Abstract
Two clinically important subspecies, Francisella tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B) are responsible for most tularaemia cases, but these isolates typically form a weak biofilm under in vitro conditions. Phase variation of the F. tularensis lipopolysaccharide (LPS) has been reported in these subspecies, but the role of variation is unclear as LPS is crucial for virulence. We previously demonstrated that a subpopulation of LPS variants can constitutively form a robust biofilm in vitro, but it is unclear whether virulence was affected. In this study, we show that biofilm-forming variants of both fully virulent F. tularensis subspecies were highly attenuated in the murine tularaemia model by multiple challenge routes. Genomic sequencing was performed on these strains, which revealed that all biofilm-forming variants contained a lesion within the wbtJ gene, a formyltransferase involved in O-antigen synthesis. A ΔwbtJ deletion mutant recapitulated the biofilm, O-antigen and virulence phenotypes observed in natural variants and could be rescued through complementation with a functional wbtJ gene. Since the spontaneously derived biofilm-forming isolates in this study were a subpopulation of natural variants, reversion events to the wbtJ gene were detected that eliminated the phenotypes associated with biofilm variants and restored virulence. These results demonstrate a role for WbtJ in biofilm formation, LPS variation and virulence of F. tularensis.
Keywords: biofilm, Francisella tularensis, LPS, O-antigen, phase variation, SNP reversion, tularaemia, virulence, WbtJ
Introduction
Francisella tularensis is an intracellular pathogen that is endemic throughout much of the northern hemisphere and is the causative agent of tularaemia [1, 2]. While only a few hundred cases occur per year in humans in the USA [3], tularaemia can be life-threatening if left untreated [4]. Two Francisella subspecies are almost exclusively responsible for causing illness in humans: F. tularensis subsp. tularensis (type A) and F. tularensis subsp. holarctica (type B). In North America, type A isolates tend to be more virulent and are most often transmitted by deer flies and tick vectors in arid environments [1, 2, 5, 6]. On the other hand, type B isolates are usually less virulent, are distributed throughout much of Eurasia, and are more often associated with aquatic environments and transmitted through mosquitoes [7–10]. From a biodefence perspective, F. tularensis poses a significant threat as a bioweapon due to its notoriously low infectious dose, aerosizable nature and high morbidity [5]. While a live vaccine strain (LVS) has previously been developed, approval by the US Food and Drug Administration (FDA) for use in the USA has not occurred due to multiple concerns, such as an unknown ancestral history, instability of colony morphology and potential reversion to a virulent form [11, 12].
Tularaemia is known to manifest clinically in several forms in humans (ulceroglandular, glandular, oculoglandular, oropharyngeal, pneumonic, typhoidal) that can often be indicative of the route of exposure [8]. The most common forms of tularaemia are ulceroglandular and glandular, which are characterized by an infection at the bacterial entry site and are often attributed to bites from insects harbouring F. tularensis or the handling of an infected animal [2, 13]. Pneumonic tularaemia, occurring from aerosol exposure or hematogenous spread, is the most severe form of the disease and is associated with higher mortality rates [4]. As an intracellular pathogen, F. tularensis is able to avoid immune detection to invade a variety of host cells, most often macrophages and hepatocytes [14, 15]. It is generally agreed that the unique structure of F. tularensis lipopolysaccharide (LPS) allows the bacteria to avoid the canonical TLR4 signalling pathway that alerts immune cells to bacterial invasion [16, 17]. Further, antigenic variation of the LPS structure, namely lipid A and the O antigen (O-Ag), has been shown to modulate host factors within macrophages, highlighting the importance LPS in the pathogenesis of F. tularensis [18, 19].
Early characterization studies on F. tularensis noted that surface factors were able to vary, which affected the colony morphology, virulence and immunogenic response [20–22]. This phenomenon was originally described as blue/grey phase variation on the basis of colony appearance under oblique lighting [21]. Blue variants were found to be the virulent form of F. tularensis, while some level of attenuation was observed in grey variants. Importantly, the reversion of grey variants to a blue form has also been reported, suggesting that variants cannot be assumed to be locked in an attenuated state [18, 21]. This phenotypic switching between blue and grey forms is readily observed in both virulent type A and B strains, as well as LVS [20, 23]. To date, it is unknown whether this variation only happens in a laboratory setting or if it is relevant to the environmental survival of F. tularensis. However, understanding this variation in F. tularensis is important because in the past, vaccine lots of LVS have displayed reduced immunogenicity in humans due to an inexplicable rise in grey variants during batch production [20, 24]. While the cellular factors that govern blue/grey variation in F. tularensis are still largely unknown, this phenotype has been attributed to alterations to the LPS molecule and/or the O-Ag that can be also incorporated into the capsule [18, 23, 25]. Culture conditions, such as growth medium, pH, inoculum density and culture duration, have been shown to influence the frequency of blue to grey variation, which has led to the working hypothesis that this variation can enable environmental survivorship for this intracellular pathogen [21]. Further supporting this hypothesis, the relative amount of O-Ag displayed on the cell surface has been reported to be influenced by growth environment [26].
In recent years, studies have convincingly shown that fully virulent type A and B isolates can produce a robust biofilm, and there is likely a link between biofilm and environmental survivorship for these isolates [27–29]. However, the ability of F. tularensis to form biofilm in a natural setting remains unknown. We previously demonstrated that in vitro biofilm formation by F. tularensis was dependent on a subpopulation of grey variants that constitutively produce a robust biofilm [28]. In most pathogens, biofilm formation is considered to be a virulence factor because adhesion to host tissues is often a prerequisite for seeding an infection. Biofilm also contributes to an active infection as the extracellular matrix provides protection from the host immune system by masking bacterial surface features and pathogen-associated molecular patterns (PAMPs) and armouring the bacteria against antimicrobial peptides (AMPs) as well as antibiotics [30]. Perhaps the most defined role of biofilm in pathogenesis is the perpetuation of an infection, leading to chronic disease or recalcitrance [31, 32]. However, it is unclear what role, if any, biofilm formation may play in pathogenesis for F. tularensis, especially considering that phase variation typically results in attenuation.
In this study, we take advantage of spontaneously laboratory-derived grey variants that constitutively form biofilm in an effort to determine the effect these events may have on pathogenesis in F. tularensis. We demonstrate that constitutive biofilm-forming variants for both type A and B isolates of F. tularensis are highly attenuated in vivo using a murine tularaemia model and multiple challenge routes. While the full genetic mechanism responsible for blue/grey variation remains elusive, we determined that the variants utilized in this study were the result of spontaneous mutations within LPS biosynthesis genes and specifically identified the mutation of the wbtJ gene as responsible for the robust biofilm production. To demonstrate the link between WbtJ with blue/grey variation and biofilm formation, an allelic exchange deletion mutant in this gene was constructed and recapitulated the robust biofilm phenotype. In addition, this mutant was shown to be completely attenuated following pneumonic challenge, suggesting that WbtJ is essential for virulence. Lastly, we provide evidence that reversion of a natural grey variant to a blue form can occur in vivo, allowing F. tularensis to revert from an avirulent to a virulent form within a host.
Methods
Bacterial strains and culture conditions
The bacterial strains used in this study are listed in Table 1. Throughout this paper, the terms ‘type A’ or ‘type B’ are used to refer to the respective subspecies, while ‘F. tularensis’ is used to refer to both subspecies (subsp. tularensis and subsp. holarctica). F. tularensis species were routinely cultured on enriched chocolate agar plates (Remel) at 37 °C. For liquid culture, either brain heart infusion (BHI) broth supplemented with 1 % IsoVitaleX (Becton Dickinson) or Chamberlain’s defined medium (CDM) at pH 6.2 [33] were used for growth.
Table 1.
Bacterial strains used in this study
|
Strain |
Strain characteristics |
Source |
|---|---|---|
|
Escherichia coli NEB5-α |
Used for routine plasmid construction in this study |
New England Biolabs |
|
FRAN244 |
Schu S4 (type A1) |
BEI (NR-10492) [20] |
|
244 BF-1 |
Grey variant; biofilm-forming isolate of FRAN244 |
[28] |
|
FRAN255 |
Isolate from human pleura in 2015 (type B2) |
BRMR* [40] |
|
255 BF-1 |
Grey variant; biofilm-forming isolate of FRAN255 |
[28] |
|
255 BF-4 |
Grey variant; biofilm-forming isolate of FRAN255 |
[28] |
|
LVS |
Live vaccine strain |
USAMRIID Repository |
|
LVS isolate #11 |
Grey variant; non-biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #13 |
Grey variant; non-biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #14 |
Grey variant; non-biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #15 |
Grey variant; biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #22 |
Grey variant; biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #26 |
Grey variant; biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #27 |
Grey variant; biofilm-forming isolate of LVS |
[28] |
|
LVS isolate #38 |
Grey variant; biofilm-forming isolate of LVS |
[28] |
|
LVS ΔwbtJ |
In-frame deletion of FTL_0602 in LVS |
This study |
|
LVS ΔwbtJ/pMP814 |
In-frame deletion of FTL_0602 in LVS with pMP814 (KanR) |
This study |
|
LVS ΔwbtJ/pKM43 |
In-frame deletion of FTL_0602 in LVS with pKM43 (KanR) |
This study |
|
LVS isolate #15/pMP814 |
LVS isolate #15 with pMP814 (KanR) |
This study |
|
LVS isolate #15/pKM43 |
LVS isolate #15 with pKM43 (KanR) |
This study |
|
#15-R1 |
Reverted colony from LVS isolate #15; blue variant, non-biofilm-forming |
This study |
|
#15-R2 |
Reverted colony from LVS isolate #15; blue variant, non-biofilm-forming |
This study |
|
#15-R3 |
Reverted colony from LVS isolate #15; blue variant, non-biofilm-forming |
This study |
|
Isolate #15 - MP-1 to 30 |
Strains isolated from mice challenged with LVS isolate #15 |
This study |
|
LVS ΔwbtJ – MP-1 to 5 |
Strains isolated from mice challenged with LVS ΔwbtJ |
This study |
*Biodefense Reference Material Repository.
All molecular cloning and plasmid construction were carried out using NEB5-α electrocompetent E. coli (New England Biolabs). When necessary, kanamycin was added to the culture medium at 50 µg ml−1 for E. coli or 10 µg ml−1 for F. tularensis to maintain selection in plasmid-containing strains. In instances where reversion or variant isolates were detected, bacteria were streak purified on chocolate agar and confirmed to be F. tularensis by PCR of wbtJ (Ft-specific gene; oKM189 and oKM192) prior to further experimentation.
Genome sequencing and SNP detection
Genomic DNA was isolated using a MasterPure isolation kit (Epicentre) following the manufacturer’s protocols. Sequencing libraries were prepared using the Illumina DNA prep kit and sequenced on the Illumina NextSeq 2000 platform using a P3 300 cycle kit. Sequencing libraries for the Oxford Nanopore Mk1C MinION were prepared using the LSK109 ligation sequencing kit and sequenced on version 9 flow cells. Raw reads were quality-trimmed using Trimmomatic v0.38 [34]. Both Illumina and Nanopore reads were used to generate de novo assemblies via Unicycler [35]. Mauve was used for whole-genome alignments [36]. Single-nucleotide polymorphism (SNP) analysis and strain consensus generation were performed using snippy v4.6.0 [37] and default settings. The reference strain used was either the Francisella holarctica LVS reference (GenBank: CP009694.1), FRAN244 (CP073128.1) or FRAN255 (CP073125.1), depending on the background of the parental isolate. Sequence data from the biofilm-forming variant genomes were uploaded under BioProject number PRJNA970844.
Construction of a ΔwbtJ mutant and complementation
Primers and plasmids used in this study are listed in Tables S1 and S2, available in the online version of this article. Flanking regions of wbtJ (FTL_0602) were amplified using primers oKM189 and oRGT_wbtJ_SOE1R to create a 520 upstream fragment and primers oRGT_wbtJ_SOE2F and oKM192 to create a 524 downstream fragment. Overlapping PCR was used to splice the two amplicons together, resulting in a 1031 bp fragment lacking the coding sequence of wbtJ. BamHI and EcoRV restriction sites engineered into the primers were used to ligate the fragment into the allelic exchange vector pEDL50 [38]. The resulting plasmid (pKM41) was confirmed by sequencing and electroporated into electrocompetent Francisella. Briefly, bacteria were grown to mid-log phase in BHI, washed several times with ice-cold 0.5M sucrose and aliquoted into 50 µl samples. Approximately 1 µg of purified plasmid (pKM41) was electroporated using a Bio-Rad Gene Pulser Xcell set to 2.5kV, 25 µF and 600 Ω. Samples were outgrown in BHI containing IsoVitalex for up to 4 h at 37 °C and plated onto chocolate agar with 10 µg ml−1 kanamycin. Colonies required 4–5 days to appear and were confirmed as co-integrates by PCR. The integrated plasmid was cured from co-integrates by overnight passage in BHI followed by serial dilution and plating on chocolate agar with 5 % sucrose. Purified colonies were screened for the ΔwbtJ allele using PCR and sequencing.
For complementing the wbtJ deletion mutant and LVS isolate #15 (a spontaneous grey variant and biofilm former), the 726 bp coding sequence of wbtJ was amplified using primers oKM192 and oKM193 that contained engineered BamHI and EcoRV restriction sites. The resulting fragment was digested and ligated into pMP814 [39], which contains a blaB promoter to drive the expression of wbtJ for trans-complementation studies. The resulting plasmid (pKM43) was electroporated into the listed F. tularensis strains and selected on chocolate agar plates containing kanamycin. Transformants were confirmed by plasmid extraction followed by PCR to confirm the presence of the intact wbtJ gene.
Static biofilm assay
Biofilm formation was assessed by crystal violet staining as previously described [28]. Briefly, F. tularensis was cultured for 24 h on chocolate agar and resuspended to an OD600 of ~0.3 in phosphate-buffered saline (PBS). Bacterial suspensions were diluted 1 to 10 into CDM (~108 c.f.u.), seeded into CoStar polystyrene 96-well plates and incubated at 37 °C for the indicated length of time. Sterility wells were included in each experiment, and peripheral wells were avoided to minimize edge effect. Prior to biofilm staining, the OD600 was measured, after which plates were aspirated, washed 3× with PBS to remove planktonic cells and fixed with 100 % ethanol for 30 min at room temperature. Following ethanol fixation, 0.1 % crystal violet (w/v) was added to each well for 15 min and washed 3× with PBS, after which the remaining crystal violet stain was solubilized in 33 % acetic acid. The OD600 was measured to quantify crystal violet staining as an indicator of biofilm formation. When necessary, samples were diluted 1 : 10 in 33 % acetic acid to ensure that OD readings were within the linear range. At least four technical replicates were averaged in each experiment. All data reported are the result of at least three individual experiments. Throughout this paper, strains that formed a robust uniform biofilm within 24–48 h are referred to as ‘biofilm formers’, while strains referred to as ‘non-biofilm formers’ did not form a biofilm within this time frame.
Macrophage assays
In assays involving macrophages, J774A.1 cells (American Type Culture Collection) were cultured at 37 °C under 5 % CO2 in Dulbecco’s modified Eagle’s medium containing glucose, l-glutamine and sodium pyruvate at 4.5 g/L and 10 % foetal bovine serum. To achieve a confluent monolayer, plates were seeded 1 day prior to experiments in 24-well plates at a density of 5×105 cells per well. Immediately prior to infection, F. tularensis was resuspended from 18 h plates to an OD600 of ~0.3 in PBS and diluted 1 : 5 into tissue culture medium to serve as a bacterial inoculum. No difference was detected for c.f.u. among strains when plating from a normalized OD600. To infect macrophages, tissue culture medium was aspirated from the J774A.1 cells and replaced with 200 µl of bacterial inoculum to achieve an m.o.i. of ~100 : 1, which was confirmed by c.f.u. enumeration of the inoculum. At 2 h postinfection, plates were washed 3× with PBS, after which tissue culture medium containing gentamicin (25 µg ml−1) was added. To obtain c.f.u. recovery at the indicated time, the infected monolayer of J774A.1 cells was washed 3× with PBS and lysed using 200 µl of sterile water and vigorous scraping. The recovered bacterial sample was diluted 1 : 5 into PBS and serial diluted to allow enumeration on chocolate agar plates. In each experiment, three replicate wells were plated in duplicate to obtain the c.f.u. recovery for an experiment. These data shown are the results of at least three independent experiments.
Detection of LPS
Whole-cell extracts were prepared from LVS grown for 18 h on chocolate agar plates by resuspending bacteria to an OD600 of ~0.5 in PBS. A 1 ml aliquot of each sample was pelleted, resuspended in 1× NuPage LDS gel loading buffer, and boiled for 15 min. c.f.u. were determined for each sample to ensure that equal amounts of bacteria were present in each sample prior to resuspension in gel loading buffer. For western analysis, samples were fractionated on a NuPage Novex 4–12 % Bis–Tris gels, transferred onto a nitrocellulouse membrane via an iBlot Gel Transfer Device and blocked overnight in 1 % skim milk in Tris-buffered saline+0.05 % Tween-20 (TBST). Samples were probed with mouse monoclonal anti-LPS antibody (FB11; MA1-7388; Invitrogen) and rabbit polyclonal anti-GroEL (Enzo Life Sciences) at 1 : 500 and 1 : 2000 dilutions, respectively. The membrane was washed 4× with TBST and the corresponding secondary antibody was applied at 1 : 10 000. After four additional TBST washes, the blot was visualized using Clarity Western ECL Substrate following the manufacturer’s protocols (Bio-Rad).
LD50 determination
In vivo testing of F. tularensis strains was performed using BALB/c mice (7–9 weeks old from Charles River Laboratories) as previously described [40]. Briefly, F. tularensis grown from an 18 h chocolate agar plate was diluted to a known concentration in PBS. To mimic vector inoculation, mice were challenged with the indicated c.f.u. using 100 µl subcutaneous inoculation (FRAN244 strains) or 50 µl intradermal (FRAN255 strains). To model pneumonic tularaemia, mice were lightly anaesthetized with ketamine, acepromazine and xylazine and inoculated intranasally with a 50 µl challenge dose. Mice were monitored for clinical symptoms several times daily and followed for 14 (FRAN244 and FRAN255 strains) or 21 days post-challenge (LVS strains) and mortality rates were recorded. Humane endpoints were used in accordance with a predetermined endpoint score sheet approved by IACUC. At the end of study, spleens and lungs were harvested and checked for c.f.u. recovery, with the exception of LVS-infected mice, as only spleens were examined.
Statistical analyses
All experiments described in this paper were performed independently at least three times. Macrophage and biofilm assays (Figs 1, 5–7) were analysed by linear mixed effects model. Analysis was implemented in the GLIMMIX procedure of SAS version 9.4 (SAS Institute, Inc., Raleigh, NC, USA). c.f.u. data were log 10-transformed prior to analysis (Figs 1 and 6). No multiplicity adjustment was applied. LD50 values were calculated using a probit model. The median time to death (TTD) was estimated using the Kaplan–Meier method and comparisons between strains were obtained from a log-normal accelerated failure time (AFT) model in the form log(TTD) = m×log(dose)+b, where m and b are strain-specific slope and intercept terms (Tables 2 and 5). A Wald chi-square test based on a Cox regression model was used to compare estimated median TTDs at the middle of the dose curve. Analysis was completed in PROC LIFEREG and PROC PROBIT, SAS version 9.4 (SAS Institute Inc, Raleigh, NC, USA).
Table 2.
LD50 and TTD for mice challenged intranasally with type A or type B biofilm-forming variants
|
244 BF-1 |
255 BF-1 |
255 BF-4 |
||
|
LD50 (c.f.u.) |
317 |
> 460 000 |
2152 |
|
|
Median TTD (Days) |
1–10 |
>14 |
>14 |
>14 |
|
10–100 |
>14 |
>14 |
>14 |
|
|
100–1000 |
>14 |
>14 |
>14 |
|
|
1000–10 000 |
8 |
>14 |
11 |
|
|
10,000–100,000 |
7 |
>14 |
9 |
|
|
>100 000 |
7 |
>14 |
8 |
|
|
Pair-Wise Comparisons (Wald-test)* |
vs. FRAN244 |
P=0.0036 |
||
|
vs. FRAN255 |
P<0.0001 |
P<0.0001 |
*Challenge dose for the parental strains was 105 c.f.u. (FRAN244) and 8 c.f.u. (FRAN255).
P values for comparisons among strains were determined by Wald Chi-square tests based on a Cox regression model.
ns, not significant (P>0.05); TTD, time to death.
Data availability
All data underlying the findings and conclusions reported in this paper are fully available within the text or the accompanying Supplementary Material. Genome sequencing is available on GenBank at the time of publication. Additional data are available upon request.
Results
Constitutive biofilm variants are impaired for intracellular replication in macrophages
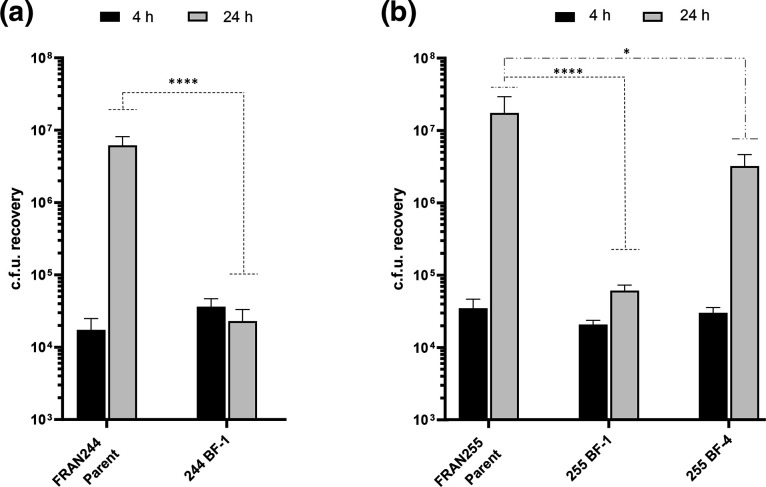
We previously showed that F. tularensis can form a robust biofilm within 24 h in vitro when blue/grey variation of the O-Ag occurred, suggesting a link between these two poorly understood events [28]. It was also previously demonstrated that grey variants of LVS with alterations to the O-Ag had a survival defect in mouse, rat and human macrophages [23, 25]. Given the importance of intracellular survival for F. tularensis pathogenesis, we sought to first assess the ability of biofilm-forming strains (as defined by forming a robust biofilm within 24–48 h) to replicate within host cells. To test this, J774A.1 macrophages were infected with the fully virulent parental wild-type strain and a corresponding spontaneous biofilm-forming variant for a type A lineage (FRAN244/Schu S4 or 244 BF-1) and a type B lineage (FRAN255, 255 BF-1 or 255 BF-4) and assayed c.f.u. recovery at 4 and 24 h post-infection. Two biofilm variants were assayed for FRAN255 as two different O-Ag profiles were previously observed [28]. At 4 h postinfection, similar levels of bacteria were recovered for both the wild-type and the biofilm-forming variants in both type A and B isolate backgrounds (Fig. 1; black bars). However, at 24 h, a significant defect in c.f.u. recovery was detected when comparing the biofilm variants to the parent isolates (Fig. 1, grey bars). More specifically, at 24 h both 244 BF-1 and 255 BF-1 displayed little to no increase compared to the 4 h time point and were significantly different when compared to the increase of the respective parent strains (P<0.0001, linear mixed effects model). Interestingly, 255 BF-4 increased approximately 2 logs when compared to the 4 h time point but was still significantly different from the FRAN255 parent for recovery at 24 h (Fig. 1b; P<0.05, linear mixed effects model). When these strains were grown extracellularly in broth media, the variants grew as well as the respective parent strain. Taken together, these data suggest that natural biofilm-forming variants are able to infect host cells but differ in their ability to replicate intracellularly.
Fig. 1.
Biofilm variants are altered in their ability to replicate within J774A.1 macrophage-like cells. Gentamicin protection assays were performed using natural biofilm forming variants of (a) FRAN244 (Type A) and (b) FRAN255 (Type B). CFUs were determined at 4 h (black bars) and 24 h (grey bars) post infection. Error bars represent the standard error of the mean from three independent experiments. Significance was assessed using linear mixed effects model on log10 transformed data. *p <0.05, ****p<0.0001.
Biofilm-forming variants display attenuation via subcutaneous and intradermal challenge in mice
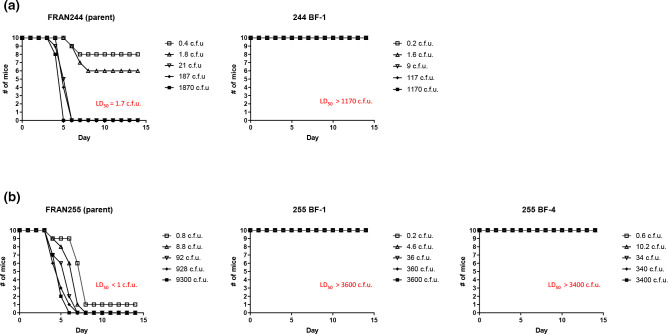
The most common route of exposure to F. tularensis occurs through a bite from an infected arthropod [2]. Given that biofilm formation has been shown to be an important mechanism of persistence and transmission for other bacterial pathogens [41], we sought to first access the virulence potential of natural biofilm-forming F. tularensis variants. LD50 analysis was performed using mice challenged either subcutaneously (type A isolates) or intradermally (type B isolates) to simulate inoculation via either a tick or mosquito, respectively (Fig. 2) [22, 42]. Using this approach, the LD50 for both parental isolates (FRAN244 and FRAN255) was determined to be approximately 1–2 c.f.u. In stark contrast, mice exposed to the biofilm-forming variants for both FRAN244 and FRAN255 survived challenge at all doses (LD50 values >1000 c.f.u.). After 14 days, spleens were harvested from three mice from the highest challenge dose group (~103) of each variant at random and plated to confirm bacterial clearance. All animals were sterile with the exception of one mouse infected with isolate 255 BF-4, from which 247 c.f.u./spleen were recovered.
Fig. 2.
Biofilm forming variants are highly attenuated by challenge mimicking vector exposure in BALB/c mice. Groups of mice (n=10) were challenged with the indicated strain either (a) subcutaneously (FRAN244) or (b) intradermally (FRAN255) and monitored for survivalfor 14 days post challenge. The LD50 values for the parent strains were determined as indicated in red. However, all mice challenged with the biofilm forming variants survived at all challenge doses; therefore, an LD50 was not able to be determined.
Biofilm-forming variants are highly attenuated in a pneumonic murine model
The most severe manifestation of tularaemia is the pneumonic form, which typically occurs following inhalation exposure [2]. With this in mind, we next assessed the virulence of the biofilm-forming variants by intranasally challenging mice and following survival for 14 days. The LD50 for BALB/c mice inoculated intranasally with wild-type F. tularensis strains is 1 c.f.u. [40], so a single challenge dose for the parental strains (FRAN244 : 105 c.f.u.; FRAN255 : 8 c.f.u.) was used as a comparison in this study (Fig. 3; red lines). For intranasal challenges, we calculated an LD50 value of 317 c.f.u. for 244 BF-1 and 2152 c.f.u. for 255 BF-4. An LD50 could not be determined for 255 BF-1, as we did not lose at least 50 % of the mice at any challenge dose. Collectively, all LD50 values for these biofilm-forming variants were still much higher than an LD50 of 1 c.f.u. for the respective parent strain. In addition, when comparing TTDs for those challenge doses that lead to the mice succumbing, the biofilm-forming variants had a significantly higher TTD when compared to the respective control strain (Table 2).
Fig. 3.
Biofilm forming variants display attenuation in a pneumonic tularemia mouse model. Groups consisting of 10 mice were challenged intranasally with doses ranging from <1 CFU up to 106 CFU for natural biofilm forming variants. As a control, one group of mice was challenged for each the parental strain (red line). Displayed are the survival curves following the mice for 14 days post challenge. The accompanying LD50 values and the median time-to-death calculated from these data and are displayed in Table 2.
To determine whether bacteria could be recovered from the organs of mice that survived challenge with the variant strains, spleens and lungs were harvested and plated from five survivors challenged with ~102 c.f.u. of 255 BF-1 (128 c.f.u.) or 255 BF-4 (680 c.f.u.). Bacterial c.f.u. were detected in both spleen and lung tissue for all animals assayed (approximately 500 and 1000 c.f.u. per organ, respectively). Notably, both large and small colony morphologies were observed on chocolate agar plates. However, further exploration of these noted select agent variants were not pursued at that time.
Genetic analysis reveals that all biofilm-forming variants contain a mutation in wbtJ, a formyltransferase involved in O-antigen synthesis
As shown above, the F. tularensis variants 244 BF-1, 255 BF-1 and 255 BF-4 were highly attenuated in both in vitro and in vivo assays. In an effort to identify a possible genetic root cause for the observed grey variant phenotype, biofilm formation and the in vivo attenuation, we performed whole-genome sequencing on these F. tularensis strains. Strains were de novo assembled and aligned to their parental strain. When compared to the respective parental strain, the genomes of these strains lacked deleterious rearrangements or large-scale (>1 kb) deletions/duplication events (Fig. S2). Two identical inversions flanked by repetitive elements were found in 255 BF-1 and 255 BF-4, but coding sequences were not affected. To us, this suggested that a more subtle mutation may be responsible for the phenotypes associated with these strains. SNP analysis was performed, which revealed that the biofilm-forming variants were highly syntenic to the parental strain, harbouring fewer than 10 mutations across the genome (Table 3). Interestingly, each variant harboured a mutation within wbtJ, a gene encoding a formyl transferase located within an operon responsible for O-antigen synthesis [43]. A missense variant of wbtJ was identified in 244 BF-1 (Ser145Phe), while frameshifts were identified in wbtJ for 255 BF1/4 variants. Combined, these data strongly suggest that wbtJ is involved in both phase variation and biofilm formation and is important for virulence in F. tularensis.
Table 3.
SNP analysis of biofilm-forming variants to identify mutated genes
|
Strain |
Position |
TYPE |
REF |
ALT |
Strand |
NT pos |
AA pos |
Effect |
Mutation |
Gene |
Product |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
244 BF-1 |
1 503 971 |
snp |
G |
A |
– |
434/726 |
145/241 |
missense |
Ser145Phe |
wbtJ |
dTDP-4-amino-4,6-dideoxyglucose formyltransferase |
|
255 BF-1 |
551 888 |
snp |
A |
T |
+ |
1368/1686 |
456/561 |
missense |
Leu456Phe |
yheS |
putative ABC transporter ATP-binding protein YheS |
|
1 016 152 |
snp |
A |
T |
– |
13/258 |
5/85 |
missense |
Leu5Ile |
hypothetical protein |
||
|
1 099 427 |
snp |
T |
A |
+ |
72/141 |
24/46 |
synonymous |
Pro24Pro |
hypothetical protein |
||
|
1 429 136 |
insert |
T |
TA |
+ |
433/447 |
145/148 |
frameshift |
Tyr145fs |
IS630 family transposase ISFtu1 |
||
|
1 503 160 |
delete |
TC |
T |
– |
370/726 |
124/241 |
frameshift |
Glu124fs |
wbtJ |
dTDP-4-amino-4,6-dideoxyglucose formyltransferase |
|
|
1 608 120 |
delete |
AT |
A |
intergenic |
|||||||
|
255 BF-4 |
460 860 |
snp |
C |
G |
+ |
383/1347 |
128/448 |
nonsense |
Ser128* |
dtpA1 |
Dipeptide and tripeptide permease A |
|
1 249 956 |
snp |
T |
G |
– |
441/465 |
147/154 |
synonymous |
Ile147Ile |
dtpD |
Dipeptide permease D |
|
|
1 503 065 |
insert |
C |
CT |
– |
465/726 |
155/241 |
frameshift |
Glu156fs |
wbtJ |
dTDP-4-amino-4,6-dideoxyglucose formyltransferase |
|
|
1 643 954 |
snp |
T |
C |
unknown |
intergenic |
||||||
|
1 736 592 |
snp |
A |
C |
unknown |
intergenic |
To bolster the sample size for biofilm and variant genomic data, we again performed whole-genome sequencing followed by SNP analysis on five additional biofilm variants isolated independently in the LVS background that were previously described (LVS isolates #15, 22, 26, 27 and 38) [28]. In agreement with the findings for biofilm-forming variants of FRAN244 and FRAN255, each additional biofilm-forming variant of the LVS isolates also harboured a unique mutation within wbtJ (Fig. 4, Table 4). As a control, we also sequenced the genomes of three grey variants isolated from the same LVS heterogenous populations (LVS #11, 13 and 14) and performed SNP analysis. We previously demonstrated that these isolates do not form biofilm within 72 h and lack reactivity to αLPS specific to F. tularensis [28]. Once again, these non-biofilm-forming strains harboured very few mutations compared to the parental LVS strain (<5). In contrast to the biofilm-forming variants, these strains lacked aberrations within wbtJ, though SNPs were identified in genes known to be involved in LPS synthesis (Table S3). Based upon these genetic observations, we hypothesized that a nonfunctional WbtJ leads to both an O-Ag variant and a biofilm-forming phenotype. Additionally, other SNPs identified caused variant alleles of wzy (O-Ag polymerase) and waaL (O-Ag ligase) and wbtA (dTDP-glucose 4,6 dehydratase). Importantly, these genes are all involved in the synthesis of the O-Ag saccharides, but the mutations do not result in a biofilm-forming phenotype, which would suggest that general O-Ag dysfunction does not result in biofilm formation.
Fig. 4.
Organization of the wbt O-antigen biosynthesis gene cluster highlighting mutations identified in grey variants used in this study. The FRAN244 (Schu S4) genome (CP0731238.1) was used as a reference to create a gene map showing organization of the wbt gene cluster. Diagram is drawn to scale; bar indicates 1 kb. Schu S4 locus tags are provided for genes in which mutations were identified. The wbtJ gene was expanded to show the relative position of each mutation identified in the gene. Triangles indicate lesions identified in isolates sequenced in this study with the color indicating biofilm status; blackfill indicates biofilm forming while white fill indicates a non-biofilm forming isolate. Strain names are provided for reference.
Table 4.
Additional SNPs in wbtJ identified in independently isolated biofilm forming variants
|
Strain |
Position |
TYPE |
REF |
ALT |
NT pos |
AA pos |
Effect |
Mutation |
|---|---|---|---|---|---|---|---|---|
|
LVS Isolate #15 |
1 315 079 |
snp |
C |
T |
459/726 |
153/241 |
nonsense |
Gln153* |
|
LVS Isolate #22 |
1 314 645 |
snp |
C |
A |
23/726 |
8/241 |
missense |
Thr8Lys |
|
LVS Isolate #26 |
1 314 764 |
deletion |
AGTGAGATTAAGCCAATAG |
A |
143/726 |
48/241 |
frameshift |
Ser48fs |
|
LVS Isolate #27 |
1 315 196 |
deletion |
ATGTG |
A |
577/726 |
193/241 |
frameshift |
Cys193fs |
|
LVS Isolate #31 |
1 314 969 |
snp |
G |
A |
347/726 |
116/241 |
missense |
Gly116Glu |
|
LVS Isolate #38 |
1 314 834 |
snp |
C |
T |
212/726 |
71/241 |
missense |
Ser71Phe |
A ΔwbtJ mutant recapitulates the phenotypes observed in natural biofilm forming variants
The wbtJ gene is a 726 bp ORF located in a locus encoding 15 genes involved in O-Ag synthesis that is flanked by insertion elements (Fig. 4). Disruption of genes (wbtA, wbtC, wbtI and wbtM) in this area via transposon mutagenesis has been reported by others to result in the loss of O-Ag on the LPS and capsule [44]. To determine whether the biofilm phenotype we observed in these naturally occurring variants was due solely to the inactivation of wbtJ, we constructed an in-frame deletion mutant (ΔwbtJ) in the wild-type background of LVS by allelic exchange. The final mutant was screened by PCR and Sanger sequencing to confirm the deletion of the wbtJ gene and ensure that no errors were generated to the upstream/downstream regions (data not shown).
When grown on a chocolate agar plate, the ΔwbtJ mutant recapitulated the colony morphology observed in the biofilm-forming variants and appears small and opaque as compared to the wild-type parent (data not shown). Additionally, the ΔwbtJ mutant appears to grow similarly to the parental LVS wild-type in CDM medium (Fig. S3). Given that we identified natural variants containing SNPs in wbtJ by their ability to quickly form biofilm, we first assayed the biofilm-forming capacity of this deletion mutant compared to the parental strain. We found that the ΔwbtJ mutant displayed the biofilm phenotype observed in a natural biofilm variant (LVS isolate #15), suggesting that lesions within the coding sequence of wbtJ in fact result in biofilm formation (Fig. 5a; P<0.0001; linear mixed effects model to compare ΔwbtJ to LVS wild-type). To demonstrate that the phenotype of the mutant was due specifically to the disruption of wbtJ, the complete coding sequence of wbtJ was cloned into shuttle vector pMP814 [39], and the resulting plasmid (pKM43) was electroporated into LVS ΔwbtJ and LVS isolate #15. In the trans-complemented strains, biofilm formation was alleviated in the transformed ΔwbtJ and LVS isolate #15 strains (Fig. 5a; P<0.0001; linear mixed effects model), consistent with mutations within wbtJ leading to the biofilm phenotype observed in natural variants and the wbtJ deletion strain.
Fig. 5.
An in-frame deletion of wbtJ recapitulates the biofilm and O-Ag phenotypes observed in natural variants and is rescued by complementation. (a) Biofilm formation of the LVS wild-type (black bar), Isolate 15 (grey bars) and a ΔwbtJ (red bars) was assayed by crystal violet staining after 1 day of growth in CDM. Biofilm was also assayed in these strains for complementation studies including the vector only (pMP814) or +wbtJ (pKM43; thatched bars) under these conditions. Significance was assessed using linear mixed effects model to compare the LVS wild-type to Isolate #15 and ΔwbtJ or strains containing pMP814 (vector only) to the isogenic strain harboring pKM43 (+wbtJ complement); ****P <0.0001. (b) Western blot analysis was performed to assess the LPS profile. Samples were suspended to an equal OD600, lysed, and separated on SDS-PAGE gels. After transferring to a membrane, blots were exposed to α-LPS (mAb FB11; top) and α-GroEL (bottom) was used as a loading control.
We previously showed that the spontaneous variants used in this study lacked reactivity to a mAb generated against the O-Ag of F. tularensis [28]. Given that WbtJ is involved in the synthesis of the dTDP-4,6-dideoxy-4-romamido-d-glucose (Qui4NFm) saccharide that is incorporated into F. tularensis O-Ag [45], we hypothesized that the LVS ΔwbtJ mutant would also display this phenotype. Western blot analysis was performed using a mAb to F. tularensis LPS to assay whole-cell extracts obtained from the parent, ΔwbtJ mutant and complemented strains. As show in Fig. 5b, the results demonstrated that the null mutant did indeed lack reactivity for antibody specific to F. tularensis O-Ag and that complementation with an intact wbtJ gene restored the hallmark laddering pattern consistent with LPS (Fig. 5b).
A ΔwbtJ mutant displays reduced fitness in macrophages
As shown above in Fig. 1, our natural biofilm-forming variants displayed an intracellular fitness defect at 24 h when compared to the parent strain. To determine whether this loss was due to the altered wbtJ gene in the variants, the ΔwbtJ mutant was tested for its ability to replicate within J774A.1 cells. Consistent with the natural variants, no differences were noted in c.f.u. recovery between the parent and deletion mutant strain at 4 h post-infection. In contrast, at the 24 h time point, the ΔwbtJ mutant did not reach a c.f.u. recovery comparable to the parent (Fig. 6a, grey bars; P<0.05, linear mixed effects model). Further, a complemented ΔwbtJ strain harbouring pKM43 was able to restore the c.f.u. recovered to wild-type levels, which was not observed in the vector-only control (pMP814; data not shown). Taken together, these results suggest that a functional copy of WbtJ is required for optimal fitness within host cells.
Fig. 6.
A ΔwbtJ mutant has a reduced fitness in J774A.1 macrophages and is rescued by complementation. Gentamicin protection assays were performed using the (a) LVS ΔwbtJ mutant and the complemented strain containing pKM43 and (b) the natural biofilm variant isolate #15. c.f.u. were determined at 4 h (black bars) and 24 h (grey bars) post infection. Error bars represent the standard error of the mean from three independent experiments. Statistical significance was determined using linear mixed effects model on log10 transformed data to compare each strain to the wild-type at each timepoint. *p<0.05; ***p<0.001. (c) J774A.1 macrophages were infected with isolate #15 and assayed over the course of 3 days to determine if replication was delayed in the biofilm forming variants.
For completeness, we also tested the ability of a natural biofilm-forming variant in the LVS background to replicate within macrophages. Consistent with the results observed in the ΔwbtJ mutant, the c.f.u. recovery of isolate #15 was not as robust as that for the parental wild-type at 24 h post-infection (Fig. 6b, grey bars; P<0.001, linear mixed effects model). To ensure that the observed fitness defect was not due to delayed replication within macrophages, we assayed isolate #15 over the course of 72 h. In agreement with other studies, wild-type c.f.u. recovery was similar between 24 and 48 h post-infection [46], with a mild decrease observed at the 72 h sampling (Fig. 6c). An increase in c.f.u. recovery was not observed for isolate #15 at 48 and 72 h, which suggests that the survival defect identified in the biofilm-forming variants likely is not due to a delay.
Natural biofilm-forming grey variants can revert to a ‘blue’ form due to repair of the wbtJ gene
While performing experiments using LVS-infected macrophages, we observed a low-level variation in the colony morphology in the natural biofilm-forming variants (<20 colonies in total per experiment, with an average of 1150 colonies counted), particularly at 24 h post-infection and beyond. Well isolated colonies were selected and streak-purified. Upon restreaking, the colony morphology appeared to resemble that of the LVS wild-type, as the colonies were larger and less opaque than those for LVS isolate #15 (data not shown). From separate experiments, three of these isolates (#15-R1, #15-R2 and #15-R3) were streak-purified and saved for further examination. Since reversion has previously been reported [18, 21], and it has been hypothesized to be the result of a secondary mutation rather than repair of the initial mutation (S→NS→S’ [21]), we performed Sanger sequencing on wbtJ for these three revertant strains to determine whether the original mutation (457C>T) was maintained. This examination revealed that not only was the original mutation present in #15 R-1 and #15 R-2, but a secondary SNP occurred directly next to this mutation that would eliminate the nonsense mutation introduced by the primary mutation (Fig. 7a). Isolate #15-R3 appeared to have the original mutation repaired to the wild-type sequence (Fig. 7a). Given that LVS isolate #15 formed a robust biofilm within 1 to 2 days, assays were performed on the reverted strains to determine whether biofilm formation would occur with these wbtJ alleles (Fig. 7b). Indeed, these strains had lost the ability to form biofilm and mirrored the LVS wild-type (P<0.0001; linear mixed effects model) Additionally, western analysis revealed that the parent O-Ag profile was restored in these isolates (Fig. 7c). Taken together, these results further suggest that the fitness of biofilm-forming variants within macrophages is impaired, and likely a selection pressure exists to favour a reversion event.
Fig. 7.
Natural reversion events within wbtJ can be detected in macrophage passaged isolate #15 samples. Heterogenous mixture of big and small colonies was observed in samples plated from macrophage assays with isolate #15 each time an experiment was conducted. Colonies that appeared larger than the parental isolate #15 were streak purified. (a) Sanger sequencing was performed of the wbtJ gene to determine the allele of the reverted isolate. Red font indicates a substituted nucleotide and the resulting codon compared to the LVS wild-type sequence for this gene. Blue and grey arrows indicate colony phenotype (b) The ability of the reverted isolates to form biofilm was assessed using crystal violet staining. For statistical analysis, a linear mixed effects model was used to compare each strain. ****p<0.0001. (c) The O-Ag was also assessed by western blotting performed with α-LPS (mAb FB11; top). α-GroEL (bottom) was included as a loading control.
Natural variants containing SNPs in wbtJ are highly attenuated via intranasal exposure but can revert to wild-type in vivo
Given that the natural LVS variant displayed a fitness defect in macrophages (though some level of replication still occurred), we sought to determine whether this natural variant (LVS isolate #15) and the reverted strain (#15 R-1) were virulent in mice via intranasal challenge. This analysis established that wild-type LVS has an LD50 of 509 c.f.u. with a median TTD of 6.5–7.5 days post-infection. Mice were challenged with LVS isolate #15 with doses ranging up to 45 000 c.f.u., and none displayed any signs of illness. Since all mice survived the challenge, we could not determine an LD50. However, virulence was restored to near wild-type levels in the reverted mutant #15 R-1 (LD50=815 c.f.u.), (Fig. 8) suggesting that virulence can be restored via phase variation.
Fig. 8.
A natural biofilm-forming variant is completely attenuated in a pneumonic tularaemia mousemodel, but a reverted isolate displays wild-type virulence. Mice were challenged intranasally with doses ranging from 1 to 104 c.f.u for the LVS wild-type (parent), isolate #15 and #15R-1. Displayed are the survival curves for groups of 10 mice following the mice for 21 days post challenge. The accompanying LD50 values and the median time-to-death calculated from these data are displayed in Table 5.
At 21 days post-challenge, spleens were harvested from survivors receiving a dose of 102 (~1 LD50 of wild-type) from both LVS wild-type and LVS isolate #15 to determine whether c.f.u. could be recovered. For the survivors challenged with the parental LVS strain (470 c.f.u.) remaining at day 21, these mice displayed no clinical signs; however, LVS (~200 c.f.u.) was recovered from all spleens. In contrast, spleens from animals challenged with LVS isolate #15 at a similar dose (450 c.f.u.) were found to be sterile. Additionally, spleens were harvested and plated from survivors challenged with 45 000 c.f.u. of LVS isolate #15. All animals within this group harboured an average of 174 c.f.u./spleen and featured a similar colony morphology to LVS wild-type. At random, 30 colonies encompassing all 10 spleens were restreaked and sequenced to analyse the wbtJ gene. As observed for macrophage studies, all restreaked isolates contained a ‘repaired’ allele of wbtJ with reversion to glutamine or leucine at residue 153 detected in these samples. Once again, biofilm formation was ablated and normal O-Ag banding was observed when western blot analysis was performed with these reverted isolates (Fig. S4). These results provide further evidence that reversion of the variants occurs in vivo.
A ΔwbtJ mutant in LVS is attenuated in a murine pneumonic model of tularaemia
We next assessed the virulence of the LVS ΔwbtJ strain and the corresponding complement ΔwbtJ+pKM43 using intranasal challenge in a mouse model. As a control for these experiments, mice were challenged with ~5 LD50 (2430 c.f.u.) of the parental LVS wild-type and a vector-only control (ΔwbtJ+pMP814) at a high dose (50 000 c.f.u.). In this experiment, we attempted to determine the intranasal LD50 for the ΔwbtJ mutant strain. However, mice challenged with a ΔwbtJ mutant displayed no clinical signs of illness even at the highest challenge dose (49 000 c.f.u.). In contrast, mice challenged with the complemented strain had an LD50 value of 933 c.f.u., demonstrating that WbtJ is required for virulence in vivo (Fig. 9, Table 5). At 21 days post-challenge, spleens from all mice challenged with ΔwbtJ were assayed for bacterial burden. For mice challenged with 490 c.f.u., only 1 animal had a detectable number of bacteria in the spleen (20 c.f.u.), while only 2 of the 10 mice from the highest challenge dose (49 000 c.f.u.) contained bacteria in the spleen (average of 25 c.f.u. per spleen). Each colony recovered from these samples were streak-purified, and the deletion of ΔwbtJ was confirmed via PCR. Notably, all mouse-passaged ΔwbtJ isolates recovered from this experiment maintained the ability to form biofilm and were grey varied by western analysis (Fig. S4, Table 6). Taken together, these data suggest that deletion of wbtJ can lock F. tularensis strains into a grey attenuated form.
Fig. 9.
A ΔwbtJ mutant is completely attenuated in a pneumonic tularaemia mouse model and virulence can be restored by complementation. Groups consisting of 10 mice were challenged intranasally with a range of doses for either a ΔwbtJ mutant (left) or the complemented strain (right). As a control for these experiments, one group of mice was challenged with the LVS parent (red line). The mice were followed for 21 days post challenge. The accompanying LD50 values and the median time-to-death calculated from these data are displayed in Table 5.
Table 5.
LD50 and TTD analysis for mice challenged intranasally with LVS variants and a ΔwbtJ mutant
|
LVS wild-type |
Isolate #15 |
#15-R1 |
Δ wbtJ |
Δ wbtJ + pKM43 |
||
|
LD50 (c.f.u.) |
509 |
>45 000 |
815 |
>49 000 |
933 |
|
|
Median TTD (Days) |
1–10 |
>21 |
>21 |
>21 |
>21 |
>21 |
|
10–100 |
>21 |
>21 |
>21 |
>21 |
>21 |
|
|
100–1000 |
>21 |
>21 |
>21 |
>21 |
>21 |
|
|
1,000–10 000 |
7.5 |
>21 |
7 |
>21 |
9 |
|
|
>10 000 |
6.5 |
>21 |
5 |
>21 |
7.5 |
|
|
Pair-Wise Comparisons (Wald-test) |
vs. LVS wild-type |
P<0.01 |
ns |
P<0.01 |
ns |
P values for comparison among strains were determined by Wald Chi-square tests based on a Cox regression model.
NS, Not Significant (P>0.05); TTD, Time to Death.
Table 6.
Phenotypic summary of isolates recovered from challenged mice at 21 days post-challenge
|
Strain |
Challenge dose (c.f.u.) |
# of mice w/ detectable LVS |
Avg c.f.u. Recovered* |
Colony morphology |
LPS status† |
Biofilm Formation |
|---|---|---|---|---|---|---|
|
LVS wild-type |
470 |
5/5 |
204 |
Normal |
nd |
nd |
|
Isolate #15 |
450 |
0/5 |
0 |
|||
|
45 000 |
10/10 |
174 |
Normal |
Blue form |
Negative |
|
|
ΔwbtJ |
490 |
1/5 |
20 |
Small |
Grey form |
Positive |
|
49 000 |
2/10 |
25 |
Small |
Grey form |
Positive |
|
*Based on plating homogenate from the complete spleen.
†As determined by western blot analysis using mAb specific to F. tularensis O-Ag.
Discussion
Biofilm formation is considered to be a virulence factor among many pathogens, but there is little information available on how biofilm and pathogenesis intersect in F. tularensis. The goal of this study was to characterize the virulence of F. tularensis variants known to produce a robust biofilm within 24 h. The major finding of this work is that each of the biofilm-producing variants for both type A and B isolates used in this study displayed high levels of attenuation via multiple exposure routes in a murine model of tularaemia. We built upon this finding as we identified that mutation of the wbtJ gene was responsible for biofilm production in the natural variants and showed that a ΔwbtJ mutant recapitulated the associated phenotypes of the spontaneous variants. Additionally, we provided evidence that grey to blue variation occurs in vivo and show that reversion from the grey to the blue form allows F. tularensis to switch from an avirulent to a virulent state.
Previous studies utilizing murine macrophages to study intracellular survival have shown that grey variants remain as infective as wild-type since similar c.f.u. are recovered post-infection (between 0–4 h) but can differ in fitness once within host cells (12–24 h). For instance, the LVSG variant studied by Soni et al. within macrophage host cells did not display a survival defect, while the variant studied by Hartley et al. showed a reduced ability to survive intracellularly [23, 25]. Each of these studies utilized J774A.1 macrophages, which is where we began assessing virulence in the natural biofilm-forming F. tularensis isolates. Within the biofilm variants we tested, variability for intracellular fitness was also observed as 244 BF-1 and 255 BF-1 showed little replication, while c.f.u. recovery for 255 BF-4 at 24 h post-infection was comparable to wild-type. An advantage of our study over previous studies is that the variant genomes have been sequenced, allowing for reasonable conclusions about affected gene(s) or disrupted enzyme(s) causing grey variation and associated attenuation to be made.
Placing these results into the context of the genomic analysis of our strains for the wbtJ gene, 255 BF-4 contained a mutation further downstream of wbtJ than these other strains, which could potentially allow some functionality of the enzyme. The location of the wbtJ mutations (Fig. 4a) could also explain the observed pneumonic LD50 differences between the two FRAN255 variants when comparing 255 BF-1 (LD50>460 000) to 255 BF-4 (LD50=2151) (Table 2). It is also worth noting that in a previous study, 255 BF-4 was identified as unique due to some residual low-molecular-weight LPS being present by western analysis [28]. The observed LD50 differences in virulence between the various described grey variants in this study could be linked to the state of the LPS. In the case of Soni et al., the LVSG mutant possessed full-length O-Ag, while the rough grey variant characterized by Hartley et al. appeared to lack O-Ag. It is likely that not all grey variants behave similarly despite a shared qualitative phenotype, as different genetic mutations can possibly result in different molecular O-Ag composition. Further analysis of the strains used in our study is currently being undertaken to better characterize the alteration of the O-Ag structure in biofilm-forming variants and the ΔwbtJ mutant.
Assessing the virulence by vector mimic showed that the variant strains were attenuated by at least a magnitude of 103 when compared to the parental strain. These data also suggest that the route of exposure impacts on the virulence in these strains, as 244 BF-1 and 255 BF-4 were completely attenuated with doses of 103 by subcutaneous and intradermal inoculation, but mice challenged intranasally were susceptible at this dose (compare Figs 2 and 3). It was also notable that all but one spleen was found to be sterile for mice challenged by vector mimic, while all mice challenged intranasally harboured bacteria within the spleen. A previous mouse challenge study with the wild-type parental isolates found that FRAN244 and FRAN255 displayed an average TTD of 4 and 6 days, respectively, when delivered at a dose between 103–104 c.f.u. intranasally [40]. Comparing those results to those of the current study, the biofilm-forming variants 244 BF-1 and 255 BF-1 TTD required nearly twice as long (8 and 11 days) at a dose between 103 and 104, respectively. Given that a low level of F. tularensis was present in the spleen homogenates, in hindsight it would be interesting to know whether these biofilm variants were reverted to the blue form. While multiple colony morphologies were observed in these spleen samples, no further characterization was performed at that time. However, since mice are exquisitely sensitive to F. tularensis, as the LD50 is normally ~1 c.f.u., reversion would presumably be lethal, which could account for the delayed TTD. To address this limitation, LVS was utilized in additional intranasal challenge studies, as this strain is attenuated at lower doses in mice, which could provide a window to detect reversion events.
In agreement with others, we have found that grey variants have a reduced fitness in mammalian host cells and in vivo [18, 23, 25]. We have extended these findings to include the link to phase variation and biofilm-forming isolates. It is still unclear how blue/grey variation of F. tularensis or biofilm formation factors into pathogenesis or survival in the environment. The blue form is the most represented isolate and considered the prototypical ‘wild-type’. Blue to grey variation has been known to occur on a scale of 10−3 and is readily observed in both type A and B isolates in a laboratory setting [21]. Perhaps variation to the grey form is an adaptation only observed under laboratory conditions, but this still remains to be determined. However, since the outcome of variation to grey leads to both an attenuated form and change in antigenicity, it is important to understand this phenomenon from a potential medical countermeasure perspective.
While certainly not exhaustive, the complete genomes of four LVS grey variants (isolates #11, #13, #14 and #15) were sequenced and were found to contain seemingly random SNPs in genes required for LPS synthesis rather than genomic rearrangements. While it was previously been observed that mutations within O-Ag synthesis genes quickly arise at a low frequency [47], it is puzzling why the variation would be at a minimum 100× more frequent than what would be expected if spontaneous mutations (rate of 10−5 to 10−8 [48]) were the sole reason for this variation. Complete genome sequencing of the eight biofilm-forming variants in this study (strains in Tables 3 and 4) detected only SNPs at a low frequency (1–6 per genome), which is consistent with the genomic stability reported for F. tularensis genomes [49, 50] and the results of studies on phase variation [51].
One possibility is that F. tularensis exists as a mixed blue/grey population in the natural environment, and this population shifts in an environment-dependent manner. For instance, robust biofilm is required for transmission of Yersinia pestis from a flea vector but is dispensable for pathogenesis within a mammalian host [41, 52]. Interestingly, the LPS of Y. pestis is also modified while maintained within a flea [53]. For F. tularensis, it is currently unknown if biofilm formation or phase variation occurs in an arthropod vector or if biofilm aids in persistence. Perhaps a mixed population is the predominant phenotype within a vector, but only blue forms are isolated from mammalian tularaemia infections as the grey form have a reduced fitness. Phenotypic plasticity is observed in Burkholderia pseudomallei, as the bacterium is thought to alter surface determinants, including the O-Ag, to facilitate survival [54, 55]. Further, variation of LPS in B. pseudomallei has been linked to persistence in an aquatic environment, ability to produce biofilm and host immune defence [56, 57]. With this in mind, a case can be made that variation of the LPS allows these Gram-negative intracellular pathogens to adapt to environmental or host reservoirs.
The emerging model from these data presented in this study is that grey variants can infect host cells but are impaired for replication or potential dissemination. This impairment then allows grey variants to persist, while reversion can occur, potentially tipping the infection toward lethality. Noteworthy, macrophages infected with our biofilm variants did not appear to undergo apoptosis as one would expect in response to F. tularensis strains having alterations to the LPS and/or O-Ag [58–60]. These data presented in this study support the theory put forth by Cowley et al. that phase variation can modulate intracellular growth to control host infection dynamics [18, 19].
To understand why these grey variants were constitutively producing biofilm, we performed whole-genome sequencing and, surprisingly, all biofilm-forming variants contained a mutation within wbtJ (eight in total). WbtJ is a N-formyltransferase that catalyses dTDP-4, 6-dideoxy-amino-d-glucose (dTDP-Qui4N) to dTDP-4, 6-dideoxy-4-formido-d-glucose (dTDP-Qui4NFm), which is incorporated into O-Ag. Within the wbt locus, wbtI, wbtJ, wbtK, wbtL and wbtM are genes unique to type A and B isolates that play a role in O-Ag synthesis [43, 61, 62] (Fig. 4). Namely, WbtL is thought to make activated dTDP-glucose, while WbtM is a dTDP-d-glucose 4,6 dehydratase; amination occurs through WbtI, resulting in dTDP-Qui4N, WbtJ formylates this sugar to become dTDP-Qui4NFm and WbtK functions as the glycosyltransferase to aid in biosynthesis of the O-Ag unit [43, 45, 61, 62]. Notably, blue/grey phase variation has not been observed in the closely related subspecies Francisella novicida, but the O-Ag unit differs in F. novicida compared to the virulent type A and B isolates [63, 64], and F. novicida lacks these five additional wbt genes [49, 65, 66]. While it is tempting to speculate that blue/grey variation is due solely to these additional genes, we detected spontaneous mutations within wbtA, wzy and waaL (Table S1), which are also encoded in the F. novicida genome. A previous study by Kurtz et al. performed genomic analysis on two different phase variants with altered LPS in an effort to understand immunogenicity, as LVS-G was moderately protective, while LVS-R provided little protection in mice [51, 67]. The exact mutation responsible for grey variation in these strains is unclear, although it is likely that a non-sense mutation in FTL_0937 (polysaccharide biosynthesis) in LVS-G and FTL_0429 (a glutamine amidotransferase) in LVS-R results in the associated phenotypes. This would suggest that an additional factor likely governs phase variation and may exert selective pressure to inactivate genes required for O-Ag synthesis or a regulatory network for LPS and cell surface antigens.
To deconstruct phase variation from biofilm formation, it is still unclear why inactivation of wbtJ specifically results in the production of a robust biofilm, and further studies are required to understand this phenomenon. One hypothesis that can be made from this work is that general dysfunction or ablation of surface polysaccharides can alter cell surface properties, such as charge, to promote attachment and/or cell-to-cell adhesion. However, if this were true, most grey variants would produce a robust biofilm. The enzymes encoded by wzy (mutated in isolate 13) and waaL (mutated in isolate 14) play a role in the polymerization and ligation of the assembled O-Ag unit, while wbtA (mutated in isolate 11) and wbtJ (mutated in isolate 15) are enzymes required to synthesize saccharides incorporated into the O-Ag [68]. It was previously shown that grey variation and O-Ag can affect the production of outer membrane vesicles (OMVs) [25, 69], which can have implications for biofilm formation, as increased OMV secretion has been linked to biofilm production [70]. LVS isolates #11, #13 and #14 do not form a biofilm, while a distinct extracellular matrix is present in biofilms produced by LVS isolate #15 [28]. The lack of biofilm formation by LVS isolate #11 (wbtA mutant) and LVS isolate #13 (wzy mutant) suggests that lack of a complete O-Ag or diminution of O-Ag repeat length does not account for the robust biofilm formation observed in wbtJ mutants. F. tularensis produces both O-antigen LPS and O-antigen capsule, both of which consists of repeating units of Qui4NFm-GalNAcAN-GalNAcAN-GuiNac. Mutations in wbtA, wzy and waaL are all thought to lack this structure [71]. While we have previously shown that stochastic biofilm formation through this specific phase variation mechanism lacks reactivity to mAb against capsule [28], it is unclear whether the capsule or capsule-like complex is present in wbtJ mutants and will require additional exploration in future studies.
We also found that a strong selective pressure exists to select for reversion events when multiple colony morphologies were identified in macrophage lysates and, more importantly, in vivo when determining the bacterial burden of spleen homogenates. While the complete genome of the reverted isolates was not sequenced, our data are the first to confirm the original hypothesis of Eigelsbach that restoration happens in the form of S → NS → S′ rather than S⇄ NS (smooth, non-smooth [21]). Simply stated, reversion occurs as a result of a secondary mutation masking the previous phenotype. A caveat to our study is that we relied upon LVS isolate #15, which harbours a SNP, to monitor reversion. We consistently identified three forms of the repaired wbtJ allele (encoding a ser, leu or gln) for reverted strains of LVS isolate #15 and wondered whether saturation was reached for the repaired alleles. However, other potential mutations that could arise at this codon would result in tyrosine (aromatic), lysine or glutamine (charged), or another stop codon. With this in mind, we suspect that the reverted isolates of LVS isolate #15 we identified are the most likely alleles that would repair WbtJ enzymatic function, especially considering codon 153 is in close proximity to the region that interacts with substrate [45].
It is likely that more deleterious mutations, such as deletion of multiple nucleotides, may be more difficult to repair and likely explain why some variants appear more stable than others [21, 33]. Thus, the in-frame deletion of ΔwbtJ LVS appears to have created a ‘locked’ grey variant that has not been observed to revert to the blue form. In addition, the ΔwbtJ mutant recapitulates the phenotypes associated with natural biofilm-forming variants. Construction of a locked variant is crucial to improving both the safety and the efficacy of live vaccine strains of F. tularensis [24]. Ideal candidates are thought to be variants locked in blue form [20, 67]; however, it is possible that a particular grey form may be able retain the immunogenic properties necessary for vaccination and display high levels of attenuation, as a systematic study of all the mutants responsible for phase variation has yet to be completed.
An additional consideration regarding biofilm formation in F. tularensis is that perhaps not all bacterial cells need to actively produce the extracellular matrix to reap the benefits offered by biofilm. Viewed in this light, stochastic variation of F. tularensis resulting in the ability to form biofilm is a novel example of division of labour in bacteria. It would be interesting, and important, to explore whether both blue and grey forms are encased within the extracellular matrix produced by biofilm-forming variants, especially considering that variation arises post-exponentially in ageing cultures [23, 28]. Given the stochastic nature of mutations causing variance, natural populations of F. tularensis may also be heterogenous to cope with changing environments, with each form differentially fitted for survival, resulting in an increase in overall fitness of the population. For instance, grey variants more readily form biofilm and stave off entering a viable but non-culturable state, which could enable better environmental persistence [28]. Grey variants, as well as a viable but non-culturable state, have been found to be attenuated in infection models, while blue variants are typically more fit within a host cell [72]. Bias towards blue fitness intracellularly has been demonstrated by using co-infection of macrophages with both blue and grey forms (1 : 10 ratio), with this resulting in the recovery of blue to grey at 100 : 1 [23]. These data presented in this current study further highlight this fitness difference as we detected reversion of grey variants to blue forms during infection.
In conclusion, the wbtJ gene has not previously been characterized in F. tularensis. We show that deletion of this gene causes F. tularensis to form robust biofilms and decreases the intracellular fitness of the bacterium. We further show that wbtJ is essential for pathogenesis in F. tularensis. The WbtJ enzyme could potentially be explored as a target for new antibiotics. Additionally, we have shown a link between the wbtJ gene and blue/grey variation that could potentially be exploited for medical countermeasure development to lock live vaccines into a particular variant form to aid in vaccine efficacy.
Supplementary Data
Funding information
This work was supported by a grant from JSTO-CBD CB10645 and CB10477 (USAMRIID project numbers 92 138 743 and 92131712).
Acknowledgements
We thank Martin Pavelka (University of Rochester) for use of the plasmid pMP814. We thank Michelle Whitehouse for technical assistance. Opinions, interpretations, conclusions and recommendations are those of the authors and not necessarily endorsed by the US Army.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This study was carried out in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals, National Research Council, 2011 and The United States Army Medical Research Institute of Infectious Diseases Institutional Animal Care and Use Committee. The protocol was approved by the United States Army of Medical Research Institute of Infectious Diseases under an Institutional Animal Care and Use Committee in compliance with the Animal Welfare Act, PHS Policy, and other Federal statutes and regulations relating to animals and experiments involving animals. The facility where this research was conducted is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Footnotes
Abbreviations: AMP, antimicrobial peptide; BHI, brain heart infusion broth; CDM, Chamberlain’s defined medium; c.f.u., colony forming unit; LD50, Lethal dose 50; LPS, lipopolysaccharide; LVS, Live Vaccine Strain; O-Ag, O-antigen; OD, optical density; PAMP, Pathogen-associated Molecular Pattern; PBS, phosphate buffered saline; PCR, polymerase chain reaction; SNP, single nucleotide polymorphism; SNP, single nucleotide polymorphism; TBST, Tris-buffered saline, Tween-20; TTD, Time to Death.
Four supplementary figures and three supplementary tables are available with the online version of this article.
References
- 1.Jellison W. Tularemia in North America. University of Montana: University of Montana Foundation; 1974. p. 276. p. [Google Scholar]
- 2.Ellis J, Oyston PCF, Green M, Titball RW. Tularemia. Clin Microbiol Rev. 2002;15:631–646. doi: 10.1128/CMR.15.4.631-646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop A, Wang H-H, Donaldson TG, Brockinton EE, Kothapalli E, et al. Tularemia cases increase in the USA from 2011 through 2019. Curr Res Parasitol Vector Borne Dis. 2023;3:100116. doi: 10.1016/j.crpvbd.2023.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hepburn MJ, Simpson AJH. Tularemia: current diagnosis and treatment options. Expert Rev Anti Infect Ther. 2008;6:231–240. doi: 10.1586/14787210.6.2.231. [DOI] [PubMed] [Google Scholar]
- 5.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, et al. Tularemia as a biological weapon: medical and public health management. JAMA. 2001;285:2763–2773. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 6.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella . Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 7.Oyston PCF, Sjostedt A, Titball RW. Tularaemia: bioterrorism defence renews interest in Francisella tularensis . Nat Rev Microbiol. 2004;2:967–978. doi: 10.1038/nrmicro1045. [DOI] [PubMed] [Google Scholar]
- 8.Sjöstedt A. Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. [DOI] [PubMed] [Google Scholar]
- 9.Hestvik G, Warns-Petit E, Smith LA, Fox NJ, Uhlhorn H, et al. The status of tularemia in Europe in a one-health context: a review. Epidemiol Infect. 2015;143:2137–2160. doi: 10.1017/S0950268814002398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maurin M, Gyuranecz M. Tularaemia: clinical aspects in Europe. Lancet Infect Dis. 2016;16:113–124. doi: 10.1016/S1473-3099(15)00355-2. [DOI] [PubMed] [Google Scholar]
- 11.Saslaw S, Eigelsbach HT, Prior JA, Wilson HE, Carhart S. Tularemia vaccine study. II. Respiratory challenge. Arch Intern Med. 1961;107:702. doi: 10.1001/archinte.1961.03620050068007. [DOI] [PubMed] [Google Scholar]
- 12.McCrumb FR. Aerosol infection of man with Pasteurella tularensis . Bacteriol Rev. 1961;25:262–267. doi: 10.1128/br.25.3.262-267.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohrbach BW, Westerman E, Istre GR. Epidemiology and clinical characteristics of tularemia in Oklahoma, 1979 to 1985. South Med J. 1991;84:1091–1096. doi: 10.1097/00007611-199109000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Conlan JW, North RJ. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect Immun. 1992;60:5164–5171. doi: 10.1128/iai.60.12.5164-5171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horzempa J, O’Dee DM, Stolz DB, Franks JM, Clay D, et al. Invasion of erythrocytes by Francisella tularensis . J Infect Dis. 2011;204:51–59. doi: 10.1093/infdis/jir221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole LE, Elkins KL, Michalek SM, Qureshi N, Eaton LJ, et al. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J Immunol. 2006;176:6888–6899. doi: 10.4049/jimmunol.176.11.6888. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Kuolee R, Shen H, Bùsa M, Conlan JW. Toll-like receptor 4 (TLR4) plays a relatively minor role in murine defense against primary intradermal infection with Francisella tularensis LVS. Immunol Lett. 2005;97:151–154. doi: 10.1016/j.imlet.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Cowley SC, Myltseva SV, Nano FE. Phase variation in Francisella tularensis affecting intracellular growth, lipopolysaccharide antigenicity and nitric oxide production. Mol Microbiol. 1996;20:867–874. doi: 10.1111/j.1365-2958.1996.tb02524.x. [DOI] [PubMed] [Google Scholar]
- 19.Cowley SC, Myltseva SV, Nano FE. Suppression of Francisella tularensis growth in the rat by co-infection with F. novicida. FEMS Microbiol Lett. 1997;153:71–74. doi: 10.1111/j.1574-6968.1997.tb10465.x. [DOI] [PubMed] [Google Scholar]
- 20.Eigelsbach HT, Downs CM. Prophylactic effectiveness of live and killed tularemia vaccines. I. Production of vaccine and evaluation in the white mouse and guinea pig. J Immunol. 1961;87:415–425. [PubMed] [Google Scholar]
- 21.Eigelsbach HT, Braun W, Herring RD. Studies on the variation of Bacterium tularense . J Bacteriol. 1951;61:557–569. doi: 10.1128/jb.61.5.557-569.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell JF, Owen CR, Larson CL. Virulence of Bacterium tularense. I. A study of the virulence of Bacterium tularense in mice, guinea pigs, and rabbits. J Infect Dis. 1955;97:162–166. doi: 10.1093/infdis/97.2.162. [DOI] [PubMed] [Google Scholar]
- 23.Hartley G, Taylor R, Prior J, Newstead S, Hitchen PG, et al. Grey variants of the live vaccine strain of Francisella tularensis lack lipopolysaccharide O-antigen, show reduced ability to survive in macrophages and do not induce protective immunity in mice. Vaccine. 2006;24:989–996. doi: 10.1016/j.vaccine.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 24.Waag DM, Sandström G, England MJ, Williams JC. Immunogenicity of a new lot of Francisella tularensis live vaccine strain in human volunteers. FEMS Immunol Med Microbiol. 1996;13:205–209. doi: 10.1111/j.1574-695X.1996.tb00238.x. [DOI] [PubMed] [Google Scholar]
- 25.Soni S, Ernst RK, Muszyński A, Mohapatra NP, Perry MB, et al. Francisella tularensis blue-gray phase variation involves structural modifications of lipopolysaccharide o-antigen, core and lipid a and affects intramacrophage survival and vaccine efficacy. Front Microbiol. 2010;1:129. doi: 10.3389/fmicb.2010.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland KM, Rosa SJ, Kristjansdottir K, Wolfgeher D, Franz BJ, et al. Differential growth of Francisella tularensis, which alters expression of virulence factors, dominant antigens, and surface-carbohydrate synthases, governs the apparent virulence of Ft SchuS4 to immunized animals. Front Microbiol. 2017;8:1158. doi: 10.3389/fmicb.2017.01158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champion AE, Catanzaro KCF, Bandara AB, Inzana TJ. Formation of the Francisella tularensis biofilm is affected by cell surface glycosylation, growth medium, and a glucan exopolysaccharide. Sci Rep. 2019;9:12252. doi: 10.1038/s41598-019-48697-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mlynek KD, Lopez CT, Fetterer DP, Williams JA, Bozue JA. Phase variation of LPS and capsule is responsible for stochastic biofilm formation in Francisella tularensis . Front Cell Infect Microbiol. 2021;11:808550. doi: 10.3389/fcimb.2021.808550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaudinn C, Rydzewski K, Meister B, Grunow R, Heuner K. Francisella tularensis subsp. holarctica wild-type is able to colonize natural aquatic ex vivo biofilms. Front Microbiol. 2023;14:1113412. doi: 10.3389/fmicb.2023.1113412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 31.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 32.James GA, Swogger E, Wolcott R, Pulcini E deLancey, Secor P, et al. Biofilms in chronic wounds. Wound Repair Regen. 2008;16:37–44. doi: 10.1111/j.1524-475X.2007.00321.x. [DOI] [PubMed] [Google Scholar]
- 33.Chamberlain RE. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl Microbiol. 1965;13:232–235. doi: 10.1128/am.13.2.232-235.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seeman T. snippy: fast bacterial variant calling from NGS reads. 2015.
- 38.LoVullo ED, Miller CN, Pavelka MS, Kawula TH. TetR-based gene regulation systems for Francisella tularensis . Appl Environ Microbiol. 2012;78:6883–6889. doi: 10.1128/AEM.01679-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LoVullo ED, Sherrill LA, Pavelka MS. Improved shuttle vectors for Francisella tularensis genetics. FEMS Microbiol Lett. 2009;291:95–102. doi: 10.1111/j.1574-6968.2008.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachert BA, Richardson JB, Mlynek KD, Klimko CP, Toothman RG, et al. Development, phenotypic characterization and genomic analysis of a Francisella tularensis panel for tularemia vaccine testing. Front Microbiol. 2021;12:725776. doi: 10.3389/fmicb.2021.725776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hinnebusch BJ, Jarrett CO, Bland DM. “Fleaing” the plague: adaptations of Yersinia pestis to its insect vector that lead to transmission. Annu Rev Microbiol. 2017;71:215–232. doi: 10.1146/annurev-micro-090816-093521. [DOI] [PubMed] [Google Scholar]
- 42.Olsufiev NG, Emelyanova OS, Dunayeva TN. Comparative study of strains of B. tularense in the old and new world and their taxonomy. J Hyg Epidemiol Microbiol Immunol. 1959;3:138–149. [PubMed] [Google Scholar]
- 43.Prior JL, Prior RG, Hitchen PG, Diaper H, Griffin KF, et al. Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J Med Microbiol. 2003;52:845–851. doi: 10.1099/jmm.0.05184-0. [DOI] [PubMed] [Google Scholar]
- 44.Apicella MA, Post DMB, Fowler AC, Jones BD, Rasmussen JA, et al. Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis . PLoS One. 2010;5:e11060. doi: 10.1371/journal.pone.0011060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zimmer AL, Thoden JB, Holden HM. Three-dimensional structure of a sugar N-formyltransferase from Francisella tularensis . Protein Sci. 2014;23:273–283. doi: 10.1002/pro.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindgren M, Bröms JE, Meyer L, Golovliov I, Sjöstedt A. The Francisella tularensis LVS ΔpdpC mutant exhibits a unique phenotype during intracellular infection. BMC Microbiol. 2013;13:20. doi: 10.1186/1471-2180-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dwibedi C, Larsson P, Ahlinder J, Lindgren P, Myrtennäs K, et al. Biological amplification of low frequency mutations unravels laboratory culture history of the bio-threat agent Francisella tularensis . Forensic Sci Int Genet. 2020;45:102230. doi: 10.1016/j.fsigen.2019.102230. [DOI] [PubMed] [Google Scholar]
- 48.Torii N, Nozaki T, Masutani M, Nakagama H, Sugiyama T, et al. Spontaneous mutations in the Helicobacter pylori rpsL gene. Mutat Res. 2003;535:141–145. doi: 10.1016/s1383-5718(02)00292-9. [DOI] [PubMed] [Google Scholar]
- 49.Rohmer L, Fong C, Abmayr S, Wasnick M, Larson Freeman TJ, et al. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 2007;8:R102. doi: 10.1186/gb-2007-8-6-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohmer L, Brittnacher M, Svensson K, Buckley D, Haugen E, et al. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect Immun. 2006;74:6895–6906. doi: 10.1128/IAI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurtz SL, Voskanian-Kordi A, Simonyan V, Elkins KL. Sequence comparison of Francisella tularensis LVS, LVS-G and LVS-R. Pathog Dis. 2018;76 doi: 10.1093/femspd/fty067. [DOI] [PubMed] [Google Scholar]
- 52.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, et al. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J Infect Dis. 2004;190:783–792. doi: 10.1086/422695. [DOI] [PubMed] [Google Scholar]
- 53.Aoyagi KL, Brooks BD, Bearden SW, Montenieri JA, Gage KL, et al. LPS modification promotes maintenance of Yersinia pestis in fleas. Microbiology (Reading) 2015;161:628–638. doi: 10.1099/mic.0.000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, et al. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei . J Bacteriol. 2007;189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wikraiphat C, Saiprom N, Tandhavanant S, Heiss C, Azadi P, et al. Colony morphology variation of Burkholderia pseudomallei is associated with antigenic variation and O-polysaccharide modification. Infect Immun. 2015;83:2127–2138. doi: 10.1128/IAI.02785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mlynek KD, Bozue JA. Why vary what’s working? Phase variation and biofilm formation in Francisella tularensis . Front Microbiol. 2022;13:1076694. doi: 10.3389/fmicb.2022.1076694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore RA, Tuanyok A, Woods DE. Survival of Burkholderia pseudomallei in water. BMC Res Notes. 2008;1:11. doi: 10.1186/1756-0500-1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindemann SR, Peng K, Long ME, Hunt JR, Apicella MA, et al. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect Immun. 2011;79:581–594. doi: 10.1128/IAI.00863-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Case EDR, Chong A, Wehrly TD, Hansen B, Child R, et al. The Francisella O-antigen mediates survival in the macrophage cytosol via autophagy avoidance. Cell Microbiol. 2014;16:862–877. doi: 10.1111/cmi.12246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chance T, Chua J, Toothman RG, Ladner JT, Nuss JE, et al. A spontaneous mutation in kdsD, A biosynthesis gene for 3 Deoxy-D-manno-Octulosonic Acid, occurred in a ciprofloxacin resistant strain of Francisella tularensis and caused a high level of attenuation in murine models of tularemia. PLoS One. 2017;12:e0174106. doi: 10.1371/journal.pone.0174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J, Ryder C, Mandal M, Ahmed F, Azadi P, et al. Attenuation and protective efficacy of an O-antigen-deficient mutant of Francisella tularensis LVS. Microbiology. 2007;153:3141–3153. doi: 10.1099/mic.0.2007/006460-0. [DOI] [PubMed] [Google Scholar]
- 62.Twine SM, Vinogradov E, Lindgren H, Sjostedt A, Conlan JW. Roles for wbtC, wbtI, and kdtA genes in lipopolysaccharide biosynthesis, protein glycosylation, virulence, and immunogenicity in Francisella tularensis strain SCHU S4. Pathogens. 2012;1:12–29. doi: 10.3390/pathogens1010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinogradov E, Conlan WJ, Gunn JS, Perry MB. Characterization of the lipopolysaccharide O-antigen of Francisella novicida (U112) Carbohydr Res. 2004;339:649–654. doi: 10.1016/j.carres.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 64.Thomas RM, Titball RW, Oyston PCF, Griffin K, Waters E, et al. The immunologically distinct O antigens from Francisella tularensis subspecies tularensis and Francisella novicida are both virulence determinants and protective antigens. Infect Immun. 2007;75:371–378. doi: 10.1128/IAI.01241-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sjödin A, Svensson K, Ohrman C, Ahlinder J, Lindgren P, et al. Genome characterisation of the genus Francisella reveals insight into similar evolutionary paths in pathogens of mammals and fish. BMC Genomics. 2012;13:268. doi: 10.1186/1471-2164-13-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLendon MK, Apicella MA, Allen L-AH. Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol. 2006;60:167–185. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Pascalis R, Chou AY, Bosio CM, Huang C-Y, Follmann DA, et al. Development of functional and molecular correlates of vaccine-induced protection for a model intracellular pathogen, F. tularensis LVS. PLoS Pathog. 2012;8:e1002494. doi: 10.1371/journal.ppat.1002494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rasmussen JA, Post DMB, Gibson BW, Lindemann SR, Apicella MA, et al. Francisella tularensis Schu S4 lipopolysaccharide core sugar and O-antigen mutants are attenuated in a mouse model of tularemia. Infect Immun. 2014;82:1523–1539. doi: 10.1128/IAI.01640-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Champion AE, Bandara AB, Mohapatra N, Fulton KM, Twine SM, et al. Further characterization of the Capsule-Like Complex (CLC) produced by Francisella tularensis subspecies tularensis: protective efficacy and similarity to outer membrane vesicles. Front Cell Infect Microbiol. 2018;8:182. doi: 10.3389/fcimb.2018.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siebert C, Lindgren H, Ferré S, Villers C, Boisset S, et al. Francisella tularensis: FupA mutation contributes to fluoroquinolone resistance by increasing vesicle secretion and biofilm formation. Emerg Microbes Infect. 2019;8:808–822. doi: 10.1080/22221751.2019.1615848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rasmussen JA, Fletcher JR, Long ME, Allen L-A, Jones BD. Characterization of Francisella tularensis Schu S4 mutants identified from a transposon library screened for O-antigen and capsule deficiencies. Front Microbiol. 2015;6:338. doi: 10.3389/fmicb.2015.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Forsman M, Henningson E, Larsson E, Johansson T, Sandström G. Francisella tularensis does not manifest virulence in viable but non-culturable state. FEMS Microbiol Ecol. 2000;31:217–224. doi: 10.1111/j.1574-6941.2000.tb00686.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data underlying the findings and conclusions reported in this paper are fully available within the text or the accompanying Supplementary Material. Genome sequencing is available on GenBank at the time of publication. Additional data are available upon request.