Abstract
Clearance of accumulated protein aggregates is one of the functions of autophagy. Recently, a clearer understanding of non-coding RNAs (ncRNAs) functions documented that ncRNAs have important roles in several biological processes associated with the development and progression of neurodegenerative disorders. Subtypes of ncRNA, including microRNA (miRNA), long noncoding RNA (lncRNA), and circular RNA (circRNA), are commonly dysregulated in neurodegenerative disorders such as Alzheimer and Parkinson diseases. Dysregulation of these non-coding RNAs has been associated with inhibition or stimulation of autophagy. Decreased miR-124 led to decreased/increased autophagy in experimental model of Alzheimer and Parkinson diseases. Increased BACE1-AS showed enhanced autophagy in Alzheimer disease by targeting miR-214-3p, Beclin-1, LC3-I/LC3-II, p62, and ATG5. A significant increase in NEAT1led to stimulated autophagy in experimental model of PD by targeting PINK1, LC3-I, LC3-II, p62 and miR-374c-5p. In addition, increased BDNF-AS and SNHG1 decreased autophagy in MPTP-induced PD by targeting miR-125b-5p and miR-221/222, respectively. The upregulation of circNF1-419 and circSAMD4A resulted in an increased autophagy by regulating Dynamin-1 and miR-29c 3p, respectively. A detailed discussion of miRNAs, circRNAs, and lncRNAs in relation to their autophagy-related signaling pathways is presented in this study.
Graphical Abstract
Autophagy-related non-coding RNAs in neurodegenerative diseases.
Keywords: Neurodegenerative disorders, Non-coding RNAs, Autophagy, microRNA, Long non-coding (lnc) RNA
Introduction
The term “neurodegenerative disease (ND)” describes the progressive loss of specific populations of neurons by toxic or metabolic processes. There are several means to classify neurodegenerative diseases by their primary clinical features, anatomic distributions of neurodegeneration, or molecular abnormalities (Dugger and Dickson 2017). Alzheimer’s disease (AD) and Parkinson’s disease (PD) are two major NDs (Checkoway et al. 2011). As the ability of the central nervous system (CNS) for regeneration is limited, it is essential to limit injury within this organ (Kuhn et al. 2001).
A cell’s autophagy process involves the transport of cytosolic components, macromolecules, viruses, bacteria, and organelles to lysosomes for destruction (Patergnani and Pinton 2015). Physiological and pathological conditions are impacted by autophagy and its specialized forms. The function of autophagy in normal conditions is to remove unnecessary material, regulate organelle turnover, and meet energy demands. Autophagy can have both beneficial and harmful effects on pathological conditions (Patergnani et al. 2020; Xue et al. 2020). The clearance of accumulated protein aggregates is one of the functions of autophagy that contributes to neuronal function and neurodegenerative disorders (Nah et al. 2015). A wide range of neurodegenerative diseases is characterized by the accumulation of proteins (Menzies et al. 2015; Ravikumar et al. 2002), with autophagy contributing to disease progression (Menzies et al. 2015) and neuronal loss (Hara et al. 2006; Komatsu et al. 2006; Levine and Kroemer 2008; Nikoletopoulou et al. 2013).
Studies have focused on evaluating specific biomarkers related to genes that encode autophagy pathways-related proteins in neurodegenerative diseases. In this regard, through their ability to adjust the translation of other RNAs, non-coding RNAs (ncRNAs) contribute to the generation of functional proteins through the effect on the expression of protein-coding transcripts (Esteller 2011). A series of cleavage events produce mature microRNAs after RNA polymerases II and III transcribe microRNA precursors. lncRNAs also contain a poly (A) terminus at 3′ and a 5′ methyl-cytosine cap which is transcribed by RNA polymerase II (Pol II). lncRNAs biogenesis is similar to that of mRNA with a few variations in the process. RNA circular molecules (circRNAs) are endogenous non-coding RNAs generated by the back-splicing process. Some circRNAs consisting of introns originate in the nucleus, while others have one or more exons with major locations in the cytoplasm. A circular RNA does not have a polyadenylated tail or a 5–3′ direction. These characteristics render ncRNAs more stable in plasma and tissues as they possess a continuous loop of covalent bonds. ncRNAs can be useful in diagnostic and prognostic goals and have roles in regulating autophagy pathways in neurodegenerative diseases (Shah et al. 2018; Xu et al. 2020).
Herein, we highlight the role of several ncRNAs associated with autophagy in AD and PD as neurodegenerative diseases. miRNAs, circRNAs, and lncRNAs will be discussed in detail. Due to the importance of RNAs for neurodegenerative diseases, multiple studies have been conducted on ncRNAs; however, in the present review, the effects of these ncRNAs on autophagy-related genes and pathways in AD and PD are discussed, distinguishing this review from others.
Autophagy and Neurodegenerative Diseases
The biochemist Christian de Duve introduced the term autophagy in late 1963, referring to cellular degradation and recycling via a self-degradative pathway (Duve and Wattiaux, 1966). In mammalian cells, macroautophagy is the best-characterized and most prevalent mode of autophagy. Macroautophagy occurs when the expanding phagophore sequesters random cytoplasm and dysfunctional organelles, leading to autophagosome formation. Following autophagosome fusion with the vacuole membrane, the autophagic body enters the vacuole lumen. Vacuolar hydrolases eventually degrade or process the sequestered cargo (Feng et al. 2014).
Autophagy initiates when two protein complexes are triggered at phagophore assembly sites, UN51-like Ser/Thr kinase (ULK) and phosphatidylinositol-3-kinase (PI3K) (Agarwal et al. 2015; Ohsumi and Mizushima 2004). ULK complexes consist of the ULK1/2 and FAK family interacted proteins, and autophagy-related gene 13 (ATG13) (Jung et al. 2009). The PI3K complex comprises Vps34, Vps15, Beclin1, and ATG14 (Fan et al. 2011). It is noteworthy that anti-apoptotic dimers BCL-XL and BCL-2 regulate Beclin1 localized on endoplasmic reticulum (ER) membranes. Beclin1 is dissociated from BCL-2. Next, it coordinates with Vps34 after autophagy is triggered (Marquez and Xu 2012; Martyniszyn et al. 2011; Pattingre et al. 2005). As a result, phosphatidylinositol 3-phosphate (PI3P) will concentrate on the phagophore’s surface (Obara and Ohsumi 2008; Puri et al. 2013). Autophagosome expansion and closure are mediated via two ubiquitin-like complexes. Initially, upon the interaction of Atg7, Atg5 covalently attaches to Atg12 (Shao et al. 2007). The complex then binds to Atg16 for Atg5-Atg12-Atg16 complex formation, which is accountable for phagophore elongation. Atg4B cleaves microtubule-associated protein 1 light chain 3 (LC3) to form LC3-I, in another ubiquitin-like complex (Fujita et al. 2008). Next, LC3-I is converted to phosphatidylethanolamine (PE)-conjugated LC3-II with assistance of the Atg5-Atg12-Atg16 complex. LC3-II is considered an important marker for autophagosomes (Kabeya et al. 2000). Subsequently, mature autophagosomes move along microtubules to fuse with lysosomes (Ravikumar et al. 2005). This involves the recruitment of multiple membrane protein complexes such as soluble NSF attachment protein receptors (SNAREs) (Itakura et al. 2012). A proteolytic reaction occurs after autolysosomes are formed to degrade the cargoes they carry (Guo et al. 2018b).
The pathophysiology of AD includes two abnormal structures: Neurofibrillary tangles (NTs) and senile plaques. Altered cleavage of amyloid precursor protein (APP) has been implicated in increased β-amyloid tangles, constituting senile plaques (Querfurth and LaFerla 2010). NTs, which are hyperphosphorylated tau proteins associated with microtubules, are pathological, insoluble aggregates. In normal conditions, microtubule stabilization and vesicle transport in neurons are controlled by tau’s interaction with tubulin (Wang et al. 2013). AD brains with autophagic vacuoles (AVs) are indicative of altered autophagy in this disease. Disruption of retrograde transport of autophagosomes along axons results in the repletion of naive AVs and Aβ-producing AVs (Ułamek-Kozioł et al. 2013; Yu et al. 2005), leading to increased production of Aβ. Autophagy contributes to clearance of soluble as well as insoluble tau aggregates in conjunction with the ubiquitin-proteasome system. Chloroquine, causes tau clearance to be delayed and tau aggregates to accumulate by inhibiting autophagosome–lysosome fusion (Hamano et al. 2008). Phosphorylated tau is more affected by autophagic failure than other forms of tau (Rodríguez-Martín et al. 2013).
Another common neurodegenerative disorder is PD, characterized by accumulation of α-synuclein and degeneration of dopaminergic neurons, and presence of ubiquitin in Lewy bodies, which are intracytoplasmic inclusions (Tan et al. 2014).
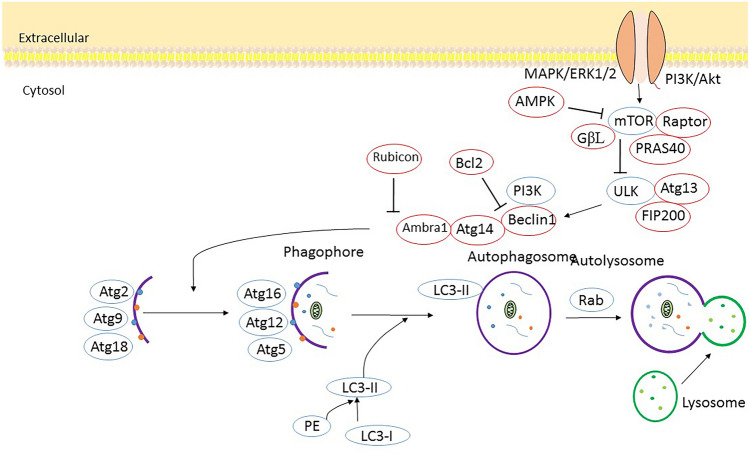
Autophagic dysfunction in the substantia nigra is inherent to PD (Alcalay et al. 2010). Autophagy pathway can be regulated by proteins encoded by PD-relevant genes. LRRK2, one of the genes whose mutation is associated with PD, may regulate macroautophagy (Bonifati 2006). Knockdown of LRRK2 stimulates autophagy and inhibits LRRK2 kinase activity, thus increasing macroautophagy, absent changes in TORC1 levels (Alegre-Abarrategui et al. 2009; Manzoni et al. 2013). Mitophagy also mediates the function of autosomal recessive genes associated with PD. Parkin, an E3-like ligase, mostly localizes to the cytosol, but once a mitochondrial uncoupler (CCCP) is applied, it is translocated to damaged mitochondria for removal (Narendra et al. 2008). As a result, Parkin mutations block the ability of mitophagy to clear damaged mitochondria in PD. In addition, studies have shown that depolarization of mitochondria recruits Parkin by accumulating PINK1 on its external membrane (Scarffe et al. 2014). Nonetheless, the link between mitophagy and the etiology of PD has yet to be fully characterized. Moreover, oxidative stress has been shown to trigger PD. In this regard, autophagy, which reduces oxidative stress, may be an effective treatment for PD (Surendran and Rajasankar 2010). The PD SNCA model of the BECN1 gene showed attenuation in neurodegenerative pathology (Spencer et al. 2009). Overexpressing RAB1A, which controls cell membrane transfer and autophagosome construct, alleviates deficiencies in dopaminergic neurons expressing SNCA (Coune et al. 2011). Dopaminergic neurodegeneration by SNCA in Drosophila has been shown to be inhibited by histone deacetylase 6, an enzyme controlling autophagosomal maturation (Du et al. 2010). Enhanced expression of transcription factor EB leads to clearance of SNCA from dopaminergic neurons through autophagy (Decressac et al. 2013). PD patients’ brains have lower expression of LAMP2A and HSC70, the constituents of CMA (Sala et al. 2014). Taken together, these studies show that autophagy has an important function in the pathogenesis of PD (Fig. 1).
Fig. 1.
A schematic representation of the autophagy pathway. This figure adapted from (Riebisch et al. 2021)
Microglia, the brain’s resident macrophages with an inherent ability to respond to CNS injury, play a crucial role in promoting repair and ensuring proper brain function. While studies examining the impact of autophagy on neurodegenerative disorders has concentrated on neurons, recent findings suggest that autophagy might also play a role in the functioning of glial cells (Dello Russo et al. 2013; Yamamoto and Yue 2014). Increased evidence purports that autophagy is regulated by both innate and adaptive responses in the peripheral immune system (Shibutani et al. 2015) and the process of phagocytosis (Green et al. 2016). Microglia are the brain’s phagocytes (Sierra et al. 2013). Autophagy and phagocytosis share striking morphological and mechanistic similarities, as both processes rely on the formation of transient vesicular structures (autophagosomes and phagosomes, respectively) that engulf and deliver cargo to the lysosomes for digestion. Interestingly, a functional cross-talk exists between autophagy and phagocytosis during the innate immune response in peripheral macrophages. The modulation of autophagy in microglia could affect both microglial phagocytosis and inflammation, potentially playing a role in the progression of neurodegeneration (Plaza-Zabala et al. 2017). Microglial autophagy has the potential to affect the activation and inflammatory responses in microglia (Su et al. 2016). In one study, the inhibition of autophagy was shown to decrease Aβ phagocytosis (Lucin et al. 2013). Microglial autophagy increased Aβ degradation and NLRP3 inflammasome in AD (Cho et al. 2014). Furthermore, the disruption of microglia autophagy exacerbated neuroinflammation and dopaminergic neuron loss via targeting NLRP3 inflammasome in animal model of PD induced by MPTP (Cheng et al. 2020; Qin et al. 2021b). Therefore, microglia autophagy plays a neuroprotective role in the progression of AD and PD.
Non-coding RNAs
A series of cleavage events produce mature microRNAs after RNA polymerases II and III transcribe microRNA precursors (Annese et al. 2020). In the canonical pathway of biogenesis, they are processed into pre-miRNAs by the microprocessor complex, including a ribonuclease III enzyme, Drosha, and an RNA binding protein DiGeorge Syndrome Critical Region 8 (DGCR8) (Gregory et al. 2004). In addition, there are two main non-canonical miRNA biogenesis pathways: Dicer-independent and Drosha/DGCR8-independent (Felekkis et al. 2010).
miRNAs expression in neurodegenerative diseases is altered, providing further evidence for their putative function in neurodegeneration. For instance, transfection of miR-7 suppresses microglial NLRP3 inflammasome activation and decreased dopaminergic neuron degeneration by decreasing microglial activation in the PD model induced by MPTP (Titze-de-Almeida and Titze-de-Almeida 2018). miR-7 inhibited neuronal apoptosis in PD cell lines model via targeting Sirt2 and Bax (Li et al. 2016). However, the upregulation of miR-7 promoted the production of extracellular Aβ in nerve cells (Fernández-de Frutos et al. 2019). miR-106b enhanced levels of secreted Aβ by regulating ABCA1 (Kim et al. 2012). miR-106b affected TGF-β signaling, thereby contributing to the pathogenesis of AD (Wang et al. 2010). Zhang et al. (2022) demonstrated that miRNA-mediated autophagy might be a putative therapeutic strategy AD.
lncRNAs also contain a poly (A) terminus at 3′ and a 5′ methyl-cytosine cap (Zhang et al. 2019), which are transcribed by RNA polymerase II (Pol II) (Nojima and Proudfoot 2022). lncRNAs biogenesis is analogous to that of mRNA with few variations. There is strong evidence that lncRNAs are capped and polyadenylated by canonical splicing in the vast majority of cases (Gourvest et al. 2019).
Information on LncRNAs target genes and signaling pathways in neurodegenerative diseases has been recently addressed. lnc-ANRIL knockdown inhibits cell inflammation via the binding of miR-125a in vitro model of AD (Zhou et al. 2020a). LncRNA H19 decreases apoptosis in neurons of MPTP-induced PD mice via regulating miR-585-3p/PIK3R3 (Zhang et al. 2020b). LncRNA-mediated autophagy may be a strategy for the treatment of neurodegenerative diseases (Jiang and Xu 2023; Xu et al. 2020).
RNA circular molecules (circRNAs) are endogenous non-coding RNA generated by the back-splicing process. Some circRNAs consisting of introns originate in the nucleus, while others have one or more exons with major locations in the cytoplasm (Guo et al. 2014). A circular RNA does not have a polyadenylated tail or a 5–3′ direction, rendering them more stable in plasma and tissues as they have a continuous loop of covalent bonds (Guo et al. 2014; Suzuki and Tsukahara 2014). For instance, circDLGAP4 has neuroprotective effects in animal models of PD by targeting the miR-134-5p and CREB (Feng et al. 2020). Circular RNA Cwc27 participates in the pathogenesis of AD by repressing Pur-α activity (Song et al. 2022).
Non-coding RNAs and Autophagy in AD and PD
miRNAs and Autophagy in AD and PD
miR-7 and miR-153
An in vitro study showed that miR-7 or miR-153 overexpression protected cortical neurons against MPP +. A small amount of miR-153 debilitated MPP + -induced initiation of p38MAPK, while miR-7 preserved that of mTOR, JNK , and SAPK protein expressions. Rapamycin co-administered with MPP + ameliorated the neuroprotective influence of miR-7 and miR-153 (Fragkouli and Doxakis 2014). An in vitro study showed that miR-7 reduced α-Syn expression and degraded α-Syn without targeting the 3′-UTR of the α-Syn mRNA. Its function is applied by increasing autophagy via the conversion of LC3-I to LC3-II and autophagosome formation (Choi et al. 2018).
miR-9-5p
miR-9-5p exacerbates MPP +—induced neurotoxicity by targeting SIRT1 (Wang et al. 2019). miR-9-5p antagomirs increased Aβ clearance and improved cognition in APPswe/PS1dE9 mice by targeting autophagy via autophagy receptor Optineurin (Optn) (Chen et al. 2021).
miR-29c
A PD mouse model has shown enhanced autophagy and down-regulation of miR-29c-3p in dopamine neurons. In a PD mice and in vitro models, increased miR-29c-3p reduced autophagy by decreasing LC3-II/I and Beclin-1 and increasing p62 expression via targeting of TET2 (Wang et al. 2021a).
miR-101
miR-101 is implicated in normal cognitive function. It has a vital role in the pathology of aging-related neurodegenerative diseases (Barbato et al. 2020). MiR-101 decreased APP in AD (Long and Lahiri 2011). Increased miR-101 led to α-syn accumulation (Valera et al. 2017). miR-101 mimic inhibited cell death in PD (Omura et al. 2021). miRNA-101a markedly down-regulated in AD patients and mouse. miRNA-101a transfection decreased autophagy in in vitro model of AD by targeting MAPK1 pathway, beclin-1 (Li et al. 2019).
miR-124
miR-124 is known as a neuron-specific miR (Lagos-Quintana et al. 2002; Mishima et al. 2007). The increased miR-124 level may be effective in decreasing neuroinflammation (Han et al. 2020). Exosomal miR-124-3p transferring into hippocampal neurons decreased neuronal injury by regulating the Rela and ApoE expressions (Ge et al. 2020). Translation of miR-124 in APP/PS1 transgenic mice showed improvements in their AD pathology and learning ability. As a consequence, the expression of p62/SQSTMl, Atg5, and LC3II was down-regulated, while Beclin-1 levels were increased (Du et al. 2017). Upon targeting LC3II/I expression, overexpression of miR-124 increased autophagy in PD induced by MPTP (Yao et al. 2019). Moreover, miR-124 was suppressed in SK-N-SH cell line treated with MPTP, resulting in enhanced autophagy by increased LC3-II/I and Beclin-1. In addition, p-AMPK levels increased in neurons upon miR-124 suppression, while p-mTOR levels decreased (Gong et al. 2016). Increased miR-124 led to decreased loss of dopamine by regulating apoptosis and autophagy in animal model of PD induced by MPTP. miR-124 decreased autophagosome accumulation and lysosomal depletion in PD (Wang et al. 2016).
miR-132/212
Tauopathies such as AD are associated with reduced microRNA (miRNA) cluster expression, miR-132/212 (Wang et al. 2017b). Decreased miR-132/212 expressions led to AD pathogenesis (Wang et al. 2017b). An in vivo study showed that a deficiency of miR-132/212 results in increased tau, along with increased tau aggregation. miR-132 directly regulated tau expression. Induction of tau aggregation by miR-132/212 deletion was associated with dysfunctional autophagy via targeting Beclin-1, ATG5–12, 9a, P62 and TMEM106b. On the other hand, miR-132 mimics were shown to restore in part cognition and tau expression in AD mice treated with miR-132 mimics. Finally, miR-132 and miR-212 were shown to be associated with cognitive dysfunction and insoluble tau in humans (Smith et al. 2015).
miR-133a
In cells treated with MPP + , a model for PD, miR-133a expression levels decreased. MiR-133a overexpression enhanced cell proliferation and autophagy (LC3II/I and Beclin-1 expression declined, while p62 expression increased), but inhibited apoptosis. The inhibition of PC12 cell autophagy and apoptosis by miR-133a was attenuated by RACY upregulation by miR-133a targeting RAC1 (Lu et al. 2020).
miR-181
Dysregulation of miR-181a is inherent to neurodegenerative diseases (Hegarty et al. 2018). miR-181a is upregulated in patients with MCI. It is involved in AD pathogenesis (Ansari et al. 2019). The inhibition of miR-181a improved synapse and cognitive function in 3xTg-AD animals (Rodriguez‐Ortiz et al. 2020). However, miR-181a was decreased in vitro model of PD (Liu et al. 2017). miR-181a/b suppressed synaptic transmission, mitochondrial respiration, and neurite outgrowth-related gene expression in MPP + -treated SK-N-SH cells (Stein et al. 2022). The upregulation of miR-181a inhibited autophagy by targeting Beclin-1, LC3II/I, p-p38, and p-JNK (Liu et al. 2017). In addition, miR-181b inhibited MPP + -induced cytotoxicity in PC12 cells via the inhibition of autophagy by targeting PTEN, Akt and mTOR expression (Li et al. 2018).
miR-185
The protective function of miR-185 was shown in controlling PD progression (Rahimmi et al. 2019). miR-185 decreased cell death in dopaminergic neuron by activating PI3K/AKT signaling in PD (Qin et al. 2021a). Increased miR-185 suppressed autophagy in cells treated with MPTP by targeting the AMPK and mTOR (Wen et al. 2018).
miR-204
miR-204 is upregulated in neurodegenerative diseases (Talepoor Ardakani et al. 2019). Enhancement in miR-204 expression was observed in AD (Zhang et al. 2021a). The upregulation of miR-204-5p induced dopaminergic cell death by targeting ER stress-mediated by DYRK1A and apoptosis (Chiu et al. 2019). Overexpression of TRPML1 led to downregulation of miR-204 and activation of STAT3, while mitochondrial autophagy was attenuated. miR-204 silencing suppressed mitochondrial autophagy by inhibiting Parkin, PINK1,,LC3II, and Beclin1 through STAT3 pathway and up-regulating TRPML1 expression in APP/PS1 transgenic AD mice and a cell model of AD induced by Aβ1-42 (Zhang et al. 2021a).
miR-214
Decreased miR-214 contributes to the neuronal injury induced by Aβ (Yu and Zhang 2013). MiR-214–5p expression decreased in AD mice. Dexmedetomidine has a neuroprotective effect on AD via targeting miR-214–5p (Hu et al. 2022). miR-214-3p inhibited autophagy by inhibiting Atg12 in SAMP8 mice (Zhang et al. 2016a).
miR-221/222
In MN9D cells treated with MPP +, miR-221/222 was targeted and bound to p27/mTOR. By downregulating p27 levels, inhibiting the mTOR pathway. Thus, miR-221/222 facilitates autophagy and protects cells from death (Qian et al. 2019). MiR-212-5p protected against cell death in dopaminergic neuron by targeting p53, SIRT2, LC3 B and p62 expression in an animal model of PD induced by MPTP (Sun et al. 2018).
The number of miRNAs associated with autophagy in AD and PD is also continually expanding as shown in Table 1.
Table 1.
Autophagy-related microRNAs in AD and PD
| Neurodegenerative disease | Model | miRNAs | Expression status | Targets | Main targets | References |
|---|---|---|---|---|---|---|
| Alzheimer disease | In vitro/SH-SY5Y cells treated by H2O2 | miR-101a | Downregulated | MAPK1 pathway, beclin-1, LC3-II | Stimulate autophagy | Li et al. (2019) |
| Alzheimer disease | In vitro/PC12 and SK-N-SH cells | let-7 | Upregulated | Atg-5, -7, LC3 II/I, beclin-1, PI3K/Akt/mTOR | Stimulate autophagy | Gu et al. (2017) |
| Alzheimer disease | In vitro/cycloheximide-treated SH-SY5Y cells | miR-9a | Upregulated | UBE4A and UBE4B | Stimulate autophagy | Subramanian et al. (2021) |
| In vivo/E64D + PEPA-treated Tau-BiFC mice | ||||||
| Alzheimer disease | in vivo/SAMP8 mice | miR-214-3p | Downregulated | Atg12, 3′-UTR, LC3βII and Beclin1 | Inhibit autophagy | Zhang et al. (2016a) |
| Alzheimer disease | In vivo/APPswe/PS1dE9 mice | miR-299-5p | Downregulated | LC3βII, p62 and Atg5 | Inhibit autophagy | Zhang et al. (2016b) |
| In vitro/N2a/SH-SY5Y cells | ||||||
| Alzheimer disease | In vivo/SH-SY5Y cell APPswe/PS1dE9 mouse | miR-9-5p and miR-331-3p | Downregulated | Aβ clearance, Sqstm1, Optn, LC3b and Becn1 | Stimulate autophagy | Chen et al. (2021) |
| Alzheimer disease | In vivo/APP/PS1 transgenic mice | miR-124 | Downregulated | BACE1, LC3II, Atg5, p62/SQSTMl, Beclin-1 | Stimulate autophagy | Du et al. (2017) |
| Alzheimer disease | In vivo/3xTg-ADWT mice | miR-132/212 | Downregulated | tau mRNA, ATG5–12, ATG9a, Beclin1, P62, TMEM106b | Inhibit autophagy | Smith et al. (2015) |
| Alzheimer disease | In vivo/Aβ1-42 and APP/presenilin-1 AD modeled mice | miR-204 | Upregulated | TRPML1 -activated STAT3 pathway | Inhibit autophagy | Zhang et al. (2021a) |
| Parkinson disease | In vitro/SK-N-SH neuroblastoma cells treated by MPP | miR-181a | downregulated | LC3II/LC3I, Beclin 1, p-p38, p-JNK | Stimulate autophagy | Liu et al. (2017) |
| Parkinson disease | In vivo/SH-SY5Y cells, BV2 cells MPTP-treated, treated with VX702 | miR-124 | downregulated | p62, p-p38 | Inhibit autophagy | Yao et al. (2019) |
| Parkinson disease | in vitro/mouse microglia cell line (BV2) treated by MPTP | miR-3473b | upregulated | TNF-α, IL-1β, TREM2 and ULK1 | Inhibit autophagy | Lv et al. (2021) |
| Parkinson disease | in vivo/C57BL/6 mice model treated by MPTP | miR-3473b | upregulated | substantia nigra pars compacta (SNpc), TREM2, ULK1 | Inhibit autophagy | Lv et al. (2021) |
| Parkinson disease | In vivo/SH-SY5Y neuroblastoma cells MPTP-treated | miR-212-5p | downregulated | SIRT2, P53 | Stimulate autophagy | Sun et al. (2018) |
| In vitro/MPP+ SH-SY5Y cells | ||||||
| Parkinson disease | In vivo/C57BL/6 mice HEK‐293T MPTP/MPP + ‐induced | miR-326 | upregulated | JNK, XBP1, c-Jun, p–c-Jun, α-Syn, p-α-Syn, iNOS and LC3-II | Stimulate autophagy | Zhao et al. (2019) |
| Parkinson disease | In vitro/HEK293A cells MPP+ | miR-7 and miR-153 | Upregulated | mTOR, SAPK/JNK, P38MAPK, Bcl-2, Caspase-3, LC3-I, LC3-II | Inhibit autophagy | Fragkouli and Doxakis (2014) |
| Parkinson disease | In vitro/SH-SY5Y cells treated by MPTP | miR-124 | Downregulated | Beclin 1, LC3 II/I, AMPK, mTOR | Stimulate autophagy | Gong et al. (2016) |
| Parkinson disease | In vivo/MPTP-treated mice | miR-221/222 | Upregulated | p27/mTOR, LC3-II, (CDKN1B/p27) | Inhibit autophagy | Qian et al. (2019) |
| In vitro/MPP + -challenged MN9D cells | ||||||
| Parkinson disease | In vitro/MPTP-treated SH-SY5Y cells | miR-185 | Upregulated | AMPK, mTOR | Inhibit autophagy | Wen et al. (2018) |
| Parkinson disease | In vivo/Bcl-2 treated 6-OHDA-induced | miR-3557 | Upregulated | CaMK2α, CaMKV, Vdac1, PI3K/mTOR and UCH-L1 | Liu et al., (2019) | |
| Parkinson disease | In vivo/SH-SY5Y cells were treated with MPP+ | miR-29c-3p | Downregulated | Beclin 1, TET2, LC3 | inhibits autophagy | Wang et al. (2021a) |
| Parkinson disease | In vivo/MPTP-treated mice | miR-124 | Upregulated | Bim, Bax | Inhibit autophagy | Wang et al. (2016) |
| In vitro/MPP+-intoxicated SH-SY5Y cells | ||||||
| Parkinson disease | In vitro/PC12 cells MPP+ | miR-199a | Upregulated | PTEN, GSK3β, Beclin1, LC3II, AKT, mTOR | Inhibit autophagy | Ba et al. (2020) |
| Parkinson disease | In vitro/MPP+ treated PC12 cell | miR-181b | Downregulated | LC3II, p-AKT, PTEN, p-mTOR, p-p70S6K | Inhibit autophagy | Li et al. (2018) |
| Parkinson disease | In vivo/MPTP | miR-106b | Downregulated | Bcl-2 and LC3II/LC3I ratio | Inhibit autophagy | Bai et al. (2021) |
| Parkinson disease | In vitro/Human neural progenitor cell line ReNcell V | miR-7 | Downregulated | LC3-I/LC3-II | Inhibit autophagy | Choi et al. (2018) |
| Parkinson disease | SH-SY5Y cells treated with MPP+ | MiR-497-5p | Upregulated | LC3-II/I, Beclin1, p62 | Inhibit autophagy | Zhu et al. (2021) |
| Parkinson disease | In vitro/PC-12 treated by MPP+ | miR-133a | Downregulated | LC3II/I, Beclin-1, p62 | Stimulate autophagy | Lu et al. (2020) |
| Parkinson disease | MPP+- treated SH-SY5Y cells | miR-132-5p | Upregulated | LC3, Beclin 1, ULK1 | Stimulate autophagy | Zhao et al. (2020c) |
| Parkinson disease | In vitro/SH-SY5Y cells | miR-103a-3p | Upregulated | Parkin,Ambra1, LC3-I/LC3-II | Stimulate autophagy | Zhou et al. (2020b) |
| In vivo/MPTP | ||||||
| Parkinson disease | 6-OHDA-treated SH-SY5Y cell model | MiR-142-5p | Downregulated | decreased P62, increased LC3-II, Beclin-1 | Stimulate autophagy | Chen et al. (2020) |
| Parkinson disease | In vivo/rotenone | MiR-24 | Upregulated | Beclin-1, LC3-II/LC3-I ratio | Inhibit autophagy | Ge et al. (2019) |
| In vitro/MN9D cells |
lncRNAs and Apoptosis in AD and PD
BACE1-AS
Increased BACE1-AS expression is inherent to AD (Zhou et al. 2021). BACE1-AS downregulation was related to decreased synuclein, iNOS and glutamate (Li et al. 2020). ATG5 expression was indirectly regulated by BACE1-AS through the interaction with miR-214-3p. Neuronal damage secondary to inhibition of autophagy is relieved by suppression of BACE1-AS in vivo (Zhou et al. 2021). In AD, it has been shown both in vitro and in vivo that BACE1-AS and miR-214-3p are overexpressed and suppressed, respectively. BACE1-AS reduction as well as miR-214-3p overexpression promote the acceleration of autophagy by targeting LC3 I/II, p62 and Beclin-1 (He et al. 2020).
NEAT1
Increased NEAT1 expression was noted in SH-SY5Y cells upon MPP + treatment. NEAT1 silencing restored the MPP + -induced effects on SH-SY5Y cells, including inflammation, cell viability, and stimulated apoptosis. NEAT1 targets miR-124 and targeting miR-124 with anti-miR-124 reversed its inhibition effects (Xie et al. 2019). NEAT1 level was elevated in vitro AD experimental model, while its depletion reversed Aβ-induced increase in apoptosis and p-Tau levels. MiR-107 is targeted by NEAT1. miR-107 expression decreased in Aβ-treated cells, and its upregulation reversed Aβ-induced damage. Neuronal injury in Aβ-treated cells was suppressed by NEAT1 knockdown and reversed by miR-107 (Ke et al. 2019). Animal models of AD have shown upregulation of NEAT1 lncRNA. Inhibition of PINK1-dependent autophagy by ubiquitinating and degrading PINK1 has been posited as a potential sequela of NEAT1 interaction with NEDD4L (Huang et al. 2020). A significant increase in NEAT1 was observed in MPTP-treated mice and PD patients, as well as in SH-SY5Y cells treated with MPP + (Dong et al. 2021; Liu et al. 2020; Xie et al. 2019). SH-SY5Y cells treated with MPP + increased proliferation upon NEAT1 inhibition, whereas apoptosis and autophagy were inhibited. MPTP-treated mice with NEAT1 inhibition showed enhanced MIR-374c-5p expression, increased cell viability, and repressed autophagy and apoptosis by targeting miR-374c-5p, LC3 II/LC3 I, P62 (Dong et al. 2021). Autophagy was suppressed in in vitro and in vivo model of MPTP- induced PD by NEAT1 knockdown via targeting PINK1, LC3-II, and LC3-I protein, decreasing dopaminergic neuronal damage (Yan et al. 2018).
SNHG1
MPP + enhanced expression of SNHG1 (Zhang et al. 2020a; Zhao et al. 2020a). Furthermore, silencing SNHG1 improved the behavior in MPTP-treated mice in vivo (Xiao et al. 2021). In addition to promoting autophagy and preventing MPP + -induced cell death, SNHG1 indirectly affects the expression of p27/mTOR by binding to the miR-221/222 cluster (Qian et al. 2019).
HOTAIR
SH-SY5Y cells treated with MPP + showed enhanced expression of lncRNA HOTAIR. PD-like symptoms were significantly alleviated in vivo when HOTAIR was knocked down. As a result of the downregulation of HOTAIR in the presence of MPP +, SH-SY5Y cells increased their viability and NLRP3-mediated pyroptotic cell death was suppressed. By targeting miR-326, HOTAIR regulates ELAVL1 expression. Downregulation of HOTAIR or ELAVL1 significantly reduced the pyroptosis-promoting effects of miR-326 inhibitor through activation of NLRP3 inflammasomes (Zhang et al. 2021c). In SK-N-SH cells treated with MPP + , increased expression of HOTAIR and ATG10 was noted (Zhao et al. 2020b). MPP + -induced neurodegeneration was decreased by the knockdown of HOTAIR. HOTAIR aggravates MPP + -mediated neuronal injury by sponging miR-874-5p (Zhao et al. 2020b). MTPT-treated mice and SH-SY5Y cells pretreated with MPP + showed upregulation of HOTAIR. In SH-SY5Y cells overexpressing HOTAIR, the expression of LRRK2 was enhanced. In SH-SY5Y cells treated by MPP +, HOTAIR knockdown improved cell viability (Kraus et al. 2017; Wang et al. 2017a). HOTAIR inhibited cell viability and stimulated autophagy by attaching to miR-221-3p and targeting LC3-II/-I, LAMP1/2, P62 expressions, and NPTX2 (Lang et al. 2020).
BDNF-AS
BDNF-AS expression increased in AD patients. The upregulation of BDNF-AS led to cognitive dysfunction in AD mice (Ding et al. 2022). Silencing BDNF-AS inhibited apoptosis and oxidative stress in PC12 cells treated by Aβ25-35 (Guo et al. 2018a). Fan et al. (Fan et al. 2020) was seen the up-regulation of BDNF-AS in both in vitro and in vivo model of PD. BDNF-AS increased autophagy in MPTP-induced PD by targeting microRNA-125b-5p, LC3II/I, and Beclin-1, p62 levels (Fan et al. 2020).
The number of lncRNAs associated with autophagy in AD and PD is also continually expanding as shown in Table 2.
Table 2.
Autophagy-related lncRNAs in AD and PD
| Neurodegenerative disease | Model | lncRNAs | Expression status | Targets | Main effect | References |
|---|---|---|---|---|---|---|
| Alzheimer disease | In vivo/transgenic mice, | BACE1-AS | Upregulated | miR-214-3p and ATG5 | Stimulate autophagy | Zhou et al. (2021) |
| In vitro/SH-SY5Y treated by Aβ1-42 | ||||||
| Alzheimer disease | In vitro/Aβ-treated SK-N-SH and SK-N-AS cells | BACE1-AS | Upregulated | miR-214-3p, Bax, Bcl-2, Beclin-1, LC3 I/LC3 II and p62 | Stimulate autophagy | He et al. (2020) |
| In vivo | ||||||
| Alzheimer disease | In vivo/transgenic mice | lncRNA RMRP | Upregulated | miR-3142/TRIB3 axis | Stimulate autophagy | Tang et al. (2022) |
| In vitro/SH-SY5Y cells | ||||||
| Parkinson disease | MPP+ | lncRNA-SNHG1 | Upregulated | miR-221/222 cluster, CDKN1B/p27/mTOR, SNHG1 | inhibit autophagy | Qian et al. (2019) |
| Parkinson disease | In vivo | NEAT1 | Upregulated | PINK1, LC3-I, LC3-II | Stimulate autophagy | Yan et al. (2018) |
| In vitro/MPP + treated-SH-SY5Y cells | ||||||
| Parkinson disease | In vitro/MPP +—treated SH-SY5Y cells | NEAT1 | Upregulated | miR-374c-5p, LC3 II/LC3 I, P62 | Stimulate autophagy | Dong et al. (2021) |
| In vivo/MPTP | ||||||
| Parkinson disease | In vitro/MN9D was treated (MPP+), In vivo/MPTP | HOTAIR | Downregulated | LC3B-II/LC3B-I, LAMP1/LAMP2, P62, NPTX2 | Inhibit autophagy | Lang et al. (2020) |
| Parkinson disease | In vivo/MPTP | BDNF-AS | Upregulated | miR-125b-5p, LC3II/I and Beclin-1, p62 levels | Inhibit autophagy | Fan et al. (2020) |
| In vitro/MPP + |
Circular RNAs and Apoptosis in AD and PD
Autophagy is regulated by CircumNF1-419 via Akt-mTOR/PI3K-I and Akt-AMPK-mTOR/PI3K-I signaling pathways. AP2B1 injection into circNF1-419 overexpressing cerebral cortex increased autophagy. Consequently, tau and p-tau were decreased, suggesting a delay in the onset of senile dementia. CirNF1-419 enhanced the activity of several signaling pathways, particularly mediators of transmission in the SAMP8 mouse (Diling et al. 2019). In in vitro model of PD, expression levels of SNCCa and circSNCA were downregulated after PPX treatment, which correlated with apoptotic gene expression, and SNCA expression was increased by CircSNCA through the downregulation of miR-7 in PD as a competitive endogenous RNA (ceRNA). Pro-apoptotic proteins were less expressed in cells with lower circSNCA expression. Downregulation of CircSNCA in PD reduced apoptosis and promoted autophagy (Sang et al. 2018). circDLGAP4 expression decreased in PD mouse model inculcated with MPTP. circDLGAP4 overexpression mitigated mitochondrial impairment, and enhanced cell viability and autophagy, decreased apoptosis and thereby attenuated neurotoxicity. A major function of circDLGAP4 is to regulate miR-134-5p. MiR-134-5p targets the transcription factor CREB, and CREB expression can be affected by the circDLGAP4/miR-134-5p axis. CircDLGAP4/miR-134-5p can also modulate the activity of CREB signaling, PGC-1α, Bcl-2, and BDNF (Feng et al. 2020). circSAMD4A was upregulated in both PD animal and cellular models. Inhibition of MPTP or MPP +-induced apoptosis and autophagy by circSAMD4A knockdown was observed. In MPTP or MPP +-induced PD models, the knockdown of circSAMD4A resulted in a reduction in phosphorylated AMPK expression and a subsequent increase in mTOR expression (Wang et al. 2021b) (Table 3).
Table 3.
Autophagy-related lncRNAs in AD and PD
| Neurodegenerative disease | Model | CircRNAs | Expression status | Targets | Main effect | References |
|---|---|---|---|---|---|---|
| Alzheimer disease | In vivo/SAMP8 mice treated with d-galactose | circNF1-419 | Upregulated | Dynamin-1, AP2B1, PI3K-I, Akt, AMPK, mTOR, Aβ1-42, Tau, p-Tau, and APOE | Stimulate autophagy | Diling et al. (2019) |
| Parkinson disease | In vitro/(MPP +) SH-SY5Y cells | CircSNCA | Downregulated | miR-7 sponge to upregulate SNCA | Induce autophagy | Sang et al. (2018) |
| Parkinson disease | In vivo/MPTP | circDLGAP4 | Downregulated | miR-134-5p/CREB, Beclin-1, LC3 II/I ratio | Inhibit autophagy | Feng et al. (2020) |
| In vitro/SH-SY5Y and MN9D cells treated with MPP+ | ||||||
| Parkinson disease | In vitro/SH-SY5Y cells MPTP- or MPP+ | circSAMD4A | Upregulated | AMPK, mTOR, miR-29c-3p | Stimulate autophagy | Wang et al. (2021b) |
Finally, dysregulation of circNF1-419, circSNCA, circHIPK3, circDLGAP4, circSAMD4A, circNF1-419, circSHOC2, circERCC2 has been associated with inhibition or stimulation of autophagy in AD and PD.
Conclusions
Increased evidence suggests that ncRNAs play a key role in autophagic homeostasis and can regulate neurodegeneration in AD and PD. ncRNAs can form complex interactions with other RNAs/DNA/proteins to regulate the autophagic process. For future perspective, the interaction between ncRNAs and autophagy in other neurodegenerative diseases and further clinical studies on the role of ncRNAs in neurodegenerative diseases progression and autophagy are required. In addition, further characterization of ncRNAs in bodily fluids is necessary, in particular, blood and CSF in patients with neurodegenerative diseases. ncRNAs (miRNAs, lncRNAs, and circRNAs) may serve as putative treatment, prognostic, and diagnostic targets for neurodegenerative diseases. The development of reliable for diagnosing early neurodegeneration and its molecular signaling will be pivotal for increased efficiency of therapies. Some studies evaluated targeting and transcriptionally repressing ncRNAs by silencing ncRNAs and RNA interference to inhibit ncRNAs expression in neurodegenerative disorders (Wang et al. 2022; Zhang et al. 2021b). Moreover, CRISPRs could be applied to delete miRNAs/lncRNAs (Ho et al. 2015) and assess the consequences for therapeutic strategies.
Author Contributions
AT-T, ZG, HZ, AA, ZF, ED, ZT, MA, HM, O-RT and FN contributed in the conception or design of the work and drafting of the manuscript. All authors confirmed the final version for submission.
Funding
No specific source of funding is associated with this work.
Data Availability
Not applicable.
Declarations
Competing Interests
The authors declare no conflict of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publish
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hamed Mirzaei, Email: h.mirzaei2002@gmail.com.
Omid Reza Tamtaji, Email: or-tamtaji@razi.tums.ac.ir.
Fatemeh Nabavizadeh, Email: nabavizadeh@tums.ac.ir.
References
- Agarwal S, Tiwari SK, Seth B, Yadav A, Singh A, Mudawal A, Chauhan LKS, Gupta SK, Choubey V, Tripathi A (2015) Activation of autophagic flux against xenoestrogen bisphenol-A-induced hippocampal neurodegeneration via AMP kinase (AMPK)/mammalian target of rapamycin (mTOR) pathways. J Biol Chem 290:21163–21184 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Alcalay RN, Caccappolo E, Mejia-Santana H, Tang MX, Rosado L, Ross BM, Verbitsky M, Kisselev S, Louis ED, Comella C (2010) Frequency of known mutations in early-onset Parkinson disease: implication for genetic counseling: the consortium on risk for early onset Parkinson disease study. Arch Neurol 67:1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegre-Abarrategui J, Christian H, Lufino MM, Mutihac R, Venda LL, Ansorge O, Wade-Martins R (2009) LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet 18:4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annese T, Tamma R, De Giorgis M, Ribatti D (2020) microRNAs biogenesis, functions and role in tumor angiogenesis. Front Oncol 10:581007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari A, Maffioletti E, Milanesi E, Marizzoni M, Frisoni GB, Blin O, Richardson JC, Bordet R, Forloni G, Gennarelli M (2019) miR-146a and miR-181a are involved in the progression of mild cognitive impairment to Alzheimer’s disease. Neurobiol Aging 82:102–109 [DOI] [PubMed] [Google Scholar]
- Ba R-Q, Liu J, Fan X-J, Jin G-L, Huang B-G, Liu M-W, Yang J-S (2020) Effects of miR-199a on autophagy by targeting glycogen synthase kinase 3β to activate PTEN/AKT/mTOR signaling in an MPP+ in vitro model of Parkinson’s disease. Neurol Res 42:308–318 [DOI] [PubMed] [Google Scholar]
- Bai X, Dong Q, Zhao L, Yao Y, Wang B (2021) microRNA-106b-containing extracellular vesicles affect autophagy of neurons by regulating CDKN2B in Parkinson’s disease. Neurosci Lett 760:136094 [DOI] [PubMed] [Google Scholar]
- Barbato C, Giacovazzo G, Albiero F, Scardigli R, Scopa C, Ciotti M, Strimpakos G, Coccurello R, Ruberti F (2020) Cognitive decline and modulation of Alzheimer’s disease-related genes after inhibition of MicroRNA-101 in Mouse hippocampal neurons. Mol Neurobiol 57:3183–3194 [DOI] [PubMed] [Google Scholar]
- Bonifati V (2006) Parkinson’s disease: the LRRK2-G2019S mutation: opening a novel era in Parkinson’s disease genetics. Euro J Human Genet: EJHG 14:1061 [DOI] [PubMed] [Google Scholar]
- Checkoway H, Lundin JI, Kelada SN (2011) Neurodegenerative diseases. IARC Sci Publ 163:407–419 [PubMed] [Google Scholar]
- Chen J, Jiang C, Du J, Xie C-L (2020) MiR-142-5p protects against 6-OHDA-induced SH-SY5Y cell injury by downregulating BECN1 and autophagy. Dose Response 18:1559325820907016 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chen M-L, Hong C-G, Yue T, Li H-M, Duan R, Hu W-B, Cao J, Wang Z-X, Chen C-Y, Hu X-K (2021) Inhibition of miR-331-3p and miR-9-5p ameliorates Alzheimer’s disease by enhancing autophagy. Theranostics 11:2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Liao Y, Dong Y, Hu H, Yang N, Kong X, Li S, Li X, Guo J, Qin L (2020) Microglial autophagy defect causes parkinson disease-like symptoms by accelerating inflammasome activation in mice. Autophagy 16:2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C-C, Yeh T-H, Chen R-S, Chen H-C, Huang Y-Z, Weng Y-H, Cheng Y-C, Liu Y-C, Cheng A-J, Lu Y-C (2019) Upregulated expression of microRNA-204-5p leads to the death of dopaminergic cells by targeting DYRK1A-mediated apoptotic signaling cascade. Front Cell Neurosci 13:399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M-H, Cho K, Kang H-J, Jeon E-Y, Kim H-S, Kwon H-J, Kim H-M, Kim D-H, Yoon S-Y (2014) Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy 10:1761–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DC, Yoo M, Kabaria S, Junn E (2018) MicroRNA-7 facilitates the degradation of alpha-synuclein and its aggregates by promoting autophagy. Neurosci Lett 678:118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coune PG, Bensadoun J-C, Aebischer P, Schneider B (2011) Rab1A over-expression prevents Golgi apparatus fragmentation and partially corrects motor deficits in an alpha-synuclein based rat model of Parkinson’s disease. J Parkinson’s Dis 1:373–387 [DOI] [PubMed] [Google Scholar]
- Decressac M, Mattsson B, Weikop P, Lundblad M, Jakobsson J, Björklund A (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc Natl Acad Sci USA 110:E1817–E1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dello Russo C, Lisi L, Feinstein DL, Navarra P (2013) mTOR kinase, a key player in the regulation of glial functions: relevance for the therapy of multiple sclerosis. Glia 61:301–311 [DOI] [PubMed] [Google Scholar]
- Diling C, Yinrui G, Longkai Q, Xiaocui T, Yadi L, Xin Y, Guoyan H, Ou S, Tianqiao Y, Dongdong W (2019) Circular RNA NF1-419 enhances autophagy to ameliorate senile dementia by binding dynamin-1 and adaptor protein 2 B1 in AD-like mice. Aging (albany NY) 11:12002 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ding Y, Luan W, Wang Z, Cao Y (2022) LncRNA BDNF-AS as ceRNA regulates the miR-9-5p/BACE1 pathway affecting neurotoxicity in Alzheimer’s disease. Arch Gerontol Geriatr 99:104614 [DOI] [PubMed] [Google Scholar]
- Dong L, Zheng Y, Gao L, Luo X (2021) lncRNA NEAT1 prompts autophagy and apoptosis in MPTP-induced Parkinson’s disease by impairing miR-374c-5p. Acta Biochim Biophys Sinica. 10.1093/abbs/gmab055 [DOI] [PubMed] [Google Scholar]
- Du G, Liu X, Chen X, Song M, Yan Y, Jiao R, Wang C-C (2010) Drosophila histone deacetylase 6 protects dopaminergic neurons against α-synuclein toxicity by promoting inclusion formation. Mol Biol Cell 21:2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Huo X, Yang Y, Hu Z, Botchway BO, Jiang Y, Fang M (2017) miR-124 downregulates BACE 1 and alters autophagy in APP/PS1 transgenic mice. Toxicol Lett 280:195–205 [DOI] [PubMed] [Google Scholar]
- Dugger BN, Dickson DW (2017) Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol 9:a028035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duve CD, Wattiaux R (1966) Functions of lysosomes. Ann Rev Physiol 28:435–492 [DOI] [PubMed] [Google Scholar]
- Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12:861–874 [DOI] [PubMed] [Google Scholar]
- Fan W, Nassiri A, Zhong Q (2011) Autophagosome targeting and membrane curvature sensing by Barkor/Atg14 (L). Proc Natl Acad Sci USA 108:7769–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Zhao X, Lu K, Cheng G (2020) LncRNA BDNF-AS promotes autophagy and apoptosis in MPTP-induced Parkinson’s disease via ablating microRNA-125b-5p. Brain Res Bull 157:119–127 [DOI] [PubMed] [Google Scholar]
- Felekkis K, Touvana E, Stefanou C, Deltas C (2010) microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia 14:236 [PMC free article] [PubMed] [Google Scholar]
- Feng Y, He D, Yao Z, Klionsky DJ (2014) The machinery of macroautophagy. Cell Res 24:24–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Zhang L, Wang S, Hong Q (2020) Circular RNA circDLGAP4 exerts neuroprotective effects via modulating miR-134-5p/CREB pathway in Parkinson’s disease. Biochem Biophys Res Commun 522:388–394 [DOI] [PubMed] [Google Scholar]
- Fernández-de Frutos M, Galán-Chilet I, Goedeke L, Kim B, Pardo-Marqués V, Pérez-García A, Herrero JI, Fernández-Hernando C, Kim J, Ramírez CM (2019) MicroRNA 7 impairs insulin signaling and regulates Aβ levels through posttranscriptional regulation of the insulin receptor substrate 2, insulin receptor, insulin-degrading enzyme, and liver X receptor pathway. Mol Cell Biol 39:e00170-e1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, Doxakis E (2014) miR-7 and miR-153 protect neurons against MPP+-induced cell death via upregulation of mTOR pathway. Front Cell Neurosci 8:182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T (2008) An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 19:4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge B, Wu H, Shao D, Li S, Li F (2019) Interfering with miR-24 alleviates rotenone-induced dopaminergic neuron injury via enhancing autophagy by upregulating DJ-1. Aging Pathobiol Ther 1:17–24 [Google Scholar]
- Ge X, Guo M, Hu T, Li W, Huang S, Yin Z, Li Y, Chen F, Zhu L, Kang C (2020) Increased microglial exosomal miR-124-3p alleviates neurodegeneration and improves cognitive outcome after rmTBI. Mol Ther 28:503–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X, Wang H, Ye Y, Shu Y, Deng Y, He X, Lu G, Zhang S (2016) miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am J Transl Res 8:2127 [PMC free article] [PubMed] [Google Scholar]
- Gourvest M, Brousset P, Bousquet M (2019) Long noncoding RNAs in acute myeloid leukemia: functional characterization and clinical relevance. Cancers (basel). 10.1038/cdd.2015.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green D, Oguin TH, Martinez J (2016) The clearance of dying cells: table for two. Cell Death Differ 23:915–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan K-P, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R (2004) The Microprocessor complex mediates the genesis of microRNAs. Nature 432:235–240 [DOI] [PubMed] [Google Scholar]
- Gu H, Li L, Cui C, Zhao Z, Song G (2017) Overexpression of let-7a increases neurotoxicity in a PC12 cell model of Alzheimer’s disease via regulating autophagy. Exp Ther Med 14:3688–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C-C, Jiao C-H, Gao Z-M (2018a) Silencing of LncRNA BDNF-AS attenuates Aβ25-35-induced neurotoxicity in PC12 cells by suppressing cell apoptosis and oxidative stress. Neurol Res 40:795–804 [DOI] [PubMed] [Google Scholar]
- Guo F, Liu X, Cai H, Le W (2018b) Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol 28:3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamano T, Gendron TF, Causevic E, Yen SH, Lin WL, Isidoro C, DeTure M, Ko L-w (2008) Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci 27:1119–1130 [DOI] [PubMed] [Google Scholar]
- Han D, Dong X, Zheng D, Nao J (2020) MiR-124 and the underlying therapeutic promise of neurodegenerative disorders. Front Pharmacol 10:1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H (2006) Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441:885–889 [DOI] [PubMed] [Google Scholar]
- He W, Chi S, Jin X, Lu J, Zheng W, Yan J, Zhang D (2020) Long non-coding RNA BACE1-AS modulates isoflurane-induced neurotoxicity to Alzheimer’s Disease through sponging miR-214-3p. Neurochem Res 45:2324–2335 [DOI] [PubMed] [Google Scholar]
- Hegarty SV, Sullivan AM, O’Keeffe GW (2018) Inhibition of miR-181a promotes midbrain neuronal growth through a Smad1/5-dependent mechanism: implications for Parkinson’s disease. Neuronal Signal. 10.1042/NS20170181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T-T, Zhou N, Huang J, Koirala P, Xu M, Fung R, Wu F, Mo Y-Y (2015) Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res 43:e17–e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Shi Z, Shao W, Xu B (2022) MicroRNA-214–5p involves in the protection effect of dexmedetomidine against neurological injury in Alzheimer’s disease via targeting the suppressor of zest 12. Brain Res Bull 178:164–172 [DOI] [PubMed] [Google Scholar]
- Huang Z, Zhao J, Wang W, Zhou J, Zhang J (2020) Depletion of LncRNA NEAT1 rescues mitochondrial dysfunction through NEDD4L-dependent PINK1 degradation in animal models of Alzheimer’s disease. Front Cell Neurosci 14:28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151:1256–1269 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xu N (2023) The emerging role of autophagy-associated lncRNAs in the pathogenesis of neurodegenerative diseases. Int J Mol Sci 24:9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro S-H, Kim Y-M, Otto NM, Cao J, Kundu M, Kim D-H (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20:1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Yang Z, Yang F, Wang X, Tan J, Liao B (2019) Long noncoding RNA NEAT1 aggravates Aβ-induced neuronal damage by targeting miR-107 in Alzheimer’s disease. Yonsei Med J 60:640–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Ramírez CM, Lee S-M, Hoe H-S, Fernández-Hernando C, Kim J (2012) MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp Neurol 235:476–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J-I, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E (2006) Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441:880–884 [DOI] [PubMed] [Google Scholar]
- Kraus TF, Haider M, Spanner J, Steinmaurer M, Dietinger V, Kretzschmar HA (2017) Altered long noncoding RNA expression precedes the course of Parkinson’s disease—a preliminary report. Mol Neurobiol 54:2869–2877 [DOI] [PubMed] [Google Scholar]
- Kuhn HG, Palmer TD, Fuchs E (2001) Adult neurogenesis: a compensatory mechanism for neuronal damage. Eur Arch Psychiatry Clin Neurosci 251:152–158 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739 [DOI] [PubMed] [Google Scholar]
- Lang Y, Li Y, Yu H, Lin L, Chen X, Wang S, Zhang H (2020) HOTAIR drives autophagy in midbrain dopaminergic neurons in the substantia nigra compacta in a mouse model of Parkinson’s disease by elevating NPTX2 via miR-221-3p binding. Aging (albany NY) 12:7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132:27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lv X, Zhai K, Xu R, Zhang Y, Zhao S, Qin X, Yin L, Lou J (2016) MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax and Sirt2. Am J Transl Res 8:993 [PMC free article] [PubMed] [Google Scholar]
- Li W, Jiang Y, Wang Y, Yang S, Bi X, Pan X, Ma A (2018) MiR-181b regulates autophagy in a model of Parkinson’s disease by targeting the PTEN/Akt/mTOR signaling pathway. Neurosci Lett 675:83–88 [DOI] [PubMed] [Google Scholar]
- Li Q, Wang Y, Peng W, Jia Y, Tang J, Li W, Zhang JH, Yang J (2019) MicroRNA-101a regulates autophagy phenomenon via the MAPK pathway to modulate Alzheimer’s-associated pathogenesis. Cell Transplant 28:1076–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Fang J, Zhou Z, Zhou Q, Sun S, Jin Z, Xi Z, Wei J (2020) Downregulation of lncRNA BACE1-AS improves dopamine-dependent oxidative stress in rats with Parkinson’s disease by upregulating microRNA-34b-5p and downregulating BACE1. Cell Cycle 19:1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song Y, Zhu X (2017) MicroRNA-181a regulates apoptosis and autophagy process in Parkinson’s disease by inhibiting p38 mitogen-activated protein kinase (MAPK)/c-Jun N-terminal kinases (JNK) signaling pathways. Med Sci Monit: Int Med J Exp Clin Res 23:1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li L, Liu S, Wang Z, Kuang H, Xia Y, Tang C, Yin D (2019) MicroRNA expression profiling screen miR-3557/324-targeted CaMK/mTOR in the rat striatum of Parkinson’s disease in regular aerobic exercise. BioMed Res Int. 10.1155/2019/7654798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu D, Zhao B, Jia C, Lv Y, Liao J, Li K (2020) Long non-coding RNA NEAT1 mediates MPTP/MPP+-induced apoptosis via regulating the miR-124/KLF4 axis in Parkinson’s disease. Open Life Sci 15:665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JM, Lahiri DK (2011) MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem Biophys Res Commun 404:889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Lin J, Zheng D, Hong C, Ke L, Wu X, Chen P (2020) Overexpression of MicroRNA-133a inhibits apoptosis and autophagy in a cell model of Parkinson’s disease by downregulating ras-related C3 botulinum toxin substrate 1 (RAC1). Med Sci Monitor: Int Med J Exp Clin Res 26:e922032–e922031 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lucin KM, O’Brien CE, Bieri G, Czirr E, Mosher KI, Abbey RJ, Mastroeni DF, Rogers J, Spencer B, Masliah E (2013) Microglial beclin 1 regulates retromer trafficking and phagocytosis and is impaired in Alzheimer’s disease. Neuron 79:873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Z, Hu B, Yan S, Yan Y, Zhang J, Shi T, Jiang L, Li W, Huang W (2021) MicroRNA-3473b regulates the expression of TREM2/ULK1 and inhibits autophagy in inflammatory pathogenesis of Parkinson’s disease. J Neurochem 157:599–610 [DOI] [PubMed] [Google Scholar]
- Manzoni C, Mamais A, Dihanich S, Abeti R, Soutar MP, Plun-Favreau H, Giunti P, Tooze SA, Bandopadhyay R, Lewis PA (2013) Inhibition of LRRK2 kinase activity stimulates macroautophagy. Biochim Biophys Acta (BBA) Mol Cell Res 1833:2900–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez RT, Xu L (2012) Bcl-2: Beclin 1 complex: multiple, mechanisms regulating autophagy/apoptosis toggle switch. Am J Cancer Res 2:214 [PMC free article] [PubMed] [Google Scholar]
- Martyniszyn L, Szulc L, Boratyńska A, Niemiałtowski MG (2011) Beclin 1 is involved in regulation of apoptosis and autophagy during replication of ectromelia virus in permissive L929 cells. Arch Immunol Ther Exp 59:463–471 [DOI] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Rubinsztein DC (2015) Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci 16:345–357 [DOI] [PubMed] [Google Scholar]
- Mishima T, Mizuguchi Y, Kawahigashi Y, Takizawa T, Takizawa T (2007) RT-PCR-based analysis of microRNA (miR-1 and-124) expression in mouse CNS. Brain Res 1131:37–43 [DOI] [PubMed] [Google Scholar]
- Nah J, Yuan J, Jung Y-K (2015) Autophagy in neurodegenerative diseases: from mechanism to therapeutic approach. Mol Cells 38:381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen D-F, Youle RJ (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 183:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N (2013) Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta (BBA) Mol Cell Res 1833:3448–3459 [DOI] [PubMed] [Google Scholar]
- Nojima T, Proudfoot NJ (2022) Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol 23:389–406 [DOI] [PubMed] [Google Scholar]
- Obara K, Ohsumi Y (2008) Dynamics and function of PtdIns (3) P in autophagy. Autophagy 4:952–954 [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Mizushima N (2004) Two ubiquitin-like conjugation systems essential for autophagy. Sem Cell Dev Biol 15:231–236 [DOI] [PubMed] [Google Scholar]
- Omura T, Nomura L, Watanabe R, Nishiguchi H, Yamamoto K, Imai S, Nakagawa S, Itohara K, Yonezawa A, Nakagawa T (2021) MicroRNA-101 regulates 6-hydroxydopamine-induced cell death by targeting suppressor/enhancer lin-12-like in SH-SY5Y cells. Front Mol Neurosci. 10.3389/fnmol.2021.748026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patergnani S, Pinton P (2015) Mitophagy and mitochondrial balance. Mitochondrial Regul. 10.1007/978-1-4939-1875-1_15 [Google Scholar]
- Patergnani S, Guzzo S, Mangolini A, dell’Atti L, Pinton P, Aguiari G (2020) The induction of AMPK-dependent autophagy leads to P53 degradation and affects cell growth and migration in kidney cancer cells. Exp Cell Res 395:112190 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122:927–939 [DOI] [PubMed] [Google Scholar]
- Plaza-Zabala A, Sierra-Torre V, Sierra A (2017) Autophagy and microglia: novel partners in neurodegeneration and aging. Int J Mol Sci 18:598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC (2013) Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154:1285–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C, Ye Y, Mao H, Yao L, Sun X, Wang B, Zhang H, Xie L, Zhang H, Zhang Y (2019) Downregulated lncRNA-SNHG1 enhances autophagy and prevents cell death through the miR-221/222/p27/mTOR pathway in Parkinson’s disease. Exp Cell Res 384:111614 [DOI] [PubMed] [Google Scholar]
- Qin X, Zhang X, Li P, Wang M, Yan L, Pan P, Zhang H, Hong X, Liu M, Bao Z (2021a) MicroRNA-185 activates PI3K/AKT signalling pathway to alleviate dopaminergic neuron damage via targeting IGF1 in Parkinson’s disease. J Drug Target 29:875–883 [DOI] [PubMed] [Google Scholar]
- Qin Y, Qiu J, Wang P, Liu J, Zhao Y, Jiang F, Lou H (2021b) Impaired autophagy in microglia aggravates dopaminergic neurodegeneration by regulating NLRP3 inflammasome activation in experimental models of Parkinson’s disease. Brain Behav Immun 91:324–338 [DOI] [PubMed] [Google Scholar]
- Querfurth HW, LaFerla FM (2010) Alzheimer’s disease. N Engl J Med 362:329–344 [DOI] [PubMed] [Google Scholar]
- Rahimmi A, Peluso I, Rajabi A, Hassanzadeh K (2019) miR-185 and SEPT5 genes may contribute to Parkinson’s disease pathophysiology. Oxid Med Cell Longev. 10.1155/2019/5019815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Duden R, Rubinsztein DC (2002) Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 11:1107–1117 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Acevedo-Arozena A, Imarisio S, Berger Z, Vacher C, O’Kane CJ, Brown SD, Rubinsztein DC (2005) Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nat Genet 37:771–776 [DOI] [PubMed] [Google Scholar]
- Riebisch AK, Mühlen S, Beer YY, Schmitz I (2021) Autophagy—a story of bacteria interfering with the host cell degradation machinery. Pathogens 10:110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Martín T, Cuchillo-Ibáñez I, Noble W, Nyenya F, Anderton BH, Hanger DP (2013) Tau phosphorylation affects its axonal transport and degradation. Neurobiol Aging 34:2146–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ortiz CJ, Prieto GA, Martini AC, Forner S, Trujillo-Estrada L, LaFerla FM, Baglietto-Vargas D, Cotman CW, Kitazawa M (2020) miR-181a negatively modulates synaptic plasticity in hippocampal cultures and its inhibition rescues memory deficits in a mouse model of Alzheimer’s disease. Aging Cell 19:e13118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala G, Stefanoni G, Arosio A, Riva C, Melchionda L, Saracchi E, Fermi S, Brighina L, Ferrarese C (2014) Reduced expression of the chaperone-mediated autophagy carrier hsc70 protein in lymphomonocytes of patients with Parkinson’s disease. Brain Res 1546:46–52 [DOI] [PubMed] [Google Scholar]
- Sang Q, Liu X, Wang L, Qi L, Sun W, Wang W, Sun Y, Zhang H (2018) CircSNCA downregulation by pramipexole treatment mediates cell apoptosis and autophagy in Parkinson’s disease by targeting miR-7. Aging (albany NY) 10:1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe LA, Stevens DA, Dawson VL, Dawson TM (2014) Parkin and PINK1: much more than mitophagy. Trends Neurosci 37:315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SZA, Zhao D, Hussain T, Sabir N, Yang L (2018) Regulation of microRNAs-mediated autophagic flux: a new regulatory avenue for neurodegenerative diseases with focus on prion diseases. Front Aging Neurosci 10:139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Gao Z, Feldman T, Jiang X (2007) Stimulation of ATG12-ATG5 conjugation by ribonucleic acid. Autophagy 3:10–16 [DOI] [PubMed] [Google Scholar]
- Shibutani ST, Saitoh T, Nowag H, Münz C, Yoshimori T (2015) Autophagy and autophagy-related proteins in the immune system. Nat Immunol 16:1014–1024 [DOI] [PubMed] [Google Scholar]
- Sierra A, Abiega O, Shahraz A, Neumann H (2013) Janus-faced microglia: beneficial and detrimental consequences of microglial phagocytosis. Front Cell Neurosci 7:6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PY, Hernandez-Rapp J, Jolivette F, Lecours C, Bisht K, Goupil C, Dorval V, Parsi S, Morin F, Planel E (2015) miR-132/212 deficiency impairs tau metabolism and promotes pathological aggregation in vivo. Hum Mol Genet 24:6721–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Zhang Y, Huang W, Shi J, Huang Q, Jiang M, Qiu Y, Wang T, Chen H, Wang H (2022) Circular RNA Cwc27 contributes to Alzheimer’s disease pathogenesis by repressing Pur-α activity. Cell Death Differ 29:393–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, Gindi R, Adame A, Wyss-Coray T, Masliah E (2009) Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in α-synuclein models of Parkinson’s and Lewy body diseases. J Neurosci 29:13578–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CS, McLendon JM, Witmer NH, Boudreau RL (2022) Modulation of miR-181 influences dopaminergic neuronal degeneration in a mouse model of Parkinson’s disease. Mol Ther Nucleic Acids 28:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P, Zhang J, Wang D, Zhao F, Cao Z, Aschner M, Luo W (2016) The role of autophagy in modulation of neuroinflammation in microglia. Neuroscience 319:155–167 [DOI] [PubMed] [Google Scholar]
- Subramanian M, Hyeon SJ, Das T, Suh YS, Kim YK, Lee J-S, Song EJ, Ryu H, Yu K (2021) UBE4B, a microRNA-9 target gene, promotes autophagy-mediated Tau degradation. Nat Commun 12:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Han X, Li X, Song Q, Lu M, Jia M, Ding J, Hu G (2018) MicroRNA-212-5p prevents dopaminergic neuron death by inhibiting SIRT2 in MPTP-induced mouse model of Parkinson’s disease. Front Mol Neurosci 11:381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendran S, Rajasankar S (2010) Parkinson’s disease: oxidative stress and therapeutic approaches. Neurol Sci 31:531–540 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Tsukahara T (2014) A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 15:9331–9342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talepoor Ardakani M, Rostamian Delavar M, Baghi M, Nasr-Esfahani MH, Kiani-Esfahani A, Ghaedi K (2019) Upregulation of miR-200a and miR-204 in MPP+-treated differentiated PC12 cells as a model of Parkinson’s disease. Mol Genet Genomic Med 7:e548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C-C, Yu J-T, Tan M-S, Jiang T, Zhu X-C, Tan L (2014) Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging 35:941–957 [DOI] [PubMed] [Google Scholar]
- Tang Z-B, Chen H-P, Zhong D, Song J-H, Cao J-W, Zhao M-Q, Han B-C, Duan Q, Sheng X-M, Yao J-L (2022) LncRNA RMRP accelerates autophagy-mediated neurons apoptosis through miR-3142/TRIB3 signaling axis in alzheimer’s disease. Brain Res 1785:147884 [DOI] [PubMed] [Google Scholar]
- Titze-de-Almeida R, Titze-de-Almeida SS (2018) miR-7 replacement therapy in Parkinson’s disease. Curr Gene Ther 18:143–153 [DOI] [PubMed] [Google Scholar]
- Ułamek-Kozioł M, Furmaga-Jabłońska W, Januszewski S, Brzozowska J, Ściślewska M, Jabłoński M, Pluta R (2013) Neuronal autophagy: self-eating or self-cannibalism in Alzheimer’s disease. Neurochem Res 38:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valera E, Spencer B, Mott J, Trejo M, Adame A, Mante M, Rockenstein E, Troncoso JC, Beach TG, Masliah E (2017) MicroRNA-101 modulates autophagy and oligodendroglial alpha-synuclein accumulation in multiple system atrophy. Front Mol Neurosci 10:329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H, Liu Y, Ma C, Huang L, Zhang L (2010) miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer’s disease targets TGF-β type II receptor. Brain Res 1357:166–174 [DOI] [PubMed] [Google Scholar]
- Wang J-Z, Xia Y-Y, Grundke-Iqbal I, Iqbal K (2013) Abnormal hyperphosphorylation of tau: sites, regulation, and molecular mechanism of neurofibrillary degeneration. J Alzheimer’s Dis 33:S123–S139 [DOI] [PubMed] [Google Scholar]
- Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, Wang H, Gong X, He X, Lu G (2016) MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to B im. Brain Pathol 26:167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang X, Guo Y, Rong H, Liu T (2017a) The long noncoding RNA HOTAIR promotes Parkinson’s disease by upregulating LRRK2 expression. Oncotarget 8:24449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Veremeyko T, Wong AH-K, El Fatimy R, Wei Z, Cai W, Krichevsky AM (2017b) Downregulation of miR-132/212 impairs S-nitrosylation balance and induces tau phosphorylation in Alzheimer’s disease. Neurobiol Aging 51:156–166 [DOI] [PubMed] [Google Scholar]
- Wang Z, Sun L, Jia K, Wang H, Wang X (2019) miR-9-5p modulates the progression of Parkinson’s disease by targeting SIRT1. Neurosci Lett 701:226–233 [DOI] [PubMed] [Google Scholar]
- Wang R, Yao J, Gong F, Chen S, He Y, Hu C, Li C (2021a) miR-29c-3p regulates TET2 expression and inhibits autophagy process in Parkinson’s disease models. Genes Cells 26:684–697 [DOI] [PubMed] [Google Scholar]
- Wang W, Lv R, Zhang J, Liu Y (2021b) circSAMD4A participates in the apoptosis and autophagy of dopaminergic neurons via the miR-29c-3p-mediated AMPK/mTOR pathway in Parkinson’s disease. Mol Med Rep 24:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Wen Z-J, Xu H-M, Zhang Y, Zhang Y-F (2022) Exosomal noncoding RNAs in central nervous system diseases: biological functions and potential clinical applications. Front Mol Neurosci 15:1004221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Zhang J, Tang P, Tu N, Wang K, Wu G (2018) Overexpression of miR-185 inhibits autophagy and apoptosis of dopaminergic neurons by regulating the AMPK/mTOR signaling pathway in Parkinson’s disease. Mol Med Rep 17:131–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Tan Z, Jia M, Zhou X, Wu K, Ding Y, Li W (2021) Long noncoding RNA SNHG1 knockdown ameliorates apoptosis, oxidative stress and inflammation in models of Parkinson’s disease by inhibiting the miR-125b-5p/MAPK1 Axis. Neuropsychiatr Dis Treat 17:1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S-P, Zhou F, Li J, Duan S-J (2019) NEAT1 regulates MPP+-induced neuronal injury by targeting miR-124 in neuroblastoma cells. Neurosci Lett 708:134340 [DOI] [PubMed] [Google Scholar]
- Xu X, Cui L, Zhong W, Cai Y (2020) Autophagy-associated lncRNAs: promising targets for neurological disease diagnosis and therapy. Neural Plast. 10.1155/2020/8881687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Patergnani S, Giorgi C, Suarez J, Goto K, Bononi A, Tanji M, Novelli F, Pastorino S, Xu R (2020) Asbestos induces mesothelial cell transformation via HMGB1-driven autophagy. Proc Natl Acad Sci 117:25543–25552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Yue Z (2014) Autophagy and its normal and pathogenic states in the brain. Annu Rev Neurosci 37:55–78 [DOI] [PubMed] [Google Scholar]
- Yan W, Chen Z-Y, Chen J-Q, Chen H-M (2018) LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem Biophys Res Commun 496:1019–1024 [DOI] [PubMed] [Google Scholar]
- Yao L, Zhu Z, Wu J, Zhang Y, Zhang H, Sun X, Qian C, Wang B, Xie L, Zhang S (2019) MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J 33:8648–8665 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zhang Y (2013) Desflurane accelerates neuronal cytotoxicity of Aβ by downregulating miR-214. Neurosci Lett 554:28–33 [DOI] [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee J-H, Mohan PS, Mercken M, Farmery MR, Tjernberg LO (2005) Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol 171:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li Q, Liu C, Gao S, Ping H, Wang J, Wang P (2016a) MiR-214-3p attenuates cognition defects via the inhibition of autophagy in SAMP8 mouse model of sporadic Alzheimer’s disease. Neurotoxicology 56:139–149 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu C, Wang J, Li Q, Ping H, Gao S, Wang P (2016b) MiR-299-5p regulates apoptosis through autophagy in neurons and ameliorates cognitive capacity in APPswe/PS1dE9 mice. Sci Rep 6:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Hong R, Chen W, Xu M, Wang L (2019) The role of long noncoding RNA in major human disease. Bioorg Chem 92:103214 [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang J, Liu Q, Xiao Z, Dai Q (2020a) Knockdown of long non-coding RNA AL049437 mitigates MPP+-induced neuronal injury in SH-SY5Y cells via the microRNA-205-5p/MAPK1 axis. Neurotoxicology 78:29–35 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xia Q, Lin J (2020b) LncRNA H19 attenuates apoptosis in MPTP-induced Parkinson’s disease through regulating miR-585-3p/PIK3R3. Neurochem Res 45:1700–1710 [DOI] [PubMed] [Google Scholar]
- Zhang L, Fang Y, Zhao X, Zheng Y, Ma Y, Li S, Huang Z, Li L (2021a) miR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol Therapy Nucleic Acids 24:822–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, He P, Bian Z (2021b) Long noncoding RNAs in neurodegenerative diseases: pathogenesis and potential implications as clinical biomarkers. Front Mol Neurosci 14:685143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Huang X-M, Liao J-X, Dong Y-K, Zhu J-L, He C-C, Huang J, Tang Y-W, Wu D, Tian J-Y (2021c) LncRNA HOTAIR promotes neuronal damage through facilitating NLRP3 mediated-pyroptosis activation in Parkinson’s disease via regulation of miR-326/ELAVL1 axis. Cell Mol Neurobiol 41:1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liang J, Chen N (2022) The potential role of miRNA-regulated autophagy in Alzheimer’s disease. Int J Mol Sci 23:7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XH, Wang YB, Yang J, Liu HQ, Wang LL (2019) MicroRNA-326 suppresses iNOS expression and promotes autophagy of dopaminergic neurons through the JNK signaling by targeting XBP1 in a mouse model of Parkinson’s disease. J Cell Biochem 120:14995–15006 [DOI] [PubMed] [Google Scholar]
- Zhao J, Geng L, Chen Y, Wu C (2020a) SNHG1 promotes MPP+-induced cytotoxicity by regulating PTEN/AKT/mTOR signaling pathway in SH-SY5Y cells via sponging miR-153-3p. Biol Res 53:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li H, Chang N (2020b) LncRNA HOTAIR promotes MPP+-induced neuronal injury in Parkinson’s disease by regulating the miR-874-5p/ATG10 axis. EXCLI J 19:1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Yang M, Li Q, Pei X, Zhu X (2020c) miR-132-5p regulates apoptosis and autophagy in MPTP model of Parkinson’s disease by targeting ULK1. NeuroReport 31:959–965 [DOI] [PubMed] [Google Scholar]
- Zhou B, Li L, Qiu X, Wu J, Xu L, Shao W (2020a) Long non-coding RNA ANRIL knockdown suppresses apoptosis and pro-inflammatory cytokines while enhancing neurite outgrowth via binding microRNA-125a in a cellular model of Alzheimer’s disease. Mol Med Rep 22:1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou J, Zhao Y, Li Z, Zhu M, Wang Z, Li Y, Xu T, Feng D, Zhang S, Tang F (2020b) miR-103a-3p regulates mitophagy in Parkinson’s disease through Parkin/Ambra1 signaling. Pharmacol Res 160:105197 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ge Y, Liu Q, Li Y-X, Chao X, Guan J-J, Diwu Y-C, Zhang Q (2021) LncRNA BACE1-AS promotes autophagy-mediated neuronal damage through the miR-214-3p/ATG5 signalling axis in Alzheimer’s disease. Neuroscience 455:52–64 [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhang H, Gao J, Xu Y (2021) Silencing of miR-497-5p inhibits cell apoptosis and promotes autophagy in Parkinson’s disease by upregulation of FGF2. Environ Toxicol 36:2302–2312 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.