Abstract
The severity of human mucopolysaccharidosis type VII (MPS VII), or Sly syndrome, depends on the relative activity of the enzyme β-glucuronidase. Loss of β-glucuronidase activity can cause hydrops fetalis, with in utero or postnatal death of the patient. In this report, we show that β-glucuronidase activity is not detectable by a standard fluorometric assay in C3H/HeOuJ (C3H) mice homozygous for a new mutation, gusmps2J. These gusmps2J/gusmps2J mice are born and survive much longer than the previously characterized β-glucuronidase-null B6.C-H-2bm1/ByBir-gusmps (gusmps/gusmps) mice. Northern blot analysis of liver from gusmps2J/gusmps2J mice demonstrates a 750-bp reduction in size of β-glucuronidase mRNA. A 5.4-kb insertion in the Gus-sh nucleotide sequence from these mice was localized by Southern blot analysis to intron 8. The ends of the inserted sequences were cloned by inverse PCR and revealed an intracisternal A-particle (IAP) element inserted near the 3′ end of the intron. The sequence of the long terminal repeat (LTR) regions of the IAP most closely matches that of a composite LTR found in transposed IAPs previously identified in the C3H strain. The inserted IAP may contribute to diminished β-glucuronidase activity either by interfering with transcription or by destabilizing the message. The resulting phenotype is much less severe than that previously described in the gusmps/gusmps mouse and provides an opportunity to study MPS VII on a genetic background that clearly modulates disease severity.
The lysosomal enzyme β-glucuronidase is present in virtually all tissues and is responsible for the degradation of glycosaminoglycans such as heparan, dermatan, and chondroitin sulfates (32). Deficiency of β-glucuronidase in humans and in the B6 mouse strain described previously (3) causes glycosaminoglycan accumulation and lysosomal distention, resulting in a progressive multisystem degenerative disease. Both humans and mice lacking β-glucuronidase suffer from decreased mobility, mental retardation, compromised functioning of most organs, and decreased life span. Death from unknown causes occurs in ∼5% of β-glucuronidase-null gusmps/gusmps mice prior to weaning, and 90% of the survivors die by 240 days of age (3). Human mucopolysaccharidosis type VII (MPS VII) is a heterogeneous disease subdivided into three categories: a severe and fatal course pre- or postnatally often associated with edema; a stable clinical course diagnosed postnatally or in early childhood; and a milder course with diagnosis near adolescence (31). β-Glucuronidase activity appears to be markedly reduced in patients with the severe disease phenotype (14, 38, 42).
In normal mice, the expression of β-glucuronidase is the product of interactions between the closely linked genes of the Gus gene complex on chromosome 5. This complex includes the structural gene, Gus-s, and regulators of both temporal expression (Gus-t) and rates of enzyme synthesis (Gus-u) (19, 23). In addition, an androgen-responsive element in intron 9 of the structural gene determines a specific allele’s ability to respond to androgen induction (18). The rigorous genetic and biochemical characterization of a number of alleles at each of these loci makes the expression of Gus one of the most interesting and well understood in mouse biology.
Here we characterize the phenotype and β-glucuronidase levels in a new MPS VII mutant mouse and identify an intracisternal A-particle (IAP) element insertion in the Gus structural gene of these animals. The IAP elements are members of a provirus-like family of sequences with approximately 2,000 copies present in the mouse genome (15) and have been shown to participate in a number of germ line mutations (2, 8, 11, 13, 16, 21, 41). Despite an absence of detectable β-glucuronidase activity, the new gusmps2J/gusmps2J mice have a pathophysiology very different from that in the previously described gusmps/gusmps mice. Normal mice from these two inbred strains differ with respect to the complex of genes that they carry at the Gus locus. B6 animals carry the thermostable Gus-sb structural allele and an allele of Gus-u that promotes a high rate of enzyme synthesis. C3H animals have a thermolabile Gus-sh gene product, the synthesis of which is decreased temporally in several tissues by Gus-t and is not strongly promoted by the associated allele of Gus-u (19). The possibility that phenotypic differences in disease expression are related to genotype or to the mutations is discussed, as is the utility of the two stocks in dissecting disease parameters.
MATERIALS AND METHODS
Animals.
The mice used in these experiments were maintained in the research colony of E. H. Birkenmeier. All genotypes on the original B6.C-H-2bm1/ByBir background (B6 +/+, +/gusmps, and gusmps/gusmps) were obtained from matings of +/gusmps animals. Normal animals of the C3H/HeOuJ line (C3H +/+) came from the production colony of The Jackson Laboratory. Both +/gusmps2J and gusmps2J/gusmps2J animals were obtained by mating homozygous mutant and heterozygous littermates. The mice were identified either by biochemical assay or by PCR (3, 29). DNA for PCR-based typing of C3H +/+, +/gusmps2J, and gusmps2J/gusmps2J animals was prepared as described previously (25) from 1- to 2-mm samples of tail tissue. Approximately 100 ng of DNA was used in a total reaction volume of 100 μl with 1.5 mM MgCl2 and a combination of three oligonucleotides, two complementary to sequences in intron 8 of the β-glucuronidase gene (5′F and 3′R, described below and in Fig. 4) and 6921, an oligonucleotide complementary to sequences in the IAP long terminal repeat (LTR). The sequence of 6921 is 5′-TGGGCTGCAGCCAATCAGGGA-3′. C3H +/+ and +/gusmps2J DNAs used as a template produce a fragment of 510 bp with oligonucleotides 5′F and 3′R. A second, 316-bp fragment is generated by oligonucleotides 6921 and 3′R on +/gusmps2J DNA. DNA from gusmps2J/gusmps2J animals produces only the 316-bp fragment. The PCR was run for 30 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 1 min.
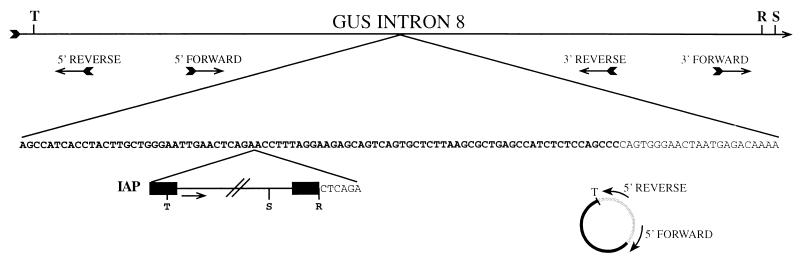
FIG. 4.
Schematic showing the insertion site of the IAP in intron 8 of the β-glucuronidase gene and the general method of inverse PCR cloning. The top line represents a fragment of intron 8, with the locations and orientations of the oligonucleotides indicated below; pertinent restriction enzyme sites are indicated above the line (R, RsaI; S, SspI; T, TaqI). Below, partial sequence of the intron shows the IAP integration site. Nucleotides in bold are part of the B2 repeat element. The remaining nucleotides represent nonrepetitive intron 8 sequence. The integrated IAP is represented at the bottom, flanked by its LTR sequences (black boxes). The direction of IAP transcription is indicated by the arrow. Restriction sites used in cloning are shown below the sequence. The six bases flanking the IAP represent the genomic sequence duplicated when the IAP integrated. An example of the mechanism by which insert sequence was obtained by inverse PCR is represented below the IAP. The circularization of the TaqI fragment at the 5′ end of the IAP element is shown. Intron 8-derived sequences are represented by the thin line, while those derived from the IAP LTR are indicated by the heavy line. Digestion of genomic DNA with TaqI cuts at the site adjacent to the oligonucleotide 5′ REVERSE (see cartoon above) and again at the site indicated in the 5′ LTR of the IAP. Subsequent circularization of that fragment via ligation produces the molecule shown, which includes sequences complementary to both 5′ REVERSE and 5′ FORWARD. Note that circularization places these oligonucleotides closer together and in an orientation suitable for PCR.
Biochemical and genetic characterization.
Brain, kidney, liver, and spleen samples from normal control animals (B6 +/+ and C3H +/+, four each), from +/gusmps2J animals (five each), and from both gusmps/gusmps and gusmps2J/gusmps2J animals (five each) were analyzed for the activity of lysosomal enzymes β-glucuronidase, α-galactosidase, and β-hexosaminidase as described elsewhere (3, 12).
To ascertain if the new mutation was allelic with gusmps, +/gusmps and gusmps2J/gusmps2J animals were mated. Offspring were scored for the facial dysmorphism characteristic of mice lacking β-glucuronidase. Kidney, liver, and spleen samples from phenotypically normal and mutant offspring were assayed for β-glucuronidase levels and evaluated for lysosomal storage by light microscopy as described previously (3).
DNA analysis.
DNA was isolated from spleens of B6 +/+ animals and of C3H +/+ and gusmps2J/gusmps2J animals by the method of Taylor and Rowe (36). DNAs were digested with restriction endonucleases, run on 0.8% agarose gels as described previously (3), transferred to nylon filters (Zetabind; AMF-Cuno, Meriden, Conn.) by the method of Southern (33), and fixed to the membrane by UV cross-linking (Stratalinker; automatic setting; Stratagene, La Jolla, Calif.). The blot was hybridized as described previously (3) to a radioactively labeled 1-kb HindIII-BamHI fragment of mouse β-glucuronidase cDNA (GUS cDNA), designated pGUS-1 (24), that includes exons 4 to 10.
Inverse PCR.
Ten-microgram samples of genomic DNA from B6 +/+, C3H +/+, and gusmps2J/gusmps2J animals were digested individually with TaqI. DNA from λ phage containing the full-length normal B6 allele (29) was digested in parallel as a positive control. Following digestion, reaction mixtures were extracted with chloroform, the DNA was precipitated with ethanol, and the pellets were washed twice with ethanol and dried under vacuum. Four micrograms of each DNA at a concentration of 6 to 7 ng per μl to favor intramolecular ligation was used in the following ligation mixture: 50 mM Tris (pH 7.5), 10 mM MgCl2, 1 mM ATP, 10 mM dithiothreitol, and 1,600 U of T4 DNA ligase (New England Biolabs) in a total volume of 600 μl. Ligation was carried out for 18 h at 16°C. Sodium acetate was added to 300 mM, and the DNAs were precipitated, washed, and dried as described above. Pellets were dissolved in 60 μl of TE (10 mM Tris [pH 7.5], 1 mM EDTA) and dialyzed against 10 ml of TE while floating on 0.025-μm-pore-size VS filters (Millipore catalogue no. VSWP 025 00) for 20 min to remove dithiothreitol. Samples were transferred to clean tubes, and 20 μl of each (approximately 1.25 μg of DNA) was used in PCR under the following conditions: 1 μM each appropriate forward and reverse oligonucleotide (for sequences, see below; The Great American Gene Company, Ramona, Calif.), 0.25 mM deoxynucleoside triphosphates, 2.5 U of Taq polymerase (Perkin-Elmer Cetus), and 1× PCR buffer with 1.5 mM MgCl2 (Perkin-Elmer Cetus) in a total volume of 50 μl. Reactions were run under the following conditions: 60 s at 95°C, followed by 30 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 65°C, and extension for 1 min at 72°C. Four 30-bp oligomers complementary to nonrepetitive intron 8 sequences of the B6 normal allele were designed for use in PCR. Two sequences 5′ and two 3′ to the insertion site were selected, the approximate locations of which are shown in Fig. 4. Oligonucleotides designated “F” (forward) read 5′ to 3′ in the direction of transcription; those designated “R” (reverse) read in the opposite orientation. Oligonucleotides designated “5′” are complementary to intronic sequences 5′ to the insertion site and were used to amplify sequences derived from TaqI-digested DNA; those designated “3′” are complementary to sequences 3′ to the insertion site and were used to amplify sequences derived from RsaI- or SspI-digested DNA. The sequences of the oligonucleotides are as follows: 5′F, 5′-CCAGAACAAGATCTCAATCGGGTAGCACAG-3′; 5′R, 5′-CCGGATGGTCGTAACACATGGCTTTCATCA-3′; 3′F, 5′-AATCGGCCGTTTTCAGCCTTCCTAACACTG-3′; and 3′R, 5′-TAGCATGCACAGGCAAGGCCCTGAACTGGA-3′.
Sequence analysis.
The nucleotide sequence of each of the PCR products was obtained by the sequencing service of The Jackson Laboratory. The oligonucleotides used in PCR served as sequencing primers. Sequence analysis was performed by using the Sequencher program, version 3.0.
Long PCR.
Long PCR of genomic DNA was carried out with the Expand Long Template PCR system (Boehringer Mannheim) and oligonucleotides (5′F and 3′R, described above) flanking the insertion site.
Northern blot analysis.
Total RNA was prepared from livers of B6 +/+, C3H +/+, gusmps/gusmps, and gusmps2J/gusmps2J animals by the method described by Chomczynski and Sacchi (6). Poly(A)+ RNA was isolated with a Poly (A) Quik mRNA isolation kit (Stratagene), and 5 μg was run on a 1% agarose gel containing formaldehyde as described previously (26). The RNA was transferred and fixed to a Nytran nylon filter (Schleicher & Schuell, Keene, N.H.) and hybridized to a 623-bp ClaI fragment containing exons 4 to 7 of the mouse GUS cDNA (7) prepared as described above. The relative amount of RNA loaded per lane was assessed by stripping and rehybridizing the blot with a 426-bp probe that detects the message encoding mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Signal intensity differences between lanes were quantified with a Molecular Dynamics (Sunnyvale, Calif.) PhosphorImager.
RT-PCR.
Combined reverse transcription-PCR (RT-PCR) was performed as described previously (27), using an annealing temperature of 60°C, a total reaction volume of 25 μl, and 1 μg of total kidney RNA template prepared as described above. Oligonucleotides were chosen to amplify exonic sequences upstream of the IAP insertion (exons 6 to 8), downstream of the insertion (exons 9 to 11), and spanning the insertion (exons 6 to 11). The oligonucleotides used in RT-PCR are listed below, with the number of the oligonucleotide corresponding to the number of the exon to which it is complementary. As before, oligonucleotides designated “F” read 5′ to 3′ in the direction of transcription and those designated “R” read in the opposite orientation. Their sequences are as follows: 6F, 5′-CCAAGGGGTCAACAAGCA-3′; 8R, 5′-GGTTTCAGAGCAGAGGAA-3′; 9F, 5′-ACACCAAAGCCCTGGACC-3′; and 11R, 5′-TTCGTCATGAAGTCGGCG-3′. The expected sizes of the products on an RNA template are as follows: exons 6 to 8, 339 bp; exons 9 to 11, 379 bp; and exons 6 to 11, 750 bp. The expected product sizes resulting from genomic DNA contamination of the RNA preparations are as follows: exons 6 to 8, 642 bp; exons 9 to 11, 5,501 bp; and exons 6 to 11, 8,426 bp. While only the 642-bp product could reasonably be expected to be produced in these reactions, it would easily be distinguishable from the RNA-derived product (339 bp) in the stained gel and would be detected by the probe during subsequent Southern hybridization. Control reactions to test the quality of the RNA templates were performed with oligonucleotides that amplify a 650-bp fragment of the Rab5b mRNA, which is expressed in the kidney. Template negative controls were run for each oligonucleotide pair. The entire 25-μl reaction volume was loaded on a 2% composite gel (1.5%/0.5% NuSieve [FMC Corp., Rockland, Maine]/agarose) containing 1× Tris-borate-EDTA and 0.75 ng of ethidium bromide per ml and separated by electrophoresis. An additional identical set of reactions was run on a 1.4% agarose gel, blotted onto Zetabind, and hybridized to an exon 6–11 fragment generated by PCR from the pGUS-1 template.
RESULTS
Genetic, histopathologic, and biochemical characterization identified a new MPS VII mutant.
The founder male mouse from the C3H/HeOuJ production colony of The Jackson Laboratory was distinguished from his littermates by a blunter face and shorter, thickened limbs and tail (Fig. 1). The male was sacrificed, and sperm was obtained for in vitro fertilization of C3H +/+ eggs. Mating of the resulting progeny produced 21 phenotypically mutant animals from 86 total offspring (24.4%), consistent with a recessive mutation.
FIG. 1.
Photograph, left to right, of gusmps/gusmps and normal B6 animals (14 weeks of age) and normal C3H and gusmps2J/gusmps2J animals (12 weeks of age). Note the overall decreased size of the mutant animals relative to their respective +/+ littermates. Also apparent are the blunted features of both mutant animals, one of the criteria initially used to identify mutant animals in their colonies of origin and to distinguish gusmps/gusmps2J progeny from +/gusmps2J in the test cross (+/gusmps × gusmps2J/gusmps2J). Note that the face of the normal C3H animal is blunted relative to that of the B6 +/+ mouse, perhaps due either to a combination of genetic differences between these two strains or specifically to the decreased constitutive levels of β-glucuronidase characteristic of C3H strains.
The mating of +/gusmps and gusmps2J/gusmps2J animals to determine whether the new mutation was allelic with gusmps produced 10 offspring of two phenotypic classes. Six appeared normal, and four had the characteristic blunted face and shortened limbs of an animal deficient in β-glucuronidase. Assay of β-glucuronidase activity in the kidney, liver, and spleen showed that phenotypically mutant animals had <1% of the activity of the phenotypically normal animals (data not shown). Tissue sections examined by light microscopy revealed no apparent storage in the phenotypically normal animals, while the mutant animals’ tissues contain distended lysosomes characteristic of β-glucuronidase deficiency (Fig. 2). Based on these data, we conclude that the C3H and B6 mutations affect the same gene. The mutant C3H allele has been designated C3H/HeOuJ-gusmps2J (gusmps2J).
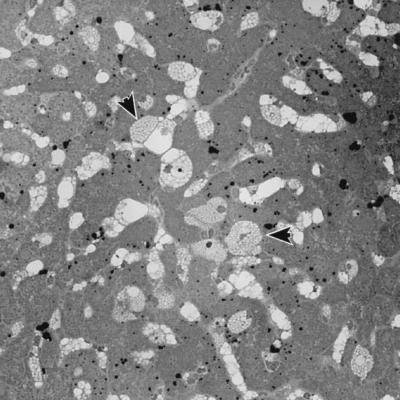
FIG. 2.
Light photomicrograph of liver tissue from a 36-week-old phenotypically mutant animal from the +/gusmps × gusmps2J/gusmps2J test cross. Note the distended lysosomes (arrows), typical of tissue from an animal with β-glucuronidase deficiency.
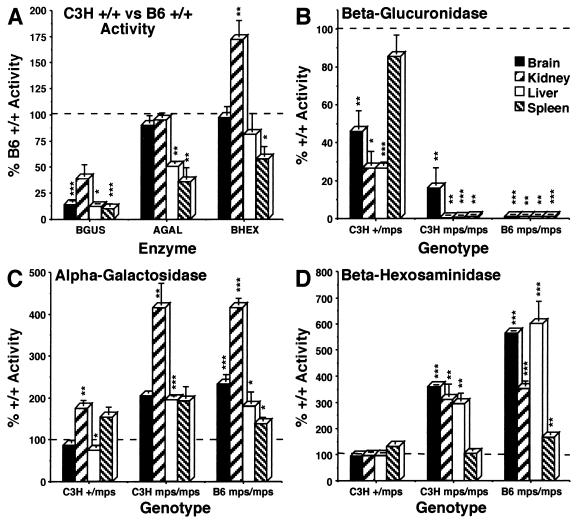
To compare the relative levels of β-glucuronidase in mutant animals and their normal littermates, lysosomal enzyme activity was analyzed in brains, kidneys, livers, and spleens of B6 +/+ and gusmps/gusmps and C3H +/+, +/gusmps2J, and gusmps2J/gusmps2J animals (Fig. 3). Specific activity was determined for β-glucuronidase and for α-galactosidase and β-hexosaminidase, two enzymes whose levels typically increase as a secondary response to diminished β-glucuronidase activity (4). Figure 3A shows the levels of these enzymes in the tissues of C3H +/+ animals, expressed as a percentage of B6 +/+ activity. Activity values for β-glucuronidase range from 10% of the B6 +/+ value in normal C3H spleen to 38% in kidney, confirming the difference in basal enzyme levels between these inbred strains. The levels of α-galactosidase and β-hexosaminidase in C3H tissues suggest that decreased β-glucuronidase activity represents a specific reduction in that enzyme and not a generalized suppression of lysosomal enzymes. The nearly twofold increase of β-hexosaminidase in C3H +/+ kidney relative to that in B6 +/+ tissue could reflect a secondary elevation of that enzyme in the normal animal in response to constitutively low β-glucuronidase activity.
FIG. 3.
Lysosomal enzyme (β-glucuronidase [BGUS], α-galactosidase [AGAL], and β-hexosaminidase [BHEX]) levels for brain, kidney, liver, and spleen. In each graph, 100% normal activity is indicated by a horizontal bar. (A) Difference in basal enzyme activities between normal animals of the C3H (bars) and B6 (horizontal line) strains. The values for C3H +/+ animals were calculated by taking the B6 +/+ level as 100% activity. (B to D) Values for tissues from animals of the indicated genotype (y axis) were calculated by reference to the normal values from the strain of origin. C3H +/+ activity was defined as 100% in calculating the values for +/gusmps2J and gusmps2J/gusmps2J animals, and the B6 +/+ level was defined as 100% for the gusmps/gusmps animals; n = 5 for all genotypes and tissues assayed, except n = 4 for all +/+ animals. The levels of significance of P values calculated by using a two-tailed Student t test are indicated as follows: P ≤ 0.001, ∗∗∗; P ≤ 0.01, ∗∗; P ≤ 0.02 to 0.05, ∗; P > 0.05, no notation.
Figures 3B to D depict β-glucuronidase activities and secondary elevations of α-galactosidase and β-hexosaminidase for +/gusmps2J animals and for both gusmps/gusmps and gusmps2J/gusmps2J animals. To reveal proportionate changes in enzyme levels, specific activities were expressed as percentages of normal activity for the strain of origin. In this way, the differences depicted represent the changes produced by each mutation within its own genetic background. Figure 3B showed that mps2J, like mps (3), has an effect in heterozygous animals, reducing β-glucuronidase activity to 26 to 85% of the normal level, depending on the tissue. Both gusmps/gusmps and gusmps2J/gusmps2J animals had β-glucuronidase activity levels less than 1% of levels for control animals. (The relatively high level of β-glucuronidase detected in gusmps2J/gusmps2J brain reflects a higher enzyme activity in one of the animals tested; the remaining animals all had levels well below 1%. Whether this represents the activity of another enzyme or compound acting on the substrate in these samples or reflects authentic β-glucuronidase enzyme activity is unknown.) Figures 3C and D show that the characteristic secondary elevations of α-galactosidase and β-hexosaminidase in gusmps/gusmps tissues are recapitulated in the tissues of the gusmps2J/gusmps2J mouse. The similarity in the degree of α-galactosidase elevation in the tissues of both mutants is striking. α-Galactosidase displays the only apparent secondary elevation in the +/gusmps2J tissues.
DNA analysis predicts an insertion within intron 8.
The probe hybridized to Southern blots to examine gusmps2J/gusmps2J DNA for gross sequence alterations detected identical restriction fragments in DNA from both B6 and C3H normal animals but an alteration in the size of some fragments from the mutant (data not shown). The sizes of the altered fragments suggested the presence of an insertion with a minimum size of 2.4 kb in the mutant DNA. Based on the altered restriction fragment pattern and the published sequence of Gus-sa (7;GenBank accession no. J02836), the insertion was localized to a 422-bp region of intron 8 between EcoRI and HindIII restriction sites. Further analysis of mutant DNA was performed with restriction enzymes selected on the basis of the location of their recognition sites relative to the putative insertion (data not shown). These analyses suggested that the minimum size of the insert was 5.4 kb.
Cloning identifies an insertion in the C3H mutant DNA.
Oligonucleotide primers 5′F and 3′R (Fig. 4) produce the predicted 510-bp fragment in PCR using DNA from normal B6 and C3H animals. No product visible by ethidium bromide staining was generated from gusmps2J/gusmps2J DNA. The failure to generate a product might be due to the increased length of the intervening DNA, although loss of the complementary primer site in the mutant DNA could not be ruled out. Because the insertion was flanked by repetitive DNA and was of unknown sequence and length, the ends of the insert were cloned by inverse PCR.
The PCR primer 5′R is located 330 bp upstream of 5′F and in the opposite orientation (Fig. 4). TaqI digestion of genomic DNA from a normal B6 or C3H animal, and from the cloned control DNA, produced a 929-bp TaqI fragment that retained sequences complementary to both of these oligonucleotides. When circularized by ligation and used as a template for PCR with 5′R and 5′F (Fig. 4), this TaqI fragment generated a product 599 bp in length. TaqI digestion of C3H mutant DNA, however, produced different results. A band of approximately 850 bp was generated, suggesting, since the intronic TaqI site remains unchanged, that the alteration in length was due to the inclusion of sequences from the insert DNA up to the first TaqI site. Sequence analysis of the isolated PCR product was performed with the PCR primers that generated the fragment. Sequence produced with primer 5′F revealed 345 bp of known intron 8 sequence followed by sequences from the 5′ end of the insert DNA. 5′R-derived sequence reads through 42 bp of intronic sequence before reaching the TaqI site of the insert and adjacent insert sequence.
A strategy similar to that outlined above was employed to obtain sequence from the 3′ end of the insert, using oligonucleotide 3′F, located 119 bp downstream of 3′R (Fig. 4). RsaI and SspI restriction enzyme sites flanking these primer sites were chosen. In total, 500 to 600 bp of DNA from the 3′ end of the insertion was obtained.
The insertion is an IAP element.
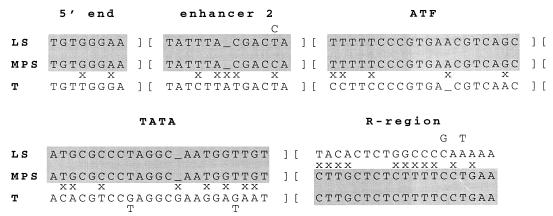
Comparison of the insert sequences with GenBank entries showed that the insertion was an IAP element. The sequences closely matched those of the LS subclass (22) of IAP elements in the U3 region of the LTR. In Fig. 5, the parts of the U3 region that are characteristic of this subclass are compared with the sequence of the element inserted in gusmps2J. The LS subclass was defined by sequencing 37 IAP cDNAs expressed in thymus and lipopolysaccharide-stimulated B cells; 34 of the clones had identical U3 regions. Differences in the R region divided the LS elements into three related families. While the U3 region of the IAP element inserted into the β-glucuronidase gene was a nearly perfect match with the LS subclass, the R-region sequences were very different from those of all three of the LS-element family R regions. They more closely matched those found in IAP elements expressed in tumor cells, which have been designated T elements (17) (Fig. 5). This type of composite LTR has been found in six other germ line mutations caused by IAP element insertions (2, 8, 9, 11, 21, 41). While the insertion in gusmps2J has a C in the 11th position in enhancer 2 where most of the LS elements have a T, it matches those in the composite LTRs in the other germ line mutations, as well as a single LS element with a C in this position.
FIG. 5.
Sequences of LTR elements derived from LS-type (22) and T-type (17) IAP elements compared to that of the IAP element inserted in intron 8 of the gusmps2J allele. The first four blocks of sequence are from the U3 region of the LTR. Enhancer 2, ATF, and TATA refer to cis-regulatory regions of the IAP promoter which have been previously described (17). The T elements contain a canonical ATF/CRE site, while the LS elements contain a variant motif. Positions at which the elements are polymorphic show two nucleotides, with the less common ones above (for LS) and below (for T) the sequence. A mismatch (x) is not marked if any member of the class has a matching nucleotide with the sequence. Similarities of the C3H-derived sequence to the others are shaded, clearly showing the composite nature of this IAP element. It is identical to the LS-type element from the 5′ end of the LTR through the U3 region, switching to share similarity to the T-type LTR in the R region.
The IAP element was inserted in a B2 repetitive element of intron 8 in the same transcriptional orientation as the β-glucuronidase gene (Fig. 4). Extended PCR of C3H +/+ and gusmps2J/gusmps2J genomic DNA with oligonucleotides 5′F and 3′R produced bands of 510 and approximately 5,400 bp, respectively (data not shown). The size of this IAP element (5.4 kb) indicates that it is an element of the IΔ1 class. IAP elements that have retrotransposed frequently have this structure, which contains a deletion of 1.9 kb of internal sequences (15). A 6-bp target site duplication characteristic of retrotransposed IAP elements is present (CTCAGA [Fig. 4]).
Northern analysis detects a shorter message in Gus mRNA from mutant animals.
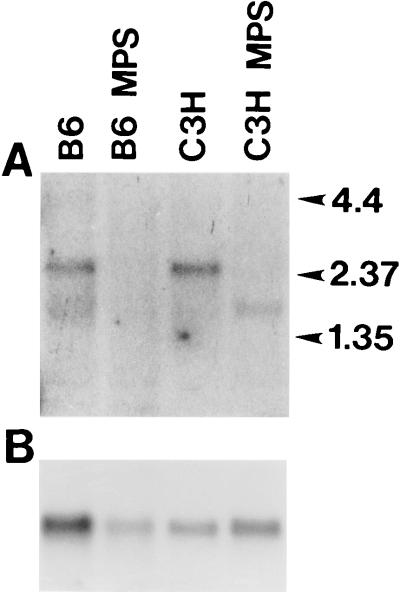
A Northern blot hybridized to a fragment of the GUS cDNA that includes exons 4 to 7 shows mRNA of the expected size in both B6 and C3H normal samples (Fig. 6). No hybridizing band of any size was detected at any exposure in the RNA derived from the B6 mutant animal. The lane containing RNA from the C3H mutant animal contains a strongly hybridizing band approximately 750 bp shorter than the normal band. Extended exposure of the blot to film shows a lightly hybridizing band that comigrates with normal message. Normalization of loading as determined by hybridization to the GAPDH probe reveals that the shorter message in the C3H mutant RNA is decreased in intensity 5- to 7-fold relative to that in the C3H normal animal, while the normal-sized band is diminished in intensity at least 20-fold.
FIG. 6.
Northern blot analysis of RNA from B6 normal (B6), B6 mutant (B6 MPS), C3H normal (C3H), and C3H mutant (C3H MPS) animals. The approximate locations of RNA molecular weight standards (Life Technologies, Inc., Gaithersburg, Md.) are indicated at the right in kilobases. (A) Results of hybridization with a fragment of the GUS cDNA including exons 4 to 7, followed by exposure to X-ray film for 96 h with intensifying screens. Note the discreteness of the shorter band relative to the normal full-length message, which suggests that this band constitutes either an equally stable or a more heavily transcribed RNA species. (B) Results of hybridization of the same blot to a mouse GAPDH probe. Data derived from this blot were used to determine the relative signal intensities of the hybridizing bands.
RT-PCR reveals a decrease in mutant transcript beyond exon 8.
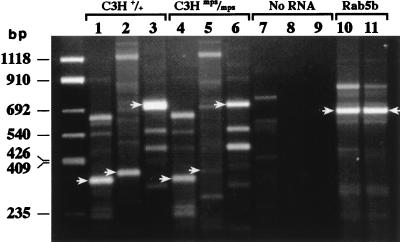
All three oligonucleotide pairs (6F-8R, 9F-11R, and 6F-11R) amplified products of the expected sizes when used in RT-PCR of total RNA from C3H +/+ kidney (Fig. 7, lanes 1 to 3) and from gusmps2J/gusmps2J kidney (lanes 4 to 6). However, products amplified by oligonucleotides from exons 9 to 11 and 6 to 11 (3′ to and spanning the insertion site, respectively) with the gusmps2J/gusmps2J RNA as the template were reduced in intensity relative to products of the +/+ RNA template (compare lanes 2 and 3 with lanes 5 and 6). Despite the presence of additional bands of various sizes in all lanes, only the bands of the sizes predicted to result from amplification of an RNA template hybridized to the cDNA probe on a Southern blot (data not shown). None of the bands staining lightly with ethidium bromide in the control lanes (no-template RNA [lanes 7 to 9]) hybridized to the probe, demonstrating that products in the other reactions were not due to low-level contamination of the reagents. The +/+ and gusmps2J/gusmps2J RNA templates produced bands of equal intensity in the control reactions using the Rab5b oligonucleotide pair (lanes 10 and 11), suggesting that the two RNA preparations were of roughly equal quality as templates. Because of the order in which products were loaded on the gel, products detected in one lane could not result from carryover during loading of the adjacent lane, as that product would be of a different size.
FIG. 7.
RT-PCR of C3H +/+ and gusmps2J/gusmps2J (mps/mps) RNA. Lanes 1 to 3 represent RT-PCR products from exons 6 to 8 (339 bp), exons 9 to 11 (379 bp), and exons 6 to 11 (750 bp) amplified from C3H +/+ RNA. Products from similar reactions loaded in the same order but amplified from C3H gusmps2J/gusmps2J RNA are shown in lanes 4 to 6. Lanes 7 to 9 represent the no-RNA template control reactions; lanes 10 and 11 show the positive control Rab5b product amplified from C3H +/+ and gusmps2J/gusmps2J RNA templates, respectively, demonstrating that the two RNA preparations serve equally well as templates. In all lanes, the arrows indicate the predicted size of fragments generated by specific oligonucleotide pairs. In lanes 1 to 6, the same arrows indicate the locations of bands corresponding in size to those detected by the GUS cDNA probe on a Southern blot of these reactions (data not shown). Left lane, pUC19 Marker (BIOSYNTHESIS, Inc., Lewisville, Tex.).
DISCUSSION
The propensity of IAP elements to transpose in the mouse genome has provided mutants useful in the elucidation of normal gene function. Many of these integrations involve a specific type of IAP element undergoing transposition on the C3H/HeOuJ inbred background. Four independent integrations in the agouti locus produced the mutations Avy, Aiy, Ahvy, and Aiapy (2, 8, 21). All of these integrations left the agouti coding sequence itself intact but caused ectopic overexpression of the gene product, providing a variety of insights into the regulation of gene expression and the role of methylation in its control. Integration of IAP sequences into the coding sequences of the genes ep and Lamb3 (11, 16) has generated mouse models for the human diseases Hermansky-Pudlak syndrome, a platelet storage deficiency disease, and Herlitz junctional epidermolysis bullosa, an often serious skin disorder characterized by severe to lethal blistering of the skin. The integration at the ep locus does not alter the quantity of mRNA produced but replaces the final 46 amino acids of the normal transcript with 78 amino acids derived from the IAP. The mutation Lamb3ap, as well as Axinkb (37) and Relnrl (28), produces changes in gene transcripts that result either in the replacement of normal coding sequence with IAP-derived sequence or in exon skipping. In the LamB3 and Relnrl mutants, message destabilization is known or postulated to result. Except for Axinkb, all of the mutations described above occurred in animals derived from the C3H/HeJ inbred background.
Mutagenic retrotransposition of IAP elements does not result exclusively from integrations into known exons or regulatory regions. A recently described mutation at the vibrator locus (13) is caused by the insertion of an IAP element into intron 4 of that gene. The insertion occurred in the same transcriptional orientation as the gene, similar to the gusmps2J insertion. Sequence analysis of the complete genomic DNA encoding the normal and mutant vibrator genes demonstrated that no other mutations distinguished these alleles. While the size of the transcript from the mutant vibrator allele appears to be unaltered, the amount of the transcript is reduced by 80 to 85%. Another mutation attributed to intronic IAP element integration involves the fused locus (37). The allele AxinFu contains an IAP insertion in intron 6 postulated to be responsible for this semidominant gain of function mutation. Normal and aberrant proteins are postulated to be produced from messages of normal length and from detected chimeric messages that contain IAP-derived sequences.
The gusmps2J allele described in this report contains an IAP element insertion in intron 8 of the sequence encoding the enzyme β-glucuronidase. Levels of the enzyme are reduced to below 1% of normal in animals homozygous for the mutation, resulting in phenotypic changes characteristic of the disease caused by β-glucuronidase deficiency, MPS VII. The disease phenotype is recapitulated in animals that are compound heterozygotes for the mutant C3H allele and a previously characterized null C57BL/6J-derived allele. While no direct evidence supports the involvement of the IAP insertion in changes in size and abundance of mRNA detected in the new mutant, several observations strongly implicate it. By Southern analysis, no gross alterations in the Gus-sh gene were detected in any fragments except those containing the insertion site in intron 8. RT-PCR detected transcript fragments of normal size from mutant mRNA from exons 5′ to the insertion (exons 6 to 8), 3′ to the insertion (exons 9 to 11), and spanning the insertion (exons 6 to 11). Similar results were obtained by RT-PCR of transcripts from the mutant AxinFu and vibrator alleles. While of normal size, the intensity of bands derived from sequences both 3′ to and spanning the insertion was consistently less than that of bands derived from sequence 5′ to the insertion in multiple replicates of these experiments. Finally, unique characteristics of the normal β-glucuronidase message may explain the possibly devastating effects of message destabilization on enzyme levels. It has been estimated that in a non-androgen-induced cell, the gene encoding β-glucuronidase is transcribed infrequently (once every 35 to 40 h) and is present on average at a concentration of one copy per cell (39). Given these numbers, it is easy to imagine that any message destabilization could result in the minimum estimated 20-fold reduction in full-length mRNA detected by Northern blot analysis.
The greatly diminished levels of full-length message in mutant RNA and the presence of a dominant shorter RNA species detected by Northern analysis suggest that either (i) IAP sequences are not spliced into the message in these cells but decrease the efficiency of transcription beyond exon 8, (ii) IAP sequences are spliced into some percentage of mutant message and are present in the shorter transcripts detected in mutant RNA, (iii) IAP sequences indistinguishable in length by agarose gel analysis are spliced into and replace normal sequences in a small percentage of mRNA molecules, or (iv) the inserted IAP has no effect on transcription of the locus. The difference in size between the normal and truncated mutant RNA bands is slightly less than expected if the products of exons 9 to 12 were simply not included (0.75 kb versus 1.0 kb). This may result from sizing inaccuracies or may represent the inclusion of β-glucuronidase gene sequences beyond exon 8 or of IAP-derived sequences. While only determination of the complete sequence of the mutant allele will prove that no additional significant sequence alterations lie hidden within that allele, given the above-described characteristics of the mutation and similarities with the AxinFu and vibrator genes, we strongly suspect that the IAP in intron 8 plays a causative role in the mutant phenotype.
Perhaps more intriguing will be the elucidation of the RNA processing mechanisms underlying the differences between gusmps2J, AxinFu, and vibrator, whose normal RNA products are nearly absent, at nearly normal levels, and diminished by 80%, respectively. The orientation of the inserted element in the intron may have differential effects. Incorporation of IAP sequences into transcripts by use of normal or cryptic splice sites in the element could lead to RNAs of reduced stability, as was postulated to be the case for the vibrator mutation (13). Recent studies have shown that RNA stability is coupled to translation, and transcripts containing premature termination codons are rapidly degraded (20). Methylation of IAP sequences affects the expression of genes into which the IAP element integrates (2, 21). Such a mechanism could affect the transcriptional machinery’s ability to recognize the IAP element in intron 8 of the β-glucuronidase gene and allow production of small amounts of normal message in some cells. Reversion of phenotype by loss of an element has not been reported.
The presence of composite LTRs in many of the IAP elements involved in germ line mutations is noteworthy since expression of such elements has not been seen in normal somatic cells. However, insertions of elements with such LTRs have caused mutations in four independently derived tumors (1, 5, 34, 35). While expression of T-type IAP elements has not been seen in normal adult somatic cells, both the LS- and T-type IAP elements are expressed early in embryogenesis (17a) as well as in tumor cells (17). Thus, sufficient levels of both types of transcripts could be present in early embryos and tumor cells to favor recombination between LTRs in transcripts that are copackaged. Such recombination would most likely occur during reverse transcription of the RNA. The reverse-transcribed products would then be available for retrotransposition.
Regardless of the precise nature of the gusmps2J mutation, the mutant animal provides a useful counterpoint to the gusmps/gusmps mouse, as it affords an opportunity to examine the effects of other genes on the severity of disease symptoms caused primarily by the deficiency of β-glucuronidase. Evidence of the effects of other loci is clearly demonstrated by the fact that the deficiency of β-glucuronidase in C3H and B6 animals does not have identical effects on the animals’ health. Both mutants have characteristically shorter, thicker limbs, accumulated storage in the lysosomes of many tissues, and secondary elevations of other lysosomal enzymes in response to decreased levels of β-glucuronidase. However, gusmps2J/gusmps2J animals are able to breed, to bear live young, and to raise litters to weaning age. Female gusmps/gusmps animals rarely conceive, have difficulty carrying litters to term, and do not lactate. In addition, preliminary evidence from our colony of gusmps2J/gusmps2J animals suggests that their average life span will be significantly longer than that of gusmps/gusmps animals, which on average live only one-third as long as their normal littermates (3).
The C3H and B6 mutations differ not only in the overall inbred genetic backgrounds in which they arose, C3H/HeOuJ and C57BL/6J, but also in the complement of alleles that they carry at the Gus gene complex, Gush and Gusb, respectively. While the normal structural alleles of C3H and B6 animals differ in thermostability, it is not the relative thermoinstability of the C3H-derived β-glucuronidase enzyme that produces the characteristic differences in enzyme levels for these two strains. Rather, it has been shown that the rate of enzyme synthesis, under the control of two other components of the Gus complex, Gus-t and Gus-u, determines the constitutive level of β-glucuronidase in these animals (10, 19). While the B6 allele of Gus-u promotes a relatively high rate of translation of Gus mRNA, the C3H Gus-u allele does not. In addition, the C3H allele of Gus-t decreases the rate of enzyme synthesis in specific tissues and at specific times in development. Although these alleles clearly help to define the normal β-glucuronidase phenotype in B6 and C3H animals, it is unclear whether they modulate the severity of the disease phenotype.
The difference in the constitutive β-glucuronidase levels in the normal C3H and B6 animals raises interesting questions regarding the different effects of β-glucuronidase deficiency on animals of these two genetic backgrounds. Perhaps specific combinations of alleles present in the C3H background enable the +/+ animals to function normally with significantly lower constitutive levels of β-glucuronidase while at the same time allowing the gusmps2J/gusmps2J animals to more easily compensate for the enzyme’s lack. Alternatively, the formal possibility remains that residual β-glucuronidase activity, below current levels of detection but significant to the animals’ health, supports the improved phenotype of gusmps2J/gusmps2J animals. As noted above, gusmps2J/gusmps2J animals are healthier than gusmps/gusmps animals, as judged by their increased capacity to breed and raise young and by their extended life span. It is reasonable to expect that as the effect of β-glucuronidase deficiency in both these strains is further characterized, specific differences in multiorgan systems other than the reproductive system may be found to contribute to this increased longevity.
The presence of mutant structural alleles in two different β-glucuronidase haplotypes has uncovered genetic interactions related to β-glucuronidase but apparently independent of the Gus gene complex. Dissection of the effect of genes linked and unlinked to Gus will be vital to assessing the efficacy of various gene transfer therapeutic measures currently being evaluated to treat this form of mucopolysaccharidosis. Because many such protocols restore only a fraction of the normal constitutive enzyme level (4, 30, 40), the role of other genes in modulating the effect of low enzyme levels may be significant to these studies and may eventually help to explain the phenotypic heterogeneity of this disease in mice and humans.
ACKNOWLEDGMENTS
We thank Jane E. Barker and Luanne L. Peters for helpful discussions and critical reading of the manuscript and Luanne Peters and Mark Mathews for superior technical assistance.
This work was funded by NIH R01 research grants DK41082 to E. H. Birkenmeier and J. E. Barker, DK49525 to J. E. Barker, and DK53920 and HD35671 to M. S. Sands.
Footnotes
This paper is dedicated to the memory of Edward Birkenmeier, a fine scientist and good friend, for whom every mutant mouse was an opportunity and every aspect of biology a joy.
REFERENCES
- 1.Algate P A, McCubrey J A. Autocrine transformation of hematopoietic cells resulting from cytokine message stabilization after intracisternal A particle transposition. Oncogene. 1993;8:1221–1232. [PubMed] [Google Scholar]
- 2.Argeson A C, Nelson K K, Siracusa L D. Molecular basis of the pleiotropic phenotype of mice carrying the hypervariable yellow (Ahvy) mutation at the agouti locus. Genetics. 1996;142:557–567. doi: 10.1093/genetics/142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birkenmeier E H, Davisson M T, Beamer W G, Ganschow R E, Vogler C A, Gwynn B, Lyford K A, Maltais L M, Wawrzyniak C J. Murine mucopolysaccharidosis type VII; characterization of a mouse with β-glucuronidase deficiency. J Clin Investig. 1989;83:1258–1266. doi: 10.1172/JCI114010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birkenmeier E H, Barker J E, Vogler C A, Kyle J W, Sly W S, Gwynn B, Levy B, Pegors C. Increased life span and correction of metabolic defects in murine mucopolysaccharidosis type VII after syngeneic bone marrow transplantation. Blood. 1991;78:3081–3092. [PubMed] [Google Scholar]
- 5.Brigle K E, Westin E H, Houghton M T, Goldman I D. Insertion of an intracisternal A particle within the 5′-regulatory region of a gene encoding folate-binding protein in L1210 leukemia cells in response to low folate selection. J Biol Chem. 1992;267:22351–22355. [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.D’Amore M A, Gallagher P M, Korfhagen T R, Ganschow R E. Complete sequence and organization of the murine β-glucuronidase gene. Biochemistry. 1988;27:7131–7140. doi: 10.1021/bi00418a070. [DOI] [PubMed] [Google Scholar]
- 8.Duhl D M, Vrieling H, Miller K A, Wolff G L, Barsh G S. Neomorphic agouti mutations in obese yellow mice. Nat Genet. 1994;8:59–64. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 9.Feng G H, Bailin T, Oh J, Spritz R A. Mouse pale ear (ep) is homologous to human Hermansky-Pudlak syndrome and contains a rare ‘AT-AC’ intron. Hum Mol Genet. 1997;6:793–798. doi: 10.1093/hmg/6.5.793. [DOI] [PubMed] [Google Scholar]
- 10.Ganschow R E. Simultaneous genetic control of the structure and rate of synthesis of murine glucuronidase. In: Markert C L, editor. Isozymes: genetics and evolution. Vol. 4. New York, N.Y: Academic Press; 1975. pp. 633–647. [Google Scholar]
- 11.Gardner J M, Wildenberg S C, Keiper N M, Novak E K, Rusniak M E, Swank R T, Puri N, Finger J N, Hagiwara N, Lehman A L, Gales T L, Bayer M E, King R A, Brilliant M H. The mouse pale ear (ep) mutation is the homologue of human Hermansky-Pudlak syndrome. Proc Natl Acad Sci USA. 1997;94:9238–9243. doi: 10.1073/pnas.94.17.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaser J H, Sly W S. β-Glucuronidase deficiency mucopolysaccharidosis: methods for enzymatic diagnosis. J Lab Clin Med. 1973;82:969–977. [PubMed] [Google Scholar]
- 13.Hamilton B A, Smith D J, Mueller K L, Kerrebrock A W, Bronson R T, van Berkel V, Daly M J, Kruglyak L, Reeve M P, Nemhauser J L, Hawkins T L, Rubin E M, Lander E S. The vibrator mutation causes neurodegeneration via reduced expression of PITPα: positional complementation cloning and extragenic suppression. Neuron. 1997;18:711–722. doi: 10.1016/s0896-6273(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 14.Islam M R, Vervoort R, Lissens W, Hoo J J, Valentino L A, Sly W S. β-Glucuronidase P408S, P415L mutations: evidence that both mutations combine to produce an MPS VII allele in certain Mexican patients. Hum Genet. 1996;98:281–284. doi: 10.1007/s004390050207. [DOI] [PubMed] [Google Scholar]
- 15.Kuff E L, Lueders K K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 16.Kuster J E, Guarnieri M H, Ault J G, Flaherty L, Swiatek P J. IAP insertion in the murine LamB3 gene results in junctional epidermolysis bullosa. Mamm Genet. 1997;8:673–681. doi: 10.1007/s003359900535. [DOI] [PubMed] [Google Scholar]
- 17.Lueders K K, Fewell J W, Morozov V E, Kuff E L. Selective expression of intracisternal A-particle genes in established mouse plasmacytomas. Mol Cell Biol. 1993;13:7439–7446. doi: 10.1128/mcb.13.12.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Lueders, K. K., and B. B. Knowles. Unpublished data.
- 18.Lund S D, Gallagher P M, Wang B, Porter S C, Ganschow R E. Androgen responsiveness of the murine β-glucuronidase gene is associated with nuclease hypersensitivity, protein binding, and haplotype-specific sequence diversity within intron 9. Mol Cell Biol. 1991;11:5426–5434. doi: 10.1128/mcb.11.11.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusis A J, Chapman V M, Wangenstein R W, Paigen K. Trans-acting temporal locus within the β-glucuronidase gene complex. Proc Natl Acad Sci USA. 1983;80:4398–4402. doi: 10.1073/pnas.80.14.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maquat L E. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1:453–465. [PMC free article] [PubMed] [Google Scholar]
- 21.Michaud E J, van Vugt M J, Bultman S J, Sweet H O, Davisson M T, Woychik R P. Differential expression of a new dominant agouti allele (Aiapy) is correlated with methylation state and is influenced by parental lineage. Genes Dev. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 22.Mietz J A, Fewell J W, Kuff E L. Selective activation of a discrete family of endogenous proviral elements in normal BALB/c lymphocytes. Mol Cell Biol. 1992;12:220–228. doi: 10.1128/mcb.12.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paigen K. Acid hydrolases as models of genetic control. Annu Rev Genet. 1979;13:417–466. doi: 10.1146/annurev.ge.13.120179.002221. [DOI] [PubMed] [Google Scholar]
- 24.Palmer R, Gallagher P M, Boyko W L, Ganschow R E. Genetic control of levels of murine kidney glucuronidase mRNA in response to androgen. Proc Natl Acad Sci USA. 1983;80:7596–7600. doi: 10.1073/pnas.80.24.7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkin-Elmer Cetus Corporation. Rapid, efficient DNA extraction for PCR from cells or blood. Amplifications. 1989;2:1–3. [Google Scholar]
- 26.Peters L L, Turtzo L C, Birkenmeier C S, Barker J E. Distinct fetal Ank1 and Ank2 related proteins and mRNA’s in normal and nb/nb mice. Blood. 1993;81:2144–2149. [PubMed] [Google Scholar]
- 27.Peters L L, John K M, Lu F M, Eicher E M, Higgins A, Yialamas M, Turtzo L C, Otsuka A J, Lux S E. Ank3 (epithelial ankyrin), a widely distributed new member of the ankyrin gene family and the major ankyrin in kidney, is expressed in alternatively spliced forms, including forms that lack the repeat domain. J Cell Biol. 1995;130:313–330. doi: 10.1083/jcb.130.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Royaux I, Bernier B, Montgomery J C, Flaherty L, Goffinet A M. Relnrl-alb2, an allele of Reeler isolated from a chlorambucil screen, is due to an IAP insertion with exon skipping. Genomics. 1997;42:479–482. doi: 10.1006/geno.1997.4772. [DOI] [PubMed] [Google Scholar]
- 29.Sands M S, Birkenmeier E H. A single-base-pair deletion in the β-glucuronidase gene accounts for the phenotype of murine mucopolysaccharidosis type VII. Proc Natl Acad Sci USA. 1993;90:6567–6571. doi: 10.1073/pnas.90.14.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sands M S, Vogler C, Kyle J W, Grubb J H, Levy B, Galvin N, Sly W S, Birkenmeier E H. Enzyme replacement therapy for murine mucopolysaccharidosis type VII. J Clin Investig. 1994;93:2324–2331. doi: 10.1172/JCI117237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sewell A C, Gehler J, Mittermaier G, Meyer E. Mucopolysaccharidosis type VII (β-glucuronidase deficiency): a report of a new case and a survey of those in the literature. Clin Genet. 1982;21:366–373. doi: 10.1111/j.1399-0004.1982.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 32.Sly W S, Quinton B A, McAlister W H, Rimoin D L. Beta-glucuronidase deficiency: report of clinical, radiologic, and biochemical features of a new mucopolysaccharidosis. J Pediatr. 1973;82:249–257. doi: 10.1016/s0022-3476(73)80162-3. [DOI] [PubMed] [Google Scholar]
- 33.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 34.Stocking C, Loliger C, Kawai M, Suciu S, Gough N, Ostertag W. Identification of genes involved in growth autonomy of hematopoietic cells by analysis of factor-independent mutants. Cell. 1988;53:869–879. doi: 10.1016/s0092-8674(88)90329-7. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka I, Ishihara H. Unusual long target duplication by insertion of intracisternal A-particle element in radiation-induced acute myeloid leukemia cells in mouse. FEBS Lett. 1995;376:146–150. doi: 10.1016/0014-5793(95)01262-2. [DOI] [PubMed] [Google Scholar]
- 36.Taylor B A, Rowe L B. Genes for serum amyloid A proteins map to chromosome 7 in the mouse. Mol Gen Genet. 1984;195:491–499. doi: 10.1007/BF00341452. [DOI] [PubMed] [Google Scholar]
- 37.Vasicek T J, Zeng L, Guan X-J, Zhang T, Constantini F, Tilghman S M. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vervoort R, Islam M R, Sly W S, Zabot M-T, Kleijer W J, Chabas A, Fenson A, Young E P, Leibaers I, Lissens W. Molecular analysis of patients with β-glucuronidase deficiency presenting as hydrops fetalis or as early mucopolysaccharidosis VII. Am J Hum Genet. 1996;58:457–471. [PMC free article] [PubMed] [Google Scholar]
- 39.Watson G, Paigen K. Genetic variations in kinetic constants that describe β-glucuronidase mRNA induction in androgen-treated mice. Mol Cell Biol. 1987;7:1085–1090. doi: 10.1128/mcb.7.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfe J H, Sands M S, Barker J E, Gwynn B, Rowe L B, Vogler C A, Birkenmeier E H. Reversal of pathology in murine mucopolysaccharidosis type VII by somatic cell gene transfer. Nature. 1992;360:749–753. doi: 10.1038/360749a0. [DOI] [PubMed] [Google Scholar]
- 41.Wu M, Rinchik E M, Wilkinson E, Johnson D K. Inherited somatic mosaicism caused by an intracisternal A particle insertion in the mouse tyrosinase gene. Proc Natl Acad Sci USA. 1997;94:890–894. doi: 10.1073/pnas.94.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamada S, Tomatsu S, Sly W S, Islam R, Wenger D A, Fukuda S, Sukegawa K, Orii T. Four novel mutations in mucopolysaccharidosis type VII including a unique base substitution in exon 10 of the β-glucuronidase gene that creates a novel 5′ splice site. Hum Mol Genet. 1995;4:651–655. doi: 10.1093/hmg/4.4.651. [DOI] [PubMed] [Google Scholar]