Abstract
The E2F transcription factors are essential regulators of cell growth in multicellular organisms, controlling the expression of a number of genes whose products are involved in DNA replication and cell proliferation. In Saccharomyces cerevisiae, the MBF and SBF transcription complexes have functions similar to those of E2F proteins in higher eukaryotes, by regulating the timed expression of genes implicated in cell cycle progression and DNA synthesis. The CDC6 gene is a target for MBF and SBF-regulated transcription. S. cerevisiae Cdc6p induces the formation of the prereplication complex and is essential for initiation of DNA replication. Interestingly, the Cdc6p homolog in Schizosaccharomyces pombe, Cdc18p, is regulated by DSC1, the S. pombe homolog of MBF. By cloning the promoter for the human homolog of Cdc6p and Cdc18p, we demonstrate here that the cell cycle-regulated transcription of this gene is dependent on E2F. In vivo footprinting data demonstrate that the identified E2F sites are occupied in resting cells and in exponentially growing cells, suggesting that E2F is responsible for downregulating the promoter in early phases of the cell cycle and the subsequent upregulation when cells enter S phase. Our data also demonstrate that the human CDC6 protein (hCDC6) is essential and limiting for DNA synthesis, since microinjection of an anti-CDC6 rabbit antiserum blocks DNA synthesis and CDC6 cooperates with cyclin E to induce entry into S phase in cotransfection experiments. Furthermore, E2F is sufficient to induce expression of the endogenous CDC6 gene even in the absence of de novo protein synthesis. In conclusion, our results provide a direct link between regulated progression through G1 controlled by the pRB pathway and the expression of proteins essential for the initiation of DNA replication.
Although E2F was originally defined as a factor that binds specifically to an element in the adenovirus E2 promoter (42), it is now evident that E2F is essential for coordinating transcription during the mammalian cell cycle (for reviews, see references 12 and 72). A number of genes are found to be regulated by E2F, particularly during the transition from G1 to S phase. To date, six members of the E2F family are known: E2F-1 through E2F-5 and the recently identified E2F-6 (11). Furthermore, two heterodimerization partners of the E2Fs, DP-1 and DP-2, have been isolated. The E2F transcription factors appear to be key downstream targets for the retinoblastoma protein pRB and two pRB-related proteins, p107 and p130 (reviewed in references 12 and 70). Binding of pRB family members (also called pocket proteins) to the E2F transcription factors results in transcriptional repression of E2F-regulated genes. Phosphorylation of the pocket proteins by cyclin-dependent kinases releases the pocket proteins from E2F, leading to derepression and/or activation of E2F-dependent genes and subsequent entry into S phase. The demonstration that deregulated E2F activity is sufficient to induce S phase in quiescent cells has provided a model for how the inactivation of the pRB pathway in human tumors leads to the development of cancer (70).
The majority of E2F-regulated genes encode proteins that are involved in DNA replication and in cell cycle progression (27, 72). These genes include DNA polymerase α (60), thymidine kinase (18), HsOrc1 (58), dihydrofolate reductase (7, 50, 71), cyclin A (35, 67), cyclin E (9, 22, 59), p107 (82), B-Myb (44), CDC2 (14, 75), E2F-1 (33, 38, 56), and E2F-2 (68). At present it is not known how different E2F-DP complexes coordinate the expression of these genes. It is also relatively unclear which gene is regulated by which heterodimer in which phase of the cell cycle. However, several data suggest that during different phases of the cell cycle, specific subsets of E2F and DP are functionally present. Importantly, the E2F family members can be divided into two subgroups based on their ability to bind pocket proteins, transactivation capacity, structure, and expression during the cell cycle (for reviews, see references 2 and 72). E2F-1, -2, and -3 (one subgroup) are potent transactivators and bind preferentially to pRB (29, 45), whereas E2F-4 and -5 (the other subgroup) are weak transactivators which are able to bind all pocket proteins (3, 23, 31, 37, 51, 66). E2F-1, -2, and -3 are expressed mainly in late G1 and early S phase, while the expression of E2F-4 is constant during the cell cycle. Recently, it was reported that the members of the two subgroups can also be distinguished functionally: whereas E2F-1, -2, and -3 are capable of inducing S phase in quiescent fibroblasts, E2F-4 and E2F-5 are not (49). The functional difference between the two subgroups is a result of different subcellular localization for the E2Fs, which appears to regulate the activity of E2F-4 and E2F-5, but not the other E2Fs that are found in the nucleus when expressed (48, 53).
Although E2Fs have been reported to regulate several genes whose products participate in DNA replication (see above), it is clear that many of these genes are only marginally upregulated by deregulated E2F expression (15, 36, 76). Moreover, since most of the proteins participating in DNA replication are very stable, it is not clear why the transcription of these genes needs to be cell cycle regulated. None of the known gene products alone is able to induce S phase, suggesting that combinations of two or more products are needed or that the responsible and limiting targets which can regulate the initiation of DNA replication have not yet been identified.
In eukaryotes, the initiation of DNA replication is a highly regulated process which implicates a large number of proteins. These proteins are assembled at the origins of replication and form the prereplication complex (for reviews, see references 8, 16, 55, and 73). Most of our knowledge regarding the regulation of eukaryotic DNA replication stems from the budding yeast Saccharomyces cerevisiae, but a substantial amount of data has also been generated for Xenopus laevis and the fission yeast Schizosaccharomyces pombe as experimental systems. In S. cerevisiae, the origin of replication is bound by a set of proteins called the origin recognition complex (ORC). ORC is bound to the origin throughout the cell cycle and consists of six polypeptides (5) that are required for DNA replication and cell division. The prereplication complex is formed at the end of mitosis, and the complex formation is initiated by the association of Cdc6p with ORC (47), followed by the binding of another set of six related proteins, Mcm2 to Mcm7 (17, 74). After the complex formation, it is thought that components in the complex are phosphorylated by the S-phase CDKs (Clb5p or Clb6p in association with Cdc28p) and the Dbf4p/Cdc7p kinase, which leads to initiation of DNA replication (73).
Recently, it was shown that the binding of Mcm proteins onto the origin is dependent on Cdc6p binding to ORC (17, 74). Although ORC remains stably attached to the origins of replication during other phases of the cell cycle, Cdc6p and Mcm proteins do not. When Cdc6p is overexpressed, it can bind to ORC throughout the cell cycle; however, the binding of Mcms to the ORC-Cdc6p complex during G2 and M phase is inhibited, most likely due to inhibitory effects from S-CDK protein kinases (74). This strongly suggests that Cdc6p plays a central and limiting role in the onset of DNA replication in S. cerevisiae. Cdc6p is a protein with a sequence similar to that of the large subunit of ORC, and it has a very short half-life, a number of consensus sites for the cyclin-dependent kinases, and an ATP-binding domain (19, 63, 83). The binding of Cdc6p to ORC in G1 requires de novo synthesis of the protein (13, 62, 63, 65). Similar findings were reported for Cdc18p, the S. pombe homolog of Cdc6p (40, 54, 57). S. cerevisiae Cdc6p is synthesized during the cell cycle in two peaks: in late mitosis, after anaphase where its expression is dependent on Swi5, and in late G1. This second peak is regulated by MBF-SBF and is sufficient to promote DNA replication.
Interestingly, the expression of cdc18+ is regulated by the transcription factor complex DSC1, the MBF homolog in S. pombe (41). Although the E2F transcription factors are not structural homologs of the transcription factors associated with the yeast MBF complex, it is believed that E2F is the functional homolog of MBF and DSC1 in mammalian cells. Thus, since the cell cycle-regulated expression of the yeast CDC6 and cdc18+ genes is dependent on MBF and DSC1, respectively, we reasoned that the expression of the mammalian homolog of CDC6 would be controlled by E2Fs. Therefore, we isolated cDNAs encoding the human and mouse homologs of Cdc6p or Cdc18p and the 5′ regulatory region of human CDC6. During the course of this work, a human cDNA for CDC6 and a partial 5′ regulatory region was published by the laboratory of Bruce Stillman (79). Our data suggest that hCDC6 (human CDC6) protein is limiting and essential for entry into the S phase of the mammalian cell cycle and that the cell cycle-regulated expression of mammalian CDC6 is E2F dependent.
MATERIALS AND METHODS
Isolation of human CDC6, mouse CDC6, and the 5′ regulatory region of hCDC6.
A search of the human expressed sequence tag (EST) database using the S. cerevisiae Cdc6p and the S. pombe Cdc18p amino acid sequences revealed several EST sequences whose translation products showed homology to Cdc6p and Cdc18p. Primers were generated to one of these clones (EST identification no. 376630), originally isolated from a Soares fetal heart NbHH19W Homo sapiens cDNA library, and PCR was used to generate a 414-bp fragment of the EST clone using a human cDNA library prepared from fetal brain (61). The PCR fragment was radiolabeled, and a human embryonic fibroblast library M426 (kindly provided by P. P. Di Fiore) was screened. One clone containing an insert of 3.0 kb was sequenced on both strands and was shown to contain an open reading frame of 560 amino acids with extended homology to Cdc6p and Cdc18p. This clone was named hCDC18 but later renamed hCDC6 in agreement with Williams et al. (79) who identified a cDNA containing an identical open reading frame.
A cDNA encoding mouse CDC6 was identified in the EST database (Image Consortium clone identification no. 477516 [46]) by searching the database with the amino acid sequence of hCDC6. This clone was obtained from Research Genetic (Huntville, Ala.) and was found to contain an insert of 2,608 bp. The cDNA was sequenced completely on both DNA strands and contained an open reading frame of 589 amino acids. Mouse CDC6 is 80% identical to human CDC6 and contains a 27-amino-acid amino-terminal extension.
The 355-bp fragment of the human promoter (79) was generated by PCR, using human genomic DNA as a template. This PCR product was used to screen a human genomic library prepared in Lambda FIX II (Stratagene). From 5 × 105 PFU, three positive phages were purified that contained three different sized inserts: 12.8, 12.9, and 16.8 kb. The results of Southern blot analysis (data not shown) identified three different SacI fragments of 1.8, 5.5, and 7.0 kb that all contained the 355-bp region. The 1.8-kb fragment was subcloned into pBSK− (Stratagene), and both strands were completely sequenced by using the Vistra DNA Sequencer 725 (Amersham).
Plasmids.
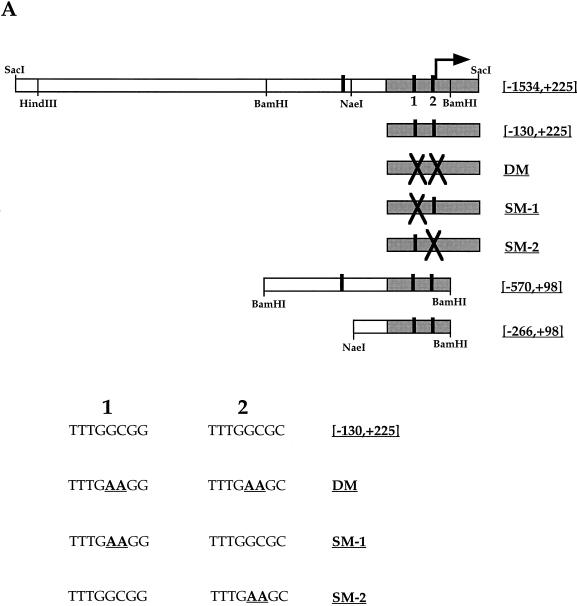
For p[−130,+225], the 355-bp PCR product described above was cloned into the NheI and HindIII sites of pGL3-Basic (Promega), a vector that contains a gene encoding the luciferase protein downstream of a multiple cloning site and which lacks any promoter or enhancer elements. Site-directed mutagenesis on p[−130,+225] was performed with the Chameleon double-stranded, site-directed mutagenesis kit (Stratagene) in accordance with the instructions of the manufacturer to produce pGL3-DM (double mutant, two E2F sites mutated, bp −43 to −36 and bp −8 to −1), pGL3-SM1 (single mutant 1, first E2F site mutated, bp −43 to −36), and pGL3-SM2 (single mutant 2, second E2F site mutated, bp −8 to −1). Except for the introduced mutations, the inserts of these three plasmids are identical to the one in p[−130,+225], as was confirmed by DNA sequencing. The primers used for the site-directed mutagenesis were 5′-GAGGCCGGGCTTTGAAGGGAGGTGGGAACG-3′ and 5′-CCATTCGGATTTGAAGCGAGCGC GGCTGG-3′ for the E2F sites and 5′-CGTGTAATTCTAGCGTCGGGGCGGCCG-3′ for the unique XbaI site in pGL3-Basic. Mutated nucleotides are underlined.
p[−1534,+225] was generated by cloning the 1.8-kb SacI fragment from the Lambda Fix phage DNA into the SacI site of pGL3-Basic. p[−570,+98] was generated by subcloning the internal BamHI/BamHI fragment from p[−1534,+225] into the BglII site of pGL3-Basic. p[−266,+98] was generated by subcloning the internal NaeI/BamHI fragment into the SmaI and BglII sites of pGL3-Basic.
pCMVHAhCDC6 was constructed by PCR amplification of the full-length open reading frame of hCDC6, and the PCR product was subsequently cloned into the BamHI site of pCMVHA (49). The entire insert was sequenced, and no mutations were introduced by PCR. pCMV-βgal was obtained from Clontech. pCMV-CD20, pCMV-E2F-1, pCMV-DP-1, and pRcCMVcyclin E have previously been described (28, 30, 32, 81). pCMV-ER-E2F-1 and pCMV-ER-E2F-1-E132, which were used to generate the Rat-1 cells, stably expressing the wild-type and a DNA-binding mutant of E2F-1 linked to the ligand-binding domain of the estrogen receptor (ER) protein, respectively, will be described elsewhere (76).
Cell culture and transfections.
U2-OS, T98G, MCF7, and Rat-1 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% South American (Gibco) bovine calf serum (BCS). Mouse Swiss 3T3 and NIH 3T3 cells were maintained in DMEM supplemented with 10% Coloradan BCS (Gibco).
Transfections were performed by the calcium phosphate method (24). NIH 3T3 cells were transiently transfected with 1.0 μg of one of the indicated human CDC6 promoter-luciferase constructs and 0.5 μg of pCMV-βgal to control for transfection efficiency. Sheared salmon sperm DNA was added to 17 μg per 10-cm-diameter dish as carrier. U2-OS cells were transiently transfected with the same amounts of promoter and βgal plasmids. pCMV-E2F-1 or pCMV-DP-1 (10 ng) or both were cotransfected for transactivation experiments to ascertain whether the different human CDC6 promoter constructs responded to E2F-1 or DP-1.
For synchronization of MCF7 cells for in vivo footprinting, fluorescence-activated cell sorting (FACS), and Northern blotting experiments, cells were starved for 36 h in DMEM lacking isoleucine and BCS. Subsequently, cells were harvested directly (0 h) or stimulated for 12, 16, and 20 h with normal medium supplemented with 10% BCS. For a G1/S block, cells were kept in DMEM–10% BCS supplemented with 1 mM hydroxyurea for 20 h and subsequently harvested.
For experiments concerning the direct regulation of hCDC6 mRNA expression by E2F, the Rat-1 cells, expressing an ER-E2F wild-type fusion and an ER-E2F-E132 DNA-binding mutant fusion, were starved for 48 h in medium containing 0.1% BCS and subsequently stimulated in the following medium: (i) fresh medium containing 10% BCS, (ii) the same starvation medium to which 1 μM 4-hydroxytamoxifen (OHT) was added, (iii) the same starvation medium to which 10 μg of cycloheximide (CHX) per ml was added, or (iv) the same starvation medium with OHT and CHX. Cells were stimulated for the times (hours) indicated in the figures and subsequently used either for reverse transcriptase PCR (RT-PCR) or FACS.
For the induction of S phase by cyclin E and hCDC6, U2-OS cells were transiently transfected with 10 μg of pCMV empty vector, 10 μg of pRcCMVcyclinE, 10 μg of pCMVHAhCDC6, or 10 μg of both plasmids. In all cases, 1 μg of pCMV-CD20 was cotransfected and amounts were adjusted to 21 μg per 10-cm-diameter dish with sheared salmon sperm DNA.
Northern blotting.
Total cellular RNA was purified from human and mouse cells described in the figure legends by using the Nonidet P-40 lysis method (64). RNA (20 to 30 μg) was electrophoresed per lane through a 1% formaldehyde agarose gel, transferred to nitrocellulose, and probed with 32P-labeled partial human or mouse CDC6 or mouse E2F-1 cDNA. Equal loading was ensured by ethidium bromide staining and/or by probing the same blot with a 32P-labeled rat glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe.
RT-PCR.
RNA from the Rat-1 cells stably expressing the wild-type ER-E2F-1 and the mutant ER-E2F-1-E132 was prepared by the guanidium thiocynate-acid phenol method (64). RNA preparations were subsequently treated with DNase, incubated for 20 min at 37°C, and phenol extracted. RNA (1 μg) was used for cDNA preparation with RT (Superscript II; Gibco) according to the instructions provided by the manufacturer. Obtained cDNA was subsequently used in a PCR using Taq polymerase (Gibco) and 32P-labeled dCTP. To be in a linear range for measuring the level of rat CDC6 GAPDH mRNA, the following PCR conditions were used. First 10 cycles were used; these cycles had 0.5°C decreases in annealing temperature from 54 to 49°C. After this, another 13 cycles were performed at 49°C.
Gel retardation assay.
Electrophoresis mobility shift assays (EMSA) were performed as described previously (29). To obtain a region of the human CDC6 promoter that harbored both E2F sites, the following oligonucleotides were used in a PCR: 5′-AATCGAGGCCGGGCTTTG-3′ and 5′-GCGGCAGCAGCAAACTCCAG-3′, giving rise to a fragment of 85 bp (bp −57 to +28). PCR was performed on p[−130,+225] and pGL3-DM as templates, and the products were end labeled with [γ-32P]ATP and analyzed for protein-binding activities. Excess (50-fold) nonlabeled wild type and mutant probes (DM fragment) were used to determine the specificity of E2F binding. MRC5 nuclear extracts were prepared as previously described (1). The following antisera were used for shifting E2F-DP-containing complexes: Monoclonal antibodies TFD10 (anti-DP1) (80), TFE41 (anti-E2F4) (53), XZ77 and 21C9 (both anti-retinoblastoma gene product pRB) (34), SD9 (anti-p107) (20), and as a negative control, M1 (anti-E1A) (26). Polyclonal antisera were anti-E2F4 (sc-866), anti-p107 (sc-318), anti-p130 (sc-317), and as a negative control, anti-farnesyl transferase (sc-137). All polyclonal sera were obtained from Santa Cruz Biotechnology.
Luciferase and β-Gal assays.
U2-OS and NIH 3T3 cells were transiently transfected in duplicate and triplicate, respectively. One dish of the NIH 3T3 transfectants or cells that were starved and then with serum stimulated was used for FACS analysis (see below). Forty-eight hours posttransfection, cells were harvested by scraping in phosphate-buffered saline (PBS) and pelleted by centrifugation. Cell pellets were subsequently resuspended in 100 μl of 100 mM Tris-HCl, pH 7.8, and cells were lysed by three freeze-thaw steps. Cell debris was spun down, and supernatant was transferred to a new tube and used for both luciferase and β-galactosidase (β-Gal) assays. Luciferase activity was typically measured by mixing 20 μl of lysate with 30 μl of 100 mM Tris-HCl [pH 7.8] and 50 μl of luciferase mix. The luciferase mix is 26 μl of luciferase buffer, 24 μl of phosphate buffer, 0.15 μl of 1 M dithiothreitol, and 1.5 μl of 0.2 M ATP. Luciferase buffer is 25 mM glycyl glycine [pH 7.8], 15 mM MgSO4, and 4 mM EGTA, and phosphate buffer is 100 mM KH2PO4/K2HPO4, pH 7.8. Subsequently, 50 μl of a 10 mM luciferin solution (in luciferase buffer) was added and activity was determined in an Anthos Lucy 1 luminometer (Labtech).
β-Gal activity was measured by mixing 2 to 20 μl of lysate with Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) to a total volume of 200 μl. To start the reaction, 40 μl of ONPG substrate (4 mg of o-nitrophenyl-β-d-galactopyranoside [Sigma] per ml of 100 mM phosphate buffer [pH 7.0]) was added and incubated at 30°C for 30 to 120 min. The reaction was stopped by adding 100 μl of 1 M Na2CO3, and the absorbance was read at 420 nm in a MRX plate reader (Dynatech Laboratories). All experiments involving luciferase and β-Gal activity were carried out at least three times.
FACS analysis.
Cells from one 10-cm-diameter dish were washed once with PBS, trypsinized, spun, and washed once more with PBS, while samples from the elutriation fractions were spun and washed once in PBS. Cells were subsequently fixed in 70% methanol and either stored at 4°C or directly used for propidium iodine (PI) staining. For PI staining, cells were washed once in PBS supplemented with 1% BCS, spun, and subsequently resuspended in 500 ml of PI buffer (10 mM Tris-HCl [pH 7.0], 5 mM MgCl2, 50 μg of PI [Sigma] per ml, 100 μg of RNase A per ml). After 30 min of incubation at 37°C, the samples were analyzed with a Becton Dickinson FACScan. CD20-positive cells were selected and treated as described previously (53).
In vivo DMS genomic footprinting.
In vivo dimethyl sulfate (DMS) genomic footprinting with the ligation-mediated PCR amplification (LMPCR) procedure was performed essentially as previously described (25, 52) with the following modifications: 2 × 106 MCF7 cells per 14-cm-diameter dish that were either incubated with hydroxyurea or that were exponentially growing or serum starved and then stimulated, were treated with the guanosine methylating agent DMS at 0.2% for 5 min at room temperature in their respective culture medium (DMEM with 0 or 10% BCS) buffered with HEPES (pH 7.4) (20 mM final concentration). After DMS treatment, cells were washed three times with cold PBS containing 2% β-mercaptoethanol and then collected in 1 ml of lysis buffer (50 mM Tris [pH 8.0], 20 mM EDTA, 1% sodium dodecyl sulfate, 2% β-mercaptoethanol). Genomic DNA was isolated by three gentle extractions with phenol (pH 8.0) followed by two precipitations in 4 M ammonium acetate with 3 volumes of ethanol. DNA was then redissolved in 1 ml of water. As a reference, MCF7 naked genomic DNA (1 mg/ml in water) was methylated in vitro with 0.5% DMS for 4 min at room temperature. Piperidine cleavage at methylated bases was performed in 1 N piperidine at 95°C for 30 min. Chemically cleaved samples were precipitated in ethanol, evaporated twice, and finally resuspended to 0.4 mg/ml in water. Portions (2 μg) of these samples were used for LMPCR.
The following primers were designed to analyze the upper strand (plus strand) and lower strand (minus strand) of the transcription start site region of the hCDC6 promoter: plus strands, S1 (5′-CTCTTCCACTGGATTGGTAGC-3′), S2 (5′-AAAGAGGCGGTGCCCAAGGCG-3′), and S3 (5′-AGAGGCGGTGCCCAAGGCGAAAGCTC-3′); for minus strands, AS1 (5′-AAGGTGGAGGAGTCACGTCC-3′), AS2 (5′-TCTGCGTCGGAGAGCCTGAGTGG-3′), and AS3 (5′-CGTCGGAGAGCCTGAGTGGTGGTGTTCGGGG-3′). LMPCRs were performed as follows: 2 μg of cleaved genomic DNA and 0.3 pmol of S1 or AS1 were denatured at 95°C for 5 min and then annealed for 30 min at 50°C in 25 μl of first-strand buffer (10 mM Tris-HCl [pH 8.9], 5 mM MgSO4, 40 mM NaCl, 20 mg of bovine serum albumin [BSA] per ml, 10% dimethyl sulfoxide [DMSO]). Vent exonuclease-negative DNA polymerase (0.5 U) (New England Biolabs) and deoxynucleoside triphosphates (dNTPs) (final concentration of each dNTP, 0.25 mM) (pH 7.0) were added to the annealed DNA mixture; elongation was performed for 15 min at 76°C. Annealed linkers (25 pmol) LINK1 and LINK2 (plus-strand LINK1, 5′-GAATTCAGATC-3′; minus-strand LINK2, 5′-GCGGTGACCCGAGAGATCTGAATTC-3′) in 45 μl of ligation mix (50 mM Tris-HCl [pH 7.5], 14 mM MgCl2, 33 mM dithiothreitol, 1.6 mM ATP [pH 7.0], 100 mg of BSA per ml, 4 U of T4 DNA ligase [Boehringer]) were then added to the elongation reaction; ligation was performed for 1.5 h at room temperature. Ligated DNA was then precipitated in ethanol, washed in 70% ethanol, and resuspended in 100 μl of amplification buffer (65 mM Tris-HCl [pH 8.8], 40 mM NaCl, 10 mM β-mercaptoethanol, 3 mM MgCl2, 50 mg of BSA per ml, 10% DMSO, 0.4 mM dNTPs). PCR amplification was then started after addition of 10 pmol of S2 or AS2 primer and 1 U of Taq DNA polymerase (Perkin Elmer). PCR was performed as follows: 1 cycle of 4 min at 95°C followed by 30 cycles consisting of 40 s at 95°C, 2 min at 62°C, and 3 min at 76°C, followed by 7 min at 76°C; this procedure was then repeated. From this amplification reaction, 15 μl were collected and mixed with 5 μl of labeling buffer (65 mM Tris-HCl [pH 8.8], 40 mM NaCl, 10 mM β-mercaptoethanol, 2 mM MgCl2, 3.75 mM dNTPs, 10% DMSO, 0.5 U of Vent exonuclease-negative DNA polymerase) containing 0.15 pmol of radioactively labeled S3 or AS3 primer (primers were 5′ end labeled with T4 polynucleotide kinase (New England Biolabs) and [μ-32P]ATP. The specific activity of the labeled primers was 3 × 106 cpm/pmol. PCR was performed as follows: 2 min at 95°C, followed by nine cycles consisting of 40 s at 95°C, 3 min at 66°C, and 5 min at 76°C. Eighty microliters of 0.3 M sodium acetate (pH 5.5) and 10 μg of tRNA were then added to stop the reaction. Labeled DNA was phenol extracted and ethanol precipitated. The pellet was washed with 70% ethanol, resuspended in 8 μl of sample loading buffer (95% formamide, 10 mM EDTA, 20 mM NaOH, 0.025% bromophenol blue, 0.025% xylene cyanol), and denatured for 3 min at 95°C. Two microliters of the sample was loaded onto a 5% sequencing gel and run at 50 W. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics Inc.).
Elutriation.
A culture of 108 asynchronously growing NIH 3T3 cells was harvested after three washes in PBS by trypsinization, spun, and resuspended in PBS containing 0.3 mM EDTA, 1 mg of glucose per ml, and 1% BCS. Subsequently, cells were separated according to cell cycle position by counter flow elutriation with Beckman equipment. Fourteen cell-containing fractions were collected and set on ice. From these fractions, samples were taken separately for FACS analysis, while the remaining cells were used for the purification of total RNA by the Nonidet P-40 lysis method. RNA from 10 fractions was used for RT-PCR analysis.
Microinjection and immunostaining.
Human glioblastoma T98G cells, attached to coverslips, were starved for 48 h in DMEM–0.1% BCS and subsequently stimulated with DMEM–10% BCS supplemented with 100 nM bromodeoxyuridine (BrdU) (Sigma). After 16 h, approximately 200 cells per coverslip were injected with either affinity-purified rabbit immunoglobulin G (IgG) as a negative control or with affinity-purified L20 (an anti-hCDC6 rabbit polyclonal antiserum) (61) in concentrations of 0.8 μg/μl in PBS. After the cells were injected, coverslips were put back in the same medium. Two hours after microinjection, cells were washed twice in PBS, fixed for 10 min in 4% formaldehyde, and subsequently treated for 10 min with Triton X-100 and for 30 s in 50 mM NaOH. Incorporated BrdU was immunostained with a mouse anti-BrdU monoclonal antibody (Becton Dickinson) and visualized with a goat anti-mouse CY3-conjugated secondary antibody. Injected cells were identified by incubation with a fluorescein isothiocyanate (FITC)-conjugated secondary goat anti-rabbit antibody recognizing the injected antibodies. Finally, cells were treated with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma), rinsed in PBS and water, and mounted before analysis with a fluorescence microscope. Noninjected cells were stimulated with DMEM–10% BCS containing 100 nM BrdU, fixed after 0, 14, 16, and 18 h, and subsequently stained for BrdU incorporation as described above.
Nucleotide sequence accession numbers.
The cDNA sequence of mouse CDC6 has been submitted to the DDBJ/EMBL/GenBank databases under accession no. AJ009559. The sequence of the 1.8-kb human CDC6 promoter has been submitted to the DDBJ/EMBL/GenBank databases under accession no. AJ009560.
RESULTS
Mammalian CDC6 is cell cycle regulated.
In order to find a human homolog of S. cerevisiae Cdc6p and S. pombe Cdc18p, a search in the EST database was performed with the amino acid sequences of the yeast proteins. One human cDNA was identified (EST identification no. 376630) that, when translated, shows homology to both Cdc6p and Cdc18p. This EST was different from human ORC1 which also has homology to Cdc6p and Cdc18p (4, 21). The cDNA coding for the homologous region was used to screen a human embryonal fibroblast library to obtain a potentially full-length cDNA. A clone containing a 3.0-kb insert was sequenced and shown to contain an open reading frame of 560 amino acids. This clone was named hCDC6 after a cDNA containing an identical open reading frame was described (79).
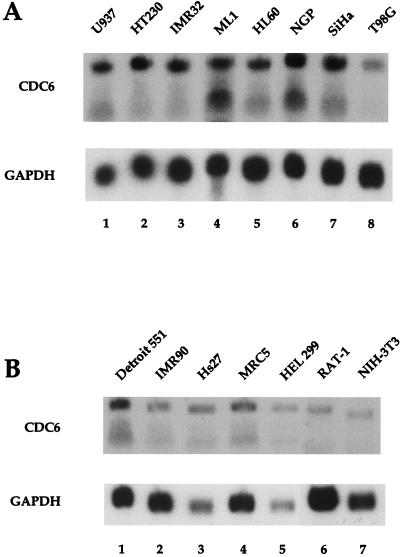
To investigate CDC6 mRNA expression, RNA was prepared from a number of human, rat, and mouse cell lines. Figure 1A shows the CDC6 mRNA expression in a number of established human tumor cell lines, while Fig. 1B shows the expression in a number of fibroblast cell lines. Two CDC6 mRNA signals (approximately 3.3 and 2.5 kb) were detected in most cell lines tested, which might be due to alternative splicing of the mRNA. The slower-migrating mRNA appeared more abundant.
FIG. 1.
Expression of endogenous human, rat, and mouse CDC6 mRNA. (A) Northern blot containing total RNA from asynchronously growing human tumor cells U937 (histiocytic lymphoma), HT230 (colon adenocarcinoma), IMR32 (neuroblastoma), ML1 (premyeloid leukemia), HL60 (promyeloid leukemia), NGP (neuroblastoma), SiHa (cervix carcinoma), and T98G (glioblastoma). The blot was probed with a partial human CDC6 cDNA probe. (B) Northern blot containing total RNA from the human fibroblast cell lines Detroit 551 (embryonic skin), IMR90 (fetal lung), Hs27 (newborn foreskin), and HEL 299 (embryonal lung), the rat cell line Rat-1, and the mouse cell line NIH 3T3. Hybridization was done with a human CDC6 cDNA probe (lanes 1 to 5) and with a mouse CDC6 cDNA probe (lanes 6 and 7). Blots were subsequently probed with a partial rat GAPDH probe, as depicted (lower blots).
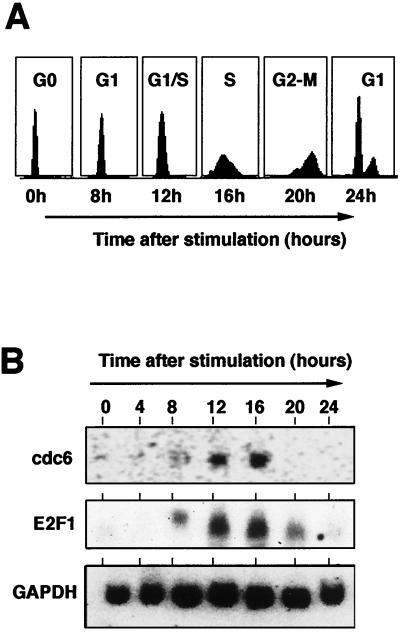
To investigate whether CDC6 expression is cell cycle regulated in immortalized murine fibroblasts, Swiss 3T3 cells were serum starved and subsequently induced to reenter the cell cycle by addition of serum. Figure 2A shows the FACS analysis of these cells that were serum starved and then stimulated, indicating that Swiss 3T3 fibroblasts reach S phase after 16 h of stimulation in a highly synchronous manner. A cDNA encoding mouse CDC6 (mCDC6) was identified in the EST database (EST identification no. 477516) by searching the database with the amino acid sequence of hCDC6. This cDNA was sequenced completely and shown to contain the complete open reading frame of mouse CDC6. The mouse protein is 80% identical to the human protein and contains a 27-amino-acid extension at the amino terminus (data not shown). A Northern blot containing total RNA from Swiss 3T3 fibroblasts, treated as described above for the FACS analysis, was probed with a radiolabeled mouse CDC6 cDNA. Figure 2B shows that the expression of CDC6 is cell cycle regulated and that the mRNA for CDC6, like E2F-1, is upregulated in Swiss 3T3 cells in mid-to-late G1. Interestingly, the CDC6 mRNA is absent in G2/M and early G1, suggesting that the expression of CDC6 is cell cycle regulated in proliferating cells as well as in cells entering the cell cycle from quiescence (discussed below). A similar expression pattern of CDC6 was observed in primary human MRC5 fibroblasts and in NIH 3T3 cells, although the downregulation of the CDC6 mRNA in G2 and early G1 was not as pronounced in these two cell lines (data not shown).
FIG. 2.
Cell cycle-regulated expression of CDC6 mRNA. (A) Swiss 3T3 cells were starved in low-serum-containing medium, subsequently stimulated with high serum, and used for FACS analysis. (B) Total RNA was isolated from Swiss 3T3 cells treated similarly and used for Northern blotting. The blot was probed first for mouse CDC6 and E2F-1 mRNAs (upper blots). Subsequently the blot was probed for GAPDH expression (lower blot).
Cell cycle expression of CDC6 in proliferating cells.
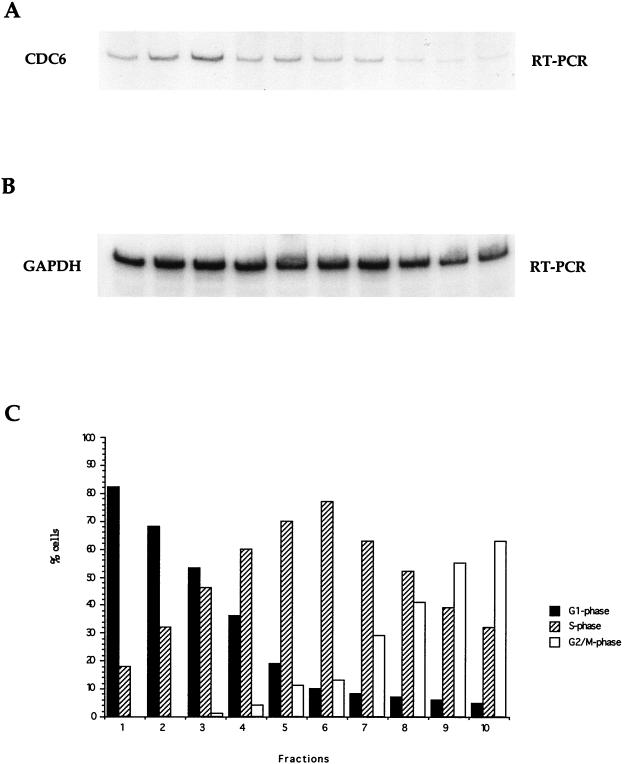
S. cerevisiae CDC6 and S. pombe cdc18+ are regulated differently during a normal cell cycle. Data obtained from synchronous populations of elutriated cells have shown that both CDC6 and cdc18+ transcription is periodic in proliferating cells; however, whereas CDC6 transcription is induced twice during the cell cycle (in M and late G1 [63]), the cdc18+ expression is induced only once (in late G1 [40]). To investigate how mammalian CDC6 is regulated in proliferating cells, exponentially growing NIH 3T3 cells were elutriated, and fractions of cells were taken for determining the cell cycle profile (Fig. 3C) and for RNA preparations. The expression of CDC6 was analyzed by RT-PCR on the obtained RNA, using primers specific for mouse CDC6 (Fig. 3A) or mouse GAPDH (Fig. 3B). The PCRs were performed under linear conditions in order to compare mRNA levels. The data shown in Fig. 3A suggest that the expression of CDC6 peaks in G1 and that the expression of CDC6 is downregulated during S and G2/M, which is in agreement with our findings shown in Fig. 2B. CDC6 mRNA in these experiments is nevertheless detectable in all analyzed fractions. This fact can most likely be attributed to the lower degree of synchrony in the NIH 3T3 cells in G2/M in the elutriation procedures compared to that in the Swiss 3T3 cells after serum starvation and stimulation. In conclusion, our data therefore suggest that the CDC6 expression is cell cycle regulated in proliferating cells as well as in cells entering the cell cycle from a state of quiescence.
FIG. 3.
Expression of CDC6 during a proliferating cell cycle. Asynchronously growing NIH 3T3 cells were fractionated by elutriation. Ten fractions are shown. (A and B) Total RNA from these elutriated cells was used for RT-PCR with sets of specific primers which amplified the cDNA obtained from mouse CDC6 mRNA (A) and mouse GAPDH mRNA (B). (C) The cell cycle distribution of cells from 10 fractions was determined by FACS.
E2F transactivates the human CDC6 promoter.
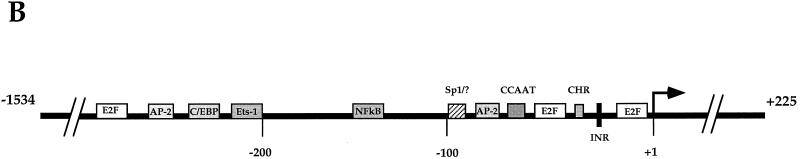
In an attempt to find the regulatory elements controlling the expression of human CDC6, the 355-bp promoter fragment of the reported gene (79) was used to screen a human genomic library. Three phages containing sequences upstream from the 355-bp promoter fragment, ranging in size from 1.8 to 7.0 kb were purified. The 1.8-kb fragment was cloned and sequenced completely (Fig. 4A). The 1,759-bp 5′ regulatory region contains three putative E2F DNA-binding sites upstream of the previously identified transcription start site, depicted in bold (bp +1). Computer analysis of the 1,759-bp sequence identified several additional potential transcription factor-binding sites (Fig. 4B). The two 3′ most E2F sites match properly with the consensus sequence TTT(G/C)(G/C)CG(G/C), while the 5′ most site does not.
FIG. 4.
Sequence and schematic representation of the human CDC6 promoter. (A) Nucleotide sequence of the 1,759-bp CDC6 promoter. The previously reported 355-bp fragment is underlined (79). Three putative E2F DNA-binding sites are boxed. A putative CHR region (bp −30 to −26) and the putative INR region (bp −16 to −11) are shown in lowercase. Restriction sites used for cloning are italic, and the transcription start site (bp +1) is bold. (B) Schematic representation of transcription factor-binding sites in the large 1,759-bp human CDC6 promoter. The transcription start site is depicted with an arrow. A CHR site, a CCAAT box, and a putative Sp1 element (Sp1/?) which are found to be protected in in vivo footprint assays, are located next to the two 3′ E2F sites close to the start site. Positions of consensus binding sites for AP-2, C/EBP, Ets-1, and NF-κB are also given. Putative recognition sites upstream of bp −325 and downstream of the start site are not shown. INR, initiator region.
Several deletion and point mutants were generated in order to study the roles of the E2F transcription factors in the regulation of mammalian CDC6 expression. These mutants are shown in Fig. 5A. The wild-type fragments, named [−1534,+225] (1,759 bp) and [−130,+225] (355 bp) were cloned upstream of the luciferase gene. In addition, three mutants harboring point mutations in the two E2F sites present in the 355-bp fragment were generated: SM1 carries a double-nucleotide mutation in the first E2F site, SM2 carries a double-nucleotide mutation in the second E2F site, while the double mutant (DM) harbors these mutations in both sites (sequences at the bottom of Fig. 5A). Finally, two deletion mutants were generated in order to measure the effect of the third (5′ most) E2F site: (i) [−570,+98], the internal BamHI/BamHI fragment (668 bp) from the 1,759-bp promoter, therefore carrying all three putative sites, and (ii) [−266,+98], the internal NaeI/BamHI fragment (364 bp), which harbors the two downstream E2F sites but not the upstream site. All mutant fragments, like the wild-type fragments, were cloned upstream of the luciferase gene.
FIG. 5.
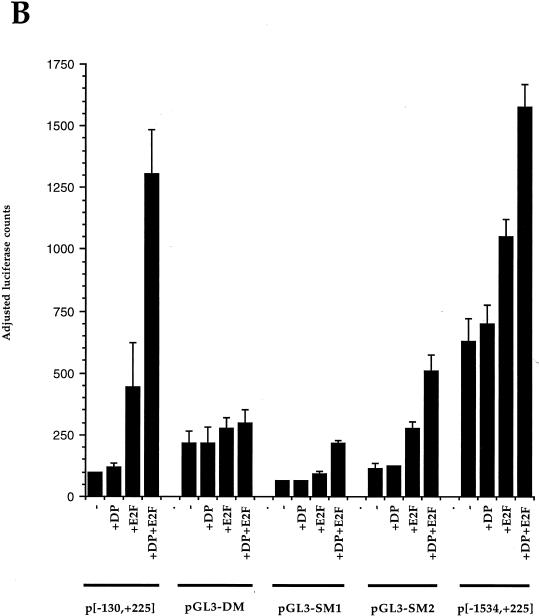
Luciferase activity mediated by different CDC6 promoter constructs. (A) Schematic representation of the large human CDC6 promoter. The transcription start site (+1) is given by an arrow. Three putative E2F sites are depicted by black bars. The two E2F sites in the 355-bp fragment (shaded) are numbered 1 and 2. The endogenous restriction sites BamHI and NaeI were used to construct p[−570,+98] and p[−266,+98]. Mutations introduced in the two most downstream E2F sites to obtain pGL3-DM, -SM1, and -SM2 are shown with big black crosses, and their sequences are given below the constructs. (B) Responses of p[−130,+225], pGL3-SM1, GL3-SM2, pGL3-DM, and p[−1534,+225] to E2F-1 and DP-1 cotransfection in U2-OS cells. The activity from the p[−130,+225] construct is upregulated approximately 13-fold by coexpression of E2F-1 and DP-1, while the mutant constructs pGL3-DM, -SM1, and -SM2 do respond significantly less to E2F-1 and DP-1. The larger p[−1534,+225] construct is upregulated approximately twofold. The p[−130,+225] construct without E2F-1 and/or DP-1 cotransfection is depicted as 100 adjusted luciferase counts after measuring β-Gal activity. All other values are given relative to this with standard deviations from the means.
To investigate the roles of the E2F sites found in the human CDC6 promoter, the activities of several of the luciferase constructs shown in Fig. 5A were tested by transient transfection in asynchronously growing human osteosarcoma U2-OS cells. To measure whether the two E2F sites in p[−130,+225] could respond to E2F and DP expression, plasmids encoding wild-type E2F-1 and DP-1 proteins were cotransfected. Figure 5B shows the luciferase activity from the different promoters, calculated after adjusting for transfection efficiency. The larger p[−1534,+225] construct alone is approximately sixfold more active than the shorter p[−130,+225] alone. It was also found that mutant versions of p[−130,+225] still harbor an activation capacity in U2-OS cells, with the double mutant (DM) being about twofold more active than the p[−130,+225] construct. This suggests that the E2F sites in this promoter are involved in repression of CDC6 expression, although we cannot exclude the possibility that the sites switch from positive to negative and back during a normal cell cycle.
Luciferase activity is upregulated significantly (13-fold) when E2F-1 and DP-1 are cotransfected with p[−130,+225]. Importantly, the effect of E2F and DP cotransfection is lost when the double mutant pGL3-DM is used, while the single mutants pGL3-SM1 and -SM2 both still respond to E2F-1 and DP-1 coexpression, although this upregulation is significantly less than found with the wild-type construct. The activity of the larger promoter (p[−1534,+225]) is upregulated about two- to threefold after E2F-1 and DP-1 coexpression, while its basal activity is much higher. A possible repressive effect by pocket proteins under more physiological conditions is lost when transcriptional activator proteins such as E2F and DP are transiently overexpressed, because endogenous pocket protein levels are too low to overcome the transient activity. Nevertheless, these data show that the E2F sites in the CDC6 promoter are E2F responsive. Moreover, we conclude that the first E2F site in p[−130,+225] appears to play a more significant role in the repression of CDC6 transcription than the second site. These results are in agreement with the findings from the in vivo footprints showing a strong protein binding to the upstream element in quiescent cells, while a weaker occupation could be detected on the downstream site (discussed below).
Roles of the E2F sites in the G1-to-S transition.
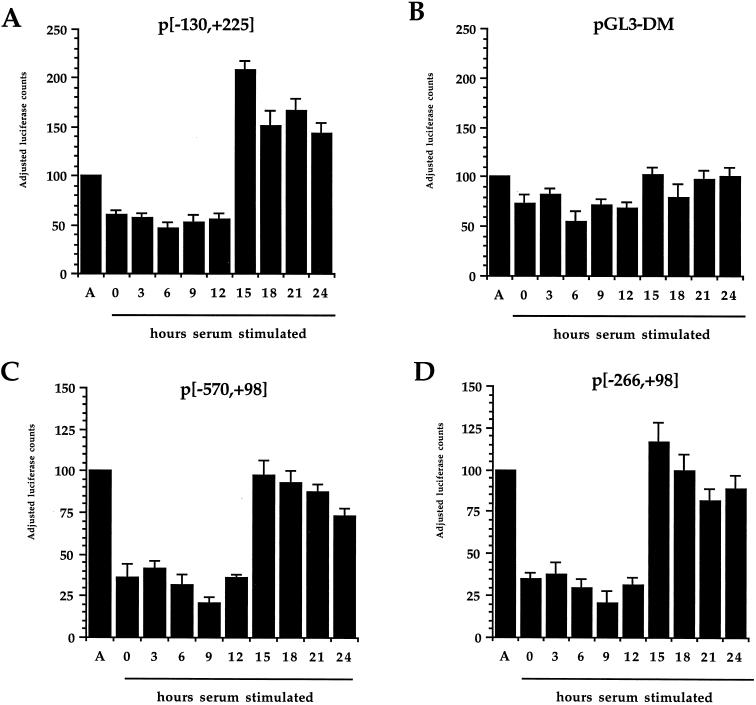
Because we found that E2F can activate transcription from the CDC6 promoter, it was interesting to determine the roles of the E2F sites in the CDC6 promoter during the cell cycle. For this, NIH 3T3 cells were transiently transfected with the wild-type construct p[−130,+225] and the mutant construct pGL3-DM. Cotransfection was performed with a β-Gal-expressing plasmid. Next, cells were starved for 24 h in a low level of serum and subsequently stimulated with a high level of serum. Lysates were prepared at certain time points, and finally all lysates were measured for luciferase activity, and compared to the activity found in asynchronously growing cells. Figure 6A shows that there is a significant upregulation of luciferase activity from the wild-type construct after 15 h of stimulation, which is around the time the cells enter S phase (FACS data not shown), while there is no upregulation by the mutant construct (Fig. 6B). From this, we conclude that the two downstream E2F sites are involved in the upregulation of the CDC6 promoter when cells reach S phase.
FIG. 6.
Cell cycle-regulated expression of the CDC6 promoter is dependent on E2F DNA-binding sites. NIH 3T3 cells were transiently transfected with p[−130,+225], pGL3-DM, p[−570,+98], and p[−266,+98]. Forty-eight hours posttransfection, cells were serum starved for 24 h and subsequently stimulated with fresh medium containing 10% BCS. Lysates were made after the depicted time spans. Asynchronous (A) samples were valued as 100 adjusted luciferase counts, while the others were calculated in comparison to this. Transfection efficiencies were determined by pCMV-βgal cotransfection. Samples were obtained in duplicate, and the presented data are representative for at least three independent experiments.
To investigate whether inclusion of the third identified E2F site upstream of the two sites in the 355-bp fragment has an impact on the regulation of the human CDC6 gene, two deletion mutants of the promoter were used: p[−570,+98] and p[−266,+98] (Fig. 5A). These were tested in NIH 3T3 cells for serum responsiveness. Figure 6C and D show that there is no detectable difference between these two constructs, but importantly, there is a comparable cell cycle regulation between these constructs and the wild-type p[−130,+225]. This suggests that inclusion of the third site does not change the regulation of the CDC6 promoter in these experiments.
E2F complexes interact with the E2F sites in the CDC6 promoter.
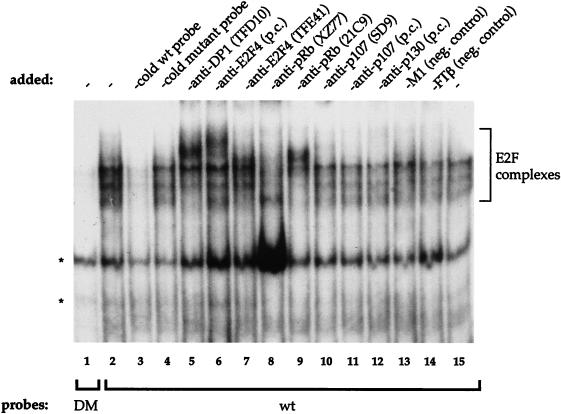
To investigate whether the two E2F sites found in the 355-bp fragment of the CDC6 promoter interact with protein complexes containing E2F and DP proteins, an EMSA was performed using radioactively labeled PCR products (85 bp) from the p[−130,+225] (wt) and pGL3-DM (double mutant) constructs shown in Fig. 5A. A set of protein complexes from MRC5 human fibroblast nuclear extracts interact with the wild-type probe (Fig. 7, lane 2), which cannot be detected when we used the DM probe (lane 1). Many different sized complexes are apparent, probably due to the fact that there are two E2F sites in the wild-type probe that are most likely bound by different E2F-containing protein complexes. This makes it difficult to distinguish complexes that contain only E2F-DP from those that contain E2F-DP bound to the different pocket proteins; also, the two sites might behave differently. We found however that both E2F sites are independently able to bind to E2F-containing complexes (data not shown).
FIG. 7.
EMSA using labeled wild-type (wt) and double mutant (DM) probes. CDC6 promoter probes were obtained by PCR on p[−130,+225] and pGL3-DM and used in combination with nuclear extract from MRC5 human fibroblasts. Specific E2F complexes that interact with the wt probe are depicted with a bracket to the right of the blot. Non-E2F containing protein-DNA complexes are given with asterisks to the left. The cold wt probe (in lane 3) and the cold mutant (derived from DM) were added in 50-fold excess over the quantity of radiolabeled fragment. Antibodies directed against DP-1, E2F-4, and pRB (lanes 5 to 9) shift different subsets of E2F-containing complexes, while the antibodies against p107 and p130 (lanes 10 to 12) do not. M1 (lane 13) is a monoclonal antibody raised against adenovirus E1A. FTβ (lane 14) is a polyclonal serum raised against farnesyl transferase. M1 and FTβ served as negative (neg.) controls. -, nothing added; PC, polyclonal antibody.
We detect complexes present on the wild-type and mutant probes, suggesting the presence of protein-DNA complexes outside (between) the E2F sites, which is in agreement with our data using in vivo footprinting (see below). Specifically bound complexes were competed by an excess of cold wild-type E2F site probe (Fig. 7, lane 3), but not by a mutant probe (lane 4). These data show that the slower-migrating bands indeed contain protein complexes that specifically interact with the E2F sites. To see which of these complexes contain E2Fs and known associated proteins was investigated by the addition of antisera against DP-1 (lane 5), E2F-4 (lanes 6 and 7), and pRB (lanes 8 and 9) which all shift or disrupt one or more E2F site-binding complexes. Antisera against p107 and p130 did not shift any of the E2F-site bound complexes (lanes 10 to 12). Using these antibodies alone with the radioactive wild-type E2F and mutant probes did not result in any detectable background shifts due to a specific binding of antisera to the DNA (data not shown). From these experiments, we conclude that the E2F sites in the 355-bp fragment of the CDC6 promoter can interact with E2F-containing complexes in vitro and that these proteins cannot bind when the E2F sites are mutated.
E2F site occupation in vivo.
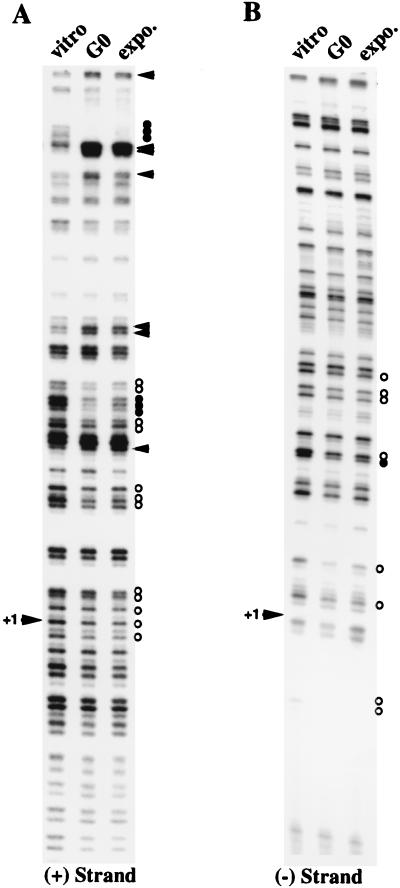
We performed a detailed analysis of the DNA-protein contacts on the hCDC6 promoter during the cell cycle by an in vivo genomic footprinting strategy. This technique allows the observation of DNA-protein contacts at single-nucleotide resolution in living cells. G0-arrested and exponentially growing MCF7 cells were incubated with DMS, a base methylating agent that reacts predominantly with the N7 position of guanines that are not protected by transcription factors. Methylated genomic DNA was extracted from DMS-treated cells. As a control, naked genomic DNA was also methylated in vitro with DMS. In vivo- or in vitro-methylated DNA was then cleaved at modified bases with hot piperidine. Using this DNA as matrix, hCDC6-specific ladders were then amplified by LMPCR and analyzed by genomic sequencing methods.
Given the locations of the E2F sites near the transcription start site, we designed our LMPCR primers to concentrate on the 200-bp sequence around the major transcription start site of the hCDC6 promoter. In vivo footprints of the sense strand (Fig. 8A) and antisense strand (Fig. 8B) demonstrate that the E2F sites located in close proximity to the transcription start site are occupied in vivo in serum-starved and exponentially growing cells. However, protection of the first E2F site (E2F/1, bp −43 to −36) is much stronger than that of the second E2F site (E2F/2, bp −8 to −1). Figure 8C shows an overview of the protected and hyperreactive sites detected. This in vivo analysis showing differences between both E2F sites strongly supports our functional analyses of the hCDC6 promoter (Fig. 5); both types of analyses clearly demonstrate that E2F proteins are directly involved in the control of hCDC6 expression.
FIG. 8.
In vivo footprinting analysis of the transcription start site region of the hCDC6 promoter. LMPCRs were performed with primer S1, S2, or [minus (−) strand] or AS1, AS2, or AS3 [plus (+) strand] on genomic DNA templates obtained from serum-starved (G0) or exponentionally growing (expo.) MCF7 cells treated in vivo with the guanosine methylating agent, DMS. Similar LMPCRs were performed with DMS-methylated naked DNA (vitro lanes). Protected residues and hyperreactive residues detected between in vitro- and in vivo-methylated DNAs are indicated as circles and arrowheads, respectively. Weak (white circles) and strong (black circles) in vivo protection is indicated. The transcription start site is indicated with an arrowhead (+1) to the left of the blots. Amplified DNA ladders that are visible correspond to guanines of the hCDC6 promoter. (A) Positive-sense strand. (B) Negative-sense strand. (C) Summary of DNA-protein contacts observed by in vivo footprinting on both strands of the hCDC6 promoter upstream of the transcription start site (black arrow, +1). Putative consensus binding sites are indicated as open boxes. A protein-bound element with a sequence similar to that of an Sp1 consensus site is depicted as Sp1/?. A protected site around the putative initiator region (INR) (bp −16 to −10) is indicated with a question mark.
These in vivo footprints also show that other transcription factors are present on this hCDC6 promoter region. An element located approximately 100 bp upstream of the start site is found to be strongly protected. It is depicted here as an Sp1 site with a question mark, because it is different from a consensus Sp1 site (GGGCGG). Constitutive protection was also observed for a putative CCAAT box and for two elements located between the two investigated E2F sites.
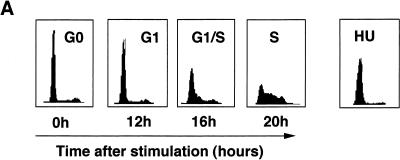
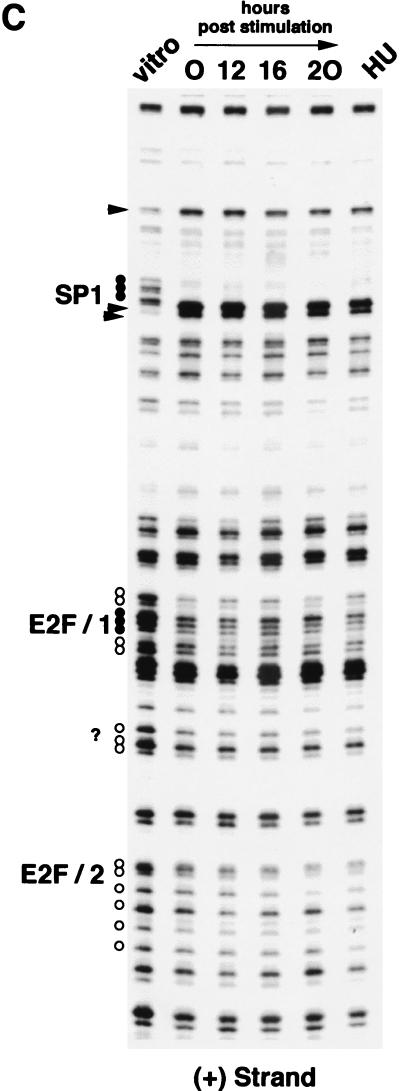
To analyze the occupation of the two E2F DNA-binding sites of the CDC6 promoter through the cell cycle in more detail, in vivo genomic footprinting was performed on MCF-7 cells that were synchronized by a combination of isoleucine and serum starvation. Cells were released into the cell cycle by addition of isoleucine-and-serum-containing medium, and cells were harvested at different times after stimulation for a FACS profile, Northern blotting, and in vivo footprinting. The FACS analysis depicted in Fig. 9A shows that these cells enter S phase approximately 16 h after serum starvation release. In agreement with the data obtained for Swiss 3T3, NIH 3T3, and MRC5 cells, the CDC6 mRNA was upregulated by the addition of serum before the onset of S phase around 12 h after release from starvation (Fig. 9B). Interestingly, the in vivo footprint of the human CDC6 promoter (Fig. 9C) shows that the two E2F DNA-binding sites are protected in quiescent cells, as well as in serum-stimulated cells after 12, 16, and 20 h of serum stimulation, while a similar protection is detected in hydroxyurea-treated samples. These findings strongly suggest that E2Fs bound to both E2F sites present in the promoter have both a negative role (when bound to pocket proteins) and a positive role in the regulation of CDC6 expression.
FIG. 9.
Cell cycle-regulated occupation of protein-binding sites in the human CDC6 promoter. Human MCF7 cells were starved in medium lacking serum and isoleucine and subsequently stimulated in normal medium with a high level of serum. (A) FACS analysis of synchronized MCF7 cells after starvation (0 h) and stimulation (12, 16, and 20 h). To block cells in G1/S, cells were also treated with hydroxyurea (HU). (B) Northern blot with total RNA from MCF7 cells that were treated as described above for panel A and then probed for human CDC6 mRNA expression (upper blot). Equal loads were ensured by ethidium bromide (EtBr) staining (lower blot). Lane A contains RNA isolated from asynchronously growing cells. (C) In vivo footprinting analysis [plus (+) strand] on genomic DNA templates from MCF7 cells that were treated as described above panels for A and B. Vitro lane contains LMPCR-treated DMS-methylated naked DNA. HU lane contains samples from hydroxyurea-treated cells. Weak (white circles) and strong (black circles) in vivo protection is indicated. Hyperactive residues, compared to in vitro-methylated DNA, are indicated with arrowheads. Sp1 refers to the putative Sp1 site upstream of the two E2F (E2F/1 and E2F/2) sites. The protected site around the putative initiator region is indicated with a question mark.
CDC6 is directly upregulated by E2F-1 activation.
It has been demonstrated that the upregulation of E2F activity in serum-deprived cells can induce S phase. Although many genes are upregulated due to E2F activity, in most cases, it is unclear whether this is a secondary effect due to the induction of S phase or whether these genes are directly targeted by E2F. To investigate this, a number of Rat-1 cell lines were generated expressing either a fusion protein between wild-type E2F-1 and the modified ligand-binding domain of the ER (Rat-1-ER-E2F-1) or a fusion between a DNA-binding mutant of E2F-1 with the same ligand-binding domain of the ER protein (Rat-1-ER-E2F-1-E132) as a negative control. Details concerning the generation, selection, and characterization of both cell lines will be described elsewhere (76). The addition of OHT (an estrogen antagonist) to cells expressing wild-type, but not mutant, E2F-1 as a fusion with ER is sufficient to induce S phase in serum-starved cells that are kept in a low level of serum (data not shown). S-phase entry occurs approximately 12 h after addition of OHT, which is 4 to 6 h before S-phase entry of serum-stimulated cells (76). Figure 10A shows that when Rat-1-ER-E2F-1 cells are starved for 48 h in low-serum-containing medium and subsequently treated with OHT, a significant upregulation of endogenous CDC6 mRNA expression occurs after 4 h (lanes 2 to 4) and that the upregulation is also detected when CHX (an inhibitor of protein synthesis) is added (lanes 6 to 9). The addition of CHX alone leads to a slight increase of CDC6 mRNA levels, which is most likely due to a minor stabilization of the RNA (lanes 10 to 13). The signal drops after 8 h of OHT addition (lane 5), suggesting that the CDC6 promoter also contains negative regulatory elements or that the stability of the ER-E2F fusion itself is regulated through downstream factors.
FIG. 10.
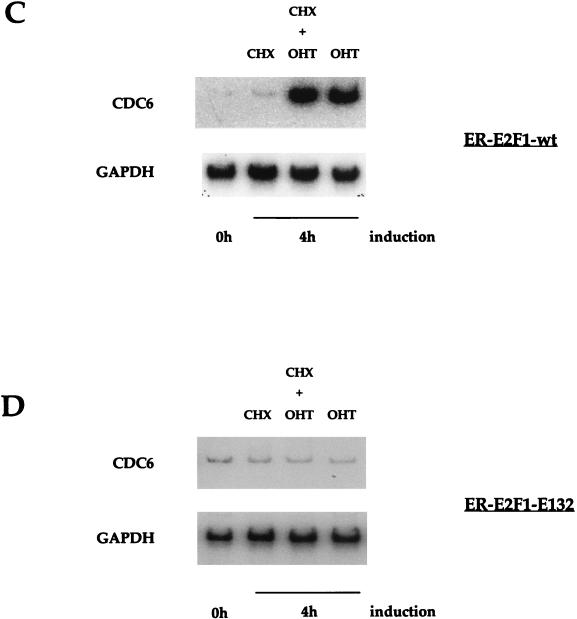
Direct regulation of CDC6 expression by E2F-1 transcription factor. (A) Northern blot containing total RNA purified from Rat-1 cells stably expressing a fusion between full-length wild-type E2F-1 and the ligand-binding domain of the ER (ER-E2F-1). E2F-1 activity was induced by the addition of OHT in the absence (lanes 3 to 5) or presence of CHX (lanes 7 to 9). The effect of CHX addition alone (lanes 11 to 13) was also monitored. The Northern blot was probed with a partial mouse CDC6 probe and subsequently hybridized with a partial rat GAPDH probe. Lane A contains the asynchronous cell sample. (B) RT-PCR on RNA obtained from the cells described above for panel A. Cells were stimulated for up to 16 h with OHT or BCS or for 4 h with OHT in the absence or presence of CHX (C). Induction of CDC6 mRNA expression was measured by a linear range radioactive RT-PCR, and the rat endogenous GAPDH mRNA served as a control (lower blot). (D) RT-PCR on total RNA purified from Rat-1 cells stably expressing a fusion between a full-length DNA-binding mutant (E132) of E2F-1 and the ligand-binding domain of ER after the addition of CHX, OHT plus CHX, or OHT alone for 4 h. The CDC6 signal was measured by an RT-PCR, and GAPDH mRNA served as a control (lower blot).
To further investigate E2F-1-regulated expression of CDC6, an RT-PCR experiment was performed using the RNA from the wild-type and E132 DNA-binding mutant cells. Expression levels of CDC6 mRNA were determined for up to 16 h of OHT addition and compared to levels after BCS addition. The RT-PCR in Fig. 10B confirms the Northern blot data shown in Fig. 10A. There is a significant upregulation of CDC6 mRNA levels after 4 h of OHT-induced activation (Fig. 10B, lane 2), which correlates with the upregulation of cyclin E, B-Myb, p107, and other putative E2F targets which we have detected in other experiments (76). After this, the level is somewhat downregulated (lanes 3 and 4). Interestingly, another upregulation of CDC6 after 16 h of OHT-induced activation was detected (lane 5), while GAPDH levels remain stable (lower blot). At present, we have no explanation for this phenomenon, but it is most likely not a direct effect of E2F activation. Lanes 6 to 9 show that the upregulation of CDC6 upon BCS addition is significantly slower.
Figure 10C shows a similar RT-PCR experiment on CDC6 and GAPDH mRNA from ER-E2F-1 wild-type cells after addition of CHX alone, CHX plus OHT, and OHT alone for 4 h. CDC6 is upregulated after addition of OHT and also when CHX is present; this shows that de novo synthesis of protein is not necessary for CDC6 transcription by ER-E2F-1. Similar results were found when an ER-E2F-3 fusion cell line was used, but not when an ER-E2F-2 fusion cell line was treated with OHT (76). These observations suggest that CDC6 expression is under the control of a specific set of E2F transcription factor complexes.
It was also tested whether the effect of OHT and CHX addition on the mutant cell line Rat-1-ER-E2F-1-E132 could influence CDC6 mRNA levels. Figure 10D shows that CDC6 expression is not upregulated after OHT and/or CHX addition in these mutant cells, which indicates that DNA binding of E2F is essential for the activation of the CDC6 promoter. From these experiments, we conclude that the upregulation of CDC6 expression in these Rat-1 cells is a rapid and direct effect of E2F-1 activation, which does not require protein synthesis and which occurs earlier than upon serum addition. These experiments also demonstrate that the upregulation of CDC6 is independent of entry into S phase, since Rat-1 cells expressing ER-E2F-1 do not enter S phase before 12 h after OHT treatment, and upregulation of CDC6 mRNA has been detected as early as 1 to 2 h after addition of OHT (data not shown). Moreover, cells treated with CHX in addition to OHT never enter S phase, probably due to a complete lack of protein synthesis.
Overexpression of hCDC6 induces S-phase entry.
E2F-induced activation leads to S-phase entry. It is unknown which of the reported E2F-regulated genes are responsible for the transition from starved cells into S phase via G1 or for the transition from G1-to-S phase in a cycling cell, since none of the reported E2F targets can mimic the E2F-induced G1-to-S transition. Here, a new E2F-regulated gene is identified, of which its yeast homologs are essential for the start of DNA replication and thus play a significant role at the G1-S boundary. To test whether human CDC6 is able to increase the number of cells in S phase, U2-OS cells were transiently transfected with a mammalian expression vector encoding an HA-tagged human CDC6 protein. Table 1 shows that expression of CDC6 alone induces a nondramatic increase in the percentage of cells in S phase and that the transient expression of cyclin E, being another E2F target, has a similar effect. However, when both human CDC6 and cyclin E are cotransfected, we observe an additive effect on the percentage of cells in S phase (54.7% increase calculated from S-phase percentage [42%] in mock-transfected cells). The percentages given are averaged from five different independent experiments. These data suggest that mammalian CDC6 is limiting for entry into the S phase of the cell cycle.
TABLE 1.
Effect on cell cycle distribution in asynchronously growing U2-OS cells after transient expression of human cyclin E and CDC6
| Transfected vector or protein | % of cells in cell cycle phase
|
% Increase in S phase | ||
|---|---|---|---|---|
| G1 | S | G2/M | ||
| Vector | 32 | 42 | 26 | |
| hCDC6 | 28 | 51 | 21 | 21.4 |
| Cyclin E | 24 | 56 | 20 | 33.3 |
| hCDC6 + cyclin E | 18 | 65 | 17 | 54.8 |
hCDC6 is required for S-phase entry.
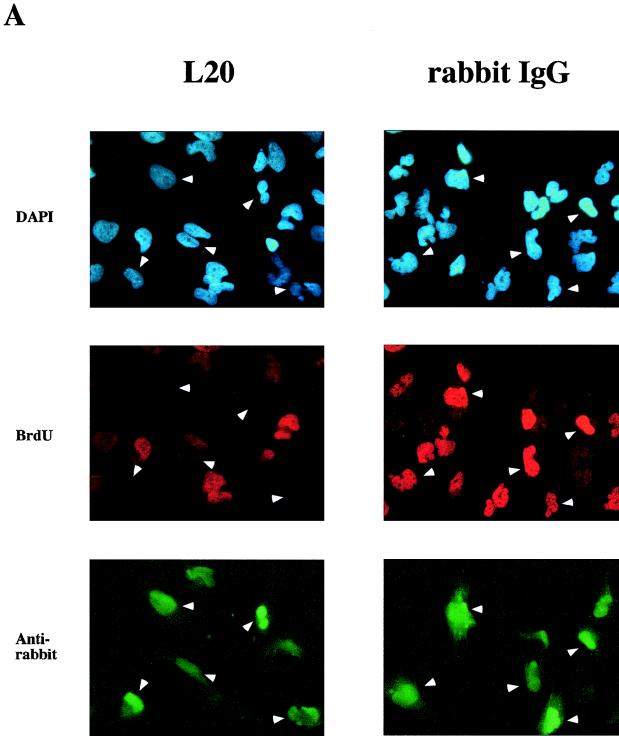
To assess whether human CDC6 is required for initiation of DNA replication, L20, an affinity-purified rabbit antiserum directed against human CDC6, was microinjected into human glioblastoma T98G cells. Based on the available data on CDC6 from other organisms, it was anticipated that human CDC6 would be required for the formation of the prereplication complex during G1 of the cell cycle. Therefore, T98G cells were serum starved and subsequently induced to enter the mammalian cell cycle by serum addition for 16 h. Then cells were microinjected with L20 or with rabbit IgG as a negative control. Figure 11A shows that a large number of L20-injected cells (green) lack BrdU incorporation (red), while in the case of the rabbit IgG injection, most cells that are injected still stain positive for BrdU. We have found that L20 recognizes endogenous levels of human CDC6 protein by immunoprecipitation and Western blotting (61). Figure 11B shows that T98G cells enter S phase 16 to 18 h after serum stimulation. Injection of anti-CDC6 antiserum after 16 h efficiently blocked BrdU incorporation, since only 25% of these cells were BrdU positive 2 h after injection (at 18 h), which corresponds to the number already committed to S phase at 16 h. In contrast, affinity-purified rabbit IgG did not prevent initiation of DNA replication, since these cells synthesized DNA as efficiently as noninjected cells up to 70% at 18 h. These data demonstrate that human CDC6, like the homologs of hCDC6 in lower eukaryotes, is essential for initiating DNA replication.
FIG. 11.
Block of DNA synthesis by microinjection of specific antibodies directed against human CDC6 protein. (A) Human T98G cells were starved for 48 h in low-serum-containing medium and subsequently stimulated with medium with high serum plus BrdU. After 16 h, cells were injected with either an affinity-purified specific anti-hCDC6 rabbit antiserum (L20) or with affinity-purified rabbit IgG. After 2 h, cells were fixed and treated with DAPI (blue; upper panels) and with antibodies for BrdU (red; middle panels) and with antirabbit antibodies to score for the injected antisera (green; lower panels). Arrowheads indicate injected cells that are not incorporating BrdU in the case of L20 or that are synthesizing DNA in the case of rabbit IgG. (B) Noninjected cells were scored for BrdU incorporation after 0, 14, 16, and 18 h following serum stimulation and compared to the total number of cells. Injected cells that were stained green because of their injected antisera were scored for BrdU incorporation and compared to the total number of injected cells. The diagram is the average of three independent experiments in which approximately 200 cells per coverslip were injected. FCS, fetal calf serum; BCS, bovine calf serum; rIgG, rabbit IgG.
DISCUSSION
Most of our knowledge concerning DNA replication is obtained from experiments using the lower eukaryotes S. cerevisiae, S. pombe, and X. laevis, while investigations in higher (multicellular) organisms are hampered by the fact that specific DNA synthesis start sites, or origins of replication, have not been identified so far. Some progress was reported by Krude et al. (43), who used extracts from HeLa cells to study the initiation of mammalian DNA replication. Numerous polypeptides involved in yeast DNA replication have been identified, and several data have provided evidence for how these proteins are implicated in the regulation of the onset of DNA replication. Cdc6p in S. cerevisiae and Cdc18p in S. pombe fulfill an essential and central role in the onset of DNA replication by regulating the timed formation of a complex known as the prereplication complex, which is necessary to start DNA synthesis with the assistance of S-CDKs (16, 73). To date, most mammalian homologs of the known yeast proteins involved in the initiation of DNA replication have been identified, although their role in DNA replication remains to be proven.
We and others have identified the mammalian homolog of Cdc6p and Cdc18p (79) and we show here that CDC6 mRNA is absent in serum-starved cells, while an upregulation of CDC6 levels is detected when cells enter S phase; these levels drop when cells progress through the cell cycle. Preliminary data show that the human CDC6 protein is absent in quiescent cells and in early G1 phase of the cell cycle, and it appears that the protein is present throughout the remaining part of the cell cycle (61).
It has been reported that the regulation of the CDC6 and cdc18+ genes in S. pombe and S. cerevisiae are under the control of the transcription factor complexes MBF/SBF and DSC1, respectively (41, 62, 73), which are the functional homologs of the mammalian E2F transcription factor family, although they are not structurally homologous. It is shown here that the E2F sites found in the 5′ regulatory region of human CDC6 are essential for the induction of hCDC6 expression during the G1-to-S transition in cycling cells. When two E2F sites found in close proximity to the transcription start site were mutated, E2F protein-containing complexes were unable to interact in band shift experiments, while the promoter lost the ability to respond to coexpression of E2F and DP proteins. Moreover, a cell cycle-regulated expression of luciferase activity was lost using a mutant promoter-luciferase gene fusion construct transfected in mouse NIH 3T3 cells that were starved and subsequently serum stimulated. In our experiments, we included a third putative E2F site (bp −279 to −271) next to the two sites that were used in the mutational analysis. Our results demonstrate that this site is neither required nor sufficient for the cell cycle-regulated expression of CDC6, presumably because the sequence does not match well with the E2F consensus binding sequence TTTSSCGS in which S is either a G or C. Although we did not analyze whether this putative E2F DNA-binding site can bind E2F, our data suggest that this site is not occupied by E2F.
To investigate to what extent the two 3′ E2F sites were bound by proteins during the mammalian cell cycle, we used an in vivo footprinting assay. Our results show that one (bp −43 to −36) of the two E2F sites is predominantly occupied by proteins in proliferating cells and that the same site is occupied by proteins in starved cells (Fig. 8), whereas the other E2F site (bp −8 to −1) is significantly less protected. Both sites, however, match well with the E2F DNA-binding consensus sequence (see above). Taken together with the fact that the E2F transcription factors are complexed with pocket proteins in G0/G1 and that pocket proteins are active repressors of transcription (10, 69, 77), our data suggest that the absence of mammalian CDC6 mRNA in G0 is due to an active repression by pRb, p107 and/or p130 bound to E2F-DP complexes via these E2F sites. Since we also found that the same sites are occupied throughout the cell cycle (Fig. 9), it seems as if these sites are crucial not only for repressing CDC6 expression but also for activating CDC6 expression in mid-to-late G1. Our data generated using promoter-luciferase constructs with wild-type and mutated E2F DNA-binding sites in synchronized cells strongly support this interpretation (Fig. 5). Similar to the CDC6 promoter, it has been reported that the E2F DNA-binding sites in the dihydrofolate reductase (78) and thymidine kinase (39) promoters are also occupied throughout the cell cycle, suggesting that the E2F sites in these promoters are both positive and negative regulatory elements.
Apart from protection of the E2F sites, we also detected protection of a putative Sp1 site, of a CCAAT region located upstream of the E2F sites and two sequences between the two 3′ E2F sites. One of these elements has homology to a region found in promoters of human cyclin A, cdc25C and cdc2 and mouse B-myb genes, called the cell cycle genes’ homology region (CHR) see below). The other element (question mark, Fig. 8C and 9C) resembles an initiator region depicted as INR in Fig. 4B. Further mutational analyses will be required to test whether this regulatory element is also critical for the cell cycle-regulated activation of the CDC6 promoter.
As noted above, the promoters of the genes encoding human cyclin A, CDC2, CDC25c, and mouse B-Myb, share a sequence of homology known as CHR, located five nucleotides downstream of a cell cycle-dependent element (CDE) (84). Some CDE sequences are involved in E2F-dependent regulation (6, 67). A CHR-homologous sequence present five nucleotides downstream of the first E2F site in the CDC6 promoter was detected (Fig. 4). It was found that nucleotide −28 in the putative CHR behaves as a hypersensitive site in in vivo footprints (Fig. 8 and 9). Therefore, we investigated whether a similar situation exists for hCDC6 by mutating the putative CHR region in the background of p[−130,+225], pGL3-SM1, pGL3-SM2, and pGL3-DM. We find that a reporter harboring only mutations in the CHR and the downstream E2F site (bp −8 to −1) behaves as pGL3-SM2 in E2F-DP cotransfection and in band shifts. We also find a comparable cell cycle regulation with this mutant promoter in starved and proliferating NIH 3T3 cells (data not shown). From this, we conclude that the putative CHR in the hCDC6 promoter does not cooperate in E2F binding and that it is not involved in E2F-dependent cell cycle regulation. However, we do not exclude the possibilities that proteins bind to the CHR and that this element has a role in hCDC6 gene regulation which is independent from E2F binding.
Next to the E2F and CHF sites we have found through computer analysis, a number of other consensus sites for transcription factor binding (Sp1, AP-2, Ets-1, NF-κB, etc. [Fig. 4B]). No E-box elements (enabling the binding of basic region–helix-loop-helix proteins) in close proximity to the transcription start site were detected. As was shown in Fig. 8 and 9, we have evidence that the CCAAT box and the putative Sp1 site are occupied in in vivo genomic footprints. To date, we have no further data indicating which of the sites present is involved in hCDC6 expression. However, we have shown that the E2F sites close to the start site are required for upregulated expression before cells reach S phase and that mutant E2F sites abolish this effect (Fig. 5B and 6).
Although it has been extensively reported that E2F activation leads to the induction of S phase, it is unclear which of the E2F regulated genes are responsible for the E2F-induced G1-to-S transition or for the entry into a new cell cycle from quiescence. We are unable to monitor the effect of hCDC6 overexpression in serum-starved cells, since the protein could not be detected by immunostaining after microinjection in quiescent cells (61). Therefore, we cannot ascertain whether hCDC6 alone or in combination with cyclin E is sufficient to induce reentry into the cell cycle. A likely explanation for the inability to detect hCDC6 in quiescent cells could be that it is an unstable protein in serum-starved cells. Nevertheless, we were able to express hCDC6 in proliferating cells and have shown that overexpression of hCDC6 can increase the number of cells in S phase. We have provided evidence that the expression of hCDC6, the mammalian homolog of the essential yeast genes CDC6 and cdc18+, is directly regulated by E2F through the E2F sites in the promoter. Significantly, hCDC6 can induce S phase in cooperation with cyclin E, another E2F target. Taken together with the demonstration that hCDC6 is required for entry into the S phase (Fig. 11), our data provide a direct link between regulation of cell cycle progression by the E2F transcription factors and the initiation of DNA replication and suggest that the function of mammalian CDC6 is similar to those of its yeast homologs.
ACKNOWLEDGMENTS
We thank Karin Holm, Alexandra Charlesworth, Stefania Lupo, Heiko Müller, and Emanuela Frittoli for technical assistance in sequencing, FACS, elutriation, and microinjection. Pier Paolo di Fiore is thanked for providing reagents, and we thank the members of the Lattanzio family who donated the microinjection setup. We thank Peter Cartwright and Simonetta Piatti for critical reading of the manuscript.
This work was supported in part by grants from the Human Frontiers Science Program and the Associazione Italiana per la Ricerca sul Cancro (AIRC) and by fellowships from the European Community (G.H.), the Fondazione Italiana per la Ricerca sul Cancro (E.V.), the Fondazione per la Formazione Oncologica (A.W.), and the Danish Research Academy (B.O.P.).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley-Interscience; 1988. [Google Scholar]
- 2.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 3.Beijersbergen R L, Kerkhoven R M, Zhu L, Carlée L, Voorhoeve P M, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–2690. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Mitchell J, Leber J, Kobayashi R, Stillman B. The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing. Cell. 1995;83:563–568. doi: 10.1016/0092-8674(95)90096-9. [DOI] [PubMed] [Google Scholar]
- 5.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 6.Bennett J D, Farlie P G, Watson R J. E2F binding is required but not sufficient for repression of B-myb transcription in quiescent fibroblasts. Oncogene. 1996;13:1073–1082. [PubMed] [Google Scholar]
- 7.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botchan M. Coordinating DNA replication DNA replication with cell division: current status of the licensing concept. Proc Natl Acad Sci USA. 1996;93:9997–10000. doi: 10.1073/pnas.93.19.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Dürr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cartwright P, Müller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–624. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 12.Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:31–60. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 13.Cocker J H, Piatti S, Santocanale C, Nasmyth K, Diffley J F. An essential role for the Cdc6 protein in forming prereplicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 14.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeGregori J, Kowalik T, Nevins J R. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–4224. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diffley J F X. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 17.Donovan S, Harwood J, Drury L S, Diffley J F X. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou Q-P, Markell P J, Pardee A B. Thymidine kinase transcription is regulated at G1/S phase by a complex that contains retinoblastoma-like protein and a cdc2 kinase. Proc Natl Acad Sci USA. 1992;89:3256–3260. doi: 10.1073/pnas.89.8.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drury L S, Perkins G, Diffley J F X. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyson N, Dembski M, Fattaey A, Ngwu C, Ewen M, Helin K. Analysis of p107-associated proteins: p107 associates with a form of E2F that differs from pRB-associated E2F-1. J Virol. 1993;67:7641–7647. doi: 10.1128/jvi.67.12.7641-7647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 22.Geng Y, Eaton E N, Picón M, Roberts J M, Lundberg A S, Gifford A, Sardet C, Weinberg R A. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene. 1996;12:1173–1180. [PubMed] [Google Scholar]
- 23.Ginsberg D, Vairo G, Chittenden T, Xiao Z-X, Cu G, Wydner K L, DeCaprio J A, Lawrence J B, Livingston D M. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–2679. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 24.Graham F L, Van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 25.Grange T, Bertrand E, Espinas M L, Fromont-Racine M, Rigaud G, Roux J, Pictet R. In vivo footprinting of the interaction of proteins with DNA and RNA. Methods. 1997;11:151–163. doi: 10.1006/meth.1996.0401. [DOI] [PubMed] [Google Scholar]
- 26.Harlow E, Franza B J, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helin K. Regulation of cell proliferation by the E2F transcription factors. Curr Opin Genet Dev. 1998;8:28–35. doi: 10.1016/s0959-437x(98)80058-0. [DOI] [PubMed] [Google Scholar]
- 28.Helin K, Harlow E, Fattaey A R. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helin K, Lees J A, Vidal M, Dyson N, Harlow E, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 30.Helin K, Wu C-L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative transactivation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 31.Hijmans E M, Voorhoeve P M, Beijersbergen R L, van’t Veer L J, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–3089. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hinds P W, Mittnacht S, Dulic V, Arnold A, Reed S I, Weinberg R A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 33.Hsiao K-M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 34.Hu Q, Bautista C, Edwards G, Defeo-Jones D, Jones R, Harlow E. Antibodies specific for the human retinoblastoma protein identify a family of related polypeptides. Mol Cell Biol. 1991;11:5792–5799. doi: 10.1128/mcb.11.11.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huet X, Rech J, Plet A, Vié A, Blanchard J M. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol. 1996;16:3789–3798. doi: 10.1128/mcb.16.7.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurford R K J, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. [Google Scholar]
- 37.Ikeda M A, Jakoi L, Nevins J R. A unique role for the Rb protein in controlling E2F accumulation during cell growth and differentiation. Proc Natl Acad Sci USA. 1996;93:3215–3220. doi: 10.1073/pnas.93.8.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 39.Karlseder J, Rothender H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelly T J, Martin G S, Forsburg S L, Stephen R J, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 41.Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 42.Kovesdi I, Reichel R, Nevins J R. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 43.Krude T, Jackman M, Pines J, Laskey R A. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 44.Lam E W-F, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lees J A, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol. 1993;13:7813–7825. doi: 10.1128/mcb.13.12.7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 47.Liang C, Weinreich M, Stillman B. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell. 1995;81:667–676. doi: 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 48.Lindeman G J, Gaubatz S, Livingston D M, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–5100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukas J, Petersen B O, Holm K, Bartek J, Helin K. Deregulated expression of E2F family members induces S-phase entry and overcomes p16INK4A-mediated growth suppression. Mol Cell Biol. 1996;16:1047–1057. doi: 10.1128/mcb.16.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Means A L, Slansky J E, McMahon S L, Knuth M W, Farnham P J. The HIP binding site is required for growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1992;12:1054–1063. doi: 10.1128/mcb.12.3.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moberg K, Starz M A, Lees J A. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol Cell Biol. 1996;16:1436–1449. doi: 10.1128/mcb.16.4.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller P R, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- 53.Müller H, Moroni M C, Vigo E, Petersen B O, Bartek J, Helin K. Induction of S-phase entry by E2F transcription factors depends on their nuclear localization. Mol Cell Biol. 1997;17:5508–5520. doi: 10.1128/mcb.17.9.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muzi-Falconi M, Brown G W, Kelly T J. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1666–1670. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muzi-Falconi M, Brown G W, Kelly T J. DNA replication: controlling initiation during the cell cycle. Curr Biol. 1996;6:229–233. doi: 10.1016/s0960-9822(02)00464-5. [DOI] [PubMed] [Google Scholar]
- 56.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. . (Author’s correction, 15:4660, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- 58.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearson B E, Nasheuer H P, Wang T S. Human DNA polymerase alpha gene: sequences controlling expression in cycling and serum-stimulated cells. Mol Cell Biol. 1991;11:2081–2095. doi: 10.1128/mcb.11.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen, B. O., and K. Helin. Unpublished data.
- 62.Piatti S, Böhm T, Cocker J H, Diffley J F X, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- 63.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 65.Santocanale C, Diffley J F X. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- 66.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, Weinberg R A. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–2407. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Dürr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sears R, Ohtani K, Nevins J R. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17:5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sherr C J. Cancer cell cycles. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 71.Slansky J, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slansky J E, Farnham P J. Introduction to the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 73.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 75.Tommasi S, Pfeifer G P. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vigo, E., and K. Helin. Unpublished data.
- 77.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 78.Wells J, Held P, Illenye S, Heintz N H. Protein-DNA interactions at the major and minor promoters of the divergently transcribed dhfr and rep3 genes during the Chinese hamster ovary cell cycle. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Williams R S, Shohet R V, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zerfass-Thome K, Schulze A, Zwerschke W, Vogt B, Helin K, Bartek J, Henglein B, Jansen-Dürr P. p27KIP1 blocks cyclin E-dependent transactivation of cyclin A gene expression. Mol Cell Biol. 1997;17:407–415. doi: 10.1128/mcb.17.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 82.Zhu L, Zhu L, Xie E, Chang L S. Differential roles of two tandem E2F sites in repression of the human p107 promoter by retinoblastoma and p107 proteins. Mol Cell Biol. 1995;15:3552–3562. doi: 10.1128/mcb.15.7.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zwerschke W, Rottjakob H W, Küntzel H. The Saccharomyces cerevisiae CDC6 gene is transcribed in late mitosis and encodes a ATP/GTPase controlling S phase initiation. J Biol Chem. 1994;269:23351–23356. [PubMed] [Google Scholar]
- 84.Zwicker J, Muller R. Cell-cycle regulation of gene expression by transcriptional repression. Trends Genet. 1997;13:3–6. doi: 10.1016/s0168-9525(96)30112-1. [DOI] [PubMed] [Google Scholar]