose beads (Amersham Pharmacia Biotech) were used in pull-down assays as described previously (21).
Transient transfection assays
NIH 3T3 fibroblasts (passage 123) (ATCC CRL 1658) were cultured in DMEM supplemented with 10% calf serum (HyClone, Logan, UT), penicillin (100 U/ml) and 100 μg/ml streptomycin (Life Technologies, Inc.). About 4 × 104 NIH 3T3 cells/well in 24-well tissue culture dishes were transfected using LipofectAmine PLUS (Invitrogen). Extracts were prepared 24 h after transfection using the Dual-Light luciferase and β-galactosidase reporter gene assay system (Tropix) and analyzed in a Labsystems Luminoskan RT dual injection luminometer. Transient transfection assays in human HeLa cells were performed as described previously (21). To probe expression levels and stability of mutant proteins after transfection, we performed western blots using the following antibodies: rabbit polyclonal anti-GFP (1:1000; Abcam), mouse monoclonal anti-HA, clone 12CA5 (1:1000, Roche), affinity-purified rabbit polyclonal anti-Pax6 C-terminal P6C (1:800) (7) antibodies and peroxidase-conjugated secondary antibodies (1:2000; Santa Cruz Biotechnology). Detection and quantification were performed using ECL chemiluminescence (Amersham Biosciences) and the LumiAnalyst imager and software (Roche Applied Sciences).
Yeast two-hybrid assays using the SOS recruitment system
Yeast two-hybrid assays using the SOS recruitment interaction system was performed essentially as described (36). The Saccharomyces cerevisiae cdc25-2 MATa strain (ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 cdc25-2 Gal+) was co-transformed with 0.3 μg of each pSOS-bait and pMYR-prey plasmids using the lithium acetate method.
Gel mobility shift assay
GST fusion proteins were purified from E.coli BL21-Star(DE3)pLysS extracts as described previously (21). The proteins were eluted from the beads using 5 mM reduced glutathione (Sigma). The GST fusion proteins were analyzed on a 10% SDS–polyacrylamide gel and quantified using SYPRO® Ruby protein stain (Bio-Rad) and LumiAnalyst imager and software (Roche Applied Sciences). Binding of GST-Pax6-HD and mutants to the 32P-labeled HDp3 probe (5′-GATCCTCTAGATAATGCGATTAGCGTAG-3′) was performed as described previously (21), with the exception that 100 mM NaCl was used instead of 30 mM KCl. The binding reactions were performed in the presence of 100 ng/μl of BSA (Sigma).
Molecular modeling of PD and HD and their interactions
The ICM Pro 3.0 program (Molsoft L.L.C., La Jolla, CA; available on the World Wide Web at www.molsoft.com) (37) was utilized for comparative modeling, computer graphics visualizations, protein–protein docking, energy calculations and calculation of molecular surfaces and electrostatic potentials. A stepwise energy refinement of the X-ray structure of human Pax6 PD [Protein Data Bank code 6pax (38)] was performed using the regul macro of ICM. Residues from S23 to Q155 of Pax6 are included in the structure. Using the A protein chain of the X-ray structure of HD from the Drosophila paired protein [Protein Data Bank code 1fjl (39)] as a structural template and the homology module of ICM, two three-dimensional models of the human Pax6 HD were built, model 1 including F8 to N64 and model 2 including L-4 to A73 (numbering of HD according to a generic HD numbering scheme). The refineModel macro of ICM was used to perform stepwise energy refinement of the HD models. Rigid body docking simulations of Pax6 PD–HD were performed using the HD model 1, and low-energy conformations were collected in a conformational stack for the docking simulation. A stack conformation having Arg44, Arg53, Arg57 and Arg58 of the HD at the PD–HD interface was selected. Extra amino acids at the N- and C-terminal end of the HD molecule were included by replacing the HD molecule with model 2 followed by manual adjustment of a few backbone dihedral angles of the N-terminal arm in order to introduce interactions between positively charged residues in HD domain (R3 and R5) and negatively charged residues of the PD domain. The model of the PD–HD complex was obtained by energy minimization using the AMBER 8 force field (http://amber.scripps.edu/).
Fluorescence resonance energy transfer experiments
For FRET microscopy, subconfluent HeLa cells (∼1 × 104 cell/well) grown in eight-chambered cover slides (Nunc), were transiently transfected with LipofectAmine PLUS (Invitrogen) using 50 ng each of the different CFP- and YFP-tagged Pax6 expression constructs. FRET analysis was performed using a Zeiss LSM510 META confocal microscope equipped with a 25 mW argon laser (Carl Zeiss GmbH, Germany). FRET was measured using the acceptor photobleaching method (40,41). Emission of CFP and YFP was detected by lambda scan from 460 to 600 nm, with 10 nm intervals, using the META detector. The YFP acceptor was bleached in the nucleus, by scanning a region of interest (ROI), by 30 iterations using 95% intensity and 45% laser power with the 514 nm Argon laser line. FRET was detected as decreased YFP-emission and a corresponding increased CFP-emission in the bleached area. The YFP acceptor was bleached between images three and four using the 458 nm laser line. We used cells expressing approximately a 1:1 ratio of the CFP- and YFP-tagged proteins for FRET experiments and at least 5 cells (nuclei) per pair of CFP and YFP fusions were analyzed.
RESULTS
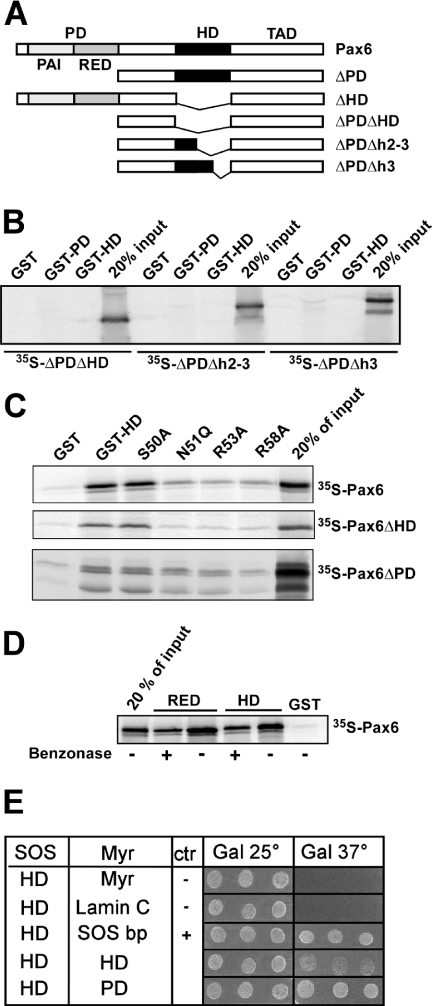
The recognition helix (helix 3) of the homeodomain of Pax6 is required for binding to both the paired domain and the homeodomain
We have previously shown that the Pax6 homeodomain is able to bind to other homeodomains, and to the C-terminal subdomain of the paired domain (RED) (21). However, the nature of the interaction surfaces involved is unknown. The homeodomain consists of three α-helices. GST pull-down assays conducted with three different deletion constructs of the HD in Pax6ΔPD implicated helix 3 either as (i) directly required for the protein–protein interactions or as (ii) required for the formation or positioning of the interaction surface located elsewhere in the domain (Figure 1A and B). We next generated four single-point mutations in helix 3. We chose to mutate surface-exposed residues conserved between homeodomains we had previously found to interact with Pax6 (21). Thus, S50, R53 and R58 were mutated to alanine while N51 was mutated to glutamine. This mutation (N51Q) is reported to abolish DNA binding by the paired class homeodomain protein Phox1 (42). GST pull-down experiments indicated that N51, R53 and R58 are important for the interaction with Pax6, while S50 mutated to alanine had no effect on the interaction between GST-HD and Pax6 (Figure 1C). The same mutations in helix 3 that reduced the interactions of GST-HD with full-length Pax6 also reduced the interaction with Pax6ΔHD and Pax6ΔPD (Figure 1C). Although the same amino acids in helix 3 seem to be important for both the interaction with the PD and the HD, there are differences between these two interactions. The HD–HD interaction (between GST-HD and Pax6ΔPD) is weaker than the HD–PD interaction (between GST-HD and Pax6ΔHD). Furthermore, the HD–PD interaction was reduced to a greater extent by the mutations, than the HD–HD interaction. R58 seems to be more important for the HD–HD interaction than N51 and R53. For the HD–PD interaction observed between GST-HD and Pax6ΔHD, the three mutants bound to Pax6ΔHD with a similarly reduced affinity (Figure 1C).
Figure 1.
The recognition helix of the homeodomain of Pax6 is important for interaction with both the PD and the HD. (A) Pax6 constructs used for in vitro translation and GST pull-downs. (B) GST pull-down assays with Pax6 HD and PD fused to GST and immobilized on glutathione–agarose beads and Pax6ΔPDΔHD, Pax6ΔPDΔh2–3 or Pax6ΔPDΔh3 produced by in vitro transcription and translation in the presence of [35S]methionine. An aliquot of 10 μl of the in vitro translation reactions was preincubated with GST immobilized on glutathione–agarose beads before incubation with the GST fusion proteins. The GST beads, GST-Pax6 HD beads and GST-Pax6 PD beads were washed several times before they were boiled in SDS loading buffer and run on a 10% SDS–polyacrylamide gel. An aliquot of 2 μl of the in vitro translated proteins was run on the same gel to visualize the signal from 20% of the input. (C) Point mutations in helix 3 of the homeodomain strongly reduce the ability of Pax6 HD to interact with the PD and the wild-type HD. The N51Q, R53A and R58A, but not S50A, mutants impede the HD–PD and HD–HD interactions. GST pull-down assays were performed with recombinant GST fusions of wild type or mutants of Pax6ΔHD against in vitro translated, [35S]methionine-labeled Pax6, Pax6ΔHD or Pax6ΔPD. (D) The interactions between full-length Pax6 and the RED subdomain and between full-length Pax6 and the HD are independent of DNA. GST pull-down assays were done with Pax6 HD and RED fused to GST as in (C). Where indicated, the pull-down experiments were performed in the presence of 500 U benzonase to degrade both DNA and RNA. The results shown are representative of three independent experiments. (E) The PD–HD and HD–HD interactions of Pax6 are also observed in the yeast-based SOS recruitment interaction system. The temperature sensitive yeast strain S.cerevisiae cdc25-2 MATa was co-transformed either with pSOS-zfPax6-HDwt and empty pMYR or pMYR-LaminC as negative controls, pMYR-SOS binding protein as a positive control, pMYR-zfPax6-HDwt, or with pMYR-zfPax6-PDwt. Three independent colonies generated from each co-transformation were replica plated onto galactose plates and grown in parallel at 25 and 37°C for 6 days. The results shown are representative of three independent experiments.
Since we have mapped the protein–protein interaction surface of the HD to the DNA-binding helix 3, it is possible that the interaction is mediated through DNA. We have earlier observed that the interaction between Pax6 and Pax6ΔHD with GST-HD is DNA-independent because it is not abolished by the addition of ethidium bromide (21). These results were confirmed using the nuclease benzonase, which destroys both DNA and RNA (Figure 1D).
In order to test the PD–HD and HD–HD interactions in another experimental setting we used the SOS-recruitment-based yeast two-hybrid system (43). This system is based on translocation of an active human SOS protein to the inner leaflet of the plasma membrane. In this system, a possible contribution of nucleic acids to the binding is completely ruled out. As shown in Figure 1E, both the PD–HD and the HD–HD interactions were verified in this system. In addition, as shown by the GST pull-down assays, the PD–HD interaction appeared stronger than the HD–HD interaction.
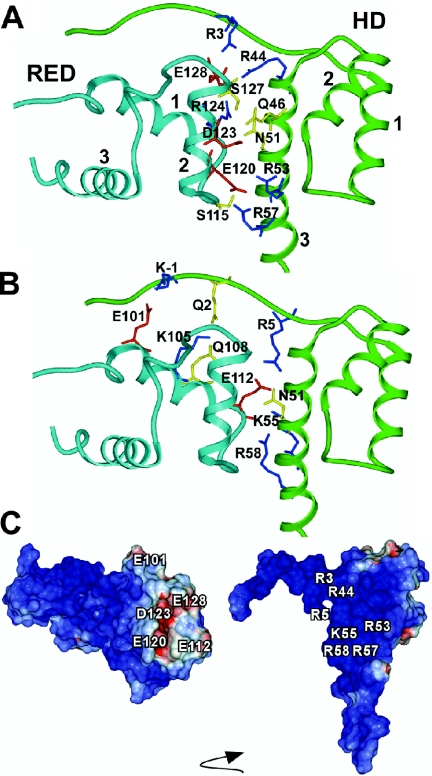
Model of the Pax6 PD–HD complex
We next constructed models of human Pax6 PD and HD using the published structures of the human Pax6 PD bound to DNA and the HD of Drosophila paired as starting points (38,39). The model of the Pax6 PD–HD complex shown in Figure 2 features salt bridge interactions between positively charged residues in helix 3 of the HD and negatively charged residues in helices 1 and 2 of RED (R44 in HD–E128 in RED, R53 in HD–E120 in RED and R57 in HD–E120 in RED). The positively charged side chain of K55 in helix 3 interacts with the carboxylate group of E112 in helix 1, whereas the side chain R58 in HD interacts with the main chain carbonyl group of E112 at the C-terminal end of helix 1 in RED. Furthermore, the side chain NH2 groups of Q46 and N51 in HD interact with the carboxylate group of D123 (helix 2) and the R124 side chain (helix 2) in RED, respectively. The side chains of Y25 in HD, W119 and F117 in RED (residues not shown in Figure 2) are localized close to salt bridges in the model. The side chain OH group of S115 in RED (loop between helices 1 and 2) interacts with the backbone CO group of A54 in HD (helix 3), whereas the side chain OH group of S127 (helix 2 of RED) forms an interaction with the backbone CO group of D123 in the same helix. Residues in the N-terminal arm of HD interact with the residues in helix 2 (R3 in HD–E128 in RED) and helix 1 (K-1 in HD–E101 in RED, Q2 in HD–K105 in RED and R5 in HD–E112 in RED) of RED.
Figure 2.
Model of the PD–HD interaction surface. (A and B) Energy minimized model of the Pax6 PD–HD complex with contact residues at the interface between the HD and the RED subdomain of the PD displayed. The side chain interactions involving helix 2 of RED are shown in (A) and those involving helix 1 in (B). Color coding of ribbon: Cyan, RED; Green, HD. Color coding of residues: red, D and E; blue, R and K; and yellow, S, Q and N. (C) Electrostatics surface potentials of the Pax6 PD (left) and HD (right) color coded according to electrostatic potentials (blue, e > 5 kcal/electron units; white, −5 ≤ e ≥ 5 kcal/electron units; and red, e < −5 kcal/electron units). Molecular surfaces and electrostatics potentials were calculated by using the REBEL (rapid exact boundary element) method of ICM for the HD and the PD. The arrow indicates that the HD has been rotated relative to the PD to orient its interaction surface towards the front. The two interacting domains were moved apart to facilitate visualization of the interacting surfaces.
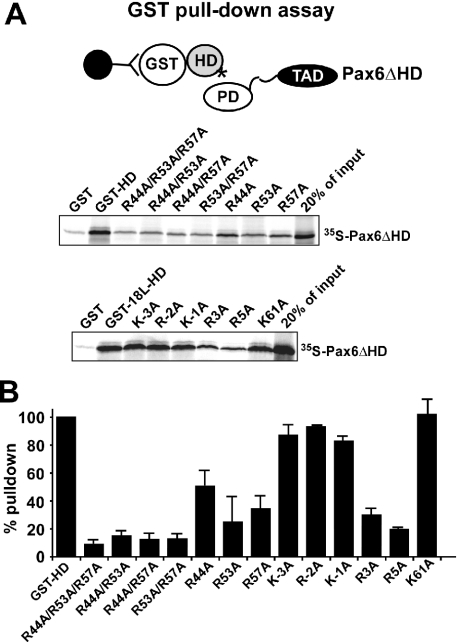
Amino acids in the homeodomain important for the PD–HD interaction
Based on the model for the PD–HD interaction (Figure 2), we mutated arginines 44 and 57 in the HD to alanine. GST pull-down experiments showed that R44, R53 and R57 were all important for the PD–HD interaction (Figure 3). We also found that the K55A mutation reduced this interaction (data not shown). The K61A mutation, affecting the basic residue immediately C-terminal to the HD, had no effect (Figure 3). Single mutations of R44, R53 and R57 to alanine resulted in 25–50% binding compared with the wild-type HD. The R44A mutant tends to interact better than the R53A and the R57A mutants. The combinations of double mutants of R44, R53 and R57 resulted in a further reduction of binding (10–15%), while the triple mutant displayed <10% binding.
Figure 3.
Arginines (R44, R53 and R57) in the recognition helix and N-terminal arm (R3 and R5) of the homeodomain of Pax6 are important for the interaction with the paired domain. GST pull-down assays with Pax6 HD, and HD mutations fused to GST and immobilized on glutathione–agarose beads and in vitro translated Pax6ΔHD. (A) The GST pull-downs were performed as described in the legend to Figure 1. The panel shows two different experiments using two different GST-HD fusions. The GST-HD protein (upper panel) contains two amino acids N-terminal to the HD while the GST-18L-HD (lower panel) contains 18 amino acids of the linker region N-terminal to the HD to study the effect of mutating also at −3 relative to the start of the HD. (B) Quantitative representation of the interaction data. A Fuji Bio-imaging analyzer (BAS5000) equipped with Image Gauge version 4.0 software was used to quantitate 35S-labeled proteins in the SDS–polyacrylamide gels. The amount 35S-labeled Pax6ΔHD pulled down by wild-type GST-HD was set to 100%. The data shown represent the mean of three independent experiments.
Basic residues in the N-terminal arm are known to be involved in protein–DNA interactions of homeodomains (39). As shown in Figure 3, R3 and R5 are both implicated by alanine substitutions as contributors to the PD–HD interaction. Single mutations of the basic residues immediately in front of the HD (KRK at positions −3 to −1) suggest that they do not contribute significantly to this interaction. Taken together, these results show that the arginine residues at positions 3, 5, 44, 53, 57 and 58 (Figure 1C), in addition to the lysine at position 55, are all important for the interaction between Pax6 HD and Pax6 PD. Furthermore, single mutations do not completely impair the interaction between GST-HD and Pax6ΔHD.
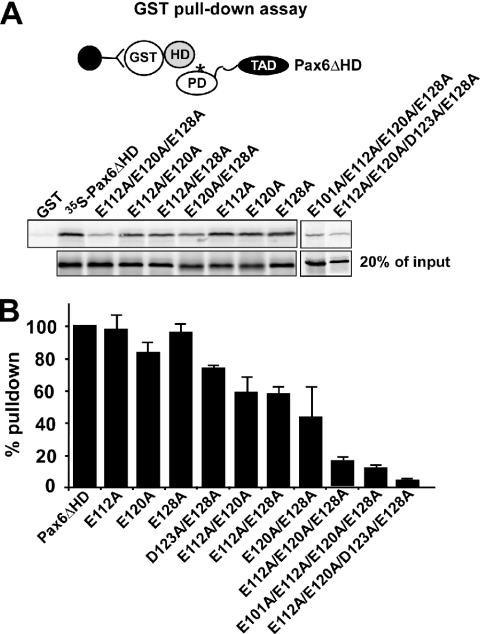
Amino acids in the paired domain important for the PD–HD interaction
The model for the PD–HD interaction implicates five acidic residues in the RED subdomain. We first mutated the glutamate residues at positions 112, 120 and 128 in the PD to alanines. Mutation of single amino acids did not result in any reduced binding between HD and PD in GST pull-down assays (Figure 4). However, when two of the Glu residues were substituted with alanines the interaction was reduced by 40–60%. Furthermore, mutating all three Glu residues resulted in only 15% residual binding (Figure 4). The D123 residue, implicated at the interaction surface in the model in Figure 2, was studied in the context of a D123A/E128A double mutant. The PD–HD interaction was inhibited more by this double mutant than the E128A single mutant suggesting that D123 also contributes to the acidic interaction surface (Figure 4B). The D123 and the E101 residues were studied in the context of two quadruple mutants, including the E112, E120 and E128 residues. The results show that E101 also contributes to the interaction and in the quadruple mutant containing D123A there is almost no residual binding activity (Figure 4). Thus, mutation of one amino acid is not sufficient to compromise the overall acidity of the interaction surface in the RED subdomain of the PD. Alternative interactions formed by one of the neighboring Glu residues probably compensate for the breakage of salt bridges occurring in single mutants. Double and triple mutations result in a substantially lowered acidity as well as breakage of multiple salt bridges. Interestingly, single mutations in the HD do impair the PD–HD interaction while single mutations in the PD do not. Double mutants in the HD give only <20% binding, whereas double mutants in the PD give 50–60% binding. The triple mutant in the PD binds with approximately the same affinity as double mutants in the HD (Figures 3 and 4).
Figure 4.
The acidic residues E101, E112, E120, D123 and E128 in the paired domain are important for the interaction with the homeodomain of Pax6. GST pull-down assays with Pax6 HD fused to GST and immobilized on glutathione–agarose beads and Pax6ΔHD protein produced by in vitro transcription and translation in the presence of [35S]methionine. (A) The single mutants of Pax6ΔHD, E112A, E120A and E128A bind to GST-HD with the same affinity as the wild type. For the double mutants the binding to the homeodomain was reduced by 40–60%. The triple mutant displays only 15% residual binding while the quadruple mutants showed 11 and 5% binding, respectively. The GST pull-downs were performed as described in the legend to Figure 1. (B) Quantitative representation of the interaction data determined as described in the legend to Figure 3. The data shown represent the mean of three independent experiments.
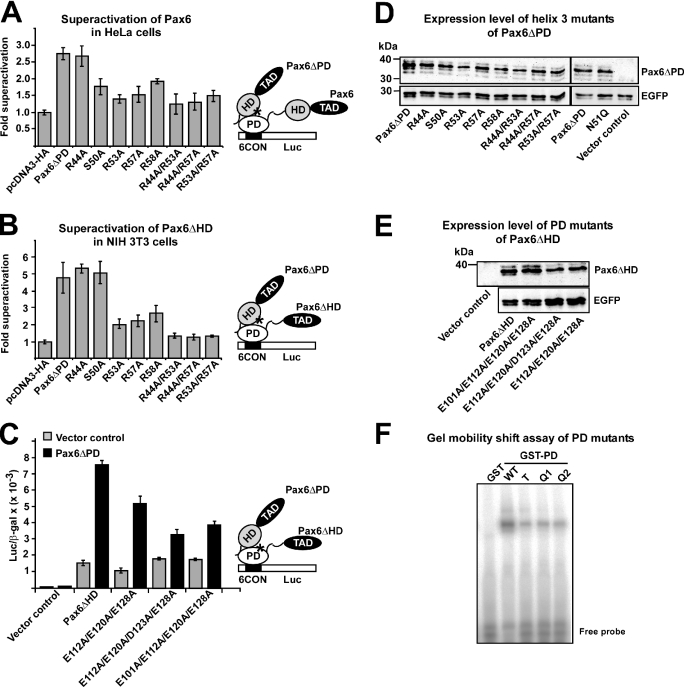
Superactivation of Pax6-mediated transactivation from paired domain-binding sites by Pax6ΔPD is dependent on the integrity of R44, R53, R57 and R58 in helix 3 of the homeodomain
We have previously shown that Pax6ΔPD as well as other homeodomain proteins, such as Rax, Chx10, HoxB1, Pbx1 and Lhx2, can enhance Pax6-mediated transactivation of a minimal promoter containing consensus Pax6 paired domain-binding sites in HeLa cells (21). Thus, we used this assay to test the effect of helix 3 mutations in vivo. HeLa cells were co-transfected with expression vectors for Pax6 and the different Pax6ΔPD constructs with mutations in helix 3 of the HD together with the reporter plasmid pP6CON-LUC (Figure 5A). The pP6CON-LUC reporter contains six consensus Pax6 paired domain binding sites upstream of the adenovirus E1b minimal promoter (12). Pax6ΔPD does not bind to the paired domain binding sites, but is able to significantly enhance (superactivate) Pax6-mediated transcriptional activation of pP6CON-LUC (21). Completely consistent with the results from the GST pull-down assays, the R53A, R57A and R58A mutations resulted in lowered superactivation of Pax6 by Pax6ΔPD (Figure 5A). The R44A mutation did not result in lowered superactivation, probably because of the higher affinity for PD than the other mutants. The S50A mutation resulted in reduced superactivation in spite of no effect on the affinity of GST-HD for Pax6 as observed by GST pull-down assays.
Figure 5.
Mutation of Arg residues in helix 3 of the homeodomain impair superactivation of Pax6-mediated transactivation from paired domain binding sites. (A) Effect of homeodomain mutants on superactivation of Pax6 in HeLa cells. HeLa cells were co-transfected with 0.25 μg of pCI-Pax6 and 0.5 μg of either pcDNA3-HA, HA-Pax6ΔPD or HA-Pax6ΔPD mutants, together with 0.5 μg of the luciferase reporter plasmid pP6CON-LUC and 0.05 μg of the control plasmid pCMVβ-gal. (B) Effect of homeodomain mutants on superactivation of Pax6ΔPD in NIH 3T3 cells. NIH 3T3 cells were co-transfected with 5 ng of Pax6ΔHD and 75 ng of either pcDNA3-HA, Pax6ΔPD or Pax6ΔPD mutants, together with 50 ng pP6CON-LUC and 5 ng pCMV β-gal. (C) Reduced superactivation of the triple and the quadruple paired domain mutants of Pax6ΔHD. NIH 3T3 cells were co-transfected with 50 ng pP6CON-LUC, 5 ng pCMVβ-gal, 5 ng of either pcDNA3-HA, Pax6ΔHD, Pax6ΔHD(E112A/E120A/E128A), Pax6ΔHD(E112A/E120A/D123A/E128A) or Pax6ΔHD(E101A/E112A/E120A/E128A) together with 75 ng pcDNA3-HA or Pax6ΔPD. (D) Western blot showing similar expression levels of wild type and all helix 3 mutants of Pax6ΔPD after transfection of HeLa cells. Transfection efficiencies were probed by co-transfecting an EGFP expression plasmid and developing the blot with an anti-GFP antibody. (E) Western blot showing similar expression levels of wild-type, triple and quadruple mutants in the PD of Pax6ΔHD. (F) Gel mobility shift assay with GST fusions of the PD of Pax6 wild type (WT), triple (T) and quadruple mutants (Q1 = E101A/E112A/E120A/E128A and Q2 = E112A/E120A/D123A/E128A) of Pax6 using a double-stranded oligonucleotide containing a single P6CON PD binding site as probe. The data shown (A–F) are representative of at least two other independent experiments.
In HeLa cells there is a background of endogenous Pax6, and Pax6ΔPD can in principle bind not only to the PD but also to the HD. We, therefore, transfected Pax6ΔHD together with mutated Pax6ΔPD into NIH 3T3 cells that do not express Pax6 (3). Reporter gene assays in NIH 3T3 cells with Pax6ΔHD and the same Pax6ΔPD wild-type and mutant constructs as in Figure 5A gave similar results as with Pax6 in HeLa cells, except for the S50A mutant, which did not give any reduced superactivation in NIH 3T3 cells (Figure 5B). Taken together, the data presented above from the superactivation experiments in cells confirm the in vitro interaction data underscoring the importance of the Arg residues (R44, R53, R57 and R58) in helix 3 of the HD for the HD–PD interaction. None of the mutations introduced in helix 3 affected expression levels and stability of the proteins (Figure 5D).
Having established the importance of the Arg residues in helix 3 of the HD for the HD–PD interaction we next examined the effect of mutating the acidic interaction surface of the RED subdomain of the PD in the superactivation assay. As shown in Figure 5C, the Pax6ΔHD(E112A/E120A/E128A) triple mutant was superactivated by Pax6ΔPD at lower efficiency than the wild-type Pax6ΔHD. For the two quadruple mutants, where the D123A or the E101A mutations are included, superactivation was severely compromised (Figure 5C). These PD mutants were expressed at similar levels as the wild-type Pax6ΔHD protein (Figure 5E). The alanine mutations in the RED subdomain do not impair the structure of the PD since the quadruple mutants transactivate the reporter gene just as well as the wild type. As analyzed by gel mobility shift assays of GST–PD fusion proteins these mutants bind to the P6CON binding site although with somewhat reduced affinity compared with wild type (Figure 5F).
The HD protein Chx10 interacts with the acidic patch of the paired domain of Pax6 via Arg residues in helix 3
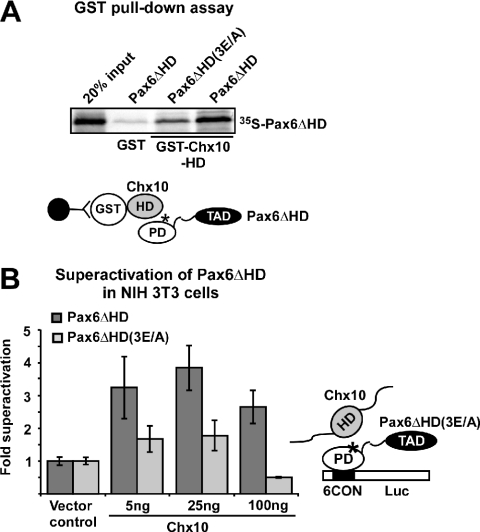
Our finding that several different homeodomain proteins with distinct TADs are able to superactivate Pax6 argues against a common interaction surface outside of the homeodomain in Pax6ΔPD. We, therefore, chose to study the interaction between the HD of the paired-type homeodomain protein Chx10 and the PD of Pax6. Chx10 and Pax6 are co-expressed in early retinogenesis and in the spinal cord (44–46). Chx10 is also able to strongly superactivate Pax6 in HeLa cells (21). As shown in Figure 6A, the HD of Chx10 interacts much more poorly with Pax6ΔHD(E112A/E120A/E128A) than with wild-type Pax6ΔHD in a GST pull-down assay. Consistently, Chx10 was not able to superactivate Pax6ΔHD(E112A/E120A/E128A) in NIH 3T3 cells while co-transfection of Chx10 enhanced wild-type Pax6ΔHD-mediated transactivation of the P6CON-LUC reporter 3.5-fold (Figure 6B). Chx10 did not activate the P6CON-LUC reporter by itself. Thus, Chx10 is clearly dependent on the acidic interaction surface in the RED subdomain to superactivate Pax6ΔPD.
Figure 6.
Reduced superactivation of the paired domain mutant Pax6ΔHD(E112A/E120A/E128A) by the paired-class homeodomain protein Chx10. (A) The Pax6ΔHD(3E/A) triple mutant show reduced binding to the homeodomain of Chx10. GST pull-down assays with the HD of murine Chx10 fused to GST and immobilized on glutathione–agarose beads and Pax6ΔHD and Pax6ΔHD(3E/A) protein produced by in vitro transcription and translation in the presence of [35S]methionine. The GST pull-downs were performed as described in the legend to Figure 1. (B) NIH 3T3 cells were co-transfected with 5 ng of either pcDNA3-HA vector, HA-Pax6ΔHD or HA-Pax6ΔHD(3E/A) expression vectors together with vector control or increasing amounts of HA-Chx10 expression vector (5, 25 and 100 ng). An aliquot of 50 ng of the pP6CON-LUC reporter vector and 5 ng of the CMV βgal vector were used. The data are shown as fold superactivation compared with Pax6ΔHD and empty vector control. The data are representative of two other independent experiments.
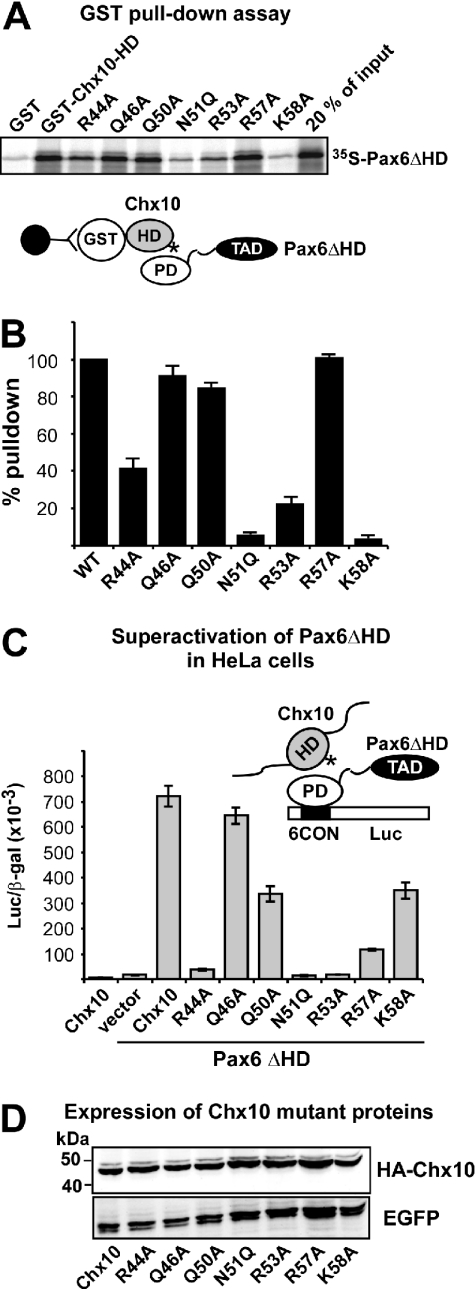
As observed for Pax6, the Chx10 homeodomain mutations, N51Q, R53A and K58A, have profound negative effects on the PD–HD interaction as observed in GST pull-down assays (Figure 7A). The R44A mutation resulted in 50% binding while Q46A and Q50A mutations did not have any significant effects. In contrast to Pax6, the R57A mutation did not reduce the interaction between the Chx10 HD and the Pax6 PD (Figure 7B). Co-transfection of Chx10 and Pax6ΔHD resulted in more than 40-fold superactivation of Pax6ΔHD-mediated transcriptional activation of the P6CON-LUC reporter in HeLa cells (Figure 7C). Chx10 alone is unable to activate P6CON-LUC. The R44A, N51Q and R53A mutations completely abolished superactivation while the R57A mutant showed a strongly reduced superactivation. The Q46A mutant does not affect the superactivation, while the Q50A and K58A mutants resulted in 50% reduction of superactivation. All mutants were expressed at similar levels as wild-type Chx10 (Figure 7D).
Figure 7.
Mutation of basic amino acids in helix 3 of the Chx10 homeodomain leads to reduced interaction with and superactivation by Pax6ΔHD. (A) GST pull-down assays with Chx10 HD wild type and mutants fused to GST and immobilized on glutathione–agarose beads and Pax6ΔHD protein produced by in vitro transcription and translation in the presence of [35S]methionine. (B) Quantitative representation of the interaction data determined as described in the legend to Figure 3. (C) Effects of mutations in the recognition helix of the HD of Chx10 on superactivation of Pax6ΔHD-mediated transactivation from paired domain-binding sites. HeLa cells were co-transfected with 0.5 μg Pax6ΔHD, 0.5 μg pP6CON-LUC and 5 ng pCMV-βgal together with either 0.25 μg pcDNA3-HA vector, HAChx10 or HA-Chx10 mutants. HA-Chx10 co-transfected with the empty Pax6ΔHD control vector shows that Chx10 alone does not activate the P6CON LUC reporter. The data in (B) and (C) represent the mean of three independent experiments. (D) Western blot showing similar expression levels of wild type and all helix 3 mutants of Chx10 following transfection of HeLa cells. EGFP served as transfection control.
The main difference between the helix 3 mutations in Pax6 and Chx10 observed in the superactivation assay is at position 44. The Chx10 R44A mutant shows complete loss of superactivation while the corresponding Pax6 mutation seemingly has no effect. However, as described above, when the Pax6 R44A mutation was combined with R53A or R57A a reduced superactivation was observed compared with the R53A or the R57A single mutants.
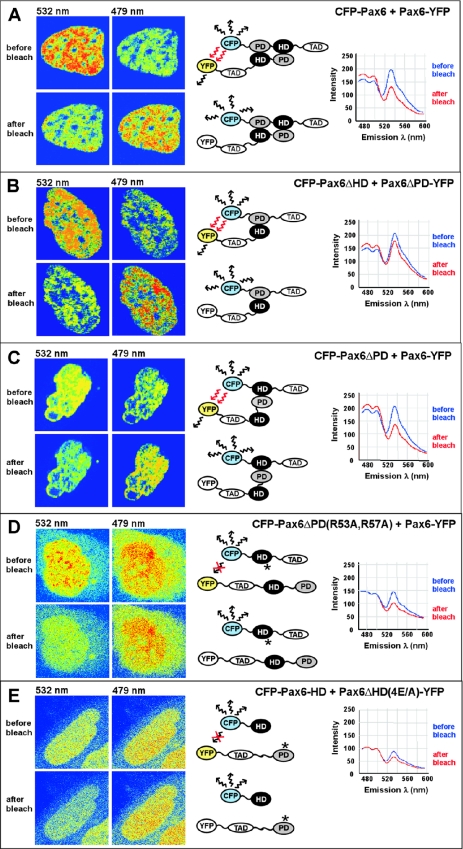
FRET between Pax6 molecules in nuclei of living cells depends on a PD–HD interaction
The results from the superactivation assays are strongly suggestive of a direct interaction between the PD and the HD of Pax6 in the nuclei of transfected cells. To test this directly in the nuclei of living cells we performed FRET experiments using acceptor photobleaching (40,41). With the cyan and yellow variants of enhanced green fluorescent protein (CFP and YFP, respectively), as donor and acceptor pair, FRET occurs only within a distance of 100 Å (47). We co-transfected expression constructs for different CFP and YFP fusions of Pax6 into HeLa cells and performed acceptor photo bleaching experiments of ROIs encompassing the nuclei of cells expressing the donor and the acceptor fluorophores at similar levels. If the CFP and the YFP fusion proteins are in close proximity to allow FRET the CFP fluorescence increases in the region where YFP is bleached. As shown in Figure 8A, FRET was readily observed in the entire nuclei of cells expressing CFP and YFP fusions of full-length Pax6 (CFP-Pax6 and Pax6-YFP). FRET was also seen between CFP-Pax6ΔHD and Pax6ΔPD-YFP and between CFP-Pax6ΔPD and Pax6-YFP (Figure 8B and C). However, no FRET could be observed when CFP-Pax6ΔPD(R53A,R57A) carrying mutations in two arginine residues in helix 3 of the HD, which are crucially involved in the PD–HD interaction, was co-transfected with Pax6-YFP (Figure 8D). The same was the case when Pax6ΔHD(4E/A)-YFP, with four of the five glutamate residues of the acidic interaction surface of the RED subdomain of the PD substituted with alanines, was co-transfected with CFP-Pax6-HD (Figure 8E). Taken together, the results from the FRET experiments strongly support the existence of a PD–HD inter-molecular interaction between Pax6 molecules in living cell nuclei involving basic residues in helix 3 and the acidic patch of the RED subdomain of the PD.
Figure 8.
The paired domain and homeodomain of Pax6 interact directly with each other, as analyzed by using FRET. The CFP- and YFP-tagged Pax6 proteins indicated in (A–E) were co-expressed in human HeLa cells. The cell images are visualized in pseudocolors before and after acceptor photobleaching to highlight changes in fluorescence intensity. Images of CFP and YFP fluorescence were obtained using the 458 nm laser line. The YFP acceptor was bleached in the whole nucleus using the 514 nm laser line. FRET was detected as decreased YFP-emission at 532 nm and a corresponding increased CFP-emission at 479 nm in the bleached area. (A) Full-length Pax6 proteins homodimerize in the nucleus of living cells. (B) The paired domain of Pax6ΔHD interacts directly with the homeodomain of the paired-less isoform Pax6ΔPD. (C) FRET analysis demonstrates interactions between the homeodomain of Pax6ΔPD and the paired domain of the full-length Pax6 isoform. (D) Mutations of arginine residues in helix 3 of the homeodomain of Pax6 impair the interaction with the paired domain, as demonstrated by no visible FRET between the HD double mutant R53A/R57A of Pax6ΔPD and the PD of full-length Pax6. (E) No FRET was detected between CFP and YFP in cells expressing a quadruple mutation in the paired domain of Pax6ΔHD-E101A/E112A/E120A/E128A and the wild-type homeodomain of Pax6. Cells expressing approximately a 1:1 ratio of the CFP- and YFP-tagged proteins were used for FRET experiments.
DNA binding of homeodomain mutants
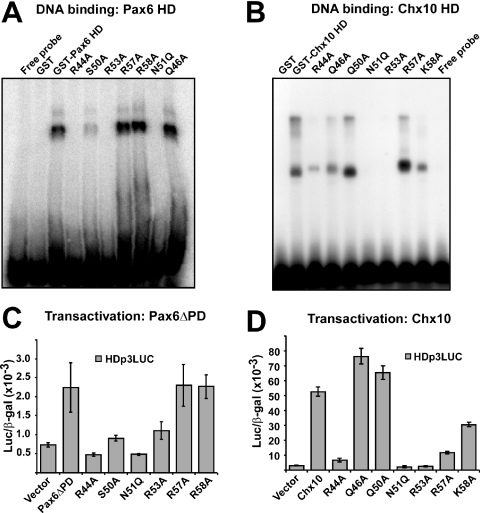
We next tested the DNA-binding activity of mutants in helix 3 of both Pax6 and Chx10 using gel mobility shift assays (Figure 9A and B). As previously reported for the paired class HD protein Phox1 the N51Q mutant did not bind DNA (42). Neither did the R53A mutant of both Pax6 and Chx10 while the R44A mutant bound weakly (Chx10) or not at all (Pax6) to the HDp3 probe (Figure 9A and B). A faint band was observed for the S50A mutant of Pax6 whereas the corresponding Q50A mutant of Chx10 bound strongly. For the basic residues at positions 57 and 58, the mutants bound strongly to DNA with K58A of Chx10 binding more weakly than the others. As can be suspected from the different mobilities of the protein–DNA complexes in Figure 9A and B, we found that, contrary to Pax6, Chx10 did not bind to the palindromic DNA-binding site as a dimer but bound as a monomer (data not shown).
Figure 9.
DNA binding and transactivation from homeodomain-binding sites of recognition helix mutants of Pax6 and Chx10. (A) Gel mobility shift assay of GST-Pax6 wild-type and helix 3 mutants. Equal amounts of GST-Pax6 HD and HD mutants were used in gel mobility shift assays as described in Materials and Methods. The Q46A mutant was included as a control of a mutant that does not affect protein–protein interaction. (B) Gel mobility shift assay of GST-Chx10 HD and HD mutants. (C) Transcriptional activation of Pax6ΔPD wild-type and HD mutants from the HDp3 homeodomain-binding site in the reporter HDp3LUC. HeLa cells were co-transfected with 0.5 μg HDp3LUC reporter, 50 ng of the control plasmid pCMV β-gal and either 0.5 μg of expression vector for HA-Pax6ΔPD wild-type, HD mutants or the empty pcDNA3-HA vector. Pax6ΔPD gives >2-fold transactivation of the HDp3LUC reporter. (D) Transcriptional activation of Chx10 wild-type and HD mutants from the HDp3 site. HeLa cells were co-transfected as in (C) except that 0.5 μg of expression vectors for HA-Chx10 or Chx10 HD mutants were used instead of expression vectors for HA-Pax6ΔPD. Chx10 shows 18-fold transactivation of the HDp3LUC reporter. The data in (A–D) are representative of two other independent experiments.
We also analyzed transactivation of the HDp3LUC reporter by mutants of Pax6ΔPD and Chx10 after transient transfection of HeLa cells (Figure 9C and D). Pax6ΔPD activated the HDp3LUC reporter >2-fold in co-transfection experiments in HeLa cells (Figure 9C). The same was the case for the DNA-binding R57A and R58A mutants whereas the R44A, S50A, N51Q and R53A mutants did not show any significant transactivation. Of these, only S50A showed some residual DNA-binding activity. These results correlate very well with the DNA-binding results.
Chx10 was able to activate the HDp3LUC reporter >15-fold when co-transfected in HeLa cells (Figure 9D). As found for Pax6, R44A, N51Q and R53A were completely unable to transactivate the reporter gene. The R57A and K58A mutants activated the reporter 4- and 10-fold, respectively (Figure 9D). The R57A mutant bound strongly to the HDp3 site whereas the K58A mutant bound a bit more weakly than the wild-type Chx10 HD. Although the same amount of protein was used in all gel mobility shift assays differential stability of the different mutant proteins could possibly affect the result. Thus, some mutants give stronger signals than the wild-type GST-HD fusion proteins.
DISCUSSION
We have now shown that the DNA recognition helices of the homeodomains of Pax6 and Chx10 are involved in mediating binding to the RED subdomain of the paired domain of Pax6. This is a novel mechanism for regulating the activity of Pax6 that can operate both intra- and inter-molecularly. In the latter case, both the paired-less isoform of Pax6 itself and other co-expressed homeodomain proteins may contribute to the regulation of gene expression by binding to the paired domain of full-length Pax6. Through this interaction Pax6 may also inhibit DNA binding by other homeodomain proteins. By combining alanine-substitution mutagenesis and molecular modeling with protein–protein interaction assays we are able to propose a model where the PD–HD interaction is largely mediated by electrostatic forces between basic residues in helix 3 of the HD and acidic residues in helices 1 and 2 of the RED subdomain of the PD. As summarized in Figure 10, the basic residues at positions 57 and 58 of homeodomain helix 3 of Pax6 are important for protein–protein interaction, but not for DNA binding. For Chx10, both the R57A and K58A mutants bound DNA. The K58A mutation reduced superactivation to 50% and showed very weak interaction in the GST pull-down assay while R57A showed strongly reduced superactivation but bound as wild type in GST pull-down assays. The basic residues R44, R53, K55, R57 and R58 of Pax6 lie on one side of helix 3 (Figures 2 and 10B). R53 is important for both DNA binding and protein–protein interaction. R44 is important for DNA binding and the R44A mutants also show some reduction in protein–protein interaction. This is consistent with both residues being involved in forming phosphate backbone contacts upon binding to DNA (39). N51 of the HD is involved in base-specific contacts in the major groove of the DNA (39). To our surprise, the N51Q mutant not only abolished DNA binding but also protein–protein interactions (Figure 10A). In contrast, the N51A mutant did not affect protein–protein interactions (data not shown and Figure 10B). Thus, steric hindrance owing to a longer side chain can explain why the PD–HD interaction is lost in the N51Q mutant.
Figure 10.
Summary of the behavior of point mutants in the recognition helix of Pax6 and Chx10 HD in protein binding (PD–HD interaction), superactivation and DNA-binding assays. (A) The results from GST pull-down assays of PD–HD interactions (see Figures 1, 3 and 7), superactivation of Pax6ΔHD-mediated transactivation of the P6CONLUC reporter (see Figures 5B and 7C) and gel mobility shift assays of DNA binding (see Figure 9A and B) are summarized. Note that except for the N51Q mutant all the other mutants are alanine substitutions. The results are scored as follows: +++, no reduction compared with wild type; ++, reduced; +, strongly reduced; −, no binding or superactivation; n.d., not determined. (B) The recognition helix of the Pax6 HD is shown with the side chains of the relevant amino acids indicated. Below, the sequence of this helix is shown with asterisks indicating positions where alanine substitutions negatively affect the PD–HD interaction and/or DNA binding. Note that the N51A mutation does not affect the PD–HD interaction in contrast to the N51Q mutation shown in (A). Open squares, no effect of alanine substitutions. The arrowhead indicates the location of the N-terminal end of helix 3.
In addition to basic residues in helix 3, the arginines at position 3 and 5 in the N-terminal arm of the HD are also involved in binding to the PD. Molecular modeling suggest that R3 may interact with E128 and R5 to E112 in the RED subdomain. In the crystal structure of the Drosophila Prd cooperative HD dimer bound to DNA both these residues are involved in DNA binding. R3 is also involved in an inter-molecular electrostatic interaction with E42 of helix 3 in the other HD (39).
Our results suggest the presence of an acidic interaction surface in the RED subdomain of the Pax6 PD consisting of the five residues E101, E112, E120, D123 and E128. When four of these residues were substituted with alanines FRET was abolished and superactivation severely inhibited. Pax6ΔHD with the triple mutation E112A/E120A/E128A in the PD still showed a significant superactivation in transient transfection assays. We also observed a weak FRET with this mutant fused to YFP and tested against CFP-Pax6 (data not shown) whereas Pax6 constructs with four of the acidic residues mutated did not display any FRET (Figure 8E).
Directly relevant to our mapping of an interaction surface in the RED subdomain is the reported binding of the basic helix–loop–helix transcription factor microphthalmia (Mitf) to the RED subdomain of Pax6 (25). A triple mutant of quail Pax6, corresponding to zebrafish S115A, F117N and E120Q, resulted in a strongly reduced interaction with Mitf. Interestingly, we have implicated E120 as part of the interaction surface in the PD–HD interaction. In our model the side chain OH group of S115 interacts with the backbone CO group of A54 in the HD and the side chain of F117 is close to the salt bridge between R57 and E120. We found that the S115A/F117N/E120Q triple mutant behaved as the double mutants E112A/E120A and E112A/E128A in GST pull-down assays (data not shown). Thus, the interaction surfaces in the RED subdomain used by the homeodomain and Mitf are at least partially overlapping.
While most HDs bind DNA as a monomer, the paired class HDs bind cooperatively as homodimers to palindromic DNA-binding sites (16,39). The HDp3 site contains two palindromic TAAT half sites separated by three nucleotides (TAATGCGATTA). Surprisingly, we found that Chx10 binds as a monomer to the HDp3-binding site (Figure 9B and data not shown). A binding site selection assay for Chx10 DNA-binding sites yielded an 8 bp consensus, TAATTAGC (48). This binding site differs from that of other paired class HDs in having only one central ATTA motif. This supports our finding of monomeric binding to the HDp3 motif.
A few residues in helix 3 of other HD proteins have been implicated in protein–protein interactions. The Axenfeld–Rieger Syndrome mutation, K50E, in the HD of PITX2a leads to increased dimerization of PITX2a in solution (49). In our case the S50A mutation did not affect the PD–HD interaction of Pax6. The Csx/Nkx2.5 homeodomain protein provides another example of a HD protein that is able to homodimerize on DNA (and in solution). K57 and R58 near the C-terminal end of helix 3 are critical for the dimerization (50). K57 was also found to be important for the interaction with the zinc finger transcription factor GATA 4 (50). Our observation that R57 and R58 are important for the PD–HD interaction of Pax6 underscores the role these residues have in protein–protein interactions. A recent report demonstrates that geminin, a protein involved in the inhibition of the licensing of DNA replication, interacts with several Hox proteins in yeast two-hybrid and GST pull-down experiments (51). Using a Hoxa11 peptide array, amino acids 54–58 in addition to the first amino acids of the HD were found to be important for this interaction. Thus, similar to our findings, both helix 3 and the N-terminal arm of the HD are implicated in a specific protein–protein interaction.
The biological relevance of the interaction studied here is as a mechanism for intra- and inter-molecular modulation of Pax6-mediated gene regulation. Evidence for an interdependence of the functionality of the PD and HD of both Pax6 and Pax3 has been reported (20,21,52–55). This interdependency could at least to a certain extent be a reflection of an intra-molecular PD–HD interaction. Additionally, co-expressed homeodomain proteins may modulate Pax6-mediated gene regulation by binding to the RED subdomain of DNA-bound Pax6 without binding to DNA themselves. Pax6 may inhibit the DNA-binding activity of co-expressed HD proteins by binding to their recognition helix as recently shown for the geminin–Hoxa11 interaction (51). In line with this notion experiments where an oligonucleotide containing the HDp3-binding site was added in increasing amounts to GST pull-down assays revealed competition between GST-HD binding to Pax6ΔHD or to the DNA-binding site (J. A. Bruun, E. I. S. Thomassen, T. Holm and T. Johansen, unpublished data). In a very recent paper Pax6 was shown to interact with the homeodomain protein Vax and with Tbx5 to establish the dorsoventral boundary of the developing eye (56). Very interestingly, Vax binds to the PD and HD of Pax6 and represses Pax6 transactivation. Furthermore, overexpression of either the PD or the HD of Pax6 is sufficient to dorsalize the eye in chicken suggesting that protein–protein interactions are involved in eye dorsalization (56). Thus, the PD–HD interaction we have characterized is highly relevant to mechanisms involved in determining gene expression boundaries. In conclusion, our studies show that helix 3 in the HD of Pax6 and other paired class HD proteins can act as a specific recognition helix for both DNA- and protein-binding.
Acknowledgments
This work was supported by grants from the ‘Top research programme’ of the Norwegian Research Council, the Norwegian Cancer Society, the Aakre Foundation, Simon Fougner Hartmanns Familiefond and the Blix Foundation to T.J. I.M. holds a grant from the Norwegian Research Council and K.K. has a postdoctoral functional genomics grant from the University of Tromsø. J.A.B. was, and E.I.S.T. is, a fellow of the Norwegian Cancer Society. Funding to pay the Open Access publication charges for this article was provided by the Norwegian Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Noll M. Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- 2.Strachan T., Read A.P. PAX genes. Curr. Opin. Genet. Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 3.Tang H.K., Singh S., Saunders G.F. Dissection of the transactivation function of the transcription factor encoded by the eye developmental gene PAX6. J. Biol. Chem. 1998;273:7210–7221. doi: 10.1074/jbc.273.13.7210. [DOI] [PubMed] [Google Scholar]
- 4.Carriere C., Plaza S., Caboche J., Dozier C., Bailly M., Martin P., Saule S. Nuclear localization signals, DNA binding, and transactivation properties of quail Pax-6 (Pax-QNR) isoforms. Cell. Growth Differ. 1995;6:1531–1540. [PubMed] [Google Scholar]
- 5.Czerny T., Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol. Cell. Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser T., Jepeal L., Edwards J.G., Young S.R., Favor J., Maas R.L. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nature Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 7.Mikkola I., Bruun J.A., Bjørkøy G., Holm T., Johansen T. Phosphorylation of the transactivation domain of Pax6 by extracellular signal-regulated kinase and p38 mitogen-activated protein kinase. J. Biol. Chem. 1999;274:15115–15126. doi: 10.1074/jbc.274.21.15115. [DOI] [PubMed] [Google Scholar]
- 8.Callaerts P., Halder G., Gehring W.J. Pax-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 9.Walther C., Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell T.N., Free S.L., Williamson K.A., Stevens J.M., Churchill A.J., Hanson I.M., Shorvon S.D., Moore A.T., Van Heyningen V., Sisodiya S.M. Polymicrogyria and absence of pineal gland due to PAX6 mutation. Ann. Neurol. 2003;53:658–663. doi: 10.1002/ana.10576. [DOI] [PubMed] [Google Scholar]
- 11.Halder G., Callaerts P., Gehring W.J. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- 12.Nornes S., Clarkson M., Mikkola I., Pedersen M., Bardsley A., Martinez J.P., Krauss S., Johansen T. Zebrafish contains two Pax6 genes involved in eye development. Mech. Dev. 1998;77:185–196. doi: 10.1016/s0925-4773(98)00156-7. [DOI] [PubMed] [Google Scholar]
- 13.Chow R.L., Altmann C.R., Lang R.A., Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- 14.Hill R.E., Favor J., Hogan B.L., Ton C.C., Saunders G.F., Hanson I.M., Prosser J., Jordan T., Hastie N.D., van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 15.van Heyningen V., Williamson K.A. PAX6 in sensory development. Hum. Mol. Genet. 2002;11:1161–1167. doi: 10.1093/hmg/11.10.1161. [DOI] [PubMed] [Google Scholar]
- 16.Wilson D., Sheng G., Lecuit T., Dostatni N., Desplan C. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 17.Epstein J.A., Glaser T., Cai J., Jepeal L., Walton D.S., Maas R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- 18.Carriere C., Plaza S., Martin P., Quatannens B., Bailly M., Stehelin D., Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol. Cell. Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaworski C., Sperbeck S., Graham C., Wistow G. Alternative splicing of Pax6 in bovine eye and evolutionary conservation of intron sequences. Biochem. Biophys. Res. Commun. 1997;240:196–202. doi: 10.1006/bbrc.1997.7623. [DOI] [PubMed] [Google Scholar]
- 20.Mishra R., Gorlov I.P., Chao L.Y., Singh S., Saunders G.F. PAX6, paired domain influences sequence recognition by the homeodomain. J. Biol. Chem. 2002;277:49488–49494. doi: 10.1074/jbc.M206478200. [DOI] [PubMed] [Google Scholar]
- 21.Mikkola I., Bruun J.A., Holm T., Johansen T. Superactivation of Pax6-mediated transactivation from paired domain-binding sites by DNA-independent recruitment of different homeodomain proteins. J. Biol. Chem. 2001;276:4109–4118. doi: 10.1074/jbc.M008882200. [DOI] [PubMed] [Google Scholar]
- 22.Plaza S., Langlois M.C., Turque N., LeCornet S., Bailly M., Begue A., Quatannens B., Dozier C., Saule S. The homeobox-containing Engrailed (En-1) product down-regulates the expression of Pax-6 through a DNA binding-independent mechanism. Cell Growth Differ. 1997;8:1115–1125. [PubMed] [Google Scholar]
- 23.Ritz-Laser B., Estreicher A., Klages N., Saule S., Philippe J. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G1 control element. J. Biol. Chem. 1999;274:4124–4132. doi: 10.1074/jbc.274.7.4124. [DOI] [PubMed] [Google Scholar]
- 24.Hussain M.A., Habener J.F. Glucagon gene transcription activation mediated by synergistic interactions of pax-6 and cdx-2 with the p300 co-activator. J. Biol. Chem. 1999;274:28950–28957. doi: 10.1074/jbc.274.41.28950. [DOI] [PubMed] [Google Scholar]
- 25.Planque N., Leconte L., Coquelle F.M., Martin P., Saule S. Specific Pax-6/microphthalmia transcription factor interactions involve their DNA-binding domains and inhibit transcriptional properties of both proteins. J. Biol. Chem. 2001;276:29330–29337. doi: 10.1074/jbc.M101812200. [DOI] [PubMed] [Google Scholar]
- 26.Nutt S.L., Morrison A.M., Dorfler P., Rolink A., Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 1998;17:2319–2333. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maitra S., Atchison M. BSAP can repress enhancer activity by targeting PU.1 function. Mol. Cell. Biol. 2000;20:1911–1922. doi: 10.1128/mcb.20.6.1911-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Hu G., Wang H., Sciavolino P., Iler N., Shen M.M., Abate-Shen C. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol. Cell. Biol. 1997;17:2920–2932. doi: 10.1128/mcb.17.5.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendall A.J., Ding J., Hu G., Shen M.M., Abate-Shen C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]
- 30.Wiggan O., Taniguchi-Sidle A., Hamel P.A. Interaction of the pRB-family proteins with factors containing paired-like homeodomains. Oncogene. 1998;16:227–236. doi: 10.1038/sj.onc.1201534. [DOI] [PubMed] [Google Scholar]
- 31.Cvekl A., Kashanchi F., Brady J.N., Piatigorsky J. Pax-6 interactions with TATA-box-binding protein and retinoblastoma protein. Invest. Ophthalmol. Vis. Sci. 1999;40:1343–1350. [PubMed] [Google Scholar]
- 32.Eberhard D., Busslinger M. The partial homeodomain of the transcription factor Pax-5 (BSAP) is an interaction motif for the retinoblastoma and TATA-binding proteins. Cancer Res. 1999;59:1716–1724. [PubMed] [Google Scholar]
- 33.Krauss S., Johansen T., Korzh V., Moens U., Ericson J.U., Fjose A. Zebrafish pax[zf-a]: a paired box-containing gene expressed in the neural tube. EMBO J. 1991;10:3609–3619. doi: 10.1002/j.1460-2075.1991.tb04927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seth A., Gonzalez F.A., Gupta S., Raden D.L., Davis R.J. Signal transduction within the nucleus by mitogen-activated protein kinase. J. Biol. Chem. 1992;267:24796–24804. [PubMed] [Google Scholar]
- 35.Simpson J.C., Wellenreuther R., Poustka A., Pepperkok R., Wiemann S. Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing. EMBO Rep. 2000;1:287–292. doi: 10.1093/embo-reports/kvd058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang C., Gonzalez F., Rothermel B., Sun L., Johnston S.A., Kodadek T. The Gal4 activation domain binds Sug2 protein, a proteasome component, in vivo and in vitro. J. Biol. Chem. 2001;276:30956–30963. doi: 10.1074/jbc.M102254200. [DOI] [PubMed] [Google Scholar]
- 37.Abagyan R., Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 38.Xu H.E., Rould M.A., Xu W., Epstein J.A., Maas R.L., Pabo C.O. Crystal structure of the human Pax6 paired domain–DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson D.S., Guenther B., Desplan C., Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 40.Karpova T.S., Baumann C.T., He L., Wu X., Grammer A., Lipsky P., Hager G.L., McNally J.G. Fluorescence resonance energy transfer from cyan to yellow fluorescent protein detected by acceptor photobleaching using confocal microscopy and a single laser. J. Microsc. 2003;209:56–70. doi: 10.1046/j.1365-2818.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- 41.Vermeer J.E., Van Munster E.B., Vischer N.O., Gadella T.W., Jr Probing plasma membrane microdomains in cowpea protoplasts using lipidated GFP-fusion proteins and multimode FRET microscopy. J. Microsc. 2004;214:190–200. doi: 10.1111/j.0022-2720.2004.01318.x. [DOI] [PubMed] [Google Scholar]
- 42.Simon K.J., Grueneberg D.A., Gilman M. Protein and DNA contact surfaces that mediate the selective action of the Phox1 homeodomain at the c-fos serum response element. Mol. Cell. Biol. 1997;17:6653–6662. doi: 10.1128/mcb.17.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aronheim A., Zandi E., Hennemann H., Elledge S.J., Karin M. Isolation of an AP-1 repressor by a novel method for detecting protein–protein interactions. Mol. Cell. Biol. 1997;17:3094–3102. doi: 10.1128/mcb.17.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu I.S., Chen J.D., Ploder L., Vidgen D., van der Kooy D., Kalnins V.I., McInnes R.R. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 45.Belecky-Adams T., Tomarev S., Li H.S., Ploder L., McInnes R.R., Sundin O., Adler R. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest. Ophthalmol. Vis. Sci. 1997;38:1293–1303. [PubMed] [Google Scholar]
- 46.Marquardt T., Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 47.Patterson G.H., Piston D.W., Barisas B.G. Forster distances between green fluorescent protein pairs. Anal. Biochem. 2000;284:438–440. doi: 10.1006/abio.2000.4708. [DOI] [PubMed] [Google Scholar]
- 48.Ferda Percin E., Ploder L.A., Yu J.J., Arici K., Horsford D.J., Rutherford A., Bapat B., Cox D.W., Duncan A.M., Kalnins V.I., et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nature Genet. 2000;25:397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- 49.Saadi I., Semina E.V., Amendt B.A., Harris D.J., Murphy K.P., Murray J.C., Russo A.F. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J. Biol. Chem. 2001;276:23034–23041. doi: 10.1074/jbc.M008592200. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara H., Usheva A., Ueyama T., Aoki H., Horikoshi N., Izumo S. Characterization of homo- and heterodimerization of cardiac Csx/Nkx2.5 homeoprotein. J. Biol. Chem. 2001;276:4570–4580. doi: 10.1074/jbc.M004995200. [DOI] [PubMed] [Google Scholar]
- 51.Luo L., Yang X., Takihara Y., Knoetgen H., Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 52.Singh S., Stellrecht C.M., Tang H.K., Saunders G.F. Modulation of PAX6 homeodomain function by the paired domain. J. Biol. Chem. 2000;275:17306–17313. doi: 10.1074/jbc.M000359200. [DOI] [PubMed] [Google Scholar]
- 53.Underhill D.A., Gros P. The paired-domain regulates DNA binding by the homeodomain within the intact Pax-3 protein. J. Biol. Chem. 1997;272:14175–14182. doi: 10.1074/jbc.272.22.14175. [DOI] [PubMed] [Google Scholar]
- 54.Fortin A.S., Underhill D.A., Gros P. Helix 2 of the paired domain plays a key role in the regulation of DNA-binding by the Pax-3 homeodomain. Nucleic Acids Res. 1998;26:4574–4581. doi: 10.1093/nar/26.20.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apuzzo S., Abdelhakim A., Fortin A.S., Gros P. Crosstalk between the paired domain and the homeodomain of PAX3: DNA binding by each domain causes a structural change in the other domain, supporting interdependence for DNA binding. J. Biol. Chem. 2004;279:33601–33612. doi: 10.1074/jbc.M402949200. [DOI] [PubMed] [Google Scholar]
- 56.Leconte L., Lecoin L., Martin P., Saule S. Pax6 interacts with cVax and Tbx5 to establish the dorsoventral boundary of the developing eye. J. Biol. Chem. 2004;279:47272–47277. doi: 10.1074/jbc.M406624200. [DOI] [PubMed] [Google Scholar]