Abstract
Antibiotic resistance is one of the greatest public health challenges of our time. International efforts to curb resistance have largely focused on drug development and limiting unnecessary antibiotic use. However, in areas where water, sanitation, and hygiene infrastructure is lacking, we propose that bacterial flow between humans and animals can exacerbate the emergence and spread of resistant pathogens. Here, we describe the consequences of poor environmental controls by comparing mobile resistance elements among Escherichia coli recovered from humans and meat in Cambodia, a middle-income country with substantial human–animal connectivity and unregulated antibiotic use. We identified identical mobile resistance elements and a conserved transposon region that were widely dispersed in both humans and animals, a phenomenon rarely observed in high-income settings. Our findings indicate that plugging leaks at human–animal interfaces should be a critical part of addressing antibiotic resistance in low- and especially middle-income countries.
Antibiotic resistance is one of the top threats facing humanity (WHO 2021). Antibiotic-resistant bacterial infections are already responsible for nearly 1.3 million deaths globally each year (ARC 2022), and some estimates suggest annual deaths from antimicrobial-resistant infections could reach 10 million by 2050 if this crisis remains unaddressed (O’Neill 2016). Due to a higher infectious disease burden, widespread informal antibiotic use, and limited resources for controlling antibiotic-resistant infections (Bebell and Muiru 2014; Klein et al. 2018), low- and middle-income countries (LMICs) are projected to experience the greatest mortality and economic fallout from this looming crisis (O’Neill 2016).
To address this challenge, global public health organizations are collaborating with countries to develop “One Health”-oriented national action plans aimed at improving antibiotic stewardship in both humans and animal production (WHO 2016; World Bank Group 2019). To date, the importance of environmental controls at the human–animal interface has not received the same level of attention as antibiotic use. This lack of prioritization may be due to high-income country bias, as studies from the UK (Day et al. 2019) and the EU (Mughini-Gras et al. 2019; Martak et al. 2021) have identified only minimal sharing of antibiotic-resistant bacteria between humans and food animals. However, the situation could be vastly different in low- and especially middle-income countries, where environmental controls at the human–animal interface are often deficient or lacking altogether (Figure 1).
Figure 1.
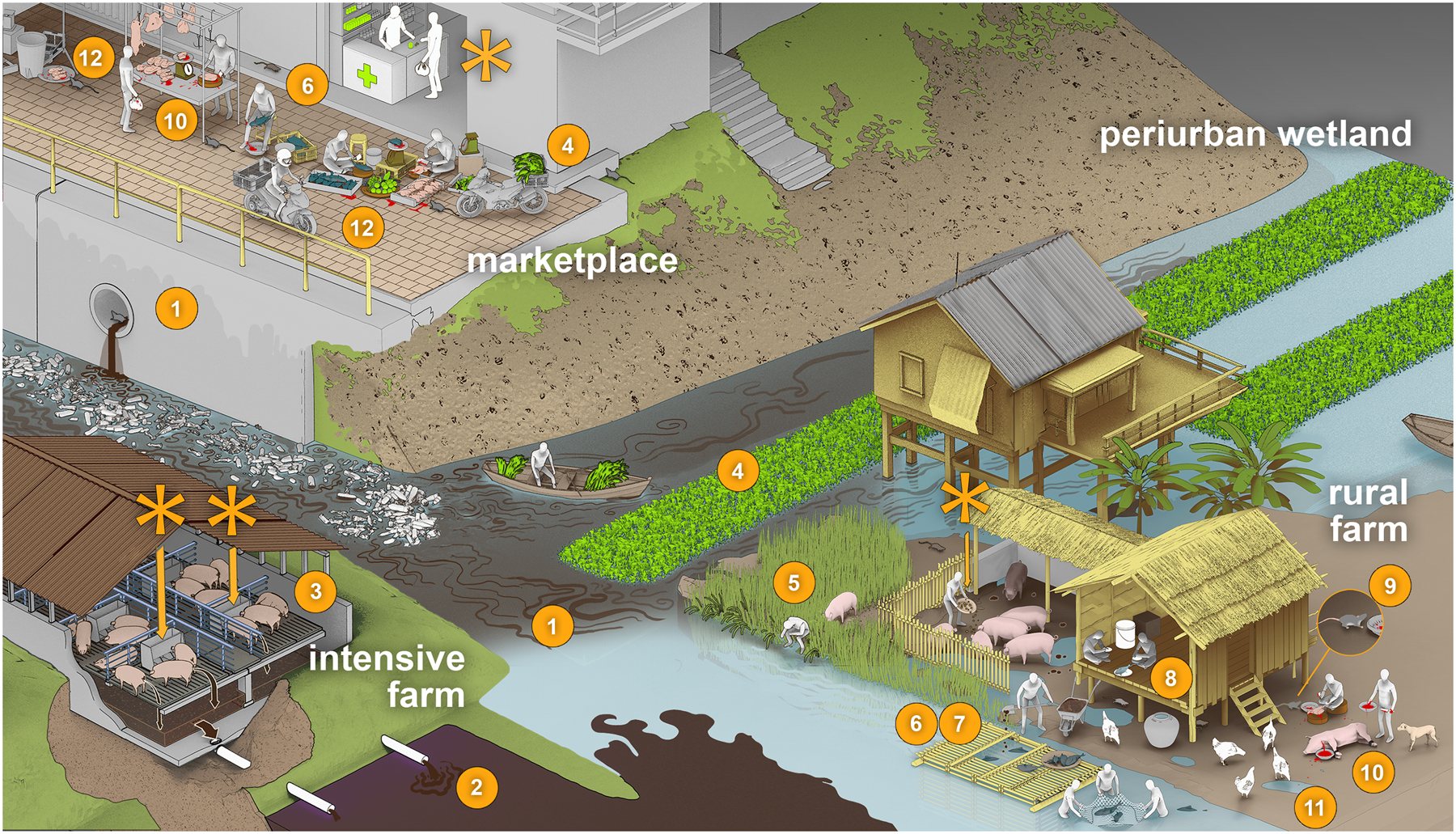
Unchecked environmental leaks and antibiotic use create myriad opportunities for the exchange of bacteria and mobile resistance elements among humans and animals. Potential transmission pathways include (1) inadequate sewage treatment infrastructure leads to contamination of human and animal drinking water; (2) animal waste contaminates drinking water; (3) raising many animals in confinement propagates disease; (4) raw sewage fertilizing crops for human consumption; (5) open defecation leads to fecal–oral transmission among humans and animals; (6) contact with animal waste; (7) farmed fish are fed pig and poultry feces; (8) consumption of undercooked chicken and fish; (9) pests contaminate food preparation areas; (10) meat contaminated by feces during processing; (11) multiple species in contact in unhygienic conditions; and (12) poor hygiene in markets. Layout stylized to indicate connectivity. Asterisks indicate antibiotic inputs.
Water, sanitation, and hygiene (WASH) in human communities and in food animal production – including on farms, in markets, and along the value chain – aim to control the flow of pathogens in and out of these systems. In settings where humans and food animals live in close proximity, the absence or breakdown of these controls – hereafter referred to as “leakiness” – has profound implications: the continuous, circular exchange of resistant bacteria selected through unregulated antibiotic use may facilitate the exchange of resistance genes among bacteria in both sectors. To our knowledge, leakiness as a potentiator of global antibiotic resistance has not been widely considered by the scientists and public health agencies who are driving policy and funding priorities in this field. Here, we provide evidence for the consequences of leakiness by describing the spread of mobile resistance elements between humans and animals in Phnom Penh, Cambodia, a highly leaky urban center in Southeast Asia.
Insufficient environmental controls and unregulated antibiotic use in Cambodia
Southeast Asia is a hotspot for emerging zoonotic infectious diseases such as highly pathogenic avian influenza H5N1, severe acute respiratory syndrome (SARS-CoV), and Nipah virus, all of which have emerged in the past few decades (Coker et al. 2011). Cambodia is among the poorest countries in Southeast Asia, and 76% of the estimated 16 million residents live in rural areas (NIS 2018). As in other Southeast Asian countries, Cambodian livestock production is rapidly increasing to support growing demand. Between 2012 and 2016, domestic poultry production increased by 53%, to 35.7 million chickens per year, and pig production by 34%, to 2.9 million pigs per year (MAFF 2017), with meat demand projected to increase by another 20% by 2024 (Sen 2018). While commercial livestock production has nearly doubled during the past decade (MAFF 2017), at least 80% of livestock continues to be raised by resource-constrained households in rural areas (MAFF 2017; NIS 2018). This distribution of livestock production between commercial farms and small-scale, household farms is similar in many LMICs in Asia and sub-Saharan Africa (Ikhimiukor et al. 2022).
WASH conditions and animal husbandry practices in Cambodia suggest ample opportunity for bidirectional exchange of fecal–oral pathogens between humans and livestock (Figures 1 and 2). First, Cambodia has the highest rate of open human defecation in the region (UNICEF Cambodia 2019), with 26.2% of rural residents and 7.7% of urban residents outside the capital continuing to defecate in fields or other open spaces (NIS 2018). Because most livestock raised in rural and urban households are allowed to roam freely during the day, these animals (especially pigs) may consume human feces (Sikasunge et al. 2007; Thomas et al. 2013). Second, nearly half of livestock-owning households report dumping untreated animal manure directly into the environment (Ström et al. 2018a), suggesting that untreated drinking water may be a source of animal fecal bacteria. Cambodia has the lowest access to piped drinking water in Southeast Asia, and nearly 1 in 5 urban residents outside the capital and 1 in 2 rural residents cannot reliably access clean water (NIS 2018). Third, poor WASH practices along the food supply chain in Cambodia, especially in antiquated slaughterhouses (Tum 2015) and crowded live-animal markets, facilitate the contamination of meat and produce by animal and human fecal bacteria. Finally, such behaviors as using untreated animal manure to fertilize crops for human consumption (Ström et al. 2018a), allowing animals to enter food preparation areas (Osbjer et al. 2015), not removing manure from the household environment (Atterby et al. 2019), handling slaughtering byproducts (Atterby et al. 2019), and eating undercooked chicken and fish (Osbjer et al. 2015; Nadimpalli et al. 2019) likely exacerbate the spread of zoonotic pathogens. With so many leaks between human communities and animal production systems, antibiotic use in both sectors could lead to the emergence and spread of antibiotic-resistant bacteria.
Figure 2.
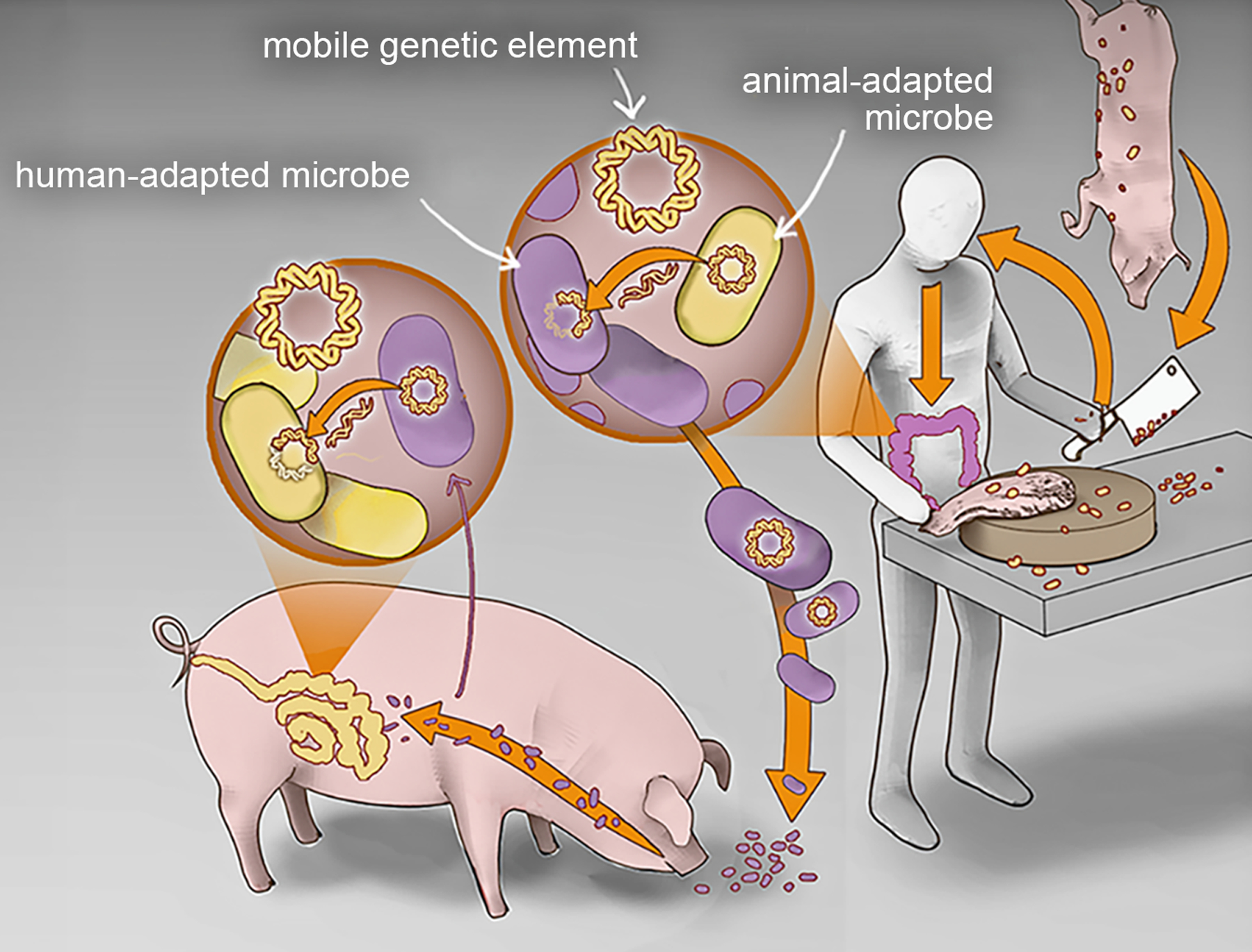
Frequent mixing of human- and animal-adapted microbes creates opportunities for the exchange of antibiotic resistance at multiple scales (ie of bacteria, plasmids, and transposable elements).
We compared published lists of antibiotics commonly used in human medicine and food animal production in Southeast Asia and found substantial overlap. Antibiotics administered to chickens and pigs included all antibiotics that are commonly used in human medicine, including third-generation cephalosporins, penicillins, fluoroquinolones, gentamicin, and co-trimoxazole (Carrique-Mas et al. 2015; Om et al. 2017; Ström et al. 2018b). Farmers in Cambodia reported purchasing human antibiotics for their animals when veterinary drugs were ineffective (Ström et al. 2018b), and antibiotics that are critically important for human medicine (eg colistin) were used extensively in poultry production (Carrique-Mas et al. 2015; Coyne et al. 2019). Such practices mean that the same antibiotic resistance genes could confer a selective advantage in multiple hosts.
Pathogen and mobile resistance element overlap across humans and food animals in Cambodia
We examined overlap among human- and animal-origin extended-spectrum β-lactamase (ESBL)-producing Escherichia coli, which are bacteria resistant to third-generation cephalosporins and other commonly used antibiotics (Panel 1). In Cambodia, gut colonization with ESBL-producing Enterobacterales has steadily increased over the past 20 years, and more than 90% of healthy community members may now be carrying these organisms in their gastrointestinal tract (Singh et al. 2020). This prevalence rate is markedly higher than what is observed in high-income countries (Bezabih et al. 2021) like the US (<5%; Islam et al. 2018), the UK (11%; Day et al. 2019), and Denmark (12%; Dall et al. 2019). Previously, we found that ESBL-producing E coli colonizing healthy community members in Phnom Penh were highly similar to strains recovered from meat and fish sold at markets (Nadimpalli et al. 2019). For example, one-third of human-origin ESBL-producing E coli strains encoded the same ESBL gene type (blaCTX-M-55 gene) that predominated among E coli recovered from meat and fish. This sharply contrasts with findings from multiple high-income countries with robust WASH infrastructure in human communities and along the food value chain (Day et al. 2019; Mughini-Gras et al. 2019); for example, a recent study from the UK found only 5% of ESBL-producing E coli strains from healthy humans harbored ESBL genes that predominated among meat (Figure 3a; Day et al. 2019). Substantial leakiness between humans and animals in Cambodia, combined with similar antibiotic pressures in both reservoirs, could result in the synergistic amplification and mutual exchange of mobile resistance elements.
Panel 1. The importance of ESBL-producing Enterobacterales.
Third-generation cephalosporins are broad-spectrum antibiotics that are generally well-tolerated, affordable, and accessible.
Bacteria that produce ESBL enzymes can degrade third-generation cephalosporins, leading to treatment failure.
Genes encoding ESBL enzymes spread among bacteria via mobile resistance elements.
Enterobacterales like Escherichia coli and Klebsiella pneumoniae have increasingly acquired these genes and now cause >50,000 deaths per year globally (ARC 2022).
ESBL-producing E coli are used as “indicator organisms” by the World Health Organization to gauge the magnitude and interconnectedness of antibiotic resistance between humans, animals, and the environment, as well as the effectiveness of potential interventions.
Figure 3.

High antibiotic use and environmental leakiness is associated with the mobilization of an extended-spectrum β-lactamase (ESBL)-encoding transposon region across different bacterial lineages and hosts in Cambodia. (a) Sankey diagram depicting the flow of ESBL genes among Escherichia coli from human and animal origin. ESBL genes flow freely among human and animal-origin E coli in the leaky Cambodian ecosystem but are largely isolated to specific hosts in the less leaky UK ecosystem. (b) Core-genome phylogenetic tree depicting diverse ESBL-encoding E coli lineages circulating in Cambodia. A blaCTX-M-encoding transposon region has been acquired by multiple E coli sub-lineages from humans and meat, underscoring rampant genetic exchange in a highly leaky system with substantial antibiotic selective pressure. Tree is mid-point rooted and scale indicates substitutions per site.
For this case study, we performed long-read DNA sequencing of ESBL-producing E coli of human and animal origin and leveraged existing E coli datasets to determine if strains from these two sources shared a common pool of mobile resistance elements. Mobile resistance elements contain repeat regions that are challenging to assemble using standard, short-read DNA sequencing technologies; using long-read sequencing allows for substantially more accurate characterization of these elements. First, we used long-read sequencing to assemble high-quality draft genomes of five ESBL-producing E coli isolates from the feces of healthy humans (n = 2) and from pork meat (n = 2) and chicken (n = 1) sold at Phnom Penh markets (WebPanel 1; WebTable 1; WebFigure 1). We identified the ESBL-encoding plasmids they harbored and annotated them. Next, we screened for these plasmids among previously published Cambodian collections of ESBL-producing E coli from healthy, gut-colonized humans (88 positive of 130 humans screened), clinical specimens (n = 15), and meat and fish from markets (93 positive of 150 samples screened) (WebTable 2).
We identified four distinct ESBL-encoding plasmids that were shared with slight variations among E coli isolates of both human and animal origin (WebPanel 2; WebFigure 2). One plasmid (pC27A-CTX-M-55) co-encoding resistance to third-generation cephalosporins, fluoroquinolones, phenicols, tetracyclines, sulfonamides, trimethoprim, and aminoglycosides was present in E coli from chicken meat (n = 3), fish (n = 12), pork meat (n = 2), and healthy humans (n = 2). A second plasmid (pP59A-CTX-M-55) co-encoding resistance to β-lactams, fluoroquinolones, macrolides, phenicols, sulfonamides, and aminoglycosides was prevalent among pork meat isolates (n = 6) and identified in a healthy human (n = 1). A third plasmid (pP225M-CTX-M-55) co-encoding resistance to third-generation cephalosporins, tetracyclines and aminoglycosides was primarily found among isolates from healthy humans (n = 6) but was also carried by E coli from a human urinary tract infection (n = 1) and from fish (n = 1). Finally, a fourth, novel plasmid type (pP276M-CTX-M-55) co-encoding resistance to third-generation cephalosporins, fluoroquinolones, phenicols, and tetracyclines was identified from a healthy human isolate (n = 1) and from chicken meat (n = 1). Although we only sequenced five ESBL-producing E coli isolates for this case study, which represent only a small fraction of the bacteria colonizing the guts of humans and food animals sampled in this setting, the multiple instances of overlap that we observed suggest that plasmid sharing among vertebrate hosts is not uncommon.
We then compared the four ESBL-encoding plasmids identified in this case study to determine whether the ESBL genes themselves shared a common origin. We identified a putative, blaCTX-M-encoding transposon region that was integrated across three of the four plasmids (WebFigure 3). This ~6-kilobase transposon region, hereafter referred to as TnCTX-M/qnrS, also encoded qnrS1, which confers low-level fluoroquinolone resistance. Among the three plasmids we sequenced, TnCTX-M/qnrS was flanked by different combinations of insertion sequences (IS26/ISEcp1/ISKpn19). Recent studies have described this same region, flanked by other combinations of insertion sequences, among E coli and Salmonella enterica recovered from retail meats, food animals, healthy humans, and clinical specimens across China and Southeast Asia (Zhang et al. 2019; Li et al. 2021; Zeng et al. 2022). This suggests that TnCTX-M/qnrS has been mobilized by diverse mobile resistance elements and is likely under high selection pressure.
Finally, we examined existing collections of ESBL-producing Enterobacterales from Cambodia as well as a global Escherichia/Shigella database (https://enterobase.warwick.ac.uk/species/index/ecoli) for the TnCTX-M/qnrS region. The blaCTX-M-55 and blaCTX-M-15 variants of this transposon region, which differed by only a single nucleotide polymorphism, were highly prevalent among Enterobacterales, including E coli, Klebsiella pneumoniae, and S enterica from human and animal products in Cambodia (Figure 3b; WebFigure 4; WebTable 2). Several strains in our existing collections were recovered from the same meat samples (Nadimpalli et al. 2019); for seven of 20 samples that were co-contaminated with different ESBL-producing species, TnCTX-M/qnrS was detected in all recovered species (WebTable 2). Examination of the global database revealed that the blaCTX-M-15 variant was broadly dispersed, while the blaCTX-M-55 variant was mainly identified in isolates from Asia, including Vietnam, Laos, Thailand, Taiwan, and China (WebPanel 2; WebTable 3). The mobilization of the blaCTX-M-55 variant across different plasmids, bacterial lineages, and hosts confirms that bacterial exchanges among humans and animals are exceptionally frequent in this region. Our findings also underscore that in settings with high leakiness, the full scope of antibiotic resistance exchange between humans and animals cannot be quantified by measuring bacterial strain overlap alone; rather, the exchange of resistance-encoding elements across bacterial lineages must also be considered.
Rebalancing priorities to combat antibiotic resistance
There is no doubt that antibiotics are the greatest selective force for the emergence of new antibiotic-resistant bacteria (Lipsitch and Samore 2002); however, poor environmental controls may be critically overlooked potentiators for such strains. In this case study, detection of the same mobile resistance elements across multiple vertebrate hosts indicates that antibiotic stewardship alone may be insufficient to stem the growing problem of antibiotic resistance in resource-poor settings. Although our analysis was limited to Cambodia, recent surveillance studies in several LMICs in Asia (Tansawai et al. 2019; Huang et al. 2020), South America (Murray et al. 2021), and Africa (Falgenhauer et al. 2019; Muloi et al. 2022) have also reported overlapping ESBL gene types among Enterobacterales from humans and food animals. Although most of these studies did not perform resource-intensive long-read sequencing to characterize the mobile resistance elements harboring these ESBL genes, as we did here, such consistent findings – again, in stark contrast to high-income settings – suggest that dissemination of mobile resistance elements across hosts is not unique to Cambodia or Southeast Asia. Overall, these data indicate that antibiotic stewardship efforts in LMICs must be complemented by implementing environmental controls at the human–animal interface (that is, “plugging the leaks”). In addition to stemming the circular flow of antibiotic-resistant bacteria, infrastructural WASH improvements in human communities and food animal production have the potential to also reduce disease and consequently antibiotic demand in both humans and animals (Pickering et al. 2019).
Higher demand for animal-based protein in low- and especially middle-income countries suggests that the need to plug leaks is becoming increasingly urgent. Specifically, as these countries develop, these dietary changes are driving up food animal production and concomitant antibiotic use (Van Boeckel et al. 2015). As a result, several countries that lack universal access to clean water and sanitation are expected to double their antibiotic use in animal production over the next 10 years (Van Boeckel et al. 2015). Currently, less than 4% of funding through the Joint Programming Initiative for Antimicrobial Resistance (https://www.jpiamr.eu) is allocated toward identifying cost-effective environmental controls that curb microbial exchange (World Bank Group 2019). Furthermore, of the 70-plus countries that have developed national antimicrobial resistance action plans in collaboration with global public health organizations, nearly all have omitted commitments to community-based WASH infrastructure (Essack 2021). This lack of prioritization may reflect a high-income country bias among scientists and policy makers who accept the generalizability of research conducted in settings with strong environmental controls, where the exchange of mobile resistance elements between food animals and the broader community is limited (Day et al. 2019; Mughini-Gras et al. 2019; Martak et al. 2021).
Conclusion
Overall, in settings with pervasive leakiness between humans and animals, a rebalancing of policy and funding priorities is needed if the efficacy of new and existing antibiotics is to be preserved. The case study presented here strongly suggests that the lack of environmental controls along with widespread antibiotic use is leading to the exchange of resistance elements and the evolution of antibiotic-resistant human pathogens in Southeast Asia. Additional studies are necessary to determine the impacts of leakiness on the dissemination of antibiotic resistance in other settings and to ascertain which leaks are the most critical and most cost-effective to plug. We propose that evaluating and implementing WASH initiatives in both the community setting and food animal production should be prioritized along with antibiotic stewardship to address the antibiotic-resistance crisis in resource-poor settings.
Supplementary Material
In a nutshell:
Antibiotic resistance is an ecological challenge, with low- and middle-income countries bearing the greatest burden
These countries have many unique interfaces at which bacteria and their resistance-encoding elements can be exchanged between humans and food animals – including on farms, at markets, and along food value chains – that are minimized or non-existent in high-income settings
We describe evidence for the consequences of poor environmental controls at these interfaces on the bidirectional spread of antibiotic resistance between humans and animals in Cambodia
Implementing water, sanitation, and hygiene infrastructure in human communities and food animal production must be prioritized to address global antibiotic resistance
Acknowledgements
This work was supported by the Dennis and Mireille Gillings Foundation, Pasteur Foundation US, MSD AVENIR, Monaco Department of International Cooperation, Institut Pasteur, and the Stuart B Levy Center for Integrated Management of Antimicrobial Resistance at Tufts (Levy CIMAR), a collaboration of Tufts Medical Center and the Tufts University Office of the Vice Provost for Research (OVPR) Research and Scholarship Strategic Plan (RSSP). MLN was supported by National Institutes of Health award KL2TR002545; FG was supported by a grant from the Région Normandie.
Footnotes
Supporting Information
Additional, web-only material may be found in the online version of this article at
Data Availability Statement
The accession numbers for the five Escherichia coli draft genomes presented in this study are available in the National Center for Biotechnology Information’s GenBank (https://www.ncbi.nlm.nih.gov/genbank) under Bioproject number PRJNA566431. The accession numbers for the Illumina sequences generated from the ESBL-producing E coli (n = 196), Salmonella enterica (n = 26), and Klebsiella pneumoniae (n = 8) isolates described in this study are available in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession numbers PRJEB25898, PRJEB27759, and PRJEB34504, respectively. Newick tree files used to create Figure 3b and WebFigure 4 are available at https://doi.org/10.15139/S3/3OMUI2.
References
- ARC (Antimicrobial Resistance Collaborators). 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399: 629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atterby C, Osbjer K, Tepper V, et al. 2019. Carriage of carbapenemase- and extended-spectrum cephalosporinase-producing Escherichia coli and Klebsiella pneumoniae in humans and livestock in rural Cambodia; gender and age differences and detection of blaOXA-48 in humans. Zoonoses Public Hlth 66: 603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebell L and Muiru A. 2014. Antibiotic use and emerging resistance – how can resource-limited countries turn the tide? Global Heart 9: 347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezabih YM, Sabiiti W, Alamneh E, et al. 2021. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemoth 76: 22–29. [DOI] [PubMed] [Google Scholar]
- Carrique-Mas JJ, Trung NV, Hoa NT, et al. 2015. Antimicrobial usage in chicken production in the Mekong Delta of Vietnam. Zoonoses Public Hlth 62 S1: 70–78. [DOI] [PubMed] [Google Scholar]
- Coker RJ, Hunter BM, Rudge JW, et al. 2011. Emerging infectious diseases in Southeast Asia: regional challenges to control. Lancet 377: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne L, Arief R, Benigno C, et al. 2019. Characterizing antimicrobial use in the livestock sector in three South East Asian countries (Indonesia, Thailand, and Vietnam). Antibiotics 8: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall LB, Lausch KR, Gedebjerg A, et al. 2019. Do probiotics prevent colonization with multi-resistant Enterobacteriaceae during travel? A randomized controlled trial. Travel Med Infect Dis 27: 81–86. [DOI] [PubMed] [Google Scholar]
- Day MJ, Hopkins KL, Wareham DW, et al. 2019. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 19: 1325–35. [DOI] [PubMed] [Google Scholar]
- Essack S 2021. Water, sanitation and hygiene in national action plans for antimicrobial resistance. B World Health Organ 99: 606–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgenhauer L, Imirzalioglu C, Oppong K, et al. 2019. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol 9: 3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zeng L, Doi Y, et al. 2020. Extended-spectrum β-lactamase-producing Escherichia coli. Lancet Infect Dis 20: 404–05. [DOI] [PubMed] [Google Scholar]
- Ikhimiukor OO, Odih EE, Donado-Godoy P, and Okeke IN. 2022. A bottom-up view of antimicrobial resistance transmission in developing countries. Nat Microbiol 7: 757–65. [DOI] [PubMed] [Google Scholar]
- Islam S, Selvarangan R, Kanwar N, et al. 2018. Intestinal carriage of third-generation cephalosporin-resistant and extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy US children. J Pediatr Infect Dis Soc 7: 234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EY, Van Boeckel TP, Martinez EM, et al. 2018. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. P Natl Acad Sci USA 115: E3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Olsen RH, Song A, et al. 2021. First report of a foodborne Salmonella enterica serovar Gloucester (4:i:l,w) ST34 strain harboring blaCTX–M–55 and qnrS genes located in IS26-mediated composite transposon. Front Microbiol 12: 646101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M and Samore MH. 2002. Antimicrobial use and antimicrobial resistance: a population perspective. Emerg Infect Dis 8: 347–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAFF (Ministry of Agriculture, Forestry and Fisheries). 2017. Annual report for agriculture, forestry, and fisheries from 2016–2017 and direction 2017–2018. Phnom Penh, Cambodia: MAFF. [Google Scholar]
- Martak D, Guther J, Verschuuren TD, et al. 2021. Populations of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae are different in human-polluted environment and food items: a multicentre European study. Clin Microbiol Infec 28: 447.e7–e14. [DOI] [PubMed] [Google Scholar]
- Mughini-Gras L, Dorado-García A, van Duijkeren E, et al. 2019. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planetary Health 3: e357–69. [DOI] [PubMed] [Google Scholar]
- Muloi DM, Wee BA, McClean DMH, et al. 2022. Population genomics of Escherichia coli in livestock-keeping households across a rapidly developing urban landscape. Nat Microbiol 7: 581–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M, Salvatierra G, Dávila-Barclay A, et al. 2021. Market chickens as a source of antibiotic-resistant Escherichia coli in a peri-urban community in Lima, Peru. Front Microbiol 12: 635871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadimpalli M, Vuthy Y, de Lauzanne A, et al. 2019. Meat and fish as sources of extended-spectrum β-lactamase-producing Escherichia coli, Cambodia. Emerg Infect Dis 25: 126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIS (National Institute of Statistics). 2018. Cambodia socio-economic survey 2017. Phnom Penh, Cambodia: NIS. [Google Scholar]
- O’Neill J 2016. Tackling drug-resistant infections globally: final report and recommendations. London, UK: Government of the United Kingdom. [Google Scholar]
- Om C, Daily F, Vlieghe E, et al. 2017. Pervasive antibiotic misuse in the Cambodian community: antibiotic-seeking behaviour with unrestricted access. Antimicrob Resist In 6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbjer K, Boqvist S, Sokerya S, et al. 2015. Household practices related to disease transmission between animals and humans in rural Cambodia. BMC Public Health 15: 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering AJ, Crider Y, Sultana S, et al. 2019. Effect of in-line drinking water chlorination at the point of collection on child diarrhoea in urban Bangladesh: a double-blind, cluster-randomised controlled trial. Lancet Glob Health 7: e1247–56. [DOI] [PubMed] [Google Scholar]
- Sen H 2018. The local chicken value chain in Cambodia: constraints and challenges of local chicken smallholder producers. Australas Agribusiness Perspect 21: 43–56. [Google Scholar]
- Sikasunge CS, Phiri IK, Phiri AM, et al. 2007. Risk factors associated with porcine cysticercosis in selected districts of eastern and southern provinces of Zambia. Vet Parasitol 143: 59–66. [DOI] [PubMed] [Google Scholar]
- Singh SR, Mao B, Evdokimov K, et al. 2020. Prevalence of MDR organism (MDRO) carriage in children and their household members in Siem Reap Province, Cambodia. JAC-Antimicrob Resist 2: dlaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ström G, Albihn A, Jinnerot T, et al. 2018a. Manure management and public health: sanitary and socio-economic aspects among urban livestock-keepers in Cambodia. Sci Total Environ 621: 193–200. [DOI] [PubMed] [Google Scholar]
- Ström G, Boqvist S, Albihn A, et al. 2018b. Antimicrobials in small-scale urban pig farming in a lower middle-income country – arbitrary use and high resistance levels. Antimicrob Resist In 7: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansawai U, Walsh TR, and Niumsup PR. 2019. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poultry Sci 98: 2622–31. [DOI] [PubMed] [Google Scholar]
- Thomas LF, de Glanville WA, Cook EA, and Fèvre EM. 2013. The spatial ecology of free-ranging domestic pigs (Sus scrofa) in western Kenya. BMC Vet Res 9: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tum S 2015. Reducing microbial contamination of meat at slaughterhouses in Cambodia. http://safetynet2008.com/wp-content/uploads/2015/11/Tum-Sothyra.pdf. Viewed 26 Oct 2022.
- UNICEF (UN Children’s Fund) Cambodia. 2019. Water, sanitation and hygiene. Phnom Penh, Cambodia: UNICEF. [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M, et al. 2015. Global trends in antimicrobial use in food animals. P Natl Acad Sci USA 112: 5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (World Health Organization). 2016. Global action plan on antimicrobial resistance. Geneva, Switzerland: WHO. [Google Scholar]
- WHO (World Health Organization). 2021. Antimicrobial resistance: fact sheet. Geneva, Switzerland: WHO. [Google Scholar]
- World Bank Group. 2019. Pulling together to beat superbugs: knowledge and implementation gaps in addressing antimicrobial resistance. Washington, DC: World Bank. [Google Scholar]
- Zeng S, Zhuo Z, Huang Y, et al. 2022. Prevalence of chromosomally located blaCTX-M-55 in Salmonella Typhimurium ST34 isolates recovered from a tertiary hospital in Guangzhou, China. Microbiol Spectrum 10: e0277121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-Z, Ding X-M, Lin X-L, et al. 2019. The emergence of chromosomally located blaCTX-M-55 in Salmonella from foodborne animals in China. Front Microbiol 10: 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the five Escherichia coli draft genomes presented in this study are available in the National Center for Biotechnology Information’s GenBank (https://www.ncbi.nlm.nih.gov/genbank) under Bioproject number PRJNA566431. The accession numbers for the Illumina sequences generated from the ESBL-producing E coli (n = 196), Salmonella enterica (n = 26), and Klebsiella pneumoniae (n = 8) isolates described in this study are available in the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession numbers PRJEB25898, PRJEB27759, and PRJEB34504, respectively. Newick tree files used to create Figure 3b and WebFigure 4 are available at https://doi.org/10.15139/S3/3OMUI2.


