Abstract
Methane is a potent greenhouse gas that contributes significantly to climate change and is primarily regulated in Nature by methanotrophic bacteria, which consume methane gas as their source of energy and carbon, first by oxidizing it to methanol. The direct oxidation of methane to methanol is a chemically difficult transformation, accomplished in methanotrophs by complex methane monooxygenase (MMO) enzyme systems. These enzymes use iron or copper metallocofactors and have been the subject of detailed investigation. While the structure, function, and active site architecture of the copper-dependent particulate methane monooxygenase (pMMO) have been investigated extensively, its putative quaternary interactions, regulation, requisite cofactors, and mechanism remain enigmatic. The iron-dependent soluble methane monooxygenase (sMMO) has been characterized biochemically, structurally, spectroscopically, and, for the most part, mechanistically. Here, we review the history of MMO research, focusing on recent developments and providing an outlook for future directions of the field. Engineered biological catalysis systems and bioinspired synthetic catalysts may continue to emerge along with a deeper understanding of the molecular mechanisms of biological methane oxidation. Harnessing the power of these enzymes will necessitate combined efforts in biochemistry, structural biology, inorganic chemistry, microbiology, computational biology, and engineering.
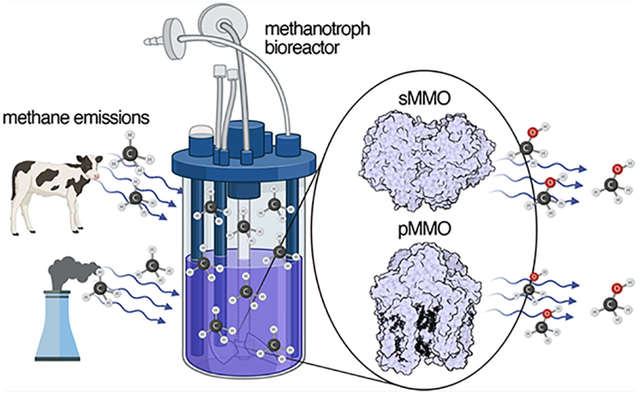
Graphical Abstract
1. INTRODUCTION
Methane is the second most abundant greenhouse gas next to carbon dioxide and has a global warming potential 84 times that of carbon dioxide over a 20-year period.1 Atmospheric methane levels have increased rapidly in recent years with global methane emissions for 2008–2017 being 576 teragrams (Tg) yr−1 (1 Tg = 1 million metric tons), exceeding those of the previous decade by 29 Tg yr−1.2 The largest yearly increase in atmospheric methane since recording began in 1983 was ~17 ppb in 2021.3,4 Approximately 60% of these record methane emissions are anthropogenic, attributable to fossil fuel production and use, livestock, rice cultivation, landfills and wastewater, and biomass burning.2,4 Of particularly high profile are frequent instances of methane leakage from oil and natural gas (composed primarily of methane) harvesting and handling systems. These increases put Earth on track for global temperature increases of >3 °C by the end of the century.2 Methane is removed from the atmosphere primarily via reaction with hydroxyl radicals to form carbon dioxide and water. Because of its short perturbation lifetime (how long it takes to decay back to the original level after an emissions increase) of ~12 years, reducing methane emissions would have an immediate impact on climate change.1,5,6
Conversion of methane to liquid fuels and chemicals would couple mitigating climate change with meeting rising energy demands, but gas-to-liquid conversion processes require steam reforming of methane to syngas (a mixture of carbon monoxide and hydrogen) followed by conversion to methanol or long chain hydrocarbons via Fischer-Tropsch synthesis. These indirect, technically demanding processes are carried out in large scale facilities and entail significant capital and operating expenses.7,8 Direct conversion of methane to methanol is highly desirable, since methanol is used to generate the gasoline additive methyl tert-butyl ether, for substitution into the gasoline pool and as a feedstock for production of olefins, formaldehyde, and acetic acid.9 However, development of high yield homogeneous or heterogeneous catalysts for direct methane conversion is challenging for two reasons.10 First, methane has an unusually high C–H bond strength of 105 kcal/mol, rendering it less reactive than other alkanes.11 Second, methanol is more reactive than methane and thus prone to further oxidation to CO2. As such, direct methane oxidation has been referred to as one of the “Holy Grails” of catalysis.12
An alternative approach to homogeneous and heterogeneous catalysis is biological catalysis using microorganisms or their isolated enzymes to oxidize methane to methanol under ambient conditions. Microbial oxidation of methane occurs in both aerobic and anaerobic environments. Aerobic methane oxidation is performed by methanotrophs,13 bacteria that consume ~30 Tg yr−1 of atmospheric methane.2,14 Methanotrophs convert methane to methanol in the first step of their metabolic pathway using methane monooxygenase (MMO) enzymes, which react with methane and dioxygen to form methanol and water. Two evolutionarily distinct MMOs can catalyze this chemically difficult reaction: a soluble enzyme (sMMO) that uses a dinuclear iron catalytic site and a membrane-bound or particulate enzyme (pMMO) whose activity is dependent on copper.15–17 In between aerobic and anaerobic methane oxidation is “intra-aerobic” methane oxidation carried out by the bacterial phylum NC10. These bacteria couple oxygen generation by nitrite reduction to methane oxidation by the pMMO system.18,19 Finally, entirely anaerobic methane oxidation occurs in anaerobic methanotrophic archaea (ANME) via reverse methanogenesis with sulfate, nitrate, or metal ions as electron acceptors.20 In contrast to aerobic methanotrophs, ANME and NC10 bacteria have not been isolated in pure culture, precluding biochemical studies. These anaerobic methane-oxidizing microbes also play a major role in offsetting methane emissions from soil and marine environments.21,22
In this review, we focus on the enzymatic oxidation of methane by aerobic methanotrophs. Recent reviews have addressed methanotroph physiology, engineering, and applications23–27 and the biochemistry, structure, and spectroscopy of MMOs.15–17,28,29 Here we address both the biology and chemistry of MMOs, spanning the history of MMO research, while highlighting recent developments and providing an outlook on unresolved questions. Progress toward understanding the molecular complexity of MMOs has required the use of a diverse scientific toolbox, involving methods in biochemistry, molecular biology, computational inorganic chemistry, spectroscopy, and structural biology. Structural approaches, in particular, have paved the way toward understanding biological methane oxidation, with recent applications of state-of-the-art methods, including cryoelectron microscopy (cryoEM) and X-ray free electron laser (XFEL) crystallography. We first address the ecology and biology of methanotrophs, focusing on the central role of copper in their physiology. We then review pMMO, addressing its molecular structure, metal centers, activity, active site, and proposed protein interaction partners. Finally, we discuss the structure, mechanism, and protein component interactions of sMMO. We also cover recent progress toward engineering both MMOs, which will be essential to their deployment in climate bioremediation and biological gas-to-liquid conversion processes.
2. BIOLOGY OF METHANOTROPHS
2.1. Taxonomy and Metabolism
Methanotrophs are gram-negative bacteria that live on methane gas as their source of carbon and energy. They were first reported in 1906,30 but initial characterization did not happen until 50 years later with the isolation of Pseudomonas (Methylomonas) methanica,31 Methanomonas methanoooxidans,32,33 and Methylococcus capsulatus,34 which would become a workhorse strain for studies of methanotroph biochemistry. It was also established early on that the oxygen atom in methanol derives from dioxygen,35,36 setting the stage for studies of MMO reaction chemistry. Methanotrophs were subsequently classified into types I, II, and X (a subset of type I) on the basis of their metabolic pathways, membrane lipid contents, cell morphologies, 16s rRNA sequences, and genomic characteristics,37,38 with multiple revisions over the years.39–41 All methanotrophs were long thought to be obligate, meaning that they can only live on one-carbon sources (primarily methane, but also possibly methanol, formate, formaldehyde and methylamines), but facultative methanotrophs that utilize multicarbon substrates such as acetate or ethanol have been isolated and characterized.42,43
Another important early observation was the presence of prominent intracytoplasmic membranes (ICMs), which were also used for classification, with the type I methanotrophs exhibiting membranes shaped like vesicular discs (Figure 1a) and the type II methanotrophs exhibiting paired membranes around the cell perimeter37,44 (Figure 1b). Thermoacidophilic methanotrophs, referred to as group III, were discovered much later45–47 and have been the subject of much interest due to their growth requirement for rare earth elements.48 The type I and type II methanotrophs are synonymous with the Gammaproteobacteria and Alphaproteobacteria classes of the Proteobacteria phylum, respectively, while the type III methanotrophs belong to the Methylacidiphilae class of the Verruocmicrobia phylum.49,50 Methanotrophs are found in diverse environments, including soil, rice paddies, freshwater lakes, oceans, tundra wetlands, landfills, and volcanic mudpots.13,51,52
Figure 1.
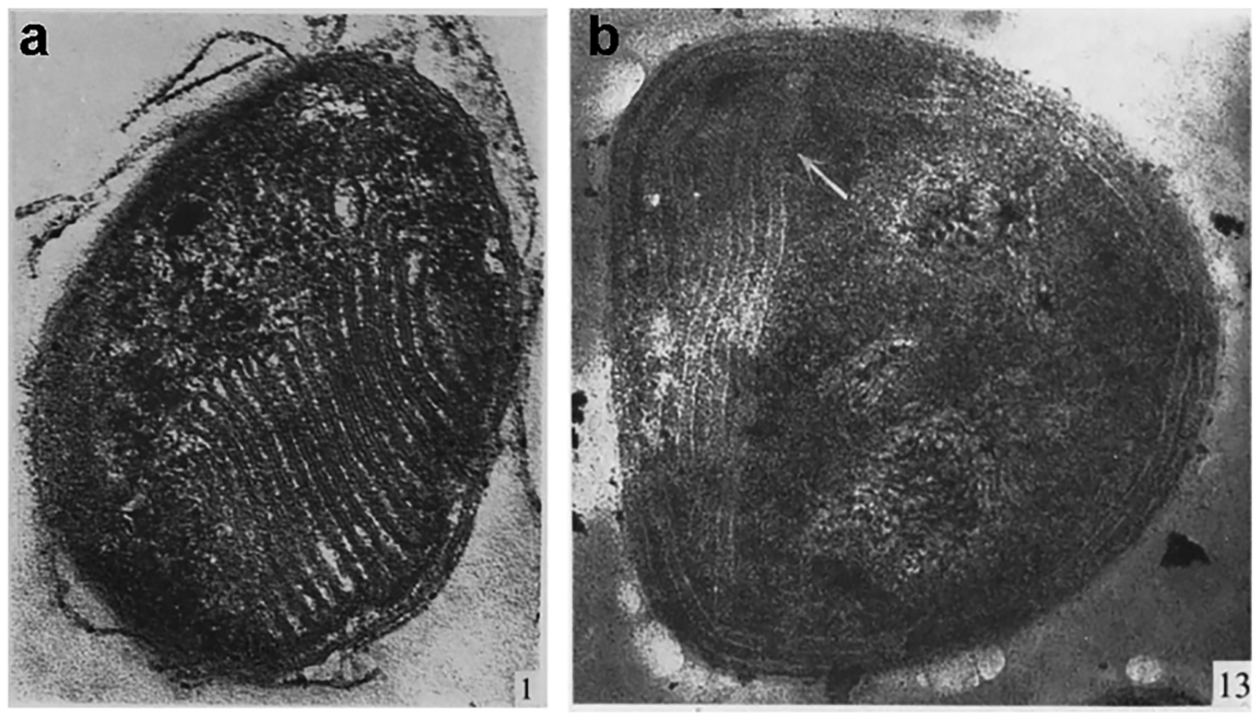
Micrographs of type I and type II methanotrophs. (a) Section of a type I Methylococcus strain magnified ×80,000. (b) Section of a type II Methylosinus strain magnified ×80,000. Adapted with permission from ref 37, copyright 1970, Society for General Microbiology.
The first step in methanotroph metabolism is the oxidation of methane to methanol by MMOs. Methanol is then oxidized to formaldehyde by methanol dehydrogenase (MDH). The next steps diverge depending on the type of methanotroph. In Gammaproteobacteria, carbon is assimilated at the stage of formaldehyde by the ribulose monophosphate pathway, whereas in Alphaproteobacteria, carbon is assimilated as formate via the serine pathway39,53,54 (Figure 2). Verrucomicrobial methanotrophs fix CO2 using the Calvin-Benson-Bassham cycle.55,56 The proteobacterial assimilatory pathways have been targeted for metabolic engineering to produce a range of fuels and chemicals, including lactate and 2,3-butanediol (reviewed in refs 23 and 26). However, further advances will require increased rates of methane conversion to methanol,28 which cannot be accomplished without a detailed understanding of MMO chemistry and regulation.
Figure 2.
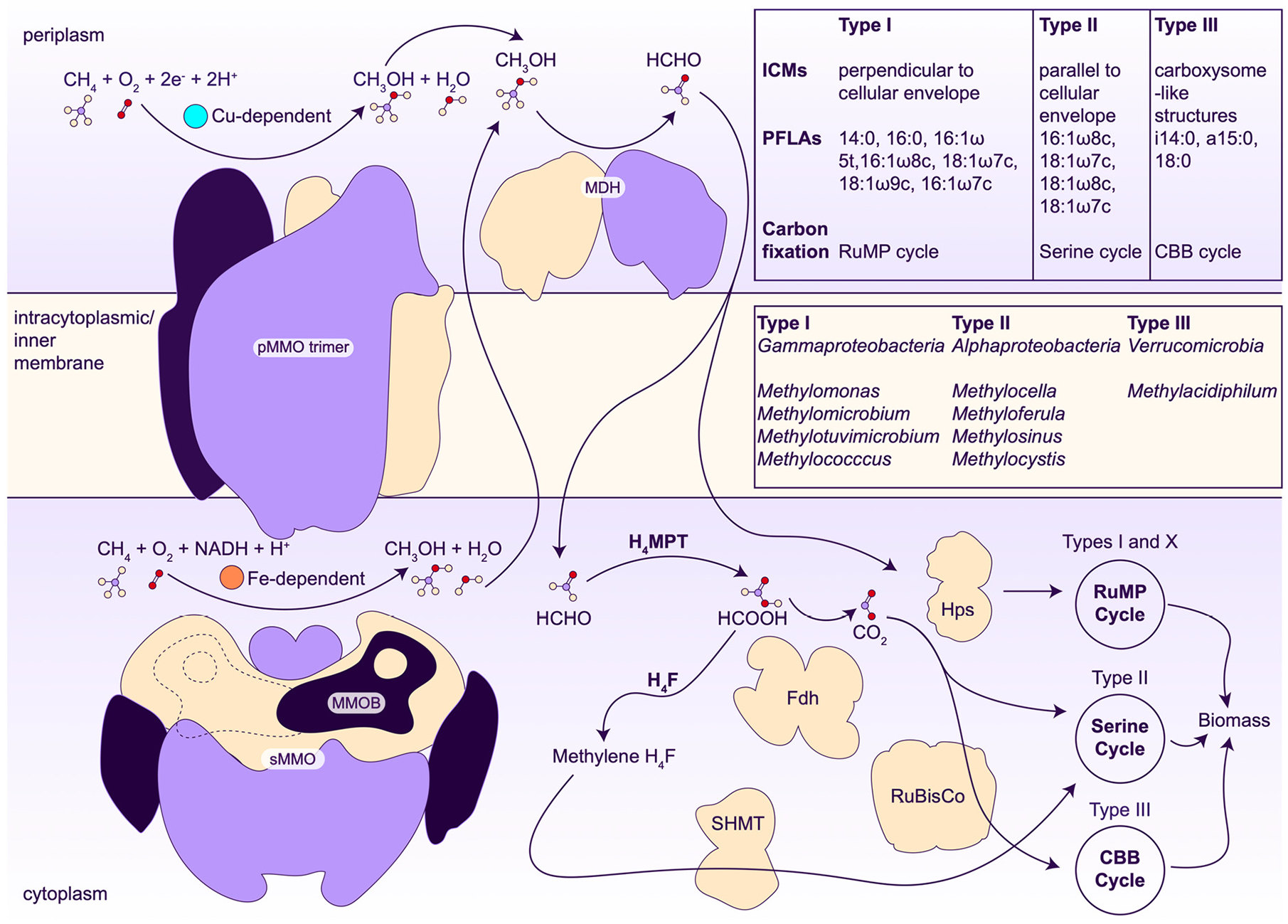
Methanotroph metabolic pathways. The pMMO trimer is colored by protomer, showing its C3 symmetrical organization. sMMO is colored by subunits that comprise the complex, along with the MMO regulatory protein B (MMOB) bound (indigo) on the front and back (dashed line) sides of sMMO. MDH, methanol dehydrogenase; ICMs, intracytoplasmic membranes; PFLAs, phospholipid fatty acids; RuMP, ribulose monophosphate; CBB, Calvin-Benson-Bassham; H4MPT, tetrahydromethanopterin pathway; H4F, tetrahydrofolate pathway; FDH, formate dehydrogenase; SHMT, serine hydroxymethyltransferase; Hps, hexulose 6-phosphate synthase; RuBisCo, ribulose 1,5-bisphosphate carboxylase.
2.2. Copper Acquisition
2.2.1. Methanobactins.
As a required cofactor for pMMO activity57–60 and an inducer of ICM formation,61–63 copper is central to methanotroph physiology. Methanotrophs have several specialized copper acquisition systems. Some methanotrophs produce natural products called methanobactins (Mbns) under conditions of copper starvation.64–66 Mbns are ribosomally synthesized, post-translationally modified peptide natural products that bind Cu(I) with particularly high affinity. The copper binding site consists of two nitrogen and two sulfur ligands provided by nitrogen-containing heterocycles and neighboring thioamide groups (Figure 3). Genes encoding the Mbn precursor peptide MbnA, biosynthetic enzymes, transporters, and other associated proteins are found in Mbn operons,67 which are coregulated with the genes encoding sMMO.68 All Mbn operons encode the MbnB/MbnC heterodimeric complex that uses a mixed valent Fe(II)Fe(III) site in MbnB to convert two cysteines in MbnA to oxazolone/thioamide groups.69,70 Additional modifying enzymes present in some Mbn operons include the aminotransferase MbnN,71 a predicted flavin-dependent oxidoreductase, MbnF, a predicted sulfotransferase, MbnS, a predicted TauD-like nonheme iron enzyme, MbnD, and a protein related to MbnB called MbnX. Variations in MbnA sequences combined with the presence of different modifying enzymes lead to a diversity of Mbn structures (Figure 3).72–76 Notably, Mbn operons are also found in a wide range of non-methanotrophic bacteria, suggesting a broader function in bacterial metal homeostasis.
Figure 3.
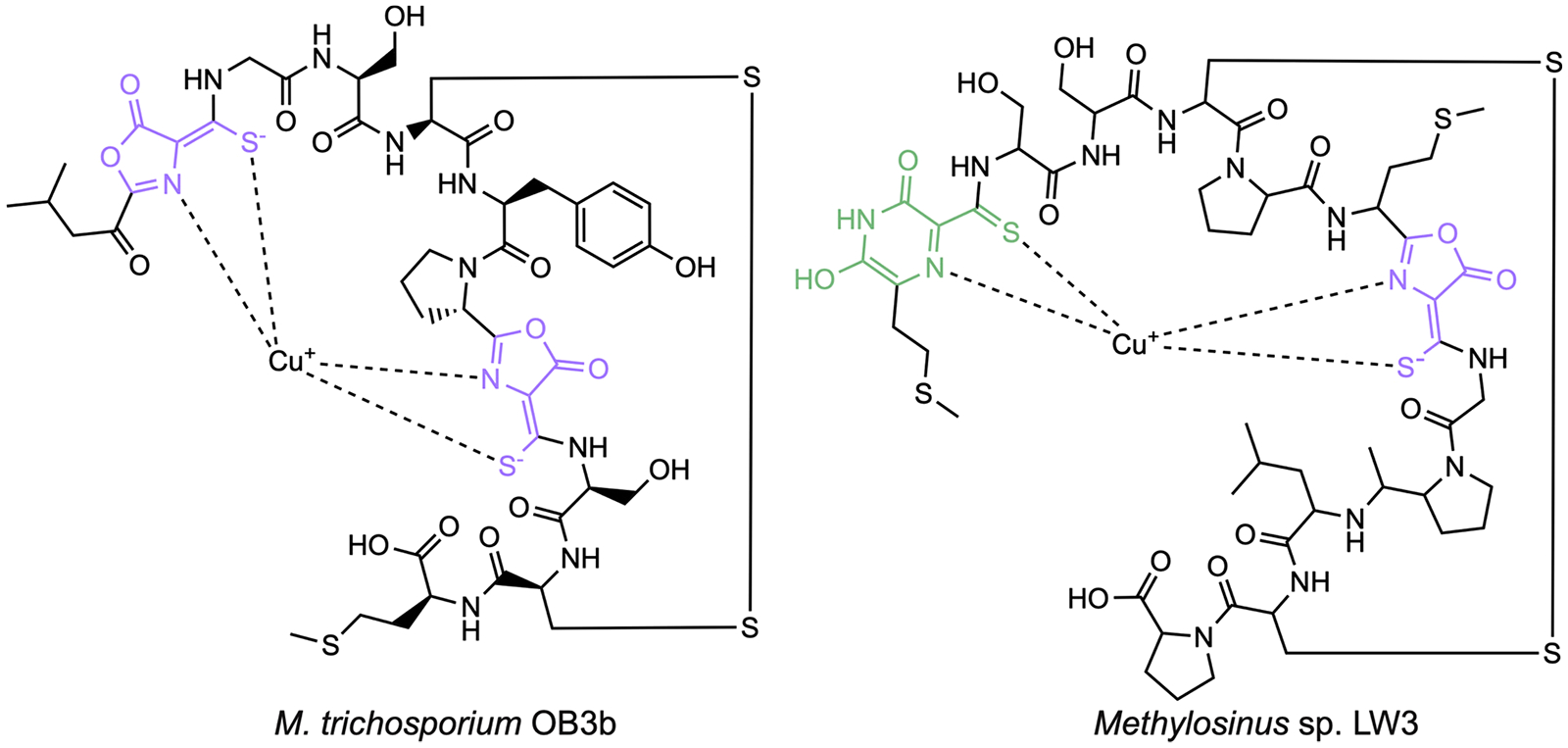
Structures of methanobactin from Methylosinus trichosporium OB3b and Methylosinus sp. LW3. The oxazolone moieties are highlighted in purple, and the pyrazinedione group is shown in green.
Under copper starvation conditions, methanotrophs secrete the apo (metal free) form of Mbn, which is then reinternalized in its copper-bound form (Figure 4).77 Addition of copper-loaded Mbn to methanotrophs can promote methane oxidation activity and the copper switch between sMMO and pMMO (section 2.3).78,79 Due to its high affinity for Cu(I), Mbn not only binds copper in solution but can also extract copper from mineral sources or glass.78,80,81 The mechanism of Mbn secretion has not been established but is proposed to involve MbnM, a member of the multidrug and toxic compound extrusion family.67,82 Uptake of copper-loaded Mbn is an active transport process mediated by the outer membrane TonB-dependent transporter MbnT, which is encoded both within Mbn operons and elsewhere in the methanotroph genomes.82–85 After intact copper-loaded Mbn enters the cell,77 it may interact with periplasmic proteins such as MbnE82 or MbnP, followed by import to the cytoplasm, perhaps by ABC transporters (Figure 4). MbnP was recently shown to bind Cu(I) using a kynurenine residue that is generated by the diheme enzyme MbnH.86–88 The MbnP and MbnH genes are typically found adjacent to genes encoding MbnT. It is not known how copper is then delivered to pMMO or cytoplasmic cellular targets, including transcription factors.
Figure 4.
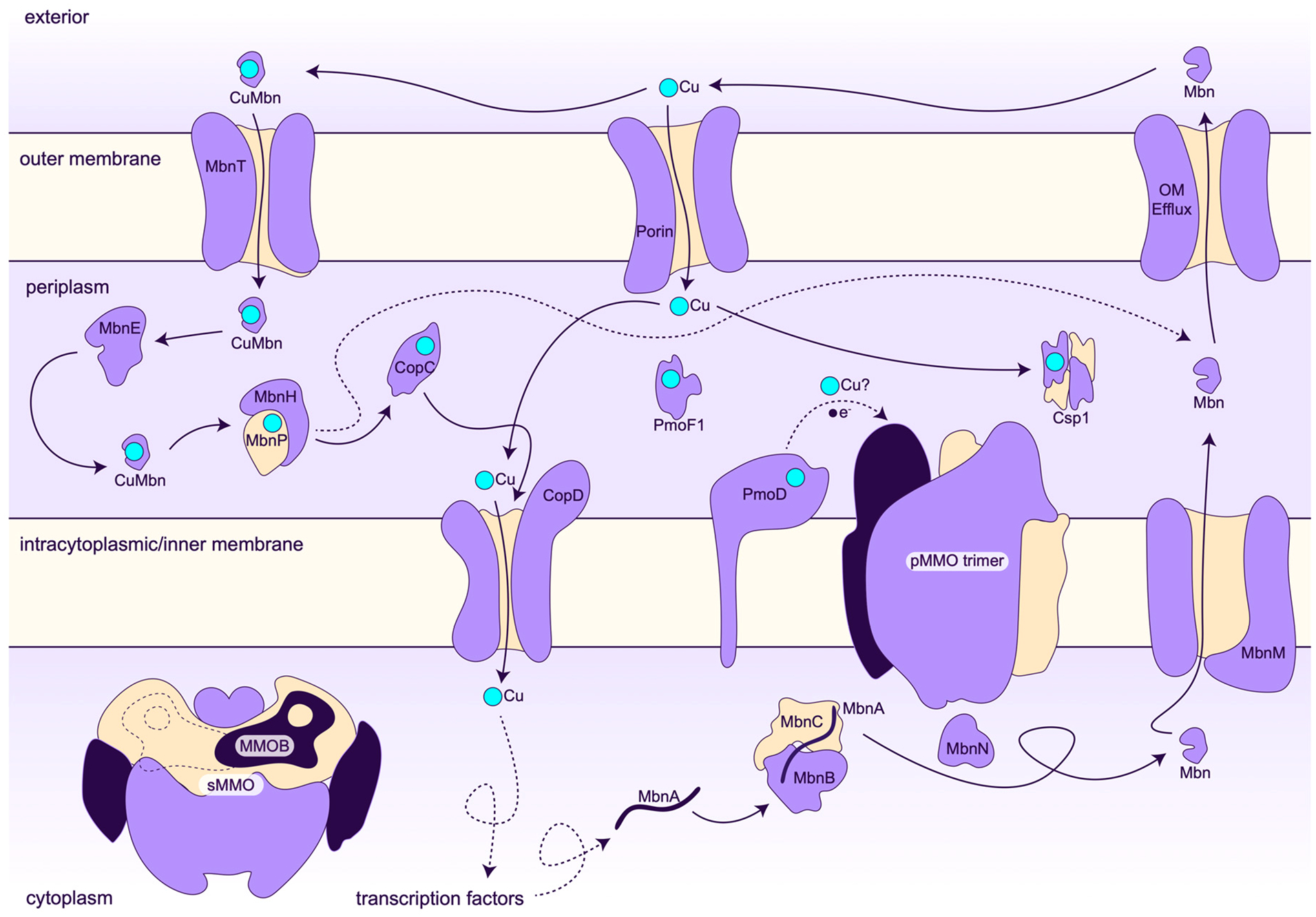
Model for copper homeostasis in the type II methanotroph Methylosinus trichosporium OB3b.
2.2.2. MopE and Csp Proteins.
Not all methanotrophs possess the ability to manufacture Mbn. There is evidence that methanotrophs can take up non-native Mbns, but so far, this Mbn piracy only involves other Mbn producers.82,84,85 Some methanotrophs instead produce copper-binding proteins belonging the MopE/CorA family. The M. capsulatus (Bath) MopE protein is truncated and modified to contain a copper-binding kynurenine residue (MopE*) and then secreted. The surface-associated CorA from Methylomicrobium album BG8 also binds Cu(I) with kynurenine.89–91 MopE* and CorA differ in overall structure and in the details of copper coordination from MbnP, which also has a kynurenine ligand.87 In MopE* and CorA, the Cu(I) is ligated by two histidines, a kynurenine, and a water molecule, whereas the Cu(I) in MbnP is coordinated by one histidine, one methionine, a kynurenine, and a water molecule. Copper downregulates expression of MopE, CorA, and a Methylotuvimicrobium alcaliphilum comb. nov. 20Z (20Z) homolog, suggesting that these proteins function in copper acquisition.91–93 How copper bound to these proteins is mobilized remains unclear.
Finally, members of the copper storage protein (Csp) family have been proposed to play a role in methanotroph copper handling.94,95 The M. trichosporium OB3b Csp1 and Csp2 proteins are predicted to be secreted from the cytoplasm to the periplasm in a copper-bound form, which for Csp1 includes binding 13 Cu(I) ions using primarily cysteine residues housed in the interior of a four-helix bundle.96 While the copper-binding properties of these proteins have been investigated in detail, their cellular localization in methanotrophs and evidence for a specific role in methane oxidation have not been reported. Disruption of the genes encoding both Csp1 and Csp2 led to a modest increase in sMMO iron-dependent activity, which could be consistent with a role in copper storage for pMMO.96 Csp3, which does not have a signal sequence and thus should reside in the cytoplasm, binds 19 Cu(I) ions, also within a four-helix bundle, and is widespread in non-methanotrophic bacteria.97,98
2.3. The Copper Switch
While the sMMO and pMMO genes were initially cloned in the late 1980s and early 1990s,99,100 numerous genomes from all types of methanotrophs are now available.23,101,102 The sMMO genes are encoded in the mmoXYBZDC operon, with mmoX, mmoY, and mmoZ encoding three subunits of the hydroxylase protein (MMOH), mmoB encoding the regulatory protein (MMOB), and mmoC encoding the reductase (MMOR) (Figure 5a).103 The pMMO genes include pmoA, pmoB, and pmoC, encoding the PmoA, PmoB, and PmoC subunits of pMMO, respectively. Methanotroph genomes contain up to three copies of the pMMO genes, depending on the species,100,104–113 along with up to two additional copies of the pmoC gene sometimes referred to as PmoC singletons.110,114 In alphaproteobacterial methanotrophs, the pmoD gene is found adjacent to the other genes (Figure 5b).115 Many methanotrophs, including the Verrucomicrobia, only contain the pMMO genes, while a few species from the Methylocella42,116 and Methyloferula117 genera only possess the sMMO genes.41 Notably, the pmo operon is similar to that encoding ammonia monooxygenase (AMO),118,119 the only enzyme besides pMMO and sMMO known to oxidize methane.120,121 AMO converts ammonia to hydroxylamine in ammonia-oxidizing bacteria and ammonia-oxidizing archaea.122–125 These nitrifying microbes also contribute to global warming by producing nitrous oxide, which is the third most important greenhouse gas next to carbon dioxide and methane.5
Figure 5.
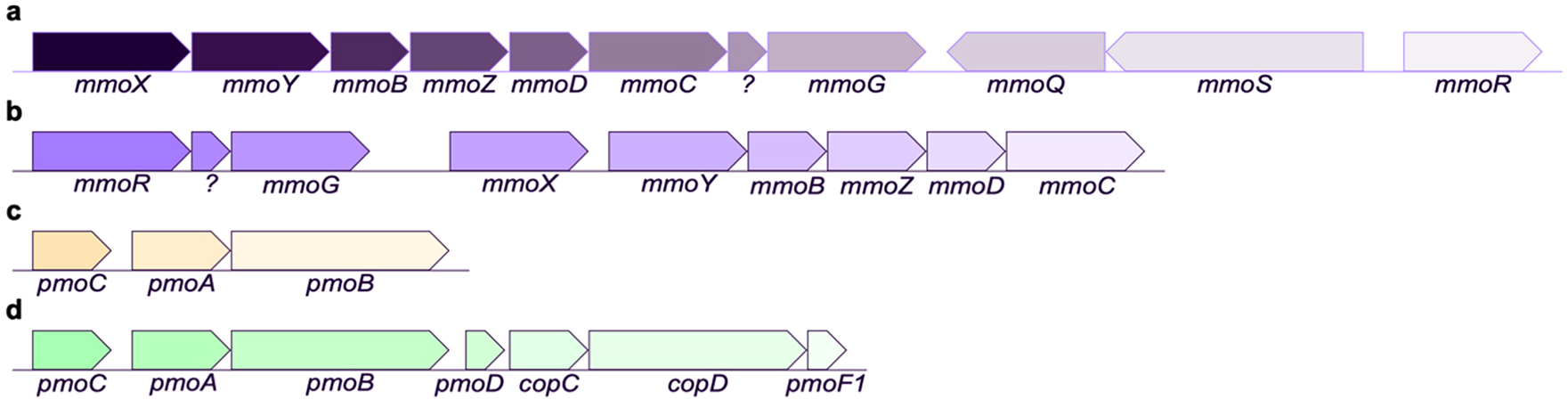
MMO operons. The operons encoding sMMO in (a) M. capsulatus (Bath) and (b) M. trichosporium OB3b and pMMO in (c) M. capsulatus (Bath) and (d) M. trichosporium OB3b are shown.
A large subset of methanotrophs encodes both sMMO and pMMO in their genomes and can switch between them depending on copper-to-biomass ratios.41 This “copper switch” was discovered ~40 years ago with the observation that MMO activity was differentially associated with the membrane (particulate) or soluble fractions as a function of copper availability and that copper and particulate fraction activity are associated with ICM formation (Figure 1).61–63 In these methanotroph strains, sMMO is prevalent at copper concentrations <1 μM, and pMMO becomes predominant at copper concentrations >5 μM. The copper-induced biogenesis of ICMs is not well understood despite their being imaged extensively by electron63,68,126,127 and fluorescence128 micros-copies as well as cryoelectron tomography (cryoET).129,130 These imaging studies indicate that the ICMs are continuous with the cytoplasmic inner membrane and form by invagination of this membrane.128,130,131
In the well-studied M. capsulatus (Bath) and M. trichosporium OB3b strains, transcription of the sMMO genes is downregulated by copper.132 While the copper switch has been referred to as “reciprocal regulation”, pMMO is in fact expressed constitutively133–135 and only mildly upregulated in the presence of copper.132,136 Recent time-dependent qRT-PCR data showed less than an order of magnitude of upregulation of pMMO expression over 24 h of exposure to copper while sMMO expression is downregulated by 2–3 orders of magnitude within 24 h of exposure to copper.68 Some studies have reported a more pronounced increase in pMMO expression,80,137 but the consensus seems to be mild upregulation. This constitutive expression of pMMO raises the questions of whether it is actually present under low copper conditions and, if so, where it is localized and whether it contains copper.
It remains unclear how copper mediates the differential expression of the two MMOs. Several proteins encoded in the mmo operon, including the transcription factor MMOR and the GroEL homolog MmoG (Figure 5a), are essential for sMMO expression.135,138 A two component system found in M. capsulatus (Bath) (Figure 5a), MmoQ/MmoS, may also play a role in sMMO regulation. Of these four proteins, only the soluble sensor domain of MmoS has been biochemically characterized, and it does not bind copper.139 No regulatory factors for pMMO have been identified. The MMOD protein, which forms a complex with and inhibits the sMMO MMOH component (section 4.1),140,141 has been proposed to bind copper and then to both repress pMMO expression and upregulate sMMO expression.137,142 This model is based on characterization of an M. trichosporium OB3b mutant in which mmoXYBZD and the first three codons of mmoC are deleted (SMDM mutant).143 For this mutant, pmoA expression decreases in the presence of copper as opposed to increasing in the wildtype strain. Since mmoD is the only disrupted gene in the SMDM mutant with an unclear function, it was suggested to mediate the copper switch.137 However, mmoD is regulated with the rest of the sMMO genes, which is inconsistent with a regulatory role. Also incompatible with this model, MMOD has no DNA binding or metal binding motifs,141 does not bind copper, and does not bind to a heparin column,68 often used as a diagnostic for DNA binding.
Several other strains of M. trichosporium OB3b have broken copper switches in that they constitutively express sMMO.127 These mutants were generated by treatment with the mutagen dichloromethane and exhibit reduced intracellular copper levels.144 Further studies of one of these mutants, the PP358 strain, showed that copper neither downregulates sMMO nor stimulates ICM formation. The PP358 genome has been sequenced, and of potential relevance to the copper switch, a frameshift deletion in the copD gene was detected.68 The copD gene neighbors (Figure 5b) and is coregulated with the pmo genes in M. trichosporium OB3b. Since CopDs are putative copper importers,145–148 a CopD disruption in Ms. trichosporium OB3b could prevent copper from reaching transcription factors in the cytoplasm (Figure 4). However, disruption of copD and the neighboring copC gene, which encodes a periplasmic copper binding protein,149 does not affect the copper switch when tested at copper concentrations of 0 and 1 μM.150 It is not known whether a partial deletion in copD, as found in the PP358 strain,68 would have the same lack of phenotype. It is also possible that a phenotype would be observed using different conditions and copper concentrations.
3. PARTICULATE METHANE MONOOXYGENASE
3.1. Enzyme Structure
pMMO comprises three subunits, PmoB, PmoA, and PmoC, arranged in an α3β3γ3 trimer (Figure 6a, b). All structurally characterized pMMOs, which include those from M. capsulatus (Bath),60,151 M. trichosporium OB3b,152 Methylocystis species strain (sp.) M,153 Methylocystis sp. Rockwell,59,60 and M. alcaliphilum 20Z60,154 (Table 1), form this trimer, and dissociation of the subunits or alternative oligomerization states have not been observed. One third of this trimer is typically referred to as the pMMO protomer. PmoB (42 kDa) consists of an N-terminal cupredoxin-like domain, two transmembrane helices, and a C-terminal cupredoxin-like domain. All PmoB subunits are predicted to have this architecture, although the related AmoB from the archaeal AMO system only contains one cupredoxin-like domain followed by a single transmembrane helix.155–157 The cupredoxin-like domains face the periplasm and constitute the only soluble regions of pMMO. PmoA (24 kDa) comprises seven transmembrane helices, along with a small β hairpin that protrudes into the PmoB periplasmic domain. PmoA has a similar fold to the S components of bacterial energy-coupling factor (ECF) ABC transporters, which are responsible for uptake of vitamins such as riboflavin, thiamin, and biotin.158,159 However, PmoA lacks a pocket equivalent to the ligand binding site in the S components.
Figure 6.
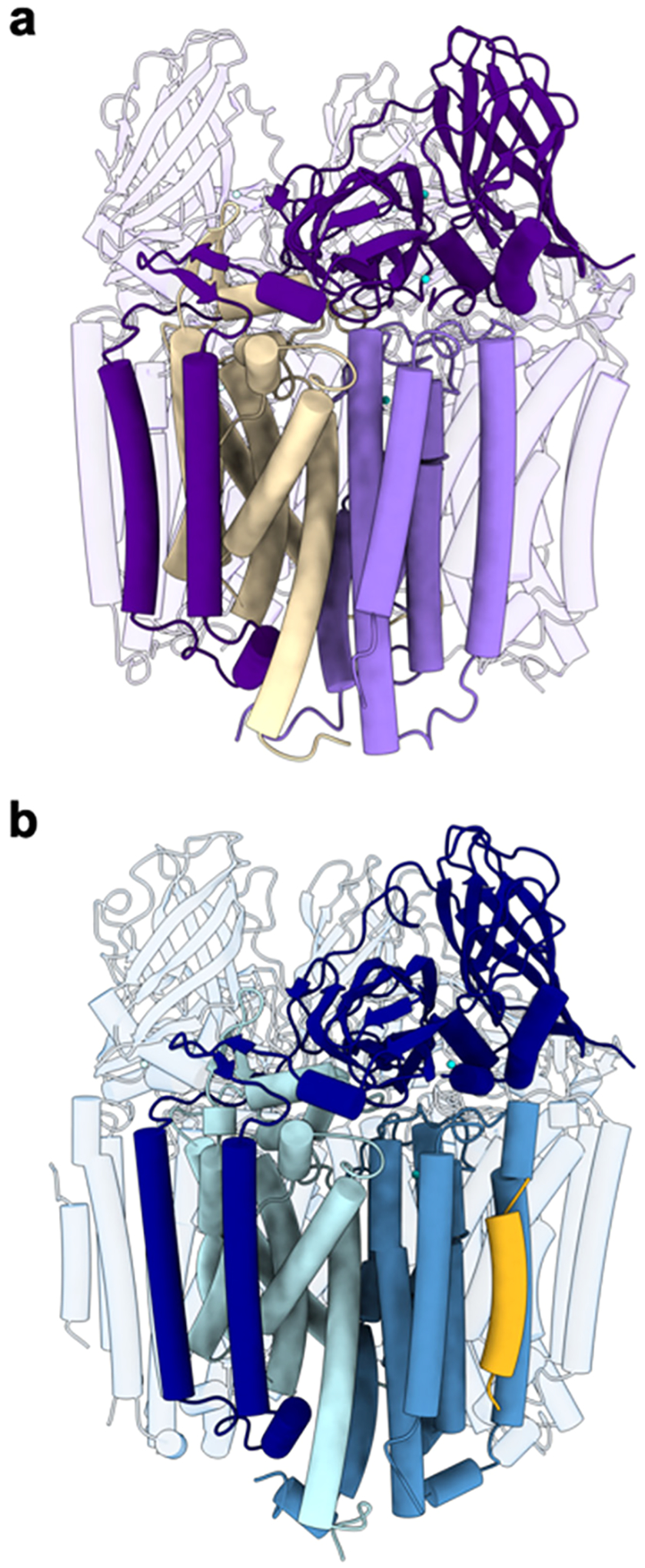
Trimeric structure of pMMO. (a) CryoEM structure of M. capsulatus (Bath) pMMO in native lipid nanodiscs (PDB ID: 7S4H). One protomer comprising PmoB (dark purple), PmoA (wheat), and PmoC (light purple) is highlighted. (b) CryoEM structure of M. sp. Rockwell pMMO in POPC nanodiscs (PDB ID: 7S4M). One protomer comprising PmoB (dark blue), PmoA (blue), PmoC (sky blue), and helix X (yellow) is highlighted.
Table 1.
pMMO Structures
| Resolution (Å) | PDB code | |
|---|---|---|
| X-ray | ||
| M. capsulatus (Bath) pMMO | 2.80 | 1YEW |
| M. capsulatus (Bath) pMMO | 2.80 | 3RGBa |
| M. trichosporium OB3b pMMO | 3.90 | 3CHX |
| M. sp. M pMMO | 2.68 | 3RFR |
| M. sp. Rockwell pMMO | 2.59 | 4PHZ |
| M. sp. Rockwell pMMO Cu(II) soaked | 3.15 | 4PI0 |
| M. sp. Rockwell pMMO Zn(II) soaked | 3.33 | 4PI2 |
| M. alcaliphilum 20Z pMMO | 2.70 | 6CXH |
| CryoEM | ||
| M. capsulatus (Bath) pMMO in native lipid nanodisc | 2.14 | 7S4H |
| M. capsulatus (Bath) pMMO in native lipid nanodisc | 2.16 | 7S4J |
| M. capsulatus (Bath) pMMO in native lipid nanodisc | 2.26 | 7S4I |
| M. capsulatus (Bath) pMMO in native lipid nanodisc | 2.34 | 7S4K |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + CN | 3.65 | 7T4O |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + CN and Cu | 3.62 | 7T4P |
| M. capsulatus (Bath) pMMO in DDM | 2.60 | 7EV9 |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + CN | 3.21 | 8SR5 |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + CN and Cu | 3.12 | 8SR4 |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + 20x TFE | 2.19 | 8OYI |
| M. capsulatus (Bath) pMMO in native lipid nanodisc xlinked + 20x TFE | 2.16 | 8SQW |
| M. capsulatus (Bath) pMMO in native lipid nanodisc + 20x TFB | 2.36 | 8SR2 |
| M. capsulatus (Bath) pMMO in native lipid nanodisc xlinked + 20x TFB | 2.18 | 8SR1 |
| M. sp. Rockwell pMMO in POPC nanodisc | 2.42 | 7S4M |
| M. alcaliphilum 20Z pMMO in POPC nanodisc | 2.46 | 7S4L |
| CryoET | ||
| M. capsulatus (Bath) pMMO | 4.80 | 7YZY |
PDB 3RGB is an improved version of structure 1YEW and should be used as the M. capsulatus (Bath) pMMO model; 1YEW is obsolete.
PmoC (22 kDa) consists of six transmembrane helices. Part of PmoC, spanning residues 225–253 in M. capsulatus (Bath) pMMO, is unmodeled in all the crystal structures due to a lack of electron density (Figure 7a). This region was finally resolved in the high resolution (up to 2.14 Å) cryoelectron microscopy (cryoEM) structures of pMMO reconstituted into nanodiscs (phospholipid bilayer discs surrounded by a membrane scaffold protein belt)160 with native methanotroph lipids (Figure 7b).60 These residues, which correspond to the most highly conserved part of the PmoC sequence, face the interior of the trimer and are stabilized by interactions with phospholipids. In the 2.6 Å resolution cryoEM structure of M. capsulatus (Bath) pMMO in n-dodecyl-β-d-maltoside (DDM) detergent, only PmoC residues 108–157 and 258–286 were modeled (Figure 7c),161 providing a less complete model than the crystal structures. The presence of lipids also stabilizes PmoA residues 192–212 (M. capsulatus (Bath) numbering).60 These residues were not modeled in the M. capsulatus (Bath) crystal structure151 or a cryoEM structure of M. capsulatus (Bath) pMMO in DDM.161
Figure 7.
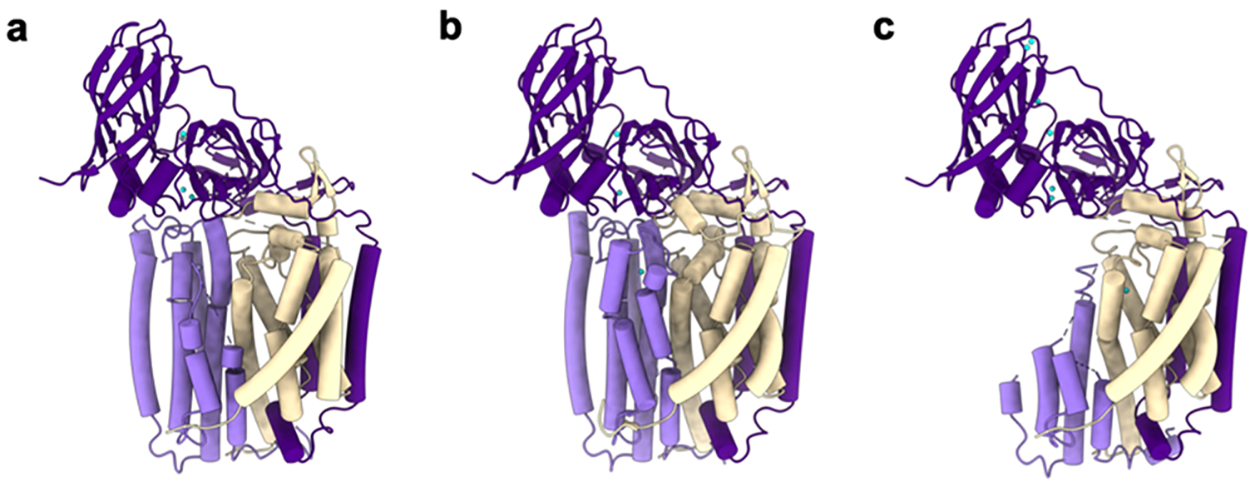
Structures of M. capsulatus (Bath) pMMO protomers showing PmoC (light purple), PmoA (wheat), PmoB (dark purple), copper ions (cyan), and zinc ions (gray) as modeled. (a) Crystal structure of pMMO showing PmoC and PmoA subunits with missing regions (PDB ID: 3RGB). (b) CryoEM structure of pMMO in native lipid nanodiscs showing the stabilized PmoC and PmoA architectures (PDB ID: 7S4H). (c) CryoEM structure of pMMO in detergent with perturbed PmoC and PmoA subunits (PDB ID: 7EV9).
In the crystal and cryoEM structures of pMMOs from the Alphaproteobacteria (M. trichosporium OB3b,152 M. sp. M,153 M. sp. Rockwell),59,60 strong density corresponding to an unidentified helix (helix X) is observed adjacent to a large groove in the surface of PmoC (Figure 6b). While helix X neighbors the PmoC N-terminus, ~15 residues of which are not modeled, its position and length are not consistent with it being connected to PmoC. Helix X could not be identified using mass spectrometry59 and has been modeled as up to 25 alanine residues, extending from the periplasm (N-terminus) toward the cytoplasm (C-terminus). In M. sp. Rockwell pMMO, lipids located between helix X and PmoC interact with two conserved arginine residues in PmoC, Arg 102 and Arg 171.59,60 Since all pMMO samples for structural characterization have been isolated directly from methanotrophs, it is likely that helix X represents a biologically relevant interaction partner. One possibility is that helix X corresponds to the transmembrane helix of PmoD (section 3.6), but attempts to model its side chain density with the PmoD sequence have not been successful. Regardless of helix X’s identity, the deep groove in the surface of PmoC is striking and is a likely binding site for either a protein partner or a large ligand. For example, an unusually shaped cryoEM density in this groove has been suggested to correspond to a quinone.60
Recent serial cryo-focused ion beam (cryoFIB) milling/scanning electron microscope (SEM) volume imaging and cryoelectron tomography (cryoET) studies of pMMO in M. capsulatus (Bath) cells have revealed that the pMMO trimers assemble into higher order array structures.130 The pMMO trimer in the intact cell was observed at 15 Å resolution in the subtomogram averaged map, and a 4.8 Å resolution map of a pMMO trimer surrounded by six lower resolution trimers was obtained by imaging isolated membranes.130 The overall architecture agrees well with the crystal and cryoEM structures, and several intertrimer contacts involving the PmoB subunit were predicted from molecular dynamics simulations. Further studies, including simulations within a lipid bilayer, are needed to assess the molecular basis for array formation and may also shed light on the mechanisms of ICM biogenesis.
3.2. Metal Centers
While a 2007 study suggested that pMMO contains a catalytic diiron center similar to that in sMMO,162 iron detected in other preparations was attributed to heme from contaminating cytochromes, identifiable by optical, electron paramagnetic resonance (EPR), and X-ray absorption spectroscopies.163 No further evidence for a diiron center has been obtained since the original report.162 Instead, pMMO is widely believed to contain copper active sites, consistent with observations that copper restores activity to pMMO samples that have been metal depleted by treatment with potassium cyanide.57–60 The copper stoichiometries of purified pMMO from M. capsulatus (Bath) (the only pMMO studied by multiple research groups) over the past 20 years are in the range of either 2–3 copper ions or 13–15 copper ions per 100 kDa pMMO protomer.41,164–166 As detailed below, 2–3 copper ions are consistent with the structural data obtained over the same time period, while the higher copper content, still favored by Chan and co-workers,167,168 is not.
3.2.1. Metal Binding Sites in the PmoB Subunit.
The structures reveal two mononuclear copper centers in the PmoB subunit, in contrast to claims that PmoB is a “copper sponge” that can bind ~10 Cu(I) ions.161,169,170 The first copper center, denoted the bis-His site, is coordinated by His48 and His72 (M. capsulatus (Bath) numbering) and is observed in the crystal151 and cryoEM60,161 structures of M. capsulatus (Bath) pMMO (Figure 8a). This site is not present in the structures of pMMO from M. trichosporium OB3b,152 M. sp. M,153 and M. sp. Rockwell59,60 because His48 is replaced with asparagine in these alphaproteobacterial PmoB sequences (Figure 8b). Notably, His48 is conserved in M. alcaliphilum 20Z PmoB, but the site is devoid of metal.154 Given that EPR spectra of pMMOs from alphaproteobacterial and gammaproteobacterial methanotrophs are virtually identical, this site in M. capsulatus (Bath) pMMO has been assigned as Cu(I).171,172 Since this site is not conserved and not always occupied, it is unlikely to play a critical functional role.
Figure 8.
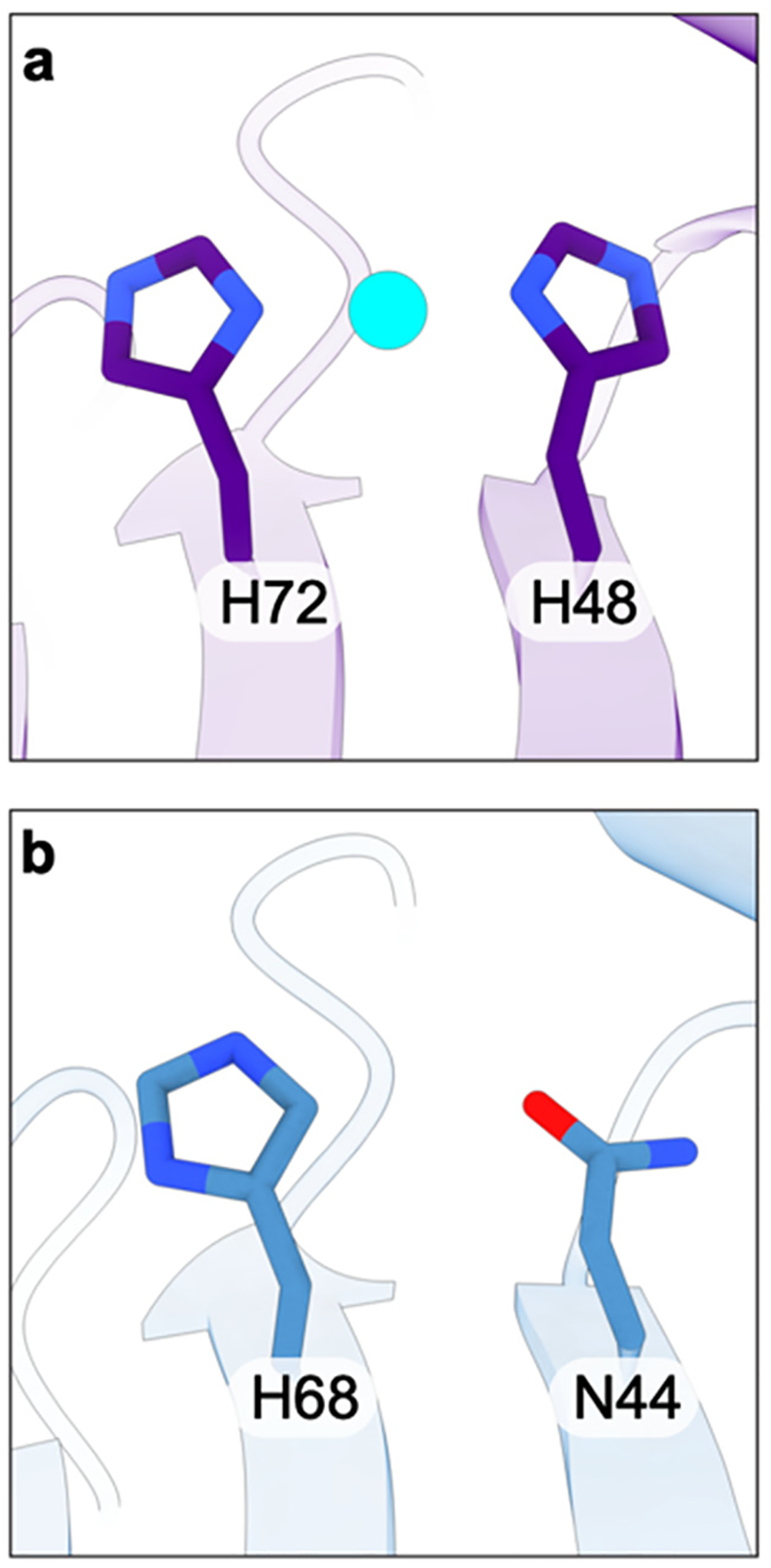
(a) The bis-His site in the PmoB subunit of M. capsulatus (Bath) (PDB ID: 7S4H) and (b) the corresponding residues in the PmoB subunit of M. sp. Rockwell pMMO (PDB ID: 7S4M).
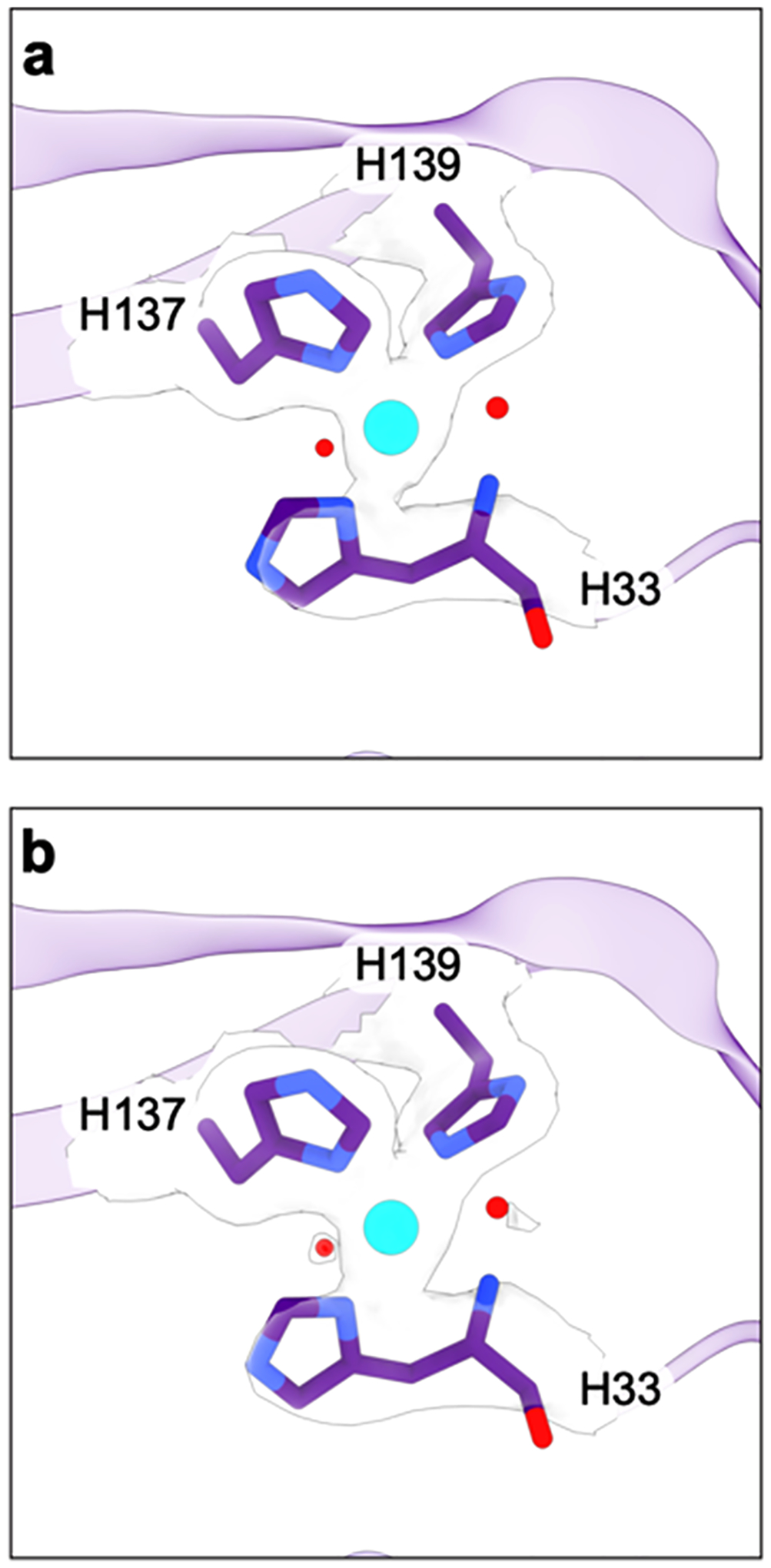
The second site in the PmoB subunit, denoted CuB, has been the subject of much discussion in the literature. In the original M. capsulatus (Bath) pMMO crystal structure, this site was modeled with two copper ions, one coordinated by His137 and His139 and the other by the side chain of His33 as well as the amino terminal group of His33, which is the first residue in the PmoB subunit.151 The first 32 residues constitute the predicted signal sequence that is presumably removed upon export to the periplasm100 and are not present in any pMMO preparation. The dicopper site model was influenced by extended X-ray absorption fine structure (EXAFS) data indicating the presence of a short (~2.5 Å) Cu–Cu interaction,163,173 and a similar model was proposed for M. trichosporium OB3b pMMO152 and for one protomer of M. sp. M pMMO.153 However, higher resolution, better quality crystallographic data obtained for pMMOs from M. sp. Rockwell59 and M. alcaliphilum 20Z154 (Table 1) were more consistent with a monocopper site in this location, as was quantum refinement174 and high-energy-resolution fluorescence detected (HERFD) EXAFS analysis.175 The crystal structure of the soluble portion of AmoB from the ammonia oxidizing archaeon Nitrosocaldus yellowstonii also revealed a single copper ion, although the amino terminal histidine was disordered in this structure.156
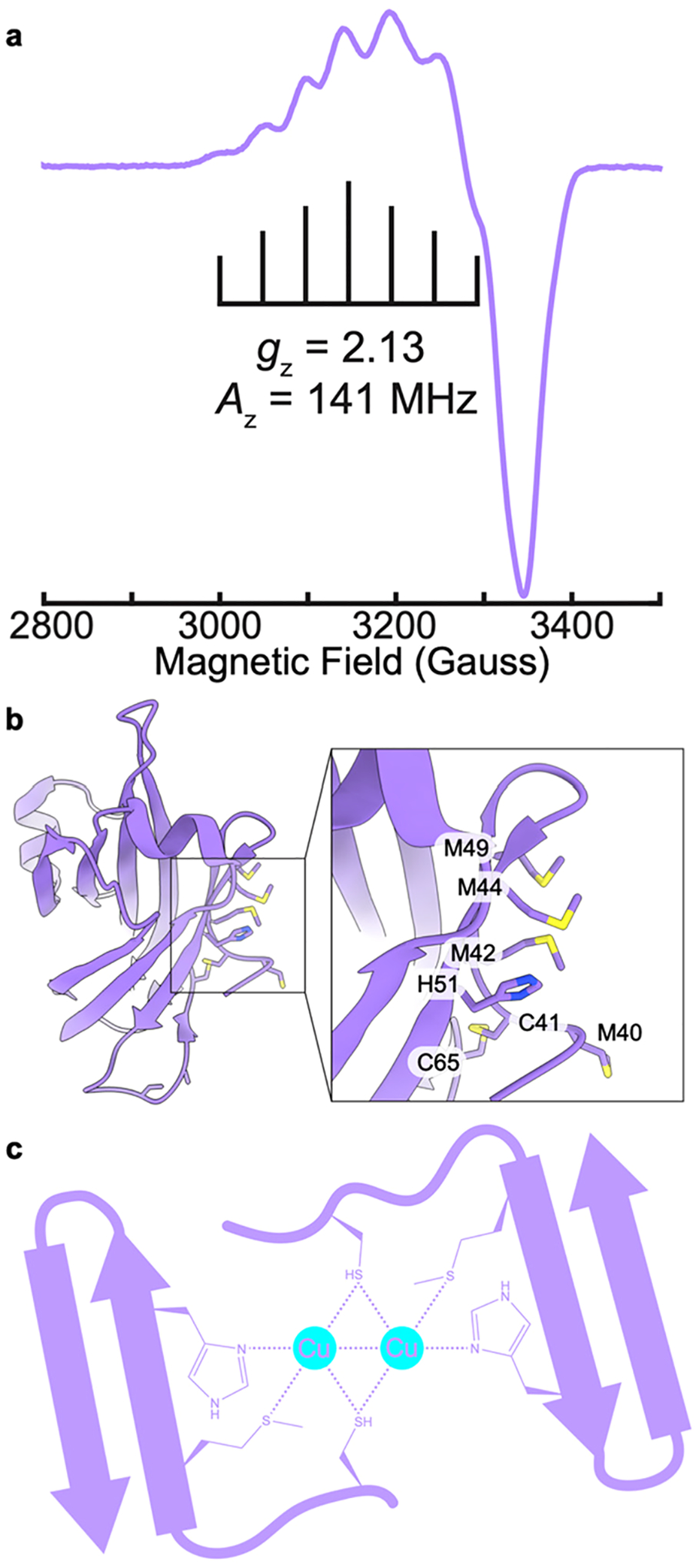
The question of the CuB nuclearity was resolved through EPR studies of M. capsulatus (Bath) whole cells cultivated in the presence of 15N and 63Cu.171 Consistent with prior whole cell EPR studies,176,177 a single type 2 Cu(II) signal was observed with superhyperfine splitting indicative of four equatorial nitrogen ligands (Figure 9). Three of these four nitrogen ligands were assigned to histidine side chains on the basis of electron nuclear double resonance (ENDOR) spectroscopic analysis. The only location in the pMMO structure with three histidines positioned to coordinate copper is the CuB site, so this EPR signal is attributable to CuB, which must be a mononuclear Cu(II) site. The same results were obtained for M. sp. Rockwell pMMO.172 In addition, 17O and 1H ENDOR data indicate the presence of an axially bound water molecule,171,172 and 1H ENDOR signals attributable to the bound amino group are observed.172 The EPR parameters of the CuB site are the same in whole cells, isolated membranes, and purified pMMO in detergent, bicelles, and nanodiscs prepared with both synthetic and native lipids.154,171,172,178
Figure 9.
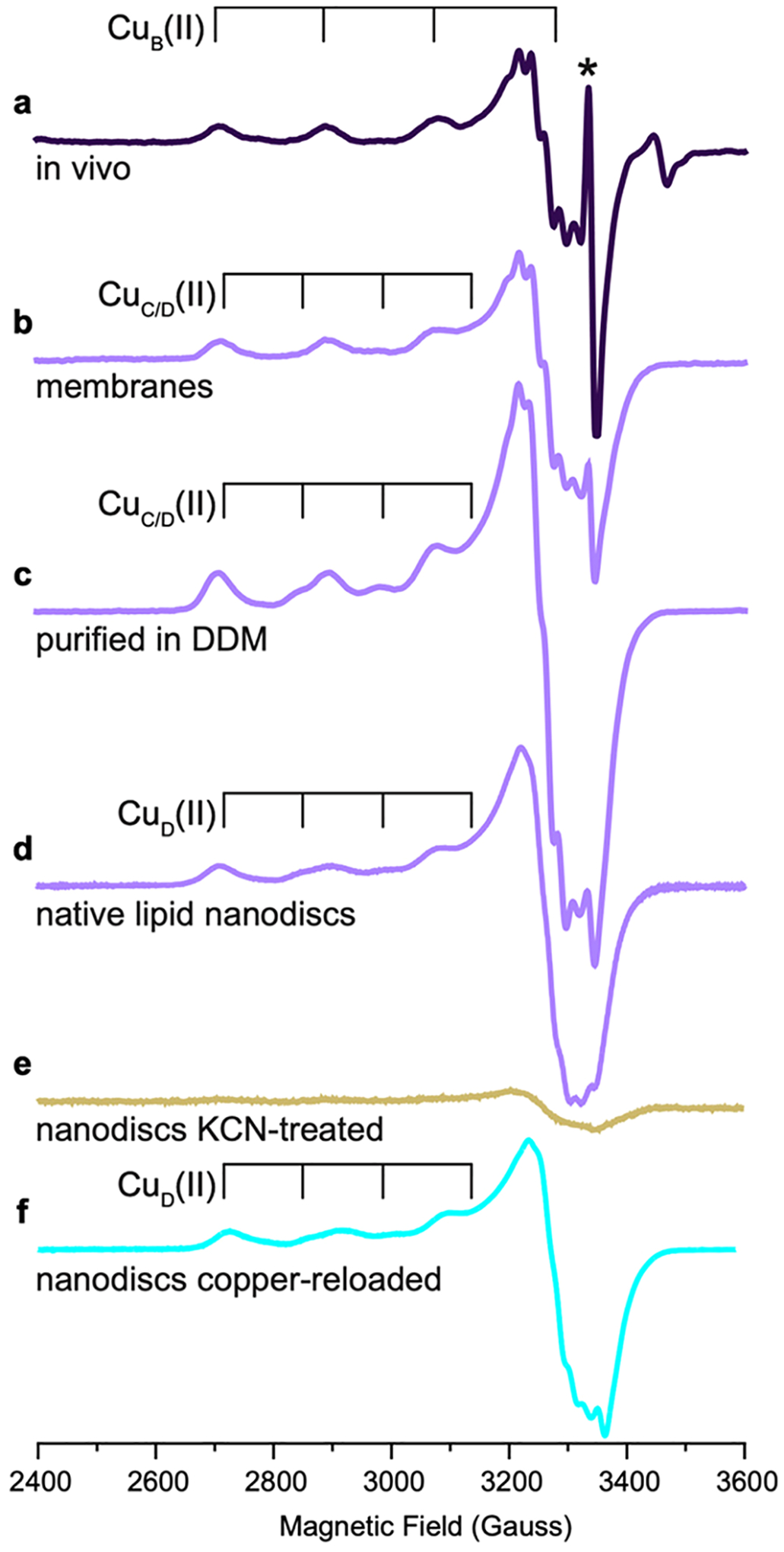
EPR spectra of pMMO from M. capsulatus (Bath) (adapted from refs 171 and 178). (a) pMMO in M. capsulatus (Bath) whole cells showing a Cu(II) EPR signature corresponding to the CuB(II) site. Asterisk indicates an organic radical signal. (b) pMMO in isolated membranes showing signals corresponding to the CuB(II) site and to the CuC/D(II) site, which exists as Cu(I) in vivo but is oxidized to Cu(II) upon membrane isolation. (c) pMMO solubilized in DDM and purified shows signals for the CuB(II) and CuC/D(II) sites. (d) pMMO in native lipid nanodiscs exhibits signals for the CuB(II) and CuD(II) sites as supported by cryoEM. (e) KCN-treated pMMO in native lipid nanodiscs shows an attenuated Cu(II) EPR spectrum with only a weak signal corresponding to partial loading of the CuB(II) site, consistent with metal depletion and supported by cryoEM. (f) KCN-treated, then copper-reloaded pMMO in native lipid nanodiscs shows recovered signals for the CuB(II) and CuD(II) sites, as supported by cryoEM.
Further support for a mononuclear CuB site is derived from native top-down mass spectrometry (nTDMS) of pMMO.179 In these studies, M. alcaliphilum 20Z PmoB ejected from a detergent micelle exhibited a mass consistent with the presence of a single Cu(II) ion, as did M. sp. Rockwell PmoB ejected from micelles. In contrast to the typical metal analysis of pMMO by inductively coupled plasma mass spectrometry (ICP-MS) or optical emission spectroscopy (ICP-OES), nTDMS enables subunit-specific localization of bound metal ions. Finally, the significantly higher resolution cryoEM structures of pMMO from M. capsulatus (Bath) (Figure 10), M. sp. Rockwell, and M. alcaliphilum 20Z (Table 1) provided unequivocal evidence for a mononuclear CuB site.60
Figure 10.
The CuB site in the cryoEM structure of M. capsulatus (Bath) pMMO in native lipid nanodiscs (PDB ID: 7S4H). The cryoEM density is shown as a transparent surface contoured at (a) 6σ and (b) 5σ.
3.2.2. Metal Binding Sites in the PmoC Subunit.
The pMMO crystal structures revealed one metal binding site in the PmoC subunit adjacent to the disordered region, denoted CuC and coordinated by Asp156, His160, and His173 (M. capsulatus (Bath) numbering) (Figure 11a). In the structures of pMMO from M. capsulatus (Bath)151 and M. sp. M,153 this site is occupied by zinc, identified by analysis of anomalous diffraction data. Both of these pMMOs were crystallized in the presence of excess ZnSO4, which not only occupies this site but also binds to the protein surface and mediates crystal lattice contacts. Zinc was not required for crystallization of M. trichosporium OB3b152 and M. sp. Rockwell59 pMMOs, and the site is occupied by copper in these structures (Figure 11b). Soaking of M. sp. Rockwell crystals in CuSO4 significantly increases the occupancy of the CuC site, while treatment with ZnSO4 results in occupancy with zinc and ordering of 10 additional residues, including a glutamic acid coordinated to the zinc ion.59
Figure 11.
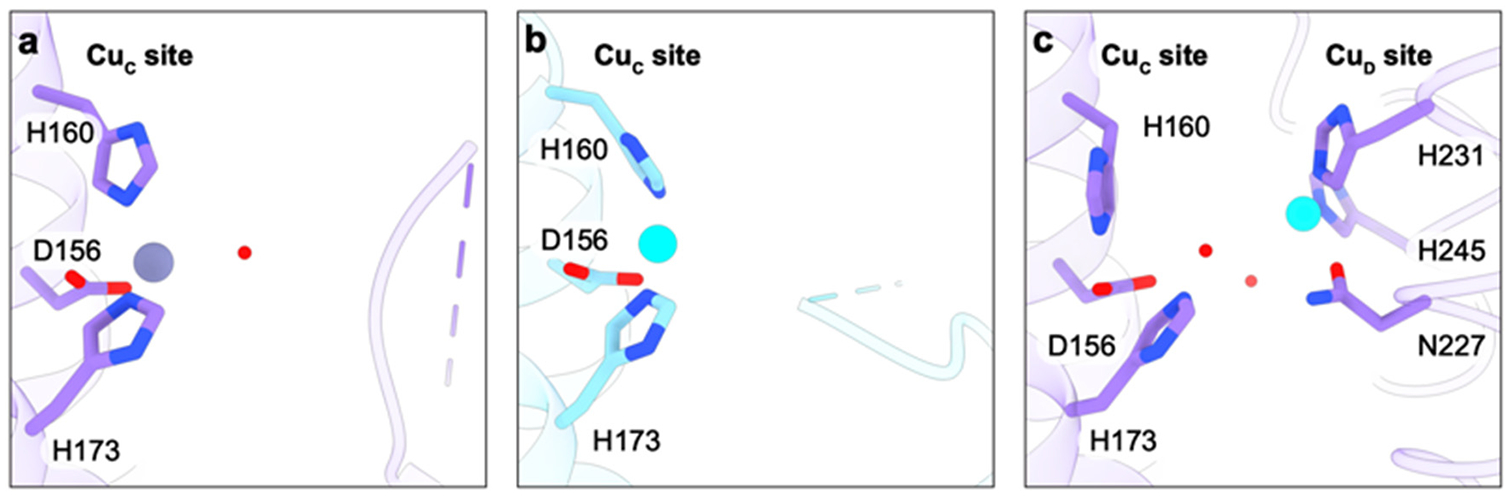
Metal binding sites in the PmoC subunit. (a) Crystal structure of M. capsulatus (Bath) pMMO (PDB ID: 3RGB) showing a zinc ion in the CuC site. (b) Crystal structure of M. sp. Rockwell pMMO (PDB ID: 4PI0) showing a copper ion in the CuC site. (c) CryoEM structure of M. capsulatus (Bath) pMMO in native lipid nanodiscs (PDB ID: 7S4H) showing an empty CuC site and a copper ion in the CuD site.
Surprisingly, the CuC site is unoccupied in the cryoEM structures of M. capsulatus (Bath) pMMO in native lipid nanodiscs. Instead, another metal binding site is apparent ~5.7 Å from the CuC site location, with ligands Asn227, His231, and His245, all derived from the PmoC region that was not observed in the crystal structures (Figure 11c).60 Unlike crystallography, there is no method to directly identify metal ions in cryoEM density maps, but structures of samples depleted of metals by potassium cyanide treatment59 and then reloaded with CuSO4 indicate that this site, denoted CuD, is indeed occupied by copper.60,178 Instead of a metal ion in the CuC site, the M. capsulatus (Bath) pMMO cryoEM maps contain density assigned as a water molecule within hydrogen bonding distance of CuC ligands Asp156, His160, and His173 (Figure 11c). The CuC site is occupied in the cryoEM map of one sample of M. capsulatus (Bath) pMMO as well as in the cryoEM maps of pMMOs from M. alcaliphilum 20Z and M. sp. Rockwell. In these maps, the residues surrounding the CuD site are poorly ordered.60 Thus, occupancy of CuC appears to correlate with disorder in the highly conserved region spanning residues 225–253 (M. capsulatus (Bath) numbering).
While whole cells exhibit a single Cu(II) EPR signal attributed to CuB (section 3.2.1), isolated membranes and purified pMMO exhibit a second Cu(II) EPR signal (Figure 9) that was initially assigned to CuC using Cu–Cu distances determined from double electron-electron resonance (DEER) spectroscopic analysis.171 15N ENDOR experiments performed at fields where this signal does not overlap with that of CuB indicate that the Cu(II) ion is coordinated by two histidine ligands, consistent with assignment to CuC. An axial water molecule was also detected by 1H ENDOR at a distance of ~3 Å from the Cu(II) ion. Similar results were obtained for pMMO reconstituted into 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) nanodiscs, although the axial water was not present.172
The cryoEM structures60 raised the question of whether this EPR signal might instead derive from the CuD site given that both CuC and CuD have two histidine nitrogen ligands and one oxygen ligand, with the only difference being the presence of asparagine instead of aspartic acid in CuD (Figure 11c). To address this question, parallel samples of M. capsulatus (Bath) pMMO in native lipid nanodiscs were interrogated by EPR and used for cryoEM structure determination. These enzymatically active samples exhibited the same two Cu(II) signals and showed occupancy only of CuD in the cryoEM structure.178 Therefore, the second EPR signal is attributable to CuD in native nanodisc samples and perhaps in isolated membranes as well. It remains unclear whether this signal in detergent-solubilized pMMO arises from CuC, CuD, or some combination of the two sites, which are separated by ~5.7 Å. Regardless, since this EPR signal is not observed in whole cells, the corresponding site must be Cu(I) in vivo.
3.3. Enzymatic Activity
3.3.1. Delivery of Electrons.
The activity of pMMO is measured by monitoring either propylene epoxidation or methane oxidation. Propylene epoxidation, which may occur by a different mechanism than that of methane oxidation, is used for whole cell activity assays, as methanol is further metabolized by downstream enzymes and thus not detectable. Methane oxidation by isolated membranes and solubilized or purified pMMO are most accurately measured using 13C-labeled methane, which ensures that all detected methanol product derives from pMMO activity.154 pMMO activity assays require a reductant, typically formate for whole cells, NADH or duroquinol for isolated membranes, and duroquinol for purified enzyme.173,180–182 Duroquinol is a synthetic analog of ubiquinol, and while ubiquinol is produced by methanotrophs,183,184 duroquinol is not a native cofactor, despite being included in some computational studies.185,186
Although these reductants are effective in vitro, the physiological source of electrons for pMMO remains unresolved, with several models under consideration. In the first model, NADH is proposed to reduce ubiquinol via a type 2 NADH:quinone oxidoreductase followed by the transfer of electrons from ubiquinol to pMMO.136,187 This scenario, which is consistent with the use of NADH and duroquinol in vitro, is referred to as the “redox arm” model (Figure 12).188 The second model, denoted “direct coupling”, involves transfer of electrons from MDH to pMMO via a cytochrome c electron shuttle (Figure 12).189 A number of metabolic modeling studies have attempted to distinguish between these pathways by correlating growth parameters and other experimental data with flux balance analysis. Depending on the methanotroph species, these studies indicate that either pathway or a combination of the two, termed “uphill electron transfer”, could be operational.188,190–195
Figure 12.
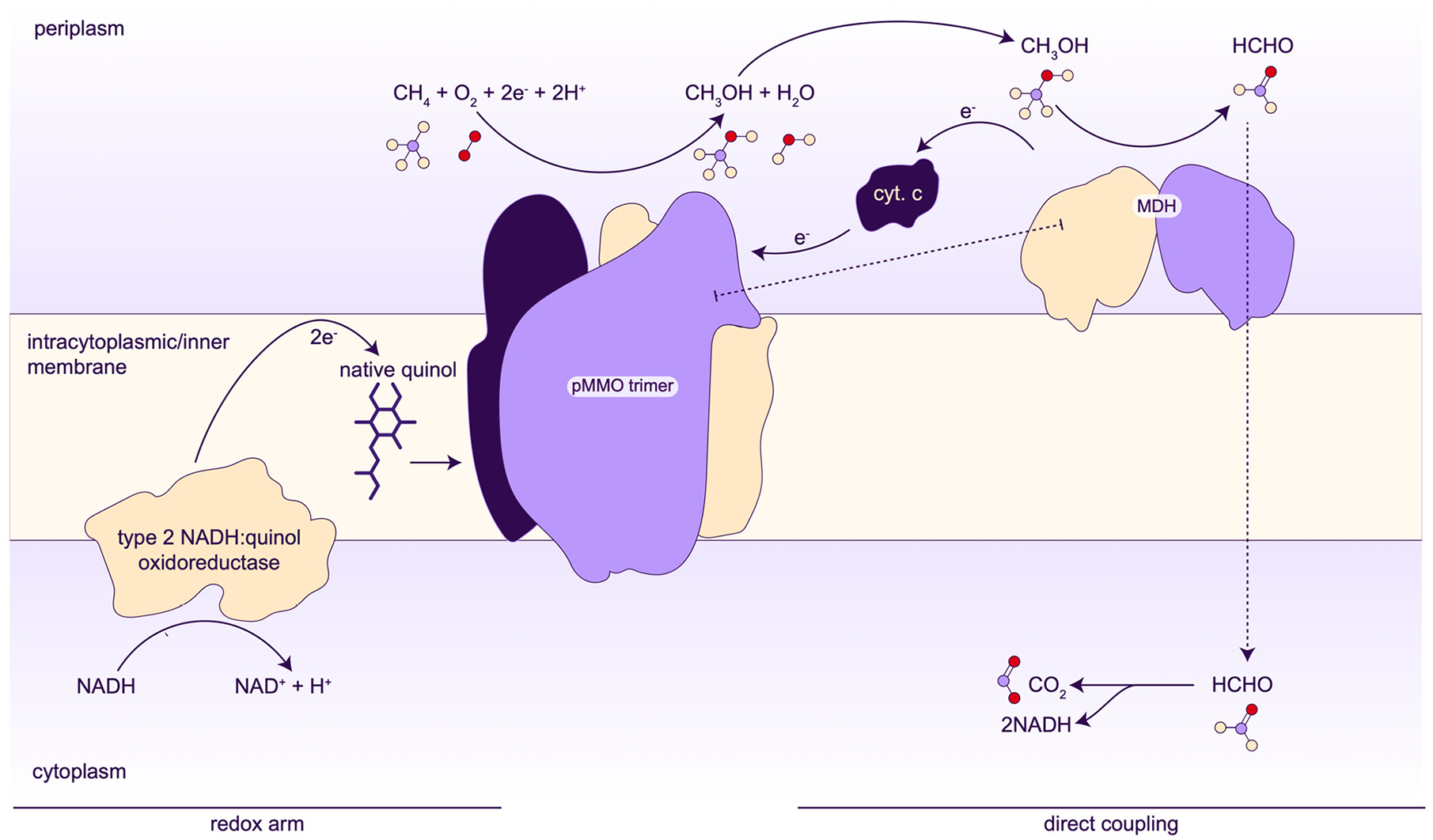
Proposed models for electron delivery to pMMO. The direct coupling model may also include the transfer of methanol from the pMMO active site to the MDH active site in a proposed supercomplex.
3.3.2. Activity of Isolated pMMO Preparations.
The specific activity of pMMO decreases upon isolation of the membranes and is significantly reduced or completely abrogated after detergent solubilization and purification (Table 2). The lack of activity upon purification for crystallization was suggested by Chan and co-workers to result from the loss of as many as 12 copper ions.168,196 However, reconstitution of pMMO into bicelles (phospholipid bilayer discs surrounded by detergent)197 recovered activity without altering the copper content or EPR spectroscopic signature, indicating that removal from the membrane, rather than massive copper loss, adversely affects pMMO activity.154 Reconstitution into nanodiscs in the presence of copper also recovers activity.60,179 The activity of M. capsulatus (Bath) pMMO nanodiscs was tested using several different lipids, including 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), POPC, and native lipids isolated directly from M. capsulatus (Bath) cells, of which the latter conferred the most activity (Table 2). The native lipids include a mixture of phosphatidylethanolamine (PE), phosphatidylcholine (PC), phosphatidylglycerol (PG), and cardiolipin (CL), as well as a significant fraction (~20%) of unidentified lipids.60 It is not clear why the native lipids confer higher activity to pMMO in nanodiscs, as lipid densities in cryoEM structures of pMMO in native lipid nanodiscs resemble those in POPC nanodiscs in the same locations, suggesting that most of the lipids observed are PCs or native lipids that remain stably bound regardless of the peripheral lipid environment.60
Table 2.
pMMO Activity Data
| Sample | Methanotroph | Substrate | Reductant | Turnover number per protomer (s−1) | Specific activity (nmol mg total protein−1 min−1) | Refs |
|---|---|---|---|---|---|---|
| Cells producing pMMO | M. capsulatus (Bath) | propylene | formate | 1.4−2.5a | 167–300 | 136,162 |
| M. trichosporium OB3b | methane | NA | 0.68–2.5b | 82–300 | 28,357 | |
| Membrane-bound pMMO | M. capsulatus (Bath) | methane | NADH | 0.083–0.146c | 40–70 | 59,154 |
| duroquinol | 0.025–0.042d | 12−20 | 59,154 | |||
| propylene | NADH | 0.044–0.246e | 21–118 | 136,173 | ||
| duroquinol | 0.033−0.179e | 16−86 | 136,173 | |||
| M. sp. Rockwell | methane | NADH | 0.017–0.024d,f | 8–11.5 | 59,179 | |
| duroquinol | 0.004–0.006d,f | 1.8–3 | 59,179 | |||
| M. alcaliphilum 20Z | methane | NADH | 0.006c,f | 3 | 154 | |
| duroquinol | 0 | 0 | 154 | |||
| Purified pMMO in DDMg | M. capsulatus (Bath) | methane | NADH | 0 | 0 | 154 |
| duroquinol | 0.002h,i | 1 | 60,154 | |||
| M. capsulatus (Bath) | propylene | NADH | 0 | 0 | 136,173 | |
| duroquinol | 0.032–0.21i,j | 18–126 | 136,173 | |||
| M. sp. Rockwell | methane | NADH | 0 | 0 | 59 | |
| duroquinol | 0 | 0 | 59 | |||
| M. alcaliphilum 20Z | methane | NADH | 0 | 0 | 154 | |
| duroquinol | 0 | 0 | 154 | |||
| Purified pMMO in bicelles | M. capsulatus (Bath) | methane | NADH | 0.009f,g | 5.2 | 154 |
| duroquinol | 0.006f,h | 3.5 | 154 | |||
| M. alcaliphilum 20Z | methane | NADH | 0 | 0 | 154 | |
| duroquinol | 0.007f,h | 4.4 | 154 | |||
| Purified pMMO in nanodiscsi | ||||||
| DMPCg | M. capsulatus (Bath) | methane | duroquinol | 0.005h | 3 | 60 |
| POPCg | M. capsulatus (Bath) | 0.009h | 5.4 | 60 | ||
| native lipids | M. capsulatus (Bath) | 0.012h | 7.2 | 60 | ||
| POPCg | M. sp. Rockwell | methane | duroquinol | 0.011g | 6.6 | 179 |
| POPCe | M. alcaliphilum 20Z | methane | duroquinol | 0 | 0 | 60 |
Calculated from rate of propylene epoxidation monitored by gas chromatography (GC) and assuming pMMO is 20% of the total protein.
Calculated from rate of methane uptake and assuming that pMMO is 20% of the total protein.
Calculated from rate of conversion of 13CH4 to 13CH3OH monitored by GC/mass spectrometry (GC/MS) and assuming that membrane-bound protein is 80% pMMO.
Calculated from rate of conversion of CH4 to CH3OH monitored by GC and assuming that membrane-bound protein is 80% pMMO. Values from ref 59 were converted to 13C values by applying a correction factor of 0.5.154
Calculated from rate of propylene epoxidation monitored by GC and assuming that membrane-bound protein is 80% pMMO.
Activity assay was performed at 30 °C. All other activity assays were performed at 45 °C.
Abbreviations used: DDM, n-dodecyl-β-D-maltoside; DMPC, 1,2-dimyristoyl-sn-glycero-3-phosphocholine; POPC, 1-palmitoyl-2-oleoylphosphatidylcholine.
Calculated from rate of conversion of 13CH4 to 13CH3OH monitored by GC/MS.
The samples used for the M. capsulatus (Bath) pMMO crystal structure determination were not assessed for methane oxidation activity and did not exhibit propylene epoxidation activity.151
Calculated from rate of propylene epoxidation monitored by GC.
The level of recovered activity in membrane mimetic systems approaches that of the isolated membranes but is still significantly less than that of whole cells (Table 2). In whole cells, pMMO is densely packed in the ICMs, forming hexagonal arrays (Figure 13).130,198 These array structures can be recapitulated to some extent in nanodiscs by altering the reconstitution conditions, and these higher order pMMO nanodisc arrays exhibit increased activity compared to single particle nanodiscs.130 Thus, pMMO activity may be enhanced in vivo by the properties of these ordered membrane structures and perhaps by protein-protein, protein-lipid, or protein-quinol interactions within these arrays. Overall, the issues with retaining activity have precluded using isolated pMMO for biotechnological applications, although one promising study demonstrated that a stable and reusable catalytic material could be generated by embedding pMMO-containing membranes in polymer hydrogels.199
Figure 13.
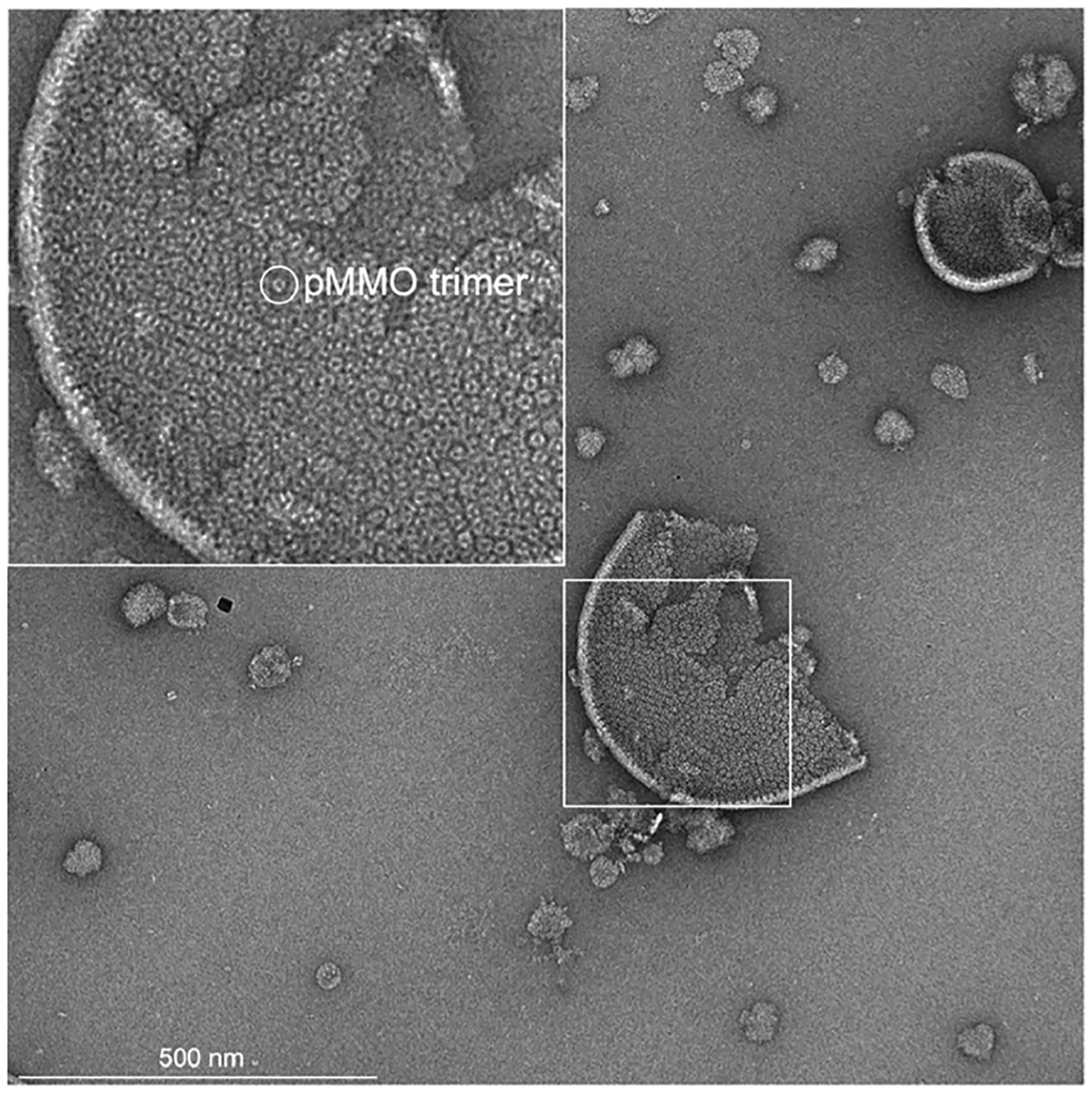
Negative stain micrograph showing pMMO in isolated membranes. The inset shows a magnified view of the isolated membranes with a single pMMO trimer circled.
Besides methane and propylene, pMMO can oxidize C1–C5 n-alkanes and terminal alkenes to 2-alcohols and 1,2-epoxides, but it does not react with aromatic and cyclic hydrocarbons.180,181,200–202 pMMO can also oxidize ammonia, the substrate of the homologous enzyme AMO, to nitrite.203 Inhibitors of pMMO activity include metal chelating agents, alkynes, and excess copper.57,203–206 Notably, inhibition by excess copper can be reversed by removal with potassium cyanide and reconstitution with stoichiometric amounts of copper.57 Zinc is also an inhibitor, with excess zinc completely inhibiting activity207 and stoichiometric amounts leading to 40–60% inhibition.59 Loading of apo pMMO with zinc almost completely abolishes activity in membranes. Zinc may occupy the copper active site and has also been proposed to interfere with proton transfer.59
3.4. Assignment of the Active Site
3.4.1. Proposed Tricopper Site.
Models for the pMMO active site have evolved as new spectroscopic and structural data have been obtained. One model proposed prior to the first crystal structures and perpetuated in the literature involves a trinuclear copper center in the PmoA subunit.165,196,208 However, no metal binding sites were observed in PmoA in any of the crystal structures. Three copper ions were modeled in PmoA in the cryoEM structure of M. capsulatus (Bath) pMMO in detergent,161 but the cryoEM structures of M. capsulatus (Bath) pMMO in native lipid nanodiscs clearly show that this region is occupied by a water molecule and a glutamate residue60 (Figure 14). While the absence of the tricopper center in the crystal structures was ascribed to the loss of activity in the crystallized samples,208 its absence in the cryoEM structure of active pMMO in native nanodiscs60 indicates that it is not a viable candidate for the active site.
Figure 14.
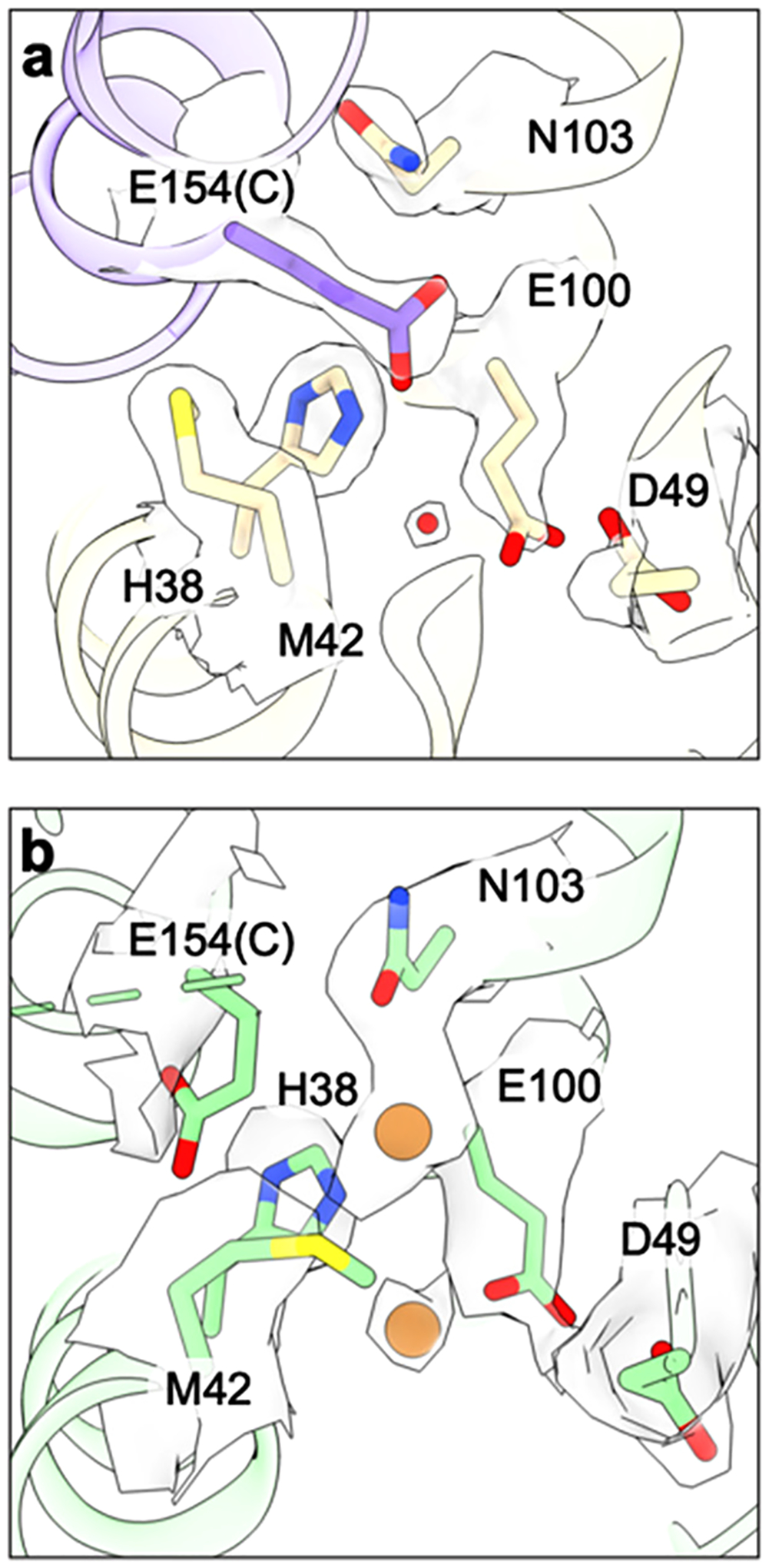
Proposed tricopper center in the PmoA subunit. (a) CryoEM structure of M. capsulatus (Bath) pMMO in native lipid nanodiscs showing the proposed tricopper center ligands and the corresponding density (PDB ID: 7S4H). (b) CryoEM structure of M. capsulatus (Bath) pMMO in DDM with copper ions and ligands shown as modeled with the corresponding density superimposed (PDB ID: 7EV9).
3.4.2. CuB Site.
The CuB site somewhat resembles the catalytic center of lytic polysaccharide monooxygenases (LPMOs), which hydroxylate and cleave glycosidic bonds of polysaccharides.209,210 The LPMO active site consists of a Cu(I) ion coordinated by the side chain and amino group of an N-terminal histidine and the side chain of a second histidine, together called a histidine brace. By contrast, the CuB site binds Cu(II) with three, not two, histidines, and differs in some details of coordination. In particular, the LPMO non-amino terminal histidine coordinates copper with its ε nitrogen atom while one of the non-amino terminal histidine residues in CuB uses its δ nitrogen atom. Nevertheless, the ability of LPMOs to activate strong C–H bonds led to the suggestion that CuB could be the site of methane oxidation.211 In support of this model, a soluble fragment of PmoB comprising the two periplasmic domains connected by a flexible linker (spmoB)57 or by monomers of apo ferritin185 ostensibly exhibited methane oxidation activity. However, further investigation of spmoB and variants thereof indicated that the activity was not from the CuB site but instead was likely attributable to reactions of the reductant duroquinol with O2.171 Consistent with this conclusion, the activity of the apo ferritin PmoB constructs was highly dependent on the presence of duroquinol.185
A number of additional observations are incompatible with CuB being the active site. First, the three histidine ligands are not conserved in the PmoB sequences of verrucomicrobial pMMOs, which instead contain methionine, proline, and glycine at these positions.113,212,213 Second, CuB is always present as Cu(II), even in whole cells,171 and is coordinatively saturated with four nitrogen ligands. Binding and activation of O2 would require reduction and the presence of an open coordination site. Relatedly, there are members of the LPMO family that have saturated copper coordination and do not exhibit LPMO activity.214 Third, CuB is exposed at the protein surface, and there is no obvious hydrophobic pocket for substrate binding. Finally, mutation of one of the CuB ligands in a related hydrocarbon monooxygenase from Mycobacterium strain NBB4 did not completely abolish activity.215
3.4.3. CuC Site.
In contrast to CuB, several lines of evidence suggest CuC as the likely active site. First, all the ligands to the CuC site are strictly conserved, including in the verrucomicrobial PmoC sequences. Second, an increase in the methane oxidation activity of M. sp. Rockwell pMMO nanodiscs observed upon copper supplementation is correlated with increased copper in the PmoC subunit as measured by nTDMS. This experiment, while not specifically pinpointing CuC, demonstrated that copper bound to PmoC is critical for activity.179 Third, the EPR signal attributed to CuC in purified pMMO is perturbed by the addition of 15N NO2−, and ENDOR data are consistent with NO2− binding to Cu(II) via its oxygen atom(s).171 This finding is significant, as NO2− inhibits methane oxidation216,217 and is therefore likely to bind at the active site. While there is no apparent substrate binding cavity near CuC in the crystal structures, a hydrophobic pocket adjacent to CuC and CuD is present in the cryoEM structures (section 3.4.4). Finally, mutation of any of the three residues corresponding to the CuC ligands in the M. strain NBB4 hydrocarbon monooxygenase completely abrogated activity.215
3.4.4. CuD Site.
When the full architecture of the region adjacent to CuC and the presence of CuD were revealed by the cryoEM structures of active pMMO in native lipid nanodiscs (sections 3.1 and 3.2.2), the CuC active site model was revised. The cryoEM maps of active samples, including M. capsulatus (Bath) pMMO in both native and POPC nanodiscs, revealed an occupied CuD site whereas the maps of samples with no activity, including M. alcaliphilum 20Z and M. sp. Rockwell pMMOs in POPC nanodiscs, exhibit an occupied CuC site and disorder at the CuD site.60 Thus, CuD occupancy appears to correlate with activity. Another key finding from the cryoEM structures in native nanodiscs is the presence of a hydrophobic cavity lined by residues from PmoA and PmoC, including three invariant phenylalanines from PmoC.60 Prior to these structures, there was no sign of a potential substrate binding cavity in pMMO.
The possibility of a CuD active site was further investigated by parallel ENDOR and cryoEM studies of M. capsulatus (Bath) pMMO in native nanodiscs in the presence of the inhibitor 2,2,2-trifluorethanol (TFE).178 Analysis of 19F ENDOR data revealed 19F couplings (Figure 15a) attributable to TFE interacting with the CuD site in an axial fashion with respect to the CuD ligand plane, with the fluorine-nuclei centroid ~5 Å away from the Cu(II) ion (Figure 15b). Modeling TFE bound with this Cu–F distance placed the TFE oxygen atom ~2 Å from CuD. CryoEM maps of the same samples showed new density connected to CuD, which was modeled well as TFE (Figure 15c). The average Cu–F distance is ~4.8 Å, consistent with the geometric information yielded by ENDOR analysis, and the TFE is situated in the aforementioned hydrophobic cavity, tilted axially out of plane with respect to the CuD-coordinating ligands. Similar experiments with 4,4,4-trifluorobutanol (TFB) showed 19F couplings to CuD via ENDOR with a larger density, modeled as TFB, connected to CuD in the cryoEM map. These combined data strongly support a model in which CuD and the surrounding cavity is the site of substrate binding and product formation. The possibility that CuC and CuD, separated by 5.7 Å, could be occupied by copper simultaneously in some form of pMMO remains open and is an important area for future investigation.
Figure 15.
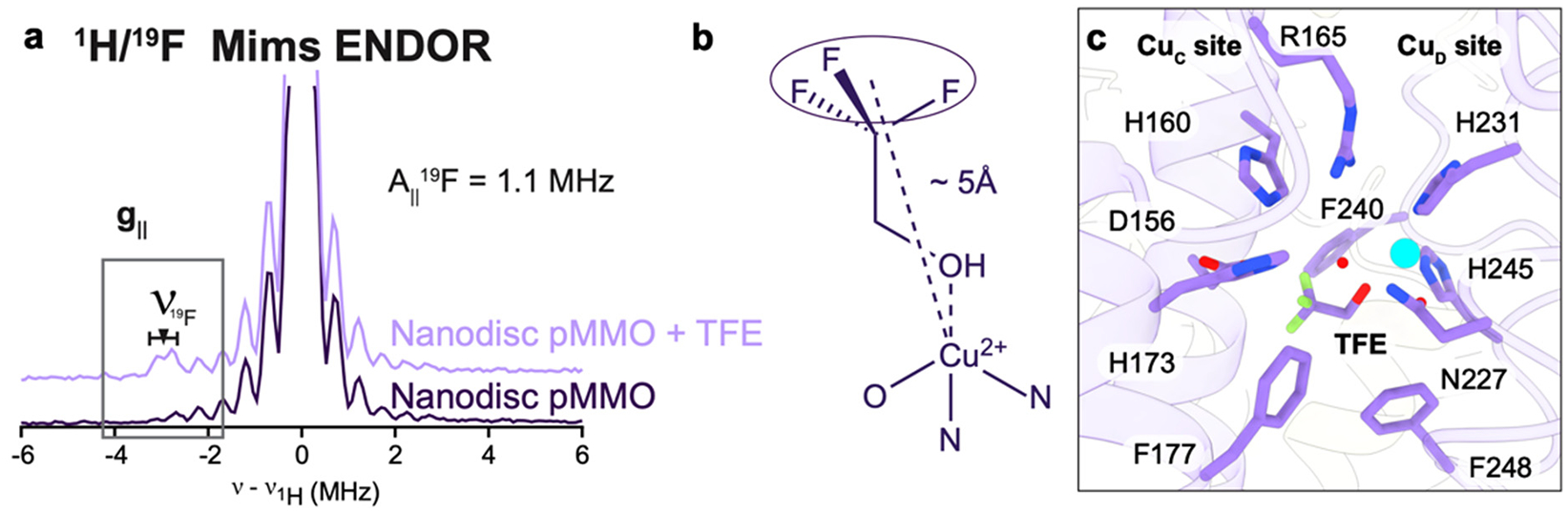
Product analog binding at the CuD site. (a) Q-band 1H/19F Mims pulsed ENDOR of M. capsulatus (Bath) pMMO in native lipid nanodiscs with (light purple) and without (dark purple) the addition of 20× TFE at g|| = 2.14 (~11200 G). (b) Model for the binding of TFE to Cu(II) based on the ENDOR-derived Cu(II)–F distance of ~5 Å. (c) Model of TFE bound at the CuD site based on the 2.19 Å resolution cryoEM map of M. capsulatus (Bath) pMMO in native lipid nanodiscs with 20× TFE added (PDB ID: 8OYI).
3.5. Interaction with Methanol Dehydrogenase
MDHs are dimeric enzymes that use a pyrroloquinoline quinone (PQQ)/calcium ion cofactor to convert methanol to formaldehyde. The MxaFI MDHs consist of two subunits, MxaF (64 kDa), which houses the PQQ cofactor, and MxaI (8.5 kDa), of which the function is not known, arranged in an α2β2 dimer (Figure 16a).218–220 A second type of MDH, XoxF, forms an α2 dimer of a single subunit and utilizes lanthanide ions instead of calcium (Figure 16b).221,222 In methanotrophs that possess both MDHs, expression is regulated by the presence of lanthanides, which repress transcription of MxaFI and activate transcription of XoxF.223–226 The verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV only possesses XoxF, and the presence of lanthanides is essential for its growth.45,48 Crystal structures of several methanotroph MDHs have been determined, including M. capsulatus (Bath) MxaFI,227 Methylotuvimicrobium buryatense 5GBC1 XoxF with lanthanum,228 and M. fumariolicum SolV XoxF in the presence of cerium,48 europium,229 and neodymium.230
Figure 16.
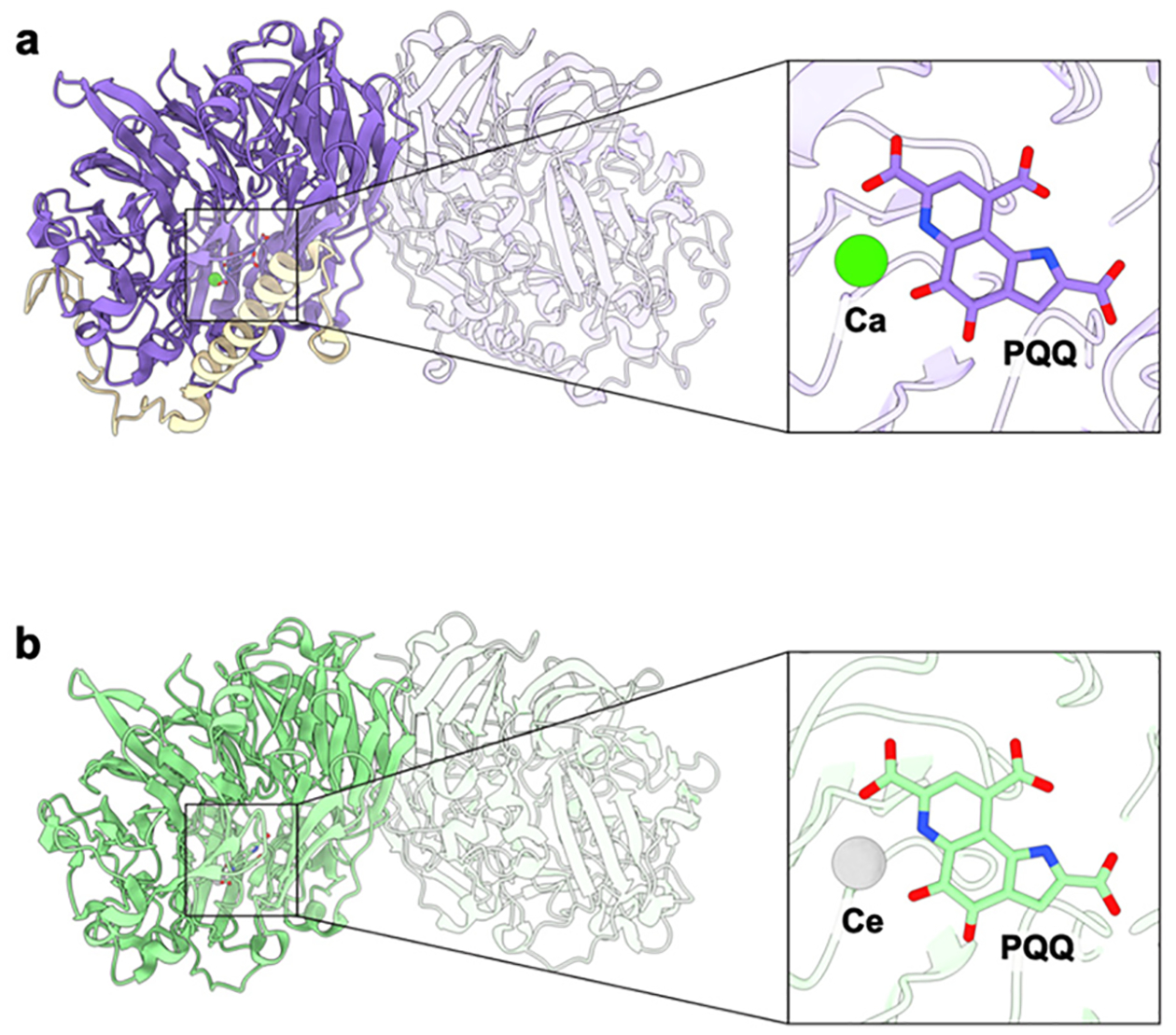
General architecture of methanol dehydrogenases and their active sites. (a) The calcium-dependent M. capsulatus (Bath) MDH shown with one αβ protomer highlighted (PDB ID: 4TQO). The MxaF subunit is shown in purple, and the MxaI subunit is shown in wheat. The inset shows the calcium (green) and the PQQ cofactor (purple) binding site. (b) The lanthanide-dependent XoxF MDH from M. fumariolicum SolV shown with one subunit of the homodimer highlighted in green (PDB ID: 4MAE). The inset shows the cerium (silver) and PQQ cofactor (green) binding site.
Several lines of evidence suggest that pMMO interacts directly with MDH. First, MDH, despite being a periplasmic enzyme, is typically associated with the ICMs.231–233 Second, transient interactions between M. capsulatus (Bath) pMMO and its cognate MxaFI as well as between M. buryatense 5GBC1 pMMO and its cognate XoxF have been detected by biolayer interferometry with KD values of ~9 and ~50 μM, respectively.227,228 An interaction between M. capsulatus (Bath) MxaFI and the spmoB protein was also detected, consistent with the location of MxaFI in the periplasm.227 Such interactions would facilitate channeling of the pMMO product methanol to the MDH active site and are consistent with the direct coupling model for electron transfer (section 3.3.1).
However, a stable pMMO-MDH complex has not been isolated by size exclusion chromatography or by reconstitution of purified proteins.227,228 A putative supercomplex between M. capsulatus (Bath) pMMO and its cognate MxaFI was reported based on a 16 Å resolution 3D volume acquired by cryoEM, but three MxaFI monomers were fit to the density,234 inconsistent with the dimeric structure of M. capsulatus (Bath) MxaFI. More recently, attempts to reproduce this result or determine the high resolution structure of a pMMO-MDH complex using improved cryoEM technology have been unsuccessful. It may be that pMMO-MDH complexes can only assemble on the membrane in the context of the pMMO arrays present in cells.130 On the basis of crystal packing interactions and the presence of multiple lysine residues, the small MxaI subunit was proposed to mediate interactions with negatively charged phospholipid headgroups in the membrane.227 This model is not generalizable to the XoxFs though, as these enzymes lack the second subunit. Nevertheless, support for the direct coupling electron transfer model, at least in type I methanotrophs,188,190,191,194 and longstanding evidence of MDH association with the membranes231–233 underscore the importance of pMMO-MDH interactions as an area for future study.
3.6. The PmoD Protein
The PmoD and AmoD/AmoE proteins belong to a unique protein family found only in methanotrophs and ammonia-oxidizing bacteria, suggesting that they are functionally linked to pMMO and AMO.115 In type II methanotrophs, including the Methylosinus and Methylocystis genera, and in gammaproteobacterial ammonia oxidizers, the pmoD/amoD gene is located directly adjacent to pmoB (Figure 5a).115,119,235 In betaproteobacterial ammonia oxidizers, amoE and amoD follow the pmo genes. Type I methanotrophs have genes encoding PmoD located elsewhere in the genome, typically adjacent to genes encoding multicopper oxidases or CopC proteins (Figure 5a).115,235 Notably, the genomes of methanotrophs and ammonia oxidizers contain multiple copies (2–11) of pmoD/amoD genes. In support of a function related to pMMO, M. trichosporium OB3b pmoD expression is coregulated with that of the pMMO subunits.68 Furthermore, its genetic disruption leads to a growth defect under pMMO-utilizing conditions, while growth under sMMO-utilizing (copper-starvation) conditions is not affected.115
PmoD proteins are predicted to comprise an N-terminal periplasmic domain followed by a transmembrane helix (Figure 4). The N-terminal domain of the PmoD protein encoded in the Mc. sp. Rockwell pmo operon has been biochemically and structurally characterized.115,236 In the presence of copper, this domain forms a dimeric species with optical and EPR spectroscopic features (Figure 17a) characteristic of a CuA site.237 Mutagenesis data indicate that unlike typical CuA sites, the ligands derive from two monomers, resulting in a symmetric site with distinct electronic properties (Figure 17b). The crystal structure of the CuA-bridged dimer has not been determined, but that of a monomeric species115 reveals key differences in the regions that provide the ligands in typical CuA domains (Figure 17c).238–240 The PmoD CuA site is also unusually unstable, decaying slowly to form two type 2 Cu(II) sites.236
Figure 17.
Structure of PmoD and a model for CuA site formation. (a) CW X-band (~9.5 GHz) EPR spectrum of the CuA of PmoD. Brackets indicate hyperfine splitting Az (adapted from ref 115). (b) Crystal structure of the PmoD soluble domain from M. sp. Rockwell (PDB ID: 6CPD) showing potential CuA-forming residues. (c) Model of CuA site formation between two PmoD proteins.
Formation of the CuA site is associated with the presence of a Cx7MxHxnC motif, which is characteristic of PmoDs encoded within pmo operons. PmoD homologs encoded in different genomic neighborhoods contain a variety of other potential metal binding motifs and also bind copper but do not form CuA sites.115 Full-length PmoD, including the C-terminal transmembrane helix, has not been biochemically characterized, so it remains unclear whether the CuA site or any type of copper site forms when PmoD is embedded in the membrane. Further, it is not known whether in vitro copper binding or CuA formation is related to the growth phenotype upon disruption of the pmo operon copy of the PmoD gene in the M. trichosporium OB3b.115 While PmoD has been proposed to play a role in pMMO copper loading, catalytic activity, and/or stabilization,115,236 further investigation is needed to elucidate its functional significance.
3.7. Mechanisms of Dioxygen and Methane C–H Bond Activation
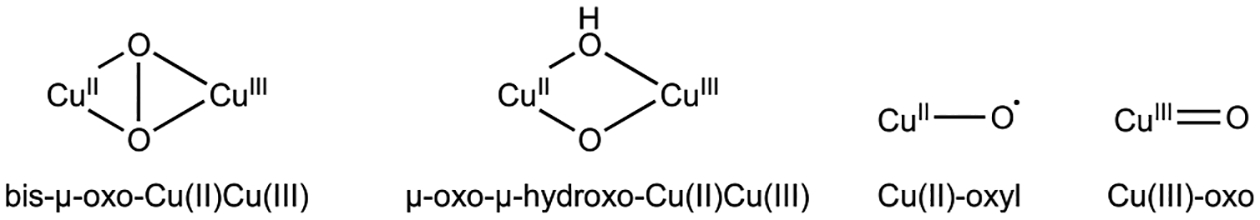
Despite the continually evolving picture of the pMMO copper active site, computational chemists have attempted to elucidate its mechanism of O2 activation, with a number of intermediates under consideration. Early studies by Yoshizawa and co-workers utilized the CuB site as a model, albeit with an oxygen atom from a nearby glutamic acid rather than the amino terminal group as a fourth ligand. Their calculated reaction pathways for M. capsulatus (Bath) pMMO involved conversion of a μ-η2:η2-peroxo-Cu(II)Cu(II) or a μ-η1:η2-peroxo-Cu(I)-Cu(II) species to a reactive bis(μ-oxo)Cu(II)Cu(III) or (μ-oxo)(μ-hydroxo)Cu(II)Cu(III) species (Figure 18) capable of methane oxidation.241–243 Formation of the latter species was proposed to occur via homolytic cleavage of the O–H bond in a nearby tyrosine residue, Tyr 374, followed by proton transfer to the μ-η2:η2-peroxo-Cu(II)Cu(II) core, yielding a μ-η1:η2 hydroperoxo-Cu(I)Cu(II) species that is converted to the (μ-oxo)(μ-hydroxo)-Cu(II)Cu(III) species.243 This mechanism was revisited more recently, this time suggesting that the nearby glutamic acid, Glu35, receives the proton from Tyr374, followed by transfer to the dicopper core.244 While Tyr374 is not strictly conserved, all the pMMO structures have a tyrosine near the CuB site. Since CuB is not dinuclear and no longer believed to be the site of methane oxidation (sections 3.2.1 and 3.4.2), these studies are likely not relevant. However, some of the proposed dicopper intermediates could be of interest in a scenario with CuC and CuD occupied simultaneously, assuming that a Cu–Cu distance significantly less than the 5.7 Å indicated by the cryoEM structures60 could be achieved via conformational changes.
Figure 18.
Copper-oxygen species proposed on the basis of computational studies to mediate methane oxidation by pMMO.
The reactivity of O2 with a monocopper center has also been investigated computationally. Using the bis-His site, which is not conserved and unlikely to bind O2, as a model, Yoshizawa and co-workers suggested that a Cu(III)-O (Cu(II)-O•) (Figure 18) species might be able to oxidize methane.241 Later studies by Ryde and co-workers using the mononuclear CuB site also suggested that a Cu(III)-O species can activate the methane C–H bond,174 though Cu(III) has yet to be detected in any biological system.245–247 A mechanism that does not invoke Cu(III) was proposed recently by Wang and co-workers.186 In this calculated mechanism, duroquinol binds at the CuC site, forming a Cu(II) duroquinol anion species. The duroquinol anion is then replaced by O2, coupled with electron transfer from O2 to the duroquinol anion to yield a Cu(II)-O2•− species and a duroquinol radical. A sequence of hydrogen atom abstraction and electron transfer steps involving a second duroquinol molecule then results in a Cu(II)-O reactive intermediate that reacts with methane. This mechanism is unlikely to be relevant to pMMO activity in vivo because duroquinol is not the physiological reductant of pMMO. Duroquinol is a synthetic analog of ubiquinol, which is too large to dock at the CuC site and thus could not participate in this mechanism. While the larger plastoquinol could be docked at the CuC site in this study, the structure used for docking lacked the crystallographically disordered region of PmoC and the CuD site, creating an artificial cavity.186 A well-folded PmoC subunit would likely preclude the binding of quinols at the CuC or CuD sites. Another recent study simulated ubiquinol binding directly at the CuD site,248 which, while more biologically plausible than the binding of synthetic duroquinol at this site, might be precluded by amino acid side chains and lipids blocking access to this cavity.
Several studies have employed different substrates to experimentally address the mechanism of C–H activation by pMMO. The reaction of membrane-bound pMMO with chiral ethane gave an intramolecular kinetic isotope effect kH/kD of 5.2–5.5 and was completely stereoselective, eliminating a mechanism involving alkyl radicals or cations and instead suggesting a concerted mechanism with a pentacoordinate hydrocarbon intermediate.249 Enantioselective hydroxylation was also observed for other substrates such as n-pentane, n-butane, and alkenes, albeit with less stereoselectivity, especially for the alkenes, as compared to ethane.200,201,250–252 No 12C/13C carbon kinetic isotope effect was detected using propane as a substrate, consistent with a concerted mechanism.253 Such studies should be interpreted cautiously, since sMMO is known to react with different substrates via different mechanisms (section 4.5).
3.8. Overexpression and Engineering
Heterologous expression of pMMO has had limited success, with no laboratory ever obtaining expression of all three subunits in E. coli. Soluble proteins corresponding to one or both of the PmoB periplasmic domains have been expressed in E. coli, but these proteins require refolding or the presence of fusion proteins and do not exhibit methane oxidation activity (section 3.4.2).57,170,171,185 The periplasmic domain of AmoB from the ammonia-oxidizing archaeon N. yellowstonii was expressed solubly without fusion tags but did not exhibit methane oxidation activity.156 There is one report of heterologous expression of M. trichosporium OB3b pMMO in Rhodococcus erythropolis LSSE8–1, but the whole cell activity is 2 orders of magnitude less than that of M. trichosporium OB3b, and the protein expression levels were not reported.28,254 Initial steps toward expression of the pMMO genes in plants have been reported, but evidence for assembly of pMMO or activity was not obtained.255 Finally, the hydrocarbon monooxygenase from M. strain NBB4, which is related to pMMO and oxidizes C2–C4 alkanes, was expressed in Mycobacterium smegmatis, conferring ethane, propane, and butane monooxygenase activity256 and allowing interrogation of several site-specific mutants,215 but further work with this system has not been reported. Thus, standard site-directed mutagenesis studies of pMMO have not been possible.
Another option for producing pMMO variants is genetic manipulation of native methanotrophs. Protocols for methanotroph gene disruption have been developed, providing insight into the functions of several proteins and facilitating metabolic engineering.26,257 Efforts to alter pMMO specifically are complicated by the presence of multiple pmo operons in most methanotroph genomes. Genetic tools exist for M. buryatense 5GB1C, which contains a single pmo operon,258,259 and site-directed mutagenesis should be possible in this strain. While growth under sMMO-utilizing conditions ought to be a viable strategy for obtaining pMMO variants, such efforts have not been successful. One possibility is that pMMO is still required for cell viability under copper-starvation conditions. In support of this idea, M. trichosporium OB3b pMMO is expressed constitutively and only mildly upregulated upon copper addition.68 Another strategy for generating point mutants is CRISPR-Cas9 genome editing, which can be performed with ~10% efficiency in M. capsulatus (Bath).257,260 The feasibility of this approach for site-directed mutagenesis of pMMO remains unproven, however.
Cell free protein synthesis (CFPS) represents a way to circumvent both difficulties with heterologous expression and the possibility that mutants with impaired pMMO viability will not grow under sMMO-utilizing conditions. In CFPS, the transcription and translation machinery is isolated from the cell,261 obviating the need for a functional pMMO for methanotroph cell viability. In recent work, M. capsulatus (Bath) pMMO was expressed in an E. coli lysate system directly into POPC nanodiscs.262 To generate the amino terminal histidine residue of the PmoB subunit, the native signal sequence was replaced by a SUMO fusion protein, and expression was conducted in the presence of SUMO protease. Remarkably, the pMMO trimer was assembled as demonstrated by negative stain EM and 2D class averaging. Activity assays on the cell-free reaction mixtures as well as on pMMO isolated from the mixture yielded no measurable methane oxidation, however. Nevertheless, this promising approach should be revisited as CFPS technology develops and more factors important for pMMO activity are elucidated.
4. SOLUBLE METHANE MONOOXYGENASE
4.1. Enzyme Structure
Three different proteins are required for methane oxidation by sMMO.16,263,264 The diiron active site is located in the multisubunit hydroxylase protein (MMOH). A reductase, MMOR, transfers two electrons from NADH to the MMOH diiron site via its two cofactors, FAD and a [2Fe-2S] cluster. The third component, MMOB, binds to MMOH and significantly increases its activity, as evidenced by a 1000-fold increase in reaction rate with dioxygen and a 150-fold increase in turnover number.265–267 A fourth protein encoded in the sMMO operon (Figure 5a), MMOD, inhibits sMMO activity.140,268 As noted above (section 2.3), MMOD has also been proposed to function in the copper switch,137,142 but its structure141 and biochemical properties68 are not consistent with this role. Another proposed role is iron loading of MMOH, but MMOD instead prevents reconstitution of apo MMOH with iron140 and reduces the rate of iron removal from MMOH.268
Components of sMMO from both M. capsulatus (Bath) and M. trichosporium OB3b have been structurally characterized (Table 3). The hydroxylase (MMOH), characterized first by crystallography269–271 and visualized 30 years later by cryoEM,272 comprises two copies each of the α, β, and γ subunits arranged in a 245 kDa α2β2γ2 dimer (Figure 19a). The α and β subunits are primarily α-helical and form a dimeric heart-shaped structure, similar to that of the R2 subunit of ribonucleotide reductase.273 The α subunit houses the diiron center in a four-helix bundle formed by helices labeled B, C, E, and F. The N-terminus of the β subunit comprises a helix that docks on the α subunit followed by a loop region that connects to the rest of the subunit. The two γ subunits, also helical, are found on opposite sides of the dimeric structure. NMR structures of MMOB (Figure 19b)274,275 and of the individual FAD/NADH binding and [2Fe-2S] cluster-containing domains of MMOR276–278 (Figure 19c) have been determined as well. The N-terminal 35 residues of MMOB are disordered in the NMR structures but were shown through NMR274 and DEER spectroscopies279 to interact with MMOH.
Table 3.
sMMO Structures
| M. capsulatus (Bath) MMOH (hydroxylase) | Resolution (Å) | PDB code |
|---|---|---|
| oxidized 4 °C | 2.20 | 1MMO |
| oxidized | 1.96 | 1FZ1 |
| oxidized | 1.70 | 1MTY |
| reduced in crystal | 2.15 | 1FYZ |
| anaerobically grown reduced | 2.40 | 1FZ5 |
| mixed valent, reduced in crystal | 2.15 | 1FZ2 |
| anaerobically grown mixed valent | 2.07 | 1FZ0 |
| methanol soaked | 2.05 | 1FZ6 |
| ethanol soaked | 1.96 | 1FZ7 |
| Xe pressurized | 3.30 | 1FZI |
| Xe pressurized | 2.60 | 1FZH |
| dibromomethane grown | 2.10 | 1FZ8 |
| iodoethane grown | 2.30 | 1FZ9 |
| pH 8.5 soaked | 2.38 | 1FZ4 |
| pH 6.2 soaked | 2.03 | 1FZ3 |
| Mn(II) soaked | 2.32 | 1XMF |
| apo (metal free) | 2.10 | 1XMG |
| Co(II) reconstituted | 2.32 | 1XMH |
| phenol soaked | 1.96 | 1XU5 |
| 6-bromohexanol soaked | 1.80 | 1XVB |
| 8-bromooctanol soaked | 2.00 | 1XVC |
| 4-fluorophenol soaked | 2.30 | 1XVD |
| 3-bromo-3-butenol soaked | 2.40 | 1XVE |
| chloropropanol soaked | 2.00 | 1XVF |
| bromoethanol soaked | 1.96 | 1XVG |
| bromophenol soaked | 2.30 | 1XU3 |
| cryoEM structure using graphene | 2.40 | 7TC8 |
| cryoEM structure using quantifoil | 2.90 | 7TC9 |
| M. trichosporium OB3b MMOH (hydroxylase) | Resolution (Å) | PDB code |
| oxidized | 2.00 | 1MHY |
| oxidized | 2.70 | 1MHZ |
| oxidized | 1.52 | 6VK6 |
| reduced in crystal | 2.12 | 6VK7 |
| MMOB, MMOR, and protein-protein complexes | Resolution (Å) | PDB code |
|---|---|---|
| M. capsulatus (Bath) MMOB NMR | 1CKV | |
| M. trichosporium OB3b MMOB NMR | 2MOB | |
| M. capsulatus (Bath) MMOR [2Fe-2S] domain NMR | 1JQ4 | |
| M. capsulatus (Bath) MMOR FAD/NADH binding domain NMR | 1TVC | |
| M. sporium MMOR FAD/NADH binding domain | 1.50 | 6L2U |
| M. capsulatus (Bath) MMOH-MMOB complex | 2.90 | 4GAM |
| M. trichosporium OB3b MMOH-MMOB with benzoate | 1.86 | 6VK5 |
| M. trichosporium OB3b MMOH-MMOB with succinate | 2.03 | 6VK8 |
| M. trichosporium OB3b MMOH-MMOB with one site reduced | 2.35 | 6VK4 |
| M. trichosporium OB3b MMOH-MMOB 5FW | 2.80 | 7M8Q |
| M. trichosporium OB3b MMOH-MMOB BTFA/K15C/5FW | 2.20 | 7M8R |
| M. trichosporium OB3b MMOH-MMOB S109A/T111A form 1 | 1.96 | 7S6Q |
| M. trichosporium OB3b MMOH-MMOB S109A/T111A form 2 | 2.40 | 7S7H |
| M. trichosporium OB3b MMOH-MMOB H5A | 1.89 | 7S6R |
| M. trichosporium OB3b MMOH-MMOB N107G/S110A | 1.98 | 7S6S |
| M. trichosporium OB3b MMOH-MMOB H33A | 1.82 | 7S6T |
| M. trichosporium OB3b diferric MMOH-MMOB XFEL | 1.95 | 6YD0 |
| M. trichosporium OB3b diferrous MMOH-MMOB XFEL | 1.95 | 6YDI |
| M. trichosporium OB3b reoxidized MMOH-MMOB XFEL | 1.95 | 6YDU |
| M. trichosporium OB3b diferrous MMOH-MMOB t = 0 XFELa | 2.00 | 6YY3 |
| M. sporium MMOH-MMOD | 2.60 | 67DK |
Treated the same as 6YDU but exposed to helium rather than O2.
Figure 19.
Structures of the sMMO proteins. (a) Overall structure of MMOH highlighting one protomer of the α2β2γ2 dimer (PDB ID: 1MTY). The α subunit is shown in light purple, the β subunit is shown in dark purple, and the γ subunit is shown in wheat. (b) Structure of MMOB (green, PDB ID: 4GAM). (c) Structure of the MMOR ferredoxin domain (light pink, PDB ID: 1JQ4) and the MMOR FAD domain (salmon, PDB ID: 1TVC).
Crystal structures of protein-protein complexes are also available (Table 3). The structures of both M. capsulatus (Bath)280,281 and M. trichosporium OB3b282 MMOH in complex with MMOB show that two molecules of MMOB bind symmetrically to the MMOH dimer, altering the conformations of α subunit helices E, F, and H (Figure 20a). The N-terminal 35 residues of MMOB order into a ring-like structure on the surface of MMOH, explaining why removal of the N-terminus obviates or significantly reduces sMMO activity280,283,284 and why mutation of specific N-terminal residues affects steps in the catalytic cycle.285 The MMOB C-terminus also becomes more ordered upon complexation, consistent with its truncation decreasing the MMOH turnover number.286 While a structure is not available for the MMOH-MMOR complex, hydrogen-deuterium exchange coupled to mass spectrometry analysis287 and chemical cross-linking data288 indicate that MMOR binds to the same region of MMOH as MMOB, specifically with its [2Fe-2S] cluster-containing domain occupying the MMOB binding site. Finally, a structure of the MMOH-MMOD complex from Methylosinus sporium strain 5 shows that MMOD binds in the same site as MMOB (Figure 20b), rationalizing its inhibitory effect in vitro.141 MMOD consists of four antiparallel β strands and a C-terminal α helix followed by an unstructured region comprising ~35 residues. Notably, MMOD displaces the N-terminal helix of the MMOH β subunit, causing helices B and C in the α subunit to shift position, thereby altering the geometry of the diiron active site.141
Figure 20.
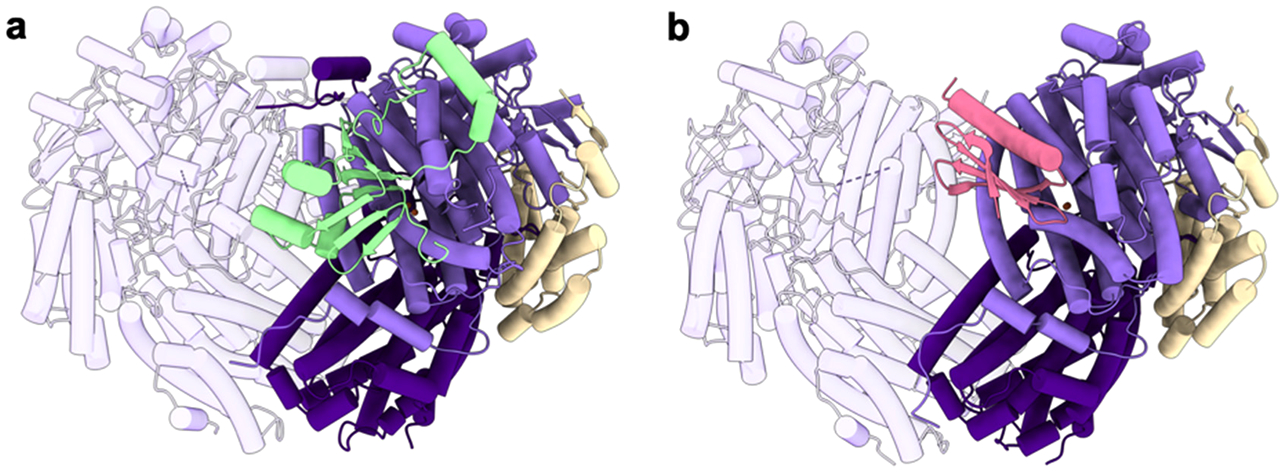
Structures of MMOH protein-protein complexes. (a) Structure of the M. capsulatus (Bath) MMOH-MMOB complex with MMOB shown in green (PDB ID: 4GAM). (b) Structure of the M. sporium MMOH-MMOD complex with MMOD shown in pink (PDB ID: 6D7K). The α subunits are shown in light purple, the β subunits are shown in dark purple, and the γ subunits are shown in wheat.
4.2. Active Site Structure
The diiron active site of sMMO, first identified by EPR, Mössbauer, and EXAFS spectroscopies, consists of two iron ions, Fe1 and Fe2, which are antiferromagnetically coupled in the Fe(III)Fe(III) state and weakly ferromagnetically coupled in the Fe(II)Fe(II) state.289–293 A mixed valent Fe(II)Fe(III) state can be generated, but it is not part of the catalytic cycle. MMOH has been crystallographically characterized in all three of these oxidation states (Table 3). In the Fe(III)Fe(III) state, the two iron ions are separated by 3.1 Å. Fe1 is coordinated by Glu114, His147, and a solvent molecule, while Fe2 is coordinated by Glu209, Glu243, and His246 (M. capsulatus (Bath) MMOH numbering). The two iron ions are bridged by two hydroxides and Glu144 (Figure 21a).269–271,294 In the Fe(II)Fe(II) state, the Fe–Fe distance increases to 3.3 Å, and Glu243 shifts to bridge Fe1 and Fe2, displacing a bridging a hydroxide and adopting a bidentate coordination to Fe2 (Figure 21b).282,295 In the Fe(II)Fe(III) state, the Fe–Fe distance increases to 3.3–3.4 Å and Glu144 no longer coordinates Fe2 (Figure 21c).294 MMOH has also been crystallized in the apo, Co(II)Co(II), and Mn(II)Mn(II) forms (Table 3).268 The latter two structures exhibit metal coordination geometries similar to that of reduced Fe(II)Fe(II) MMOH. In the MMOH-MMOB complex from M. capsulatus (Bath), Glu243 adopts coordination more similar to that of reduced MMOH.280 By contrast, the M. trichosporium OB3b MMOH-MMOB X-ray free electron laser (XFEL) structure determined at room temperature reveals a coordination similar to that of oxidized MMOH, suggesting that photoreduction occurred in the M. capsulatus (Bath) structure and that MMOB binding does not perturb Glu243.282
Figure 21.
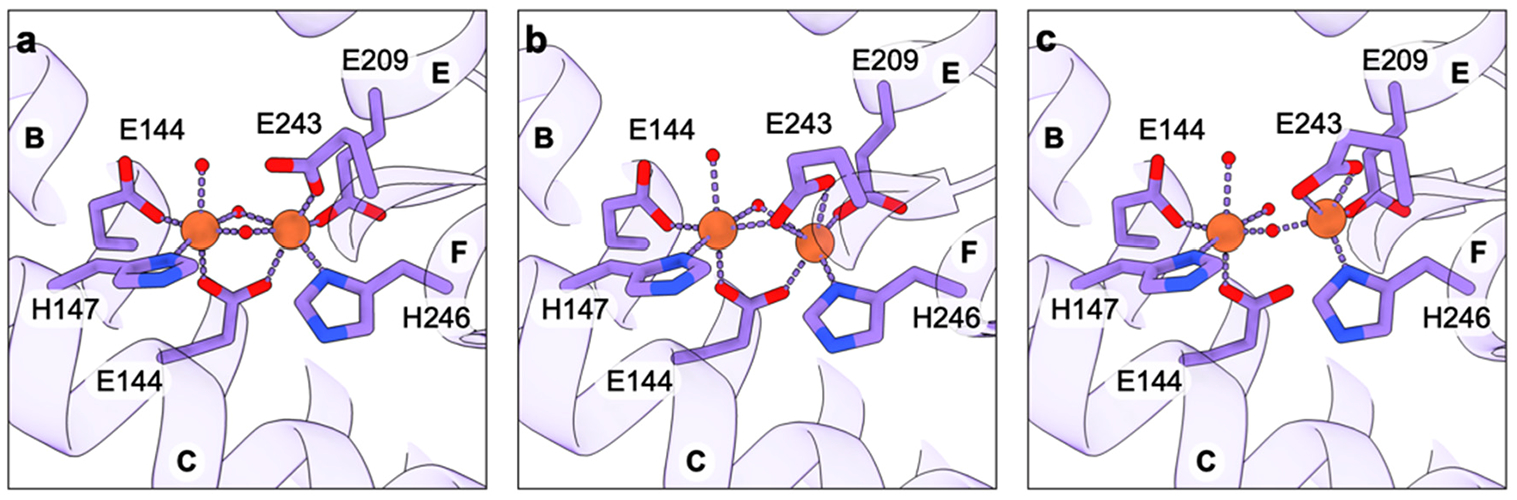
Active site of sMMO from M. capsulatus (Bath) with helices B, C, E, and F labeled. (a) The diiron cluster in the oxidized Fe(III)Fe(III) state (PDB ID: 1MTY). (b) The diiron cluster in the reduced Fe(II)Fe(II) state (PDB ID: 1FYZ). (c) The diiron cluster in the mixed valent Fe(II)Fe(III) state (PDB ID: 1FZ0).
Multiple structures of MMOH with substrates, substrate analogs, products, and product analogs bound at the diiron site are available (Table 3). The substrates dibromomethane and iodomethane and the substrate mimic xenon, often used to probe for O2 binding sites, bind in cavities extending from the diiron site to the surface (section 4.3),296 as do a range of halogenated product analogs.297 The products methanol, ethanol, 2-bromoethanol, 3-chloropropanol, 6-bromohexanol, and 3-bromo-3-butenol bind at the diiron site with the oxygen atom bridging the two iron ions.297,298 These structures are consistent with EPR and ENDOR data showing the binding of methanol, ethanol, DMSO, and TFE to the diiron center.291,299–301
4.3. Substrate Access to the Active Site
4.3.1. Chain of Cavities.
Possible pathways for substrate access to the MMOH diiron site have been investigated extensively. There are three hydrophobic cavities extending from the active site to the protein surface, denoted cavities 1, 2, and 3, as well as a pore connecting cavity 1 directly to the surface.269,302 Binding of substrate and product molecules in cavities 2 and 3 as well as at the diiron site-housing cavity 1 suggested that these pockets provide a route for methane and O2 entry.296–298 In particular, residues Phe188 and Leu110 form a gate, which is closed in oxidized M. capsulatus (Bath) MMOH and was proposed to control access to the diiron site from cavities 2 and 3.270,297 In support of this gating model, these two residues shift in the M. capsulatus (Bath) MMOH-MMOB complex, connecting the two cavities (Figure 22a).280
Figure 22.
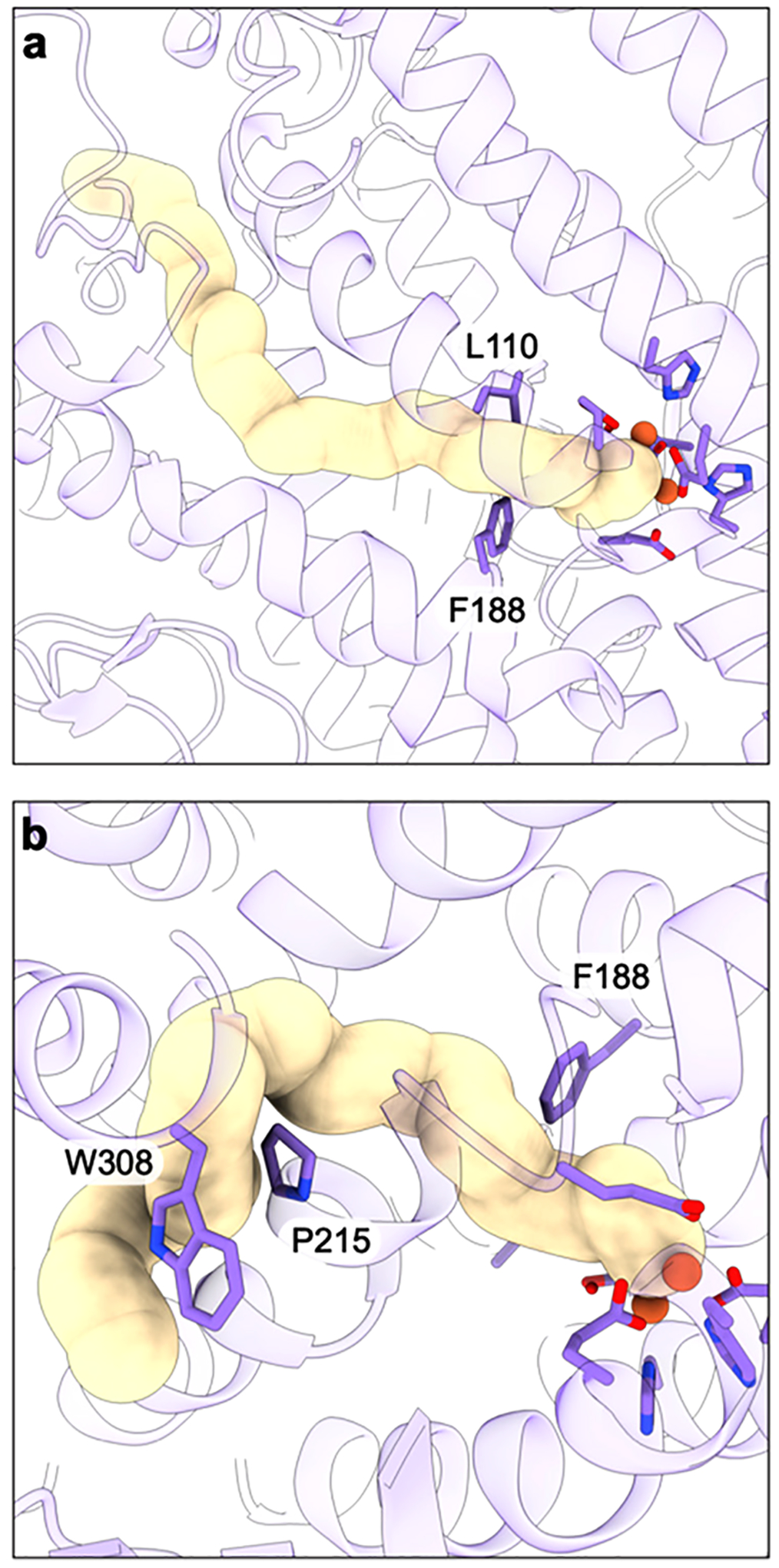
Proposed channels to and from the sMMO active site (PDB ID: 6YDI). (a) Substrate delivery channel to the hydrophobic pocket. (b) W308 tunnel 1 shown with key gating residues.
Different results were obtained for M. trichosporium OB3b MMOH: the gate is open in both oxidized MMOH and the oxidized MMOH-MMOB complex.303 Further complicating the interpretation, the gate is closed in both the oxidized and the reduced M. trichosporium OB3b MMOH-MMOB XFEL structures determined at room temperature.282 Thus, it seems that MMOB may serve to close, rather than open, the gate. Interestingly, the gate is open in structures of M. trichosporium OB3b MMOH-MMOB with bound benzoate and succinate, and further examination of the M. capsulatus (Bath) MMOH-MMOB complex electron density map suggests that an unmodeled substrate molecule might be present and perturb the gate in that structure.303 Another issue with this pathway is that the reaction kinetics are not consistent with methane accessing the diiron site from the 35–40 Å chain of cavities 1–3.16,304,305 In particular, the linear decay rate of reactive intermediate Q (section 4.4) with substrate concentration266,306 is inconsistent with the cavities filling with methane prior to reaction.
4.3.2. W308 Tunnels.
There are two possible alternatives to the cavity path. First, direct entry to the active site might be available through the pore region. However, binding of MMOB covers this region and blocks the diiron center.280,303 Second, a narrow tunnel, denoted W308 tunnel 1 (Figure 22b), has been identified recently using a probe with a solvent radius of 1.1 Å as opposed to the typically used water solvent radius of 1.4 Å. This tunnel is gated by residues Pro215 and Trp208 and is lined with conserved hydrophobic residues. The tunnel is closed in the structure of reduced MMOH but open in the reduced MMOH-MMOB complex from M. trichosporium OB3b.303,307 The binding of MMOB leads to organization of a dome of hydrophobic residues at the tunnel entrance, proposed to facilitate O2 entry.303 The tunnel is also adjacent to a number of MMOB residues shown by mutagenesis to be important for catalysis.284,304 Notably, replacement of MMOB residue Val41, located at the tunnel entry to the α subunit, with arginine and other bulky residues almost completely abrogated enzymatic activity.303 While both the chain of cavities (section 4.3.1) and W308 tunnel 1 have been proposed as access routes for both methane and O2, recent work suggests that a different, related path exists for methane access. This path, denoted W308 tunnel 2, is widened in the complex between MMOH and a double mutant of MMOB, S109A/T111A,308 consistent with this MMOB variant exhibiting increased rates of reactivity with larger substrates.309
4.4. Mechanism of Dioxygen Activation
Activation of O2 by sMMO has been studied extensively, with the first iron-oxygen intermediates reported 30 years ago.266,310 Single-turnover kinetic and spectroscopic studies of reduced MMOH with O2 in the presence of MMOB have established a detailed reaction cycle (Figure 23).15,16,285,311 The first intermediate, O, is an Fe(II)Fe(II) species that is proposed to have O2 bound to the protein but not at the diiron center, as it exhibits the same optical and EPR spectroscopic features as reduced MMOH.266,267,312,313 Intermediate O forms irreversibly, and its existence explains why formation of the subsequent intermediates does not depend on the O2 concentration.
Figure 23.
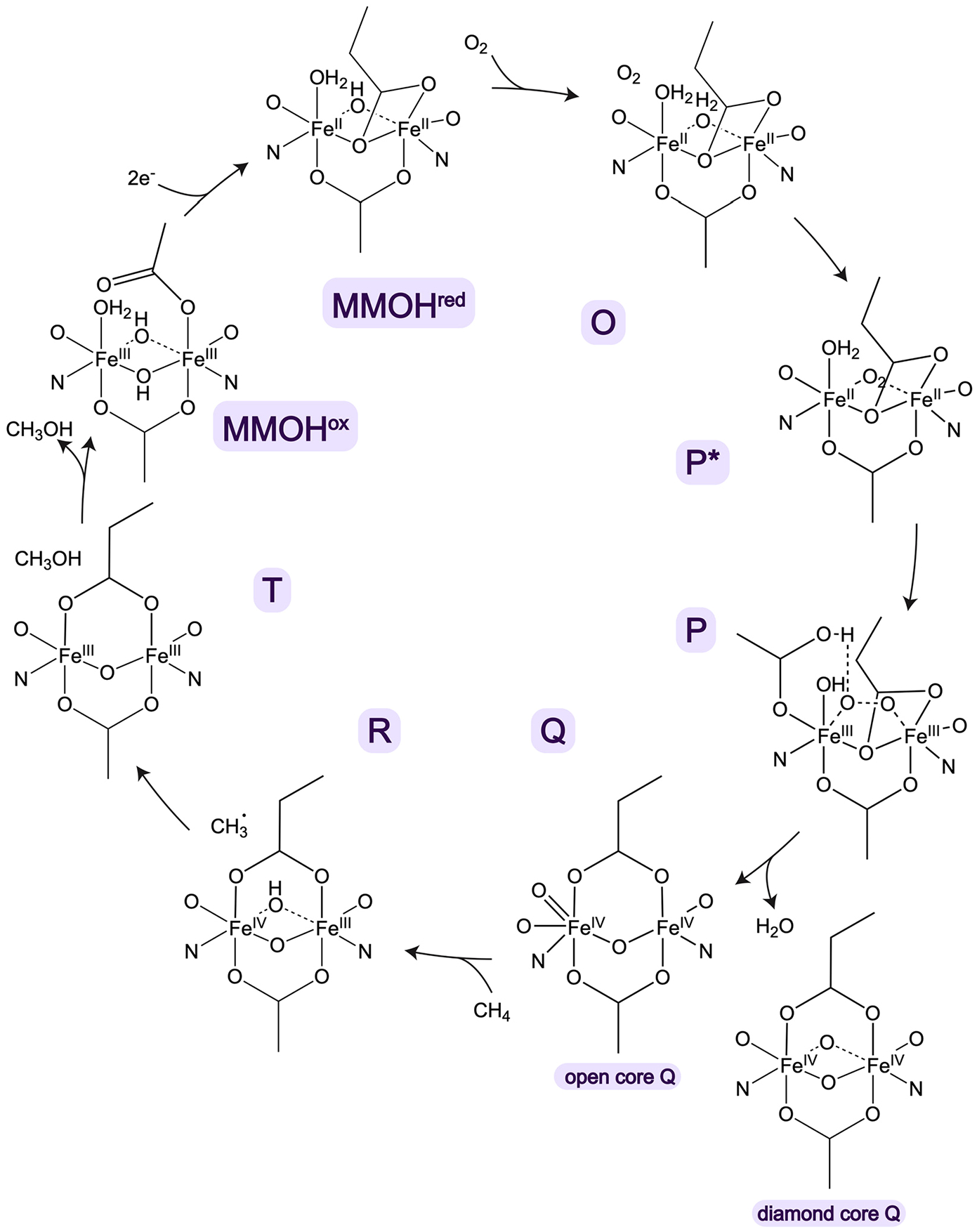
sMMO reaction cycle. All of the intermediates, with the exception of R, have been detected directly.
The binding of O2 to the diiron center then yields intermediate P* followed by intermediate P. For M. capsulatus (Bath) MMOH, intermediate P* was proposed to be an Fe(III)Fe(III) species differing from the subsequent intermediate P in the protonation of a coordinating ligand or solvent molecule.314 Intermediate P* in the M. trichosporium MMOH reaction cycle was also originally proposed to be an Fe(III)Fe(III) species, but Mössbauer data indicate that it is actually an Fe(II)Fe(II) species.315,316 These studies of M. trichosporium MMOH P* were facilitated by using the MMOB His33Ala variant, which slows the decay of P*.304,316 Intermediate P is an Fe(III)Fe(III) peroxo species, identified by its optical and Mössbauer spectra, which are consistent with a cis or trans μ−1,2 bridging coordination.306,311,315 The formation and decay of P depends on pH, and kinetic solvent isotope effects are observed in D2O, indicating that proton transfer, likely involving a bound solvent molecule or one of the carboxylate ligands, is involved.314,315
In the next step, the O–O bond is cleaved to form Q, the intermediate that reacts directly with methane. Intermediate Q is an antiferromagnetically coupled Fe(IV)Fe(IV) species,266,312,317 of which the exact structure has been the subject of ongoing debate (Figure 23). On the basis of Mössbauer parameters and EXAFS data fit with a short 2.46 Å Fe–Fe interaction, Q was proposed to have a diamond core structure,317,318 an assignment later supported by time-resolved resonance Raman data.319,320 However, difficulties reproducing the short Fe–Fe distance computationally321 and reactivity comparisons of biomimetic diamond and open core model compounds322 suggested that alternative structures might be plausible. Using HERFD XAS, it was possible to compare the pre-edge energy of Q with those of a range of Fe(IV)Fe(IV) model complexes. Combined with calculations, these data led to the conclusion that Q is better described as an open core structure.323,324 A comparison of newly acquired HERFD EXAFS data with the prior partial fluorescent yield (PFY) EXAFS results further indicated that the 2.46 Å Fe–Fe distance could derive from background contamination, and gave a revised Fe–Fe distance of 3.30–3.34 Å, which is more consistent with an open core.325 The tide then turned back, with a systematic nuclear resonance vibrational spectroscopic (NRVS) study supporting only closed core models.326 In addition, DFT calculations predict that concerted motions of the two oxo bridges in the closed core structure provide the reactivity necessary to break the methane C–H bond.326 Once Q reacts with methane (section 4.5), an oxo-bridged Fe(III)Fe(III) product complex with an oxygen atom derived from O2,319 intermediate T, is formed. Finally, methanol is released, regenerating the resting Fe(III)Fe(III) state (Figure 23).
4.5. Mechanism of C–H Activation
The reaction of intermediate Q with methane has been studied by a range of experimental and computational approaches. In contrast to pMMO (section 3.7), reactions with chiral ethane and chiral butane yield some inversion of stereochemistry, consistent with hydrogen abstraction by Q producing a short-lived radical intermediate.327–331 Radical clock substrates have also been employed as probes, including substituted cyclopropanes, methylcubane, and norcarane.332–337 In these studies, rearrangement of the probe substrate upon reaction with Q can inform upon the existence and lifetime of transient intermediates. The overall results are consistent with the involvement of a short-lived radical but suggest that different substrates are oxidized by different mechanisms, rendering it difficult to draw conclusions regarding methane oxidation.16,321,338,339
Kinetic isotope effect (KIE) measurements have also provided insight into the sMMO mechanism. A remarkably large KIE is obtained for the reaction of deuterated methane with intermediate Q, 50 for M. trichosporium OB3b sMMO and 28 for M. capsulatus (Bath) sMMO, while no KIE is measured using other substrates.306,340,341 Analysis of the temperature dependence of the KIE for methane is consistent with significant quantum tunneling,309 which is facilitated by interactions with MMOB. Using the MMOB quad variant (N107G/S109A/S110A/T111A), which increases the decay rate of Q with larger substrates presumably by increasing the size of the entry pathway (pore or other),304 reduces the methane KIE to 6. This result indicates that conformational changes upon interaction with MMOB are not only relevant to substrate access and methane specificity but also important for tunneling.341 Thus, MMOB facilitates selectivity for methane, despite it having the highest C–H bond strength, both by modulating substrate access and by enabling quantum tunneling.
4.6. Interplay between MMOR and MMOB Binding
The role of MMOR in providing electrons to MMOH is clear and the ways in which MMOB regulates substrate access and steps in the catalytic cycle have emerged over the years, but how the binding of these two proteins is orchestrated has been the subject of debate. MMOB is believed to prevent further reduction of intermediate Q by MMOR before it can react with methane. In support of this role, reduced MMOH in the presence of MMOR, but not MMOB, exhibits significantly less activity.342 As summarized recently, several distinct models for regulation of electron transfer from MMOR to MMOH have been proposed.343 In one scenario, MMOR and MMOB bind to MMOH simultaneously using separate binding sites, consistent with cross-linking data suggesting formation of a ternary complex.344 Alternatively, only one component can interact with MMOH at a time. This model is supported by hydrogen-deuterium exchange coupled to mass spectrometry analysis showing that the binding sites overlap and fluorescence anisotropy measurements indicating that the MMOR [2Fe-2S] domain can displace MMOB from MMOH.287 In this scenario, MMOR reduces MMOH and is then replaced by MMOB, which might remain loosely associated via its N-terminal region or might dissociate completely.
Displacement of MMOR from reduced MMOH by MMOB is consistent with fluorescence anisotropy data showing that M. capsulatus (Bath) MMOB has a higher affinity for reduced MMOH than for oxidized MMOH.279 The increased affinity of MMOB for reduced MMOH contradicts early reports that MMOB decreases the MMOH redox potential,345,346 which would mean it binds oxidized MMOH with higher affinity. Initial studies of M. trichosporium OB3b MMOB and MMOH using fluorescent probes did indicate a higher affinity for the oxidized form,347 but recent reinvestigation of the affinity of M. trichosporium OB3b MMOB for MMOH using 19F NMR gave different results.343 Instead of attaching large probes to cysteine residues in MMOB as done previously, two tryptophan residues in MMOB and one in MMOR were replaced with 5-fluorotryptophan. In addition, an MMOB variant labeled with 3-bromo-1,1,1-trifluoroacetone was generated. The combined use of these less disruptive labels and the sensitivity of 19F NMR provided new insight into the interactions between the components. In particular, the same binding constants were measured for the interactions of both oxidized and reduced M. trichosporium OB3b MMOH with MMOB. The observed effect of MMOB on the MMOH redox potential345,346 is not consistent with this finding but may be due to experimental considerations in the redox titration.343
The 19F NMR study also showed that the affinity of MMOR for MMOH is similar to that of MMOB for MMOH, regardless of the MMOH oxidation state. These observations suggest that there is an equilibrium and that MMOR can completely displace MMOB, ruling out the simultaneous binding mechanism. Thus, a third model was proposed in which MMOB and MMOR compete for the binding site on MMOH regardless of oxidation state (Figure 24).343 In this dynamic equilibrium model, irreversible reaction steps, including reduction of MMOH while in complex with MMOR and subsequent reactions with O2 and methane while in complex with MMOB, pull the reaction forward. The slow kinetics of MMOB dissociation from MMOH are proposed to protect intermediate Q from unproductive reduction by MMOR before it can react with methane.
Figure 24.
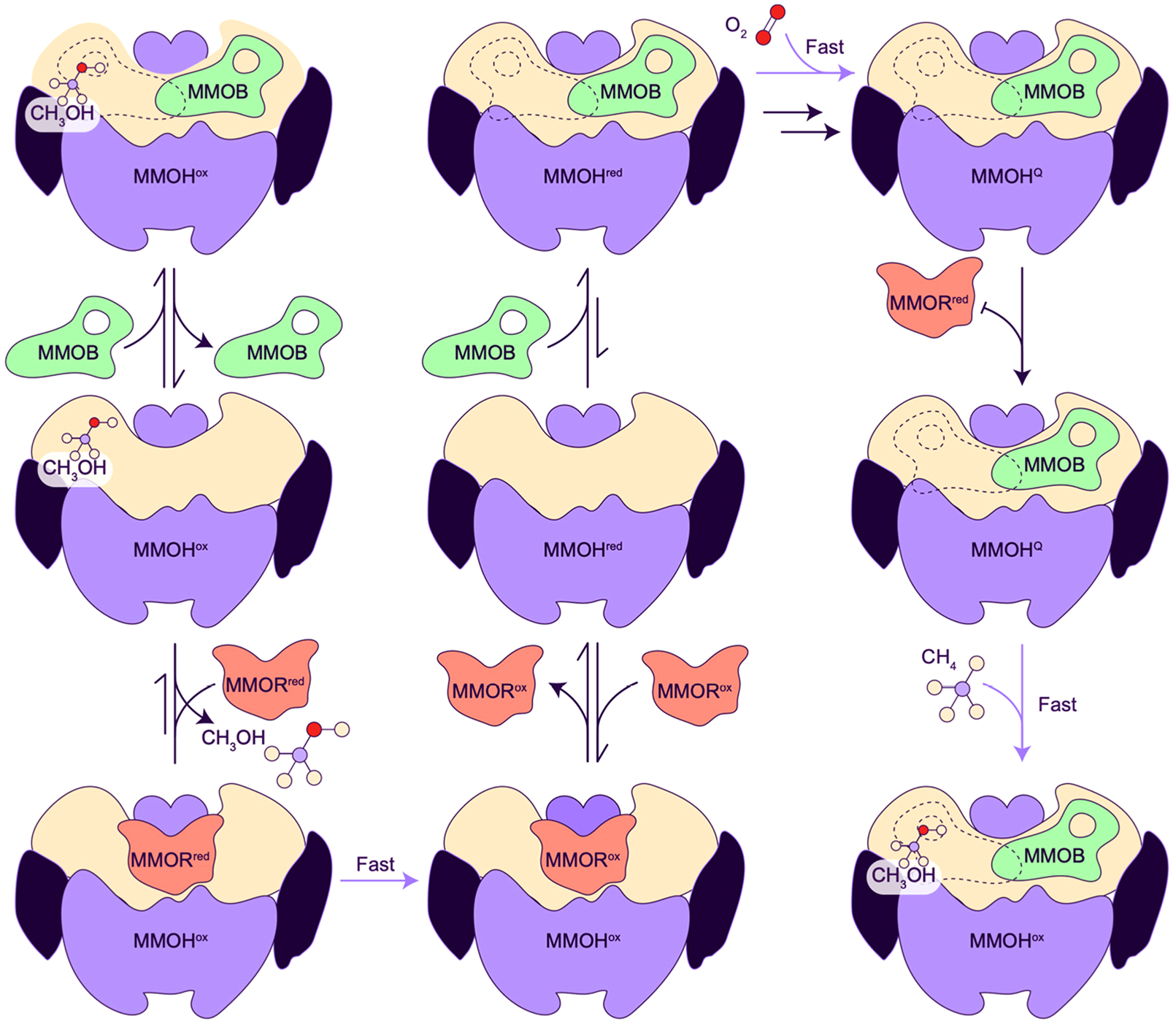
Model for regulation of electron transfer and substrate binding in sMMO adapted with permission from ref 343.
4.7. Overexpression and Engineering
Many studies of the MMOH mechanism and its interactions with MMOB and MMOR were facilitated by the ability to heterologously express MMOB in E. coli and produce site-specific and truncated variants. It has proven much more difficult to generate variants of MMOH. Expression of MMOH, MMOB, and MMOR in Pseudomonas strains has been reported, and Pseudomonas putida expressing sMMO degrades trichloroethylene (TCE) at 12.5% of the rate of TCE degradation by M. trichosporium OB3b.28,348,349 TCE oxidation was also observed for strains of Agrobacterium tumefaciens and Rhizobium meliloti expressing sMMO,350 but none of these systems were reported to oxidize methane.
More recently, attempts to express sMMO have focused on coexpression with the E. coli chaperone proteins GroES and GroEL. Coexpression of the Methylomonas methanica MC09 sMMO operon with E. coli GroEL and GroES led to assembly of the MMOH dimer as detected by native PAGE.351 This MMOH exhibited nitrobenzene oxidation activity at about half the level of M. trichosporium OB3b sMMO and an EPR signal consistent with the presence of a mixed valent Fe(II)Fe(III) center. While the sMMO operon encodes a GroEL homolog, MmoG (Figure 5a), it is unclear whether it interacts with a GroES homolog and MmoG alone is not sufficient to yield soluble MMOH. In a preprint report, screening and directed evolution yielded soluble M. capsulatus (Bath) MMOH upon coexpression with a GroEL/GroES pair from M. capsulatus (Bath). Methane conversion to methanol was observed in the E. coli cells expressing sMMO, and further metabolic engineering to produce acetone was successful.352 In addition, mutations that enhance activity, likely by enhancing solubility, were identified using directed evolution. Overall, this approach is promising both for biochemical studies and for biotechnological applications.
Another strategy for generating MMOH variants is homologous expression in methanotrophs. Site-directed mutagenesis of M. trichosporium OB3b MMOH has been performed by introducing genes with mutations into a strain lacking part of the sMMO operon. Unlike pMMO (section 3.8), sMMO is not required for cell viability, and these strains can be cultivated under pMMO-utilizing conditions followed by expression of sMMO variants as copper levels are lowered.143,353–355 This system has been used to alter several residues near the diiron center, including Cys151 and Thr213,353 as well as a leucine, Leu110, separating the active site from cavity 2.143 Two Cys151 variants, Cys151Glu and Cys151Tyr, could not be produced at high levels, while a Thr213Ser variant was purified and exhibited diminished propylene oxidation activity.353 Mutation of Leu110 to glycine, cysteine, arginine, and tyrosine resulted in differences in regioselectivity,143 as did alterations to Phe192, which resides close to the diiron center and to Arg98, which is part of a hydrogen bonding network proposed to modulate access to the cavity pathway.356 Despite these promising results over the past 20 years, this approach has yet to be deployed on large scale, likely due to limitations in working with methanotrophs. Finally, as noted above (section 3.8), CRISPR-Cas9 gene editing can be performed with ~10% efficiency in M. capsulatus (Bath),257,260 but its efficacy in producing point mutants has not yet been demonstrated.
5. CONCLUSIONS AND OUTLOOK
Within the time of preparing this article, the climate crisis has intensified, as manifested in dangerous air and water temperatures, wildfires and accompanying air pollution, and extreme weather. As an abundant yet short-lived greenhouse gas, methane is a prime target for immediate mitigation efforts, and methanotrophs and MMOs present a promising route forward. While significant progress has been made toward an atomic level understanding of both pMMO and sMMO function, important questions remain unanswered. The picture of pMMO has been revised multiple times, with the most recent studies indicating that the active site is located at the PmoC CuD site, with the possibility that CuD and CuC can be occupied simultaneously still on the table. Activity and structural studies of pMMO in membranes and native lipid nanodiscs have underscored the importance of studying the enzyme in its native environment, and future work should prioritize in situ characterization. Protein-protein interactions with candidates such as PmoD and MDH may only be detectable in situ and have the potential to shed light on the physiological reductant(s). The state of the field is more advanced for sMMO, with an established catalytic cycle and a detailed model of interactions between MMOH and its partner proteins MMOR and MMOB. Regulation of intermediate Q formation, methane selectivity, and active site access are also well understood. The nature of Q remains controversial, but the continual application of advanced techniques should resolve this debate in the near future. While computational studies have indicated that a pMMO monocopper site could oxidize methane, experimental evidence for reactive intermediates analogous to sMMO intermediate Q has not yet been obtained. It is likely that pMMO also has specific mechanisms for preventing inactivation of intermediates and the enzyme subunits themselves. Engineering of both MMOs is nascent, with progress in heterologous and homologous expression, cell free protein synthesis, and CRISPR-Cas9 gene editing over the past few years. Further efforts will be required to establish robust systems that will ultimately be scalable to a commercially viable level. Such systems will require a deep understanding of methanotroph physiology, particularly as pertains to the copper switch, metal acquisition, and ICM formation. In the future, biochemists and structural biologists will need to interface closely with microbiologists, synthetic biologists, and engineers to leverage the full potential of these remarkable bacteria and enzymes for the sake of our planet.
ACKNOWLEDGMENTS
Research in the Rosenzweig laboratory on pMMO is supported by NIH grant R35GM118035. Research in the Rosenzweig laboratory on PmoD is supported by DOE grant DE-SC0016284. F.J.T. was supported by NIH grants T32GM105538 and F31ES034283.
Biographies
Frank J. Tucci, from Hartford, CT, received his B.A. in Chemistry and in Neuroscience from Wesleyan University, where he conducted research in the laboratory of Professor Erika A. Taylor, studying an enzyme involved in lipopolysaccharide biosynthesis, Heptosyltransferase I. Frank is now a Ph.D. candidate in the laboratory of Professor Amy C. Rosenzweig at Northwestern University, where he is using cryoEM to study pMMO, with particular interests in its copper centers, its interactions with the native membrane, and electron microscopy methods. He is a former trainee of the NIH-funded Chemistry of Life Processes Training Program and is currently supported by a Ruth L. Kirschstein Predoctoral Individual National Research Service Award and the Northwestern University Rappaport Award for Research Excellence.
Amy C. Rosenzweig, originally from Pittsburgh, PA, is the Weinberg Family Distinguished Professor of Life Sciences in the Departments of Molecular Biosciences and Chemistry at Northwestern University. She received a B.A. in Chemistry from Amherst College and a Ph.D. in Inorganic Chemistry from Massachusetts Institute of Technology. The Rosenzweig laboratory uses structural, biochemical, and biophysical approaches to attack problems at the forefront of bioinorganic chemistry. Their work has been honored recently by the American Chemical Society Alfred Bader Award in Bioinorganic or Bioorganic Chemistry and the Protein Society Hans Neurath Award. Rosenzweig, a member of the National Academy of Sciences and a fellow of the American Academy of Arts and Sciences, is currently chair of the Department of Molecular Biosciences.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.chemrev.3c00727
Special Issue Paper
This paper is an additional review for Chem. Rev. 2023, volume 123, issue 9, “Bridging the Gaps: Learning from Catalysis across Boundaries”.
The authors declare no competing financial interest.
Contributor Information
Frank J. Tucci, Departments of Molecular Biosciences and of Chemistry, Northwestern University, Evanston, Illinois 60208, United States
Amy C. Rosenzweig, Departments of Molecular Biosciences and of Chemistry, Northwestern University, Evanston, Illinois 60208, United States
REFERENCES
- (1).Mar KA; Unger C; Walderdorff L; Butler T Beyond CO2 equivalence: The impacts of methane on climate, ecosystems, and health. Environ. Sci. Policy 2022, 134, 127–136. [Google Scholar]
- (2).Saunois M; Stavert AR; Poulter B; Bousquet P; Canadell JG; Jackson RB; Raymond PA; Dlugokencky EJ; Houweling S; Patra PK; et al. The global methane budget 2000–2017. Earth Syst. Sci. Data 2020, 12, 1561–1623. [Google Scholar]
- (3).Lan X; Thoning KW; Dlugokencky EJ Trends in globally-averaged CH4, N2O, and SF6 determined from NOAA Global Monitoring Laboratory measurements, Version 2023–06; 2023. DOI: 10.15138/P8XG-AA10. [DOI] [Google Scholar]
- (4).Lan X; Nisbet EG; Dlugokencky EJ; Michel SE What do we know about the global methane budget? Results from four decades of atmospheric CH4 observations and the way forward. Philos. Trans. R. Soc. A 2021, 379, 20200440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Dhakal S; Minx JC; Toth FL; Abdel-Aziz A; Meza MJF; Hubacek K; Jonckheere IGC; Kim Y-G; Nemet GF; Pachauri S; et al. IPCC, 2022: Climate Change 2022: Mitigation of Climate Change. Contribution of Working Group III to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press, 2022. [Google Scholar]
- (6).Abernethy S; O’Connor F; Jones C; Jackson R Methane removal and the proportional reductions in surface temperature and ozone. Philos. Trans. Royal Soc. A 2021, 379, 20210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Haynes CA; Gonzalez R Rethinking biological activation of methane and conversion to liquid fuels. Nat. Chem. Biol 2014, 10, 331–339. [DOI] [PubMed] [Google Scholar]
- (8).Ail SS; Dasappa S Biomass to liquid transportation fuel via Fischer-Tropsch synthesis - Technology review and current scenario. Renewable Sustainable Energy Rev 2016, 58, 267–286. [Google Scholar]
- (9).Dummer NF; Willock DJ; He Q; Howard MJ; Lewis RJ; Qi GD; Taylor SH; Xu J; Bethell D; Kiely CJ; et al. Methane oxidation to methanol. Chem. Rev 2023, 123, 6359–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ravi M; Ranocchiari M; van Bokhoven JA The direct catalytic oxidation of methane to methanol-a critical assessment. Angew. Chem., Int. Ed. Engl 2017, 56, 16464–16483. [DOI] [PubMed] [Google Scholar]
- (11).Crabtree RH Aspects of methane chemistry. Chem. Rev 1995, 95, 987–1007. [Google Scholar]
- (12).Arndtsen BA; Bergman RG; Mobley TA; Peterson TH Selective intermolecular carbon-hydrogen bond activation by synthetic metal complexes in homogeneous solution. Acc. Chem. Res 1995, 28, 154–162. [Google Scholar]
- (13).Hanson RS; Hanson TE Methanotrophic bacteria. Microbiol. Rev 1996, 60, 439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Murguia-Flores F; Arndt S; Ganesan AL; Murray-Tortarolo G; Hornibrook ERC Soil Methanotrophy Model (MeMo v1.0): a process-based model to quantify global uptake of atmospheric methane by soil. Geosci. Model Dev 2018, 11, 2009–2032. [Google Scholar]
- (15).Ross MO; Rosenzweig AC A tale of two methane monooxygenases. J. Biol. Inorg. Chem 2017, 22, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Banerjee R; Jones JC; Lipscomb JD Soluble methane monooxygenase. Annu. Rev. Biochem 2019, 88, 409–431. [DOI] [PubMed] [Google Scholar]
- (17).Koo CW; Rosenzweig AC Biochemistry of aerobic biological methane oxidation. Chem. Soc. Rev 2021, 50, 3424–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wu ML; Ettwig KF; Jetten MSM; Strous M; Keltjens JT; van Niftrik L A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera’. Biochem. Soc. Trans 2011, 39, 243–248. [DOI] [PubMed] [Google Scholar]
- (19).Welte CU; Rasigraf O; Vaksmaa A; Versantvoort W; Arshad A; Op den Camp HJM; Jetten MSM; Luke C; Reimann J Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ. Microbiol. Rep 2016, 8, 941–955. [DOI] [PubMed] [Google Scholar]
- (20).Cai C; Zhang XQ; Wu MX; Liu T; Lai CY; Frank J; He BQ; Marcellin E; Guo JH; Hu SH; et al. Roles and opportunities for microbial anaerobic oxidation of methane in natural and engineered systems. Energy Environ. Sci 2021, 14, 4803–4830. [Google Scholar]
- (21).Thauer RK Functionalization of methane in anaerobic microorganisms. Angew. Chem., Int. Ed 2010, 49, 6712–6713. [DOI] [PubMed] [Google Scholar]
- (22).Segarra KEA; Schubotz F; Samarkin V; Yoshinaga MY; Hinrichs KU; Joye SB High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat. Commun 2015, 6, 7477. [DOI] [PubMed] [Google Scholar]
- (23).Kalyuzhnaya MG; Puri AW; Lidstrom ME Metabolic engineering in methanotrophic bacteria. Metabolic Engineering 2015, 29, 142–152. [DOI] [PubMed] [Google Scholar]
- (24).Strong PJ; Kalyuzhnaya M; Silverman J; Clarke WP A methanotroph-based biorefinery: Potential scenarios for generating multiple products from a single fermentation. Bioresour. Technol 2016, 215, 314–323. [DOI] [PubMed] [Google Scholar]
- (25).Semrau JD; DiSpirito AA Methanotrophy - environmental, industrial and medical applications. Curr. Issues Mol. Biol 2019, 33, 1–22. [DOI] [PubMed] [Google Scholar]
- (26).Nguyen AD; Lee EY Engineered methanotrophy: a sustainable solution for methane-based industrial biomanufacturing. Trends Biotechnol 2021, 39, 381–396. [DOI] [PubMed] [Google Scholar]
- (27).Pham DN; Nguyen AD; Lee EY Outlook on engineering methylotrophs for one-carbon-based industrial biotechnology. Chem. Eng. J 2022, 449, 137769. [Google Scholar]
- (28).Lawton TJ; Rosenzweig AC Methane-oxidizing enzymes: An upstream problem in biological gas-to-liquids conversion. J. Am. Chem. Soc 2016, 138, 9327–9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Sirajuddin S; Rosenzweig AC Enzymatic oxidation of methane. Biochemistry 2015, 54, 2283–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Söhngen N Über bakterien, welche methane als Kohlen-stoffnahrung und energiequelle gebrauchen. Centr. Bakt. Parasitenk, Abt. II 1906, 15, 513–517. [Google Scholar]
- (31).Dworkin M; Foster JW Studies on Pseudomonas methanica (Söhngen) nov. comb. J. Bacteriol 1956, 72, 646–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Brown LR; Strawinski RJ; McCleskey CS Isolation and characterization of Methanomonas methanooxidans Brown and Strawinski. Can. J. Microbiol 1964, 10, 791–799. [DOI] [PubMed] [Google Scholar]
- (33).Stocks PK; McCleskey CS Morphology and phyisology of Methanomonas methanooxidans. J. Bacteriol 1964, 88, 1071–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Foster JW; Davis RH A methane-dependent coccus with notes on classification and nomenclature of obligate methane-utilizing bacteria. J. Bacteriol 1966, 91, 1924–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Leadbetter ER; Foster JW Incorporation of molecular oxygen in bacterial cells utilizing hydrocarbons for growth. Nature 1959, 184, 1428–1429. [DOI] [PubMed] [Google Scholar]
- (36).Higgins IG; Quayle JR Oxygenation of methane by methane-utilizing bacteria. Biochem. J 1970, 118, 28P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Davies SL; Whittenbury R Fine structure of methane and other hydrocarbon-utilizing bacteria. J. Gen. Microbiol 1970, 61, 227–232. [DOI] [PubMed] [Google Scholar]
- (38).Makula RA Phospholipid composition of methane-oxidizing bacteria. J. Bacteriol 1978, 134, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Bowman JP; Sly LI; Nichols PD; Hayward AC Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol 1993, 43, 735–753. [Google Scholar]
- (40).Bowman JP; Sly LI; Stackebrandt E The phylogenetic position of the family Methylococcaceae. Int. J. Syst. Bacteriol 1995, 45, 182–185. [DOI] [PubMed] [Google Scholar]
- (41).Semrau JD; Dispirito AA; Yoon S Methanotrophs and copper. FEMS Microbiol. Lett 2010, 34, 496–531. [DOI] [PubMed] [Google Scholar]
- (42).Dedysh SN; Knief C; Dunfield PF Methylocella species are facultatively methanotrophic. J. Bacteriol 2005, 187, 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dedysh SN; Dunfield PF Facultative methane oxidizers. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis KN, Ed.; Springer, 2010; pp 1967–1976. [Google Scholar]
- (44).Whittenbury R; Dalton H The methylotrophic bacteria. In The Procaryotes; Starr MP, Stolp H, Truper HG, Balowes A, Schlegel HG, Eds.; Springer-Verlag, 1981; pp 894–902. [Google Scholar]
- (45).Pol A; Heijmans K; Harhangi HR; Tedesco D; Jetten MSM; Op den Camp HJM Methanotrophy below pH1 by a new Verrucomicrobia species. Nature 2007, 450, 874–878. [DOI] [PubMed] [Google Scholar]
- (46).Dunfield PF; Yuryev A; Senin P; Smirnova AV; Stott MB; Hou S; Ly B; Saw JH; Zhou Z; Ren Y; et al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 2007, 450, 879–882. [DOI] [PubMed] [Google Scholar]
- (47).Islam T; Jensen S; Reigstad LJ; Larsen O; Birkeland NK Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U.S.A 2008, 105, 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Pol A; Barends TRM; Dietl A; Khadem AF; Eygensteyn J; Jetten MSM; Op den Camp HJM Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ. Microbiol 2014, 16, 255–264. [DOI] [PubMed] [Google Scholar]
- (49).Knief C Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol 2015, 6, 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Knief C Diversity of methane-cycling microorganisms in soils and their relation to pxygen. Curr. Issues Mol. Biol 2019, 33, 23–56. [DOI] [PubMed] [Google Scholar]
- (51).Trotsenko YA; Khmelenina VN Biology of extremophilic and extremotolerant methanotrophs. Arch. Microbiol 2002, 177, 123–131. [DOI] [PubMed] [Google Scholar]
- (52).Sharp CE; Smirnova AV; Graham JM; Stott MB; Khadka R; Moore TR; Grasby SE; Strack M; Dunfield PF Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol 2014, 16, 1867–1878. [DOI] [PubMed] [Google Scholar]
- (53).Trotsenko YA; Murrell JC Metabolic aspects of aerobic obligate methanotrophy. In Advances in Applied Microbiology; Laskin AL, Sariaslani S, Eds.; Elsevier Academic Press Inc., 2008; Vol 63, pp 183–229. [DOI] [PubMed] [Google Scholar]
- (54).Kalyuzhnaya MG; Yang S; Rozova ON; Smalley NE; Clubb J; Lamb A; Gowda GAN; Raftery D; Fu Y; Bringel F Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun 2013, 4, 2785. [DOI] [PubMed] [Google Scholar]
- (55).Khadem AF; Pol A; Wieczorek A; Mohammadi SS; Francoijs KJ; Stunnenberg HG; Jetten MSM; Op den Camp HJM Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J. Bacteriol 2011, 193, 4438–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).van Teeseling MC; Pol A; Harhangi HR; van der Zwart S; Jetten MS; Op den Camp HJ; van Niftrik L Expanding the verrucomicrobial methanotrophic world: description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol 2014, 80, 6782–6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Balasubramanian R; Smith SM; Rawat S; Stemmler TL; Rosenzweig AC Oxidation of methane by a biological dicopper centre. Nature 2010, 465, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Smith SM; Balasubramanian R; Rosenzweig AC Metal reconstitution of particulate methane monooxygenase and heterologous expression of the pmoB subunit. Methods Enzymol 2011, 495, 195–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Sirajuddin S; Barupala D; Helling S; Marcus K; Stemmler TL; Rosenzweig AC Effects of zinc on particulate methane monooxygenase activity and structure. J. Biol. Chem 2014, 289, 21782–21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Koo CW; Tucci FJ; He Y; Rosenzweig AC Recovery of particulate methane monooxygenase activity in a lipid bilayer. Science 2022, 375, 1287–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Scott D; Brannan J; Higgins IJ The effect of growth conditions on intracytoplasmic membranes and methane monooxygenase activities in Methylosinus trichosporium OB3b. J. Gen. Microbiol 1981, 125, 63–72. [Google Scholar]
- (62).Stanley SH; Prior SD; Leak DJ; Dalton H Copper stress underlies the fundamental change in intracellular location of methane monooxygenase in methane oxidizing organisms: studies in batch and continuous cultures. Biotechnol. Lett 1983, 5, 487–492. [Google Scholar]
- (63).Prior SD; Dalton H The effect of copper ions on membrane content and methane monooxygenase activity in methanol-grown cells of Methylococcus capsulatus (Bath). J. Gen. Microbiol 1985, 131, 155–163. [Google Scholar]
- (64).Kenney GE; Rosenzweig AC Methanobactins: maintaining copper homeostasis in methanotrophs and beyond. J. Biol. Chem 2018, 293, 4606–4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Kenney GE; Rosenzweig AC Chalkophores. Annu. Rev. Biochem 2018, 87, 645–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Semrau JD; DiSpirito AA; Obulisamy PK; Kang-Yun CS Methanobactin from methanotrophs: genetics, structure, function and potential applications. FEMS Microbiol. Lett 2020, 367, fnaa045. [DOI] [PubMed] [Google Scholar]
- (67).Kenney GE; Rosenzweig AC Genome mining for methanobactins. BMC Biol 2013, 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Kenney GE; Sadek M; Rosenzweig AC Copper-responsive gene expression in the methanotroph Methylosinus trichosporium OB3b. Metallomics 2016, 8, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Kenney GE; Dassama LMK; Pandelia ME; Gizzi AS; Martinie RJ; Gao P; DeHart CJ; Schachner LF; Skinner OS; Ro SY; et al. The biosynthesis of methanobactin. Science 2018, 359, 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Park YJ; Jodts RJ; Slater JW; Reyes RM; Winton VJ; Montaser RA; Thomas PM; Dowdle WB; Ruiz A; Kelleher NL; et al. A mixed-valent Fe(II)Fe(III) species converts cysteine to an oxazolone/thioamide pair in methanobactin biosynthesis. Proc. Natl. Acad. Sci. U.S.A 2022, 119, No. e2123566119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Park YJ; Kenney GE; Schachner LF; Kelleher NL; Rosenzweig AC Repurposed HisC aminotransferases complete the biosynthesis of some methanobactins. Biochemistry 2018, 57, 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Behling LA; Hartsel SC; Lewis DE; Dispirito AA; Choi DW; Masterson LR; Veglia G; Gallagher WH NMR, mass spectrometry and chemical evidence reveal a different chemical structure for methanobactin that contains oxazolone rings. J. Am. Chem. Soc 2008, 130, 12604–12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Krentz BD; Mulheron HJ; Semrau JD; DiSpirito AA; Bandow NL; Haft DH; Vuilleumier S; Murrell JC; McEllistrem MT; Hartsel SC; et al. A comparison of methanobactins from Methylosinus trichosporium OB3b and Methylocystis strain SB2 predicts methanobactins are synthesized from diverse peptide precursors modified to create a common core for binding and reducing copper ions. Biochemistry 2010, 49, 10117–10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).El Ghazouani A; Basle A; Gray J; Graham DW; Firbank SJ; Dennison C Variations in methanobactin structure influences copper utilization by methane-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A 2012, 109, 8400–8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Kenney GE; Goering AW; Ross MO; DeHart CJ; Thomas PM; Hoffman BM; Kelleher NL; Rosenzweig AC Characterization of methanobactin from Methylosinus sp. LW4. J. Am. Chem. Soc 2016, 138, 11124–11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Park YJ; Roberts GM; Montaser R; Kenney GE; Thomas PM; Kelleher NL; Rosenzweig AC Characterization of a copper-chelating natural product from the methanotroph Methylosinus sp. LW3. Biochemistry 2021, 60, 2845–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Balasubramanian R; Kenney GE; Rosenzweig AC Dual pathways for copper uptake by methanotrophic bacteria. J. Biol. Chem 2011, 286, 37313–37319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Kulczycki E; Fowle DA; Knapp C; Graham DW; Roberts JA Methanobactin-promoted dissolution of Cu-substituted borosilicate glass. Geobiology 2007, 5, 251–263. [Google Scholar]
- (79).Kulczycki E; Fowle DA; Kenward PA; Leslie K; Graham DW; Roberts JA Stimulation of methanotroph activity by Cu-substituted borosilicate glass. Geomicrobiol. J 2011, 28, 1–10. [Google Scholar]
- (80).Knapp CW; Fowle DA; Kulczycki E; Roberts JA; Graham DW Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc. Natl. Acad. Sci. U.S.A 2007, 104, 12040–12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).El Ghazouani A; Basle A; Firbank SJ; Knapp CW; Gray J; Graham DW; Dennison C Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorg. Chem 2011, 50, 1378–1391. [DOI] [PubMed] [Google Scholar]
- (82).Dassama LM; Kenney GE; Ro SY; Zielazinski EL; Rosenzweig AC Methanobactin transport machinery. Proc. Natl. Acad. Sci. U.S.A 2016, 113, 13027–13032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Gu W; Farhan Ul Haque M; Baral BS; Turpin EA; Bandow NL; Kremmer E; Flatley A; Zischka H; DiSpirito AA; Semrau JD A TonB-dependent transporter is responsible for methanobactin uptake by Methylosinus trichosporium OB3b. Appl. Environ. Microbiol 2016, 82, 1917–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Peng P; Kang-Yun CS; Chang J; Gu WY; DiSpirito AA; Semrau JD Two TonB-dependent transporters in Methylosinus trichosporium OB3b are responsible for uptake of different forms of methanobactin and are involved in the canonical “copper switch″. Appl. Environ. Microbiol 2022, 88, No. e0179321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Peng P; Gu WY; DiSpirito AA; Semrau JD Multiple mechanisms for copper uptake by Methylosinus trichosporium OB3b in the presence of heterologous methanobactin. mBio 2022, 13, No. e0223922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Manesis AC; Slater JW; Cantave K; Martin Bollinger J Jr.; Krebs C; Rosenzweig AC Capturing a bis-Fe(IV) State in Methylosinus trichosporium OB3b MbnH. Biochemistry 2023, 62, 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Manesis AC; Jodts RJ; Hoffman BM; Rosenzweig AC Copper binding by a unique family of metalloproteins is dependent on kynurenine formation. Proc. Natl. Acad. Sci. U.S.A 2021, 118, No. e2100680118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Kenney GE; Dassama LMK; Manesis AC; Ross MO; Chen S; Hoffman BM; Rosenzweig AC MbnH is a diheme MauG-like protein associated with microbial copper homeostasis. J. Biol. Chem 2019, 294, 16141–16151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Helland R; Fjellbirkeland A; Karlsen OA; Ve T; Lillehaug JR; Jensen HB An oxidized tryptophan facilitates copper binding in Methylococcus capsulatus-secreted protein MopE. J. Biol. Chem 2008, 283, 13897–13904. [DOI] [PubMed] [Google Scholar]
- (90).Ve T; Mathisen K; Helland R; Karlsen OA; Fjellbirkeland A; Røhr AK; Andersson KK; Pedersen RB; Lillehaug JR; Jensen HB The Methylococcus capsulatus (Bath) secreted protein, MopE*, binds both reduced and oxidized copper. PLoS One 2012, 7, No. e43146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Johnson KA; Ve T; Larsen O; Pedersen RB; Lillehaug JR; Jensen HB; Helland R; Karlsen OA CorA Is a copper repressible surface-associated copper(I)-binding protein produced in Methylomicrobium album BG8. PLoS One 2014, 9, No. e87750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Berson O; Lidstrom ME Cloning and characterization of corA, a gene encoding a copper-repressible polypeptide in the type I methanotroph, Methylomicrobium albus BG8. FEMS Microbiol. Lett 1997, 148, 169–174. [DOI] [PubMed] [Google Scholar]
- (93).Shchukin VN; Khmelenina VN; Eshinimayev BT; Suzina NE; Trotsenko YA Primary characterization of dominant cell surface proteins of halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Microbiology 2011, 80, 608–618. [PubMed] [Google Scholar]
- (94).Dennison C; David S; Lee J Bacterial copper storage proteins. J. Biol. Chem 2018, 293, 4616–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Dennison C The coordination chemistry of copper uptake and storage for methane oxidation. Chem.Eur. J 2019, 25, 74–86. [DOI] [PubMed] [Google Scholar]
- (96).Vita N; Platsaki S; Basle A; Allen SJ; Paterson NG; Crombie AT; Murrell JC; Waldron KJ; Dennison C A four-helix bundle stores copper for methane oxidation. Nature 2015, 525, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Vita N; Landolfi G; Baslé A; Platsaki S; Lee J; Waldron KJ; Dennison C Bacterial cytosolic proteins with a high capacity for Cu(I) that protect against copper toxicity. Sci. Rep 2016, 6, 39065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Lee JC; Dalton RA; Basle A; Vita N; Dennison C; Subtilis B Important structural features of thiolate-rich four-helix bundles for Cu(I) uptake and removal. Inorg. Chem 2023, 62, 6617–6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Murrell JC Genetics and molecular biology of methanotrophs. FEMS Microbiol. Lett 1992, 88, 233–248. [DOI] [PubMed] [Google Scholar]
- (100).Semrau JD; Chistoserdov A; Lebron J; Costello A; Davagnino J; Kenna E; Holmes AJ; Finch R; Murrell JC; Lidstrom ME Particulate methane monooxygenase genes in methanotrophs. J. Bacteriol 1995, 177, 3071–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Kelly DP; Anthony C; Murrell JC Insights into the obligate methanotroph Methylococcus capsulatus. Trends Microbiol 2005, 13, 195–198. [DOI] [PubMed] [Google Scholar]
- (102).Kalyuzhnaya MG; Gomez OA; Murrell JC The methane-oxidizing bacteria (methanotrophs). In Taxonomy, Genomics and Ecophysiology of Hydrocarbon-Degrading Microbes; McGenity TJ, Ed.; Springer International Publishing AG, 2019; pp 245–278. [Google Scholar]
- (103).Murrell JC Molecular genetics of methane oxidation. Biodeg 1994, 5, 145–159. [DOI] [PubMed] [Google Scholar]
- (104).Stolyar S; Costello AM; Peeples TL; Lidstrom ME Role of multiple gene copies in particulate methane monooxygenase activity in the methane-oxidizing bacterium Methylococcus capsulatus Bath. Microbiol 1999, 145, 1235–1244. [DOI] [PubMed] [Google Scholar]
- (105).Ward N; Larsen O; Sakwa J; Bruseth L; Khouri H; Durkin AS; Dimitrov G; Jiang L; Scanlan D; Kang KH; et al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol 2004, 2, No. e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Hou SB; Makarova KS; Saw JHW; Senin P; Ly BV; Zhou ZM; Ren Y; Wang JM; Galperin MY; Omelchenko MV; et al. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol. Direct 2008, 3, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Stein LY; Yoon S; Semrau JD; DiSpirito AA; Crombie A; Murrell JC; Vuilleumier S; Kalyuzhnaya MG; Op den Camp HJ; Bringel F; et al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J. Bacteriol 2010, 192, 6497–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Stein LY; Bringel F; DiSpirito AA; Han S; Jetten MSM; Kalyuzhnaya MG; Kits KD; Klotz MG; Op den Camp HJM; Semrau JD; et al. Genome sequence of the methanotrophic alphaproteobacterium Methylocystis sp strain Rockwell (ATCC 49242). J. Bacteriol 2011, 193, 2668–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Svenning MM; Hestnes AG; Wartiainen I; Stein LY; Klotz MG; Kalyuzhnaya MG; Spang A; Bringel F; Vuilleumier S; Lajus A; et al. Genome sequence of the arctic methanotroph Methylobacter tundripaludum SV96. J. Bacteriol 2011, 193, 6418–6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).del Cerro C; García JM; Rojas A; Tortajada M; Ramón D; Galán B; Prieto MA; García JL Genome sequence of the methanotrophic poly-β-hydroxybutyrate producer Methylocystis parvus OBBP. J. Bacteriol 2012, 194, 5709–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Vuilleumier S; Khmelenina VN; Bringel F; Reshetnikov AS; Lajus A; Mangenot S; Rouy Z; Op den Camp HJM; Jetten MSM; Dispirito AA; et al. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol 2012, 194, 551–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Anvar SY; Frank J; Pol A; Schmitz A; Kraaijeveld K; den Dunnen JT; Op den Camp HJ The genomic landscape of the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. BMC Genomics 2014, 15, 914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Kruse T; Ratnadevi CM; Erikstad HA; Birkeland NK Complete genome sequence analysis of the thermoacidophilic verrucomicrobial methanotroph ″Candidatus Methylacidiphilum kamchatkense″ strain Kam1 and comparison with its closest relatives. BMC Genomics 2019, 20, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Dam B; Dam S; Kube M; Reinhardt R; Liesack W Complete genome sequence of Methylocystis sp strain SC2, an aerobic methanotroph with high-affinity methane oxidation potential. J. Bacteriol 2012, 194, 6008–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Fisher OS; Kenney GE; Ross MO; Ro SY; Lemma BE; Batelu S; Thomas PM; Sosnowski VC; DeHart CJ; Kelleher NL; et al. Characterization of a long overlooked copper protein from methane- and ammonia-oxidizing bacteria. Nat. Commun 2018, 9, 4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Dunfield PF; Khmelenina VN; Suzina NE; Trotsenko YA; Dedysh SN Methylocella silvestris sp. nov., a novel methanotroph isolated from an acidic forest cambisol. Int. J. Syst. Evol. Microbiol 2003, 53, 1231–1239. [DOI] [PubMed] [Google Scholar]
- (117).Vorobev AV; Baani M; Doronina NV; Brady AL; Liesack W; Dunfield PF; Dedysh SN Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol 2011, 61, 2456–2463. [DOI] [PubMed] [Google Scholar]
- (118).Norton JM; Alzerreca JJ; Suwa Y; Klotz MG Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol 2002, 177, 139–149. [DOI] [PubMed] [Google Scholar]
- (119).Arp DJ; Chain PSG; Klotz MG The impact of genome analyses on our understanding of ammonia-oxidizing bacteria. Annu. Rev. Microbiol 2007, 61, 503–528. [DOI] [PubMed] [Google Scholar]
- (120).Hyman MR; Wood PM Methane oxidation by Nitrosomonas europaea. Biochem. J 1983, 212, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (121).Arp DJ; Sayavedra-Soto LA; Hommes NG Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch. Microbiol 2002, 178, 250–255. [DOI] [PubMed] [Google Scholar]
- (122).Arp DJ; Stein LY Metabolism of inorganic N compounds by ammonia-oxidizing bacteria. Crit. Rev. Biochem. Mol. Biol 2003, 38, 471–495. [DOI] [PubMed] [Google Scholar]
- (123).Klotz MG; Stein LY Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol. Lett 2008, 278, 146–156. [DOI] [PubMed] [Google Scholar]
- (124).Stein LY Insights into the physiology of ammonia-oxidizing microorganisms. Curr. Opin. Chem. Biol 2019, 49, 9–15. [DOI] [PubMed] [Google Scholar]
- (125).Stein LY The long-term relationship between microbial metabolism and greenhouse gases. Trends Microbiol 2020, 28, 500–511. [DOI] [PubMed] [Google Scholar]
- (126).Collins MLP; Buchholz LA; Remsen CC Effect of copper on Methylomonas albus BG8. Appl. Environ. Microbiol 1991, 57, 1261–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (127).Phelps PA; Agarwal SK; Speitel GE Jr.; Georgiou G Methylosinus trichosporium OB3b mutants having constitutive expression of soluble methane monooxygenase in the presence of high levels of copper. Appl. Environ. Microbiol 1992, 58, 3701–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (128).Whiddon KT; Gudneppanavar R; Hammer TJ; West DA; Konopka MC Fluorescence-based analysis of the intracytoplasmic membranes of type I methanotrophs. Microb. Biotechnol 2019, 12, 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (129).Tavormina PL; Kellermann MY; Antony CP; Tocheva EI; Dalleska NF; Jensen AJ; Valentine DL; Hinrichs KU; Jensen GJ; Dubilier N; et al. Starvation and recovery in the deep-sea methanotroph Methyloprofundus sedimenti. Mol. Microbiol 2017, 103, 242–252. [DOI] [PubMed] [Google Scholar]
- (130).Zhu YA; Koo CW; Cassidy CK; Spink MC; Ni T; Zanetti-Domingues LC; Bateman B; Martin-Fernandez ML; Shen J; Sheng YW; et al. Structure and activity of particulate methane monooxygenase arrays in methanotrophs. Nat. Commun 2022, 13, 5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (131).Brantner CA; Buchholz LA; Remsen CC; Collins MLP Isolation of intracytoplasmic membrane from methanotrophic bacterium Methylomicrobium album BG8. Curr. Microbiol 2000, 40, 132–134. [DOI] [PubMed] [Google Scholar]
- (132).Nielsen AK; Gerdes K; Murrell JC Copper-dependent transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol 1997, 25, 399–409. [DOI] [PubMed] [Google Scholar]
- (133).Gilbert B; McDonald IR; Finch R; Stafford GP; Nielsen AK; Murrell JC Molecular analysis of pmo (particulate methane monooxygenase) operons from two type II methanotrophs. Appl. Environ. Microbiol 2000, 66, 966–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (134).Stolyar S; Franke M; Lidstrom ME Expression of individual copies of Methylococcus capsulatus Bath particulate methane monooxygenase genes. J. Bacteriol 2001, 183, 1810–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (135).Stafford GP; Scanlan J; McDonald IR; Murrell JC rpoN, mmoR and mmoG, genes involved in regulating the expression of soluble methane monooxygenase from Methylosinus trichosporium OB3b. Microbiol 2003, 149, 1771–1784. [DOI] [PubMed] [Google Scholar]
- (136).Choi DW; Kunz RC; Boyd ES; Semrau JD; Antholine WE; Han JI; Zahn JA; Boyd JM; de la Mora AM; DiSpirito AA The membrane-associated methane mono-oxygenase pMMO and pMMO-NADH:quinone oxidoreductase complex from Methylococcus capsulatus Bath. J. Bacteriol 2003, 185, 5755–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (137).Semrau JD; Jagadevan S; DiSpirito AA; Khalifa A; Scanlan J; Bergman BH; Freemeier BC; Baral BS; Bandow NL; Vorobev A; et al. Methanobactin and MmoD work in concert to act as the ‘copper-switch’ in methanotrophs. Environ. Microbiol 2013, 15, 3077–3086. [DOI] [PubMed] [Google Scholar]
- (138).Csáki R; Bodrossy L; Klem J; Murrell JC; Kovács KL Genes involved in the copper-dependent regulation of soluble methane monooxygenase of Methylococcus capsulatus (Bath): cloning, sequencing and mutational analysis. Microbiol 2003, 149, 1785–1795. [DOI] [PubMed] [Google Scholar]
- (139).Ukaegbu UE; Henery S; Rosenzweig AC Biochemical characterization of MmoS, a sensor protein involved in copper-dependent regulation of soluble methane monooxygenase. Biochemistry 2006, 45, 10191–10198. [DOI] [PubMed] [Google Scholar]
- (140).Merkx M; Lippard SJ Why OrfY? Characterization of mmoD, a long overlooked component of the soluble methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem 2002, 277, 5858–5865. [DOI] [PubMed] [Google Scholar]
- (141).Kim H; An S; Park YR; Jang H; Yoo H; Park SH; Lee SJ; Cho US MMOD-induced structural changes of hydroxylase in soluble methane monooxygenase. Sci. Adv 2019, 5, No. eaax0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (142).DiSpirito AA; Semrau JD; Murrell JC; Gallagher WH; Dennison C; Vuilleumier S Methanobactin and the link between copper and bacterial methane oxidation. Microbiol. Mol. Biol. Rev 2016, 80, 387–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (143).Borodina E; Nichol T; Dumont MG; Smith TJ; Murrell JC Mutagenesis of the ″leucine gate″ to explore the basis of catalytic versatility in soluble methane monooxygenase. Appl. Environ. Microbiol 2007, 73, 6460–6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (144).Fitch MW; Graham DW; Arnold RG; Agarwal SK; Phelps P; Speitel GE Jr.; Georgiou G Phenotypic characterization of copper-resistant mutants of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol 1993, 59, 2771–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (145).Cha JS; Cooksey DA Copper hypersensitivity and uptake in Pseudomonas syringae containing cloned components of the copper resistance operon. Appl. Environ. Microbiol 1993, 59, 1671–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (146).Zhang XX; Rainey PB Regulation of copper homeostasis in Pseudomonas fluorescens SBW25. Environ. Microbiol 2008, 10, 3284–3294. [DOI] [PubMed] [Google Scholar]
- (147).Chillappagari S; Miethke M; Trip H; Kuipers OP; Marahiel MA Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J. Bacteriol 2009, 191, 2362–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (148).Serventi F; Youard ZA; Murset V; Huwiler S; Buhler D; Richter M; Luchsinger R; Fischer HM; Brogioli R; Niederer M; et al. Copper starvation-inducible protein for cytochrome oxidase biogenesis in Bradyrhizobium japonicum. J. Biol. Chem 2012, 287, 38812–38823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (149).Lawton TJ; Kenney GE; Hurley JD; Rosenzweig AC The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry 2016, 55, 2278–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (150).Gu W; Farhan Ul Haque M; Semrau JD Characterization of the role of copCD in copper uptake and the ‘copper-switch’ in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett 2017, 364, fnx094. [DOI] [PubMed] [Google Scholar]
- (151).Lieberman RL; Rosenzweig AC Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 2005, 434, 177–182. [DOI] [PubMed] [Google Scholar]
- (152).Hakemian AS; Kondapalli KC; Telser J; Hoffman BM; Stemmler TL; Rosenzweig AC The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 2008, 47, 6793–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Smith SM; Rawat S; Telser J; Hoffman BM; Stemmler TL; Rosenzweig AC Crystal structure and characterization of particulate methane monooxygenase from Methylocystis species strain M. Biochemistry 2011, 50, 10231–10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Ro SY; Ross MO; Deng YW; Batelu S; Lawton TJ; Hurley JD; Stemmler TL; Hoffman BM; Rosenzweig AC From micelles to bicelles: Effect of the membrane on particulate methane monooxygenase activity. J. Biol. Chem 2018, 293, 10457–10465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Walker CB; de la Torre JR; Klotz MG; Urakawa H; Pinel N; Arp DJ; Brochier-Armanet C; Chain PSG; Chan PP; Gollabgir A; et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U.S.A 2010, 107, 8818–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Lawton TJ; Ham J; Sun T; Rosenzweig AC Structural conservation of the B subunit in the ammonia monooxygenase/particulate methane monooxygenase superfamily. Proteins 2014, 82, 2263–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (157).Hodgskiss LH; Melcher M; Kerou M; Chen W; Ponce-Toledo RI; Savvides SN; Wienkoop S; Hartl M; Schleper C Unexpected complexity of the ammonia monooxygenase in archaea. ISME J 2023, 17, 588–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (158).Zhang P; Wang J; Shi Y Structure and mechanism of the S component of a bacterial ECF transporter. Nature 2010, 468, 717–720. [DOI] [PubMed] [Google Scholar]
- (159).Rempel S; Stanek WK; Slotboom DJ ECF-type ATP-binding cassette transporters. Annu. Rev. Biochem 2019, 88, 551–576. [DOI] [PubMed] [Google Scholar]
- (160).McLean MA; Gregory MC; Sligar SG Nanodiscs: A controlled bilayer surface for the study of membrane proteins. Annu. Rev. Biophys 2018, 47, 107–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (161).Chang W-H; Lin H-H; Tsai I-K; Huang S-H; Chung S-C; Tu I-P; Yu SS-F; Chan SI Copper centers in the cryo-EM Structure of particulate methane monooxygenase reveal the catalytic machinery of methane oxidation. J. Am. Chem. Soc 2021, 143, 9922–9932. [DOI] [PubMed] [Google Scholar]
- (162).Martinho M; Choi DW; DiSpirito AA; Antholine WE; Semrau JD; Münck E Mössbauer studies of the membrane-associated methane monooxygenase from Methylococcus capsulatus Bath: evidence for a diiron center. J. Am. Chem. Soc 2007, 129, 15783–15785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (163).Lieberman RL; Kondapalli KC; Shrestha DB; Hakemian AS; Smith SM; Telser J; Kuzelka J; Gupta R; Borovik AS; Lippard SJ; et al. Characterization of the particulate methane monooxygenase metal centers in multiple redox states by X-ray absorption spectroscopy. Inorg. Chem 2006, 45, 8372–8381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (164).Lieberman RL; Rosenzweig AC Biological methane oxidation: regulation, biochemistry, and active site structure of particulate methane monooxygenase. Crit. Rev. Biochem. Mol. Biol 2004, 39, 147–164. [DOI] [PubMed] [Google Scholar]
- (165).Chan SI; Chen KH-C; Yu SS-F; Chen C-L; Kuo SS-J Toward delineating the structure and function of the particulate methane monooxygenase from methanotrophic bacteria. Biochemistry 2004, 43, 4421–4430. [DOI] [PubMed] [Google Scholar]
- (166).Hakemian AS; Rosenzweig AC The biochemistry of methane oxidation. Annu. Rev. Biochem 2007, 76, 223–241. [DOI] [PubMed] [Google Scholar]
- (167).Chan SI; Chang WH; Huang SH; Lin HH; Yu SSF Catalytic machinery of methane oxidation in particulate methane monooxygenase (pMMO). J. Inorg. Biochem 2021, 225, 111602. [DOI] [PubMed] [Google Scholar]
- (168).Chan SI; Wang VCC; Chen PPY; Yu SSF Methane oxidation by the copper methane monooxygenase: Before and after the cryogenic electron microscopy structure of particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Chin. Chem. Soc 2022, 69, 1147–1158. [Google Scholar]
- (169).Yu SSF; Ji CZ; Wu YP; Lee TL; Lai CH; Lin SC; Yang ZL; Wang VCC; Chen KHC; Chan SI The C-terminal aqueous-exposed domain of the 45 kDa subunit of the particulate methane monooxygenase in Methylococcus capsulatus (Bath) is a Cu(I) sponge. Biochemistry 2007, 46, 13762–13774. [DOI] [PubMed] [Google Scholar]
- (170).Lu YJ; Hung MC; Chang BTA; Lee TL; Lin ZH; Tsai IK; Chen YS; Chang CS; Tsai YF; Chen KHC; et al. The PmoB subunit of particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath): The CuI sponge and its function. J. Inorg. Biochem 2019, 196, 110691. [DOI] [PubMed] [Google Scholar]
- (171).Ross MO; MacMillan F; Wang J; Nisthal A; Lawton TJ; Olafson BD; Mayo SL; Rosenzweig AC; Hoffman BM Particulate methane monooxygenase contains only mononuclear copper centers. Science 2019, 364, 566–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (172).Jodts RJ; Ross MO; Koo CW; Doan PE; Rosenzweig AC; Hoffman BM Coordination of the copper centers in particulate methane monooxygenase: comparison between methanotrophs and characterization of the Cuc site by EPR and ENDOR spectroscopies. J. Am. Chem. Soc 2021, 143, 15358–15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Lieberman RL; Shrestha DB; Doan PE; Hoffman BM; Stemmler TL; Rosenzweig AC Purified particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a dimer with both mononuclear copper and a copper-containing cluster. Proc. Natl. Acad. Sci. U.S.A 2003, 100, 3820–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Cao LL; Caldararu O; Rosenzweig AC; Ryde U Quantum refinement does not support dinuclear copper sites in crystal structures of particulate methane monooxygenase. Angew. Chem., Int. Ed 2018, 57, 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (175).Cutsail GE III; Ross MO; Rosenzweig AC; DeBeer S Towards a unified understanding of the copper sites in particulate methane monooxygenase: an X-ray absorption spectroscopic investigation. Chem. Sci 2021, 12, 6194–6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Yuan H; Collins MLP; Antholine WE Low-frequency EPR of the copper in particulate methane monooxygenase from Methylomicrobium albus BG8. J. Am. Chem. Soc 1997, 119, 5073–5074. [Google Scholar]
- (177).Lemos SS; Collins MLP; Eaton SS; Eaton GR; Antholine WE Comparison of EPR-visible Cu2+ sites in pMMO from Methylococcus capsulatus (Bath) and Methlyomicrobium album BG8. Biophys. J 2000, 79, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (178).Tucci FJ; Jodts RJ; Hoffman BM; Rosenzweig AC Product analog binding identifies the copper active site of particulate methane monooxygenase. Nat. Catal 2023, 6, 1194–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Ro SY; Schachner LF; Koo CW; Purohit R; Remis JP; Kenney GE; Liauw BW; Thomas PM; Patrie SM; Kelleher NL; et al. Native top-down mass spectrometry provides insights into the copper centers of membrane-bound methane monooxygenase. Nat. Commun 2019, 10, 2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (180).Burrows KJ; Cornish A; Scott D; Higgins IJ Substrate specificities of the soluble and particulate methane monooxygenases of Methylosinus trichosporium OB3b. J. Gen. Microbiol 1984, 130, 3327–3333. [Google Scholar]
- (181).Smith DDS; Dalton H Solubilisation of methane monooxygenase from Methylococcus capsulatus (Bath). Eur. J. Biochem 1989, 182, 667–671. [DOI] [PubMed] [Google Scholar]
- (182).Shiemke AK; Cook SA; Miley T Quinols as electron donors for detergent solubilized and membrane-bound methane monooxygenase. J. Inorg. Biochem 1995, 59, 385. [DOI] [PubMed] [Google Scholar]
- (183).Collins MD; Green PN Isolation and characterization of a novel coenzyme Q from some methane-oxidizing bacteria. Biochem. Biophys. Res. Commun 1985, 133, 1125–1131. [DOI] [PubMed] [Google Scholar]
- (184).Urakami T; Komagata K Occurrence of isoprenoid compounds in gram-negative methanol-utilizing, methane-utilizing, and methylamine-utilizing bacteria. J. Gen. Appl. Microbiol 1986, 32, 317–341. [Google Scholar]
- (185).Kim HJ; Huh J; Kwon YW; Park D; Yu Y; Jang YE; Lee BR; Jo E; Lee EJ; Heo Y; et al. Biological conversion of methane to methanol through genetic reassembly of native catalytic domains. Nat. Catal 2019, 2, 342–353. [Google Scholar]
- (186).Peng W; Qu X; Shaik S; Wang B Deciphering the oxygen activation mechanism at the CuC site of particulate methane monooxygenase. Nat. Catal 2021, 4, 266–273. [Google Scholar]
- (187).Cook SA; Shiemke AK Evidence that a type-2 NADH:quinone oxidoreductase mediates electron transfer to particulate methane monooxygenase in Methylococcus capsulatus. Arch. Biochem. Biophys 2002, 398, 32–40. [DOI] [PubMed] [Google Scholar]
- (188).de la Torre A; Metivier A; Chu F; Laurens LML; Beck DAC; Pienkos PT; Lidstrom ME; Kalyuzhnaya MG Genome-scale metabolic reconstructions and theoretical investigation of methane conversion in Methylomicrobium buryatense strain 5G(B1). Microb. Cell Fact 2015, 14, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (189).Leak DJ; Dalton H Growth yields of methanotrophs. 2. A Theoretical analysis. Appl. Microbiol. Biotechnol 1986, 23, 477–481. [Google Scholar]
- (190).Lieven C; Petersen LAH; Jorgensen SB; Gernaey KV; Herrgard MJ; Sonnenschein N A genome-scale metabolic model for Methylococcus capsulatus (Bath) suggests reduced efficiency electron transfer to the particulate methane monooxygenase. Front. Microbiol 2018, 9, 2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (191).Akberdin IR; Thompson M; Hamilton R; Desai N; Alexander D; Henard CA; Guarnieri MT; Kalyuzhnaya MG Methane utilization in Methylomicrobium alcaliphilum 20ZR: a systems approach. Sci. Rep 2018, 8, 2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (192).Bordel S; Rodriguez Y; Hakobyan A; Rodriguez E; Lebrero R; Munoz R Genome scale metabolic modeling reveals the metabolic potential of three Type II methanotrophs of the genus Methylocystis. Metab. Eng 2019, 54, 191–199. [DOI] [PubMed] [Google Scholar]
- (193).Bordel S; Rojas A; Munoz R Reconstruction of a genome scale metabolic model of the polyhydroxybutyrate producing methanotroph Methylocystis parvus OBBP. Microb. Cell Fact 2019, 18, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Nguyen AD; Park JY; Hwang IY; Hamilton R; Kalyuzhnaya MG; Kim D; Lee EY Genome-scale evaluation of core one-carbon metabolism in gammaproteobacterial methanotrophs grown on methane and methanol. Metab. Eng 2020, 57, 1–12. [DOI] [PubMed] [Google Scholar]
- (195).Naizabekov S; Lee EY Genome-scale metabolic model reconstruction and in silico investigations of methane metabolism in Methylosinus trichosporium OB3b. Microorganisms 2020, 8, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (196).Chan SI; Yu SSF Controlled oxidation of hydrocarbons by the membrane-bound methane monooxygenase: The case for a tricopper cluster. Acc. Chem. Res 2008, 41, 969–979. [DOI] [PubMed] [Google Scholar]
- (197).Dörr JM; Scheidelaar S; Koorengevel MC; Dominguez JJ; Schäfer M; van Walree CA; Killian JA The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J 2016, 45, 3–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Kitmitto A; Myronova N; Basu P; Dalton H Characterization and structural analysis of an active particulate methane monooxygenase trimer from Methylococcus capsulatus (Bath). Biochemistry 2005, 44, 10954–10965. [DOI] [PubMed] [Google Scholar]
- (199).Blanchette CD; Knipe JM; Stolaroff JK; DeOtte JR; Oakdale JS; Maiti A; Lenhardt JM; Sirajuddin S; Rosenzweig AC; Baker SE Printable enzyme-embedded materials for methane to methanol conversion. Nat. Commun 2016, 7, 11900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (200).Elliott SJ; Zhu M; Tso L; Nguyen H-H; Yip JH-K; Chan SI Regio- and stereoselectivity of particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc 1997, 119, 9949–9955. [Google Scholar]
- (201).Miyaji A; Miyoshi T; Motokura K; Baba T The substrate binding cavity of particulate methane monooxygenase from Methylosinus trichosporium OB3b expresses high enantioselectivity for n-butane and n-pentane oxidation to 2-alcohol. Biotechnol. Lett 2011, 33, 2241–2246. [DOI] [PubMed] [Google Scholar]
- (202).Jiang H; Chen Y; Jiang PX; Zhang C; Smith TJ; Murrell JC; Xing XH Methanotrophs: Multifunctional bacteria with promising applications in environmental bioengineering. Biochem. Eng. J 2010, 49, 277–288. [Google Scholar]
- (203).Bedard C; Knowles R Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev 1989, 53, 68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (204).Zahn JA; DiSpirito AA Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath). J. Bacteriol 1996, 178, 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (205).Cook SA; Shiemke AK Evidence that copper is a required cofactor for the membrane-bound form of methane monooxygenase. J. Inorg. Biochem 1996, 63, 273–284. [Google Scholar]
- (206).Basu P; Katterle B; Andersson KK; Dalton H The membrane-associated form of methane monooxygenase from Methylococcus capsulatus (Bath) is a copper/iron protein. Biochem. J 2003, 369, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Chen CL; Chen KHC; Ke SC; Yu SSF; Chan SI Preparation and characterization of a (Cu, Zn)-pMMO from Methylococcus capsulatus (Bath). J. Inorg. Biochem 2004, 98, 2125–2130. [DOI] [PubMed] [Google Scholar]
- (208).Chan SI; Wang VCC; Lai JCH; Yu SSF; Chen PPY; Chen KHC; Chen CL; Chan MK Redox potentiometry studies of particulate methane monooxygenase: support for a trinuclear copper cluster active site. Angew. Chem., Int. Ed 2007, 46, 1992–1994. [DOI] [PubMed] [Google Scholar]
- (209).Walton PH; Davies GJ; Diaz DE; Franco-Cairo JP The histidine brace: nature’s copper alternative to haem? FEBS Lett 2023, 597, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (210).Ipsen JO; Hallas-Møller M; Brander S; Lo Leggio L; Johansen KS Lytic polysaccharide monooxygenases and other histidine-brace copper proteins: structure, oxygen activation and biotechnological applications. Biochem. Soc. Trans 2021, 49, 531–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (211).Ciano L; Davies GJ; Tolman WB; Walton PH Bracing copper for the catalytic oxidation of C-H bonds. Nat. Catal 2018, 1, 571–577. [Google Scholar]
- (212).Op den Camp HJ; Islam T; Stott MB; Harhangi HR; Hynes A; Schouten S; Jetten MSM; Birkeland N-K; Pol A; Dunfield PF Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep 2009, 1, 293–306. [DOI] [PubMed] [Google Scholar]
- (213).Koo CW; Rosenzweig AC Particulate methane monooxygenase and the PmoD protein. In Encylopedia of Inorganic and Bioinorganic Chemistry; John Wiley & Sons, Ltd., 2020. DOI: 10.1002/9781119951438.eibc2740. [DOI] [Google Scholar]
- (214).Labourel A; Frandsen KEH; Zhang F; Brouilly N; Grisel S; Haon M; Ciano L; Ropartz D; Fanuel M; Martin F; et al. A fungal family of lytic polysaccharide monooxygenase-like copper proteins. Nat. Chem. Biol 2020, 16, 345–350. [DOI] [PubMed] [Google Scholar]
- (215).Liew EF; Tong DC; Coleman NV; Holmes AJ Mutagenesis of the hydrocarbon monooxygenase indicates a metal centre in subunit-C, and not subunit-B, is essential for copper-containing membrane monooxygenase activity. Microbiology 2014, 160, 1267–1277. [DOI] [PubMed] [Google Scholar]
- (216).Stein LY; Arp DJ Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol 1998, 64, 4098–4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (217).Nyerges G; Stein LY Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol. Lett 2009, 297, 131–136. [DOI] [PubMed] [Google Scholar]
- (218).Anthony C; Williams P The structure and mechanism of methanol dehydrogenase. Biochim. Biophys. Acta 2003, 1647, 18–23. [DOI] [PubMed] [Google Scholar]
- (219).Anthony C The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys 2004, 428, 2–9. [DOI] [PubMed] [Google Scholar]
- (220).Gvozdev AR; Tukhvatullin IA; Gvozdev RI Quinonedependent alcohol dehydrogenases and FAD-dependent alcohol oxidases. Biochemistry-Moscow 2012, 77, 843–856. [DOI] [PubMed] [Google Scholar]
- (221).Picone N; Op den Camp HJM Role of rare earth elements in methanol oxidation. Curr. Op. Chem. Biol 2019, 49, 39–44. [DOI] [PubMed] [Google Scholar]
- (222).Keltjens JT; Pol A; Reimann J; Op den Camp HJM PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl. Microbiol. Biotechnol 2014, 98, 6163–6183. [DOI] [PubMed] [Google Scholar]
- (223).Gu WY; Farhan Ul Haque M; DiSpirito AA; Semrau JD Uptake and effect of rare earth elements on gene expression in Methylosinus trichosporium OB3b. FEMS Microbiol. Lett 2016, 363, fnw129. [DOI] [PubMed] [Google Scholar]
- (224).Farhan Ul Haque M; Kalidass B; Bandow N; Turpin EA; DiSpirito AA; Semrau JD Cerium regulates expression of alternative methanol dehydrogenases in Methylosinus trichosporium OB3b. Appl. Environ. Microbiol 2015, 81, 7546–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (225).Chu F; Lidstrom ME XoxF acts as the predominant methanol dehydrogenase in the type I methanotroph Methylomicrobium buryatense. J. Bacteriol 2016, 198, 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (226).Crombie AT The effect of lanthanum on growth and gene expression in a facultative methanotroph. Environ. Microbiol 2022, 24, 596–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (227).Culpepper MA; Rosenzweig AC Structure and protein-protein interactions of methanol dehydrogenase from Methylococcus capsulatus (Bath). Biochemistry 2014, 53, 6211–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (228).Deng YW; Ro SY; Rosenzweig AC Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C. J. Biol. Inorg. Chem 2018, 23, 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (229).Jahn B; Pol A; Lumpe H; Barends TRM; Dietl A; Hogendoorn C; Op den Camp HJM; Daumann LJ Similar but not the same: first kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. Chembiochem 2018, 19, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (230).Schmitz RA; Picone N; Singer H; Dietl A; Seifert KA; Pol A; Jetten MSM; Barends TRM; Daumann LJ; Op den Camp HJM Neodymium as metal cofactor for biological methanol oxidation: structure and kinetics of an XoxF1-type methanol dehydrogenase. mBio 2021, 12, No. e0170821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (231).Wadzinski AM; Ribbons DW Oxidation of C1 compounds by particulate fractions from Methylococcus capsulatus: properties of methanol oxidase and methanol dehydrogenase. J. Bacteriol 1975, 122, 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (232).Fassel TA; Buchholz LA; Collins MLP; Remsen CC Localization of methanol dehydrogenase in two strains of methylotrophic bacteria detected by immunogold labeling. Appl. Environ. Microbiol 1992, 58, 2302–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (233).Brantner CA; Remsen CC; Owen HA; Buchholz LA; Collins MLP Intracellular localization of the particulate methane monooxygenase and methanol dehydrogenase in Methylomicrobium album BG8. Arch. Microbiol 2002, 178, 59–64. [DOI] [PubMed] [Google Scholar]
- (234).Myronova N; Kitmitto A; Collins RF; Miyaji A; Dalton H Three-dimensional structure determination of a protein supercomplex that oxidizes methane to formaldehyde in Methylococcus capsulatus (Bath). Biochemistry 2006, 45, 11905–11914. [DOI] [PubMed] [Google Scholar]
- (235).El Sheikh AF; Poret-Peterson AT; Klotz MG Characterization of two new genes, amoR and amoD, in the amo operon of the marine ammonia oxidizer Nitrosococcus oceani ATCC 19707. Appl. Environ. Microbiol 2008, 74, 312–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (236).Ross MO; Fisher OS; Morgada MN; Krzyaniak MD; Wasielewski MR; Vila AJ; Hoffman BM; Rosenzweig AC Formation and electronic structure of an atypical CuA Site. J. Am. Chem. Soc 2019, 141, 4678–4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (237).Liu J; Chakraborty S; Hosseinzadeh P; Yu Y; Tian S; Petrik I; Bhagi A; Lu Y Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev 2014, 114, 4366–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (238).Williams PA; Blackburn NJ; Sanders D; Bellamy H; Stura EA; Fee JA; McRee DE The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 Å resolution. Nat. Struct. Biol 1999, 6, 509–516. [DOI] [PubMed] [Google Scholar]
- (239).Brown KR; Djinovic-Carugo K; Haltia T; Cabrito I; Saraste M; Moura JJG; Moura I; Tegoni M; Cambillau C Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase. Evidence of a bridging inorganic sulfur. J. Biol. Chem 2000, 275, 41133–41136. [DOI] [PubMed] [Google Scholar]
- (240).Robinson H; Ang MC; Gao YG; Hay MT; Lu Y; Wang AHJ Structural basis of electron transfer modulation in the purple CuA center. Biochemistry 1999, 38, 5677–5683. [DOI] [PubMed] [Google Scholar]
- (241).Yoshizawa K; Shiota Y Conversion of methane to methanol at the mononuclear and dinuclear copper sites of particulate methane monooxygenase (pMMO): a DFT and QM/MM study. J. Am. Chem. Soc 2006, 128, 9873–9881. [DOI] [PubMed] [Google Scholar]
- (242).Shiota Y; Yoshizawa K Comparison of the reactivity of bis(μ-oxo)CuIICuIII and CuIIICuIII species to methane. Inorg. Chem 2009, 48, 838–845. [DOI] [PubMed] [Google Scholar]
- (243).Shiota Y; Juhasz G; Yoshizawa K Role of tyrosine residue in methane activation at the dicopper site of particulate methane monooxygenase: a density functional theory study. Inorg. Chem 2013, 52, 7907–7917. [DOI] [PubMed] [Google Scholar]
- (244).Miyanishi M; Abe T; Hori Y; Shiota Y; Yoshizawa K Role of amino acid residues for dioxygen activation in the second coordination sphere of the dicopper site of pMMO. Inorg. Chem 2019, 58, 12280–12288. [DOI] [PubMed] [Google Scholar]
- (245).Solomon EI; Heppner DE; Johnston EM; Ginsbach JW; Cirera J; Qayyum M; Kieber-Emmons MT; Kjaergaard CH; Hadt RG; Tian L Copper active sites in biology. Chem. Rev 2014, 114, 3659–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (246).Keown W; Gary JB; Stack TDP High-valent copper in biomimetic and biological oxidations. J. Biol. Inorg. Chem 2017, 22, 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (247).DiMucci IM; Lukens JT; Chatterjee S; Carsch KM; Titus CJ; Lee SJ; Nordlund D; Betley TA; MacMillan SN; Lancaster KM The myth of d8 copper(III). J. Am. Chem. Soc 2019, 141, 18508–18520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (248).Peng W; Wang Z; Zhang Q; Yan S; Wang B Unraveling the valence state and reactivity of copper centers in membrane-bound particulate methane monooxygenase. J. Am. Chem. Soc 2023, 145, 25304–25317. [DOI] [PubMed] [Google Scholar]
- (249).Wilkinson B; Zhu M; Priestley ND; Nguyen H-HT; Morimoto H; Williams PG; Chan SI; Floss HG A concerted mechanism for ethane hydroxylation by the particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc 1996, 118, 921–922. [Google Scholar]
- (250).Ono M; Okura I On the reaction mechanism of alkene epoxidation with Methylosinus trichosporium OB3b. J. Mol. Catal 1990, 61, 113–122. [Google Scholar]
- (251).Yu SS-F; Wu L-Y; Chen KH-C; Luo W-I; Huang D-S; Chan SI The stereospecific hydroxylation of [2,2-2H2] butane and chiral dideuteriobutanes by the particulate methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem 2003, 278, 40658–40669. [DOI] [PubMed] [Google Scholar]
- (252).Ng K-Y; Tu L-C; Wang Y-S; Chan SI; Yu SSF Probing the hydrophobic pocket of the active site in the particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) by variable stereoselective alkane hydroxylation and olefin epoxidation. ChemBioChem 2008, 9, 1116–1123. [DOI] [PubMed] [Google Scholar]
- (253).Huang DS; Wu SH; Wang YS; Yu SS; Chan SI Determination of the carbon kinetic isotope effects on propane hydroxylation mediated by the methane nonooxygenases from Methylococcus capsulatus (Bath) by using stable carbon isotopic analysis. ChemBioChem 2002, 3, 760–765. [DOI] [PubMed] [Google Scholar]
- (254).Gou Z; Xing X-H; Luo M; Jiang H; Han B; Wu H; Wang L; Zhang F Functional expression of the particulate methane monooxygenase gene in recombinant Rhodococcus erythropolis. FEMS Microbiol. Lett 2006, 263, 136–141. [DOI] [PubMed] [Google Scholar]
- (255).Spatola Rossi T; Tolmie AF; Nichol T; Pain C; Harrison P; Smith TJ; Fricker M; Kriechbaumer V Recombinant expression and subcellular targeting of the particulate methane monooxygenase (pMMO) protein components in plants. Sci. Rep 2023, 13, 15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (256).Coleman NV; Le NB; Ly MA; Ogawa HE; McCarl V; Wilson NL; Holmes AJ Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J 2012, 6, 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (257).Nath S; Henard JM; Henard CA Optimized tools and methods for methanotroph genome editing. Methods Mol. Biol 2022, 2489, 421–434. [DOI] [PubMed] [Google Scholar]
- (258).Puri AW; Owen S; Chu F; Chavkin T; Beck DAC; Kalyuzhnaya MG; Lidstrom ME Genetic tools for the industrially promising methanotroph Methylomicrobium buryatense. Appl. Environ. Microbiol 2015, 81, 1766–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (259).Yan X; Chu F; Puri AW; Fu Y; Lidstrom ME Electroporation-based genetic manipulation in type I methanotrophs. Appl. Environ. Microbiol 2016, 82, 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (260).Tapscott T; Guarnieri MT; Henard CA Development of a CRISPR/Cas9 system for Methylococcus capsulatus in vivo gene editing. Appl. Environ. Microbiol 2019, 85, No. e00340–00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (261).Silverman AD; Karim AS; Jewett MC Cell-free gene expression: an expanded repertoire of applications. Nat. Rev. Genet 2020, 21, 151–170. [DOI] [PubMed] [Google Scholar]
- (262).Koo CW; Hershewe JM; Jewett MC; Rosenzweig AC Cell-free protein synthesis of particulate methane monooxygenase into nanodiscs. ACS Synth. Biol 2022, 11, 4009–4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (263).Lipscomb JD Biochemistry of the soluble methane monooxygenase. Annu. Rev. Microbiol 1994, 48, 371–399. [DOI] [PubMed] [Google Scholar]
- (264).Merkx M; Kopp DA; Sazinsky MH; Blazyk JL; Müller J; Lippard SJ Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: a tale of two irons and three proteins. Angew. Chem., Int. Ed 2001, 40, 2782–2807. [DOI] [PubMed] [Google Scholar]
- (265).Fox BG; Froland WA; Dege JE; Lipscomb JD Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J. Biol. Chem 1989, 264, 10023–10033. [PubMed] [Google Scholar]
- (266).Lee S-K; Nesheim JC; Lipscomb JD Transient intermediates of the methane monooxygenase catalytic cycle. J. Biol. Chem 1993, 268, 21569–21577. [PubMed] [Google Scholar]
- (267).Liu Y; Nesheim JC; Lee SK; Lipscomb JD Gating effects of component B on oxygen activation by the methane monooxygenase hydroxylase component. J. Biol. Chem 1995, 270, 24662–24665. [DOI] [PubMed] [Google Scholar]
- (268).Sazinsky MH; Merkx M; Cadieux E; Tang SY; Lippard SJ Preparation and X-ray structures of metal-free, dicobalt and dimanganese forms of soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath). Biochemistry 2004, 43, 16263–16276. [DOI] [PubMed] [Google Scholar]
- (269).Rosenzweig AC; Frederick CA; Lippard SJ; Nordlund P Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 1993, 366, 537–543. [DOI] [PubMed] [Google Scholar]
- (270).Rosenzweig AC; Brandstetter H; Whittington DA; Nordlund P; Lippard SJ; Frederick CA Crystal structure of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) at 1.7 Å resolution: implications for substrate gating and component interactions. Proteins 1997, 29, 141–152. [PubMed] [Google Scholar]
- (271).Elango N; Radhakrishnan R; Froland WA; Wallar BJ; Earhart CA; Lipscomb JD; Ohlendorf DH Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci 1997, 6, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (272).Ahn E; Kim B; Park S; Erwin AL; Sung SH; Hovden R; Mosalaganti S; Cho US Batch production of high-quality graphene grids for cryo-EM: cryo-EM structure of Methylococcus capsulatus soluble methane monooxygenase hydroxylase. ACS Nano 2023, 17, 6011–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (273).Nordlund P; Sjöberg B-M; Eklund H Three dimensional structure of the free radical protein of ribonucleotide reductase. Nature 1990, 345, 593–598. [DOI] [PubMed] [Google Scholar]
- (274).Chang SL; Wallar BJ; Lipscomb JD; Mayo KH Solution structure of component B from methane monooxygenase derived through heteronuclear NMR and molecular modeling. Biochemistry 1999, 38, 5799–5812. [DOI] [PubMed] [Google Scholar]
- (275).Walters KJ; Gassner GT; Lippard SJ; Wagner G Structure of the soluble methane monooxygenase regulatory protein B. Proc. Natl. Acad. Sci. U. S. A 1999, 96, 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (276).Müller J; Lugovskoy AA; Wagner G; Lippard SJ NMR structure of the [2Fe-2S] ferredoxin domain from soluble methane monooxygenase reductase and interaction with its hydroxylase. Biochemistry 2002, 41, 42–51. [DOI] [PubMed] [Google Scholar]
- (277).Chatwood LL; Muller J; Gross JD; Wagner G; Lippard SJ NMR structure of the flavin domain from soluble methane monooxygenase reductase from Methylococcus capsulatus (Bath). Biochemistry 2004, 43, 11983–11991. [DOI] [PubMed] [Google Scholar]
- (278).Lee C; Ha SC; Rao Z; Hwang Y; Kim DS; Kim SY; Yoo H; Yoon C; Na JG; Park JH; et al. Elucidation of the electron transfer environment in the MMOR FAD-binding domain from Methylosinus sporium 5. Dalton Trans 2021, 50, 16493–16498. [DOI] [PubMed] [Google Scholar]
- (279).Wang WX; Lippard SJ Diiron oxidation state control of substrate access to the active site of soluble methane monooxygenase mediated by the regulatory component. J. Am. Chem. Soc 2014, 136, 2244–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (280).Lee SJ; McCormick MS; Lippard SJ; Cho U-S Control of substrate access to the active site in methane monooxygenase. Nature 2013, 494, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (281).Rosenzweig AC Metalloenzymes: Put a ring on it. Nat. Chem. Biol 2013, 9, 220–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (282).Srinivas V; Banerjee R; Lebrette H; Jones JC; Aurelius O; Kim IS; Pham CC; Gul S; Sutherlin K; Bhowmick A; et al. High resolution XFEL structure of the soluble methane monooxygenase hydroxylase complex with its regulatory component at ambient temperature in two oxidation states. J. Am. Chem. Soc 2020, 142, 14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (283).Brandstetter H; Whittington DA; Lippard SJ; Frederick CA Mutational and structural analyses of the regulatory protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Chem. Biol 1999, 6, 441–449. [DOI] [PubMed] [Google Scholar]
- (284).Chang SL; Wallar BJ; Lipscomb JD; Mayo KH Residues in Methylosinus trichosporium OB3b methane monooxygenase component B involved in molecular interactions with reduced- and oxidized-hydroxylase component: A role for the N-terminus. Biochemistry 2001, 40, 9539–9551. [DOI] [PubMed] [Google Scholar]
- (285).Wallar BJ; Lipscomb JD Oxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev 1996, 96, 2625–2657. [DOI] [PubMed] [Google Scholar]
- (286).Zhang JY; Lipscomb JD Role of the C-terminal region of the B component of Methylosinus trichosporium OB3b methane monooxygenase in the regulation of oxygen activation. Biochemistry 2006, 45, 1459–1469. [DOI] [PubMed] [Google Scholar]
- (287).Wang WX; Iacob RE; Luoh RP; Engen JR; Lippard SJ Electron transfer control in soluble methane monooxygenase. J. Am. Chem. Soc 2014, 136, 9754–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (288).Kopp DA; Berg EA; Costello CE; Lippard SJ Structural features of covalently cross-linked hydroxylase and reductase proteins of soluble methane monooxygenase as revealed by mass spectrometric analysis. J. Biol. Chem 2003, 278, 20939–20945. [DOI] [PubMed] [Google Scholar]
- (289).Ericson A; Hedman B; Hodgson KO; Green J; Dalton H; Bentsen JG; Beer RH; Lippard SJ Structural Characterization by EXAFS Spectroscopy of the Binuclear Iron Center in Protein A of Methane Monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc 1988, 110, 2330. [Google Scholar]
- (290).Fox BG; Surerus KK; Münck E; Lipscomb JD Evidence for a m-oxo-bridged binuclear iron cluster in the hydroxylase component of methane monooxygenase. J. Biol. Chem 1988, 263, 10553–10556. [PubMed] [Google Scholar]
- (291).Hendrich MP; Fox BG; Andersson KK; Debrunner PG; Lipscomb JD Ligation of the diiron site of the hydroxylase component of methane monooxygenase. J. Biol. Chem 1992, 267, 261–269. [PubMed] [Google Scholar]
- (292).DeWitt JG; Bentsen JG; Rosenzweig AC; Hedman B; Green J; Pilkington S; Papaefthymiou GC; Dalton H; Hodgson KO; Lippard SJ X-ray absorption, Mössbauer, and EPR studies of the dinuclear iron center in the hydroxylase component of methane monooxygenase. J. Am. Chem. Soc 1991, 113, 9219–9235. [Google Scholar]
- (293).Fox BG; Hendrich MP; Surerus KK; Andersson KK; Froland WA; Lipscomb JD; Münck E Mössbauer, EPR, and ENDOR studies of the hydroxylase and reductase components of methane monooxygenase from Methylosinus trichosporium OB3b. J. Am. Chem. Soc 1993, 115, 3688–3701. [Google Scholar]
- (294).Whittington DA; Lippard SJ Crystal structures of soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site. J. Am. Chem. Soc 2001, 123, 827–838. [DOI] [PubMed] [Google Scholar]
- (295).Rosenzweig AC; Nordlund P; Takahara PM; Frederick CA; Lippard SJ Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem. Biol 1995, 2, 409–418. [PubMed] [Google Scholar]
- (296).Whittington DA; Rosenzweig AC; Frederick CA; Lippard SJ Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase. Biochemistry 2001, 40, 3476–3482. [DOI] [PubMed] [Google Scholar]
- (297).Sazinsky MH; Lippard SJ Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the α-subunit. J. Am. Chem. Soc 2005, 127, 5814–5825. [DOI] [PubMed] [Google Scholar]
- (298).Whittington DA; Sazinsky MH; Lippard SJ X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase. J. Am. Chem. Soc 2001, 123, 1794–1795. [DOI] [PubMed] [Google Scholar]
- (299).Davydov R; Kuprin S; Graslund A; Ehrenberg A Electron-paramagnetic-resonance study of the mixed-valent diiron center in Escherichia coli ribonucleotide reductase produced by reduction of radical-free protein R2 at 77 K. J. Am. Chem. Soc 1994, 116, 11120–11128. [Google Scholar]
- (300).DeRose VJ; Liu KE; Lippard SJ; Hoffman BM Investigation of the dinuclear Fe center of methane monooxygenase by advanced paramagnetic resonance techniques: on the geometry of DMSO binding. J. Am. Chem. Soc 1996, 118, 121–134. [Google Scholar]
- (301).Smoukov SK; Kopp DA; Valentine AM; Davydov R; Lippard SJ; Hoffman BM Product binding to the diiron(III) and mixed-valence diiron centers of methane monooxygenase hydroxylase studied by 1,2H and 19F ENDOR spectroscopy. J. Am. Chem. Soc 2002, 124, 2657–2663. [DOI] [PubMed] [Google Scholar]
- (302).Wang W; Liang AD; Lippard SJ Coupling oxygen consumption with hydrocarbon oxidation in bacterial multicomponent monooxygenases. Acc. Chem. Res 2015, 48, 2632–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (303).Jones JC; Banerjee R; Shi K; Aihara H; Lipscomb JD Structural studies of Methylosinus trichosporium OB3b soluble methane monooxygenase hydroxylase and regulatory component complex reveal a transient substrate tunnel. Biochemistry 2020, 59, 2946–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (304).Wallar BJ; Lipscomb JD Methane monooxygenase component B mutants alter the kinetics of steps throughout the catalytic cycle. Biochemistry 2001, 40, 2220–2233. [DOI] [PubMed] [Google Scholar]
- (305).Brazeau BJ; Lipscomb JD Key amino acid residues in the regulation of soluble methane monooxygenase catalysis by component B. Biochemistry 2003, 42, 5618–5631. [DOI] [PubMed] [Google Scholar]
- (306).Valentine AM; Stahl SS; Lippard SJ Mechanistic studies of reduced methane monooxygenase hydroxylase with dioxygen and substrates. J. Am. Chem. Soc 1999, 121, 3876–3887. [Google Scholar]
- (307).Banerjee R; Lipscomb JD Small-molecule tunnels in metalloenzymes viewed as extensions of the active site. Acc. Chem. Res 2021, 54, 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (308).Jones JC; Banerjee R; Semonis MM; Shi K; Aihara H; Lipscomb JD X-ray crystal structures of methane monooxygenase hydroxylase complexes with variants of its regulatory component: correlations with altered reaction cycle dynamics. Biochemistry 2022, 61, 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (309).Zheng H; Lipscomb JD Regulation of methane monooxygenase catalysis based on size exclusion and quantum tunneling. Biochemistry 2006, 45, 1685–1692. [DOI] [PubMed] [Google Scholar]
- (310).Liu KE; Wang D; Huynh BH; Edmondson DE; Salifoglou A; Lippard SJ Spectroscopic detection of intermediates in the reaction of dioxygen with the reduced methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc 1994, 116, 7465–7466. [Google Scholar]
- (311).Tinberg CE; Lippard SJ Dioxygen activation in soluble methane monooxygenase. Acc. Chem. Res 2011, 44, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (312).Liu KE; Valentine AM; Wang D; Huynh BH; Edmondson DE; Salifoglou A; Lippard SJ Kinetic and spectroscopic characterization of intermediates and component interactions in reactions of methane monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc 1995, 117, 10174–10185. [Google Scholar]
- (313).Brazeau BJ; Lipscomb JD Kinetics and activation thermodynamics of methane monooxygenase compound Q formation and reaction with substrates. Biochemistry 2000, 39, 13503–13515. [DOI] [PubMed] [Google Scholar]
- (314).Tinberg CE; Lippard SJ Revisiting the mechanism of dioxygen activation in soluble methane monooxygenase from M. capsulatus (Bath): evidence for a multi-step, proton-dependent reaction pathway. Biochemistry 2009, 48, 12145–12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (315).Lee S-K; Lipscomb JD Oxygen activation catalyzed by methane monooxygenase hydroxylase component: proton delivery during the O-O bond cleavage steps. Biochemistry 1999, 38, 4423–4432. [DOI] [PubMed] [Google Scholar]
- (316).Banerjee R; Meier KK; Münck E; Lipscomb JD Intermediate P* from soluble methane monooxygenase contains a diferrous cluster. Biochemistry 2013, 52, 4331–4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (317).Lee S-K; Fox BG; Froland WA; Lipscomb JD; Münck E A transient intermediate of the methane monooxygenase catalytic cycle containing an FeIVFeIV cluster. J. Am. Chem. Soc 1993, 115, 6450–6451. [Google Scholar]
- (318).Shu L; Nesheim JC; Kauffmann K; Münck E; Lipscomb JD; Que L Jr. An Fe2IVO2 diamond core structure for the key intermediate Q of methane monooxygenase. Science 1997, 275, 515–518. [DOI] [PubMed] [Google Scholar]
- (319).Banerjee R; Proshlyakov Y; Lipscomb JD; Proshlyakov DA Structure of the key species in the enzymatic oxidation of methane to methanol. Nature 2015, 518, 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (320).Rosenzweig AC Biochemistry: Breaking methane. Nature 2015, 518, 309–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (321).Baik M-H; Newcomb M; Friesner RA; Lippard SJ Mechanistic studies on the hydroxylation of methane by methane monooxygenase. Chem. Rev 2003, 103, 2385–2419. [DOI] [PubMed] [Google Scholar]
- (322).Xue G; De Hont R; Munck E; Que L Jr. Million-fold activation of the [Fe2(μ-O)2] diamond core for C-H bond cleavage. Nat. Chem 2010, 2, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (323).Castillo RG; Banerjee R; Allpress CJ; Rohde GT; Bill E; Que L; Lipscomb JD; DeBeer S High-energy-resolution fluorescence-detected X-ray absorption of the Q intermediate of soluble methane monooxygenase. J. Am. Chem. Soc 2017, 139, 18024–18033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (324).Schulz CE; Castillo RG; Pantazis DA; DeBeer S; Neese F Structure-spectroscopy correlations for intermediate Q of soluble methane monooxygenase: insights from QM/MM calculations. J. Am. Chem. Soc 2021, 143, 6560–6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (325).Cutsail GE 3rd; Banerjee R; Zhou A; Que L Jr.; Lipscomb JD; DeBeer S High-resolution extended X-ray absorption fine structure analysis provides evidence for a longer Fe···Fe distance in the Q intermediate of methane monooxygenase. J. Am. Chem. Soc 2018, 140, 16807–16820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (326).Jacobs AB; Banerjee R; Deweese DE; Braun A; Babicz JT Jr.; Gee LB; Sutherlin KD; Böttger LH; Yoda Y; Saito M; et al. Nuclear resonance vibrational spectroscopic definition of the Fe(IV)2 intermediate Q in methane monooxygenase and its reactivity. J. Am. Chem. Soc 2021, 143, 16007–16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (327).Priestley ND; Floss HG; Froland WA; Lipscomb JD; Williams PG; Morimoto H Cryptic stereospecificity of methane monooxygenase. J. Am. Chem. Soc 1992, 114, 7561–7562. [Google Scholar]
- (328).Valentine AM; Wilkinson B; Liu KE; Komar-Panicucci S; Priestley ND; Williams PG; Morimoto H; Floss HG; Lippard SJ Tritiated chiral alkanes as substrates for soluble methane monooxygenase from Methylococcus capsulatus (Bath): probes for the mechanism of hydroxylation. J. Am. Chem. Soc 1997, 119, 1818–1827. [Google Scholar]
- (329).Gherman BF; Dunietz BD; Whittington DA; Lippard SJ; Friesner RA Activation of the C-H bond of methane by intermediate Q of methane monoozygenase: A theoretical study. J. Am. Chem. Soc 2001, 123, 3836–3837. [DOI] [PubMed] [Google Scholar]
- (330).Guallar V; Gherman BF; Miller WH; Lippard SJ; Friesner RA Dynamics of alkane hydroxylation at the non-heme diiron center in methane monooxygenase. J. Am. Chem. Soc 2002, 124, 3377–3384. [DOI] [PubMed] [Google Scholar]
- (331).Baik MH; Gherman BF; Friesner RA; Lippard SJ Hydroxylation of methane by non-heme diiron enzymes: molecular orbital analysis of C-H bond activation by reactive intermediate Q. J. Am. Chem. Soc 2002, 124, 14608–14615. [DOI] [PubMed] [Google Scholar]
- (332).Liu KE; Johnson CC; Newcomb M; Lippard SJ Radical clock studies and kinetic isotope effect studies of the hydroxylation of hydrocarbons by methane monooxygenase. J. Am. Chem. Soc 1993, 115, 939–947. [Google Scholar]
- (333).Choi S-Y; Eaton PE; Kopp DA; Lippard SJ; Newcomb M; Shen R Cationic species can be produced in soluble methane monooxygenase-catalyzed hydroxylation reactions; radical intermediates are not formed. J. Am. Chem. Soc 1999, 121, 12198–12199. [Google Scholar]
- (334).Jin Y; Lipscomb JD Probing the mechanism of C-H activation: oxidation of methylcubane by soluble methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 1999, 38, 6178–6186. [DOI] [PubMed] [Google Scholar]
- (335).Valentine AM; LeTadic-Biadatti MH; Toy PH; Newcomb M; Lippard SJ Oxidation of ultrafast radical clock substrate probes by the soluble methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem 1999, 274, 10771–10776. [DOI] [PubMed] [Google Scholar]
- (336).Jin Y; Lipscomb JD Mechanistic insights into C-H activation from radical clock chemistry: oxidation of substituted methylcyclopropanes catalyzed by soluble methane monooxygenase from Methylosinus trichosporium OB3b. Biochim. Biophys. Acta 2000, 1543, 47–59. [DOI] [PubMed] [Google Scholar]
- (337).Brazeau BJ; Austin RN; Tarr C; Groves JT; Lipscomb JD Intermediate Q from soluble methane monooxygenase hydroxylates the mechanistic substrate probe norcarane: evidence for a stepwise reaction. J. Am. Chem. Soc 2001, 123, 11831–11837. [DOI] [PubMed] [Google Scholar]
- (338).Kopp DA; Lippard SJ Soluble methane monooxygenase: activation of dioxygen and methane. Curr. Opin. Chem. Biol 2002, 6, 568–576. [DOI] [PubMed] [Google Scholar]
- (339).Sazinsky MH; Lippard SJ Methane monooxygenase: functionalizing methane at iron and copper. Met. Ions Life Sci 2015, 15, 205–256. [DOI] [PubMed] [Google Scholar]
- (340).Nesheim JC; Lipscomb JD Large kinetic isotope effects in methane oxidation catalyzed by methane monooxygenase - evidence for C-H bond cleavage in a reaction cycle intermediate. Biochemistry 1996, 35, 10240–10247. [DOI] [PubMed] [Google Scholar]
- (341).Brazeau BJ; Wallar BJ; Lipscomb JD Unmasking of deuterium kinetic isotope effects on the methane monooxygenase compound Q reaction by site-directed mutagenesis of component B. J. Am. Chem. Soc 2001, 123, 10421–10422. [DOI] [PubMed] [Google Scholar]
- (342).Liu Y; Nesheim JC; Paulsen KE; Stankovich MT; Lipscomb JD Roles of the methane monooxygenase reductase component in the regulation of catalysis. Biochemistry 1997, 36, 5223–5233. [DOI] [PubMed] [Google Scholar]
- (343).Jones JC; Banerjee R; Shi K; Semonis MM; Aihara H; Pomerantz WCK; Lipscomb JD Soluble methane monooxygenase component interactions monitored by 19F NMR. Biochemistry 2021, 60, 1995–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (344).Fox BG; Liu Y; Dege JE; Lipscomb JD Complex formation between the protein components of methane monooxygenase from Methylosinus trichosporium OB3b. J. Biol. Chem 1991, 266, 540–550. [PubMed] [Google Scholar]
- (345).Liu KE; Lippard SJ Redox properties of the hydroxylase component of methane monooxygenase from Methylococcus capsulatus (Bath). J. Biol. Chem 1991, 266, 12836–12839. [PubMed] [Google Scholar]
- (346).Paulsen KE; Liu Y; Fox BG; Lipscomb JD; Münck E; Stankovich MT Oxidation-reduction potentials of the methane monooxygenase hydroxylase component from Methylosinus trichosporium OB3b. Biochemistry 1994, 33, 713–722. [DOI] [PubMed] [Google Scholar]
- (347).Zhang J; Wallar BJ; Popescu CV; Renner DB; Thomas DD; Lipscomb JD Methane monooxygenase hydroxylase and B component interactions. Biochemistry 2006, 45, 2913–2926. [DOI] [PubMed] [Google Scholar]
- (348).Oldenhuis R; Oedzes JY; van der Waarde JJ; Janssen DB Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl. Environ. Microbiol 1991, 57, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (349).Jahng D; Wood TK Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol 1994, 60, 2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (350).Jahng D; Kim CS; Hanson RS; Wood TK Optimization of trichloroethylene degradation using soluble methane monooxygenase of Methylosinus trichosporium OB3b expressed in recombinant bacteria. Biotechnol. Bioeng 1996, 51, 349–359. [DOI] [PubMed] [Google Scholar]
- (351).Zill D; Lettau E; Lorent C; Seifert F; Singh PK; Lauterbach L Crucial role of the chaperonin GroES/EL for heterologous production of the soluble methane Monooxygenase from Methylomonas methanica MC09. Chembiochem 2022, 23, No. e202200195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (352).Bennett RK; Dzvova N; Dillon M; Jones S; Hestmark K; Zhu B; Helman N; Greenfield D; Clarke E; Papoutsakis ET Expression of soluble methane monooxygenase in Escherichia coli enables methane conversion. bioRxiv preprint 2021, DOI: 10.1101/2021.08.05.455234, (accessed 8/6/21). [DOI] [Google Scholar]
- (353).Smith TJ; Slade SE; Burton NP; Murrell JC; Dalton H Improved system for protein engineering of the hydroxylase component of soluble methane monooxygenase. Appl. Environ. Microbiol 2002, 68, 5265–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (354).Smith TJ; Murrell JC Mutagenesis of soluble methane monooxygenase. Methods Enzymol 2011, 495, 135–147. [DOI] [PubMed] [Google Scholar]
- (355).Smith TJ; Nichol T Engineering soluble methane monooxygenase for biocatalysis. In Methane biocatalysis: paving the way to sustainability; Kalyuzhnaya MG; Xing X-H, Eds.; Springer International Publishing, 2018; pp 153–168. [Google Scholar]
- (356).Lock M; Nichol T; Murrell JC; Smith TJ Mutagenesis and expression of methane monooxygenase to alter regioselectivity with aromatic substrates. FEMS Microbiol. Lett 2017, 364. fnx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (357).Lontoh S; Semrau JD Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl. Environ. Microbiol 1998, 64, 1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]


