Abstract
Salmonella and E. coli synthesize, import, and export cadaverine, putrescine, and spermidine to maintain physiological levels and provide pH homeostasis. Both low and high intracellular levels of polyamines confer pleiotropic phenotypes or lethality. Here, we demonstrate that the previously uncharacterized inner membrane protein PaeA (YtfL) is required for reducing cytoplasmic cadaverine and putrescine concentrations. We identified paeA as a gene involved in stationary phase survival when cells were initially grown in acidic medium, in which they produce cadaverine. The paeA mutant is also sensitive to putrescine, but not to spermidine or spermine. Sensitivity to external cadaverine in stationary phase is only observed at pH >8, suggesting that the polyamines need to be deprotonated to passively diffuse into the cell cytoplasm. In the absence of PaeA, intracellular polyamine levels increase and the cells lose viability. Degradation or modification of the polyamines is not relevant. Ectopic expression of the known cadaverine exporter, CadB, in stationary phase partially suppresses the paeA phenotype, and overexpression of PaeA in exponential phase partially complements a cadB mutant grown in acidic medium. These data support the hypothesis that PaeA is a cadaverine/putrescine exporter, reducing potentially toxic levels under certain stress conditions.
Keywords: polyamines, cadaverine, putrescine, Salmonella, stationary phase
Plain Language Summary
Polyamines are critical for survival in most life forms. We have identified a putative polyamine exporter that prevents toxic buildup of certain polyamines in the bacterium Salmonella.
Graphical Abstract
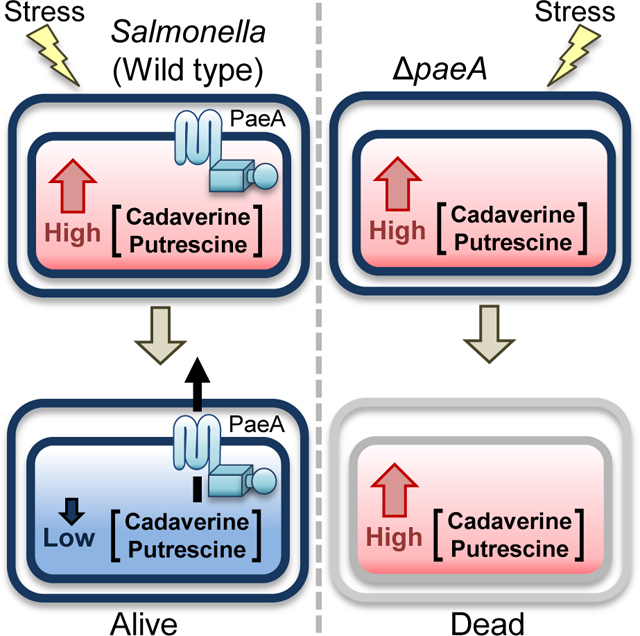
Certain stresses or conditions induce production or import of putrescine and/or cadaverine. Once stress is relieved or conditions change, the polyamines need to be exported via PaeA. Failure to export leads to high polyamine concentrations that are lethal under certain conditions.
1. INTRODUCTION
Salmonella enterica is a major food-borne pathogen causing an estimated 1.2 million infections each year in the United States (Scallan et al., 2011). Ingested Salmonella colonize the small intestinal epithelium and induce both inflammatory diarrhea and bacterial invasion, leading to systemic spread (Darwin and Miller, 1999; Tsolis et al., 1999; Wallis and Galyov, 2000; Zhou and Galan, 2001). The most serious Salmonella disease results from this extraintestinal infection (Pegues and Miller, 2010; Feasey et al., 2012), the hallmark of which is the ability of Salmonella to survive in macrophages (Burton et al., 2014; Fenlon and Slauch, 2014). Macrophages engulf bacteria into phagosomes, which subsequently acidify and mature into phagolysosomes upon delivery or production of various antimicrobial effectors, including superoxide and nitric oxide (Slauch, 2011). We do not have a complete understanding of the additional stress tolerance systems required for Salmonella to survive and replicate within macrophages.
Polyamines are found in all three domains of life. They interact with negatively charged molecules, such as nucleic acids and phospholipids. Indeed, it is estimated that over half of the polyamines in the cell exist in polyamine–RNA complexes (Miyamoto et al., 1993). Physiologically, polyamines have been implicated in numerous cellular processes in bacteria, such as growth under anaerobic conditions, siderophore synthesis, peptidoglycan synthesis, biofilm formation, swarming motility, oxidative stress resistance, and acid stress resistance (Park et al., 1996; Chattopadhyay et al., 2003; Chattopadhyay et al., 2009; Kurihara et al., 2009a; Michael, 2018). Polyamines also affect ribosome assembly, translation initiation, and translation fidelity (Igarashi and Kashiwagi, 2015; Igarashi and Kashiwagi, 2018). Indeed, it has been proposed that alterations in polyamine levels affect translation of a subset of proteins critical to overall cell physiology and stress response (Igarashi and Kashiwagi, 2015; Igarashi and Kashiwagi, 2018; Sakamoto et al., 2020).
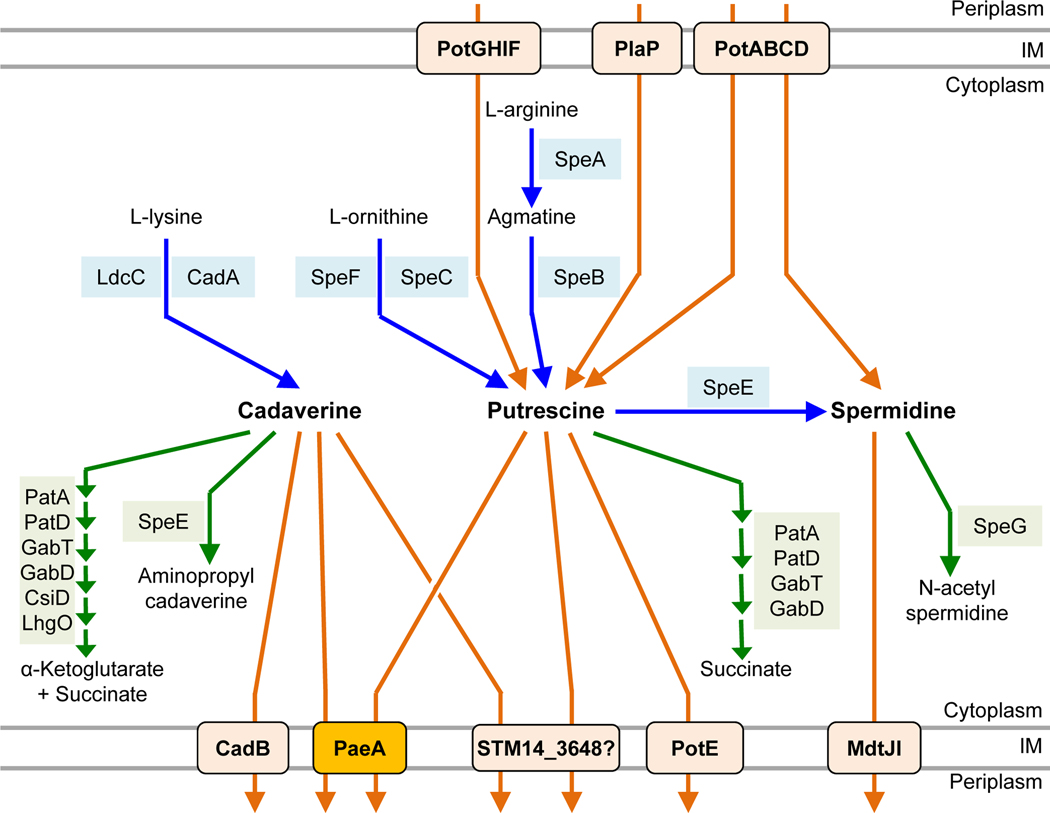
Spermidine and putrescine are broadly found in bacteria (Michael, 2018). Salmonella enterica and E. coli are among the bacteria that also produce cadaverine (Gale and Epps, 1944; Park et al., 1996; Chattopadhyay et al., 2009). Salmonella has systems for uptake, export, synthesis, and degradation of polyamines (Fig. 1). Salmonella synthesizes cadaverine from L-lysine by the constitutively expressed lysine decarboxylase LdcC (Yamamoto et al., 1997), or the acid-inducible CadA (Gale and Epps, 1944; Park et al., 1996)(see below). Putrescine is synthesized from L-arginine by SpeA and SpeB or from L-ornithine by SpeC or the inducible SpeF (Tabor and Tabor, 1985; Michael, 2016). Spermidine is synthesized from decarboxylated S-adenosyl-L-methionine and putrescine by SpeE (Bowman et al., 1973; Tabor and Tabor, 1987). Salmonella cannot synthesize spermine (Tabor and Tabor, 1985). Cadaverine and putrescine can be degraded to succinate via the lysine degradation pathway (Schneider and Reitzer, 2012; Knorr et al., 2018). In contrast to E.coli, Salmonella enterica does not have the second putrescine degradation pathway mediated by PuuABCD (Schneider and Reitzer, 2012). Spermidine is converted to N-acetylspermidine by SpeG (Fukuchi et al., 1995), but no further mechanism of degradation is known.
FIGURE 1. Known pathways of polyamine metabolism in Salmonella enterica.
Blue lines represent synthetic pathways, green lines represent degradation or modification pathways, and orange lines represent transport pathways.
Cadaverine is exported by the lysine:cadaverine antiporter CadB (Soksawatmaekhin et al., 2004), putrescine is excreted by the ornithine:putrescine antiporter PotE (Kashiwagi et al., 1997), and spermidine is excreted by MdtJI (Higashi et al., 2008). More recently, Hassan et al. identified AceI in Acinetobacter baumannii as a cadaverine/putrescine exporter (Hassan et al., 2019). The putative homolog in Salmonella (STM14_3648) has not been characterized. The ABC transporter PotABCD preferentially imports spermidine, but can also transport putrescine (Igarashi et al., 2001), while the ABC transporter PotFGHI is apparently specific for putrescine (Igarashi et al., 2001). PlaP is a putrescine:proton symporter (Kurihara et al., 2011). Salmonella does not encode the PuuP putrescine importer identified in E. coli (Kurihara et al., 2009b).
Under acidic conditions in vitro, the lysine-dependent acid stress resistance system CadAB is transcriptionally induced. CadA decarboxylates lysine to produce cadaverine, which is secreted by CadB (Park et al., 1996). During this process, one proton is consumed and one carbon dioxide is produced, and the protonated diamine is secreted, helping to neutralize the cytoplasmic pH (Park et al., 1996). Inside the macrophage phagosome, Salmonella is exposed to acid stress, pH 5 to 5.5. However, the cadBA genes remain repressed and the cytoplasm becomes acidic (Chakraborty et al., 2015). Keeping the cytoplasm acidic is important for enhancing prolonged up-regulation of the ssrAB two component system, which regulates the SPI2 type III secretion system and its effector genes (Chakraborty et al., 2015), required for survival in the phagosome (Jennings et al., 2017). It is unclear if there are the other reasons for Salmonella to repress the genes responsible for polyamine production in the phagosome, and whether this further impacts Salmonella virulence.
We are interested in metabolic pathways important for survival of Salmonella in the phagosome. We focused on the genomic region that encodes ytfL. Previous studies in E.coli demonstrated that ytfK and msrA, encoded on either side of ytfL are important for nitrate and nitrite-related stress tolerances, respectively (St John et al., 2001; Iwadate and Kato, 2017). The addition of nitrite is known to enhance acid stress (Muhlig et al., 2014). We deleted these genes and investigated the survival of mutants in acidic medium with nitrite. Although none of the mutants showed defects in growth, we found that ytfL is responsible for stationary phase survival when cells are grown in acidic medium with nitrite. Subsequent analyses showed that, in the ytfL mutant, the polyamines cadaverine or putrescine can build up to high concentration in the cytoplasm, causing loss of viability in stationary phase. The simplest explanation is that YtfL acts as a polyamine exporter, although we cannot rule out some indirect effect on cadaverine/putrescine concentrations. We have renamed the ytfL gene paeA, for Polyamine Export.
2. RESULTS
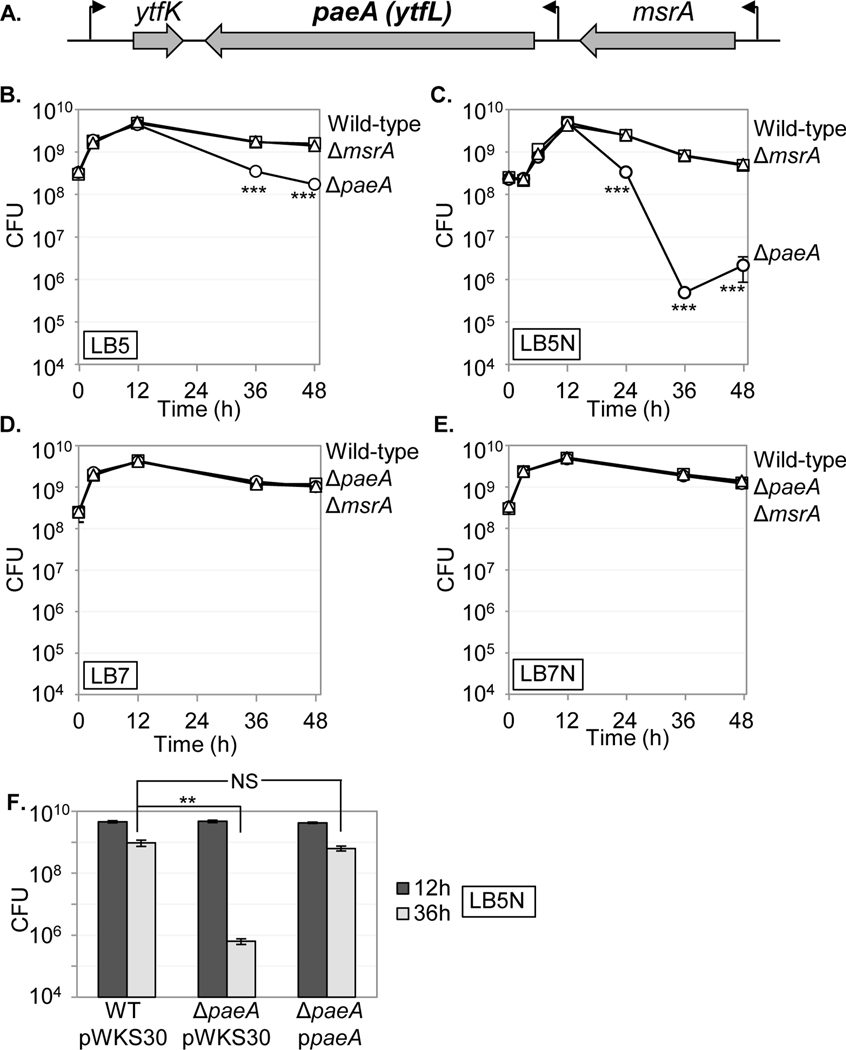
2.1. A mutant loses viability in stationary phase when grown in pH 5 LB medium with nitrite.
Nitric oxide is produced by macrophages (MacMicking et al., 1997) and known to be important in controlling Salmonella infection (Vazquez-Torres et al., 2000; Chakravortty et al., 2002). Both ytfK and msrA have been implicated in resistance to reactive nitrogen species (St John et al., 2001; Iwadate and Kato, 2017). The paeA (ytfL) gene is located between ytfK and msrA (Fig 2A). Transcriptomic data suggest that paeA is transcribed from both its own promoter and via read-through from the msrA promoter, with an increase in transcription when Salmonella is replicating in macrophages (Kroger et al., 2013; Srikumar et al., 2015). Under acidic conditions, nitrite is converted to nitric oxide in vitro (St John et al., 2001). To see the effect of the deletions of paeA or mrsA on the sensitivity to nitrite under acidic conditions, we grew wild-type, a ::Cm strain, or an in-frame strain in unbuffered pH 5 LB medium with nitrite (LB5N) or without nitrite (LB5) and measured their growth and survival. As shown in Fig. 2B and C, all strains showed an increased lag when grown LB5N, but there was no overall difference in growth to stationary phase among the strains. After the entry into stationary phase, the mutant lost viability over 24 hours when cultured in LB5 medium. The effect was greater when cells were grown in LB5N. There was no effect on viability in stationary phase when the paeA mutant was grown in pH 7 LB medium (LB7), with or without nitrite (Fig. 2D and E). Introduction of a low-copy plasmid encoding PaeA (pWKS30-paeA) into the paeA mutant complemented the survival defect in LB5N (Fig. 2F). These observations revealed that paeA, which encodes an uncharacterized putative inner membrane protein, is important for the survival in stationary phase when cells are grown in pH 5 LB medium, especially in the presence of nitrite.
FIGURE 2. The mutant loses viability in stationary phase when grown in pH 5 LB medium.
(A) The paeA locus (to scale). (B-E) Cells were grown in pH 7 LB (LB7), pH 5 LB (LB5), or each medium with 2 mM nitrite (LB7N, LB5N). After 12, 24, 36, and 48 hours, appropriate dilutions were plated on LB glucose plates to determine CFUs. Symbols: squares, wild-type; circles, ; triangles, . (F) Stationary phase survival of the indicated strains containing the vector or pWKS30-paeA plasmid grown in LB5N. Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare wild-type strain with strain or as indicated in panel F. (NS Not significant, *p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: 14028, JS2430, JS2431, JS2452, JS2453, and JS2454.
Although the loss of viability in stationary phase in the paeA mutant is dependent on initial growth at pH 5, this phenotype is enhanced by nitrite, suggesting that nitric oxide might be involved in the sensitivity. To test this hypothesis, we determined if nitric oxide scavengers or methionine sulfoxide reductases, important for tolerance to nitric oxide stress (St John et al., 2001; Vine and Cole, 2011), had any effect on the phenotype. As shown in Fig. S1A and B, none of the single or quintuple deletion mutants of nitric oxide scavenger genes, hmpA, norW, hcp, nrfA, and nirBD, nor the single or quadruple deletion mutants of methionine sulfoxide reductase genes, msrA, msrB, msrC, and msrPQ, showed any loss of viability in stationary phase under these conditions, nor did the mutations affect the phenotype caused by deletion of paeA in these backgrounds. The strains lacking the nitric oxide dioxygenase HmpA, the primary nitric oxide scavenger in the presence of oxygen (Gardner and Gardner, 2002), grew to a lower CFU at 12h after incubation in LB5N medium, and thus the CFU ratio (CFU at 36h after incubation / CFU at 12h after incubation) was higher than that of wild-type, but the paeA derivative still lost viability in stationary phase. These results suggested that nitric oxide scavengers and methionine sulfoxide reductases are not responsible for the survival in stationary phase when strains are grown in LB5N. Thus, although addition of nitrite to the medium enhances the paeA phenotype in stationary phase (Fig. 2B and C), it appears to be an indirect effect and not dependent on nitric oxide per se.
2.2. The mutant is sensitive to supernatant of stationary phase cultures grown in pH5 LB medium with nitrite.
We next addressed the question of why the loss of viability at stationary phase in the strain is observed only when the cells are grown in LB5 but not LB7. First, we asked if the initial difference in pH of the media alters the pH in stationary phase, and thus survival of the strain. As shown in Fig. S2A-D, all of the unbuffered LB cultures reached a pH of ~8.8 independent of the strain, the starting pH, or the presence of nitrite. This is as expected in LB, given the fact that peptides are the primary carbon source, resulting in the release of ammonia (Sezonov et al., 2007). Thus, the effect of the initial medium pH on the survival of the strain in stationary phase is not due to any difference in the pH of the stationary phase medium.
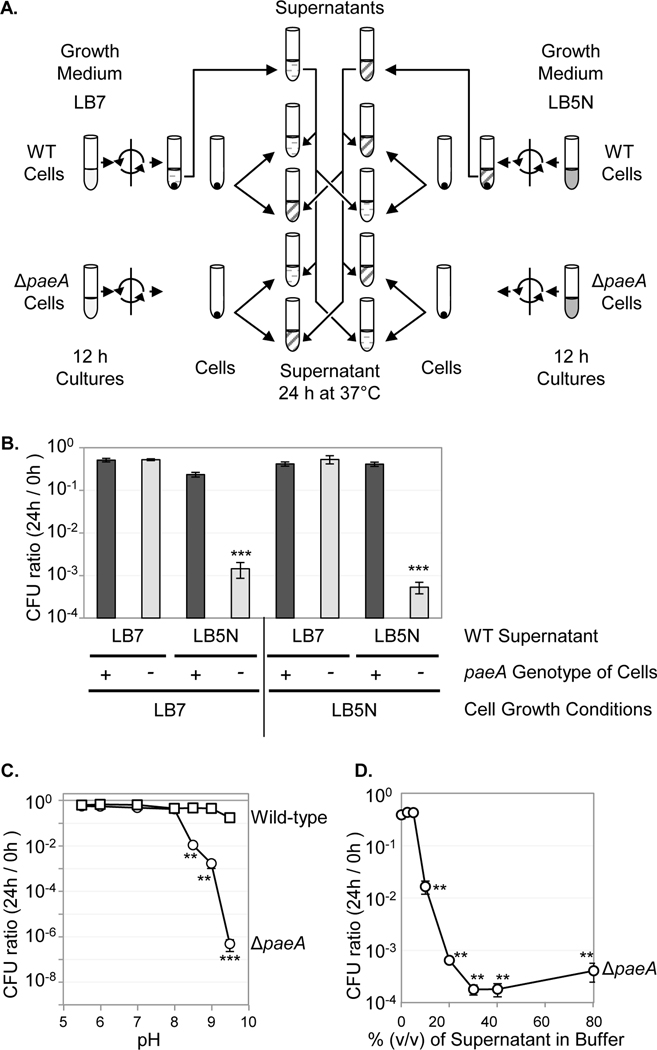
Second, we asked whether it was the growth of the strain in LB5N or the resulting medium that conferred loss of viability in stationary phase. To do so, we switched the cells and media from cultures that were grown in LB7 or LB5N (Fig. 3A). As shown in Fig. 3B, the cells lost viability when suspended in conditioned medium from cells grown in LB5N, independent of whether the cells were initially grown in LB5N or LB7. These results indicated that the strain is sensitive to some compound in the culture medium produced when cells are grown in LB5N.
FIGURE 3. The mutant is sensitive to supernatant of stationary phase cultures grown in pH 5 LB medium with nitrite.
(A) The wild type and strains were grown for 12 h in either LB7 or LB5N. The cells were removed by centrifugation and the supernatant from the wild-type cultures was filtered. The cells were washed and suspended in the indicated supernatant (time 0) and incubated at 37℃ for 24 hours. CFUs were determined at 0 and 24 h with the results shown in panel B. (C) Cells are the wild type or strain grown 12 h in LB7. Supernatant is from a wild type LB5N culture that was mixed 1:1 with saline containing 100 mM MES buffer (pH 5.5 or 6.0), MOPS buffer (pH 7.0, 8.0, or 8.5), or CAPS buffer (pH 9.0 or 9.5). CFUs were determined at 0 and 24 h. (C) The cells were suspended in the pH 9.0 CAPS buffered saline containing the indicated percent of wild-type LB5N supernatant. Symbols: squares, wild-type; circles, . Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare the wild-type strain with the strain. For panel D, paired t tests compare each condition to 0% supernatant (*p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: 14028 and JS2430.
To further understand the toxicity of the supernatant of stationary phase LB5N cultures, we examined the effect of the pH of the supernatant during stationary phase incubation. Wild-type or stationary phase cells grown in LB7 were suspended in the pH-adjusted LB5N supernatant solutions and incubated for 24 hours at 37°C. As shown in Fig. 3C, the toxic effect of the supernatant on the strain was observed only when the pH was between 8.5 and 9.5. We also checked if the toxicity of the supernatant is dose dependent. As shown in Fig. 3D, the toxic effect of LB5N supernatant was observed with little loss in activity when diluted to 20% in pH 9 buffered saline. Activity was significantly reduced at 10% supernatant and completely lost at 5% supernatant.
2.3. The toxicity of supernatant is dependent on acid-inducible cadaverine production.
In an attempt to determine what compound in the supernatant was responsible for toxicity, we asked if supplementation with any particular amino acid in the original culture would affect production of the toxic compound. We grew wild-type cells to stationary phase in LB5N in which we added one of the twenty amino acids to 0.4%. We then tested the effect of the resulting supernatants, diluted 1:20 in pH 9 buffer, on stationary phase cells. As shown in Fig. S3, addition of lysine to the starting culture significantly increased the toxicity of resulting supernatant. None of the other 19 amino acids had any effect. When lysine was added to the conditioned medium, rather than the starting culture, the enhancement of the toxicity of supernatant was not observed (Fig. S3). These results implied that a compound made from or with lysine is responsible for the toxicity of the supernatant.
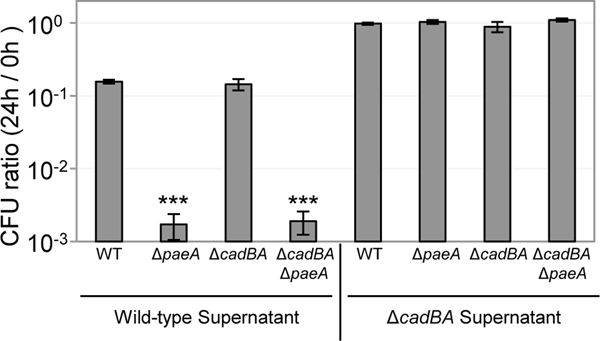
In Salmonella, lysine plays an important role in acid stress tolerance; CadA-mediated decarboxylation of lysine into cadaverine neutralizes cytoplasmic pH (Park et al., 1996). We hypothesized that the addition of lysine in LB5N enhances the production of cadaverine and this process is required or responsible for the accumulation of the toxic compound in the supernatant. Therefore, we made a strain and checked the toxicity of the supernatant after growth of this strain in LB5N. As shown in Fig. 4, the supernatant of the culture had no effect on the survival of the strain in stationary phase. Note that the strain remained sensitive to the supernatant from the wild type culture. Thus, the CadAB system is required for production of the toxic compound, but is not involved in the sensitivity of a mutant per se.
FIGURE 4. The cadBA genes are required for the toxicity of supernatant.
The supernatants are from the wild-type or strain grown in LB5N. The indicated strains were grown in LB7, then cells were suspended in 5 mL total of CAPS buffer (pH 9.0) containing the indicated supernatant (4 ml) and incubated at 37℃ for 24 hours. Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare the wild-type and indicated strains in the same condition (*p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: 14028, JS2430, JS24304, and JS2456.
The above results suggest that cadaverine, produced and exported by CadAB, is toxic to the mutant in stationary phase. Therefore, we measured polyamine levels in the cultures via LC/MS (Fig. S4A). Cells produced very little spermidine, independent of culture conditions or time of incubation. Putrescine production ranged from 0.3 to 0.6 mM, again largely independent of the pH of the medium. In contrast, cadaverine was less than 0.5 mM in LB7, but increased to >2 mM in LB5 with a further increase when cells were grown in LB5N for 24 hrs. This increased production was dependent on CadAB.
2.4. The strain is sensitive to cadaverine and putrescine but not to spermine or spermidine at pH 9.
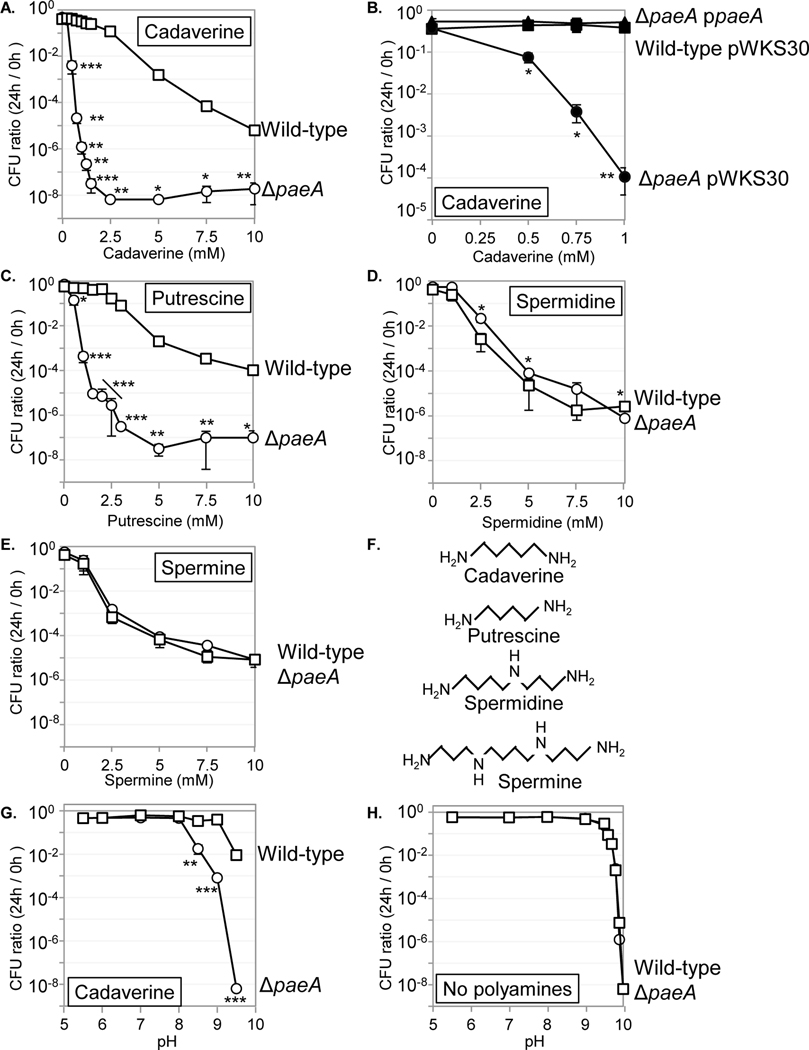
To directly test the hypothesis that strains are sensitive to cadaverine in stationary phase, we added cadaverine at various concentrations to pH 9 buffer and monitored the viability of the wild type and strain. As shown in Fig. 5A, the strain was sensitive to 0.5 mM cadaverine. Sensitivity increased in a concentration-dependent manner, peaking at concentrations at or above 2.5 mM cadaverine with an 8-log loss in viability. This phenotype was complemented when PaeA was expressed from a plasmid (Fig. 5B). The wild-type strain also showed sensitivity to cadaverine in stationary phase, but only at higher concentrations. Indeed, the strain is ~10-fold more sensitive compared to the wild type strain; the survival of strain in the presence of 0.75 mM cadaverine equaled that of the wild-type strain in the presence of 7.5 mM cadaverine. These results indicated that the deletion of paeA dramatically increases sensitivity to cadaverine in stationary phase. These results also support the supposition that it is cadaverine produced by wild type or paeA strains grown in LB5N that is the toxic substance in the culture supernatant.
FIGURE 5. The mutant is sensitive to cadaverine and putrescine, but not to spermidine and spermine.
The indicated strains were grown for 12 h in LB7, then suspended in 5 mL of CAPS buffer (pH 9.0) containing the indicated amount of (A and B) cadaverine, (C) putrescine, (D) spermidine, or (E) spermine, and incubated at 37℃ for 24 h. (F) Structure of cadaverine, putrescine, spermidine, and spermine. (G) The effect of pH on survival of the wild-type and mutant strains in the presence of 0.625 mM cadaverine and (H) in the absence of cadaverine. Symbols: open squares, wild-type; open circles, ; closed squares, wild-type harboring pWKS30 vector; closed circles, harboring pWKS30 vector; closed triangles, harboring paeA pWKS30-placZ-paeA. Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare wild type and strains (*p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: 14028, JS2430, JS2452, JS2453, and JS2454.
Salmonella synthesizes putrescine and spermidine, as well as cadaverine (Tabor and Tabor, 1985). Neither Salmonella nor E. coli synthesize spermine (Tabor and Tabor, 1985), but they might encounter spermine in the environment. We determined if a strain is sensitive specifically to cadaverine or more broadly to polyamines. As shown in Fig. 5C, when stationary phase wild-type and strains were treated with putrescine at pH 9, the strain showed a dramatic loss of viability relative to wild type in a concentration-dependent manner. Analogous to what was observed with cadaverine, the wild type strain was also sensitive to putrescine, but only at higher concentrations. These results indicate that the paeA mutant shows increased sensitivity to both cadaverine and putrescine at similar concentrations and to similar extents.
In contrast to what was observed above, the wild-type and strains were equally sensitive to spermidine and spermine in a concentration-dependent manner, with sensitivity being approximately equal in degree to what is observed with the wild-type strain treated with cadaverine (Figs. 5D and E). These observations correlate with the structural similarities and differences in these four compounds (Fig. 5F), and suggest that the paeA mutant is not simply sensitive to polyamines, but more specifically to cadaverine and putrescine.
The toxic effect of supernatant on was pH-dependent (Fig. 3B). We tested whether the toxicity of cadaverine is also pH dependent. As shown in Fig. 5G, the strain is sensitive to cadaverine only when the pH of the solution ranged from 8.5 to 9.5. The much reduced sensitivity of the wild type strain was also pH dependent, only observed at pH 9.5. In the absence of cadaverine, survival rates of the and wild-type strains in solutions of increasing pH were indistinguishable. Thus, as was seen for the toxic supernatants, sensitivity to cadaverine is pH dependent.
We also measured the sensitivities of wild type E. coli strain MG1655 and a strain to cadaverine and putrescine. As shown in supplementary Fig. S5, the deletion of paeA in E. coli resulted in increased sensitivity to cadaverine and putrescine, but not to spermidine and spermine. These results suggest that the function of PaeA in polyamine tolerance is conserved.
We propose that the pH dependence is related to the pKa of the amine groups of cadaverine (pKa= 10.25, and 9.13). Under these basic conditions, a fraction of the amine groups of cadaverine are deprotonated, making it uncharged and membrane permeable. Theoretically, at pH 9, 2.7% of cadaverine is deprotonated. As cadaverine enters the bacterial cytoplasm, it would become protonated and charged. If true, this implies that 1) toxicity is caused by cadaverine in the cytoplasm, and 2) transport into the cytoplasm is passive. Putrescine would behave in a similar fashion.
2.5. Evidence for high cytoplasmic cadaverine or putrescine as the basis for toxicity.
To further distinguish whether PaeA protects the cell from cytoplasmic cadaverine or extra-cytoplasmic cadaverine, we checked whether the exogenous expression of the cadaverine efflux pump CadB suppresses the sensitivity. As shown in Fig. S6A, the introduction of a pWKS30-cadB plasmid into a strain partially suppressed its sensitivity to cadaverine compared to the strain harboring the pWKS30 vector. The deletion of the cadBA genes in the mutant resulted in increased sensitivity to cadaverine (Fig. S6B). Introduction of the pWKS30-cadB plasmid suppressed this additional sensitivity in the strain (Fig. S6B). These results support the conclusion that the cell is sensitive to intracellular cadaverine. Loss of the cadaverine exporter CadB enhances sensitivity, while ectopic expression of CadB partially suppresses sensitivity.
PaeA protects the cytoplasm from cadaverine and putrescine. We hypothesized various mechanisms for cadaverine tolerance. PaeA could 1) prevent influx, 2) promote efflux, 3) promote degradation or modification, 4) or affect toxicity per se. We set out to test these various hypotheses.
The pH dependence of the sensitivity to both cadaverine and putrescine implies that deprotonation of the compound is required. The simplest interpretation is that deprotonation of the polyamines is required for their passive entry into the cell through the cytoplasmic membrane. Although it is theoretically possible that PaeA somehow blocks this passive entry, it is difficult to imagine a molecular mechanism for such action.
Once inside the cell, the polyamines would become protonated. They could then be pumped out of the cytoplasm. A potential consequence is an increase in cytoplasmic pH, which could be toxic. Indeed, raising the cytoplasmic pH is why cadaverine is normally made and pumped out under acidic conditions (Park et al., 1996). An argument against this mechanism of toxicity is the fact that overproduction of the cadaverine exporter CadB suppresses sensitivity in the paeA mutant (Fig S6). If loss of protons from the cytoplasm was toxic, then CadB should exacerbate sensitivity. To more directly test the effects of cadaverine, we monitored intracellular pH of wild-type and strains during cadaverine treatment. We used a green fluorescent protein (GFP) that responds to pH change (Arce-Rodriguez et al., 2019). We confirmed the response to pH by incubating cells in saline solutions buffered at various pH in the presence of cell-permeable acids and bases, thereby equilibrating intracellular and extracellular pH. As shown in Fig. S7A, the pH-responsive GFP expressed in wild-type and strains uniformly changes fluorescence in response to pH. As shown in Fig. S7B, incubation of wild-type or strains in pH 9 buffer for 24 hrs increased the intracellular pH from ~7.4 to 7.6. The cells remained viable over this time period. Addition of 0.2 mM cadaverine did not affect viability or the degree of alkalization of the cytoplasm during this time (Fig. S7B and C). When wild-type and strains were incubated in the presence of 0.5 mM cadaverine for 12 hours, the strain showed ~75% loss in viability, while the intracellular pH was ~7.7, compared to a pH of ~7.6 seen in the wild type under the same conditions. The intracellular pH of the strain ultimately reached ~7.8 after twenty-four hours. Note that it is difficult to determine the contribution of the dead cells to this overall change in fluorescence. However, given that the intracellular pH in most of the cultures was ultimately above 7.6 without any significant loss in viability, it seems unlikely that the minor increase in intracellular pH seen in the strain could account for the toxicity of cadaverine. Moreover, in the wild type culture in which the cells remained viable, the addition of cadaverine did not significantly affect the intracellular pH compared to buffer alone. Given all of our results, we conclude that sensitivity to cadaverine is not due to any effect of the polyamine on intracellular pH.
PaeA could play some role in degradation of cadaverine. The Salmonella genome encodes orthologs to E. coli proteins PatA, PatD, GabT, GabD, CsiD, and LhgO, known to be involved in the degradation of cadaverine to succinate (Knorr et al., 2018)(Fig. 1). We constructed a strain () that lacks all the genes involved in cadaverine degradation and monitored cadaverine sensitivity in this strain and in an isogenic paeA mutant. As shown in Fig. S8, the strain was resistant to 1 mM cadaverine. This shows that it is not simply degradation of cadaverine that protects wild-type cells. Deletion of paeA in the degradation-defective background conferred sensitivity. It is noteworthy that strain exhibited higher sensitivity to cadaverine than strain (Fig. S8). This is presumably because the concentration of cadaverine is higher in the absence of degradation.
Next, we examined if the modification of cadaverine or putrescine is involved in PaeA-dependent tolerance. It is known that putrescine is converted into spermidine by the propylamine transferase SpeE (Bowman et al., 1973); SpeE can also convert cadaverine to aminopropylcadaverine (Bowman et al., 1973). The propylamine donor, decarboxylated S-adenosylmethionine, is made by SpeD (Markham et al., 1982; Tabor and Tabor, 1987). SpeG can further modify spermidine to N-acetylspermidine (Matsui et al., 1982; Fukuchi et al., 1994). We constructed a strain that lacks this polyamine modification pathway and determined the effect of deleting the paeA gene in this background on cadaverine sensitivity. As shown in Fig. S8, the deletion of paeA in the strain still increased the sensitivity to cadaverine, which suggested that the modification pathway of cadaverine and putrescine is not required. All together, these results show that PaeA acts independently of the known degradation or modification pathways, and suggest that cadaverine (or putrescine) per se is toxic.
2.6. PaeA is involved in cadaverine and putrescine efflux.
The STM14_3648 locus encodes an uncharacterized putative inner membrane protein homologous to the PACE family of multidrug efflux pumps. Hassan et al. recently reported that polyamines were the physiological substrate for the Acinetobacter homolog AceI (Hassan et al., 2019). Although they showed that the Salmonella protein did not reduce cadaverine levels when expressed in E. coli, we deleted STM14_3648 and tested the effect in our assay. As shown in Fig. S9, deletion of STM14_3648 has no effect on cadaverine tolerance in either the or backgrounds.
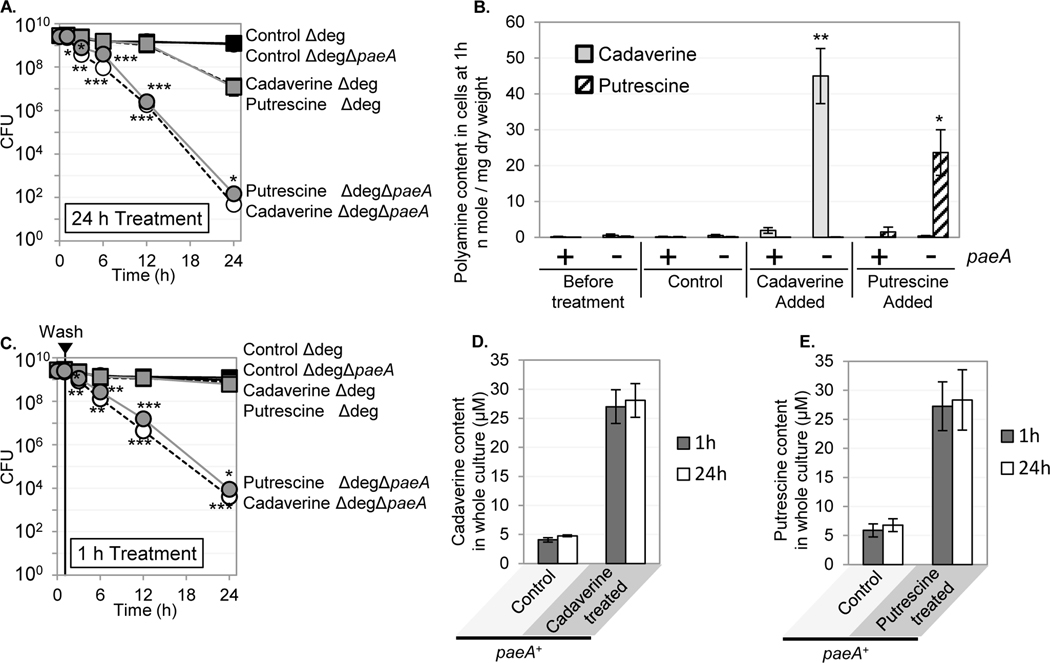
To test if PaeA is involved in polyamine efflux, we measured the intracellular level of cadaverine or putrescine after a short period of treatment. To avoid the complication of degradation, we used the strain. When the and strains were treated with 5 mM cadaverine or putrescine, the paeA mutant started to lose viability after 3 hours. However, there was little loss of viability after one hour (Fig. 6A). Therefore, we collected cells one hour after treatment. The treated cells were washed and total intracellular cadaverine and putrescine were measured by LC/MS. We also continued to monitor viability in these cells after removing extracellular polyamine. As shown in Fig. 6B, the paeA strain had a 24-fold higher level of cadaverine compared to the isogenic strain. In the putrescine treated cells, the strain showed a 16-fold higher level of putrescine compared to the isogenic strain (Fig. 6B). Fig S4B shows that the strain also had higher levels of intracellular cadaverine after growth in LB5 or LB5N. As shown in Fig. 6D and E, both the and strains showed similar overall levels of cadaverine and putrescine in the total cultures after washing and suspending the cells in pH 9 buffer. These results suggest that PaeA is not involved in degradation or modification of cadaverine and putrescine, but rather enhances efflux of cadaverine and putrescine from the cytoplasm.
FIGURE 6. The intracellular cadaverine and putrescine concentrations are increased in the mutant after short treatment with cadaverine or putrescine.
The or strains lacking all known genes for degradation of cadaverine and putrescine ( = ) were grown for 12 h in LB7, then suspended in 5 mL of CAPS buffer (pH 9.0) with or without 5 mM of either cadaverine or putrescine and incubated at 37℃. (A) CFUs were determined at the indicated time points. (B) The intracellular content of cadaverine and putrescine were determined at the 1 h time point. Spermidine was <0.15 nmoles mg−1 and spermine was <0.25 nmoles mg−1 in all cells. These data are re-plotted in Fig. S10. (C) One hour after initial treatment, cells were washed and re-suspended in 5 mL of CAPS buffer (pH 9.0), and CFUs were determined at the indicated time points. (D) Cadaverine contents in whole wild-type cultures (medium and cells) immediately after removing cadaverine or (E) putrescine, and after 24 h incubation. Symbols: Squares, ; circles, ; black filled markers with black lines, control; open markers with dashed lines, cadaverine treatment; gray filled markers with gray lines, putrescine treatment. Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare the indicated strains to the parent strain in the same treatment (*p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: JS2464 and JS2465.
Fig. 6C shows that the treated cells continued to lose viability after removing extracellular cadaverine or putrescine. Either the lethal damage had occurred by one hour or the intracellular concentration remained high. We monitored intracellular cadaverine over time after treating or strains with various concentrations of cadaverine and washing the cells after one hour. As shown in Fig. S11C and consistent with Fig. 6, treatment with 5 mM cadaverine resulted in a 16-fold increase in cell-associated cadaverine in the versus cells. Even when we treated the cells with 50 mM cadaverine, the levels were below that observed at 5 mM in the strain. Levels in the strains showed an initial drop in cellular cadaverine concentrations immediately after washing the cells. However, at three hours, the intracellular levels remained constant and significantly higher than those in the paeA+ cells treated with 5 mM cadaverine. Even at 50 mM cadaverine, levels in the cells dropped to an apparent baseline level. We propose that the drop in intracellular cadaverine contents immediately after washing is due to non-specific loss. The fact that intracellular concentrations remains high in the paeA mutant is consistent with a role of PaeA in cadaverine efflux. The fact that the cells treated with 50 mM cadaverine continued to lose viability at the same rate as the cells treated with 5 mM suggests that the initial high concentrations can cause damage that is ultimately lethal.
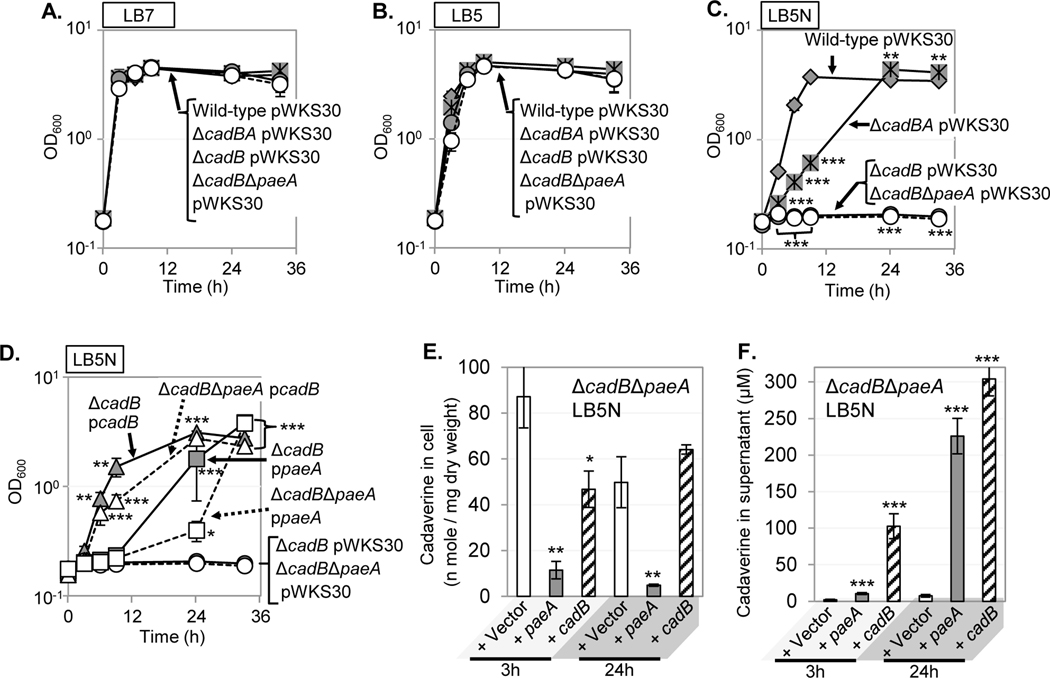
We next asked if ectopic expression of PaeA suppresses the growth defect conferred by endogenously produced cadaverine. As shown in Fig. 7, strains deleted for cadAB, cadB, or cadB paeA grew essentially like the wild-type control in LB7 and LB5, with no significant loss of viability in stationary phase. However, when cells were grown in LB5N, the cadAB mutant showed a significant growth defect, although the strain ultimately reached the same final . These results suggest that nitrite exacerbates the acid stress normally counteracted by CadAB, as previously noted (Muhlig et al., 2014). In contrast, neither the nor the were able to grow in LB5N. The fact that the cadB mutant showed a more severe growth defect in LB5N than the mutant suggests that the defect is not only the lack of appropriate acid stress tolerance but also the result of intracellular CadA-mediated cadaverine production, which is increased when cells are grown in LB5N versus LB5 (Fig. S4B).
FIGURE 7. Ectopic expression of paeA partially suppresses the growth defect of a mutant in acidic medium.
The wild-type, , , and strains harboring the pWKS30 vector were grown in (A) LB7, (B) LB5, or (C) LB5N (1.5 mM ). (D) The and strains harboring the pWKS30, pWKS30 -cadB, or pWKS30 -paeA plasmid were grown in LB5N (1.5 mM ). Symbols: closed diamonds, wild-type pWKS30; closed asterisks, pWKS30; closed circles, pWKS30; opened circles, pWKS30; closed triangles, pWKS30 -cadB; opened triangles, pWKS30 -cadB; filled squares, pWKS30 -paeA; opened squares, pWKS30 -paeA. (E) Cadaverine content in cell pellet or (F) supernatant of the strain containing indicated plasmids grown for 3h or 24h in LB5N (1.5 mM ). Values are reported as mean ± standard deviation, where n = 3. Unpaired t tests compare the indicated strains to the wild-type pWKS30 strain (A-C) or a corresponding isogenic strain with pWKS30 (D-E) (*p < 0.05, **p < 0.005, ***p < 0.0005). Strains used: JS2452, JS2457, JS2458, JS2459, JS2460, JS2461, JS2462, and JS2463.
As shown in Fig. 7D, the introduction of pWKS30 PlacZ-cadB into and strains restored their growth in LB5N. The introduction of pWKS30 PlacZ-paeA partially restored the growth of both and strains. These results suggested that PaeA reduces stress caused by the accumulation of cadaverine that is endogenously produced by CadA in the acidic environment, consistent with PaeA acting in cadaverine efflux. Indeed, as shown in Fig. 7E and F, the amount of intracellular cadaverine in the strain expressing paeA ectopically is 7- or 9-fold lower than in the strain harboring the vector after 3 and 24 hours of growth in LB5N, respectively, with a concomitant increase in cadaverine found in the supernatant. These results suggest that PaeA is involved in cadaverine efflux under physiological conditions. Note that the total concentration of cadaverine produced in the complemented strains is higher, likely due to the fact that CadB is a lysine:cadaverine antiporter, thus increasing the amount of substrate for cadaverine production.
3. DISCUSSION
In the current study, we identified paeA (ytfL) as a gene involved in cadaverine and putrescine tolerance in stationary phase. The simplest explanation for our data is that PaeA is required for cadaverine and putrescine efflux under certain conditions that would otherwise lead to a toxic buildup. Our results confirm that Salmonella, grown in low pH LB medium, produces significant concentrations of cadaverine via the CadAB pathway and excretes it into the medium. This production is independent of PaeA and is exacerbated by adding nitrite to the growth medium, for reasons that are not understood. A mutant lacking PaeA becomes sensitive to cadaverine in stationary phase. The mutant is also sensitive to putrescine, but not to spermidine or spermine. The sensitivity is pH dependent and only observed at pH >8. This is presumably because the external polyamine needs to be deprotonated so that it can passively diffuse into the cell cytoplasm. It is cytoplasmic cadaverine that is conferring sensitivity. This is supported by several lines of evidence. First, we can directly measure the significant increase in intracellular cadaverine or putrescine in the paeA mutant. Second, a cadB mutant is sensitive to growth in pH 5 LB with nitrite. Deleting cadA suppresses this phenotype, as does ectopic expression of PaeA. Thus, CadA is producing intracellular cadaverine that cannot be excreted, conferring sensitivity. Third, ectopic expression of the cadaverine exporter, CadB, in stationary phase partially suppresses the paeA phenotype. It is cadaverine or putrescine per se that are toxic, in that degradation or modification are not relevant. Note that, although we uncovered the function of PaeA under specific conditions, it seems likely that it has a broader role is cellular homeostasis. Polyamines, produced in response to any number of stress conditions or other physiological need could build up to potentially lethal levels when the need has been met or conditions change. These excess polyamines need to be removed from the cytoplasm.
In contrast to mammalian cells (Mandal et al., 2013), in Salmonella and E.coli, polyamine synthesis is not essential for cell growth and survival (Chattopadhyay et al., 2009). However, under certain conditions, such as growth in an anaerobic environment or in 95% oxygen, mutants unable to synthesize polyamines cannot grow or even lose viability (Chattopadhyay et al., 2003; Chattopadhyay et al., 2009). As shown in Fig. 1, Salmonella has multiple redundant pathways for polyamine synthesis, import, and export, suggesting that Salmonella controls polyamine levels in response to changes in the external environment and/or conditions inside the cell. Despite the existence of the known import/export systems, our data suggest that import of cadaverine and putrescine in our experiments was the result of passive diffusion of the uncharged species into the cell, explaining the pH dependence, and that export was dependent largely on PaeA. These results can be explained by the fact that none of the known polyamine import or export systems are expressed in late stationary phase in Salmonella (Kroger et al., 2013).
Polyamines have pleiotropic effects on cells. Many studies have addressed the need for polyamines using E. coli mutants defective for polyamine synthesis (Chattopadhyay et al., 2009; Chattopadhyay et al., 2013; Chattopadhyay and Tabor, 2013; Chattopadhyay et al., 2015; Igarashi and Kashiwagi, 2018; Keller et al., 2019). The effects of excess polyamines on cell physiology are less well understood. In E.coli, polyamines inhibit pore function in the outer membrane porins OmpF and OmpC by binding within the pores from the periplasmic side (Dela Vega and Delcour, 1996; Iyer and Delcour, 1997). Though homologous, OmpF is 2-fold more susceptible to polyamines than OmpC (Dela Vega and Delcour, 1996). Among the four polyamines, spermine is the most potent, followed by spermidine and cadaverine, with putrescine being the least effective (Dela Vega and Delcour, 1996). However, loss of porin function is not lethal (Hall and Silhavy, 1981). Excess polyamines, like low levels, affect RNA-mediated processes, particularly translation initiation of certain mRNAs, defining the “polyamine modulon” (Sakamoto et al., 2020). Whether the result of changes in expression of specific genes or more general defects in translation, this could certainly be lethal. At sublethal concentrations, excess polyamines confer particular phenotypes, including sensitivity to hydrogen peroxide, high temperature, and normally sub-inhibitory concentrations of kanamycin, in strains that cannot degrade putrescine or cadaverine (Schneider et al., 2013), and growth arrest and cell lysis in low temperature in strains with excess spermidine (Limsuwun and Jones, 2000). Note that the sensitivity to polyamines is dependent on overall physiology of the cell. For example, in our experiments, intracellular putrescine concentrations were relatively high in exponential phase, yet cells grew normally, whereas similar concentrations of cadaverine in stationary phase were lethal (Fig. S4).
Our data suggest that PaeA facilitates efflux of both of cadaverine and putrescine, but the molecular mechanism is unclear. The 447 amino acid PaeA protein contains three conserved domains that are often found together. The N-terminal 197 amino acids comprise a four-transmembrane domain in the DUF21 (PF01595) family, associated with transport proteins (Chen et al., 2020). The N terminus and C-terminal domains are predicted to be in the cytoplasm. The transmembrane domain is followed by two tandem CBS repeat domains (PF00571), which are known to bind ligands with an adenosyl group, including ATP (Hirata et al., 2014). The C-terminal 100 amino acids constitute a CorC_HlyC (PF03471) domain. This overall structure is certainly consistent with PaeA being a transporter. However, homologous proteins containing all three domains are primarily implicated in / transport, such as MpfA from Bacillus subtilis (Pi et al., 2020), and MpfA from Staphylococcus aureus (Armitano et al., 2016). and polyamines both have important roles as cations binding to nucleic acids, although they are not completely interchangeable (Tabor and Tabor, 1985; Lightfoot and Hall, 2014). Thus, we cannot rule out that PaeA is an ion transporter that is indirectly affecting polyamine efflux. However, it is interesting that PaeA seems to be specific for cadaverine and putrescine, having no effect on spermine or spermidine toxicity, and that it can partially complement loss of CadB in exponential phase. Further biochemical analyses of PaeA will be required to prove that it is an exporter, define the substrate, and identify the putative energy source. We also need to more fully understand the physiological role of PaeA. Transcriptomic data suggest that paeA is expressed under most conditions, but is most highly induced when Salmonella is replicating in macrophages (Kroger et al., 2013; Srikumar et al., 2015). More detailed analysis of paeA regulation is required.
Polyamines are also implicated in pathogenesis. The PaeA/YtfL of Klebsiella pneumonia, which is 91% identical to PaeA of S. enterica, is reported to be responsible for making spent medium that induces disassembly of host microtubules in lung epithelial cells. Expressing the Klebsiella protein in an E. coli strain conferred the same phenomenon (Chua et al., 2019). Although they did not identify the active compound, polyamines are involved in microtubule assembly (Savarin et al., 2010). Interestingly, most Staphylococci neither synthesize nor metabolize polyamines (Anzaldi and Skaar, 2011; Joshi et al., 2011). Indeed, these organisms are highly sensitive to spermine and spermidine, but not cadaverine or putrescine, and showed increased sensitivity at alkaline pH (Joshi et al., 2011). The epidemic S. aureus MRSA strain USA300 possesses an “arginine catabolic mobile element” that encodes a SpeG spermine/spermidine acetyl transferase that can detoxify polyamines found in skin and inflamed tissues, thought to contribute to its increased pathogenicity (Anzaldi and Skaar, 2011; Joshi et al., 2011). We need to determine if PaeA plays any role in Salmonella pathogenesis.
In conclusion, we found that PaeA is involved in stationary phase survival when Salmonella is exposed to cadaverine or putrescine, but not to spermidine and spermine. Further, we showed that PaeA reduces intracellular cadaverine and putrescine level during treatment. These results suggest that PaeA is involved in selective efflux of cadaverine and putrescine and this is important for survival under specific stress conditions.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and plasmids
Strains and plasmids are described in Table S1. All Salmonella strains used are derivatives of Salmonella enterica serovar Typhimurium strain 14028. Deletions with simultaneous insertion of antibiotic resistance cassettes were constructed using Red-mediated recombination as previously described (Datsenko and Wanner, 2000; Ellermeier et al., 2002), with the indicated endpoints. Deletions were verified by PCR analysis and then transduced into unmutagenized backgrounds using phage P22 HT105/1 int-201 (Maloy et al., 1996). All plasmids were passaged through a restriction-minus modification-plus Pi+ Salmonella strain (JS198) (Ellermeier et al., 2002) prior to transformation into derivatives of strain 14028. E. coli K12 strains used are derivatives of strain MG1655. Deletion mutations were constructed by using Red-mediated recombination (Datsenko and Wanner, 2000) and the mutations were transferred to clean backgrounds by P1 transduction (Silhavy et al., 1984).
The pWKS30- plasmid was constructed by amplifying the paeA gene using primers ytfLF and ytfLR (Table S3). Both the PCR product and pWKS30 plasmid were digested with EcoRI and XbaI restriction enzymes (New England Biolabs) according to the manufacturer’s protocol. The resulting fragments were ligated with T4 DNA ligase (New England Biolabs) according to manufacturer’s protocol and transformed into E. coli strain . The plasmid was confirmed by DNA sequence.
Plasmid pWKS30- was constructed using primers YIS_275–17 and YIS_275–18 to amplify the cadB gene, which was introduced into a pWKS30 fragment amplified with YI_pWKS30–3 and YI_pWKS30–10 by Gibson assembly (NEBuilder® HiFi DNA Assembly). The resulting product was transformed into . The plasmid was confirmed by DNA sequencing.
4.2. Stationary phase survival assay
Cells from overnight LB medium cultures were harvested by centrifugation and suspended in the same volume of 0.85% NaCl. The suspensions were diluted 1:25 into 5 mL of unbuffered pH 5 LB medium with 2 mM (LB5N) or without (LB5) or pH 7 LB medium (LB7) and incubated for 48 hours at 37°C on a roller drum. For the strains harboring a pWKS30 plasmid, 50 μg ml− of ampicillin was added to the medium. At indicated times, the cultures were diluted and plated on LB agar supplemented with 0.1 % glucose and incubated overnight at 37°C to determine CFU. The pH of spent medium was determined using a pH meter. For growth curves, of the culture was measured in a BioTek ELx808 Absorbance Reader at the indicated time points. When necessary, the cultures were diluted in the original growth medium.
4.3. Spent medium tolerance assay
To prepare spent medium, wild type cells from overnight LB cultures were harvested by centrifugation and suspended in the same volume of 0.85% NaCl. The suspensions were then diluted 1:25 into 5 mL of LB5N or LB7 and incubated for 12 hours at 37°C in a roller drum. Cells were then removed by centrifugation (7,000 g) and the supernatant was filtered through a 0.22 μm Millex-GP syringe filter (Millipore). For preparation of stationary phase cells, overnight LB medium cultures were centrifuged and the cells were suspended in the same volume of 0.85% NaCl. The suspensions were diluted 1:25 into 5 mL of LB5N or LB7. After 12 hours of incubation at 37°C in a roller drum, the cells were collected by centrifugation (7,000 g) and washed twice with 0.85% NaCl. The stationary phase cells were suspended in the original volume of prepared spent medium with or without 100 mM MES buffer (pH5.5 or 6.0), MOPS buffer (pH7.0, 8.0, or 8.5), or CAPS buffer (pH9.0 or 9.5). When necessary, prepared spent media were diluted with 0.85% NaCl. The resulting 5 mL cultures were incubated for 24 hours at 37°C in a roller drum. After serial dilution, the cultures, before and after incubation, were plated on LB agar plates supplemented with 0.1 % glucose and incubated overnight at 37 C to determine CFUs. Spent medium tolerance was calculated by dividing the CFU after 24 h incubation by the CFU before incubation.
4.4. Polyamine tolerance assay
Stationary phase cells, prepared as above, were suspended in the original volume of 0.85% NaCl with CAPS buffer (pH 9.0) with the indicated amount of cadaverine, putrescine, spermidine, or spermine. Polyamine stocks were first prepared at 20 mM in 0.85% NaCl with CAPS buffer (pH 9.0). To perform the assay at varied pH, cells were suspended in original volume of 0.85% NaCl with 100 mM MES buffer (pH5.5 or 6.0), MOPS buffer (pH7.0, 8.0, or 8.5), or CAPS buffer (pH 9.5). The cultures were incubated for 24 hours at 37°C in a roller drum. After serial dilution, the cultures, before and after incubation, were plated on LB agar plates supplemented with 0.1 % glucose and incubated overnight at 37°C to determine CFUs. Polyamine tolerance was calculated by dividing the CFU after 24 h incubation by the CFU before incubation.
4.5. Measurement of intracellular pH
To obtain a calibration curve, pH 7 LB medium grown stationary phase wild-type or cells harboring pS2513-PHP were suspended to = 0.4 to 0.5 in saline containing 50 mM of MES buffer (pH 6.0 or 6.5) or MOPS buffer (pH 7.0, 7.5, 8.0, or 8.5), each with 50 mM sodium benzoate and 50 mM methylamine HCl and incubated at 30°C for 10 min. The fluorescence emission of PHP () was recorded at of either 405 nm or 485 nm using a BioTek Synergy H1 Hybrid Multi-Mode Reader. The relative excitation (R 405/485) was plotted versus the pH of the buffer. To measure cytoplasmic pH during cadaverine treatment, stationary phase cells harboring pS2513-PHP were suspended in 5 mL of CAPS buffer (pH9.0) with or without 0.2 or 0.5 mM of cadaverine. At the indicated time the fluoresce emission of PHP () was recorded at of either 405 nm or 485 nm and cytoplasmic pH was calculated using calibration curve. CFUs were determined by plating appropriate dilutions on LB glucose plates containing 50 μg ml- of kanamycin.
4.6. Measurement of polyamine contents in cells, culture, and spent medium
To measure intracellular polyamine levels, cells were collected by centrifugation (7,000 g) at the indicated time points and washed with 0.85% NaCl 100 mM CAPS buffer (pH 9.0) twice. After carefully and completely removing the supernatant, the cell pellet was frozen at −80°C. Cells were suspended in 1 mL methanol and disrupted by sonication. The samples were cleared by centrifugation and the concentration of polyamines in the solvent was determined by LC-MS at the Metabolomics Center, Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign. The concentration is reported per 1 mg of dry weight. Where indicated, the concentration of polyamines in the supernatant was determined after removal of cells by centrifugation. For determination of polyamine concentrations in whole cultures, cells were disrupted by sonication and the supernatant was cleared by centrifugation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Thomas E. Kehl-Fie, Jacqueline Morey, and Katie Frye for help with measurements of intracellular pH using the pH-responsive GFP, and to Cari Vanderpool and Jim Imlay for valuable discussions. This study was supported by NIH grant R01 AI123381 to J.M.S. Y.I. is supported by a JSPS Postdoctoral Fellowship for Research Abroad.
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Anzaldi LL and Skaar EP (2011) The evolution of a superbug: how Staphylococcus aureus overcomes its unique susceptibility to polyamines. Mol Microbiol 82: 1–3. 10.1111/j.1365-2958.2011.07808.x [DOI] [PubMed] [Google Scholar]
- Arce-Rodriguez A, Volke DC, Bense S, Haussler S and Nikel PI (2019) Non-invasive, ratiometric determination of intracellular pH in Pseudomonas species using a novel genetically encoded indicator. Microb Biotechnol 12: 799–813. 10.1111/1751-7915.13439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitano J, Redder P, Guimaraes VA and Linder P (2016) An Essential Factor for High Mg(2+) Tolerance of Staphylococcus aureus. Front Microbiol 7: 1888. 10.3389/fmicb.2016.01888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman WH, Tabor CW and Tabor H (1973) Spermidine biosynthesis. Purification and properties of propylamine transferase from Escherichia coli. J Biol Chem 248: 2480–2486. [PubMed] [Google Scholar]
- Burton NA, Schurmann N, Casse O, Steeb AK, Claudi B, Zankl J, et al. (2014) Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection in mice. Cell Host Microbe 15: 72–83. 10.1016/j.chom.2013.12.006 [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Mizusaki H and Kenney LJ (2015) A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13: e1002116. 10.1371/journal.pbio.1002116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravortty D, Hansen-Wester I and Hensel M (2002) Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med 195: 1155–1166. 10.1084/jem.20011547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Chen W and Tabor H (2013) Escherichia coli glutathionylspermidine synthetase/amidase: phylogeny and effect on regulation of gene expression. FEMS Microbiol Lett 338: 132–140. 10.1111/1574-6968.12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Keembiyehetty CN, Chen W and Tabor H (2015) Polyamines Stimulate the Level of the sigma38 Subunit (RpoS) of Escherichia coli RNA Polymerase, Resulting in the Induction of the Glutamate Decarboxylase-dependent Acid Response System via the gadE Regulon. J Biol Chem 290: 17809–17821. 10.1074/jbc.M115.655688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW and Tabor H (2003) Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc Natl Acad Sci U S A 100: 2261–2265. 10.1073/pnas.2627990100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK, Tabor CW and Tabor H (2009) Polyamines are not required for aerobic growth of Escherichia coli: preparation of a strain with deletions in all of the genes for polyamine biosynthesis. J Bacteriol 191: 5549–5552. 10.1128/JB.00381-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay MK and Tabor H (2013) Polyamines are critical for the induction of the glutamate decarboxylase-dependent acid resistance system in Escherichia coli. J Biol Chem 288: 33559–33570. 10.1074/jbc.M113.510552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, Kozlov G, Fakih R, Yang M, Zhang Z, Kovrigin EL, et al. (2020) Mg(2+)-ATP Sensing in CNNM, a Putative Magnesium Transporter. Structure 28: 324–335 e324. 10.1016/j.str.2019.11.016 [DOI] [PubMed] [Google Scholar]
- Chua MD, Liou CH, Bogdan AC, Law HT, Yeh KM, Lin JC, et al. (2019) Klebsiella pneumoniae disassembles host microtubules in lung epithelial cells. Cell Microbiol 21: e12977. 10.1111/cmi.12977 [DOI] [PubMed] [Google Scholar]
- Darwin KH and Miller VL (1999) Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin Microbiol Rev 12: 405–428. 10.1128/CMR.12.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America 97: 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Vega AL and Delcour AH (1996) Polyamines decrease Escherichia coli outer membrane permeability. J Bacteriol 178: 3715–3721. 10.1128/jb.178.13.3715-3721.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Janakiraman A and Slauch JM (2002) Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161. 10.1016/S0378-1119(02)00551-6 [DOI] [PubMed] [Google Scholar]
- Feasey NA, Dougan G, Kingsley RA, Heyderman RS and Gordon MA (2012) Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379: 2489–2499. 10.1016/S0140-6736(11)61752-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenlon LA and Slauch JM (2014) Phagocyte roulette in Salmonella killing. Cell Host Microbe 15: 7–8. 10.1016/j.chom.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi J, Kashiwagi K, Takio K and Igarashi K (1994) Properties and structure of spermidine acetyltransferase in Escherichia coli. J Biol Chem 269: 22581–22585. [PubMed] [Google Scholar]
- Fukuchi J, Kashiwagi K, Yamagishi M, Ishihama A and Igarashi K (1995) Decrease in cell viability due to the accumulation of spermidine in spermidine acetyltransferase-deficient mutant of Escherichia coli. J Biol Chem 270: 18831–18835. 10.1074/jbc.270.32.18831 [DOI] [PubMed] [Google Scholar]
- Gale EF and Epps HM (1944) Studies on bacterial amino-acid decarboxylases: 1. l(+)-lysine decarboxylase. Biochem J 38: 232–242. 10.1042/bj0380232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM and Gardner PR (2002) Flavohemoglobin detoxifies nitric oxide in aerobic, but not anaerobic, Escherichia coli. Evidence for a novel inducible anaerobic nitric oxide-scavenging activity. J Biol Chem 277: 8166–8171. 10.1074/jbc.M110470200 [DOI] [PubMed] [Google Scholar]
- Hall MN and Silhavy TJ (1981) Genetic analysis of the ompB locus in Escherichia coli K-12. J. Mol. Biol 151: 1–15. [DOI] [PubMed] [Google Scholar]
- Hassan KA, Naidu V, Edgerton JR, Mettrick KA, Liu Q, Fahmy L, et al. (2019) Short-chain diamines are the physiological substrates of PACE family efflux pumps. Proc Natl Acad Sci U S A 116: 18015–18020. 10.1073/pnas.1901591116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Ishigure H, Demizu R, Uemura T, Nishino K, Yamaguchi A, et al. (2008) Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J Bacteriol 190: 872–878. 10.1128/JB.01505-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y, Funato Y, Takano Y and Miki H (2014) Mg2+-dependent interactions of ATP with the cystathionine-beta-synthase (CBS) domains of a magnesium transporter. J Biol Chem 289: 14731–14739. 10.1074/jbc.M114.551176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K, Ito K and Kashiwagi K (2001) Polyamine uptake systems in Escherichia coli. Res Microbiol 152: 271–278. 10.1016/s0923-2508(01)01198-6 [DOI] [PubMed] [Google Scholar]
- Igarashi K and Kashiwagi K (2015) Modulation of protein synthesis by polyamines. IUBMB Life 67: 160–169. 10.1002/iub.1363 [DOI] [PubMed] [Google Scholar]
- Igarashi K and Kashiwagi K (2018) Effects of polyamines on protein synthesis and growth of Escherichia coli. J Biol Chem 293: 18702–18709. 10.1074/jbc.TM118.003465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwadate Y and Kato JI (2017) Involvement of the ytfK gene from the PhoB regulon in stationary-phase H2O2 stress tolerance in Escherichia coli. Microbiology 163: 1912–1923. 10.1099/mic.0.000534 [DOI] [PubMed] [Google Scholar]
- Iyer R and Delcour AH (1997) Complex inhibition of OmpF and OmpC bacterial porins by polyamines. J Biol Chem 272: 18595–18601. 10.1074/jbc.272.30.18595 [DOI] [PubMed] [Google Scholar]
- Jennings E, Thurston TLM and Holden DW (2017) Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences. Cell Host Microbe 22: 217–231. 10.1016/j.chom.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Joshi GS, Spontak JS, Klapper DG and Richardson AR (2011) Arginine catabolic mobile element encoded SpeG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol Microbiol 82: 9–20. 10.1111/j.1365-2958.2011.07809.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Shibuya S, Tomitori H, Kuraishi A and Igarashi K (1997) Excretion and uptake of putrescine by the PotE protein in Escherichia coli. J Biol Chem 272: 6318–6323. 10.1074/jbc.272.10.6318 [DOI] [PubMed] [Google Scholar]
- Keller C, Chattopadhyay M and Tabor H (2019) Absolute requirement for polyamines for growth of Escherichia coli mutants (mnmE/G) defective in modification of the wobble anticodon of transfer-RNA. FEMS Microbiol Lett 366. 10.1093/femsle/fnz110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr S, Sinn M, Galetskiy D, Williams RM, Wang C, Muller N, et al. (2018) Widespread bacterial lysine degradation proceeding via glutarate and L-2-hydroxyglutarate. Nat Commun 9: 5071. 10.1038/s41467-018-07563-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, et al. (2013) An Infection-Relevant Transcriptomic Compendium for Salmonella enterica Serovar Typhimurium. Cell Host. Microbe 14: 683–695. 10.1016/j.chom.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Kurihara S, Suzuki H, Oshida M and Benno Y (2011) A novel putrescine importer required for type 1 pili-driven surface motility induced by extracellular putrescine in Escherichia coli K-12. J Biol Chem 286: 10185–10192. 10.1074/jbc.M110.176032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara S, Suzuki H, Tsuboi Y and Benno Y (2009a) Dependence of swarming in Escherichia coli K-12 on spermidine and the spermidine importer. FEMS Microbiol Lett 294: 97–101. 10.1111/j.1574-6968.2009.01552.x [DOI] [PubMed] [Google Scholar]
- Kurihara S, Tsuboi Y, Oda S, Kim HG, Kumagai H and Suzuki H (2009b) The putrescine Importer PuuP of Escherichia coli K-12. J Bacteriol 191: 2776–2782. 10.1128/JB.01314-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot HL and Hall J (2014) Endogenous polyamine function--the RNA perspective. Nucleic Acids Res 42: 11275–11290. 10.1093/nar/gku837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limsuwun K and Jones PG (2000) Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J Bacteriol 182: 5373–5380. 10.1128/jb.182.19.5373-5380.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J, Xie QW and Nathan C (1997) Nitric oxide and macrophage function. Annu Rev Immunol 15: 323–350. 10.1146/annurev.immunol.15.1.323 [DOI] [PubMed] [Google Scholar]
- Maloy SR, Stewart VJ and Taylor RK (1996) Genetic Analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- Mandal S, Mandal A, Johansson HE, Orjalo AV and Park MH (2013) Depletion of cellular polyamines, spermidine and spermine, causes a total arrest in translation and growth in mammalian cells. Proc Natl Acad Sci U S A 110: 2169–2174. 10.1073/pnas.1219002110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham GD, Tabor CW and Tabor H (1982) S-adenosylmethionine decarboxylase of Escherichia coli. Studies on the covalently linked pyruvate required for activity. J Biol Chem 257: 12063–12068. [PubMed] [Google Scholar]
- Matsui I, Kamei M, Otani S, Morisawa S and Pegg AE (1982) Occurrence and induction of spermidine-N1-acetyltransferase in Escherichia coli. Biochem Biophys Res Commun 106: 1155–1160. 10.1016/0006-291x(82)91233-5 [DOI] [PubMed] [Google Scholar]
- Michael AJ (2016) Biosynthesis of polyamines and polyamine-containing molecules. Biochem J 473: 2315–2329. 10.1042/BCJ20160185 [DOI] [PubMed] [Google Scholar]
- Michael AJ (2018) Polyamine function in archaea and bacteria. J Biol Chem 293: 18693–18701. 10.1074/jbc.TM118.005670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Kashiwagi K, Ito K, Watanabe S and Igarashi K (1993) Estimation of polyamine distribution and polyamine stimulation of protein synthesis in Escherichia coli. Arch Biochem Biophys 300: 63–68. 10.1006/abbi.1993.1009 [DOI] [PubMed] [Google Scholar]
- Muhlig A, Behr J, Scherer S and Muller-Herbst S (2014) Stress response of Salmonella enterica serovar Typhimurium to acidified nitrite. Appl Environ Microbiol 80: 6373–6382. 10.1128/AEM.01696-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Bearson B, Bang SH, Bang IS and Foster JW (1996) Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol Microbiol 20: 605–611. 10.1046/j.1365-2958.1996.5441070.x [DOI] [PubMed] [Google Scholar]
- Pegues DA and Miller SI (2010) Salmonella species, including Salmonella Typhi. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Mandell GL, Bennett JE and Dolin R (eds). Philadelphia, PA: Churchill Livingstone, pp. 2887–2903. [Google Scholar]
- Pi H, Wendel BM and Helmann JD (2020) Dysregulation of Magnesium Transport Protects Bacillus subtilis against Manganese and Cobalt Intoxication. J Bacteriol 202. 10.1128/JB.00711-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A, Sahara J, Kawai G, Yamamoto K, Ishihama A, Uemura T, et al. (2020) Cytotoxic Mechanism of Excess Polyamines Functions through Translational Repression of Specific Proteins Encoded by Polyamine Modulon. Int J Mol Sci 21. 10.3390/ijms21072406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarin P, Barbet A, Delga S, Joshi V, Hamon L, Lefevre J, et al. (2010) A central role for polyamines in microtubule assembly in cells. Biochem J 430: 151–159. 10.1042/BJ20091811 [DOI] [PubMed] [Google Scholar]
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. (2011) Foodborne Illness Acquired in the United States-Major Pathogens. Emerging Infectious Diseases 17: 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Hernandez VJ and Reitzer L (2013) Putrescine catabolism is a metabolic response to several stresses in Escherichia coli. Mol Microbiol 88: 537–550. 10.1111/mmi.12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL and Reitzer L (2012) Pathway and enzyme redundancy in putrescine catabolism in Escherichia coli. J Bacteriol 194: 4080–4088. 10.1128/JB.05063-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D and D’Ari R (2007) Escherichia coli physiology in Luria-Bertani broth. J Bacteriol 189: 8746–8749. 10.1128/JB.01368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman ML and Enquist LW (1984) Experiments with gene fusions. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Slauch JM (2011) How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol. Microbiol 80: 580–583. 10.1111/j.1365-2958.2011.07612.x [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soksawatmaekhin W, Kuraishi A, Sakata K, Kashiwagi K and Igarashi K (2004) Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol Microbiol 51: 1401–1412. 10.1046/j.1365-2958.2003.03913.x [DOI] [PubMed] [Google Scholar]
- Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, et al. (2015) RNA-seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella Typhimurium. PLoS Pathog 11: e1005262. 10.1371/journal.ppat.1005262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, et al. (2001) Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A 98: 9901–9906. 10.1073/pnas.161295398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW and Tabor H (1985) Polyamines in microorganisms. Microbiol Rev 49: 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor CW and Tabor H (1987) The speEspeD operon of Escherichia coli. Formation and processing of a proenzyme form of S-adenosylmethionine decarboxylase. J Biol Chem 262: 16037–16040. [PubMed] [Google Scholar]
- Tsolis RM, Adams LG, Ficht TA and Baumler AJ (1999) Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect Immun 67: 4879–4885. 10.1128/IAI.67.9.4879-4885.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H and Fang FC (2000) Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192: 227–236. 10.1084/jem.192.2.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vine CE and Cole JA (2011) Unresolved sources, sinks, and pathways for the recovery of enteric bacteria from nitrosative stress. FEMS Microbiol Lett 325: 99–107. 10.1111/j.1574-6968.2011.02425.x [DOI] [PubMed] [Google Scholar]
- Wallis TS and Galyov EE (2000) Molecular basis of Salmonella-induced enteritis. Mol Microbiol 36: 997–1005. 10.1046/j.1365-2958.2000.01892.x [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Miwa Y, Miyoshi K, Furuyama J and Ohmori H (1997) The Escherichia coli ldcC gene encodes another lysine decarboxylase, probably a constitutive enzyme. Genes Genet Syst 72: 167–172. 10.1266/ggs.72.167 [DOI] [PubMed] [Google Scholar]
- Zhou DG and Galan J (2001) Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes and Infection 3: 1293–1298. 10.1016/S1286-4579(01)01489-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.