Abstract
Mutations in the human mismatch repair protein hMSH2 have been found to cosegregate with hereditary nonpolyposis colorectal cancer (HNPCC). Previous biochemical and physical studies have shown that hMSH2 forms specific mispair binding complexes with hMSH3 and hMSH6. We have further characterized these protein interactions by mapping the contact regions within the hMSH2-hMSH3 and the hMSH2-hMSH6 heterodimers. We demonstrate that there are at least two distinct interaction regions of hMSH2 with hMSH3 and hMSH2 with hMSH6. Interestingly, the interaction regions of hMSH2 with either hMSH3 or hMSH6 are identical and there is a coordinated linear orientation of these regions. We examined several missense alterations of hMSH2 found in HNPCC kindreds that are contained within the consensus interaction regions. None of these missense mutations displayed a defect in protein-protein interaction. These data support the notion that these HNPCC-associated mutations may affect some other function of the heterodimeric complexes than simply the static interaction of hMSH2 with hMSH3 or hMSH2 with hMSH6.
Mismatch repair involves the recognition and repair of incorrectly paired nucleotides that result from misincorporation errors during DNA replication; from physical or chemical damage to DNA, such as the deamination of methyl cytosine which results in a G-T mispair; and from genetic recombination between DNA strands which lack perfect homology (for a review, see reference 13). Postreplication mismatch repair of polymerase misincorporation errors has been shown to increase the overall fidelity of DNA synthesis by up to a 1,000-fold (for reviews see references 19 and 24 to 26). The Escherichia coli DNA adenine methylation-instructed pathway is the best characterized postreplication mismatch repair system and has been shown to be genetically dependent on mutS, mutH, and mutL. These gene products have been purified and the bacterial mismatch repair reaction has been reconstituted in vitro. The MutS protein binds preferentially to mismatched DNA substrates as a homodimer (33). The MutH protein displays an intrinsic endonuclease activity that cleaves the unmethylated strand of a hemimethylated GATC sequence (34). Since a newly synthesized strand of DNA is transiently undermethylated, it is the MutH protein which targets mismatch repair to the nascent strand of DNA. Although the role of MutL is not entirely understood, it appears to interact with MutS and participates in the activation of the endonuclease activity of MutH (5, 15). The removal of the DNA strand containing the mispaired nucleotide as well as synthesis and ligation of this excised strand appears to be mediated by proteins which are not specific to the mismatch repair pathway.
Homologs of the bacterial mismatch repair proteins have been identified in virtually every organism examined to date with the exception of the Archaea (12). In the yeast Saccharomyces cerevisiae, several homologs of the bacterial MutS protein have been identified: MSH1 (for MutS homolog) is a nuclear-encoded protein which functions in mitochondrial mismatch repair (31, 32); MSH2, MSH3, and MSH6 function in nuclear mismatch repair (22, 31, 32); and MSH4 and MSH5 perform an as yet undetermined essential function in meiosis (7, 17, 30). A nearly identical set of human MutS homologs (hMSH2, hMSH3, hMSH4, hMSH5, and hMSH6) have been identified, the exception being MSH1 (for a review, see reference 12). Similarly, three MutL homologs (MLH1 [hMLH1], MLH2 or MLH3 [hPMS1], and PMS1 [hPMS2]) have been identified in yeast and humans (12).
Both in humans and yeast, MSH2 and MSH6 form a heterodimer which specifically binds single-mispaired nucleotides and a subset of nucleotide insertion-deletion mismatches, while MSH2 and MSH3 form a heterodimer which is specific for an overlapping subset of nucleotide insertion-deletion mismatches as well as larger insertion-deletion loops (1, 22). Insertion-deletion structures have been proposed to arise when DNA polymerase “slips” while replicating through DNA containing simple repeat sequences (20).
Germ line mutations in hMSH2 and hMLH1 account for the majority of hereditary nonpolyposis colon cancer (HNPCC) families, implicating mismatch repair in the etiology of disease (6, 11, 29). Interestingly, mutations of hMSH6, hPMS1, and hPMS2 appear to be rare, and there have been no reported mutations of hMSH3 in HNPCC (2, 23, 27). While the functional consequences of mutations that result in protein truncations might be anticipated (approximately 75% of the germ line mutations), the functional consequences of missense mutations that predispose to HNPCC is unknown. Furthermore, the missense mutations of hMSH2 and hMLH1 do not appear to be clustered but are rather spread throughout the coding sequence of both genes (29).
Mutation analysis has determined some of the functional regions of the MutS homologs. Amino acid sequence comparisons have revealed a highly conserved adenine nucleotide binding-hydrolysis region associated with a helix-turn-helix region (10). Furthermore, a number of MutS homologs have been shown to possess a weak ATPase activity (4, 8, 14, 16). Site-directed mutagenesis of this region revealed that although the ATPase region is not essential for mismatch binding activity, it is essential for the completion of a repair event (4, 16). This conclusion is underlined by the observation that several of these mutants display a dominant-negative phenotype (4, 16, 35).
Based on structural comparisons to the lambda repressor, Cro, and Cap proteins, it was suggested that the helix-turn-helix region might be involved in mispair recognition by the MutS proteins (32). However, site-directed mutagenesis of the helix-turn-helix region in the yeast MSH2 appeared to produce no effect on mismatch binding activity (3). Thus, it appears unlikely that the helix-turn-helix region plays a singular role in mismatch recognition. In addition, the carboxy-terminal 114 amino acids (aa) of yeast MSH2 were found to be essential for interaction with MSH6 (3). Interestingly, several reports have detailed frame-shift and/or truncation mutations in hMSH3 and hMSH6 in up to 60% of microsatellite unstable sporadic tumors (21), even though germ line mutations of these genes are rare or absent. These results suggest that a more thorough understanding of the structure-function regions of the human MutS homologs hMSH2, hMSH3, and hMSH6 might provide a biochemical foundation for their role(s) in carcinogenesis.
In this study, we have defined the hMSH2-hMSH3 and hMSH2-hMSH6 interaction regions. We found that there are two distinct interaction regions for both the hMSH2-hMSH3 and hMSH2-hMSH6 heterodimers. The interaction regions of hMSH2 with either hMSH3 or hMSH6 appear to be identical. We have constructed several missense mutations of hMSH2 that were reported to cosegregate with HNPCC. Although the interaction assay is nonquantitative, we found that none of these alterations affected the contacts within these protein heterodimers.
MATERIALS AND METHODS
Reagents and enzymes.
Restriction endonucleases were purchased from New England Biolabs (Beverly, Mass.). PCRs were performed with the High Fidelity PCR Kit from Boehringer Mannheim (Mannheim, Germany). Oligonucleotides were synthesized on a 3948 Nucleic Acid Synthesis and Purification System (Applied Biosystems; Foster City, Calif.). DNA plasmid constructs were purified by using Qiagen (Hilden, Germany) DNA purification kits. In vitro transcription and translation (IVTT) reactions were performed by using the TNT Coupled Rabbit Reticulocyte Lysate System (Promega; Madison, Wis.). Radiolabeled [35S]methionine used to label proteins was obtained from Dupont NEN (Wilmington, Del.). Glutathione-linked agarose beads were purchased from Sigma (St. Louis, Mo.).
Subcloning of hMSH2 and hMSH3.
The cloning of hMSH2, hMSH3, and hMSH6 cDNAs and subcloning into pET expression vectors (Novagen) has been previously described (1). In this study, we used a HeLa cDNA clone of hMSH3 (GenBank accession no. U61981).
Glutathione S-transferase (GST) fusion proteins were made by using the pGEX system (Pharmacia, Uppsala, Sweden). For ease of cloning, pGEX-4T-2 was modified as follows. The vector DNA was digested with EcoRI and BamHI and gel purified. The following linker was introduced by ligation (top strand, 5′-GAT CCG AGA ACC TGT ACT TCC AGG GAC ATA TGG CCA TGG GTA CCG-3′; bottom strand, 5′-AAT TCG GTA CCC ATG GCC ATA TGT CCC TGG AAG TAC AGG TTC TCG-3′); this vector is referred to as pGEX-SG1 and allows for subcloning with NdeI and NcoI restriction endonuclease sites in which the ATG initiation codon within each site is in-frame with the GST moiety. This vector also contains a tobacco etch virus (TEV) protease site just upstream of the NdeI and NcoI sites.
Construction of hMSH2 truncation mutations.
The hMSH2 deletion mutants were constructed by PCR truncation mutagenesis. Forward primers were designed by using a codon which had a guanine in the first position and adding the subsequent 17 nucleotides to the following sequence: 5′-GCG GAT CCC ATG G-3′. Reverse primers were designed by adding the first 18 nucleotides of the complementary strand to the following sequence: 5′-GGA GGA TCC CTA-3′. By using a forward and reverse primer, PCR was performed with pET3d-hMSH2 as template. The PCR product and pET24d were digested with NcoI and BamHI, gel purified, and ligated together.
To make truncated peptides containing an internal deletion, pET 24d-hMSH2 (aa 700 to 800 deleted with NdeI) was constructed by performing a PCR on hMSH2 with the primers 5′-GCG GAT CCC ATG GCA GAA GTG TCC ATT GTG-3′ and 5′-GGA GGA TCC CAT ATG TAG ATT ATT AAC AGT TGG-3′, digesting this product and pET24d with NcoI and BamHI, gel purifying each, and ligating them together. This vector allowed for the ligation of fragments with NdeI and BamHI. Forward primers were designed with the first 18 nucleotides preceded by the sequence 5′-GGC GGT ATC CAT ATG-3′. The reverse primer was the same as described above. PCR fragments were ligated into this vector with NdeI and BamHI. Site-directed mutagenesis of hMSH2 was done by using overlap PCR (18). All of the site-directed mutations were completely sequenced with a Perkin-Elmer ABI Sequencer with XL upgrade.
Construction of hMSH3 and hMSH6 truncation mutations.
The hMSH3 and hMSH6 truncation constructs were created similarly to how the hMSH2 deletion mutants were constructed. The forward primers were designed exactly the same. The reverse primers were designed by using the sequence 5′-GGC ATA CTC GAG CTA-3′. The PCR product was subcloned into either pET24d or pGEX-SG1. pET24d-hMSH3 (aa 800 to 990 deleted with AgeI) was constructed by PCR with the primers 5′-GCG GAT CCC ATG GAT TTT CTA GAG AAA TTC-3′ and 5′-GGA CGC GTC GTC GAC CTA ACC GGT ATC TCT GAT GAA ATA CTC-3′. The product from this PCR and pET24d were then digested with NcoI and SalI and subcloned. This vector then allowed for the ligation of inserts with AgeI and XhoI. Forward primers used the sequence 5′-GCG GTG ACC GGT-3′. PCR was performed and the product was ligated into pET24d-hMSH3 (aa 800 to 990 deleted with AgeI). In order to avoid errors introduced by random PCR mutagenesis, all constructs made by PCR were either completely sequenced or the experiments were conducted by using two separately isolated PCR products. A complete list of primers used in these constructions is available upon request.
GST-fusion protein interaction assay.
An overnight culture of pGEX-hMSH(X) was grown in Luria broth (LB) with 50 μg of ampicillin/ml. A total of 50 ml of LB with ampicillin was inoculated with 1 ml of overnight culture and grown to an optical density at 600 nm of 0.5. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mM, and the mixture was placed in a shaker at 30°C for 2 h. Induced cells were pelleted and resuspended in 800 μl of phosphate-buffered saline (Boehringer Mannheim) plus protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 0.8 mg of leupeptin/ml, 0.8 mg of pepstatin/ml, and 0.1 mM EDTA). Lysozyme was added to a concentration of 1 mg/ml, and the mixture was left on ice for 30 min. Triton X-100 and dithiothreitol (DTT) were then added to final concentrations of 0.2% and 2 mM, respectively, and the lysate was frozen and thawed two times to completely lyse the cells. DNase (Boehringer Mannheim) was added to a final concentration of 20 μg/ml, and the lysate was incubated on ice for 20 min. Cell debris was cleared by centrifugation at 14,000 rpm with a refrigerated Eppendorf (model 5402) centrifuge for 30 min, and the supernatant was transferred to a new microcentrifuge tube with rehydrated GST beads such that approximately 10 to 50 ng of protein were bound to each 25 μl of beads (see below for quantitation of GST-fusion protein levels). The lysate-GST beads were incubated at 4°C on a rocking platform.
After being rocked at 4°C for 1 to 2 h, the lysate-GST beads were spun at 1,000 rpm in the Eppendorf centrifuge (model 5415C) for 30 s, the supernatant was removed, and the beads were gently resuspended in 500 μl of Binding Buffer (20 mM Tris [pH 7.5], 10% glycerol, 150 mM NaCl, 5 mM EDTA, 1 mM DTT, 0.1% Tween 20, 0.75 mg of bovine serum albumin [BSA], 0.5 mM phenylmethylsulfonyl fluoride, 0.8 mg of leupeptin/ml and 0.8 mg of pepstatin/ml). The centrifugation-resuspension was repeated three times to wash the beads free of most nonspecific lysate proteins. The slurry was then added to a 14-ml sterile polypropylene tube, diluted with Binding Buffer to approximately 50 μl of packed glutathione beads per ml, and incubated at 4°C on a rocking platform for 30 min in order to allow BSA to coat the beads. A total of 500 μl (10 to 50 ng of bound GST-fusion protein) of these coated GST-fusion protein-associated glutathione beads was then aliquoted into 1.5-ml microcentrifuge tubes. GST-fusion protein expression levels were determined by binding the lysate to glutathione beads (as described above), followed by three washes and quantitation of protein on Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels with BSA as a standard (14).
IVTT (Promega) reactions with [35S]methionine were performed with pET-hMSH(Y) by using purified DNA (Qiagen) according to the manufacturer’s recommendations. IVTT reactions were prerun to determine the relative molar concentration of each construct. This was calculated by using the specific activity of the [35S]methionine, correcting for the number of methionines in each IVTT construct, and using SDS-PAGE and a Molecular Dynamics PhosphorImager with ImageQuant software (Sunnyvale, Calif.). Up to 10 μl of the IVTT protein was added to each tube such that each sample had a relatively equimolar concentration of IVTT protein. An IVTT reaction which used pET24d as the vector was added to normalize the total amount of IVTT mixture in each tube. The tubes were incubated for at least 1 h at 4°C on a rocker. The beads were washed three times with Binding Buffer and then resuspended in 50 μl of SDS loading buffer (0.25 Tris [pH 6.8], 5% sucrose, 2% SDS, 5% 2-mercaptoethanol, and 0.005% bromophenol blue). The samples were resolved on an SDS-PAGE gel and then imaged by using a PhosphorImager. The GST-IVTT interaction assay system is not quantitative and may depend on the relative association constant (kassoc), which is related to the concentration of interacting peptides. Thus, subtle changes in the relative concentrations of the peptides may obscure potentially altered interactions. In order to provide a modest control for such concentration-dependent processes between experiments we determined the molar concentration of the GST-fusion protein and the molar concentration of the IVTT sample (see above). Furthermore, we have observed clear changes in interaction between hMLH1 and hPMS2 by using a similar assay system that correlates with alterations known to be mutations versus polymorphisms (14a).
RESULTS
GST interaction assay.
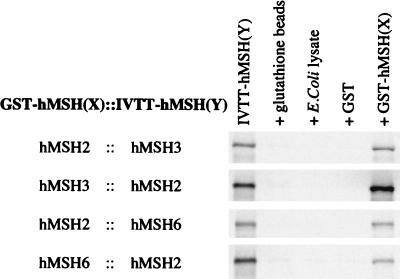
We have previously demonstrated the physical interaction between hMSH2 with hMSH3 and hMSH2 with hMSH6 by using protein-protein cross-linking and immunoprecipitation (IP) with anti-hMSH2 antibodies (1). However, interaction region mapping experiments with this same system, using truncation mutants of hMSH3 and hMSH6, resulted in elevated background signal as a result of anti-hMSH2 antibody binding to the truncated probes. In addition, this IP assay did not appear to be sensitive enough to detect weak interactions. Therefore, we developed an alternative assay that relies on the use of a GST-fusion protein expressed in E. coli as “bait” and IVTT protein as “prey.” This assay proved to be effective for all of the combinations that would be necessary for this study: GST-hMSH2::IVTT-hMSH3, GST-hMSH3::IVTT-hMSH2, GST-hMSH2::IVTT-hMSH6, and GST-hMSH6::IVTT-hMSH2 (Fig. 1). The interaction for each of these IVTT full-length peptides was specific for the GST-hMSH(X) fusion proteins, since we observed nearly undetectable background nonspecific binding as demonstrated by incubation and centrifugal precipitation of the IVTT-MSH(Y) with (i) the glutathione beads alone; (ii) E. coli lysate and glutathione beads; and (iii) pGEX (the GST moiety alone) and glutathione beads as controls (Fig. 1, lanes 2 to 4). Furthermore, densitometric comparison of the pGEX (only) lane with the GST-hMSH(X) lane has demonstrated that the signal to background ratio in this assay approaches 100-fold. These results suggested that this bait-prey system is sufficient to map the interaction regions of the hMSH2-hMSH3 and the hMSH2-hMSH6 heterodimers. In these studies we assign a clear interaction by comparison of the GST alone plus IVTT-MSH(Y) and GST-MSH(X) plus IVTT-MSH(Y) as “pairs.” Furthermore, this assay provides a qualitative measure of interaction efficiency since each experiment contains a nearly identical molar ratio of GST-MSH(X) and IVTT-MSH(Y) (see Materials and Methods). In addition, we have shown that the GST-hMSH3 and GST-hMSH6 fusion proteins are active for mispair binding when they are combined with purified hMSH2 (data not shown). These results suggest that the structures of the hMSH3 and hMSH6 proteins are not substantially altered by fusion to GST. The reverse experiment (GST-hMSH2 with purified hMSH3 or hMSH6) could not be performed because of difficulties in purifying homogeneous hMSH3 and hMSH6.
FIG. 1.
GST-fusion protein assay to study the interaction regions between hMSH2 and hMSH3 and between hMSH2 and hMSH6. Radiolabeled ([35S]methionine) IVTT protein [IVTT-hMSH(Y)] was precipitated with a GST-fusion protein [GST-hMSH(X)]. The first lane contains 10% of the total IVTT mixture used in each experiment. Three controls are shown for each experiment. The second lane contains the IVTT protein which was added to glutathione beads (only). The third lane contains the glutathione beads which were pretreated with a control E. coli lysate which was similar to the lysates from which the GST moiety and the GST-MSH(X) proteins were isolated. The fourth lane contains the glutathione beads which were pretreated with an E. coli lysate which had been induced for expression of GST moiety (alone) from the vector pGEX-SG1. The fifth lane contains the IVTT-MSH(Y) protein incubated with glutathione beads which had been pretreated with lysates that had been made from E. coli which had been induced for expression of a GST-MSH(X) fusion protein. After stringent washing, the samples were resolved on an SDS–8% PAGE gel and imaged with a PhosphorImager. No other bands were visible on the gels other than those shown.
Interaction regions of hMSH2 and hMSH3.
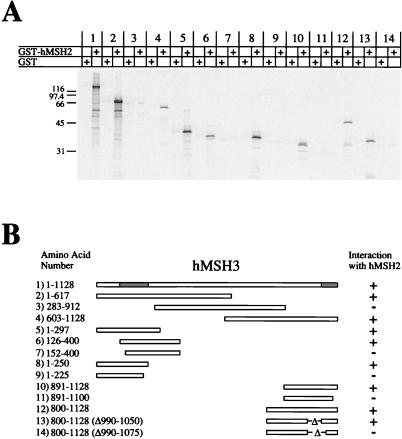
We determined the hMSH3 interaction region(s) with hMSH2 (Fig. 2). Truncated peptides of hMSH3 were constructed such that the protein was divided into three overlapping sections (Fig. 2, pairs 2 to 4). Interestingly, there appeared to be two separate interaction regions: an amino-terminal region and a carboxy-terminal region (Fig. 2, pairs 5 and 10, respectively). The amino-terminal region was resolved between aa 126 and 250 (Fig. 2, pairs 6 to 9). It is important to note that we found the level of IVTT expression to be insufficient with polypeptides that contained less than 100 aa (data not shown). Thus, in order to fully map the carboxy-terminal region we adopted an internal deletion strategy. Using this strategy, the carboxyl interaction region was localized between aa 1050 and 1128 (Fig. 2, pairs 10 to 14).
FIG. 2.
Interaction regions of hMSH3 with hMSH2. 35S-labeled full-length and truncation mutants of IVTT-hMSH3 were added to glutathione beads which had been pretreated with either GST (alone) or GST-hMSH2. (A) Phosphorimage of samples resolved on an SDS–12% PAGE gel. (B) Illustration of the constructs that were used in this experiment and corresponding region locations. The numbers correspond to the pairs shown in panel A. The hMSH2 interaction regions are shaded gray in the full-length hMSH3.
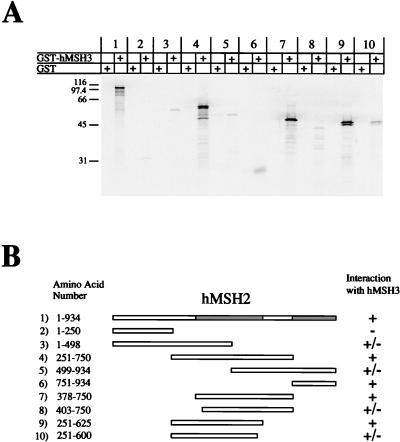
The interaction region of hMSH2 with hMSH3 was similarly determined. We found that hMSH2 also contained two interaction regions with hMSH3 (Fig. 3, pairs 1 to 6). A potential third interaction region from aa 1 to 250 (Fig. 3, pair 2) appears to have been ruled out by constructing a GST-hMSH2 (aa 1 to 250) and observing no interaction with IVTT-hMSH3 (data not shown). We further resolved the amino-terminal interaction region between aa 378 and 625 (Fig. 3, pairs 7 to 10). However, we were unable to adequately resolve the carboxyl interaction region of hMSH2 with full-length GST-hMSH3 because the signal was insufficient (Fig. 3, pair 6).
FIG. 3.
Interaction regions of hMSH2 with hMSH3. 35S-labeled full-length and truncation mutants of IVTT-hMSH2 were added to glutathione beads which had been pretreated with either GST (alone) or GST-hMSH3. (A) Phosphorimage of samples resolved on an SDS–12% PAGE gel. (B) Illustration of the constructs that were used in this experiment and corresponding region locations. The numbers correspond to the pairs shown in panel A. The hMSH2 interaction regions are shaded gray in the full-length hMSH2.
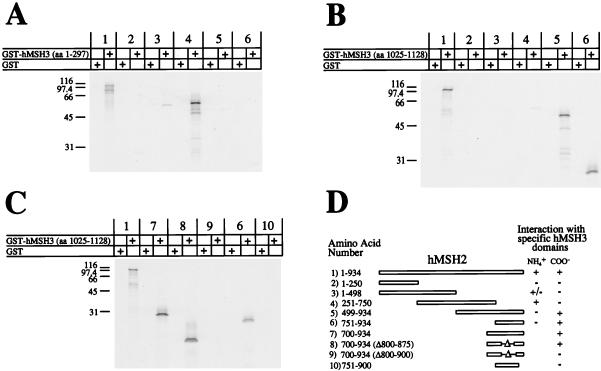
Because there appeared to be two interaction regions between hMSH2 and hMSH3 we designed a system to determine the linear orientations of the two regions. GST fusion proteins containing truncations of hMSH3 were constructed. GST-hMSH3 (aa 1 to 297) contained the consensus amino-terminal interaction region and GST-hMSH3 (aa 1025 to 1128) contained the consensus carboxy-terminal interaction region. These two constructs were used as bait against a series of hMSH2 prey truncation mutants. We found that the full-length hMSH2 interacted with both the GST-hMSH3 truncation constructs (Fig. 4A to C, pair 1). GST-hMSH3 (aa 1 to 297) interacted most strongly with aa 251 to 750 of hMSH2 (Fig. 4A, pair 4). GST-hMSH3 (aa 1025 to 1128) interacted with aa 751 to 934 of hMSH2 (Fig. 4B and C, pairs 5 to 8). These results suggest that the amino-terminal interaction region of hMSH3 contacts the amino-terminal interaction region of hMSH2 and the carboxyl region of hMSH3 contacts the carboxyl region of hMSH2. The GST-hMSH3 (aa 1025 to 1128) truncation also allowed us to further resolve the carboxy-terminal interaction region of hMSH2 to aa 875 to 934 (Fig. 4C).
FIG. 4.
Linear orientations of the hMSH2-hMSH3 interaction regions. 35S-labeled full-length and truncation mutants of IVTT-hMSH2 were added to a GST construct which contained the amino-terminal interaction region of hMSH3 (aa 1 to 297) (A) or a GST construct which contained the carboxy-terminal interaction region of hMSH3 (aa 1025 to 1129) (B). The carboxy-terminal interaction region of hMSH2 with hMSH3 was further resolved by using GST-hMSH3 (aa 1025 to 1129) and carboxy-terminal fragments containing internal deletions of IVTT-hMSH2 (C). Each of these experiments was resolved on an SDS–15% PAGE gel and visualized with a PhosphorImager. (D) Illustration of the constructs and consensus interactions with amino- and carboxy-terminal regions shown in panels A to C.
Interaction regions of hMSH2 and hMSH6.
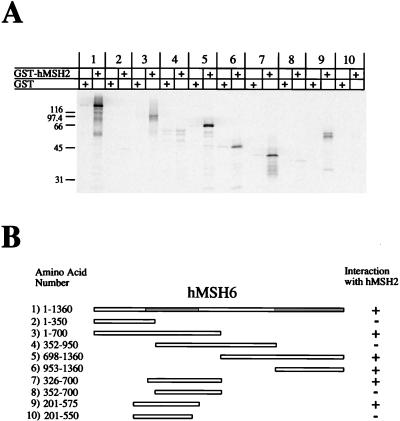
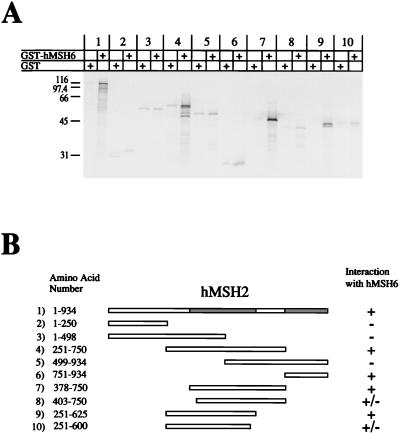
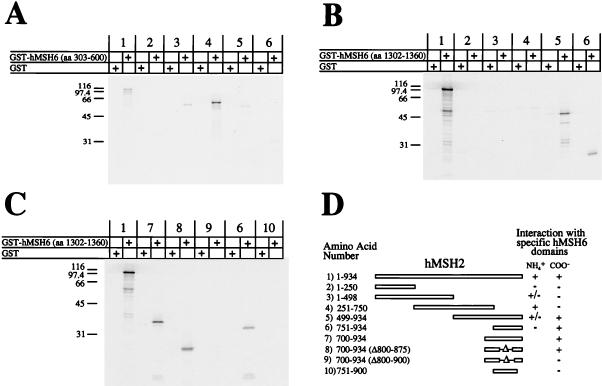
Using a similar strategy, we also determined the interaction regions of hMSH2 and hMSH6. As was observed with hMSH3, hMSH6 contained two interaction regions with hMSH2 (Fig. 5, pairs 1 to 6). The amino-terminal interaction region was mapped from aa 326 to 575 (Fig. 5, pairs 7 to 10). The carboxy-terminal interaction region lies between aa 953 and 1360 (Fig. 5, pair 6). We also mapped the interaction region of hMSH2 with hMSH6 (Fig. 6). We found that hMSH2 also had two interaction regions with hMSH6 (Fig. 6, pairs 1 to 6). The amino-terminal region was mapped to aa 378 to 625 of hMSH2 (Fig. 6, pairs 7 to 10). By using a GST fusion protein which contained a truncation mutant of hMSH6 containing aa 1302 to 1360 (similar to the GST-hMSH3 truncation system described above), the carboxy-terminal interaction region of hMSH2 was localized to aa 875 to 934 (Fig. 7B and C). These results suggest that the same amino acid regions of hMSH2 are employed in the interaction with either hMSH3 or hMSH6.
FIG. 5.
Interaction regions of hMSH6 with hMSH2. 35S-labeled full-length and truncation mutants of IVTT-hMSH6 were added to glutathione beads which had been pretreated with either GST (alone) or GST-hMSH2. (A) Phosphorimage of samples resolved on an SDS–12% PAGE gel. (B) Illustration of the constructs that were used in this experiment and corresponding region locations. The numbers correspond to the pairs shown in panel A. The hMSH2 interaction regions are shaded gray in the full-length hMSH6.
FIG. 6.
Interaction regions of hMSH2 with hMSH6. 35S-labeled full-length and truncation mutants of IVTT-hMSH2 were added to glutathione beads which had been pretreated with either GST (alone) or GST-hMSH6. (A) Phosphorimage of samples resolved on an SDS–12% PAGE gel. (B) Illustration of the constructs that were used in this experiment and corresponding region locations. The numbers correspond to the pairs shown in panel A. The hMSH6 interaction regions are shaded gray in the full-length hMSH2.
FIG. 7.
Linear orientations of the hMSH2-hMSH6 interaction regions. 35S-labeled full-length and truncation mutants of IVTT-hMSH2 were added to a GST construct which contained the amino-terminal interaction region of hMSH6 (aa 303 to 600) (A) or a GST construct which contained the carboxy-terminal interaction region of hMSH6 (aa 1302 to 1360) (B). The carboxy-terminal interaction region of hMSH2 with hMSH6 was further resolved by using GST-hMSH6 (aa 1302 to 1360) and carboxy-terminal fragments containing internal deletions of IVTT-hMSH2 (C). Each of these experiments was resolved on an SDS–15% PAGE gel and visualized with a PhosphorImager. (D) Illustration of the constructs and consensus interactions with amino- and carboxy-terminal regions shown in panels A to C.
The linear orientations of the hMSH2-hMSH6 interaction regions were also determined. Using IVTT N- and C-terminal hMSH2 interaction regions and GST N- and C-terminal interaction regions of hMSH6, we found that the N-terminal interaction region of hMSH6 interacted with the N-terminal interaction region of hMSH2 (Fig. 7A) and the C-terminal interaction region of hMSH6 interacted with the C-terminal interaction region of hMSH2 (Fig. 7B). Thus, the linear orientations of the interaction regions of the hMSH2-hMSH6 heterodimer are identical to those observed with the hMSH2-hMSH3 heterodimer.
Interaction regions of hMSH2 with itself.
We have previously shown that hMSH2 binds mismatched nucleotides and appears to form a homodimer (1). Using a GST-hMSH2 (aa 751 to 934) construct we found that this C-terminal region interacted with the carboxy terminus of hMSH2 (data not shown). This construct displayed specific interaction with the hMSH2 truncation mutants aa 751 to 934 and aa 700 to 934 (Δ800 to 875). However, it did not interact with either the hMSH2 truncation mutant aa 751 to 900 or aa 700 to 934 (Δ800 to 900). Thus, the hMSH2 homodimer appears to display the same C-terminal interaction pattern that we observed with hMSH2 binding to hMSH3 or hMSH6 (Fig. 4D and 7D, pairs 6 to 10), implicating aa 875 to 934 of hMSH2 in self-association. We also attempted to map the amino-terminal homodimer interaction region of hMSH2. However, we were unable to clearly identify distinct regions in the amino-terminal region with the full-length GST-hMSH2 construct (data not shown).
The effect of hMSH2 mutations found in HNPCC kindreds on protein-protein interaction.
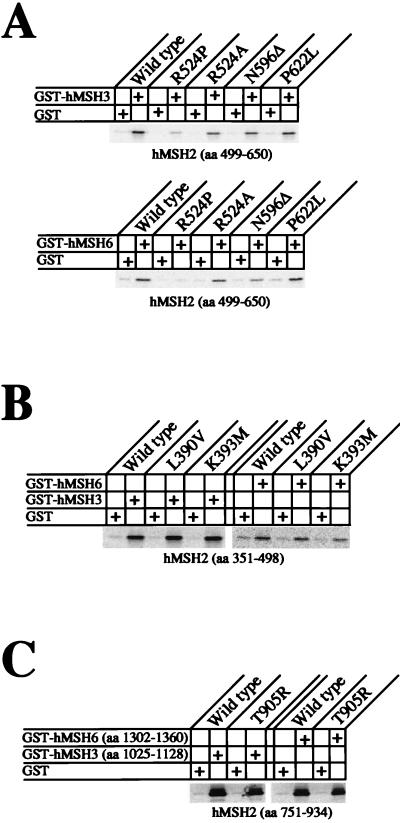
Once the interaction regions of hMSH2 were identified, we determined that several HNPCC missense mutations were located within these regions. Six of these HNPCC mutations were constructed and tested for their interaction properties: L390V, K393M, R524P, N596Δ, P622L, and T905R. Each interaction experiment was performed such that we only tested the interaction with the consensus N- or C-terminal interaction region to eliminate any confusion that multiple interaction regions might display. These IVTT mutant consensus interaction regions were tested for interaction with full-length GST-hMSH3 and GST-hMSH6. There was no discernible difference in the level of interaction between that of any of the mutant hMSH2 constructs and that of the wild-type hMSH2 consensus interaction region (data not shown).
However, close examination of the amino-terminal interaction region of hMSH2 suggested that both the aa 403 to 750 and the aa 251 to 600 truncation proteins are not completely devoid of interaction with hMSH3 and hMSH6 (Fig. 3 and 6, pairs 8 and 10). This result appeared to suggest that the consensus amino-terminal interaction region might be composed of at least two mini-interaction regions. Therefore, we further divided the amino-terminal region into two subregions: aa 351 to 498 and aa 499 to 650. We found that by allowing a slightly longer incubation time and exposing the SDS-PAGE gel for a longer period of time, a sufficient signal could be garnered with these truncated fragments (Fig. 8). Interestingly, one of these HNPCC mutant constructs, R524P (aa 499 to 650), appeared to show reduced interaction (Fig. 8A). However, when we replaced the proline residue found in the HNPCC kindred with an alanine, this reduced interaction was not evident. Thus, it is likely that the proline residue merely disrupted the local peptide structure to obscure the remaining interaction(s) in this N-terminal region. There was no difference in interaction when the N596Δ and P622L mutant peptides were compared to the wild type (Fig. 8A). We also tested L390V and K393M in the context of the aa 351 to 498 consensus truncations and found no significant effect with either mutations (Fig. 8B), nor did we observe a difference with the T905R truncation (Fig. 8C). These results suggest that altered static interaction between hMSH2 and either hMSH3 or hMSH6 is unlikely to cause functional defects resulting in HNPCC. Furthermore, because there are two interaction regions, it is unlikely that the loss of function in one region would result in the complete loss of heterodimer interaction.
FIG. 8.
Effects of HNPCC mutations on hMSH2 interaction with hMSH3 or hMSH6. Analysis of the interaction of hMSH2 missense mutations, found in well-defined HNPCC kindreds, with hMSH3 or hMSH6. The binding association of GST-MSH3 or GST-MSH6 with each of these mutations was determined by using IVTT-hMSH2 truncated peptides known to contain the consensus interaction region. Peptides were resolved on SDS–15% PAGE gels and examined by PhosphorImager.
DISCUSSION
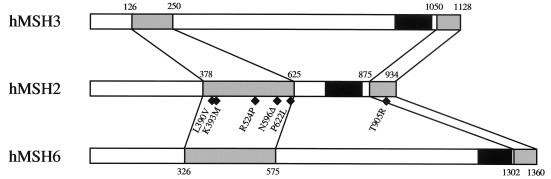
The biochemical copurification of hMSH2 with hMSH3 and of hMSH2 with hMSH6 suggested that hMSH2-hMSH3 and hMSH2-hMSH6 might exist as heterodimeric proteins (9, 28). This interaction was confirmed by Acharya et al. (1) when they demonstrated physical interaction via protein-protein cross-linking within hMSH2-hMSH3 and within hMSH2-hMSH6. These latter results suggested that the regions of interaction could be identified. Based on the data presented here, we propose a model for the regional interactions of hMSH2 with hMSH3 and hMSH6 (Fig. 9). Our results suggest that hMSH2 employs the same interaction region for either hMSH3 or hMSH6. These interactions occur through two distinct regions of hMSH2: aa 378 to 625 and 875 to 934. The adenine nucleotide binding region and the putative helix-turn-helix motif of hMSH2 are not contained within either of these regions. These results suggest that it is unlikely that the helix-turn-helix motif is essential for interaction with hMSH3 and hMSH6. Also shown in our model are the linear orientations of the N-terminal and C-terminal interaction regions of hMSH2. The N-terminal interaction region of hMSH2 (aa 378 to 625) was found to contact aa 126 to 250 of hMSH3 and aa 326 to 575 of hMSH6. The C-terminal interaction region of hMSH2 (aa 875 to 934) was found to contact aa 1050 to 1128 of hMSH3 and aa 1302 to 1360 of hMSH6.
FIG. 9.
Model of the hMSH2 consensus interaction with hMSH3 or hMSH6. The interaction regions of hMSH2 with hMSH3 and of hMSH2 with hMSH6 are shown in gray and they are connected with lines that illustrate the specificity of each region to their pairing partner. The nucleotide binding regions are shown as black boxes. The location of the HNPCC mutations tested in these studies are illustrated as black diamonds.
Since hMSH3 and hMSH6 appear to contact hMSH2 within the same binding regions, we aligned both the amino-terminal and carboxyl-terminal regions of hMSH3 and hMSH6. We found that amino-terminal regions of hMSH3 and hMSH6 had little identifiable homology while the carboxyl-terminal interaction regions suggested moderate homology with 16 of 60 residues being identical (data not shown). It is possible that the carboxy-terminal regions of hMSH3 and hMSH6 provide some undetermined conserved function for these proteins that includes, but is not limited to, protein-protein interaction.
It had previously been postulated that S. cerevisiae MSH2 interacted with MSH6 through a single region containing the carboxy-terminal 114 aa of MSH2. We have made a similar observation in that we have mapped an interaction region in the carboxy-terminal 70 aa of hMSH2 (aa 875 to 934). However, our results have suggested a second interaction region toward the N terminus of the hMSH2 protein (aa 378 to 625). One possible explanation for this discrepancy is that the human MutS homologs have acquired this extra interaction region. Alternatively, the S. cerevisiae MutS homologs may also have two interaction regions and the system that was used to map these regions trapped the interaction in a conformation which only required the carboxyl terminus. The likelihood of this explanation is increased when one considers the observation that the hMSH2-hMSH6 heterodimer undergoes a conformational transition as a result of nucleotide binding and/or exchange (14, 34a).
The identification of two separate interaction regions offers several intriguing possibilities. The simplest possibility is that the two protein heterodimers merely interact through two regions and do not undergo any dynamic changes. However, it is also possible that one or both of the interaction regions is altered both in conformation and/or activity when either of the MutS homologs is bound to an ADP or ATP molecule (10, 14). This idea is further supported when one considers that the two interaction regions flank the adenine nucleotide binding region. It is not hard to envision a hinge mechanism in which the binding regions change configuration depending on whether an ADP or ATP molecule is bound.
Previous studies of the yeast MSH2-MSH6 interaction have suggested that the carboxy-terminal interaction region of MSH2 interacted with MSH6 through a hepta-hydrophobic repeat motif (3). We do not find any sequence which resembles a hepta-hydrophobic repeat in the carboxy-terminal interaction region of hMSH2. Nor do we find any corresponding hepta-hydrophobic repeat sequence in the carboxy-terminal interaction regions of hMSH3 and hMSH6. Thus, based on the interaction region that we mapped, the carboxyl termini of the human MutS proteins are unlikely to interact through a hepta-hydrophobic motif.
A biochemical study of the human mismatch repair proteins has clearly aided our understanding of how hMSH2 and hMLH1 contribute to the pathogenesis of cancer. Mutations in hMSH2 have been reported in approximately 45% of HNPCC patients. Of these mutations, approximately 15% have been reported to be missense mutations which appear to be spread throughout the coding sequence (reference 29 and unpublished data). Thus far, hMSH2 has been observed to have several biochemical activities: mismatch binding, nucleotide binding-hydrolysis, and interaction with hMSH3 and hMSH6. By examining HNPCC mutations constructed in the hMSH2 protein, we have begun to address the question of whether altered interaction with hMSH3 and/or hMSH6 might play a role in tumorigenesis. We did not find any significant effect of six missense mutations, found in well-characterized HNPCC kindreds, on the static interaction between hMSH2 and hMSH3 or hMSH6. While the GST-IVTT interaction assay is clearly not quantitative and we cannot rule out undetermined in vivo effects of these mutations on hMSH2 interaction with hMSH3 and hMSH6, these results would appear to suggest that altered static interactions within the hMSH2-hMSH3 or the hMSH2-hMSH6 heterodimers is unlikely to be causative of disease. Additional support for this hypothesis is evident when one considers that clear identification of these protein-protein interactions only occurs when the N- and C-terminal domains are physically separated from one another (i.e., if either domain is present, then there is strong interaction between the peptides). Finally, while proof that we could identify interaction defects in the system described here is not possible at present, it should be noted that similar studies with the hMLH1 and hPMS2 proteins have clearly detailed interaction defects associated with missense mutations (15a). We entertain the possibility that the missense amino acid changes of hMSH2 affect some other function than interaction with their heterodimeric partner. For example, these alterations might affect the dynamic function of hMSH2 by altering important conformational transitions associated with the heterodimers.
A role for hMSH2 in cancer has been firmly established in HNPCC. Our data suggest that truncation mutations of hMSH2, hMSH3, or hMSH6 may retain the amino-terminal interaction region. One potential mechanism for interfering with mismatch repair would be if the truncated peptides interfered with the wild-type function of the hMSH2-hMSH3 and hMSH2-hMSH6 heterodimers. Elucidation of the role of the human mismatch repair proteins in cancer depends on a complete understanding of the biochemical and functional properties of these proteins. The function(s) of the interaction regions detailed here is under study.
ACKNOWLEDGMENTS
We thank Hansjuerg Alder and the employees of the Sidney Kimmel Nucleic Acid Facility for nucleotide synthesis and sequencing; Christoph Schmutte for helping to prepare the figures for this study; and Jason Krupnick, Tim Roth, Samir Acharya, and Greg Tombline for helpful discussions.
This work was supported by NIH grants CA56542 and CA67007.
REFERENCES
- 1.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, Yuasa Y. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 3.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alani E, Sokolsky T, Studamire B, Miret J J, Lahue R S. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 6.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 7.Burns N, Grimwade B, Ross-Macdonald P B, Choi E, Finberg K, Roeder G S, Snyder M S. Large-scale analysis of gene expression, protein localization and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- 8.Chi N W, Kolodner R D. Purification and characterization of MSH1, a yeast mitochondrial protein that binds to DNA mismatches. J Biol Chem. 1994;269:29984–29992. [PubMed] [Google Scholar]
- 9.Drummond J T, Li G-M, Longley M J, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 10.Fishel R. Mismatch repair, molecular switches, and signal transduction. Genes Dev. 1998;12:2096–2101. doi: 10.1101/gad.12.14.2096. [DOI] [PubMed] [Google Scholar]
- 11.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 12.Fishel R, Wilson T. MutS homologs in mammalian cells. Curr Opin Genet Dev. 1997;7:105–113. doi: 10.1016/s0959-437x(97)80117-7. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 14.Gradia S, Acharya S, Fishel R. The Human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 15.Grilley M, Welsh K M, Su S S, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 15a.Guerrette, S., S. Acharya, and R. Fishel. Submitted for publication.
- 16.Haber L T, Walker G C. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollingsworth N M, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 18.Kallal L, Kurjan J. Analysis of the receptor binding domain of Gpa1p, the Gα subunit involved in the yeast pheromone response pathway. Mol Cell Biol. 1997;17:2897–2907. doi: 10.1128/mcb.17.5.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T. Slippery DNA and diseases. Nature. 1993;365:207–208. doi: 10.1038/365207a0. [DOI] [PubMed] [Google Scholar]
- 21.Malkhosyan S, Rampino N, Yamamoto H, Perucho M. Frameshift mutator mutations. Nature. 1996;382:499–500. doi: 10.1038/382499a0. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 22.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 23.Miyaki M, Konishi M, Tanaka K, Kikuchi Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T. Germ line mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 24.Modrich P. Methyl-directed DNA mismatch correction. J Biol Chem. 1989;264:6597–6600. [PubMed] [Google Scholar]
- 25.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 26.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 27.Nicolaides N C, Papadopoulos N, Liu B, Wei Y F, Carter K C, Ruben S M, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, et al. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 28.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 29.Peltomaki P, Vasen H F. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. Gastroenterology. 1997;113:1146–1158. doi: 10.1053/gast.1997.v113.pm9322509. [DOI] [PubMed] [Google Scholar]
- 30.Pochart P, Woltering D, Hollingsworth N M. Conserved properties between functionally distinct Muts homologs in yeast. J Biol Chem. 1997;272:30345–30349. doi: 10.1074/jbc.272.48.30345. [DOI] [PubMed] [Google Scholar]
- 31.Reenan R A G, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reenan R A G, Kolodner R D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su S-S, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Welsh K M, Lu A L, Clark S, Modrich P. Isolation and characterization of the Escherichia coli mutH gene product. J Biol Chem. 1987;262:15624–15629. [PubMed] [Google Scholar]
- 34a.Wilson, T., S. Gradia, and R. Fishel. Unpublished data.
- 35.Wu T H, Marinus M G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]