Abstract
The INK4a/ARF locus encodes two proteins involved in tumor suppression in a manner virtually unique in mammalian cells. Distinct first exons, driven from separate promoters, splice onto a common exon 2 and 3 but utilize different reading frames to produce two completely distinct proteins, both of which play roles in cell cycle control. INK4a, a critical element of the retinoblastoma gene pathway, binds to and inhibits the activities of CDK4 and CDK6, while ARF, a critical element of the p53 pathway, increases the level of functional p53 via interaction with MDM2. Here we clone and characterize the promoter of the human ARF gene and show that it is a CpG island characteristic of a housekeeping gene which contains numerous Sp1 sites. Both ARF and INK4a are coordinately expressed in cells except when their promoter regions become de novo methylated. In one of these situations, ARF transcription could be reactivated by treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine, and the reactivation kinetics of ARF and INK4a were found to differ slightly in a cell line in which both genes were silenced by methylation. The ARF promoter was also found to be highly responsive to E2F1 expression, in keeping with previous results at the RNA level. Lastly, transcription from the ARF promoter was down-regulated by wild-type p53 expression, and the magnitude of the effect correlated with the status of the endogenous p53 gene. This finding points to the existence of an autoregulatory feedback loop between p53, MDM2, and ARF, aimed at keeping p53 levels in check.
The INK4a/ARF (alternative reading frame) cell cycle regulatory locus has proven to be a unique and interesting experimental system. The INK4a (p16 INK4a, MTS1, CDKN2, p16α) gene was implicated as a tumor suppressor gene by its frequent mutation, deletion, or promoter hypermethylation in a variety of human tumors (14, 39; reviewed in reference 64). The role of the INK4a locus became more complicated with the finding of a second gene, ARF, which shares a portion of the INK4a coding region and has a unique first exon (termed exon 1β) originating approximately 20 kb centromeric to INK4a exon 1 (now termed exon 1α) (reviewed in references 31, 36, and 68). This exon, under the control of its own promoter, splices onto exon 2 of INK4a in an alternative reading frame, allowing for the production of two totally unrelated proteins, both of which are cell cycle regulators and tumor suppressors in mouse models (27, 33, 52, 61). The critical importance of this locus may lie in the involvement of its two gene products in two of the most important cell cycle regulatory pathways: ARF in the p53 pathway and INK4a in the retinoblastoma gene product (pRb) pathway (reviewed in references 16 and 27).
Of the four known members of the INK4 family (p15 INK4b, p16 INK4a, p18 INK4c, and p19 INK4d), only INK4a has a firmly established role in human tumorigenesis (23, 26). In addition to INK4a mutations being associated with familial melanoma, mutation or deletion of INK4a occurs with a frequency ranging from approximately 30% in esophageal tumors to nearly 100% in pancreatic and non-small-cell lung carcinomas (4; reviewed in references 17, 57, and 59). The INK4 proteins can block cyclin D-dependent kinase (cdk) activities by preventing their association with the D-type cyclins, causing cells to arrest in G1 (60; reviewed in reference 64). Arrest is mediated primarily through hypophosphorylation of pRb, a substrate of the cyclin D-cdk complex. pRb binds to and inhibits a subset of transcriptional regulatory proteins termed the E2Fs, which then act as repressors of E2F target genes. Phosphorylation of pRb in these complexes allows the E2F proteins to transactivate their target genes, and their products then promote cell cycle progression (reviewed in reference 64).
Another mechanism of INK4a inactivation that has been observed in cases in which no deletion or mutation could be found is promoter region hypermethylation. The promoter of INK4a resides within a CpG island, and abnormal, tumor-associated hypermethylation has been observed in many tumor types and found to result in silencing of the gene (14, 39). CpG islands are regions rich in the CpG dinucleotide which are often associated with genes and are normally kept unmethylated in cells by an unknown mechanism but which may involve binding of the Sp1 transcription factor (35).
The newer member of the INK4a locus, ARF (murine p19 ARF and human p14 ARF [63, 65] and p16β [33]), is predicted to encode a basic polypeptide with no homology to known proteins and which shows approximately 50% identity between mouse and human proteins (compared to 65% for INK4a). ARF is ubiquitously expressed and is elevated in cells lacking functional p53. ARF does not bind to cdks or inhibit the activities of cyclin-cdk complexes; however, overexpression of ARF results in cell cycle arrest in both G1 and G2 (52). Mutations within exon 1β have not been reported; however, mutations within the shared exon 2 region, which inactivate INK4a, also have the potential to affect ARF, but no definitive mutations have yet been found in ARF that do not also disrupt INK4a (11, 51, 71). Large deletions of this region of chromosome 9p that remove INK4a would also likely knock out ARF. The role of mutations in exon 2 of ARF is questionable in any case, since the cell cycle inhibitory functions of ARF are encoded entirely by sequences within the unique exon 1β (51). The role of ARF in tumorigenesis was firmly established, at least in mice, by recent work involving a targeted disruption of exon 1β. ARF-null mice developed lymphomas and sarcomas at an early age, a phenotype indistinguishable from that of a previous INK4a exon 2 knockout which effectively disrupted both INK4a and ARF (61). Furthermore, cell cycle arrest mediated by ARF was abolished in cells lacking functional p53, indicating that ARF may act upstream of p53 (reviewed in references 16, 27, and 48).
Recent work has shown that the role of ARF in the p53 pathway is to bind to MDM2 (48, 78). MDM2, a proto-oncogene itself, binds to p53 and targets it for degradation in the ubiquitin pathway, resulting in abrogation of its antiproliferative and apoptosis-promoting effects (19, 30, 32, 42). MDM2 lso masks the p53 transcriptional activation domain (42, 43). In one study, ARF was shown to bind to and induce the degradation of the MDM2 proto-oncogene, resulting in a stabilization of p53 (78). In a second study, ARF was shown to suppress oncogenic transformation in a p53-dependent manner, block MDM2’s ability to mask the transcriptional activating function of p53, and be necessary for the efficient execution of the p53-dependent apoptotic response (48). Regulation of cell division and apoptosis by p53 is thought to be due to the ability of p53 to modulate transcription of genes involved in controlling both processes. p53, by virtue of its interaction with many cellular proteins, has been shown to act as a sequence-specific DNA binding protein and transcriptional activator (8, 46) and also repress transcription from promoters which do not contain p53 binding sites (6, 13, 34, 38, 58, 69) through sequestration of the TATA binding protein (TBP) and inhibition of transcriptional initiation (62).
The focus of this work was to gain a better understanding of the factors involved in regulating the INK4a/ARF locus. Specifically, we cloned and sequenced the promoter region of the human ARF gene and found that it possesses many of the features of a housekeeping gene. Gene expression studies indicated that ARF is ubiquitously expressed in cell lines, with only two notable exceptions. In these two cases, promoter region hypermethylation was demonstrated by Southern blotting and ARF expression could be reactivated after treatment with the methylation inhibitor 5-aza-2′-deoxycytidine (5-aza-CdR). Promoter deletion analysis revealed the location of potentially important regulatory regions, including numerous Sp1 sites. Potential E2F binding sites were also detected, and the ARF promoter was found to be highly responsive to E2F1 as has been observed previously at the RNA level (9). Transfection studies with ARF and INK4a reporter constructs indicated that the transcription factors necessary for promoter activity were ubiquitously expressed and that the activities of the endogenous genes were related to their methylation status. Lastly, studies comparing the effects of wild-type p53 overexpression on ARF and INK4a promoter constructs indicated that transcription from the ARF promoter could be potently repressed, in keeping with previous observations at the protein level that ARF levels are elevated in cells lacking functional p53. Thus, we propose the existence of an autoregulatory feedback loop involving p53, MDM2, and ARF which functions to keep p53 levels tightly controlled in normal cells.
MATERIALS AND METHODS
Cell lines, tissue culture, and drug treatments.
The colon cancer-derived cell lines HCT116, HCT15, SW48, LoVo, SW837, and HT-29 were maintained in McCoy’s 5-a medium. The bladder cancer lines 5637 and J82 and the cervical cancer line C-33A were maintained in minimal essential medium supplemented with sodium pyruvate and nonessential amino acids (Life Technologies). The lymphoid lines Raji, CA46, and HL-60 were maintained in RPMI 1640. The hepatocellular carcinoma line Hep 3B and the bladder cell line T24 were maintained in Dulbecco modified Eagle medium, and the pancreatic line CFPac-1 was maintained in Iscove modified Dulbecco medium. All media were supplemented with 10% fetal bovine serum (20% for the HL-60 line). All cell lines were purchased from the American Type Culture Collection. For 5-aza-CdR (Sigma) treatments, cells were diluted to 3.0 × 105/ml and allowed to grow overnight; then freshly prepared 5-aza-CdR was added to a final concentration of 1.0 μM, and the cells were allowed to grow for the times indicated. For DNA damage transfection experiments, camptothecin (CMT; Sigma) was used at a final concentration of 5 μM in dimethyl sulfoxide (DMSO) and cytosine arabinoside (Ara-C; Oncogene Science) was used at a final concentration of 50 μM in water. DMSO was also added to Ara-C cultures at the same concentration as that for the CMT treatments to control for nonspecific solvent effects on cells.
Reverse transcription (RT)-PCR analysis of ARF and INK4a expression.
Total RNA (2.5 μg) was isolated with Trizol (Life Technologies) and reverse transcribed by using gene-specific 3′ primers, deoxynucleoside triphosphates (Boehringer Mannheim), and Superscript II reverse transcriptase (Life Technologies) in a 20-μl volume. The primers used were specific for INK4a and ARF (common antisense 5′-TTC CCG AGG TTT CTC AGA G-3′) and for PCNA (proliferating cell nuclear antigen) (antisense 5′-GCT AGG ATC CTA AGA TCC TTC TTC ATC CTC GAT C-3′). cDNA was amplified by PCR with 5′ primers specific for ARF, INK4a, and PCNA with the same antisense primer used for creation of cDNA. PCR was performed in a 50-μl volume as follows: for ARF and INK4a, 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 58°C for 1 min, and 72°C for 1 min; for PCNA, 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min. 5′ primers were 5′-CAT GGT GCG CAG GTT CTT G-3′ for ARF, 5′-AAC GCA CCG AAT AGT TAC G-3′ for INK4a, and 5′-GAT CGG ATC CGT ATG TTC GAG GCG CGC CTG GTC-3′ for PCNA. PCR products were resolved on 1.2% agarose gels, transferred to nylon membranes (NEN), and probed as described previously (56) with the following end-labeled primers: for ARF, 5′-TAC TGA GGA GCC AGC GTC TAG-3′ (exon 1β); for INK4a, 5-TAC TGA GGA GCC AGC GTC TAG-3′ (exon 1α); and for PCNA, 5-CTA GCG CCA AGG TAT CCG CG-3′. Quantitation was performed on a Molecular Dynamics PhosphorImager.
Cloning and sequencing of the human ARF promoter.
A lambda genomic library (human placenta) was probed with an exon 1β-specific probe according to instructions of the manufacturer (Stratagene). The probe was generated by PCR using the sense primer 5′-GAT CGC ATG CTC CCA GTC TGC AGT TAA GG-3′ and antisense primer 5′-GAT CGT CGA CGT CTA AGT CGT TGT AAC CCG-3′, based on the previously published exon 1β sequence (36), and cloned into the TA cloning vector (Invitrogen). PCR conditions were similar to those used for ARF/INK4a RT-PCR, with 10% DMSO added and 50 ng of human placental DNA (Sigma) as the template. A single phage clone was isolated, the insert was characterized by restriction analysis, and an approximately 8.5-kb SacI fragment was subcloned into the SacI site of pBluescript II (Stratagene) to create pKR19. The SacI site at the 3′ end of this fragment corresponded to the SacI site at +49 of the previously published sequence. All numbering is relative to the previously published transcription start site (36). This fragment was further subcloned, and the region from +49 to −5650 was sequenced. A detailed description of subcloning procedures and sequencing primers will be furnished upon request; note that only a portion of the region sequenced is shown in Fig. 2B. All DNA sequencing was performed at the USC DNA Sequencing Core Facility.
FIG. 2.
(A) Plots comparing the frequency of the CpG dinucleotide in the ARF promoter/exon 1β region (top) and the INK4a/exon 1α region (bottom) derived from GenBank accession no. AC000048. Bent arrows are the transcription start sites, and open boxes are transcribed regions. Analysis of the regions of each promoter denoted by the brackets in terms of their CpG contents yields the following: G/C content = 0.64, observed/expected for CpG = 0.85 for ARF and G/C content = 0.54, observed/expected for CpG = 0.68 for INK4a over approximately 2,400 bp. (B) Sequence of a portion of the ARF promoter region (GenBank accession no. AF082338). The previously mapped transcription start site (36) is indicated by a bent arrow above the italicized “G” and is defined here as position +1. The positions of several potential transcription factor binding sites are underlined, as is a region homologous to other known initiator elements (Inr). Potential E2F binding sites are denoted with a line above the sequence; (−) indicates that the consensus binding site is 5′ to 3′ on the bottom strand. Positions of restriction enzyme sites used in subsequent cloning steps for promoter deletion analysis are underlined, and their positions relative to the transcription start site are indicated. Downward arrows denote 5′ ends of deletion constructs generated by PCR. A subset of the repetitive elements described in the text (Alu and purine-pyrimidine [Pur-Pyr]) is also shown.
Plasmid constructs.
All ARF CAT promoter constructs were derived from pKR19. Initially, the SphI (−5502)-PstI (−18) fragment from pKR19 was cloned into these same sites of pCAT-Basic (Promega) to create pKR21-3. This construct lacked the native transcription start site, which was added back by digesting pKR21-3 with SalI and cloning the SalI fragment derived from pKR19 (containing the ARF sequences from −735 to +49 and a SalI site derived from pBluescript II) into this same site and screening for proper orientation to create p(−5502)19ARF. All subsequent deletion constructs had the same 3′ end at the +49 SacI site. 5′-end deletions were then made by digesting p(−5502)19ARF with HindIII (within the pCAT-Basic vector polylinker) and a second enzyme within the ARF sequence, blunting the ends with T4 DNA polymerase (Boehringer Mannheim), and recircularizing with T4 DNA ligase (Boehringer Mannheim). The second restriction enzyme sites and the deletion constructs created were as follows: BstXI for p(−4690)19ARF, KpnI for p(−3407)19ARF, SpeI for p(−2465)19ARF, Eco47III for p(−925)19ARF, SmaI for p(−776)19ARF, SacII for p(−331)19ARF, BglII for p(−151)19ARF, and BssHII for p(−67)19ARF. p(−44)19ARF and p(−19)19ARF were created by PCR with p(−151)19ARF as the template (10 ng), a common 3′ primer located within the vector sequence (5′-CAA CGG TGG TAT ATC CAG TG-3′), and 5′ primer 5′-AGT CGG CAT GCG CAG GGG GCG GTG CGT GGG-3′ for the −44 construct or 5′-AGC TAG CAT GCT CTG CAG TTA AGG GGG CAG G-3′ for the −19 construct. PCR conditions were the same as those used for ARF/INK4a RT-PCR analysis except that Pfu DNA polymerase (Stratagene) was used. The PCR product was digested with SphI (incorporated into 5′ primer) and SalI (derived from pCAT-Basic vector sequences) and cloned into these same sites of pCAT-Basic. All sequences were confirmed. All INK4a promoter constructs were created by PCR with Pfu DNA polymerase and the previously described (15) INK4a promoter-containing lambda phage clone as the template. All constructs were named relative to the 5′-most transcription start site for INK4a, which corresponds to −306 relative to the translation start site and which contains all of the previously mapped transcription start sites (18). A common 3′ primer was used for all constructs at +100 (5′-GCT AGT CGA CGG AGG AGG TGC TAT TAA CTC-3′), and 5′ primers were 5′-GAT CGC ATG CCA AAC ACG CCT TTG CTG GCA-3′ for p(−118)16INK4a, 5′-GAT CGC ATG CGG GGC TCT CAC AAC TAG GAA-3′ for p(−293)16INK4a, 5′-GAT CGC ATG CCC AGA CAG CCG TTT TAC ACG-3′ for p(−474)16INK4a, 5′-GAT CGC ATG CAG CAC TTT TTC TGG TCT AGG A-3′ for p(−654)16INK4a, and 5′-GAT CAA GCT TGA ACT TTT ACC TCC TTG CGC-3′ for p(−1729)16INK4a. The sequence of each construct was confirmed. Each PCR product was digested with SphI [incorporated into the 5′ primer; HindIII was used for p(−1729)16INK4a] and SalI (incorporated into the 3′ primer) and cloned into these same sites of pCAT-Basic. The control consensus p53 binding site chloramphenicol acetyltransferase (CAT) plasmid [2X(p53)BSCAT] was created by annealing complementary oligonucleotides (top strand, 5′-GAT CTA GGC ATG CCT AGG CAT GCC TAA AGG CAT GCC TAG GCA TGC CTA-3′) containing two copies of a consensus p53 binding site (46). When annealed, BglII site overhangs were created and used to clone the oligonucleotide into the BglII site of pGH262 (provided by Gary Hayward, The Johns Hopkins University) to create 2X(p53)BSCAT. The p53 promoter-CAT plasmid (p53proCAT) was created by PCR as described for the INK4a reporter constructs with 5′ primer 5′-ACT GAG CAT GCG GGA GAA AAC GTT AGG GTG TGG-3′, 3′ primer 5′-GTG GCT CTA GAC TTT TGA GAA GCT C-3′, and human placental DNA as the template. The product was digested with SphI and XbaI and ligated into these same sites of pCAT-Basic to create p53proCAT, which contains the region from −96 to −534, relative to the translation start site, of the human p53 promoter (73).
Transfection, CAT assay, and in vitro methylation.
Cells were transfected with Lipofectamine as instructed by the manufacturer (Life Technologies). Briefly, 6 × 105 cells were incubated in the presence of 2 μg of CAT plasmid and 8 μl of Lipofectamine in the absence of serum for 7 h. Cell extracts were prepared 48 h later for CAT and β-galactosidase assays as described previously (15, 56). Most CAT plasmid vectors were cotransfected with pSV40LacZ (15), and CAT activity was normalized relative to β-galactosidase activity to control for differences in transfection efficiency. For the p53 dose-response transfections, 2 μg of CAT plasmid was used along with a total of 2 μg of expression vector. The amount of wild-type or mutant p53 expression vector was kept constant at 2 μg by addition of parental expression vector containing no cDNA insert. Wild-type p53 (pC53-SN3), mutant p53 (pC53-SCX3), and parental expression vector (pCMVNeoBam) were kindly provided by Bert Vogelstein (The Johns Hopkins University). For DNA damage transfections, cells were transfected identically except that drug was added with serum-containing medium 7 h after transfection, the cells were incubated overnight, and fresh medium was added the following morning. The total transfection time remained the same, 48 h. CAT activities were normalized to the protein concentration to account for small toxicity differences. Protein concentrations were determined with the Bio-Rad protein assay reagent according to the manufacturer’s instructions. For E2F1 response element mapping experiments, 2.0 μg of reporter plasmid was cotransfected with 2.0 μg of E2F1 expression vector (provided by Joseph Nevins, Duke University) or empty parental expression vector [pcDNA 3.1(+); Invitrogen]. In vitro methylation reactions were carried out as described previously (55) with methylases purchased from New England Biolabs. The completeness of the methylation reaction was confirmed by digestion of an aliquot of the reaction with an appropriate methylation-sensitive restriction enzyme (HpaII or HhaI). Quantitation of CAT activity was performed on a Molecular Dynamics PhosphorImager.
Methylation analysis.
Ten micrograms of genomic DNA was isolated from cell lines by standard procedures, digested with 100 U of each of the restriction enzymes described in Results, resolved on a 1% agarose gel, transferred to a nylon membrane (NEN), and probed with ARF promoter-derived fragments as described previously (25, 55). Double digestions were performed sequentially, so that each restriction enzyme was in the optimal incubation buffer with a precipitation step in between. All enzymes were purchased from New England Biolabs, and digestions were performed according to the manufacturer’s instructions.
Nucleotide sequence accession number.
The DNA sequence of the human ARF promoter has been deposited in GenBank under accession no. AF082338.
RESULTS
Analysis of ARF and INK4a expression patterns in cell lines and evidence for suppression by DNA methylation.
We initially screened a panel of tumor cell lines derived from a variety of different tissues for both ARF and INK4a expression by RT-PCR (Table 1 and Fig. 1A). Expression of ARF was observed in all tumor cell lines examined except the colon cancer cell lines HCT15 and LoVo (Table 1; note that cell lines with known deletions of this region were excluded from this analysis). Expression of INK4a was more restricted, and the data summarized in Table 1 indicated that there were three expression patterns from the INK4a/ARF locus: (i) ARF+ INK4a+, (ii) ARF+ INK4a−, and (iii) ARF− INK4a−. Interestingly we found no cell line in which ARF− INK4a+ was the pattern. A large body of work has shown that the promoters of INK4a and p15 INK4b can become de novo methylated and silenced in tumor cells (14, 20, 39). It was therefore possible that ARF, which resides between these two genes, might be subject to such silencing.
TABLE 1.
Analysis of INK4a and ARF expression in cell lines by RT-PCRa
| Tissue source | Cell line | 5-Aza-CdR treatment | Expression of:
|
|
|---|---|---|---|---|
| ARF | INK4a | |||
| Colon | HCT116 | − | + | + |
| + | + | + | ||
| SW48 | − | + | − | |
| + | + | + | ||
| HCT15 | − | − | − | |
| + | + | + | ||
| LoVo | − | − | − | |
| SW837 | − | + | − | |
| HT-29 | − | + | − | |
| Cervix | C-33A | − | + | + |
| Bladder | 5637 | − | + | + |
| T24 | − | + | − | |
| + | + | + | ||
| J82 | − | + | + | |
| Lymphoid | Raji | − | + | − |
| + | + | + | ||
| CA46 | − | + | − | |
| HL-60 | − | + | − | |
| Liver | Hep 3B | − | + | + |
| Pancreas | CFPac-1 | − | + | − |
| + | + | + | ||
Expression of the transcript indicated was scored as present (+) or absent (−) after Southern hybridization with oligonucleotide probes specific for the first exon of each gene. 5-Aza-CdR treatments were at 1.0 μM for 72 h.
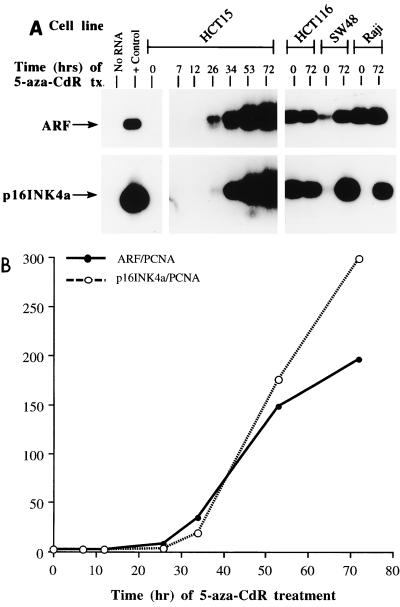
FIG. 1.
(A) RT-PCR analysis of ARF (top) and p16 INK4a (bottom) expression patterns in the HCT15 cell line after treatment with 1.0 μM 5-aza-CdR for the times indicated. Also shown is a single time point (72 h) for the SW48, HCT116, and Raji cell lines. Hep 3B RNA was used as the positive (+) control. PCR products were probed with oligonucleotides specific for the unique first exon of each transcript, and RNA integrity was verified by amplification of transcripts for β-actin and PCNA (not shown). Note that a longer exposure is shown for the HCT15 5-aza-CdR time course experiment in order to emphasize differences at the earlier time points. (B) Quantitation of the results in panel A for the HCT15 cell line relative to the ubiquitously expressed transcript for PCNA.
Several of the lines in Table 1 were treated with 5-aza-CdR (Fig. 1A) to obtain evidence that the ARF promoter could be silenced by DNA methylation. The SW48, HCT116, and Raji lines were used as controls, and ARF was expressed before drug treatment in all three lines, although SW48 expressed more ARF RNA after treatment. INK4a was expressed before drug treatment in HCT116 cells and was activated following 5-aza-CdR treatment in the SW48 and Raji lines (Fig. 1A). HCT15 cells, which expressed neither INK4a nor ARF, were monitored at various times after treatment to determine if there was differential reactivation kinetics for the two tandemly linked genes. Induction of both transcripts occurred in a time-dependent fashion, with ARF expression slightly preceding INK4a activation, an effect most noticeable at the 34-h time point (Fig. 1A). The results were quantitated relative to the ubiquitously expressed transcript for PCNA (Fig. 1B). Although ARF transcripts appeared earlier than INK4a transcripts, the INK4a transcript levels were ultimately higher than those of ARF transcripts, as shown by the crossover of the two lines at approximately 40 h posttreatment. Our results clearly demonstrate that both transcripts can be reactivated in tandem. Although we cannot rule out the possibility that each transcript is expressed from a different allele, such a situation has not been observed in other, similar tandem promoter systems (7, 49, 74, 75).
Cloning and characterization of the human ARF promoter.
We used the known sequence of exon 1β (36) as a probe to screen a lambda genomic library to further characterize both the role of DNA methylation in suppressing ARF promoter-driven transcription and its regulation in general. A clone containing all of exon 1β and at least 10 kb of upstream sequence (data not shown) was identified, and an approximately 8.5-kb SacI fragment subcloned from the phage clone and the 3′ end corresponded to a SacI site at +49 (relative to the transcription start site) of the previously published exon 1β sequence (36). This fragment was subcloned further (see Materials and Methods) and sequenced to bp −5650 relative to the previously published transcription start site (36). The sequence indicated that ARF is a TATA-less promoter and a CpG island like that seen for many other housekeeping genes. The region extending from +49 to −2678, the most CpG-rich region (encompassed by brackets in Fig. 5B), had a G/C content of 0.59 and an observed-over-expected ratio of CpG of 0.78 and contained 183 CpG sites, clearly meeting the established criteria for a CpG island (3, 12). Figure 2A compares the ARF promoter region reported here with the previously analyzed INK4a promoter, also a CpG island (15, 18).
FIG. 5.
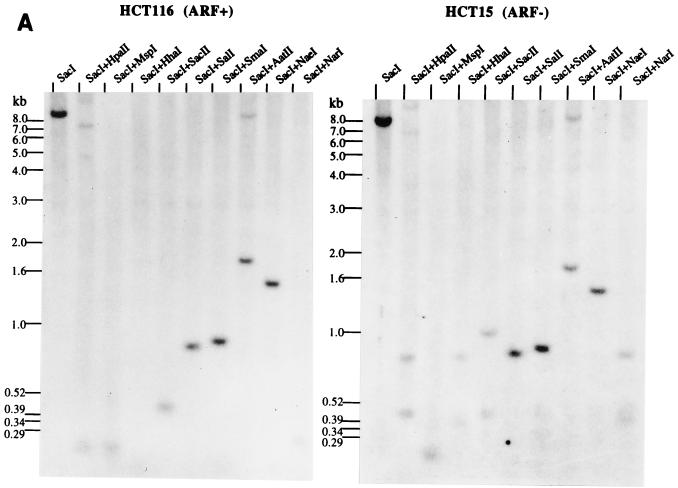
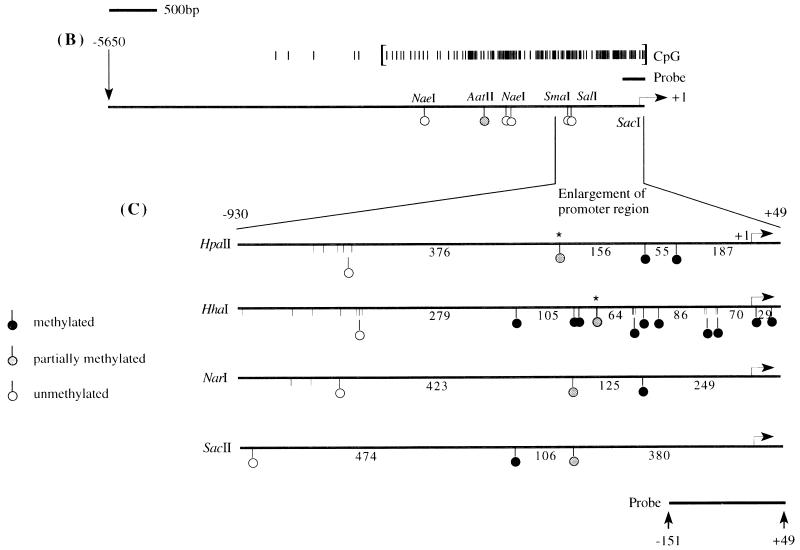
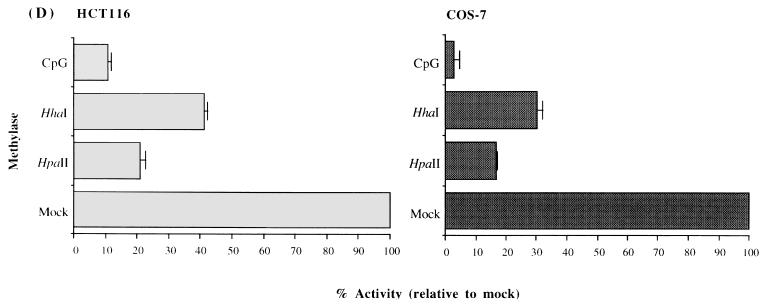
Methylation analysis of the ARF promoter. (A) Representative Southern blots after digestion of HCT116 and HCT15 genomic DNAs with the enzymes indicated and probing with the fragment shown in panels B and C. The presence of higher-molecular-weight bands in the HCT15 digests compared to the HCT116 digests is indicative of methylation. The low level of hybridization after digestion with enzymes like HpaII and HhaI is a result of the large number of such sites, creating many small restriction fragments which hybridize poorly. (B) Schematic of the location of several of the rare-cutting methylation-sensitive restriction enzyme sites, a CpG plot of the entire sequenced region, and the transcription start site (+1). Brackets in the CpG plot indicate the boundaries used for the calculation of CpG island status. The location of the probe is indicated by the thick bar, and the methylation status at the CpG sites analyzed by restriction digest is indicated by the lollipops. (C) Blowup of the region immediately adjacent to the ARF promoter (−930 to +49) and summary of the methylation status of CpG sites in this region as determined from blots in panel A for the HCT15 cell line. Asterisks indicate that these particular CpG sites, while partially methylated in HCT15 cells, were completely methylated in the LoVo cell line (not shown). The sizes of the fragments are indicated below the lines. A lollipop displaced below a group of restriction enzyme sites indicates that the sites were too close together to accurately determine which site was digested. (D) Analysis of the methylation sensitivity of the p(−151)19ARF promoter CAT construct after in vitro methylation and transfection into HCT116 or COS-7 cells. Results after treatment with the various methylases are presented as the mean percent activity relative to the mock-methylated control (no methylase) for triplicate transfections. Error bars represent the standard deviations.
The promoter region of the INK4a gene had been previously characterized (18). It is negatively regulated by pRb; however, the ARF promoter and its regulation have not been previously reported. Figure 2B indicates the locations of numerous potential transcription factor binding sites. In particular, there are seven potential binding sites for the Sp1 transcription factor and a region homologous to the initiator element often present in TATA-less promoters such as this. The placement of the putative initiator relative to the transcription start site and potential Sp1 binding sites is similar to that for other previously described promoters (37; reviewed in reference 67). Also notable are several repetitive elements, such as an Alu element at positions −2942 to −2695, a thymine-cytosine run from −3109 to −3056 (not shown), a guanine-thymine run from −125 to −92, three potential AP-1 sites at −4977, −4829, and −3394 (not shown) (reviewed in reference 24), and one potential YY1 site at −3152 (not shown) (66).
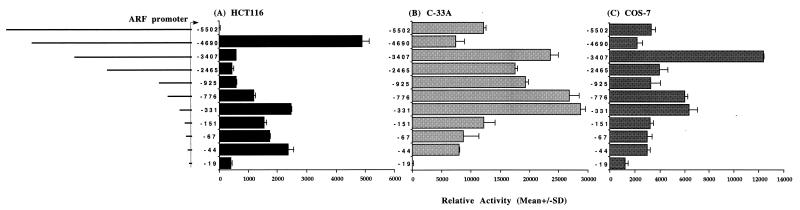
To determine if this region possessed promoter activity and to define important regulatory regions, 5′-end deletions were made by using naturally occurring restriction enzyme sites or PCR and fused to the CAT reporter gene, and the effects were assayed in several cell lines. The results, shown in Fig. 3, indicated that the largest construct, to −5502 (all numbering is relative to the transcription start site), ranged from slightly inhibitory in COS-7 and C-33A cells to potently inhibitory in HCT116 cells. Deletion to −4690 restored high level activity in HCT116 cells but had a relatively minor effect in the other two lines. The binding site for a tissue-specific repressor protein or a silencer may reside within this region. Deletion to −3407 dramatically reduced ARF-driven promoter activity in HCT116 cells; however, a large increase in promoter-driven CAT activity was observed in C-33A and COS-7 cells. Deletion to −2465 reduced activity to some extent, and further deletion to −925 caused a very modest reduction in activity in COS-7 cells but resulted in slight increases in activity in HCT116 and C-33A cells despite deletion of two potential Sp1 binding sites (Fig. 2B). Deletion to −776 and −331 resulted in significant increases in CAT activity in all three cell lines, indicating that a repressive element may reside within this region. Fine deletions nearer the transcription start site to −151, −67, −44, and −19, which delete two, three, four, and five potential Sp1 binding sites respectively, resulted in a gradual decline of activity in COS-7 and C-33A cells. There was a slight increase in activity upon deletion to −44 in HCT116 cells, the reason for which is unclear; however, further deletion to −19 resulted in low-level promoter activity in HCT116 cells. Overall, the effects of the various promoter deletion constructs were similar in COS-7 and C-33A cells but differed significantly from effects in the HCT116 line. This finding may indicate a difference in the transcription factor milieu in the colon cancer line HCT116; however, this analysis does provide clear evidence that the region we have cloned upstream of ARF acts as a promoter.
FIG. 3.
ARF promoter deletion analysis. The extent of each of the 5′ deletions fused to the CAT reporter gene, as well as the extent of each construct relative to the transcription start site (indicated by the bent arrow), is indicated schematically at the left. Results are presented as the mean relative activity (percent acetylation/β-galactosidase activity) for triplicate transfections into the HCT116 (A), C-33A (B), and COS-7 (C) cell lines. Error bars indicate the standard deviations (SD) from the means.
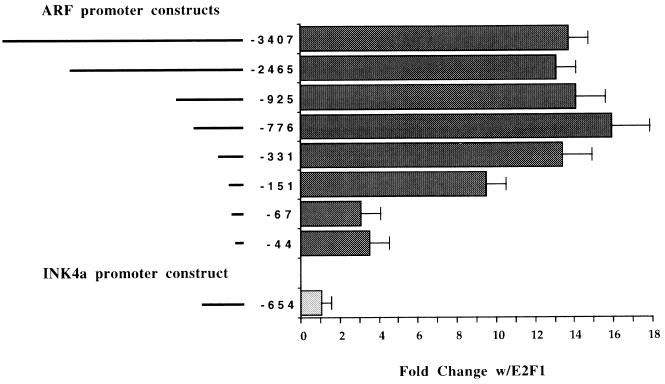
A recent study using adenoviral vectors expressing the five members of the E2F family of transcription factors indicated that ARF mRNA levels were elevated after infection with adenoviruses expressing E2F1 and E2F2 but not E2F3 to E2F5 (9), and we have noted several potential E2F binding sites in Fig. 2B. Good matches to the binding site consensus TTTCCCGCC(A/T)(A/T)(A/T), found to be optimal binding sites for E2F1 to E2F4 in a binding site selection assay (72), were detected at −265 and +27, while poorer matches to this and the standard E2F consensus binding site TTTCGCGC (reviewed in reference 24) were detected at −249 and −69. Figure 3 also indicated that these regions were important for transcription in all cell lines. To determine if the previously observed increase in ARF mRNA after E2F1 overexpression was modulated at the transcriptional level, we cotransfected various ARF reporter deletion constructs with an E2F1 expression vector or empty parental expression vector. Figure 4 indicates that the ARF promoter is highly responsive to E2F1 (nearly 15-fold) and that the regions mediating this response correlate with the locations of potential E2F binding sites. We are at present unsure if the E2F site at +27 would be utilized in the intact natural promoter since it is downstream of the transcription start site. This will require further fine mapping experiments. An INK4a reporter construct served as a negative control and was unaffected by E2F1 overexpression.
FIG. 4.
The ARF promoter is upregulated by E2F1 expression. Each of the ARF or INK4a promoter constructs denoted schematically at the left was cotransfected with an equal amount of an expression vector for E2F1 or empty parental expression vector. The results of duplicate transfections in the HCT116 cell line are presented as the mean fold activation with E2F1 cotransfection (activity with E2F1/activity with empty expression vector). Error bars indicate the standard deviations (SD) from the means.
Comparison of the overall activities of the ARF promoter constructs in Fig. 3 showed that the cell lines could be ordered as C-33A > COS-7 > HCT116 in terms of promoter strength, which correlated with the p53 status of the lines. HCT116 contains a wild-type p53 gene (47), while C-33A has a mutant p53 gene (22), and COS-7 cells express the simian virus 40 large T antigen (T-Ag), which complexes with and inactivates both p53 and pRb (reviewed in references 29 and 45). This is particularly relevant since it has been noted that ARF levels are elevated in cells lacking functional p53 (27, 52), the implication being that p53 may suppress ARF transcription. Results of the transfections studies were consistent with this idea, and further direct evidence for the role of p53 in suppressing ARF at the transcriptional level will be presented later.
Suppression of ARF promoter activity by CpG methylation.
The previous results indicated that the ARF promoter was silent in only two cell lines (HCT15 and LoVo) and that ARF transcription could be activated in HCT15 cells after treatment with 5-aza-CdR. HCT15 and LoVo cells were investigated in more detail to determine if this transcriptional silence might be mediated by hypermethylation of the CpG island associated with the ARF promoter. Genomic DNA from these lines was digested with various methylation-sensitive restriction enzymes and analyzed by Southern blotting (Fig. 5). The digestion pattern obtained with restriction enzymes sensitive to CpG methylation with DNA obtained from the ARF-expressing HCT116 cells was consistent with complete hypomethylation of the CpG sites examined since the high-molecular-weight SacI band of approximately 8.5 kb was absent in the double digests. The exception was the AatII site, which was partially methylated in this line (Fig. 5A). A similar result was obtained with normal leukocyte DNA except that the AatII site was completely hypomethylated (not shown), showing that the CpG island of ARF was unmethylated in normal tissues and in HCT116 cells which expressed the ARF transcript (Table 1 and Fig. 1A). Many of the CpG sites within the promoter were, however, hypermethylated in HCT15 cells, as indicated by the presence of higher-molecular-weight bands, relative to the HCT116 digests, indicating the the 8.5-kb SacI fragment was only partially cut by the methylation-sensitive enzymes. The results, summarized in Fig. 5B and C, indicated that there was a relatively small region of hypermethylation at the 3′ end of the ARF CpG island which did not extend significantly beyond −450 relative to the transcription start site.
Sensitivity of the ARF promoter to CpG methylation was further examined by in vitro methylation and transfection. The ARF construct containing sequences from +49 to −151 fused to the CAT gene was chosen for this study since the Southern blotting studies (Fig. 5A to C) indicated that hypermethylation was confined to the region close to the transcription start site and that this construct possessed a significant degree of promoter activity in its unmethylated state. The −151 construct was methylated in vitro with HpaII methylase (1 recognition site), HhaI methylase (4 recognition sites), and CpG methylase (16 recognition sites) and transfected into HCT116 and COS-7 cells. Figure 5D shows that CpG methylase had the most repressive effect, reducing reporter gene activity to 10% or less of that of the mock-methylated control in both lines. Interestingly, methylation at the single HpaII site had a larger repressive effect than methylation at the four HhaI sites. This finding may indicate that the HpaII site resides in or near the binding site of a critical transcription factor, although no matches to consensus binding sites of known transcription factors were detected in this region.
ARF and INK4a promoter constructs are expression competent in all cell lines.
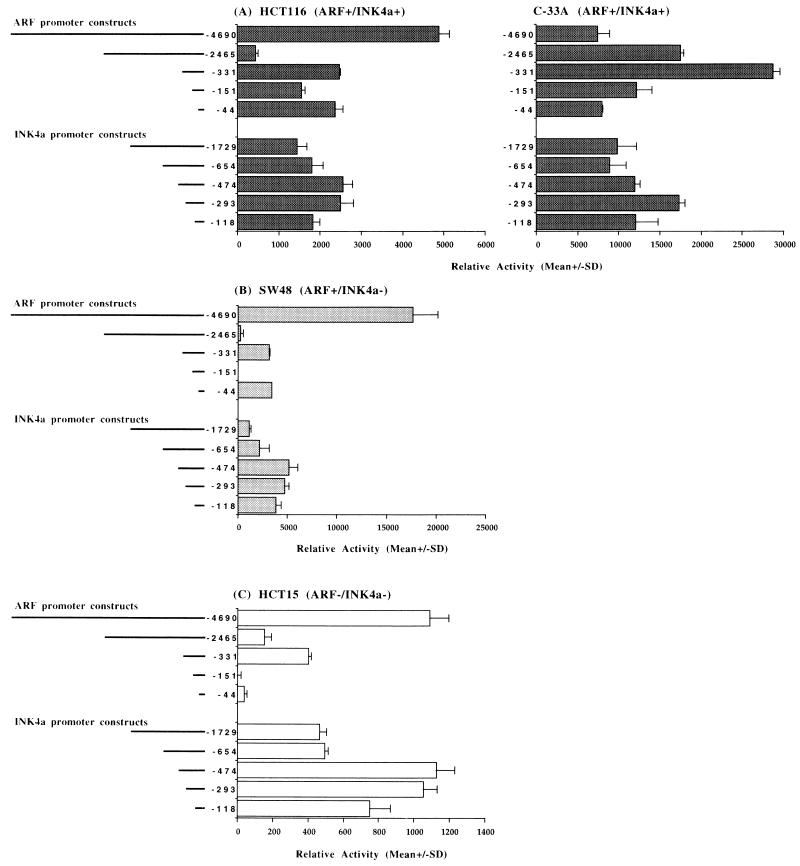
We next determined if the expression patterns for INK4a and ARF shown in Table 1 were a result of differences in the transcription factor milieu or differences in the methylation status of the endogenous gene promoter sequences. A series of ARF and INK4a promoter reporter constructs were transfected into cells with each of the previously mentioned expression patterns. Both the ARF promoter and the INK4a promoter were capable of driving expression of the CAT gene in all cell lines regardless of whether the endogenous ARF or INK4a gene was transcribed (Fig. 6). Both promoters were also nearly equal in their overall activities when the most active construct for each promoter was compared, with the exception of the ARF+ INK4a+ and ARF+ INK4a− cell lines, in which the ARF promoter gave a slightly higher level of expression. This effect was most notable in the ARF+ INK4a− SW48 line and may indicate that there is some differential regulation of these two promoters in some situations. There was, however, some correlation between the absolute CAT activity levels and the activity levels of the endogenous promoters. For example, the overall activities of the ARF and INK4a CAT constructs were highest in the ARF+ INK4a+ and ARF+ INK4a− lines (Fig. 6A and B), while the ARF− INK4a− HCT15 cell line had the lowest overall promoter activity (Fig. 6C). Thus, the transcription factors necessary for ARF and INK4a promoter activity are present in all cell lines, although the relative levels of these factors may vary to some extent. The inactivity of the endogenous promoter was related to its methylation status. It has been shown that the ARF promoter is hypermethylated and inactive in the HCT15 line (Fig. 1A and 5A) and the INK4a promoter is inactive and hypermethylated in the HCT15 and SW48 lines (Fig. 1A; references 2 and 21). It should also be noted that the INK4a promoter analysis presented here differed in some respects from previously published data in that deletions beyond −654 retained relatively high activity levels (18). The reasons for this are unclear but may be due to differences in promoter construction, since our constructs contained less sequence at the 3′ end than in the previous analysis.
FIG. 6.
Comparison of activities of ARF and INK4a promoter-CAT constructs after transfection into ARF+ INK4a+ (A), ARF+ INK4a− (B), and ARF− INK4a− (C) cell lines. The map of each of the reporter constructs relative to the transcription start site of each promoter (for INK4a, the 5′-most initiation site was defined as +1) is indicated at the left and the mean relative activity (percent acetylation/β-galactosidase activity) for triplicate (duplicate for SW48 and HCT15) transfections is shown at the right. Error bars indicate the standard deviations (SD) from the means.
Regulation of the INK4a/ARF locus by p53.
The above studies showed that the ARF promoter region can become de novo methylated and transcriptionally silenced; however, this appeared to be a low-frequency event in cell lines, making it unlikely that this phenomenon would occur frequently in primary tumors. Another potential regulatory control mechanism of ARF expression may be mediated by the tumor suppressor gene p53, since ARF levels are elevated in cell lines lacking functional p53, implying that p53 may suppress ARF. While not rigorously quantitated, the increase in ARF expression in previous reports has ranged from 5- to 10-fold (18, 27, 52). Since we now had available the ARF promoter, we were able to test the hypothesis that p53 may suppress ARF at the transcriptional level.
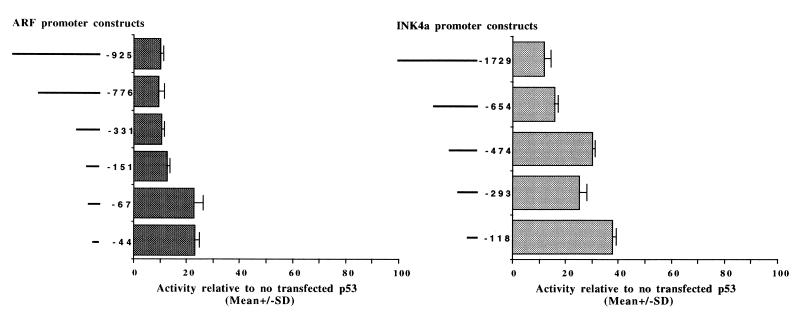
Initially we transfected a series of ARF and INK4a reporter deletion constructs with a fixed amount of wild-type p53 expression vector or empty parental expression vector into the p53-mutant cell line C-33A. The results (Fig. 7) indicated that both the ARF and INK4a promoters were significantly repressed and that the majority of the repressive effect was mediated by the region encompassing the transcription initiation sites. A small proportion of the repressive effect also appeared to be mediated by additional regions from −151 to −67 of the ARF promoter and −654 to −474 of the INK4a promoter. The majority of the repressive effect being mediated by the region containing the transcription start site is entirely consistent with the proposed mechanism of p53 repression, that is, interaction with the TBP component of the TFIID complex, causing a reduction in transcriptional initiation of certain promoters which do not contain p53 binding sites (62). We did not detect any p53 binding sites within the sequenced region of ARF or the reported INK4a promoter sequence. It should also be noted that although both promoters are TATA-less, the TBP-TFIID complex has been shown to be required for transcriptional initiation at TATA-less promoters (5).
FIG. 7.
Mapping p53-responsive regions of the ARF and INK4a promoters. Each of the reporter constructs indicated schematically at the left of each graph was cotransfected with 1.0 μg of wild-type p53 expression vector or empty parental expression vector into the C-33A cell line. Results are presented as the mean activity for triplicate transfections relative to the transfection containing no p53 expression vector, set at 100%. Error bars represent the standard deviations (SD).
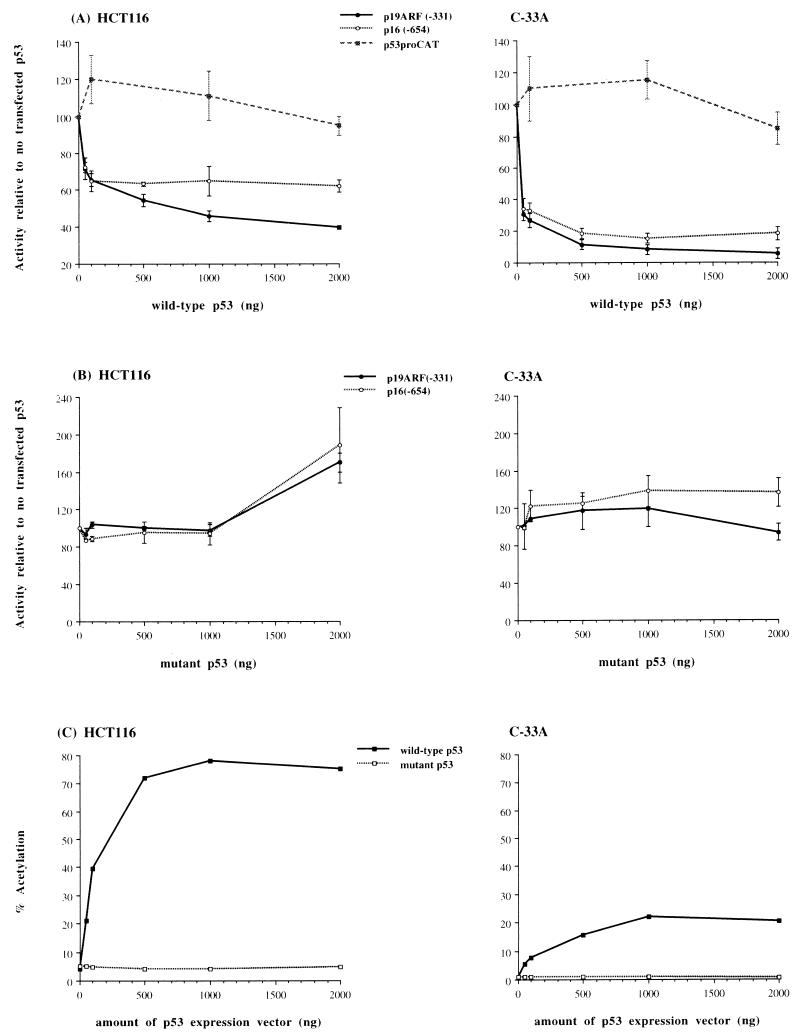
To further study the effects of p53 overexpression on the ARF and INK4a promoters, we cotransfected reporter constructs demonstrating a high degree of repression in the previous experiment (−331 for ARF and −654 for INK4a) with increasing amounts of an expression vector encoding wild-type p53 or mutant p53 (DNA binding domain, Val-143→Ala) into both the HCT116 cell line (endogenous p53 wild type) (47) and the C-33A cell line (endogenous p53 mutant) (22). The total amount of expression vector was held constant by addition of parental expression vector containing no cDNA (Fig. 8). Promoter activities were repressed approximately 1.6-fold for INK4a and 2.5-fold for ARF at the highest levels of wild-type p53 in the HCT116 cell line (Fig. 8A). Transfection into the C-33A line with wild-type p53 resulted in an even greater degree of repression, approximately 5.5-fold for INK4a and 17.2-fold for ARF at the highest levels of p53 (Fig. 8A). Clear differences in the sensitivities of the two promoters to p53 could been seen (1.6- and 3.1-fold for HCT116 and C-33A, respectively), indicating that the repression was unlikely to be a nonspecific effect of p53 overexpression. The degree of repression seen with wild-type p53 overexpression, particularly in C-33A, appears sufficient to account for the previously observed differences in ARF expression between p53-expressing and -nonexpressing cell lines (18, 27, 52). Transfection with increasing amounts of mutant p53 resulted in an approximately twofold increase in the activities of both the INK4a and ARF promoters in the HCT116 cell line, with no clear difference between the two promoters; however, in C-33A cells a very slight increase in INK4a promoter activity was observed with increasing amounts of mutant p53, while the ARF promoter appeared not to be significantly affected (Fig. 8B). The differences in the effect of overexpression of mutant p53 in HCT116 and C-33A cells were most likely due to differences in the status of the endogenous p53 gene. Two additional CAT reporter constructs were used as comparison and control. A CAT reporter vector driven by the human p53 gene promoter (p53proCAT) served as a negative control for the effects of p53 overexpression (Fig. 8A), and a CAT reporter plasmid containing two copies of a consensus p53 binding site driving an E1b TATA minimal promoter [2X(p53)BSCAT] was used as a positive control for a promoter containing p53 binding sites (46) and therefore able to be activated by p53. Cotransfection of increasing amounts of wild-type p53 with the latter CAT reporter plasmid showed that the repressive effects were not due to nonspecific inhibition of all transcription in these cells. Reporter gene activity (expressed as the percent acetylation) was dramatically (approximately 20-fold) increased in both cell lines, while cotransfection with mutant p53 had little effect (Fig. 8C).
FIG. 8.
Effects of p53 on ARF and INK4a promoter-CAT constructs. (A) Dose-response cotransfection of wild-type p53 expression vector and p(−331)19ARF, p(−654)16INK4a, or p53proCAT promoter-CAT constructs into the HCT116 and C-33A cell lines. (B) The identical CAT reporter constructs cotransfected with increasing amounts of a mutant (Val-143→Ala) p53 expression vector. Results in panels A and B are presented as the mean activity for triplicate transfections relative to the transfection containing no p53 expression vector, set at 100%. Error bars represent the standard deviations. The total amount of expression vector was held constant by addition of parental vector containing no cDNA. (C) Representative results after transfection of a CAT reporter construct containing two copies of a consensus p53 binding site with increasing amounts of wild-type or mutant p53 expression vector into the HCT116 and C-33A cell lines. Results are presented as percent acetylation; independent experiments yielded similar results. Note that the endogenous p53 gene is wild type in HCT116 and mutant in C-33A.
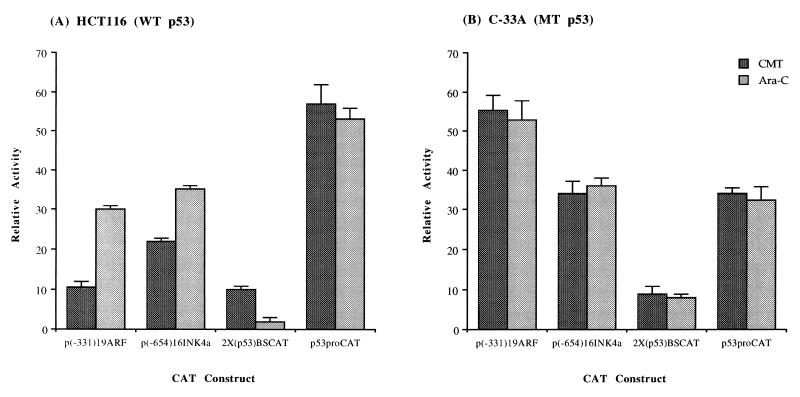
The previously described experiments relied on overexpression of p53, and so we next wished to determine if similar effects on transcription could be observed under physiologic conditions in which p53 is known to be upregulated. We chose to use the DNA-damaging drugs CMT, an inhibitor of topoisomerase I known to induce p53 in a time- and dose-dependent manner (41), and Ara-C, an S-phase-specific antimetabolite that does not directly damage DNA (53) and which has been shown not to induce p53 (28). The latter was used as a comparison with the CMT treatment to control for nonspecific effects on transcription due to drug cytotoxicity and cellular insult. The HCT116 and C-33A cell lines were used again to compare effects on ARF (−331) and INK4a (−654) reporter constructs. Figure 9 shows that CAT activity driven by the ARF and INK4a promoters was less in HCT116 cells treated with CMT than in HCT116 cells treated with Ara-C. The degree of repression was similar to that observed previously with the wild-type p53 expression vector in HCT116 cells (Fig. 8A). No effect on ARF and INK4a reporter activity was observed in C-33A cells treated in a similar manner, also consistent with previous experiments (Fig. 8B), indicating that repression of the ARF and INK4a promoters can occur at physiologic levels of p53. The 2X(p53)BSCAT reporter plasmid served as a positive control to show that the CMT treatment did indeed result in an increase in functional wild-type p53 protein. The activity of this construct was activated nearly sixfold in CMT-treated HCT116 cells relative to Ara-C-treated HCT116 cells but unaffected in similarly treated C-33A cells (Fig. 9). An additional negative control for the effects of drug treatment, employing the p53proCAT vector used previously as a negative control for p53 overexpression (Fig. 8A), was unchanged by drug treatment in both cell lines (Fig. 9).
FIG. 9.
ARF and INK4a promoters are repressed after induction of p53 by DNA-damaging agents. The reporter constructs indicated below each graph were transfected into HCT116 (A) or C-33A (B) cells, after which cells were exposed to the indicated DNA-damaging agent (5 μM CMT or 50 μM Ara-C). After 48 h, cells were harvested for CAT assay and the protein concentration was determined for each. Results are presented as the mean relative activity (percent acetylation normalized to the protein concentration to account for small differences in toxicity) of triplicate transfections. Error bars are the standard deviations from the means. WT, wild-type; MT, mutant.
DISCUSSION
We have cloned and characterized the promoter region of the ARF putative tumor suppressor gene and investigated suppression of this CpG island promoter by DNA methylation. Two colon cancer-derived cell lines had hypermethylation of the ARF promoter, although this occurred infrequently in cell lines and is therefore unlikely to be common in uncultured tumors. We have also provided functional evidence that hypermethylation can suppress ARF promoter activity. Transfection of ARF and INK4a promoter constructs revealed that they were active in all cell lines tested. Activity of the endogenous genes, however, was correlated with promoter methylation status. Transfection studies also revealed that the ARF promoter was highly responsive to E2F1 overexpression, in keeping with previous results (9). Studies with overexpression of both wild-type and mutant forms of p53 indicated that the ARF promoter was repressed by wild-type p53 and, unexpectedly, so was its downstream neighbor INK4a, although there were clear differences in the degree of repression. This repression was also observable under physiologic conditions in which p53 levels are increased due to DNA damage.
Sequence analysis of the ARF promoter region revealed that it possessed many of the characteristics of a housekeeping gene in that it was a CpG island and a TATA-less promoter containing a region homologous to the initiator element which is responsible for correctly positioning the site of transcriptional initiation (5). Deletion analysis revealed that several of the potential Sp1 binding sites, as well as potential E2F binding sites, were important; however, other regions not containing recognizable binding sites were also important. It was interesting that the largest ARF construct was less active than smaller constructs in the HCT116 cell line and that this inhibitory activity appeared to be somewhat cell type dependent. Whether a repressor protein binding site is present in this region or an active silencer element is present is currently under investigation, but such elements have been noted previously in other promoters (77). This region, however, clearly possesses many of the features of a promoter and is capable of driving significant levels of reporter gene activity.
Treatment of the HCT15 cell line with 5-aza-CdR combined with RT-PCR expression data was interesting in several respects. First, ARF activation preceded INK4a activation after drug treatment, and second, both transcripts could clearly be expressed at the same time. Based on previous work and the structure of the INK4a/ARF locus, we had predicted that a transcriptional interference model may be in operation and that transcripts originating from the upstream ARF promoter might reduce INK4a transcriptional initiation. It was in fact observed that INK4a levels were elevated in ARF-null mice (27). Our results do not support a transcriptional interference mechanism such as has been observed in other situations containing two promoters in tandem, where it has been shown that when the upstream promoter was active the downstream one was not, and vice versa (7, 49, 50, 74, 75). It is not clear how the INK4a/ARF situation differs from these examples except that the two promoters are very widely spaced (estimated at 20 kb in humans [36, 68]). We cannot rule out the possibility of allele-specific transcription of each gene, as has been observed for imprinted genes (reviewed in reference 10). We have no evidence for differential methylation of this region and have not yet detected a polymorphism in the transcribed regions of the ARF and INK4a genes in the HCT15 line to allow for investigation of this possibility.
Methylation-mediated silencing of INK4a has been reported in numerous situations (14, 21, 39, 59), and so it is interesting that its neighbor ARF appeared to only rarely be subject to such regulation. It is believed that CpG islands may be protected from de novo methylation by binding of the Sp1 transcription factor (35). A comparison of the ARF and INK4a CpG islands revealed that the CpG density of the ARF CpG island (number of CpG sites/bp = 0.085) was significantly higher than the INK4a CpG island (number of CpG sites/bp = 0.048) over a similarly sized region (bracketed regions in Fig. 2A). A more dense, or larger, CpG island may confer a greater protective effect against de novo methylation. The ARF promoter contains seven potential Sp1 sites whereas the INK4a promoter contains four potential Sp1 sites upstream of the transcription initiation sites, and this may also help explain the differences in the propensity of the two promoters to become de novo methylated. An additional influence on de novo methylation of the INK4a CpG island may arise from transcription through the INK4a promoter region from the upstream ARF gene. The INK4a/ARF locus provides a naturally occurring system in which to study such possible effects.
Studies of the effect of E2F expression on the ARF promoter indicated a high level of responsiveness to E2F1, an effect that was unlikely to be nonspecific because the stimulation correlated with deletion of potential E2F binding sites and had no effect on an INK4a promoter construct. This stimulation at the transcriptional level clearly correlated with the increased levels of ARF mRNA observed previously with E2F overexpression (9). The significance of this finding implies that increased levels of cellular E2F, resulting from pRb phosphorylation or mutations in the pRb pathway, would increase ARF levels, which would in turn act to increase the levels of functional p53 (48). We speculate that in normal cells, this might account for the observed increase in p53 levels as cells enter the cell cycle from quiescence (54). In a malignant or premalignant cell resulting from mutation in the pRb pathway, this transient increase in the level of p53 may be a mechanism to induce apoptosis since pRb dysfunction, would, unlike DNA damage, be irreparable. Such potential relationships will be the subject of future study.
Investigation of the role of p53 in regulating ARF expression at the transcriptional level was motivated by the findings in previous work that ARF levels were elevated approximately 5- to 10-fold in p53-deficient cell lines at the protein level (27, 52). Our results from the wild-type p53 dose-response transfections show that the ARF promoter is repressed 3- to 17-fold at the highest levels of wild-type p53 and that the degree of repression correlated with the status of the endogenous p53 gene. We attribute the muted effect in HCT116 cells compared to C-33A cells to the presence of wild-type endogenous p53 in the former, which would be expected to reduce the initial levels of reporter gene activity in this line. Further support for the role of p53 came from dose-response studies utilizing a mutant form of p53. Cotransfection of mutant p53 with the ARF and INK4a reporter constructs into HCT116 resulted in an increase in activity, presumably due to heterotetramerization with the endogenous wild-type p53 interfering with its repressive abilities, while in C-33A, cotransfection of the mutant form of p53 had little to no effect.
Is the repression observed with p53 overexpression a nonspecific effect, and is it physiologically relevant? Overexpression of p53 has been shown to repress a variety of other promoters (for the c-fos, c-jun, hsc70 [13], PCNA [38], and interleukin-6 [58] genes), while other promoters are unaffected (the human p53 gene promoter in our studies; the β-actin, c-Ha-ras, epidermal growth factor [6], and β2-microglobulin [38] gene promoters). We feel that the repression of the ARF and INK4a promoters is meaningful for several reasons. First, the degree of repression seen with the ARF promoter was reproducibly greater than that observed with the INK4a promoter even though the two promoters had similar activities in the absence of added p53 and were within the same plasmid backbone. Second, the effects were reproducible in two different cell lines and correlated with the status of the endogenous p53 gene in each line. Third, the effect of mutant p53 was selective in that promoter activity actually increased significantly in the cell line in which the endogenous p53 gene was wild type, while little to no effect of mutant p53 overexpression was observed in the cell line containing a mutant endogenous p53 gene. Lastly, a similar degree of transcriptional repression of the ARF and INK4a promoters was observed after induction of physiologic levels of p53 by DNA-damaging agents in a cell line with wild-type p53 function but not in a cell line with a mutant p53 gene compared to the p53 overexpression studies.
We were initially surprised by the effects of p53 on the INK4a reporter construct. It had been reported that the INK4a promoter was repressed three- to fivefold by pRb overexpression and that p53 status had no effect in a cell line containing a temperature-sensitive simian virus 40 T-Ag. T-Ag binds to and inactivates both pRb and p53 (reviewed in reference 40), and this study concluded that the repressive effect observed when the T-Ag was inactivated after shift to the nonpermissive temperature was due solely to the pRb component. The effect of p53 was not directly tested (18), and so reinterpretation of both this study and others is consistent with the notion that p53 overexpression may have a repressive effect on INK4a levels. For example, INK4a levels were elevated after transfection of the E6 oncoprotein (which specifically inactivates p53 [reviewed in reference 40]) into a cell line with otherwise wild-type pRb and p53 function (70). Thus, we propose that both pRb and p53 may regulate INK4a and ARF. The regulation is, however, differential, with p53 being the dominant component for ARF and pRb being the dominant component for INK4a. The effect of p53 on INK4a may in fact be mediated through pRb as has been previously proposed (70).
The nearly 20-fold decrease in ARF-driven reporter gene activity in the C-33A cell line when wild-type p53 was overexpressed may well be sufficient to account for the corresponding increase in ARF levels in p53-deficient cell lines that has been reported previously (27, 52); however, two additional factors need to be considered: (i) the stability of the RNA and (ii) the stability of the protein. It has been shown that the INK4a RNA is very stable (18), although similar information is not available for the ARF RNA. INK4a protein has a long half-life of at least 3 h (44), while ARF has a significantly shorter half-life of approximately 90 min (78). This has important implications for our results because in order to rapidly change the level of a protein by controlling the rate of transcription, a short-lived RNA or protein is essential. The short half-life of ARF indicates that control at the transcriptional level would allow for rapid changes in protein levels when p53 levels are elevated under physiological situations and why less of an effect of p53 overexpression is seen on the INK4a protein level with its long half-life. We cannot rule out the possibility that there are other levels of regulation between ARF and p53; however, it is clear from these studies that the ARF promoter can be regulated by p53 at the transcriptional level.
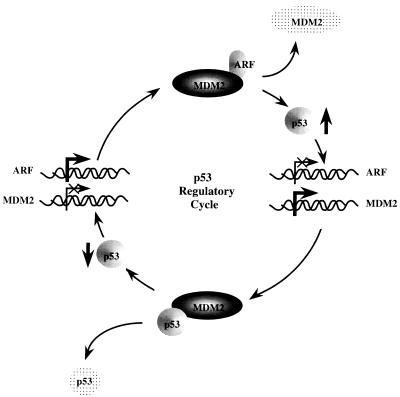
Recent work has shown that the role of ARF in p53-dependent cell cycle checkpoint control is mediated by its binding to the MDM2 proto-oncogene accompanied by an increase in functional p53 levels and an enhancement of the capability of p53 to transactivate promoters containing p53 binding sites (48). Based on this study and the work presented here, we propose the existence of an autoregulatory feedback loop which allows for tight controls over the levels of p53 in normal cells (Fig. 10). In this model, ARF expression and binding to MDM2 results in an increase in functional p53 levels, allowing for either cell cycle arrest or induction of apoptosis. The increased levels of p53 will then feed back on the ARF promoter, resulting in a down-regulation of ARF transcription and concomitant decreases in ARF protein. This will then free MDM2 to bind to p53 and reduce its levels by targeting it for degradation. Additional elements of this pathway, such as the factors regulating MDM2 and ARF levels, and the role of E2F and pRb, will no doubt be the focus of much future study given that the INK4a/ARF locus now stands firmly at the crossroads of two of the most important cell cycle regulatory pathways.
FIG. 10.
Proposed regulatory cycle controlling cellular p53 levels mediated by p53, MDM2, and ARF. Interaction between MDM2 and ARF results in increased degradation of MDM2 (78) (shown by light stipple) and an increase in functional p53 (upward arrow) (48). This then represses ARF (×) and activates MDM2 (large arrow) transcription (1, 76). Elevated levels of MDM2 and decreased levels of ARF then promote degradation of p53 (shown by light stipple) and continue the cycle as shown. E2F levels may be one of the outside influences on the cycle.
ACKNOWLEDGMENTS
This work was supported by grants R35 CA 49758-09 and T32 CA 09320-15 from the National Institutes of Health.
We thank Felicidad Gonzales for technical assistance and Richard Ambinder for providing cell lines and plasmids.
REFERENCES
- 1.Barak Y, Juven T, Haffner R, Oren M. Mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bender C M, Pao M M, Jones P A. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. [PubMed] [Google Scholar]
- 3.Bird A. CpG-Rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 4.Caldas C, Hahn S A, daCosta L T, Redston M S, Schutte M, Seymour A B, Weinstein C L, Hruban R H, Yeo C J, Kern S E. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 1994;8:27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 5.Carcamo J, Buckbinder L, Reinberg D. The initiator directs the assembly of a transcription factor IID-dependent transcription complex. Proc Natl Acad Sci USA. 1991;88:8052–8056. doi: 10.1073/pnas.88.18.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin K-V, Ueda K, Pastan I, Gottesman M M. Modulation of activity of the promoter of the human MDR1 gene by ras and p53. Nature. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 7.Corbin V, Maniatis T. Role of transcriptional interference in the Drosophila melanogaster Adh promoter switch. Nature. 1989;337:279–282. doi: 10.1038/337279a0. [DOI] [PubMed] [Google Scholar]
- 8.Crook T, Marston N J, Sara E A, Vousden K H. Transcriptional activation by p53 correlates with suppression of growth but not transformation. Cell. 1994;79:817–827. doi: 10.1016/0092-8674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 9.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994;4:265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald M G, Harkin D P, Silva-Arrieta S, MacDonald D J, Lucchina L C, Unsal H, O’Neill E, Koh J, Finkelstein D M, Isselbacher K J, Sober A J, Haber D A. Prevalence of germ-line mutations in p16, p19ARF, and CDK4 in familial melanoma: analysis of a clinic-based population. Proc Natl Acad Sci USA. 1996;93:8541–8545. doi: 10.1073/pnas.93.16.8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 13.Ginsberg D, Mechta F, Yaniv M, Oren M. Wild-type p53 can down-regulate the activity of various promoters. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Zulueta M, Bender C M, Yang A S, Nguyen T-D, Beart R W, VanTornout J M, Jones P A. Methylation of the CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 15.Gonzalgo M L, Hayashida T, Bender C M, Pao M M, Tsai Y C, Gonzales F A, Nguyen H D, Nguyen T-D, Jones P A. The role of DNA methylation in expression of the p19/p16 locus in human bladder cancer cell lines. Cancer Res. 1998;58:1245–1252. [PubMed] [Google Scholar]
- 16.Haber D A. Splicing into senescence: the curious case of p16 and p19ARF. Cell. 1997;91:555–558. doi: 10.1016/s0092-8674(00)80441-9. [DOI] [PubMed] [Google Scholar]
- 17.Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- 18.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 20.Herman J G, Jen J, Merlo A, Baylin S B. Hypermethylation associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;54:722–727. [PubMed] [Google Scholar]
- 21.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 22.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 23.Hussussian C J, Struewing J P, Goldstein A M, Higgins P A T, Ally D S, Sheahan M D, Clark W H, Tucker M A, Dracopoli N C. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 24.Jones N C, Rigby P W, Ziff E B. Trans-acting protein factors and the regulation of eukaryotic transcription: lessons from studies on DNA tumor viruses. Genes Dev. 1988;2:267–281. doi: 10.1101/gad.2.3.267. [DOI] [PubMed] [Google Scholar]
- 25.Jones P A, Wolkowicz M J, Rideout III W M, Gonzales F A, Marziasz C M, Coetzee G, Tapscott S J. De novo methylation of the MyoD1 CpG island during the establishment of immortal cell lines. Proc Natl Acad Sci USA. 1990;87:6117–6121. doi: 10.1073/pnas.87.16.6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamb A, Shattuck-Eidens D, Eeles R, Liu Q, Gruis N A, Ding W, Hussey C, Tran T, Miki Y, Weaver-Feldhaus J, McClure M, Aitken J F, Anderson D E, Bergman W, Frants R, Goldgar D E, Green A, MacLennan R, Martin N G, Meyer L J, Youl P, Zone J J, Skolnick M H, Cannon-Albright L A. Analysis of the p16 gene (CDKN2) as a candidate for the chromosome 9p melanoma susceptibility locus. Nat Genet. 1994;8:22–26. doi: 10.1038/ng0994-22. [DOI] [PubMed] [Google Scholar]
- 27.Kamijo T, Zindy F, Roussel M F, Quelle D Q, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 28.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 29.Ko L J, Prives C. p53: Puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 30.Kubbutat M H G, Jones S N, Vousden K H. Regulation of p53 stability by mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 31.Larsen C-J. p16INK4a: a gene with a dual capacity to encode unrelated proteins that inhibit cell cycle progression. Oncogene. 1996;12:2041–2044. [PubMed] [Google Scholar]
- 32.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 33.Liggett W H, Sewell D A, Rocco J, Ahrendt S A, Koch W, Sidransky D. p16 and p16β are potent growth suppressors of head and neck squamous carcinoma cells in vitro. Cancer Res. 1996;56:4119–4123. [PubMed] [Google Scholar]
- 34.Mack D H, Vartikar J, Pipas J M, Laimins L A. Specific repression of TATA-mediated but not initiator-mediated transcription repression by wild-type p53. Nature. 1993;363:281–283. doi: 10.1038/363281a0. [DOI] [PubMed] [Google Scholar]
- 35.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse Aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 36.Mao L, Merlo A, Bedi G, Shapiro G I, Edwards C D, Rollins B J, Sidransky D. A novel p16INK4A transcript. Cancer Res. 1995;55:2995–2997. [PubMed] [Google Scholar]
- 37.Means A L, Farnham P J. Transcriptional initiation from the dihydrofolate reductase promoter is positioned by HIP-1 binding at the initiation site. Mol Cell Biol. 1990;10:653–661. doi: 10.1128/mcb.10.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer W E, Shields M T, Lin D, Appella E, Ullrich S J. Growth suppression induced by wild-type p53 protein is accompanied by selective down-regulation of proliferating-cell nuclear antigen expression. Proc Natl Acad Sci USA. 1991;88:1958–1962. doi: 10.1073/pnas.88.5.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumor suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 40.Moran E. DNA tumor virus transforming proteins and the cell cycle. Curr Opin Genet Dev. 1993;3:63–70. doi: 10.1016/s0959-437x(05)80342-9. [DOI] [PubMed] [Google Scholar]
- 41.Nelson W G, Kastan M B. DNA strand breaks: the DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 43.Oliner J D, Pietenpol J A, Thiagalingam S, Gyuris J, Kinzler K W, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumor suppressor p53. Nature. 1993;362:857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 44.Parry D, Bates S, Mann D J, Peters G. Lack of cyclin D-cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumor suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry M E, Levine A J. Tumor-suppressor p53 and the cell cycle. Curr Opin Genet Dev. 1993;3:50–54. doi: 10.1016/s0959-437x(05)80340-5. [DOI] [PubMed] [Google Scholar]
- 46.Pietenpol J A, Tokino T, Thiagalingam S, El-Deiry W, Kinzler K W, Vogelstein B. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc Natl Acad Sci USA. 1994;91:1998–2002. doi: 10.1073/pnas.91.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polyak K, Waldman T, He T-C, Kinzler K W, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 48.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R A. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 49.Proudfoot N J. Transcriptional interference and termination between duplicated α-globin gene constructs suggests a novel mechanism for gene regulation. Nature. 1986;322:562–565. doi: 10.1038/322562a0. [DOI] [PubMed] [Google Scholar]
- 50.Puglielli M T, Desai N, Speck S H. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp) J Virol. 1997;71:120–128. doi: 10.1128/jvi.71.1.120-128.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quelle D E, Cheng M, Ashmun R A, Sherr C J. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not the alternative reading frame protein p19ARF. Proc Natl Acad Sci USA. 1997;94:669–673. doi: 10.1073/pnas.94.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 53.Rashbaum S A, Cozzarelli N R. Mechanism of DNA synthesis inhibition by arabinosyl cytosine and arabinosyl adenine. Nature. 1976;264:679. doi: 10.1038/264679a0. [DOI] [PubMed] [Google Scholar]
- 54.Reich N C, Levine A J. Growth regulation of a cellular tumor antigen, p53, in nontransformed cells. Nature. 1984;308:199–201. doi: 10.1038/308199a0. [DOI] [PubMed] [Google Scholar]
- 55.Robertson K D, Ambinder R F. Mapping promoter regions that are hypersensitive to methylation-mediated inhibition of transcription: application of the methylation cassette assay to the Epstein-Barr virus major latency promoter. J Virol. 1997;71:6445–6454. doi: 10.1128/jvi.71.9.6445-6454.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robertson K D, Hayward S D, Ling P D, Samid D, Ambinder R F. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: evidence that demethylation at a single CpG site is crucial. Mol Cell Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rozenblum E, Schutte M, Goggins M, Hahn S A, Panzer S, Zahurak M, Goodman S N, Sohn T A, Hruban R H, Yeo C J, Kern S E. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57:1731–1734. [PubMed] [Google Scholar]
- 58.Santhanam U, Ray A, Sehgal P B. Repression of the interleukin 6 gene promoter by p53 and the retinoblastoma susceptibility gene product. Proc Natl Acad Sci USA. 1991;88:7605–7609. doi: 10.1073/pnas.88.17.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schutte M, Hruban R H, Geradts J, Maynard R, Hilgers W, Rabindran S K, Moskaluk C A, Hahn S A, Schwarte-Waldhoff I, Schmiegel W, Baylin S B, Kern S E, Herman J G. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 60.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 61.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 62.Seto E, Usheva A, Zambetti G P, Momand J, Horikoshi N, Weinmann R, Levine A J, Shenk T. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc Natl Acad Sci USA. 1992;89:12028–12032. doi: 10.1073/pnas.89.24.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sewell D A, Liggett W H, Jr, Yarbrough W G, Sidransky D. Human p16β encodes a novel 14 kD protein and suppresses growth in vitro independently from pRb. Proc Am Assoc Cancer Res. 1997;38:80. [Google Scholar]
- 64.Sherr C J. Cancer cell cycles. Science. 1996;276:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 65.Shores C, Liu E, Yarbrough W. Expression of p16β in human cells. Proc Am Assoc Cancer Res. 1997;38:277. [Google Scholar]
- 66.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smale S T, Baltimore D. The “initiator” as a transcriptional control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 68.Stone S, Jiang P, Dayananth P, Tavtigian S V, Katcher H, Parry D, Peters G, Kamb A. Complex structure and regulation of the p16 (MTS1) locus. Cancer Res. 1995;55:2988–2994. [PubMed] [Google Scholar]
- 69.Subler M A, Martin D W, Deb S. Inhibition of viral and cellular promoters by human wild-type p53. J Virol. 1992;66:4757–4762. doi: 10.1128/jvi.66.8.4757-4762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tam S W, Shay J W, Pagano M. Differential expression and cell cycle regulation of the cyclin-dependent kinase 4 inhibitor p16INK4. Cancer Res. 1994;54:5816–5820. [PubMed] [Google Scholar]
- 71.Tanaka H, Shimada Y, Imamura M, Shabagaki I, Ishizaki K. Multiple types of aberrations in the p16(INK4a) and the p15(INK4b) genes in 30 esophageal squamous-cell-carcinoma cell lines. Int J Cancer. 1997;70:437–442. doi: 10.1002/(sici)1097-0215(19970207)70:4<437::aid-ijc11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 72.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tuck S P, Crawford L. Characterization of the human p53 gene promoter. Mol Cell Biol. 1989;9:2163–2172. doi: 10.1128/mcb.9.5.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vales L D, Darnell J E. Promoter occlusion prevents transcription of Adenovirus polypeptide IX mRNA until after DNA replication. Genes Dev. 1989;3:49–59. doi: 10.1101/gad.3.1.49. [DOI] [PubMed] [Google Scholar]
- 75.Wu J, Grindlay G J, Bushel P, Mendelsohn L, Allan M. Negative regulation of the human ɛ-globin gene by transcriptional interference: role of an Alu repetitive element. Mol Cell Biol. 1990;10:1209–1216. doi: 10.1128/mcb.10.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu X, Bayle J H, Olsen D, Levine A J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 77.Xu Z-P, Saunders G F. Transcriptional regulation of the human PAX6 gene promoter. J Biol Chem. 1997;272:3430–3436. doi: 10.1074/jbc.272.6.3430. [DOI] [PubMed] [Google Scholar]
- 78.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]